- 1College of Life Sciences, Nanjing Agricultural University, Nanjing, China
- 2Department of Criminal Science and Technology, Nanjing Police University, Nanjing, China
N6-methyladenosine (m6A), a well-characterized RNA modification, is involved in regulating multiple biological processes; however, genome-wide identification and functional characterization of the m6A modification in legume plants, including soybean (Glycine max (L.) Merr.), remains lacking. In this study, we utilized bioinformatics tools to perform comprehensive analyses of molecular writer candidates associated with the RNA m6A modification in soybean, characterizing their conserved domains, motifs, gene structures, promoters, and spatial expression patterns. Thirteen m6A writer complex genes in soybean were identified, which were assigned to four families: MT-A70, WTAP, VIR, and HAKAI. It also can be identified that multiple cis elements in the promoters of these genes, which were classified into five distinct groups, including elements responsive to light, phytohormone regulation, environmental stress, development, and others, suggesting that these genes may modulate various cellular and physiological processes in plants. Importantly, the enzymatic activities of two identified m6A writers, GmMTA1 and GmMTA2, were confirmed in vitro. Furthermore, we analyzed the expression patterns of the GmMTAs and GmMTBs under different abiotic stresses, revealing their potential involvement in stress tolerance, especially in the response to alkalinity or darkness. Overexpressing GmMTA2 and GmMTB1 in soybean altered the tolerance of the plants to alkalinity and long-term darkness, further confirming their effect on the stress response. Collectively, our findings identified the RNA m6A writer candidates in leguminous plants and highlighted the potential roles of GmMTAs and GmMTBs in the response to abiotic stress in soybean.
Introduction
Over 150 distinct chemical modifications of eukaryotic RNA molecules have been identified, including methylation, acetylation, and glycosylation (Delaunay et al., 2023). Many exist in noncoding RNAs, particularly transfer RNAs and ribosomal RNAs. Massage RNA (mRNA) can also carry several base modifications, such as N6-methyladenosine (m6A), N1-methyladenosine, 5-methylcytidine, N4-acetylcytidine, N7-methylguanosine, and pseudouridine (Frye et al., 2016, 2018). These chemical modifications influence gene expression by regulating the structure, splicing, transport, stability, and translation efficiency of the target transcripts (Roundtree et al., 2017). Of all known modifications, m6A is the most abundant in mRNA (Jia et al., 2013). It is installed by a “writer” protein complex and dynamically removed by “eraser” proteins in the nucleus (Shi et al., 2019). Often, “reader” proteins are responsible for decoding the m6A signature (Han et al., 2021).
In plants, m6A modifications in mRNA are decorated by a methyltransferase complex comprising MRNA ADENOSINE METHYLASE A (MTA; the ortholog of human METHYLTRANSFERASE-LIKE 3 (METTL3), MRNA ADENOSINE METHYLASE B (MTB; the ortholog of human METTL14), FKBP12 INTERACTING PROTEIN 37 kDa (FIP37), VIRILIZER (VIR), and the E3 ubiquitin ligase HAKAI (Zhong et al., 2008; Shen et al., 2016). Mutations of MTA, MTB, FIP37, or VIR in Arabidopsis (Arabidopsis thaliana (L.) Heynh.) are embryo-lethal, indicating that m6A RNA modifications are essential for plant development (Vespa et al., 2004; Zhong et al., 2008; Shen et al., 2016; Růžička et al., 2017). The absence of HAKAI decreases the abundance of m6A modifications but does not result in obvious growth defects (Růžička et al., 2017). In cotton (Gossypium hirsutum (L.)), m6A modifications enhance the stability of GhMYB44 mRNA, contributing to fiber elongation and secondary cell wall thickening (Xing et al., 2023). m6A modifications in plants are also involved in the stress response. In Arabidopsis, transcripts involved in the salt and osmotic stress responses showed an increased abundance of m6A modifications when the plants were grown under high salinity (Anderson et al., 2018). In apple (Malus domestica (Suckow) Borkh.), MdMTA-mediated m6A modifications improved drought tolerance by promoting the mRNA stability and translation efficiency of genes associated with lignin deposition and oxidative stress (Hou et al., 2022). The overexpression of the m6A reader Malus hupehensis YTH-domain family protein 2 (MhYTP2) can elevate the mRNA stability of its target M. domestica allantoinase-like gene (MdALN) in apple (Guo et al., 2023). Together, this evidence highlights the significant roles of m6A modifications in developmental regulation and stress tolerance in plants.
Soybean (Glycine max (L.) Merr.) is an economically important leguminous crop, as it contributes a huge proportion of the total global oilseed and biodiesel and provides vital protein and oil sources for human food and animal fodder (Qi and Lee, 2014; Kim et al., 2017). Soybean cultivation faces challenges from abiotic stresses, such as high or low temperatures and soil salinity or alkalinity. Over the course of its long evolutionary history, soybean has developed intricate strategies to withstand abiotic stresses, which involve modifications across multiple dimensions, including its metabolism, physiology, and transcriptome (Feng et al., 2021; Sheikh et al., 2024). One such abiotic stress, soil salinity, causes ion toxicity and osmotic stress, and thus induces the generation of reactive oxygen species (ROS) (van Zelm et al., 2020). Enzymatic antioxidants, such as superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT), can remove ROS to reduce oxidative stress and protect plants from damage (Wang et al., 2016a; Fu et al., 2017). Additionally, ion channel proteins play a crucial role in the response to salt stress; for instance, Na+/H+ ANTIPORTER 1 (GmNHX1) sequesters Na+ into the vacuole, thereby reducing its level in the cytoplasm (Yang et al., 2017). Similarly, CATION/PROTON EXCHANGER 1 (GmCHX1) facilitates the exclusion of Na+ from leaf tissues, mitigating the toxic effects associated with excessive salt accumulation (Qu et al., 2022). Moreover, stress-inducible transcription factors (TFs), including NACs, bZIPs, MYBs, and WRKYs (Katiyar et al., 2012; Ji et al., 2013; Zhu et al., 2014; Phukan et al., 2016; Lim et al., 2022), enhance plant stress tolerance by regulating the expression of target genes during the stress response. Gene expression is also modulated by epigenetic modifications, including m6A, which play important roles in plant responses to abiotic stress; however, studies on the involvement of m6A in the soybean response to abiotic stress are limited, and the key components involved have yet to be identified.
In this study, we employed genome-wide analyses to identify the m6A writer proteins in soybean and analyzed the gene structure and evolutionary aspects of each family member. We particularly focused on elucidating the expression patterns and subcellular localization of the core members of the m6A writer complex, the GmMTAs and GmMTBs, which were determined to be involved in abiotic stress responses. Our study provides a foundation for further exploring the function of the m6A modification in soybean.
Materials and methods
Plant materials and stress treatments
Soybean (Glycine max, Williams 82) seeds were sterilized using chlorine and then planted on moist vermiculite. The plants were cultivated in a growth chamber under long-day conditions (16 h light of 100 μmol m-2 s-1 intensity provided by white LED lamps at 25°C and 8 h dark at 23°C, 70% relative humidity). 15-day-old seedlings were used for the different abiotic stress treatment. For the cold or heat treatments, seedlings were grown at 8°C or 42°C for 24 h, respectively. For the drought stress treatment, soybean seedlings were transferred to Hoagland liquid culture containing 20% polyethylene glycol (PEG) and incubated for 1 day. For the salinity or alkalinity stress treatments, soybean plants were grown in Hoagland liquid culture containing 150 mM NaCl or 100 mM NaHCO3, respectively, for 1 day. For the darkness treatment, soybean seedlings were grown in Hoagland liquid culture under the dark condition for 3 days. Following these abiotic stress treatments, the leave and roots samples were collected separately and frozen at -80°C for subsequent experiments.
Soybean leaf transient transformation
To generate transgenic soybean lines overexpressing GmMTA2 or GmMTB1, a modified soybean transient transformation was utilized based on a previously study (Wang et al., 2023a). In brief, the transformed Agrobacterium cells were incubated with infiltration buffer (OD600 = 1) containing 10 mM MES (pH 5.6) and 200 μM acetosyringone (Sigma) and infiltrated into the lower epidermis of the leaves from 7-day-old soybean seedlings by employing a vacuum pump until the leaves were thoroughly wetted. Following a recovery period of one day under the continuous darkness, the transformed soybean seedlings were transferred to the normal growth condition for another five days before being subjected to the stress treatment. For alkalinity treatment, the seedlings were grown in Hoagland liquid culture containing 100 mM NaHCO3 for 36 hours. Histochemical detection of hydrogen peroxide (H2O2) and superoxide anion (O2.-) and the enzymatic activity measurement of CAT, POD, and SOD from the infiltrated leaves were performed according to previous studies (Cheng et al., 2019; Pi et al., 2023). For darkness treatment, the seedlings were grown under the continuous darkness for 10 days. Chlorophyll content of soybean leaves from each sample was measured according to a previous description (Li et al., 2024).
RNA extraction, cloning, and quantitative RT-PCR analyses
Total RNA was isolated from 15-day-old soybean leaves and cDNA was prepared according to previous studies (Anderson et al., 2018; Gao et al., 2023; Wang et al., 2023b). The following primers were used for cloning: for GmMTA1 (Glyma.07G067100), Cp994 and Cp995; for GmMTA2 (Glyma.16G033100), Cp1293 and Cp1294, for GmMTB1 (Glyma.10G232300), Cp988 and Cp989; for GmMTB2 (Glyma.20G161800), Cp1291 and Cp1292 (Supplementary Table S3). GmMTA1, GmMTA2, GmMTB1, and GmMTB2 were cloned into pBA002-flag-HA-StrepII (VN21) and pXCS-YFP (V36) (Chen and Witte, 2020), respectively, for the protein purification and subcellular localization analysis. The VN21 was generated by introducing a 135-bp fragment encoding the flag-HA-StrepII tag using the primer pair Cp795/Cp796 into pBA002 vector (Kost et al., 2008).
To analyze mRNA abundance, reverse transcription quantitative PCR (RT-qPCR) was performed with QuantStudio 1 (Thermo Fisher Scientific) using Hieff qPCR SYBR Green Master Mix (Yeasen Biotechnology) according to the previous description (Wang et al., 2023b). Total RNA from each sample was isolated using the RNA isolater Total RNA Extraction Reagent (Vazyme Biotech Co., Ltd). The quantity and concentration of the resulted RNA were evaluated using a Thermo Scientific NanoDrop™ spectrophotometer. A total of 800 ng of RNA was used to synthesize the first-strand cDNA using an Oligo (dt) primer. Transcript abundance of GmMTA1, GmMTA2, GmMTB1, and GmMTB2 was analyzed by employing the primer pairs Cp633/Cp634 or Cp631/Cp632, Cp637/Cp638, and Cp635/Cp636, respectively. GmF-BOX (Glyma.12G051100) was amplified with the primer pairs Cp363/Cp364 as an internal reference gene in mRNA. The calculation was based on the 2−ΔΔCT method (Livak and Schmittgen, 2001). All primer sequences are detailed in Supplementary Table S3.
Phylogenetic and characteristics analyses of the components m6A writer proteins in soybean
The protein sequences of Arabidopsis m6A writers were obtained from the TAIR database (https://www.arabidopsis.org) and used as reference sequences. Subsequently, the m6A writer protein sequences from Glycine max, Glycine soja, Phaseolus vulgaris, Medicago truncatula, and Lotus japonicus were identified by using the BlastP method from Phytozome database (https://phytozome-next.jgi.doe.gov). Multiple sequence alignment was performed with the ClustalW method (Larkin et al., 2007). To compare evolutionary relationships, the m6A writer protein sequences from A. thaliana, G. max, G. soja, P. vulgaris, M. truncatula, and L. japonicus were used to construct a phylogenetic tree using MEGA11 (Tamura et al., 2021) with the Neighbor-Joining (NJ) method and 1000 bootstrap replications. Thereafter, the phylogenetic tree was visualized using ChiPlot (https://www.chiplot.online/) (Xie et al., 2023).
Analyses of gene structure, conserved motifs, collinearity relationship, and cis-elements analyses of the m6A writer genes
Gene structures were visualized using the TBtools software with GFF files provided as input. Conserved motifs within the m6A writer protein sequences were analyzed using the MEME online tool (https://meme-suite.org/meme/tools/meme). The parameters were set as following: the site distribution was designated as ‘any number of repetitions’ (anr), the number of motifs was specified as 10, and all other optional parameters were kept at their default settings. Results of the conserved domains were visualized by using TBtools. To investigate the collinearity relationships among m6A writer genes in G. max, the One Step MCScanX-Super Fast program integrated into TBtools was employed. For cis-elements analysis, 2000-bp region upstream of the start codons of m6A writer genes were obtained from the Phytozome database. Subsequently, the promoter sequences were submitted to the PlantCARE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Prediction from the PlantCARE database were visualized using TBtools (Chen et al., 2020).
Tissue-specific expression of the m6A writer proteins in soybean
To analyze the tissue-specific expression of m6A writer proteins, transcriptome sequencing data were obtained from the SoyOmics database (Liu et al., 2023) for various soybean tissues, including cotyledon, stem, leaf bud, leaf, flower, seed, shoot, and root. These data were graphically represented and visualized using the ChiPlot.
Subcellular localization
GmMTAs-eYFP or GmMTBs-eYFP was transiently co-expressed with the nucleus marker protein RFP-H2B (RFP fused to histone 2B) (Martin et al., 2009) in Nicotiana benthamiana leaves for 5 days. The samples were analyzed using a ZEISS LSM 980 with Airyscan2 microscope equipped with an HC PLAPO CS2 40 × 1.0 water immersion objective (ZEISS Microsystems) according to the previous description (Wang et al., 2023b).
Protein purification and enzymatic activity measurement
Recombinant soybean GmMTA1 and GmMTA2 were affinity purified after transient expression in N. benthamiana as described before (Chen et al., 2016; Baccolini and Witte, 2019). Purified protein was quantified by employing the Bradford reagent from Tiangen with bovine serum albumin (BSA) as the standard.
To assess the enzymatic activity, 0.15 nmol of recombinant GmMTA1 or GmMTA2 was incubated with a 50 µl substrate solution, containing 0.8 mM S-adenosylmethionine (SAM; Sigma), 0.15 nmol RNA probe (UACACUCGAUCUGGACUAAAGCUGCUC, synthesized by Genscript), 80 mM KCl, 1.5 mM MgCl2, 0.2 U μl−1 RNasin, 10 mM dithiothreitol (DTT), 4% glycerol and 15 mM HEPES (pH 7.9), at 28°C for 1 h. Then the reaction was terminated by incubating at 95°C for 15min. The resulting RNA product (800 ng) were fully digested into single nucleosides in a 50μl reaction buffer containing 10 mM Tris–HCl, pH 7.9, 1 mM MgCl2, 0.1 mg mL−1 BSA, 0.4 units benzonase (Sigma-Aldrich), 0.004 units phosphodiesterase I (Sigma-Aldrich) and 0.04 units shrimp alkaline phosphatase (NEB) according to previous description with minor revision (Chen et al., 2018; Chen and Witte, 2020; Gao et al., 2023; Wang et al., 2023b). After incubation at 37°C for 10 h, enzyme reaction was terminated and the sample was filtered by an ultrafiltration tube (3 kDa cutoff; Pall). 2μl aliquots were analyzed by an Agilent 1290 HPLC system coupled with a Sciex 6500 Qtrap mass spectrometer. The following mass transitions were monitored: m/z 268.1 to 136 (A, adenosine); m/z 282.12 to 150 (m6A, N6-methyladenosine). Standard solutions of A: 1, 5, 25 50, 100, 200, 400, 2000 and 10 000 ng/ml; m6A: 0.1, 0.5, 2.5, 5, 10, 20, 40, 200, and 1000 ng/ml were used for quantification. The ratio of m6A to A were calculated based on the calibrated concentrations.
Statistics
Statistical analysis was performed by GraphPad Prism 9.5.1 software. Statistical methods and sample sizes are shown in the figure legends. All replicates are biological replicates or experimental replicates.
Results
Genome-wide identification and evolutionary analysis of mRNA m6A writer genes in legume plants
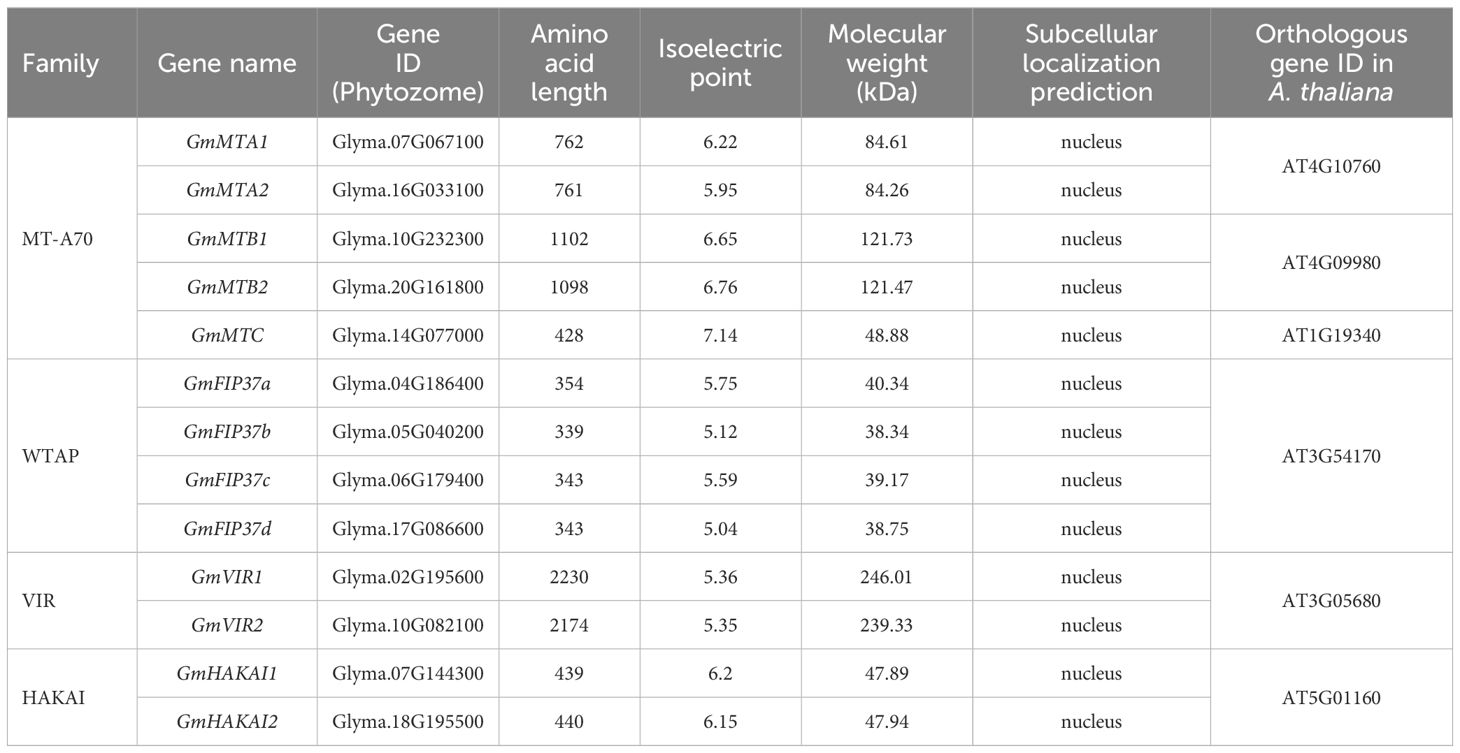
To identify the m6A writer candidate genes in legume plants, we used the sequences of the known writer proteins MTA, MTB, MTC, FIP37, VIR, and HAKAI from the model plant Arabidopsis as queries in BLASTp searches against the genomes of soybean (G. max), wild soybean (G. soja Siebold & Zucc.), common bean (Phaseolus vulgaris L.), Medicago truncatula Gaertn., and Lotus japonicus (Regel) K. Larsen in the Phytozome V13 database (https://phytozome-next.jgi.doe.gov/blast-search). We identified thirteen candidate m6A writer homologs in G. max, thirteen in G. soja, six in P. vulgaris, eight in M. truncatula, and six in L. japonicus (Figure 1; Supplementary Table S1). All identified proteins were divided into four families (MT-A70, WTAP, VIR, and HAKAI) according to their topological structure. At least one homolog for each family was identified in each of the plant species, with the MT-A70 family representing the most identified (22 candidates in total) and the least identified for the VIR family (eight in total). Among the five analyzed legumes, the G. max and G. soja genomes contained the highest number of m6A writer candidate genes (Supplementary Table S1). In G. max, five candidate genes were identified in the MT-A70 family, including Glyma.14G077000, Glyma.10G232300, Glyma.20G161800, Glyma.07G067100, and Glyma.16G033100. Four candidate genes were identified for the WTAP family, and two each were revealed for the VIR or HAKAI families (Figure 1; Table 1). The members of each writer protein family were named sequentially according to their order on the chromosomes (Table 1).
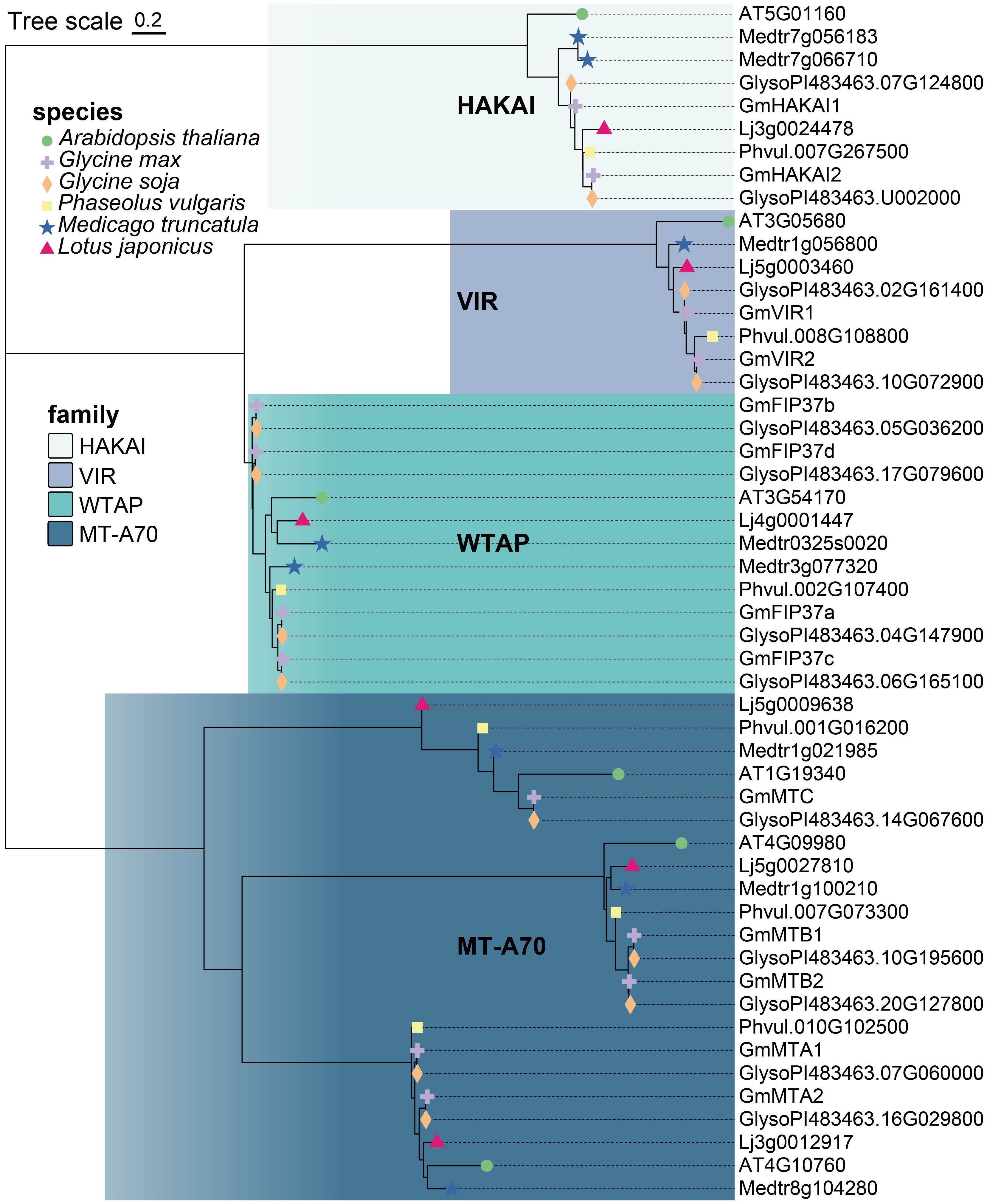
Figure 1 Phylogenetic analysis of m6A writer proteins in Arabidopsis thaliana, Glycine max, Glycine soja, Phaseolus vulgaris, Medicago truncatula, and Lotus japonicus. The phylogenetic tree was constructed using MEGA11 software with the Neighbor-Joining algorithm and 1,000 bootstrap replicates.
We identified similar characteristics in the candidate proteins from each family in G. max. The lengths of the MT-A70 family candidate proteins ranged from 428 to 1102 amino acids. The predicted molecular weights were 48.88 to 121.73 kDa, with theoretical isoelectric points (pIs) in the range of 5.95 to 7.14. The two VIR family candidates had the longest sequences, which were 2230 and 2174 aa with molecular weights of 246.01 and 239.33 kDa and pIs of 5.36 and 5.35. The HAKAI and WTAP candidates were shorter, ranging from 439 to 440 aa and 343 to 354 aa, respectively. Their molecular weights ranged from 47.89 to 47.94 kDa (HAKAI family) and 38.34 to 40.34 kDa (WTAP family), and their pIs were 6.15 to 6.2 and 5.04 to 5.75, respectively.
Conserved motifs and gene exon–intron structures of the mRNA m6A writer genes
We constructed a phylogenetic tree using the Neighbor-Joining method to reconstruct the evolutionary relationships among all writer candidates from G. max (Figure 2). The result was consistent with that of the phylogenetic analysis constructed using the proteins from all five legume plants and Arabidopsis (Figure 1). We employed a motif analysis using the MEME program to identify the conserved motifs present within the m6A writer candidates in G. max. In total, 10 distinct and highly conserved motifs were predicted (Figure 2A; Supplementary Table S2). Each candidate protein contained 2–6 motifs except GmMTC (Figure 2A), hinting that most are likely m6A writer proteins. Motif 4, motif 7, and motif 10 were common to many members, suggesting that they may be important for methyltransferase activity. Additionally, motif 1, motif 2, and motif 3 were unique to the WTAP/FIP37 subfamily, while motif 5 and motif 8 exclusively existed in the MT-A70 subfamily. The homologous proteins of each family share identical conserved motifs, suggesting they may be functionally redundant.
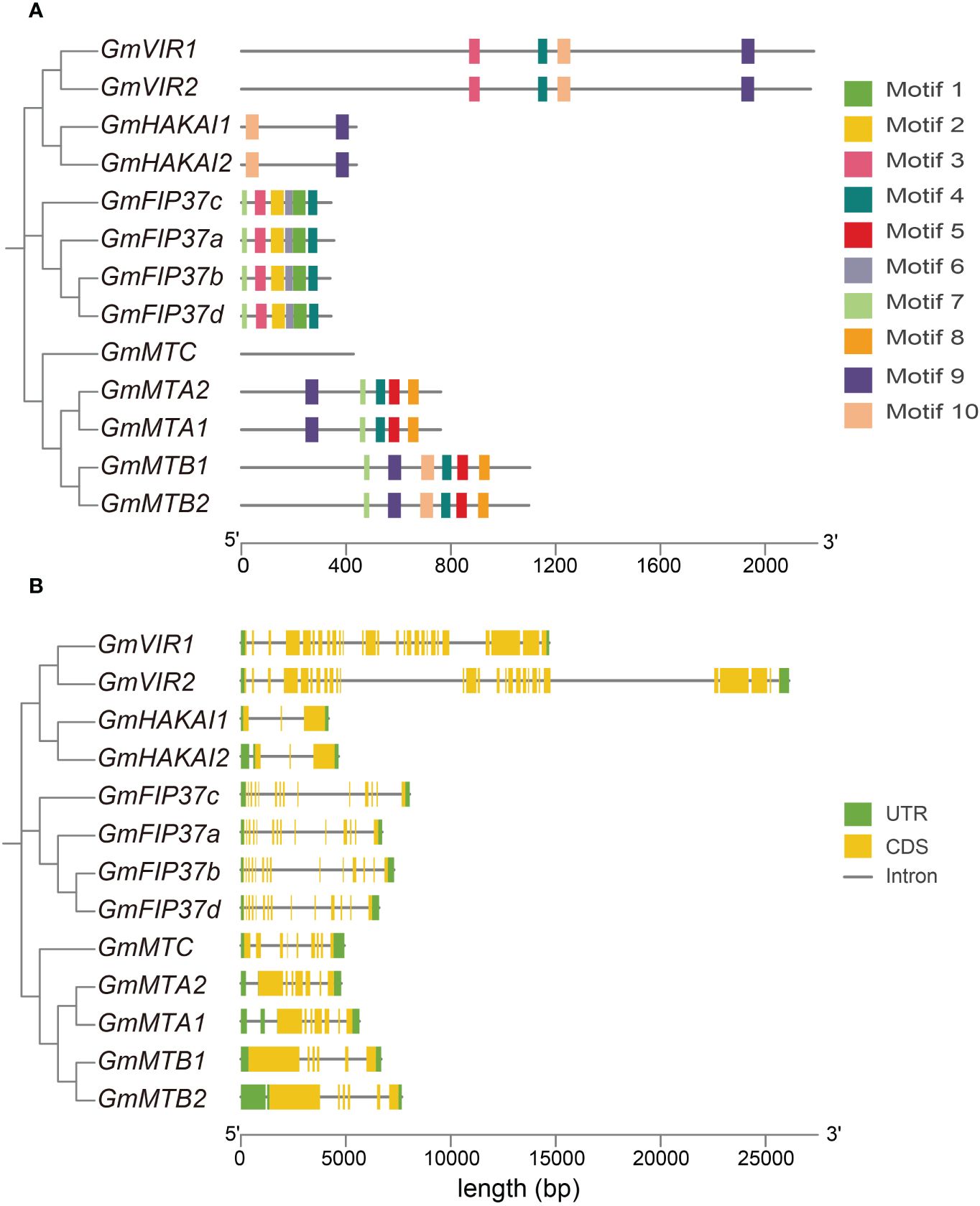
Figure 2 The conserved motifs and gene structure of m6A writer genes in G max. (A) Phylogenetic analysis of m6A writer candidates from G max and the organization and distribution of the conserved motifs in the m6A writer genes. (B) Phylogenetic analysis of m6A writer candidates from G max and the exon–intron structures of the m6A writer genes. Untranslated regions (UTRs) are represented by green boxes, coding sequences (CDSs) are represented by yellow boxes, and introns are represented by gray lines.
We analyzed the exon–intron distribution to investigate the genetic structural diversity. Notably, m6A writer candidates from each subfamily had a similar exon–intron pattern, although across all writer candidates the exon number varied substantially, from 3 to 28. The two GmVIRs had the most exons (27 and 28 exons), while two GmHAKAIs had three exons each. Seven and six exons, respectively, were identified in the core methyltransferase genes, GmMTAs and GmMTBs. Generally, closely related candidate writers tended to have similar conserved motifs (Figure 2A) and exon–intron structure patterns (Figure 2B), suggesting their relative conservation during the evolutionary process and thereby substantiating the accuracy of the clustering analysis presented in Figure 1.
Chromosomal distribution of mRNA m6A writer genes
We investigated the chromosomal distribution of the m6A writer genes in soybean and identified associated gene duplication events. In total, 13 genes were randomly distributed on 11 of the 20 chromosomes of G. max (Figure 3). Chromosomes 7 and 10 each possess two m6A writer genes, whereas chromosomes 2, 4, 5, 6, 14, 16, 17, 18, and 20 each contain one such gene. Different gene replication events, such as tandem duplication and fragment duplication, occur in plant genomes, resulting in the expansion of gene families (Cannon et al., 2004). The collinearity analysis revealed no tandem duplication between the m6A writer genes; however, nine pairs arising from fragment duplications (GmMTA1/GmMTA2, GmMTB1/GmMTB2, GmMTC/Glyma.17G248801, GmVIR1/GmVIR2, GmHAKAI1/GmHAKAI2, GmFIP37a/GmFIP37b, GmFIP37b/GmFIP37c, GmFIP37c/GmFIP37d, and GmFIP37a/GmFIP37d) were observed, showing that members from each m6A writer family originated from gene duplication in soybean. A duplication partner from one such pair, Glyma.17G248801, is an uncharacterized gene without an MT-A70 domain, suggesting that it possesses an unknown function other than m6A methyltransferase activity.
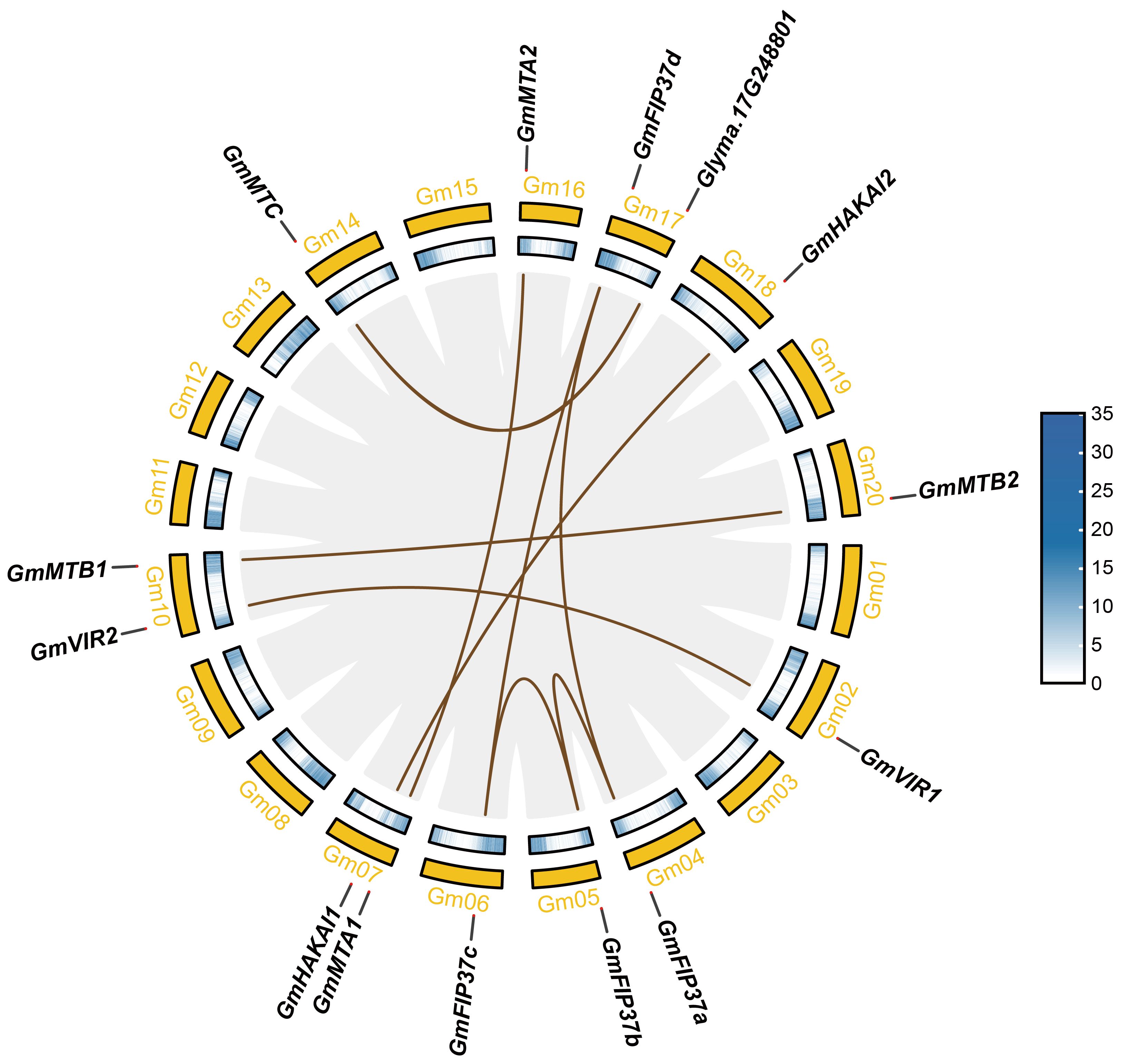
Figure 3 Collinearity analysis of the m6A writer genes in soybean. The tan lines represent collinear pairs of m6A writer genes, and the gray lines represent the collinearity results of the soybean genome.
Spatial expression patterns and subcellular localization of the mRNA m6A writer genes and proteins
Employing the publicly available data from the SoyOmics website (https://ngdc.cncb.ac.cn/soyomics/index), we examined the expression profiles of all m6A writer genes across various tissues from G. max, including the cotyledon, stem, leaf bud, leaf, flower, pod_seed (pod plus seed), pod, seed, shoot meristem, and root. Our analyses revealed distinct expression patterns among these genes across different tissue types (Supplementary Figure S1). Notably, we observed a pronounced upregulation of the expression of m6A writer genes in the leaf bud, flower, and shoot, indicating their potential involvement in orchestrating floral development. By contrast, we were surprised to discover relatively low expression levels of these genes in the leaves.
Furthermore, we used WoLF PSORT (https://wolfpsort.hgc.jp/) to predict the subcellular localization of all members, which were all predicted to be localized in the nucleus (Table 1). To test this prediction, we fused the full-length amino acid sequences of the four core members, GmMTA1, GmMTA2, GmMTB1, and GmMTB2, with a C-terminal yellow fluorescent protein (YFP) tag each and then independently transiently co-expressed them with the nucleus marker (H2B) fused with mCherry (RFP) in N. benthamiana leaves. The results showed that all four proteins were indeed localized to the nucleus (Figure 4), which is consistent with the prediction.
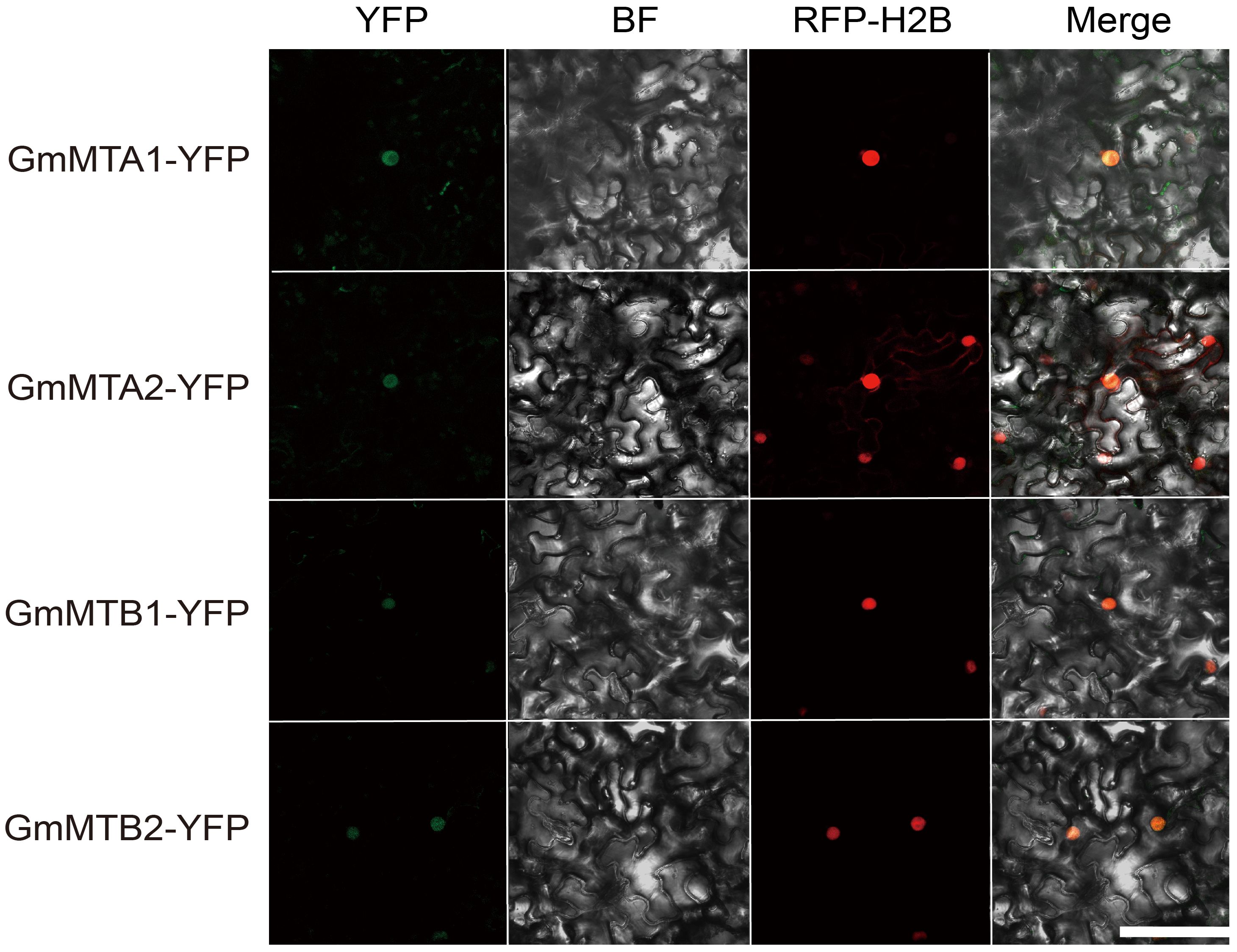
Figure 4 Subcellular localization analysis of GmMTAs and GmMTBs in N. benthamiana leaves. RFP-H2B was used as a nuclear marker. Bar, 100 μM. BF, brightfield.
Methyltransferase activity analysis of the GmMTAs
In mammals, an MTase complex comprising METTL3 and METTL14 efficiently catalyzes the transfer of a methyl group to m6A on RNA. METTL3 primarily functions as the catalytic core, while METTL14 acts as an RNA-binding platform (Wang et al., 2016b). To investigate the enzymatic activities of GmMTAs, which are the homologs of mammalian METTL3 in soybean, we independently expressed GmMTA1 and GmMTA2 recombinantly in N. benthamiana and performed methylation assays with the purified proteins using an unmodified RNA probe and SAM as the methyl donor (Figure 5A). Subsequently, the RNA product was digested into nucleosides and the abundance of m6A was analyzed using liquid chromatography–tandem mass spectrometry (LC-MS/MS). Remarkably, our findings revealed a significant increase in m6A levels upon adding GmMTA1 or GmMTA2 proteins compared with the control. These experimental results strongly support our hypothesis that GmMTA1 and GmMTA2 possess similar methyltransferase activities, indicating their ability to catalyze mRNA m6A modifications (Figure 5).
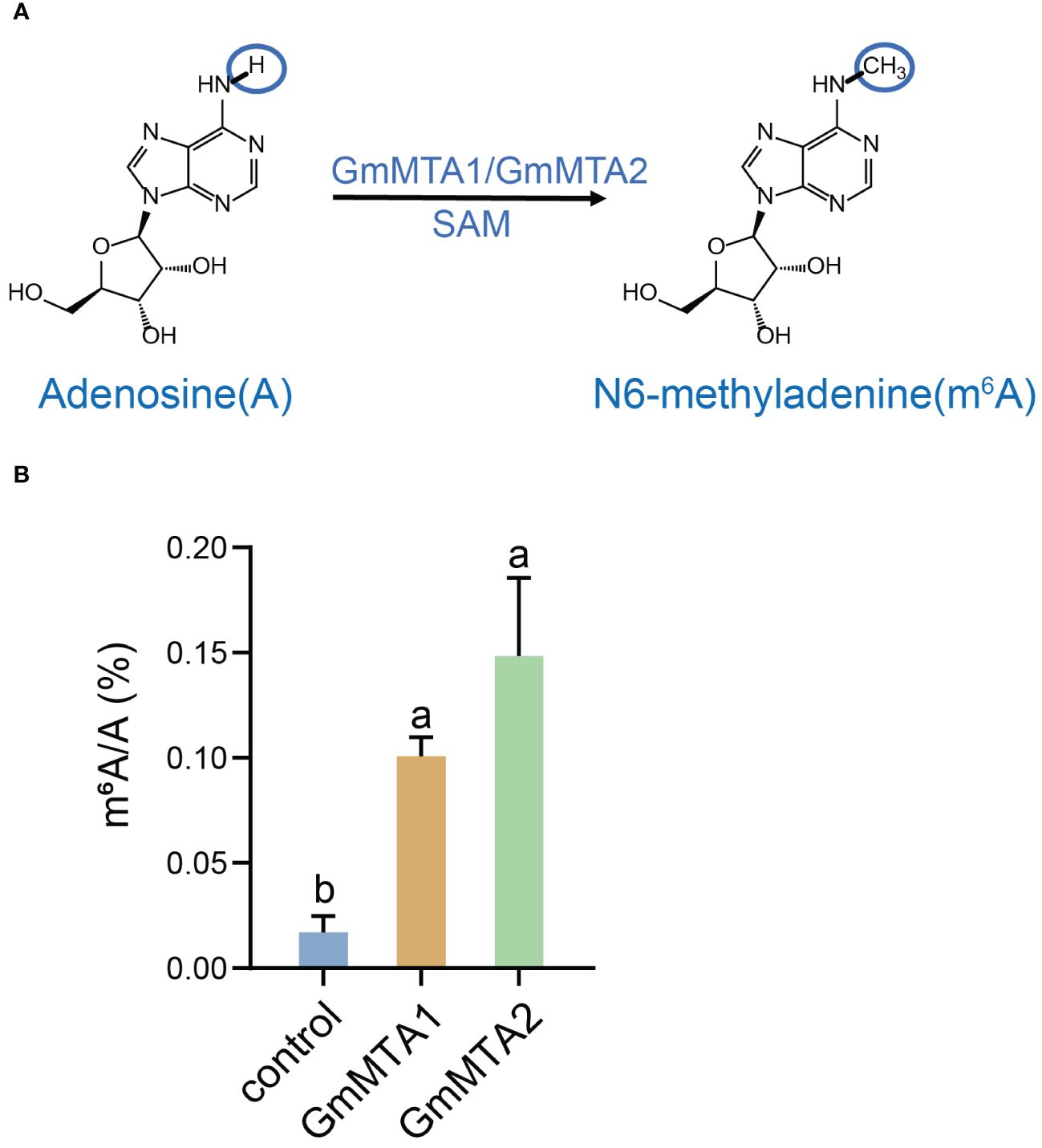
Figure 5 Biochemical analyses of GmMTA1 and GmMTA2. (A) A proposed reaction mechanism of adenosine methylation producing the N6-methyladenosine (m6A) RNA modification performed by GmMTAs in soybean. (B) In vitro methylation assay of GmMTA1 and GmMTA2. Error bars represent SD (n = 3). Different letters indicate significant differences at P < 0.05.
Cis-element analyses of mRNA m6A writer genes
To gain deeper insights into the transcriptional regulatory activity of RNA m6A writer genes, we predicted the cis elements within the 2000-bp promoter regions of all candidates using the PlantCARE web server (http://bioinformatics.psb.ugent.be/webtools/). The identified cis-acting elements were involved in 25 functional categories, which could be classified into five groups: light-responsive elements, phytohormone-responsive elements, environmental stress-related elements, development-responsive elements, and other elements (Figures 6A, B). Remarkably, light-responsive elements could be identified in all promoters. Among the phytohormone-responsive elements, those involved in the abscisic acid (ABA) response and methyl jasmonate (MeJA) response were the most abundant. The most frequent environmental stress-related elements were anaerobic induction, followed by the low temperature-responsive ones. Furthermore, cis elements related to the developmental response were also identified (Figure 6B). The detection of light-responsive elements in all promoters (Figure 6C) suggests that the expression levels of m6A writer genes are probably influenced by light, which may affect the development and environmental stimuli responses of soybean.
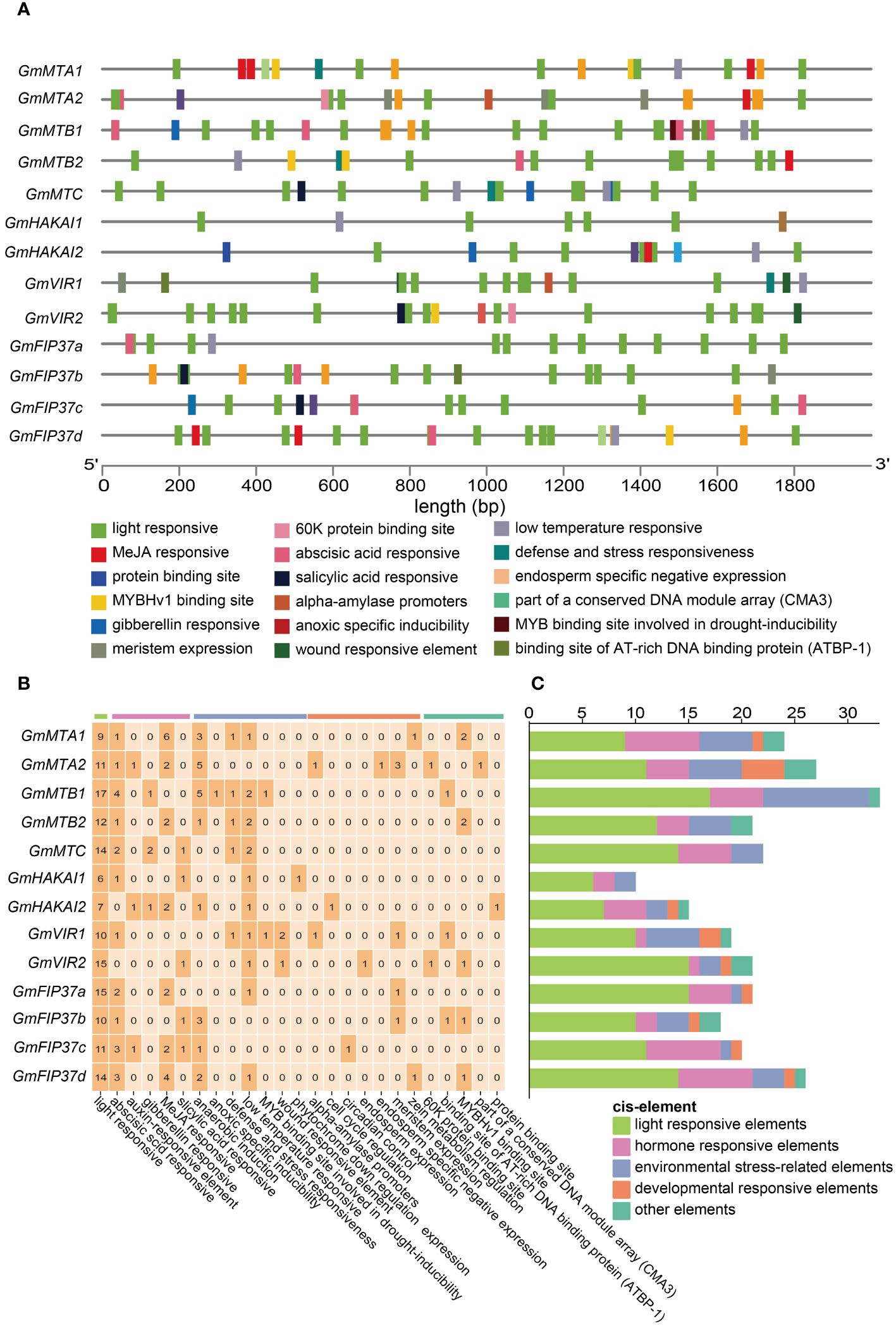
Figure 6 Cis elements in the promoters of m6A writer genes. (A) The distribution of cis elements in the promoters of the m6A writer genes. Different cis elements were depicted in different colored boxes. (B) The number of each category of cis element in m6A writer genes. (C) Cis elements were classified into those responsive to light, phytohormones, development, environmental stress, and other regulated categories.
Expression patterns of GmMTAs and GmMTBs in response to abiotic stress
The results of the cis-element analysis suggest that m6A RNA modifications in soybean play crucial roles in the response to different abiotic stresses. We therefore analyzed the expression pattern of the m6A writer components GmMTA1, GmMTA2, GmMTB1, and GmMTB2 in the leaf and root of 15-day-old soybean plants grown under normal conditions or different stresses, including cold, heat, drought, salinity, alkalinity, or darkness, over 24 h. Generally, the abiotic stress treatments significantly altered the expression patterns of the GmMTAs and GmMTBs compared with the control (Figure 7; Supplementary Figure S2). Notably, each gene exhibited distinct responses to different stress conditions in the leaf; for instance, GmMTA1 was initially suppressed after 2 h of cold treatment but induced after 6 h, whereas the expression level of GmMTA2 remained unchanged until 12 h. By contrast, the expression levels of GmMTB1 and GmMTB2 were both enhanced during the cold treatment (Figure 7B).
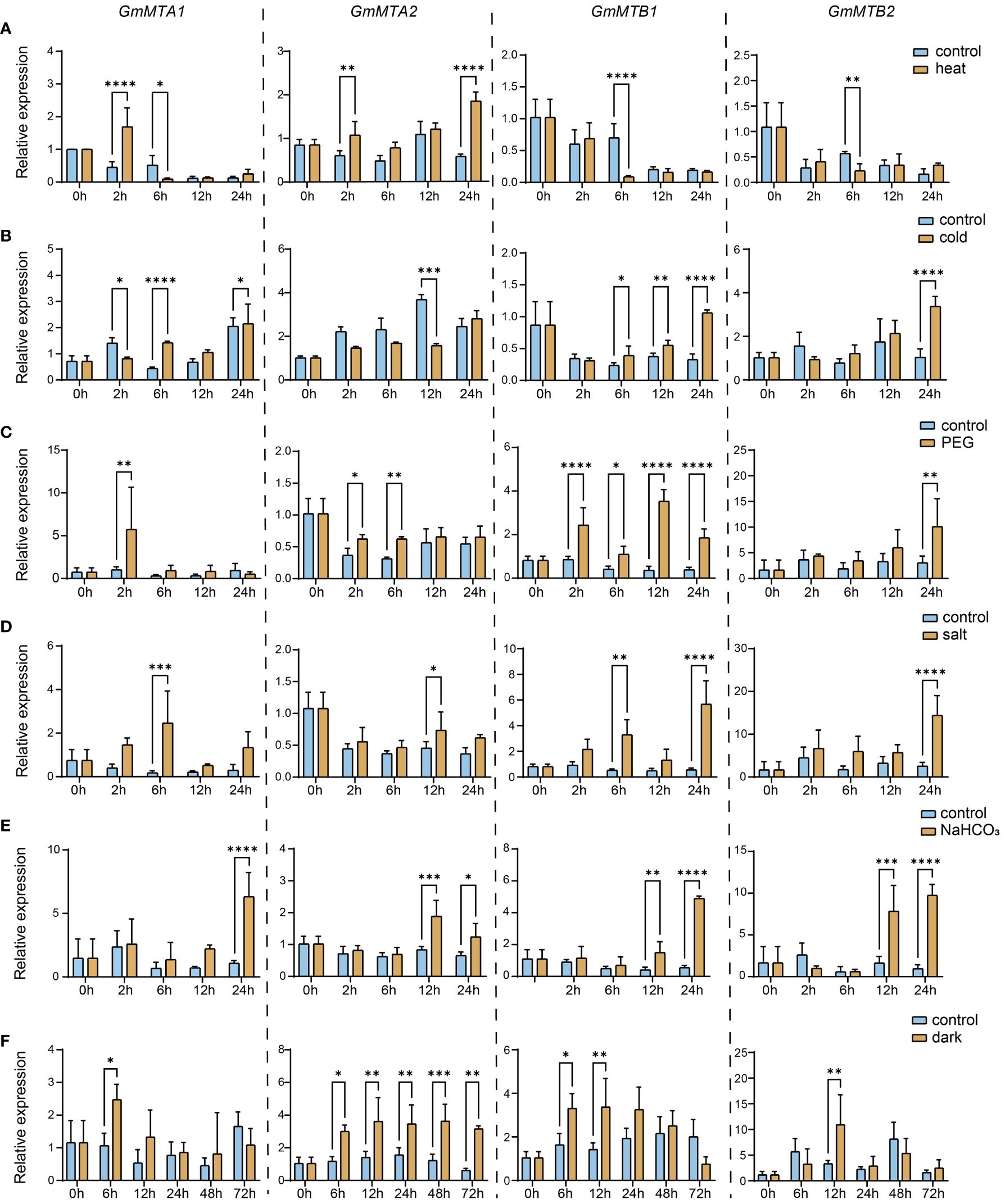
Figure 7 Relative expression levels of GmMTAs and GmMTBs in leaf under different abiotic stresses detected using reverse transcription quantitative PCR (RT-qPCR). 15-day-old soybean seedings were subjected to heat (A), cold (B), polyethylene glycol (PEG) (C), salt (D), alkalinity (NaHCO3) (E), or darkness (F) treatments. GmF-BOX (Glyma.12G051100) was used as the internal control. Error bars represent SD (n = 3, *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ***P ≤ 0.0001).
In addition, we also observed that GmMTAs and GmMTBs exhibit different response times to various stresses in the leaf. The expression of all four genes was increased after 12 to 24 h of alkalinity stress (NaHCO3) treatment (Figure 7E), while darkness induced these changes after 6 to 12 h (Figure 7F). These data provide evidence that GmMTA- and GmMTB-mediated m6A modifications generally participate in the abiotic stress response in soybean. Moreover, our findings also suggest that GmMTA2 and GmMTB1 are the dominant genes involved in the cellular response to these environmental stressors.
Overexpression of GmMTA2 and GmMTB1 in soybean enhances plant tolerance to alkalinity and darkness
To further verify the roles of GmMTAs and GmMTBs in the response to abiotic stress, we overexpressed GmMTA2 and GmMTB1 in the leaves of 7-day-old wild-type soybean plants, respectively, which were then subjected to alkalinity or darkness treatment. Reverse transcription quantitative PCR (RT-qPCR) and immunoblot analyses revealed that pBA002-GmMTA2-HA (GmMTA2-OE) and pBA002-GmMTB1-HA (GmMTB1-OE) were all overexpressed in the transformed soybean (Figure 8). Compared with the control expressing the empty pBA002 vector (EV), the leaves overexpressing GmMTB1 but not GmMTA2 exhibited significantly increased tolerance to the alkalinity treatment (Figure 9A). In addition, we detected the H2O2 content by Diaminobenzidine staining (Figure 9B) and measured the catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD) activities (Figures 9C-E) in the leaves from each genotype after the NaHCO3 treatment. These results confirmed that the leaves harboring GmMTB1 overexpression were indeed more resistant to the alkalinity. During the darkness treatment, the leaves overexpressing GmMTA2 showed the highest tolerance, while leaves overexpressing GmMTB1 also performed better than the control EV leaves (Figure 10A). We confirmed these findings by measuring the chlorophyll contents (Figure 10B). These results, together with RT-qPCR analyses (Figure 7), demonstrated that GmMTB1 played a role in the tolerance to alkalinity treatment while both GmMTA2 and GmMTB1 were involved in dark stress.
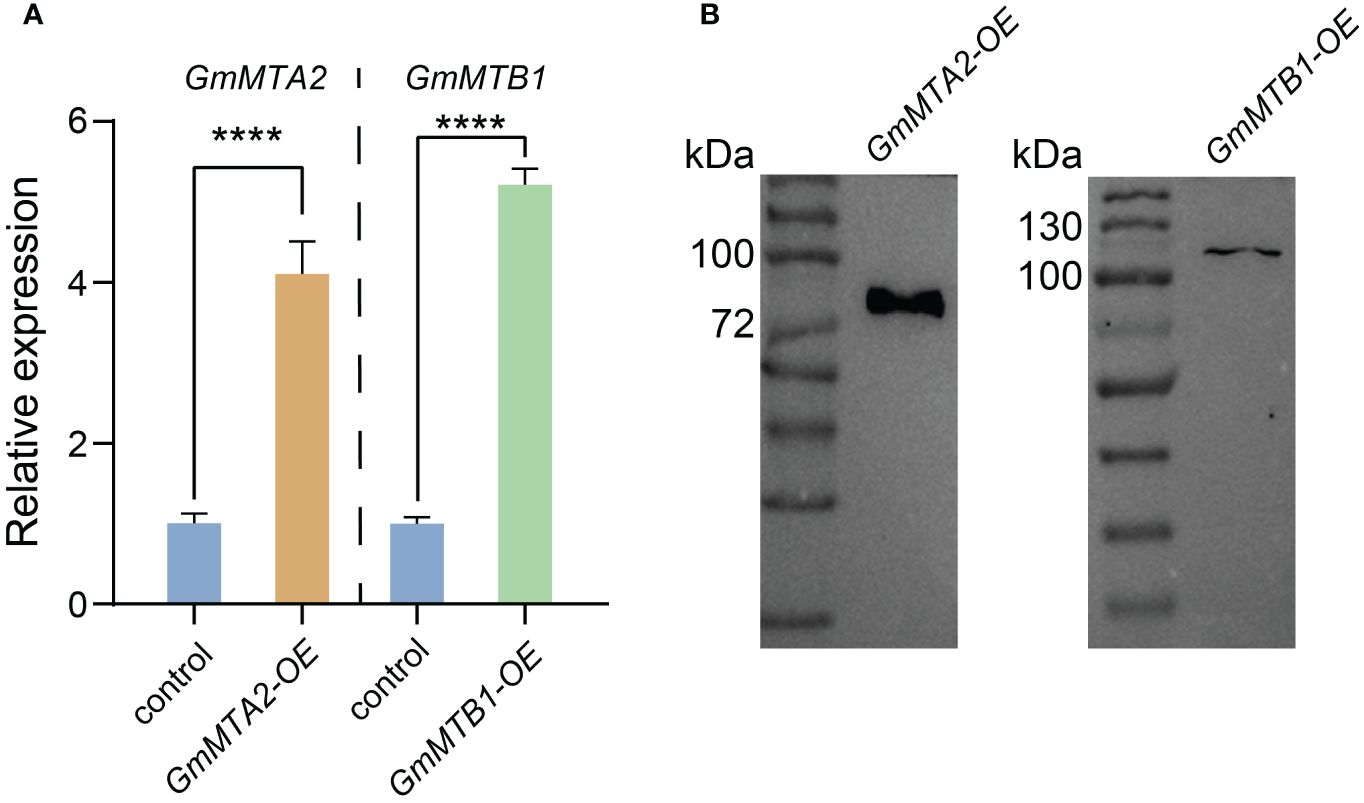
Figure 8 Confirmation of GmMTA2 and GmMTB1 overexpression in soybean. (A) Relative expression of GmMTA2 and GmMTB1 quantified using RT-qPCR in the wild type and the lines overexpressing GmMTA2 (GmMTA2-OE) or GmMTB1 (GmMTB1-OE). Error bars represent SD (n = 3, ****P ≤ 0.0001). (B) Immunoblot analyses of leaf extracts from the soybean lines overexpressing GmMTA2 or GmMTB1. An anti-HA antibody was used for detection. Control = plants expressing the empty vector.
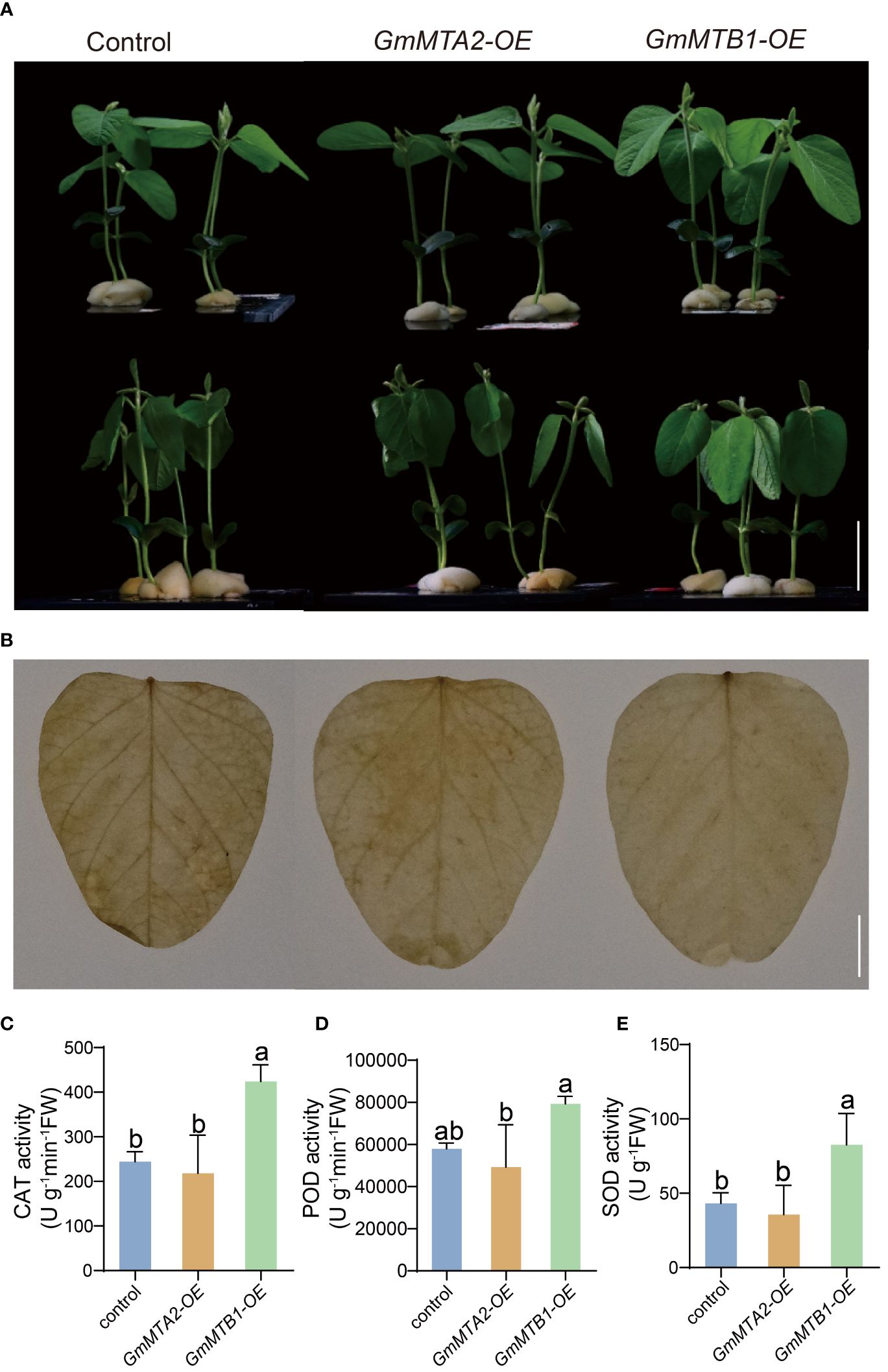
Figure 9 Leaf phenotypes of the empty vector-expressing control, GmMTA2-OE, and GmMTB1-OE plants under the NaHCO3 treatment. (A) Plant documents before (top panel) and after (bottom panel) a 36-h NaHCO3 treatment. Bar, 3 cm. (B) Diaminobenzidine staining of leaves after the NaHCO3 treatment. Bar, 1 cm. (C-E) The catalase (CAT; C), peroxidase (POD; D), and superoxide dismutase (SOD; E) activities in the leaves after the NaHCO3 treatment. Error bars represent SD (n = 3). Different letters indicate significant differences at P < 0.05.
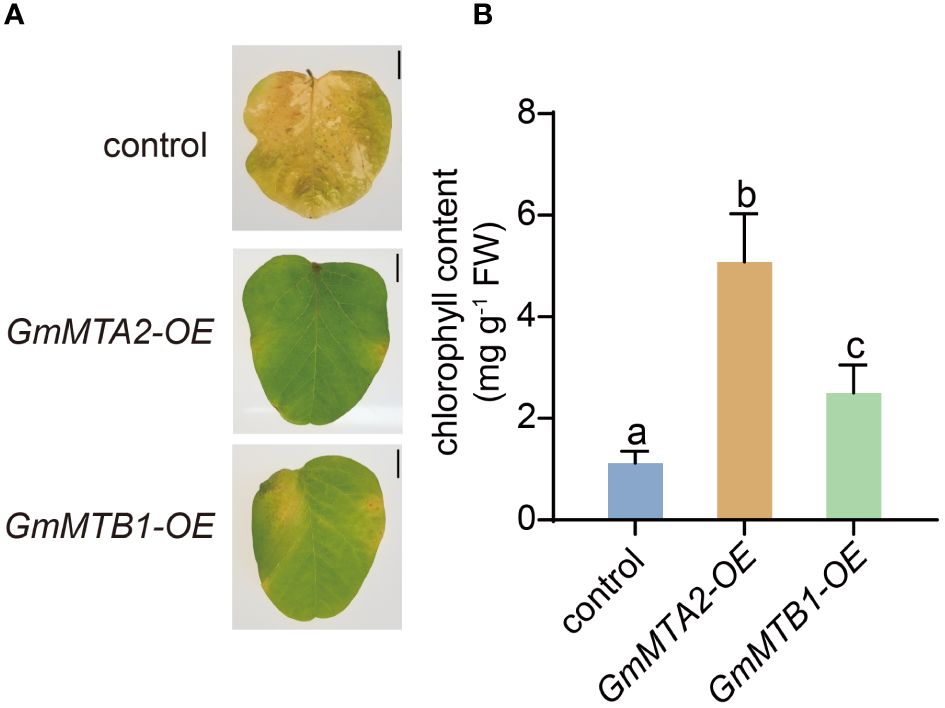
Figure 10 Leaf phenotypes of the control, GmMTA2-OE, and GmMTB1-OE plants upon darkness treatment. (A) The phenotypic changes of the leaves subjected to darkness for 10 days. Bar, 1 cm. (B) Chlorophyll contents of leaves from the dark-treated plants. Error bars represent SD (n = 3). Different letters indicate significant differences at P < 0.05.
Discussion
Here, we identified and characterized the gene family members of the m6A writer complex in legume plants. They were assigned to four families: MT-A70, WTAP, VIR, and HAKAI. In soybean, the MT-A70 family comprised five members, the WTAP subfamily had four members, and both the HAKAI and VIR families were represented by only two members each (Figure 1). Among these m6A writer candidates, the enzymatic activities of GmMTA1 and GmMTA2 were confirmed in an in vitro assay (Figure 5). In addition, the gene structures of all candidates in G. max were analyzed, with similar structures and conserved motifs present among the members of each family. The gene structures varied between members of the different families; however, the VIR members exhibited more exons per gene (average of 27), while members of the HAKAI family had the fewest (only three exons per gene, on average; Figure 2). Upon analyzing the promoter regions of the m6A writer complex genes, several cis elements were identified. These cis elements were subsequently categorized into five functional clusters: responsive to light, phytohormone signaling (such as MeJA and ABA), environmental stress (such as low temperature or drought), development and other regulation (Figure 6). Simultaneously, we performed a subcellular localization analysis of all members of the MT-A70 family from G. max, revealing them all to be located within the nucleus, in line with our prediction (Table 1).
Abiotic stress reduces crop yields and can even kill the plant; therefore, it is critical to understand how soybean responds to environmental stresses, such as saline–alkali stress, cold stress, drought stress, and darkness (Hu et al., 2021; Zheng et al., 2021). There is mounting evidence that m6A modifications play crucial roles in regulating plant responses to these stresses. In Arabidopsis, salt stress was reported to significantly affect the m6A methylation levels on mRNA (Hu et al., 2021). Additionally, VIR-mediated m6A methylation modulates ROS homeostasis by negatively regulating the mRNA stability of several negative regulators of the salt stress response, such as Arabidopsis NAC transcription factor (AtATAF1), GIGANTEA (AtGI), and glutathione S-transferaset U17 (AtGSTU17) (Zheng et al., 2021). In apple, MdMTA enhances lignin deposition and ROS scavenging under drought conditions. In poplar (Populus trichocarpa), plants overexpressing PtrMTA had a higher trichome density and a more developed root system (Lu et al., 2020). Moreover, they exhibited better drought tolerance (Lu et al., 2020). These findings show that the m6A modification plays a pivotal role in the abiotic stress responses across diverse plant species, but our understanding of these mechanisms in soybean remains very limited. Here, we identified key m6A component genes in soybean and analyzed their roles in different abiotic stress conditions, including cold, heat, drought, salinity, alkalinity, and darkness. Our data suggest that GmMTAs and GmMTBs, as core components of the m6A writer complex, are altered in response to various stressors, especially induced upon drought, alkalinity, and darkness treatment; however, their expression patterns and response times differ in accordance with the specific stress conditions (Figure 7).
Leaf senescence has a crucial effect on crop quality and yield. It is an age-dependent process that can be regulated by several factors, including leaf age, phytohormones, temperature, and light. Upon entering the senescence stage, a leaf’s cells undergo a sequential disorganization of cellular organelles, accompanied by systematic changes in metabolism and gene expression (Woo et al., 2019; Guo et al., 2021). Under darkness, the key transcription factors PHYTOCHROME-INTERACTING FACTOR 4/5 (PIF4/5) in the light signaling pathway are activated and regulate the expression of chlorophyll-catabolic genes through the ABA or ethylene pathways, accelerating chlorophyll degradation and promoting leaf yellowing and senescence (Sakuraba et al., 2014; Song et al., 2014). Long-term darkness has commonly been used as a tool to investigate the process of leaf senescence (Guo et al., 2021). In addition to transcriptional regulation, epigenetic modification also plays an important role in the regulation of leaf senescence. In Arabidopsis, the mutation of MTA resulted in a more pronounced aging phenotype than was observed in the wild type (Sheikh et al., 2024). The m6A levels in the mRNA were shown to increase during a darkness treatment, preventing premature aging by destabilizing transcripts of age-related genes (Sheikh et al., 2024). We assessed the expression levels of the GmMTAs and GmMTBs under a darkness treatment and observed a significant upregulation after 6-12 h (Figure 7). In particular, GmMTA2 showed continuous induction and maintained high expression levels after 72 h of treatment (Figure 7). Furthermore, we overexpressed GmMTA2 and GmMTB1 in soybean leaves, which resulted in a more prominent anti-aging phenotype and higher chlorophyll content (Figure 10). These findings suggest that soybean GmMTA2 and GmMTB1 play essential roles in dark-induced leaf senescence, providing additional evidence for m6A involvement in crop or fruit ripening (Zhou et al., 2022). In animal studies related to m6A, METTL3 (the human homolog of MTA) is also vital for slowing the aging of human mesenchymal stem cells (Wu et al., 2020); hence, further research into the role of m6A in aging regulation is warranted.
Conclusion
In this study, a total of thirteen m6A writer genes in G. max, thirteen in G. soja, six in P. vulgaris, eight in M. truncatula, and six in L. japonicus were identified. The phylogenetic analysis divided these genes into four families based on their topological structure. In soybean, the collinearity analysis revealed members from each m6A writer family originated from gene duplication. WoLF PSORT prediction coupled with subcellular localization analysis suggested that these m6A writer genes all localized in nucleus. Furthermore, enzymatic analysis showed that both GmMTA1 and GmMTA2 possessed the methyltransferase activities toward adenosine on RNA. The cis-acting elements of 2000 bp promoter regions of all m6A writer genes were investigated and the expression pattern of four MT-A70 family members upon abiotic stress treatment were determined. The results suggested that all members, especially GmMTA2 and GmMTB1, were involved in the cellular response to various abiotic stress. Notably, soybean leaves overexpressing GmMTB1 exhibited more resistant to the alkalinity, while overexpressing GmMTA2 or GmMTB1 both showed the highest tolerance to darkness treatment. These results will provide a basis for further exploring the biological functions of the m6A writer genes from legume plants in growth regulation and stress responses.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
PL: Data curation, Formal analysis, Methodology, Writing – original draft. HL: Writing – original draft, Data curation, Formal analysis. JZ: Data curation, Formal analysis, Methodology, Writing – original draft. TY: Data curation, Writing – original draft. SG: Data curation, Writing – original draft. LC: Data curation, Writing – original draft. TX: Data curation, Writing – original draft. AX: Data curation, Writing – original draft. XL: Formal analysis, Methodology, Writing – original draft. CZ: Formal analysis, Methodology, Writing – original draft. LG: Formal analysis, Methodology, Writing – original draft. MC: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (grant number 32371365), the Natural Science Foundation of Jiangsu Province, China (grant number BK20230101), the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the start-up fund for advanced talents from Nanjing Agricultural University to MC.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1446591/full#supplementary-material
References
Anderson, S. J., Kramer, M. C., Gosai, S. J., Yu, X., Vandivier, L. E., Nelson, A. D. L., et al. (2018). N6-methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in Arabidopsis. Cell Rep. 25, 1146–1157. doi: 10.1016/j.celrep.2018.10.020
Baccolini, C., Witte, C.-P. (2019). AMP and GMP catabolism in Arabidopsis converge on xanthosine, which is degraded by a nucleoside hydrolase heterocomplex. Plant Cell 31, 734–751. doi: 10.1105/tpc.18.00899
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4. doi: 10.1186/1471-2229-4-10
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, M., Herde, M., Witte, C.-P. (2016). Of the nine cytidine deaminase like genes in Arabidopsis thaliana eight are pseudogenes and only one is required to maintain pyrimidine homeostasis in vivo. Plant Physiol. 171, 799–809. doi: 10.1104/pp.15.02031
Chen, M., Urs, M. J., Sánchez-González, I., Olayioye, M. A., Herde, M., Witte, C.-P. (2018). m6A RNA degradation products are catabolized by an evolutionarily conserved N6-Methyl-AMP deaminase in plant and mammalian cells. Plant Cell 30, 1511–1522. doi: 10.1105/tpc.18.00236
Chen, M., Witte, C.-P. (2020). A kinase and a glycosylase catabolize pseudouridine in the peroxisome to prevent toxic pseudouridine monophosphate accumulation. Plant Cell 32, 722–739. doi: 10.1105/tpc.19.00639
Cheng, C., Liu, Y., Liu, X., An, J., Jiang, L., Yu, B. (2019). Recretohalophyte Tamarix TrSOS1 confers higher salt tolerance to transgenic plants and yeast than glycophyte soybean GmSOS1. Environ. Exp. Bot. 165, 196–207. doi: 10.1016/j.envexpbot.2019.06.006
Delaunay, S., Helm, M., Frye, M. (2023). RNA modifications in physiology and disease: towards clinical applications. Nat. Rev. Genet. 25, 104–122. doi: 10.1038/s41576-023-00645-2
Feng, N., Yu, M., Li, Y., Jin, D., Zheng, D. (2021). Prohexadione-calcium alleviates saline-alkali stress in soybean seedlings by improving the photosynthesis and up-regulating antioxidant defense. ECOTOX Environ. SAFE 220, 112369. doi: 10.1016/j.ecoenv.2021.112369
Frye, M., Harada, B. ,. T., Behm, M., He, C. (2018). RNA Modifications modulate gene expression during development. Science 361, 1346–1349. doi: 10.1126/science.aau1646
Frye, M., Jaffrey, S. R., Pan, T., Rechavi, G., Suzuki, T. (2016). RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet. 17, 365–372. doi: 10.1038/nrg.2016.47
Fu, Y., Guo, C., Wu, H., Chen, C. (2017). Arginine decarboxylase ADC2 enhances salt tolerance through increasing ROS scavenging enzyme activity in Arabidopsis thaliana. Plant Growth Regul. 83, 253–263. doi: 10.1007/s10725-017-0293-0
Gao, S., Sun, Y., Chen, X., Zhu, C., Liu, X., Wang, W., et al. (2023). Pyrimidine catabolism is required to prevent the accumulation of 5-methyluridine in RNA. Nucleic Acids Res. 51, 7451–7464. doi: 10.1093/nar/gkad529
Guo, Y., Ren, G., Zhang, K., Li, Z., Miao, Y., Guo, H. (2021). Leaf senescence: progression, regulation, and application. Mol. Hortic. 1, 5. doi: 10.1186/s43897-021-00006-9
Guo, T., Yang, Z., Bao, R., Fu, X., Wang, N., Liu, C., et al. (2023). The m6A reader MhYTP2 regulates the stability of its target mRNAs contributing to low nitrogen tolerance in apple (Malus domestica). Hortic 10, uhad094. doi: 10.1093/hr/uhad094
Han, B., Wei, S., Li, F., Zhang, J., Li, Z., Gao, X. (2021). Decoding m6A mRNA methylation by reader proteins in cancer. Cancer Lett. 518, 256–265. doi: 10.1016/j.canlet.2021.07.047
Hou, N., Li, C., He, J., Liu, Y., Yu, S., Malnoy, M., et al. (2022). MdMTA-mediated m6A modification enhances drought tolerance by promoting mRNA stability and translation efficiency of genes involved in lignin deposition and oxidative stress. New Phytol. 234, 1294–1314. doi: 10.1111/nph.18069
Hu, J., Cai, J., Park, S. J., Lee, K., Li, Y., Chen, Y., et al. (2021). N6-Methyladenosine mRNA methylation is important for salt stress tolerance in Arabidopsis. Plant J. 106, 1759–1775. doi: 10.1111/tpj.15270
Ji, X., Liu, G., Liu, Y., Zheng, L., Nie, X., Wang, Y. (2013). The bZIP protein from Tamarix hispida, ThbZIP1, is ACGT elements binding factor that enhances abiotic stress signaling in transgenic Arabidopsis. BMC Plant Biol. 13, 151. doi: 10.1186/1471-2229-13-151
Jia, G., Fu, Y., He, C. (2013). Reversible RNA adenosine methylation in biological regulation. Trends Genet. 29, 108–115. doi: 10.1016/j.tig.2012.11.003
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., Bansal, K. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom 13, 544. doi: 10.1186/1471-2164-13-544
Kim, E., Hwang, S., Lee, I. (2017). SoyNet: a database of co-functional networks for soybean Glycine max. Nucleic Acids Res. 45, D1082–D1089. doi: 10.1093/nar/gkw704
Kost, B., Spielhofer, P., Chua, N. H. (2008). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. doi: 10.1046/j.1365-313x.1998.00304.x
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, Z., Lyu, X., Li, H., Tu, Q., Zhao, T., Liu, J., et al. (2024). The mechanism of low blue light-induced leaf senescence mediated by GmCRY1s in soybean. Nat. Commun. 15, 798. doi: 10.1038/s41467-024-45086-5
Lim, C., Kang, K., Shim, Y., Yoo, S. C., Paek, N. C. (2022). Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 188, 1900–1916. doi: 10.1093/plphys/kiab492
Liu, Y., Zhang, Y., Liu, X., Shen, Y., Tian, D., Yang, X., et al. (2023). SoyOmics: A deeply integrated database on soybean multi-omics. Mol. Plant 16, 794–797. doi: 10.1016/j.molp.2023.03.011
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, L., Zhang, Y., He, Q., Qi, Z., Zhang, G., Xu, W., et al. (2020). MTA, an RNA m6A Methyltransferase, enhances drought tolerance by regulating the development of trichomes and roots in Poplar. Int. J. Mol. Sci. 21, 2462. doi: 10.3390/ijms21072462
Martin, K., Kopperud, K., Chakrabarty, R., Banerjee, R., Brooks, R., Goodin, M. M. (2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59, 150–162. doi: 10.1111/j.1365-313X.2009.03850.x
Phukan, U. J., Jeena, G. S., Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00760
Pi, B., Liu, X., Huang, Q., Zhang, T., Yu, B. (2023). Comparative transcriptomic analysis of Glycine soja and G. max and functional identification of GsCNGC20-d interacted with GsCDPK29 under salt stress. Environ. Exp. Bot. 206, 105185. doi: 10.1016/j.envexpbot.2022.105185
Qi, D. H., Lee, C. F. (2014). Influence of soybean biodiesel content on basic properties of biodiesel-diesel blends. J. Taiwan Inst. Chem. Eng. 45, 504–507. doi: 10.1016/j.jtice.2013.06.021
Qu, Y., Guan, R., Yu, L., Berkowitz, O., David, R., Whelan, J., et al. (2022). Enhanced reactive oxygen detoxification occurs in salt-stressed soybean roots expressing GmSALT3. Physiol. Plant 174, e13709. doi: 10.1111/ppl.13709
Roundtree, I. A., Evans, M. E., Pan, T., He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi: 10.1016/j.cell.2017.05.045
Růžička, K., Zhang, M., Campilho, A., Bodi, Z., Kashif, M., Saleh, M., et al. (2017). Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215, 157–172. doi: 10.1111/nph.14586
Sakuraba, Y., Jeong, J., Kang, M.-Y., Kim, J., Paek, N.-C., Choi, G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5, 4636. doi: 10.1038/ncomms5636
Sheikh, A. H., Tabassum, N., Rawat, A., Almeida, T. M., Nawaz, K., Hirt, H. (2024). m6A RNA methylation counteracts dark-induced leaf senescence in Arabidopsis. Plant Physiol. 194, 2663–2678. doi: 10.1093/plphys/kiad660
Shen, L., Liang, Z., Gu, X., Chen, Y., Teo, Z. W. N., Hou, X., et al. (2016). N6-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev. Cell 38, 186–200. doi: 10.1016/j.devcel.2016.06.008
Shi, H., Wei, J., He, C. (2019). Where, When, and How: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650. doi: 10.1016/j.molcel.2019.04.025
Song, Y., Yang, C., Gao, S., Zhang, W., Li, L., Kuai, B. (2014). Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 7, 1776–1787. doi: 10.1093/mp/ssu109
Tamura, K., Stecher, G., Kumar, S., Battistuzzi, F. U. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
van Zelm, E., Zhang, Y., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Vespa, L., Vachon, G., Berger, F., Perazza, D., Faure, J.-D., Herzog, M. (2004). The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 134, 1283–1292. doi: 10.1104/pp.103.028050
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016b). Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 534, 575–578. doi: 10.1038/nature18298
Wang, T., Guo, J., Peng, Y., Lyu, X., Liu, B., Sun, S., et al. (2023a). Light-induced mobile factors from shoots regulaterhizobium-triggered soybean root nodulation. Science 374, 65–71. doi: 10.1126/science.abh2890
Wang, W., Liu, H., Wang, F., Liu, X., Sun, Y., Zhao, J., et al. (2023b). N4-acetylation of cytidine in mRNA plays essential roles in plants. Plant Cell 35, 3739–3756. doi: 10.1093/plcell/koad189
Wang, F., Liu, J., Zhou, L., Pan, G., Li, Z., Zaidi, S.-H.-R., et al. (2016a). Senescence-specific change in ROS scavenging enzyme activities and regulation of various SOD isozymes to ROS levels in psf mutant rice leaves. Plant Physiol. Biochem. 109, 248–261. doi: 10.1016/j.plaphy.2016.10.005
Woo, H. R., Kim, H. J., Lim, P. O., Nam, H. G. (2019). Leaf senescence: systems and dynamics aspects. Annu. Rev. Plant Biol. 70, 347–376. doi: 10.1146/annurev-arplant-050718-095859
Wu, Z., Shi, Y., Lu, M., Song, M., Yu, Z., Wang, J., et al. (2020). METTL3 counteracts premature aging via m6A-dependent stabilization of MIS12 mRNA. Nucleic Acids Res. 48, 11083–11096. doi: 10.1093/nar/gkaa816
Xie, J., Chen, Y., Cai, G., Cai, R., Hu, Z., Wang, H. (2023). Tree Visualization By One Table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 51, W587–W592. doi: 10.1093/nar/gkad359
Xing, K., Liu, Z., Liu, L., Zhang, J., Qanmber, G., Wang, Y., et al. (2023). N6-methyladenosine mRNA modification regulates transcripts stability associated with cotton fiber elongation. Plant J. 115, 967–985. doi: 10.1111/tpj.16274
Yang, L., Han, Y., Wu, D., Yong, W., Liu, M., Wang, S., et al. (2017). Salt and cadmium stress tolerance caused by overexpression of the Glycine Max Na+/H+ Antiporter (GmNHX1) gene in duckweed (Lemna turionifera 5511). Aquat. Toxicol. 192, 127–135. doi: 10.1016/j.aquatox.2017.08.010
Zheng, H., Sun, X., Li, J., Song, Y., Song, J., Wang, F., et al. (2021). Analysis of N6-methyladenosine reveals a new important mechanism regulating the salt tolerance of sweet sorghum. Plant Sci. 304, 110801. doi: 10.1016/j.plantsci.2020.110801
Zhong, S., Li, H., Bodi, Z., Button, J., Vespa, L., Herzog, M., et al. (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288. doi: 10.1105/tpc.108.058883
Zhou, L., Gao, G., Tang, R., Wang, W., Wang, Y., Tian, S., et al. (2022). m6A-mediated regulation of crop development and stress responses. Plant Biotechnol. 20, 1447–1455. doi: 10.1111/pbi.13792
Keywords: m6A, soybean, RNA methylation, abiotic stress, MTA, MTB
Citation: Liu P, Liu H, Zhao J, Yang T, Guo S, Chang L, Xiao T, Xu A, Liu X, Zhu C, Gan L and Chen M (2024) Genome-wide identification and functional analysis of mRNA m6A writers in soybean under abiotic stress. Front. Plant Sci. 15:1446591. doi: 10.3389/fpls.2024.1446591
Received: 10 June 2024; Accepted: 25 June 2024;
Published: 11 July 2024.
Edited by:
Huatao Chen, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaCopyright © 2024 Liu, Liu, Zhao, Yang, Guo, Chang, Xiao, Xu, Liu, Zhu, Gan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingjia Chen, bWpjaGVuQG5qYXUuZWR1LmNu
†These authors have contributed equally to this work
 Peng Liu1†
Peng Liu1† Mingjia Chen
Mingjia Chen