- 1Wheat Research Institute, School of Life Sciences and Engineering, Southwest University of Science and Technology, Mianyang, Sichuan, China
- 2College of Tobacco Science of Guizhou University, Key Laboratory of Plant Resource Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education), Guizhou Key Lab of Agro-Bioengineering, Guiyang, China
- 3US Department of Agriculture, Agricultural Research Service, Wheat Health, Genetics, and Quality Research Unit, and Department of Plant Pathology, Washington State University, Pullman, WA, United States
- 4State Key Laboratory of Crop Stress Biology in Arid Areas and College of Plant Protection, Northwest A&F University, Xianyang, Shaanxi, China
Stripe rust, caused by Puccinia striiformis f. sp. tritici (Pst), is a serious disease that affects wheat worldwide. There is a great need to develop cultivars with combinations of all-stage resistance (ASR) and adult-plant resistance (APR) genes for sustainable control of the disease. QYrsv.swust-1BL in the Italian durum wheat (Triticum turgidum ssp. durum) cultivar Svevo is effective against Pst races in China and Israel, and the gene has been previously mapped to the long arm of chromosome 1B. The gene is flanked by SNP (single nucleotide polymorphism) markers IWB5732 and IWB4839 (0.75 cM). In the present study, we used high-density 660K SNP array genotyping and the phenotypes of 137 recombinant inbred lines (RILs) to fine map the QYrsv.swust-1BL locus within a 1.066 Mb region in durum wheat Svevo (RefSeq Rel. 1.0) on chromosome arm 1BL. The identified 1.066 Mb region overlaps with a previously described map of Yr29/QYr.ucw-1BL, a stripe rust APR gene. Twenty-five candidate genes for QYrsv.swut-1BL were identified through comparing polymorphic genes within the 1.066 Mb region in the resistant cultivar. SNP markers were selected and converted to Kompetitive allele-specific polymerase chain reaction (KASP) markers. Five KASP markers based on SNP were validated in a F2 and F2:3 breeding population, providing further compelling evidence for the significant effects of QYrsv.swut-1BL. These markers should be useful in marker-assisted selection for incorporating Yr29/QYrsv.swust-1BL into new durum and common wheat cultivars for resistance to stripe rust.
Introduction
Wheat is a widely cultivated grain crop and is critical for global food security. Thus, it is important to limit pathogen-caused losses in wheat yield. Stripe rust (Yellow rust, Yr), caused by the fungal pathogen Puccinia striiformis f. sp. tritici (Pst), is a growing threat to global wheat production (Chen, 2005, 2020). Although fungicides can control stripe rust, this approach can be costly and harmful to humans, animals, and the environment. The most economical, effective, and environmentally friendly approach to controlling stripe rust is to develop resistant cultivars. However, because of co-evolution pressures, race-specific resistance can be ultimately overcome by new races of the pathogen, often within just a few years (Beddow et al., 2015). There is an urgent need to identify effective stripe rust-resistant genes and develop new resistant cultivars.
At present, 86 formally named resistance genes, more than 70 temporarily named genes, and more than 380 quantitative trait loci (QTL) for resistance to stripe rust have been identified from common wheat, durum wheat, and wild relatives (Bansal et al., 2014; Wang and Chen, 2017; Feng et al., 2018; Nsabiyera et al., 2018; Gessese et al., 2019; Maccaferri et al., 2019; Pakeerathan et al., 2019; Li et al., 2020; Singh et al., 2021; Klymiuk et al., 2022; Yao et al., 2022; Feng et al., 2023; Zhu et al., 2023). However, most of the genes were overcome by the pathogen shortly after being introduced to commercial cultivars. Race-specific (mostly all-stage resistance genes) and non-race-specific [mostly adult-plant resistance (APR) genes] are two major classes of resistance to wheat stripe rust (Chen et al., 2013). Race-specific resistance genes typically encode nucleotide-binding site-leucine-rich repeat (NBS-LRR) resistance proteins. These proteins can detect the presence of pathogen effectors or modify the defense proteins they produce in the host, thereby triggering hypersensitive reactions and reducing the growth of pathogens (Periyannan et al., 2013; Zhang et al., 2017; Chen et al., 2018; Marchal et al., 2018). However, many race-specific resistance genes in commercial cultivars are no longer effective. This is because amino acid changes in the effectors can help the pathogen evade recognition by the nucleotide-binding leucine-rich repeat (NB-LRR) genes (Chen et al., 2017; Salcedo et al., 2017; Cobo et al., 2019). By contrast, APR genes typically show more durable but less strong resistance than all-stage race-specific genes (Krattinger et al., 2009; Singh et al., 2011; Chen et al., 2013). Three APR genes against wheat stripe rust have been cloned, encoding proteins that are more complex and diverse than NBS-LRR. Yr18 encodes an ATP-binding cassette (ABC) transporter (Krattinger et al., 2009); Yr36 encodes a kinase and a putative START lipid-binding domain (Fu et al., 2009; Gou et al., 2015); and Yr46 encodes a hexose transporter (Moore et al., 2015). Using APR is a demonstrated approach to developing wheat cultivars with durable resistance to stripe rust (Chen et al., 2013; Liu et al., 2018, 2019).
Stripe rust resistance QTL QYrsv.swust-1BL was previously mapped between the linked SNP markers IWB5732 and IWB4839 in a 0.75 cM (1.74 Mb) region derived from durum wheat cultivar Svevo (Zhou et al., 2021), which overlaps the map of Yr29, a long-term utilized gene for APR to stripe rust (William et al., 2003; Kolmer, 2015). The known pleiotropic APR gene Yr29/Lr46/Sr58/Pm39 has been reported in several studies (Ren et al., 2017; Cobo et al., 2018, 2019; Herrera-Foessel et al., 2011; Ponce-Molina et al., 2018; Li et al., 2020). Yr29/Lr46 (Singh et al., 1998; William et al., 2003) provides partial APR to rust diseases (Herrera-Foessel et al., 2015) and powdery mildew (Lillemo et al., 2008, 2013) in wheat. Yr29/Lr46 is also associated with the leaf tip necrosis (Ltn) gene Ltn2 (William et al., 2003; Rosewarne et al., 2006). QYrsv.swust-1BL explained 11.0% to 34.4% of the phenotypic variation and was effective across all tested environments in China and Israel. The phenotypic variance explained by QYrsv.swust-1BL and Yr29 was very similar (Zhou et al., 2021). However, it is not certain whether the two genes are the same gene, allelic genes, or closely linked genes. In addition, the flanking markers were not sufficiently close for efficient marker-assisted selection.
The objectives of this study were to genotype recombinant inbred lines (RILs) used in the previous study (Zhou et al., 2021) using the wheat 660K SNP iSelect array, produce a high-resolution map for the resistance gene QYrsv.swust-1BL in Svevo with more markers, identify candidates for this resistance gene, transfer QYrsv.swust-1BL into common wheat, and develop user-friendly markers for validation to hasten the use of this gene in breeding programs.
Materials and methods
Plant materials
To fine-map the locus of QYrsv.swust-1BL for stripe rust APR in Svevo, 137 RILs developed from Svevo/Zavitan (Avni et al., 2014; Zhou et al., 2021) were used as a mapping population. To transfer and validate stripe rust resistance from durum wheat Svevo to the Chinese common wheat (T. aestivum) background, Svevo was crossed with Mianmai 37 (MM 37), a widely grown common wheat cultivar developed by the Wheat Research Institute, Mianyang Academy of Agricultural Sciences, Sichuan, China. However, this cultivar has become susceptible to stripe rust in recent years. Twelve F1 plants derived from MM 37/Svevo were selfed to generate 474 F2 plants. Due to sterility, only 318 F2:3 families were successfully harvested from 474 F2 plants. F2 plants correspond to 318 F2:3 lines, which were phenotyped for stripe rust response and used to detect the availability of markers. Common wheat cultivars Jinmai 47 (JM 47) and Mingxian 169 (MX 169) were used in the phenotyping experiments as stripe rust-susceptible checks.
Field evaluation
In 2019 and 2020, the 137 RILs of Svevo/Zavitan were phenotyped for their responses to stripe rust in Yangling (YL, 34.292N, 108.077E), Shaanxi Province, and Mianyang (MY, 31.682N, 104.663E), Sichuan Province, China. A randomized complete block design was used, with two replicates in each year at each location. For each line in each replicate, 25 to 30 seeds were sown in a 1-m row, with 25 cm separating the rows. As susceptible checks, JM 47 was planted in every 20th row and MX 169 around the nursery to increase the pathogen inoculum. The YL site was planted on 6 October 2019 and 10 October 2020, and the MY site on 12 November 2019 and 15 November 2020. In Yangling, when flag leaves emerged [Zadoks growth stage (GS) 40] (Zadoks et al., 1974) in late March, the plants were inoculated with a mixture of equal amounts of urediniospores of Pst races CYR32, CYR33, and CYR34, which were predominant races in China. The fields in Mianyang were tested for natural Pst infection in these two growing seasons, as this region is one of the major Pst inoculum sources in China (Chen et al., 2014). The parents, 12 F1, and 474 F2 plants of MM 37/Svevo were grown at the research farm of Southwest University of Science and Technology in Mianyang during the 2017–2018 and 2018–2019 growing seasons, respectively. Three hundred eighteen F2:3 family seeds were harvested from 474 F2 plants and were planted, together with the parental lines, at Mianyang on 12 November 2019 in a randomized complete block design with two replications. Infection type (IT) based on the 0–9 scale (Line and Qayoum, 1992) and disease severity (DS) as percentage of infected foliage were recorded for each parent and RIL when JM 47 and MX 169 had 80% or higher DS at Zadoks growth stages (GS) 65–69.
Phenotypic analysis
Relative to the phenotypic scores of the parents and susceptible checks, the RILs of Svevo/Zavitan and the F2:3 families of MM 37/Svevo were classified into the groups of homozygous resistant (HR) and homozygous susceptible (HS). In addition, the segregating F2:3 families were classified as segregating (Seg.). An analysis of variance (ANOVA) was conducted based on the IT and DS data of the populations in the tests of different years and fields (considered as different environments) using the “AOV” tool in QTL IciMapping V4.2 software (Wang, 2009; Meng et al., 2015).
DNA extraction, KASP marker development, and genotyping the RIL and F2 populations
Total genomic DNA was extracted from the parents, 137 RILs of Svevo/Zavitan, and 318 F2 plants from MM 37/Svevo using the cetyltrimethylammonium bromide (CTAB) method (Anderson et al., 1993). To identify closer SNP markers and saturate the targeted QTL region, the genetic maps of the RIL mapping population produced in the previous studies with the 90K SNP array (Avni et al., 2014; Zhou et al., 2021) and the sequences of closely linked polymorphic SNPs were used to BLAST search version 1.0 of the Chinese Spring and Svevo genome sequences from the International Wheat Genome Consortium (IWGSC) and at the GrainGenes Svevo Genome Browser (Durum Wheat Svevo RefSeq Rel. 1.0), respectively. Svevo, Zavitan, and MM 37 were first genotyped with the 660K SNP array by China Golden Marker Biotech Co., Ltd. (Beijing, China). The Kompetitive allele-specific polymerase chain reaction (KASP) markers were developed from the 660K SNP map (http://wheat.pw.usda.gov/ggpages/topics/Wheat660_SNP_array_developed_by_CAAS.pdf). Based on chromosome positions in the 660K SNP map, SNPs within a more conservative target region between IWB8812 and IWB4839 were selected for conversion to KASP markers (Figure 1). The KASP markers were used to screen the Svevo and Zavitan to confirm polymorphisms before genotyping the entire RIL population. Similarly, before screening the 318 F2 plants, MM 37 and Svevo were tested with the KASP markers to confirm the polymorphisms before the markers were used to genotype the 318 F2 plants from the MM 37/Svevo cross. The design of KASP primers, PCR amplification, and marker detection were conducted following the PolyMarker method (Ramirez-Gonzalez et al., 2015; Wu et al., 2018).
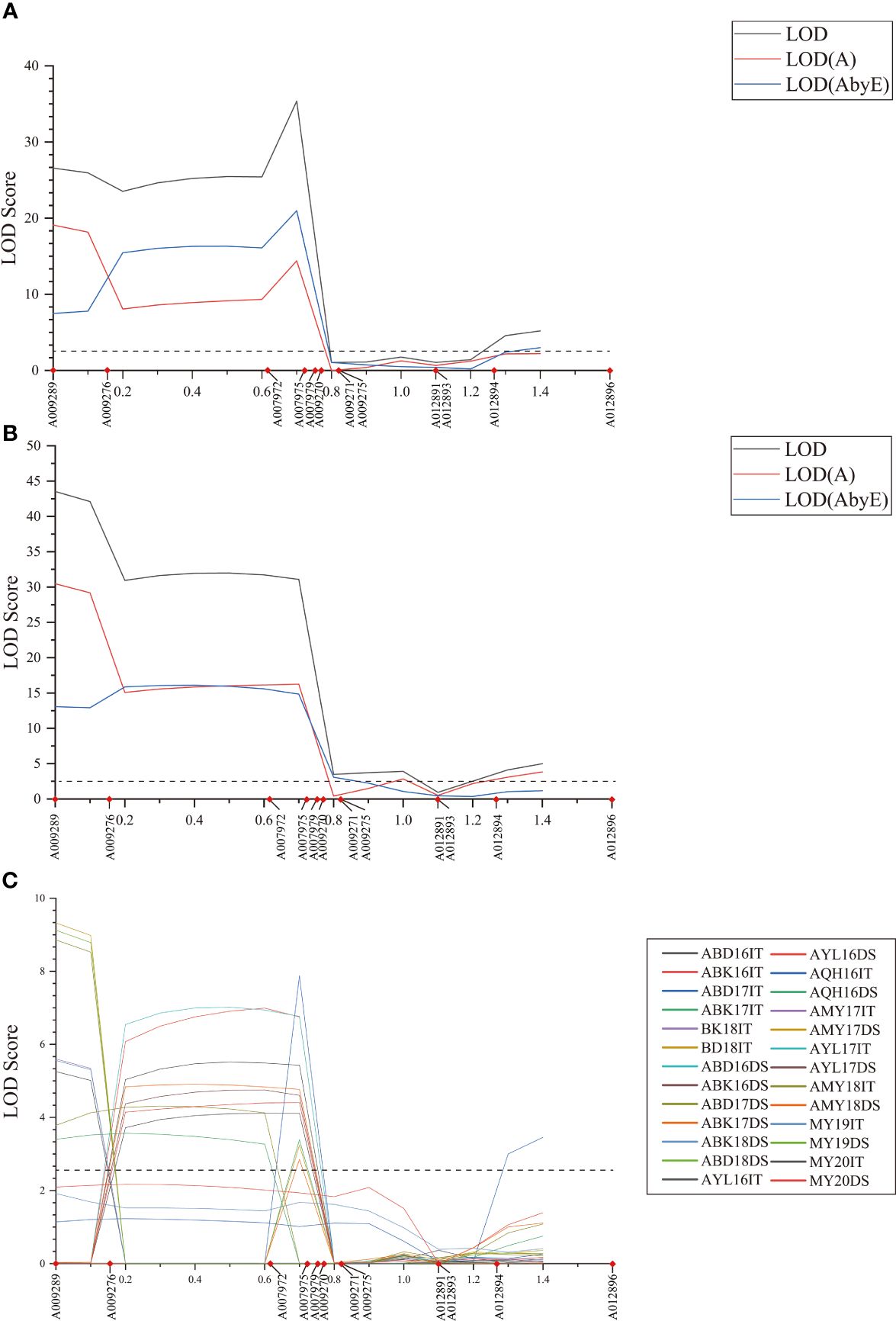
Figure 1 LOD scores of the MET (A, B) and BIP (C) analyses on infection type (IT) and disease severity (DS) values in stripe rust resistance QYrsv.swust-1BL. LOD, the total Lod score; LOD (A), the additive effect component of the LOD; LOD (AbyE), the environmental by additive effect component of the LOD. The X-axis represents the genetic distance of the chromosomes. The Y-axis represents the threshold LOD score.
Construction of genetic maps and QTL mapping
Genome Studio Polyploid Clustering v1.0 (Illumina, New York, USA) was used for genotype calling and clustering. Markers with >20% of missing data were excluded from further analyses. Marker binning was done using pattern segregation methods (Wang, 2009; Meng et al., 2015). The linkage groups were ordered, and non-binned markers were placed in the groups based on their genome positions. The Chi-squared test was used to detect any segregation distortions. Markers with a p-value of < 0.01 in the Chi-squared test were removed prior to generating the genetic map.
Genetic linkage maps of the KASP markers were constructed using the previously described procedures (Zhou et al., 2019; 2021). The likelihood of an odd score of 2.5 was set as the threshold for linkages between loci, and QTL mapping was conducted based on the phenotyping and genotyping data using the software IciMapping V4.2 (Wang, 2009; Meng et al., 2015). Mapping was conducted first using the BIP (biparental population) module and the ICIM-ADD model in the years 2019–2020 and by combining the data from the previous years 2016–2018 from all environments. The MET (multi-environmental trials) module was also employed to identify the QTL loci in the years 2016–2020 in all environments. The methodology used for the BIP and MET analyses closely follows the approach described in Zhou et al. (2021). But the mapping parameters were step=0.1 cM and PIN=0.001 (probability of SNP to be included in the model).
Identification of candidate genes
The probes of the SNP identified in the QTL region were aligned to the newly released Chinese Spring sequences (IWGSC RefSeq V1.0, annotation V1.1, the International Wheat Genome Consortium (IWGSC), https://www.wheatgenome.org/about/iwgsc-2.0) through BLAST searching. This provided physical positions, reference sequences, annotations, and alignments of the polymorphic SNP markers. The SNP probes were also BLAST searched against the Svevo genome sequences at the GrainGenes Svevo Genome Browser (Durum Wheat Svevo RefSeq Rel. 1.0). Annotated genes in the target region were bioinformatically analyzed using the NCBI (https://bl-ast.ncbi.nlm.nih.gov/Blast.cgi) and Genoscope (https://www.cea.fr/drf/ifr-ancoisjacob/Pages/Departements/Genoscope.aspx) databases and the Web site-based software FGENSH (http://www.softberry.com/berry.phtml?topic=fgenesh&group=he-lp&subgroup=gfind) for their putative functions and possible involvement in plant disease resistance to identify candidates of the stripe rust resistance genes.
Results
Developing KASP markers
Using the initial markers IWB8812 and IWB4839 from the 90K wheat SNP array as the conservative flanking loci, QYrsv.swust-1BL from Svevo was mapped in the 1.855 Mb (physical position: 669,649,666 ~ 671,505,587 bp) region corresponding to the Chinese Spring IWGSC RefSeq v1.0 1BL chromosome, which includes the segments previously reported and considered as QYr.ucw-1BL/Yr29 (Cobo et al., 2019; Zhou et al., 2021). By comparing the genome sequences of durum wheat Svevo RefSeq Rel. 1.0 (the GrainGenes Svevo Genome Browser) and Chinese Spring IWGSC RefSeq v1.0 (https://urgi.versailles.inra.fr/download/iwgsc/), a nucleic acid database was constructed with the genome sequences of Svevo. The sequences of 630,517 SNP probes from the 660K SNP array were compared to the genome sequences of Svevo by BLASTn. All SNP probes that matched the expected sequences of the 660,683,255 ~ 663,334,719 bp region of the Svevo chromosome 1BL were selected. Similarly, in the Chinese Spring genome, SNPs in the 669,649,666 ~ 671,505,587 bp region were identified. Based on both physical positions of the commonly polymorphic SNP loci on 1BL, a total of 42 SNP markers were selected from the wheat 660K SNP array in the 669,649,666 ~ 671,505,587 bp region, from which 21 KASP markers were successfully developed. After confirming the polymorphisms between the parental lines (Svevo and Zavitan), thirteen KASP markers (A009289, A009276, A009272, A007975, A007979, A009270, A009271, A007972, A009275, A012891, A012893, A012894, and A012896) successfully distinguished Svevo and Zavitan (Table 1). A total of 47 wheat DNA samples, including the 2 parents and 23 resistant and 22 susceptible RILs from the Svevo/Zavitan cross, were selected to verify these markers, which were then tested on 137 RILs from the Svevo/Zavitan cross. The sequences of the 13 KASP markers are shown in Table 1. The SNP sequences associated with QYrsv.swust-1BL identified from the wheat 660K SNP array are shown in Table 2, which were used to design the KASP primer sequence. When these 13 KASP markers were used to genotype MM 37, Svevo, five markers—A009289, A012891, A012893, A012894, and A012896—were polymorphic between the parents (MM 37, Svevo) and among the tested F2 plants from their cross. Indicating that the markers A009289, A012891, A012893, A012894, and A012896 are useful in marker-assisted selection for introducing the resistance gene into common wheat.

Table 1 Marker name and primer sequences of polymorphic KASP markers developed from SNP markers in the genome region between the 669,955,772 and 672,333,864 bp positions on chromosome 1BL of the Chinese Spring reference genome (IWGSC Ref Seq v1.0).
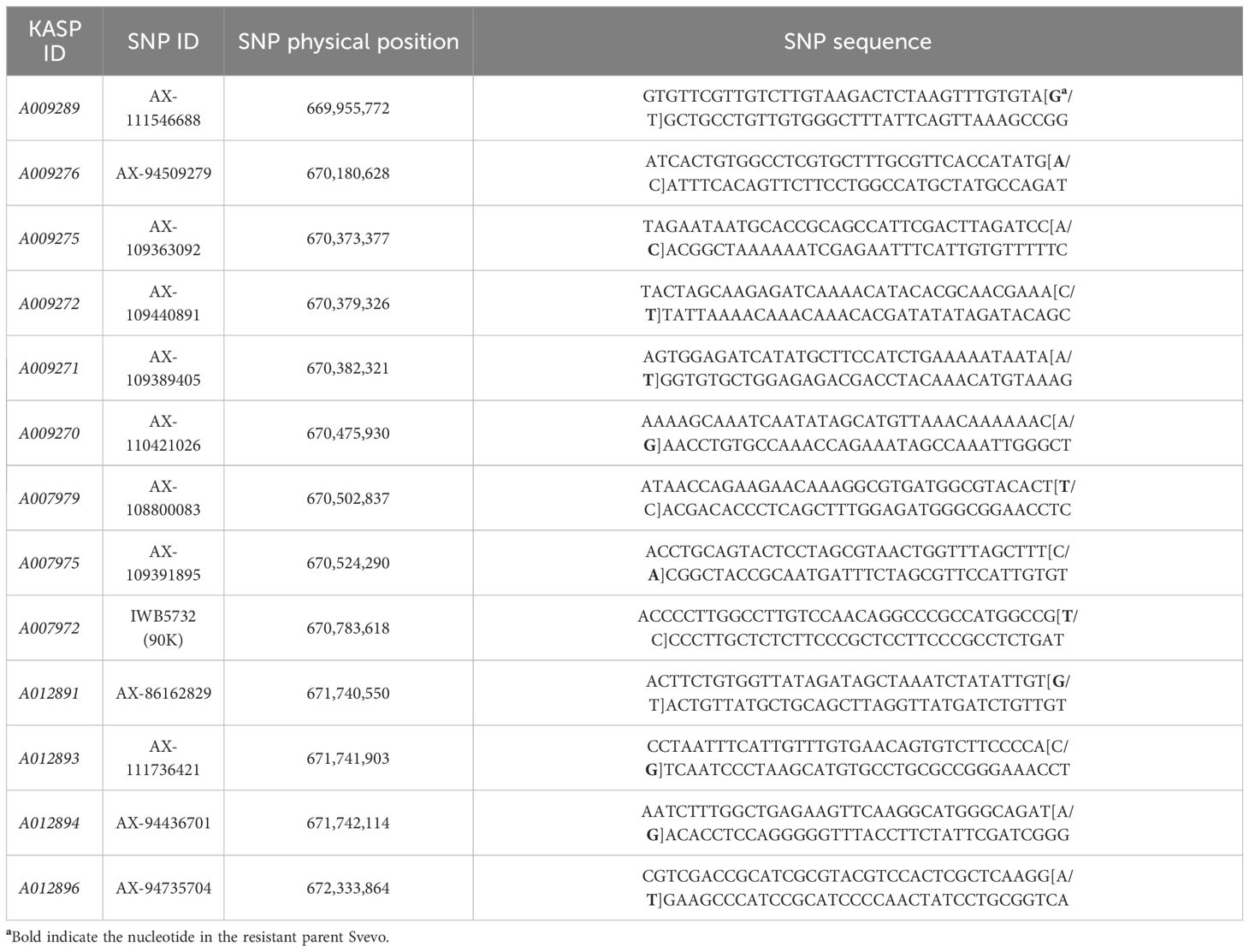
Table 2 Primer sequences were designed for the KASP markers derived from single nucleotide polymorphisms (SNPs) associated with QYrsv.swust-1BL, identified from the wheat 660K SNP array.
Multi-environment analysis of QYrsv.swust-1BL stripe rust resistance loci
QTL for stripe rust resistance was scanned through all 13 KASP markers by the inclusive composite interval mapping (ICIM) method implemented in Ici-Mapping V4.2 software. QYrsv.swust-1BL was discovered using the BIP and MET methods (LOD > 2.5, P < 0.01). QYrsv.swust-1BL was significant for both IT and DS, with the highest peak in a 0.72 cM region. Figure 1 represents a summary of the analysis effects of QYrsv.swust-1BL using the BIP and MET models for IT and DS in multiple seasons from 2016–2020. On the BIP analysis, it has a PVE (percent of explained variation) of 10.43–24.12% in IT and a PVE of 9.32–27.34% in DS across 2016–2020 environments (Table 3). In the MET analysis, it has a high LOD score of 35.38 and a PVE of 11.68% in IT between markers A007972 and A007975. Additionally, it has a high LOD score of 43.53 and a PVE of 24.20% in DS between markers A009289 and A009276 (Table 4).
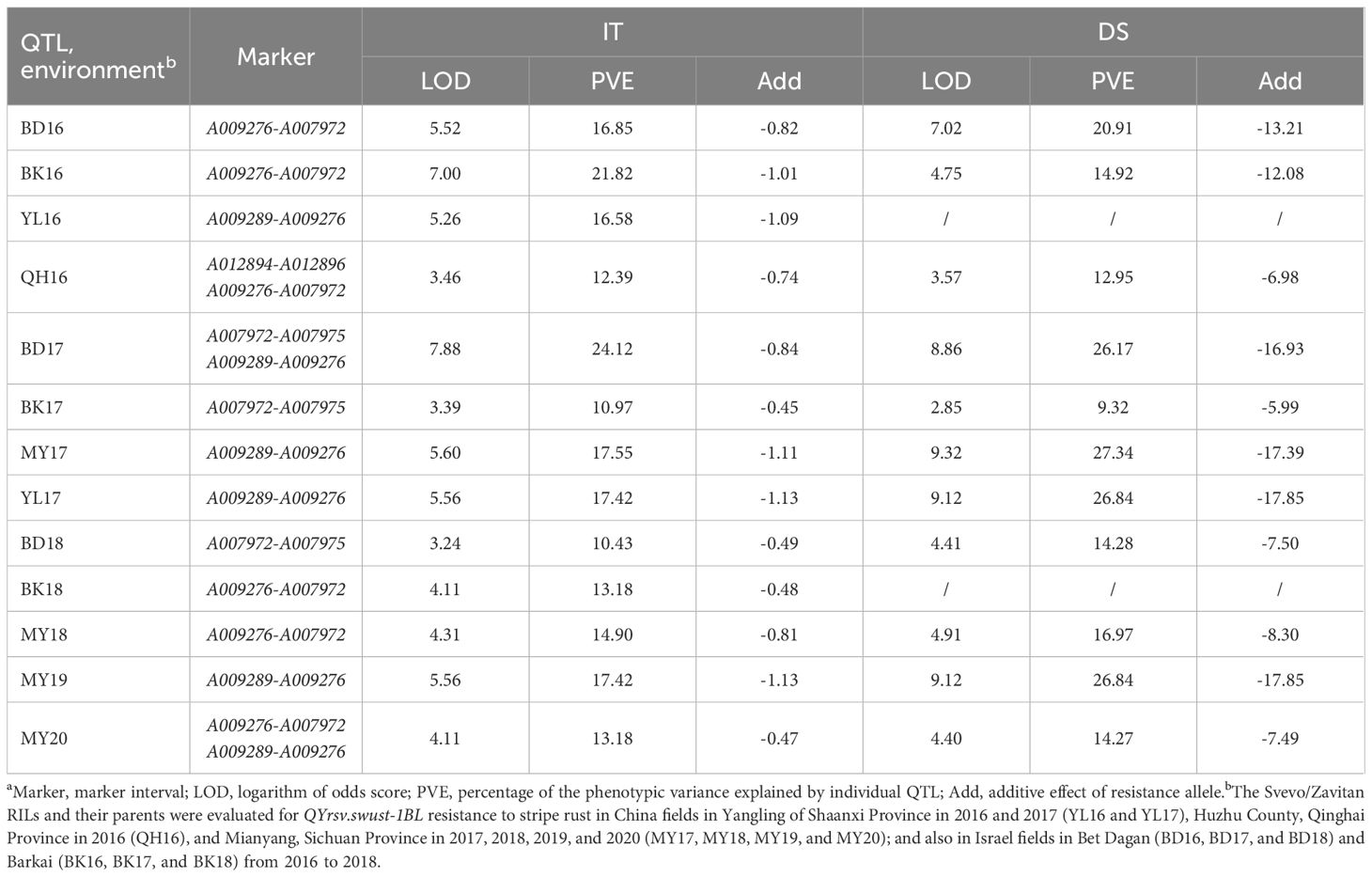
Table 3 Summary of stripe rust resistance gene QYrsv.swust-1BL identified using the biparental population model (BIP) based on mean disease severity (DS) and infection type (IT) of the Svevo/Zavitan recombinant inbred line (RIL) population tested in China and Israel from 2016 to 2020a.
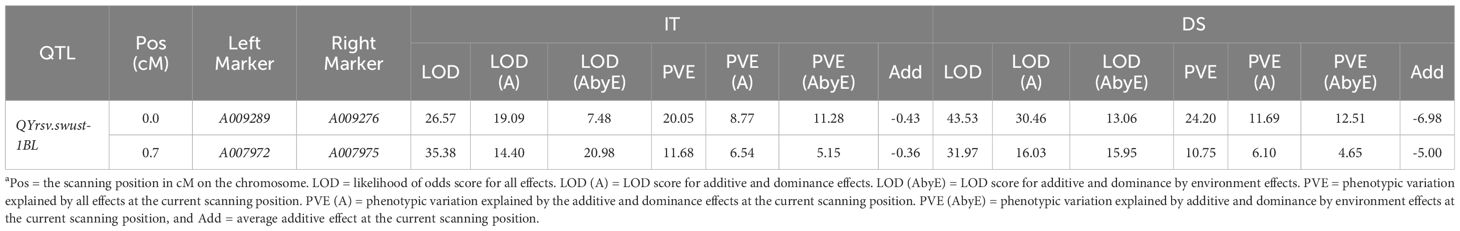
Table 4 Summary of stripe rust resistance gene QYrsv.swust-1BL identified using the multi-environmental trials (MET) model based on the mean infection type (IT) and disease severity (DS) of the Svevo/Zavitan recombinant inbred line (RIL) population tested in China and Israel from 2016 to 2020a.
Identification of recombination events
These 13 markers were used to genotype the 137 RILs, from which 4 RILs were found to have only one recombination event proximal to markers A009289, A009276, A007972, and A007975, respectively. Two markers (A009289 and A007975) were most closely linked to the target gene on two sides and flanked the 1,065,719 bp region between the 660,683,255 bp and 661,748,974 bp positions on the durum wheat Svevo RefSeq Rel. 1.0 (Table 2) and the 568,518 bp region between the 669,955,772 and 670,524,290 bp positions in Chinese Spring IWGSC RefSeq v1.0 (Table 3) on chromosome 1BL. It is clear that QYrsv.swust-1BL overlaps with QYr.ucw-1BL/Yr29. Although the physical mapping region surpasses that of QYr.ucw-1BL/Yr29 (669.902 ~ 670.234 Mp), it is noteworthy that only three candidate genes have been identified within the Chinese spring reference genome (Figure 2).
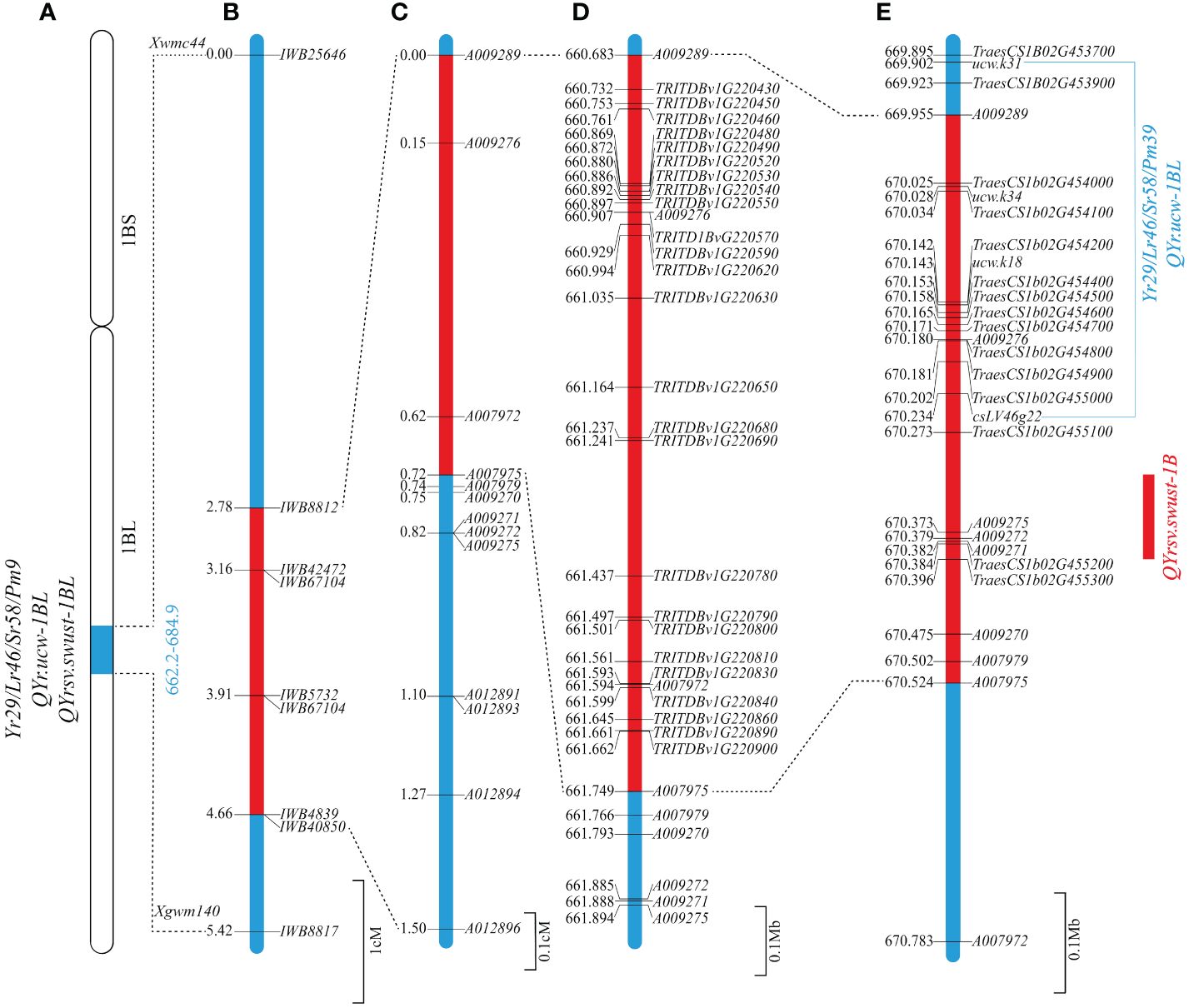
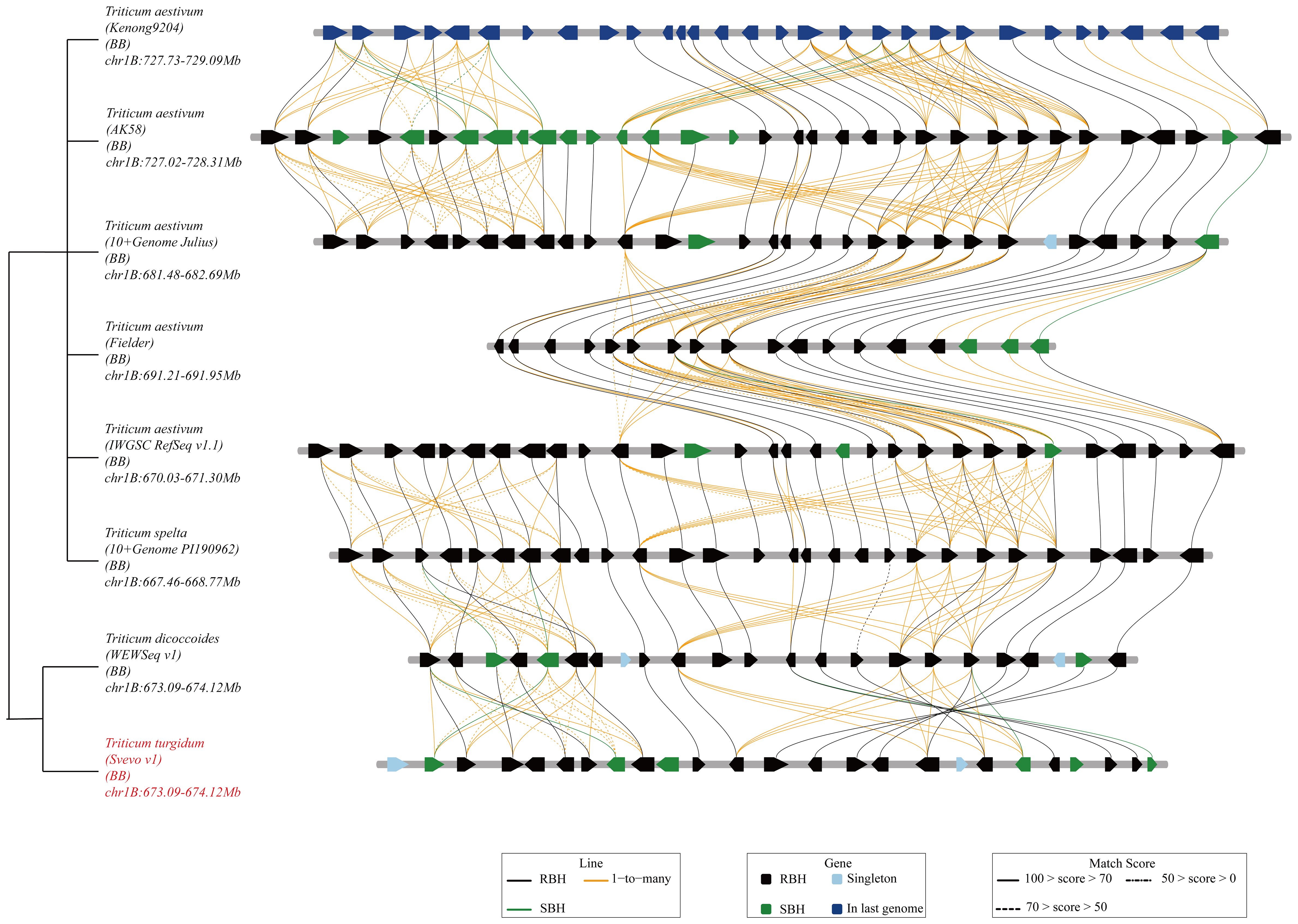
Figure 2 Genetic and comparative genomic linkage map of the stripe rust resistance QTL QYrsv.swust-1BL. (A) Stripe rust resistance gene/QTL: Yr29/Lr46/Sr58/Pm38, QYr.ucw-1BL, and QYrsv.swust-1BL share the same distal region in chromosome 1BL. (B) Genetic map constructed using a 90K SNP chip showing QYrsv.swust-1BL (red) within different flanked markers in comparison with Yr29/Lr46/Sr58/Pm38 and QYr.ucw-1BL (blue). (C) A fine map of QYrsv.swust-1BL was constructed using newly developed KASP markers. (D) The Svevo genome region from the 660.372 Mb to 663.335 Mb position on chromosome 1B. (E) The Chinese Spring (RefSeq V1.0) genome region from the 669.956 Mb to 672.334 Mb position on chromosome 1B.
Candidates for QYrsv.swust-1BL
Based on the analysis of BIP and MET, as well as the examination of recombination events in RILs, QYrsv.swust-1BL was delimited by the linked markers A009289 and A007975 (0.72 cM) (Figure 2). The genetic distance between the two flanking markers in this candidate region was found to be conservative in the durum wheat Svevo genome and 1,065,719 bp in the Svevo RefSeq Rel. 1.0 (Figure 2). The 1.066 Mb region was found to contain 25 annotated genes (Figure 2; Table 5). These 25 high-confidence genes in Svevo were found to be related to disease resistance. The protein encoded by five of these genes (TRITD1Bv1G220450, TRITD1Bv1G220460, TRITD1Bv1G220520, TRITD1Bv1G220540, and TRITD1Bv1G220550) belongs to the disease resistance protein receptor-like protein kinase, which is involved in pathogen recognition and plays an important role in effector-induced protein status monitoring (Mchale et al., 2006). Four of these genes are S-type anion channels (TRITD1Bv1G220620, TRITD1Bv1G220780, TRITD1Bv1G220800, and TRITD1Bv1G220810). Two of these genes are glucan endo-1,3-beta-glucosidase (TRITD1Bv1G220480 and TRITD1Bv1G220530). The other fourteen genes are: Acid beta-fructofuranosi-dase (TRITD1Bv1G220430), Sugar transporter (TRITD1Bv1G220570), WRKY family transcription factor (TRITD1Bv1G220590), Band 7 stomatin family protein (TRITD1Bv1G220630), FAD-binding Berberine family protein (TRITD1Bv1G220650), Calcium-dependent protein kinase (TRITD1Bv1G220680), Protein ABIL1 (TRITD1Bv1G220690), Divalention symporter (TRITD1Bv1G220790), Late embryogenesis abundant protein Lea14 (TRITD1Bv1G220830), F-box family protein (TRITD1Bv1G220840), Late embryogenesis abundant hydroxyproline-rich glycoprotein family (TRITD1Bv1G220860), Thrombospondin type-1-domain-containing protein (TRITD1Bv1G220890), RING/FYVE/PHD zinc finger superfamily protein (TRITD1Bv1G220900), and the other is an unannotated gene (TRITD1Bv1G220490) (Table 5). There are a total of 13 candidate genes in the 568,518 bp region between positions 669,955,772 and 670,524,290 bp of the Chinese Spring IWGSC RefSeq v1.0 (Table 5) on chromosome 1BL. Five of the receptor-like protein kinases (TraesCS1B02G454000, TraesCS1B02G454100, TraesCS1B02G454400, TraesCS1B02G454600, and TraesCS1B02G454700), two of the glucan endo-1,3-beta-glucosidase (TraesCS1B02G454200 and TraesCS1B02G454500), one of the sugar transporter (TraesCS1B02G454800), one of the WRKY family transcription factor (TraesCS1B02G455000), and one of the S-type anion channel (TraesCS1B02G455100) are candidate genes that share common genes with the Svevo genome. The other three different genes are: protein kinase (TraesCS1B02G454900), ATP-dependent zinc metalloprotease FtsH (TraesCS1B02G455200), and carboxypeptidase (TraesCS1B02G455300). These four classes of genes are shared between the Chinese spring genome and the Svevo genome and will be used to verify candidate genes for QYrsv.swust-1BL in the future. A collinearity analysis of these genes was performed using the Zavitan (WEWSeq v1.0), Triticum spelta (spelta PI190962 v1.0), and other 5 Triticum aestivum genomes (http://wheat.cau.edu.cn/TGT/) data (Figure 3). The results of collinearity analysis indicate that there have been multiple chromosomal structural variations in this region. An intrachromosomal translocation exists within the range of approximately 673.1 MB to 673.8 MB, while there is a paracentric inversion within the range of 673.8 MB to 674.1 MB. Through candidate gene functional annotation. Most of the candidate genes in the Svevo reference genome have high match scores, which refer to high confidence.
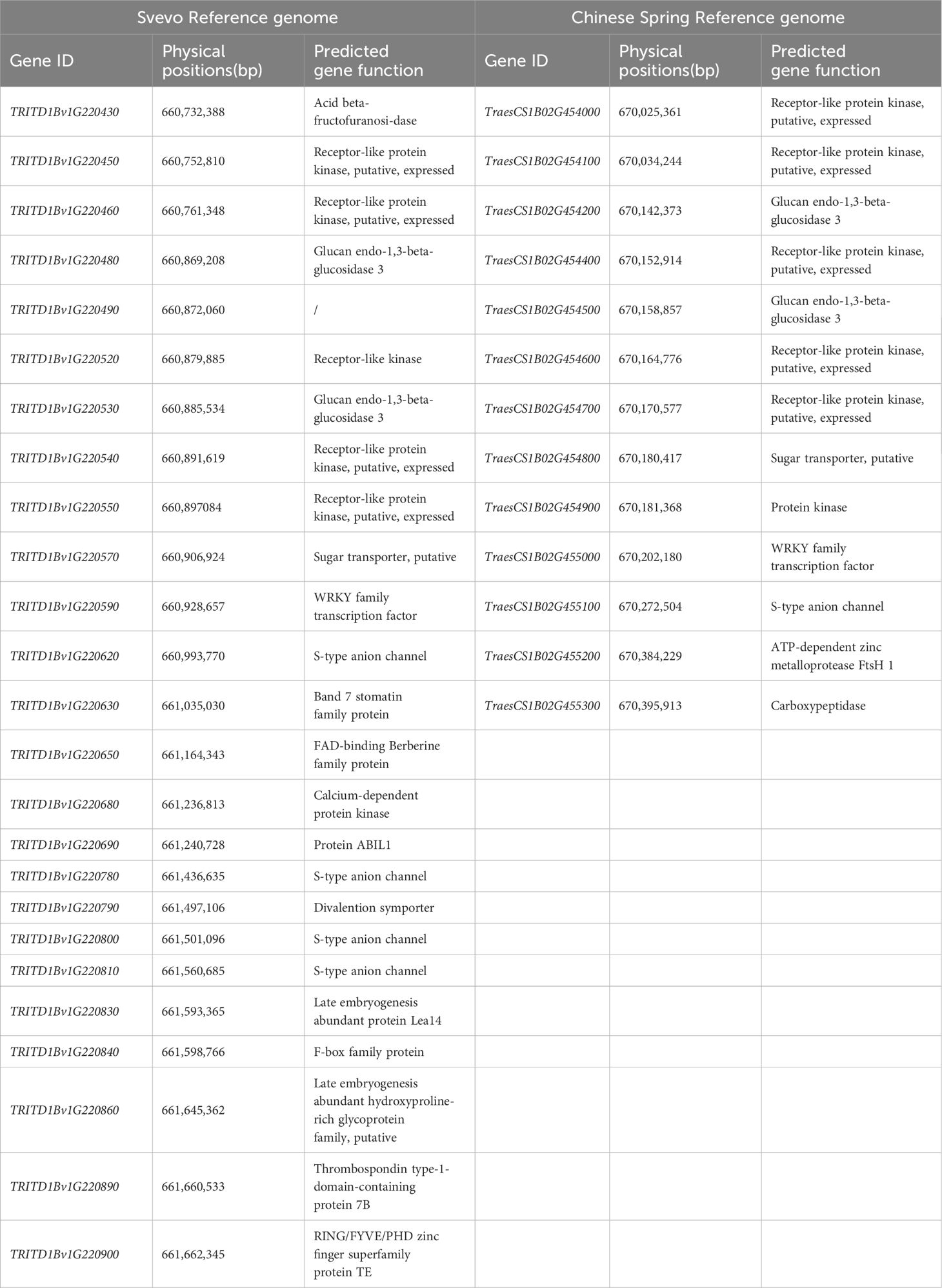
Table 5 The putative genes identified in the genomic regions of the durum wheat Svevo genome (Ref Seq Rel. 1.0.) and the Chinese Spring genome (IWGSC v1.0) covering QYrsv.swust-1BL.
QYrsv.swust-1BL KASP marker validation
To assess the applicability of the newly developed KASP marker in breeding programs, an F2 breeding population derived from a cross between the Chinese hexaploid wheat variety Mianmai 37 and the durum wheat variety Svevo was utilized. The developed KASP markers A009289, A012891, A012893, A012894, and A012896 effectively distinguished genotypes within the F2 breeding population into three distinct clusters (Figure 4). The allele cluster nucleotide of homozygote associated with resistance for KASP markers A009289, A012891, A012893, A012894, and A012896 were identified as ‘G’, ‘G’, ‘G’, ‘G’, ‘T’, respectively, while the alternative allele had a nucleotide cluster of ‘T’, ‘T’, ‘C’, ‘A’, ‘A’, respectively (Table 2; Figure 4). F2 plants and corresponds to 318 F2:3 lines, which were phenotyped for stripe rust response. Among homozygous alleles associated with resistance for marker A009289, F2 plants carrying positive alleles from QYrsv.swust-1BL exhibited a reduction in IT and DS by 34.14% and 45.24% compared with those of the alternative allele ‘T’, respectively. Progenies of F2:3 carrying positive alleles from QYrsv.swust-1BL exhibited a reduction in IT and DS by 54.29% and 41.63% compared with those of the alternative allele ‘T’, respectively. Progenies with heterozygous genotypes for marker A009289 also demonstrated a decrease in IT and DS by 47.27 and 57.71 in F2 plants and 53.80% and 46.92% in F2:3 lines compared with those of the alternative allele ‘T’, respectively. Conversely, progenies carrying alternative alleles from Mianmai 37 showed an increase in IT and DS. The phenotypes of F2 plants and F2:3 families carrying positive alleles and heterozygous genotypes with A012891, A012893, and A012894 markers were significantly different from those carrying alternative alleles, with a difference of 0.005 level, similar to the results of the A009289 marker. The results of marker A012896 in F2 generation were not significant in IT and DS, while the IT differences in F2:3 generation were significant at the levels of 0.05 and 0.01. These findings highlight the effective utilization of KASP markers A009289, A012891, A012893, A012894, and A012896 for marker-assisted selection in breeding programs, confirming that QYrsv.swust-1BL is indeed a significant and stable major gene locus controlling resistance to stripe rust disease.
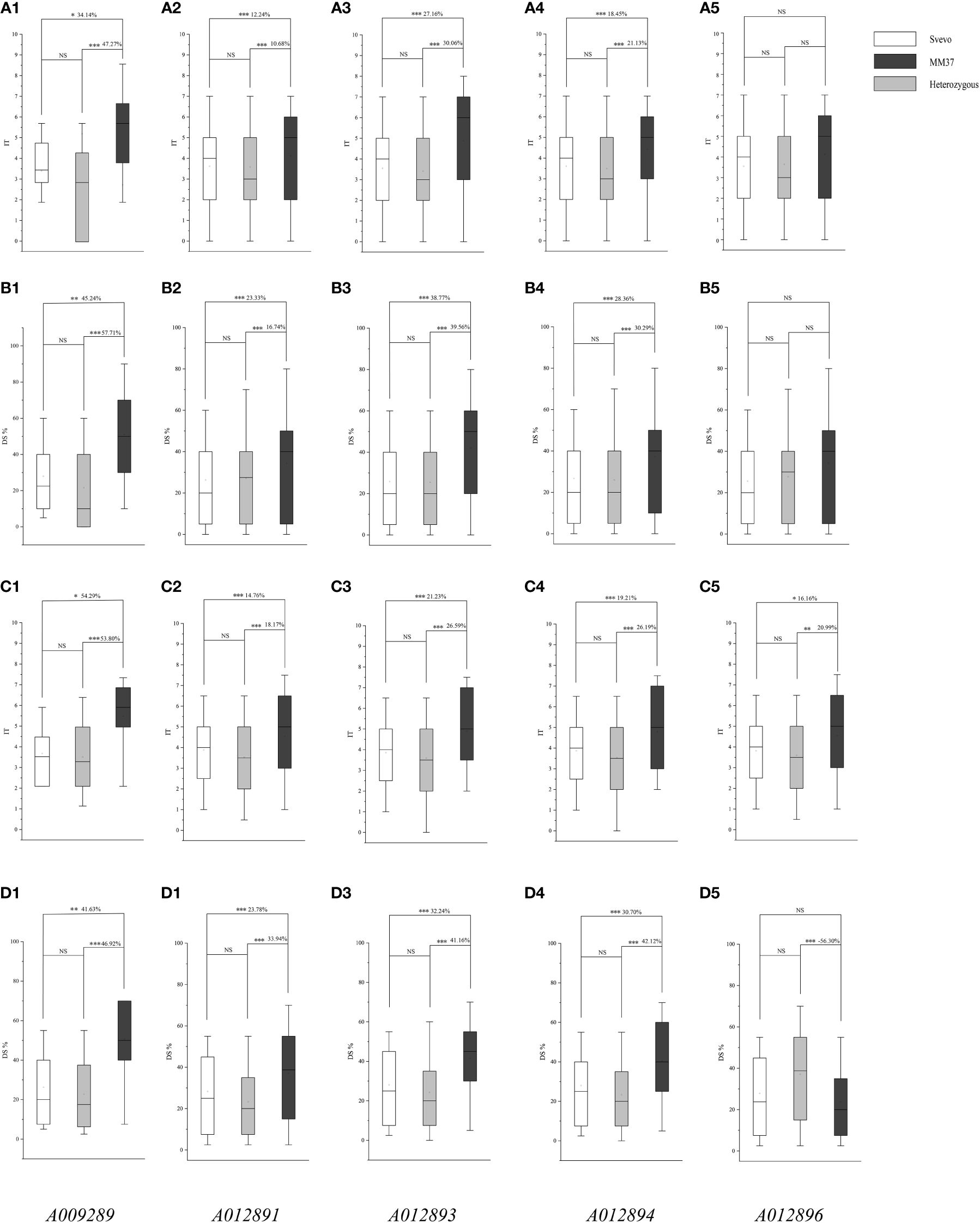
Figure 4 Effects of QYrsv.swust-1BL in the F2 and F2:3 validation populations. The lines with QYrsv.swust-1BL had lower IT and DS than the lines carrying the allele gene of MM37, and the difference between Svevo and heterozygous is not significant. (A1-A5, B1-B5) are based on the IT and DS data of the F2 population, and (C1-C5, D1-D5) are based on the IT and DS data of the F2:3 lines. Numbers 1-5 refer to the validation by five KASP markers A009289, A012891, A012893, A012894 and A012896. * Significance level at P < 0.05; ** Significance level at P <0.01; *** Significance level at P < 0.005. “ns” refers to the none significance.
Discussion
Conventional phenotypic screening approaches have been used to develop cultivars with long-lasting disease resistance, but the breeding process takes many years. Molecular markers associated with rust resistance can be used to speed up the process and stack different resistance genes into a single cultivar. In a previous study, a QTL, QYrsv.swust-1BL, for APR to stripe rust was identified and mapped in chromosome 1BL in durum wheat Svevo (Zhou et al., 2021). By comparing the genetic and physical positions of linked molecular markers, QYrsv.swust-1BL was found to be in the same position as Yr29 on chromosome 1BL.
In the present study, we used the high-density wheat 660K SNP array to find more SNP markers in the QYrsv.swust-1BL region and successfully converted the SNP markers into KASP markers. By re-mapping QYrsv.swust-1BL using the same Svevo/Zavitan RIL population as used in the previous study (Zhou et al., 2021), we added 13 KASP markers to the target region with a much higher resolution. These KASP markers were validated using an F2 population of 318 plants from a cross of Svevo with the common elite wheat cultivar MM 37. The genetic distance of the QYrsv.swust-1BL interval was reduced to 0.72 cM (Figure 2). Finally, we mapped this stripe rust resistance locus within a 1.066 Mb region of chromosome 1BL in durum wheat Svevo RefSeq Rel. 1.0, which overlaps with the previously reported genes, Yr29/Lr46/Sr58/Pm39, for resistance to stripe rust (yellow rust), leaf rust, stem rust, and powdery mildew, respectively (William et al., 2003; Lillemo et al., 2008; Singh et al., 2013). The genomic region between markers A009289 and A007975 in the Chinese Spring reference genome was found to be 0.569 Mb, comparable to the size of the region in the durum wheat genome.
Based on the results of high-resolution mapping using a large population, exome capture, and allelism analysis of a cross between common wheat RIL55 (with the Klein Chajá allele of QYr.ucw-1BL) and Lalbahadur (with the Pavon-1B allele of Yr29), Cobo et al. (2019) reported that QYr.ucw-1BL and Yr29 represent the same gene. According to the comparative maps and annotated genes, we hypothesized that QYrsv.swust-1BL should be the same as QYr.ucw-1BL/Yr29. The genomic interval between KASP markers A009289 and A007975 in the Chinese Spring genome (IWGSC v1.0) spans a smaller region of 0.569 Mb and shares four complete classes of genes. However, the 1.066 Mb region between KASP markers A009289 (at the 660,683,255 bp position) and A007975 (at the 661,748,974 bp position) contains 25 high-confidence genes annotated in the Svevo reference genome, including the QYr.ucw-1BL region (Cobo et al., 2018, 2019). These 25 and 13 genes in Svevo RefSeq Rel. 1.0 and Chinese Spring IWGSC RefSeq v1.0 are listed in Table 5, respectively. Ten annotated genes were reported in the QYr.ucw-1BL region between markers ucw.k31 and csLV46G22 (Cobo et al., 2019). As QYrsv.swust-1BL and QYr.ucw-1BL/Yr29 were mapped to the same regions of the Chinese Spring genome, we found four classes of identical genes in these genomic regions. Thus, QYrsv.swust-1BL and QYr.ucw-1BL/Yr29 are likely the same gene. More conclusive evidence for these two QTL as the same gene needs future studies of direct comparison of both QTL in common wheat backgrounds through allelism testing, gene cloning, and expression. The F2:3 lines or later generation lines with 42 chromosomes from the MM 37/Svevo cross can be used together with the QYrsv.swust-1BL/QYr.ucw-1BL/Yr29 near-isogenic lines in the Avocet S background for such experiments.
In the present study, we showed that the common wheat Chinese Spring and durum wheat Svevo genome sequences can be used to hasten high-resolution mapping of traits of interest in wheat. We encountered difficulties, such as sterile plants caused by aneuploids from the common wheat and durum wheat crosses, when attempting to clone QYrsv.swust-1BL because of the ploid differences between the parental lines and the reference genomes. Therefore, regions like QYrsv.swust-1BL from durum wheat in this study should be assembled using the donor genome to enable the cloning of causal genes and to develop diagnostic markers that can be used in breeding programs.
For breeding wheat cultivars with stripe rust resistance, QYrsv.swust-1BL can be used together with other effective resistance genes. The progeny lines with 42 chromosomes and QYrsv.swust-1BL derived from cross MM 37/Svevo should be more efficient than the original donor of durum wheat for incorporating the APR QTL into other common wheat backgrounds. The KASP markers tightly linked to QYrsv.swust-1BL can be used in marker-assisted selection for speeding up its incorporation and pyramiding with other effective genes for developing wheat cultivars with high-level, durable resistance to stripe rust.
Conclusion
In this study, using high-density 660K SNP array genotyping, we fine-mapped the QYrsv.swust-1BL APR locus as a starting point to develop a diagnostic marker for use in breeding and to clone this gene. We mapped QYrsv.swust-1BL to within a 1.066 Mb region in Triticum turgidum durum wheat Svevo (RefSeq Rel. 1.0) on the chromosome arm 1BL, which overlaps with a previously described map of QYr.ucw-1BL/Yr29, a Pst resistance gene. The four gene families within the identified 1.066 Mb region, identical to the Chinese spring reference genome, have been implicated in disease resistance. SNP markers were then used to select high-throughput KASP markers that could be used in wheat breeding programs to hasten the deployment of these Pst resistance loci.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
XZ: Writing – review & editing, Writing – original draft, Visualization, Investigation, Formal analysis, Data curation. GJ: Writing – review & editing, Writing – original draft, Visualization, Investigation, Formal analysis, Data curation. YL: Writing – review & editing. XL: Writing – review & editing, Supervision. LC: Writing – review & editing, Visualization, Supervision, Data curation. XC: Writing – review & editing, Visualization, Supervision, Investigation, Data curation. ZK: Writing – review & editing, Visualization, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the International S&T Cooperation Program of China (2015DFG32340) and was partially funded by the Key Research and Development Program of International Science and Technology Innovation Cooperation of Science and Technology Department of Sichuan Province, China (No. 22YFH0032), the National Natural Science Foundation of China (32101707), the Longshan Academic Talent Research Support Program of SWUST (No. 17LZX5), and the PhD Foundation of Southwest University of Science and Technology (No. 16zx7162).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anderson, J. A., Churchill, G. A., Autrique, J. E., Tanksley, S. D., Sorrells, M. E. (1993). Optimizing parental selection for genetic-linkage maps. Genome 36, 181–186. doi: 10.1139/g93-024
Avni, R., Nave, M., Eilam, T., Sela, H., Alekperov, C., Peleg, Z., et al. (2014). Ultra-dense genetic map of durum wheat × wild emmer wheat developed using the 90K iSelect SNP genotyping assay. Mol. Breed. 34, 1549–1562. doi: 10.1007/s11032-014-0176-2
Bansal, U. K., Kazi, A. G., Singh, B., Hare, R. A., Basiana, H. S. (2014). Mapping of durable stripe rust resistance in a durum wheat cultivar Wollaroi. Mol. Breed. 33, 51–59. doi: 10.1007/s11032-013-9933-x
Beddow, J. M., Pardey, P. G., Chai, Y., Hurley, T. M., Kriticos, D. J., Braun, H. J., et al. (2015). Research investment implications of shifts in the global geography of wheat stripe rust. Nat. Plants 1, 1–5. doi: 10.1038/nplants.2015.132
Chen, X. M. (2005). Epidemiology and control of stripe rust (Puccinia striiformis f. sp. tritici) on wheat. Can. J. Plant Pathol. 27, 314–337. doi: 10.1080/07060660509507230
Chen, X. M., Coram, T., Huang, X. L., Wang, M. N., Dolezal, A. (2013). Understanding molecular mechanisms of durable and non-durable resistance to stripe rust in wheat using a transcriptomics approach. Cur. Genomics 14, 111–126. doi: 10.2174/1389202911314020004
Chen, W. Q., Wellings, C., Chen, X. M., Kang, Z. S., Liu, T. G. (2014). Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 15, 433–446. doi: 10.1111/mpp.12116
Chen, J., Upadhyaya, N. M., Ortiz, D., Sperschneider, J., Li, F., Bouton, C., et al. (2017). Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science 358, 1607–1610. doi: 10.1126/science.aao4810
Chen, S., Zhang, W., Bolus, S., Rouse, M. N., Dubcovsky, J. (2018). Identification and characterization of wheat stem rust resistance gene Sr21 effective against the Ug99 race group at high temperature. PloS Genet. 14, e1007287. doi: 10.1371/journal.pgen.1007287
Chen, X. M. (2020). Pathogens which threaten food security: Puccinia striiformis, the wheat stripe rust pathogen. Food Secur. 12, 239–251. doi: 10.1007/s12571-020-01016-z
Cobo, N., Pflüger, L., Chen, X. M., Dubcovsky, J. (2018). Mapping QTL for resistance to new virulent races of wheat stripe rust from two argentinean wheat cultivars. Crop Sci. 58, 2470–2483. doi: 10.2135/cropsci2018.04.0286
Cobo, N., Wanjugi, H., Lagudah, E., Dubcovsky, J. (2019). A high-resolution map of wheat QYr.ucw-1BL, an adult plant stripe rust resistance locus in the same chromosomal region as Yr29. Plant Genome 12, 180055. doi: 10.3835/plantgenome2018.08.0055
Feng, J. Y., Wang, M. N., See, D. R., Chao, S. M., Zheng, Y. L., Chen, X. M. (2018). Characterization of novel gene Yr79 and four additional QTL for all-stage and high-temperature adult-plant resistance to stripe rust in spring wheat PI 182103. Phytopathology 108, 737–747. doi: 10.1094/PHYTO-11-17-0375-R
Feng, J., Yao, F., Wang, M., See, D. R., Chen, X. (2023). Molecular mapping of Yr85 and comparison with other genes for resistance to stripe rust on wheat chromosome 1B. Plant Dis. 0, null. doi: 10.1094/PDIS-11-22-2600-RE
Fu, D., Uauy, C., Distelfeld, A., Blechl, A., Epstein, L., Chen, X. M., et al. (2009). A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323, 1357–1360. doi: 10.1126/science.1166289
Gessese, M., Bariana, H., Wong, D., Hayden, M., Bansal, U. (2019). Molecular mapping of stripe rust resistance gene Yr81 in a common wheat landrace Aus27430. Plant Dis. 103, 1166–1171. doi: 10.1094/PDIS-06-18-1055-RE
Gou, J. Y., Li, K., Wu, K., Wang, X., Lin, H., Cantu, D., et al. (2015). Wheat stripe rust resistance protein WKS1 reduces the ability of the thylakoid-associated ascorbate peroxidase to detoxify reactive oxygen species. Plant Cell 27, 1755–1770. doi: 10.1105/tpc.114.134296
Herrera-Foessel, S. A., Singh, R. P., Huerta-Espino, J., Salazar, V. C., Lagudah, E. S. (2011). “First report of slow rusting gene Lr46 in durum wheat,” in Poster Abstracts of the Borlaug Global Rust Initiative Technical Workshop. Ed. McIntosh, M. (Saint Paul, Minnesota, USA), 191.
Herrera-Foessel, S. A., Singh, R. P., Lan, C. X., Huerta-Espino, J., Calvo-Salazar, V., Bansal, U. K., et al. (2015). Yr60, a gene conferring moderate resistance to stripe rust in wheat. Plant Dis. 99, 508–511. doi: 10.1094/PDIS-08-14-0796-RE
Klymiuk, V., Chawla, H. S., Wiebe, K., Ens, J., Fatiukha, A., Govta, L., et al. (2022). Discovery of stripe rust resistance with incomplete dominance in wild emmer wheatusing bulked segregant analysis sequencing. Commun. Biol. 5, 826. doi: 10.1038/s42003–022-03773–3
Kolmer, J. A. (2015). A QTL on chromosome 5BL in wheat enhances leaf rust resistance of Lr46. Mol. Breed. 35, 74–81. doi: 10.1007/s11032-015-0274-9
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Li, J., Dundas, I., Dong, C., Li, G., Trethowan, R., Yang, Z., et al. (2020). Identification and characterization of a new stripe rust resistance gene Yr83 on rye chromosome 6R in wheat. Theor. Appl. Genet. 133, 1095–1107. doi: 10.1007/s00122-020-03534-y
Lillemo, M., Asalf, B., Singh, R. P., Huerta-Espino, J., Chen, X. M., He, Z. H., et al. (2008). The adult plant rust resistance loci Lr34/Yr18 and Lr46/Yr29 are important determinants of partial resistance to powdery mildew in bread wheat line Saar. Theor. Appl. Genet. 116, 1155–1166. doi: 10.1007/s00122-008-0743-1
Lillemo, M., Joshi, A. K., Prasad, R., Chand, R., Singh, R. P. (2013). QTL for spot blotch resistance in bread wheat line Saar co-locateto the biotrophic disease resistance loci Lr34 and Lr46. Theor. Appl. Genet. 126, 711–719. doi: 10.1007/s00122-012-2012-6
Line, R. F., Qayoum, A. (1992). Virulence, aggressiveness, evolution and distribution of races of Puccinia striiformis (the cause of stripe rust of wheat) in North America 1968–1987 (Washington, DC: US Department of Agriculture).
Liu, L., Wang, M. N., Feng, J. Y., See, D. R., Chao, S. M., Chen, X. M. (2018). Combination of all-stage and high-temperature adult-plant resistance QTL confers high level, durable resistance to stripe rust in winter wheat cultivar Madsen. Theor. Appl. Genet. 131, 1835–1849. doi: 10.1007/s00122-018-3116-4
Liu, L., Yuan, C. Y., Wang, M. N., See, D. R., Zemetra, R. S., Chen, X. M. (2019). QTL analysis of durable stripe rust resistance in the North American winter wheat cultivar Skiles. Theor. Appl. Genet. 132, 1677–1691. doi: 10.1007/s00122-019-03307-2
Maccaferri, M., Harris, N. S., Twardziok, S. O., Pasam, R. K., Gundlach, H., Spannagl, M., et al. (2019). Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 51, 885–895. doi: 10.1038/s41588-019-0381-3
Marchal, C., Zhang, J., Zhang, P., Fenwick, P., Steuernagel, B., Adamski, N. M., et al. (2018). BED-domain containing immune receptors confer diverse resistance spectra to yellow rust. Nat. Plants 4, 662–668. doi: 10.1038/s41477-018-0236-4
Mchale, L., Tan, X., Koehl, P., Michelmore, R. W. (2006). Plant NBS-LRR proteins: adaptable guards. Genome Biol. 7, 212. doi: 10.1186/gb-2006-7-4-212
Meng, L., Li, H. H., Zhang, L. Y., Wang, J. K. (2015). QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Moore, J. W., Herrera-Foessel, S., Lan, C., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi: 10.1038/ng.3439
Nsabiyera, V., Bariana, H. S., Qureshi, N., Wong, D., Hayden, M. J., Bansal, U. K. (2018). Characterization and mapping of adult plant stripe rust resistance in wheat accession Aus27284. Theor. Appl. Genet. 131, 1–9. doi: 10.1007/s00122-018-3090-x
Pakeerathan, K., Bariana, H., Qureshi, N., Wong, D., Hayden, M., Bansal, U. (2019). Identification of a new source of stripe rust resistance Yr82 in wheat. Theor. Appl. Genet. 132, 3169–3176. doi: 10.1007/s00122-019-03416-y
Periyannan, S., Moore, J., Ayliffe, M., Bansal, U., Wang, X., Huang, L., et al. (2013). The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341, 786–788. doi: 10.1126/science.1239028
Ponce-Molina, L. J., Huerta-Espino, J., Singh, R., Basnet, B. R., Lagudah, E. S., Aguilar-Rincón, V. H., et al. (2018). Characterization of adult plant resistance to leaf rust and stripe rust in Indian wheat cultivar ‘New Pusa 876’. Crop Sci. 58, 630–638. doi: 10.2135/cropsci2017.06.0396
Ramirez-Gonzalez, R. H., Uauy, C., Caccamo, M. (2015). PolyMarker: a fast polyploid primer design pipeline. Bioinformatics 31, 2038–2039. doi: 10.1093/bioinformatics/btv069
Ren, Y., Singh, R. P., Basnet, B. R., Lan, C. X., Huerta-Espino, J., Lagudah, E. S., et al. (2017). Identification and mapping of adult plant resistance loci to leaf rust and stripe rust in common wheat cultivar Kundan. Plant Dis. 101, 456–463. doi: 10.1094/PDIS-06-16-0890-RE
Rosewarne, G. M., Singh, R. P., Huerta-Espino, J., William, H. M., Bouchet, S., Cloutier, S., et al. (2006). Leaf tip necrosis, molecular markers and β1-proteasome subunits associated with the slow rusting resistance genes Lr46/Yr29. Theor. Appl. Genet. 112, 500–508. doi: 10.1007/s00122-005-0153-6
Salcedo, A., Rutter, W., Wang, S., Akhunova, A., Bolus, S., Chao, S., et al. (2017). Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science 358, 1604–1606. doi: 10.1126/science.aao7294
Singh, R. P., Herrera-Foessel, S. A., Huerta-Espino, J., Lan, C. X., Basnet, B. R., Bhavani, S., et al. (2013). “Pleiotropic gene Lr46/Yr29/Pm39/Ltn2 confers slow rusting, adult plant resistance to wheat stem rust fungus,” in Proceedings of the Borlaug Global Rust Initiative Technical Workshop, 19–22 Aug (Indian Council of Agricultural Research, New Delhi), 17.1.
Singh, R. P., Huerta-Espino, J., Bhavani, S., Herrera-Foessel, S. A., Singh, D., Singh, P. K., et al. (2011). Race non-specific resistance to rust diseases in CIMMYT spring wheats. Euphytica 179, 175–186. doi: 10.1007/s10681-010-0322-9
Singh, R. P., Mujeeb-Kazi, A., Huerta-Espino, J. (1998). Lr46: A gene conferring slow-rusting resistance to leaf rust in wheat. Phytopathology 88, 890–894. doi: 10.1094/PHYTO.1998.88.9.890
Singh, A. K., Zhang, P., Dong, C., Li, J., Singh, S., Trethowan, R., et al. (2021). Generation and molecular marker and cytological characterization of wheat - Secale strictum subsp. anatolicum derivatives. Genome 64, 29–38. doi: 10.1139/gen-2020–0060
Wang, J. K. (2009). Inclusive composite interval mapping of quantitative trait genes. Acta Phytopathol. Sin. 35, 239–245. doi: 10.3724/SP.J.1006.2009.00239
Wang, M. N., Chen, X. M. (2017). “Stripe rust resistance,” in Stripe Rust. Eds. Chen, X. M., Kang, Z. S. (Springer, Dordrecht), 353–558. doi: 10.1007/978–94-024–1111-9
William, H. M., Singh, R. P., Huerta-Espino, J., Islas, S. O., Hoisington, D. (2003). Molecular marker mapping of leaf rust resistance gene Lr46 and its association with stripe rust resistance gene Yr29 in wheat. Phytopathology 93, 153–159. doi: 10.1094/PHYTO.2003.93.2.153
Wu, J. H., Zeng, Q. D., Wang, Q. L., Liu, S. J., Yu, S. Z., Mu, J. M., et al. (2018). SNP−based pool genotyping and haplotype analysis accelerate fine−mapping of the wheat genomic region containing stripe rust resistance gene Yr26. Theor. Appl. Genet. 131, 1481–1496. doi: 10.1007/s00122-018-3092-8
Yao, E., Blake, V. C., Cooper, L., Wight, C. P., Michel, S., Cagirici, H. B. (2022). “Graingenes: a data-rich repository for small grains genetics and genomics”. Database, 2022, baac034. doi: 10.1093/database/baac034
Zadoks, J. C., Chang, T. T., Konzak, C. F. (1974). A decimal code for the growth stages of cereals. Weed Res. 14, 415–421. doi: 10.1111/j.1365-3180.1974.tb01084.x
Zhang, W., Chen, S., Abate, Z., Nirmala, J., Rouse, M. N., Dubcovsky, J. (2017). Identification and characterization of Sr13, a tetraploid wheat gene that confers resistance to the Ug99 stem rust race group. Proc. Natl. Acad. Sci. U.S.A. 114, E9483–E9492. doi: 10.1073/pnas.1706277114
Zhou, X. L., Hu, T., Li, X., Yu, M., Li, Y., Yang, S., et al. (2019). Genome-wide mapping of adult plant stripe rust resistance in wheat cultivar Toni. Theor. Appl. Genet. 132, 1693–1704. doi: 10.1007/s00122-019-03308-1
Zhou, X. L., Zhong, X., Roter, J., Li, X., Yao, Q., Yan, J. H., et al. (2021). Genome-wide mapping of adult plant stripe rust resistance locus derived from durum wheat Svevo by 90K SNP array. Plant Dis. 105, 879–888. doi: 10.1094/PDIS-09–20-1933-RE
Keywords: stripe rust, resistance gene, durum wheat, fine mapping, quantitative trait locus
Citation: Zhou X, Jia G, Luo Y, Li X, Cai L, Chen X and Kang Z (2024) Fine mapping of QYrsv.swust-1BL for resistance to stripe rust in durum wheat Svevo. Front. Plant Sci. 15:1395223. doi: 10.3389/fpls.2024.1395223
Received: 03 March 2024; Accepted: 06 May 2024;
Published: 12 June 2024.
Edited by:
Changlin Liu, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Francesca Desiderio, Council for Agricultural and Economics Research, ItalyJindong Liu, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Zhou, Jia, Luo, Li, Cai, Chen and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhensheng Kang, a2FuZ3pzQG53c3VhZi5lZHUuY24=; Lin Cai, bGluY2FpQGd6dS5lZHUuY24=
†These authors have contributed equally to this work
 Xinli Zhou
Xinli Zhou Guoyun Jia1†
Guoyun Jia1† Xin Li
Xin Li Lin Cai
Lin Cai Zhensheng Kang
Zhensheng Kang