
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 15 February 2024
Sec. Aquatic Photosynthetic Organisms
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1346759
This article is part of the Research TopicPhotosynthetic Regulation and Carbon/Nitrogen Metabolism in Genetically Engineered AlgaeView all 5 articles
The carboxysome is a bacterial microcompartment (BMC) which plays a central role in the cyanobacterial CO2-concentrating mechanism. These proteinaceous structures consist of an outer protein shell that partitions Rubisco and carbonic anhydrase from the rest of the cytosol, thereby providing a favorable microenvironment that enhances carbon fixation. The modular nature of carboxysomal architectures makes them attractive for a variety of biotechnological applications such as carbon capture and utilization. In silico approaches, such as molecular dynamics (MD) simulations, can support future carboxysome redesign efforts by providing new spatio-temporal insights on their structure and function beyond in vivo experimental limitations. However, specific computational studies on carboxysomes are limited. Fortunately, all BMC (including the carboxysome) are highly structurally conserved which allows for practical inferences to be made between classes. Here, we review simulations on BMC architectures which shed light on (1) permeation events through the shell and (2) assembly pathways. These models predict the biophysical properties surrounding the central pore in BMC-H shell subunits, which in turn dictate the efficiency of substrate diffusion. Meanwhile, simulations on BMC assembly demonstrate that assembly pathway is largely dictated kinetically by cargo interactions while final morphology is dependent on shell factors. Overall, these findings are contextualized within the wider experimental BMC literature and framed within the opportunities for carboxysome redesign for biomanufacturing and enhanced carbon fixation.
Photoautotrophic microbes, such as algae and cyanobacteria, have shown promise as biomanufacturing platforms which can use CO2 as their sole carbon source. At the core of this process lies the enzyme ribulose bisphosphate carboxylase/oxygenase (Rubisco), recognized as the most influential and abundant enzyme in our planet’s carbon cycle (Phillips and Milo, 2009; Raven, 2013; Bar-On and Milo, 2019). Rubisco is used by plants, algae, cyanobacteria, and even some non-photosynthetic chemoautotrophs (Andersson and Backlund, 2008) to assimilate nearly 250 billion tons of carbon from the atmosphere every year (Field et al., 1998). Despite its widespread importance, Rubisco is a catalytically inefficient enzyme in today’s oxygen-rich environment, achieving CO2 fixation rates on the order of 1-10 s-1 (Flamholz et al., 2019; Davidi et al., 2020) while also capable of photorespiration [Rubisco-catalyzed oxygenation of ribulose bisphosphate (Busch, 2020; Savir et al., 2010)]. Carbon assimilation in aquatic organisms is further complicated by the often low availability of dissolved CO2 under ambient conditions (Maberly and Gontero, 2017).
To overcome these challenges, certain aquatic microorganisms evolved ornate CO2-concentrating mechanisms (CCMs) (Iñiguez et al., 2020; Badger et al., 1998), which consist of inorganic carbon pumps and Rubisco-filled compartments (pyrenoids and carboxysomes) (Badger and Price, 1992) that work by selectively increasing the CO2 concentration around Rubisco (Price et al., 1998). Carboxysomes are a part of a larger class of structurally related protein organelles called bacterial microcompartments (BMC). As a class, BMCs are associated with an array of programmable, modular characteristics that can be leveraged to support biomanufacturing and carbon sequestration applications.
The deployment of CCMs as “modules” for CO2 fixation has been suggested as a promising target to bolster the productivity of biomanufacturing platforms that utilize CO2 as their primary precursor to produce biomass and biofuels/bioproducts. However, heterologous expression and redesign of CCMs requires mechanistic insights elusive to current high resolution experimental methods. To alleviate this limitation, molecular dynamics (MD) simulations are quickly gaining attention for revealing atomic-detailed processes underpinning CCM assembly and function. MD provides spatio-temporal information which can potentially facilitate rational modifications and in silico prototyping. This review will present the current state of MD and other computational applications towards studying and redesigning the core of the cyanobacterial CCM, the carboxysome. Since carboxysome-specific simulations are limited, this review draws on and contextualizes the wider experimental BMC literature with implications for their synthetic adaptation for enhanced carbon fixation.
The cyanobacterial CCM has been a focus of multiple studies for the elucidation of structure, function, and its integration into cellular metabolism (Kupriyanova et al., 2023). Carbon assimilation in cyanobacteria begins with the uptake and accumulation of inorganic carbon sources within the cytoplasm (Figure 1A). CO2 can simply diffuse through the outer cellular membrane while charged bicarbonate must be actively pumped into the cell coupled with Na+ or in an ATP-dependent fashion with BicA/SbtA and BCT1, respectively (Shibata et al., 2002a; Shibata et al., 2001; Shibata et al., 2002b). Internalized CO2 can be converted to bicarbonate by an NADPH-dependent reduction by the complexes NDH-13 and NDH-14, which are coupled to CO2-uptake proteins (Cup) (Artier et al., 2018). CO2 and the resulting bicarbonate pool feed into the carboxysome – a bacterial protein-derived organelle that houses Rubisco and carbonic anhydrase within a semi-permeable protein shell (Kerfeld and Melnicki, 2016; Rae et al., 2013) – where CO2 and ribulose-bisphosphate react to form the central metabolite 3-phospoglycerate (3-PGA).
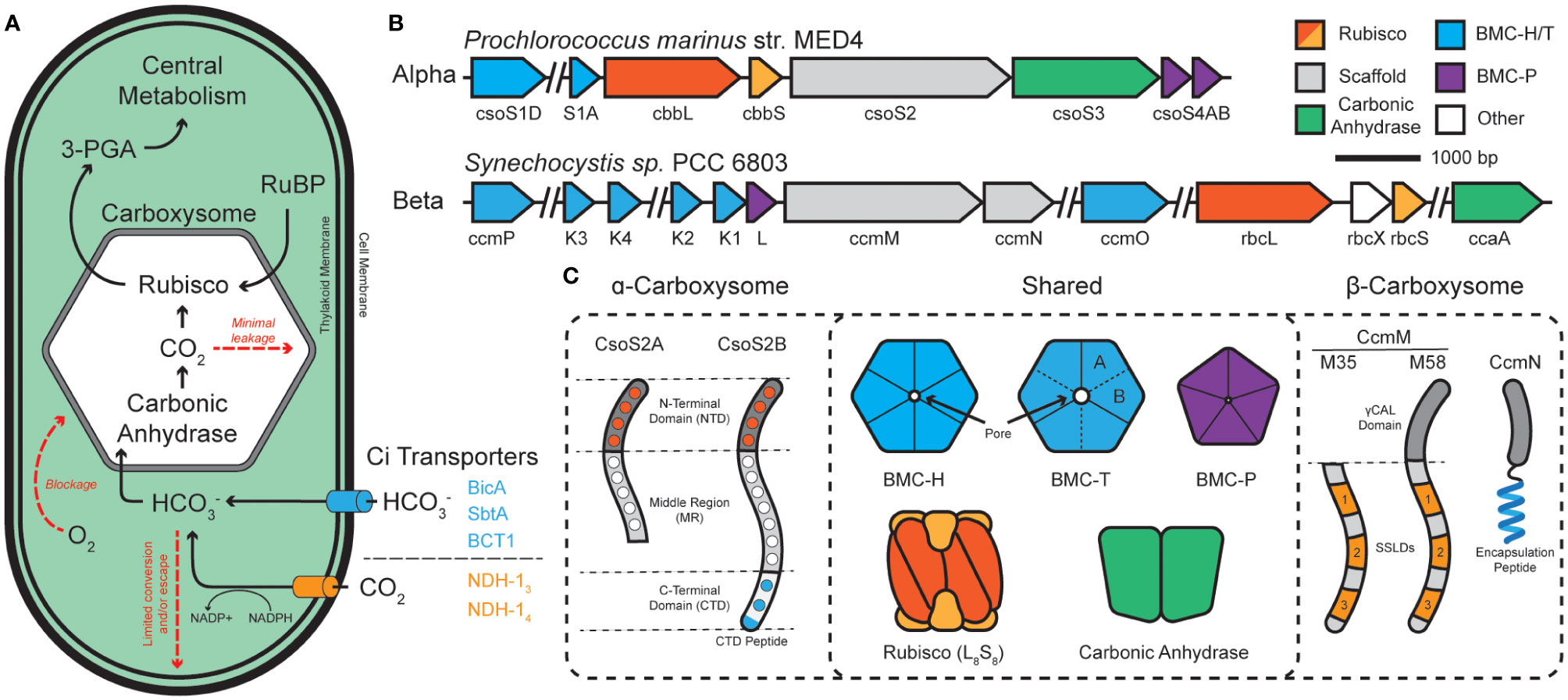
Figure 1 The cyanobacterial carbon concentrating mechanism (CCM) is centered around the carboxysome. (A) Cyanobacterial inorganic carbon (Ci) sequestration begins with CO2 and HCO3- transporters. Bicarbonate enters the carboxysome and is converted into CO2 and combined with ribulose bisphosphate (RuBP) to form central metabolite 3-phosphoglycerate (3-PGA). (B) The two classes of carboxysome, ɑ- and β-, differ in their genetic organization. ɑ-carboxysomes tend to organize into distinct operons while β-carboxysomes tend to be more disjointed among numerous satellite loci. (C) ɑ- and β-carboxysomes share many components but differ in their use of scaffold proteins. ɑ-types use two forms of CsoS2, which is composed of a multivalent, Rubisco-binding N-terminal domain (NTD; orange dots specify repeats), a multivalent middle region (MR; white dots specify repeats), and a multivalent, shell-binding C-terminal domain (CTD; blue dots specify repeats). CsoS2 is expressed in two forms which differ in their C-termini. β-types use two forms of CcmM to aggregate Rubisco through 3-repeats of small subunit-like domains (SSLDs). The M58 form includes an N-terminal γCAL domain that also binds carbonic anhydrase and CcmN. CcmN contains a C-terminal encapsulation peptide which enables interactions with the shell. Components are colored according to which other components they interact with.
Carboxysomes are comprised of an outer protein shell and an inner enzymatic cargo, and their primary function is to (i) decrease photorespiration by avoiding high [O2] from reaching Rubisco (Li et al., 2020; Ferlez et al., 2019), (ii) concentrate CO2 around Rubisco >1000x (Badger and Price, 2003), and (iii) physically compartmentalize the cytosolic bicarbonate pool from the wide majority of carbonic anhydrase activity to prevent carbon loss (Cai et al., 2009; Dou et al., 2008; Price and Badger, 1989) (Figure 1A). Functional carboxysomes are essential for cyanobacterial growth at low (ambient) CO2 environments (Abernathy et al., 2019), therefore, we must fully understand their underpinning mechanisms for their ultimate manipulation as modules for carbon fixation. Structural features of cyanobacterial carboxysomes.
Carboxysomes are part of a larger class of protein-bounded organelles in bacteria called bacterial microcompartments (BMCs). All BMC shells, including those of carboxysomes, are built from an array of structurally conserved hexameric (BMC-H), pseudo-hexameric/trimeric (BMC-T), and pentameric (BMC-P) proteins (Kerfeld et al., 2018; Melnicki et al., 2021). These proteins natively self-assemble into icosahedral shells which form a barrier between the bacterial cytosol and the interior of the BMC. BMC-H proteins consist of a single Pfam00936 domain and, together with BMC-T and their various permutations (Sutter et al., 2021), make up the bulk of the facets by tessellating tightly into a honeycomb-like lattice (Sutter et al., 2016). BMC-P, on the other hand, consists of a Pfam03319 domain and exists more fleetingly within the shell (Yang et al., 2020; Sun et al., 2022; Sutter et al., 2017), serving to only cap the vertex positions (Cai et al., 2009; Sutter et al., 2017; Tanaka et al., 2008). All BMC shell proteins have characteristic concave (cytosol facing) and convex (luminally facing) surfaces (Sutter et al., 2017; Trettel et al., 2022). Together, these proteins assemble into a barrier that enables selective influx/efflux of metabolites (Dou et al., 2008) thanks to central pores located at their central axis of symmetry (Kerfeld et al., 2005).
Carboxysomes are categorized into 2-classes; α- and β-carboxysomes housed in α- and β-cyanobacterial lineages (using cso or ccm gene nomenclature), respectively. While structurally conserved, experimental evidence suggests that ɑ- and β-carboxysomes differ in their evolution, operon structure, components (Rubisco type, scaffolds, carbonic anhydrases), and modes of assembly (Kerfeld and Melnicki, 2016; Rae et al., 2013) (Figures 1B, C). Current models propose that ɑ-carboxysomes assemble concomitantly (Kerfeld and Melnicki, 2016; Iancu et al., 2010) with the disordered scaffold protein CsoS2 acting as an essential hub (Cai et al., 2015a) that supports the co-condensation of the Rubisco holoenzyme and carbonic anhydrase (Oltrogge et al., 2020; Blikstad et al., 2023) with mosaicked shell subunits (Ni et al., 2023), eventually maturing into a concentration-dependent paracrystalline/fibril array of Rubisco packaged within the shell (Ni et al., 2023; Metskas et al., 2022; Evans et al., 2023). In contrast, β-carboxysome assemble core-first (Cameron et al., 2013) with the essential scaffold protein CcmM initiating the condensation of Rubisco (Ludwig et al., 2000; Wang et al., 2019; Zang et al., 2021; Wang and Hayer-Hartl, 2023; Ryan et al., 2019), carbonic anhydrase (Long et al., 2007; Long et al., 2011), and CcmN into a ‘pro-carboxysome’. The encapsulation peptide (EP) of CcmN (Kinney et al., 2012) promotes shell envelopment of the pro-carboxysome resulting in a mature particle (Cameron et al., 2013; Chen et al., 2013) where Rubisco also exists in a paracrystalline lattice (Faulkner et al., 2017). Despite their functional differences, BMC particles [carboxysomes and metabolosomes (Yang et al., 2022)] rely on the liquid-liquid phase separation (LLPS) (Azaldegui et al., 2021) of their internal components to trigger their assembly. Despite recent experimental insights, engineering aspects such as size, morphology, and multiplexed cargo packaging remain a challenge.
Carboxysome shells enhance carbon assimilation by concentrating CO2 and limiting O2 diffusion within the luminal space while enabling the influx of bicarbonate and blocking CO2 leakage outwards (Rae et al., 2013; Cai et al., 2009; Dou et al., 2008). Permeation is understood to primarily occur at the central pores in the cyclic axis of symmetry in BMC-H shell proteins (Kerfeld et al., 2005) (Figure 2A). These pores, being typically ~4-7 Å in diameter (Tanaka et al., 2008; Kerfeld et al., 2005; Tanaka et al., 2009), have been experimentally attributed as gates for substrate passage. For example, mutagenized pore-adjacent residues on BMC-H proteins alter the biochemical activity for the entire BMC particle in vitro as well as cell growth when tested in vivo (Chowdhury et al., 2015); it is hypothesized that these mutations at the pore constriction change the rate at which the interior enzymes access substrates that diffuse through those pores.
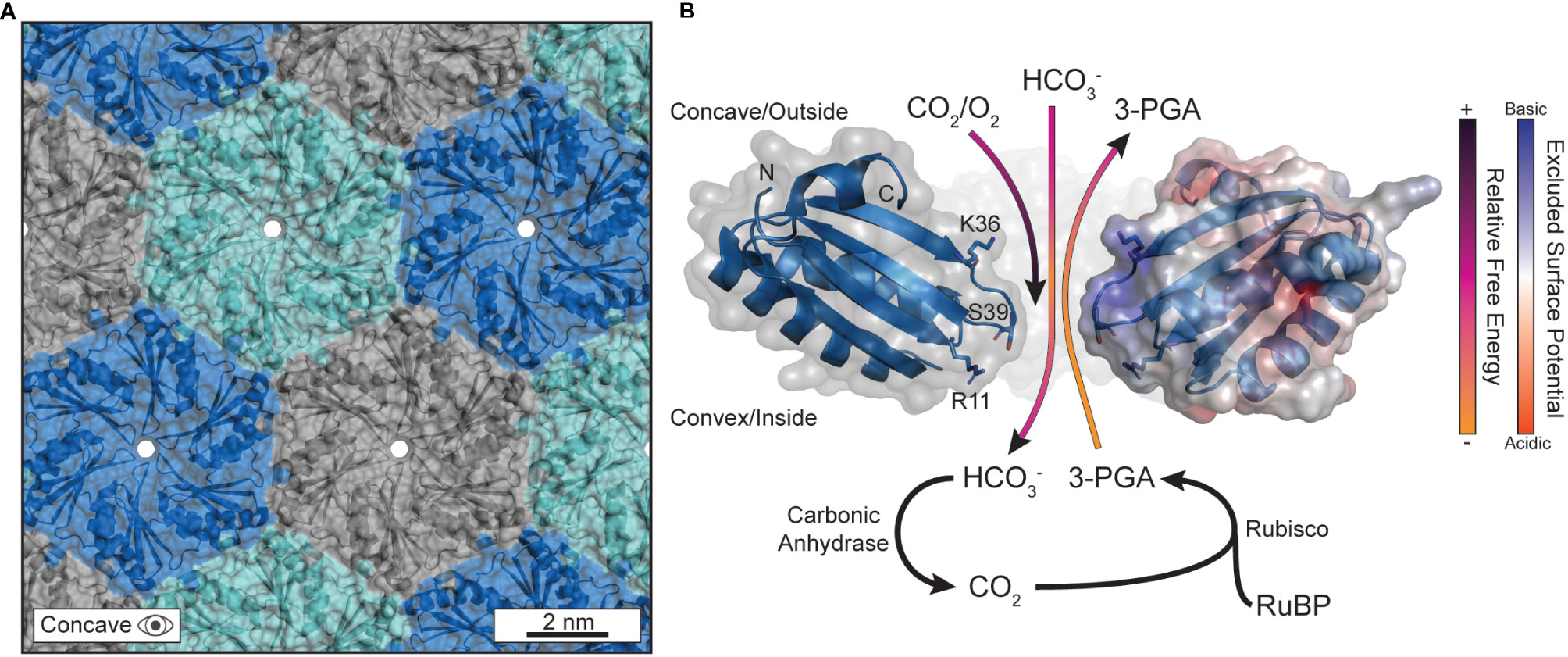
Figure 2 The pores in the protein shell are responsible for gating substrate permeation. (A) The outer protein shell is composed of hexagonally arranged shell proteins that tesselate into a tight-knit honeycomb-like lattice that forms a barrier against the cytosol. Currently, the only understood path for substrate diffusion is through pores that form at the central axis of symmetry in the hexameric quaternary structure. The PDB structure for CsoS1A (2G13) was used to generate this panel in PyMOL. (B) The biophysical properties that differentiate substrates, and their permeation through the central pore, are sequence encoded. Several substrates are visualized traversing the pore with their anticipated trajectories colored by their approximate relative free energy at that location as informed by Faulkner et al. The PDB structure for CcmK2 (2A1B) was used to generate this panel in PyMOL. The C-termini were clipped at residue 90 for clarity. The right-most monomer has its surface colored according to its excluded surface potential as calculated by the ABPS plugin in PyMOL.
MD approaches have affirmed and expanded upon the evidential importance of the central pore for substrate gating. Pioneering work came from modeling the major BMC-H protein (Yang et al., 2020) of the propanediol metabolosome in Salmonella enterica, PduA, using biased potentials (e.g. umbrella sampling, metadynamics) (Park et al., 2017). Calculations indicated a more favorable passage of 1,2-propanediol compared to the higher free-energy barrier for propionaldehyde (Park et al., 2017), confirming earlier assumptions that some intermediates, like the reactive propionaldehyde, are selectively sequestered within the BMC lumen to prevent toxicity (Sampson and Bobik, 2008) or carbon loss due to volatility (Penrod and Roth, 2006). The higher free-energy barrier does not wholly block substrates, like propionaldehyde, from diffusing but does impede the process kinetically. The mechanism was attributed simply to the higher hydrogen-bonding capacity of the central pore constriction, lined with a serine residue (S40) (Crowley et al., 2010), for 1,2-propanediol over propionaldehyde due to the extra hydroxyl group which acts as an additional hydrogen bond donor. Effectively, the serine-lined pore is a better binding site for 1,2-propanediol than propionaldehyde which promotes the formers passage so long as the binding is not overly strong (Bauer and Nadler, 2006). Such features from the Pdu BMC can reasonably be applied to carboxysomes, due to the high sequence and structural conservation between all BMC shell proteins (Melnicki et al., 2021). Indeed, experimental work in the propanediol metabolosome has further attributed the pore-lining S40 of PduA as critical to influencing permeation (Chowdhury et al., 2015) This result was later confirmed via simulation and experimentation on CcmK2 of the Synechocystis sp. PCC 6803 β-carboxysome with its S39 pore (Faulkner et al., 2020).
The residues lining the central constriction alone are not sufficient to explain substrate permeation across ɑ- and β-carboxysome shells as many BMC-H proteins common encode residues like serine and glycine at the pore constriction. Surface electrostatic density around the central pore has also been observed to be an effective mechanism of attracting/repelling substrates in several studies focusing on BMC-H proteins (Faulkner et al., 2020; Mahinthichaichan et al., 2018). In all cases, the pore-adjacent concave surface exhibits a high extent of positive charge effectively turning the outward facing surface into an electrostatic funnel (Figure 2B). This may explain the lower free-energy profile for bicarbonate just outside of the pore within the concavity, essentially attracting the negatively charged bicarbonate anions while conversely impeding the passage of neutral CO2 and O2 from reaching the interior [or, in the case of CO2, escaping once bicarbonate is converted luminally by carbonic anhydrase (Cai et al., 2009)] (Figure 2B). These works importantly provided the groundwork to understand not just CO2 concentration within the shell, but also the inhibition of O2 diffusion too. MD observations that O2 diffusion is biophysically impeded is substantiated by the presence of bioinformatically identified glycl-radical enzyme associated microcompartments (GRMs) (Zarzycki et al., 2015). GRMs have been speculated to help extend the range of environments that glycyl-radical enzymes can reasonably act in, as oxygen exposure inactivates these enzymes (Zarzycki et al., 2017; Zhang et al., 2001). This notion is further supported by engineered carboxysome shells that package oxygen-sensitive hydrogenases can impart enhanced activity in an aerobic environment (Li et al., 2020).
The specific residues (corresponding to R11, K36, and the pore S39 in CcmK2) responsible for forming a substrate barrier are largely conserved among CcmK2 proteins (Faulkner et al., 2020) and emphasize the importance of both the pore and concave surface overall in substrate gating (Figure 2B). The alignment of β-strands proximal to the pore on the concave surface (L31 to K36 in CcmK2) also expose their backbone amines and contribute to this effect. We note, however, that these specifics will differ between shell proteins. For example, BMC-H even among the same class will differ in surface electrostatics (Schmidt-Dannert et al., 2018) and therefore should not be taken as a one-size-fits-all rule (i.e. concave surface always being positive to the same degree) but instead as another layer of consideration when assessing permeation. While Rubisco and the carboxysome shell may not be able to sufficiently differentiate between CO2 and O2 (Poudel et al., 2020), the outer shell can enhance the passage bicarbonate and, in combination with the encapsulated carbonic anhydrase, locally increase the CO2 concentration around Rubisco. MD simulations have helped explain the molecular basis for carbon concentration in the carboxysomal CCM and will be an essential methodology to predictively modify the shell for augmented substrate specificities moving forward.
The proclivity of BMC shell pores to bind anionic species is not limited to solely bicarbonate. Other anions, like sulfate, have been found in crystal structures of a broad range of BMC-H proteins including CcmK1, CsoS1A, and EutM to list a few (Tanaka et al., 2008; Tsai et al., 2007; Takenoya et al., 2010). MD have also revealed structural aspects of ion coordination. For instance, chloride ions have been observed to occupy pore-adjacent positions (Faulkner et al., 2020; Mahinthichaichan et al., 2018) and coordinate with either backbone amides or basic residues, such as arginine. Similar results were recently found for the metabolosome BMC-H PduA, where chloride ions were found to coordinate with the backbone amide of the pore-lining S40 (Trettel et al., 2023). This study also found that chloride itself also acts competitively with 1,2-propanediol, the intended substrate, for pore access thereby hindering permeation rates. Altogether, simulation data from both carboxysome and metabolosome models both agree on the ability of anions to coordinate with and occupy shell protein pores via non-specific backbone interactions (Faulkner et al., 2020; Trettel et al., 2023). While only currently reported for metabolosome shells, this suggests that ion coordination may be a widespread phenomenon which can also regulate permeation events in carboxysomes. The role of other physiologically relevant anions, such as inorganic phosphate which can regulate Rubisco activity (Marcus and Gurevitz, 2000), has yet to be explored in this context.
Bacterial microcompartments, including carboxysomes, can vary in size and regularly do not demonstrate a singular defined structure. This differs greatly from similarly icosahedral, although evolutionarily unrelated (Krupovic and Koonin, 2017), viral capsids and complicates the direct structural assessment of native BMC complexes. Understanding the dynamics of carboxysome self-assembly can shed light on the polydispersity and factors that control it and thereby tune factors which directly contribute to carbon fixation like surface-to-volume ratios, Rubisco organization, and Rubisco packaging efficiency. While inspired by simulations that explain viral capsid assembly that typically form around nucleic acids (Perlmutter et al., 2013; Lynch et al., 2023), new models pertaining to BMC assembly specifically needed to be developed to explain the subtle differences that trigger biogenesis and heterogeneous assemblies.
Initial attempts at modeling BMC assembly were inspired by carboxysomes where evidence has been found for both concomitant and core-first assembly pathways (Perlmutter et al., 2016) as observed in both ɑ- and β- lineages (Kerfeld and Melnicki, 2016) (Figure 3). The principle differentiating factor was the relative strength of attraction cargo had for other cargo, where weaker interactions led to ‘one-step’ or concomitant assembly (observed in ɑ-carboxysomes) (Figures 3A, B) while stronger interactions led to ‘two-step’ or core-first (observed in β-carboxysomes) (Figures 3A, C). Specifically, for ɑ-carboxysomes, modeling (Mahalik et al., 2016) and atomic-force microscopy (Sutter et al., 2016; Garcia-Alles et al., 2017) have both suggested that shell facets form by nucleation, which can further provide an area to locally concentrate cargo (Oltrogge et al., 2020) and nucleate ɑ-carboxysome formation, since cargo-cargo interactions are predicted to not be strong enough drivers on their own (Perlmutter et al., 2016) (Figure 3B). β-carboxysome cargo (Rubisco and CcmM M35) in two-step assembly modes coalesce strongly enough on their own without the need of a shell-templated trigger (Figure 3C). Interestingly, these simulations predicted that cargo would become organized into concentric layers, observed prior in both ɑ- (Iancu et al., 2007; Shively et al., 1973; Schmid et al., 2006) and β-carboxysomes (Kaneko et al., 2006; Iancu et al., 2005). Paracrystalline order was not a prerequisite for forming complete particles in these simulations and in fact would inhibit budding (Figure 3A). These observations have held up to additional recent higher-resolution experimental scrutiny, where Rubisco in both ɑ- and β-carboxysomes is now understood to assemble into concentric layers (Evans et al., 2023; Faulkner et al., 2017; Ni et al., 2022) when the internal concentration is sufficiently high (Metskas et al., 2022). While just the first of many follow-up studies, Perlmutter et al.’s above work demonstrated the utility of computational modeling to understand carboxysome assembly. However, the system employed at the time, albeit elegant, only investigated one shell geometry (T = 3), one BMC-H and BMC-P, and one cargo. This initial model has been greatly expanded to include considerations like the impact of cargo packaging on BMC size (Mohajerani and Hagan, 2018), the role of scaffolds (Mohajerani et al., 2021), and even multiple cargos (Tsidilkovski et al., 2022) on microcompartment size, assembly pathway, and packaging efficiency (Figure 4A).
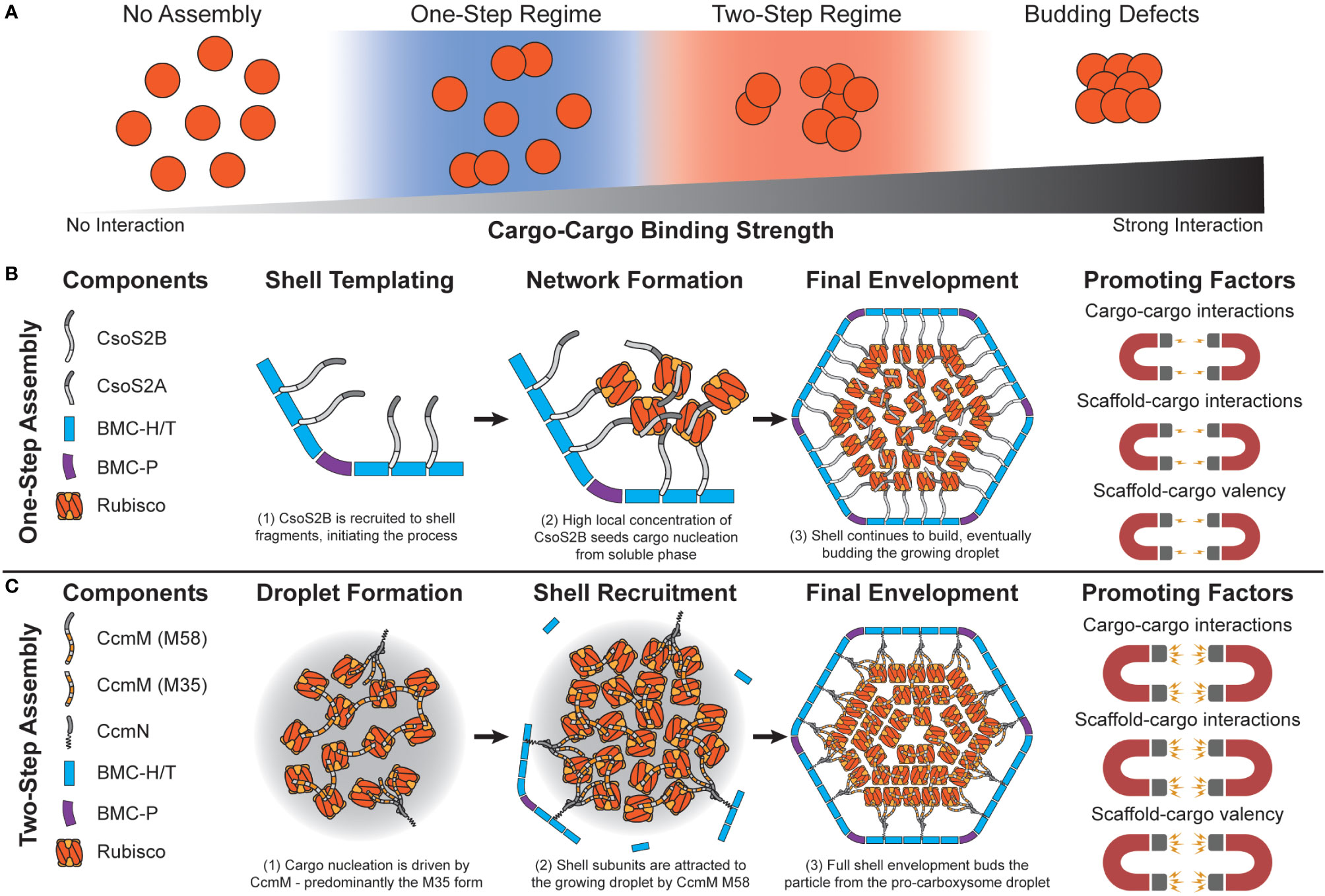
Figure 3 Carboxysome assembly pathways depend on the relative propensity of cargo to aggregate. (A) Simulations predict that carboxysome assembly pathway exists on a continuum dependent on cargo-cargo binding strengths. Very weak or no binding propensity inhibits assembly of filled shells. Weak/moderate binding strengths results in one-step assembly pathways, as either high concentrations of cargo or shell components that locally increase cargo concentration are needed. Moderate/strong binding strengths lead to a two-step pathway, where cargo can coalesce independent of a shell. Overly strong binding strengths inhibits budding of the cargo droplet by shell components. (B) In the one-step pathway (also called concamitant), cargo proteins Rubisco, CsoS2A, and CsoS2B do not interact strongly enough to inititate phase separation from the bulk. CsoS2B must first bind shell facets/vertices. This creates a local environment with a high concentration of CsoS2B N-terminal repeats that attract Rubisco and CsoS2A. The droplet growth cascades until a critical mass of shell proteins envelope it, resulting in a mature alpha carboxysome. Molecular simulations reveal that this pathway is promoted by relatively weaker cargo-cargo interactions/valency. (C) In the two-step pathway (also called core-first), cargo proteins Rubisco and both forms of CcmM together coalesce a pro-carboxysome droplet. CcmN allows for shell components to begin templating around the growing droplet, eventually budding a complete particle. Molecular simulations reveal that this pathway is promoted by relatively strong cargo-cargo interactions/valencies. The carbonic anhydrase component in both examples is omitted for clarity.
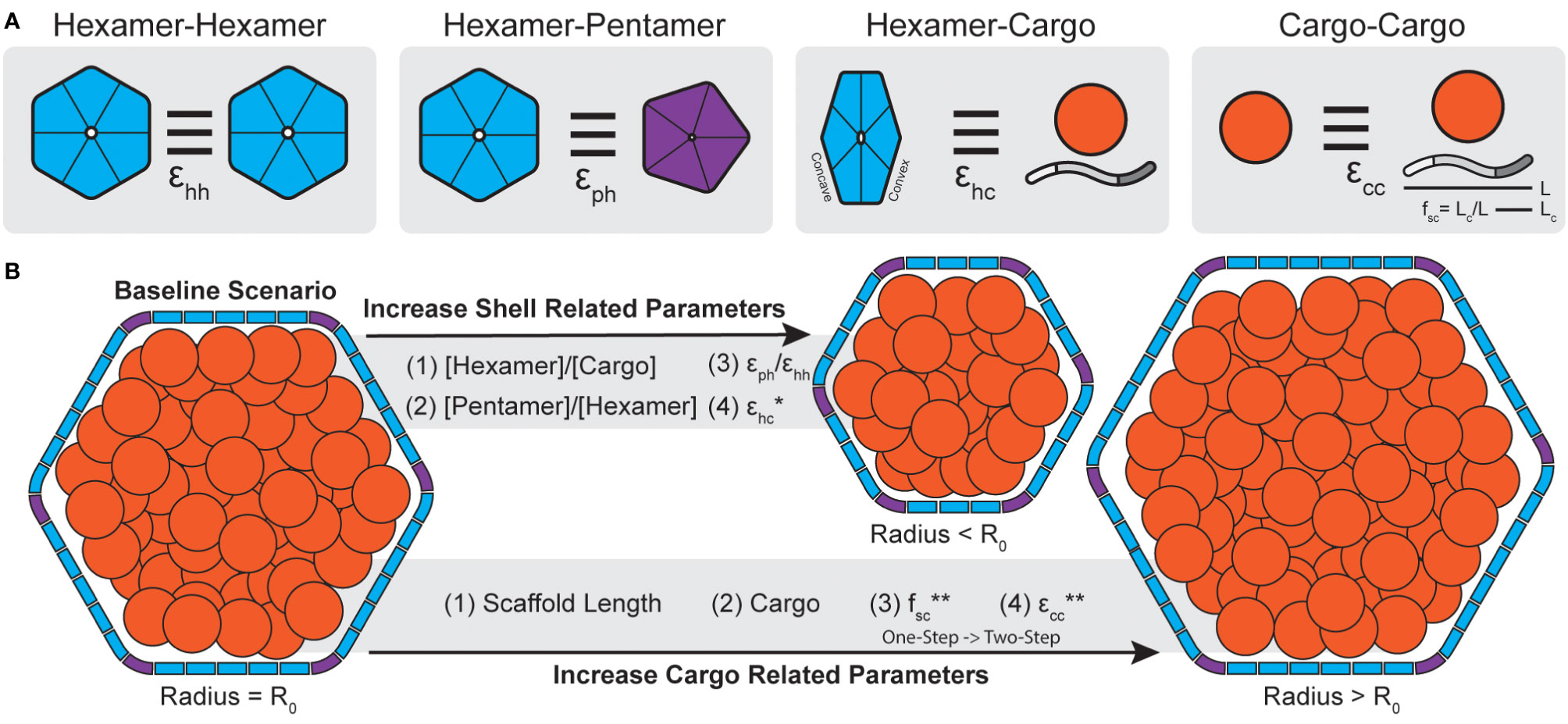
Figure 4 Carboxysome morphology is determined by a combination of shell and cargo related parameters. (A) The interactions strengths between different components of a BMC can be parameterized (ϵ). In simulations, hexamers can interact edge-to-edge while having matching surfaces oriented in parallel with interactions strength ϵhh. Hexamers and pentamers can likewise interact with strength ϵph. Cargo components, defined as both scaffolds like CsoS2 (shown) or general cargo (orange circle) can interact with the convex surface of shell hexamers with strength ϵhc. Lastly, cargo, in the form of general cargo or scaffolds, can form self interactions of strength ϵcc. In the case of scaffolds, the fraction of the protein that bind cargo (fsc) is defined as the length of the cargo binding domain (Lc, akin to valency or number of binding sites) divided by the total scaffold length (L). (B) Molecular simulations reveal parameters that alter morphology during assembly. Generally, increasing parameters (stoichiometry, interaction strengths) related to the shell lead to smaller particles. Conversely, increasing parameters related to cargo aggregation, or including cargo at all, leads to larger particles. One except is that increasing ϵhc for general cargo will result in smaller particles but for scaffolds will not (*, see Mohajerani et al., 2021). Note, assembly pathways in simulations shift from one-step to two-step with increasing fsc and ecc (**). Accordingly, the two-step pathways are generally associated with larger particles.
In the context of microcompartment size, dynamical simulations reveal that shells packaged with cargo, generally, tend to be larger than non-packaged shells (Mohajerani and Hagan, 2018). Further, these simulations showed that BMC size also correlates with assembly pathways where core-first assembly modes, dominated kinetically by relatively stronger cargo-cargo interactions, led to larger particles and up to 5-fold more packaged cargo than concomitant modes (Mohajerani and Hagan, 2018). These results similarly apply to multi-component systems, where assembly pathway is primarily delineated by the sum of the cargo interaction strengths and the strength of self-cargo interactions (Figure 4B) can dictate packaging stoichiometry (Tsidilkovski et al., 2022). This has been likewise observed in real BMC systems where β-carboxysomes, which follow a core-first assembly pathway, tend to be larger than their ɑ- counterparts (Whitehead et al., 2014). Empty, synthetic structural models reported thus far are also always far smaller than native BMCs (<40 nm diameter) (Sutter et al., 2017; Ni et al., 2023; Tan et al., 2021; Sutter et al., 2019a; Kalnins et al., 2020; Greber et al., 2019; Sutter et al., 2019b).
Dynamical simulations have also revealed that shell components, while not the predominant factor, can also influence final morphology. For instance, simply increasing the ratio of shell proteins to cargo can lead to overnucleation (Mohajerani and Hagan, 2018) and thus smaller particles (Figure 4B). This has also been found in simulations which assume shell proteins demonstrate no spontaneous curvature of their own [motivated by atomic force microscopy studies on shell subunits (Sutter et al., 2016; Garcia-Alles et al., 2017)] and can essentially trap a growing cargo droplet out of equilibrium (Rotskoff and Geissler, 2018). This, however, may depend on the system of study as shell proteins have been observed to form sheets, nanotubes, and empty icosahedra among other morphologies, sometimes within the same sample (Ferlez et al., 2023), without the need of cargo templating to induce curvature (Ferlez et al., 2023; Trettel and Winkler, 2023; Uddin et al., 2018; Hagen AR. et al., 2018; Noël et al., 2015). The presence of excess pentamers or stronger pentamer-hexamer interactions can likewise lead to more pentamer insertion and thus overnucleation into smaller particles (Mohajerani and Hagan, 2018) (Figure 4B). This latter point is interesting since many BMC operons encode for more than one BMC-P (Sutter et al., 2021). BMC-P proteins appear to play different roles in different contexts, where in some metabolosomes they can directly influence BMC morphology (Mills et al., 2022) and in others they are completely dispensable and can be added exogenously to “cap” the icosahedron (Sutter et al., 2019b; Hagen A. et al., 2018; Kirst et al., 2022). Observations from simulations further emphasize the importance of studying the effects of BMC-P and how they can influence morphology, packaging efficiency and permeability.
The above studies ascribed the connection of homogenous cargo to end morphology. However, the models used may be more applicable to metabolosomes, where cargo directly interacts with the shell (Fan et al., 2010; Aussignargues et al., 2015), than carboxysomes, where scaffolds act as an intermediary connecting the shell and cargo domains (Oltrogge et al., 2020; Wang et al., 2019). Accordingly, Mohajerani et al. have also conducted a study, motivated by ɑ-carboxysomes specifically, on the role of a CsoS2-inspired scaffold proteins in BMC assembly (Mohajerani et al., 2021) (Figure 4A). Scaffolds proteins, as a type of cargo themselves, can potentially affect shell size and assembly pathway in a much more programmable manner than typical cargo due to their modular nature (Chaijarasphong et al., 2016) (Figure 1C). Simulations parametrized these physical aspects by defining the length of the CsoS2-inspired scaffold (L), the length of the cargo binding domain [Lc, with longer Lc meaning more cargo binding sites, akin to more CsoS2 N-terminal domains (NTDs)] and the fractional length of the cargo binding domain (fsc = Lc/L) as shown in Figure 4A. Importantly, simulations reveal that there is a critical interplay between the total length of the scaffold and its valency with cargo. By fixing the overall scaffold length (L) and increasing Lc (and therefore fsc), simulations showed that cargo packaging likewise increases. Moreover, increasing fsc, analogous to the number of cargo binding sites, transitioned systems from a one-step to a two-step assembly pathway (Figure 4B) where two-step pathways are again associated with more cargo packaging (Mohajerani et al., 2021). Similarly to cargo packaging alone (Mohajerani and Hagan, 2018), as the scaffold is itself a type of cargo, physically longer scaffolds also generally result in larger shells to a point as they increase volume requirements (Mohajerani et al., 2021). These simulations are supported by work in the model H. neapolitanus α-carboxysome that demonstrates a requirement for a minimal threshold of NTDs in CsoS2 to be met to achieve carboxysome formation (Oltrogge et al., 2020). Further, more recent work by Oltrogge and colleagues likewise agree that increasing CsoS2 length by increasing the number of middle region (MR) repeats leads to larger α-carboxysomes (Oltrogge et al., 2023). However, they ascribe this phenomenon to the MR repeats of CsoS2 stabilizing the low-curvature regions (i.e. the facets) of the carboxysome shell, enabling their extension, while Mohajerani et al. argue for the need to meet increased volume requirements. We note that these arguments are not mutually exclusive.
MD simulations of shell permeation to date have focused on a small subset of model BMC-H. While impactful, future permeation studies may wish to sample a greater diversity of BMC-H to develop a deeper understanding of the natural biophysical diversity shell proteins can accommodate. For instance, sampling a wider array of carboxysomal BMC-H may highlight subtle differences that influence bicarbonate, O2, and 3-PGA diffusion. Similar methodologies can and should be applied towards describing permeation in mixed heterohexamer systems, like those reported for CcmK3/K4 (Sommer et al., 2019; Garcia-Alles et al., 2019) or purely synthetic systems with the potential for asymmetric pore designs (Česle et al., 2023) that may further regulate substrate diffusion in ways that homo-hexameric BMC-H cannot. The various classes of BMC-T should also be considered to better grasp their hypothesized connection to substrate gating (Klein et al., 2009; Tanaka et al., 2010). Simulation scale also needs to be accounted for, and future studies may wish to engage with physiologically relevant systems with multiple components like the small synthetically-derived BMC shells (Sutter et al., 2017; Ni et al., 2023; Tan et al., 2021; Sutter et al., 2019a; Kalnins et al., 2020; Greber et al., 2019; Sutter et al., 2019b), as a proxy for larger native-like systems. Investigating more complex shells will progress our understanding of how chemical gradients, a physiologically critical component, behave and evolve within a BMC context. For instance, differences in density and packing of Rubisco within carboxysomes (Kaneko et al., 2006; Ni et al., 2022) may result in CO2/O2 gradients proportional to the enzymes’ proximity to the shell. Detailed permeation studies could discern the packaging attributes within BMCs that would result in more efficient catalytic properties in engineered architectures. Simulating whole-BMC shell models can also limit pore-centric bias and explore if flux exists in non-porous areas such as the hexamer-hexamer interfaces or corner junctions where three hexamers meet. Similarly, permeability studies can be expanded to study the diffusion of a wider swath of metabolites and cofactors through BMC shell structures. Current research suggests that these cofactors, like NAD(P)H, are maintained as private pools that are internally recycled and do not appreciably diffuse through the shell barrier (Huseby and Roth, 2013; Cheng et al., 2012). Regardless, novel BMCs may be sought to transform metabolites far larger than those found in current model systems. MD simulations of permeation, therefore, will continue to facilitate rapid in silico prototyping of permeation through protein shells for altered substrate specificities or enhanced carbon concentration within the carboxysome lumen.
Specific structural components of the shell, such as the extended C-termini on many BMC-H, should also be addressed. While typically ignored due to missing crystallographic data, these termini can now be predicted and integrated into computational models thanks to emerging computational tools. These outward-facing, flexible/disordered (Faulkner et al., 2020) termini have been implicated in functions such as assembly (Trettel et al., 2022; Klein et al., 2009) like in viruses (Xue et al., 2014), but some data suggests they also reach into the concavity of adjacent subunits (Trettel et al., 2022) which may impact permeation or fine-tune assembly in environmentally responsive ways.
The collective knowledge on carboxysome systems continually expands and reinvents our understandings of these complex systems. Incorporating simulations to complement emerging experimental insights will lead to more meaningful outputs to inform design choices. For instance, future modeling may wish to explore evidence-informed shell-cargo interaction sites that form from predominantly (i) the edge-edge interaction surface of two adjoining shell proteins (Ni et al., 2023) and, in some cases, (ii) interactions with specific interior-oriented domains such as the N-terminus of the PduB BMC-T (Trettel et al., 2022; Lehman et al., 2017; Kennedy et al., 2022). This is further underpinned by the multitude of different shell proteins BMCs can encode and their synthetic interchangeability (Cai et al., 2015b; Slininger Lee et al., 2017) which certainly influence shell-shell (including curvature) and shell-cargo/scaffold interactions. For example, many BMCs encode for BMC-T proteins where every other edge may be better attuned for specific shell interactions on adjacent subunits (Trettel et al., 2022; Waltmann et al., 2023) and influence factors like shell curvature and/or shell-cargo interactions. Many others bioinformatically identified BMC loci entirely lack these factors for unknown reasons (Sutter et al., 2021).
The luminal organization of Rubisco is also now known to differ between related carboxysomes and may be tied to overall carboxysome activity, For instance, Halothiobacillus α-carboxysomes exhibit ~2-fold higher activity than Cyanobium α-carboxysomes and have different modes of Rubisco organization (Ni et al., 2022). Future simulations may have an opportunity to explain how these subtle structural differences arise (i.e. Rubisco surface charge difference, CsoS2 binding affinity, internal Rubisco concentration), ascribe functional consequences, and reveal how to program desired internal conformations.
Shell-focused assembly simulations can also help better define and explain the mechanisms behind the varied supramolecular structures BMC shell proteins can form in vivo and in vitro, such as nanotubes, for designer protein scaffolds (Young et al., 2017). Recent work suggests that BMC-H curvature trends can be inferred by their crystal structural arrangements (Garcia-Alles et al., 2023) and that these trends can be modulated rationally with computationally-informed amino acid substitutions (Li et al., 2021). However, factors like buffer/environmental composition (Faulkner et al., 2019), shell protein class and stoichiometry, and protein disorder undoubtedly also factor into supramolecular, and native-like, structures in unknown ways. In particular, the disordered termini many BMC-H proteins carry, predominantly on their outward facing C-terminus (Sutter et al., 2017; Trettel et al., 2022) have been speculated to fine-tune both shell-shell and shell-cargo interactions (Fan et al., 2012). Further, currently described simulation systems may already be attuned to ascribe the role of the multiple pentamers BMCs can encode for by tuning their relative stoichiometry and interaction strengths. Computational studies will undoubtedly continue to address these considerations and many more for custom carbon-fixing scaffolds.
Assembly-focused simulations teach us that assembly pathway is chiefly governed by cargo interaction strengths (Figure 3A) while final morphology is determined by both cargo and shell contributions (Figure 4B). In terms of assembly pathway, stronger cargo-cargo (including scaffold) interactions or higher cargo stoichiometries are typically associated with two-step assembly pathways that lead to larger shells with more cargo (Mohajerani and Hagan, 2018; Mohajerani et al., 2021; Tsidilkovski et al., 2022) (Figure 4B). Conversely, weaker cargo-cargo interactions or higher shell stoichiometries are associated with one-step assembly pathways and smaller shells (Mohajerani and Hagan, 2018; Mohajerani et al., 2021; Tsidilkovski et al., 2022) (Figure 4B). These findings carry direct carboxysome design implications related to assembly kinetics that manifest physically in the forms of (i) expression system design and (ii) scaffold design.
Cargo and shell constructs can be designed in both a continuous synthetic operon or discontinuously into different plasmids with different modes of induction for testing (Lee and Tullman-Ercek, 2017). Single-vector/operon designs have been successful using a variety of induction approaches (Bonacci et al., 2012; Graf et al., 2018; Flamholz et al., 2020; Jiang et al., 2023). Notably, similar strategies also result in morphologically and functionally sound carboxysomes when genomically integrated and expressed in plants (Chen et al., 2023; Long et al., 2018). Double-vector systems, which independently express shell and cargo components, have also been described (Jiang et al., 2023; Jakobson et al., 2016; Wagner et al., 2017) although they do need to be tuned and timed appropriately (Lee and Tullman-Ercek, 2017; Jakobson et al., 2016; Nichols et al., 2019) likely due to kinetic effects of aggregation described by simulations. In one case, researchers redesigned a carboxysome for hydrogen production by serially inducing hydrogenase cargo followed by a β-carboxysome shell (Li et al., 2020). BMCs with concomitant assembly pathways, like those commonly employed for heterologous α-carboxysomes formation, may benefit from single vector designs which promote co-expression of both shell and cargo components under native-like controls (i.e. ribosomal binding sites). Similarly, two-step pathways may be promoted by a well-tuned cargo-preaggregation step proceeded by shell expression. Researchers should consider the kinetics of interactions and expression to prevent off-target assemblies.
Scaffold choice and design is also an emerging route for modification. In ɑ-carboxysomes, modifying CsoS2 and the ratios of CsoS2A and CsoS2B (analogous to fsc in simulations) or the number of NTD/MR repeats (Oltrogge et al., 2023) are approachable routes to alter morphology and Rubisco packaging for CCM augmentation. Similarly, in β-carboxysomes, modifications of CcmM and CcmN may also be sufficient routes for modification. However, both classes of scaffolds act as specific adaptors between the Rubisco cargo and the shell domains and therefore cannot coalesce a more diverse range of cargo by themselves without extensive modification. Heterologous encapsulation and assembly methods may wish to rely on carboxysome-inspired fusions (Gonzalez-Esquer et al., 2015) or metabolosome EPs which trigger both shell-cargo (Fan et al., 2010) and cargo-cargo (Lawrence et al., 2014). Cargo fused with metabolosome EPs may act more akin to the assembly models produced in several assembly simulation works to date (Perlmutter et al., 2016; Mohajerani and Hagan, 2018; Tsidilkovski et al., 2022).
One bottleneck with biomass productivity lies in the connection between photosynthetic efficiency and carbon fixation. To alleviate these bottlenecks, some groups have installed various CCM components into plant chloroplasts including Nicotiana benthamiana (Lin et al., 2014), Rhodosprillum rubrum (Long et al., 2018), and Nicotiana tabacum (Chen et al., 2023). These studies have been able to generate carboxysomes nearly structurally and catalytically equivalent to native carboxysomes and support photosynthesis (Chen et al., 2023). Further additional factors like the incorporation of bicarbonate transporters, removal of the stromal carbonic anhydrase, and including Rubisco activates (Chen et al., 2022) may be needed to significantly enhance growth under ambient CO2 conditions. A deeper fundamental understanding of carboxysome assembly offered by computational simulations may assist in full implementation of cyanobacterial CCMs into C3 plants.
Simulations have played a critical role in exploring the physical phenomena that underpin carboxysome assembly. However, many conclusions remain explored at low resolution, partially due to the technics used. More investment in multi-resolution calculations is required for incorporating high accuracy detailed mechanisms at commensurate computational investment. Such methodologies may only be possible after exercising high fidelity energy landscape reconstruction based on accelerated MD or AI assisted methodologies for the fast interconversion between low resolution models (e.g. supra coarse-grained) and fully atomic detailed structures. Only this approach would be able to lead to a more fine-tuned and robust rational carboxysome manipulation.
The integration of in silico predictive and analytical methods with in vivo structure/function studies of BMCs is essential to advance BMC-based biotechnologies. MD simulations have been critical in describing the fundamental principles underlying permeation events through protein shells and fundamental principles that underpin carboxysome assembly. MD simulations reveal that substrate permeation is controlled by a series of biophysical properties, encoded by residues mainly along the outer concave surface, and substrate competition. Simulations studying BMC assembly demonstrate that assembly pathway is controlled kinetically by cargo accumulation and morphology is dictated by a combination of shell and cargo parameters. The ever-increasing access to computational power, and methodologies (i.e., machine-learning algorithms), will undoubtably expand these findings and allow for a higher-throughput exploration of the BMC diversity and the redesign of these architectures for specific non-native biochemical traits. Such advancements will continue to impact how we think and tinker with these architectures and help implement programmable BMCs for biomanufacturing and enhanced CO2 sequestration roles.
DT: Conceptualization, Formal Analysis, Investigation, Writing – original draft, Writing – review & editing. SP: Writing – original draft, Writing – review & editing. AL: Writing – original draft, Writing – review & editing. CG: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. DT, SP, AL, and CG acknowledge funding by LANL’s Laboratory Directed Research and Development (LDRD) grant 20220387ER.
We would like to thank Dr.’s S. (Gnana) Gnanakaran, Cesar Lopez, and Hung Do for helpful discussions and their expertise on computational simulations while preparing this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abernathy, M. H., Czajka, J. J., Allen, D. K., Hill, N. C., Cameron, J. C., Tang, Y. J. (2019). Cyanobacterial carboxysome mutant analysis reveals the influence of enzyme compartmentalization on cellular metabolism and metabolic network rigidity. Metab. Eng. 54, 222–231. doi: 10.1016/j.ymben.2019.04.010
Andersson, I., Backlund, A. (2008). Structure and function of Rubisco. Plant Physiol. Biochem. 46, 275–291. doi: 10.1016/j.plaphy.2008.01.001
Artier, J., Holland, S. C., Miller, N. T., Zhang, M., Burnap, R. L. (2018). Synthetic DNA system for structure-function studies of the high affinity CO. Biochim. Biophys. Acta Bioenerg. 1859, 1108–1118. doi: 10.1016/j.bbabio.2018.06.015
Aussignargues, C., Paasch, B. C., Gonzalez-Esquer, R., Erbilgin, O., Kerfeld, C. A. (2015). Bacterial microcompartment assembly: The key role of encapsulation peptides. Commun. Integr. Biol. 8, e1039755. doi: 10.1080/19420889.2015.1039755
Azaldegui, C. A., Vecchiarelli, A. G., Biteen, J. S. (2021). The emergence of phase separation as an organizing principle in bacteria. Biophys. J. 120, 1123–1138. doi: 10.1016/j.bpj.2020.09.023
Badger, M. R., Andrews, T. J., Whitney, S. M., Ludwig, M., Yellowlees, D. C., Leggat, W., et al. (1998). The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO 2 -concentrating mechanisms in algae. Can. J. Bot 76 (6), 1052–1071. doi: 10.1139/cjb-76-6-1052
Badger, M. R., Price, G. D. (1992). The CO2 concentrating mechanism in cyanobacteria and microalgae. Physiol. Plantarum 84, 606–615. doi: 10.1111/j.1399-3054.1992.tb04711.x
Badger, M. R., Price, G. D. (2003). CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J. Exp. Bot. 54, 609–622. doi: 10.1093/jxb/erg076
Bar-On, Y. M., Milo, R. (2019). The global mass and average rate of rubisco. Proc. Natl. Acad. Sci. U. S. A. 116, 4738–4743. doi: 10.1073/pnas.1816654116
Bauer, W. R., Nadler, W. (2006). Molecular transport through channels and pores: effects of in-channel interactions and blocking. Proc. Natl. Acad. Sci. U. S. A. 103, 11446–11451. doi: 10.1073/pnas.0601769103
Blikstad, C., Dugan, E. J., Laughlin, T. G., Turnšek, J. B., Liu, M. D., Shoemaker, S. R., et al. (2023). Identification of a carbonic anhydrase-Rubisco complex within the alpha-carboxysome. Proc. Natl. Acad. Sci. U. S. A. 120, e2308600120. doi: 10.1073/pnas.2308600120
Bonacci, W., Teng, P. K., Afonso, B., Niederholtmeyer, H., Grob, P., Silver, P. A., et al. (2012). Modularity of a carbon-fixing protein organelle. Proc. Natl. Acad. Sci. U. S. A. 109, 478–483. doi: 10.1073/pnas.1108557109
Busch, F. A. (2020). Photorespiration in the context of Rubisco biochemistry, CO2 diffusion and metabolism. Plant J. 101, 919–939. doi: 10.1111/tpj.14674
Cai, F., Dou, Z., Bernstein, S. L., Leverenz, R., Williams, E. B., Heinhorst, S., et al. (2015a). Advances in understanding carboxysome assembly in prochlorococcus and synechococcus implicate CsoS2 as a critical component. Life (Basel) 5, 1141–1171. doi: 10.3390/life5021141
Cai, F., Menon, B. B., Cannon, G. C., Curry, K. J., Shively, J. M., Heinhorst, S. (2009). The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PloS One 4, e7521. doi: 10.1371/journal.pone.0007521
Cai, F., Sutter, M., Bernstein, S. L., Kinney, J. N., Kerfeld, C. A. (2015b). Engineering bacterial microcompartment shells: chimeric shell proteins and chimeric carboxysome shells. ACS Synth. Biol. 4, 444–453. doi: 10.1021/sb500226j
Cameron, J. C., Wilson, S. C., Bernstein, S. L., Kerfeld, C. A. (2013). Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155, 1131–1140. doi: 10.1016/j.cell.2013.10.044
Česle, E. E. L., Ta Rs, K., Jansons, J., Kalniņš, G. (2023). Modulation of hybrid GRM2-type bacterial microcompartment shells through BMC-H shell protein fusion and incorporation of non-native BMC-T shell proteins. ACS Synth. Biol. 12, 3275–3286. doi: 10.1021/acssynbio.3c00281
Chaijarasphong, T., Nichols, R. J., Kortright, K. E., Nixon, C. F., Teng, P. K., Oltrogge, L. M., et al. (2016). Programmed ribosomal frameshifting mediates expression of the α-carboxysome. J. Mol. Biol. 428, 153–164. doi: 10.1016/j.jmb.2015.11.017
Chen, T., Fang, Y., Jiang, Q., Dykes, G. F., Lin, Y., Price, G. D., et al. (2022). Incorporation of functional Rubisco activases into engineered carboxysomes to enhance carbon fixation. ACS Synth. Biol. 11, 154–161. doi: 10.1021/acssynbio.1c00311
Chen, T., Hojka, M., Davey, P., Sun, Y., Dykes, G. F., Zhou, F., et al. (2023). Engineering α-carboxysomes into plant chloroplasts to support autotrophic photosynthesis. Nat. Commun. 14, 2118. doi: 10.1038/s41467-023-37490-0
Chen, A. H., Robinson-Mosher, A., Savage, D. F., Silver, P. A., Polka, J. K. (2013). The bacterial carbon-fixing organelle is formed by shell envelopment of preassembled cargo. PloS One 8, e76127. doi: 10.1371/journal.pone.0076127
Cheng, S., Fan, C., Sinha, S., Bobik, T. A. (2012). The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PloS One 7, e47144. doi: 10.1371/journal.pone.0047144
Chowdhury, C., Chun, S., Pang, A., Sawaya, M. R., Sinha, S., Yeates, T. O., et al. (2015). Selective molecular transport through the protein shell of a bacterial microcompartment organelle. Proc. Natl. Acad. Sci. U. S. A. 112, 2990–2995. doi: 10.1073/pnas.1423672112
Crowley, C. S., Cascio, D., Sawaya, M. R., Kopstein, J. S., Bobik, T. A., Yeates, T. O. (2010). Structural insight into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell. J. Biol. Chem. 285, 37838–37846. doi: 10.1074/jbc.M110.160580
Davidi, D., Shamshoum, M., Guo, Z., Bar-On, Y. M., Prywes, N., Oz, A., et al. (2020). Highly active rubiscos discovered by systematic interrogation of natural sequence diversity. EMBO J. 39, e104081. doi: 10.15252/embj.2019104081
Dou, Z., Heinhorst, S., Williams, E. B., Murin, C. D., Shively, J. M., Cannon, G. C. (2008). CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J. Biol. Chem. 283, 10377–10384. doi: 10.1074/jbc.M709285200
Evans, S. L., Al-Hazeem, M. M. J., Mann, D., Smetacek, N., Beavil, A. J., Sun, Y., et al. (2023). Single-particle cryo-EM analysis of the shell architecture and internal organization of an intact α-carboxysome. Structure 31, 677–88.e4. doi: 10.1016/j.str.2023.03.008
Fan, C., Cheng, S., Liu, Y., Escobar, C. M., Crowley, C. S., Jefferson, R. E., et al. (2010). Short N-terminal sequences package proteins into bacterial microcompartments. Proc. Natl. Acad. Sci. U. S. A. 107, 7509–7514. doi: 10.1073/pnas.0913199107
Fan, C., Cheng, S., Sinha, S., Bobik, T. A. (2012). Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc. Natl. Acad. Sci. U. S. A. 109, 14995–15000. doi: 10.1073/pnas.1207516109
Faulkner, M., Rodriguez-Ramos, J., Dykes, G. F., Owen, S. V., Casella, S., Simpson, D. M., et al. (2017). Direct characterization of the native structure and mechanics of cyanobacterial carboxysomes. Nanoscale 9, 10662–10673. doi: 10.1039/C7NR02524F
Faulkner, M., Szabó, I., Weetman, S. L., Sicard, F., Huber, R. G., Bond, P. J., et al. (2020). Molecular simulations unravel the molecular principles that mediate selective permeability of carboxysome shell protein. Sci. Rep. 10, 17501. doi: 10.1038/s41598-020-74536-5
Faulkner, M., Zhao, L., Barrett, S., Lu-Ning, L. (2019). Self-assembly stability and variability of bacterial microcompartment shell proteins in response to the environmental change. Nanoscale Res. Lett. 14, 1–8. doi: 10.1186/s11671-019-2884-3
Ferlez, B. H., Kirst, H., Greber, B. J., Nogales, E., Sutter, M., Kerfeld, C. A. (2023). Heterologous assembly of pleomorphic bacterial microcompartment shell architectures spanning the nano- to microscale. Adv. Mater. 35, e2212065. doi: 10.1002/adma.202212065
Ferlez, B., Sutter, M., Kerfeld, C. A. (2019). Glycyl radical enzyme-associated microcompartments: redox-replete bacterial organelles. mBio 10, e02327–18. doi: 10.1128/mBio.02327-18
Field, C. B., Behrenfeld, M. J., Randerson, J. T., Falkowski, P. (1998). Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240. doi: 10.1126/science.281.5374.237
Flamholz, A. I., Dugan, E., Blikstad, C., Gleizer, S., Ben-Nissan, R., Amram, S., et al. (2020). Functional reconstitution of a bacterial CO2 concentrating mechanism in Escherichia coli. Elife 9, e59882. doi: 10.7554/eLife.59882.sa2
Flamholz, A. I., Prywes, N., Moran, U., Davidi, D., Bar-On, Y. M., Oltrogge, L. M., et al. (2019). Revisiting trade-offs between Rubisco kinetic parameters. Biochemistry 58, 3365–3376. doi: 10.1021/acs.biochem.9b00237
Garcia-Alles, L. F., Fuentes-Cabrera, M., Truan, G., Reguera, D. (2023). Inferring assembly-curving trends of bacterial micro-compartment shell hexamers from crystal structure arrangements. PloS Comput. Biol. 19, e1011038. doi: 10.1371/journal.pcbi.1011038
Garcia-Alles, L. F., Lesniewska, E., Root, K., Aubry, N., Pocholle, N., Mendoza, C. I., et al. (2017). Spontaneous non-canonical assembly of CcmK hexameric components from β-carboxysome shells of cyanobacteria. PloS One 12, e0185109. doi: 10.1371/journal.pone.0185109
Garcia-Alles, L. F., Root, K., Maveyraud, L., Aubry, N., Lesniewska, E., Mourey, L., et al. (2019). Occurrence and stability of hetero-hexamer associations formed by β-carboxysome CcmK shell components. PloS One 14, e0223877. doi: 10.1371/journal.pone.0223877
Gonzalez-Esquer, C. R., Shubitowski, T. B., Kerfeld, C. A. (2015). Streamlined construction of the cyanobacterial CO2-fixing organelle via protein domain fusions for use in plant synthetic biology. Plant Cell. 27, 2637–2644. doi: 10.1105/tpc.15.00329
Graf, L., Wu, K., Wilson, J. W. (2018). Transfer and analysis of Salmonella pdu genes in a range of Gram-negative bacteria demonstrate exogenous microcompartment expression across a variety of species. Microb. Biotechnol. 11, 199–210. doi: 10.1111/1751-7915.12863
Greber, B. J., Sutter, M., Kerfeld, C. A. (2019). The plasticity of molecular interactions governs bacterial microcompartment shell assembly. Structure 27, 749–63.e4. doi: 10.1016/j.str.2019.01.017
Hagen, A. R., Plegaria, J. S., Sloan, N., Ferlez, B., Aussignargues, C., Burton, R., et al. (2018). In vitro assembly of diverse bacterial microcompartment shell architectures. Nano Lett. 18, 7030–7037. doi: 10.1021/acs.nanolett.8b02991
Hagen, A., Sutter, M., Sloan, N., Kerfeld, C. A. (2018). Programmed loading and rapid purification of engineered bacterial microcompartment shells. Nat. Commun. 9, 2881. doi: 10.1038/s41467-018-05162-z
Huseby, D. L., Roth, J. R. (2013). Evidence that a metabolic microcompartment contains and recycles private cofactor pools. J. Bacteriol. 195, 2864–2879. doi: 10.1128/JB.02179-12
Iancu, C. V., Ding, H. J., Morris, D. M., Dias, D. P., Gonzales, A. D., Martino, A., et al. (2007). The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J. Mol. Biol. 372, 764–773. doi: 10.1016/j.jmb.2007.06.059
Iancu, C. V., Morris, D. M., Dou, Z., Heinhorst, S., Cannon, G. C., Jensen, G. J. (2010). Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells. J. Mol. Biol. 396, 105–117. doi: 10.1016/j.jmb.2009.11.019
Iancu, C. V., Wright, E. R., Benjamin, J., Tivol, W. F., Dias, D. P., Murphy, G. E., et al. (2005). A “flip-flop” rotation stage for routine dual-axis electron cryotomography. J. Struct. Biol. 151, 288–297. doi: 10.1016/j.jsb.2005.07.004
Iñiguez, C., Capó-Bauçà, S., Niinemets, Ü, Stoll, H., Aguiló-Nicolau, P., Galmés, J. (2020). Evolutionary trends in RuBisCO kinetics and their co-evolution with CO2 concentrating mechanisms. Plant J. 101, 897–918. doi: 10.1111/tpj.14643
Jakobson, C. M., Chen, Y., Slininger, M. F., Valdivia, E., Kim, E. Y., Tullman-Ercek, D. (2016). Tuning the catalytic activity of subcellular nanoreactors. J. Mol. Biol. 428, 2989–2996. doi: 10.1016/j.jmb.2016.07.006
Jiang, Q., Li, T., Yang, J., Aitchison, C. M., Huang, J., Chen, Y., et al. (2023). Synthetic engineering of a new biocatalyst encapsulating [NiFe]-hydrogenases for enhanced hydrogen production. J. Mater. Chem. B. 11, 2684–2692. doi: 10.1039/D2TB02781J
Kalnins, G., Cesle, E. E., Jansons, J., Liepins, J., Filimonenko, A., Tars, K. (2020). Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles. Nat. Commun. 11, 388. doi: 10.1038/s41467-019-14205-y
Kaneko, Y., Danev, R., Nagayama, K., Nakamoto, H. (2006). Intact carboxysomes in a cyanobacterial cell visualized by hilbert differential contrast transmission electron microscopy. J. Bacteriol. 188, 805–808. doi: 10.1128/JB.188.2.805-808.2006
Kennedy, N. W., Mills, C. E., Abrahamson, C. H., Archer, A. G., Shirman, S., Jewett, M. C., et al. (2022). Linking the Salmonella enterica 1,2-Propanediol Utilization Bacterial Microcompartment Shell to the Enzymatic Core via the Shell Protein PduB. J. Bacteriol. 204, e0057621. doi: 10.1128/jb.00576-21
Kerfeld, C. A., Aussignargues, C., Zarzycki, J., Cai, F., Sutter, M. (2018). Bacterial microcompartments. Nat. Rev. Microbiol. 16, 277–290. doi: 10.1038/nrmicro.2018.10
Kerfeld, C. A., Melnicki, M. R. (2016). Assembly, function and evolution of cyanobacterial carboxysomes. Curr. Opin. Plant Biol. 31, 66–75. doi: 10.1016/j.pbi.2016.03.009
Kerfeld, C. A., Sawaya, M. R., Tanaka, S., Nguyen, C. V., Phillips, M., Beeby, M., et al. (2005). Protein structures forming the shell of primitive bacterial organelles. Science 309, 936–938. doi: 10.1126/science.1113397
Kinney, J. N., Salmeen, A., Cai, F., Kerfeld, C. A. (2012). Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J. Biol. Chem. 287, 17729–17736. doi: 10.1074/jbc.M112.355305
Kirst, H., Ferlez, B. H., Lindner, S. N., Cotton, C. A. R., Bar-Even, A., Kerfeld, C. A. (2022). Toward a glycyl radical enzyme containing synthetic bacterial microcompartment to produce pyruvate from formate and acetate. Proc. Natl. Acad. Sci. U. S. A. 119, e2116871119. doi: 10.1073/pnas.2116871119
Klein, M. G., Zwart, P., Bagby, S. C., Cai, F., Chisholm, S. W., Heinhorst, S., et al. (2009). Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J. Mol. Biol. 392, 319–333. doi: 10.1016/j.jmb.2009.03.056
Krupovic, M., Koonin, E. V. (2017). Cellular origin of the viral capsid-like bacterial microcompartments. Biol. Direct. 12, 25. doi: 10.1186/s13062-017-0197-y
Kupriyanova, E. V., Pronina, N. A., Los, D. A. (2023). Adapting from Low to High: An Update to CO2-Concentrating Mechanisms of Cyanobacteria and Microalgae. Plants (Basel) 12, 1569. doi: 10.3390/plants12071569
Lawrence, A. D., Frank, S., Newnham, S., Lee, M. J., Brown, I. R., Xue, W. F., et al. (2014). Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth. Biol. 3, 454–465. doi: 10.1021/sb4001118
Lee, M., Tullman-Ercek, D. (2017). Practical considerations for the encapsulation of multi-enzyme cargos within the bacterial microcompartment for metabolic engineering. Curr. Opin. Syst. Biol. 5, 16–22. doi: 10.1016/j.coisb.2017.05.017
Lehman, B. P., Chowdhury, C., Bobik, T. A. (2017). The N terminus of the PduB protein binds the protein shell of the Pdu microcompartment to its enzymatic core. J. Bacteriol. 199, e00785–16. doi: 10.1128/JB.00785-16
Li, T., Jiang, Q., Huang, J., Aitchison, C. M., Huang, F., Yang, M., et al. (2020). Reprogramming bacterial protein organelles as a nanoreactor for hydrogen production. Nat. Commun. 11, 5448. doi: 10.1038/s41467-020-19280-0
Li, Y., Kennedy, N. W., Li, S., Mills, C. E., Tullman-Ercek, D., Olvera de la Cruz, M. (2021). Computational and experimental approaches to controlling bacterial microcompartment assembly. ACS Cent. Sci. 7, 658–670. doi: 10.1021/acscentsci.0c01699
Lin, M. T., Occhialini, A., Andralojc, P. J., Devonshire, J., Hines, K. M., Parry, M. A., et al. (2014). β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J. 79, 1–12. doi: 10.1111/tpj.12536
Long, B. M., Badger, M. R., Whitney, S. M., Price, G. D. (2007). Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J. Biol. Chem. 282, 29323–29335. doi: 10.1074/jbc.M703896200
Long, B. M., Hee, W. Y., Sharwood, R. E., Rae, B. D., Kaines, S., Lim, Y. L., et al. (2018). Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat. Commun. 9, 3570. doi: 10.1038/s41467-018-06044-0
Long, B. M., Rae, B. D., Badger, M. R., Price, G. D. (2011). Over-expression of the β-carboxysomal CcmM protein in Synechococcus PCC7942 reveals a tight co-regulation of carboxysomal carbonic anhydrase (CcaA) and M58 content. Photosynth. Res. 109, 33–45. doi: 10.1007/s11120-011-9659-8
Ludwig, M., Sültemeyer, D., Price, G. (2000). Isolation of ccmKLMN genes from the marine cyanobacterium, Synechococcus sp. PCC7002 (Cyanophyceae), and evidence that CcmM is essential for carboxysome assembly. J. Phycol. 36 (6), 1109–1119. doi: 10.1046/j.1529-8817.2000.00028.x
Lynch, D. L., Pavlova, A., Fan, Z., Gumbart, J. C. (2023). Understanding virus structure and dynamics through molecular simulations. J. Chem. Theory Comput. 19, 3025–3036. doi: 10.1021/acs.jctc.3c00116
Maberly, S. C., Gontero, B. (2017). Ecological imperatives for aquatic CO2-concentrating mechanisms. J. Exp. Bot. 68, 3797–3814. doi: 10.1093/jxb/erx201
Mahalik, J. P., Brown, K. A., Cheng, X., Fuentes-Cabrera, M. (2016). Theoretical study of the initial stages of self-assembly of a carboxysome’s facet. ACS Nano. 10, 5751–5758. doi: 10.1021/acsnano.5b07805
Mahinthichaichan, P., Morris, D. M., Wang, Y., Jensen, G. J., Tajkhorshid, E. (2018). Selective permeability of carboxysome shell pores to anionic molecules. J. Phys. Chem. B. 122, 9110–9118. doi: 10.1021/acs.jpcb.8b06822
Marcus, Y., Gurevitz, M. (2000). Activation of cyanobacterial RuBP-carboxylase/oxygenase is facilitated by inorganic phosphate via two independent mechanisms. Eur. J. Biochem. 267, 5995–6003. doi: 10.1046/j.1432-1327.2000.01674.x
Melnicki, M. R., Sutter, M., Kerfeld, C. A. (2021). Evolutionary relationships among shell proteins of carboxysomes and metabolosomes. Curr. Opin. Microbiol. 63, 1–9. doi: 10.1016/j.mib.2021.05.011
Metskas, L. A., Ortega, D., Oltrogge, L. M., Blikstad, C., Lovejoy, D. R., Laughlin, T. G., et al. (2022). Rubisco forms a lattice inside alpha-carboxysomes. Nat. Commun. 13, 4863. doi: 10.1038/s41467-022-32584-7
Mills, C. E., Waltmann, C., Archer, A. G., Kennedy, N. W., Abrahamson, C. H., Jackson, A. D., et al. (2022). Vertex protein PduN tunes encapsulated pathway performance by dictating bacterial metabolosome morphology. Nat. Commun. 13, 3746. doi: 10.1038/s41467-022-31279-3
Mohajerani, F., Hagan, M. F. (2018). The role of the encapsulated cargo in microcompartment assembly. PloS Comput. Biol. 14, e1006351. doi: 10.1371/journal.pcbi.1006351
Mohajerani, F., Sayer, E., Neil, C., Inlow, K., Hagan, M. F. (2021). Mechanisms of scaffold-mediated microcompartment assembly and size control. ACS Nano. 15, 4197–4212. doi: 10.1021/acsnano.0c05715
Ni, T., Jiang, Q., Ng, P. C., Shen, J., Dou, H., Zhu, Y., et al. (2023). Intrinsically disordered CsoS2 acts as a general molecular thread for α-carboxysome shell assembly. Nat. Commun. 14, 5512. doi: 10.1038/s41467-023-41211-y
Ni, T., Sun, Y., Burn, W., Al-Hazeem, M. M. J., Zhu, Y., Yu, X., et al. (2022). Structure and assembly of cargo Rubisco in two native α-carboxysomes. Nat. Commun. 13, 4299. doi: 10.1038/s41467-022-32004-w
Nichols, T. M., Kennedy, N. W., Tullman-Ercek, D. (2019). Cargo encapsulation in bacterial microcompartments: Methods and analysis. Methods Enzymol. 617, 155–186. doi: 10.1016/bs.mie.2018.12.009
Noël, C., Cai, F., Kerfeld, C. (2015). Purification and characterization of protein nanotubes assembled from a single bacterial microcompartment shell subunit. Adv. Mater. Interf 3 (1), 1500295. doi: 10.1002/admi.201500295
Oltrogge, L. M., Chaijarasphong, T., Chen, A. W., Bolin, E. R., Marqusee, S., Savage, D. F. (2020). Multivalent interactions between CsoS2 and Rubisco mediate α-carboxysome formation. Nat. Struct. Mol. Biol. 27, 281–287. doi: 10.1038/s41594-020-0387-7
Oltrogge, L. M., Chen, A. W., Chaijarasphong, T., Turnšek, J. B., Savage, D. F. (2023). α-carboxysome size is controlled by the disordered scaffold protein CsoS2. Biochemistry 63 (2), 219–229. doi: 10.1021/acs.biochem.3c00403
Park, J., Chun, S., Bobik, T. A., Houk, K. N., Yeates, T. O. (2017). Molecular dynamics simulations of selective metabolite transport across the propanediol bacterial microcompartment shell. J. Phys. Chem. B. 121, 8149–8154. doi: 10.1021/acs.jpcb.7b07232
Penrod, J. T., Roth, J. R. (2006). Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J. Bacteriol. 188, 2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006
Perlmutter, J. D., Mohajerani, F., Hagan, M. F. (2016). Many-molecule encapsulation by an icosahedral shell. Elife 5, e14078. doi: 10.7554/eLife.14078.029
Perlmutter, J. D., Qiao, C., Hagan, M. F. (2013). Viral genome structures are optimal for capsid assembly. Elife 2, e00632. doi: 10.7554/eLife.00632.024
Phillips, R., Milo, R. (2009). A feeling for the numbers in biology. Proc. Natl. Acad. Sci. U. S. A. 106, 21465–21471. doi: 10.1073/pnas.0907732106
Poudel, S., Pike, D. H., Raanan, H., Mancini, J. A., Nanda, V., Rickaby, R. E. M., et al. (2020). Biophysical analysis of the structural evolution of substrate specificity in RuBisCO. Proc. Natl. Acad. Sci. U. S. A. 117, 30451–30457. doi: 10.1073/pnas.2018939117
Price, G. D., Badger, M. R. (1989). Expression of human carbonic anhydrase in the cyanobacterium synechococcus PCC7942 creates a high CO(2)-requiring phenotype: evidence for a central role for carboxysomes in the CO(2) concentrating mechanism. Plant Physiol. 91, 505–513. doi: 10.1104/pp.91.2.505
Price, G. D., Sültemeyer, D., Klughammer, B., Ludwig, M., Badger, M. (1998). The functioning of the CO2 concentrating mechanism in several cyanobacterial strains: a review of general physiological characteristics, genes, proteins, and recent advances. Can. J. Bot. 76 (6), 973–1002. doi: 10.1139/b98-081
Rae, B. D., Long, B. M., Badger, M. R., Price, G. D. (2013). Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol. Mol. Biol. Rev. 77, 357–379. doi: 10.1128/MMBR.00061-12
Raven, J. A. (2013). Rubisco: still the most abundant protein of Earth? New Phytol. 198, 1–3. doi: 10.1111/nph.12197
Rotskoff, G. M., Geissler, P. L. (2018). Robust nonequilibrium pathways to microcompartment assembly. Proc. Natl. Acad. Sci. U. S. A. 115, 6341–6346. doi: 10.1073/pnas.1802499115
Ryan, P., Forrester, T. J. B., Wroblewski, C., Kenney, T. M. G., Kitova, E. N., Klassen, J. S., et al. (2019). The small RbcS-like domains of the β-carboxysome structural protein CcmM bind RubisCO at a site distinct from that binding the RbcS subunit. J. Biol. Chem. 294, 2593–2603. doi: 10.1074/jbc.RA118.006330
Sampson, E. M., Bobik, T. A. (2008). Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J. Bacteriol. 190, 2966–2971. doi: 10.1128/JB.01925-07
Savir, Y., Noor, E., Milo, R., Tlusty, T. (2010). Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape. Proc. Natl. Acad. Sci. U. S. A. 107, 3475–3480. doi: 10.1073/pnas.0911663107
Schmid, M. F., Paredes, A. M., Khant, H. A., Soyer, F., Aldrich, H. C., Chiu, W., et al. (2006). Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J. Mol. Biol. 364, 526–535. doi: 10.1016/j.jmb.2006.09.024
Schmidt-Dannert, S., Zhang, G., Johnston, T., Quin, M. B., Schmidt-Dannert, C. (2018). Building a toolbox of protein scaffolds for future immobilization of biocatalysts. Appl. Microbiol. Biotechnol. 102, 8373–8388. doi: 10.1007/s00253-018-9252-6
Shibata, M., Katoh, H., Sonoda, M., Ohkawa, H., Shimoyama, M., Fukuzawa, H., et al. (2002a). Genes essential to sodium-dependent bicarbonate transport in cyanobacteria: function and phylogenetic analysis. J. Biol. Chem. 277, 18658–18664. doi: 10.1074/jbc.M112468200
Shibata, M., Ohkawa, H., Kaneko, T., Fukuzawa, H., Tabata, S., Kaplan, A., et al. (2001). Distinct constitutive and low-CO2-induced CO2 uptake systems in cyanobacteria: genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc. Natl. Acad. Sci. U. S. A. 98, 11789–11794. doi: 10.1073/pnas.191258298
Shibata, M., Ohkawa, H., Katoh, H., Shimoyama, M., Ogawa, T. (2002b). Two CO2 uptake systems in cyanobacteria: four systems for inorganic carbon acquisition in Synechocystis sp. strain PCC6803. Funct. Plant Biol. 29, 123–129. doi: 10.1071/PP01188
Shively, J. M., Ball, F. L., Kline, B. W. (1973). Electron microscopy of the carboxysomes (polyhedral bodies) of Thiobacillus neapolitanus. J. Bacteriol. 116, 1405–1411. doi: 10.1128/jb.116.3.1405-1411.1973
Slininger Lee, M. F., Jakobson, C. M., Tullman-Ercek, D. (2017). Evidence for improved encapsulated pathway behavior in a bacterial microcompartment through shell protein engineering. ACS Synth. Biol. 6, 1880–1891. doi: 10.1021/acssynbio.7b00042
Sommer, M., Sutter, M., Gupta, S., Kirst, H., Turmo, A., Lechno-Yossef, S., et al. (2019). Heterohexamers formed by CcmK3 and CcmK4 increase the complexity of beta carboxysome shells. Plant Physiol. 179, 156–167. doi: 10.1104/pp.18.01190
Sun, Y., Harman, V. M., Johnson, J. R., Brownridge, P. J., Chen, T., Dykes, G. F., et al. (2022). Decoding the absolute stoichiometric composition and structural plasticity of α-carboxysomes. mBio 13, e0362921. doi: 10.1128/mbio.03629-21
Sutter, M., Faulkner, M., Aussignargues, C., Paasch, B. C., Barrett, S., Kerfeld, C. A., et al. (2016). Visualization of bacterial microcompartment facet assembly using high-speed atomic force microscopy. Nano Lett. 16, 1590–1595. doi: 10.1021/acs.nanolett.5b04259
Sutter, M., Greber, B., Aussignargues, C., Kerfeld, C. A. (2017). Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 356, 1293–1297. doi: 10.1126/science.aan3289
Sutter, M., Laughlin, T. G., Sloan, N. B., Serwas, D., Davies, K. M., Kerfeld, C. A. (2019a). Structure of a Synthetic β-Carboxysome Shell. Plant Physiol. 181, 1050–1058. doi: 10.1104/pp.19.00885
Sutter, M., McGuire, S., Ferlez, B., Kerfeld, C. A. (2019b). Structural characterization of a synthetic tandem-domain bacterial microcompartment shell protein capable of forming icosahedral shell assemblies. ACS Synth. Biol. 8, 668–674. doi: 10.1021/acssynbio.9b00011
Sutter, M., Melnicki, M. R., Schulz, F., Woyke, T., Kerfeld, C. A. (2021). A catalog of the diversity and ubiquity of bacterial microcompartments. Nat. Commun. 12, 3809. doi: 10.1038/s41467-021-24126-4
Takenoya, M., Nikolakakis, K., Sagermann, M. (2010). Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli. J. Bacteriol. 192, 6056–6063. doi: 10.1128/JB.00652-10
Tan, Y. Q., Ali, S., Xue, B., Teo, W. Z., Ling, L. H., Go, M. K., et al. (2021). Structure of a minimal α-carboxysome-derived shell and its utility in enzyme stabilization. Biomacromolecules 22, 4095–4109. doi: 10.1021/acs.biomac.1c00533
Tanaka, S., Kerfeld, C. A., Sawaya, M. R., Cai, F., Heinhorst, S., Cannon, G. C., et al. (2008). Atomic-level models of the bacterial carboxysome shell. Science 319, 1083–1086. doi: 10.1126/science.1151458
Tanaka, S., Sawaya, M. R., Phillips, M., Yeates, T. O. (2009). Insights from multiple structures of the shell proteins from the beta-carboxysome. Protein Sci. 18, 108–120. doi: 10.1002/pro.14
Tanaka, S., Sawaya, M. R., Yeates, T. O. (2010). Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 327, 81–84. doi: 10.1126/science.1179513
Trettel, D. S., Neale, C., Zhao, M., Gnanakaran, S., Gonzalez-Esquer, C. R. (2023). Monatomic ions influence substrate permeation across bacterial microcompartment shells. Sci. Rep. 13, 15738. doi: 10.1038/s41598-023-42688-9
Trettel, D. S., Resager, W., Ueberheide, B. M., Jenkins, C. C., Winkler, W. C. (2022). Chemical probing provides insight into the native assembly state of a bacterial microcompartment. Structure 30, 537–50.e5. doi: 10.1016/j.str.2022.02.002
Trettel, D. S., Winkler, W. C. (2023). Analysis of bacterial microcompartments and shell protein superstructures by confocal microscopy. Microbiol. Spectr. 11, e0335722. doi: 10.1128/spectrum.03357-22
Tsai, Y., Sawaya, M. R., Cannon, G. C., Cai, F., Williams, E. B., Heinhorst, S., et al. (2007). Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PloS Biol. 5, e144. doi: 10.1371/journal.pbio.0050144
Tsidilkovski, L., Mohajerani, F., Hagan, M. F. (2022). Microcompartment assembly around multicomponent fluid cargoes. J. Chem. Phys. 156, 245104. doi: 10.1063/5.0089556
Uddin, I., Frank, S., Warren, M. J., Pickersgill, R. W. (2018). A generic self-assembly process in microcompartments and synthetic protein nanotubes. Small 14, e1704020. doi: 10.1002/smll.201704020
Wagner, H. J., Capitain, C. C., Richter, K., Nessling, M., Mampel, J. (2017). Engineering bacterial microcompartments with heterologous enzyme cargos. Eng. Life Sci. 17, 36–46. doi: 10.1002/elsc.201600107
Waltmann, C., Kennedy, N. W., Mills, C. E., Roth, E. W., Ikonomova, S. P., Tullman-Ercek, D., et al. (2023). Kinetic growth of multicomponent microcompartment shells. ACS Nano. 17, 15751–15762. doi: 10.1021/acsnano.3c03353
Wang, H., Hayer-Hartl, M. (2023). Phase separation of rubisco by the folded SSUL domains of CcmM in beta-carboxysome biogenesis. Methods Mol. Biol. 2563, 269–296. doi: 10.1007/978-1-0716-2663-4_14
Wang, H., Yan, X., Aigner, H., Bracher, A., Nguyen, N. D., Hee, W. Y., et al. (2019). Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature 566, 131–135. doi: 10.1038/s41586-019-0880-5
Whitehead, L., Long, B. M., Price, G. D., Badger, M. R. (2014). Comparing the in vivo function of α-carboxysomes and β-carboxysomes in two model cyanobacteria. Plant Physiol. 165, 398–411. doi: 10.1104/pp.114.237941
Xue, B., Blocquel, D., Habchi, J., Uversky, A. V., Kurgan, L., Uversky, V. N., et al. (2014). Structural disorder in viral proteins. Chem. Rev. 114, 6880–6911. doi: 10.1021/cr4005692
Yang, M., Simpson, D. M., Wenner, N., Brownridge, P., Harman, V. M., Hinton, J. C. D., et al. (2020). Decoding the stoichiometric composition and organisation of bacterial metabolosomes. Nat. Commun. 11, 1976. doi: 10.1038/s41467-020-15888-4
Yang, M., Wenner, N., Dykes, G. F., Li, Y., Zhu, X., Sun, Y., et al. (2022). Biogenesis of a bacterial metabolosome for propanediol utilization. Nat. Commun. 13, 2920. doi: 10.1038/s41467-022-30608-w
Young, E. J., Burton, R., Mahalik, J. P., Sumpter, B. G., Fuentes-Cabrera, M., Kerfeld, C. A., et al. (2017). Engineering the bacterial microcompartment domain for molecular scaffolding applications. Front. Microbiol. 8, 1441. doi: 10.3389/fmicb.2017.01441
Zang, K., Wang, H., Hartl, F. U., Hayer-Hartl, M. (2021). Scaffolding protein CcmM directs multiprotein phase separation in β-carboxysome biogenesis. Nat. Struct. Mol. Biol. 28, 909–922. doi: 10.1038/s41594-021-00676-5
Zarzycki, J., Erbilgin, O., Kerfeld, C. A. (2015). Bioinformatic characterization of glycyl radical enzyme-associated bacterial microcompartments. Appl. Environ. Microbiol. 81, 8315–8329. doi: 10.1128/AEM.02587-15
Zarzycki, J., Sutter, M., Cortina, N. S., Erb, T. J., Kerfeld, C. A. (2017). In vitro characterization and concerted function of three core enzymes of a glycyl radical enzyme - associated bacterial microcompartment. Sci. Rep. 7, 42757. doi: 10.1038/srep42757
Keywords: carbon fixation, bacterial microcompartments, carboxysome, molecular dynamics, permeation, phase separation
Citation: Trettel DS, Pacheco SL, Laskie AK and Gonzalez-Esquer CR (2024) Modeling bacterial microcompartment architectures for enhanced cyanobacterial carbon fixation. Front. Plant Sci. 15:1346759. doi: 10.3389/fpls.2024.1346759
Received: 29 November 2023; Accepted: 30 January 2024;
Published: 15 February 2024.
Edited by:
Jianping Yu, National Renewable Energy Laboratory (DOE), United StatesReviewed by:
Harvey J.M. Hou, Alabama State University, United StatesCopyright © 2024 Trettel, Pacheco, Laskie and Gonzalez-Esquer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel S. Trettel, ZHRyZXR0ZWxAbGFubC5nb3Y=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.