- College of Life Sciences, Xinyang Normal University, Xinyang, China
Interactions between rice plants (Oryza sativa L.) and brown planthoppers (Nilaparvata lugens Stål, BPHs) are used as a model system to study the molecular mechanisms underlying plant-insect interactions. Small RNAs (sRNAs) regulate growth, development, immunity, and environmental responses in eukaryotic organisms, including plants and insects. Recent research suggests that sRNAs play significant roles in rice-BPH interactions by mediating post-transcriptional gene silencing. The focus of this review is to explore the roles of sRNAs in rice-BPH interactions and to highlight recent research progress in unraveling the mechanism of cross-kingdom RNA interference (ckRNAi) between host plants and insects and the application of ckRNAi in pest management of crops including rice. The research summarized here will aid in the development of safe and effective BPH control strategies.
Introduction
Rice (Oryza sativa L.) is a globally-important staple food which is susceptible to damage from hundreds of insect herbivores throughout its lifecycle (Du et al., 2020). One of the most destructive of these insect herbivores is the brown planthopper (Nilaparvata lugens Stål, BPH), which is responsible for severely reduced rice yields and substantial economic losses each year (Shi et al., 2021; Shi et al., 2023). Once outbreaks, the insects can completely destroy crops, an effect called “hopperburn” (Backus et al., 2005).
Plants have evolved an intricate, double-layered defense system to effectively resist and respond to herbivorous pests. The first layer is referred to as pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) (Jing et al., 2017). PTI activates downstream defense-related signaling cascades, such as the phytohormone-mediated defense response pathway (Erb and Reymond, 2019; Wang et al., 2023). The second layer is known as effector-triggered immunity (ETI), which is a robust resistance (R) protein-mediated defense response (Jones and Dangl, 2006; Takken and Tameling, 2009; Rodriguez et al., 2017). Recent research suggests that plants respond to herbivory through a series of defense-related processes, including phytohormone signaling and secondary metabolite biosynthesis, many of which are regulated by small RNAs (sRNAs) (Sattar and Thompson, 2016).
sRNAs are eukaryotic non-coding RNA molecules, approximately 20-30 nucleotides (nt) in length, which regulate gene expression via RNA silencing (Zamore and Haley, 2005; Chapman and Carrington, 2007). According to their precursor structures and associated genetic pathways, plant sRNAs are classified into two major classes: microRNAs (miRNAs) and small interfering RNAs (siRNAs) (Bartel, 2009; Chen, 2009; Katiyar-Agarwal and Jin, 2010). Likewise, insect sRNAs are divided into three major classes: miRNAs, endogenous-siRNAs (endo-siRNAs), and piwi-interacting RNAs (piRNAs) (Golden et al., 2008). In both plants and animals, miRNAs are 20-24 nt single-stranded non-coding RNAs which mediate post-transcriptional gene silencing by binding to mRNAs containing specific complementary base pairs (Zhang et al., 2006; Bartel, 2009; Ghini et al., 2018). Global sRNA sequence profiling of rice and BPH has enabled the identification and characterization of many sRNAs, particularly miRNAs, involved in rice-BPH interactions (Zha et al., 2016; Wu et al., 2017; Nanda et al., 2020). The focus of this review is to explore the roles of sRNAs in rice-BPH interactions and to highlight recent research progress in unraveling the mechanism of cross-kingdom RNA interference (RNAi) between host plants and insects. The research summarized here will aid in the development of safe and effective BPH control strategies.
Rice-derived sRNAs involved in BPH resistance
In plants, sRNAs play significant roles in growth, development, abiotic and biotic stress responses (Khraiwesh et al., 2012; Duan et al., 2015; Yue et al., 2017; Chen et al., 2019; Kryovrysanaki et al., 2022). Several studies have utilized RNA and sRNA profiling to identify sRNAs in rice. Functional validation experiments indicate that these sRNAs fine-tune plant innate immunity by integrating R gene-mediated resistance, phytohormone signaling, callose deposition, reactive oxygen species (ROS) production, and secondary metabolite biosynthesis (Wu et al., 2017; Ge et al., 2018; Dai et al., 2019; Tan et al., 2020; Lü et al., 2022; Shen et al., 2023).
To date, approximately 17 BPH-resistance (R) genes have been identified in both wild and cultivated rice (Wang et al., 2023). Considerable research has been conducted to characterize the mechanism by which R genes confer BPH resistance (Jing et al., 2017; Zheng et al., 2021). Through miRNA sequencing, Wu et al. (2017) identified 23 and 674 differentially expressed miRNAs (DEMs) (including 464 known and 183 novel miRNAs) between resistant (carrying BPH-resistance gene Bph15) and susceptible rice varieties before and after BPH infestation, respectively. The identified DEMs were primarily involved in basal defense and BPH-specific resistance. Similarly, an integrated miRNA and mRNA analysis identified 217 DEMs between Bph6-carrying transgenic rice lines and wild type plants after BPH infestation (Tan et al., 2020). Of these, nine miRNAs were specifically expressed in transgenic rice lines, suggesting their involvement in Bph6-mediated resistance to the BPH. In addition, both Nanda et al. (2020) and Lü et al. (2022) identified an array of BPH-responsive miRNAs between resistant and susceptible rice varieties. Although these findings suggest that miRNAs likely participate in the BPH defense response, the involvement of only a few miRNAs has been experimentally verified (Table 1).
It is well known that the phytohormone signaling plays an important role in rice defense against BPH (Zhou et al., 2009). Recent research suggests that miRNAs regulate rice resistance to BPH by post-transcriptionally regulating the expression of target genes involved in phytohormone signaling. For example, Osa-miR156 negatively regulates BPH resistance by modulating jasmonic acid (JA) signaling (Ge et al., 2018). Osa-miR156-silenced plants (MIM156) exhibited increased resistance to BPH via upregulated expression of OsMPK3 and OsMPK6 and downregulated expression of OsWRKY70, a transcription factor which positively regulates JA signaling. Furthermore, the expression of the JA biosynthesis gene OsHI-LOX and the contents of JA and bioactive jasmonoyl-isoleucine (JA-Ile) were significantly reduced in MIM156 plants. Altogether, it appears that Osa-miR156 regulates JA biosynthesis and BPH resistance via the MAPK cascade in rice. In addition, Osa-miR162a is strongly induced by BPH herbivory in rice seedlings (Chen et al., 2023). Functional verification indicated that Osa-miR162a regulates BPH resistance in rice by inhibiting the α-linolenic acid metabolism pathway, which itself regulates JA biosynthesis (Chen et al., 2023).
In rice, secondary metabolites have been shown to inhibit both the feeding and development of BPH. Furthermore, miRNAs can regulate the expression of genes involved in secondary metabolite biosynthesis to modulate BPH resistance. For example, OsmiR396 was found to negatively regulate BPH resistance via the OsmiR396–growth-regulating factor 8 (OsGRF8)–OsF3H–flavonoid module (Dai et al., 2019). Transgenic plants over-expressing growth-regulating factor 8 (OsGRF8), the target gene of OsmiR396, exhibit enhanced BPH resistance due to downregulation of OsmiR396. Overall, it appears that OsmiR396-OsGRF8 modulates BPH resistance by regulating the expression of the flavanone 3-hydroxylase (OsF3H) gene, which is involved in flavonoid biosynthesis (Dai et al., 2019). More recent research indicated that OsmiR159 negatively regulates BPH resistance through the OsmiR159–OsGA-MYBL2 module and the OsmiR159–OsGAMYBL2–GS3 signaling pathway (Shen et al., 2023). Despite these advancements, the molecular mechanism underlying miRNA-mediated BPH resistance in rice is still poorly understood.
The roles of sRNAs in BPH physiology
Advances in genomics have greatly expanded our understanding of the roles sRNAs play in BPH physiology and environmental response (Sattar and Thompson, 2016; Zha et al., 2016). Emerging evidence suggests that sRNAs participate in BPH metamorphosis, wing polyphenism, molting, and reproductive development (Chen et al., 2013; Xu et al., 2013; Chen et al., 2018; Ye et al., 2019; Xu et al., 2020; Li et al., 2021; Wang et al., 2022). Combing transcriptomic and genomic data, Xu et al. (2013) identified key genes involved in the BPH siRNA and miRNA pathways. RNAi knockdown of these genes severely affected BPH development and morphology, suggesting that siRNAs and miRNAs may play a crucial role in BPH development and metamorphosis (Xu et al., 2013).
In BPH, wing polyphenism is determined by environmental cues such as the nutritional status of host rice plants, population density, and photoperiod (Xu et al., 2020; Li et al., 2021). These environmental cues affect wing polyphenism by way of several complex regulatory pathways, including insulin/IGF-1 signaling (IIS), juvenile hormone (JH), and 20-hydroxyecdysone (20E) signaling (Xu et al., 2015). Research suggests that these signaling pathways are modulated by an array of miRNAs. For example, RNA sequencing of long wing (LW) and short wing (SW) BPH strains identified a complicated miRNA network which may modulate wing morphological plasticity in a growth-stage dependent manner (Xu et al., 2020). Three miRNAs (Nlu-miR-14-3p, Nlu-miR-9a-5p, and Nlu-miR-315-5p) have been confirmed to interact with NlInR genes, which are the part of IIS signaling pathway (Xu et al., 2020). In addition, Nlu-miR-34 has been shown to modulate wing polyphenism by targeting NlInR1 and mediating the cross-talk between the IIS, JH, and 20E signaling pathways via a positive autoregulatory feedback loop (Ye et al., 2019). Both Nlu-miR-1000-1-3p (Xu et al., 2020) and Nlu-mir-9a (Li et al., 2021) were predicted to target the wing development regulatory gene Ultrabithorax (NlUbx), and both were found to be differentially expressed between LW and SW BPH. Finally, the NlInRs/Nlu-mir-9a/NlUbx regulatory cascade appears to control wing dimorphism by regulating the host’s nutritional status (Li et al., 2021).
Molting is crucial to normal insect development, and is at least partially controlled by the chitin biosynthesis pathway and 20E signaling (Chen et al., 2013). Through deep miRNA sequencing of BPH instars at specific stages and during four molting periods, 21 (Chen et al., 2013) and 36 (Chen et al., 2018) specific mature miRNAs were identified, respectively. Among them, Nlu-miR-8-5p, Nlu-miR-2a-3p, and Nlu-miR-173 were found to target genes in the chitin biosynthesis pathway, as well as transcription factor NlFtz-F1. All three miRNAs appear to regulate molting and chitin biosynthesis through 20E signaling (Chen et al., 2013; Chen et al., 2018). The expression of chitin synthase gene A was downregulated when its specific siRNA and its regulated miRNA (Nlu-miR-2703) were injected into BPH, reducing both chitin biosynthesis and molting success (Li et al., 2017).
sRNAs have also been found to regulate BPH fecundity by modulating the expression of genes associated with reproductive development. For example, injecting Nlu-miR-34-5p mimics can decrease BPH fecundity by reducing vitellogenin (Vg) expression (Wang et al., 2022). The biosynthesis of Vg is crucial for oocyte accumulation and successful reproduction (Wang et al., 2022). Glutamine synthetase (NlGS), a protein involved in ovary development which regulates Vg accumulation, has been identified as a target of Nlu-miR-4868b (Zhai et al., 2013; Fu et al., 2015). NlGS expression was downregulated following injection of the Nlu-miR-4868b mimic, but upregulated following injection of the Nlu-miR-4868b inhibitor. Additionally, overexpression of Nlu-miR-4868b reduced both insect fecundity and Vg expression.
Finally, miRNAs play important regulatory roles in environmental responses such as the adaptation to resistant rice varieties. Zha et al. (2016) constructed and sequenced two sRNA libraries using two BPH populations exhibiting different levels of virulence: biotype 1, which only survives on the susceptible rice variety ‘Taichung Native 1 (TN1)’, and biotype Y, which is able to survive on the resistant rice variety ‘YHY15’ (carrying BPH-resistance gene Bph15). The researchers identified 26 DEMs between these two BPH populations, suggesting that these BPH miRNAs may regulate adaptability to resistant rice varieties. However, the precise functions of these miRNAs require further confirmation.
Cross-kingdom RNAi in the rice-BPH interaction
Research suggests that sRNAs can be transferred between host plants and interacting organisms, thereby inducing gene silencing via a mechanism known as “cross-kingdom RNAi” (Huang et al., 2019). This scenario was first reported in the interaction between plants and fungi. For example, gray mold (Botrytis cinerea)-derived sRNAs were found to be able to control the Arabidopsis thaliana RNAi system by binding to AGO1, ultimately silencing genes involved in plant immunity (Weiberg et al., 2013). Cross-kingdom RNAi has also been observed in the rice-BPH interaction (Shen et al., 2021; Lü et al., 2022). Rice-derived sRNAs may be ingested when BPH feed on rice plants, allowing them to regulate BPH gene expression.
Recently, rice-derived Osa-miR162a, a conserved plant miRNA, was found to effectively silence NlTOR (Target of rapamycin) expression in BPH through the cross-kingdom RNAi mechanism (Shen et al., 2021). Both ingestion and injection of Osa-miR162a mimics result in reduced female BPH fecundity and Vg activity, which is regulated by the TOR signaling pathway. In addition, allowing BPH adults to feed on Osa-miR162a- or Osa-miR162a-m1 (a modified derivative of Osa-miR162a)-overexpressing transgenic rice lines consistently resulted in reduced egg production and hatching success. These results suggest that these miRNAs confer resistance to BPH in rice, and that Osa-miR162a may be a potential target for BPH control (Shen et al., 2021; Chen et al., 2023).
Another rice-derived miRNA, Osa-miR5795, has also been found to impact BPH fecundity (Lü et al., 2022). By sequencing and analyzing sRNAs from six rice varieties exhibiting variable BPH resistance, 45 resistance-related DEMs were identified between BPH-susceptible and BPH-resistant rice varieties prior to BPH infestation, as well as 144 feeding-induced DEMs. Twenty-five of these DEMs were shared between both groups and were found to be directly involved in the rice-BPH interaction. In addition, seven potential cross-kingdom miRNAs were identified, and their targets were primarily involved in fecundity, feeding, digestion, and detoxification. Based on their predicted binding sites, two of these cross-kingdom miRNAs were selected to verify their function in BPH fecundity. Consequently, BPH oviposition was significantly reduced following injection with Osa-miR5795 mimics targeting the fecundity marker gene NlVg (Lü et al., 2022).
Both of these rice-derived miRNAs (Osa-miR162a and Osa-miR5795) appear to play an important role in rice-BPH interactions through cross-kingdom regulation of NlTOR and NlVg expression, both of which regulate fecundity in BPH (Table 1). However, to date no sRNAs, particularly BPH-derived miRNAs, appear to be involved in rice-BPH interactions through cross-kingdom RNAi trafficking.
Application of cross-kingdom RNAi in crop protection
miRNA-mediated gene regulation has emerged as a novel strategy to improve insect resistance in crop plants, including rice. Host-induced gene silencing (HIGS) is a novel concept based on the cross-kingdom RNAi mechanism. HIGS involves overexpressing insect-targeted double-stranded RNAs (dsRNAs) or artificial miRNAs in host plants to specifically block the expression of feeding- and survival-related genes in target pests and pathogens (Huang et al., 2019; Jiang et al., 2023; Mahanty et al., 2023) (Figure 1A). A growing number of studies have demonstrated the successful application of HIGS in crop protection (Escobar et al., 2001; Seemanpillai et al., 2003; Zha et al., 2011; Van et al., 2014; Coleman et al., 2015; Shivakumara et al., 2017; Panwar et al., 2018). In this context, we will use the application of HIGS to manage BPH as an example. Two salivary proteins secreted by BPH are mucin-like protein (NlMLP) and salivary protein 1 (NlSP1). Ectopic expression of these genes in tobacco (Nicotiana benthamiana) leaves induced the expression of defense-related genes and callose deposition, suggesting that these two proteins function as elicitors (Shangguan et al., 2018; Huang et al., 2020). Compared to controls which received no injection or were injected with dsGFP, insects injected with dsNlMLP or dsNlSP1 exhibited significantly reduced weight gain and survival rates, suggesting that NlMLP and NlSP1 are essential for BPH survival (Shangguan et al., 2018; Huang et al., 2020). Similarly, BPH feeding on transgenic plants constitutively expressing dsNlMLP or dsNlSP1 also exhibited reduced weight gain and survival rates compared to insects feeding on wild type plants (Shangguan et al., 2018; Huang et al., 2020). Although allowing insects to feed on plants overexpressing exogenous dsRNA was not as effective as injecting insects directly, HIGS remains a promising pest control strategy. However, the implementation of HIGS depends on the generation of transgenic plants, which is both time-intensive and costly (Jiang et al., 2023; Mahanty et al., 2023). These limitations have so far hampered the application of HIGS to BPH control in rice.
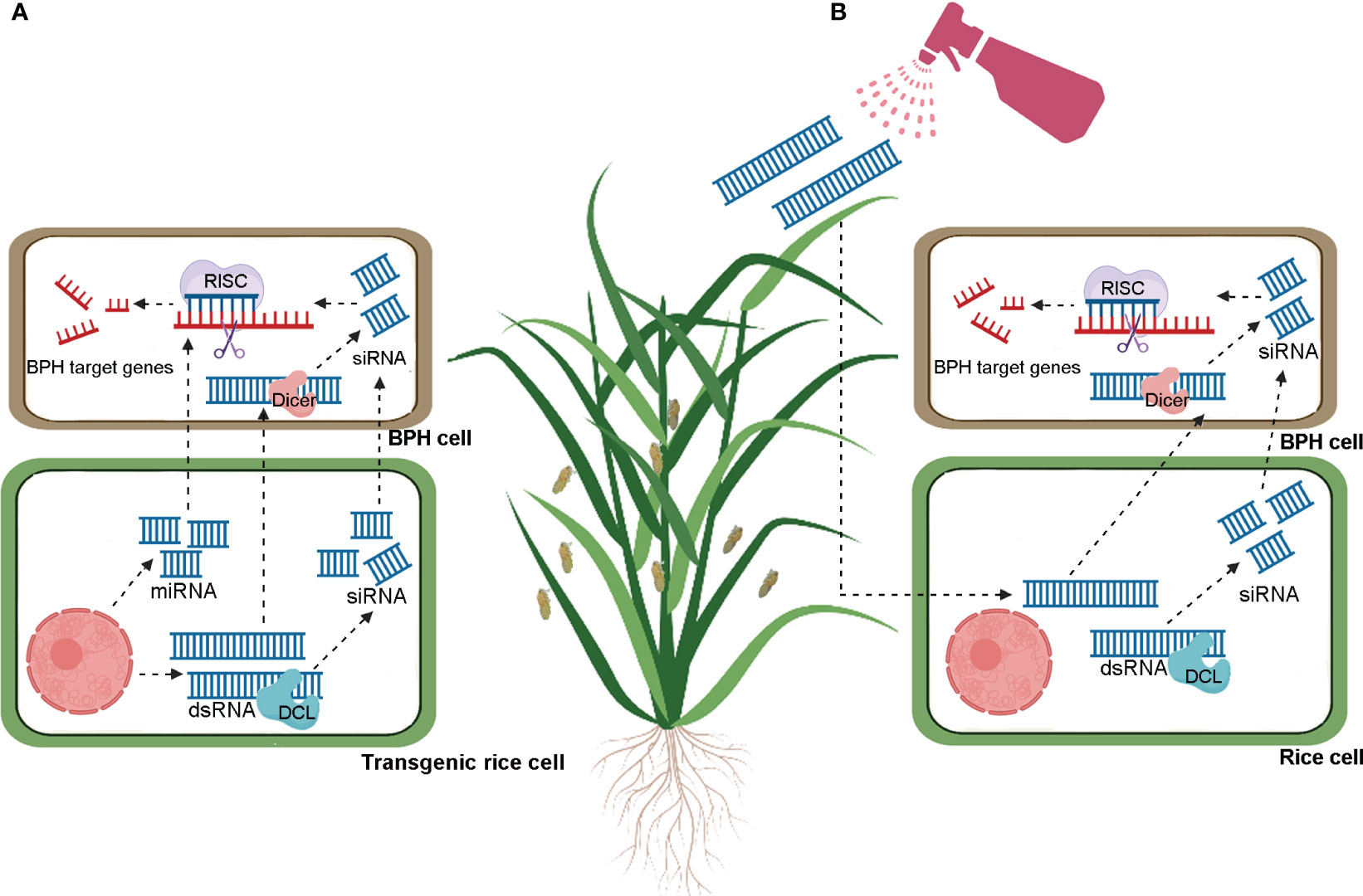
Figure 1 Schematic models for brown planthoppers control through host induced gene silencing (HIGS, A) and spray induced gene silencing (SIGS, B). In HIGS (A), transgenic plants produce exogenous dsRNA or miRNA, or external spraying (B) delivers exogenous dsRNA. These dsRNA are processed into small interfering RNAs (siRNAs) by rice Dicer-like (DCL) proteins. The siRNAs are then transferred to brown planthopper (BPH) cells and bind to complementary sequences on BPH target mRNA. Through the assistance of the RNA-induced silencing complex (RISC), the target transcripts are silenced. Additionally, exogenous dsRNA and miRNAs produced by transgenic plants can be directly absorbed by BPH, resulting in gene silencing.
Recently, a novel RNAi-based crop protection strategy called “spray-induced gene silencing (SIGS)” has been developed (Jiang et al., 2023; Mahanty et al., 2023). As the name implies, SIGS does not require genetic modification and instead involves simply spraying crop plants with synthesized exogenous dsRNA to selectively knock down insect or pathogen genes (Figure 1B). This technology has been successfully used to control rice blast disease (Magnaporthe oryzae) by spraying dsRNA targeting the fungal pathogenicity gene MoDES1 (Sarkar and Roy-Barman, 2021). Recently, a nanocarrier-dsRNA spray delivery system was developed to control the white-backed planthopper (WBPH) (Sogatella furcifera) under laboratory conditions (Guo et al., 2023a, and Guo et al., 2023b). The results demonstrated the efficacy of the nanocarrier spray system for inducing RNAi-mediated knockdown of WBPH genes, including SfTH, SfEGFR, Sfzfh-2, SfAbd-A, and SfAbd-B. In addition, the treatment resulted in significant phenotypic defects and increased mortality in WBPH (Guo et al., 2023a, and Guo et al., 2023b). These promising results lay a foundation for the further development and application of SIGS to control rice pests, including BPH.
Perspectives and challenges
A growing body of research has revealed the involvement of sRNAs in the interaction between rice and BPH. The majority of these sRNAs have been predicted and/or identified through multi-omics analyses, and their targets have been predicted computationally. However, many of these results still require experimental validation. Moreover, the molecular mechanisms underlying sRNA-mediated rice-BPH interactions remain poorly understood. The pathways of sRNA transfer between rice and BPH should also be comprehensively evaluated. Our growing understanding of cross-kingdom RNAi has paved the way for the development of promising agricultural pest control strategies, including HIGS and SIGS. Nevertheless, HIGS and SIGS face several technical challenges. The stability and uptake efficiency of dsRNA and sRNA need to be strengthened and off-target activities must be avoided. We predict that the development and application of environmentally-friendly RNAi-based technology will become an agronomic research focus, and that the communication of cross-kingdom sRNAs will emerge as a hot research topic.
Author contributions
SJ: Project administration, Writing – original draft, Writing – review & editing. JX: Writing – review & editing. HT: Writing – review & editing. PL: Writing – review & editing. BY: Writing – original draft, Writing – review & editing. QL: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This review was supported by grants from the National Natural Science Foundation of China (U1704111 and 31401732), ZHONGYUAN YINGCAI JIHUA (ZYYCYU202012165), and Henan Province Science and Technology Research Project (222102110116).
Acknowledgments
We acknowledge the BioRender tool that we used to create Figure 1. We appreciate the linguistic assistance provided by TopEdit (www.topeditsci.com) during the preparation of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Backus, E., Serrano, M., Ranger, C. (2005). Mechanisms of hopperburn: an overview of insect taxonomy, behavior, and physiology. Annu. Rev. Entomol. 50, 125–151. doi: 10.1146/annurev.ento.49.061802
Bartel, D. (2009). MicroRNAs: Target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Chapman, E., Carrington, J. (2007). Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 8, 884–896. doi: 10.1038/nrg2179
Chen, X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25, 21–44. doi: 10.1146/annurev.cellbio.042308.113417
Chen, X., Jiang, L., Zheng, J., Chen, F., Wang, T., Wang, M., et al. (2019). A missense mutation in Large Grain Size 1 increases grain size and enhances cold tolerance in rice. J. Exp. Bot. 70, 3851–3866. doi: 10.1093/jxb/erz192
Chen, J., Li, T., Pang, R., Yue, X., Hu, J., Zhang, W. (2018). Genome-wide screening and functional analysis reveal that the specific microRNA nlu-miR-173 regulates molting by targeting Ftz-F1 in Nilaparvata lugens. Front. Physiol. 9. doi: 10.3389/fphys.2018.01854
Chen, J., Liang, Z., Liang, Y., Pang, R., Zhang, W. (2013). Conserved microRNAs miR-8-5p and miR-2a-3p modulate chitin biosynthesis in response to 20-hydroxyecdysone signaling in the brown planthopper, Nilaparvata lugens. Insect Biochem. Mol. Biol. 43, 839–848. doi: 10.1016/j.ibmb.2013.06.002
Chen, J., Liu, Q., Yuan, L., Shen, W., Shi, Q., Qi, G., et al. (2023). Osa-miR162a enhances the resistance to the brown planthopper via α-linolenic acid metabolism in rice (Oryza sativa). J. Agric. Food Chem. 71, 11847–11859. doi: 10.1021/acs.jafc.3c02637
Coleman, A., Wouters, R., Mugford, S., Hogenhout, S. (2015). Persistence and transgenerational effect of plant-mediated RNAi in aphids. J. Exp. Bot. 66, 541–548. doi: 10.1093/jxb/eru450
Dai, Z., Tan, J., Zhou, C., Yang, X., Yang, F., Zhang, S., et al. (2019). The OsmiR396-OsGRF8-OsF3H-flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa). Plant Biotechnol. J. 17, 1657–1669. doi: 10.1111/pbi.13091
Du, B., Chen, R., Guo, J., He, G. (2020). Current understanding of the genomic, genetic, and molecular control of insect resistance in rice. Mol. Breed. 40, 24. doi: 10.1007/s11032-020-1103-3
Duan, P., Ni, S., Wang, J., Zhang, B., Xu, R., Wang, Y., et al. (2015). Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2, 15203. doi: 10.1038/nplants.2015.203
Erb, M., Reymond, P. (2019). Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 29, 527–557. doi: 10.1146/annurev-arplant-050718-095910
Escobar, M., Civerolo, E., Summerfelt, K., Dandekar, A. (2001). RNAi-mediated oncogene silencing confers resistance to crown gall tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 98, 13437–13442. doi: 10.1073/pnas.241276898
Fu, X., Li, T., Chen, J., Dong, Y., Qiu, J., Kang, K., et al. (2015). Functional screen for microRNAs of Nilaparvata lugens reveals that targeting of glutamine synthase by miR-4868b regulates fecundity. J. Insect Physiol. 83, 22–29. doi: 10.1016/j.jinsphys.2015.11.003
Ge, Y., Han, J., Zhou, G., Xu, Y., Ding, Y., Shi, M., et al. (2018). Silencing of miR156 confers enhanced resistance to brown planthopper in rice. Planta 248, 813–826. doi: 10.1007/s00425-018-2942-6
Ghini, F., Rubolino, C., Climent, M., Simeone, I., Marzi, M., Nicassio, F. (2018). Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nat. Commun. 9, 3119. doi: 10.1038/s41467-018-05182-9
Golden, D., Gerbasi, V., Sontheimer, E. (2008). An inside job for siRNAs. Mol. Cell 31, 309–312. doi: 10.1016/j.molcel.2008.07.008
Guo, H., Liu, X. Z., Long, G. J., Gong, L. L., Zhang, M. Q., Ma, Y. F., et al. (2023a). Functional characterization of developmentally critical genes in the white-backed planthopper: Efficacy of nanoparticle-based dsRNA sprays for pest control. Pest Manage. Sci. 79, 1048–1061. doi: 10.1002/ps.7271
Guo, H., Lona, G. J., Liu, X. Z., Ma, Y. F., Zhang, M. Q., Gong, L. L., et al. (2023b). Functional characterization of tyrosine melanin genes in the white-backed planthopper and utilization of a spray-based nanoparticle-wrapped dsRNA technique for pest control. Int. J. Biol. Macromol. 230, 123123. doi: 10.1016/j.ijbiomac.2022.123123
Huang, C., Wang, H., Hu, P., Hamby, R., Jin, H. (2019). Small RNAs-big players in plant-microbe interactions. Cell Host Microbe 26, 173–182. doi: 10.1016/j.chom.2019.07.021
Huang, J., Zhang, N., Shan, J., Peng, Y., Guo, J., Zhou., C., et al. (2020). Salivary protein 1 of brown planthopper is required for survival and induces immunity response in plants. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.571280
Jiang, C., Li, Z., Zheng, L., Yu, Y., Niu, D. (2023). Small RNAs: Efficient and miraculous effectors that play key roles in plant-microbe interactions. Mol. Plant Pathol. 24, 999–1013. doi: 10.1111/mpp.13329
Jing, S., Zhao, Y., Du, B., Chen, R., Zhu, L., He, G. (2017). Genomics of interaction between the brown planthopper and rice. Curr. Opin. Insect Sci. 19, 82–87. doi: 10.1016/j.cois.2017.03.005
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Katiyar-Agarwal, S., Jin, H. (2010). Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 48, 225–246. doi: 10.1146/annurev-phyto-073009-114457
Khraiwesh, B., Zhu, J., Zhu, J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819, 137–148. doi: 10.1016/j.bbagrm.2011.05.001
Kryovrysanaki, N., James, A., Tselika, M., Bardani, E., Kalantidis, K. (2022). RNA silencing pathways in plant development and defense. Int. J. Dev. Biol. 66, 163–175. doi: 10.1387/ijdb.210189kk
Li, T., Chen, J., Fan, X., Chen, W., Zhang, W. (2017). MicroRNA and dsRNA targeting chitin synthase A reveal a great potential for pest management of the hemipteran insect Nilaparvata lugens. Pest Manage. Sci. 73, 1529–1537. doi: 10.1002/ps.4492
Li, X., Zhao, M., Tian, M., Zhao, J., Cai, W., Hua, H. (2021). An InR/mir-9a/NlUbx regulatory cascade regulates wing diphenism in brown planthoppers. Insect Sci. 28, 1300–1313. doi: 10.1111/1744-7917.12872
Lü, J., Liu, J., Chen, L., Sun, J., Su, Q., Li, S., et al. (2022). Screening of brown planthopper resistant miRNAs in rice and their roles in regulation of brown planthopper fecundity. Rice Sci. 29, 559–568. doi: 10.1016/j.rsci.2022.05.003
Mahanty, B., Mishra, R., Joshi, R. K. (2023). Cross-kingdom small RNA communication between plants and fungal phytopathogens-recent updates and prospects for future agriculture. RNA Biol. 20, 109–119. doi: 10.1080/15476286.2023.2195731
Nanda, S., Yuan, S., Lai, F., Wang, W., Fu, Q., Wan, P. (2020). Identification and analysis of miRNAs in IR56 rice in response to BPH infestations of different virulence levels. Sci. Rep. 10, 19093. doi: 10.1038/s41598-020-76198-9
Panwar, V., Jordan, M., McCallum, B., Bakkeren, G. (2018). Host-induced silencing of essential genes in Puccinia triticina through transgenic expression of RNAi sequences reduces severity of leaf rust infection in wheat. Plant Biotechnol. J. 16, 1013–1023. doi: 10.1111/pbi.12845
Rodriguez, P. A., Escudero-Martinez, C., Bos, J. I. (2017). An aphid effector targets trafficking protein VPS52 in a host-specific manner to promote virulence. Plant Physiol. 173, 1892–1903. doi: 10.1104/pp.16.01458
Sarkar, A., Roy-Barman, S. (2021). Spray-induced silencing of pathogenicity gene MoDES1 via exogenous double-stranded RNA can confer partial resistance against fungal blast in rice. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.733129
Sattar, S., Thompson, G. A. (2016). Small RNA regulators of plant-hemipteran interactions: micromanagers with versatile roles. Front. Plant Sci. 7, 1241. doi: 10.3389/fpls.2016.01241
Seemanpillai, M., Dry, I., Randles, J., Rezaian, A. (2003). Transcriptional silencing of geminiviral promoter-driven transgenes following homologous virus infection. Mol. Plant Microbe Interact. 16, 429–438. doi: 10.1094/MPMI.2003.16.5.429
Shangguan, X., Zhang, J., Liu, B., Zhao, Y., Wang, H., Wang, Z., et al. (2018). A mucin-like protein of planthopper is required for feeding and induces immunity response in plants. Plant Physiol. 176, 552–565. doi: 10.1104/pp.17.00755
Shen, W., Cao, S., Liu, J., Zhang, W., Chen, J., Li, J. (2021). Overexpression of an Osa-miR162a derivative in rice confers cross-kingdom RNA interference-mediated brown planthopper resistance without perturbing host development. Int. J. Mol. Sci. 22, 12652. doi: 10.3390/ijms222312652
Shen, Y., Yang, G., Miao, X., Shi, Z. (2023). OsmiR159 modulate BPH resistance through regulating G-protein γ subunit GS3 gene in rice. Rice 16, 30. doi: 10.1186/s12284-023-00646-z
Shi, S., Wang, H., Nie, L., Tan, D., Zhou, C., Zhang, Q., et al. (2021). Bph30 confers resistance to brown planthopper by fortifying sclerenchyma in rice leaf sheaths. Mol. Plant 14, 1714–1732. doi: 10.1016/j.molp.2021.07.004
Shi, S., Zha, W., Yu, X., Wu, Y., Li, S., Xu, H., et al. (2023). Integrated transcriptomics and metabolomics analysis provide insight into the resistance response of rice against brown planthopper. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1213257
Shivakumara, T., Chaudhary, S., Kamaraju, D., Dutta, T., Papolu, P., Banakar, P., et al. (2017). Host-induced silencing of two pharyngeal gland genes conferred transcriptional alteration of cell wall-modifying enzymes of Meloidogyne incognita vis-a-vis perturbed nematode infectivity in eggplant. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00473
Takken, F. L. W., Tameling, W. I. L. (2009). To nibble at plant resistance proteins. Science 324, 744–746. doi: 10.1126/science.1171666
Tan, J., Wu, Y., Guo, J., Li, H., Zhu, L., Chen, R., et al. (2020). A combined microRNA and transcriptome analyses illuminates the resistance response of rice against brown planthopper. BMC Genom. 21, 144. doi: 10.1186/s12864-020-6556-6
Van, E., Powell, C., Shatters, R., Borovsky, D. (2014). Control of larval and egg development in Aedes aegypti with RNA interference against juvenile hormone acid methyl transferase. J. Insect Physiol. 70, 143–150. doi: 10.1016/j.jinsphys.2014.08.001
Wang, H., Shi, S., Hua, W. (2023). Advances of herbivore-secreted elicitors and effectors in plant-insect interactions. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1176048
Wang, N., Zhang, C., Chen, M., Shi, Z., Zhou, Y., Shi, X., et al. (2022). Characterization of MicroRNAs associated with reproduction in the brown planthopper, Nilaparvata lugens. Int. J. Mol. Sci. 23, 7808. doi: 10.3390/ijms23147808
Weiberg, A., Wang, M., Lin, F., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Wu, Y., Lv, W., Hu, L., Rao, W., Zeng, Y., Zhu, L., et al. (2017). Identification and analysis of brown planthopper-responsive microRNAs in resistant and susceptible rice plants. Sci. Rep. 7 (1), 8712. doi: 10.1038/s41598-017-09143-y
Xu, H. J., Chen, T., Ma, X. F., Xue, J., Pan, P. L., Zhang, X. C., et al. (2013). Genome-wide screening for components of small interfering RNA (siRNA) and microRNA (miRNA) pathways in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Mol. Biol. 22, 635–647. doi: 10.1111/imb.12051
Xu, H., Xue, J., Lu, B., Zhang, X., Zhuo, J., He, S., et al. (2015). Two insulin receptors determine alternative wing morphs in planthoppers. Nature 519, 464–467. doi: 10.1038/nature14286
Xu, L., Zhang, J., Zhan, A., Wang, Y., Ma, X., Jie, W., et al. (2020). Identification and analysis of microRNAs associated with wing polyphenism in the brown planthopper, Nilaparvata lugens. Int. J. Mol. Sci. 21, 9754. doi: 10.3390/ijms21249754
Ye, X., Xu, L., Li, X., He, K., Hua, H., Cao, Z., et al. (2019). miR-34 modulates wing polyphenism in planthopper. PloS Genet. 15, e1008235. doi: 10.1371/journal.pgen.1008235
Yue, E., Li, C., Li, Y., Liu, Z., Xu, J. (2017). MiR529a modulates panicle architecture through regulating squamosa promoter binding-like genes in rice (Oryza sativa). Plant Mol. Biol. 94, 469–480. doi: 10.1007/s11103-017-0618-4
Zamore, P., Haley, B. (2005). Ribo-gnome: The big world of small RNAs. Science 309, 1519–1524. doi: 10.1126/science.1111444
Zha, W., Peng, X., Chen, R., Du, B., Zhu, L., He, G. (2011). Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PloS One 6, e20504. doi: 10.1371/journal.pone.0020504
Zha, W., Zhou, L., Li, S., Liu, K., Yang, G., Chen, Z., et al. (2016). Characterization and comparative profiling of the small RNA transcriptomes in the Hemipteran insect Nilaparvata lugens. Gene 595, 83–91. doi: 10.1016/j.gene.2016.09.042
Zhai, Y., Zhang, J., Sun, Z., Dong, X., He, Y., Kang, K., et al. (2013). Proteomic and transcriptomic analyses of fecundity in the brown planthopper Nilaparvata lugens (Stål). J. Proteome. Res. 12, 5199–5212. doi: 10.1021/pr400561c
Zhang, B., Pan, X., Cobb, G., Anderson, T. (2006). Plant microRNA: a small regulatory molecule with big impact. Dev. Biol. 289, 3–16. doi: 10.1016/j.ydbio.2005.10.036
Zheng, X., Zhu, L., He, G. (2021). Genetic and molecular understanding of host rice resistance and Nilaparvata lugens adaptation. Curr. Opin. Insect Sci. 45, 14–20. doi: 10.1016/j.cois.2020.11.005
Keywords: sRNAs, Oryza sativa, brown planthopper, RNAi, resistance
Citation: Jing S, Xu J, Tang H, Li P, Yu B and Liu Q (2023) The roles of small RNAs in rice-brown planthopper interactions. Front. Plant Sci. 14:1326726. doi: 10.3389/fpls.2023.1326726
Received: 23 October 2023; Accepted: 10 November 2023;
Published: 23 November 2023.
Edited by:
Jihong Hu, Northwest A&F University, ChinaCopyright © 2023 Jing, Xu, Tang, Li, Yu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Yu, eXViaW5fMjAxNUAxMjYuY29t; Qingsong Liu, bGl1cWluZ3NvbmdAeHludS5lZHUuY24=
 Shengli Jing
Shengli Jing Jingang Xu
Jingang Xu Hengmin Tang
Hengmin Tang Peng Li
Peng Li Bin Yu
Bin Yu Qingsong Liu
Qingsong Liu