- 1North Florida Research and Education Center, University of Florida, Quincy, FL, United States
- 2Plant Pathology Department, University of Florida, Gainesville, FL, United States
- 3Indian Council of Agricultural Research (ICAR) - National Centre for Integrated Pest Management, New Delhi, India
Plant disease classification is quite complex and, in most cases, requires trained plant pathologists and sophisticated labs to accurately determine the cause. Our group for the first time used microscopic images (×30) of tomato plant diseases, for which representative plant samples were diagnostically validated to classify disease symptoms using non-coding deep learning platforms (NCDL). The mean F1 scores (SD) of the NCDL platforms were 98.5 (1.6) for Amazon Rekognition Custom Label, 93.9 (2.5) for Clarifai, 91.6 (3.9) for Teachable Machine, 95.0 (1.9) for Google AutoML Vision, and 97.5 (2.7) for Microsoft Azure Custom Vision. The accuracy of the NCDL platform for Amazon Rekognition Custom Label was 99.8% (0.2), for Clarifai 98.7% (0.5), for Teachable Machine 98.3% (0.4), for Google AutoML Vision 98.9% (0.6), and for Apple CreateML 87.3 (4.3). Upon external validation, the model’s accuracy of the tested NCDL platforms dropped no more than 7%. The potential future use for these models includes the development of mobile- and web-based applications for the classification of plant diseases and integration with a disease management advisory system. The NCDL models also have the potential to improve the early triage of symptomatic plant samples into classes that may save time in diagnostic lab sample processing.
Introduction
Plants contribute up to 80% of food for human consumption and feed for livestock. Numerous pests and diseases cause approximately 20%–40% losses in crop yield, with diseases resulting in $220 billion in losses annually (FAO, 2020). In 2021, tomato was a major vegetable crop grown worldwide with an annual production of 189 million tons from 5.2 million hectares (FAO, 2022). Tomato is affected by almost 200 diseases, and these frequently create bottlenecks for optimum tomato production (Jones et al., 2014). The accurate identification of disease symptoms is the first step in the management of plant diseases. In many cases, incorrect diagnosis of plant diseases leads to mismanagement and economic losses (Riley et al., 2002). Currently, disease identification is done by a combination of visual observations and lab-based techniques (e.g., microscopy, culturing, antibody, and molecular methods). This process can be time-consuming and needs highly trained personnel, who are available in developed countries while limited or not present in developing and less-developed countries. Added to this challenge, some plant diseases can spread like “wildfire” and can destroy a crop in days (Fry et al., 2013; Chen, 2020). Thus, there is a need for faster, cheaper, and reliable disease classification approaches to identify potential causes of disease and selection of management strategies.
Deep learning is a subfield of artificial intelligence (AI). Deep learning refers to the use of artificial neural network architectures that contain a substantial number of internal processing layers to provide processed output from raw data (Lecun et al., 2015). Deep learning tools are used in a wide variety of tasks in agriculture (e.g., disease classification, yield estimation and forecasting, crop genomics, and modeling). In the last decade, several researchers used machine learning-based tools for the classification of plant diseases (Mohanty et al., 2016; Sladojevic et al., 2016; Amara et al., 2017; Brahimi et al., 2017; Ferentinos, 2018; Lee et al., 2020; Liu et al., 2021). For tomato, researchers used various deep learning models like AlexNet, GoogleNet, Inception V3, Residual Network (ResNet) 18, and ResNet 50 (Tran et al., 2019; Maeda-Gutiérrez et al., 2020; Chong et al., 2023); Yolo V3 convolutional neural network (Liu and Wang, 2020); or conditional generative adversarial network (Abbas et al., 2021). Many of these models had long training times (up to 2 weeks) with modest datasets (Picon et al., 2019) and low accuracy with test datasets (Liu et al., 2021). In addition, the models had a lower performance matrix on external field captured images and required resources including 1) high-performance computer with a costly graphic processing unit (GPU) or access to cloud computing and 2) expertise with skills in deep learning.
Researchers working directly with farmers have limited resources and lack robust AI talent pool, especially in developing and underdeveloped countries (Gwagwa et al., 2021). One potential solution to these challenges is to use no coding or minimum coding deep learning models called automated machine learning. This is a process of applying machine learning models to real-world problems by streamlining raw data processing, feature, and model selection and hyperparameter optimization with minimal human inferences (Kanti Karmaker et al., 2021). Recently automated machine learning approaches were tested in the medical field for the classification of images of the eye (Korot et al., 2021), video of cataract (Touma et al., 2022), and carotid injury (Unadkat et al., 2022). Automated machine learning facilitates users to build state-of-the-art models without writing codes, making them accessible to a wide range of individuals without coding experience.
The accurate classification of diseases is extremely challenging as most disease symptoms are randomly distributed within plant parts (Riley et al., 2002). Disease symptoms can vary with cultivar, crop stage, environment, and other biotic and abiotic factors necessitating that the image databases used in building models are updated regularly with new sets of images. Deep learning models already used in plant disease classification have the same image database such as PlantVillage or Kaggle and have led to non-ideal robustness and overfitting, resulting in a reduction in testing accuracy (Mohanty et al., 2016; Darwish et al., 2020). In some cases, classification performance on external data evaluation was reduced below 50% (Mohanty et al., 2016; Ferentinos, 2018). AlexNet and GoogLeNet deep learning model performance was reduced to 31% from 99% with the PlantVillage database (Mohanty et al., 2016). Deep learning VGG model success rate on testing with real field images reduced to 33.3% on laboratory-tested models despite a large dataset of over 80,000 images used for training of the models (Ferentinos, 2018).
Most of the existing database images are taken using a mobile phone or camera. These macroscopic images capture more background features (pixels) beyond the focus on the diseased section of the plant parts. Lab disease diagnosis is based on both visual symptom examination, using a stereoscopic microscope often followed by a molecular diagnostic assay (Riley et al., 2002). Stereoscopic microscopes provide ×25–70 magnification, which helps in better visualization of fungal symptoms and signs like sclerotia, mycelial growth, and bacterial oozing, all of which can help in the diagnosis of plant diseases. Images taken with a higher magnification than normal mobile phone cameras may be a potential solution to improve the accuracy of the models in field conditions as the magnified view of the disease symptoms could help in better training with little or no noise from the background features.
Most deep learning models are data-centric and require a well-curated dataset (Kanti Karmaker et al., 2021). Based on the limitations in the accuracy of models for field disease classification and the potential for using non-coding deep learning (NCDL) platforms to improve a better system of classification, this present study was undertaken. We built a new tomato image database that consisted of field-collected microscopic images using a ×30 mobile-mounted camera lens. These images were trained and tested using NCDL platforms including Amazon Rekognition Custom Label (Custom Label), Teachable Machine from Google (Teachable Machine), Microsoft Custom Azure Vision (Custom Vision), Google AutoML Vision (AutoML), Clarifai, and Apple CreateML (CreateML). The goal of this study is to demonstrate the potential use of microscopic images of plant disease with NCDL platforms for improved classification. The specific objectives were i) to develop a tomato biotic stress microscopic images database and ii) to analyze the potential of non-coding machine learning models for disease classification using microscopic images.
Materials and methods
Collection of images and diagnostic validation
The study was designed to collect diverse images from major tomato-growing regions in Florida and Georgia. Images were collected during 2020 and 2021 in 10 different field surveys. The images were collected using a mobile-mounted ×30 camera (GoMicro). The android smartphone Xiaomi Redmi Note 5 Pro 12 MP camera was used for image capture by a single individual (Supplementary Figure 1). Images were captured using the GoMicro Detect application in natural sunlight in field conditions.
Images were collected from all symptomatic parts, i.e., leaves (upper and lower sides) and fruits. To reduce background noise and effects from shaking, samples were placed on a flat cardboard surface for image capture. Disease symptoms at different stages were captured to have a broad and diverse image collection. In addition to the images of the symptomatic samples, healthy samples were also collected. When symptoms were too large to be captured in a single microscopic image, multiple images were taken of the symptom region. While capturing the images, the optical zoom of the mobile camera was not used. Representative samples during the surveys were diagnostically identified using microscopic, microbiological, and molecular tools to ensure images were classified correctly (Supplementary Table 1). All images were visually examined to remove any that were out-of-focus. Images were labeled based on the diagnostic report provided by the plant disease diagnostic lab at the University of Florida, and expert knowledge of an experienced horticulturist was used when relevant to abiotic issues. A small subset of representative images for each class is presented in Supplementary Figure 2.
Non-coding deep learning platform selection
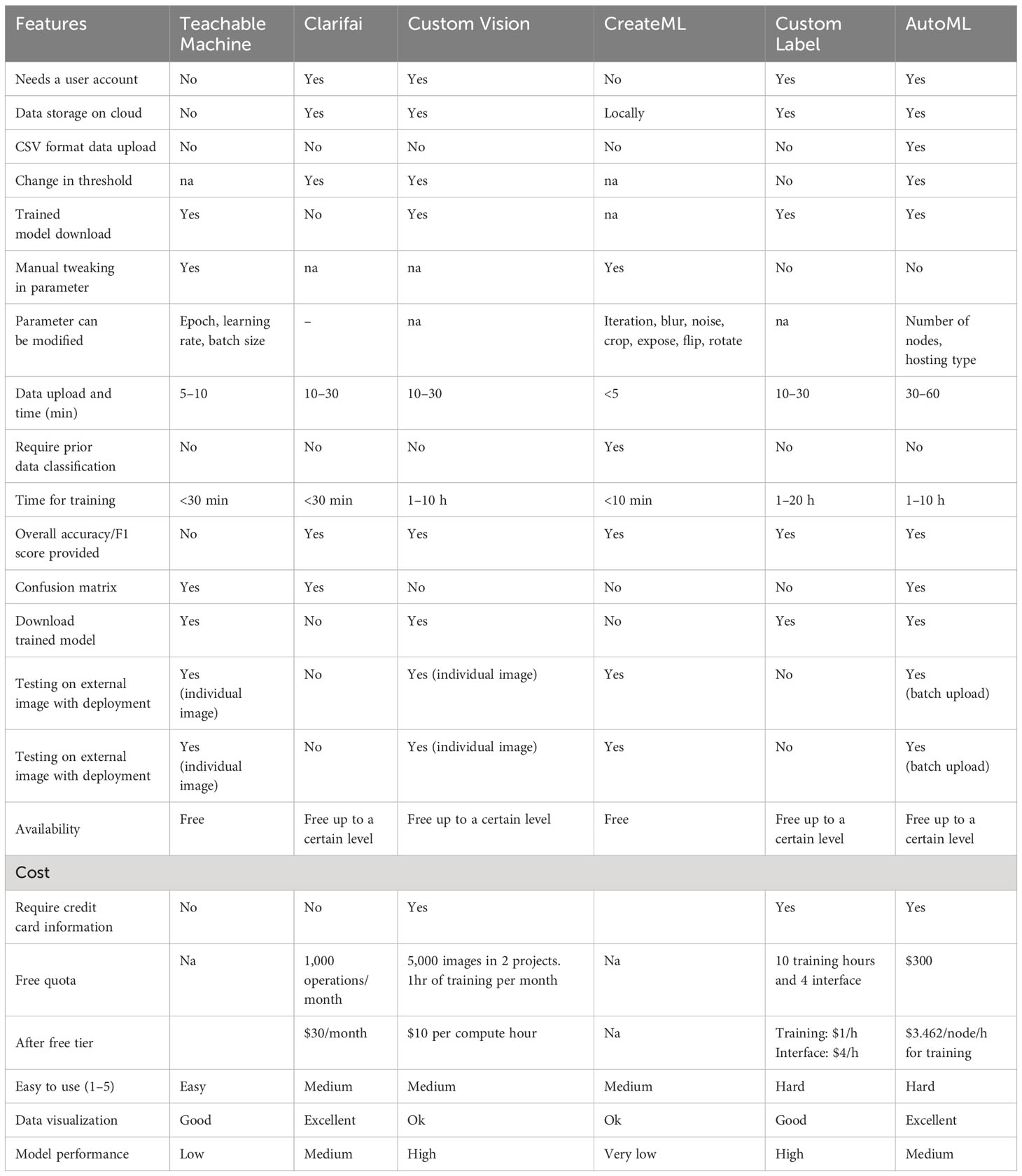
Six different NCDL platforms were selected based on prior literature and information available through Internet searches. Each platform was reviewed for available features, including access for use in the U.S., cost, type of output, graphical user interface, time taken for analysis, and ease of use by a person with minimal coding experience. A summary of the NCDL platforms and reasons for not using a particular platform is provided in Supplementary Table 2. Platforms included in this study were Amazon Rekognition Custom Label, Teachable Machine from Google, Microsoft Custom Vision, Google AutoML Vision, Clarifai, and Apple CreateML. All platforms except CreateML use a Microsoft Windows-based system on the web without downloading locally. CreateML was downloaded in a Mac-based system and run locally using the XCode developer program without active Internet. Teachable Machine and CreateML were freely available without any limitations. For the other platforms, the free tier (the use of the model without making any payment) was used first, and once the free limit was exhausted, the basic tier (cheapest plan) was used with payment.
Data upload and labeling
All dataset images were labeled according to their diagnostic result/expert knowledge. Each NCDL platform has a distinct way of uploading and storage of the dataset. In the case of the Teachable Machine platform, images had to be uploaded each time for training, and the data were not stored locally. For the other platforms, the uploaded data were stored for further use. Only the AutoML platform allows image upload in CSV format. In all the NCDL platforms except CreateML, data had to be labeled manually after upload. Images were labeled separately as training and testing for the CreateML NCDL platform. A detailed description of each NCDL platform is provided in Table 1.
Data processing
Training with the supervised NCDL platform needs images to be split into training, validation, and testing sets. All except CreateML did not use prior splitting of the image dataset. Each NCDL platform default setting was used for the splitting of each image dataset, and if manual setting was required, 80:20 (training:testing) split was used. For CreateML, python library split-folders (V 0.5.1) (Split-folders:PyPI, 2022) were used to split the dataset into training and testing datasets of 80:20 ratio. Teachable Machine and AutoML have 85:15 and 90:10 split ratios, respectively. Duplicate images were automatically detected by AutoML and Custom Vision, and these images, if any, were removed from the training databases in all the other NCDL platforms. Custom Vision advanced (no time for training) option was used for training.
Model training
NCDL platform models were trained using image databases described in Table 2 using a web interface except in the case of CreateML. These models did not require any specific CPU or GPU uses. CreateML platform was run locally on the Mac-based system that has 256 GB SSD and 8 GB RAM. For the default threshold value of each model, or where a manual option was available, 50% (0.5) was used as the threshold value for classification. The parameters that can be modified within each platform are described in Table 1. Unless otherwise specified, we did not use hyperparameters or change the default setting of the NCDL platform models.
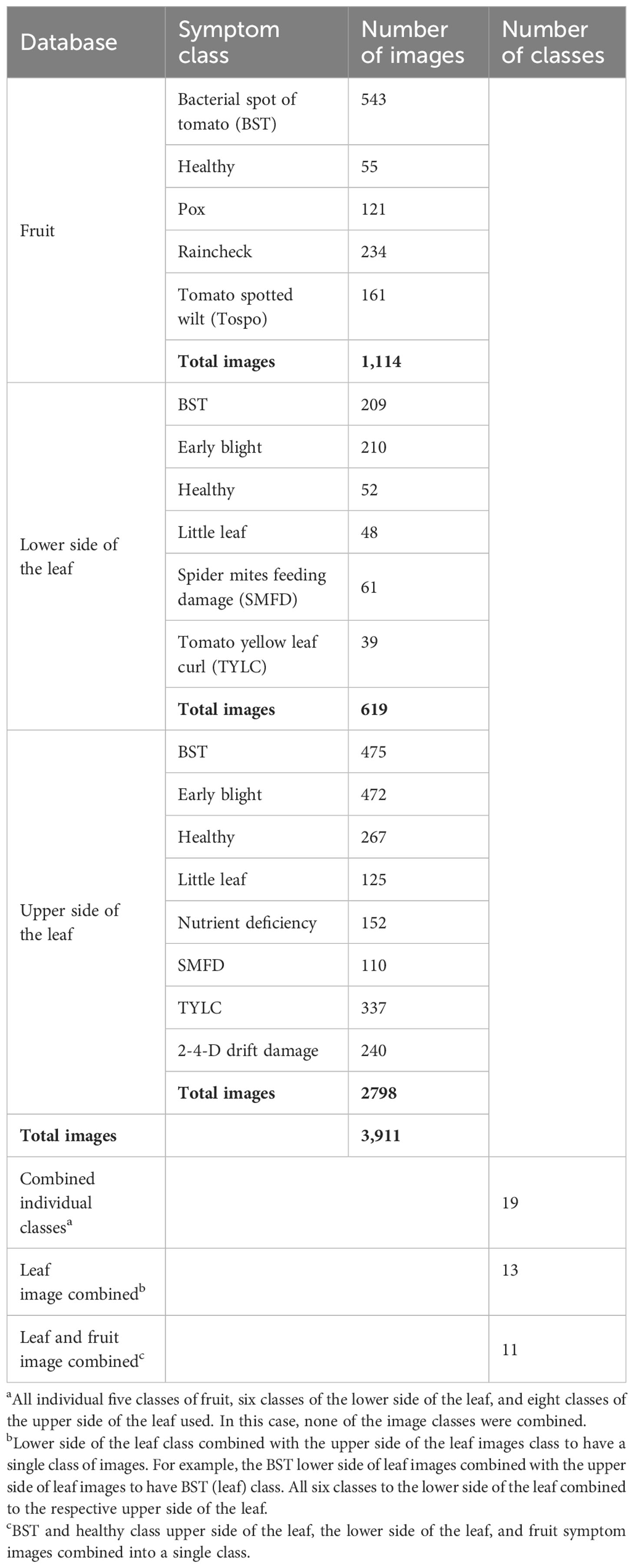
Table 2 Databases for the training of the non-coding deep learning (NCDL) platform models used in this study.
Metrics and statistical analysis
Each model graphic user interface provides different types of outputs. To compare the models, we used precision/positive predictive value (PPV), negative predictive value (NPV), recall/sensitivity, specificity, accuracy, and F1 score. To evaluate these parameters, we extracted the following: true positive (TP), which is the number of positive images that were classified as the same as the labeled class; true negative (TN), which is the number of negative images that were classified as the same as the labeled class; false positive (FP), which is the number of images classified as positive but do not belong to that labeled class; and false negative (FN), which is the number of images that belong to the labeled class but were classified in a different class. A summary of the approach is given below.
Some models provided a confusion matrix, which is an easy way to understand models in terms of each class and how many misclassified images belong to which class in a table format. Graphs were created with precision and recall value, and the area under this graph line referred to as the area under precision–recall curve (AUPRC) was only available in AutoML, mentioned as the average precision.
Each of the NCDL platform model outputs was distinct. The Teachable Machine provides only accuracy and a confusion matrix. Clarifai produces one K spilt value of cross-validation report along with a confusion matrix. Custom Vision provides only precision, recall, and average precision (AUPRC/AP) values. CreateML provides precision and recall for each labeled class and overall accuracy for validation and test data. Custom Label provides the threshold used for the classification of each labeled class and the corresponding precision, recall, and F1 score. The AutoML model provides a confusion matrix and accuracy. The AutoML model precision and recall curve can be modified with an interactive threshold level. A default threshold value of 0.5 was used for all NCDL platforms.
Results
Image collection and database
Image databases were created for fruit images (symptomatic and healthy) and for leaf images [symptomatic and healthy adaxial (upper) and abaxial (lower) surfaces]. Overall, a total of 3,911 images were collected and labeled in 19 different classes (Table 2). For NCDL platform model analyses, these images were further classified into three different categories: 1) all 19 individual class labeled images were used for analysis and referred as “combined individual class”; 2) individual classes of adaxial and abaxial leaf symptoms were pooled into one class, making 13 total classes, and referred as “leaf images combined”; and 3) finally, images of classes with fruit and abaxial and adiaxial leaf symptoms were pooled in single class making a total of 11 classes and referred as “leaf and fruit images combined.” In total, six different types of image databases were analyzed using different models.
Deep learning model performance
The F1 score (Standard Deviation (S.D.)) of the different NCDL platforms across all models-databases were as follows: Custom Label, 98.5 (1.6); Clarifai, 93.9 (2.5); Teachable Machine, 91.6 (3.9); AutoML, 95.0 (1.9); and Custom Vision, 97.5 (2.7) (Figure 1A and Supplementary Table 3). The F1 score was significantly different between the models (F(4, 25) = 6.729, P = 0.0008). The Custom Label F1 score was significantly higher than Teachable Machine and Clarifai with Tukey’s multiple post-hoc comparisons. The Custom Vision F1 score was significantly different than only Teachable Machine with a mean difference (95% CI) of 5.9 (1.5–10.4) (Supplementary Table 4). All the other NCDL models showed statistically similar F1 scores.
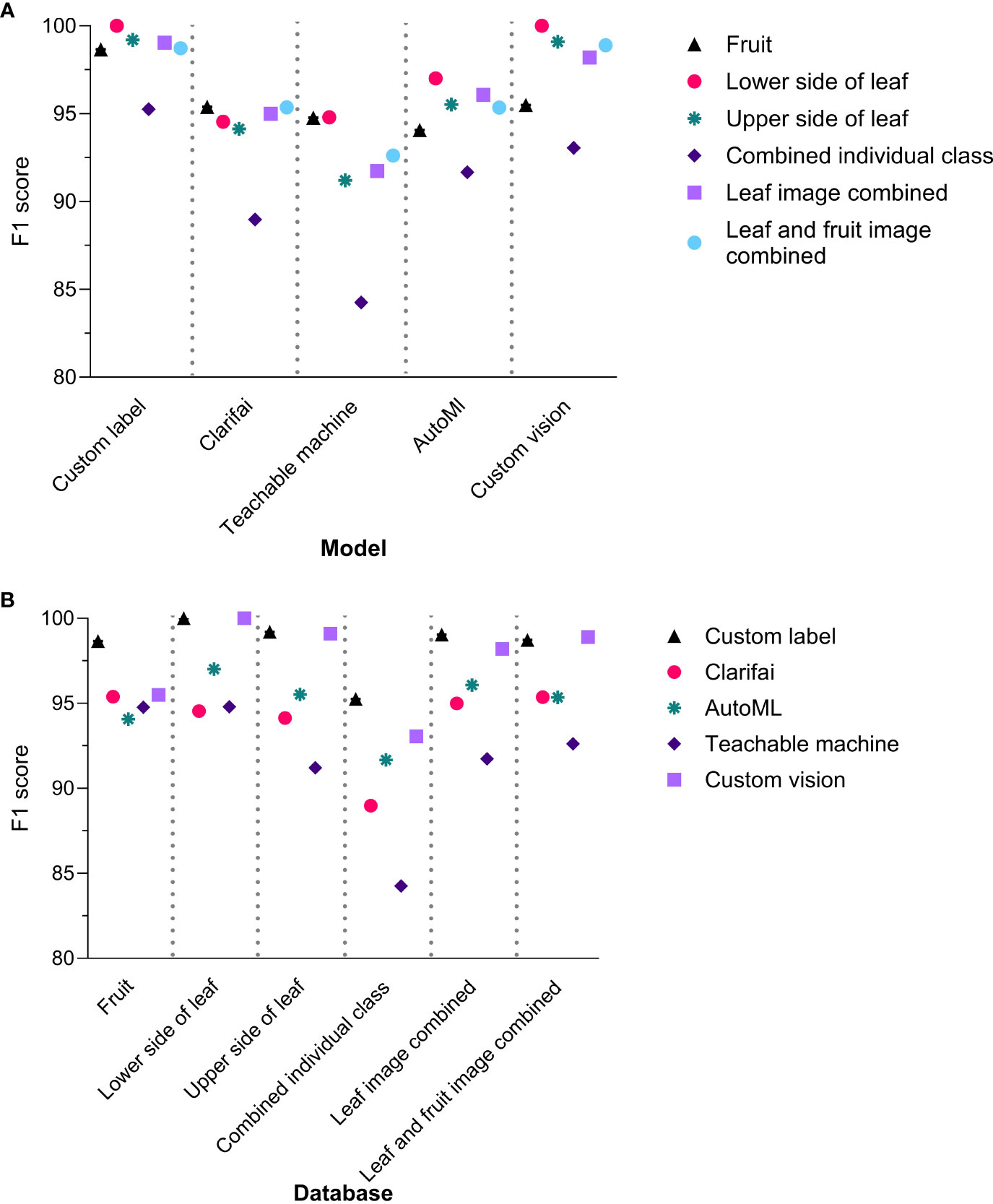
Figure 1 F1 scores of the non-coding deep learning (NCDL) models grouped by (A) models and (B) database.
Accuracy (S.D.) denotes the percentage of images identified correctly and were as follows: Custom Label, 99.8% (0.2); Clarifai, 98.7% (0.5); Teachable Machine, 98.3% (0.4); AutoML, 98.9% (0.6); and CreateML, 87.3% (4.3) (F(4, 25) = 42.29, P = 8.412e−11) (Supplementary Table 5). CreateML had a significantly lower accuracy among all the NCDL platform models tested with the following mean difference (95% CI): Custom Label 12.4 (9.1–15.7), Clarifai 11.4 (8.0–14.6), Teachable Machine 10.9 (7.7–14.3), and AutoML 11.6 (8.2–14.85) (Supplementary Table 6).
Among the different image database–model pairs, F1 scores were as follows: fruit, 95.7 (1.6); lower side of the leaf, 97.3 (2.7); upper side of the leaf, 95.8 (3.4); combined individual class, 90.6 (4.2); leaf images combined, 96.0 (2.9); and leaf and fruit images combined, 96.2 (2.6) (Figure 1B). A database-wide comparison resulted in only the lower side of the leaf image showing a higher F1 score than the combined individual class database (F(5, 24) = 2.79, P = 0.0316) (Supplementary Table 7).
Evaluation based on platform type
CreateML and Custom Vision were not used for all comparisons, as their web-based interface did not provide the number of true positive, false positive, false negative, and true negative that are required for precision, recall, NPV, specificity, accuracy, and F1 value calculations. Clarifai provides individual classes and overall and ROC curve (ROC AUC), but no other model provides a similar output in their interface, therefore was not used for comparison.
Individual classes
In general, all NCDL platform models had very good disease image classification outputs in all the databases (Table 3). The fruit image database performance across five classes was as follows: F1 score (precision, NPV, recall, specificity): Custom Label, 98.7 (98.7%, 99.7%, 98.7%, 99.7%); Clarifai, 95.4 (95.4%, 98.8%, 95.4%, 98.8%); Teachable Machine, 94.8 (94.8%, 98.7%, 94.8%, 98.7%); AutoML, 94.1 (94.1%, 98.5%, 94.1%, 98.5%); and Custom Vision, 95.5 (95.9%, na, 95.1%, na). All NCDL platforms except Clarifai had numerically higher performance with the lower side of the leaf database than the upper side of the leaf database in the respective models. The Clarifai model had a lower accuracy of 98.2% on the lower side of the leaf database compared with 98.5% on the upper side of the leaf database.
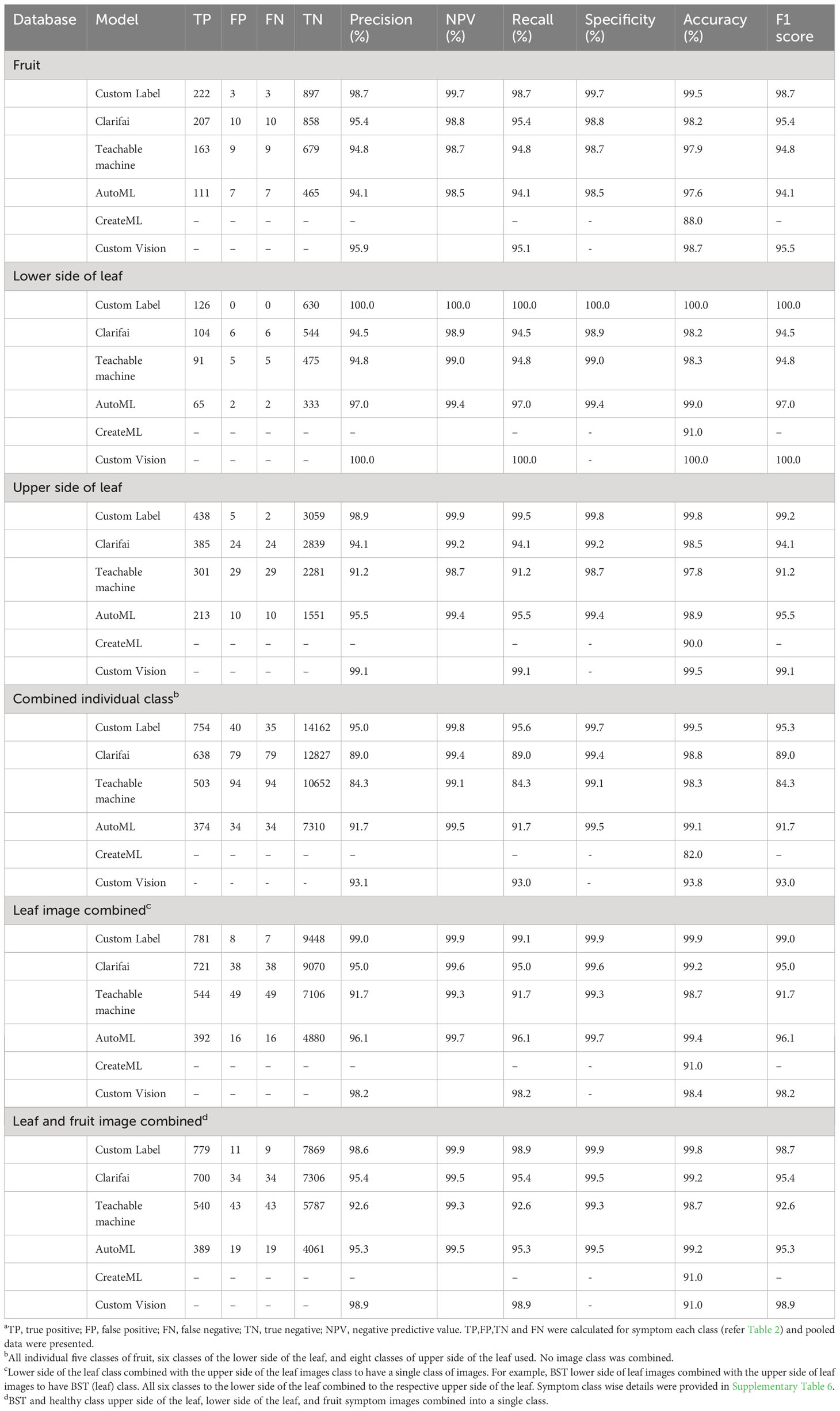
Table 3 Comparison of the model–database classification performance matrices for non-coding deep learning (NCDL) platformsa.
Combined classes
The NCDL platform models trained on a diverse combined individual class database had relatively lower model-all classes classification performance with F1 score (precision, NPV, recall, specificity) as follows: Custom Label, 95.3 (95.0%, 99.8%, 95.6%, 99.7%); Clarifai, 89.0 (89.0%, 99.4%, 89.0%, 99.4%); Teachable Machine, 84.3 (84.3%, 99.1%, 84.3%, 99.1%); AutoML, 91.7 (91.7%, 99.5%, 91.7%, 99.4%); and Custom Vision, 93.0 (93.1%, na, 93.0%, na) (Table 3, Supplementary Table 8). For example, for bacterial spot (BST), out of 91 test set images from the upper side of the leaf, 73 images were classified correctly, while 16 images were classified as false negative for the lower side of BST symptoms, and two false negatives were identified as early blight symptom in the Clarifai model (Supplementary Table 9).
Classification matrices improved when the upper and lower sides of the leaf images were combined into one class comparison to all combined individual classes database (Table 3). The Custom Label F1 score increased from 95.3 in the combined individual class to 99.0. The F1 score of the lowest performing model, the Teachable Machine, increased from 84.3 to 91.7. In most of the individual classes, the F1 score was above 90, except for the fruit pox symptom (Supplementary Table 8). Pox had an F1 score of 90.6, 70.6, 78.9, 76.2, 69.5, and 79.2 with Custom Label, Clarifai, Teachable Machine, AutoML, CreateML, and Custom Vision, respectively (Supplementary Table 8). Other classes with low F1 score in CreateML were little leaf (84.7), BST on leaf (83.4), and 2-4-D (2,4-dichlorophenoxyacetic acid) drift (88.8) symptoms.
To explore further, symptoms of the same disease on fruit and leaf symptom image classes were combined into one class (BST and healthy class fruit and leaf images combined). Overall, this image database had no impact on either accuracy or F1 score (Figures 1, 2). The F1 score of the leaf and fruit image combined database of Custom Label (99.0 to 98.7) and AutoML (96.1 to 95.3) dropped slightly, while Clarifai (95.0 to 95.4), Teachable Machine (91.7 to 92.6), and Custom Vision (98.2 to 98.9) increased slightly compared with the leaf images combined database.
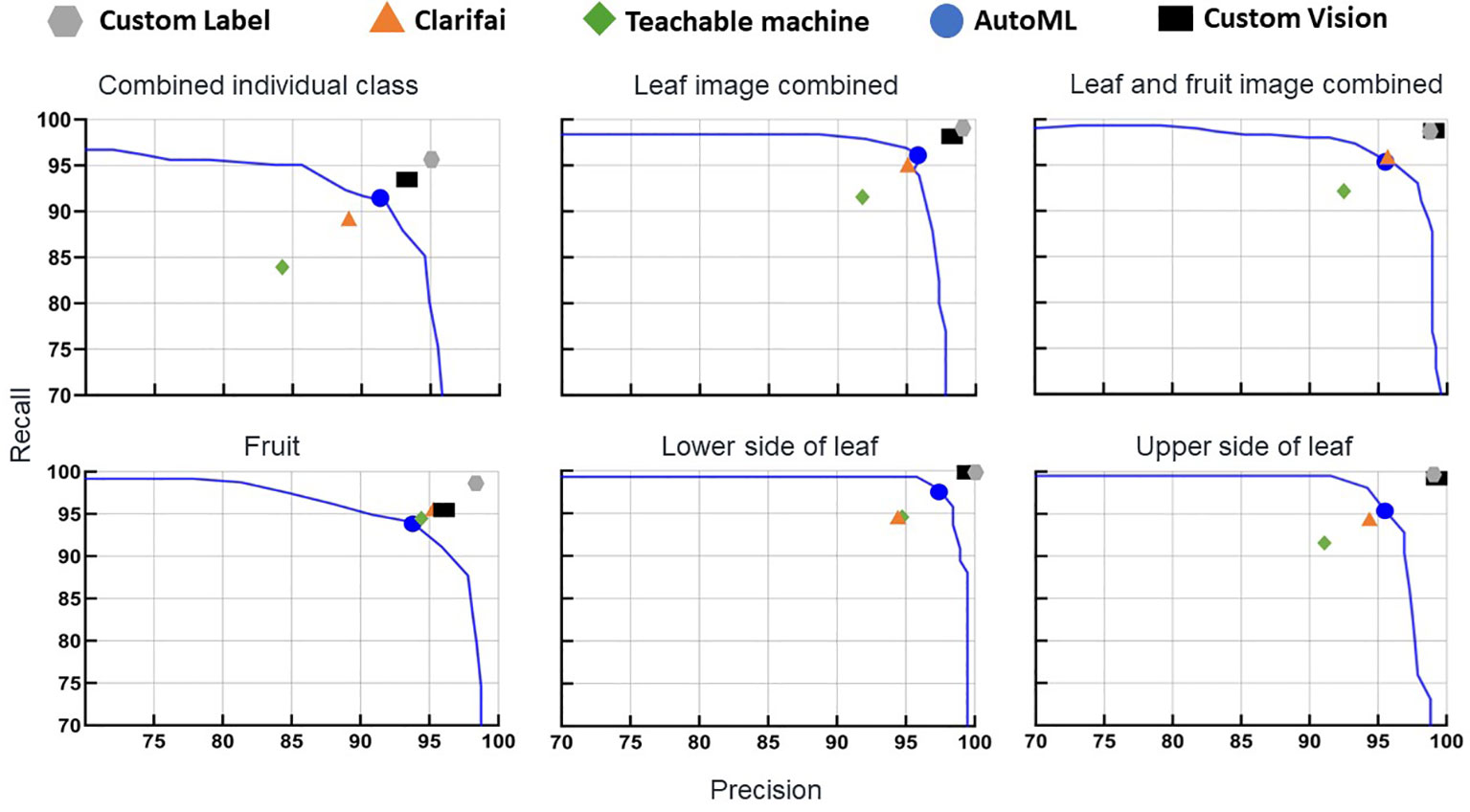
Figure 2 Precision and recall of the non-coding deep learning (NCDL) platform models, with plots grouped by image database. Each point is an individual model’s precision and recall at the default threshold, plotted against the AutoML platform precision–recall curves.
External validation of data
External validation of the data was conducted for images collected using ×30 microscopic images by another independent user (Sentil et al., unpublished) using an iOS-based mobile phone on the leaf image combined database models for three NCDLs (i.e., Teachable Machine, Custom Vision, and AutoML). Clarifai and Custom Label do not have an option of testing without deployment of the model and, therefore, were not used. Teachable Machine and Custom Vision only allow testing of individual images, while batch prediction was done with the AutoML model. From a random selection of 200 images, classification performance F1 scores (precision, NPV, recall, specificity) were as follows: Teachable Machine, 76.3 (76.3%, 95.3%, 76.3%, 95.3%); AutoML, 86.6 (86.6%, 97.3%, 86.6%, 97.3%); and Custom Vision, 88.3 (88.3%, 97.7%, 88.3%, 99.7%) (Figure 3, Supplementary Table 10). The overall accuracy of CreateML in testing data with 50 iterations was 69% (data not shown).
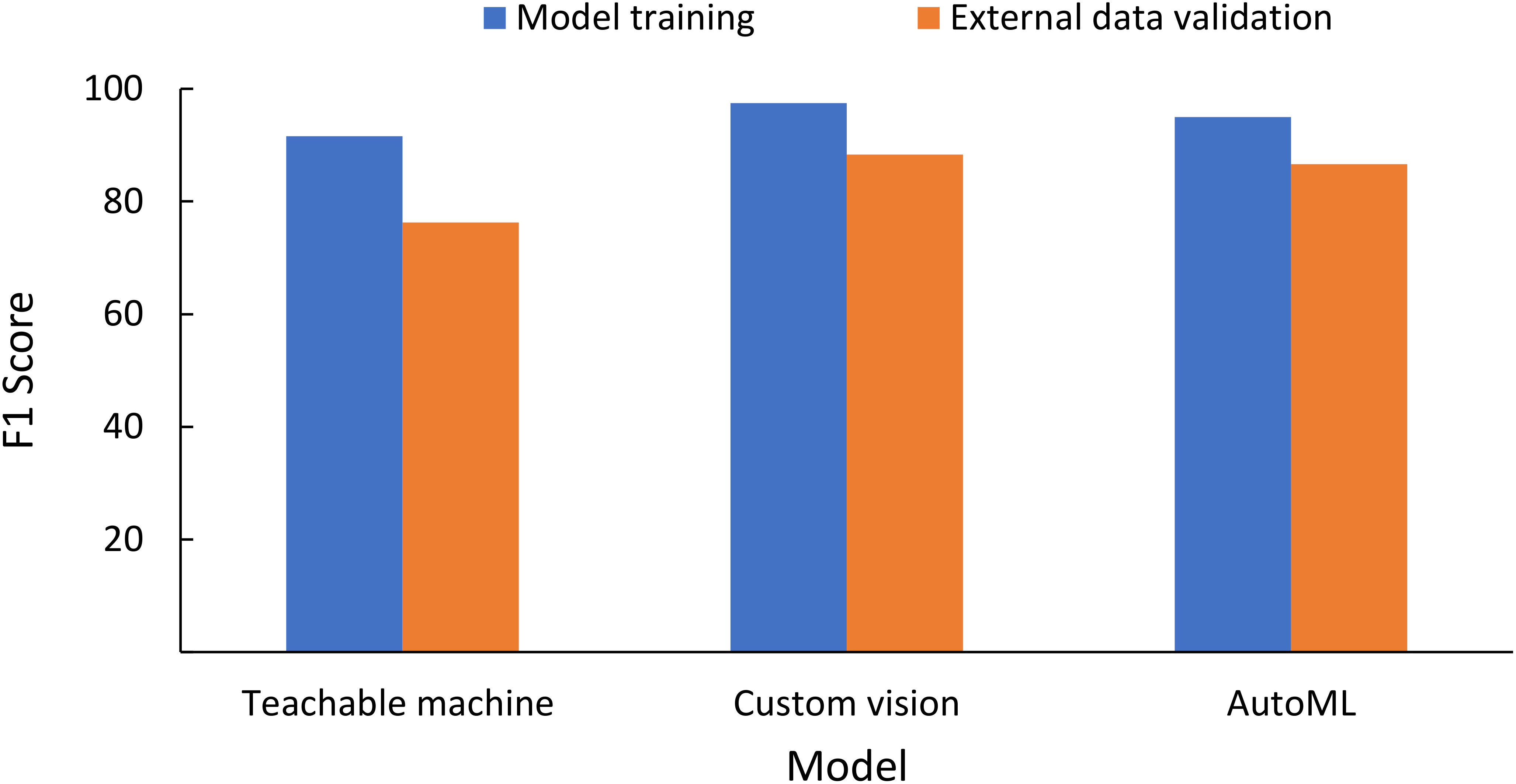
Figure 3 Testing classification matrices of the non-coding deep learning (NCDL) platform models on the external image database (image collected by an independent user and not used in training, testing and validation of the models) using the leaf image combined database model.
Repeatability of model
The leaf image combined database was trained on five models three times. The F1 scores (SD) of the models were 89.9 (0.2) for Custom Label, 95.2 (0.4) for Clarifai, 91.4 (3.6) for Teachable Machine, 96.2 (1.2) for AutoML, and 98.6 (0.6) for Custom Vision. The standard deviation was low except for the Teachable Machine, indicating the repeatability of model classification performance.
Model features
The use of NCDL platforms for plant disease classification is a relatively new area, so each model was examined for various user features from a plant pathologist’s point of view (Table 1). Each model has its own set of advantages and technical specifications that need to be worked out. Teachable Machine has the simplest graphic user inference that allows three changeable parameter epochs, batch size, and learning rate. Teachable Machine is the only model in which data need to be uploaded from a local computer or Google Drive, each time the model is trained. The Teachable Machine and CreateML platforms do not require a user account. CreateML models run on a local computer and allow image augmentation like blur, focus, flip, and rotate, which none of the other models have. Modification of the classification threshold allows the user to train models for higher and lower confidence with each classification. Clarifai, Custom Vision, and AutoML allow tweaking of the threshold values. In Custom Label, each class had a different threshold for disease classification at a system-chosen threshold, but this cannot be selected by the user. After uploading, images need to be manually class-wise-labeled in all models. Only AutoML allows data upload in CSV format which makes the process faster than the other models. All NCDL platforms except CreateML automatically separate the testing and training datasets. CreateML requires preclassified testing and training datasets.
CreateML and Teachable Machine were free without any restrictions in use. Clarifai allows 1,000 operations per month in the community tier; after this, monthly charges are required. There were no mentioned criteria for operation, but in this study, we did not run out of free tier quota. Custom Vision has a free limit of 5,000 training images per project and a maximum of 2 projects with a speed of 2 transactions per second (TPS). After free usage, it charges computing hours. The Custom Label free tier allows 10 training hours and 4 interface hours after, hourly training, and interface charges. Custom Label also provides up to 5 GB of image storage in the free tier. AutoML provides initial credit in the free tier, and it charges per node per hour basis for training. Each model has other associated costs for deployment and prediction, which were not used in this study. The use of all the NCDL platform models comprised of three steps: upload and labeling of data, training, and output visualization. Based upon these parameters, Custom Label, AutoML, and Custom Vision models look promising for future uses.
Discussion
There are many models already available in the web domain, but issues persist with their practical use and field-based validation. Siddiqua et al. (2022) evaluated 17 different mobile applications for disease classification and noted that only “Plantix–your crop doctor” has the capability of disease identification to recommend a suitable management option to users (CGIAR, 2017, https://plantix.net/en/). Some of these models are available for specific crops like “Cassava Plant Disease Identify” for cassava disease and “Tumaini” for banana disease (Selvaraj et al., 2019). However, these mobile application models cannot be customized by local pathologists or extension agents on a local database of choice to improve disease classification (Siddiqua et al., 2022). Microscopic images used in this study have a magnified view of disease symptoms that captures minute characteristics in symptoms than images taken by a normal camera (macroscopic images). Most macroscopic images have the noise of background features in addition to the disease symptoms that make feature extraction more complicated and, thus, reduce accuracy (Lu et al., 2021; Tugrul et al., 2022).
In this study, we evaluated models and multiple combinations of databases using the same number of identical training images using freely available resources in the NCDL platform. Most models (Custom Label, Custom Vision, and AutoML) have similar higher classification performance. Teachable Machine was the least performing model among all web-based cloud computing models tested. If we include locally run models also, then CreateML had significantly lower classification accuracy among all the tested models. Similar findings were also noted when using ophthalmology images with multimodality databases on the CreateML platform (Korot et al., 2021). The exact reason why these two models have poorer performance compared with others for disease classification is unknown as most platforms neither provide information on their configuration and nor allow users to change parameters. Teachable Machine which was built using Tensorflow.js works mostly on transfer learning approaches (Teachable machine, 2022) and may have computational limits or may exclude some image data. We used different hyperparameters like epoch and learning rate for Teachable Machine and number of iterations, blur, and focus for CreateML to improve the performance, but it was not significantly improved compared with the default setting (data now shown). As both models (Teachable Machine and CreateML) were freely available without any limit, it is possible that the developers were not routinely improve these models.
The average F1 score of NCDL models ranged from 91.6 to 98.5, while accuracy ranged from 87.3% to 99.8%. Earlier researchers used mostly macroscopic image databases from the PlantVillage database for plant disease classification using the CNN model. Most of the earlier models (AlexNet, GoogLeNet, ResNet, MobiNet, R-CNN, D-CNN) were trained on the same set of images with different augmentations from scratch, or the transfer learning approach has classification matrices from 76% to 99.8% (Abbas et al., 2021; Tugrul et al., 2022). Very few models were tested using external images, and the ones that were tested had a disease classification performance of below 50% (Mohanty et al., 2016; Ferentinos, 2018). Performance reduction on external validation is a major obstacle to the practical application of deep learning models. In our study, only five NCDL models were capable of external validation of the model, in which the F1 score of AutoML and Custom Vision was greater than 86, while Teachable Machine had an F1 score of 76 on a leaf image combined dataset. For the NCDL models, accuracy was reduced by a maximum of 7% in the case of Teachable Machine (the lowest performing model in this study) in external data testing. This indicates that the NCDL platform models were not overfitting or leaking data during the training of the models. In the medical field, models used with eye images have a similar F1 score of as high as 93.9 to the Custom Label models (Korot et al., 2021).
Most leaf and fruit databases had similar classification performance. Only the lower side of the leaf had statistically higher classification performance than the “all individual class” database. This may be due to two reasons: 1) when individual classes were combined, the same type of symptom from the upper and lower side of the leaf images had more false positives and negatives than other databases. The bacterial spot of tomato (BST) upper side of the leaf had 18% (16 out 91) false-positive images as lower side of the leaf. This may be due to early disease symptoms being observed first on the lower side of the leaf and expanding to the upper side of the leaf (Osdaghi et al., 2021). 2) It has also been observed that the lower side of the leaf has more characteristic early and uniform symptoms than other plant parts (Schor et al., 2016). In our study, tomato fruit pox, a genetic disorder (Crill et al., 1973) that produces an incipient oval-shaped lesion, had the lowest classification performance in all databases. Pox was misclassified with fruit BST symptoms and in some cases of healthy samples. This exemplifies issues with the background of images that can reduce classification accuracy. This type of Image-based classification can be further improved using a convex lens that can take the curvature of the plant stem and fruit into consideration and reduce the blurriness of the image.
In terms of the evaluation performance, the top three models (Custom Label, AutoML, and Custom Vision) had no significant differences. Most of the NCDL platforms took less than 1 h for the training of the models. CreateML was the fastest, taking<5 min to create the models. Other models (Custom Label, AutoML, and Custom Vision) took approximately 10 h with the largest dataset. Most models are freely available; however, these models are free only up to certain levels which can be quickly drained. In our analysis, we ran out of free usage in all models except Clarifai. The cost associated with these models is not cheap: Custom Vision charges as high as $10/h/node, which is comparatively costly for a developing nation model developer. Although cloud-based NCDL platforms are easy to use and infinity-scalable, plant pathologists must think about cost requirements compared with the already available traditional methods or outsourcing using machine experts with local resources.
Limitations with microscopic image collection and models
Even though image collection is quite simple with a microscopic lens, there are no public databases available as of now, and it will take time to build a database of the nature used in this study. A database of such nature that is being built at the University of Florida for tomato, cucurbits, and pepper diseases currently has >30,000 images and is expected to be available for public use by next year (Paret, personal communication). Quality images are necessary for the successful implementation of any AI model. In a recent survey, 96% of the respondents (scientists, AI experts) faced challenges with quality data for the training of ML algorithms (Research Dimensional, 2019). Given that microscopic images provide a magnified view, some diseases like early blight of tomato symptoms may not be captured in a single image, which would increase the number of images to be taken of each class. In diseases like tomato Fusarium wilt, which shows dropping of the leaves and yellowing symptoms, whole-plant images in addition to microscopic images may be necessary. In the ideal scenario, a combined approach would be to use both magnified and whole-plant images. However, the machine learning process toward integrating macro- and microscopic images first needs to be established. Another aspect for consideration is the different shapes of the fruits that make it difficult to capture images. The convex shape of tomato fruit may lead to distorted and blurred images. A flexible outing cover for the lens reduces blur due to the shape of the leaf and fruit and facilitates easy adjustment of the camera/lens.
Threshold is an important parameter to have classification with more confidence (Ferri et al., 2019), but the high-performing model such as —Custom Label—did allow modifying this value. Most models do not have an option to change any hyperparameter that makes these models opaque. We do not have information on what kind of parameters and underlying architecture was used by NCDL platform models that reduce the option for improvement in classification performance (Liang et al., 2022). In external evaluation, cloud-based models were lagging a lot. Custom Label and Clarifai do not have the option for external validation before deployment. Teachable Machine and Custom Vision allow individual images for testing that make testing cumbersome and time-consuming. AutoML allows batch processing, but preparing data again requires a laborious process.
Potential of microscope image coupled NCDL platform
Globally, 95% of the population has mobile network coverage that makes mobile use accessible everywhere (ITU, 2021). In 2021, 5.34 billion people had smartphones which increased by 73% within 5 years (2016–2021) (DataReportal, 2022). This study demonstrates the potential use of mobile phones integrated with deep learning models for plant disease classification. Currently, a major challenge in agriculture in remote places of the world is real-time advisory to endpoint stakeholders. Various down-term applications can be easily integrated with the NCDL platform. Trained models can be utilized as a research tool, deployed on new databases or on prospectively curated data, to collate images that fit a criterion. These potential use cases are not comprehensive, and more will be revealed as agriculture researchers gain an understanding of ML principles through the exploration of NCDL platforms.
AI progression is similar to what has occurred in other technologies (sequencing, mobile, etc.) first by a small core group of scientists, later by experts that navigate the technical nuance of new development, and finally by the general public. Slowly, AI is coming to the domain of “citizen science” where it can be used to reap benefits (Craig, 2022). We explored only a limited number of publicly available NCDL platforms. In coming times, advancement in NCDL platforms and more use of these platforms likely will persuade other big AI companies to improve these models. For now, though, most no-code AI users are business professionals who want to streamline the way things are done without having to involve a programmer.
Conclusion
We evaluated the classification matrices and usability of six NCDL platforms using microscopic-magnified ×30 images captured using a mobile phone camera-mounted lens. The outcomes from the study demonstrate that the NCDL platform-developed models improved disease classification. Our finding illustrated that plant disease classification using microscopic-magnified images in combination with NCDL models can be used by any plant pathologist and diagnostician for the recommendation of disease management measures to farmers. This study demonstrates the potential use of mobile phones integrated with deep learning models in remote corners of the world without the help of computer personnel. The methods used in this work can speed up disease identification and save a lot of resources (chemicals, lab space, and personnel). The NCDL platforms have a broad scope for agriculture, in which it can potentially be used for understanding nutrient limitations, weed identification, and insect diagnosis. Currently, a major challenge in agriculture in remote places of the world is real-time advisory to endpoint stakeholders. This technology should improve real-time advisory capabilities in remote places.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://github.com/manoj044/Tomato_microscopic_images.git.
Author contributions
MC: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. SS: Investigation, Writing – review & editing. JJ: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing, Project administration. MP: Conceptualization, Funding acquisition, Investigation, Methodology, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by University of Florida Informatics Institute seed funding. Manoj Choudhary acknowledges the Fellowship awarded through the Netaji Subhas - Indian Council of Agricultural Research International Fellowship for his doctoral studies.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1292643/full#supplementary-material
References
Abbas, A., Jain, S., Gour, M., Vankudothu, S. (2021). Tomato plant disease detection using transfer learning with C-GAN synthetic images. Comput. Electron Agric. 187, 106279. doi: 10.1016/J.COMPAG.2021.106279
Amara, J., Bouaziz, B., Algergawy, A. (2017). A deep learning-based approach for banana leaf diseases classification. in. Lecture Notes Inf. (LNI) Gesellschaft für Informatik 79–89.
Brahimi, M., Boukhalfa, K., Moussaoui, A. (2017). Deep learning for tomato diseases: classification and symptoms visualization. Appl. Artif. Intell. 31, 299–315. doi: 10.1080/08839514.2017.1315516
CGAIR (2017) Plant disease diagnosis using artificial intelligence: a case study on Plantix - CGIAR Platform for Big Data in Agriculture. Available at: https://bigdata.cgiar.org/digital-intervention/plant-disease-diagnosis-using-artificial-intelligence-a-case-study-on-plantix/ (Accessed October 23, 2022).
Chen, X. (2020). Pathogens which threaten food security: ,Puccinia striiformis, the wheat stripe rust pathogen. Food Secur 12, 239–251. doi: 10.1007/s12571-020-01016-z
Chong, H. M., Yap, X. Y., Chia, K. S. (2023). Effects of different pretrained deep learning algorithms as feature extractor in tomato plant health classification. Pattern Recognition Image Anal. 33, 39–46. doi: 10.1134/S1054661823010017/FIGURES/4
Craig, S.S. (2022) No-code’ Brings the power of A.I. to the masses. The New York times. Available at: https://www.nytimes.com/2022/03/15/technology/ai-no-code.html (Accessed September 14, 2022).
Crill, P., Jones, J. P., Strobel, J. W. (1973). The fruit pox and gold fleck syndromes of tomato. Phytopathology 63, 1285. doi: 10.1094/Phyto-63-1285
Darwish, A., Ezzat, D., Hassanien, A. E. (2020). An optimized model based on convolutional neural networks and orthogonal learning particle swarm optimization algorithm for plant diseases diagnosis. Swarm Evol. Comput. 52, 100616. doi: 10.1016/j.swevo.2019.100616
DataReportal (2022) Digital around the world — DataReportal – global digital insights. Available at: https://datareportal.com/global-digital-overview (Accessed September 14, 2022).
FAO (2020) International Year of Plant Health 2020 (FAO | Food and Agriculture Organization of the United Nations). Available at: https://www.fao.org/plant-health-2020/about/en/ (Accessed September 13, 2022).
FAO (2022) FAOSTAT. Available at: http://www.fao.org/faostat/en/#data/RP (Accessed September 6, 2023).
Ferentinos, K. P. (2018). Deep learning models for plant disease detection and diagnosis. Comput. Electron Agric. 145, 311–318. doi: 10.1016/j.compag.2018.01.009
Ferri, C., Hernández-Orallo, J., Flach, P. (2019). Setting decision thresholds when operating conditions are uncertain. Data Min Knowl. Discovery 33, 805–847. doi: 10.1007/s10618-019-00613-7
Fry, W. E., McGrath, M. T., Seaman, A., Zitter, T. A., McLeod, A., Danies, G., et al. (2013). The 2009 late blight pandemic in the eastern United States - Causes and results. Plant Dis. 97, 296–306. doi: 10.1094/PDIS-08-12-0791-FE
Gwagwa, A., Kazim, E., Kachidza, P., Hilliard, A., Siminyu, K., Smith, M., et al. (2021). Road map for research on responsible artificial intelligence for development (AI4D) in African countries: The case study of agriculture. Patterns 2, 100381. doi: 10.1016/j.patter.2021.100381
ITU (2021). Measuring digital development - Facts and figures 2021. https://www.itu.int/itu-d/reports/statistics/facts-figures-2021/.
Jones, J. B., Zitter, T. A., Momol, M. T., Miller, S. A. (2014). Compendium of Tomato Diseases and Pests. 2nd ed. Eds. Jones, J. B., Zitter, T. A., Momol, M. T., Miller, S. A. (St. Paul,MN, USA: The American Phytopathological Society).
Kanti Karmaker, S., Hassan, M., Smith, M. J., Mahadi Hassan, M., Xu, L., Zhai, C., et al. (2021). AutoML to date and beyond: challenges and opportunities. ACM Comput. Surv 54, 175. doi: 10.1145/3470918
Korot, E., Guan, Z., Ferraz, D., Wagner, S. K., Zhang, G., Liu, X., et al. (2021). Code-free deep learning for multi-modality medical image classification. Nat. Mach. Intell. 3, 288–298. doi: 10.1038/s42256-021-00305-2
Lecun, Y., Bengio, Y., Hinton, G. (2015). Deep learning. Nature 521, 436–444. doi: 10.1038/nature14539
Lee, S. H., Goëau, H., Bonnet, P., Joly, A. (2020). New perspectives on plant disease characterization based on deep learning. Comput. Electron Agric. 170, 105220. doi: 10.1016/j.compag.2020.105220
Liang, W., Tadesse, G. A., Ho, D., Li, F. F., Zaharia, M., Zhang, C., et al. (2022). Advances, challenges and opportunities in creating data for trustworthy AI. Nat. Mach. Intell. 4, 669–677. doi: 10.1038/s42256-022-00516-1
Liu, J., Wang, X. (2020). Tomato diseases and pests detection based on improved yolo V3 convolutional neural network. Front. Plant Sci. 11. doi: 10.3389/FPLS.2020.00898/BIBTEX
Liu, X., Min, W., Mei, S., Wang, L., Jiang, S. (2021). Plant disease recognition: a large-scale benchmark dataset and a visual region and loss reweighting approach. IEEE Trans. Image Process. 30, 2003–2015. doi: 10.1109/TIP.2021.3049334
Lu, J., Tan, L., Jiang, H. (2021). Review on convolutional neural network (CNN) applied to plant leaf disease classification. Agriculture 11, 707. doi: 10.3390/AGRICULTURE11080707
Maeda-Gutiérrez, V., Galván-Tejada, C. E., Zanella-Calzada, L. A., Celaya-Padilla, J. M., Galván-Tejada, J. I., Gamboa-Rosales, H., et al. (2020). Comparison of convolutional neural network architectures for classification of tomato plant diseases. Appl. Sci. 10, 1245. doi: 10.3390/APP10041245
Mohanty, S. P., Hughes, D. P., Salathé, M. (2016). Using deep learning for image-based plant disease detection. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01419
Osdaghi, E., Jones, J. B., Sharma, A., Goss, E. M., Abrahamian, P., Newberry, E. A., et al. (2021). A centenary for bacterial spot of tomato and pepper. Mol. Plant Pathol. 22, 1500–1519. doi: 10.1111/mpp.13125
Picon, A., Seitz, M., Alvarez-Gila, A., Mohnke, P., Ortiz-Barredo, A., Echazarra, J. (2019). Crop conditional Convolutional Neural Networks for massive multi-crop plant disease classification over cell phone acquired images taken on real field conditions. Comput. Electron Agric. 167, 105093. doi: 10.1016/j.compag.2019.105093
Research Dimensional. (2019). Artificial intelligence and machine learning projects are obstructed by data issues. Available at: https://cdn2.hubspot.net/hubfs/3971219/Survey%20Assets%201905/Dimensional%20Research%20Machine%20Learning%20PPT%20Report%20FINAL.pdf. (Accessed September 14, 2022).
Riley, M., Williamson, M., Maloy, O. (2002). Plant disease diagnosis. Plant Health Instructor. doi: 10.1094/PHI-I-2002-1021-01
Schor, N., Bechar, A., Ignat, T., Dombrovsky, A., Elad, Y., Berman, S. (2016). Robotic disease detection in greenhouses: combined detection of powdery mildew and tomato spotted wilt virus. IEEE Robot Autom Lett. 1, 354–360. doi: 10.1109/LRA.2016.2518214
Selvaraj, M. G., Vergara, A., Ruiz, H., Safari, N., Elayabalan, S., Ocimati, W., et al. (2019). AI-powered banana diseases and pest detection. Plant Methods 15, 1–11. doi: 10.1186/S13007-019-0475-Z/FIGURES/6
Siddiqua, A., Kabir, M. A., Ferdous, T., Ali, I. B., Weston, L. A. (2022). Evaluating plant disease detection mobile applications: quality and limitations. Agronomy 12, 1869. doi: 10.3390/agronomy12081869
Sladojevic, S., Arsenovic, M., Anderla, A., Culibrk, D., Stefanovic, D. (2016). Deep neural networks based recognition of plant diseases by leaf image classification. Comput. Intell. Neurosci. 2016. doi: 10.1155/2016/3289801
Split-folders:PyPI (2022). Available at: https://pypi.org/project/split-folders/ (Accessed September 15, 2022).
Teachable machine (2022) GitHub - googlecreativelab/teachablemachine-community: Example code snippets and machine learning code for Teachable Machine. Available at: https://github.com/googlecreativelab/teachablemachine-community (Accessed September 14, 2022).
Touma, S., Antaki, F., Duval, R. (2022). Development of a code-free machine learning model for the classification of cataract surgery phases. Sci. Rep. 12, 1–7. doi: 10.1038/s41598-022-06127-5
Tran, T. T., Choi, J. W., Le, T. T. H., Kim, J. W. (2019). A comparative study of deep CNN in forecasting and classifying the macronutrient deficiencies on development of tomato plant. Appl. Sci. 9, 1601. doi: 10.3390/APP9081601
Tugrul, B., Elfatimi, E., Eryigit, R. (2022). Convolutional neural networks in detection of plant leaf diseases: A review. Agriculture 12, 1192. doi: 10.3390/agriculture12081192
Keywords: diseases, code-free models, machine learning, tomato, deep learning, biotic stress, abiotic stress, microscopic images
Citation: Choudhary M, Sentil S, Jones JB and Paret ML (2023) Non-coding deep learning models for tomato biotic and abiotic stress classification using microscopic images. Front. Plant Sci. 14:1292643. doi: 10.3389/fpls.2023.1292643
Received: 11 September 2023; Accepted: 24 November 2023;
Published: 08 January 2023.
Edited by:
Mario Cunha, University of Porto, PortugalReviewed by:
Michail D. Kaminiaris, Agricultural University of Athens, GreeceSuneet Kr Gupta, Bennett University, India
Peng Chen, Anhui University, China
Copyright © 2023 Choudhary, Sentil, Jones and Paret. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mathews L. Paret, cGFyZXRAdWZsLmVkdQ==
 Manoj Choudhary
Manoj Choudhary Sruthi Sentil1
Sruthi Sentil1 Jeffrey B. Jones
Jeffrey B. Jones Mathews L. Paret
Mathews L. Paret