- Shandong Provincial Key Laboratory of Plant Stress, College of Life Sciences, Shandong Normal University, Ji’nan, Shandong, China
In recent years, numerous genes that encode proteins with specific domains that participate in different biological processes or have different molecular functions have been identified. A class of genes with typical domains whose function has rarely been identified and another type of genes with no typical domains have attracted increasing attentions. As many of these so-called as unknown/uncharacterized (U/U) genes are involved in important processes, such as plant growth and plant stress resistance, there is much interest in deciphering their molecular roles. Here, we summarize our current understanding of these genes, including their structures, classifications, and roles in plant growth and stress resistance, summarize progress in the methods used to decipher the roles of these genes, and provide new research perspectives. Unveiling the molecular functions of unknown/uncharacterized genes may suggest strategies to fine-tune important physiological processes in plants, which will enrich the functional network system of plants and provide more possibilities for adaptive improvement of plants.
Introduction
The development of modern molecular biology tools has accelerated the discovery of genes involved in various biological processes. Many genes have known functions in regulating various physiological processes and mechanisms in plants, such as vegetative growth that the overexpressing of Lb1G04899 from Limonium bicolor improved the salt tolerance of transgenic Arabidopsis under NaCl environment (Liu et al., 2022a; Wang et al., 2022b); flowering time that CYLIN - DEPENDENT KINASE G2 (CDKG2) gene affected flowering time in Arabidopsis (Ma and Chen, 2007; Ma et al., 2015; Zhou et al., 2019); changes in phytohormone status that the NHL family genes of wild soybeans can regulate ABA and MeJA, laying the foundation for potential roles in signal transduction mechanisms (Xu et al., 2020; Zhang et al., 2022b; Zhao et al., 2023a); anther and pollen development that BcMF19 inhibited pollen development in Chinese cabbage-pak-choi (Huang et al., 2011) and resistance to harsh environments including drought that TaDTG6-BDel574 regulates the transcription of TaPIF1 to enhance drought resistance in wheat (Mei et al., 2022); salt that CycC1 controlled salt tolerance in Arabidopsis by regulating transcriptional regulation of SOS1 (Lu et al., 2023; Ma et al., 2023); diseases that adult-plant resistance (APR) genes played roles in inhibiting the occurrence of wheat rust (Dinglasan et al., 2022) and insect pests that three genes (Cry1Ac-Cry2Ab-EPSPS) in cotton have resistance to lepidopteran insect (Siddiqui et al., 2022). Based on conserved structural domains in their encoded proteins, various gene families are known to regulate different physiological processes, including development, reproduction, and environmental adaptation. For example, members of the SWEET family (containing an MtN3/saliva transmembrane domain) promote ion and sugar transport (Guan et al., 2008; Chen et al., 2010; Slewinski, 2011; Fang et al., 2022; Liu et al., 2022c); members of the WRKY family (containing a WRKY domain) participate in plant defense and aging processes (Silke and Somssich, 2001; Miao et al., 2004; Besseau et al., 2012); and members of the MYB family (containing an MYB domain) are widely involved in development and stress responses.
Genes with established functions are annotated based on the domains in their encoded proteins (defined as structural annotation) and their functions are verified by deletion or overexpression analysis (functional annotation). Genome annotation is primarily based on gene structure, that is, the boundaries of exons/introns and CDS (coding sequences)/UTRs (untranslated regions), at protein-coding loci (Zhang et al., 2022). With the advent of high-throughput sequencing technology, numerous genes have been sequenced and found to encode proteins with unknown/uncharacterized domains. The localizations of these proteins are also uncertain based on structural annotation. Thus, these genes are defined as unknown/uncharacterized (U/U) genes. Although the biological functions of proteins encoded by U/U genes are unclear, these genes occupy a large proportion of genes reported to date (Imtiaz, 2022).
Do U/U genes matter? More and more of these genes have been shown to play important roles in plants, such as controlling growth and development (Wang et al., 2020) and stress resistance (Soda et al., 2013; Qi et al., 2023). It is challenging to classify U/U genes. Here, we focus on recent progress in our understanding U/U genes, including their classifications, methods and functions. We also discuss research methods used to further study U/U genes.
Classification of U/U genes
Genes with typical domains whose function has rarely been identified and genes with no typical domains whose roles are uncertain were named unknown/uncharacterized (U/U) genes. Here, we classify U/U genes into two types based on the presence or absence of conserved domains.
One type is genes with domains but functions have not been identified. Many U/U genes have been identified in food crops and uncultivated plants. Many genes of unknown function contain conserved domains, allowing them to be classified into gene families that encode proteins with known functions. The presence of conserved domains helps researchers predict the roles of U/U genes and provides direction for the functional research of unknown genes. Exogenously overexpressing MbMYBC1 and MbMYB108 from Malus baccata enhanced the cold and drought resistance of transgenic Arabidopsis (Arabidopsis thaliana) (Yao et al., 2022; Liu et al., 2023). Exogenously overexpressing FvMYB82 from strawberry (Fragaria vesca) and the R1-MYB transcription factor gene LcMYB1 from sheepgrass (Leymus chinensis (Trin.) Tzvel.) enhanced the salt tolerance of transgenic Arabidopsis (Cheng et al., 2013; Li et al., 2022a). AgMYB5, an unknown gene from celery (Apium graveolens L.), enhanced β-carotene synthesis in transgenic Arabidopsis (Sun et al., 2023)(Figure 1A). Among NAC family transcription factor genes, overexpressing CaNAC46 from pepper (Capsicum annuum) and SlNAC10 from Suaeda liaotungensis enhanced the salt and drought resistance of transgenic Arabidopsis (Ma et al., 2021; Du et al., 2022) (Figure 1B). Among genes in the WRKY transcription factor family, OsWRKY54 is associated with salt tolerance in rice; heterologous expression of VvWRKY28 from grapevine (Vitis vinifera) and PcWRKY11 from Polygonum cuspidatum in Arabidopsis enhanced salt tolerance (Liu et al., 2022b; Wang et al., 2022a) (Figure 1C).
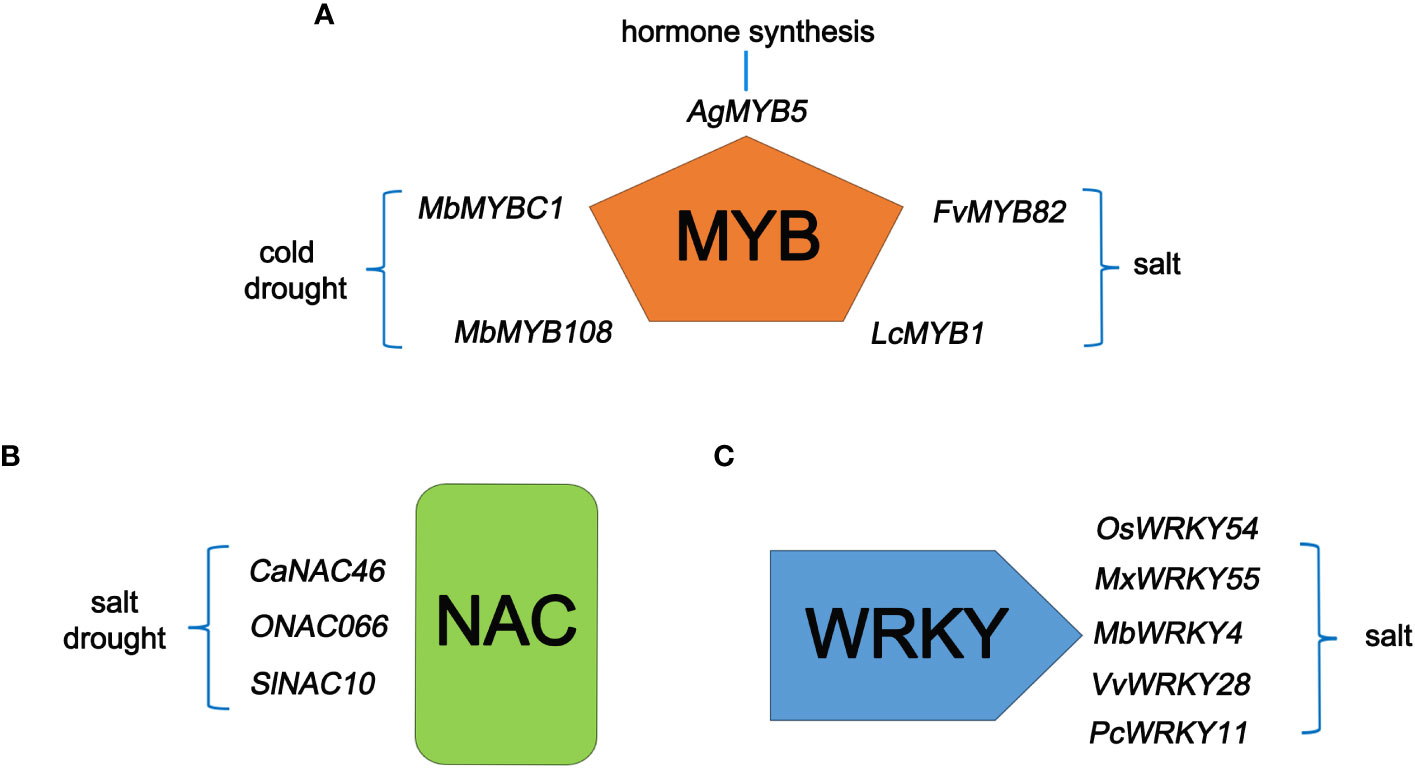
Figure 1 U/U genes with typical conserved domains and their roles in plants. (A) Five newly identified MYB type genes and their related functions. (B) Two newly discovered NAC family genes and their related functions. (C) Three newly isolated WRKY genes and their related functions.
The other type is genes without a domain and with unknown functions. Several unknown genes that lack typical conserved domains also function in plant growth, development, and resistance to stress. The U/U gene Lb1G04202 from the halophyte Limonium bicolor, which lacks known structural domains or special structures, encodes a protein that functions in the nucleus and enhanced NaCl tolerance in Arabidopsis by alleviating osmotic stress. The RNA of Lb1G04202 localizes to the salt gland (a unique salt-secreting structure) of Limonium bicolor, suggesting that this gene plays a role in salt gland development (Wang et al., 2022b). In a word, U/U genes with and without conserved domains play significant roles in plants.
Methods to study U/U genes
U/U genes are almost always discovered in non-model plants, making functional studies quite challenging due to unstable transformation systems. Whole genome sequencing and comparative genomics are essential techniques for performing functional studies of these genes (Geng et al., 2022; Liu and Zhang, 2022; Yuan et al., 2022). U/U genes are always identified by RNA-seq, but their assembled sequences are not always accurate. Therefore, transcriptome sequencing techniques such as Iso-seq that yield full-length reference sequences are recommended in studies examining U/U function (Yuan et al., 2015; Yuan et al., 2016; Jia et al., 2022). Iso-seq can directly obtain complete transcripts without disrupting splicing, in order to accurately analyze structural information such as variable splicing and fusion genes of reference genome species. This technology can also promote the optimization of genome annotation and quantification of transcriptome abundance, providing opportunities for the discovery of new genes (Rhoads and Au, 2015; Li et al., 2017; Beiki et al., 2019; Jia et al., 2022).
Map-based cloning can be used to isolate and clone plant genes and to localize genes on chromosomes (Lee et al., 2019). This technology is particularly suitable for situations where the expression products of genes are unknown, functional information for unknown genes is lacking, or no suitable phenotypes are observed (Jin et al., 2022; Zhan et al., 2023). However, the complete sequence of new genes cannot be fully mastered, which undoubtedly poses difficulties for the full-length cloning and isolation. RACE (rapid amplification of cDNA ends) is an effective method for studying new genes, which based on PCR and RNA reverse transcription. It rapidly amplifies the unknown sequence regions of the 3’ or 5’ ends of cDNA through partial known gene sequences to obtain full-length cDNA (Groot Kormelink and Luyten, 1997; Lindberg et al., 1997; Cheng et al., 2006; Yeku and Frohman, 2011).
Bioinformatics analysis of candidate genes is crucial, as it provides a rough understanding of the possible range of gene action through domain prediction (SMART), hydrophilicity analysis (Expasy-ProtScale) (Dong et al., 2022), transmembrane region display (TMHMM 2.0) (Zhao et al., 2022), and subcellular localization prediction (WoLF PSORT) (Wang et al., 2021; Song et al., 2022). Bioinformatics analysis can lay a solid foundation for further in-depth research of U/U genes (Chen et al., 2022).
RNA interference (RNAi) is an efficient tool for studying the effects of gene deletions (Koeppe et al., 2023; Traber and Yu, 2023). Gene silencing mediated by double-stranded RNA (dsRNA) is widely used to study gene functions in various plants (Akond et al., 2022; Bharathi et al., 2023). Another efficient method to identify gene function is clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated gene knockout (Hu et al., 2023). The phenotypes obtained using these two methods can be compared to phenotypes obtained via overexpression to analyze the biological function of the target gene or protein (Yuan et al., 2022). Of course, it is not sufficient to conduct research solely in the species harboring U/U genes. The transfer of candidate genes into model plants (Arabidopsis) or prokaryotic bacteria is extensively used for further functional research (Leng et al., 2021; Wang et al., 2022b).
The completion of various life functions in plant cells relies on interactions between proteins (Beihammer et al., 2023). Typically, functional proteins combine with other proteins (known or unknown) to form complexes and function in specific pathways (Zhao et al., 2008). Therefore, it is necessary to identify genes that are involved upstream or downstream of the U/U gene of interest and validate the interactions between their encoded proteins. U/U proteins and candidate proteins that may interact with each other identified by screening yeast libraries can be validated by examining in vitro and in vivo interactions using yeast-two hybrid assays (Cao et al., 2022), bimolecular fluorescence complementation (BiFC) (Choi et al., 2022), co-Immunoprecipitation (CoIP), and GST-pulldown (Du et al., 2023) in order to elucidate the associated signaling pathways (Liu et al., 2022a).
The roles of U/U genes in regulating plant growth
U/U genes that regulate plant growth and development are distributed across a variety of species, particularly soybean (Glycine max), rice (Oryza sativa), and non-model plants. The functions of reported U/U genes throughout the lifecycles of soybean and rice are shown in Figure 2.
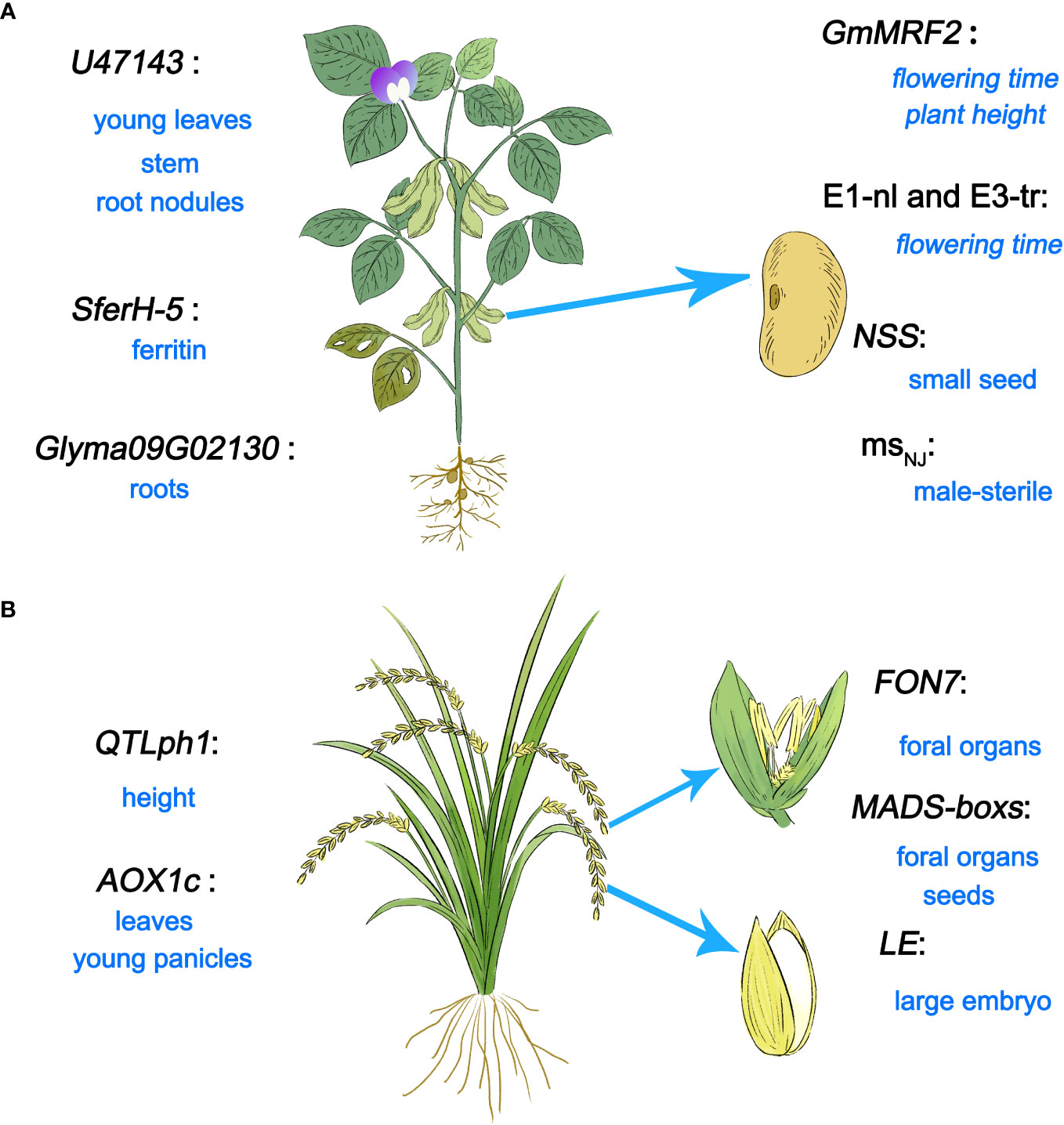
Figure 2 U/U genes related to plant growth regulation. (A) U/U genes related to growth and development identified in soybean. (B) Newly identified genes in rice and their functions in growth and development.
U/U genes in soybean
To date, many U/U genes that participate in reproductive growth have been identified in soybean (Figure 2A). The hemoglobin gene U47143 from soybean shares a protein sequence similarity of only 58% with another hemoglobin gene in soybean. As soybean is a non-symbiotic leguminous plant, hemoglobin is expressed in various tissues of this plant, such as cotyledons, seedling stems, roots, young leaves, and root nodules (Anderson et al., 1996). Glyma09G02130, a new NHX (Na+/H+ antiporter gene family) gene, was identified from the soybean genome, which is related to root growth. Under salt stress, the expression level of Glyma09G02130 in roots is significantly upregulated (Chen et al., 2015). A new ferritin gene SferH-5 has been cloned from soybean seedlings, which is related to the production of ferritin in soybean (Dong et al., 2007). The MORN motif type gene GmMRF2 has also been isolated from soybean. Soybean lines overexpressing GmMRF2 exhibited earlier flowering under long day (LD) conditions and showed an increase in plant height under both LD and short day (SD) conditions. In addition, gibberellin pathway genes which positively regulate plant height and promote flowering, were significantly upregulated in GmMRF2-overexpression lines (Zhang et al., 2023a), reflecting the important roles of GmMRF2 in regulating flowering time and plant height. In addition, E1-nl and E3-tr were identified as related to flowering time by AmpliSeq technology (Ogiso-Tanaka et al., 2019). By analyzing T-DNA mutant (S006) seeds, a gene related to seed development was discovered, named New Seed Size (NSS). Seeds of the CRISPR/Cas9-generated nss1 mutant were small with brown seed coats, which is consistent with the phenotypes of S006 seeds (Zhang et al., 2023b). What’s more, a novel male-stelile gene msNJ has been discovered (Nie et al., 2019). In a word, U/U genes are distributed at various locations in soybean and participate in different life activities.
U/U genes in rice
A gene underlying a quantitative trait locus (QTL) controlling plant height on chromosome 1 (QTLph1) was identified in rice; this gene encodes a protein that promotes sucrose transport to the leaves (Ishimaru et al., 2004) (Figure 2B). Ten new MADS-box homologous genes were identified in rice using pan-genome, all of which were expressed in flower tissue and six were highly expressed during seed development (Li et al., 2023a). A novel gene encoding alternating oxidase (AOX1c) was isolated from rice, mainly expressed in leaves and young panicles (Saika et al., 2002). The U/U gene Os08g0299000, named FLORAL ORGAN NUMBER7 (FON7), was identified in a mutant with altered floral organ number (generated by ethyl methanesulfonate treatment of Korean japonica rice cultivar Ilpum); this gene controls the number of floral organs. The fon7 mutant shows an increased number of stamens and pistils. The number of floral organs plays crucial roles in fruit development and grain maturity (Maung et al., 2023). In rice, LARGE EMBRYO (LE), a U/U gene that controls embryo size, was identified and characterized. In le mutants and RNA interference lines, the embryo size is increased, indicating that LE plays a decisive role in controlling embryo size (Lee et al., 2019). Therefore, U/U genes in rice play a major role in growth and reproduction.
U/U genes in other plants
Most other U/U genes have been reported in Arabidopsis, poplar, and sweet potato (Ipomoea batatas Lam). Four Arabidopsis mutants (rem1.2, orc1a, ppd1, and mcm4), exhibit varying degrees of reduction rosette size, confirming the novel role of these U/U genes in effective leaf surface area (ELSA) (Gonzalez et al., 2020). In Arabidopsis, the line expressing COBRA gene family showed a significant decrease in cellulose content, and the new member was identified related to the secondary cell wall formation (Brown et al., 2005). The protein encoded by the U/U transcription factor gene PebHLH35 (from Populus euphratica) enhances drought resistance by regulating stomatal development and photosynthesis, as demonstrated in transgenic Arabidopsis plants heterologously expressing this gene (Dong et al., 2014). The protein encoded by the U/U sucrose transporter gene IbSUT4 from sweet potato participates in plant growth by intervening in the abscisic acid signaling pathway (Wang et al., 2020). The U/U BrSCC1 gene BraA03g040800.3C identified in Brassica rapa L has been verified to be related to the seed coat color (Zhang et al., 2023c). The U/U gene GhMPK7 has been isolated from cotton, whose overexpression in transgenic tobacco promoted the transcription level of SA pathway quickly and efficiently and showed earlier germination compared to WT (Shi et al., 2010). It can be seen that there are numerous U/U genes distributed in different plants and participated in various life activities.
The roles of U/U genes in abiotic stress resistance
Most U/U genes identified in different plant play function in responses to different types and degrees of stress, including high salt (Figure 3A), water scarcity (Figure 3B), and harsh temperatures (high or low) (Figure 3C).
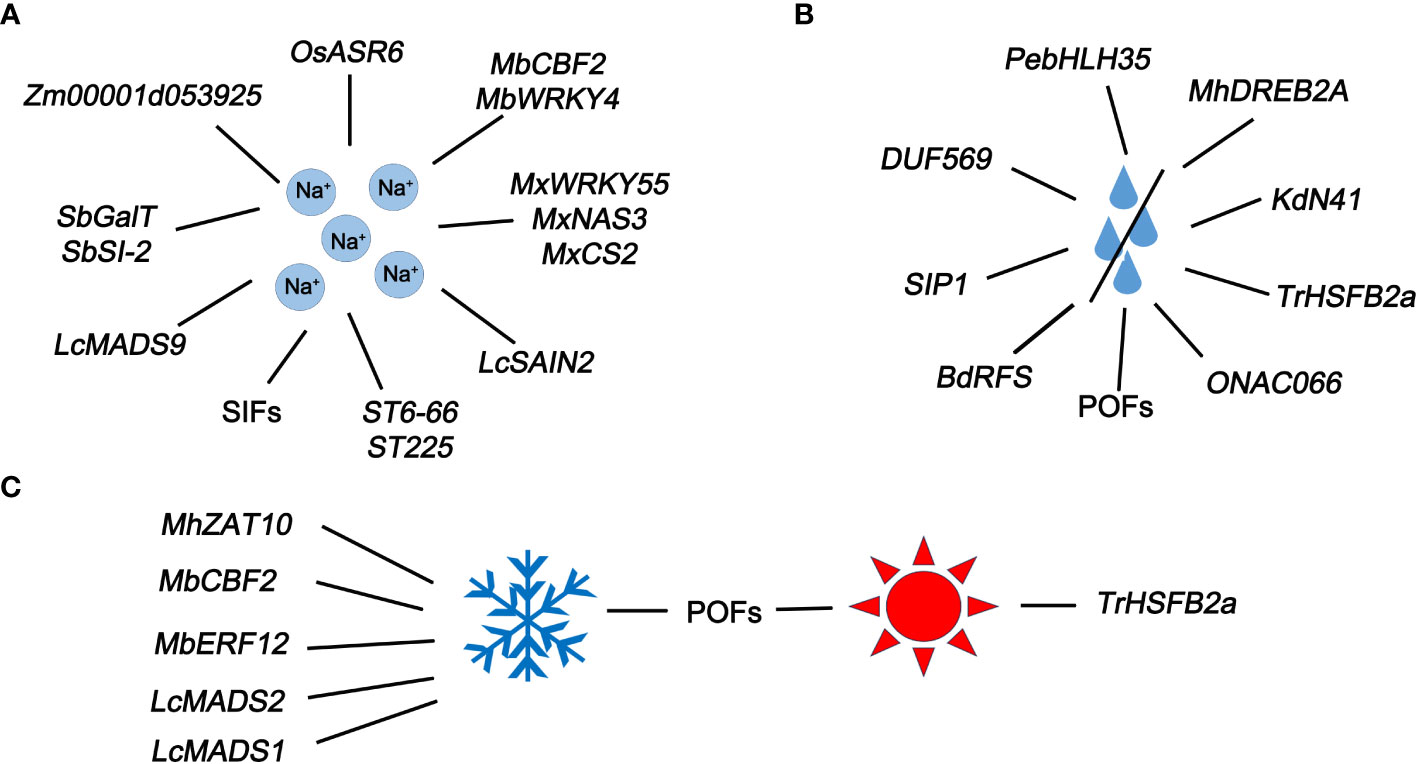
Figure 3 U/U genes closely related to abiotic stress. (A) U/U genes that respond to Na+ in plants. (B) U/U genes that respond to water deficiency in plants. (C) U/U genes that respond to extreme temperatures.
Salt stress
In rice, OsASR6 (a newly identified salt-induced ASR gene) is upregulated under salt stress. OsASR6 RNAi transgenic lines showed poorer salt tolerance and oxidative stress capacity than the untransformed control plants, while genetically modified rice lines with OsASR6 overexpression showed excellent performance (Zhang et al., 2022a). A U/U SIF gene in rice might be involved in the plant response to salinity stress (Soda et al., 2013). A new WRKY gene named MxWRKY55 was isolated from Malus xiaojinensis and introduced into Arabidopsis to significantly improve its salt tolerance and biomass (Han et al., 2020). What’s more, overexpression of MxNAS3 cloned from M. xiaojinensis in transgenic Arabidopsis improve biomass and root length. Importantly, the high expression of MxNAS3 in transgenic Arabidopsis is associated with the formation of malformed flowers (Han et al., 2018b). Also, a new gene MxCS2 encoding citrate synthase promotes the synthesis of citrate synthase and increases the content of CA in Arabidopsis. Overexpression of MxCS2 also increased the fresh weight, root length, CS activity, as well as chlorophyll and citric acid content (Han et al., 2014). MbCBF2, an uncharacterized gene from Malus baccata (L.) Borkh, increased its expression in young tissues under high salt induction. In heterologous overexpressed Arabidopsis lines, it can enhance the adaptation to high salt environment and change physiological indicators related to stress, including proline, malondialdehyde (MDA) and superoxide dismutase (SOD), which reflects the tolerance of MbCBF2 to salt stress (Li et al., 2022b). Overexpression of a novel WRKY gene MbWRKY4 in transgenic tobacco enhances salt tolerance (Han et al., 2018a).
Functional analysis of the salt cress (Thellungiella halophila) genes ST6-66 and ST225 in Arabidopsis revealed their importance in salt resistance (Du et al., 2008). The U/U gene LcMADS9 was significantly upregulated in sheepgrass (Leymus chinensis (Trin.) Tzvel) under NaCl treatment, highlighting the response of this gene to NaCl (Jia et al., 2018). LcSAIN2, another salt-induced U/U gene from sheepgrass, enhanced salt tolerance in transgenic Arabidopsis plants (Li et al., 2013). Transcriptome sequencing of two maize (Zea mays) inbred lines revealed the U/U gene Zm00001d053925, whose expression level was significantly higher in AS5 (salt tolerant line) than in NX420 (salt intolerant line), indicating that Zm00001d053925 functions in the plant response to salt stress (Zhu et al., 2023) (Figure 3A). A U/U gene galactosyl transferase-like (SbGalT) from Salicornia brachiata alleviates osmotic and salt stress in transgenic tobacco (Dubey et al., 2021). Also, another new salt induced gene SbSI-2 (Salicornia brachiata salt-inducible-2) has been functionally identified to have the same function as SbGalT (Pandey et al., 2014). In one word, U/U genes exercise significant functions in response to salt stress.
Drought stress
In Populus euphratica, the transcription factor PebHLH35 confers drought resistance by regulating various developmental and physiological processes (Dong et al., 2014). DEHYDRATION RESPONSE ELEMENT-BINDING PROTEIN 2A (DREB2A) in apple (Malus domestica) responds to drought stress and plants overexpressing MhDREB2A exhibited enhanced tolerance to drought (Li et al., 2023b). An uncharacterized KdNOVEL41 (KdN41) gene from Kalanchoe (K.) daigremontiana confers drought resistance on K. daigremontiana and tobacco (Nicotiana tabacum) by playing a role in clearing reactive oxygen species and reducing osmotic damage (Wang et al., 2018). The unique proteins with obscure features (POFs) of Arabidopsis enhance tolerance to oxidative stress, including osmotic, salinity, and temperature stress (Luhua et al., 2008). The U/U gene BdRFS of Brachypodium distachyon has been identified to be functionally conserved, together with improve the drought resistance of Brachypodium and Arabidopsis (Ying et al., 2023). The inactivation of SIP1, encoding an unknown protein in Arabidopsis, decreased drought tolerance (Anderson and Kohorn, 2001). Furthermore, a novel gene DUF569 (AT1G69890) with “domain of unknown function” positively regulates drought stress in Arabidopsis (Nabi et al., 2021). The HSF transcription factor gene TrHSFB2a (B-type HSF), which was recently identified in drought-sensitive white clover (Trifolium repens), negatively regulates drought resistance (Iqbal et al., 2022) (Figure 3B). Under drought stress conditions, overexpression of the ONAC066 gene (a novel gene whose function has been newly determined) enhances the tolerance of rice to drought stress and sensitivity to ABA (Yuan et al., 2019). Numerous U/U genes responding to drought stress undoubtedly bring new possibilities for improving plant drought resistance.
Extreme temperature stress
MbCBF2, a U/U CBF transcription factor gene from Malus baccata (L.) Borkh, shows elevated expression at low temperatures. Exogenously overexpressing MbCBF2 enhanced the adaptability of transgenic Arabidopsis to cold conditions (Li et al., 2022b). MbERF12, an ERF gene, enhances its ability to scavenge reactive oxygen species through ethylene signaling, playing a crucial role in the response of salt and low temperature stress (Han et al., 2021). ZINC FINGER OF ARABIDOPSIS THALIANA 10 (ZAT10), a U/U gene in Malus domestica, is activated under low temperature stress. Apple lines overexpressing MhZAT10 showed increased tolerance to low temperature stress, indicating that this gene plays an important role in cold resistance (Li et al., 2023b). Low temperature significantly induced LcMADS1 and LcMADS2 expression in sheepgrass (Jia et al., 2018).
TrHSFB2a expression in white clover was strongly induced by exposure to high temperature (35°C) and the encoded protein negatively regulates heat tolerance (Iqbal et al., 2022). The POFs in Arabidopsis enhance plant tolerance to oxidative stress under both cold and heat stress (Luhua et al., 2008) (Figure 3C). Under harsh temperatures, in addition to previously characterized genes, there are also these uncharacterized genes, which enriches the large category of genes that have resistance to extreme temperature.
Though different new genes were identified in various stress, plants usually experience fluctuations in several key hormone levels during their early stress response, such as ABA, SA and JA (Verma et al., 2016). ABA is a regulatory factor for many plants under environmental stress, including drought, low temperature, and salinity. Abiotic stress generates osmotic signals, leading to ABA accumulation (Danquah et al., 2014). SnRKs are involved in osmotic stress and ABA signal transduction, and both SnRKs and ABA pathways involve MAPK responses (Zhu, 2016). Under extreme stress conditions, ROS is overproduced and causes oxidative damage to plants (Verma et al., 2016). After ROS signal transduction, anthocyanins are produced, which are used for antioxidant activities by clearing excess ROS (Naing and Kim, 2021). In short, plants have a similar fate after being subjected to abiotic stresses, which can trigger a series of homologous stress tolerance activities.
The roles of U/U genes in biological stress responses
Plant diseases such as powdery mildew, bacterial blight, and leaf rust frequently occur in plants (especially food crops), which greatly reduces crop quality and yields. Many U/U genes in crops are related to diseases responses. Here we summarize progress in identifying genes involved in biological stress resistance. Among them, most do not have obvious domains, except for family genes such as NAC.
Leaf rust resistance
Wheat leaf rust, a disease caused by Puccinia triticina, mainly damages the leaves of common wheat (Triticum aestivum) and causes serious losses in wheat production (Qi et al., 2023). Currently, the most effective control measure involves breeding and using resistant wheat varieties.
The U/U gene Lr68 in common wheat confers slow-rusting resistance to wheat rust, as demonstrated in the field (Herrera-Foessel et al., 2012). Lr46 is also associated with slow-rusting resistance to leaf rust in wheat (Singh et al., 1998). A leaf rust resistance gene named Lr81 was identified in wheat line PI470121, which is a unique leaf rust resistance locus (Xu et al., 2022a). A stable APR gene, named LrYang16G216, was detected in wheat and identified as a new and effective gene for leaf rust resistance (Zhao et al., 2023b). A gene Pc54 with leaf rust resistance has been identified in oat (Avena sativa) (Admassu-Yimer et al., 2022).These newly identified genes all have excellent activity in inhibiting rust (Figure 4A), which could contribute to the breeding of rust resistant wheat varieties.
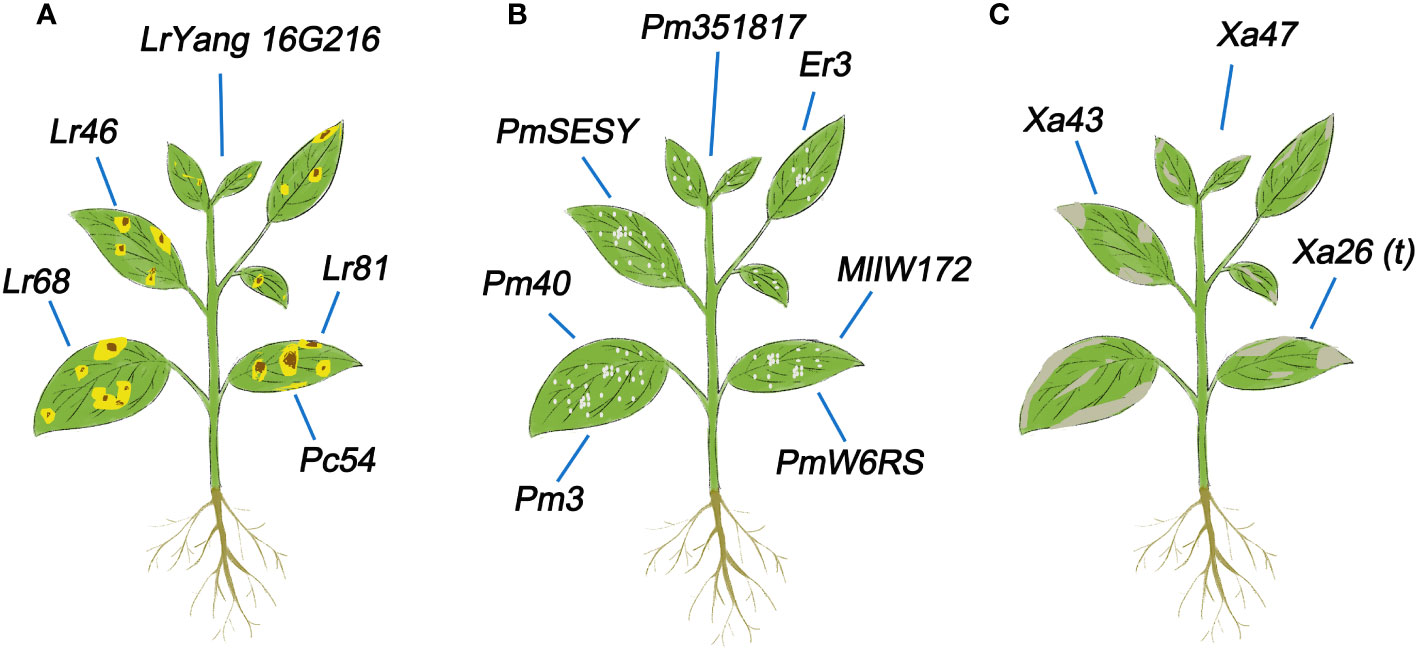
Figure 4 U/U genes related to resistance to biological stress in plants. (A) U/U genes involved in leaf rust resistance in plants. (B) U/U genes involved in powdery mildew resistance in plants. (C) U/U genes involved in bacterial blight resistance in plants.
Powdery mildew resistance
Powdery mildew is a crop disease induced by Blumeria graminis f. sp. Tritici that is extremely destructive to common wheat (Mapuranga et al., 2022). Identifying powdery mildew resistance genes could suggest strategies to improve growth and yield in wheat and other crops.
The U/U powdery mildew resistance gene Pm40 of Elytrigia intermedium confers resistance to this disease and has been transferred to wheat to enhance its resistance to powdery mildew (Luo et al., 2009). A gene Pm3 with powdery mildew resistance was identified in oat (Avena sativa) (Admassu-Yimer et al., 2022). PmW6RS has been identified as a powdery mildew resistant gene in rye (Secale cereale L., RR), providing a new gene selection for wheat disease resistance breeding (Wang et al., 2023). Pm351817, a new Pm65 allele in wheat, exhibits resistance to powdery mildew (Xu et al., 2023). PmSESY in rye (Secale sylvestre) also confers resistance to powdery mildew and can significantly improve resistance to this disease (He et al., 2021). The U/U gene Er3, which was identified in Pisum fulvum, markedly improves the resistance to powdery mildew (Sara et al., 2007). The U/U allele MlIW172 of Pm60 was shown to enhance resistance to powdery mildew in wheat by transgenic complementation (Wu et al., 2022). These genes provide genetic diversity for breeding wheat with enhanced resistance to powdery mildew (Figure 4B).
Bacterial blight resistance
Bacterial blight (BB), a disease caused by Xanthomonas oryzae pv. oryzae (Xoo), is a serious rice disease worldwide (Javed et al., 2022). Therefore, identifying and isolating BB resistance genes from different rice resources is of great significance. Different rice varieties have multiple different BB resistance genes. The BB resistance gene Xa43 was recently identified in Zhangpu wild rice (Oryza rufipogon) (Huang et al., 2023). A new NLR disease resistance gene Xa47 has long-term resistance to rice BB disease (Lu et al., 2022b). Xa26(t), which was identified in rice variety Minghui 63, has a dominant effect on the Chinese Xoo strain JL691 at both the seedling and adult stages (Yang et al., 2003) (Figure 4C). It can be seen that the identification and utilization of U/U genes are of great significance for resisting BB.
Resistance to other diseases
Multiple resistance genes to downy mildew exist in wild Lactuca, 11 of which were introduced into lettuce (Lactuca sativa L.) to facilitate the development of multi-gene downy mildew resistant lines (Parra et al., 2020). The resistance gene Rsg3 was recently discovered in Chinese barley landrace PI 565676 (a landrace from China). This gene, which provides strong resistance to greenbug (Schizaphis graminum Rondani), should help alleviate the major threat of this insect pest to global food production (Xu et al., 2022b). The resistance gene bph42, which confers resistance to brown planthopper (BPH), was identified in wild rice line Oryza rufipogon (Griff.) and transferred to cultivated rice (Oryza sativa), laying the foundation for the production of high-quality rice with enhanced insect resistance (Kaur et al., 2022). Brassica rapa shows obvious resistance to turnip mosaic virus (TuMV). Through genetic analysis, a uncharacterized TuMV resistance gene, BraA06g035130.3C, was recently identified, paving the way for improving TuMV resistance and agricultural production (Lu et al., 2022a). The U/U gene GbNAC1 from Gossypium barbadense L. has been identified to be positively involved in the regulation of Verticillium Wilt resistance (Wang et al., 2016).
Perspectives
More than a quarter of genes in the genomes of both crops and halophytes encode proteins of unknown function (Luhua et al., 2008). Some of these genes encode at least one previously defined domain or motif, but most lack previously defined features. Although transcriptome, metabolome, and proteome data show that many of these genes play important roles in plant growth, metabolism, physiology, and other life processes, their functions remain to be identified.
Nowadays the model organism Arabidopsis can be used to verify the functions of these genes via heterologous transformation and other experimental techniques, but studies of unknown genes should focus on their functions in the species harboring these genes and establishing genetic transformation systems for these species. Generating overexpression and silencing lines of the target gene of the species of interest via genetic transformation and observing the phenotypes of the transgenic lines would enable the analysis of gene function more directly and accurately. The functional study of unknown genes is not limited to the genes themselves. Genes are usually regulated by key upstream factors, and they encode proteins that regulate downstream genes. Therefore, clarifying the functions of the upstream and downstream factors of U/U genes and establishing a complete gene regulatory network are important aspects of functional studies of these genes.
U/U genes not only encode proteins that perform various biological functions in plants, but they also play important roles in the life activities of animals, microorganisms, and especially humans. We can also find inspiration from the study of U/U genes in animals. FREPs, a recently identified gene family in mussels (Mytilus edulis), are related to immune recognition in mollusks (Gorbushin and Iakovleva, 2011). A recently identified Ig kappa gene in sea star (Asterias rubens) confers specific resistance to horseradish peroxidase (Vincent et al., 2014). Previously unidentified genes obtained from chromosome replication promoted the study of the Neuropeptide Y family in vertebrates (Sundström et al., 2008). Innexin 3, a gene involved in dorsal closure in embryos, has also been identified in Drosophila (Fanning et al., 2013). A newly discovered gene that confers resistance to influenza virus H5N1 was identified in duck (Anas platyrhynchos) through transcriptome analysis (Huang et al., 2019). A new human membrane-associated mucin of the ocular surface was recently identified, which could contribute to the protection of human eyes (Fini et al., 2020). The discovery of a series of new genes in males revealed a new pathway for the production of testosterone (Flück and Pandey, 2014). New genes that function in osmotic stress resistance in the yeast Saccharomyces cerevisiae have also been identified (Gonzalez et al., 2016).
The study of U/U genes faces challenges because it is often unclear to which pathways these genes contribute. We can also uncover the unexpected functions of U/U genes in plant development and resistance, providing essential information to supplement our knowledge of known functional genes and improve our understanding of the connections between biological molecules.
In summary, numerous uncharacterized genes in living organisms have yet to be discovered, isolated, analyzed, cloned, and functionally identified. Some of these genes play key roles in the lifecycles of living organisms. Exploring these genes may enrich our understanding of existing physiological processes, metabolic pathways, and functional networks and offer new strategies to modulate them.
Author contributions
XW: Writing – original draft. BW: Writing – review & editing. FY: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Research Foundation of China (NSFC, project nos. 32370304 and 32170301).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Admassu-Yimer, B., Klos, K. E., Griffiths, I., Cowan, A., Howarth, C. (2022). Mapping of Crown Rust (Puccinia coronata f. sp. avenae) Resistance Gene Pc54 and a Novel Quantitative Trait Locus Effective Against Powdery Mildew (Blumeria graminis f. sp. avenae) in the Oat (Avena sativa) Line Pc54. Phytopathology® 112 (6), 1316–1322. doi: 10.1094/phyto-10-21-0445-r
Akond, Z., Rahman, H., Ahsan, M. A., Mosharaf, M. P., Alam, M., Mollah, M. N. H. (2022). Comprehensive in silico analysis of RNA silencing-related genes and their regulatory elements in wheat (Triticum aestivum L.). BioMed. Res. Int. 17(1). doi: 10.1155/2022/4955209
Anderson, C. M., Kohorn, B. D. (2001). Inactivation of Arabidopsis SIP1 leads to reduced levels of sugars and drought tolerance. J. Plant Physiol. 158 (9), 1215–1219. doi: 10.1078/s0176-1617(04)70149-2
Anderson, C. R., Jensen, E. O., Llewellyn, D. J., Dennis, E. S., Peacock, W. J. (1996). A new hemoglobin gene from soybean: a role for hemoglobin in all plants. Proc. Natl. Acad. Sci. 93 (12), 5682–5687. doi: 10.1073/pnas.93.12.5682
Beihammer, G., Romero-Pérez, A., Maresch, D., Figl, R., Mócsai, R., Grünwald-Gruber, C., et al. (2023). Pseudomonas syringae DC3000 infection increases glucosylated N-glycans in Arabidopsis thaliana. Glycoconj J. 40 (1), 97–108. doi: 10.1007/s10719-022-10084-6
Beiki, H., Liu, H., Huang, J., Manchanda, N., Nonneman, D., Smith, T. P. L., et al. (2019). Improved annotation of the domestic pig genome through integration of Iso-Seq and RNA-seq data. BMC Genomics 20 (1), 344. doi: 10.1186/s12864-019-5709-y
Besseau, S., Li, J., Palva, E. T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 63 (7), 2667–2679. doi: 10.1093/jxb/err450
Bharathi, J., Anandan, R., Benjamin, L. K., Muneer, S., Prakash, M. A. S. (2023). Recent trends and advances of RNA interference (RNAi) to improve agricultural crops and enhance their resilience to biotic and abiotic stresses. Plant Physiol. Biochem. 194, 600–618. doi: 10.1016/j.plaphy.2022.11.035
Brown, D. M., Zeef, L. A. H., Ellis, J., Goodacre, R., Turner, S. R. (2005). Identification of novel genes in arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17 (8), 2281–2295. doi: 10.1105/tpc.105.031542
Cao, Y., Dou, D., Zhang, D., Zheng, Y., Ren, Z., Su, H., et al. (2022). ZmDWF1 regulates leaf angle in maize. Plant Sci 325, 111459. doi: 10.1016/j.plantsci.2022.111459
Chen, H.-t., Chen, X., Wu, B.-y., Yuan, X.-x., Zhang, H.-m., Cui, X.-y., et al. (2015). Whole-genome identification and expression analysis of K+ efflux antiporter (KEA) and Na+/H+ antiporter (NHX) families under abiotic stress in soybean. J. Integr. Agric. 14 (6), 1171–1183. doi: 10.1016/s2095-3119(14)60918-7
Chen, L. Q., Hou, B. H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468 (7323), 527–532. doi: 10.1038/nature09606
Chen, H., Li, X., Li, F., Li, D., Dong, Y., Fan, Y. (2022). Bioinformatics analysis of WRKY family genes in erianthus fulvus ness. Genes (Basel) 13 (11), 2102. doi: 10.3390/genes13112102
Cheng, L., Li, X., Huang, X., Ma, T., Liang, Y., Ma, X., et al. (2013). Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 70, 252–260. doi: 10.1016/j.plaphy.2013.05.025
Cheng, W., Tung, Y., Chiou, T., Chen, J. (2006). Cloning and characterisation of mitochondrial manganese superoxide dismutase (mtMnSOD) from the giant freshwater prawn Macrobrachium rosenbergii. Fish Shellfish Immunol. 21 (4), 453–66. doi: 10.1016/j.fsi.2006.02.005
Choi, J., Kim, J. W., Oh, M. H. (2022). Identification of Feronia-interacting proteins in Arabidopsis thaliana. Genes Genomics 44 (12), 1477–1485. doi: 10.1007/s13258-022-01292-3
Danquah, A., de Zelicourt, A., Colcombet, J., Hirt, H. (2014). The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32 (1), 40–52. doi: 10.1016/j.bioteChadv.2013.09.006
Dinglasan, E., Periyannan, S., Hickey, L. T. (2022). Harnessing adult-plant resistance genes to deploy durable disease resistance in crops. Essays Biochem. 66 (5), 571–580. doi: 10.1042/EBC20210096
Dong, X., Sun, Q., Wei, D., Li, J., Li, J., Tang, B., et al. (2007). A novel ferritin gene, SferH-5, reveals heterogeneity of the 26.5-kDa subunit of soybean (Glycine max) seed ferritin. FEBS Lett. 581 (30), 5796–5802. doi: 10.1016/j.febslet.2007.11.049
Dong, Y., Wang, C., Han, X., Tang, S., Liu, S., Xia, X., et al. (2014). A novel bHLH transcription factor PebHLH35 from Populus euphratica confers drought tolerance through regulating stomatal development, photosynthesis and growth in Arabidopsis. Biochem. Biophys. Res. Commun. 450 (1), 453–458. doi: 10.1016/j.bbrc.2014.05.139
Dong, X., Zhu, H., Hao, X., Wang, Y., Ma, X., Zhao, J., et al. (2022). Genome-Wide identification of common bean pvLTP family genes and expression profiling analysis in response to drought stress. Genes (Basel) 13 (12), 2394. doi: 10.3390/genes13122394
Du, X., Fang, L., Li, J., Chen, T., Cheng, Z., Zhu, B., et al. (2023). The TabHLH094-TaMYC8 complex mediates the cadmium response in wheat. Mol. Breed 43 (7), 57. doi: 10.1007/s11032-023-01404-1
Du, J., Huang, Y. P., Xi, J., Cao, M. J., Ni, W. S., Chen, X., et al. (2008). Functional gene-mining for salt-tolerance genes with the power of Arabidopsis. Plant J. 56 (4), 653–664. doi: 10.1111/j.1365-313X.2008.03602.x
Du, X., Su, M., Jiao, Y., Xu, S., Song, J., Wang, H., et al. (2022). A transcription factor slNAC10 gene of suaeda liaotungensis regulates proline synthesis and enhances salt and drought tolerance. Int. J. Mol. Sci. 23 (17), 9625. doi: 10.3390/ijms23179625
Dubey, A. K., Khatri, K., Jha, B., Rathore, M. S. (2021). The novel galactosyl transferase-like (SbGalT) gene from Salicornia brachiata maintains photosynthesis and enhances abiotic stress tolerance in transgenic tobacco. Gene 786. doi: 10.1016/j.gene.2021.145597
Fang, T., Rao, Y., Wang, M., Li, Y., Liu, Y., Xiong, P., et al. (2022). Characterization of the SWEET gene family in longan (Dimocarpus longan) and the role of dlSWEET1 in cold tolerance. Int. J. Mol. Sci. 23 (16), 8914. doi: 10.3390/ijms23168914
Fanning, A. S., Giuliani, F., Giuliani, G., Bauer, R., Rabouille, C. (2013). Innexin 3, a new gene required for dorsal closure in drosophila embryo. PloS One 8 (7). doi: 10.1371/journal.pone.0069212
Fini, M. E., Jeong, S., Gong, H., Martinez-Carrasco, R., Laver, N. M. V., Hijikata, M., et al. (2020). Membrane-associated mucins of the ocular surface: New genes, new protein functions and new biological roles in human and mouse. Prog. Retinal Eye Res. 75. doi: 10.1016/j.preteyeres.2019.100777
Flück, C. E., Pandey, A. V. (2014). Steroidogenesis of the testis – new genes and pathways. Annales d'Endocrinologie 75 (2), 40–47. doi: 10.1016/j.ando.2014.03.002
Geng, Y., Zhang, S., Yang, N., Qin, L. (2022). Whole-Genome Sequencing and Comparative Genomics Analysis of the Wild Edible Mushroom (Gomphus purpuraceus) Provide Insights into Its Potential Food Application and Artificial Domestication. Genes (Basel) 13 (9), 1628. doi: 10.3390/genes13091628
Gonzalez, R., Butkovic, A., Rivarez, M. P. S., Elena, S. F. (2020). Natural variation in Arabidopsis thaliana rosette area unveils new genes involved in plant development. Sci. Rep. 10 (1), 17600. doi: 10.1038/s41598-020-74723-4
Gonzalez, R., Morales, P., Tronchoni, J., Cordero-Bueso, G., Vaudano, E., Quirós, M., et al. (2016). New genes involved in osmotic stress tolerance in saccharomyces cerevisiae. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01545
Gorbushin, A. M., Iakovleva, N. V. (2011). A new gene family of single fibrinogen domain lectins in Mytilus. Fish Shellfish Immunol. 30 (1), 434–438. doi: 10.1016/j.fsi.2010.10.002
Groot Kormelink, P., Luyten, W. (1997). Cloning and sequence of full-length cDNAs encoding the human neuronal nicotinic acetylcholine receptor (nAChR) subunits beta3 and beta4 and expression of seven nAChR subunits in the human neuroblastoma cell line SH-SY5Y and/or IMR-32. FEBS Lett. 400 (3), 309–314. doi: 10.1016/s0014-5793(96)01383-x
Guan, Y. F., Huang, X. Y., Zhu, J., Gao, J. F., Zhang, H. X., Yang, Z. N. (2008). RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 147 (2), 852–863. doi: 10.1104/pp.108.118026
Han, D., Han, J., Xu, T., Li, X., Yao, C., Li, T., et al. (2021). Overexpression of MbERF12, an ERF gene from Malus baccata (L.) Borkh, increases cold and salt tolerance in Arabidopsis thaliana associated with ROS scavenging through ethylene signal transduction. In Vitro Cell. Dev. Biol. - Plant 57 (5), 760–770. doi: 10.1007/s11627-021-10199-9
Han, D., Hou, Y., Ding, H. (2018a). Isolation and preliminary functional analysis of MbWRKY4 gene involved in salt tolerance in transgenic tobacco. Int. J. Agric. Biol. 20 (9), 433–441. doi: 10.1080/17429145.2018.1499145
Han, D.-G., Shi, Y., Wang, B., Liu, W., Yu, Z.-Y., Lv, B.-Y., et al. (2014). Isolation and preliminary functional analysis of mxCS2: a gene encoding a citrate synthase in malus xiaojinensis. Plant Mol. Biol. Rep. 33 (1), 133–142. doi: 10.1007/s11105-014-0735-z
Han, D., Zhang, Z., Ni, B., Ding, H., Liu, W., Li, W., et al. (2018b). Isolation and functional analysis of MxNAS3 involved in enhanced iron stress tolerance and abnormal flower in transgenic Arabidopsis. J. Plant Interact. 13 (1), 433–441. doi: 10.1080/17429145.2018.1499145
Han, D., Zhou, Z., Du, M., Wu, X., Yu, J., Zhang, P., et al. (2020). Overexpression of a Malus xiaojinensis WRKY transcription factor gene (MxWRKY55) increased iron and high salinity stress tolerance in Arabidopsis thaliana. In Vitro Cell. Dev. Biol. Plant 56 (5). doi: 10.1007/s11627-021-10241-w
He, H., Du, H., Liu, R., Liu, T., Yang, L., Gong, S., et al. (2021). Characterization of a new gene for resistance to wheat powdery mildew on chromosome 1RL of wild rye Secale sylvestre. Theor. Appl. Genet. 134 (3), 887–896. doi: 10.1007/s00122-020-03739-1
Herrera-Foessel, S. A., Singh, R. P., Huerta-Espino, J., Rosewarne, G. M., Periyannan, S. K., Viccars, L., et al. (2012). Lr68: a new gene conferring slow rusting resistance to leaf rust in wheat. Theor. Appl. Genet. 124 (8), 1475–1486. doi: 10.1007/s00122-012-1802-1
Hu, L., Li, Y., Wang, J., Zhao, Y., Wang, Y. (2023). Controlling CRISPR-Cas9 by guide RNA engineering. Wiley Interdiscip Rev. RNA 14, (1). doi: 10.1002/wrna.1731
Huang, Y.-h., Feng, H.-p., Huang, L.-r., Yi, K., Rong, E.-g., Chen, X.-y., et al. (2019). Transcriptomic analyses reveal new genes and networks response to H5N1 influenza viruses in duck (Anas platyrhynchos). J. Integr. Agric. 18 (7), 1460–1472. doi: 10.1016/s2095-3119(19)62646-8
Huang, F., He, N., Yu, M., Li, D., Yang, D., Pastor, V. (2023). Identification and fine mapping of a new bacterial blight resistance gene, Xa43(t), in Zhangpu wild rice (Oryza rufipogon). Plant Biol. 25 (3), 433–439. doi: 10.1111/plb.13502
Huang, L., Liu, Y., Yu, X., Xiang, X., Cao, J. (2011). A polygalacturonase inhibitory protein gene (BcMF19) expressed during pollen development in Chinese cabbage-pak-choi. Mol. Biol. Rep. 38 (1), 545–552. doi: 10.1007/s11033-010-0139-6
Imtiaz, A. (2022). ARNSHL gene identification: past, present and future. Mol. Genet. Genomics 297 (5), 1185–1193. doi: 10.1007/s00438-022-01926-x
Iqbal, M. Z., Jia, T., Tang, T., Anwar, M., Ali, A., Hassan, M. J., et al. (2022). A heat shock transcription factor TrHSFB2a of white clover negatively regulates drought, heat and salt stress tolerance in transgenic arabidopsis. Int. J. Mol. Sci. 23 (21), 12769. doi: 10.3390/ijms232112769
Ishimaru, K., Ono, K., Kashiwagi, T. (2004). Identification of a new gene controlling plant height in rice using the candidate-gene strategy. Planta 218 (3), 388–395. doi: 10.1007/s00425-003-1119-z
Javed, M. A., Ali, S. W., Ashfaq, M., Tabassam, J., Ali, M., IhsanUllah, M., et al. (2022). Molecular profiling of bacterial blight resistance in Malaysian rice cultivars. Braz. J. Biol. 82. doi: 10.1590/1519-6984.256189
Jia, D., Jin, C., Gong, S., Wang, X., Wu, T. (2022). RNA-seq and iso-seq reveal the important role of COMT and CCoAOMT genes in accumulation of scopoletin in noni (Morinda citrifolia). Genes (Basel) 13 (11), 1993. doi: 10.3390/genes13111993
Jia, J., Zhao, P., Cheng, L., Yuan, G., Yang, W., Liu, S., et al. (2018). MADS-box family genes in sheepgrass and their involvement in abiotic stress responses. BMC Plant Biol. 18 (1), 42. doi: 10.1186/s12870-018-1259-8
Jin, X., Tsago, Y., Lu, Y., Sunusi, M., Khan, A. U. (2022). Map-based cloning and transcriptome analysis of the more-tiller and small-grain mutant in rice. Planta 256 (5), 98. doi: 10.1007/s00425-022-04011-0
Kaur, P., Neelam, K., Sarao, P. S., Babbar, A., Kumar, K., Vikal, Y., et al. (2022). Molecular mapping and transfer of a novel brown planthopper resistance gene bph42 from Oryza rufipogon (Griff.)To cultivated rice (Oryza sativa L.). Mol. Biol. Rep. 49 (9), 8597–8606. doi: 10.1007/s11033-022-07692-8
Koeppe, S., Kawchuk, L., Kalischuk, M. (2023). RNA interference past and future applications in plants. Int. J. Mol. Sci. 24 (11), 9755. doi: 10.3390/ijms24119755
Lee, G., Piao, R., Lee, Y., Kim, B., Seo, J., Lee, D., et al. (2019). Identification and characterization of LARGE EMBRYO, a new gene controlling embryo size in rice (Oryza sativa L.). Rice (N Y) 12 (1), 22. doi: 10.1186/s12284-019-0277-y
Leng, B., Wang, X., Yuan, F., Zhang, H., Lu, C., Chen, M., et al. (2021). Heterologous expression of the Limonium bicolor MYB transcription factor LbTRY in Arabidopsis thaliana increases salt sensitivity by modifying root hair development and osmotic homeostasis. Plant Sci. 302, 110704. doi: 10.1016/j.plantsci.2020.110704
Li, Y., Dai, C., Hu, C., Liu, Z., Kang, C. (2017). Global identification of alternative splicing via comparative analysis of SMRT- and Illumina-based RNA-seq in strawberry. Plant J. 90 (1), 164–176. doi: 10.1111/tpj.13462
Li, X., Gao, Q., Liang, Y., Ma, T., Cheng, L., Qi, D., et al. (2013). A novel salt-induced gene from sheepgrass, LcSAIN2, enhances salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 64, 52–59. doi: 10.1016/j.plaphy.2012.12.014
Li, X., Liang, X., Li, W., Yao, A., Liu, W., Wang, Y., et al. (2022b). Isolation and Functional Analysis of MbCBF2, a Malus baccata (L.) Borkh CBF Transcription Factor Gene, with Functions in Tolerance to Cold and Salt Stress in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 23 (17), 9827. doi: 10.3390/ijms23179827
Li, X. L., Meng, D., Li, M. J., Zhou, J., Yang, Y. Z., Zhou, B. B., et al. (2023b). Transcription factors MhDREB2A/MhZAT10 play a role in drought and cold stress response crosstalk in apple. Plant Physiol. 192 (3), 2203–2220. doi: 10.1093/plphys/kiad147
Li, W., Wang, D., Hong, X., Shi J, H. J., Su, S., Loaiciga, C. R., et al. (2023a). Identification and validation of new MADS-box homologous genes in 3010 rice pan-genome. Plant Cell Rep. 42 (6), 975–988. doi: 10.1007/s00299-023-03006-9
Li, W., Zhong, J., Zhang, L., Wang, Y., Song, P., Liu, W., et al. (2022a). Overexpression of a Fragaria vesca MYB Transcription Factor Gene (FvMYB82) Increases Salt and Cold Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 23 (18). doi: 10.3390/ijms231810538
Lindberg, A., Polacek, C., Johansson, S. (1997). Amplification and cloning of complete enterovirus genomes by long distance PCR. J. Virol. Methods 65 (2), 191–199. doi: 10.1016/s0166-0934(97)02178-2
Liu, Z., Fan, H., Ma, Z. (2022c). Comparison of SWEET gene family between maize and foxtail millet through genomic, transcriptomic, and proteomic analyses. Plant Genome 15 (3), e20226. doi: 10.1002/tpg2.20226
Liu, W., Liang, X., Cai, W., Wang, H., Liu, X., Cheng, L., et al. (2022b). Isolation and Functional Analysis of VvWRKY28, a Vitis vinifera WRKY Transcription Factor Gene, with Functions in Tolerance to Cold and Salt Stress in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 23 (21). doi: 10.3390/ijms232113418
Liu, S., Wang, Y., Shi, M., Maoz, I., Gao, X., Sun, M., et al. (2022a). SmbHLH60 and SmMYC2 antagonistically regulate phenolic acids and anthocyanins biosynthesis in Salvia miltiorrhiza. J. Adv. Res. 42, 205–219. doi: 10.1016/j.jare.2022.02.005
Liu, W., Wang, T., Wang, Y., Liang, X., Han, J., Han, D. (2023). MbMYBC1, a M. baccata MYB transcription factor, contribute to cold and drought stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1141446
Liu, W., Zhang, B. (2022). The landscape of genome sequencing and assembling in plants. Funct. Integr. Genomics 22 (6), 1147–1152. doi: 10.1007/s10142-022-00916-x
Lu, X., Li, Z., Huang, W., Wang, S., Zhang S, L. F., Zhang, H., et al. (2022a). Mapping and identification of a new potential dominant resistance gene to turnip mosaic virus in Brassica rapa. Planta 256 (4), 66. doi: 10.1007/s00425-022-03981-5
Lu, K., Song, R., Guo, J. X., Zhang, Y., Zuo, J. X., Chen, H. H., et al. (2023). CycC1;1-WRKY75 complex-mediated transcriptional regulation of SOS1 controls salt stress tolerance in Arabidopsis. Plant Cell 35 (7), 2570–2591. doi: 10.1093/plcell/koad105
Lu, Y., Zhong, Q., Xiao, S., Wang, B., Ke, X., Zhang, Y., et al. (2022b). A new NLR disease resistance gene Xa47 confers durable and broad-spectrum resistance to bacterial blight in rice. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1037901
Luhua, S., Ciftci-Yilmaz, S., Harper, J., Cushman, J., Mittler, R. (2008). Enhanced tolerance to oxidative stress in transgenic Arabidopsis plants expressing proteins of unknown function. Plant Physiol. 148 (1), 280–292. doi: 10.1104/pp.108.124875
Luo, P. G., Luo, H. Y., Chang, Z. J., Zhang, H. Y., Zhang, M., Ren, Z. L. (2009). Characterization and chromosomal location of Pm40 in common wheat: a new gene for resistance to powdery mildew derived from Elytrigia intermedium. Theor. Appl. Genet. 118 (6), 1059–1064. doi: 10.1007/s00122-009-0962-0
Ma, C., Chen, L. (2007). Research progress on isolation and cloning of functional genes in tea plants. Front. Agric. China 1 (4), 449–455. doi: 10.1007/s11703-007-0074-z
Ma, L., Han, R., Yang, Y., Liu, X., Li, H., Zhao, X., et al. (2023). Phytochromes enhance SOS2-mediated PIF1 and PIF3 phosphorylation and degradation to promote Arabidopsis salt tolerance. Plant Cell. 35 (8), 2997–3020. doi: 10.1093/plcell/koad117
Ma, X., Qiao, Z., Chen, D., Yang, W., Zhou, R., Zhang, W., et al. (2015). CYCLIN-DEPENDENT KINASE G2 regulates salinity stress response and salt mediated flowering in Arabidopsis thaliana. Plant Mol. Biol. 88 (3), 287–299. doi: 10.1007/s11103-015-0324-z
Ma, J., Wang, L.-Y., Dai, J.-X., Wang, Y., Lin, D. (2021). The NAC-type transcription factor CaNAC46 regulates the salt and drought tolerance of transgenic Arabidopsis thaliana. BMC Plant Biol. 21 (1). doi: 10.1186/s12870-020-02764-y
Mapuranga, J., Chang, J., Yang, W. (2022). Combating powdery mildew: Advances in molecular interactions between Blumeria graminis f. sp. tritici and wheat. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1102908
Maung, P. P., Kim, B., Jin, Z., Jang, S., Lee, Y. K., Koh, H. J. (2023). Identification and characterization of a novel gene controlling floral organ number in rice (Oryza sativa L.). PloS One 18 (1), e0280022. doi: 10.1371/journal.pone.0280022
Mei, F., Chen, B., Du, L., Li, S., Zhu, D., Chen, N., et al. (2022). A gain-of-function allele of a DREB transcription factor gene ameliorates drought tolerance in wheat. Plant Cell 34 (11), 4472–4494. doi: 10.1093/plcell/koac248
Miao, Y., Laun, T., Zimmermann, P., Zentgraf, U. (2004). Targets of the wrky53 transcription factor and its role during leaf senescence in arabidopsis. Plant Mol. Biol. 55 (6), 853–867. doi: 10.1007/s11103-005-2142-1
Nabi, R. B. S., Tayade, R., Hussain, A., Adhikari, A., Lee, I.-J., Loake, G. J., et al. (2021). A novel DUF569 gene is a positive regulator of the drought stress response in arabidopsis. Int. J. Mol. Sci. 22 (10), 5316. doi: 10.3390/ijms22105316
Naing, A. H., Kim, C. K. (2021). Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiologia Plantarum 172 (3), 1711–1723. doi: 10.1111/ppl.13373
Nie, Z., Zhao, T., Liu, M., Dai, J., He, T., Lyu, D., et al. (2019). Molecular mapping of a novel male-sterile gene msNJ in soybean [Glycine max (L.) Merr.]. Plant Reprod. 32 (4), 371–380. doi: 10.1007/s00497-019-00377-6
Ogiso-Tanaka, E., Shimizu, T., Hajika, M., Kaga, A., Ishimoto, M. (2019). Highly multiplexed AmpliSeq technology identifies novel variation of flowering time-related genes in soybean (Glycine max). DNA Res. 26 (3), 243–260. doi: 10.1093/dnares/dsz005
Pandey, G. K., Yadav, N. S., Singh, V. K., Singh, D., Jha, B. (2014). A novel gene sbSI-2 encoding nuclear protein from a halophyte confers abiotic stress tolerance in E. coli Tobacco. PloS One 9 (7), 31–8. doi: 10.1371/journal.pone.0101926
Parra, L., Nortman, K., Sah, A., Truco, M. J., Ochoa, O., Michelmore, R. (2020). Identification and mapping of new genes for resistance to downy mildew in lettuce. Theor. Appl. Genet. 134 (2), 519–528. doi: 10.1007/s00122-020-03711-z
Qi, Y., Li, J., Mapuranga, J., Zhang, N., Chang, J., Shen, Q., et al. (2023). Wheat leaf rust fungus effector Pt13024 is avirulent to TcLr30. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1098549
Rhoads, A., Au, K. F. (2015). PacBio sequencing and its applications. Genomics Proteomics Bioinf. 13 (5), 278–289. doi: 10.1016/j.gpb.2015.08.002
Saika, H., Ohtsu, K., Hamanaka, S., Nakazono, M., Tsutsumi N, H. A. (2002). AOX1c, a novel rice gene for alternative oxidase; comparison with rice AOX1a and AOX1b. Genes Genet. Syst. 77 (1), 1–17. doi: 10.1266/ggs.77.31
Sara, F., Torres, A. M, Moreno, M.T, Diego, R. (2007). Identification of a new gene for resistance to powdery mildew in pisum fulvum, a wild relative of pea. Breed. ence 57 (2), 181–184. doi: 10.1270/jsbbs.57.181
Shi, J., An, H. L., Zhang, L., Gao, Z., Guo, X. Q. (2010). GhMPK7, a novel multiple stress-responsive cotton group C MAPK gene, has a role in broad spectrum disease resistance and plant development. Plant Mol. Biol. 74. doi: 10.1007/s11103-010-9661-0
Siddiqui, H. A., Asad, S., Naqvi, R. Z., Asif, M., Liu, C., Liu, X., et al. (2022). Development and evaluation of triple gene transgenic cotton lines expressing three genes (Cry1Ac-Cry2Ab-EPSPS) for lepidopteran insect pests and herbicide tolerance. Sci. Rep. 12 (1), 18422. doi: 10.1038/s41598-022-22209-w
Silke, R., Somssich, I. E. (2001). A new member of the arabidopsis wrky transcription factor family, atwrky6, is associated with both senescence- and defence-related processes. Plant J. 28, (2). doi: 10.1046/j.1365-313x.2001.01131.x
Singh, R., Mujeeb-Kazi, A., Huerta-Espino, J. (1998). Lr46: a gene conferring slow-rusting resistance to leaf rust in wheat. Phytopathol. 88, (9). doi: 10.1094/PHYTO.1998.88.9.890
Slewinski, T. L. (2011). Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: a physiological perspective. Mol. Plant 4 (4), 641–662. doi: 10.1093/mp/ssr051
Soda, N., Kushwaha, H. R., Soni, P., Singla-Pareek, S. L., Pareek, A. (2013). A suite of new genes defining salinity stress tolerance in seedlings of contrasting rice genotypes. Funct. Integr. Genomics 13 (3), 351–365. doi: 10.1007/s10142-013-0328-1
Song, H., Duan, Z., Wang, Z., Li, Y., Wang, Y., Li, C., et al. (2022). Genome-wide identification, expression pattern and subcellular localization analysis of the JAZ gene family in Toona ciliata. Ind. Crops Products 178, 151. doi: 10.1016/j.indcrop.2022.114582
Sun, M., Xu, Q.-Y., Zhu, Z.-P., Liu, P.-Z., Yu, J.-X., Guo, Y.-X., et al. (2023). AgMYB5, an MYB transcription factor from celery, enhanced β-carotene synthesis and promoted drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 23 (1), 133–154. doi: 10.1186/s12870-023-04157-3
Sundström, G., Larsson, T. A., Brenner, S., Venkatesh, B., Larhammar, D. (2008). Evolution of the neuropeptide Y family: New genes by chromosome duplications in early vertebrates and in teleost fishes. Gen. Comp. Endocrinol. 155 (3), 705–716. doi: 10.1016/j.ygcen.2007.08.016
Traber, G., Yu, A. (2023). RNAi-based therapeutics and novel RNA bioengineering technologies. J. Pharmacol. Exp. Ther. 384 (1). doi: 10.1124/jpet.122.001234
Verma, V., Ravindran, P., Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16 (1), 179. doi: 10.1186/s12870-016-0771-y
Vincent, N., Osteras, M., Otten, P., Leclerc, M. (2014). A new gene in A. rubens: a sea star Ig kappa gene. Meta Gene 2, 320–322. doi: 10.1016/j.mgene.2014.03.005
Wang, J., Han, G., Liu, H., Yan, H., Jin, Y., Cao, L., et al. (2023). Development of novel wheat-rye 6RS small fragment translocation lines with powdery mildew resistance and physical mapping of the resistance gene PmW6RS. Theor. Appl. Genet. 136 (9), 4357. doi: 10.1007/s00122-023-04433-8
Wang, D., Liu, H., Wang, H., Zhang, P., Shi, C. (2020). A novel sucrose transporter gene IbSUT4 involves in plant growth and response to abiotic stress through the ABF-dependent ABA signaling pathway in Sweetpotato. BMC Plant Biol. 20 (1), 157. doi: 10.1186/s12870-020-02382-8
Wang, G., Wang, X., Ma, H., Fan, H., Lin, F., Chen, J., et al. (2022a). PcWRKY11, an II-d WRKY Transcription Factor from Polygonum cuspidatum, Enhances Salt Tolerance in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 23 (8), 5401. doi: 10.3390/ijms23084357
Wang, X., Wang, B., Yuan, F. (2022b). Lb1G04202, an uncharacterized protein from recretohalophyte limonium bicolor, is important in salt tolerance. Int. J. Mol. Sci. 23 (10). doi: 10.3390/ijms23105401
Wang, W., Yuan, Y., Yang, C., Geng, S., Sun, Q., Long, L., et al. (2016). Characterization, expression, and functional analysis of a novel NAC gene associated with resistance to verticillium wilt and abiotic stress in cotton. G3 Genes|Genomes|Genetics 6 (12), 3951–3961. doi: 10.1534/g3.116.034512
Wang, X., Zhou, Y., Xu, Y., Wang, B., Yuan, F. (2021). A novel gene LbHLH from the halophyte Limonium bicolor enhances salt tolerance via reducing root hair development and enhancing osmotic resistance. BMC Plant Biol. 21 (1), 284. doi: 10.1186/s12870-021-03094-3
Wang, L., Zhu, C., Jin, L., Xiao, A., Duan, J., Ma, L. (2018). A novel gene of Kalanchoe daigremontiana confers plant drought resistance. Sci. Rep. 8 (1), 2547. doi: 10.1038/s41598-018-20687-5
Wu, Q., Chen, Y., Li, B., Li J, Z. P., Xie, J., Zhang, H., et al. (2022). Functional characterization of powdery mildew resistance gene MlIW172, a new Pm60 allele and its allelic variation in wild emmer wheat. . J. Genet. Genomics 49, (8). doi: 10.1016/j.jgg.2022.01.010
Xu, Y., Jiao, X., Wang, X., Zhang, H., Wang, B., Yuan, F. (2020). Importin-beta From the Recretohalophyte Limonium bicolor Enhances Salt Tolerance in Arabidopsis thaliana by Reducing Root Hair Development and Abscisic Acid Sensitivity. Front. Plant Sci. 11, 4846. doi: 10.3389/fpls.2020.582459
Xu, X., Kolmer, J., Li, G., Tan, C., Carver, B. F., Bian, R., et al. (2022a). Identification and characterization of the novel leaf rust resistance gene Lr81 in wheat. Theor. Appl. Genet. 135 (8), 2725–2734. doi: 10.1007/s00122-022-04145-5
Xu, X., Li, G., Cowger, C., Bai, G., Carver, B. F., Bian, R., et al. (2023). Identification of a novel Pm65 allele conferring a wide spectrum of resistance to powdery mildew in wheat accession PI 351817. Phytopathology. doi: 10.1094/PHYTO-01-23-0032-R
Xu, X., Mornhinweg, D., Bai, G., Li, G., Bian, R., Bernardo, A., et al. (2022b). Characterization of Rsg3, a novel greenbug resistance gene from the Chinese barley landrace PI 565676. Plant Genome 16 (1). doi: 10.1002/tpg2.20287
Yang, Z., Sun, X., Wang, S., Zhang, Q. (2003). Genetic and physical mapping of a new gene for bacterial blight resistance in rice. Theor. Appl. Genet. 106 (8), 1467–1472. doi: 10.1007/s00122-003-1205-4
Yao, C., Li, W., Liang, X., Ren, C., Liu, W., Yang, G., et al. (2022). Molecular Cloning and Characterization of MbMYB108, a Malus baccata MYB Transcription Factor Gene, with Functions in Tolerance to Cold and Drought Stress in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 23 (9), 107–22. doi: 10.3390/ijms23094846
Yeku, O., Frohman, M. (2011). Rapid amplification of cDNA ends (RACE). Methods Mol. Biol. 703, 278. doi: 10.1007/978-1-59745-248-9_8
Ying, S., Scheible, W. R., Lundquist, P. K. (2023). A stress-inducible protein regulates drought tolerance and flowering time in Brachypodium and Arabidopsis. Plant Physiol. 191 (1), 643–659. doi: 10.1093/plphys/kiac486
Yuan, F., Lyu, M. J., Leng, B. Y., Zheng, G. Y., Feng, Z. T., Li, P. H., et al. (2015). Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 38 (8), 1637–1657. doi: 10.1111/pce.12514
Yuan, F., Lyu, M. J., Leng, B. Y., Zhu, X. G., Wang, B. S. (2016). The transcriptome of NaCl-treated Limonium bicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Mol. Biol. 91 (3), 241–256. doi: 10.1007/s11103-016-0460-0
Yuan, X., Wang, H., Cai, J., Bi, Y., Li, D., Song, F. (2019). Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 19 (1), 4189. doi: 10.1186/s12870-019-1883-y
Yuan, F., Wang, X., Zhao, B., Xu, X., Shi, M., Leng, B., et al. (2022). The genome of the recretohalophyte Limonium bicolor provides insights into salt gland development and salinity adaptation during terrestrial evolution. Mol. Plant 15 (6), 1024–1044. doi: 10.1016/j.molp.2022.04.011
Zhan, J., Zhong, J., Cheng, J., Wang, Y., Hu, K. (2023). Map-based cloning of the APRR2 gene controlling green stigma in bitter gourd (Momordica charantia). Front. Plant Sci. doi: 10.3389/fpls.2023.1128926
Zhang, J., Chen, L., Cai, Y., Su, Q., Chen, Y., Li, M., et al. (2023a). A novel MORN-motif type gene GmMRF2 controls flowering time and plant height of soybean. Int. J. Biol. Macromol 245, 125464. doi: 10.3390/ijms24044189
Zhang, M., Dong, R., Huang, P., Lu, M., Feng, X., Fu, Y., et al. (2023b). Novel seed size: a novel seed-developing gene in glycine max. Int. J. Mol. Sci. 24 (4), 245. doi: 10.3390/ijms24044189
Zhang, Q., Liu, Y., Jiang, Y., Li, A., Cheng, B., Wu, J. (2022a). OsASR6 enhances salt stress tolerance in rice. Int. J. Mol. Sci. 23 (16), 11. doi: 10.3390/ijms23169340
Zhang, Y., Qin, Y., Li, D., Wang, W., Gao, X., Hao, C., et al. (2023c). Fine mapping and cloning of a novel BrSCC1 gene for seed coat color in Brassica rapa L. Theor. Appl. Genet. 136 (1), 9340. doi: 10.1007/s00122-023-04287-0
Zhang, X., Xue, Y., Wang, H., Nisa, Z. U., Jin, X., Yu, L., et al. (2022b). Genome-wide identification and characterization of NHL gene family in response to alkaline stress, ABA and MEJA treatments in wild soybean (Glycine soja). PeerJ 10, e14451. doi: 10.7717/peerj.14451
Zhang, T., Zhou, J., Gao, W., Jia, Y., Wei, Y., Wang, G. (2022). Complex genome assembly based on long-read sequencing. Brief Bioinform. 23 (5), 1. doi: 10.1093/bib/bbac305
Zhao, R., Liu, B., Wan, W., Jiang, Z., Chen, T., Wang, L., et al. (2023b). Mapping and characterization of a novel adult-plant leaf rust resistance gene LrYang16G216 via bulked segregant analysis and conventional linkage method. Theor. Appl. Genet. 136 (1). doi: 10.1007/s00122-023-04270-9
Zhao, M., Morohashi, K., Hatlestad, G., Grotewold, E., Lloyd, A. (2008). The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135 (11), 1991–1999. doi: 10.1242/dev.016873
Zhao, Y., Qin, Q., Chen, L., Long, Y., Song, N., Jiang, H., et al. (2022). Characterization and phylogenetic analysis of multiple C2 domain and transmembrane region proteins in maize. BMC Plant Biol. 22 (1), 388. doi: 10.1186/s12870-022-03771-x
Zhao, B., Zhou, Y., Jiao, X., Wang, X., Wang, B., Yuan, F. (2023a). Bracelet salt glands of the recretohalophyte Limonium bicolor: Distribution, morphology, and induction. J. Integr. Plant Biol. 65 (4), 950–966. doi: 10.1111/jipb.13417
Zhou, Y., Hu, L., Song, J., Jiang, L., Liu, S. (2019). Isolation and characterization of a MADS-box gene in cucumber (Cucumis sativus L.) that affects flowering time and leaf morphology in transgenic Arabidopsis. Biotechnol. Biotechnol. Equip. 33 (1), 54–63. doi: 10.1080/13102818.2018.1534556
Zhu, J.-K. (2016). Abiotic stress signaling and responses in plants. Cell 167 (2), 313–324. doi: 10.1016/j.cell.2016.08.029
Keywords: function, growth, stress resistance, structure, unknown/uncharacterized genes
Citation: Wang X, Wang B and Yuan F (2023) Deciphering the roles of unknown/uncharacterized genes in plant development and stress responses. Front. Plant Sci. 14:1276559. doi: 10.3389/fpls.2023.1276559
Received: 12 August 2023; Accepted: 08 November 2023;
Published: 23 November 2023.
Edited by:
Muthusamy Ramakrishnan, Nanjing Forestry University, ChinaReviewed by:
Mingzhe Sun, Heilongjiang Bayi Agricultural University, ChinaDeguo Han, Northeast Agricultural University, China
Copyright © 2023 Wang, Wang and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baoshan Wang, YnN3YW5nQHNkbnUuZWR1LmNu; Fang Yuan, eXVhbmZhbmdAc2RudS5lZHUuY24=
 Xi Wang
Xi Wang Baoshan Wang
Baoshan Wang Fang Yuan
Fang Yuan