- 1Key Laboratory of Specialty Agri-products Quality and Hazard Controlling Technology of Zhejiang Province, College of Life Sciences, China Jiliang University, Hangzhou, Zhejiang, China
- 2Institute of Crop Science and Zhejiang Key Laboratory of Crop Germplasm, Zhejiang University, Hangzhou, China
- 3Mizuda Group Co., Ltd., Huzhou, Zhejiang, China
- 4Institute of Vegetables, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
The Brassica napus (B. napus) LOR (Lurp-One-Related) gene family is a little-known gene family characterized by a conserved LOR domain in the proteins. Limited research in Arabidopsis showed that LOR family members played important roles in Hyaloperonospora parasitica (Hpa) defense. Nevertheless, there is a paucity of research investigating the role of the LOR gene family towards their responses to abiotic stresses and hormone treatments. This study encompassed a comprehensive survey of 56 LOR genes in B. napus, which is a prominent oilseed crop that holds substantial economic significance in China, Europe, and North America. Additionally, the study evaluated the expression profiles of these genes in response to salinity and ABA stress. Phylogenetic analysis showed that 56 BnLORs could be divided into 3 subgroups (8 clades) with uneven distribution on 19 chromosomes. 37 out of 56 BnLOR members have experienced segmental duplication and 5 of them have undergone tandem repeats events with strong evidence of purifying selection. Cis-regulatory elements (CREs) analysis indicated that BnLORs involved in process such as light response, hormone response, low temperature response, heat stress response, and dehydration response. The expression pattern of BnLOR family members revealed tissue specificity. RNA-Seq and qRT-PCR were used to validate BnLOR gene expression under temperature, salinity and ABA stress, revealing that most BnLORs showed inducibility. This study enhanced our comprehension of the B. napus LOR gene family and could provide valuable information for identifying and selecting genes for stress resistant breeding.
Introduction
Abiotic stress can reduce plant growth and development via inhibiting cell division and elongation, reducing leaf expansion, and slowing down metabolic processes (Suzuki et al., 2014). Consequently, plants may exhibit stunted growth and a decrease in biomass. Among them, salt stress could lead to osmotic stress, ion toxicity and nutrient imbalance (Raza et al., 2022; Sheteiwy et al., 2022). Abscisic acid (ABA) treatment could lead to the inhibition of growth, stomatal closure, and senescence (Waadt et al., 2022). Salt stress and ABA are known to have significant effects on rapeseed (Brassica napus). Salt stress induces oxidative stress and disrupts ion homeostasis in rapeseed, resulting in reduced growth, photosynthesis, and yield (Shu et al., 2018). ABA plays a crucial role in mediating the response of rapeseed to salt stress, by regulating stomatal closure, antioxidant defense, and osmotic adjustment (Zhu and Assmann, 2017). Salt stress and ABA also modulate the expression of stress-related genes, including those involved in ion transport, osmotic regulation, and stress signaling pathways in rapeseed (Raza et al., 2023). Furthermore, salt stress and ABA have been shown to interact with other signaling molecules, such as calcium and reactive oxygen species (ROS), to modulate the physiological and molecular responses of rapeseed to stress (Raza et al., 2023).
The LOR (Lurp-One-Related) gene family is widely presented in various organisms, including plants, fungi, bacteria, and certain archaea. LOR family members are characterized by a conserved LOR domain in the protein (Knoth and Eulgem, 2008). Members of this gene family typically exhibit a 12-stranded β-barrel structure that encompasses a central C-terminal α-helix, similar to the C-terminal domain observed in Tubby proteins (Bateman et al., 2009). β-barrel and α-helix are structural motifs commonly found in proteins that play important roles in gene structure organization. β-barrel structures are characterized by a cylindrical shape formed by β-strands arranged in a closed loop, while α-helices refer to α-helices located at the C-terminus of a protein. These structural elements are involved in protein-protein interactions, protein stability, and protein-DNA interactions, influencing gene expression, transcriptional regulation, and chromatin organization (Yin et al., 2017). The LOR gene family was initially named after the first member discovered in Arabidopsis thaliana, LURP1 (Late up-regulated in response to Hyaloperonospora parasitica). LURP1 belongs to the LURP cluster and plays a pivotal role in plant defense against pathogenic bacteria. Knoth and Eulgem (2008) demonstrated that LURP1 is indispensable in the defense response against Hyaloperonospora parasitica (Hpa) in Arabidopsis. Moreover, the promoter of AtLURP1 contain a W box and two TGA boxes, which could mediate gene expression in response to Hpa and salicylic acid (SA) (Knoth and Eulgem, 2008). Later Caillaud et al. (2016) proved that AtLURP1 could respond to SA and function as a marker gene of SA triggered immunity. Gupta and Senthil-Kumar (2017) conducted a global transcriptome profiling of Arabidopsis investigating their response to Pseudomonas syringae infection during drought recovery stage. They found that the transcription level of AtLURP1 was up-regulated in response to pathogen infection, but down-regulated during drought stress or at the time of drought recovery. Lee et al. (2017) found that AtLURP1 was the target gene of NAC4 transcription factor (TF) and could negatively regulate cell death caused by plant pathogen infection. AtLURP1 also participated in plant defense against the widespread pest Myzus persicae (Bricchi et al., 2012). Apart from AtLURP1, AtLOR1 is another gene that has been extensively studied. Baig (2018) reported that AtLOR1 displayed constitutive expression and played a significant role in basal defense against Hpa. These findings highlight the potential important roles of LOR gene family in plant in response to environmental stimuli. However, apart from the AtLURP1 and AtLOR1, the expression pattern and biological function of other members are still unknown.
Brassica napus (B. napus, AACC, 2n=4x=38), also called rapeseed, is a prominent oilseed crop that holds substantial economic significance in China, Europe, and North America, with extensive applications as oilseeds, vegetable, biofuel, and fodder (Yang et al., 2022). As a representative species of Brassicaceae, B. napus was originated from the spontaneous hybridization between Brassica rapa (B. rapa, AA, 2n=2x=20) and Brassica oleracea (B. oleracea, CC, 2n=2x=18) at nearly 7500 years ago, followed by allopolyploidization (Yang et al., 2016; Yang et al., 2018; Yang et al., 2020). With the continuous development of sequencing technology, the reference genomes of Darmor-bzh (winter ecotype), Zhongshuang 11 and NY7 (semi-winter ecotype) had been successfully sequenced and assembled (Chalhoub et al., 2014; Sun et al., 2017; Zou et al., 2019), but no systematic analysis and function prediction about the LOR gene family in B. napus had been reported yet. A comprehensive investigation of the BnLOR gene family could shed light on the evolutionary mechanisms of allopolyploidization between B. rapa and B. oleracea and provide a theoretical foundation for future studies on the roles of BnLOR genes.
In this study, 56 LOR family members were identified in the B. napus reference genome. The phylogenetic relationship, chromosomal distributions, protein conserved motifs, gene structures, whole genome duplication (WGD) events, synteny relationship, and putative cis-regulatory elements (CREs) in promoter regions were systematically analyzed. Besides, the expression pattern of BnLORs in different tissue as well as under salt stress and ABA treatment was analyzed. Results showed that most BnLOR genes could be induced by salt stress and ABA treatment, suggesting their potential function on salinity and ABA stress responsiveness. These results could provide useful information for further investigation about the function and molecular mechanisms of BnLOR genes towards abiotic salinity and ABA stress.
Materials and methods
Sequence extraction and domain identification
The reference genome and protein sequences of B. napus were downloaded from http://yanglab.hzau.edu.cn/BnIR/ while the reference genome of B. rapa, B. oleracea and Arabidopsis was obtained from http://plants.ensembl.org/index.html. The Hidden Markov Model (HMM) file of the LOR domain was acquired from Pfam protein family database (http://pfam.xfam.org/) (El-Gebali et al., 2019). LOR genes were searched from B. napus reference genome using HMMER 3.3 (Potter et al., 2018). The preliminarily identified LOR family members from B. napus (BnLOR) were validated by blasting with LOR gene family from A. thaliana as queries and all candidate genes were further validated at CDD (https://www.ncbi.nlm.nih.gov/cdd/), Pfam and SMART (http://smart.embl.de/) to verify the existence of LOR domain. The length of the BnLOR protein sequences as well as the molecular weight (Mw) and the isoelectric points (pI) were predicted by the pI/Mw tool in ExPASy (http://www.expasy.ch/tools/pi_tool.html). Besides, the subcellular location of BnLOR genes were predicted by the Cell-PLoc web-server (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc/).
Multiple sequence alignment and phylogenetic analysis
To identify the evolutionary relationship among LOR gene family from different species, the protein sequences of LORs from Arabidopsis, B. rapa, B. oleracea, and B. napus were aligned by the Clustal W algorithm with default parameters (Larkin et al., 2007). The ALN file was converted into MEGA 6.0 to build a neighbor-joining (NJ) phylogenetic tree with following parameters: Poisson model, and pairwise deletion for the reliability of interior branches (Tamura et al., 2013). The phylogenetic tree was modified and displayed using the iTol tool (https://itol.embl.de/).
Gene structures and protein conserved motifs analysis
The gene structures and protein conserved motifs of the BnLOR family members were analyzed in this study. The exon-intron structures of each BnLOR gene were extracted using gene structure view (advanced) in TBtools (Chen et al., 2020), which provided a graphical representation of the gene structures. The conserved motifs within the BnLOR protein sequences were identified using the Multiple Em for Motif Elicitation program (MEME, https://meme-suite.org/meme/) with default parameters set for maximum number of motifs, distribution of motif occurrences, and optimum motif width (Bailey et al., 2009). The identified exon-intron structures and conserved motifs were then visualized by TBtools (Chen et al., 2020).
Gene distribution and WGD visualization
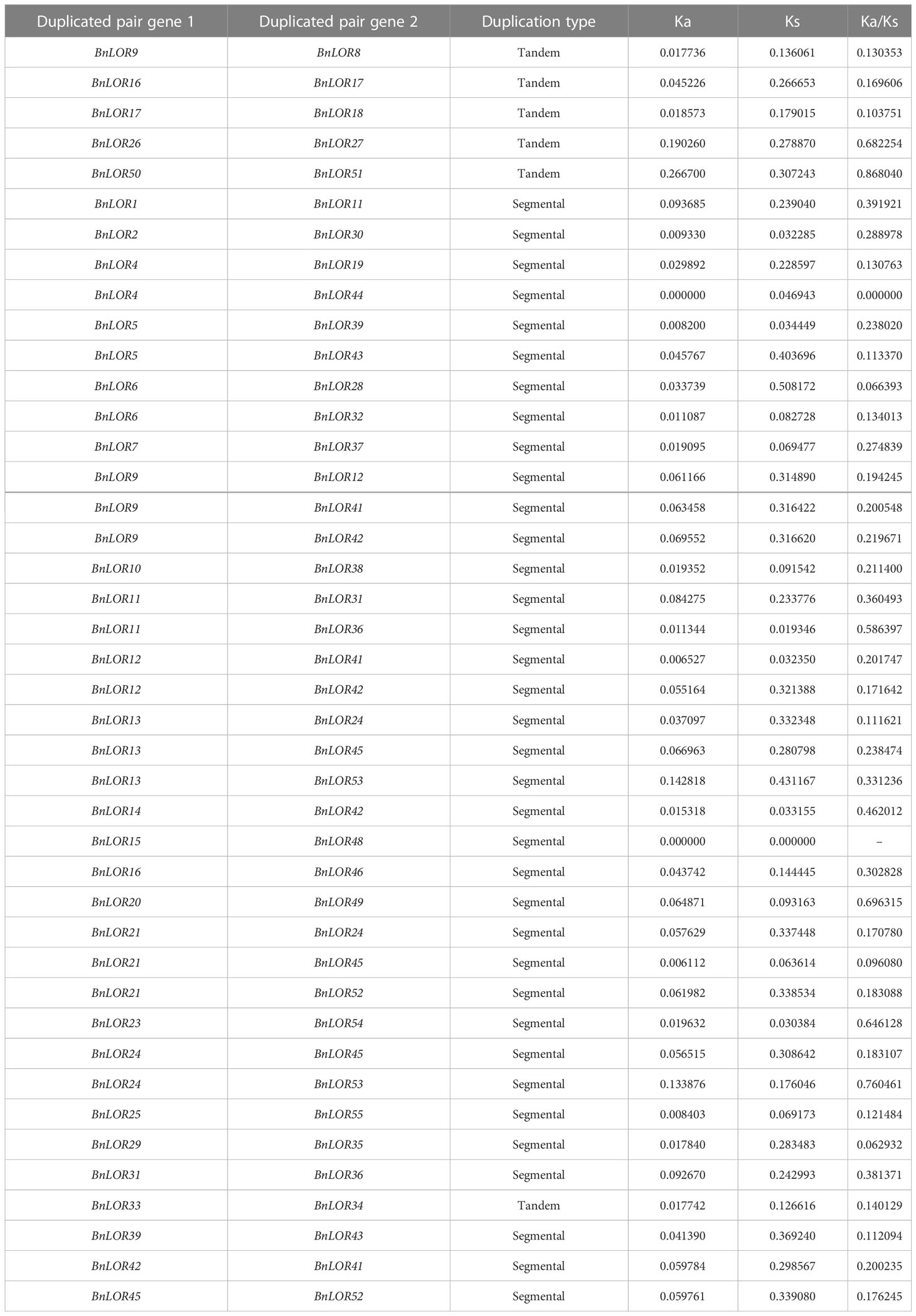
The chromosomal positions and relative distances of the BnLOR family members were drafted using TBtools (Chen et al., 2020). The WGD landscape within B. napus was generated by MCScanX (Wang et al., 2012). To confirm WGD events, the shorter aligned sequence was required to cover over 70% of the longer sequence and the similarity of the aligned regions had to be over 70% (Gu et al., 2002; Yang et al., 2008). WGD events includes segmental duplication and tandem repeats. Tandem repeats are defined as two genes located in the same chromosomal fragment with a length of less than 100 kb (Wang et al., 2010b). Segmental duplications refer to two genes experienced polyploidization followed by chromosome rearrangements (Yu et al., 2005). The synteny map was created using Circos (Krzywinski et al., 2009). Additionally, the MCScanX was used to show the collinearity relationship between B. napus, B. rapa and B. oleracea. Finally, the KaKs_Calculator 2.0 program was utilized to calculate the non-synonymous (ka) and synonymous (ks) substitutions of each duplicated LOR gene (Wang et al., 2010a).
CREs prediction in the promoter regions
The promoter sequence (about 2000 bp upstream of the translation start codon) of BnLOR family members were extracted from the B. napus reference genome. PlantCARE program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict the putative CRE responsive to transcription binding sites, hormone treatments, biotic and abiotic stresses. PlantRegMap program (http://plantregmap.gao-lab.org/) was used to predict the potential TFs using B. napus as the target species (Jin et al., 2017).
Orthologous gene clusters among multiple species
To identify orthologous gene clusters among multiple species, the reference genomes of eight commonly studied species including Arabidopsis, B. oleracea, B. rapa, B. napus, Glycine max (G. max), Oryza sativa (O. sativa), Triticum aestivum (T. aestivum) and Zea mays (Z. mays) were analyzed using the Ortho Venn 2.0 tool (https://orthovenn2.bioinfotoolkits.net/home). The e-value threshold and the inflation value were set at 1e-5 and 1.5, respectively. The protein sequences of LORs in these species were uploaded, analyzed, and visualized. (Wang et al., 2022; Wang et al., 2023).
Gene expression patterns analysis
To determine the gene expression pattern of BnLOR gene family, transcriptome file of various tissues including blossomy pistil, flower, leaf, ovule, pericarp, pistil, root, silique, sepal, stamen, stem, and wilting pistil were obtained from a previous study by Sun et al. (2017) under the project ID of PRJNA394926 in the NCBI. Moreover, transcriptome data of B. napus under dehydration, salt stress, ABA treatment and cold stress conditions were obtained from Zhang et al. (2019) under the project ID of CRA001775. Differential expression analysis of BnLOR genes was performed using the DSEeq2 R package and the heatmaps were created by TBtools software (Chen et al., 2020).
Comparative modeling of BnLOR proteins
Homology modeling was used to create the three-dimensional (3D) structures of BnLOR proteins from their amino acid sequences. The amino acid sequences of BnLORs were submitted to SWISS-MODEL program (https://swissmodel.expasy.org/) for model generating. The quality of the models was assessed using ERRAT and PROCHECK tests through SAVES program (https://saves.mbi.ucla.edu/). The resulting 3D structures of BnLOR proteins were visualized using the Pymol software (Karnes and Shaikh, 2021).
Plant materials and treatments
One leading cultivar of B. napus (ZD622) was chosen in this research to validate the relative expression levels of BnLORs under salinity and ABA stress (Ma et al., 2022). Mature and healthy seeds of ZD622 underwent sterilization by submersion in 1% NaClO for 15 minutes. The seeds were thoroughly rinsed with distilled water before being germinated in a petri dish for 24 hours with a damp filter paper placed underneath. After germination, the seedlings were transferred to a 12 cm2 growth box containing full-strength Hoagland’s solution, along with a sponge placed inside. Germinated plants were shifted to growth chamber with 16-hr photoperiod, 24/16 day/night temperature, 300 μmol/m2/s light intensity and 60% to 70% humidity. After one week, uniformly developed seedlings were subjected to salt stress (200 mM) and ABA treatment (25 μM). Leaf samples were immediately frozen using liquid nitrogen and stored in a -80°C refrigerator in preparation for RNA isolation at 0 h, 4 h, and 24 h after treatment. Each sample had at least three replicates.
qRT-PCR validation
The MiniBEST Plant RNA Extraction Kit (Takara, Japan) was employed to isolate total RNA from leaf samples. The PrimeScriptTM RT reagent Kit (Takara, Japan) was used to carry out reverse transcription reactions with 1 μg RNA. The synthesized cDNAs were used as templates for qRT-PCR by employing the SuperReal PreMix Plus/SYBR Green (Tiangen, China) on a Bio-Rad CFX96 (BIO RAD, USA). The qRT-PCR conditions were elucidated in our recently published research (Wang et al., 2022). B. napus actin-7 was used as the internal control gene. All qRT-PCR primers used were listed in Table S1. The relative expression levels were calculated using the 2–ΔΔCt method with three replicates each (Livak and Schmittgen, 2001). Statistical analysis of the data was performed using the SPSS 20.0 statistical package (SPSS, Chicago, IL, USA). One-way variance analysis (ANOVA) was conducted followed by Duncan’s multiple range test (p < 0.05) to determine significant differences. Figures were generated using GraphPad Prism 7.0 (Yang et al., 2022).
Results
Systematic identification and classification of BnLORs
According to the HMM file (PF04525) of LOR gene family, 72 candidate BnLOR genes were identified through model search of the B. napus reference genome. After filtering out the redundant genes and alternative splicing, the existence of LOR domain in candidate genes was further verified by CDD, Pfam and SMART program. As a result, 56 LOR genes in B. napus with complete open reading frame and LOR domain were identified. Their details, along with their encoded proteins, were listed in Tables S2, S3. The predicted length of BnLOR proteins range from 90 amino acids (BnLOR15/48) to 257 amino acids (BnLOR17) (Tables S2, S3). The MWs of BnLOR proteins ranged from 10.49 kDa (BnLOR48) to 29.28 kDa (BnLOR17), suggesting structural variance and functional diversity among members of this gene family (Table S2). The pIs of 15 BnLOR proteins were less than 7, indicating that they were acidic proteins (Table S2). The rest 41 proteins were alkalinity proteins with pIs higher than 7 (Table S2). The largely variance of pIs indicated the possibility of the proteins existing in different parts of cells. The grand average of hydropathicity (GRAVY) prediction results showed that all proteins were hydrophilic (Table S2). The instability index of 25 proteins were lower than 40, indicating that these proteins were relatively stable, while the rest 31 proteins were relatively unstable (Table S2). The aliphatic index of 56 BnLOR proteins fluctuated from 71.48 (BnLOR11) to 100.88 (BnLOR28). Aliphatic index is positively correlated with the thermal stability of protein, suggesting that the thermal stability of the BnLOR proteins was largely variant. Besides, the prediction of subcellular localization of BnLOR proteins exhibited significant variation. Among the 56 identified BnLOR proteins, 25 were exclusively localized in a single compartment such as nucleus, cytoplasm, or chloroplast, while the remaining 31 proteins were found in at least two different subcellular locations. Notably, chloroplast was the most frequent localization site for BnLOR proteins (75.00%), followed by nucleus (48.21%), cytoplasm (33.93%), cell membrane (14.29%), mitochondrion (14.29%), peroxisome (12.50%), cell wall (10.71%), and vacuole (3.57%).
Phylogenetic analysis of Brassicaceae species
The phylogenetic tree among B. napus (56 BnLORs), model plants Arabidopsis (20 AtLORs), ancestral species B. rapa (33 BrLORs) and B. oleracea (30 BoLORs) was constructed by NJ method. According to its topological structures, BnLORs could be divided into three subgroups (Figure 1). Subgroup I was the largest group with 27 BnLOR genes and could be further divided into four clades (Clade A to D) (Figure 1). Subgroup II (Clade E) was the smallest group with only 3 BnLOR genes (Figure 1). Subgroup III had 26 BnLOR genes and could be further divided into three clades (Clade F to H) (Figure 1). Clade A, C and H were relatively large, each contains 10 or more family members. Clade D and E had only 2 or 3 members each while Clade B, F and G had 4 to 8 family members, respectively. Both Subgroup I and III had plenty of members and branches, indicating that the structures and function of these two subgroups were relatively diverse. Each clade had LOR family members from four species (B. napus, Arabidopsis, B. rapa and B. oleracea), indicating that the phylogenic relationship among these four species were relatively close (Figure 1). At the end of the phylogenetic tree branch, there were 46 homologous gene pairs, among which 22 were Bn-Bo homologous gene pairs, 17 were Bn-Br homologous gene pairs and 2 were Br-Bo homologous gene pairs (Figure 1). Bn-Bo and Bn-Br homologous gene pairs could be found in every clade, but their distribution rates were inconsistent. It is noticed that all homologous gene pairs in Clade B, C, D and F belonged to Bn-Bo or Bn-Br homologous gene pairs (Figure 1).
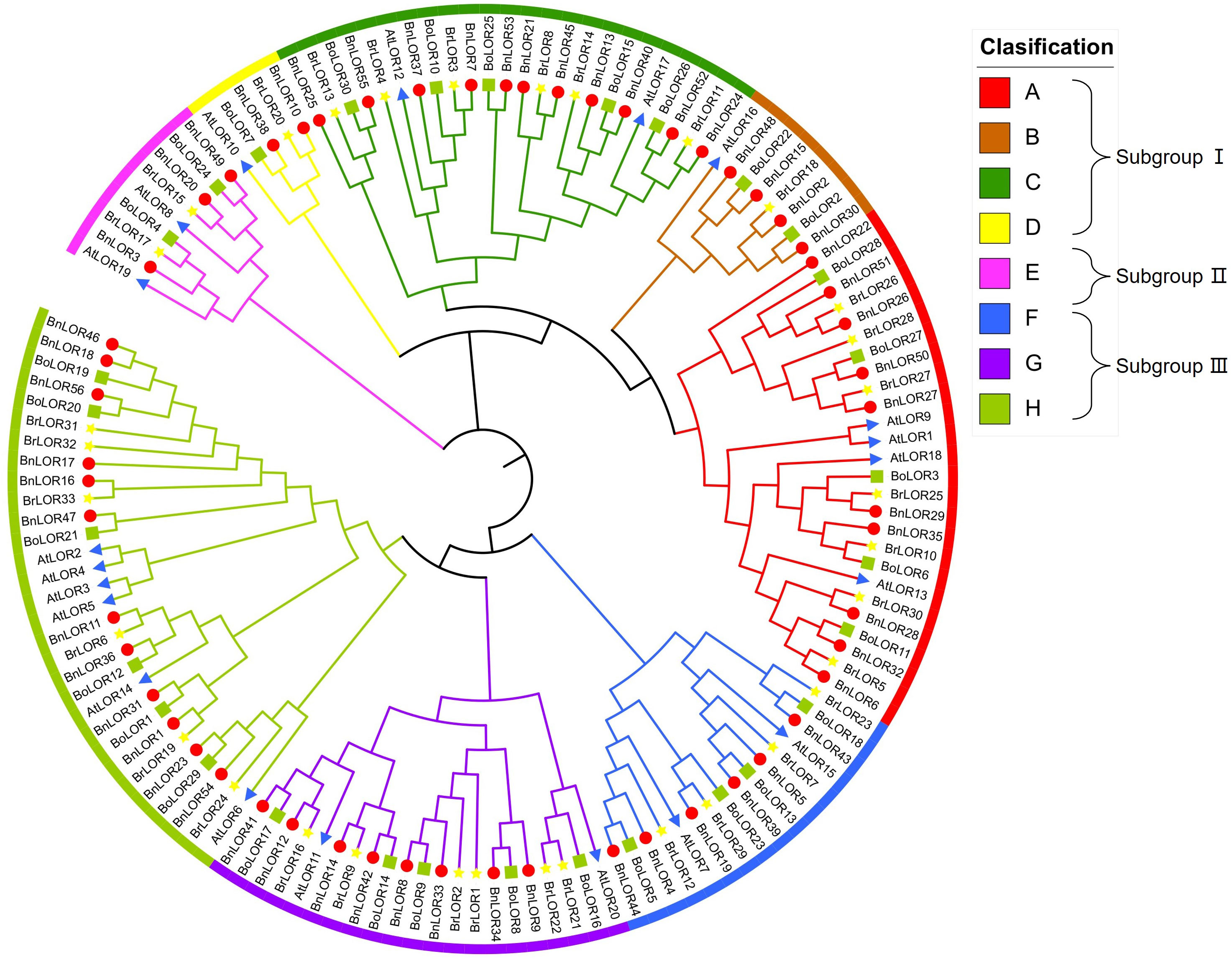
Figure 1 A neighbor-joining (NJ) phylogenetic tree of 142 LOR proteins from B. napus, B. oleracea, B.rapa, and A. thaliana. Overall, 56 BnORs (red circle), 33 BrLORs (yellow star), 30 BoLORs (green box), and 20 AtLORs (blue triangle) were grouped into 3 subgroups (8 clades) based on domain and 1000 bootstrap values.
Gene structures and protein conserved motif analysis
The reference genome sequences of BnLORs were submitted to GSDS server to display their gene structures. Results showed that the exon-intron composition from Clade A to Clade D was similar (Figure 2). Each of them owned 3 exons separated by 2 introns apart from BnLOR48/15/53, which had one intro inside two exons (Figure 2). Meanwhile, the motif structures from Clade E to H were also similar (Figure 2). Most of them contained two exons and one intron with a few exceptions (Figure 2). BnLOR3 and BnLOR54 had three exons separated by two introns; BnLOR47/16/17 only had one exon; BnLOR20 had two exons and two introns (Figure 2). Among all the 56 BnLORs, 19 BnLOR family members had no upstream or downstream non-coding regions regulating the transcription of coding regions (Figure 2).
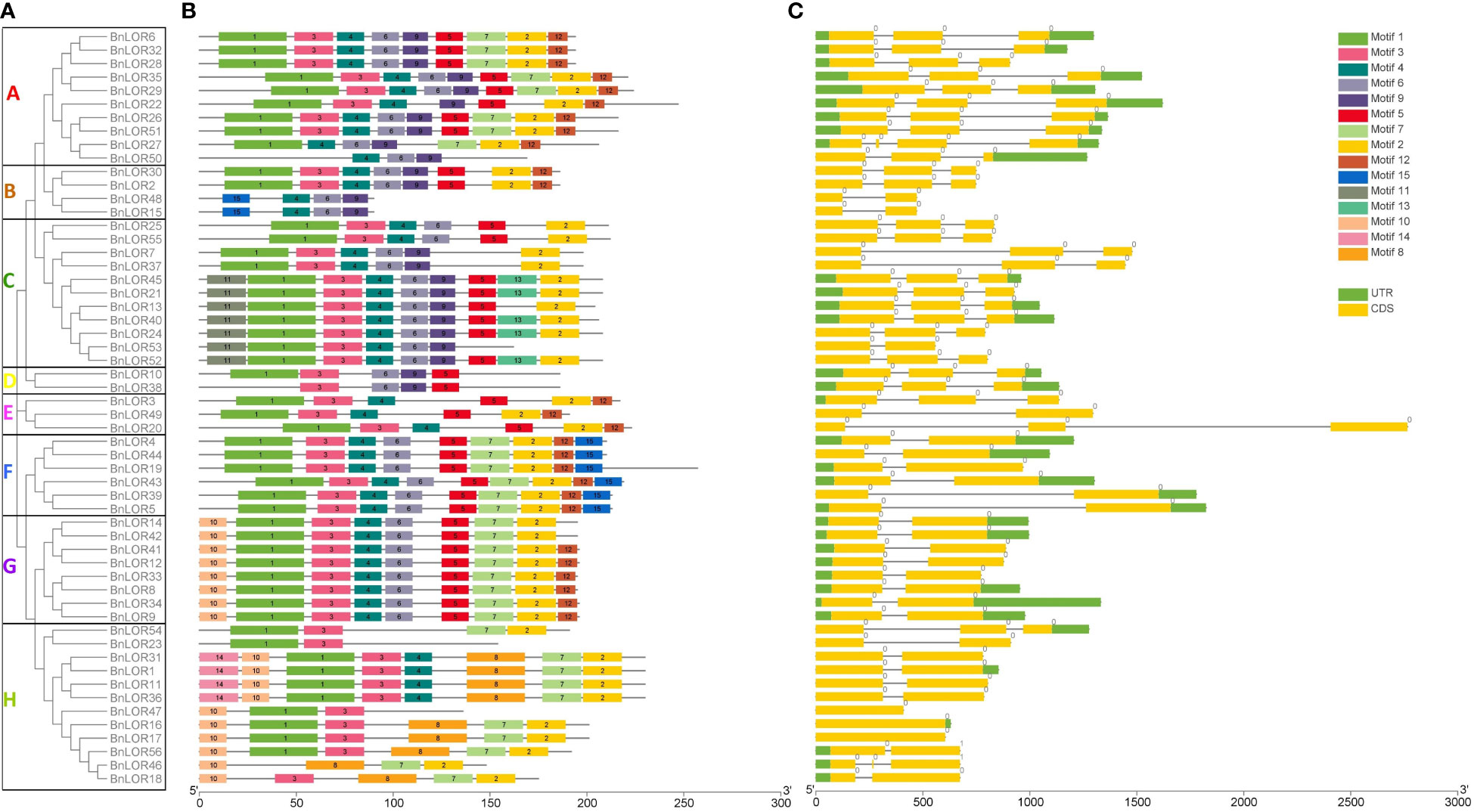
Figure 2 The gene structures and motif analysis of LOR family in B. napus. (A) Based on phylogenetic relationship and domain identification, the BnLORs were grouped into eight clades. (B) Conserved motif compositions identified in BnLOR proteins. 15 Different color boxes denote different motifs. (C) Gene structures of BnLORs. The yellow rectangle represents CDS or exons, and the grey line represents introns. Different numbers indicate the phase of introns.
The reference protein sequences of BnLORs were uploaded to MEME Suite to identify their conserved motif. As a result, the MEME program identified 15 conserved motifs (p<0.05) with the length varied from 11 to 36 amino acids (Figure 2; Supplementary Figure S1; Table S4). The range of conserved motifs in BnLOR proteins was from 4 to 52 (Figure 2). Most of the BnLOR proteins contained motif 3 while only 4 proteins (BnLOR31/1/11/36) contained motif 14 (Figure 2). It was noticed that the motif structures of these BnLOR proteins showed high similarities. Most proteins had motif 1 at the head and motif 2 at the tail (Figure 2). Besides, BnLOR proteins on the same clade or adjacent clade had similar motif structures. For example, all proteins in Clade A have the same motif composition except for BnLOR50 (Figure 2). Each protein in Clade F shared the same motif composition (Figure 2). In Clade G, every protein had the same motif distribution except BnLOR14 and 42, which lacked motif 12 (Figure 2). Most proteins from Clade A to G had motif 6 (Figure 2). At the same time, we also noticed that the distribution of motifs in BnLOR proteins showed specificity. For instance, motif 11 only existed in Clade C while motif 10 only existed in Clade G and H (Figure 2). Motif 15 only appeared in Clade F while motif 13 only appeared in Clade C (Figure 2). Motif 14 only existed in Clade H and there was no motif 12 in Clade C or H (Figure 2).
Gene distribution and WGD visualization
The position of 56 BnLOR gene were retrieved from the B. napus reference genome and their position was visualized and mapped on chromosomes with TBtools (Chen et al., 2020). To facilitate identification, these genes were named as BnLOR1 to BnLOR56 based on their chromosomal location (Figure 3). Our analysis revealed that the distribution of BnLORs across the A and C genomes of B. napus was uneven, with 29 genes located on the A genome and 27 genes located on the C genome (Figure 3). Notably, chromosomes A3 and C3 exhibited the highest number of BnLOR genes, with 7 and 8 genes, respectively (Figure 3). There were 4 genes on chromosome C6 and C8 and 3 genes on chromosome A6, A7, A9, C4 (Figure 3). Two genes were located on chromosome A1, A2, A4, A5, C1, C5 and C9, one BnLOR gene was located on chromosome A10 and C7 while no BnLOR gene was found on chromosome A8 and C2 (Figure 3). For random chromosomes, there were 0 to 3 BnLOR genes, respectively (Figure 3).
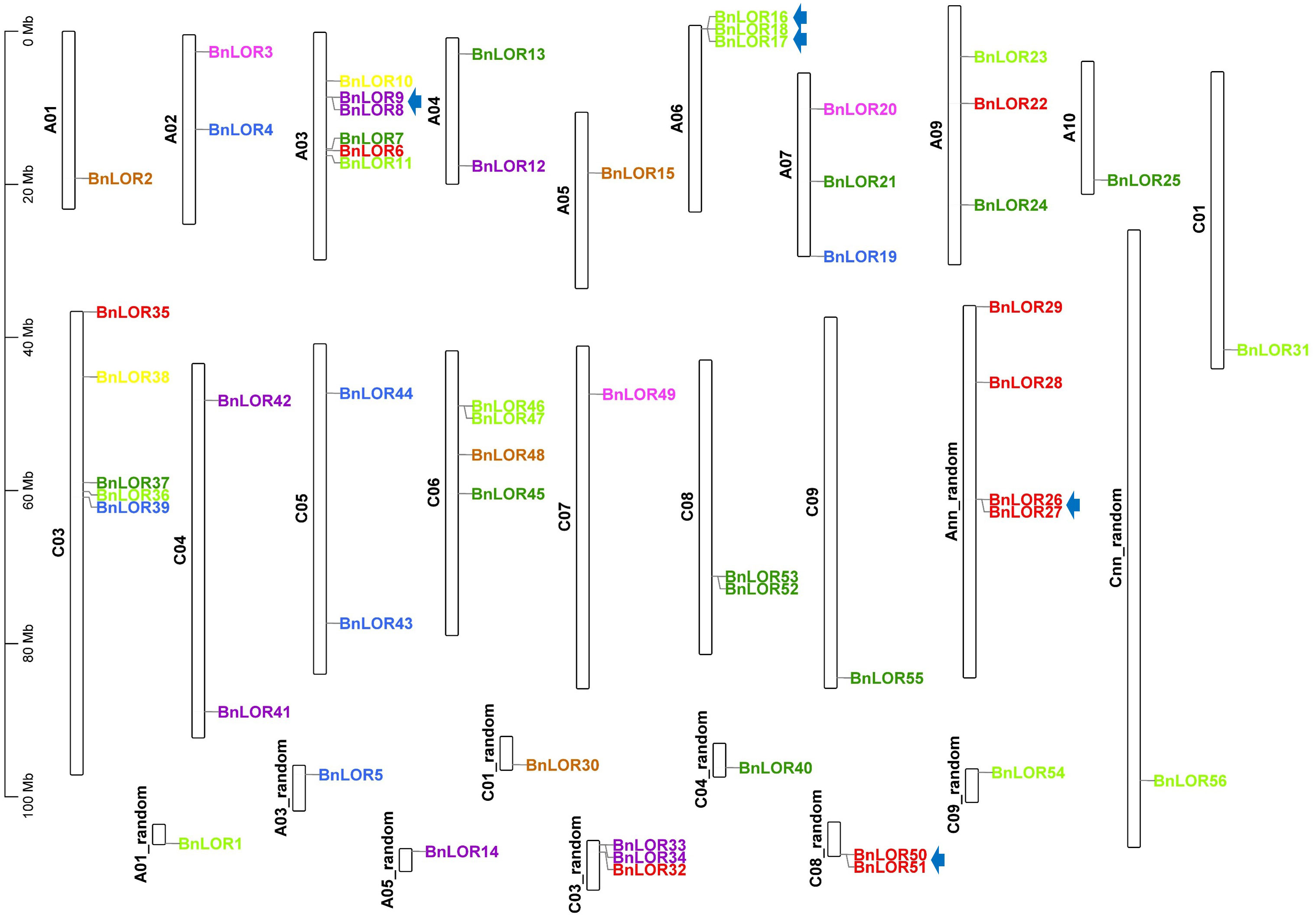
Figure 3 The distribution of LOR genes on B. napus chromosomes. The tandem repeats are highlighted with arrow. Different color of genes is accordance with their group distribution in Figure 1.
WGD has been widely recognized as a crucial impetus underlying the evolution of species, providing the foundation for morphological and physiological changes in plants. WGD events include tandem repeats and segmental duplication. B. napus was originated from the spontaneous hybridization between B. rapa and B. oleracea followed by allopolyploidization, leading to the prevalence of paralogous genes with multiple copies in rapeseed. Thus, in this study, we aimed to gain insights into the expansion of BnLOR genes by analyzing the WGD. As a result, 42 pairs of gene duplication events occurred in 28 BnLOR family members on 19 chromosomes (Figure 4; Table 1). Among them, there were five tandem repeats (BnLOR9/8, BnLOR16/17, BnLOR17/18, BnLOR26/27, BnLOR50/51) (Table 1). The other 37 were all segmental duplication (Table 1). Our findings suggest that BnLOR genes have undergone a considerable number of duplication events throughout the evolutionary process, thereby contributing to the rapid expansion of the BnLOR gene family.
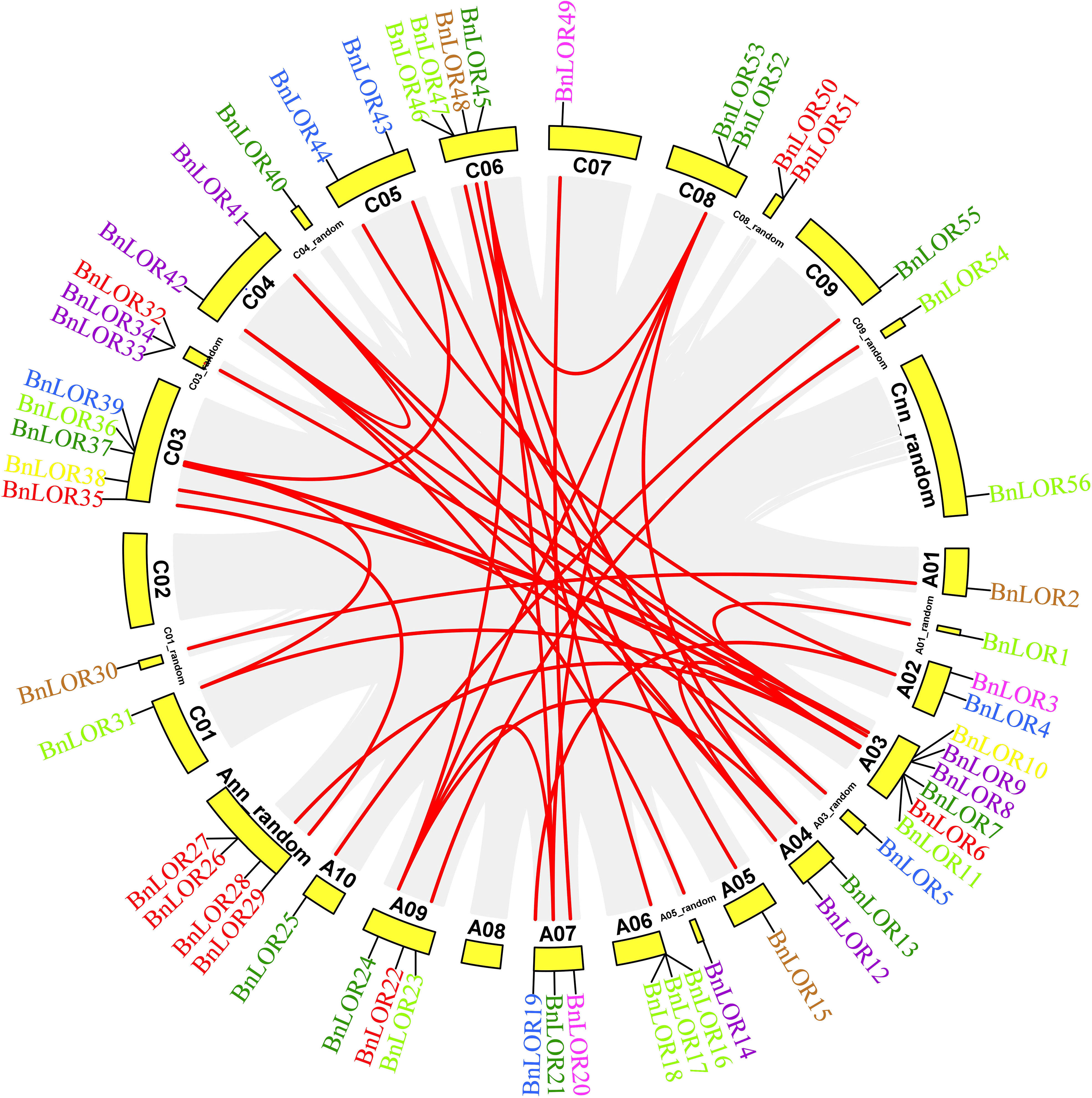
Figure 4 Chromosome locations and inter-chromosomal associations of BnLOR genes. Grey lines in the background display all syntenic blocks in the B. napus genome and the red lines display syntenic BnLOR gene pairs. Different color of genes is accordance with their group distribution in Figure 1.
To elucidate the phylogenetic relationship among LOR family members, we employed MCScanX to conduct collinearity analysis across B. napus, B. rapa, and B. oleracea genomes. Our analysis identified 148 sets of collinearity relationships, including 78 pairs of collinear genes between B. napus and B. rapa and 70 pairs of collinear genes between B. napus and B. oleracea (Figure 5 and Table S5). The collinear genes on each chromosome were distributed on two to several chromosomes. For example, the collinear genes of B. napus on Chromosome A2 were located on Chromosome A2, A7 of B. rapa and on Chromosome C2 and C8 of B. oleracea (Figure 5). Notably, chromosomes A3 and C3 exhibited the largest number of collinear gene pairs.
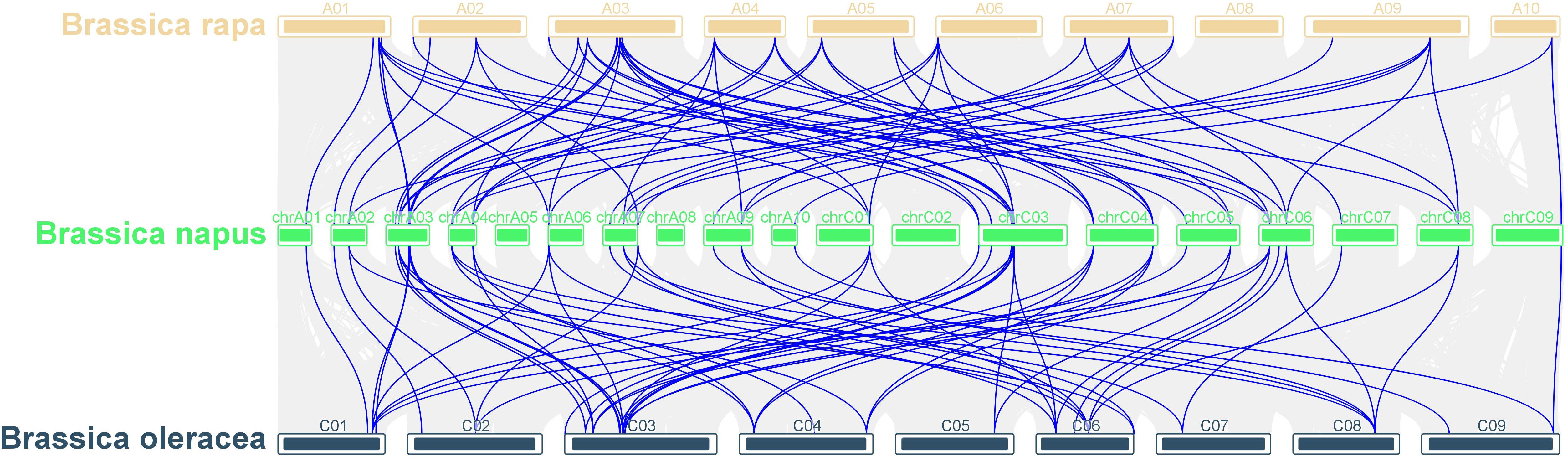
Figure 5 Collinearity analysis of LOR gene family in B. napus, B. rapa and B. oleracea. The grey lines in the background indicate the collinearity blocks within B. napus, B. rapa and B. oleracea, whereas the blue lines indicate the syntenic LOR gene pairs.
To further explore the diversity of duplicated gene pairs during evolution, we calculated the Ka/Ks value. Except for one gene (BnLOR15) that could not be calculated in B. napus, the Ka/Ks value of all the segmental duplications and tandem repeats of BnLOR genes were less than 1, indicating that BnLOR genes were under the pressure of purifying selection during the evolutionary process of B. napus (Table 1).
Putative CREs analysis of BnLOR genes
After uploading the promoter sequence of BnLORs onto PlantCARE web server, a total of 1333 CREs with annotations were discovered. Nearly half (41.34%) of the CREs were associated with light responsiveness, such as AE-box, Box 4, ATCT-motif, GATA-motif, G-Box, GT1-motif, ACE, MRE, GA-motif, TCT-motif, I-box, AT1-motif, etc. 32.11% of them were involved in hormone responsiveness, such as ABRE, P-box, CGTCA-motif, TCA-element, TGA-element, TATC-box and TGACG-motif. Among them, ABRE was an ABA responsive CRE. 49 out of the 56 BnLOR promoters had ABRE element, indicating that most of BnLORs might be inducted by ABA treatment. 18.38% of the CREs were related to stress responsiveness, such as LTR, ARE, TC-rich repeats, GC-motif, MBS, and WUN-motif. About 80% of the CREs in BnLOR promoter regions were associated with anaerobic induction, implying that BnLOR gene family might confer the ability to survive under inundation stress or high-altitude environments. CREs related to circadian control were also detected in the BnLOR promoter regions, suggesting that this gene family might be regulated by light induction and circadian rhythms. Besides, CREs involved in metabolic regulation and meristem expression were also discovered.
This study focused on CREs in response to hormone treatments such as auxin responsive elements (TGA element and AuxRR core), ethylene responsive element (ERE), gibberellin (GA) responsive elements (GARE motif and TATC box), SA responsive elements (SARE), ABA responsive elements (ABRE) and methyl jasmonate (MeJA) related motif (TGACG motif and CGTCA motif) (Song et al., 2020). Besides, CREs associated with abiotic stress were also our focus, such as low temperature responsive element (LTR), heat stress responsive element (HSE), anaerobic induced elements (ARE), drought-induced elements (MBS), etc. (Song et al., 2020). Results showed that ABA and MeJA responsive CREs were the most abundant in the promoter regions of BnLOR genes, with 135 and 154 elements respectively, followed by auxin (47), GA (41) and SA (38) responsive elements (Figure 6A; Table S6). Among the stress responsive elements, the anaerobic induced elements were the most (123). Moreover, a relatively high number of low-temperature responsive elements, defense responsive elements and stress responsive elements were also detected, with 40, 39 and 37 CREs respectively. Generally speaking, BnLOR genes should be able to respond to various abiotic stresses and hormone treatments. However, we noticed that some genes contained very limited CREs such as BnLOR17/14/31, indicating that their expression might weakly or couldn’t respond to abiotic stresses and hormone treatments.
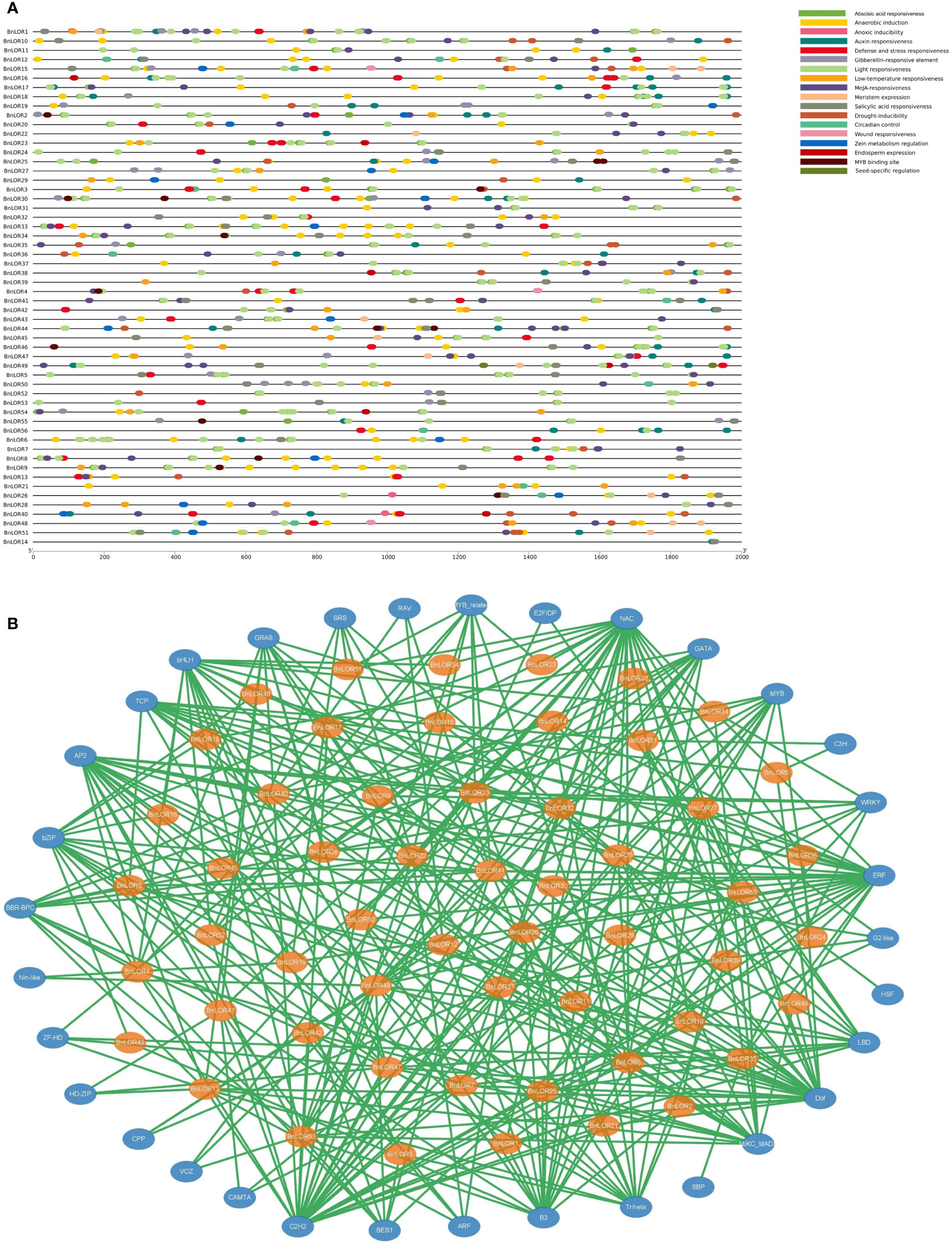
Figure 6 Analysis of cis-regulatory elements (CREs) in the BnLORs promoter regions and the predicted transcription factors (TFs) that regulate BnLORs. (A) CREs in the BnLORs promoter regions. Different CREs with functional similarity is denoted by similar colors. (B) Putative TFs that regulate BnLORs. The orange ovals indicate the BnLORs, and the yellow ovals indicate the predicted TFs. The green lines indicate the regulation relationship between TFs and BnLOR genes.
To further understand the regulation network of BnLORs, the putative TFs regulating BnLOR family members were also predicted. As a result, 56 BnLORs were regulated by 347 TFs (Figure 6B; Table S7). Except for BnLOR56 which couldn’t be retrieved, the rest 55 BnLOR genes were regulated by 1 (BnLOR38) to 15 (BnLOR49) TFs. Among the 347 TFs, NAC had the largest amount (31), followed by C2H2 (30), Dof (25), ERF (25), AP2 (19), B3(18), bHLH (17) and MIKC_MADS (17). These eight types of TFs occupied more than 50% of the total numbers (Figure 6B; Table S7). Most of these TFs are related to abiotic stresses and hormone treatments. For instance, The NAC Transcription factor family is unique to plants and plays a crucial role in plant growth and development as well as biotic and abiotic stress resistance (Wu et al., 2020). Dof proteins are involved in various biological processes, including but not limited to endosperm development, plant defense, seed germination, gibberellin response and auxin response (Yu et al., 2019). AP2/ERF proteins play vital roles in regulating plant growth and development and various abiotic stress response (Wu et al., 2021). It has noticed that BnLORs were mainly regulated by these TFs, further supporting the prediction that BnLORs perform crucial functions in the regulation of abiotic stresses and hormone treatments.
Orthologous clusters comparisons among multiple species
To deepen our understanding about the evolutionary history and the orthologous relationship among LOR genes, orthologous gene clusters in different species were compared and annotated. The quantity of orthologous proteins in each cluster exhibited variability, with the number ranging from 5 to 37 (Figure 7; Table S8). Twenty-nine orthologous proteins in B. oleracea and 28 orthologous proteins in B. rapa were predicted to have similar conserved domains to BnLOR proteins. Only 6 orthologous proteins were discovered in O. sativa, showing a large difference, this might be due to their early evolutionary differentiation. For the other six species (Arabidopsis, B. oleracea, B. rapa, G. max, T. aestivum and Z. mays), the number of orthologous proteins ranged from 8 (Z. mays) to 14 (Arabidopsis). Ten BnLORs proteins only had orthologous proteins in B. oleracea while another 10 BnLOR proteins only had orthologous proteins in B. rapa. The orthologous proteins of BnLOR5 could only be found in G. max. None of the BnLORs had orthologous proteins in every species. BnLOR49 had orthologous proteins in up to 6 species except B. rapa. Additionally, BnLOR4/29/44 proteins had orthologous proteins in 5 different kinds of species. Two orthologous clusters, comprising a total of 31 proteins, were found to be overlapping in each species (Figure 7; Table S8). Furthermore, 9 singleton proteins (BnLOR8/18/22/23/30/45/46/50/56) didn’t have any orthologous proteins in any of the studied species.
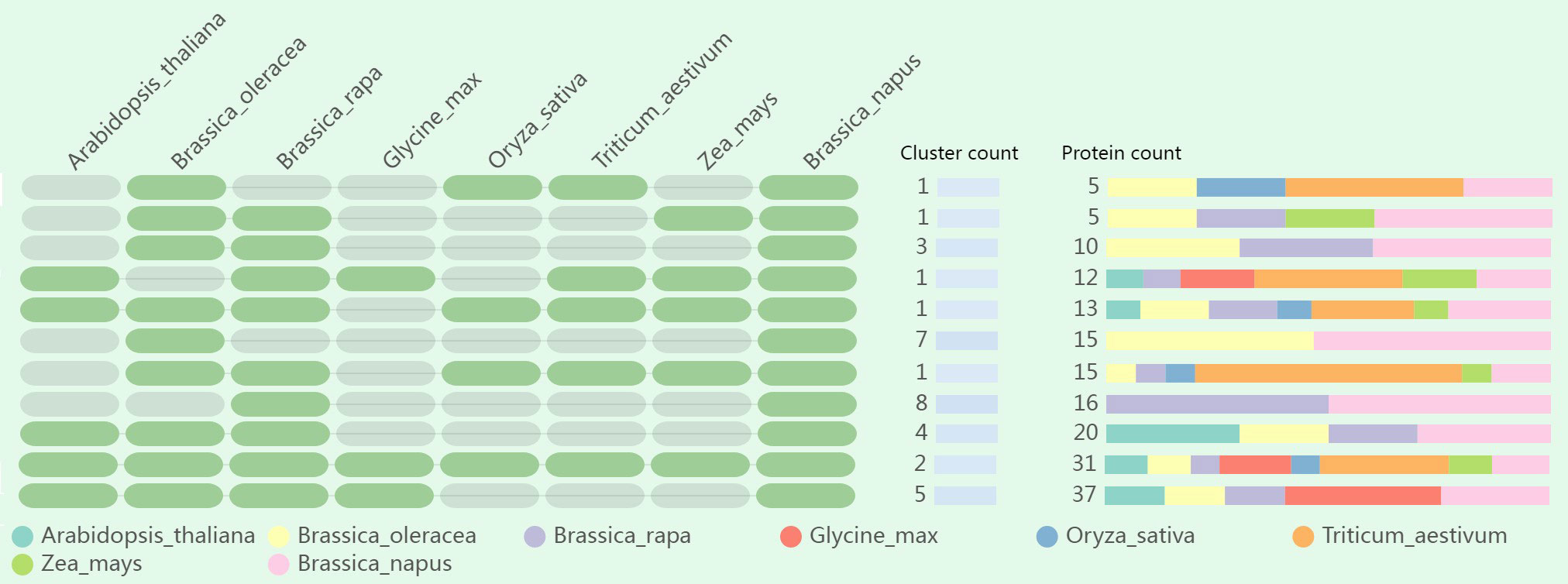
Figure 7 Comparative analysis of orthologous gene clusters among eight species (Arabidopsis thaliana, Brassica oleracea, Brassica rapa, Glycine max, Oryza sativa, Triticum aestivum and Zea mays). Green ovals on the left represent the presence of orthologous genes while grey ovals represent the absence of orthologous genes. Different colors bars on the right indicate the number and ratio of orthologous proteins in eight species.
Expression profiling of BnLORs in different tissues
The expression profiles of 56 BnLOR genes among 12 distinct tissues including blossomy pistil, flower, leaf, ovule, pericarp, pistil, root, silique, sepal, stamen, stem, wilting pistil were obtained by Sun et al. (2017) under the project ID of PRJNA394926. Results indicated that some members of the BnLOR gene family exhibited closely resembling expression patterns across diverse tissues of B. napus (Figure 8A; Table S9). For example, BnLOR28/29/35 had high expression levels in almost every tissue while BnLOR4/5/33/39/43 had relatively high expression levels in each tissue. However, the expression levels of BnLOR1/16/18/22/46/54/56 were relatively low in every tissue. BnLOR22 were found not expressed in any tissue. These 8 genes with low relative expression levels were all belonged to Clade H, suggesting that genes in this clade might be pseudogenes or act in response to biotic/abiotic stresses or hormone treatments. Certain genes in the BnLOR family displayed tissue-specific expression patterns. For instance, BnLOR13/21/40/45 were mainly expressed in stamen. These four genes also had a small amount of expression in blossomy pistil, wilting pistil and flower, but they didn’t express in other tissues, suggesting that these genes might have a potential association with reproductive growth. BnLOR6 and BnLO32 were also highly expressed in reproductive organs, but their distribution was different. Both of them were largely expressed in stamen and flower, followed by wilting pistil and blossomy pistil. BnLOR10 and BnLOR12 had high expression levels in ovule and moderate expression levels in other tissues. BnLOR14 and BnLOR42 demonstrated elevated expression levels in blossomy pistil with moderate expression levels in other tissues, indicating that these two genes might involve in plant reproductive growth, but might regulate reproductive growth through different pathways. BnLOR19 was highly expressed in pericarp, suggesting that it might take part in the vegetative growth. The remaining genes were slightly expressed in different tissues.
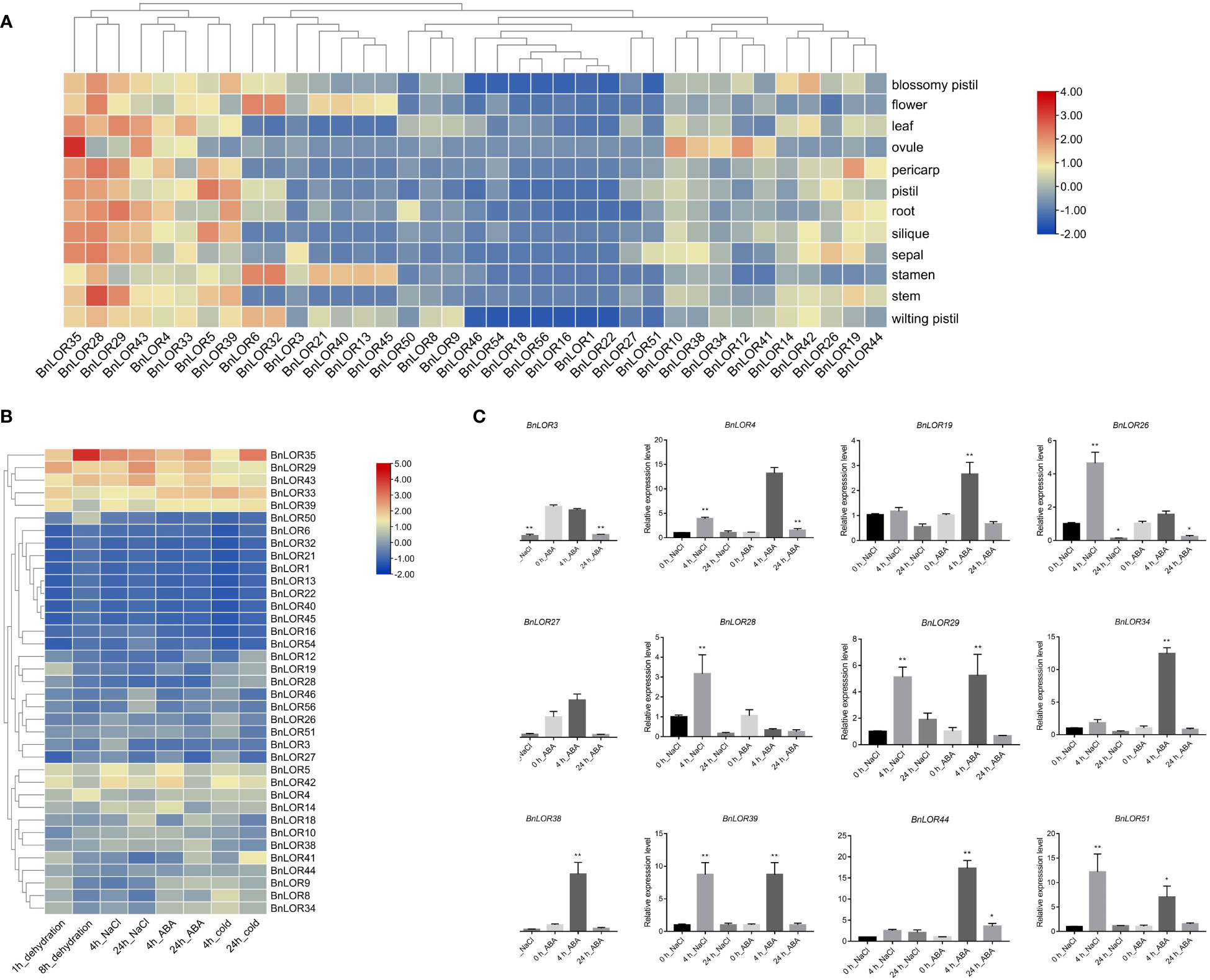
Figure 8 Expression profile of BnLOR genes under normal condition and abiotic stress. (A) Expression analysis of BnLOR genes in different tissues (blossomy pistil, flower, leaf, ovule, pericarp, pistil, root, silique, sepal, stamen, stem, wilting pistil) under normal condition. Data were transformed with a log2(TPM+1) transformation. Red square means the gene is up-regulated while blue square means the gene is down-regulated. (B) The expression levels of differentially expressed genes in B. napus (ZS11) under dehydration, salt stress, ABA treatment and cold stress. Data were transformed with a log2(TPM+1) transformation. Red color means the gene is up-regulated while blue color means the gene is down-regulated. (C) Relative expression levels of 12 randomly selected BnLOR genes in response to salt stress and ABA treatment in B. napus (ZD622). Mean ± SE of three replicates represent the significant difference with LSD test. * and ** indicate significance at the 5% and 1% level, respectively.
Expression profiling of BnLORs under temperature, salinity, and ABA stress
To examine the initial changes in gene expression, we obtained the transcriptional profile of B. napus under temperature, salinity and ABA stress under the project ID of CRA001775 (Zhang et al., 2019). The stress treatments consisted of drought stress, salt stress (200 mM), ABA treatment (25 μM), and low temperature stress (4 °C). The differentially expressed genes (DEGs) with a false rate less than 0.5 and an absolute fold change over 2 were chosen and displayed in Figure 8B. In general, the relative expression levels of most BnLOR gene at 4 h was higher than that at 24 h under salt stress, ABA treatment and cold stress while and the situation was similar under dehydration stress, indicating that the response speed of mRNA was very fast in BnLOR gene family (Figure 8B; Table S10). The relative expression levels of BnLOR5/29/33/35/39/42/43 genes showed a significant up-regulation while BnLOR1/6/13/16/21/22/32/40/45/50/54 were significantly down-regulated under four different treatments. Among them, BnLOR33 exhibited significant upregulation in response to cold stress, indicating its potential significance in facilitating adaptation to such stress. For the remaining 19 BnLOR genes, their relative expression levels varied under different stress. Almost all of these 19 genes could increase their expression levels under cold stress, but the degree was different, and the relative expression levels at 4 h were larger than 24 h. Meanwhile, the expression of BnLOR4/8/9/14/34/41 was partially up regulated at 4 h and decreased at 24 h under dehydration stress. Several other genes demonstrated slight up or down regulation in response to dehydration, salt stress, and ABA treatment.
To further verify the expression pattern under salinity and ABA stress, twelve BnLOR family members were chosen at random for qRT-PCR analysis. The relative expression levels of these genes had altered in varying degrees under salt stress or ABA treatment, indicating that these genes were capable of being induced by salinity and ABA stress (Figure 8C; Table S10). Under salt stress, the relative expression levels of BnLOR4/29/39/44/51 increased dramatically at 4 h and went down a little bit at 24 h, but still showed an overall upward trend. The remaining seven genes increased at the initial stage of salt stress but decreased dramatically compared to control after 24 h, indicating an exceptionally swift response under salt stress. Under ABA treatment, the relative expression levels of BnLOR4/39/44/51 exhibited an upward trend, with a larger increase observed at 4 h compared to 24 h. Conversely, the relative expression levels of BnLOR3 and BnLOR28 decreased at both 4 h and 24 h after treatment. The relative expression levels of the remaining six genes increased at the initial stage and then decreased over time, as depicted in Figure 8C and Table S10. Overall, the expression pattern trends of qRT-PCR and transcriptome file were relatively consistent, but there were some differences in the specific values.
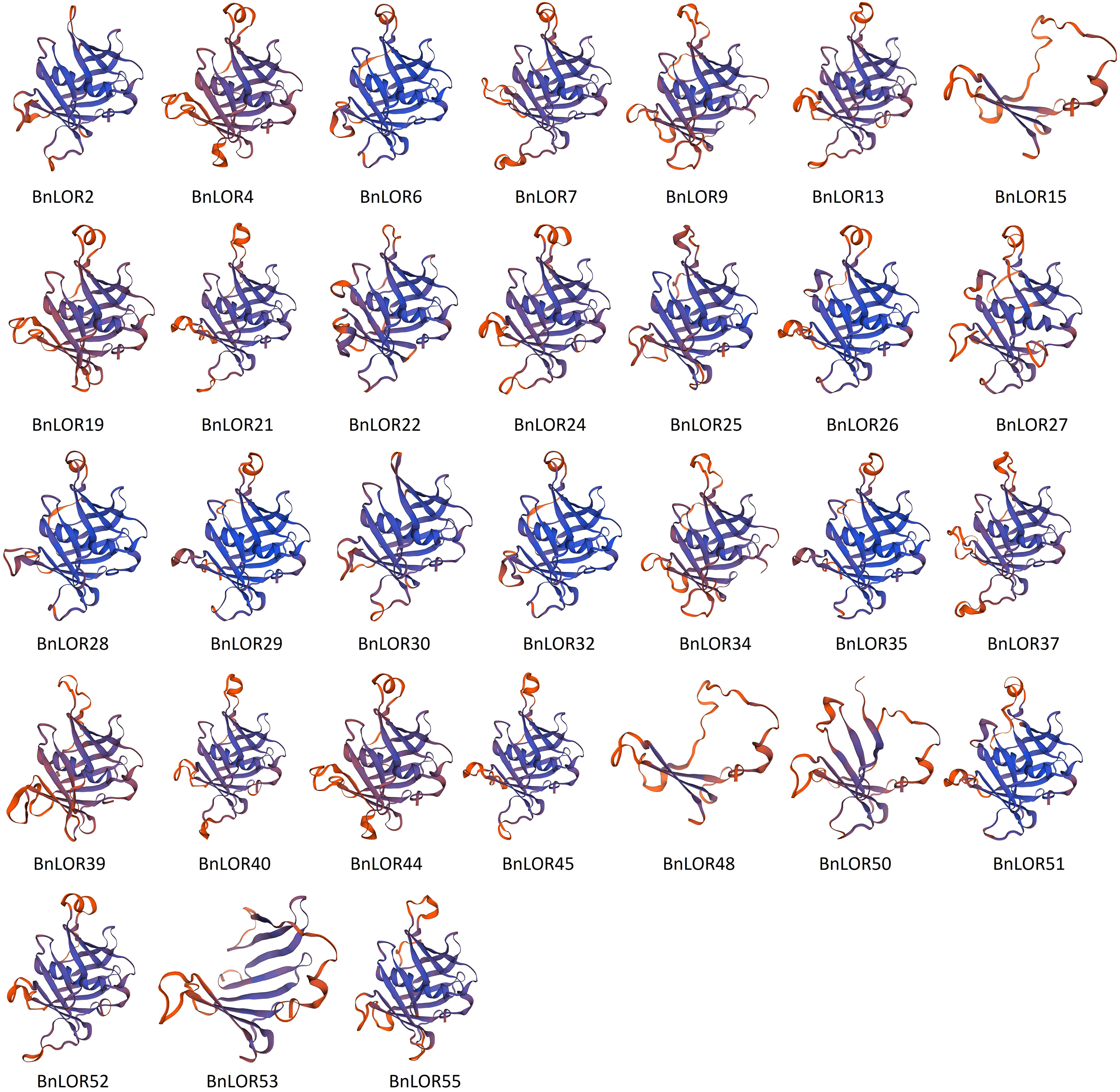
Comparative modeling of BnLOR proteins
To deepen our understanding about the structures of BnLOR proteins, homology modeling was carried out to predict the 3D structures of BnLOR proteins. The predicted models were generated based on reported templates and optimized to maximize alignment coverage, percentage identity, global model quality estimate (GMQE), and overall quality factor. Models were considered reliable if the sequence identity between the template and the BnLOR protein was greater than 30%. Results showed that the 3D structures of 31 BnLOR proteins could be predicted by SWISS-MOLDEL (Figure 9; Table S11). The LOR protein family was discovered to exhibit structural similarities to the mammalian PLSCR (Phospholipid scramlase) protein family (Baig, 2018). X-ray crystallography technique was employed to determine the 3D structures of AtLOR1 and found that this protein comprised a 12-stranded β-barrel structure that encompassed a central C-terminal α-helix (Bateman et al., 2009). According to this structural model, sequence conservation was found to be mainly located within the secondary structures and β-hairpin turns between strands 2 to 3 and 4 to 5 (Bateman et al., 2009). All 31 proteins shared the same template 2q4m.1.A. The tertiary structures of the BnLOR proteins were relatively similar and complex with a few exceptions. For the 31 BnLOR proteins with predicted 3D structures, BnLOR15/48/50/53 were significantly different from the other 27 members, with no central C-terminal α-helix and less than 12 stranded β-barrels. Among these four genes, the expression pattern of BnLOR15/48/53 couldn’t be detected by RNA-seq, indicating that they were either low expressive genes or pseudogene. The relative expression level of BnLOR50 could be induced by abiotic stress suggests that it may be involved in facilitating adaptation to such stress.
Discussion
With the availability of reference genomes for numerous plant species, many gene families have been identified, yet the LOR gene family remains relatively unknown. In this research, the LOR gene family in B. napus was identified by investigating its phylogenetic relationship, chromosomal locations, protein conserved motifs, gene structures, WGD events, synteny relationship, putative CREs in promoter regions, expression profiles, and responses to temperature, salinity and ABA stress. The LOR gene family, been discovered in plants, fungi, eubacteria, and some archaea, is distinguished by its conserved LOR domain. Our phylogenetic analysis revealed that BnLORs, BoLORs, BrLORs, and AtLORs could be divided into three subgroups and these three subgroups can be further divided into eight clades (Clade A to H) (Figure 1). Notably, the BnLOR family comprises a relatively large number of family members (56), which is approximately three times that of Arabidopsis (20), Solanum lycopersicum (17), and Solanum tuberosum (16). Basically, plants with larger genome sizes tend to possess a greater number of gene family members (Wang et al., 2020). B. napus was derived from the spontaneous hybridization between B. rapa and B. oleracea followed by allopolyploidization (Chalhoub et al., 2014). It has not only experienced genome wide triplication events specific to Brassica species, but also experienced allopolyploidization, resulting in a larger genome size and larger gene family members. Additionally, due to homoeologous recombination and chromosomal structural variation between the A and C genomes of B. napus during the domestication process, a small number of genes were lost or gained (Chalhoub et al., 2014). Consequently, the number of BnLORs is not simply the sum of BoLORs and BrLORs. Previous studies found that AtLURP1 and AtLOR1 were both located in Clade A and demonstrated significant involvement in the resistance to Hpa, suggesting that genes in Clade A (BnLOR6/32/28/35/29/27/50/26/51/22) might be associated with plant defense.
During the process of evolution, multiple gene families underwent differential adaptation (Xu et al., 2012), with introns being a crucial component in the acquisition of new gene functions (Roy et al., 2003). Among the 56 BnLOR members, 27 contained two introns, 26 had one intron, and 3 had no introns (Figure 2). It is commonly believed that the presence of introns is more prevalent during the earlier stages of gene expansion, and gradually diminishing during the evolutionary process (Roy and Gilbert, 2006; Roy and Penny, 2007; Rose, 2019), as they play a crucial role in activating genes by providing compact gene structures with fewer introns, thereby facilitating timely responses to various stresses (Chung et al., 2006; Jeffares et al., 2008). Consistent with these findings, the majority of the BnLOR genes could be rapidly and significantly induced under salt stress and ABA treatment (Figures 8B, C, Table S10), indicating their important roles towards abiotic stresses and hormone treatments. With the help of MEME online tools, up to 15 putative conserved motifs were predicted within the BnLOR proteins (Figure 2). The majority of these proteins contained 6 to 9 conserved motifs, with motifs 1 to 6 being highly conservative and could be found in most BnLOR proteins (Figure 2). The other motifs had an obvious preference in certain subgroups or clades. The rest motifs demonstrated distinct preferences within specific subgroups or clades. For instance, motif 10 only existed in Clade G and H, while motif 11 and 13 were merely presented in Clade C. The functions of these conserved motifs in the BnLOR proteins were not well understood yet, but they might play a role in the interaction between LOR proteins and their substrates (Fang et al., 2021). Motifs 1 to 6 may be integral in determining essential molecular functions among BnLOR genes, while the preference for certain motifs may contribute to the structural foundation for the diversity in gene functions. Similarities in intron numbers and motif arrangements were observed within the same subgroups or clades, further supporting the phylogenetic relationship of the BnLOR gene family.
The syntenic relationship of BnLORs, BrLORs, BoLORs, and AtLORs family members were investigated in this research. A total of 78 collinear gene pairs between B. napus and B. rapa and 70 collinear gene pairs between B. napus and B. oleracea were discovered (Figure 5; Table S5). Chromosomes A3 and C3 showed the highest number of collinear gene pairs. We also identified 14 pairs of orthologous genes between B. napus and Arabidopsis, 28 pairs of orthologous genes between B. napus and B. rapa, and 29 pairs of orthologous genes between B. napus and B. oleracea (Table S8). Notably, one AtLOR gene showed two syntenic orthologous gene pairs (AT1G80120-BnLOR4/44), indicating that these two genes originated after the divergence of Arabidopsis. The remaining 12 orthologous gene pairs were one-to-one syntenic orthologs. The high conservation of collinearity among LORs in B. napus, B. rapa, and B. oleracea suggests that these genes have been evolutionarily conserved. Gene duplication is a crucial impetus for expanding gene families and evolving new functions, such as stress adaptation and disease resistance (Vision et al., 2000; Panchy et al., 2016). Previous studies have suggested that tandem repeats and segmental duplications are the primary patterns of gene family expansion, with segmental duplications occurring more frequently due to the retention of duplicated chromosomal blocks in polyploidy plants followed by chromosome rearrangements (Yu et al., 2005; Kong et al., 2010). Our analysis showed that 56 BnLOR genes underwent 37 segmental duplications and five tandem repeat events under purifying selection, suggesting that BnLOR gene family had undergone gene expression during the evolutionary process.
CREs has been reported to play crucial roles in regulating the transcription of genes involved in environmental stress responses (Yamaguchi-Shinozaki and Shinozaki, 2005; Chen et al., 2016). Knoth et al. (2007) identified a region in the AtLURP1 promoter that contained a key pathogen response element W box (a binding site for WRKY TFs). After binding with W box, AtWRKY70 could regulate the expression of AtLURP1 (Knoth et al., 2007). In this study, we analyzed the CREs presented in the promoter regions of BnLOR genes and classified them into three main categories including TF binding sites, hormone responsive elements, and stress responsive elements. CREs analysis revealed that BnLORs were mainly involved in light response, hormone response, and stress response processes. Specifically, we identified 123 anaerobic induction related CREs, 40 low temperature responsive elements, 39 defense responsive elements, and 37 stress responsive elements among the 56 BnLOR genes. These elements might contribute to the resistance of B. napus to various abiotic stresses, such as cold, heat, drought, and salt stress, and could explain the differential expression patterns and stress responses of duplicated genes. The presence of a wide range of CREs distributed throughout the promoter regions of BnLOR genes suggested that these genes might have complex expression profiles and played important roles in regulating stress resistance in rapeseed. These CREs provided valuable genetic information for breeding rapeseed varieties with improved stress resistance.
Transcription regulatory networks play a crucial role in the regulation of gene expression in plants. Among these networks, NAC, WRKY, MYB, and GATA transcription factors have been identified as important players in abiotic stress responses. NAC transcription factors are known to be involved in various plant developmental processes as well as responses to abiotic stress. They have been reported to regulate the expression of stress-responsive genes and play a role in enhancing plant tolerance to abiotic stresses such as drought, salinity, and temperature extremes (Nuruzzaman et al., 2013). WRKY transcription factors are another important group of regulators that are involved in plant responses to abiotic stress. In B. napus, WRKY transcription factors have been shown to play a role in response to abiotic stresses such as drought, cold, and salt stress (Jiang et al., 2016). MYB and GATA transcription factors have been implicated in the regulation of genes involved in abiotic stress responses, including drought and salt stress in B. napus (Hajiebrahimi et al., 2017; Zhu et al., 2020). These transcription factors have been implicated in the regulation of the majority of the BnLOR genes., showing consistency with the predicted function of BnLORs on abiotic stress resistance. However, the regulatory mechanisms involving NAC, WRKY, MYB, and GATA transcription factors in modulating BnLOR expression and its role in abiotic stress responses in B. napus are still being elucidated and represent a promising area of research.
The expression profile of BnLORs exhibited gene specificity, whereby BnLOR35/28/29/43/4/33/5/39/6/32 exhibited relatively high expression levels across various tissues, suggesting their indispensable functions. Certain BnLOR genes may perform specialized functions in specific tissues, thereby potentially participating in the life cycle of B. napus. Conversely, other BnLOR genes displayed negligible or non-existent expression levels. Moreover, the expression levels of the same gene differed across diverse tissues, thus indicating tissue-specificity. Notably, the majority of the BnLOR members displayed low expression levels in 12 tissues under normal conditions, whereas their relative expression levels were significantly induced by abiotic stress.
Previous studies in Arabidopsis have reported the participation of AtLURP1 and AtLOR1 in biotic stress (Knoth and Eulgem, 2008; Coker et al., 2015; Lee et al., 2017; Baig, 2018). Nevertheless, it remains uncertain whether the LOR family in plants responds to abiotic stress or not. In this study, we investigated the transcriptional responses of 56 B. napus LOR genes under dehydration, salt stress, ABA treatment, and cold stress. Our findings revealed that BnLOR5/29/33/35/39/42/43 exhibited significantly up-regulated expression levels, whereas BnLOR1/6/13/16/21/22/32/40/45/50/54 displayed significant down-regulation under diverse stress conditions. These results implied that these genes may have pivotal roles in response to dehydration, salt stress, ABA treatment, and cold stress. Additionally, qRT-PCR was employed to validate the expression pattern of BnLORs under salinity and ABA stress. Results showed that the expression trends of qRT-PCR and transcriptome profile were relatively consistent, although differed in specific values. Therefore, these genes might serve as promising candidates for genetic engineering to enhance plant fitness in response to salt stress and ABA treatment.
To gain a better understanding of the interplay between BnLOR proteins, we generated 3D structures via homology modeling. Interestingly, all 31 predicted proteins shared the same template, 2q4m.1.A, which was derived from the ensemble refinement of the crystal structure of At5g01750 from the DUF567 family in Arabidopsis (Table S11). Notably, the DUF567 family solely comprises uncharacterized proteins that are found in plants, fungi, eubacteria, and some archaea. In plants, DUF567 is extensively expanded, with Arabidopsis harboring 21 family members. Gene ontology analysis revealed that At5g01750 is involved in protein domain-specific binding in the plasma membrane. Wang et al. (2022) reported that NBS-LRR proteins in Lagenaria siceraria can be classified into four groups, indicating that NBS-LRR proteins form a complex protein network to recognize multiple effectors. Similar results were also observed in Yer et al. (2018). Nonetheless, our study demonstrated that the tertiary structures of all predicted BnLOR proteins were remarkably consistent, sharing the same template. This finding implied that BnLORs might be functionally consistent, which was consistent with our observation that most BnLORs genes could respond to abiotic stresses and hormone treatments.
Conclusion
This study conducted a genome-wide survey of B. napus LOR gene family and analyzed their phylogenetic relationships, chromosomal distributions, protein conserved motifs, gene structures, WGD events, synteny relationship, putative CREs in promoter regions, expression profile and response to temperature, salinity and ABA stress through bioinformatics analysis and qRT-PCR. Results showed that there was a high degree of homology within BnLOR members. 56 BnLOR genes could be divided into 3 subgroups (8 clades) and have undergone 37 segmental duplication and 5 tandem repeats events with purifying selection. The LORs in B. napus had a high level of collinearity with that of B. rapa and B. oleracea, indicating they were quite conservative during the evolutionary process. The analysis of conserved domain, motif structures and WGD events of BnLORs were consistent with the phylogenetic analysis. BnLOR family genes had tissue specific expression patterns and most of them could be induced by salt stress and ABA treatment, suggesting their potential function on stress and hormone responsiveness. These results provided a reference for comprehensive understanding and utilization of BnLOR genes in B. napus in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
SY and YD analyzed the data. JC and QH grew the plants and conducted the qRT-PCR analysis. JWang drew the figures, SY and ZU edited the manuscript. GC, JWei, JC, and QH contributed regents/materials/analysis tool. SY and JWang conceived and designed the study, obtained funds, and critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (32200251) and the Science and Technology Department of Zhejiang Province (2021C02064-2).
Conflict of interest
Author JWei was employed by the Mizuda Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1197781/full#supplementary-material
Supplementary Figure 1 | The motif logos of 15 motifs among BnLOR family proteins.
References
Baig, A. (2018). Role of arabidopsis LOR1 (LURP-one related one) in basal defense against Hyaloperonospora arabidopsidis. Physiol. Mol. Plant Pathol. 103, 71–77. doi: 10.1016/j.pmpp.2018.05.003
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi: 10.1093/nar/gkp335
Bateman, A., Finn, R. D., Sims, P. J., Wiedmer, T., Biegert, A., Soding, J. (2009). Phospholipid scramblases and tubby-like proteins belong to a new superfamily of membrane tethered transcription factors. Bioinformatics 25, 159–162. doi: 10.1093/bioinformatics/btn595
Bricchi, I., Bertea, C. M., Occhipinti, A., Paponov, I. A., Maffei, M. E. (2012). Dynamics of membrane potential variation and gene expression induced by spodoptera littoralis, myzus persicae, and Pseudomonas syringae in Arabidopsis. PloS One 7, e46673. doi: 10.1371/journal.pone.0046673
Caillaud, M. C., Asai, S., Rallapalli, G., Piquerez, S., Fabro, G., Jones, J. D. G. (2016). A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PloS Biol. 14, e1002408. doi: 10.1371/journal.pbio.1002408
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A. P., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H. Y., Hsieh, E. J., Cheng, M. C., Chen, C. Y., Hwang, S. Y., Lin, T. P. (2016). ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element. New Phytol. 211, 599–613. doi: 10.1111/nph.13914
Chung, B. Y., Simons, C., Firth, A. E., Brown, C. M., Hellens, R. P. (2006). Effect of 5’UTR introns on gene expression in Arabidopsis thaliana. BMC Genomics 7, 120. doi: 10.1186/1471-2164-7-120
Coker, T. L. R., Cevik, V., Beynon, J. L., Gifford, M. L. (2015). Spatial dissection of the Arabidopsis thaliana transcriptional response to downy mildew using fluorescence activated cell sorting. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00527
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The pfam protein families database in 2019. Nucleic Acids Res. 47, 427–432. doi: 10.1093/nar/gky995
Fang, Y., Cao, D., Yang, H., Guo, W., Ouyang, W., Chen, H., et al. (2021). Genome-wide identification and characterization of soybean GmLOR gene family and expression analysis in response to abiotic stresses. Int. J. Mol. Sci. 22, 12515. doi: 10.3390/ijms222212515
Gu, Z., Cavalcanti, A., Chen, F. C., Bouman, P., Li, W. H. (2002). Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 19, 256–262. doi: 10.1093/oxfordjournals.molbev.a004079
Gupta, A., Senthil-Kumar, M. (2017). Transcriptome changes in Arabidopsis thaliana infected with Pseudomonas syringae during drought recovery. Sci. Rep. 7, 9124. doi: 10.1038/s41598-017-09135-y
Hajiebrahimi, A., Owji, H., Hemmati, S. (2017). Genome-wide identification, functional prediction, and evolutionary analysis of the R2R3-MYB superfamily in Brassica napus. Genome 60 (10), 797–814. doi: 10.1139/gen-2017-0059
Jeffares, D. C., Penkett, C. J., Bahler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Jiang, J., Ma, S., Ye, N., Jiang, M., Gao, J., Zhang, J. (2016). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59 (2), 86. doi: 10.1111/jipb.12513
Jin, J., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, 1040–1045. doi: 10.1093/nar/gkw982
Karnes, K., Shaikh, K. (2021). Characterization and structure-function analysis of autophagy factor ATG7 from Trypanosoma brucei (TbATG7) as a potential drug target. FASEB J. 35, 03861. doi: 10.1096/fasebj.2021.35.S1.03861
Knoth, C., Eulgem, T. (2008). The oomycete response gene LURP1 is required for defense against Hyaloperonospora parasitica in Arabidopsis thaliana. Plant J. 55, 53–64. doi: 10.1111/j.1365-313X.2008.03486.x
Knoth, C., Ringler, J., Dangl, J. L., Eulgem, T. (2007). Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol. Plant Microbe Interact. 20, 120–128. doi: 10.1094/MPMI-20-2-0120
Kong, H., Landherr, L. L., Frohlich, M. W., Leebens-Mack, J., Depamphilis, C. W. (2010). Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 50, 873–885. doi: 10.1111/j.1365-313X.2007.03097.x
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Larkin, M. A., Blackshields, G., Brown, N., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, M. H., Jeon, H. S., Kim, H. G., Park, O. K. (2017). An Arabidopsis NAC transcription factor NAC4 promotes pathogen-induced cell death under negative regulation by microRNA164. New Phytol. 214, 343–360. doi: 10.1111/nph.14371
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, J., Islam, F., Ayyaz, A., Fang, R., Hannan, F., Farooq, M. A., et al. (2022). Wood vinegar induces salinity tolerance by alleviating oxidative damages and protecting photosystem II in rapeseed cultivars. Ind. Crop Prod. 189, 115763. doi: 10.1016/j.indcrop.2022.115763
Nuruzzaman, M., Sharoni, A. M., Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00248
Panchy, N., Lehti-Shiu, M., Shiu, S. H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316. doi: 10.1104/pp.16.00523
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, 200–204. doi: 10.1093/nar/gky448
Raza, A., Tabassum, J., Fakhar, A. Z., Sharif, R., Chen, H., Zhang, C., et al. (2022). Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit. Rev. Biotechnol. 1–28. doi: 10.1080/07388551.2022.2093695
Raza, A., Charagh, S., Najafi-Kakavand, S., Abbas, S., Shoaib, Y., Anwar, S., et al. (2023). Role of phytohormones in regulating cold stress tolerance: physiological and molecular approaches for developing cold-smart crop plants. Plant Stress. 8, 100152. doi: 10.1016/j.stress.2023.100152
Rose, A. B. (2019). Introns as gene regulators: a brick on the accelerator. Front. Genet. 9. doi: 10.3389/fgene.2018.00672
Roy, S. W., Fedorov, A., Gilbert, W. (2003). Large-Scale comparison of intron positions in mammalian genes shows intron loss but no gain. P. Natl. Acad. Sci. 100 (12), 7158–7162. doi: 10.1073/pnas.1232297100
Roy, S. W., Gilbert, W. (2006). The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 7, 211–221. doi: 10.1038/nrg1807
Roy, S. W., Penny, D. (2007). A very high fraction of unique intron positions in the intron-rich diatom Thalassiosira pseudonana indicates widespread intron gain. Mol. Biol. Evol. 24, 1447–1457. doi: 10.1093/molbev/msm048
Sheteiwy, M. S., Ulhassan, Z., Qi, W., Lu, H., AbdElgawad, H., Minkina, T., et al. (2022). Association of jasmonic acid priming with multiple defense mechanisms in wheat plants under high salt stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.886862
Shu, K., Qi, Y., Chen, F., Meng, Y., Luo, X., Shuai, H., et al. (2018). Salt stress represses soybean seed germination by negatively regulating GA biosynthesis while positively mediating ABA biosynthesis. Front. Plant Sci. 9. doi: 10.3389/fpls.2017.01372
Song, C., Zhang, D., Zheng, L., Shen, Y., Zuo, X., Mai, J., et al. (2020). Genome-wide identification and expression profiling of the YUCCA gene family in Malus domestica. Sci. Rep. 10, 10866. doi: 10.1038/s41598-020-66483-y
Sun, F., Fan, G., Hu, Q., Zhou, Y., Guan, M., Tong, C., et al. (2017). The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J. 92, 452–468. doi: 10.1111/tpj.13669
Suzuki, N., Rivero, R. M., Shulaev, V., Blumwald, E., Mittler, R. (2014). Abiotic and biotic stress combinations. New Phytol. 203, 32–43. doi: 10.1111/nph.12797
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Vision, T. J., Brown, D. G., Tanksley, S. D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. doi: 10.1126/science.290.5499.2114
Waadt, R., Seller, C. A., Hsu, P. K., Takahashi, Y., Munemasa, S., Schroeder, J. I. (2022). Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 23, 680–694. doi: 10.1038/s41580-022-00479-6
Wang, L., Ding, X., Gao, Y., Yang, S. (2020). Genome-wide identification and characterization of GRAS genes in soybean (Glycine max). BMC Plant Biol. 20, 415. doi: 10.1186/s12870-020-02636-5
Wang, L., Guo, K., Li, Y., Tu, Y., Hu, H., Wang, B., et al. (2010a). Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 10, 282. doi: 10.1186/1471-2229-10-282
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010b). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinf. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, J., Yang, C., Wu, X., Wang, Y., Wang, B., Wu, X., et al. (2022). Genome-wide characterization of NBS-LRR family genes and expression analysis under powdery mildew stress in Lagenaria siceraria. Physiol. Mol. Plant Pathol. 118, 101798. doi: 10.1016/j.pmpp.2022.101798
Wang, J., Wang, Y., Wu, X., Wang, B., Lu, Z., Zhong, L., et al. (2023). Insight into the bZIP gene family in Lagenaria siceraria: genome and transcriptome analysis to understand gene diversification in cucurbitaceae and the roles of LsbZIP gene expression and function under cold stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1128007
Wu, X., Lai, Y., Lv, L., Ji, M., Han, K., Yan, D., et al. (2020). Fasciclin-like arabinogalactan gene family in Nicotiana benthamiana: genome-wide identification, classification and expression in response to pathogens. BioMed. Central. 20, 305. doi: 10.1186/s12870-020-02501-5
Wu, X., Lai, Y., Rao, S., Lv, L., Ji, M., Han, K., et al. (2021). Genome-wide identification reveals that Nicotiana benthamiana hypersensitive response (HR)-like lesion inducing protein 4 (NbHRLI4) mediates cell death and salicylic acid-dependent defense responses to turnip mosaic virus. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.627315
Xu, G., Guo, C., Shan, H., Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1187–1192. doi: 10.1073/pnas.1109047109
Yamaguchi-Shinozaki, K., Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Yang, S., Chen, S., Geng, X. X., Yan, G., Li, Z. Y., Meng, J. L., et al. (2016). The first genetic map of a synthesised allohexaploid Brassica with a, b and c genomes based on simple sequence repeat markers. Theor. Appl. Genet. 129, 689–701. doi: 10.1007/s00122-015-2657-z
Yang, S., Chen, S., Zhang, K., Li, L., Yin, Y., Gill, R. A. (2018). A high-density genetic map of an allohexaploid Brassica doubled haploid population reveals quantitative trait loci for pollen viability and fertility. Front. Plant Sci. 9, 1161. doi: 10.3389/fpls.2018.01161
Yang, S., Gill, R. A., Zaman, Q. U., Ulhassan, Z., Zhou, W. (2020). Insights on SNP types, detection methods and their utilization in Brassica species: recent progress and future perspectives. J. Biotechnol. 324, 11–20. doi: 10.1016/j.jbiotec.2020.09.018
Yang, S., Zhang, K., Lu, C., Chen, G., Huang, Q., Ulhassan, Z., et al. (2022). Genetic variation and stability analysis of an artificially synthesized allohexaploid Brassica for breeding innovations. Agronomy 12, 2843. doi: 10.3390/agronomy12112843
Yang, S., Zhang, X., Yue, J. X., Tian, D., Chen, J. Q. (2008). Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genom. 280, 187–198. doi: 10.1007/s00438-008-0355-0
Yer, E. N., Baloglu, M. C., Ayan, S. (2018). Identification and expression profiling of all hsp family member genes under salinity stress in different poplar clones. Gene 678, 324–336. doi: 10.1016/j.gene.2018.08.049
Yin, Y., Morgunova, E., Jolma, A., Kaasinen, E., Sahu, B., Khund-Sayeed, S., et al. (2017). Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356 (6337), eaaj2239. doi: 10.1126/science.aaj223
Yu, Y. H., Bian, L., Wan, Y. T., Jiao, Z. L., Yu, K. K., Zhang, G. H., et al. (2019). Grape (Vitis vinifera) VvDOF3 functions as a transcription activator and enhances powdery mildew resistance. Plant Physiol. Biochem. 143, 183–189. doi: 10.1016/j.plaphy.2019.09.010
Yu, J., Wang, J., Lin, W., Li, S., Li, H., Zhou, J., et al. (2005). The genomes of Oryza sativa: a history of duplications. PloS Biol. 3, e38. doi: 10.1371/journal.pbio.0030038
Zhang, Y., Ali, U., Zhang, G., Yu, L., Fang, S., Iqbal, S., et al. (2019). Transcriptome analysis reveals genes commonly responding to multiple abiotic stresses in rapeseed. Mol. Breed. 39, 158. doi: 10.1007/s11032-019-1052-x
Zhu, M., Assmann, S. M. (2017). Metabolic signatures in response to abscisic acid (ABA) treatment in Brassica napus guard cells revealed by metabolomics. Sci. Rep. 7, 12875. doi: 10.1038/s41598-017-13166-w
Zhu, W., Guo, Y., Chen, Y., Wu, D., Jiang, L. (2020). Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in Brassica napus. BMC Plant Biol. 20, 543. doi: 10.21203/rs.3.rs-30607/v1
Keywords: Brassica napus, LOR gene family, phylogenetic tree, expression profiles, abiotic stress
Citation: Yang S, Chen J, Ding Y, Huang Q, Chen G, Ulhassan Z, Wei J and Wang J (2023) Genome-wide investigation and expression profiling of LOR gene family in rapeseed under salinity and ABA stress. Front. Plant Sci. 14:1197781. doi: 10.3389/fpls.2023.1197781
Received: 31 March 2023; Accepted: 02 May 2023;
Published: 31 May 2023.
Edited by:
Meng Jiang, Zhejiang University, ChinaReviewed by:
Muhammad Azhar Hussain, Hainan University, ChinaMuhammad Waseem, Hainan University, China
Ali Raza, Fujian Agriculture and Forestry University, China
Muhammad Zulfiqar Ahmad, Gomal University, Pakistan
Copyright © 2023 Yang, Chen, Ding, Huang, Chen, Ulhassan, Wei and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Wang, d2FuZ2ppYW5AemFhcy5hYy5jbg==
†These authors have contributed equally to this work
 Su Yang
Su Yang Jialuo Chen
Jialuo Chen Yonghe Ding
Yonghe Ding Qian Huang
Qian Huang Guangna Chen
Guangna Chen Zaid Ulhassan
Zaid Ulhassan Ji’an Wei
Ji’an Wei Jian Wang
Jian Wang