
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Plant Sci., 08 February 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1152603
This article is a correction to:
Integrated analysis of small RNAs, transcriptome and degradome sequencing reveal the drought stress network in Agropyron mongolicum Keng
 Bobo Fan1
Bobo Fan1 Fengcheng Sun2
Fengcheng Sun2 Zhuo Yu1
Zhuo Yu1 Xuefeng Zhang1
Xuefeng Zhang1 Xiaoxia Yu1
Xiaoxia Yu1 Jing Wu1
Jing Wu1 Xiuxiu Yan1
Xiuxiu Yan1 Yan Zhao3
Yan Zhao3 Lizhen Nie2
Lizhen Nie2 Yongyu Fang2
Yongyu Fang2 Yanhong Ma1*
Yanhong Ma1*A Corrigendum on:
Integrated analysis of small RNAs, transcriptome and degradome sequencing reveal the drought stress network in Agropyron mongolicum Keng.
By Fan B, Sun F, Yu Z, Zhang X, Yu X, Wu J, Yan X, Zhao Y, Nie L, Fang Y and Ma Y (2022) Front. Plant Sci. 13:976684. doi: 10.3389/fpls.2022.976684
In the published article, there was an error in Figure 9. Figure 9 should have been a bar graph of RT-qPCR, but since the relative expression trends of RT-qPCR are the same to that of RNA-seq, we unintentionally put the bar graph of gene expression from RNA-seq in Figure 9. We have corrected Figure 9 to a combined graph from the gene expression of RT-qPCR and RNA-seq.
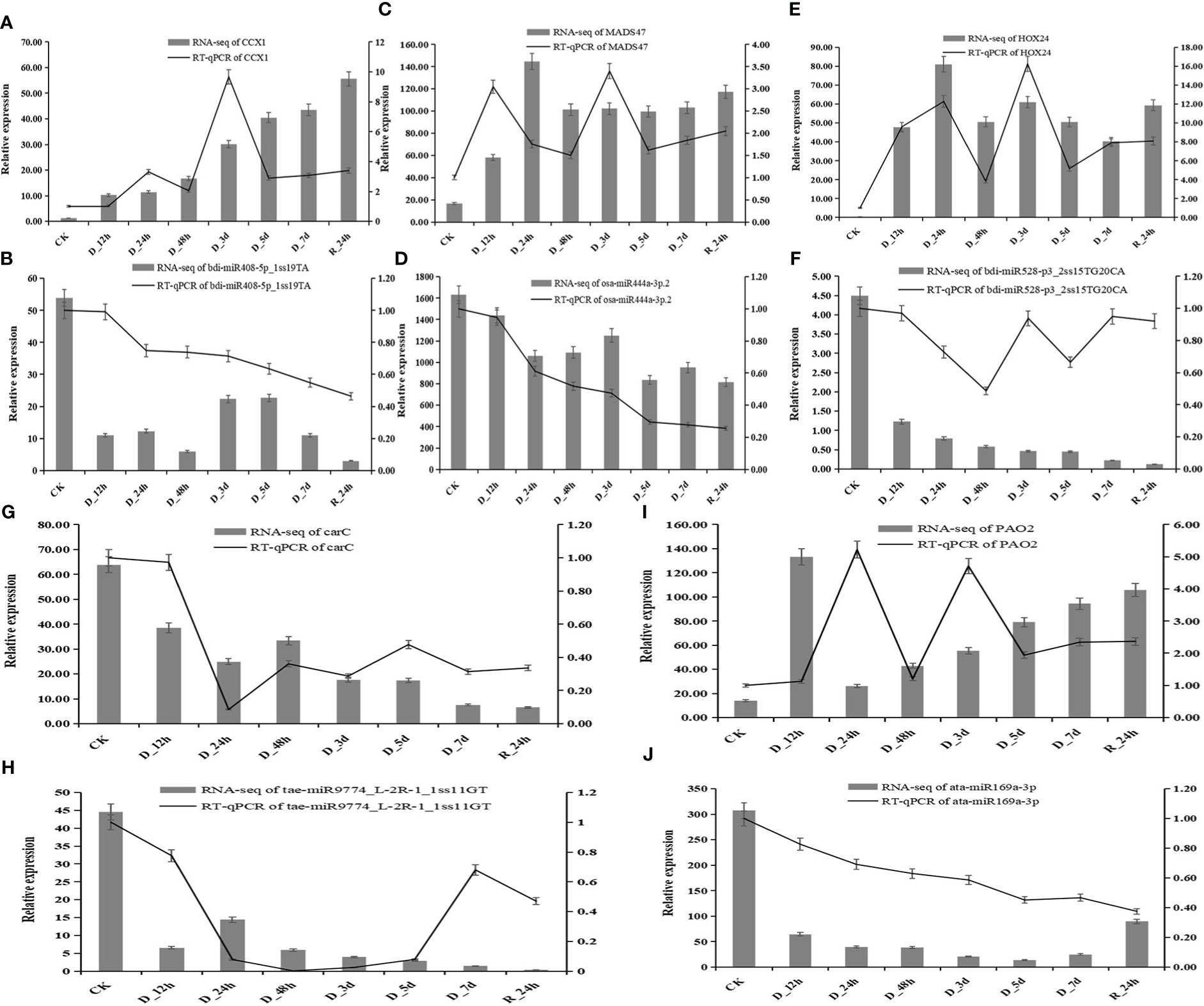
Figure 9 Correlation of the expression between miRNAs and the hub genes at eight durations of different drought treatments (25% PEG solution PEG6000). bdi-miR408-5p_1ss19TA target CCX1(A, B); osa-miR444a-3p.2 target MADS47 (C, D); bdi-miR528-p3_2ss15TG20CA target HOX24 (E, F); tae-miR9774_L-2R-1_1ss11GT target carC (G, H); and ata-miR169a-3p target PAO2 (I, J).
In the last sentence of “Correlation analysis of miRNAs and their candidate hub genes for drought resistance”, “The relative expression of tae-miR9774_ L-2R-1_ 1ss11GT increased overall, but the relative expression of its target genes decreased”, the first “increased” should be “decreased”, and “but the relative expression of its target genes decreased” should be removed.
The corrected sentence appears below:
“The relative expression of tae-miR9774_ L-2R-1_ 1ss11GT and target gene carC decreased overall”.
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: Agropyron mongolicum Keng, drought resistance, microRNAs, transcriptome, degradome, integration analysis, co-expression network
Citation: Fan B, Sun F, Yu Z, Zhang X, Yu X, Wu J, Yan X, Zhao Y, Nie L, Fang Y and Ma Y (2023) Corrigendum: Integrated analysis of small RNAs, transcriptome and degradome sequencing reveal the drought stress network in Agropyron mongolicum Keng. Front. Plant Sci. 14:1152603. doi: 10.3389/fpls.2023.1152603
Received: 28 January 2023; Accepted: 30 January 2023;
Published: 08 February 2023.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2023 Fan, Sun, Yu, Zhang, Yu, Wu, Yan, Zhao, Nie, Fang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanhong Ma, bWF5YW5ob25nODBAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.