- 1Key Laboratory of Agricultural Biotechnology of Liaoning Province, College of Bioscience and Biotechnology, Shenyang Agricultural University, Shenyang, Liaoning, China
- 2Collaborative Innovation Center for Genetic Improvement and High Quality and Efficiency Production of Northeast Japonica Rice in China, Rice Research Institute, Shenyang Agricultural University, Shenyang, Liaoning, China
Introduction: β-Mannanase is a plant cell wall remodeling enzyme involved in the breakdown of hemicellulose and plays an important role in growth by hydrolyzing the mannan-like polysaccharide, but its function in adaptation to salt stress has been less studied.
Methods: Based on cloned the mannanase (MAN) gene from Mirabilis jalapa L., the study was carried out by heterologously expressing the gene in Arabidopsis thaliana, and then observing the plant phenotypes and measuring relevant physiological and biochemical indicators under 150 mM salt treatment.
Results and discussion: The results indicate that MirMAN is a protein with a glycohydrolase-specific structural domain located in the cell wall. We first found that MirMAN reduced the susceptibility of transgenic Arabidopsis thaliana to high salt stress and increased the survival rate of plants by 38%. This was corroborated by the following significant changes, including the reduction in reactive oxygen species (ROS) levels, increase in antioxidant enzyme activity, accumulation of soluble sugars and increase of the expression level of RD29 in transgenic plants. We also found thatthe heterologous expression of MirMAN promoted root growth mainly by elongating the primary roots and increasing the density of lateral roots. Meanwhile, the expression of ARF7, ARF19, LBD16 and LBD29 was up-regulated in the transgenic plants, and the concentration of IAA in the roots was increased. Those results indicate that MirMAN is involved in the initiation of lateral root primordia in transgenic plants through the IAA-ARF signalling pathway. In conclusion, MirMAN improves plant salt tolerance not only by regulating ROS homeostasis, but also by promoting the development of lateral roots. Reflecting the potential of the MirMAN to promote root plastic development in adaptation to salt stress adversity.
1 Introduction
Soil salinity is one of the major issues limiting plant growth and agricultural production, affecting an estimated 50% of arable land in China by 2050 (Jiang et al., 2019; Lu et al., 2019). High salinity reduces water absorption and ultimately leads to osmotic stress in the plant cell. Moreover, root uptake of large amounts of Na+ and Cl− can harm metabolic processes and reduce photosynthetic efficiency (Oka et al., 2013). Oxidative stress, mediated by reactive oxygen species (ROS), is typically accompanied by osmotic stress and ionic toxicity. The effective control of the oxidative damage induced by osmotic and toxic is still critical for plant tolerance to stress.
The root is the first plant organ to encounter salt stress and sense the salt signal. Root activity and biomass are the main indicators to assess the tolerance of plants to stress due to directly affecting plant growth (Li et al., 2018). Root cells respond to adverse environments and boost endogenous defense mechanisms, involving the enhanced processes of synthesis of compatible osmolytes, antioxidants, hormones, transporters, and regulatory proteins. The upregulated expression gene, the response to dehydration 29A (AtRD29A), regarded as an indicator of stress tolerance, performs a protective and regulatory mechanism in plants (Jiang et al., 2019). These physiological processes further influence root system conformation via distinct mechanisms, including control of cell cycle progression, cell size and shape, and cell wall biosynthesis (Zelm et al., 2020).
Root cell proliferation, elongation, and differentiation all involve cell wall remodeling. As we know, the hemicellulose of the cell wall consists of many different polysaccharides that play a significant role in the highly dynamic changes of the cell wall (Cosgrove, 2005; Mohnen, 2008; Moreira and Filho, 2008). Mannans are major components of polysaccharides present within the plant cell wall and endosperm and are highly difficult to degrade (Jin et al., 2019). The enzymatic cleavage of mannans is achieved by several glycoside hydrolase (GH) enzymes, which act on concert to deconstruct the β-1,4-mannan backbone, ultimately releasing d-mannose and/or mannan oligosaccharides (Wang et al., 2013; Malgas et al., 2015), including mannan endo-1,4-β-mannosidase (EC 3.2.1.25) and 1,4-β-d-mannanase (EC 3.2.1.78), commonly named β-mannanase (MAN) (Sak-Ubol et al., 2016). According to the Carbohydrate-Active enZYmes (CAZY) database, MAN belongs to the glycoside hydrolase family 5 (GH5), a multigene family (Guillon et al., 2012). Previous studies have shown that endo-β-1,4-mannanases regulates a variety of plant growth and developmental processes including embryogenesis, leaf formation, stem elongation, shoot branching, fruit development, pollen development, and lateral root emergence (Nonogaki et al., 2000; Peters and Tomos, 2000; Buckeridge, 2010; Konozy et al., 2012; Yan et al., 2012; Wang et al., 2013; Carrillo-Barral et al., 2018; Yamamoto, 2019; Zang et al., 2019).
PtrMAN6 in poplar is specifically expressed in xylem duct cells and catalyzes hydrolysis for the production of mannan oligosaccharides (Zhao et al., 2013). It confirms that MAN coordinates xylem cell expansion and secondary cell wall deposition. It has been determined that AtMAN3 gene plays an important role in the regulation of heavy metal Cd tolerance in Arabidopsis (Chen et al., 2015). Notably, MAN2/6 gene expression significantly increased in Carex rigescens following salt treatment from transcriptome data and then may have contributed to salt tolerance in C. rigescens (Zhang et al., 2020). In plants, MAN participates in a large number of developmental processes. Nevertheless, its function in abiotic stress tolerance remains unknown.
Mirabilis jalapa L., a kind of ornamental plant, has a strong tolerance to petroleum hydrocarbon-contaminated soil and the ability to promote the degradation of pollutant organic molecules (Peng et al., 2009). In this study, we successfully isolated the β-mannanases gene (MirMAN); MirMAN encodes a protein of 411 amino acids that contains a domain showing 68.13% sequence homology to the glycosyl hydrolase motif with Spinacia oleracea (XP_021852790.1), but its exact biological function has not yet been characterized. Complete loss of MirMAN function in plants suggests that MirMAN may be important for cell wall development and plant-related defense systems following salt stress. Accordingly, we generated Arabidopsis plants by heterologous expression of MirMAN and compared phenotypic features and responses to salt stress with wild-type plants. Then, we identified the role of MirMAN in responding to salt stress with an evaluation of the antioxidant status, soluble sugar accumulation, and lipid peroxidation, as well as an analysis of the expression levels of stress resistance-related genes. These results provide important new insights into the function of MirMAN gene in plant stress tolerance.
2 Materials and methods
2.1 Total RNA extraction and PCR amplification of MirMAN gene
M. jalapa RNA was extracted from plants grown with petroleum contamination for 45 days using the RNAprep Pure Plant Total RNA Extraction Kit (TianGen, Beijing, China) following the manufacturer’s instructions. After detection by 1% gel electrophoresis, the total RNA was first treated to remove the genomic DNA and then transferred into the first strand of cDNA using the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The first strand of synthesized cDNA was used as a template. The 2× PCR mix (TaKaRa, Dalian, China) was used to perform the PCR (Supplementary Table 1). The amplified product was recovered, directly ligated to the pMD-19T vector using the TA cloning method according to the operating instruction of pMD™ 19-T Vector Cloning Kit (TaKaRa, Dalian, China), and then transformed into TOP 10 competent cells. The positive clones, identified by bacterial liquid PCR using the sequencing primers M13 (M13F: TGTAAAACGACGGCCAGT, M13R: CAGGAAACAGCTATGACC) in the pMD-19T vector, were sent to Biobios (Shanghai, China) for sequencing.
2.2 MirMAN gene bioinformatics analysis
The molecular mass and isoelectric point of MirMAN were performed with ExPASy (http://web.expasy.org). The sequences of MAN in Arabidopsis thaliana, L. esculentum, and Brachypodium distachyon were obtained from the National Center for Biotechnology Information (NCBI) database (https://blast.ncbi.nlm.nih.gov/Blast). The sequence secondary and tertiary structures were predicted using SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) and SWISSMODEL (https://www.swissmodel.expasy.org/interactive/DTDUCp/models/), respectively. The sequence was analyzed for potential nuclear localization signals using cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi), transmembrane regions using TMHMM (Services- DTU Health Tech), and signal peptides using seqNLS (http://mleg.cse.sc.edu/seqNLS/). The predicted amino acid sequences of MirMAN and homologous genes were compared using DNAMAN, and a molecular phylogenetic tree was constructed using the neighbor-joining method in MEGA ver. 7.0.
2.3 Subcellular localization of MirMAN: GFP
Primer 5.0 software was used to design the primers pCAMBIA1303-MirMAN-F: GAAGATCTAAAGAAAGAAAAAATGAAAATAAAT and pCAMBIA1303-MirMAN-R: CGGACTAGTTCACCTCCTTAACTTTCTAATAATCCTA. With the use of the double-enzyme (BgIII and BamHI) digestion method, the target gene was inserted into the plant overexpression vector pCAMBIA1303 to construct the pCAMBIA1303-MirMAN : GFP vector. The recombinant plasmid and PAD62 plasmid were transformed into competent Agrobacterium tumefaciens GV3101 cells, which were injected into tobacco leaves. Plants were grown in the dark for 1 day and then transferred to long-day conditions (16-h light/8-h dark) for 2 days. The leaf epidermal cells were incubated in 10% sucrose solution for 10 min to induce plasmolysis. The pCAMBIA1303-35S-MirMAN : GFP protein localization was observed under a laser confocal microscope (Leica, TCSSP8, Wetzlar, Germany).
2.4 Genetic transformation and identification of MirMAN overexpression transgenic lines
Agrobacterium LBA4404 containing the recombinant pCAMBIA1303-MirMAN overexpression vector was transfected into Arabidopsis Col-0 inflorescences, and the infected mature seeds (T0) were collected and cultured on 0.5× Murashige and Skoog (MS) (containing 20 mg/L of hygromycin) solid medium to screen for positive transformants. The Arabidopsis seedlings of various lines capable of rooting on the hygromycin-containing medium were identified by leaf PCR with a 2× PCR mix (Aidlab, Beijing, China) and the detection primers. A total of 33 transgenic lines with positive PCR results were selected for subsequent experiments. PCR conditions were 35 cycles of amplification (98°C for 10 s, 55°C for 5 s, and 72°C for 30 s). The total RNA was isolated from the transgenic Arabidopsis lines OE#1-33 and wild-type Arabidopsis using the RNA plant Plus reagent (TianGen, Beijing, China) based on the manufacturer’s instructions. The quantitative PCR (qPCR) was carried out using a FastKing RT Kit with gDNase (KR116, TianGen, Beijing, China) and SYBR Green qPCR Mix (Monad, Suzhou, China) according to the manufacturer’s protocols. qPCR was performed using Monad Selected q225MX (Monad Biotech Co., Ltd., Suzhou, China) real-time detection system. We used the reference gene AtACTIN2 as endogenous controls for Arabidopsis (Supplementary Table 1). Three replicate reactions were routinely performed per sample.
2.5 Western blotting assays
For the Western blotting test of the Arabidopsis leaves, the protein content was measured using the method of Bradford (1976). Proteins were separated according to molecular weight by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) on a 10%–14% gradient gel and then transferred to an immobilon-P polyvinylidene difluoride (PVDF) membrane (Solarbio, Beijing, China). Proteins were conventionally detected with the MirMAN antibody (custom-made by GenScript, Piscataway, NJ, USA). Further, an anti-rabbit horseradish peroxidase (HRP) antibody was used as a secondary antibody. The blots were incubated with ECL Western Blotting Substrate (Solarbio, Beijing, China) and imaged using a Bio-Rad image analyzer (Bio-Rad, Hercules, CA, USA).
2.6 Morphological characterization of plant
Surface-sterilized seeds were plated on 0.5× MS medium with 1% sucrose and 1% agar (pH 5.7), and the plates were incubated in the dark at 4°C for 3 days. Then, plants were grown in the artificial growing room at a temperature of 22°C, 16-h light and 8-h dark (60% humidity and 120 μmol m–2 s–1 during a 16-h daily light period). We conducted digital images of root system morphology using a desktop scanner and determined the root length and the number of tips, forks, and crossings by using WinRHIZO. The seedlings were grown on 0.5× MS medium for 4 days, and then the agar plates were rotated 90°, causing the gravistimulus for 48 h. Data were compiled from three independent experiments with each line. Methods for the determination of IAA contents in the seedlings by mass spectrometry were described (Jia et al., 2020).
2.7 Salt stress treatment and physiological index determination of transgenic plants
For salt stress treatment, wild-type (WT) Arabidopsis and transgenic lines were planted in the same black plastic pots in the soil, and the plants were grown in a greenhouse. The 3-week-old seedlings in soil were irrigated with water and 150 mM of NaCl solution every 3 days and sampled at 0, 3, 5, and 7 days. Soil-grown Arabidopsis plants that were 24 days old were continuously watered with or without 150 mM of NaCl for 7 days, followed by an analysis of the survival rate after recovery for 1 week.
Catalase (CAT) activity was assayed on the basis of its ability to decompose H2O2 and measured at 240 nm. Superoxide dismutase (SOD) activity was determined by measuring its ability to inhibit the reduction of nitro blue tetrazolium chloride (NBT). Peroxidase (POD) activity was measured as the oxidation of guaiacol in the presence of H2O2, during which tetraguaiacol was formed. Lipid peroxidation in the plants was measured by the thiobarbituric acid-reacting substance and was expressed in terms of malondialdehyde (MDA) content. Diaminobenzidine (DAB) and NBT staining were used to confirm the detection of and H2O2, respectively. Glutathione (GSH) levels were measured according to the instructions supplied with the GSH Assay Kit (Suzhou Comin Biotechnology, Suzhou, China). Total soluble sugar was assayed by the anthrone method, which is widely used for the determination of soluble sugars. In brief, approximately 2 mL of the anthrone chemical reagent was added to 1 mL of the sample and then measured at 630 nm, and total soluble sugar content was calculated. Endo-β-mannanase activity was measured by a spectrophotometric method (Determination of activity β-mannanase as feed additives—Spectrophotometric method, GB/T 36861-2018, China). Briefly, the reducing oligosaccharides and monosaccharides can react with the 3,5-dinitrosalicylic acid reagent in a boiling water bath to yield a color change indicative of mannose hydrolysis. Each measurement was repeated at least three times, with data collection repeated at least three times.
2.8 Statistical analysis
All the data were presented as averaged values of three independent replicates. Statistical analyses of the data were carried out using Student’s t-tests, and multiple comparisons of means were analyzed using Tukey’s test. Differences were considered statistically significant at p< 0.05.
3 Results
3.1 Cloning and characterization of MirMAN in Mirabilis jalapa
The coding sequence of MirMAN was predicted from transcriptome sequencing results (data not published). The open reading frame (ORF) sequence of MirMAN is 1,233 bp long and contains 411 amino acids, with a predicted molecular weight of 47.08 kDa and an isoelectric point of 7.16 (pI 7.16). The calculated instability coefficient and hydrophobicity are 38.81, and −0.46, respectively, which are indicative of stable hydrophilic protein. Multiple sequence comparisons showed that MirMAN was 46.96%, 45.85%, 43.99%, 42.65%, and 42.31% consistent with LeMAN1, AtMAN7, LeMAN4, AtMAN3, and AtMAN1, respectively (Figure 1). In the secondary structure of MirMAN, α-helix accounted for 38.2%, β-sheets for 10.7%, extension chain for 20.9%, and random structures for 30.17%. Computational analysis predicted that MirMAN folds in a typical (β/α)8-TIM barrel structure, with multiple β-sheets forming a hydrophobic domain (Supplementary Figure 2).
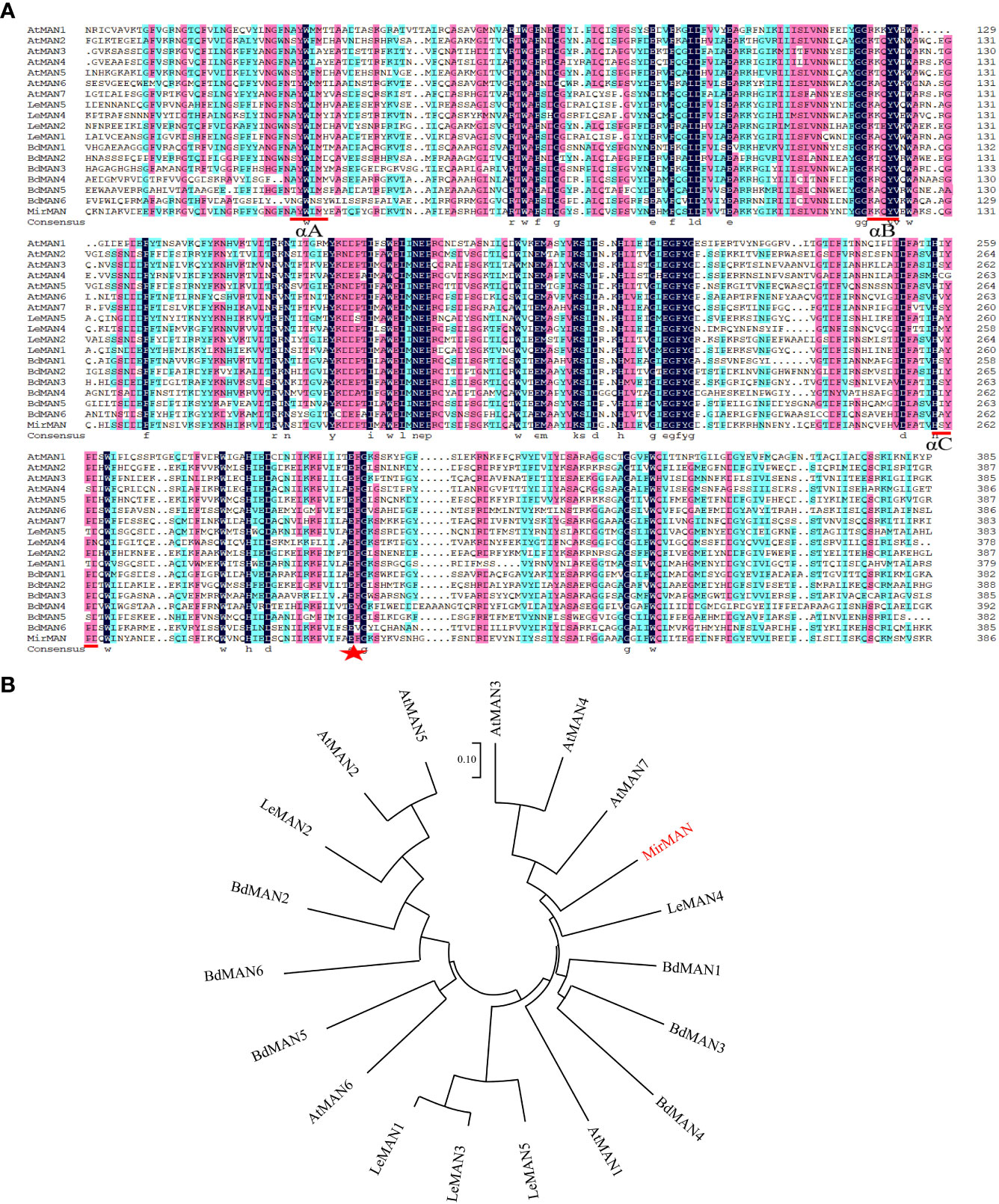
Figure 1 Alignment of sequences and phylogenetic tree of MirMAN proteins. (A) Alignment of sequences of MirMAN proteins and homologs from others species. Blue inverted triangles indicate signal peptide sites, red lines indicate the MAN conserved sequence, and red star indicates the catalytic glutamate residue structural motif. Multiple sequence comparisons showed that MirMAN was 46.96%, 45.85%, 43.99%, 42.65%, 42.31%, 41.79%, 39.72%, 39.15%, 39.08%, 35.71%, 35.03%, 34.98%, 34.57%, 34.41%, 33.64%, 31.98%, 31.57%, and 29.13%, consistent with LeMAN1, AtMAN7, LeMAN4, AtMAN3, AtMAN1, AtMAN4, LeMAN5, BdMAN1, LeMAN3, AtMAN6, BdMAN5, BdMAN2, AtMAN5, AtMAN2, BdMAN3, LeMAN2, BdMAN4 and AtMAN6, respectively. (B) Phylogenetic tree of MirMAN proteins from Mirabilis jalapa and other species. At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Le, Lycopersicon esculentum.
3.2 Subcellular localization of MirMAN protein
The marker for plasma membrane (PM), PAD62-mcherry, was co-expressed with pCAMBIA1303-35S-MirMAN-GFP by Agrobacterium-mediated transient expression in Nicotiana benthamiana leaves. After 16 h of agroinfiltration, we observed fluorescence in the green channel with a confocal laser scanning microscope and compared its position. Red PAD62 fluoresced in membrane epithelial cells (Figure 2A). pCAMBIA1303-35S-MirMAN-GFP green fluorescence was clearly distributed in a dotted pattern in the cell (Figure 2A). To investigate whether these signals were outside or inside the membrane, 30% sucrose was added to induce plasmolysis of N. benthamiana leaf cells, and images were acquired using a confocal microscope. Upon plasmolysis, the pCAMBIA1303-35S-MirMAN-GFP signal remained predominantly associated with the cell wall, although the PAD62-labeled PM retracted from the cell wall (Figure 2B).
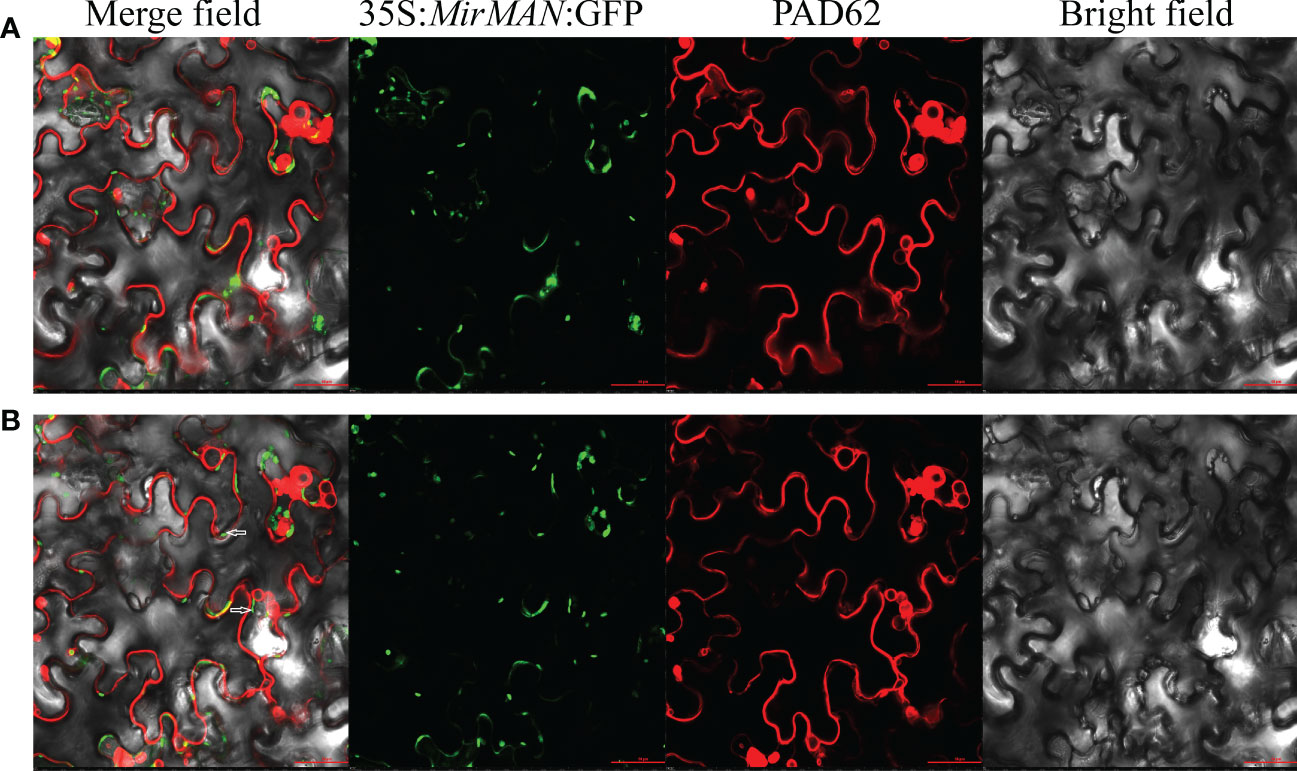
Figure 2 Subcellular localization of MirMAN. (A) Subcellular localization of 35S:MirMAN: GFP in tobacco cells. (B) Subcellular localization of 35S:MirMAN: GFP following 30% sucrose treatment. Bar = 50 μm. After plasmolysis, GFP signal observed in the cell wall is indicated by white arrows.
3.3 Generation of transgenic Arabidopsis lines
The MirMAN ORF sequence was conducted into a plant binary vector control of the constitutive CaMV 35S promoter and transformed into Arabidopsis Columbia-0 (Col-0) plants. Thirty-three independent transgenic Arabidopsis lines were generated, their identity from 33 lines had already been confirmed by PCR analysis, and these lines were designated OE#1–33 (Figure 3A). Expression of MirMAN protein in the transgenic lines was demonstrated by Western blotting. Figure 3B shows the result of Western blotting after polyclonal MirMAN antibody detection. As shown in Figure 3B, a clear band was observed at approximately 47 kDa and was specifically precipitated by the antibody, but not in the control group. These results indicate that MirMAN has been successfully integrated into the Arabidopsis genome. Therefore, T3 mutants (#20, #26, and #33) were selected for further analysis. We identified MirMAN gene expression levels in T3 generation transgenic lines under normal conditions (Supplementary Figure 1).
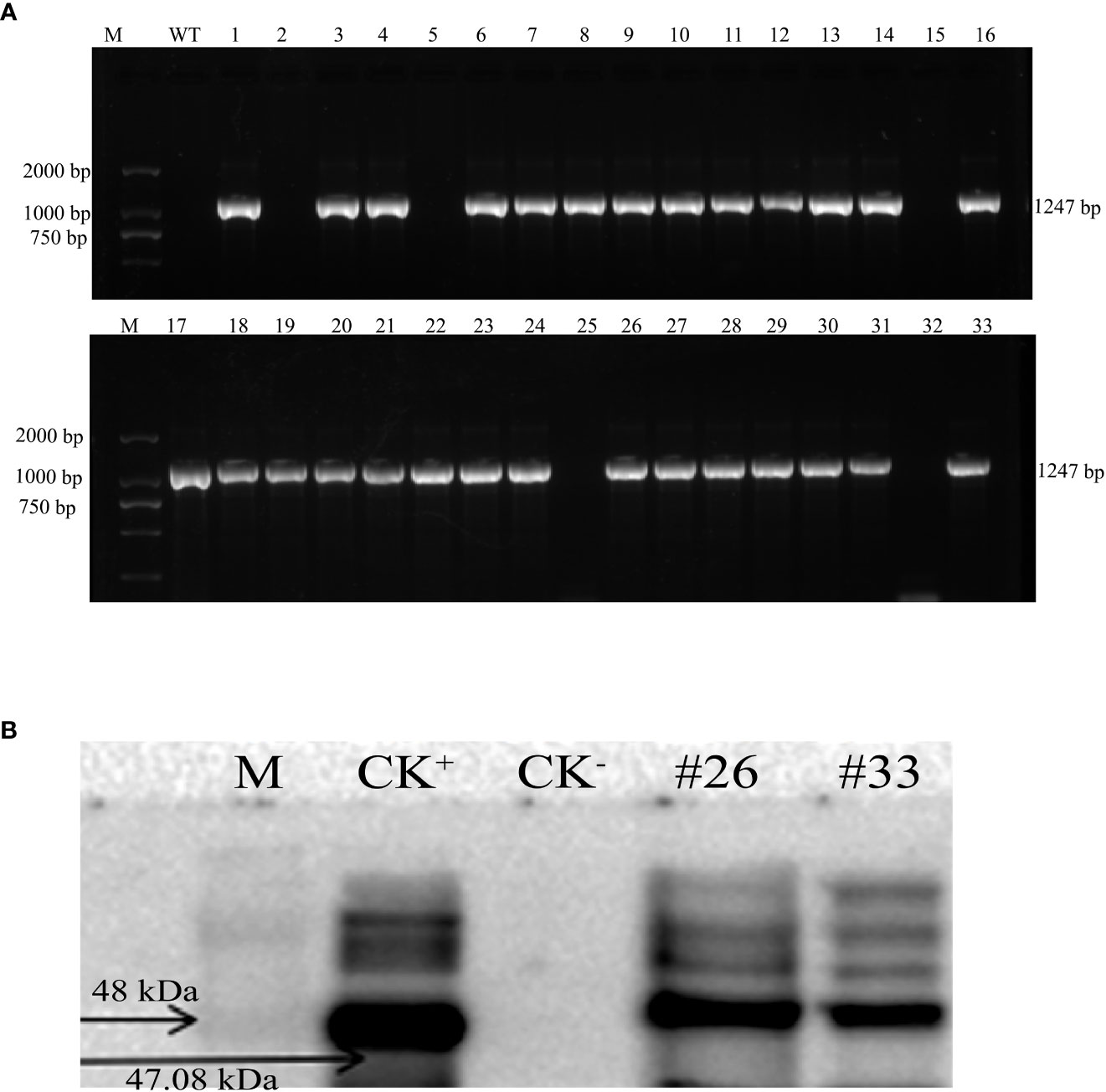
Figure 3 Characterization of recombinant MirMAN gene in Arabidopsis. (A) Cloning of MirMAN gene and PCR identification of T2 transgenic Arabidopsis. (B) The specific antibody was used for Western blotting. (A) M: DL marker 2,000; WT: wild type (negative control); the others are the 33 transgenic lines. (B) M: marker; CK+, positive control; CK−, negative control; the others are the transgenic lines.
3.4 Effects of root system architecture in MirMAN heterologous-expressing Arabidopsis
Compared to WT Arabidopsis, transgenic Arabidopsis lines #20/26/33 display an increased total number of lateral roots and primary root lengths in the 0.5× MS medium (Figure 4). Indeed, we noticed that the OE#20/26/33 lines with numerous lateral roots were up to 3.5–4-fold. To assess this possibility, we used gravitropic stimulation to synchronize lateral root primordia initiation in WT and OE#20/26/33 seedlings (Figure 5A). The period of the seedlings’ lateral root primordia emergence was defined as eight time periods, and their temporal developmental axis is shown in Figure 5B. We observed that transgenic Arabidopsis #20/26/33 lines display increased lateral root density and lateral root primordia per developmental stage III–VIII (Figure 5C). To better understand the relationship between IAA content and the numerous lateral roots, transgenic Arabidopsis lines’ IAA content was tested. IAA content was more accumulated in transgenic Arabidopsis line #26 (Figure 5D). We focused on the expression of root development-related genes, which was upregulated by 3–10-fold in transgenic lines compared to that of control plants, including LBD16 (lateral organ boundaries domain), LBD29, ARF7 (auxin response factor), and ARF19 (Figure 5E).
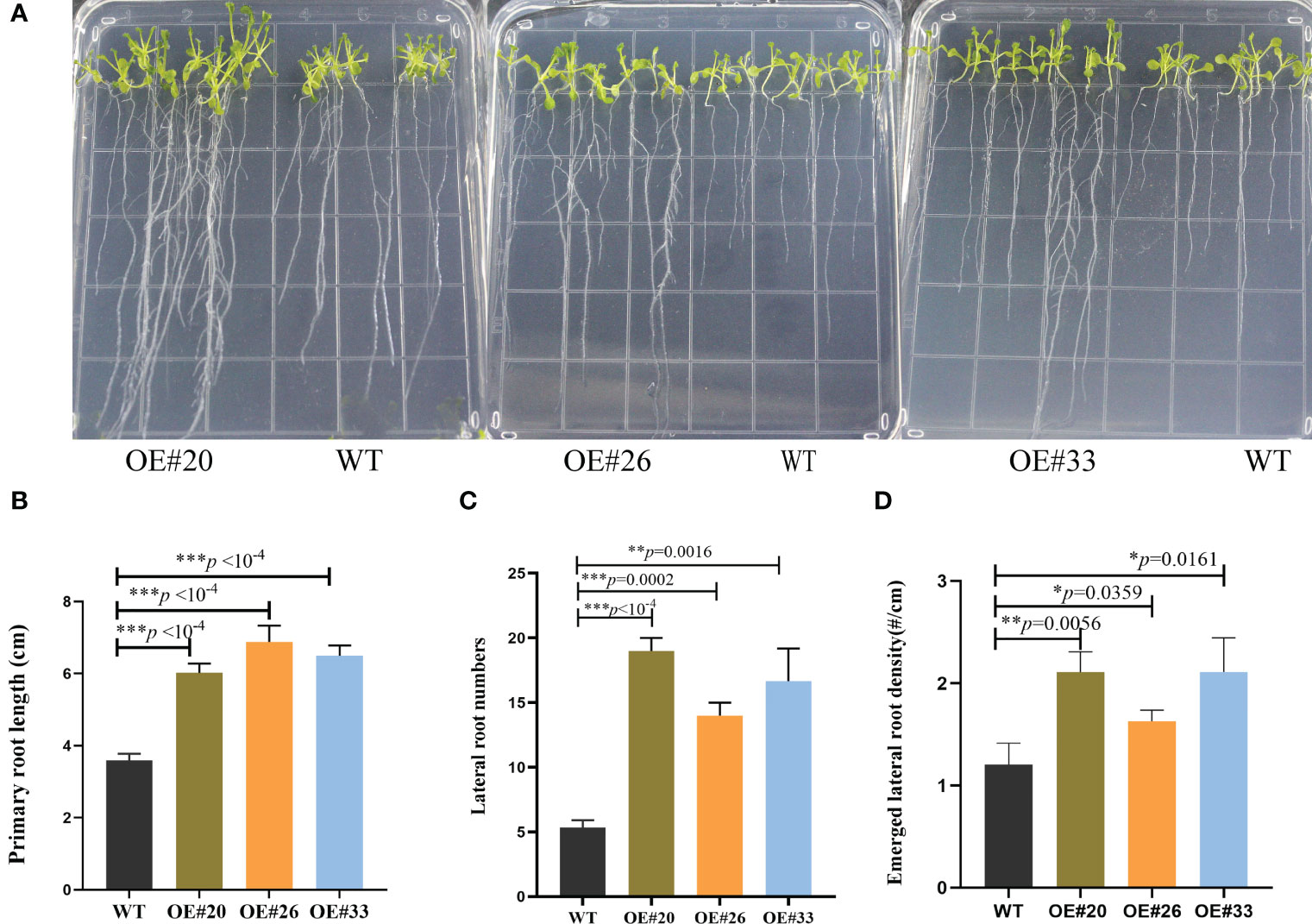
Figure 4 MirMAN regulation of root development on transgenic Arabidopsis plants. (A) The phenotypic root observation of wild type (WT) and MirMAN-OE lines. (B) Length of the primary root. (C) Number of lateral roots. (D) Density of lateral root primordia. The seedlings were cultured for 12 days. Data are means ± SD (n = 10). *p< 0.05, **p< 0.01, ***p< 0.001, independent-samples t-test.
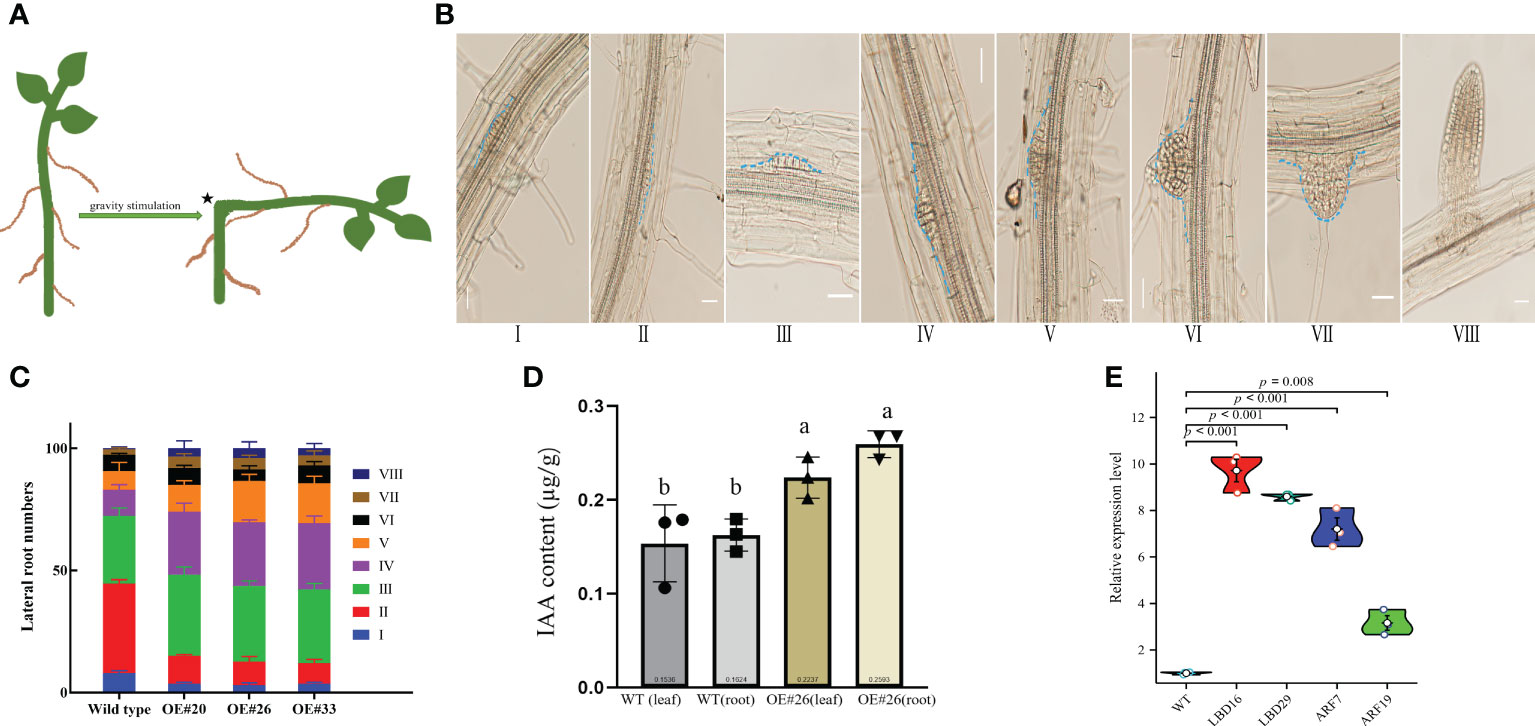
Figure 5 Comparison of lateral root density of wild type (WT) and OE lines Arabidopsis plants at various developmental stages. (A) Diagram showing a classic approach that was used to induce lateral root initiation and formation. Bending the horizontally placed primary root tip 90° and allowing the root tip to grow in the direction of gravity can induce the initiation and formation of lateral roots at the convex side of the bending site, as indicated by an asterisk. (B) Lateral root formation at different developmental stages detected from a root of single plant. (C) Measurements of the percentage of lateral root formation of WT and OE lines Arabidopsis at various developmental stages (n = 100). Measurements were carried out after gravity stimulation for 48 h. (D) IAA content in WT and OE#26 seedlings. (E) Expression level of LED16, LBD29, ARF7, and ARF19 genes. At least three independent biological replicates were carried out, and consistent results were obtained. Results from one representative experiment are shown. Ten-day-old seedlings were used for analysis. Data shown represent average and SD (n = 100). Letters indicate the results for comparisons across groups using Tukey’s post hoc tests.
3.5 Salt tolerance analysis of MirMAN heterologous-expressing transgenic Arabidopsis
In the comparison of phenotypic differences between the WT and the OE#26 plants under salt stress, the treatment of WT Arabidopsis with 150 mM of NaCl strongly inhibited root growth, and OE#26 showed less growth inhibition when treated with 150 mM of NaCl in soil (Figure 6). To determine salt tolerance in soil, we treated 24-day-old soil-grown plants with 150 mM of NaCl for additional 7 days, followed by recovery for 1 week. Most of the leaves of WT Arabidopsis gradually turned yellow, shrunk in size, and wilted, but the wilting symptoms of OE#26 plants were significantly slower than those of the control (Figure 6A). In addition, after rehydration, 61% of OE#26 plants and only 23.0% of control plants resumed their growth (Figure 6B). Salt stress significantly inhibited root and shoot fresh weight in WT and transgenic Arabidopsis. Specifically, the OE#26 plants showed a significantly higher root fresh weight, shoot fresh weight, and root/shoot ratio than those in WT plants under salt stress (p ≤ 0.05) (Figures 6C, D). While WT plants responded to salt stress by decreasing their root-to-shoot ratio, the ratio in transgenic Arabidopsis remained unchanged (Figure 6E).
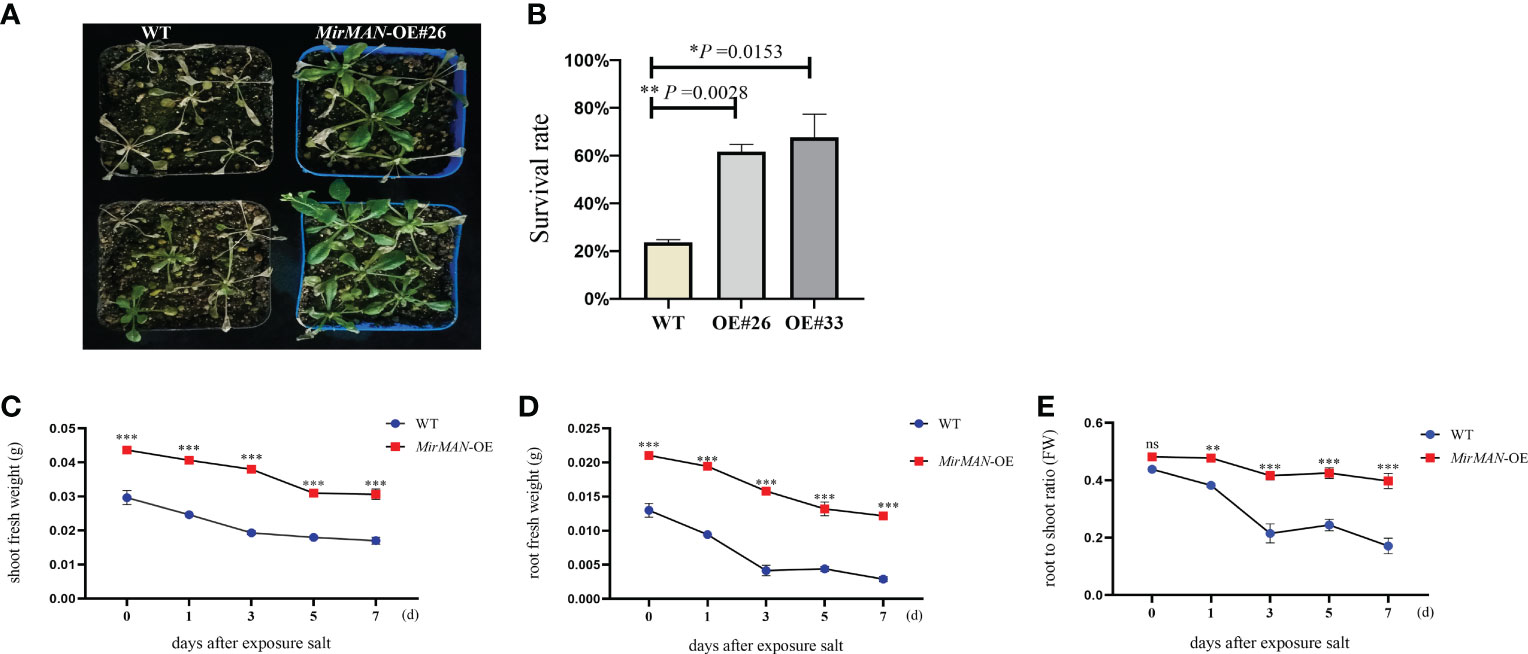
Figure 6 Effect of salt stress on MirMAN-overexpressed Arabidopsis plants. (A) Plants were grown on soil for 3 weeks and then exposed to 150 mM NaCl stress. (B) Survival rate under 150 mM NaCl stress. (C) Shoot fresh weight of plants. (D) Root fresh weight of plants. (E) Root-to-shoot ratio. n = 5. Error bars indicate ± SD. Student’s t-tests were used to compare the measurements of OE lines with those from wild type (WT). *p< 0.05, **p< 0.01, ***p< 0.001. ns represents no significance at p > 0.05.
3.6 MirMAN heterologous expression improves free radical scavenging activity and antioxidant status
We next tested the physiological alterations in WT and OE#20/26 plants under salt stress conditions. We also measured the H2O2 and characteristics by chemical staining and analyzed the H2O2 and contents of WT and OE#26 plants after salt treatment. We found that WT plants accumulated more H2O2 and than OE#26 plants (Figures 7A, B). Upon NaCl treatment, the average MDA content in WT plants was 4.49 times higher than in MirMAN heterologous expression plants (Figure 7C). The OE#20/26 plants increased the levels of GSH compared to the values of the WT Arabidopsis during the normal conditions (Figure 7D). Upon salt treatment, the values of GSH and GSH/GSSH in both the OE lines and WT plants increased significantly, with the OE lines exhibiting significantly higher levels than WT Arabidopsis (Figures 7D, F). The opposite was true for oxidized GSH (GSSG) concentrations, whose levels were 25.64 and 24.67 significantly lower for the OE lines compared to the WT (52.09), respectively (Figure 7E). In addition, GSH/GSSG values in the OE lines under salt stress increased to a greater extent compared to those in the WT Arabidopsis (Figure 7F). The total soluble sugar content in the OE#20/26 plants was 1.6 times that in the WT plants (Figure 8A). Under control conditions, total mannanase activity in both OE lines had significant increases compared to that in the WT plants (Figure 8B), showing that increased MirMAN overexpression resulted in more mannanase production. Moreover, salt stress increased the transcript levels of RD29A in transgenic plants by at least twice compared to those in WT plants (Figure 8C). Furthermore, our data show that MirMAN substantially increases CAT, POD, and SOD activities (Figures 8D–F).
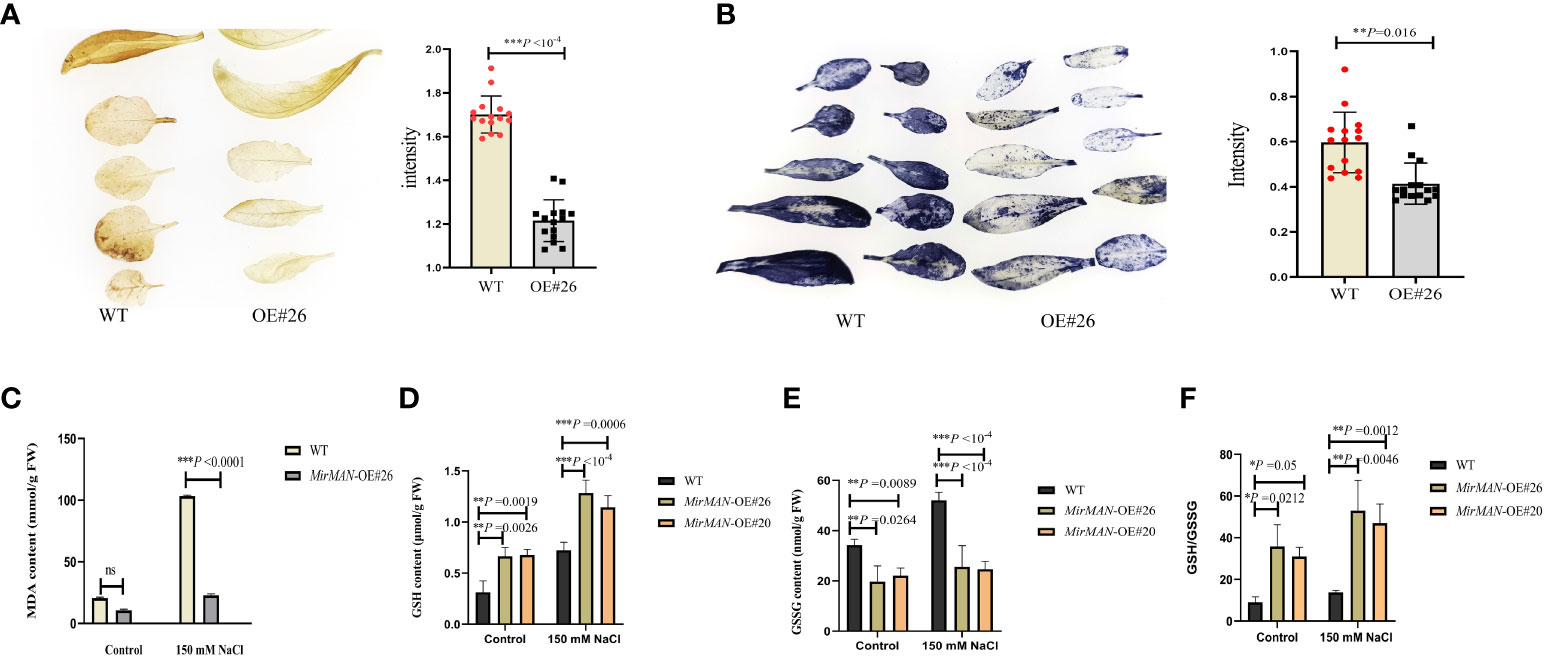
Figure 7 Evaluation of salt resistance in WT and OE lines Arabidopsis plants. (A) H2O2 contents. (B) contents. (C) MDA contents. (D) GSH contents. (E) GSSG contents. (F) GSH/GSSG. MDA, malondialdehyde; GSH, glutathione; GSSG, oxidized glutathione; WT, wild type. Error bars indicate ± SD. Student’s t-tests were used to compare the measurements of OE lines with those from WT. *p< 0.05, **p< 0.01, *** p< 0.001. ns represents no significance at p > 0.05.
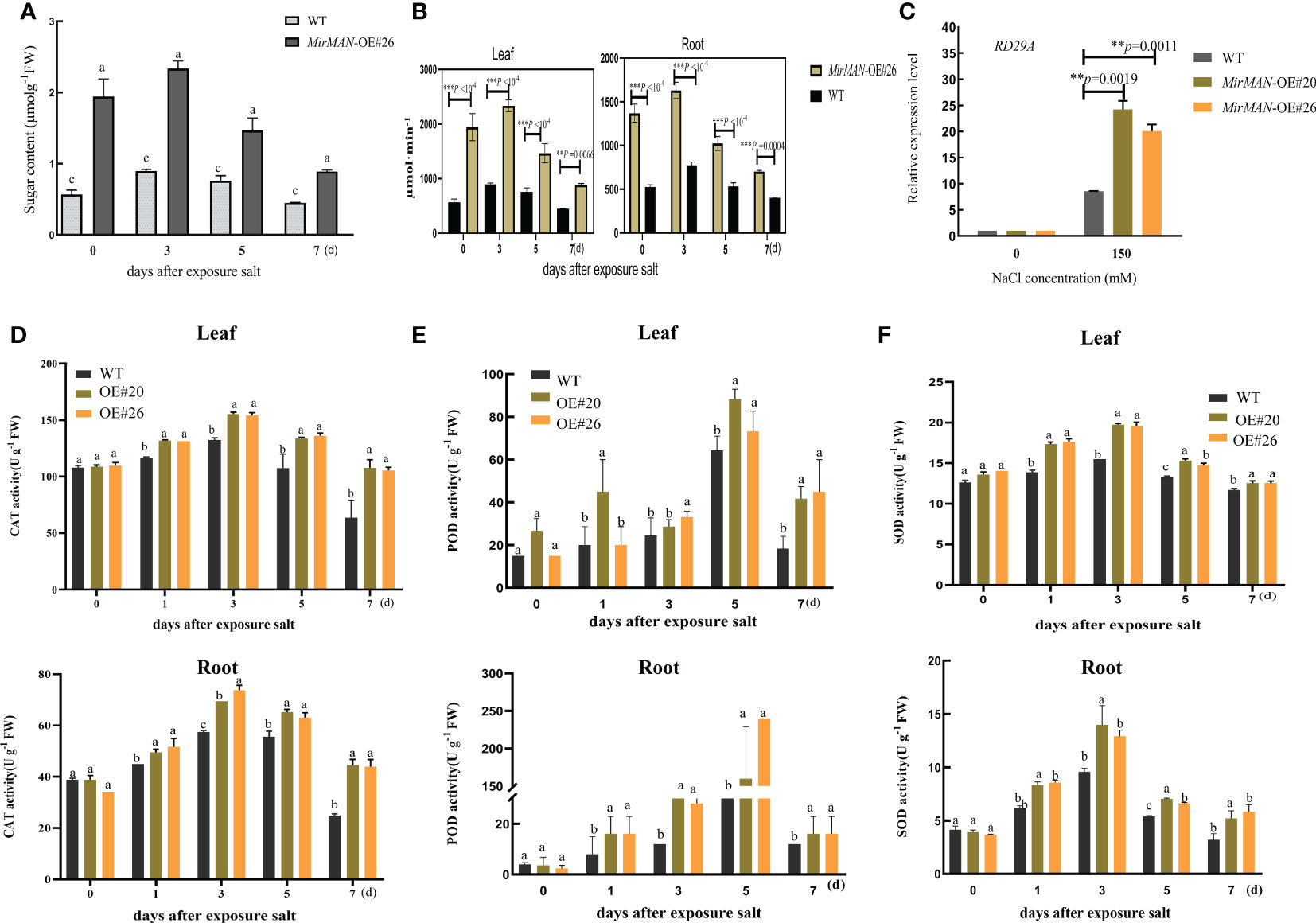
Figure 8 Salt resistance analysis of WT and transgenic lines of Arabidopsis plants. (A) Soluble sugar contents. (B) Endo-β-mannanase activity. (C) RD29A gene expressions after salt experiment. (D) CAT activity. (E) POD activity. (F) SOD activity. SOD, superoxide dismutase; POD, peroxidase; CAT, catalase; WT, wild type. Error bars indicate ± SD. Student’s t-tests were used to compare the measurements of OE lines with those from WT. **p< 0.01, *** P < 0.001. Letters indicate the results for comparisons across group using Tukey’s post hoc test.
4 Discussion
Although the orthologues of MAN genes have been identified in many plant species, such as L. esculentum, B. distachyon, and Brassica rapa, the influence of MirMAN on the regulation of growth remains almost unknown (Nonogaki et al., 2000; Iglesias-Fernández et al., 2011; González-Calle et al., 2015; Carrillo-Barral et al., 2018). Here, MirMAN was cloned from M. jalapa L., and its function of improving the growth and salt tolerance of plants was analyzed in Arabidopsis.
4.1 Characteristic and subcellular localization analyses of MirMAN
Sequence and phylogenetic analyses showed that MirMAN is homologous to AtMAN7, but its function has not been fully resolved (Figure 1). Here, we demonstrated that the heterologous expression of MirMAN can substantially alter relevant root development without abiotic stress in Arabidopsis. Like other MANs, MirMAN has a putative signal peptide and an active site with the expected conserved amino acids. The MirMAN is composed of a sole GH5 catalytic module without any attached carbohydrate-binding module (CBM). Judging from the subcellular distribution of MirMAN : GFP protein fluorescence signal in Nicotiana tabacum cells, MirMAN was localized to the cell wall (Figure 2). The subcellular localization of MirMAN on the cell wall might play an important role in cell-wall loosening, as many cell expansion requires primary cell-wall loosening and incorporation of newly synthesized cell-wall material.
4.2 Effects of heterologous expression of MirMAN on plant growth in Arabidopsis
Heterologous expression of MirMAN gene in Arabidopsis enhances plant growth. The effect of 35S: MirMAN transgenic plants on leaf development was particularly striking in rosette leaves. In MirMAN transgenic plants, the rosette leaves were large and elongated with increases in the compactness of the branches and leaves of the plants (Supplementary Figure 3). MirMAN causes a series of metabolic changes in transgenic plants, among which the increased cell growth rate is one of them. Moreover, the heterologous expression of MirMAN obviously changed root system architecture in transgenic plants, including longer primary roots and more numbers of lateral roots (Figures 4, 5). This indicates that MirMAN ultimately affects lateral root formation at different developmental stages. Lateral roots are important not only for determining plants’ root system phenotypes in soil but also for absorbing water and nutrients (Lucob-Agustin et al., 2020). Several studies have set to demonstrate cell wall composition and extensibility mediated by cell wall modification enzymes necessary for root cell elongation, thereby controlling root elongation and, consequently, root development (Tabuchi and Matsumoto, 2001; Ma et al., 2004). In our study, the number of lateral roots of transgenic plants increased, which may enable them to store more water. Transgenic plants showed an enhanced survival capacity compared with the control plants in salt treatment.
Exogenous application of auxin or improved endogenous auxin synthesis leads to a significant increase in the number of lateral roots (Laskowski et al., 1995). In these experimental results, the increase of lateral roots of overexpression MirMAN is closely related to the increase of endogenous auxin (Figure 5D). It is speculated that there may mechanisms, as follows: 1) the increase of auxin concentration promoted the early arrival of auxin oscillation peak. After detection of the initial signal auxin, rhizosphere meristem cells rapidly select lateral root cells, accelerate lateral root primordium development, and further develop lateral root organs. 2) When the lateral root primordium breaks through the epidermis, the cell wall is more likely to change; that is, cells are more likely to detach and form new cells, thereby speeding up the overall lateral root development. 3) The polysaccharide composition of the cell wall changed. Mannanase cleaves mannan or mannan oligosaccharide, and its products mannan oligosaccharide and mannose are speculated to be involved in upstream signal control hormone signal, which may have a positive development feedback mechanism, but the relationship between them needs to be further proved.
Like other MANs, C has putative glutamate catalytic residues for breaking the mannosidic bond (http://smart.embl-heidelberg.de/). Typical MAN proteins are composed of three helices: αA, αB, and αC (Figure 1). MirMAN is mainly predicted to control the extensibility of the cell wall by causing crosslinking or cleaving of cell wall polysaccharides and thus promote cell wall loosening and therefore growth. We speculated that MirMAN promotes hydrolysis of hemicellulose by endo-β-1,4-mannanases, thereby enhancing cell wall expansion. Endo-β-mannanase hydrolyzes mannan oligosaccharide or mannans to produce mannose and/or mannan oligosaccharide (Wang et al., 2013). The research found that galactoglucomannan oligosaccharide (GGMO) treatment, while promoting the primary root elongation and stimulating the emergence of lateral roots, influences Zea mays L. growth development (Kollárová et al., 2018). Previous studies have focused on the downstream effects of lateral root formation during root development, as glucose acts upstream of the BR signal transduction pathway to regulate the lateral root process by dose-dependently. Similarly, glucose also acts on the fiber elongation in cotton directly by BR signal transduction (Gupta et al., 2015; Li et al., 2022).
4.3 Heterologous expression MirMAN most likely leads to salt tolerance in Arabidopsis
Salt stress is dramatically imposing severe constraints on plant adaptation, crop productivity, and quality. The search for molecule regulation of plant stress responses and plant development remains a key priority in the fight against abiotic stress resistance. As plants are exposed to abiotic stresses, most of the root system architectures (RSAs) altered according to the environment contribute to tolerance and recovery, which involves gene expression of root cell wall remodeling (Motte and Beeckman, 2019; Zhang et al., 2019). Overexpression of MirMAN causes suitable root phenotypes in transgenic plants within salt stress, including main root elongation and promoting lateral root density (Supplementary Figure 4), which positively regulates the salt stress resistance. This positive effect may have partially offset the growth inhibition induced by salt stress in transgenic Arabidopsis.
Adversity can cause the overproduction of ROS and thereby affects oxidative stress, endoplasmic reticulum (ER) stress, and consequent damage to proteins and subcellular organelles in plants (Mittler et al., 2011). Several cellular mechanisms have been shown to counterbalance the production of ROS, including enzymatic and non-enzymatic pathways. In plants undergoing salt stress responses, antioxidant enzyme activities are important for suppressing ROS-induced oxidative stress. At 150 mM NaCl-treated groups, H2O2 and were overproduced in wild-type plants (Figures 7A, B). Considering SOD, CAT, and ascorbate peroxidase (APX) activity data in transgenic lines of Arabidopsis (Figures 8D–F), it is concluded that both antioxidant enzymes successfully eliminate the excess of H2O2 and , avoiding the oxidative stress damage caused by salt stress. By contrast, the heterologous expression in MirMAN plants shows a lower MDA content than in wild-type plants (Figure 7C). Similarly, glutathione is an endogenous antioxidant that exists in either reduced (GSH) or oxidized (GSSG) form that protects various cellular structures from oxidative damage. Under salt stress, the GSH levels of both WT and MirMAN transgenic lines of Arabidopsis were significantly upregulated, but the GSH level of OE#26 was more significant (Figure 7D), indicating that there are more GSH in the OE#26 against more oxidative stress. Also, in the transgenic plants, more glutathione was used for the reduction reaction, and less glutathione was produced by oxidation (Figure 7E). Glutathione accumulated in higher concentrations in MirMAN transgenic lines, which in turn helped plants to withstand oxidative stress mediated by the H2O2 and during episodes of salt stress. These results suggest that overexpression of MirMAN plants reduces the accumulation of oxidative H2O2 and by enhancing antioxidant defenses to oxidative stress. The MirMAN transgenic Arabidopsis lines have optimized an antioxidant defense system to increase salt tolerance. Soluble sugar fraction levels in these lines are significantly increased compared with wild-type levels (Figure 8A). Plants can use metabolites to reduce the osmotic potential of plants, which prevents the imbalance of ROS production and ensures water balance for the normal growth of plants under salt stress. The metabolites are bioactive molecules that play a key role in stress, but they are also osmotic regulators of many intracellular processes including preventing membrane fusion and stabilizing enzymes and other intracellular components to cope with salt stress. The relationship between these metabolites interacting with salt tolerance needs further studies (Ismail and Horie, 2017; Liu et al., 2021).
Moreover, we believe that the proposed root development strategy could also substantially improve their performance in salt stress and recovery efficiency. Studies showed that the BR signal transduction component BES1 directly affects the expression of cell wall remodeling genes XTH19 and XTH23 as a positive regulator of salt stress, which promoted the development of lateral roots and thus improved the salt adaptability (Xu et al., 2020). Mannose acts as a signal molecule to regulate plant growth and development (Richterová-Kučerová et al., 2012). It suggests that these sugar molecules can specifically integrate various responses to endogenous and environmental signals to modulate root development in Arabidopsis (Valifard et al., 2021). Plant abiotic stress resistance is improved by the application of the small molecule sugar and interferes with the transcriptional dynamics of downstream signal transduction, indicating that it plays an important role in stress-related pathways. Most of the underlying molecular mechanisms remain unclear (Liu et al., 2021). Understanding how the MirMAN hydrolysate perceives, integrates, and responds to environmental signals is critical for improving plant resistance to stress and will be the focus of our future research efforts.
The experiment aimed to study the response mechanism of MirMAN to salt stress. Little is known about the physiological functions of these enzymes in planta. Although the causal relationship between hydrolysis products of mannanase and lateral root development is yet to be established by designing further experiments, this provides us a new insight into the accumulation of endo-β-mannanase in plants, which causes a change in plant growth and development under salt stress. It also provides further intriguing evidence suggesting that the cell wall remodeling gene may have widespread and as yet largely unrecognized functions in plant growth, development, and plant stress tolerance. At present, we cannot distinguish between direct effects (e.g., specific binding to a receptor) and indirect effects (e.g., hydrolysate). Nevertheless, elucidation of the mechanistic basis of these effects is likely to provide new insights into the factors that govern plant growth and stress tolerance.
In summary, we successfully isolated MirMAN gene from M. jalapa and defined its location and function (Figure 9). The heterologous expression of MirMAN enhanced Arabidopsis growth and development and improved the resistance of transgenic Arabidopsis to salt stress by increasing soluble sugar contents and enhancing antioxidant defenses. MirMAN plays an important role in plants by enhancing root growth. It can be a candidate gene for plant molecular breeding and provide valuable insights into the physiological significance of MAN in plant abiotic stress.
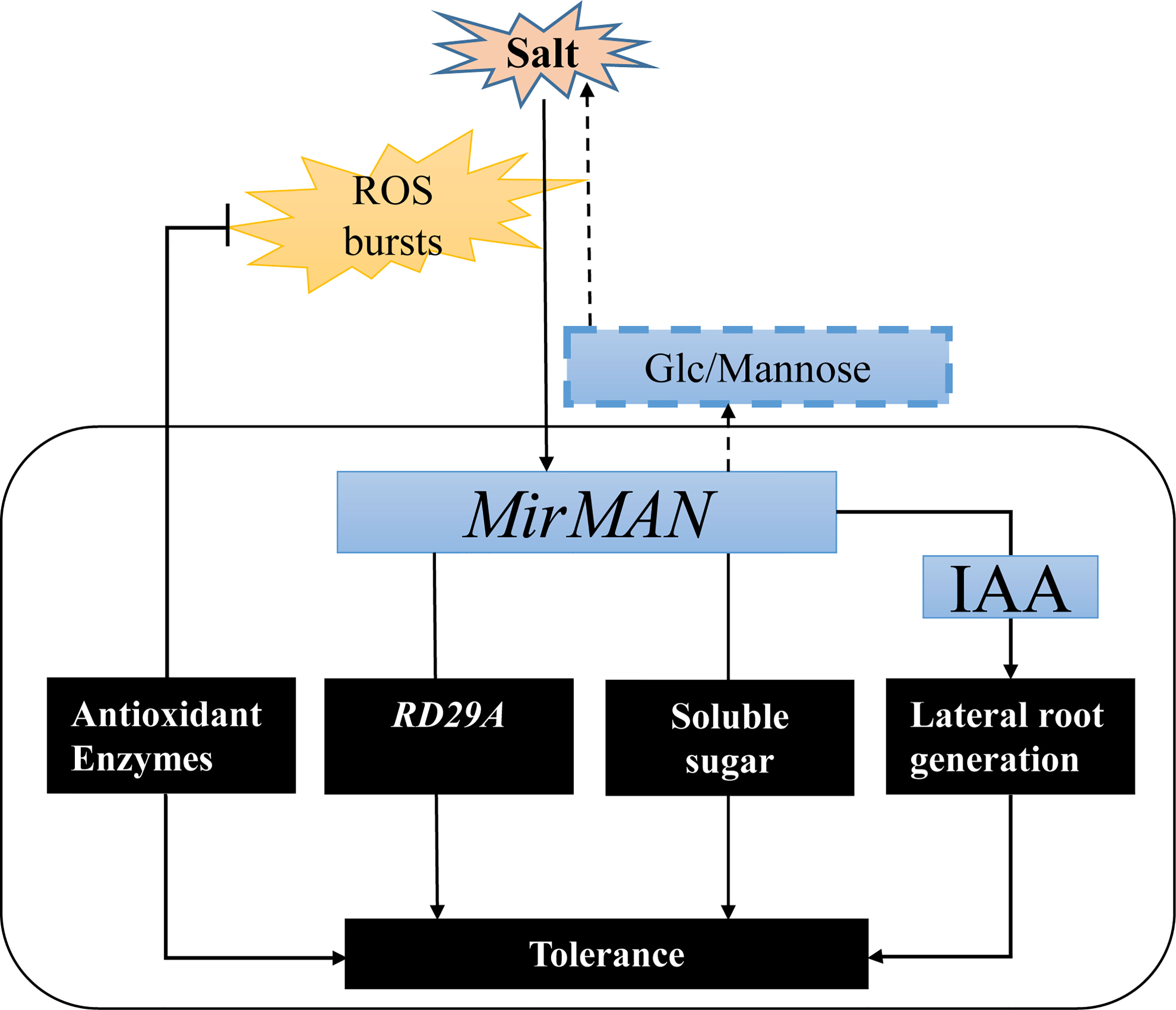
Figure 9 A hypothetical model showing the roles of MirMAN in regulating salt tolerance. MirMAN affects the expression of its downstream stress genes such as RD29A, mediating plant tolerance during development and salt stress. MirMAN can hydrolyze mannans and increase soluble sugar content to alleviate osmotic stress. It can also regulate plant tolerance by causing changes in the antioxidant enzyme system.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Author contributions
MZ and HX were responsible for the study design. JX, CY, SC and HL drafted the manuscript. JX, CY and SJ performed the experiments. SJ, CY, HM and JL performed the statistical analyses. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Special funds of “Collaborative Innovation Center for Genetic Improvement and High Quality and Efficiency Production of Northeast Japonica Rice In China (2022)” (Grant No. KF2022-07).
Acknowledgments
We thank the team of researcher Lu Xiaochun from Liaoning Province Key Laboratory of Crop Molecular Improvement and Sorghum Institute of Liaoning Academy of Agricultural Sciences for kindly helping us with Arabidopsis cultivation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1118548/full#supplementary-material
References
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Buckeridge, M. S. (2010). Seed cell wall storage polysaccharides: Models to understand cell wall biosynthesis and degradation. Plant Physiol. 154, 1017–1023. doi: 10.1104/pp.110.158642
Carrillo-Barral, N., Matilla, A. J., Rodríguez-Gacio, M., del, C., Iglesias-Fernández, R. (2018). Mannans and endo-β-mannanase transcripts are located in different seed compartments during brassicaceae germination. Planta 247, 649–661. doi: 10.1007/s00425-017-2815-4
Chen, J., Yang, L., Gu, J., Bai, X., Ren, Y., Fan, T., et al. (2015). MAN3 gene regulates cadmium tolerance through the glutathione-dependent pathway in Arabidopsis thaliana. New Phytol. 205, 570–582. doi: 10.1111/nph.13101
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. doi: 10.1038/nrm1746
González-Calle, V., Barrero-Sicilia, C., Carbonero, P., Iglesias-Fernández, R. (2015). Mannans and endo-β-mannanases (MAN) in brachypodium distachyon: Expression profiling and possible role of the BdMAN genes during coleorhiza-limited seed germination. J. Exp. Bot. 66, 3753–3764. doi: 10.1093/jxb/erv168
Guillon, F., Larré, C., Petipas, F., Berger, A., Moussawi, J., Rogniaux, H., et al. (2012). A comprehensive overview of grain development in Brachypodium distachyon variety Bd21. J. Exp. Bot. 63, 739–755. doi: 10.1093/jxb/err298
Gupta, A., Singh, M., Laxmi, A. (2015). Interaction between glucose and brassinosteroid during the regulation of lateral root development in arabidopsis. Plant Physiol. 168, 307–320. doi: 10.1104/pp.114.256313
Iglesias-Fernández, R., Rodríguez-Gacio, M. C., Barrero-Sicilia, C., Carbonero, P., Matilla, A. (2011). Three endo-β-mannanase genes expressed in the micropylar endosperm and in the radicle influence germination of Arabidopsis thaliana seeds. Planta 233, 25–36. doi: 10.1007/s00425-010-1257-z
Ismail, A. M., Horie, T. (2017). Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 68, 405–434. doi: 10.1146/annurev-arplant-042916-040936
Jia, M., Li, Q., Hua, J., Liu, J., Zhou, W., Qu, B., et al. (2020). Phytohormones regulate both “Fish scale” galls and cones on picea koraiensis. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.580155
Jiang, Z., Zhou, X., Tao, M., Yuan, F., Liu, L., Wu, F., et al. (2019). Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572, 341–346. doi: 10.1038/s41586-019-1449-z
Jin, M., Guo, X., Zhang, R., Qu, W., Gao, B., Zeng, R. (2019). Diversities and potential biogeochemical impacts of mangrove soil viruses. Microbiome 7, 1–15. doi: 10.1186/s40168-019-0675-9
Kollárová, K., Kamenická, V., Vatehová, Z., Lišková, D. (2018). Impact of galactoglucomannan oligosaccharides and cd stress on maize root growth parameters, morphology, and structure. J. Plant Physiol. 222, 59–66. doi: 10.1016/j.jplph.2017.12.017
Konozy, E. H. E., Causse, M., Faurobert, M. (2012). Cell wall glycosidase activities and protein content variations during fruit development and ripening in three texture contrasted tomato cultivars. Saudi J. Biol. Sci. 19, 277–283. doi: 10.1016/j.sjbs.2012.04.006
Laskowski, M. J., Williams, M. E., Nusbaum, H. C., Sussex, I. M., Biology, P., Hall, K. (1995). Formation of lateral root meristems is a two-stage process. Development 3310, 3303–3310. doi: 10.1242/dev.121.10.3303
Li, X., Chen, R., Chu, Y., Huang, J., Jin, L., Wang, G., et al. (2018). Overexpression of RCc3 improves root system architecture and enhances salt tolerance in rice. Plant Physiol. Biochem. 130, 566–576. doi: 10.1016/j.plaphy.2018.08.008
Li, X., Liu, W., Ren, Z., Wang, X., Liu, J., Yang, Z., et al. (2022). Glucose regulates cotton fiber elongation by interacting with brassinosteroid. J. Exp. Bot 73 (3), 711–726. doi: 10.1093/jxb/erab451
Liu, L., Wu, X., Sun, W., Yu, X., Demura, T., Li, D., et al. (2021). Galactinol synthase confers salt-stress tolerance by regulating the synthesis of galactinol and raffinose family oligosaccharides in poplar. Ind. Crops Prod. 165, 113432. doi: 10.1016/j.indcrop.2021.113432
Lu, P., Magwanga, R. O., Kirungu, J. N., Hu, Y., Dong, Q., Cai, X., et al. (2019). Overexpression of cotton a DTX/MATE gene enhances drought, salt, and cold stress tolerance in transgenic arabidopsis. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00299
Lucob-Agustin, N., Kawai, T., Takahashi-Nosaka, M., Kano-Nakata, M., Wainaina, C. M., Hasegawa, T., et al. (2020). WEG1, which encodes a cell wall hydroxyproline-rich glycoprotein, is essential for parental root elongation controlling lateral root formation in rice. Physiol. Plant 169 (2), 214–227. doi: 10.1111/ppl.13063
Ma, J. F., Shen, R., Nagao, S., Tanimoto, E. (2004). Aluminum targets elongating cells by reducing cell wall extensibility in wheat roots. Plant Cell Physiol. 45, 583–589. doi: 10.1093/pcp/pch060
Malgas, S., van Dyk, J. S., Pletschke, B. I. (2015). A review of the enzymatic hydrolysis of mannans and synergistic interactions between β-mannanase, β-mannosidase and α-galactosidase. World J. Microbiol. Biotechnol. 31, 1167–1175. doi: 10.1007/s11274-015-1878-2
Mittler, R., Vanderauwera, S., Suzuki, N., Miller, G., Tognetti, V. B., Vandepoele, K., et al. (2011). ROS signaling: The new wave? Trends Plant Sci. 16, 300–309. doi: 10.1016/j.tplants.2011.03.007
Mohnen, D. (2008). Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11, 266–277. doi: 10.1016/j.pbi.2008.03.006
Moreira, L. R. S., Filho, E. X. F. (2008). An overview of mannan structure and mannan-degrading enzyme systems. Appl. Microbiol. Biotechnol. 79, 165–178. doi: 10.1007/s00253-008-1423-4
Motte, H., Beeckman, T. (2019). The evolution of root branching: Increasing the level of plasticity. J. Exp. Bot. 70 (3), 785–793. doi: 10.1093/jxb/ery409
Nonogaki, H., Gee, O. H., Bradford, K. J. (2000). A germination-specific endo-β-mannanase gene is expressed in the micropylar endosperm cap of tomato seeds. Plant Physiol. 123, 1235–1245. doi: 10.1104/pp.123.4.1235
Oka, Y., Butnaru, M., Von Buchholtz, L., Ryba, N. J. P., Zuker, C. S. (2013). High salt recruits aversive taste pathways. Nature 494, 472–475. doi: 10.1038/nature11905
Peng, S., Zhou, Q., Cai, Z., Zhang, Z. (2009). Phytoremediation of petroleum contaminated soils by mirabilis jalapa l. in a greenhouse plot experiment. J. Hazard Mater. 168, 1490–1496. doi: 10.1016/j.jhazmat.2009.03.036
Peters, W. S., Tomos, A. D. (2000). The mechanic state of “inner tissue” in the growing zone of sunflower hypocotyls and the regulation of its growth rate following excision. Plant Physiol. 123, 605–612. doi: 10.1104/pp.123.2.605
Richterová-Kučerová, D., Kollárová, K., Zelko, I., Vatehová, Z., Lišková, D. (2012). How do galactoglucomannan oligosaccharides regulate cell growth in epidermal and cortical tissues of mung bean seedlings? Plant Physiol. Biochem. 57, 154–158. doi: 10.1016/j.plaphy.2012.05.014
Sak-Ubol, S., Namvijitr, P., Pechsrichuang, P., Haltrich, D., Nguyen, T. H., Mathiesen, G., et al. (2016). Secretory production of a beta-mannanase and a chitosanase using a lactobacillus plantarum expression system. Microb. Cell Fact 15, 1–12. doi: 10.1186/s12934-016-0481-z
Tabuchi, A., Matsumoto, H. (2001). Changes in cell-wall properties of wheat (Triticum aestivum ) roots during aluminum-induced growth inhibition. Physiol. Plant 112, 353–358. doi: 10.1034/j.1399-3054.2001.1120308.x
Valifard, M., Le Hir, R., Müller, J., Scheuring, D., Neuhaus, H. E., Pommerrenig, B. (2021). Vacuolar fructose transporter SWEET17 is critical for root development and drought tolerance. Plant Physiol. 187 (4), 2716–2730. doi: 10.1093/plphys/kiab436
Wang, Y., Vilaplana, F., Brumer, H., Aspeborg, H. (2013). Enzymatic characterization of a glycoside hydrolase family 5 subfamily 7 (GH5-7) mannanase from Arabidopsis thaliana. Planta 239, 653–665. doi: 10.1007/s00425-013-2005-y
Xu, P., Fang, S., Chen, H., Cai, W. (2020). The brassinosteroid-responsive xyloglucan endotransglucosylase/hydrolase 19 (XTH19) and XTH23 genes are involved in lateral root development under salt stress in Arabidopsis. Plant J. 104 (1), 59–75. doi: 10.1111/tpj.14905
Yamamoto, Y. (2019). Aluminum toxicity in plant cells: Mechanisms of cell death and inhibition of cell elongation. Soil Sci. Plant Nutr. 65, 41–55. doi: 10.1080/00380768.2018.1553484
Yan, M., Zhang, Y., Guo, W., Wang, X. (2012). Soybean endo-β-Mannanase GmMAN1 is not associated with leaf abscission, but might be involved in the response to wounding. PloS One 7, 1–9. doi: 10.1371/journal.pone.0049197
Zhao, Y, Song, D, Sun, J, Li, L. (2013). Populus endo-beta-mannanase PtrMAN6 plays a role in coordinating cell wall remodeling with suppression of secondary wall thickening through generation of oligosaccharide signals. Plant J. 74 (3), 473–485. doi: 10.1111/tpj.12137
Zang, H., Xie, S., Zhu, B., Yang, X., Gu, C., Hu, B., et al. (2019). Mannan oligosaccharides trigger multiple defence responses in rice and tobacco as a novel danger-associated molecular pattern. Mol. Plant Pathol. 20, 1067–1079. doi: 10.1111/mpp.12811
Zelm, E., Zhang, Y., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Zhang, T., Tang, H., Vavylonis, D., Cosgrove, D. J. (2019). Disentangling loosening from softening: Insights into primary cell wall structure. Plant J. 100, 1101–1117. doi: 10.1111/tpj.14519
Keywords: mannanase, salt stress, antioxidant enzyme system, transgenic Arabidopsis, Mirabilis jalapa L
Citation: Xu J, Yang C, Ji S, Ma H, Lin J, Li H, Chen S, Xu H and Zhong M (2023) Heterologous expression of MirMAN enhances root development and salt tolerance in Arabidopsis. Front. Plant Sci. 14:1118548. doi: 10.3389/fpls.2023.1118548
Received: 07 December 2022; Accepted: 20 March 2023;
Published: 14 April 2023.
Edited by:
Meng Jiang, Zhejiang University, ChinaCopyright © 2023 Xu, Yang, Ji, Ma, Lin, Li, Chen, Xu and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Zhong, bWluZ3pob25nQHN5YXUuZWR1LmNu; Hai Xu, Y2hpbmF4dWhhaUAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Juanjuan Xu1†
Juanjuan Xu1† Caiyu Yang
Caiyu Yang Shuisen Chen
Shuisen Chen Ming Zhong
Ming Zhong