
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 01 March 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1112354
This article is part of the Research TopicGene Regulatory Networks involved in the Molecular Response to Drought, Salt and Osmotic Stresses in CropsView all 7 articles
Laccases are plant enzymes with essential functions during growth and development. These monophenoloxidases are involved in lignin polymerization, and their expression respond to environmental stress. However, studies of laccases in some plants and fungi have highlighted that many structural and functional aspects of these genes are still unknown. Here, the laccase gene family in Aeluropus littoralis (AlLAC) is described based on sequence structure and expression patterns under abiotic stresses and ABA treatment. Fifteen non-redundant AlLACs were identified from the A. littoralis genome, which showed differences in physicochemical characteristics and gene structure. Based on phylogenetic analysis, AlLACs and their orthologues were classified into five groups. A close evolutionary relationship was observed between LAC gene family members in rice and A. littoralis. According to the interaction network, AlLACs interact more with proteins involved in biological processes such as iron incorporation into the metallo-sulfur cluster, lignin catabolism, regulation of the symbiotic process and plant-type primary cell wall biogenesis. Gene expression analysis of selected AlLACs using real-time RT (reverse transcription)-PCR revealed that AlLACs are induced in response to abiotic stresses such as cold, salt, and osmotic stress, as well as ABA treatment. Moreover, AlLACs showed differential expression patterns in shoot and root tissues. Our findings indicate that AlLACs are preferentially involved in the late response of A. littoralis to abiotic stress.
Laccases (EC 1.10.3.2), named after their presence in the Japanese lacquer tree (Toxicodendron vernicifluum), are multi-copper oxidases (LMCOs). These enzymes are involved in catalyzing the oxidation of one electron of various cellular compounds such as arylamines, phenols, and aromatic thiols (Reiss et al., 2013). Members of the multi-copper oxidases superfamily include ceruloplasmin, nitrite reductase, ascorbate oxidase, and ferroxidase. Laccases have been identified in many organisms, including plants, fungi, insects, and bacteria, and are involved in a wide range of cellular processes (Wang et al., 2015a; Janusz et al., 2020; Lu et al., 2021). Plant laccases are copper-containing glycoproteins that act as key regulators for lignin polymerization and deposition in the plant cell wall (Liu et al., 2017). Laccases contain three conserved copper (Cu)-oxidase domains (Mot and Silaghi-Dumitrescu, 2012), which can be used to identify laccase family members. Due to their importance, members of this gene family have been identified and analyzed in plants, including Lolium perenne (Gavnholt et al., 2002), Oryza Sativa (Liu et al., 2017), Sorghum bicolor (Wang et al., 2017), Arabidopsis thaliana (Turlapati et al., 2011), Brachypodium distachyon (Wang et al., 2015b), Zea mays (Caparrós-Ruiz et al., 2006), Setaria viridis (Simões et al., 2020), Brassica napus (Zhang et al., 2013), Gossypium arboretum (Zhang et al., 2019), Saccharum officinarum (Cesarino et al., 2013), Eucalyptus grandis (Arcuri et al., 2020) and Pyrus bretschneideri (Lu et al., 2021).
According to previous studies, laccases can be involved in various cellular processes. For instance, Liu and coworkers, reported that laccases are mostly induced in the early development stage of rice seedlings (Liu et al., 2017), and Simões and coworkers, showed that laccases from Setaria viridis are potentially involved in monolignol oxidation (Simões et al., 2020). Moreover, the study of laccases in loblolly pine revealed that these genes are expressed mainly in immature secondary xylem, the tissue for synthesized lignin (Sato et al., 2001). Lignin is critical in increasing plant vigor and resistance to biotic and abiotic stresses. Accordingly, the lignin content in the mutant line for laccases genes, LAC17 and LAC14, in Arabidopsis was decreased compared to wild-type lines (Berthet et al., 2011), suggesting that laccases are involved in lignin polymerization and decomposition. Also the involvement of laccases in Cleome seed coat lignification was described (Zhuo et al., 2022). Other roles have also been suggested for laccases in plants, highlighting that the members of this gene family are involved in responding to environmental stresses. For instance, the overexpression of OsChI1, a laccase gene from rice, sharply increased the salt and drought tolerance in transgenic Arabidopsis (Cho et al., 2014). In addition, copper tolerance was improved in Arabidopsis transgenic lines for the overexpressed OsLAC10, a laccase10 gene from rice (Liu et al., 2017). Similarly, Cai and coworkers reported that the laccase2 gene in Arabidopsis is involved in response to abiotic stresses such as drought (Cai et al., 2006), and Xu et al. showed that laccase genes are induced by abiotic stresses such as low/high temperature, drought, and hormone application (ABA, MeJA, and SA) in citrus (Xu et al., 2019). Based on their broad substrate spectrum and ability to depolymerize lignin, laccases moved into the focus of current research. Furthermore they became of economic importance (Riva, 2006; Arregui et al., 2019) due to their use as bioactive component of ascorbic acid (vitamin C) sensors (Lee et al., 2018).
Aeluropus littoralis is a halophyte monocot model that can grow under salt and drought conditions (Saad et al., 2011; Hashemi et al., 2016). A. littoralis does not have the ability to be cultivated, but as a valuable source of resistance genes, it can be used in breeding and genetic engineering programs. The genomic resources for A. littoralis provide the opportunity to identify the key genes and molecular mechanisms of response to abiotic stresses (Hashemi et al., 2016). Motivated by the importance of laccase genes in increasing the resistance of plants against environmental conditions, the present study analyzed the sequence structure of laccase genes as well as their regulatory systems. In addition, the expression pattern of these genes in response to environmental stress and ABA hormone was investigated in root and shoot tissues.
The protein sequences of Arabidopsis thaliana laccase family genes (17 members) were retrieved from the Arabidopsis information resource TAIR10 (TAIRv10, http://www.arabidopsis.org/). The sequences were cross-checked with the Pfam database for the presence of three domains of laccases including CuRO_1_LCC_plant (cd13849), CuRO_2_LCC_plant (cd13875), and CuRO_3_LCC_plant (cd13897). Arabidopsis laccase protein sequences were aligned as queries against the Aeluropus Proteome and genome version 1 (Hashemi-Petroudi et al., 2022) using the local BLASTP and TBLASTN program, respectively. The resulting peptide sequences were verified with the blastP tool in NCBI and the Pfam database for the presence of conserved laccase (TIGR03389). In order to reduce the redundancy, % identity of all sequences was computed with the Decrease Redundancy program (https://web.expasy.org/decrease_redundancy), and any pairs of sequences with more than 99% identity were removed from the analysis. The use of the Decrease Redundancy program resulted in reduction from 21 putative AlLACs to 15 defined protein sequences. The naming of each laccase gene in Aeluropus (AlLAC) was based on the closest known orthologue of Arabidopsis.
To construct a phylogenetic tree, AlLACs with their orthologous in Arabidopsis (TAIR10) and rice (IRGSP-1.0) were analyzed using the multiple sequence alignment Muscle tool of MEGA v. 11 software (Tamura et al., 2021) with the default parameters. A phylogenetic tree was constructed using the maximum-likelihood (ML) method with 1000 bootstrap replicates.
The MEME database was used to predict the conserved motifs of AlLAC protein sequences. The number of conserved motifs was adjusted to 10, and other parameters were set as the default. The gene structure of AlLAC genes was constructed using Tbtools (Chen et al., 2020) based on GFF format file for exon and intron location information known from Arabidopsis orthologs. The physical and chemical properties, including number of amino acids, molecular weight (kDa), theoretical pI, grand average of hydropathicity (GRAVY), total number of negatively charged residues (Asp + Glu), total number of positively charged residues (Arg + Lys), and instability index of AlLACs, were calculated using the online ExPASy-ProtParam tool (Gasteiger et al., 2005). The subcellular localization of AlLACs were predicted using WoLF PSORT (Horton et al., 2007) based on default settings.
The Phyre2 server was applied to predict the 3D structure of AlLAC proteins (Kelley et al., 2015). Similar structures were analyzed using the Phyre investigator tool to recognize the pocket site related to the binding region.
PlantCare (Lescot et al., 2002) was used to study cis-regulatory elements in the 1000 bp promoter region. The sequence information was retrieved from Aeluropus genome version 1 (Hashemi-Petroudi et al., 2022). The identified cis-regulatory elements were classified based on their function, and drawn as a graph.
The STRING v11.5 database was used to identify the interactions of AlLAC proteins based on their orthologues in Arabidopsis. The first shell of the network was adjusted to ≤ 20 and the second shell was fixed to no more than 10. Gene ontology (GO) enrichment analysis was used to identify the significant (FDR ≤ 0.05) molecular function, biological process, and cellular component terms presented in the LAC-interaction network using the STRING. Cytoscape v3.8.2 (Shannon et al., 2003) was used to construct the LAC-interaction network.
Seeds of Aeluropus were grown at 25 ± 2°C under 16 hours of light and 8 hours of darkness in the greenhouse at Sari Agricultural Sciences and Natural Resources University. After three weeks, the samples were transferred in groups of three to plastic containers, each containing five liters of Hoagland’s nutrient solution under hydroponic culture (Hoagland and Arnon, 1950). After two months, plants of similar size were selected for exposure to stress conditions. For salinity stress, plants were treated gradually by adding 100 mM salt (NaCl) every 48 hours to a final concentration of 600 mM NaCl. For osmotic stress, plants were treated with 20% PEG 6000 of -0.80 MPa. PEG was added to a plastic container and samples (roots and leaves) were collected at different exposure periods of 0 hour (h), as control sample, 6 hours, 48 hours and one week. For cold stress, the plants were exposed to a temperature of 4°C. Sampling of roots and leaves tissues was done 6 h, 48 h and one week after stress exposure, in leaves three biological replicates. The abscisic acid (ABA) treatment was also done by spraying 100 micromolar hormones on the leaves; two leaf and root tissues were sampled at 3, 6, 24 and 48 hours after applying the treatment, in three biological replicates. Three untreated vases were used as controls.
Total RNA was extracted from leaf and root tissue of all three biological replicates using the Threezol kit (Threezol, Riragene, Iran). The quantity and quality of the RNA samples were measured by spectrophotometry and 1.5% agarose gel electrophoresis, respectively. DNase I treatment (DNase I RNase-free, Thermo Scientific) was used to remove genomic DNA. After combining the RNA of the biological replicates, cDNA synthesis was performed using a kit (Thermo Scientific) according to the company’s instructions, and diluted five times.
The expression levels of the target genes were measured with a Bio-Rad CFX96 machine, using The Maxima SYBR Green/ROX qPCR Master Mix kit (Thermo Scientific) in three technical replicates. The cycling profile started at 95°C for 15 seconds, then 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds. At least one negative control (NTC) was considered for each primer. Melting curve analysis of samples and threshold cycle were calculated with CFX software (Bio-Rad). Normalization of gene expression was done with the geometric mean method using specific reference genes of each tissue. Selecting the appropriate internal control gene is very important when determining gene expression, opting for genes that show the least amount of gene expression changes in different stress conditions (either different tissues or sampled at different times). Specific reference genes for leaf tissue were GTF and U2SnRNP, and specific reference genes for root tissue were PRS3 and EF1a (Hashemi et al., 2016). Five AlLAC genes, AlLAC5, AlLAC12.2, AlLAC14, AlLAC16.1, and AlLAC17.1, were selected based on bioinformatics analyses and primers were designed using AlleleID software (version 7.5, Table S1). Quantitative analysis of the data related to the relative expression level of the studied genes was done using the 2-△△CT method (Livak and Schmittgen, 2001).
In total, 15 LAC proteins in Aeluropus littoralis (AlLAC) were identified (Tables 1, S2). According to their physicochemical properties, the AlLACs encoded proteins ranging from 175 (AlLAC16.1) to 648 aa (AlLAC7.3). The pI values were predicted to be between 5.17 (AlLAC11.1) and 8.93 (AlLAC17.3); eight of the 15 AlLACs showed a pI greater than 7.0 (Table 1). Based on the instability index, four of the 15 AlLACs could be introduced as unstable proteins, while the other 11 were predicted to be stable. In addition, most AlLAC proteins (10 of the 15) showed negative values for the GRAVY index, indicating that AlLACs are more hydrophilic. Based on the prediction of the subcellular localization, AlLACs are predominantly located in organelles such as chloroplasts (Chlo.) and vacuoles (Vacu.) (Table 1). Overall, AlLACs showed variation based on their physicochemical properties, suggesting that they may have diverse functions.
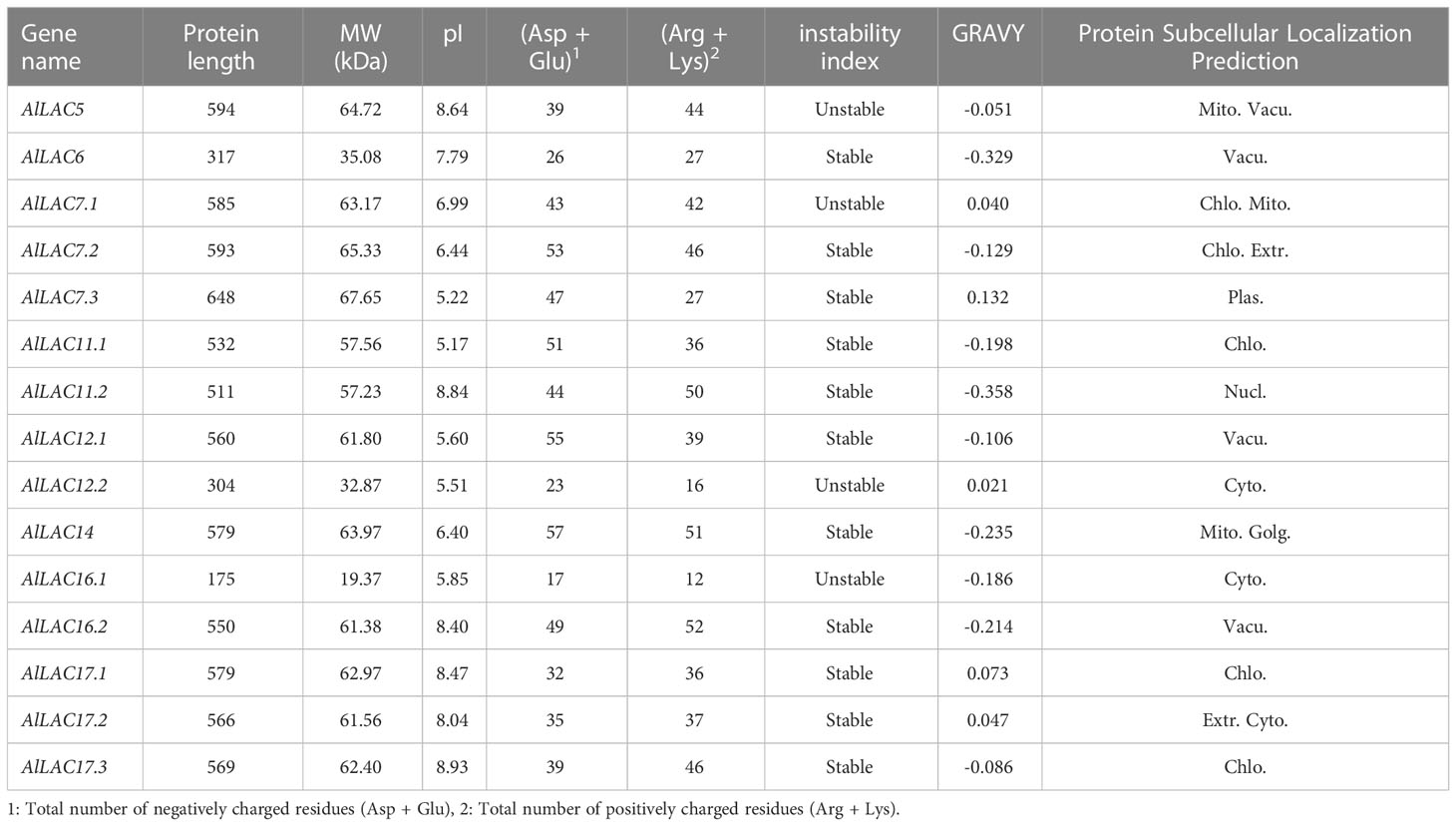
Table 1 Physicochemical properties of AlLAC gene family members in A.littoralis.Accession number and sequences (gene, protein, and CDS) of AlLAC gene family members are provided in Table S2.
The phylogenetic tree for members of the LAC gene family in Arabidopsis (AtLACs), rice (OsLACs), and A. littoralis (AlLACs) separated LACs into five main groups (Figure 1). Furthermore, AlLACs were more similar to their orthologs in rice, suggesting that a close evolutionary process may have occurred in this gene family in rice and A. littoralis. Moreover, it seems that the diversity of LAC family has more occurred after derivation of monocots and dicots. AlLACs from group I, including AlLAC11.1, AlLAC6, AlLAC16.2, and AlLAC11.2 showed a greater genetic distance than other members, suggesting that these members may have a higher potential for further molecular functional investigation. In addition, the gene structure, conserved motifs, and encoded domain distribution of AlLACs were analyzed (Figure 2 and Table S3). Ten conserved motifs and three encoded domains were identified in AlLACs. Motif 4 was in the Cu-oxidase 1 domain region, while motifs 6, 1, and 3 were predicted in Cu-oxidase 2 domain, and motifs 5 and 2 were located in Cu-oxidase 3 domain (Figure 2). Some of the conserved motifs (motif 9 and motif 10) were identified in the extra-domain region that can be used to identify the AlLACs. Among the various gene structures observed in AlLACs, the maximum exon/intron number was observed in AlLAC11.1.
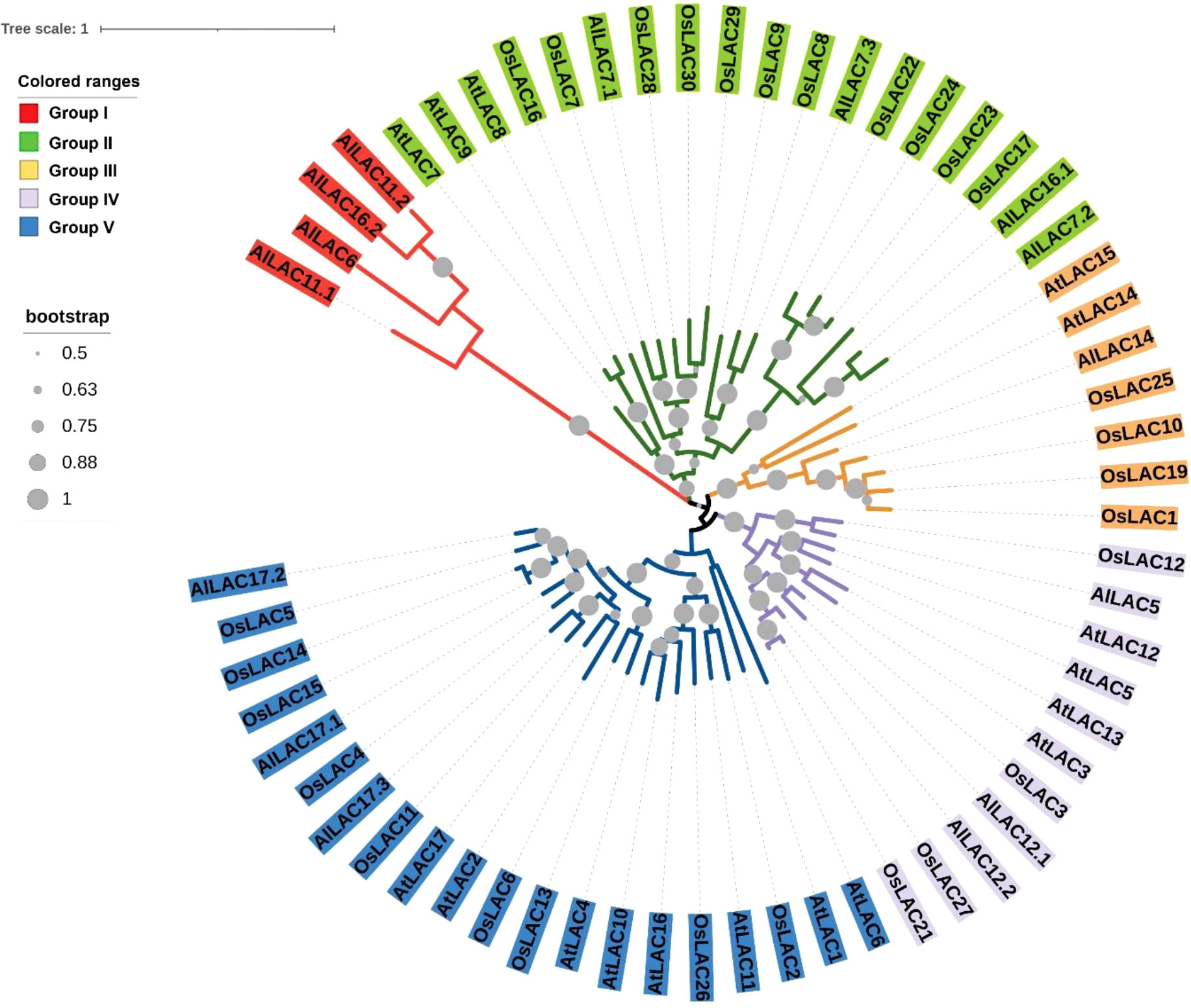
Figure 1 Phylogenetic tree of LAC gene members in Arabidopsis (AtLAC), rice (OsLAC), and A.littoralis (AlLAC).
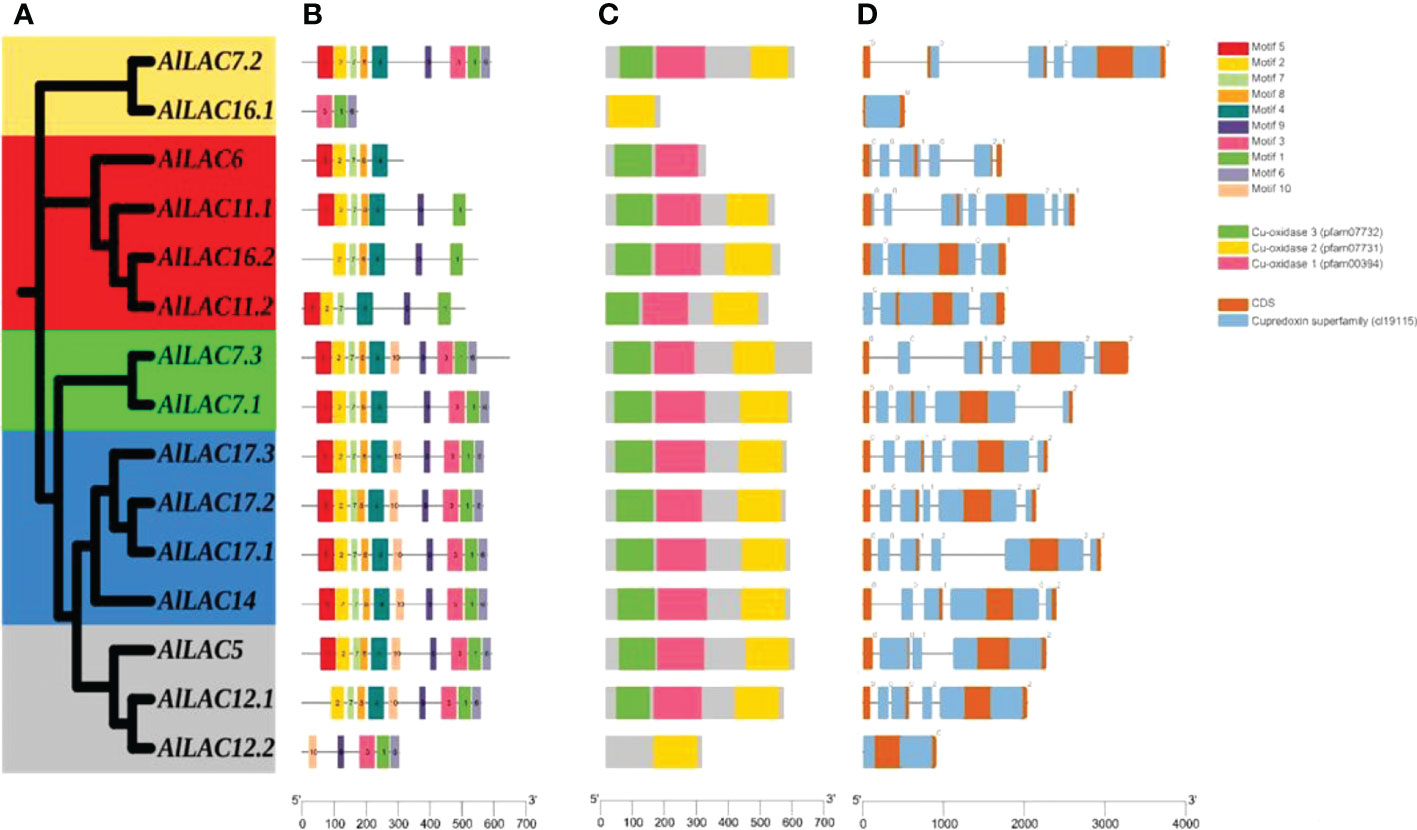
Figure 2 Phylogenetic tree of AlLAC proteins (A), conserved motif distribution of AlLAC proteins (B), conserved domains of AlLAC proteins (C), and gene structure of AlLAC genes (D). The logo and sequence of conserved motifs are provided in Table S3.
The predicted 3D structure and binding sites of AlLACs revealed diverse structures among members (Figure 3). Moreover, leucine (L), proline (P), valine (V), phenylalanine (F), glycine (G), and alanine (A) were frequently predicted in the binding site region of AlLACs (Figure 4). The residues detected in the pocket sites of all AlLACs demonstrate the key positions of the important amino acids likely related to their function and interaction points.
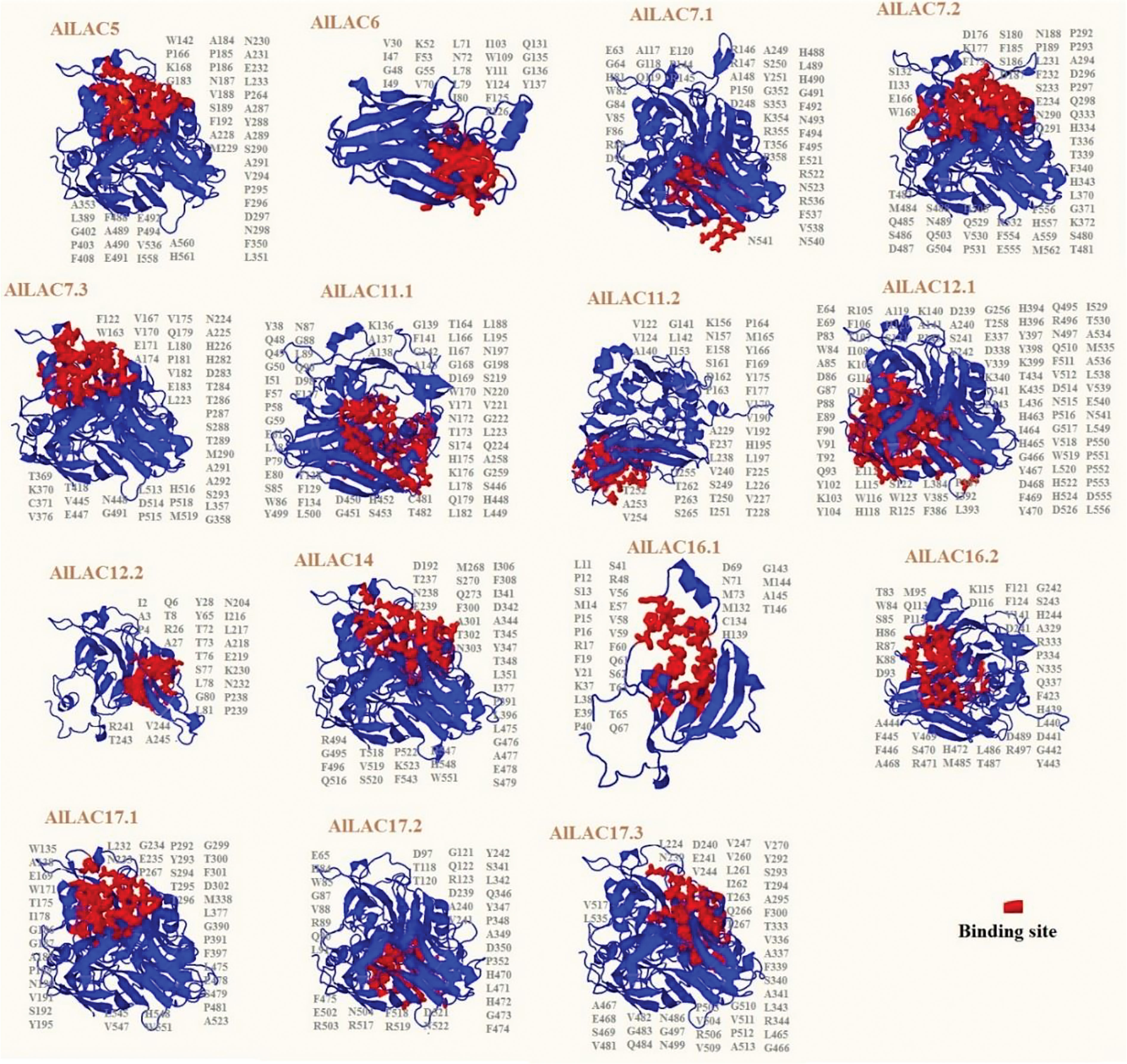
Figure 3 Three-dimensional docking analysis of AlLACs. The binding residues are shown on protein structure.
The interaction network for AlLAC proteins was drawn based on their orthologs in the model plant, Arabidopsis. Sks11, sks13, and SKU5 proteins, which have oxidoreductase activity, were the most similar to AlLAC11.2, AlLAC16.2, and AlLAC6 proteins, respectively (Figure 5). There were no direct interactions between the laccases themselves. Three interaction groups were observed: the first group contained Sks11 (AT3G13390), sks13 (AT3G13400), and SKU5 (AT4G12420); the second group contained LAC7 and LAC17; and the third group contained LAC5 and LAC6. LAC5 showed the most and strongest interactions compared to the other laccases. Gene ontology (GO) enrichment analysis revealed several biological process terms; iron incorporation into metallo-sulfur cluster, lignin catabolism, regulation of symbiotic processes, plant-type primary cell wall biogenesis, and L-ascorbic acid biosynthesis were significantly linked with the LAC-interaction network (Table S4). In addition, molecular function terms including cysteine desulfurase activity, glucose-6-phosphate isomerase activity, ferrochelatase activity, copper ion binding, oxidoreductase activity, cellulose synthase (udp-forming) activity, and antioxidant activity were significantly associated with the LAC-interaction network (Table S4).
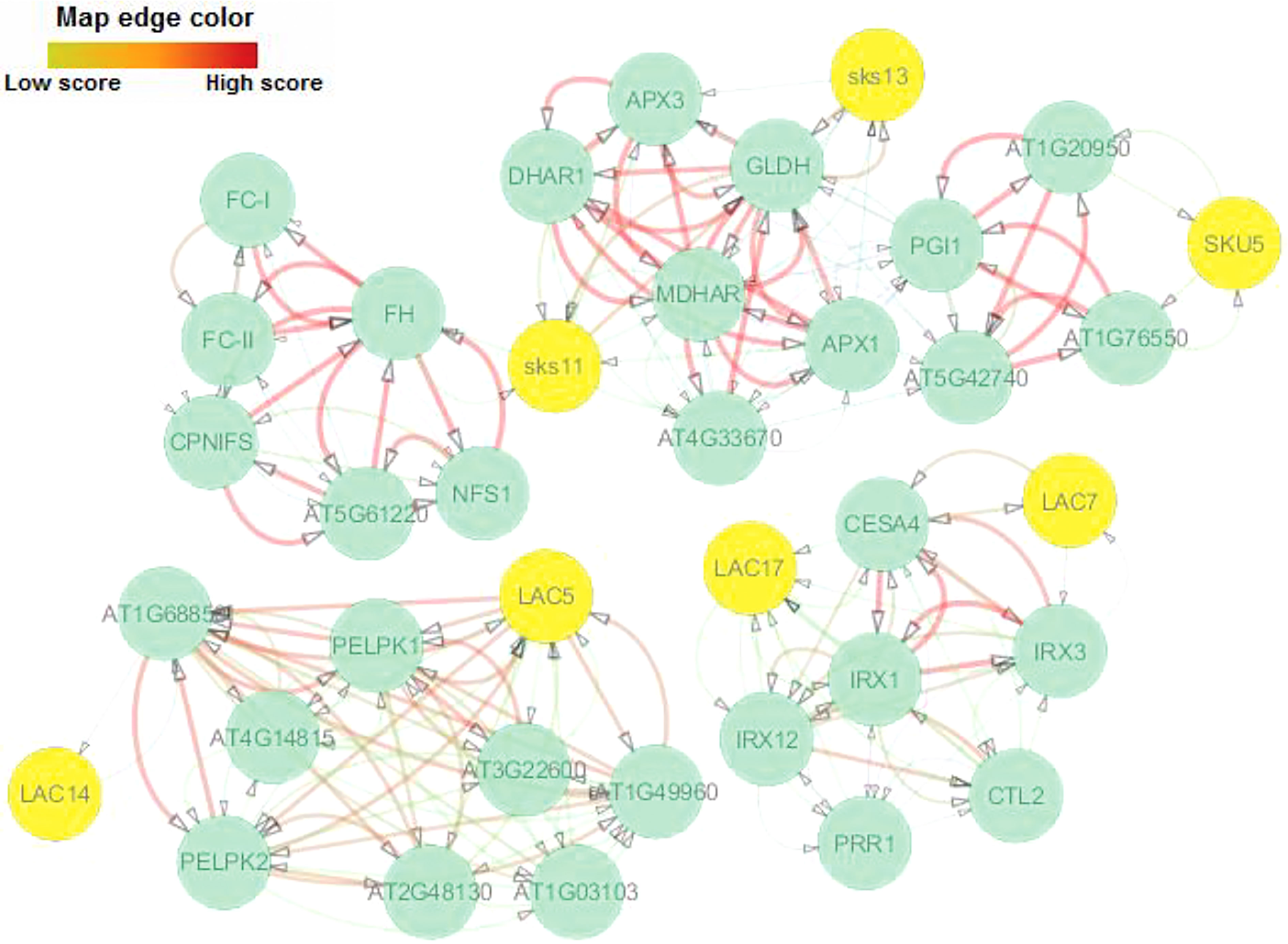
Figure 5 Protein–protein interaction network of AlLACs using STRING server v11 based on Arabidopsis interactome data.
The upstream region of AlLAC genes was analyzed to detect putative cis-regulatory elements. The cis-regulatory elements that are involved with stress, growth and development, phytohormones, and transcription factor (TF) site were observed in the promoter region (Figure 6). The common and unknown function of cis-regulatory elements were frequently observed upstream of AlLACs (Figure 6A). Cis-regulatory elements to methyl jasmonate (MeJA) and ABA responsiveness (ABRE) were frequently detected in the promoter region (Figure 6B). Moreover, acting elements related to ethylene (ERE), salicylic acid (SA), gibberellic acid (GA), and auxin responsiveness were also observed in the promoter region (Figure 6B), as were stress-responsive MYB elements such as as-1, an acting element involved in oxidative stress-responsive, and STRE (stress-controlled transcription factors) elements (Figure 6C). Dehydration-responsive element (DRE) and MBS (MYB binding site involved in drought-inducibility) elements were also observed upstream of AlLACs (Figure 6C). These results suggest that laccases probably cooperate with factors involved in stress response and are probably present in stress-dependent signaling pathways.
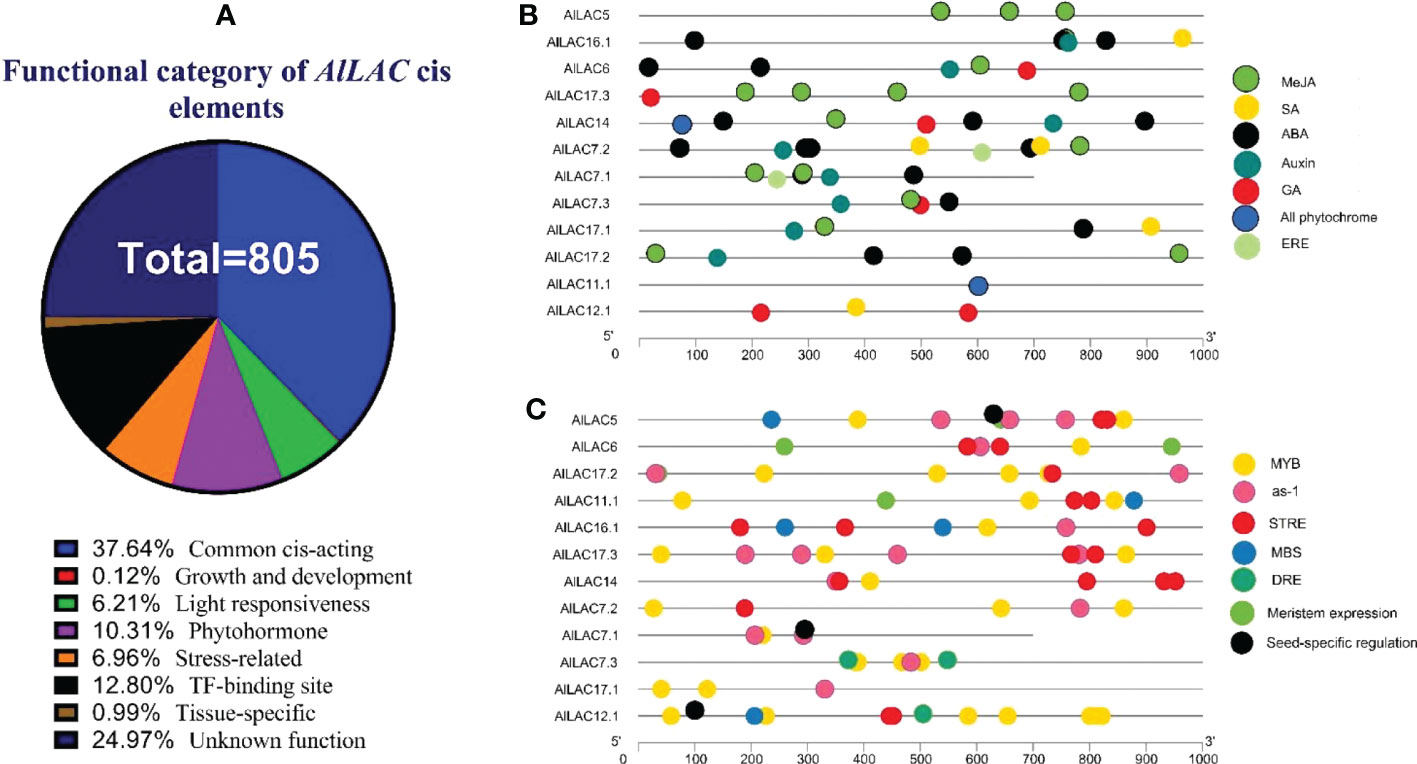
Figure 6 Cis-regulatory distribution in the promoter region of AlLAC genes. Percentage of cis-regulatory elements based on their functions (A), distribution of cis-regulatory elements related to phytohormone (B), and distribution of cis-regulatory elements related to growth and development, TF binding site, and stress responsiveness (C). More details of Cis-regulatory distribution in the promoter region of AlLAC genes are provided in Table S5.
The expression levels of five candidate AlLAC genes (AlLAC5, AlLAC14, AlLAC16.1, AlLAC17.1, and AlLAC12.2) were investigated under ABA application in shoot and root tissues of A. littoralis (Figure 7). The AlLAC genes for real time RT-PCR analysis were selected on their mRNA abundance and salt stress inducibility reported in (Younesi-Melerdi et al., 2020). Based on their expression profile, all selected genes illustrated tissue-specific expression, and they were highly expressed in shoot tissues. Interestingly, all studied AlLACs were sharply upregulated after 48 hours of ABA application in shoot tissues, while AlLAC14 was upregulated at all-time points of ABA application in shoot tissues. Besides, all AlLACs showed a down-regulation after 3 h of ABA application in root tissues. Overall, ABA may indirectly affect the expression of AlLACs in the shoot.
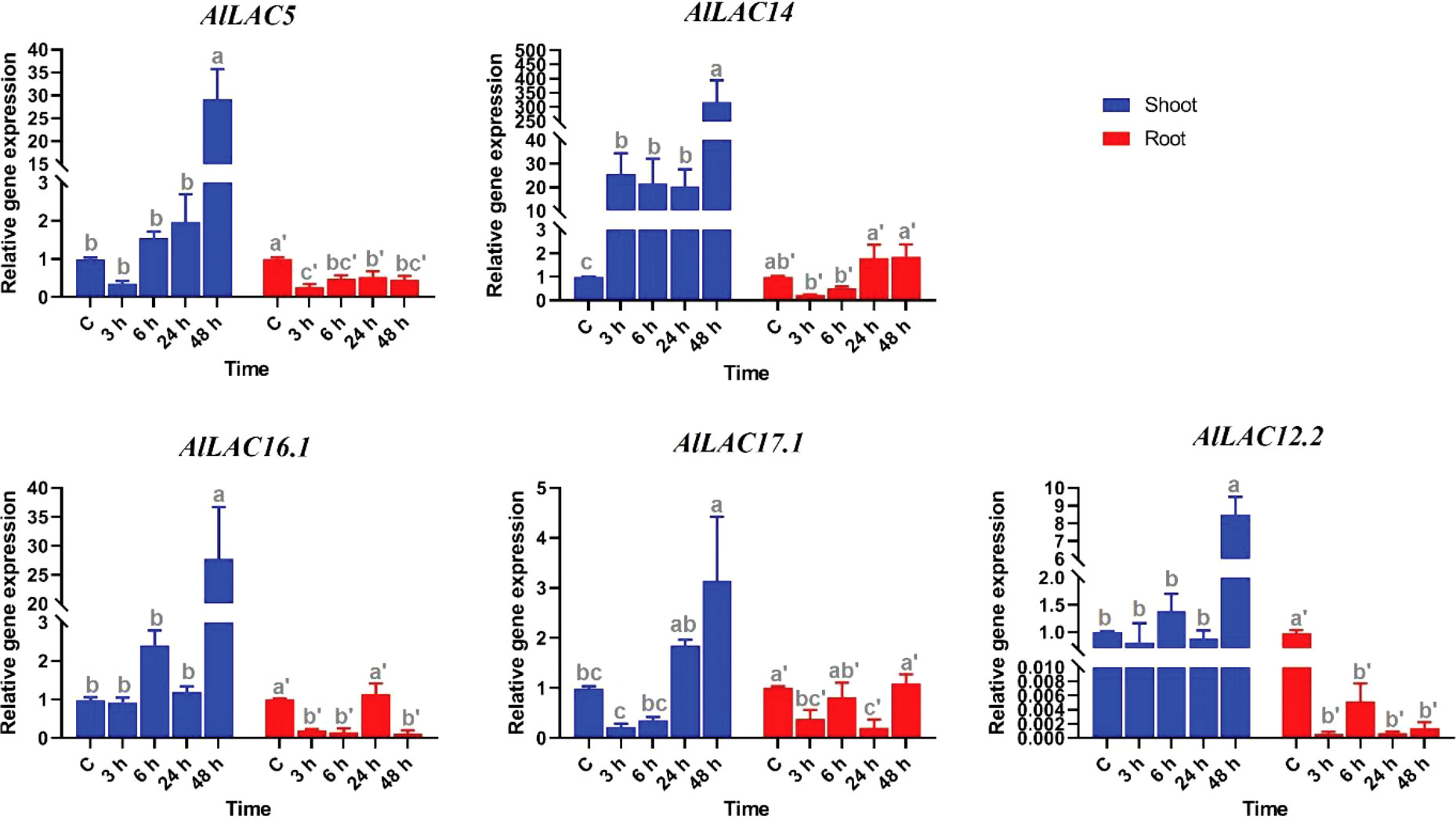
Figure 7 Expression profile of AlLAC genes under ABA treatments. Different letters above each bar indicate a significant difference (p < 0.05) based on Tukey’s range test.
Expression patterns of five selected AlLAC genes in response to abiotic stresses – cold, osmotic (using PEG application), and salt stress – were evaluated in shoot and root tissues (Figures 8-10). In response to cold stress, AlLAC5 was upregulated after seven days in shoot tissues, while AlLAC14 was highly induced after 48 hours and seven days in both shoot and root tissues (Figure 8). AlLAC16.1 and AlLAC17.1 were less induced in response to cold stress, and AlLAC12.2 showed upregulation at all studied time points, especially after seven days, in root tissues. In response to salt stress, AlLAC genes showed diverse expression patterns. For instance, AlLAC5, AlLAC17.1, and AlLAC12.2 were upregulated in roots, while AlLAC14 showed a high upregulation in the shoot (Figure 9). Furthermore, AlLAC5, and AlLAC17.1 were upregulated in both root and shoot, suggesting that these genes are directly associated with response to salt stress. In addition, AlLAC16.1 showed a down-regulation in root tissues at all-time points in response to salinity (Figure 9). The expression profile of AlLACs was analyzed under PEG application for induction of osmotic stress. Interestingly, all studied genes showed an upregulation in root tissues, and high expression was recorded after 48 h (Figure 10). In shoot tissues, AlLAC14 was sharply induced after 48 hours in response to drought stress. Overall, AlLAC14 appears to be more expressed in shoot tissues, and AlLACs are more involved in the late response of A. littoralis to abiotic stress.
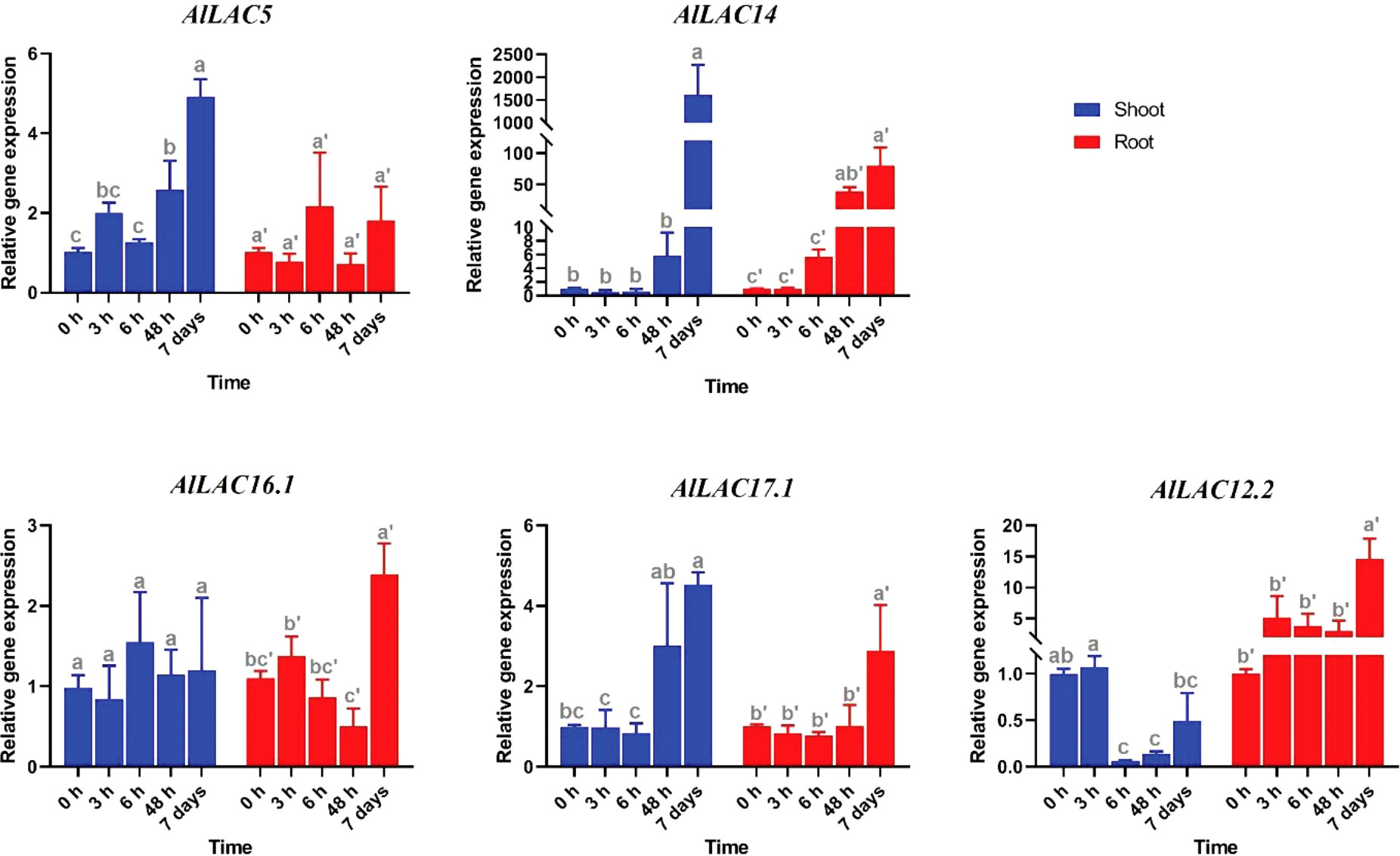
Figure 8 Expression profile of AlLAC genes in response to cold stress. Different letters above each bar indicate a significant difference (p < 0.05) based on Tukey’s range test.
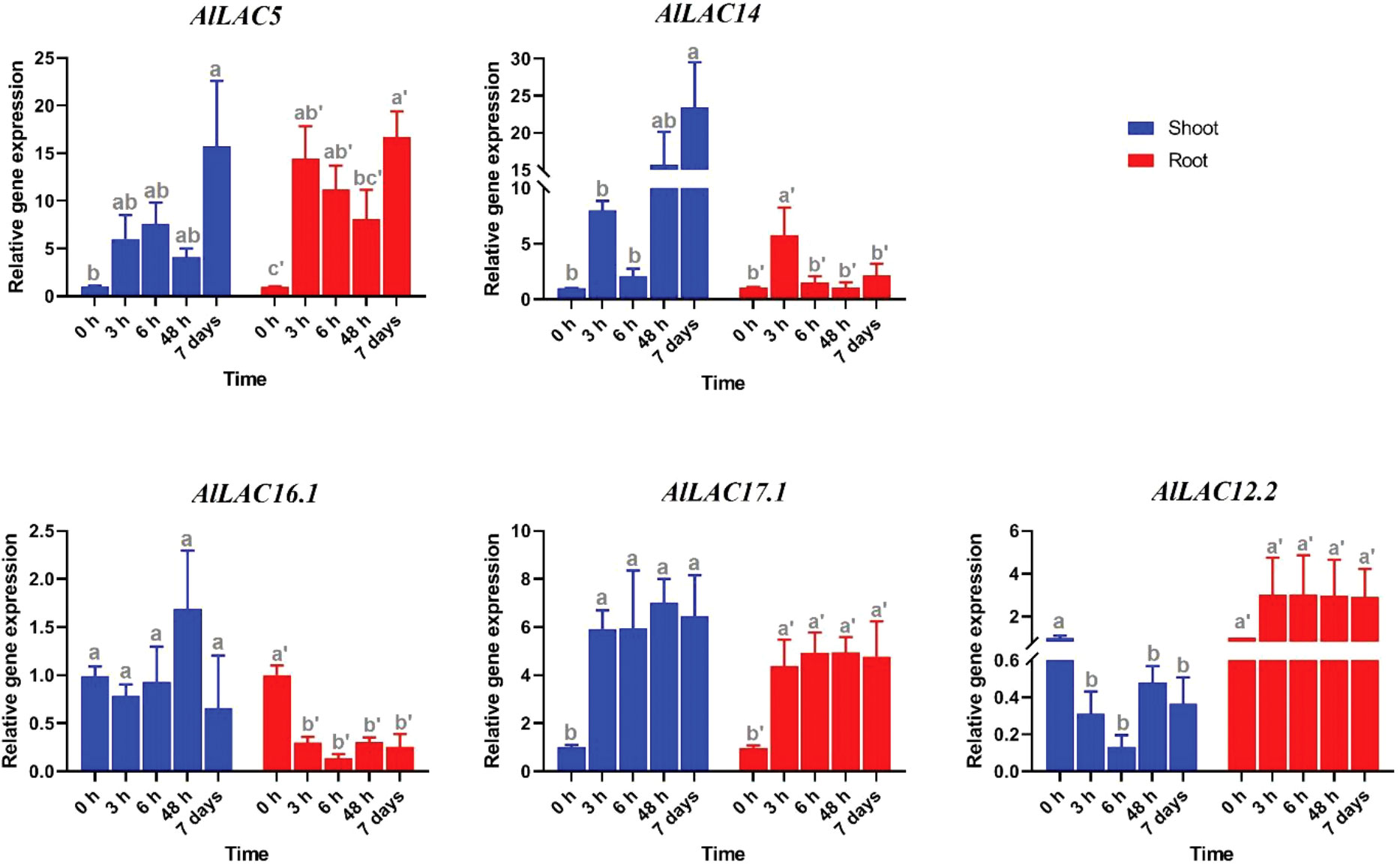
Figure 9 Expression profile of AlLAC genes in response to salt stress. Different letters above each bar indicate a significant difference (p < 0.05) based on Tukey’s range test.
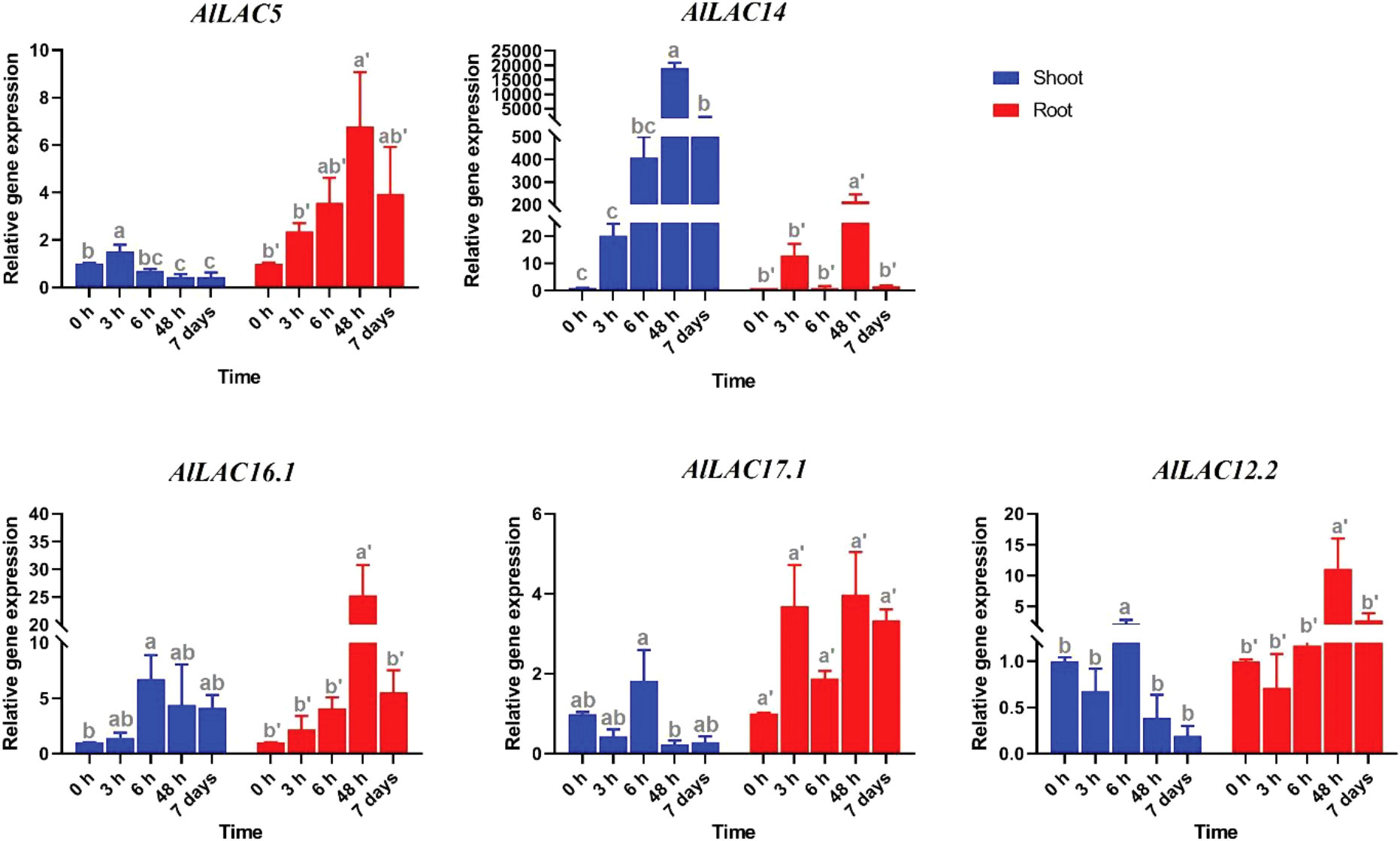
Figure 10 Expression profile of AlLAC genes in response to osmotic stress (PEG application). Different letters above each bar indicate a significant difference (p < 0.05) based on Tukey’s range test.
Laccases have important roles such as lignin polymerization in plants, and also contribute to resistance to environmental stresses (Bao et al., 1993; Sato et al., 2001; Berthet et al., 2011). Enzymes of this gene family moved into focus due to their ability to catalyze steps in bio-economy (Arregui et al., 2019; Agustin et al., 2021) and their utilization as biosensors (Lee et al., 2018). Thus far, laccases have not been studied in Aeluropus littoralis, a model plant that is highly resistant to salinity stress (Saad et al., 2011; Hashemi et al., 2016). A. littoralis has a high potential in the field of resistance to abiotic stresses and serves to identify and isolate new stress-adapted genes. In the current study, 15 AlLACs were identified from the A. littoralis genome. Previous studies have identified 30 LACs from rice (Liu et al., 2017), 40 LACs from the pear genome (Lu et al., 2021), 52 LACs from the Setaria viridis genome (Simões et al., 2020), 54 LACs from the Eucalyptus grandis genome (Arcuri et al., 2020), 29 LACs from the Brachypodium distachyon genome (Wang et al., 2015b), 17 LACs from the Arabidopsis thaliana genome (Turlapati et al., 2011), and 27 LACs from the Sorghum bicolor genome (Wang et al., 2017). The lower number of laccase genes found here suggests that the laccase gene family is probably less extended in A. littoralis during evolutionary processes. Based on their physicochemical characteristics and gene structure, AlLACs were diverse, indicating that the members of this gene family are involved in various cellular processes (Ahmadizadeh et al., 2020; Heidari et al., 2021a). Intron/exon number was also diverse among AlLACs. It was stated that the number of exons/introns can affect the expression levels; genes with fewer exons can be rapidly activated (Jeffares et al., 2008; Koralewski and Krutovsky, 2011; Iñiguez and Hernández, 2017; Heidari et al., 2022). According to phylogenetic analysis, LAC gene family members from A. littoralis, Arabidopsis, and rice were grouped into five classes, and a close evolutionary process was observed between AlLACs and their orthologues in rice. This finding suggests that mutations have occurred in coding sequence regions after the divergence between dicots and monocots (Faraji et al., 2021a; Musavizadeh et al., 2021). In addition, the distribution of conserved motifs might be associated with the diversity and function of genes from a family (Faraji et al., 2021b). Some conserved motifs were observed in the extra-domain site – these regions can be used to identify AlLACs and their role of these proteins in stress response.
AlLACs also showed diverse 3D structures. Leucine, proline, valine, phenylalanine, glycine, and alanine were identified as the amino acids that affect the interaction and function of AlLACs. The interaction network for AlLAC proteins based on their orthologues in the model plant, Arabidopsis, suggested that laccases are probably involved in diverse cellular processes related to cellulose synthase activity and oxidoreductase activity, including iron incorporation into metallo-sulfur cluster, lignin catabolism, regulation of symbiotic processes, plant-type primary cell wall biogenesis, and L-ascorbic acid biosynthesis. Previous studies have also stated that laccases are associated with processes related to plant cell wall components (Cesarino et al., 2013; Liu et al., 2017). Notably, the weak interaction between the laccases indicates that they likely work in independent pathways.
Several classes of cis-acting elements related to response to hormones, light, abiotic and biotic stresses, growth and development processes were identified upstream of AlLAC genes, suggesting that the members of this gene family have multifunctional roles in A. littoralis (Ahmadizadeh and Heidari, 2014; Rezaee et al., 2020). We investigated the expression profile of five AlLAC genes in response to ABA and PEG application, cold, and salt stress using qRT-PCR. Notably, AlLACs showed differential expression patterns in shoot and root tissues, indicating that these genes have different functions. Our findings demonstrated that AlLAC14 is more induced in shoot tissues after 48 hours of exposure to stresses, however further molecular functional studies of this gene are recommended. In addition, the selected AlLAC genes were upregulated in response to application of ABA hormone, a stress-dependent regulator that many signaling pathways use in response to adverse environmental conditions (Heidari et al., 2021b). The induction of AlLAC genes after 48 hours of ABA application raises that these genes probably have some interaction with signaling pathway related to ABA. Furthermore, the selected AlLAC genes illustrated diverse expression in response to abiotic stresses, including cold, salt, and osmotic stress. Interestingly, these AlLACs were induced after 48 hours of exposure to stress conditions, revealing that AlLACs are associated with late response pathways of A. littoralis. Several studies have mentioned the role of laccases in the process of lignification (Sato et al., 2001; Berthet et al., 2011). Laccases involves in lignin polymerization and can affects the lignin synthesis (Berthet et al., 2011). Lignin is a component of plant cell walls that plays an important role in increasing the resistance and strength of plants. Besides, it was reported that LACs can be regulated by transcription factor MYB and be induced by abiotic stresses (Xu et al., 2022). Our results suggest that laccases are involved in response to adverse conditions from direct and indirect pathways in A. littoralis.
The present study identified 15 AlLACs in the Aeluropus littoralis genome, which formed five groups based on phylogenetic analysis. AlLACs showed high diversity in structure and physicochemical properties, suggesting that these gene family members were influenced by evolutionary pressure. Various cis-regulatory elements were observed upstream of AlLACs, revealing that AlLACs are involved in different cell signaling pathways. Moreover, AlLACs showed tissue-specific expression, suggesting that AlLAC genes might be associated with growth and development processes. All of the selected AlLAC genes were induced in response to salt, osmotic, and cold stress, indicating that they can play a role in increasing the tolerance to adverse conditions.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SH and MA performed the experiments, SH MA and MK analyzed the data and wrote the manuscript. PH contributed to the data analysis and preparation of the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by Genetics and Agricultural Biotechnology Institute of Tabarestan (GABIT), Sari Agricultural Sciences and Natural Resources University (SANRU) (Grant number: GABIT-98/D/PI271). Costs for open access publishing were partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, grant 491250510).
We thank Dr. Peter Poczai (University of Helsinki, Finland) for the helpful discussion and suggestion.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1112354/full#supplementary-material
Supplementary Table 1 | List of AlLAC genes primers used in real-time PCR.
Supplementary Table 2 | Accession number and sequences (gene, protein, and CDS) of AlLAC genes.
Supplementary Table 3 | Sequence and logo of conserved motifs distributed in AlLAC proteins.
Supplementary Table 4 | Gene ontology (GO) enrichment of AlLAC-interaction network.
Supplementary Table 5 | Functional category of AlLAC promoter element.
Agustin, M. B., de Carvalho, D. M., Lahtinen, M. H., Hilden, K., Lundell, T., Mikkonen, K. S. (2021). Laccase as a tool in building advanced lignin‐based materials. ChemSusChem, 14(21), 4615–4635.
Ahmadizadeh, M., Heidari, P. (2014). Bioinformatics study of transcription factors involved in cold stress. Biharean Biol. 8, 83–86.
Ahmadizadeh, M., Rezaee, S., Heidari, P. (2020). Genome-wide characterization and expression analysis of fatty acid desaturase gene family in camelina sativa. Gene Rep. 21, 100894. doi: 10.1016/j.genrep.2020.100894
Arcuri, M. L. C., Fialho, L. C., Vasconcellos Nunes-Laitz, A., Fuchs-Ferraz, M. C. P., Wolf, I. R., Valente, G. T., et al. (2020). Genome-wide identification of multifunctional laccase gene family in eucalyptus grandis: potential targets for lignin engineering and stress tolerance. Trees 34, 745–758. doi: 10.1007/s00468-020-01954-3
Arregui, L., Ayala, M., Gomez-Gil, X., Gutierrez-Soto, G., Hernandez-Luna, C. E., Herrera de Los Santos, M., et al. (2019). Laccases: Structure, function, and potential application in water bioremediation. Microb. Cell Fact 18 (1), 200. doi: 10.1186/s12934-019-1248-0
Bao, W., O’Malley, D. M., Whetten, R., Sederoff, R. R. (1993). A laccase associated with lignification in loblolly pine xylem. Science 260, 672–674. doi: 10.1126/science.260.5108.672
Berthet, S., Demont-Caulet, N., Pollet, B., Bidzinski, P., Cézard, L., Le Bris, P., et al. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of arabidopsis thaliana stems. Plant Cell 23, 1124–1137. doi: 10.1105/tpc.110.082792
Cai, X., Davis, E. J., Ballif, J., Liang, M., Bushman, E., Haroldsen, V., et al. (2006). Mutant identification and characterization of the laccase gene family in arabidopsis. J. Exp. Bot. 57, 2563–2569. doi: 10.1093/jxb/erl022
Caparrós-Ruiz, D., Fornalé, S., Civardi, L., Puigdomènech, P., Rigau, J. (2006). Isolation and characterisation of a family of laccases in maize. Plant Sci. 171, 217–225. doi: 10.1016/j.plantsci.2006.03.007
Cesarino, I., Araújo, P., Sampaio Mayer, J. L., Vicentini, R., Berthet, S., Demedts, B., et al. (2013). Expression of SofLAC, a new laccase in sugarcane, restores lignin content but not s: G ratio of arabidopsis lac17 mutant. J. Exp. Bot. 64, 1769–1781. doi: 10.1093/jxb/ert045
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cho, H. Y., Lee, C., Hwang, S.-G., Park, Y. C., Lim, H. L., Jang, C. S. (2014). Overexpression of the OsChI1 gene, encoding a putative laccase precursor, increases tolerance to drought and salinity stress in transgenic arabidopsis. Gene 552, 98–105. doi: 10.1016/j.gene.2014.09.018
Faraji, S., Ahmadizadeh, M., Heidari, P. (2021a). Genome-wide comparative analysis of mg transporter gene family between triticum turgidum and camelina sativa. BioMetals 4, 34, 639–660. doi: 10.1007/s10534-021-00301-4
Faraji, S., Heidari, P., Amouei, H., Filiz, E., Poczai, P. (2021b). Investigation and computational analysis of the sulfotransferase (SOT) gene family in potato (Solanum tuberosum): Insights into sulfur adjustment for proper development and stimuli responses. Plants 10, 2597. doi: 10.3390/plants10122597
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). “Protein identification and analysis tools on the ExPASy server,” in The proteomics protocols handbook (Totowa, NJ: Humana Press), 571–607. doi: 10.1385/1-59259-890-0:571
Gavnholt, B., Larsen, K., Rasmussen, S. K. (2002). Isolation and characterisation of laccase cDNAs from meristematic and stem tissues of ryegrass (Lolium perenne). Plant Sci. 162, 873–885. doi: 10.1016/S0168-9452(02)00035-3
Hashemi, S. H., Nematzadeh, G., Ahmadian, G., Yamchi, A., Kuhlmann, M. (2016). Identification and validation of aeluropus littoralis reference genes for quantitative real-time PCR normalization. J. Biol. Res. 23, 1–13. doi: 10.1186/s40709-016-0053-8
Hashemi-Petroudi, S. H., Arab, M., Dolatabadi, B., Kuo, Y. T., Baez, M. A., Himmelbach, A., et al. (2022). Initial description of the genome of aeluropus littoralis, a halophile grass. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.906462
Heidari, P., Abdullah, Faraji, S., Poczai, P. (2021a). Magnesium transporter gene family: Genome-wide identification and characterization in theobroma cacao, corchorus capsularis and gossypium hirsutum of family malvaceae. Agronomy 11, 1651. doi: 10.3390/agronomy11081651
Heidari, P., Entazari, M., Ebrahimi, A., Ahmadizadeh, M., Vannozzi, A., Palumbo, F., et al. (2021b). Exogenous EBR ameliorates endogenous hormone contents in tomato species under low-temperature stress. Horticulturae 7, 84. doi: 10.3390/horticulturae7040084
Heidari, P., Puresmaeli, F., Mora-Poblete, F. (2022). Genome-wide identification and molecular evolution of the magnesium transporter (MGT) gene family in citrullus lanatus and cucumis sativus. Agronomy 12, 2253. doi: 10.3390/agronomy12102253
Hoagland, D. R., Arnon, D. I. (1950). The water-culture method for growing plants without soil. Circ. Calif. Agric. Exp. Stn. 347.
Horton, P., Park, K.-J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor 347 (2nd edit). Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Iñiguez, L. P., Hernández, G. (2017). The evolutionary relationship between alternative splicing and gene duplication. Front. Genet. 8, 14.
Janusz, G., Pawlik, A., Świderska-Burek, U., Polak, J., Sulej, J., Jarosz-Wilkołazka, A., et al. (2020). Laccase properties, physiological functions, and evolution. Int. J. Mol. Sci. 21, 966. doi: 10.3390/ijms21030966
Jeffares, D. C., Penkett, C. J., Bähler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Koralewski, T. E., Krutovsky, K. V. (2011). Evolution of exon-intron structure and alternative splicing. PloS One 6, e18055. doi: 10.1371/journal.pone.0018055
Lee, Y. G., Liao, B. X., Weng, Y. C. (2018). Ascorbic acid sensor using a PVA/laccase-Au-NPs/Pt electrode. RSC Adv. 8 (66), 37872–37879. doi: 10.1039/c8ra06280c
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Liu, Q., Luo, L., Wang, X., Shen, Z., Zheng, L. (2017). Comprehensive analysis of rice laccase gene (OsLAC) family and ectopic expression of OsLAC10 enhances tolerance to copper stress in arabidopsis. Int. J. Mol. Sci. 18, 209. doi: 10.3390/ijms18020209
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, C., Yang, T., Zhang, Y., Miao, X., Jin, C., Xu, X. (2021). Genome-wide analyses and expression patterns under abiotic stress of LAC gene family in pear (Pyrus bretschneideri). Plant Biotechnol. Rep. 15, 403–416. doi: 10.1007/s11816-021-00675-4
Mot, A. C., Silaghi-Dumitrescu, R. (2012). Laccases: complex architectures for one-electron oxidations. Biochem 77, 1395–1407.
Musavizadeh, Z., Najafi-zarrini, H., Kazemitabar, S. K., Hashemi, S. H. (2021). Genome-wide analysis of potassium channel genes in Rice : Expression of the OsAKT and OsKAT genes under salt stress. Genes 12, 784. doi: 10.3390/Genes12050784
Reiss, R., Ihssen, J., Richter, M., Eichhorn, E., Schilling, B., Thöny-Meyer, L. (2013). Laccase versus laccase-like multi-copper oxidase: A comparative study of similar enzymes with diverse substrate spectra. PloS One 8, e65633. doi: 10.1371/journal.pone.0065633
Rezaee, S., Ahmadizadeh, M., Heidari, P. (2020). Genome-wide characterization, expression profiling, and post-transcriptional study of GASA gene family. Gene Rep. 20, 100795. doi: 10.1016/j.genrep.2020.100795
Riva, S. (2006). Laccases: blue enzymes for green chemistry. Trends Biotechnol. 24 (5), 219–226. doi: 10.1016/j.tibtech.2006.03.006
Saad, R.B., Romdhan, W.B., Zouari, N., Azaza, J., Mieulet, D., Verdeil, J.-L., et al. (2011). Promoter of the AlSAP gene from the halophyte grass aeluropus littoralis directs developmental-regulated, stress-inducible, and organ-specific gene expression in transgenic tobacco. Transgenic Res. 20, 1003–1018. doi: 10.1007/s11248-010-9474-6
Sato, Y., Wuli, B., Sederoff, R., Whetten, R. (2001). Molecular cloning and expression of eight laccase cDNAs in loblolly pine (Pinus taeda). J. Plant Res. 114, 147–155. doi: 10.1007/PL00013978
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Simões, M. S., Carvalho, G. G., Ferreira, S. S., Hernandes-Lopes, J., de Setta, N., Cesarino, I. (2020). Genome-wide characterization of the laccase gene family in setaria viridis reveals members potentially involved in lignification. Planta 251, 1–18. doi: 10.1007/s00425-020-03337-x
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Turlapati, P. V., Kim, K.-W., Davin, L. B., Lewis, N. G. (2011). The laccase multigene family in arabidopsis thaliana: towards addressing the mystery of their gene function (s). Planta 233, 439–470. doi: 10.1007/s00425-010-1298-3
Wang, Y., Bouchabke-Coussa, O., Lebris, P., Antelme, S., Soulhat, C., Gineau, E., et al. (2015b). LACCASE5 is required for lignification of the brachypodium distachyon culm. Plant Physiol. 168, 192–204. doi: 10.1104/pp.114.255489
Wang, J., Feng, J., Jia, W., Chang, S., Li, S., Li, Y. (2015a). Lignin engineering through laccase modification: a promising field for energy plant improvement. Biotechnol. Biofuels 8, 1–11. doi: 10.1186/s13068-015-0331-y
Wang, J., Feng, J., Jia, W., Fan, P., Bao, H., Li, S., et al. (2017). Genome-wide identification of sorghum bicolor laccases reveals potential targets for lignin modification. Front. Plant Sci. 8, 714. doi: 10.3389/fpls.2017.00714
Xu, X., Zhou, Y., Wang, B., Ding, L., Wang, Y., Luo, L., et al. (2019). Genome-wide identification and characterization of laccase gene family in citrus sinensis. Gene 689, 114–123. doi: 10.1016/j.gene.2018.12.015
Xu, N., Han, X. M., Xue, Y., Zhuge, X. L., Guan, C. N., Yang, H. L., et al. (2022). Genome-wide comprehensive analysis of PtLACs: Prediction and verification of the functional divergence of tandem-duplicated genes. Forests, 13(2), 157.
Younesi-Melerdi, E., Nematzadeh, G. A., Pakdin-Parizi, A., Bakhtiarizadeh, M. R., Motahari, S. A. (2020). De novo RNA sequencing analysis of Aeluropus littoralis halophyte plant under salinity stress. Scientific Reports, 10(1), 9148.
Zhang, K., Lu, K., Qu, C., Liang, Y., Wang, R., Chai, Y., et al. (2013). Gene silencing of BnTT10 family genes causes retarded pigmentation and lignin reduction in the seed coat of brassica napus. PloS One 8, e61247. doi: 10.1371/journal.pone.0061247
Zhang, Y., Wu, L., Wang, X., Chen, B., Zhao, J., Cui, J., et al. (2019). The cotton laccase gene GhLAC15 enhances verticillium wilt resistance via an increase in defence-induced lignification and lignin components in the cell walls of plants. Mol. Plant Pathol. 20, 309–322. doi: 10.1111/mpp.12755
Keywords: ABA treatment, salt stress, cold stress, gene expression, gene structure, plant gene family
Citation: Hashemipetroudi SH, Arab M, Heidari P and Kuhlmann M (2023) Genome-wide analysis of the laccase (LAC) gene family in Aeluropus littoralis: A focus on identification, evolution and expression patterns in response to abiotic stresses and ABA treatment. Front. Plant Sci. 14:1112354. doi: 10.3389/fpls.2023.1112354
Received: 30 November 2022; Accepted: 23 January 2023;
Published: 01 March 2023.
Edited by:
Elena Baldoni, National Research Council (CNR), ItalyCopyright © 2023 Hashemipetroudi, Arab, Heidari and Kuhlmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seyyed Hamidreza Hashemipetroudi, c2hyLmhhc2hlbWlAc2FucnUuYWMuaXI=; Parviz Heidari, aGVpZGFyaXBAc2hhaHJvb2R1dC5hYy5pcg==; Markus Kuhlmann, a3VobG1hbm5AaXBrLWdhdGVyc2xlYmVuLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.