
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 19 May 2023
Sec. Plant Breeding
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1061803
 Anuj Sharma1
Anuj Sharma1 Jian Li2,3
Jian Li2,3 Rebecca Wente2,3
Rebecca Wente2,3 Gerald V. Minsavage1
Gerald V. Minsavage1 Upinder S. Gill2,3†
Upinder S. Gill2,3† Arturo Ortega4,5
Arturo Ortega4,5 C. Eduardo Vallejos3
C. Eduardo Vallejos3 John P. Hart6
John P. Hart6 Brian J. Staskawicz4,5
Brian J. Staskawicz4,5 Michael R. Mazourek6
Michael R. Mazourek6 Robert E. Stall1
Robert E. Stall1 Jeffrey B. Jones1*‡
Jeffrey B. Jones1*‡ Samuel F. Hutton2,3*‡
Samuel F. Hutton2,3*‡Bacterial spot caused by Xanthomonas euvesicatoria is a major disease of pepper (Capsicum annuum L.) in warm and humid production environments. Use of genetically resistant cultivars is an effective approach to manage bacterial spot. Two recessive resistance genes, bs5 and bs6, confer non-race-specific resistance against bacterial spot. The objective of our study was to map these two loci in the pepper genome. We used a genotyping-by-sequencing approach to initially map the position of the two resistances. Segregating populations for bs5 and bs6 were developed by crossing susceptible Early CalWonder (ECW) with near-isogenic lines ECW50R (bs5 introgression) or ECW60R (bs6 introgression). Following fine-mapping, bs5 was delimited to a ~535 Kbp interval on chromosome 3, and bs6 to a ~666 Kbp interval in chromosome 6. We identified 14 and 8 candidate resistance genes for bs5 and bs6, respectively, based on predicted protein coding polymorphisms between ECW and the corresponding resistant parent. This research enhances marker-assisted selection of bs5 and bs6 in breeding programs and is a crucial step towards elucidating the molecular mechanisms underlying the resistances.
Pepper (Capsicum annuum L.) is an important solanaceous crop that is cultivated throughout the world. Bacterial spot of pepper (BSP) is a major disease responsible for loss of marketable yield in many pepper-growing regions (Osdaghi et al., 2021). The disease is manifested as dark brown necrotic lesions in all aerial parts of the plant. Foliar infection can lead to defoliation, which in turn leads to yield loss. The marketability of fresh fruits is also affected by the presence of scab-like symptoms or due to sun-scalding resulting from extensive defoliation (Ritchie, 2000). The disease is caused by three species of Xanthomonas — X. vesicatoria, X. euvesicatoria (Xe), and X. gardneri (Xg) (Osdaghi et al., 2021). The management of BSP often relies on application of copper-based bactericides; however, the emergence of copper-tolerant strains has rendered this strategy unsustainable (Stall et al., 2009). Alternatively, host plant resistance has been deployed as an effective, economical, and environmentally friendly way of mitigating economic damage caused by BSP.
Most of the resistances deployed in modern agriculture are conditioned by dominant resistance (R) genes which often belong to Nucleotide-Binding Leucine Rich Repeats (NLR) or Receptor-Like Kinase (RLK) protein families (Sharma et al., 2022a). Five dominant resistances have been reported against BSP — Bs1 from C. annuum accession PI 163192 (Cook and Stall, 1963), Bs2 from C. chacoense PI 260435 (Cook and Guevara, 1984), Bs3 from C. annuum PI 271322 (Kim and Hartmann, 1985), Bs4C from C. pubescens PI 235047 (Sahin and Miller, 1998), and Bs7 from C. baccatum var. pendulum UENF 1556 (Potnis et al., 2011). Among them, only Bs2 and Bs3, and to some extent Bs1, have been commercially deployed. Based on gene-for-gene interactions between R genes and their corresponding avirulence genes, BSP causing Xe has been classified into eleven races (P0 – P10) (Stall et al., 2009). Bs1 provides resistance against races P0, P2, and P5; Bs2 against races P0, P1, P2, P3, P7, and P8; and Bs3 against races P0, P1, P4, P7, and P9. Dominant resistance following infection often results in elicitation of a hypersensitive response (HR) and programmed cell death which creates high selection pressure for emergence and enrichment of pathogen races that overcome such resistance through loss/modification of avirulence genes (Gassmann et al., 2000). As a result, R genes are usually short-lived as exemplified by emergence and increased prevalence of races P6 and P10 in bell pepper cultivation, which are insensitive to the deployed R-genes (Kousik and Ritchie, 1996a; Kousik and Ritchie, 1996b; Kousik and Ritchie, 1998; Pernezny et al., 1999; Stall et al., 2009).
In contrast to R genes, recessive resistances typically result from the loss or modification of host susceptibility (S) factors that are exploited by bacteria to initiate a disease response (Sharma et al., 2022a). Recessive resistances are not race-specific and, following infection, do not elicit an HR — the lower selection pressure reduces the chance of emergence of resistance-breaking virulent strains (Parlevliet, 2002; Poland et al., 2009). This makes recessive resistance, despite the breeding challenges, highly desirable for management of rapidly evolving bacterial pathogens, such as Xe. Currently, three recessive resistances have been identified against BSP — bs5 derived from C. annuum PI 271322, bs6 from C. annuum PI 163192 or PI 264281, and bs8 from C. annuum PI 163192 (Jones et al., 2002; Sharma et al., 2022b). Two of these genes, bs5 and bs6, confer resistance to all known Xe races, including race P6 and P10 (Jones et al., 2002; Vallejos et al., 2010). Although bs8 has been demonstrated to suppress Xg, its effect on Xe is not known (Sharma et al., 2022b). Only bs5 has been commercially deployed (McCarthy, 2011; McCarthy, 2012), and there have been no reports of its suppression by Xe.
Both bs5 and bs6 were first reported as monogenic, recessive, non-HR resistances against Xe race P6 (Jones et al., 2002). Both resistance genes were derived from hot pepper accessions collected from India and maintained at the USDA Plant Genetic Resources Conservation Unit, GA (npgsweb.ars-grin.gov/gringlobal). bs5 was reported to originate from C. annuum PI 271322 (Russell, 1955), which had previously been reported to carry field resistance against BSP (Sowell and Dempsey, 1977). Although bs6 is described as originating from either PI 163192 or PI 264281, the most probable source is PI 163192 (Hyland, 1967), which Dempsey et al. (1981) utilized to incorporate bacterial spot resistance into the C44 series of pepper breeding lines; included in this series is the Pep13 line which was used as bs6 donor by Jones et al. (2002) (Lane et al., 1997). Jones et al. (2002) transferred bs5 to the bell pepper C. annuum Early CalWonder (ECW) background by repeated backcrosses to ultimately generate ECW-50R line (Vallejos et al., 2010). A similar strategy was used to develop an ECW NIL containing bs6, which has been named ECW-60R. Recent literature has uncovered that bs5 is also present in PI 163192 (Szarka et al., 2022).
In order to understand the mechanism of resistance, it is often necessary to identify the underlying resistance gene. This is accomplished by gene mapping, which is the process of determining the physical location of a gene in the genome. Mapping of a resistance gene locus also enables the development and use of linked molecular markers (in addition to phenotypic selection) to accelerate the breeding process through marker-assisted selection. Genotyping-by-sequencing (GBS) is a robust sequencing-based method of surveying genome-wide polymorphisms which can be utilized to discover molecular markers (such as SNPs and InDels) and genotypes the samples with those markers in a single step (Elshire et al., 2011). As a large number of small genomic variations from all chromosomes can be utilized in mapping, GBS often provides higher resolution than traditional genotyping methods. In this paper, we (i) identified the genomic localization of bs5 and bs6 resistance genes in pepper genome using GBS, (ii) fine mapped the respective resistance regions and identified flanking markers, and (iii) identified and analyzed candidate resistance genes.
The phenotypic differences between ECW and ECW50R (bs5) were clear and easily distinguishable following inoculation at a relatively low bacterial concentration (105 CFU/ml) (Figure 1). The ECW leaf tissue developed necrotic lesions surrounded by yellow halos while the ECW50R tissue remained mostly green. In the GBS F2 population, 91 out of 100 F2s (19 resistant and 72 susceptible) were phenotyped with high confidence and thus were used for GBS step. The ratio of resistant to susceptible F2s (1:3.8) was slightly lower than the expected ratio of 1:3 for recessive monogenic inheritance, however the difference was not statistically significant (X2 = 0.824 at 1 degree of freedom; p=0.364).
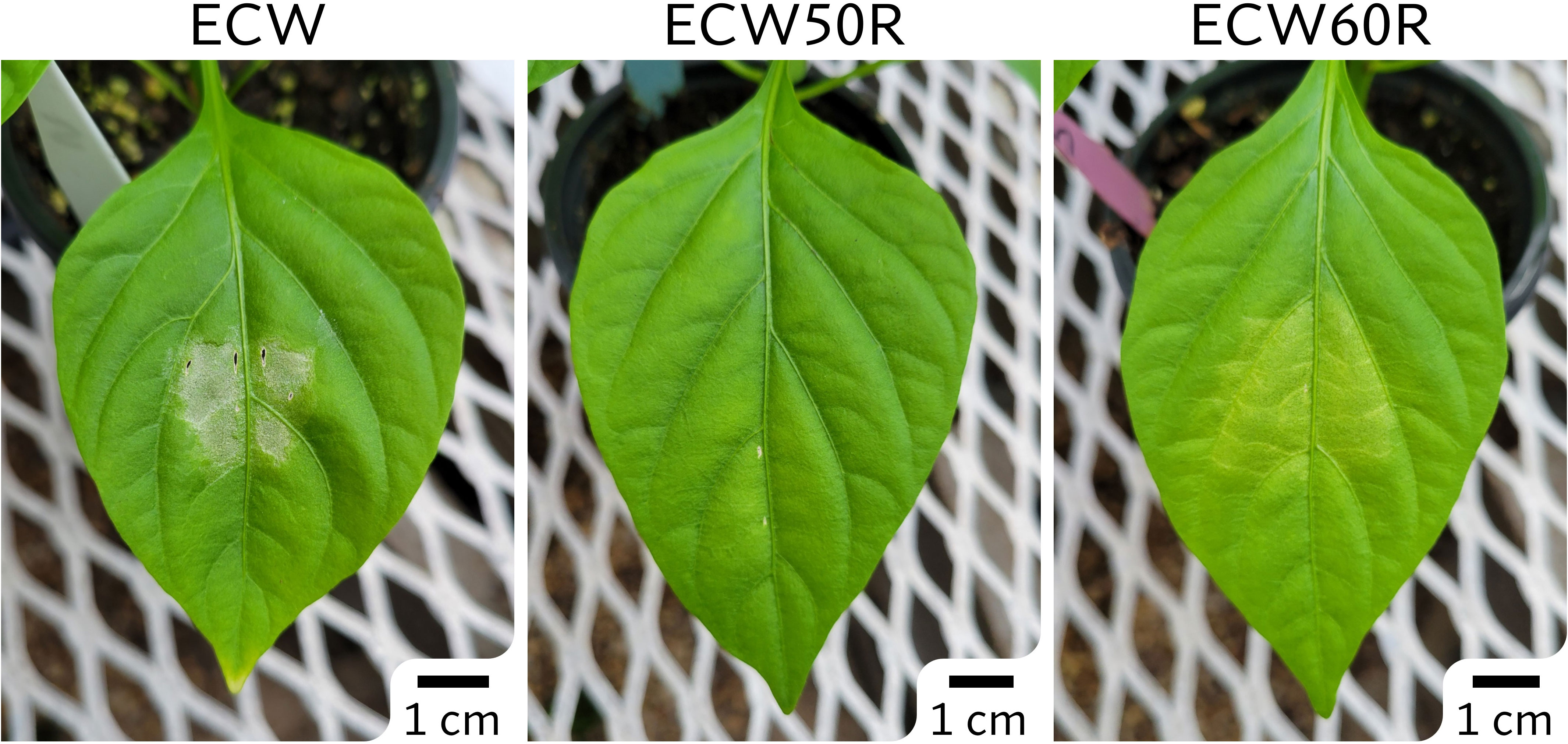
Figure 1 Phenotypes of ECW, ECW50R (bs5), and ECW60R (bs6) pepper 5 days after inoculation of Xanthomonas euvesicatoria strain Xv157 at 105 CFU/ml.
The phenotype of ECW60R (bs6) resistance was not as distinct as bs5 (Figure 1). As expected, bs6 resistance was characterized by extensive chlorosis. Out of 120 F2s, 92 most clearly phenotyped individuals (29 resistant and 63 susceptible) were selected for GBS analysis. The ratio of resistant to susceptible F2s (1:2.2) was not statistically different (X2 = 2.087 at 1 degree of freedom; p=0.1486) from the expected 1:3 ratio.
A total of 169,398,995 reads were generated from the bs5 GBS library (Supplementary Table 1). The GBS pipeline discovered 101 high quality SNPs that were polymorphic between the two parents, and those SNPs were selected for further analysis. The linkage analysis of 88 F2s that could be genotyped identified thirteen linkage groups, and the bs5 resistance mapped to linkage group 1 in chromosome 3 with highest significance (Figure 2; Supplementary Tables 2, 3). SNPs between positions 134,620 and 1,098,542 of chromosome 3 were the most significantly associated with bs5 (p<0.0001). Genotyping of the F2 population with CAPS markers spanning the linkage region confirmed 100% marker-trait co-segregation in the mapping population (Supplementary Tables 4, 5). The results indicate that bs5 is located towards the distal end of the short arm of chromosome 3, within a ~1 Mbp interval between 0.1 and 1.1 Mbp position.
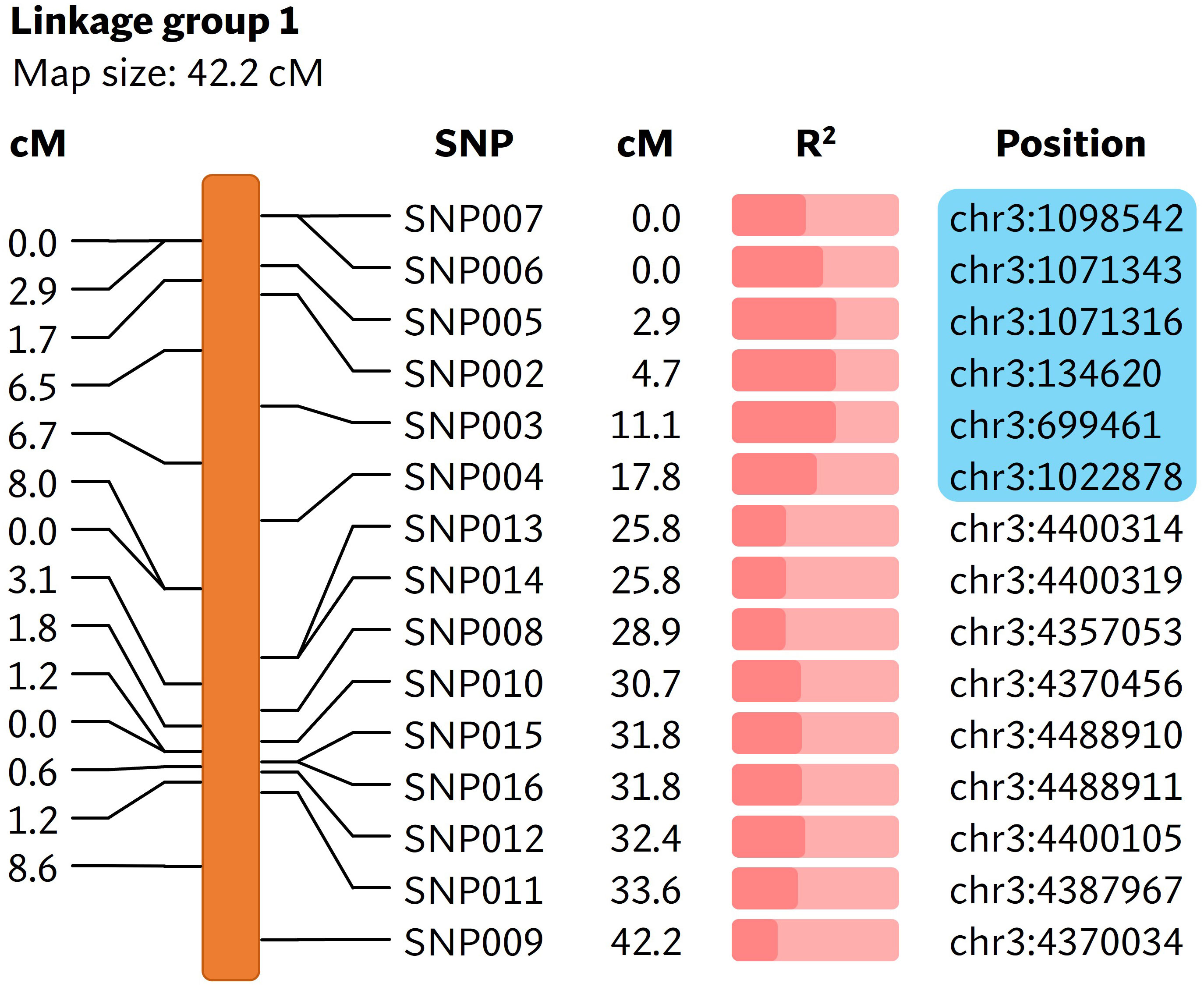
Figure 2 Linkage map showing markers associated with bs5. The cM values in the left represent the linkage distance between the markers and the cM values in the middle represent the positions of markers in the linakge group. The R2 value (Supplementary Table 3) is represented by the fraction of light salmon background filled by darker color. The physical positions of markers are based on C. annuum UCD10X genome, release 1.1. Blue box encloses genomic area that was further investigated by fine-mapping. cM, centimorgan; R2, coefficient of determination.
A larger ECW × ECW50R F2 population was developed to fine-map the position of bs5. Out of 1270 F2s genotyped with flanking markers 3g_C0.134 and 3g_C1.11 (Supplementary Table 4), 16 individuals were identified as recombinants and were phenotyped. Ten informative recombinants and F3 RILs developed from six non-informative recombinants placed bs5 into an ~546 Kb interval between markers 3g_C0.134 (~0.4 cM) and 3g_C0.68 (~0.95 cM) with tight linkage with marker 3g_C0.26. (Figure 3; Supplementary Tables 6, 7).
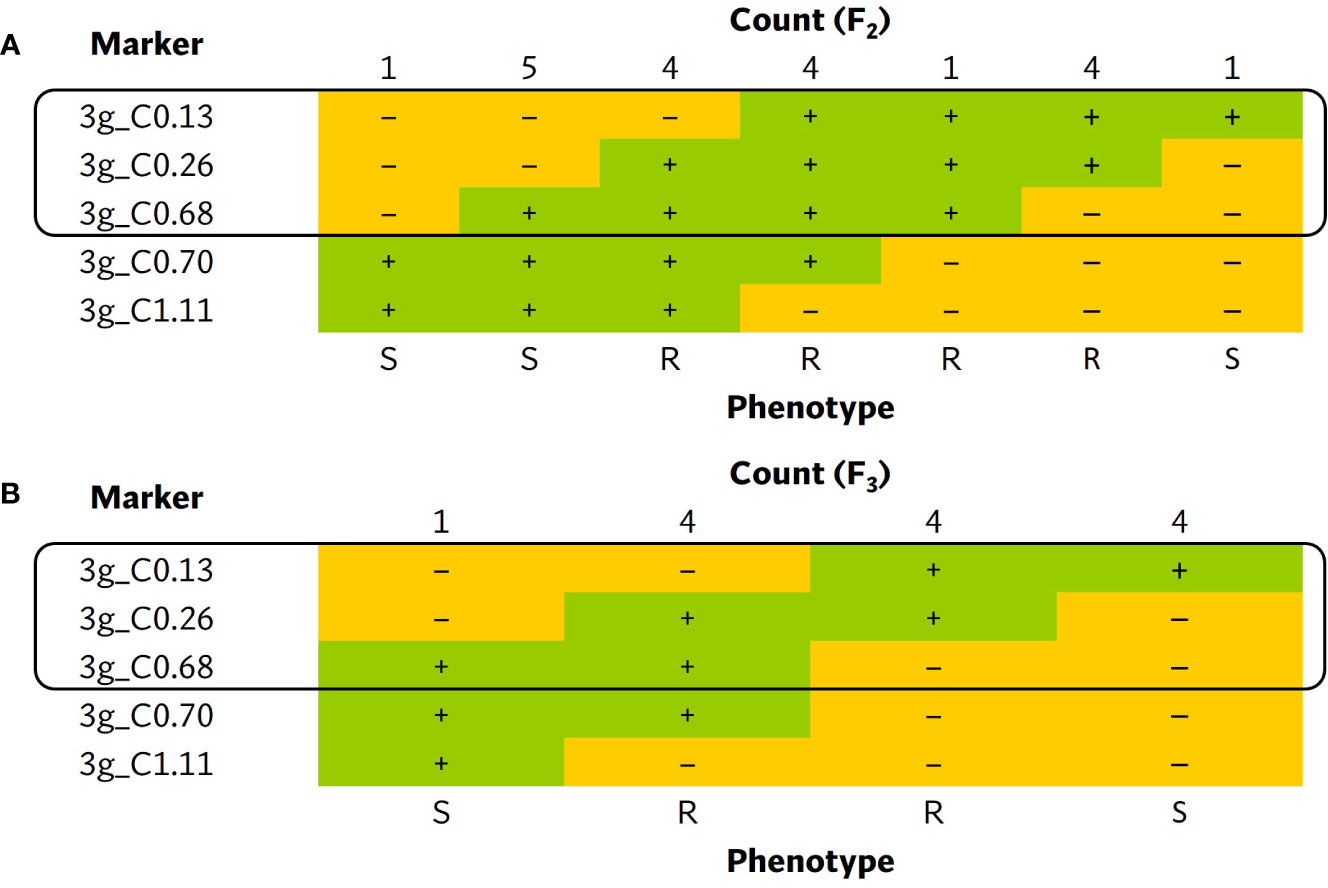
Figure 3 Tabulation of genotypes of the (A) F2 and (B) F3 progenies from bs5 fine-mapping population that recombine within the bs5 mapped region, together with their phenotypes. The black boxes enclose the closest markers flanking the new resistance interval. The numeric portion of marker names following “C” represent their approximate position (in megabases) in chromosome 3 of C. annuum UCD10X genome, release 1.1. +, homozygous for the resistant/ECW50R allele; –, heterozygous or homozygous for the susceptible/ECW allele; R, resistant phenotype; S, susceptible phenotype.
An ECW bs5 super-scaffold was developed by concatenating C. annuum ECW scaffolds that align with in C. annuum UCD10X bs5 interval. This super-scaffold consisted of 535 Kbp sequence including gaps and flanking region and provided complete coverage of UCD10X bs5 interval (Supplementary Table 8). Comparison of whole genome polymorphisms between bs5-fixed line (PI 163192 × ECW50R) and ECW identified a total of 1,718 variants in this region under stringent filtration (data not shown). However, only 28 variants were found to alter the protein sequences, which resulted in 14 putative candidate genes for bs5 resistance (Table 1; Alignment S1-S14).
As the reference-based GBS pipeline only identified a small number of polymorphic markers, the reference-free UNEAK pipeline was used for mapping bs6. This pipeline discovered 133 SNPs from a total of 173,074,228 reads generated from sequencing (Supplementary Table 9). Nine linkage groups were generated from the linkage analysis using genotyping information from 92 F2 plants (Supplementary Table 10), out of which the bs6 resistance phenotype was significantly (p < 0.0001) linked to SNPs on linkage group 3 (Figure 4; Supplementary Tables 10, 11). The linkage group was determined to be physically located in chromosome 6. CAPS markers were developed in the bs6-mapped region, and genotyping of the F2 population validated the linkage between those markers and the resistance phenotype (Supplementary Tables 12, 13). The results indicated that bs6 was located within an ~21 Mbp interval between positions 168–189 Mbp in C. annuum UCD10X genome.
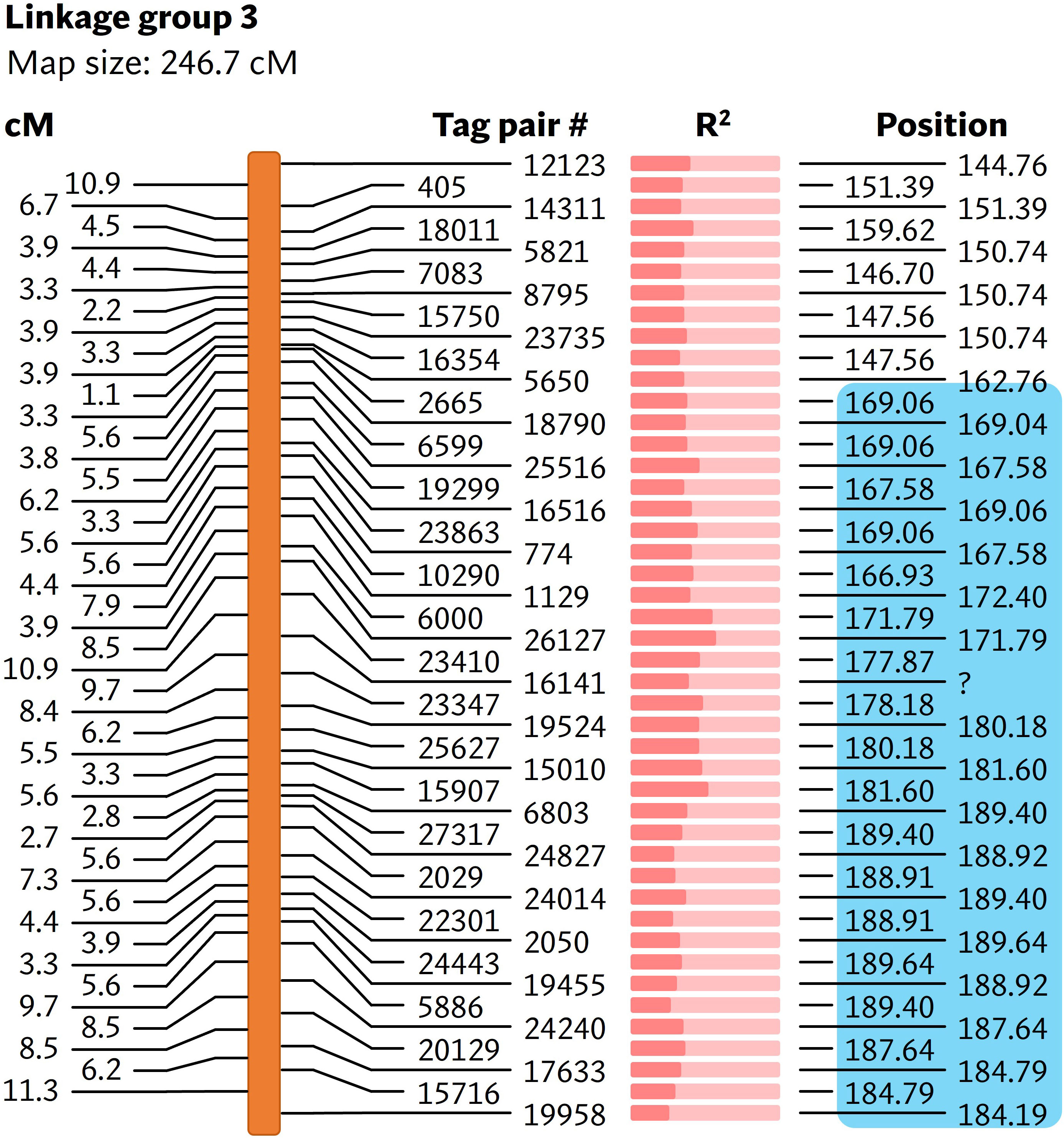
Figure 4 Linkage map showing markers associated with bs6. The cM values on the left represent the linkage distance between the markers. The R2 value (Supplementary Table 11) is represented by the fraction of light salmon background filled by darker color. The physical positions of the markers are based on chromosome 6 of C. annuum UCD10X genome, release 1.1. Blue box encloses genomic area that was further investigated by fine-mapping. cM, centimorgan; R2, coefficient of determination. #, number; ?, unmapped / unknown position.
Five of the CAPS markers within the ~21 Mbp bs6 interval were initially used to more precisely determine the position of bs6. In a fine mapping F2 population of 940 plants, 277 plants were identified as recombinants, 123 of which were homozygous for 60R alleles throughout part of the recombined region and were phenotyped as F2 plants; genotyping of these F2s delimited the resistance locus to an ~9.8 Mbp region between markers 6g_C171.79 and 6g_C181.60 (Figure 5A; Supplementary Table 14). F3 RILs developed from 61 F2s that recombined within the region were genotyped with eight new CAPS markers within the interval (Supplementary Table 12); this delimited bs6 within an ~5.1 Mbp interval between markers 6g_C175.02 and 6g_C180.10 (Figure 5B; Supplementary Table 15). A second ECW60R × ECW F2 population of 940 plants was developed and genotyped with new HRM markers (Supplementary Table 12), and 41 recombinants between flanking markers 6g_H171.54 and 6g_H183.16 were identified and developed into F3 RILs. All 41 RILs were phenotyped and were genotyped with markers in the 5.1 Kbp interval, thereby delimiting bs6 to an ~656 Kbp region between markers 6g_H178.44 (~0.11 cM) and 6g_H179.10 (~0.11 cM) (Figure 5C; Supplementary Table 16).

Figure 5 Tabulation of genotypes of the (A) F2 plants from first bs6 fine-mapping population, (B) F3 plants from first bs6 fine-mapping population, and (C) F3 plants from second bs6 fine-mapping population that recombine within the bs6 interval, together with their phenotypes. The black boxes enclose the closest markers flanking the new resistance interval. Each blue and white block in the scale bar on the left represents 1 Mbp region. The numeric portion of marker names following “C” or “H” represent their approximate position (in megabases) in chromosome 6 of C. annuum UCD10X genome, release 1.1. +, homozygous for the resistant/ECW60R allele; –, heterozygous or homozygous for the susceptible/ECW allele; R, resistant phenotype; S, susceptible phenotype.
The ECW bs6 super-scaffold spanned three C. annuum ECW scaffolds with a total size of 681 Kb, providing complete coverage of UCD10X bs6 interval (Supplementary Table 8). A total of 1,718 variants were identified between ECW and ECW60R genome in this region after filtration. Annotation of those variants identified protein coding changes in eight genes, which are candidates for bs6 (Table 2; Alignment S15-S22). Interestingly, four of those candidates are functionally annotated as ZED1-related serine/threonine kinases, and three have protein polymorphisms within the putative kinase domain (Table 2).
In this paper, we determined the genomic localization of two recessive BSP resistance genes: bs5 and bs6. bs5 was mapped to the telomeric region of chromosome 3 and bs6 to chromosome 6. The genomic position of bs5 is in discordance with a previous report on the position of bs5, which had mapped it to the centromeric region of chromosome 6 (Vallejos et al., 2010). However, the chromosomal position in the previous study was based upon two populations of 60 F2 and 88 F3 progenies and only utilized 64 markers for screening the entire pepper genome. In contrast, the bs5 locations identified in the present study benefited from a much larger number of markers identified through GBS, and has been validated in large fine mapping populations. Furthermore, the recent availability of a high-quality pepper reference genome enabled us to cross-validate our mapping results with the physical positions in the pepper chromosomes.
Several pepper lines have been reported to have varying degrees of recessive resistance against BSP. One of the earliest discoveries of recessive resistance was made by Dempsey (1953) in the pepper cultivar, Santanka. Hibberd et al. (1988) reported quantitative non-race-specific resistance in PI 163189. Poulos et al. (1992) reported that the quantitative, non-HR, non-race-specific resistance in CNPH 703 is controlled by at least two genes. Both PI 163189 and PI 183441 (parent of CNPH 703) were imported together with PI 163192, and thus the resistances in those accessions could also be due to bs5/bs6. A monogenic, recessive, non-HR and non-race-specific resistance in PI 163192 was identified by Szarka and Csilléry (2001) and named gds (general defense system); gds has since been shown to be the same as bs5 (Timár et al., 2019). Riva et al. (2004) reported recessive resistance in UENF 1381 that may be governed by multiple genes. Furthermore, several genes have been identified in pepper which are required for complete virulence; reduced expression of such genes resulted in reduced susceptibility to BSP. Some notable examples include GLIP1 (Hong et al., 2008), MRP1 (An et al., 2008), MLO2 (Kim and Hwang, 2012), and GRP1 (Kim et al., 2015).
A patent filed in 2013 and granted in the US in 2021 describes a recessive, non-race-specific resistance gene in pepper called “xcv-1”, which encodes a cysteine-rich transmembrane region with the resistant allele containing a double leucine deletion (Kiss et al., 2021). Interestingly, one of the polymorphic genes located towards the center of the bs5 fine mapped interval (GeneID: 107864425) encodes a cysteine-rich transmembrane domain-containing protein (CYSTM) and has a double leucine deletion in the resistant allele (Table 1). The genomic localization of xcv-1 has not been reported; however, out of 6 cysteine-rich transmembrane genes annotated in the C. annuum UCD10X genome, two are present in the bs5 region (Supplementary Table 17), and only 107864425 is polymorphic between ECW and ECW50R with a double leucine deletion (Table 1). Thus, it is likely that xcv-1 and bs5 are identical resistances (Szarka et al., 2022) and are encoded by gene 107864425. CYSTM proteins are known to have a role in stress tolerance and disease resistance. Ectopic overexpression of a group of pathogen-induced CYSTM proteins in Arabidopsis reduced in-planta population of Pseudomonas syringae pv. tomato (Pereira Mendes et al., 2021).
A number of bs6 candidate resistance genes are ZED1-related kinases (ZRKs), which are members of the broad receptor-like kinase/Pelle family of protein kinases (Shiu et al., 2004). ZRKs belong to family RLCK-XII, which includes several pseudokinases that can participate in biotic defense response (Lewis et al., 2013; Wang et al., 2015; Seto et al., 2017). A tomato ZRK, JIM2 (RxopJ4), provides resistance against bacterial spot of tomato by serving as a decoy target for the type III effector, XopJ4, and consequently activates a ZAR1-mediated defense response (Schultink et al., 2019). Surprisingly, RxopJ4 is one of several ZRKs located in the syntenic region of bs6 in tomato genome (data not shown) (Sharlach, 2013). Since ZRKs can be targeted by bacterial effectors, and since recessive resistances such as bs6 often result from modification of bacterial susceptibility targets, four ZRKs in the bs6 interval are also intriguing candidates for bs6.
bs5 and bs6 act synergistically and provide resistance against all races of Xe. Together with bs8, which provides resistance against Xg, they enable development of pepper varieties carrying long-lasting recessive resistance to all known BSP pathogens. Pyramiding of resistance genes also increases stability of resistance, both in terms of durability, and against unfavorable conditions. As an example, bs5 or bs6, alone, provides lower levels of resistance at high temperatures (Vallejos et al., 2010). The next steps are to functionally characterize the candidate genes to identify bs5/bs6. Identification of the resistance genes will facilitate understanding of the mechanism of resistance, which in turn can contribute to the development of novel disease control strategies. Apart from pepper, development of bacterial spot-resistant tomatoes is highly desirable, and identification of the bs5/bs6 genes will be a crucial step for identifying tomato homologs which can be targeted by gene-editing technologies.
For developing populations segregating for resistance, ECW50R and ECW60R were used as resistant parents for bs5 and bs6, respectively. ECW was used as susceptible parent for both populations. For both resistances, ECW was crossed with respective resistant parent to produce an F1 population, which was self-pollinated to generate F2 seeds. F3 populations were generated by selfing of F2s when necessary. F2 recombinant individuals were self-pollinated, and progeny were genotyped to identify plants fixed for the recombined chromosomal segments (recombinant inbred lines (RILs)). A complete outline of all populations is presented in Supp. Image 1. For all plants, seeds were sown in a seedling flat, and fourteen-day-old seedlings were transplanted to 10-cm pots containing Fafard Mix 4 (Fafard, Inc., Agawam, MA). For fine-mapping F2 populations, the plants were grown in 242-well trays (Speedling Inc., Sun City, FL) containing Speedling peat-lite soilless media (Speedling Inc., Sun City, FL). The transplants were grown in a greenhouse at temperatures ranging between 20-30 °C.
As the resistant responses due to bs5 and bs6 do not result in HR induction, they are differentiated from the susceptible response by infiltration of bacterial suspension into pepper leaves at a low concentration (Stall, 1981). In contrast to the development of necrotic lesion in susceptible pepper, the bs5 resistance only causes a slight yellowing of the infiltrated area and the bs6 resistant response is characterized by a more intense chlorosis (Vallejos et al., 2010). Xe race P6 strain Xv157 was grown in nutrient broth (BBL, Cockeysville, MD) overnight at 28 °C with constant shaking. Bacterial cells were pelleted by centrifugation, the supernatant was discarded, and the cells were re-suspended in sterile tap water. The bacterial suspension was adjusted using Spectronic 20 Genesys spectrophotometer (Spectronic Instruments, Rochester, NY) to OD600 = 0.3, which is approximately 108 CFU/ml, then diluted to 105 CFU/ml in sterile tap water. The resulting bacterial suspension was infiltrated with a syringe and hypodermic needle into the mesophyll of the first and second true leaf of five- to six-week-old pepper plants. Inoculated plants were maintained in a greenhouse for disease development, and the plants were evaluated three weeks after inoculation. Plants showing confluent necrosis were rated as susceptible, else they were rated as resistant for the respective resistance. For bs6 resistance, the disease screen of each RIL was repeated multiple times to obtain accurate phenotypic result.
Foliar tissue from young leaves was lyophilized and used for DNA extraction. Genomic DNA was extracted using the Qiagen Plant DNeasy Mini Kit (Qiagen, Germantown, MD) according to the manufacturer’s instructions. The DNA was normalized to 5 ng/µL based on quantification with a Synergy 2 multimode microplate reader (Biotek Instruments, Winooski, VT) with the Quant-iT PicoGreen double-stranded DNA quantification assay (Thermo Fisher Scientific, Waltham, MA). A 96-plex (ninety one F2s, a single F1, and two each of ECW and respective resistant parent) ApeKI GBS library was constructed using a previously published protocol (Elshire et al., 2011). Barcode-adapter titration indicated that 0.9 ng µL-1 of each barcode-adapter per 50 ng of genomic DNA produced satisfactory libraries without dimer formation. The barcode-adapter titration mixture and the final GBS library were analyzed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) to ensure acceptable fragment size distribution and quantities. The GBS library was diluted to 3.6 pM and sequenced on one lane (single end, 101 base pair read length) of an Illumina HiSeq 2500 (Illumina Inc, San Diego, CA) at the Genomics Resources Core Facility (Weill Cornell Medicine, NY).
The raw sequencing reads were processed in TASSEL version 3.0 (Bradbury et al., 2007) using either the reference genome-reliant TASSEL-GBS pipeline (Glaubitz et al., 2014) or the reference-free UNEAK pipeline (for bs6) (Lu et al., 2013). For both pipelines, high quality sequencing reads that contained a barcode-adapter, an ApeKI restriction site, and an inserted genomic sequence (hereafter termed GBS tags) were identified and selected based on polymorphism between parents. In TASSEL-GBS pipeline, the reads were aligned with the bwa v0.7.8 (Li and Durbin, 2009) to the C. annuum UCD10X reference genome, release 1.1 (Hulse-Kemp et al., 2018) to identify polymorphisms (Supplementary Table 1). For the UNEAK pipeline, reference genome information was not necessary, and SNPs were identified by pairwise alignment of all unique sequence tags across the entire dataset (Supplementary Table 2). Raw read files from sequencing of GBS libraries are deposited in NCBI SRA under bioproject PRJNA863731.
Polymorphic SNPs identified between the parental lines were employed for linkage analyses using MapDisto v1.7 (implemented within Microsoft Excel 2007), (Lorieux, 2012). The parameters in linkage analyses were a minimum LOD=5, a maximum r=0.3, and the ‘Kosambi’ mapping function. The loci were ordered within each linkage map using the auto-order function. QTL analysis was conducted for each population to determine the association between the SNPs within a linkage group and resistance to race P6. Single marker analysis was performed using the R/qtl package in R v3.3.1 (Broman et al., 2003).
Cleaved Amplified Polymorphic Sequence (CAPS) markers were designed for validating the mapping results from GBS and for fine mapping. Primers for the markers were designed using Primer 3 software (Untergasser et al., 2007) utilizing SNPs identified from GBS. DNA was extracted using a Cetyltrimethylammonium Bromide (CTAB) method (Doyle and Doyle, 1987) and polymerase chain reaction (PCR) was carried out with Phire Hot Start II DNA polymerase (Thermo Fisher Scientific, Waltham, MA) in a 10 μl volume, which consisted of 2 μl of DNA (adjusted to ~20 ng/μl), 4.89 μl of HPLC-H2O, 2 μl of 5X Phire Reaction Buffer, 1 μl of dNTPs, 0.03 μl each of forward and reverse primers, and 0.05 μl of polymerase. The amplicons were digested with appropriate restriction enzymes according to the manufacturer’s recommendations (New England Biolabs, Ipswich, MA). Results were detected using electrophoresis on 3% agarose gels stained with ethidium bromide.
High Resolution Melting curve (HRM) markers were developed from SNPs identified from GBS. Primers were developed using the IDT PrimerQuest (idtdna.com/Primerquest). DNA was extracted using a NaOH rapid DNA extraction method (Lee et al., 2017). The 5 μl PCR reactions were mixed with 2x AccuStart II PCR SuperMix (Quantabio, Beverly, MA), 0.5 μM of each primer, and 20x EvaGreen Dye (Biotium, Hayward, CA) and run as follows: (95 °C @ 60s) + 40 × ((94 °C @ 5s) + (Tm @ 10s) + (72 °C @ 15s)) + (72 °C for 60s), where Tm is the annealing temperature. For allele determination, melting curve analysis was performed by scanning the PCR product in a LightCycler 480 Instrument II (Roche, Pleasanton, CA).
A modified microprep protocol was used for DNA extraction for whole genome sequencing of ECW60R (Fulton et al., 1995; Sharma et al., 2022b). DNA concentration and purity was verified using NanoDrop (Thermo Fisher Scientific, Waltham, MA). Subsequently, DNA was cleaned using DNeasy PowerClean Pro Cleanup Kit (Qiagen, Germantown, MD) following the manufacturer’s recommendations. Illumina sequencing library was prepared using a Nextera DNA Flex Library Prep Kit (Illumina Inc, San Diego, CA) using the protocol recommended by the manufacturer. The DNA was sequenced to produce 100 base-pairs (bp) paired end reads in one lane of Illumina HiSeq 3000 at University of Florida Interdisciplinary Center for Biotechnology Research.
The C. annuum ECW whole genome sequence (GCA_011745845.1) was only assembled to scaffold level at the time of analysis (Kim et al., 2017). To produce contiguous sequence, the bs5 or bs6 fine mapped intervals were blasted against the reference genome C. annuum UCD10X (GCF_002878395.1). All ECW scaffolds with query coverage greater than 2% and matching to unique regions were identified and concatenated together in correct order and orientation to produce ECW super-scaffolds for bs5 and bs6. The super-scaffolds also consisted of 5 Kbp region up- and down–stream from flanking markers and 3 Kbp gap between stitched scaffolds. The super-scaffolds were aligned with C. annuum UCD10X resistance intervals to verify complete coverage.
The ECW genes were predicted de-novo to overcome differences in gene annotations between reference genomes. ECW gene prediction model was developed using BRAKER v2.1.6 (Brůna et al., 2021). Within BRAKER, three publicly available ECW RNAseq sequences (SRR13488414, SRR13488423, and SRR13488424) were aligned to C. annuum ECW genome sequence (GCA_011745845.1) and supplied to Genemark-ET v4.68 (Lomsadze et al., 2014) to generate hints for training Augustus v3.4.0 (Stanke et al., 2008). The resulting ECW gene prediction model was used to identify potential protein coding regions in the bs5 and bs6 super-scaffolds. The genes were validated based on their posterior probability and annotation of homologous regions in C. annuum UCD10X or C. annuum CM334 annotation.
Polymorphisms for bs5 were identified using whole genome bulk sequences of PI 163192 × ECW50R F2 population, which is fixed for bs5 gene (Sharma et al., 2022b). For bs6, the whole genome sequence of ECW60R was used. The sequences were analyzed using an in-house pipeline. The quality of the reads was verified with FastQC 0.11.7 (bioinformatics.babraham.ac.uk/projects/fastqc) and the adapters were trimmed using trim_galore v0.6.5 (Krueger et al., 2021). The trimmed reads were aligned to C. annuum ECW genome using Bwa-mem2 v2.2.1 (Vasimuddin et al., 2019). The resulting alignment file was used for variant calling with the HaplotypeCaller tool in GATK 4 (DePristo et al., 2011). The variants were filtered under high stringency as follows: depth ≥ 12, quality-normalized depth ≥ 10, mapping quality ≥ 50, and reference allele depth ≤ 0.1 × alternate allele depth. The sequencing data for PI 163192 × ECW50R F2s has previously been deposited in NCBI/ENA/DDBJ database under bioproject PRJNA789991. ECW60R whole genome sequence is deposited under bioproject PRJNA863893.
The coordinates and allelic sequence of high-quality polymorphisms in bs5/bs6 super-scaffolds were derived from variant calling of C. annuum ECW scaffolds with an in-house script. The polymorphism were annotated with snpEff v5.0 (Cingolani et al., 2012) using a custom super-scaffold variant annotation database built using previously described sequences and protein coding regions. Only the variations that result in protein coding changes were selected to identify potential candidate genes. Potential homologs of candidate genes in other C. annuum genomes were identified by blasting the predicted amino acid sequences of those genes, which also provided the functional annotations of the candidates. Finally, protein domains containing the polymorphisms between ECW and ECW50R/ECW60R were identified by Pfam (Mistry et al., 2021) and InterPro search (Blum et al., 2021).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
GM, JJ, and RS initially developed ECW50R and ECW60R lines. JL, JH, and MM performed GBS and contributed to its analysis. Fine mapping was conducted by JL, RW and SH (genotyping) and GM and JJ (phenotyping). UG generated the whole genome sequences. AS contributed to manuscript writing, fine-mapping, sequence analysis, and identification of candidate genes. All authors contributed to the article and approved the submitted version.
The research was supported by USDA NIFA Specialty Crop Research Initiative Grants Program Grant Number: 2015-51181-24312 and 2019-51181-30010. We would also like to thank 2blades foundation for providing support for initial stages of mapping. John Hart was supported by a Seed Matters Postdoctoral Fellowship.
Xanthomonas euvesicatoria strain Xv157 (race 6) was kindly provided by S. A. Miller (The Ohio State University, Wooster).
AO and BS are applicants for patent WO2021011348A1/US20210071193A1. MM is a cofounder of Row 7 Seeds, but neither receives compensation nor holds equity.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1061803/full#supplementary-material
An, S. H., Choi, H. W., Hwang, I. S., Hong, J. K., Hwang, B. K. (2008). A novel pepper membrane-located receptor-like protein gene CaMRP1 is required for disease susceptibility, methyl jasmonate insensitivity and salt tolerance. Plant Mol. Biol. 67, 519. doi: 10.1007/s11103-008-9337-1
Blum, M., Chang, H.-Y., Chuguransky, S., Grego, T., Kandasaamy, S., Mitchell, A., et al. (2021). The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354. doi: 10.1093/nar/gkaa977
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Broman, K. W., Wu, H., Sen, Ś., Churchill, G. A. (2003). R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890. doi: 10.1093/bioinformatics/btg112
Brůna, T., Hoff, K. J., Lomsadze, A., Stanke, M., Borodovsky, M. (2021). BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR. Genomics Bioinf. 3, lqaa108. doi: 10.1093/nargab/lqaa108
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 6, 80–92. doi: 10.4161/fly.19695
Cook, A., Guevara, Y. (1984). Hypersensitivity in Capsicum chacoense to race 1 of the bacterial spot pathogen of pepper. Plant Dis. 68, 329–330. doi: 10.1094/PD-69-329
Cook, A., Stall, R. (1963). Inheritance of resistance in pepper to bacterial spot. Phytopathology 53, 1060–1062.
Dempsey, A. (1953) Inheritance studies of certain fruit and plant characters in capsicum frutescens. Available at: http://rave.ohiolink.edu/etdc/view?acc_num=osu1486473800771257 (Accessed March 29, 2021).
Dempsey, A., Demski, J., Sowell, G. (1981). Breeding pimiento. Ed. Charleston, S. C. (Charleston, South Carolina: US Vegetable Laboratory, USDA-ARS).
DePristo, M. A., Banks, E., Poplin, R., Garimella, K. V., Maguire, J. R., Hartl, C., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. doi: 10.1038/ng.806
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-Sequencing (GBS) approach for high diversity species. PloS One 6, e19379. doi: 10.1371/journal.pone.0019379
Fulton, T. M., Chunwongse, J., Tanksley, S. D. (1995). Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol. Biol. Rep. 13, 207–209. doi: 10.1007/BF02670897
Gassmann, W., Dahlbeck, D., Chesnokova, O., Minsavage, G. V., Jones, J. B., Staskawicz, B. J. (2000). Molecular evolution of virulence in natural field strains of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 182, 7053–7059. doi: 10.1128/JB.182.24.7053-7059.2000
Glaubitz, J. C., Casstevens, T. M., Lu, F., Harriman, J., Elshire, R. J., Sun, Q., et al. (2014). TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PloS One 9, e90346. doi: 10.1371/journal.pone.0090346
Hibberd, A., Stall, R., Bassett, M. (1988). Quantitatively assessed resistance to bacterial leaf spot in pepper that is simply inherited. Phytopathology 78, 607–612. doi: 10.1094/Phyto-78-607
Hong, J. K., Choi, H. W., Hwang, I. S., Kim, D. S., Kim, N. H., Choi, D. S., et al. (2008). Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta 227, 539–558. doi: 10.1007/s00425-007-0637-5
Hulse-Kemp, A. M., Maheshwari, S., Stoffel, K., Hill, T. A., Jaffe, D., Williams, S. R., et al. (2018). Reference quality assembly of the 3.5-Gb genome of Capsicum annuum from a single linked-read library. Hortic. Res. 5, 1–13. doi: 10.1038/s41438-017-0011-0
Hyland, H. L. (1967) Plant material introduced January 1 to December 31, 1961 (Nos. 270535 to 277783) (Beltsville, MD: United States Department of Agriculture). Available at: https://handle.nal.usda.gov/10113/39196 (Accessed January 4, 2023).
Jones, J. B., Minsavage, G. V., Roberts, P. D., Johnson, R. R., Kousik, C. S., Subramanian, S., et al. (2002). A non-hypersensitive resistance in pepper to the bacterial spot pathogen is associated with two recessive genes. Phytopathology 92, 273–277. doi: 10.1094/phyto.2002.92.3.273
Kim, B. S., Hartmann, R. W. (1985). Inheritance of a gene (Bs3) conferring hypersensitive resistance to Xanthomonas campestris pv. vesicatoria in pepper (Capsicum annuum). Plant Dis. 69, 233–235.
Kim, D. S., Hwang, B. K. (2012). The pepper MLO gene, CaMLO2, is involved in the susceptibility cell-death response and bacterial and oomycete proliferation. Plant J. 72, 843–855. doi: 10.1111/tpj.12003
Kim, D. S., Kim, N. H., Hwang, B. K. (2015). GLYCINE-RICH RNA-BINDING PROTEIN1 interacts with RECEPTOR-LIKE CYTOPLASMIC PROTEIN KINASE1 and suppresses cell death and defense responses in pepper (Capsicum annuum). New Phytol. 205, 786–800. doi: 10.1111/nph.13105
Kim, S., Park, J., Yeom, S.-I., Kim, Y.-M., Seo, E., Kim, K.-T., et al. (2017). New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 18, 210. doi: 10.1186/s13059-017-1341-9
Kiss, G. B., Szabó, Z., Iliescu, C. E., Balogh, M. (2021) Identification of a xanthomonas euvesicatoria resistance gene from pepper (Capsicum annuum) and method for generating plants with resistance. Available at: https://patents.google.com/patent/US11041165B2/en (Accessed April 5, 2022).
Kousik, C. S., Ritchie, D. F. (1996a). Disease potential of pepper bacterial spot pathogen races that overcome the Bs2 gene for resistance. Phytopathology 86, 1336–1343.
Kousik, C. S., Ritchie, D. F. (1996b). Race shift in Xanthomonas campestris pv. vesicatoria within a season in field-grown pepper. Phytopathology 86, 952–958. doi: 10.1094/Phyto-86-952
Kousik, C. S., Ritchie, D. F. (1998). Response of bell pepper cultivars to bacterial spot pathogen races that individually overcome major resistance genes. Plant Dis. 82, 181–186. doi: 10.1094/PDIS.1998.82.2.181
Krueger, F., James, F., Ewels, P., Afyounian, E., Schuster-Boeckler, B. (2021). FelixKrueger/TrimGalore: v0.6.7 (Charleston, South Carolina: Centro Internacional de Agricultura Tropical). doi: 10.5281/zenodo.5127899
Lane, R., McCarter, S., Kuhn, C., Deom, C. (1997). “Dempsey”, a virus-and bacterial spot-resistant bell pepper. Hortscience 32, 333–334. doi: 10.21273/HORTSCI.32.2.333
Lee, S., Noh, Y.-H., Roach, J. A., Mangandi, J., Verma, S., Whitaker, V. M., et al. (2017). A high-throughput genotyping system combining rapid DNA extraction and high-resolution melting analysis in allo-octoploid strawberry. Acta Hortic. 1156, 89–94. doi: 10.17660/ActaHortic.2017.1156.12
Lewis, J. D., Lee, A. H.-Y., Hassan, J. A., Wan, J., Hurley, B., Jhingree, J. R., et al. (2013). The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc. Natl. Acad. Sci. U.S.A. 110, 18722–18727. doi: 10.1073/pnas.1315520110
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Lomsadze, A., Burns, P. D., Borodovsky, M. (2014). Integration of mapped RNA-seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res. 42, e119. doi: 10.1093/nar/gku557
Lorieux, M. (2012). MapDisto: fast and efficient computation of genetic linkage maps. Mol. Breed. 30, 1231–1235. doi: 10.1007/s11032-012-9706-y
Lu, F., Lipka, A. E., Glaubitz, J., Elshire, R., Cherney, J. H., Casler, M. D., et al. (2013). Switchgrass genomic diversity, ploidy, and evolution: Novel insights from a network-based SNP discovery protocol. PloS Genet. 9, e1003215. doi: 10.1371/journal.pgen.1003215
McCarthy, W. (2011) Sweet pepper hybrid 9954288. Available at: https://patents.google.com/patent/US8013222B2 (Accessed October 11, 2021).
McCarthy, W. (2012) Sweet pepper hybrid 9942815. Available at: https://patents.google.com/patent/US8138398B2 (Accessed September 12, 2022).
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Osdaghi, E., Jones, J. B., Sharma, A., Goss, E. M., Abrahamian, P., Newberry, E. A., et al. (2021). A centenary for bacterial spot of tomato and pepper. Mol. Plant Pathol. 22, 1500–1519. doi: 10.1111/mpp.13125
Parlevliet, J. E. (2002). Durability of resistance against fungal, bacterial and viral pathogens; present situation. Euphytica 124, 147–156. doi: 10.1023/A:1015601731446
Pereira Mendes, M., Hickman, R., Van Verk, M. C., Nieuwendijk, N. M., Reinstädler, A., Panstruga, R., et al. (2021). A family of pathogen-induced cysteine-rich transmembrane proteins is involved in plant disease resistance. Planta 253, 102. doi: 10.1007/s00425-021-03606-3
Pernezny, K., Collins, J., Stall, R. E., Shuler, K., Datnoff, L. E. (1999). A serious outbreak of race 6 of xanthomonas campestris pv. vesicatoria on pepper in southern Florida. Plant Dis. 83, 79–79. doi: 10.1094/PDIS.1999.83.1.79C
Poland, J. A., Balint-Kurti, P. J., Wisser, R. J., Pratt, R. C., Nelson, R. J. (2009). Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 14, 21–29. doi: 10.1016/j.tplants.2008.10.006
Potnis, N., Minsavage, G., Smith, J. K., Hurlbert, J. C., Norman, D., Rodrigues, R., et al. (2011). Avirulence proteins AvrBs7 from Xanthomonas gardneri and AvrBs1.1 from Xanthomonas euvesicatoria contribute to a novel gene-for-Gene interaction in pepper. Mol. Plant Microbe Interact. 25, 307–320. doi: 10.1094/MPMI-08-11-0205
Poulos, J. M., Reifschneider, F. J.B., Coffmann, W. R. (1992). Inheritance of quantitative components of resistance to Xanthomonas campestris pv. vesicatoria in pepper line ‘CNPH 703’. In: Proceedings of the VIIIth EUCARPIA Meeting on Genetics and Breeding on Capsicum and Eggplant. Rome, Italy. pp 166–171.
Ritchie, D. (2000). Bacterial spot of pepper and tomato. Plant Health Instr. doi: 10.1094/PHI-I-2000-1027-01
Riva, E., Rodrigues, R., Pereira, M., Sudré, C., Karasawa, M. (2004). Inheritance of bacterial spot disease in Capsicum annuum l. Crop Breed. Appl. Biotechnol. 4, 490–494. doi: 10.12702/1984-7033.v04n04a18
Russell, P. G. (1955) Plant material introduced by the section of plant introduction, horticultural crops research branch, agricultural research service, January 1 to December 31, 1948 (Nos. 161667 to 172310) (Beltsville, MD: United States Department of Agriculture). Available at: https://handle.nal.usda.gov/10113/39236 (Accessed January 4, 2023).
Sahin, F., Miller, S. A. (1998). Resistance in capsicum pubescens to Xanthomonas campestris pv. vesicatoria pepper race 6. Plant Dis. 82, 794–799. doi: 10.1094/PDIS.1998.82.7.794
Schultink, A., Qi, T., Bally, J., Staskawicz, B. (2019). Using forward genetics in Nicotiana benthamiana to uncover the immune signaling pathway mediating recognition of the Xanthomonas perforans effector XopJ4. New Phytol. 221, 1001–1009. doi: 10.1111/nph.15411
Seto, D., Koulena, N., Lo, T., Menna, A., Guttman, D. S., Desveaux, D. (2017). Expanded type III effector recognition by the ZAR1 NLR protein using ZED1-related kinases. Nat. Plants 3, 1–4. doi: 10.1038/nplants.2017.27
Sharlach, M. (2013) Mapping and identification of the RXopJ4 resistance gene and the search for new sources of durable resistance to bacterial spot disease of tomato. Available at: https://escholarship.org/uc/item/1zx6v1gg (Accessed July 27, 2022).
Sharma, A., Abrahamian, P., Carvalho, R., Choudhary, M., Paret, M. L., Vallad, G. E., et al. (2022a). Future of bacterial disease management in crop production. Annu. Rev. Phytopathol. 60, 259–282. doi: 10.1146/annurev-phyto-021621-121806
Sharma, A., Minsavage, G. V., Gill, U. S., Hutton, S. F., Jones, J. B. (2022b). Identification and mapping of bs8, a novel locus conferring resistance to bacterial spot caused by Xanthomonas gardneri. Phytopathology 112, 1640–1650. doi: 10.1094/PHYTO-08-21-0339-R
Shiu, S.-H., Karlowski, W. M., Pan, R., Tzeng, Y.-H., Mayer, K. F. X., Li, W.-H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220–1234. doi: 10.1105/tpc.020834
Sowell, G., Dempsey, A. (1977). Additional sources of resistance to bacterial spot of pepper. Plant Dis. Reptr. 61, 684–686.
Stall, R. E. (1981). “Selection for component of horizontal resistance to bacterial spot of pepper,” In: Lozano, J.C. (ed). Proceedings of international conference on plant pathogenic bacteria, Centro Internacional de Agricultura Tropical, Cali, Colombia. 511–517.
Stall, R. E., Jones, J. B., Minsavage, G. V. (2009). Durability of resistance in tomato and pepper to xanthomonads causing bacterial spot. Annu. Rev. Phytopathol. 47, 265–284. doi: 10.1146/annurev-phyto-080508-081752
Stanke, M., Diekhans, M., Baertsch, R., Haussler, D. (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644. doi: 10.1093/bioinformatics/btn013
Szarka, J., Csilléry, G. (2001). General defense system in the plant kingdom. Int. J. Hortic. Sci. 7, 79–84. doi: 10.31421/IJHS/7/1/254
Szarka, J., Timár, Z., Hári, R., Palotás, G., Péterfi, B. (2022). General defense response under biotic stress and its genetics at pepper (Capsicum annuum l.). Sustainability 14, 6458. doi: 10.3390/su14116458
Timár, Z., Palotás, G., Csilléry, G., Szarka, J. (2019). “Study of recessive bacterial leaf spot resistance genes in capsicum annuum l,” in Innovations in genetics and breeding of capsicum and eggplant. Eds. Lefebvre, V., Daunay, M.-C. (Avignon, France: Institut National de la Recherche Agronomique), 132–133. Available at: https://symposium.inrae.fr/eucarpiacapsicumeggplant2019/content/download/5824/79309/version/1/file/Proceedings_17thCapsEggEUCARPIA_Avignon2019.pdf.
Untergasser, A., Nijveen, H., Rao, X., Bisseling, T., Geurts, R., Leunissen, J. A. M. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–W74. doi: 10.1093/nar/gkm306
Vallejos, C. E., Jones, V., Stall, R. E., Jones, J. B., Minsavage, G. V., Schultz, D. C., et al. (2010). Characterization of two recessive genes controlling resistance to all races of bacterial spot in peppers. Theor. Appl. Genet. 121, 37–46. doi: 10.1007/s00122-010-1289-6
Vasimuddin, Md., Misra, S., Li, H., Aluru, S. (2019). “Efficient architecture-aware acceleration of BWA-MEM for multicore systems,” in 2019 IEEE International parallel and distributed processing symposium (IPDPS) (New York: IEEE), 314–324. doi: 10.1109/IPDPS.2019.00041
Keywords: capsicum annuum, genotyping-by-sequencing, xanthomonas euvesicatoria, disease resistance, marker-assisted selection, recessive resistance
Citation: Sharma A, Li J, Wente R, Minsavage GV, Gill US, Ortega A, Vallejos CE, Hart JP, Staskawicz BJ, Mazourek MR, Stall RE, Jones JB and Hutton SF (2023) Mapping of the bs5 and bs6 non-race-specific recessive resistances against bacterial spot of pepper. Front. Plant Sci. 14:1061803. doi: 10.3389/fpls.2023.1061803
Received: 05 October 2022; Accepted: 22 March 2023;
Published: 19 May 2023.
Edited by:
Dilip R. Panthee, North Carolina State University, United StatesReviewed by:
Jelli Venkatesh, Seoul National University, Republic of KoreaCopyright © 2023 Sharma, Li, Wente, Minsavage, Gill, Ortega, Vallejos, Hart, Staskawicz, Mazourek, Stall, Jones and Hutton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Samuel F. Hutton, c2ZodXR0b25AdWZsLmVkdQ==; Jeffrey B. Jones, amJqb25lc0B1ZmwuZWR1
†Present address: Upinder S. Gill, Department of Plant Pathology, North Dakota State University, Fargo, ND, United States
‡These authors share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.