- 1China Tobacco Gene Research Center, Zhengzhou Tobacco Research Institute of China National Tobacco Corporation (CNTC), Zhengzhou, China
- 2National Tobacco Genetic Engineering Research Center, Yunnan Academy of Tobacco Agricultural Sciences, Kunming, China
Proteins of the Nitrate Transporter 1/Peptide Transporter (NPF) family transport a diverse variety of substrates, such as nitrate, peptides, hormones and chloride. In this study, a systematic analysis of the tobacco (Nicotiana tabacum) NPF family was performed in the cultivated ‘K326’. In total, 143 NtNPF genes were identified and phylogenetically classified into eight subfamilies, NPF1 to NPF8, based on the classification of NPF families in other plant species. The chromosomal locations and structures of the NtNPF genes were analyzed. The expression profiles of NtNPF genes under NaCl stress were analyzed to screen the possible NPF genes involving in chloride regulation in tobacco. Most NtNPF6 genes responded to salt stress in the roots and leaves. The expression of NtNPF6.13 was significantly down-regulated after salt stress for 12h. The chloride content was reduced in the roots of ntnpf6.13 mutant. These findings support the participation of NtNPF6.13 in chloride uptake. Several other NtNPF genes that play potential roles in chloride metabolism of tobacco require further study.
Introduction
As an important macronutrient, nitrogen (N) plays an essential role in plant growth and development (Crawford, 1995; Xu et al., 2012). Among the different forms of nitrogen, nitrate () is predominant in plant roots. The four most reported nitrate transport protein families are the Nitrate Transporter 1/Peptide Transporter family (NPF), the Nitrate Transporter 2 family (NRT2), the Chloride Channel family (CLC), and the slow anion channel-associated homologues family (SLAC/SLAH) (Krapp et al., 2014).The NPF family typically comprises a large number of members within a species, for example, 53 NPFs in Arabidopsis and 93 NPFs in rice are known and have been well studied (Leran et al., 2014; Wang et al., 2018b). The NPF proteins can be classified into eight subfamilies and named by a standard rule. In Arabidopsis, 20 AtNPF genes are involved in nitrate uptake or efflux from the soil. AtNPF6.3, also named NRT1.1 or CHL1, was the first nitrate transporter to be cloned and has dual-affinity nitrate transport activity (Tsay et al., 1993). Regardless of their participation in nitrate uptake, root-to-shoot transport, or leaf nitrate allocation, AtNPF genes all have important functions. In other species, NPF genes have been reported to contribute to nitrate transportation, for example, NPF2.2 in rice (Li et al., 2015), NPF6 in maize (Wen et al., 2017), and NPF6.5 in grape (He et al., 2020).
In addition to nitrate transport, NPF proteins perform diverse functions (Anfang and Shani, 2021; Prabhala et al., 2021).An increasing number of other substrates of NPF proteins have been reported (Corratge-Faillie and Lacombe, 2017; Payne et al., 2017; Wulff et al., 2019), of which one is chloride. It is well known that CLC and SLAC/SLAH genes encode chloride transporters or channels that participate in chloride transport (Barbier-Brygoo et al., 2011; Zhang et al., 2018).The micronutrient Cl- shows strongly dynamic interaction with (Wege et al., 2017). Given the similar physical properties in solution, the selectivity of proteins for these two monovalent anions is often ambiguous. In Arabidopsis, two NPF members are involved in chloride transport, namely AtNPF2.4 (Li et al., 2016a), and AtNPF2.5 (Li et al., 2016b). AtNPF2.4is highly expressed in the root stele and facilitates the transfer of chloride from root to the shoot. Accumulation of Cl- increases with overexpression of AtNPF2.4and is reduced with knockdown of AtNPF2.4. AtNPF2.5, which is the closest homolog to AtNPF2.4, is expressed predominantly in the root and modulates chloride efflux from the root. Both AtNPF2.4 and AtNPF2.5may modulate Cl- transportation without affecting accumulation in the shoot. In maize, ZmNPF6.4 and ZmNPF6.6 transport both chloride and nitrate (Wen et al., 2017). With regard to chloride transport, ZmNPF6.4 shows high-affinity chloride transport activity, whereas ZmNPF6.6 shows low-affinity chloride transport activity. In contrast, with respect to nitrate transport, ZmNPF6.4 exhibits low-affinity nitrate transport activity, whereas ZmNPF6.6 shows high-affinity nitrate transport activity. Recently, it has been reported that MtNPF6.5 and MtNPF6.7 mediate chloride uptake and nitrate preference in Medicago roots (Xiao et al., 2021). Apart from these reported NPF genes, little information is available on the participation of NPF genes in chloride transport in other species.
To date, the NPF gene family has been identified and characterized in many plant species, including Arabidopsis (Chiba et al., 2015), wheat (Wang et al., 2020), rice (Drechsler et al., 2018), apple (Wang et al., 2018a), rapeseed (Zhang et al., 2020), spinach (Wang et al., 2021), potato (Zhang et al., 2021) and poplar (Zhao et al., 2021). No previous study has investigated the NPF gene family in Nicotiana genus. In the current study, we identified the NPF family members of tobacco (Nicotiana tabacum) and analyzed the phylogenetic relationships, gene structure and chromosomal location. To screen NtNPF genes involved in chloride transportation, the expression profiles of NtNPF genes were evaluated using an RNA sequencing (RNA-seq) data from plants exposed to salt stress. The results revealed that 80% of NtNPF6genes were responsive to salt stress either in the roots or the leaves. Using the CRISPR/Cas9 gene-editing system to knockout NtNPF6.13, one of most responsive NtNPF6 members in roots under salt stress, the chloride content of roots was significantly decreased in the mutants compared to the wild type. The findings indicate that NtNPF6.13 might play an important role in chloride uptake.
Materials and methods
Identification of NPF genes in tobacco
Genome sequences of ‘K326’ (a commonly cultivated tobacco cultivar) were downloaded from the Sol Genomics Network (https://solgenomics.net/). Fifty-three AtNPF seqences were obtained from The Arabidopsis Information Resource database (https://www.arabidopsis.org/). The AtNPF protein sequences were used to perform a BLASTP search in the K326 database with E-value<1e-5. The PFAM domain (PF00854.18: PTR2) was used to identify NtNPFs. The online ExPASy tool (https://www.expasy.org/) was used to predict the molecular weights and isoelectric points of each NtNPF protein.
Phylogenetic analysis of NtNPF proteins
Protein sequence alignments for Arabidopsis (53 AtNPFs) and tobacco (143 NtNPFs) NPF family members were generated using ClustalW (https://www.genome.jp/tools-bin/clustalw). A phylogenetic tree was constructed with RAxML (version 8.2.10) under the PROTGAMMAGTR model with 100 bootstrap replications. The phylogenetic tree was edited and visualized in Evolview V3 (https://www.evolgenius.info/evolview/) (Subramanian et al., 2019).
Chromosomal localization and syntenic analysis of NtNPF genes
The chromosomal location of the NtNPF genes was determined from the Sol Genomics Network database. Syntenic blocks were identified using MCScanX (Wang et al., 2012). The results were visualized with Circos software (Krzywinski et al., 2009), with syntenic blocks involving NtNPF gene pairs connected by lines.
Gene structure and conserved motifs analysis of NtNPF gene family
The genomic DNA and CDS information for the NtNPF genes were downloaded from Sol Genomics Network database. To explore the structure of NtNPF genes, the Gene Structure Display Server 2.0online tool (GSDS, http://gsds.gao-lab.org/index.php; Hu et al., 2015) was used to display the genomic length and organization of introns/exons. The conserved motifs of NtNPFs were analyzed using the MEME online tool (version 5.3.0, http://meme-suite.org/tools/meme; Bailey et al., 2015). The maximum number of motifs was set 10.
Cis-regulatory element analysis of NtNPFs
To predict the function of NtNPFs, putative cis-reguatory elements were analyzed. The 2-kb upstream sequence from the translation start site of all 143 NtNPF genes was obtained and set as the promoter. The sequences were analyzed using the plantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; Lescot et al., 2002).
Plant materials and salt treatment
Seedlings of tobacco’K326’ were cultivated in plastic pots under a 16-h photoperiod at 28 and 23 °C (day/night). For salt treatment, plants at the six-leaf stage were transferred to a nutrient solution for 1 week, and then NaCl (final concentration 300 mM) was added to the solution to initiate salt treatment. After treatment for 12 h, 3d or 7 d, leaves and roots were sampled and used for RNA extraction and RNA-seq. Three biological replicates were used and the nutrient solution without NaCl was used as control (CK). All samples were immediately frozen in liquid nitrogen and stored at -80 °C, except that the roots were washed with water before freezing. The nutrient solution contains 5mM potassium nitrate (KNO3), 1 mM magnesium sulfate heptahydrate (MgSO4·7H2O), 1 mM monopotassium phosphate (KH2PO4), 4 mM calcium nitrate tetrahydrate (Ca(NO3)2·4H2O), 1mM ammonium nitrate (NH4NO3), 0.1 mM ferric sodium EDTA (Fe-Na-EDTA), 0.1mM boric acid (H3BO3), 30 µM zinc sulfate (ZnSO4·7H2O), 100 µM manganese monosulfate (MnSO4·H2O), 0.1 µM copper sulfate pentahydrate (CuSO4·5H2O), 0.1 µM cobalt chloride hexahydrate (CoCl2·6H2O) and 1 µM sodium molybdate dihydrate (Na2MoO4·2H2O) per liter, with about 0.2 μM of background chloride concentration.
RNA isolation and qPCR analysis
Total RNA was isolated using the RNAprep Pure Plant Kit (Tiangen, Beijing, China) in accordance with the manufacturer’s protocol. The RNA quality and concentration were determined using a Nano Drop 2000 spectrophotometer. The cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche). Quantitative real-time PCR (qPCR) analysis was performed as described previously (Zhang et al., 2018). All specific primers used are listed in Supplementary Table S1.
RNA-sequencing and transcriptomic analysis
For RNA-Seq, total RNA was extracted from the K326 root and leaf samples (CK and treated with 300mM NaCl for 12h, 3d or 7d) with the RNAprep Pure Plant Kit. Each sample, comprising three seedlings, was analyzed in triplicate under the same conditions. A total of 24 samples were used for RNA-seq by the Novogene Company (Beijing, China) on an Illumina HiSeq4000 platform. After filtering for quality control, the clean reads were mapped to the K326 genome (ftp://ftp.solgenomics.net/genomes/Nicotiana_tabacum/edwards_et_al_2017/assembly/) with Hisat2 (Kim et al., 2019). Gene abundances were estimated using StringTie (Pertea et al., 2015). The expression levels of NtNPF genes were then analyzed.
Subcellular localization of NtNPF6.13
The subcellular localization of NtNPF6.13 was first predicated with the online tools Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/; Chou and Shen, 2010), WoLF PSORT (https://wolfpsort.hgc.jp; Horton et al., 2007), YLoc (https://abi-services.informatik.uni-tuebingen.de/yloc/webloc.cgi; Briesemeister et al., 2010) and PSORT (https://www.genscript.com/tools/psort; Nakai and Horton, 1999). The online tool TMHMM2.0 (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0) was used to predict the transmembrane helices of NtNPF6.13.To verify the predicted localization, NtNPF6.13 was cloned without the termination codon using a pair of gene-specific primers, (5’-ATGGCACTTCCTGAGACAC-3’ and 5’-ACAAACCGGTCCATCATC-3’). The fragment was purified and inserted into the vector pFFCS1300 to fuse the C-terminus with the green fluorescent protein (GFP) under the constitutive control of the CaMV35S promoter. After sequencing, the recombinant plasmid 35S: NtNPF6.13-GFP was transferred to Agrobacterium tumefaciens strain GV3101. The fusion construct was infected into the leaf of Nicotiana benthamiana and the GFP signal was observed using a TCS STED CW confocal laser microscope (LEICA, Wetzlar, Germany).
Generation of NtNPF6.13 knockout lines and salt treatment
The CRISPR/Cas9-based genome editing method was used to generateNtNPF6.13 knockout lines. A 20-bp coding sequence (5’-GCTCTTAGATCCTCCTCCGG-3’) of NtNPF6.13 was inserted into the sgRNA-Cas9 expression vector and transformed into tobacco K326. A 252-bp DNA fragment was amplified from the transgenic lines by using a primer pair (5’-CTTCCTGAGACACAGCAAG-3’ and 5’-TTCCCATCTAATCTCGCC-3’) bordering the target region. The PCR products were sequenced to determine the mutation sites of NtNPF6.13. The existence of Cas9 sequence in T1 transgenic plants was examined using a Cas9-sepecific primer pair (5’-GGGACCCTAAGAAGTACGGC-3’ and 5’-TATTCTCGGCCTGCTCTCTG-3’).Plants free of Cas9 were retained and theNtNPF6.13 genotype was determined.
Using the NaCl treatment method described above, ntnpf6.13 mutants and K326 control plants were treated with 300 mM NaCl for 7 d. The roots were collected for ion determination. For phenotyping of plants under the salt stress, ntnpf6.13 mutants and K326seeds were geminated on 1/2 MS plates for 1 week, then the seedlings were transferred to fresh MS plates supplemented with 150 mM NaCl for additional1 week of growth before photographing and measurement of the root length.
Determination of chloride content in roots
The concentration of Cl- was determined as described previously (Zhang et al., 2018). Briefly, the roots were dried and ground into powder. Then, 50 mg powder was digested with 10 ml of 5% acetic acid. After incubation at 30 °C for 30min, the digested solution was filtered and diluted, and analyzed with an AA3 Continuous Flow Analytical System (Germany).
Results
Genome-wide identification and characterization of tobacco NPF genes
To comprehensively identify the NtNPF genes in tobacco, NPF protein sequences of Arabidopsis were used to perform a BLASTP search of the genome database of Nicotiana tabacum (K326) from Sol Genomics Network. As shown in Supplementary Table S2, 143 NtNPF genes were identified and named in accordance with the nomenclatural rules for the gene family (Leran et al., 2014). The gene sequence length ranged from 375bp (NtNPF4.18) to 18,162bp (NtNPF7.10). The corresponding proteins were predicted ranging from 103aa (NtNPF2.8) to 1,158aa (NtNPF7.10). The length of more than 65% of NtNPF proteins was 500 aa -600 aa. The isoelectric point ranged from 4.31 (NtNPF5.9) to 10.99 (NtNPF2.15), and in most NtNPFs (>85%) was greater than7.
Phylogenetic tree of NtNPF gene family in tobacco
To gain insight into the phylogenetic relationship of the 143NtNPFs, a phylogenetic tree was constructed using RAxML version 8.2.10 under the PROTGAMMAGTR model together with 53 AtNPF proteins. Based on the relationships with AtNPFs, the NtNPFs were classified into eight subfamilies (Figure 1), which was consistent with the classifications of NPFs from other species, such as Brassica napus (Wen et al., 2020), apple (Malus × domestica Borkh.) (Wang et al., 2018a), and spinach (Wang et al., 2021). The eight subfamilies were designatedNtNPF1 to NtNPF8, and each contained 20, 25, 4, 24, 25, 20, 11 and 14 members, respectively.
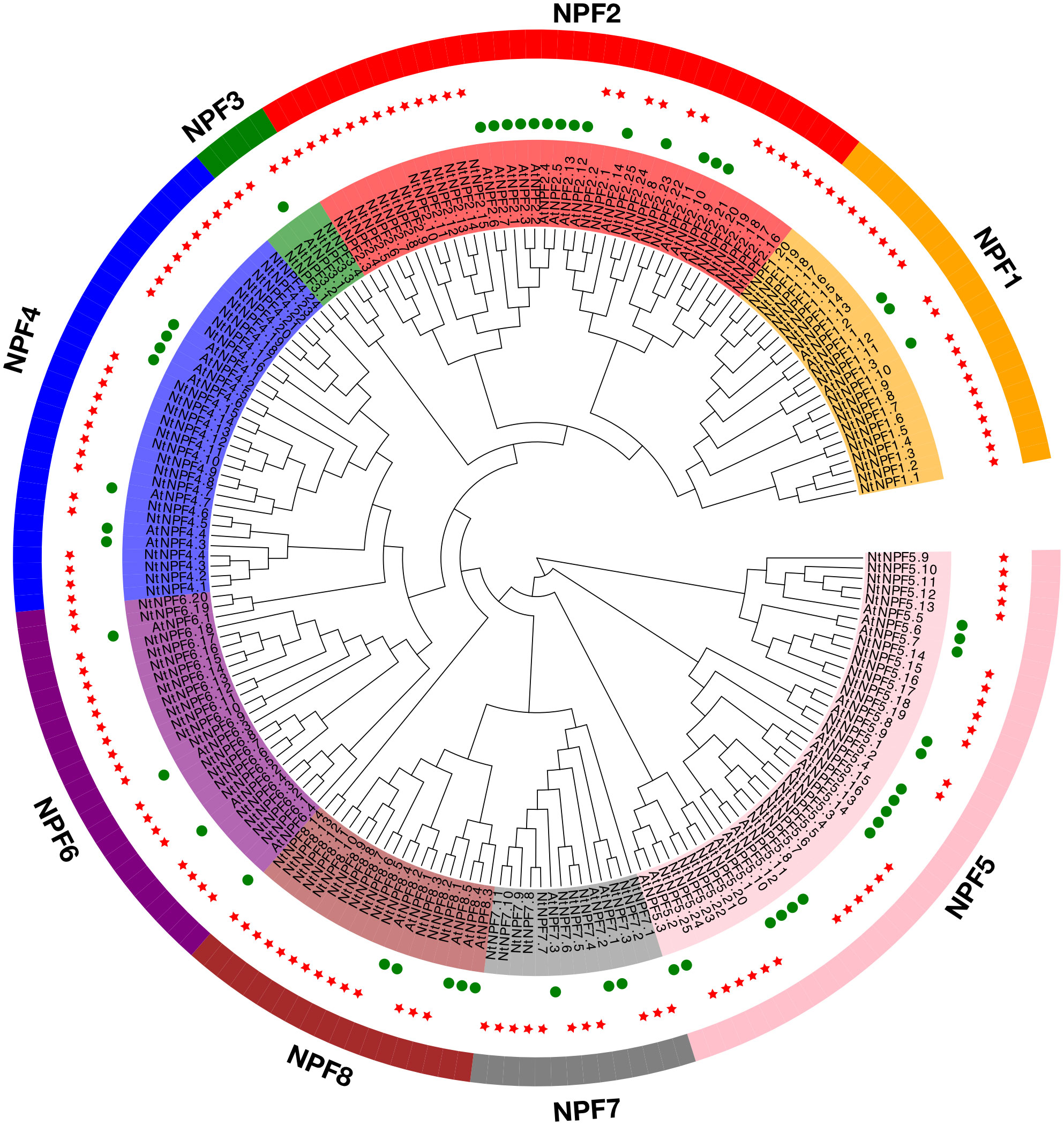
Figure 1 Phylogenetic tree of NtNPF genes of tobacco and AtNPF genes of Arabidopsis. The clades representing different subfamilies (groups 1–8) are indicated by different colors. NtNPF genes are marked with a green circle and AtNPF genes are marked with a red star.
Chromosomal locations and synteny analysis of NtNPF genes
The chromosomal distribution showed that 88 of the 143 NtNPF genes were mapped on 20 of 24 tobacco chromosomes (Figure 2). Five chromosomes (Nt04, Nt09, Nt18, Nt20 and Nt24) carried seven NtNPF genes each, one chromosome (Nt02) carried six NtNPF genes, five chromosomes (Nt08, Nt12, Nt15, Nt22 and Nt23) harbored five NtNPF genes each, two chromosomes (Nt05 and Nt07) contained four NtNPF genes each, two chromosomes (Nt06 and Nt10)carried three genes each, three chromosomes (Nt01, Nt19 and Nt21) harbored two NtNPF genes each, and two chromosomes (Nt03 and Nt17) each carried a single NtNPF gene. With regard to the four NtNPF3 genes, only one was mapped to a chromosome; the remaining three were mapped to scaffolds. In addition, it was noted that most NtNPF genes were located on the chromosome arms, whereas several NtNPF genes, such as NtNPF5.4 and NtNPF5.6, were positioned near the telomere.
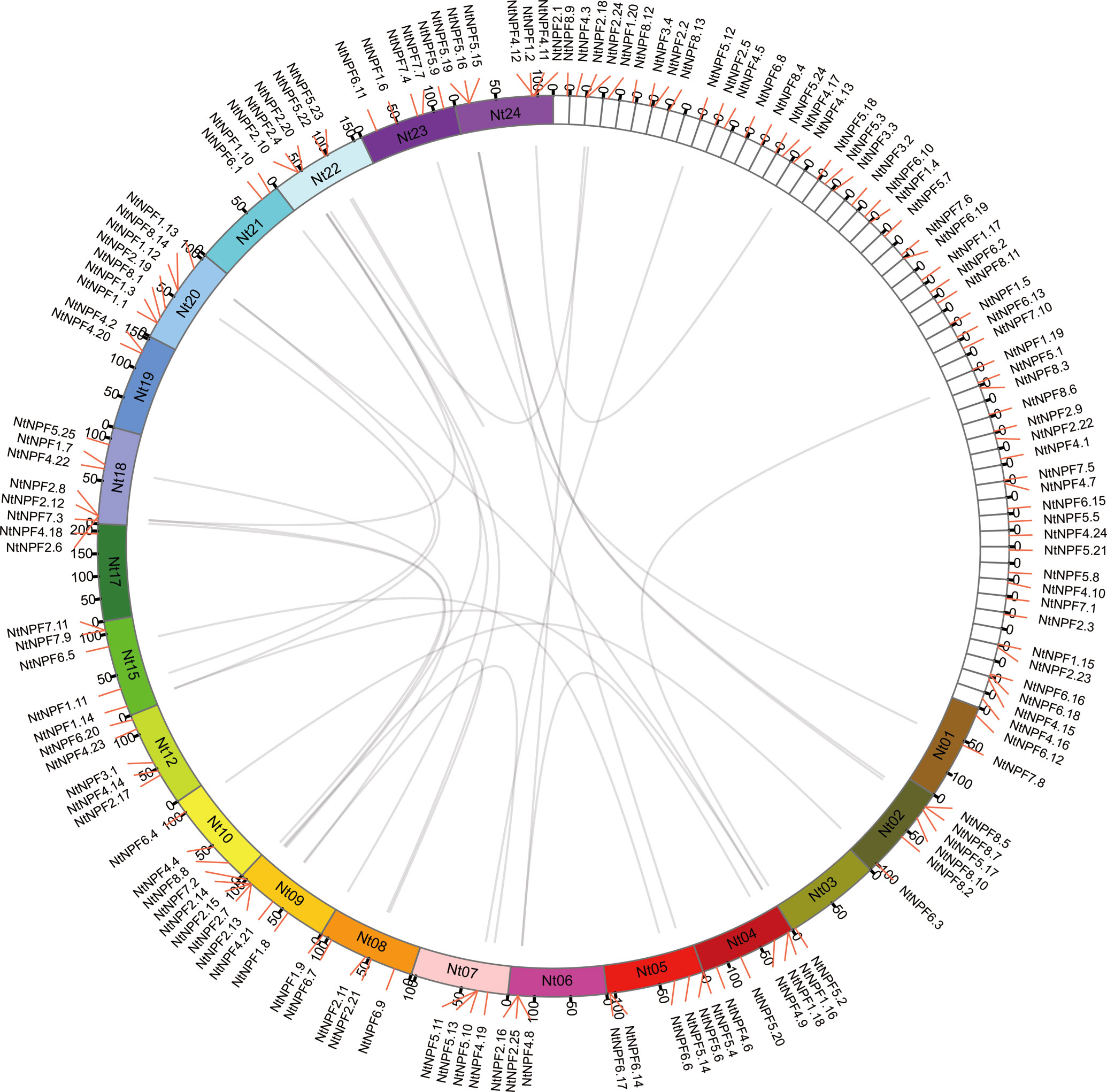
Figure 2 Chromosomal distribution and segmental duplication of NtNPF genes in tobacco. The panel shows the 20 chromosomes and unassigned scaffolds using a circle; gray lines connect homologous genes.
Using the predicted NtNPF protein sequences, a snyteny analysis was performed with MCScanX. Thirty-nine NtNPF genes were included in syntenic blocks, forming 26 syntenic NtNPF gene pairs (Figure 2). The 39 NtNPF genes belonged to seven NtNPF subfamilies excluding the NtNPF3 subfamily. The NtNPF5 subfamily contributing nine genes, NtNPF1, NtNPF2, and NtNPF4 contributed seven genes each, NtNPF6 and NtNPF7 contributed four genes each, andNtNPF8 contributed one gene.
Gene structure and conserved motifs of NtNPF genes in tobacco
The NtNPF gene structure was determined using the GSDS online tools. The exon number of the 143 NtNPFs ranged from 1 to 13, and almost 110NtNPFs contained 3-5 exons (Figure 3). The longest NtNPF gene, NtNPF7.10, contained eight exons, whereasNtNPF1.10included the highest exon number (13) among all NtNPF genes. Seven genes, includingNtNPF1.2, NtNPF1.16 and NtNPF2.9, contained no introns. The diversity of the gene structure might imply an abundance of gene functions. Using the multiple sequence alignment of tobacco NPF proteins, the MEME tool was used to investigate the sequence features and functional motifs. Ten conserved motifs for NtNPF proteins were identified, which were designated motif 1 to motif 10 (Figure 3).

Figure 3 Phylogenetic relationships, gene structure, and architecture of conserved protein motifs of NtNPF genes in tobacco.
Cis-acting elements in the promoters of NtNPF genes
The promoter (2 kb upstream) of each gene was analyzed using the PlantCARE database to explore the cis-acting regulatory elements of the NtNPF genes. Multiple cis-elements in the NtNPF gene promoters were detected, such as plant hormone response elements (auxin, methyl jasmonate [MeJA], gibberellin, salicylic acid and abscisic acid), abiotic stress response elements (light, low-temperature, wound, hypoxia and drought), biotic stress response elements and circadian control elements (Figure 4).The G-box, Box 4, ABRE and ARE elements were most frequently enriched in the NtNPF promoter regions (Figure 4A and Supplementary Table S3 & Figure S1). The CGTCA-motif, TGACG-motif and GT1-motif elements, which are associated with MeJA response, were mostly enriched in the promoters of NtNPF3 subfamily genes (Figure 4B), which indicated that NtNPF3genes might function in MeJA regulation.
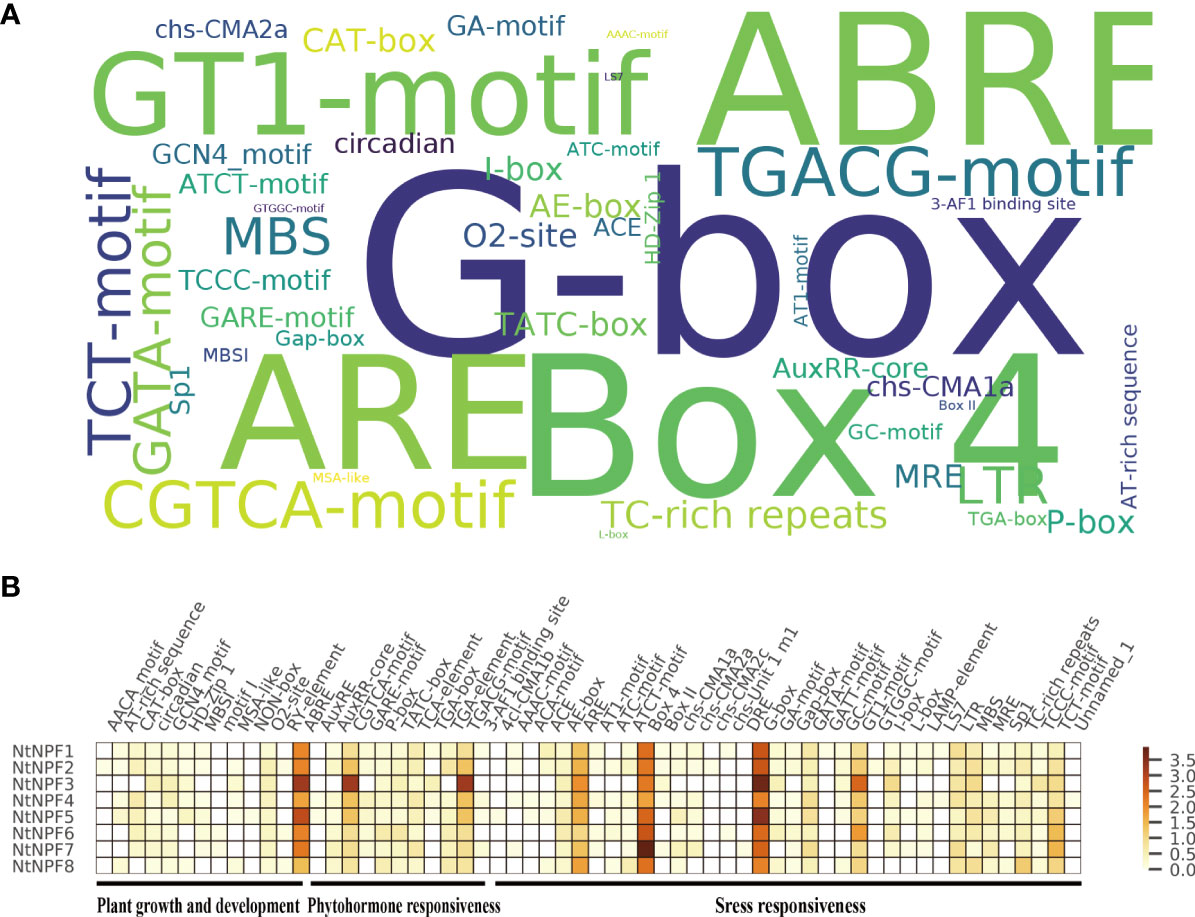
Figure 4 Cis-acting elements in the promoter of NtNPF genes of tobacco. (A) Over-presentation of the cis-acting elements in the promoter of all NtNPF genes. The font size is positively associated with the occurrence number of corresponding cis-acting elements. (B) Heatmap of average number of cis-acting elements in different NtNPF subfamilies. Brown represents a high number and white represents a low number of cis-acting elements.
Transcriptional profiles of NtNPF genes under salt stress
In order to screen the potential NtNPF genes in response to salt stress, the transcriptional profile was analyzed using RNA-seq data sets for the roots and leaves derived from tobacco K326 seedlings exposed to 300mM NaCl stress for 0h, 12h, 3d and 7d.Among the 143 NtNPF genes, more than 70% showed extremely low or undetectable transcript levels before and after exposure to salt stress (Supplementary Figure S2).In contrast, several NtNPF6genes, namelyNtNPF6.16, NtNPF6.18, NtNPF6.13, NtNPF6.12, and NtNPF6.11, showed the highest expression levels in the root prior to salt stress (Figure 5 and Supplementary Table S4). Thirteen NtNPF genes responded to salt stress in the leaves and 30 NtNPF genes responded to salt stress in the roots. Two NtNPF genes, NtNPF2.22 and NtNPF2.23, responded to salt stress in the roots and leaves. Almost all NtNPF6subfamily members responded to salt stress either in the leaves or the roots, except NtNPF6.1, NtNPF6.2, NtNPF6.19 and NtNPF6.20. Interestingly, AtNPF6.3 homologous NtNPF6 genes usually responded in the roots, whereasAtNPF6.2 homologous NtNPF6 genes usually responded in the leaves. Notably, most of the responsive NtNPF genes were down-regulated at 12h under salt stress both in the roots and leaves. The root-specific highly expressed NtNPF6.13, NtNPF6.16 and NtNPF6.18 showed strongly similar expression patterns in response to salt stress: transcription was down-regulated after salt stress for 12h; and thereafter was mostly recovered at 3d and 7d under salt stress. The expression changes of the aforementioned NtNPF genes indicated that these NtNPF genes might play important roles in tolerance to salt stress in tobacco.
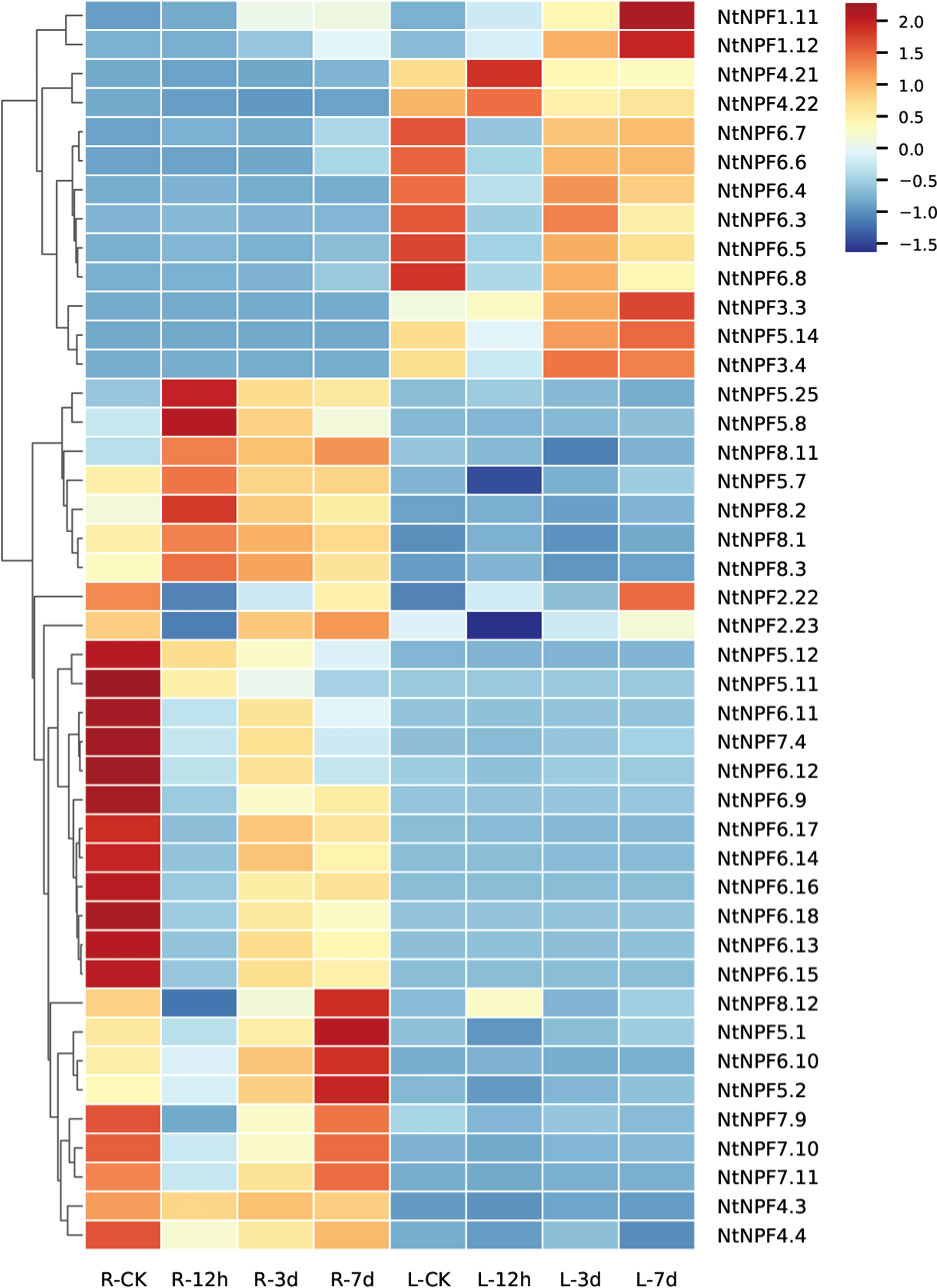
Figure 5 Expression profiles of NtNPF genes responsive to salt stress in roots and leaves of tobacco. R-CK, R-12h, R-3d and R-7d represent root samples, and L-CK, L-12h, L-3d and L-7d represent leaf samples. The expression values are shown as the z score of the RPKM values. The scale bar is shown on the right, and high expression levels are indicated by a red color.
Additionally, the expression levels of 16 selected NtNPF6 genes, NtNPF6.3 to NtNPF6.18, were evaluated by qRT-PCR. Two distinct expression patterns were identified within 7 d of salt treatment (Supplementary Figure S3), except for NtNPF6.5, which was not detected in all samples. NtNPF6.4was the only gene that was initially up-regulated, and thereafter the expression level continuously decreased with prolonged salt treatment. The expression levels of the remaining 14 NtNPF6 genes showed a similar pattern with the duration of salt treatment, namely significantly reduced expression after salt stress for 12 h, and thereafter recovery to different extents at 3 d of salt treatment. These results were essentially consistent with the transcriptome data. It was noted that NtNPF6.13, a highly expressed gene in the root, showed the greatest decline in expression level at 12hof salt treatment and recovered the least at 3d of salt treatment. Therefore, NtNPF6.13 was selected to detect its function in response to salt stress.
Subcellular localization of NtNPF6.13
The NtNPF6.13 protein was inconsistently predicted to be localized in the vacuole, plasma membrane, cytoplasm or endoplasmic reticulum, respectively, by four online tools (Figure 6A). To clarify its subcellular localization, NtNPF6.13fused with GFP was transformed in Agrobacterium and inoculated in N. benthamiana leaves. As shown in Figure 6B, NtNPF6.13 protein co-localized with the plasma membrane marker FM4-64 (Bolte et al., 2004), which indicated that NtNPF6.13 was localized to the plasma membrane. This result is consistent with the prediction by WoLF PSORT and the predicted existence of transmembrane region by TMHMM.
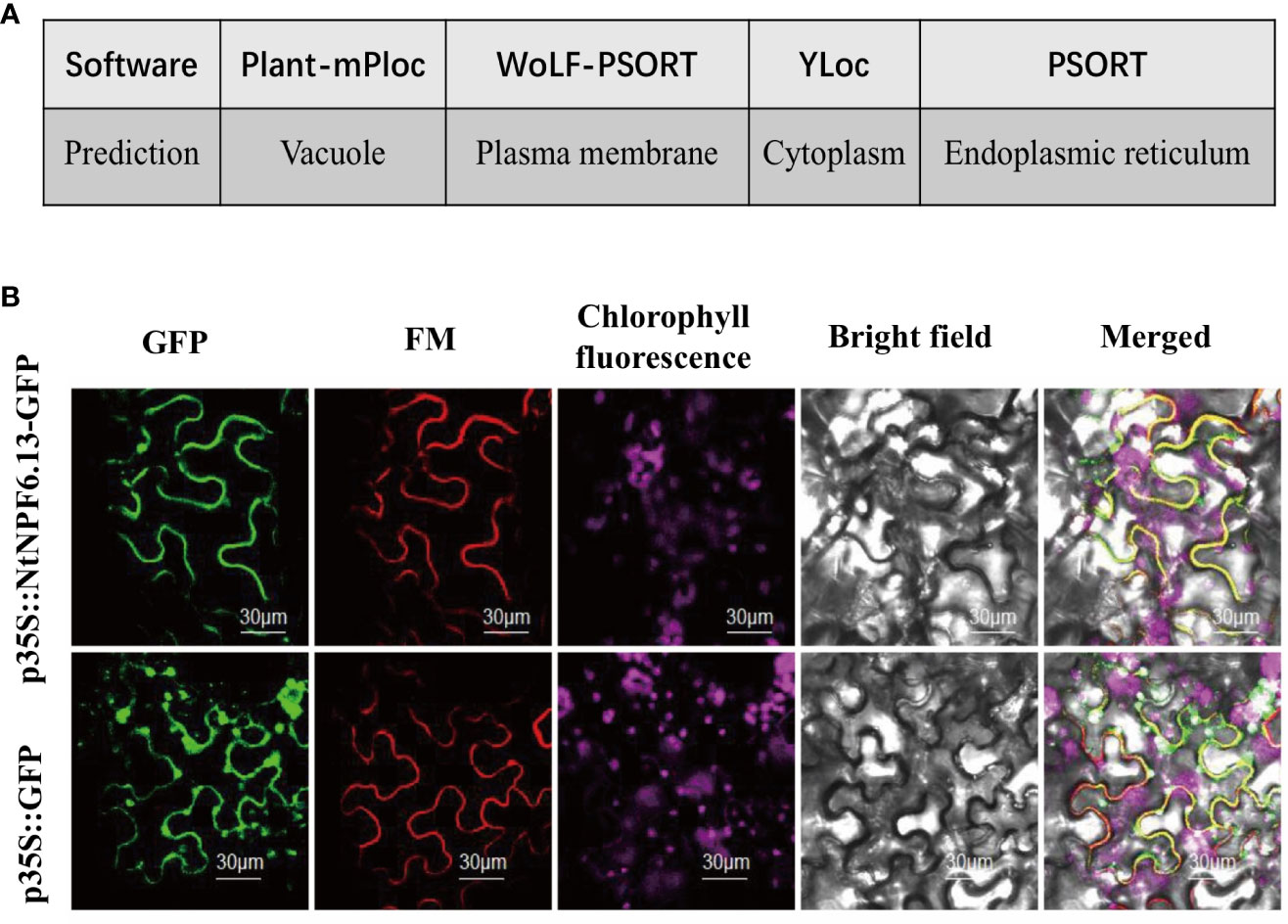
Figure 6 Subcellular localization of NtNPF6.13. (A) Localization predicted by different online tools. (B) Co-localization of NtNPF6.13-GFP with the plasma membrane marker FM in N.benthamiana leaves. Confocal images were captured the day after agroinfiltration. NtNPF6.13-GFP represents the gene and GFP represents the control with an empty vector.
Knockout of NtNPF6.13 reduces Cl- content in root
To further evaluate the role of NtNPF6.13 in chloride absorption and transportation, NtNPF6.13 knockout lines were generated using CRISPR/Cas9-mediated genome-editing technology. Three mutants, ntnpf6.13-1, ntnpf6.13-2 and ntnpf6.13-3, were generated in the K326 background (Figure 7A and Supplementary Figure S4). Sequencing revealed that, in the target region of the first exon, ntnpf6.13-1 and ntnpf6.13-2harbored a 1-bp insertion, whereas ntnpf6.13-3 had a 1-bp deletion, resulting in frame shift mutations ofNtNPF6.13 for all three mutants. By selfing and genotyping, we selected homozygous ntnpf6.13-1 mutants lacking the Cas9 transgene for further analyses.
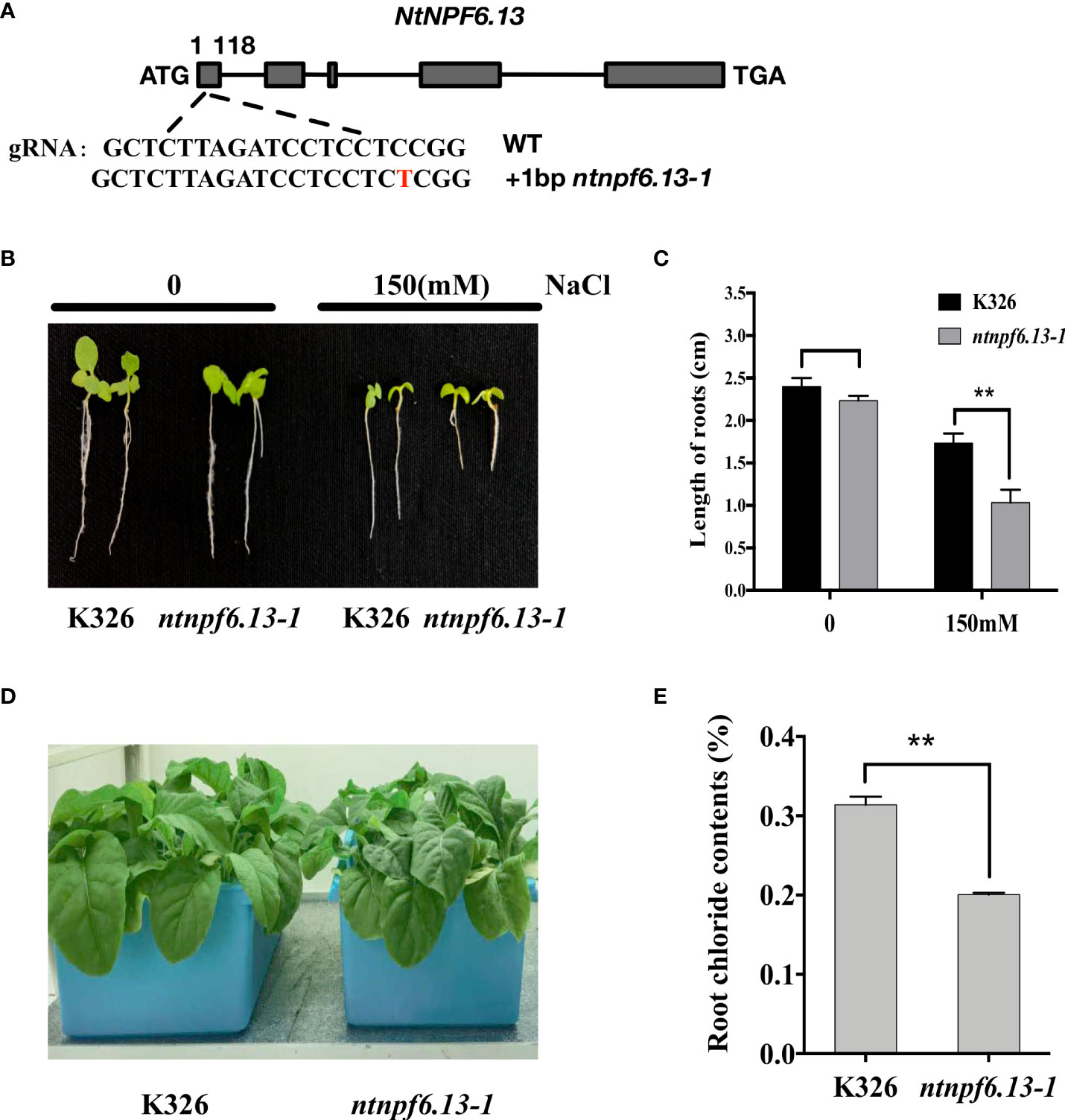
Figure 7 CRISPR/Cas9-mediated knockout of NtNPF6.13 reduces the Cl− content in roots of tobacco. (A) Diagram of gene structure and mutation sites of NtNPF6.13. (B) Phenotype of ntnpf6.13-1 mutant under salt stress compared with K326. (C) Root length of ntnpf6.13-1 mutant and K326 under salt stress. The error bars and asterisks indicate the SD and statistical significance of three biological replicates (Student’s t-test; ** P< 0.01), respectively. (D) Hydroponic culture of ntnpf6.13-1 mutant and K326. (E) Root chloride contents of K326 and ntnpf6.13-1 mutant.
The growth of ntnpf6.13-1 mutants was significantly suppressed under salt stress in comparison with K326 control plants (Figure 7B). Under the non-stress condition, almost no difference in plant growth was observed between the mutant and K326. Under salt stress (induced by 150mM NaCl treatment), the root length of the mutant was significantly shorter than that of K326 (Figure 7C). The Cl- concentration in the roots was reduced by 36.1% in ntnpf6.13-1 mutants compared with that of the K326 control plants (Figure 7D, E). Under treatment with NaCl (150 and 300 mM, respectively), the Cl- concentration in the roots was also reduced by 24% and 15.5%, respectively, in the ntnpf6.13-1 mutants (Supplementary Figure S5A). These results indicated that NtNPF6.13 might play an important role in Cl- absorption in tobacco. Compared to K326 control plants, the chloride contents were significantly (p<0.01) increased in the leaves of ntnpf6.13 mutant both before and after 150mM NaCl stress, while no significant difference was observed under 300mM NaCl stress (Supplementary Figure S5B). Even though the down-regulated chloride contents in root of ntnpf6.13 was observed in comparison with K326, the chloride content did significantly increase in root of ntnpf6.13 after NaCl stress. Therefore, it should be reasonable to conclude that NtNPF6.13 might affect the transportation of chloride from roots to shoots, but such effect would be neglectable small under higher Cl- content in root.
Discussion
In this study, 143 NtNPF genes in tobacco (K326) were identified and classified into eight groups consistent with the gene families of other species. As NPF gene families are characterized in an increasing number of plant species, the number of NPF members might be determined to be associated with ploidy. For example, hexaploidy wheat (Triticum aestivum) has the largest known NPF family (331 members) (Wang et al., 2020), and allotetraploid rapeseed (Brassica napus) has 193 NPF genes (Zhang et al., 2020). In contrast, diploid plants have dozens of NPF genes, such as Arabidopsis (53) (Krapp et al., 2014), rice (82) (Yang et al., 2020), apple (73) (Wang et al., 2018a) and spinach (57) (Wang et al., 2021).Tetraploid tobacco was determined to have 143 NtNPF genes in the present study. In addition, the phylogenetic classification of the NtNPF members was similar to other species accessed with the NPF genes classified into eight subfamilies. Compared with AtNPF5, which comprised the most members among AtNPF subfamilies, NtNPF2 and NtNPF5 were the largest subfamilies, with 25memberseach, among the eight NtNPF subfamilies. The intragenomic snyteny analysis revealed that 39 NtNPF genes formed syntenic blocks, indicating that the NtNPF gene family might have experienced whole-genome duplication and/or segmental duplications.
Almost 38% (55 genes) of the NtNPF genes were not mapped to tobacco chromosomes. The genome assembly used in this study was the latest version for N. tabacum, which was assembled by Edwards et al. (2017). Although it has been improved, anchorage of the tobacco genome to chromosomal locations was increased to 64%. It is expected that additional NtNPF genes will be mapped to chromosomes with further improvements in the tobacco genome assembly.
Considering previous NPF family analyses in different plant species, although the main focus has been nitrogen use efficiency (Wang et al., 2018a; Wang et al., 2020; Yang et al., 2020; Wang et al., 2021; Zhang et al., 2021; Zhao et al., 2021), the participation of certain NPF genes in chloride transport has been noted (Wen et al., 2017; Xiao et al., 2021).In the present study, we explored the NtNPF genes that contribute to chloride transportation in tobacco. The RNA-seq data for NtNPF genes under salt stress showed that most NtNPF6 genes were down-regulated after salt stress exposure for 12 h. Ten NtNPF6s (NtNPF6.9-NtNPF6.18) that were homologous to AtNPF6.3 responded in the roots to salt stress (Supplementary Figure S3).In AtNPF6.3-like proteins, His356 is the structural element crucial for nitrate transportation (Wen and Kaiser, 2018). Mutation of this conserved His residue to Tyr conferred the ability to transport chloride (Xiao et al., 2021). Alignment of the conserved residues showed that six of the ten NtNPF6s, including NtNPF6.13, contained a Tyr residue at the position corresponding to AtNPF6.3:H356 (Supplementary Table S5), which indicated that these NtNPF6s might function in chloride metabolism. Recently, it was reported that the AtNPF6.3 subclade in rosids could be grouped into three haplotypes (A-, B-, C-type)based on four residues corresponding to T101, H356, T360 and F511 of AtNPF6.3 (Xiao et al., 2021). In addition, the haplotypes of ten NtNPF6s of AtNPF6.3 orthologs were analyzed (Supplementary Figure S6 and Supplementary Table S5). NtNPF6.13 to NtNPF6.18 was unable to be classified to any subtype owing to the varying deficiencies among THTF residues. The other four AtNPF6.3 orthologs, NtNPF6.9 to NtNPF6.12, were all classified to the A-type (TYTF). It was also noted that NtNPF6.3 and 6.4 (AtNPF6.4 orthologs) and NtNPF6.5 to 6.8 (AtNPF6.2 orthologs) were responsive to salt stress in the leaves (Figure 5 and Supplementary Figure S7).Whether these NtNPF6s play roles in leaf chloride metabolism requires additional experimental evidence. In addition toNtNPF6 genes, other NtNPF genes were responsive to salt stress, such as NtNPF7.11, NtNPF5.11 and NtNPF4.21. Previously, AtNPF7.3 and AtNPF7.2 were reported to play roles in chloride transportation (Li et al., 2016a). It would be promising to elucidate the function of NtNPF7.11in tobacco chloride metabolism.
In this study, we showed that NtNPF6.13 may function in Cl- uptake and transportation, but the exact molecular mechanism of NtNPF6.13 remains unknown. As ZmNPF6.4 and ZmNPF6.6 could transport both chloride and nitrate in maize, it was interesting to explore whether NtNPF6.13 is involved in transportation. As shown in Supplementary Figure S8, the root of ntnpf6.13 mutant showed a significantly (p<0.01) down-regulated nitrate content only under normal condition (control), while in the leaves, the significantly decreased nitrate content of ntnpf6.13 mutant was observed under both control and 300mM NaCl conditions. The lower nitrate content in ntnpf6.13 mutant suggested that NtNPF6.13 did affect nitrate transport besides chloride in tobacco. That is, both nitrate and chloride could be the substrate of NtNPF6.13, and further studies are needed to identify their characters on affinity and efficiency of uptake and transport.
Meanwhile, Na+, brought in with Cl-, was also of interest and was determined. As shown in Supplementary Figure S9, ntnpf6.13 mutant showed a significant (p<0.05) higher Na+ concentration in root under control, and a significant (p<0.05) higher Na+ concentration in leaves under salt treatment. Obviously, the changes of Na+ and Cl- were converse at roots. Such contrast changes upon cation and anion were also observed on other transportation proteins (such as NtSLAH) related transgenic lines (unpublished data). All of these indicated the complexity of ion transport mechanism, and need to be further studied and clarified.
As a stress signal of higher amount of NaCl, ntnpf6.13 mutant responded this salt stress with changes on gene expression level, Na+ content, Cl- content, content, root length, etc. In addition, several light responsive elements such as Box 4, GT1-motif and TCT motif, were detected in the NtNPF6.13 promoter (Supplementary Table S3), which indicated NtNPF6.13 might also be regulated by light signal.
Conclusion
A total of 143 NtNPFs were identified and the phylogenetic relationships, chromosomal distribution and conserved domain structure were analyzed. RNA-seq data showed that the expression level of most NtNPF6 genes (10 in roots and 6 in leaves) was reduced after treatment with 300mM NaCl for 12h. The geneNtNPF6.13 was responsive to salt stress in the roots and was localized to the plasma membrane. After knockout by CRISPR/Cas9, ntnpf6.13 mutants showed a significantly reduced Cl- content in the roots compared with that of the control K326. In addition, the root length of the mutants was significantly shorter than that of K326 after exposure to salt stress. These results showed that NtNPF6.13 might be involved in the absorption of Cl- in tobacco. Further studies are needed to explore the function of other NtNPF6s that responded to salt stress, and their molecular mechanisms in chloride absorption and transportation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA827645.
Author contributions
HZ and HnZ conceived the experiments. ZH drafted the manuscript. ZL analyzed the RNA-seq data. GX, GB, PZ and NZ participated in the main experiments in this work, with assistance from QZ, QC and PL. LJ and HNZ contributed the revise of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the CNTC Research Program [grant no. 110202001018(JY-01)] and YNTC Research Program (grant no. 2021530000241016).
Acknowledgments
We thank Towin Biotechnology (www.towinbio.com) for providing the pCS1300 vector. We also thank Charlesworth Author Services (https://www.cwauthors.com.cn) for language editing.
Conflict of interest
Author HZ, ZL, GX, PZ, NZ, QZ, QC, PL, LJ, HnZ are employed by Zhengzhou Tobacco Research Institute of CNTC. This study received funding from CNTC and YNTC. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.999403/full#supplementary-material
Supplementary Table 1 | Sequences of NtNPF6-specificqPCR primers.
Supplementary Table 2 | Characterization of the NtNPF family in tobacco.
Supplementary Table 3 | Cis-acting elements of NtNPF genes.
Supplementary Table 4 | Expression analysis of NtNPF genes responsive to salt stress according to RNA-seq data.
Supplementary Table 5 | Haplotype of AtNPF6.3 subclade.
Supplementary Figure 1 | Numbers of different cis-acting elements in NtNPF genes.
Supplementary Figure 2 | Expression profiles of NtNPF genes under salt stress.
Supplementary Figure 3 | Expression pattern analysis of all NtNPF genes under salt stress.
Supplementary Figure 4 | Schematic diagram of gene structure and mutation sites of NtNPF6.13.
Supplementary Figure 5 | Cl− contents in root (A) and leave (B) of K326 and ntnpf6.13-1 mutant under salt stress.
Supplementary Figure 6 | AtNPF6.3 subclade in different plant species.
Supplementary Figure 7 | Phylogenetic analysis of NPF proteins.
Supplementary Figure 8 | contents in root (A) and leave (B) of K326 and ntnpf6.13-1 mutant under salt stress.
Supplementary Figure 9 | Na+ contents in root (A) and leave (B) of K326 and ntnpf6.13-1 mutant under salt stress.
References
Anfang, M., Shani, E. (2021). Transport mechanisms of plant hormones. Curr. Opin. Plant Biol. 63, 102055. doi: 10.1016/j.pbi.2021.102055
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43 (W1), W39–W49. doi: 10.1093/nar/gkv416
Barbier-Brygoo, H., De Angeli, A., Filleur, S., Frachisse, J. M., Gambale, F., Thomine, S., et al. (2011). Anion channels/transporters in plants: from molecular bases to regulatory networks. Annu. Rev. Plant Biol. 62, 25–51. doi: 10.1146/annurev-arplant-042110-103741
Bolte, S., Talbot, C., Boutte, Y., Catrice, O., Read, N. D., Satiat-Jeunemaitre, B. (2004). FM-Dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc 214 (Pt 2), 159–173. doi: 10.1111/j.0022-2720.2004.01348.x
Briesemeister, S., Rahnenfuhrer, J., Kohlbacher, O. (2010). YLoc–an interpretable web server for predicting subcellular localization. Nucleic Acids Res. 38 (Web Server issue), W497–W502. doi: 10.1093/nar/gkq477
Chiba, Y., Shimizu, T., Miyakawa, S., Kanno, Y., Koshiba, T., Kamiya, Y., et al. (2015). Identification of arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J. Plant Res. 128 (4), 679–686. doi: 10.1007/s10265-015-0710-2
Chou, K. C., Shen, H. B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PloS One 5, ), e11335. doi: 10.1371/journal.pone.0011335
Corratge-Faillie, C., Lacombe, B. (2017). Substrate (un)specificity of arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 68 (12), 3107–3113. doi: 10.1093/jxb/erw499
Crawford, N. M. (1995). Nitrate: nutrient and signal for plant growth. Plant Cell 7 (7), 859–868. doi: 10.1105/tpc.7.7.859
Drechsler, N., Courty, P. E., Brule, D., Kunze, R. (2018). Identification of arbuscular mycorrhiza-inducible nitrate transporter 1/Peptide transporter family (NPF) genes in rice. Mycorrhiza 28 (1), 93–100. doi: 10.1007/s00572-017-0802-z
Edwards, K. D., Fernandez-Pozo, N., Drake-Stowe, K., Humphry, M., Evans, A. D., Bombarely, A., et al (2017). A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics. 19 18 (1), 448. doi: 10.1186/s12864-017-3791-6
He, Y., Xi, X., Zha, Q., Lu, Y., Jiang, A. (2020). Ectopic expression of a grape nitrate transporter VvNPF6.5 improves nitrate content and nitrogen use efficiency in arabidopsis. BMC Plant Biol. 20 (1), 549. doi: 10.1186/s12870-020-02766-w
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35 (Web Server issue), W585–W587. doi: 10.1093/nar/gkm259
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37 (8), 907–915. doi: 10.1038/s41587-019-0201-4
Krapp, A., David, L. C., Chardin, C., Girin, T., Marmagne, A., Leprince, A. S., et al. (2014). Nitrate transport and signalling in arabidopsis. J. Exp. Bot. 65 (3), 789–798. doi: 10.1093/jxb/eru001
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 (9), 1639–1645. doi: 10.1101/gr.092759.109
Leran, S., Varala, K., Boyer, J. C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., et al. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19 (1), 5–9. doi: 10.1016/j.tplants.2013.08.008
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Li, B., Byrt, C., Qiu, J., Baumann, U., Hrmova, M., Evrard, A., et al. (2016a). Identification of a stelar-localized transport protein that facilitates root-to-Shoot transfer of chloride in arabidopsis. Plant Physiol. 170 (2), 1014–1029. doi: 10.1104/pp.15.01163
Li, Y., Ouyang, J., Wang, Y. Y., Hu, R., Xia, K., Duan, J., et al. (2015). Disruption of the rice nitrate transporter OsNPF2.2 hinders root-to-shoot nitrate transport and vascular development. Sci. Rep. 5, 9635. doi: 10.1038/srep09635
Li, B., Qiu, J., Jayakannan, M., Xu, B., Li, Y., Mayo, G. M., et al. (2016b). AtNPF2.5 modulates chloride (Cl(-)) efflux from roots of arabidopsis thaliana. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.02013
Nakai, K., Horton, P. (1999). PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24 (1), 34–36. doi: 10.1016/s0968-0004(98)01336-x
Payne, R. M., Xu, D., Foureau, E., Teto Carqueijeiro, M. I., Oudin, A., Bernonville, T. D., et al. (2017). An NPF transporter exports a central monoterpene indole alkaloid intermediate from the vacuole. Nat. Plants 3, 16208. doi: 10.1038/nplants.2016.208
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33 (3), 290–295. doi: 10.1038/nbt.3122
Prabhala, B. K., Rahman, M., Nour-Eldin, H. H., Jorgensen, F. S., Mirza, O. (2021). PTR2/POT/NPF transporters: what makes them tick? Adv. Protein Chem. Struct. Biol. 123, 219–240. doi: 10.1016/bs.apcsb.2020.10.002
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., Chen, W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47 (W1), W270–W275. doi: 10.1093/nar/gkz357
Tsay, Y. F., Schroeder, J. I., Feldmann, K. A., Crawford, N. M. (1993). The herbicide sensitivity gene CHL1 of arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72 (5), 705–713. doi: 10.1016/0092-8674(93)90399-b
Wang, X., Cai, X., Xu, C., Wang, Q. (2021). Identification and characterization of the NPF, NRT2 and NRT3 in spinach. Plant Physiol. Biochem. 158, 297–307. doi: 10.1016/j.plaphy.2020.11.017
Wang, Y. Y., Cheng, Y. H., Chen, K. E., Tsay, Y. F. (2018b). Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69, 85–122. doi: 10.1146/annurev-arplant-042817-040056
Wang, Q., Liu, C., Dong, Q., Huang, D., Li, C., Li, P., et al. (2018a). Genome-wide identification and analysis of apple NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) genes reveals MdNPF6.5 confers high capacity for nitrogen uptake under low-nitrogen conditions. Int. J. Mol. Sci. 19 (9), 2761. doi: 10.3390/ijms19092761
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi: 10.1093/nar/gkr1293
Wang, H., Wan, Y., Buchner, P., King, R., Ma, H., Hawkesford, M. J. (2020). Phylogeny and gene expression of the complete NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER FAMILY in triticum aestivum. J. Exp. Bot. 71 (15), 4531–4546. doi: 10.1093/jxb/eraa210
Wege, S., Gilliham, M., Henderson, S. W. (2017). Chloride: not simply a 'cheap osmoticum', but a beneficial plant macronutrient. J. Exp. Bot. 68 (12), 3057–3069. doi: 10.1093/jxb/erx050
Wen, Z., Kaiser, B. N. (2018). Unraveling the functional role of NPF6 transporters. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00973
Wen, J., Li, P. F., Ran, F., Guo, P. C., Zhu, J. T., Yang, J., et al. (2020). Genome-wide characterization, expression analyses, and functional prediction of the NPF family in brassica napus. BMC Genomics 21 (1), 871. doi: 10.1186/s12864-020-07274-7
Wen, Z., Tyerman, S. D., Dechorgnat, J., Ovchinnikova, E., Dhugga, K. S., Kaiser, B. N. (2017). Maize NPF6 proteins are homologs of arabidopsis CHL1 that are selective for both nitrate and chloride. Plant Cell 29 (10), 2581–2596. doi: 10.1105/tpc.16.00724
Wulff, N., Ernst, H. A., Jorgensen, M. E., Lambertz, S., Maierhofer, T., Belew, Z. M., et al. (2019). An optimized screen reduces the number of GA transporters and provides insights into nitrate transporter 1/Peptide transporter family substrate determinants. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01106
Xiao, Q., Chen, Y., Liu, C. W., Robson, F., Roy, S., Cheng, X., et al. (2021). MtNPF6.5 mediates chloride uptake and nitrate preference in medicago roots. EMBO J. 40 (21), e106847. doi: 10.15252/embj.2020106847
Xu, G., Fan, X., Miller, A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182. doi: 10.1146/annurev-arplant-042811-105532
Yang, X., Xia, X., Zeng, Y., Nong, B., Zhang, Z., Wu, Y., et al. (2020). Genome-wide identification of the peptide transporter family in rice and analysis of the PTR expression modulation in two near-isogenic lines with different nitrogen use efficiency. BMC Plant Biol. 20 (1), 193. doi: 10.1186/s12870-020-02419-y
Zhang, J., Han, Z., Lu, Y., Zhao, Y., Wang, Y., Zhang, J., et al. (2021). Genome-wide identification, structural and gene expression analysis of the nitrate transporters (NRTs) family in potato (Solanum tuberosum l.). PloS One 16 (10), e0257383. doi: 10.1371/journal.pone.0257383
Zhang, H., Jin, J., Jin, L., Li, Z., Xu, G., Wang, R., et al. (2018). Identification and analysis of the chloride channel gene family members in tobacco (Nicotiana tabacum). Gene 676, 56–64. doi: 10.1016/j.gene.2018.06.073
Zhang, H., Li, S., Shi, M., Wang, S., Shi, L., Xu, F., et al. (2020). Genome-wide systematic characterization of the NPF family genes and their transcriptional responses to multiple nutrient stresses in allotetraploid rapeseed. Int. J. Mol. Sci. 21 (17), 5947. doi: 10.3390/ijms21175947
Keywords: Nicotiana tabacum, nitrate transporter 1/peptide transporter family, NPF, salt stress, gene expression, chloride
Citation: Zhang H, Li Z, Xu G, Bai G, Zhang P, Zhai N, Zheng Q, Chen Q, Liu P, Jin L and Zhou H (2022) Genome-wide identification and characterization of NPF family reveals NtNPF6.13 involving in salt stress in Nicotiana tabacum. Front. Plant Sci. 13:999403. doi: 10.3389/fpls.2022.999403
Received: 21 July 2022; Accepted: 27 September 2022;
Published: 13 October 2022.
Edited by:
Awadhesh Kumar, National Rice Research Institute (ICAR), IndiaReviewed by:
Hongfang Jia, Henan Agricultural University, ChinaJawahar Singh, National Institute of Plant Genome Research (NIPGR), India
Copyright © 2022 Zhang, Li, Xu, Bai, Zhang, Zhai, Zheng, Chen, Liu, Jin and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huina Zhou, am9hbnpobkAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
 Hui Zhang
Hui Zhang Zefeng Li2†
Zefeng Li2† Guoyun Xu
Guoyun Xu Ge Bai
Ge Bai Qiansi Chen
Qiansi Chen Pingping Liu
Pingping Liu Lifeng Jin
Lifeng Jin Huina Zhou
Huina Zhou