
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 15 December 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.984702
This article is part of the Research TopicEpigenetic Modifications Associated with Abiotic and Biotic Stresses in Plants: An Implication for Understanding Plant Evolution, Volume IIView all 5 articles
Plants have developed multiple mechanisms as an adaptive response to abiotic stresses, such as salinity, drought, heat, cold, and oxidative stress. Understanding these regulatory networks is critical for coping with the negative impact of abiotic stress on crop productivity worldwide and, eventually, for the rational design of strategies to improve plant performance. Plant alterations upon stress are driven by changes in transcriptional regulation, which rely on locus-specific changes in chromatin accessibility. This process encompasses post-translational modifications of histone proteins that alter the DNA-histones binding, the exchange of canonical histones by variants that modify chromatin conformation, and DNA methylation, which has an implication in the silencing and activation of hypervariable genes. Here, we review the current understanding of the role of the major epigenetic modifications during the abiotic stress response and discuss the intricate relationship among them.
Chromatin is a highly organized eukaryotic complex of DNA and proteins, where DNA is packaged into regularly spaced nucleosomes, assembled as beads on a string. Each nucleosome is formed by ∼147 bp of DNA wrapped around a core histone octamer (Olins and Olins, 1974; Thomas and Kornberg, 1975; Libertini et al., 1988; Luger et al., 1997; Wolffe and Hayes, 1999). Throughout evolution, histone proteins have gradually evolved from archaeal ancestors into the four distinct subunits that compose the common octamer of the nucleosome. The core histones H2A, H2B, H3, and H4 are structured in two H2A-H2B dimers and an H3-H4 tetramer. The linker histone H1 helps to condense the chromatin by binding to the DNA between nucleosomes (Campos and Reinberg, 2009; Henikoff and Smith, 2015; Talbert and Henikoff, 2017).
The chromatin landscape is in constant reorganization to guarantee the transcriptomic reprogramming required during developmental processes (Baulcombe and Dean, 2014; Kawashima and Berger, 2014; Xiao and Wagner, 2015; Lee and Seo, 2018; Gehring, 2019), such as germline differentiation (Borg et al., 2009; Feng et al., 2010; Baroux et al., 2011; Borg et al., 2021b) or leaf senescence (Brusslan et al., 2015). Alterations in chromatin structure have been associated to different states of DNA accessibility (Sequeira-Mendes et al., 2014). Functionally, chromatin is divided into two conformational states: heterochromatin, in which DNA is strongly condensed, and euchromatin, where the DNA is more accessible and less compacted. The molecular mechanisms regulating the switch between euchromatin and heterochromatin include complex epigenetic regulatory networks (Adam et al., 2001; Nakayama et al., 2001; Ahmad and Henikoff, 2002; Francis et al., 2004; Castellano-Pozo et al., 2013; Sequeira-Mendes et al., 2014; Yelagandula et al., 2014; Morrison and Thakur, 2021). We have included several excellent and recent reviews that discuss and detail the function of the major drivers of chromatin restructuration: histone variants (Loppin and Berger, 2020; Probst et al., 2020; Foroozani et al., 2022), histone post translational modifications (Antunez-Sanchez et al., 2020; Jiang et al., 2020b), and DNA methylation (Zhang et al., 2018; Mattei et al., 2022).
Abiotic factors such as salinity, limited water availability, extreme temperature, low-light and chemical composition of the soil severely impact plant growth and developmental programs. Thus, variations in any of these conditions lead to an alteration in the homeostasis, known as abiotic stress (Singh and Laxmi, 2015). Each form of abiotic stress contains a unique signaling pathway. Nevertheless, there are conserved cellular responses orchestrated by a complex regulatory network involving (1) upstream signaling molecules, such as ROS, NO, Ca2+ or ABA, and (2) downstream regulation, in which transcription factors and epigenetic regulators intervene (He et al., 2018). Here, we will focus on the downstream regulation and summarize the mechanisms of these epigenetic agents, which redefine the plant chromatin landscape when exposed to external stimuli.
There are different scales —global and local— at which these modifications happen in the context of abiotic stressors (Figure 1). Global changes in response to abiotic stress include an increase in histone acetylation (Pandey et al., 2002; Earley et al., 2007), a loss in the chromocenter organization —typical of plant heterochromatin— (Pecinka et al., 2010; reviewed in Probst and Mittelsten Scheid, 2015) and a reduction in nucleosome occupancy (Brzezinka et al., 2016; Park et al., 2018; reviewed in Bäurle and Trindade, 2020). These modifications occur globally in the sense that they are not directed to a particular genomic region but are genome-wide instead. On the contrary, there are local changes particular of stress-responsive areas of the genome characterized by an increase in methylation of the residues K4/K36 of H3 histone tails (Lee et al., 2016) and changes in nucleosome composition (Rutowicz et al., 2015). Additionally, there are gene-specific changes unique of each type of stress [e. g. P5CS2 is upregulated upon salt stress and dehydration, whereas HSP17.4 responds to heat (Port et al., 2004; Székely et al., 2008)]. However, their local epigenetic regulation shares identical features: an increase in histone modifications associated with an increase in DNA accessibility and a reduction in marks associated with less accessibility. During this review, we have decided not to focus on the specific changes that occur in presence of each abiotic factor, but instead on the general mechanisms involved in chromatin reorganization during the stress response —commonly shared between the different abiotic agents—. Deciphering how the expression of stress-responsive genes occurs is fundamental in unravelling the hidden details of the abiotic stress response.
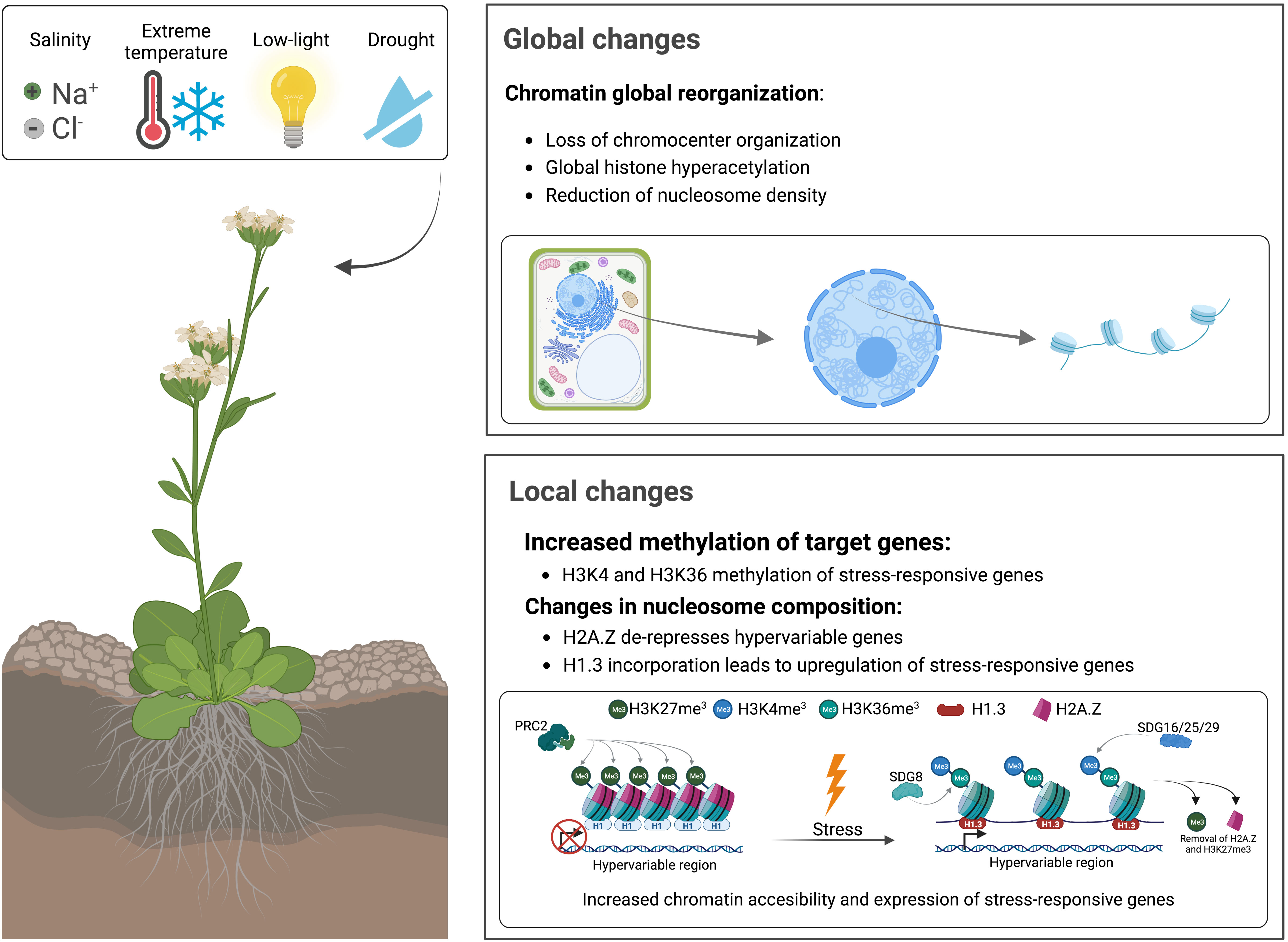
Figure 1 Global and local changes in chromatin structure in response to abiotic factors, such as salt, limited water availability, cold, heat, and low-light. Global changes are distributed genome-wide, whereas local changes are directed to specific genomic regions, commonly associated with stress-responsive genes.
How histone post translational modifications impact on the transcriptional changes required for plant survival during the stress response has been previously discussed (Kim et al., 2015; Ueda and Seki, 2020; Bhadouriya et al., 2021). However, the implication of histone variants during the abiotic stress response has not been discussed in depth. Moreover, most of the reviews about epigenetic regulation include either histone variants or histone modifications, but it is rare to see a combination of both. Hence, this review aims to furnish deeper insights into the transient coordination between histone variants and histone modifications in response to abiotic stress in plants.
Arabidopsis thaliana serves as an excellent model organism in plant research due to (1) its small, fully sequenced and well-annotated genome, (2) its short life cycle, (3) its tolerance to mutations in chromatin key genes, generally lethal in other organisms and (4) its post-embryonic organogenesis process characteristic of plants (Perianez-Rodriguez et al., 2014). These circumstances create an ideal scenario to study epigenetic changes during growth and morphogenesis in response to developmental and environmental cues. For that purpose, most of the epigenetic research in plants uses this organism as a model. In this review, we will focus on the major epigenetic modifications in the plant Arabidopsis thaliana as an approach to plant epigenetics.
The paralogous genes of a histone family encode identical isoforms, but also non-allelic protein isoforms commonly referred to as histone variants. These variants differ in their amino acid sequence from the canonical form and play critical roles in diverse processes such as transcription, chromatin remodelling, and DNA packaging, conferring unique characteristics of chromatin (Talbert et al., 2012; reviewed in Talbert and Henikoff, 2017; Probst, 2022).
Canonical histones, also known as replicative histones, are predominantly expressed during the S-phase and deposited in a DNA synthesis-dependent manner. Conversely, histone variants, or replacement histones, are expressed throughout the cell cycle, are incorporated in a DNA synthesis-independent manner and have sequence divergence and specific genomic localization (Chaubet et al., 1992; reviewed in Henikoff and Smith, 2015).
Histone variants have been described in all model organisms studied, from the unicellular yeast Saccharomyces cerevisiae and algae to plants and animals, and for all histones but histone H4, with only a few exceptions (Long et al., 2019; reviewed in Probst et al., 2020). Some histone variants, like H3.3 and H2A.Z, are conserved in eukaryotes, while others are lineage-specific, such as the flowering plant-specific H2A.W variant (Yelagandula et al., 2014; Giaimo et al., 2019; Bourguet et al., 2021; Lei et al., 2021). There are tissue-specific variants, such as the Arabidopsis H3.10 and H2B.8, that function in sperm cells (Jiang et al., 2020a; Borg et al., 2021a; Buttress et al., 2022).
Given the important role that histone variants have in chromatin regulation, their deposition needs to be temporally orchestrated. Histone chaperones promote nucleosome assembly and disassembly during replication, transcription and repair (Daniel Ricketts et al., 2015; Hammond et al., 2017).
The diversity of nucleosome composition provided by canonical histones and variants is associated with different chromatin states. Depending on the histone variant incorporated into the nucleosome, chromatin adopts a more open —accessible to transcriptional machinery— or closed chromatin conformation. Thus, H3.3, H2A.Z, and H2A.X variants are abundant in euchromatic regions, along with histone marks in active chromatin, e.g., H3K4me3, H3K36me3 and H2B ubiquitylation, and coincide with high RNA Pol II occupancy (Stroud et al., 2012; Wollmann et al., 2012). These features form a chromatin state typical of active transcription (Sequeira-Mendes et al., 2014; Borg et al., 2021a). On the contrary, H2A.W and H1 histones colocalize with heterochromatin marks —like H3K9me2, H3K27me3, H3K27me1— and DNA methylation in silent genomic regions, favoring the compaction of the chromatin (Grewal and Jia, 2007; Vaillant and Paszkowski, 2007; Roudier et al., 2009; Stroud et al., 2012; Zemach et al., 2013; Rutowicz et al., 2019; Choi et al., 2020). The histone variants that play a role during the stress response are incorporated into nucleosomes in specific regions of the genome —stress-responsive genes— that are crucial for the upstream signaling stress response (Coleman-Derr and Zilberman, 2012; Rutowicz et al., 2015).
An intriguing feature of histone variants is the organization within one family. In histone families where there are several histone variant proteins with similar functions, there is usually a prevalence in the abundance of one or two among the others (reviewed in Martire and Banaszynski, 2020). This suggests either specific pathways ensure deposition of these variants or that the slight differences between the proteins lead to a favored deposition of some of them against the rest, developing into a specific role of certain variants. In Arabidopsis, the histones —canonical or variants— are organized into the four families (Table 1) discussed below.
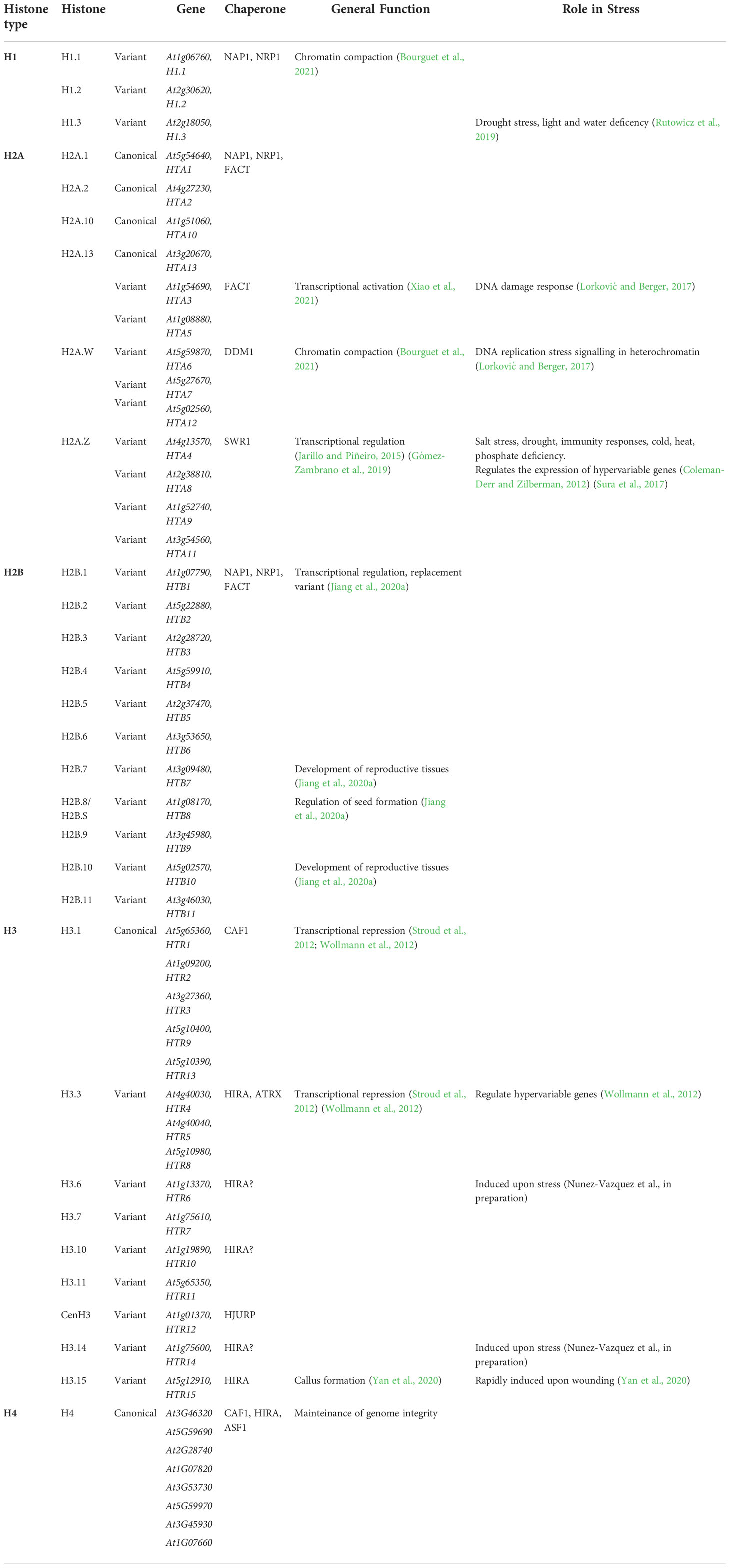
Table 1 Classification of histone families in Arabidopsis: genes, variants, proteins, chaperones, general function and role in stress.
H1 histones are known as “linker histones” because they bind to the linker DNA between nucleosomes, further facilitating chromatin compaction. These histones consist of a globular domain, which binds the DNA at the dyad axis of the nucleosome to the core histones, a short N-terminal chain, and a C-terminal tail that binds to the DNA between nucleosomes (Zhou et al., 2013; Zhou et al., 2015). The tight bound between the nucleosome and the linker DNA results in higher nucleosome density. Histone H1, together with H2A.W, coordinates heterochromatin accessibility and DNA methylation (Bourguet et al., 2021). In vertebrates, several evolutionarily conserved subfamilies of H1 can be distinguished, and they play redundant and specific roles during development and cellular differentiation (Mcbryant et al., 2010; Talbert and Henikoff, 2017). In humans and mice, 11 different H1 variants have been identified (Fyodorov et al., 2018), while the Arabidopsis H1 family is formed by the H1.1, H1.2, and H1.3 histones (Table 1).
H1.1 and H1.2 —the replicative histones H1— are highly similar, whereas the H1.3 variant is shorter and lacks the (S/T)PXK motifs required for DNA binding (Kotliński et al., 2016). Consequently, the H1.3 variant has higher mobility within chromatin. H1.1 and H1.2 are enriched in heterochromatin, anti-correlate with gene expression (Rutowicz et al., 2015), and are also necessary for H3K27me3 deposition (Rutowicz et al., 2019). Alternatively, H1.3, although it is not abundant in the histone H1 pool, plays a specific role in the abiotic stress response. Under normal conditions, it is exclusively expressed in guard cells, but when the plant is exposed to a stimulus, such as light deficency, drought, and abscisic acid (ABA), H1.3 competes with H1.1 and H1.2 for the incorporation into the nucleosome (Rutowicz et al., 2015). Physiological and transcriptomic analyses of h1.3 null mutants demonstrate that H1.3 is required for proper stomatal functioning under normal growth conditions and adaptive developmental responses to combined light and water deficiency (Rutowicz et al., 2015). The putative differences in the deposition patterns of H1.3 in different tissues in response to stress have not been explored.
The H2A histone family in Arabidopsis comprises four replicative H2A, four H2A.Z, three H2A.X, and two H2A.W (Table 1), composed of ~130 amino acid residues. H2A variants differ in the C-terminal motifs of their primary amino acid sequences (Kawashima et al., 2015). Some of these variants’ properties are conserved throughout the kingdoms. For instance, H2A.Z diverged from the canonical H2A early in eukaryotic evolution. H2A.Z properties have been thoroughly described in humans, mice, yeast, and plants. In all these kingdoms, H2A.Z histone is a replacement variant with similar roles in transcriptional regulation and DNA repair (Jarillo and Piñeiro, 2015; Giaimo et al., 2019; Gómez-Zambrano et al., 2019). In fact, H2A.Z sequences from different organisms show a higher similarity level than the H2A.Z and H2A within the same organism. The diverse relationship between H2A variants and gene expression explains histone variants’ impact on chromatin structure. H2A.X is distributed along the whole Arabidopsis genome, whilst H2A.W is enriched in pericentromeric regions, colocalizing with heterochromatin and transposable elements (TEs) (Lei and Berger, 2020; Bourguet et al., 2021). On the other hand, replicative H2A and H2A.Z are excluded from pericentromeric heterochromatin (Zilberman et al., 2008; Yelagandula et al., 2014). The exclusion of H2A.Z from pericentromeric heterochromatin has been linked to its shortened C-terminal tail, which is thought to limit the binding of the linker histone H1 to the core nucleosome particle (Osakabe et al., 2018).
Histone variants mediate the nucleosome adaptability to different stimuli. Changes in nucleosome composition directly reports on nucleosome stability (Osakabe et al., 2018). For instance, H2A.Z-H2B dimers are replaced more rapidly than H2A-H2B dimers (Brahma et al., 2017), conferring the genes covered by H2A.Z-H2B nucleosomes the ability to respond quickly to a stimulus. An intriguing plant H2A feature is that, in contrast to animals and yeast, H2A-containing nucleosomes are homotypic, since each variant associates only with itself (Osakabe et al., 2018).
The distribution of H2A.Z in the Arabidopsis genome is puzzling because of its dual, and perhaps interconvertible, deposition patterns. H2A.Z can be deposited either at the transcription start site (TSS) of a large set of constitutively expressed genes across cell types or at the gene-body of repressed genes (Coleman-Derr and Zilberman, 2012) associated with repressive H3K27me3. When incorporated at the TSS, it is thought to maintain genome integrity with stable transcription rates by facilitating the transcription of genes essential for plant survival (Mahrez et al., 2016). This process is thought to occur by reducing the energy required by the RNA polymerase II to overcome the first nucleosomal barrier (Sura et al., 2017). Over a decade ago, the involvement of the H2A.Z histone variant in gene responsiveness during environmental stress was elucidated by showing that H2A.Z is deposited within gene bodies in genes categorized as “hypervariable” (Coleman-Derr and Zilberman, 2012). Furthermore, transcriptome data of h2a.z knock-out mutant plants revealed a deregulation of Arabidopsis genes with high responsiveness scores, which correlates with those that have H2A.Z deposited on their gene body in the absence of stress. Notably, under normal conditions, gene-body H2A.Z deposition participates in the repression of genes involved in response to wounding, drought, ABA, salinity, UV light, heat, cold, immune response, defense response, and phosphate in Arabidopsis (Coleman-Derr and Zilberman, 2012). Since then, several authors have reported the implication of H2A.Z not only as a transcriptional regulator but also as a key player in gene repression under biotic and abiotic stress conditions (Cortijo et al., 2017; Sura et al., 2017; Nguyen and Cheong, 2018; Gómez-Zambrano et al., 2019; Bieluszewski et al., 2022).
The role of H2A.Z in stress resembles the function of the histone mark H3K27me3, as they both actively regulate the expression of hypervariable genes. Due to the similarities in the regulation of their targets, it was hypothesized that H2A.Z and H3K27me3 could functionally interact. In mouse embryonic stem cells, H2A.Z promotes chromatin compaction, favoring H3K27me3 deposition by the POLYCOMB REPRESSIVE COMPLEX 2 (PRC2) (Wang et al., 2018). Consistent with this, H3K27me3 is dependent on H2A.Z deposition in Arabidopsis (Dai et al., 2017; Carter et al., 2018). SWI2/SNF2-Related 1 Chromatin Remodeling Complex (SWR1), the complex incorporating the H2A.Z variant, is required for H3K27 trimethylation (Luo et al., 2020; Liu et al., 2021). However, the variant H2A.Z and the Polycomb modification H3K27me3 do not share most of their targets, as shown by the limited overlap of upregulated genes between hta9-hta11, defective in H2A.Z protein, and mutants of the Polycomb repressive complex 2 (PRC2) catalytic subunit curly leaf (clf) (Gómez-Zambrano et al., 2019). These differences suggest that the repression of targets via H2A.Z gene body-deposition targets a wide range of hypervariable genes and is not limited to stress-responsive genes. These data suggest an exciting timeframe in the repression of responsive genes, where deposition of H2A.Z by SWR1 is first needed to achieve PRC2 repression of hypervariable genes. H3K27me3 usually works in bivalent genes and is released from the environment of the gene it represses shortly after the stress stimulus (Zhang et al., 2011; Molitor et al., 2014; Zhang et al., 2020). ChIP-seq data of H2A.Z after stress are not available so far, and, consequently, it is not possible to conclude whether the H2AZ is evicted from the gene body or if it is deposited in a different region of the same locus —although it has been proposed that there is H2A.Z depletion from the gene body upon transcriptional activation (Sura et al., 2017). Establishing a timeline to clarify further the role of these critical actors in activating these repressed genes during the stress response remains unclear and needs further investigation.
The H2A.X variant, which only differs from the replicative H2A in the additional SQEF amino acid motif that H2A.X contains in its C-terminal tail, has been described to regulate the DNA damage response (DDR) (Dantuma and van Attikum, 2016; Lorković and Berger, 2017). In replicative stress, the ATR and ATM kinases phosphorylate H2A.X by a mechanism that is conserved in both animals and plants. The H2A.W.7 variant is necessary for DNA replication stress signaling in heterochromatin, which shows there might be an interaction between these H2A.X and H2A.W by the joint action of kinases to act in response to DNA damage in different regions of the Arabidopsis genome (Yelagandula et al., 2014; Lorković and Berger, 2017).
Compared with the extensive published studies defining H2A variants, only a handful of H2B variants have been characterized. Despite the similarities between H2A and H2B histones and the conserved status of their dimers, the Arabidopsis histone H2B family is formed by 11 genes that encode proteins of high sequence divergence (Jiang et al., 2020a). The expression of Arabidopsis H2B varies across development. Defining the role of H2B.8 —also known as H2B.S—is particular intriguing since this histone specifically accumulates during chromatin compaction of dry seed embryos (Jiang et al., 2020a; Buttress et al., 2022). The potential response of H2B proteins to abiotic stress has not been explored so far.
The Arabidopsis histone H3 family is one of the most studied and complex. It is composed of fifteen genes encoding nine H3 proteins with unique roles. The canonical form, the protein H3.1, is encoded by five intronless genes: HTR1, HTR2, HTR3, HTR9, and HTR13. This protein is only deposited during DNA replication and DNA repair. The histone H3.3, the best-characterized histone H3 variant, is encoded by the HTR4, HTR5, and HTR8 genes and is incorporated throughout the whole cell cycle constitutively, in a DNA replication-independent manner, allowing a rapid chromatin adaptation to different environmental stimuli (March-Díaz and Reyes, 2009; reviewed in Talbert and Henikoff, 2017).
The differences between the variants in the H1 and H2A histone families are driven by the distinct amino acid motifs, even domains, that they include in their sequence. Instead, the H3 family maintains a high amino acid homology degree. The differences in the H3 variants consist of changes of a relatively small number of aminoacids (Figure 2). H3.1 and H3.3 have unique properties, despite that their amino acid sequences differ only in 4 amino acid residues at positions 31, 41, 87, 90 (Figure 2). The substitution at position 41 of H3.1 is specific to dicotyledon plants (Lu et al., 2018). The differences in amino acid sequences between H3.1 and H3.3 are almost identical in plants and animals. Their distribution patterns are also highly similar across species (Ingouff and Berger, 2010; Stroud et al., 2012; Wollmann et al., 2012; Müller and Almouzni, 2014; reviewed in Loppin and Berger, 2020; Foroozani et al., 2022). This evidence of convergent evolution strongly points toward the importance of those specific residues in the function of the eukaryotic genome. Regarding histone H3 distribution along the Arabidopsis genome, ChIP-seq studies showed that H3.1 is enriched in heterochromatin, specifically in TEs and pericentromeric heterochromatin, colocalizing with histone modifications associated with gene repression such as H3K9me2, H3K27me1, H3K27me3 or DNA methylation, H2B ubiquitination, and RNA polymerase II occupancy (Stroud et al., 2012; Wollmann et al., 2012). In contrast, H3.3 is associated with active chromatin marks, including H3K4me3, H3K9me3, and H3K36me3. Therefore, H3.3 is associated with euchromatic regions, being deposited preferentially at the 3’ UTR end of constitutively expressed genes (Shi et al., 2011; Stroud et al., 2012; Wollmann et al., 2012; Shu et al., 2014).

Figure 2 Differences in the amino acid sequence of the members of the histone H3 family in Arabidopsis thaliana. The length of each variant’s amino acid sequence is indicated in black.
Histone variants often play a role in the activation of certain groups of inducible genes. For example, H3.3 specifically regulates the expression of genes involved in environmental responses (Wollmann et al., 2017). Also, a recent study showed that H3.3 inhibits flowering by increasing the levels of H3K4me3 and H3K36me3 marks at the FLOWERING LOCUS C (FLC) gene (Zhao et al., 2021), although the specific mechanisms underlying the relationship between histone H3.3 and stress responses have not yet been clarified.
Genome architecture can be structurally shaped with the help of histone variants. A H3 variant known as CENH3 in plants —and CENP-A in mammals— is specifically incorporated in the centromere region (Malik and Henikoff, 2009; Fukagawa and Earnshaw, 2014; Müller and Almouzni, 2014). CENH3 is an essential protein that function in centromere organization and chromosome segregation (Ravi et al., 2010). The CENH3 amino acid sequence strongly diverges from that of the rest of H3 family members. A clear role of CENH3 in stress response has not been described. However, its expression was drastically reduced in the mutant background of MUT9-LIKE KINASE1 and 2 (MLK1 and 2). These kinases are in charge of H3.3 phosphorylation in a process that is dependent of the ABA pathway (Wang et al., 2015).
There is a group of atypical plant-specific H3 variants with specific substitutions in their N-terminal tail, encoded by the genes HTR6 and HTR14 that share features with both H3.1 and H3.3, although they are thought to be more similar to H3.3, as they contain the four critical amino acids (T31, Y41, H87, L90) in which H3.3 differ from H3.1 (Figure 2). Furthermore, H3.14 and H3.6 have been described to contain an enrichment of transcription factor binding sites implicated in salinity and drought stress responses in their respective promoter regions (Nunez-Vazquez et al., in preparation). Further differences are present in these atypical H3 variants, but the functional impact of these changes has yet to be explored. The atypical H3.15 has a distinguishing feature due to its lack of the K27 residue, which prevents the trimethylation of this residue by the Polycomb PRC2 complex and has been reported to be induced after wounding and has a role in cell fate reprogramming during plant regeneration (Yan et al., 2020). The sperm-specific H3.10 variant has an intricate role in heterochromatin formation and gene silencing, as it reprograms H3K27me3 during Arabidopsis spermatogenesis (Okada et al., 2005).
Several histone chaperones have been described to incorporate H3-H4 dimers in the nucleosome. CAF1 is the typical H3.1 chaperone, whereas HIRA commonly incorporates H3.3 by binding to its C-terminal tail’s H87 and L90 amino acids (Daniel Ricketts et al., 2015) (Figure 2). As many atypical histone variants (H3.6, H3.14, H3.10) contain the H87 and L90 residues, we hypothesize HIRA is likely to be responsible for their deposition, although further research is needed to demonstrate this assumption.
Chromatin stability is favored by the interaction of the negatively charged phosphate groups of DNA with the positively charged amino acids of histone proteins. The post-translational modifications (PTMs) of both histone tails and histone fold domains contribute to chromatin control and accessibility. The histone PTMs environment is founded and maintained by a set of highly coordinated enzymes (Kouzarides, 2007). PTMs are considered to favor the oscillation between relaxed or packaged chromatin configurations. However, whether histone PTMs are a cause or a consequence of changes in transcriptional regulation is a controversial and puzzling topic (Millán-Zambrano et al., 2022; Policarpi et al., 2022; Wang et al., 2022). On one hand, some evidence indicates that active histone modifications support transcription in an informative manner rather than serving as an essential regulatory function (Wang et al., 2022). On the other hand, a different study points towards de novo H3K4me3 deposition can induce major transcription activation (Policarpi et al., 2022). Here, we will discuss recent discoveries and summarize the current understanding of the regulation and function of histone post translational modifications in response to abiotic stress.
Histone PTMs include methylation, acetylation, phosphorylation, ubiquitination, and sumoylation, among others. These reactions are catalyzed by histone-modifying enzymes recruited to specific genomic regions (Kouzarides, 2007). The chromatin landscape of active genes is preferentially associated with highly acetylated histones, whereas inactive genes are associated with hypoacetylated histones (Hebbes et al., 1988). The general assumption is that acetylation of lysine and arginine residues reduces the DNA-histone interactions and relaxes the chromatin structure, resulting in increasing accessibility to the DNA of the transcriptional machinery (Allis and Jenuwein, 2016). The association between histones and DNA is also regulated by histone methylation. Due to the neutral character of this modification, methylation of amino acids does not directly perturb nucleosome stability (Xiao et al., 2016), although it affects the local hydrophobicity. Hence, it appears in association with actively transcribed or repressed genes, depending on the methylated amino acid residue (Xiao et al., 2016; Yung et al., 2021). In contrast, the phosphorylation of threonine, serine, and tyrosine adds an extra negative charge to the chain, weakening the DNA-histone interaction. Ubiquitination of lysines, consisting of the addition of small amino acid chains to the histone tail, also compromises nucleosome stability. H2Aub has been associated with gene silencing, whereas H2Bub is linked to transcriptional activation. The specific mechanism of transcription regulation by ubiquitination has not yet been clarified (Zhou et al., 2017; Zhou et al., 2018).
Although nucleosomes are present in all eukaryotic cells, the role of specific PTMs varies between animals and plants. For example, H3K9me3, a constitutive heterochromatin mark in mammals, is present in plant euchromatin, whereas the dimethylated state, H3K9me2, is associated with plant heterochromatin (Lippman et al., 2004; Zhang et al., 2008). The monomethylation of H3K27 is a plant-specific heterochromatin mark, although it also appears at lower level in repressed genes of euchromatin (Jacob et al., 2009). H3K27me3 regulates facultative heterochromatin —specific regions of the genome that behave as heterochromatin in some cells or developmental stages but as euchromatin in others— both in plants and animals (Schuettengruber et al., 2007; Zheng and Chen, 2011; Makarevitch et al., 2013), as it is the case for H3K4me3 in actively transcribed genes (Zhang et al., 2009).
Several studies have reported that PTMs are involved in seed formation, flowering, and biotic and abiotic stress responses (Cao et al., 2008; Zou and Mallampalli, 2014; Huang et al., 2016; Zhou and Zeng, 2017). In the presence of stress, the plant needs to reorganize and optimize its resources. (Atkinson and Urwin, 2012). For that purpose, it pauses different ongoing processes, such as protein translation and cell elongation, and prioritizes those that are strictly necessary for plant survival (Muñoz and Castellano, 2012; Yamamoto, 2019). Modifying the local chromatin landscape during the stress response does not comprise a specific PTM. Instead, it involves globally induced changes that could be summed up as (1) an increase in histone acetylation in the promoters and gene bodies of drought-inducible genes and (2) derepression of hyperresponsive targets by histone and DNA demethylation (To and Kim, 2014) (Figures 1, 3).
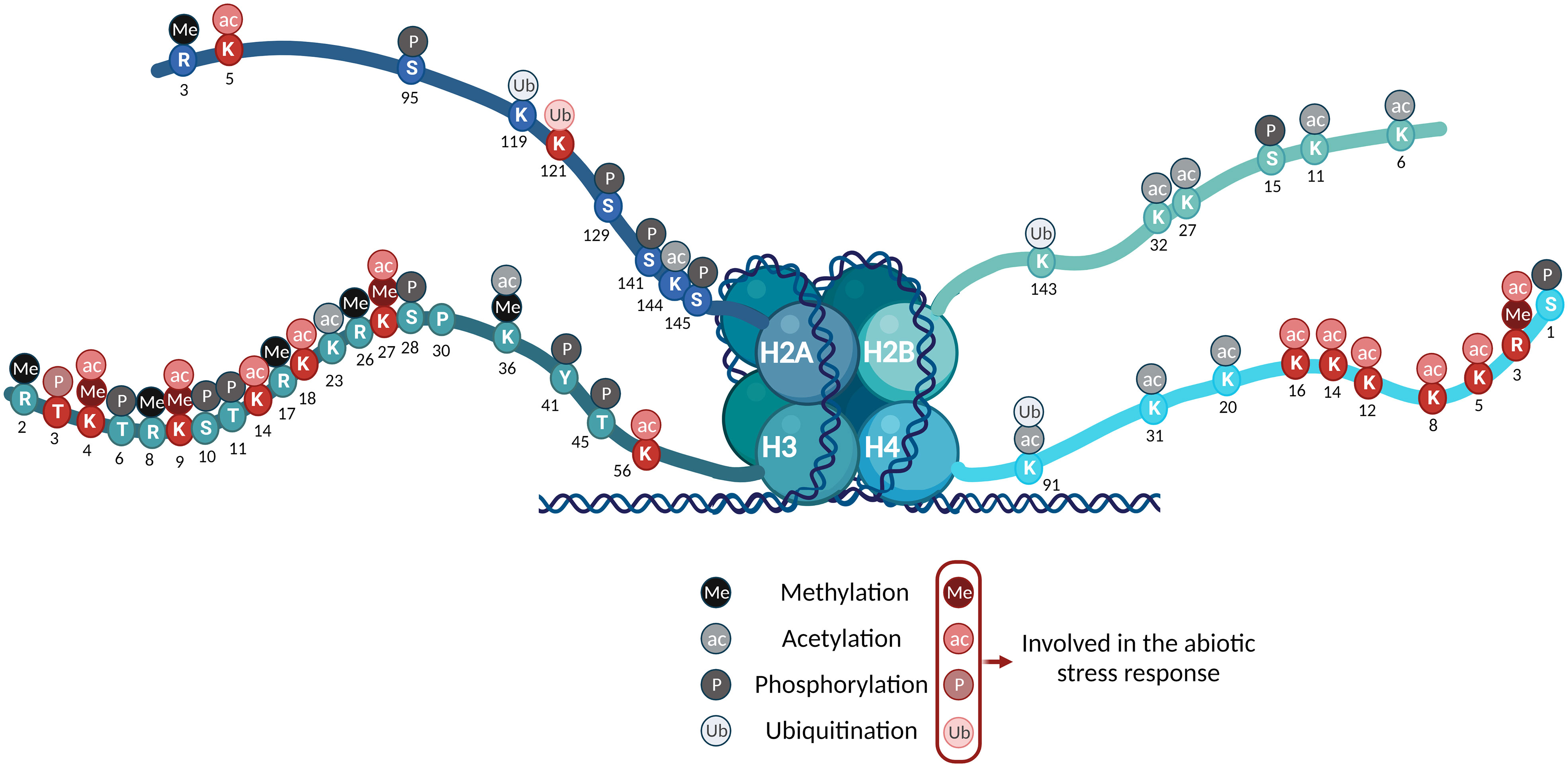
Figure 3 Summary of the histone PTMs in the nucleosome’s core histones in Arabidopsis. The PTMs detected are color-coded as indicated. Those involved in the abiotic stress response are highlighted in red.
The induction of the abiotic stress-responsive genes is independent of the mechanism of the stress-memory (Ding et al., 2012). Consequently, we consider that the regulation of stress memory is out of the scope of this review article. We have selected a list of recent review articles that detail the stress memory process (Oberkofler et al., 2021; Liu et al., 2022; Perrella et al., 2022).
Histone acetyltransferases (HATs) catalyze the transfer of the acetyl group from acetyl-CoA to the amino group of the lysine residues at the N-terminal tail of the histones. This reaction results in an acetylated lysine that compromise the interaction of the histone with the negatively charged DNA, leading to an open status of the chromatin (Smith and Denu, 2009). In presence of diverse abiotic agents —heat, salt, limited water availability— there is a global increase in histone acetylation (Pandey et al., 2002; Earley et al., 2007), (Figure 1). Acetylation marks allow the binding of stress-specific transcription factors —such as ABRE or DREB— during the stress response to areas of the genome that are generally silent (Kim et al., 2014; Widiez et al., 2014).
In Arabidopsis, 12 different HATs belong to four families: the GNAT/HAG, the MYST/HAM, the p300/CBP/HAC and the TAFII250/HAF families (Pandey et al., 2002; Fina et al., 2017) (Table 2). They regulate plant development, flowering time, and some specific processes of abiotic stress response that include response to light, salt tolerance, DNA damage and hormonal pathways (Earley et al., 2007; Xu et al., 2012; Xiao et al., 2013).
The GNAT superfamily member histone acetyltransferases GENERAL CONTROL NONDEREPRESSIBLE 5 (GCN5), encoded by HAG1, has been positively linked to cold and heat stress (Pavangadkar et al., 2010; Hu et al., 2015) and with the positive regulation of salt tolerance (Zheng et al., 2019). GCN5 was the first HAT identified in Arabidopsis. Transcriptomic analyses of gcn5 mutant show pleiotropic defects in the expression of genes involved in plant development and adaptation to environmental conditions (Cohen et al., 2009; Servet et al., 2010; Kim et al., 2018; Wang et al., 2019; Zheng et al., 2019). Importantly, under salt stress, gcn5 plants present inhibited growth compared to wild type plants (Zheng et al., 2019). The preferential GCN5 acetylation sites are the lysine residues of histone H2B and H3, with a lower preference for histone H4 (Fan et al., 2017; Mutlu and Puigserver, 2020). In fact, a decrease in the H3K9ac and H3K14ac marks has been reported in gcn5 mutants under salt stress (Li et al., 2022).
HAC1 and HAC5, two members of the p300/CBP family, participate in the ethylene response. The transcriptional levels of the ethylene response factors (ERFs) ERF1, ERF4, ERF6 and ERF11 significantly increase in the hac1hac5 double mutant (Li et al., 2014). It is possible that HAC1 and HAC5 might as well be involved in salinity stress response, as there is a close relationship between ethylene and salinity tolerance (Tao et al., 2015). Nevertheless, further research is needed to demonstrate it.
The Arabidopsis MYST family includes homologs of the catalytic subunit of the Nucleosome Acetyltransferase of the yeast H4 (NuA4) complex. Its components, HAG4/HAM1 and HAG5/HAM2, regulate general developmental processes in the plant, such as flowering, gametogenesis, chlorophyll synthesis, cell growth, and ploidy (Latrasse et al., 2008; Zacharaki et al., 2012; Crevillén et al., 2019). HAG4/HAM1 and HAG5/HAM2 also take part in ABA and UVB light responses, and other cell functions such as transcriptional activation and DNA damage repair (Campi et al., 2012; Umezawa et al., 2013).
The opposite action of HATs is conducted by histone deacetylases (HDAC). These enzymes catalyze the hydrolysis of the acetyl group from the amine of acetyl-lysine residues within histone tails. The 16 HDACs encoded in the Arabidopsis genome (Table 3) are organized into three families (RPD3/HDA1, HD2, and SIR2).
In Arabidopsis, HDA6 and HDA19 are the most extensively studied HDACs (Mehdi et al., 2016). They belong to the RPD3/HDA1 family. HDA6 and HDA19 have similar developmental functions. Both participate in pathogen defense systems, JA, and salicylic acid-mediated defense responses (Zhou et al., 2005; Wu et al., 2008; Choi et al., 2012), regulation of flowering, senescence (Wu et al., 2008; Yu et al., 2011; Mehdi et al., 2016), and abiotic stress responses (Chen and Wu, 2010). The fundamental difference between HDA6 and HDA19 is the antagonistic function, positive and negative, respectively, they have in the regulation of salt stress (reviewed in Luo et al., 2017).
HDA19 represses gene expression upon ABA and drought treatments by four different ways: 1) interaction with the ethylene response factor ERF7 and the transcriptional repressor SIN3, originating a repressive complex that silences stress-responsive genes (Song et al., 2005); 2) binding to SIN3-LIKE1 (SNL1) and SIN3-LIKE2 (SNL2), homologs of SIN3, to form a repressive complex that prevents ABA biosynthesis via the deacetylation of H3K9/14/18 (Wang et al., 2013); 3) formation of a complex with MSI1 that represses expression of genes in the ABA pathway such as the ABA receptors PYL4, PYL5, and PYL6 (Mehdi et al., 2016); and 4) binding to HDA6 and HDC1 and deacetyl K3K9/K14 in response to drought stress (Perrella et al., 2013).
HDA6 is upregulated by cold stress. This enzyme regulates cold-responsive (COR) genes during freezing tolerance (Park et al., 2018). HDA6 forms complexes with MSI4 and MSI5, that cause histone deacetylation in specific target loci, leading to transcriptional gene silencing (Gu et al., 2011; Mehdi et al., 2016).
The histone deacetylase HDA9, another member of the RPD3/HDA1 family, is a negative regulator of the ABA pathway. hda9 loss-of-function mutant displays increased tolerance to dehydration and upregulation of drought-responsive genes. During drought conditions, HDA9 interacts with critical components of the ABA pathway, such as ABI4 (Baek et al., 2020), and results in the induction of critical enzymes in the ABA catabolic pathways like ABA 8’-hydroxylases, encoded by CYP707A1 and CYP707A2 (Baek et al., 2020). HDA9 is also particularly important because it collaborates with the PRC2 complex by deacetylating H3K27 prior to its trimethylation (Qian et al., 2012).
HDA15 participates in the regulation of several warm temperature genes, including HEAT SHOCK PROTEIN 20 (HSP20), INDOLE-3-ACETIC ACID INDUCIBLE 19 (IAA19), and IAA29 (Shen et al., 2019). HDA15 participates in the ABA pathway. On one hand, it interacts with the transcription factor MYB96 to repress the expression of RHO GTPASE OF PLANTS in response to ABA (Lee and Seo, 2019). On the other hand, HDA15 interacts with MAC3A and MAC3B, subunits of the MAC complex, by a process enhanced by ABA. Moreover, hda15 and mac3a/mac3b mutants are ABA insensitive in seed germination and hyposensitive to salinity (Tu et al., 2022).
The expression of HD2A, HD2B, HD2C, and HD2D —members of the HD2A deacetylase family— is repressed by ABA and NaCl, which indicates their potential role in stress response. Overexpression of HD2D and HD2C results in increased drought tolerance (Sridha and Wu, 2006; Han et al., 2016). The expression of the ABA-responsive genes ABI1 and ABI2 increase in hda6, hd2c, and hda6/hd2c-1 mutant backgrounds, which was associated with increased histone H3K9/K14 acetylation (Luo et al., 2012). In the regulation of salinity tolerance, HD2C, together with HDA6 and HD2D, act as positive regulators (Luo et al., 2012).
In summary, HDA9 and HDA19 negatively regulate salt stress tolerance (Mehdi et al., 2016; Zheng et al., 2016; Ueda et al., 2017), while HDA6, HD2C, and HD2D positively regulate salinity tolerance (Chen and Wu, 2010; Chen et al., 2010; Luo et al., 2012; Han et al., 2016). These roles are further supported by the phenotypes of the previously mentioned HDAC mutants (reviewed in Ueda and Seki, 2020).
Histone methyltransferases catalyze mono-, di- and trimethylation of the amino group of lysines and arginines. This process is dependent on S-adenosyl-L-methionine (Smith and Denu, 2009). In plants, there are only eight histone lysine methylation sites: H3K4, H3K9, H3K26, H3K27, H3K36, H3K79, H4K20 and H1K26, and six arginine methylation sites: H3R2, H3R8, H3R17, H3R26, H4R3 and H4R17 (Zhang and Reinberg, 2001; Springer et al., 2003; Liu et al., 2010; Pontvianne et al., 2010; reviewed in Ueda and Seki, 2020).
The Arabidopsis genome contains 49 genes encoding SET domain-containing (SDG) methyltransferases (Baumbusch et al., 2001; Ng et al., 2007). Out of the 49 SDG proteins, 31 have histone lysine methyltransferase (HKMT) activity and are divided into five classes (I to V) based on their domain architectures (Table 4) (Baumbusch et al., 2001; Springer et al., 2003; Ng et al., 2007). In addition, there is an additional HKMT family, known as telomeric silencing 1-like (DOT1), which does not contain a SET domain and specifically adds methyl groups at the telomeric regions of H3K79 (Ng et al., 2002). Protein arginine methyltransferases (PRMTs) are classified as Type I or Type II, depending on the position of the methyl group on the guanidine of the methylated arginine (Hernando et al., 2015).
Plant SET proteins are classified into five classes: E(Z), ASH1, TRX (trithorax), PHD and SU(VAR) (Table 4).
The most common forms of methylation consist of the trimethylation of H3K27 and H3K4. H3K27me3 increases chromatin condensation and limits the recruitment of the transcriptional machinery and transcription factors to genes. Thus, H3K27me3 is associated with gene repression (Aranda et al., 2015; Zhao et al., 2021). In contrast, H3K4me3 colocalizes with actively transcribed genes, where it promotes the recruitment of transcription initiation factors to promoters of target genes (Lauberth et al., 2013; Zhao et al., 2021).
The leading writers of H3K27me3 in plants and animals are the PRC2 complexes. In Arabidopsis, the histone methyl transferase of the PRC2 complex and EZ homologs are MEDEA (MEA), CURLY LEAF (CLF) and SWINGER (SWN) (Makarevitch et al., 2013). The H3K27me3 mark has an intricate relationship with stress. It is a mark of facultative heterochromatin, involved primarily in the repression of developmentally regulated genes (Füßl et al., 2018). The H3K27me3 mark gives a more plastic structure to chromatin than constitutive heterochromatin. This structure allows condensation or decondensation of regions and permits transcription in temporal and spatial contexts, such as the derepression of genes involved in the abiotic stress response. Together with H3K4me3, it can have an implication on bivalent and responsive genes (Zhao et al., 2021).
The members of the trithorax family are responsible of H3K4 trimethylation. ATX1 drives H3K4me3 methylation in response to drought and osmotic stress (Ding et al., 2011). ATX1 together with ATRX7 regulate the expression of heat stress-responsive genes, not only during heat stress but also during stress recovery (Song et al., 2021). atx4 and atx5 mutants, also members of the trithorax family, showed increased tolerance to drought and salt stresses (Liu et al., 2018).
The loss of function of CAU1/PRMT5/SKB1, a member of the Type II PRMT family, results in salt hypersensitivity (Zhang et al., 2011). This enzyme catalyzes the addition of methyl groups to H4R3. In the presence of the salt stimulus, SKB1 dissociates from chromatin, leading to demethylation of arginine residues, and the transcription of stress-responsive genes (Fu et al., 2013).
Histone demethylases perform the antagonistic reaction to histone methyltransferases. It consists of the removal of the methyl group of the lysines and arginine residues of H3 and H4 tails. The nature of histone demetylation is intriguing due to the irreversible nature of the C-N bond. The first histone demethylase activity was identified in 1973 (Paik and Kim, 1973). There are 25 histone demethylase genes encoded in the Arabidopsis genome (Table 5) organized into two families: the FAD-dependent LSD/LDL/FLD and Jumonji C JMJ (Tsukada et al., 2006; Gu et al., 2016).
LSD1 was the first isolated demethylase (David Allis et al., 1980). This enzyme catalyzes the reduction of FAD to FADH2 and oxidizes the methylated lysine, resulting in an imine intermediate (Smith and Denu, 2009). The mechanism of histone demethylation by LSD1 is highly conserved among most eukaryotes (Lan et al., 2007b; Lan et al., 2007a; Liu et al., 2007; Opel et al., 2007; Rudolph et al., 2007; Katz et al., 2009). Another potential mechanism is the conversion of methylarginine to citrulline by a peptidyl arginine deiminase (Wang et al., 2004).
The histone demethylases included in the JMJ family contain a JmjC domain, which catalyzes the histone demethylation through the oxidation of ferrous ions Fe (II) and α- ketoglutarate (α-kg) (Lu et al., 2010). JMJ15 and JMJ17 demethylases take part in salinity and dehydration stress response, respectively (Liu et al., 2010; Shen et al., 2014; Xiao et al., 2016; Huang et al., 2019). There is an accumulation of lignin in the jmj15 mutant, although the regulation of lignin biosynthetic genes by JMJ15 still remains uncertain. It would be interesting to study whether JMJ15 and the HAT GCN5 participate in a common regulatory pathway in cell wall modification (Yung et al., 2021). jmj15 exhibits increased sensitivity to salinity stress. Similarly, overexpression of JMJ15 increases salinity tolerance in the plant and enhance seed germination under salt treatment (Yang et al., 2012; Shen et al., 2014). The loss-of-function mutants of JMJ17 display dehydration stress tolerance and ABA hypersensitivity regarding stomatal closure. During high temperature conditions, JMJ14 and JMJ15 remove H3K4me3 from transcriptional repressors of responsive genes in response to trigger thermomorphogenesis (Cui et al., 2021).
A recent study details the role in abiotic stress of JMJ27 (Wang et al., 2021). They revealed that JMJ27 positively regulates both ABA and drought-responsive genes and establishes a permissive chromatin state to enable an efficient transcriptional induction upon drought stress conditions (Chen et al., 2010; van Dijk et al., 2010). This is achieved by the demethylation of H3K9me2. Likewise, JMJ27 may function together with drought stress-activated H3K4 methyltransferase and histone acetyltransferase to co-activate their target genes under drought stress conditions.
Although acetylation and methylation are the most studied PTMs, there are additional modifications that influence chromatin accessibility. Histone ubiquitination comprises the incorporation of a 76-amino acid polypeptide into lysine residues of histones. This modification occurs mainly in H2A and H2B histones and is catalyzed by the formation of an isopeptide bond between the carboxy-terminal glycine of ubiquitin and the ϵ-group of a lysine residue on the carboxy-terminal tail of the histones. Substrates can be both poly- and monoubiquitinated. Polyubiquitination creates an irreversible signal for proteasomal-mediated degradation, whereas monoubiquitination generates a regulatory signal, which can be reversed by the action of ubiquitin-specific proteases (USPs/UBPs) called deubiquitinating enzymes (DUBs) (Zhou et al., 2017).
Arabidopsis E3 ubiquitin ligases (HUB1 and HUB2) and E2 ubiquitin conjugases (UBC1 and UBC2) are responsible for histone H2B mono-ubiquitination (H2Bub) (Cao et al., 2008). H2Aub is preferentially linked to transcriptional repression by counteracting H3K4me3. Specifically, the PRC1 complex catalyzes the monoubiquitination of H2A.ZK129 in a process linked to transcriptional repression (Gómez-Zambrano et al., 2019). On the other hand, H2Bub is a significant regulator of transcriptional activation, as it is required for H3K4me3 and H3K36me3. The monoubiquitination of H2B leads to the activation of responsive genes involved in abiotic and biotic stress that includes drought, salt, fungal pathogens, cold, heat and immune responses (Cao et al., 2008; Dhawan et al., 2009; Zou et al., 2014; Zhou and Zeng, 2017; Chen et al., 2019; Sun et al., 2020).
Histone phosphorylation consists of the addition of a phosphate group, and thus, of a negative charge, to serine, threonine, or tyrosine residues of the N-terminal tail of histones (Figure 3). This modification is involved in response to DNA damage, extracellular signals, and mitosis, where it leads to chromatin condensation in prophase (Dai et al., 2005; Houben et al., 2007; Rossetto et al., 2012; Wang and Higgins, 2013; Wang et al., 2015). The phosphorylation process is conserved along eukaryotes (Bi et al., 2011; Pirrotta, 2015).
There is also induction of H3 phosphorylation in response to abiotic stress, although the specific molecular mechanisms of the response are not clearly understood. H3S10ph is encompassed with acetylation in response to salt stress and cold (Sokol et al., 2007), suggesting that H3 phosphorylation is associated with transcription reprogramming after stress inducement. The phosphorylation of H3T3ph increases in pericentromeric regions after drought stress treatments (Wang et al., 2015) and is thought to be important in the maintenance of heterochromatin, suggesting that phosphorylation is implicated in gene silencing upon abiotic stress.
How chromatin marks affect transcription is a hot topic that brings the attention of epigenetics researchers from diverse backgrounds and fields. Thus, whether chromatin changes cause or correlate with the changes in gene expression is an area of active debate (Millán-Zambrano et al., 2022; Policarpi et al., 2022; Wang et al., 2022). Over the last decade, different studies have addressed this question with different technologies including ChIP-seq, CUT&RUN, CUT&Tag and TADs (Barski et al., 2007; Lieberman-Aiden et al., 2009; Dixon et al., 2012; Brind’Amour et al., 2015; Skene and Henikoff, 2017; Hainer and Fazzio, 2019; Kaya-Okur et al., 2019; Deng et al., 2022). On one hand, some evidence support that the marks are not instructing the activation/silencing, but instead they are informative of DNA processes (Wang et al., 2022). On the other hand, recent data support that some histone PTMs, such as H3K4me3, directly drive changes in gene expression (Policarpi et al., 2022). We consider that the answer to this question is far from being simple. It is likely that some histone PTMs may cause the initiation of DNA processes, such as transcriptional activation. In these cases, the histone PTM could be responsible for driving the genomic response. Similarly, there is a good chance that other histone PTMs are written as a consequence of these processes. For example, as the trace of a polymerase in a specific region of the genome. The mechanism that determines which marks acts as instructors or consequences of a genomic response depends on the context provided by the tissue, the developmental stage and the genomic landscape.
This topic is particularly intriguing during abiotic stress conditions, where the timing and order of the histone modifications is a crucial step to decipher the mechanism guiding the stress response. For that purpose, it is essential to establish a timeline of the epigenetic reprogramming in this scenario. During the early response, which ranges from the first to the fifth hour, a signaling network drives the binding of stress-specific transcription factors —DREB, ABRE— and the transcriptional machinery (Geng et al., 2013). Deciphering which epigenetic mark or histone variant is deposited/removed first after the abiotic stress would be the basis to understand better the intricate relationship between the histone variants and modifications. New studies focused on the temporal analysis of loss of and gain of function mutants of the enzymes that drive the major epigenetic regulators (H3K27me3, H3K4me3 and H2A.Z) will be crucial to establish the temporal epigenetic dynamics.
Bivalent chromatin is composed of epigenetic marks that play opposite roles on gene expression and co-localize in the same genomic regions (Voigt et al., 2013; Zhao et al., 2021). The H3K27me3/H3K4me3 pair of marks is the most usual form of bivalent chromatin. The first bivalent genes identified participate in cell differentiation in human embryonic stem cells (Bernstein et al., 2006). Since then, bivalent genes were identified in distinct species. An example of bivalent gene in Arabidopsis is the FLOWERING LOCUS C (FLC) (Jiang et al., 2008). The main hypothesis is that bivalent chromatin serves as a fast mechanism inducing developmentally regulated genes during differentiation (Bernstein et al., 2006). As the early stress response requires a rapid activation of responsive genes, a hypothesis is that poising of genes for transcriptional activation could be a mechanism for a fast gene regulation in response to abiotic stress. However, the role of bivalency marks has not been properly characterized in whole organisms nor during stress responses —with the exception of cold stress in potato tuber (Zeng et al., 2019)— so we consider it is an interesting topic of research.
There are histone variants that behave differently depending on the tissue they are deposited. If we take the chromatin organization within the sperm cells as an example, we find a regulatory network of histone PTMs and variants that define the accessibility to the transcriptional machinery. Histone variants H3.10 and H2B.8 are specific of sperm cells and represent the major pool of histones H3 and H2B, respectively (Okada et al., 2005; Jiang et al., 2020a; Buttress et al., 2022). Additionally, H3K27 is demethylated in sperm nuclei in a well-coordinated system in which the loss of H3K27me3 is associated to an increase of H3K4me3 in those genes required for embryo patterning, seed dormancy and flowering (Borg et al., 2020). Altogether, this suggest there is an intricate and well-conserved relationship between histone variants and modifications in specific tissues during development. Therefore, the tissue specificity of some epigenetic players rise the question of whether there are tissue-specific epigenetic changes during the abiotic stress response. Due to the nature of the stress response, it is possible that external organs and tissues reprogram their epigenetic landscape differently from internal tissues.
There is a need to perform studies that not only study the role of standard PTMs but also of those that are only present in specific histone variants. It makes sense that epigenetic marks involved in the abiotic stress response coincide with stress-specific histone variants in the same nucleosome. An example of specific modifications in histone variants is the ubiquitination of H2A.Z in its K129 residue. This mark specifically controls transcriptional repression by a group of genes silenced by PRC1, suggesting the possible function that the K129Ub might have in the dual role of H2A.Z (Gómez-Zambrano et al., 2019). Moreover, a recent preprint suggest that the K27 residue in the histone variant H3.3 is indispensable for many developmental processes that ranges from flowering to callus formation (Fal et al., 2022). Additionally, it has been reported that the SQ motif present in H2A.W.7 prevents the phosphorylation of the KSPK motif, a mark associated with DNA damage response (Schmücker et al., 2021), which indicates that the differences in the variants’ sequences result in diversity in transcriptional regulation. Consequently, further analyses are needed to fine-tune the relationship between these epigenetic players during the abiotic stress response.
In addition to histone variants and histone modifications, there is a need to unveil the role of DNA methylation, another major epigenetic regulator, in the chromatin landscape of stress-responsive genes. DNA methylation has been broadly described to regulate gene expression and silencing (Robertson, 2005; Slotkin and Martienssen, 2007; Zhang et al., 2018; He et al., 2022). Its relationship in the abiotic stress response as salinity, heat stress, cold, drought, heavy metals or nutrient deficit has been proposed earlier (Villagómez-Aranda et al., 2021; Reddy et al., 2022), although no significant conclusion has been made yet. As this modification colocalizes with heterochromatic regions and transposable elements (TEs) (Henderson and Jacobsen, 2007; Zhang et al., 2018), it makes sense to hypothesize that the CG, CHG and CHH regions of the genome can be methylated and demethylated to alter the transcription of specific stress-responsive genes. So far, it is known that NaCl exposure of Arabidopsis DECREASE IN DNA METHYLATION 1 mutant ddm1, a chromatin remodeler that facilitates DNA methylation, led to structural chromatin alterations (Yao et al., 2012; Sahu et al., 2013). Also, changes in DNA methylation in response to drought were not only Arabidopsis specific but are also observed in rice, which under salt stress showed altered DNA methylation levels (Zhang et al., 2013), maize (Sallam and Moussa, 2021), tomato (Huang et al., 2016), cotton (Wang and Qiao, 2020) and soy (Chen et al., 2019). However, more work needs to be done to explore the in-depth mechanisms and effect of DNA methylation on abiotic stress responses in plants.
To sum up, we consider that the epigenetic changes during the abiotic stress response should not be studied individually but, as the fundamental components of a complex network that provides a regulatory potential. Future insights into how the histone variants and modifications define chromatin organization and impact plant development during the abiotic stress response hold a great potential.
RN-V contributed most of the writing, that was supervised and revised by BD and CG. All authors contributed to the article and approved the submitted version.
Research in the laboratory is supported by grants RTI2018-094793-B-I00 (Ministerio de Ciencia e Innovación and Fondo Europeo de Desarrollo Regional FEDER), and 2018-AdG_833617 (European Research Council, EU), and by institutional grants from Banco de Santander and Fundación Ramon Areces to the CBMSO. RN-V is recipient of FPI contract PRE2019-087501 (Ministerio de Ciencia e Innovación).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adam, M., Robert, F., Larochelle, M., Gaudreau, L. (2001). H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 21, 6270–6279. doi: 10.1128/MCB.21.18.6270-6279.2001
Ahmad, K., Henikoff, S. (2002). The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200. doi: 10.1016/S1097-2765(02)00542-7
Allis, C. D., Jenuwein, T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500. doi: 10.1038/nrg.2016.59
Antunez-Sanchez, J., Naish, M., Ramirez-Prado, J. S., Ohno, S., Huang, Y., Dawson, A., et al. (2020). A new role for histone demethylases in the maintenance of plant genome integrity. eLife 9, e58533. doi: 10.7554/eLife.58533
Aranda, S., Mas, G., Di Croce, L. (2015). Regulation of gene transcription by polycomb proteins. Sci. Adv. 1, e1500737. doi: 10.1126/sciadv.1500737
Atkinson, N. J., Urwin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. doi: 10.1093/jxb/ers100
Baek, D., Shin, G., Kim, M. C., Shen, M., Lee, S. Y., Yun, D.-J. (2020). Histone deacetylase HDA9 with ABI4 contributes to abscisic acid homeostasis in drought stress response. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00143
Baroux, C., Raissig, M. T., Grossniklaus, U. (2011). Epigenetic regulation and reprogramming during gamete formation in plants. Curr. Opin. Genet. Dev. 21, 124–133. doi: 10.1016/j.gde.2011.01.017
Barski, A., Cuddapah, S., Cui, K., Roh, T.-Y., Schones, D. E., Wang, Z., et al. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. doi: 10.1016/j.cell.2007.05.009
Baulcombe, D. C., Dean, C. (2014). Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 6, a019471. doi: 10.1101/cshperspect.a019471
Bäurle, I., Trindade, I. (2020). Chromatin regulation of somatic abiotic stress memory. J. Exp. Bot. 71 (17), 5269–5279. doi: 10.1093/jxb/eraa098
Baumbusch, L. O., Thorstensen, T., Krauss, V., Fischer, A., Naumann, K., Assalkhou, R., et al. (2001). The arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29, 4319–4333. doi: 10.1093/nar/29.21.4319
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. doi: 10.1016/j.cell.2006.02.041
Bhadouriya, S. L., Mehrotra, S., Basantani, M. K., Loake, G. J., Mehrotra, R. (2021). Role of chromatin architecture in plant stress responses: An update. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.603380
Bieluszewski, T., Sura, W., Dziegielewski, W., Bieluszewska, A., Lachance, C., Kabza, M., et al. (2022). NuA4 and H2A.Z control environmental responses and autotrophic growth in arabidopsis. Nat. Commun. 13, 277. doi: 10.1038/s41467-021-27882-5
Bi, Y.-D., Wang, H.-X., Lu, T.-C., Li, X., Shen, Z., Chen, Y.-B., et al. (2011). Large-Scale analysis of phosphorylated proteins in maize leaf. Planta 233, 383–392. doi: 10.1007/s00425-010-1291-x
Borg, M., Brownfield, L., Twell, D. (2009). Male Gametophyte development: a molecular perspective. J. Exp. Bot. 60, 1465–1478. doi: 10.1093/jxb/ern355
Borg, M., Jacob, Y., Susaki, D., LeBlanc, C., Buendía, D., Axelsson, E., et al. (2020). Targeted reprogramming of H3K27me3 resets epigenetic memory in plant paternal chromatin. Nat. Cell Biol. 22, 621–629. doi: 10.1038/s41556-020-0515-y
Borg, M., Jiang, D., Berger, F. (2021a). Histone variants take center stage in shaping the epigenome. Curr. Opin. Plant Biol. 61, 101991. doi: 10.1016/j.pbi.2020.101991
Borg, M., Papareddy, R. K., Dombey, R., Axelsson, E., Nodine, M. D., Twell, D., et al. (2021b). Epigenetic reprogramming rewires transcription during the alternation of generations in arabidopsis. eLife 10, e61894. doi: 10.7554/eLife.61894
Bourguet, P., Picard, C. L., Yelagandula, R., Pélissier, T., Lorković, Z. J., Feng, S., et al. (2021). The histone variant H2A.W and linker histone H1 co-regulate heterochromatin accessibility and DNA methylation. Nat. Commun. 12, 2683. doi: 10.1038/s41467-021-22993-5
Brahma, S., Udugama, M. I., Kim, J., Hada, A., Bhardwaj, S. K., Hailu, S. G., et al. (2017). INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat. Commun. 8, 15616. doi: 10.1038/ncomms15616
Brind’Amour, J., Liu, S., Hudson, M., Chen, C., Karimi, M. M., Lorincz, M. C. (2015). An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat. Commun. 6, 6033. doi: 10.1038/ncomms7033
Brusslan, J. A., Bonora, G., Rus-Canterbury, A. M., Tariq, F., Jaroszewicz, A., Pellegrini, M. (2015). A genome-wide chronological study of gene expression and two histone modifications, H3K4me3 and H3K9ac, during developmental leaf senescence. Plant Physiol. 168, 1246–1261. doi: 10.1104/pp.114.252999
Brzezinka, K., Altmann, S., Czesnick, H., Nicolas, P., Gorka, M., Benke, E., et al. (2016). Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. eLife 5, e17061. doi: 10.7554/eLife.17061
Buszewicz, D., Archacki, R., Palusiński, A., Kotliński, M., Fogtman, A., Iwanicka-Nowicka, R., et al. (2016). HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant, Cell & Environmen 39, 2108–2122. doi: 10.1111/pce.12756
Buttress, T., He, S., Wang, L., Zhou, S., Saalbach, G., Vickers, M., et al. (2022). Histone H2B.8 compacts flowering plant sperm through chromatin phase separation. Nature 611, 614–622. doi: 10.1038/s41586-022-05386-6
Campi, M., D’Andrea, L., Emiliani, J., Casati, P. (2012). Participation of chromatin-remodeling proteins in the repair of ultraviolet-B-Damaged DNA. Plant Physiol. 158, 981–995. doi: 10.1104/pp.111.191452
Campos, E. I., Reinberg, D. (2009). Histones: Annotating chromatin. Annu. Rev. Genet. 43, 559–599. doi: 10.1146/annurev.genet.032608.103928
Cao, Y., Dai, Y., Cui, S., Ma, L. (2008). Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS c regulates flowering time in arabidopsis. Plant Cell 20, 2586–2602. doi: 10.1105/tpc.108.062760
Carter, B., Bishop, B., Ho, K. K., Huang, R., Jia, W., Zhang, H., et al. (2018). The chromatin remodelers PKL and PIE1 act in an epigenetic pathway that determines H3K27me3 homeostasis in arabidopsis. Plant Cell 30, 1337–1352. doi: 10.1105/tpc.17.00867
Castellano-Pozo, M., Santos-Pereira, J. M., Rondón, A. G., Barroso, S., Andújar, E., Pérez-Alegre, M., et al. (2013). R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol. Cell 52, 583–590. doi: 10.1016/j.molcel.2013.10.006
Chaubet, N., Clement, B., Gigot, C. (1992). Genes encoding a histone H3.3-like variant in arabidopsis contain intervening sequences. J. Mol. Biol. 225, 569–574. doi: 10.1016/0022-2836(92)90943-E
Chen, H., Feng, H., Zhang, X., Zhang, C., Wang, T., Dong, J. (2019). An arabidopsis E3 ligase HUB2 increases histone H2B monoubiquitination and enhances drought tolerance in transgenic cotton. Plant Biotechnol. J. 17, 556–568. doi: 10.1111/pbi.12998
Chen, L.-T., Luo, M., Wang, Y.-Y., Wu, K. (2010). Involvement of arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. doi: 10.1093/jxb/erq154
Chen, L.-T., Wu, K. (2010). Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 5, 1318–1320. doi: 10.4161/psb.5.10.13168
Choi, J., Lyons, D. B., Kim, M. Y., Moore, J. D., Zilberman, D. (2020). DNA Methylation and histone H1 jointly repress transposable elements and aberrant intragenic transcripts. Mol. Cell 77, 310–323.e7. doi: 10.1016/j.molcel.2019.10.011
Choi, S.-M., Song, H.-R., Han, S.-K., Han, M., Kim, C.-Y., Park, J., et al. (2012). HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in arabidopsis. Plant J. Cell Mol. Biol. 71, 135–146. doi: 10.1111/j.1365-313X.2012.04977.x
Cohen, R., Schocken, J., Kaldis, A., Vlachonasios, K. E., Hark, A. T., McCain, E. R. (2009). The histone acetyltransferase GCN5 affects the inflorescence meristem and stamen development in arabidopsis. Planta 230, 1207. doi: 10.1007/s00425-009-1012-5
Coleman-Derr, D., Zilberman, D. (2012). Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PloS Genet. 8, e1002988. doi: 10.1371/journal.pgen.1002988
Cortijo, S., Charoensawan, V., Brestovitsky, A., Buning, R., Ravarani, C., Rhodes, D., et al. (2017). Transcriptional regulation of the ambient temperature response by H2A.Z nucleosomes and HSF1 transcription factors in arabidopsis. Mol. Plant 10, 1258–1273. doi: 10.1016/j.molp.2017.08.014
Crevillén, P., Gómez-Zambrano, Á., López, J. A., Vázquez, J., Piñeiro, M., Jarillo, J. A. (2019). Arabidopsis YAF9 histone readers modulate flowering time through NuA4-complex-dependent H4 and H2A.Z histone acetylation at FLC chromatin. New Phytol. 222, 1893–1908. doi: 10.1111/nph.15737
Cui, X., Zheng, Y., Lu, Y., Issakidis-Bourguet, E., Zhou, D.-X. (2021). Metabolic control of histone demethylase activity involved in plant response to high temperature. Plant Physiol. 185, 1813–1828. doi: 10.1093/plphys/kiab020
Dai, X., Bai, Y., Zhao, L., Dou, X., Liu, Y., Wang, L., et al. (2017). H2A.Z represses gene expression by modulating promoter nucleosome structure and enhancer histone modifications in arabidopsis. Mol. Plant 10, 1274–1292. doi: 10.1016/j.molp.2017.09.007
Dai, J., Sultan, S., Taylor, S. S., Higgins, J. M. G. (2005). The kinase haspin is required for mitotic histone H3 thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 19, 472–488. doi: 10.1101/gad.1267105
Daniel Ricketts, M., Frederick, B., Hoff, H., Tang, Y., Schultz, D. C., Singh Rai, T., et al. (2015). Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat. Commun. 6 (1), 8711. doi: 10.1038/ncomms8711
Dantuma, N. P., van Attikum, H. (2016). Spatiotemporal regulation of posttranslational modifications in the DNA damage response. EMBO J. 35, 6–23. doi: 10.15252/embj.201592595
David Allis, C., Bowen, J. K., Abraham, G. N., Glover, C. V. C., Gorovsky, M. A. (1980). Proteolytic processing of histone H3 in chromatin: a physiologically regulated event in tetrahymena micronuclei. Cell 20, 55–64. doi: 10.1016/0092-8674(80)90234-2
Deng, Y., Bartosovic, M., Kukanja, P., Zhang, D., Liu, Y., Su, G., et al. (2022). Spatial-CUT&Tag: Spatially resolved chromatin modification profiling at the cellular level. Science 375, 681–686. doi: 10.1126/science.abg7216
Dhawan, R., Luo, H., Foerster, A. M., AbuQamar, S., Du, H.-N., Briggs, S. D., et al. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in arabidopsis. Plant Cell 21, 1000–1019. doi: 10.1105/tpc.108.062364
Ding, Y., Avramova, Z., Fromm, M. (2011). The arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J. 66, 735–744. doi: 10.1111/j.1365-313X.2011.04534.x
Ding, Y., Fromm, M., Avramova, Z. (2012). Multiple exposures to drought “train” transcriptional responses in arabidopsis. Nat. Commun. 3, 740. doi: 10.1038/ncomms1732
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. doi: 10.1038/nature11082
Earley, K. W., Shook, M. S., Brower-Toland, B., Hicks, L., Pikaard, C. S. (2007). In vitro specificities of arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J. Cell Mol. Biol. 52, 615–626. doi: 10.1111/j.1365-313X.2007.03264.x
Fal, K., Tomkova, D., Masson, M. L., Faigenboim, A., Pano, E., Ishkhneli, N., et al. (2022). Lysine 27 of histone H3.3 is a fine modulator of developmental gene expression and stands as an epigenetic checkpoint for lignin biosynthesis in arabidopsis. bioRxiv. doi: 10.1101/2022.06.08.495374
Fan, A., Mi, W., Liu, Z., Zeng, G., Zhang, P., Hu, Y., et al. (2017). Deletion of a histone acetyltransferase leads to the pleiotropic activation of natural products in metarhizium robertsii. Org. Lett. 19, 1686–1689. doi: 10.1021/acs.orglett.7b00476
Feng, S., Jacobsen, S. E., Reik, W. (2010). Epigenetic reprogramming in plant and animal development. Science 330, 622–627. doi: 10.1126/science.1190614
Fina, J. P., Masotti, F., Rius, S. P., Crevacuore, F., Casati, P. (2017). HAC1 and HAF1 histone acetyltransferases have different roles in UV-b responses in arabidopsis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01179
Foroozani, M., Holder, D. H., Deal, R. B. (2022). Histone variants in the specialization of plant chromatin. Annu. Rev. Plant Biol. 73, 149–172. doi: 10.1146/annurev-arplant-070221-050044
Francis, N. J., Kingston, R. E., Woodcock, C. L. (2004). Chromatin compaction by a polycomb group protein complex. Science 306, 1574–1577. doi: 10.1126/science.1100576
Füßl, M., Lassowskat, I., Née, G., Koskela, M. M., Brünje, A., Tilak, P., et al. (2018). Beyond histones: New substrate proteins of lysine deacetylases in arabidopsis nuclei. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00461
Fukagawa, T., Earnshaw, W. C. (2014). The centromere: Chromatin foundation for the kinetochore machinery. Dev. Cell 30, 496–508. doi: 10.1016/j.devcel.2014.08.016
Fu, Y.-L., Zhang, G.-B., Lv, X.-F., Guan, Y., Yi, H.-Y., Gong, J.-M. (2013). Arabidopsis histone methylase CAU1/PRMT5/SKB1 acts as an epigenetic suppressor of the calcium signaling gene CAS to mediate stomatal closure in response to extracellular calcium. Plant Cell 25, 2878–2891. doi: 10.1105/tpc.113.113886
Fyodorov, D. V., Zhou, B.-R., Skoultchi, A. I., Bai, Y. (2018). Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 19, 192–206. doi: 10.1038/nrm.2017.94
Gehring, M. (2019). Epigenetic dynamics during flowering plant reproduction: evidence for reprogramming? New Phytol. 224, 91–96. doi: 10.1111/nph.15856
Geng, Y., Wu, R., Wee, C. W., Xie, F., Wei, X., Chan, P. M. Y., et al. (2013). A spatio-temporal understanding of growth regulation during the salt stress response in arabidopsis. Plant Cell 25, 2132–2154. doi: 10.1105/tpc.113.112896
Giaimo, B. D., Ferrante, F., Herchenröther, A., Hake, S. B., Borggrefe, T. (2019). The histone variant H2A.Z in gene regulation. Epigenet. Chromatin. 12, 37. doi: 10.1186/s13072-019-0274-9
Gómez-Zambrano, Á., Merini, W., Calonje, M. (2019). The repressive role of arabidopsis H2A.Z in transcriptional regulation depends on AtBMI1 activity. Nat. Commun. 10, 2828. doi: 10.1038/s41467-019-10773-1
Grewal, S. I. S., Jia, S. (2007). Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46. doi: 10.1038/nrg2008
Gu, T., Han, Y., Huang, R., McAvoy, R. J., Li, Y. (2016). Identification and characterization of histone lysine methylation modifiers in fragaria vesca. Sci. Rep. 6, 23581. doi: 10.1038/srep23581
Gu, X., Jiang, D., Yang, W., Jacob, Y., Michaels, S. D., He, Y. (2011). Arabidopsis homologs of retinoblastoma-associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PloS Genet. 7, e1002366. doi: 10.1371/journal.pgen.1002366
Hainer, S. J., Fazzio, T. G. (2019). High-resolution chromatin profiling using CUT&RUN. Curr. Protoc. Mol. Biol. 126, e85. doi: 10.1002/cpmb.85
Hammond, C. M., Strømme, C. B., Huang, H., Patel, D. J., Groth, A. (2017). Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18, 141–158. doi: 10.1038/nrm.2016.159
Han, Z., Yu, H., Zhao, Z., Hunter, D., Luo, X., Duan, J., et al. (2016). AtHD2D gene plays a role in plant growth, development, and response to abiotic stresses in arabidopsis thaliana. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00310
Hebbes, T. R., Thorne, A. W., Crane-Robinson, C. (1988). A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7, 1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x
He, M., He, C.-Q., Ding, N.-Z. (2018). Abiotic stresses: General defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01771
He, L., Huang, H., Bradai, M., Zhao, C., You, Y., Ma, J., et al. (2022). DNA Methylation-free arabidopsis reveals crucial roles of DNA methylation in regulating gene expression and development. Nat. Commun. 13, 1335. doi: 10.1038/s41467-022-28940-2
Henderson, I. R., Jacobsen, S. E. (2007). Epigenetic inheritance in plants. Nature 447, 418–424. doi: 10.1038/nature05917
Henikoff, S., Smith, M. M. (2015). Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 7, a019364. doi: 10.1101/cshperspect.a019364
Hernando, C. E., Sanchez, S. E., Mancini, E., Yanovsky, M. J. (2015). Genome wide comparative analysis of the effects of PRMT5 and PRMT4/CARM1 arginine methyltransferases on the arabidopsis thaliana transcriptome. BMC Genomics 16, 192. doi: 10.1186/s12864-015-1399-2
Houben, A., Demidov, D., Caperta, A. D., Karimi, R., Agueci, F., Vlasenko, L. (2007). Phosphorylation of histone H3 in plants–a dynamic affair. Biochim. Biophys. Acta BBA Gene Struct. Expr. 1769, 308–315. doi: 10.1016/j.bbaexp.2007.01.002
Huang, K.-Y., Su, M.-G., Kao, H.-J., Hsieh, Y.-C., Jhong, J.-H., Cheng, K.-H., et al. (2016). dbPTM 2016: 10-year anniversary of a resource for post-translational modification of proteins. Nucleic Acids Res. 44, D435–D446. doi: 10.1093/nar/gkv1240
Huang, S., Zhang, A., Jin, J. B., Zhao, B., Wang, T.-J., Wu, Y., et al. (2019). Arabidopsis histone H3K4 demethylase JMJ17 functions in dehydration stress response. New Phytol. 223, 1372–1387. doi: 10.1111/nph.15874
Hu, Z., Song, N., Zheng, M., Liu, X., Liu, Z., Xing, J., et al. (2015). Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in arabidopsis. Plant J. Cell Mol. Biol. 84, 1178–1191. doi: 10.1111/tpj.13076
Ingouff, M., Berger, F. (2010). Histone3 variants in plants. Chromosoma 119, 27–33. doi: 10.1007/s00412-009-0237-1
Jacob, Y., Feng, S., LeBlanc, C. A., Bernatavichute, Y. V., Stroud, H., Cokus, S., et al. (2009). ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat. Struct. Mol. Biol. 16, 763–768. doi: 10.1038/nsmb.1611
Jarillo, J. A., Piñeiro, M. (2015). H2A.Z mediates different aspects of chromatin function and modulates flowering responses in arabidopsis. Plant J. 83, 96–109. doi: 10.1111/tpj.12873
Jiang, D., Borg, M., Lorković, Z. J., Montgomery, S. A., Osakabe, A., Yelagandula, R., et al. (2020a). The evolution and functional divergence of the histone H2B family in plants. PloS Genet. 16, e1008964. doi: 10.1371/journal.pgen.1008964
Jiang, J., Ding, A. B., Liu, F., Zhong, X. (2020b). Linking signaling pathways to histone acetylation dynamics in plants. J. Exp. Bot. 71, 5179–5190. doi: 10.1093/jxb/eraa202
Jiang, D., Wang, Y., Wang, Y., He, Y. (2008). Repression of FLOWERING LOCUS c and FLOWERING LOCUS T by the arabidopsis polycomb repressive complex 2 components. PloS One 3, e3404. doi: 10.1371/journal.pone.0003404
Jung, J.-H., Park, J.-H., Lee, S., To, T. K., Kim, J.-M., Seki, M., et al. (2013). The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS c transcription via chromatin remodeling under short-term cold stress in arabidopsis. Plant Cell 25, 4378–4390. doi: 10.1105/tpc.113.118364
Katz, D. J., Edwards, T. M., Reinke, V., Kelly, W. G. (2009). A c. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137, 308–320. doi: 10.1016/j.cell.2009.02.015
Kawashima, T., Berger, F. (2014). Epigenetic reprogramming in plant sexual reproduction. Nat. Rev. Genet. 15, 613–624. doi: 10.1038/nrg3685
Kawashima, T., Lorković, Z. J., Nishihama, R., Ishizaki, K., Axelsson, E., Yelagandula, R., et al. (2015). Diversification of histone H2A variants during plant evolution. Trends Plant Sci. 20, 419–425. doi: 10.1016/j.tplants.2015.04.005
Kaya-Okur, H. S., Wu, S. J., Codomo, C. A., Pledger, E. S., Bryson, T. D., Henikoff, J. G., et al. (2019). CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10 (1), 1930. doi: 10.1038/s41467-019-09982-5
Kim, K., Jang, Y.-J., Lee, S.-M., Oh, B.-T., Chae, J.-C., Lee, K.-J. (2014). Alleviation of salt stress by enterobacter sp. EJ01 in tomato and arabidopsis is accompanied by up-regulation of conserved salinity responsive factors in plants. Mol. Cells 37, 109–117. doi: 10.14348/molcells.2014.2239
Kim, J.-M., Sasaki, T., Ueda, M., Sako, K., Seki, M. (2015). Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00114
Kim, J.-M., To, T. K., Matsui, A., Tanoi, K., Kobayashi, N. I., Matsuda, F., et al. (2017). Acetate-mediated novel survival strategy against drought in plants. Nat. Plants 3, 1–7. doi: 10.1038/nplants.2017.97
Kim, J., Yang, W., Forner, J., Lohmann, J. U., Noh, B., Noh, Y. (2018). Epigenetic reprogramming by histone acetyltransferase HAG1/AtGCN5 is required for pluripotency acquisition in arabidopsis. EMBO J. 37, e98726. doi: 10.15252/embj.201798726
Kotliński, M., Rutowicz, K., Kniżewski, Ł., Palusiński, A., Olędzki, J., Fogtman, A., et al. (2016). Histone H1 variants in arabidopsis are subject to numerous post-translational modifications, both conserved and previously unknown in histones, suggesting complex functions of H1 in plants. PloS One 11, e0147908. doi: 10.1371/journal.pone.0147908
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi: 10.1016/j.cell.2007.02.005
Lan, F., Collins, R. E., De Cegli, R., Alpatov, R., Horton, J. R., Shi, X., et al. (2007a). Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 448, 718–722. doi: 10.1038/nature06034
Lan, F., Zaratiegui, M., Villén, J., Vaughn, M. W., Verdel, A., Huarte, M., et al. (2007b). S. pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription. Mol. Cell 26, 89–101. doi: 10.1016/j.molcel.2007.02.023
Latrasse, D., Benhamed, M., Henry, Y., Domenichini, S., Kim, W., Zhou, D.-X., et al. (2008). The MYST histone acetyltransferases are essential for gametophyte development in arabidopsis. BMC Plant Biol. 8, 121. doi: 10.1186/1471-2229-8-121
Lauberth, S. M., Nakayama, T., Wu, X., Ferris, A. L., Tang, Z., Hughes, S. H., et al. (2013). H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152, 1021–1036. doi: 10.1016/j.cell.2013.01.052
Lee, S., Fu, F., Xu, S., Lee, S. Y., Yun, D.-J., Mengiste, T. (2016). Global regulation of plant immunity by histone lysine methyl transferases. Plant Cell 28, 1640–1661. doi: 10.1105/tpc.16.00012
Lee, K., Seo, P. J. (2018). Dynamic epigenetic changes during plant regeneration. Trends Plant Sci. 23, 235–247. doi: 10.1016/j.tplants.2017.11.009
Lee, H. G., Seo, P. J. (2019). MYB96 recruits the HDA15 protein to suppress negative regulators of ABA signaling in arabidopsis. Nat. Commun. 10, 1713. doi: 10.1038/s41467-019-09417-1
Lei, B., Berger, F. (2020). H2A variants in arabidopsis: Versatile regulators of genome activity. Plant Commun. 1, 100015. doi: 10.1016/j.xplc.2019.100015
Lei, B., Capella, M., Montgomery, S. A., Borg, M., Osakabe, A., Goiser, M., et al. (2021). A synthetic approach to reconstruct the evolutionary and functional innovations of the plant histone variant H2A.W. Curr. Biol. 31, 182–191.e5. doi: 10.1016/j.cub.2020.09.080
Libertini, L. J., Ausió, J., van Holde, K. E., Small, E. W. (1988). Histone hyperacetylation. its effects on nucleosome core particle transitions. Biophys. J. 53, 477–487. doi: 10.1016/S0006-3495(88)83126-6
Lieberman-Aiden, E., van Berkum, N. L., Williams, L., Imakaev, M., Ragoczy, T., Telling, A., et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. doi: 10.1126/science.1181369
Lippman, Z., Gendrel, A.-V., Black, M., Vaughn, M. W., Dedhia, N., McCombie, W. R., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476. doi: 10.1038/nature02651
Liu, H., Able, A. J., Able, J. A. (2022). Priming crops for the future: rewiring stress memory. Trends Plant Sci. 27, 699–716. doi: 10.1016/j.tplants.2021.11.015
Liu, L., Chai, M., Huang, Y., Qi, J., Zhu, W., Xi, X., et al. (2021). SDG2 regulates arabidopsis inflorescence architecture through SWR1-ERECTA signaling pathway. iScience 24, 103236. doi: 10.1016/j.isci.2021.103236
Liu, C., Lu, F., Cui, X., Cao, X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61, 395–420. doi: 10.1146/annurev.arplant.043008.091939
Liu, F., Quesada, V., Crevillén, P., Bäurle, I., Swiezewski, S., Dean, C. (2007). The arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol. Cell 28, 398–407. doi: 10.1016/j.molcel.2007.10.018
Liu, Y., Zhang, A., Yin, H., Meng, Q., Yu, X., Huang, S., et al. (2018). Trithorax-group proteins ARABIDOPSIS TRITHORAX4 (ATX4) and ATX5 function in abscisic acid and dehydration stress responses. New Phytol. 217, 1582–1597. doi: 10.1111/nph.14933
Li, C., Xu, J., Li, J., Li, Q., Yang, H. (2014). Involvement of arabidopsis histone acetyltransferase HAC family genes in the ethylene signaling pathway. Plant Cell Physiol. 55, 426–435. doi: 10.1093/pcp/pct180
Li, Z., Zhang, H., Cai, C., Lin, Z., Zhen, Z., Chu, J., et al. (2022). Histone acetyltransferase GCN5-mediated lysine acetylation modulates salt stress aadaption of trichoderma. Appl. Microbiol. Biotechnol. 106, 3033–3049. doi: 10.1007/s00253-022-11897-z
Long, M., Sun, X., Shi, W., Yanru, A., Leung, S. T. C., Ding, D., et al. (2019). A novel histone H4 variant H4G regulates rDNA transcription in breast cancer. Nucleic Acids Res. 47, 8399–8409. doi: 10.1093/nar/gkz547
Loppin, B., Berger, F. (2020). Histone variants: The nexus of developmental decisions and epigenetic memory. Annu. Rev. Genet. 54, 121–149. doi: 10.1146/annurev-genet-022620-100039
Lorković, Z. J., Berger, F. (2017). Heterochromatin and DNA damage repair: Use different histone variants and relax. Nucleus 8, 583–588. doi: 10.1080/19491034.2017.1384893
Lu, L., Chen, X., Qian, S., Zhong, X. (2018). The plant-specific histone residue Phe41 is important for genome-wide H3.1 distribution. Nat. Commun. 9, 630. doi: 10.1038/s41467-018-02976-9
Lu, F., Cui, X., Zhang, S., Liu, C., Cao, X. (2010). JMJ14 is an H3K4 demethylase regulating flowering time in arabidopsis. Cell Res. 20, 387–390. doi: 10.1038/cr.2010.27
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F., Richmond, T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 a resolution. Nature 389, 251–260. doi: 10.1038/38444
Luo, M., Cheng, K., Xu, Y., Yang, S., Wu, K. (2017). Plant responses to abiotic stress regulated by histone deacetylases. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02147
Luo, Y.-X., Hou, X.-M., Zhang, C.-J., Tan, L.-M., Shao, C.-R., Lin, R.-N., et al. (2020). A plant-specific SWR1 chromatin-remodeling complex couples histone H2A.Z deposition with nucleosome sliding. EMBO J. 39, e102008. doi: 10.15252/embj.2019102008
Luo, M., Wang, Y.-Y., Liu, X., Yang, S., Lu, Q., Cui, Y., et al. (2012). HD2C interacts with HDA6 and is involved in ABA and salt stress response in arabidopsis. J. Exp. Bot. 63, 3297–3306. doi: 10.1093/jxb/ers059
Mahrez, W., Arellano, M. S. T., Moreno-Romero, J., Nakamura, M., Shu, H., Nanni, P., et al. (2016). H3K36ac is an evolutionary conserved plant histone modification that marks active genes. Plant Physiol. 170, 1566–1577. doi: 10.1104/pp.15.01744
Makarevitch, I., Eichten, S. R., Briskine, R., Waters, A. J., Danilevskaya, O. N., Meeley, R. B., et al. (2013). Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27[W]. Plant Cell 25, 780–793. doi: 10.1105/tpc.112.106427
Malik, H. S., Henikoff, S. (2009). Major evolutionary transitions in centromere complexity. Cell 138, 1067–1082. doi: 10.1016/j.cell.2009.08.036
March-Díaz, R., Reyes, J. C. (2009). The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol. Plant 2, 565–577. doi: 10.1093/mp/ssp019
Martire, S., Banaszynski, L. A. (2020). The roles of histone variants in fine-tuning chromatin organization and function. Nat. Rev. Mol. Cell Biol. 21, 522–541. doi: 10.1038/s41580-020-0262-8
Mattei, A. L., Bailly, N., Meissner, A. (2022). DNA Methylation: a historical perspective. Trends Genet. 38, 676–707. doi: 10.1016/j.tig.2022.03.010
Mcbryant, S., Lu, X., Hansen, J. (2010). Multifunctionality of the linker histones: An emerging role for protein-protein interactions. Cell Res. 20, 519–528. doi: 10.1038/cr.2010.35
Mehdi, S., Derkacheva, M., Ramström, M., Kralemann, L., Bergquist, J., Hennig, L. (2016). The WD40 domain protein MSI1 functions in a histone deacetylase complex to fine-tune abscisic acid signaling. Plant Cell 28, 42–54. doi: 10.1105/tpc.15.00763
Millán-Zambrano, G., Burton, A., Bannister, A. J., Schneider, R. (2022). Histone post-translational modifications - cause and consequence of genome function. Nat. Rev. Genet. 23, 563–580. doi: 10.1038/s41576-022-00468-7
Molitor, A. M., Bu, Z., Yu, Y., Shen, W.-H. (2014). Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PloS Genet. 10, e1004091. doi: 10.1371/journal.pgen.1004091
Morrison, O., Thakur, J. (2021). Molecular complexes at euchromatin, heterochromatin and centromeric chromatin. Int. J. Mol. Sci. 22, 6922. doi: 10.3390/ijms22136922
Müller, S., Almouzni, G. (2014). A network of players in H3 histone variant deposition and maintenance at centromeres. Biochim. Biophys. Acta 1839, 241–250. doi: 10.1016/j.bbagrm.2013.11.008
Muñoz, A., Castellano, M. M. (2012). Regulation of translation initiation under abiotic stress conditions in plants: Is it a conserved or not so conserved process among eukaryotes? Comp. Funct. Genomics 2012, 406357. doi: 10.1155/2012/406357
Mutlu, B., Puigserver, P. (2020). GCN5 acetyltransferase in cellular energetic and metabolic processes. Biochim. Biophys. Acta BBA Gene Regul. Mech. 1864, 194626. doi: 10.1016/j.bbagrm.2020.194626
Nakayama, J., Rice, J. C., Strahl, B. D., Allis, C. D., Grewal, S. I. S. (2001). Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113. doi: 10.1126/science.1060118
Ng, H. H., Feng, Q., Wang, H., Erdjument-Bromage, H., Tempst, P., Zhang, Y., et al. (2002). Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and sir protein association. Genes Dev. 16, 1518–1527. doi: 10.1101/gad.1001502
Nguyen, N. H., Cheong, J.-J. (2018). H2A.Z-containing nucleosomes are evicted to activate AtMYB44 transcription in response to salt stress. Biochem. Biophys. Res. Commun. 499, 1039–1043. doi: 10.1016/j.bbrc.2018.04.048
Ng, D. W.-K., Wang, T., Chandrasekharan, M. B., Aramayo, R., Kertbundit, S., Hall, T. C. (2007). Plant SET domain-containing proteins: Structure, function and regulation. Biochim. Biophys. Acta BBA Gene Struct. Expr. 1769, 316–329. doi: 10.1016/j.bbaexp.2007.04.003
Oberkofler, V., Pratx, L., Bäurle, I. (2021). Epigenetic regulation of abiotic stress memory: maintaining the good things while they last. Curr. Opin. Plant Biol. 61, 102007. doi: 10.1016/j.pbi.2021.102007
Okada, T., Endo, M., Singh, M. B., Bhalla, P. L. (2005). Analysis of the histone H3 gene family in arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 44, 557–568. doi: 10.1111/j.1365-313X.2005.02554.x
Olins, A. L., Olins, D. E. (1974). Spheroid chromatin units (v bodies). Science 183, 330–332. doi: 10.1126/science.183.4122.330
Opel, M., Lando, D., Bonilla, C., Trewick, S. C., Boukaba, A., Walfridsson, J., et al. (2007). Genome-wide studies of histone demethylation catalysed by the fission yeast homologues of mammalian LSD1. PloS One 2, e386. doi: 10.1371/journal.pone.0000386
Osakabe, A., Lorković, Z. J., Kobayashi, W., Tachiwana, H., Yelagandula, R., Kurumizaka, H., et al. (2018). Histone H2A variants confer specific properties to nucleosomes and impact on chromatin accessibility. Nucleic Acids Res. 46, 7675–7685. doi: 10.1093/nar/gky540
Paik, W. K., Kim, S. (1973). Enzymatic demethylation of calf thymus histones. Biochem. Biophys. Res. Commun. 51, 781–788. doi: 10.1016/0006-291X(73)91383-1
Pandey, R., Müller, A., Napoli, C. A., Selinger, D. A., Pikaard, C. S., Richards, E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. doi: 10.1093/nar/gkf660
Park, J., Lim, C. J., Shen, M., Park, H. J., Cha, J.-Y., Iniesto, E., et al. (2018). Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. 115, E5400–E5409. doi: 10.1073/pnas.1721241115
Pavangadkar, K., Thomashow, M. F., Triezenberg, S. J. (2010). Histone dynamics and roles of histone acetyltransferases during cold-induced gene regulation in arabidopsis. Plant Mol. Biol. 74, 183–200. doi: 10.1007/s11103-010-9665-9
Pecinka, A., Dinh, H. Q., Baubec, T., Rosa, M., Lettner, N., Scheid, O. M. (2010). Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis[W][OA]. Plant Cell 22, 3118–3129. doi: 10.1105/tpc.110.078493
Perianez-Rodriguez, J., Manzano, C., Moreno-Risueno, M. A. (2014). Post-embryonic organogenesis and plant regeneration from tissues: two sides of the same coin? Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00219
Perrella, G., Bäurle, I., van Zanten, M. (2022). Epigenetic regulation of thermomorphogenesis and heat stress tolerance. New Phytol. 234, 1144–1160. doi: 10.1111/nph.17970
Perrella, G., Lopez-Vernaza, M. A., Carr, C., Sani, E., Gosselé, V., Verduyn, C., et al. (2013). Histone deacetylase Complex1 expression level titrates plant growth and abscisic acid sensitivity in arabidopsis. Plant Cell 25, 3491–3505. doi: 10.1105/tpc.113.114835
Pirrotta, V. (2015). Histone marks direct chromosome segregation. Cell 163, 792–793. doi: 10.1016/j.cell.2015.10.043
Policarpi, C., Munafo, M., Tsagkris, S., Carlini, V., Hackett, J. A. (2022). Systematic epigenome editing captures the context-dependent instructive function of chromatin modifications. bioRxiv. doi: 10.1101/2022.09.04.506519
Pontvianne, F., Blevins, T., Pikaard, C. S. (2010). Arabidopsis histone lysine methyltransferases. Adv. Bot. Res. 53, 1–22. doi: 10.1016/S0065-2296(10)53001-5
Popova, O. V., Dinh, H. Q., Aufsatz, W., Jonak, C. (2013). The RdDM pathway is required for basal heat tolerance in arabidopsis. Mol. Plant 6, 396–410. doi: 10.1093/mp/sst023
Port, M., Tripp, J., Zielinski, D., Weber, C., Heerklotz, D., Winkelhaus, S., et al. (2004). Role of Hsp17.4-CII as coregulator and cytoplasmic retention factor of tomato heat stress transcription factor HsfA2. Plant Physiol. 135, 1457–1470. doi: 10.1104/pp.104.042820
Probst, A. V. (2022). Deposition and eviction of histone variants define functional chromatin states in plants. Curr. Opin. Plant Biol. 69, 102266. doi: 10.1016/j.pbi.2022.102266
Probst, A. V., Desvoyes, B., Gutierrez, C. (2020). Similar yet critically different: the distribution, dynamics and function of histone variants. J. Exp. Bot. 71, 5191–5204. doi: 10.1093/jxb/eraa230
Probst, A. V., Mittelsten Scheid, O. (2015). Stress-induced structural changes in plant chromatin. Curr. Opin. Plant Biol. 27, 8–16. doi: 10.1016/j.pbi.2015.05.011
Qian, W., Miki, D., Zhang, H., Liu, Y., Zhang, X., Tang, K., et al. (2012). A histone acetyltransferase regulates active DNA demethylation in arabidopsis. Science 336, 1445–1448. doi: 10.1126/science.1219416
Ravi, M., Kwong, P. N., Menorca, R. M. G., Valencia, J. T., Ramahi, J. S., Stewart, J. L., et al. (2010). The rapidly evolving centromere-specific histone has stringent functional requirements in arabidopsis thaliana. Genetics 186, 461–471. doi: 10.1534/genetics.110.120337
Reddy, D., Bhattacharya, S., Shah, S., Rashid, M., Gupta, S. (2022). DNA Methylation mediated downregulation of histone H3 variant H3.3 affects cell proliferation contributing to the development of HCC. Biochim. Biophys. Acta BBA Mol. Basis Dis. 1868, 166284. doi: 10.1016/j.bbadis.2021.166284
Robertson, K. D. (2005). DNA Methylation and human disease. Nat. Rev. Genet. 6, 597–610. doi: 10.1038/nrg1655
Roca Paixão, J., Gillet, F.-X., Ribeiro, T., Bournaud, C., Lourenço-Tessutti, I., Noriga, D., et al. (2019). Improved drought stress tolerance in arabidopsis by CRISPR/dCas9 fusion with a histone AcetylTransferase. Sci. Rep. 9, 8080. doi: 10.1038/s41598-019-44571-y
Rossetto, D., Avvakumov, N., Côté, J. (2012). Histone phosphorylation. Epigenetics 7, 1098–1108. doi: 10.4161/epi.21975
Roudier, F., Teixeira, F. K., Colot, V. (2009). Chromatin indexing in arabidopsis: an epigenomic tale of tails and more. Trends Genet. 25, 511–517. doi: 10.1016/j.tig.2009.09.013
Rudolph, T., Yonezawa, M., Lein, S., Heidrich, K., Kubicek, S., Schäfer, C., et al. (2007). Heterochromatin formation in drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol. Cell 26, 103–115. doi: 10.1016/j.molcel.2007.02.025
Rutowicz, K., Lirski, M., Mermaz, B., Teano, G., Schubert, J., Mestiri, I., et al. (2019). Linker histones are fine-scale chromatin architects modulating developmental decisions in arabidopsis. Genome Biol. 20, 157. doi: 10.1186/s13059-019-1767-3
Rutowicz, K., Puzio, M., Halibart-Puzio, J., Lirski, M., Kotliński, M., Kroteń, M. A., et al. (2015). A specialized histone H1 variant is required for adaptive responses to complex abiotic stress and related DNA methylation in arabidopsis. Plant Physiol. 169, 2080–2101. doi: 10.1104/pp.15.00493
Sahu, P. P., Pandey, G., Sharma, N., Puranik, S., Muthamilarasan, M., Prasad, M. (2013). Epigenetic mechanisms of plant stress responses and adaptation. Plant Cell Rep. 32, 1151–1159. doi: 10.1007/s00299-013-1462-x
Sallam, N., Moussa, M. (2021). DNA Methylation changes stimulated by drought stress in ABA-deficient maize mutant vp10. Plant Physiol. Biochem. 160, 218–224. doi: 10.1016/j.plaphy.2021.01.024
Schmücker, A., Lei, B., Lorković, Z. J., Capella, M., Braun, S., Bourguet, P., et al. (2021). Crosstalk between H2A variant-specific modifications impacts vital cell functions. PloS Genet. 17, e1009601. doi: 10.1371/journal.pgen.1009601
Schuettengruber, B., Chourrout, D., Vervoort, M., Leblanc, B., Cavalli, G. (2007). Genome regulation by polycomb and trithorax proteins. Cell 128, 735–745. doi: 10.1016/j.cell.2007.02.009
Sequeira-Mendes, J., Aragüez, I., Peiró, R., Mendez-Giraldez, R., Zhang, X., Jacobsen, S. E., et al. (2014). The functional topography of the arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 26, 2351–2366. doi: 10.1105/tpc.114.124578
Servet, C., Conde e Silva, N., Zhou, D.-X. (2010). Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in arabidopsis. Mol. Plant 3, 670–677. doi: 10.1093/mp/ssq018
Shen, Y., Conde e Silva, N., Audonnet, L., Servet, C., Wei, W., Zhou, D.-X. (2014). Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in arabidopsis. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00290
Shen, Y., Lei, T., Cui, X., Liu, X., Zhou, S., Zheng, Y., et al. (2019). Arabidopsis histone deacetylase HDA15 directly represses plant response to elevated ambient temperature. Plant J. Cell Mol. Biol. 100, 991–1006. doi: 10.1111/tpj.14492
Shi, L., Wang, J., Hong, F., Spector, D. L., Fang, Y. (2011). Four amino acids guide the assembly or disassembly of arabidopsis histone H3.3-containing nucleosomes. Proc. Natl. Acad. Sci. 108, 10574–10578. doi: 10.1073/pnas.1017882108
Shu, H., Nakamura, M., Siretskiy, A., Borghi, L., Moraes, I., Wildhaber, T., et al. (2014). Arabidopsisreplacement histone variant H3.3 occupies promoters of regulated genes. Genome Biol. 15, R62. doi: 10.1186/gb-2014-15-4-r62
Singh, D., Laxmi, A. (2015). Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00895
Skene, P. J., Henikoff, S. (2017). An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856. doi: 10.7554/eLife.21856
Slotkin, R. K., Martienssen, R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285. doi: 10.1038/nrg2072
Smith, B. C., Denu, J. M. (2009). Chemical mechanisms of histone lysine and arginine modifications. Biochim. Biophys. Acta 1789, 45–57. doi: 10.1016/j.bbagrm.2008.06.005
Sokol, A., Kwiatkowska, A., Jerzmanowski, A., Prymakowska-Bosak, M. (2007). Up-regulation of stress-inducible genes in tobacco and arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 227, 245–254. doi: 10.1007/s00425-007-0612-1
Song, C.-P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., et al. (2005). Role of an arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384–2396. doi: 10.1105/tpc.105.033043
Song, Z.-T., Zhang, L.-L., Han, J.-J., Zhou, M., Liu, J.-X. (2021). Histone H3K4 methyltransferases SDG25 and ATX1 maintain heat-stress gene expression during recovery in arabidopsis. Plant J. 105, 1326–1338. doi: 10.1111/tpj.15114
Springer, N. M., Napoli, C. A., Selinger, D. A., Pandey, R., Cone, K. C., Chandler, V. L., et al. (2003). Comparative analysis of SET domain proteins in maize and arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 132, 907–925. doi: 10.1104/pp.102.013722
Sridha, S., Wu, K. (2006). Identification of AtHD2C as a novel regulator of abscisic acid responses in arabidopsis. Plant J. 46, 124–133. doi: 10.1111/j.1365-313X.2006.02678.x
Stroud, H., Otero, S., Desvoyes, B., Ramírez-Parra, E., Jacobsen, S. E., Gutierrez, C. (2012). Genome-wide analysis of histone H3.1 and H3.3 variants in arabidopsis thaliana. Proc. Natl. Acad. Sci. 109, 5370–5375. doi: 10.1073/pnas.1203145109
Sun, Y., Zhao, J., Li, X., Li, Y. (2020). E2 conjugases UBC1 and UBC2 regulate MYB42-mediated SOS pathway in response to salt stress in arabidopsis. New Phytol. 227, 455–472. doi: 10.1111/nph.16538
Sura, W., Kabza, M., Karlowski, W. M., Bieluszewski, T., Kus-Slowinska, M., Pawełoszek, Ł., et al. (2017). Dual role of the histone variant H2A.Z in transcriptional regulation of stress-response Genes[OPEN]. Plant Cell 29, 791–807. doi: 10.1105/tpc.16.00573
Székely, G., Ábrahám, E., Cséplő, Á., Rigó, G., Zsigmond, L., Csiszár, J., et al. (2008). Duplicated P5CS genes of arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53, 11–28. doi: 10.1111/j.1365-313X.2007.03318.x
Talbert, P. B., Ahmad, K., Almouzni, G., Ausió, J., Berger, F., Bhalla, P. L., et al. (2012). A unified phylogeny-based nomenclature for histone variants. Epigenet. Chromatin. 5, 7. doi: 10.1186/1756-8935-5-7
Talbert, P. B., Henikoff, S. (2017). Histone variants on the move: substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol. 18, 115–126. doi: 10.1038/nrm.2016.148
Tao, J.-J., Chen, H.-W., Ma, B., Zhang, W.-K., Chen, S.-Y., Zhang, J.-S. (2015). The role of ethylene in plants under salinity stress. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01059
Thomas, J. O., Kornberg, R. D. (1975). An octamer of histones in chromatin and free in solution. Proc. Natl. Acad. Sci. U. S. A. 72, 2626–2630. doi: 10.1073/pnas.72.7.2626
To, T., Kim, J.-M. (2014). Epigenetic regulation of gene responsiveness in arabidopsis. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00548
To, T. K., Nakaminami, K., Kim, J.-M., Morosawa, T., Ishida, J., Tanaka, M., et al. (2011). Arabidopsis HDA6 is required for freezing tolerance. Biochem. Biophys. Res. Commun. 406, 414–419. doi: 10.1016/j.bbrc.2011.02.058
Tsukada, Y., Fang, J., Erdjument-Bromage, H., Warren, M. E., Borchers, C. H., Tempst, P., et al. (2006). Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816. doi: 10.1038/nature04433
Tu, Y.-T., Chen, C.-Y., Huang, Y.-S., Chang, C.-H., Yen, M.-R., Hsieh, J.-W. A., et al. (2022). HISTONE DEACETYLASE 15 and MOS4-associated complex subunits 3A/3B coregulate intron retention of ABA-responsive genes. Plant Physiol. 190, 882–897. doi: 10.1093/plphys/kiac271
Ueda, M., Matsui, A., Tanaka, M., Nakamura, T., Abe, T., Sako, K., et al. (2017). The distinct roles of class I and II RPD3-like histone deacetylases in salinity stress response. Plant Physiol. 175, 1760–1773. doi: 10.1104/pp.17.01332
Ueda, M., Seki, M. (2020). Histone modifications form epigenetic regulatory networks to regulate abiotic stress Response1 [OPEN]. Plant Physiol. 182, 15–26. doi: 10.1104/pp.19.00988
Umezawa, T., Sugiyama, N., Takahashi, F., Anderson, J. C., Ishihama, Y., Peck, S. C., et al. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in arabidopsis thaliana. Sci. Signal. 6, rs8–rs8. doi: 10.1126/scisignal.2003509
Vaillant, I., Paszkowski, J. (2007). Role of histone and DNA methylation in gene regulation. Curr. Opin. Plant Biol. 10, 528–533. doi: 10.1016/j.pbi.2007.06.008
van Dijk, K., Ding, Y., Malkaram, S., Riethoven, J.-J. M., Liu, R., Yang, J., et al. (2010). Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in arabidopsis thaliana. BMC Plant Biol. 10, 238. doi: 10.1186/1471-2229-10-238
Villagómez-Aranda, A. L., García-Ortega, L. F., Torres-Pacheco, I., Guevara-Gonzalez, R. G. (2021). Whole-genome DNA methylation analysis in hydrogen peroxide overproducing transgenic tobacco resistant to biotic and abiotic stresses. Plants 10, 178. doi: 10.3390/plants10010178
Voigt, P., Tee, W.-W., Reinberg, D. (2013). A double take on bivalent promoters. Genes Dev. 27, 1318–1338. doi: 10.1101/gad.219626.113
Wang, Z., Cao, H., Sun, Y., Li, X., Chen, F., Carles, A., et al. (2013). Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid–ethylene antagonism mediated by histone deacetylation. Plant Cell 25, 149–166. doi: 10.1105/tpc.112.108191
Wang, Z., Casas-Mollano, J. A., Xu, J., Riethoven, J.-J. M., Zhang, C., Cerutti, H. (2015). Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of arabidopsis thaliana. Proc. Natl. Acad. Sci. 112, 8487–8492. doi: 10.1073/pnas.1423325112
Wang, Z., Chivu, A. G., Choate, L. A., Rice, E. J., Miller, D. C., Chu, T., et al. (2022). Prediction of histone post-translational modification patterns based on nascent transcription data. Nat. Genet. 54, 295–305. doi: 10.1038/s41588-022-01026-x
Wang, F., Higgins, J. M. G. (2013). Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol. 23, 175–184. doi: 10.1016/j.tcb.2012.11.005
Wang, Q., Liu, P., Jing, H., Zhou, X. F., Zhao, B., Li, Y., et al. (2021). JMJ27-mediated histone H3K9 demethylation positively regulates drought-stress responses in arabidopsis. New Phytol. 232, 221–236. doi: 10.1111/nph.17593
Wang, Y., Long, H., Yu, J., Dong, L., Wassef, M., Zhuo, B., et al. (2018). Histone variants H2A.Z and H3.3 coordinately regulate PRC2-dependent H3K27me3 deposition and gene expression regulation in mES cells. BMC Biol. 16, 107. doi: 10.1186/s12915-018-0568-6
Wang, L., Qiao, H. (2020). Chromatin regulation in plant hormone and plant stress responses. Curr. Opin. Plant Biol. 57, 164–170. doi: 10.1016/j.pbi.2020.08.007
Wang, Y., Wysocka, J., Sayegh, J., Lee, Y.-H., Perlin, J. R., Leonelli, L., et al. (2004). Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306, 279–283. doi: 10.1126/science.1101400
Wang, T., Xing, J., Liu, Z., Zheng, M., Yao, Y., Hu, Z., et al. (2019). Histone acetyltransferase GCN5-mediated regulation of long non-coding RNA At4 contributes to phosphate starvation response in arabidopsis. J. Exp. Bot. 70, 6337–6348. doi: 10.1093/jxb/erz359
Widiez, T., Symeonidi, A., Luo, C., Lam, E., Lawton, M., Rensing, S. A. (2014). The chromatin landscape of the moss physcomitrella patens and its dynamics during development and drought stress. Plant J. Cell Mol. Biol. 79, 67–81. doi: 10.1111/tpj.12542
Wolffe, A. P., Hayes, J. J. (1999). Chromatin disruption and modification. Nucleic Acids Res. 27, 711–720. doi: 10.1093/nar/27.3.711
Wollmann, H., Holec, S., Alden, K., Clarke, N. D., Jacques, P.-É., Berger, F. (2012). Dynamic deposition of histone variant H3.3 accompanies developmental remodeling of the arabidopsis transcriptome. PloS Genet. 8, e1002658. doi: 10.1371/journal.pgen.1002658
Wollmann, H., Stroud, H., Yelagandula, R., Tarutani, Y., Jiang, D., Jing, L., et al. (2017). The histone H3 variant H3.3 regulates gene body DNA methylation in arabidopsis thaliana. Genome Biol. 18, 94. doi: 10.1186/s13059-017-1221-3
Wu, K., Zhang, L., Zhou, C., Yu, C.-W., Chaikam, V. (2008). HDA6 is required for jasmonate response, senescence and flowering in arabidopsis. J. Exp. Bot. 59, 225–234. doi: 10.1093/jxb/erm300
Xiao, J., Lee, U.-S., Wagner, D. (2016). Tug of war: adding and removing histone lysine methylation in arabidopsis. Curr. Opin. Plant Biol. 34, 41–53. doi: 10.1016/j.pbi.2016.08.002
Xiao, J., Wagner, D. (2015). Polycomb repression in the regulation of growth and development in arabidopsis. Curr. Opin. Plant Biol. 23, 15–24. doi: 10.1016/j.pbi.2014.10.003
Xiao, J., Zhang, H., Xing, L., Xu, S., Liu, H., Chong, K., et al. (2013). Requirement of histone acetyltransferases HAM1 and HAM2 for epigenetic modification of FLC in regulating flowering in arabidopsis. J. Plant Physiol. 170, 444–451. doi: 10.1016/j.jplph.2012.11.007
Xiao, S., Jiang, L., Wang, C., Ow, D. W.. (2021). Arabidopsis OXS3 family proteins repress ABA signaling through interactions with AFP1 in the regulation of ABI4 expression. J. Exp. Botany 72, 5721–5734. doi: 10.1093/jxb/erab237
Xu, D., Huang, W., Li, Y., Wang, H., Huang, H., Cui, X. (2012). Elongator complex is critical for cell cycle progression and leaf patterning in arabidopsis. Plant J. Cell Mol. Biol. 69, 792–808. doi: 10.1111/j.1365-313X.2011.04831.x
Yamamoto, Y. (2019). Aluminum toxicity in plant cells: Mechanisms of cell death and inhibition of cell elongation. Soil Sci. Plant Nutr. 65, 41–55. doi: 10.1080/00380768.2018.1553484
Yan, A., Borg, M., Berger, F., Chen, Z. (2020). The atypical histone variant H3.15 promotes callus formation in arabidopsis thaliana. Development 147, dev184895. doi: 10.1242/dev.184895
Yang, H., Mo, H., Fan, D., Cao, Y., Cui, S., Ma, L. (2012). Overexpression of a histone H3K4 demethylase, JMJ15, accelerates flowering time in arabidopsis. Plant Cell Rep. 31, 1297–1308. doi: 10.1007/s00299-012-1249-5
Yao, C., Li, H., Shen, X., He, Z., He, L., Guo, Z. (2012). Reproducibility and concordance of differential DNA methylation and gene expression in cancer. PloS One 7, e29686. doi: 10.1371/journal.pone.0029686
Yelagandula, R., Stroud, H., Holec, S., Zhou, K., Feng, S., Zhong, X., et al. (2014). The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in arabidopsis. Cell 158, 98–109. doi: 10.1016/j.cell.2014.06.006
Yu, C.-W., Liu, X., Luo, M., Chen, C., Lin, X., Tian, G., et al. (2011). HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS d and regulates flowering in arabidopsis. Plant Physiol. 156, 173–184. doi: 10.1104/pp.111.174417
Yung, W., Li, M., Sze, C., Wang, Q., Lam, H. (2021). Histone modifications and chromatin remodelling in plants in response to salt stress. Physiol. Plant 173, 1495–1513. doi: 10.1111/ppl.13467
Zacharaki, V., Benhamed, M., Poulios, S., Latrasse, D., Papoutsoglou, P., Delarue, M., et al. (2012). The arabidopsis ortholog of the YEATS domain containing protein YAF9a regulates flowering by controlling H4 acetylation levels at the FLC locus. Plant Sci. 196, 44–52. doi: 10.1016/j.plantsci.2012.07.010
Zemach, A., Kim, M. Y., Hsieh, P.-H., Coleman-Derr, D., Eshed-Williams, L., Thao, K., et al. (2013). The arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153, 193–205. doi: 10.1016/j.cell.2013.02.033
Zeng, Z., Zhang, W., Marand, A. P., Zhu, B., Buell, C. R., Jiang, J. (2019). Cold stress induces enhanced chromatin accessibility and bivalent histone modifications H3K4me3 and H3K27me3 of active genes in potato. Genome Biol. 20, 123. doi: 10.1186/s13059-019-1731-2
Zhang, X., Bernatavichute, Y. V., Cokus, S., Pellegrini, M., Jacobsen, S. E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in arabidopsis thaliana. Genome Biol. 10, R62. doi: 10.1186/gb-2009-10-6-r62
Zhang, H., Lang, Z., Zhu, J.-K. (2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506. doi: 10.1038/s41580-018-0016-z
Zhang, K., Mosch, K., Fischle, W., Grewal, S. I. S. (2008). Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 15, 381–388. doi: 10.1038/nsmb.1406
Zhang, Y., Reinberg, D. (2001). Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360. doi: 10.1101/gad.927301
Zhang, C. Y., Wang, N. N., Zhang, Y. H., Feng, Q. Z., Yang, C. W., Liu, B. (2013). DNA Methylation involved in proline accumulation in response to osmotic stress in rice (Oryza sativa). Genet. Mol. Res. GMR 12, 1269–1277. doi: 10.4238/2013.April.17.5
Zhang, Y.-Z., Yuan, J., Zhang, L., Chen, C., Wang, Y., Zhang, G., et al. (2020). Coupling of H3K27me3 recognition with transcriptional repression through the BAH-PHD-CPL2 complex in arabidopsis. Nat. Commun. 11, 6212. doi: 10.1038/s41467-020-20089-0
Zhang, Z., Zhang, S., Zhang, Y., Wang, X., Li, D., Li, Q., et al. (2011). Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 23, 396–411. doi: 10.1105/tpc.110.081356
Zhao, K., Kong, D., Jin, B., Smolke, C. D., Rhee, S. Y. (2021). A novel bivalent chromatin associates with rapid induction of camalexin biosynthesis genes in response to a pathogen signal in arabidopsis. eLife 10, e69508. doi: 10.7554/eLife.69508
Zheng, B., Chen, X. (2011). Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr. Opin. Plant Biol. 14, 123–129. doi: 10.1016/j.pbi.2011.01.001
Zheng, Y., Ding, Y., Sun, X., Xie, S., Wang, D., Liu, X., et al. (2016). Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in arabidopsis. J. Exp. Bot. 67, 1703–1713. doi: 10.1093/jxb/erv562
Zheng, M., Liu, X., Lin, J., Liu, X., Wang, Z., Xin, M., et al. (2019). Histone acetyltransferase GCN5 contributes to cell wall integrity and salt stress tolerance by altering the expression of cellulose synthesis genes. Plant J. Cell Mol. Biol. 97, 587–602. doi: 10.1111/tpj.14144
Zhou, B.-R., Feng, H., Kato, H., Dai, L., Yang, Y., Zhou, Y., et al. (2013). Structural insights into the histone H1-nucleosome complex. Proc. Natl. Acad. Sci. 110, 19390–19395. doi: 10.1073/pnas.1314905110
Zhou, B.-R., Jiang, J., Feng, H., Ghirlando, R., Xiao, T. S., Bai, Y. (2015). Structural mechanisms of nucleosome recognition by linker histones. Mol. Cell 59, 628–638. doi: 10.1016/j.molcel.2015.06.025
Zhou, J., Liu, D., Wang, P., Ma, X., Lin, W., Chen, S., et al. (2018). Regulation of arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc. Natl. Acad. Sci. U. S. A. 115, E1906–E1915. doi: 10.1073/pnas.1712251115
Zhou, Y., Romero-Campero, F. J., Gómez-Zambrano, Á., Turck, F., Calonje, M. (2017). H2A monoubiquitination in arabidopsis thaliana is generally independent of LHP1 and PRC2 activity. Genome Biol. 18, 69. doi: 10.1186/s13059-017-1197-z
Zhou, B., Zeng, L. (2017). Conventional and unconventional ubiquitination in plant immunity. Mol. Plant Pathol. 18, 1313–1330. doi: 10.1111/mpp.12521
Zhou, C., Zhang, L., Duan, J., Miki, B., Wu, K. (2005). HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in arabidopsis. Plant Cell 17, 1196–1204. doi: 10.1105/tpc.104.028514
Zilberman, D., Coleman-Derr, D., Ballinger, T., Henikoff, S. (2008). Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456, 125–129. doi: 10.1038/nature07324
Zou, C., Mallampalli, R. K. (2014). Regulation of histone modifying enzymes by the ubiquitin–proteasome system. Biochim. Biophys. Acta BBA Mol. Cell Res. 1843, 694–702. doi: 10.1016/j.bbamcr.2013.12.016
Keywords: chromatin, epigenetics, histone, histone variant, histone modification, acetylation, methylation, abiotic stress
Citation: Nunez-Vazquez R, Desvoyes B and Gutierrez C (2022) Histone variants and modifications during abiotic stress response. Front. Plant Sci. 13:984702. doi: 10.3389/fpls.2022.984702
Received: 02 July 2022; Accepted: 28 September 2022;
Published: 15 December 2022.
Edited by:
Mahmoud Yaish, Sultan Qaboos University, OmanReviewed by:
Namisha Sharma, Institute of Life Sciences (ILS), IndiaCopyright © 2022 Nunez-Vazquez, Desvoyes and Gutierrez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Crisanto Gutierrez, Y2d1dGllcnJlekBjYm0uY3NpYy5lcw==; Bénédicte Desvoyes, YmRlc3ZveWVzQGNibS5jc2ljLmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.