
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 25 August 2022
Sec. Plant Metabolism and Chemodiversity
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.959698
This article is part of the Research Topic Plant Chemoecology: Integrating Micro- and Macrolevel Approaches in Regulating Secondary Metabolism View all 8 articles
A correction has been applied to this article in:
Corrigendum: Tartary buckwheat FtF3′H1 as a metabolic branch switch to increase anthocyanin content in transgenic plant
Tartary buckwheat (TB) is a pseudocereal rich in flavonoids, mainly including flavonols and anthocyanins. The flavonoid 3′-hydroxylase (F3′H) is a key enzyme in flavonoid biosynthesis and is encoded by two copies in TB genome. However, its biological function and effects on flavonol and anthocyanin synthesis in TB have not been well validated yet. In this study, we cloned the full-length FtF3′H1 gene highly expressed in all tissues (compared with FtF3′H2) according to TB flowering transcriptome data. The corresponding FtF3′H1 protein contains 534 amino acids with the molecular properties of the typical plant F3′H and belongs to the CYP75B family. During the flowering stage, the FtF3′H1 expression was highest in flowers, and its expression pattern showed a significant and positive correlation with the total flavonoids (R2 > 0.95). The overexpression of FtF3′H1 in Arabidopsis thaliana, Nicotiana tabacum and TB hairy roots resulted in a significant increase in anthocyanin contents (p < 0.05) but a decrease in rutin (p < 0.05). The average anthocyanin contents were 2.94 mg/g (fresh weight, FW) in A. thaliana (about 135% increase), 1.18 mg/g (FW) in tobacco (about 17% increase), and 1.56 mg/g (FW) TB hairy roots (about 44% increase), and the rutin contents were dropped to about 53.85, 14.99, 46.31%, respectively. However, the expression of genes involved in anthocyanin (DFRs and ANSs) and flavonol (FLSs) synthesis pathways were significantly upregulated (p < 0.05). In particular, the expression level of DFR, a key enzyme that enters the anthocyanin branch, was upregulated thousand-fold in A. thaliana and in N. tabacum. These results might be attributed to FtF3′H1 protein with a higher substrate preference for anthocyanin synthesis substrates. Altogether, we identified the basic biochemical activity of FtF3′H1 in vivo and investigated its involvement in anthocyanin and flavonol metabolism in plant.
Tartary buckwheat (Fagopyrum tataricum Gaertn.) belongs to the Fagopyrum genus (Polygonaceae), mainly being cultivated in Asian countries. It is a brilliant cereal with edible and medicinal characteristics. Enriched flavonoids provide the pharmacological activity for TB (Li et al., 2012b; Dong et al., 2020; Park et al., 2021). As a focus in the secondary metabolism of plants, flavonoids not only endow various colors including red, blue, and purple as pigmentation resources in plants, but also are associated with health as active compounds in vegetables and fruits (Grotewold, 2006). In addition, flavonoids are well-known antioxidants that have numerous pharmacologic properties such as anti-inflammatory, anticarcinogenic, antimetastatic, and other effects (Li et al., 2012a).
Since the mid-1980s, biochemical information about flavonoid enzymes has proliferated, and the biosynthesis, accumulation, core enzymes, and genes involved in flavonoid biosynthesis are now widely described in different species through the flavonoid biosynthesis pathway, such as Arabidopsis thaliana, Malus, Fragaria vesca, and so on (Prescott and John, 1996; Han et al., 2010; Thill et al., 2013; Wu et al., 2020). In the model plant A thaliana, at least 54 flavonoid molecules are found and more than 30 genes are involved in their biosynthesis (Routaboul et al., 2006). The enzyme-coding genes Phenylalanine ammonia-lyase (PAL; Shufflebottom et al., 1993), Cinnamic acid 4-hydroxylase (C4H; Mizutani et al., 1997), 4-coumarate CoA ligase (4CL; Hamberger and Hahlbrock, 2004), Chalcone synthase (CHS; Austin and Noel, 2003), Chalcone isomerase (CHI; Winkel-Shirley, 2001), Flavanone 3-hydroxylase (F3H; Pelletier and Shirley, 1996), Flavonoid 3′-hydroxylase (F3′H; Schoenbohm et al., 2000), Flavonol synthase (FLS; Prescott et al., 2002), Dihydroflavonol 4-reductase (DFR; Shirley et al., 1992), Leucoanthocyanidin dioxygenase/anthocyanidin synthase (LDOX/ANS; Saito et al., 1999; Abrahams et al., 2003), Anthocyanidin reductase (ANR; Devic et al., 1999), and Polyphenol oxidase (PPO; Pourcel et al., 2007) constitute the central flavonoid biosynthetic pathway in A thaliana. The flavonoid 3′-hydroxylase (F3′H) is a vital enzyme in the flavonoid biosynthesis pathway (Figure 1). It belongs to a member of cytochrome P450 monooxygenase (CYP) superfamily, which catalyzes NADPH-dependent hydroxylation of substrates and associates to various metabolic pathways, including flavonoid biosynthesis (Awasthi et al., 2016). The main function of F3′H is to increase water solubility by catalyzing the hydroxylation of C atoms in the B-ring (Grotewold, 2006). CYP encoded by F3′H has been well explored in a variety of plants and its function has been demonstrated. For example, F3′H overproduction induced the F3′H-and F3′5′H-deficient pale-pink petunia to produce fuchsia flowers due to the accumulated anthocyanidins (Tanaka, 2006). In Actinidia (Peng et al., 2019), Arachis hypogaea (Xue et al., 2021), Brassica rapa (Park et al., 2021), and other plants, the expression of F3′H was strongly correlated with the content of different flavonoids, respectively. Obviously, F3′H is involved in multiple actions and has a significant effect on flavonoid metabolism in plants.
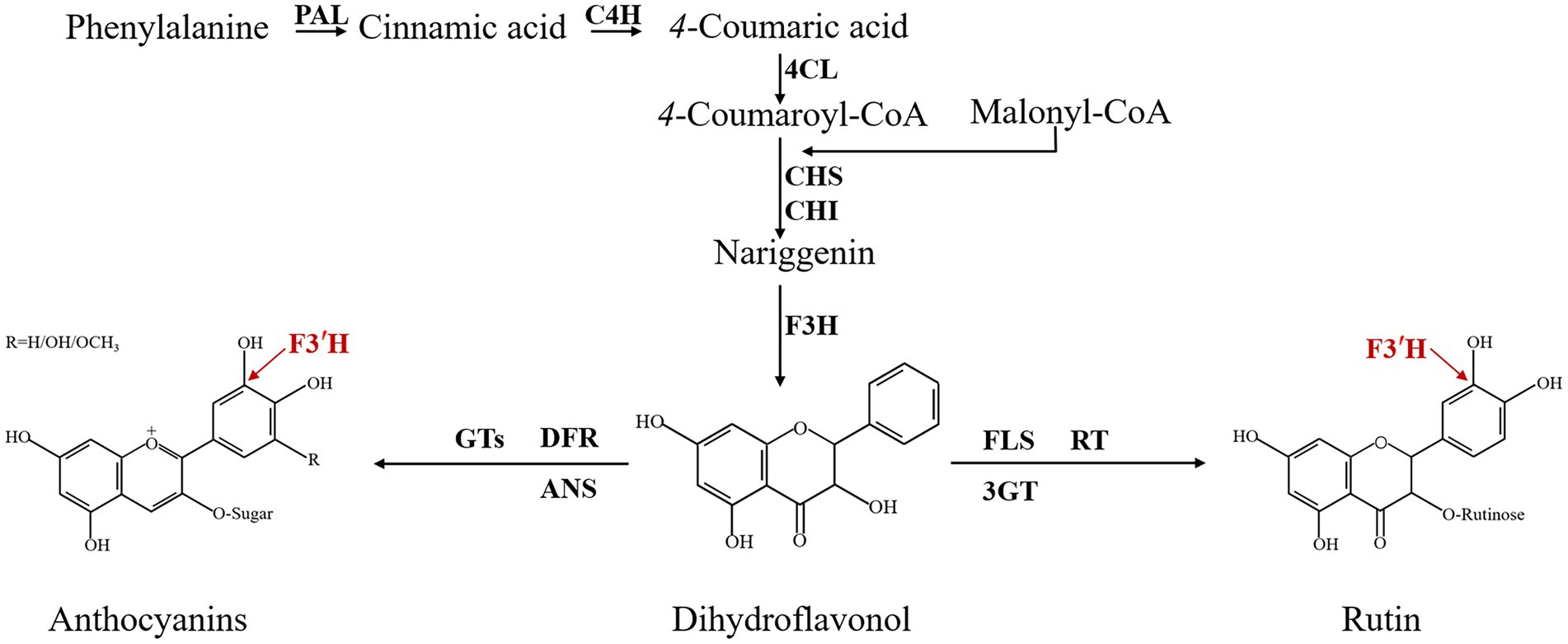
Figure 1. Flavonoid biosynthesis pathway of TB. PAL: phenylalanine ammonialyase; C4H: cinnamate 4-hydroxylase; 4CL: 4-coumarate CoA ligase; CHS: chalcone synthase; CHI: chalcone isomerase; F3H: flavanone 3-hydroxylase; F3′H: flavonoid 3′-hydroxylase; DFR: dihydroflavonol 4-reductase; FLS: flavonol synthase; ANS: anthocyanidin synthase; GTs: glucosyl transferases; 3GT: flavonoid 3-O-glucosyltransferase; RT: 3-O-rhamnosyltransferase.
Tartary buckwheat contains distinct types of flavonoids, the most abundant active biological compounds (Peng et al., 2015; Huda et al., 2021; Li et al., 2022). Flavonols, represented by rutin, are the main type of TB flavonoids, accounting for 1.7% of the dry weight of TB grains. Besides, anthocyanins largely accumulate in the TB stems and leaves, thereby strengthening its external stress-resistance (Fabjan et al., 2003; Park et al., 2011). Hence, it is an indispensable process to promote TB flavonoid accumulation and identify the associated key enzyme genes in the biosynthetic flavonoid pathway. To date, studies on TB genome, transcriptome, and metabolome, have provide a new perspective on molecular regulation of flavonoid synthesis in TB (Gao et al., 2016; Yao et al., 2016; Huang et al., 2019; Liu et al., 2019; Hou et al., 2021). However, the biological function of some genes such as FtF3′H1 has not been identified, which impedes to clarify the metabolic branches of flavonoids synthesis in TB. In our previous work, FtF3′H1 gene containing the conserved motif (GGER/K) was successfully cloned by RACE. However, the corresponding sequence lacked about 40 amino acids at the N-terminal, thereby resulting in unidentified biological activity (Li et al., 2014). In this study, the complete FtF3′H coding sequence was isolated from TB, and its activity and function were analyzed by performing homologous and heterologous expression experiments, respectively. These results will improve the acknowledge of the flavonoid metabolic pathways in TB, subsequently promote the accumulation of natural medicinal metabolites and contribute to the molecular breeding of TB.
The TB cultivar., Xiqiao 2, was provided by Professor An’hu Wang from Xichang University. TB seeds were grown in farm field of Sichuan Agriculture University at Ya’an, Sichuan. TB roots, stems, leaves, flowers, and seeds during plant flowering were collected to investigate the total flavonoid content and relative expression level of FtF3′H1. The corresponding samples were frozen in liquid nitrogen, and stored at –80°C.
Nicotiana tabacum “NC89” and Arabidopsis thaliana ecotype Columbia-0 (Col-0), tt7 mutant an Arabidopsis line lacking F3′H gene function were provided by Professor Jinwen Zhang from Gansu Agricultural University and Professor Yi Cai from Sichuan Agricultural University, respectively. N. tabacum and A thaliana were germinated in a greenhouse with 16 h photoperiod. Additionally, TB seeds were sterilized with 70% ethanol for 90 s and HgCl solution for 10 min and then rinsed several times in sterile water. These seeds were grown on agar plates containing sucrose-free half Murashige and Skoog (½ MS) medium with pH5.8 and 0.8% agar at 25°C with 16 h photoperiod for subsequent hair roots preparation.
Based on TB genome sequence and transcriptome data (Yao et al., 2017a; Zhang et al., 2018), we obtained the full-length cDNA sequence of FtF3′H1 by RT-PCR. The amplified PCR fragments were cloned into pMD@19-T vector and then sequenced. The amino acid sequence of FtF3′H1 was deduced by DNAman software (version 9.0), following with the homology analysis performed using MEGA software (version 7.0). Continually, the genetic structure was predicted by the online software GSD2.0.1 The physicochemical characteristics of the deduced FtF3′H1 protein, including molecular formula, relative molecular weight, and isoelectric point were characterized using the online website ExPASy ProtParam tool.2 Predicted molecular docking models of FtF3′H1 with substrates were obtained with the MOE (version 2019) software.
The ORF of FtF3′H1 was amplified and inserted in to pGEX-4T. Then the recombinant plasmid of pGEX-4 T-FtF3′H1 was transformed into E. coli BL21(DE3). A single colony was inoculated and cultivated in LB medium supplemented with kanamycin (50 μg/ml) at 37°C in the incubator with 220 rpm. When the OD600 of the bacterial culture reached to 0.6, 0.2 mmol/l isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce the expression of FtF3′H1 fused with a GST-tag at 18°C for 8 h.
To investigate the potential function of the FtF3′H1 recombinant protein, we performed affinity chromatography to purify it with a BeyoGold™ GST labeling Resin kit. Continually, the purified FtF3′H1 was supplemented with 100 mmol/l KH2PO4-NaOH buffer (pH = 0.8), 20 mmol/l β-mercaptoethanol, 1.25 mmol/l NADPH, and then mixed with 100 mmol/l kaempferol and 100 mmol/l dihydrokaempferol as the substrate. Enzyme activity was determined under conditions at 30°C for 1 h, and an equal volume of ethyl acetate was used to extract reaction compounds. Subsequently, the extracted precipitation was dissolved in 100 ml of 80% methanol solution and then determined by High-performance liquid chromatography (HPLC).
The FtF3′H1 ORF was cloned into the constitutive expression vector pCHF3-eYFP driven by 35S promoter. The recombinant plasmid pCHF3-FtF3′H1-eYFP was transformed into the strain Agrobacterium tumefaciens strains GV3101 and A4. GV3101 was used to transform Arabidopsis and N. tabacum, and A4 for TB to generate transgenic plants. The transgenic Arabidopsis T3 lines were selected on MS medium with 50 mg/ml kanamycin (Huang et al., 2019), and the N. tabacum T3 lines were obtained on MS medium with 100 mg/ml kanamycin. Moreover, the FtF3′H-overexpressing TB hairy root lines were selected in ½ MS liquid medium with 50 mg/ml kanamycin (Huang et al., 2016).
The flavonoid and anthocyanin were extracted from TB tissues (root, stem, leaf, flower, and seed), transgenic Arabidopsis lines, N. tabacum flowers and TB hairy roots, respectively, using the method as described previously (Liu and Zhu, 2007; Yao et al., 2017b). The total flavonoid and anthocyanin contents were determined by a spectrophotometry as previously reported (Liu and Zhu, 2007; Zhang et al., 2009). HPLC was performed to determine rutin content according to the reported method (Yao et al., 2020). Three biological repeats were set for each experimental group.
To determine the expression levels of flavonoid-associated genes in different samples, total RNA was extracted using RNAout Kit (TIANGEN, China). Quantitative real-time PCR (qRT-PCR) was performed using the PreMix Ex Taq II kit (Tli RNAseH Plus) and with CFX96 RT-PCR machine (Bio-Rad, United States). The PCR amplification procedure was as follows: 95°C for 10 s, 40 cycles at 95°C for 5 s, 61°C for 30 s. The housekeeping gene FtH3 (GenBank ID: HM628903 for TB), β-Actin2 (GenBank ID: AF149413 for Arabidopsis), and β-actin (GenBank ID: AB158612 for N. tabacum) were used as the internal standards, respectively. The analysis of each sample was repeated three times. Relative expression level was calculated using the 2−ΔΔCt method.
All the primer sequences used in this study are shown in Supplementary Table S1. Data were analyzed using the SPSS 19.0 statistical software. Significant differences were indicated when p < 0.05.
Based on TB transcriptome data, two different transcripts were annotated as F3′H, named FtF3′H1 and FtF3′H2. To investigate the role of two FtF3′Hs in flavonoid biosynthesis, we performed a correlation analysis between the expression level and flavonoid content in different tissues of flowering TB. Results illustrated that FtF3′H2 revealed traces of expression in all TB tissues except roots, however, FtF3′H1 showed a high expression pattern (Supplementary Figure S1). Thus, FtF3′H1 was selected as the candidate gene for further characterization.
Analysis of chromosome location showed that the FtF3′H1 was located on chromosome 8 in TB (Supplementary Figure S2A). Sequence analysis revealed that FtF3′H1 contained three exons and two introns, encoding a peptide containing 534 amino acids (Supplementary Figure S2C). The deduced protein molecular formula is C2655H4212N724O755S23, with a predicted relative molecular weight of 59.09 kDa and the theoretical pI of 7.69. Compared with the P450 proteins of Arabidopsis, results revealed that FtF3′H1 closely related to AtCYP75B1, which was the only gene encoding F3′H protein and regulated anthocyanin accumulation in Arabidopsis (Figure 2A). Basing on the characterized plant F3′Hs, the evolutionary tree showed that FtF3′H1 was classified into a member of the CYP75B subfamily (Supplementary Figure S2B). Furthermore, sequence alignment exhibited that FtF3′H1 had a high homology (about 80%) with F3′H sequence from A. thaliana, sweet potato, Malus domestica, and Petunia hybrida (Figure 2B). In addition, FtF3′H1 contains four typically conserved motifs of cytochrome P450 and three specifically conserved motifs of F3′H protein, suggesting that FtF3′H1 might have similar biological function in other species. Importantly, the interaction analysis emphasized that FtF3′H1 successfully docked to the substrates of dihydrokaempferol and kaempferol (Figure 2C). Altogether, the above data systematically revealed the molecular features of FtF3′H1 and suggested that FtF3′H1 might be involved in the regulation of flavonoid biosynthesis in TB.
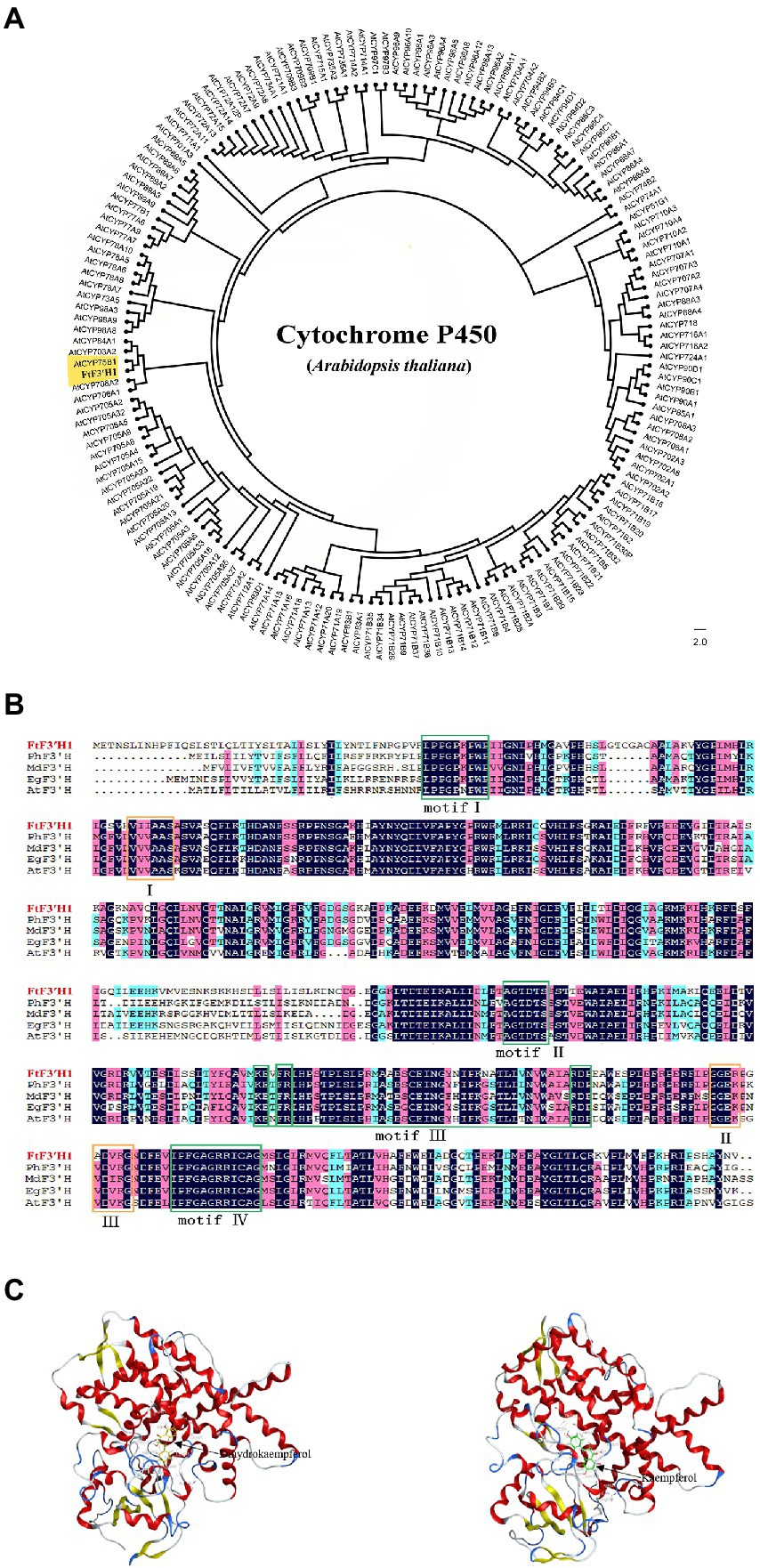
Figure 2. Structural features and phylogenetic analysis of the FtF3′H1 protein. (A) Molecular phylogenetic tree of F3′H proteins by the maximum likelihood method. (B) Sequence alignment of the deduced FtF3′H1 with F3′H proteins from other species, motifI (PPGPTPWP), motifII (FGAGRRICAG), motifIII (E-R-R), motifIV (AGTDTS), I (VVVAAS), II (GGEK), III (VDVKG). GenBank accession numbers for the proteins in the alignment are as follows: Petunia (AAD56282.1), Malus (ACR14867.1), Eustoma grandiflorum (BAP94456.1), Arabidopsis thaliana (CAB62611.1). (C) Docking prediction for FtF3′H1 protein with Dihydrokaempferol and Kaempferol.
To further understand the relationship between FtF3′H1 expression and flavonoid content in TB. The flavonoids were extracted and measured from roots, stems, leaves, flowers, and seeds of TB. Correspondingly, the expression pattern of FtF3′H1 in five tissues was analyzed using qRT-PCR (Figure 3). The results showed a positive correlation between flavonoid content and FtF3′H1 expression pattern (R2 > 0.95), which were highest in flowers, followed by roots, leaves, seeds, and stems in a decreasing order. Our results were also strongly correlated with the data obtained previously (Yao et al., 2017a; R2 > 0.95).

Figure 3. Relationship between flavonoid content and FtF3′H1 transcript level in different tissues of TB at flowering stage. The expression of FtF3′H1 was determined by qPCR. FtH3 gene was used as the reference gene. Determination of total flavonoids of TB by UV spectrophotometer. The average value is calculated repeatedly by three times of technology. Error bars indicate standard deviations (± SD).
To directly observe the effect of FtF3′H1 on flavonoid biosynthesis, FtF3′H1-overproducing lines of Arabidopsis, and tt7 mutant, N. tabacum and TB hair roots were generated (Supplementary Figure S4).
In the transgenic Arabidopsis lines (OE1 ~ 3), the seeds visually showed deeper pigmentation than WT (Figure 4A), which might be attributed to the increased anthocyanin from its active biosynthesis. Data showed that the average anthocyanin content was 2.94 mg/g (fresh weight, FW) and significantly increased than WT (p < 0.05), while the rutin content was significantly reduced (p < 0.05; Figure 4B). Accordingly, the expression levels of flavonoid synthesis related gene, including AtCHS, AtCHI, AtF3′H, AtFLS, AtDFR, and AtANS, were upregulated (p < 0.05). Interestingly, the expression levels of AtDFR and AtANS, two key enzyme genes in the anthocyanin synthesis branch, were upregulated by thousand-folds, much higher than that of AtFLS in the flavonol synthesis pathway. The result might be the main reason for the increase in anthocyanin and decrease in rutin in OE1 ~ 3 (Figure 4C; p < 0.05). Similarly, in the tt7 mutant Arabidopsis lines (tt7::FtF3′H1-3), FtF3′H1 overexpression restored the seeds color of tt7 mutants (Figure 5A), increased anthocyanin and reduced rutin levels (Figure 5B), and upregulated expression of 5 genes involved in flavonoid synthesis (Figure 5C). Furthermore, the downregulation of AtFLS’s expression in tt7 mutant demonstrated that FtF3′H1 significantly enhanced the metabolic strength of anthocyanin synthesis branch in Arabidopsis (Figure 5C).
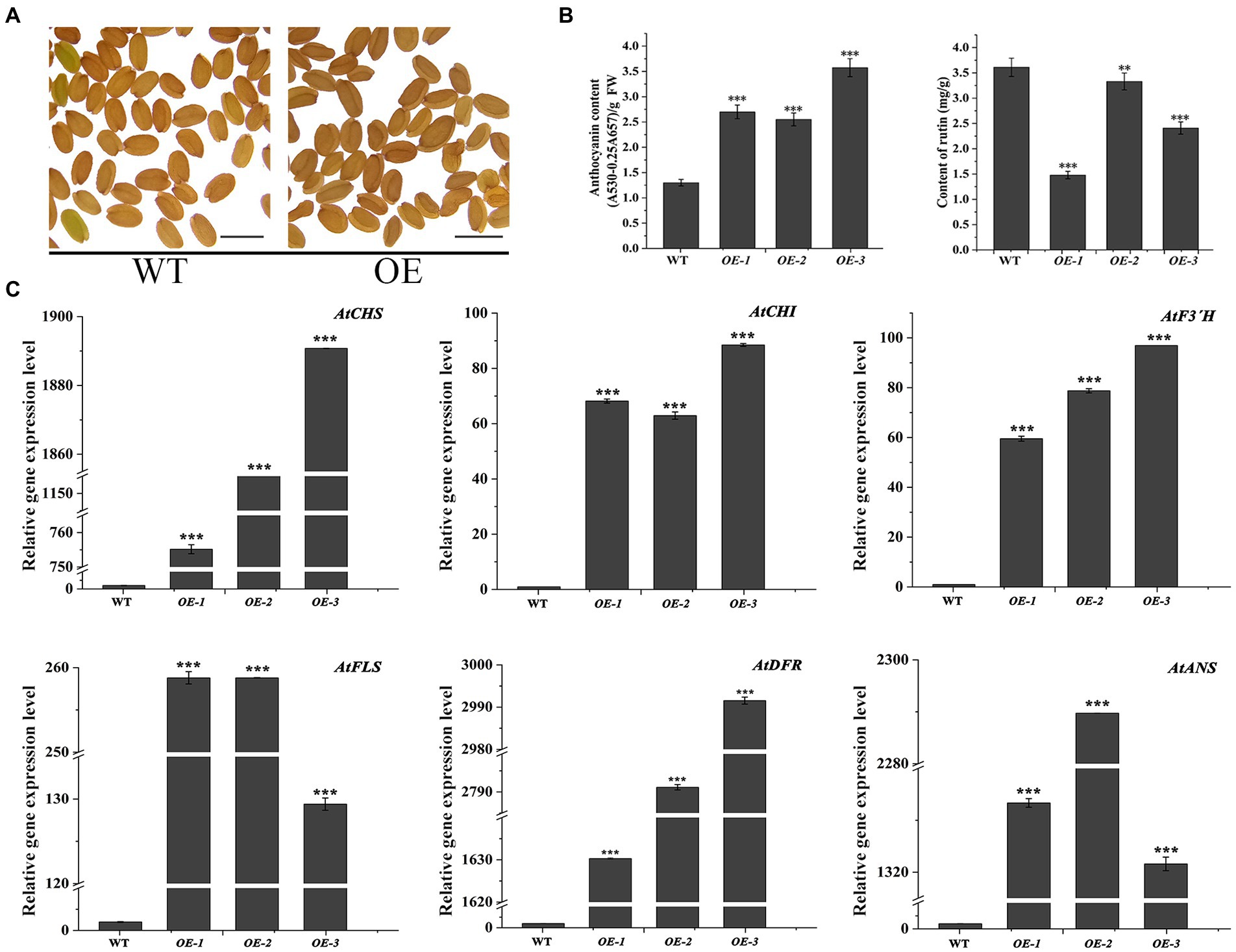
Figure 4. Overexpression of FtF3′H1 gene in Arabidopsis thaliana. (A) Seed phenotypes of transgenic A. thaliana overexpressing FtH3′H1 (OE) and WT (scale bar = 0.05 cm). (B) Determination of anthocyanin and rutin content in transgenic Arabidopsis lines OE-1, OE-2, and OE-3. WT is the control. Each value represents the mean of three replicates, and error bars indicate ± SD. **p < 0.01 and ***p < 0.001. (C) qPCR analysis of the expression levels of anthocyanin biosynthesis genes in A. thaliana with the FtF3′H1 overexpression. Atactin2 was used as the reference gene. Results represent mean values ± SD from three biological replicates. *p < 0.05, **p < 0.01 and ***p < 0.001.
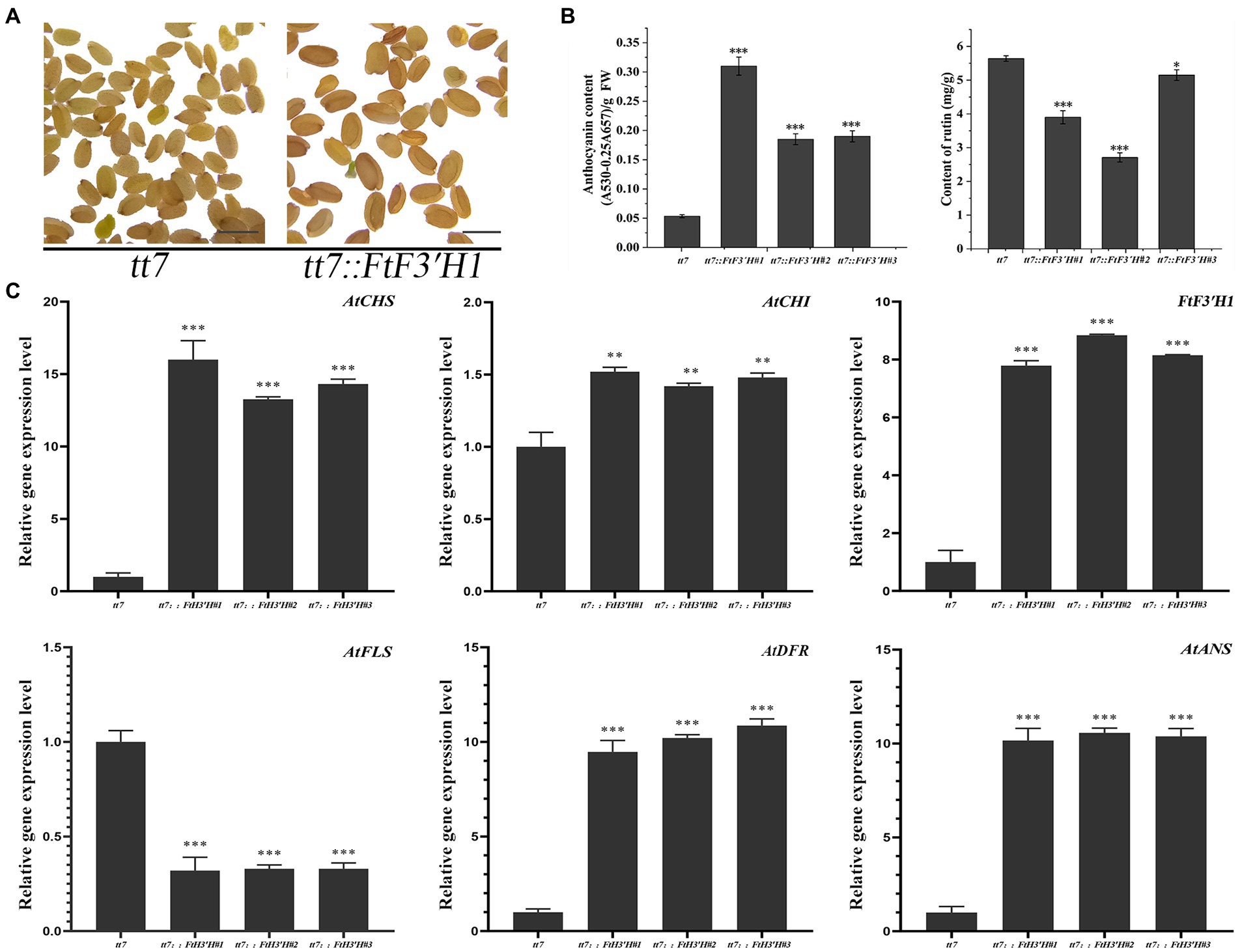
Figure 5. The original phenotype was restored after complementation of FtH3′H1 in tt7. (A) The seed coat color of A. thaliana tt7 with or without FtH3′H1 (scale bar = 0.05 cm). (B) Determination of anthocyanin and rutin content in the corresponding Arabidopsis as indicated. Each value represents the mean of three replicates, and error bars indicate ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001. (C) Transcription level of genes related to the anthocyanin biosynthesis in transgenic materials. Atactin2 was used as the reference gene. Results represent mean values ± SD from three biological replicates. *p < 0.05, **p < 0.01 and ***p < 0.001.
The transgenic N. tabacum lines (OE1 ~ 3) were used to confirm the biological feature of FtF3′H1. Compared to WT, FtF3′H1 transgenic N. tabacum leaves were longer and narrower, and the flowers color were redder (Figure 6A). The core enzyme genes of flavonoid biosynthesis were significantly upregulated (p < 0.05), and the NtDFR gene was most strongly expressed (Figure 6C; p < 0.05). To accurately validate the effects of FtF3′H1 in TB, TB hairy root lines overexpressing FtF3′H1 (OE1 ~ 3) were generated (Figure 7A). Consistent with the results in transgenic A. thaliana (Figure 5B) and N. tabacum (Figure 6B), pigmentation in TB hairy roots derived from anthocyanin accumulation, accompanied by a decrease in rutin content (Figure 7B; p < 0.05). qRT-PCR assays showed an increased expression levels of FtCHS, FtF3′H1, FtDFR, and FtANS, whereas decreased levels of FtCHI and FtFLS (Figure 7C). Taken together, FtF3′H1 is a key enzyme gene and branch switch for the metabolic pathway of flavonoids synthesis, and its overexpression can lead the metabolic flow to anthocyanin synthesis.
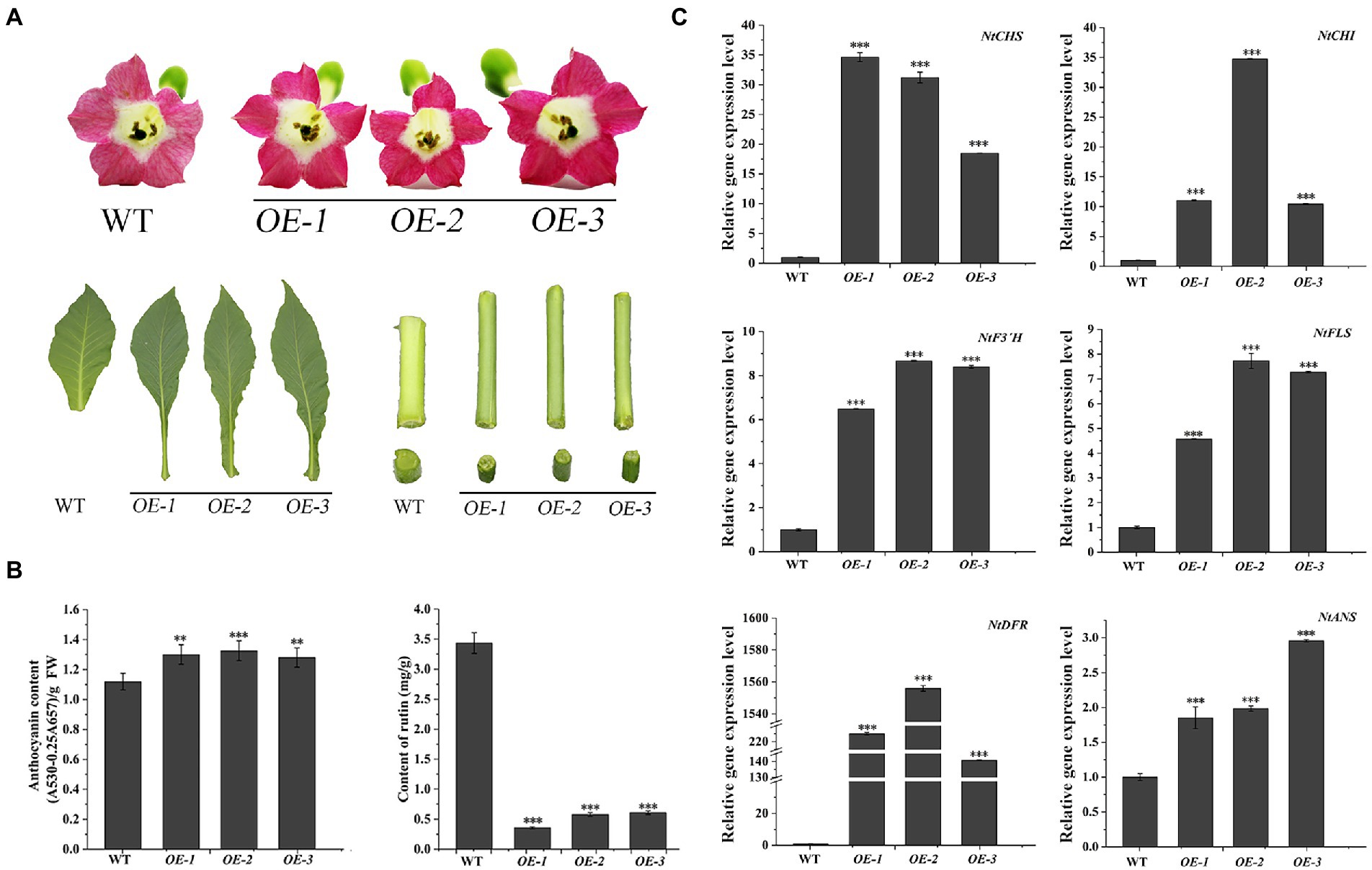
Figure 6. Overexpression of FtF3′H1 gene promotes the accumulation of anthocyanins and inhibits the accumulation of rutin in N. tabacum. (A) Flowers petal pigmentation, leaf and stem growth in transgenic N. tabacum lines OE-1, OE-2 and OE-3. WT is the control. (B) Anthocyanins and rutin contents in transgenic N. tabacum. The average value is calculated repeatedly by three times of technology, and error bars indicate ± SD. **p < 0.01 and ***p < 0.001. (C) Detection of transcriptional levels of flavonoid biosynthetic genes in transgenic N. tabacum. Ntβ-actin gene are used as a reference in N. tabacum. The average value is calculated repeatedly by three times of technology, and error bars indicate ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001.
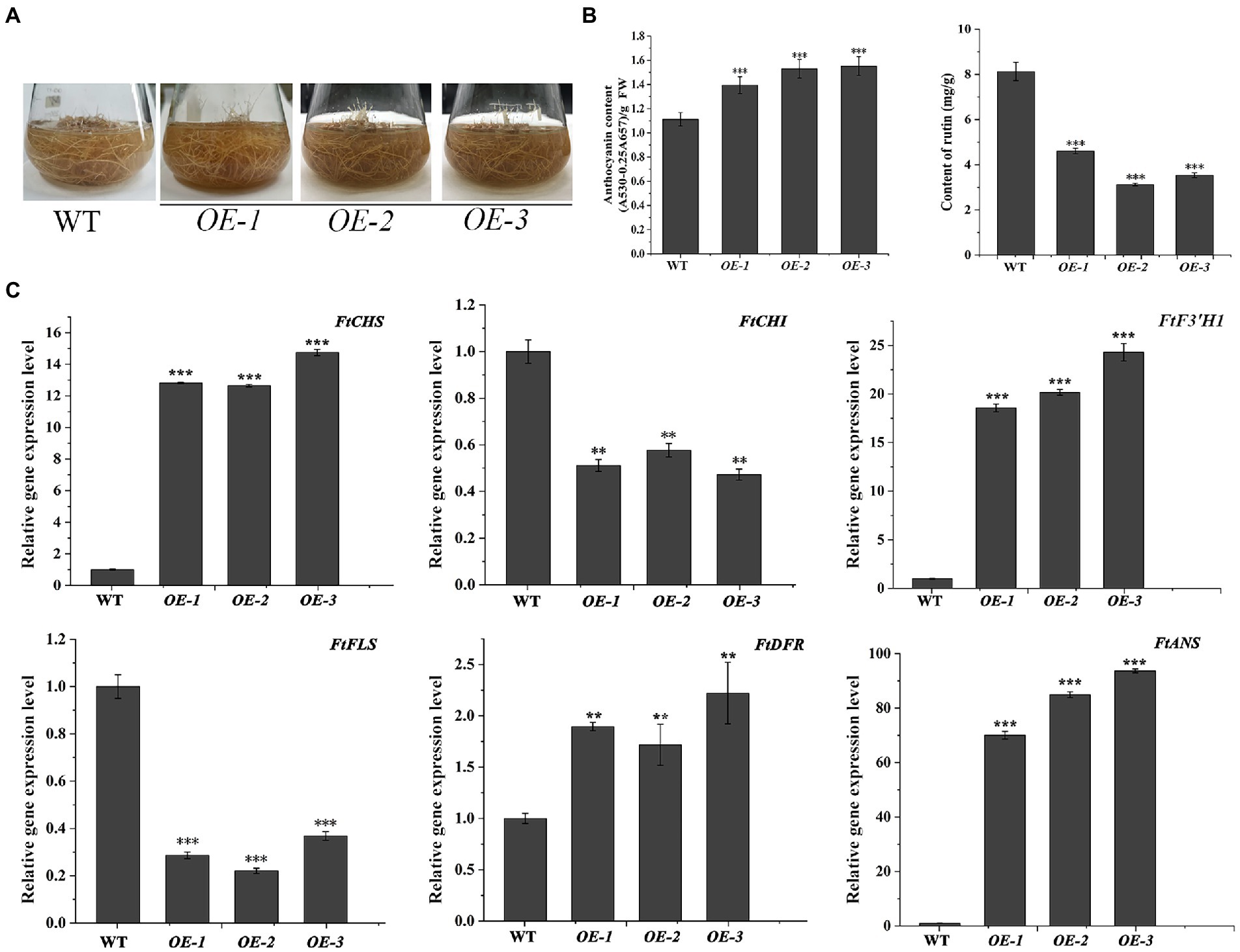
Figure 7. Overexpression of FtF3′H1 in TB hairy roots. (A) Phenotypic comparison between WT and transgenetic TB hairy roots overproducing FtF3′H1 (OE-1, OE-2 and OE-3). (B) Determination of anthocyanin and rutin contents in hairy roots. The average value is calculated repeatedly by three times of technology. Error bars represent ± SD. **p < 0.01 and ***p < 0.001. (C) Expression analysis of genes associated with the anthocyanin biosynthesis in TB hairy roots by qPCR. FtH3 gene is the reference gene. The average value is calculated repeatedly by three times of technology. Error bars indicate ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001.
Tartary buckwheat is an annual herb of the dicotyledonaceae family and as a homology resource for medicine and food because of its enriched flavonoids. Flavonols and anthocyanins are the main flavonoids in TB, and they are produced from different metabolic branches in the same metabolism pathway. The distribution of anthocyanin and flavonol in TB has always been a complicated issue. Therefore, it is a key task to identify the switch gene functioning in flavonoid metabolism pathway from TB. In this study, FtF3′H1 was shown to increase the content of anthocyanin and decrease the rutin levels in TB. It strongly indicates that FtF3′H1 might be a perspective gene for the quality improvement of TB by increasing anthocyanins via genetic engineering.
In the CYP75 subgroup of plant P450 family, molecular characterization is an effective way to identify the F3′H gene. In this study, FtF3′H1 protein are highly homologous to the F3′H proteins in Vitis vinifera (Jeong et al., 2006; Jochen Bogs et al., 2006), Malus (Hutabarat and Halbwirth, 2020), and Petunia (Brugliera et al., 1999). Unsurprisingly, the FtF3′H1 contained four cytochrome P450 conserved motifs (PPGPTPWP, FGAGRRICAG, AGTDTS, and E-R-R) and three characteristic motifs of F3′H (VVVAAS, GGER/K, and VDVKG; Figure 2B). Among them, PPGPTPWP is a proline-rich region, which is the main conserved domain and responsible for signal anchoring (Murakami et al., 1994). FGAGRRICAG, a heme-binding region containing conserved cysteine residues, can bind iron ions as well as carbon monoxide (Chika et al., 2001). AGTDTS is a molecular oxygen-binding domain that is essential for catalytic reactions (Baba and Ashraf, 2019). E-R-R, a domain that stabilizes protein structures (Zabala and Vodkin, 2003; Seitz et al., 2006). Each of the three characteristic motifs can be the F3′H marker, such as GGER/K, which can be used as an important marker to distinguish F3′H from F3′5′H and other P450s (Zabala and Vodkin, 2003; Seitz et al., 2006).
In order to explore how FtF3′H1 affects the biosynthesis of flavonoids, a soluble recombinant protein FtF3′H1 tagged with GST was induced in E. coli and purified. Subsequently, the enzyme activity was determined by feeding substrates (kaempferol and dihydrokaempferol) and analyzed by HPLC. Unfortunately, the theoretical products including quercetin and dihydroquercetin were not able to be detected in the reaction (Supplementary Figure S3). This failure may be due to the lack of cytochrome P450 reductase in the prokaryotic expression system, which is essential for the delivery of electrons (Sevrioukova et al., 1999). Additionally, the deficiency of endoplasmic reticulum membrane structures in the system also prevents the FtF3′H1 from fulfilling its activity (Sevrioukova et al., 1999). Differently, in Catharanthus roseus (Hotze et al., 1995) and Camellia sinensis (Guo et al., 2019), the F3′H enzyme activities were measured by excising the membrane-bound region of cytochrome P450 enzyme and cytochrome P450 reductase. In the future, we will use different strategies to further analyze the catalytic activity of FtF3′H1 in vitro.
Previous studies have shown that the transcription levels of F3′Hs are significantly related to the synthesis of flavonoids and accumulation of pigments in plants. During fruit development and coloring of Vitis vinifera, it was found that the activation of the flavonoid pathway resulting in the total flavonoid and anthocyanin accumulation had a strong correlation with the F3′H expression (Jeong et al., 2006). Similar conclusions were drawn in Arabidopsis, Camellia sinensis, Sorghum bicolor, and other plants (Forkmann and Martens, 2001; He et al., 2020a; Cai et al., 2022; Hildreth et al., 2022). Interestingly, in Korean varieties of white, black, and red rice (Oryza sativa), 2 F3′Hs (CYP75B3 and CYP75B4) showed different expression patterns. CYP75B3 and CYP75B4 were mainly expressed in the developing seeds of black rice, but not in those of white and red rice. The expression levels of CYP75B4 were much higher than those of CYP75B3 in the developing seeds, leaves, and roots of white rice. The results suggested that different copies of the F3′Hs might regulate the synthesis and accumulation of different types of flavonoids in plants through changes in expression pattern. Consistently, the same phenomenon was also observed in this study. Based on the transcriptome and qRT-PCR data of flowering TB, the accumulation of total flavonoids may be mainly due to the high expression of FtF3′H1 rather than its homologous copy FtF3′H2 (Supplementary Figure S1), and it shows a significantly positive correlation with the FtF3′H1 expression (R2 = 0.95; Figure 3).
In the metabolic pathway of flavonoids, the function of F3′H enzyme is to catalyze the hydroxylation of the 3rd C atom in different flavonoid subtracts on the B ring, such as dihydrokaempferol, kaempferol, pelargonidin and so on. In this biochemical process, FLS, DFR, ANS, and F3′H may share the same subtracts and compete with each other, leading to metabolic flow favoring anthocyanin or flavonol branch (Martínez-Lüscher et al., 2014; Sakuta et al., 2021; Shi et al., 2021). In some cases, increased expression of F3′Hs have been shown to improve anthocyanin accumulation in some plants via shifting flavonoid metabolism toward the anthocyanin branch. In apple (Malus × domestica), expression of MdFLS is down-regulated during late stages of fruit development, which can be attributed to high levels of expression of MdF3′H genes and the competition of MdF3′H and MdFLS for the same substrate (Han et al., 2010). Wu reported that overexpression of the GbF3′H (Ginkgo biloba) could enhance the anthocyanin synthesis and accumulate more red-colored pigments in leaves of transgenic Populus (Wu et al., 2020). In another cases, silencing or mutation of the F3′H gene may lead to the enhancement of flavonol synthesis branch and the reduction of anthocyanin content. In Paeonia suffruticosa, an indel in F3′H caused the upregulation of FLS and drastically reduced the anthocyanin content in acyanic petals (Zhang et al., 2020). Our data from transgenic lines, including Arabidopsis, Arabidopsis tt7 mutants, N. tabacum, and TB hairy roots, showed that the overexpression of FtF3′H1 caused an increase in anthocyanin synthesis and decreased the content of rutin. These results are consistent with the reports in Chinese Cabbage (Brassica rapa L. subsp. pekinensis; He et al., 2020a,b, Park et al., 2021), O. sativa (Shih et al., 2008), and Paeonia suffruticosa (Shih et al., 2006; Zhang et al., 2020; Nitarska et al., 2021). In our experiments, FtF3′H1 increased the accumulation of anthocyanin, which may due to the higher expression abundance of FtF3′H1 than FtFLSs or FtF3′H1 has a stronger substrate preference for anthocyanin branch.
In conclusion, we successfully cloned FtF3′H1 with a complete domain from TB, characterized its molecular features and biological function in vivo. Notably, this study validated that FtF3′H1 is not only a key enzyme gene, but more essentially acting as a metabolic branch switch for TB flavonoid synthesis.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
CL and KY wrote the first draft of the paper. JY performed most of the experiments. HW helped the writing language of the draft. HC provided modification information in the revised version of manuscript. HZ and QW participated in the preparation of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key R&D Program of China (2021YFD1200105) and National Natural Science Foundation of China (31871698).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.959698/full#supplementary-material
Abrahams, S., Lee, E., Walker, A. R., Tanner, G. J., Larkin, P. J., and Ashton, A. R. (2003). The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J. 35, 624–636. doi: 10.1046/j.1365-313x.2003.01834.x
Austin, M. B., and Noel, J. P. (2003). The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 20, 79–110. doi: 10.1039/b100917f
Awasthi, P., Gupta, A. P., Bedi, Y. S., Vishwakarma, R. A., and Gandhi, S. G. (2016). Mannitol stress directs flavonoid metabolism toward synthesis of flavones via differential regulation of two cytochrome p 450 monooxygenases in coleus forskohlii. Front. Plant Sci. 7:985. doi: 10.3389/fpls.2016.00985
Baba, S. A., and Ashraf, N. (2019). Functional characterization of flavonoid 3′-hydroxylase, CsF3′H, from Crocus sativus L: insights into substrate specificity and role in abiotic stress. Arch. Biochem. Biophys. 667, 70–78. doi: 10.1016/j.abb.2019.04.012
Brugliera, F., Barri-Rewell, G., Holton, T. A., and Mason, J. G. (1999). Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J. 19, 441–451. doi: 10.1046/j.1365-313x.1999.00539.x
Cai, J., Lv, L. T., Zeng, X. F., Zhang, F., Chen, Y. L., Tian, W. L., et al. (2022). Integrative analysis of metabolomics and transcriptomics reveals molecular mechanisms of anthocyanin metabolism in the zikui tea plant (Camellia sinensis cv. Zikui). Int. J. Mol. Sci. 23:4780. doi: 10.3390/jmsa094780
Chika, K., Gong, Z., Yoshikazu, T., Mami, Y., and Kazuki, S. (2001). Differential expression of two cytochrome p450s involved in the biosynthesis of flavones and anthocyanins in chemo-varietal forms of Perilla frutescens. Plant Cell Physiol. 42, 1338–1344. doi: 10.1093/pcp/pce169
Devic, M., Guilleminot, J., Debeaujon, I., Bechtold, N., Bensaude, E., and Delseny, M. (1999). The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J. 19, 387–398. doi: 10.1046/j.1365-313x.1999.00529.x
Dong, Q. X., Zhao, H. X., Huang, Y. J., Chen, Y., Wan, M., Wu, Q., et al. (2020). FtMYB18 acts as a negative regulator of anthocyanin/proanthocyanidin biosynthesis in Tartary buckwheat. Plant Mol. Biol. 104, 309–325. doi: 10.1007/s11103-020-01044-5
Fabjan, N., Rode, J., Košir, I. J., Wang, Z. H., Zhang, Z., Kreft, I., et al. (2003). Tartary buckwheat (Fagopyrum tataricum Gaertn.) as a source of dietary rutin and quercitrin. J. Agric. Food Chem. 51, 6452–6455. doi: 10.1021/jf034543e
Forkmann, G., and Martens, S. (2001). Metabolic engineering and applications of flavonoids. Curr. Opin. Biotechnol. 12, 155–160. doi: 10.1016/s0958-1669(00)00192-0
Gao, F., Zhao, H. X., Yao, H. P., Li, C. L., Chen, H., Wu, Q., et al. (2016). Identification, isolation and expression analysis of eight stress-related R2R3-MYB genes in tartary buckwheat (Fagopyrum tataricum). Plant Cell Rep. 35, 1385–1396. doi: 10.1007/s00299-016-1971-5
Guo, L. N., Gao, L. P., Ma, X. B., Guo, F. R., Ruan, H. X., Bao, Y., et al. (2019). Functional analysis of flavonoid 3′-hydroxylase and flavonoid 3′, 5′-hydroxylases from tea plant (Camellia sinensis), involved in the B-ring hydroxylation of flavonoids. Gene 717:144046. doi: 10.1016/j.gene.2019.144046
Hamberger, B., and Hahlbrock, K. (2004). The 4-coumarate: CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc. Natl. Acad. Sci. 101, 2209–2214. doi: 10.1073/pnas.0307307101
Han, Y. P., Vimolmangkang, S., Soria-Guerra, R. E., Rosales-Mendoza, S., Zheng, D. M., Lygin, A. V., et al. (2010). Ectopic expression of apple F3′H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiol. 153, 806–820. doi: 10.1104/pp.109.152801
He, Q., Lu, Q. Q., He, Y. T., Wang, Y. X., Zhang, N. N., Zhao, W. B., et al. (2020a). Dynamic changes of the anthocyanin biosynthesis mechanism during the development of heading Chinese cabbage (Brassica rapa L.) and Arabidopsis under the control of BrMYB2. Front. Plant Sci. 11:593766. doi: 10.3389/fpls.2020.593766
He, Q., Wu, J. Q., Xue, Y. H., Zhao, W. B., Li, R., Zhang, L. G., et al. (2020b). The novel gene BrMYB2, located on chromosome A07, with a short intron 1 controls the purple-head trait of Chinese cabbage (Brassica rapa L.). Hortic. Res. 7, 97. doi: 10.1038/s41438-020-0319-z
Hildreth, S. B., Littleton, E. S., Clark, L. C., Puller, G. C., Kojima, S., and Winkel, B. S. (2022). Mutations that alter Arabidopsis flavonoid metabolism affect the circadian clock. Plant J., [Epub ahead of preprint]. doi: 10.1111/tpj.15718, 110, 932–945.
Hotze, M., Schröder, G., and Schröder, J. (1995). Cinnamate 4-hydroxylase from catharanthus roseus and a strategy for the functional expression of plant cytochrome p 450 proteins as translational fusions with p 450 reductase in Escherichia coli. FEBS Lett. 374, 345–350. doi: 10.1016/0014-5793(95)01141-z
Hou, S. Y., Du, W., Hao, Y. R., Han, Y. H., Li, H. Y., Liu, L. L., et al. (2021). Elucidation of the regulatory network of flavonoid biosynthesis by profiling the metabolome and transcriptome in Tartary buckwheat. J. Agric. Food Chem. 69, 7218–7229. doi: 10.1021/acs.jafc.1c00190
Huang, Y. J., Wu, Q., Wang, S., Shi, J. Q., Dong, Q. X., Yao, P. F., et al. (2019). FtMYB8 from Tartary buckwheat inhibits both anthocyanin/proanthocyanidin accumulation and marginal trichome initiation. BMC Plant Biol. 19, 263. doi: 10.1186/s12870-019-1876-x
Huang, X., Yao, J. W., Zhao, Y. Y., Xie, D. F., Xie, D. F., Xu, Z. Q., et al. (2016). Efficient rutin and quercetin biosynthesis through flavonoids-related gene expression in Fagopyrum tataricum Gaertn. Hairy root cultures with UV-B irradiation. Front. Plant Sci. 7:63. doi: 10.3389/fpls.2016.00063
Huda, M. N., Lu, S., Jahan, T., Ding, M. Q., Jha, R., Zhou, M. L., et al. (2021). Treasure from garden: bioactive compounds of buckwheat. Food Chem. 335:127653. doi: 10.1016/j.foodchem.2020.127653
Hutabarat, O. S., and Halbwirth, H. (2020). Enzymatic characterization of apple (Malus Domestica). IOP Conference Series: Earth and Environmental Science. 486:012058. doi: 10.1088/1755-1315/486/1/012058
Jeong, S. T., Goto-Yamamoto, N., Hashizume, K., and Esaka, M. (2006). Expression of the flavonoid 3′-hydroxylase and flavonoid 3′, 5′-hydroxylase genes and flavonoid composition in grape (Vitis vinifera). Plant Sci. 170, 61–69. doi: 10.1016/j.plantsci.2005.07.025
Jochen Bogs, A. E., Mcdavid, D., and Robinson, S. P. (2006). Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiol. 140, 279–291. doi: 10.1104/pp.105.073262
Li, C. L., Bai, Y. C., Chen, H., Zhao, H. X., Shao, J. R., and Wu, Q. (2012a). Cloning, characterization and functional analysis of a phenylalanine ammonia-lyase gene (FtPAL) from Fagopyrum tataricum Gaertn. Plant Mol. Biol. Report. 30, 1172–1182. doi: 10.1007/s11105-012-0431-9
Li, C. L., Bai, Y. C., Li, S. J., Chen, H., Han, X. Y., Zhao, H. X., et al. (2012b). Cloning, characterization, and activity analysis of a flavonol synthase gene FtFLS1 and its association with flavonoid content in tartary buckwheat. J. Agric. Food Chem. 60, 5161–5168. doi: 10.1021/jf205192q
Li, H. Y., Lv, Q. Y., Liu, A., Wang, J. R., Sun, X. Q., Deng, J., et al. (2022). Comparative metabolomics study of tartary (Fagopyrum tataricum (L.) Gaertn) and common (Fagopyrum esculentum Moench) buckwheat seeds. Food Chem. 371:131125. doi: 10.1016/j.foodchem.2021.131125
Li, S. J., Zhu, D. Y., Dong, Q. X., and Li, C. L. (2014). Cloning of flavonoid 3′-hydroxylase gene from Fagopyrum tataricum and its tissue-specific expression under cold stress. Chin. Tradit. Herb. Drug 45, 1300–1306. doi: 10.7501/j.issn.0253-2670.2014.09.020
Liu, M. Y., Wen, Y. D., Sun, W. J., Ma, Z. T., Huang, L., Wu, Q., et al. (2019). Genome-wide identification, phylogeny, evolutionary expansion and expression analyses of bZIP transcription factor family in tartary buckwheat. BMC Genom. 20, 483–418. doi: 10.1186/s12864-019-5882-z
Liu, B. G., and Zhu, Y. Y. (2007). Extraction of flavonoids from flavonoid-rich parts in tartary buckwheat and identification of the main flavonoids. J. Food Eng. 78, 584–587. doi: 10.1016/j.jfoodeng.2005.11.001
Martínez-Lüscher, J., Sánchez-Díaz, M., Delrot, S., Aguirreolea, J., Pascual, I., and Gomes, E. (2014). Ultraviolet-B radiation and water deficit interact to alter flavonol and anthocyanin profiles in grapevine berries through transcriptomic regulation. Plant Cell Physiol. 55, 1925–1936. doi: 10.1093/pcp/pcu121
Mizutani, M., Ohta, D., and Sato, R. (1997). Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol. 113, 755–763. doi: 10.1104/pp.113.3.755
Murakami, K., Mihara, K., and Omura, T. (1994). The transmembrane region of microsomal cytochrome P 450 identified as the endoplasmic reticulum retention signal. J. Biochem. 116, 164–175. doi: 10.1093/oxfordjournals.jbchem.a124489
Nitarska, D., Boehm, R., Debener, T., Lucaciu, R. C., and Halbwirth, H. (2021). First genome edited poinsettias: targeted mutagenesis of flavonoid 3′-hydroxylase using CRISPR/Cas9 results in a colour shift. Plant Cell Tissue Organ Cult. (PCTOC) 147, 49–60. doi: 10.1007/s11240-021-02103-5
Park, S., Lee, H., Min, M. K., Ha, J., Song, J., Lim, C. J., et al. (2021). Functional characterization of BrF3′H, which determines the typical flavonoid profile of purple Chinese cabbage. Front. Plant Sci. 12, 793589. doi: 10.3389/fpls.2021.793589
Park, N. I., Li, X. H., Suzuki, T., Kim, S. J., Woo, S. H., Park, C. H., et al. (2011). Differential expression of anthocyanin biosynthetic genes and anthocyanin accumulation in tartary buckwheat cultivars ‘Hokkai T8’and ‘Hokkai T10’. J. Agric. Food Chem. 59, 2356–2361. doi: 10.1021/jf200020b
Pelletier, M. K., and Shirley, B. W. (1996). Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings (coordinate regulation with chalcone synthase and chalcone isomerase). Plant Physiol. 111, 339–345. doi: 10.1104/pp.111.1.339
Peng, Y. Y., Lin-Wang, K., Cooney, J. M., Wang, T. C., Espley, R. V., and Allan, A. C. (2019). Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species). HORTICULTURE RESEARCH 6, 3. doi: 10.1038/s41438-018-0076-4
Peng, L. X., Zou, L., Su, Y. M., Fan, Y., and Zhao, G. (2015). Effects of light on growth, levels of anthocyanin, concentration of metabolites in Fagopyrum tataricum sprout cultures. Int. J. Food Sci. Technol. 50, 1382–1389. doi: 10.1111/ijfs.12780
Pourcel, L., Routaboul, J.-M., Cheynier, V., Lepiniec, L., and Debeaujon, I. (2007). Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci. 12, 29–36. doi: 10.1016/j.tplants.2006.11.006
Prescott, A. G., and John, P. (1996). Dioxygenases: molecular structure and role in plant metabolism. Annu. Rev. Plant Biol. 47, 245–271. doi: 10.1146/annurev.arplant.47.1.245
Prescott, A. G., Stamford, N. P., Wheeler, G., and Firmin, J. L. (2002). In vitro properties of a recombinant flavonol synthase from Arabidopsis thaliana. Phytochemistry 60, 589–593. doi: 10.1016/s0031-9422(02)00155-3
Routaboul, J.-M., Kerhoas, L., Debeaujon, I., Pourcel, L., Caboche, M., Lepiniec, L., et al. (2006). Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224, 96–107. doi: 10.1007/s00425-005-0197-5
Saito, K., Kobayashi, M., Gong, Z., Tanaka, Y., and Yamazaki, M. (1999). Direct evidence for anthocyanidin synthase as a 2-oxoglutarate-dependent oxygenase: molecular cloning and functional expression of cDNA from a red forma of Perilla frutescens. Plant J. 17, 181–189. doi: 10.1046/j.1365-313x.1999.00365.x
Sakuta, M., Tanaka, A., Iwase, K., Miyasaka, M., Ichiki, S., and Hatai, M. (2021). Anthocyanin synthesis potential in betalain-producing Caryophyllales plants. J. Plant Res. 134, 1335–1349. doi: 10.1007/s10265-021-01341-0
Schoenbohm, C., Martens, S., Eder, C., Forkmann, G., and Weisshaar, B. (2000). Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P 450 enzyme. Biol. Chem. 381, 749–753. doi: 10.1515/BC.2000.095
Seitz, C., Eder, C., Deiml, B., Kellner, S., Martens, S., and Forkmann, G. (2006). Cloning, functional identification and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3′, 5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′, 5′-hydroxylase in the asteraceae family. Plant Mol. Biol. 61, 365–381. doi: 10.1007/s11103-006-0012-0
Sevrioukova, I. F., Li, H., Zhang, H., Peterson, J. A., and Poulos, T. L. (1999). Structure of a cytochrome p 450–redox partner electron-transfer complex. Proc. Natl. Acad. Sci. 96, 1863–1868. doi: 10.1073/pnas.96.5.1863
Shi, Y. F., Jiang, X. L., Chen, L. B., Li, W. W., Lai, S. Y., Fu, Z. P., et al. (2021). Functional analyses of flavonol synthase genes from camellia sinensis reveal their roles in anther development. Front. Plant Sci. 12:753131. doi: 10.3389/fpls.2021.753131
Shih, C. H., Chu, H., Tang, L. K., Sakamoto, W., Maekawa, M., and Lo, C. (2008). Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 228, 1043–1054. doi: 10.1007/s00425-008-0806-1
Shih, C. H., Chu, I. K., Yip, W. K., and Lo, C. (2006). Differential expression of two flavonoid 3′-hydroxylase cDNAs involved in biosynthesis of anthocyanin pigments and 3-deoxyanthocyanidin phytoalexins in sorghum. Plant Cell Physiol. 47, 1412–1419. doi: 10.1093/pcp/pcl003
Shirley, B. W., Hanley, S., and Goodman, H. M. (1992). Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 4, 333–347. doi: 10.1105/tpc.4.3.333
Shufflebottom, D., Edwards, K., Schuch, W., and Bevan, M. (1993). Transcription of two members of a gene family encoding phenylalanine ammonia-lyase leads to remarkably different cell specificities and induction patterns. Plant J. 3, 835–845. doi: 10.1111/j.1365-313x.1993.00835.x
Tanaka, Y. (2006). Flower colour and cytochromes P 450. Phytochem. Rev. 5, 283–291. doi: 10.1098/rstb.2012.0432
Thill, J., Miosic, S., Gotame, T. P., Mikulic-Petkovsek, M., Gosch, C., Veberic, R., et al. (2013). Differential expression of flavonoid 3′-hydroxylase during fruit development establishes the different B-ring hydroxylation patterns of flavonoids in Fragaria × ananassa and Fragaria vesca. Plant Physiol. Biochem. 72, 72–78. doi: 10.1016/j.plaphy.2013.03.019
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Wu, Y. Q., Wang, T. L., Xin, Y., Wang, G. B., and Xu, L. A. (2020). Overexpression of the GbF3′H1 gene enhanced the epigallocatechin, gallocatechin, and catechin contents in transgenic populus. J. Agric. Food Chem. 68, 998–1006. doi: 10.1021/acs.jafc.9b07008
Xue, Q. Q., Zhang, X. R., Yang, H., Li, H. D., Lv, Y. Y., Zhang, K., et al. (2021). Transcriptome and metabolome analysis unveil anthocyanin metabolism in pink and red testa of peanut (Arachis hypogaea L.). Int. J. Genom. 2021:5883901. doi: 10.1155/2021/5883901
Yao, P. F., Huang, Y. J., Dong, Q. X., Wan, M., Wang, A. H., Chen, Y. W., et al. (2020). FtMYB6, a light-induced SG7 R2R3-MYB transcription factor, promotes flavonol biosynthesis in tartary buckwheat (Fagopyrum tataricum). J. Agric. Food Chem. 68, 13685–13696. doi: 10.1021/acs.jafc.0c03037
Yao, H. P., Li, C. L., Zhao, H. X., Zhao, J. N., Chen, H., Bu, T. L., et al. (2017a). Deep sequencing of the transcriptome reveals distinct flavonoid metabolism features of black tartary buckwheat (Fagopyrum tataricum Garetn.). Prog. Biophys. Mol. Biol. 124, 49–60. doi: 10.1016/j.pbiomolbio.2016.11.003
Yao, P. F., Zhao, H. X., Luo, X. P., Gao, F., Li, C. L., Yao, H. P., et al. (2017b). Fagopyrum tataricum FtWD40 functions as a positive regulator of anthocyanin biosynthesis in transgenic tobacco. J. Plant Growth Regul. 36, 755–765. doi: 10.1007/s00344-017-9678-6
Yao, P. F., Zhao, H. X., Luo, X. P., Gao, F., Yao, H. P., Li, C. L., et al. (2016). Chalcone synthase homologous genes cloning and expression pattern in flowering Fagopyrum tataricum Gaertn. Russ. J. Plant Physiol. 63, 790–799. doi: 10.1134/S1021443716060169
Zabala, G., and Vodkin, L. (2003). Cloning of the pleiotropic T locus in soybean and two recessive alleles that differentially affect structure and expression of the encoded flavonoid 3′ hydroxylase. Genetics 163, 295–309. doi: 10.1093/genetics/163.1.295
Zhang, Y. Z., Cheng, Y. W., Xu, S. Z., Ma, H. P., Han, J. M., and Zhang, Y. (2020). Tree peony variegated flowers show a small insertion in the F3′H gene of the acyanic flower parts. BMC Plant Biol. 20, 1–13. doi: 10.1186/s12870-020-02428-x
Zhang, L., J, L., X, X., Ma, B., Gao, Q., Du, H. L., et al. (2018). The tartary buckwheat genome provides insightsinto rutin biosynthesis and abiotic stresstolerance. Mol. Plant 10, 1224–1237. doi: 10.1016/j.molp.2017.08.013
Keywords: Tartary buckwheat (Fagopyrum tataricum), metabolism, flavonoid biosynthesis, flavonoid 3′-hydroxylase, transgenic plants
Citation: Li C, Yang J, Yang K, Wu H, Chen H, Wu Q and Zhao H (2022) Tartary buckwheat FtF3′H1 as a metabolic branch switch to increase anthocyanin content in transgenic plant. Front. Plant Sci. 13:959698. doi: 10.3389/fpls.2022.959698
Received: 02 June 2022; Accepted: 08 August 2022;
Published: 25 August 2022.
Edited by:
Koichi Sugimoto, University of Tsukuba, JapanReviewed by:
Jiang Chen, Chengdu University of Traditional Chinese Medicine, ChinaCopyright © 2022 Li, Yang, Yang, Wu, Chen, Wu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haixia Zhao, emhhb2hhaXhpYUBzaWNhdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.