- Department of Agriculture Biotechnology, National Agri-Food Biotechnology Institute, SAS Nagar, India
Intrinsically disorder regions or proteins (IDRs or IDPs) constitute a large subset of the eukaryotic proteome, which challenges the protein structure–function paradigm. These IDPs lack a stable tertiary structure, yet they play a crucial role in the diverse biological process of plants. This study represents the intrinsically disordered nature of a plant-specific DNA binding with one finger transcription factor (DOF-TF). Here, we have investigated the role of OsDOF27 and characterized it as an intrinsically disordered protein. Furthermore, the molecular role of OsDOF27 in thermal stress tolerance has been elucidated. The qRT-PCR analysis revealed that OsDOF27 was significantly upregulated under different abiotic stress treatments in rice, particularly under heat stress. The stress-responsive transcript induction of OsDOF27 was further correlated with enriched abiotic stress-related cis-regulatory elements present in its promoter region. The in vivo functional analysis of the potential role of OsDOF27 in thermotolerance was further studied in yeast and in planta. Ectopic expression of OsDOF27 in yeast implicates thermotolerance response. Furthermore, the rice transgenic lines with overexpressing OsDOF27 revealed a positive role in mitigating heat stress tolerance. Collectively, our results evidently show the intrinsically disorderedness in OsDOF27 and its role in thermal stress response in rice.
Introduction
Rice is one of the most important cereal crops as it is consumed by one-third of the global population (Fornasiero et al., 2022). Despite an enormous rise in rice cultivation, its productivity is severely compromised by different environmental stresses, including extreme temperature, water scarcity, salinity stress, heavy metal stress, etc. Besides, the dwindling agrarian land and continuously growing population further escalated the gap between its demand and supply (Rasheed et al., 2020). To combat the stressful conditions, diverse molecular responses are instigated at the cellular level, which include signaling cascade, phytohormonal regulation, altered gene expression, transcriptional regulation, and post-transcriptional and post-translational modifications (Singh and Jwa, 2013; Cohen and Leach, 2019; Kaur et al., 2021; Manna et al., 2021; Salvi et al., 2021b, 2022). Transcriptional control involves the regulation mediated via transcription factors (TFs) and transcriptional regulators (TRs). TF bind to cis-elements present in the promoter region of the target gene while TR indirectly regulates the gene expression by modulating the DNA—protein interaction through chromatin remodeling (Zheng et al., 2016; Chandran et al., 2019; Manna et al., 2021). In rice, about 2,048 gene are predicted to encode for TFs, whereas 328 as transcriptional regulators (Kikuchi et al., 2003; Pérez-Rodríguez et al., 2010). Similarly, about 6% of the total Arabidopsis genome is assigned to encode for TF (Riechmann et al., 2000). Overall, the TFs are ascribed to perform diverse cellular functions. Amongst them, several TF families are known to play general functions, while some of the TF families belong to a plant-specific clade. Generally, the plant-specific TFs orchestrate the regulation of gene networks associated with processes that are specific to plants, such as seed germination and maturation, vasculature formation, photosynthesis, stomatal regulation, etc. (Hrmova and Hussain, 2021; Wani et al., 2021; Strader et al., 2022). Amongst a large number of TFs families, TCP, AP2, and DOF-TF are documented as plant-specific TFs (Manna et al., 2021).
The DNA binding with one finger transcription factor family belongs to a plant-specific clade of TF which is structured as a Cys2/Cys2 zinc finger-TF (Yanagisawa, 2002). As DOF-TFs are exclusively present in plants, they are widely known to regulate diverse biological events that are specific to the plant system, such as abiotic stress, vasculature formation, photoperiodic flowering, seed germination, seed development, circadian cycle, phytochrome signaling, nitrogen use efficiency, etc. (Shigyo et al., 2007; Zou et al., 2013; Gupta et al., 2015; Corrales et al., 2017; Manna et al., 2021). The number of DOF-TF varies greatly across the species, for instance, there are 37 DOF-TF encoding genes in Arabidopsis (Lijavetzky et al., 2003), 34 in tomato (Cai et al., 2013), 31 in wheat (Shaw et al., 2009), 60 in apple (Zhang et al., 2018), 74 in banana (Dong et al., 2016), 41 in poplar (Yang et al., 2006), and 78 in soybean (Guo and Qiu, 2013) to name a few. In general, the DOF-TF is a 200–400 aa long protein with a highly conserved DNA binding motif that usually resides in the N-terminal and greatly divergent C-terminal region. The N-terminal DNA binding domain of all DOF-TF specifically binds to the 5'-[T/A]AAAG-3' (except AGTA in pumpkin) cis-regulatory element present in the promoter region of its target gene, while its C-terminal interacts with the RNA polymerase to modulate the gene (Kisu et al., 1995, 1998; Noguero et al., 2013). Previous reports have also shown that apart from the DNA-binding potential of the DOF-domain, it also participates in the protein–protein interaction and thus indicates its functional versatility (Yanagisawa, 2004; Waqas et al., 2020; Manna et al., 2021). DOF-TF interacts with different proteins, especially other TFs such as TCP, MYB, and bZIP, which hold a crucial role in many biological pathways. The first DOF-TF encoding gene was identified in maize in 1995 which binds to the CaMV35S promoter sequence (Yanagisawa, 1995). Several reports indicated the contribution of DOF-TFs in biotic and abiotic stress responses. In tomatoes, SlDOF22 negatively regulates the ascorbic acid content. Besides, in RNAi lines of SlDOF22, the SlSOS1 gene was down-regulated, which results in decreased salinity stress tolerance (Cai et al., 2016). The overexpression of GhDOF1 enhances the salt and cold stress response in cotton (Su et al., 2017). Corrales et al. (2017) showed that the expression level of cycling DOF factor 3 (CDF3) is triggered by abiotic stress conditions including salt, heat, drought, and ABA treatment, and its overexpression lines exhibited improved stress tolerance while RNAi lines displayed reduced stress tolerance. Several reports highlighted that the transcription factors are profuse with the intrinsically disordered region (IDR), which offers functional plasticity and versatility to the TFs (Liu et al., 2006; Brodsky et al., 2020; Salladini et al., 2020). Intrinsically disordered proteins are characterized as proteins with an enriched disordered region and devoid of an ordered three-dimension structure, yet they are capable to execute diverse cellular functions (Jung et al., 2011; Sun et al., 2013). As compared to prokaryotes, eukaryotic proteomes are known to possess higher disordered proteins. This aspect of abundant IDR in eukaryotes also indicated the correlation of IDR with complexity at the functional and organism levels (DeForte and Uversky, 2017). These IDRs emerged as a new field in structural biology which are largely studied in the animal system. However, due to their central role in the regulation of metabolic and cell signaling processes, they are attaining much attention in plant science as well (Zamora-Briseño et al., 2021). Here, we aimed to explore the role of an OsDOF27 using diverse in-silico molecular and functional analyses. In our analysis, we found the OsDOF27 protein to be highly disordered in nature and appeared to play an important role in thermal stress response in rice.
Materials and methods
Plant materials, treatment, and bacterial strain
Rice (Oryza sativa) seeds (var. Taipei-309) grown at the National Agri-Food Biotechnology Institute, Mohali (Punjab) were used in this study for genetic transformation and stress experiments. For the genetic transformation of rice, embryogenic calli were used (Thakur et al., 2022). For stress imposition on rice seedlings, 7-day-old rice seedlings were challenged to different abiotic stressors such as heat (42°C), salt (200 mM), cold (4°C), methyl viologen (20 μM), polyethylene glycol (PEG; 20%) for 12 h. After 12 h of stress, the seedling was immediately used for expression profiling or snap frozen and stored at −80°C for later use (Yokotani et al., 2009). For cloning experiments, the Escherichia coli DH5α strain was used, while Agrobacterium tumefaciens LBA-4404 was used for rice transformation. Yeast strain (INVSc1) was used as a host strain for the yeast expression system to study thermotolerance response in yeast.
Molecular cloning
For molecular cloning of OsDOF27 (LOC_Os10g35300), we used gateway-based cloning as per the manufacturer's instructions. Initially, the OsDOF27 was cloned in pENTR™/D-TOPO™ vector and the positive constructs were confirmed by Sanger sequencing. The sequenced confirmed entry clone was then used for its subcloning to the destination vectors like pANIC6B (CD3-1708), pSITEYFP3CA (CD3-1638), and pDEST52 (pYES2-DEST52 Gateway™ destination vector) using LR Clonase-II. The primers used for this study are enlisted in Supplementary Table 1.
Agroinfiltration of N. benthamiana for subcellular localization study
The subcellular localization study was conducted by agroinfiltration according to our method described previously (Thakur et al., 2021). Briefly, Agrobacterium cells harboring positive transformant of pSITEYFP3CA were grown in LB medium with selection markers at 28°C for 24 h. The plasmid with 35S-YFP was used as the positive control. The Agrobacterium cells were then pelleted down by centrifugation, mixed with resuspension buffer, and incubated for 6 h prior to infiltration. The mix was then infiltrated into the abaxial surface of 6-week-old N. benthamiana leaves with a needleless syringe (1 ml). The plants were covered and kept in dark for 12 h and then transferred to the growth chamber at 24°C ± 1 with a 16/8 h photoperiod. The leaf sample was collected after 48 h for microscopy.
DAPI staining and confocal microscopy
The nuclear localization of OsDOF27 protein was confirmed by colocalizing it with 4, 6-diamidino-2-phenylindole (DAPI) that specifically stains the nucleus. Briefly, the leaf sample was cut and washed by dipping (for 5 s) with 1X phosphate-buffered saline (PBS, pH-7.5) containing 0.5% Triton-X (which aids the DAPI staining). After a gentle wash (for 5–10 s) in 1X phosphate-buffered saline (PBS, pH-7.5; without Triton-X), the leaf sample was incubated in DAPI solution (15 μg mL−1 DAPI prepared in 1 × PBS) for 20 min. The fluorescent signals were detected using a Carl Zeiss confocal laser scanning microscope (LSM880; Karl Zeiss, Jena, Germany) with an oil immersion objective (Thakur et al., 2021).
QPCR analysis
Total RNA was isolated using the TRIzol reagent (Sigma) and reverse transcribed into cDNA using iScript™ cDNA Synthesis Kit (BioRad) according to the manufacturer's instruction. A no template control was incorporated as a negative control in each assay. For normalization, two endogenous reference genes (Ubiquitin-5 and EF1α) were used in each assay. The gene-specific primers and reference genes used for the qRT-PCR were previously described and validated.
Yeast thermo-tolerance assay
To assess the thermotolerance assay in the yeast system, we cloned OsDOF27 in the yeast expression vector (pDEST52 vector) using gateway-based cloning (Invitrogen). The construct pDEST52:OsDOF27 was then mobilized to yeast strain (INVSc1) through PEG-lithium acetate-based transformation. Yeast spot assay and growth curve analysis were performed to determine the thermotolerance of the yeast cell harboring pDEST52:OsDOF27. The yeast cells were allowed to grow in a YEB medium till the mid-exponential phase with A600 0.5 (about 1 × 107 cells ml−1). The cell density was then adjusted to 0.2 (A600) for spot assay. Spot assay was conducted by spotting a serial dilution yeast culture with equal cell density (A600 0.2), and a 5 μl spot was placed on the plate. An empty-vector-transformed INVsc1 strain was used in each case as a control for the stress experiment. For unstressed conditions, the plate was incubated at 30°C, while to determine the thermotolerance potential, the plate was incubated at 42°C for 8 h and then moved to 30°C. The yeast cell growth on the plate was monitored closely and photographed. For growth curve analysis, the initial cell density of the yeast cell harboring pDEST52:OsDOF27 and the empty vector was kept similar (OD = 0.2 A), and the growth was observed and plotted against time. Each assay was repeated three times, with at least four biological replicates (Salvi et al., 2021a).
In silico analysis of OsDOF27 for IDR analysis and stability curve
The OsDOF27 sequence was obtained from the Rice Genome Annotation Project database (http://rice.uga.edu/). The disorder propensity was determined with different software such as http://www.pondr.com/, https://iupred.elte.hu/, and http://sparks-lab.org/server/SPOT-disorder/index.php. The compositional profiler was used to assess the enrichment or depletion of the amino acid composition of OsDOF27 against SwissProt52. The amino acid composition was color-coded for disorders and sorted by the differences observed (http://www.cprofiler.org/cgi-bin/profiler.cgi; Vacic et al., 2007). For generating the sequence, annotated Das-Pappu phase diagram Classification of Intrinsically Disordered Ensemble Regions, CIDER, http://pappulab.wustl.edu/CIDER/analysis/) was used to evaluate the parameters associated with disordered protein sequences (Kyte and Doolittle, 1982; Das and Pappu, 2013; Holehouse et al., 2015). The prediction of secondary structures such as alpha helix, beta turns, extended strand, and the random coil was conducted by the online tool Self-Optimized Prediction Method with Alignment (SOPMA, https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html; Geourjon and Deléage, 1995). To assess parameters like the standard heat capacity of folding (ΔCp), the standard enthalpy of folding (ΔHm) at melting temperature, Tm (melting temperature), the protein sequence of OsDOF27 was subjected to an online tool SCooP tool (http://babylone.3bio.ulb.ac.be/SCooP/index.php). The curve for the change in standard free energy of folding with respect to the temperature was assessed by protein stability curve prediction (Pucci and Rooman, 2014; Pucci et al., 2017).
Agrobacterium-mediated rice transformation
Agrobacterium-mediated rice transformation was performed as per our previous report (Thakur et al., 2022). Briefly, Agrobacterium tumefaciens strain LBA4404 harboring pANIC6COsDOF27 (LOC_Os10g35300) was used for the genetic transformation of rice. For rice transformation, embryogenic calli were co-cultivated with Agrobacterium culture for 36 h and further washed and selected for hygromycin resistance. After three rounds of selection, the callus was transferred to regeneration media. Plants with developed roots and shoots were transferred to a hydroponics medium and subsequently hardened and were grown to maturity. The offspring of the primary transformants were propagated by selfing and T2 generation seeds were used for further experiments.
Stress treatment
For stress experiments, the seeds of three independent transgenic lines (Ubi:OsDOF27-OE1, OE-2, and OE-5), wild type (WT), and vector control (VC) were exposed to high temperature. Seed germination was assessed on two layers of filter paper (Whatman No. 1) moistened with autoclaved double-distilled water (ddH2O). For non-stressed conditions, seed germination was evaluated at 28 ± 1°C while for heat stress, the seeds were pre-exposed to high temperature (45°C) for 12 h. The germination rate was determined by monitoring the germination score after 72 h. Gemination assay was conducted in triplicate with n=30 seeds per treatment in each experimental replicate. Seeds that exhibited radicle protrusion were scored as germinated. For stress imposition at the seedling stage, rice transgenic and control plants were germinated under non-stressed/control conditions at 28 ± 1°C with a photoperiod of 16 h-light/8 h-dark cycle (300–400 μmol photons m−2s−1). Two-week-old plants were challenged to heat stress by transferring them to 45°C for 24 h and then transferred to 28 °C, while for non-stressed (NS) conditions, plants were continuously grown at 28°C.
Results
Gene expression analysis of rice OsDOF27 during abiotic stress conditions
To understand the biological function of DOF-TF in rice plant growth and development, we have investigated the expression of DOF27 under abiotic stress treatments. For this, rice seedlings were challenged with abiotic stressors including heat, salt, cold, PEG, and methyl viologen, and the transcript level of OsDOF27 was assessed using qRT-PCR. OsDOF27 showed maximum induction in heat stress followed by oxidative and osmotic stress imposed by methyl viologen (paraquat) and PEG treatment, respectively (Figure 1). To get a molecular insight into the gene regulation of OsDOF27, we inspected the cis-regulatory elements present in the promoter region of OsDOF27. For this, the 1.5 KB promoter sequences of OsDOF27 were analyzed using web-based tools i.e., PLACE database. The results revealed that the promoter regions were enriched with various stress and phytohormonal-related cis-regulatory elements indicating the stress-responsive expression of OsDOF27. Supplementary Table 2 showed the detailed cis-element present in the promoter region of OsDOF27.
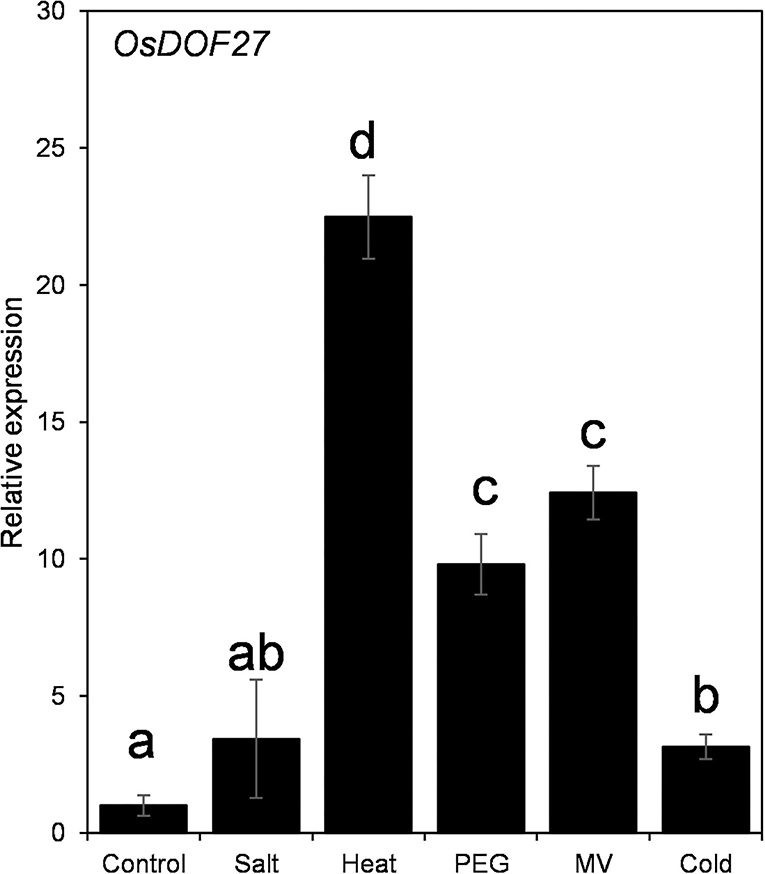
Figure 1. qRT-PCR for expression analysis of OsDOF27 under different abiotic stress conditions including salt, heat, PEG, methyl viologen, and cold. Seven-day-old rice seedlings were treated with abiotic stressors (20% PEG, 200 mM NaCl, heat (42°C), cold (4°C), and methyl viologen (20 μM). Total RNA was extracted, and cDNA was synthesized, followed by qPCR. Two endogenous controls (Ubiquitin-5 and EF1α) were used to normalize the relative expression value of OsDOF27 followed by a calculation using the ΔΔCT method. The error bars represent the standard deviation of triplicate analysis. The significant difference in the mean of the values is indicated by different letters (α = 0.01).
DOF27 is an intrinsically disorder protein
Furthermore, while designing the primers for OsDOF27, we observed multiple repeats in its sequence. It prompted us to inspect the protein sequence of OsDOF27, which showed a biased composition for amino acids as around 40% of the total sequence comprises three amino acids i.e., proline (15.4%), glycine (14.7%), and alanine (11%). The amino acid composition of OsDOF27 suggested the presence of the intrinsically disordered region. Therefore, to understand the sequence–structure relationship of OsDOF27, we inspected the protein sequence of OsDOF27 for the presence of an intrinsically disordered region (IDR). The OsDOF27 appears to be a highly disordered protein that was enriched in the IDR region, especially in N- and C- terminals. However, the DNA binding DOF-domain was found to be deficient in IDR sequences (Figure 2A). The compositional bias of OsDOF27 was assessed and compared against the Swissport 52 database, which revealed that OsDOF27 possessed an abundant subset of disordered promoting amino acids such as proline, glycine, alanine, serine, and arginine, while the order promoting residues, such as isoleucine, phenylalanine, valine, etc., were dramatically low in OsDOF27 (Figure 2B). Additionally, we also conducted an evaluation of the OsDOF27 protein sequence for parameters like FCR (fraction of charged residue), NCPR (net charge per residue), Hydropathy plot (based on Kyte-Doolittle hydrophobicity scale), and diagram of states which represent the position of protein in graph drawn between the fraction of positive and negative charge. As shown in Figure 2, the different characterized IDPs were observed as κ (kappa) (0.310), FCR (0.139), NCPR (0.015), and Hydropathy (4.184) as per Das et al. (2015; Supplementary Table 3). Furthermore, the secondary structure prediction of OsDOF27 also showed that it comprises 24.91% of alpha-helix, 12.09% extended-strand, 7.69% beta-turn, and more than 50%, i.e., 55.31%, is random-coil (Supplementary Figure 1).
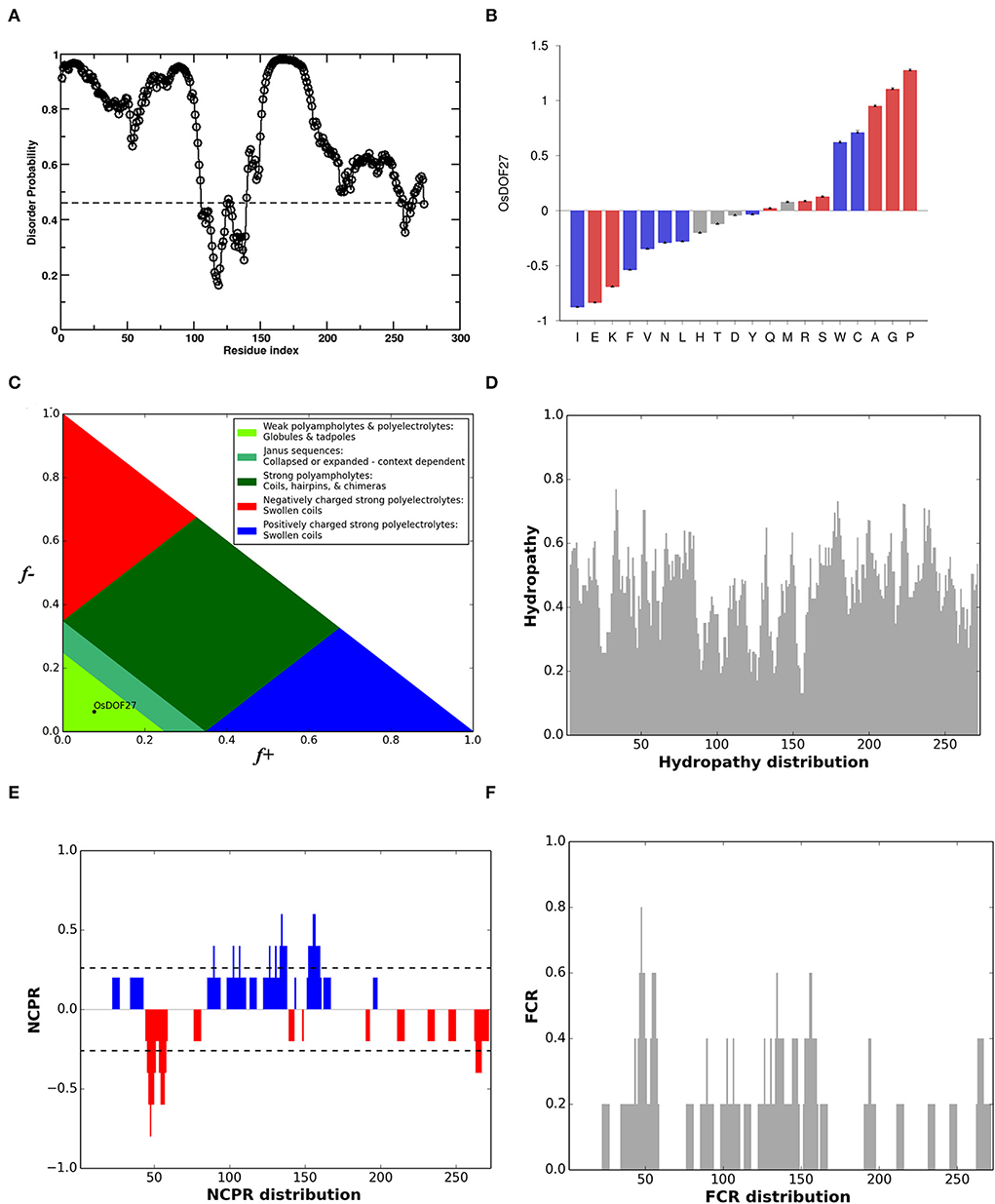
Figure 2. (A) Graph represents a prediction of disorder regions present in the protein sequence of OsDOF27 based on the amino acid composition. Calculations were performed using the database of Protein Disorder (DisProt) software. A score over 0.5 indicates a high probability of disorder. (B) Depiction of compositional bias of OsDOF27 to show enriched or depleted aa of OsDOF27 against the SwissProt52 database. The amino acid composition was color-coded for disorder and sorted by the difference observed. The red and blue color bars indicate the disordered and ordered promoting amino acid, respectively, while the gray color-coded bar represents the neutral amino acid. (C) Analysis for diagram of states which represent the position of protein in graph drawn between the fraction of positive and negative charge, (D) Hydropathy plot (based on Kyte-Doolittle hydrophobicity scale), (E) NCPR (net charge per residue), (F) FCR (fraction of charged residue) of OsDOF27 protein sequence.
Subcellular localization of OsDOF27
An in silico assessment of subcellular localization suggested the nuclear localization of OsDOF27 (Supplementary Figure 2A). To examine the subcellular localization of OsDOF27 in planta, the CDS was cloned in the pSITE-YFP, and after sequence confirmation, the construct was transformed to Agrobacterium. The Agrobacterium strain harboring OsDOF27 was used for agroinfiltration of transient transfection in Nicotiana leaves. After 2 days of agroinfiltration, the YFP signal was visualized using a confocal laser scanning microscope to track the subcellular localization of OsDOF27. The microscopic analysis revealed that OsDOF27 was localized to the nucleus. The nuclear localization was further confirmed using DAPI staining, which specifically stains the nucleus. The YFP signal colocalized with DAPI, which confirms the nuclear localization of OsDOF27 (Figure 3). However, in the control, fluorescence was detected in the entire cell, including the cytoplasm and nucleus (Supplementary Figure 2B).
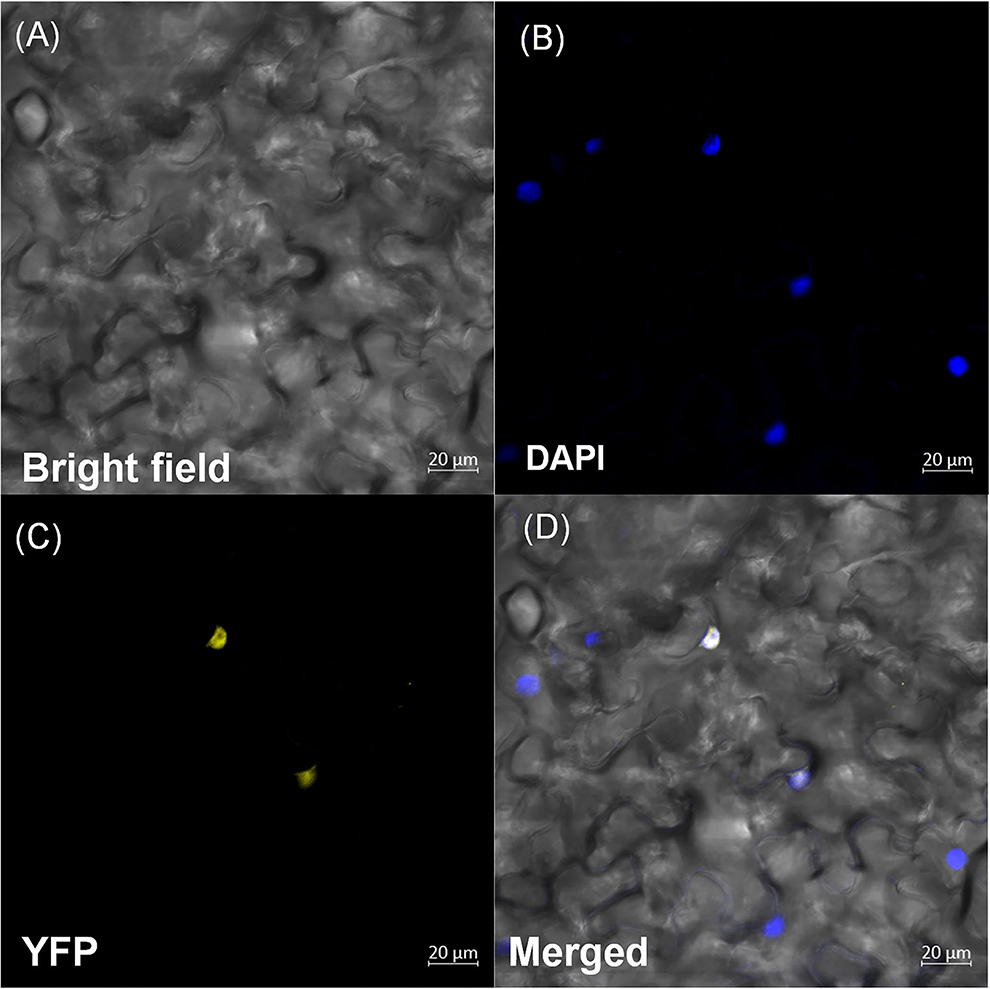
Figure 3. Subcellular localization of OsDOF27 protein. Six-week-old Nicotiana leaves were syringe-infiltrated at the abaxial surface with Agrobacterium strain harboring pSITEYFP3CA:OsDOF27 constructs. YFP fluorescence and differential interference contrast images and visible/GFP merged images are visualized by confocal microscopy. Scale bars = 20 μm. Confocal images of nicotiana leaves showing OsDOF27 targeted to the nucleus, with (A) bright field; (B) DAPI; (C) YFP; (D) merged or overlay.
OsDOF27 imparts thermotolerance in yeast
As OsDOF27 was highly induced under heat stress, we examined the thermotolerance potential of yeast cells harboring OsDOF27. For this, the OsDOF27 was cloned in pDEST-52 in which the OsDOF27 was regulated by gal4 promoter. After verifying the sequence of construct by Sanger sequencing, the construct (pDEST-52-DOF27 or pDEST-52-VC) was mobilized to the yeast strain (INVSc1). We then examined the thermotolerance potential of yeast transformants using growth kinetics and the spot assay. The growth analysis indicated that OsDOF27 ameliorates the thermal stress response of yeast cells expressing OsDOF27 (Figures 4A,B). Additionally, in an in silico analysis based on the Gibbs-Helmholtz equation ΔG(T)= ΔHm[1–(T/Tm)]–ΔCp[Tm–T+T Log(T/Tm)], the OsDOF27 exhibited high Tm (melting temperature) of 69.5°C, which indicates its thermostability (Figure 4C). A Tm value indicates the temperature for a protein at which it gets denatured resulting in its functional impairment. For a protein to be thermally stable, it needs a more negative value for ΔH, which makes the ΔG value negative and thus more thermally stable.
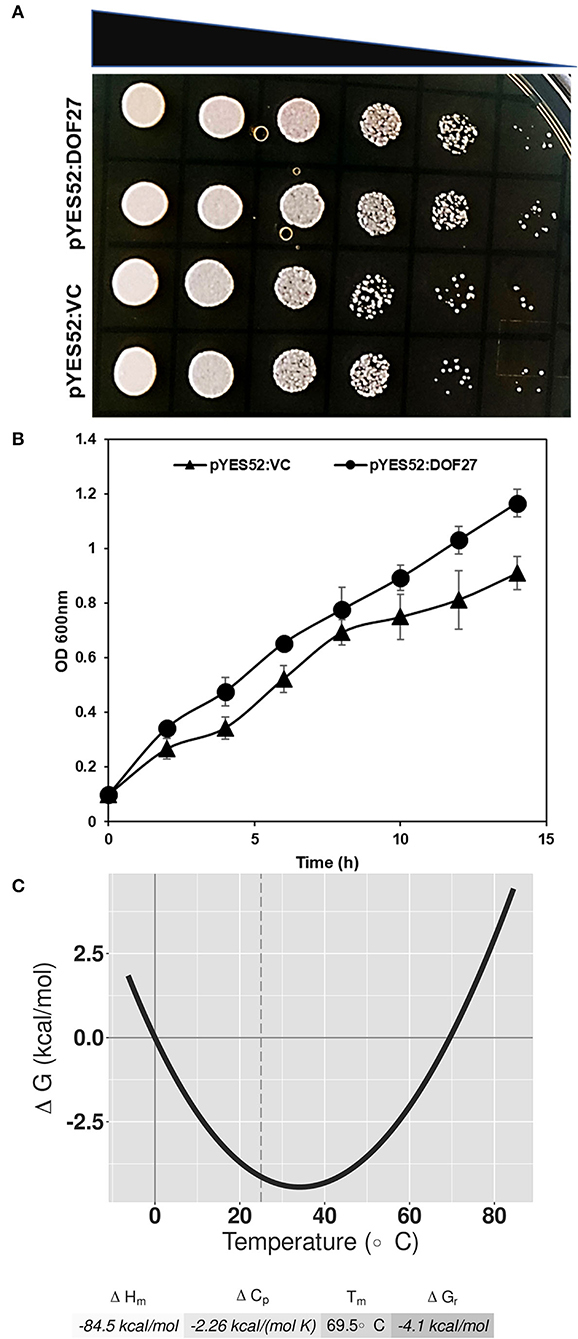
Figure 4. Thermotolerance potential of yeast is shown by (A) Spot assay which represents the comparative growth of yeast cells harboring pDEST52:OsDOF27 and empty vector. Five microliters of each yeast sample (two colonies) were used for spot assay with 10-fold serial dilutions. (B) the curve depicts the growth of yeast cells harboring pDEST52:OsDOF27 and empty vector under thermal stress. (C) The thermal stability curve represents the standard free energy of protein folding against the temperature.
Overexpression of OsDOF27 improves seed germination and stress response in rice transgenics
For functional analysis of OsDOF27, we generated rice transgenic lines overexpressing OsDOF27 using Agrobacterium-mediated rice transformation (Figure 5A). The enhanced transcript level of OsDOF27 in transgenic lines was assessed through qRT-PCR analysis (Figure 5B). Three independent transgenic lines (T2 lines) with enhanced expression of OsDOF27 were then assessed for heat stress tolerance as qRT results of OsDOF27, and yeast thermotolerance assay indicated its role in heat stress. For this, the transgenic lines were challenged with heat stress at the germination stage and the seedling stage. First, the germination potential of three independent lines of T2 transgenic seed of OsDOF27-OE with enhanced OsDOF27 expression along with the WT counterpart was examined. The transgenic seeds of the overexpression line showed better germination potential as compared to wild-type during heat stress (Figure 6A). While in the non-stressed condition, the transgenic and WT seeds did not show any significant difference in the germination assay. At the vegetative stage, when the seedlings were exposed to 45°C for 24 h and then transferred to 28°C, all three transgenic lines of OsDOF27-OE recovered better in comparison to their control counterpart (WT and VC; Figures 6B,C). The survival rate of transgenic plants was almost double than that of wild-type counterparts (Figure 6D). Besides, under non-stressed conditions, OsDOF27-OE lines behaved similar to WT and VC and did not exhibit any significant differences.
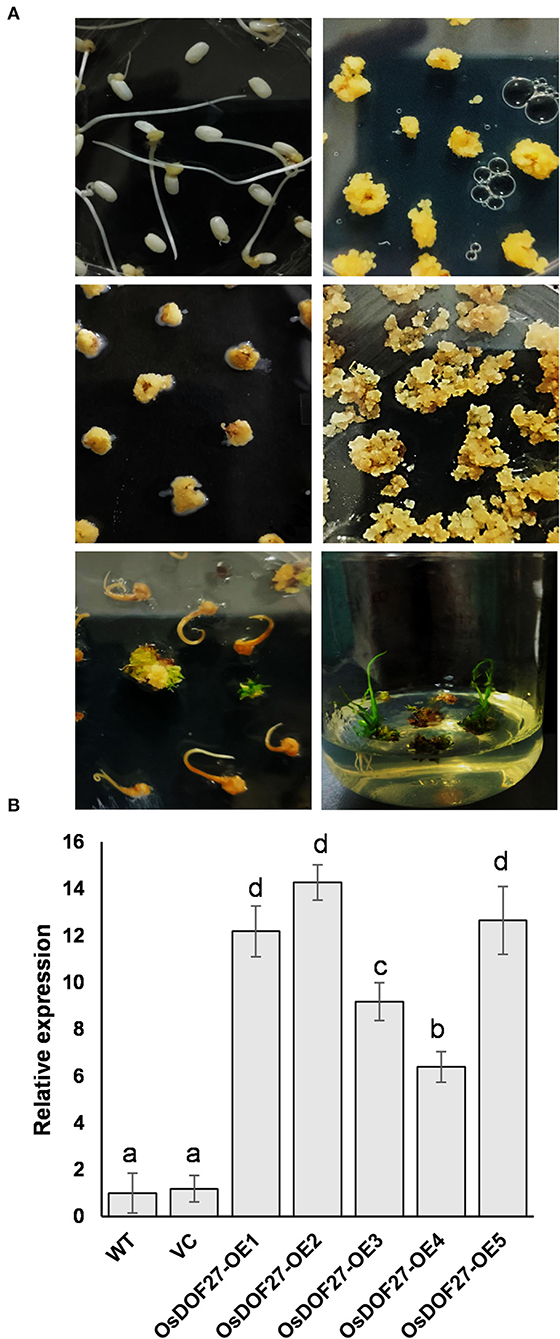
Figure 5. (A) Schematic representation of the Agrobacterium-mediated rice transformation of OsDOF27. (B) qPCR analysis of positive transformants. Total RNA was extracted from independent transgenic lines, and cDNA was synthesized followed by qPCR. Two endogenous controls (Ubiquitin-5 and EF1α) were used to normalize the relative expression value of OsDOF27 followed by calculation using the ΔΔCT method. The error bars represent the standard deviation of triplicate analysis. The significant difference in the mean of the values is indicated by different letters (α = 0.01).
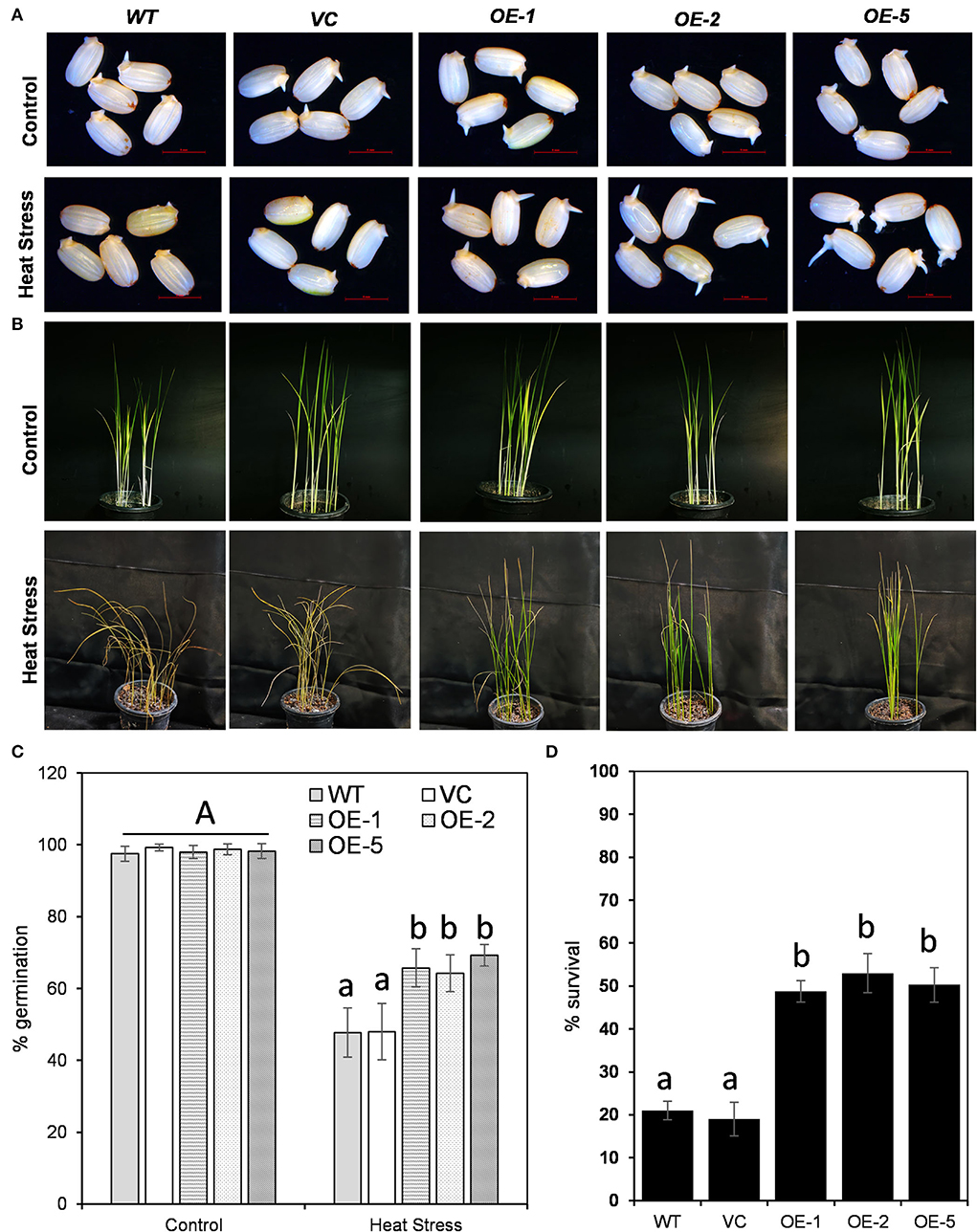
Figure 6. Comparative stress tolerance potential of transgenic lines overexpressing OsDOF27 and its control counterpart (WT, wildtype plants, and VC, vector control plants) at (A) germination stage and (B) seedling stages. (C) quantitative analysis of percent germination and (D) percent survival of transgenic lines overexpressing OsDOF27 in comparison to WT and VC under heat stress treatment. The error bars represent the standard deviation of triplicate analysis. The significant difference in the mean of the values is indicated by different letters (α = 0.01).
Discussion
In general, plants possess incredible molecular plasticity, which supports their growth and development during their inevitable exposure to abiotic stresses. Some plants have an intrinsic ability for higher stress tolerance than others, such tolerance is dependent on different genetic and environmental factors of the plant. The ability of plants to tolerate the stressful milieu is largely controlled by their highly complex yet regulated transcriptional process (Manna et al., 2021; Strader et al., 2022). The transcription factors play a crucial role by regulating the expression of their target gene and controlling almost all the biological processes, including seed maturation, growth and development, stress tolerance, and senescence to name a few (Yanagisawa, 2002; Dubos et al., 2010; Manna et al., 2021). Being a DNA binding protein, the role of TFs is largely known for regulating the transcription by binding to the promoter region of their target gene. However, in light of recent research developments in protein biology, intrinsically disorder proteins are attaining much focus. The IDPs challenge the sequence–structure–function paradigm, as IDPs do not possess a stable three-dimension structure. The IDP with molecular flexibility possesses the ability to interact with different interacting partners due to the altered conformation under different physiological conditions (Sun et al., 2013; Zamora-Briseño et al., 2021). IDP or the presence of IDRs in the protein facilitates functional versatility. Apparently, the presence of abundant IDP or IDR in plants explains the molecular plasticity that the plant might have acquired during the course of evolution to support their adequate growth and development (Pazos et al., 2013).
Here, in this study, we have the intrinsically disorder nature of DOF-TF. We characterize the molecular role of OsDOF27 under heat stress using two important systems, that is yeast and rice. The in silico analysis showed that the OsDOF27 is involved in different biological processes, which indicated its functional versatility (Supplementary Figure 3). The promoter region of OsDOF27 was enriched in abiotic stress-responsive cis-regulatory elements, and the expression profile tested also showed that OsDOF27 induces under abiotic stress. Besides, stress response, its promoter region is also enriched in cis-elements related to the response of phytohormones such as ABA, ethylene, and JA. The promoter region also showed a high propensity for the DOF-TF binding site, indicating its self-regulation or regulation by other members of the DOF-TF family (Supplementary Table 2). The ponder fit analysis indicated that OsDOF27 is an IDP; however, the region encompassing the DOF domain (109–158 aa; Supplementary Figure 4) has a very low disorder region. Since the DOF domain is a characteristic domain of the DOF family with a highly conserved region, which is known as the DNA binding region, this domain needs to have an ordered promoting amino acid. The abundance of arginine (R), serine (S), alanine (A), glycine (G), and proline (P) amino acids in the OsDOF27 indicated its discorded nature and thus its structural flexibility. Also, the predicted secondary structure of OsDOF27 encompasses more than 50% protein as a random coil (55%) supporting the disorder region of the protein (Smith et al., 1996). The abundance of glycine and proline in the OsDOF27 drives it entropically unpropitious to form ordered secondary structures and makes the polypeptide more disordered and flexible. As glycine (lack side chain) caters to flexibility, proline (constrained phi angle) is too rigid to facilitate the adequate secondary structure (Eyles and Gierasch, 2000; Rauscher et al., 2006; Muiznieks and Keeley, 2010) Being a transcription factor, OsDOF27 localizes to the nucleus, also the similar subcellular localization for DOF proteins are reported previously (Corrales et al., 2017; Su et al., 2017). The OsDOF27 imparts yeast thermotolerance, which was revealed by spot assay as well as growth curve analysis. The thermal stability curve also showed that OsDOF27 possesses negative ΔG between temperatures of 20° and 40°C, indicating its stability index within the temperature range (Pucci et al., 2017; Yadav et al., 2021). Besides, the high negative value of Tm and less negative ΔH with negative ΔG signifies the thermal stability of OsDOF27. In planta analysis for thermotolerance also depicted that rice transgenic lines with enhanced expression of OsDOF27 were more tolerant to heat stress exposure as compared to their wild-type counterpart. Transgenic lines displayed improved tolerance potential at germination and the seedling stage, which indicated the important role of OsDOF27 under heat stress challenges. Similarly, JrDOF3 was found to have a positive role in heat stress tolerance by binding to the DOFCOREZM motif of JrGRAS2 to regulate its expression. JrGRAS2 in turn regulates the expression of heat shock proteins and thereby improves high temperature stress tolerance (Yang et al., 2018). In a recent transcriptomics study in cucumber, DOF transcription factor was found to be upregulated (Yu et al., 2022). A transcriptomic and proteomic analysis would be conducted to understand the candidate molecular targets of OsDOF27, which will allow us to get deeper insight and delineate the detailed molecular mechanisms of heat stress tolerance imparted by DOF27 in rice. Besides, the protein–protein interaction analysis of OsDOF27 is also underway to comprehend the integrated network of this intrinsically disorder protein and other biological functions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
PS conceptualized the manuscript, supervised the authors, wrote the manuscript, analyzed the data, and edited the manuscript. NG, K, and PS performed the experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the research funding to PS from the Department of Science and Technology (DST), Government of India under the scheme of DST-INSPIRE Faculty Award (DST/INSPIRE/04/2018/003425), and SERB Core-Research-Grant (CRG/2021/000949), Government of India.
Acknowledgments
The authors gratefully acknowledge the Executive Director, NABI, Mohali for constant support. The authors also thank the DBT-e-Library Consortium (DeLCON) at National Agri-Food Biotechnology Institute for providing e-resource facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.956299/full#supplementary-material
References
Brodsky, S., Jana, T., Mittelman, K., Chapal, M., Kumar, D. K., Carmi, M., et al. (2020). Intrinsically disordered regions direct transcription factor in vivo binding specificity. Mol. Cell 79, 459–471.e4. doi: 10.1016/j.molcel.2020.05.032
Cai, X., Zhang, C., Shu, W., Ye, Z., Li, H., and Zhang, Y. (2016). The transcription factor SlDof22 involved in ascorbate accumulation and salinity stress in tomato. Biochem. Biophys. Res. Commun. 474, 736–741. doi: 10.1016/j.bbrc.2016.04.148
Cai, X., Zhang, Y., Zhang, C., Zhang, T., Hu, T., Ye, J., et al. (2013). Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J. Integr. Plant Biol. 55, 552–566. doi: 10.1111/jipb.12043
Chandran, A. K. N., Moon, S., Yoo, Y.-H., Gho, Y.-S., Cao, P., Sharma, R., et al. (2019). A web-based tool for the prediction of rice transcription factor function. Database 2019:baz061. doi: 10.1093/database/baz061
Cohen, S. P., and Leach, J. E. (2019). Abiotic and biotic stresses induce a core transcriptome response in rice. Sci. Rep. 9, 1–11. doi: 10.1038/s41598-019-42731-8
Corrales, A.-R., Carrillo, L., Lasierra, P., Nebauer, S. G., Dominguez-Figueroa, J., Renau-Morata, B., et al. (2017). Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant. Cell Environ. 40, 748–764. doi: 10.1111/pce.12894
Das, R. K., and Pappu, R. V. (2013). Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. U. S. A. 110, 13392–13397. doi: 10.1073/pnas.1304749110
Das, R. K., Ruff, K. M., and Pappu, R. V. (2015). Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 32, 102–112. doi: 10.1016/j.sbi.2015.03.008
DeForte, S., and Uversky, V. N. (2017). Not an exception to the rule: the functional significance of intrinsically disordered protein regions in enzymes. Mol. Biosyst. 13, 463–469. doi: 10.1039/C6MB00741D
Dong, C., Hu, H., and Xie, J. (2016). Genome-wide analysis of the DNA-binding with one zinc finger (Dof) transcription factor family in bananas. Genome 59, 1085–1100. doi: 10.1139/gen-2016-0081
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Eyles, S. J., and Gierasch, L. M. (2000). Multiple roles of prolyl residues in structure and folding. J. Mol. Biol. 301, 737–747. doi: 10.1006/jmbi.2000.4002
Fornasiero, A., Wing, R. A., and Ronald, P. (2022). Rice domestication. Curr. Biol. 32, R20–R24. doi: 10.1016/j.cub.2021.11.025
Geourjon, C., and Deléage, G. (1995). SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 11, 681–684. doi: 10.1093/bioinformatics/11.6.681
Guo, Y., and Qiu, L.-J. (2013). Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 8:e76809. doi: 10.1371/journal.pone.0076809
Gupta, S., Malviya, N., Kushwaha, H., Nasim, J., Bisht, N. C., Singh, V. K., et al. (2015). Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta. 241, 549–562. doi: 10.1007/s00425-014-2239-3
Holehouse, A. S., Ahad, J., Das, R. K., and Pappu, R. V. (2015). CIDER: classification of intrinsically disordered ensemble regions. Biophys. J. 108:228a. doi: 10.1016/j.bpj.2014.11.1260
Hrmova, M., and Hussain, S. S. (2021). Plant transcription factors involved in drought and associated stresses. Int. J. Mol. Sci. 22:5662. doi: 10.3390/ijms22115662
Jung, J.-H., Seo, P. J., Kang, S. K., and Park, C.-M. (2011). miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Mol. Biol. 76, 35–45. doi: 10.1007/s11103-011-9759-z
Kaur, H., Manna, M., Thakur, T., Gautam, V., and Salvi, P. (2021). Imperative role of sugar signaling and transport during drought stress responses in plants. Physiol. Plant. 171, 833–848. doi: 10.1111/ppl.13364
Kikuchi, S., Satoh, K., Nagata, T., Kawagashira, N., Doi, K., Kishimoto, N., et al. (2003). Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301, 376–379. doi: 10.1126/science.1081288
Kisu, Y., Esaka, M., and Suzuki, M. (1995). Putative zinc-binding domain of plant transcription factor, AOBP, is related to DNA-binding domains of steroid hormone receptors and GATA1. Proc. Japan Acad. Ser. B 71, 288–292. doi: 10.2183/pjab.71.288
Kisu, Y., Ono, T., Shimofurutani, N., Suzuki, M., and Esaka, M. (1998). Characterization and expression of a new class of zinc finger protein that binds to silencer region of ascorbate oxidase gene. Plant cell Physiol. 39, 1054–1064. doi: 10.1093/oxfordjournals.pcp.a029302
Kyte, J., and Doolittle, R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. doi: 10.1016/0022-2836(82)90515-0
Lijavetzky, D., Carbonero, P., and Vicente-Carbajosa, J. (2003). Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 3, 1–11. doi: 10.1186/1471-2148-3-17
Liu, J., Perumal, N. B., Oldfield, C. J., Su, E. W., Uversky, V. N., and Dunker, A. K. (2006). Intrinsic disorder in transcription factors. Biochemistry 45, 6873–6888. doi: 10.1021/bi0602718
Manna, M., Thakur, T., Chirom, O., Mandlik, R., Deshmukh, R., and Salvi, P. (2021). Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiol. Plant. 172, 847–868. doi: 10.1111/ppl.13268
Muiznieks, L. D., and Keeley, F. W. (2010). Proline periodicity modulates the self-assembly properties of elastin-like polypeptides. J. Biol. Chem. 285, 39779–39789. doi: 10.1074/jbc.M110.164467
Noguero, M., Atif, R. M., Ochatt, S., and Thompson, R. D. (2013). The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 209, 32–45. doi: 10.1016/j.plantsci.2013.03.016
Pazos, F., Pietrosemoli, N., García-Martín, J. A., and Solano, R. (2013). Protein intrinsic disorder in plants. Front. Plant Sci. 4:363. doi: 10.3389/fpls.2013.00363
Pérez-Rodríguez, P., Riano-Pachon, D. M., Corrêa, L. G. G., Rensing, S. A., Kersten, B., and Mueller-Roeber, B. (2010). PlnTFDB: updated content and new features of the plant transcription factor database. Nucl. Acids Res. 38, D822–D827. doi: 10.1093/nar/gkp805
Pucci, F., Kwasigroch, J. M., and Rooman, M. (2017). SCooP: an accurate and fast predictor of protein stability curves as a function of temperature. Bioinformatics 33, 3415–3422. doi: 10.1093/bioinformatics/btx417
Pucci, F., and Rooman, M. (2014). Stability curve prediction of homologous proteins using temperature-dependent statistical potentials. PLoS Comput. Biol. 10:e1003689. doi: 10.1371/journal.pcbi.1003689
Rasheed, R., Ashraf, M. A., Iqbal, M., Hussain, I., Akbar, A., Farooq, U., et al. (2020). “Major constraints for global rice production: changing climate, abiotic and biotic stresses,” in Rice Research for Quality Improvement: Genomics and Genetic Engineering, ed A. Roychoudhury (Singapore: Springer). 15–45. doi: 10.1007/978-981-15-4120-9_2
Rauscher, S., Baud, S., Miao, M., Keeley, F. W., and Pomes, R. (2006). Proline and glycine control protein self-organization into elastomeric or amyloid fibrils. Structure 14, 1667–1676. doi: 10.1016/j.str.2006.09.008
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C.-Z., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Salladini, E., Jørgensen, M. L. M., Theisen, F. F., and Skriver, K. (2020). Intrinsic disorder in plant transcription factor systems: functional implications. Int. J. Mol. Sci. 21:9755. doi: 10.3390/ijms21249755
Salvi, P., Agarrwal, R., Kajal Gandass, N., Manna, M., Kaur, H., et al. (2022). Sugar transporters and their molecular tradeoffs during abiotic stress responses in plants. Physiol. Plant. 2022:e13652. doi: 10.1111/ppl.13652
Salvi, P., Kumar, B., Kamble, N. U., Hazra, A., and Majee, M. (2021a). A conserved NAG motif is critical to the catalytic activity of galactinol synthase, a key regulatory enzyme of RFO biosynthesis. Biochem. J. 478, 3939–3955. doi: 10.1042/BCJ20210703
Salvi, P., Manna, M., Kaur, H., Thakur, T., Gandass, N., Bhatt, D., et al. (2021b). Phytohormone signaling and crosstalk in regulating drought stress response in plants. Plant Cell Rep. 40, 1305–1329. doi: 10.1007/s00299-021-02683-8
Shaw, L. M., McIntyre, C. L., Gresshoff, P. M., and Xue, G.-P. (2009). Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct. Integr. Genomics 9, 485–498. doi: 10.1007/s10142-009-0130-2
Shigyo, M., Tabei, N., Yoneyama, T., and Yanagisawa, S. (2007). Evolutionary processes during the formation of the plant-specific Dof transcription factor family. Plant Cell Physiol. 48, 179–185. doi: 10.1093/pcp/pcl044
Singh, R., and Jwa, N.-S. (2013). Understanding the responses of rice to environmental stress using proteomics. J. Proteome Res. 12, 4652–4669. doi: 10.1021/pr400689j
Smith, L. J., Fiebig, K. M., Schwalbe, H., and Dobson, C. M. (1996). The concept of a random coil: Residual structure in peptides and denatured proteins. Fold. Des. 1, R95–R106. doi: 10.1016/S1359-0278(96)00046-6
Strader, L., Weijers, D., and Wagner, D. (2022). Plant transcription factors—being in the right place with the right company. Curr. Opin. Plant Biol. 65:102136. doi: 10.1016/j.pbi.2021.102136
Su, Y., Liang, W., Liu, Z., Wang, Y., Zhao, Y., Ijaz, B., et al. (2017). Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum. J. Plant Physiol. 218, 222–234. doi: 10.1016/j.jplph.2017.07.017
Sun, X., Rikkerink, E. H. A., Jones, W. T., and Uversky, V. N. (2013). Multifarious roles of intrinsic disorder in proteins illustrate its broad impact on plant biology. Plant Cell 25, 38–55. doi: 10.1105/tpc.112.106062
Thakur, T., Gandass, N., Mittal, K., Jamwal, P., Muthamilarasan, M., and Salvi, P. (2021). A rapid, efficient, and low-cost BiFC protocol and its application in studying in vivo interaction of seed-specific transcription factors, RISBZ and RPBF. Funct. Integr. Genomics 21, 593–603. doi: 10.1007/s10142-021-00801-z
Thakur, T., Sinha, K., Kaur, T., Kapoor, R., Kumar, G., Bhunia, R. K., et al. (2022). Efficient genetic transformation of rice for CRISPR/Cas9 mediated genome-editing and stable overexpression studies: a case study on rice lipase 1 and galactinol synthase encoding genes. Agronomy 12:179. doi: 10.3390/agronomy12010179
Vacic, V., Uversky, V. N., Dunker, A. K., and Lonardi, S. (2007). Composition Profiler: a tool for discovery and visualization of amino acid composition differences. BMC Bioinformat. 8, 1–7. doi: 10.1186/1471-2105-8-211
Wani, S. H., Anand, S., Singh, B., Bohra, A., and Joshi, R. (2021). WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep. 40, 1071–1085. doi: 10.1007/s00299-021-02691-8
Waqas, M., Shahid, L., Shoukat, K., Aslam, U., Azeem, F., and Atif, R. M. (2020). “Role of DNA-binding with one finger (Dof) transcription factors for abiotic stress tolerance in plants,” in Transcription Factors for Abiotic Stress Tolerance in Plants, ed S. H. Wani (Cambridge, MA: Academic Press), 1–14. doi: 10.1016/B978-0-12-819334-1.00001-0
Yadav, S., Pandey, A. K., and Dubey, S. K. (2021). Molecular modeling, docking and simulation dynamics of β-glucosidase reveals high-efficiency, thermo-stable, glucose tolerant enzyme in Paenibacillus lautus BHU3 strain. Int. J. Biol. Macromol. 168, 371–382. doi: 10.1016/j.ijbiomac.2020.12.059
Yanagisawa, S. (1995). A novel DNA-binding domain that may form a single zinc finger motif. Nucl. Acids Res. 23, 3403–3410. doi: 10.1093/nar/23.17.3403
Yanagisawa, S. (2002). The Dof family of plant transcription factors. Trends Plant Sci. 7, 555–560. doi: 10.1016/S1360-1385(02)02362-2
Yanagisawa, S. (2004). Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 45, 386–391. doi: 10.1093/pcp/pch055
Yang, G., Gao, X., Ma, K., Li, D., Jia, C., Zhai, M., et al. (2018). The walnut transcription factor JrGRAS2 contributes to high temperature stress tolerance involving in Dof transcriptional regulation and HSP protein expression. BMC Plant Biol. 18:367. doi: 10.1186/s12870-018-1568-y
Yang, X., Tuskan, G. A., and Cheng, Z.-M. (2006). Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol. 142, 820–830. doi: 10.1104/pp.106.083642
Yokotani, N., Ichikawa, T., Kondou, Y., Matsui, M., Hirochika, H., Iwabuchi, M., et al. (2009). Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta 229, 1065–1075. doi: 10.1007/s00425-009-0895-5
Yu, B., Ming, F., Liang, Y., Wang, Y., Gan, Y., Qiu, Z., et al. (2022). Heat stress resistance mechanisms of two cucumber varieties from different regions. Int. J. Mol. Sci. 23:31817. doi: 10.3390/ijms23031817
Zamora-Briseño, J. A., Pereira-Santana, A., Reyes-Hernández, S. J., Cerqueda-García, D., Castaño, E., and Rodríguez-Zapata, L. C. (2021). Towards an understanding of the role of intrinsic protein disorder on plant adaptation to environmental challenges. Cell Stress Chaperones 26, 141–150. doi: 10.1007/s12192-020-01162-5
Zhang, Z., Yuan, L., Liu, X., Chen, X., and Wang, X. (2018). Evolution analysis of Dof transcription factor family and their expression in response to multiple abiotic stresses in Malus domestica. Gene 639, 137–148. doi: 10.1016/j.gene.2017.09.039
Zheng, Y., Jiao, C., Sun, H., Rosli, H. G., Pombo, M. A., Zhang, P., et al. (2016). iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 9, 1667–1670. doi: 10.1016/j.molp.2016.09.014
Keywords: intrinsically disorder proteins, heat stress response, DOF-transcription factor, crop improvement, transcriptional regulation
Citation: Gandass N, Kajal and Salvi P (2022) Intrinsically disordered protein, DNA binding with one finger transcription factor (OsDOF27) implicates thermotolerance in yeast and rice. Front. Plant Sci. 13:956299. doi: 10.3389/fpls.2022.956299
Received: 30 May 2022; Accepted: 30 June 2022;
Published: 29 July 2022.
Edited by:
Iftikhar Ali, Institute of Genetics and Developmental Biology (CAS), ChinaCopyright © 2022 Gandass, Kajal and Salvi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prafull Salvi, c2FsdmkucHJhZnVsbEBnbWFpbC5jb20=; cHJhZnVsbC5zYWx2aUBuYWJpLnJlcy5pbg==
†ORCID: Prafull Salvi https://orcid.org/0000-0002-7834-8803
 Nishu Gandass
Nishu Gandass Kajal
Kajal Prafull Salvi
Prafull Salvi