- State Key Laboratory of Crop Biology, College of Agronomy, Shandong Agricultural University, Tai’an, China
Powdery mildew, caused by Blumeria graminis f. sp. tritici (Bgt), is a devastating fungal disease that seriously damages the yield and quality of wheat in many regions of the world. Identifying new resistance genes and breeding new resistant varieties are effective methods to control this disease. The breeding line SN15218 shows good resistance against powdery mildew. We, therefore, developed an F2 population and 287 F2:3 families crossed between SN15218 and the powdery mildew susceptible cultivar Huixianhong (HXH). Genetic analysis indicated that a single dominant gene, designated herein PmSN15218, conferred resistance to the Bgt isolate E09 in SN15218. Bulked segregant RNA-Seq (BSR-Seq) analysis revealed that PmSN15218 is located in a ∼25-Mb interval on chromosome 2AL. Using the polymorphism information between SN15218 and HXH, we developed 13 polymerase chain reaction (PCR) markers and mapped this gene to a 0.5-cM genetic interval between the two flanking markers PmM12 and PmM14, corresponding to a 6.01-Mb physical region in the Chinese Spring reference genome. The results of molecular marker analysis, allelic tests of resistance spectrum, and DNA resequencing indicated that PmSN15218 is distinct from the known resistance gene Pm4b on 2AL.
Introduction
Wheat (Triticum aestivum L.) is an important food crop worldwide, providing about 20% of human calories (Ni et al., 2017). However, wheat quality and yield stability are frequently affected by fungal diseases. For example, powdery mildew caused by Blumeria graminis f. sp. tritici (Bgt), is a devastating wheat foliar disease. In recent decades, powdery mildew has led to severe yield losses of 5–8% during the years of average infection, and as high as 40% in some years, in China and other parts of the world (Singh et al., 2016). In addition to fungicides and other biological agents, the deployment of disease-resistant cultivars is the most economical and environmentally friendly method for managing this foliar disease (Fu et al., 2013; Qie et al., 2019). However, because frequent changes in pathogen populations often overcome the effects of available resistance genes (Yu et al., 2018), most reported powdery mildew resistance genes in wheat are race-specific and susceptible to resistance loss once they are widely deployed in commonly planted cultivars. Therefore, it is necessary for breeders to discover additional sources of powdery mildew resistance genes in breeding programs.
At present, more than 100 powdery mildew-resistance genes or alleles mapping to 63 different loci (Pm1–Pm68, Pm18/Pm1c, Pm22/Pm1e, Pm23/Pm4c, Pm31/Pm21, Pm48/Pm46, and Pm17/Pm8) have been reported, with the loci Pm1, Pm3, Pm4, Pm5, and Pm24 having 5, 17, 8, 5, and 2 alleles, respectively (Li et al., 2020; McIntosh et al., 2020; He et al., 2021; Zhang et al., 2021; Wu et al., 2022). These include five resistance alleles, Pm4a–Pm4e, reported at the locus Pm4, located on the long arm of chromosome 2A. Pm4a was first identified in the emmer wheat cultivar Khapli and the durum wheat cultivar Yuma and was linked to marker Xgwm356 (Ma et al., 2004). Pm4b was derived from the French common wheat cultivar VPM1, a lineage derived from a cross between Aegilops ventricosa, Triticum turgidum var. carthlicum (Triticum persicum), and the common wheat variety Marne (Bariana and McIntosh, 1994), and has been mapped to a 3.0-cM genetic interval between flanking markers Xics43 and Xics13 in chromosome 2AL (Wu et al., 2018). Over time, wheat cultivars carrying Pm4a have gradually lost resistance to powdery mildew in many countries and regions, such as the United States, the Middle East, and China, while Pm4b, which was identified over 30 years ago, still provides effective resistance in parts of China and the United States (El-Shamy et al., 2016; Cowger et al., 2017; Wu et al., 2018). Pm4c (also called Pm23) comes from the common wheat line 81-7241, which exhibits high resistance to powdery mildew and has a broad resistance spectrum. Pm4c is located at a 4.9-cM genetic interval between Xbarc122 and Xgwm356 on chromosome 2AL (Hao et al., 2008). Pm4d and Pm4e were identified from the common wheat introgression line Tm27d2, which inherited its resistance from the Triticum monococcum accession Tm27 and the Chinese landrace D29, respectively. Pm4d was mapped to a 6.7-cM genetic interval, while Pm4e was precisely mapped to a 0.19-cM interval corresponding to a 360-kb physical region (Schmolke et al., 2012; Ullah et al., 2018). Recently, researchers successfully cloned the Pm4a-Fed and Pm4b-Fed genes by sequencing related mutants from the wheat lines Fed-Pm4a and Fed-Pm4b (Fed refers to the common wheat cultivar “Federation”), respectively, and identified multiple resistant or susceptible haplotypes of Pm4-Fed simultaneously (Sánchez-Martín et al., 2021).
Novel powdery mildew resistance genes identified from modern cultivars are easier to use in breeding than those from wheat relatives or wild species. However, the powdery mildew resistance genes so far identified are almost from landraces or wild relatives and often have adverse factors or linkage drag of undesirable genes. Only a few, such as Pm2a, Pm4a, Pm6, Pm8, and Pm21, originated from modern cultivars and can be directly used in breeding programs (Qie et al., 2019). Moreover, of the 18 resistance genes (Pm51–Pm68) newly identified in recent years, only Pm52, Pm54, and Pm65 originated from adapted cultivars (Liangxing99, Pioneer 26R61, and Xinmai208, respectively) (Zhao et al., 2013; Hao et al., 2015; Li et al., 2019; He et al., 2021; Zhang et al., 2021).
SN15218 is derived from a natural mutant plant of the cultivar Yannong 19 (YN19) in the seed production field. It shows a high level of resistance to powdery mildew at both seedling and adult stages, suggesting that it harbors a powdery mildew resistance gene that could be valuable for wheat breeding. In this study, we characterized the resistance gene in SN15218, temporarily named PmSN15218, through genetic analysis, molecular mapping, spectrum analysis, and allelic relationship comparison.
Materials and Methods
Plant Materials
The materials used in this study included the winter wheat cultivars YN19 and Huixianhong (HXH) and powdery mildew resistant line, SN15218. YN19 and HXH are widely planted commercial cultivars and are susceptible to most Bgt isolates. SN15218 is a breeding line with a YN19 background. It is similar to common wheat and shows resistance to many powdery mildew races. To analyze the inheritance of powdery mildew resistance, SN15218 was crossed with HXH to construct a segregating population, and F1, F2, and F2:3 materials were tested for resistance to the Bgt isolate E09, which is avirulent on SN15218 but virulent on HXH.
Tests of Powdery Mildew Resistance
SN15218, HXH, and their F1 hybrids, F2 populations, and F2:3 families (25 seedlings for each line), as well as YN19 as a susceptible control, were tested for powdery mildew resistance using the Bgt isolate E09. Meanwhile, 23 single-spore-derived Bgt isolates and several wheat lines carrying known Pm genes were grown in a greenhouse at the Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, to compare the reaction patterns of SN15218 and the other wheat lines with the different Bgt isolates. At the two-leaf stage, seedlings were inoculated with fresh spores and then transferred to a plant growth chamber at 15–18°C with 60% relative humidity and a 12 h light/12 h dark cycle to allow symptom development. Two weeks after the susceptible control materials HXH and YN19 were heavily infected, the plants were scored into infection types (ITs) according to a 0–4 scale (Tan et al., 2018). We classified the plants into two groups: resistant (R, IT = 0–2) and susceptible (S, IT = 3–4). Observed and expected segregation ratios were compared using the Chi-squared (χ2) test for goodness of fit.
Bulked Segregant RNA-Seq Analysis
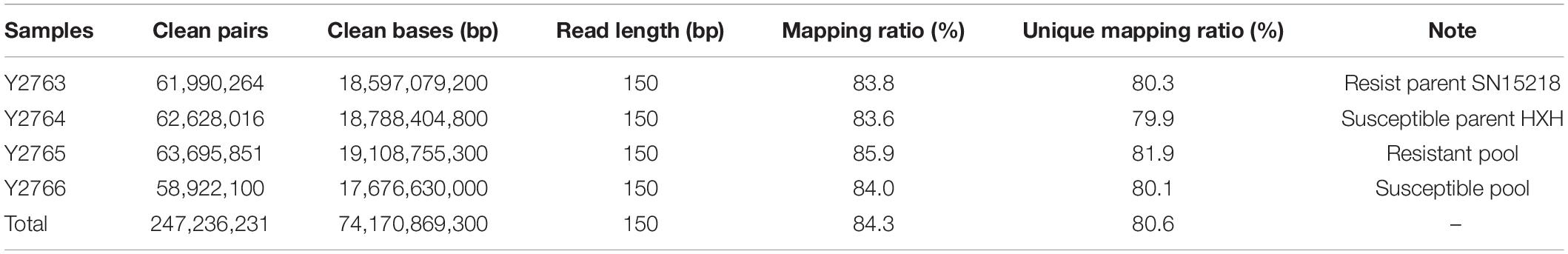
The F2:3 families with contrasting resistance phenotypes against isolate Bgt E09 (30 homozygous resistant and 30 susceptible families) were used to construct sample pools. Leaves of similar quality were collected from five plants of each family. Resistant and susceptible leaves were pooled for RNA isolation. Two parents, SN15218 and HXH, were also processed as parental checks. Total RNA was extracted using TRIzol reagent (Tiangen Biotech Co., Beijing, China) according to the manufacturer’s protocol. Library construction, high-throughput sequencing (Illumina NovaSeq 6000), and sequencing data qualification were performed by Berry Genomics Company (Beijing, China). Adapter sequences and low-quality bases were trimmed using fastp v0.19 with default paraments (Chen et al., 2018). High-quality reads were mapped to the Chinese Spring reference genome, RefSeq V1.0 (International Wheat Genome Sequencing Consortium, IWGSC), using Hisat2 v2.1.0 (Kim et al., 2019). Variant calling was performed using the HaplotypeCaller module in the GATK v3.8 toolkit (Poplin et al., 2018). Single-nucleotide polymorphism (SNP) filtration, sliding window analysis, and SNP-index plotting were conducted following a previously described protocol (Takagi et al., 2013). We excluded SNPs with read depth <5 from resistant and susceptible pools and two parent samples. SNP index values in the two pools were calculated using a custom perl script with SNPs of susceptible parent HXH as the reference, and the Δ(SNP-index) = (SNP-index of the resistant pool) − (SNP-index of the susceptible pool) was calculated for each SNP. Sliding window analysis was applied to ΔSNP-index plots with 1-Mb window size and 10-kb increment, and the average ΔSNP-index of SNPs was used for the sliding window plot (Takagi et al., 2013).
Genotyping and Linkage Analysis
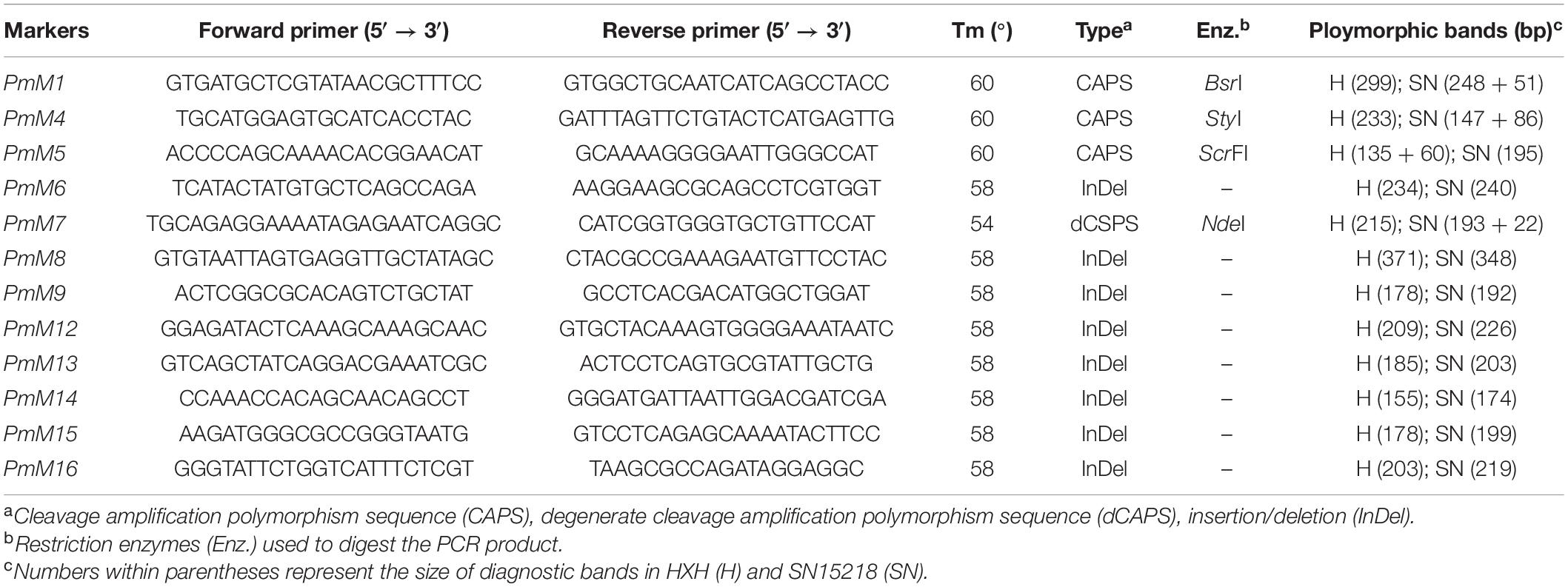
Leaf tissue was collected from 2-week-old plants, and total genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method (Porebski et al., 1997). According to the variants data from bulked segregant RNA-Seq (BSR-Seq) analysis, four polymerase chain reaction (PCR) markers were developed based on SNPs (Table 1). These markers were used to genotype the 285 F2 plants crossed with SN15218 and HXH and confirm the mapping result of BSR-Seq data. To develop more PCR markers, DNA resequencing and variants calling with two parents, SN15218 and HXH, were performed. The method of variant calling was similar to that described above for BSR-Seq, except that BWA v0.7.171 was used instead of Hisat2 for mapping. Based on DNA resequencing data and the Chinese Spring RefSeq v1.0 reference map, seven insertion/deletion (InDel) markers were newly developed (Table 1) for further genotyping. χ2 tests of the F2 and F2:3 populations were used to evaluate the goodness of fit between observed data and expected segregation ratios. A genetic linkage map was constructed using the software JoinMap 4.0.2 The recombination values were converted to genetic distance (cM) using the Kosambi mapping function.
De novo Assembly
DNA resequencing reads of the powdery mildew resistant line SN15218 were first aligned to the Triticeae repeat database mipsREdat 9.3p (PGSB Repeat Database) using BWA-mem v0.7.17 in order to filter out repeat noises. Non-mapped reads (which come from the genome region of non-repeat sequences) were fished using Samtools v1.93 and then used for de novo assembly using SPAdes v3.134 with default parameters.
Results
SN15218 Powdery Mildew Resistance Is a Monogenic Inherited Trait
When inoculated with Bgt isolate E09, SN15218 was highly resistant (IT = 0) and HXH was highly susceptible to the fungus (IT = 4) (Figure 1). F1 plants of SN15218 × HXH showed the same resistant phenotype as the parent SN15218, indicating the dominance of the powdery mildew resistance of SN15218. In the F2 population, 216 individuals showed resistance (IT = 0–2) and 69 individuals showed susceptibility (IT = 3–4), indicating segregation of a single resistance gene (χ23:1 = 0.09, P1df > 0.05). All the 285 F2:3 families derived from these two parents were also tested using the same Bgt isolate E09 in the climate chamber at the seedling stage. A total of 82 and 69 families were homozygous-resistant and homozygous-susceptible, respectively, and the remaining 134 families were segregated, confirming that the powdery mildew resistance of SN15218 was inherited as a single gene (χ21:2:1 = 2.2, P2df > 0.05). We temporarily named this resistance gene PmSN15218.

Figure 1. Powdery mildew infection of seedling from susceptible materials Huixianhong (HXH), Yannong 19 (YN19), and resistant parent SN15218 at 15 dpi inoculated with Blumeria graminis f. sp. tritici (Bgt) isolate E09.
PmSN15218 Gene Is Localized on Chromosome 2AL
We sequenced the two parents (SN15218 and HXH) and two resistant and susceptible descendant pools in 150-bp paired-end mode, generating 58.9–63.7 million clean paired reads; the average read depth was more than 60 for each SNP after filtration (Table 1). Approximately, 84.3% of the clean reads from each parent or pool could be mapped to the reference, and 80.6% of them were uniquely mapped and used for variant calling. We identified 46,895 high-quality homozygous SNPs between the two parents. Meanwhile, there were 1,027 and 1,791 homozygous SNPs in the resistant and susceptible pools, respectively, most of which were located on chromosome 2A (255 SNPs in the resistant pool, 645 SNPs in the susceptible pool). Only a single sharp peak (ΔSNP-index > 0.8) was identified on the long arm of wheat chromosome 2A, and these SNPs were enriched in a ∼25-Mb interval (Chr2A: Mb 751-776) (Figure 2), suggesting that the powdery mildew resistance gene of SN15218 was located on chromosome 2AL.
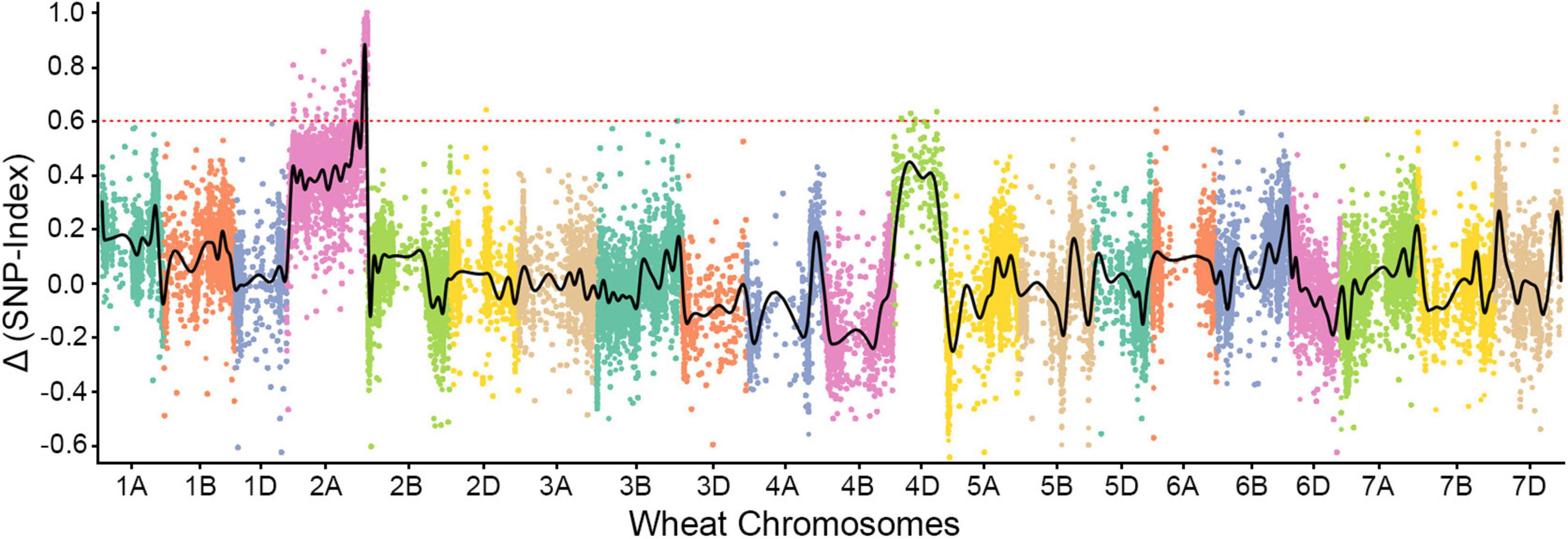
Figure 2. Bulked segregant RNA-Seq (BSR-Seq) analysis of PmSN15218. PmSN15218 was localized at the distal end of the long arm of chromosome 2A.
PmSN15218 Maps to a 0.5-cM Genetic Interval
In the ∼25-Mb target region, we dispersedly selected five co-segregating SNPs/InDels identified from BSR-Seq data and converted them into cleaved amplified polymorphic sequences (CAPSs)/derived cleaved amplified polymorphic sequences (dCAPSs) markers (PmM1, PmM4, PmM5, and PmM7) or an InDel marker (PmM6) (Table 2). Using 90 F2 plants from the offspring of hybridization between SNP15218 and HXH, we genotyped these five markers and found that they were all closely linked to PmSN15218, which confirmed the preliminary BSR-Seq mapping result. Based on DNA resequencing data, 6,172 SNPs/InDels were called in the ∼25-Mb target region, including 67 InDel sites larger than 10 bp between the 2 parents. We further created seven new InDel markers (Table 2) using the polymorphic information and reference data of Chinese Spring RefSeq v1.0 and mapped PmSN15218 to the 0.5-cM PmM12–PmM14 interval using 287 F2 plants, which corresponded to a 6.01-Mb physical region (chr2A: Mb 764.093–770.104) (Figure 3).
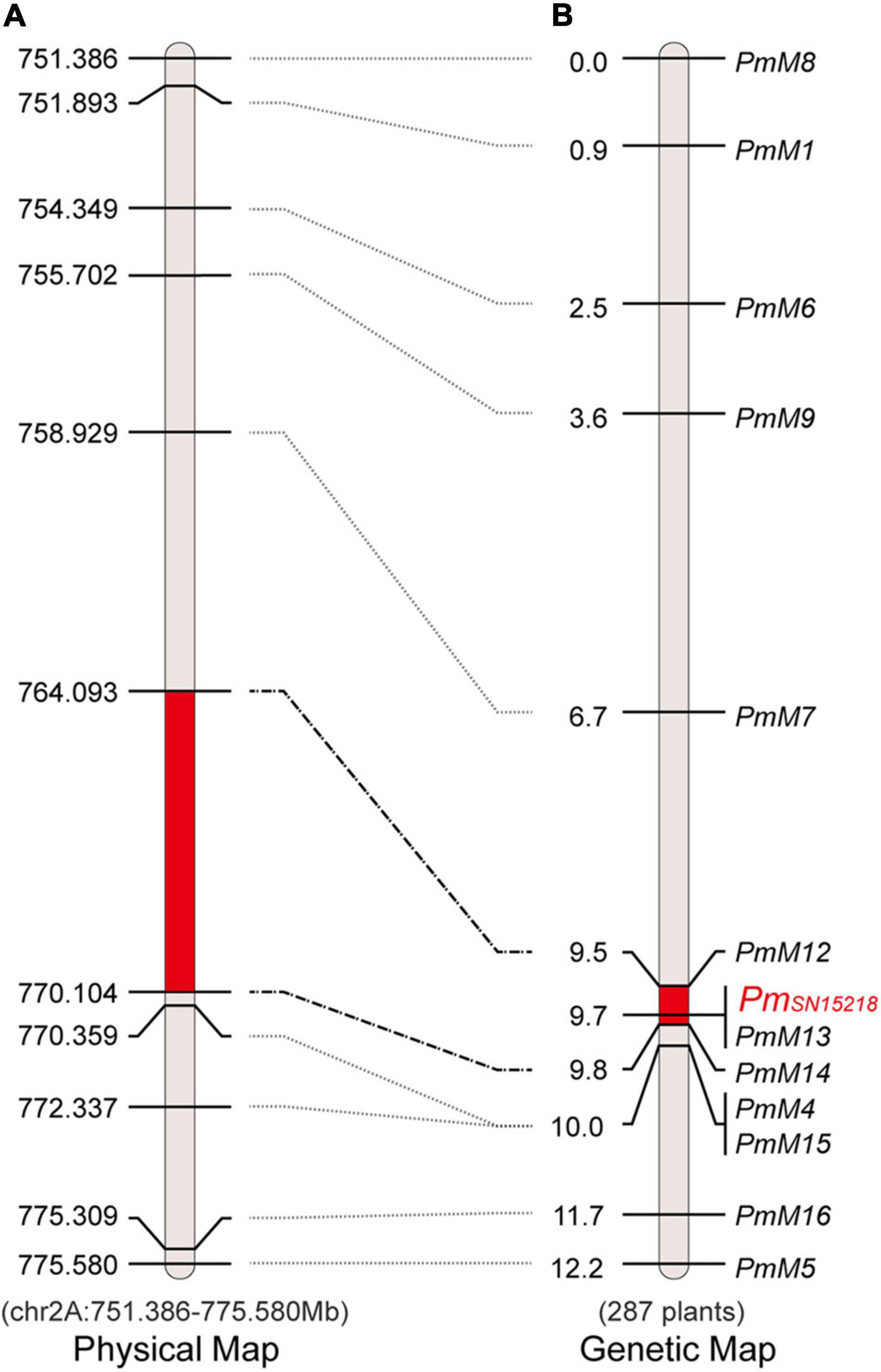
Figure 3. Physical and genetic map of PmSN15218 region. (A) Physical map of chromosome 2AL 751.4–775.6 Mb. (B) The precision genetic map with 287 F2 individuals. The map units were physical positions (Mb) or centimorgan (cM) that was labeled on the left sides of the map.
Resistance Spectra
In addition to PmSN15218 from SN15218, several powdery mildew resistance genes, Pm4a, Pm4b, and Pm4c, that were previously identified from different wheat varieties or lines (Khapli, Armada, and 81-7241, respectively) were also mapped on chromosome 2AL. We evaluated the disease reactions of SN15218 and the above resistant landraces to 23 Bgt isolates. Line 81-7241, carrying Pm4c, showed high resistance to all of the isolates. Khapli, carrying Pm4a, showed resistance to most powdery mildew isolates but was susceptible to seven isolates. Armada, carrying Pm4b, showed a similar resistance spectrum to Khapli except for two isolates, E05 and E15 (Table 3). Line SN15218 exhibited better resistance than Khapli and Armada and was susceptible to only four Bgt isolates, E18, E20, E31, and E32 (Table 3). The results demonstrated that the resistance spectrum of PmSN15218 was distinct from that of Pm4a, Pm4b, or Pm4c.
Allelic Relationship
To compare the physical locations of Pm genes at the Pm4 locus, including Pm4a (Hao et al., 2008; Fu et al., 2013), Pm4b-Fed (Sánchez-Martín et al., 2021), Pm4c (Pm23) (Hao et al., 2008), Pm4d (Schmolke et al., 2012), Pm4e (Ullah et al., 2018), Pm65 (Li et al., 2019), and PmSN15218, we integrated these tightly linked markers into chromosome 2AL of the Chinese Spring reference genome (IWGSC RefSeq v1.0) (Supplementary Table 1). These seven genes were located at the same physical interval: Pm4a was 4.8cM from marker Xgwm365, which was located in a region around Mb 762 of chromosome 2A; Pm4b-Fed, Pm4c (Pm23), Pm4d, Pm4e, Pm65, and PmSN15218 were located in the physical interval of Mb 760–770. Among them, the Pm4b-Fed gene, originating from tetraploid T. carthlicum, is not in the “Chinese Spring” genome reference but is most closely related to the gene TraesCS2A01G557900 near Mb 761 of chromosome 2AL, the ancestor of which was duplicated and fused to form the kinase structural domain of Pm4b-Fed (Sánchez-Martín et al., 2021). To further confirm the relationship between PmSN15218 and Pm4b-Fed, we performed a PCR assay using three Pm4-specific PCR markers, JS717 × JS718, Pm4.1, and Pm4b-Fed-S (newly developed in this study), and the results showed that SN15218 did not contain Pm4. We also performed de novo assembly using DNA resequencing data of SN15218 and found no Pm4b-Fed gene or fragment in the assembly database. In addition, we compared five other Pm genes, PmX (Fu et al., 2013), PmLK906 (Niu et al., 2008), PmPS5A (Niu et al., 2010), PmMl92145E8-9 (Yu et al., 2018), and PmXMM (Yao et al., 2022), which were also located on chromosome 2AL (Supplementary Table 1). Three of these, PmX, PmLK906, and PmPS5A, shared similar physical locations with PmSN15218, but PmMl92145E8-9 and PmXMM were located in the proximal region of the Pm4 locus.
Discussion
Powdery mildew is a widespread fungal disease of wheat, which can infect wheat at both the seedling stage and adult stage and causes significant yield losses. Loss of resistance in wheat cultivars caused by the rapid evolution of this pathogenic fungus has prompted a continuing search for new sources of resistance. Here, we identified a single dominant resistance gene (PmSN15218) near the Pm4 loci derived from the resistant mutant SN15218 through genetic analysis, molecular mapping, resistance spectrum investigation, and allelic relationship analysis. We tested different wheat genotypes against 23 Bgt isolates. The SN15218, carrying PmSN15218, displayed a different resistance pattern from wheat carrying Pm4a, Pm4b, or Pm4c. SN15218 was highly resistant to powdery mildew in fields and has the genetic background of common wheat. Therefore, it could serve as a good intermediate material for powdery mildew resistance breeding.
The PmSN15218 was mapped to a 0.5-cM genetic interval flanked by markers PmM12 and PmM14, responding to the physical region of Mb 764.1–770.1 on chromosome 2A in the Chinese Spring RefSeq V1.0. One hundred and twenty-three genes were annotated in this region. Among them, 12 encode for NBS-LRR-like resistance proteins or disease-resistance proteins, and 11 encode for receptor-like kinases or protein kinases. Comparative genomics analysis indicated that most genes in the PmSN15218 region were conserved between the common wheat Chinese Spring (IWGSC), wild emmer wheat Zavitan (Avni et al., 2017), and durum wheat Seveo (Maccaferri et al., 2019; Figure 4). However, there were some inversions between Chinese Spring and Svevo or Zavitan and small chromosome fragment translocations between Chinese Spring and Zavitan in the upstream flanking region at the proximal side (Figure 4). These results indicate that the colinear genes located upstream of the target region should be considered when filtering candidate genes. In addition, more molecular markers and a larger segregation population need to be developed to enable further fine mapping and cloning of PmSN15218.
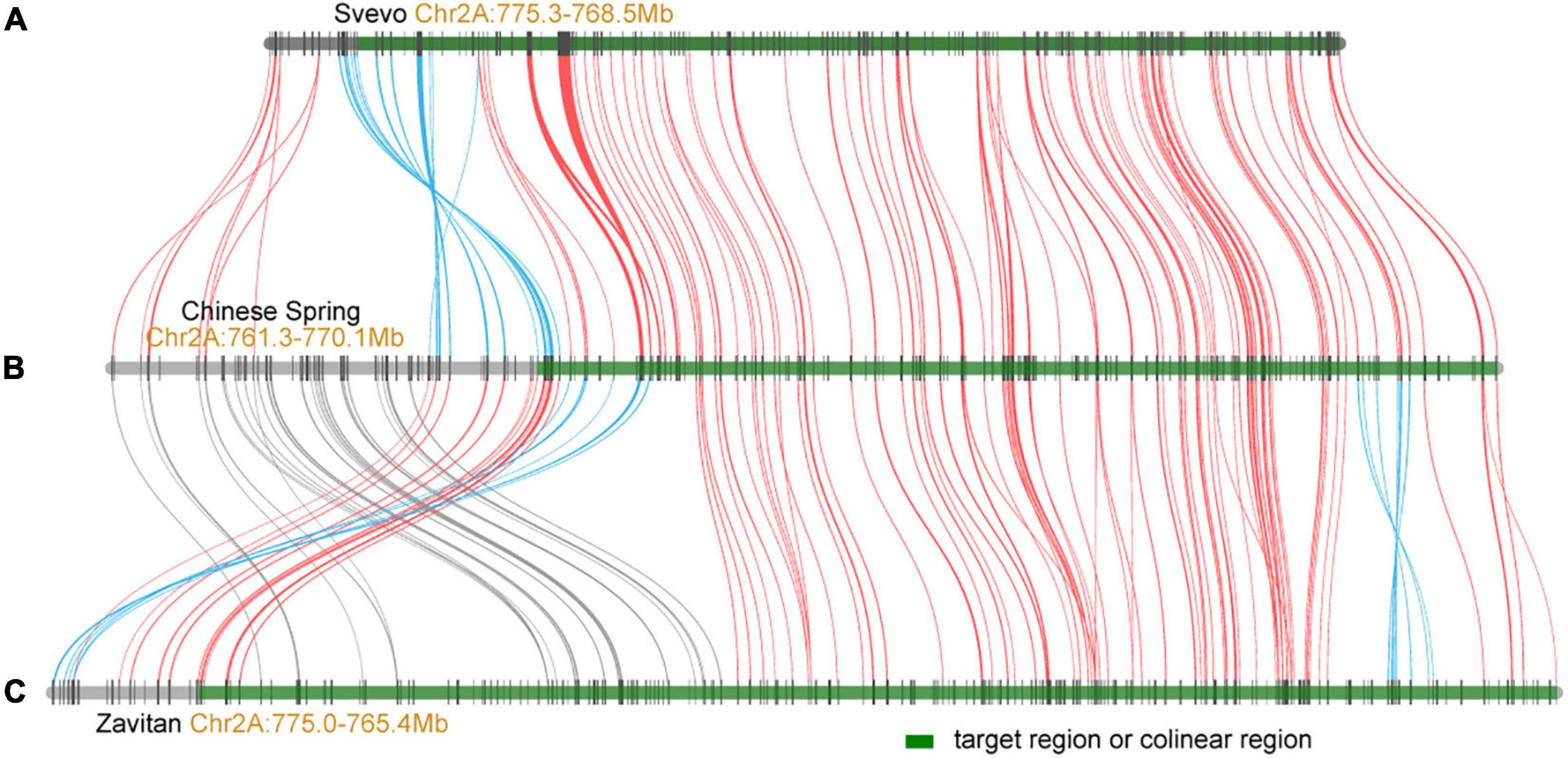
Figure 4. Collinearity of PmSN15218 region between durum wheat Svevo (A), common wheat Chinese Spring (B), and wild emmer wheat Zavitan (C). The green regions represent the colinear intervals of PmSN15218 gene.
The Pm4 loci for powdery mildew resistance in wheat are complex, and much research has focused on its localization, cloning, and breeding applications. Previous studies have shown that Pm4b is localized in the physical interval of Mb 771.887–779.732 on chromosome 2A (Wu et al., 2018; Supplementary Table 1). However, Sánchez-Martín et al. (2021) cloned the Pm4b-Fed gene from the wheat cultivar Federation using the MutChromSeq approach, and their analysis revealed that this gene was localized near Mb 761 on chromosome 2A. Pm4b is derived from the French cultivar VPM1, and VPM1 drives from a cross between A. ventricosa, T. turgidum var. carthlicum, and the common wheat (T. aestivum) variety Marne (Bariana and McIntosh, 1994). It was introduced to China from Europe in the 1980s for genetic improvement of wheat disease resistance breeding. Pm4b-Fed is derived from three wheat lines, Weihenstephan M1, ELS, and TP229, and was first named Mle, which was identified as an allele of Pm4a and then transferred to the Australian spring wheat variety Federation by backcrossing (Briggle, 1966; Wolfe, 1967; McIntosh and Bennett, 1979). Since the two genes have different donors, the contradictory localization results could be due to ectopic recombination that occurred during transfer to the common wheat background. To determine the allelic relationship of PmSN15218 and Pm4b-Fed, we tested JS717 × JS718 and Pm4.1, two specific markers for Pm4b-Fed (Sánchez-Martín et al., 2021; Yao et al., 2022), and a newly developed marker, Pm4b-Fed-S (Supplementary Table 1), and found that this gene was not present in HXH, YN19, or SN15218. In addition, we analyzed the closely linked markers of other Pm genes, including PmX, Pm65, PmLK906, PmPS5A, and PmMl92145E8-9, on chromosome 2AL using in silico PCR (Supplementary Table 2). These markers are either undetectable or differ in product size in the de novo assembly database of SN15218. Combining the results of the genetic map, resistance spectra, allelic relationship, and DNA resequencing data, we speculate that PmSN15218 may be a potential new powdery mildew resistance gene, distinct from Pm4, on chromosome 2AL.
Conclusion
Herein, we identified a wheat powdery mildew resistance gene, PmSN15218, located on chromosome 2AL. It is distinct from the previously cloned Pm4b. This gene was derived from the breeding line SN15218 and could serve as a valuable genetic resource for wheat powdery mildew resistance breeding without any adverse factors or linkage drag of undesirable genes.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
YB, FN, and JW designed the project. MS, QL, YH, GL, and JQ performed the experiments. YB, FN, and JQ analyzed the data and wrote the manuscript. All authors discussed the results and reviewed the manuscript.
Funding
This research was funded by the Key R&D Program of Shandong Province (Major Science and Technology Innovation Project 2021LZGC009) and the Shandong Provincial Scientific Innovation Project for Young Scholars of Universities (2019KJF026).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.931778/full#supplementary-material
Supplementary Table 1 | Closely linked markers for several reported powdery mildew resistance genes on chromosome 2AL.
Supplementary Table 2 | In silico polymerase chain reaction (PCR) analysis of selected powdery mildew resistance genes on chromosome 2AL.
Footnotes
- ^ https://github.com/lh3/bwa
- ^ https://www.kyazma.nl
- ^ http://www.htslib.org
- ^ https://github.com/ablab/spades
References
Avni, R., Nave, M., Barad, O., Baruch, K., Twardziok, S. O., Gundlach, H., et al. (2017). Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97. doi: 10.1126/science.aan0032
Bariana, H., and McIntosh, R. (1994). Characterisation and origin of rust and powdery mildew resistance genes in VPM1 wheat. Euphytica 76, 53–61.
Briggle, L. W. (1966). Transfer of resistance to Erysiphe graminis f. sp. tritici from khapli emmer and yuma durum to hexaploid wheat. Crop Sci. 6:crosci1966.
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cowger, C., Mehra, L., Arellano, C., Meyers, E., and Murphy, J. P. (2017). Virulence differences in Blumeria graminis f. sp. tritici from the central and eastern united states. Phytopathology 108, 402–411. doi: 10.1094/PHYTO-06-17-0211-R
El-Shamy, M. M., Emara, H. M., and Mohamed, M. E. (2016). Virulence analysis of wheat powdery Mildew (Blumeria graminis f. sp. tritici) and effective genes in middle delta, egypt. Plant Dis. 100, 1927–1930.
Fu, B., Chen, Y., Li, N., Ma, H., Kong, Z., Zhang, L., et al. (2013). pmX: a recessive powdery mildew resistance gene at the Pm4 locus identified in wheat landrace Xiaohongpi. Theor. Appl. Genet. 126, 913–921. doi: 10.1007/s00122-012-2025-1
Hao, Y., Liu, A., Wang, Y., Feng, D., Gao, J., Li, X., et al. (2008). Pm23: a new allele of Pm4 located on chromosome 2AL in wheat. Theor. Appl. Genet. 117, 1205–1212. doi: 10.1007/s00122-008-0827-y
Hao, Y., Parks, R., Cowger, C., Chen, Z., Wang, Y., Bland, D., et al. (2015). Molecular characterization of a new powdery mildew resistance gene Pm54 in soft red winter wheat. Theor. Appl. Genet. 128, 465–476. doi: 10.1007/s00122-014-2445-1
He, H., Liu, R., Ma, P., Du, H., Zhang, H., Wu, Q., et al. (2021). Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor. Appl. Genet. 134, 53–62. doi: 10.1007/s00122-020-03681-2
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Li, G., Cowger, C., Wang, X., Carver, B. F., and Xu, X. (2019). Characterization of Pm65, a new powdery mildew resistance gene on chromosome 2AL of a facultative wheat cultivar. Theor. Appl. Genet. 132, 2625–2632. doi: 10.1007/s00122-019-03377-2
Li, H., Dong, Z., Ma, C., Xia, Q., Tian, X., Sehgal, S., et al. (2020). A spontaneous wheat-Aegilops longissima translocation carrying Pm66 confers resistance to powdery mildew. Theor. Appl. Genet. 133, 1149–1159. doi: 10.1007/s00122-020-03538-8
Ma, Z., Wei, J., and Cheng, S. (2004). PCR-based markers for the powdery mildew resistance gene Pm4a in wheat. Theor. Appl. Genet. 109, 140–145. doi: 10.1007/s00122-004-1605-0
Maccaferri, M., Harris, N. S., Twardziok, S. O., Pasam, R. K., Gundlach, H., Spannagl, M., et al. (2019). Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 51, 885–895. doi: 10.1038/s41588-019-0381-3
McIntosh, R., and Bennett, F. G. (1979). Cytogenetical studies in wheat. IX. monosomic analyses, telocentric mapping and linkage relationships of genes Sr21, Pm4 and Mle. Austr. J. Biol. Sci. 32, 115–126.
McIntosh, R., Dubcovsky, J., Rogers, J. W., Xia, X., and Raupp, W. (2020). Catalogue of Gene Symbols for Wheat. Available online at: https://wheat.pw.usda.gov/GG3/WGC (accessed December 31, 2021).
Ni, F., Qi, J., Hao, Q., Lyu, B., Luo, M. C., Wang, Y., et al. (2017). Wheat Ms2 encodes for an orphan protein that confers male sterility in grass species. Nat. Commun. 8:15121. doi: 10.1038/ncomms15121
Niu, J., Jia, H., Yin, J., Wang, B., Ma, Z., and Shen, T. (2010). Development of an STS marker linked to powdery mildew resistance genes PmLK906 and Pm4a by gene chip hybridization. Agric. Sci. China 9, 331–336.
Niu, J. S., Wang, B. Q., Wang, Y. H., Cao, A. Z., Qi, Z. J., and Shen, T. M. (2008). Chromosome location and microsatellite markers linked to a powdery mildew resistance gene in wheat line ‘Lankao 90(6)’. Plant Breed. 127, 346–349.
Poplin, R., Ruano-Rubio, V., Depristo, M. A., Fennell, T. J., Carneiro, M. O., Van Der Auwera, G. A., et al. (2018). Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv [Perprint]. bioRxiv, 201178,
Porebski, S., Bailey, L. G., and Baum, B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15.
Qie, Y., Sheng, Y., Xu, H., Jin, Y., Ma, F., Li, L., et al. (2019). Identification of a new powdery mildew resistance gene pmDHT at or closely linked to the Pm5 locus in the Chinese wheat landrace dahongtou. Plant Dis. 103, 2645–2651. doi: 10.1094/PDIS-02-19-0401-RE
Sánchez-Martín, J., Widrig, V., Herren, G., Wicker, T., Zbinden, H., Gronnier, J., et al. (2021). Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants 7, 327–341. doi: 10.1038/s41477-021-00869-2
Schmolke, M., Mohler, V., Hartl, L., Zeller, F. J., and Hsam, S. L. K. (2012). A new powdery mildew resistance allele at the Pm4 wheat locus transferred from einkorn (Triticum monococcum). Mol. Breed. 29, 449–456.
Singh, R. P., Singh, P. K., Rutkoski, J., Hodson, D. P., He, X., Jørgensen, L. N., et al. (2016). Disease impact on wheat yield potential and prospects of genetic control. Ann. Rev. Phytopathol. 54, 303–322. doi: 10.1146/annurev-phyto-080615-095835
Takagi, H., Abe, A., Yoshida, K., Kosugi, S., Natsume, S., Mitsuoka, C., et al. (2013). QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74, 174–183. doi: 10.1111/tpj.12105
Tan, C., Li, G., Cowger, C., Carver, B. F., and Xu, X. (2018). Characterization of Pm59, a novel powdery mildew resistance gene in afghanistan wheat landrace PI 181356. Theor. Appl. Genet. 131, 1145–1152. doi: 10.1007/s00122-018-3067-9
Ullah, K. N., Li, N., Shen, T., Wang, P., Tang, W., Ma, S., et al. (2018). Fine mapping of powdery mildew resistance gene Pm4e in bread wheat (Triticum aestivum L.). Planta 248, 1319–1328.
Wolfe, M. S. (1967). Physiologic specialization of Erysiphe graminis f. sp. tritici in the united kingdom, 1964–5. Trans. Br. Mycol. Soc. 50, 631–640.
Wu, L., Zhu, T., He, H., Cao, X., Li, H., Xu, H., et al. (2022). Genetic dissection of the powdery mildew resistance in wheat breeding line LS5082 using BSR-Seq. Crop J. doi: 10.1094/PDIS-12-21-2771-RE
Wu, P., Xie, J., Hu, J., Qiu, D., Liu, Z., Li, J., et al. (2018). Development of molecular markers linked to powdery mildew resistance gene Pm4b by combining SNP discovery from transcriptome sequencing data with bulked segregant analysis (BSR-Seq) in wheat. Front. Plant Sci. 9:95. doi: 10.3389/fpls.2018.00095
Yao, D., Ijaz, W., Liu, Y., Hu, J., Peng, W., Zhang, B., et al. (2022). Identification of a Pm4 allele as a powdery mildew resistance gene in wheat line xiaomaomai. Int. J. Mol. Sci. 2022:23. doi: 10.3390/ijms23031194
Yu, X., Ren, S., Zhao, L., Guo, J., Bao, Y., Ma, Y., et al. (2018). Molecular mapping of a novel wheat powdery mildew resistance gene Ml92145E8-9 and its application in wheat breeding by marker-assisted selection. Crop J. 6, 621–627.
Zhang, R., Xiong, C., Mu, H., Yao, R., Meng, X., Kong, L., et al. (2021). Pm67, a new powdery mildew resistance gene transferred from Dasypyrum villosum chromosome 1V to common wheat (Triticum aestivum L.). Crop J. 9, 882–888.
Keywords: wheat, powdery mildew, BSR-Seq, Pm4 loci, PmSN15218
Citation: Sun M, Liu Q, Han Y, Liu G, Wu J, Qi J, Ni F and Bao Y (2022) PmSN15218: A Potential New Powdery Mildew Resistance Gene on Wheat Chromosome 2AL. Front. Plant Sci. 13:931778. doi: 10.3389/fpls.2022.931778
Received: 29 April 2022; Accepted: 19 May 2022;
Published: 15 June 2022.
Edited by:
Handong Su, Huazhong Agricultural University, ChinaReviewed by:
Haiyan Jia, Nanjing Agricultural University, ChinaLiu Dengcai, Sichuan Agricultural University, China
Copyright © 2022 Sun, Liu, Han, Liu, Wu, Qi, Ni and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Ni, bmlmZWkxOTk4QHNkYXUuZWR1LmNu; Yinguang Bao, YmFveWluZ3VhbmdAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Meng Sun†
Meng Sun† Jiajie Wu
Jiajie Wu Fei Ni
Fei Ni Yinguang Bao
Yinguang Bao