- 1College of Agronomy, Sichuan Agricultural University, Chengdu, China
- 2Provincial Key Laboratory for Plant Genetics and Breeding, Sichuan Agricultural University, Chengdu, China
- 3Center for Informational Biology, University of Electronic Science and Technology of China, Chengdu, China
It was reported that the chromosome 6R of rye (Secale cereale L.) carries stripe rust resistance gene Yr83, and the region with the candidate resistance gene(s) still needs to be narrowed down. This study confirmed that the chromosome 6RLAr derived from rye AR106BONE contains stripe rust resistance gene(s). A wheat-rye T6BS.6RLAr translocation chromosome, a wheat-rye small-segment translocation T6RLAr-6AS.6AL, and three kinds of deleted T6BS.6RLAr translocations, T6BS.6RLAr-1, T6BS.6RLAr-2, and T6BS.6RLAr-3, were identified. Translocations T6BS.6RLAr, T6BS.6RLAr-2, and T6RLAr-6AS.6AL were highly resistant to stripe rust and T6BS.6RLAr-1 and T6BS.6RLAr-3 were highly susceptible. The molecular markers specific to 6RL determined that the three regions of the 6RLAr arm from 732,999,830 bp to the telomere, from 735,010,030 to 848,010,414 bp, and from 848,011,262 bp to the telomere were deleted from T6BS.6RLAr-1, T6BS.6RLAr-2, and T6BS.6RLAr-3, respectively. T6BS.6RLAr-2 and T6RLAr-6AS.6AL contained the segment that was deleted in T6BS.6RLAr-3. Therefore, it can be concluded that about 37 Mb segment from 848,011,262 bp to the telomere carried stripe rust resistance gene(s), and it was smaller than that with the Yr83 gene. Gene annotation indicated that about 37 Mb region contains 43 potential resistance genes, and 42 of them are nucleotide-binding site and leucine-rich repeat (NBS-LRR)-like resistance protein genes. The results in this study narrowed down the size of the region with candidate stripe rust resistance gene(s) on the 6RL arm, and the T6RLAr-6AS.6AL is a promising small-segment translocation for improvement of wheat cultivars.
Introduction
Wheat stripe rust is caused by Puccinia striiformis f. sp. tritici (Pst) and is one of the most serious diseases in wheat. Developing wheat cultivars with resistance to stripe rust is the most practical way to control this disease. More than 80 Yr (yellow rust resistance) genes were officially named; however, most of them lost their resistance because of variations in the prevalence of virulent pathotypes (Ren et al., 2022). The resistance to stripe rust of 103 wheat lines was tested using pathogenic races CYR32, CYR33, and CYR34, which are currently prevalent in China, and only Yr5, Yr15, and Yr45 exhibited all-stage resistance, and only Yr41, Yr47, and Yr50 were adult plant resistant (Hu et al., 2022). The replacement of historically clonal Pst races by new ones occurred continually (Jamil et al., 2020; Bouvet et al., 2022). Therefore, it is important to explore new resistance genes to stripe rust and enrich the resource pool for wheat resistance breeding.
Wheat-related species contain abundant resistance genes. For example, rye (Secale cereale L.) is an important gene source for wheat disease resistance breeding (Spetsov and Daskalova, 2022). Stripe rust gene Yr 9 located on the 1RS arm was successfully used in commercial wheat cultivars. Additionally, 2R, 4R, 5R, 6R, and 7R chromosomes also carry stripe rust-resistant genes (Lei et al., 2011; Li et al., 2016a, 2020a,b; Schneider et al., 2016; An et al., 2019; Xi et al., 2019; Johansson et al., 2020; Ren et al., 2020). Four of these reports indicated that rye chromosome 6R carried stripe rust resistance genes, and they were derived from different rye sources (Schneider et al., 2016; Johansson et al., 2020; Li et al., 2020a,b). A new stripe rust resistance gene on chromosome 6R was named Yr83, and it was located in a bin FL0.73-1.00 of 6RL (Li et al., 2020b). To clone the stripe rust resistance gene(s) on 6RL, it is necessary to narrow the segment with resistant gene(s). In this study, a wheat-rye 6RAr monosomic addition line (MA6RAr) with resistance to stripe rust, a 6RSAr monotelosomic addition line (MTA6RSAr), a 6RLAr monotelosomic addition line (MTA6RLAr), a T6BS.6RLAr translocation line, and three kinds of deleted T6BS.6RLAr translocation lines were used to locate the stripe rust resistance gene(s) on a smaller segment of 6RLAr arm, and some resistance protein genes in this segment were found.
Materials and Methods
Plant Materials
Octoploid triticale lines Mianyang 11/AR106BONE-4 (MAR4) and Mianyang 11/Kustro (MK) were derived from common wheat (Triticum aestivum L.) Mianyang 11 (MY11) × Secale cereale L. AR106BONE and MY11 × Secale cereale L. Kustro, respectively. Crossing between MAR4 and a wheat line J1025 (T. aestivum L.) and between MK and MY11 was carried out. From the progenies of MAR4 × J1025 and MK × MY11, wheat-rye 6R monosomic addition lines MA6RAr and MA6RKu were identified, respectively. These monosomic addition lines were used to investigate the stripe rust resistance of the two kinds of 6R chromosomes. MTA6RSAr and MTA6RLAr were identified from the selfed progeny of MA6RAr. The seeds of MA6RAr were irradiated with 60Co-γ rays at a dosage of 200 Gy at Biotechnology and Nuclear Technology Research Institute, Sichuan Academy of Agricultural Sciences, China. From the progeny of irradiated MA6RAr, a wheat-rye T6BS.6RLAr translocation line was obtained, and the seeds derived from this translocation line were also irradiated with 60Co-γ rays and some deleted T6BS.6RLAr translocation chromosomes were detected. The rye Kustro and AR106BONE and the common wheat MY11 and J1025 were kept in the seed store in our laboratory.
Cytological Analysis
The root-tip metaphase chromosomes of the materials used in this study were analyzed using non-denaturing fluorescence in situ hybridization (ND-FISH) technology. Oligo-pSc200, Oligo-pSc250 (Vershinin et al., 1995; Fu et al., 2015), Oligo-Ku (Xiao et al., 2017), Oligo-pSc119.2-1 (McIntyre et al., 1990; Tang et al., 2014), and Oligo-pTa535-1 (Komuro et al., 2013; Tang et al., 2014) were used as probes. Probes Oligo-pSc200, Oligo-pSc250, and Oligo-Ku were used to distinguish rye chromosomes from wheat (Fu et al., 2015; Xiao et al., 2017). Probes Oligo-pSc119.2-1 and Oligo-pTa535-1 were used to identify individual wheat chromosomes (Tang et al., 2014). Additionally, the different cytological structure of rye chromosomes was displayed by the combination of probes Oligo-pSc200, Oligo-pSc250, and Oligo-pSc119.2-1 (Fu et al., 2015). These oligonucleotide (oligo) probes were 5′ end-labeled with 6-carboxyfluorescein (6-FAM) or 6-carboxytetramethylrhodamine (TAMRA). The root-tip metaphase chromosomes were prepared according to the methods described by Han et al. (2006). The ND-FISH analysis was carried out following the methods described by Fu et al. (2015) and Xiao et al. (2017). Additionally, the genomic DNA of rye AR106BONE was used as a probe for the genomic in situ hybridization (GISH) analysis of the T6RLAr-6AS.6AL small-segment translocation. The GISH analysis was carried out according to the methods described by Fu et al. (2015).
Development of 6RL-Specific Markers
Primers were designed according to the 6RL sequence of rye Lo7 (Rabanus-Wallace et al., 2021) using Primer 3 software (version 4.0), and the optimal melting temperature and size values were set to 60°C and to 20 bases, respectively. In total, 423 pairs of primers were designed. Additionally, 124 developed 6RLKu-specific length amplified fragment sequencing (SLAF-seq) markers (Li et al., 2016b) were also used. The PCR amplification and agarose gel electrophoresis were performed following the procedure described by Li et al. (2016b). Chinese Spring (CS), MY11, T6BS.6RLAr translocation line, and rye AR106BONE were used to test the 6RLAr specificity of the 423 newly designed primer pairs and the 124 6RLKu-specific SLAF-seq markers. All the 6RLAr-specific markers were physically mapped to specific regions of the 6RLAr arm using deleted T6BS.6RLAr translocation lines. The sequences of the 124 SLAF-seq primer pairs were used for nucleotide Basic Local Alignment Search Tool (BLAST) searches against the 6R sequence of rye Lo7 (Rabanus-Wallace et al., 2021) using a BLAST tool in the Triticeae Multi-omics Center (http://202.194.139.32/), and the positions of these markers on 6RL arm were determined according to the BLAST results.
Cloning Partial Sequences of the Candidate Resistance Genes From 6RLAr
To confirm that the candidate resistance genes on the 6RL arm of Lo7 are similar to that of 6RLAr, the sequences of two candidate resistance genes SECCE6Rv1G0449960.1 and SECCE6Rv1G0453070.1 of Lo7 were randomly selected for designing primer pairs. Primer pairs of P60 (5′ TGGGG AATAG CTGGC ATTGG3′, 5′ TCGGT AGGGT AGACG GTGAG3′) and P70 (5′ AATGG GAGGA CTCTT GCGTG3′, 5′ CTGGG AATGA ACCGA CAGCT3′) were used to amplify the partial sequences of the two genes from 6RLAr arm. The PCR reactions and product separation were also performed according to the methods described by Li et al. (2016b), and the annealing temperature of the primer pairs was 60°C. The target products amplified by the two primer pairs, P60 and P70, were cloned and then sequenced by the Tsingke Biotechnology Co., Ltd. (Chengdu, China). Sequence alignment was performed using the software DNAMAN (Ver. 4.0, Lynnon Corp., Quebec, QC, Canada).
Stripe Rust Response Test
The response of parental wheat J1025, MY11, lines MA6RKu, MA6RAr, MTA6RSAr, MTA6RLAr, T6BS.6RLAr, and the deleted T6BS.6RLAr to stripe rust was evaluated. The mixed stripe rust prevalent isolates CYR32, CYR33, and CYR34 were used to inoculate seedlings in field according to the method described by Xi et al. (2019). A 0–9 numerical scale of infection types (IT) was scored according to the standard described by Wan et al. (2017) at the adult stage. The disease resistance of J1025, MY11, the lines MA6RKu, MA6RAr, MTA6RSAr, MTA6RLAr, and T6BS.6RLAr was evaluated in 2018–2019 in Qionglai, Sichuan Province, China and in 2019–2020, 2020–2021, and 2021–2022 in Wenjiang and Dayi, Sichuan Province, China. The disease resistance of the deleted T6BS.6RLAr and the T6RLAr-6AS.6AL translocation was evaluated in 2020–2021 and 2021–2022 in Wenjiang and Dayi. When the segment of 6RLAr with stripe rust resistance was identified, the candidate resistance genes were found according to the gene annotation of rye Lo7 (http://202.194.139.32/jbrowse.html) (Rabanus-Wallace et al., 2021).
Results
Identification of Wheat-Rye 6R Addition and Translocation Lines
The ND-FISH based on oligo probes was used to analyze the progeny of wheat × rye. Wheat-rye 6R monosomic addition lines MA6RAr and MA6RKu were identified (Figures 1A–D). Both the short arms of the 6RAr and 6RKu chromosomes (6RSAr and 6RSKu) contained signals of Oligo-pSc119.2-1 and Oligo-pSc250, and no signals of Oligo-pSc250 were observed on their long arms (6RLAr and 6RLKu) (Figure 1E). The probe Oligo-pSc119.2-1 produced three and four signal bands on 6RLAr and 6RLKu, respectively (Figure 1E). Two and three signal bands of probe Oligo-pSc200 were observed on 6RLAr and 6RLKu arms, respectively (Figure 1E). The results indicated that the structure of the long arms of the 6RAr and 6RKu chromosomes is different. Line MA6RAr was highly resistant to stripe rust (IT = 1), whereas line MA6RKu was susceptible (IT = 9; Figure 1E).
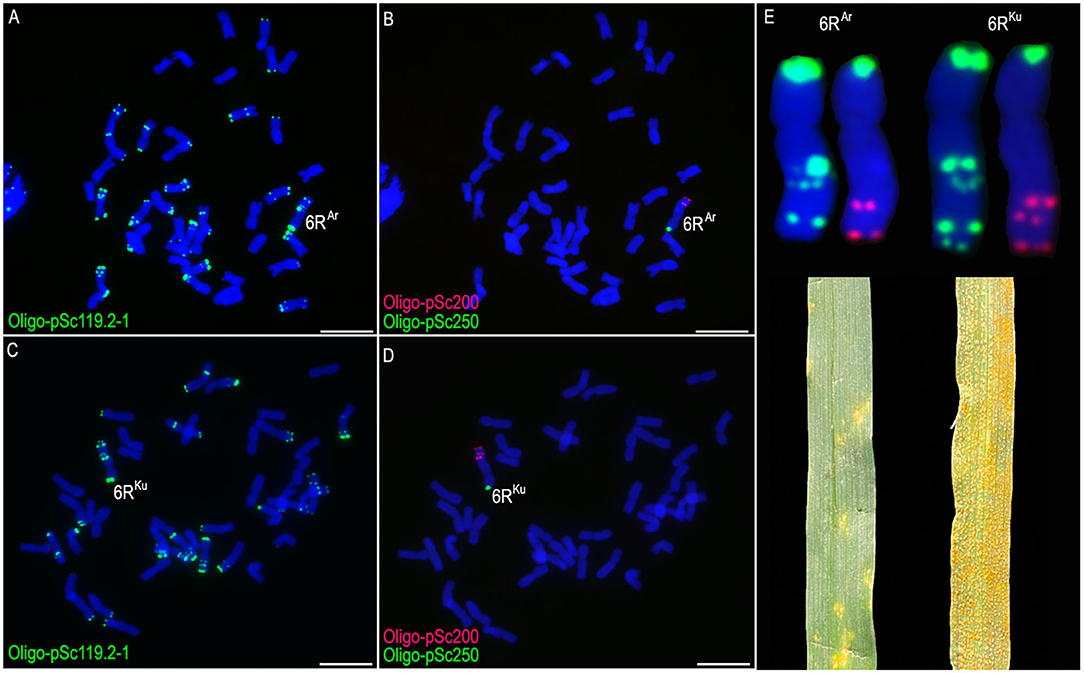
Figure 1. The non-denaturing fluorescence in situ hybridization (ND-FISH) analysis of lines MA6RAr and MA6RKu using probes Oligo-pSc119.2-1 (green), Oligo-pSc200 (red), Oligo-pSc250 (green), and their stripe rust resistance test. (A,B) A root-tip metaphase cell of line MA6RAr. (C,D) A root-tip metaphase cell of line MA6RKu. (E) Cut-and-paste 6RAr and 6RKu chromosomes. 6RAr was highly resistant to stripe rust and 6RKu was susceptible. Scale bar: 10 μm.
From the selfed progeny of line MA6RAr, the 6RSAr and the 6RLAr monotelosomic addition lines, MTA6RSAr and MTA6RLAr, were identified (Figures 2A,B). Line MTA6RSAr was susceptible to stripe rust (IT = 9), and line MTA6RLAr displayed high resistance (IT = 1; Figure 2C). In total, 200 seeds of MA6RAr were irradiated. A plant 18T231-46 containing a wheat-rye T6BS.6RLAr translocation chromosome was identified from 1,182 M1 seeds, and a line 19T177-21 containing a pair of T6BS.6RLAr translocations was identified from 30 seeds of the selfed progeny of 18T231-46 (Figures 3A,B). Additionally, 104 seeds of the selfed progeny of 18T231-46 were also irradiated. Four lines, 21F1, 21F3, 21F7, and 21F11, were identified from the 1,090 seeds (M2) of the progeny of irradiated 18T231-46. In these lines, three kinds of deleted T6BS.6RLAr translocation chromosomes and a small-segment translocation T6RLAr-6AS.6AL were found (Figures 3C–I). Line 21F1 contained two deleted translocation chromosomes T6BS.6RLAr-1, on which the Oligo-pSc200 signals and the distal Oligo-pSc119.2-1 signals on the 6RLAr arms were disappeared (Figures 3C,I). Line 21F3 contained two deleted translocation chromosomes T6BS.6RLAr-2, carrying one Oligo-pSc200 signal band and two intercalary Oligo-pSc119.2-1 signal bands (Figures 3D,I). Line 21F7 contained a pair of deleted translocation chromosomes T6BS.6RLAr-3, and the signals of Oligo-pSc200 on telomeric regions were disappeared (Figures 3E,I). In line 21F11, probe Oligo-pSc200 produced signals on the telomeric regions of the short arms of chromosomes 6A (Figures 3F–I). Therefore, line 21F11 contained a pair of small-segment translocations T6RLAr-6AS.6AL and a pair of T6BS.6RLAr-3 (Figures 3F–I). The weak GISH signals on the chromosomes T6RLAr-6AS.6AL indicated that the segment of 6RLAr involved in translocation is small (Figures 3H,I).
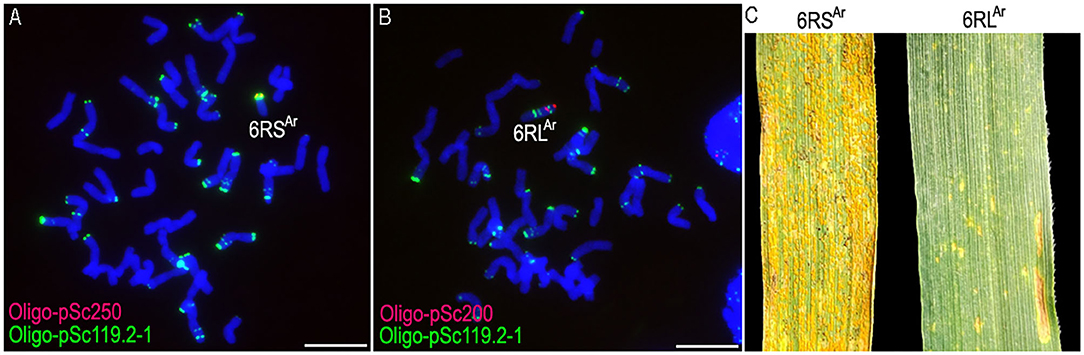
Figure 2. The non-denaturing fluorescence in situ hybridization (ND-FISH) analysis of lines MTA6RSAr and MTA6RLAr using probes Oligo-pSc119.2-1 (green), Oligo-pSc200 (red), Oligo-pSc250 (red), and their stripe rust resistance test. (A) A root-tip metaphase cell of line MTA6RSAr. (B) A root-tip metaphase cell of line MTA6RLAr. (C) Line MTA6RSAr was susceptible to stripe rust and line MTA6RLAr was highly resistant. Scale bar: 10 μm.
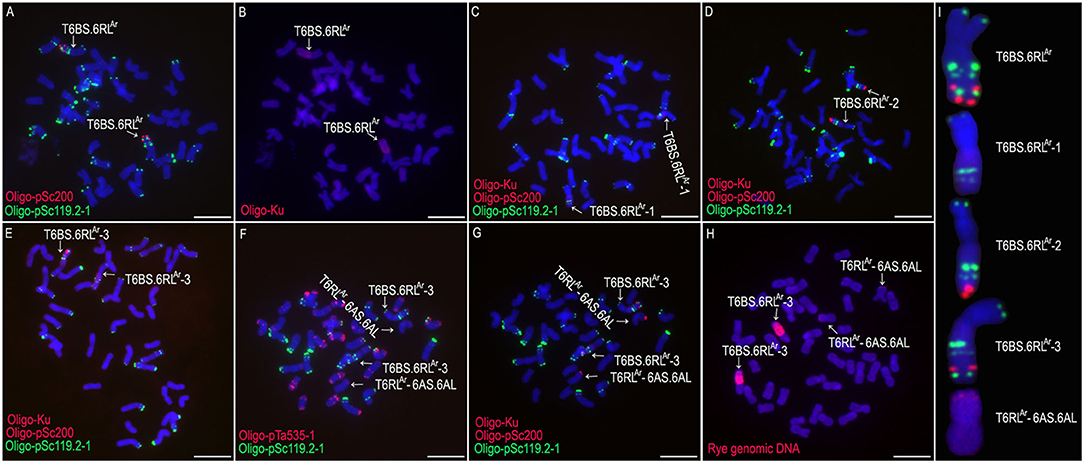
Figure 3. The non-denaturing fluorescence in situ hybridization (ND-FISH) and genomic in situ hybridization (GISH) analyses of T6BS.6RLAr translocations and its deleted T6BS.6RLAr using probes Oligo-pSc119.2-1 (green), Oligo-pTa535-1 (red), Oligo-pSc200 (red), Oligo-Ku (red) and rye genomic DNA (red). (A,B) A root-tip metaphase cell of line 19T177-21. (C) A root-tip metaphase cell of line 21F1. (D) A root-tip metaphase cell of line 21F3. (E) A root-tip metaphase cell of line 21F7. (F,G) A root-tip metaphase cell of line 21F11. (H) GISH analysis of root-tip metaphase cell of line 21F11. (I) Cut-and-paste chromosomes of T6BS.6RLAr and deleted T6BS.6RLAr. Scale bar: 10 μm.
The Physical Location of 6RLAr-Specific Markers
Among the 423 newly designed primer pairs, 204 amplified specific bands from T6BS.6RLAr translocation line and rye AR106BONE, but not from CS and MY11, indicated that these 204 primer pairs are 6RLAr-specific markers (Figure 4 and Supplementary Table S1). Additionally, 95 of the 124 SLAF-seq markers amplified target bands from both T6BS.6RLAr and AR106BONE. A total of 299 (204 + 95) 6RLAr-specific markers were obtained, and their target amplification regions on the 6RL arm are listed in Supplementary Table S1. The breakpoints on the 6RLAr arms of T6BS.6RLAr-1, T6BS.6RLAr-2, and T6BS.6RLAr-3 were determined using the 299 markers. In total, 103 of the 299 markers amplified 6RLAr-specific bands from translocation T6BS.6RLAr-1 (Figure 4A), and these markers covered the region of 6RL from 333,302,151 to 732,900,689 bp (Supplementary Table S1). The first distal marker (Lo7.6RL-71) that did not amplify products from T6BS.6RLAr-1 occupied the region from 732,999,830 to 733,000,692 bp (Supplementary Table S1). Therefore, the breakpoint in T6BS.6RLAr-1 was located in the region of the 6RLAr arm between 732,900,689 and 732,999,830 bp (Figure 5A and Supplementary Table S1). Totally, 118 and 52 markers that were distributed into two regions of 6RL from 333,302,151 to 733,480,409 bp and from 848,011,262 to 885,003,466 bp, respectively amplified target bands from T6BS.6RLAr-2, and 129 markers that distributed the region of 6RL from 735,010,030 to 848,010,414 bp did not produce amplicons from this translocation chromosome (Figures 4B–D and Supplementary Table S1). It can be determined that the two breakpoints in T6BS.6RLAr-2 occurred in the two regions of 6RLAr arm between 733,480,409 and 735,010,030 bp and between 848,010,414 and 848,011,262 bp (Figure 5A and Supplementary Table S1). For T6BS.6RLAr-3 translocation, 247 markers that were distributed on the 6RL region from 333,302,151 to 848,010,414 bp amplified target bands, and the rest 52 markers, which were the same as those amplified products from T6BS.6RLAr-2, did not amplify amplicons (Figure 4D and Supplementary Table S1). According to the amplification of these markers, the breakpoint regions on 6RLAr in the three deleted T6BS.6RLAr translocations can be determined (Figure 5A and Supplementary Table S1). For the T6BS.6RLAr-3 chromosome, the breakpoint between 848,010,414 and 848,011,262 bp was the same as the second breakpoint in the T6BS.6RLAr-2 chromosome (Figure 5A and Supplementary Table S1). Therefore, the segments from 732,999,830 bp to telomere, from 735,010,030 to 848,010,414 bp, and from 848,011,262 bp to telomere of 6RLAr were deleted from T6BS.6RLAr-1, T6BS.6RLAr-2, and T6BS.6RLAr-3, respectively (Figure 5A). The 6RLAr segment deleted from T6BS.6RLAr-3 was transferred to T6RLAr-6AS.6AL (Figures 4, 5A).
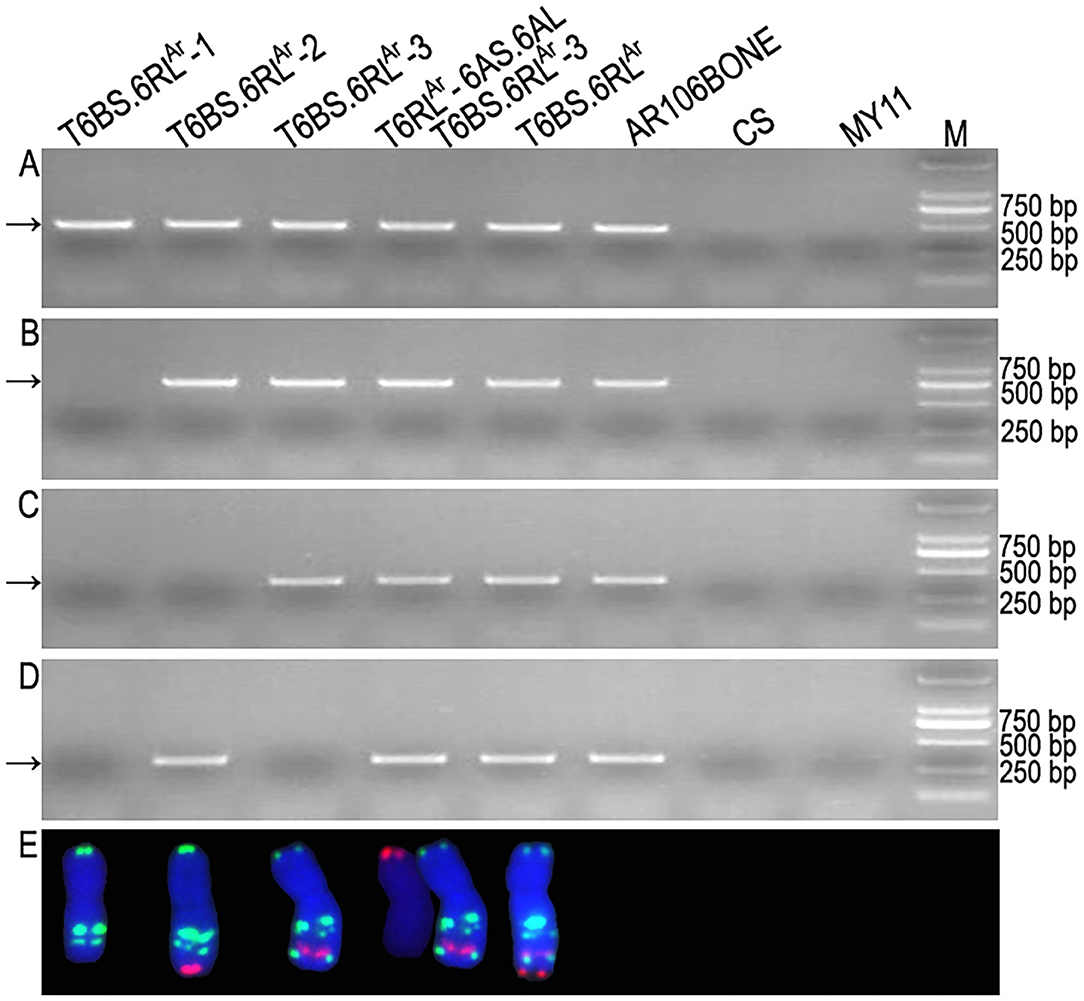
Figure 4. Physically mapped 6RLAr-specific markers using deleted T6BS.6RLAr translocations. (A) Products amplified by primer pair Lo7.6RL-2 represent the markers located on T6BS.6RLAr and all the deleted T6BS.6RLAr. (B) Products amplified by primer pair Lo7.6RL-74 represent the markers located on T6BS.6RLAr, T6BS.6RLAr-2, and T6BS.6RLAr-3. (C) Products amplified by primer pair Ku.6RL-260 represent the markers located on T6BS.6RLAr and T6BS.6RLAr-3. (D) Products amplified by primer pair Lo7.6RL-183 represent the markers located on T6BS.6RLAr, T6BS.6RLAr-2, and T6RLAr-6AS.6AL. (E) Cut-and-paste translocation chromosomes corresponding to their amplified products in each electrophoresis lane. Arrows indicate the target bands amplified by 6RLAr-specific markers. Common wheat Chinese Spring (CS), Mianyang11 (MY11), and rye AR106BONE were used as controls. M: DNA marker.
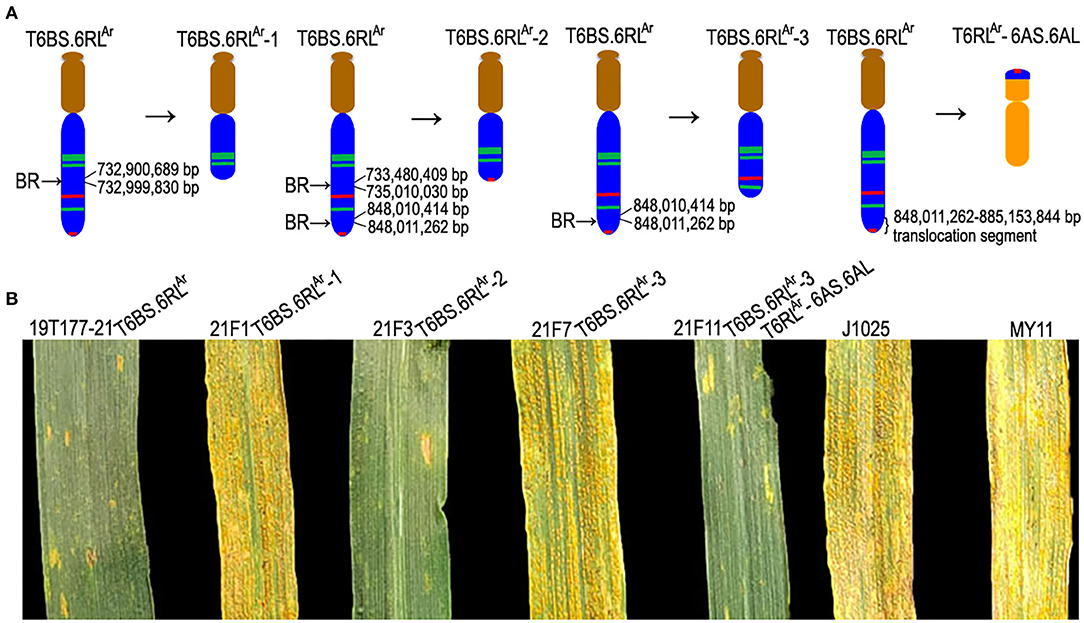
Figure 5. Ideograms of wheat-rye 6RL translocation chromosomes and stripe rust response test. (A) Ideograms of T6BS.6RLAr, deleted T6BS.6RLAr, and the small-segment translocation T6RLAr-6AS.6AL. Brown, blue, and orange indicate segments of 6B, 6R, and 6A chromosomes, respectively. Green and red bands indicate the signals of Oligo-pSc119.2-1 and Oligo-pSc200, respectively. “BR” indicates a break point. The right brace indicates the 6RLAr segment that was transferred on the 6A chromosome. (B) Parental wheat J1025 and MY11 and lines 21F1 and 21F7 were highly susceptible to stripe rust and lines 19T177-21, 21F3, and 21F11 were highly resistant.
Location of Stripe Rust Resistance Genes on Segment of 6RLAr
Stripe rust response test indicated that translocation lines T6BS.6RLAr, 21F3 (T6BS.6RLAr-2), and 21F11 (T6BS.6RLAr-3 and T6RLAr-6AS.6AL) were highly resistant to stripe rust (IT = 1), and translocation lines 21F1 (T6BS.6RLAr-1) and 21F7 (T6BS.6RLAr-3) were highly susceptible (IT = 9; Figure 5B). According to the resistance of 21F3, 21F7, and 21F11, and the 6RLAr segments existed in T6BS.6RLAr-2, T6BS.6RLAr-3, and T6RLAr-6AS.6AL, it was determined that the 6RLAr segment in T6RLAr-6AS.6AL carried the stripe rust resistance gene. Therefore, it was determined that the 6RLAr region from 848,371,946 bp to the telomere (885,153,844 bp) carried the stripe rust resistance gene. Gene annotation indicated that there are 43 potential resistance genes located in the region from 848,011,885 to 885,153,844 bp of chromosome 6R (Supplementary Table S2). Only one of the 43 genes belongs to the kinase family protein gene and the other 42 genes are nucleotide-binding site and leucine-rich repeat (NBS-LRR)-like resistance protein genes (Supplementary Table S2).
The Similarity of the Candidate Genes on 6RL Arms of Lo7 and AR106BONE
The two primer pairs P60 and P70 amplified specific bands from translocation chromosomes T6BS.6RLAr, T6BS.6RLAr-2, and T6RLAr-6AS.6AL but not from T6BS.6RLAr-1 and T6BS.6RLAr-3 translocations (Figure 6). The two primer pairs also did not amplify products from CS and MY11 (Figure 6). The sequences amplified from T6RLAr-6AS.6AL were cloned and sequenced. The sequences amplified by primer pairs P60 and P70 were named Ar-60.1 (1,857 bp) and Ar-70.1 (2,423 bp), respectively (Supplementary Figures S1, S2). Sequences Ar-60.1 and Ar-70.1 had 99.03 and 99.50% similarity to the corresponding segments of genes SECCE6Rv1G0449960.1 and SECCE6Rv1G0453070.1, respectively (Supplementary Figures S1, S2). These results indicated the conservation of resistance genes on the 6RL arms of Lo7 and AR106BONE.
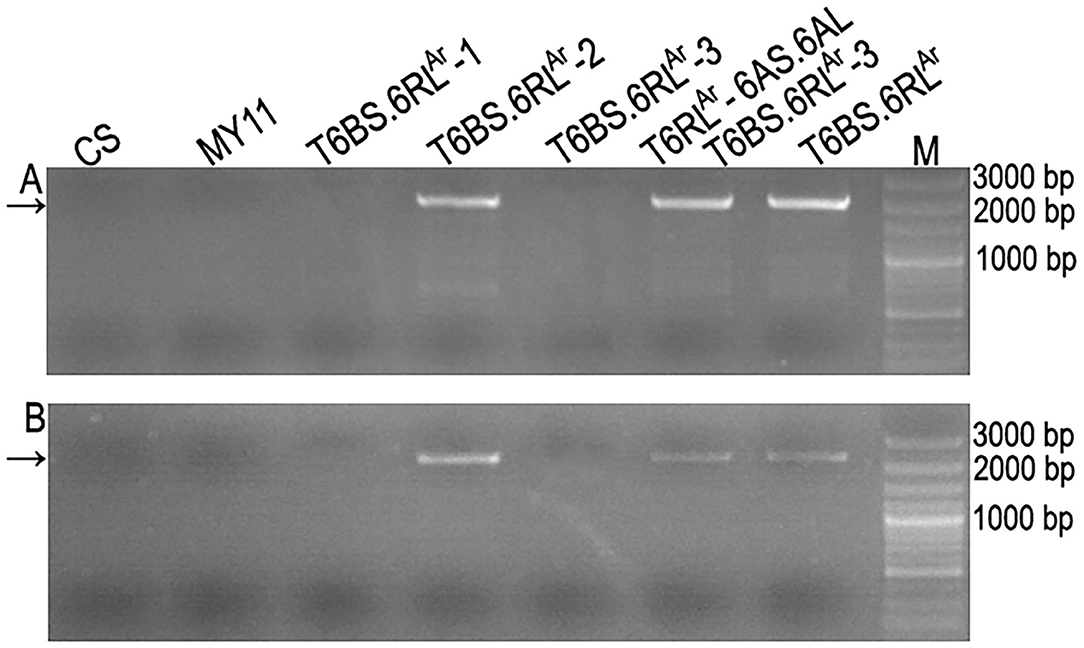
Figure 6. Amplification of partial segments of genes SECCE6Rv1G0449960.1 and SECCE6Rv1G0453070.1. (A) Products amplified by primer pair P60. (B) Products amplified by primer pair P70. Arrows indicate the target bands amplified by the two primer pairs. Common wheat Chinese Spring (CS) and Mianyang11 (MY11) were used as controls. M: DNA marker.
Discussion
Polymorphism of 6R Chromosomes
The complex cytological structure of the 6R chromosome has been reported. It was known that the long arm of the rye 6R chromosome contains homologous groups 3 and 7 segments in the distal region (Devos et al., 1993; Li et al., 2013). FISH signal patterns of tandem repeats pSc119.2 on S. cereale 6R and S. africanum 6Rafr were different, and this indicated the structural alteration between the two kinds of 6R chromosomes (Li et al., 2020a). In this study, the cytological structure of 6R chromosomes derived from rye AR106BONE and Kustro is different. Some 6RLKu-specific SLAF-seq markers that did not amplify products from 6RAr also exhibited the diversity between chromosomes 6RKu and 6RAr. The 6RL arm with stripe rust resistance gene Yr83 carried weak pSc119.2 signals in the intercalary and telomeric regions (Li et al., 2020b), and it is different from that of the 6RLAr arm. However, the pSc119.2 signal pattern of 6RLAr is similar to that on the 6RL arm of rye cv. Qinling (Hao et al., 2018). These results exhibited the abundant genetic diversity of chromosome 6R in the genus Secale. Therefore, it is necessary to continue to study the allelic variations of 6R chromosomes as they may enrich the genetic diversity of the resistance genes on them. Especially, the difference among the stripe rust resistance genes on different 6R chromosomes needs to be confirmed. Additionally, the results in this study make us think about whether the stripe rust resistance of the 6RL segment is controlled by a single gene or multiple genes.
Using 6R Deletions to Locate Stripe Rust Resistance Genes
Rye chromosome deletion lines are useful for the location of resistance genes. The powdery mildew resistance gene Pm56 was located in the subtelomeric region of the 6RS arm using 6R deletion lines (Hao et al., 2018). The stripe rust resistance gene Yr83 was mapped to the bin of FL0.73-1.00 of 6RL using deletion mapping (Li et al., 2020b). Similarly, 6R chromosome deletion and translocation lines were used to locate the powdery mildew resistance gene on the 6RL arm at FL0.85-1.00 and the stripe rust resistance gene on the 6RS arm at FL0.95-1.00 (Li et al., 2020a). However, the detailed information about the physical location of these reported resistance genes on 6R chromosomes is still unclear. In this study, 6RL-specific markers have been anchored to the exact physical positions on the 6R chromosome using reference genomic sequences of rye, and these markers combined with deleted T6BS.6RL translocation chromosomes were used to reveal that the 6RL segment with stripe rust resistance gene(s) is about 37 Mb. The primer sequences of the 33 markers that were located in the bin with the Yr83 gene were used for nucleotide BLAST searches against the 6R sequence of rye Lo7 (Rabanus-Wallace et al., 2021) using a BLAST tool in the Triticeae Multi-omics Center (http://202.194.139.32/), and these markers covered the 6R segment between 743,894,430 and 880,064,740 bp. Therefore, in this study, the 6RL segment with stripe rust resistance is smaller than that reported by Li et al. (2020b), and this narrows down the candidate genes with stripe rust resistance on 6R chromosome. The high similarity of the candidate resistance gene sequences between rye Lo7 and AR106BONE indicated that the physical mapping based on the reference genome of rye Lo7 can be applied to 6RLAr arm. Additionally, the new markers developed in this study enrich the 6R-specific molecular markers. It is a pity that the effect of the T6RLAr-6AS.6AL chromosome on agronomic traits is unclear because it exists in a plant together with the T6BS.6RLAr-3 chromosome. To get rid of the T6BS.6RLAr-3, the cross between line 21F11 and some wheat lines that are susceptible to stripe rust has already been carried out. The T6RLAr-6AS.6AL chromosome is a promising small-segment translocation for the improvement of wheat cultivars.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
ZT and SF conceived and designed the study, created the materials, analyzed the data, and wrote the manuscript. YD and JL performed the experiments and analyzed the data. ZY and GL created the materials. All authors read and approved the final manuscript.
Funding
This manuscript was provided by the National Natural Science Foundation of China (no. 31770373), the Open Project Program of the State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, China (no. PCCE-KF-2021-02), and the Sichuan Science and Technology Program (no. 2022ZDZX0014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are thankful to Jie Zhang and Yun Jiang (Biotechnology and Nuclear Technology Research Institute, Sichuan Academy of Agricultural Sciences) for that they help us to irradiate the seeds used in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.928014/full#supplementary-material
Supplementary Figure S1. Alignment between the sequence Ar-60.1 and Lo7-60.1. Ar-60.1 represents the sequence amplified by P60 from T6RLAr-6AS.6AL translocation. Lo7-60.1 represents the partial sequence of SECCE6Rv1G0449960.1 of rye Lo7. Red arrows indicate the primer sequences.
Supplementary Figure S2. Alignment between the sequence Ar-70.1 and Lo7-70.1. Ar-70.1 represents the sequence amplified by P70 from T6RLAr-6AS.6AL translocation. Lo7-70.1 represents the partial sequence of SECCE6Rv1G0453070.1 of rye Lo7. Red arrows indicate the primer sequences.
References
An, D., Ma, P., Zheng, Q., Fu, S., Li, L., Han, F., et al. (2019). Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot. Theor. Appl. Genet. 132, 257–272. doi: 10.1007/s00122-018-3214-3
Bouvet, L., Percival-Alwyn, L., Berry, S., Fenwick, P., Mantello, C. C., Sharma, R., et al. (2022). Wheat genetic loci conferring resistance to stripe rust in the face of genetically diverse races of the fungus Puccinia striiformis f. sp. tritici. Theor. Appl. Genet. 135, 301–319. doi: 10.1007/s00122-021-03967-z
Devos, K. M., Atkinson, M. D., Chinoy, C. N., Francis, H. A., Harcourt, R. L., Koebner, R. M., et al. (1993). Chromosomal rearrangements in the rye genome relative to that of wheat. Theor. Appl. Genet. 85, 673–680. doi: 10.1007/BF00225004
Fu, S. L., Chen, L., Wang, Y. Y., Li, M., Yang, Z. J., Qiu, L., et al. (2015). Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci. Rep. 5, 10552. doi: 10.1038/srep10552
Han, F. P., Lamb, J. C., and Birchler, J. A. (2006). High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. U. S. A. 103, 3238–3243. doi: 10.1073/pnas.0509650103
Hao, M., Liu, M., Luo, J., Fan, C., Yi, Y., Zhang, L., et al. (2018). Introgression of powdery mildew resistance gene Pm56 on rye chromosome arm 6RS into wheat. Front. Plant. Sci. 9, 1040. doi: 10.3389/fpls.2018.01040
Hu, C. Y., Wang, F. T., Lang, X. W., Feng, J., Li, K. J., Lin, R. M., et al. (2022). Resistance analyses on wheat stripe rust resistance genes to the predominant races of Puccinia striiformis f. sp. tritici in China. Sci. Agr. Sin. 55, 491–502. https://doi.org/10.3864/j.issn.0578-1752.2022.03.006
Jamil, S., Shahzad, R., Ahmad, S., Fatima, R., Zahid, R., Anwar, M., et al. (2020). Role of genetics, genomics, and breeding approaches to combat stripe rust of wheat. Front. Nutr. 7, 580715. doi: 10.3389/fnut.2020.580715
Johansson, E., Henriksson, T., Prieto-Linde, M. L., Andersson, S., Ashraf, R., and Rahmatov, M. (2020). Diverse wheat-alien introgression lines as a basis for durable resistance and quality characteristics in bread wheat. Front. Plant. Sci. 11, 1067. doi: 10.3389/fpls.2020.01067
Komuro, S., Endo, R., Shikata, K., and Kato, A. (2013). Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56, 31–137. doi: 10.1139/gen-2013-0003
Lei, M., Li, G., Zhang, S., Liu, C., and Yang, Z. (2011). Molecular cytogenetic characterization of a new wheat Secale africanum 2Ra(2D) substitution line for resistance to stripe rust. J. Genet. 90, 283–287. doi: 10.1007/s12041-011-0081-y
Li, G. R., Gao, D., La, S. X., Wang, H. J., Li, J. B., He, W. L., et al. (2016a). Characterization of wheat-Secale africanum chromosome 5R(a) derivatives carrying Secale specific genes for grain hardness. Planta 243, 1203–1212. doi: 10.1007/s00425-016-2472-z
Li, G. R., Tang, L. R., Yin, Y., Zhang, A. H., Yu, Z. H., Yang, E. N., et al. (2020a). Molecular dissection of Secale africanum chromosome 6Rafr in wheat enabled localization of genes for resistance to powdery mildew and stripe rust. BMC. Plant. Biol. 20, 134. doi: 10.1186/s12870-020-02351-1
Li, J. B., Dundas, I., Dong, C. M., Li, G. R., Trethowan, R., Yang, Z. J., et al. (2020b). Identification and characterization of a new stripe rust resistance gene Yr83 on rye chromosome 6R in wheat. Theor. Appl. Genet. 133, 1095–1107. doi: 10.1007/s00122-020-03534-y
Li, J. J., Endo, T. R., Saito, M., Ishikawa, G., Nakamura, T., and Nasuda, S. (2013). Homoeologous relationship of rye chromosome arms as detected with wheat PLUG markers. Chromosoma 122, 555–564. doi: 10.1007/s00412-013-0428-7
Li, M., Tang, Z. X., Qiu, L., Wang, Y. Y., Tang, S. Y., and Fu, S. L. (2016b). Identification and physical mapping of new PCR-based markers specific for the long arm of rye (Secale cereale L.) chromosome 6. J. Genetics. Genomics. 43, 209–216. doi: 10.1016/j.jgg.2015.11.005
McIntyre, C. L., Pereira, S., Moran, L. B., and Appels, R. (1990). New Secale cereale (rye) DNA derivatives for the detection of rye chromosome segments in wheat. Genome 33, 635–640. doi: 10.1139/g90-094
Rabanus-Wallace, M. T., Hackauf, B., Mascher, M., Lux, T., Wicker, T., Gundlach, H., et al. (2021). Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat. Genet 53, 564–573. doi: 10.1038/s41588-021-00807-0
Ren, T., Li, Z., Tan, F., Jiang, C., and Luo, P. (2022). “Advances in identifying stripe rust resistance genes in cereals,” in Achieving Durable Disease Resistance in Cereals, ed R. Oliver (Cambridge: Burleigh Dodds Science Publishing), 39–80. doi: 10.19103/AS.2021.0092.04
Ren, T. H., Sun, Z. X., Ren, Z. L., Tan, F. Q., Luo, P. G., Tang, Z. X., et al. (2020). Molecular and cytogenetic characterization of a wheat-rye 7BS.7RL translocation line with resistance to stripe rust, powdery mildew, and fusarium head blight. Phytopathology 110, 1713–1720. doi: 10.1094/PHYTO-02-20-0061-R
Schneider, A., Rakszegi, M., Molnár-Láng, M., and Szakács, É. (2016). Production and cytomolecular identification of new wheat-perennial rye (Secale cereanum) disomic addition lines with yellow rust resistance (6R) and increased arabinoxylan and protein content (1R, 4R, 6R). Theor. Appl. Genet. 129, 1045–1059. doi: 10.1007/s00122-016-2682-6
Spetsov, P., and Daskalova, N. (2022). Resistance to pathogens in wheat-rye and triticale genetic stocks. J. Plant. Pathol. 104, 99–114. https://doi.org/10.1007/s42161-021-01019-5
Tang, Z. X., Yang, Z. J., and Fu, S. L. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Vershinin, A. V., Schwarzacher, T., and Heslop-Harrison, J. S. (1995). The large-scale genomic organization of repetitive DNA families at the telomeres of rye chromosomes. Plant. Cell. 7, 1823–1833. doi: 10.1105/tpc.7.11.1823
Wan, A., Wang, X., Kang, Z., and Chen, X. (2017). “Variability of the stripe rust pathogen,” in The Stripe Rust, ed X. Chen (Van Godewijckstraat: Springer Nature), 35–154. doi: 10.1007/978-94-024-1111-9_2
Xi, W., Tang, Z., Luo, J., and Fu, S. (2019). Physical location of new stripe rust resistance gene(s) and PCR-based markers on rye (Secale cereal L.) chromosome 5 using 5R dissection lines. Agronomy 9, 498. doi: 10.3390/agronomy9090498
Keywords: wheat, rye, stripe rust, 6R chromosome, small-segment translocation
Citation: Duan Y, Luo J, Yang Z, Li G, Tang Z and Fu S (2022) The Physical Location of Stripe Rust Resistance Genes on Chromosome 6 of Rye (Secale cereale L.) AR106BONE. Front. Plant Sci. 13:928014. doi: 10.3389/fpls.2022.928014
Received: 25 April 2022; Accepted: 23 May 2022;
Published: 29 June 2022.
Edited by:
Fangpu Han, Institute of Genetics and Developmental Biology (CAS), ChinaReviewed by:
Peng Zhang, The University of Sydney, AustraliaAndreas Houben, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Germany
Hongjie Li, Institute of Crop Sciences (CAAS), China
Copyright © 2022 Duan, Luo, Yang, Li, Tang and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zongxiang Tang, enh0YW5nQHNpY2F1LmVkdS5jbg==; Shulan Fu, ZnVzaHVsYW5Ac2ljYXUuZWR1LmNu
 Yanling Duan
Yanling Duan Jie Luo
Jie Luo Zujun Yang
Zujun Yang Guangrong Li
Guangrong Li Zongxiang Tang
Zongxiang Tang Shulan Fu
Shulan Fu