
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 14 July 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.918202
This article is part of the Research Topic Physiological and Molecular Basis of Fruit Ripening and Development and its Applications for Quality Improvement View all 5 articles
 Wenjun Liu1,2†
Wenjun Liu1,2† Zijing Chen1,2†
Zijing Chen1,2† Shenghui Jiang3
Shenghui Jiang3 Yicheng Wang4
Yicheng Wang4 Hongcheng Fang5
Hongcheng Fang5 Zongying Zhang1,2
Zongying Zhang1,2 Xuesen Chen1,2*
Xuesen Chen1,2* Nan Wang1,2*
Nan Wang1,2*Identifying the genetic variation characteristics of phenotypic traits is important for fruit tree breeding. During the long-term evolution of fruit trees, gene recombination and natural mutation have resulted in a high degree of heterozygosity. Apple (Malus × domestica Borkh.) shows strong ecological adaptability and is widely cultivated, and is among the most economically important fruit crops worldwide. However, the high level of heterozygosity and large genome of apple, in combination with its perennial life history and long juvenile phase, complicate investigation of the genetic basis of fruit quality traits. With continuing augmentation in the apple genomic resources available, in recent years important progress has been achieved in research on the genetic variation of fruit quality traits. This review focuses on summarizing recent genetic studies on apple fruit quality traits, including appearance, flavor, nutritional, ripening, and storage qualities. In addition, we discuss the mapping of quantitative trait loci, screening of molecular markers, and mining of major genes associated with fruit quality traits. The overall aim of this review is to provide valuable insights into the mechanisms of genetic variation and molecular breeding of important fruit quality traits in apple.
Apple (Malus × domestica Borkh.) shows strong ecological adaptability and is widely cultivated around the world. In 2020, the total global planting area of apple was 4.622 million ha, with total production output of 86.443 million tonnes, distributed in almost 100 countries on six continents (FAO and WFP, 2021). Consequently, apple is among the most economically important fruit crops worldwide. Most apple fruit are crisp, juicy, sweet, and delicious, and are rich in vitamins, dietary fiber, polyphenols, and mineral elements. Apple has many benefits to human health and is a favorite fruit among consumers (Eberhardt et al., 2000; Hyson, 2011; Oyenihi et al., 2022). Apple fruit quality is determined by many individual traits, most of which are quantitative traits controlled by minor polygenes or oligogenes (Conner et al., 1998; Cãtãlina et al., 2015; Zheng et al., 2020). According to the research objective, fruit quality can be divided into appearance, flavor, nutritional, storage and transportation, resistance, and processing qualities (Chen et al., 2015). The determinants of the different quality attributes include single fruit weight, fruit shape, color, texture, flavor, aroma, and functional ingredients (Chen et al., 2015).
Exploring the genetic basis and molecular mechanism of phenotypic traits is of great significance for fruit tree breeding. The inheritance of traits can be divided into qualitative traits controlled by major genes and quantitative traits controlled by minor genes. The former are inherited simply and follow Mendelian inheritance principles (Mendel, 1965), whereas the latter are affected by the environment and gene interactions, and hybrid offspring often deviate from Mendelian inheritance patterns. Over the course of long-term evolution, gene recombination and natural mutation have resulted in a high degree of heterozygosity in apple (Sun et al., 2020). In addition, owing to extensive artificial selection, non-additive genetic effects are important sources of genetic variation for apple phenotypes (Kumar et al., 2015). These genetic characteristics, in conjunction with a perennial life history and long juvenile phase, complicate research on apple genetics.
Previously, reverse genetics was used to explore the genes that influence apple fruit quality. For example, the V-myb myeloblastosis viral oncogene homolog transcription factors (TFs) MdMYB1 and MdMYB10 regulate anthocyanin synthesis and coloration in apple fruit (Takos et al., 2006; Espley et al., 2007). The sucrose transporter MdSUT1 and sorbitol transporter MdSOT1 regulate the sugar uptake and transport mechanism (Fan et al., 2009). The 1-aminocyclopropane-1-carboxylic acid synthase gene (MdACS1) regulates ethylene synthesis and apple fruit ripening (Sunako et al., 1999). The successful assembly of the complete genome sequence of “Golden Delicious” apple in 2010 has enabled whole-genome analysis of apple. In addition, availability of a reference genome has permitted related research on apple fruit quality, including genome association analysis, and the cloning and identification of key functional genes. A total of 57,386 genes, including 4,021 TFs, 178 microRNAs (miRNAs), 992 resistance genes, and 1,246 biosynthetic genes, were annotated in the “Golden Delicious” genome (Velasco et al., 2010). Subsequently, additional high-quality apple genome assemblies have been generated using the latest sequencing technologies or more homozygous and diverse samples (Li et al., 2016; Daccord et al., 2017; Zhang et al., 2019; Sun et al., 2020). In addition, substantial progress has been achieved in genome-resequencing analysis and genome-wide association analysis (GWAS) (Duan et al., 2017; Liao et al., 2021). These achievements are essential for the genetic analyses of apple fruit traits, for example, for the construction of high-density linkage maps, mapping of quantitative trait loci (QTLs), and identification of genes crucial for apple trait development.
This review focuses on the genetic variation characteristics of important fruit quality traits in apple. We summarize recent genetic studies on apple fruit quality traits, including appearance, flavor, nutritional, ripening, and storage qualities. In addition, we discuss the mapping of QTLs, screening of molecular markers, and mining of major genes associated with each quality trait. The overall aim of this review is to provide valuable insights into the mechanisms of genetic variation and molecular breeding of important fruit quality traits in apple.
The appearance quality of apple fruit, mainly comprising fruit size (single fruit weight), fruit shape, and fruit color, is an important aesthetic attribute that strongly determines the commercial value of the fruit. For example, the breeding of the large-fruit cultivar “World No. 1” (a single fruit weighs more than 600 g), the long-oval shaped cultivar “Starkrimson” (fruit shape index of 0.98; Westwood and Blaney, 1963), and various red-colored cultivars all represent the genetic improvement of apple fruit appearance and quality (Chen et al., 2021). Here, we present an overview of the genetic characteristics of fruit appearance quality, such as single fruit weight, fruit shape, coloration, and fruit russeting, and discuss the QTLs, molecular markers, and main-effect genes associated with each appearance quality trait.
Fruit weight is an economically important trait in apple and is a quantitative trait controlled by multiple genes (Brown, 1960). As a quantitative indicator of apple fruit size, it directly determines the market price of the fruit. Almost 100 QTLs associated with apple fruit weight have been reported, with logarithm of the odds (LODs) ranging from 2.97 to 10.98, and include major and minor loci distributed on almost all 17 chromosomes (Liebhard et al., 2003; Kenis et al., 2008; Devoghalaere et al., 2012; Chang et al., 2014; Potts et al., 2014). Devoghalaere et al. (2012) detected six QTL regions for fruit weight. Two of the QTLs were conserved across both segregating populations on linkage group (LG) 8 (designated fw2) and LG 15 (designated fw1). One QTL was mapped to a region containing an auxin response factor gene (ARF106). This gene is expressed during cell division and cell expansion, consistent with a potential role in the control of fruit size (Devoghalaere et al., 2012). Consistently, transcriptome analysis of “Longfeng” apple and its large-fruited bud sport variety “Grand Longfeng” showed that most of the differential genes were related to auxin signaling, including the auxin synthetic genes MdTAR1 and MdYUCCA6 (Bu et al., 2020). These results suggested that auxin played a critical role in trait development of single fruit weight in apple. Recent apple genome resequencing studies identified one cell division regulatory gene and two β-galactosidase genes in fw1 by GWAS analysis. In addition, the miRNAs miR172g and miR172h are indicated to be important for fruit enlargement during domestication of cultivated apples (Duan et al., 2017). Similarly, overexpression of miR172p in transgenic “Royal Gala” apple significantly reduces fruit size by targeting the expression of AP2 TFs (Yao et al., 2015). The QTLs, molecular markers, major-effect genes, and miRNAs associated with apple fruit weight have been extensively mined, but for the majority, except miR172p, their functions remain to be elucidated (Figure 1).
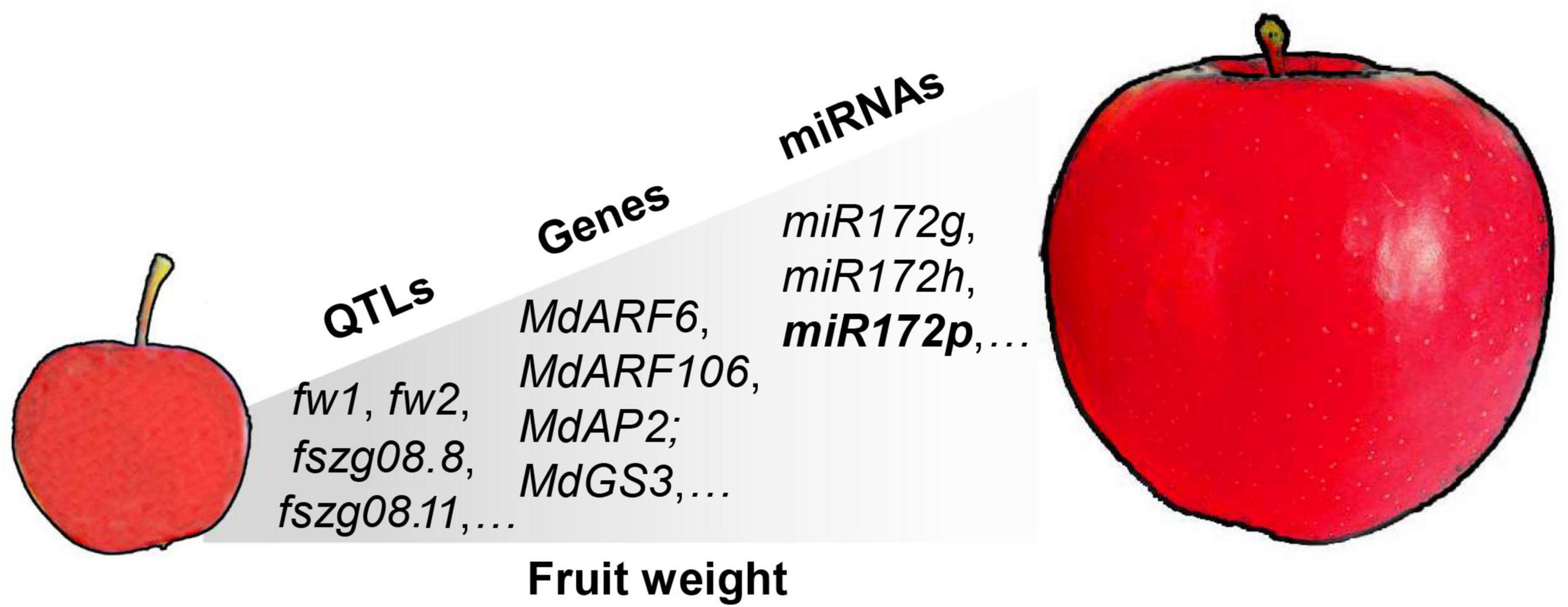
Figure 1. Quantitative trait loci (QTLs), molecular markers, major-effect genes, and miRNAs associated with apple fruit weight. The main QTLs associated with fruit weight include fw1, fw2, and fszg08.8 (Devoghalaere et al., 2012; Chang et al., 2014). The major-effect genes include the auxin response factors MdARF6 and MdARF106, and APETALA 2 family TF MdAP2 (Devoghalaere et al., 2012; Yao et al., 2015). The main miRNAs include miR172g, miR172h, and miR172p (Yao et al., 2015; Duan et al., 2017). The function of miR172p has been elucidated in transgenic “Royal Gala” apple.
The fruit color of apple can be divided into skin color and flesh color. The skin color can be subdivided into presence or absence of red pigmentation, striped red, or blushed red. The flesh color is mainly subdivided into red fleshed and non-red fleshed. The red pigmentation of apple fruit is determined mainly by anthocyanins, which are color-producing secondary metabolites that accumulate in different tissues and organs of plants (Jaakola, 2013). The major gene MdMYB1 was the first gene identified to control the presence or absence of red pigmentation in the skin (Takos et al., 2006) and associated molecular markers were explored subsequently (Kumar et al., 2012). More recently, GWAS analysis using single-nucleotide polymorphism (SNP) markers confirmed the association between fruit color and the MdMYB1 locus on chromosome 9 (Migicovsky et al., 2016; McClure et al., 2019). Comparative genomics analysis of 148 apple populations and a segregated hybrid population revealed that a gypsy-like long terminal repeat retrotransposon (designated redTE) was inserted 3297 bp upstream of MdMYB1, thereby activating the expression of MdMYB1 and controlling the redness of the skin (Figure 2A; Zhang et al., 2019). In Japanese plums, high levels of variability in the intronic and intergenic regions of the MYB10 LG3 cluster were also closely associated with polymorphisms in their skin color (Fiol et al., 2021). These results reveal the important function of MYB1 as a major gene in regulating fruit coloration, and its function is affected by extensive variation in gene regions and intergenic regions.
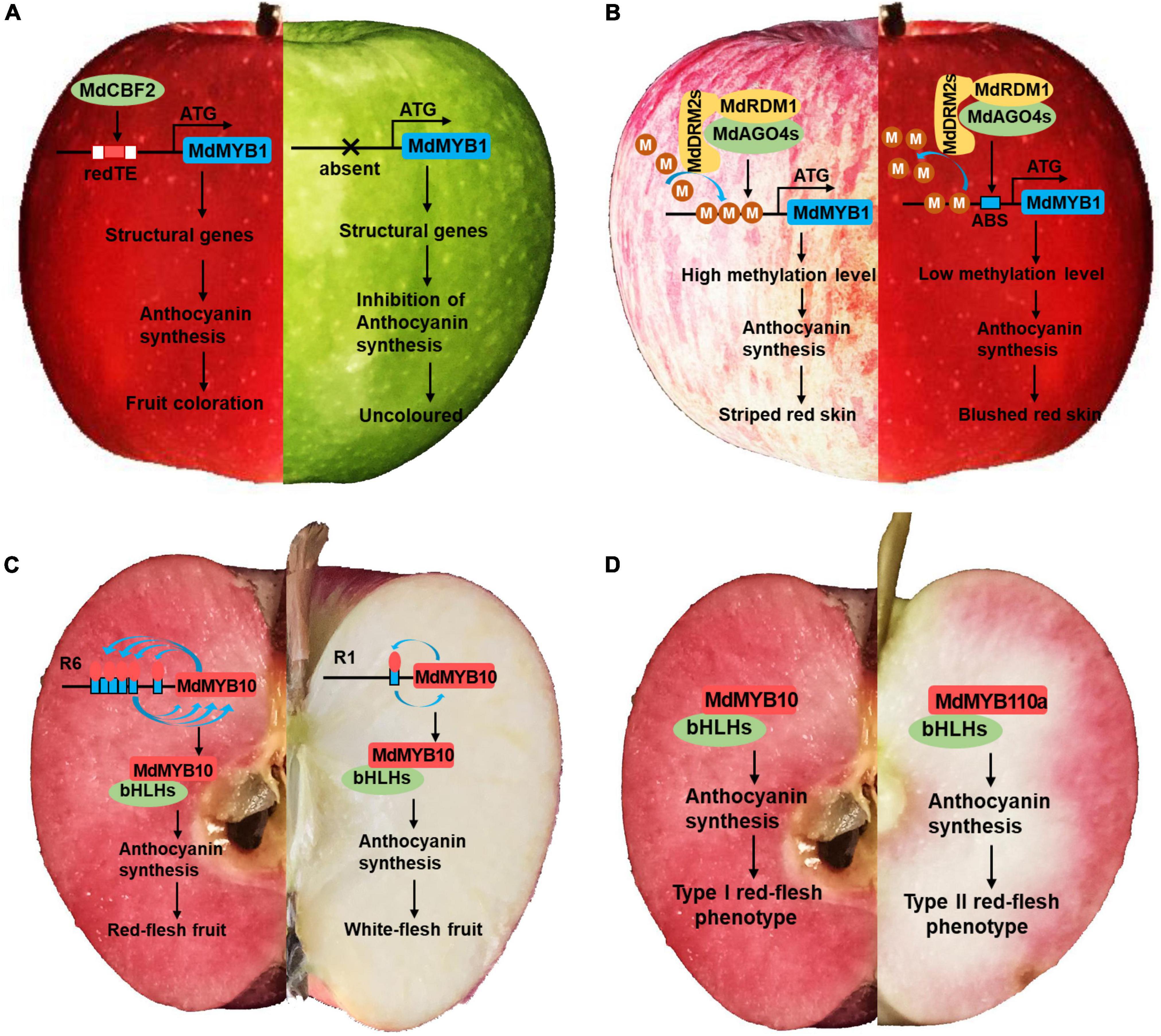
Figure 2. Red pigmentation of apple fruit skin and flesh. (A) A gypsy-like long terminal repeat retrotransposon (designated redTE) was inserted 3297 bp upstream of MdMYB1, thereby activating the expression of MdMYB1 and controlling the redness of the fruit skin. (B) MdAGO4s, MdDRM2s, and MdRDM1 interact with each other and form an effector complex. MdAGO4s recruit MdDRM2s, which catalyze CHH methylation of the MdMYB1 promoter. MdMYB1 then regulates anthocyanin accumulation to determine the coloration. M, a -CH3 (methyl); ABS, AGO4 binding sequence. (C) Model showing autoregulation of the R6 and R1 promoters by MdMYB10. The MdMYB10 promoter in red-fleshed apple contains six 23 bp repeating microsatellite sequences (R6), which confer MdMYB10 with self-activation. The MdMYB10 promoter in white-fleshed apple contains only one 23 bp repeating microsatellite sequence. (D) MdMYB10 and its homolog MdMYB110a are involved in the red pigmentation of type I and type II red-fleshed apples, respectively.
In contrast with the presence or absence of red pigmentation in the fruit ground color, the color patterns of the fruit overcolor, such as blushed red and striped red, are mostly associated with methylation modification, which represents a type of epigenetic inheritance. Differences in anthocyanin levels between the red and green stripes can be explained by differences in methylation levels of the MdMYB10 promoter (Telias et al., 2011). Whole-genome bisulfite sequencing of “Red Delicious” and its four-generation red sport mutants “Starking Red,” “Starkrimson,” “Campbell Redchief,” and “Vallee Spur” showed that differences in DNA methylation levels were responsible for genetic variation of red sport mutants from “Red Delicious” (Li et al., 2019). Subsequent detailed methylation modification studies using striped red or blushed red fruit of “Fuji” apple as the study material showed that Argonaute 4 (MdAGO4s) methylated the MdMYB1 promoter, thereby regulating anthocyanin biosynthesis (Figure 2B; Jiang et al., 2020).
Red-fleshed apples have received widespread attention from apple breeders and consumers owing to their more attractive color and higher functional nutritional content (Wang et al., 2018a). Red-fleshed apples are mainly divided into type I (red pigmentation in the fruit flesh, stems, flowers, and young leaves) and type II (red flesh only in the outer cortex, no red pigmentation in the leaves, stems, or other tissues) (Volz et al., 2009; Sekido et al., 2010). The red-flesh phenotype of type I apples is controlled by MdMYB10, which contains six 23 bp repeating microsatellite sequences in its promoter that confer it with self-activation, resulting in anthocyanin synthesis (Figure 2C; Espley et al., 2009). The red-flesh phenotype of type II apples is controlled by MdMYB110, a MdMYB10 homolog located in LG 17, which is not expressed in type I red-fleshed fruit (Figure 2D; Chagné et al., 2013). In red-fleshed kiwifruit, both MYB10 and MYB110 could upregulate anthocyanin biosynthesis in fruit, while MYB10 resulted in anthocyanin accumulation limited to the inner pericarp (Wang W. Q. et al., 2022a). These results suggest that the differential expression patterns of MYB10 and MYB110 contribute to the variation of the red flesh phenotype.
Fruit shape is a quantitative trait controlled by minor polygenes. The fruit shape index is the ratio of the fruit height to width. Using multiple hybrid populations, Brown (1960) observed that the fruit shape index of most combinations was close to the parent median value. Previous studies have shown that the fruit shape index of apple is a quantitative trait controlled by five pairs of genes (Sun et al., 2012). Four simple sequence repeat markers and one amplified fragment length polymorphism marker linked to the fruit shape index were screened in a “Jonathan” × “Golden Delicious” hybrid population using bulked segregant analysis (Sun et al., 2012). Using the same hybrid population, QTL mapping analysis showed that LG11 and LG15 contained QTLs associated with fruit shape index, and the QTL loci in LG11 were stable in different years (Chang et al., 2014). Interestingly, there are not only overlapping QTLs for single-fruit weight and fruit shape, but also many independent QTLs. However, to date, the crucial genes that regulate apple fruit shape have not been identified.
Fruit russeting occurs in many apple cultivars, such as “Egremont Russet,” and seriously affects the appearance quality and commercial value of the fruit. Fruit russeting in apple is the result of formation of a plastic periderm following microscopic cracking of the cuticle (Khanal et al., 2013). It was initially considered that fruit russeting in apple might be controlled solely by the Ru gene. However, evaluation of the offspring of crosses of non-fully russeted and less-russeted cultivars with fully russeted cultivars revealed that the non-fully russeted phenotype was controlled by multiple factors (Alston and Watkins, 1973). A high-density genetic map was developed using a F1 segregating population derived from a cross between the fully russeted cultivar “Renetta Grigia di Torriana” and “Golden Delicious,” and a major QTL associated with fruit russeting, designated Ru_RGT, was detected on chromosome 12. In addition, a putative ATP-BINDING CASSETTE G family transporter ABCG has been identified as a candidate gene that controls russet development in apple (Falginella et al., 2015). Overexpression of MdMYB93 in tobacco leaves leads to accumulation of a large amount of lignin monomers, suberin, and their precursors (Legay et al., 2016), suggesting that MdMYB93 may promote the formation of apple fruit russeting. In contrast, MdLIM1 can directly bind to the CCACTTGAGTAC element in the phenylalanine ammonia-lyase promoter, thereby inhibiting lignin synthesis and reducing the formation of fruit russeting (Yuan et al., 2019). However, the functional verification of the genes regulating fruit russeting so far is only related to the lignin synthesis, and the subsequent direct relationship with the fruit russeting phenotype remains to be verified.
With economic development and improvement of living standards, increasing attention is paid to the flavor quality, nutritional value, and health benefits of fruit. While consumers are influenced by aesthetic traits, they also are paying greater attention to taste attributes. Here, we introduce the genetic characteristics of apple fruit flavor and nutritional components, such as soluble sugars, organic acids, aromatic substances, and polyphenols, and discuss the QTLs, molecular markers, and main-effect genes associated with each quality trait.
In apple, sugar and acid contents and the sugar:acid ratio are important indicators of fruit flavor quality (Jayasena and Cameron, 2008). The soluble sugars in apple fruit mainly comprise sucrose, fructose, glucose, and sorbitol, among which glucose and fructose are almost entirely localized to vacuoles (Yamaki, 1984). The sugar content of apple fruit is a typical quantitative trait controlled by minor genes (Liao et al., 2021). The sugar content in hybrid offspring shows a normal distribution and the average sugar content in the population is close to the parent median (Ma et al., 2015), which indicates that the of sugar content is mainly controlled by additive genetic effects. More than 70 QTLs associated with fruit sugar content have been detected in different populations, distributed on at least 15 chromosomes with LODs ranging from 1.9 to 12.6 (Table 1). Taken together, the focal LGs for sugar content in apple fruit are LG1 and LG3, which have been located frequently in multiple hybrid populations.
Whole-genome resequencing of 497 apple accessions revealed that apple breeding has resulted in a reduction in the degree of population-level genetic polymorphisms, and that sweet apples and wine apples were domesticated independently (Liao et al., 2021). GWAS for soluble sugar contents showed that a SNP mutation of the main-effect gene MdSOT2 significantly reduced accumulation of sorbitol in apple fruit (Liao et al., 2021). In addition, a stable QTL was detected in LG01 in “Honeycrisp.” The SNP A/G variation of the sorbitol dehydrogenase gene MdSDH2 promoter affects its binding to the TF MdABI3, thereby downregulating the expression level of MdSDH2 and reducing the fructose content in the fruit (Wang Z. et al., 2022b). Thus, SNP mutations in genes that encode key enzymes for sugar synthesis have significant effects on the accumulation of various soluble sugars.
The unloading of sugars from the phloem to the fruit via the apoplast and symplast pathways, and the transport of sugars to the plasma and tonoplast membranes of fruit flesh cells are regulated by various genes, such as the sucrose transporters MdSUT1, MdSUT2, and MdSUT4, monosaccharide transporter MdTMT1, glucose transporter MdVGT1, and multiple MdSWEET genes (Fan et al., 2009; Ma et al., 2017; Zhen et al., 2018; Peng et al., 2020; Xu et al., 2020). The genetics, synthesis, metabolism, and transport of soluble sugars in apple fruit are complex and further research is needed.
The predominant organic acid in apple fruit is malic acid, which accounts for more than 80% of the total acid content and is the main contributor to tartness in the fruit (Scherer et al., 2012). The content of malic acid is jointly regulated by the major gene Ma and a minor gene (Verma et al., 2019). The recessive homozygous mama genotype exhibits low acidity, whereas the dominant homozygous MaMa and heterozygous Mama genotypes are controlled by minor polygenes and exhibit high to moderate acidity (Xu et al., 2012). Using a molecular marker linkage map, the major gene associated with malic acid was localized to LG16 and designated the Ma locus (Maliepaard et al., 1998). Subsequently, two QTLs associated with fruit acidity were detected, which were located in LG8 and LG16, respectively, and the QTL in LG16 was identical to that detected previously (Liebhard et al., 2003; Kumar et al., 2012; Ma et al., 2016).
On this basis, through further precise mapping, the major Ma locus in LG16 was localized to a 65–82 kb segment, containing 12–19 candidate genes, one of which encodes an aluminum-activated malate transporter, designated MdALMTII (or Ma1) (Bai et al., 2012; Xu et al., 2012). Ma1 is considered to be the primary gene that determines the Ma locus and fruit acidity, which has been verified in a subsequent genome-wide linkage analysis (Liao et al., 2021). A SNP mutation (A/G) in the coding sequence of Ma1 leads to premature termination of translation and reduces the malic acid content (Bai et al., 2012). The premature termination of translation changes the subcellular localization of Ma1 and, consequently, its malate transport function is lost (Ma et al., 2015). Similarly, deletion of a coding region at the C-terminus of MdALMT9, a homolog of Ma1, leads to premature termination of its translation and reduction in malic acid content (Li et al., 2020). Therefore, SNPs or indels mutations in Ma1 gene coding sequences significantly affect its transport function and malate content. In addition, recent studies have showed that MdWRKY126 and MdMa12 can lead to the accumulation of malate by regulating the activity of malate dehydrogenase (Gao et al., 2022; Zhang et al., 2022).
The aroma of apple fruit is composed of more than 300 different volatile substances, mainly including alcohols, aldehydes, esters, ketones, and ethers. The genetic mechanism of aroma caused by diverse volatile substances, different synthetic pathways, and multiple regulatory genes is extremely difficult to study. Except for LG8, LG10, and LG13, QTLs associated with fruit aroma may be localized to the remaining 14 LGs (Zini et al., 2005; Dunemann et al., 2009; Costa et al., 2013). Among them, LG2 and LG15 are the focal LGs for apple fruit aroma. In contrast, the main QTL associated with fruit aroma compounds in peach was detected in LG4 (Eduardo et al., 2013), and the QTL associated with γ-decalactone content in strawberry was detected in LG3 (Sánchez-Sevilla et al., 2014). Given the distinct aroma components in different fruits, the genetic linkage associations are diverse. In apple, genes have been cloned and identified for several enzymes crucial for aroma synthesis, such as MdLOX1 encoding lipoxygenases (Schiller et al., 2015), MdAAT1 and MdAAT2 encoding alcohol acyltransferases (Li et al., 2006; Dunemann et al., 2012), and MdADH1 and MADH2 encoding alcohol dehydrogenases (Echeverrıa et al., 2004). However, the genetic mechanism and crucial regulatory genes associated with apple fruit aroma require further study.
Apple fruit are rich in a variety of vitamins, dietary fiber, polyphenols, and mineral elements. Among these components, functional nutrients such as polyphenols, flavonoids, chlorogenic acid, and L-ascorbic acid (vitamin C) are effective in antioxidation, prevention of cardiovascular and cerebrovascular diseases, and anti-tumor effects in the human body (Eberhardt et al., 2000; Naveed et al., 2018; Bondonno et al., 2019). The polyphenols in apple fruit comprise mainly flavonoids, tannins, phenolic acids, and anthocyanins, which are quantitative traits controlled by multiple genes. Using a “Royal Gala” × “Braeburn” hybrid population, QTLs associated with the contents of polyphenols, such as chlorogenic acid, quinic acid, anthocyanin, catechin, epicatechin, quercetin, and phloridzin, were located in LG1, LG6, LG7, LG9, LG13, LG14, LG15, LG16, and LG17 with LODs ranging from 3.03 to 41.28 (Chagné et al., 2012). Among these QTLs, a crucial gene encoding leucoanthocyanidin reductase, MdLAR1, involved in flavonol synthesis was screened from the QTL located in LG16, and genes encoding hydroxy cinnamate transferase/hydroxy quinate transferase, MdHCT/HQT, involved in chlorogenic acid synthesis were screened from the QTL located in LG17 (Chagné et al., 2012). Using a “Prima” × “Fiesta” hybrid population, QTLs associated with polyphenols were located in LG1, LG8, and LG13, of which 33 metabolite quantitative trait loci (mQTLs) associated with skin phenolic compounds and 17 mQTLs associated with flesh phenolic compounds were detected in LG16 (Khan et al., 2012). Extensive progress has been achieved in understanding the genetic mechanism and regulation of polyphenol accumulation in apple fruit. TFs of the MYB, bZIP, WRKY, HSF, ERF, and other families are widely reported to be involved in apple polyphenol synthesis (An et al., 2017; Wang et al., 2017, 2018b,2020; Zhang et al., 2018). In comparison, research on other nutritional components of apple fruit has lagged behind.
L-ascorbic acid is an important antioxidant (Davey et al., 2000). The content of L-ascorbic acid in apple fruit is a quantitative trait controlled by multiple genes (Mellidou et al., 2012). Using “Telamon” × “Braeburn” hybrid populations, QTLs associated with apple fruit L-ascorbic acid were located in LG10, LG11, LG16, and LG17, among which genes for crucial enzymes, such as GDP-L-galactose phosphorylase (GGP) and dehydroascorbate reductase (DHAR), were identified. Allelic variation of MdGGP1 and MdGGP3 in “Braeburn” is associated with high L-ascorbic acid content in the fruit (Mellidou et al., 2012). A recent study has shown that the transcript and protein abundances of Asc Mannose pathway Regulator 1 Like 1 (MdAMR1L1), a regulator involved in the L-ascorbic acid synthesis pathway, are negatively correlated with L-ascorbic acid content during apple fruit development (Ma et al., 2022). Up to now, the research on apple L-ascorbic acid is still preliminary, and its regulatory mechanism and key genes still need to be further studied.
The respiration of apple fruit is typical of a climacteric fruit. The fruit ripening and storability periods strongly determine the commercial value of different apple cultivars (Johnston et al., 2002). Inevitably, ethylene, as an important senescence-related endogenous hormone in plants, can initiate fruit ripening and synergistically complete the entire storage process, degrade cell walls, and lead to fruit softening (Giovannoni, 2004; Chaves and Mello-Farias, 2006). Here, we review the genetic characteristics of apple fruit ripening and storage quality, and discuss the relevant regulatory functions of ethylene and other factors.
Apple fruit ripening is a quantitative trait controlled by multiple genes, which basically conforms to the characteristics of mesotrophic variation. Using “Fiesta” × “Discovery” hybrid populations, one major-effect QTL associated with fruit ripening was located in LG3 (LOD = 4.7); the QTL originated from the early ripening parent “Discovery” and explained 16% of the phenotypic variability (Liebhard et al., 2003). Using a “Telamon” × “Braeburn” hybrid population, QTLs associated with fruit ripening were located in LG3, LG9, LG10, and LG16, with LODs ranging from 3.2 to 8.6 (Kenis et al., 2008). Through GWAS of 1168 samples of different apple genotypes and phenotypic analysis of their phenological traits, six SNPs (four on chromosome 3, one on chromosome 10, and one on chromosome 16) were retained as cofactors for ripening period at the whole-population level, which accounted for 17.2% of the phenotypic variance. Among the candidate genes, NAC family TFs and AP2/ERF family TFs were widely identified, indicating that they play key roles in apple fruit ripening (Urrestarazu et al., 2017). Consistent with this finding, the involvement of NAC and AP2/ERF family TFs in fruit ripening has been extensively demonstrated in other fruits, such as tomato, strawberry, peach, and kiwifruit (Wang et al., 2019; Gao et al., 2020; Fu et al., 2021; Kou et al., 2021; Martín-Pizarro et al., 2021).
Ethylene is an important regulator of fruit ripening, and ethylene biosynthesis, ethylene receptors, and ethylene response-related genes all affect fruit ripening (Sunako et al., 1999; Varanasi et al., 2011; Tan et al., 2013). In apple, MdSnRK2-1 can phosphorylate MdHB1 and MdHB2 to enhance their protein stability and transcriptional activity toward MdACO1, thereby promoting ethylene synthesis and fruit ripening (Jia et al., 2022). Besides, the TF MdMYC2 is responsive to jasmonate treatment and directly binds to the promoters of MdACS1, MdACO1, and MdERF3, thereby activating the ethylene signaling pathway during apple fruit ripening (Li et al., 2017). The auxin response factor MdARF5 is responsive to naphthaleneacetic acid treatment and directly binds to the promoters of MdERF2, MdACS1, MdASC3a, and MdACO1, and thereby induces ethylene biosynthesis during apple fruit ripening (Yue et al., 2020). These findings indicate that different hormonal signals are closely associated with ethylene synthesis and fruit ripening.
Fruit storability is a commercially important trait of apple, which directly determines the shelf life and commodity value. Fruit storability can be evaluated by retention of firmness and crispness, which are quantitative traits controlled by multiple genes (King et al., 2001; Soglio et al., 2009; Longhi et al., 2013). Ethylene plays an important role in the formation and maintenance of fruit firmness and crispness. The transcriptional activity of the ethylene synthesis gene MdACS1-2 is greatly reduced by insertion in the promoter of a specific retrotransposon, namely a short interspersed nuclear element, resulting in significant reduction in ethylene synthesis and improved storage stability (Sunako et al., 1999). MdACS1-2/-2 homozygous apple cultivars show lower ethylene production, higher fruit firmness during storage, and are generally storable, whereas MdACS1-1/-2 heterozygous and MdACS1-1/-1 homozygous cultivars show higher ethylene production, the fruit softens readily, and is generally intolerant of storage (Sunako et al., 1999; Harada et al., 2000; Oraguzie et al., 2004). Recent research has shown that genetic variation of the ethylene-responsive factors MdERF3 and MdERF118 is involved in regulating flesh firmness and crispness retention of apple fruit (Wu et al., 2021).
Differences in the activities of various cell wall metabolic enzymes, such as polygalacturonase (PG) and β-galactosidase (β-Gal), also contribute to variation in fruit firmness and crispness among apple cultivars (Nybom et al., 2020). Comparison of the MdPG1 sequence between “Fuji” and “Mondial Gala” with significantly different storage traits revealed a SNP (T/T in “Fuji” and G/T in “Mondial Gala”) in the exon, which resulted in an amino acid change from valine to phenylalanine (Costa et al., 2010). Subsequently, 22 SNPs (10 in exons and 12 in introns) in MdPG1 were identified by genome-whole resequencing analysis of 77 apple cultivars, of which six SNPs lead to changes of MdPG1 amino acid and polygalacturonase activity (Longhi et al., 2013). QTLs associated with fruit firmness have been detected on chromosomes 12, 16, and 17 by GWAS analysis, and three polygalacturonases, one pectinesterase, and one glucan endo-1,3-β-glucosidase have been identified that are associated with cell wall modifications (Duan et al., 2017).
Genetic characteristics research is an important basis for crop breeding. This review has summarized research on the genetic variation characteristics of important quality traits in apple fruit in recent decades. Compared with annual and biennial self-compatible field or vegetable crops, perennial fruit trees mainly show the following characteristics of genetic variation: First, most perennial fruit trees are self-incompatible, highly heterozygous, and have a long juvenile phase. Consequently, it is difficult to design specific experiments to conduct genetic research on flowering and fruits. Second, most quality traits of fruit trees are quantitative traits controlled by multiple genes. This is the case for fruit quality traits such as fruit size, color, sugar and acid contents, aroma, and polyphenol content. However, in self-compatible peach genotypes, more than 20 fruit quality traits are controlled by one or two genes, such as white flesh/yellow flesh (Y/y), freestone/clingstone (F/f), and melting/non-melting (M/m) (Hesse, 1975). Finally, the prevalence of self-incompatibility and heterozygosity leads to non-additive genetic effects as an important source of genetic variation in apple phenotypes (Kumar et al., 2015).
In recent years, with the development of omics technologies, such as genomics, transcriptomics, proteomics, metabolomics, and phenomics, as well as GWAS, metabolic GWAS, structural variation analysis, molecular marker-assisted selection, and other emerging molecular technologies, substantial progress has been achieved in the genetic research of fruit traits. Many crucial genes associated with fruit quality traits have also been identified. However, owing to limitations in fruit tree gene editing and the generally longer juvenility period, the function of most fruit quality trait-related genes has not been fully validated (Lobato-Gómez et al., 2021). Furthermore, the heritable variation of the genome caused by epigenetics, such as histone modification, DNA methylation modification, chromatin remodeling, and non-coding RNA regulation, complicates the study of quality traits in fruit trees. Consequently, there remains much to do to comprehensively elucidate the genetic mechanism of fruit quality traits.
NW, WL, and ZC conceived the idea and prepared the figures. WL, ZC, SJ, YW, HF, and ZZ contributed to the writing. XC and NW reviewed and edited the manuscript. All authors have read and approved the final version of the manuscript.
This work was supported by the National Natural Science Foundation of China (CN) (32172533) and the Agricultural Variety Improvement Project of Shandong Province (2021LZGC024).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Robert McKenzie, Ph.D., from Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing the English text of a draft of this manuscript.
Alston, F., and Watkins, R. (1973). Apple breeding at east malling. Proc. Eucarpia Fruit Breed Symp. 11, 14–29.
Amyotte, B., Bowen, A. J., Banks, T., Rajcan, I., and Somers, D. J. (2017). Mapping the sensory perception of apple using descriptive sensory evaluation in a genome wide association study. PLoS One 12:e0171710. doi: 10.1371/journal.pone.0171710
An, J., Qu, F., Yao, J., Wang, X., You, C., Wang, X., et al. (2017). The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 4:17023. doi: 10.1038/hortres.2017.23
Bai, Y., Dougherty, L., Li, M., Fazio, G., Cheng, L., and Xu, K. (2012). A natural mutation led truncation in one of the two aluminum activated malate transporter like genes at the Ma locus is associated with low fruit acidity in apple. Mol. Genet. Genomics 287, 663–678. doi: 10.1007/s00438-012-0707-7
Bondonno, N., Dalgaard, F., Kyrø, C., Murray, K., Bondonno, C., Lewis, J., et al. (2019). Flavonoid intake is associated with lower mortality in the Danish Diet Cancer and Health Cohort. Nat. Commun. 10:3651. doi: 10.1038/s41467-019-11622-x
Brown, A. (1960). The inheritance of shape, size and season of ripening in progenies of the cultivated apple. Euphytica 9, 327–337. doi: 10.1007/BF00029485
Bu, H., Yu, W., Yuan, H., Yue, P., Wei, Y., and Wang, A. (2020). Endogenous auxin content contributes to larger size of apple fruit. Front. Plant Sci. 11:592540. doi: 10.3389/fpls.2020.592540
Cãtãlina, D., Adriana, S., Bozdog, C., and Radu, S. (2015). Estimation of genetic effects implied in apple inheritance of quantitative traits. J. Hortic. Forestry Biotechnol. 19, 85–90. doi: 10.1007/bf00029485
Chagné, D., Krieger, C., Rassam, M., Sullivan, M., Fraser, J., André, C., et al. (2012). QTL and candidate gene mapping for polyphenolic composition in apple fruit. BMC Plant Biol. 12:12. doi: 10.1186/1471-2229-12-12
Chagné, D., Lin-Wang, K., Espley, R., Volz, R., How, N., Rouse, S., et al. (2013). An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 161, 225–239. doi: 10.1104/pp.112.206771
Chang, Y., Sun, R., Sun, H., Zhao, Y., Han, Y., Chen, D., et al. (2014). Mapping of quantitative trait loci corroborates independent genetic control of apple size and shape. Sci. Hortic. 174, 126–132. doi: 10.1016/j.scienta.2014.05.019
Chaves, A., and Mello-Farias, P. (2006). Ethylene and fruit ripening: from illumination gas to the control of gene expression, more than a century of discoveries. Genet. Mol. Biol. 29, 508–515. doi: 10.1590/S1415-47572006000300020
Chen, Z., Yu, L., Liu, W., Zhang, J., Wang, N., and Chen, X. (2021). Research progress of fruit color development in apple (Malus domestica Borkh.). Plant Physiol. Biochem. 162, 267–279. doi: 10.1016/j.plaphy.2021.02.033
Chen, X., Guo, W., Xu, J., Cong, P., Wang, L., Liu, C., et al. (2015). Genetic improvement and promotion of fruit quality of main fruit trees. Sci. Agric. Sin. 48, 3524–3540.
Conner, P., Brown, S., and Weeden, N. (1998). Molecular-marker analysis of quantitative traits for growth and development in juvenile apple trees. Theor. Appl. Genet. 96, 1027–1035. doi: 10.1007/s001220050835
Costa, F., Peace, C., Stella, S., Serra, S., Musacchi, S., Bazzani, M., et al. (2010). QTL dynamics for fruit firmness and softening around an ethylene-dependent polygalacturonase gene in apple (Malus×domestica Borkh.). J. Exp. Bot. 61, 3029–3039. doi: 10.1093/jxb/erq130
Costa, F., Cappellin, L., Zini, E., Patocchi, A., Kellerhals, M., Komjanc, M., et al. (2013). QTL validation and stability for volatile organic compounds (VOCs) in apple. Plant Sci. 211, 1–7. doi: 10.1016/j.plantsci.2013.05.018
Costa, F. (2015). MetaQTL analysis provides a compendium of genomic loci controlling fruit quality traits in apple. Tree Genet. Genomes 11:819. doi: 10.1007/s11295-014-0819-9
Daccord, N., Celton, J., Linsmith, G., Becker, C., Choisne, N., Schijlen, E., et al. (2017). High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 49, 1099–1106. doi: 10.1038/ng.3886
Davey, M., Montagu, M., Inze, D., Sanmartin, M., Kanellis, A., Smirnoff, N., et al. (2000). Plant L-ascorbic acid: chemistry, function, metabolism, bioavailability and effects of processing. J. Sci. Food Agric. 80, 825–860.
Devoghalaere, F., Doucen, T., Guitton, B., Keeling, J., Payne, W., Ling, T., et al. (2012). A genomics approach to understanding the role of auxin in apple (Malus x domestica) fruit size control. BMC Plant Biol. 12:7. doi: 10.1186/1471-2229-12-7
Duan, N., Bai, Y., Sun, H., Wang, N., Ma, Y., Li, M., et al. (2017). Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nat. Commun. 8:249. doi: 10.1038/s41467-017-00336-7
Dunemann, F., Ulrich, D., Boudichevskaia, A., Grafe, C., and Weber, W. (2009). QTL mapping of aroma compounds analysed by headspace solid-phase microextraction gas chromatography in the apple progeny ‘Discovery’בPrima’. Mol. Breed. 23, 501–521. doi: 10.1007/s11032-008-9252-9
Dunemann, F., Ulrich, D., Malysheva-Otto, L., Weber, W., Longhi, S., Velasco, R., et al. (2012). Functional allelic diversity of the apple alcohol acyl-transferase gene MdAAT1 associated with fruit ester volatile contents in apple cultivars. Mol. Breed. 29, 609–625. doi: 10.1007/s11032-011-9577-7
Eberhardt, M., Chang, Y., and Rui, H. (2000). Antioxidant activity of fresh apples. Nature 405, 903–904. doi: 10.1038/35016151
Echeverrıa, G., Graell, J., López, M., and Lara, I. (2004). Volatile production, quality and aroma-related enzyme activities during maturation of ‘Fuji’ apples. Postharvest Biol. Technol. 31, 217–227. doi: 10.1016/j.postharvbio.2003.09.003
Eduardo, I., Chietera, G., Pirona, R., Pacheco, I., Troggio, M., Banchi, E., et al. (2013). Genetic dissection of aroma volatile compounds from the essential oil of peach fruit: QTL analysis and identification of candidate genes using dense SNP maps. Tree Genet. Genomes 9, 189–204. doi: 10.1007/s11295-012-0546-z
Espley, R. V., Brendolise, C., Chagne, D., Kutty-Amma, S., Green, S., Volz, R., et al. (2009). Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. plant cell 21, 168–183. doi: 10.1105/tpc.108.059329
Espley, R., Hellens, R., Putterill, J., Stevenson, D., Kutty-Amma, S., and Allanet, A. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Falginella, L., Cipriani, G., Monte, C., Gregori, R., Testolin, R., Velasco, R., et al. (2015). A major QTL controlling apple skin russeting maps on the linkage group 12 of ‘Renetta Grigia di Torriana’. BMC Plant Biol. 15:150. doi: 10.1186/s12870-015-0507-4
Fan, R., Peng, C., Xu, Y., Wang, X., Li, Y., Shang, Y., et al. (2009). Apple sucrose transporter SUT1 and sorbitol transporter SOT6 interact with cytochrome B5 to regulate their affinity for substrate sugars. Plant Physiol. 150, 1880–1901. doi: 10.1104/pp.109.141374
Fiol, A., García-Gómez, B. E., Jurado-Ruiz, F., Alexiou, K., Howad, W., and Aranzana, M. J. (2021). Characterization of Japanese Plum (Prunus salicina) PsMYB10 alleles reveals structural variation and polymorphisms correlating with fruit skin color. Front. Plant Sci. 12:655267. doi: 10.3389/fpls.2021.655267
Fu, B. L., Wang, W. Q., Liu, X. F., Duan, X. W., Allan, A. C., Grierson, D., et al. (2021). An ethylene-hypersensitive methionine sulfoxide reductase regulated by NAC transcription factors increases methionine pool size and ethylene production during kiwifruit ripening. New Phytol. 232, 237–251. doi: 10.1111/nph.17560
Gao, J., Zhang, Y., Li, Z., and Liu, M. (2020). Role of ethylene response factors (ERFs) in fruit ripening. Food Qual. Saf. 4, 15–20. doi: 10.1093/fqsafe/fyz042
Gao, M., Zhao, H., Zheng, L., Zhang, L., Peng, Y., Ma, W., et al. (2022). Overexpression of apple Ma12, a mitochondrial pyrophosphatase pump gene, leads to malic acid accumulation and the upregulation of malate dehydrogenase in tomato and apple calli. Hortic. Res. [Epub ahead of print]. doi: 10.1093/hr/uhab053
Giovannoni, J. (2004). Genetic regulation of fruit development and ripening. Plant Cell 16, S170–S180. doi: 10.1105/tpc.019158
Guan, Y., Peace, C., Rudell, D., Verma, S., and Evans, K. (2015). QTLs detected for individual sugars and soluble solids content in apple. Mol. Breed. 35:135. doi: 10.1007/s11032-015-0334-1
Harada, T., Sunako, T., Wakasa, Y., Soejima, J., Satoh, T., and Niizeki, M. (2000). An allele of the 1-aminocyclopropane-1-carboxylate synthase gene (Md-ACS1) accounts for the low level of ethylene production in climacteric fruits of some apple cultivars. Theor. Appl. Genet. 101, 742–746. doi: 10.1007/s001220051539
Hesse, C. (1975). “Peaches,” in Advanced in Fruit Breeding, eds J. Janick and J. N. Moore (West Lafayette, IN: Purdue University Press), 285–335.
Hyson, D. (2011). A comprehensive review of apples and apple components and their relationship to human health. Adv. Nutr. 2, 408–420. doi: 10.3945/an.111.000513
Jaakola, L. (2013). New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18, 477–483. doi: 10.1016/j.tplants.2013.06.003
Jayasena, V., and Cameron, I. (2008). °Brix/acid ratio as a predictor of consumer acceptability of Crimson Seedless table grapes. J. Food Qual. 31, 736–750. doi: 10.1111/j.1745-4557.2008.00231.x
Jia, M., Li, X., Wang, W., Li, T., Dai, Z., Chen, Y., et al. (2022). SnRK2 subfamily I protein kinases regulate ethylene biosynthesis by phosphorylating HB transcription factors to induce ACO1 expression in apple. New Phytol. 234, 1262–1277. doi: 10.1111/nph.18040
Jiang, S., Wang, N., Chen, M., Zhang, R., Sun, Q., Xu, H., et al. (2020). Methylation of MdMYB1 locus mediated by RdDM pathway regulates anthocyanin biosynthesis in apple. Plant Biotechnol. J. 18, 1736–1748. doi: 10.1111/pbi.13337
Johnston, J., Hewett, E., and Hertog, M. (2002). Postharvest softening of apple (Malus domestica) fruit: a review. New Zealand J. Crop Hortic. Sci. 30, 145–160. doi: 10.1080/01140671.2002.9514210
Kenis, K., Keulemans, J., and Davey, M. (2008). Identification and stability of QTLs for fruit quality traits in apple. Tree Genet. Genomes 4, 647–661. doi: 10.1007/s11295-008-0140-6
Khan, S., Chibon, P., De-Vos, R., Schipper, B., Walraven, E., Beekwilder, J., et al. (2012). Genetic analysis of metabolites in apple fruits indicates an mQTL hotspot for phenolic compounds on linkage group 16. J. Exp. Bot. 63, 2895–2908. doi: 10.1093/jxb/err464
Khanal, B., Grimm, E., and Knoche, M. (2013). Russeting in apple and pear: a plastic periderm replaces a stiff cuticle. AoB Plants 5:pls048. doi: 10.1093/aobpla/pls048
King, G., Lynn, J., Dover, C., Evans, K., and Seymour, G. (2001). Resolution of quantitative trait loci for mechanical measures accounting for genetic variation in fruit texture of apple (Malus pumila Mill.). Theor. Appl. Genet. 102, 1227–1235. doi: 10.1007/s001220000530
Kou, X., Zhou, J., Wu, C. E., Yang, S., Liu, Y., Chai, L., et al. (2021). The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening: a review. Plant Mol. Biol. 106, 223–238. doi: 10.1007/s11103-021-01128-w
Kumar, S., Chagné, D., Bink, M., Volz, R., Whitworth, C., and Carlisle, C. (2012). Genomic selection for fruit quality traits in apple (Malus×domestica Borkh.). PLoS One 7:e36674. doi: 10.1371/journal.pone.0036674
Kumar, S., Molloy, C., Muñoz, P., Daetwyler, H., Chagné, D., and Volz, R. (2015). Genome-enabled estimates of additive and nonadditive genetic variances and prediction of apple phenotypes across environments. G3 5, 2711–2718. doi: 10.1534/g3.115.021105
Kunihisa, M., Moriya, S., Abe, K., Okada, K., Haji, T., Hayashi, T., et al. (2014). Identification of QTLs for fruit quality traits in Japanese apples: QTLs for early ripening are tightly related to preharvest fruit drop. Breed. Sci. 64, 240–251. doi: 10.1270/jsbbs.64.240
Larsen, B., Migicovsky, Z., Jeppesen, A. A., Gardner, K. M., Toldam-Andersen, T. B., Myles, S., et al. (2019). Genome-wide association studies in apple reveal loci for aroma volatiles, sugar composition, and harvest date. Plant Genome 12:180104. doi: 10.3835/plantgenome2018.12.0104
Legay, S., Guerriero, G., André, C., Guignard, C., Cocco, E., Charton, S., et al. (2016). MdMYB93 is a regulator of suberin deposition in russeted apple fruit skins. New Phytol. 212, 977–991. doi: 10.1111/nph.14170
Li, D., Xu, Y., Xu, G., Gu, L., Li, D., and Shu, H. (2006). Molecular cloning and expression of a gene encoding alcohol acyltransferase (MdAAT2) from apple (cv. Golden Delicious). Phytochemistry 67, 658–667. doi: 10.1016/j.phytochem.2006.01.027
Li, X., Kui, L., Zhang, J., Xie, Y., Wang, L., Yan, Y., et al. (2016). Improved hybrid de novo genome assembly of domesticated apple (Malus x domestica). GigaScience 5:35. doi: 10.1186/s13742-016-0139-0
Li, T., Xu, Y., Zhang, L., Ji, Y., Tan, D., Yuan, H., et al. (2017). The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29, 1316–1334. doi: 10.1105/tpc.17.00349
Li, W. F., Ning, G. X., Mao, J., Guo, Z. G., Zhou, Q., and Chen, B. H. (2019). Whole-genome DNA methylation patterns and complex associations with gene expression associated with anthocyanin biosynthesis in apple fruit skin. Planta 250, 1833–1847. doi: 10.1007/s00425-019-03266-4
Li, C., Dougherty, L., Coluccio, A., Meng, D., El-Sharkawy, I., Borejsza-Wysocka, E., et al. (2020). Apple ALMT9 requires a conserved C-terminal domain for malate transport underlying fruit acidity. Plant Physiol. 182, 992–1006. doi: 10.1104/pp.19.01300
Liao, L., Zhang, W., Zhang, B., Fang, T., Wang, X., Cai, Y., et al. (2021). Unraveling a genetic roadmap for improved taste in the domesticated apple. Mol. Plant 14, 1454–1471. doi: 10.1016/j.molp.2021.05.018
Liebhard, R., Kellerhals, M., Pfammatter, W., Jertmini, M., and Gessler, C. (2003). Mapping quantitative physiological traits in apple (Malus × domestica Borkh.). Plant Mol. Biol. 52, 511–526. doi: 10.1023/A:1024886500979
Liu, Z., Bao, D., Liu, D., Zhang, Y., Ashraf, M. A., and Chen, X. (2016). Construction of a genetic linkage map and QTL analysis of fruit-related traits in an F1 Red Fuji x Hongrou apple hybrid. Open Life Sci. 11, 487–497. doi: 10.1515/biol-2016-0063
Lobato-Gómez, M., Hewitt, S., Capell, T., Christou, P., Dhingra, A., and Girón-Calva, P. S. (2021). Transgenic and genome-edited fruits: background, constraints, benefits, and commercial opportunities. Hortic. Res. 8:166. doi: 10.1038/s41438-021-00601-3
Longhi, S., Hamblin, M., Trainotti, L., Peace, C., Velasco, R., and Costa, F. (2013). A candidate gene based approach validates Md-PG1 as the main responsible for a QTL impacting fruit texture in apple (Malus x domestica Borkh). BMC Plant Biol. 13:37. doi: 10.1186/1471-2229-13-37
Ma, B., Liao, L., Zheng, H., Chen, J., Wu, B., Ogutu, C., et al. (2015). Genes encoding aluminum-activated malate transporter II and their association with fruit acidity in apple. Plant Genome 8:plantgenome2015.03.0016. doi: 10.3835/plantgenome2015.03.0016
Ma, B., Zhao, S., Wu, B., Wang, D., Peng, Q., Owiti, A., et al. (2016). Construction of a high density linkage map and its application in the identification of QTLs for soluble sugar and organic acid components in apple. Tree Genet. Genomes 12:1. doi: 10.1007/s11295-015-0959-6
Ma, Q., Sun, M., Lu, J., Liu, Y., Hu, D., and Hao, Y. (2017). Transcription factor AREB2 is involved in soluble sugar accumulation by activating sugar transporter and amylase genes. Plant Physiol. 174, 2348–2362. doi: 10.1104/pp.17.00502
Ma, S., Li, H., Wang, L., Li, B., Wang, Z., Ma, B., et al. (2022). F-box protein MdAMR1L1 regulates ascorbate biosynthesis in apple by modulating GDP-mannose pyrophosphorylase. Plant Physiol. 188, 653–669. doi: 10.1093/plphys/kiab427
Maliepaard, C., Alston, F., Van-Arkel, G., Brown, L., Chevreau, E., Dunemann, F., et al. (1998). Aligning male and female linkage maps of apple (Malus pumila Mill.) using multi-allelic markers. Theor. Appl. Genet. 97, 60–73. doi: 10.1007/s001220050867
Martín-Pizarro, C., Vallarino, J. G., Osorio, S., Meco, V., Urrutia, M., Pillet, J., et al. (2021). The NAC transcription factor FaRIF controls fruit ripening in strawberry. Plant Cell 33, 1574–1593. doi: 10.1093/plcell/koab070
McClure, K., Gong, Y., Song, J., Vinqvist-Tymchuk, M., Palmer, L., Fan, L., et al. (2019). Genome-wide association studies in apple reveal loci of large effect controlling apple polyphenols. Hortic. Res. 6:107. doi: 10.1038/s41438-019-0190-y
Mellidou, I., Chagné, D., Laing, W. A., Keulemans, J., and Davey, M. W. (2012). Allelic variation in paralogs of gdp-l-galactose phosphorylase is a major determinant of vitamin c concentrations in apple fruit. Plant Physiol. 160, 1613–1629. doi: 10.1104/pp.112.203786
Mendel, G. (1965). Experiments in Plant Hybridisation, Vol. 2. Cambridge, MA: Harvard University Press, 694. doi: 10.1136/bmj.2.5463.694-a
Migicovsky, Z., Gardner, K., Money, D., Sawler, J., Bloom, J., Moffett, P., et al. (2016). Genome to phenome mapping in apple using historical data. Plant Genome 9:plantgenome2015.11.0113. doi: 10.3835/plantgenome2015.11.0113
Naveed, M., Hejazi, V., Abbas, M., Kamboh, A., Khan, G., Shumzaid, M., et al. (2018). Chlorogenic acid (CGA): a pharmacological review and call for further research. Biomed. Pharmacother. 97, 67–74. doi: 10.1016/j.biopha.2017.10.064
Nybom, H., Ahmadi-Afzadi, M., Rumpunen, K., and Tahir, I. (2020). Review of the impact of apple fruit ripening, texture and chemical contents on genetically determined susceptibility to storage rots. Plants 9:831. doi: 10.3390/plants9070831
Oraguzie, N., Iwanami, H., Soejima, J., Harada, T., and Hall, A. (2004). Inheritance of the Md-ACS1 gene and its relationship to fruit softening in apple (Malus × domestica Borkh.). Theor. Appl. Genet. 108, 1526–1533. doi: 10.1007/s00122-003-1574-8
Oyenihi, A. B., Belay, Z. A., Mditshwa, A., and Caleb, O. J. (2022). “An apple a day keeps the doctor away”: the potentials of apple bioactive constituents for chronic disease prevention. J. Food Sci. 87, 2291–2309. doi: 10.1111/1750-3841.16155
Peng, Q., Cai, Y., Lai, E., Nakamura, M., Liao, L., Zheng, B., et al. (2020). The sucrose transporter MdSUT4.1 participates in the regulation of fruit sugar accumulation in apple. BMC Plant Biol. 20:191. doi: 10.1186/s12870-020-02406-3
Potts, S., Khan, M., Han, Y., Kushad, M., and Korban, S. (2014). Identification of quantitative trait loci (QTLs) for fruit quality traits in apple. Plant Mol. Biol. Rep. 32, 109–116. doi: 10.1007/s11105-013-0634-8
Sánchez-Sevilla, J. F., Cruz-Rus, E., Valpuesta, V., Botella, M. A., and Amaya, I. (2014). Deciphering gamma-decalactone biosynthesis in strawberry fruit using a combination of genetic mapping, RNA-Seq and eQTL analyses. BMC Genomics 15:218. doi: 10.1186/1471-2164-15-218
Scherer, R., Rybka, A., Ballus, C., Meinhart, A., Filho, J., and Godoy, H. (2012). Validation of a HPLC method for simultaneous determination of main organic acids in fruits and juices. Food Chem. 135, 150–154. doi: 10.1016/j.foodchem.2012.03.111
Schiller, D., Contreras, C., Vogt, J., Dunemann, F., Defilippi, B., Beaudry, R., et al. (2015). A dual positional specific lipoxygenase functions in the generation of flavor compounds during climacteric ripening of apple. Hortic. Res. 2:15003. doi: 10.1038/hortres.2015.3
Sekido, K., Hayashi, Y., Yamada, K., Shiratake, K., Matsumoto, S., Maejima, T., et al. (2010). Efficient breeding system for red-fleshed apple based on linkage with s3-RNase allele in ‘Pink Pearl’. Hortscience 45, 534–537. doi: 10.1007/BF01645299
Soglio, V., Costa, F., Molthoff, J., Weemen-Hendriks, W., Schouten, H., and Gianfranceschi, L. (2009). Transcription analysis of apple fruit development using cDNA microarrays. Tree Genet. Genomes 5, 685–698. doi: 10.1007/s11295-009-0219-8
Sun, H., Zhao, Y., Li, C., Chen, D., Wang, Y., Zhang, X., et al. (2012). Identification of markers linked to major gene loci involved in determination of fruit shape index of apples (Malus domestica). Euphytica 185, 185–193. doi: 10.1007/s10681-011-0515-x
Sun, R., Chang, Y., Yang, F., Wang, Y., Li, H., Zhao, Y., et al. (2015). A dense SNP genetic map constructed using restriction site-associated DNA sequencing enables detection of QTLs controlling apple fruit quality. BMC Genomics 16:747. doi: 10.1186/s12864-015-1946-x
Sun, X., Jiao, C., Schwaninger, H., Chao, C., Ma, Y., Duan, N., et al. (2020). Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 52, 1423–1432. doi: 10.1038/s41588-020-00723-9
Sunako, T., Sakuraba, W., Senda, M., Akada, S., Ishikawa, R., Niizeki, M., et al. (1999). An allele of the ripening-specific 1-aminocyclopropane-1-carboxylic acid synthase gene (ACS1) in apple fruit with a long storage life. Plant Physiol. 119, 1297–1304. doi: 10.1104/pp.119.4.1297
Takos, A., Jaffé, F., Jacob, S., Bogs, J., Robinson, S., and Walker, A. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi: 10.1104/pp.106.088104
Tan, D., Li, T., and Wang, A. (2013). Apple 1-aminocyclopropane-1-carboxylic acid synthase genes, MdACS1 and MdACS3a, are expressed in different systems of ethylene biosynthesis. Plant Mol. Biol. Rep. 31, 204–209. doi: 10.1007/s11105-012-0490-y
Telias, A., Lin-Wang, K., Stevenson, D., Cooney, J., Hellens, R., Allan, A., et al. (2011). Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 11:93. doi: 10.1186/1471-2229-11-93
Urrestarazu, J., Muranty, H., Denancé, C., Leforestier, D., Ravon, E., Guyader, A., et al. (2017). Genome-wide association mapping of flowering and ripening periods in apple. Front. Plant Sci. 8:1923. doi: 10.3389/fpls.2017.01923
Varanasi, V., Shin, S., Mattheis, J., Rudell, D., and Zhu, Y. (2011). Expression profiles of the MdACS3 gene suggest a function as an accelerator of apple (Malus× domestica) fruit ripening. Postharvest Biol. Technol. 62, 141–148. doi: 10.1016/j.postharvbio.2011.05.005
Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A., Kalyanaraman, A., et al. (2010). The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 42, 833–839. doi: 10.1038/ng.654
Verma, S., Evans, K., Guan, Y., Luby, J. J., Rosyara, U. R., Howard, N. P., et al. (2019). Two large-effect QTLs, Ma and Ma3, determine genetic potential for acidity in apple fruit: breeding insights from a multi-family study. Tree Genet. Genomes 15:18. doi: 10.1007/s11295-019-1324-y
Volz, R., Oraguzie, N., Whitworth, C., How, N., Chagné, D., Carlisle, C., et al. (2009). Breeding for red flesh colour in apple: progress and challenges. Acta Hortic. 814, 337–342. doi: 10.17660/ActaHortic.4.54
Wang, N., Xu, H., Jiang, S., Zhang, Z., Lu, N., Qiu, H., et al. (2017). MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90, 276–292. doi: 10.1111/tpj.13487
Wang, N., Jiang, S., Zhang, Z., Fang, H., Xu, H., Wang, Y., et al. (2018a). Malus sieversii: the origin, flavonoid synthesis mechanism, and breeding of red-skinned and red-fleshed apples. Hortic. Res. 5:70. doi: 10.1038/s41438-018-0084-4
Wang, N., Liu, W., Zhang, T., Jiang, S., Xu, H., Wang, Y., et al. (2018b). Transcriptomic analysis of red-fleshed apples reveals the novel role of MdWRKY11 in flavonoid and anthocyanin biosynthesis. J. Agric. Food Chem. 66, 7076–7086. doi: 10.1021/acs.jafc.8b01273
Wang, X., Zeng, W., Ding, Y., Wang, Y., Niu, L., Yao, J. L., et al. (2019). PpERF3 positively regulates ABA biosynthesis by activating PpNCED2/3 transcription during fruit ripening in peach. Hortic. Res 6:19. doi: 10.1038/s41438-018-0094-2
Wang, N., Liu, W., Yu, L., Guo, Z., Chen, Z., Jiang, S., et al. (2020). HEAT SHOCK FACTOR A8a modulates flavonoid synthesis and drought tolerance. Plant Physiol. 184, 1273–1290. doi: 10.1104/pp.20.01106
Wang, W. Q., Moss, S. M., Zeng, L., Espley, R. V., Wang, T., Lin-Wang, K., et al. (2022a). The red flesh of kiwifruit is differentially controlled by specific activation–repression systems. New Phytol. 235, 630–645. doi: 10.1111/nph.18122
Wang, Z., Ma, B., Yang, N., Jin, L., Wang, L., Ma, S., et al. (2022b). Variation in the promoter of the sorbitol dehydrogenase gene MdSDH2 affects binding of the transcription factor MdABI3 and alters fructose content in apple fruit. Plant J. 109, 1183–1198. doi: 10.1111/tpj.15624
Westwood, M., and Blaney, L. (1963). Non-climatic factors affecting the shape of apple fruits. Nature 200, 802–803. doi: 10.1038/200802a0
Wu, B., Shen, F., Wang, X., Zheng, W., Xiao, C., Deng, Y., et al. (2021). Role of MdERF3 and MdERF118 natural variations in apple flesh firmness/crispness retainability and development of QTL-based genomics-assisted prediction. Plant Biotechnol. J. 19, 1022–1037. doi: 10.1111/pbi.13527
Xu, K., Wang, A., and Brown, S. (2012). Genetic characterization of the Ma locus with pH and titratable acidity in apple. Mol. Breed. 30, 899–912. doi: 10.1007/s11032-011-9674-7
Xu, H., Zou, Q., Yang, G., Jiang, S., Fang, H., Wang, Y., et al. (2020). MdMYB6 regulates anthocyanin formation in apple both through direct inhibition of the biosynthesis pathway and through substrate removal. Hortic. Res. 7:72. doi: 10.1038/s41438-020-0294-4
Yamaki, S. (1984). Isolation of vacuoles from immature apple fruit flesh and copartmentation of sugars, organic acids, phenolic compounds and amino acids. Plant Cell Physiol. 25, 151–166. doi: 10.1093/oxfordjournals.pcp.a076688
Yao, J., Xu, J., Cornille, A., Tomes, S., Karunairetnam, S., Luo, Z., et al. (2015). A micro RNA allele that emerged prior to apple domestication may underlie fruit size evolution. Plant J. 84, 417–427. doi: 10.1111/tpj.13021
Yuan, G., Bian, S., Han, X., He, S., Liu, K., Zhang, C., et al. (2019). An integrated transcriptome and proteome analysis reveals new insights into russeting of bagging and non-bagging “Golden Delicious” apple. Int. J. Mol. Sci. 20:4462. doi: 10.3390/ijms20184462
Yue, P., Lu, Q., Liu, Z., Lv, T., Li, X., Bu, H., et al. (2020). Auxin-activated MdARF5 induces the expression of ethylene biosynthetic genes to initiate apple fruit ripening. New Phytol. 226, 1781–1795. doi: 10.1111/nph.16500
Zhang, J., Xu, H., Wang, N., Jiang, S., Fang, H., Zhang, Z., et al. (2018). The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple. Plant Mol. Biol. 98, 205–218. doi: 10.1007/s11103-018-0770-5
Zhang, L., Hu, J., Han, X., Li, J., Gao, Y., and Richards, C. (2019). A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 10:1494. doi: 10.1038/s41467-019-09518-x
Zhang, L., Ma, B., Wang, C., Chen, X., Ruan, Y. L., Yuan, Y., et al. (2022). MdWRKY126 modulates malate accumulation in apple fruit by regulating cytosolic malate dehydrogenase (MdMDH5). Plant Physiol. 188, 2059–2072. doi: 10.1093/plphys/kiac023
Zhen, Q., Fang, T., Peng, Q., Liao, L., Zhao, L., Owiti, A., et al. (2018). Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hortic. Res. 5:14. doi: 10.1038/s41438-018-0024-3
Zheng, W., Shen, F., Wang, W., Wu, B., Wang, X., Xiao, C., et al. (2020). Quantitative trait loci-based genomics-assisted prediction for the degree of apple fruit cover color. Plant Genome 13:e20047. doi: 10.1002/tpg2.20047
Keywords: apple, genetic characteristics, quality traits, QTLs, genes
Citation: Liu W, Chen Z, Jiang S, Wang Y, Fang H, Zhang Z, Chen X and Wang N (2022) Research Progress on Genetic Basis of Fruit Quality Traits in Apple (Malus × domestica). Front. Plant Sci. 13:918202. doi: 10.3389/fpls.2022.918202
Received: 12 April 2022; Accepted: 23 June 2022;
Published: 14 July 2022.
Edited by:
Rebecca Grumet, Michigan State University, United StatesReviewed by:
Sujeet Verma, University of Florida, United StatesCopyright © 2022 Liu, Chen, Jiang, Wang, Fang, Zhang, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuesen Chen, Y2hlbnhzQHNkYXUuZWR1LmNu; Nan Wang, bmFud2FuZ2ppbmd6aUAxNjMuY29t
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.