- 1College of Horticulture, Vegetable Genetics and Breeding Laboratory, Anhui Agricultural University, Hefei, China
- 2Guangdong Technology Research Center for Marine Algal Bioengineering, Guangdong Key Laboratory of Plant Epigenetics, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, China
- 3School of Agronomy, Anhui Agricultural University, Hefei, China
- 4Department of Plant Pathology, College of Plant Protection, Anhui Agricultural University, Hefei, China
Heat shock protein 90 (Hsp90) plays an important role in plant developmental regulation and defensive reactions. Several plant species have been examined for the Hsp90 family gene. However, the Hsp90 gene family in cabbage has not been well investigated to date. In this study, we have been discovered 12 BoHsp90 genes in cabbage (Brassica oleracea var. capitata L.). These B. oleracea Hsp90 genes were classified into five groups based on phylogenetic analysis. Among the five groups, group one contains five Hsp90 genes, including BoHsp90-1, BoHsp90-2, BoHsp90-6, BoHsp90-10, and BoHsp90-12. Group two contains three Hsp90 genes, including BoHsp90-3, BoHsp90-4, and BoHsp90. Group three only includes one Hsp90 gene, including BoHsp90-9. Group four were consisting of three Hsp90 genes including BoHsp90-5, BoHsp90-7, and BoHsp90-8, and there is no Hsp90 gene from B. oleracea in the fifth group. Synteny analysis showed that a total of 12 BoHsp90 genes have a collinearity relationship with 5 Arabidopsis genes and 10 Brassica rapa genes. The promoter evaluation revealed that the promoters of B. oleracea Hsp90 genes included environmental stress-related and hormone-responsive cis-elements. RNA-seq data analysis indicates that tissue-specific expression of BoHsp90-9 and BoHsp90-5 were highly expressed in stems, leaves, silique, and flowers. Furthermore, the expression pattern of B. oleracea BoHsp90 exhibited that BoHsp90-2, BoHsp90-3, BoHsp90-7, BoHsp90-9, BoHsp90-10, and BoHsp90-11 were induced under cold stress, which indicates these Hsp90 genes perform a vital role in cold acclimation and supports in the continual of normal growth and development process. The cabbage Hsp90 gene family was found to be differentially expressed in response to cold stress, suggesting that these genes play an important role in cabbage growth and development under cold conditions.
Introduction
Crop plants during their growth cycle often confronted with various biotic and abiotic stresses (Food and Agriculture Organization [FAO], 2020). Recent climatic changes continuously trigger extreme temperature events and disrupt optimal plant growth (IPCC, 2021; Masson-Delmotte et al., 2021). Increased global warming instigated the risk of cold damage to crop plants (Gu et al., 2008; Moreno and Orellana, 2011; Hassan et al., 2021). Plants contain regulatory mechanisms that enable them to withstand harsh environmental conditions, called acclimatization (Liu et al., 2019). To acclimatize the extreme temperature (low and high) conditions, plants exhibit a number of protein expressions either to promote biosynthesis of new proteins or to protect existing proteins; cold acclimation process carried through expressions of highly conserved stress proteins called heat shock proteins (Hsps; Queitsch et al., 2002). Various types of Hsps are particularly found in all plant species (Xiong and Ishitani, 2006). Based on molecular mass, Hsps were categorized into five groups, i.e., the sHsp family, the chaperonin (Hsp60/GroEL) family, the 70-kDa heat shock protein (Hsp70/DnaK) family, the Hsp90 family, and the Hsp100/ClpB family (Pratt and Toft, 2003; Wang et al., 2004; Al-Whaibi, 2011).
The Hsp90 protein family is a renowned and extremely conserved class of molecular chaperones in eukaryotic cytoplasm (Chen et al., 2006).” For instance, in Arabidopsis, there are 7 Hsp90 proteins, of which AtHsp90-1, AtHsp90-2, AtHsp90-3 and AtHsp90-4 are present in the cytoplasm, while AtHsp90-5, AtHsp90-6 and AtHsp90-7 are present in the mitochondria, chloroplast, and endoplasmic reticulum, respectively (Krishna and Gloor, 2001; Yamada et al., 2007). It is a part of the ATPase superfamily (GHKL: gyrase, histidine kinase, and MutL; Dutta and Inouye, 2000); It functions as an ATP-regulated dimer and chaperone protein with three highly conserved domains: a 25 kDa C-terminal substrate-binding domain, a 12 kDa intermediate ATP-binding domain, and a 12 kDa N-terminal ATP-binding domain (Prodromou et al., 1997; Pearl and Prodromou, 2000).” The protein channel that binds to ATP in Hsp90 is frequently closed (Sung et al., 2016) which results in the poor activity of N-terminal ATPase (Raman and Suguna, 2015). The cytoplasm of eukaryotic cells contains the intermediate and N-terminal domains of Hsp90; the charged portions are varied in length between species (Pearl and Prodromou, 2000). It is commonly known that a protein’s function is determined by its ability to fold into a three-dimensional structure. The Hsp90 proteins together with other molecular chaperones provide a course that facilitates protein folding (Wang et al., 2004). The Hsp90 gene family suppress protein aggregation and enable the ubiquitination of inactive proteins (Picard, 2002); further, it belongs to a group of molecular chaperones that are involved in the creation of the spatial structure of kinase substrates, DNA repair and substrate activation, early stress signaling, and transcription factor spatial structure maintenance (Jackson et al., 2004; Lachowiec et al., 2015).” The expression of stressor Hsp90 gene is upregulated when plants are stressed; it interacts and repairs the deformed proteins with support of non-proteinaceous compounds (Mukhopadhyay et al., 2003).
This protein (Hsp90) has been found in a number of plant species such as 7 in Arabidopsis (Krishna and Gloor, 2001), 7 in Solanum lycopersicum and 7 in Asteridae and tobacco (Liu et al., 2014; Song et al., 2019) and pepper (Jing et al., 2020). Nine and ten Hsp90 genes were identified in rice (Hu et al., 2009) and Populus (Zhang et al., 2013), respectively. Recent studies have shown that Hsp90 has a vital role in plant’ acclimation to various biotic and abiotic stresses (Di Donato and Geisler, 2019). The HSP gene family were considered to be involved or induced in cold environments (Krishna et al., 1995); it has the potential to protect plant biological systems from cold damage by preventing freezing-induced protein denaturation (Yan et al., 2006). Many studies reported that the low-temperature stress activates the gene expressions of both chaperonins and Hsps, particularly Hsp90, Hsp70, and the smHsps (Wang et al., 2004). In Brassica napus, the expression of Hsp90 was upregulated under cold stress (Krishna et al., 1995; Gill et al., 2015). Likewise, rice also exhibited the up-regulated expression of OsHsp90-2 and OsHsp90-4 under drought, cold, and heat stress (Hu et al., 2009). Liu et al. (2022) reported that in rubber trees (Hevea brasiliensis Müll. Arg.) the expression of the HbHsp90.1 gene was significantly up-regulated at 4°C for 6 h of cold stress treatment. Similarly, Song et al. (2019) observed an identical trend in the tobacco plant, where NtHsp90-4, NtHsp90-5 and NtHsp90-9 showed high levels of transcription at 4°C for 6 to 12 h duration under cold stress. Increased Hsp90 protein accumulation was observed in winter wheat under cold stress (Vítámvás et al., 2012).
Brassica oleracea var. capitata (Cabbage) is a popular vegetable crop cultivated all over the world and contains a variety of nutrients and health-promoting compounds (Lv et al., 2020). Temperature required for optimal cabbage growth is 17°C, while its normal growth and development decreased when temperature falls below 10°C; further it can withstand up to –5°C for short phase of cold exposure, prolonged exposure leads to cell death (Food and Agricultural Organization of the United Nations (FAO), 2007). This investigation executed a genome-wide analysis of the Brassica Genome Network database using Hsp90 protein sequences of Arabidopsis thaliana. We analyzed the gene structure, phylogenetic tree, chromosomal location, homologous gene pairs, conserved motifs, collinearity analysis, sectioning pressure and cis-elements of the cabbage Hsp90 gene promoter by utilizing bioinformatics approaches. Moreover, this study explored the relevant roles of cabbage Hsp90 gene, provides a vibrant approach for molecular breeding and develop a better understanding for its tolerance responses under cold conditions.
Materials and Methods
Hsp90 Gene Family Identification in Brassica oleracea
To extract the Hsp90 gene from the B. oleracea genome, “the Arabidopsis Hsp90 gene was used as a query in the online database Phytozome 11.0 (Goodstein et al., 2012).1 The genome, CDS, protein and 1.5 kb upstream promoter region of the Hsp90 gene were extracted and used as a query against the NCBI database2 to identify conserved domains and discard Hsp90 domains missing genes. To ensure the presence of Hsp90 specific domains, all non-redundant protein sequences were checked using manual and Pfam databases, and 12 BoHsp90 genes were finally found.
Physical Properties and Position on Chromosomes
To predict the protein physical properties of B. oleracea Hsp90 (BoHsp90), “the ExPASy tool is utilized.3 The phytozome was used to obtain information about gene start-end position, the number of amino acids, and the chromosome location of the BoHsp90 family. Plot phenograms4 indicate the location of each BoHsp90 gene on its chromosome. Using standard parameters in the ExPASy program, the protein sequence of the BoHsp90 gene was determined5“(Gasteiger et al., 2003).”
Structural Analysis and Phylogenetic Relationship and Motif Analysis
Using the CDS and genome sequences of the related BoHsp90 genes, construct gene structures in the Gene Structure Display Server to find exons-introns within genes (Hu et al., 2015)6. Multiple alignments of Hsp90 protein sequences of B. oleracea, Brassica rapa, B. napus, cucumber, tomato, potato, soybean, Vitis vinifera, Zea mays, Arabidopsis, rice, tobacco, cotton, Medicago truncatula, Elaeis guineensis, Nelumbo nucifera, and P. bretschneideri were performed by MEGA 7.0 with default parameters. Then, based on the alignment results, a phylogenetic tree was constructed using the maximum likelihood method with the following parameters: partial deletion, and 1,000 bootstrap tests. MEME is used to carry out the conserved sections of the BoHsp90 gene (Bailey et al., 2006)7 and the Pfam database was used to screen their genomic assemblies (Punta et al., 2012).8
Synteny Analysis of Hsp90 Genes in Brassica oleracea, Arabidopsis, and Brassica rapa
By using a software MCScanX, a syntenic study of the Hsp90 gene in B. oleracea, Arabidopsis, and B. rapa was performed to better understand the syntenic interactions between A. thaliana, B. oleracea, and B. rapa (Wang et al., 2012). It was used to check the Hsp90 genes synteny in, Arabidopsis, B. oleracea and B. rapa.
Cis-Regulatory Element Prediction for Brassica oleracea Hsp90 Gene Promoters
The promoter sequence (1.5 kb DNA sequence in the start codon stream) of each Hsp90 gene was derived from the Brassica genome. “Using the PlantCARE database, the cis-acting regulatory element then examines the promoter of each B. oleracea Hsp90 gene9 (Plantcare; Lescot et al., 2002). The anticipated cis-acting regulatory elements were categorized based on their regulatory functions.”
Gene Duplication, Ka/Ks Calculation and Positive Selection Analysis
Segmental and Tandem duplication occurred when two closely related BoHsp90 genes were discovered on the same or different chromosomes.“Additionally, the gene duplication events BoHsp90 genes and also synonymous substitution rate (Ka) and the non-synonymous substitution rate (Ks) were calculated using an online tool and computed their ratios.10 The divergence time (T) for B. oleracea was calculated by using T = ks/2r where R 1.5 × 10−8; Koch et al., 2000).” The ratio of Ka/Ks evaluates the selection pressure of BoHsp90 repeated genes. When the ratio of Ka/Ks is>1, <1, or = 1, it is considered a positive, negative, or neutral selection. “The amino acids in the BoHsp90 protein that are under selective pressure are then clarified using the Selection 2.2 tool.11 The maximum likelihood test from the Bayesian inference method is used to calculate the offset ratio between codons in an aligned sequence (Yang, 1997).” Different kinds the selection type appears in the selection result, and evaluate by scale color.
Transcriptomic Data Analysis
Gene expression patterns of cabbage in different tissues (roots, stems, flowers, leaves, scales, bulbs and callus) were retrieved from the NCBI database12 based on FPKM expression data with accession number GSE42891 (Yu et al., 2014). Finally, an expression heatmap was created using the TBtools software.13
Plant Materials and Stress Treatments
The cabbage (cultivar Yingchuan) seeds were planted in plug trays in moistened mixed soil (peat moss) were provided by School of Horticulture, Anhui Agricultural University, Hefei, China. Seedlings were germinated and cultured for 5 to 6 weeks in a glass chamber at 22°C on a 16/8 h. (light/dark) cycle. Then, they are treated at low temperatures (−2°C, 0°C, and 4°C) for 6, 12 and 24 h (Ahmed et al., 2015). Plants that had not been treated were cultivated normally. At 6, 12, and 24 h following treatment, all true leaves were sampled and quickly put into liquid nitrogen and stored at −80°C for RNA extraction and synthesis of cDNA.
RNA Isolation, cDNA Synthesis and qRT-PCR Analysis to Validate the BoHsp90 Genes Expression
The Novaprotein reagent or RNA isolation kit for RNA extraction from cabbage leaves and quantification using a NanoDrop spectrophotometer (Thermo Scientific). “The superscript III Reverse Transcriptase (Novaprotein) according to the manufacturer’s guidelines, and the cDNA was dilute to 100 ng/l with ddH2O for further analysis. Quantitative RT-PCR was performed with the CFX96 Real Time System (Bio-Rad) with 96 well plates were used, and each well contained a total reaction mixture (20 μl) consisting of 10 μl of Power SYBR Master mix, 1 μl primer mix each F/R primers +6 μl ddH2O) and 2 μl cDNA.” The transcript level was normalized with a housekeeping gene BolActin. The 3 biological replicates were used for each BoHsp90s. The primers sequence for each gene were designed by using Primer5 software as shown in Supplementary Table S2.
Results
Identification of the Hsp90 Gene Family in Brassica oleracea
Using the A. thaliana Hsp90 protein sequence (BAB09283.1) as a query sequence, by using the online database phytozome14 to find Hsp90 genes in the Brassica genome. “The blast outcomes of A. thaliana in Brassica gave 18 hits in phytozome.” The signature BoHsp90 domain for each protein was confirmed using SMART Pfam15 (Punta et al., 2012). Additionally, NCBI web-based conserved domain search tool16 (Marchler-Bauer et al., 2015) was used to verify the conserved domain, and the protein sequences lacking the BoHsp90 domain were removed. During an extensive search of the Hsp90 gene, 12 putative genes were found. The annotation of known Brassica BoHsp90 family members were carried out, based on their chromosomal wise arrangements (BoHsp901-12). The protein sequences of BoHsp90 genes extended from 44 (BoHsp90-12) to 1819 (BoHsp90-11) amino (aa) residue with molecular weight (MW) range of 46.93–201.60 kDa. All 12 Hsp90 members have an average length of 773.4 aa and an average MW of 87.92 KDa furthermore, 41.66% (5 out of 12 genes) of BoHsp90 genes had high pI values (more than 5), while 58.33% (7 out of 12 genes) had pI values below 5. The predicted subcellular localization includes nuclear, endoplasmic reticulum and cytoplasmic. These results revealed that the maximum number of genes are found in the cytoplasm and percentage found in the nuclear (Supplementary Table S1).
Phylogenetic Relationship of the BoHsp90 Genes in Brassica oleracea
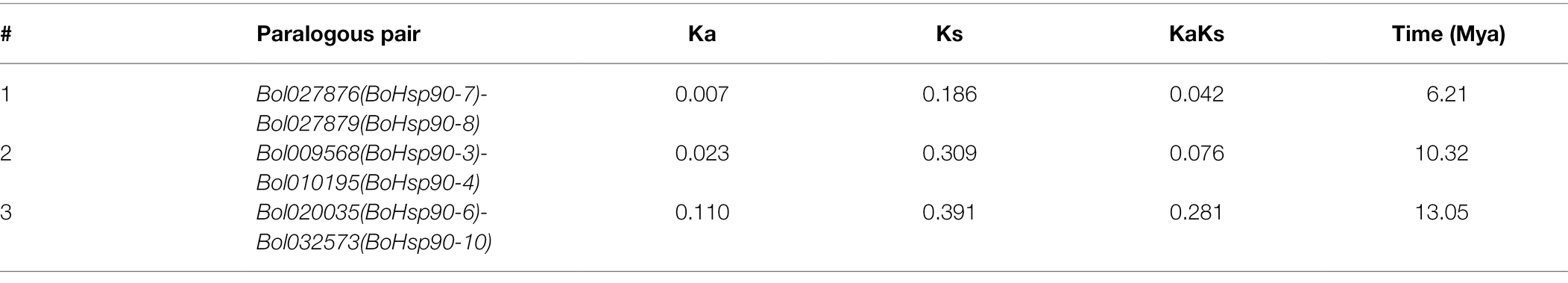
To study their evolutionary relationship, the predicted protein sequence of BoHsp90 was employed to create a rootless phylogenetic tree. The phylogenetic study of the Hsp90 genes distributed into five groups (G1, G2, G3, G4, and G5) containing B. oleracea, B. rapa, B. napus, tomato, cucumber, potato, V. vinifera, Sorghum bicolor, cotton, tobacco, Arabidopsis, rice, Z. mays, soybean, M. truncatula, E. guineensis, Chinese white pear (Pyrus bretschneideri) and N. nucifera. Among them G1 is the largest group with a total of 25 members, having B. oleracea (4 genes), B. napa (4 genes), B. rapa (3 genes), rice (3 genes), cucumbers (2 genes), potatoes (2 genes), A. thaliana (2 genes), 1 V. vinifera (1 gene), E. guineensis (1 gene), N. nucifera (1 gene) and one from sorghum. The second largest group, G4, comprises of 22 members, having B. oleracea (3 genes), B. rapa (3 genes), B. napus (1 gene), A. thaliana (3 genes), soybeans (2 genes), tomato (2 genes), rice (2 genes), Z. mays (2 genes), potato (1 gene), tobacco (1 gene), cotton (1 gene), and one P. bretschneideri. The G2 group contain 15 members, among which 3 in B. oleracea, 3 in B. rapa, one in A. thaliana, 2 in Z. mays, 2 in rice, 2 in tomato, one in cucumber, and one in soybean. While G3 comprised of 10 members as 2 in cotton, 2 in soybean, one in B. oleracea, B. rapa, A. thaliana, cucumber, tomato, and M. truncatula. G5 composed of 9 members and considered as a small group, having 3 in cotton, 2 in soybean, 2 in M. truncatula, one in cucumber and rice member (Figure 1).
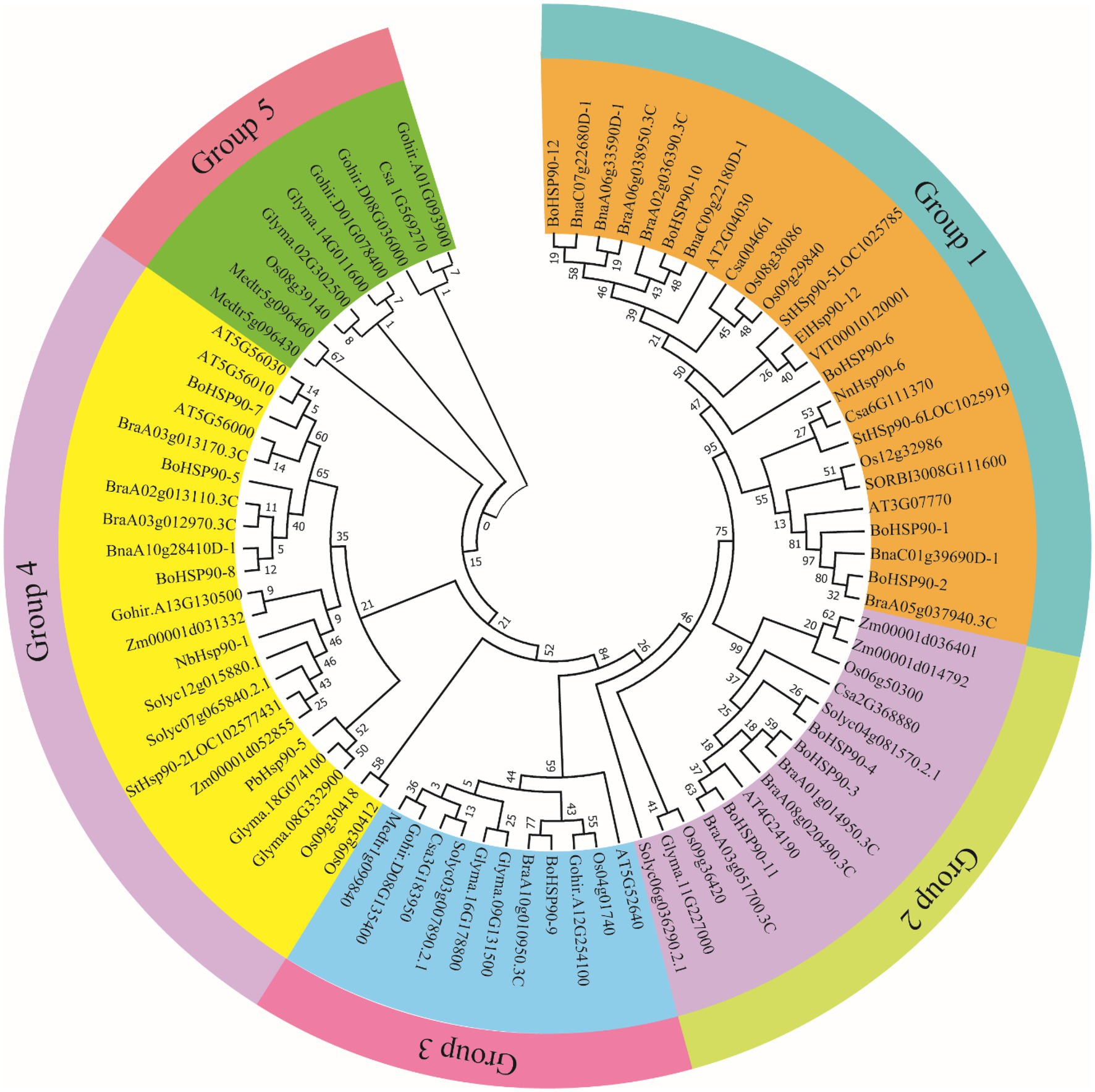
Figure 1. Phylogenetic tree of the Hsp90 proteins from Brassica oleracea, Brassica rapa, Brassica napus, tomato, cucumber, potato, Vitis vinifera, Sorghium bicolor, cotton, tobacco, Arabidopsis, rice, Zea mays, soybean, Medicago truncatula, Elaeis guineensis, Chinese white pear (Pyrus bretschneideri) and Nelumbo nucifera”
Structural Analysis of the BoHsp90 Genes
The arrangement of the BoHsp90 genes was plotted using the online tool GSDS based on their coding sequence. Within each BoHsp90 gene, “(Figure 2) displays the comparative lengths of introns and the conservation of corresponding exon sequences.” All these genes have between 1 and 19 introns. However, half of the BoHsp90 genes contain 1 to 14 introns, while another half of BoHsp90 genes contain different numbers of introns likewise BoHsp90-2 has 19 introns, BoHsp90–10 contains 17 introns, 12 in BoHsp90-11, 9 in BoHsp90-6, BoHsp90-12 has 8 introns and 3 introns in the BoHsp90–9. The position and length of introns also vary widely. In order to better understand the structural properties of BoHsp90 protein, 10 conserved motifs in the protein were identified using the online program MEME motif search tool and the distribution of these conserved motifs in BoHsp90 protein was explored.” The findings revealed that the majority of the genes comprised 10 conserved motifs. There are similar motifs in closely related genes, indicating similar functions in the BoHsp90 gene family. The BoHsp90-3 and BoHsp90-12 contain six motifs, BoHsp90-7 has eight and 9 motifs were found in BoHsp90-9. According to these findings, structural motif alignment varies between members of the BoHsp90 family genes but is similar within closely related genes (Supplementary Table S5).
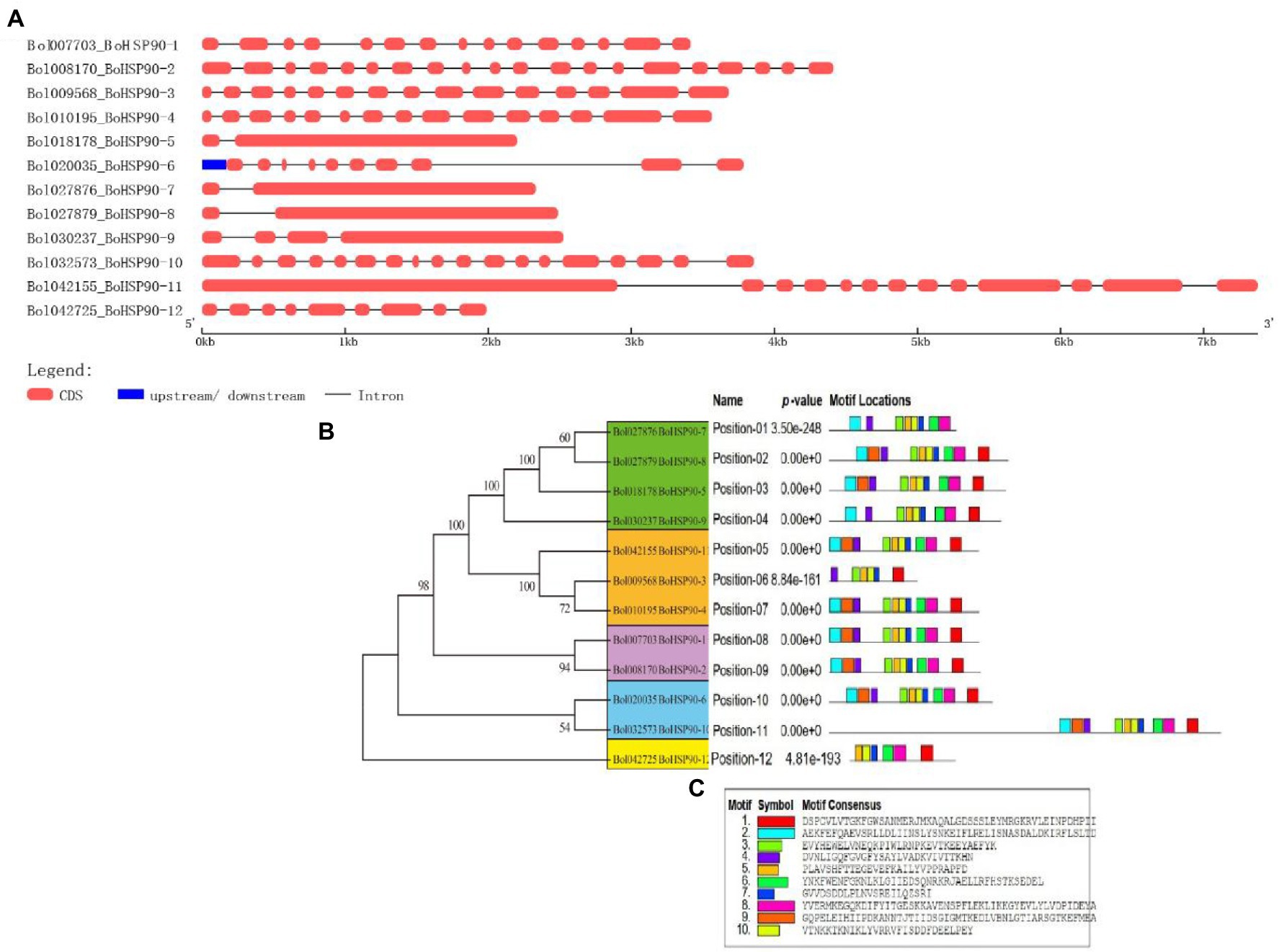
Figure 2. Gene structure and conserved motif analysis of BoHsp90 genes. (A) Gene structure of the BoHsp90 genes in cabbage. (B) Motif composition of BoHsp90. (C) A conserved motif in BoHsp90 gene was detected with MEME. Ten different motifs are represented by variously colored boxes.
Chromosomal Distribution, Collinearity Analysis of the Hsp90 Gene Family in Brassica oleracea
The chromosomal location map shown in (Figure 3) was created to specify the chromosomal orientation and deletion site of the BoHsp90 gene. The findings revealed that a total of 12 genes were linked to four chromosomes in cabbage. Of these, 83% of the BoHsp90 gene (10 loci) are present on three chromosomes in cabbage. Chromosomes 3, 2, and the more repressed Hsp90 members each have three and four genes, while chromosome 4 contains two genes. Furthermore, we also explored the collinearity relationships between the Cabbage BoHsp90 genes and related genes from 2 representative species including A. thaliana and B. rapa. A total of 12 cabbage Hsp90 genes have collinearity relationships with 5 A. thaliana genes. 10 B. rapa genes.
Figure 3. Chromosomal location and duplication of Cabbage (B. oleracea) Hsp90 genes. (A) Red boxes indicate tandem duplications, and black lines indicate segmental duplications. (B) Collinearity relationship or Synteny analysis of Hsp90 genes in B. oleracea, B. rapa, and Arabidopsis.
Duplication, Ka/Ks and Evolutionary Analysis of the BoHsp90 Genes in Brassica oleracea
The Ka/Ks ratio can be used to practice the selection process history of coding sequences “(Li et al., 1981). To explore the bias of duplicated BoHsp90 members, the Ka, Ks and Ka/Ks ratios of Hsp90 paralogous members were determined.” The Ks values were used to estimate each gene’s fraction. “All of the paralogous Hsp90 pairs had a Ks value ranging from 0.1 to 0.3, which was made up of cabbage duplication events. In addition, the third pair of BoHspP90 genes obtained duplications from 0.28 to 13.05 Mya (Table 1). The Ka/Ks ratio can also be used to determine selection pressure. The value of Ka/Ks1, greater than or equal to one, shows that the paralogous pair was subjected to, positive, purifying and neutral selection, or rare selection, respectively (Juretic et al., 2005).” Consequently, all the values were smaller than one, the representative that the BoHsp90 genes changed largely under the influence of purifying selection (Table 1). “Consistent evolution of amino acid positions is imperative for conservation of function and protein structure.” Certain amino acid identifications may be useful in elucidating selection pressures and assessing the relevance of specific amino acid interactions in Hsp90 protein interactions. The MEC model’s positive selection test for Hsp90 protein estimates the selection pressure of a given coding region. Our research shows that 38.76% (271 out of 699) amino acids are purifying selected, while the rest are affected by the neutral process (Figure 4). The results clarify the evolution all gene pairs are affected by purification selection.

Figure 4. Selection pressures among Hsp90 gene sequences using mechanistic-empirical combination (MEC) model of selection online tool. Gray and white highlights represent neutral selection, and purple highlight represents purifying or negative selection on codons.”
Analysis of the Promoter Region of Brassica oleracea Hsp90 genes
Each BoHsp90 gene was isolated from the Brassica plant genome, and the cis-regulatory region (1,500 bp upstream) was assessed using PlantCARE to better understand its possible functions and regulatory processes. The 12 BoHsp90 promoter region of contain 50% of light-responsive elements, responsive to hormones 29%, environmental responsive elements 7%, development-related elements contain 11% and site binding elements 2% (Figure 5A). “There are various transcription factor binding sites that govern hormones such as gibberellic acid (GA), abscisic acid (ABA), jasmonic acid, auxin, silicic acid, and ethylene. In addition to the previously mentioned regions (Figure 5B) or by stress response (defense, heat, low-temperature stress, etc). The BoHsp90-3 promoter region contains a maximum of 11 ABRE-related elements and 3 for jasmonic acid-responsive elements (Supplementary Table S3).
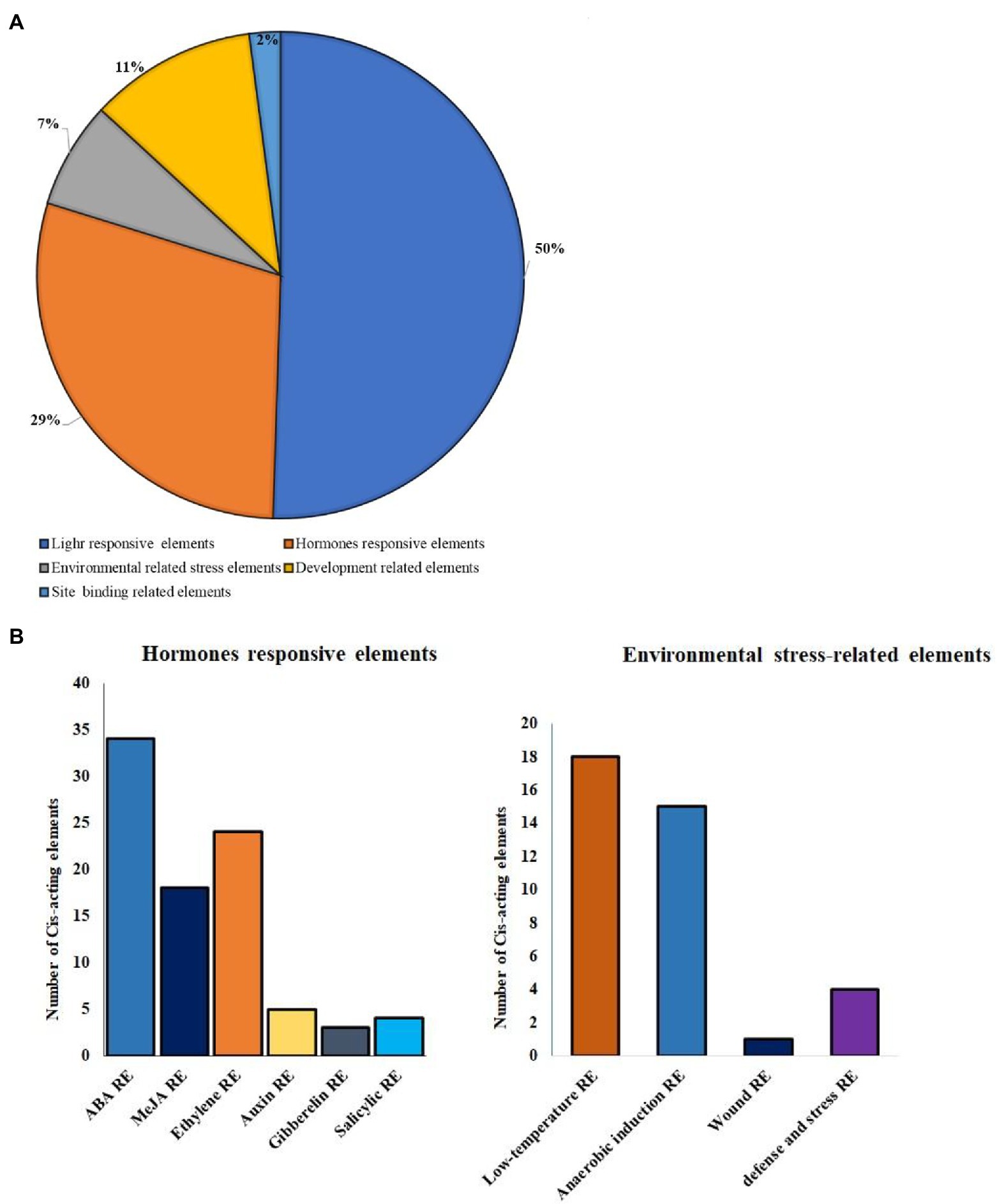
Figure 5. Distribution of cis-elements in the promoter of B. oleracea Hsp90 genes. (A) The proportion of cis-elements in the promoters of B. oleracea Hsp90 genes are are shown in a pie chart. (B) Different hormones responsive elements and environmental stress-related elements in 12 BoHsp90 genes cis-elements regions.
Tissue-Specific Expression Profile Analysis of Brassica oleracea the Hsp90 Genes
The expression profile of the BoHsp90 gene was generated using RNA-seq data from seven tissues (root, stem, leaf, shoot, flower, callus, and silique). The expression pattern was derived from publicly accessible Illumina RNA-Seq data for Cabbage at NCBI (Liu et al., 2014).17 The expression of the BoHsp90 gene was found to be higher in stem and silique tissues than in other tissues (Figure 6). The 12 predicted BoHsp90 genes were found to be active in at least one of the six tissues. The expression of BoHsp90–9 and BoHsp90–5 was found to be high in the stem, silique, leaves, and flowers. Two genes were upregulated in stem, silique, leaves, and flowers (BoHsp90-9 and BoHsp90-5). “Target genes involved in plant growth and development have been shown to be affected by genes that are highly expressed in plant tissues or organs. In this study, we found development-related BoHsp90 genes in the cabbage genome at the transcription level, and two genes (BoHsp90–5, and 9) were selected in stem, leaves, silique, and flowers that were upregulated, downregulated, or not different in other tissues according to normalized FPKM values. (Supplementary Table S4).
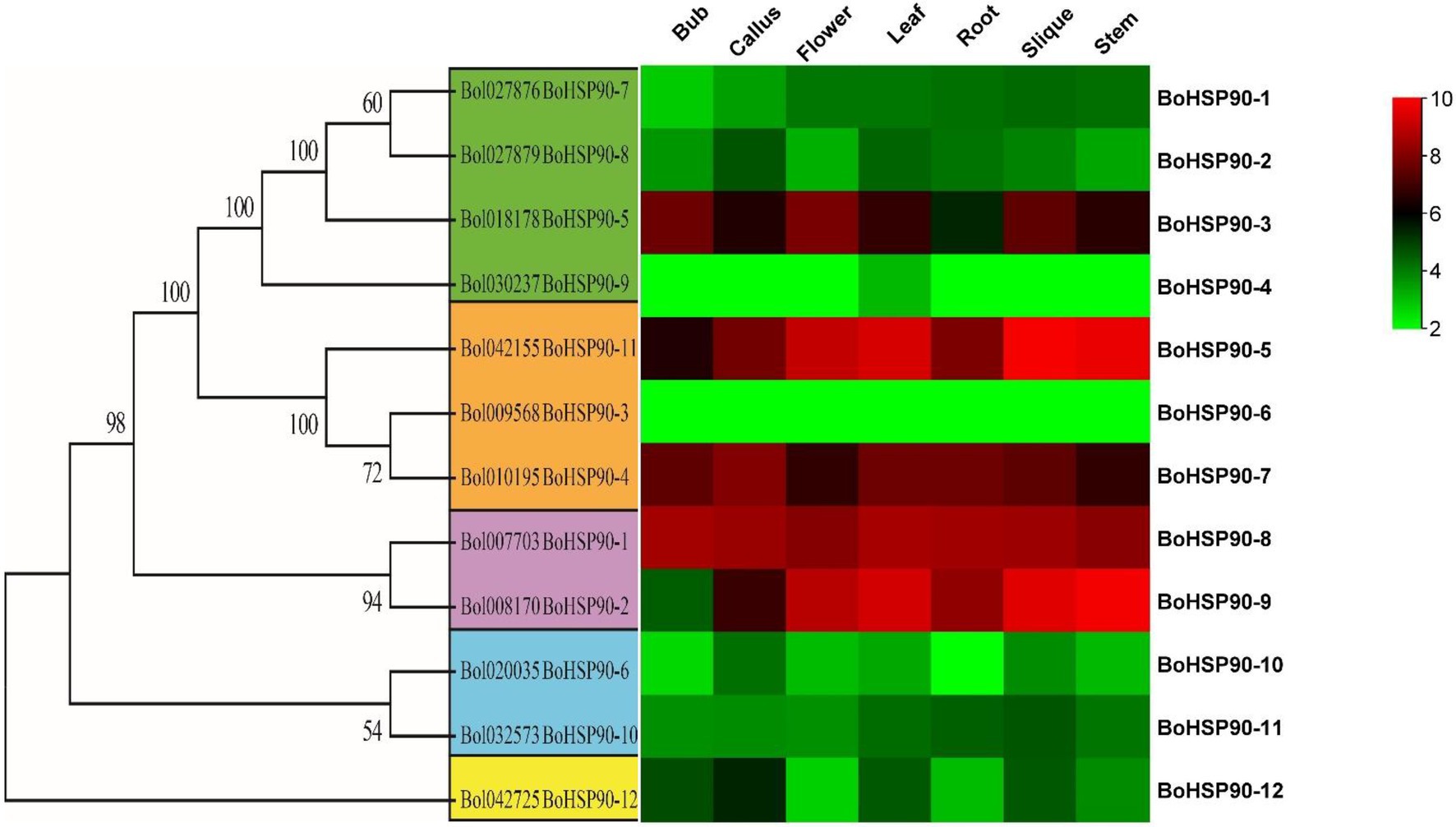
Figure 6. The heat map of 12 BoHsp90 genes was constructed using Tbtools software. The FPKM values were log2 transformed and mean-centered by genes using the scale 10 for minimum expression (red color) and 2 (green color) for maximum to make a heat map visible for each cabbage tissue.
Expression Profiles Analysis of Brassica oleracea Hsp90 Genes Under Cold Stress
To confirm the expression patterns determined by the qRT-PCR was employed to analyze the expression patterns of 12 different BoHsp90 genes under cold stress conditions. Different expression profiles of 12 BoHsp90 genes in response to cold stress were identified. (Figure 7). The result showed that most of the genes showed high expression levels at 4°C for 12 h intervals. Besides the expression level of BoHsp90-2, BoHsp90-3, BoHsp90-7, BoHsp90-9, BoHsp90-2, and BoHsp90-11 were highly upregulated at 4°C after 12 h exposure of cold stress. we have further noticed that expression patterns of genes, including, BoHsp90-2, BoHsp90-3, BoHsp90-4, BoHsp90-5, BoHsp90-7, BoHsp90-8, BoHSP90-10, and BoHSP90-12 were gradually downregulated as time passed. Similarly, BoHSP90-1, BoHsp90-4, BoHsp90-6, BoHsp90-9 were highly upregulated at −2 and 0°C for 24 h intervals, but these genes showed downregulation trend at 6 h. The findings from this study exhibited that the B.oleracea Hsp90 genes family, which are responded to cold stress promptly and with a long response time.
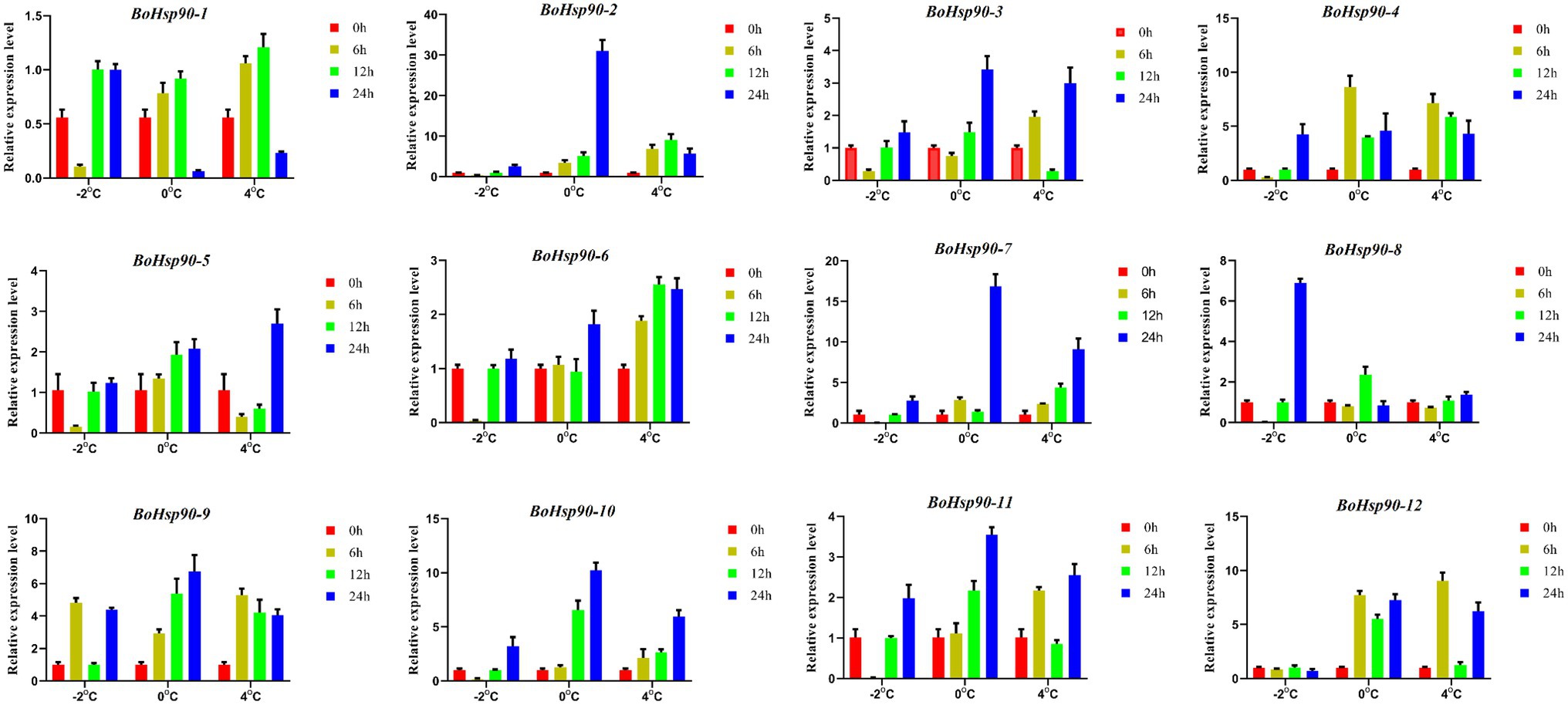
Figure 7. Relative expression levels of 12 Hsp90 genes in B. oleracea leaves by qRT-PCR after 0 h (control), 6 h, 12 h and 24 h after treatment of cold stress at −2°C, 0°C, and 4°C. The expression levels of Hsp90 genes were normalized to BoActin using as an internal control. Error bars indicate the standard error (SE) between three replicates.
Discussion
The unpredictability of global temperatures is followed by an increase in the frequency of cold spells, which is harmful for crop growth and effected the yields, and understanding the response of plants to low-temperature stress is very important for plant growth (Ding et al., 2019). When the temperature fluctuates (rises and falls) in comparison to normal, the physiological and biochemical reactions in the organism are inhibited, and cold stress inhibits the majority of protein and mRNA transcription. However, temperature stress (high and low) typically alters gene expression in plants, and an important type of highly conserved protein known as HSPs is rapidly synthesized. The Hsp90 is a chaperone protein that is highly conserved in prokaryotes and all eukaryotes and is involved in the cell cycle, signaling, and other key biological processes (Pearl and Prodromou, 2006). HSPs were first identified in the salivary gland chromosomes of Drosophila larvae (Ritossa, 1962). The function of Hsp90 genes has been extensively studied in animal and fungal systems to date, but not widely studied in plants. However, with the continuous advancement and wider application of genome sequencing technologies, more and more plant genome data for Hsp90 have been released recently (Kersey, 2019). The Hsp90 gene was found to be involved not only in stress signal transduction, receptor folding, transcription factors and kinases, and physiological processes in plants (Chory and Wu, 2001; Mach, 2009; Cha et al., 2013; Han et al., 2015) but also in assisting cell survival in stressful situations (Jackson et al., 2004). The Hsp90 gene family has been identified in two model plants, Arabidopsis and rice (Hu et al., 2009). Subsequently, the Hsp90 gene family was discovered in some major vegetable crops, such as tomato (Liu et al., 2014) and pepper (Jing et al., 2020). Cabbage is a very important vegetable crop cultivated in many areas worldwide. B. oleracea seedlings must be vernalized at 0–10°C and then licked, blossomed, and bore fruit under long sunshine and moderate temperature. Due to its characteristics of different maturity, suitable for different seasons, wide adaptability, and strong stress resistance, B. oleracea occupies an important position in the annual production of vegetables (Zhu et al., 2017). But the low temperature was one of the most important limiting factors for the wintering cultivation of B. oleracea in open fields; during winter B. oleracea often suffers from low temperatures and even sub-zero temperatures. The low-temperature conditions make its yield and quality decline. However, Hsp90 gene family has not been characterized in cabbage (B. oleracea). Therefore, we investigated the whole genome of the BoHsp90 gene family in cabbage, including phylogeny, chromosomal location, gene structure, conserved motifs, and expression profiles. A total of 12 Hsp90 genes in cabbage were identified in this study, which are greater than other plant species such as 7 in Arabidopsis (Krishna and Gloor, 2001), 9 in rice (Hu et al., 2009), 10 in poplar (Zhang et al., 2013), 7 in tomato (Liu et al., 2014), and 7 in pepper (Jing et al., 2020). This could be the one of the main reasons for the difference in cold tolerance ability among different plant species. In this study, using the Arabidopsis Hsp90 protein sequence, 12 Hsp90 genes were retrieved from the Brassica plant genome. These Hsp90 family proteins are vital in acclimatizing unfavorable environmental conditions through regulation and maintenance of physiological mechanism of cabbage plant (Zhang et al., 2021). The various physicochemical properties of the Hsp90 family proteins imply that there is a great deal of diversity among members, which will help in further research into the functions of Hsp90 genes. Different features of the Hsp90 protein family, such as molecular weight, isoelectric point, and the number of exon-introns, were investigated in cabbage in this work. “Furthermore, all Hsp90 proteins in B. oleracea were acidic nature, which was similar with findings from A. thaliana, tomato, and other plants (Liu et al., 2014). These results suggested that Hsp90 proteins were conserved across plant species. The amino acid sequences of Hsp90 family proteins provided phylogenetic information based on their basic structures. In different BoHsp90 gene sequences, we found different numbers of introns (1–19 introns). The fewer introns a plant has, the better its capability to respond to various environmental stimuli and developmental processes (Jin et al., 2014). Long-term evolution has resulted in a high number of introns in BoHsp90s. Song et al. (2019) revealed that conserved motif analysis of Hsp90 genes in tobacco, Nitab4.5_0001622g0050 and Nitab4.5_0003328g0120 consist of fewer motifs, hinting that it had lost sequencing parts during evolution. Referring to previous investigations, the cabbage Hsp90 genes’ conserved motif analysis, Bol009568.BoHsp90-3 and Bol42725.BoHsp90-12 shown fewer motifs, which might be due to misplaced sequence sections during evolution. In addition, BoHsp90-3 and BoHsp90-12 contain 6 motifs, BoHsp90-7 has 8 motifs, and 9 motifs were found in BoHsp90-9. These findings suggested that the structural motifs are aligned differently in various members of the BoHsp90 family genes but are similar within closely related genes.
Gene duplication is a crucial mechanism in the evolution of gene families (Yang et al., 2008). In cabbage, at least three pairs of duplicated genes were discovered, representing that gene replication may have occurred throughout the evolution of the Hsp90 genes. Phylogenetic analysis is frequently used to learn more about species’ evolutionary changes and to discover the orthologs and paralogs within species (Moreira and Philippe, 2000). An unrooted phylogenetic tree was constructed using protein sequences from B. oleracea, B. rapa, B. napus, tomato, cucumber, potato, V. vinifera, Sorghium bicolor, cotton, tobacco, Arabidopsis, rice, Z. mays, soya bean, E. guineensis, Chinese white pear (Pyrus bretschneideri) and N. nucifera. Hsp90s can be split into five groups based on evolutionary study. Protein function is determined by its structure (Skolnick and Fetrow, 2000). According to the collinearity relationships between the B. orleacea BoHsp90 family genes and 2 representative plant species including A. thalian and B. rapa, only three pair of paralogous pair (Bol027876BoHsp90-7-Bol027879.BoHsp90-8, Bol01019568.BoHsp90-3-Bol010195Hsp90.4 and Bol020035.BoHsp90-6-Bol032573.BolHsp90-10) were perceived and segmental duplication were not found, representing that enlargement of Hsp90 gene in B. oleracea was mainly caused by tandem duplication genes. Only four B. oleracea Hsp90 genes Bol7703-BoHsp90-1, Bol008170.BolHsp90-2, Bol018178.BoHsp90-5 and Bol027879.BoHsp90-8 was conservative and not collinearity relation with Hsp90 genes in B. rapa and Arabidopsis. However, eight B. oleracea Hsp90 genes have collinearity relationships with 5 genes of A. thaliana and 10 genes of B. rapa. These findings suggested that the Hsp90 gene family expands differently in each species, as this pattern was also noticed in other plant gene families (Huang et al., 2016). Furthermore, promoter analysis revealed that the promoter of the B. oleracea Hsp90 gene contained various regulatory elements related to light-responsive elements, responsive to hormones, environmental responsive elements, development-related elements, and site binding elements, among others, implying that these regulatory elements actively engage in a variety of biotic and abiotic stress responses in plants and also indicating that Hsp90 genes are important in cell-cycle control, signal transduction, genomic silencing, protein degradation, and protein trafficking.
Recent studies have shown that Hsp90 has a vital role in plant acclimation to various biotic and abiotic stresses (Di Donato and Geisler, 2019). The Hsp gene family was considered to be involved or induced under cold environments (Krishna et al., 1995); it has the potential to protect plant biological systems from cold damage by preventing freezing-induced protein denaturation (Yan et al., 2006). In this study, the publicly online tissue specific expression analysis showed that the BoHsp90 gene was found to be higher in stem and silique tissues than other tissues. Similarly, the expression of BoHsp90–9 and BoHsp90–5 was found to be high in the stem, silique, leaves, and flowers. While upregulated in stem, silique, leaves and flowers. These findings indicated that the functions of B. oleracea Hsp90 genes varied across tissues. Many studies indicated that Hsp90 gene family induced under low-temperature stress (Krishna et al., 1995; Di Donato and Geisler, 2019).
Gene expression patterns were highly correlated with their functions. The tissue-specific expression patterns at a given developmental stage are crucial for determining the gene functions in which they are involved. The expression pattern of different genes was studied in our work under diverse stresses and distinct cabbage tissues to better understand their possible significance during cold stress and development responses. The majority of the genes investigated exhibited expression in developmental tissues, indicating their function in these tissues (Figure 7). However, the Hsp90 gene family in cabbage has not been investigated to date. Thus, to understand the functions of Hsp90 genes under cold stress, we accomplish qRT-PCR of 12 different genes under cold stress. These results indicated that expressions of Hsp90 genes family, BoHsp90-1/6/8/9/11, BoHsp90-1, and BoHsp90-9 were highly upregulated at 4°C, 0°C, and -2°C respectively, while remaining were down-regulated at respective low temperatures for 6 h duration. In the same manner, gene expressions of BoHsp90-2/3/5/6/7/9/10/11/12, BoHsp90-1/4/9/10/11/12, and BoHsp90-1 were significantly upregulated at 4°C, 0°C, and -2°C, respectively; while BoHsp90-1/4/8, BoHsp90-2/3/6/7/8, BoHsp90-2/3/4/5/6/7/8/9/10/11/12 were downregulated at 4°C, 0°C, and -2°C, respectively, for 12 h duration. Lastly for 24 h duration, gene expressions of BoHsp90-3/5/6/7/10/11/12, BoHsp90-1/4/6/9, and BoHsp90-1/3/4/6/9/11/12 were upregulated at 4°C, 0°C, and -2°C, respectively; while BoHsp90-1/2/4/8/9, BoHsp90-2/3/5/7/8/10/11/12, and BoHsp90-2/5/7/8/10 were downregulated at 4°C, 0°C, and -2°C, respectively. Previous studies also reported that Hsp90 protein accumulation was found to be altered in cold-treated winter wheat followed cold treatment (Vítámvás et al., 2012). Overall, the cabbage Hsp90 gene family was found to be differentially expressed under cold stress, suggesting that these genes play an important role in cabbage growth and development under cold conditions. We found the evolution of Hsp90 genes in cabbage using comparative genomics analysis, and we screened putative regulatory genes associated with cold stress. The biological activities of these genes will be examined in the subsequent study, which will give an essential theoretical foundation for increasing cabbage quality.
Conclusion
In this study, Hsp90 family genes were identified and characterized in the Brassica oleracea. A total of 12 Hsp90 genes of Brassica were identified and characterized using phylogenetic tree, gene structure, physicochemical characteristics, conserved motif analysis, chromosomal location, synteny, selection pressure, homologous gene pairs, and cis-elements in the promoters, which revealed a rich evolutionary history for this Brassica family. The expression pattern analysis of B. oleracea BoHsp90 exhibited that BoHsp90-2, BoHsp90-3, BoHsp90-7, BoHsp90-9, BoHsp90-10, and BoHsp90-11 were induced under cold stress, which indicates these Hsp90 genes perform a vital role in cold acclimation and supports in the continual of normal growth and development process. Our findings provide a scientific foundation for more comprehensive understanding of the cabbage Hsp90 gene family, and it will also contribute to the development of new high yielding and stress tolerant cultivars of B. oleracea.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/gds/?term=GSE42891.
Author Contributions
SS and SJ conceived and designed the work. MA, QD, YL, MH, and CT performed the experiments and conduct the bioinformatics analysis. SS and JS wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by the National Natural Science Foundation of China (31872108, 31272170) and the Key Research and Development Projects in Anhui Province (No. 202104b11020006).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.908511/full#supplementary-material
Footnotes
1. ^https://phytozome-next.jgi.doe.gov/
2. ^www.ncbi.nlm.nih.gov/Structure/
3. ^http://web.expasy.org/protparam
4. ^http://visualization.ritchielab.org/phenograms/plot
5. ^http://www.expasy.org/tools
6. ^http://gsds.cbi.pku.edu.cn/
7. ^https://www.meme-suite.org/meme/
8. ^https://pfam.sanger.ac.uk/
9. ^http://bioinformatics.psb.ugent.be/
10. ^https://services.cbu.uib.no/tools/kaks
11. ^http://selecton.tau.ac.il
12. ^https://www.ncbi.nlm.nih.gov/
14. ^http://phytozome.jgi.doe.gov/pz/portal.html
References
Ahmed, N. U., Jung, H.-J., Park, J.-I., Cho, Y.-G., Hur, Y., and Nou, I.-S. (2015). Identification and expression analysis of cold and freezing stress responsive genes of Brassica oleracea. Gene 554, 215–223. doi: 10.1016/j.gene.2014.10.050
Al-Whaibi, M. H. (2011). Plant heat-shock proteins: a mini review. J. King Saud Univ.-Sci. 23, 139–150. doi: 10.1016/j.jksus.2010.06.022
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Cha, J.-Y., Ahn, G., Kim, J. Y., Kang, S. B., Kim, M. R., Su'udi, M., et al. (2013). Structural and functional differences of cytosolic 90-kDa heat-shock proteins (Hsp90s) in Arabidopsis thaliana. Plant Physiol. Biochem. 70, 368–373. doi: 10.1016/j.plaphy.2013.05.039
Chen, B., Zhong, D., and Monteiro, A. (2006). Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genomics 7, 1–19. doi: 10.1186/1471-2164-7-156
Chory, J., and Wu, D. (2001). Weaving the complex web of signal transduction. Plant Physiol. 125, 77–80. doi: 10.1104/pp.125.1.77
Di Donato, M., and Geisler, M. (2019). HSP 90 and co-chaperones: a multitaskers’ view on plant hormone biology. FEBS Lett. 593, 1415–1430. doi: 10.1002/1873-3468.13499
Ding, Y., Shi, Y., and Yang, S. (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222, 1690–1704. doi: 10.1111/nph.15696
Dutta, R., and Inouye, M. (2000). GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 25, 24–28. doi: 10.1016/S0968-0004(99)01503-0
Food and Agriculture Organization [FAO] (2020). Responding to the Impact of the COVID-19 Outbreak on Food Value Chains Through Efficient Logistics. Rome: Food and Agriculture Organization.
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gill, R. A., Ali, B., Islam, F., Farooq, M. A., Gill, M. B., Mwamba, T. M., et al. (2015). Physiological and molecular analyses of black and yellow seeded Brassica napus regulated by 5-aminolivulinic acid under chromium stress. Plant Physiol. Biochem. 94, 130–143. doi: 10.1016/j.plaphy.2015.06.001
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Gu, L., Hanson, P. J., Post, W., Mac Kaiser, D. P., Yang, B., Nemani, R., et al. (2008). The 2007 eastern US spring freeze: increased cold damage in a warming world? Bioscience 58, 253–262. doi: 10.1641/B580311
Han, Y., Chen, Y., Yin, S., Zhang, M., and Wang, W. (2015). Over-expression of TaEXPB23, a wheat expansin gene, improves oxidative stress tolerance in transgenic tobacco plants. J. Plant Physiol. 173, 62–71. doi: 10.1016/j.jplph.2014.09.007
Hassan, M. A., Xiang, C., Farooq, M., Muhammad, N., Yan, Z., Hui, X., et al. (2021). Cold stress in wheat: plant acclimation responses and management strategies. Front. Plant Sci. 12:676884. doi: 10.3389/FPLS.2021.676884
Hu, W., Hu, G., and Han, B. (2009). Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 176, 583–590. doi: 10.1016/j.plantsci.2009.01.016
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, Z., Duan, W., Song, X., Tang, J., Wu, P., Zhang, B., et al. (2016). Retention, molecular evolution, and expression divergence of the auxin/indole acetic acid and auxin response factor gene families in Brassica rapa shed light on their evolution patterns in plants. Genome Biol. Evol. 8, 302–316. doi: 10.1093/gbe/evv259
Jackson, S. E., Queitsch, C., and Toft, D. (2004). Hsp90: from structure to phenotype. Nat. Struct. Mol. Biol. 11, 1152–1155. doi: 10.1038/nsmb1204-1152
Jin, Z., Chandrasekaran, U., and Liu, A. (2014). Genome-wide analysis of the Dof transcription factors in castor bean (Ricinus communis L.). Genes Genomics 36, 527–537. doi: 10.1007/s13258-014-0189-6
Jing, W., Fangjun, T., Chengliang, L., Xilu, Z., Lijun, O., Juntawong, N., et al. (2020). Genome-wide identification and analysis of HSP90 gene family in pepper. Acta Horticult. Sin. 47, 665–674. doi: 10.16420/j.issn.0513-353x.2019-0470
Juretic, N., Hoen, D. R., Huynh, M. L., Harrison, P. M., and Bureau, T. E. (2005). The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 15, 1292–1297. doi: 10.1101/gr.4064205
Kersey, P. J. (2019). Plant genome sequences: past, present, future. Curr. Opin. Plant Biol. 48, 1–8. doi: 10.1016/j.pbi.2018.11.001
Koch, M. A., Haubold, B., and Mitchell-Olds, T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17, 1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248
Krishna, P., and Gloor, G. (2001). The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 6, 238–246. doi: 10.1379/1466-1268(2001)006<0238:THFOPI>2.0.CO;2
Krishna, P., Sacco, M., Cherutti, J. F., and Hill, S. (1995). Cold-induced accumulation of hsp90 transcripts in Brassica napus. Plant Physiol. 107, 915–923. doi: 10.1104/pp.107.3.915
Lachowiec, J., Lemus, T., Borenstein, E., and Queitsch, C. (2015). Hsp90 promotes kinase evolution. Mol. Biol. Evol. 32, 91–99. doi: 10.1093/molbev/msu270
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, W.-H., Gojobori, T., and Nei, M. (1981). Pseudogenes as a paradigm of neutral evolution. Nature 292, 237–239. doi: 10.1038/292237a0
Liu, Y., Dang, P., Liu, L., and He, C. (2019). Cold acclimation by the CBF-COR pathway in a changing climate: Lessons from Arabidopsis thaliana. Plant Cell Rep 38, 511–519. doi: 10.1007/s00299-019-02376-3
Liu, Y., Wan, H., Yang, Y., Wei, Y., Li, Z., Ye, Q., et al. (2014). Genome-wide identification and analysis of heat shock protein 90 in tomato. Yi chuan=Hereditas 36, 1043–1052. doi: 10.3724/SP.J.1005.2014.1043
Liu, M., Wang, L., Ke, Y., Xian, X., Wang, J., Wang, M., et al. (2022). Identification of HbHSP90 gene family and characterization HbHSP90. 1 as a candidate gene for stress response in rubber tree. Gene :827, 146475. doi: 10.1016/j.gene.2022.146475
Lv, H., Wang, Y., Han, F., Ji, J., Fang, Z., Zhuang, M., et al. (2020). A high-quality reference genome for cabbage obtained with SMRT reveals novel genomic features and evolutionary characteristics. Sci. Rep. 10, 1–9. doi: 10.1038/s41598-020-69389-x
Mach, J. (2009). Alternative splicing produces a JAZ protein that is not broken down in response to jasmonic acid. Plant Cell 21:14. doi: 10.1105/tpc.108.210111
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Masson-Delmotte, V., Zhai, P., Pirani, A., Connors, S., Péan, C., Berger, S., et al. (2021). “Climate change 2021: the physical science basis,” in Working Group I contribution to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. ambridge: Cambridge University Press.
Moreira, D., and Philippe, H. (2000). Molecular phylogeny: pitfalls and progress. Int. Microbiol. 3, 9–16.
Moreno, A. A., and Orellana, A. (2011). The physiological role of the unfolded protein response in plants. Biol. Res. 44, 75–80. doi: 10.4067/S0716-97602011000100010
Mukhopadhyay, I., Nazir, A., Saxena, D., and Chowdhuri, D. K. (2003). Heat shock response: hsp70 in environmental monitoring. J. Biochem. Mol. Toxicol. 17, 249–254. doi: 10.1002/jbt.10086
Pearl, L. H., and Prodromou, C. (2000). Structure and in vivo function of Hsp90. Curr. Opin. Struct. Biol. 10, 46–51. doi: 10.1016/S0959-440X(99)00047-0
Pearl, L. H., and Prodromou, C. (2006). Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 75, 271–294. doi: 10.1146/annurev.biochem.75.103004.142738
Picard, D. (2002). Heat-shock protein 90, a chaperone for folding and regulation. Cell. Mol. Life Sci. CMLS 59, 1640–1648. doi: 10.1007/pl00012491
Pratt, W. B., and Toft, D. O. (2003). Regulation of signaling protein function and trafficking by the Hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228, 111–133. doi: 10.1177/153537020322800201
Prodromou, C., Roe, S. M., O’brien, R., Ladbury, J. E., Piper, P. W., and Pearl, L. H. (1997). Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90, 65–75. doi: 10.1016/S0092-8674(00)80314-1
Punta, M., Coggill, P. C., Eberhardt, R. Y., Mistry, J., Tate, J., Boursnell, C., et al. (2012). The Pfam protein families database. Nucleic Acids Res. 40, D290–D301. doi: 10.1093/nar/gkr1065
Queitsch, C., Sangster, T. A., and Lindquist, S. (2002). Hsp90 as a capacitor of phenotypic variation. Nature 417, 618–624. doi: 10.1038/nature749
Raman, S., and Suguna, K. (2015). Functional characterization of heat-shock protein 90 from Oryza sativa and crystal structure of its N-terminal domain. Acta Crystallograph. Sect. F: Struct. Biol. Commun. 71, 688–696. doi: 10.1107/S2053230X15006639
Ritossa, F. (1962). A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia 18, 571–573. doi: 10.1007/BF02172188
Skolnick, J., and Fetrow, J. S. (2000). From genes to protein structure and function: novel applications of computational approaches in the genomic era. Trends Biotechnol. 18, 34–39. doi: 10.1016/S0167-7799(99)01398-0
Song, Z., Pan, F., Yang, C., Jia, H., Jiang, H., He, F., et al. (2019). Genome-wide identification and expression analysis of HSP90 gene family in Nicotiana tabacum. BMC Genet. 20, 1–12. doi: 10.1186/s12863-019-0738-8
Sung, N., Lee, J., Kim, J.-H., Chang, C., Joachimiak, A., Lee, S., et al. (2016). Mitochondrial Hsp90 is a ligand-activated molecular chaperone coupling ATP binding to dimer closure through a coiled-coil intermediate. Pro. Nati. Acad. Sci. 113, 2952–2957. doi: 10.1073/pnas.1516167113
Vítámvás, P., Prášil, I. T., Kosová, K., Planchon, S., and Renaut, J. (2012). Analysis of proteome and frost tolerance in chromosome 5A and 5B reciprocal substitution lines between two winter wheats during long-term cold acclimation. Proteomics 12, 68–85. doi: 10.1002/pmic.201000779
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Xiong, L., and Ishitani, M. (2006). “Stress signal transduction: components, pathways and network integration.” in Abiotic Stress Tolerance in Plants. eds. M. Way and K. J. Green. (Berlin: Springer), 3–29.
Yamada, K., Fukao, Y., Hayashi, M., Fukazawa, M., Suzuki, I., and Nishimura, M. (2007). Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J. Biol. Chem. 282, 37794–37804. doi: 10.1074/jbc.M707168200
Yan, S.-P., Zhang, Q.-Y., Tang, Z.-C., Su, W.-A., and Sun, W.-N. (2006). Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol. Cell. Proteom. 5, 484–496. doi: 10.1074/mcp.M500251-MCP200
Yang, Z. (1997). PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556. doi: 10.1093/bioinformatics/13.5.555
Yang, Z., Zhou, Y., Wang, X., Gu, S., Yu, J., Liang, G., et al. (2008). Genomewide comparative phylogenetic and molecular evolutionary analysis of tubby-like protein family in Arabidopsis, rice, and poplar. Genomics 92, 246–253. doi: 10.1016/j.ygeno.2008.06.001
Yu, J., Tehrim, S., Zhang, F., Tong, C., Huang, J., Cheng, X., et al. (2014). Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genom. 15, 1–18. doi: 10.1186/1471-2164-15-3
Zhang, K., He, S., Sui, Y., Gao, Q., Jia, S., Lu, X., et al. (2021). Genome-wide characterization of HSP90 gene family in cucumber and their potential roles in response to abiotic and biotic stresses. Front. Genet. 12, 95. doi: 10.3389/fgene.2021.584886
Zhang, J., Li, J., Liu, B., Zhang, L., Chen, J., and Lu, M. (2013). Genome-wide analysis of the Populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC Genomics 14, S1–S14. doi: 10.1186/1471-2164-14-S8-S1
Keywords: cabbage, genome wide, Hsp90, expression pattern, phylogenetic analysis
Citation: Sajad S, Jiang S, Anwar M, Dai Q, Luo Y, Hassan MA, Tetteh C and Song J (2022) Genome-Wide Study of Hsp90 Gene Family in Cabbage (Brassica oleracea var. capitata L.) and Their Imperative Roles in Response to Cold Stress. Front. Plant Sci. 13:908511. doi: 10.3389/fpls.2022.908511
Edited by:
Parviz Heidari, Shahrood University of Technology, IranReviewed by:
Abdullah, Quaid-i-Azam University, PakistanMostafa Ahmadizadeh, University of Hormozgan, Iran
Copyright © 2022 Sajad, Jiang, Anwar, Dai, Luo, Hassan, Tetteh and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianghua Song, amhzb25nQGFoYXUuZWR1LmNu
 Shoukat Sajad
Shoukat Sajad Shuhan Jiang
Shuhan Jiang Muhammad Anwar
Muhammad Anwar Qian Dai
Qian Dai Yuxia Luo
Yuxia Luo Muhammad A. Hassan
Muhammad A. Hassan Charles Tetteh
Charles Tetteh Jianghua Song
Jianghua Song