- 1Institute of Forestry and Pomology, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 2Key Laboratory of Biology and Genetic Improvement of Horticultural Crops (North China), Ministry of Agriculture and Rural Affairs, Beijing, China
- 3Beijing Engineering Research Center of Functional Floriculture, Beijing, China
- 4Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China
- 5Institute of Botany, University of Chinese Academy of Sciences, Beijing, China
- 6Beijing Engineering Research Center for Deciduous Fruit Trees, Beijing, China
Rose (Rosa sp.) flowers have a rich diversity of colors resulting from the differential accumulation of anthocyanins, flavonols, and carotenoids. However, the genetic and molecular determinants of the red-petal trait in roses remains poorly understood. Here we report that a transposable element-like insertion (Rosa1) into RcMYB114, a R2R3-MYB transcription factor’s promoter region causes its transcription, resulting in red petals. In red-petal varieties, RcMYB114 is expressed specifically in flower organs, but is absent from non-red varieties. Sequencing, yeast two-hybrid, transient transformation, and promoter activity assays of RcMYB114 independently confirmed the role of Rosa1 in altering RcMYB114’s transcription and downstream effects on flower color. Genetic and molecular evidence confirmed that the Rosa1 transposable element-like insertion, which is a previously unknown DNA transposable element, is different from those in other plants and is a reliable molecular marker to screen red-petal roses.
Introduction
Rose (Rosa sp.) is a widely cultivated plant prized for its ornamental, therapeutic, and cosmetic properties (Marmol et al., 2017; Hibrand Saint-Oyant et al., 2018; Raymond et al., 2018). Flower color is of primary importance in ornamental plants and rose exhibits a diverse array of petal colors. Many studies have sought to uncover the molecular and genetic basis of petal coloration in order to accelerate the selective breeding process. In general, differences in color patterns among flowers are determined by the differential regulation of pigment biosynthesis genes during flower development (Martin and Gerats, 1993). For example, red flower color is conferred by the anthocyanin pigment, the biosynthesis of which has been widely studied (Grotewold, 2006; Carbone et al., 2009; Schaart et al., 2013b; Hsu et al., 2015). Anthocyanin biosynthesis involves many structural genes encode essential biosynthetic enzymes including phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate:coenzyme A ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), anthocyanin synthase (ANS), and UDP flavonoid glucosyltransferase (UFGT) (Winkel-Shirley, 2001). In particularly, the enzymes DFR, ANS, and UFGT are markers of the late anthocyanin biosynthetic pathway (Winkel-Shirley, 2001; Grotewold, 2006; Tanaka et al., 2008; Wang et al., 2020). The genes encoding these late-stage enzymes are regulated by various R2R3-MYB transcription factor genes such as AtMYB123(TT2) (Baudry et al., 2004), FaMYB9/FaMYB11 (Schaart et al., 2013a), FaMYB10 (Wang et al., 2020), MlPELAN and MlNEGAN (Yuan et al., 2014), PavMYB10.1 (Jin et al., 2016), PsMYB12 (Gu et al., 2019), VvMYBA2r and VvMYBA2w (Jiu et al., 2021), CgsMYB6, and CgsMYB11 (Lin and Rausher, 2021).
The total numbers of R2R3-MYB transcription factors were different in different plant species. For example, a 70 R2R3-MYB transcription factors were identified in sugar beet (Stracke et al., 2014). In grapevine, 108 R2R3-type MYBs were described and classified (Matus et al., 2008). In Arabidopsis thaliana, there are 126 MYBs of the R2R3-type described (Stracke et al., 2001). A total of 285 R2R3-MYB transcription factors were identified in banana (Pucker et al., 2020). According to its phylogenetic relationships and short signature motifs the R2R3-MYBs were classified into different subgroups (Kranz et al., 1998; Dubos et al., 2010). The anthocyanin activating R2R3-MYB transcription factors which belong to subgroup 6, had the signature motif “[R/K]PRPRx[F/L].” The first anthocyanin activating R2R3-MYB transcription factors were identified by Paz-Ares et al. (1987). Then many newly R2R3-MYB transcription factors activating anthocyanin were identified in other plants such as snapdragon, lily, petunia, monkey-flower, peony, moth orchid, strawberry, cheery, apple (Goodrich et al., 1992; Quattrocchio et al., 1999; Borevitz et al., 2000; Spelt et al., 2000; Schwinn et al., 2006; Jin et al., 2016; Wang et al., 2020). In snapdragons (Antirrhinum majus), the genes Rosea1, Rosea2, and Venosa regulate petal color intensity and anthocyanin pigmentation (Schwinn et al., 2006). In Asiatic hybrid lily (Lilium asiatica hybrid), the genes LhMYB12 and LhMYB6 regulate anthocyanin pigmentation in tepals, filaments, and styles, and LhMYB6 also regulates light-induced pigmentation in leaves (Yamagishi et al., 2010). In monkey-flower (Mimulus spp.), the genes PELAN and NEGAN regulate anthocyanin pigmentation in the petal lobe and nectar guide, respectively. NEGAN is activated by the NEGAN-MlANbHLH1-WD40 complex via autocatalytic feedback, which is required to generate anthocyanin spots. The abnormal expression of PELAN leads to yellow petals (Yuan et al., 2014). In the orchid Phalaenopsis equestris, the differential expression profiles of R2R3-MYB transcription factors regulate the formation of red flowers, which in the orchid Phalaenopsis Aphrodite, overexpression of the PeMYB2 gene causes anthocyanin accumulation in sepals and petals of white-flowered species, suggesting that in this species, PeMYB2 is responsible for the fully red flower trait (Hsu et al., 2015).
Gene transcription is influenced by several mechanisms including silencing, rearrangement, and insertion of transposable elements (McClintock, 1950; Brown, 1981; Signor and Nuzhdin, 2018; Nakayama and Kataoka, 2019; Gil and Ulitsky, 2020). Barbara McClintock first predicts that transposable elements (Ac/Ds system), which was a mobile piece of DNA, were present in eukaryotic genomes and her studies showed that transposable elements influenced the color of kernels in maize (McClintock, 1950). Transposable elements can replicate and integrate into different positions of the genome, altering the expression of adjacent genes (McClintock, 1950; Butelli et al., 2012; Drongitis et al., 2019; Niu et al., 2019). Transposable elements can be quite volatile and are able to insert themselves into intergenic regions, promoters, exons, introns, and both the 5′ and 3′ untranslated regions (UTRs) of genes. These insertions can lead to both genetic and phenotypic variation (McClintock, 1950; Elbarbary et al., 2016; Hirsch and Springer, 2017; Niu et al., 2019). Often, environmental factors, such as increased temperatures, alter the number and activity of transposable elements, potentially leading to adaptation (Niu et al., 2019). Therefore, transposable elements have potential to quickly create genetic and phenotypic diversity within a population.
During a field survey, we found an interesting red rose (Rosa chinensis ‘Semperflorens’ cv. ‘Slater’s Crimson China’) specimen that contained both red and white flowers on the same branch. The flowers had no other obvious differences, having the same number of petals, pistils, stamens, and sepals. We suspected a transposable element may be affecting the expression of one or more R2R3-MYB transcription factor genes. In this research, we sought to understand the genetic basis of this phenomenon.
Materials and Methods
Plant Materials and Growth Conditions
Samples of a red rose (R. chinensis ‘Semperflorens’ cv. ‘Slater’s Crimson China’) containing both red and white (mutant) flowers on the same plant were identified and collected. Beijing (116°20′ N, 39°56′ E) has a warm temperate semi humid and semi-arid monsoon climate. Summer is hot and rainy, winter is cold and dry, and spring and autumn are short. The annual average temperature is 10∼12°C. All these rose plants were grown outside under nature cultivation conditions at the Institute of Forestry and Pomology, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China. The plants are pruned each December. Samples of the leaves, stems, styles, and petals were used for gene expression analysis. Petals at different developmental stages were used to assess anthocyanin content (Guterman et al., 2002). The leaves, stems, styles, and petals of the other rose varieties were sampled and quick frozen using liquid nitrogen, and stored in the freezer (−70°C).
Petal Anthocyanin Quantification and Identification
A pH differential method was used to identify and quantify the total anthocyanin content of petals of both red and white (mutant) ‘Slater’s Crimson China’ roses at stage 4 (Cheng and Breen, 1991; Benvenuti et al., 2004). In order to quantify anthocyanin content, rose petals were ground in liquid nitrogen and 10 mg of petal powder was extracted with 0.1% HCl-methanol solution for 4 h, in the dark, at room temperature. All petal extractions were centrifuged for 20 min at 8500 g and all supernatants were filtered using a 0.45-μm membrane. A pH differential method was used to estimate the total content of anthocyanins (Cheng and Breen, 1991; Benvenuti et al., 2004). The absorbances at 510 and 700 nm were determined. The anthocyanin content was calculated using anthocyanin content A = (a510-a700) pH 1.0–(a510-a700) pH 4.5. All analyses were performed using three biological replicates.
The anthocyanin contents were quantified and calculated as cyanidin-3,5-O-diglucoside equivalents in μg per g FW. In order to identify the anthocyanin present, the extracts were assayed using ultra-high-performance liquid chromatography–mass spectrometry (UPLC-MS/MS) using a Acquity UPLC system (Waters, MA, United States) coupled to a XEVO-TQ triple-quadrupole mass spectrometer (Waters, Milford, MA, United States) with electrospray ionization (ESI). The analysis conditions were as follows: a 0.4 mL/min flow rate and positive ion ESI modes, 3.0 kV capillary voltages, and 16 L/h nebulization nitrogen flow. The chromatographs were plotted and analyzed using Origin software (OriginLabs, Northampton, MA, United States). The characteristics of UV-Vis spectra of peaks and the mass spectrometric information of the petal anthocyanin compounds were analyzed according to the difference of the retention times of standards. The anthocyanins content’s relative quantification was analyzed by calculating the peak areas of samples according to the corresponding standard compound’s intensity. All analyses were performed using three biological replicates.
cDNA and Genomic Sequence Amplification and Sequencing
Genomic DNA was isolated from 250 mg fresh leaf samples from red and white (mutant) with a Super Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China). Total RNA extractions of both red and white (mutant) ‘Slater’s Crimson China’ roses at stage 4 were performed using an RNA isolation kit (Tiangen Biotech Co., Beijing, China). After the RNA extract had been treated with DNase I, first-strand cDNA was synthesized with a Revert Aid First-Strand cDNA synthesis kit (Thermo Scientific Inc., Waltham, MA, United States). Primer synthesis was performed by Shanghai Sangon (Sangon, Shanghai, China) (Supplementary Table 3). cDNA sequence RcMYB114 was cloned using the red petal cDNA as template. The genomic sequence of RcMYB114 was cloned using the Genomic DNA as template from wild type and mutant petals. PCR was conducted in a 50 μL volume containing 5 μL 10 × buffer, 5 μL dNTPs (2 mmol), 3 μL MgSO4 (25 mmol), 1.5 μL of each primer (10 pmol), 1 U KOD plus polymerase (Toyoboco, Ltd. Life Science Department, Osaka, Japan), and 3 μL genomic DNA (100 ng) or cDNA (100 ng). The cycling conditions were as follows: 1 cycle at 94°C for 4 min, 35 cycles at 98°C for 30 s, 55 ∼58°C for 30 s, and 68°C for 2 min; followed by a final cycle at 68°C for 5 min. PCR products were separated. The amplifying fragments were ligated into pLB-Simple Vector (Tiangen Biotech Co., Beijing, China), transformed into Escherichia coli strain, and sequenced.
Sequence Alignment and Phylogenetic Analysis
The evolutionary analysis of RcMYB114Red gene was carried out using its protein sequence. The other MYB transcription factor’s protein sequences were acquired from GenBank,1 including A. thaliana (AtMYB4, AtMYB75, AtMYB90, AtMYB105, AtMYB114) (Kranz et al., 1998; Borevitz et al., 2000; Jin et al., 2000; Stracke et al., 2001), Solanum pennellii (SpMYB114) (Kiferle et al., 2015), A. majus (AmROSEA1, AmROSEA2, AmVENOSA) (Schwinn et al., 2006), Malus domestica (MdMYB16, MdMYB17, MdMYB111, MdMYB114) (Lin-Wang et al., 2010; Xu et al., 2017; Song et al., 2019), Prunus avium (PaMYB114) (Jin et al., 2016), Prunus persica (PpMYB114) (XP_020420992), Prunus mume (PmMYB114) (XP_016652295), Pyrus × bretschneideri (PbMYB114) (Yao et al., 2017), Rosa rugosa (RrMYB114) (QEV87523), Fragaria vesca (FvMYB114) (XP_004288854), Fragaria × ananassa (FaMYB1, FaMYB5, FaMYB9, FaMYB11) (Paolocci et al., 2011; Schaart et al., 2013a), R. chinensis (RcMYB23, RcMYB308, RcMYB4, RcMYB113, RcMYB105), Rosa hybrid (RhMYB10) (Lin-Wang et al., 2010). The MYB protein alignment was performed using CLUSTALW2 (Chenna et al., 2003; Larkin et al., 2007). An evolutionary tree was produced using MEGA X3 by the neighbor-joining approach executing 1000 bootstrap replicates (Tamura et al., 2011; Kumar et al., 2018).
RNA-Seq Analysis of Transcription Levels
Total RNA extractions of both red and white (mutant) ‘Slater’s Crimson China’ roses at stage 4 were performed using an RNA isolation kit (Tiangen Biotech Co., Beijing, China). After the total RNA extraction and DNase I treatment, magnetic beads with Oligo (dT) are used to isolate mRNA. Mixed with the fragmentation buffer, the mRNA is fragmented into short fragments. Than the cDNA was synthesized using the mRNA fragments as templates by reverse transcriptase (Invitrogen, Carlsbad, CA, United States). The sequencing libraries were prepared using the Library Prep Kit (New England BioLabs, Rowley, MA, United States) Short fragments are purified and resolved with EB buffer for end reparation and single nucleotide A (adenine) addition. After that, the short fragments are connected with adapters. After agarose gel electrophoresis, the suitable fragments are selected for the PCR amplification as templates. At last, the library could be sequenced using the HiSeqTM 2000 system (Illumina, San Diego, CA, United States) by Novogene (Novogene Biotech Co., Ltd., Beijing, China). Primary sequencing data that produced by Illumina HiSeqTM 2000, called as raw reads. Raw data (raw reads) of fastq format were firstly processed through in-house perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing N base and low quality reads from raw data. At the same time, Q20, Q30, and GC content the clean data were calculated. All the downstream analyses were based on the clean data with high quality. After QC, clean reads was aligned to the reference sequences with SOAPaligner/SOAP2. The alignment data is utilized to calculate distribution of reads on reference genes and perform coverage analysis. The gene expression level is calculated by using RPKM method (Mortazavi et al., 2008). The RPKM method is able to eliminate the influence of different gene length and sequencing discrepancy on the calculation of gene expression. Therefore, the calculated gene expression can be directly used for comparing the difference of gene expression among samples. Differential expression analysis was performed using the edgeR (Robinson et al., 2010). The P values were adjusted using the Benjamini and Hochberg method. Corrected P-value 0.05 and absolute fold change of 2 were set as the threshold for significantly different expression.
Real-Time Quantitative PCR Assay of Genes Related to Anthocyanin Biosynthesis
Total RNA of each tissue sample was extracted and first-strand cDNA was synthesized with a cDNA synthesis kit (Thermo Scientific, Waltham, MA, United States). RT-qPCR was performed using a Bio-Rad CFX96 system (Bio-Rad, California, CA, United States). For RT-qPCR conditions, 10 μL reaction mixture included 5 μL 2 × SYBR Premix, 1 μL forward primer (10 μM), 1 μL reverse primer (10 μM), 1 μL cDNA template (20 ng) and 2 μL ddH2O; the PCR conditions were as follows: 1 cycle at 95°C for 30 s, 40 cycles at 95°C for 5 s, and 1 cycle at 60°C for 30 s. Primers for various regulatory and structural genes related to the anthocyanin biosynthesis pathway are shown in Supplementary Table 3. Data were analyzed using the 2–ΔΔCT method as outlined by Livak and Schmittgen (2001). Expression of specific genes were normalized to actin (KC514920) (Meng et al., 2013). All analyses were performed using three biological replicates.
Yeast Two-Hybrid Assay of Gene Function
Yeast Two-Hybrid (Y2H) experiments were performed according to the method used in a previous study. Briefly, AH109-competent cells were co-transformed according to the manufacturer’s instructions (Clontech Laboratories, California, CA, United States). RcMYB114Red and RcWD40 were introduced into pGADT7 to produce fusion proteins using the GAL4 activation domain (AD). RcMYB114Red and RcWD40 were separately cloned into pGBKT7 to make recombinants with the GAL4 DNA binding domain (BD). The vector of RcbHLH fused with the GAL4 AD and BD was kept in our laboratory (Li et al., 2017). All primers are listed in Supplementary Table 3. All constructs were confirmed by enzyme digestion and sequencing. The various combinations of BD and AD vectors were co-transformed into yeast strain AH109 using the lithium acetate method (Gietz et al., 1995) and selected on SD/–Leu–Trp media under 30°C culture conditions for 3–4 days. To assay the interaction, these clones were then incubated on SD/–Ade–His–Leu–Trp culture media under 30°C culture conditions for 7 days. β-galactosidase tests were performed on the same plate and positive clones were dyeing by using 3–5 μL 4 mg/mL X-α-gal, and false-positive activation was excluded using the P53 plus SV40 vector.
Nicotiana benthamiana Expression Assay of Gene Function
Transient expression of RcMYB114Red constructs was performed using a Hyper Trans system (Sainsbury et al., 2009; Butelli et al., 2017). Briefly, RcMYB114Red was isolated from the genomic DNA of both the white (mutant) and red flowers of ‘Slater’s Crimson China.’ RcMYB114Red was placed into the pEAQ686HT-DEST1 vector. The RcMYB114Red plasmid was transformed into Agrobacterium tumefaciens GV3101. The transformation protocol was conducted as previously described (Sparkes et al., 2006). Leaves were sampled 7 days after injection. We then photographed the leaves and carried out measurement of the total anthocyanin content, high-performance liquid chromatography analysis of anthocyanin compounds, and RT-qPCR analysis of the expression of the RcMYB114Red transcription factor and other genes.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assays were performed according to Bowler’s methods (Bowler et al., 2004). Briefly, we used a rabbit (New Zealand) to produce an IgG antibody to RcMYB114Red. The rabbit IgG was purified using Pan’s protocol (Pan et al., 2005). The chromatin immunoprecipitation experiments were carried out as described by Bowler (Bowler et al., 2004) using a Pierce Agarose ChIP kit (No. 26156, Thermo Scientific, Waltham, MA, United States). Primers were designed according to the promoter sequences of RcPAL, RcC4H, RcCHS, RcCHI, RcF3H, RcFLS, RcLAR, RcDFR, RcANS, and RcUFGT.
pRcMYB114Red and pRcMYB114White Promoter-β-Glucuronidase Fusion Gene Transformation and Histochemical β-Glucuronidase Assay
We fused the pRcMYB114Red and pRcMYB114White promoters to the β-Glucuronidase (GUS) gene vector, which were subsequently injected into Nicotiana benthamiana leaves (Sainsbury et al., 2009; Butelli et al., 2017). Leaves were sampled 7 days after injection and soaked in X-Gluc buffer (12 mM potassium ferricyanide, 12 mM potassium ferrocyanide, 0.3% (v/v) Triton X-100, and 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-glucuronide). The buffer was infiltrated into the samples under a vacuum. The leaves were stained overnight at 37°C, washed in 70% (v/v) ethanol several times, and then photographed (Koo et al., 2007).
Results
RcMYB114 Shares High Homology With Anthocyanin-Regulating Genes in Many Plants
During a field survey, we found a red rose (R. chinensis ‘Semperflorens’ cv. ‘Slater’s Crimson China’) specimen that contained both red and white (mutant) flowers on the same branch (Figure 1A). We collected both red and white flowers from this specimen and performed an RNA-seq analysis. We found that several key genes upstream of the anthocyanin biosynthesis pathway, including RcPAL, RcCHI, RcCHS, and RcC4H, had significantly higher expression levels in red flowers compared to white flowers. Additionally, several key downstream genes, including RcDFR, RcANS, and RcUFGT, also had significantly higher expression levels in red flowers compared to white flowers (Figure 1B). We further confirmed these results by real-time quantitative PCR (RT-qPCR). Overall, we found 125 MYB genes expressed in red flowers and 118 MYB genes expressed in white flowers. Among these MYB genes, RcMYB113, RcMYB308, RcMYB75, RcMYB90, RcMYB114, RcMYB4, RcMYB105, and RcMYB23 encode R2R3-type MYB transcription factors. The RNA-seq heatmap indicated that the relative transcription levels of these genes showed two distinct expression patterns between red and white flowers. The expression levels of RcMYB113, RcMYB308, and RcMYB4 were lower in red flowers compared to white flowers. Conversely, the expression levels of RcMYB75, RcMYB114, RcMYB105, and RcMYB23 were higher in red flowers compared to white flowers. Notably, RcMYB114 was highly expressed in red flowers, but was absent in white flowers (Figure 1B). Based on gene annotation analysis, RcMYB114 was mapped to chromosome 7 (GenBank accession: MW239569). BLAST similarity analysis indicated that RcMYB114 was identical to the RchiOBHmChr7g0235271.4 Further, SMART analysis showed that RcMYB114 encoded an R2R3-MYB transcription factor.5
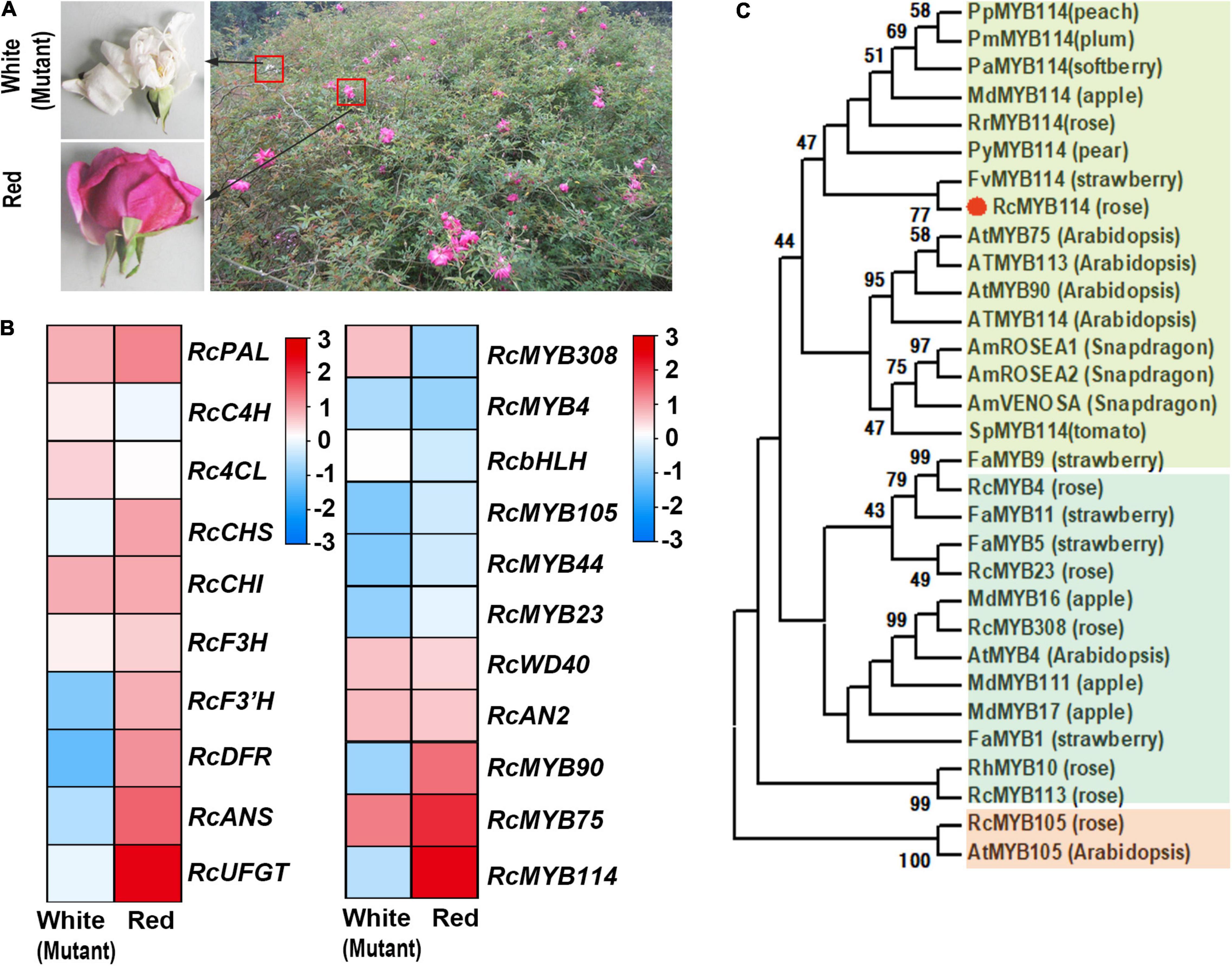
Figure 1. Identification of the gene (RcMYB114) regulating anthocyanin biosynthesis in our mutant rose. (A) A red rose (R. chinensis ‘Semperflorens’ cv. ‘Slater’s Crimson China’) specimen was found to contain both red and white (mutant) flowers. (B) RNA-seq heat map of genes related to anthocyanin biosynthesis. (C) Evolutionary analysis of MYB genes.
To determine the phylogenetic relationship of RcMYB114 to other known R2R3-MYB transcription factors, we constructed a tree of about 30 closely related transcription factors by the neighbor-joining approach (Figure 1C). We found that the protein product of RcMYB114 (RcMYB114) clustered with strawberry (Fragaria spp.) FvMYB114. The related proteins are known to primarily regulate anthocyanin biosynthesis, including FvMYB114 (F. vesca), and it was suspected that RCMYB114 does the same (Figure 1C).
RcMYB114 Regulates Anthocyanin Biosynthesis in Roses
We cloned RcMYB114 using the genomic DNA as template from both white mutant petals and red wild-type petals. The results found that the sequences of RcMYB114red and RcMYB114white were identical. To confirm whether RcMYB114 is responsible for regulating anthocyanin biosynthesis, a Hyper Trans expression vector was used to transiently deliver the RcMYB114Red plus RcbHLH into N. benthamiana leaves. We found that, after 5 days, leaves successfully infiltrated with RcMYB114Red plus RcbHLH showed red color (Figures 2A,B). We found that the pigmentation was mainly comprised of cyanidin-3-O-sophoroside, and the total anthocyanin content of these leaves was 57.92 mg/100 g fresh weight (FW).
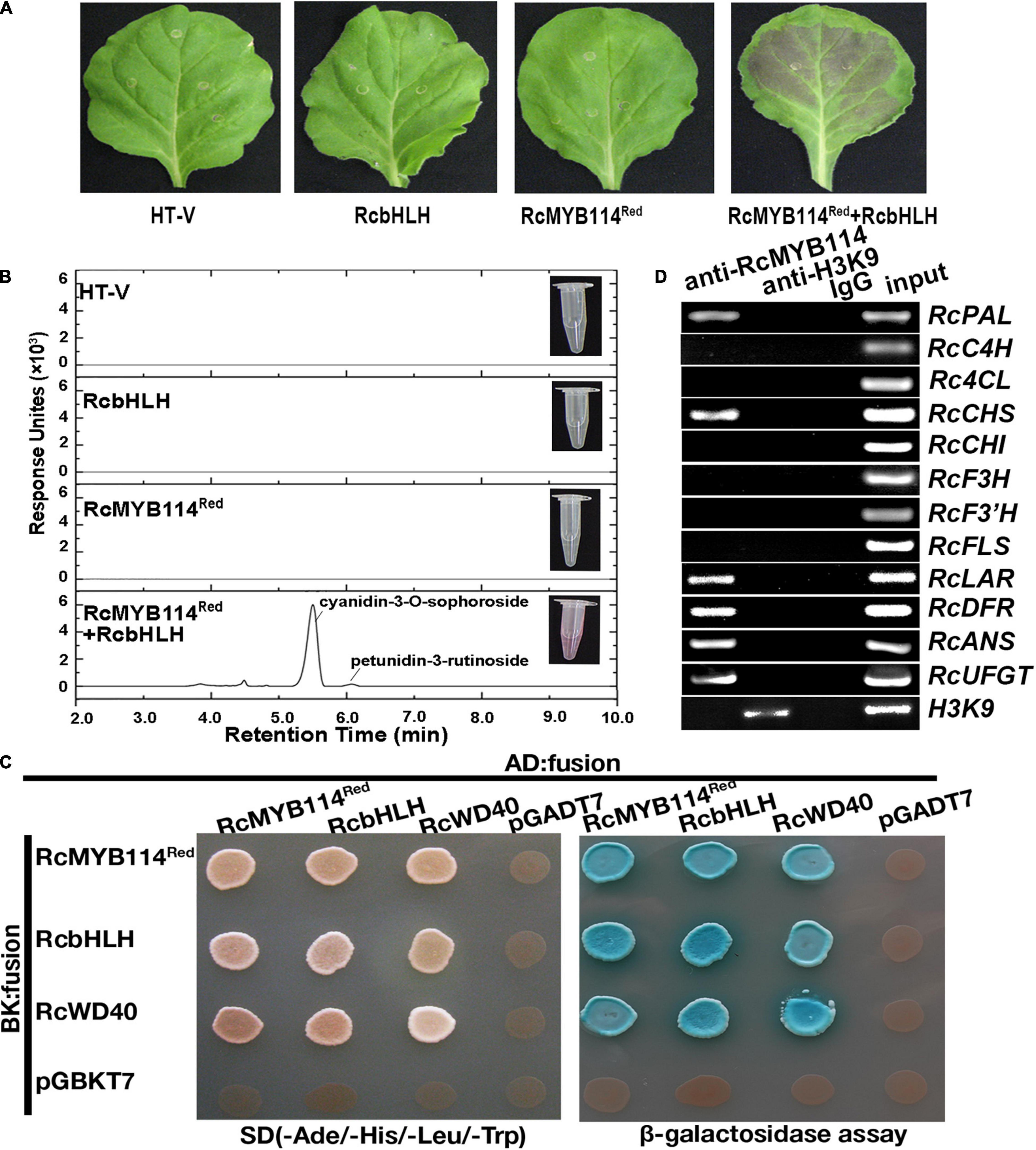
Figure 2. Confirmation of the role of RcMYB114 in regulating anthocyanin biosynthesis. (A) Leaf phenotypes of N. benthamiana after being infiltrated with different constructs or the empty vector (HT-V). (B) Anthocyanins from the infiltrated N. benthamiana leaves were assayed by UPLC-MS/MS. (C) The yeast two-hybrid (Y2H) assay to probe the interaction of RcMYB114 with RcWD40 and RcbHLH within the MBW complex. The co-transformants were screened on media (−Ade/−His/−Leu/−Trp). β-galactosidase tests were performed on the same plate and positive clones were dyeing by using 3–5 μL 4 mg/mL X-α-gal, and false-positive activation was excluded using the P53 plus SV40 vector. (D) RcMYB114 was selectively recruited to RcPAL, RcCHS, RcLAR, RcDFR, RcANS, and RcUFGT promoter regions as determined by the ChIP assay. Anti-H3K9 was used as positive control and Anti-IgG was negative control.
To further verify the role of RcMYB114, yeast two-hybrid (Y2H) and chromatin immunoprecipitation (ChIP) assays were carried out. The classical MBW complex, which acts as a regulatory hub for the anthocyanin biosynthesis and other processes, consists of MYB, basic helix-loop-helix (bHLH), and WD40 proteins. In the Y2H assay, yeast system vectors were constructed using RcMYB114Red, RcWD40, and RcbHLH. We observed that all three MBW complex proteins interacted with each other in yeast cells (Figure 2C). ChIP analysis illustrated that RcMYB114 selectively bound the RcPAL, RcCHS, RcLAR, RcDFR, RcANS, and RcUFGT promoter regions containing the MYB binding site (AACCTAA) for light-responsive elements (Figure 2D). These results indicate that RcMYB114 encodes a transcription factor protein that interacts with RcWD40 and RcbHLH and is selectively recruited to RcPAL, RcCHS, RcLAR, RcDFR, RcANS, and RcUFGT promoter regions to regulate anthocyanin biosynthesis and accumulation.
A Fragment Insertion (Rosa1) in the Promoter Region of Rcmyb114 Causes Its Expression in Red Rose
We found that RcMYB114Red and RcMYB114White were identical in both form and function. We also found that red roses expressed RcMYB114Red in high quantities while white roses did not express RcMYB114White. To determine the cause of this dramatic difference in transcript abundance between red and white roses on the same plant, we isolated the upstream promoters of RcMYB114Red and RcMYB114White. The promoter sequence of RcMYB114Red (pRcMYB114Red) was approximately 3 kb, but that of RcMYB114White was only 2866 bp (Figures 3A,B). After cloning and sequencing these fragments, we found a 148-bp fragment inserted at −758 bp upstream of the ATG start codon of RcMYB114Red, which was absent in RcMYB114White. The 148-bp fragment was named Rosa1 and contained a cis-acting element binding site for transcription factors. For example, the sense chain had bZIP, TCP, and bHLH domains and the antisense chain had GRF, WRKY, E2F/PD, NAC, and SBP domains (Figure 3C). The element CATTCATACGGAAGTG of SBP is the binding site for MYB transcription factors, which are involved in regulation of flavonoid biosynthesis (Solano et al., 1995). There are seven chromosomes in haploid roses, and we found that the Rosa1 fragment is found in 5–8 locations, distributed across all chromosomes. Rosa1 is mainly inserted in the promoter, 5′UTR, 3′UTR, intron, and intergenic regions of transcription factor genes, including MYB, TIFY, and WD40, and other genes related to growth and development, including zinc finger, wuschel family, adenyltransferase, and CoA reductase (Supplementary Table 1).
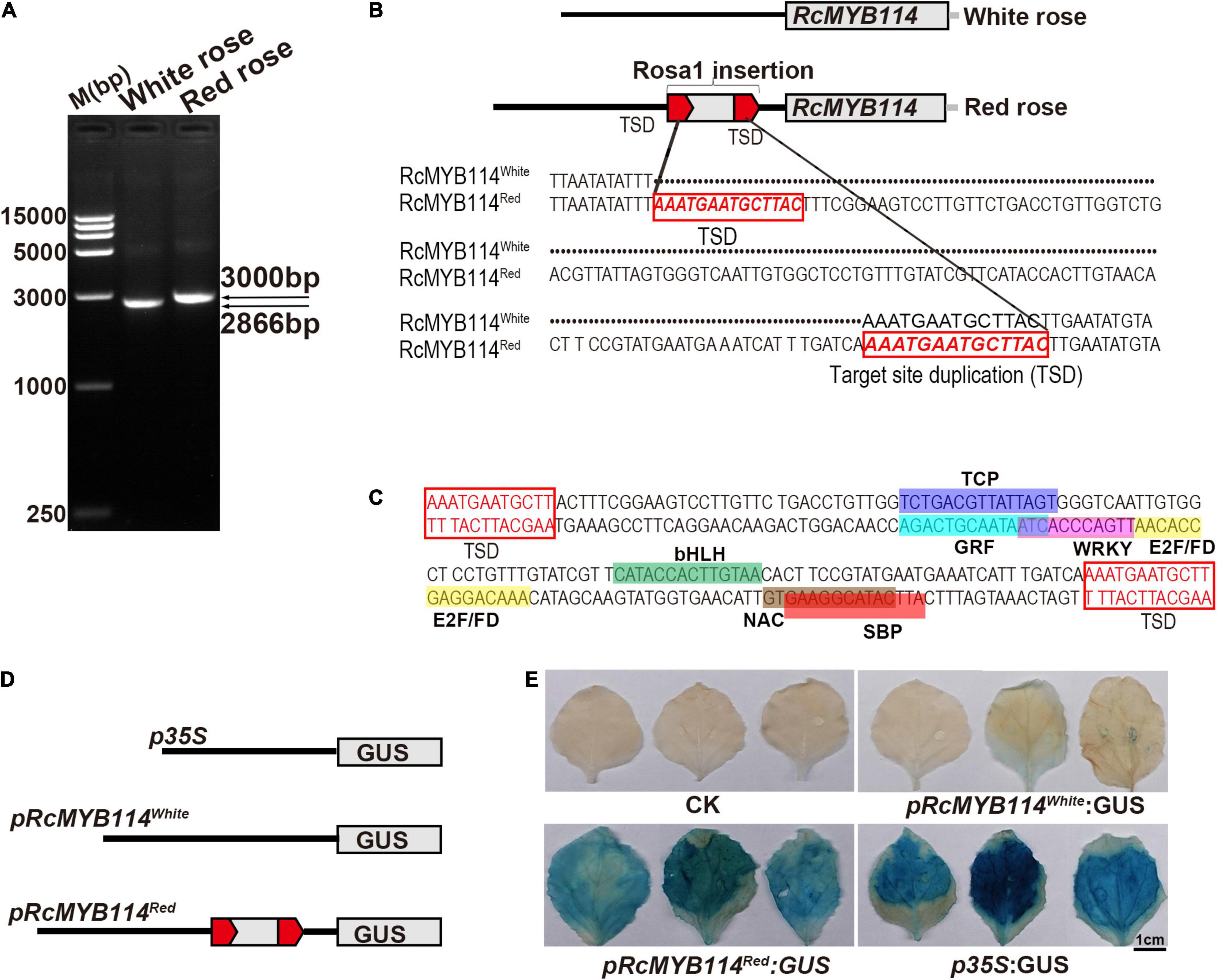
Figure 3. Confirmation that the Rosa1 transposable element-like sequence inserted into the promoter region of RcMYB114 alters its expression. (A) Promoters, pRcMYB114Red and pRcMYB114White, were cloned by PCR amplification. (B) Sequence analysis of Rosa1, a transposable element-like sequence in the promoter of RcMYB114Red. Red frame and letters indicate the target site duplication (TSD) and sequence. (C) Domain analysis of the Rosa1 transposable element-like sequence. (D) Schematic diagram of the pRcMYB114Red and pRcMYB114White promoter-β-Glucuronidase (GUS) fusion vectors. The RcMYB114 promoters replaced the CaMV35S promoter in pBI121. (E) Histochemical assay of GUS activity in N. benthamiana leaves after being infiltrated with the p35S:GUS, pRcMYB114Red:GUS, and pRcMYB114White:GUS constructs.
To investigate whether the Rosa1 inserted in the promoter region of RcMYB114Red alters its expression, we used the GUS reporter system. We constructed pRcMYB114Red (3 kb, including Rosa1) and pRcMYB114White (2866 bp, without Rosa1) promoter-GUS fusion constructs (Figure 3D). The RcMYB114 promoter-GUS fusion vectors were infiltrated into the abaxial surface of N. benthamiana leaves. After GUS staining, the 35S:GUS construct showed the strongest expression, followed by the pRcMYB114Red GUS construct, with the pRcMYB114White GUS construct having the weakest expression. This experiment confirmed that Rosa1 does alter gene expression (Figure 3E).
Next, we analyzed the RcMYB114 gene and its upstream sequence in different rose varieties (red, yellow, white, and green) (Supplementary Table 2), to confirm the universal existence of this transposable element-like sequence and how it is related to RcMYB114 transcript levels and petal color across phenotypes. RcMYB114 was amplified from the genomic DNA of 51 rose varieties (Figure 4). All the tested red-petal varieties showed high expression of RcMYB114, including ‘Slater’s Crimson China,’ ‘Blue River,’ ‘Betty Prior,’ ‘Dortmund,’ ‘Uncle Walter,’ ‘Pierre de Ronasard,’ ‘Hiohgi,’ ‘Hohoemi,’ ‘Red Success,’ ‘Terrazza Voila,’ ‘Wonderful Wife,’ ‘Crimson Glory,’ ‘Red Cap,’ ‘Carola,’ ‘Seba,’ ‘Black Lady,’ ‘Gold Carriage,’ ‘Zajibiaoyan,’ ‘Hana-Busa,’ ‘Huangjiabaxinuo,’ ‘2018-08-3,’ ‘Burgundy Iceberg,’ ‘Xiangchun,’ ‘Dongfanghong,’ ‘Cherry Bonica,’ ‘Red Lace,’ ‘Rhapsody in Blue,’ and ‘Ingrid Bergman.’ RcMYB114 was not expressed in the non-red varieties (green, yellow, and white), including the green varieties ‘Viridiflora,’ ‘Green Star,’ ‘Lvye,’ ‘Éclair,’ and ‘Creamy Eden’; the yellow varieties ‘Golden Celebration,’ ‘Yellow Meilove,’ ‘Adolf Horstmann,’ ‘Golden Scepter,’ ‘Kent Princess,’ ‘Oregold,’ ‘Gold Bunny,’ ‘Australian Gold,’ and ‘Golden Shower’; and the white varieties ‘Baihe,’ ‘2018-31-117,’ ‘White Ohara,’ ‘Bridal White,’ ‘Lvyun,’ ‘Beizhi,’ ‘Snowflake,’ ‘White Satin,’ and ‘Tiantanbai’ (Figure 4). All the red-petal varieties contained the Rosa1 sequence in the promoter of their RcMYB114 gene, and all non-red varieties lacked Rosa1 (Figure 4). This suggests that the Rosa1 insertion in the upstream regulatory sequence of RcMYB114 altered RcMYB114 expression, resulting in anthocyanin biosynthesis accumulation, and was therefore responsible for the red-petal phenotype in rose.
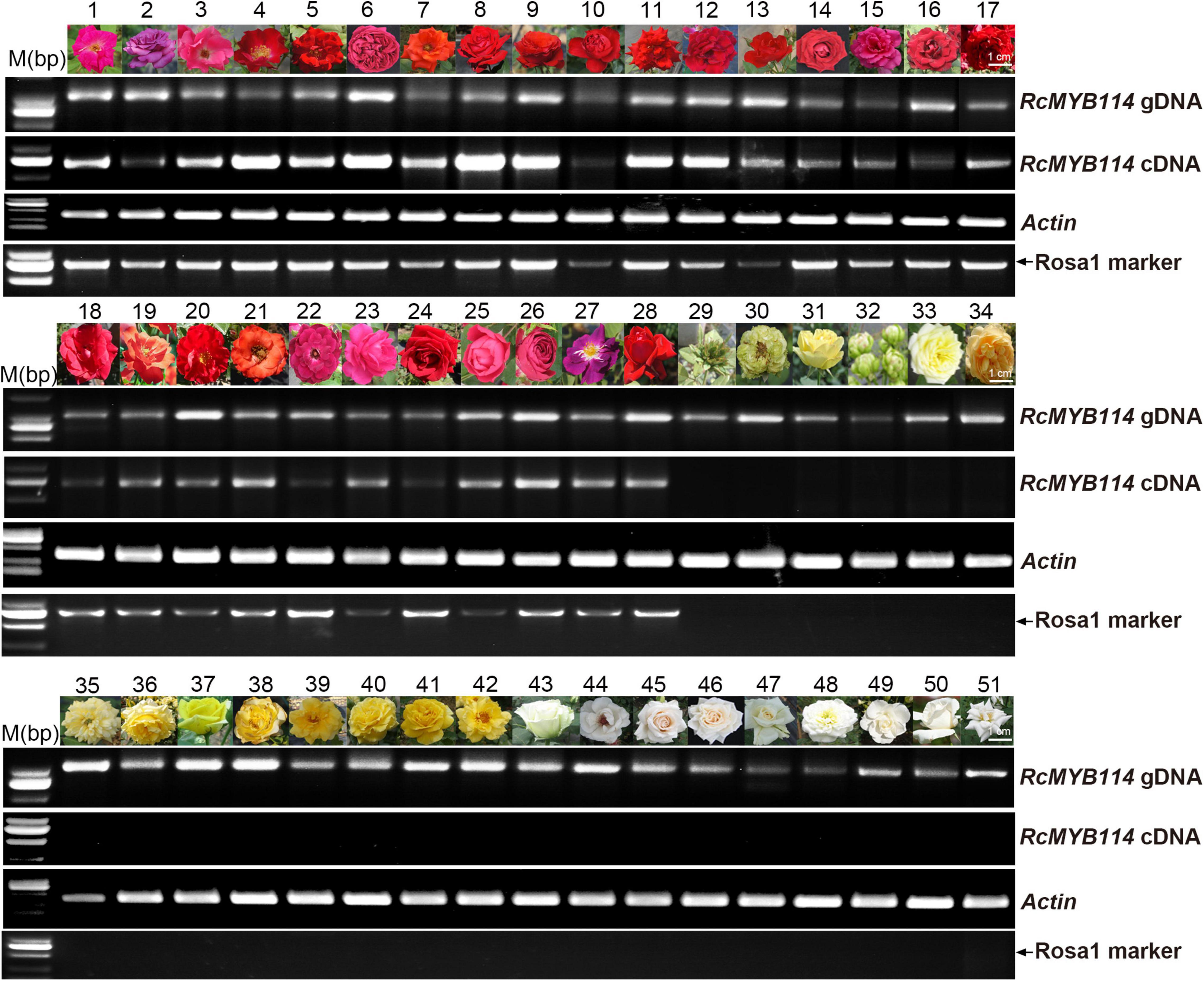
Figure 4. Expression of RcMYB114 across different petal-color phenotypes is dependent on the presence of Rosa1 transposable element-like sequence within the upstream regulatory sequence of RcMYB114. PCR amplification of RcMYB114 from genomic DNA and petal cDNA of 51 rose varieties. Actin was used as a loading control. Only the red-petal varieties contained the Rosa1 transposable element-like sequence.
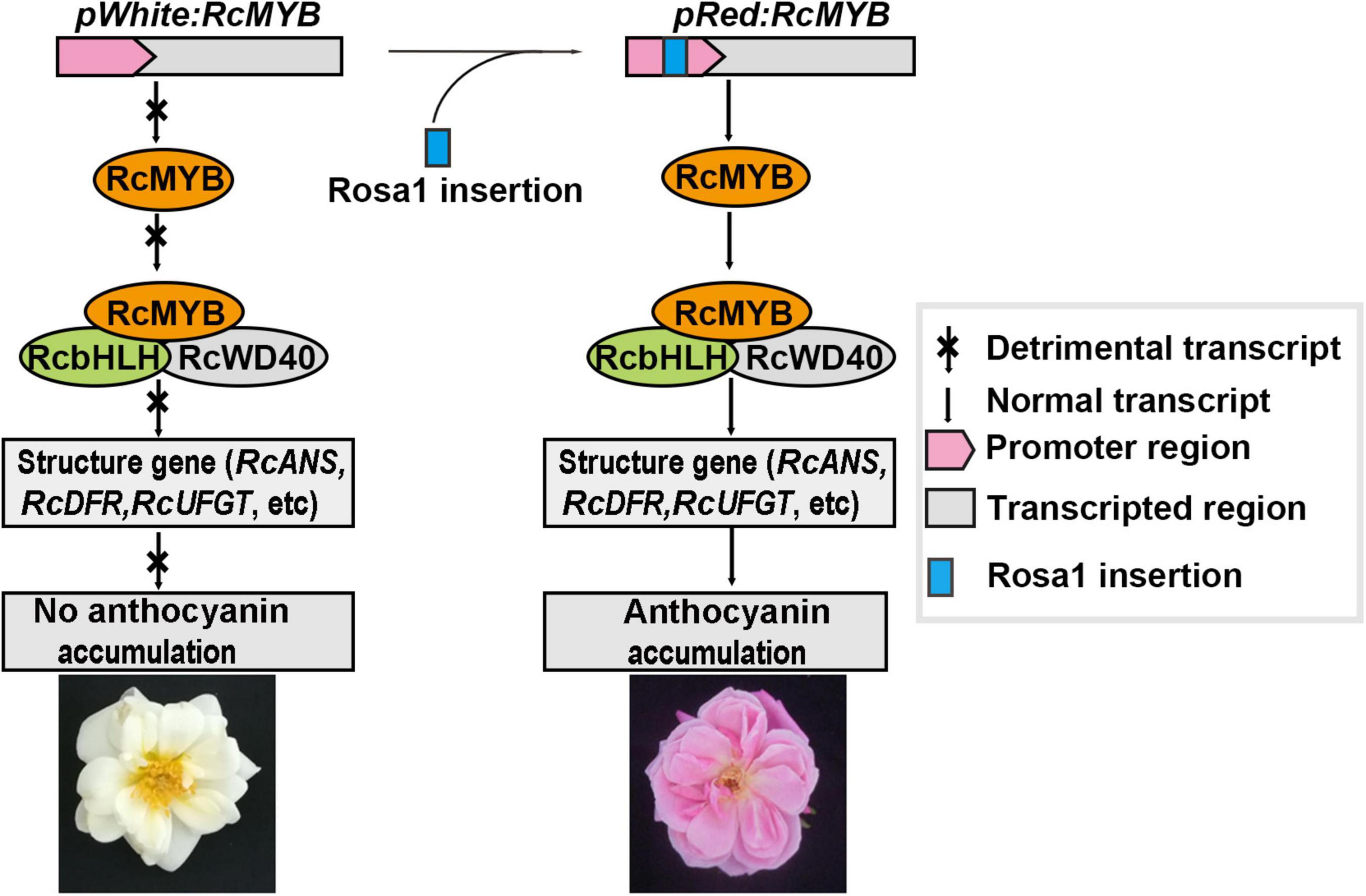
A Regulatory Model for Anthocyanin Biosynthesis and Accumulation in Rose Petals
Rose plants show a rich diversity of flower colors. The formation of petal color depends on the differential activation of pigment biosynthesis genes and accumulation of pigments during flower development. Red petal color is conferred by anthocyanins, and the RcMYB114 transcription factor regulates anthocyanin biosynthesis and accumulation in rose. A Rosa1 fragment insertion can change gene expression by altering transcription. In white, green, and yellow rose varieties, RcMYB114 is not expressed without the Rosa1 fragment insertion during flower development. Without RcMYB114 transcription and translation, a putative canonical MBW complex can’t form, which is responsible for the expression of the key anthocyanin structural genes, such as RcDFR, RcANS, and RcUFGT. Therefore, anthocyanin biosynthesis and accumulation are blocked, resulting in white-, green-, and yellow-petal roses. However, the Rosa1 fragment insertion in the upstream regulatory sequence of RcMYB114 causes its expression during flower development. RcMYB114 is then available to form the canonical MBW complex with RcWD40 and RcbHLH. This promotes the expression of RcDFR, RcANS, RcUFGT, and other downstream anthocyanin structural genes, eventually leading to the accumulation of anthocyanins and red petal color in rose flowers (Figure 5).
Discussion
R2R3-MYB Family Genes Are Critical Regulators of Anthocyanin Accumulation in Plants
Anthocyanins are secondary metabolites that have multiple biological functions in plants (Falcone Ferreyra et al., 2012). The R2R3-MYB transcription factors, regulate the anthocyanin biosynthesis pathway, thus effecting anthocyanin biosynthesis and accumulation (Hichri et al., 2011; Jin et al., 2016; Andersen et al., 2019; Gu et al., 2019; Li et al., 2019; Wang et al., 2020; Jiu et al., 2021; Lin and Rausher, 2021; Yin et al., 2021). The addition or deletion of sequences in the coding region of R2R3-MYB genes can affect their transcription and protein synthesis and ultimately alter the accumulation of anthocyanins. For example, in strawberry (Fragaria × ananassa Duch.), FaMYB10 plays an important role in controlling anthocyanin biosynthesis. An 8-bp ACTTATAC insertion at the C terminus genomic region, leads to a code shift mutation and produces the white octoploid strawberry (Wang et al., 2020). In sweet cherry (P. avium L.), the inheritance of cherry fruit skin color is regulated by a single gene involved in anthocyanin biosynthesis, PavMYB10.1. This gene has three different alleles: PavMYB10.1a, PavMYB10.1b, and PavMYB10.1c. PavMYB10.1a contains an intact 672-bp cDNA sequence, conferring red skin color. A 1-bp deletion in PavMYB10.1b confers blush skin color and an insertion/deletion (indel) in the same sequence region of PavMYB10.1c confers yellow skin color (Jin et al., 2016).
Although different genes control fruit coloration and flower coloration, their regulation mechanisms are very similar. In petunia (Petunia × atkinsiana), the anthocyanin2 (an2) locus, which encodes an R2R3-MYB regulator, is a significant regulator of petal limb color. Other petunia species, including P. integrifolia and P. axillaris, possess several color variants. The an2 allele has two alternative code shifts through insertion into one position, which cause an an2 functorial defect and alter the flower color (Quattrocchio et al., 1999). In snapdragons (Antirrhinum spp.), the Rosea locus, which includes Rosea1, Rosea2, and Venosa MYB-related transcription factors, has three different alleles that regulate the intensities and patterns of magenta anthocyanin pigmentation in petal. Wild-type petals are nearly wholly colored and contain a high concentration of magenta anthocyanin in the corolla. Two mutant alleles (roscol and rosdor) at the Rosea locus are created by indels, resulting in a low level of anthocyanin presentation confined to the petals’ inner epidermis or a low level anthocyanin presentation toward tube’s base and anthocyanin presentation on dorsal lobes’ outer epidermis, respectively (Schwinn et al., 2006). We found that the RcMYB114 gene can affect the accumulation of anthocyanins in roses. According to the analysis results of different varieties with different petal colors, at the genomic level, RcMYB114 gene was amplified in the genomic DNA of all rose varieties. Meanwhile, at the level of gene transcription, the RcMYB114 was only expressed in all red petal variety, but not in non-red rose varieties such as yellow, green and white petals. Also, we found that the transcription level of RcMYB114 was different in all red varieties, and some varieties had very low expression. These results indicated that there are other MYB genes that cooperate with RcMYB114 and form MYBs regulation networks to determine the red color of rose petals.
Rosa1 Plays a Critical Role in Determining Flower Color by Altering the Transcription of R2R3-MYB Genes
Insertion incidents influence nearby gene transcription and raise the mutation rate near the insertion site, leading to the diversification of plant traits (Bennetzen, 2000; Elbarbary et al., 2016; Chuong et al., 2017; Niu et al., 2019; Jo and Kim, 2020). For example, in apple (M. domestica Borkh.), MdMYB1, which is related to the anthocyanin biosynthesis pathway, regulates fruit red skin phenotype. The MdMYB1 gene possesses three different alleles: MdMYB1-1, MdMYB1-2, and MdMYB1-3. The MdMYB1-1 allele is dominant and leads to anthocyanin biosynthesis and red fruit skin. The other two alleles, MdMYB1-2 and MdMYB1-3, lead to limited anthocyanin biosynthesis, producing non-red fruit skin. There is a 4097-bp retrotransposon insertion with two target site duplications upstream of the MdMYB1 promoter region, which controls the development of red skin color in apple (Zhang et al., 2019). Whether this insertion is present or not is stably transferred from one generation to the next (Zhang et al., 2019). In citrus, Ruby, an R2R3-MYB gene, regulates fruit color. A Copia-like retrotransposon inserted into Ruby induces its expression, resulting in the striking red color of Sicilian blood oranges (Citrus sinensis). In addition, the differential expression of Ruby due to various site mutations, deletions, and insertions of transposable elements gives rise to variations in leaf and petal colors in different Citrus species and domesticated cultivars (Butelli et al., 2012, 2017). In grape (Vitis vinifera L.), red fruit color is closely associated with the expression of VvMYBA1, which regulates anthocyanin biosynthesis. Black-skinned cultivars contain a retrotransposon, Gret1, inserted into the upstream region of VvMYBA1. Because no VvMYBA1 transcripts are detected in white-skinned grapes, it can be concluded that the insertion drives expression of VvMYBA1, presenting as dark-skinned grapes (Kobayashi et al., 2004; Walker et al., 2007).
The Rosa1 transposable element we found is a previously unknown DNA transposon, and different from those in apple, citrus, and grape. For example, Rosa1 contains cis-acting element binding sites for transcription factors: the sense chain contains bZIP, TCP, and bHLH domains, and the antisense chain contains GRF, WRKY, E2F/PD, NAC, and SBP domains. In rose, the Rosa1 transposable element, which inserts into RcMYB114’s promoter region, may be considered as an enhancer, promoting the development of red flower color. Cultivars without the Rosa1 transposable element do not effectively produce anthocyanin pigments. Thus, the Rosa1 transposable element can serve as a DNA molecular marker to distinguish red petal roses.
Conclusion
RcMYB114, a R2R3-MYB transcription factor, shares high homology with anthocyanin-regulating genes in many plants. RcMYB114 is part of an MBW complex and selectively recruited to structure gene’s promoter regions to regulate anthocyanin biosynthesis and accumulation in rose. Rosa1, a148-bp transposable element-like, insertion in the promoter region of RcMYB114, enhanced RcMYB114 transcript level and resulted in upregulation of anthocyanin biosynthesis genes, accumulated anthocyanins. Thus, the Rosa1 can alter gene transcription and produce rose red petal.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The raw RNA-seq data were deposited in the CNGB Nucleotide Sequence Archive (https://db.cngb.org/cnsa/) of China National GenBank (CNGB) database (accession number: CNP0001468). The RcMYB114, RcWD40 cDNA, RcbHLH, and Rosa1 are available in the GenBank database under the accession numbers MW239568, MW239571, KY783912, and MW430097.
Author Contributions
WJ designed research. ML, YY, HW, HoZ, XZ, and ZX performed research. ML, HuZ, YF, and PS analyzed data. WJ and ML wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Science and Technology Innovation Ability Construction Projects of Beijing Academy of Agriculture and Forestry Science (KJCX20200205, KJCX202000425, and KJCX20200109) and the National Natural Science Foundation China (32172608).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Li Guanglin from College of Life Sciences, Shanxi Normal University, for providing assistance with data analysis. We also thank George Lomonossoff from John Innes Centre for providing Hyper Trans vector system.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.857684/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/genbank/
- ^ https://www.genome.jp/tools-bin/clustalw
- ^ https://www.megasoftware.net/
- ^ https://lipm-browsers.toulouse.inra.fr/pub/RchiOBHm-V2/
- ^ http://smart.embl-heidelberg.de/
References
Andersen, O. M., Chen, K., Du, L., Liu, H., and Liu, Y. (2019). A novel R2R3-MYB from grape hyacinth, MaMybA, which is different from MaAN2, confers intense and magenta anthocyanin pigmentation in tobacco. Proc. Natl. Acad. Sci. U S A. 19, 20232–20239. doi: 10.1073/pnas.1912741116
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Bennetzen, J. L. (2000). Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 42, 251–269. doi: 10.1023/A:1006344508454
Benvenuti, S., Pellati, F., Melegari, M., and Bertelli, D. (2004). Polyphenols, anthocyanins, ascorbic acid, and radical scavenging activity of Rubus, Ribes, and Aronia. J. Food Sci. 69, 164–169. doi: 10.1111/j.1365-2621.2004.tb13352.x
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394. doi: 10.1105/tpc.12.12.2383
Bowler, C., Benvenuto, G., Laflamme, P., Molino, D., Probst, A., Tariq, M., et al. (2004). Chromatin techniques for plant cells. Plant J. 39, 776–789. doi: 10.1111/j.1365-313X.2004.02169.x
Brown, D. D. (1981). Gene expression in eukaryotes. Science 211, 667–674. doi: 10.1126/science.6256857
Butelli, E., Garcia-Lor, A., Licciardello, C., Las Casas, G., Hill, L., Recupero, G. R., et al. (2017). Changes in Anthocyanin Production during Domestication of Citrus. Plant Physiol. 173, 2225–2242. doi: 10.1104/pp.16.01701
Butelli, E., Licciardello, C., Zhang, Y., Liu, J., Mackay, S., Bailey, P., et al. (2012). Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24, 1242–1255. doi: 10.1105/tpc.111.095232
Carbone, F., Preuss, A., De Vos, R. C., D’Amico, E., Perrotta, G., Bovy, A. G., et al. (2009). Developmental, genetic and environmental factors affect the expression of flavonoid genes, enzymes and metabolites in strawberry fruits. Plant Cell Environ. 32, 1117–1131. doi: 10.1111/j.1365-3040.2009.01994.x
Cheng, G. W., and Breen, P. J. (1991). Activity of phenylalanine ammonia-lyase (Pal) and concentrations of anthocyanins and phenolics in developing strawberry fruit. J. Am. Soc. Horticult. Sci. 116, 865–869. doi: 10.21273/JASHS.116.5.865
Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. G., et al. (2003). Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 31, 3497–3500. doi: 10.1093/nar/gkg500
Chuong, E. B., Elde, N. C., and Feschotte, C. (2017). Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18, 71–86. doi: 10.1038/nrg.2016.139
Drongitis, D., Aniello, F., Fucci, L., and Donizetti, A. (2019). Roles of transposable elements in the different layers of gene expression regulation. Int. J. Mol. Sci. 20, 5755–5766. doi: 10.3390/ijms20225755
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Elbarbary, R. A., Lucas, B. A., and Maquat, L. E. (2016). Retrotransposons as regulators of gene expression. Science 351:aac7247. doi: 10.1126/science.aac7247
Falcone Ferreyra, M. L., Rius, S., and Casati, P. (2012). Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3:222. doi: 10.3389/fpls.2012.00222
Gietz, R. D., Schiestl, R. H., Willems, A. R., and Woods, R. A. (1995). Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360. doi: 10.1002/yea.320110408
Gil, N., and Ulitsky, I. (2020). Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 21, 102–117. doi: 10.1038/s41576-019-0184-5
Goodrich, J., Carpenter, R., and Coen, E. S. (1992). A common gene regulates pigmentation pattern in diverse plant species. Cell 68, 955–964. doi: 10.1016/0092-8674(92)90038-e
Grotewold, E. (2006). The genetics and biochemistry of floral pigments. Ann. Rev. Plant Bol. 57, 761–780. doi: 10.1146/annurev.arplant.57.032905.105248
Gu, Z., Zhu, J., Hao, Q., Yuan, Y. W., Duan, Y. W., Men, S., et al. (2019). A novel R2R3-MYB transcription factor contributes to petal bblotch formation by regulating organ-specificexpression of PsCHS in tree peony (Paeonia suffruticosa). Plant Cell Physiol. 60, 599–611. doi: 10.1093/pcp/pcy232
Guterman, I., Shalit, M., Menda, N., Piestun, D., Dafny-Yelin, M., Shalev, G., et al. (2002). Rose scent: genomics approach to discovering novel floral fragrance-related genes. Plant Cell 14, 2325–2338. doi: 10.1105/tpc.005207
Hibrand Saint-Oyant, L., Ruttink, T., Hamama, L., Kirov, I., Lakhwani, D., Zhou, N. N., et al. (2018). A high-quality genome sequence of Rosa chinensis to elucidate ornamental traits. Nat. Plants 4, 473–484. doi: 10.1038/s41477-018-0166-1
Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S., and Lauvergeat, V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62, 2465–2483. doi: 10.1093/jxb/erq442
Hirsch, C. D., and Springer, N. M. (2017). Transposable element influences on gene expression in plants. Biochim. Biophys. Acta Gene. Regul. Mech. 1860, 157–165. doi: 10.1016/j.bbagrm.2016.05.010
Hsu, C. C., Chen, Y. Y., Tsai, W. C., Chen, W. H., and Chen, H. H. (2015). Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 168, 175–191. doi: 10.1104/pp.114.254599
Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. Embo J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Jin, W., Wang, H., Li, M., Wang, J., Yang, Y., Zhang, X., et al. (2016). The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 14, 2120–2133. doi: 10.1111/pbi.12568
Jiu, S., Guan, L., Leng, X., Zhang, K., Haider, M. S., Yu, X., et al. (2021). The role of VvMYBA2r and VvMYBA2w alleles of the MYBA2 locus in the regulation of anthocyanin biosynthesis for molecular breeding of grape (Vitis spp.) skin coloration. Plant Biotechnol. J. 19, 1216–1239. doi: 10.1111/pbi.13543
Jo, C., and Kim, S. (2020). Transposition of a non-autonomous DNA transposon in the gene coding for a bHLH transcription factor results in a white bulb color of onions (Allium cepa L.). Theor. Appl. Genet. 133, 317–328. doi: 10.1007/s00122-019-03460-8
Kiferle, C., Fantini, E., Bassolino, L., Povero, G., Spelt, C., Buti, S., et al. (2015). Tomato R2R3-MYB Proteins SlANT1 and SlAN2: Same Protein Activity, Different Roles. PLoS One 10:e0136365. doi: 10.1371/journal.pone.0136365
Kobayashi, S., Goto-Yamamoto, N., and Hirochika, H. (2004). Retrotransposon-induced mutations in grape skin color. Science 304:982. doi: 10.1126/science.1095011
Koo, J., Kim, Y., Kim, J., Yeom, M., Lee, I. C., and Nam, H. G. (2007). A GUS/Luciferase fusion reporter for plant gene trapping and for assay of promoter activity with luciferin-dependent control of the reporter protein stability. Plant Cell Physiol. 48, 1121–1131. doi: 10.1093/pcp/pcm081
Kranz, H. D., Denekamp, M., Greco, R., Jin, H., Leyva, A., Meissner, R. C., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276. doi: 10.1046/j.1365-313x.1998.00278.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, C., Qiu, J., Huang, S., Yin, J., and Yang, G. (2019). AaMYB3 interacts with AabHLH1 to regulate proanthocyanidin accumulation in Anthurium andraeanum (Hort.)-another strategy to modulate pigmentation. Mol. Genet. Genom. 294, 469–478. doi: 10.1007/s00438-018-1525-3
Li, M., Yang, Y., Wang, H., Liu, J., and Wanmei, J. (2017). Cloning and expression of bHLH Gene from Rosa chinensis‘Slater’s Crimson China’and its interaction with transcription factors MYB and WD40. Acta Horticult. Sin. 44, 1949–1958. doi: 10.16420/j.issn.0513-353x.2017-0183
Lin, R. C., and Rausher, M. D. (2021). R2R3-MYB genes control petal pigmentation patterning in Clarkia gracilis ssp. sonomensis (Onagraceae). New Phytol. 229, 1147–1162. doi: 10.1111/nph.16908
Lin-Wang, K., Bolitho, K., Grafton, K., Kortstee, A., Karunairetnam, S., McGhie, T. K., et al. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10:50. doi: 10.1186/1471-2229-10-50
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Marmol, I., Sanchez-de-Diego, C., Jimenez-Moreno, N., Ancin-Azpilicueta, C., and Rodriguez-Yoldi, M. J. (2017). Therapeutic applications of rose hips from different Rosa species. Int. J. Mol. Sci. 18:1137. doi: 10.3390/ijms18061137
Martin, C., and Gerats, T. (1993). Control of pigment biosynthesis genes during petal development. Plant Cell 5, 1253–1264. doi: 10.1105/tpc.5.10.1253
Matus, J. T., Aquea, F., and Arce-Johnson, P. (2008). Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 8:83. doi: 10.1186/1471-2229-8-83
McClintock, B. (1950). The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. U S A. 36, 344–355. doi: 10.1073/pnas.36.6.344
Meng, Y. L., Li, N., Tian, J., Gao, J. P., and Zhang, C. Q. (2013). Identification and validation of reference genes for gene expression studies in postharvest rose flower (Rosa hybrida). Sci. Horticult. 158, 16–21. doi: 10.1016/j.scienta.2013.04.019
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nakayama, K., and Kataoka, N. (2019). Regulation of gene expression under hypoxic conditions. Int. J. Mol. Sci. 20, 3278–3293. doi: 10.3390/ijms20133278
Niu, X. M., Xu, Y. C., Li, Z. W., Bian, Y. T., Hou, X. H., Chen, J. F., et al. (2019). Transposable elements drive rapid phenotypic variation in Capsella rubella. Proc. Natl. Acad. Sci. U S A. 116, 6908–6913. doi: 10.1073/pnas.1811498116
Pan, Q.-H., Zou, K.-Q., Peng, C.-C., Wang, X.-L., and Zhang, D.-P. (2005). Purification, biochemical and immunological characterization of acid invertases from apple fruit. J. Integrat. Plant Biol. 47, 50–59. doi: 10.1111/j.1744-7909.2005.00018.x
Paolocci, F., Robbins, M. P., Passeri, V., Hauck, B., Morris, P., Rubini, A., et al. (2011). The strawberry transcription factor FaMYB1 inhibits the biosynthesis of proanthocyanidins in Lotus corniculatus leaves. J. Exp. Bot. 62, 1189–1200. doi: 10.1093/jxb/erq344
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P. A., and Saedler, H. (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6, 3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x
Pucker, B., Pandey, A., Weisshaar, B., and Stracke, R. (2020). The R2R3-MYB gene family in banana (Musa acuminata): Genome-wide identification, classification and expression patterns. PLoS One 15:e0239275. doi: 10.1371/journal.pone.0239275
Quattrocchio, F., Wing, J., van der Woude, K., Souer, E., de Vetten, N., Mol, J., et al. (1999). Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11, 1433–1444. doi: 10.1105/tpc.11.8.1433
Raymond, O., Gouzy, J., Just, J., Badouin, H., Verdenaud, M., Lemainque, A., et al. (2018). The Rosa genome provides new insights into the domestication of modern roses. Nat. Genet. 50, 772–777. doi: 10.1038/s41588-018-0110-3
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Sainsbury, F., Thuenemann, E. C., and Lomonossoff, G. P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693. doi: 10.1111/j.1467-7652.2009.00434.x
Schaart, J. G., Dubos, C., Romero De La Fuente, I., van Houwelingen, A., de Vos, R. C. H., et al. (2013a). Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 197, 454–467. doi: 10.1111/nph.12017
Schaart, J. G., Dubos, C., Romero De La Fuente, I., van Houwelingen, A. M., de Vos, R. C., et al. (2013b). Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria x ananassa) fruits. New Phytologist. 197, 454–467. doi: 10.1111/nph.12017
Schwinn, K., Venail, J., Shang, Y., Mackay, S., Alm, V., Butelli, E., et al. (2006). A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18, 831–851. doi: 10.1105/tpc.105.039255
Signor, S. A., and Nuzhdin, S. V. (2018). The evolution of gene expression in cis and trans. Trends Genet. 34, 532–544. doi: 10.1016/j.tig.2018.03.007
Solano, R., Nieto, C., and Paz-Ares, J. (1995). MYB.Ph3 transcription factor from Petunia hybrida induces similar DNA-bending/distortions on its two types of binding site. Plant J. 8, 673–682. doi: 10.1046/j.1365-313x.1995.08050673.x
Song, T., Li, K., Wu, T., Wang, Y., Zhang, X., Xu, X., et al. (2019). Identification of new regulators through transcriptome analysis that regulate anthocyanin biosynthesis in apple leaves at low temperatures. PLoS One 14:e0210672. doi: 10.1371/journal.pone.0210672
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Spelt, C., Quattrocchio, F., Mol, J. N., and Koes, R. (2000). anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12, 1619–1632. doi: 10.1105/tpc.12.9.1619
Stracke, R., Holtgrawe, D., Schneider, J., Pucker, B., Sorensen, T. R., and Weisshaar, B. (2014). Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol. 14:249. doi: 10.1186/s12870-014-0249-8
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/s1369-5266(00)00199-0
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tanaka, Y., Nakamura, N., and Togami, J. (2008). Altering flower color in transgenic plants by RNAi-mediated engineering of flavonoid biosynthetic pathway. Methods Mol. Biol. 442, 245–257. doi: 10.1007/978-1-59745-191-8_17
Walker, A. R., Lee, E., Bogs, J., McDavid, D. A., Thomas, M. R., and Robinson, S. P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49, 772–785. doi: 10.1111/j.1365-313X.2006.02997.x
Wang, H., Zhang, H., Yang, Y., Li, M. F., Zhang, Y. T., Liu, J., et al. (2020). The control of red colour by a family of MYB transcription factors in octoploid strawberry (Fragaria x ananassa) fruits. Plant Biotechnol. J. 18, 1169–1184. doi: 10.1111/pbi.13282
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Xu, H., Wang, N., Liu, J., Qu, C., Wang, Y., Jiang, S., et al. (2017). The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 94, 149–165. doi: 10.1007/s11103-017-0601-0
Yamagishi, M., Shimoyamada, Y., Nakatsuka, T., and Masuda, K. (2010). Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of asiatic hybrid lily. Plant Cell Physiol. 51, 463–474. doi: 10.1093/pcp/pcq011
Yao, G., Ming, M., Allan, A. C., Gu, C., Li, L., Wu, X., et al. (2017). Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 92, 437–451. doi: 10.1111/tpj.13666
Yin, X., Zhang, Y., Zhang, L., Wang, B., Zhao, Y., Irfan, M., et al. (2021). Regulation of MYB transcription factors of anthocyanin synthesis in Lily flowers. Front. Plant Sci. 12:761668. doi: 10.3389/fpls.2021.761668
Yuan, Y. W., Sagawa, J. M., Frost, L., Vela, J. P., and Bradshaw, H. D. Jr. (2014). Transcriptional control of floral anthocyanin pigmentation in monkeyflowers (Mimulus). New Phytologist. 204, 1013–1027. doi: 10.1111/nph.12968
Keywords: Rosa1 transposable element-like, RcMYB114, transcription, petal color, rose, anthocyanin
Citation: Li M, Zhang H, Yang Y, Wang H, Xue Z, Fan Y, Sun P, Zhang H, Zhang X and Jin W (2022) Rosa1, a Transposable Element-Like Insertion, Produces Red Petal Coloration in Rose Through Altering RcMYB114 Transcription. Front. Plant Sci. 13:857684. doi: 10.3389/fpls.2022.857684
Received: 18 January 2022; Accepted: 11 April 2022;
Published: 29 April 2022.
Edited by:
Jian Wu, China Agricultural University, ChinaReviewed by:
Wenjun Huang, Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden (CAS), ChinaYaping Kou, Institute of Vegetables and Flowers (CAAS), China
Copyright © 2022 Li, Zhang, Yang, Wang, Xue, Fan, Sun, Zhang, Zhang and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanmei Jin, andtMDgwOUAxNjMuY29t
 Maofu Li
Maofu Li Hui Zhang
Hui Zhang Yuan Yang
Yuan Yang Hua Wang
Hua Wang Zhen Xue
Zhen Xue Youwei Fan
Youwei Fan Pei Sun
Pei Sun Hong Zhang
Hong Zhang Xinzhu Zhang
Xinzhu Zhang Wanmei Jin
Wanmei Jin