
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 25 March 2022
Sec. Plant Development and EvoDevo
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.820439
This article is part of the Research Topic Recent Advances in Flower and Fruit Development in Perennial plants View all 22 articles
 Qinchang Chen1,2
Qinchang Chen1,2 Xiaodan Zhang3
Xiaodan Zhang3 Yaxue Fang4
Yaxue Fang4 Baiyu Wang4
Baiyu Wang4 Shaosi Xu1,2
Shaosi Xu1,2 Kai Zhao1,2
Kai Zhao1,2 Jisen Zhang4*
Jisen Zhang4* Jingping Fang1,2*
Jingping Fang1,2*
Longan (Dimocarpus longan Lour.) is a productive fruit crop with high nutritional and medical value in tropical and subtropical regions. The MYB gene family is one of the most widespread plant transcription factor (TF) families participating in the flowering regulation. However, little is known about the MYB TFs involved in the flowering process in longan and its regulatory network. In this study, a total of 119 DlR2R3-MYB genes were identified in the longan genome and were phylogenetically grouped into 28 subgroups. The groupings were supported by highly conserved gene structures and motif composition of DlR2R3-MYB genes in each subgroup. Collinearity analysis demonstrated that segmental replications played a more crucial role in the expansion of the DlR2R3-MYB gene family compared to tandem duplications, and all tandem/segmental duplication gene pairs have evolved under purifying selection. Interspecies synteny analysis among longan and five representative species implied the occurrence of gene duplication events was one of the reasons contributing to functional differentiation among species. RNA-seq data from various tissues showed DlR2R3-MYB genes displayed tissue-preferential expression patterns. The pathway of flower development was enriched with six DlR2R3-MYB genes. Cis-acting element prediction revealed the putative functions of DlR2R3-MYB genes were related to the plant development, phytohormones, and environmental stresses. Notably, the orthologous counterparts between Arabidopsis and longan R2R3-MYB members tended to play conserved roles in the flowering regulation and stress responses. Transcriptome profiling on off-season flower induction (FI) by KClO3 indicated two up-regulated and four down-regulated DlR2R3-MYB genes involved in the response to KClO3 treatment compared with control groups. Additionally, qRT-PCR confirmed certain genes exhibited high expression in flowers/flower buds. Subcellular localization experiments revealed that three predicted flowering-associated MYB proteins were localized in the nucleus. Future functional studies on these potential candidate genes involved in the flowering development could further the understanding of the flowering regulation mechanism.
Myeloblastosis (MYB) proteins were first identified in the avian myeloblastosis virus (AMV) (Klempnauer et al., 1982), but have since been found in all eukaryote genomes and belong to one of the largest transcription factor (TF) gene families in plants. The first plant MYB gene identified from Zea mays, known as COLORED1 (C1), was involved in the regulation of anthocyanin biosynthesis (Paz-Ares et al., 1987). The MYB TF contains a highly conserved MYB DNA-binding domain (DBD) in the N-terminal region that is usually composed of 1–4 serial imperfect repeat sequences. Each repeat sequence covers about 50–55 amino acid residues including three regularly spaced tryptophan (W) residues [-W-(X19)-W-(X19)-W…-F/I-(X-18)-W–(X-18)-W-], which fold into three α-helices (Kanei-Ishii et al., 1990; Kranz et al., 2000). The second and third helices form a helix-turn-helix (HTH) structure, and the third α-helix directly participates in binding with DNA (Ogata et al., 1992; Williams and Grotewold, 1997; Jia et al., 2004). MYB TFs can be divided into four categories based on the number of MYB repeats in their DBDs: 4R-MYB (four repeats), R1R2R3-MYB (three repeats), R2R3-MYB (two repeats), and 1RMYB (one repeat). The R2R3-MYB genes have received significant attention because they are massively expanded in the plant lineage and involved in regulating a wide array of plant-specific biological processes, such as primary and secondary metabolism, response to plant hormones and environmental stresses, disease resistance and leaf morphogenesis (Roy, 2016; Millard et al., 2019a; Jiang and Rao, 2020). The evolutionary history of the R2R3-MYB (2R-MYBs) gene family throughout the eukaryotic kingdom remains unknown (Du et al., 2015). The “loss” model proposes that R2R3-MYB genes potentially evolved from R1R2R3-MYB progenitor genes through the loss of the R1 repeat sequence (Rosinski and Atchley, 1998; Braun and Grotewold, 1999). In contrast, the more convincing “gain” model hypothesizes that R1R2R3-MYB genes evolved from the R2R3-MYBs via intragenic structural domain duplication (Jiang et al., 2004; Du et al., 2015).
With the rapid development of high-throughput sequencing technologies, genome-wide identification of R2R3-MYB proteins has been achieved in many plant species, for instance, Arabidopsis thaliana (Stracke et al., 2001; Dubos et al., 2010), Medicago truncatula (Li et al., 2019), Casuarina equisetifolia (Wang et al., 2021), Solanum tuberosum (Sun et al., 2019), Oryza sativa (Katiyar et al., 2012), and Capsicum annuum (Wang et al., 2020). In general, the number of R2R3-MYBs in plants ranged in size from 70 to 285, and these MYB genes evolved mainly based on the natural evolution of organisms and the recombination and amplification of genomes (Chang and Duda, 2012). The highest number of R2R3-MYBs (285) was found in the banana genome (Pucker et al., 2020). Respectively, 244, 192, 157, and 134 R2R3-MYB genes have been identified in the soybean (Du et al., 2012b), populus (Wilkins et al., 2009), maize (Du et al., 2012a), and grape (Wong et al., 2016) based on previous studies. In addition, the lowest number of R2R3-MYB genes, a total of 70, were identified in the sugar beet genome (Stracke et al., 2014).
R2R3-MYB TFs as regulatory proteins play crucial roles in plant growth and development (Kranz et al., 2000; Dubos et al., 2010). With regards to the flower development, AtMYB33 and AtMYB65 were microRNA-regulated genes that could promote anther and pollen development (Millar and Gubler, 2005). AtMYB80 regulated pollen development and programed cell death in the chorioallantoic layer (Phan et al., 2011). AtMYB106 regulated the flowering time of plants by inhibiting the expression of the Flowering Locus T (AtFT) gene, which is a negative regulator in plant flowering development (Hong et al., 2021). In chrysanthemum, CmMYB2 interacted with CmBBX24 to influence the synthesis of gibberellin and ultimately regulated flowering (Zhu et al., 2020). In addition, the R2R3-MYB TFs are known to be widely involved in the regulation of plant specialized metabolism. Previous studies showed that several AtR2R3-MYB genes such as AtMYB11/PFG1, AtMYB12/PFG1, AtMYB111/PFG3, AtMYB21, and AtMYB24 were involved in regulating the flavonol biosynthesis pathway in Arabidopsis (Stracke et al., 2007; Dubos et al., 2010; Shan et al., 2020). Overexpression of a flavonol regulator, MtMYB134, in hairy roots of Medicago truncatula could enhance the biosynthesis of various flavonol derivatives (Naik et al., 2021). AtMYB75/PAP1, AtMYB90/PAP2, AtMYB113, and AtMYB114 were identified as regulators of anthocyanin biosynthesis (Teng et al., 2005; Gonzalez et al., 2008). R2R3-MYB family genes are also widely involved in the plant responses to biotic and abiotic stress besides participating in various physiological activities. AtMYB15 and AtMYB44 could enhance drought and salt tolerance by regulating stomatal closure under abscisic acid (ABA) treatments (Jung et al., 2008; Ding et al., 2009). AtMYB96 increased resistance to low temperature through promoting the expression of C-repeat binding factor (CBF) (Lee and Seo, 2015). The overexpression of AtMYB96 also enhanced drought tolerance by mediating ABA signaling and regulating the target gene Lipid transfer protein 3 (LTP3) (Seo et al., 2009). In other species, LcMYB2 promoted seed germination and root growth under drought stress (Zhao et al., 2019). Overexpression of ZmMYB3R improved tolerance to drought and salt stress in transgenic Arabidopsis plants (Wu et al., 2019).
Longan (Dimocarpus longan Lour., 2n = 2x = 30) is an important tropical and subtropical fruit tree of the Sapindaceae family in the Sapindales order, widely grown in southern China and Southeast Asia (Matsumoto, 2006). As one of the cradles of longan, China currently ranks first in the longan cultivation acreage and yields in the world accounting for 50% of global longan production. Longan has high nutritional and medicinal values and mainly uses its palatable and juicy fruit for commercial purposes (Rangkadilok et al., 2005; Zhang et al., 2020). Flower induction (FI) is an important regulatory process in the transition from vegetative to reproductive growth in plants. Environmental factors like low temperature, drought, and oxidative stress can regulate the flowering of many plants. Potassium chlorate (KClO3) treatment is one of the most effective ways that widely apply in longan production to induce off-season flowering (Huang et al., 2021). To the best of our knowledge, KClO3-induced flowering has only been utilized in longan (Huang et al., 2021). Studies on the physiological mechanism of KClO3-mediated FI showed that KClO3 could increase the content of hormones such as cytokinin (CK) and salicylic acid (SA) in longan, which are closely related to plant flower formation (Sringarm et al., 2009). The KClO3 treatment up-regulated CK concentration together with DlFT1 expression in mature leaves (Winterhagen et al., 2020). In addition, KClO3 treatment caused changes in the carbon to nitrogen ratio by decreasing the starch content and increasing the soluble sugar, sucrose and soluble amino acid content, which provided nutrients for the flower bud differentiation (Chang, 2010). A regulatory model of FI by KClO3 in longan has been proposed: KClO3 may be reduced to chlorite and hypochlorite under the action of nitrate reductase (NR) and nitrite reductase (NiR) to directly induce stress responses, leading to an increase in CK content and expression of flowering-related genes (Borges et al., 2004; Cho et al., 2017). Thus far, several flowering-related genes have been identified and analyzed in longan such as flowering locus T (DlFT), gigantea (DlGI), early flowering 4 (DlELF4), flavin-binding, kelch repeat, F-box 1 (DlFKF1), short vegetative phase (DlSVP) and APETALA1-like (DlAP1) genes (Winterhagen et al., 2013; Jia et al., 2014; Huang F. N. et al., 2017; Waheed et al., 2020) but the molecular mechanism underlying longan FI and floral organ development is largely unknown.
The R2R3-MYB genes comprise one of the largest TF families in longan. Previous studies have identified 35 (Zheng et al., 2020) and 98 (Lin et al., 2017) R2R3-MYB genes based on transcriptome data and the Next Generation Sequencing (NGS)-based assembled genome sequences, respectively. Recently, a high-quality chromosome-level genome of longan “Shixia” (“SX”) was assembled by our team (NCBI GenBank: JAIFKA000000000.1), which will help uncover the distribution, evolution and function of genome-wide R2R3-MYB TFs. By combining PacBio long reads, HiC data and genetic maps, the longan genome was assembled into 483.32 Mb with a contig N50 size of 763.91 kb, which were further anchored onto 15 pseudochromosomes with a 99.30% anchored rate. Benchmarking Universal Single-Copy Orthologs (BUSCO) assessment of the genome was 94.6%, which was an indicator of good quality assembly. Transcriptome dynamics revealed that six associated genes including one MYB transcript factor (Dil.04g005810.2.t1) were closely related to the flowering time under the KClO3 treatment (unpublished data). Studying the MYB gene expression profiles associated with off-season FI by KClO3 in longan will provide a deeper understanding of the molecular regulation mechanism of the flowering process in woody plants.
In this present study, we identified 119 DlMYB genes from the “Shixia” longan genome sequences and divided them into 28 subgroups. The DlMYB gene sequences were further investigated for their characterization, phylogenetic relationship, gene structure, subcellular localization, conserved motifs, cis-acting elements, chromosome distribution, synteny analysis, selection pressure, and gene duplication. In addition, the RNA-seq data showed expression patterns of DlMYBs in nine different tissues and in flower bud tissues treated with KClO3 at different stages which assisted us in determining the potential role of DlMYBs in longan flowering. The qRT-PCR method was used to verify the expression levels of the selected MYB genes in longan tissues. Moreover, three potential flowering-associated MYB genes were further experimentally validated by their subcellular localization in Arabidopsis protoplasts. This is the first systematic report on the genome-wide characterization of the MYB gene family in longan. This study will help investigate members of the R2R3-MYB family involved in the flowering process in longan and the regulatory network, which will pave the way for future functional studies and candidate gene selection for genetic improvement in longan.
Longan cultivar trees (“Shixia,” abbreviated “SX”) about 4− to 6− year-olds in Maoming, Guangdong, China, were selected for treatment with white KClO3 powder of >99% purity in 2016. The apical buds were collected under controlled conditions and KClO3 treatment from longan trees at ten time points with three biological replications including 0 day after treatment (DAT) (November 18th, 2016), 5 DAT (November 23th, 2016), 10 DAT (November 28th, 2016), 15 DAT (December 3rd, 2016), 20 DAT (December 8th, 2016), 25 DAT (December 13th, 2016), 30 DAT (December 18th, 2016), 35 DAT (December 23th, 2016), 41 DAT (December 29th, 2016) and 54 DAT (January 11th, 2017). Immediately after harvesting, all tissue samples were frozen in liquid nitrogen and then stored at –80°C prior to the RNA isolation.
Total RNAs were extracted from collected samples with three biological replicates using the RNAprep Pure Plant Kit (DP432, TIANGEN Biotech, Beijing, China) according to the manufacturer’s protocol. The quality of total RNAs was verified on an Agilent 2100 Bioanalyzer (Plant RNA Nano Chip, Agilent, CA, United States). After quantity and quality determination, RNA-Seq libraries were constructed using the Illumina® TruSeq™ RNA Sample Preparation Kit [RS-122-2001(2), Illumina] and then subjected to Illumina transcriptome short reads sequencing on Illumina X Ten platform with paired-end (PE) 150 nt mode (San Diego, CA, United States).
The genomic data of Dimocarpus longan “Shixia” (GenBank accession No.: JAIFKA000000000.1), Litchi chinensis (GenBank accession No.: JAIUGE000000000.1), Oryza sativa (GenBank accession No.: RPSM01000000), and Zea mays (GenBank accession No.: CABHLF010000000) were downloaded from the National Center for Biotechnology Information (NCBI). The data for Xanthoceras sorbifolium was downloaded from GigaDB (Sneddon et al., 2012). The data for Arabidopsis thaliana R2R3-MYB genes were retrieved from The Arabidopsis Information Resource (TAIR) database. The RNA-seq data of “Sijimi” (“SJM”) longan were downloaded from the NCBI (GenBank accession No.: PRJNA329283), including nine tissues (roots, stems, leaves, flower buds, flowers, young fruits, pulps, pericarps, and seeds) from a previous study (Lin et al., 2017).
The Hidden Markov Model (HMM) profiles for the Myb_DNA-binding domain (PF00249) and SWIRM domain (PF04433) were downloaded from the Pfam protein family database. The specific identification method of R2R3-MYB genes was as follows. The largest number of R2R3-MYB proteins were identified based on the HMM profiles of the MYB domain and SWIRM domain by using HMMER 3.0 with E-values ≤ 0.01 threshold (Potter et al., 2018). Gene sequences containing MYB domains but no SWIRM domain were considered as putative MYB genes. All longan R2R3-MYB genes were verified using AtR2R3-MYB gene sequences as target sequences by BLASTP, with the following parameters: E-value cut-off of 1e–5 and 30% identity. These sequences were manually inspected by the NCBI Conserved Domain Database (NCBI-CDD) and the SMART website to ensure that the genes contained two MYB domains. The full-length amino acid sequences of DlMYB proteins were submitted to the Expasy website (Gasteiger et al., 2003) to predict their molecular weights (MW), isoelectric points (PIs), hydrophilia, and instability. Detailed information on the DlMYB genes is listed in Supplementary Table 1. Multiple sequence alignments of the MYB domains were performed by Clustal X (2.1) (Larkin et al., 2007) with default parameters. The amino acid sequences in MYB domains were visualized by GeneDoc (Nicholas et al., 1997). Weblogo (Crooks et al., 2004) was used to display the sequence logos of R2 and R3 MYB domain repeats based on the multiple alignment files.
The multiple sequence alignments of the full-length R2R3-MYB protein sequences from longan and Arabidopsis were performed by Muscle v3.8.1 (Edgar, 2004) with default parameters. The phylogenetic tree of R2R3-MYB proteins from both genomes was constructed using the Maximum Likelihood (ML) method of RAxML (Stamatakis, 2014), with the following parameters: JTT model and 1,000 bootstrap replications. The Evolview (Subramanian et al., 2019) was used to visualize the ML phylogenetic tree. Longan R2R3-MYB proteins were grouped according to the previously reported AtR2R3-MYB family classification (S1–S25) (Stracke et al., 2001; Dubos et al., 2010). Another ML phylogenetic tree of R2R3-MYB protein sequences from the longan genome was constructed using the same method.
The MEME v5.3.3 (Multiple expectation maximization for Motif Elicitation) (Bailey et al., 2009) online tool was used to determine the conserved motifs in DlMYB amino acid sequences, with the following parameters: zero or one occurrence per sequence; maximum number of motif to find, 20; minimum width of motif, 6; and maximum width of motif, 200. Weblogo (Crooks et al., 2004) was used to display these conserved motifs.
The exon/intron structure patterns of DlMYB genes, including intron distribution patterns and intron-exon boundaries, were graphically visualized by the TBtools v1.087 (Chen et al., 2020), using the nucleotide sequences of DlMYB genes and CDSs (coding sequences). The 2-kb sequences upstream of each longan R2R3-MYB gene sequence were submitted to the PlantCARE database (Lescot et al., 2002) to predict cis-acting elements. The predicted results of gene structures and cis-acting elements were visualized by TBtools program (Chen et al., 2020).
The detailed information of the physical locations of DlMYB genes were generated based on the longan genome sequences (NCBI GenBank: JAIFKA000000000.1). The TBtools was used to exhibit chromosomal locations of DlMYB genes using the gene location visualization program. The Multiple Collinearity Scan toolkit (MCScanX) (Wang et al., 2012) and TBtools were used to analyze and visualize DlMYB gene replication events with E-value cut-off of < 1e–10. The Dual Systeny Plot program in TBtools (Chen et al., 2020) was adopted to elucidate the synteny relationship of orthologous R2R3-MYB genes between longan and other five species, including three dicots Arabidopsis, litchi and yellow horn, and two monocots rice and maize. The Ka/Ks (non-synonymous/synonymous substitution ratio) values were used to determine the selection pressure after replication. Ka/Ks > 1 signifies purified selection, Ka/Ks = 1 indicates neutral selection, and Ka/Ks < 1 represents positive selection (Hurst, 2002). The MS model in KaKs_calculator 2.0 (Wang et al., 2010) was used to calculate the values of Ka, Ks, and Ka/Ks of MYB duplicated gene pairs.
Subcellular localization was used to analyze the specific location of the DlMYB genes within the cell. The subcellular localization and nuclear localization signal (NLS) were predicted using the YLoc (Briesemeister et al., 2010) and NLStradamus online tool (Nguyen Ba et al., 2009), respectively. Three flower-preferential expressed DlMYB genes, DlMYB16, DlMYB72, and DlMYB116 were selected for further experimental validation of their subcellular localizations. Total RNAs were extracted from leaves, flowers and flower buds of “Shixia” longan plants using the RNAprep pure Plant Kit (Tiangen, #DP432) according to the manufacturer’s protocol. The full-length coding sequences excluding stop codon of these three R2R3-MYB genes were amplified and cloned into the BsaI/Eco31I restriction sites of the pBWA(V)HS-GLosgfp vector (Supplementary Figure 5; Sun et al., 2018). Briefly, the pBWA(V)HS-DlMYB-GFP (35S:GFP) recombinant vector and the nucleus marker vector pBWA(V)HS-Nucleus-mKATE (Shcherbo et al., 2007) were co-transformed in Arabidopsis protoplasts with PEG solution (40% W/V PEG 4000, 0.6 mol/L Mannitol, 0.1 mol/L calcium chloride, pH 5.8). The pBWA(V)HS-GFP empty vector was used as a control. After incubation in darkness at 28°C for 18–24 h, the subcellular distribution and localization of DlMYB-GFP were captured in protoplasts using the Nikon C2-ER laser scanning confocal microscope (LSCM) at a 488 nm excitation wavelength and a 510 nm emission wavelength. Meanwhile, colocalization of DlMYB-GFP and Nucleus-mKATE (NLS-mKate) was used to confirm the nuclear localization of MYB proteins with the mKATE protein at 561 nm excitation wavelength and 580 nm emission wavelength (Shcherbo et al., 2007; Zhao et al., 2017). The chlorophyll fluorescence was captured to confirm the localization of chloroplasts at 640 nm excitation wavelength and 675 nm emission wavelength. The primers used for PCR amplification are listed in Supplementary Table 2.
RNA-seq data from nine tissues of “Sijimi” (“SJM”) longan (See more details in section “Public Data Retrieval”) were used to explore the expression patterns of R2R3-MYB genes at different developmental stages of the longan plant (Lin et al., 2017). The Fastp v0.20.1 software (Chen et al., 2018) was used to filter low-quality reads and adaptor sequences. High-quality clean reads were mapped to the “SX” longan reference genome sequences using HISAT2 v2.2.1 (Kim et al., 2015). The trimmed mean of M values (TMM) method was used to normalize the expression levels of R2R3-MYB genes in longan using edgR software (Robinson et al., 2010). The expression profiles of the R2R3-MYB genes in longan tissues were visualized by TBtools and the ratios were log2-transformed. Gene ontology (GO) annotations were conducted in the eggNOG-mapper (Huerta-Cepas et al., 2019) for the DlMYB genes. All 37,142 genes of longan were used as the reference set, and 119 DlMYB genes were taken as the test set. The annotation results of DlMYB genes were enriched and analyzed using the R package clusterProfiler (Yu et al., 2012), including three categories, cellular component (CC), biological process (BP), and molecular function (MF).
The RNA-seq data of “SX” under KClO3 treatment was also processed using the same method. The differentially expressed genes (DEGs) under KClO3 treatments and controlled conditions in longan were identified using the R package DESeq2 v3.11 (Love et al., 2014), with the threshold of |log2(fold-change)| ≥ 1 and a false discovery rate (FDR) ≤ 0.05. The DESeq2’s Likelihood Ratio Test was used to conduct the time course analysis to find genes which were significantly upregulated/downregulated over time in different treatments. The weighted gene co-expression network analysis (WGCNA) was conducted using the WGCNA v4.0.2 package (Langfelder and Horvath, 2008) based on DEGs with TMM ≥ 1 at least in one sample. Cytoscape v3.7.1 software (Shannon et al., 2003) was used to visualize the gene interaction networks of longan R2R3-MYB genes in modules. FLOweRing Interactive Database (FLOR-ID) of Arabidopsis (Bouché et al., 2016) was used to search flowering-related genes that may interact with DlR2R3-MYB genes. GO and KEGG enrichment analysis were performed using the R package clusterProfiler.
Eight DlMYB genes potentially involved in the flower development were selected for the quantitative real-time polymerase chain reaction (qRT-PCR) in five different tissues (leaves, pericarps, pulps, flowers, and flower buds) of “Shixia.” The total RNA in the samples were isolated using the TRNzol Universal (DP424, TIANGEN Biotech, Beijing, China) reagent according to the manufacturer’s instructions. The concentration and quality of the extracted RNAs was determined using Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, MA, United States) and gel electrophoresis. For cDNA synthesis, 0.5 μg RNA was reverse-transcribed using the M-MLV 4 First-Strand cDNA Synthesis Kit (MT403-01, Biomed, Beijing, China) following the supplier’s instructions. The cDNA was stored at –20°C for further use. Gene-specific primers were designed using the online tool primer3plus (Untergasser et al., 2007; Supplementary Table 2). The longan actin gene (Dil.06g016430.1) was used as the internal reference gene. The qRT-PCR amplification was conducted on a CFX Connect Real-Time PCR detection system (Bio-Rad, Hercules, CA, United States) with 2 × SYBR Green qPCR MasterMix (MT521, Biomed, Beijing, China). Three independent biological replicates and three technical repeats were taken. The reaction was performed using the protocol for this kit with the following minor modifications: 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 30 s. The relative expression levels of DlMYB genes were further calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
A total of 219 non-redundant DlMYB genes were identified in the longan reference genome, including 119 R2R3-MYB genes (2R-MYBs), 95 MYB-like repeats genes (1R-MYBs), three R1R2R3-MYB genes (3R-MYBs), and two 4RMYB genes (4R-MYBs). The R2R3-MYB genes constituted the largest group of the MYB gene family in longan. The R2R3-MYB genes were renamed DlMYB1 to DlMYB119 based on their chromosomal locations in longan genome (Supplementary Table 1). The detailed information of DlMYB gene features, including coding sequences, protein sequences, chromosomal locations, protein sizes, molecular weights (MWs), isoelectric points (PIs), subcellular localizations, instability, and hydrophilia, were listed in Supplementary Tables 1, 3. The length of DlMYB proteins ranged from 134 (DlMYB38) to 661 (DlMYB55) amino acids (aa) with an average of 306 aa, and the MWs had a range of sizes from 15.7 (DlMYB55) to 75.8 (DlMYB38) KDa (mean 34.6). The PIs of the DlMYB proteins ranged from 4.91 (DlMYB32) to 10.60 (DlMYB23). All DlMYB protein hydrophilia values were negative, indicating that longan MYBs are hydrophilic.
The 119 R2R3-MYB amino acid sequence logos of R2 and R3 repeats were produced to show the sequence characterization and frequency of the most widespread amino acids at each position (Figure 1 and Supplementary Figure 1). In general, the R2 and R3 repeats of MYB domains had ∼108 basic residues (including the linker region). Critical amino acid insertions were detected in both R repeats, especially in the Helix2 (H2) segment of R2 and R3. The R2R3-MYB domains showed amino acid insertion points for DlMYB23 (24aa), DlMYB27 (8aa), DlMYB98 (3aa), and DlMYB101 (3aa) in R2, DlMYB4 (2aa), DlMYB53 (2aa), and DlMYB54 (2aa) in R3 (Figure 1 and Supplementary Figure 1). The results confirmed the conservation of the MYB domain, but amino acid compositions and lengths of the regions outside the MYB domain were highly variable.
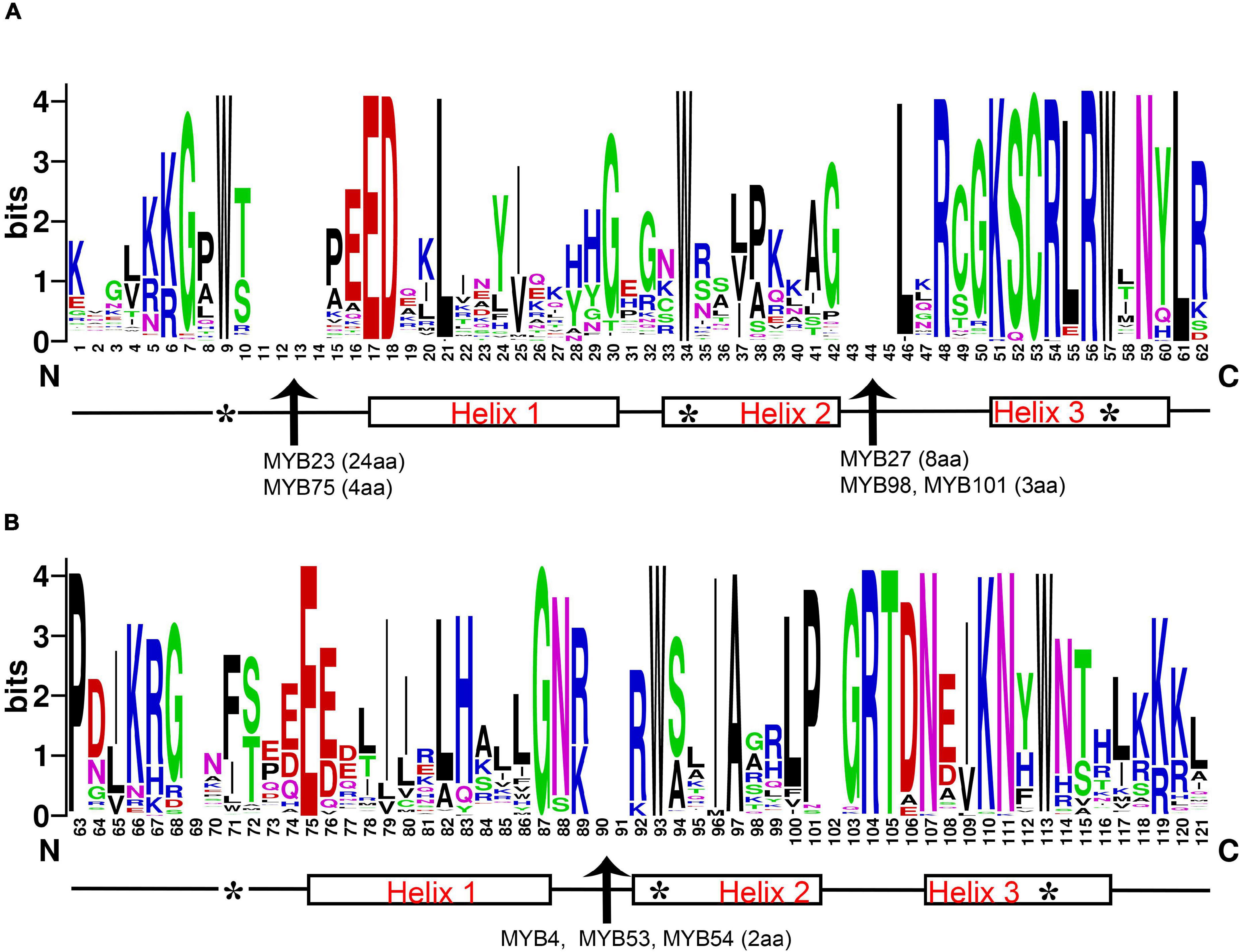
Figure 1. Characterization of R2 (A) and R3 (B) MYB repeats of longan R2R3-MYB proteins. The position score indicated the degree of conservation of each amino acid at the specific position within sequences. The positions of the three α-helices are marked (helices 1–3). Arrows represent amino acid insertion sites detected in the R2 repeat of genes DlMYB23, DlMYB27, DlMYB75, and DlMYB98 and the R3 repeat of DlMYB53, DlMYB54, and DlMYB93. The length of each insertion (amino acid, aa) is highlighted in parentheses. Characteristic tryptophan (W) residues are marked with the symbol “*”.
A series of highly conserved and regularly distributed Trp (W) residues were detected in R2 and R3 repeats of DlMYBs, which are noted for landmarks of plant MYB proteins. Interestingly, only the second and third Trp residues were found to be conserved in the R3 repeat and the first Trp was generally replaced by a hydrophobic amino acid, such as Phe (F) or Ile (I). Apart from Phe and Ile, the substitution with the amino acids Met (M), Leu (L), and Try (Y) were also found in the first Trp residue in the R3 repeat at Trp-62 position of DlMYB proteins (DlMYB6, DlMYB10, DlMYB51, DlMYB57, DlMYB72, DlMYB81, DlMYB82, DlMYB98, DlMYB101, and DlMYB114). In addition, the Trp residues, Cys-45, Arg-48 in the R2 repeat, Leu-53 in the linker region, and Glu-66, Gly-78 in the R3 repeat were conserved in all DlMYB proteins (Figure 1 and Supplementary Figure 1). These highly conserved amino acid residues were mainly distributed in the third helix and the helix-turn-helix (HTH) motif. In DlMYB32 and DlMYB75, the Pro-55 was replaced with Ala and Ser in the R3 repeat, respectively, implying they may have different evolutionary divergence rates.
The maximum likelihood (ML) phylogenetic tree of 119 DlMYB and 126 AtMYB proteins showed that all R2R3-MYBs were categorized into 28 subgroups (designed as S1–S28) based on sequence similarity and topology (Figure 2). Each clade in the ML tree contained R2R3-MYB proteins from longan and Arabidopsis. Five AtMYBs and thirteen DlMYBs that could not be retrieved in the previously constructed AtMYB phylogenetic tree were rearranged in this study, in S26–S28, according to the phylogenetic analysis of Arabidopsis and longan MYBs. Eventually, all 119 longan R2R3-MYB proteins were divided into 28 subgroups, including S1 (three members), S2 (seven members), S3 (one member), S4 (seven members), S5 (nine members), S6 (eight members), S7 (one member), S8 (four members), S9 (four members), S10 (two members), S11 (three members), S12 (one member), S13 (seven members), S14 (seven members), S15 (four members), S16 (three members), S17 (four members), S18 (four members), S19 (one member), S20 (four members), S21 (five members), S22 (five members), S23 (one member), S24 (five members), S25 (six members), S26 (four members), S27 (four members), and S28 (five members) (Figure 2 and Supplementary Table 1). Not only the gene duplications but gene loss events seem to have an impact on the expansion and contraction of MYB members after the divergence of longan and Arabidopsis genomes. As shown in Figure 2, some subgroups (S2, S5, S6, S24, S27, S28) of the phylogenetic tree harbored twice as many longan MYB members as Arabidopsis MYBs. The opposite was also observed in subgroups 7, 10, 12, 16, 19, and 23, where the number of DlMYB proteins was less than half that of AtMYB proteins.
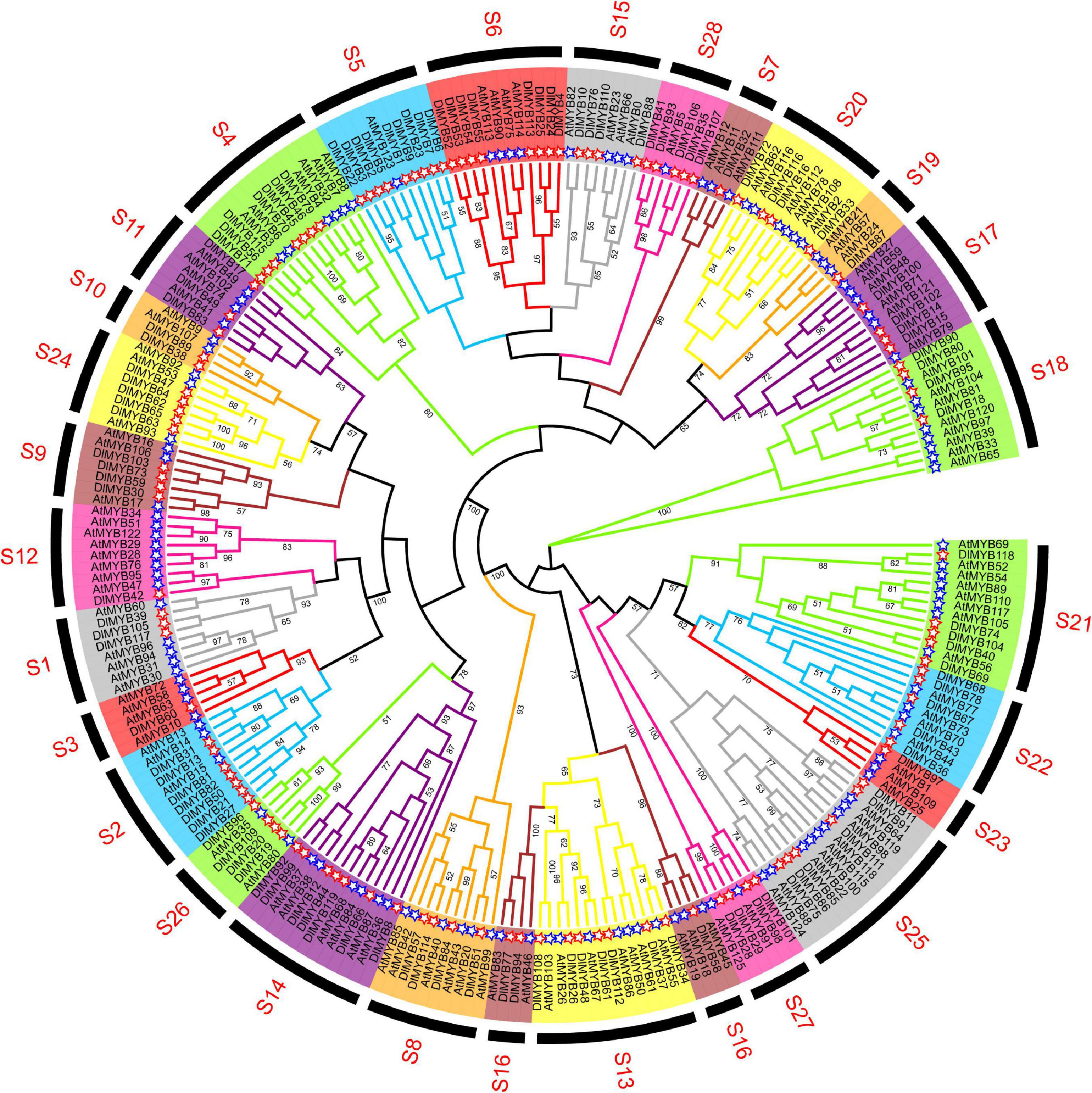
Figure 2. Phylogenetic analysis of longan and Arabidopsis MYB proteins. The full-length amino acid sequences of longan (119) and Arabidopsis (126) MYB proteins were compared using RAxML software to generate ML phylogenetic trees with 1,000 bootstrap values. The Evolview3 was used to visualize and display > 50 bootstrap values at the nodes. Red and blue stars represent DlMYB and AtMYB proteins, respectively. All MYB proteins were divided into 28 subgroups (S1–S28).
The ML phylogenetic tree constructed for 119 DlMYB proteinsalone also revealed that all DlMYB proteins were divided into 28 subgroups (Figure 3A). In order to have a more comprehensive understanding of the diversity and conserved domains of these R2R3-MYB genes in longan, conserved motifs of these DlMYB protein sequences were examined. A total of 20 conserved motifs were examined and designated as motif 1 to 20 (Figure 3B, Supplementary Figure 2, and Supplementary Table 4). The majority of R2R3-MYB protein sequences had four significantly similar conserved motifs and motif orders. Ninety-seven out of 119 R2R3-MYB protein sequences contained motif 3, motif 4, motif 1, and motif 2 present in that order, and these motifs were composed of 21, 11, 40, and 29 amino acids, respectively (Figure 3B and Supplementary Table 4). Those four motifs were found to encode the MYB DNA-binding domain. Motifs 3, 4, and the left part of motif 1 were composed of the R2 repeat, and the right part of motif 1 and motif 2 were found in the R3 repeat. In other 17 R2R3-MYB proteins, motif 4 in the R2 repeat was replaced by motif 18. In addition, motif 4 and motif 1 of DlMYB28 and DlMYB29 were replaced by motif 8. Three R2R3-MYB proteins, DlMYB98, DlMYB100, and DlMYB101 only contained motif 1, motif 2, and motif 3. Almost all DlMYB proteins (118/119) contained motif 2 and motif 3 with the exception of DlMYB 42 that did not have motif 2. Most DlMYB proteins from the same clade had a similar motif composition. For instance, motifs 9 and 17 were shared by the members in S6, motifs 8 and 13 were exclusively in S27, and motif 15 was unique to S25, suggesting that these unique motifs may contribute to the differentiation in specific functions.
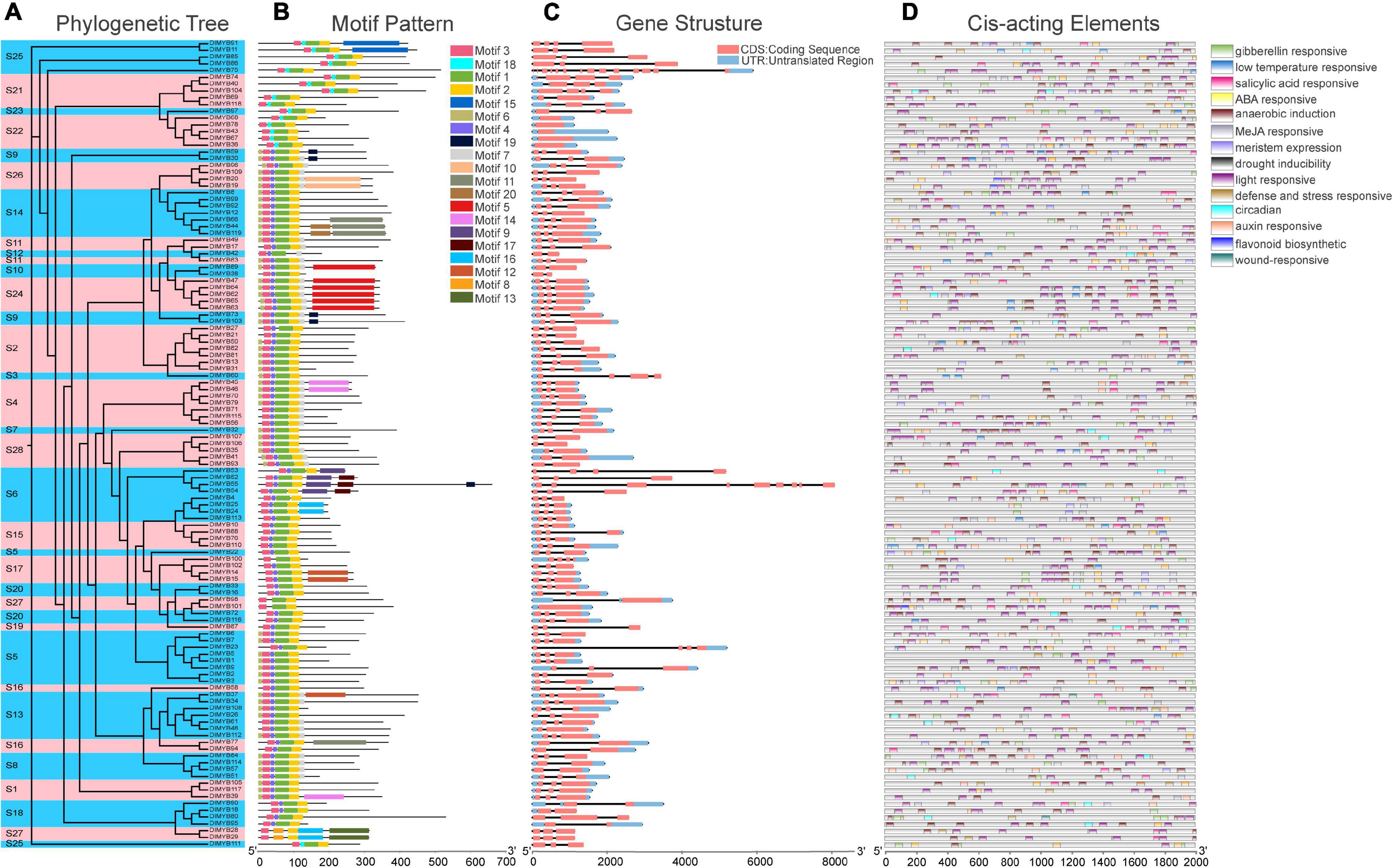
Figure 3. Phylogenetic tree, conserved motifs, gene structural, and cis-acting element analysis of DlMYBs. (A) An Maximum Likelihood (ML) tree of 119 DlMYBs proteins with 1,000 bootstrap replicates. (B) The conserved motifs in DlMYBs were predicted using the MEME Suite web server. (C) Exon/intron structures of the longan R2R3-MYB genes. The introns, exons, and UTRs are indicated by orange boxes, black lines, and blue boxes, respectively. (D) The cis-acting elements of the longan R2R3MYB proteins were predicted using PlantCARE.
The exon/intron organization was analyzed to investigate the structural diversity of DlR2R3-MYB genes. The number of introns in longan R2R3-MYB genes ranged from zero to thirteen (Figure 3C). Most (111/119) of the coding sequences of R2R3-MYB genes in longan were disrupted by one to three introns, with the exception of six DlMYB genes (DlMYB36, DlMYB43, DlMYB67, DlMYB68, DlMYB78, and DlMYB101) that completely lacked introns, indicating the conserved intron number in the R2R3-MYB genes. Only two DlMYB genes had an excess of introns, including DlMYB75 (13 introns) and DlMYB55 (8 introns). Most genes clustered in the same subgroup exhibited highly similar gene structures and intron numbers, which could strongly support the accuracy of the classification of subgroups.
The predicted results for cis-regulatory elements showed that a total of fourteen types of cis-regulatory elements were predicted in DlR2R3-MYB genes. The most widespread cis-acting elements in the promoter region of the DlMYB genes were light responsiveness (119/119), anaerobic induction (102/119), and ABA responsiveness elements (101/119) (Figure 3D and Supplementary Tables 5, 6), while the least common ones were wound-responsive (4/119), flavonoid biosynthetic genes regulation (13/119) and circadian (23/119). Moreover, the promoters of 79 (66.39%), 70 (58.82%), and 56 (47.06%) DlMYB genes were predicted to contain cis-acting elements associated with MeJA (methyl jasmonate), GA (gibberellin), and SA (salicylic acid), respectively. The detailed information regarding the cis-regulatory elements in longan DlMYB genes is given in Supplementary Tables 5, 6. All 14 types of cis-acting elements could be divided into three major functional categories, including cellular development or photoresponsive elements (two types of elements), phytohormone (five types of elements), and environmental stress (seven types of elements). In general, DlR2R3-MYB gene promoters containing environmental stress-related elements (398) were the most common followed by phytohormone-related ones (358).
Genome chromosomal location analysis showed that 119 R2R3-MYB genes were unevenly distributed on 15 chromosomes (Figure 4A), and each chromosome had at least two DlR2R3-MYB genes. DlChr1 harbored the largest group of R2R3-MYB genes (16/119), followed by DlChr15 with 14 R2R3-MYB genes and DlChr12 with 11 genes, while only two genes were located on DlChr11. DlR2R3-MYB genes tend to cluster to the proximal end of some chromosomes, including DlChr1, DlChr2, DlChr3, DlChr4, DlChr8, DlChr10, and DlChr12.
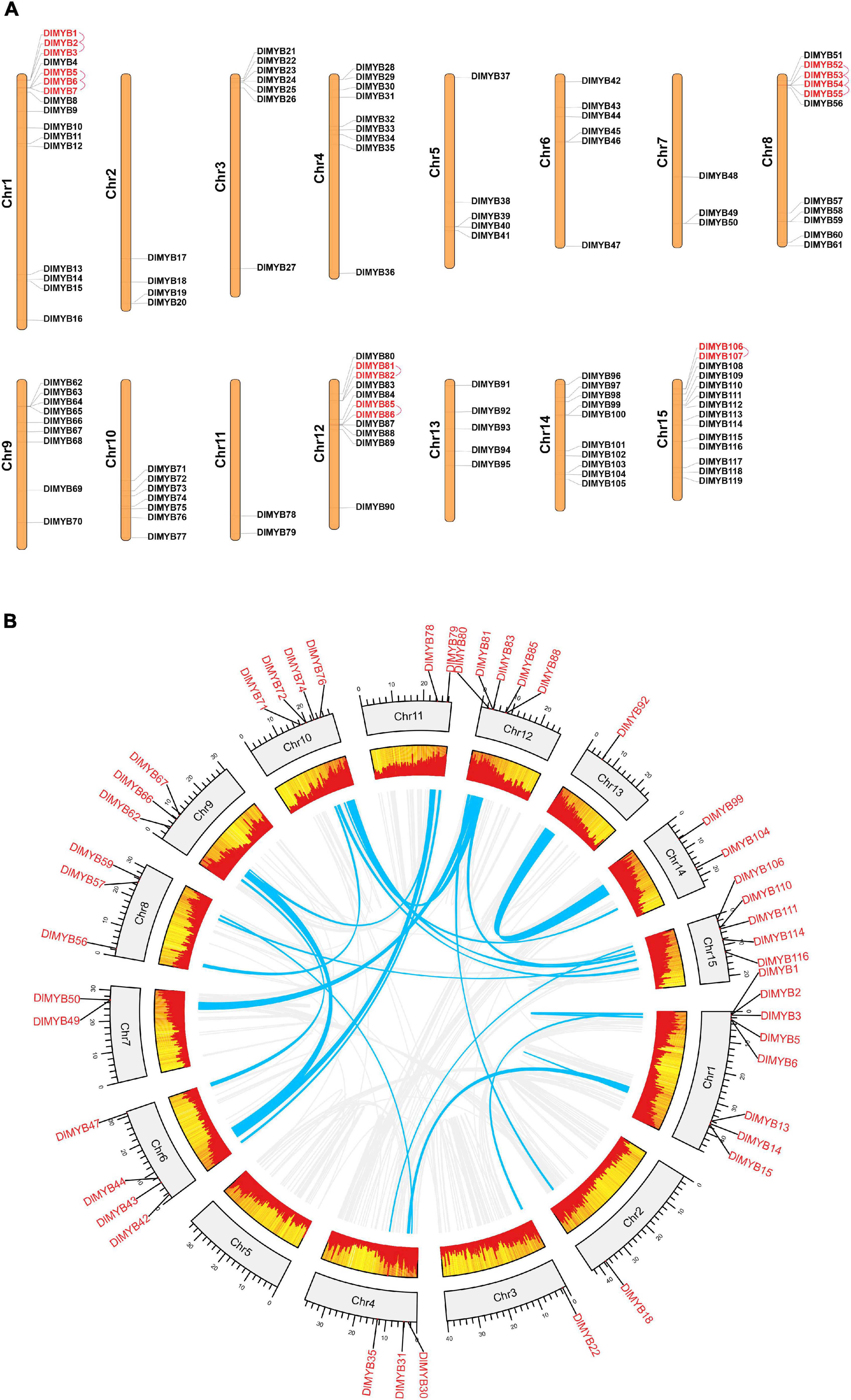
Figure 4. Chromosomal location, tandem duplication, and segmental gene duplication analysis of the DlMYB genes. (A) The distribution map of DlMYB genes on the 15 chromosomes of longan (genes with tandem clusters are marked in red). (B) Circos plot of DlMYB genes in the longan genome. Gene pairs with segmental duplications (SDs) are indicated by blue lines, and gene density was highlighted by red rectangular boxes.
In our study, a total of 25 segmental duplications (SD) involving 44 MYB genes were identified on different chromosomes (Figure 4B and Supplementary Tables 7, 8). DlMYB genes were located within synteny blocks on almost all chromosomes except for DlChr5. The SD events occurred mostly on chromosomes 1, 12, and 15 (Supplementary Table 8). Some DlR2R3-MYB genes, such as DlMYB2, DlMYB78, and DlMYB110, were involved in multiple duplications. In total, six tandem duplications (TD) clusters consisting of 16 DlR2R3-MYB genes were identified in DlChr1, DlChr8, DlChr12, and DlChr15 (Figure 4A). The distribution of TD genes on chromosomes were consistent with aforementioned chromosomes with a high-density of MYB genes. These results indicated that approximately 43.7% of DlMYB genes might be generated by duplication events, which played a vital role in the expansion of the MYB gene family in longan (Supplementary Table 7). The Ka/Ks values (non-synonymous/synonymous substitution ratios) were calculated to understand the selection mode of segmentally and tandemly duplicated R2R3-MYB gene pairs in longan. Ka had a range of values from 0.06 to 0.52 while Ks values varied from 0.14 to 4.36 (Supplementary Table 8). In particular, one duplicated gene pair DlMYB14-DlMYB15 had no Ka and Ks value (NaN). Through manual checking, we found that mutations between DlMYB14 and DlMYB15 sequences occurred at the nucleic acid level but their amino acid sequences remained unchanged. The Ka/Ks ratios of 35 DlMYB gene duplicated pairs (including 10 tandem and 25 segmental duplication gene pairs) ranged from 0.05 to 0.84 (Supplementary Table 8) (all less than 1), indicating that longan R2R3-MYB duplicated genes had undergone negative purifying selection in the process of evolution and contribute largely to the maintenance of function in the longan R2R3-MYB gene family. The average Ka/Ks value of tandem duplication genes was 0.34, two-fold that of segmented duplication genes (0.17) (Supplementary Figure 6). Furthermore, the Ka/Ks value of each orthologous DlMYB gene pair was also calculated in the Sapindaceae family (Supplementary Figure 6 and Supplementary Table 9). Nearly all of the 289 (289/291) orthologous gene pairs between longan and litchi, longan and yellow horn had Ka/Ks values of < 1, except for two gene pairs DlMYB32-LITCHI026531.m1 (Ka/Ks = 1.20) and DlMYB100-LITCHI018670.m1 (Ka/Ks = 1.78) between longan and litchi, implying that purifying selection may be the dominant force driving the evolution of R2R3-MYB genes in the lineage of Sapindaceae. The Ka/Ks mean values of the R2R3-MYB syntenic gene pairs between longan and Arabidopsis, longan and litchi, longan and yellow horn orthologs were 0.15, 0.32, and 0.23, respectively (Supplementary Figure 6 and Supplementary Table 9).
To gain more insight into the evolutionary model of the longan R2R3-MYB gene family, the comparative syntenic maps of longan along with three representative dicots including Arabidopsis, litchi (Litchi chinensis) and yellow horn (Xanthoceras sorbifolium Bunge), and two monocots, rice (Oryza sativa L.) and maize (Zea mays) were generated (Figure 5A). Litchi and yellow horn were phylogenetically closer to longan, which were also in the family of Sapindaceae. The results revealed that 99 DlMYB genes were involved in the formation of a syntenic relationship with those in litchi, followed by yellow horn (94 genes involved), Arabidopsis (61), maize (17), and rice (16) (Figure 5B). The maximum number of ortholog DlR2R3-MYB gene pairs were found in DlMYB-XsMYB (148) and DlMYB-LcMYB (143) followed by DlMYB-AtMYB (102), while the minimum number were found in DlMYB-ZmMYB (25) and DlMYB-OsMYB (17) (Supplementary Table 9), which is consistent with their taxonomic positions. Notably, a total of nine DlMYB genes were predicted to form collinear pairs with genes from all the other five species (Supplementary Table 10). Eighty-five syntenic gene pairs were only found between longan and other dicots (Figure 5B). For example, the gene pair DlMYB38-AT3G02940.1 was only detected between longan and Arabidopsis, DlMYB51-EVM0018212.1-AT1G66230.1 was only found among longan, Arabidopsis, and yellow horn. A total of 26 MYB genes were only found in longan, litchi, and yellow horn (Supplementary Table 10). Additionally, 18 AtMYB genes were predicted to be paired with at least two longan genes (Supplementary Table 9), suggesting the newly evolved functions of redundant DlMYB genes. The detailed information of syntenic gene pairs is provided in Supplementary Tables 9, 10.
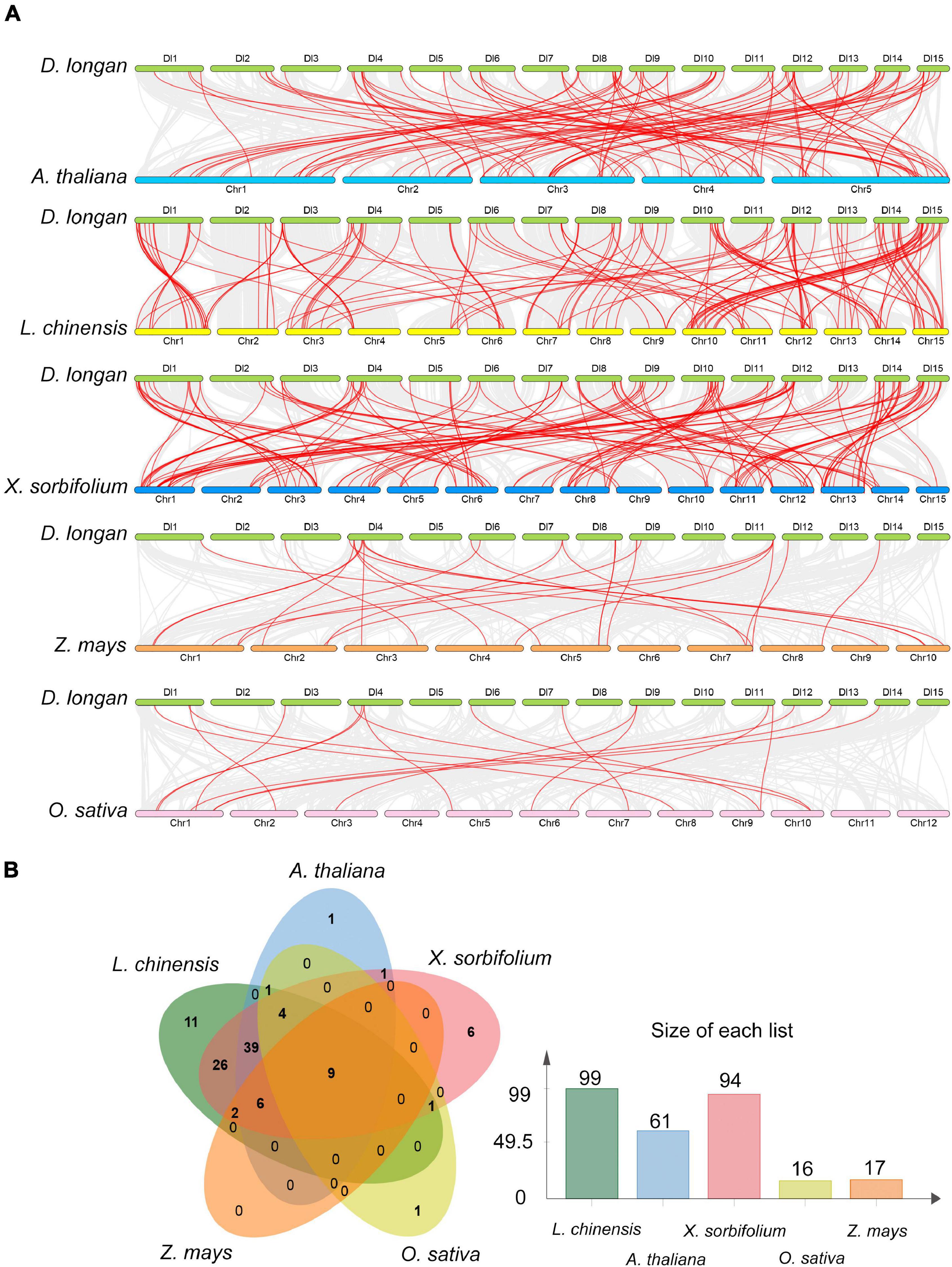
Figure 5. Collinearity analysis of the DlMYB genes. (A) Synteny relationship of R2R3MYB genes between longan and five other representative plant species: Arabidopsis, litchi, yellow horn, maize, and rice. Gray lines in the background indicate the collinear blocks within two genomes, and red lines highlight the syntenic MYB gene pairs. (B) The R2R3MYB genes formed the colinear pairs between longan and the other five species. Bar graphs indicate the number of genes in colinear pairs.
Subcellular localization predictions revealed that all 119 DlMYB proteins were located in the nucleus (Supplementary Figure 7 and Supplementary Tables 1, 3). To experimentally validate the predicted subcellular localization, we selected three potential flowering-associated DlMYB genes (DlMYB16, DlMYB72, and DlMYB116) for analysis in Arabidopsis protoplasts. These three genes were expressed at significantly high levels in flower or flower bud tissues compared with other tissues. The NLS-mKATE and Cy5 were applied in each transformed design as a marker for nuclear and chloroplast localization, respectively. The results showed that DlMYB16, DlMYB72, and DlMYB116 proteins were localized to the nucleus (Figure 6), which was consistent with subcellular localization predictions.
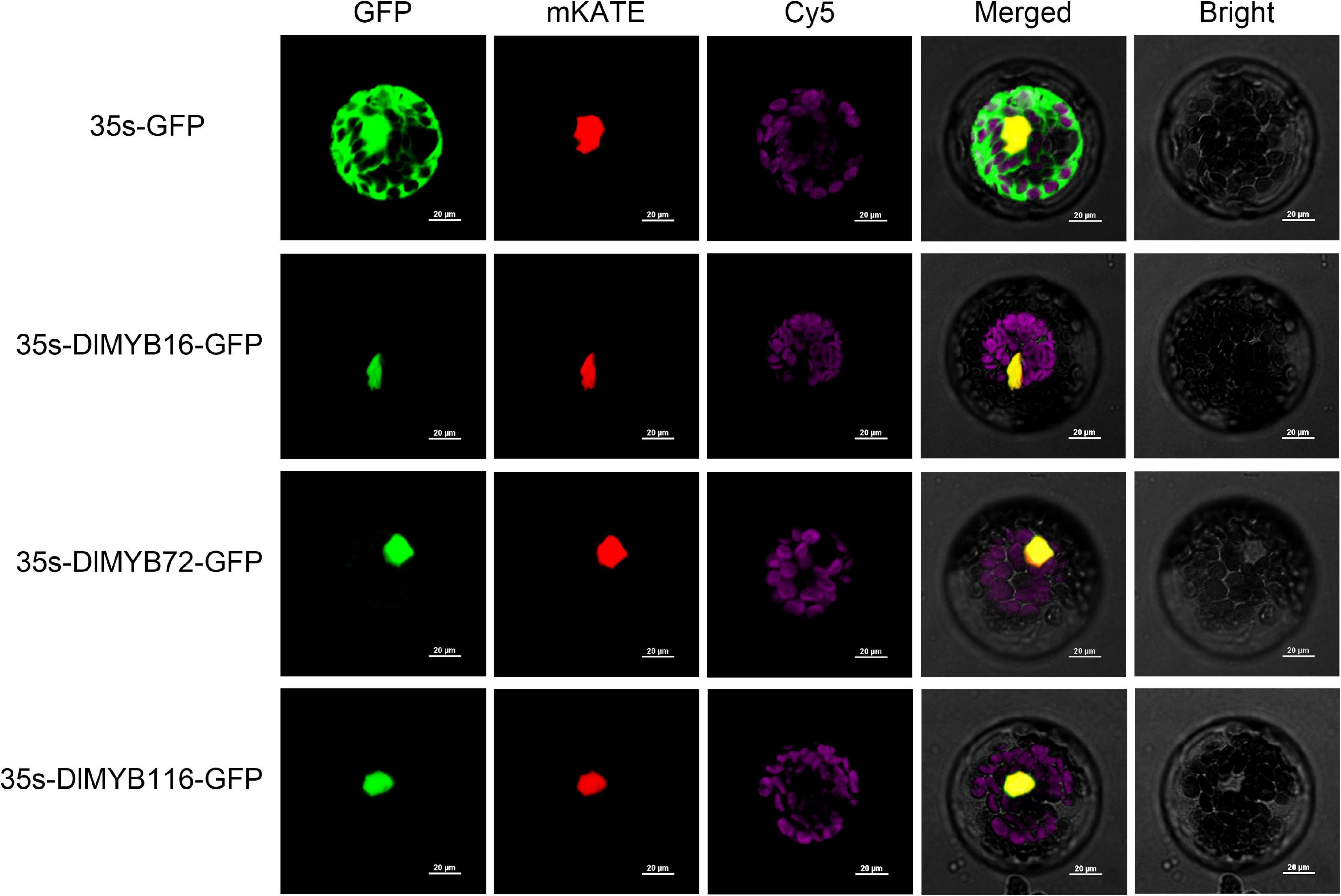
Figure 6. Nuclear localization of DlMYB16, DlMYB72, and DlMYB116 proteins in Arabidopsis protoplasts. GFP: green fluorescent protein; mKATE: nuclear marker; Cy5: chloroplast; Bright: bright field; merged: combined mKATE and GFP and bright field. Bars, 20 μm.
The differential expression patterns of DlR2R3-MYB genes in nine tissues were analyzed using RNA-seq data (Figure 7A, Supplementary Figures 3, 4, and Supplementary Tables 11, 12). The results revealed that the DlR2R3-MYB genes showed varying expression patterns in different tissues and most genes displayed tissue-specific expressions. In general, 119 R2R3-MYB genes were clustered into seven groups according to the preferential expression tissues, which were designated as group I to group VII (Figure 7A). Group I genes were highly expressed in flowers and flower buds. Group II genes were widely expressed in different tissues, which indicated that they were involved in various organ formation processes in longan. Group III genes were mainly expressed in the roots, suggesting that they might execute certain functions in the root development. Genes in group IV-VII exhibited preferential expression in seeds, young fruits, pericarps, and pulps, respectively. In total, 74 (62.18%), 58 (48.74%), 47 (39.50%), 62 (52.10%), 57 (47.90%), 69 (57.98%), 30 (25.21%), 62 (52.10%), 50 (42.02%) DlR2R3-MYB genes were expressed (TMM > 1) in roots, stems, leaves, flower buds, flowers, young fruits, pulps, pericarps and seeds, respectively (Supplementary Figure 3 and Supplementary Table 12). Flowers and flower buds were observed to have the most DlR2R3-MYB genes that had TMM ≥ 100, with 10.08 and 7.56%, respectively, suggesting that more R2R3-MYB genes were highly expressed in flower tissues than in other tissues. Of 119 DlMYB genes, 98 genes (82.35%) were expressed in at least one tissue (Supplementary Table 11). By contrast, five genes (DlMYB11, DlMYB21, DlMYB38, DlMYB43, and DlMYB54) in longan were not expressed (TMM = 0) in any of the examined tissues, possibly because they could only be expressed under given conditions, or detected in certain tissues that were not examined in our study, or they were pseudogenes. Three DlMYB genes (DlMYB67, DlMYB70, and DlMYB79) were observed to have high expression levels (TMM > 10) in all nine tissues examined. It is worth noting that fourteen genes (DlMYB8, DlMYB16, DlMYB17, DlMYB33, DlMYB35, DlMYB67, DlMYB70, DlMYB72, DlMYB78, DlMYB79, DlMYB87, DlMYB93, DlMYB115, and DlMYB116) showed extremely high expression levels (TMM > 100) in flowers and flower buds. Some genes exhibited flower-specific expressions. Four genes (DlMYB28, DlMYB29, DlMYB42, and DlMYB109) were expressed exclusively in flower buds, and DlMYB87 was expressed only in flowers and flower buds.
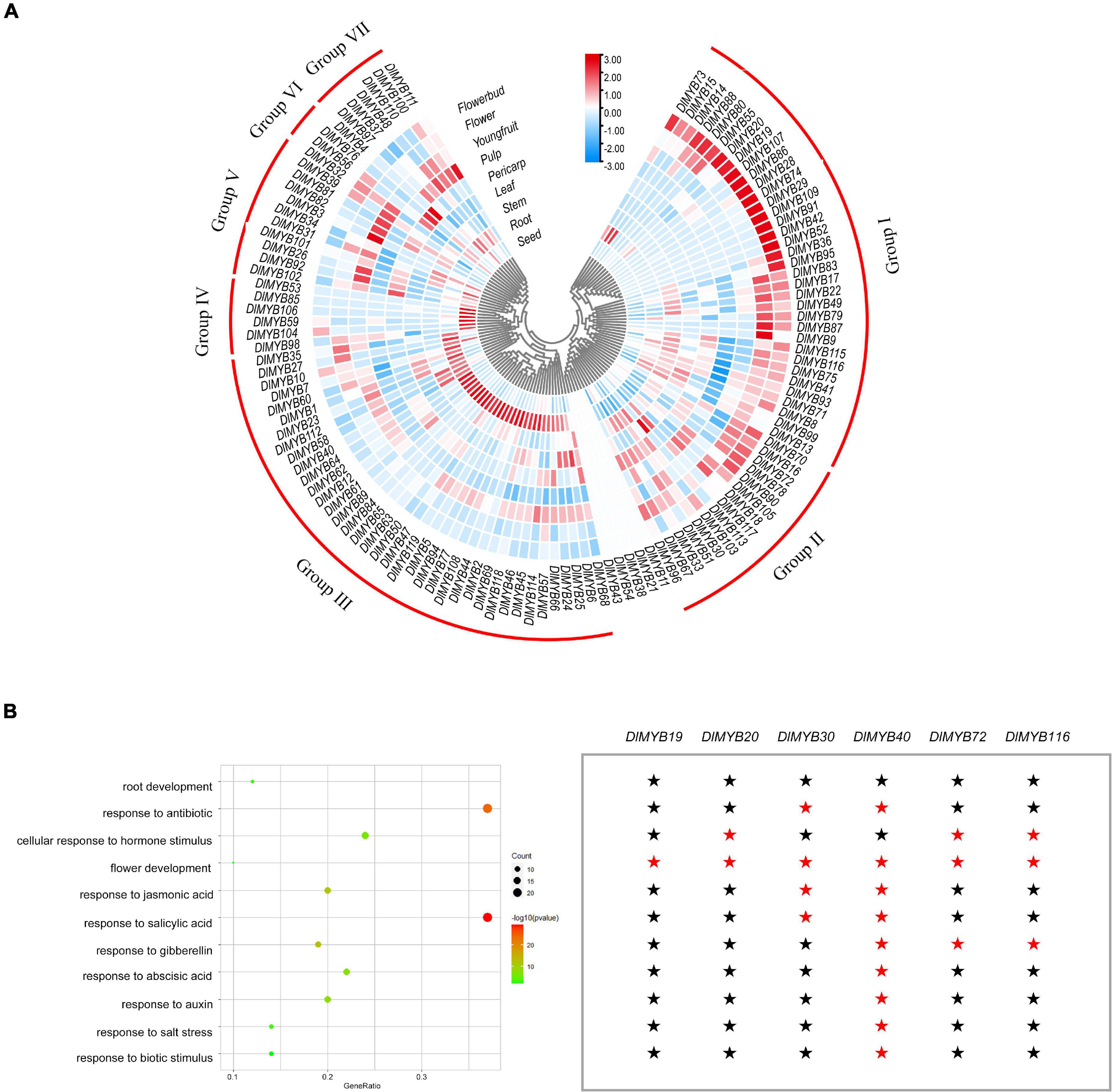
Figure 7. Tissue-specific expression and GO enrichment analysis of DlMYB genes. (A) The expression levels of DlMYB genes in nine tissues were classified into groups I–VII. Different colors indicate log2 transformed expression levels, with red indicating high expression levels and blue indicating low expression levels. (B) Some DlMYB genes were classified as related to stress response, development, and hormones. The size of the circle indicates the number of DlMYB genes, and the color gradient indicates the size of the P-value. Six DlMYB genes are categorized into “flower development” function. A red pentagram represents that the gene is relevant, while a black pentagram indicates that the gene is not involved in the category.
A total of 153 GO items were enriched by the GO annotation with a cut-off value of P ≤ 0.05, among which 146 GO terms were classified into the biological process category and only seven into the molecular function category (Supplementary Table 13). Fifty-eight of all DlMYB genes (48.74%) were assigned to categories related to hormones, stress response, and tissue development (Figure 7B). Interestingly, six DlMYB genes (DlMYB19, DlMYB20, DlMYB30, DlMYB40, DlMYB72, and DlMYB116) were assigned to the “flower development” category. Among them, DIMYB19 belonged exclusively to the “flower development” category and DIMYB20 was only associated with two categories “flower development” and “cellular response to hormone stimulus,” while DlMYB40 was also related to other several categories such as development and salt stress response (Figure 7B and Supplementary Table 13). Regarding their expression levels, however, DlMYB40 was lowly expressed in all examined tissues except for roots and stems. DIMYB19 and DIMYB20 showed slightly higher expression (with TMM of 10∼12) in flower buds. Two genes, DlMYB72 and DlMYB116, that had extremely high expression levels in flowers or flower buds were given more attention in the following analysis.
To further investigate the biological function of DlMYB genes responding to KClO3 treatment, ten time points with untreated control and KClO3 treatment were used to detect the expression profiles of DlMYB genes in apical buds. The hierarchical cluster analysis was performed based on the expression (TMM values) of DlMYB genes treated with KClO3 at different periods (Figure 8A and Supplementary Table 14). In the TMM analysis results, 20.17% (24/119) of all DlMYB genes were observed to have no expression (TMM < 1) at different time points (Supplementary Table 14). A total of 18 DlMYB genes (15.13%) were highly expressed (TMM > 10) at all ten time points, among which, DlMYB35, DlMYB93, DlMYB98, and DlMYB101 were extremely highly expressed (TMM > 100). The time course analysis showed that six genes were significantly upregulated or downregulated (Padj < 0.05) under KClO3 treatments at different time points compared with control. For instance, the gene expression of DlMYB31 and DlMYB68 were inhibited after KClO3 treatment compared to the control, indicating they were negatively regulated by KClO3 (Figure 8B and Supplementary Table 14). DlMYB22 and DlMYB71 were inhibited in the later stages after KClO3 treatment. In contrast, the expression of DlMYB23 and DlMYB99 were promoted in the initial stages by the KClO3 treatment. In addition, Some genes in the same subfamilies tended to exhibit similar expression patterns. For example, DlMYB33 and DlMYB116 in subfamily 20 were up-regulated at day 5 and day 10, and then commenced to decline at later time points (Figure 2 and Supplementary Table 14).
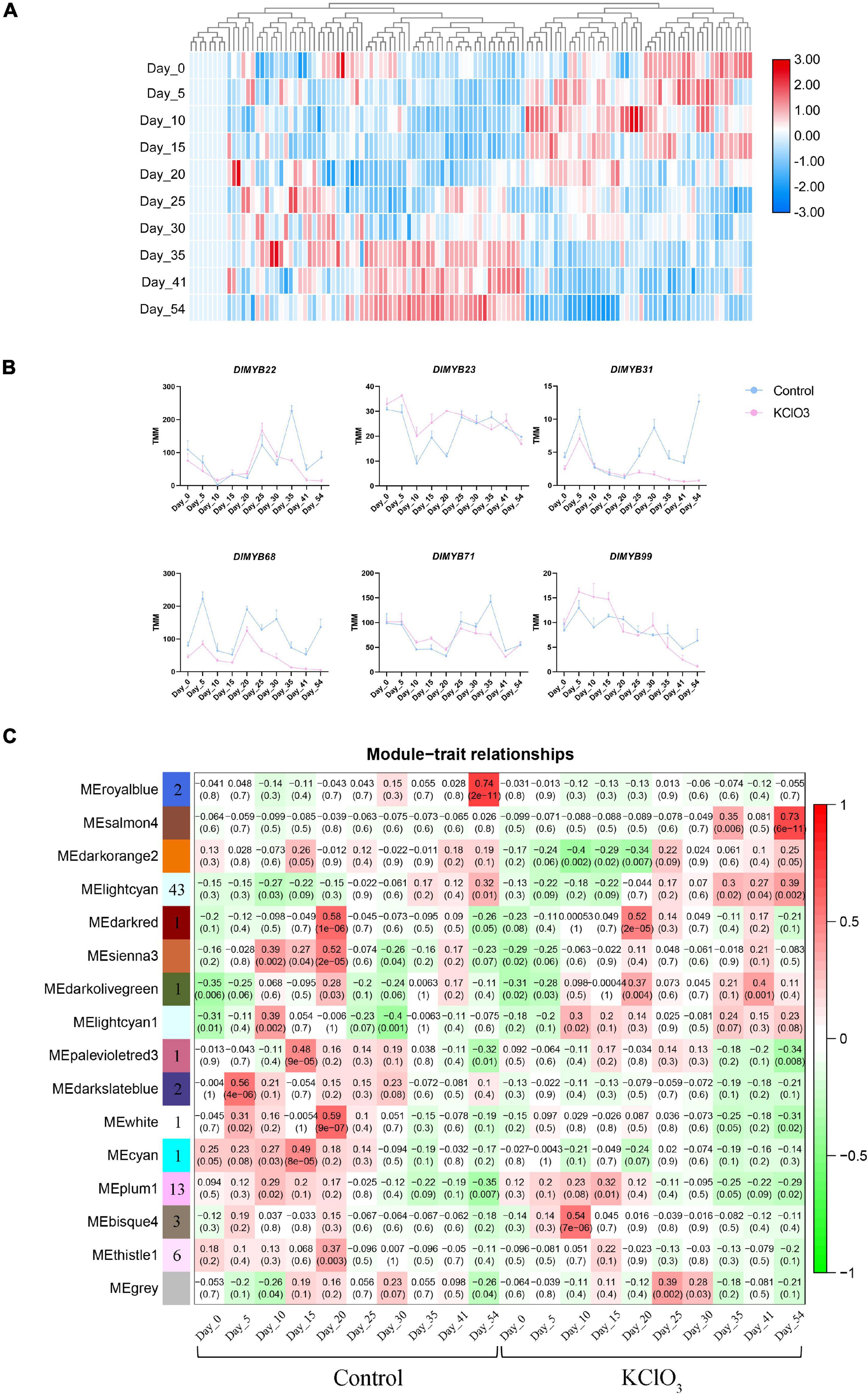
Figure 8. Expression patterns of DlMYB genes under KClO3 treatment. (A) The heat map displays the hierarchical clustering of the 119 DlMYB genes at different time points, with red indicating high expression levels and blue indicating low expression. (B) The line graphs display the effect of KClO3 treatment on the expression of DlMYB genes. The blue line represents the control and the pink line represents KClO3 treatment. (C) The heatmap from WGCNA of differentially expressed genes (DEGs) in off-season flower induction at different treatments and developmental stages. Each row corresponds to one module resulting from an WGCNA eigengene value. Each column corresponds to the control treatment and the KClO3 treatment at different time-points. The color and number of each cell at the row-column intersection indicate the correlation coefficient between one specific module and one time point under control or KClO3 treatment. Darker red represents a higher degree of correlation. The number in brackets indicate the significance (p-value) of the correlation coefficient. The number of DlMYB genes in each module is shown in the leftmost column.
To elucidate the regulatory pathway of off-season flowering among DlMYBs and other genes, coexpression networks were generated incorporating 16 modules via WGCNA. The heatmap of ten time points under control and KClO3 treatment showed that 74 differentially expressed R2R3-MYB genes were distributed in eleven modules represented by different colors (MEbisque4, MEcyan, MEdarkolivegreen, MEdarkred, MEdarkslateblue, MElightcyan1, MEpalevioletred3, MEplum1, MEroyalblue, MEthistle1, and MEwhite) (Figure 8C). After filtering out the low-interacting genes based on the weight values reflecting the connectivity between genes in each module (the thresholds of weight values were shown in Supplementary Table 15), 35 R2R3-MYB genes remained that potentially interact with 567 genes. KEGG pathway enrichment analysis revealed that the “plant hormone signal transduction” pathway (ko04075) was enriched with 18 genes (Supplementary Figure 9A), and GO enrichment showed that these genes were mainly associated with “DNA binding TF activity” (Supplementary Figure 9B).
Among the eleven distinct modules, the MElightcyan module with 211 genes had an expression pattern correlated with the late period of KClO3 (35-54 DAT) (Figure 8C and Supplementary Table 15). A gene expression network was developed to further mine candidate DlMYB genes (Supplementary Figure 8). Fifteen DlMYB genes were contained in the 211 genes. DlMYB113 was identified as hub genes, with several TF genes such as HISTONE H2A 11 (HTA11, Dil.04g007090.1), EARLY IN SHORT DAYS 7/TILTED 1 (TIL1, Dil.13g008690.1), and MITOTIC ARREST-DEFICIENT 2 (MAD2, Dil.10g018750.1). The MEplum1 module was found to be correlated with the early stage (15 DAT) of KClO3 treatment. In total, 133 genes were involved in this module, including DlMYB30, DlMYB41 and DlMYB60, and other genes such as TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR1-20 (TCP20, Dil.12g008890.1) and HISTIDINE-CONTAINING PHOSPHOTRANSMITTER 2 (AHP2, Dil.04g009050.1). The MEdarkolivegreen module with 7 genes were associated with the middle stage (20 DAT) of KClO3 treatment. DlMYB13 was identified as a hub gene in the central location, closely linked to HUMAN WDR5 HOMOLOG A (WDR5a, Dil.04g005430.1). The MEbisque4 module was comprised of 58 genes that were specifically and highly associated with the early period (10 DAT) of KClO3 treatment. This module included three R2R3-MYB TF genes (DlMYB8, DlMYB33, and DlMYB78). DlMYB8 was identified as the top hub gene and was directly connected to WRKY75 and ERF114.
To validate the expression of DlMYB genes in flower development, eight representative DlMYB genes (DlMYB16, DlMYB17, DlMYB19, DlMYB20, DlMYB35, DlMYB72, DlMYB78, and DlMYB116) were selected for qRT-PCR analysis (Figure 9). Overall, the trends of the qRT-PCRs for the eight DlMYB genes were consistent with the results of RNA-seq analysis. However, a slight difference existed due to differences between longan cultivars used, the cultivation conditions of plants, and the geographical location of tissue collection.
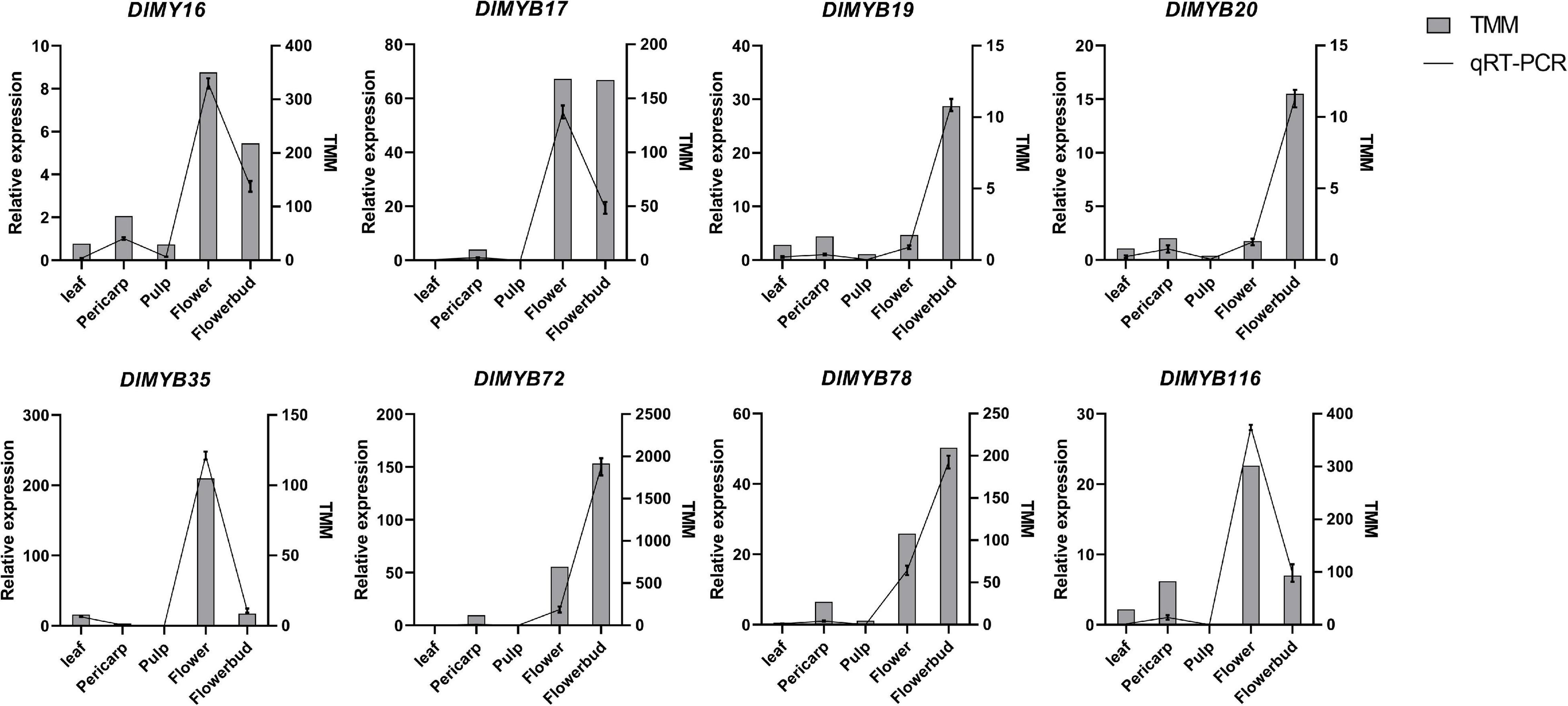
Figure 9. qRT-PCR and RNA-seq analyses of eight selected DlMYB genes. The samples from flowers, flower buds, leaves, pulps, and pericarps show a range of expression changes of selected DlMYB genes. The left and right sides of the Y-axis indicate relative expression levels and TMM, respectively. To confirm the tissue-specific expression of the DlMYB genes, the expression pattern of selected DlMYB genes was calculated as a fold relative to the expression level of the pericarp.
As shown in Figure 9, DlMYB17 and DlMYB72 were observed to be expressed in the pericarps, flowers, and flower buds specifically. DlMYB16 and DlMYB116 were detected to have similar expression patterns and high expression in flowers and flower buds, suggesting a potential redundancy in their functions. Similar trends were found for DlMYB19 and DlMYB20 in subfamily 26. In addition, DlMYB35 was specifically expressed in leaves, flowers, and flower buds. Strongly in line with our theorized expectations, the eight selected genes were significantly highly expressed in both flowers and flower buds compared with other tissues in longan.
The MYB gene family constitutes one of the largest TFs implicated in the regulation of a wide array of physiological processes in plants. Thus far, this present study is the first to conduct in silico systematic and genome-wide characterization of the R2R3-MYB gene family in the longan genome and reveal their potential roles in longan flower development.
In this study, a total of 119 R2R3-MYB proteins were identified in the “Shixia” longan genome, which were classified into 28 subgroups (Figure 2 and Supplementary Table 1). A considerable fraction of subgroups (S1–S25) in our classification were consistent with the subgroupings in other plants (Dubos et al., 2010). Division of R2R3-MYBs into subgroups based on conserved sequence motifs would identify clades whose members commonly exhibited similar biological functions (Millard et al., 2019a). The numbers of R2R3-MYB genes in longan and Arabidopsis were remarkedly different in some subgroups within the phylogenetic tree (Figure 2). This phenomenon was also observed in many other species such as potato (Sun et al., 2019), maize (Du et al., 2012a), grape (Wong et al., 2016), and flax (Tombuloglu, 2020). For one reason, the number of R2R3-MYB genes may be related to the quality of the assembled genome. The marked improvement of the chromosome-level longan genome assembly would expand and enrich the predicted DlMYB gene pool. In comparison with our result, only 35 (Zheng et al., 2020) and 98 (Lin et al., 2017) DlMYB genes were identified based on transcriptome data and the previous scaffold-level longan assembly using Illumina data, respectively. For another, the variation in the total number of R2R3-MYB genes and subgroup genes among plants may have arisen from evolutionary divergence during which the expansion/contraction of MYB genes occurred in different species (Jiang and Rao, 2020). TD, SD and whole genome duplication (WGD) events have long been known to occur throughout plant evolution and perform a significant role in new gene generation and gene family expansion (Van de Peer, 2004). The results showed that tandem duplication and segmental duplication events were unevenly distributed throughout all chromosomes, with 16 DlMYB genes identified as TDs and nearly three times more DlMYB genes (44) classified as SDs (Figure 4 and Supplementary Tables 7, 8). Similar trends of more SDs were observed in Arabidopsis (Cannon et al., 2004) and potato (Sun et al., 2019), suggesting a more crucial role of SD events in the expansion of MYB genes compared to TD events. More SD events and fewer TDs have commonly been present in gene families involved in housekeeping or core regulatory functions (Cannon et al., 2004). Ka/Ks was used to reflect the selection pressure on plants during evolution. The Ka/Ks ratios of all 35 tandemly and segmentally duplicated DlR2R3-MYB gene pairs were less than 1 (Supplementary Table 8), indicating that they might have been subject to purifying selection with constraint functional divergence after species-specific duplication events. This result corresponds well with previous observations in many plant species (Abbas et al., 2021; Wang et al., 2021). Although R2R3-MYB TFs contain “non-MYB regions” that are highly variable, disordered and compositionally biased, they have been evolving at a neutral rate and under the pressure of purifying selection with evolutionary constraints (Millard et al., 2019a).
Interspecies synteny analysis demonstrated that DlR2R3-MYB genes had higher collinearity with R2R3-MYB genes of three dicot species, especially Litchi, and yellow horn in the same family of Sapindaceae (Figures 5A,B and Supplementary Tables 9, 10), suggesting their divergence from the common dicot ancestor. In addition, some syntenic gene pairs were only present between longan and other dicots. For instance, 39 syntenic gene pairs were identified between longan and all examined dicots. Those pairs may appear after the divergence of dicotyledon and monocotyledon species. There were 26 syntenic DlMYB genes exclusively found in litchi and yellow horn, implying those genes may be Sapindaceae specific MYB genes, which have arisen after the divergence of Sapindaceae and Brassicaceae. Eighteen AtMYB genes paired with at least two longan genes indicate that additional longan genes would be generated due to duplication events and play vital roles during the evolution of longan. Nine DlMYB genes were identified to have synteny with their orthologs in five plants analyzed, suggesting that these genes may have existed prior to the divergence of these plant species and are highly conserved in evolution (Figure 5B and Supplementary Tables 10, 11).
We constructed a separate ML phylogenetic tree of 119 DlMYB proteins, which showed that either tandem replication or segmental replication gene pairs were classified into the same subgroup, except for the DlMYB42-DlMYB79 segmental replication gene pair (Figure 3A). In addition, the motif composition and the distribution of exon/intron structures varied among DlMYB genes (Figures 3B,C). For example, DlMYB98, DlMYB100, and DlMYB101 contained only three motifs, while 16 DlMYB genes harbored the largest number of 7 motifs (Figure 3B). In general, a high degree of conservation in the MYB structural domain (R2 and R3 repeats) were found and members of the same subgroup tended to have similar motif combinations. The motifs that made up R2 and R3 repetitive sequences were highly conserved and mainly distributed at the N terminus, while specific motifs were present at the C terminus (Figure 1 and Supplementary Figure 1). The motifs 9 and 17 belonged exclusively to S6, motifs 8 and 13 were only found in S27, and motif 15 was unique in S25. Previous researches revealed that unique motifs may be associated with the functional divergence of DlMYBs (Millard et al., 2019b). Gene structure analysis showed that members in the same subgroup were also inclined to have similar exon/intron structures and intron numbers. Four genes in subgroup 6, DlMYB52, DlMYB53, DlMYB54, and DlMYB55, however, showed significantly different gene structures from each other although they formed a tandem gene cluster in Chr8. Consistent with previous studies (Abbas et al., 2021), most R2R3-MYB genes were found to contain only two introns with the exception of DlMYB75 and DlMYB55 that contained 13 and 8 introns, respectively. Introns were found to have functions in promoting cellular resistance to starvation by enhancing the repression of ribosomal protein genes (Parenteau et al., 2019).
The cis-acting element analysis results showed that DlMYB proteins were predicted to have various elements related to phytohormone, environmental stress, and cellular development (Figure 3D and Supplementary Tables 5, 6). Phytohormones such as ABA, ethylene, GA, and MeJA are involved in the regulation of plant flower development and senescence (Bao et al., 2020; Huang et al., 2021). The promoters of the DlMYB genes were predicted to contain several cis-acting elements associated with hormone responses, implying the potential role of DlMYB genes in the regulation of flower development. GO enrichment analysis also indicated that some DlMYB proteins have functions in response to hormones, tissue development and environmental stress (Figure 7B and Supplementary Table 13). Among them, the “flower development” pathway was enriched with six genes, indicating their potential functions in flowering (Figure 7B and Supplementary Table 13). In agreement with previous research results in Arabidopsis (Dubos et al., 2010), the MYB binding sites involved in flavonoid biosynthetic regulation (MBSI) elements were found in promoters of thirteen longan R2R3-MYB genes. Another 61 DlMYB genes contained the drought inducibility element known for its role in response to drought stress, suggesting that more than half of the DlR2R3-MYB genes might be involved in the drought stress response.
Many R2R3-MYB members are involved in regulating plant development (Millar and Gubler, 2005; Cheng et al., 2009; Phan et al., 2011; Song et al., 2011; Huang H. et al., 2017). The expression analysis allowed us to further estimate the functions of DlMYB genes in longan. Our results revealed that varying expression patterns were observed among longan tissues and most genes exhibited tissue-preferential expression (Figure 7A), suggesting their tissue-specific roles in the development of differentiated tissues. Five genes were not expressed in all examined tissues, which were probably pseudogenes or could only be detected in other tissues that we did not collect. The functions of DlMYB genes were closely linked to their orthologous counterparts in Arabidopsis. Four longan genes, DlMYB18, DlMYB80, DlMYB81, and DlMYB90 clustered with AtMYB33 and AtMYB65 in subgroup 18, which could promote anther and pollen development in Arabidopsis (Millar and Gubler, 2005). In addition, DlMYB18 and DlMYB80 formed segmental replication gene pairs in the longan genome and had synteny with AtMYB97/AtMYB101/AtMYB120 that were found to be synergistic pollen tube factors and could control the fertilization process (Liang et al., 2013). These four longan MYB genes abovementioned were found to be active in flowers or young fruits, suggesting their potential roles in pollen development. Similarly, DlMYB19, DlMYB20, DlMYB96, and DlMYB109, which grouped with AtMYB80 in subgroup 26, also showed high expression in flowers, indicating they may participate in flower development, especially in regulating pollen development (Phan et al., 2011).
Moreover, AtMYB21 and AtMYB24 that take part in the regulation of anther development (Cheng et al., 2009; Song et al., 2011) grouped with DlMYB87 in subgroup 19. DlMYB87 was extremely highly expressed in flowers, suggesting its possible involvement in regulating anther development. AtMYB108 in subfamily 20 can regulate stamen maturation in Arabidopsis (Mandaokar and Browse, 2009). Another gene AtMYB62 in S20 is a regulator of phosphorus stress in Arabidopsis. Overexpression of AtMYB62 inhibits the synthesis of gibberellic acid (GA) and the expression of the flowering regulators SCO1 and SUPERMAN, ultimately leading to delayed flowering in Arabidopsis (Devaiah et al., 2009). Their homologs in longan, DlMYB16, DlMYB33, DlMYB72, and DlMYB116 were observed to have high expression in flowers. Among them, DlMYB72 and DlMYB116 had similar expression patterns as segmental replication genes and formed syntenic gene pairs with AtMYB62/AtMYB116 and AtMYB62, respectively (Figure 7A and Supplementary Tables 10, 11). Therefore, these genes may have similar functional characteristics in the regulation of flower development. Beyond its function in flowering, AtMYB108 also participated in the regulation of the leaf senescence process, acting by binding to a specific region of ANAC003 promoter to form a MYB-NAC regulatory complex (Chou et al., 2018). Its homologs DlMYB16 and DlMYB116 were also highly expressed in leaves, suggesting that genes in the same subgroup may play different biological functions. In subgroup 14, AtMYB68 was demonstrated to specifically regulate the root growth (Jiang et al., 2004), and its homologs DlMYB8, DlMYB12, DlMYB44, DlMYB66, DlMYB99, and DlMYB119 were also found to be highly expressed in the roots, presumably having an important role in longan root development. The high levels of association between longan MYB expression patterns and the roles of their orthologs in Arabidopsis may be evidence of functional conservation between homologous R2R3-MYB members of Arabidopsis and longan.
The qRT-PCR experiments demonstrated the high expression levels of DlMYB19, DlMYB20, DlMYB72, and DlMYB116 in flowers and flower buds, which were also annotated in the “flower development” pathway by GO enrichment analysis, further suggesting that these genes may play an active part in the regulation of flower development (Figure 9).
Multiple R2R3MYB proteins were found to be capable of abiotic and biotic stress tolerance in plants (Dubos et al., 2010). AtMYB15, which clustered with the segmental replication gene pairs DlMYB13 and DlMYB31 in subgroup 2, was demonstrated to possess cold stress tolerance and drought resistance (Ding et al., 2009; Kim et al., 2017). AtMYB15 enhances the susceptibility to freezing by negatively regulating CBF genes which are essential for freezing resistance in Arabidopsis, whereas MPK6-mediated phosphorylation reduces the affinity of AtMYB15 to bind to the CBF3 promoter to enhance cold stress tolerance (Kim et al., 2017). Meanwhile, the overexpression of AtMYB15 enhanced ABA-mediated drought resistance and salt tolerance in Arabidopsis (Ding et al., 2009). Interestingly, DlMYB13 and DlMYB31 happened to have low-temperature responsiveness (LTR) elements and several ABA-responsive (ABRE) elements, and DlMYB31 was assigned to the “response to cold” pathway in GO enrichment analysis (Supplementary Tables 5, 6, 9–13), indicating their potential role in frost and drought resistance. Also, lignin biosynthesis is regulated by AtMYB15, which plays a key role in regulating plant growth and development as well as in plant innate immunity by promoting the synthesis of secondary cell walls (Kim et al., 2020). These two homologous DlMYB genes may also have similar biological functions that remain to be uncovered. In subgroup 22, five longan genes (DlMYB36, DlMYB43, DlMYB67, DlMYB68, and DlMYB78) formed a single cluster with four orthologous AtMYB genes (AtMYB44, AtMYB70, AtMYB73, and AtMYB77), which have been reported to be involved in the abiotic stress response (Jung et al., 2008; Zhao et al., 2014; Yang et al., 2020). For example, AtMYB44 among them functions in the regulation of ABA-mediated stomatal closure as well as drought tolerance (Jung et al., 2008). The cis-acting element prediction showed that these four longan genes all have drought inducibility elements except for DlMYB68 (Supplementary Tables 5, 6). In particularly, DlMYB78 contained four drought inducibility elements, indicating it may have greater potential in response to drought induction. In terms of transcript expression, DlMYB67 was extremely highly expressed in all tissues. qRT-PCR experiments showed that DlMYB78 was extremely highly expressed in flowers and flower buds, which may promote flowering under drought induction (Figure 9 and Supplementary Tables 9–11).
R2R3-MYB proteins are involved in different primary and secondary metabolic regulations in plants (Dubos et al., 2010). In Arabidopsis subgroup5, AtMYB123 ensures a high accumulation of proanthocyanidin (PAs) by forming the MYB-BHLH-WDR (MBW) complex (Baudry et al., 2004). The AtMYB123 homolog, DlMYB7, was highly expressed in roots, stems, leaves, flowers and young fruits, with a possible function in proanthocyanidin synthesis (Figure 7A and Supplementary Figure 4). Four AtMYB genes, AtMYB75/PAP1, AtMYB90/PAP2, AtMYB113, and AtMYB114 have been reported to control anthocyanin biosynthesis (Teng et al., 2005; Gonzalez et al., 2008), which formed a subgroup (S6) with the tandem replication gene cluster DlMYB52/DlMYB53/DlMYB54/DlMYB55 in longan (Figures 2, 4A). DlMYB32, AtMYB11/PFG2, ATMYB12/PFG1, and AtMYB111/PFG3 were classified as subgroup 7. The AtMYBs in this group mainly participate in the regulation of flavonol biosynthesis (Dubos et al., 2010). Previous studies have found AtMYB21 and AtMYB24 in subgroup 19, that cluster with DlMYB87, played conserved regulatory roles in flavonol biosynthesis in Arabidopsis anthers and pollen by activating the flavonol synthase 1 (AtFLS 1) gene (Stracke et al., 2010; Shan et al., 2020). The flower-specific expressed gene DlMYB42 clustered with AtMYB28 (HAG1), AtMYB29 (HAG2), and AtMYB86 (HAG2) with functions involved in aliphatic glucosinolate biosynthesis (Gigolashvili et al., 2008) in subgroup 12.
The transcriptome dynamics of 119 DlMYB genes in apical buds at different periods under KClO3 treatment were analyzed(Figure 8A and Supplementary Table 14). The time course analysis of 119 DlMYB genes at different periods under control and KClO3 treatment promoted the expression of DlMYB23 and DlMYB99 at the initial stage, which may be activated in response to the KClO3 induction (Figure 8B and Supplementary Table 14). Compared with the gene expression under control without KClO3 treatment, we observed that the expression of four genes (DlMYB22, DlMYB31, DlMYB68, and DlMYB71) were inhibited under KClO3 treatment, indicating that they may have a negative effect on flowering by KClO3 treatment. In general, DlMYB genes in the same subfamily tended to have similar expression patterns.
Co-expression network analysis via WGCNA was used to identify the role of DlMYB genes in KClO3-mediated FI (Figure 8C). Based on the FLOR-ID of Arabidopsis (Bouché et al., 2016), we found seven flower-related genes out of these 567 genes, including HTA11, WDR5a, TIL1, SENSITIVE TO FREEZING 6 (SFR6), HISTONE ACETYLTRANSFERASE OF THE MYST FAMILY 1 (HAM1), CURLY LEAF (CLF), and CYCLING DOF FACTOR 3 (CDF3). Some DlMYB hub genes were closely linked to other TF genes. For example, three DlMYB genes (DlMYB8, DlMYB33, and DlMYB78) were directly linked to TF genes such as WRKY75 and ERF114 in the MEbisque4 module (Supplementary Figure 8). The WRKY75 could positively regulate flowering in Arabidopsis by GA-mediated binding to the promoter of the FT gene (Zhang et al., 2018). WRKY2 and WRKY34 repressed AtMYB97, AtMYB101, and AtMYB120 expression during male gametogenesis (Lei et al., 2018). DlMYB8 and WRKY75 were up-regulated in the early stages of KClO3 treatment and presumably they may positively coregulate FT gene expression and eventually induce flowering in longan. In the MEroyalblue module, on the contrary, DlMYB31, DlMYB51 and ERF1 were down-regulated after KClO3 treatment. In Arabidopsis, ERF1 delayed flowering by repressing the expression of the FT gene (Chen et al., 2021). The downregulation of those genes under KClO3 may attenuate the repression of FT expression and ultimately promoted flowering. In the MEplum1 module, DlMYB41, TCP20, and AHP2 were all up-regulated in the earlier stages after KClO3 treatment (Supplementary Figure 8 and Supplementary Table 14). In Arabidopsis, interestingly, AHP2 functioned in the cytokinin pathway to promote female gametophyte development and TCP20 promoted the absorption of nitrate (Guan et al., 2014; Liu et al., 2017). Chlorate was a nitrate analog that could be absorbed by the nitrate transporters (Borges et al., 2004). Thus, DlMYB41 and TCP20 may together facilitate the utilization of KClO3, which was subsequently reduced to chlorite and hypochlorite and directly induced the stress response. Meanwhile, DlMYB41 and AHP2 were interacted to stimulate CK-mediated signal transduction. Eventually, they acted together to influence the expression of flowering-related genes and induce flowering. Whether and how these DlMYB genes function in longan flowering induction and development requires further experiments to elucidate.
In the current study, a systematic identification of R2R3-MYB gene family members in longan was carried out. We identified a total of 119 full-length R2R3-MYB genes in longan, which were further divided into 28 distinct subgroups by phylogenetic classification. A comprehensive bioinformatics analysis was performed to investigate the protein physicochemical properties, motif composition, gene structure, and cis-element prediction of the DlMYB genes. An uneven distribution of DlMYB genes on 15 chromosomes was observed with at least two genes per chromosome. The synteny analysis indicated segmental replication events were major contributors to the expansion of the R2R3-MYB gene family in longan. Comparative syntenic mapping among longan and other five representative species provided insights into the evolutionary relationship of orthologous MYB members and implied the presence of functional differentiation led by duplication events during evolution. Subcellular localization analysis revealed that the selected flowering-associated DlMYB genes mainly function in the nucleus as predicted. Expression profiling of DlMYB genes indicates their tissue-preferential expression pattern and KClO3-induced off-season flowering effect. GO enrichment analysis revealed that six DlMYB genes were assigned to the “flower development” category. The selected eight genes were further proved to be highly expressed in flowers and flower buds by qRT-PCR, indicating their possible involvement in the flower development process. Expression patterns in KClO3-mediated FI indicated two up-regulated and four down-regulated DlR2R3-MYB genes were involved. These results will open up untapped avenues for further functional analysis of the R2R3-MYB genes to elucidate their roles in longan FI and floral organ development.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JF and JZ conceived the study and designed the experiments. QC, JF, XZ, YF, BW, SX, and KZ carried out the experiments and analyzed the data. QC and JF wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by the Natural Science Foundation of Fujian Province, China under Grant Number 2019J0102; the Natural Science Foundation of China under Grant Number 41906096; the Middle-aged Teachers from Fujian Provincial Department of Education Grant Number JT180082; and the Scientific Research Innovation Program “Xiyuanjiang River Scholarship” of College of Life Sciences, Fujian Normal University under Grant Number 22FSSK011.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the editor and reviewers for their constructive comments on this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.820439/full#supplementary-material
Abbas, F., Ke, Y., Zhou, Y., Yu, Y., Waseem, M., Ashraf, U., et al. (2021). Genome-wide analysis reveals the potential role of MYB transcription factors in floral scent formation in Hedychium coronarium. Front. Plant Sci. 12:623742. doi: 10.3389/fpls.2021.623742
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bao, S., Hua, C., Shen, L., and Yu, H. (2020). New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 62, 118–131. doi: 10.1111/jipb.12892
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Borges, R., Miguel, E. C., Dias, J. M. R., Da Cunha, M., Bressan-Smith, R. E., De Oliveira, J. G., et al. (2004). Ultrastructural, physiological and biochemical analyses of chlorate toxicity on rice seedlings. Plant Sci. 166, 1057–1062. doi: 10.1016/j.plantsci.2003.12.023
Bouché, F., Lobet, G., Tocquin, P., and Périlleux, C. (2016). FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 44, D1167–D1171. doi: 10.1093/nar/gkv1054
Braun, E. L., and Grotewold, E. (1999). Newly discovered plant c-myb-like genes rewrite the evolution of the plant myb gene family. Plant Physiol. 121, 21–24.
Briesemeister, S., Rahnenführer, J., and Kohlbacher, O. (2010). YLoc–an interpretable web server for predicting subcellular localization. Nucleic Acids Res. 38, W497–W502. doi: 10.1093/nar/gkq477
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Chang, D., and Duda, T. F. Jr. (2012). Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol. Biol. Evol. 29, 2019–2029. doi: 10.1093/molbev/mss068
Chang, Q. (2010). Study on Relationship between off-Season Floral Induction in Longan and Nutrition of the Carbon and Nitrogen. Fuzhou: Fujian Agriculture & Forestry University. Master’s thesis.
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chen, Y., Zhang, L., Zhang, H., Chen, L., and Yu, D. (2021). ERF1 delays flowering through direct inhibition of FLOWERING LOCUS T expression in Arabidopsis. J. Integr. Plant Biol. 63, 1712–1723. doi: 10.1111/jipb.13144
Cheng, H., Song, S., Xiao, L., Soo, H. M., Cheng, Z., Xie, D., et al. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5:e1000440. doi: 10.1371/journal.pgen.1000440
Cho, L. H., Yoon, J., and An, G. (2017). The control of flowering time by environmental factors. Plant J. 90, 708–719. doi: 10.1111/tpj.13461
Chou, M. L., Liao, W. Y., Wei, W. C., Li, A. Y., Chu, C. Y., Wu, C. L., et al. (2018). The direct involvement of dark-Induced Tic55 protein in chlorophyll catabolism and its indirect role in the MYB108-NAC signaling pathway during leaf senescence in Arabidopsis thaliana. Int. J. Mol. Sci. 19:1854. doi: 10.3390/ijms19071854
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Devaiah, B. N., Madhuvanthi, R., Karthikeyan, A. S., and Raghothama, K. G. (2009). Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant 2, 43–58. doi: 10.1093/mp/ssn081
Ding, Z., Li, S., An, X., Liu, X., Qin, H., and Wang, D. (2009). Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genomics 36, 17–29. doi: 10.1016/s1673-8527(09)60003-5
Du, H., Yang, S. S., Liang, Z., Feng, B. R., Liu, L., Huang, Y. B., et al. (2012b). Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 12:106. doi: 10.1186/1471-2229-12-106
Du, H., Feng, B. R., Yang, S. S., Huang, Y. B., and Tang, Y. X. (2012a). The R2R3-MYB transcription factor gene family in maize. PLoS One 7:e37463. doi: 10.1371/journal.pone.0037463
Du, H., Liang, Z., Zhao, S., Nan, M. G., Tran, L. S., Lu, K., et al. (2015). The evolutionary history of R2R3-MYB proteins across 50 eukaryotes: new insights into subfamily classification and expansion. Sci. Rep. 5:11037. doi: 10.1038/srep11037
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gigolashvili, T., Engqvist, M., Yatusevich, R., Müller, C., and Flügge, U. I. (2008). HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 177, 627–642. doi: 10.1111/j.1469-8137.2007.02295.x
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Guan, P., Wang, R., Nacry, P., Breton, G., Kay, S. A., Pruneda-Paz, J. L., et al. (2014). Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 111, 15267–15272. doi: 10.1073/pnas.1411375111
Hong, L., Niu, F., Lin, Y., Wang, S., Chen, L., and Jiang, L. (2021). MYB106 is a negative regulator and a substrate for CRL3BPM E3 ligase in regulating flowering time in Arabidopsis thaliana. J Integr. Plant Biol. 63, 1104–1119. doi: 10.1111/jipb.13071
Huang, F. N., Fu, Z. Y., Zeng, L. H., and Morley-Bunker, M. (2017). Isolation and characterization of GI and FKF1 homologous genes in the subtropical fruit tree Dimocarpus longan. Mol. Breed. 37:13. doi: 10.1007/s11032-017-0691-z
Huang, H., Gao, H., Liu, B., Qi, T., Tong, J., Xiao, L., et al. (2017). Arabidopsis MYB24 regulates jasmonate-mediated stamen sevelopment. Front. Plant Sci. 8:1525. doi: 10.3389/fpls.2017.01525
Huang, S., Han, D., Wang, J., Guo, D., and Li, J. (2021). Floral induction of longan (Dimocarpus longan) by potassium chlorate: application, mechanism, and future perspectives. Front. Plant Sci. 12:670587. doi: 10.3389/fpls.2021.670587
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Hurst, L. D. (2002). The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18:486. doi: 10.1016/s0168-9525(02)02722-1
Jia, L., Clegg, M. T., and Jiang, T. (2004). Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol. 134, 575–585. doi: 10.1104/pp.103.027201
Jia, T. Q., Wei, D. F., Meng, S., Allan, A. C., and Zeng, L. H. (2014). Identification of regulatory genes implicated in continuous flowering of longan (Dimocarpus longan L.). PLoS One 9:e114568. doi: 10.1371/journal.pone.0114568
Jiang, C., Gu, J., Chopra, S., Gu, X., and Peterson, T. (2004). Ordered origin of the typical two- and three-repeat Myb genes. Gene 326, 13–22. doi: 10.1016/j.gene.2003.09.049
Jiang, C. K., and Rao, G. Y. (2020). Insights into the diversification and evolution of R2R3-MYB transcription factors in plants. Plant Physiol. 183, 637–655. doi: 10.1104/pp.19.01082
Jung, C., Seo, J. S., Han, S. W., Koo, Y. J., Kim, C. H., Song, S. I., et al. (2008). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 146, 623–635. doi: 10.1104/pp.107.110981
Kanei-Ishii, C., Sarai, A., Sawazaki, T., Nakagoshi, H., He, D. N., Ogata, K., et al. (1990). The tryptophan cluster: a hypothetical structure of the DNA-binding domain of the myb protooncogene product. J. Biol. Chem. 265, 19990–19995. doi: 10.1016/S0021-9258(17)45472-X
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., and Bansal, K. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:544. doi: 10.1186/1471-2164-13-544
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, S. H., Kim, H. S., Bahk, S., An, J., Yoo, Y., Kim, J. Y., et al. (2017). Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res. 45, 6613–6627. doi: 10.1093/nar/gkx417
Kim, S. H., Lam, P. Y., Lee, M. H., Jeon, H. S., Tobimatsu, Y., and Park, O. K. (2020). The Arabidopsis R2R3 MYB transcription factor MYB15 is a key regulator of lignin biosynthesis in effector-triggered immunity. Front. Plant Sci. 11:583153. doi: 10.3389/fpls.2020.583153
Klempnauer, K. H., Gonda, T. J., and Bishop, J. M. (1982). Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell 31, 453–463. doi: 10.1016/0092-8674(82)90138-6
Kranz, H., Scholz, K., and Weisshaar, B. (2000). c-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J. 21, 231–235. doi: 10.1046/j.1365-313x.2000.00666.x
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, H. G., and Seo, P. J. (2015). The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 82, 962–977. doi: 10.1111/tpj.12866
Lei, R., Ma, Z., and Yu, D. (2018). WRKY2/34-VQ20 Modules in Arabidopsis thaliana negatively regulate expression of a trio of related MYB transcription factors during pollen development. Front. Plant Sci. 9:331. doi: 10.3389/fpls.2018.00331
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, W., Liu, Y., Zhao, J., Zhen, X., Guo, C., and Shu, Y. (2019). Genome-wide identification and characterization of R2R3-MYB genes in Medicago truncatula. Genet. Mol. Biol. 42, 611–623. doi: 10.1590/1678-4685-gmb-2018-0235
Liang, Y., Tan, Z. M., Zhu, L., Niu, Q. K., Zhou, J. J., Li, M., et al. (2013). MYB97, MYB101 and MYB120 function as male factors that control pollen tube-synergid interaction in Arabidopsis thaliana fertilization. PLoS Genet. 9:e1003933. doi: 10.1371/journal.pgen.1003933
Lin, Y., Min, J., Lai, R., Wu, Z., Chen, Y., Yu, L., et al. (2017). Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics. Gigascience 6, 1–14. doi: 10.1093/gigascience/gix023
Liu, Z., Yuan, L., Song, X., Yu, X., and Sundaresan, V. (2017). AHP2, AHP3, and AHP5 act downstream of CKI1 in Arabidopsis female gametophyte development. J. Exp. Bot. 68, 3365–3373. doi: 10.1093/jxb/erx181
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative pcr and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Mandaokar, A., and Browse, J. (2009). MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol. 149, 851–862. doi: 10.1104/pp.108.132597
Matsumoto, T. K. (2006). Genes uniquely expressed in vegetative and potassium chlorate induced floral buds of Dimocarpus longan. Plant Sci. 170, 500–510. doi: 10.1016/j.plantsci.2005.09.016
Millar, A. A., and Gubler, F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17, 705–721. doi: 10.1105/tpc.104.027920
Millard, P. S., Kragelund, B. B., and Burow, M. (2019a). R2R3 MYB transcription factors - functions outside the DNA-binding domain. Trends Plant Sci. 24, 934–946. doi: 10.1016/j.tplants.2019.07.003
Millard, P. S., Weber, K., Kragelund, B. B., and Burow, M. (2019b). Specificity of MYB interactions relies on motifs in ordered and disordered contexts. Nucleic Acids Res. 47, 9592–9608. doi: 10.1093/nar/gkz691
Naik, J., Rajput, R., Pucker, B., Stracke, R., and Pandey, A. (2021). The R2R3-MYB transcription factor MtMYB134 orchestrates flavonol biosynthesis in Medicago truncatula. Plant Mol. Biol. 106, 157–172. doi: 10.1007/s11103-021-01135-x
Nguyen Ba, A. N., Pogoutse, A., Provart, N., and Moses, A. M. (2009). NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 10:202. doi: 10.1186/1471-2105-10-202
Nicholas, K. B., Nicholas, H., and Deerfield, D. (1997). GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14.
Ogata, K., Hojo, H., Aimoto, S., Nakai, T., Nakamura, H., Sarai, A., et al. (1992). Solution structure of a DNA-binding unit of Myb: a helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc. Natl. Acad. Sci. U. S. A. 89, 6428–6432. doi: 10.1073/pnas.89.14.6428
Parenteau, J., Maignon, L., Berthoumieux, M., Catala, M., Gagnon, V., and Abou Elela, S. (2019). Introns are mediators of cell response to starvation. Nature 565, 612–617. doi: 10.1038/s41586-018-0859-7
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P. A., and Saedler, H. (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. Embo J. 6, 3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x
Phan, H. A., Iacuone, S., Li, S. F., and Parish, R. W. (2011). The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23, 2209–2224. doi: 10.1105/tpc.110.082651
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Pucker, B., Pandey, A., Weisshaar, B., and Stracke, R. (2020). The R2R3-MYB gene family in banana (Musa acuminata): genome-wide identification, classification and expression patterns. PLoS One 15:e0239275. doi: 10.1371/journal.pone.0239275
Rangkadilok, N., Worasuttayangkurn, L., Bennett, R. N., and Satayavivad, J. (2005). Identification and quantification of polyphenolic compounds in longan (Euphoria longana Lam.) fruit. J. Agric. Food Chem. 53, 1387–1392. doi: 10.1021/jf0403484
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rosinski, J. A., and Atchley, W. R. (1998). Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J. Mol. Evol. 46, 74–83. doi: 10.1007/pl00006285
Roy, S. (2016). Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 11:e1117723. doi: 10.1080/15592324.2015.1117723
Seo, P. J., Xiang, F., Qiao, M., Park, J. Y., Lee, Y. N., Kim, S. G., et al. (2009). The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151, 275–289. doi: 10.1104/pp.109.144220
Shan, X., Li, Y., Yang, S., Yang, Z., Qiu, M., Gao, R., et al. (2020). The spatio-temporal biosynthesis of floral flavonols is controlled by differential phylogenetic MYB regulators in Freesia hybrida. New Phytol. 228, 1864–1879. doi: 10.1111/nph.16818
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shcherbo, D., Merzlyak, E. M., Chepurnykh, T. V., Fradkov, A. F., Ermakova, G. V., Solovieva, E. A., et al. (2007). Bright far-red fluorescent protein for whole-body imaging. Nat. Methods 4, 741–746. doi: 10.1038/nmeth1083
Sneddon, T. P., Li, P., and Edmunds, S. C. (2012). GigaDB: announcing the GigaScience database. Gigascience 1:11. doi: 10.1186/2047-217x-1-11
Song, S., Qi, T., Huang, H., Ren, Q., Wu, D., Chang, C., et al. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23, 1000–1013. doi: 10.1105/tpc.111.083089
Sringarm, K., Potchanasin, P., Sruamsiri, P., and Bangerth, K. F. (2009). Floral induction (FI) in longan (Dimocarpus longan, Lour.) trees-The possible participation of endogenous hormones II. Low temperature and potassium chlorate effects on hormone concentrations in and their export out of leaves. Sci. Hortic. 122, 295–300. doi: 10.1016/j.scienta.2008.11.031
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stracke, R., Holtgräwe, D., Schneider, J., Pucker, B., Sörensen, T. R., and Weisshaar, B. (2014). Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol. 14:249. doi: 10.1186/s12870-014-0249-8
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Stracke, R., Jahns, O., Keck, M., Tohge, T., Niehaus, K., Fernie, A. R., et al. (2010). Analysis of production of flavonol glycosides-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol. 188, 985–1000. doi: 10.1111/j.1469-8137.2010.03421.x
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/s1369-5266(00)00199-0
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., and Chen, W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Sun, J., Chen, M., Zhu, M., Jiang, Y., Meng, J., Zhao, D., et al. (2018). Cloning, characterization, and expression analysis of three FAD8 genes encoding a fatty acid desaturase from seeds of Paeonia ostii. Molecules 23:929. doi: 10.3390/molecules23040929
Sun, W., Ma, Z., Chen, H., and Liu, M. (2019). MYB gene family in potato (Solanum tuberosum L.): genome-wide identification of hormone-responsive reveals their potential functions in growth and development. Int. J. Mol. Sci. 20:4847. doi: 10.3390/ijms20194847
Teng, S., Keurentjes, J., Bentsink, L., Koornneef, M., and Smeekens, S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139, 1840–1852. doi: 10.1104/pp.105.066688
Tombuloglu, H. (2020). Genome-wide identification and expression analysis of R2R3, 3R- and 4R-MYB transcription factors during lignin biosynthesis in flax (Linum usitatissimum). Genomics 112, 782–795. doi: 10.1016/j.ygeno.2019.05.017
Untergasser, A., Nijveen, H., Rao, X., Bisseling, T., Geurts, R., and Leunissen, J. A. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–W74. doi: 10.1093/nar/gkm306
Van de Peer, Y. (2004). Computational approaches to unveiling ancient genome duplications. Nat. Rev. Genet. 5, 752–763. doi: 10.1038/nrg1449
Waheed, S., Peng, Y., and Zeng, L. H. (2020). Identification and characterization of DlGI promoter Involved in photoperiod, light Intensity, hormone, and DlELF4 response from longan. J. Am. Soc. Hortic. Sci. 145, 340–348. doi: 10.21273/jashs04946-20
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics 8, 77–80. doi: 10.1016/s1672-0229(10)60008-3
Wang, J., Liu, Y., Tang, B., Dai, X., Xie, L., Liu, F., et al. (2020). Genome-wide identification and capsaicinoid biosynthesis-related expression analysis of the R2R3-MYB gene family in Capsicum annuum L. Front. Genet. 11:598183. doi: 10.3389/fgene.2020.598183
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wang, Y., Zhang, Y., Fan, C., Wei, Y., Meng, J., Li, Z., et al. (2021). Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 21:328. doi: 10.1186/s12870-021-03083-6
Wilkins, O., Nahal, H., Foong, J., Provart, N. J., and Campbell, M. M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 149, 981–993. doi: 10.1104/pp.108.132795
Williams, C. E., and Grotewold, E. (1997). Differences between plant and animal Myb domains are fundamental for DNA binding activity, and chimeric Myb domains have novel DNA binding specificities. J. Biol. Chem. 272, 563–571. doi: 10.1074/jbc.272.1.563
Winterhagen, P., Hegele, M., Tiyayon, P., and Wunsche, J. N. (2020). Cytokinin accumulation and flowering gene expression are orchestrated for floral meristem development in longan (Dimocarpus longan Lour.) after chemical flower induction. Sci. Hortic. 270:109467. doi: 10.1016/j.scienta.2020.109467
Winterhagen, P., Tiyayon, P., Samach, A., Hegele, M., and Wunsche, J. N. (2013). Isolation and characterization of FLOWERING LOCUS T subforms and APETALA1 of the subtropical fruit tree Dimocarpus longan. Plant Physiol. Biochem. 71, 184–190. doi: 10.1016/j.plaphy.2013.07.013
Wong, D. C. J., Schlechter, R., Vannozzi, A., Höll, J., Hmmam, I., Bogs, J., et al. (2016). A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Res. 23, 451–466. doi: 10.1093/dnares/dsw028
Wu, J., Jiang, Y., Liang, Y., Chen, L., Chen, W., and Cheng, B. (2019). Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol. Biochem. 137, 179–188. doi: 10.1016/j.plaphy.2019.02.010
Yang, Y., Zhang, L., Chen, P., Liang, T., Li, X., and Liu, H. (2020). UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. Embo J. 39:e101928. doi: 10.15252/embj.2019101928
Yu, G., Wang, L. G., Han, Y., and He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287. doi: 10.1089/omi.2011.0118
Zhang, L., Chen, L., and Yu, D. (2018). Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 176, 790–803. doi: 10.1104/pp.17.00657
Zhang, X. F., Guo, S., Ho, C. T., and Bai, N. S. (2020). Phytochemical constituents and biological activities of longan (Dimocarpus longan Lour.) fruit: a review. Food Sci. Hum. Wellness 9, 95–102. doi: 10.1016/j.fshw.2020.03.001
Zhao, F., Zhao, T., Deng, L., Lv, D., Zhang, X., Pan, X., et al. (2017). Visualizing the essential role of complete virion assembly machinery in efficient hepatitis c virus cell-to-cell transmission by a viral infection-activated split-intein-mediated reporter system. J. Virol. 91:e1720-16. doi: 10.1128/jvi.01720-16
Zhao, P., Hou, S., Guo, X., Jia, J., Yang, W., Liu, Z., et al. (2019). A MYB-related transcription factor from sheepgrass, LcMYB2, promotes seed germination and root growth under drought stress. BMC Plant Biol. 19:564. doi: 10.1186/s12870-019-2159-2
Zhao, Y., Xing, L., Wang, X., Hou, Y. J., Gao, J., Wang, P., et al. (2014). The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 7:ra53. doi: 10.1126/scisignal.2005051
Zheng, W., Dong, X. M., Zhang, Q. Y., Yu, X. F., Li, W. L., Ji, Y. B., et al. (2020). Identification and characterization of myb genes in Dimocarpus Longan Lour. Bangladesh J. Bot. 49, 97–104. doi: 10.3329/bjb.v49i1.49100
Keywords: Dimocarpus longan, R2R3-MYB, flower development, KClO3, qRT-PCR
Citation: Chen Q, Zhang X, Fang Y, Wang B, Xu S, Zhao K, Zhang J and Fang J (2022) Genome-Wide Identification and Expression Analysis of the R2R3-MYB Transcription Factor Family Revealed Their Potential Roles in the Flowering Process in Longan (Dimocarpus longan). Front. Plant Sci. 13:820439. doi: 10.3389/fpls.2022.820439
Received: 23 November 2021; Accepted: 02 March 2022;
Published: 25 March 2022.
Edited by:
Shunquan Lin, South China Agricultural University, ChinaReviewed by:
Fanchang Zeng, Shandong Agricultural University, ChinaCopyright © 2022 Chen, Zhang, Fang, Wang, Xu, Zhao, Zhang and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingping Fang, amlucGhpYUAxNjMuY29t; Jisen Zhang, emppc2VuQGZhZnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.