
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 10 January 2023
Sec. Plant Genetics, Epigenetics and Chromosome Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1101912
This article is part of the Research TopicRecent Advances of Epigenetics in Crop Biotechnology, Volume IIView all 7 articles
Regulation of gene expression underpins gene function and is essential for regulation of physiological roles. Epigenetic modifications regulate gene transcription by physically facilitating relaxation or condensation of target loci in chromatin. Transcriptional corepressors are involved in chromatin remodeling and regulate gene expression by establishing repressive complexes. Genetic and biochemical studies reveal that a member of the Groucho/Thymidine uptake 1 (Gro/Tup1) corepressor family, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 15 (HOS15), is recruited via the evening complex (EC) to the GIGANTEA (GI) promoter to repress gene expression, and modulating flowering time. Therefore, HOS15 connects photoperiodic pathway and epigenetic mechanism to control flowering time in plants. In addition, growing body of evidence support a diverse roles of the epigenetic regulator HOS15 in fine-tuning plant development and growth by integrating intrinsic genetic components and various environmental signals.
Plants are the most fundamental organisms on earth that sustain global ecosystem and food production (Raven, 2019; Eckardt et al., 2022). The changing environmental conditions, however, has threatened the plants. Unfortunately, land plants as sessile organisms are unable to choose or change environment for their development and growth. Plants as multicellular organisms are strikingly plastic and have evolved the ability to coordinate their growing phenotype in response to the changing environment (Pierik et al., 2021). For instance, the timing of flowering is a highly plastic phenomenon. A variety of external signals, such as day length and temperature, may induce endogenous physiological changes and accelerate flowering, the critical switch from vegetative to reproductive growth (Srikanth and Schmid, 2011; Freytes et al., 2021). Cells of a complex multicellular organism are homogeneous at genomic level but are physiologically and functionally heterogeneous due to the dynamic expression of genes. The spatiotemporal regulation of gene expression, which determines the timing and pattern of gene function, supports the phenotypic plasticity of plant development and growth (Nicotra et al., 2010; Jong and Leyser, 2012). Epigenetic regulation of chromatin status has become one of the most exciting frontiers of gene expression research in plant science. Epigenetic modification influences chromatin conformation and related transcriptional states by means of DNA methylation and histone modifications (Jaenisch and Bird, 2003; Gibney and Nolan, 2010; Eriksson et al., 2020).
Corepressors are transcriptional regulators (Courey and Jia, 2001). These transcriptional corepressors epigenetically repress target genes by forming a multi-protein complex with other transcription factors, adapters and accessory proteins. HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 15 (HOS15) belongs to the most prominent and evolutionarily conserved Groucho/Thymidine uptake 1 (Gro/Tup1) family corepressors, which include the Gro protein in nematodes, the Tup1 protein in yeasts, and the transducin beta-like protein 1 (TBL1) protein in mammals. They contain a N-terminal glutamine (Q)-rich motif and a C-terminal region comprising multiple-repeat of approximately forty amino acids enriched for tryptophan-aspartic acid dipeptide (WD40). In Arabidopsis, at least 13 members of the Gro/Tup1 family corepressor have been identified, including TOPLESS (TPL) and four TPL-related proteins (TPRs), LEUNIG (LUG) and LUG HOMOLOG (LUH), HOS15 and other uncharacterized members (Liu and Karmarkar, 2008; Lee and Golz, 2012). Among them, the TPL/TPR sub-family proteins selectively interact with HDA19. The TPL-HDA19 module has been implicated in regulating a broad range of developmental and environmental processes by association with different transcription factors (Long et al., 2006; Causier et al., 2012; Krogan et al., 2012; Wang et al., 2013; Oh et al., 2014; Plant et al., 2021). Thus, it was not surprising when HOS15 was found to be involved in a variety of biological processes, like TPL/TPRs, through connecting with histone deacetylases. However, neither HOS15 nor TPL/TPRs contains a DNA binding domain, therefore, the corepressors require additional patterners to reach promoter regions of target genes. Recent study revealed that HOS15 was recruited by a protein complex consisting of transcription factors to the promoter region of GIGANTEA (GI) to repress photoperiodic pathway which accelerates Arabidopsis flowering under long days (Hammond, 2019; Park et al., 2019). This review focus on the molecular mechanism of HOS15-mediated epigenetic regulation of flowering and highlights the role of HOS15 in integration of both endogenous physiological cues and exogenous environmental stimuli stresses.
Previously, an elegant high-throughput genetic screening system was designed to decipher the complex osmotic and cold stress signaling cascade in Arabidopsis using the chimeric gene construct as reliable reporter, in which the stress-responsive RD29A (also known as COR78 for cold regulated 78) gene promoter was used to control the expression of the firefly luciferase gene that quantitatively and faithfully reflecting the RD29A promoter activity (Ishitani et al., 1997). A large number of different categorized mutants with altered expression of the reporter gene was identified, such as cos, los, and hos, named after constitutive, low, and high expression of osmotically responsive genes, respectively. Interestingly, some of those mutants also displayed a plethora of developmental phenotypes, for examples, the hos1 mutant with defect in a RING finger E3 ubiquitin ligase coding gene (gene locus: AT2G39810) flowers remarkably earlier than wild-type (Jung et al., 2012; Lee et al., 2012); while the hos2 [also designated fiery1 (fry1) for strong expression of firefly luciferase reporter activity] mutant with defect in a bifunctional phosphoadenosine phosphatase coding gene (AT5G63980) has fewer lateral roots, shorter petioles, crinkly leaves, and flowers later (Xiong et al., 2001; Chen and Xiong, 2010). The pleiotropic developmental phenotypes of the HOS family mutants manifest that the developmental and stress response signaling pathways are genetically interactive.
The casual gene responsive for hos15 mutant phenotype was identified to encode a WD40-repeat protein belonging to the Gro/Tup1 family corepressor (Zhu et al., 2008), which includes a dozen of members in Arabidopsis (Figure 1). However, neither HOS15 interacting proteins nor its role in plant development has been described. While studying the molecular function of HOS15, Park et al. noticed, by coincidence, that HOS15 loss-of-function (hos15) plants flower earlier than wild-type (WT) plants. By contrast, the flowering time of HOS15 gain-of-function plants become delayed. Hooked by this robust and interesting phenotype, the authors set out to elucidate the molecular mechanism underlying HOS15-mediated control of flowering time.
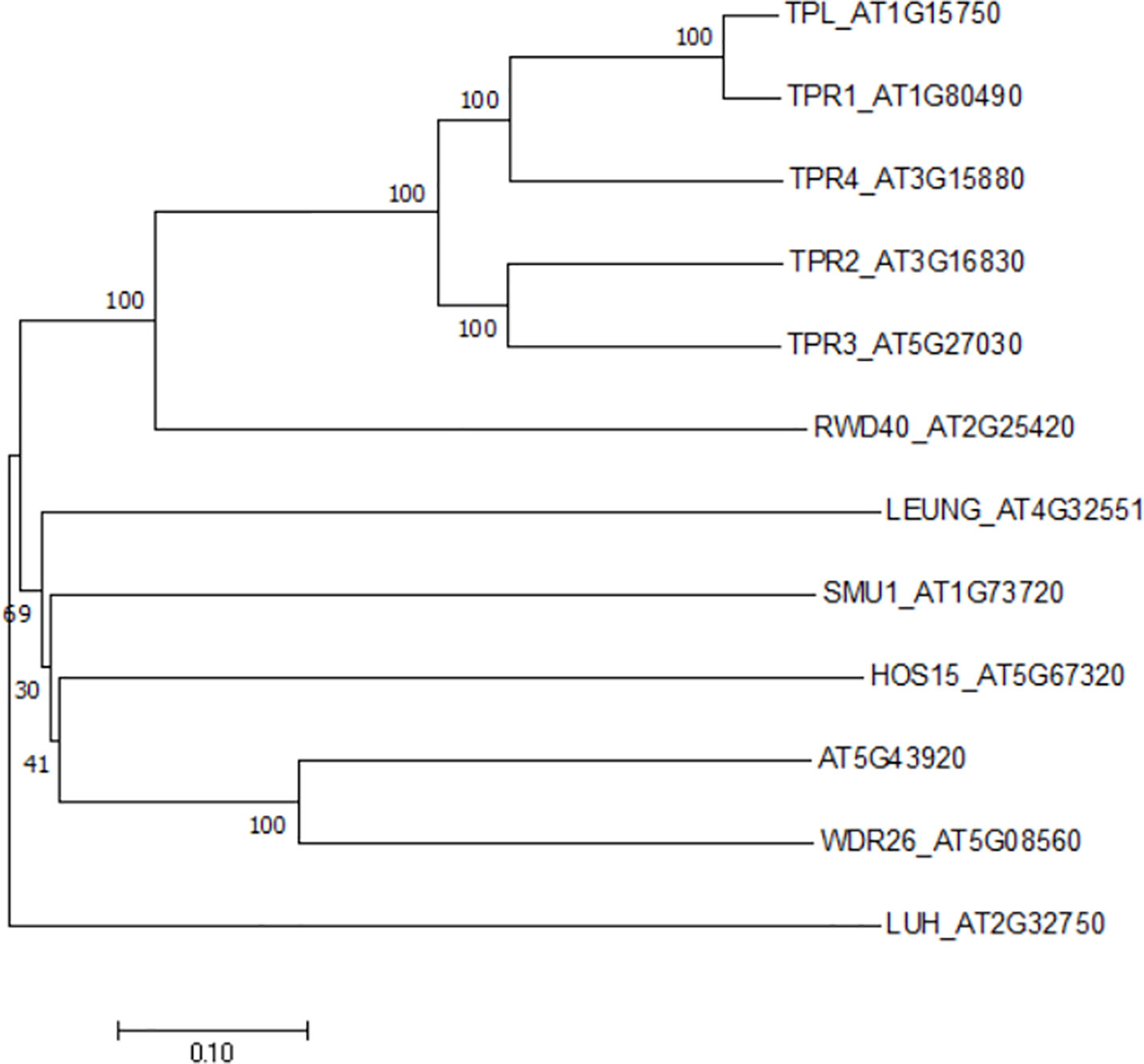
Figure 1 A phylogenetic tree of the Gro/Tup1 corepressor gene family in Arabidopsis. The gene names were followed by TAIR locus numbers. An unrooted neighbor- joining tree was generated by MEGA X with full-length sequences (1, 000 bootstrap replicates).
Regulation of flowering time has been a hot topic studied intensively over the past 100 years. These studies implicated that the initiation of flowering was controlled by multiple genetically defined pathways that integrate both exogenous environmental cues and endogenous developmental signals (Andrés and Coupland, 2012). The vernalization pathway refers to the establishment of competence to flower after a prolonged exposure of plants to cold. The gibberellic pathway refers to the requirement of the phytohormone, gibberellin (GA), to promote normal flowering patterns. The photoperiodic pathway refers to the regulation of flowering by day length and light quality (Shim et al., 2017). CONSTANS (CO) plays a central role in the photoperiodic flowering pathway, in which the rhythmic expression pattern of CO is regulated by GIGANTEA (GI) (Fowler et al., 1999; Park et al., 1999; Mizoguchi et al., 2005; Park et al., 2013). The endogenous regulators that promote flowering in a photoperiod- and GA-independent way often referred to as the autonomous pathway. To first crack the mystery by determining which pathway was affected in hos15 mutants, a high-throughput RNA sequencing experiment was performed to identify if any genes of specific pathway were differentially expressed in hos15 mutants when compared to WT. The expression of key floral pathway integrator genes, such as FLOWERING LOCUS T (FT) (Corbesier et al., 2007), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) (Samach et al., 2000; Yoo et al., 2005), APETALA1 (AP1) (Mandel et al., 1992), AGAMOUS-LIKE 19 (AGL19) and AGL24 (Yu et al., 2002; Schönrock et al., 2006), was higher in hos15 mutants. This unbiased analysis also revealed that the expression of photoperiodic flowering pathway genes, including GI, CO, and FLAVIN-BINDING KELCH REPEAT F-BOX 1 (FKF1) (Ito et al., 2012), was specifically upregulated; whereas the expression of other flowering pathway genes was unchanged in hos15 mutants. The results strongly indicate that the photoperiodic pathway is provoked in hos15 mutants leading to early flowering.
As mentioned above, genes such as GI and CO are involved in promoting flowering in response to photoperiod. GI is a key circadian integrator and a master regulator of CO expression. Under long-day condition, light promotes the expression of GI and a ubiquitin-ligase FKF1 peak at the same time, leading to the optimal formation of the GI–FKF1 complex, which, in turn, promotes the degradation of transcriptional repressors of CO (Imaizumi et al., 2005; Sawa et al., 2007). CO is a floral activator that directly activates FT expression. Therefore, GI controls flowering time through regulating the CO-FT module. Indeed, GI protein stability is controlled by light and circadian clock. The clock-associated protein EARLY FLOWERING 3 (ELF3) acts as a substrate adaptor, enabling a RING-type E3 ubiquitin-ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) to target GI for the ubiquitin-26S proteasome-mediated proteolysis (Yu et al., 2008). Interestingly, GI transcript levels were consistently increased in elf3 mutants, which displays an early-flowering phenotype (Kim et al., 2005). Therefore, ELF3 controls GI protein abundancy at both transcriptional and post-transcriptional levels. But how ELF3 represses GI expression remain mysterious. ELF3 is a constituent component of a protein complex (called the evening complex [EC]), consisting of ELF3, ELF4 and a transcription factor, LUX ARRHYTHMO (LUX, also termed PHYTOCLOCK1), which directly binds to the GI promoter through LUX binding site (LBS) to repress GI transcription in the night (Mizuno et al., 2014).
Since GI transcription is repressed by both the evening complex and the transcriptional corepressor HOS15, the authors wondered whether the evening complex brings HOS15 to the GI promoter. A pilot chromatin Immunoprecipitation (ChIP) assay revealed that like LUX, HOS15 associated with the GI promoter via binding to the same LBS regions (Park et al., 2019). This result promoted the author to further investigate whether HOS15 can bind to the evening complex. Both in vitro Y2H and in vivo experiments confirmed that HOS15 interacted with the evening complex components, LUX and ELF3 as well. A gel-filtration analysis showed the presence of a high-order complex containing HOS15-LUX-ELF3 (Park et al., 2019). Further ChIP assays with loss-of-function mutants indicated that HOS15 and LUX were interdependent in terms of binding to GI promoter; HOS15 was unable to bind to GI promoter in lux mutants, and vice versa. Thus, it become clear that the transcriptional corepressor HOS15 was recruited to the GI promoter via direct association with the evening complex to silence GI expression. Now, there comes another question where is the real core repressor of the repression-complex established on the target promoter.
HOS15 contains a WD40 repeat motif and is ortholog to TBL1 in mammals (Zhu et al., 2008). TBL1 coordinates the formation of transcriptional repression complexes comprising either nuclear receptor corepressor (N-CoR, also known as NCOR1) or silencing mediator for retinoic acid and thyroid receptor (SMRT, also known as NCOR2), which function in chromatin modification of target gene promoter regions. TBL1 selectively deploys histone deacetylases (HDACs) of the Reduced potassium dependency 3 (Rpd3)-like superfamily that is broadly conserved in all eukaryotes (Yoon et al., 2005). HDACs are responsible for catalyzing histone deacetylation modification. In Arabidopsis genome the class I Rpd3-like HDAC family is composed of HDA6, HDA7, HDA9, and HDA19. Interestingly, HDA9 and HDA19 were found in a mass spectrometry-based proteomic approach aimed to identify HOS15-interacting proteins (Park et al., 2018a).
Genetic evidence that both hda9 single and hda9 hos15 double mutants phenocopied the early-flower phenotype of hos15 mutants, suggested that HDA9 was a strong candidate that functioned in association with HOS15. The authors biochemically confirmed that HDA9 interacted with HOS15 and HDA9 bond to GI promoter regions in a HOS15-dependent manner, by harnessing co-IP and ChIP analysis, respectively. Consistently, the Histone H3 on the GI locus was hyper-acetylated in hos15 mutants. Therefore, it appears evident that the EC complex recruits the HOS15-HDA9 module to the GI promoter and represses GI transcription by modulating the epigenetic status, and thereby delaying flowering time (Figure 2). The corepressor HOS15 connects photoperiodic pathway regulated by GI and epigenetic modification at histones catalyzed by HDA9 to control flowering time in plants.
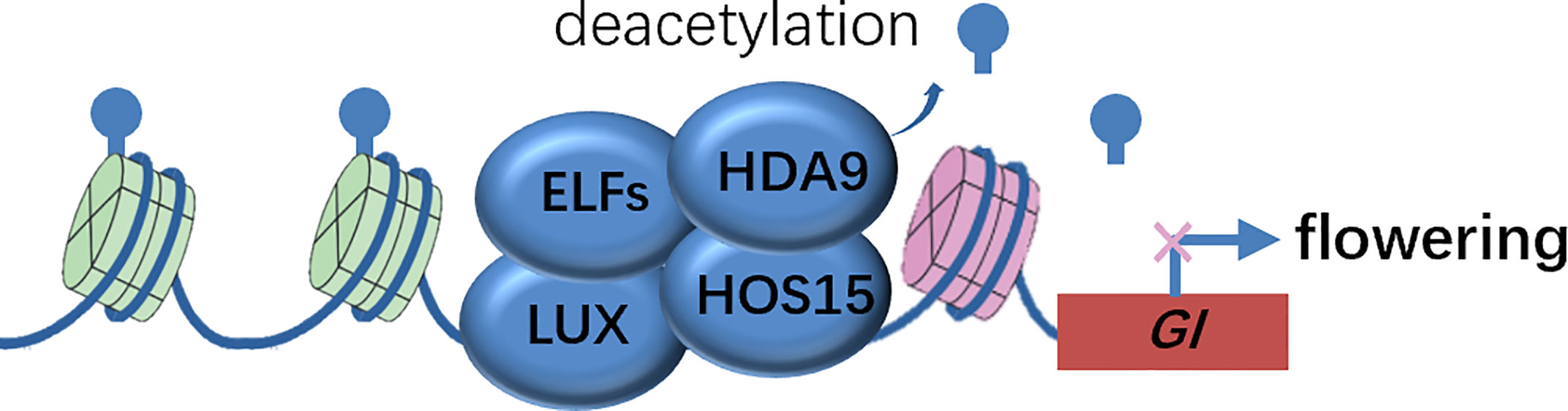
Figure 2 Molecular model of HOS15-mediated higher-order repressive complex in GI promoter region to inhibit GI expression via histone deacetylation. Inhibition of GI expression delays flowering time. EFLS, EARLY FLOWERINGS; GI, GIGANTEA; HDA9, HISTONE DEACETYLASE 9; HOS15, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 15; LUX, LUX ARRHYTHMO.
As broached above, the Arabidopsis Rpd3-like HDACs in general were believed to be involved in different plant biological processes by regulating gene expression through histone deacetylation. The mechanisms by which HDACs are selectively recruited to specific target gene loci in chromosome are not well understood. Notably, HDACs may have non-histone substrates (Zheng et al., 2020; Xu et al., 2021). In an immunoaffinity purification assay to identify protein complex associated with HDA9 revealed that the WRKY53 transcription factor could co-purified with HDA9 (Chen et al., 2016). A domain-swapping experiment further demonstrated that the deacetylase domain of HDA9 mediated the interaction with WRKY53. Biochemical analysis using an anti-acetylated lysine antibody revealed that WRKY53 lysine acetylation levels were increased and decrease in HDA9 loss- and gain-of-function mutants, respectively, as compared to the wild-type background (Chen et al., 2016). HDA9 might inhibit WRKY53 transcriptional activity through deacetylation modification, thereby regulating downstream gene expression. Interestingly, through a large-scale protein acetylome analysis, ribosomal proteins were identified as substrates of HDA714, a member of Rpd3-like HDACs in rice (Xu et al., 2021). Contrary to the long-held belief that histone deacetylases act only on the tails of histone proteins, HDACs might form distinct protein complexes and regulate gene expression through various mechanisms in different physiological contexts, which offers the possibility of complicate crosstalk and delicate balance among diverse signaling pathways.
The role of HOS15-mediated regulation of flowering time has been clearly elucidated in Arabidopsis. However, as a universal corepressor, it is not unexpected that HOS15 is involved in a myriad of physiological processes through interacting with corresponding transcription factors.
Mutation of HOS15 massively affects histone acetylation and methylation levels, which subsequently alters expression of a large number of genes involved in developmental programs and environmental responses (Mayer et al., 2019). In addition to regulate flowering time, Suzuki et al. reported that HOS15 plays a critical role in the specification and definition of aerial lateral organ size by promoting cell proliferation during leaf developmental program (Fujikura et al., 2009; Suzuki et al., 2018). HOS15 also promotes age-associated leaf senescence via differential regulation of senescence- and photosynthesis-related genes (Zareen et al., 2022).
HOS15 not only regulates plant development and growth but also participates in plant response and adaptation to environmental changes. Several lines of evidence supported that HOS15 interacts with histone deacetylase 2C (HD2C) and induces HD2C degradation via the CULLIN4-based E3 ubiquitin ligase in a cold-dependent manner, which releases the expression of cold responsive genes to enhance cold tolerance (Zhu et al., 2008; Park et al., 2018b; Lim et al., 2020; Lim et al., 2021). HOS15 negatively regulates Snf1-Related Kinases (SnRKs) activity of the abscisic acid (ABA) signaling pathway (Ali and Yun, 2020; Khan et al., 2022). In addition to be involved in abiotic stress response, HOS15 also regulates plants resistance to pathogen infections. Shen et al. revealed that HOS15 sophisticatedly manipulates plant defense response by destabilizing the transcriptional activity of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1), the master regulator of plant immunity networks (Shen et al., 2020). Yang et al. revealed that HOS15 intercepts plant defense response by repressing the expression of nucleotide-binding leucine rich repeat/NOD-like receptors, which recognize invading pathogen effectors to provoke effector-triggered immunity; therefore, mutation of HOS15 confers plants more resistant to pathogen infection (Yang et al., 2020). They also noticed that mutation of HOS15 results in enlarged siliques, the seed capsules of cruciferous plants (Mayer et al., 2019; Shen et al., 2020). In addition to acetylation, histone methylation, another type of post-translational modification may also be involved in fine-tuning the trade-off between growth and defense in plants. For example, the members of the histone methyltransferase family, SET DOMAIN GROUP 8 (SDG8) and ARABIDOPSIS TRITHORAX 1 (ATX1, also known as SDG27), positively regulate plant immune response to phytopathogens while actively repress plant flowering time (Berr et al., 2010; S., Y., Kim et al., 2005; Zhao et al., 2005; Pien et al., 2008; Lee et al., 2016). Regulation of histone modification is a gigantic platform for different pathways convergence and interaction that deserves further exploration.
The so far identified biological roles of Arabidopsis HOS15 were summarized in Figure 3. Studies of HOS15 function will elucidate the molecular mechanism underlying how plants epigenetically regulate development and environmental adaptations and trade-off between these two processes, which will eventually provide insights on how to develop novel high quality, high yield and stress resilient crops by innovative bottom-up molecular breeding technologies to sustain agriculture under global climate change.
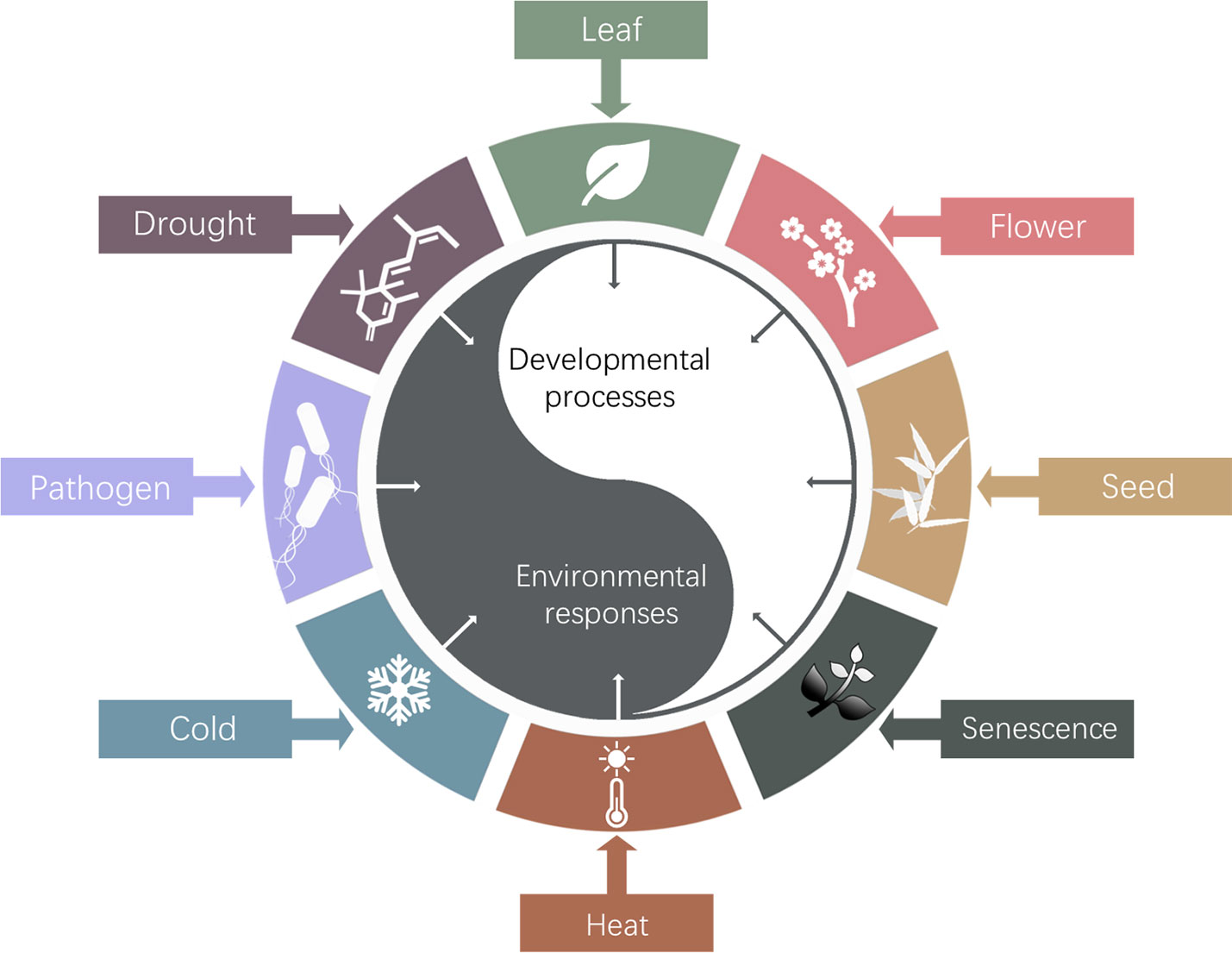
Figure 3 Schematic diagram of HOS15 regulated developmental processes and response to environmental stresses. So far, HOS15 was found to play multiple roles in plant organ development, such as leaf, flower, seed and senescence; and plant response to various abiotic and biotic stresses, such as drought, temperature, and pathogens.
L-JH, YW, and NL: conceptualization. L-JH, DJ, and NL: literature review. L-JH, ZL, and NL: writing—original preparation. NL and YL: writing—review and editing. L-JH, YW, DJ, and NL: design and revision of the images. All authors have read and agreed to the final version of the manuscript.
This work was supported by Training Program for Excellent Young Innovators of Changsha (kq2009016), National Natural Science Foundation of China (31901345), Natural Science Foundation of Hunan Province (2020JJ5970 and 2021JJ31141), the Education Department of Hunan Province (20A517 and 22B0260), and Postgraduate Scientific Research Innovation Project (CX20210885).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ali, A., Yun, D.-J. (2020). HOS15: A missing link that fine-tunes ABA signaling and drought tolerance in arabidopsis. Plant Signaling Behav. 15, 1770964. doi: 10.1080/15592324.2020.1770964
Andrés, F., Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Berr, A., McCallum, E. J., Alioua, A., Heintz, D., Heitz, T., Shen, W.-H. (2010). Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154, 1403–1414. doi: 10.1104/pp.110.161497
Causier, B., Ashworth, M., Guo, W., Davies, B. (2012). The TOPLESS interactome: a framework for gene repression in arabidopsis. Plant Physiol. 158, 423–438. doi: 10.1104/pp.111.186999
Chen, X., Lu, L., Mayer, K. S., Scalf, M., Qian, S., Lomax, A., et al. (2016). POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in arabidopsis. eLife 5, e17214. doi: 10.7554/eLife.17214
Chen, H., Xiong, L. (2010). The bifunctional abiotic stress signalling regulator and endogenous RNA silencing suppressor FIERY1 is required for lateral root formation. Plant Cell Environ. 33, 2180–2190. doi: 10.1111/j.1365-3040.2010.02218.x
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of arabidopsis. Sci. (New York N.Y.) 316, 1030–1033. doi: 10.1126/science.1141752
Courey, A. J., Jia, S. (2001). Transcriptional repression: the long and the short of it. Genes Dev. 15, 2786–2796. doi: 10.1101/gad.939601
Eckardt, N. A., Ainsworth, E. A., Bahuguna, R. N., Broadley, M. R., Busch, W., Carpita, N. C., et al. (2022). Climate change challenges, plant science solutions. Plant Cell, koac303. doi: 10.1093/plcell/koac303
Eriksson, M. C., Szukala, A., Tian, B., Paun, O. (2020). Current research frontiers in plant epigenetics: an introduction to a virtual issue. New Phytol. 226, 285–288. doi: 10.1111/nph.16493
Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., et al. (1999). GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688. doi: 10.1093/emboj/18.17.4679
Freytes, S. N., Canelo, M., Cerdán, P. D. (2021). Regulation of flowering time: When and where? Curr. Opin. Plant Biol. 63, 102049. doi: 10.1016/j.pbi.2021.102049
Fujikura, U., Horiguchi, G., Ponce, M. R., Micol, J. L., Tsukaya, H. (2009). Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of arabidopsis thaliana. Plant J.: For Cell Mol. Biol. 59, 499–508. doi: 10.1111/j.1365-313X.2009.03886.x
Gibney, E. R., Nolan, C. M. (2010). Epigenetics and gene expression. Heredity 105, 4–13. doi: 10.1038/hdy.2010.54
Hammond, R. K. (2019). HOS15 coregulates photoperiodic flowering with the evening complex via transcriptional repression of GIGANTEA. Plant Cell 31, 3–4. doi: 10.1105/tpc.19.00039
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A., Kay, S. A. (2005). FKF1 f-box protein mediates cyclic degradation of a repressor of CONSTANS in arabidopsis. Sci. (New York N.Y.) 309, 293–297. doi: 10.1126/science.1110586
Ishitani, M., Xiong, L., Stevenson, B., Zhu, J. K. (1997). Genetic analysis of osmotic and cold stress signal transduction in arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9, 1935–1949. doi: 10.1105/tpc.9.11.1935
Ito, S., Song, Y. H., Imaizumi, T. (2012). LOV domain-containing f-box proteins: light-dependent protein degradation modules in arabidopsis. Mol. Plant 5, 573–582. doi: 10.1093/mp/sss013
Jaenisch, R., Bird, A. (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33 (Suppl), 245–254. doi: 10.1038/ng1089
Jong, M., Leyser, O. (2012). Developmental plasticity in plants. Cold Spring Harbor Symp. Quant. Biol. 77, 63–73. doi: 10.1101/sqb.2012.77.014720
Jung, J.-H., Seo, P. J., Park, C.-M. (2012). The E3 ubiquitin ligase HOS1 regulates arabidopsis flowering by mediating CONSTANS degradation under cold stress. J. Biol. Chem. 287, 43277–43287. doi: 10.1074/jbc.M112.394338
Khan, I. U., Ali, A., Zareen, S., Khan, H. A., Lim, C. J., Park, J., et al. (2022). Non-expresser of PR-genes 1 positively regulates abscisic acid signaling in arabidopsis thaliana. Plants (Basel Switzerland) 11, 815. doi: 10.3390/plants11060815
Kim, S. Y., He, Y., Jacob, Y., Noh, Y.-S., Michaels, S., Amasino, R. (2005). Establishment of the vernalization-responsive, winter-annual habit in arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17, 3301–3310. doi: 10.1105/tpc.105.034645
Kim, W.-Y., Hicks, K. A., Somers, D. E. (2005). Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol. 139, 1557–1569. doi: 10.1104/pp.105.067173
Krogan, N. T., Hogan, K., Long, J. A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Dev. (Cambridge England) 139, 4180–4190. doi: 10.1242/dev.085407
Lee, S., Fu, F., Xu, S., Lee, S. Y., Yun, D.-J., Mengiste, T. (2016). Global regulation of plant immunity by histone lysine methyl transferases. Plant Cell 28, 1640–1661. doi: 10.1105/tpc.16.00012
Lee, J. E., Golz, J. F. (2012). Diverse roles of Groucho/Tup1 co-repressors in plant growth and development. Plant Signaling Behav. 7, 86–92. doi: 10.4161/psb.7.1.18377
Lee, J. H., Kim, J. J., Kim, S. H., Cho, H. J., Kim, J., Ahn, J. H. (2012). The E3 ubiquitin ligase HOS1 regulates low ambient temperature-responsive flowering in arabidopsis thaliana. Plant Cell Physiol. 53, 1802–1814. doi: 10.1093/pcp/pcs123
Lim, C. J., Ali, A., Park, J., Shen, M., Park, K. S., Baek, D., et al. (2021). HOS15-PWR chromatin remodeling complex positively regulates cold stress in arabidopsis. Plant Signaling Behav. 16, 1893978. doi: 10.1080/15592324.2021.1893978
Lim, C. J., Park, J., Shen, M., Park, H. J., Cheong, M. S., Park, K. S., et al. (2020). The histone-modifying complex PWR/HOS15/HD2C epigenetically regulates cold tolerance. Plant Physiol. 184, 1097–1111. doi: 10.1104/pp.20.00439
Liu, Z., Karmarkar, V. (2008). Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci. 13, 137–144. doi: 10.1016/j.tplants.2007.12.005
Long, J. A., Ohno, C., Smith, Z. R., Meyerowitz, E. M. (2006). TOPLESS regulates apical embryonic fate in arabidopsis. Sci. (New York N.Y.) 312, 1520–1523. doi: 10.1126/science.1123841
Mandel, M. A., Gustafson-Brown, C., Savidge, B., Yanofsky, M. F. (1992). Molecular characterization of the arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. doi: 10.1038/360273a0
Mayer, K. S., Chen, X., Sanders, D., Chen, J., Jiang, J., Nguyen, P., et al. (2019). HDA9-PWR-HOS15 is a core histone deacetylase complex regulating transcription and development. Plant Physiol. 180, 342–355. doi: 10.1104/pp.18.01156
Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., et al. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in arabidopsis. Plant Cell 17, 2255–2270. doi: 10.1105/tpc.105.033464
Mizuno, T., Kitayama, M., Oka, H., Tsubouchi, M., Takayama, C., Nomoto, Y., et al. (2014). The EC night-time repressor plays a crucial role in modulating circadian clock transcriptional circuitry by conservatively double-checking both warm-night and night-time-light signals in a synergistic manner in arabidopsis thaliana. Plant Cell Physiol. 55, 2139–2151. doi: 10.1093/pcp/pcu144
Nicotra, A. B., Atkin, O. K., Bonser, S. P., Davidson, A. M., Finnegan, E. J., Mathesius, U., et al. (2010). Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15, 684–692. doi: 10.1016/j.tplants.2010.09.008
Oh, E., Zhu, J.-Y., Ryu, H., Hwang, I., Wang, Z.-Y. (2014). TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat. Commun. 5, 4140. doi: 10.1038/ncomms5140
Park, H. J., Baek, D., Cha, J.-Y., Liao, X., Kang, S.-H., McClung, C. R., et al. (2019). HOS15 interacts with the histone deacetylase HDA9 and the evening complex to epigenetically regulate the floral activator GIGANTEA. Plant Cell 31, 37–51. doi: 10.1105/tpc.18.00721
Park, H. J., Kim, W.-Y., Yun, D.-J. (2013). A role for GIGANTEA. Plant Signaling Behav. 8, e24820. doi: 10.4161/psb.24820
Park, J., Lim, C. J., Khan, I. U., Jan, M., Khan, H. A., Park, H. J., et al. (2018a). Identification and molecular characterization of HOS15-interacting proteins in arabidopsis thaliana. J. Plant Biol. 61, 336–345. doi: 10.1007/s12374-018-0313-2
Park, J., Lim, C. J., Shen, M., Park, H. J., Cha, J.-Y., Iniesto, E., et al. (2018b). Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. United States America 115, E5400–E5409. doi: 10.1073/pnas.1721241115
Park, D. H., Somers, D. E., Kim, Y. S., Choy, Y. H., Lim, H. K., Soh, M. S., et al. (1999). Control of circadian rhythms and photoperiodic flowering by the arabidopsis GIGANTEA gene. Sci. (New York N.Y.) 285, 1579–1582. doi: 10.1126/science.285.5433.1579
Pien, S., Fleury, D., Mylne, J. S., Crevillen, P., Inzé, D., Avramova, Z., et al. (2008). ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS c activation via histone 3 lysine 4 trimethylation. Plant Cell 20, 580–588. doi: 10.1105/tpc.108.058172
Pierik, R., Fankhauser, C., Strader, L. C., Sinha, N. (2021). Architecture and plasticity: optimizing plant performance in dynamic environments. Plant Physiol. 187, 1029–1032. doi: 10.1093/plphys/kiab402
Plant, A. R., Larrieu, A., Causier, B. (2021). Repressor for hire! the vital roles of TOPLESS-mediated transcriptional repression in plants. New Phytol. 231, 963–973. doi: 10.1111/nph.17428
Raven, P. H. (2019). Saving plants, saving ourselves. PLANTS PEOPLE PLANET 1, 8–13. doi: 10.1002/ppp3.3
Samach, A., Onouchi, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of arabidopsis. Sci. (New York N.Y.) 288, 1613–1616. doi: 10.1126/science.288.5471.1613
Sawa, M., Nusinow, D. A., Kay, S. A., Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in arabidopsis. Sci. (New York N.Y.) 318, 261–265. doi: 10.1126/science.1146994
Schönrock, N., Bouveret, R., Leroy, O., Borghi, L., Köhler, C., Gruissem, W., et al. (2006). Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev. 20, 1667–1678. doi: 10.1101/gad.377206
Shen, M., Lim, C. J., Park, J., Kim, J. E., Baek, D., Nam, J., et al. (2020). HOS15 is a transcriptional corepressor of NPR1-mediated gene activation of plant immunity. Proc. Natl. Acad. Sci. United States America 117, 30805–30815. doi: 10.1073/pnas.2016049117
Shim, J. S., Kubota, A., Imaizumi, T. (2017). Circadian clock and photoperiodic flowering in arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 173, 5–15. doi: 10.1104/pp.16.01327
Srikanth, A., Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life sci.: CMLS 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Suzuki, M., Shinozuka, N., Hirakata, T., Nakata, M. T., Demura, T., Tsukaya, H., et al. (2018). OLIGOCELLULA1/HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES15 promotes cell proliferation with HISTONE DEACETYLASE9 and POWERDRESS during leaf development in arabidopsis thaliana. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00580
Wang, L., Kim, J., Somers, D. E. (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. United States America 110, 761–766. doi: 10.1073/pnas.1215010110
Xiong, L., Bh, L., Ishitani, M., Lee, H., Zhang, C., Zhu, J. K. (2001). FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in arabidopsis. Genes Dev. 15, 1971–1984. doi: 10.1101/gad.891901
Xu, Q., Liu, Q., Chen, Z., Yue, Y., Liu, Y., Zhao, Y., et al. (2021). Histone deacetylases control lysine acetylation of ribosomal proteins in rice. Nucleic Acids Res. 49, 4613–4628. doi: 10.1093/nar/gkab244
Yang, L., Chen, X., Wang, Z., Sun, Q., Hong, A., Zhang, A., et al. (2020). HOS15 and HDA9 negatively regulate immunity through histone deacetylation of intracellular immune receptor NLR genes in arabidopsis. New Phytol. 226, 507–522. doi: 10.1111/nph.16380
Yoo, S. K., Chung, K. S., Kim, J., Lee, J. H., Hong, S. M., Yoo, S. J., et al. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in arabidopsis. Plant Physiol. 139, 770–778. doi: 10.1104/pp.105.066928
Yoon, H.-G., Choi, Y., Cole, P. A., Wong, J. (2005). Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol. Cell. Biol. 25, 324–335. doi: 10.1128/MCB.25.1.324-335.2005
Yu, J.-W., Rubio, V., Lee, N.-Y., Bai, S., Lee, S.-Y., Kim, S.-S., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32, 617–630. doi: 10.1016/j.molcel.2008.09.026
Yu, H., Xu, Y., Tan, E. L., Kumar, P. P. (2002). AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc. Natl. Acad. Sci. United States America 99, 16336–16341. doi: 10.1073/pnas.212624599
Zareen, S., Ali, A., Lim, C. J., Khan, H. A., Park, J., Xu, Z.-Y., et al. (2022). The transcriptional corepressor HOS15 mediates dark-induced leaf senescence in arabidopsis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.828264
Zhao, Z., Yu, Y., Meyer, D., Wu, C., Shen, W.-H. (2005). Prevention of early flowering by expression of FLOWERING LOCUS c requires methylation of histone H3 K36. Nat. Cell Biol. 7, 1256–1260. doi: 10.1038/ncb1329
Zheng, Y., Ge, J., Bao, C., Chang, W., Liu, J., Shao, J., et al. (2020). Histone deacetylase HDA9 and WRKY53 transcription factor are mutual antagonists in regulation of plant stress response. Mol. Plant 13, 598–611. doi: 10.1016/j.molp.2019.12.011
Keywords: epigenetics, transcriptional corepressor, HOS15, GIGANTEA, flowering
Citation: Huang L-J, Wang Y, Lin Z, Jiang D, Luo Y and Li N (2023) The role of corepressor HOS15-mediated epigenetic regulation of flowering. Front. Plant Sci. 13:1101912. doi: 10.3389/fpls.2022.1101912
Received: 18 November 2022; Accepted: 21 December 2022;
Published: 10 January 2023.
Edited by:
Ming Luo, South China Botanical Garden (CAS), ChinaReviewed by:
Alexandre Berr, UPR2357 Institut de biologie moléculaire des plantes (IBMP), FranceCopyright © 2023 Huang, Wang, Lin, Jiang, Luo and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ning Li, bmxpQGNzdWZ0LmVkdS5jbg==
†ORCID: Li-Jun Huang, orcid.org/0000-0001-8072-5180
Ning Li, orcid.org/0000-0001-5297-8551
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.