
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 29 November 2022
Sec. Plant Metabolism and Chemodiversity
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1054586
This article is part of the Research Topic Natural Products from Plants and Applications for Pest Management View all 10 articles
Bacterial fruit blotch, caused by the seedborne gram-negative bacterium Acidovorax citrulli, is one of the most destructive bacterial diseases of cucurbits (gourds) worldwide. Despite its prevalence, effective and reliable means to control bacterial fruit blotch remain limited. Transcriptomic analyses of tissue culture-based regeneration processes have revealed that organogenesis-associated cellular reprogramming is often associated with upregulation of stress- and defense-responsive genes. Yet, there is limited evidence supporting the notion that the reprogrammed cellular metabolism of the regenerated tissued confers bona fide antimicrobial activity. Here, we explored the anti-bacterial activity of protocorm-like-bodies (PLBs) of Phalaenopsis aphrodite. Encouragingly, we found that the PLB extract was potent in slowing growth of A. citrulli, reducing the number of bacteria attached to watermelon seeds, and alleviating disease symptoms of watermelon seedlings caused by A. citrulli. Because the anti-bacterial activity can be fractionated chemically, we predict that reprogrammed cellular activity during the PLB regeneration process produces metabolites with antibacterial activity. In conclusion, our data demonstrated the antibacterial activity in developing PLBs and revealed the potential of using orchid PLBs to discover chemicals to control bacterial fruit blotch disease.
Bacterial fruit blotch (BFB) is a serious seedborne pathogen for watermelon and melon worldwide and its outbreaks have caused severe fruit loss in the Americas, Asia, Europe, the Middle and Far East, and Australia (Burdman et al., 2005; Bahar and Burdman, 2010; Burdman and Walcott, 2012). In Taiwan, BFB disease was first reported in 1992-1993 (Tzeng et al., 2010). The recurrence of BFB in 1994 caused more than 60% loss of the watermelon crop nationwide (Tang, 1997; Cheng et al., 2000; Cheng, 2009; Tzeng et al., 2010). The primary source of inoculum is often contaminated seeds (Assouline et al., 1997). Because of the destructive nature of BFB disease, evaluation of pathogen contamination in seed lots prior to their sale and distribution has become a critical practice for seed companies. Based on the National Seed Health System USDA standard (http://www.seedhealth.org/cb1-1), one infected seedling in 30,000 tested seeds can be sufficient to lead to rejection of the entire seed lot. However, seed disinfestations and chemical control only have limited efficacy in controlling BFB (Burdman and Walcott, 2012). Even though seedling grow-out assay is widely used to evaluate seed health, infected seedlings may or may not show disease symptoms. It has been reported that the amount of bacteria present on a seed, environmental conditions, virulence levels of the strains, and the susceptibility of host plants as a whole affect BFB outbreaks (Schaad et al., 2003; Bahar and Burdman, 2010). If infected seedlings are not detected, they might be transplanted into the field and become the primary source of inoculum for field outbreaks. To date, no BFB disease resistant plants have been developed and BFB management remains a major challenge to global watermelon and melon agriculture (Bahar and Burdman, 2010; Islam et al., 2020). Hence, an innovative plan for a BFB management program is a pressing need.
The Orchidaceae represents one of the largest angiosperm families comprising more than 25,000 species that grow in wide range of habitats ranging from rainforest and mountain, to swamp and arctic tundra (Stokstad, 2015). Considering the rich diversity of the orchid species, it is likely that orchids provide a substantial resource of novel compounds for potential application. In fact, orchids have been utilized by humans for thousand years. For example, vanilla orchid Vanilla planifolia, probably endemic to tropical forests in Eastern Mexico, is a major source of vanilla (Bory et al., 2008). Additionally, orchids such as Gastrodia elata, Bletilla striata, and Dendrobium species have been used for medicinal purposes in China and other Asian countries for thousands of years (Bulpitt et al., 2007). Despite this background, only a few phytochemicals have been characterized from orchids and the potential of orchid derived-phytochemicals has not been fully explored.
Protocorm-like bodies (PLBs) morphologically resemble the orchid germinated structures, protocorms, but are derived from somatic explants via a de novo regeneration pathway (Jones and Tisserat, 1990; Chugh et al., 2009; Fang et al., 2022). Because each PLB has the ability to regenerate into an individual plant, PLB-based micropropagation is often used to produce clonal plantlets in the orchid industry (Yam and Arditti, 2009). Our previous comparative transcriptomic studies to dissect the developmental origin of PLB supported that PLB and protocorm share similar molecular signatures and unexpectedly revealed that many genes involved in plant defense responses are specifically enriched in the developing PLB (Fang et al., 2016; Fang, 2021; Fang et al., 2022). Considering the potential functions of PLB-enriched defense related genes, we explored the antimicrobial activity of PLB extract and tested its effects on BFB of watermelon. Our results demonstrate the antibacterial activity of PLB extract and suggest the potential of using the orchid PLBs for developing a reagent to control BFB.
Acidovorax citrulli Aac153 (A. citrulli Aac153), isolated by the Laboratory of Phytopathogenic Bacteria, Department of Plant Pathology, National Chung Hsing University, Taiwan was a kind gift from Dr. Yi-Hsien Lin from National Pingtung University of Science and Technology. A. citrulli Aac153 has been shown to cause watermelon fruit blotch disease (Chang et al., 2019). A. citrulli Aac153 was stored at -75°C in tryptic soy broth (TSB) supplemented with 15% glycerol (v/v) and allowed to grow on selective medium AacG containing 0.5g/l KH2PO4, 2g/l Na2HPO4·12H2O, 2 g/l (NH4)2SO4, 5 g/l L-glutamic acid, 12.5 mg/l bromothymol blue, 15g/l agar, 20 mg/l ampicillin, 25 ppm/l cycloheximde as described previously (Chen, 2014). The single colony was used as inoculum for the primary culture. For subculture, single colony of primary culture was used to inoculate TSB and allowed to grow overnight at 28°C. The overnight culture was then grown on a selective AacG agar plate and the single colony was used as inoculum for the secondary culture. Only the primary and secondary cultures were used for inoculation in all the experiments. This strain produces reproducible, severe symptoms on the commercialized watermelon cultivar China Baby (Chang et al., 2019).
PLB tissues were homogenized by pestle and mortar in the presence of liquid nitrogen. One gram of pulverized PLB tissues was sonicated in the presence of 5 ml of ethyl acetate (EtOAc) for 30 mins (Branson 8510 DTH). The EtOAc-based PLB extract was incubated at 55°C for 10 min. Large tissue debris was removed by centrifugation at 3220 x g for 10 min. Supernatant was transferred to a new tube and concentrated by a rotary evaporator (EYELA, USA). The pellet was resuspended in 1 ml 100% MeOH. The PLB extract was then filtered by a 0.2 μm filter (13 mm Acrodisc Syringe filter, Pall) followed by concentration using a CentriVap Vacuum Concentrator (Labconco). The concentrated PLB-extract was flash frozen and stored at -80°C.
Frozen PLB extract was resuspended in 2 ml 30% MeOH. The 1 cc 50 mg Sep-PaK C18 cartridge (Waters) was first equilibrated with 1 ml of 100% MeOH followed by 1 ml of H2O once and then 1 ml of 30% MeOH once. For fractionation, solid phase extraction (SPE) was carried out by applying 1 ml PLB-based extract onto the equilibrated Sep-PaK C18 cartridge using a step gradient of MeOH-water mixture at a concentration of 30%, 45%, 60%, 80%, and 100% MeOH PLB and the eluents were collected individually. Methanol was allowed to evaporate by CentriVap Vacuum Concentrator (Labconco) and the fractionated PLB extract from 1 g tissues was pooled and resuspended in 100 μl 100% MeOH for seed infestation and pathogenicity assays as described below.
Frozen PLB extract from 1 g of PLB tissues (see above) was resuspended directly in 1 ml 100% MeOH. The overnight A. citrulli Aac153 culture was pelleted by centrifugation and washed once with 5 ml TSB medium, and diluted to OD600 = ~0.05 with TSB medium. The diluted culture was mixed with crude PLB extract (933 μl bacterial culture + 67 μl PLB extract) and aliquoted into 6 technical replicates (100 μl each) to a 96-well microtiter plate and allowed to incubate at 28°C. The OD600 was recorded at 0, 15, 19 hours after incubation. For each biological replicate, the absorbance measurements of OD600 were recorded in three technical replicates. This experiment was repeated three times.
Disease symptoms of seedlings at 12 days after transplantation (DAT) were recorded. Normally, 12 DAT watermelon seedlings have two expanded true leaves. Disease index was rated as follows (Figure 1): 0, no symptoms; 1, slight (< 20%) water-soaking or necrotic spots on cotyledons or hypocotyls; 2, increased water-soaking or necrotic spots (>20%) on cotyledons or hypocotyls; 3, expanded water-soaking and necrosis (>50%) on cotyledons or hypocotyls, true leaves often failed to emerge from infected seedlings, for seedlings with emerging true leaves, leaves failed to expand and were often distorted; 4, bent seedlings with necrotic cotyledons and hypocotyls, no true leaves were observed; 5, falling-over seedlings with complete necrotic cotyledons and hypocotyls. Disease severity was calculated as DS (%) = [sum (class frequency × score of rating class)]/[(total number of plants) × (maximal disease index)] × 100.

Figure 1 The represented disease severity index used to categorize disease symptoms of watermelon seedlings infected with A. citrulli Aac153. The numbers indicate the disease index described in Materials and Methods.
Watermelon seeds (China Baby) were purchased from Known-You Seed Company (Taiwan). For seed sterilization, seeds were imbibed in distilled water supplemented with 0.1% Triton X100 and gently shaken for 30 min. Imbibed seeds were sterilized by incubating with 75% ethanol for 5 min followed by washing with sterilized water 5 times. Seeds were allowed to dry in a laminar hood overnight.
For bacterial culture preparation, A. citrulli Aac153 culture that grew in 4 ml TSB overnight was pelleted by centrifugation at 3220 x g at 25°C for 10 min, washed once with 5 ml 0.5% carboxymethyl cellulose (CMC), resuspended in 0.5% CMC, and adjusted to OD600 to ~0.3. This preparation was used as the bacterial stock for seed infestation and seedling pathogenicity assays.
For seed infestation assay, five sterilized seeds were incubated with A. citrulli Aac153 culture in the presence (933 μl diluted bacterial culture + 67 μl fractionated PLB extract) or absence (933 μl diluted bacterial culture + 67 μl methanol) of PLB extract with gentle shaking (200 rpm) at 28°C for 2 h. The infested seeds were allowed to dry in a laminar flow hood overnight. The infected seeds were then resuspended in 1 ml AacG selective medium, incubated at 4°C for 30 min followed by incubation at 37°C for 1 h as described previously (Chen, 2014). Bacteria were concentrated by centrifugation at 15871 x g at 4°C for 10 min. The supernatant was carefully removed and the bacterial pellet was resuspended in 1 ml distilled water. The bacterial suspension was diluted 10 or 100 times by distilled water, plated on AacG selective plates, and allowed to grow at 28°C for three days. For each biological repeat, measurement of colony forming units was performed in three technical replicates. This experiment was repeated three times. A two-tailed Student’s t-test was applied. *, 0.05 > p > 0.01; **, p < 0.01. Only the treatments showing statistically significant reduction of bacteria were marked.
For seedling pathogenicity assay, the diluted A. citrulli Aac153 culture (OD600 to ~0.3) was further diluted 500 times with 0.5% CMC immediately before infection experiment. Eight sterilized seeds were incubated with 1 ml of diluted A. citrulli Aac153 culture in the presence (933µl diluted A. citrulli Aac153 + 67µl fractionated PLB extract, treatment) or absence (933 µl diluted A. citrulli Aac153 + 67µl MeOH, control) of PLB extract at 28°C with gentle shaking (200 rpm) for 24 h. The infested seeds were allowed to germinate in a humidity chamber at 32°C in the dark for 72 h. The germinated seedlings were then transferred to soil and allowed to grow in a growth chamber with a 16-h:8-h light:dark cycle under illumination of ~300 μmol photons m-2s-1 at 32°C. Plastic wrap was used to cover soil pots to maintain humidity and removed 5 days after transplantation. Disease symptoms were rated and recorded based on disease index scale (Figure 1) as described previously. The experiment was conducted three times. A two-tailed Student’s t-test was applied. **, 0.01 > p > 0.001; ***, p < 0.001.
RNA was extracted as described previously (Fang et al., 2016). Three micrograms of DNA-free RNA were reverse transcribed in the presence of a mixture of oligo(dT) and random primers in a 9:1 ratio using the GoScript Reverse Transcription System (Promega) based on the manufacturer’s instructions. Ten microliters of reverse transcription-PCR reaction contained 2.5 μL of 1:20 diluted cDNA, 0.2 μM of primers, and 5 μL of 2x KAPA SYBR FAST master mix (KAPA Biosystems). The amplification program was as follows: 95°C for 1 min, and 40 cycles at 95°C for 5 s and 58°C to 60°C for 20 s. PCR was performed in triplicate. Data are from technical triplicates and the error bars are presented as standard error of the mean. The RNA samples used for RT-qPCR analysis were independent from those for RNA-seq analyses. Primer pairs and the specified annealing temperature used for quantitative PCR are listed in Supplementary Table 1. UBIQUITIN was used as an internal control (Lin et al., 2014). The nomenclature of chalcone synthases was based on the previous study (Kuo et al., 2019). List of gene IDs used in this study and their corresponding IDs in the P. aphrodite databases are listed in Supplementary Table 2.
All experiments were performed three times or as otherwise mentioned in the figure legends. The data are presented as means and standard deviations obtained from at least three replicates of a single experiment. The significant difference between the treatments was analyzed by running a Student’s t-test in IBM SPSS v.20.
Our previous RNA-seq study investigating the developmental origin of PLBs revealed that Gene Ontology (GO) terms such as oxidation-reduction process, terpene synthase activity, and stress responses are overrepresented in developing PLBs (Fang et al., 2016). The biochemical and biological properties of these GO terms are generally associated with plant defense responses (Field et al., 2006; Almagro et al., 2009; Ton et al., 2009; Gonzalez et al., 2010; Denance et al., 2013; Yang et al., 2013; Savatin et al., 2014). Among them, Phalaenopsis chalcone synthase (CHS) and flavonoid 3’ hydroxylase (F3’ H) genes, PaCHS4, PaCHS5, and PaF3’ H1, were preferentially upregulated in developing PLBs (Table 1). Chalcone synthase and flavonoid 3’ hydroxylase act at the initial steps to produce flavonoids- and isoflavonoid-type phytoalexins (Bak et al., 2011; Dao et al., 2011; Ahuja et al., 2012) that are part of plant defense responses (Ahuja et al., 2012). A PLB-enriched PaCYC71A1, which is related to Arabidopsis CYP71A12 (Bak et al., 2011), encodes a putative cytochrome P450 monooxygenase (Table 1). Arabidopsis CYP71A12 takes part in biosynthesis of camalexin, a major phytoalexin important for disease resistance (Millet et al., 2010; Klein et al., 2013; Pastorczyk et al., 2020). Additionally, two PLB-enriched WRKY transcription factors, PaWRKY3 and PaWRKY4, were identified. PaWRKY3 belongs to the group III WRKY transcription factors (Supplementary Figure 1) and is related to Arabidopsis WRKY70 (Wu et al., 2005). Arabidopsis WRKY70 modulates salicylic acid (SA)- and jasmonic acid (JA)-mediated defense pathways to regulate plant immunity against bacterial pathogens (Li et al., 2004; Li et al., 2006; Zhou et al., 2018). PaWRKY4, on the other hand, encodes a group I WRKY transcription factor that is related to Arabidopsis WRKY33 (Supplementary Figure 1), which is an important regulator for biosynthesis of camalexin and pathogen-associated molecular patterns (PAMP)/pathogen-triggered reactive oxygen species (ROS) and ethylene production (Qiu et al., 2008; Mao et al., 2011; Li et al., 2012; Zhao et al., 2020; Zhou et al., 2020).
These PLB-enriched genes (Table 1) are also associated with other aspects of plant defense responses. For example, PaECR1 encodes a potential enoyl-coA reductase that has been shown to play roles in plant defense responses in cotton and P. amabilis orchid (Fu et al., 2012; Mustafa et al., 2017). A potential PLANT NATRIURETIC PEPTIDE (PEP) encoded by PaPEP1A belongs to a family of peptides involved in regulation of defense responses and ion and water homeostasis (Gehring and Irving, 2013; Ficarra et al., 2018). MAJOR LATEX PROTEIN1 (PaMLP1) and NORCOCLAURINE SYNTHASE1 (PaNCS1) belong to pathogen-related 10 (PR10) and Bet v1 proteins (Radauer et al., 2008). MLP proteins are known to be involved in various abiotic and biotic responses (Yang et al., 2015; Wang et al., 2016; Holmquist et al., 2021). NCS proteins catalyze the first committed step of biosynthesis of benzylisoquinoline alkaloids (BIAs) that possess antimicrobial activity (Lee and Facchini, 2010). PaRALF1, on the other hand, encodes a protein that is related to Arabidopsis RAPID ALKALINIZATION FACTOR (RALF) peptides, which are found to interact with receptor-like kinase FERONIA to modulate ROS production and plant immune responses (Stegmann et al., 2017; Li et al., 2018; Abarca et al., 2021). The PLB-enriched genes, PaCOMT1 and PaCOMT2, encode putative caffeic acid O-methyltransferases. Rice COMT is reported to have N-acetylserotonin O-methyltransferase (ASMT) activity that converts N-acetylserotonin to melatonin (Byeon et al., 2015), which modulates ROS and SA levels to improve plant responses to various abiotic and biotic stresses (Lee et al., 2015; Khan et al., 2020). Additionally, PaPRX1 encodes a PLB-enriched peroxidase, which may take part in plant defense responses (O’Brien et al., 2012). The expression patterns of the described PLB-enriched genes have been documented in an independent RNA-seq dataset (Fang et al., 2022) and validated by RT-qPCR analysis in a separate set of samples (Figure 2).
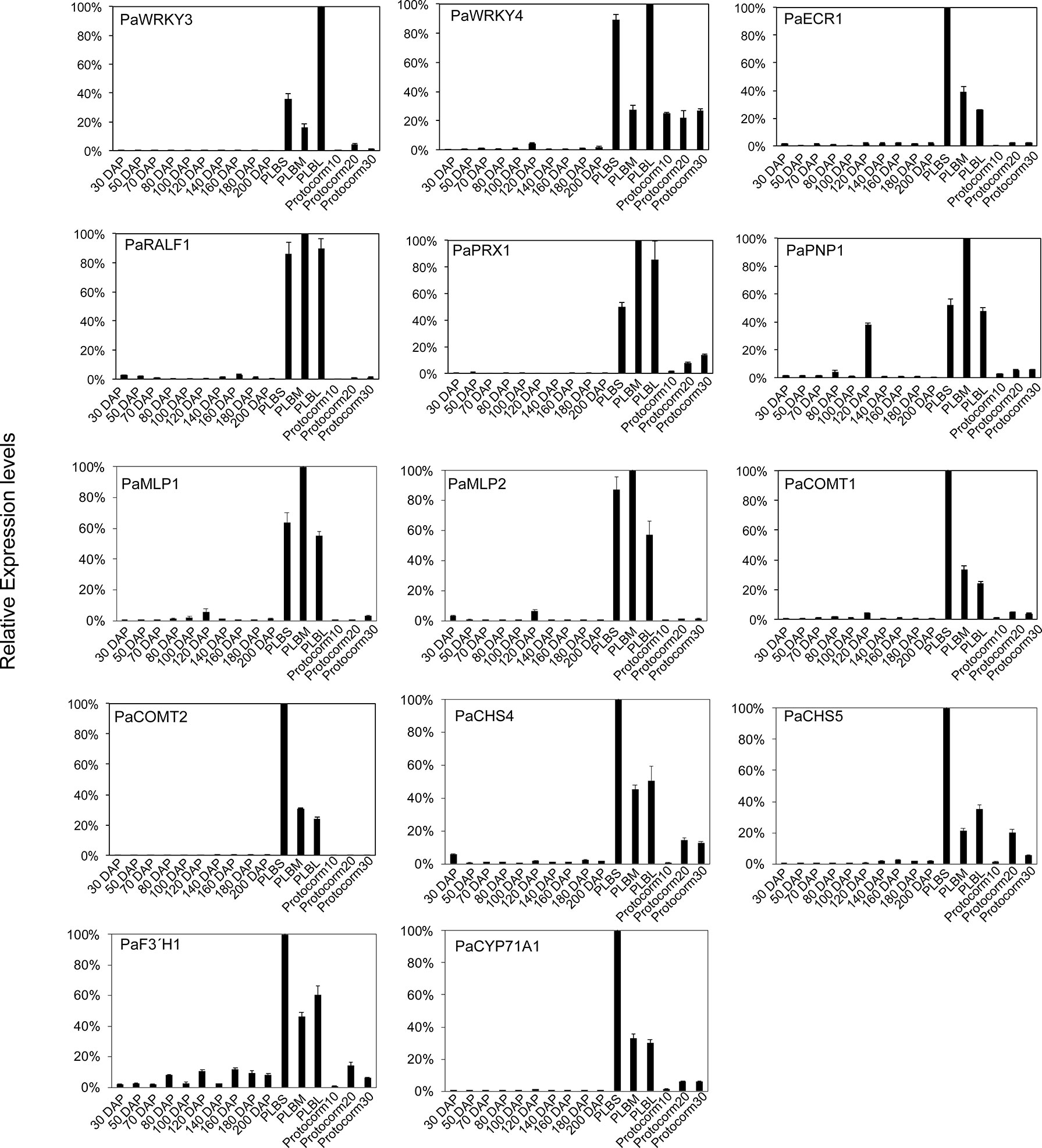
Figure 2 Expression profiles of the selected PLB-enriched genes in developing ovaries of P. aphrodite collected at 30 to 200 days after pollination (DAP), and developing PLBs and protocorms. Small PLBs (PLBS), medium PLBs (PLBM), large PLBs (PLBL), 10-d-old protocorms (protocorm10), 20-d-old protocorms (protocorm20), and 30-d-old protocorms (protocorm30) are defined as previously described (Fang et al., 2016) by RT-qPCR analysis. Expression was normalized to the Ubiquitin (PaUBI) signal. Data are from technical triplicates and the error bars are presented as standard error of the mean. Similar expression pattern was observed in RNA-seq data from two independently collected samples (Fang et al., 2016; Fang et al., 2022).
Because many plant defense-relates genes were specifically induced in developing PLBs, we hypothesized that the dynamic metabolomic reprogramming of developing PLBs leads to synthesis of antimicrobial metabolites. Ethyl acetate (EtOAc), which is commonly used for plant metabolite extraction (Tamokou et al., 2012; Oliveira et al., 2013; Lien et al., 2014; Yang et al., 2022), was used to prepare PLB crude extract. The crude extract was then tested for its effect on growth of three plant pathogens including Acidovorax citrulli Aac153 (watermelon bacterial fruit blotch disease), Pectobacterium carotovorum subsp. carotovorum (bacterial soft rot disease), and Xanthomonas citri pv. mangiferaeindicae (mango bacterial black spot disease). Because PLB crude extract showed consistent inhibitory effect on growth of A. citrulli Aac153 in the preliminary test (data not shown), we decided to focus on A. citrulli Aac153. To confirm this inhibitory effect, bacterial growth was monitored over 19 hours. In three separate experiments, PLB crude extract slowed the growth of A. citrulli Aac153 15 hours after treatment (Figure 3), indicating the presence of anti-bacterial activity in the developing PLBs. However, growth of A. citrulli Aac153 eventually caught up 19 hours after incubation, suggesting other substances may interfere with the inhibitory activity.
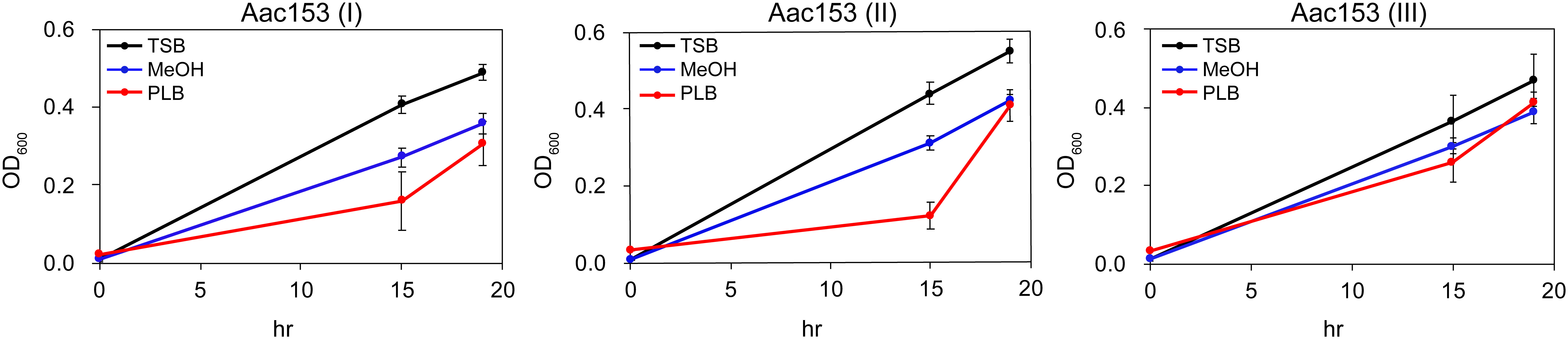
Figure 3 Crude PLB extract affected growth of A. citrulli Aac153. Aac153, A. citrulli Aac153 culture. TSB, cells were allowed to grow in TSB medium. MeOH, cells were allowed to grow in TSB medium containing 6.7% MeOH. PLB, cells were allowed to grow in TSB medium supplemented with 6.7% PLB extract. hr, hour after incubation. I, II, and III represent three independent experiments.
To enrich and separate the active metabolites from interfering substances, PLB crude extract was fractionated by solid phase extraction (SPE) based on the chemical polarity. Chemicals eluted with different concentrations of MeOH were collected for seed infestation test. Watermelon seeds were inoculated with A. citrulli Aac153 in the presence of different PLB eluents (see Materials and Methods). Interestingly, 30% MeOH PLB eluent was shown to be most effective in reducing the number of bacteria associated with watermelon seeds (Figure 4). Only 2.1% to 6.5% of bacterial cells remained after co-incubation with 30% MeOH-water PLB eluent. Co-incubating seeds with the 100% MeOH PLB eluents was also effective in reducing bacterial number but to a lesser extent (between 7.9% to 53.2%) with relatively large variations as compared to the 30% MeOH-water PLB eluents. This indicates that compounds with the potent antibacterial activity were enriched in 30% MeOH-water PLB eluent.
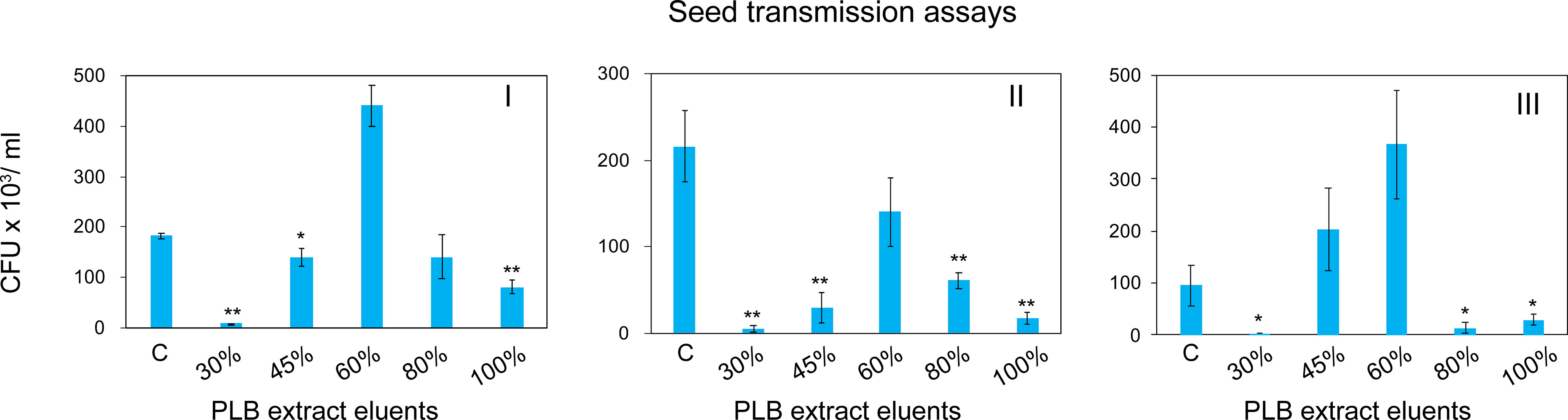
Figure 4 Watermelon seed infestation assay. CFU, colony forming unit. CFU Data were collected from 5 pooled watermelon seeds. C, no PLB extract control. 30%, PLB extract eluted by 30% MeOH-H2O. 45%, PLB extract eluted by 45% MeOH-H2O. 60%, PLB extract eluted by 60% MeOH-H2O. 80%, PLB extract eluted by 80% MeOH-H2O. 100%, PLB extract eluted by 100% MeOH-H2O. The experiment was repeated three times with similar results. I, II, and III represent three independent experiment. A two-tailed Student’s t-test was applied. *0.05 > p > 0.01; **p < 0.01. Only the treatments showing statistically significant reduction of bacteria were marked.
Because 30% MeOH PLB eluent was effective in reducing the number of bacteria attached to the watermelon seeds, we then investigated whether it can protect watermelon seedlings from A. citrulli Aac153 infection. To this end, watermelon seeds were incubated with A. citrulli Aac153 culture in the presence of 1x or 1/5x of 30% MeOH PLB eluent.
Watermelon seedlings from seeds incubated with CMC or CMC + 6.7% MeOH were used as controls, and no disease symptoms were observed on these seedlings (Figure 5A; Supplementary Figure 2). On the other hand, watermelon seedlings inoculated with A. citrulli Aac153 inoculation, showed water-soaking spots and necrotic lesions on the hypocotyl or cotyledons, typical BFB symptoms (Walcott, 2008; Bahar and Burdman, 2010), at 12 days after transplantation (DAP). Co-treatment of 1x 30% MeOH PLB eluent alleviated disease symptoms on the A. citrulli Aac153-infected seedlings. Moreover, 1/5 x 30% MeOH PLB eluent was also potent in protecting the A. citrulli Aac153-infected seedlings. This experiment was conducted three times and similar results were obtained each time. Disease assessment was quantified by the disease index scale as detailed in Materials and Methods. Disease severity of the A. citrulli Aac153-infected seedlings ranged from 65.5% to 81% (Figure 5B). The disease severity of the A. citrulli Aac153-infected seedlings treated with 1x 30% MeOH PLB eluent was reduced to 38.0% to 46%. Even 1/5 x 30% MeOH PLB eluent was able to protect the A. citrulli Aac153-infected seedlings and disease severity was reduced to 46% to 48.2%. Importantly, PLB treatment at the higher concentration only slightly affected seed germination (p value = 0.03, Table 2). Together, we conclude that PLB-derived metabolites possess the antibacterial activity that protects watermelon seedlings from A. citrulli Aac153 infection.
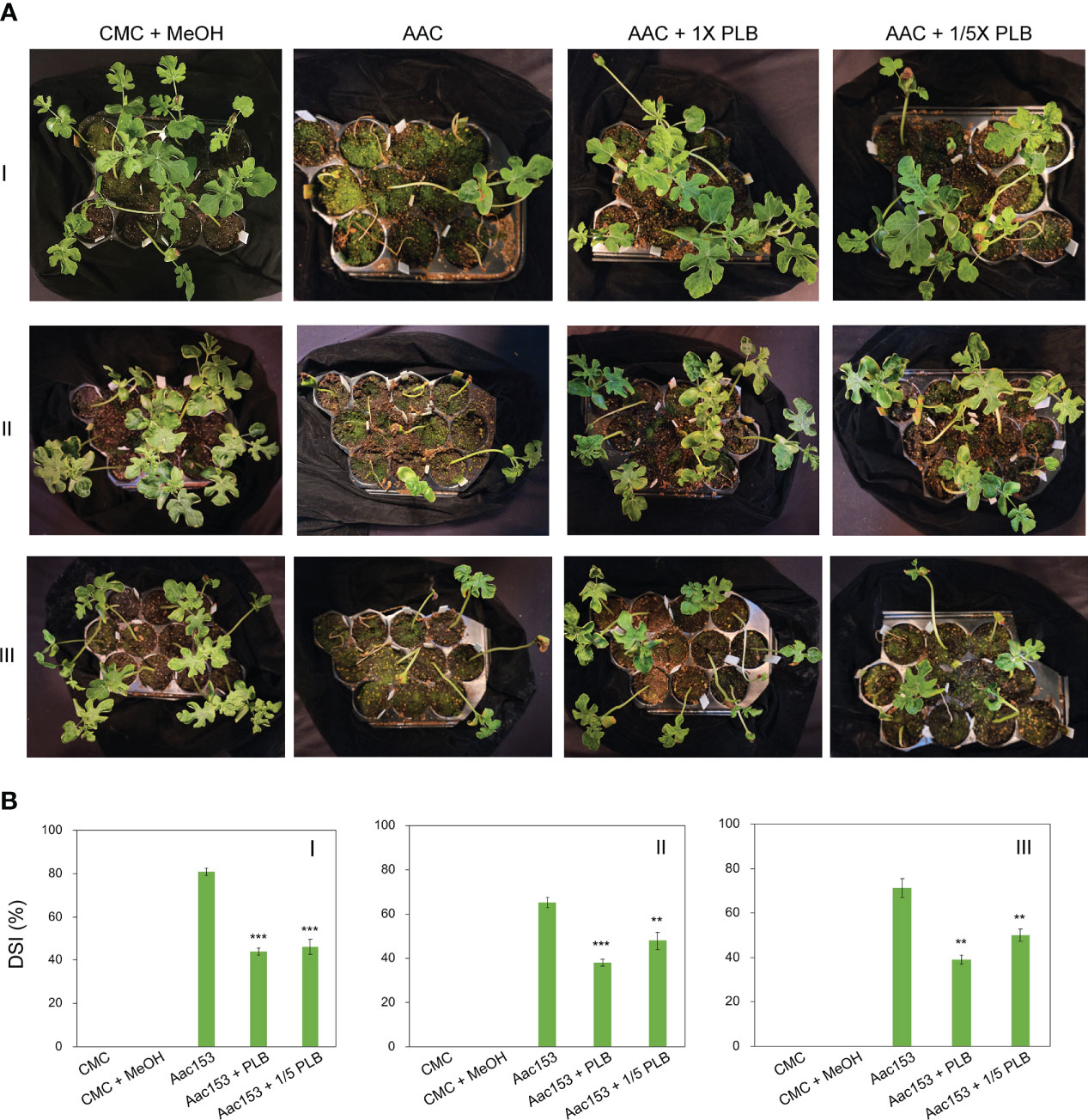
Figure 5 Thirty percent MeOH PLB eluent alleviates disease symptoms of watermelon bacterial blotch disease. (A) Disease symptoms of A citrulli Aac153-infested seedlings at 12 days after transplantion (DAT). (B) Disease severity index (DSI) of 12 DAT watermelon seedlings after treatments. CMC + MeOH, seeds were incubated with 6.7% MeOH in CMC medium. Aac153, seeds were incubated with A citrulli Aac153 in the presence of 6.7% MeOH. Aac153 + PLB, seeds were incubated with A citrulli Aac153 in the presence of 30% MeOH PLB extract. Aac153 + 1/5 PLB, seeds were incubated with A citrulli Aac153 in the presence of one-fifth 30% MeOH PLB extract. The experiment was repeated three times. I, II, and III represent three independent experiments. Twenty-two seeds were used in each experiment except CMC and CMC + MeOH treatments. A two-tailed Student’s t-test was applied. **0.01 > p > 0.001; ***p < 0.001.
Accumulated studies have provided molecular evidence linking pluripotency acquisition of plant regeneration processes to activation of defense responses (Chupeau et al., 2013; Ikeuchi et al., 2017; Iwase et al., 2021; Li et al., 2021). Furthermore, defense- or stress-associated pathways are proposed to be part of a gene regulatory network for cell proliferation and organ regeneration (Heyman et al., 2018; Wu et al., 2020a; Zeng et al., 2021). However, there is little evidence to support the notion that the rewired gene regulatory network of regenerated tissues is capable of synthesizing antimicrobial metabolites. Here, we showed that orchid PLB contains antibacterial substances that are potent in slowing the growth of A. citrulli Aac153, reducing the number of A. citrulli Aac153 associated with watermelon seeds, and protecting watermelon seedlings from severe infection by A. citrulli Aac153. Why would the developing PLBs possess the antibacterial activity? PLB is a regenerated structure induced by cutting during tissue culture (Yam and Arditti, 2009). Tissue injury caused by cutting during tissue culture triggers wounding-induced responses that mimic mechanical wounding triggered by herbivores and insects. It is therefore possible that wounding activates responses that contribute to production of antibacterial activity in PLBs. Wounding induced by herbivores and insects is known to trigger de novo synthesis of ethylene, jasmonic acid, and abscisic acid (Hyodo et al., 1983; Peña-Cortés et al., 1995; Bergey et al., 1996; Bouquin et al., 1997) that subsequently induce plant immunity responses to protect plants from infection by microbial pathogens (Savatin et al., 2014). Wounding also induces an array of immunity-related transcription factors such as WRKY transcription factors whose functions are to activate plant defense responses and prevent bacterial and fungal infection (Li et al., 2004; Li et al., 2006; Zheng et al., 2006; Pandey et al., 2010; Sarris et al., 2015; Zhou et al., 2018). Coincidently, some of the WRKY transcription factors also play a role in tissues regeneration (Che et al., 2006; Xu et al., 2012; Iwase et al., 2021). In this study, we showed that PaWRKY3 and PaWRKY4 (Table 1, Figure 2) are PLB-enriched transcription factors. PaWRKY3 and PaWRKY4 are the homologs of Arabidopsis defense response regulators, WRKY70 and WRKY33, respectively (Li et al., 2017; Zhou et al., 2018). Arabidopsis WRKY70 is directly regulated by the key immunity signaling regulator NONEXPRESSOR OF PR GENES1 (NPR1) and wrky70 mutant displayed reduced resistance to the oomycte Hyaloperonospora parasitica (Wang et al., 2006; Knoth et al., 2007). Arabidopsis WRKY33, on the other hand, is required for pathogen-induced biosynthesis of camalexin and ethylene response (Mao et al., 2011; Li et al., 2012). Considering the antimicrobial activity of PLB extract, we hypothesize that PaWRKY3 and PaWRKY4 may be part of cell reprogramming networks in developing PLBs that contribute to the defense activation and accumulation of antimicrobial metabolites. The functions of these PaWRKY3 and PaWRKY4 transcription factors remain to be determined.
Phytoalexins are reported to play important roles in combating a broad range of bacterial and fungal pathogens (Glazebrook and Ausubel, 1994; Graham et al., 2007; Ahuja et al., 2012; Schmelz et al., 2014). Because wounding has been reported to induce phytoalexin biosynthesis (Guillet and De Luca, 2005; Naoumkina et al., 2007; Farag et al., 2008), we speculated that phytoalexins make up part of the PLB-based antibacterial metabolites. Since chalcone synthases (PaCHS4 and PaCHS5) and flavonoid 3’ hydroxylase (PaF3’ H1) were specifically upregulated in PLBs, it is possible that flavonoids- and isoflavonoid-type phytoalexins were accumulated to provide the antimicrobial activity. However, we cannot exclude the possibility that other types of phytoalexins or cellular processes provide the inhibitory effect against A. citrulli Aac153. The active substance(s) remain to be purified and identified.
Plants possess a rich repertoire of phytochemicals that are important for plants to combat pathogens and predators, and adapt to biotic and abiotic stresses in the natural environment (Pichersky and Lewinsohn, 2011; Moghe and Last, 2015). The fact that the stress-activated plant regeneration program is often associated with defense-associated cellular activity suggests that the molecular wiring of development and plant immunity processes is overlapping. This notion is supported by a recent study showing that plant development and immunity share a signaling network (Wu et al., 2020b). In addition to a signaling network, we hypothesize that the cellular metabolism is also reprogrammed to accommodate stress-associated organogenesis and plant immunity functions. The available genome and transcriptome databases of P. aphrodite (Chao et al., 2017; Chao et al., 2018) provide the molecular basis to discover pathways and to decode the molecular wiring of PLB-associated antimicrobial metabolites (Owen et al., 2017). We suggest that PLBs may be used as a metabolite tap for identifying novel antibacterial compounds. Identification and characterization of the PLB-associated antimicrobial metabolites may provide a new route to harness the chemical diversity of orchid species.
Bacterial fruit blotch disease is a serious threat to the cucurbit (gourd) industry. Even though seed sanitation with hydrochloric acid or peroxyacetic acid have been proven to be effective at eradicating pathogens from infested seeds, seed quality is affected substantially (Hopkins et al., 1996; Hopkins et al., 2003). Moreover, seed disinfestation treatments and chemical control in the field are limited in their ability to reduce the yield losses associated with BFB (Burdman and Walcott, 2012), most likely because the applied chemicals cannot reach bacteria that are associated with developing embryos and seed coats (Rane and Latin, 1992; Hopkins and Thompson, 2002; Dutta et al., 2016). Since PLB-based extract did not affect the viability or germination rate of the watermelon seeds (Table 2), it may serve as an alternative to control watermelon fruit blotch disease.
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding authors.
S-CF conceived the idea and coordinated the study. S-CF, B-LH, and T-PH designed the experiments. S-CF, B-LH, and T-PH analyzed the data. S-CF and T-PH wrote the manuscript. B-LH and J-CC conducted the experiments. All the authors have read and approved the final manuscript.
This work was supported by the Ministry of Science and Technology (MOST 106-2313-B- 001-004-MY3 and MOST 109-2313-B-001 -015 -MY3 to SCF).
We thank Dr. Yi-Hsien Lin for providing us Acidovorax citrulli Aac153 strain, Mr. Fu-Chieh Yang for technical support, and Ms. Miranda Loney for English editing.
S-CF and B-LH are named inventors on a patent application pertaining to the technology that was filed by Academia Sinica.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1054586/full#supplementary-material
Abarca, A., Franck, C. M., Zipfel, C. (2021). Family-wide evaluation of RAPID ALKALINIZATION FACTOR peptides. Plant Physiol. 187 (2), 996–1010. doi: 10.1093/plphys/kiab308
Ahuja, I., Kissen, R., Bones, A. M. (2012). Phytoalexins in defense against pathogens. Trends Plant Sci. 17 (2), 73–90. doi: 10.1016/j.tplants.2011.11.002
Almagro, L., Gomez Ros, L. V., Belchi-Navarro, S., Bru, R., Ros Barcelo, A., Pedreno, M. A. (2009). Class III peroxidases in plant defence reactions. J. Exp. Bot. 60 (2), 377–390. doi: 10.1093/jxb/ern277
Assouline, I., Milshtein, H., Mizrahi, M., Levy, E., Ben-Ze’ev, I. S. (1997). Acidovorax avenae subsp.citrulli transmitted by solanaceous seeds. Phytoparasitica 25 (2), 117–118. doi: 10.1007/bf02981192
Bahar, O., Burdman, S. (2010). Bacterial fruit blotch: A threat to the cucurbit industry. Israel J. Plant Sci. 58 (1), 19–31. doi: 10.1560/Ijps.58.1.19
Bak, S., Beisson, F., Bishop, G., Hamberger, B., Hofer, R., Paquette, S., et al. (2011). Cytochromes p450. Arabidopsis Book 9, e0144. doi: 10.1199/tab.0144
Bergey, D. R., Howe, G. A., Ryan, C. A. (1996). Polypeptide signaling for plant defensive genes exhibits analogies to defense signaling in animals. Proc. Natl. Acad. Sci. U.S.A. 93 (22), 12053–12058. doi: 10.1073/pnas.93.22.12053
Bory, S., Grisoni, M., Duval, M. F., Besse, P. (2008). Biodiversity and preservation of vanilla: present state of knowledge. Genet. Resour. Crop Evol. 55 (4), 551–571. doi: 10.1007/s10722-007-9260-3
Bouquin, T., Lasserre, E., Pradier, J., Pech, J. C., Balague, C. (1997). Wound and ethylene induction of the ACC oxidase melon gene CM-ACO1 occurs via two direct and independent transduction pathways. Plant Mol. Biol. 35 (6), 1029–1035. doi: 10.1023/a:1005902226054
Bulpitt, C. J., Li, Y., Bulpitt, P. F., Wang, J. (2007). The use of orchids in Chinese medicine. J. R Soc. Med. 100 (12), 558–563. doi: 10.1258/jrsm.100.12.558
Burdman, S., Kots, N., Kritzman, G., Kopelowitz, J. (2005). Molecular, physiological, and host-range characterization of Acidovorax avenae subsp. citrulli isolates from watermelon and melon in Israel. Plant Dis. 89 (12), 1339–1347. doi: 10.1094/PD-89-1339
Burdman, S., Walcott, R. (2012). Acidovorax citrulli: generating basic and applied knowledge to tackle a global threat to the cucurbit industry. Mol. Plant Pathol. 13 (8), 805–815. doi: 10.1111/j.1364-3703.2012.00810.x
Byeon, Y., Choi, G. H., Lee, H. Y., Back, K. (2015). Melatonin biosynthesis requires n-acetylserotonin methyltransferase activity of caffeic acid O-methyltransferase in rice. J. Exp. Bot. 66 (21), 6917–6925. doi: 10.1093/jxb/erv396
Chang, J.-J., Wu, P.-Y., Lin, Y.-N., Deng, W.-., Lin, Y.-H. (2019). Intensification of PAMP-triggered immunity in watermelon by Bacillus spp. strains as a strategy for controlling bacterial fruit blotch disease. J. Plant Med. 61 (1), 39–48. doi: 10.6716/JPM.201903_61(1).0004
Chao, Y. T., Chen, W. C., Chen, C. Y., Ho, H. Y., Yeh, C. H., Kuo, Y. T., et al. (2018). Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 16 (12), 2027–2041. doi: 10.1111/pbi.12936
Chao, Y. T., Yen, S. H., Yeh, J. H., Chen, W. C., Shih, M. C. (2017). Orchidstra 2.0-a transcriptomics resource for the orchid family. Plant Cell Physiol. 58 (1), 1–13. doi: 10.1093/pcp/pcw220
Che, P., Lall, S., Nettleton, D., Howell, S. H. (2006). Gene expression programs during shoot, root, and callus development in arabidopsis tissue culture. Plant Physiol. 141 (2), 620–637. doi: 10.1104/pp.106.081240
Chen, Y.-J. (2014). Development of a new selective medium for acidovorax citrulli. master thesis (Taichung, Taiwan: National Chung Hsing University).
Cheng, A.-S. (2009). “Bacterial fruit blotch disease,” in Compendium of melon diseases and pests (Taiwan: Bureau of Animal and Plant Health Inspection and Quarantine, Council of Agriculture).
Cheng, A. H., Hsu, Y. L., Huang, T. C., Wang, H. L. (2000). Susceptibility of cucurbits to acidovorax avenae subsp. citrulli and control of fruit bloth on melon. Plant Pathol. Bull. 9, 151–156. doi: 10.6649/PPB.200012_9(4).0004
Chugh, S., Guha, S., Rao, I. U. (2009). Micropropagation of orchids: a review on the potential of different explants. Sci. Hortic. 122 (4), 507–520. doi: 10.1016/J.Scienta.2009.07.016
Chupeau, M. C., Granier, F., Pichon, O., Renou, J. P., Gaudin, V., Chupeau, Y. (2013). Characterization of the early events leading to totipotency in an arabidopsis protoplast liquid culture by temporal transcript profiling. Plant Cell 25 (7), 2444–2463. doi: 10.1105/tpc.113.109538
Dao, T. T., Linthorst, H. J., Verpoorte, R. (2011). Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 10 (3), 397–412. doi: 10.1007/s11101-011-9211-7
Denance, N., Sanchez-Vallet, A., Goffner, D., Molina, A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00155
Dutta, B., Schneider, R. W., Robertson, C. L., Walcott, R. R. (2016). Embryo localization enhances the survival of Acidovorax citrulli in watermelon seeds. Phytopathology 106 (4), 330–338. doi: 10.1094/PHYTO-09-15-0232-R
Fang, S. C. (2021). “Comparative transcriptomics study provides new insights into the specialized reproductive developmental programs of phalaenopsis aphrodite,” in Orchid biotechnology IV, vol. 173-205 . Eds. Chen, W. H., CHen, H. H. (Singapore: World Scientific).
Fang, S. C., Chen, J. C., Chang, P. Y., Lin, H. Y. (2022). Co-Option of the SHOOT MERISTEMLESS network regulates protocorm-like body development in Phalaenopsis aphrodite. Plant Physiol. 190 (1), 127–145. doi: 10.1093/plphys/kiac100
Fang, S. C., Chen, J. C., Wei, M. J. (2016). Protocorms and protocorm-like bodies are molecularly distinct from zygotic embryonic tissues in Phalaenopsis aphrodite. Plant Physiol. 171 (4), 2682–2700. doi: 10.1104/pp.16.00841
Farag, M. A., Huhman, D. V., Dixon, R. A., Sumner, L. W. (2008). Metabolomics reveals novel pathways and differential mechanistic and elicitor-specific responses in phenylpropanoid and isoflavonoid biosynthesis in medicago truncatula cell cultures. Plant Physiol. 146 (2), 387–402. doi: 10.1104/pp.107.108431
Ficarra, F. A., Grandellis, C., Garavaglia, B. S., Gottig, N., Ottado, J. (2018). Bacterial and plant natriuretic peptides improve plant defence responses against pathogens. Mol. Plant Pathol. 19 (4), 801–811. doi: 10.1111/mpp.12560
Field, B., Jordan, F., Osbourn, A. (2006). First encounters–deployment of defence-related natural products by plants. New Phytol. 172 (2), 193–207. doi: 10.1111/j.1469-8137.2006.01863.x
Fu, S. F., Tsai, T. M., Chen, Y. R., Liu, C. P., Haiso, L. J., Syue, L. H., et al. (2012). Characterization of the early response of the orchid, Phalaenopsis amabilis, to Erwinia chrysanthemi infection using expression profiling. Physiol. Plant 145 (3), 406–425. doi: 10.1111/j.1399-3054.2012.01582.x
Gehring, C., Irving, H. (2013). Plant natriuretic peptides: systemic regulators of plant homeostasis and defense that can affect cardiomyoblasts. J. Investig. Med. 61 (5), 823–826. doi: 10.2310/JIM.0b013e3182923395
Glazebrook, J., Ausubel, F. M. (1994). Isolation of phytoalexin-deficient mutants of arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 91 (19), 8955–8959. doi: 10.1073/pnas.91.19.8955
Gonzalez, A. M., Marcel, T. C., Kohutova, Z., Stam, P., van der Linden, C. G., Niks, R. E. (2010). Peroxidase profiling reveals genetic linkage between peroxidase gene clusters and basal host and non-host resistance to rusts and mildew in barley. PloS One 5 (8), e10495. doi: 10.1371/journal.pone.0010495
Graham, T. L., Graham, M. Y., Subramanian, S., Yu, O. (2007). RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues. Plant Physiol. 144 (2), 728–740. doi: 10.1104/pp.107.097865
Guillet, G., De Luca, V. (2005). Wound-inducible biosynthesis of phytoalexin hydroxycinnamic acid amides of tyramine in tryptophan and tyrosine decarboxylase transgenic tobacco lines. Plant Physiol. 137 (2), 692–699. doi: 10.1104/pp.104.050294
Heyman, J., Canher, B., Bisht, A., Christiaens, F., De Veylder, L. (2018). Emerging role of the plant ERF transcription factors in coordinating wound defense responses and repair. J. Cell Sci. 131 (2), jcs208215. doi: 10.1242/jcs.208215
Holmquist, L., Dolfors, F., Fogelqvist, J., Cohn, J., Kraft, T., Dixelius, C. (2021). Major latex protein-like encoding genes contribute to Rhizoctonia solani defense responses in sugar beet. Mol. Genet. Genomics 296 (1), 155–164. doi: 10.1007/s00438-020-01735-0
Hopkins, D. L., Cucuzza, J. D., Watterson, J. C. (1996). Wet seed treatments for the control of bacterial fruit blotch of watermelon. Plant Dis. 80 (5), 529–532. doi: 10.1094/Pd-80-0529
Hopkins, D. L., Thompson, C. M. (2002). Seed transmission of Acidovorax avenae subsp citrulli in cucurbits. Hortscience 37 (6), 924–926. doi: 10.21273/Hortsci.37.6.924
Hopkins, D. L., Thompson, C. M., Hilgren, J., Lovic, B. (2003). Wet seed treatment with peroxyacetic acid for the control of bacterial fruit blotch and other seedborne diseases of watermelon. Plant Dis. 87 (12), 1495–1499. doi: 10.1094/Pdis.2003.87.12.1495
Hyodo, H., Tanaka, K., Watanabe, K. (1983). Wound-induced ethylene production and 1-aminocyclopropane-1-carboxylic acid synthase in mesocarp tissue of winter squash fruit. Plant Cell Physiol. 24 (6), 963–969. doi: 10.1093/oxfordjournals.pcp.a076626
Ikeuchi, M., Iwase, A., Rymen, B., Lambolez, A., Kojima, M., Takebayashi, Y., et al. (2017). Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol. 175 (3), 1158–1174. doi: 10.1104/pp.17.01035
Islam, M. R., Hossain, M. R., Jesse, D. M. I., Jung, H. J., Kim, H. T., Park, J. I., et al. (2020). Characterization, identification and expression profiling of genome-wide r-genes in melon and their putative roles in bacterial fruit blotch resistance. BMC Genet. 21 (1), 80. doi: 10.1186/s12863-020-00885-9
Iwase, A., Kondo, Y., Laohavisit, A., Takebayashi, A., Ikeuchi, M., Matsuoka, K., et al. (2021). WIND transcription factors orchestrate wound-induced callus formation, vascular reconnection and defense response in arabidopsis. New Phytol. 232 (2), 734–752. doi: 10.1111/nph.17594
Jones, D., Tisserat, B. (1990). Clonal propagation of orchids. Methods Mol. Biol. 6, 181–191. doi: 10.1385/0-89603-161-6:181
Khan, A., Numan, M., Khan, A. L., Lee, I. J., Imran, M., Asaf, S., et al. (2020). Melatonin: Awakening the defense mechanisms during plant oxidative stress. Plants (Basel) 9 (4), 407. doi: 10.3390/plants9040407
Klein, A. P., Anarat-Cappillino, G., Sattely, E. S. (2013). Minimum set of cytochromes P450 for reconstituting the biosynthesis of camalexin, a major arabidopsis antibiotic. Angew Chem. Int. Ed Engl. 52 (51), 13625–13628. doi: 10.1002/anie.201307454
Knoth, C., Ringler, J., Dangl, J. L., Eulgem, T. (2007). Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol. Plant Microbe Interact. 20 (2), 120–128. doi: 10.1094/MPMI-20-2-0120
Kuo, Y. T., Chao, Y. T., Chen, W. C., Shih, M. C., Chang, S. B. (2019). Segmental and tandem chromosome duplications led to divergent evolution of the chalcone synthase gene family in Phalaenopsis orchids. Ann. Bot. 123 (1), 69–77. doi: 10.1093/aob/mcy136
Lee, H. Y., Byeon, Y., Tan, D. X., Reiter, R. J., Back, K. (2015). Arabidopsis serotonin n-acetyltransferase knockout mutant plants exhibit decreased melatonin and salicylic acid levels resulting in susceptibility to an avirulent pathogen. J. Pineal Res. 58 (3), 291–299. doi: 10.1111/jpi.12214
Lee, E. J., Facchini, P. (2010). Norcoclaurine synthase is a member of the pathogenesis-related 10/Bet v1 protein family. Plant Cell 22 (10), 3489–3503. doi: 10.1105/tpc.110.077958
Li, J., Brader, G., Kariola, T., Palva, E. T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46 (3), 477–491. doi: 10.1111/j.1365-313X.2006.02712.x
Li, J., Brader, G., Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16 (2), 319–331. doi: 10.1105/tpc.016980
Lien, H. M., Tseng, C. J., Huang, C. L., Lin, Y. T., Chen, C. C., Lai, Y. Y. (2014). Antimicrobial activity of Antrodia camphorata extracts against oral bacteria. PloS One 9 (8), e105286. doi: 10.1371/journal.pone.0105286
Li, C., Liu, X., Qiang, X., Li, X., Li, X., Zhu, S., et al. (2018). EBP1 nuclear accumulation negatively feeds back on FERONIA-mediated RALF1 signaling. PloS Biol. 16 (10), e2006340. doi: 10.1371/journal.pbio.2006340
Li, G., Meng, X., Wang, R., Mao, G., Han, L., Liu, Y., et al. (2012). Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in arabidopsis. PloS Genet. 8 (6), e1002767. doi: 10.1371/journal.pgen.1002767
Lin, H. Y., Chen, J. C., Wei, M. J., Lien, Y. C., Li, H. H., Ko, S. S., et al. (2014). Genome-wide annotation, expression profiling, and protein interaction studies of the core cell-cycle genes in Phalaenopsis aphrodite. Plant Mol. Biol. 84 (1-2), 203–226. doi: 10.1007/s11103-013-0128-y
Li, D., Shu, X., Zhu, P., Pei, D. (2021). Chromatin accessibility dynamics during cell fate reprogramming. EMBO Rep. 22 (2), e51644. doi: 10.15252/embr.202051644
Li, J., Zhong, R., Palva, E. T. (2017). WRKY70 and its homolog WRKY54 negatively modulate the cell wall-associated defenses to necrotrophic pathogens in arabidopsis. PloS One 12 (8), e0183731. doi: 10.1371/journal.pone.0183731
Mao, G., Meng, X., Liu, Y., Zheng, Z., Chen, Z., Zhang, S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in arabidopsis. Plant Cell 23 (4), 1639–1653. doi: 10.1105/tpc.111.084996
Millet, Y. A., Danna, C. H., Clay, N. K., Songnuan, W., Simon, M. D., Werck-Reichhart, D., et al. (2010). Innate immune responses activated in arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22 (3), 973–990. doi: 10.1105/tpc.109.069658
Moghe, G. D., Last, R. L. (2015). Something old, something new: conserved enzymes and the evolution of novelty in plant specialized metabolism. Plant Physiol. 169 (3), 1512–1523. doi: 10.1104/pp.15.00994
Mustafa, R., Hamza, M., Kamal, H., Mansoor, S., Scheffler, J., Amin, I. (2017). Tobacco rattle virus-based silencing of enoyl-coA reductase gene and its role in resistance against cotton wilt disease. Mol. Biotechnol. 59 (7), 241–250. doi: 10.1007/s12033-017-0014-y
Naoumkina, M., Farag, M. A., Sumner, L. W., Tang, Y., Liu, C. J., Dixon, R. A. (2007). Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 104 (46), 17909–17915. doi: 10.1073/pnas.0708697104
O’Brien, J. A., Daudi, A., Finch, P., Butt, V. S., Whitelegge, J. P., Souda, P., et al. (2012). A peroxidase-dependent apoplastic oxidative burst in cultured arabidopsis cells functions in MAMP-elicited defense. Plant Physiol. 158 (4), 2013–2027. doi: 10.1104/pp.111.190140
Oliveira, D. A., Salvador, A. A., Smania, A., Jr., Smania, E. F., Maraschin, M., Ferreira, S. R. (2013). Antimicrobial activity and composition profile of grape (Vitis vinifera) pomace extracts obtained by supercritical fluids. J. Biotechnol. 164 (3), 423–432. doi: 10.1016/j.jbiotec.2012.09.014
Owen, C., Patron, N. J., Huang, A., Osbourn, A. (2017). Harnessing plant metabolic diversity. Curr. Opin. Chem. Biol. 40, 24–30. doi: 10.1016/j.cbpa.2017.04.015
Pandey, S. P., Roccaro, M., Schon, M., Logemann, E., Somssich, I. E. (2010). Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of arabidopsis. Plant J. 64 (6), 912–923. doi: 10.1111/j.1365-313X.2010.04387.x
Pastorczyk, M., Kosaka, A., Pislewska-Bednarek, M., Lopez, G., Frerigmann, H., Kulak, K., et al. (2020). The role of CYP71A12 monooxygenase in pathogen-triggered tryptophan metabolism and arabidopsis immunity. New Phytol. 225 (1), 400–412. doi: 10.1111/nph.16118
Peña-Cortés, H., Fisahn, J., Willmitzer, L. (1995). Signals involved in wound-induced proteinase inhibitor II gene expression in tomato and potato plants. Proc. Natl. Acad. Sci. U.S.A. 92 (10), 4106–4113. doi: 10.1073/pnas.92.10.4106
Pichersky, E., Lewinsohn, E. (2011). Convergent evolution in plant specialized metabolism. Annu. Rev. Plant Biol. 62, 549–566. doi: 10.1146/annurev-arplant-042110-103814
Qiu, J. L., Fiil, B. K., Petersen, K., Nielsen, H. B., Botanga, C. J., Thorgrimsen, S., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27 (16), 2214–2221. doi: 10.1038/emboj.2008.147
Radauer, C., Lackner, P., Breiteneder, H. (2008). The bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol. Biol. 8, 286. doi: 10.1186/1471-2148-8-286
Rane, K. K., Latin, R. X. (1992). Bacterial fruit blotch of watermelon - association of the pathogen with seed. Plant Dis. 76 (5), 509–512. doi: 10.1094/Pd-76-0509
Sarris, P. F., Duxbury, Z., Huh, S. U., Ma, Y., Segonzac, C., Sklenar, J., et al. (2015). A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161 (5), 1089–1100. doi: 10.1016/j.cell.2015.04.024
Savatin, D. V., Gramegna, G., Modesti, V., Cervone, F. (2014). Wounding in the plant tissue: the defense of a dangerous passage. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00470
Schaad, N. W., Postnikova, E., Randhawa, P. (2003). Pseudomonas syringae “Emergence of Acidovorax avenae subsp. citrulli as a crop threatening disease of watermelon and melon,” (Netherlands: Springer), 573–581.
Schmelz, E. A., Huffaker, A., Sims, J. W., Christensen, S. A., Lu, X., Okada, K., et al. (2014). Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J. 79 (4), 659–678. doi: 10.1111/tpj.12436
Stegmann, M., Monaghan, J., Smakowska-Luzan, E., Rovenich, H., Lehner, A., Holton, N., et al. (2017). The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355 (6322), 287–289. doi: 10.1126/science.aal2541
Stokstad, E. (2015). BOTANY. orchids’ dazzling diversity explained. Science 349 (6251), 914. doi: 10.1126/science.349.6251.914
Tamokou, J. D., Simo Mpetga, D. J., Keilah Lunga, P., Tene, M., Tane, P., Kuiate, J. R. (2012). Antioxidant and antimicrobial activities of ethyl acetate extract, fractions and compounds from stem bark of Albizia adianthifolia (Mimosoideae). BMC Complement Altern. Med. 12, 99. doi: 10.1186/1472-6882-12-99
Tang, C. J. (1997). Studies on bacterial fruit blotch of watermelon caused by acidovorax avenae subsp. citrulli (National Chung Hsing University: Master Thesis).
Ton, J., Flors, V., Mauch-Mani, B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14 (6), 310–317. doi: 10.1016/j.tplants.2009.03.006
Tzeng, K.-C., Lu, Y.-S., Cheng, A.-S., Huang, T.-C., Hsu, S.-T. (2010). “瓜類細菌性果斑病:病原菌檢測與病害管理,” in Special publication TARI (Taiwan: Taiwan Agricultural Research Institute).
Walcott, R. R. (2008). “Integrated pest management of bacterial fruit blotch of cucurbits,” in Integrated management of diseases caused by fungi, phytoplasma and bacteria. Eds. Ciancio, A., Mukerji, K. G.(Springer Netherlands: Dordrecht).
Wang, D., Amornsiripanitch, N., Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PloS Pathog. 2 (11), e123. doi: 10.1371/journal.ppat.0020123
Wang, Y., Yang, L., Chen, X., Ye, T., Zhong, B., Liu, R., et al. (2016). Major latex protein-like protein 43 (MLP43) functions as a positive regulator during abscisic acid responses and confers drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 67 (1), 421–434. doi: 10.1093/jxb/erv477
Wu, K. L., Guo, Z. J., Wang, H. H., Li, J. (2005). The WRKY family of transcription factors in rice and arabidopsis and their origins. DNA Res. 12 (1), 9–26. doi: 10.1093/dnares/12.1.9.
Wu, H., Qu, X., Dong, Z., Luo, L., Shao, C., Forner, J., et al. (2020a). WUSCHEL triggers innate antiviral immunity in plant stem cells. Science 370 (6513), 227–231. doi: 10.1126/science.abb7360
Wu, Q., Xu, F., Liu, L., Char, S. N., Ding, Y., Je, B. I., et al. (2020b). The maize heterotrimeric G protein beta subunit controls shoot meristem development and immune responses. Proc. Natl. Acad. Sci. U.S.A. 117 (3), 1799–1805. doi: 10.1073/pnas.1917577116
Xu, K., Liu, J., Fan, M., Xin, W., Hu, Y., Xu, C. (2012). A genome-wide transcriptome profiling reveals the early molecular events during callus initiation in arabidopsis multiple organs. Genomics 100 (2), 116–124. doi: 10.1016/j.ygeno.2012.05.013
Yam, T. W., Arditti, J. (2009). History of orchid propagation: a mirror of the history of biotechnology. Plant Biotechnol. Rep. 3 (1), 1–56. doi: 10.1007/S11816-008-0066-3
Yang, C. L., Liang, S., Wang, H. Y., Han, L. B., Wang, F. X., Cheng, H. Q., et al. (2015). Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticillium dahliae. Mol. Plant 8 (3), 399–411. doi: 10.1016/j.molp.2014.11.023
Yang, Z., Li, L., Chen, C. H., Zhang, Y. Y., Yang, Y., Zhang, P., et al. (2022). Chemical composition and antibacterial activity of 12 medicinal plant ethyl acetate extracts using LC-MS feature-based molecular networking. Phytochem. Anal. 33 (3), 473–489 doi: 10.1002/pca.3103
Yang, D. L., Yang, Y., He, Z. (2013). Roles of plant hormones and their interplay in rice immunity. Mol. Plant 6 (3), 675–685. doi: 10.1093/mp/sst056
Zeng, J., Li, X., Ge, Q., Dong, Z., Luo, L., Tian, Z., et al. (2021). Endogenous stress-related signal directs shoot stem cell fate in Arabidopsis thaliana. Nat. Plants 7 (9), 1276–1287. doi: 10.1038/s41477-021-00985-z
Zhao, J., Chen, Q., Zhou, S., Sun, Y., Li, X., Li, Y. (2020). H2Bub1 regulates RbohD-dependent hydrogen peroxide signal pathway in the defense responses to Verticillium dahliae toxins. Plant Physiol. 182 (1), 640–657. doi: 10.1104/pp.19.00913
Zheng, Z., Qamar, S. A., Chen, Z., Mengiste, T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48 (4), 592–605. doi: 10.1111/j.1365-313X.2006.02901.x
Zhou, M., Lu, Y., Bethke, G., Harrison, B. T., Hatsugai, N., Katagiri, F., et al. (2018). WRKY70 prevents axenic activation of plant immunity by direct repression of SARD1. New Phytol. 217 (2), 700–712. doi: 10.1111/nph.14846
Zhou, J., Wang, X., He, Y., Sang, T., Wang, P., Dai, S., et al. (2020). Differential phosphorylation of the transcription factor WRKY33 by the protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulates camalexin biosynthesis in arabidopsis. Plant Cell 32 (8), 2621–2638. doi: 10.1105/tpc.19.00971
Keywords: Acidovorax citrulli, bacterial fruit blotch (BFB), protocorm-like-body, Phalaenopsis orchids, antibacterial activity
Citation: Ho B-L, Chen J-C, Huang T-P and Fang S-C (2022) Protocorm-like-body extract of Phalaenopsis aphrodite combats watermelon fruit blotch disease. Front. Plant Sci. 13:1054586. doi: 10.3389/fpls.2022.1054586
Received: 27 September 2022; Accepted: 10 November 2022;
Published: 29 November 2022.
Edited by:
Minmin Li, Institute of Food Science and Technology (CAAS), ChinaReviewed by:
Zhiqiang Kong, Chinese Academy of Agricultural Sciences (CAAS), ChinaCopyright © 2022 Ho, Chen, Huang and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tzu-Pi Huang, dHBodWFuZ0BuY2h1LmVkdS50dw==; Su-Chiung Fang, c2NmYW5nQGdhdGUuc2luaWNhLmVkdS50dw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.