- 1Center for Agricultural Resources Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Shijiazhuang, China
- 2The National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 3Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing, China
Developing effective and durable host plant resistance is crucial for controlling powdery mildew, a devastating disease caused by Blumeria graminis f. sp. tritici (Bgt). In the present study, we dissected the genetic basis of the adult plant resistance to powdery mildew using a recombinant inbred line (RIL) composed of 176 F9 RILs population derived from a cross between PuBing 3228 (P3228) and susceptible cultivar Gao 8901. P3228 exhibits stable adult-plant resistance to powdery mildew in the field over consecutive years. We identified two QTLs on chromosomes 7DS (QPm.cas-7D) and 1AL (QPm.cas-1A) contributed by P3228, and one QTL on 3DS (QPm.cas-3D) contributed by Gao 8901, which could explain 65.44%, 3.45%, and 2.18% of the phenotypic variances, respectively. By analyzing the annotated genes in the 1.168 Mb physical interval of the major QTL QPm.cas-7D, we locked a previously cloned adult-plant resistance gene Pm38 that was most probably the candidate gene of QPm.cas-7D. Sequence alignment analysis revealed that the candidate gene of QPm.cas-7D in P3228 was identical to the reported Pm38 sequence. Two haplotypes QPm-7D-R and QPm-7D-S were identified in the whole Pm38 genomic regions between P3228 and Gao 8901. To apply QPm.cas-7D in wheat breeding, we developed a kompetitive allele-specific PCR (KASP) marker Kasp5249 that is closely linked with these haplotypes. It is worth mentioning that the QPm-7D-R haplotype significantly decreased TKW and underwent negative selection for higher yields in China wheat breeding. In this study, we identified a major QTL QPm.cas-7D and revealed the relationship between its resistance and yield, which could be beneficial for further applications in wheat disease resistance and high-yield breeding.
Introduction
Common wheat (Triticum aestivum) is an important contributor to national food security and sustains one-third of humankind (IWGSC, 2018). With an estimated global population of more than nine billion over the next 30 years, wheat production is facing an approximately 70% growth challenge to meet the food demands (IWGSC, 2014). However, wheat powdery mildew, a globally epidemic wheat disease caused by the biotrophic fungus Blumeria graminis f. sp. tritici (Bgt), can severely reduce wheat yields and affect grain quality (Yahiaoui et al., 2004; Singh et al., 2016). In recent decades, the planting area of winter wheat in China affected annually by powdery mildew has reached 6 m ha, resulting in 300,000 tons of yield loss each year (Jia et al., 2020).
Developing effective and durable host plant resistance is crucial for controlling powdery mildew epidemics. Resistance to disease in crops is typically classified into two main patterns: qualitative resistance and quantitative resistance (Spielmeyer et al., 2005; Lillemo et al., 2008; McIntosh et al., 2019; He et al., 2021). Qualitative resistance is mostly race-specific where resistance (R) gene based and confers strong and life-long immunity at all stages (Kang et al., 2020; Sánchez-Martín et al., 2021). However, Bgt isolates have complex and highly variable virulence structures, so their constant evolution causes the constant breakdown of R genes, particularly in areas where R genes were widely used (An et al., 2019). Different from qualitative resistance, quantitative resistance which conferred by polygenes is mostly non-race-specific, of which adult plant resistance (APR) is one of the main types and exhibits effectiveness at the post-seedling stages. APR usually cannot display complete immunity, to great extent, which reduces selection pressure on pathogens (Li et al., 2014). Together, these two forms of resistance have provided the genetic basis of powdery mildew resistance in wheat.
To date, more than 100 powdery mildew (Pm) genes/alleles at 63 loci (Pm1-Pm68, noting that Pm8 = Pm17, Pm18 = Pm1c, Pm22 = Pm1e, Pm23 = Pm4c, and Pm31 = Pm21) have been found from common wheat and its wild relatives (He et al., 2018; McIntosh et al., 2020). Most of the 68 formally designated Pm genes provided qualitative resistance which showed all stage resistance (ASR), only Pm38 (Lagudah et al., 2009), Pm39 (Lillemo et al., 2008), Pm46 (Moore et al., 2015), and Pm62 (Zhang et al., 2018) showed APR. Despite numerous Pm genes having been reported, most of them cannot be directly applied in wheat production due to unexpected linkage drag or longer breeding cycles required for genes that were from wheat relatives or landraces. For instance, the broad-spectrum powdery mildew resistance gene Pm16 led to 15% yield loss when it was introgressed into the wheat backgrounds (Summers and Brown, 2013). In the current wheat breeding programs in China, only a few Pm genes have been extensively used in wheat improvement (Jin et al., 2021), which are facing huge selective pressure. It is therefore necessary for resistance durability to unceasingly identify and rationally deploy various types of Pm genes/alleles from various germplasm resources.
Once the effective gene was identified, the next challenge is its accurate and rapid transfer in breeding programs. Compared with the conventional breeding, marker-assisted selection (MAS) is more accurate for it also combines genotypic identification. The targeted genes could be selected or excluded in fewer generations by using the powerful diagnostic markers (Jiang et al., 2016). Therefore, cloning of target genes/loci and their tightly-linked molecular markers are key points for MAS. Recent progress in whole-genome sequencing and data-processing strategies have greatly promoted the isolation of the resistance genes. Up to now, 13 Pm genes, including Pm1a (Hewitt et al., 2021), Pm2 (Sánchez-Martín et al., 2016), Pm3 (Yahiaoui et al., 2004), Pm4b (Sánchez-Martín et al., 2021), Pm5e (Xie et al., 2020), Pm8 (Hurni et al., 2013), Pm17 (Singh et al., 2018), Pm21 (He et al., 2018; Xing et al., 2018), Pm24 (Lu et al., 2020), Pm38/Yr18/Lr34/Sr57 (Krattinger et al., 2009), Pm41 (Li et al., 2020), Pm46/Yr46/Lr67/Sr55 (Moore et al., 2005) and Pm60 (Zou et al., 2018), have been cloned by multiple strategies. Many diagnostic markers based on variations in the functional gene sequence have been consequently developed, such as the functional kompetitive allele-specific PCR (KASP) marker Pm5e-KASP for Pm5e (Xie et al., 2020), and STS-Pm24 for Pm24 (Lu et al., 2020). Those markers have no recombination with the target genes, which highlighted the extremely precise and efficient genotyping results.
PuBing3228 (P3228) is a wheat-Agropyron cristatum introgression line (Liu et al., 2020). It exhibits stable resistance to powdery mildew in wheat-growing regions over consecutive years, indicating that it should be a promising resource for durable powdery mildew resistance. To better clarify and use the resistance against powdery mildew in P3228, the objectives of this study were to (i) assess its resistance to powdery mildew, (ii) map the major QTL for powdery mildew resistance and predict candidate gene(s), (iii) reveal the relationship between Pm gene(s) and yield traits, (iv) determine the geographical distribution of the major Pm gene(s), and (v) develop KASP marker of the candidate gene(s) for MAS breeding.
Materials and methods
Plant materials and field trials
A mapping population composed of 176 F9 RILs derived from ‘P3228 × Gao 8901’ was developed by single seed descent method. The wheat line P3228 exhibited resistance to powdery mildew at the adult stage whereas Gao 8901 was highly susceptible (Figure 1). The 176 RILs, and two parents were grown at the Center for Agricultural Resources Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences during 2021-2022 growing seasons (2022SJZ). Hill-drop (20 seeds per hill) were used and all trials used a randomized block design with three replicates. The water, fertilizer and other management of all field trials were carried out in accordance with local standard practices. Two natural populations including 157 landraces of the Chinese wheat mini-core collection, and 348 modern cultivars from ten ecological zones of China were used for marker screening and association analysis as previously described (Zhao et al., 2019; Liu et al., 2020).
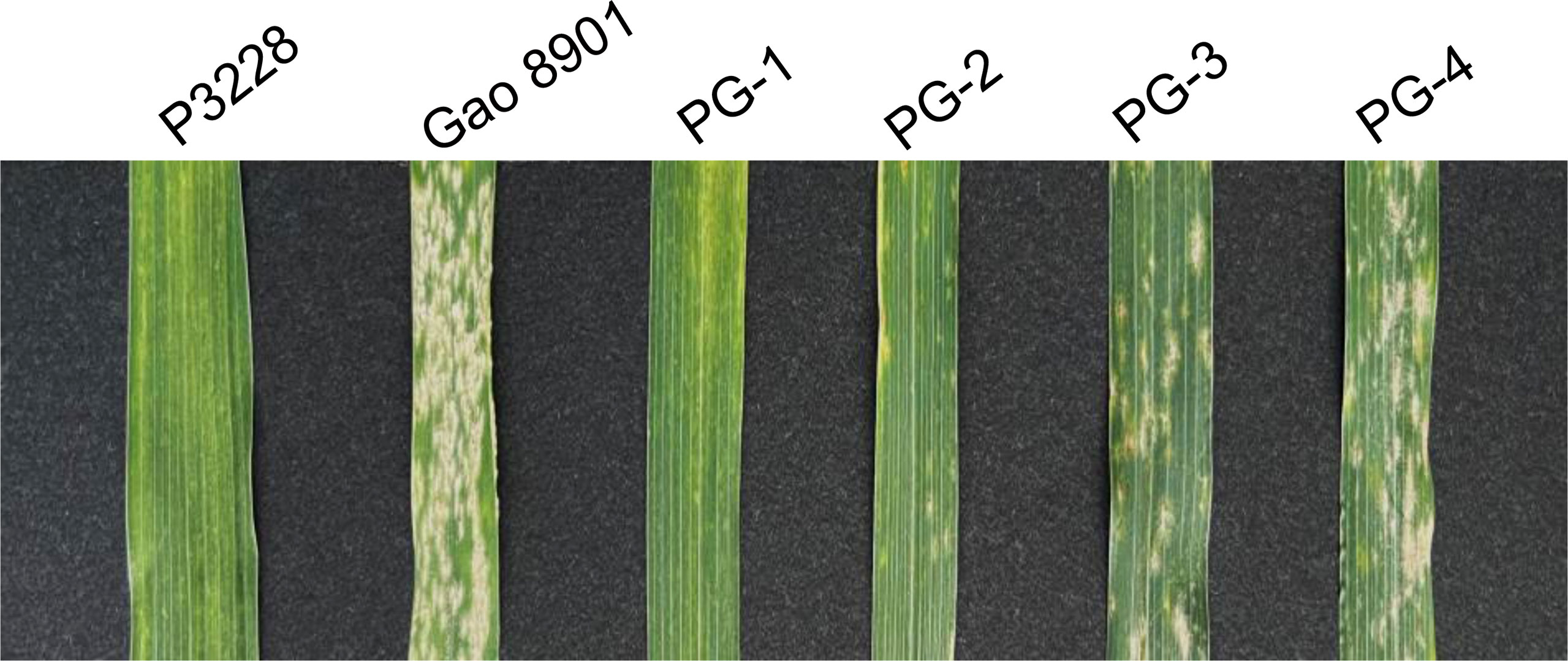
Figure 1 Phenotypic characterization of resistance to powdery mildew at the adult stage in two parents and some representative RILs. P3228, PuBing 3228; PG-1, PG-2, PG-3, and PG-4 are representative lines in PG-RIL population.
Phenotypic assessment of powdery mildew
Seedling stage reactions of P3228 and Gao 8901 to virulent Bgt isolates E09, E11, and E20 were separately tested with three replicates at a greenhouse as previously described (Qie et al., 2019). Heng 4399 was used as the susceptible control. For each line, 20 seeds were planted in rectangular trays (54 cm × 28 cm × 4 cm) with 128 wells (3 cm × 3 cm × 4 cm). After 15 days after inoculation, when sporulation was observed on the first leaf of Heng 4399, the tested plants were scored using a 0-4 scale, in which infection types (ITs) 0-2 considered resistant, while ITs 3-4 considered susceptible (Si et al., 1992).
At the jointing stage, the plants were inoculated with a mixture of Bgt isolates E09, E11, and E20. Adult plant reactions to powdery mildew were scored with mean maximum disease severity (MDS). When the susceptible control (Heng 4399) reached 80%, the MDS scores were calculated based on the Cobb scale (Peterson et al., 1948). According to the actual percentage of powdery mildew covered area (0-100%), the severity of infection in the secondary leaves (leaf below flag leaf) of five randomly selected plants in each hill was scored. Disease severities of five selected plants in each line were averaged to obtain a mean severity for each line. Each plant was assessed twice for confirmation.
QTL mapping
A whole-genome genetic map of the PG-RIL population to analysis the genetics of MDS was previously constructed from Wheat 660 K SNP array data (Liu et al., 2020). QTL mapping was conducted in IciMapping v4.1 software by the inclusive composite interval mapping of additive and dominant QTL (ICIM-ADD, Meng et al., 2015). The logarithm of odds (LOD) score≥2.5 (Sun et al., 2013). The MapChart 2.2 was used to draw the genetic map (Voorrips, 2002). The QTLs were named based on McIntosh et al. (2019) and ‘cas’ represents the Chinese Academy of Sciences.
Comparison of the identified QTLs with the known powdery mildew resistance genes
The physical position of the QTL was identified using the flanking SNP markers sequence of QTL to BLAST against the genome sequences of Chinese Spring v2.1 (Zhu et al., 2021). Candidate genes in the QTL interval was acquired based on coding sequence of Chinese Spring v2.1 and gene function annotations were manually using NCBI Non-redundant protein sequences.
Development of KASP markers
To develop markers that can efficiently trace the powdery mildew resistance genes in P3228 in MAS, KASP marker Kasp5249 (QPm-7D-FAM: gaaggtgaccaagttcatgctATGGGAGCATTATTTTTTTCCATCT, QPm-7D-HEX: gaaggtcggagtcaacggattATGGGAGCATTATTTTTTTCCATCA, QPm-7D-R: TGCTCATCTCTGGTATGCCATTTAA) was developed based on the distinctive insertions/deletions (InDels) in the targeted interval. KASP assays were performed in 96-well format in 5 μl mixture comprising 2.81 μl of 2 × KASP mix (LGC Genomics, UK), 1μl of DNA template, 1.11 μl of ddH2O and 0.08 μl of primer mixture. KASP reactions were carried out on an Applied Biosytems™ Veriti™ 96 PCR system (Thermo Fisher, USA). PCR amplification procedure was performed as previously described (Liu et al., 2020). The fluorescence value was read using FLUOstar Omega SNP (LGC Genomics, UK). The KASP genotyping results were read using KlusterCaller genotyping software (LGC Genomics, UK).
Phenotypic evaluation of agronomic traits
Phenotypic traits of 157 landraces and 348 modern cultivars, including thousand kernel weight (TKW), kernel number per spike (KNS), spikelet number per spike (TSS), spike length (SL), effective tiller number (ETN) and plant height (PH), were investigating from plants grown in 2002 and 2005 at Luoyang, Henan province and 2010 at Shunyi, Beijing.
Statistical analyses
The frequency distribution of powdery mildew responses and analysis of variance (ANOVA) was calculated in performed with SPSS Statistics v20.0 software (SPSS, USA). The broad-sense heritability (H2) was calculated using the QGAStation 2.0 (http://ibi.zju.edu.cn/software/qga/v2.0/indexc.htm) and the following formula H2 = VG/VP; where VG and VP are the genetic variance and phenotypic variance, respectively. Two-tailed t test was performed with SPSS Statistics v20.0 software (SPSS, USA).
Results
Evaluation of powdery mildew resistance and correlation analysis
At the seedling stage, both P3228 and Gao 8901 developed abundant sporulation on the leaves with an IT 4 when inoculated with Bgt isolate E09, E11 and E20, respectively. At the adult plant stage, when the MDS of the susceptible control Heng 4399 ranged from 80% to 100%, the P3228 and Gao 8901 showed 1.00% and 67.67%, respectively, showing significant differences on MDS (Table 1). For the RIL population, the frequency distributions of MDS showed continuous variation (Supplemental Figure 1). The MDS scores showed broad-sense heritability (H2) at 0.63 (Table 1).
QTL mapping
Based on the results of powdery mildew reaction evaluation, two QTLs from P3228 on chromosomes 1A and 7D, and one from Gao 8901 on chromosomes 3D, respectively, were detected in 2022SJZ environment (Table 2 and Figure 2), and were designated as QPm.cas-1A, QPm.cas-3D, and QPm.cas-7D, respectively. The QTL QPm.cas-1A was located in the marker interval AX-109816727–AX-10877999 on the short arm of chromosome 1A and explained 3.45% of the phenotypic variance with an additive effect of -10.58 (Table 2 and Figure 2). The QTL QPm.caas-3DS was mapped on chromosome 3DS and flanked by markers AX-94989783 and AX-109499958, which accounted for 2.18% of the phenotypic variance with an additive effect of 5.33 (Table 2 and Figure 2). The major QTL QPm.cas-7D was mapped on marker interval AX-111197303–AX-89471347 on the short arm of chromosome 7D and explained 65.44% of the phenotypic variance with an additive effect of -29.38 (Table 2 and Figure 2).
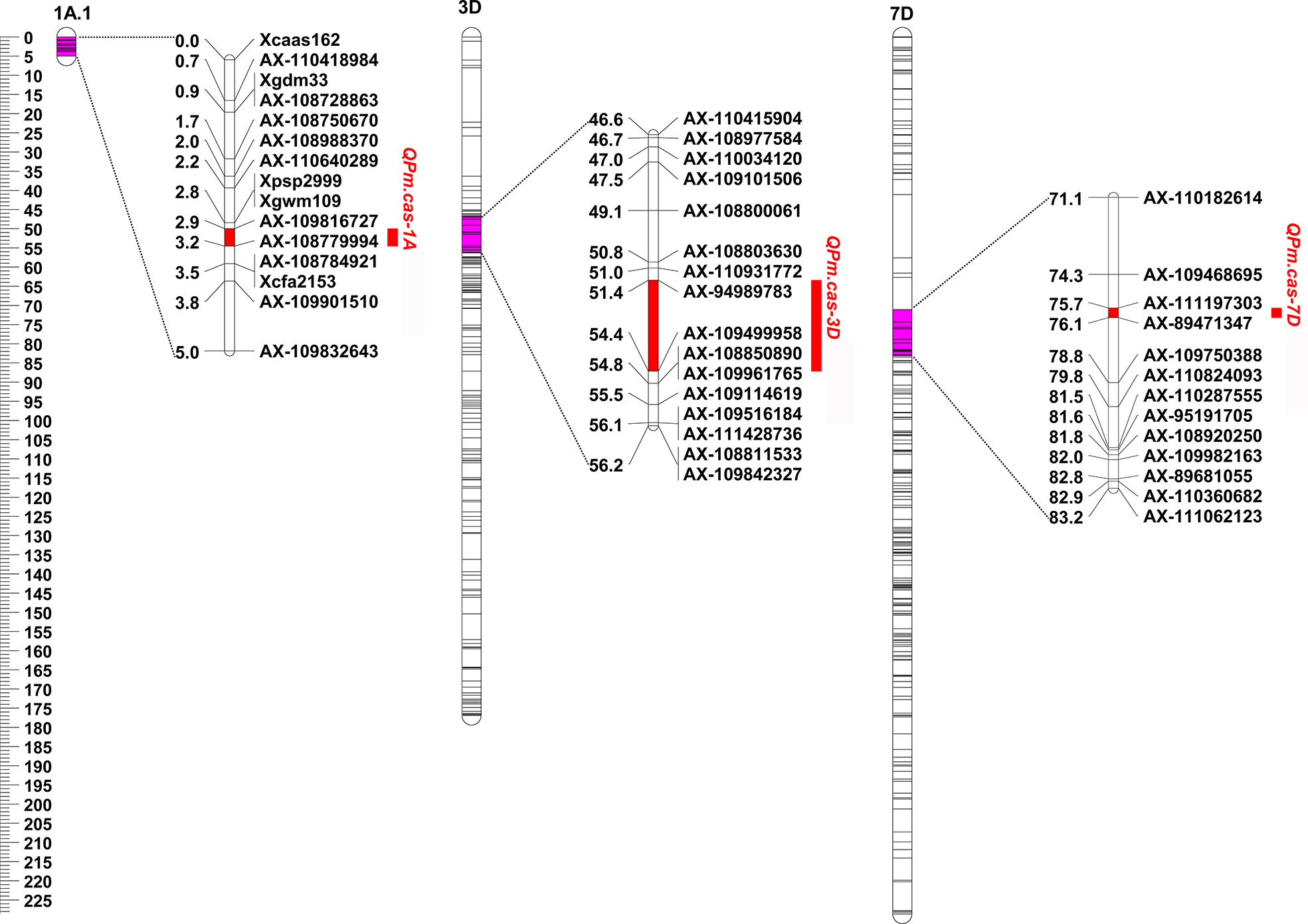
Figure 2 Genetic locations of QTL intervals associated with maximum disease severities detected in 2022SJZ environment. Uniform centimorgan (cM) scales are shown on the left. QTLs are indicated on the right side of each chromosome.
Predicting of candidate gene Pm38 for QTL QPm.cas-7D
The peak interval of the major QTL QPm.cas-7D was collocated between the markers AX-111197303 and AX-89471347. Combined with the physical position of Bin markers Bin-AX-111197303 (including AX-111197303 and AX-89378255) and Bin-AX-89471347 (including AX-89471347) based on the Chinese Spring reference genome v2.1 (Zhu et al., 2021), the QTL QPm.cas-7D was mapped to the 48.917–50.085 Mb position on chromosome arm 7DS. We found 17 high confidence annotated genes in the 1.168 Mb region using Chinese Spring reference genome v2.1 (Table 3). Among them, Pm38 (TraesCS7D03G0183600), a previously cloned adult-plant resistance gene to powdery mildew, was considered as the preferred candidate gene for QPm.cas-7D. Then, we analyzed the genomic sequence of Pm38 from Gao 8901 and P3228 using two pairs of genome-specific primers as reported to amplify the gene separately from the start codon to exon 14 (ExpF1 and Cssfr6-MR1), and from exon 11 to the stop codon (Cssfr6-MF2 and Lr34-ExpR1) (Fang et al., 2020). The results showed that the Pm38 allele in P3228 was identical to the previously reported Pm38. Moreover, two SNPs (A1654T and C5597T) and two InDels (1-bp InDels at 4996th and 3-bp InDels at 5249th) were identified between P3228 and Gao 8901 in the whole Pm38 genomic regions, which formed two haplotypes: QPm-7D-R (resistant haplotype) and QPm-7D-S (susceptible haplotype) (Figure 3A). Meanwhile, sequence alignment revealed that the 1-bp deletion at 4996th caused frameshift mutation, resulting in a loss-of-function Pm38 protein in Gao 8901 (Figure 3A).

Table 3 Candidate genes identified for QTL QPm.cas-7D with putative functions of interest and their functional annotation.
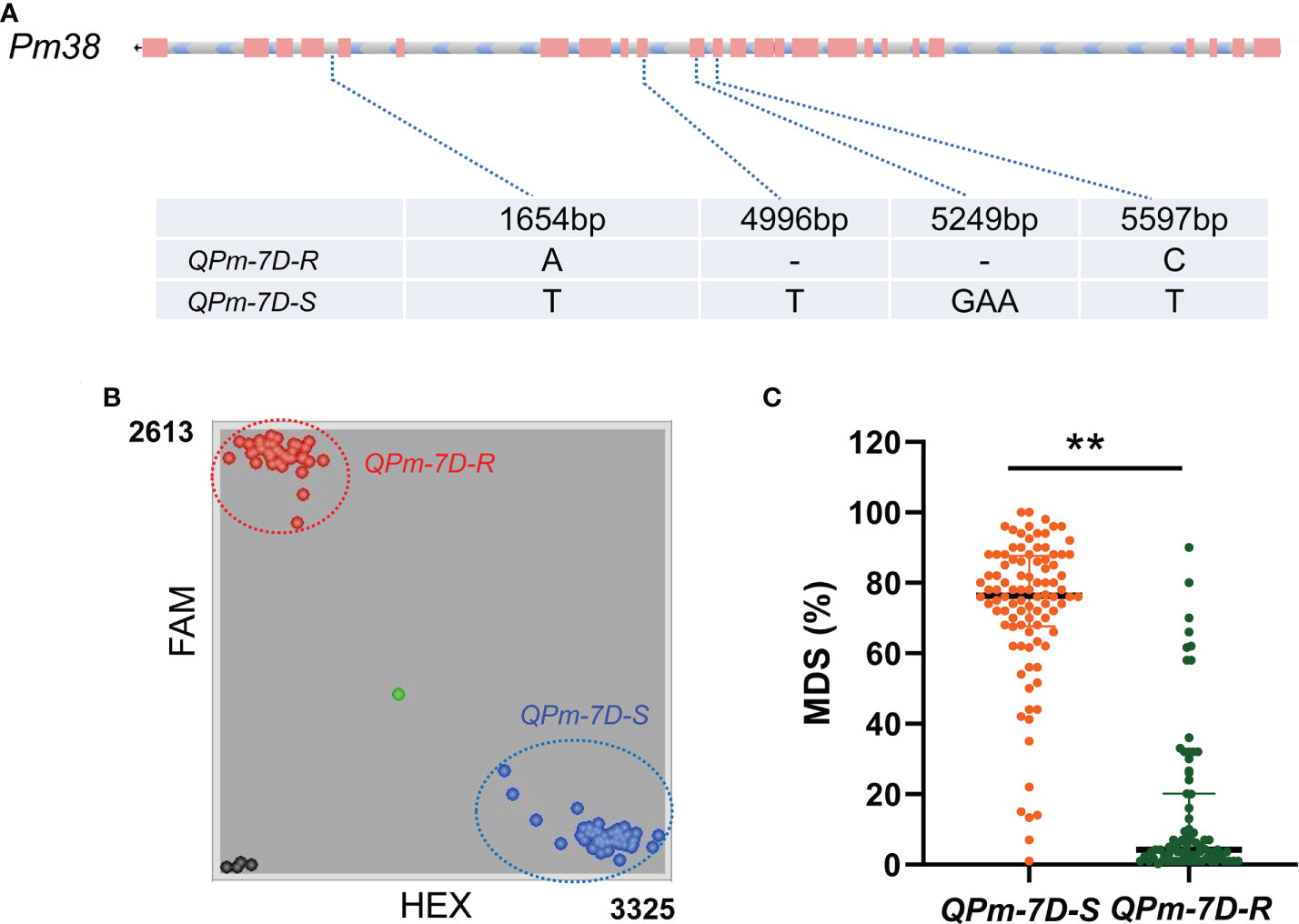
Figure 3 Haplotypes analysis with maximum disease severities (MDS) of QPm.cas-7D in PG-RIL population. (A) Haplotypes of QPm.cas-7D based on its genome regions in P3228 and Gao 8901. (B) Genotyping results of the diagnostic marker Kasp5249 developed for QPm-7D-R and QPm-7D-S haplotypes in the PG-RIL population. (C) Haplotypes analysis with MDS of Pm38 in the PG-RIL population. **P < 0.01 (two-tailed t test).
Development of KASP markers and analysis for Pm38 alleles
Based on the 3-bp InDel on the Pm38- homologous sequence between P3228 and Gao 8901, we developed a KASP marker Kasp5249 (Figure 3B). After screening the PG-RIL population using marker Kasp5249, a two-tailed t test was performed between the InDel of Kasp5249 and MDS. The results showed that Kasp5249 was significantly correlated with MDS in the PG-RIL population (Figure 3C). These results further demonstrated that the candidate gene for QPm.cas-7D was most likely Pm38.
Association analysis of QPm-7D-R haplotype with yield-related traits in common wheat
After screening 157 landraces of the Chinese wheat mini-core collection and 348 Chinese modern cultivars using the diagnostic marker Kasp5249, we performed haplotype association analysis of six agronomic traits (TKW, KNS, TSS, SL, ETN, PH) in multiple environments. The resistance haplotype QPm-7D-R was significantly correlated with TKW and SL in the 157 landraces of the Chinese wheat mini-core collection (Figures 4A-F). The mean TKW of QPm-7D-R plants was significantly lower than those of the QPm-7D-S plants (4.47 g lower in 2002, 5.71 g lower in 2005, and 3.55 g lower in 2010) (Figure 4A). Similar results were found in 348 Chinese modern cultivars. Significant differences were also detected in TKW and PH between QPm-7D-R and QPm-7D-S haplotypes (Figures 5A-F). The mean TKW of QPm-7D-R plants in three environments was also significantly lower than that of QPm-7D-S plants (5.28 g lower in 2002, 4.58 g lower in 2005, and 3.49 g lower in 2010) (Figure 5A). In summary, the above results indicate that QPm-7D-R is the lower TKW haplotype than QPm-7D-S.
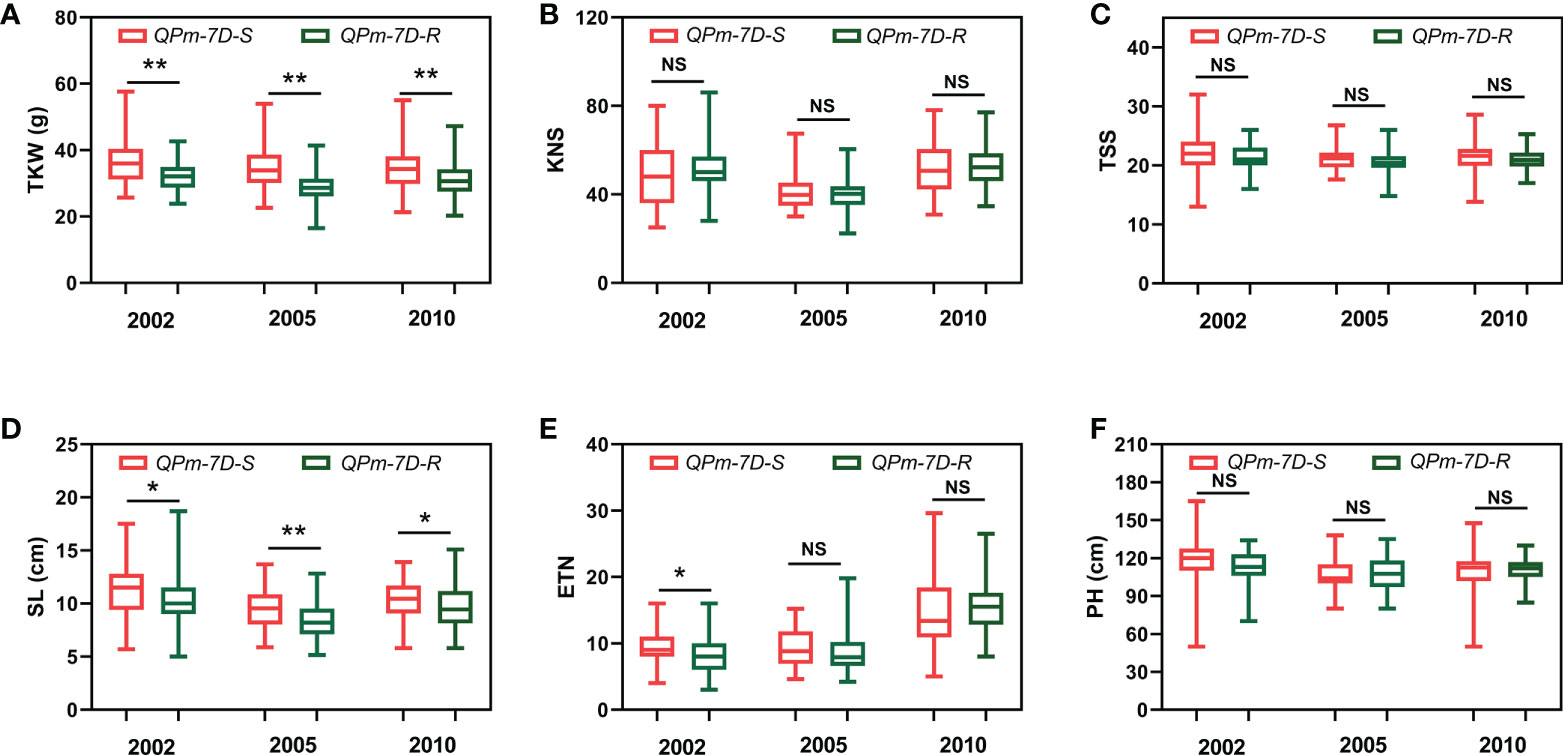
Figure 4 Haplotypes analysis with agronomic traits of QPm.cas-7D in the landraces of the Chinese wheat mini-core collection. Comparison analysis of QPm.cas-7D haplotypes with the TKW (A), KNS (B), TSS (C), SL (D), ETN (E), and PH (F) of the landraces of the Chinese wheat mini-core collection in three environments. **P < 0.01 and *P < 0.05 (two-tailed t test) indicates a significant difference to the two haplotypes. NS, no significant difference.
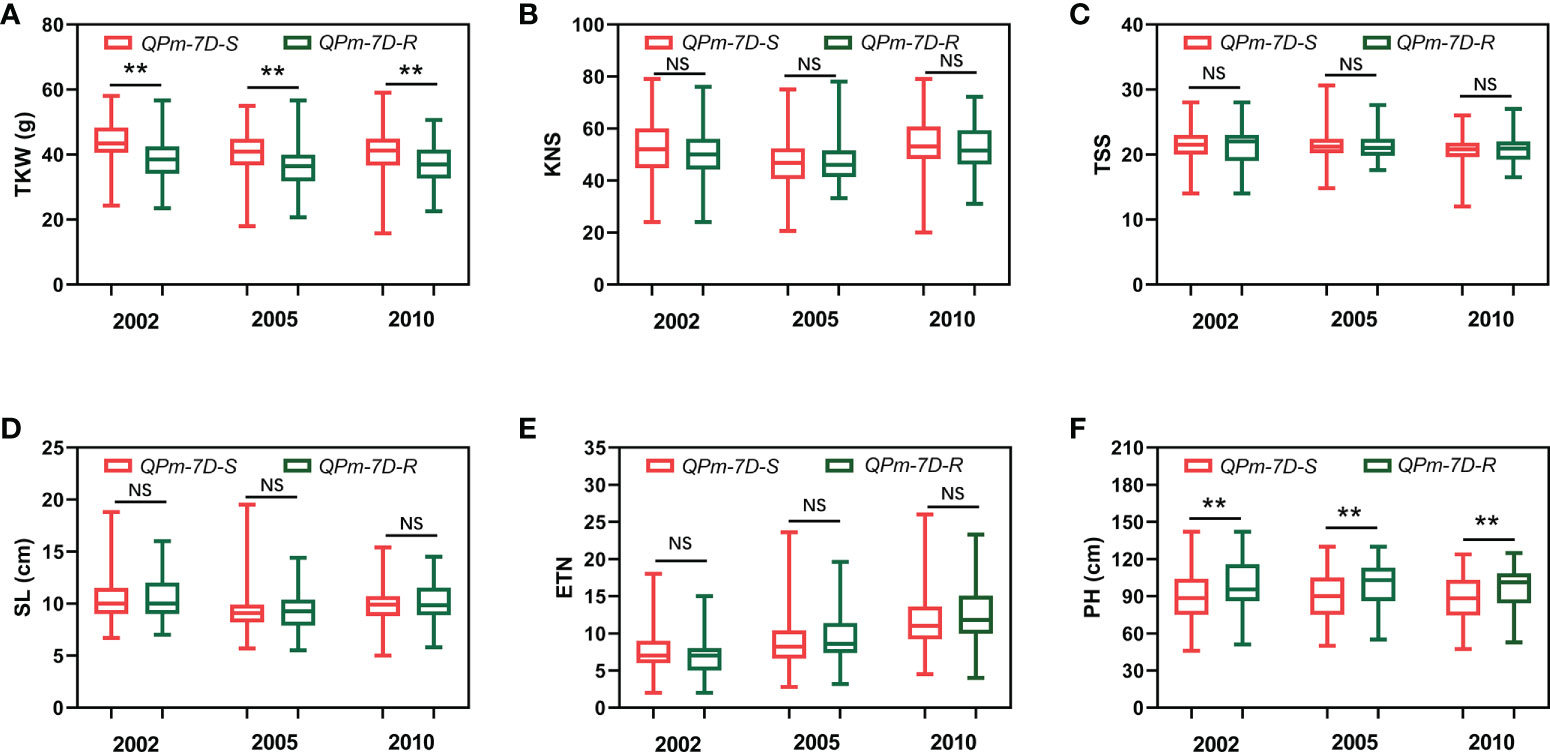
Figure 5 Haplotypes analysis with agronomic traits of QPm.cas-7D in the Chinese modern cultivars. Comparison analysis of QPm.cas-7D haplotypes with the TKW (A), KNS (B), TSS (C), SL (D), ETN (E), and PH (F) of the Chinese modern cultivars in three environments. **P < 0.01 and *P < 0.05 (two-tailed t test) indicates a significant difference to the two haplotypes. NS, no significant difference.
QPm.cas-7D-R haplotype underwent negative selection during Chinese wheat breeding
The geographic distribution of the QPm-7D-R and QPm-7D-S haplotypes was evaluated in both landraces and modern cultivars from ten ecological zones of China. The frequency of the QPm-7D-R haplotype declined in the modern cultivars relative to the landraces in the major Chinese production zones (Figures 6A, B). By contrast, the frequency of the QPm-7D-S haplotype with high TKW was increased during the transition from landraces to modern cultivars (Figures 6A, B). These results suggested that QPm-7D-R underwent negative selection in China wheat breeding for higher yields.
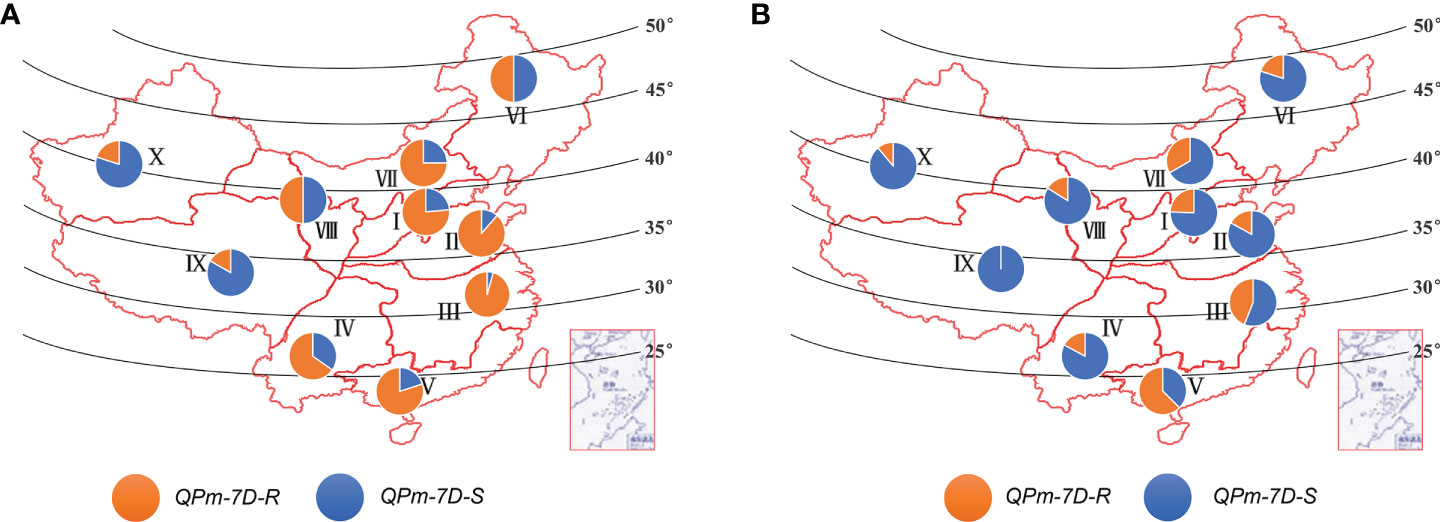
Figure 6 Geographic distribution of the QPm.cas-7D haplotypes among ten Chinese ecological regions. Distribution of QPm.cas-7D haplotypes in landraces (A) and modern cultivars (B) among ten Chinese ecological regions. I, northern winter wheat region; II, Yellow and Huai River valley winter wheat region; III, low and middle Yangtze River valley winter wheat region; IV, southwestern winter wheat region; V, southern winter wheat region; VI, northeastern spring wheat region; VII, northern spring wheat region; VIII, northwestern spring wheat region; IX, Qinghai–Tibet spring–winter wheat region; X, Xinjiang winter–spring wheat region.
Discussion
P3228 is a valuable wheat germplasm line carrying multiple and stable QTLs related to yield traits (Wang et al., 2011; Liu et al., 2020). In the present study, we found that P3228 was susceptible to Bgt isolates E09, E11, and E20 at the seedling stage but highly resistant to a mixture of those Bgt isolates at the adult plant stage, suggesting that P3228 conferred APR against powdery mildew (Figure 1). Subsequently, using a RIL population of ‘P3228 × Gao 8901’, we identified a major QTL QPm.cas-7D contributed by P3228 in an interval flanked by AX-111197303–AX-89471347 on the short arm of chromosome 7D, which explained 65.44% of the phenotypic variance with an additive effect of -29.38 (Table 2 and Figure 2). Meanwhile, a minor QTL QPm.cas-1A also from P3228 was detected in the marker interval AX-109816727–AX-10877999 on the short arm of chromosome 1A and explained 3.45% of the phenotypic variance with an additive effect of -10.58 (Table 2 and Figure 2). Pm38 (TraesCS7D03G0183600), a previously cloned Pm gene conferring APR against powdery mildew (Krattinger et al., 2009) was included in the targeted physical interval of the major QTL QPm.cas-7D. Sequencing showed that P3228 had an identical sequence to the reported Pm38 (Krattinger et al., 2009), while Gao 8901 had two different SNPs and two InDels in the genomic regions, resulting a frameshift mutation, the findings indicated that QPm.cas-7D was most likely Pm38 (Figure 3A). Of course, we cannot completely exclude the possibility that QPm.cas-7D might be one of the other 17 candidate genes that cooperated with Pm38 in the targeted physical interval. Further fine mapping and functional validation of the candidate genes should be performed to confirm the Pm38 as the caused gene for QPm.cas-7D in the future.
Due to the ease of selection and phenotypic evaluation, most disease resistance studies have focused on ASR genes, which are known as qualitative or race-specific resistance genes (Wu et al., 2022). However, overuse of single race-specific resistance genes can lead to the rapid evolution of new virulent Bgt races and consequent massive economic losses (Singh et al., 2016). In this case, developing durable resistance conferred by APR genes is emphasized. Many APR genes appear to provide broad-spectrum resistance to one or multiple diseases at the same locus, such as Pm38/Lr34/Yr18/Sr57 (Krattinger et al., 2009), Pm39/Lr46/Yr29/Sr58 (Singh et al., 2013), Pm46/Lr67/Yr46/Sr55 (Moore et al., 2015), and Pmx/Lr27/Yr30/Sr2 (Mago et al., 2011). Previous studies revealed that the pyramiding of multiple APR genes enabled near-immunity of the plants (Singh and Trethowan, 2007). Therefore, the QTL for powdery mildew resistance identified in P3228 could enrich the available wheat genetic resources in breeding for durable and multiple resistance.
The balance of resistance and yield is the main concern for breeders when using a resistance gene in wheat breeding programs, but often, disease resistance is at the expense of some agronomic traits and reduces plant adaptation (Deng et al., 2017; Han et al., 2022; Mu et al., 2022). For example, mildew resistance locus O (MLO), is a durable and broad-spectrum resistance to powdery mildew in various plant species including common wheat, however, it also leads to growth penalties and yield losses, thereby limiting its widespread use in wheat breeding (Li et al., 2022). So, investigation of the corresponding yield traits tends to be an important index for evaluating the use of resistance gene(s). Recent research shows that Pm5e has no yield penalty by investigating the agronomic performance in a pair of near-isogenic lines H962R with Pm5e and H962S without Pm5e (Qiu et al., 2022). In this study, we found that QPm.cas-7D significantly decreased TKW in the 157 landraces of the Chinese wheat mini-core collection and 348 Chinese modern cultivars (Figures 4A-F and Figures 5A-F). An additional interesting phenomenon was also observed that the frequency of QPm.cas-7D in wheat landraces was higher than in breeding lines, which might attribute to the artificial selection of high TKW trait in wheat major production regions. Thus, QPm.cas-7D could be designed to transfer into the various wheat cultivars exhibiting desirable performance on TKW.
To better transfer QPm.cas-7D in MAS, we developed a KASP marker Kasp5249 (Figure 3B) based on the differences in the Pm38- homologous sequence between P3228 and Gao 8901. It is worth mentioning that Kasp5249 was developed according to the 3-bp deletion that causes the loss of protein function, which could also be used as the functional marker of Pm38. We believe that after the suitable selection for resistance and agronomic performance, QPm.cas-7D will release its full potential in wheat breeding programs.
Conclusion
We performed QTL analysis using the PG-RIL population for MDS, and three QTLs QPm.cas-1A, QPm.cas-3D, and QPm.cas-7D were identified in 2022SJZ environment (Table 2 and Figure 2). Notably, the major QTL QPm.cas-7D contributed by P3228, could explain 65.44% of the phenotypic variances (Table 2). Furthermore, the QTL QPm.cas-7D was delimited to the physical interval of approximately 1.168 Mb, and Pm38 was considered as the candidate gene (Table 3). Based on a 3-bp InDel of Pm38 genomic sequence between the two parents, a KASP marker Kasp5249 of Pm38 allele was developed and verified by PG-RIL population (Figures 3B, C). Furthermore, the QPm-7D-R haplotype significantly decreased TKW and underwent negative selection in China wheat breeding for higher yields (Figures 4A-F and Figures 5A-F). Our finding identified a major QTL QPm.cas-7D and analyzed its effects for yield-related traits, which could be helpful in improving wheat disease resistance and high-yield breeding.-
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
DA and MZ conceived the study. TG, YJ, LX, HY, JW, and CH evaluated the phenotype. HL, GH and ZS carried out QTL mapping, predicted candidate gene, and developed the KASP markers. HL, GH and TG analyzed the data and wrote the manuscript. DA and MZ supervised and revised the writing of the article. All authors approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24030102), the National Natural Science Foundation of China (32101686), and the Hebei Province Key Research and Development Program (22326306D). The funding bodies were not involved in the design of the study, and collection, analysis, and interpretation of data, and manuscript writing.
Acknowledgments
The authors are grateful to Prof. Xueyong Zhang for providing 157 landraces of the Chinese wheat mini-core collection and 348 Chinese modern cultivars.
Conflict of interests
The reviewer JL declared a shared affiliation with the author CH to the handling editor at the time of review.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1042399/full#supplementary-material
References
An, D. G., Ma, P. T., Zheng, Q., Fu, S. L., Li, L. H., Han, F. P., et al. (2019). Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot. Theor. Appl. Genet. 132, 257–272. doi: 10.1007/s00122-018-3214-3
Deng, Y. W., Zhai, K. R., Xie, Z., Yang, D. Y., Zhu, X. D., Liu, J. Z., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, eaai8898. doi: 10.1126/science.aai8898
Fang, T., Lei, L., Li, G., Powers, C., Hunger, R. M., Carver, B. F., et al. (2020). Development and deployment of KASP markers for multiple alleles of Lr34 in wheat. Theor. Appl. Genet. 133, 2183–2195. doi: 10.1007/s00122-020-03589-x
Han, G. H., Yan, H. W., Gu, T. T., Cao, L. J., Zhou, Y. L., Wei, L., et al. (2022). Identification of a wheat powdery mildew dominant resistance gene in the Pm5 locus for high-throughput marker-assisted selection. Plant Dis. doi: 10.1094/PDIS-07-22-1545-RE
He, H., Liu, R., Ma, P., Du, H., Zhang, H., Wu, Q., et al. (2021). Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor. Appl. Genet. 134, 53–62. doi: 10.1007/s00122-020-03681-2
Hewitt, T., Müller, M. C., Molnár, I., Mascher, M., Holušová, K., Šimková, H., et al. (2021). A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. New Phytol. 229, 2812–2826. doi: 10.1111/nph.17075
He, H. G., Zhu, S. Y., Zhao, R. H., Jiang, Z. N., Ji, Y. Y., Ji, J., et al. (2018). Pm21, encoding a typical CC-NBS-LRR protein, confers broad spectrum resistance to wheat powdery mildew disease. Mol. Plant 11, 879–882. doi: 10.1016/j.molp.2018.03.004
Hurni, S., Brunner, S., Buchmann, G., Herren, G., Jordan, T., Krukowski, P., et al. (2013). Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 76, 957–996. doi: 10.1111/tpj.12345
IWGSC (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788. doi: 10.1126/science.1251788
IWGSC (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Jia, M. S., Xu, H. X., Liu, C., Mao, R. X., Li, H. S., Liu, J. J., et al. (2020). Characterization of the powdery mildew resistance gene in the elite wheat cultivar Jimai 23 and its application in marker-assisted selection. Front. Genet. 11, 241. doi: 10.3389/fgene.2020.00241
Jiang, Y., Schulthess, A. W., Rodemann, B., Ling, J., Plieske, J., Kollers, S., et al. (2016). Validating the prediction accuracies of marker-assisted and genomic selection of fusarium head blight resistance in wheat using an independent sample. Theor. Appl. Genet. 130, 471–482. doi: 10.1007/s00122-016-2827-7
Jin, Y. L., Shi, F. Y., Liu, W. H., Fu, X. Y., Gu, T. T., Han, G. H., et al. (2021). Identification of resistant germplasm and detection of genes for resistance to powdery mildew and leaf rust from 2,978 wheat accessions. Plant Dis. 105, 3900–3908. doi: 10.1094/PDIS-03-21-0532-RE
Kang, Y. C., Zhou, M. X., Merry, A. M., Barry, K. M. (2020). Mechanisms of powdery mildew resistance of wheat-a review of molecular breeding. Plant Pathol. 69, 601–617. doi: 10.1111/ppa.13166
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Lagudah, E. S., Krattinger, S. G., Herrera-Foessel, S., Singh, R. P., Huerta-Espino, J., Spielmeyer, W., et al. (2009). Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theor. Appl. Genet. 119, 889–898. doi: 10.1007/s00122-009-1097-z
Li, M., Dong, L., Li, B., Wang, Z., Xie, J., Qiu, D., et al. (2020). A CNL protein in wild emmer wheat confers powdery mildew resistance. New Phytol. 228, 1027–1037. doi: 10.1111/nph.16761
Li, Z., Lan, C., He, Z., Singh, R. P., Rosewarne, G. M., Chen, X., et al. (2014). Overview and application of QTL for adult plant resistance to leaf rust and powdery mildew in wheat. Crop Sci. 54, 1907–1925. doi: 10.2135/cropsci2014.02.0162
Li, S., Lin, D., Zhang, Y., Deng, M., Chen, Y., Lv, B., et al. (2022). Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602, 455–460. doi: 10.1038/s41586-022-04395-9
Lillemo, M., Asalf, B., Singh, R. P., Huerta-Espino, J., Chen, X. M., He, Z. H., et al. (2008). The adult plant rust resistance loci Lr34/Yr18 and Lr46/Yr29 are important determinants of partial resistance to powdery mildew in bread wheat line Saar. Theor. Appl. Genet. 116, 1155–1166. doi: 10.1007/s00122-008-0743-1
Liu, H., Li, H., Hao, C., Wang, K., Wang, Y., Qin, L., et al. (2020). TaDA1, a conserved negative regulator of kernel size, has an additive effect with TaGW2 in common wheat (Triticum aestivum l.). Plant Biotechnol. J. 18, 1330–1342. doi: 10.1111/pbi.13298
Liu, H., Zhang, X., Xu, Y., Ma, F., Zhang, J., Cao, Y., et al. (2020). Identification and validation of quantitative trait loci for kernel traits in common wheat (Triticum aestivum l.). BMC Plant Biol. 20, 529. doi: 10.1186/s12870-020-02661-4
Lu, P., Guo, L., Wang, Z., Li, B., Li, J., Li, Y., et al. (2020). A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun. 11, 680. doi: 10.1038/s41467-020-14294-0
Mago, R., Tabe, L., McIntosh, R. A., Pretorius, Z., Kota, R., Paux, E., et al. (2011). A multiple resistance locus on chromosome arm 3BS in wheat confers resistance to stem rust (Sr2), leaf rust (Lr27) and powdery mildew. Theor. Appl. Genet. 123, 615–623. doi: 10.1007/s00122-011-1611-y
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Xia, X. C., Raupp, W. J. (2019) Catalogue of gene symbols for wheat 2019 supplement. Available at: https://wheat.pw.usda.gov/GG3/WGC.
Jia, M. S., Xu, H. X., Liu, C., Mao, R. X., Li, H. S., Liu, J. J., et al (2020). Characterization of the powdery mildew resistance gene in the elite wheat cultivar Jimai 23 and its application in marker-assisted selection. Front. Genet. 11, 241. doi: 10.3389/fgene.2020.00241
Meng, L., Li, H., Zhang, L., Wang, J. (2015). QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Moore, J. W., Herrera-Foessel, S., Lan, C., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–8. doi: 10.1038/ng.3439
Moore, J. W., Herrera-Foessel, S., Lan, C., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi: 10.1038/ng.3439
Mu, Y. J., Gong, W. P., Qie, Y. M., Liu, X. Q., Li, L. Z., Sun, N. N., et al. (2022). Identification of the powdery mildew resistance gene in wheat breeding line yannong 99102-06188 via bulked segregant exome capture sequencing. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1005627
Peterson, R. F., Campbell, A. B., Hannah, A. E. (1948). A diagrammatic scale for estimating rust intensity of leaves and stem of cereals. Can. J. Res. Sect. C. 26, 496–500. doi: 10.1139/CJR48C-033
Qie, Y., Sheng, Y., Xu, H., Jin, Y., Ma, F., Li, L., et al. (2019). Identification of a new powdery mildew resistance gene pmDHT at or closely linked to the Pm5 locus in the Chinese wheat landrace dahongtou. Plant Dis. 103, 2645–2651. doi: 10.1094/PDIS-02-19-0401-RE
Qiu, D., Huang, J., Guo, G. H., Hu, J. H., Li, Y. H., Zhang, H. J., et al. (2022). The Pm5e gene has no negative effect on wheat agronomic performance: evidence from newly established near-isogenic lines. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.918559
Sánchez-Martín, J., Steuernagel, B., Ghosh, S., Herren, G., Hurni, S., Adamski, N., et al. (2016). Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17, 221. doi: 10.1186/s13059-016-1082-1
Sánchez-Martín, J., Widrig, V., Herren, G., Wicker, T., Zbinden, H., Gronnier, J., et al. (2021). Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants 7, 327–341. doi: 10.1038/s41477-021-00869-2
Singh, R. P., Herrera-Foessel, S. A., Huerta-Espino, J., Lan, C. X., Basnet, B. R., Bhavani, S., et al. (2013). Pleiotropic gene Lr46/Yr29/Pm39/Ltn2 confers slow rusting, adult plant resistance to wheat stem rust fungus. proceedings borlaug global rust initiative. New Delhi India: 2013 Tech. Workshop, 17.1.
Singh, S. P., Hurni, S., Ruinelli, M., Brunner, S., Sanchez-Martin, J., Krukowski, P., et al. (2018). Evolutionary divergence of the rye Pm17 and Pm8 resistance genes reveals ancient diversity. Plant Mol. Biol. 98, 249–260. doi: 10.1007/s11103-018-0780-3
Singh, R. P., Singh, P. K., Rutkoski, J., Hodson, D. P., He, X., Jørgensen, L. N., et al. (2016). Disease impact on wheat yield potential and prospects of genetic control. Annu. Rev. Phytopathol. 54, 303–322. doi: 10.1146/annurev-phyto-080615-095835
Singh, R. P., Trethowan, R. (2007). Breeding major food staples. Eds. Kang, M., Priyadarshan, P. M. (Ames, IA: Blackwell), 109–140, 349-355.
Si, Q. M., Zhang, X. X., Duan, X. Y., Sheng, B. Q., Zhou, Y. L. (1992). On gene analysis and classification of powdery mildew (Erysiphe graminis f. sp. tritici) resistant wheat varieties. Acta Phytopathol. Sinica. 22, 349–355. doi: 10.13926/j.cnki.apps.1992.04.021
Spielmeyer, W., McIntosh, R. A., Kolmer, J., Lagudah, E. S. (2005). Powdery mildew resistance and Lr34/Yr18 genes for durable resistance to leaf and stripe rust cosegregate at a locus on the short arm of chromosome 7D of wheat. Theor. Appl. Genet. 111, 731–735. doi: 10.1007/s00122-005-2058-9
Summers, R. W., Brown, J. K. M. (2013). Constraints on breeding for disease resistance in commercially competitive wheat cultivars. Plant Phothol. 62, 115–121. doi: 10.1111/ppa.12165
Sun, Z., Li, H., Zhang, L., Wang, J. (2013). Properties of the test statistic under null hypothesis and the calculation of LOD threshold in quantitative trait loci (QTL) mapping. Acta Agronom. Sin. 39, 1–11. doi: 10.3724/SP.J.1006.2013.00001
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, J., Liu, W., Wang, H., Li, L., Gao, A. (2011). QTL mapping of yield-related traits in the wheat germplasm 3228. Euphytica 177, 277–292. doi: 10.1007/s10681-010-0267-z
Wu, L. R., Zhu, T., He, H. G., Cao, X. Y., Li, H. S., Xu, H. X., et al. (2022). Genetic dissection of the powdery mildew resistance in wheat breeding line LS5082 using BSR-seq. Crop J. doi: 10.1016/j.cj.2021.12.008
Xie, J., Guo, G., Wang, Y., Hu, T., Wang, L., Li, J., et al. (2020). A rare single nucleotide variant in Pm5e confers powdery mildew resistance in common wheat. New Phytol. 228, 1011–1026. doi: 10.1111/nph.16762
Xing, L., Hu, P., Liu, J., Witek, K., Zhou, S., Xu, J., et al. (2018). Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol. Plant 11, 874–878. doi: 10.1016/j.molp
Yahiaoui, N., Srichumpa, P., Dudler, R., Keller, B. (2004). Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 37, 528–538. doi: 10.1046/j.1365-313x.2003.01977.x
Zhang, R. Q., Fan, Y. L., Kong, L. N., Wang, Z. J., Wu, J. Z., Xing, L. P., et al. (2018). Pm62, an adult-plant powdery mildew resistance gene introgressed from Dasypyrum villosum chromosome arm 2VL into wheat. Theor. Appl. Genet. 131, 2613–2620. doi: 10.1007/s00122-018-3176-5
Zhao, J., Wang, Z., Liu, H., Zhao, J., Li, T., Hou, J., et al. (2019). Global status of 47 major wheat loci controlling yield, quality, adaptation and stress resistance selected over the last century. BMC Plant Biol. 19, 5. doi: 10.1186/s12870-018-1612-y
Zhu, T., Wang, L., Rimbert, H., Rodriguez, J. C., Deal, K. R., De Oliveira, R., et al. (2021). Optical maps refine the bread wheat Triticum aestivum cv Chinese spring genome assembly. Plant J. 107, 303–314. doi: 10.1111/tpj.15289
Keywords: adult plant resistance, powdery mildew, Pm38, haplotypes, molecular markers, common wheat
Citation: Liu H, Han G, Gu T, Jin Y, Shi Z, Xing L, Yan H, Wang J, Hao C, Zhao M and An D (2022) Identification of the major QTL QPm.cas-7D for adult plant resistance to wheat powdery mildew. Front. Plant Sci. 13:1042399. doi: 10.3389/fpls.2022.1042399
Received: 12 September 2022; Accepted: 06 October 2022;
Published: 19 October 2022.
Edited by:
Xiaoli Fan, Chengdu Institute of Biology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Jindong Liu, Institute of Crop Sciences, (CAAS), ChinaLiqiang Song, Hebei Agricultural University, China
Copyright © 2022 Liu, Han, Gu, Jin, Shi, Xing, Yan, Wang, Hao, Zhao and An. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meicheng Zhao, bWN6aGFvQHNqemlhbS5hYy5jbg==; Diaoguo An, YW5kaWFvZ3VvQDE2My5jb20=
†These authors have contributed equally to this work
 Hong Liu
Hong Liu Guohao Han
Guohao Han Tiantian Gu1†
Tiantian Gu1† Yuli Jin
Yuli Jin Hanwen Yan
Hanwen Yan Chenyang Hao
Chenyang Hao Diaoguo An
Diaoguo An
