- 1Hunan Provincial Engineering and Technology Research Center for Agricultural Microbiology Application, Hunan Institute of Microbiology, Changsha, China
- 2College of Plant Protection, Hunan Agricultural University, Changsha, China
Edeines are a group of non-ribosomal antibacterial peptides produced by Brevibacillus brevis. Due to the significant antibacterial properties of edeines, increasing edeine yield is of great interest in biomedical research. Herein, we identified that EdeB, a member of the ParB protein family, significantly improved edeine production in B. brevis. First, overexpression of edeB in B. brevis X23 increased edeine production by 92.27%. Second, in vitro bacteriostasis experiment showed that edeB-deletion mutant exhibited less antibacterial activity. Third, RT-qPCR assay demonstrated that the expression of edeA, edeQ, and edeK, which are key components of the edeine biosynthesis pathway, in edeB-deletion mutant X23(ΔedeB) was significantly lower than that in wild-type B. brevis strain X23. Finally, electrophoretic mobility shift assay (EMSA) showed that EdeB directly bound to the promoter region of the edeine biosynthetic gene cluster (ede BGC), suggesting that EdeB improves edeine production through interaction with ede BGC in B. brevis.
Introduction
B. brevis is a Gram-positive, rod-shaped, and spore-producing bacterium, which produces several antibacterial substances, but not toxins (Cook, 1993; Che et al., 2013; Hou et al., 2015; Jianmei et al., 2015). Its antibacterial products are widely used in biological control of plant diseases caused by pathogenic bacteria (Yang and Yousef, 2018; Saxena et al., 2020). Edeines, which are non-ribosomal antibacterial peptides produced by B. brevis, consist of four unconventional amino acid residues, a glycine, and a polyamine (Figure 1A) (Hettinger and Craig, 1970; Czerwinski et al., 1983; Johnson et al., 2020). As non-ribosomal antibacterial peptides, edeines are synthesized by the multifunctional enzyme complex system of “sulfur template polymerase mechanism”. The unique molecular structure of edeines contributes to a variety of biological function, including antimicrobial and antitumor activities, which can greatly inhibit the growth of numerous bacteria, fungi, mycoplasma, and tumor cells (Kurylo-Borowska and Szer, 1973; Cozzarelli, 1977; Czajgucki et al., 2010). Edeines inhibit bacterial growth through a number of different targets, including DNA, RNA, and ribosomes (protein synthesis), at a dose-dependent manner (Chen and Lu, 2020). At low concentration (<15 μg/mL), edeines reversibly constrain the activity of DNA polymerase II and III to inhibit DNA synthesis, however, the synthesis of protein is not affected. Edeines at high concentration (>150 μg/mL) bind to the P site of small ribosomal subunit (30S subunit), which competitively blocks the binding of fMet-tRNA onto ribosomes and simultaneously inhibits the initiation of transcription and protein translation (Szer and Kurylo-Borowska, 1972; Dinos et al., 2004). In addition, edeines can be used as a transcriptional inhibitor to study ribosome function and protein synthesis. Moreover, edeines can also eliminate bacterial resistance caused by plasmid genes and inhibit the division of Bacillus subtilis cells (Shimotohno et al., 2010; Semenchenko et al., 2016).
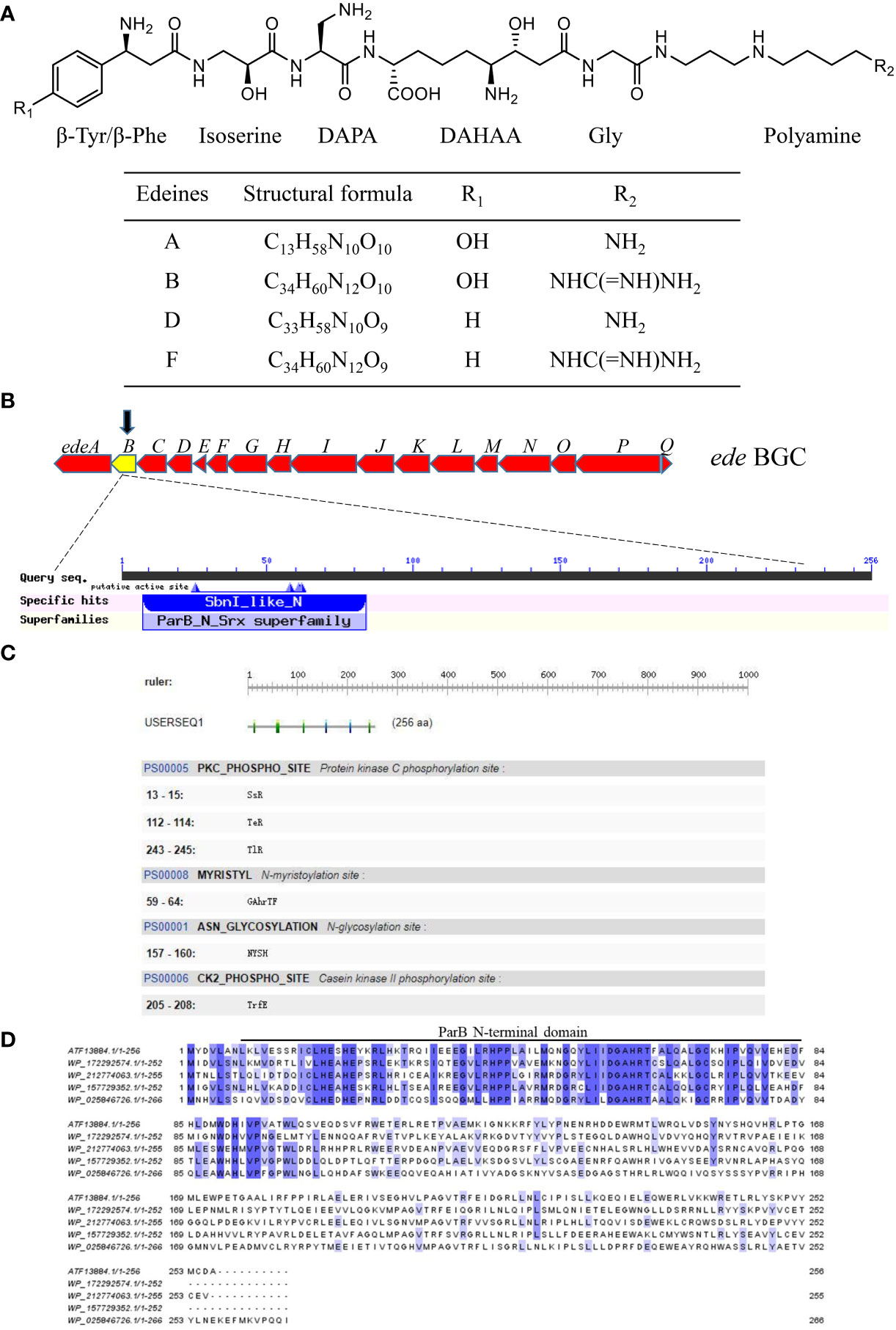
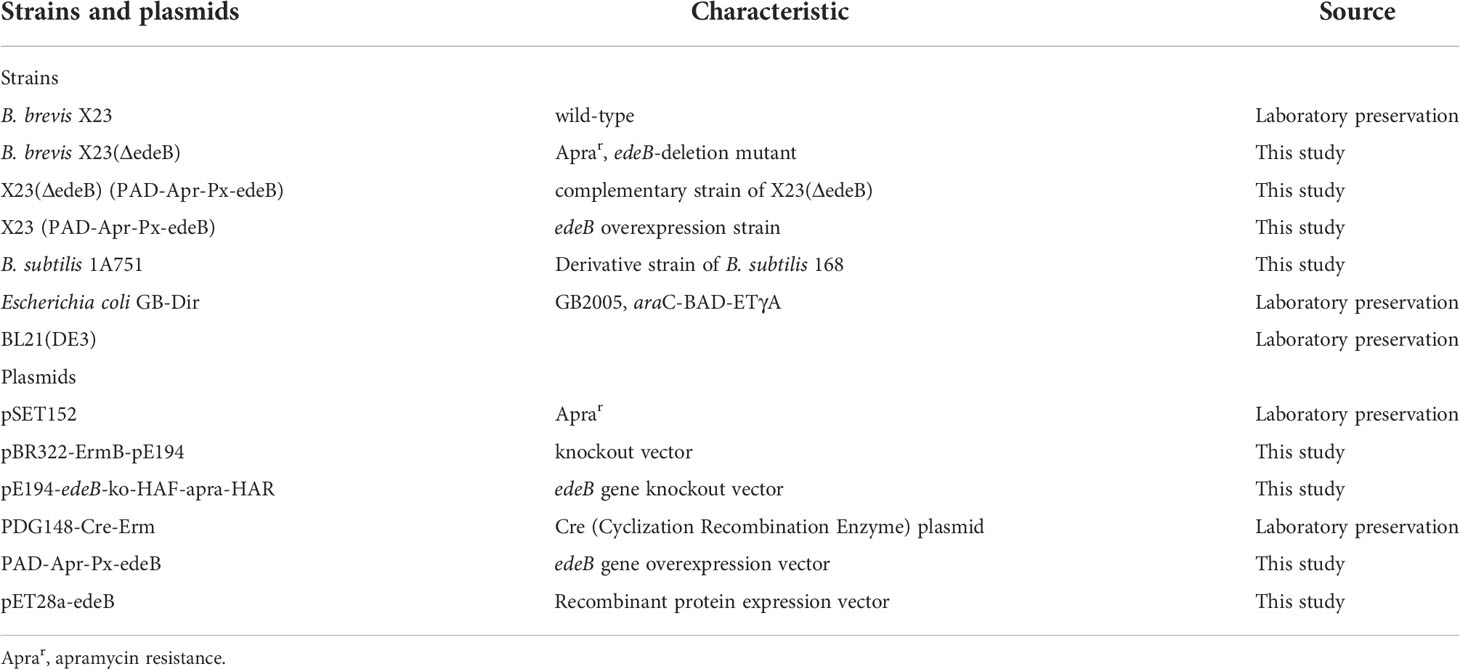
Figure 1 Characteristics of the edeB gene and EdeB protein. (A) Chemical structure of edeine A, B, D, and F. DAPA, 2,3-diaminopropionic acid; DAHAA, 2,6-diamino-7-hydroxyazaleic acid. (B) Bioinformatic analysis of edeB gene. The edeB gene on the edeines gene cluster (ede BGC) was shown in the yellow box. Conserved domain analysis of EdeB protein was performed by NCBI-CDD and Pfam database search. (C) Functional sites of EdeB. (D) Sequence alignment of EdeB and other ParB family proteins from Bacillus sp. WMMC1349 (GenBank accession: WP_172292574.1), Polycladomyces abyssicola (GenBank accession: WP_212774063.1), Tumebacillus algifaecis (GenBank accession: WP_157729352.1) and Brevibacillus agri (GenBank accession: WP_025846726.1).
Regardless of the biological activities of edeines mentioned above, low production level of edeines by edeine-producing bacteria is a major issue limiting their applications in medicine and agriculture. Edeine A and B produced by B. brevis X23 exhibit wide antibacterial spectrum and have been used as biocontrol agents against bacterial wilt in solanaceae crops. The complete genomic sequence of B. brevis X23 allows the identification of edeine biosynthetic gene cluster (ede BGC) (Wu et al., 2012; Liu et al., 2016). In this study, we identified and explored the biological function of regulatory proteins involved in biosynthesis of edeines, which will provide a theoretical basis for the construction of B. brevis strains with high-yield of edeines through genetic manipulation.
A potential transcriptional factor gene edeB on the ede BGC was identified on the basis of bioinformatic analysis of B. brevis X23 genome. Protein EdeB contains a typical helix-turn-helix (HTH) domain, which is conserved in ParB family transcriptional factors. Although ParB homolog proteins have been annotated from many genomes of Bacillus species, their function has not been fully understood yet. In this study, we constructed a B. brevis mutant X23(ΔedeB) by disrupting the edeB gene through Red/ET homologous recombination. Furthermore, a complementary strain and edeB-overexpression strain were constructed using the expression plasmid PAD-Apr-Px-edeB vector. Subsequently, we compared edeine production among those different B. brevis strains to explore the effect of edeB on the yield of edeines. The binding ability between EdeB protein and the promoter region of ede BGC was also evaluated. Our results demonstrated that EdeB significantly improved edeine production in B. brevis X23 through binding to ede BGC.
Materials and methods
Strains, plasmids, culture medium, and antibiotics
The strains and plasmids used in this study are shown in Table 1. E. coli and B. brevis were cultured in LB medium (tryptone 10.0 g/L, yeast extract 5.0 g/L, NaCl 1.0 g/L, pH 7.0 ~ 7.5, and 1.5% of agar powder for solid culture). NB medium (tryptone 10.0 g/L, beef extract 3.0 g/L, sodium chloride 5.0 g/L, glucose 10.0 g/L, pH 7.0 ~ 7.5) was used for fermentation cultivation. Apramycin (20 μg/mL) and Kanamycin (30 μg/mL) were added into the media for E. coli as required. Gene-engineered B.brevis strains were selected on LB media containing apramycin (10 μg/mL).
Reagents
PrimerSTAR Max DNA Polymerase and DNA molecular weight standard were purchased from TaKaRa Co., Ltd (Dalian, China). All restriction endonucleases were purchased from New England Biolabs (Beijing, China). Genomic DNA, RNA, and plasmid extraction kits, reverse transcription kits, and DNA gel recovery kits were purchased from TransGen Biotech (Beijing, China). Antibiotics and other biochemical reagents were purchased from Sangon Biotech Co., Ltd. (Shanghai, China).
Primers, genomic DNA extraction, and DNA sequencing
Primers used in this study are shown in Supplementary Table 1. All primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The genomic DNA of B. brevis X23 was extracted using the phenol chloride method according to previous study (Mcorist et al., 2002). DNA sequencing was conducted at Hunan Tsingke Biotechnology Co., Ltd. (Changsha, China).
Identification of the regulatory gene edeB on ede BGC
The genome of B. brevis strain X23 was sequenced and deposited at NCBI with the accession number NZ_CP023474.1. Regulatory genes around ede BGC in B. brevis X23 genome were reviewed to identify edeB. The conserved domain of identified EdeB protein (Genebank accession: ATF13884.1) was analyzed by the NCBI’s conserved domain database (https://www.ncbi.nlm.nih.gov/cdd/) and Pfam (http://pfam.xfam.org/). The function sites of EdeB were analyzed by PROSITE (https://prosite.expasy.org/).The homologs of EdeB were compared and analyzed using Jalview software.
Construction of edeB-deletion, complementation, and overexpression mutants
The edeB-knockout vectors were constructed based on Red/ET homologous recombination (Yu et al., 2000; Zhang et al., 2000; Muyrers et al., 2001; Sharan et al., 2009). Genomic DNA of B. brevis X23 was used as a template to amplify the upstream and downstream homologous arms of edeB with primers edeB-F1/edeB-F2 and edeB-R1/edeB-R2 using PCR assay. Subsequently, the apramycin resistance gene (Aprar) was amplified from plasmid pSET152 with primers Apra-F/Apra-R using PCR. The pBR322-ErmB-ori194 vector skeleton was amplified plasmid pE194 with primers pE194-F/pE194-R. Then, the above four DNA fragments were mixed and electroshocked into E. coli GB-Dir containing the Red/ET recombination system to be connected into a circular plasmid through homologous recombination. Then proper knockout vectors were selected and sequenced through enzyme digestion and the selected plasmid with correct sequence was named as pE194-edeB-ko-HAF-apra-HAR. The knockout vector was transferred into B. brevis X23, and undergone homologous recombination to replace the edeB gene by Aprar. The mutant was further confirmed through PCR assay with primers P1/P2 and P3/P4, and named as B. brevis X23(ΔedeB)-Apra. Finally, the Cre plasmid was transferred into B. brevis X23(ΔedeB)-Apra to generate B. brevis X23(ΔedeB) strain by removing Aprar.
Next, the edeB gene was amplified by PCR with primer pairs edeB-Px-F and edeB-Px-R. The amplicons were recovered from agarose gel and purified. The edeB gene fragments and vector PAD-Apr-Px were digested with BamH I and Pst I endonucleases, respectively, and then connected by T4 DNA ligase to obtain the expression plasmid PAD-Apr-Px-edeB. Finally, the expression plasmid was transferred into wild-type B. brevis X23 strain and X23(ΔedeB) by electro-transfer. The edeB-complementary and overexpression strains were obtained through apramycin antibiotic resistance screening, and confirmed by PCR with primers xylR-5-out/pAD123-end.
Assessment of bacterial growth
The growth of wild-type B. brevis strain and its mutants were evaluated according to the previously published report (Lan et al., 2017). Briefly, the wild-type B. brevis X23 strain or mutants were preconditioned overnight in LB medium, and then transferred to fresh NB liquid medium (initial OD600 of 0.1), cultured at 30°C and 180 r/min. Then, the bacterial solution concentration (OD600) was measured on a spectrophotometer after 12, 24, 36, and 48 h of cultivation in the NB liquid medium. The fermentation experiments were repeated for three times, and each measurement was also conducted for three times in parallel.
Evaluation of antibacterial activity
B. subitlis 1A751 strain, which is sensitive to edeines, was used to evaluate the antibacterial activity of edeines produced by B. brevis. The antibacterial activity of fermentation broth was evaluated according to the previous study (Dang et al., 2013). Briefly, B. brevis was cultivated in liquid NB for 48 h, and centrifugated at 10,000 g for 10 min at 4°C to collect the supernatant, which was then filtrated through a filter (0.22 μm) to obtain sterile filtrate. B. subtilis 1A751 was spread on LB agar plates after dilution to OD600 of 0.1. A total of 100 μL filtrated B. brevis supernatant was added into each hole (diameter of 8 mm) of B. subtilis culture. Sterile LB medium was used as blank control (CK). After incubation at 30°C for 48 h, the antibacterial activity was evaluated by measuring the radius of antibacterial circle. The experiments were repeated for three times with three replicates each.
RNA isolation and RT-qPCR
The relatively transcriptional level of edeA, edeQ, and edeK on ede BGC was determined through RT-qPCR assay. The primers used on qPCR assay are shown in Supplementary Table 1. Total RNA was extracted from B. brevis X23 and mutants after 48h of fermentation according to the instructions of EasyPure ® RNA Kit. Reverse transcription was conducted using the reverse transcription kit (EasyScript®One-Step gDNA Removal and cDNA Synthesis SuperMix, TransGen). 2X Universal SYBR Green Fast For qPCR Mix kit (Abclone) was used in real-time fluorescent quantitative PCR assay on an ABI-QS5 real-time PCR experiment platform. Three parallel experiments were designed and each sample was repeated three times. The 16S rRNA gene in B. brevis was used as a control, and the comparative cycle threshold method was used for relative quantification (Livak and Schmittgen, 2013).
Analysis of the promoter sequence of ede BGC
The binding sites of ede BGC promoter were predicted through BPROM (http://linux1.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfingf) and BDGP (https://www.fruitfly.org/seq_tools/promoter.html). WeBlogo 3.0 (http://weblogo.threeplusone.com/) was used to create sequence logos for the -10 and -35 regions of the promoter.
Electrophoretic mobility shift assay (EMSA)
EMSA was conducted to explore whether EdeB directly binds to the promoter region of ede BGC according to the previous method (Hellman and Fried, 2007). Briefly, the edeB gene was amplified from genomic DNA of wild-type B. brevis X23 using primers edeB-F and edeB-R. The vector pET28a was digested by Xba I and Xho I, and then incubated with purified amplicons of edeB. Then, the mixture was electroshocked into Escherichia coli GB05-dir containing the recombinant system through homologous recombination. The extracted plasmid was identified and transformed into competent cells BL21(DE3) to obtain recombinant strain BL21/pET28a-edeB. The EdeB protein was purified from the culture after IPTG induction. The promoter region edePro of ede BGC was amplified by PCR using primer pairs edePro-F/edePro-R with 6-FAM at the 5’end (Supplementary Table 1). Subsequently, the labeled DNA fragments were mixed with purified EdeB protein. The binding reaction system (20 uL) consists of 10 mM Tris (pH 7.5), 5 mM MgCl2, 50 mM EDTA, 60 mM KCl, 10 mM DTT, 10% glycerol, indicated amounts of EdeB protein and purified edePro composition. Randomly interrupted salmon sperm DNA was added to the binding reaction system to prevent nonspecific competition. After incubation on ice for 10 minutes, the reactants were run on 8% of TBE polyacrylamide gel (Bio-Rad) with 0.5×TBE running buffer for 1 hour at 30 mA.
Evaluation of edeine production through HPLC–MS
Edeines produced by wild-type B. brevis X23 and its mutants were quantitatively analyzed by HPLC-MS. Briefly, HPLC-MS was performed on the HPLC system (Agilent 1260) in an ODS column (Shimadzu AQ-C18 HP, 2.1 mm×100 mm×3μm) with the following parameters: solvent A, acetonitrile; solvent B, 0.1% formic acid-water; elution procedures:5% A,10 min, 100% A, 2 min; flow rate was 0.3 mL/min; column temperature 40°C. Mass spectrometry conditions: MRM scanning mode, positive ion, molecular weight scanning range of 100 m/z. 1000 m/z, nebulizer flow rate of 3 L/min, heating gas flow rate of 10 L/min, interface temperature of 200°C, DL temperature of 200°C, heating block temperature of 350°C, and dryer flow rate of 10 L/min. The peak areas of edeine A (m/z 755.444 [M+H]+) and B (m/z 797.465 [M+H]+) were compared by HPLC-MS.
Results
edeB encodes a putative ParB family protein
The protein EdeB (Genebank accession: ATF13884.1) encoded by the edeB gene consists of 256 amino acids with a molecular weight of 30.4 kDa. Bioinformatics analyses showed that EdeB protein was a member of the ParB family proteins, and the N-terminal of EdeB contained a SnbI_like domain (Figure 1B). PROSITE analysis identified a number of functional sites in EdeB, including Protein kinase C phosphorylation, N-myristoylation, and N-glycosylation, and Casein kinase II phosphorylation sites (Figure 1C). The amino acid sequences of EdeB and three homologs of ParB family proteins from other bacterial species were aligned and reviewed using the Jalview software. The results showed that EdeB was a member of the ParB family proteins with highly conserved sequences (Figure 1D).
Disruption of edeB significantly reduced edeine biosynthesis and inhibited the antibacterial activity of B. brevis X23
In order to explore the role of edeB in edeine biosynthesis, the edeB gene was replaced by apramycin resistance gene through Red/ET homologous recombination, and the mutant was named as X23 (ΔedeB)-apra. Then the Cre enzyme plasmid was transferred into X23 (ΔedeB)-apra to finally obtain B. brevis X23(ΔedeB) strain through removing the Aprar (Figure 2A). We compared the cell growth curves of wild-type strain B. brevis X23, edeB-deletion mutant X23(ΔedeB), complementary strain X23(ΔedeB)(PAD-Apr-Px-edeB) in NB fermentation medium. The results showed that no significant difference in the cell growth curves was detected among the wild-type B. brevis X23, X23 (ΔedeB), and X23(ΔedeB)(PAD-Apr-Px-edeB). (Figure 2B).
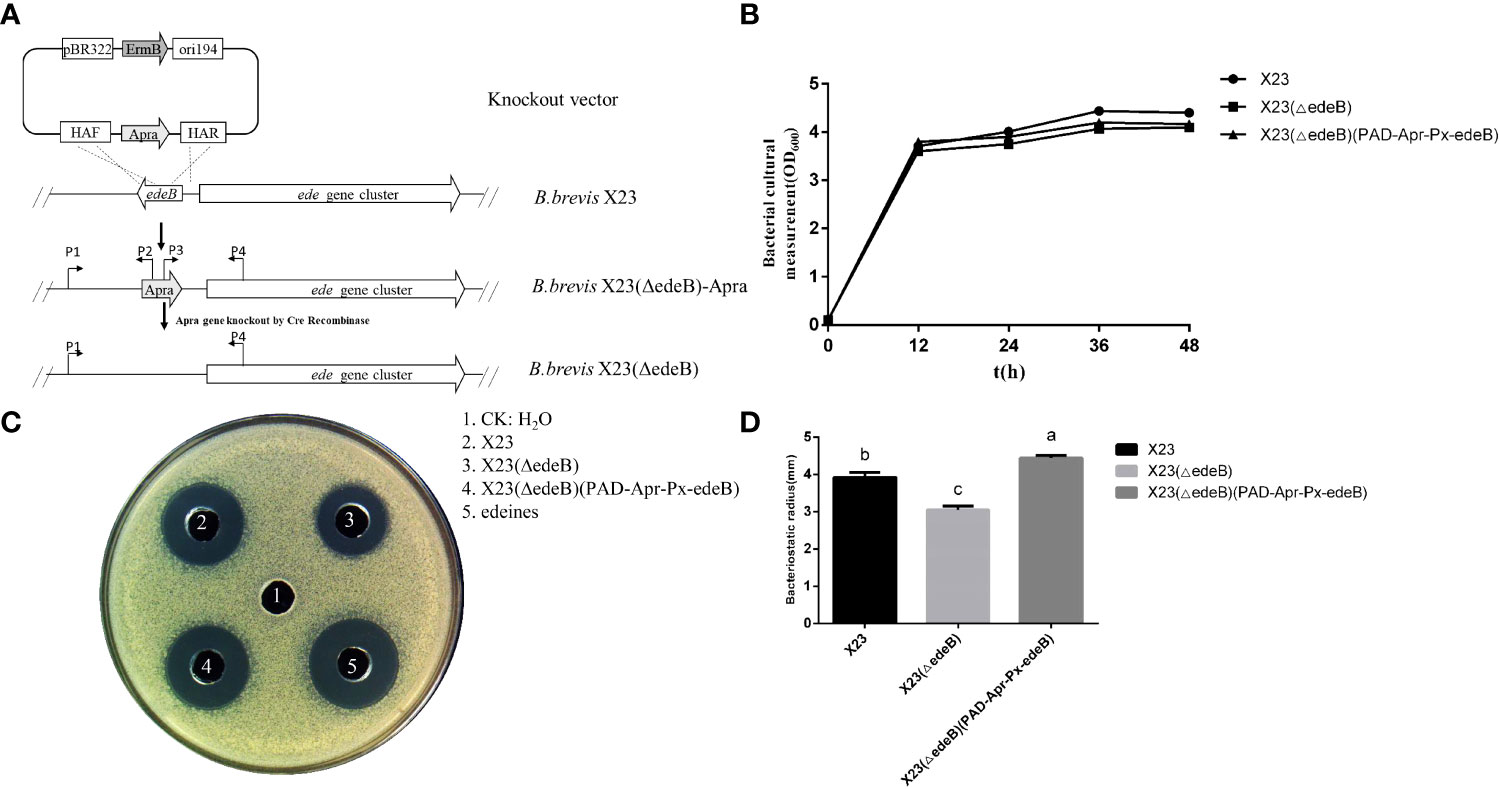
Figure 2 Comparison of antibacterial activity between edeB -deletion mutant and complementation strains. (A) edeB gene knockout flow chart. (B) Growth curve comparison. (C) Antibacterial ability comparison. (D)Comparison of radius of the inhibition zone. Different letters indicate statistically significant differences (P < 0.05).
The antibacterial activity of fermentation products was compared among wild-type B. brevis X23, X23 (ΔedeB), and X23(ΔedeB)(PAD-Apr-Px-edeB) cultured in NB medium for 48 hours. The results showed that the inhibition zone of X23 (ΔedeB) was significantly smaller than that of the wild-type B. brevis X23. The inhibition zone of the X23 (ΔedeB) (PAD-Apr-Px-edeB) was significantly larger than that of wild-type B. brevis X23 (Figures 2C, D). Taken together, edeB disruption significantly inhibited the antibacterial activity of B. brevis X23.
To further explore the effect of EdeB on edeine biosynthesis, HPLC-MS was used to compare edeine production among wild-type B. brevis strain X23, X23 (ΔedeB), and X23 (ΔedeB) (PAD-Apr-Px-edeB). The results showed that X23 (ΔedeB) still produced edeines, but the yield of edeine A and B decreased by 17.67% and 16.16%, respectively, compared with wild-type B. brevis strain X23. edeB gene complementation increased the yields of edeine A and B in X23 (ΔedeB) (PAD-Apr-Px-edeB) by 20.88% and 13.53%, respectively, compared with wild-type B. brevis X23 (Figure 3). These results confirmed that EdeB improved edeines biosynthesis, which is consistent with the above results of antibacterial activity evaluation.
Figure 3 Comparison of edeine production among wild-type B. brevis X23, X23 (ΔedeB), and X23 (ΔedeB) (PAD-Apr-Px-edeB) by HPLC-MS. (A) Comparison of edeine yield by HPLC-MS analysis. The retention time of edeine A and B was indicated by the dotted lines. (B) The mass spectrogram of edeine A and B. The molecular ions [M+H]+ were 755.444 m/z and 797.465 m/z, respectively. (C) Change of edeine A and B yield after 48 h fermentation.
Deletion of edeB gene significantly inhibited the expression of ede BGC
To study the effect of EdeB on the transcription of ede BGC, we compared the expression of genes edeA (encoding a transporter protein), edeQ (encoding a resistance enzyme), and edeK (encoding a non-ribosomal peptide synthase, NPRS) between X23(ΔedeB) and wild-type B. brevis X23. And the 16S rRNA gene with stable transcriptional expression was selected as the internal reference.
The value of the target gene measured by the ΔΔCt formula. Greater than 2.0 or less than 0.5 means that the transcription level of the target gene is significantly increased or decreased. The RT-qPCR results showed that the transcription level of edeA, edeQ, and edeK in X23(ΔedeB) significantly decreased compared with that in the wild-type B. brevis X23, suggesting that EdeB was a positive regulator in edeine biosynthesis (Figure 4).
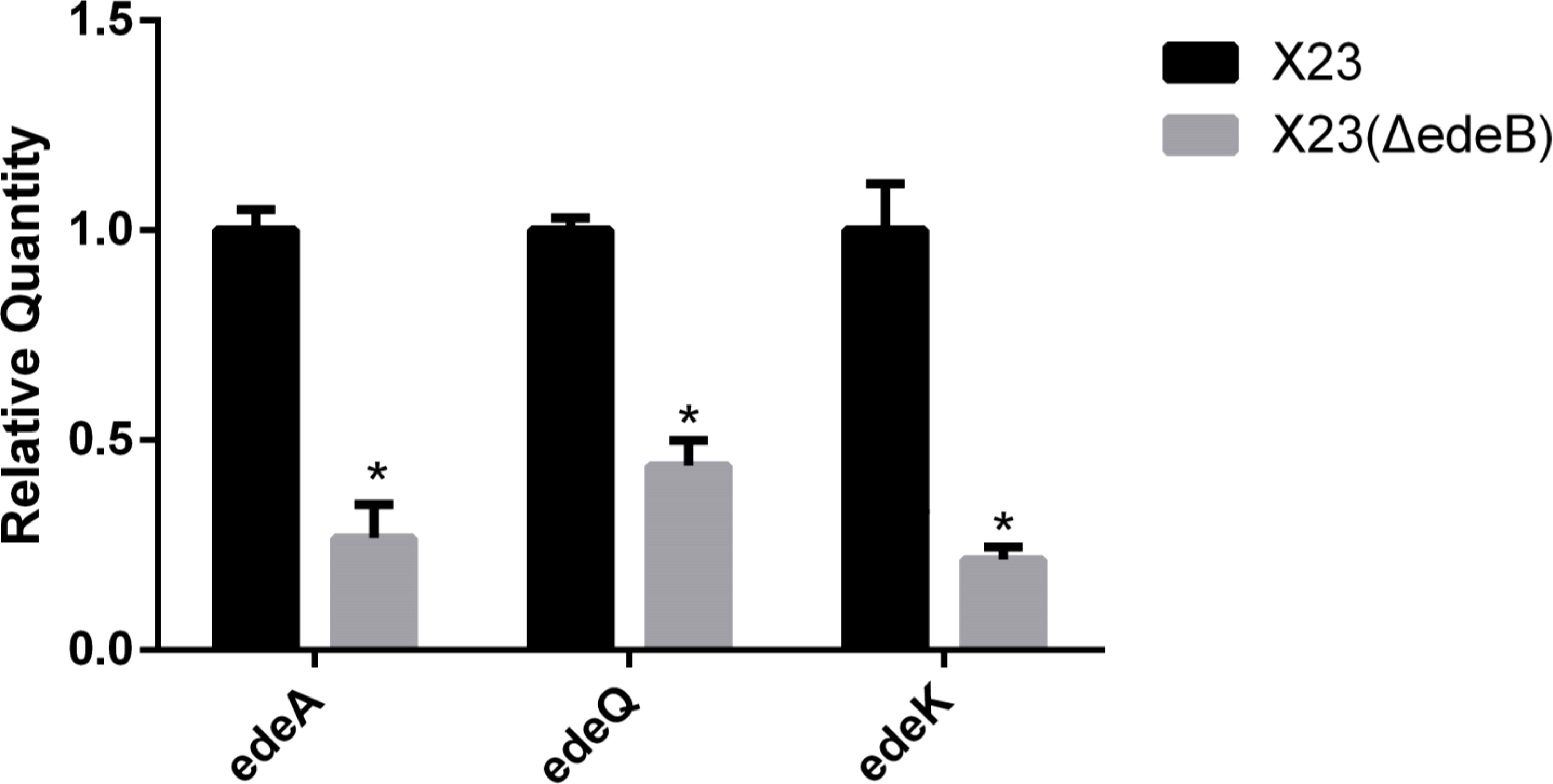
Figure 4 Effects of edeB gene knockout on the transcription levels of edeA, edeQ, and edeK. * statistically significant difference.
Bioinformatics analysis of the main elements of promoter edepro in ede BGC
The ede BGC promoter (~200 bp), namely edePro, has been previously identified and characterized (Liu et al., 2022). Bioinformatic analyses identified a -10 and -35 region in the sequence of ede BGC promoter edePro (Figure 5A). The transcriptional factor binding site (TFBS) sequences were involved in binding with transcription factors Crp (cAMP receptor protein) and arcA (Arginine deiminase). WebLogo 3.0 conservative identification program showed that the 6-nucleotide sequence in the -10 region was similar to the classical -10 region (TATAAT), and the 6-nucleotide sequence in the -35 region was identical to the classical -35 region (TTGACA) (Figure 5B).
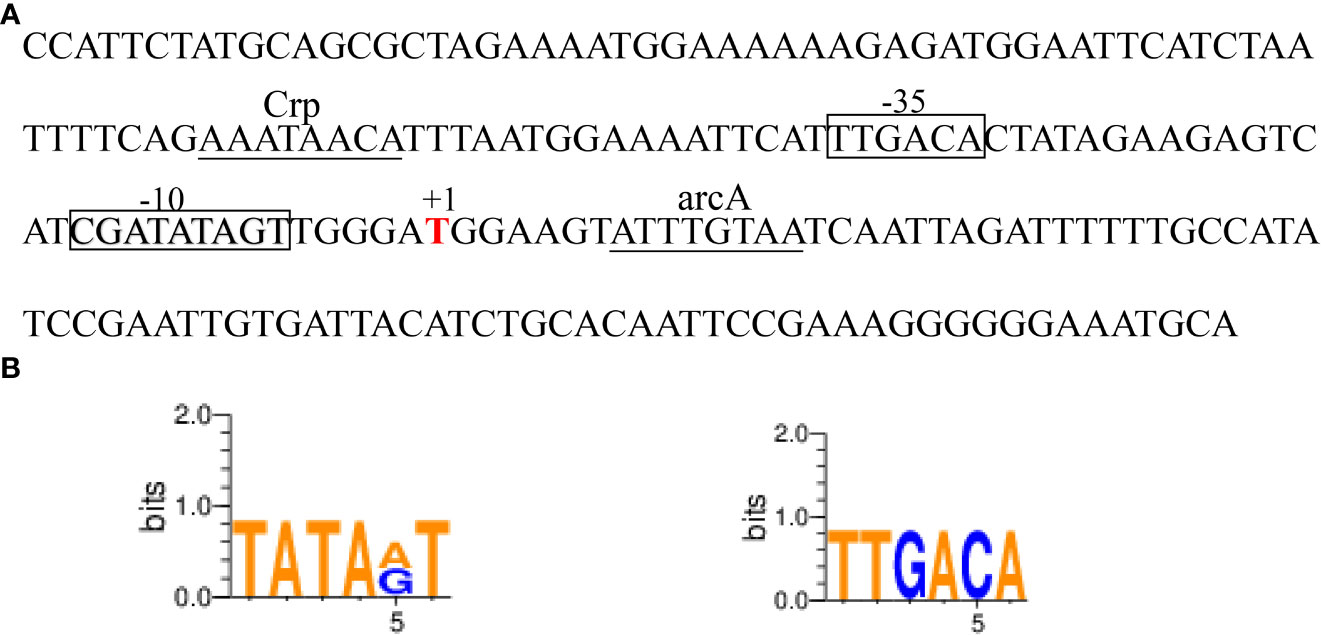
Figure 5 The edeine promoter edePro. (A) Bioinformatic analysis of edePro by BPROM and BDGP. (B) Conservative identification of -10 and-35 region in edePro.
EdeB directly regulates the promoter region of ede BGC
Transcriptional regulators usually control transcription by specifically binding to the promoter region of target genes (Leslie and Justin, 2013). In order to study whether EdeB regulates edeine biosynthesis through the same way, EMSA was used to evaluate the binding ability of EdeB with the promoter region of ede BGC. The results showed that EdeB inhibited the migration of promoter fragment edePro, suggesting that EdeB directly interact with ede BGC. The interaction between EdeB and ede BGC was further confirmed (Figure 6). Taken together, EdeB improved the transcription of ede BGC and edeine production in B. brevis by directly binding to its promoter region.
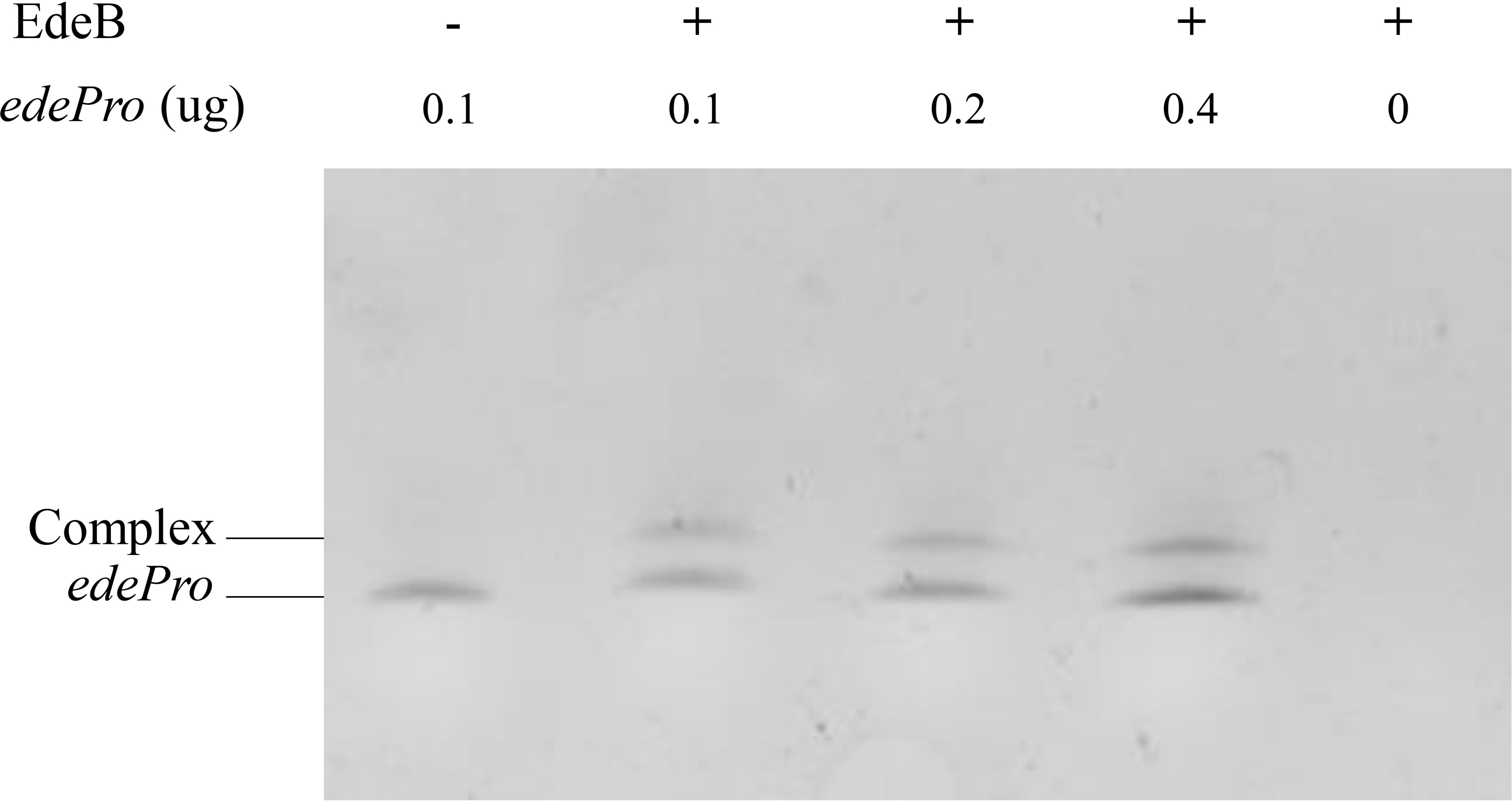
Figure 6 The binding ability of EdeB to ede BGC analyzed by EMSA. – : 0 μg EdeB protein; + : 1 μg EdeB protein.
edeB overexpression enhanced edeine production
The yield of edeine A and B in X23 (PAD-Apr-Px-edeB) was increased by 96.60% and 27.26%, respectively, by edeB gene overexpression and the total yield of edeines was increased by 92.27% (Figure 7). These results confirmed that EdeB improved edeine biosynthesis.
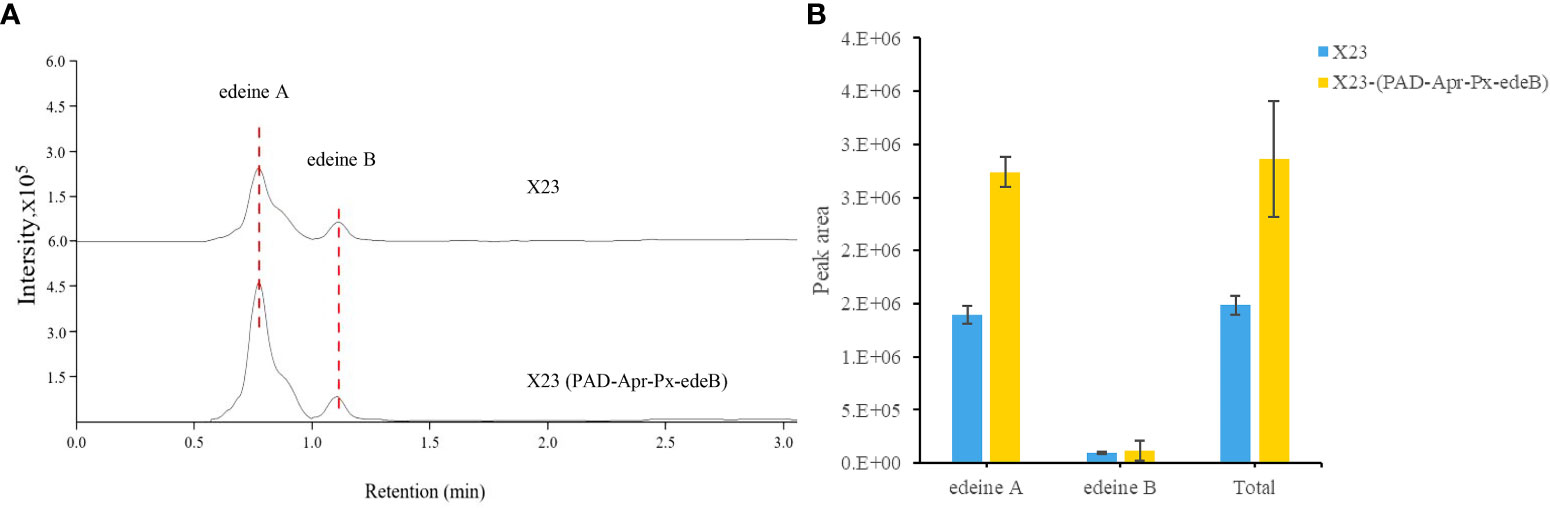
Figure 7 Edeine production in wild-type B. brevis X23 and X23 (PAD-Apr-Px-edeB). (A) Comparison of edeine yield by HPLC-MS analysis. The retention time of edeine A and B was indicated by the dotted lines. (B) Change of edeine A and B yield after 48 h fermentation.
Discussion
Edeines are natural nonribosomal peptides with antibacterial, antifungal, and antitumor activities (Johnson et al., 2020). Edeines are produced by B. brevis under appropriate condition, however, only a few reports focusing on the molecular pathway and regulatory factors involved in edeine production in B. brevis. Westman first identified the edeine biosynthetic gene cluster in B. brevis Vm4, and confirmed that edeQ was a resistance gene. Heterologous expression of edeQ enhanced resistance of edeines in Bacillus subtilis (Westman et al., 2013). In addition, Zhang et al. reported that AbrB downregulated the biosynthesis of edeines in B. brevis X23, and abrB knockout improved the yield of edeines (Zhang et al., 2020).
Currently, our understanding of the molecular pathway and regulation of edeine biosynthesis is still limited. In this study, we identified that the regulatory gene edeB on ede BGC was involved in edeine biosynthesis of B. brevis X23. Bioinformatics analysis showed that EdeB protein is a member of the ParB family, which are plasmid active distribution proteins involved in the distribution of plasmids and chromosomes, as well as embodying DNA-binding activity (Jalal et al., 2020). Recent studies have shown that many ParB family proteins functioned as transcription factors in numerous biological processes (Khare et al., 2004). In addition, EdeB protein contains a typical HTH domain, which can bind with DNA, suggesting that EdeB protein plays a regulatory role in transcription (Kusiak1 et al., 2011). While ParB family proteins are common in Bacillus, their role in the regulation of antibiotic biosynthesis has not been clearly studied (Webb et al., 2010). SbnI, a protein of the ParB superfamily, is a heme-inducible transcriptional regulator involved in Staphyloferrin B (SB) production (Laakso et al., 2016). SbnI also exhibited enzymatic function in SB biosynthesis (Skaar et al., 2004). It has been shown that Glu20 and Asp58 were required for the kinase activity of SbnI (Verstraete et al., 2018). The N-terminal of EdeB protein contains a SnbI_like domain, suggesting that it may has similar function as SnbI.
The important role of gene edeB in the edeine biosynthesis was confirmed by edeB knockout and complementation experiments. The inhibition zone of B. brevis strain with edeB knockout significantly smaller, and the antibacterial activity was significantly reduced compared with the wild-type B. brevis strain X23. Gene edeB complementation restored and improved its antibacterial ability. HPLC-MS results showed that edeB knockout reduced edeine production by 18%, but edeB complementation increased edeine production by 20.43%. Taken together, these results suggest that edeB was essential for edeine biosynthesis by upregulating edeine production.
The expression of edeA, edeQ, and edeK on ede BGC was analyzed by RT-qPCR. The results showed that the expression of these three genes were significantly reduced in edeB-deletion mutant. The genes with decreased transcription were involved in antibiotic transport, core skeleton formation, and resistance in edeine biosynthesis. Reduced transcription of these genes may be regulated by EdeB, which is consistent with decreased production of edeines in edeB-deletion mutant, suggesting that edeB is a positive regulator in the biosynthesis of edeines.
EMSA analysis showed that EdeB could directly bound to the promoter region of ede BGC. Based on the direct interaction between EdeB and edePro, and edeB deletion and complementation experiments discussed above, we propose a molecular mechanism underlying the regulatory role of EdeB in edeine biosynthesis. EdeB upregulates the expression of the genes on ede BGC to improve the production of edeines through binding to the promoter region of ede BGC. However, whether other proteins are involved in this process or interact with EdeB is still not known. In summary, EdeB is a major component in edeine biosynthesis. Overexpressing edeB can significantly improve edeine production in B. brevis, which may have wide applications in medicine and agriculture.
Conclusion
In this study, we identified and characterized a ParB family transcription factor, EdeB, which significantly increased the expression of several genes on ede BGC, and significantly improved the production of edeine A and B. In addition, our results demonstrated that EdeB directly bound to the promoter region of ede BGC to improve the transcription and translation of edeine genes. Our study elucidated the molecular mechanism underlying the biosynthesis of edeines, and can be used to improve the production of edeines in industry in the future.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number NZ_CP023474.1 and ATF13884.1.
Author contributions
JD, QLi and WC designed the study; JD, CZ, LZ and QLo conducted the experiments. JD and LZ performed data collection and analyses. JD, QLi and WC drafted the manuscript. All authors have read, edited, and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (31772216, 32000047, and 31501698), the Science and Technology Project of Hunan Province, China (2021NK1040), and High-tech Industry Science and Technology Innovation Leading Project of Hunan province, China (2020NK2006).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1022476/full#supplementary-material
References
Che, J., Bo, L., Lin, Y., Tang, W., Tang, J. (2013). Draft genome sequence of biocontrol bacterium Brevibacillus brevis strain FJAT-0809-GLX. Genome Announc. 1, e0016013. doi: 10.1128/genomeA.00160-13
Chen, C. H., Lu, T. K. (2020). Development and challenges of antimicrobial peptides for therapeutic applications. Antibiotics (Basel) 9 (1), 24. doi: 10.3390/antibiotics9010024
Cook, R. J. (1993). Making greater use of introduced microorganisms for biological control of plant pathogens. Annual review of phytopathology. Annu. Rev. Phytopathol. 31, 53–80. doi: 10.1146/annurev.phyto.31.1.53
Cozzarelli, N. R. (1977). The mechanism of action of inhibitors of DNA synthesis. Annu. Rev. Biochem. 46 (1), 641–668. doi: 10.1146/annurev.bi.46.070177.003233
Czajgucki, Z., Andruszkiewicz, R., Kamysz, W. J. (2010). Structure activity relationship studies on the antimicrobial activity of novel edeine a and d analogues. J. Pept. Sci. 12, 653–662. doi: 10.1002/psc.775
Czerwinski, A., Wojciechowskra, H., Andruszkiewicz, R., Grzybowska, J., Gumieniak, J., Borowski, E., et al. (1983). Total synthesis of edeine d. J. Antibiot (Tokyo). 36 (8), 1001–1006. doi: 10.7164/antibiotics.36.1001
Dang, Y., Zhao, F., Liu, X., Fan, X., Huang, R., Gao, W., et al. (2013). Enhanced production of antifungal lipopeptide iturin a by Bacillus amyloliquefaciens LL3 through metabolic engineering and culture conditions optimization. Microb. Cell Fact. 18, 68. doi: 10.1186/s12934-019-1121-1
Dinos, G., Wilson, D. N., Teraoka, Y., Szaflarski, W., Fucini, P., Kalpaxis, D., et al. (2004). Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: The universally conserved residues G693 and C795 regulate p-site RNA binding. Mol. Cell. 13, 113–124. doi: 10.1016/S1097-2765(04)00002-4
Hellman, L. M., Fried, M. G. (2007). Electrophoretic mobility shift assay (EMSA) for detecting protein–nucleic acid interactions. Nat. Protoc. 2, 1849–1861. doi: 10.1038/nprot.2007.249
Hettinger, T. P., Craig, L. C. (1970). Edeine. IV. structures of the antibiotic peptides edeines A1 and B1. Biochemistry. 9, 1224–1232. doi: 10.1021/bi00807a025
Hou, Q., Wang, C., Hou, X., Xia, Z., Ye, J., Liu, K., et al. (2015). Draft genome sequence of Brevibacillus brevis DZQ7, a plant growth-promoting rhizobacterium with broad-spectrum antimicrobial activity. Genome Announc. 3, e00831-15. doi: 10.1128/genomeA.00831-15
Jalal, A., Tran, N. T., Stevenson, C. E., Chan, E. W., Le, T. J. (2020). Diversification of DNA-binding specificity by permissive and specificity-switching mutations in the ParB/Noc protein family. Cell Rep. 32, 107928. doi: 10.1016/j.celrep.2020.107928
Jianmei, C., Bo, L., Zheng, C., Huai, S., Guohong, L., Cibin, G. J. (2015). Identification of ethylparaben as the antimicrobial substance produced by Brevibacillus brevis FJAT-0809-GLX. Microbiol. Res. 172, 48–56. doi: 10.1016/j.micres.2014.11.007
Johnson, E. T., Bowman, M. J., Dunlap, C. A. (2020). Brevibacillus fortis NRS-1210 produces edeines that inhibit the in vitro growth of conidia and chlamydospores of the onion pathogen fusarium oxysporum f. sp. cepae. Antonie Van Leeuwenhoek. 113 (7), 973–987. doi: 10.1007/s10482-020-01404-7
Khare, D., Ziegelin, G., Lanka, E., Heinemann, U. J. (2004). Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat. Struct. Mol. Biol. 11, 656–663. doi: 10.1038/nsmb773
Kurylo-Borowska, Z., Szer, W. J. (1973). Inhibition of bacterial DNA synthesis by edeine. effect on Escherichia coli mutants lacking DNA polymerase I. Biochim. Biophys. Acta 287, 236–245. doi: 10.1016/0005-2787(72)90373-5
Kusiak1, M., Gapczyska, A., Pochocka, D., Jagura-Burdzy, G. (2011). Binding and spreading of ParB on DNA determine its biological function n Pseudomonas aeruginosa. J. Bacteriol. 193 (13), 3342–3355. doi: 10.1128/JB.00328-11
Laakso, H. A., Marolda, C. L., Pinter, T. B., Stillman, M. J., Heinrichs, D. E. (2016). A heme-responsive regulator controls synthesis of staphyloferrin b in Staphylococcus aureus. J. Biol. Chem. 291, 29–40. doi: 10.1074/jbc.M115.69662
Lan, H. E., Wang, L., Zhu, Q., Qiao, J., Wang, W. J. (2017). Comparison of two methods for drawing the growth curve of Bacillus subtilis and Escherichia coli. Tianjin Agric. Sci. 23, 14–18. doi: 10.3969/j.issn.1006-6500.2017.05.004
Leslie, C., Justin, R. (2013). The TetR family of regulators. Microbiol. Mol. Biol. Rev. 77 (3), 440–475. doi: 10.1128/MMBR.00018-13
Liu, Q., Shen, Q., Bian, X., Chen, H., Fu, J., Wang, H., et al. (2016). Simple and rapid direct cloning and heterologous expression of natural product biosynthetic gene cluster in Bacillus subtilis via Red/ET recombineering. Sci. Rep. 6, 34623. doi: 10.1038/srep34623
Liu, Q., Zhang, L., Wang, Y., Zhang, C., Liu, T., Duan, C., et al. (2022). Enhancement of edeine production in brevibacillus brevis X23 via in situ promoter engineering. Microb. Biotechnol. 15 (2), 577–589. doi: 10.1111/1751-7915.13825
Livak, K. J., Schmittgen, T. D. (2013). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Mcorist, A. L., Jackson, M., Bird, A. R. (2002). A comparison of five methods for extraction of bacterial DNA from human faecal samples. J. Microbiol. Methods 50, 131–139. doi: 10.1016/s0167-7012(02)00018-0
Muyrers, J., Zhang, Y., Stewart, A. F. (2001). Techniques: Recombinogenic engineering–new options for cloning and manipulating DNA. Trends Biochem. Sci. 26, 325–331. doi: 10.1016/s0968-0004(00)01757-6
Saxena, A. K., Kumar, M., Chakdar, H., Anuroopa, N., Bagyaraj, D. J. (2020). Bacillus species in soil as a natural resource for plant health and nutrition. J. Appl. Microbiol. 128, 1583–1594. doi: 10.1111/jam.14506
Semenchenko, A., Oliveira, G., Atman, A. P. (2016). Hybrid agent-based model for quantitative in-silico cell-free protein synthesis. Biosystems. 150, 22–34. doi: 10.1016/j.biosystems.2016.07.008
Sharan, S. K., Thomason, L. C., Kuznetsov, S. G., Court, D. L. (2009). Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206–223. doi: 10.1038/nprot.2008.227
Shimotohno, K. W., Kawamura, F., Natori, Y., Nanamiya, H., Magae, J., Ogata, H., et al. (2010). Inhibition of septation in Bacillus subtilis by a peptide antibiotic, edeine B(1). Biol. Pharm. Bull. 33, 568–571. doi: 10.1248/bpb.33.568
Skaar, E. P., Humayun, M., Bae, T., Debord, K. L., Schneewind, O. J. (2004). Iron-source preference of Staphylococcus aureus infections. Science. 305, 1626–1628. doi: 10.1126/science.1099930
Szer, W., Kurylo-Borowska (1972). Interactions of edeine with bacterial ribosomal subunits. selective inhibition of aminoacyl-tRNA binding sites. Biochim. Biophys. Acta 259, 357–368. doi: 10.1016/0005-2787(72)90310-3
Verstraete, M., Perez-Borrajero, M., Brown, C., Heinrichs, D. E, Murphy, M. M. (2018). SbnI is a free serine kinase that generates O-phospho-L-serine for staphyloferrin b biosynthesis in Staphylococcus aureus. J. Biol. Chem. 93, 6147–6160. doi: 10.1074/jbc.RA118.001875
Webb, C. D., Graumann, P. L., Kahana, J. A., Teleman, A. A., Losick, R. (2010). Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol. Microbiol. 28, 883–892. doi: 10.1046/j.1365-2958.1998.00808.x
Westman, E., Yan, M., Waglechner, N., Koteva, K., Wright, G. (2013). Self resistance to the atypical cationic antimicrobial peptide edeine of Brevibacillus brevis Vm4 by the n-acetyltransferase EdeQ. Chem. Biol. 20, 983–990. doi: 10.1016/j.chembiol.2013.06.010
Wu, C., Yunsheng, W., Dingjun, L., Lin, L., Qiming, X., Qingming, Z., et al. (2012). Draft genome sequence of brevibacillus brevis strain X23, a biocontrol agent against bacterial wilt. J. Bacteriol. 194, 6634–66355. doi: 10.1128/JB.01312-12
Yang, X., Yousef, A. E. (2018). Antimicrobial peptides produced by brevibacillus spp.: Structure, classification and bioactivity: A mini review. World J. Microbiol. Biotechnol. 34, 57. doi: 10.1007/s11274-018-2437-4
Yu, D., Ellis, H. M., Lee, E. C., Jenkins, N.A., Copeland, N.G., Court, D.L., et al. (2000). An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U S A. 97, 5978–5983. doi: 10.1073/pnas.100127597
Zhang, L., Lv, C., Duan, C., Wang, Y., Huang, J., Du, J., et al. (2020). Effect of transcription factor AbrB on the biosynthesis of antibiotic edeines in biocontrol bacteria Brevibacillus brevis X23. Chin. J. Biol. Control. 36, 564–567. doi: 10.16409/j.cnki.2095-039x.2020.04.017
Keywords: Brevibacillus brevis, edeines, ParB protein family, EdeB, gene overexpression
Citation: Du J, Zhang C, Long Q, Zhang L, Chen W and Liu Q (2022) Characterization of a pathway−specific activator of edeine biosynthesis and improved edeine production by its overexpression in Brevibacillus brevis. Front. Plant Sci. 13:1022476. doi: 10.3389/fpls.2022.1022476
Received: 18 August 2022; Accepted: 27 September 2022;
Published: 25 October 2022.
Edited by:
Yonghua Zhu, Hunan University, ChinaReviewed by:
Dixit Sharma, Central University of Himachal Pradesh, IndiaYan Li, Institute of Vegetables and Flowers (CAAS), China
Fu Yan, Shandong University, China
Jiangshan Ma, Central South University Forestry and Technology, China
Copyright © 2022 Du, Zhang, Long, Zhang, Chen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingshu Liu, bGl1cWluZ3NodTIwMTJAMTI2LmNvbQ==; Wu Chen, Y2hlbnd1d2FycmlvckAxNjMuY29t
 Jie Du
Jie Du Cuiyang Zhang1
Cuiyang Zhang1 Qingshu Liu
Qingshu Liu