
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 06 September 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1005627
This article is part of the Research Topic Genomics and disease resistance in wheat and maize View all 16 articles
 Yanjun Mu1†
Yanjun Mu1† Wenping Gong2†
Wenping Gong2† Yanmin Qie3†
Yanmin Qie3† Xueqing Liu4
Xueqing Liu4 Linzhi Li4
Linzhi Li4 Nina Sun4
Nina Sun4 Wei Liu4
Wei Liu4 Jun Guo2
Jun Guo2 Ran Han2
Ran Han2 Ziyang Yu1
Ziyang Yu1 Luning Xiao1
Luning Xiao1 Fuyu Su1
Fuyu Su1 Wenjing Zhang1
Wenjing Zhang1 Jiangchun Wang4*
Jiangchun Wang4* Guohao Han5*
Guohao Han5* Pengtao Ma1*
Pengtao Ma1*Powdery mildew of wheat (Triticum aestivum), caused by Blumeria graminis f.sp. tritici (Bgt), is a destructive disease that seriously threatens the yield and quality of its host. Identifying resistance genes is the most attractive and effective strategy for developing disease-resistant cultivars and controlling this disease. In this study, a wheat breeding line Yannong 99102-06188 (YN99102), an elite derivative line from the same breeding process as the famous wheat cultivar Yannong 999, showed high resistance to powdery mildew at the whole growth stages. Genetic analysis was carried out using Bgt isolate E09 and a population of YN99102 crossed with a susceptible parent Jinhe 13–205 (JH13–205). The result indicated that a single recessive gene, tentatively designated pmYN99102, conferred seedling resistance to the Bgt isolate E09. Using bulked segregant exome capture sequencing (BSE-Seq), pmYN99102 was physically located to a ~33.7 Mb (691.0–724.7 Mb) interval on the chromosome arm 2BL, and this interval was further locked in a 1.5 cM genetic interval using molecular markers, which was aligned to a 9.0 Mb physical interval (699.2–708.2 Mb). Based on the analysis of physical location, origin, resistant spectrum, and inherited pattern, pmYN99102 differed from those of the reported powdery mildew (Pm) resistance genes on 2BL, suggesting pmYN99102 is most likely a new Pm gene/allele in the targeted interval. To transfer pmYN99102 to different genetic backgrounds using marker-assisted selection (MAS), 18 closely linked markers were tested for their availability in different genetic backgrounds for MAS, and all markers expect for YTU103-97 can be used in MAS for tracking pmYN99102 when it transferred into those susceptible cultivars.
Common wheat (Triticum aestivum) is one of the most important crops to food security for it provides ~20% of the calories consumed by humans (Isham et al., 2021). With an estimated global population of more than nine billion by 2050, wheat production is needed an ~70% growth to meet the food demands (International Wheat Genome Sequencing Consortium (IWGSC), 2014). However, powdery mildew, a global devastating wheat disease caused by Blumeria graminis f. sp. tritici (Bgt), can significantly reduce wheat yield and affect flour quality (Li et al., 2019; Wang et al., 2021). It typically decreases wheat yield by 10–15% and up to 62% in severe cases (Singh et al., 2016).
To control this disease, utilization of host resistance is regarded as the most effective, and environmentally friendly way (Chen, 2013; Ma et al., 2015). Nevertheless, it cannot be ignored that Bgt isolates have complex and highly variable virulence structures, so their frequent evolution will lead to the continuous breakdown of resistance genes, particularly in the areas where race-specific resistance genes were widely used. A well-known example was the “boom-bust” of Pm8, which led to severe epidemics after extended periods of use in the main wheat production regions of China (He et al., 2011, 2015; An et al., 2019). On the other hand, despite more than 80 formally designated Pm genes (Pm1-Pm68, noting that Pm8=Pm17, Pm18=Pm1c, Pm22=Pm1e, Pm23=Pm4c, and Pm31=Pm21) having been reported (Li H. H. et al., 2020; McIntosh et al., 2020; He et al., 2021), most of them cannot be directly applied in wheat production due to undesirable linkage drag, such as a broad-spectrum gene Pm16 which caused up to 15% yield loss when introduced into wheat backgrounds (Summers and Brown, 2013; Tan et al., 2018). In the modern wheat breeding programs of China, only a few Pm genes including Pm2, Pm4, Pm5, Pm8, and Pm21 have been extensively applied in wheat improvement (Jia et al., 2020; Jin et al., 2021), making them face huge selective pressure. Therefore, unceasingly exploring and utilizing the novel Pm genes/alleles that could balance the broad resistance and comprehensive agronomic performance is an ongoing and essential process.
Once the effective gene was identified, its accurate and rapid transfer or pyramiding is the key point in breeding practice. In comparison to conventional breeding based on phenotypic selection, marker-assisted selection (MAS) is more effective because it combines both genotypic and phenotypic identification. Using the tightly linked or diagnostic markers, the targeted genes could be selected or excluded in fewer generations and thus promote the breeding process (Jiang et al., 2016). Therefore, the isolation of target genes/loci and the development of their tightly linked markers are the two key factors for MAS. Recent advances in the whole-genome sequencing of wheat and corresponding high-throughput sequencing techniques have significantly accelerated the identification and isolation of the resistance genes (Zhu et al., 2020; Ma et al., 2021). A newly-developed strategy bulked segregant exome capture sequencing (BSE-Seq), which combines bulked segregant analysis (BSA) and the exome sequence strategy, has great potential to accelerate gene mapping, particularly in polyploid species with large and complex genome properties such as common wheat. BSE-Seq could effectively identify the linked interval which is not limited to the multiple gene copies, high similarity among the homoeologs, and various types of mapping segregant populations. More importantly, most of the variations obtained from BSE-Seq are existed in the coding regions, making it an economical but effective method for constructing linkage maps and also analyzing the differentially expressed genes associated with the targeted traits (Dong et al., 2020).
Wheat genotypes carrying high-resistance genes but with poor agronomic performance will be greatly limited in breeding because of multigeneration of backcrossing needed, which is not preferred by breeders (Summers and Brown, 2013; Yu et al., 2022). Genes identified in wheat cultivars/breeding lines can be more easily applied in breeding practice compared with those originated from wheat relatives or landraces (Xu et al., 2015). In this case, wheat breeding lines are of high breeding priority as the donor to improve powdery mildew resistance. Yannong 99102-06188 (YN99102), is an elite wheat breeding line developed by the Yantai Academy of Agricultural Sciences (Yantai, China). It exhibited both high resistance to powdery mildew and elite agronomic traits for consecutive years of observation in the field. To better clarify and use the powdery mildew resistance in YN99102, the objectives of this study were to (i) assess the powdery mildew resistance of YN99102 and determine its inheritance; (ii) rapidly map the Pm gene(s) using BSE-Seq; (iii) evaluate and develop the tightly linked markers suitable for MAS.
Wheat breeding line YN99102, a derivative line from the same breeding process as the elite wheat cultivar Yannong 999, was derived from the multiple crosses of Lumai 14 and Lin 9,511 through space mutation breeding (Supplementary Figure 1). YN99102 showed high resistance to powdery mildew at both seedling and adult plant stages. To determine the genetic analysis and map the Pm gene(s) at the seedling stage in YN99102, the wheat line Jinhe 13–205 (JH13–205) was used as the susceptible parent to cross with YN99102 to generate F1 hybrids, F2 populations, and F2:3 families. Wheat cultivar Mingxian 169 without any known Pm gene, was used as the susceptible control for phenotypic evaluation and served as the Bgt inoculum spreader. Eight wheat genotypes with known Pm genes on chromosome 2BL, Coker 747 (with Pm6) (Wan et al., 2020), Am9/3 (with Pm33) (Zhu et al., 2005), CH7086 (with Pm51) (Zhan et al., 2014), Liangxing 99 (with Pm52) (Zhao et al., 2013), WE35 (with Pm64) (Zhan et al., 2014), LS5082 (with PmLS5082) (Wu et al., 2019), KN0816 (with PmKN0816) (Wang et al., 2021) and Qingxinmai (with PmQ) (Li Y. H. et al., 2020) were tested with different Bgt isolates in order to compare their reactions with that of YN99102 (Table 1). Forty-two susceptible wheat cultivars from different regions of China were used to evaluate the usefulness of the closely linked markers for MAS of the Pm gene(s) in YN99102 (Supplementary Table 1).
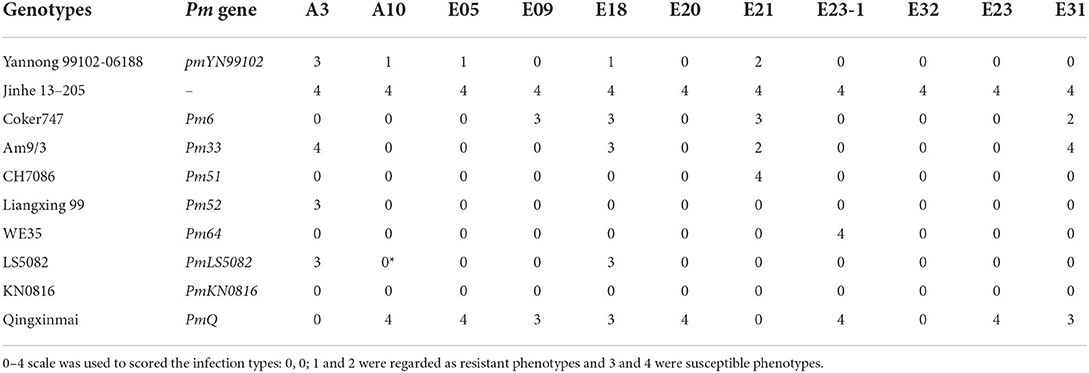
Table 1. Comparative responses of Yannong 99102-06188 and wheat genotypes with known powdery mildew resistance genes on chromosome arm 2BL to 11 isolates of Blumeria graminis f. sp. tritici (Bgt) with different virulence.
At the adult stage, YN99102 was inoculated with a mixture of 11 Bgt isolates including A3, A10, E05, E09, E18, E20, E21, E23–1, E32, E23, and E31 in the field nurseries with three replicates. The assessments were performed from 2018 to 2021 at Yantai University, Yantai City, Shandong Province, China (121.39'E, 37.52'N). For each replicate, YN99102 was planted with 30 seeds per row in four 1.2-m rows, with Mingxian 169 planted on each side of YN99102 as susceptible control and inoculum spreader. When Mingxian 169 showed severe disease symptoms, the disease reaction of YN99102 was assessed using a 0–9 scale for infection types (ITs), in which 0–4 were considered resistant and 5–9 were susceptible (Sheng and Duan, 1991). Each plant was assessed twice for confirmation.
To determine the inheritance of the powdery mildew resistance in YN99102 at the seedling stage, the Bgt isolate E09, which is prevalent in the main wheat producing regions of North China (Zhou et al., 2005), was used to inoculate YN99102, susceptible parent JH13–205, and their F1 hybrids, F2 population, and F2:3 families at the one-leaf stage. Each of the F2:3 families was tested with 30 seeds to confirm the phenotypic reaction of the F2 plants. The resistance assessment was carried out in a greenhouse in a high humidity environment with a daily cycle of 14 h of light at 22°C and 10 h of darkness at 18°C. The tested seeds were planted in rectangular trays (54 × 28 × 4.2 cm) with 128 wells (3.2 × 3.2 × 4.2 cm) and then inoculated at one leaf stage by dusting the fresh conidia of Bgt isolate E09, and Mingxian 169 was planted randomly in the trays as the susceptible control. When the pustules were fully developed on the first leaf of Mingxian 169 about 14–15 days after inoculation, each plant was assessed on a 0–4 scale, plants with ITs 0–2 were regarded as resistant and those with ITs 3 and 4 as susceptible (Sheng, 1988; Wang et al., 2005). Goodness-of-fit was analyzed using the chi-square (χ2) test to investigate deviations of the observed phenotypic data of F2 populations and F2:3 families from theoretically expected segregation ratios.
BSE-Seq was used to rapidly located Pm gene(s) in the targeted interval by Oebiotech (Shanghai, China). After the susceptible control Mingxian 169 showed serious powdery mildew symptoms on the first leaf, equal leaf tissues from 30 homozygous resistant and 30 homozygous susceptible F2:3 plants of YN99102 × JH13–205, respectively, were randomly collected to construct resistant and susceptible bulks. These two DNA bulks were subjected to exome capture sequencing with deep coverage (~70 ×). The construction, assessment, and sequencing of the libraries were performed as described by (Dong et al., 2020).
Raw sequence reads were filtered using Fastp (v0.12.4) to remove the low-quality reads and adapters used. The high-quality reads were then aligned to IWGSC RefSeq v1.0 genome. After that, raw cohort vcf was worked out with GATK (v4.0.10.1) (McCormick et al., 2015). The minimum-mapping-quality parameter was set as 30 for only high-quality alignment reads used to call variants. SNP calling and density analysis were carried out using sliding window calculation based on the reference of Takagi et al. (2013) The data filtering parameters were set as AF (Allele Frequence) <0.3 or >0.7. Bcftools (v1.9) (Narasimhan et al., 2016) was performed for variants quality filtering with “QUAL > 30” and “DP ≥5.” The statistical model varBScore was carried out to determine the candidate interval. SnpEff (v4.3T) (Cingolani et al., 2012) was used to generate customized databases containing IWGSC v1.1 HC/LC genes for the annotation of the variants.
Based on the candidate interval obtained from BSE-Seq, 98 molecular markers linked to the known Pm genes in the candidate interval were firstly used to test for polymorphisms between resistant and susceptible parents and bulks (Table 2). Then, the polymorphic markers between the parents and the bulks were used to genotype the F2:3 families of YN99102 × JH13–205 for a preliminary mapping of the Pm gene(s) in YN99102. Moreover, 70 new markers in the target interval were developed based on the simple sequence repeat (SSR) and small insertion-deletion (InDel) that were discovered by BSE-Seq (Supplementary Table 2).
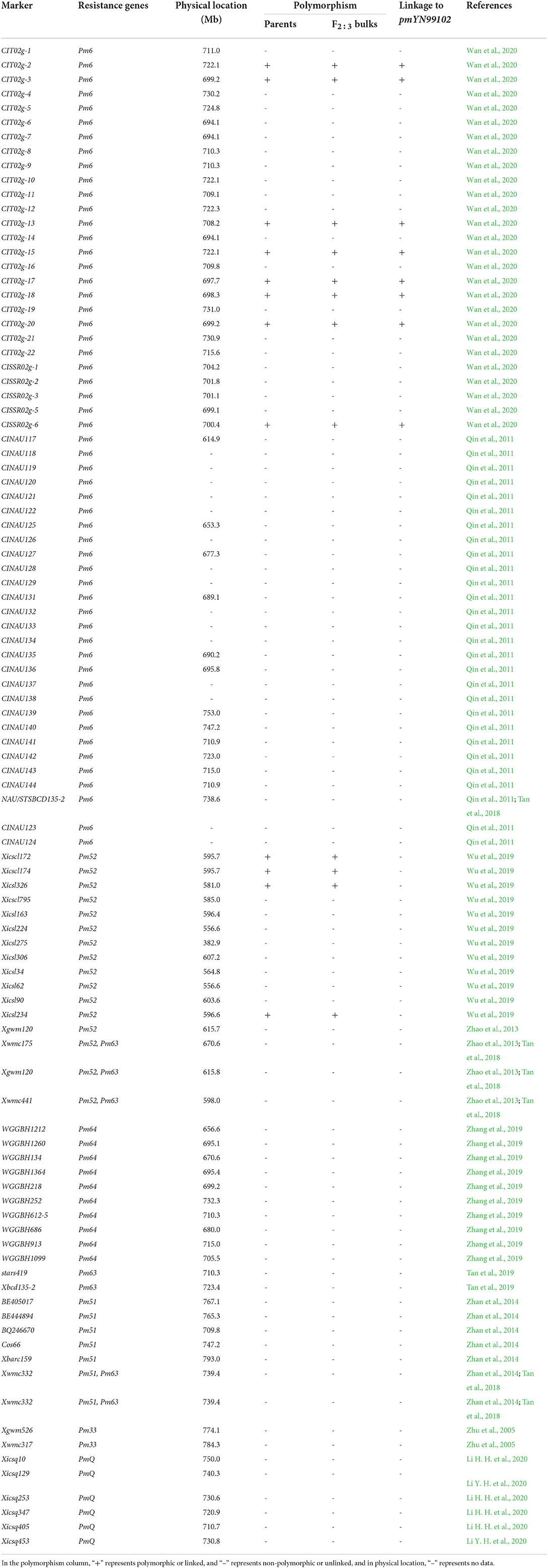
Table 2. Polymorphic and linkage analyses of the markers linked to the powdery mildew resistance genes located on chromosome arm 2BL using the mapping population derived from the cross of Yannong 99102-06188 × Jinhe 13–205.
PCR amplification was performed as described by Han et al. (2022a,b) with minor modification. The PCR products were then separated in 8% non-denaturing polyacrylamide gels with a 29:1 ratio of acrylamide and bisacrylamide, finally visualized by silver staining.
After obtaining phenotyping data from the evaluation of disease resistance and the genotyping data of the F2:3 families from molecular marker analysis, the linkage map of the Pm gene in YN99102 was constructed using MAPMAKER 3.0 (Lincoln et al., 1992) and the Kosambi function as reported previously (Kosambi, 1944). Functional annotation was performed based on the information from IWGSC RefSeq [version 1.0; The International Wheat Genome Sequencing Consortium (IWGSC, 2018)].
Considering that the Pm gene in YN99102 was assigned to the chromosome 2BL, YN99102 and eight wheat genotypes also carrying known Pm genes on chromosome 2BL, including Coker 747 (with Pm6), Am9/3 (with Pm33), CH7086 (with Pm51), Liangxing 99 (with Pm52), WE35 (with Pm64), LS5082 (with PmLS5082), KN0816 (with PmKN0816) and Qingxinmai (with PmQ), were tested against 11 Bgt isolates that were collected from the diseased wheat fields in different wheat growing areas of China to compare their resistance spectrum (Table 1). Each isolate was developed through single-spore purification and separately stored in glass tubes with three layers of gauzes. The methods of inoculation and incubated conditions were described previously (Wu et al., 2019).
To further distinguish the pmYN99102 from the documented Pm genes on chromosome arm 2BL at the level of genetic diversity, 98 markers closely linked to those Pm genes were tested for polymorphisms between resistant and susceptible parents and bulks derived from the F2:3 families of YN99102 × JH13–205 to investigate the genetic diversity of the candidate interval of Pm gene in YN99102 and the known Pm genes in chromosome arm 2BL (Table 2).
To evaluate the applicability of the markers for MAS breeding, 42 susceptible wheat cultivars from different regions of China were tested with the closely linked or co-segregated markers. The markers which were able to consistently amplify polymorphic band(s) between YN99102 and these susceptible cultivars were regarded as effective for MAS in those genetic backgrounds (Supplementary Table 1). To transfer the Pm gene(s) in YN99102 to applicable backgrounds, these cultivars were crossed with YN99102 to construct BC1F2 and F3 segregation populations for MAS.
For the adult plant investigations with powdery mildew in the field, YN99102 showed high resistance with ITs 0–1 to the Bgt mixture including Bgt isolates A3, A10, E05, E09, E18, E20, E21, E23–1, E32, E23 and E31 over the consecutive growing seasons from 2018 to 2021.
Then, the Bgt isolate E09 was used to determine the inheritance of powdery mildew resistance in YN99102 at the seedling stage. When inoculated with this isolate, YN99102 was highly resistant with IT 0, whereas JH13–205 was highly susceptible with IT 4. All the 10 F1 plants of the cross YN99102 × JH13–205 were susceptible with IT 4, indicating the resistance of YN99102 to Bgt isolate E09 was controlled by recessive Pm gene(s). The F2 populations segregated in 31 resistant plants scored as IT 0, and 77 susceptible plants scored as IT 4, which fits a theoretical ratio of 1:3 for the monogenic segregation (χ2 = 0.79; P = 0.37). Subsequently, all the 108 F2 plants were transplanted in the field to generate F2:3 families for the confirmation of the homozygous or heterozygous genotype of the susceptible F2 plants. The 108 F2:3 families segregated with 31 homozygous resistant (rr), 52 segregating (Rr), and 25 homozygous susceptible (RR), and the phenotypic result of F2:3 families further confirmed the ratio of monogenic inheritance of the powdery mildew resistance 1:2:1 (χ2 = 0.48; P = 0.49) (Table 3). Therefore, it suggested that the resistance to Bgt isolate E09 in YN99102 was controlled by a single recessive gene, tentatively designated as pmYN99102.

Table 3. Segregation ratios of F2 and F2:3 generations of Yannong 99102-06188 (YN) and Jinhe 13-205 (JH) following inoculation with Blumeria graminis f. sp. tritici (Bgt) isolate E09 at the seedling stage.
To confirm the genetic position of pmYN99102, the resistant and susceptible DNA bulks were genotyped using BSE-Seq. Based on the results of BSE-Seq, a total of 32,711 high-quality SNPs were identified between the resistant and susceptible bulks by ΔSNP index analysis, which was distributed on all of the wheat chromosomes (Figure 1). Among them, 10,731 (32.8%) SNPs were detected on chromosome arm 2B, 2,978 (27.8%) SNPs enriched on chromosome arm 2BL 691.0–724.7 Mb with varBScore analysis (Figure 2), indicating that the pmYN99102 was likely located in this 33.7 Mb interval on chromosome arm 2BL.
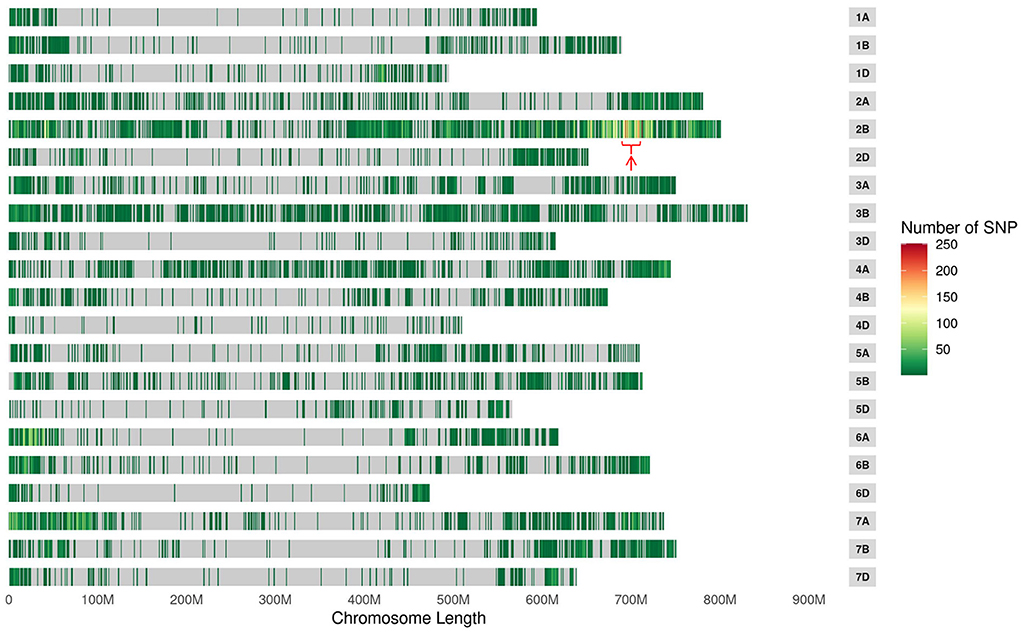
Figure 1. The distribution of single nucleotide polymorphisms (SNPs) and significant candidate intervals (indicated with red arrow) across the 21 wheat chromosomes.
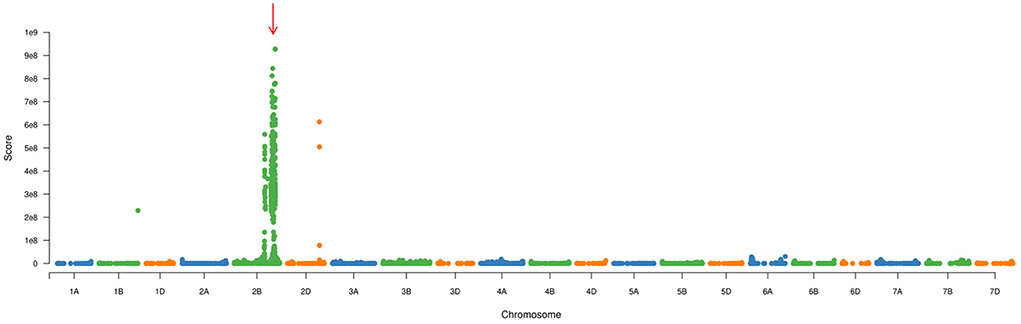
Figure 2. Manhattan plot of the varBScore across the 21 wheat chromosomes for analyzing the candidate region (indicated with red arrow).
With the candidate interval confirmed, 98 previously reported markers linked to the candidate interval on 2BL (Table 2) and 70 newly developed markers based on the results of BSE-Seq (Supplementary Table 2) were used to screen polymorphism between the resistant and susceptible parents and bulks. Among them, 19 markers, including 10 newly developed markers (Table 4), amplified consistent polymorphisms between the resistant and susceptible parents and bulks, which were used to genotype the F2:3 families of the cross of YN99102 × JH13–205 to construct the linkage map and further narrow down the candidate interval of pmYN99102 (Figure 3). The result showed that pmYN99102 was flanked by markers CIT02g-3/CIT02g-20 and CIT02g-13/CIT02g-2/CIT02g-15 with genetic distances of 0.5 and 1.0 cM, corresponding to 699.2–708.2 Mb physical interval, and co-segregated with CISSR02g-6 (700.4 Mb) according to the IWGSC Chinese Spring reference genome v1.0 (Figure 4). In this interval, we obtained a total of 76 high confidence genes based on the gene annotation results. Among them, four genes TraesCS2B01G505200, TraesCS2B01G507000, TraesCS2B01G507100 and TraesCS2B01G509000 were related to disease resistance and were regarded as the candidate genes of pmYN99102 (Supplementary Table 3).
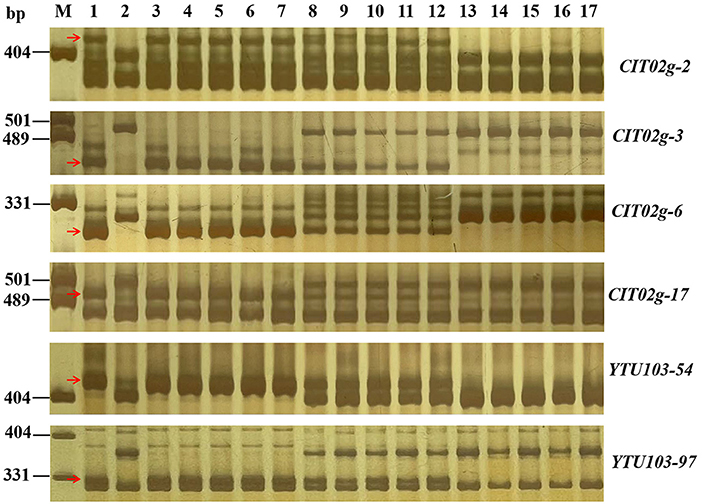
Figure 3. Amplification patterns of pmYN99102-linked markers CIT02g-2 (A), CIT02g-3 (B), CIT02g-6 (C), CIT02g-17 (D), CIT02g-54 (E) and YTU103-97 in genotyping resistant parent Yannong 99102-06188 (YN99102), susceptible parent Jinhe13–205 (JH13–205) and randomly selected F2:3 families of YN99102 × JH13–205. Lane M, pUC18 Msp I; lanes 1–2, YN99102 and JH13–205; lanes 3–7, homozygously resistant F2:3 families; lanes 8–12, heterozygously F2:3 families; lanes 13–17, homozygously susceptible F2:3 families. The red arrows were used to indicate the polymorphic bands linked to pmYN99102.
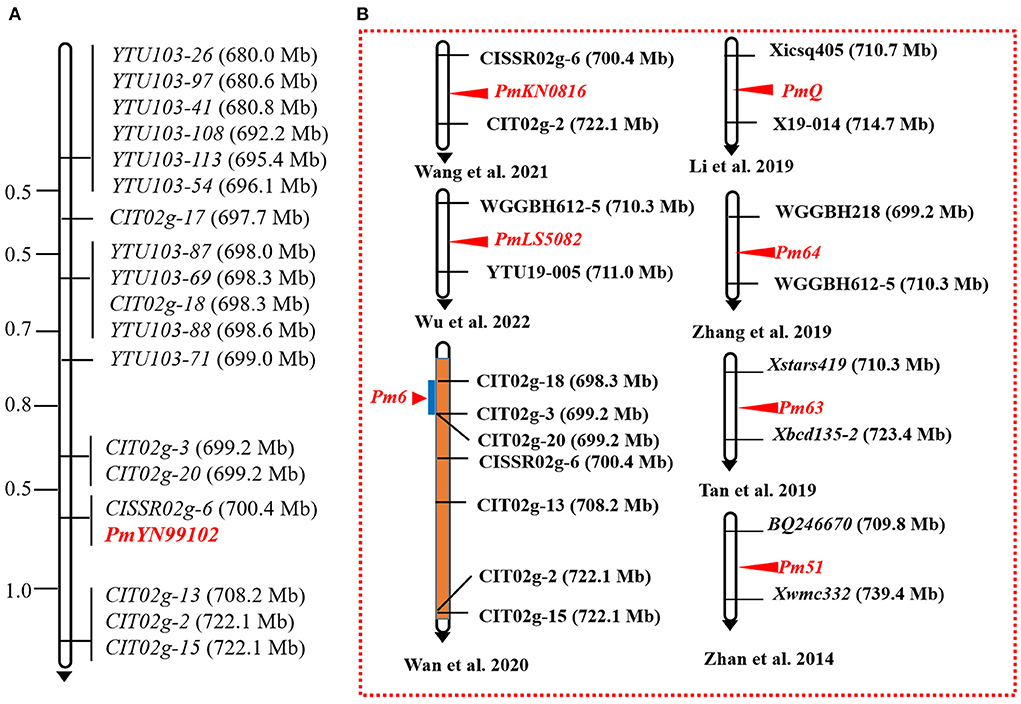
Figure 4. Linkage map of pmYN99102 using the F2:3 families of Yannong 99102-06188 × Jinhe13–205 (A) and the physical locations of cataloged Pm genes on chromosome arm 2BL (B). Genetic distances in cM are showed to the left. The black arrows point to the centromere.
To identify the relationship between pmYN99102 and the known Pm genes on chromosome arm 2BL, YN99102 (with pmYN99102), Coker747 (with Pm6), Am9/3 (with Pm33), CH7086 (with Pm51), Liangxing99 (with Pm52), WE35 (with Pm64), LS5082 (with PmLS5082), KN0816 (with PmKN0816) and Qingxinmai (with PmQ) were tested against 11 Bgt isolates to evaluate their resistance spectrum. The results showed that YN99102 was resistant to 10 of 11 (90.9%) isolates, while Coker747, Am9/3, CH7086, Liangxing99, WE35, LS5082, KN0816 and Qingxinmai was resistant to 8 of 11 (72.7%), 8 of 11 (72.7%), 10 of 11 (90.9%), 10 of 11 (90.9%), 10 of 11 (90.9%), 9 of 11 (81.8%), 11 of 11 (100%), 3 of 11 (27.2%), respectively (Figure 5 ; Table 1). Even though YN99102, CH7086, and Liangxing 99 were all resistant to 10 out of 11, they showed significantly different phenotypes against different isolates. Therefore, YN99102 had different resistance spectrum from the known Pm genes on 2BL.
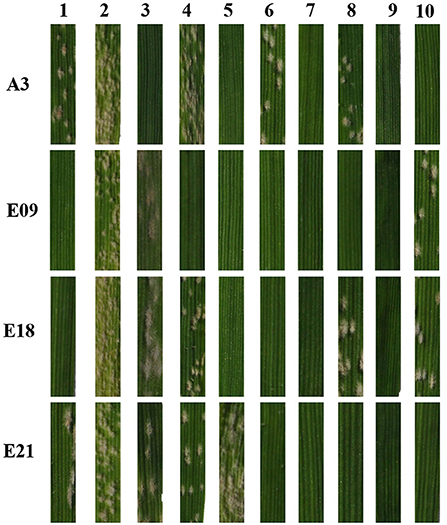
Figure 5. Reaction patterns of wheat breeding line Yannong 99102-06188 (YN99102) and the resistant donors with cataloged Pm genes on 2BL chromosome to selected Blumaria graminis f. s. tritici isolates A3, E09, E18, and E21. 1: YN99102; 2: susceptible parent Jinhe13-205 (JH13–205); 3: Coker747 (Pm6); 4: Am9/3 (Pm33); 5: CH7086 (Pm51); 6: Liangxing 99 (Pm52); 7: WE35 (Pm64); 8: LS5082 (PmLS5082); 9: KN0816 (PmKN0816); 10: Qingxinmai (PmQ).
To further distinguish pmYN99102 from the known Pm genes on 2BL, 98 previously reported markers that were closely linked to the documented Pm genes on chromosome arm 2BL were test the polymorphisms between the resistant and susceptible bulks derived from the F2:3 families of YN99102 × JH13–205 (Table 2). Among them, 12 markers, including eight markers for Pm6 (CIT02g−2, CIT02g−3, CIT02g−13, CIT02g−15, CIT02g−17, CIT02g−18, CIT02g−20, CISSR02g−2) and four markers for Pm52 (Xicscl172, Xicscl174, Xicsl326, and Xicsl234) amplified polymorphisms between the resistant and susceptible parents and bulks, while other 86 markers showed no polymorphism. Four markers closely linked to Pm52 were not linked to pmYN99102 (Table 2). Molecular markers analysis combined with different resistance spectrum demonstrated that pmYN99102 is most likely different from the known Pm genes on chromosome arm 2BL.
To better use pmYN99102 in MAS, 18 markers closely linked to pmYN99102 were tested for their availability in the 42 susceptible wheat cultivars for MAS (Figure 6; Supplementary Table 1). All markers except for YTU103–97 could amplify polymorphic bands between YN99102 and most of the 42 susceptible cultivars, suggesting that these markers can be used in MAS for tracking pmYN99102 when transferred into those cultivars. YTU103–97 amplified consistent bands in 38 out of 42 susceptible cultivars, meaning that these two markers were less appropriate for MAS of pmYN99102.
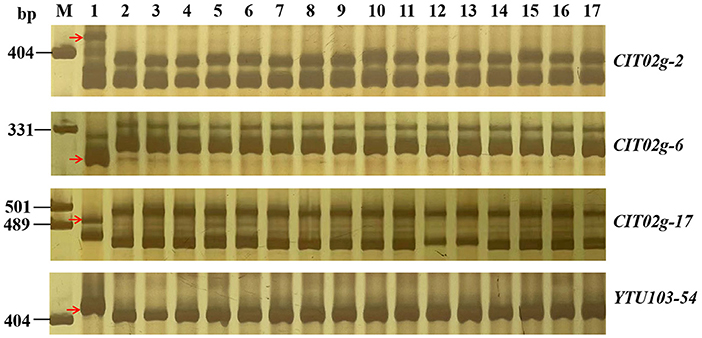
Figure 6. Amplification patterns of pmYN99102-linked markers CIT02g-2 (A) and CIT02g-6 (B), CIT02g-17 (C) and YTU103-54 (D) in Yannong 99102-06188 (YN99102), Jinhe13–205 (JH13–205), and 15 wheat cultivars/breeding lines susceptible to powdery mildew. M, DNA marker pUC18 Msp I; lanes 1 and 2, YN99102 and JH13–205; and lanes 3–17: Shannong 1,538, Hanmai 13, Huaimai 0,226, Zhoumai 27, Xinong 979, Lumai 185, Zhongyu 1,311, Jimai 268, Tainong 1,014, Jimai 229, Jimai 21, Jimai 20, Daimai 2,173, Zhoumai 1,751, Jinan 17. The white arrows indicate the polymorphic bands in YN99102.
In the present study, using genetic analysis, BSE-Seq, and molecular marker analysis, we accurately and rapidly identified a recessive Pm gene pmYN99102 in YN99102 and localized it to a 0.9 Mb interval (699.2–708.2 Mb) on chromosome arm 2BL, an enrichment region carrying many Pm genes (Wu et al., 2019). Eleven Pm genes have been reported to be located on chromosome arm 2BL from various gene donors, including Pm6 from Triticum timopheevii (Wan et al., 2020), Pm33 from T. persicum Vav. (Zhu et al., 2005), Pm51 from a Thinopyrum ponticum introgression line (Zhan et al., 2014), Pm52 from Chinese wheat cultivar Liangxing 99 (Wu et al., 2019), Pm63 from Iranian wheat landrace PI 628024 (Tan et al., 2019), Pm64 from wild emmer (Zhang et al., 2019), PmQ from Chinese wheat landrace Qingxinmai (Li H. H. et al., 2020), MlZec1 and MlAB10 both from T. dicoccoides (Mohler et al., 2005; Maxwell et al., 2010), PmKN0816 from the Chinese wheat breeding line KN0816 (Wang et al., 2021) and PmLS5082 from the Chinese wheat breeding line LS5082 (Wu et al., 2019), suggesting chromosome arm 2BL is a potential R gene-rich region and complex molecular modules and mechanisms may be involved in the chromosome arm 2BL. Compared with those documented genes, pmYN99102 (699.2–708.2 Mb) could be clearly distinguished from eight of them: Pm33 (773.2–784.3 Mb), Pm51 (709.8–739.4 Mb), Pm52 (581.0–585.0 Mb), Pm63 (710.3–723.4 Mb), PmQ (710.7–714.7 Mb), MlZec1 and MlAB10 (both 796.7–780.0 Mb), and PmLS5082 (710.3–711.0 Mb) based on their physical locations and/or origins. However, the physical intervals of three genes Pm6 (698.3–699.2 Mb), Pm64 (699.2–710.3 Mb), and PmKN0816 (700.4–710.3 Mb) overlapped that of pmYN99102 (699.2–708.2 Mb) and hence it is necessary to clarify their relationships.
T. timopheevii derived Pm6 was the first Pm gene identified on chromosome 2BL and was transferred into the wheat genetic background in the form of wheat-T. timopheevii 2B/2G introgression lines (Jorgensen and Jensen, 1973; Bennett, 1984; Ji et al., 2008). The 2G chromosome introgression segment carrying Pm6 has strong recombination suppression in the wheat genome, so seven of the eight Pm6-linked markers used in this study had no recombination between the 2G chromosome introgression segment carrying Pm6 and the corresponding wheat segment, while they showed normal recombination frequency as common wheat in the pmYN99102 interval. Moreover, only 8 of 55 Pm6-linked markers showed polymorphisms between YN99102 and JH13–205, and their derivative F2:3 resistant and susceptible bulks (Table 2), which revealed a distinct genetic diversity between the intervals of pmYN99102 and Pm6. Meanwhile, YN99106 had a broader resistance spectrum than Coker 747 (with Pm6). Critically, YN99106 was resistant to Bgt isolate E09 and pmYN99102 was identified by inoculating the Bgt isolate E09, whereas Coker 747 (with Pm6) was susceptible to the Bgt isolate E09 (Table 1). It was reported that Coker 747 was moderate effectiveness at the one-leaf stage to the two-leaf stage, but showed gradually increased resistance from the third leaf stage and reached complete resistance at the fourth leaf stage and later (Qin et al., 2011). Whereas, YN99102 exhibited high resistance to Bgt isolates from the one-leaf stage and continued through all growth stages. Therefore, the combined evidence indicated that pmYN99102 was different from Pm6. Pm64, derived from a wheat-T. dicoccoides introgression line showed a different resistance spectrum from pmYN99102 (Table 1). When tested with 10 Pm64-linked markers, none of them amplified polymorphisms in YN99102 and JH03–125 and their mapping population. PmKN0816 was a broad-spectrum resistance gene and also discovered in a Chinese wheat breeding line, and its donor KN0816 was resistant to all the 11 tested Bgt isolates, but YN99102 was susceptible to the Bgt isolate A03. In addition, all of the three Pm genes Pm6, Pm64, and PmKN0816 followed dominant inheritance pattern, while pmYN99102 was distinctively recessive. Taken together, pmYN99102 is most likely a new gene different from the cataloged Pm genes on chromosome 2BL based on their origins, chromosome intervals, resistance spectrum, and inheritance pattern. Of course, allelism tests and cloning of these genes are necessary in the future to finally determine their relationships in such a mysterious interval containing multiple and complex resistance genes. Further validation on the four candidate genes of pmYN99102 that was directly related to disease resistance (Supplementary Table 3) will be our focus in the near future.
When a resistance gene was identified, less linkage drag is the critical factor associated with its easy use in wheat breeding programs, but often, disease resistance is at the expense of some agronomic traits and reduced plant adaptation (Deng et al., 2017; Ma et al., 2018; Han et al., 2022b). Fortunately, YN99102 is an elite derivative line from the same breeding process as a famous wheat cultivar Yannong 999 in China, which is the first wheat cultivar to exceed 800 kilograms per mu yield in China. In the breeding process of Yannong 999, two prominent breeding lines, Yannong 99102-06072 and Yannong 99102-06188 (YN99102) were selected as the candidate lines. Of the two lines, Yannong 99102-06072 has favorable synergy between yield and quality, making it the current Yannong 999 official registration which met the breeding goal of high yield and quality; whereas YN99102 has the advantages of both powdery mildew resistance and high yield, making it a valuable resistance resource for both wheat breeding and genetic study. To facilitate the transfer of pmYN99102 in MAS, we evaluated the applicability of 17 markers including 10 newly developed markers and seven reported markers in 42 susceptible cultivars (Supplementary Table 1). The results showed that all markers except for YTU103–97 were polymorphic between YN99102 and most of the 42 susceptible cultivars, suggesting that these markers can be used in MAS for detecting pmYN99102 once it was introduced to those susceptible cultivars. In fact, we have obtained the BC1F2 and F3 segregation populations from the cross of some applicable cultivars and YN99102 currently. We believe that pmYN99102 will release its full potential following the selection for resistance and agronomic performance in wheat breeding programs.
In conclusion, using BSE-Seq and molecular markers, we identified a powdery mildew resistance gene pmYN99102 in the wheat breeding line YN99102. Based on the analysis of physical location, origin, resistant spectrum, and inherited pattern, pmYN99102 is most likely a new Pm gene. Molecular markers available for marker-assisted selection were also selected for tracking pmYN99102 in breeding. Our study can be valuable for enhancing the genetic diversity of powdery mildew resistance in breeding.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
PM, GH, and JW conceived the research. YM, WG, YQ, ZY, LX, FS, and WZ performed the experiments. JW and XL developed the experimental materials. NS and WL performed the phenotypic assessment. GH, JG, and RH analyzed the data. PM wrote the manuscript. All authors read and approved the final manuscript.
This research was financially supported by the National Natural Science Foundation of China (32072053, 31971874) and the Key Research and Development Project of Shandong Province (2020CXGC010805, 2021LZGC009).
We are grateful to Prof. Emerita Paula E. Jameson, University of Canterbury, financially supported by Double Hundred Plan for Foreign Experts in Shandong Province, China, for critically reviewing and editing this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1005627/full#supplementary-material
Supplementary Figure 1. Genealogy of the wheat breeding line Yannong 99102-06188 and its relationship with the wheat cultivar Yannong 999.
Supplementary Table 1. Validation of pmYN99102-linked markers on 42 Chinese wheat cultivars/breeding lines in marker-assisted selection (MAS) breeding.
Supplementary Table 2. Molecular markers developed by bulked segregant exome capture sequencing (BSE-Seq).
Supplementary Table 3. Gene annotation in the candidate interval of wheat powdery mildew resistance gene pmYN99102.
An, D. G., Ma, P. T., Zheng, Q., Fu, S. L., Li, L. H., Han, F. P., et al. (2019). Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot. Theor. Appl. Genet. 132, 257–272. doi: 10.1007/s00122-018-3214-3
Bennett, F. G. A. (1984). Resistance to powdery mildew in wheat: a review of its use in agriculture and breeding programmes. Plant Pathol. 33, 279–300. doi: 10.1111/j.1365-3059.1984.tb01324.x
Chen, X. M. (2013). Review article: high-temperature adult-plant resistance, key for sustainable control of stripe rust. Am. J. Plant Sci. 4, 608–627. doi: 10.4236/ajps.2013.43080
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6, 80–92. doi: 10.4161/fly.19695
Deng, Y. W., Zhai, K. R., Xie, Z., Yang, D. Y., Zhu, X. D., Liu, J. Z., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965. doi: 10.1126/science.aai8898
Dong, C. H., Zhang, L. C., Chen, Z. X., Xia, C., Gu, Y. Q., Wang, J. R., et al. (2020). Combining a new exome capture panel with an effective varBScore algorithm accelerates BSA-based gene cloning in wheat. Front. Plant Sci. 11, 1249. doi: 10.3389/fpls.2020.01249
Han, G. H., Yan, H. W., Gu, T. T., Cao, L. J., Zhou, Y. L., Wei, L., et al. (2022a). Identification of a wheat powdery mildew dominant resistance gene in the Pm5 locus for high-throughput marker-assisted selection. Plant Dis. doi: 10.1094/PDIS-07-22-1545-RE. [Epub ahead of print].
Han, G. H., Yan, H. W., Wang, J., Cao, L. J., Liu, S. Y., Li, X. Q., et al. (2022b). Molecular cytogenetic identification of a new wheat-rye 6R addition line and physical localization of its powdery mildew resistance gene. Front. Plant Sci. 13, 889494. doi: 10.3389/fpls.2022.889494
He, H. G., Liu, R. K., Ma, P. T., Du, H. N., Zhang, H. H., Wu, Q. H., et al. (2021). Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of greek durum wheat TRI 1796. Theor. Appl. Genet. 134, 53–62. doi: 10.1007/s00122-020-03681-2
He, Z. H., Xia, X. C., Chen, X. M., Zhang, Y., Zhang, Y., Yan, J., et al. (2015). “Application of molecular markers in plant quality and disease resistance breeding”. In the Proceedings of the Seventh National Symposium on Wheat Genetics and Breeding. Zhengzhou.
He, Z. H., Xia, X. C., Chen, X. M., and Zhuang, Q. S. (2011). Progress of wheat breeding in China and the future perspective. Acta Agron. Sin. 37, 202–215. doi: 10.3724/SP.J.1006.2011.00202
International Wheat Genome Sequencing Consortium (IWGSC) (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788. doi: 10.1126/science.1251788
Isham, K., Wang, R., Zhao, W., Wheeler, J., Klassen, N., Akhunov, E., et al. (2021). QTL mapping for grain yield and three yield components in a population derived from two high-yielding spring wheat cultivars. Theor. Appl. Genet. 134, 2079–2095. doi: 10.1007/s00122-021-03806-1
IWGSC. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Ji, J. H., Qin, B., Wang, H. Y., Cao, A. Z., Wang, S. L., Chen, P. D., et al. (2008). STS markers for powdery mildew resistance gene Pm6 in wheat. Euphytica 163, 159–165. doi: 10.1007/s10681-007-9578-0
Jia, M. S., Xu, H. X., Liu, C., Mao, R. X., Li, H. S., Liu, J. J., et al. (2020). Characterization of the powdery mildew resistance gene in the elite wheat cultivar Jimai 23 and its application in marker-assisted selection. Front. Genet. 11, 241. doi: 10.3389/fgene.2020.00241
Jiang, Y., Schulthess, A. W., Rodemann, B., Ling, J., Plieske, J., Kollers, S., et al. (2016). Validating the prediction accuracies of marker-assisted and genomic selection of fusarium head blight resistance in wheat using an independent sample. Theor. Appl. Genet. 130, 471–482. doi: 10.1007/s00122-016-2827-7
Jin, Y. L., Shi, F. Y., Liu, W. H., Fu, X. Y., Gu, T. T., Han, G. H., et al. (2021). Identification of resistant germplasm and detection of genes for resistance to powdery mildew and leaf rust from 2,978 wheat accessions. Plant Dis. 105, 3900–3908. doi: 10.1094/PDIS-03-21-0532-RE
Jorgensen, J. H., and Jensen, C. J. (1973). Gene Pm6 for resistance to powdery mildew in wheat. Euphytica 22, 423–423. doi: 10.1007/BF00022656
Kosambi, D. D. (1944). The estimation of map distances from recombination values. Ann. Hum. Genet. 12, 172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x
Li, H. H., Dong, Z. J., Ma, C., Xia, Q., Tian, X. B., Sehgal, S., et al. (2020). A spontaneous wheat-aegilops longissima translocation carrying Pm66 confers resistance to powdery mildew. Theor. Appl. Genet. 133, 1149–1159. doi: 10.1007/s00122-020-03538-8
Li, H. J., Murray, T. D., Mcintosh, R. A., and Zhou, Y. (2019). Breeding new cultivars for sustainable wheat production. Crop J. 7, 715–717. doi: 10.1016/j.cj.2019.11.001
Li, Y. H., Shi, X. H., Hu, J. H., Wu, P. P., Qiu, D., Qu, Y. F., et al. (2020). Identification of a recessive gene PmQ conferring resistance to powdery mildew in wheat landrace Qingxinmai using BSR-Seq analysis. Plant Dis. 104, 743–751. doi: 10.1094/pdis-08-19-1745-re
Lincoln, S. E., Daly, M. J., Lander, E. S., Lincoln, S., Daly, M., and Lander, E. (1992). Constructing Genetics Maps With MAPMAKER/EXP 3.0. Whitehead Institute Technical Report, 3rd Edn. Cambridge, MA: Whitehead Institute.
Ma, P. T., Wu, L. R., Xu, Y. F., Xu, H. X., Xu, Z., Zhang, X., et al. (2021). Bulked segregant RNA-Seq provides distinctive expression profile against powdery mildew in the wheat genotype YD588. Front. Plant Sci. 12, 764978. doi: 10.3389/fpls.2021.764978
Ma, P. T., Xu, H. X., Xu, Y. F., Li, L. H., Qie, Y. M., Luo, Q. L., et al. (2015). Molecular mapping of a new powdery mildew resistance gene Pm2b in Chinese breeding line KM2939. Theor. Appl. Genet. 128, 613–622. doi: 10.1007/s00122-015-2457-5
Ma, P. T., Xu, H. X., Xu, Y. F., Song, L. P., Liang, S. S., Sheng, Y., et al. (2018). Characterization of a powdery mildew resistance gene in wheat breeding line 10V-2 and its application in marker-assisted selection. Plant Dis. 102, 925–931. doi: 10.1094/pdis-02-17-0199-re
Maxwell, J. J., Lyerly, J. H., Srnic, G., Parks, R., Cowger, C., Marshall, D., et al. (2010). MlAB10: A Triticum turgidum subsp. Dicoccoides derived powdery mildew resistance gene identified in common wheat. Crop Sci. 50, 2261. doi: 10.2135/cropsci2010.04.0195
McCormick, R. F., Truong, S. K., and Mullet, J. E. (2015). RIG: recalibration and interrelation of genomic sequence data with the GATK. G3 (Bethesda). 5, 655–665. doi: 10.1534/g3.115.017012
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Xia, X. C., and Raupp, W. J. (2020). Catalogue of Gene Symbols For Wheat 2020 Supplement. Available online at: https://wheat.pw.usda.gov/GG3/WGC
Mohler, V., Zeller, F. J., Wenzel, G., and Hsam,. S. L. K. (2005). Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 9. Gene MlZec1 from the Triticum dicoccoides-derived wheat line Zecoi-1. Euphytica 142, 161–167. doi: 10.1007/s10681-005-1251-x
Narasimhan, V., Danecek, P., Scally, A., Xue, Y., Tyler-Smith, C., and Durbin, R. (2016). BCFtools/RoH: a hidden markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 32, 1749–1751. doi: 10.1093/bioinformatics/btw044
Qin, B., Cao, A., Wang, H., Chen, T., You, F. M., Liu, Y., et al. (2011). Collinearity-based marker mining for the fine mapping of Pm6, a powdery mildew resistance gene in wheat. Theor. Appl. Genet. 123, 207–218. doi: 10.1007/s00122-011-1577-9
Sheng, B. Q. (1988). Wheat powdery mildew was recorded using infection type in seedling stage. Plant Prot. 14:49.
Sheng, B. Q., and Duan, X. Y. (1991). Improvement of scale 0-9 method for scoring adult plant resistance to powdery mildew of wheat. Beijing Agr. Sci. 1, 38–39.
Singh, R. P., Singh, P. K., Rutkoski, J., Hodson, D. P., He, X., Jørgensen, L. N., et al. (2016). Disease impact on wheat yield potential and prospects of genetic control. Annu. Rev. Phytopathol. 54, 303–322. doi: 10.1146/annurev-phyto-080615-095835
Summers, R. W., and Brown, J. K. M. (2013). Constraints on breeding for disease resistance in commercially competitive wheat cultivars. Plant Phothol. 62, 115–121. doi: 10.1111/ppa.12165
Takagi, H., Uemura, A., Yaegashi, H., Tamiru, M., Abe, A., Mitsuoka, C., et al. (2013). MutMap-Gap: whole-genome resequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol. 200, 276–283. doi: 10.1111/nph.12369
Tan, C. C., Li, G. Q., Cowger, C., Carver, B. F., and Xu, X. Y. (2018). Characterization of Pm59, a novel powdery mildew resistance gene in Afghanistan wheat landrace PI 181356. Theor. Appl. Genet. 131, 1145–1152. doi: 10.1007/s00122-018-3067-9
Tan, C. C., Li, G. Q., Cowger, C., Carver, B. F., and Xu, X. Y. (2019). Characterization of Pm63, a powdery mildew resistance gene in Iranian landrace PI 628024. Theor. Appl. Genet. 132, 1137–1144. doi: 10.1007/s00122-018-3265-5
Wan, W. T., Xiao, J., Li, M. L., Tang, X., Wen, M. X., Cheruiyot, A. K., et al. (2020). Fine mapping of wheat powdery mildew resistance gene Pm6 using 2B/2G homoeologous recombinants induced by the ph1b mutant. Theor. Appl. Genet. 133, 1265–1275. doi: 10.1007/s00122-020-03546-8
Wang, W. R., He, H. G., Gao, H. M. , Xu, H. X., Song, W. Y., Zhang, X., et al. (2021). Characterization of the powdery mildew resistance gene in wheat breeding line KN0816 and its evaluation in marker-assisted selection. Plant Dis. 105, 4042–4050. doi: 10.1094/pdis-05-21-0896-re
Wang, Z. L., Li, L. H., He, Z. H., Duan, X. Y., Zhou, Y. L., Chen, X. M., et al. (2005). Seedling and adult plant resistance to powdery mildew in Chinese bread wheat cultivars and lines. Plant Dis. 89, 457–463. doi: 10.1094/pd-89-0457
Wu, P. P., Hu, J. H., Zou, J. W., Qiu D., Qu, Y. F., Li, Y. H., et al. (2019). Fine mapping of the wheat powdery mildew resistance gene Pm52 using comparative genomics analysis and the Chinese Spring reference genomic sequence. Theor. Appl. Genet. 132, 1451–1461. doi: 10.1007/s00122-019-03291-7
Xu, H. X., Yi, Y. J., Ma, P. T., Qie, Y. M., Fu, X. Y., Xu, Y. F., et al. (2015). Molecular tagging of a new broad-spectrum powdery mildew resistance allele Pm2c in Chinese wheat landrace Niaomai. Theor. Appl. Genet. 128, 2077–2084. doi: 10.1007/s00122-015-2568-z
Yu, Z. Y., Xiao, L. N., Su, F. Y., Liu, W., Luo, F. Y., Han, R., et al. (2022). Mining of wheat Pm2 alleles for goal-oriented marker-assisted breeding. Front. Plant Sci. 13, 912589. doi: 10.3389/fpls.2022.912589
Zhan, H. X., Li, G. R., Zhang, X. J., Li, X., Guo, H. J., Gong, W. P., et al. (2014). Chromosomal location and comparative genomics analysis of powdery mildew resistance gene Pm51 in a putative wheat-Thinopyrum ponticum introgression line. PLoS ONE. 9, e113455. doi: 10.1371/journal.pone.0113455
Zhang, D. Y., Zhu, K. Y., Dong, L. L., Liang, Y., Li, G. Q., Fang, T. L., et al. (2019). Wheat powdery mildew resistance gene Pm64 derived from wild emmer (Triticum turgidum var. dicoccoides) is tightly linked in repulsion with stripe rust resistance gene Yr5. Crop J. 7, 761–770. doi: 10.1016/j.cj.2019.03.003
Zhao, Z. H., Sun, H. G., Song, W., Lu, M., Huang, J., Wu, L. F., et al. (2013). Genetic analysis and detection of the gene MlLX99 on chromosome 2BL conferring resistance to powdery mildew in the wheat cultivar Liangxing 99. Theor. Appl. Genet. 126, 3081–3089. doi: 10.1007/s00122-013-2194-6
Zhou, R. H., Zhu, Z. D., Kong, X. Y., Huo, N. X., Tian, Q. Z., Li, P., et al. (2005). Development of wheat near-isogenic lines for powdery mildew resistance. Theor. Appl. Genet. 110, 640–648. doi: 10.1007/s00122-004-1889-0
Zhu, T., Wu, L. R., He, H. G., Song, J. C., Jia, M. S., Liu, L. C., et al. (2020). Bulked segregant RNA-Seq reveals distinct expression profiling in Chinese wheat cultivar Jimai 23 responding to powdery mildew. Front. Genet. 11, 474. doi: 10.3389/fgene.2020.00474
Keywords: wheat powdery mildew, pmYN99102, MAS, BSE-Seq, molecular mapping
Citation: Mu Y, Gong W, Qie Y, Liu X, Li L, Sun N, Liu W, Guo J, Han R, Yu Z, Xiao L, Su F, Zhang W, Wang J, Han G and Ma P (2022) Identification of the powdery mildew resistance gene in wheat breeding line Yannong 99102-06188 via bulked segregant exome capture sequencing. Front. Plant Sci. 13:1005627. doi: 10.3389/fpls.2022.1005627
Received: 28 July 2022; Accepted: 15 August 2022;
Published: 06 September 2022.
Edited by:
Handong Su, Huazhong Agricultural University, ChinaReviewed by:
Jindong Liu, Institute of Crop Sciences (CAS), ChinaCopyright © 2022 Mu, Gong, Qie, Liu, Li, Sun, Liu, Guo, Han, Yu, Xiao, Su, Zhang, Wang, Han and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengtao Ma, cHRtYUB5dHUuZWR1LmNu; Guohao Han, Z2hoYW5AbXMuc2p6aWFtLmFjLmNu; Jiangchun Wang, eXQtd2pjQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.