- 1Triticeae Research Institute, Sichuan Agricultural University, Chengdu, China
- 2State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Sichuan Agricultural University, Chengdu, China
- 3College of Resources, Sichuan Agricultural University, Chengdu, China
- 4College of Grassland Science and Technology, Sichuan Agricultural University, Chengdu, China
Early maturation is an important objective in wheat breeding programs that could facilitate multiple-cropping systems, decrease disaster- and disease-related losses, ensure stable wheat production, and increase economic benefits. Exploitation of novel germplasm from wild relatives of wheat is an effective means of breeding for early maturity. Psathyrostachys huashanica Keng f. ex P. C. KUO (2n=2x=14, NsNs) is a promising source of useful genes for wheat genetic improvement. In this study, we characterized a novel wheat-P. huashanica line, DT23, derived from distant hybridization between common wheat and P. huashanica. Fluorescence in situ hybridization (FISH) and sequential genomic in situ hybridization (GISH) analyses indicated that DT23 is a stable wheat-P. huashanica ditelosomic addition line. FISH painting and PCR-based landmark unique gene markers analyses further revealed that DT23 is a wheat-P. huashanica 7Ns ditelosomic addition line. Observation of spike differentiation and the growth period revealed that DT23 exhibited earlier maturation than the wheat parents. This is the first report of new earliness per se (Eps) gene(s) probably associated with a group 7 chromosome of P. huashanica. Based on specific locus-amplified fragment sequencing technology, 45 new specific molecular markers and 19 specific FISH probes were developed for the P. huashanica 7Ns chromosome. Marker validation analyses revealed that two specific markers distinguished the Ns genome chromosomes of P. huashanica and the chromosomes of other wheat-related species. These newly developed FISH probes specifically detected Ns genome chromosomes of P. huashanica in the wheat background. The DT23 line will be useful for breeding early maturing wheat. The specific markers and FISH probes developed in this study can be used to detect and trace P. huashanica chromosomes and chromosomal segments carrying elite genes in diverse materials.
Introduction
Early maturation is an important target in wheat breeding programs. In recent years, breeding efforts have targeted increase in wheat yield potential to meet the growing food demand resulting from global population explosion, climate change, and reduction in arable land area (Mondal et al., 2013; Chen et al., 2016). Certain adverse factors, such as Fusarium head blight, powdery mildew, and rust infection, high temperature, frost damage, excessive rainfall, and preharvest sprouting, are liable to occur during the mid to late wheat-growing season and comprise the primary causes of reduction in wheat yield and quality (Iqbal et al., 2006; Mahdiyeh and Bahram, 2012; Sheehan and Bentley, 2020). Exploitation of new early maturation genes and the breeding of early maturing wheat cultivars are strategies proven to be effective against these emerging threats to agricultural production and are suitable for a double-cropping management system, in which earlier sowing of the following crop improves the probability of its success (Hunger et al., 2014; Su et al., 2018). Such an approach promotes effective utilization of the limited arable land and enhances grain yield.
The wheat heading stage is jointly controlled by vernalization (Vrn), photoperiod (Ppd), and earliness per se (Eps) genes (Shi et al., 2019). Given their involvement in essential growth and development processes, these genes are crucial in determining the length of the wheat growth period (Zhao et al., 1989). The Ppd and Vrn genes show significant interactions with the environment, whereas Eps genes participate at various stages of wheat growth and function independently (Ochagavía et al., 2018; Shi et al., 2019). To date, a number of Eps quantitative trait loci (QTLs) have been mapped on different wheat and barley chromosomes (Laurie et al., 1995; Kamran et al., 2014; Lombardo et al., 2019). Only a small number of Eps genes originating from wheat relatives have been exploited, such as LUX and ELF3 derived from Triticum monococcum (Gawroński and Schnurbusch, 2012; Gawroński et al., 2014; Alvarez et al., 2016), and HvCEN derived from Hordeum vulgare, respectively (Comadran et al., 2012). Therefore, the discovery and identification of additional sources of early maturation in wheat-related germplasm are an important and long-term objective for breeding early maturing wheat cultivars.
Psathyrostachys huashanica Keng f. ex P. C. KUO (2n=2x=14, NsNs) is a diploid, perennial, outcrossing graminaceous species restricted to the Huashan section of the Qinling Mountains, Shaanxi Province, China (Baden, 1991). As a wild relative of common wheat, P. huashanica is favored by many wheat breeders because it possesses numerous agronomically beneficial traits, such as early maturity, dwarf stature, disease resistance (to scab, stripe rust, powdery mildew, and take-all), and tolerance to abiotic factors (cold, drought, salinity, and infertile soil; Chen et al., 1991; Kang et al., 2016). To transfer these desirable genes from P. huashanica to wheat, distant hybridization between common wheat and P. huashanica has been performed since the 1980s, from which a heptaploid hybrid H8911 (2n=7x=49, AABBDNs) was successfully obtained by embryo culture (Chen et al., 1991). Subsequently, derivative lines with P. huashanica chromosome(s) incorporated into the common wheat background were generated, such as an amphiploid line (PHW-SA, 2n=8x=56, AABBDDNsNs; Kang et al., 2009), and chromosome addition and substitution lines (Zhao et al., 2004, 2010; Kishii et al., 2010). In the last decade, a series of wheat-P. huashanica derivative lines were developed and identified using molecular cytological methods, including wheat-P. huashanica 1Ns-7Ns disomic addition lines (Du et al., 2013a,b,c, 2014a,b,c,d), 1Ns(1D), 2Ns(2D), 3Ns(3D), and 5Ns(5D) disomic substitution lines (Li et al., 2019 2021a Bai et al., 2020; Qu et al., 2021), and several translocation lines (Kang et al., 2016; Li et al., 2020a; Liu et al., 2021). These progeny lines outperformed their wheat parents with regard to disease resistance and agronomic traits, demonstrating that P. huashanica is a superior wild relative useful to wheat breeding programs. However, the transfer of elite Eps genes carried by P. huashanica to common wheat has been neglected to date.
Our research team has performed wide hybridization between common wheat and P. huashanica since 2004 and successfully obtained F1 hybrids without adoption of an embryo rescue technique (Kang et al., 2008). Subsequently, wheat-P. huashanica derivative lines were generated by backcrossing and selfing (Wang et al., 2011). In the present study, we identified a novel wheat-P. huashanica 7Ns ditelosomic addition line, DT23, from offspring lines that exhibited earlier maturation than the wheat parents by fluorescence in situ hybridization (FISH), genomic in situ hybridization (GISH), FISH painting, and PCR-based landmark unique gene (PLUG) marker analyses. Furthermore, we developed and validated new specific molecular markers and specific FISH probes, based on specific locus amplified fragment sequencing (SLAF-seq) technology, to efficiently trace P. huashanica 7Ns chromatin in wheat breeding programs and identify chromosomes of P. huashanica and other wheat-related species.
Materials and Methods
Plant Materials
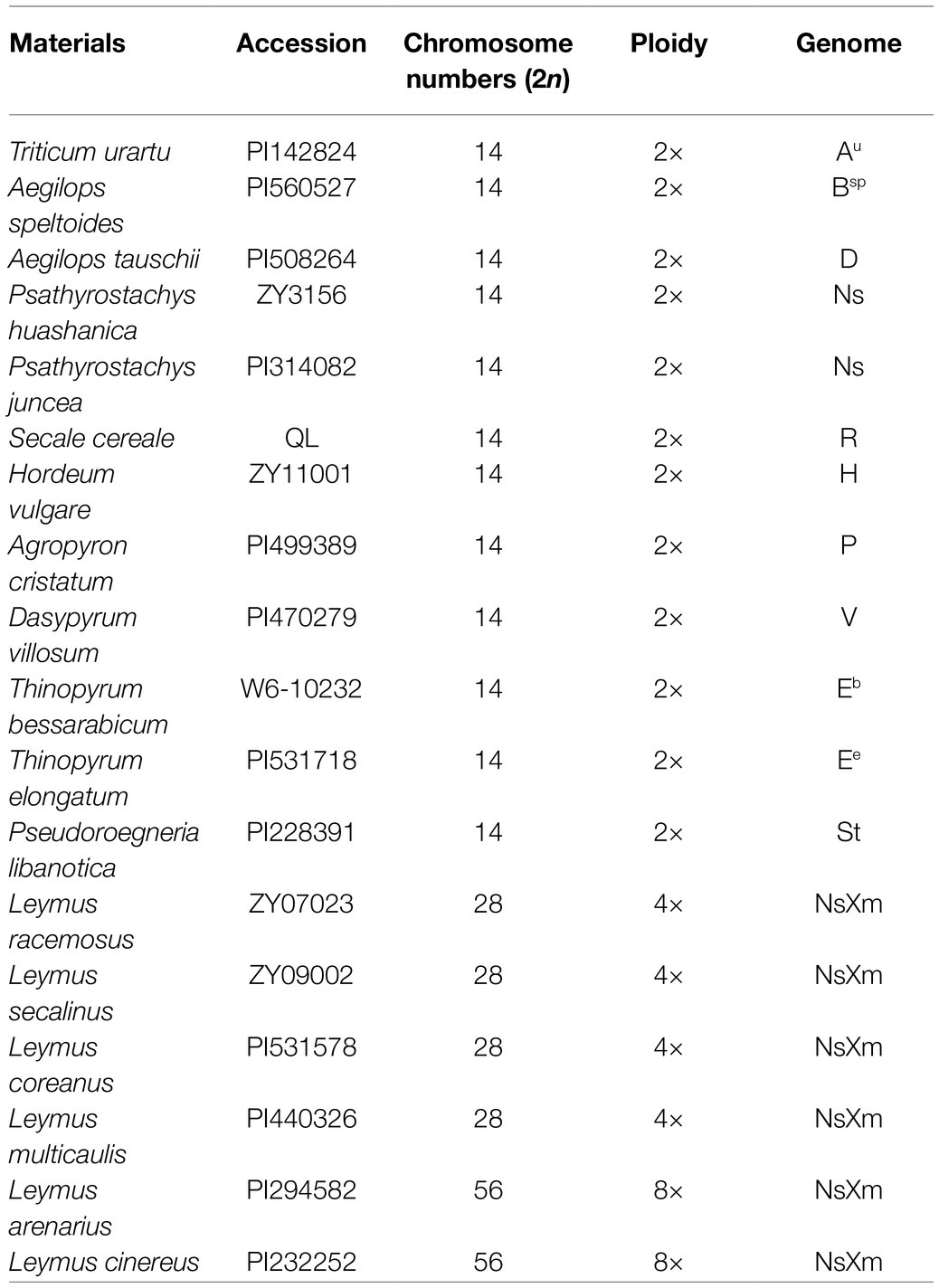
Wheat (Triticum aestivum L.) “Chinese Spring” (CS; 2n=6x=42, AABBDD) is a Sichuan white-grained cultivar. Chinese Spring ph2b (CSph2b), a chemically induced mutant, was produced by a terminal segment deletion or point mutation of ph2 gene on the short arm of CS chromosome 3D (Wall et al., 1971; Sears, 1982). Psathyrostachys huashanica accession ZY3156 (2n=2x=14, NsNs) was collected from the Huashan Mountains, Shaanxi Province, China, by Profs. C. Yen and J.L. Yang of Sichuan Agricultural University. We first crossed CSph2b with P. huashanica, and thereafter, the F1 hybrids were crossed with CS, as the male parent, to obtain the BC1F1 generation (Kang et al., 2008). Subsequently, a wheat-P. huashanica 7Ns ditelosomic addition line, designated DT23, was developed through three consecutive cycles of self-pollination. Finally, molecular markers and FISH probes were validated in other wheat-related species listed in Table 1. All plant materials used are preserved in the herbarium of the Triticeae Research Institute, Sichuan Agricultural University, China.
FISH and Sequential GISH Analyses
Actively growing root tips from seeds germinated at 22°C in a constant temperature incubator were treated with nitrous oxide gas for 2.5h and 90% glacial acetic acid for at least 10min and then digested with pectinase and cellulase (Komuro et al., 2013). Mitotic chromosome spreads from root tip cells were prepared and observed for the FISH and sequential GISH analyses using a previously described method (Han et al., 2006), with minor modifications. For the FISH experiment, a pair of fluorescent-modified probes comprising oligo-pSc119.2 (6-FAM-5′) and oligo-pTa535 (TAMRA-5′), synthesized by Sangon Biotech (Chengdu, China; Tang et al., 2014), were used to distinguish each wheat chromosome of DT23. The FISH procedure was performed as described by Han et al. (2006) and Gong et al. (2019). The chromosomes were counterstained with 4,6-diamino-2-phenylindole solution (Vector Laboratories, Burlingame, CA, United States). Mitotic chromosome number counts and fluorescent signals were visualized and captured using a fluorescence microscope (Olympus BX63) equipped with a Photometric SenSys DP-70 CCD camera (Olympus, Tokyo, Japan). The images were optimized for contrast and brightness using Adobe Photoshop software.
The slides photographed were eluted sequentially in 75% alcohol for 10min, 2× SSC in boiling water for 5min, 75% alcohol for 20min, and 100% alcohol for 20min, then exposed to bright light for 48h, and finally were prepared for sequential GISH. In the sequential GISH analysis, total genomic DNA of P. huashanica and T. aestivum “J-11” was extracted from fresh leaves using the improved cetyltrimethylammonium bromide method (Cota-Sánchez et al., 2006). Genomic DNA of the former species was labeled with Texas Red-12-dUTP (Red) using the nick translation method (Thermo Fisher Scientific, Eugene, OR, United States) and served as a probe, whereas genomic DNA of the latter cultivar was used as blocking DNA with a probe:block ratio of 1:150. The sequential GISH protocol was performed in accordance with that of Han et al. (2006) and Gong et al. (2019). Detection and visualization of GISH signals were performed as described above.
FISH Painting Analysis
Seven bulked oligonucleotide-based FISH probes (Synt1 to Synt7) derived from single-copy sequences on chromosomes 1 to 7 of barley and corresponding to each of the seven Triticeae linkage groups (Li et al., 2021b), which were kindly provided by Dr. ZJ Yang, University of Electronic Science and Technology of China, Chengdu, China, were used to determine the homoeologous group relationships of the introduced P. huashanica chromosomes in line DT23. FISH painting with the bulked oligo probes was performed as described previously by Han et al. (2015) and Bi et al. (2020). After the bulked oligo-based FISH, sequential FISH and GISH were similarly conducted as described above.
PLUG Marker Analysis
Primer pairs for 135 PLUG markers distributed evenly among the seven wheat homoeologous groups (Ishikawa et al., 2009) were employed to determine the homoeologous group relationships of the added P. huashanica chromosomes in line DT23. CS and CSph2b were used as negative controls, whereas P. huashanica was used as a positive control. PCR amplification was performed as described previously by Yang et al. (2020) with slight modifications.
Observation of Spike Differentiation and Growth Period
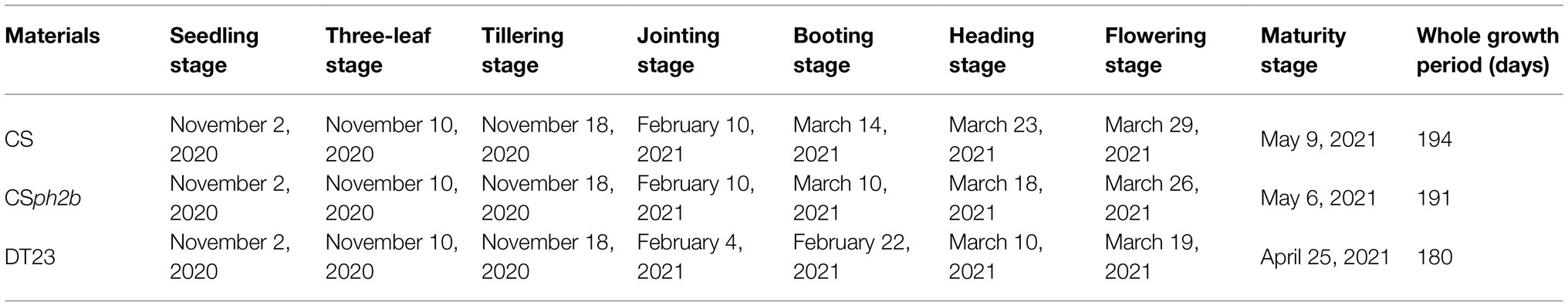
Plants of CS, CSph2b, and DT23 were grown in the field in Wenjiang district, Sichuan Province, China, during the 2020–2021 growing season. The field layout consisted of plots 1.5m in width with 30cm rows and a sowing density of 15 plants per row. In accordance with the criteria described by Cui et al. (2008), wheat spike differentiation was categorized into eight stages, comprising apex elongation, single-ridge, double-ridge, glume primordia differentiation, floret primordia differentiation, stamen and pistil differentiation, anther separation, and tetrad stages. During the three-leaf stage to the heading stage, spike differentiation was observed with a stereomicroscope (ZEISS SteREO Discovery.V20) at seven-day intervals. Ten randomly selected individuals from each material were observed at each time point. In addition, the timing of stages in the growth period of each material was assessed, comprising the seedling, three-leaf, tillering, jointing, booting, heading, flowering, and maturity stages (Guo and Fan, 2011).
Evaluation of Agronomic Performance
Morphological traits of DT23 and its wheat parents were evaluated in the field in Wenjiang district, Sichuan Province, China, with three replications in the 2020–2021 growing season. At the physiological maturity stage, 20 randomly selected plants of each line were harvested to evaluate their morphological traits, comprising plant height, tiller number, spike length, number of spikelets per spike, number of kernels per spike, and 1,000-grain weight. The IBM SPSS Statistics 24.0 software package was used for statistical analysis of the data.
Molecular Marker Development
Genomic DNA of CSph2b, P. huashanica, and DT23 was sequenced using the SLAF-seq technique (Biomarker, Beijing, China). Genomic DNA digestion, PCR fragment amplification, fragment selection, and SLAF-seq library construction were conducted as previously described by Sun et al. (2013) with slight modifications. Amplicons with appropriate sizes of 464–494bp were excised and diluted for sequencing using an Illumina HiSeq 2,500 platform (Illumina, Inc., San Diego, CA, United States). The SLAFs were identified, filtered, and clustered following the methods described by Chen et al. (2013). Psathyrostachys huashanica 7Ns chromosome-specific sequences were generated as follows. The high-quality DT23 sequences were first compared with the CS reference genome sequence1 using the Burrows-Wheeler Alignment software. The sequences with 0% similarity to CS were selected. These sequences were then compared with the CSph2b sequences acquired using SLAF-seq in this study, and the sequences with identities less than 23% were selected. Finally, the retained DT23 sequences were compared with the sequences of P. huashanica and the sequences with identities greater than 90% were selected, which were regarded to be the P. huashanica 7Ns chromosome-specific sequences.
Based on these specific sequences, PCR primers were designed using the Primer3Plus online tool2 and synthesized by Sangon Biotech (Chengdu, China). The amplified products were electrophoresed in 3% agarose gel. The markers detected in DT23 and P. huashanica but absent in CS were identified as 7Ns chromosome-specific molecular markers. The stability, repeatability, and specificity of these markers were validated in CS, CSph2b, P. huashanica, and DT23 as well as in 17 wheat-related species. The PCR amplification mixture (in a final volume of 20μl) contained 1μl template DNA (200ng/μL), 10μl of 2× Taq Master Mix for PAGE (Dye Plus), 1.0μl of each primer (10μM), and 7.0μl ddH2O. The PCR procedure was as follows: 94°C for 5min, followed by 35cycles of 94°C for 30s, an appropriate annealing temperature of 60–70°C for 30s, and 72°C for 30s, and a final extension at 72°C for 10min.
FISH Probes Development
The verified molecular markers specific to the Ns genome of P. huashanica, P. juncea, and Leymus species were amplified by PCR from P. huashanica genomic DNA. An aliquot (5μl) of the amplicons was electrophoresed in 3% agarose gel to check the product length and yield. The PCR products were extracted with sodium acetate and precipitated with ethanol. The pellets were rinsed with 70% ethanol and dissolved in 30μl ddH2O. The extracted DNA fragments were labeled with fluorescein-12-dUTP (Green) using the nick translation method (Thermo Fisher Scientific) and served as probes in the FISH analysis of CS, DT23, P. huashanica, and 11 wheat-related species.
Results
Chromosomal Constitution of DT23
FISH and sequential GISH analyses were performed to determine the chromosomal composition of the wheat-P. huashanica line DT23. According to the standard FISH karyotype of Chinese Spring (Tang et al., 2014), FISH analysis with the probes Oligo-pSc119.2 (green) and Oligo-pTa535 (red) revealed that DT23 carried 42 wheat chromosomes and two telocentric chromosomes that lacked fluorescent signals (Figure 1A). When P. huashanica genomic DNA was used as the probe and J-11 genomic DNA as the blocker, sequential GISH analysis further revealed that the two telocentric chromosomes with strong red hybridization signals were Ns chromosomes from P. huashanica (Figure 1B). To confirm the cytological stability of the line DT23, GISH was used to identify 40 randomly selected seeds from selfed progeny of DT23. Thirty-six seeds carried a pair of Ns telocentric chromosomes (Figure 1C), and four seeds carried one Ns telocentric chromosome (Figure 1D). Therefore, these findings suggested that DT23 was a cytogenetically relatively stable wheat-P. huashanica ditelosomic addition line.
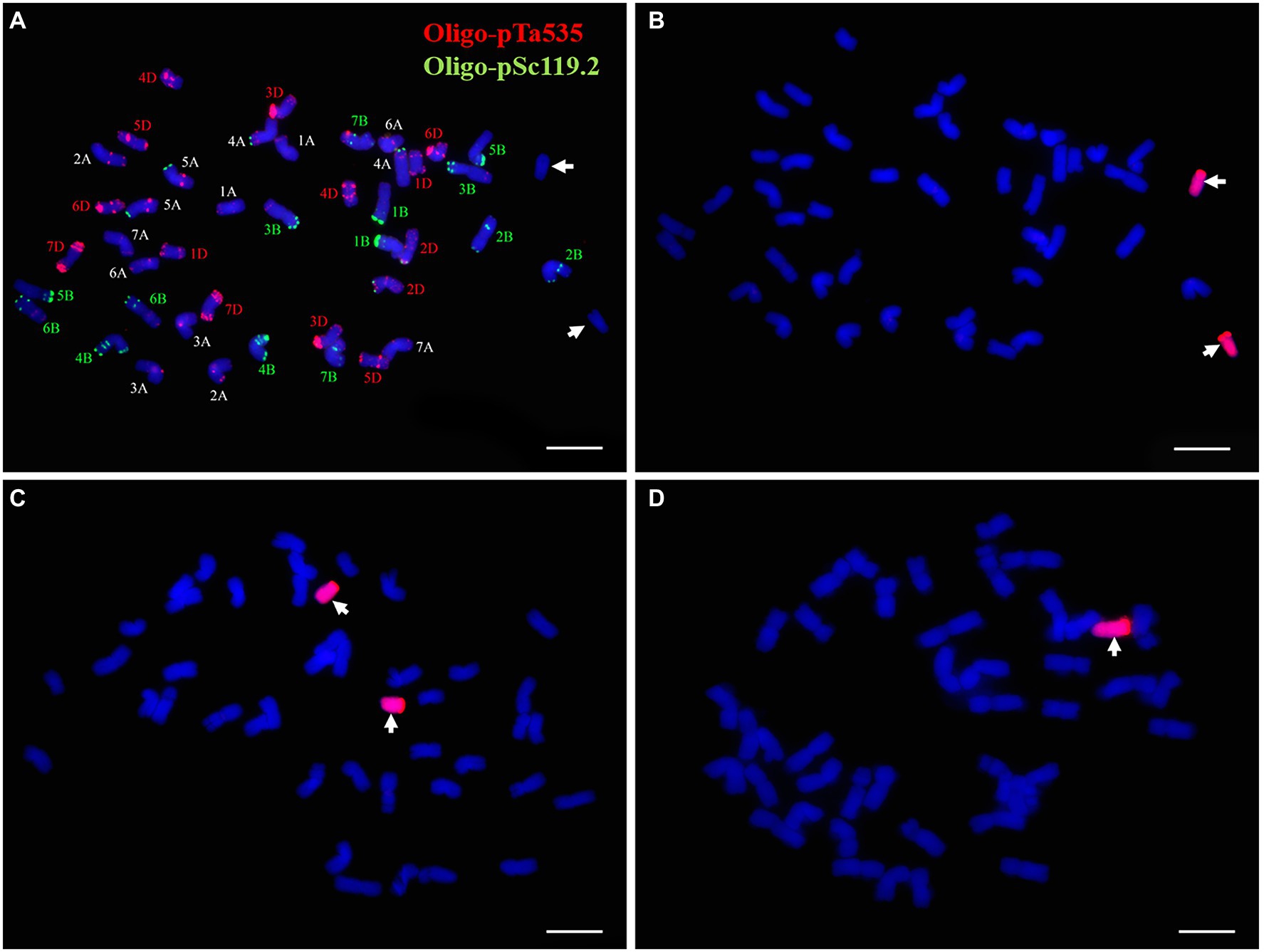
Figure 1. FISH and sequential GISH identification of the wheat-P. huashanica ditelosomic addition line DT23. (A) FISH identification of DT23 using Oligo-pSc119.2 (green) and Oligo-pTa535(red). (B) Sequential GISH analysis on the same metaphase cell of DT23 using P. huashanica genomic DNA as a probe (red). (C) GISH identification of the selfed progeny of DT23 which carried a pair of Ns telocentric chromosomes. (D) GISH identification of the selfed progeny of DT23 which carried one Ns telocentric chromosome. Arrows indicate the introduced P. huashanica chromosomes in DT23. Scale bar: 10μm.
FISH Painting Analysis of DT23
FISH painting with the probes (Synt1 to Synt7) corresponding to the first to seventh linkage group was performed on DT23. Probe Synt7 painted the six complete chromosomes and two telocentric chromosomes (Figure 2A). Sequential FISH using Oligo-pSc119.2 and Oligo-pTa535 indicated that three pairs of complete chromosomes were painted, comprising the common wheat chromosomes 7A, 7B, and 7D, whereas the two telocentric chromosomes did not show hybridization signals (Figure 2B). GISH analysis revealed that the two telocentric chromosomes with strong red hybridization signals were Ns chromosomes of P. huashanica (Figure 2C). Therefore, these results convincingly indicated that the alien chromosomal segments in DT23 belonged to the seventh group and that DT23 is a wheat–P. huashanica 7Ns ditelosomic addition line.
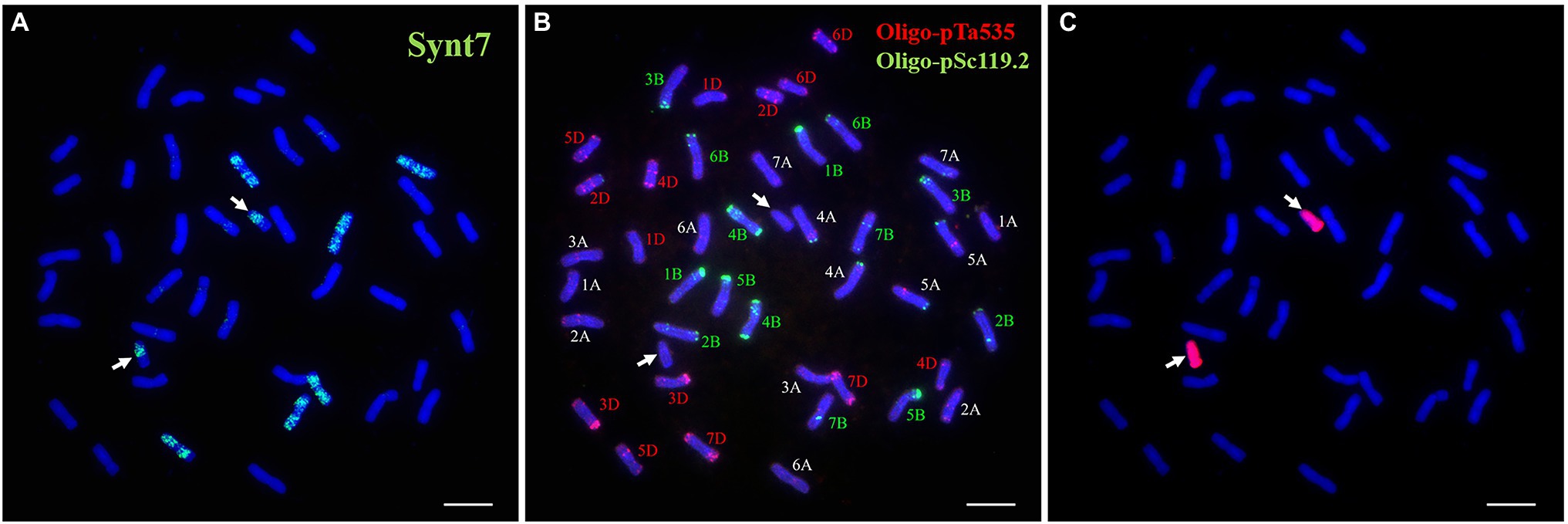
Figure 2. Sequential FISH and GISH analyses of the wheat-P. huashanica ditelosomic addition line DT23. (A) FISH painting analysis of DT23 using probe Synt7 (green). (B) FISH identification of DT23 using Oligo-pSc119.2 (green) and Oligo-pTa535(red). (C) P. huashanica genomic DNA was used as a probe for GISH (red). Arrows indicate the introduced P. huashanica chromosomes in DT23. Scale bar: 10μm.
PLUG Marker Analysis of DT23
As expected, among 135 PLUG markers, two markers (TNAC1782-7AS 7BS 7DS and TNAC1845-7AL 7BL 7DL) distributed on the group 7 chromosomes of wheat amplified the same specific bands in DT23 and P. huashanica but not in CS and CSph2b (Figure 3). This result also suggested that P. huashanica chromosomal segments introduced into DT23 belonged to the seventh group.
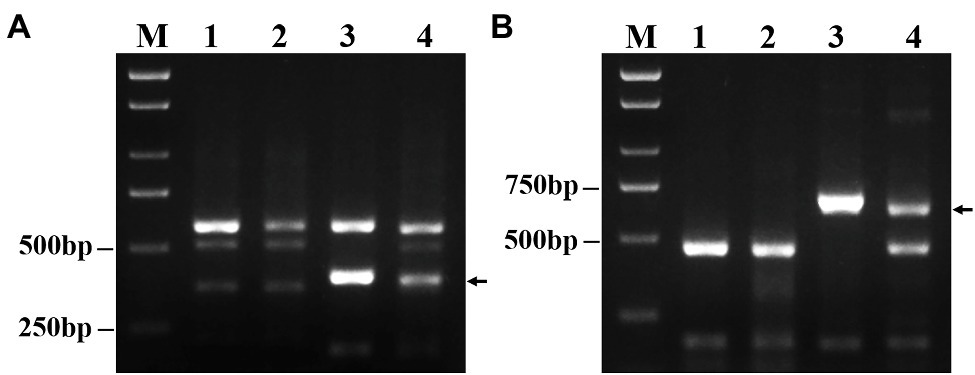
Figure 3. Amplification patterns of wheat PLUG markers. (A) TNAC1782-TaqI, (B) TNAC1845-TaqI. M: Marker (2000bp); 1: CS; 2: CSph2b; 3: P. huashanica; 4: DT23 (wheat-P. huashanica ditelosomic addition line). Arrows indicate the diagnostic amplification products for Ns genome.
Observation of Spike Differentiation and Growth Period
During the 2020–2021 growing season, the growth period was investigated in the field for DT23, CS, and CSph2b. Statistical analysis revealed that the timing of the seedling, three-leaf, and tillering stages was consistent among the three materials, but the timing of the jointing to maturity stages in DT23 was strongly accelerated compared with those of CS and CSph2b (Table 2). These findings were identical to those observed under greenhouse conditions (Supplementary Table 1). The entire growth period of DT23 was 14 and 11days shorter than that of CS and CSph2b, respectively (Table 2).
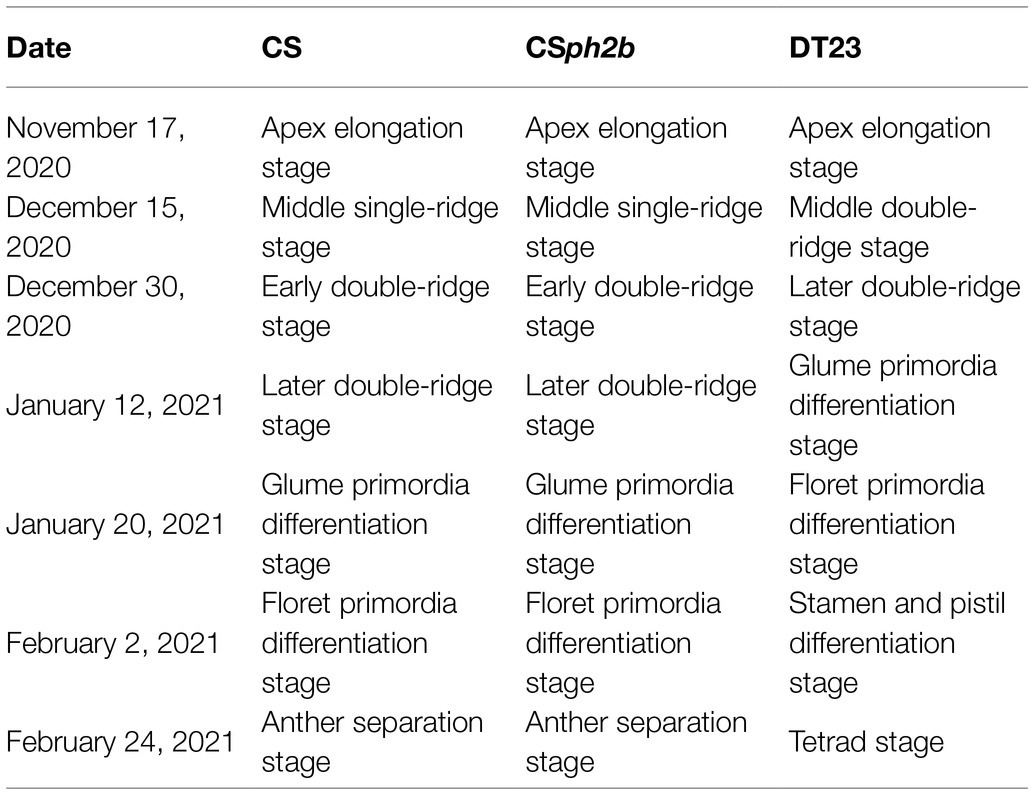
To further clarify the phenotypic differences between DT23 and its wheat parents CS and CSph2b, spike differentiation from the three-leaf stage to the heading stage in the field was observed by stereomicroscopy during the 2020–2021 growing season. On November 17, 2020, DT23, CS, and CSph2b plants were all at the apex elongation stage (Table 3, Figure 4A), indicating that no obvious difference in development was observed between DT23 and its wheat parents up to this stage. However, a clear developmental difference was observed on December 15, 2020, when CS and CSph2b plants were at the mid-single-ridge stage, whereas DT23 plants were at the mid-double-ridge stage (Table 3, Figure 4B). Subsequently, DT23 developed more rapidly than CS and CSph2b from the late-double-ridge stage to the tetrad stage (Table 3, Figures 4C–G). When CS and CSph2b were at the booting stage, DT23 plants were already at the heading stage and had developed distinctly larger spikes (Figures 4H,I).

Figure 4. Morphological observation of spike differentiation in different dates under field conditions. Different uppercase letters indicate the date of observation. (A) November 17, 2020. (B) December 15, 2020. (C) December 30, 2020. (D) January 12, 2021. (E) January 20, 2021. (F) February 2, 2021. (G) February 24, 2021. (H,I) March 10, 2021. Images of each panel from left to right are CS, CSph2b and DT23, respectively. Scale bars: A, 200μm; B-F, 150μm; G, 50μm; H, 2cm; I, 1cm.
Morphology of DT23
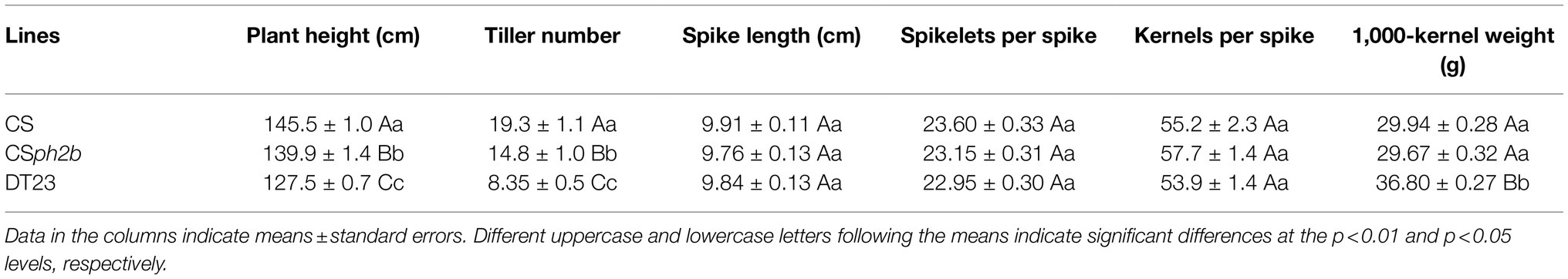
DT23 showed stability in morphological traits, which were similar to those of the wheat parents CS and CSph2b (Table 4, Figure 5). The average plant height and tiller number of DT23 were significantly lower than those of CS and CSph2b. The 1,000-kernel weight of DT23 was significantly higher than that of CS and CSph2b. No significant differences between DT23 and either CS or CSph2b were observed with regard to spike length, number of spikelets per spike, and number of kernels per spike.

Figure 5. Plant morphology of the wheat-P. huashanica ditelosomic addition line DT23 and its wheat parents. (A) Adult plants, (B) Spikes, and (C) Spikelets and grains. 1: CS; 2: CSph2b; 3: DT23.
Molecular Marker Development
A total of 6,963,342, 3,723,959, and 16,237,871 reads were, respectively, obtained for CSph2b, P. huashanica ZY3156, and DT23 using the SLAF-seq approach. The average Q30 score was 94.89%, and the GC content was 48.56%. After filtering out low-quality, repeat, and ambiguous reads, a total of 399,489, 120,418, and 462,445 effective SLAFs were generated for CSph2b, P. huashanica, and DT23, respectively. The average sequencing depth was 13.95×. These results were optimal and fulfilled the expected requirements. Sequence comparison revealed that 54 DT23 sequences showed 0% homology with the CS reference genome and less than 23% homology with CSph2b sequences, but more than 90% homology with P. huashanica sequences. These sequences were considered to be candidates for specific sequences of chromosome 7Ns from P. huashanica.
To develop P. huashanica 7Ns chromosome-specific molecular markers, 52 primer pairs were designed based on the candidate specific sequences and used to amplify sequences from CS, CSph2b, P. huashanica, and DT23 (Supplementary Table 2). In total, 45 primer pairs amplified specific bands from DT23 and P. huashanica but not from CS and CSph2b, such as PH7Ns-12 and PH7Ns-38 (Figures 6A,B). Therefore, these markers were regarded as P. huashanica 7Ns chromosome-specific molecular markers, with a success rate of up to 86.54%.
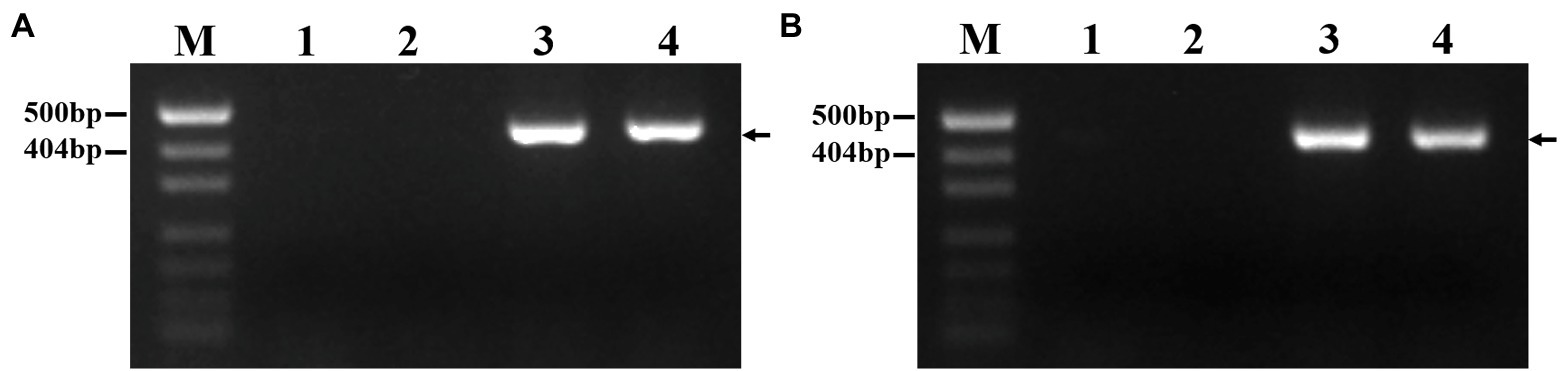
Figure 6. PCR amplification of specific molecular markers. (A) PH7Ns-12, (B) PH7Ns-38. M: Marker (500bp); 1: CS; 2: CSph2b; 3: P. huashanica; 4: DT23 (wheat-P. huashanica ditelosomic addition line). Arrows show the diagnostic amplification products of P. huashanica 7Ns chromosome.
To verify their specificity and stability, the 45 molecular markers were used to amplify sequences from 17 wheat-related species. The PCR amplification results are presented in Supplementary Table 3. Among these markers, two markers amplified sequences only from P. huashanica but not from the wheat-related species (Figure 7A). In contrast, one and two markers amplified specific sequences not only from P. huashanica, but also from L. racemosus and P. juncea, respectively (Figures 7B,C). In addition, one marker amplified a sequence common to P. huashanica, P. juncea, L. arenarius, and L. cinereus, but not from any other wheat-related species (Figure 7D). Thirteen markers amplified specific sequences not only from P. huashanica and P. juncea, but also from six Leymus species (Figure 7E). Furthermore, specific bands were amplified for the other wheat-related species. Five, 3, 4, 5, 6, 9, 5, 11, 15, and 4 markers amplified specific sequences from T. urartu, Aegilops speltoides, Ae. tauschii, Secale cereale, H. vulgare, Agropyron cristatum, Dasypyrum villosum, Th. bessarabicum, Thinopyrum elongatum, and Pseudoroegneria libanotica, respectively.
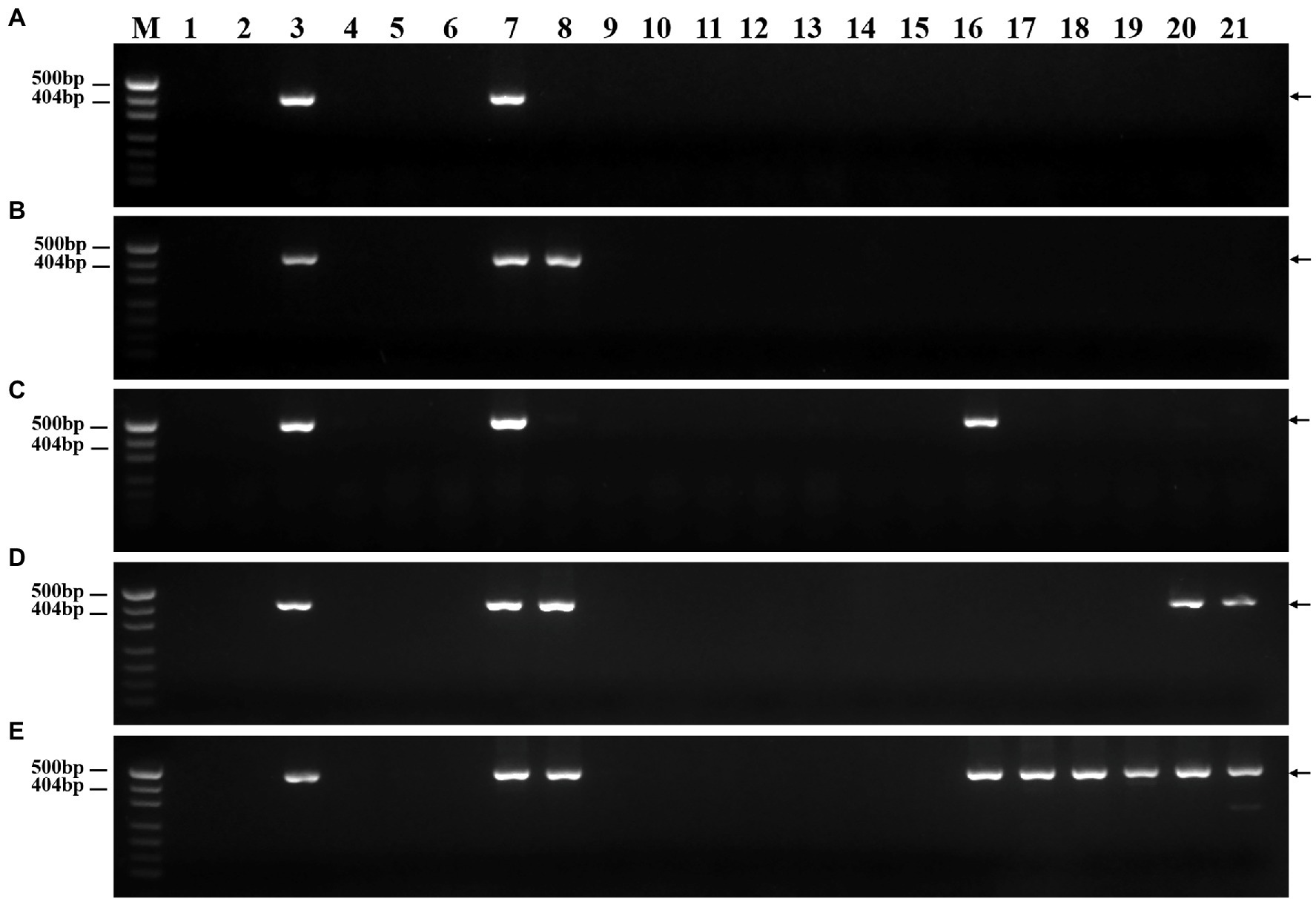
Figure 7. Specificity and stability of specific molecular markers in other wheat-related species. (A) PH7Ns-15, (B) PH7Ns-43, (C) PH7Ns-12, (D) PH7Ns-14, and (E) PH7Ns-5. M: Marker (500bp); 1: CS; 2: CSph2b; 3: DT23 (wheat-P. huashanica ditelosomic addition line); 4: T. urartu; 5: Ae. speltoides; 6: Ae. tauschii; 7: P. huashanica; 8: P. juncea; 9: S. cereale; 10: H. vulgre; 11: Ag. cristatum; 12: Das. villosum; 13: Pse. libanotica; 14: Th. elongatum; 15: Th. bessarabicum; 16: L. racemosus; 17: L. secalinus; 18: L. coreanus; 19: L. multicaulis; 20: L. arenarius; and 21: L. cinereus. Arrows show the diagnostic amplification products of P. huashanica 7Ns chromosome.
FISH Probes Development
To develop P. huashanica-specific FISH probes, PCR products of 19 markers, comprising two markers specific to P. huashanica, two markers specific to P. huashanica and P. juncea, and 15 markers specific to the Ns genome-containing species, were fluorescently labeled as probes for FISH analysis of P. huashanica. A total of 19 P. huashanica-specific FISH probes were developed (Supplementary Table 4). All probes produced strong and distinct hybridization signals in telomeric regions of every chromosome for P. huashanica (Supplementary Figure 1), comprising 10 chromosomes with two telomeric signals and four chromosomes with one telomeric signal, such as pPh15 and pPh37 (Figures 8A,B).
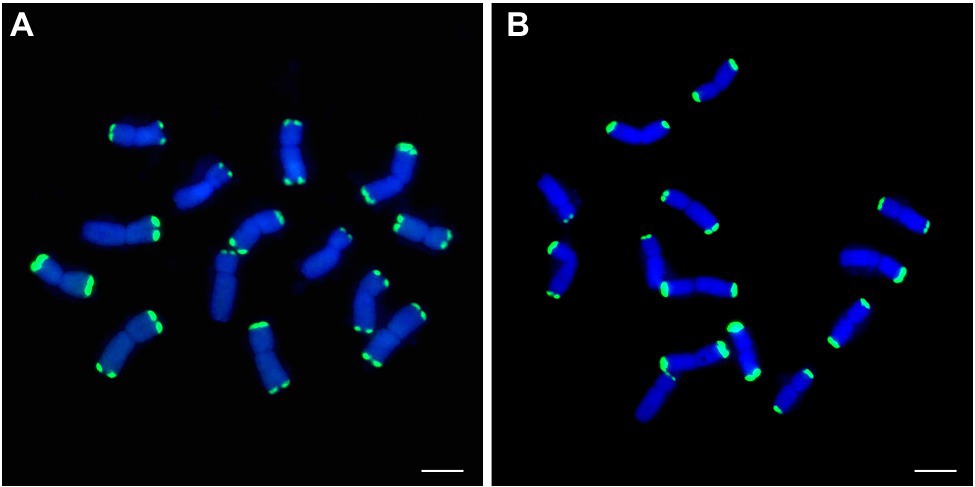
Figure 8. FISH patterns of repetitive DNA probes in P. huashancia. (A) pPh15 (green), (B) pPh37 (green). Scale bar: 10μm.
To assess the utility of these probes, further FISH analysis with probe pPh37 was performed on CS and 11 wheat-related species. P. juncea carried two chromosomes with distinct hybridization signals in telomeric and middle chromosomal arm regions (Figure 9B), Das. villosum, Pse. libanotica, and L. multicaulis carried five, two, and two chromosomes, respectively, with strong hybridization signals in telomeric regions (Figures 9F,G,J). In addition, L. racemosus and L. cinereus had 12 and 50 chromosomes, respectively, with strong hybridization signals in telomeric and subtelomeric regions (Figures 9I,L). No hybridization signals were observed on chromosomes of CS, S. cereale, H. vulgare, Ag. cristatum, Th. elongatum, and L. arenarius (Figures 9A,C–E,H,K).
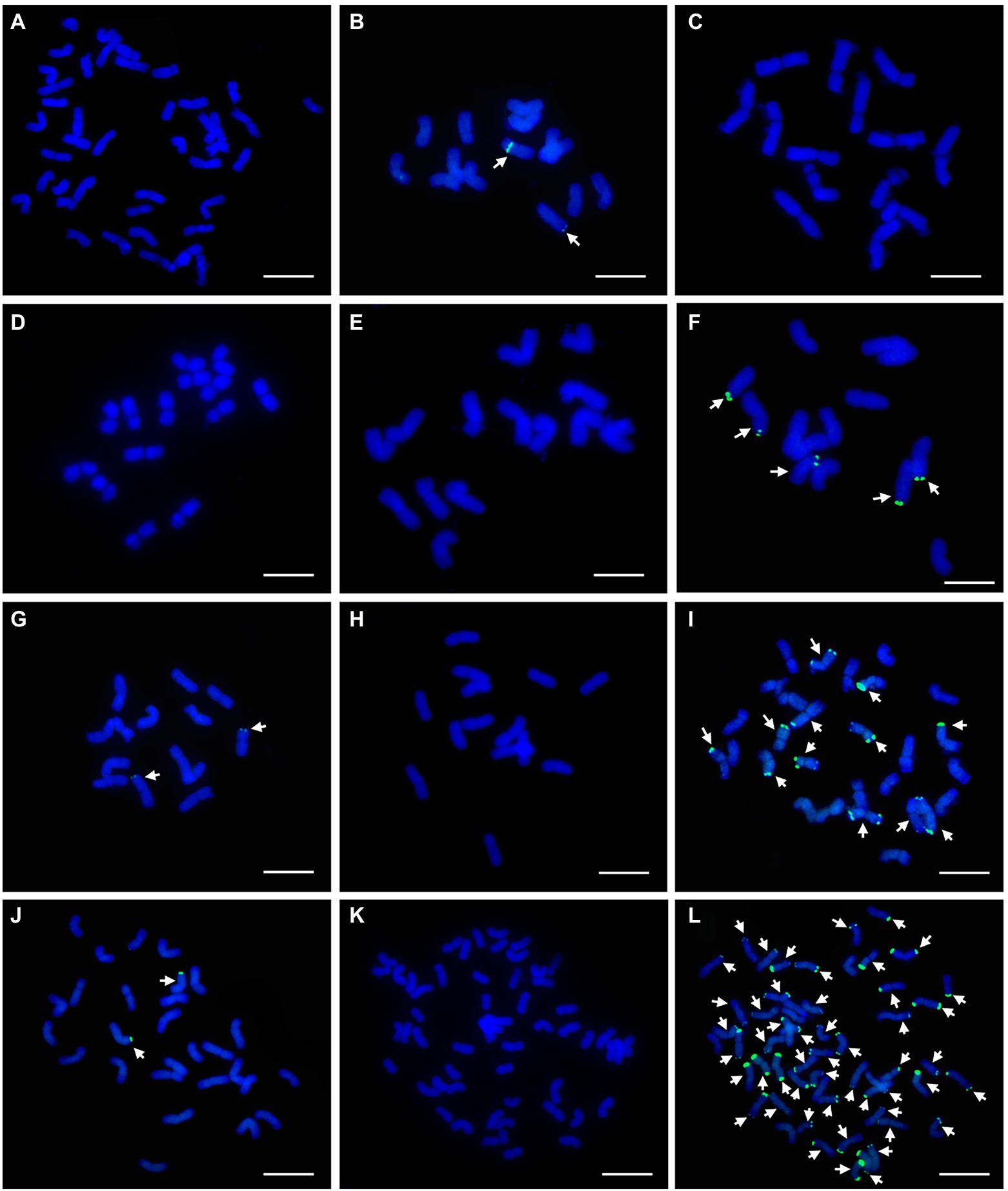
Figure 9. FISH patterns of repetitive DNA probe pPh37 (green) in CS and 11 wheat-related species. (A) CS; (B) P. juncea; (C) S. cereal; (D) H. vulgre; (E) Ag. cristatum; (F) Das. villosum; (G) Pse. libanotica; (H) Th. elongatum; (I) L. racemosus; (J) L. multicaulis; (K) L. arenarius; and (L) L. cinereus. Arrows indicate partial chromosomes with strong hybridization signals. Scale bar: 10μm.
In comparison with the GISH pattern of P. huashanica genomic DNA as a probe (Figure 10A), the probe pPh37 generated strong hybridization signals in telomeric regions of the two alien chromosomal segments in line DT23, but not on common wheat A-, B-, and D-genome chromosomes (Figure 10B). This result indicated that the probes developed in this study could be applied to detect P. huashanica chromosomes or chromosomal segments in a wheat background.
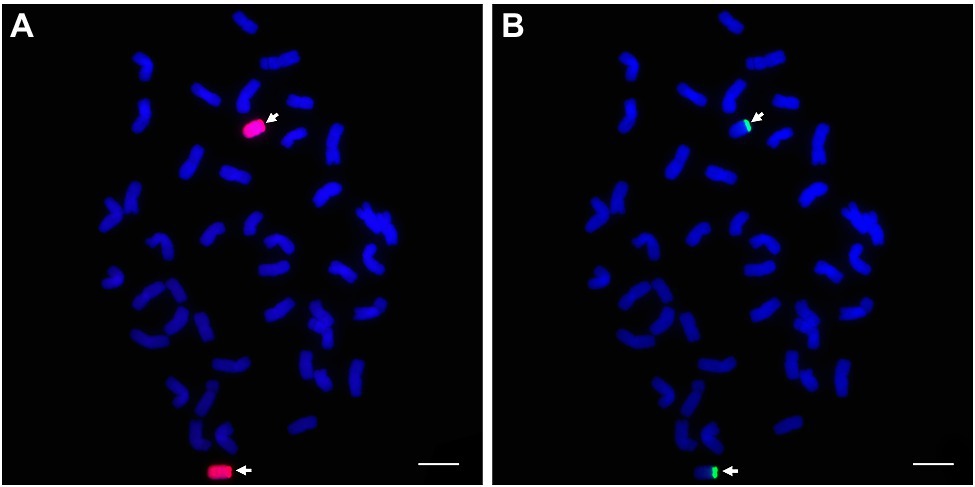
Figure 10. GISH and sequential FISH identification of the wheat-P. huashanica ditelosomic addition line DT23. (A) P. huashanica genomic DNA was used as a probe for GISH (red). (B) FISH identification of DT23 using pPh37 (green). Arrows indicate the introduced P. huashanica chromosomes in DT23. Scale bar: 10μm.
Discussion
Wild relatives of wheat are important for the breeding of new early maturing genotypes and to broaden the genetic base in wheat breeding programs. Early maturing germplasm has previously been generated by crossing common wheat with wild relatives. For instance, Efremova et al. (1996) developed 12 alien 5R(5A) disomic substitution lines with different wheat genetic backgrounds using the alien substitution line “Saratovskaya 29” 5R(5A) as a donor. Efremova et al. (2006) demonstrated that these lines differed in timing of ear emergence and response to vernalization. Bao et al. (2009) developed a new dwarf and early maturing germplasm line Shannong 0057–2 derived from the BC3F6 progeny of the cross between common wheat “Yannong 15” and Th. intermedium. Liu et al. (2011) produced two early maturing disomic addition lines, WB0528 and WB0647, selected from the hybrid progenies between common wheat and cultivated barley. Farkas et al. (2013) suggested that the addition of different barley chromosomes significantly influenced the flowering time of wheat both in controlled environment tests and in the field. The authors reported that the wheat-barley 7H addition line was consistently the earliest flowering, whereas the wheat-barley 4H addition line was the latest to flower in all treatments. Wang et al. (2015) identified a novel wheat-P. huashanica 6Ns disomic addition line that exhibited earlier maturation than its wheat parent and suggested that P. huashanica 6Ns chromosomes significantly affected the spike primordium development and flowering time of wheat in the field. Wang et al. (2017) developed a new germplasm line with early maturation derived from hybridization between common wheat “Yannong 15” and Ae. ventricosa × Ae. cylindrica amphiploid SDAU18. However, few early maturing germplasm lines derived from wide hybrids between common wheat and P. huashanica have been reported previously. In the present study, we developed and characterized a new wheat-P. huashanica 7Ns ditelosomic addition line DT23 derived from the P. huashanica/CSph2b/CS F4 progeny. The maturity stage of DT23 was 14 and 11days earlier than that of the parents CS and CSph2b, respectively. Morphological observation revealed that spike differentiation of DT23 and the parents CS and CSph2b displayed distinct differences after the apex elongation stage. The main reason for the differences was that spike differentiation of DT23 was more rapid than that of CS and CSph2b during the single-ridge to double-ridge stages, thereby giving rise to the subsequent discrepancy in timing of maturity. The pedigree provided the only evidence that DT23 carried novel Eps gene(s) from the P. huashanica 7Ns chromosome. All seven Ns chromosomes of P. huashanica have been verified to be useful on account of the vast number of beneficial genes carried: Stripe rust resistance genes are located on 1Ns, 2Ns, 3Ns, 4Ns, and 5Ns (Du et al., 2014a,c,d; Li et al., 2019; Qu et al., 2021); leaf rust resistance genes are located on 1Ns and 7Ns (Du et al., 2013c, 2014b); powdery mildew resistance genes are located on 1Ns, 3Ns, 4Ns, and 5Ns (Han et al., 2020 Li et al., 2020a 2021a Liu et al., 2021); a take-all resistance gene is located on 2Ns (Bai et al., 2020); gluten and gliadin-related genes are located on 1Ns and 6Ns (Du et al., 2013b; Qu et al., 2021); and early maturity-related genes are located on 6Ns (Wang et al., 2015). The present study is the first report of new Eps gene(s) probably associated with a group 7 chromosome of P. huashanica. Furthermore, the additional chromosomal segments have no obvious genetic linkage drag affecting agronomic performance. Accordingly, the ditelosomic addition line DT23, which exhibits an early maturation phenotype, may represent valuable germplasm for breeding early maturity wheat cultivars.
Molecular markers for detecting and tracking alien chromosomes and/or chromosomal segments carrying elite genes are of vital importance in wheat breeding programs (Fedak, 1999). Conventional methods previously used to develop markers for P. huashanica are time-consuming, imprecise, and expensive, such as random-amplified polymorphic DNA (RAPD) and sequence-characterized amplified region (SCAR) markers. For example, Chen et al. (2010) screened three P. huashanica genome-specific repetitive sequences using 200 RAPD primers. Subsequently, five Ns-specific and 11 5Ns-specific SCAR markers for P. huashanica were developed (Wang et al., 2013; Du et al., 2013d; Wang et al., 2014a,b; Zhang et al., 2017; Li et al., 2020b). The SLAF-seq approach, a high-throughput, high accuracy, low cost, and next-generation sequencing-based technology, has been applied successfully to develop large numbers of highly accurate molecular markers in a variety of wild relatives of wheat, such as Th. elongatum (Chen et al., 2013), Th. intermedium (Li et al., 2016), Th. ponticum (Yang et al., 2021), S. cereale (Du et al., 2018), and Ae. biuncialis (Song et al., 2020). To date, no P. huashanica-specific SLAF-based markers have been reported. In the current study, we developed 45 specific markers for chromosome 7Ns of P. huashanica in the ditelosomic addition line DT23 based on SLAF-seq data, with a success rate of up to 86.54%. These markers may be potentially useful not only for tracking P. huashanica 7Ns chromosomal segments harboring Eps gene(s) in a wheat background, but also for distinguishing the Ns genome of P. huashanica and other closely related genomes from Triticeae species. In addition, marker validation analyses indicated that the marker amplification frequencies in P. juncea (Ns) and Leymus species (NsXm) were much higher than that in other wheat-related species. These findings suggested that the relationship between Psathyrostachys and Leymus is closer than that with other wild relatives, which supported evidence that the Ns genome of Leymus was donated by Psathyrostachys (Zhang and Dvořák, 1991; Sha et al., 2017).
FISH analysis with repetitive DNA probes has been widely used to identify alien chromosomes and/or chromosomal segments integrated into a common wheat background (Cseh et al., 2011; González-García et al., 2011). For instance, Li et al. (2016) produced a novel FISH probe pSt122 representing terminal repeats from Th. intermedium using SLAF-seq. Liu et al. (2018a) generated a blue grain-related FISH probe pThp12.19 from the wheat-Th. ponticum 4Ag (4D) disomic substitution line Blue 58 by SLAF-seq. Liu et al. (2018b) developed eight Th. ponticum-specific FISH probes based on SLAF-seq. In the present study, we developed 19 P. huashanica-specific FISH probes by SLAF-seq, which produced strong and identical fluorescent signals in one or both telomeric regions of all P. huashanica chromosomes, but not in wheat chromosomes. Compared with GISH using genomic DNA of P. huashanica as a probe, FISH analysis with these probes was successfully applied to detect P. huashanica chromosomal segments in DT23. To the best of our knowledge, this is the first report of P. huashanica-specific FISH probes. Furthermore, the probes provide the possibility of distinguishing different P. huashanica chromosomes together with other FISH probes. Therefore, these probes will be convenient and applicable for discrimination of P. huashanica chromosomes and/or chromosomal segments in a wheat background and for identification of chromosomes from other wheat-related species. The present specificity analysis suggested that probe pPh37 generated strong hybridization signals from 12 L. racemosus chromosomes and two L. multicaulis chromosomes, but not L. arenarius chromosomes. These findings revealed that the donor species of the Ns genome to Leymus was not P. huashanica, which was consistent with the inferences of Bödvarsdóttir and Anamthawat-Jónsson (2003) and Wang et al. (2006) based on DNA hybridization and FISH patterns.
Conclusion
Herein, a novel wheat-P. huashanica 7Ns ditelosomic addition line, DT23, was identified by FISH, GISH, PLUG marker, and FISH painting analyses. Compared with the wheat parents, DT23 exhibits earlier maturation. Hence, it can be employed as a valuable intermediate material for breeding early maturing wheat cultivars. In addition, 45 P. huashanica 7Ns chromosome-specific markers and 19 P. huashanica-specific FISH probes were developed based on SLAF-seq. The newly developed markers and probes will be useful for accurate detection of P. huashanica chromosomes and/or chromosomal segments in a wheat background as well as chromosomes from other closely related species.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
BT, LZ, LL, and HK conducted the experiment, analyzed the data, and drafted the manuscript. HZ, WZ, LX, YW, and JZ developed addition line and evaluated morphological traits. XF, LS, HZ, DW, YC, and GC provided technique guidance. YZ and HK designed the experiment and formulated the questions. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 31771781 and 31971883), and the Applied Basic Research Programs of the Science and Technology Bureau of Sichuan Province (2020YJ0348), and the Science and Technology Bureau of Chengdu City (2021-YF05-00681-SN).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. ZJ Yang, University of Electronic Science and Technology of China, Chengdu, China, for kindly providing FISH probes and supplying technique guidance of FISH painting.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.784001/full#supplementary-material
Supplementary Table 1 | Growth period statistics of CS, CSph2b, and DT23 in the greenhouse.
Supplementary Table 2 | PCR amplification results of P. huashanica molecular markers.
Supplementary Table 3 | Specific amplification of 7Ns chromosome markers in wheat-related species.
Supplementary Table 4 | SLAF sequences of the P. huashanica-specific FISH probes.
Supplementary Figure 1 | FISH patterns of 19 repetitive DNA probes in P. huashancia. (A) pPh1, (B) pPh3, (C) pPh5, (D) pPh12, (E) pPh13, (F) pPh14, (G) pPh15, (H) pPh18, (I) pPh19, (J) pPh24, (K) pPh27, (L) pPh32, (M) pPh33, (N) pPh37, (O) pPh38, (P) pPh43, (Q) pPh48, (R) pPh49, and (S) pPh50. All probes are green. Scale bar: 10μm.
Footnotes
References
Alvarez, M. A., Tranquilli, G., Lewis, S., Kippes, N., and Dubcovsky, J. (2016). Genetic and physical mapping of the earliness per se locus Eps-Am1 in Triticum monococcum identifies EARLY FLOWERING 3 (ELF3) as a candidate gene. Funct. Integr. Genomics 16, 365–382. doi: 10.1007/s10142-016-0490-3
Baden, C. (1991). A taxonomic revision of Psathyrostachys (Poaceae). Nord. J. Bot. 11, 3–26. doi: 10.1111/j.1756-1051.1991.tb01790.x
Bai, S. S., Yuan, F. P., Zhang, H. B., Zhang, Z. Y., Zhao, J. X., Yang, Q. H., et al. (2020). Characterization of the wheat-Psathyrostachys huashania Keng 2Ns/2D substitution line H139: A novel germplasm with enhanced resistance to wheat take-all. Front. Plant Sci. 11:233. doi: 10.3389/fpls.2020.00233
Bao, Y. G., Cui, F., Li, X. F., Feng, D. S., Wang, L., Gao, J. R., et al. (2009). Breeding and identification of a dwarf and earliness Trititrigia germplasm Shannong 0057-2. J. Agric. Biotechnol. 17, 684–689. doi: 10.3969/j.issn.1674-7968.2009.04.024
Bi, Y. F., Zhao, Q. Z., Yan, W. K., Li, M. X., Liu, Y. X., Cheng, C. Y., et al. (2020). Flexible chromosome painting based on multiplex PCR of oligonucleotides and its application for comparative chromosome analyses in Cucumis. Plant J. 102, 178–186. doi: 10.1111/tpj.14600
Bödvarsdóttir, S. K., and Anamthawat-Jónsson, K. (2003). Isolation, characterization, and analysis of Leymus-specific DNA sequences. Genome 46, 673–682. doi: 10.1139/g03-029
Chen, S. Q., Huang, Z. F., Dai, Y., Qin, S. W., Gao, Y. Y., Zhang, L. L., et al. (2013). The development of 7E chromosome-specific molecular markers for Thinopyrum elongatum based on SLAF-seq technology. PLoS One 8:e65122. doi: 10.1371/journal.pone.0065122
Chen, L. G., Wu, J., Zhao, J. X., Liu, S. H., Yang, Q. H., Du, W. L., et al. (2010). Screening, cloning and southern blotting analysis of specific repetitive DNA sequences of Psathyrostachys huashanica. J. Triticeae Crops 30, 23–28. doi: 10.1080/00949651003724790
Chen, S. Y., Zhang, A. J., and Fu, J. (1991). The hybridization between Triticum aestivum and Psathyrotachys huashanica. Acta Genet. Sin. 18, 508–512.
Chen, D., Zhang, J. P., Liu, W. H., Wu, X. Y., Yang, X. M., Li, X. Q., et al. (2016). Gene and protein expression profiling analysis of young spike development in large spike wheat germplasms. J. Integr. Agric. 15, 744–754. doi: 10.1016/S2095-3119(15)61179-0
Comadran, J., Kilian, B., Russell, J., Ramsay, L., Stein, N., Ganal, M., et al. (2012). Natural variation in a homolog of antirrhinum CENTRORADIALIScontributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 44, 1388–1392. doi: 10.1038/ng.2447
Cota-Sánchez, J. H., Remarchuk, K., and Ubayasena, K. (2006). Readyto-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Mol. Biol. Report. 24, 161–167. doi: 10.1007/BF02914055
Cseh, A., Kruppa, K., Molnár, I., Rakszegi, M., Doležel, J., and Molnár-Láng, M. (2011). Characterization of a new 4BS.7HL wheat-barley translocation line using GISH, FISH, and SSR markers and its effect on the β-glucan content of wheat. Genome 54, 795–804. doi: 10.1139/g11-044
Cui, J. M., Wang, Y. H., and Wang, H. C. (2008). “Differentiation and formation of winter wheat spikes,” in The Spike of Wheat. eds. Cui, J. M., and Guo, T. C.. (Beijing: China Agriculture Press), 22–33.
Du, H. M., Tang, Z. X., Duan, Q., Tang, S. Y., and Fu, S. L. (2018). Using the 6RLKu minichromosome of rye (Secale cereale L.) to create wheat-rye 6D/6RLKu small segment translocation lines with powdery mildew resistance. Int. J. Mol. Sci. 19:3933. doi: 10.3390/ijms19123933
Du, W. L., Wang, J., Lu, M., Sun, S. G., Chen, X. H., Zhao, J. X., et al. (2013a). Molecular cytogenetic identification of a wheat- Psathyrostachys huashanica Keng 5Ns disomic addition line with stripe rust resistance. Mol. Breed. 31, 879–888. doi: 10.1007/s11032-013-9841-0
Du, W. L., Wang, J., Lu, M., Sun, S. G., Chen, X. H., Zhao, J. X., et al. (2014a). Characterization of a wheat-Psathyrostachys huashanica Keng 4Ns disomic addition line for enhanced tiller numbers and stripe rust resistance. Planta 239, 97–105. doi: 10.1007/s00425-013-1957-2
Du, W. L., Wang, J., Pang, Y. H., Li, Y. L., Chen, X. H., Zhao, J. X., et al. (2013b). Isolation and characterization of a Psathyrostachys huashanica Keng 6Ns chromosome addition in common wheat. PLoS One 8:e53921. doi: 10.1371/journal.pone.0053921
Du, W. L., Wang, J., Pang, Y. H., Wu, J., Zhao, J. X., Liu, S. H., et al. (2014b). Development and application of PCR markers specific to the 1Ns chromosome of Psathyrostachys huashanica Keng with leaf rust resistance. Euphytica 200, 207–220. doi: 10.1007/s10681-014-1145-x
Du, W. L., Wang, J., Pang, Y. H., Wu, J., Zhao, J. X., Yang, Q. H., et al. (2014c). Isolation and characterization of a wheat-Psathyrostachys huashanica Keng 3Ns disomic addition line with resistance to stripe rust. Genome 57, 37–44. doi: 10.1139/gen-2013-0199
Du, W. L., Wang, J., Wang, L. M., Pang, Y. H., Wu, J., Zhao, J. X., et al. (2013d). A novel SCAR marker for detecting Psathyrostachys huashanica Keng chromatin introduced in wheat. Genet. Mol. Res. 12, 4797–4806. doi: 10.4238/2013.October.18.16
Du, W. L., Wang, J., Wang, L. M., Wu, J., Zhao, J. X., Liu, S. H., et al. (2014d). Molecular characterization of a wheat-Psathyrostachys huashanica Keng 2Ns disomic addition line with resistance to stripe rust. Mol. Gen. Genomics. 289, 735–743. doi: 10.1007/s00438-014-0844-2
Du, W. L., Wang, J., Wang, L. M., Zhang, J., Chen, X. H., Zhao, J. X., et al. (2013c). Development and characterization of a Psathyrostachys huashanica Keng 7Ns chromosome addition line with leaf rust resistance. PLoS One 8:e70879. doi: 10.1371/journal.pone.0070879
Efremova, T. T., Maystrenko, O. I., Arbuzova, V. S., Laykova, L. I., Panina, G. M., Popova, O. M., et al. (2006). Effect of alien 5R(5A) chromosome substitution on ear-emergence time and winter hardiness in wheat-rye substitution lines. Euphytica 151, 145–153. doi: 10.1007/s10681-006-9135-2
Efremova, T. T., Maystrenko, O. I., and Laykova, L. I. (1996). Development of alien substitution lines of wheat with rye chromosome 5R. Cereal Res. Commun. 24, 33–39. doi: 10.1007/bf02849301
Farkas, A., Molnár, I., Kiss, T., Karsai, I., and Molnár-Láng, M. (2013). Effect of added barley chromosomes on the flowering time of new wheat/winter barley addition lines in various environments. Euphytica 195, 45–55. doi: 10.1007/s10681-013-0970-7
Fedak, G. (1999). Molecular aids for integration of alien chromatin through wide crosses. Genome 42, 584–591. doi: 10.1139/g99-046
Gawroński, P., Ariyadasa, R., Himmelbach, A., Poursarebani, N., Kilian, B., Stein, N., et al. (2014). A distorted circadian clock causes early flowering and temperature-dependent variation in spike development in the Eps-3Am mutant of einkorn wheat. Genetics 196, 1253–1261. doi: 10.1534/genetics.113.158444
Gawroński, P., and Schnurbusch, T. (2012). High-density mapping of the earliness per se-3Am (Eps-3Am) locus in diploid einkorn wheat and its relation to the syntenic regions in rice and Brachypodium distachyon L. Mol. Breed. 30, 1097–1108. doi: 10.1007/s11032-011-9697-0
Gong, B. R., Zhu, W., Li, S. Y., Wang, Y. Q., Xu, L. L., Wang, Y., et al. (2019). Molecular cytogenetic characterization of wheat-Elymus repens chromosomal translocation lines with resistance to Fusarium head blight and stripe rust. BMC Plant Biol. 19:590. doi: 10.1186/s12870-019-2208-x
González-García, M., Cuacos, M., González-Sánchez, M., Puertas, M. J., and Vega, J. M. (2011). Painting the rye genome with genome-specific sequences. Genome 54, 555–564. doi: 10.1139/g11-003
Guo, W. S., and Fan, G. Q. (2011). “Wheat,” in Treatise on Crop Cultivation eds. Yang, W. Y., and Tu, N. M., (Beijing: China Agriculture Press), 78.
Han, F. P., Lamb, J. C., and Birchler, J. A. (2006). High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. PNAS 103, 3238–3243. doi: 10.1073/pnas.0509650103
Han, J., Liu, Y. X., Hou, C. C., Li, J. C., Wang, J. L., Zhang, Q. Y., et al. (2020). A 1Ns disomic addition from Psathyrostachys huashanica Keng confers resistance to powdery mildew in wheat. Agronomy 10, 312. doi: 10.3390/agronomy10020312
Han, Y. H., Zhang, T., Thammapichai, P., Weng, Y. Q., and Jiang, J. M. (2015). Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 200, 771–779. doi: 10.1534/genetics.115.177642
Hunger, R. M., Edwards, J. T., Bowden, R. L., Yan, L., Rayas-Duarte, P., Bai, G., et al. (2014). ‘Billings’ wheat combines early maturity, disease resistance, and desirable grain quality for the southern Great Plains, USA. J. Plant Regist. 8, 22–31. doi: 10.3198/jpr2012.11.0053crc
Iqbal, M., Navabi, A., Salmon, D. F., Yang, R. C., and Spaner, D. (2006). A genetic examination of early flowering and maturity in Canadian spring wheat. Can. J. Plant Sci. 86, 995–1004. doi: 10.4141/P06-002
Ishikawa, G., Nakamura, T., Ashida, T., Saito, M., Nasuda, S., Endo, T. R., et al. (2009). Localization of anchor loci representing five hundred annotated rice genes to wheat chromosomes using PLUG markers. Theor. Appl. Genet. 118, 499–514. doi: 10.1007/s00122-008-0916-y
Kamran, A., Iqbal, M., and Spaner, D. (2014). Flowering time in wheat (Triticum aestivum L.): a key factor for global adaptability. Euphytica 197, 1–26. doi: 10.1007/s10681-014-1075-7
Kang, H. Y., Wang, Y., Sun, G. L., Zhang, H. Q., Fan, X., and Zhou, Y. H. (2009). Production and characterization of an amphiploid between common wheat and Psathyrostachys huashanica Keng ex Kuo. Plant Breed. 128, 36–40. doi: 10.1111/j.1439-0523.2008.01542.x
Kang, H. Y., Zhang, H. Q., Fan, X., and Zhou, Y. H. (2008). Morphological and cytogenetic studies on the hybrid between bread wheat and Psathyrostachys huashanica Keng ex Kuo. Euphytica 162, 441–448. doi: 10.1007/s10681-007-9608-y
Kang, H. Y., Zhang, Z. J., Xu, L. L., Qi, W. L., Wang, H., Zhu, W., et al. (2016). Characterization of wheat-Psathyrostachys huashanica small segment translocation line with enhanced kernels per spike and stripe rust resistance. Genome 59, 221–229. doi: 10.1139/gen-2015-0138
Kishii, M., Dou, Q. W., Garg, M., Ito, M., Tanaka, H., and Tsujimoto, H. (2010). Production of wheat-Psathyrostachys huashanica chromosome addition lines. Genes Genet. Syst. 85, 281–286. doi: 10.1266/ggs.85.281
Komuro, S., Endo, R., Shikata, K., and Kato, A. (2013). Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56, 131–137. doi: 10.1139/gen-2013-0003
Laurie, D. A., Pratchett, N., Snape, J. W., and Bezant, J. H. (1995). RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter × spring barley (Hordeum vulgare L.) cross. Genome 38, 575–585. doi: 10.1139/g95-074
Li, J. C., Li, J. J., Cheng, X. N., Zhao, L., Yang, Z. J., Wu, J., et al. (2021a). Molecular cytogenetic and agronomic characterization of the similarities and differences between wheat-Leymus mollis Trin. And wheat-Psathyrostachys huashanica Keng 3Ns(3D) substitution lines. Front. Plant Sci. 12:644896. doi: 10.3389/fpls.2021.644896
Li, J. C., Liu, Y., Cheng, X. N., Yao, X. N., Yang, Z. J., Wu, J., et al. (2020b). Molecular characteristics and inheritance of a chromosome segment from Psathyrostachys huashanica Keng in a wheat background. Genet. Resour. Crop. Evol. 67, 1245–1257. doi: 10.1007/s10722-020-00908-5
Li, G. R., Wang, H. J., Lang, T., Li, J. B., La, S. X., Yang, E. N., et al. (2016). New molecular markers and cytogenetic probes enable chromosome identification of wheat-Thinopyrum intermedium introgression lines for improving protein and gluten contents. Planta 244, 865–876. doi: 10.1007/s00425-016-2554-y
Li, J. C., Yao, X. N., Yang, Z. J., Cheng, X. N., Yuan, F. P., Liu, Y., et al. (2019). Molecular cytogenetic characterization of a novel wheat-Psathyrostachys huashanica Keng 5Ns (5D) disomic substitution line with stripe rust resistance. Mol. Breed. 39:109. doi: 10.1007/s11032-019-1014-3
Li, G. R., Zhang, T., Yu, Z. H., Wang, H. J., Yang, E. N., and Yang, Z. J. (2021b). An efficient Oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae. Plant J. 105, 978–993. doi: 10.1111/TPJ.15081
Li, J. C., Zhao, L., Cheng, X. N., Bai, G. H., Li, M., Wu, J., et al. (2020a). Molecular cytogenetic characterization of a novel wheat-Psathyrostachys huashanica Keng T3DS-5NsL•5NsS and T5DL-3DS•3DL dual translocation line with powdery mildew resistance. BMC Plant Biol. 20:163. doi: 10.1186/s12870-020-02366-8
Liu, S. H., Chen, X. H., Cai, Z. H., Wu, J., Zhao, J. X., and Yang, Q. H. (2011). Selection and identification of early mature derive lines from common wheat× cultivated barley hybrid progenies. J. Shanxi Agric. Univ. 31, 142–145. doi: 10.3969/j.issn.1671-8151.2011.02.011
Liu, Y. X., Huang, S. H., Han, J., Hou, C. C., Zheng, D. S., Zhang, Z. M., et al. (2021). Development and molecular cytogenetic identification of a new wheat-Psathyrostachys huashanica Keng translocation line resistant to powdery mildew. Front. Plant Sci. 12:689502. doi: 10.3389/fpls.2021.689502
Liu, L. Q., Luo, Q. L., Li, H. W., Li, B., Li, Z. S., and Zheng, Q. (2018a). Physical mapping of the blue-grained gene from Thinopyrum ponticumchromosome 4Ag and development of blue-grain-related molecular markers and a FISH probe based on SLAF-seq technology. Theor. Appl. Genet. 131, 2359–2370. doi: 10.1007/s00122-018-3158-7
Liu, L. Q., Luo, Q. L., Teng, W., Li, B., Li, H. W., Li, Y. W., et al. (2018b). Development of Thinopyrum ponticum-specific molecular markers and FISH probes based on SLAF-seq technology. Planta 247, 1099–1108. doi: 10.1007/s00425-018-2845-6
Lombardo, L. A., Ghione, C. E., Nisi, M. M., Tranquilli, G. E., and Helguera, M. (2019). Earliness per se variation in argentine bread wheat varieties and mapping of QTLs in a recombinant inbred line population. Euphytica 215, 1–17. doi: 10.1007/s10681-019-2474-6
Mahdiyeh, Z. K., and Bahram, H. (2012). Estimation of genetic parameters for maturity and grain yield in diallel crosses of five wheat cultivars using two different models. J. Agric. Sci. 4:74. doi: 10.5539/jas.v4n8p74
Mondal, S., Singh, R. P., Crossa, J., Huerta-Espino, J., Sharma, I., Chatrath, R., et al. (2013). Earliness in wheat: a key to adaptation under terminal and continual high temperature stress in South Asia. Field Crop Res. 151, 19–26. doi: 10.1016/j.fcr.2013.06.015
Ochagavía, H., Prieto, P., Savin, R., Griffiths, S., and Slafer, G. A. (2018). Earliness per se effects on developmental traits in hexaploid wheat grown under field conditions. Eur. J. Agron. 99, 214–223. doi: 10.1016/j.eja.2018.07.007
Qu, X. J., Zhang, D. S., Zhang, X. Y., Wang, S. W., Wang, C. Y., Wang, Y. Z., et al. (2021). Cytogenetic and marker assisted identification of a wheat-Psathyrostachys huashanica Keng f. ex P.C.Kuo alien substitution line conferring processing quality and resistance to stripe rust. Genet. Resour. Crop. Evol., 1–12. doi: 10.1007/s10722-021-01253-x
Sears, E. R. (1982). A wheat mutation conditioning an intermediate level of homoeologous chromosome pairing. Can. J. Genet. Cytol. 24, 715–719. doi: 10.1139/g82-076
Sha, L. N., Fan, X., Li, J., Liao, J. Q., Zeng, J., Wang, Y., et al. (2017). Contrasting evolutionary patterns of multiple loci uncover new aspects in the genome origin and evolutionary history of Leymus (Triticeae; Poaceae). Mol. Phylogenet. Evol. 114, 175–188. doi: 10.1016/j.ympev.2017.05.015
Sheehan, H., and Bentley, A. (2020). Changing times: opportunities for altering winter wheat phenology. Plants People Planet 3, 113–123. doi: 10.1002/ppp3.10163
Shi, C. N., Zhao, L., Zhang, X. F., Lv, G. G., Pan, Y. B., and Chen, F. (2019). Gene regulatory network and abundant genetic variation play critical roles in heading stage of polyploidy wheat. BMC Plant Biol. 19:6. doi: 10.1186/s12870-018-1591-z
Song, L. Q., Zhao, H., Zhang, Z., Zhang, S., Liu, J. J., Zhang, W., et al. (2020). Molecular cytogenetic identification of wheat-Aegilops Biuncialis 5Mb disomic addition line with tenacious and black glumes. Int. J. Mol. Sci. 21:4053. doi: 10.3390/ijms21114053
Su, J. J., Ma, Q., Li, M., Hao, F. S., and Wang, C. (2018). Multi-locus genome-wide association studies of fiber-quality related traits in Chinese early-maturity upland cotton. Front. Plant Sci. 9:1169. doi: 10.3389/fpls.2018.01169
Sun, X. W., Liu, D. Y., Zhang, X. F., Li, W. B., Liu, H., Hong, W. G., et al. (2013). SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One 8:e58700. doi: 10.1371/journal.pone.0058700
Tang, Z. X., Yang, Z. J., and Fu, S. L. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Wall, A. M., Riley, R., and Chapman, V. (1971). Wheat mutants permitting homoeologous meiotic chromosome pairing. Genet. Res. 18, 311–328. doi: 10.1017/S0016672300012714
Wang, J., Du, W. L., Wu, J., Chen, X. H., Liu, C. J., Zhao, J. X., et al. (2014a). Development of a specific SCAR marker for the Ns genome of Psathyrostachys huashanica Keng. Can. J. Plant Sci. 94, 1441–1447. doi: 10.4141/cjps-2014-076
Wang, Y. H., Han, Q. D., He, F., Bao, Y. G., Ming, D. F., and Wang, H. G. (2017). Characterization of a Triticum aestivum-Aegilops germplasm line presenting reduced plant height and early maturation. Crop J. 5, 185–194. doi: 10.1016/j.cj.2016.10.004
Wang, L. M., Liu, Y., Du, W. L., Jing, F., Wang, Z. H., Wu, J., et al. (2015). Anatomy and cytogenetic identification of a wheat-Psathyrostachys huashanica Keng line with early maturation. PLoS One 10:e0131841. doi: 10.1371/journal.pone.0131841
Wang, J., Lu, M., Du, W. L., Zhang, J., Dong, X. Y., Wu, J., et al. (2013). A novel PCR-based marker for identifying Ns chromosomes in wheat-Psathyrostachys huashanica Keng derivative lines. Span. J. Agric. Res. 11, 1094–1100. doi: 10.5424/sjar/2013114-4245
Wang, J., Wang, L. M., Du, W. L., Chen, L. G., Liu, S. H., Wu, J., et al. (2014b). Development of 5Ns chromosome-specific SCAR markers for utilization in future wheat breeding programs. Russ. J. Genet. 50, 606–612. doi: 10.1134/s1022795414060131
Wang, Y., Yu, K., Xie, Q., Kang, H. Y., Lin, L., Fan, X., et al. (2011). The 3Ns chromosome of Psathyrostachys huashanica carries the gene(s) underlying wheat stripe rust resistance. Cytogenet. Genome Res. 134, 136–143. doi: 10.1159/000324928
Wang, R. R., Zhang, J. Y., Lee, B. S., Jensen, K. B., Kishii, M., and Tsujimoto, H. (2006). Variations in abundance of 2 repetitive sequences in Leymus and Psathyrostachys species. Genome 49, 511–519. doi: 10.1139/g05-126
Yang, G. T., Boshoff, W. H. P., Li, H. W., Pretorius, Z. A., Luo, Q. L., Li, B., et al. (2021). Chromosomal composition analysis and molecular marker development for the novel Ug99-resistant wheat-Thinopyrum ponticum translocation line WTT34. Theor. Appl. Genet. 134, 1587–1599. doi: 10.1007/s00122-021-03796-0
Yang, X. F., Wang, C. Y., Chen, C. H., Tian, Z. R., and Ji, W. Q. (2020). Development and characterization of a wheat-Leymus mollis lm#7Ns disomic addition line with resistance to stripe rust. Cereal Res. Commun. 48, 467–476. doi: 10.1007/s42976-020-00058-5
Zhang, H. B., and Dvořák, J. (1991). The genome origin of tetraploid species of Leymus (Poaceae: Triticeae) inferred from variation in repeated nucleotide sequences. Am. J. Bot. 78, 871–884. doi: 10.1002/j.1537-2197.1991.tb14491.x
Zhang, J., Jiang, Y., Guo, Y. L., Li, Y. H., Wang, Y., and Xuan, P. (2017). Cloning of Ns genome-specific sequence of Psathyrostachys huashanica and construction of molecular markers. J. Agric. Biotechnol. 25, 1391–1399. doi: 10.3969/j.issn.1674-7968.2017.09.002
Zhao, J. X., Chen, X. H., Wang, X. L., Wu, J., Fu, J., He, B. R., et al. (2004). Molecular cytogenetic study on the alien substitution lines of Triticum-Psathyrostachys. Acta Bot. Boreal. Occident. Sin. 24, 2277–2281. doi: 10.3321/j.issn:1000-4025.2004.12.015
Zhao, J. X., Ji, W. Q., Wu, J., Chen, X. H., Cheng, X. N., Wang, J. W., et al. (2010). Development and identification of a wheat-Psathyrostachys huashanica addition line carrying HMW-GS, LMW-GS and gliadin genes. Genet. Resour. Crop. Evol. 57, 387–394. doi: 10.1007/s10722-009-9477-4
Keywords: Psathyrostachys huashanica, ditelosomic addition line, early maturation, SLAF-seq, molecular marker, FISH probes
Citation: Tan B, Zhao L, Li L, Zhang H, Zhu W, Xu L, Wang Y, Zeng J, Fan X, Sha L, Wu D, Cheng Y, Zhang H, Chen G, Zhou Y and Kang H (2021) Identification of a Wheat-Psathyrostachys huashanica 7Ns Ditelosomic Addition Line Conferring Early Maturation by Cytological Analysis and Newly Developed Molecular and FISH Markers. Front. Plant Sci. 12:784001. doi: 10.3389/fpls.2021.784001
Edited by:
Vijay Kumar Tiwari, University of Maryland, College Park, United StatesCopyright © 2021 Tan, Zhao, Li, Zhang, Zhu, Xu, Wang, Zeng, Fan, Sha, Wu, Cheng, Zhang, Chen, Zhou and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Houyang Kang, aG91eWFuZy5rYW5nQHNpY2F1LmVkdS5jbg==
†These authors have contributed equally to this work
 Binwen Tan1,2†
Binwen Tan1,2† Yi Wang
Yi Wang Jian Zeng
Jian Zeng Lina Sha
Lina Sha Haiqin Zhang
Haiqin Zhang Yonghong Zhou
Yonghong Zhou Houyang Kang
Houyang Kang