- 1Independent Researcher, Bangalore, India
- 2Department of Botany, St. Joseph’s College Autonomous, Bangalore, India
- 3Sericultural Research Institute, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
Salt stress affects the plant growth and productivity worldwide and NHX is one of those genes that are well known to improve salt tolerance in transgenic plants. It is well characterized in several plants, such as Arabidopsis thaliana and cotton; however, not much is known about NHXs in tea plant. In the present study, NHX genes of tea were obtained through a genome-wide search using A. thaliana as reference genome. Out of the 9 NHX genes in tea, 7 genes were localized in vacuole while the remaining 2 genes were localized in the endoplasmic reticulum (ER; CsNHX8) and plasma membrane (PM; CsNHX9), respectively. Furthermore, phylogenetic relationships along with structural analysis which includes gene structure, location, and protein-conserved motifs and domains were systematically examined and further, predictions were validated by the expression analysis. The dN/dS values show that the majority of tea NHX genes is subjected to strong purifying selection under the course of evolution. Also, functional interaction was carried out in Camellia sinensis based on the orthologous genes in A. thaliana. The expression profiles linked to various stress treatments revealed wide involvement of NHX genes from tea in response to various abiotic factors. This study provides the targets for further comprehensive identification, functional study, and also contributed for a better understanding of the NHX regulatory network in C. sinensis.
Background
Excessive use of inorganic fertilizers is making the land infertile and unavailable for agriculture due to over accumulation of salts in it (Kovda, 1983). Moreover, abiotic stresses, such as drought and heat stress, cause an additive effect and overall decrease the crop yield and quality (Zhu, 2001). Therefore, to keep up with growing demands of population, there is a pressing need to identify and characterize more salt tolerant genes from different plant species and use them for improvement of salt tolerance in crop plants.
Sodium chloride is one of major salts present in the soil and most of the salt tolerance mechanism focuses on the transport and compartmentalization of sodium ions. Na+ influx is controlled by either sodium/hydrogen antiporter (NHX) family of cation/H+ transporters (Apse et al., 1999) or nonselective cation channels (NSCCs), or high-affinity K+ transporters (HKTs; Waters et al., 2013). HKT can regulate the long-distance transport of Na+ (Rubio et al., 1995) while Na+/H+ antiporter (NHX) is involved in the transport of Na+ ions from cytoplasm to vacuole or outside of the cell. To achieve this, it utilizes the H+ electrochemical gradient formed by two proton pumps, i.e., H+-ATPase and H+-PPase thereby avoiding the cell from the toxic effects of sodium ions (Apse et al., 1999).
NHX proteins belong to the cation/proton antiporter 1 (CPA1) superfamily and most of NHX proteins possess 10 transmembrane helices (Yamaguchi et al., 2003; Brett et al., 2005; Chanroj et al., 2012; Wu et al., 2019b). Localization of NHX proteins is mainly restricted to plasma membranes, vacuoles, and endosomes (Aharon et al., 2003; Pehlivan et al., 2016). The first plant NHX gene was recognized in barley root tips (Ratner and Jacoby, 1976) followed by its identification and characterization in Arabidopsis thaliana (At; Roberto et al., 1999), and a total of 8 NHX genes have been reported in A. thaliana till date. Out of 8 NHXs in A. thaliana, 2 genes (AtNHX7 and AtNHX8) belong to PM-class (plasma membranes), 2 genes (AtNHX5 and AtNHX6) belong to Endo-class (endosomes), and 4 genes (AtNHX1-4) belong to Vac-class (vacuoles). This classification is done on the basis of their subcellular localization (Shi et al., 2000; Aharon et al., 2003; Brett et al., 2005; Bassil et al., 2011b). Apart from the involvement of these genes in salt tolerance, NHX antiporters are involved in the regulation of wide variety of physiological processes, such as vesicle trafficking, pH regulation, K+ homeostasis, protein transport, and growth/development (Pardo et al., 2006; Rodriguez Rosales et al., 2009; Bassil et al., 2012; Reguera et al., 2014).
Camellia sinensis is native to East Asia, the Indian Subcontinent, and Southeast Asia, but it is today cultivated across the world in tropical and subtropical regions. Tea plant (C. sinensis L.) is an important economic crop, leaves of which are an important source of non-alcoholic beverage. As a leaf-harvested crop, tea plant is unavoidably threatened with various adverse environment stresses throughout the whole life cycle, such as drought (Xie et al., 2019), salt (Wan et al., 2018), and cold (Li et al., 2018) stresses, which critically hinders the development of the tea industry. With drastic environmental changes leading to a decline in the cultivated land area, like many other economic crops, tea planting fields are moving to salinity and drought-affected areas. In this study, we performed a genome-wide analysis of NHX genes in C. sinensis including the phylogenetic relationships, a motif analysis, promoter analysis, gene expression pattern, and the gene structures. Through a systematic analysis of all the members of the NHX gene, we can understand the gene regulation, expression pattern, and eventually their biological functions in tea.
Materials and Methods
Identification of NHX Genes of Tea Plant
The tea plant genome sequence was recovered from the Tea Plant Information Archive, TPIA (Xia et al., 2019).1 The NHX genes from A. thaliana and rice were retrieved from TAIR database (Berardini et al., 2015)2 and Rice Genome Annotation Project database (Kawahara et al., 2013)3, respectively. These sequences were then used as a query sequences to scan the tea genome database using the BLASTp algorithm with an e-value of 1e-5 and an identity match of 50% as the threshold. To further confirm the presence of Na+/H+_Exchanger domain, the NHX genes were submitted to SMART (Letunic et al., 2015)4 and Pfam web tool. ProtParam tool integrated in ExPASy database was used to predict the physicochemical properties of the NHX peptides (Gasteiger et al., 2005).5 BaCello (Balanced subcellular localization predictor) online server was used to predict the subcellular localization of the protein sequences (Pierleoni et al., 2006).6 Additionally, TMHMM server v2.07 was used to predict the transmembrane helices in NHX peptide sequences (Sonnhammer et al., 1998).
Phylogenetic Analysis of NHX Genes
The NHX peptide sequences from C. sinensis (Cs), A. thaliana (At), Oryza sativa (Os), Solanum lycopersicum (Sl), Solanum tuberosum (St), Medicago truncatula (Mt), Populus trichocarpa (Pt), Gossypium hirsutum (Gh), Sorghum bicolor (Sb), Zea mays (Zm), and Glycine max (Gm) were aligned by using MUSCLE (Robert, 2004), with default parameters. The aligned sequences were then used to generate the phylogenetic tree using MEGA7.0.14 software (Kumar et al., 2016). The tree was constructed using Neighbor-Joining (NJ) algorithm with default parameters. The reliability of the phylogenetic tree was analyzed by the bootstrap method and replicates were set to 1,000.
Conserved Motif and Gene Structure Analysis
In order to identify the conserved motifs, the MEME (Bailey et al., 2009)8 suite was used with default parameters. The intron/exon distribution pattern of NHX genes was obtained and then analyzed using the gene structure display server V2.0 (Hu et al., 2015).9
Analysis of Cis-Regulatory Elements
The promoter sequences of 2,000 bp of the tea NHX genes were retrieved from the TPIA database to analyze the cis-acting regulatory elements (CAREs). The PlantCARE program10 (Rombauts et al., 1999; Lescot et al., 2002) was used for identifying and analyzing the CAREs.
Genomic Distribution of NHX Genes and Ka/Ks Ratios
Due to the incomplete genome assembly information available in the TPIA database, the NHX genes were mapped into their corresponding scaffolds. MapGene2chromosome web v2 (MG2C) server (Jiangtao et al., 2015)11 was used to map the genes into their scaffolds based on their positional information in the TPIA database, which includes scaffold length, number, gene ID, starting and ending position of the genes, and scaffold ID. Further, the dN (Ka) and dS (Ks) ratios were evaluated using the SNAP v.2.1.1 online tool (Korber, 2000)12 to assess the synonymous and non-synonymous groups. The dS values represent the time of divergence of duplication events and the dN/dS values represent the selective pressure of duplicate genes.
GO Ontology Annotation and Functional Interaction Network
QuickGO13 was used to perform GO Ontology (GO) analysis for all the 9 tea NHX genes. Furthermore, the network of functionally interacting homologous genes between tea and A. thaliana was identified and constructed using STRING online tool (Szklarczyk et al., 2019)14 with default parameters.
Expression Profile of Tea NHXs
The tissue-specific expression profiles in 8 plant tissues, which include expression levels in apical bud, flower, fruit, young leaf, mature leaf, old leaf, root, and stem, were retrieved from TPIA database and analyzed (Xia et al., 2019). Furthermore, gene expression data under cold, drought, and salt stresses were analyzed to understand the potential role of tea NHXs in response to the abiotic stress factors. Additionally, to check the effects of methyl-jasmonate (MeJA) treatment, its expression data were retrieved from TPIA database and analyzed for the 9 tea NHXs. Respective graphs for the gene expression for all the tea NHX genes were generated. Heat maps for the same were generated using heatmapper online server (Babicki et al., 2016; Heatmapper).
Results
Genome-Wide Identification of NHX Genes in C. sinensis
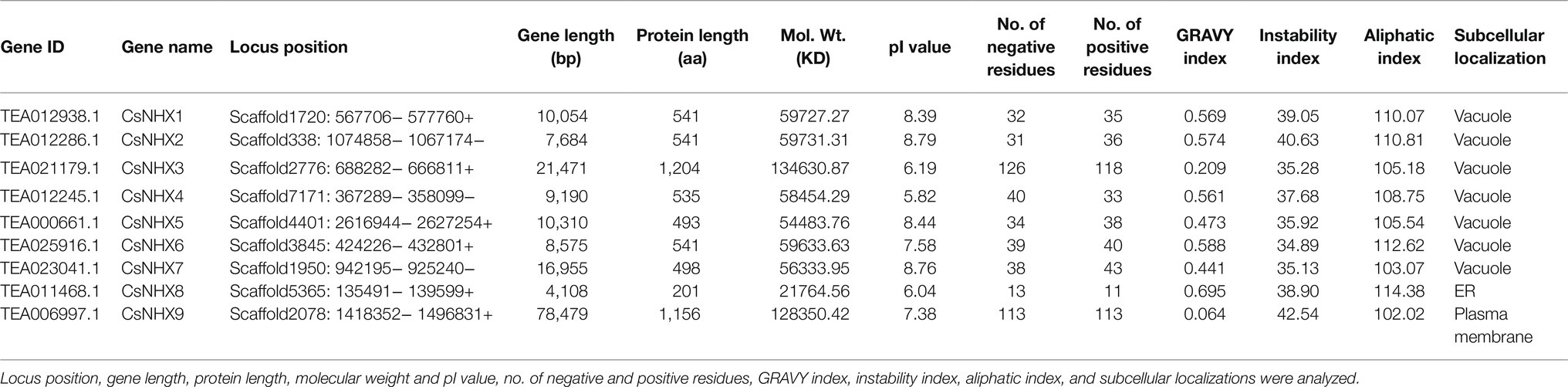
In order to retrieve the members of the NHX gene family in tea, the published NHX protein sequences of A. thaliana (8) and rice (7) were retrieved from TAIR database (see footnote 2) and Rice Genome Annotation Project database (see footnote 3), respectively. These peptide sequences were then used as queries to search against the genome database of tea, Tea Plant Information Archive (TPIA; see footnote 1) by making use of the BLASTp algorithm with e-value and identity percentages set to 1e-5 and 50% as threshold, respectively (Supplementary Table S1). The tea NHX peptide sequences identified were further screened using the Hidden Markov Model (PF00999) to confirm the presence of the Na+/H+ _Exchanger domain. Based on the results, 9 putative tea NHX genes were incorporated into the final dataset.
The physicochemical properties of the identified tea NHX protein sequences were evaluated and analyzed by the ExPASy ProtParam tool (Table 1). The length of the NHX peptide sequences ranged from 201 (CsNHX8) to 1,204 (CsNHX3) amino acid residues while the molecular weights varied from 21764.56 (CsNHX8) to 134630.87 (CsNHX3) kDa. The predicted isoelectric points (pI) values ranged from 5.82 (CsNHX4) to 8.79 (CsNHX2). 5 out of the 9 NHX peptide sequences had more positive residues than negative ones, 3 had more of negative residues and remaining one (CsNHX9) had equal number of positive and negative residues. All the 9 NHX peptide sequences had positive grand average of hydropathy (GRAVY index) values, ranging from 0.209 (CsNHX3) to 0.695 (CsNHX8). This indicated that all the 9 NHX peptides identified are hydrophobic in nature. The instability index scores revealed that 2 out of 9 NHX peptides (CsNHX2 and CsNHX9) were above 40 while the rest 7 had scores below the given level, indicating that most of the screened peptides had a stable nature (Wang et al., 2018). The aliphatic index of the peptides ranged from 102.02 (CsNHX9) to 114.38 (CsNHX8). The subcellular localization revealed that most of the NHX genes in tea were localized in vacuole (7 out of 9), while the remaining 2 genes were localized in the endoplasmic reticulum (ER; CsNHX8) and plasma membrane (PM; CsNHX9), respectively. Additionally, the presence of transmembrane helices was also analyzed and it revealed that all the NHX peptides had a considerable number of transmembrane helices, ranging from a minimum of 6 in CsNHX8 to a maximum of 12 in CsNHX9 (Supplementary Figure S1).
Phylogenetic Analysis of Tea NHXs
To explore the evolutionary relationships of the NHX genes among the different plant species, a phylogenetic analysis was conducted comparing the identified tea NHX genes along with NHXs from 10 other plants. For this study, we retrieved the NHX peptide sequences from A. thaliana (At), O. sativa (Os), S. lycopersicum (Sl), S. tuberosum (St), M. truncatula (Mt), P. trichocarpa (Pt), G. hirsutum (Gh), S. bicolor (Sb), Z. mays (Zm), and G. max (Gm) from their respective genome databases. The NHX peptide sequences from A. thaliana were used as query sequences to search for the NHX genes in all these plants. The sizes of all the NHX gene family from the 11 members ranged from a minimum of 5 in S. tuberosum to a maximum of 23 in G. hirsutum (Table 2).
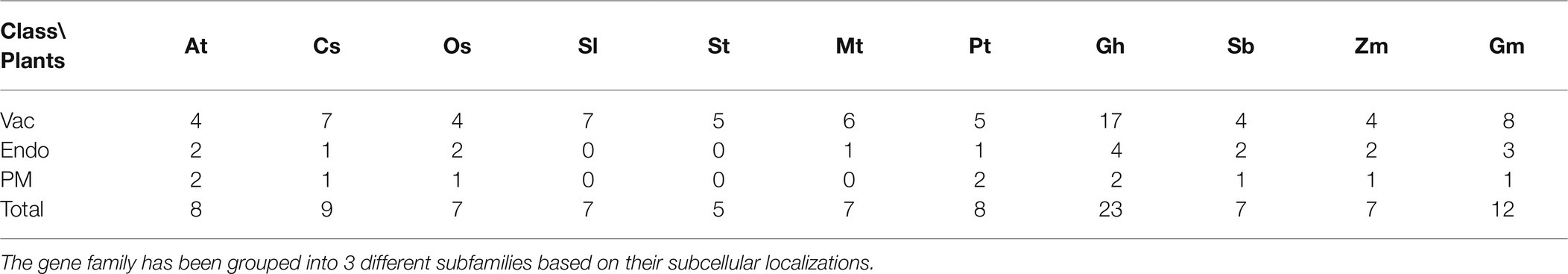
Table 2. NHX gene family members from A. thaliana (At), C. sinensis (Cs), O. sativa (Os), S. lycopersicum (Sl), S. tuberosum (St), M. truncatula (Mt), P. trichocarpa (Pt), G. hirsutum (Gh), S. bicolor (Sb), Z. mays (Zm), and G. max (Gm).
The phylogenetic tree was then constructed using all the 100 NHX peptide sequences from the 11 species. MEGA 7.0.14 was used to generate the phylogenetic trees, using the Neighbor-Joining (NJ) algorithm, at default parameters and 1,000 bootstrap replicates. The phylogenetic tree shows a direct relation with the subcellular localization as all the NHX peptides clustered into 3 different clades based on their localizations (Figure 1). The 3 different clades were the Vac-class (Vacuole), Endo-class (Endosomal), and PM-class (Plasma membrane). Among these 3 classes, the Vac-class was the most abundantly present class of NHXs in all the 11 species with 71 genes, followed by the Endo-class and PM-class with 18 and 11 genes, respectively.
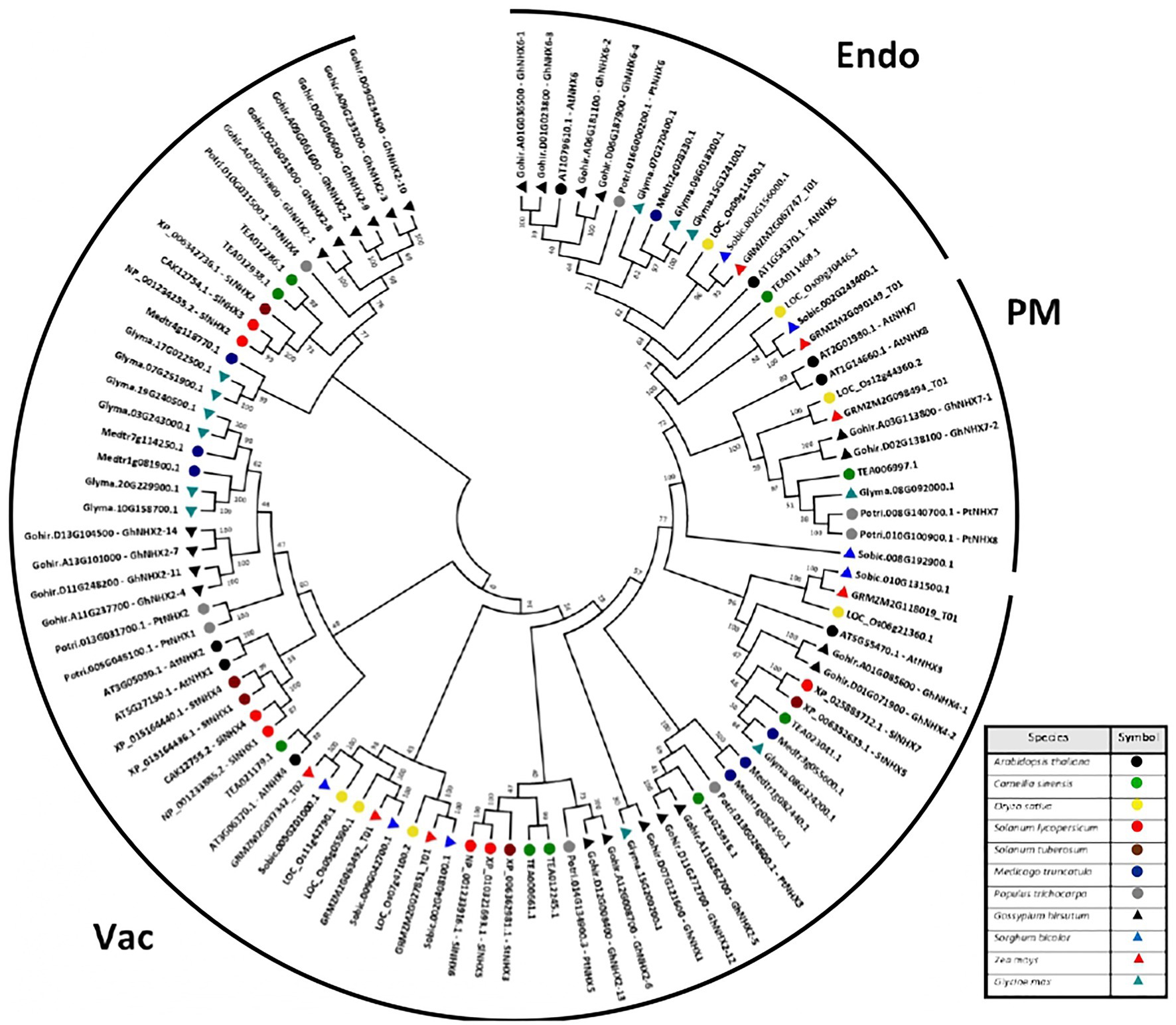
Figure 1. Phylogenetic tree of NHX genes from Arabidopsis thaliana, Camellia sinensis, Oryza sativa, Solanum lycopersicum, Solanum tuberosum, Medicago truncatula, Populus trichocarpa, Gossypium hirsutum, Sorghum bicolor, Zea mays, and Glycine max. The full-length NHX protein sequences were aligned using MUSCLE, and the phylogenetic tree was constructed using MEGA 7.0.14 by the Neighbor-Joining (NJ) method with default parameters and 1,000 bootstrap replicates. The tree is divided into three major classes of NHX genes, consisting of the Vac-, Endo-, and PM-classes.
Motif Composition of Tea NHXs
To evaluate the structural characteristics and diversity of the tea NHXs, a correlative study of the conserved motifs from the NHX peptides of A. thaliana, C. sinensis, and O. sativa was conducted using the MEME suite (Figure 2). 15 motifs were identified from 24 NHXs used out of which 2 (Motif 8 and Motif 14) were conserved across all the genes. Motif 1, 5, and 11 were each present in 18 NHXs. These 3 motifs existed only in the Vac- and Endo-classes. The amiloride-binding site (FFIYLLPPI) is a characteristic feature of NHX proteins. It was detected in Motif 3 and was found in 16 NHXs, existing only in the Vac- and Endo-classes. Motif 2, 4, 6, 10, and 12 existed only in the Vac-class and was present in 15, 15, 10, 14, and 12 NHXs, respectively. Motif 13 and 15 were present in 8 and 7 NHXs correspondingly. These 2 motifs existed only in the PM- and Endo-classes. The remaining motif 7 and 9 were present in all the classes and were harbored by 22 and 21 NHXs correspondingly. Additionally, the motif logos of all the 15 motifs were also obtained and are presented in the supplemental information (Supplementary Figure S2). The NHXs present in the same class had similar conserved motifs except Endo-class, which showed partial conservancy. These results provided noteworthy evidence that the NHX genes were highly conserved.
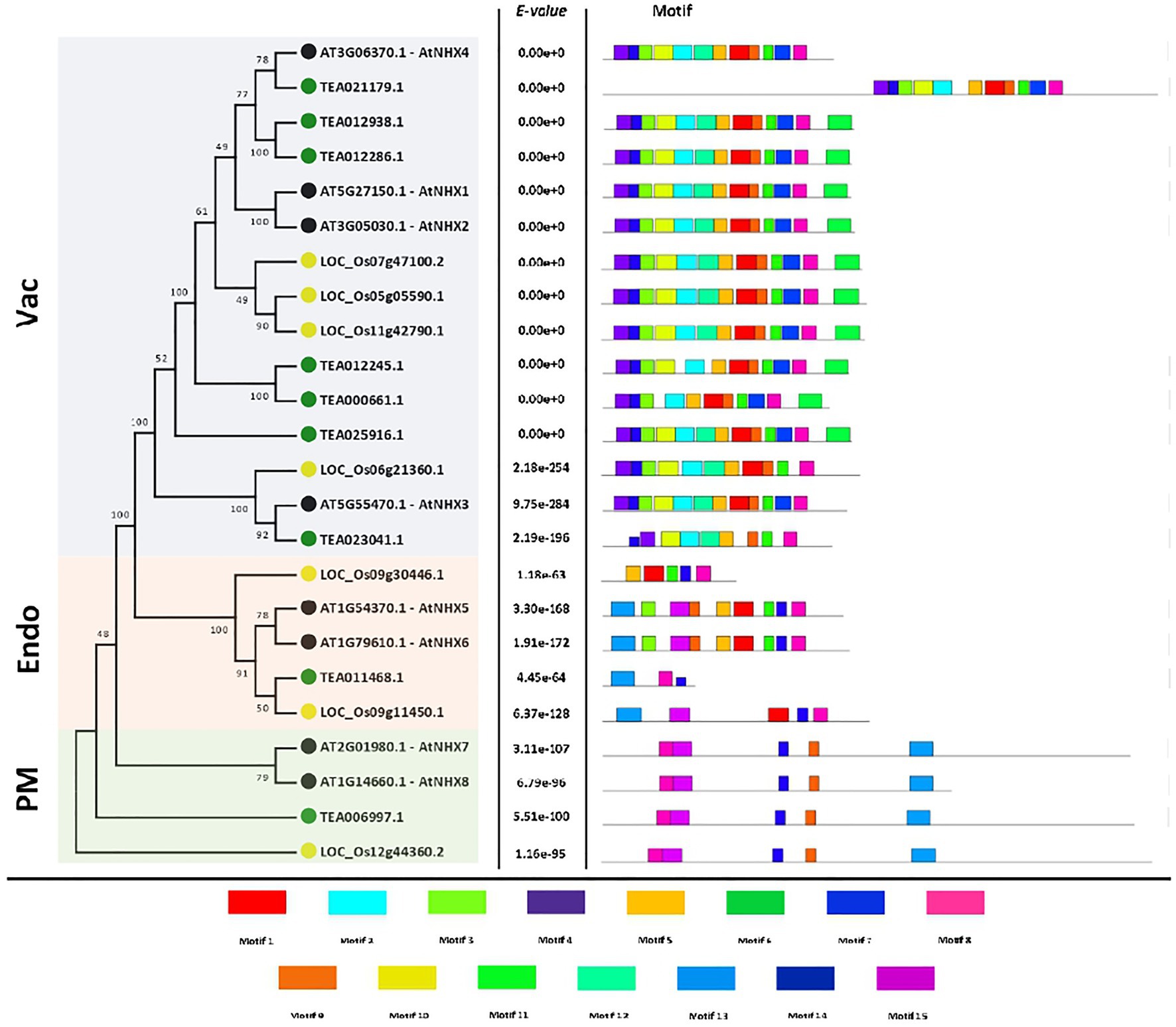
Figure 2. The motif analysis of NHX genes in A. thaliana, C. sinensis, and O. sativa. The motif figures were generated by MEME suite. A total of 15 motifs were identified and are marked individually.
Gene Structure Analysis of Tea NHXs
To identify the structural characteristics of the tea NHXs, the intron/exon architecture of the genes were analyzed using Gene Structure Display Server v2.0. Study of the intron/exon patterns revealed some significant differences concerning the number of introns and exons, which further contributes to the variation in gene lengths. Abundant presence of non-coding sequences within a genome is regarded to be an indicator of genome complexity (Taft et al., 2007; Goyal et al., 2018; Chatterjee et al., 2020). Analyzing these intron arrangements thereby provides significant information regarding the evolution, regulation, and function of the NHXs (Deutsch and Long, 1999; Fedorova and Fedorov, 2003; Zhang et al., 2014; Liu et al., 2015). The analysis of the tea NHX gene structures indicated considerable differences with respect to the number of introns and exons across the 3 classes (Figure 3A). Among the 9 tea NHXs, only CsNHX1 (TEA012938.1) possessed UTR (Untranslated Regions) segments at both 5′ and 3′ ends. 5 out the 7 Vac-class NHXs had 14 exons and 13 introns. CsNHX3 (TEA021179.1) possessed 19 exons and 18 introns while CsNHX7 (TEA023041.1) had 13 exons and 12 introns, respectively. The Endo-class NHX CsNHX8 (TEA011468.1) had the least share of IEs (Introns-exons) among the 3 classes with only 6 exons and 5 intron segments. However, the PM-class NHX CsNHX9 (TEA006997.1) had the most share of IEs with 25 exons and 24 introns. It was observed that the genes belonging to the same clade had a similar distribution of introns and exons. The intron segments and exon lengths were relatively conserved among the genes of the same class. Additionally, analyzing the amino acid sequence identity also supported the sequence conservation among the tea NHXs (Figure 3B). Two paralogous pairs of NHX in Vac-class displayed high amino acid sequence identities (TEA012286.1/TEA012938.1 = 86.14% and TEA012245.1/TEA000661.1 = 79.05%). On the flip side, tea NHXs in different classes displayed lower levels of sequence identities (TEA012245.1/TEA006997.1 = 21.98%; TEA012245.1/TEA011468.1 = 31.22%; and TEA011468.1/TEA006997.1 = 22.63%).
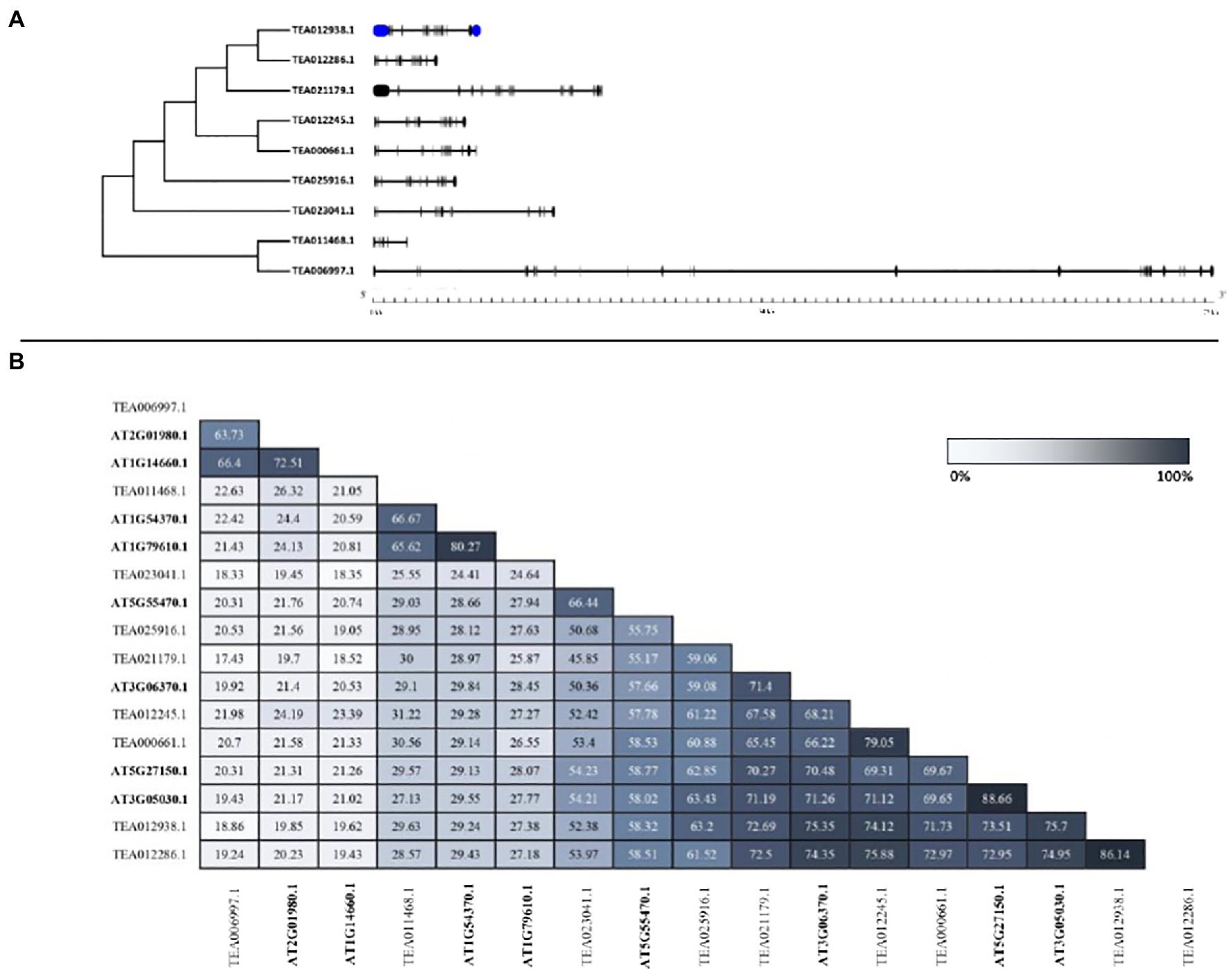
Figure 3. The intron/exon architecture and pairwise sequence identity among NHX proteins of A. thaliana and C. sinensis. (A) Gene structure maps were drawn using the Gene Structure Display Server 2.0. Black boxes represent exons, blue boxes represent the UTRs, and black lines represent introns. The gene length can be estimated by using the scale (in kb) given at the bottom. (B) The full-length protein sequences were aligned using MUSCLE tool (https://www.ebi.ac.uk/Tools/msa/muscle/) with default settings. The table has been marked based on a gradient with lighter shades representing minimum percentage identity and darker shades representing maximum identity between the sequences.
Retrieval of Tea NHX Promoter Regions and Analysis of CAREs
Cis-acting regulatory elements (CAREs) play a key role in determining gene regulation, function, transcription, and gene expression (Wu et al., 2019a; Liu et al., 2021). Analysis of these regulatory elements helps in defining the plant responses to various environmental stimuli, stress factors, thereby affecting the growth regulation (Akram et al., 2020). To explore the transcriptional potential of the tea NHX genes, the promoter sequences of 2,000 bp upstream of the transcriptional start codon “ATG” were retrieved from the TPIA database. These promoter sequences were then used to predict and analyze the CAREs using the PlantCARE database. 41 total CAREs were identified randomly distributed across the promoter regions of the 9 tea NHXs (Supplementary Table S2). Based on the specific biological functions of the identified CAREs, they were grouped together into a pie chart under 20 different sections (Figure 4A). Most of the CAREs had sequence lengths of 6 and 9 bp, while the others ranged between 5 to 13 bp (Figure 4B). Analyzing the 41 CAREs, it was observed that 18 elements were involved in light responsiveness, 9 elements in phyto-hormonal as well as plant growth and regulation each, and 5 elements in stress response. The light responsive elements had the largest share of CAREs and were present in all the 9 tea NHXs. Among these 18 light responsive elements, the Box-4 and G-box elements were abundantly present in 8 and 6 NHXs, respectively. Few of the other light responsive elements were TCCC-motif, AE-box, AT1-motif, Box-II, TCT-motif, chs-CMA1a, and chs-CMA2a in 4, 2, 2, 1, 1, 3, and 3 tea NHXs, respectively. NHXs are mostly involved in response to various environmental stresses and regulation (Akram et al., 2020). The stress responsiveness elements comprised of elements responding to drought stress (MBS), low temperature (LTR), defense and stress (TC-rich repeats), and anaerobic induction (ARE) in 1, 2, 3, and 7 tea NHXs correspondingly. Another element was involved in maximal elicitor mediated activation (AT-rich sequences) was harbored by 1 tea NHX (TEA023041.1). The CAREs involved in phytohormone responses mainly comprised of abscisic acid responsive element (ABRE), gibberellin responsive elements (GARE-motif, TATC-box, and P-box), and salicylic acid responsive element (TCA-element) in 6, 5, and 5 genes, respectively. Other phytohormone response elements included elements responsive to MeJA (CGTCA-motif and TGACG-motif) in 4 genes and auxin (TGA-element and AuxRR-core) in 2 genes. The elements associated with plant growth and development mainly comprised of MYBvH1-binding site (CCAAT-box), zein metabolism regulatory element (O2-site), endosperm expression element (GCN4_motif), and palisade mesophyll differentiation element (HD-Zip 1) in 5, 4, 3, and 3 tea NHXs, respectively. A regulatory element (A-box) was present in CsNHX1 (TEA012938.1) and CsNHX9 (TEA006997.1), while AT-rich DNA-binding site (ATCT-motif) was present in CsNHX8 (TEA011468.1) and CsNHX9 (TEA006997.1). Some of the other growth-related CAREs included elements involved in meristem expression (CAT-box) and circadian control, both present in CsNHX5 (TEA000661.1) and cell cycle regulation (MSA-like) in CsNHX8. The results obtained from the analysis of these CAREs suggests the involvement of the tea NHXs in various phytohormone, light, and stress responses.
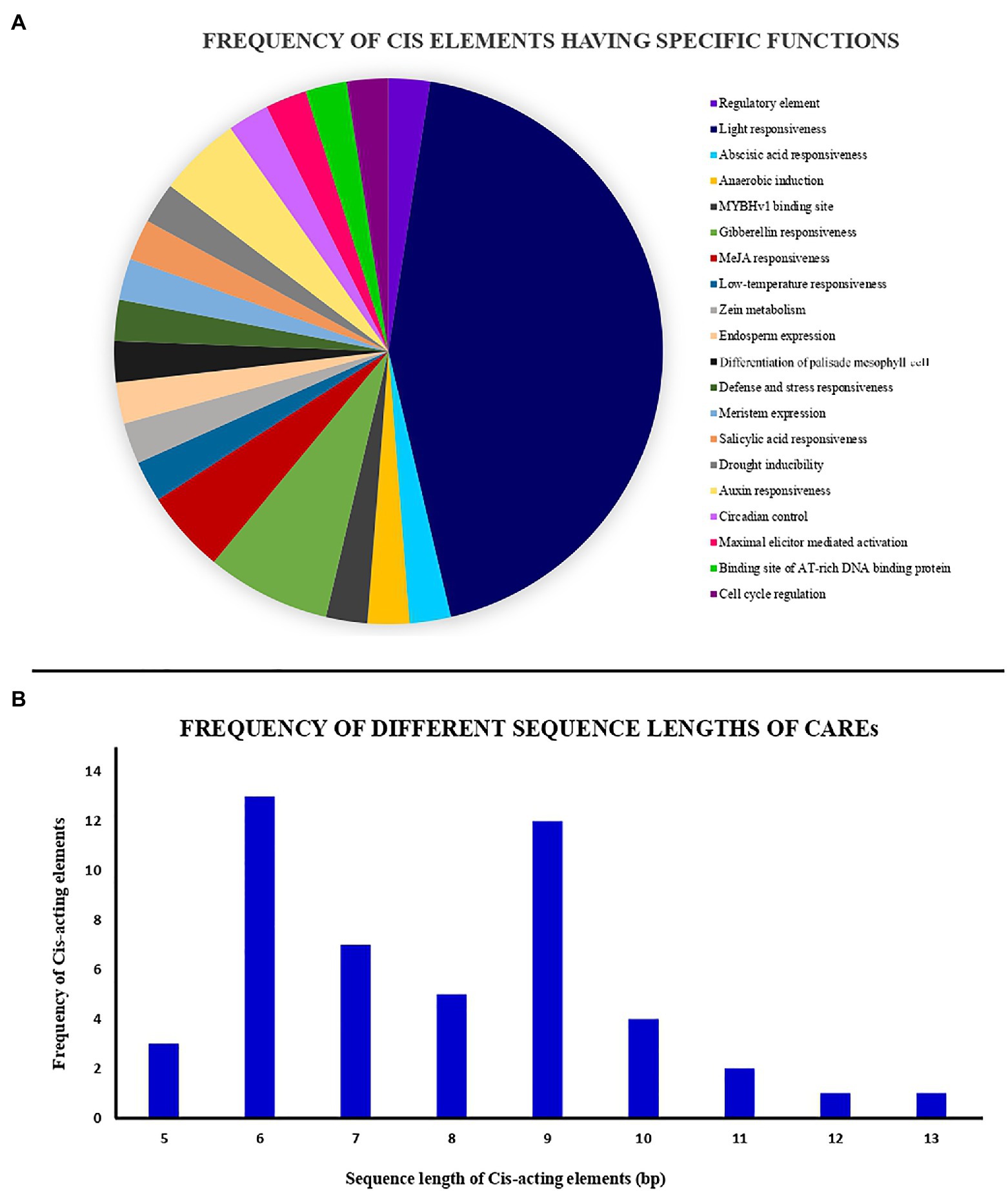
Figure 4. Analysis of cis-acting elements identified from the NHX genes of C. sinensis. All cis-acting elements have been identified using PlantCARE database. (A) Pie chart showing the frequency of different cis-acting elements based on their specific biological activities. (B) Histogram showing the frequency of different sequence lengths of the cis-acting elements.
Genomic Distribution Map and Evolutionary Pressures on Tea NHXs
In an attempt to understand the genome distribution pattern of the tea NHXs, the genes were mapped into their genomic scaffolds. Due to the lack of chromosome-level assembly data in the TPIA database, the genes had to be mapped into their scaffolds instead of the chromosomes. The 9 tea NHXs were distributed evenly across 9 different scaffolds (Figure 5). The genes were positioned such that a single scaffold housed individual genes. Additionally, the Ka/Ks or dN/dS (non-synonymous substitution rate/synonymous substitution rate) ratios were calculated in order to understand the evolutionary pressures and gene divergence mechanisms (Supplementary Table S3). The dN/dS ratio helps determine whether Darwinian selection pressures were involved in the duplication events (Tian et al., 2017; Chatterjee et al., 2020). If the value of the dN/dS ratio is >1, it implies a positive or Darwinian selection. If the ratio is equal to 1, it implies a neutral selection and if the ratio is <1, it determines a purifying selection (Bowers et al., 2003; Liu et al., 2014). Pairwise comparisons of the 9 tea NHXs revealed 13 gene pairs having their dN/dS ratios >1, indicating a positive selection. The rest 23 gene pairs had their ratios <1, indicating a negative or purifying selection. Additionally, a cumulative graph of the tea NHXs was also generated (Supplementary Figure S3). The results from the gene distribution pattern and dN/dS ratios showed that the NHXs were extensively distributed across the C. sinensis genome. Tandem duplication events were however absent across the tea NHXs. The dN/dS ratios are conclusive proof that strong purifying selection pressures had occurred during the evolution thereby enabling a number of different factors to regulate the NHXs in tea genome.
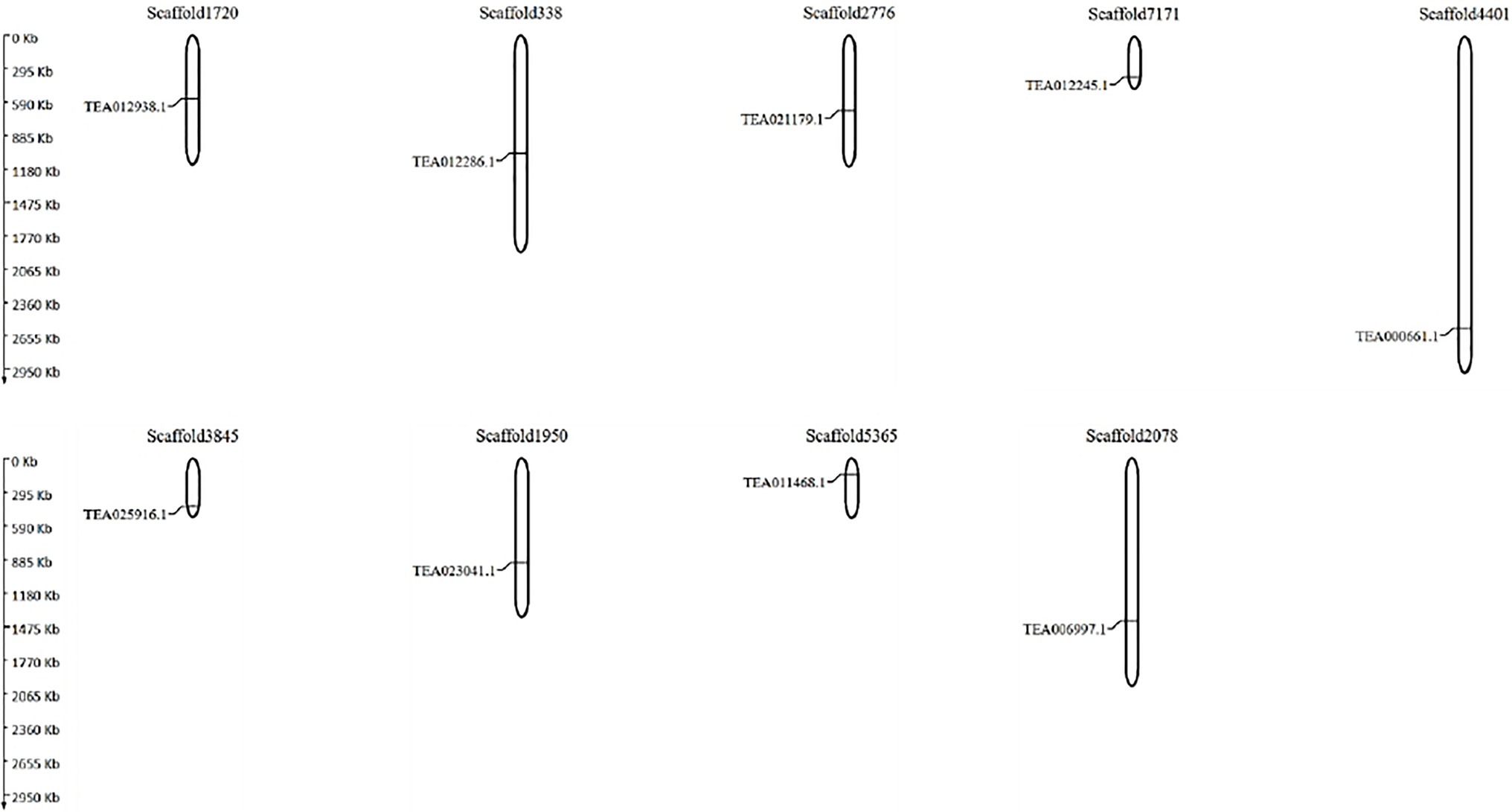
Figure 5. The scaffold distribution 9 NHX genes in C. sinensis. MapGene2chromosome web v2 (MG2C) software tool (http://mg2c.iask.in/mg2c_v2.1/) was used to map genes into their respective scaffolds. The scaffolds are drawn to scale and the scaffold numbers are indicated on the top.
GO Ontology Analysis of Tea NHXs
In order to predict the functions of the 9 tea NHXs, GO ontology analysis was done. It was observed that tea NHXs were enriched in 24 GO terms (Supplementary Table S4). The 9 NHX genes were divided into 3 major groups, which included biological process, cellular component, and molecular function. The first group featured 13 different GO terms with “proton transmembrane transport” (GO:1902600; 6 sequences; 66.67%) having the highest representation (Figure 6). It was followed by “Na+ transmembrane transport” (GO:0035725; 5 sequences; 55.56%), “K+ homeostasis” (GO:0055075; 5 sequences; 55.56%), “regulation of pH” (GO:0006885; 5 sequences; 55.56%), and “response to salt” (GO:0009651; 5 sequences; 55.56%). Few of the other GO terms included “monovalent inorganic cation homeostasis” (GO:0055067), “metal ion transport” (GO:0030001), and “RNA splicing” (GO:0008380). The first group was followed by the cellular component group that featured 6 different GO terms. Among these 6, “integral component of membrane” (GO:0016021; 6 sequences; 66.67%) was featured the most and was closely followed by “vacuolar membrane” (GO:0005774; 5 sequences; 55.56%) and “plasma membrane” (GO:0005886; 5 sequences; 55.56%). The rest was “intrinsic component of membrane” (GO:0031224; 3 sequences; 33.33%), “plastid” (GO:0009536; 1 sequence; 11.11%), and “mitochondria” (GO:0005739; 1 sequence; 11.11%). The remaining 5 of the 24 identified GO terms were featured in the molecular function group. Among these 5, “Na:proton antiporter activity” (GO:0015386; 5 sequences; 55.56%) was represented the most. It was followed by “monovalent cation:proton antiporter activity” (GO:0005451; 3 sequences; 33.33%), “solute:proton antiporter activity” (GO:0015299; 2 sequences; 22.22%), “double stranded DNA binding” (GO:0003690; 1 sequence; 11.11%), and “antiporter activity” (GO:0015297; 1 sequence; 11.11%).
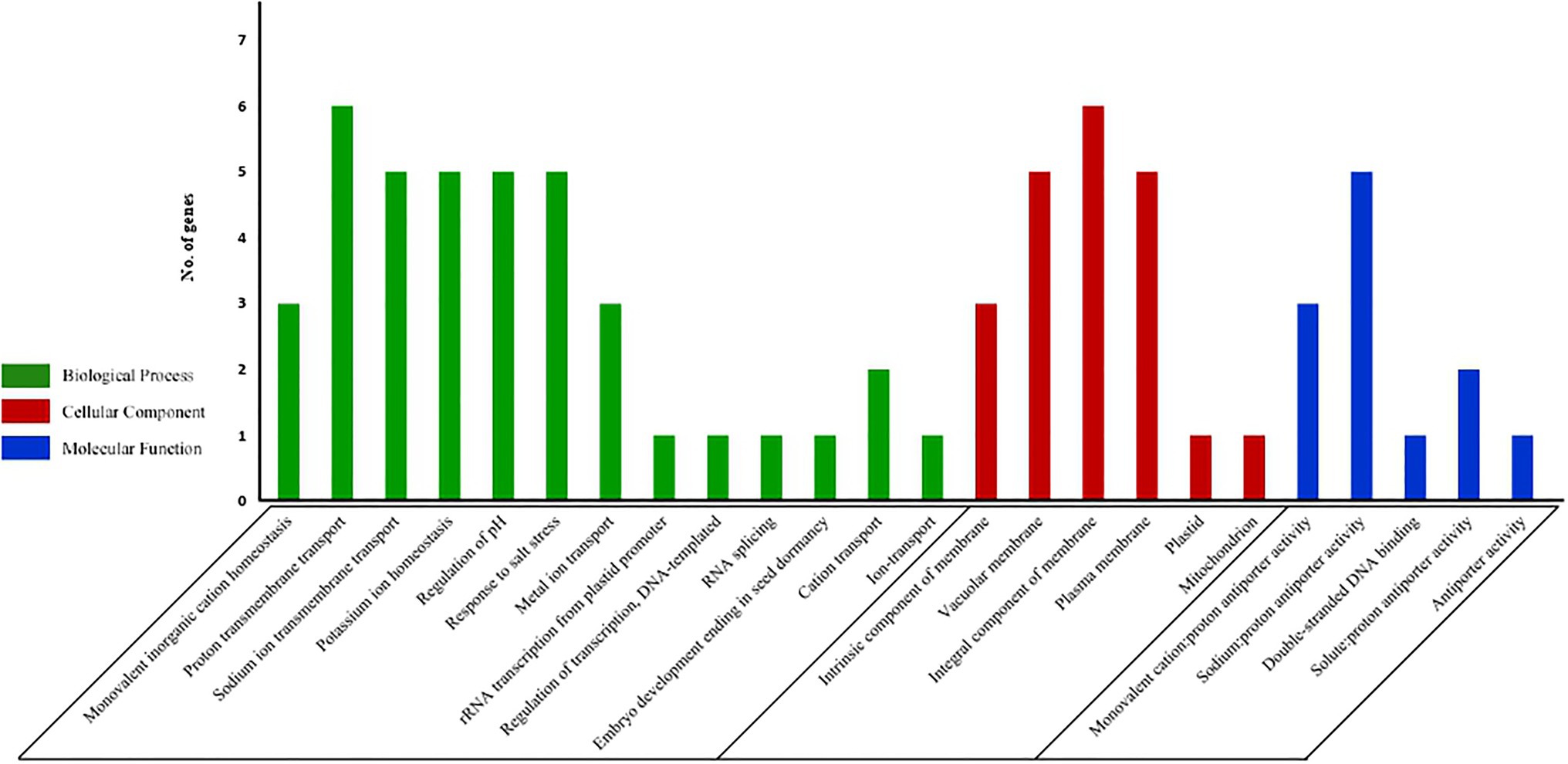
Figure 6. GO analysis of all the 9 NHX genes in C. sinensis. The results have been grouped into three main categories: Biological Process, Cellular Component, and Molecular function. The y-axis represents the frequency of genes while the x-axis represents the potential functions.
Functional Interaction Network of Tea NHX Proteins
To understand and explore the interaction pattern of NHX genes in tea, a protein interaction network was constructed using the STRING server based on an Arabidopsis association model (Figure 7). The Arabidopsis model had to be employed due to the absence of tea database in the STRING server. NHXs are largely involved in a variety of biological processes of which response to salt stress is one of the prominent ones (Tian et al., 2017). The interaction network was therefore constructed based on 5 tea NHXs (CsNHX1, CsNHX2, CsNHX3, CsNHX4, and CsNHX5), involved in response to salt stress according to GO ontology. The A. thaliana homolog for these 5 tea NHXs, AtNHX2 (AT3G05030), was used as the central node to build the full network. The tea proteins, homologs to the Arabidopsis proteins participating in the network, were also added. These homologous proteins were designated as STRING proteins and were selected based on high bit scores in BLAST results (Chatterjee et al., 2020). Similarity search program, such as BLAST, is frequently used to produce accurate statistical estimates that help ensuring protein sequences with significant similarity to have similar structures (Pearson, 2013). In addition, proteins sharing higher degree of sequence and structural similarities often tend to have similar functions as well (Gan et al., 2002). AtNHX2 (AT3G05030) is involved in active K+ uptake at the tonoplast and involved in regulating stomatal closure (Berardini et al., 2015). AtNHX1 (AT5G27150) encodes a vacuolar sodium-proton antiporter involved in salt tolerance, ion homeostasis, and leaf development. Two of the tea NHXs (CsNHX1 and CsNHX2) which are homologous to AtNHX1 are massively involved in response to salt stress. AVP1 (AT1G15690) is involved in regulation of apoplastic pH and auxin transport (Berardini et al., 2015). CLC-C (AT5G49890) is a chloride channel protein and is involved in the Cl− transmembrane transporter activity. CLC-F (AT1G55620) is another chloride channel protein, localized in chloroplast and Golgi apparatus, and is involved in voltage-gated Cl− channel activity (Berardini et al., 2015). HKT1 (AT4G10310) encodes for a sodium transporter expressed in xylem parenchyma cells and is involved in response to osmotic stress and salt stress. SOS2 (AT5G35410) encodes a member of the CBL-interacting protein kinase family and is a regulatory component controlling plant K+ nutrition (Berardini et al., 2015). TEA006066.1, which is homologous to SOS2, also shows response to nutrient and water deprivation according to GO ontology. SOS3 (AT5G24270) encodes for a calcium sensor that is essential for K+ nutrition, K+/Na+ selectivity, and salt tolerance. CHX18 (AT5G41610), CHX17 (AT4G23700), and CHX15 (AT2G13620) are all involved in regulation of pH and are members of the putative Na+/H+ antiporter family (Berardini et al., 2015).
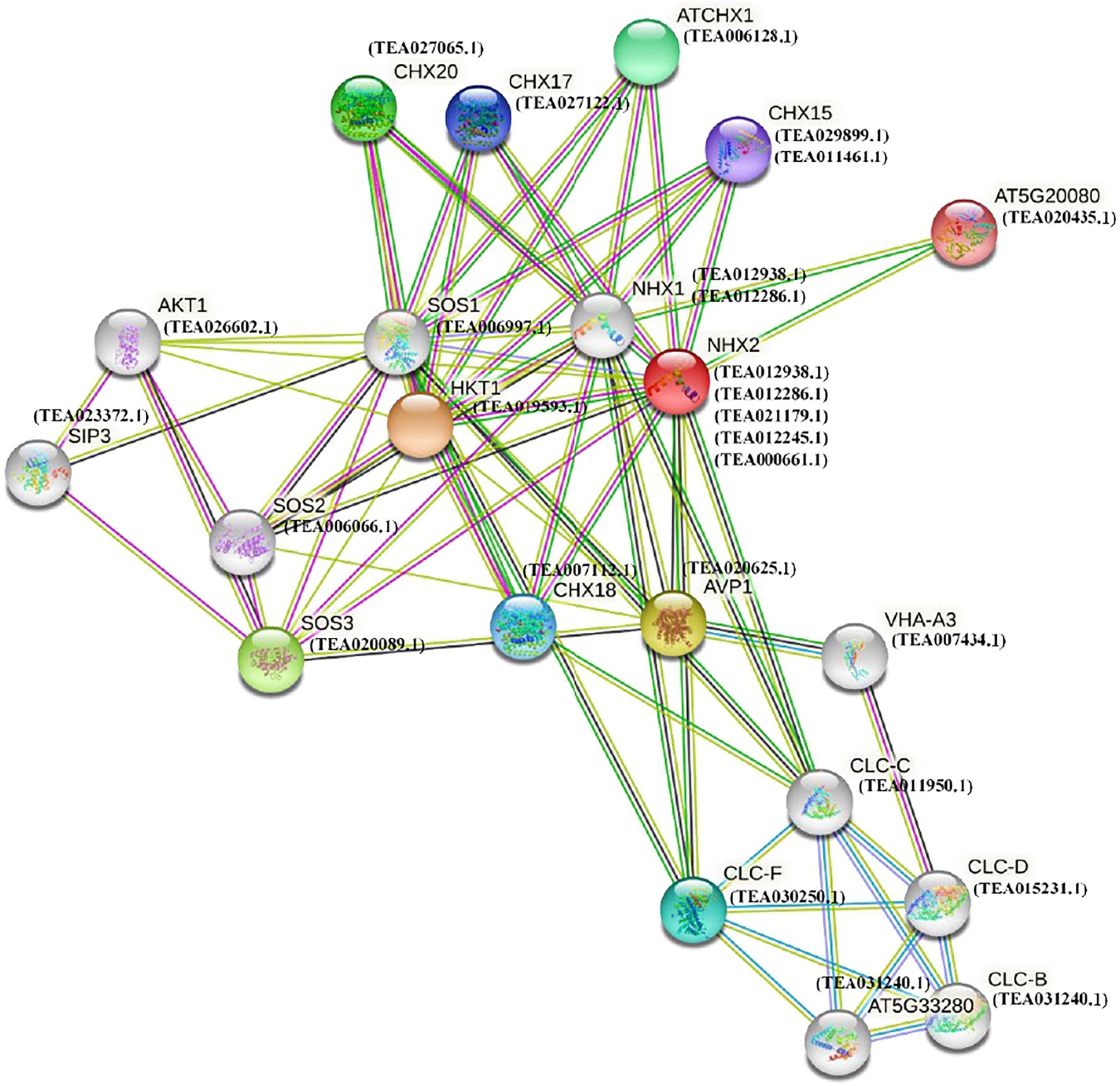
Figure 7. Functional interaction networks of NHX proteins in C. sinensis. The interaction network was formed according to homologs in A. thaliana.
Tissue-Specific Gene Expression of Tea NHXs
The tissue-specific expression levels of the 9 tea NHXs in 8 different tissues were retrieved from the TPIA database wherein the levels of expressions were evaluated in transcripts per million (TPM). The database has the expression profile data of all the C. sinensis genes, which have been experimentally validated (Wei et al., 2018). The plant tissues that have been assessed in the study involved apical bud, flower, fruit, young leaf, mature leaf, old leaf, root, and stem (Supplementary Table S5). All the 9 tea NHXs exhibited varying levels of expression in these 8 different tissues. Few of the genes had high levels of expression while the rest had negligible transcript levels (Figure 8). CsNHX1 (TEA012938.1) was expressed the most in apical bud, closely followed by CsNHX5 (TEA000661.1). This similar pattern was observed when the expression levels were checked in flower, young leaf, root, and stem. The highest expression level was recorded by CsNHX1 in fruit, followed by CsNHX2 (TEA012286.1) and CsNHX5 (TEA000661.1). In mature leaf and old leaf, CsNHX2 was expressed the most, followed by CsNHX1 and CsNHX5. These results suggested that 3 out of the 9 tea NHXs (CsNHX1, CsNHX2, and CsNHX5) were significantly expressed in all the 8 tissues. The rest of the 6 NHXs were minimally expressed in these 8 tissues with CsNHX7 (TEA023041.1) and CsNHX4 (TEA012245.1) being the least.
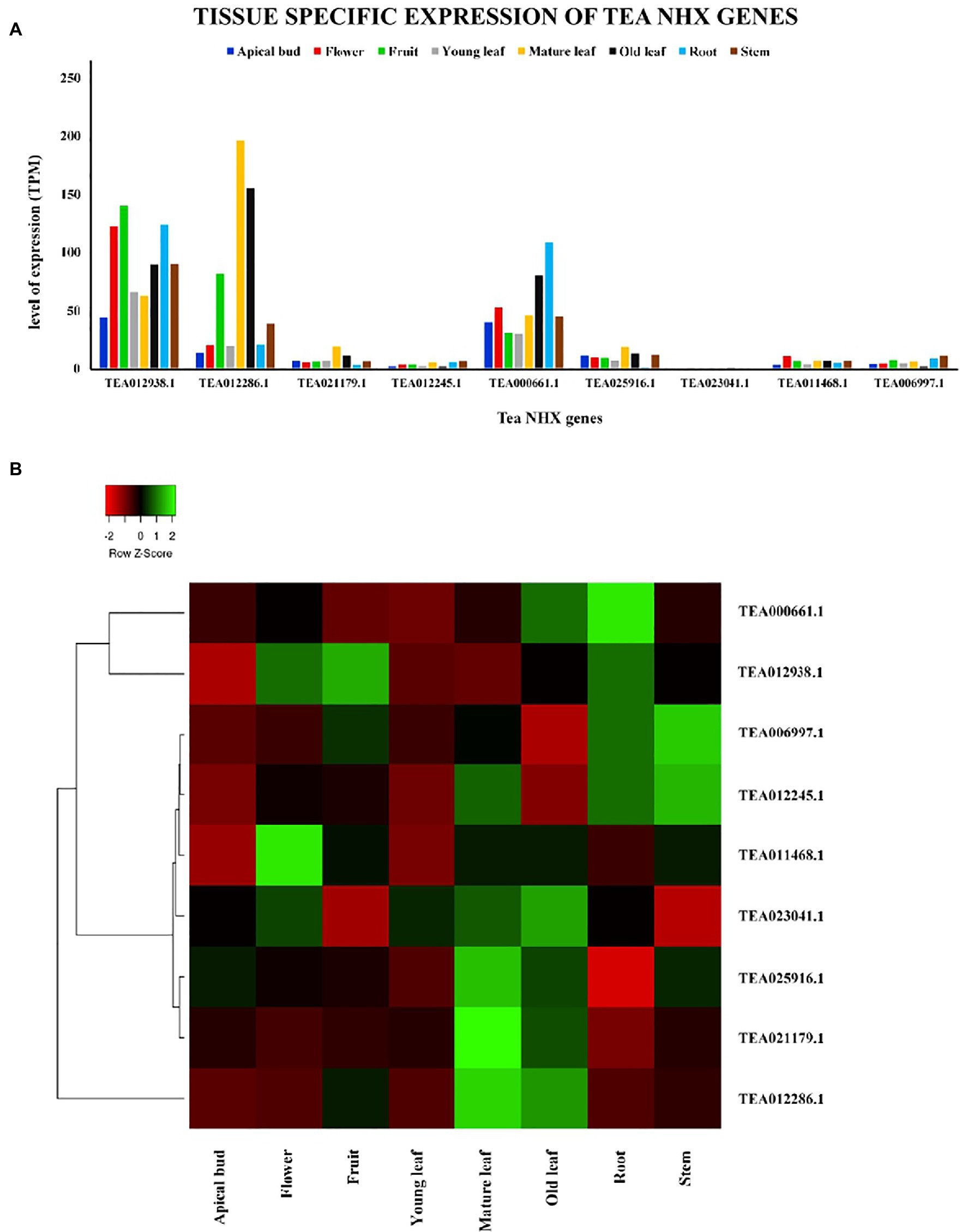
Figure 8. Tissue-specific expression patterns of NHX genes in C. sinensis in 8 different plant tissues. (A) The relative expression of Tea NHX genes represented graphically by analyzing the transcriptome data. (B) Relative expression represented as a heatmap, generated using heatmapper online server. The color bar on the top represents the normalized transcript per million (TPM) values. Green and red colors represent the up- and downregulation values while black represents no expression.
Expression Profiles of Tea NHXs Under Cold and Drought Stress
In order to check how the 9 tea NHXs respond to varying levels of cold and drought stress, their expression data were retrieved from the TPIA database. The TPIA database has experimentally verified expression data for all the C. sinensis genes under cold (Wang et al., 2013) and drought stress (Zhang et al., 2017). The cold acclimated data comprised of 5 stages of expression: (1) 25~20°C (CK), (2) Fully acclimated at 10°C for 6 h (CA1-6 h), (3) 10~4°C for 7 days (CA1-7 d), (4) Cold response at 4~0°C for 7 days (CA2-7 d), and (5) Recovering under 25~20°C for 7 days (DA-7 d; Xia et al., 2019), where CK is the control (Supplementary Table S6). Expression levels for CA1-6 h showed that 7 out of 9 tea NHXs were upregulated while the rest 2 were downregulated. Out of these 7 upregulated genes, CsNHX1 (TEA012938.1), CsNHX5 (TEA000661.1), and CsNHX2 (TEA012286.1) were upregulated the most. When the cold stress was increased to the next stage (CA1-7 d), again 7 genes showed upregulation with CsNHX2, CsNHX1, and CsNHX5 being the highest. CsNHX9 (TEA006997.1), which was initially upregulated in the first condition, was downregulated in this present condition. Further increasing the cold stress levels at CA2-7 d, expression data revealed 5 NHXs being upregulated. CsNHX6 (TEA025916.1) was slightly upregulated at CA1-7 d but was downregulated at CA2-7 d. CsNHX2 was also upregulated for the previous two levels of cold stress, but at CA2-7 d, it was downregulated. CsNHX5 was expressed the most followed by TEA12938.1. Expression levels under the recovery phase (DA-7 d) showed that only 3 NHXs were upregulated (Figure 9). Throughout the cold stress conditions, 2 genes namely, CsNHX1 and CsNHX5 consistently maintained high levels of expression, followed by CsNHX2 and CsNHX3. These results indicated the active participation of these 4 tea NHX genes in response to cold stress. The expression levels were further checked under drought stress. The expression data in the TPIA database with respect to 25% polyethylene glycol (PEG) treatment include four stages: (1) 0 h, (2) 24 h, (3) 48 h, and (4) 72 h (Zhang et al., 2017), where 0 h was taken as the control (Supplementary Table S7). Under the first drought stress period of 24 h, 5 of the 9 tea NHXs were upregulated with CsNHX1 being expressed the most. The same set of genes was upregulated when the drought stress condition was extended to a period of 48 h and then for 72 h (Figure 10). These 5 genes showed upregulated levels of expression throughout the experimental conditions and thereby suggest their roles in response to drought stress.
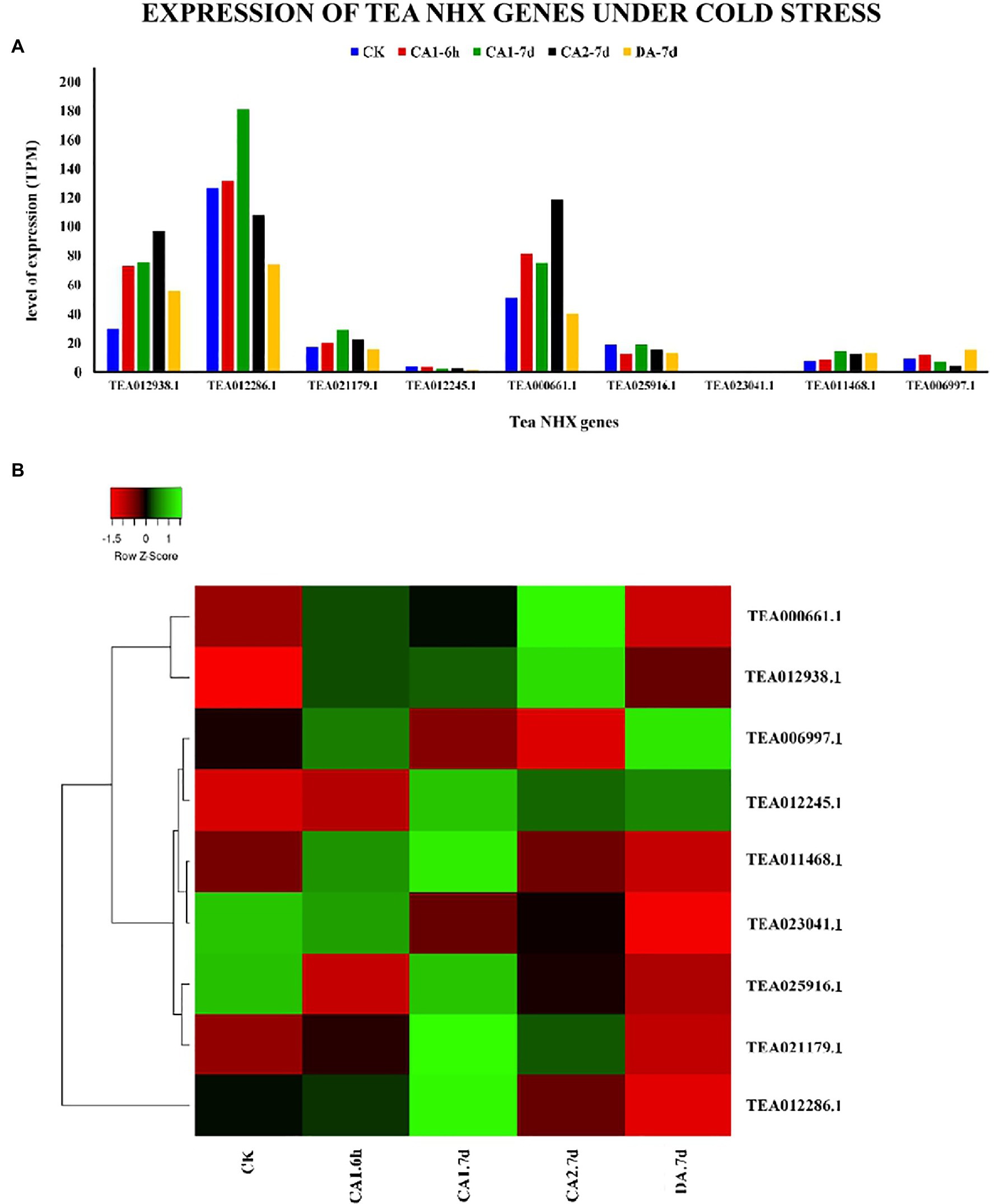
Figure 9. Expression of NHX genes in C. sinensis under cold stress. (A) The relative expression of Tea NHX genes represented graphically by analyzing the transcriptome data. (B) Relative expression represented as a heatmap, generated using heatmapper online server. The color bar on the top represents the normalized TPM values. Green and red colors represent the up- and downregulation values while black represents no expression.
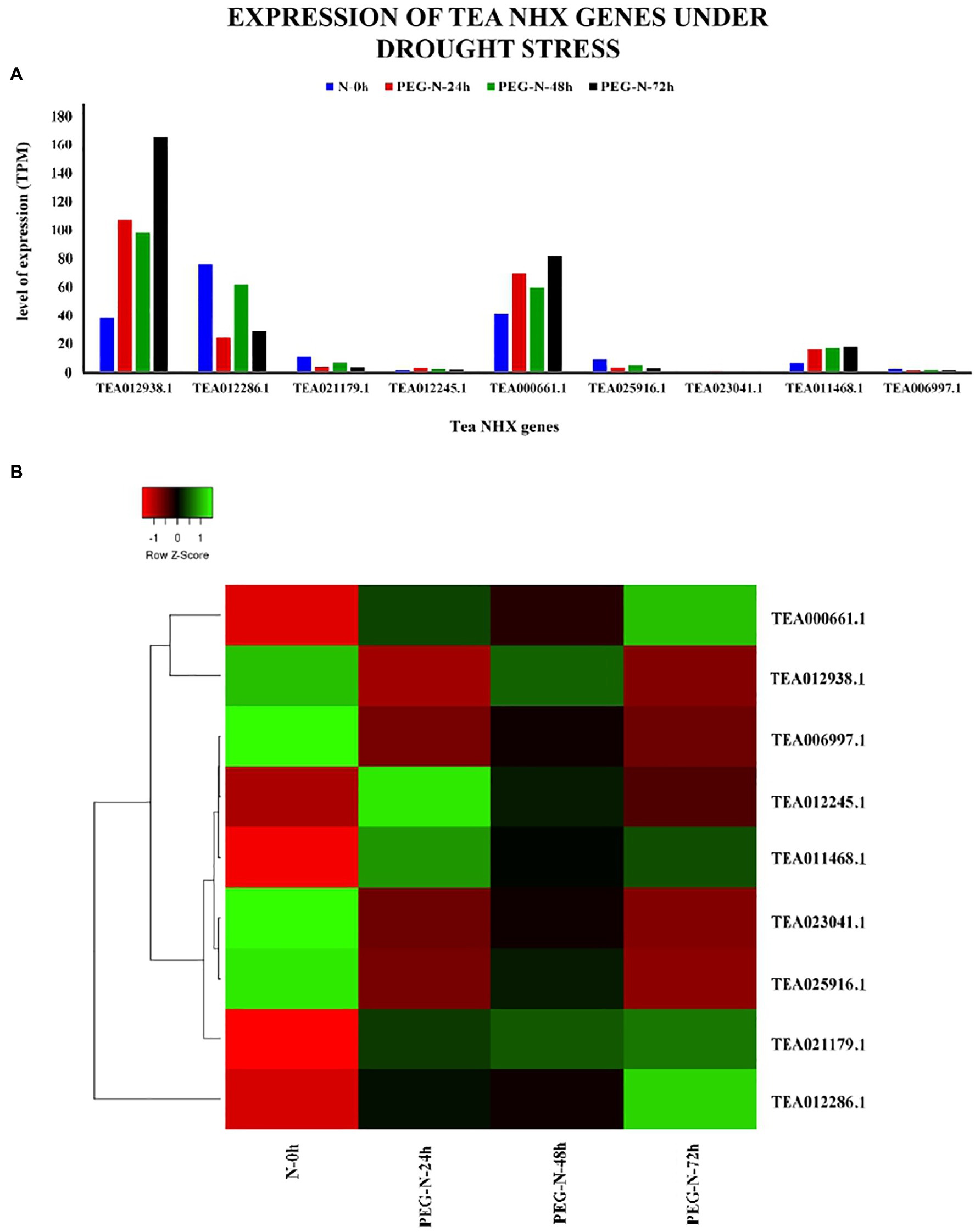
Figure 10. Expression of NHX genes in C. sinensis under drought stress. (A) The relative expression of Tea NHX genes, represented graphically by analyzing the transcriptome data. (B) Relative expression represented as a heatmap, generated using heatmapper online server. The color bar on the top represents the normalized TPM values. Green and red colors represent the up- and downregulation values, respectively, while black represents no expression.
Expression Profiles of Tea NHXs Under Salt Stress
The primary role of the NHX genes is response to salt stress (Tian et al., 2017). To understand the potential role of the 9 tea NHXs in response to high levels of salinity, the expression data were analyzed. The salt stress data in TPIA database are recorded based on treatment with 200 mm NaCl under 4 stages: (1) 0 h, (2) 24 h, (3) 48 h, and (4) 72 h (Zhang et al., 2017) where 0 h was taken as the control (Supplementary Table S8). Expression data under the 24 h salt stress condition revealed 3 genes being upregulated. Among these 3 tea NHXs, CsNHX1 (TEA012938.1) was expressed the most. A similar pattern was observed when the salt stress conditions were extended for periods of 48 h and 72 h (Figure 11). CsNHX7 (TEA023041.1) and CsNHX8 (TEA011468.1) were upregulated to a fair extent while CsNHX1 maintained very high levels of expression throughout the experimental condition, with increasing transcript levels at each stage. GO ontology data too suggested the involvement of CsNHX1 in response to salt stress. These results clearly indicate the active role of these tea NHXs in response to prolonged levels of salt stress.
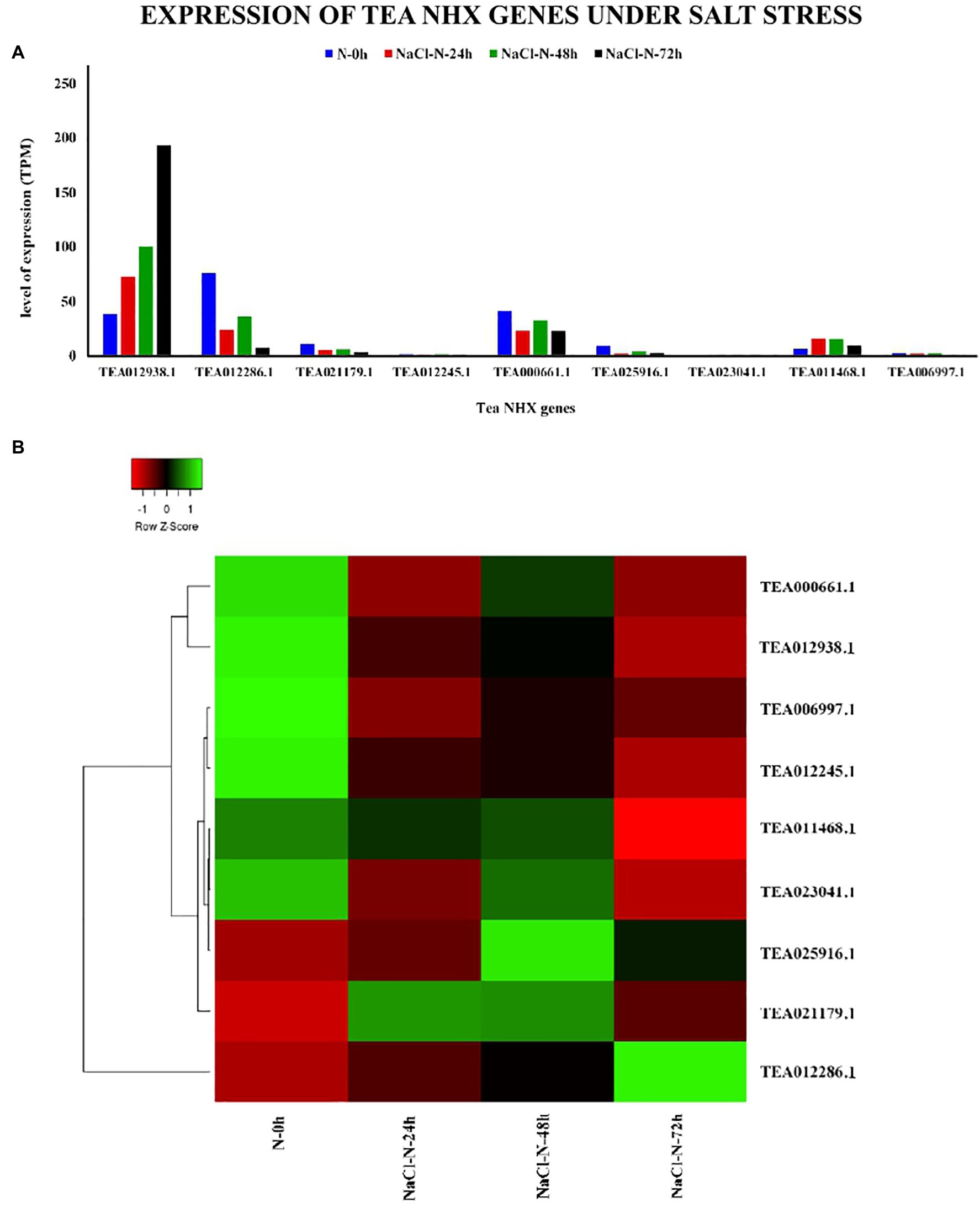
Figure 11. Expression of NHX genes in C. sinensis under salt stress. (A) The relative expression of Tea NHX genes, represented graphically by analyzing the transcriptome data. (B) Relative expression represented as a heatmap, generated using heatmapper online server. The color bar on the top represents the normalized TPM values. Green and red colors represent the up- and downregulation values while black represents no expression.
Response of Tea NHXs to MeJA Treatment
The analysis of the cis-acting elements in the promoter regions of the 9 tea NHXs had revealed the presence of 2 MeJA responsive elements (CGTCA-motif and TGACG-motif; Supplementary Table S2). To further understand the effect of MeJA on the 9 tea NHXs, their expression data were retrieved from the TPIA database and analyzed. This data is recorded based on the results of exposing the plant parts to aqueous solution of MeJA, under 4 stages: (1) 0 h, (2) 12 h, (3) 24 h, and (4) 48 h (Shi et al., 2015) where, 0 h was used as the control (Supplementary Table S9). 7 out of 9 tea NHXs showed upregulation in expression levels when exposed to the MeJA treatment for a period of 12 h. Extending the duration of the experiment to 24 h showed a few minor changes in the genes showing upregulation. CsNHX2 (TEA012286.1), which was showing upregulation initially, now was slightly downregulated. On the other hand, CsNHX6 (TEA025916.1) was downregulated in the initial phase but showed upregulated levels of expression in the present condition. 7 genes were upregulated in total at 24 h of MeJA treatment. Further extending the experiment to 48 h revealed that 5 genes were upregulated while the rest 4 were downregulated (Figure 12). CsNHX1 (TEA012938.1) and CsNHX5 (TEA000661.1) consistently maintained high levels of expression throughout the 48 h of the exposure to MeJA. These results suggested that the transcription levels of the tea NHXs might have a close relation to the regulation of MeJA.
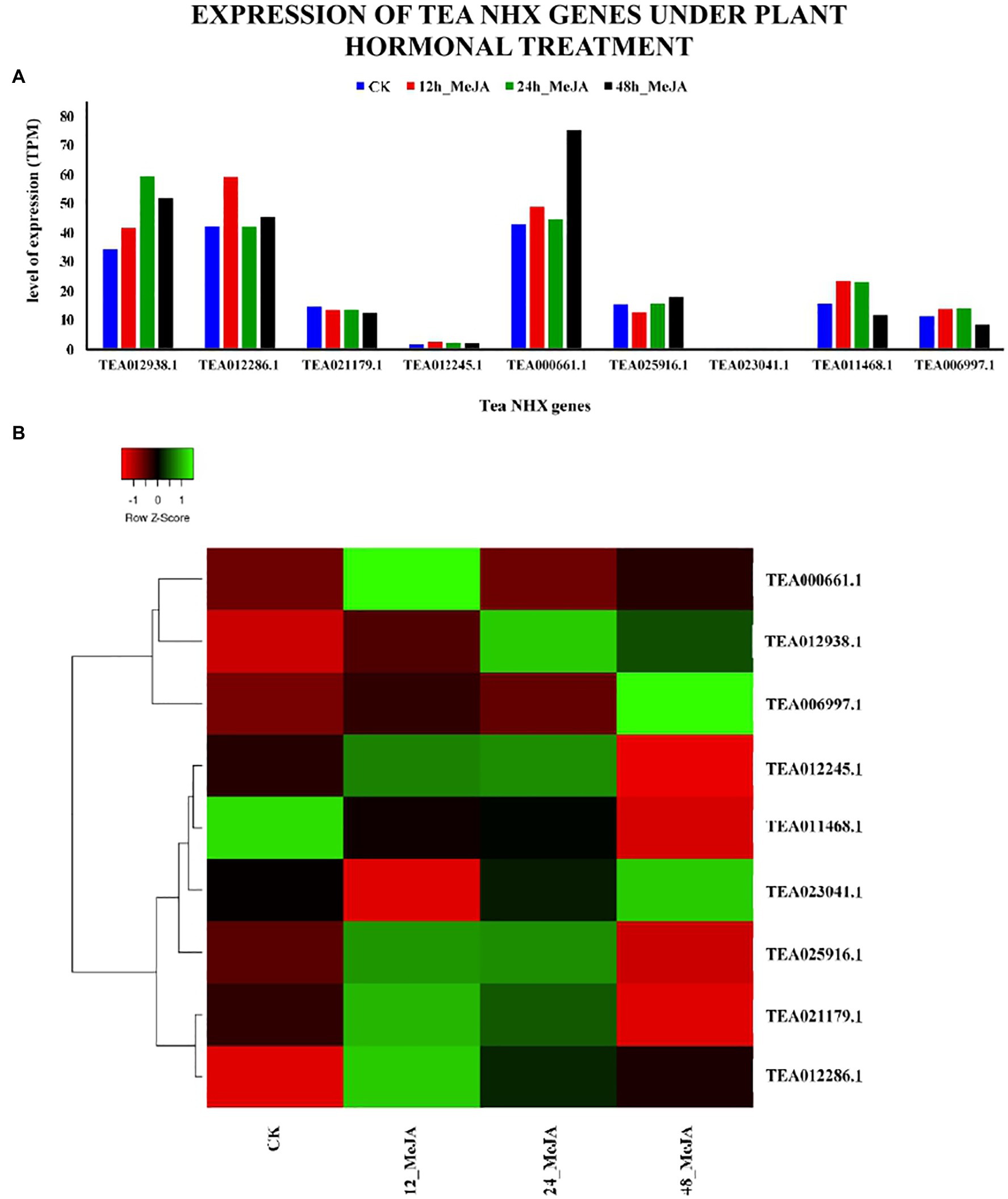
Figure 12. Expression levels of NHX genes in C. sinensis under plant hormonal treatment. The plant hormone under study was Methyl-jasmonate (MeJA). (A) The relative expression of Tea NHX genes, represented graphically by analyzing the transcriptome data. (B) Relative expression represented as a heatmap, generated using heatmapper online server. The color bar on the top represents the normalized TPM values. Green and red colors represent the up- and downregulation values while black represents no expression.
Discussion
NHX gene families have already been identified and functionally characterized for several plants, including A. thaliana, rice, wheat, sweet beet, cotton, and other (Kumari et al., 2018; Yarra, 2019; Wu et al., 2019b; Fu et al., 2020; Khare et al., 2021). However, the NHX genes in C. sinensis have not been studied yet. In this study, the gene structure, phylogenetic relationship, genomic distribution, and expression of NHX genes in C. sinensis were all analyzed at the genomic level. A diverse no. of NHX genes have been identified in various plant species. Gene duplication and loss specific to different subfamilies of NHX over the course of evolution could explain these differences in the number of NHX genes in plants. A total of 9 NHX genes have been identified in C. sinensis based on the Na+/H+ exchanger domain (Table 1).
In-silico studies based on subcellular localizations showed that NHXs are grouped into three classes (Vac-, Endo-, and PM-class). In A. thaliana, both NHX7 and NHX8 are localized in the plasma membrane (Shi et al., 2002), whereas in tea, CsNHX9 (TEA006997.1) localized in the plasma membrane, CsNHX8 (TEA011468.1) is localized in endosome, and the others in the vacuole (Table 1). Members in each of the classes from algae to higher plants showed that the NHX families were fairly similar, indicating that NHXs had conserved functions throughout the evolutionary process (Chanroj et al., 2012). The function of NHX transporters may be influenced by their subcellular localization. Members of the NHX family, which are found on both the plasma membrane and tonoplast, help to maintain ionic homeostasis by excluding and compartmentalizing excess Na+. Furthermore, endomembrane-bound NHX members have been discovered to be important for cellular cargo trafficking, growth development, and protein processing regulation (Bassil et al., 2011a,b). The exon/intron structural diversity, which plays an important role in the evolution of gene families, brings to the evidence for phylogenetic groupings. In C. sinensis, CsNHX3 (TEA021179.1) possesses a greater number of introns (18) and exons (19) while CsNHX1 (TEA012938.1) has lesser number of introns (12) and exons (18) than the rest of the 5 genes present in Vac-class. However, in P. trichocarpa, Vac-class NHXs (PtNHX1-5) contain 14 exons and the Endo-class NHX (PtNHX6) has 22 exons, while the PM-class NHXs (PtNHX7 and PtNHX8) displays 23 exons (Tian et al., 2017). Similarly, for NHX genes in G. max (Gm), seven members of GmNHX contain 14–15 exons, whereas the rest three members have 20 exons (Chen et al., 2015). These findings suggested that NHX gene families in plants have a fair share of structural diversity.
The putative amiloride-binding site and membrane-spanning pore in the NHX gene families, which contain the amino acid sequence “FFIYLLPPI” (Bassil et al., 2012), have been found to be highly conserved (Brett et al., 2005; Rodriguez Rosales et al., 2009; Bassil et al., 2012). In the presence of the drug amiloride and/or its derivatives, this domain inhibits the cation/H+ exchange (Wu et al., 2011). In the motif study, the amiloride-binding site was found to be located in the N-terminus of motif 3 and it is found in 6 NHX genes of C. sinensis (Figure 2). The C-terminus of NHX proteins was diverse in contrast to the conserved N-terminus. Studies have shown that the deletion of the C-terminal hydrophilic region results in increased Na+/H+ transport activity, implying that the C-terminus is important not only for subcellular localization but also for transport activity regulation (Yamaguchi et al., 2003; Orlowski and Grinstein, 2007). The phylogenetic analysis indicated that the NHXs in P. trichocarpa (Tian et al., 2017), S. bicolor (Kumari et al., 2018), and Beta vulgaris (Wu et al., 2019b) showed three phylogenetic clusters based on their location in the cell; we found the same results for tea NHX transporters. So according to these findings, the NHX family genes have remained relatively conserved throughout evolution.
Cis-acting regulatory elements function as key molecular switches in transcriptional regulation of gene activities that control a variety of biological processes, such as hormonal response, abiotic stress response, and development (Ding et al., 2018; Verma et al., 2019). Hormones including ABA, ethylene, SA, and IAA play significant roles in plants development and stress response (Mishra et al., 2014; Li et al., 2019; Wang and Huang, 2019; Zhang and Li, 2019). In this study, cis-acting regulatory elements related to transcription factors were identified to be randomly distributed across the promotor region of the 9 tea NHXs (Supplementary Table S2). One ABA-responsive element (ABRE) has been discovered in 6 NHXs (CsNHX1, CsNHX2, CsNHX4, CsNHX5, CsNHX6, and CsNHX8) of C. sinensis (Supplementary Table S2). Whereas in poplar, one or two ABREs were observed (Tian et al., 2017). This analysis showed that NHX genes may play a role in the ABA signaling pathway. Furthermore, ARE (anaerobic induction), DRE (drought-responsive cis-acting element), LTR (low-temperature responsive element), MBS (drought response), and STRE (stress response) were identified as stress responsive regulatory elements in tea. Similarly, in PtNHXs from poplar and SbNHXs from S. bicolor are also found to contain similar elements (Kumari et al., 2018). The results indicated that the identified regulatory elements in this study aid in understanding their roles in various abiotic and biotic stress-related pathways.
Further to understand the distribution pattern of the tea NHXs, the genomic distribution mapping was performed. Tandem duplication events were absent across the tea NHXs (Figure 5). The duplication of genes increases the functional divergence, which is an essential factor in adaptability under changing environmental conditions (Conant and Wolfe, 2008). The dN/dS ratio indicates different selection pressure on genes throughout the evolutionary changes. Wang et al. (2019) found that positive selection of a gene during evolution increases its potential and transcription levels under stress conditions in Triticum aestivum and TaBT1. Whereas in tea, the dN/dS ratios provided conclusive evidence that strong purifying selection pressure existed during evolution, allowing a variety of factors to regulate the genes (Supplementary Table S3).
In plants, sodium-proton antiporters facilitate both Na+/H+ and K+/H+ exchanges, contributing to stress tolerance as well as K+ nutrition (Venema et al., 2002; Apse et al., 2003; Leidi et al., 2010). NHXs have been reported to enhance salinity tolerance in different species, such as A. thaliana (Shi et al., 2000), B. vulgaris (Xia et al., 2002), S. lycopersicum (Zhang and Blumwald, 2001; Rodríguez-Rosales et al., 2008), Hordeum vulgare (Vasekina et al., 2005), Z. mays (Zörb et al., 2005), T. aestivum (Brini et al., 2005), G. max (Li et al., 2006), O. sativa (Fukuda et al., 2011; Zeng et al., 2018), and S. bicolor (Kumari et al., 2018). The expression data for various tissues and stress conditions showed that the tea NHXs may be involved in developmental processes and abiotic stress responses. Our study revealed that in C. sinensis, the NHX genes express differentially in 8 different tissues (Supplementary Table S5). The different expression patterns in various tissues (Figure 8) indicated that the NHX gene family provides opportunities to break the functional constraint from the original gene during the course of evolution.
Based on data from other species, functional annotation and interaction analysis of NHX proteins can help us predict their potential regulatory roles. The electrochemical gradient of protons across tonoplasts, generated by two vacuolar H+-pumps, H+-ATPase, and H+-PPase, has been shown to drive the Vac-class NHXs (Brett et al., 2005; Bao et al., 2009; Wu et al., 2011). In this analysis, all the tea genes considered for building the interaction network, belongs to the Vac-class. By increasing cation accumulation, co-expression of ZxNHX and ZxVP1 genes can improve salt tolerance in transgenic plant species, such as Lotus corniculatus (Bao et al., 2014), Alfalfa (Bao et al., 2015), and sugar beet (Wu et al., 2015). These finding suggested that when plants were exposed to salt stress, Vac-class NHXs might work together to transport Na+ across tonoplasts. Calcineurin B-like (CBL) is well known for its ability to interact and modulate CBL-interacting protein kinases (CIPK), which then mediate Ca+ signal transduction (Yu et al., 2014; Tian et al., 2017). During the salinity response, CBL regulates NHX7 (SOS1) and CIPK mediates the Ca2+ signaling pathway (Miranda et al., 2017). A salt stress elicited Ca2+ signal activates a protein kinase complex consisting of CBL4 (SOS3) and CIPK24 (SOS2), and the complex then phosphorylates and activates the SOS1 protein to extrude Na+ out of the cell in A. thaliana under salt stress (Quintero et al., 2011). In transgenic tobacco, overexpression of SOS1 gene increased salt tolerance by maintaining a higher K+/Na+ ratio (Yue et al., 2012). In the current study, CLBs are hypothesized to interact with TEA006066.1, CsNHX1 (TEA012938.1), CsNHX2 (TEA012286.1), CsNHX3 (TEA021179.1), and CsNHX4 (TEA012245.1) but not with CIPK (Figure 7). Similarly, NHX7 (SOS1) interactions with CBLs were predicted in poplar (Tian et al., 2017) and S. bicolor (Bassil et al., 2011a). However, in the future, yeast two hybrid research will need to confirm these proteins interactions.
Ion transporters are important in many biological processes, including ion uptake and sequestration, energy provision, and cell expansion (Bassil and Blumwald, 2014). Previous studies in plants found that Na+/H+ antiporters as important members in transporters mediate the coupled exchange of Na+ or K+ for H+ in all cellular compartments (Qiu, 2012; Bassil and Blumwald, 2014). The NHX genes primarily use two proton pumps, the H+-ATP enzyme and H+-PPase, to produce H+ electrochemical gradients that transport Na+ from the cytoplasm to vacuoles or outside the cell, thereby maintain Na+ ion stability and avoiding the toxic effect of Na+ accumulation in cells (Munns, 2002; Sze and Chanroj, 2018).
Stress response analysis showed that each tea NHX genes were responsive to abiotic stresses of drought, cold, and salt. Under PEG treatment, the expression of CsNHX1 (TEA012938.1), CsNHX4 (TEA012245.1), TEA00066.1, CsNHX7 (TEA023041.1), CsNHX8 (TEA011468.1) reached the highest level at 12 h (Figure 10), and CsNHX1, CsNHX7 and CsNHX8 also responded to salt stress in varying degree, demonstrating these genes may be associated with salt and drought stress. MeJA was found to be linked to salt tolerance in few studies (Zhang et al., 2019; Zhao et al., 2019) and the expression data suggested the close relation of the NHXs toward the regulation of MeJA. Further in the study, the expression levels of CsNHX1, CsNHX7, and CsNHX8 were significantly upregulated by various concentrations of NaCl over a 48 h period and 72 h period (Figure 11), and their expression levels under high-salt stress were relatively higher than those under either mild or moderate-salt stress. In Reaumuria trigyna, the expression levels of RtNHX1 in leaves showed an increase and reached a high level at 3 h, and then reduced after 6 h when exposed to high-salt stress (200 mm NaCl; Li et al., 2017). A similar expression pattern was found in sweet potato, where IbNHX2 was significantly upregulated at 4 h after treatment of 200 mm NaCl (Wang et al., 2016). Another study (Lu et al., 2014) found that the transcription level of TaNHX3 in both leaves and roots sharply increased at 24 h and then gradually decreased after 48 h over a 96 h period in different wheat cultivars subjected to salt stress. Moreover, CsNHX1 belonging to the Vac-class NHX showed the highest level of expression for all the salt stress condition. The study showed that the expression levels of Vac-class NHXs are significantly higher than other class genes thereby confirming that Vac-class NHXs might play critical roles in salt tolerance. The study also notices that CsNHX1 and CsNHX7 showed significant expression levels under all abiotic stress conditions thereby providing a comprehensive understanding of the functions of NHXs in C. sinensis.
Conclusion
Among the numerous transporters in monovalent cation/proton antiporter (CAP1) family, the Na+/H+ antiporters (NHXs) are secondary ion transporters to exchange H+ and transfer the Na+ or K+ across membrane. The objective of this study was to identify, characterize, and determine the role of NHX genes in tea at the genomic stage. Using phylogenetic relationship, the 9 tea NHXs were grouped into three major classes (Vac-, Endo-, and PM-classes). The amiloride-binding site (FFIYLLPPI) is a characteristic feature of NHX proteins and it was found in the N-terminus of motif 3. The lack of tandem duplication events was a result of the close distribution pattern of the NHX genes in tea. ABA-responsive element (ABRE) was found in 6 genes, implying NHX gene might be involved in ABA signaling pathway as well. Furthermore, responses of tea NHX to drought, cold, and salinity indicated that the genes were involved in single or multiple stress responses. The study also confirms the active role of Vac-class NHXs in response to salt stress over the other two classes. CsNHX1 (TEA012938.1) and CsNHX7 (TEA023041.1) maintained high expression levels for all the abiotic stress conditions thereby giving us a comprehensive understanding of the role of NHX genes. Adding to the tally, the responses to MeJA treatment also suggested the involvement of the tea NHXs in MeJA regulation. Tea is a commercial crop, grown all over the world and abiotic stresses are one of the major factors that limit the crop productivity worldwide. This work will therefore serve as a basis to provide valuable information for future studies and exploration of the role of NHX genes in various developmental process, as well as the elucidation of other potential functions in C. sinensis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
AP and AC designed and performed the experiments. SS and NM devised the experiments. GS helped in data analysis and writing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by the Key Technologies R&D Program for Crop Breeding of Zhejiang Province (2016C02054-19 and 2017C02010), the Natural Science Foundation of China (31670303), and the Joint Laboratory of Olive Oil Quality and Nutrition among China, Australia, and Spain.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are thankful to DBT-eLibrary Consortium (DeLCON) for providing access to e-resources.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.777884/full#supplementary-material
Abbreviations
ABA, Abscisic acid; ABRE, ABA-responsive element; ARE, Anaerobic-responsive element; CBL, Calcineurin B-like proteins; CDC2, Cell division cycle protein 2 CDS Coding sequences; CIPK, CBL-interacting protein kinases; CMP, Calcium-binding protein; CYB5R1, NADH-cytochrome b5 reductase 1; DER, Drought-responsive element; ERE, Ethylene-responsive element; GSDS, Gene structure display serve; GSK3, Glycogen synthase kinase 3; HKT, High-affinity K+ transporter; IAA, Indole Acetic Acid; LTR, Low-temperature responsiveness; ORF, Open reading frame; pI, Isoelectric point; PM, Plasma membrane; PPI, Protein–protein interaction; SA, Salicylic acid; SOS1, Salt overly sensitive 1; 3-D, Three-dimension; TM, Transmembrane helical domain; VP, Vacuolar H+-PPase; MEME, Multiple expectation maximization for motif elicitation; MW, Molecular weight; NHX, Na+/H+ antiporter; ROS, Reactive-oxygen species; TPIA, Tea Plant Information Archive; CS/Cs, Camellia sinensis; TAIR, The Arabidopsis Information Resource; HMM, Hidden Markov Model; dN, non-synonymous substitution; dS, Synonymous substitution; NJ, Neighbor Joining; AT/At, Arabidopsis thaliana; CK, Non acclimated; CA1, Fully acclimated; CA3, De-acclimated; TPM, Transcripts per million; UTR, Untranslated Region.
Footnotes
2. ^https://www.arabidopsis.org/
3. ^http://rice.plantbiology.msu.edu/
4. ^http://smart.embl-heidelberg.de/
6. ^http://gpcr.biocomp.unibo.it/bacello/index.htm
7. ^http://www.cbs.dtu.dk/services/TMHMM/
9. ^http://gsds.cbi.pku.edu.cn/
10. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
11. ^http://mg2c.iask.in/mg2c_v2.0/
12. ^https://www.hiv.lanl.gov/content/sequence/SNAP/SNAP.html
References
Aharon, G. S., Apse, M. P., Duan, S., Hua, X., and Blumwald, E. (2003). Characterization of a family of vacuolar Na+/H+ antiporters in Arabidopsis thaliana. Plant Soil 253, 245–256. doi: 10.1023/A:1024577205697
Akram, U., Song, Y., Liang, C., Abid, M. A., Askari, M., Myat, A. A., et al. (2020). Genome-wide characterization and expression analysis of NHX gene family under salinity stress in Gossypium barbadense and its comparison with Gossypium hirsutum. Genes 11:803. doi: 10.3390/genes11070803
Apse, M. P., Aharon, G. S., Snedden, W. A., and Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258. doi: 10.1126/science.285.5431.1256
Apse, M. P., Sottosanto, J. B., and Blumwald, E. (2003). Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 36, 229–239. doi: 10.1046/j.1365-313X.2003.01871.x
Babicki, S., Arndt, D., Marcu, A., Liang, Y., Grant, J. R., Maciejewski, A., et al. (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, W147–W153. doi: 10.1093/nar/gkw419
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bao, A. K., Du, B. Q., Touil, L., Kang, P., Wang, Q. L., and Wang, S. M. (2015). Co-expression of tonoplast cation/H+ antiporter and H+-pyrophosphatase from xerophyte Zygophyllum xanthoxylum improves alfalfa plant growth under salinity, drought and field conditions. Plant Biotechnol. J. 14, 964–975. doi: 10.1111/pbi.12451
Bao, A. K., Wang, S. M., Wu, G. Q., Xi, J. J., Zhang, J. L., and Wang, C. M. (2009). Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.). Plant Sci. 176, 232–240. doi: 10.1016/j.plantsci.2008.10.009
Bao, A. K., Wang, Y. W., Xi, J. J., Liu, C., Zhang, J. L., and Wang, S. M. (2014). Co-expression of xerophyte Zygophyllum xanthoxylum ZxNHX and ZxVP1-1 enhances salt and drought tolerance in transgenic Lotus corniculatus by increasing cations accumulation. Funct. Plant Biol. 41, 203–214. doi: 10.1071/FP13106
Bassil, E., and Blumwald, E. (2014). The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr. Opin. Plant Biol. 22, 1–6. doi: 10.1016/j.pbi.2014.08.002
Bassil, E., Coku, A., and Blumwald, E. (2012). Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 63, 5727–5740. doi: 10.1093/jxb/ers250
Bassil, E., Ohto, M. A., Esumi, T., Tajima, H., Zhu, Z., Cagnac, O., et al. (2011a). The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. Plant Cell 23, 224–239. doi: 10.1105/tpc.110.079426
Bassil, E., Tajima, H., Liang, Y., Ohto, M., Ushijima, K., Nakano, R., et al. (2011b). The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23, 3482–3497. doi: 10.1105/tpc.111.089581
Berardini, T. Z., Reiser, L., Li, D., Mezheritsky, Y., Muller, R., Strait, E., et al. (2015). The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53, 474–485. doi: 10.1002/dvg.22877
Bowers, J. E., Chapman, B. A., Rong, J., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. doi: 10.1038/nature01521
Brett, C. L., Donowitz, M., and Rao, R. (2005). Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 288, C223–C239. doi: 10.1152/ajpcell.00360.2004
Brini, F., Gaxiola, R. A., Berkowitz, G. A., and Masmoudi, K. (2005). Cloning and characterization of a wheat vacuolar cation/proton antiporter and pyrophosphatase proton pump. Plant Physiol. Biochem. 43, 347–354. doi: 10.1016/j.plaphy.2005.02.010
Chanroj, S., Wang, G., Venema, K., Zhang, M. W., Delwiche, C. F., and Sze, H. (2012). Conserved and diversified gene families of monovalent cation/H(+) antiporters from algae to flowering plants. Front. Plant Sci. 3:25. doi: 10.3389/fpls.2012.00025
Chatterjee, A., Paul, A., Unnati, G. M., Rajput, R., Biswas, T., Kar, T., et al. (2020). MAPK cascade gene family in Camellia sinensis: in-silico identification, expression profiles and regulatory network analysis. BMC Genomics 21:613. doi: 10.1186/s12864-020-07030-x
Chen, H., Chen, X., Wu, B., Yuan, X., Zhang, H., Cui, X., et al. (2015). Whole-genome identification and expression analysis of K+ efflux antiporter (KEA) and Na+/H+ antiporter (NHX) families under abiotic stress in soybean. J. Integr. Agric. 14, 1171–1183. doi: 10.1016/S2095-3119(14)60918-7
Conant, G. C., and Wolfe, K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi: 10.1038/nrg2482
Deutsch, M., and Long, M. (1999). Intron-exon structures of eukaryotic model organisms. Nucleic Acids Res. 27, 3219–3228. doi: 10.1093/nar/27.15.3219
Ding, X., Li, J., Pan, Y., Zhang, Y., Ni, L., Wang, Y., et al. (2018). Genome-wide identification and expression analysis of the UGlcAE gene family in tomato. Int. J. Mol. Sci. 19:1583. doi: 10.3390/ijms19061583
Fedorova, L., and Fedorov, A. (2003). “Introns in gene evolution,” in Origin and Evolution of New Gene Functions. ed. M. Long (Dordrecht, Netherlands: Springer), 123–131.
Fu, X., Lu, Z., Wei, H., Zhang, J., Yang, X., Wu, A., et al. (2020). Genome-wide identification and expression analysis of the NHX (sodium/hydrogen antiporter) gene family in cotton. Front. Genet. 11:964. doi: 10.3389/fgene.2020.00964
Fukuda, A., Nakamura, A., Hara, N., Toki, S., and Tanaka, Y. (2011). Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 233, 175–188. doi: 10.1007/s00425-010-1289-4
Gan, H. H., Perlow, R. A., Roy, S., Ko, J., Wu, M., Huang, J., et al. (2002). Analysis of protein sequence/structure similarity relationships. Biophys. J. 83, 2781–2791. doi: 10.1016/s0006-3495(02)75287-9
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). “Protein identification and analysis tools on the ExPASy server,” in The Proteomics Protocols Handbook. ed. J. M. Walker (Springer: Humana Press), 571–607.
Goyal, R. K., Tulpan, D., Chomistek, N., Fundora, D. G., West, C., Ellis, B. E., et al. (2018). Analysis of MAPK and MAPKK gene families in wheat and related Triticeae species. BMC Genomics 19:178. doi: 10.1186/s12864-018-4545-9
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jiangtao, C., Yingzhen, K., Qian, W., Yuhe, S., Daping, G., Jing, L., et al. (2015). Mapgene2chrom, a tool to draw gene physical map based on Perl and SVG languages. Hereditas 37, 91–97. doi: 10.16288/j.yczz.2015.01.013
Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. doi: 10.1186/1939-8433-6-4
Khare, T., Joshi, S., Kaur, K., Srivastav, A., Shriram, V., Srivastava, A. K., et al. (2021). Genome-wide in silico identification and characterization of sodium-proton (Na+/H+) antiporters in Indica rice. Plant Gene 26:100280. doi: 10.1016/j.plgene.2021.100280
Korber, B. (2000). “HIV signature and sequence variation analysis,” in Computational Analysis of HIV Molecular Sequences. eds. G. R. Allen and H. L. Gerald (Dordrecht, Netherlands: Kluwer), 55–72.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kumari, P. H., Kumar, S. A., Ramesh, K., Reddy, P. S., Nagaraju, M., Prakash, A. B., et al. (2018). Genome-wide identification and analysis of Arabidopsis sodium proton antiporter (NHX) and human sodium proton exchanger (NHE) homologs in Sorghum bicolor. Genes 9:236. doi: 10.3390/genes9050236
Leidi, E. O., Barragán, V., Rubio, L., El-Hamdaoui, A., Ruiz, M. T., Cubero, B., et al. (2010). The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 61, 495–506. doi: 10.1111/j.1365-313X.2009.04073.x
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Li, K., Liu, Z., Xing, L., Wei, Y., Mao, J., Meng, Y., et al. (2019). miRNAs associated with auxin signaling, stress response, and cellular activities mediate adventitious root formation in apple rootstocks. Plant Physiol. Biochem. 139, 66–81. doi: 10.1016/j.plaphy.2019.03.006
Li, N., Wang, X., Ma, B., Du, C., Zheng, L., and Wang, Y. (2017). Expression of a Na+/H+ antiporter RtNHX1 from recretohalophyte Reaumuria trigyna improved salt tolerance of transgenic Arabidopsis thaliana. J. Plant Physiol. 218, 109–120. doi: 10.1016/j.jplph.2017.07.015
Li, W. Y. F., Wong, F. L., Tsai, S. N., Phang, T. H., Shao, G., and Lam, H. M. (2006). Tonoplast-located GmCLC1 and GmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow (BY)-2 cells. Plant Cell Environ. 29, 1122–1137. doi: 10.1111/j.1365-3040.2005.01487.x
Li, N. N., Yue, C., Cao, H. L., Qian, W. J., Hao, X. Y., Wang, Y. C., et al. (2018). Transcriptome sequencing dissection of the mechanisms underlying differential cold sensitivity in young and mature leaves of the tea plant (Camellia sinensis). J. Plant Physiol. 224, 144–155. doi: 10.1016/j.jplph.2018.03.017
Liu, Z., An, C., Zhao, Y., Xiao, Y., Bao, L., Gong, C., et al. (2021). Genome-wide identification and characterization of the CsFHY3/FAR1 gene family and expression analysis under biotic and abiotic stresses in tea plants (Camellia sinensis). Plan. Theory 10:570. doi: 10.3390/plants10102186
Liu, W., Li, W., He, Q., Daud, M. K., Chen, J., and Zhu, S. (2014). Genome-wide survey and expression analysis of calcium-dependent protein kinase in Gossypium raimondii. PLoS One 9:e98189. doi: 10.1371/journal.pone.0116352
Liu, Z., Shi, L., Liu, Y., Tang, Q., Shen, L., Yang, S., et al. (2015). Genome wide identification and transcriptional expression analysis of mitogen-activated protein kinase and mitogen-activated protein kinase kinase genes in Capsicum annuum. Front. Plant Sci. 6:780. doi: 10.3389/fpls.2015.00780
Lu, W., Guo, C., Li, X., Duan, W., Ma, C., Zhao, M., et al. (2014). Overexpression of TaNHX3, a vacuolar Na+/H+ antiporter gene in wheat, enhances salt stress tolerance in tobacco by improving related physiological processes. Plant Physiol. Biochem. 76, 17–28. doi: 10.1016/j.plaphy.2013.12.013
Miranda, R. S., Alvarez-Pizarro, J. C., Costa, J. H., Paula, S. O., Prisco, J. T., and Gomes-Filho, E. (2017). Putative role of glutamine in the activation of CBL/CIPK signaling pathways during salt stress in sorghum. Plant Signal. Behav. 12:e1361075. doi: 10.1080/15592324.2017.1361075
Mishra, S., Shukla, A., Upadhyay, S., Sharma, P., Singh, S., Phukan, U. J., et al. (2014). Identification, occurrence, and validation of DRE and ABRE cis-regulatory motifs in the promoter regions of genes of Arabidopsis thaliana. J. Integr. Plant Biol. 56, 388–399. doi: 10.1111/jipb.12149
Munns, R. (2002). Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Orlowski, J., and Grinstein, S. (2007). Emerging roles of alkali cation/proton exchangers in organellar homeostasis. Curr. Opin. Cell Biol. 19, 483–492. doi: 10.1016/j.ceb.2007.06.001
Pardo, J. M., Cubero, B., Leidi, E. O., and Quintero, F. J. (2006). Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J. Exp. Bot. 57, 1181–1199. doi: 10.1093/jxb/erj114
Pearson, W. R. (2013). An introduction to sequence similarity (“homology”) searching. Curr. Protoc. Bioinformatics 42:3.1. doi: 10.1002/0471250953.bi0301s42
Pehlivan, N., Sun, L., Jarrett, P., Yang, X., Mishra, N., Chen, L., et al. (2016). Cooverexpressing a plasma membrane and a vacuolar membrane sodium/proton antiporter significantly improves salt tolerance in transgenic Arabidopsis plants. Plant Cell Physiol. 57, 1069–1084. doi: 10.1093/pcp/pcw055
Pierleoni, A., Martelli, P. L., Fariselli, P., and Casadio, R. (2006). BaCelLo: a balanced subcellular localization predictor. Bioinformatics 22:e408-16. doi: 10.1093/bioinformatics/btl222
Qiu, Q. S. (2012). Plant and yeast NHX antiporters: roles in membrane trafficking. J. Integr. Plant Biol. 54, 66–72. doi: 10.1111/j.1744-7909.2012.01097.x
Quintero, F. J., Martinez-Atienza, J., Villalta, I., Jiang, X. Y., Kim, W. Y., Ali, Z., et al. (2011). Activation of the plasma membrane Na/H antiporter salt-overly-sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. U. S. A. 108, 2611–2616. doi: 10.1073/pnas.1018921108
Ratner, A., and Jacoby, B. (1976). Effect of K, its counter anion, and pH on sodium efflux from barley root tips. J. Exp. Bot. 27, 843–852. doi: 10.1093/jxb/27.5.843
Reguera, M., Bassil, E., and Blumwald, E. (2014). Intracellular NHX type cation/H+ antiporters in plants. Mol. Plant 7, 261–263. doi: 10.1093/mp/sst091
Robert, C. E. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Roberto, A., Gaxiola, R. R., Amir, S., Paula, G., and Seth, L. A. (1999). The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. U. S. A. 96, 1480–1485. doi: 10.1073/pnas.96.4.1480
Rodriguez Rosales, M. P., Galvez, F. J., Huertas, R., Aranda, M. N., Baghour, M., Cagnae, O., et al. (2009). Plant NHX cation/proton antiporters. Plant Signal. Behav. 4, 265–276. doi: 10.4161/psb.4.4.7919
Rodríguez-Rosales, M. P., Jiang, X., Gálvez, F. J., Aranda, M. N., Cubero, B., and Venema, K. (2008). Overexpression of the tomato K+/H+ antiporter LeNHX2 confers salt tolerance by improving potassium compartmentalization. New Phytol. 179, 366–377. doi: 10.1111/j.1469-8137.2008.02461.x
Rombauts, S., Déhais, P., Van Montagu, M., and Rouzé, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Rubio, F., Gassmann, W., and Schroeder, J. I. (1995). Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270, 1660–1663. doi: 10.1126/science.270.5242.1660
Shi, H., Ishitani, M., Kim, C., and Zhu, J. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U. S. A. 97, 6896–6901. doi: 10.1073/pnas.120170197
Shi, J., Ma, C., Qi, D., Lv, H., Yang, T., Peng, Q., et al. (2015). Transcriptional responses and flavor volatiles biosynthesis in methyl jasmonate-treated tea leaves. BMC Plant Biol. 15:233. doi: 10.1186/s12870-015-0609-z
Shi, H., Quintero, F. J., Pardo, J. M., and Zhu, J. K. (2002). The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell 14, 465–477. doi: 10.1105/tpc.010371
Sonnhammer, E. L. L., von Heijne, G., and Krogh, A. (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182.
Sze, H., and Chanroj, S. (2018). Plant endomembrane dynamics: studies of K(+)/H(+) antiporters provide insights on the effects of pH and ion homeostasis. Plant Physiol. 177, 875–895. doi: 10.1104/pp.18.00142
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Cepas, J. H., et al. (2019). STRING v11: proteinprotein association networks with increased coverage, supporting functional discovery in genome wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Taft, R. J., Pheasant, M., and Mattick, J. S. (2007). The relationship between non-proteincoding DNA and eukaryotic complexity. BioEssays 29, 288–299. doi: 10.1002/bies.20544
Tian, F., Chang, E., Li, Y., Sun, P., Hu, J., and Zhang, J. (2017). Expression and integrated network analyses revealed functional divergence of NHX-type Na+/H+ exchanger genes in poplar. Sci. Rep. 7:2607. doi: 10.1038/s41598-017-18149-5
Vasekina, A., Yershov, P., Reshetova, O., Tikhonova, T., Lunin, V., Trofimova, M., et al. (2005). Vacuolar Na+/H+ antiporter from barley: identification and response to salt stress. Biochemistry 70, 100–107. doi: 10.1007/s10541-005-0057-8
Venema, K., Quintero, F. J., Pardo, J. M., and Donaire, J. P. (2002). The Arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. J. Biol. Chem. 277, 2413–2418. doi: 10.1074/jbc.M105043200
Verma, D., Lakhanpal, N., and Singh, K. (2019). Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. BMC Genomics 20:227. doi: 10.1186/s12864-019-5593-5
Wan, S. Q., Wang, W., Zhou, T., Zhang, Y., Chen, J., Xiao, B., et al. (2018). Transcriptomic analysis reveals the molecular mechanisms of Camellia sinensis in response to salt stress. Plant Growth Regul. 84, 481–492. doi: 10.1007/s10725-017-0354-4
Wang, Y., Hou, J., Liu, H., Li, T., Wang, K., Hao, C., et al. (2019). TaBT1, affecting starch synthesis and thousand kernel weight, underwent strong selection during wheat improvement. J. Exp. Bot. 70, 1497–1511. doi: 10.1093/jxb/erz032
Wang, J., and Huang, R. (2019). Modulation of ethylene and ascorbic acid on reactive oxygen species scavenging in plant salt response. Front. Plant Sci. 10:319. doi: 10.3389/fpls.2019.01782
Wang, Y., Liu, Z., Wu, Z., Li, H., Wang, W. L., Cui, X., et al. (2018). Genome-wide identification and expression analysis of GRAS family transcription factors in tea plant (Camellia sinensis). Sci. Rep. 8:3949. doi: 10.1038/s41598-018-36676-7
Wang, B., Zhai, H., He, S., Zhang, H., Ren, Z., and Zhang, D. A. (2016). Vacuolar Na+/H+ antiporter gene, IbNHX2, enhances salt and drought tolerance in transgenic sweetpotato. Sci. Hortic. 201, 153–166. doi: 10.1016/j.scienta.2016.01.027
Wang, X., Zhao, Q., Ma, C., Zhang, Z. H., Cao, H. L., Kong, Y. M., et al. (2013). Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics 14:415. doi: 10.1186/1471-2164-14-415
Waters, S., Gilliham, M., and Hrmova, M. (2013). Plant high-affinity potassium (HKT) transporters involved in salinity tolerance: structural insights to probe differences in ion selectivity. Int. J. Mol. Sci. 14, 7660–7680. doi: 10.3390/ijms14047660
Wei, C. L., Yang, H., Wang, S. B., Zhao, J., Liu, C., Gao, L. P., et al. (2018). Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. U. S. A. 115, E4151–E4158. doi: 10.1073/pnas.1719622115
Wu, G. Q., Feng, R. J., Wang, S. M., Wang, C. M., Bao, A. K., Wei, L., et al. (2015). Co-expression of xerophyte Zygophyllum xanthoxylum ZxNHX and ZxVP1-1 confers enhanced salinity tolerance in chimeric sugar beet (Beta vulgaris L.). Front. Plant Sci. 6:581. doi: 10.3389/fpls.2015.00581
Wu, A., Hao, P., Wei, H., Sun, H., Cheng, S., Chen, P., et al. (2019a). Genome-wide identification and characterization of glycosyltransferase family 47 in cotton. Front. Genet. 10:824. doi: 10.3389/fgene.2019.00824
Wu, G. Q., Wang, Q., Bao, A. K., and Wang, S. M. (2011). Amiloride reduces sodium transport and accumulation in the succulent xerophyte Zygophyllum xanthoxylum under salt conditions. Biol. Trace Elem. Res. 139, 356–367. doi: 10.1007/s12011-010-8662-9
Wu, G. Q., Wang, J. L., and Li, S. J. (2019b). Genome-wide identification of Na(+)/H(+) antiporter (NHX) genes in sugar beet (Beta vulgaris L.) and their regulated expression under salt stress. Genes 10:401. doi: 10.3390/genes10050401
Wu, G. Q., Xi, J. J., Wang, Q., Ma, Q., Bao, A. K., Zhang, J. L., et al. (2011). The ZxNHX gene encoding tonoplast Na+/H+ antiporter in the xerophyte Zygophyllum xanthoxylum plays important roles in response to salt and drought. J. Plant Physiol. 168, 758–767. doi: 10.1016/j.jplph.2010.10.015
Xia, T., Ape, M. P., Aharon, G. S., and Blumwald, E. (2002). Identification and characterization of a NaCl-inducible vacuolar Na+/H+ antiporter in Beta vulgaris. Physiol. Plant. 116, 206–212. doi: 10.1034/j.1399-3054.2002.1160210.x
Xia, E., Li, F., Tong, W., Li, P. H., Wu, Q., Zhao, H. J., et al. (2019). Tea plant information archive (TPIA): a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol. J. 17, 1938–1953. doi: 10.1111/pbi.13111
Xie, H., Wang, Y., Ding, Y., Qiu, C., Sun, L., Gai, Z., et al. (2019). Global ubiquitome profiling revealed the roles of ubiquitinated proteins in metabolic pathways of tea leaves in responding to drought stress. Sci. Rep. 9:4286. doi: 10.1038/s41598-019-54958-6
Yamaguchi, T., Apse, M. P., Shi, H., and Blumwald, E. (2003). Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc. Natl. Acad. Sci. U. S. A. 100, 12510–12515. doi: 10.1073/pnas.2034966100
Yarra, R. (2019). The wheat NHX gene family: potential role in improving salinity stress tolerance of plants. Plant Gene 18:100178. doi: 10.1016/j.plgene.2019.100178
Yu, Q., An, L., and Li, W. (2014). The CBL-CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 33, 203–214. doi: 10.1007/s00299-013-1507-1
Yue, Y., Zhang, M., Zhang, J., Duan, L., and Li, Z. (2012). SOS1 gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio. J. Plant Physiol. 169, 255–261. doi: 10.1016/j.jplph.2011.10.007
Zeng, Y., Li, Q., Wang, H., Zhang, J., Du, J., Feng, H., et al. (2018). Two NHX-type transporters from Helianthus tuberosus improve the tolerance of rice to salinity and nutrient deficiency stress. Plant Biotechnol. J. 16, 310–321. doi: 10.1111/pbi.12773
Zhang, H. X., and Blumwald, E. (2001). Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat. Biotechnol. 19, 765–768. doi: 10.1038/90824
Zhang, Q., Cai, M., Yu, X., Wang, L., Guo, C., Ming, R., et al. (2017). Transcriptome dynamics of Camellia sinensis in response to continuous salinity and drought stress. Tree Genet. Genomes 13:78. doi: 10.1007/s11295-017-1161-9
Zhang, Y., and Li, X. (2019). Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 50, 29–36. doi: 10.1016/j.pbi.2019.02.004
Zhang, G., Li, C., Li, Q., Li, B., Larkin, D. M., Lee, C., et al. (2014). Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1320. doi: 10.1126/science.1251385
Zhang, W., Liu, S., Li, C., Zhang, P., and Zhang, P. (2019). Transcriptome sequencing of Antarctic moss under salt stress emphasizes the important roles of the ROS-scavenging system. Gene 696, 122–134. doi: 10.1016/j.gene.2019.02.037
Zhao, M., Yin, L., Ma, J., Zheng, J., Wang, Y., Lan, J., et al. (2019). GmERF135 the roles of in improving salt tolerance and decreasing ABA sensitivity in soybean. Front. Plant Sci. 10:940. doi: 10.3389/fpls.2019.00940
Zhu, J. K. (2001). Plant salt tolerance. Trends Plant Sci. 6, 66–71. doi: 10.1016/S1360-1385(00)01838-0
Keywords: salt tolerance, Arabidopsis, genome-wide search, expression profiles, Camellia sinensis, NHXs
Citation: Paul A, Chatterjee A, Subrahmanya S, Shen G and Mishra N (2021) NHX Gene Family in Camellia sinensis: In-silico Genome-Wide Identification, Expression Profiles, and Regulatory Network Analysis. Front. Plant Sci. 12:777884. doi: 10.3389/fpls.2021.777884
Edited by:
Xiyin Wang, Agricultural University of Hebei, ChinaReviewed by:
Yuhua Wang, Nanjing Agricultural University, ChinaShoupu He, National Key Laboratory of Cotton Biology, Institute of Cotton Research (CAAS), China
Copyright © 2021 Paul, Chatterjee, Subrahmanya, Shen and Mishra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoxin Shen, Z3VveGluLnNoZW5AdHR1LmVkdQ==; Neelam Mishra, bmVlbGFtaWl0a2dwQGdtYWlsLmNvbQ==; bmVlbGFtbWlzaHJhQHNqYy5hYy5pbg==
†These authors have contributed equally to this work
 Abhirup Paul
Abhirup Paul Archita Chatterjee
Archita Chatterjee Shreya Subrahmanya
Shreya Subrahmanya Guoxin Shen
Guoxin Shen Neelam Mishra
Neelam Mishra