
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 01 October 2021
Sec. Plant Development and EvoDevo
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.760041
This article is part of the Research Topic Growth Patterns Underlying Plant Development View all 20 articles
 Chen-li Zhu1†
Chen-li Zhu1† Bao Xing1†
Bao Xing1† Shou-zhen Teng1
Shou-zhen Teng1 Chen Deng1
Chen Deng1 Zhen-yong Shen3
Zhen-yong Shen3 Peng-fei Ai3
Peng-fei Ai3 Tie-gang Lu1
Tie-gang Lu1 Sheng-wei Zhang2*
Sheng-wei Zhang2* Zhi-guo Zhang1*
Zhi-guo Zhang1*Leaf angle is one of the most important agronomic traits in rice, and changes in leaf angle can alter plant architecture to affect photosynthetic efficiency and thus determine grain yield. Therefore, it is important to identify key genes controlling leaf angle and elucidate the molecular mechanisms to improve rice yield. We obtained a mutant rela (regulator of leaf angle) with reduced leaf angle in rice by EMS mutagenesis, and map-based cloning revealed that OsRELA encodes a protein of unknown function. Coincidentally, DENSE AND ERECT PANICLE 2 (DEP2) was reported in a previous study with the same gene locus. RNA-seq analysis revealed that OsRELA is involved in regulating the expression of ILI and Expansin family genes. Biochemical and genetic analyses revealed that OsRELA is able to interact with OsLIC, a negative regulator of BR signaling, through its conserved C-terminal domain, which is essential for OsRELA function in rice. The binding of OsRELA can activate the expression of downstream genes repressed by OsLIC, such as OsILI1, a positive regulator of leaf inclination in rice. Therefore, our results suggest that OsRELA can act as a transcriptional regulator and is involved in the regulation of leaf inclination by regulating the transcriptional activity of OsLIC.
As one of the agronomic traits closely related to grain yield, leaf angle not only controls plant architecture but also moderate leaf erectness is essential for improving rice yield (Wang and Li, 2011). Leaf erectness improves light capture and CO2 diffusion efficiency (Sakamoto et al., 2006), thus increasing photosynthetic efficiency. The leaf angle in rice is determined by the lamina joint that connects the leaf blade and leaf sheath. Compared with the leaf tissue, mesophyll cells were replaced by parenchyma cells, and more sclerenchyma cells were gathered around the vascular bundles on the adaxial and abaxial sides. This structure not only provides mechanical strength to the lamina joint but also a determinant factor of leaf angle (Zhou et al., 2017). Many genes have been reported to be involved in the regulation of lamina joint development in rice, and many of these mutants have shown great potential to increase yield. Therefore, it has become the goal of breeders to find and elucidate the molecular mechanism of genes in regulating leaf angle in rice.
In rice, many transcription factors have been reported to be involved in the regulation of lamina joint development; therefore, control of leaf angle can be achieved through genetic manipulation. Altering the expression of some specific transcription factors resulted in increased leaf angle; for example, overexpression of the Lax panicle1 (LAX1) gene resulted in increased leaf angle (Komatsu et al., 2003), while the OsLIC (LEAF and TILLER ANGLE INCREASED CONTROLLER) mutant exhibited a significant increase in leaf angle and tiller angle (Wang et al., 2008). Overexpression of auxin response Factor 19 (OsARF19) can promote cell division on the adaxial side of the lamina joint, which results in an increased leaf angle (Zhang et al., 2015). Interestingly, overexpression of OsIAA1, a repressor of OsARF, also led to increased leaf angle (Song et al., 2009). However, some mutants of transcription factors exhibit reduced leaf angle, such as the OsLIGULELESS1 (OsLG1) mutant, which has a complete absence of the lamina joint, auricle and ligule tissues, resulting in erect leaves (Lee et al., 2007). The reduced leaf angle 1 (RLA1) and BRASSINAZOLE RESISTANT1 (OsBZR1) protein complex is a positive regulator of the BR signaling pathway, and their mutants have an obvious erect leaf phenotype (Bai et al., 2007; Qiao et al., 2017). INCLINATION1 (ILI1) and ILI1 binding bHLH (IBH1) are direct downstream target genes of the transcription factors OsBZR1 and OsLIC, which are antagonistically involved in regulating the elongation of parenchyma cells in the lamina joint (Zhang L.Y. et al., 2009). It has also been reported that REGULATOR OF LEAF INCLINATION 1 (RLI1) can directly activate the expression of BRASSINISTEROID UPREGULATED1 (BU1) and BUL1 COMPLEX1 (BC1), thereby promoting elongation of lamina joint cells (Ruan et al., 2018). Although multiple types of transcription factors are involved in the regulation of the leaf angle, most of them are related to the metabolism and signaling of plant hormones, such as auxin and brassinosteroids.
In addition to transcription factors, some enzymes related to the transport, metabolism, or signaling of auxin and brassinosteroids are also involved in the development of the lamina joint. For example, LAZY1 regulates leaf angle by affecting the gravitropism of aboveground tissues of rice (Li et al., 2007), while Loose Plant Architecture 1 (LPA1) regulates the expression of OsPIN1a and thus affects the polar transport of auxin (Sun et al., 2019). The GH3 family gene LEAF INCLINATION1 (LC1) and an increased number of tillers, enlarged leaf angles, and dwarfism (TLD1) can promote cell elongation by decreasing auxin content at the lamina joint (Zhang S.W. et al., 2009; Zhao et al., 2013). Inhibition of auxin receptor OsTIR1 expression also resulted in increased leaf angle, suggesting that auxin can negatively regulate leaf angle by inhibiting cell division and cell elongation (Bian et al., 2012). Mutations in the cytochrome P450 genes D2, D11 and OsDWARF, which are involved in BR biosynthesis (Mori et al., 2002; Hong et al., 2003; Tanabe et al., 2005), and the receptor for BR signaling OsBRI1 cause shortening of parenchyma cells of the lamina joint and result in an erect leaf phenotype (Yamamuro et al., 2000). The expression and protein stability of the U-type cyclin CYC U4;1 is negatively regulated by BR, which positively regulates leaf erectness by promoting the division of sclerenchyma cells on the abaxial side of the lamina joint (Sun et al., 2015). From these analyses, it is clear that auxin and BRs are of great importance in the regulation of leaf angle; however, whether other molecules regulate leaf angle by directly or indirectly integrating the metabolism or physiological process of hormones in rice needs to be further evaluated.
In this study, we obtained a mutant rela (regulator of leaf angle) with reduced leaf angle in rice by EMS mutagenesis. RNA-seq analysis revealed that OsRELA is involved in the regulation of OsEXPA5, OsEXPB6, and OsEXPB11 and OsILI1, OsILI4 (OsBU1), and OsILI5 (OsBUL1) expression. Biochemical and genetic analyses revealed that OsRELA interacts with OsLIC through the conserved C-terminal domain, and the binding of OsRELA can activate the expression of downstream genes repressed by OsLIC, thus affecting the leaf angle by positively regulating the expression of OsILI1 in rice.
To screen for genes involved in the regulation of leaf angle in rice, we created a library of 60Co-γ irradiation-induced mutants from which we isolated a mutant with semidwarf and leaf erectness (Figures 1A,B), reduced panicle and leaf length phenotypes (Supplementary Figures 1B,C), based on which the mutant was named regulator of leaf angle (rela). We crossed the rela mutant with the wild-type and found that the ratio of wild-type to mutant in the segregating population was 415:134, close to 3:1, indicating that rela is a recessive mutation controlled by a single nuclear gene. To further reveal the reason for the reduced leaf angle of the rela mutant, we performed a transverse section of the lamina joint and found that the number of sclerenchyma cell layers and the cell area on the abaxial side of the rela mutant were increased compared to those of the wild-type (Figures 1C,F,G). Longitudinal section revealed that the length of parenchyma cells on the adaxial side of the rela mutant was shorter than that of the wild-type (Figures 1D,E). In addition, longitudinal section of the uppermost internode revealed that the cell length in the rela mutant was significantly shorter than that of the wild-type (Supplementary Figures 1A,D–F). These results suggest that OsRELA promotes the elongation of cells in the culm and parenchyma cells on the adaxial side and inhibits the division and expansion of sclerenchyma cells on the abaxial side of the lamina joint, thus positively regulating the leaf angle and plant height.
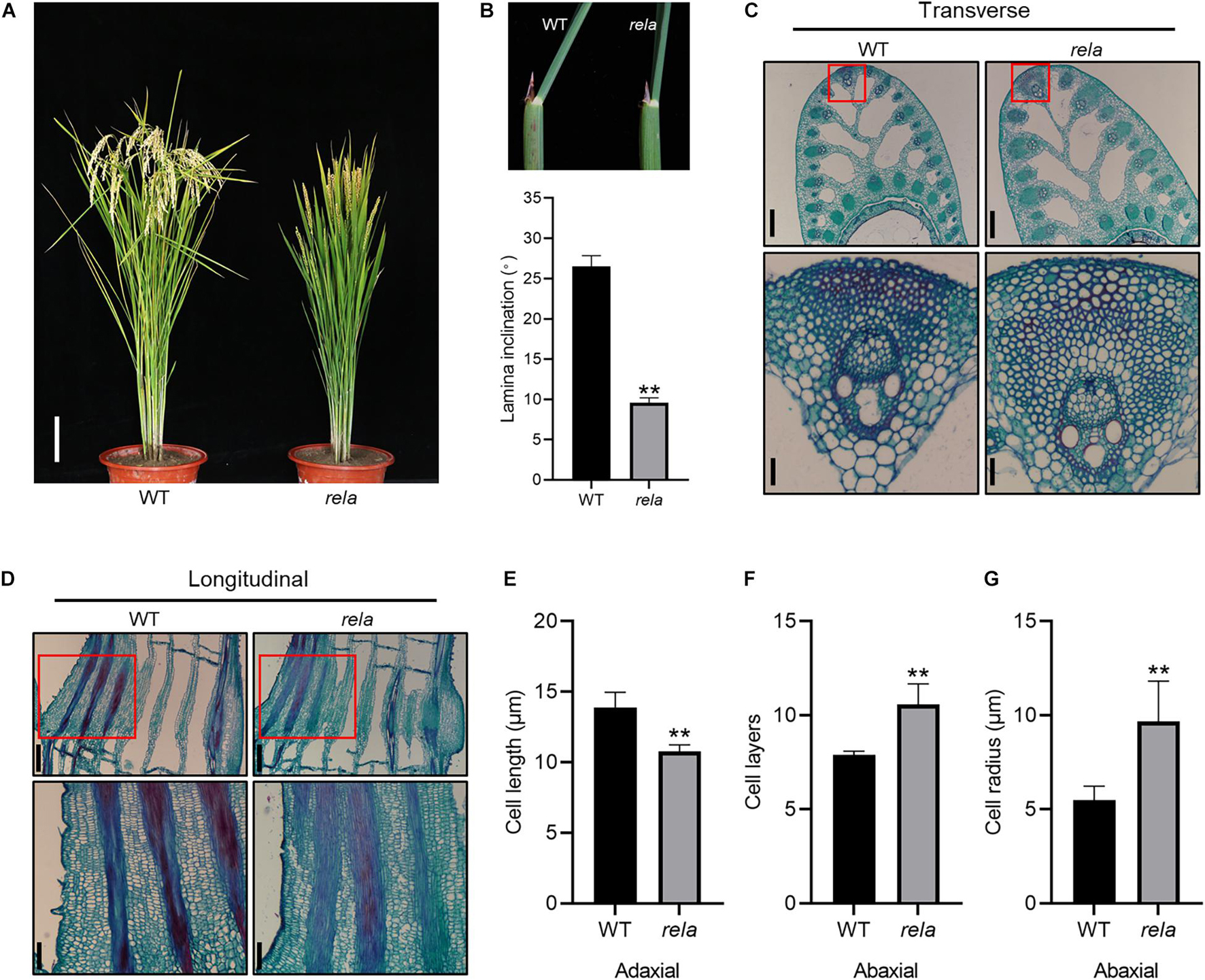
Figure 1. Phenotypic characterization of the rela mutant. (A) The morphological phenotypes of the wild-type and rela at the grain-filling stage. Bar = 20 cm. WT, wild-type. (B) Quantification of the leaf inclination of the second lamina joint of wild-type and rela. The picture at top shows the second lamina joints. Error bars are SD (n = 30). Asterisks indicate significant differences from the WT (**P < 0.01, Student’s t-test). (C) Transverse section of the lamina joints of wild-type and rela. The red box indicates the enlarged abaxial sides of the lamina joint. Bars = 250 μm in the upper panel and 30 μm in the lower panel. (D) Longitudinal section of the lamina joints of wild-type and rela. The red box indicates the enlarged adaxial sides of the lamina joint. Bars = 250 μm in the upper panel and 30 μm in the lower panel. (E) Measurement of lamina joint adaxial cell lengths of wild-type and rela [shown in (D)]. Error bars are SD (n = 60). Asterisks indicate significant differences from the WT (**P < 0.01, Student’s t-test). (F) The number of sclerenchyma cell layers in the adaxial sides of wild-type and rela [shown in (C)]. Error bars are SD (n = 30). Asterisks indicate significant differences from the WT (**P < 0.01, Student’s t-test). (G) Measurement of sclerenchyma cell radius in the abaxial sides of wild-type and rela [shown in (C)]. Error bars are SD (n = 30). Asterisks indicate significant differences from the WT (**P < 0.01, Student’s t-test).
To clone the gene encoding OsRELA, we crossed the rela mutant with Dular and obtained a segregating population of the F2 generation. By linkage analysis, OsRELA was narrowed between two genetic markers, Indel 7–10 and Indel 7–12, located on the long arm of chromosome 7. By further fine mapping using 2871 F2 generation plants, we targeted the OsRELA gene between SNP 25.338 M, and Indel 25.402 M, and six genes were contained in this 64-kb interval: Os07g0615200, Os07g0615400, Os07g0615500, Os07g0615800, Os07g0616000, and Os07g0616200 (Figure 2A). Further genome sequencing revealed a 28-bp deletion in the seventh exon of the Os07g0616000 gene, which shifted the reading frame and eventually led to a premature stop codon, while the rest of the genes were sequenced without mutations (Figure 2B). Our immunoblotting experiments using a specific polyclonal antibody that recognizes the protein encoded by Os07g0616000 showed that the expression of the protein encoded by Os07g0616000 was not detected in the rela mutant (Figure 2C), indicating that the phenotypes of the rela mutant are most likely due to a 28-bp deletion in the Os07g0616000 gene. Coincidentally, OsRELA is located at the same gene locus as the previously reported DENSE AND ERECT PANICLE 2. DEP2 was revealed to be associated with panicle development (Li et al., 2010), and we also found this phenotype in the rela mutant; however, the biological function of OsRELA involved in the regulation of leaf angle has not been elucidated. Phylogenetic tree analysis revealed that OsRELA belongs to a Spermatophyta-specific gene family, and its homologs are distributed in Gymnosperm, Amborella, Eudicots, and Monocots. However, the gene family has fewer members in Eudicots, and there are three copies in Arabidopsis: AT3G14172, AT1G72410, and AT1G17360. Most of the family members are distributed in the Monocots, and there is only one copy in rice (Supplementary Figure 2).
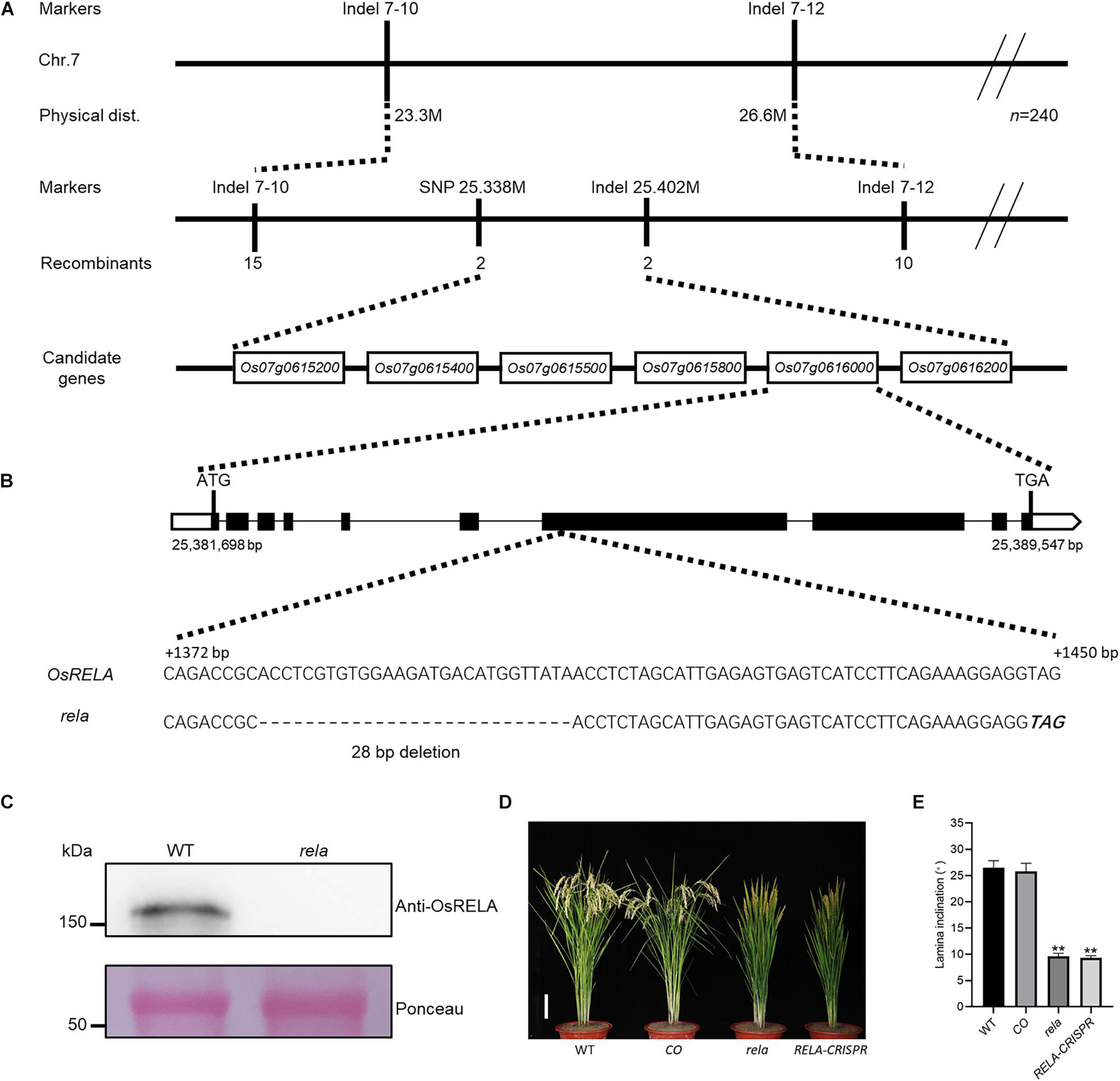
Figure 2. Map-based cloning of OsRELA and complementation test. (A) Linkage map of the OsRELA locus. OsRELA is located on the long arm of chromosome 7, between the molecular marker SNP 25.338 M and Indel 25.402 M, a genomic region of ∼64 kb containing six ORFs. The markers, numbers of recombinants and candidate genes are indicated. (B) Schematic diagram of the OsRELA locus. Comparison with the wild-type sequence revealed a 28-bp deletion (dashed) in the seventh exon, generating a premature stop codon (bold italics). (C) Protein levels of OsRELA in the seedlings of wild-type and rela detected by immunoblot using anti-OsRELA–specific polyclonal antibodies. Ponceau staining of the Rubisco large subunit is shown as a loading control. Molecular masses of proteins (kDa) are shown on the left. (D) Plant morphology of wild-type, rela/proOsRELA:OsRELA(CO), rela and RELA-CRISPR at the heading stage. Bar = 10 cm. (E) Quantification of the leaf inclination of the second lamina joint of wild-type, rela/proOsRELA:OsRELA (CO), rela and RELA-CRISPR plants. Error bars are SD (n = 20). Asterisks indicate significant differences from the WT (**P < 0.01, Student’s t-test).
To further clarify the function of OsRELA in regulating leaf inclination, we constructed the pOsRELA:OsRELA vector and transformed it into the rela mutant. The leaf angle of transgenic lines expressing OsRELA was restored to the wild-type state (Figure 2D). We also obtained genome-edited lines of Os07g0616000 using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) genome editing tool (Supplementary Figure 3). In all genome-edited homozygous lines obtained, the leaf angle was significantly smaller than that of the wild-type line, consistent with the rela mutant phenotype (Figure 2E). These genetic results suggest that OsRELA is the key gene that regulates leaf angle in rice.
To determine the localization of OsRELA in rice cells, we transformed the 35S:OsRELA-GFP vector into rice protoplasts for transient expression. Confocal laser scanning microscopy revealed that the fluorescence of the OsRELA-GFP fusion protein merged with the nuclear marker mCherry-NLS signal (Figure 3A). Consistently, GFP fluorescence of the GFP-OsRELA fusion protein was colocalized with the nuclear stain DAPI in the rela/pOsRELA:GFP-OsRELA lines (Figure 3B), suggesting that OsRELA functions mainly in the nucleus.
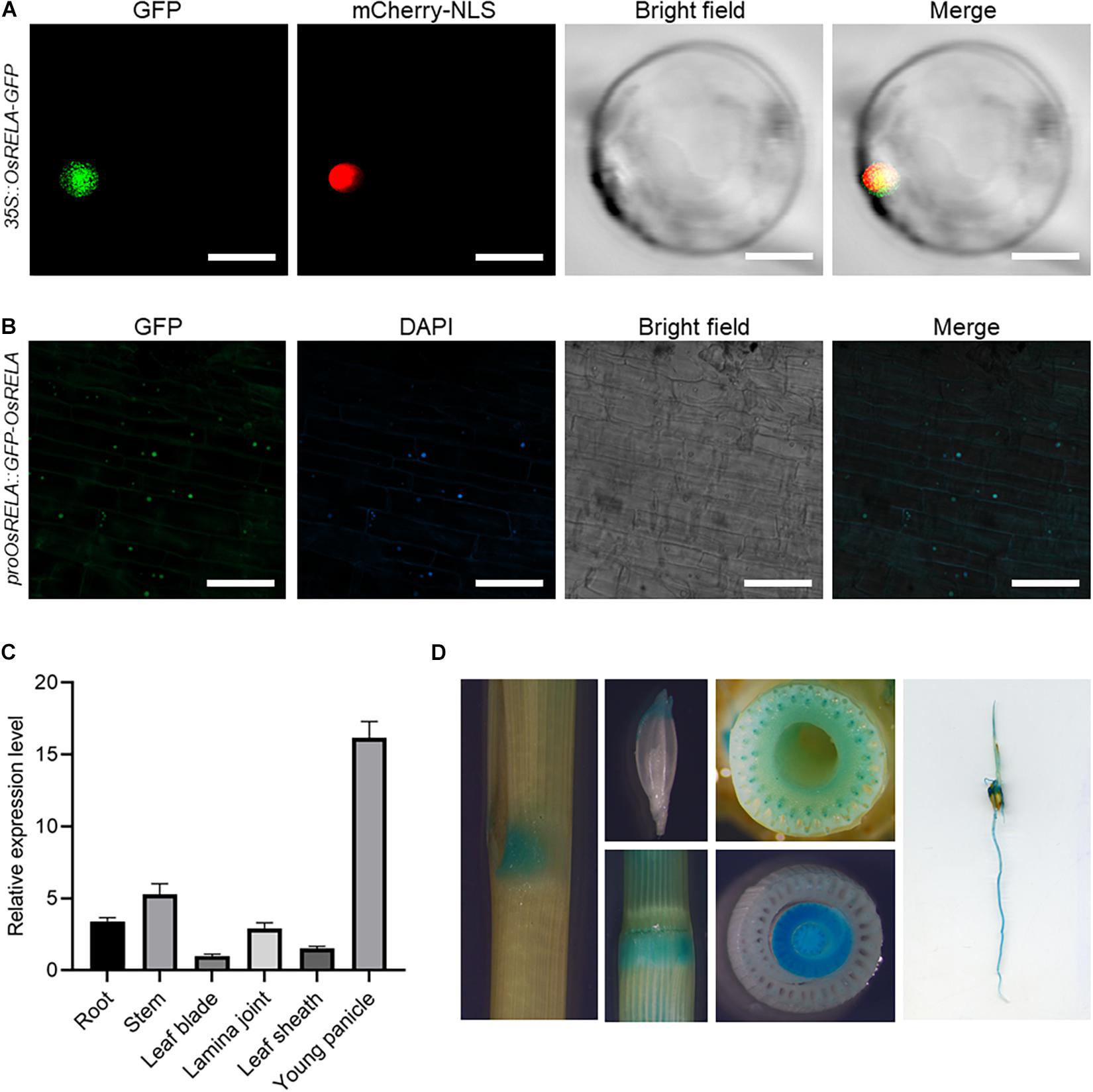
Figure 3. Subcellular localization and tissue expression patterns of OsRELA. (A) Subcellular localization of the OsRELA-GFP fusion protein in rice protoplasts. mCherry-NLS was used as the nuclear marker. Bar = 20 μm. (B) Fluorescent signals in transgenic rice plants expressing GFP-OsRELA. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Bar = 80 μm. (C) RT–qPCR analysis of the expression patterns of OsRELA in various tissues, including root, stem, leaf blade, lamina joint, leaf sheath and young panicle. The rice UBQ gene was amplified as the internal control. (D) Examination of GUS activity in transgenic plants expressing proOsRELA:GUS. GUS activity is found in the lamina joint, young florets, stem, vascular bundles, leaf sheath and root.
To further analyze the expression pattern of OsRELA, we performed real-time quantitative PCR (RT–qPCR) to measure the OsRELA expression level, and the results showed that OsRELA was expressed in various tissues of rice (Figure 3C). GUS staining experiments performed on pOsRELA:GUS transgenic plants revealed strong GUS staining in the lamina joint, young florets, stem, vascular bundles, leaf sheath and root (Figure 3D). All these tissues were associated with the phenotypes of the rela mutant.
To analyze OsRELA-regulated genes that participate in the regulation of leaf angle, we performed RNA-seq analysis on the rela mutant and identified 285 genes with significantly different expression levels compared to the wild-type, including 156 upregulated genes and 129 downregulated genes (Figure 4A). We next performed a hierarchical clustering analysis of all genes that were significantly differentially expressed, and the representative top 50 most significant genes are shown (Figure 4B). In addition, gene ontology (GO) enrichment analysis demonstrated that the molecular functions of the differentially expressed genes were mainly involved in DNA binding (GO:0043566), suggesting that OsRELA is likely to be involved in transcriptional regulation by regulating DNA binding (Figure 4C). We then examined the genes with significant expression changes in the wild type and rela by RT–qPCR. Consistent with the RNA-seq results, the expression of OsEXPA5, OsEXPB6, and OsEXPB11 was significantly upregulated, while the expression of OsmiR408, OsmiR528, ILA1 interacting protein 3, OsOFP14, OsILI1, OsILI4 (OsBU1), and OsILI5 (OsBUL1) was significantly downregulated in the rela mutant (Figure 4D), and the expression of seven genes related to cell wall synthesis was also significantly suppressed in rela (Figure 4E). It is worth noting that ILA1 interacting protein 3 (OsIIP3), OsILI1, OsILI4 (OsBU1), and OsILI5 (OsBUL1) were reported to positively regulate leaf angle (Tanaka et al., 2009; Zhang L.Y. et al., 2009; Ning et al., 2011; Jang et al., 2017). Expansin family genes and cell wall synthesis genes were involved in the process of cell wall loosening and cell extension (Cho and Kende, 1997; Kuluev et al., 2014). The phenotypes of the rela mutant are similar to those of BR-deficient and BR-insensitive mutants (Yamamuro et al., 2000; Hong et al., 2003), and both Expansins and ILI family genes have been reported to be downstream of BR signaling and involved in the regulation of leaf angle (Wang et al., 2018), implying that OsRELA may intersect with the BR signaling pathway to regulate leaf inclination in rice.
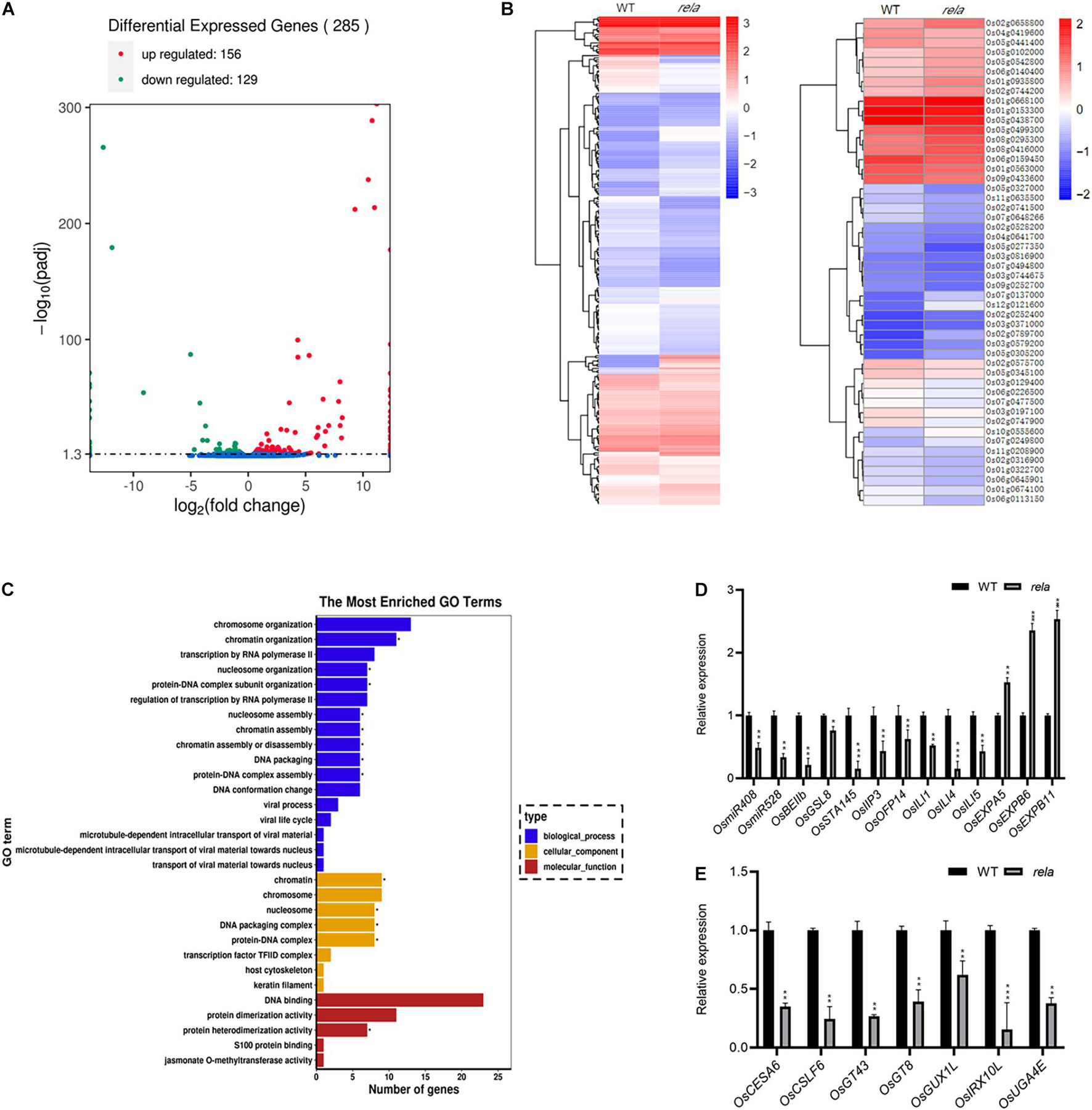
Figure 4. RNA-seq analysis of OsRELA-modulated genes. (A) A volcano plot illustrating differentially expressed genes from RNA-seq analysis between wild-type and rela. Genes upregulated and downregulated are shown in red and green, respectively. Values are presented as the log2 of tag counts. (B) Heat map of the RNA-seq analysis results shows all genes that were significantly differentially expressed (left panel) and the representative top 50 genes that were differentially expressed (right panel). (C) Gene ontology (GO) functional clustering of all genes that were differentially expressed. (D) RT–qPCR validation analysis of the gene expression levels between wild-type and rela revealed by RNA-seq. The rice UBQ gene was amplified as the internal control. Asterisks indicate significant differences from the WT (*P < 0.05, **P < 0.01, ***P < 0.001, Student’s t-test). (E) RT–qPCR analysis of the genes involved in cell wall synthesis. The rice UBQ gene was amplified as the internal control. Asterisks indicate significant differences from the WT (**P < 0.01, ***P < 0.001, Student’s t-test).
To further reveal the relationship between OsRELA and the BR signaling pathway, we obtained OsLIC from a rice cDNA library by a yeast two-hybrid approach using full-length OsRELA as bait (Figure 5A). OsLIC negatively regulates leaf angle and acts as an antagonistic transcription factor of OsBZR1 (Zhang et al., 2012). However, OsRELA does not interact with other BR signaling components, such as OsGSK2, DLT, OsBZR1, and RLA1, in yeast (Supplementary Figure 4). Bimolecular fluorescence complementation (BiFC) assays showed that OsRELA and OsLIC interact in rice cells (Figure 5B), and the semi-in vivo pull-down assay results also indicated that the OsLIC-GST fusion protein interacts with OsRELA (Figure 5C).
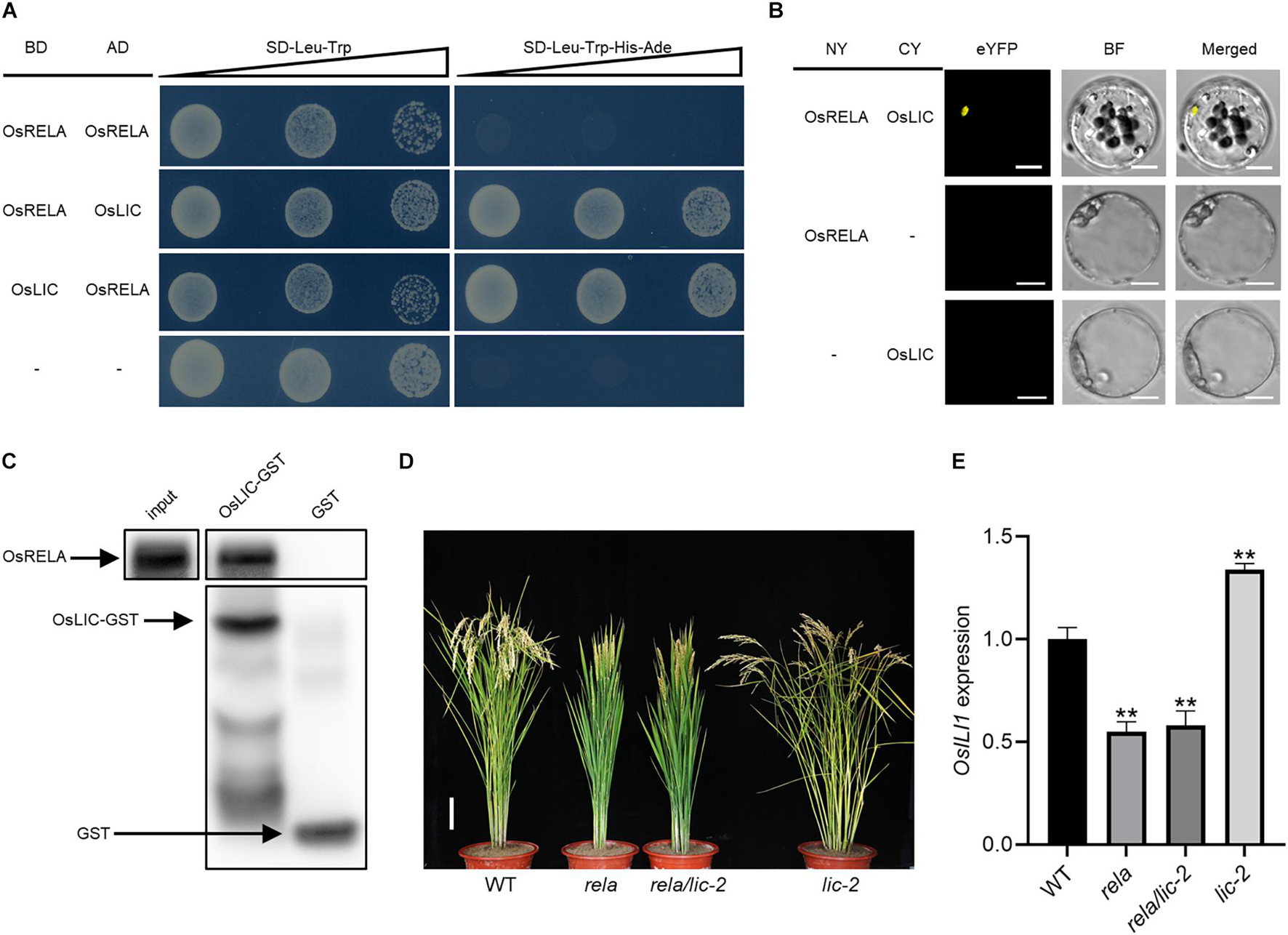
Figure 5. OsRELA physically interacts with and acts downstream of OsLIC. (A) Interactions between OsRELA and OsLIC in yeast two-hybrid assays. OsRELA could not form homodimer in yeast. AD, activation domain; BD, binding domain; SD, synthetic dropout; the gradients indicate tenfold serial dilutions. (B) BiFC analysis of OsRELA and OsLIC interactions in rice protoplasts. BF, brightfield. Scale bars, 5 μm. (C) LIC-GST proteins can pull down OsRELA from total protein extracts of 1-week-old wild-type plants. OsRELA was detected by immunoblotting using anti-OsRELA-specific polyclonal antibodies. (D) Plant morphologies of wild-type, rela, rela/lic-2 and lic-2 at the heading stage. Bar = 10 cm. (E) RT–qPCR analysis of OsILI1 expression levels in wild-type, rela, rela/lic-2 and lic-2 lamina joints. The rice UBQ gene was amplified as the internal control. Asterisks indicate significant differences from the WT (**P < 0.01, Student’s t-test).
To further clarify the genetic relationship between OsRELA and OsLIC, using the CRISPR/Cas9 system, a lic-2 mutant line was obtained by targeting the second exon of the OsLIC gene, and DNA sequencing showed that a 1 bp insertion in the second exon of OsLIC resulted in premature termination of translation (Supplementary Figure 5). The lic-2 mutant showed an increased leaf and tiller angle (Wang et al., 2008). We then crossed rela with the lic-2 mutant and found that the phenotypes of the rela/lic-2 double mutant were similar to those of the rela mutant (Figure 5D), implying that the function of OsLIC is dependent on OsRELA. Quantitative real-time PCR results showed that the expression level of OsILI1, the direct downstream target of OsLIC, was reduced in both the rela and rela/lic-2 mutants (Figure 5E). The elevated expression level of OsILI1 and the increased leaf angle in the lic-2 mutant are consistent with what has been reported previously, suggesting that the regulation of OsILI1 expression by OsLIC is dependent on OsRELA.
To clarify the minimal domain of OsRELA that interacts with OsLIC, we performed sequence and bioinformatics analysis on OsRELA. We found a speckle-type BTB/POZ domain at the N-terminus of OsRELA by Bioinformatics Toolkit analysis, which is a multifunctional protein–protein interaction motif (Bardwell and Treisman, 1994). Two COILED-COIL domains were predicted by the ExPASy tool kit: 697–724 aa and 798–849 aa. The COILED-COIL domain plays an important role in mediating protein–protein interactions (Burkhard et al., 2001). A nuclear localization sequence was found in its conserved C-terminal domain by SeqNLS software: 1240–1256 aa (Supplementary Figures 6, 7).
We next created different truncated forms of OsRELA attached to the GAL4-BD domain named BD-OsRELA (1–1365 aa), BD-OsRELA-T1 (1–199 aa), BD-OsRELA-T2 (200–643 aa), BD-OsRELA-T3 (1173–1365 aa) and BD-OsRELA-T4 (1–1172 aa) (Figure 6A). After cotransforming yeast with the OsLIC-AD vector, we found that both OsRELA and OsRELA-T3 interacted strongly with OsLIC, while OsRELA-T1, OsRELA-T2, and OsRELA-T4 did not interact with OsLIC (Figure 6B). The above results suggest that OsRELA interacts with OsLIC through its conserved C-terminal domain. We further used the promoter of OsILI1 to drive luciferase (LUC) as a reporter and OsRELA-FLAG, OsRELAΔC-FLAG, OsLIC-FLAG, and FLAG as effectors for the transient expression assay (Figure 6C). The results showed that OsLIC represses OsILI1 expression, and when OsRELA-FLAG was cotransformed with OsLIC-FLAG into rice protoplasts, the expression level of OsILI1 was elevated. When OsRELAΔC-FLAG was cotransformed with OsLIC-FLAG, OsILI1 expression was repressed (Figure 6D). These results indicate that OsRELA interacts with OsLIC through its conserved C-terminal domain and thus abolishes the transcriptional repression of OsLIC on OsILI1.
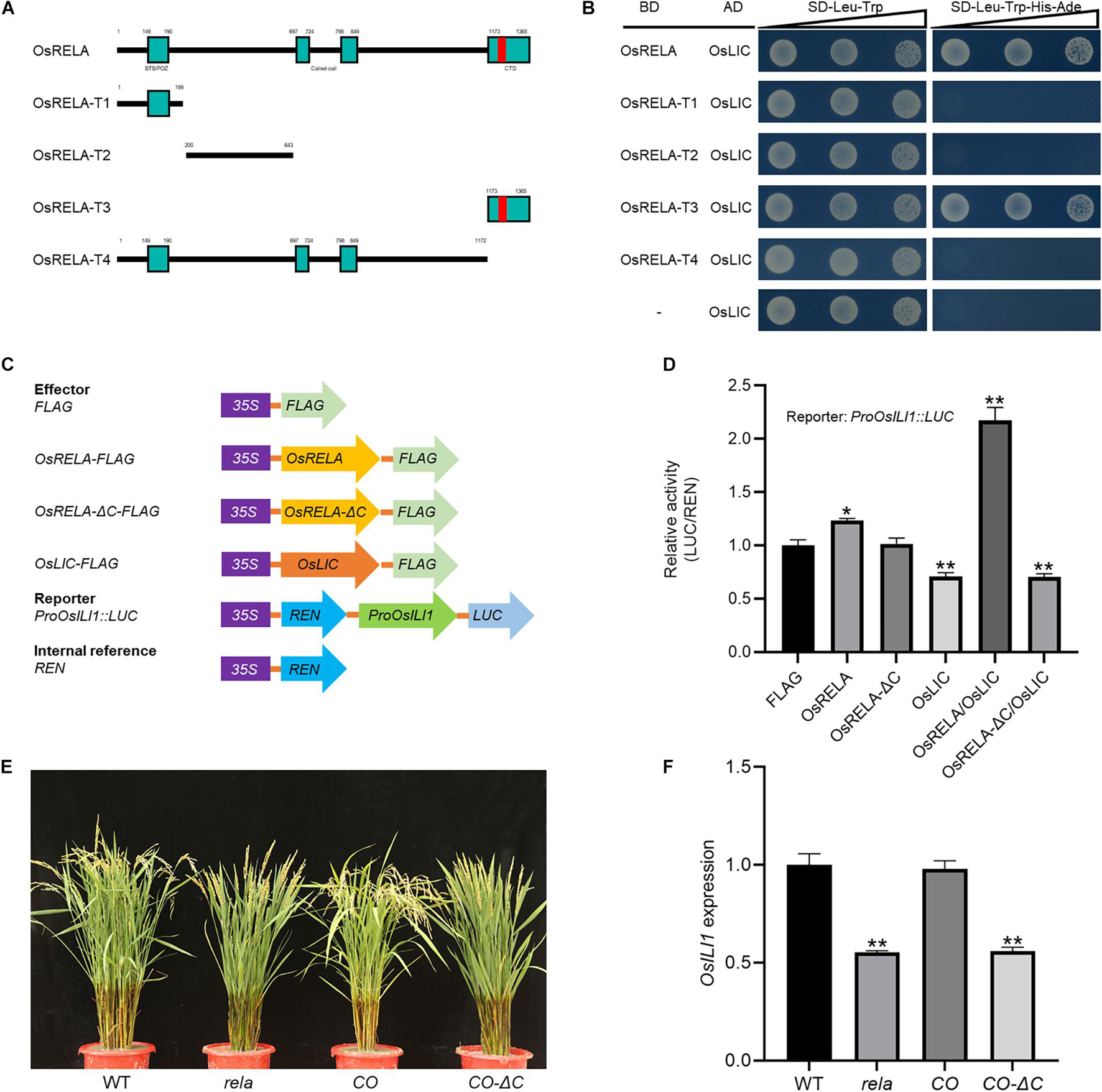
Figure 6. The OsRELA-OsLIC interaction occurs via the conserved C-terminal of OsRELA. (A) Schematic representation of wild-type and different truncated forms of OsRELA. (B) Yeast two-hybrid assays show that OsLIC interacts with OsRELA and OsRELA-T3. AD, activation domain; BD, binding domain; SD, synthetic dropout; the gradients indicate tenfold serial dilutions. (C) Schematic diagrams of the reporter and effector constructs. The firefly luciferase (LUC) gene driven by the OsILI1 promoter and the Renilla luciferase (REN) reporter gene driven by the 35S promoter were used as the reporter and internal control, respectively. For the effectors, OsRELA, OsRELAΔC and OsLIC were fused with FLAG. (D) Transient gene expression assays in rice protoplasts. The LUC reporter gene was cotransfected with OsRELA, OsRELAΔC and OsLIC or both. Asterisks indicate significant differences from the control (*P < 0.05, **P < 0.01, Student’s t-test). (E) Plant morphologies of wild-type, rela, rela/proOsRELA:OsRELA(CO) and rela/proOsRELA:OsRELAΔC(CO–ΔC) at the heading stage. Bar = 10 cm. (F) RT–qPCR analysis of OsILI1 expression levels in wild-type, rela, rela/proOsRELA:OsRELA(CO) and rela/proOsRELA:OsRELAΔC(CO–ΔC) lamina joints. The rice UBQ gene was amplified as the internal control. Asterisks indicate significant differences from the WT (**P < 0.01, Student’s t-test).
Finally, we genetically analyzed the effect of the C-terminal domain of the OsRELA protein on its function and found that deletion of the C-terminal domain could not rescue the phenotypes of the rela mutant (Figure 6E). Moreover, quantitative real-time PCR results revealed that the expression level of OsILI1 was not recovered in the CO-ΔC lines (Figure 6F), indicating that the regulation of OsRELA on OsILI1 expression through OsLIC also depends on its C-terminal domain.
Brassinosteroids are important plant hormones that regulate leaf angle in rice (Tong and Chu, 2018). BR deficiency inhibits the elongation of parenchyma cells on the adaxial side and promotes the division of sclerenchyma cells on the abaxial side of the lamina joint (Zhang L.Y. et al., 2009; Sun et al., 2015); therefore, many BR-deficient and BR-insensitive mutants exhibit an erect leaf phenotype. In this study, we obtained a mutant rela with a reduced leaf angle by EMS mutagenesis and found that the cell length in the internode and adaxial parenchyma cells in the lamina joint was reduced compared with that in the wild type. The number of sclerenchyma cell layers and the cell area on the abaxial side were increased, and these phenotypes were similar to those of BR-related mutants. The expression of OsEXPA5, OsEXPB6, and OsEXPB11 was significantly upregulated in the rela mutant, while the expression of OsILI1, OsILI4 (OsBU1), OsILI5 (OsBUL1), and genes related to cell wall synthesis were significantly downregulated in the rela mutant, and all of these genes were reported to be downstream of the BR signaling pathway. As the only transcription factor in the BR signaling pathway that specifically interacts with OsRELA, OsLIC acts as an antagonistic transcription factor of OsBZR1 to regulate the expression of OsILI1 and OsIBH1, thus affecting leaf inclination. Our results indicate that OsRELA interacts with OsLIC and that OsRELA can act as an important regulator of OsLIC to affect its transcriptional regulation of downstream genes, suggesting that OsRELA intersects with the BR signaling pathway by regulating the transcriptional activity of OsLIC and is thus involved in the regulation of leaf inclination in rice.
OsRELA is located in the same gene locus as previously reported DENSE AND ERECT PANICLE 2 (DEP2) (Li et al., 2010), ERECT PANICLE2 (EP2) (Zhu et al., 2010) and SMALL AND ROUND SEED1 (SRS1) (Abe et al., 2010). OsRELA/DEP2/SRS1 encodes a protein of unknown function. Through the analysis of the evolutionary relationship between OsRELA and its homologs, we found that it is an ancient gene that is conserved among angiosperms, and there are conserved motifs at the N-terminus and C-terminus. Through the GUS reporter gene and RT–qPCR results, we found that OsRELA is strongly expressed in young tissues, such as young spikelets, florets and lamina joints, and the amino acid sequence at its N-terminus changed when the gene evolved from gymnosperms to angiosperms. The large difference between gymnosperms and angiosperms is that angiosperms produce seeds within an enclosure, suggesting that gene evolution may be related to this. In addition, no functional analysis has been reported for its three homologs in Arabidopsis, and future research should verify whether these three genes are involved in the regulation of Arabidopsis architecture, siliques or seed size by similar mechanisms.
Notably, the N-terminal amino acid sequences of OsRELA homologs in Arabidopsis share high similarity to that of COP1-interacting protein 7 (CIP7). It has been reported that CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) acts as an E3 ubiquitin ligase and is able to inhibit photomorphogenesis in the dark by mediating the ubiquitinated degradation of light-inducible transcription factors, such as HY5, HYH, and AtMYB21 (Holm et al., 2002; Shin et al., 2002; Srivastava et al., 2015). CIP7 is also localized in the nucleus and may act as a transcription factor to activate light-induced gene expression as well as anthocyanin synthesis and chlorophyll accumulation. However, CIP7 RNA-interfering plants do not have defects in light-inhibited hypocotyl elongation (Yamamoto et al., 1998). To further investigate the possible mechanism by which OsRELA exerts transcriptional regulatory functions, we performed sequence alignment analysis of OsRELA and CIP7 and found that their N-terminal sequences may encode a speckle-type BTB/POZ domain. The BTB domain (Broad-Complex, Tramtrack, and Bric a brac) is a multifunctional protein–protein interaction domain involved in many biological processes, including transcriptional regulation (Deweindt et al., 1995), ion channel assembly (Aravind and Koonin, 1999) and ubiquitinated protein degradation (Julian et al., 2019). The BTB domain often combines with other structural domains to form multifunctional proteins, BTB-zinc finger proteins were reported to mediate transcriptional repression by interacting with histone deacetylase complexes N-CoR or SMRT (Huynh and Bardwell, 1998). ZBTB24 can specifically bind to the 12-bp DNA motif [CT(G/T)CCAGGACCT] to activate downstream gene expression (Ren et al., 2019), and BACH1 and BACH2 are transcriptional regulators that bind to Maf recognition elements to coordinate transcription activation and repression in cooperate with MAFK (Oyake et al., 1996). LPA1 has also been reported to be involved in a similar process to regulate PIN1a expression in rice (Sun et al., 2019). While our results suggest that OsRELA functions as a transcriptional activator, whether OsRELA has specific DNA binding activity is unclear, and whether its promoter selection for target genes depends on the interacting transcription factors or related proteins needs further validation. In this study, whether the regulation of ILI family gene expression by OsRELA requires the binding of the transcription factor OsLIC is unclear. In addition, the specific mechanism of OsRELA in regulating gene expression needs to be further evaluated. Whether OsRELA regulates gene expression by changing the three-dimensional conformation of chromosomes similar to that of chromatin remodeling factors or regulates the activation or repression of gene expression by recruiting histone modification complexes needs to be further evaluated. Further studies are needed to reveal the specific mechanism of OsRELA.
In this study, we identified OsRELA through forward genetics, which is an important regulator of leaf angle in rice. As summarized in Figure 7, OsRELA regulates several lamina inclination-related gene expressions, which have been demonstrated as the positive regulators in rice leaf inclination (Tong and Chu, 2018). Furthermore, OsRELA affects leaf angle by influencing the transcriptional regulation of OsILI1 via interacting and inhibiting OsLIC, which antagonizes with OsBZR1 each other (Zhang et al., 2012), the key transcriptional factor in BR signaling. However, OsRELA does not interact with OsBZR1, indicating they response to different environmental or developmental cues to regulate lamina inclination via suppressing the OsLIC in rice. Collectively, our data not only clarify a new mechanism of leaf inclination regulation but also provide a reference and new genetic resources for future crop improvement.
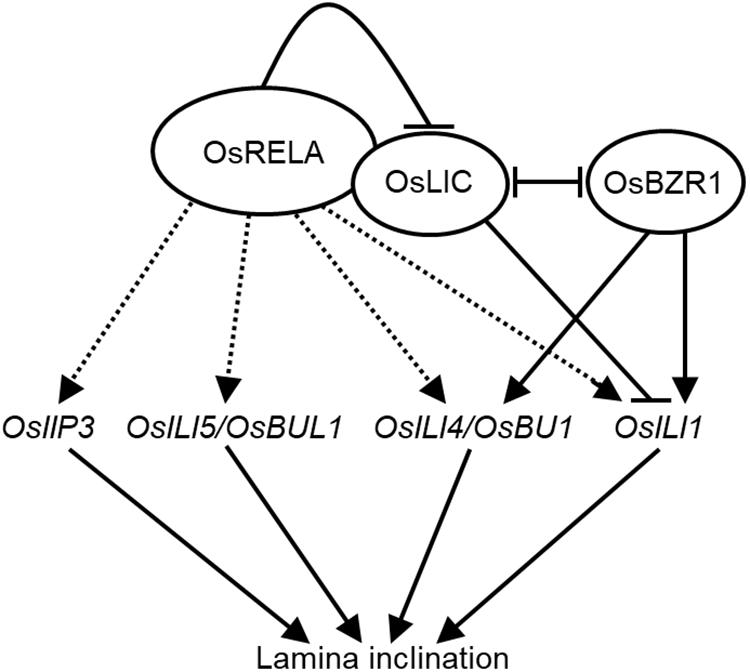
Figure 7. A model for OsRELA in regulating the lamina inclination. OsRELA positively regulates the expression of OsIIP3, OsILI5/OsBUL1, OsILI4/OsBU1, and OsILI1, which are positive regulators involved in the lamina inclination. Since it has been reported that OsBZR1 and OsLIC are pair of antagonistic transcription factors that involved in the lamina inclination by regulate the expression of OsILI1. The binding of OsRELA can activate the expression of OsILI1 repressed by OsLIC as well. Whether the OsRELA directly or indirectly activates the listed gene expressions (dashed arrows) should be elucidated in the future.
In this study, Nipponbare rice (Oryza sativa subsp. japonica) and Dular rice (Oryza sativa L. ssp. indica) were used as wild-type controls, and the rela mutant was obtained from EMS-induced mutant populations. The F2 segregation population used for genetic segregation analysis was obtained by crossing the rela mutant with Nipponbare. The F2 segregant population used for map-based cloning was obtained by crossing the rela mutant with Dular. The rice materials used in this study were grown in the fields of the Chinese Academy of Agricultural Sciences under natural conditions.
After the mature lamina joints of the wild-type and rela mutant were separated, they were rapidly fixed in formaldehyde-acetic acid-alcohol (FAA) at 4°C for 72 h. Subsequently, the samples were dehydrated through a gradient ethanol solution. The dehydrated samples were subjected to a xylene:ethanol clear solution of 1:2, 1:1, and 2:1 and finally transitioned to xylene. The samples were then incubated in a xylene:Paraplast Plus solution (Sigma–Aldrich) solution of 1:1 at 37°C for 48 h. The samples were embedded in Paraplast Plus for 3 days. The samples were then sectioned into 8-μm-thick sections by an RM2245 rotary microtome (Leica). After the removal of Paraplast Plus with xylene:ethanol ratios of 2:1, 1:1, and 1:2, the sections were rehydrated with gradient ethanol, followed by safranin-fixed green staining. Images were taken with an ICC50 HD microscope (Leica), and the cell length and cell area were subsequently measured using ImageJ.
A total of 154 primer pairs exhibiting polymorphisms between Nipponbare and Dular were identified from our primer library. We targeted OsRELA between two genetic markers, Indel 7–10 and Indel 7–12, located on the long arm of chromosome 7. By further fine-mapping using the F2 generation population of 2871 progeny, we targeted the OsRELA gene between two newly developed genetic markers. We then performed PCR amplification and sequencing of the genomic sequences of all genes within this 64-kb interval in wild-type and rela mutants.
For complementation assays, the putative 2.2-kb promoter region of OsRELA was amplified using the OsRELA-Pro primer pairs, and the coding region was amplified using the OsRELA-CDS and OsRELA-CDSΔC primers and inserted into the pCAMBIA1300 binary vector using EcoRI/HindIII restriction sites.
To knock out the OsRELA and OsLIC genes, we selected 20-bp gene-specific spacer sequences in the coding region and cloned them into the sgRNA-Cas9 vector to obtain OsRELA-CRISPR and OsLIC-CRISPR vectors.
To obtain OsRELA overexpression lines, the promoter of OsRELA was amplified by primer OsRELA-Pro, and the full-length CDS was amplified by using primer OsRELA-GFP and inserted into the pCAMBIA1300 binary vector using EcoRI/HindIII restriction sites to obtain proOsRELA:GFP-OsRELA.
To determine the expression pattern of OsRELA, a 2.2-kb region upstream of the ATG start codon was amplified using OsRELA-GUS primers, and the PCR product was cloned into the pCAMBIA1391z binary vector using HindIII/EcoRI restriction sites to obtain the proOsRELA:GUS vector.
The above vectors were subsequently transformed into wild-type and rela mutant via Agrobacterium-mediated transformation.
An in vitro purified OsRELA polypeptide (Os07g0616000, 500–740 aa) fragment was injected into rabbits to generate the corresponding polyclonal antibody at ABclonal Technology. The specificity of the polyclonal antibody against OsRELA was subsequently verified by immunoblotting assays using wild-type rice seedlings. Samples were well ground in liquid nitrogen and then incubated with protein extraction buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, and protease inhibitor cocktail) at 4°C for 15 min. After centrifugation at 4°C for 10 min, the supernatant was boiled for 10 min and separated on a 6% SDS–PAGE gel. The anti-OsRELA polyclonal antibody was diluted at a concentration of 1:2000 and detected by chemiluminescence reagent (GE Health care), and the Rubisco large subunit was used as the endogenous control.
To determine the subcellular localization of the OsRELA protein, the 4095-bp coding region was cloned into the pAN580 vector using the XbaI restriction site. The 35S:OsRELA-GFP vector was subsequently introduced into rice protoplasts as previously described (Zhang et al., 2011), and the fluorescence of GFP was visualized using a confocal laser scanning microscope (LSM 980; Zeiss).
Different tissues of proOsRELA:GUS transgenic plants were incubated in GUS staining buffer (1 M sodium phosphate, pH 7.0, 0.5 M EDTA, pH 8.0, 10% Triton X-100, 50 mM K3Fe(CN)6, 0.1 M X-Gluc) at 37°C after isolation. After 12–16 h, the samples were decolorized using 75% ethanol. The pictures were photographed using an ICC50 HD microscope (Leica).
The sequences of OsRELA and OsLIC were obtained from The Rice Annotation Project1. Homologous of OsRELA in plants were obtained from the NCBI BLAST server2. Multiple sequence alignment was performed using DNAMAN9 with default settings. A neighbor-joining tree of OsRELA and its homologs was generated by MEGA7 with 1000 bootstrap replicates. The protein structure analysis was performed using the following software:
HHpred: https://toolkit.tuebingen.mpg.de/tools/hhpred
COILS: https://embnet.vital-it.ch/software/COILS_form.html
SeqNLS: http://mleg.cse.sc.edu/seqNLS/
Total RNA was extracted from lamina joints of rice seedlings using TRIzol reagent (Invitrogen). First-strand cDNA synthesis was performed using the PrimeScript 1st strand cDNA Synthesis Kit (6110A). RT–qPCR was performed as previously described (Zhou et al., 2008), and the rice ubiquitin gene was used as an internal control.
The differential gene expression analysis of 2-week-old rice seedlings of wild-type and rela mutant were performed by Allwegene Technology Inc3.
The full-length coding region of OsRELA was cloned into the pGBKT7 vector as bait, and then yeast two-hybrid screening was performed with a rice cDNA library. The positive clones were subsequently identified by sequencing. The full-length coding regions of OsRELA, OsLIC, OsBZR1, RLA1, DLT, and OsGSK2 were cloned into the corresponding pGBKT7 and pGADT7 vectors using EcoRI/BamHI restriction sites. The corresponding bait and prey constructs were cotransformed into the yeast strain AH109 (Clontech) and grown on the selective medium SD-Trp/-Leu. The positive clones were then transferred to the selective medium SD-Trp/-Leu/-His/-Ade.
The full-length coding region of OsRELA was amplified and cloned into the pSPYNE vector using EcoRI/SalI to obtain NY-OsRELA, and the full-length coding region of OsLIC was amplified and cloned into the pSPYCE vector using EcoRI/SalI to obtain OsLIC-CY. OsRELA-NY/OsLIC-CY, OsRELA-NY/pSPYCE, and pSPYNE/OsLIC-CY were cotransformed into rice protoplasts by PEG-mediated transformation. The fluorescence of YFP was visualized using a confocal laser scanning microscope (LSM 980; Zeiss).
To test the interaction between OsRELA and OsLIC, the full-length coding region of OsLIC was amplified and cloned into pGEX-4T-1 using EcoRI/XhoI, and the vector was subsequently transformed into E. coli strain BL21 to obtain the OsLIC-GST fusion protein. Two-week-old wild-type seedlings were ground in liquid nitrogen and solubilized with 2X protein extraction buffer [100 mM Tris-HCl, pH 7.5, 300 mM NaCl, 2 mM EDTA, pH 8.0, 1% TrionX-100, 10% (v/v) glycerol, and protease inhibitor mixtures (M307; AMRESCO)]. The semi-in vivo pull-down assay was performed as previously described (Qiao et al., 2017).
In the dual-luciferase assay, the 2.2-kb promoter region of OsILI1 was cloned upstream of the firefly luciferase gene (LUC) in pGreenII 0800-LUC to generate the OsILI1-LUC reporter vector. OsLIC-FLAG, OsRELA-FLAG, OsRELAΔC-FLAG, and FLAG were cloned into pGreenII 62-SK as effectors using XbaI/HindIII restriction sites. The combined reporter and effector vectors were cotransformed into rice protoplasts by PEG-mediated transformation. Renilla reniformis (Ren) driven by the 35S promoter was used as an internal control. The LUC activity was quantified with a Dual-Luciferase Assay Kit (Promega) following the manufacturer’s recommendations, and the relative LUC activity was calculated as the ratio of LUC/Ren.
Sequence data from this article can be found in the database of The Rice Annotation Project (RAP) under the following accession numbers: OsRELA,Os07g0616000;OsLIC,Os06g0704300;OsmiR408,Os01g0322700;OsmiR528,Os03g0129400;OsBEIIb,Os02g0528200;OsG SL8,Os06g0113150;OsSTA145,Os05g0327000;OsIIP3,Os02g0575700;OsOFP14,Os05g0441400;OsILI1,Os04g0641700;OsILI4(OsB U1),Os06g0226500;OsILI5(OsBUL1),Os02g0747900;OsEXPA5, Os02g0744200;OsEXPB6,Os10g0555600;OsEXPB11,Os02g0658800;OsCESA6,Os07g0252400;OsCSLF6,Os08g0160500;OsGT43,Os04g0650300;OsGT8,Os02g0739400;OsGUX1L,Os01g0880200;OsI RX10L,Os01g0926700;OsUGA4E,Os08g0526100 and UBIQUI TIN, Os03g0234200.
The primers used are shown in Supplementary Table 1 and Supplementary Table 2.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
T-gL conceived the project and designed the research. Z-gZ and P-fA conceived the original screening, constructed the mapping population. C-lZ and BX carried out most of the experiments, map-based cloning of OsRELA gene, paraffin section, genetic analysis, and chemical parameters. CD completed the protein purification. Z-yS and S-zT performed rice transformation. S-wZ and Z-gZ designed the experiments, analyzed, and wrote the data with C-lZ. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This research was supported by the National Key Research and Development Program of China (2020YFA0907603), National Natural Science Foundation of China (32070283), the Agricultural Science and Technology Innovation Program (CAAS-ZDXT2019003), and the Science Foundation of Hebei Province (C2020205014).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.760041/full#supplementary-material
Abe, Y., Mieda, K., Ando, T., Kono, I., Yano, M., Kitano, H., et al. (2010). The SMALL AND ROUND SEED1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice. Genes Genet. Syst. 85, 327–339. doi: 10.1266/ggs.85.327
Aravind, L., and Koonin, E. V. (1999). Fold prediction and evolutionary analysis of the POZ domain: structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285, 1353–1361. doi: 10.1006/jmbi.1998.2394
Bai, M. Y., Zhang, L. Y., Gampala, S. S., Zhu, S. W., Song, W. Y., Chong, K., et al. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. U. S. A. 104, 13839–13844. doi: 10.1073/pnas.0706386104
Bardwell, V. J., and Treisman, R. (1994). The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 8, 1664–1677. doi: 10.1101/gad.8.14.1664
Bian, H., Xie, Y., Guo, F., Han, N., Ma, S., Zeng, Z., et al. (2012). Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol. 196, 149–161. doi: 10.1111/j.1469-8137.2012.04248.x
Burkhard, P., Stetefeld, J., and Strelkov, S. V. (2001). Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11, 82–88. doi: 10.1016/s0962-8924(00)01898-5
Cho, H. T., and Kende, H. (1997). Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell 9, 1661–1671. doi: 10.1105/tpc.9.9.1661
Deweindt, C., Albagli, O., Bernardin, F., Dhordain, P., Quief, S., Lantoine, D., et al. (1995). The LAZ3/BCL6 oncogene encodes a sequence-specific transcriptional inhibitor: a novel function for the BTB/POZ domain as an autonomous repressing domain. Cell Growth Differ. 6, 1495–1503.
Holm, M., Ma, L. G., Qu, L. J., and Deng, X. W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16, 1247–1259. doi: 10.1101/gad.969702
Hong, Z., Ueguchi-Tanaka, M., Umemura, K., Uozu, S., Fujioka, S., Takatsuto, S., et al. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15, 2900–2910. doi: 10.1105/tpc.014712
Huynh, K. D., and Bardwell, V. J. (1998). The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene 17, 2473–2484. doi: 10.1038/sj.onc.1202197
Jang, S., An, G., and Li, H. Y. (2017). Rice leaf angle and grain size are affected by the OsBUL1 transcriptional activator complex. Plant Physiol. 173, 688–702. doi: 10.1104/pp.16.01653
Julian, J., Coego, A., Lozano-Juste, J., Lechner, E., Wu, Q., Zhang, X., et al. (2019). The MATH-BTB BPM3 and BPM5 subunits of Cullin3-RING E3 ubiquitin ligases target PP2CA and other clade A PP2Cs for degradation. Proc. Natl. Acad. Sci. U. S. A. 116, 15725–15734. doi: 10.1073/pnas.1908677116
Komatsu, K., Maekawa, M., Ujiie, S., Satake, Y., Furutani, I., Okamoto, H., et al. (2003). LAX and SPA: major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. U. S. A. 100, 11765–11770. doi: 10.1073/pnas.1932414100
Kuluev, B. R., Kniazev, A. V., Nikonorov Iu, M., and Chemeris, A. V. (2014). [Role of the expansin genes NtEXPA1 and NtEXPA4 in the regulation of cell extension during tobacco leaf growth]. Genetika 50, 560–569.
Lee, J., Park, J. J., Kim, S. L., Yim, J., and An, G. (2007). Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol. Biol. 65, 487–499. doi: 10.1007/s11103-007-9196-1
Li, F., Liu, W., Tang, J., Chen, J., Tong, H., Hu, B., et al. (2010). Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res. 20, 838–849. doi: 10.1038/cr.2010.69
Li, P. J., Wang, Y. H., Qian, Q., Fu, Z. M., Wang, M., Zeng, D. L., et al. (2007). LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 17, 402–410. doi: 10.1038/cr.2007.38
Mori, M., Nomura, T., Ooka, H., Ishizaka, M., Yokota, T., Sugimoto, K., et al. (2002). Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol. 130, 1152–1161. doi: 10.1104/pp.007179
Ning, J., Zhang, B., Wang, N., Zhou, Y., and Xiong, L. (2011). Increased leaf angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the Lamina joint of rice. Plant Cell 23, 4334–4347. doi: 10.1105/tpc.111.093419
Oyake, T., Itoh, K., Motohashi, H., Hayashi, N., Hoshino, H., Nishizawa, M., et al. (1996). Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol. 16, 6083–6095. doi: 10.1128/MCB.16.11.6083
Qiao, S., Sun, S., Wang, L., Wu, Z., Li, C., Li, X., et al. (2017). The RLA1/SMOS1 transcription factor functions with OsBZR1 to regulate brassinosteroid signaling and rice architecture. Plant Cell 29, 292–309. doi: 10.1105/tpc.16.00611
Ren, R., Hardikar, S., Horton, J. R., Lu, Y., Zeng, Y., Singh, A. K., et al. (2019). Structural basis of specific DNA binding by the transcription factor ZBTB24. Nucleic Acids Res. 47, 8388–8398. doi: 10.1093/nar/gkz557
Ruan, W., Guo, M., Xu, L., Wang, X., Zhao, H., Wang, J., et al. (2018). An SPX-RLI1 module regulates leaf inclination in response to phosphate availability in rice. Plant Cell 30, 853–870. doi: 10.1105/tpc.17.00738
Sakamoto, T., Morinaka, Y., Ohnishi, T., Sunohara, H., Fujioka, S., Ueguchi-Tanaka, M., et al. (2006). Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 24, 105–109. doi: 10.1038/nbt1173
Shin, B., Choi, G., Yi, H., Yang, S., Cho, I., Kim, J., et al. (2002). AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. Plant J. 30, 23–32. doi: 10.1046/j.1365-313x.2002.01264.x
Song, Y., You, J., and Xiong, L. (2009). Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol. Biol. 70, 297–309. doi: 10.1007/s11103-009-9474-1
Srivastava, A. K., Senapati, D., Srivastava, A., Chakraborty, M., Gangappa, S. N., and Chattopadhyay, S. (2015). Short hypocotyl in white light1 interacts with elongated Hypocotyl5 (HY5) and Constitutive Photomorphogenic1 (COP1) and promotes COP1-mediated degradation of HY5 during arabidopsis seedling development. Plant Physiol. 169, 2922–2934. doi: 10.1104/pp.15.01184
Sun, Q., Li, T. Y., Li, D. D., Wang, Z. Y., Li, S., Li, D. P., et al. (2019). Overexpression of loose plant architecture 1 increases planting density and resistance to sheath blight disease via activation of PIN-FORMED 1a in rice. Plant Biotechnol. J. 17, 855–857. doi: 10.1111/pbi.13072
Sun, S., Chen, D., Li, X., Qiao, S., Shi, C., Li, C., et al. (2015). Brassinosteroid signaling regulates leaf erectness in Oryza sativa via the control of a specific U-type cyclin and cell proliferation. Dev. Cell 34, 220–228. doi: 10.1016/j.devcel.2015.05.019
Tanabe, S., Ashikari, M., Fujioka, S., Takatsuto, S., Yoshida, S., Yano, M., et al. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17, 776–790. doi: 10.1105/tpc.104.024950
Tanaka, A., Nakagawa, H., Tomita, C., Shimatani, Z., Ohtake, M., Nomura, T., et al. (2009). BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 151, 669–680. doi: 10.1104/pp.109.140806
Tong, H., and Chu, C. (2018). Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci. 23, 1016–1028. doi: 10.1016/j.tplants.2018.08.007
Wang, B., Smith, S. M., and Li, J. (2018). Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69, 437–468. doi: 10.1146/annurev-arplant-042817-040422
Wang, L., Xu, Y., Zhang, C., Ma, Q., Joo, S. H., Kim, S. K., et al. (2008). OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS One 3:e3521. doi: 10.1371/journal.pone.0003521
Wang, Y., and Li, J. (2011). Branching in rice. Curr. Opin. Plant Biol. 14, 94–99. doi: 10.1016/j.pbi.2010.11.002
Yamamoto, Y. Y., Matsui, M., Ang, L. H., and Deng, X. W. (1998). Role of a COP1 interactive protein in mediating light-regulated gene expression in arabidopsis. Plant Cell 10, 1083–1094. doi: 10.1105/tpc.10.7.1083
Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1606. doi: 10.1105/tpc.12.9.1591
Zhang, C., Xu, Y., Guo, S., Zhu, J., Huan, Q., Liu, H., et al. (2012). Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet. 8:e1002686. doi: 10.1371/journal.pgen.1002686
Zhang, L. Y., Bai, M. Y., Wu, J., Zhu, J. Y., Wang, H., Zhang, Z., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21, 3767–3780. doi: 10.1105/tpc.109.070441
Zhang, S., Wang, S., Xu, Y., Yu, C., Shen, C., Qian, Q., et al. (2015). The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 38, 638–654. doi: 10.1111/pce.12397
Zhang, S. W., Li, C. H., Cao, J., Zhang, Y. C., Zhang, S. Q., Xia, Y. F., et al. (2009). Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 151, 1889–1901. doi: 10.1104/pp.109.146803
Zhang, Y., Su, J., Duan, S., Ao, Y., Dai, J., Liu, J., et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7:30. doi: 10.1186/1746-4811-7-30
Zhao, S. Q., Xiang, J. J., and Xue, H. W. (2013). Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol. Plant 6, 174–187. doi: 10.1093/mp/sss064
Zhou, J., Jiao, F., Wu, Z., Li, Y., Wang, X., He, X., et al. (2008). OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 146, 1673–1686. doi: 10.1104/pp.107.111443
Zhou, L. J., Xiao, L. T., and Xue, H. W. (2017). Dynamic cytology and transcriptional regulation of rice lamina joint development. Plant Physiol. 174, 1728–1746. doi: 10.1104/pp.17.00413
Keywords: leaf inclination, OsRELA, OsLIC, phytohormone, rice
Citation: Zhu C-l, Xing B, Teng S-z, Deng C, Shen Z-y, Ai P-f, Lu T-g, Zhang S-w and Zhang Z-g (2021) OsRELA Regulates Leaf Inclination by Repressing the Transcriptional Activity of OsLIC in Rice. Front. Plant Sci. 12:760041. doi: 10.3389/fpls.2021.760041
Received: 17 August 2021; Accepted: 09 September 2021;
Published: 01 October 2021.
Edited by:
Yuling Jiao, Peking University, ChinaReviewed by:
Meng Yuan, Huazhong Agricultural University, ChinaCopyright © 2021 Zhu, Xing, Teng, Deng, Shen, Ai, Lu, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng-wei Zhang, c3d6aGFuZ0BoZWJ0dS5lZHUuY24=; Zhi-guo Zhang, emhhbmd6aGlndW9AY2Fhcy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.