- 1Hunan Province Key Laboratory of Crop Sterile Germplasm Resource Innovation and Application, College of Life Sciences, Hunan Normal University, Changsha, China
- 2Department of Plant and Microbial Biology, University of California, Berkeley, Berkeley, CA, United States
Potassium (K+) levels in the soil often limit plant growth and development. As a result, crop production largely relies on the heavy use of chemical fertilizers, presenting a challenging problem in sustainable agriculture. To breed crops with higher K+-use efficiency (KUE), we must learn how K+ is acquired from the soil by the root system and transported to the rest of the plant through K+ transporters. In this study, we identified the function of the rice K+ transporter OsHAK8, whose expression level is downregulated in response to low-K+ stress. When OsHAK8 was disrupted by CRISPR/Cas9-mediated mutagenesis, Oshak8 mutant plants showed stunted growth, especially under low-K+ conditions. Ion content analyses indicated that K+ uptake and root-to-shoot K+ transport were significantly impaired in Oshak8 mutants under low-K+ conditions. As the OsHAK8 gene was broadly expressed in different cell types in the roots and its protein was targeted to the plasma membrane, we propose that OsHAK8 serves as a major transporter for both uptake and root-to-shoot translocation in rice plants.
Highlights
- Potassium transporter OsHAK8 mediates K+ uptake and translocation under low-K+ stress.
Introduction
Potassium (K+) is an essential macronutrient for plant growth and development. Therefore, sufficient K+ supplies are necessary to promote crop growth and development (Clarkson and Hanson, 1980; Leigh and Wyn Jones, 1984). Although the K+ concentration in the cytoplasm often reaches 100 mM, the K+ concentration in the soil is much lower, usually below 1 mM (Maathuis, 2009). Therefore, plants often suffer low-K+ stress under natural conditions, limiting their growth and development (Schroeder et al., 1994). However, plants can sense potassium levels in soil and adapt to low-K+ stress by altering root morphology, modifying K+ acquisition, and changing the K+ utilization strategy they utilize (Schachtman and Shin, 2007; Luan et al., 2009; Luan et al., 2017; Tang et al., 2020).
K+ absorption by plant roots and K+ transport throughout the plant are mediated by transport proteins including K+ channels and carriers. The activities of these K+ channels and carriers are highly regulated, allowing plants to respond and adapt to low-K+ stress (Schachtman and Shin, 2007; Luan et al., 2009; Luan et al., 2017; Wang and Wu, 2017; Tang et al., 2020). To date, many channels and carriers involved in K+ absorption and translocation have been identified and their functions characterized in the model plant Arabidopsis (Adams and Shin, 2014; Luan et al., 2017; Tang et al., 2020). For example, AtAKT1 encodes an inward-rectifying K+ channel that is expressed highly in the roots and contributes to K+ uptake (Lagarde et al., 1996; Hirsch et al., 1998). Under low-K+ conditions, AtAKT1 is activated in plant root hair by a regulatory pathway, which entails calcineurin B-like calcium sensors (CBL1 and CBL9) functioning together with their interacting kinase (CIPK23) (Li et al., 2006; Xu et al., 2006; Cheong et al., 2007; Luan, 2009; Luan et al., 2009). Moreover, AtHAK5 from the HAK/KUP/KT family is characterized as a high-affinity K+ transporter in Arabidopsis roots. The transcription of AtHAK5 is induced under K+-deficient conditions (Rubio et al., 2000; Gierth et al., 2005), and transport activity is also enhanced by CBL1/9-CIPK23-dependent phosphorylation (Ragel et al., 2015). AtHAK5 and AtAKT1 are considered as the two major components that contribute to K+ uptake in Arabidopsis roots (Gierth et al., 2005; Pyo et al., 2010; Nieves-Cordones et al., 2014). After being absorbed into the root hairs and epidermis, K+ is transported into root stele tissues and then translocated from roots toward shoots via xylem vessels which serve as the highway for long-distance K+ translocation. The outward-rectifying Shaker K+ channel AtSKOR is involved in this process. AtSKOR is expressed in stele tissues and is responsible for K+ release into the xylem sap (Gaymard et al., 1998). In addition to AtSKOR, the K+ transporter AtKUP7 may also participate in K+ transport into the xylem sap, affecting K+ translocation from roots toward shoots, especially under K+-limited conditions (Han et al., 2016). It is noteworthy that other transporters that are not K+-specific, such as Nitrate Transporter 1/Peptide Transporter AtNRT1.5, Cation Chloride Co-transporter AtCCC, and Barley Non-selective Outwardly Rectifying Current channel NORC have also been reported to facilitate K+ efflux into the xylem sap (Colmenero-Flores et al., 2007; Pottosin and Dobrovinskaya, 2014; Li et al., 2017).
Rice is a staple food for half of the global population and serves as a critical model for studying the basic biology of cereal crops. The growth of rice plants is highly susceptible to K+ deficiency; as a result, rice production requires a large amount of K+-fertilizers (Wanasuria et al., 1981; Dobermann et al., 1996; Li et al., 2014; Zhang et al., 2018). Therefore, effective K+ uptake and translocation within the organism will be the key to KUE, and dissecting the mechanism of K+ transport will be crucial to understanding the molecular basis for KUE. However, comparing with Arabidopsis, much less is known about the mechanism underlying K+ absorption and translocation in rice plants. Although OsAKT1 and OsHAK5, like AtAKT1 and AtHAK5, mediate K+ uptake in rice roots (Li et al., 2014; Yang et al., 2014), K+ channels such as OsKATs and OsAKTs may be involved in K+ accumulation in the cytoplasm in response to salt stress (Musavizadeh et al., 2021). Thus, more transporters related to K+ absorption and translocation need to be explored.
Compared with Arabidopsis HAK members, the rice genome features a large gene expansion in the HAK family. According to an earlier study (Yang et al., 2009) and up-to-date genomic information, we have confirmed that the genomes of the Nipponbare rice subspecies encode 27 OsHAK family members vs. 13 in Arabidopsis. The Arabidosis AtHAK family consists of four major clades (I–IV), whereas the rice OsHAK family has five clades (I–V). In particular, the singleton of AtHAK5 clade (clade I) in Arabidopsis is expanded to eight members in rice (Yang et al., 2009). Therefore, mechanisms in rice are more complex, reflecting the more elaborate anatomy of the plant.
To date, 4 out of 27 OsHAK genes have been characterized in rice. In addition to OsHAK5 (Li et al., 2014), OsHAK1 and OsHAK16 function in K+ uptake and translocation under a broad range of external K+ levels (0.05–1 mM), and OsHAK21 confers significant potassium uptake and growth in yeasts at low K+ concentrations (Chen et al., 2015; Shen et al., 2015; Feng et al., 2019). In order to fully address the mechanism of K+ uptake and translocation in rice, we took a systematic approach in studying the function of OsHAK genes in rice using CRISPR/Cas9-mediated mutagenesis. Here, we showed that OsHAK8 plays crucial role in K+ uptake and K+ translocation from root to shoot, especially under low-K+ conditions.
Materials and Methods
Plant Growth Conditions
Rice (Oryza sativa ssp. japonica “Nipponbare”) was used as the wild type in this study and for the development of all transgenic plant lines. Hoagland hydroponic cultivation of rice was performed according to an earlier procedure (Li et al., 2014) with modification in the concentrations of K+ as specified in the figure legends. Nutrient solutions were replaced once per week or as needed by experimental treatments.
Yeast Complementation Assay
For the constructs used in functional complementation in CY162 cells, the coding sequences of OsHAK8 were amplified using the specific primers listed in Supplementary Table 1 from rice genomic cDNA. The PCR product was digested with HindIII and XbaI and ligated into the pYES2 vector, and transformed into the yeast strain CY162, a K+ uptake-deficient strain of Saccharomyces cerevisiae (MATα, Δtrk1, trk2:pCK64, his3, leu2, ura3, trp1, and ade2) (Anderson et al., 1992). We performed yeast transformations using the LiAC/ssDNA/PEG method, and the growth assays on the arginine phosphate (AP) medium were performed as described previously (Horie et al., 2011).
qRT-PCR Analysis
Total RNA was extracted using the TRIzol reagent (Thermo Fisher Scientifiec Baltics UAB, United States) (Invitrogen) following the instructions of the manufacturer. Samples were treated with DNase (Promega) (Accurate Biotechnology (Hunan) Co., Ltd.) to eliminate genomic DNA contamination. Two micrograms of DNA-free RNA were used for reverse transcription using a Moloney murine leukemia virus reverse transcriptase (Promega) with anchored oligo (dT18). qRT-PCR was performed on an ABI PRISM 7,500 real-time PCR instrument (Thermo Fisher Scientific) (Applied Biosystems) using Power SYBR Green (TaKaRa) (Qiagen; CFX96 system). All the primer pairs used for the expression assays or subcloning are listed in Supplementary Table 1. Primer specificity was confirmed by analysis of the melting curves. The 2–△△CT method (Livak and Schmittgen, 2001) was used to quantify the value of every sample using Actin as an internal reference.
GUS Assays
The 2,000-bp fragment before the ATG codon of OsHAK8 was amplified from rice genomic DNA (cv Nipponbare) with the specific primers listed in Supplementary Table 1. This fragment was used as the OsHAK8 promoter in this study, digested with BamHI and HindIII, and then constructed into the pCAMBIA1301 vector. The plasmid was transformed into Nipponbare rice via callus transformation as described previously (Upadhyaya et al., 2000), which was mediated by Agrobacterium EHA105. The transgenic lines were initially screened by hygromycin resistance and GUS staining. The histochemical analysis of GUS staining in different tissues of rice was performed as described previously (Ai et al., 2009). The samples were examined and photographed with an Olympus SZX12 microscope (Olympus, Olympus Corporation, Tokyo, Japan) equipped with a camera. The quantification of GUS activity was performed using the Image J software provided by the microscope manufacturer.
Subcellular Localization Analysis
For subcellular localization analysis, the OsHAK8 cDNA was amplified without its stop codon using the specific primers listed in Supplementary Table 1, subsequently digested with XbaI, and then inserted into the multicloning sites in the plant binary vector pCAMBIA2300 to make the 35S:OsHAK8-GFP fusion construct. Transient expression in rice of OsHAK8:GFP in the pCAMBIA2300 vector and SP1:RFP in the modified pCAMBIA1300 vector were conducted using the polyethyleneglycol method (Li et al., 2009). For the plasma membrane localization of OsHAK8, we used a confocal laser scanning microscope (LSM880, Carl Zeiss, Jena, Germany) for detection.
Generation of OsHAK8 CRISPR/Cas9 Knockout Transgenic Plants
The CRISPR/Cas9 system (the Cas9 vector pYLCRISPR/Cas9 Pubi-H provided by Prof. Yaoguang Liu) was applied to generate Oshak8-1 mutants. The pYLCRISPR/Cas9 Pubi-H is a plant binary vector in which Cas9p was driven by the maize ubiquitin promoter (pUBI) with the hygromycin selectable marker gene having two BsaI sites flanked by a toxic negative selectable marker (ccd B gene) for the cloning of the sgRNA expression cassette. The sgRNA1 sequences (GTGTGCTCTCTTTCGTGTTCTGG) which targeted exons2 of the OsHAK8 locus and the sgRNA2 sequences (CCCTGGTGATCATGCTGTGCTGG) which targeted exons9 of the OsHAK8 locus (Supplementary Figure 1) were designed with the web tool CRISPR Plant (Xie et al., 2014). The 20-bp target sequence had to be immediately followed by an NGG motif (PAM), which is essential for Cas9 binding to the target DNA. sgRNA1 and sgRNA2 cloning into pYLCRISPR/Cas9Pubi-H was performed as described (Usman et al., 2020).
Transgenic plants were obtained by the co-culture of seed embryo-derived calli (Oryza sativa L. cv. Nipponbare) with A. tumefaciens strain EHA105 (harboring the sgRNAs in the pYLCRISPR/Cas9 Pubi-H plasmids) following the procedure of Upadhyaya et al. (2000). To identify mutations in regenerated plants, genomic DNA was amplified by using mutation detection primers which were designed to flank the designated target site. Then, the sequence chromatograms were analyzed using the DSDecode tool1 to check the genotype of the transgenic plants. The two independent homozygous mutant lines from the T1 generation, named Oshak8-1 and Oshak8-2, were used for further study. The primers used for CRISPR/Cas9 construction and mutant detection are listed in Supplementary Table 1.
Measurement of Chlorophyll
We used the equations developed by Porra et al. (1989) to estimate chlorophyll contents. Chlorophyll was extracted from the rice shoots with 10 ml of 80% (v/v) acetone containing 2.5 mM of sodium phosphate buffer (pH 7.8) and incubated in darkness for 30 h at room temperature. After centrifugation (4°C, 15 min, 13,000 × g), we measured the absorbances A663.6 and A646.6 with a spectrophotometer (UV-1700, Shimadzu, Kyoto, Japan).
Ion Content Analysis
For ion content analysis (K+), samples were washed thoroughly with double-distilled water three times and dried at 80°C for 48 h to constant weight. The samples were treated in a muffle furnace at 300°C for 1 h, 575°C for 7 h, and finally dissolved in 0.1-N HNO3. K+ concentrations were then measured by ICP-AES (Varian 715-ES) at the 766.49-nm wavelength (for K+).
Measurement of Root K+ Net Uptake Rate
The measurement of the K+ uptake rate followed previously described procedures (Nieves-Cordones et al., 2010; Yang et al., 2014) with some modifications. Five-day-old rice seedlings were grown in hydroponic solutions containing 10 mM of K+ for 14 days and then transferred to 0 mM-K+ solutions for 5 days. For the K+ uptake assay, K+-starved plants were incubated in the medium containing 0.01 or 10 mM of K+ for 3 days, and the roots and shoots were collected for K+ content measurements using inductively coupled plasma (ICP) analysis. The net K+ uptake rate into the plant was calculated according to the equation (Nieves-Cordones et al., 2010): Net uptake rate = (C2 – C1)/[(t2– t1) × (R2 + R1)/2], where C is the total K+ content, R is the root dry weight, and t is the time; the subscripts 1 and 2 indicate the start and end of the period for which the uptake rate is calculated: t2 – t1 = 3 days, and (R2 + R1)/2 = the mean root dry weight.
Measurement of K+ Export Rate in Xylem Sap
Seven-day-old rice seedlings were grown in the hydroponic solutions containing 10 mM of K+ for 14 days and then transferred to the medium containing 0.01 or 10 mM of K+ for 14 days. The detailed protocols for the shoot excision, collection of the xylem sap by cotton ball, determination of K+ concentration by ICP, and calculation of the xylem K+ export rate were described previously by Yang et al. (2014).
Results
Rice Oshak8 Mutants Are Sensitive to Low K+ Stress
To explore the possible role of HAK family transporters in rice, we analyzed expression pattern and subcellular localization and generated gene-edited loss-of-function lines for each of the HAK members. Here, we focused on the functional analysis of OsHAK8. We produced two knockout lines of OsHAK8 using the CRISPR/Cas9 system in the Nipponbare background. The Oshak8-1 had a 1-bp insertion in the second exon of LOC_Os03g21890, resulting in frameshift mutations at the 68th amino acid and premature translation termination at the 87th amino acid. The Oshak8-2 mutant had a 2-bp deletion in the 9th exon of LOC_Os03g21890, resulting in a frameshift mutation at the 444th amino acid (Supplementary Figure 1). No off-target cleavage was detected by the web-based tool CRISPR-P22 (Liu et al., 2017). We examined the growth of the two independent Oshak8 mutants (Oshak8-1, Oshak8-2) under K+-sufficient and K+-deficient conditions. When grown in the K+-sufficient hydroponic solution (10-mM K+), the Oshak8 mutants and the wild-type plants (WT) grew well without discernible differences (Figure 1A). When treated in K+-deficient hydroponic conditions (0-mM K+) however, the Oshak8 mutants showed more stunted statures than the wild-type plants. Along with the increase of external K+ concentrations, the differences between the Oshak8 mutants and the wild-type plants became smaller and eventually disappeared at 10-mM K+ concentration (Figure 1A).
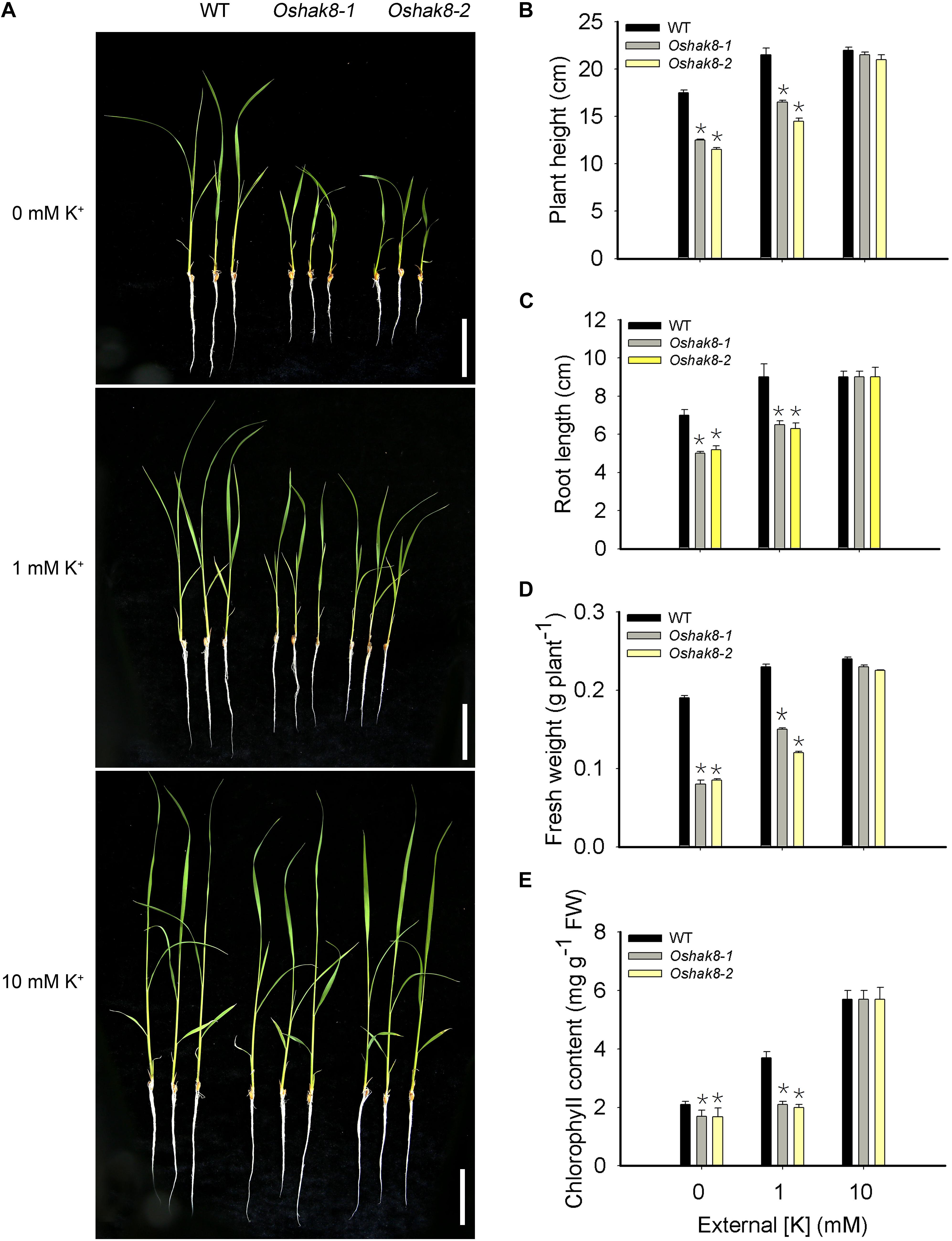
Figure 1. Growth retardation of the Oshak8 mutants under K+-deficient conditions. (A) Growth retardation of the Oshak8 mutants under K+-deficient conditions. The seeds of the wild-type plants (WT) and Oshak8 mutants (Oshak8-1, Oshak8-2) germinated in water for 5 days, grown in hydroponic solutions containing 0, 1, or 10 mM of K+, and then photographed after 14 days. Oshak8 mutants (Oshak8-1, Oshak8-2) displayed growth retardation under the K+-deficient conditions. Bars = 4 cm. (B) Plant height of WT and Oshak8 mutants under various K+ concentrations. Root lengths of 14-day-old seedlings were measured. Growth conditions were as described in (A). (C) Root lengths of WT and Oshak8 mutants. Growth conditions were as described in (A). (D) Fresh weight of WT and Oshak8 mutants. Growth conditions were as described in (A). (E) Chlorophyll contents of WT and Oshak8 mutants. Growth conditions were as described in (A). Significant differences were found between WT and Oshak8 knockout plants (Oshak8-1, Oshak8-2) (*P < 0.01 by Student’s t-test). Data are means of three replicates of one experiment. The experiment was repeated three times with similar results. Error bars represent ± SD. Asterisks represent significant differences.
As a more quantitative measure, we assayed plant height and root length in the wild-type and mutant plants. The Oshak8 mutants were more severely inhibited compared with the wild-type plants when grown in hydroponic conditions containing less than 10-mM K+ (Figures 1B,C). Consistent with the observed size difference, the fresh weights of the Oshak8 mutants were significantly lower compared with the wild-type plants (Figure 1D). The Oshak8 mutants also had significantly lower levels of chlorophyll than the wild-type plants (Figure 1E).
In addition, we examined the growth of the Oshak8 mutants in soil (with 0.18 mg/g of external K+) (Supplementary Figure 2Aa). The Oshak8 mutants showed necrotic brown spots and burnt tips on older leaves, which are typical K+-deficient symptoms of rice plants (Supplementary Figure 2Ab). The mutants were also shorter than the wild-type plants (Supplementary Figure 2B). The seed-setting rate and 1,000-grain weight in the mutants were reduced as compared to the wild-type plants (Supplementary Figures 2D,E). The main panicle length of the Oshak8 mutants and the wild-type plants showed no discernible differences (Supplementary Figures 2Ac,C). The pollen grains in the mutants contained less starch and were less viable than those in the wild-type plants (Supplementary Figures 3A,B). These results suggest that the knockout of the OsHAK8 gene affected rice growth and development, especially under low-K+ conditions, possibly due to changes in K+ accumulation in the mutant plants.
Knockout of OsHAK8 Impaired K+ Accumulation in Rice Plants
To test whether the growth retardation phenotype of Oshak8 was due to the reduction of K+ content, we measured the K+ content in the wild-type plants and the Oshak8 mutants. As shown in Figure 2A, there was no significant difference in K+ content among the different plants when grown in the K+-sufficient hydroponic solution (10-mM K+). As the K+ level in the medium became lower, all plants accumulated less K+ as expected. However, the Oshak8 mutants were affected more severely. For example, under 0 mM of K+, the Oshak8 mutants contained 67% less K+ compared with the wild-type plants. Under 1 mM of K+, the Oshak8 mutants accumulated 44% less K+ than the wild-type plants.
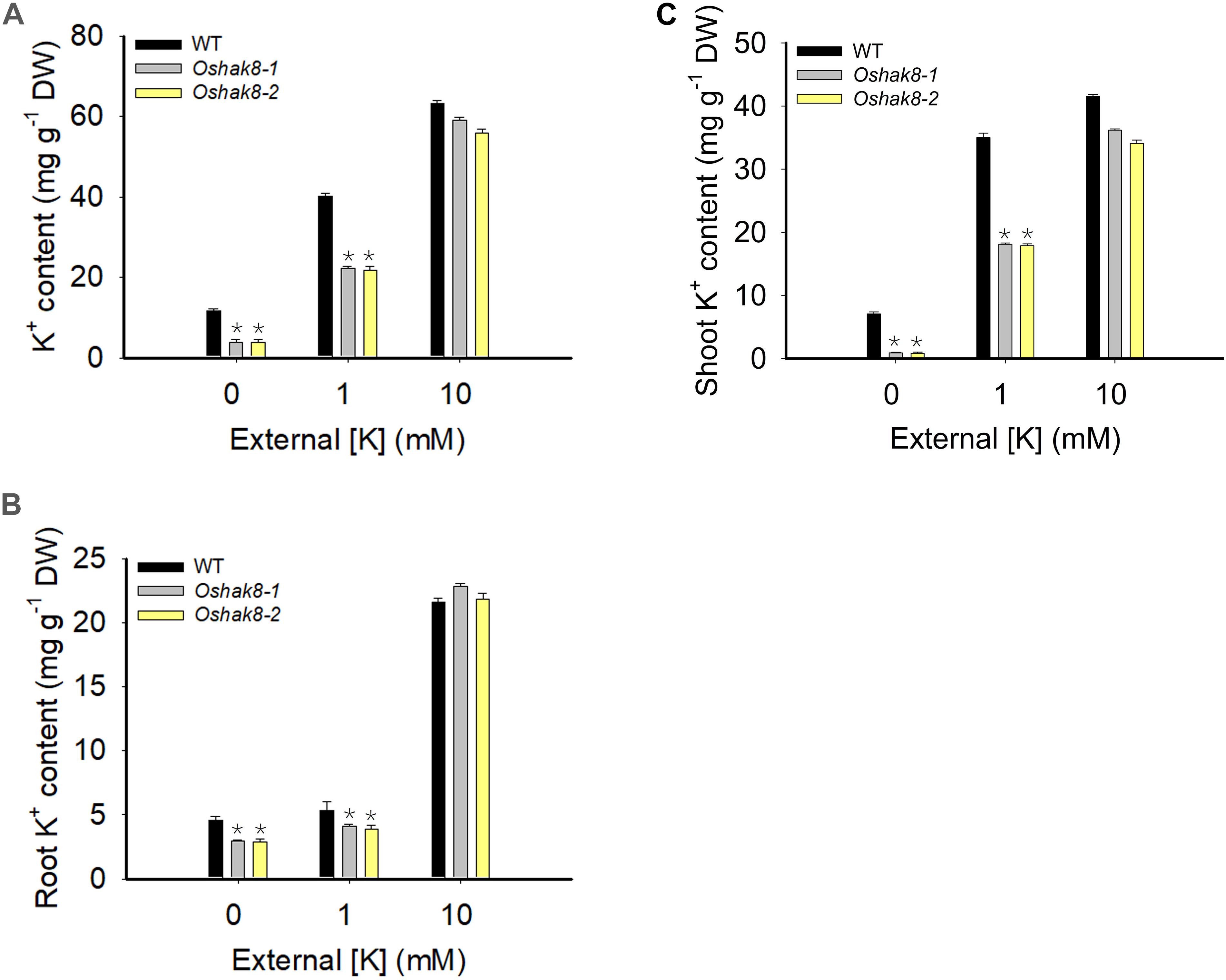
Figure 2. Growth retardation of the Oshak8 mutants under K+-deficient conditions. The seeds of the wild-type plants (WT) and Oshak8 mutants (Oshak8-1, Oshak8-2) germinated in water for 5 days, then grown in hydroponic solutions containing 0, 1, or 10 mM of K+ for 14 days. The whole plants, roots, and shoots were collected for K+ content measurements using inductively coupled plasma analysis. (A) K+ contents of WT and Oshak8 mutant (Oshak8-1, Oshak8-2) plants. (B) K+ contents in the roots of WT and Oshak8 mutant (Oshak8-1, Oshak8-2) plants. (C) K+ contents in the shoots of WT and Oshak8 mutant (Oshak8-1, Oshak8-2) plants. Significant differences were found between WT and Oshak8 mutant plants (Oshak8-1, Oshak8-2) (*P < 0.01 by Student’s t-test). Data are means of three replicates of one experiment. The experiment was repeated three times with similar results. Error bars represent ± SD. Asterisks represent significant differences.
Then, we measured the K+ contents in roots and shoots separately to examine the distribution of K+ in the plants. As shown in Figures 2B,C, there is no difference in K+ content among different plants under K+-sufficient conditions. When grown in low-K+ conditions, the Oshak8 mutants contained less K+ in both roots and shoots compared with the wild-type plants. We conclude from the above data that the knockout of OsHAK8 impaired K+ accumulation in the plants, especially under the low-K+ conditions.
OsHAK8 Mediates K+ Transport in Yeast
To determine whether OsHAK8 encodes a functional K+ transporter, we performed complementation tests using the yeast strain CY162 (MATα, Δtrk1, trk2:pCK64, his3, leu2, ura3, trp1, and ade2), which lacks the high-affinity K+-transporters Trk1/2 and, thus, is defective in K+ uptake (Anderson et al., 1992). When grown on an arginine phosphate (AP) medium containing 10 mM of K+, no significant differences were scored among transformants harboring either the empty vector or the OsHAK8 construct (Figure 3). When the concentration of K+ in the AP medium was decreased to 0.5 mM, the yeast mutant transformed with OsHAK8 grew better as compared to the mutant transformed with the empty pYES2 vector (Figure 3). The results indicated that OsHAK8 is capable of mediating K+ uptake.
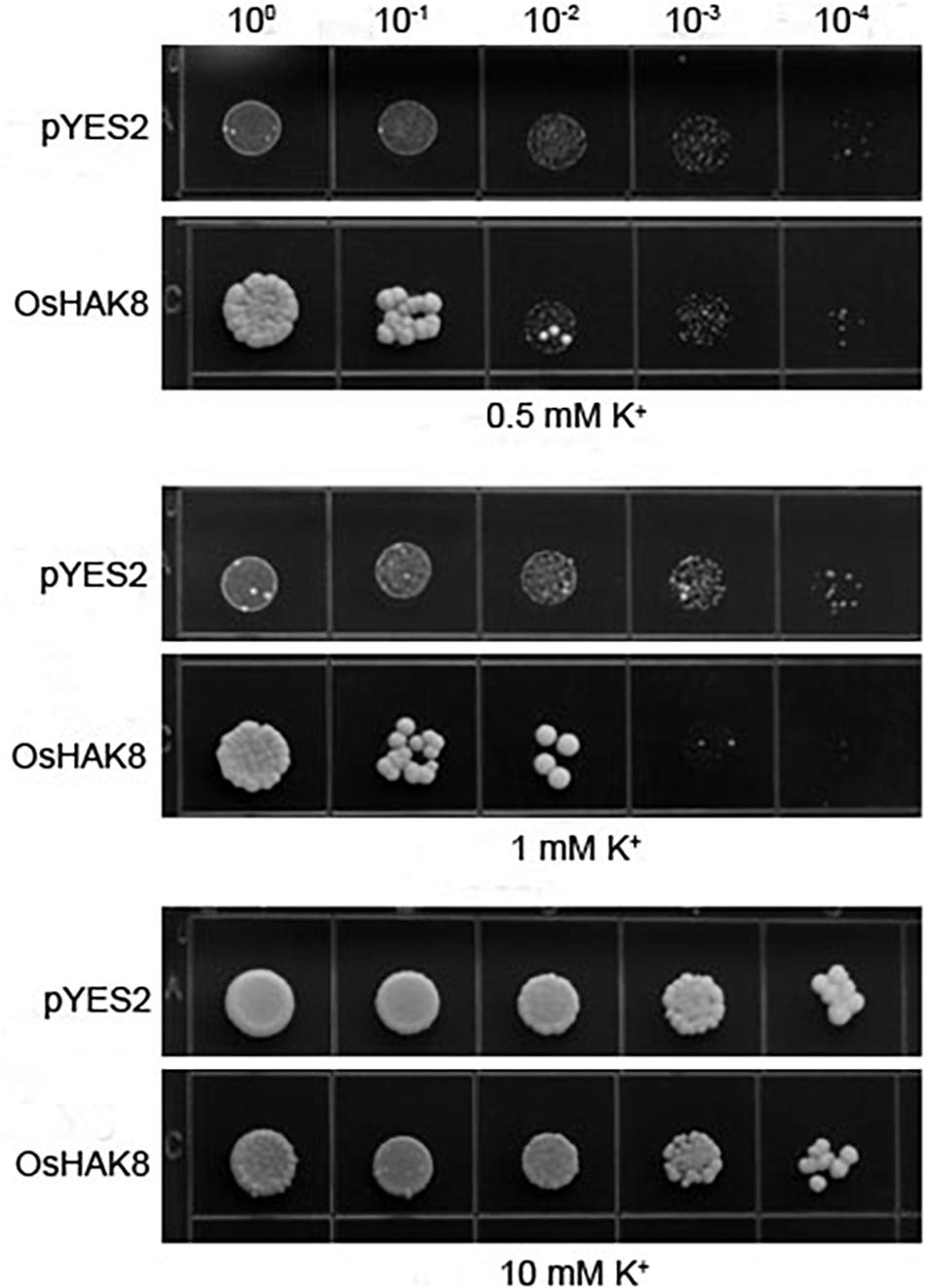
Figure 3. Functional complementation of OsHAK8 in the K+ uptake-deficient mutant strain CY162 of Saccharomyces cerevisiae. Growth of yeast strains on arginine phosphate (AP) medium containing different K+ concentrations. The strains are CY162 cells transformed with the pYES2 vector only (pYES2), OsHAK8 cDNA in the pYES2 vector (OsHAK8). CY162 cells which harbored pYES2 or expressed OsHAK8, were grown on the AP medium containing 0.5, 1, or 10 mM of K+. No significant differences were found between pYES2 and OsHAK8 when grown on the AP medium containing 10 mM of K+ (P>0.05 by Student’s t-test). Significant differences were found between pYES2 and OsHAK8 when grown on the AP medium containing 0.5 mM K+ (P<0.01 by Student’s t-test). The experiment was repeated three times with similar results.
OsHAK8 Encodes a Plasma Membrane Protein
The expression of OsHAK8 facilitated K+ uptake in yeast, which suggested that OsHAK8 was likely to be targeted to the plasma membrane. To test the subcellular localization of OsHAK8 in planta, a green fluorescence protein (GFP) reporter construct was generated to express an OsHAK8-GFP fusion protein driven by the cauliflower mosaic virus 35S promoter. The same vector expressing GFP only was used as a control. The GFP-only control and the OsHAK8-GFP fusion construct were each introduced into the protoplasts from rice stem cells, and GFP signals were examined using confocal microscopy. While the GFP-only signal was observed mainly in the cytoplasm and nucleus, the OsHAK8-GFP fluorescence was predominantly localized at the plasma membrane. To confirm the plasma membrane localization of OsHAK8, we co-transformed the OsHAK8-GFP construct with a construct containing a known plasma membrane protein fused to red fluorescent protein (SP1-RFP) (Li et al., 2009) into the protoplasts from rice stem cells. We found that GFP signals produced by the OsHAK8-GFP fusion overlapped well with RFP signals generated by SP1-RFP (Figure 4). Based on these results, we concluded that OsHAK8 is localized to the plasma membrane in rice cells.
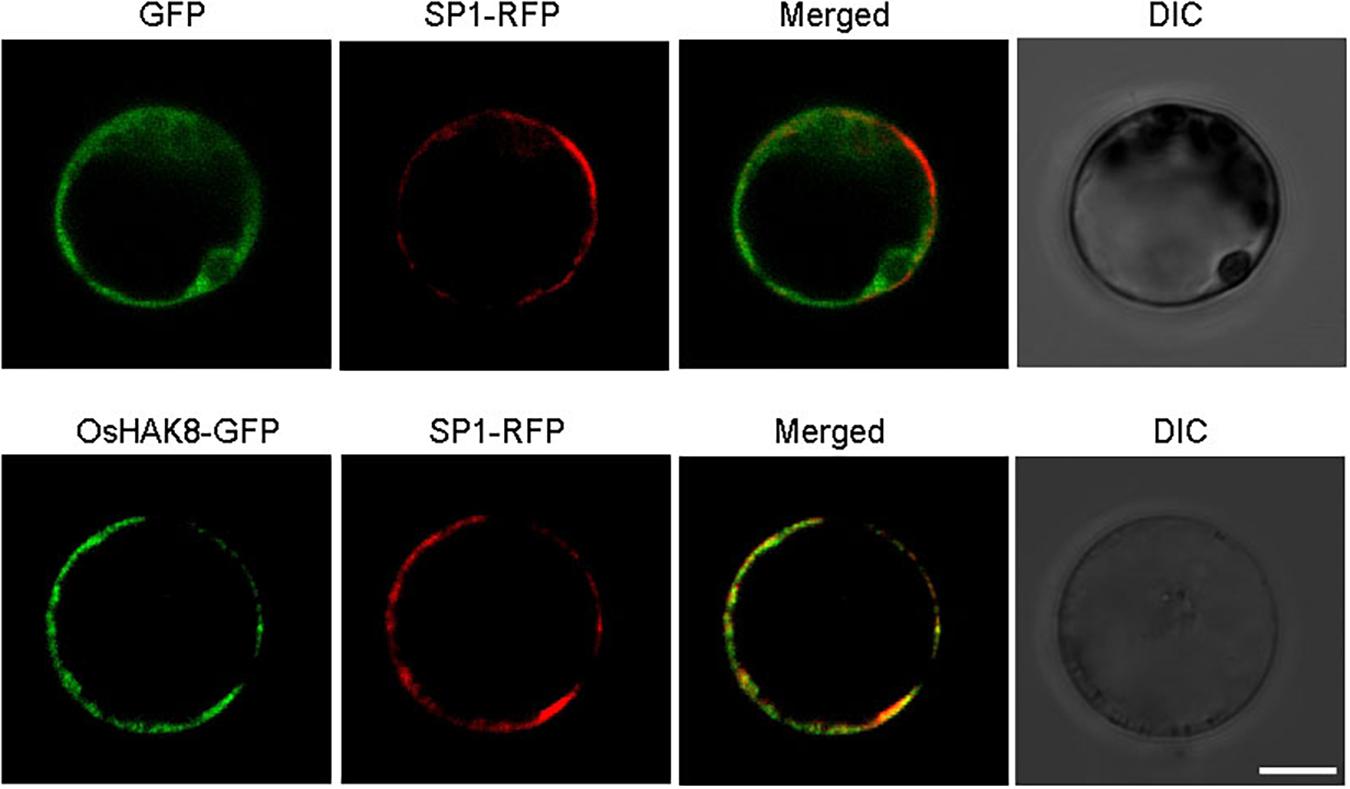
Figure 4. Plasma membrane localization of OsHAK8. The GFP coding sequence was fused to the C-terminus of the OsHAK8 coding region in the pCambia2300 vector and then transformed into rice mesophyll protoplasts. GFP alone in the same vector was used as a control. Signals from GFP, OsHAK8-GFP, and SP1-RFP (a plasma membrane localization marker) were imaged using a Zeiss LSM880 confocal laser scanning microscope. Columns 1–4 show GFP signals, SP1-RFP signals, merged images of GFP and RFP signals, and bright-field differential interference contrast (DIC), respectively. For each localization experiment, ≥ 30 individual cells were analyzed using a Zeiss LSM880 confocal laser scanning microscope (Carl Zeiss). Bar = 10 μm.
OsHAK8 Is Expressed Mainly in Rice Root
Analyses of the OsHAK8 expression pattern are critical to explaining the physiological functions of the gene in plants. We first determined the expression pattern of the OsHAK8 gene in different tissues of rice plants. The qRT-PCR analysis of the RNA samples isolated from various organs of the Nipponbare plants showed that OsHAK8 was expressed mainly in the roots. Transcripts of OsHAK8 were also present in lower amounts in stems, leaves, anthers, and glumes (Figure 5A). To investigate the expression pattern of OsHAK8 in more detail, we performed GUS activity staining of the transgenic rice plants harboring the OsHAK8 promoter-GUS fusion construct. Consistent with the qRT-PCR results, strong GUS signals were detected in the roots (Figures 5Ba,b). In addition, GUS activity was also detected in the stem (Figure 5Bc), mature anthers (Figure 5Bd), and glume (Figure 5Be). We chose three independent transgenic lines (lines 2, 5, and 8) to analyze the results in more detail through the examination of root cross-sections. The results showed that OsHAK8-GUS was expressed mainly in the vascular tissues, cortex, and exodermis of the roots (Figure 5Bf).
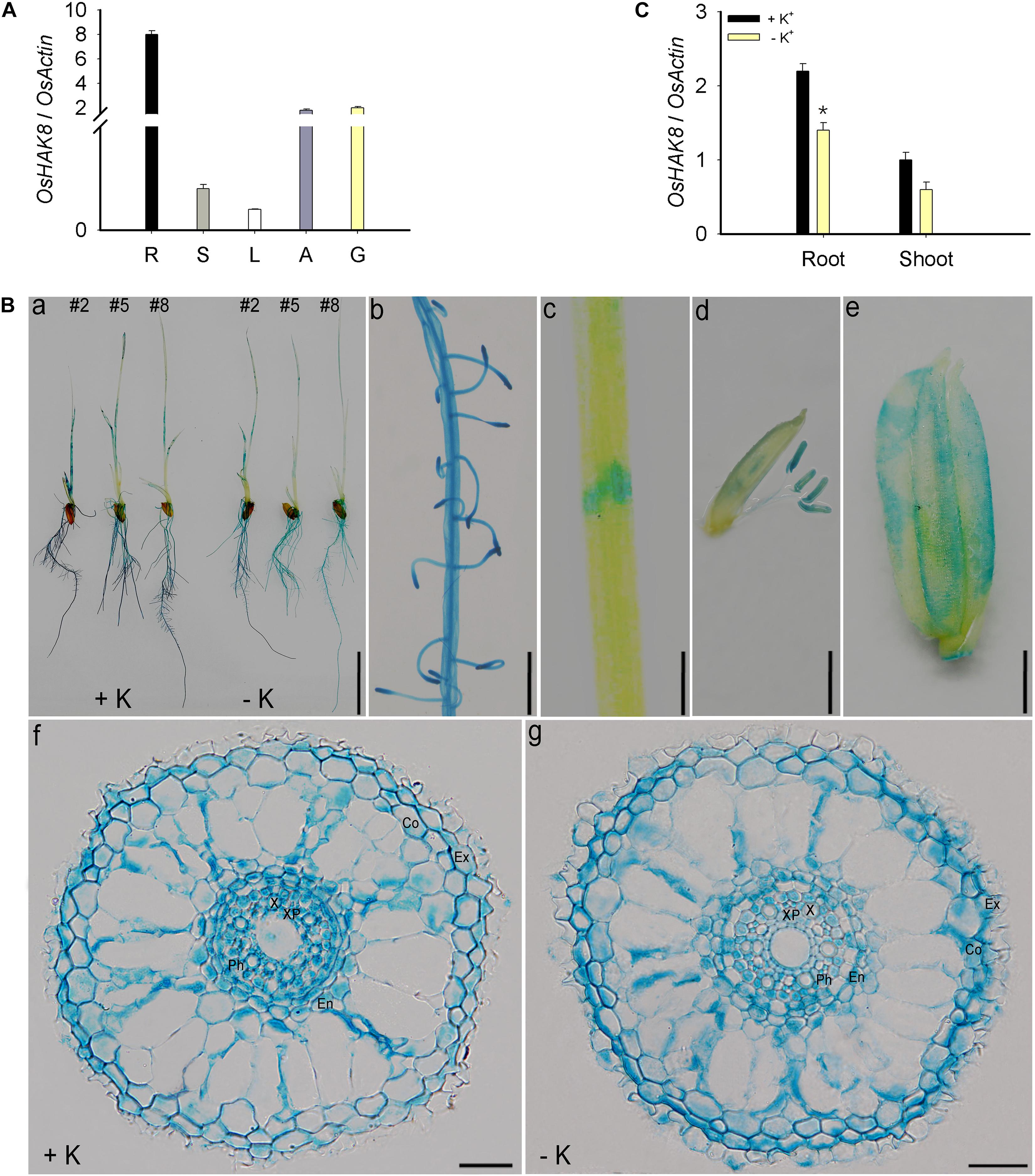
Figure 5. Expression pattern of OsHAK8. qRT-PCR analyses showing OsHAK8 mRNA accumulation in roots, stems, leaves, anthers, and glumes. Rice seedlings (Nipponbare) were grown in soil for 10 weeks. R, root; S, stem; L, leaf; A, anther; G, glume. (A) Histochemical analysis of OsHAK8 promoter-driven GUS reporter expression in transgenic rice plants. The transgenic plants were grown as described in (B), and GUS activities (indicated in blue) were examined using histochemical staining. (a) GUS staining of 7-days-old plants (Line 2) grown in hydroponic solutions with 10-mM K+ (+ K) or 0-mM K+ (-K). (b) Enlarged image of the root hair zones in (a). (c) Enlarged image of the node in (a). (d) GUS staining of anthers. The transgenic plants were grown as described in (A). (e) GUS staining of glume. The transgenic plants were grown as described in (A). (f,g) Cross-section images of the elongation zone in (a). Ex, exodermis; Co, cortex; En, endodermis; Ph, phloem; X, xylem; XP, xylem parenchyma. Bar in (a) = 1 cm, bar in (b) = 5 mm, bar in (c) = 1 mm, bar in (d) = 1 mm, bar in (e) = 1 mm, and bars in (f) to (g) = 100 μm. (C) Effect of K+ deficiency on the transcriptional expression of OsHAK8 in rice. 3-day-old rice seedlings (Nipponbare) were grown in a hydroponic solution containing 10 mM K+ for 3 days and then transferred to 10-mM K+ (+ K) and 0-mM K+ (−K) solutions for 24 h, respectively. Total RNAs were isolated from the roots and shoot of the seedlings, and OsHAK8 transcript levels were determined by qRT-PCR. Actin was used as an internal standard. Significant difference was found between 0- and 10-mM K+ samples indicated in root tissue (P<0.01 by Student’s t-test). Data are means of three replicates of one experiment. The experiment was repeated three times with similar results. Error bars represent ± SD. Asterisks represent significant differences.
Expression analysis showed that the expression of OsHAK8 in the roots was downregulated when seedlings were transferred from the K+-sufficient to the K+-deficient condition (Figure 5C). In addition, the OsHAK8-GUS signal was decreased by K+ deficiency in stelar cells when transferred to K+-deficient conditions (Figures 5Ba,f,g and Supplementary Figure 4A). We quantified the GUS signals in the root stele using the Image J software in three plants from lines (lines 2, 5, and 8) treated under K+-sufficient or K+-deficient conditions. There was a 2-fold decrease in the GUS signal in the stele under K+-deficient conditions (Supplementary Figure 4B). This expressed pattern suggests the potential role of OsHAK8 in K+ transport in response to the low-K+ stress in the roots.
Root Uptake and Root-to-Shoot K+ Transport Are Reduced in Oshak8 Mutants Under K+-Deficient Condition
The expression analysis indicated that OsHAK8 showed strong expression in root tissues (Figure 5A). Direct K+ measurements indicated that the roots and shoots of the two Oshak8 mutants both accumulated less K+ than that of the wild-type plants under low-K+ conditions (Figures 2B,C). These results suggest that OsHAK8 may be involved in K+ acquisition and/or root-to-shoot translocation in root tissues.
To test whether OsHAK8 participated in K+ uptake, we performed the K+ uptake assay. The seeds of the wild-type plants and Oshak8 mutants were germinated in water for 5 days, transferred to the hydroponic solutions containing 10 mM of K+ for 14 days, and then transferred to the hydroponic solutions containing 0 mM of K+ for 5 days. For the K+ uptake and translocation assay, K+-starved plants were incubated in the medium containing 0.01 or 10 mM of K+ for 3 days, and the roots and shoots were collected separately for K+ content measurements using ICP analysis. As shown in Figure 6A, the K+ uptake rate was reduced in the Oshak8 mutants compared with the wild-type plants. The results showed that the knockout of OsHAK8 impaired K+ uptake in rice roots.
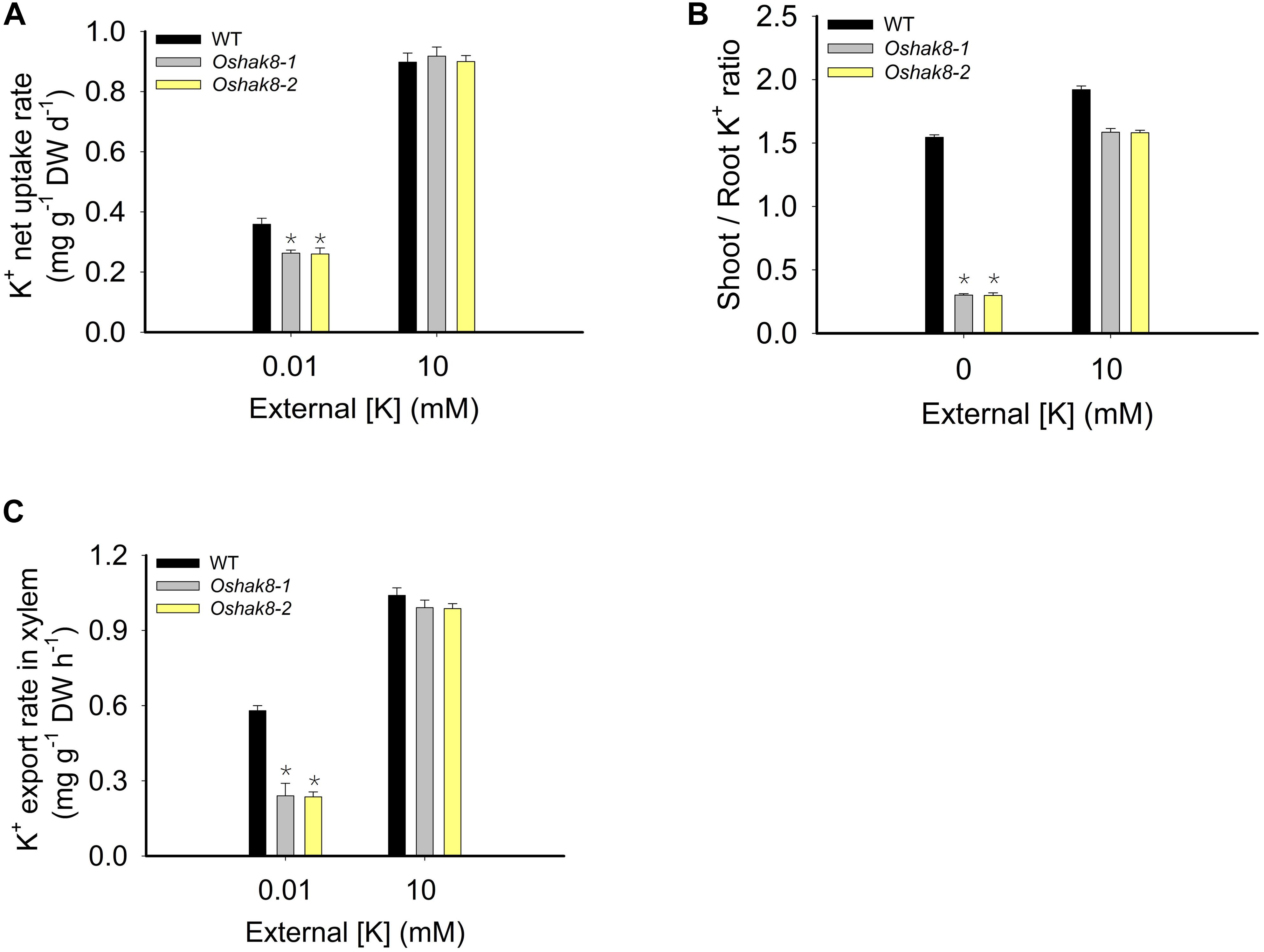
Figure 6. Effects of Oshak8 knockout on K translocation under different K+-supply conditions. (A) K+ net uptake rate of WT and Oshak8 mutant (Oshak8-1, Oshak8-2) plants. The seeds of the WT and Oshak8 mutants were germinated in water for 5 days, transferred to the hydroponic solutions containing 10 mM of K+ for 14 days, and then transferred to the hydroponic solutions containing 0 mM of K+ for 5 days. For the K+ uptake assay, K+-starved plants were incubated in the medium containing 0.01 or 10 mM of K+ for 3 days and the roots and shoots were collected for K+ content measurements using inductively coupled plasma analysis. (B) The shoot/root ratios of K+ in various plant materials. Growth conditions were as described in Figure 2. The roots were collected for K+ content measurements using inductively coupled plasma analysis. (C) K+ export rate in the xylem. Seven-day-old rice seedlings were grown in the International Rice Research Institute (IRRI) solution for 2 weeks and then transferred to 10- or 0.01-mM K+ solutions for 2 weeks before the sampling. The detailed protocols for the shoot excision, collection of the xylem sap by cotton ball, determination of K+ concentration by ICP, and calculation of the xylem K+ export rate were described previously by Yang et al. (2014). K+ contents were determined by ICP-AES. Significant differences were found between WT and Oshak8 mutants (Oshak8-1, Oshak8-2) (*P < 0.01 by Student’s t-test). Data are means of three replicates of one experiment. The experiment was repeated three times with similar results. Error bars represent ± SD. Asterisks represent significant differences. DW, dry weight.
We calculated the shoot/root ratio of K+ content to determine the K+ distribution between shoots and roots. The shoot/root K+ ratio in the Oshak8 mutants was lower than that of the wild-type plants under K+-deficient conditions (Figure 6B), indicating that K+ translocation from roots to shoots was affected in Oshak8 mutants. As a critical parameter for root-to-shoot transport, K+ concentration in the xylem sap was also measured. Under K+-sufficient conditions (10 mM of K+), there was no difference in K+ levels in the xylem sap between mutants and wild-type plants (Figure 6C). Under low-K+ conditions (0.01 mM of K+), the K+ levels in the xylem sap were lower in Oshak8 compared to the wild-type (Figure 6C), consistent with the reduced root-to-shoot K+ transport in the Oshak8 mutants. Taken together, these results demonstrated that OsHAK8 is involved in K+ uptake and root-to-shoot K+ transport in roots, especially under low-K+ conditions.
Discussion
Although OsAKT1 and OsHAK5, like AtAKT1 and AtHAK5 in Arabidopsis, mediate K+ uptake in rice roots (Li et al., 2014; Yang et al., 2014), more transporters related to rice K+ absorption and translocation need to be explored. Here, we showed that the OsHAK family gene OsHAK8 plays a crucial role in K+ uptake and root-to-shoot K+ transport in rice roots.
The GUS reporter analysis indicated that OsHAK8 was broadly expressed in different cell types in the roots (Figure 5Cf), especially in the exodermis, cortex, and stele (Figures 5Cb,f), which support the possible function of the gene in K+ uptake and root-to-shoot K+ translocation in rice roots. Several lines of genetic data connected growth defects of the Oshak8 mutant plants with the K+ transport function of the OsHAK8. The K+ contents in both the roots and shoots of the Oshak8 mutants were significantly reduced (Figures 2B,C), suggesting that the growth retardation of Oshak8 may be due to the reduction of K+ uptake and root-to-shoot K+ transport. Furthermore, the results from K+ depletion assays in which both root uptake and root-to-shoot K+ transport were measured again indicated that the Oshak8 mutants performed worse as compared to the wild-type plants. As K+ is a critical nutrient that supports plant growth, reduced K+ uptake and root-to-shoot K+ transport in the Oshak8 mutants may result in the inhibition of plant growth in both young seedlings and adult plants in the field (Figure 1A and Supplementary Figures 2A,B). Another process where OsHAK8 may function is in gametogenesis associated with pollen viability and seed setting. We observed reduced pollen viability and 1,000-grain weight associated with the mutant plants (Supplementary Figures 2E, 3A,B). Together, these results show that OsHAK8 is expressed and functions in multiple tissues and developmental stages in rice plants. Because OsHAK8 is a plasma membrane transporter, its function in K+ uptake may be due to its expression and activity in the root epidermal cells. In addition, its expression in the stelar cells may contribute to K+ transport into the vasculature and, eventually, secretion into the xylem vessels. Detailed analyses of K+ contents in various cell types of vascular tissues may help further dissect the role of OsHAK8 in a specific step of root-to-shoot long-distance transport.
Plants can sense K+ levels in the soil and accordingly regulate root-to-shoot translocation to balance K+ distribution between roots and shoots (Du et al., 2019). Under K+-sufficient conditions, the root-to-shoot K+ transport is enhanced to support plant growth. In contrast, when facing K+ deficiency in the soil, the long-distance transport activity is often reduced to maintain root growth as a priority. In other words, root-to-shoot K+ transport needs to be fine-tuned according to external K+ levels (Du et al., 2019). Transcriptional regulation provides an essential strategy in plant regulation of root-to-shoot translocation. Previous studies have revealed that the NRT1/PTR family member NPF7.3/NRT1.5 is regulated at the transcriptional level in response to external low-K+ levels. During low-K+ stress, NPF7.3/NRT1.5 transcripts are downregulated to inhibit root-to-shoot K+ transport in Arabidopsis (Du et al., 2019), which suggests that root-to-shoot K+ translocation can be achieved via the transcriptional regulation of related transporters such as NPF7.3/NRT1.5. In the present study, we found that OsHAK8 functions in root-to-shoot transport, especially under low-K+ conditions. Interestingly, OsHAK8 transcript level was reduced in response to K+ deficiency in root stelar cells (Figures 5Ba,g,C and Supplementary Figures 4A,B), consistent with the idea that downregulation of root-to-shoot K+ translocation may be achieved through reduced OsHAK8 expression. However, downregulation of OsHAK8 expression under low-K+ conditions does not mean plants do not need them. In loss-of-function mutants of OsHAK8, root-to-shoot K+ transport was largely abolished (Figure 6C), causing stunted growth (Figure 1A and Supplementary Figure 2Aa). Therefore, the transcript levels of OsHAK8 need to be fine-tuned in response to external K+ levels. These transcriptional levels of OsHAK8 may be regulated by some yet unknown components in the low-K+ response pathway. This hypothesis should be investigated in future experiments.
Compared with Arabidopsis HAK members, the rice genome features a large gene expansion in the HAK family. The dramatic gene expansion events in the rice HAK family imply that K+ uptake and transport mechanisms in rice are more complex. Indeed, according to the expression patterns in rice roots, different OsHAK transporters are expressed and responsible for K+ transport in different root cell types in response to external low K+ levels. For example, OsHAK1 is abundantly expressed in entire roots, particularly at the root tips. OsHAK5 is much less abundant in roots as compared to shoots. The expression of OsHAK16 is detected at the root epidermal and stelar cells. OsHAK8 was abundantly expressed in almost all cell types of roots. K+ deficiency steadily upregulated the expression of OsHAK1 and OsHAK16 in the roots, while enhancing OsHAK5 expression only transiently (Yang et al., 2014; Chen et al., 2015; Feng et al., 2019). In contrast, we found that K+ deficiency downregulated the expression of OsHAK8 in the steles. Such unique expression patterns of different OsHAK genes may have determined their functions in plants. OsHAK5 is involved in the root K+ acquisition and root-shoot translocation only at low (0.1–0.3 mM), but not at high (1 mM) external K+ mediums (Yang et al., 2014). OsHAK1 and OsHAK16 contribute to K+ acquisition and root-shoot translocation in the 0.05–1 mM range. Here, we showed that OsHAK8 mediated K+ acquisition and root-shoot translocation at a wide range of external K+ levels (0.01–10 mM). For further dissection of the distinct roles of OsHAK8, OsHAK1, OsHAK5, and OsHAK16 in rice, it will be important to reveal the regulatory mechanisms of OsHAK1, OsHAK5, OsHAK16, and OsHAK8 at both the transcriptional and post-transcriptional levels in the future.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XW, DM, LC, and SL conceived, designed the experiments, and analyzed the data. XW, JL, FL, DC, and YP performed the experiments. XW, DM, and SL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (Grant No. 2016y FD0101107), the National Science Foundation of China (Grant No. NSFC-31500200), and the Research Foundation of Education Bureau of Hunan Province of China (Grants Nos. 20A295, 20C1124, and 17B165).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Yaoguang Liu (SouthChina Agricultural University, Guangzhou, China.) and Lijia Qu (College of Life Science, Peking University, China) for providing the CRISPR/Cas9 system.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.730002/full#supplementary-material
Supplementary Figure 1 | Verification of the Oshak8 mutants. Verification of the Oshak8 mutants by PCR-based sequencing. Two representative transgenic lines (abbreviated as Oshak8-1 and Oshak8-2, respectively) for Oshak8 mutants are generated from the Nipponbare genetic background.
Supplementary Figure 2 | Phenotype analysis of Oshak8 mutants grown in soil. (A) Phenotype analysis of plants grown in soil. (a) Phenotype of plants grown in soil. WT and Oshak8 mutants (Oshak8-1, Oshak8-2) grown in soil (0.18-mg/g K+) for 95 days. Bar = 20 cm. (b) Comparison of brown spots, which is a typical K+-deficient symptom of rice on old leaves of plants. Growth conditions were as described in (A). Oshak8-1 and Oshak8-2 displayed brown spots on old leaves. WT displayed no brown spots on old leaves. Bar = 1 cm. (c) The main panicle lengths of plants. WT and Oshak8 mutants (Oshak8-1, Oshak8-2) grown in soil (0.18-mg/g K+) for 105 days. Main panicle lengths were measured. Bars = 8 cm. Data are presented as means ± SD. Bar = 10 cm. (B–E) Statistical analysis of plant heights (B), main panicle length (C), seed setting rate (D), and 1,000-grain weight (E) of plants. Growth conditions were as described in (A). Significant differences were found between WT and Oshak8 mutants (Oshak8-1, Oshak8-2) (^*P < 0.01 by Student’s t-test). n ≥ 20 for each data point. Asterisks represent significant differences.
Supplementary Figure 3 | Pollen viability analysis of Oshak8 mutants. (A) I2-KI staining of pollen grains from the wild type and Oshak8 mutants (Oshak8-1, Oshak8-2). Bar = 100 μm. (B) Pollen viability of the wild-type and Oshak8 mutants (Oshak8-1, Oshak8-2). The experiment was repeated three times with similar results. Error bars represent ± SD. Significant difference was found between wild-type and Oshak8 mutants (Oshak8-1, Oshak8-2) (P < 0.01 by Student’s t-test). The experiment was repeated three times with similar results. Error bars represent ± SD.
Supplementary Figure 4 | Quantitative analysis of OsHAK8 promoter activity represented by GUS signal. Histochemical analysis of OsHAK8 promoter-driven GUS reporter expression in transgenic rice plants. Three-day-old rice seedlings (Nipponbare) were grown in hydroponic solution containing 10 mM K+ for 3 days and then transferred to 10 mM K+ (+ K) and 0 mM K+ (-K) solutions for 24 h, respectively. GUS activity was examined by staining with GUS substrate (X-Gluc) in three independent lines (#2, #5, #8) of transgenic plants harboring OsHAK8-GUS construct. Plants were stained in each line and a representative plant was shown in each group. (A) Cross-section images of the root hair zone of the transgenic plants harboring OsHAK8-GUS construct (#2, #5, #8) in (A). Bars = 100 μm. (B) Quantification of GUS activity in the cortex region of transgenic rice plants containing the OsHAK8 promoter-GUS construct in (A). Image J software was used to quantify the GUS signals in the root cortex areas of the transgenic lines (#2, #5, #8) treated with K+-sufficient (10-mM K+) or low-K+ conditions (0-mM K+). Three plants from each line were used for the analysis in one experiment. The experiment was repeated three times with similar results. Error bars represent ± SD. Significant difference was found between 0.01- and 10-mM K+ samples are indicated in root tissues (P < 0.01 by Student’s t-test). The experiment was repeated three times with similar results. Error bars represent ± SD.
Supplementary Table 1 | List of PCR primers.
Footnotes
References
Adams, E., and Shin, R. (2014). Transport, signaling, and homeostasis of potassium and sodium in plants. J. Integ. Plant Biol. 56, 231–249. doi: 10.1111/jipb.12159
Ai, P., Sun, S., Zhao, J., Fan, X., Xin, W., Guo, Q., et al. (2009). Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 57, 798–809.
Anderson, J. A., Huprikar, S. S., Kochian, L. V., Lucas, W. J., and Gaber, R. F. (1992). Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 89, 3736–3740. doi: 10.1073/pnas.89.9.3736
Chen, G., Hu, Q., Luo, L., Yang, T., Zhang, S., Hu, Y., et al. (2015). Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 38, 2747–2765. doi: 10.1111/pce.12585
Cheong, Y. H., Pandey, G. K., Gran, J. J., Batistic, O., Li, L. G., Kim, B. G., et al. (2007). Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 52, 223–239. doi: 10.1111/j.1365-313x.2007.03236.x
Clarkson, D. T., and Hanson, J. B. (1980). The mineral nutrition of higher plants. Annu. Rev. Plant Physiol. 31, 239–298.
Colmenero-Flores, J. M., Martinez, G., Gamba, G., Vázquez, N., Iglesias, D. J., Brumós, J., et al. (2007). Identification and functional characterization of cation-chloride cotransporters in plants. Plant J. 50, 278–292. doi: 10.1111/j.1365-313x.2007.03048.x
Dobermann, A., StaCruz, P. C., and Cassman, K. G. (1996). Fertilizer inputs, nutrient balance, and soil nutrient-supplying power in intensive, irrigated rice systems. I. Potassium uptake and K balance. Nutr. Cycl. Agroecosyst. 46:110.
Du, X. Q., Wang, F. L., Li, H., Jing, S., Yu, M., Li, J. G., et al. (2019). The transcription factor MYB59 regulates K+/NO3– translocation in the Arabidopsis response to Low K+ stress. Plant Cell. 31, 699–714. doi: 10.1105/tpc.18.00674
Feng, H. M., Tang, Q., Cai, J., Xu, B. C., Xu, G. H., and Yu, L. (2019). Rice OsHAK16 functions in potassium uptake and translocation in shoot, maintaining potassium homeostasis and salt tolerance. Planta 250, 549–561. doi: 10.1007/s00425-019-03194-3
Gaymard, F., Pilot, G., Lacombe, B., Bouchez, D., Bruneau, D., Boucherez, J., et al. (1998). Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94, 647–655. doi: 10.1016/s0092-8674(00)81606-2
Gierth, M., Mäser, P., and Schroeder, J. I. (2005). The potassium transporter HAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 137, 1105–1114. doi: 10.1104/pp.104.057216
Han, M., Wu, W., Wu, W. H., and Wang, Y. (2016). Potassium transporter KUP7 is involved in K+ acquisition and translocation in Arabidopsis root under K+-limited conditions. Mol. Plant 9, 437–446. doi: 10.1016/j.molp.2016.01.012
Hirsch, R. E., Lewis, B. D., Spalding, E. P., and Sussman, M. R. (1998). Arole for the AKT1 potassium channel in plant nutrition. Science 280, 918–921. doi: 10.1126/science.280.5365.918
Horie, T., Sugawara, M., Okada, T., Taira, K., Nakayama, P. K., Katsuhara, M., et al. (2011). Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J. Biosci. Bioeng. 111, 346–356. doi: 10.1016/j.jbiosc.2010.10.014
Lagarde, D., Basset, M., Lepetit, M., Conejero, G., Gaymard, F., Astruc, S., et al. (1996). Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 9, 195–203. doi: 10.1046/j.1365-313x.1996.09020195.x
Leigh, R. A., and Wyn Jones, R. G. (1984). A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytologist. 97, 1–13. doi: 10.1111/j.1469-8137.1984.tb04103.x
Li, H., Yu, M., Du, X. Q., Wang, Z. F., Wu, W. H., Quintero, F. J., et al. (2017). NRT1.5/NPF7.3 functions as a protoncoupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis. Plant Cell. 29, 2016–2026. doi: 10.1105/tpc.16.00972
Li, J., Yu, L., Guo, N. Q., Li, J., Xu, Z. J., Wu, W. H., et al. (2014). OsAKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell. 26, 3387–3402. doi: 10.1105/tpc.114.123455
Li, L. G., Kim, B. G., Cheong, Y. H., Pandey, G. K., and Luan, S. (2006). A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 12625–12630. doi: 10.1073/pnas.0605129103
Li, S. B., Qian, Q., Fu, Z. M., Zeng, D. L., Meng, X. B., Kyozuka, J., et al. (2009). Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 58, 592–605. doi: 10.1111/j.1365-313x.2009.03799.x
Liu, H., Ding, Y., Jin, W., Xie, K., and Chen, L. L. (2017). CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 10, 530–532. doi: 10.1016/j.molp.2017.01.003
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-DeltaDeltaC(T)). Method Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luan, M., Tang, R. J., Tang, Y., Tian, W., Hou, C., Zhao, F., et al. (2017). Transport and homeostasis of potassium and phosphate: limiting factors for sustainable crop production. J. Exp. Bot. 68, 3091–3105.
Luan, S. (2009). The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 14, 37–42. doi: 10.1016/j.tplants.2008.10.005
Luan, S., Lan, W. Z., and Lee, S. C. (2009). Potassium nutrition, sodium toxcity, and calcium signaling: connections through the CBL-CIPK network. Curr. Opin. Plant Biol. 12, 339–346. doi: 10.1016/j.pbi.2009.05.003
Maathuis, F. J. M. (2009). Physiological functions of mineral macronutrients. Curr. Opin. Plant Biol. 12, 250–258. doi: 10.1016/j.pbi.2009.04.003
Musavizadeh, Z., Najafi-Zarrini, H., Kazemitabar, S. K., Hashemi, S. H., Faraji, S., Barcaccia, G., et al. (2021). Genome-wide analysis of potassium channel genes in rice: expression of the OsAKT and OsKAT genes under salt stress. Genes 12:784. doi: 10.3390/genes12050784
Nieves-Cordones, M., Alemán, F., Martínez, V., and Rubio, F. (2010). The Arabidopsis thaliana HAK5 K+ transporter is required for plant growth and K+ acquisition from low K+ solutions under saline conditions. Mol. Plant 3, 326–333. doi: 10.1093/mp/ssp102
Nieves-Cordones, M., Alemán, F., Martínez, V., and Rubio, F. (2014). K+ uptake in plant roots. The systems involved, their regulation and parallels in other organisms. J. Plant Physiol. 171, 688–695. doi: 10.1016/j.jplph.2013.09.021
Porra, R. J., Thompson, W. A., and Kriedemann, P. E. (1989). Determination of accurate extinction coefficients and simultaneous-equations for assaying chlorophyll-a and b extracted with 4 different solvents-verification of the concentration of chlorophyll standards by atomic-absorption spectroscopy. Biochimica et Biophysica Acta 975, 384–394. doi: 10.1016/S0005-2728(89)80347-0
Pottosin, I., and Dobrovinskaya, O. (2014). Non-selective cation channels in plasma and vacuolar membranes and their contribution to K+ transport. J. Plant Physiol. 171, 732–742. doi: 10.1016/j.jplph.2013.11.013
Pyo, Y. J., Gierth, M., Schroeder, J. I., and Cho, M. H. (2010). High-affinity K+ transport in Arabidopsis: HAK5 and AKT1 are vital for seedling stablishment and postgermination growth under low-potassium conditions. Plant Physiol. 153, 863–875. doi: 10.1104/pp.110.154369
Ragel, P., Rodenas, R., Garcia-Martin, E., Andres, Z., Villalta, I., Nieves-Cordones, M., et al. (2015). The CBL-Interacting Protein Kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol. 169, 2863–2873.
Rubio, F., Santa-María, G. E., and Rodríguez-Navarro, A. (2000). Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol. Plant. 109, 34–43. doi: 10.1034/j.1399-3054.2000.100106.x
Schachtman, D. P., and Shin, R. (2007). Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol. 58, 47–69. doi: 10.1146/annurev.arplant.58.032806.103750
Schroeder, J. I., Ward, J. M., and Gassmann, W. (1994). Perspectives on the physiology and structure of inward-rectifying K+ channels in higher plants: biophysical implications for K+ uptake. Annu. Rev. Biophys. Biomol. Struct. 23, 441–471. doi: 10.1146/annurev.bb.23.060194.002301
Shen, Y., Shen, L., Shen, Z. X., Jing, W., Ge, H. L., Zhao, J. Z., et al. (2015). The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stressin rice. Plant Cell Environ. 38, 2766–2779. doi: 10.1111/pce.12586
Tang, R. J., Zhao, F. G., Yang, Y., Wang, C., Li, K. L., Kleist, T. J., et al. (2020). A calcium signalling network activates vacuolar K+ remobilization to enable plant adaptation to low-K environments. Nat. Plants 6, 384–393. doi: 10.1038/s41477-020-0621-7
Upadhyaya, N., Surin, B., Ramm, K., Gaudron, J., Schunmann, P. H. D., Taylor, W., et al. (2000). Agrobacterium-mediated transformation of Australian rice cultivars Jarrah and Amaroo using modified promoters and selectable markers. Aust. J. Plant Physiol. 27, 201–210. doi: 10.1071/pp99078
Usman, B., Nawaz, G., Zhao, N., Liu, Y. G., and Li, R. B. (2020). Generation of high yielding and fragrant rice (Oryza sativa L.) lines by CRISPR/Cas9 targeted mutagenesis of three homoeologs of cytochrome P450 gene family and OsBADH2 and transcriptome and proteome profiling of revealed changes triggered by mutations. Plants 9:788. doi: 10.3390/plants9060788
Wanasuria, S., De Datta, S. K., and Mengel, K. (1981). Rice yield in relation toelectro-ultrafiltrat -ion extractable soil potassium. Plant Soil 59, 23–31. doi: 10.1007/bf02183589
Wang, Y., and Wu, W. H. (2017). Regulation of potassium transport and signaling in plants. Curr. Opin. Plant Biol. 39, 123–128. doi: 10.1016/j.pbi.2017.06.006
Xie, K., Zhang, J., and Yang, Y. (2014). Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol. Plant 7, 923–926. doi: 10.1093/mp/ssu009
Xu, J., Li, H. D., Chen, L. Q., Wang, Y., Liu, L. L., He, L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360. doi: 10.1016/j.cell.2006.06.011
Yang, T., Zhang, S., Hu, Y., Wu, F., Hu, Q., Chen, G., et al. (2014). The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 166, 945–959. doi: 10.1104/pp.114.246520
Yang, Z. F., Gao, Q. S., Sun, C. S., Li, W. J., Gu, S. L., and Xu, C. W. (2009). Molecular evolution and functional divergence of HAK potassium transporter gene family in rice (Oryza sativa L.). J. Genet. Genomics 36, 161–172. doi: 10.1016/s1673-8527(08)60103-4
Keywords: potassium, rice, OsHAK8, K+ uptake, K+ translocation, low K+ stress
Citation: Wang X, Li J, Li F, Pan Y, Cai D, Mao D, Chen L and Luan S (2021) Rice Potassium Transporter OsHAK8 Mediates K+ Uptake and Translocation in Response to Low K+ Stress. Front. Plant Sci. 12:730002. doi: 10.3389/fpls.2021.730002
Received: 24 June 2021; Accepted: 13 July 2021;
Published: 03 August 2021.
Edited by:
Jon Pittman, The University of Manchester, United KingdomReviewed by:
Ashish Kumar Srivastava, Bhabha Atomic Research Centre (BARC), IndiaParviz Heidari, Shahrood University of Technology, Iran
Copyright © 2021 Wang, Li, Li, Pan, Cai, Mao, Chen and Luan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dandan Mao, bWRkMDMwM0AxNjMuY29t; Liangbi Chen, Y2hlbmxpYW5nYmlAMTI2LmNvbQ==; Sheng Luan, c2x1YW5AYmVya2VsZXkuZWR1
†These authors have contributed equally to this work
 Xiaohui Wang1†
Xiaohui Wang1† Dandan Mao
Dandan Mao Sheng Luan
Sheng Luan