- 1Crop Research Institute, Shandong Academy of Agricultural Sciences, Jinan, China
- 2Key Laboratory of Wheat Biology and Genetic Improvement in the North Huang and Huai River Valley, Ministry of Agriculture, Jinan, China
- 3National Engineering Laboratory for Wheat and Maize, Jinan, China
- 4School of Life Sciences and Technology, University of Electronic Science and Technology of China, Chengdu, China
Aegilops sharonensis, a wild relative of wheat, harbors diverse disease and insect resistance genes, making it a potentially excellent gene source for wheat improvement. In this study, we characterized and evaluated six wheat-A. sharonensis derivatives, which included three disomic additions, one disomic substitution + monotelosomic addition and two disomic substitution + disomic additions. A total of 51 PLUG markers were developed and used to allocate the A. sharonensis chromosomes in each of the six derivatives to Triticeae homoeologous groups. A set of cytogenetic markers specific for A. sharonensis chromosomes was established based on FISH using oligonucleotides as probes. Molecular cytogenetic marker analysis confirmed that these lines were a CS-A. sharonensis 2Ssh disomic addition, a 4Ssh disomic addition, a 4Ssh (4D) substitution + 5SshL monotelosomic addition, a 6Ssh disomic addition, a 4Ssh (4D) substitution + 6Ssh disomic addition and a 4Ssh (4D) substitution + 7Ssh disomic addition line, respectively. Disease resistance investigations showed that chromosome 7Ssh of A. sharonensis might harbor a new powdery mildew resistance gene, and therefore it has potential for use as resistance source for wheat breeding.
Introduction
Aegilops sharonensis Eig (Sharon goatgrass, SshSsh, 2n = 2x = 14), a wild relative of wheat, is endemic to the coastal plains of Israel and southern Lebanon (Slageren, 1994), and its genome is closely related to the B genome of common wheat (Olivera and Steffenson, 2009). A. sharonensis is a diverse source of genes for disease and insect resistance (Gill et al., 1985; Olivera et al., 2007). It has been reported that A. sharonensis carries resistance to leaf rust (Snyman et al., 2004; Olivera et al., 2007), stem rust (Valkoun et al., 1985; Olivera et al., 2007), stripe rust (Anikster et al., 2005; Olivera et al., 2007), powdery mildew (Dhaliwal et al., 1993; Olivera et al., 2007), and greenbug (Gill et al., 1985). Moreover, A. sharonensis has high tolerance to salt, drought, aluminum, boron, and nutrient deficiencies (Manyowa, 1989; Waines et al., 1993; Xu et al., 1993; Gorny and Garczynski, 2008). Consequently, A. sharonensis is potentially an excellent gene source for wheat improvement.
Miller et al. (1982) succeeded in producing and identifying a wheat-A. sharonensis addition line, which was due to the preferential transmission of one chromosome from A. sharonensis. Subsequently, this chromosome was identified as a gametocidal chromosome 4Ssh by cytological methods such as chromosome observation, C-banding and in situ hybridization. They also produced a wheat-A. sharonensis 4Ssh (4D) substitution line (Miller et al., 1982). Later, Xu et al. (1992) also reported that they had succeeded in producing a wheat-A. sharonensis 4Ssh (4D) substitution line by using a nullisomic backcrossing procedure. Millet (2007) developed a tetraploid wheat-A. sharonensis amphiploid (genome AABBS1S1). Yu et al. (2017) identified two novel wheat stem rust resistance genes in A. sharonensis. Antonyuk et al. (2009) studied 26 wheat-A. sharonensis introgression lines. Recently, both Zhao et al. (2014) and Jiang et al. (2014) developed tetraploid wheat-A. sharonensis amphidiploids. However, there are very few reports on the isolation of wheat plants carrying individual A. sharonensis chromosomes. Li X. Y. et al. (2019); Li et al. (2020) reported 24 HMW-GSs homozygous lines derived from progenies of cross wheat/A. sharonensis, and produced three 1Ssh (1A) substitution lines, two 1Ssh (1B) substitution lines, three 1Ssh (1D) substitution lines and two 1Ssh (5D) substitution lines. Therefore, the set of wheat-A. sharonensis chromosome lines is still not complete, which greatly limits the mapping and utilization of excellent genes derived from this species in wheat.
In this study, six wheat-A. sharonensis chromosome derivatives, including three disomic addition lines, one disomic substitution + monotelosomic addition line, and two disomic substitution + disomic addition lines, were identified by (Polymerase Chain Reaction) PCR-based landmark unique gene (PLUG) markers and fluorescence in situ hybridization (FISH) analysis. In addition, the infection types (ITs) of disease resistance, spike and grain characteristics of these wheat-A. sharonensis chromosome lines were also investigated to provide useful information for the possible subsequent development of wheat-A. sharonensis translocations for wheat genetic improvement.
Materials and Methods
Plant Materials
Triticum aestivum cv. Jinan17 (JN17) and Jimai22 (JM22) were maintained at the Crop Research Institute, Shandong Academy of Agricultural Sciences in Jinan. T. aestivum cv. Chinese Spring (CS) was provided by Prof. Z. J. Yang, School of Life Science and Technology, University of Electronic Science and Technology of China, Chengdu. The diploid A. sharonensis accession (TA1995) was provided by Mr. J. Raupp, Wheat Genetic and Genomic Resources Center, Kansas State University, United States. The CS-A. sharonensis amphiploid (JIC-31) and six unidentified CS-A. sharonensis chromosome lines (JIC-32, JIC-33, JIC-34, JIC-35, JIC-36, and JIC-37) were kindly provided by Prof. S. M. Reader, John Innes Centre, United Kingdom.
Fluorescence in situ Hybridization Analysis
Root tip treatments, chromosome slide preparations, and chromosome counting were according to Liu et al. (2011). Fifteen seeds of each of the materials were germinated for collection of root tips for FISH analysis and fifteen cells of each of the materials were studied. Probes Oligo-pTa535-1, Oligo-pSc119.2-1, and Oligo-(GAA)8 were synthesized by Chengdu Ruixin Biological Technology Co., Ltd. Probe sequences, the fluorochromes for probe labeling, FISH protocols and labeled DNA signal detection methods were according to Danilova et al. (2012) and Tang et al. (2014) after comparison with the CS standard FISH map. FISH using Oligo-(GAA)8 as a probe could be used to identify wheat chromosomes except 1A, 3D, 4D, 5D, and 6D, as described by Danilova et al. (2012). FISH using Oligo-pSc119.2-1 and Oligo-pTa535-1 probes could identify all 42 wheat chromosomes simultaneously as described by Tang et al. (2014). pTa71 (45S rDNA) contains a 9-kb EcoRI fragment isolated from bread wheat (Gerlach and Bedbrook, 1979), which could be used to identify homoeologous groups 1 and 6 of Triticum and Aegilops. Photomicrographs of FISH chromosomes were taken using an Olympus BX-51 microscope.
DNA Isolation and PLUG-PCR
Total genomic DNA isolation was according to the protocol of Liu et al. (2006). A total of 526 PLUG primer pairs were synthesized according to Ishikawa et al. (2009). All primer pairs were synthesized by Chengdu Ruixin Biological Technology Co., Ltd., and PCR protocol was according to Ishikawa et al. (2009). In order to obtain high levels of polymorphism, the PCR products were digested with the four-base cutter enzymes HaeIII or TaqI according to Ishikawa et al. (2009) and were separated on 2% agarose gels.
Disease Resistance Testing
The resistance reactions to stripe rust, leaf rust, stem rust, and powdery mildew of the six suspected CS-A. sharonensis derivatives were tested. We investigated the disease resistance data for two consecutive years in 2015 and 2016, and 20 individual plants of each line were investigated each year. CS is highly susceptible to all four pathogens, hence the disease response scoring did not begin until CS was fully infected. According to Wang et al. (2014), the disease responses were scored on a 0–4 rating scale, 0 means immune, 0; indicates nearly immune but showing a small fleck on the leaf, 1 means highly resistant, 2 indicates moderately resistant, 3 means moderately susceptible, and 4 indicates highly susceptible. Scores of 0–2 were classified as resistant and 3–4 as susceptible. The pathogenic race selection and disease response rating scale of the four diseases were all according to Gong et al. (2017). The pathogen inoculation methods for stripe rust, leaf rust and powdery mildew were according to Liu et al. (2013), while stem rust inoculation was according to Han et al. (2018). Stripe rust resistance was determined on adult plants using mixed isolates of races CY32, CY33, and Su-4 in the experimental farmland of School of Life Science and Technology, University of Electronic Science and Technology of China, Chengdu, Sichuan Province. Stem rust resistance was determined on seedlings using mixed isolates of pathotypes 34MKGQM and 21C3CTHSM in the greenhouse of College of Plant Protection, Shenyang Agricultural University, Shenyang, Liaoning Province. Leaf rust resistance was determined on seedlings using mixed isolates of THTT, PHTT, THKS, THTS, and THKT (these isolates are prevalent and highly damaging on wheat crops throughout China) in the greenhouse of College of Plant Protection, Hebei Agricultural University, Baoding, Hebei Province. Powdery mildew resistance was determined on seedlings (in the greenhouse) and also on adult plants (in the field) following inoculation with mixed powdery mildew races collected from four different cities including Jinan, Linyi, Dezhou, and Heze of Shandong Province.
Spike and Grain Characteristics
Chinese Spring and the six suspected CS-A. sharonensis chromosome lines were planted in the field at Jinan in Shandong Province on October 25, 2015 and harvested on June 5, 2016. The spikes were collected for photographs on May 10, 2016 then threshed and the grain extracted when fully mature. The spike and grain characters of these materials were investigated and described according to Li et al. (2006).
Results
Cytogenetic Identification of Wheat-A. sharonensis Chromosome Lines
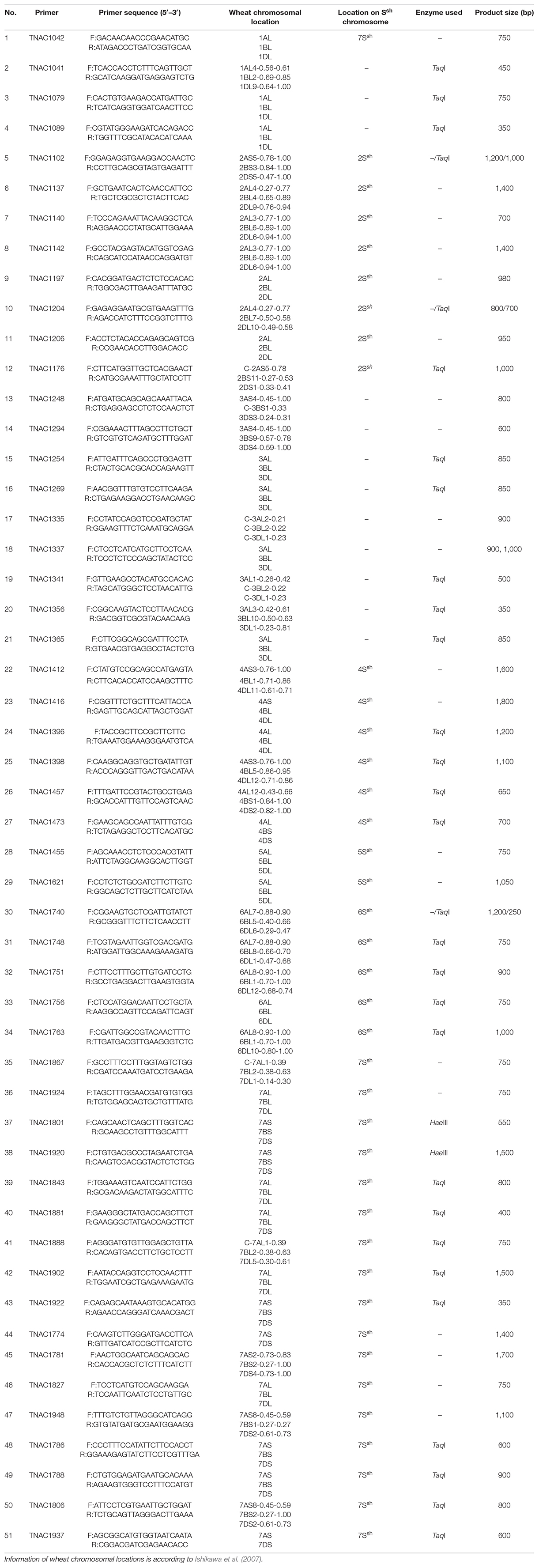
Sequential FISH with probes Oligo-pSc119.2-1, Oligo-pTa535-1, and Oligo-(GAA)8 was used to detect the chromosome constitution of wheat-A. sharonensis amphiploid JIC-31 (Figure 1). The karyotype of the seven pairs of A. sharonensis chromosomes in JIC-31 is shown in Figure 1C. Cytological studies revealed that the chromosome numbers of JIC-32 to JIC-37 were 44, 44, 42 + monotelosomic, 42, 44, and 44, respectively. FISH on mitotic metaphase chromosomes of these lines showed that the lines JIC-32 and JIC-33 had 42 wheat chromosomes, while the 4D chromosomes in JIC-34 to JIC-37 were missing. A pair of A. sharonensis chromosomes with distinct FISH signals different from wheat chromosomes was detected in JIC-32, JIC-33, and JIC-35. Two different pairs of A. sharonensis chromosomes were found in JIC-36 and JIC-37, while disomic and monotelosomic additions of A. sharonensis chromosomes were detected in JIC-34. Therefore, JIC-32, JIC-33, and JIC-35 are CS-A. sharonensis disomic addition lines, JIC-36 and JIC-37 are CS-A. sharonensis disomic substitution + disomic addition lines. JIC-34 is CS-A. sharonensis disomic substitution + monotelosomic addition line. FISH patterns of JIC-32 and JIC-33 are shown in Figure 2 (FISH patterns of JIC-34 to JIC-37 are shown in Supplement Figure 1).
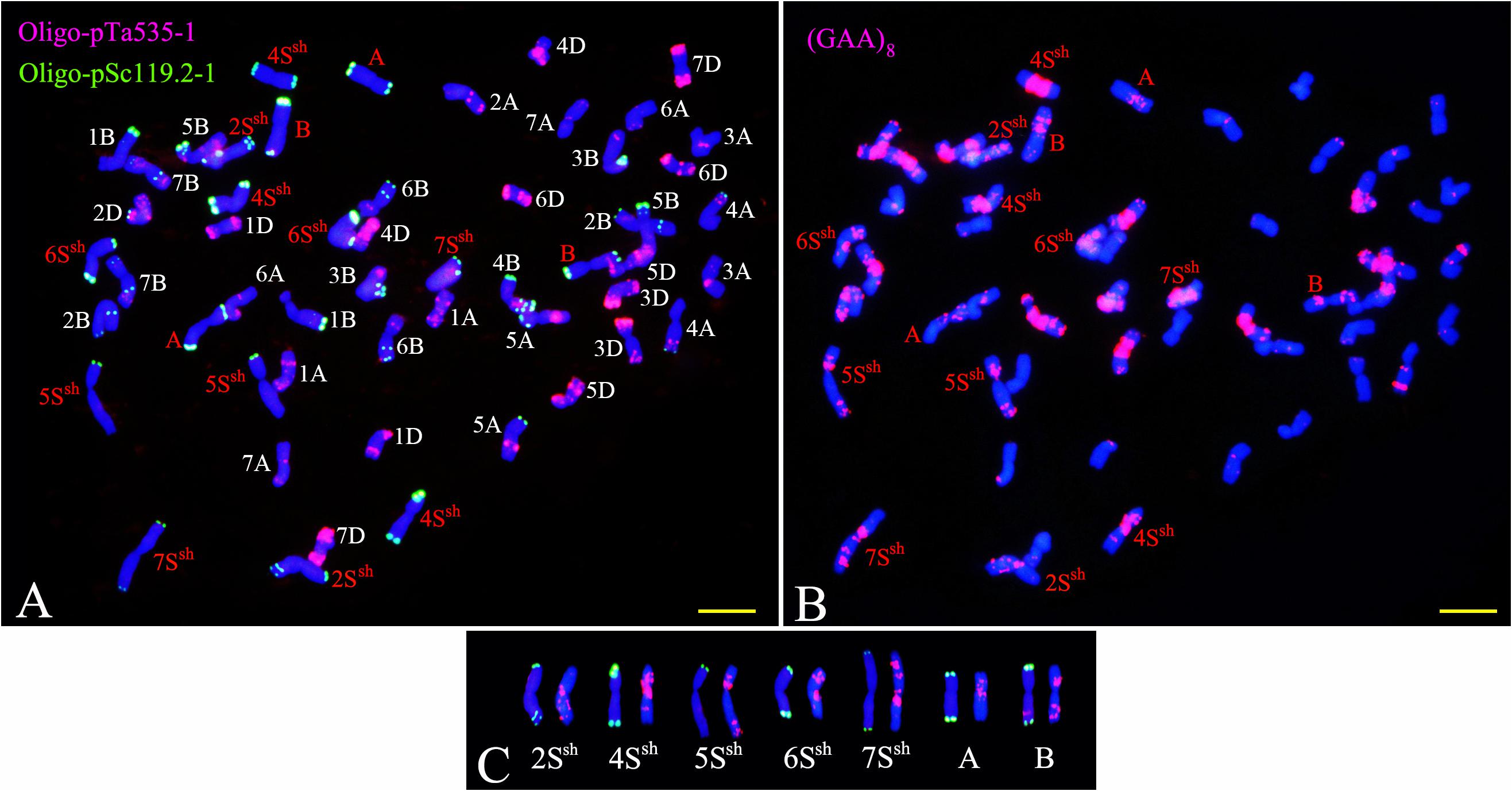
Figure 1. FISH identification of the chromosome constitution of wheat-Aegilops sharonensis amphiploid JIC-31. Panel (A) shows the probes were Oligo-pSc119.2-1 (green) and Oligo-pTa535-1 (red). Panel (B) shows the probe was (GAA)8 (red). (C) shows the karyotype of the seven pairs of A. sharonensis chromosomes in JIC-31; Panel (A,B) indicates unidentified chromosome. Bar indicates 10 μm.
Figure 2. FISH using Oligo-nucleotides as probes on the CS-A. sharonensis 2Ssh disomic addition (A,B) and the 4Ssh disomic addition (C,D). Panels (A,C) are double-color FISH patterns using Oligo-pTa535-1 (red) and Oligo-pSc119.2-1 (green) as probes; (B,D) are double-color FISH patterns using pTa71 (red) and (GAA)8 (green) as probes. Bar indicates 10 μm.
FISH using Oligo-pTa535-1 onto A. sharonensis chromosomes in lines JIC-32 to JIC-37 showed no signals associated with that probe. However, slightly different Oligo-pSc119.2-1 signals were found on both terminal regions of all A. sharonensis chromosomes (Figure 3). In addition to signals on terminal regions of chromosomes, hybridization associated with Oligo-pSc119.2-1 was observed on sub-terminal regions on the A. sharonensis chromosomes in line JIC-32 (Figure 3). Moreover, probe Oligo-(GAA)8 with different signal positions and signal strengths, together with chromosome arm ratios, could clearly characterize all A. sharonensis chromosomes in lines JIC-32 to JIC-37. The pTa71 signals on the sub-terminal regions could be detected on the short arms of A. sharonensis chromosomes in JIC-35 and JIC-36 (Figure not shown), indicating that this pair of A. sharonensis chromosomes might be 1Ssh or 6Ssh.
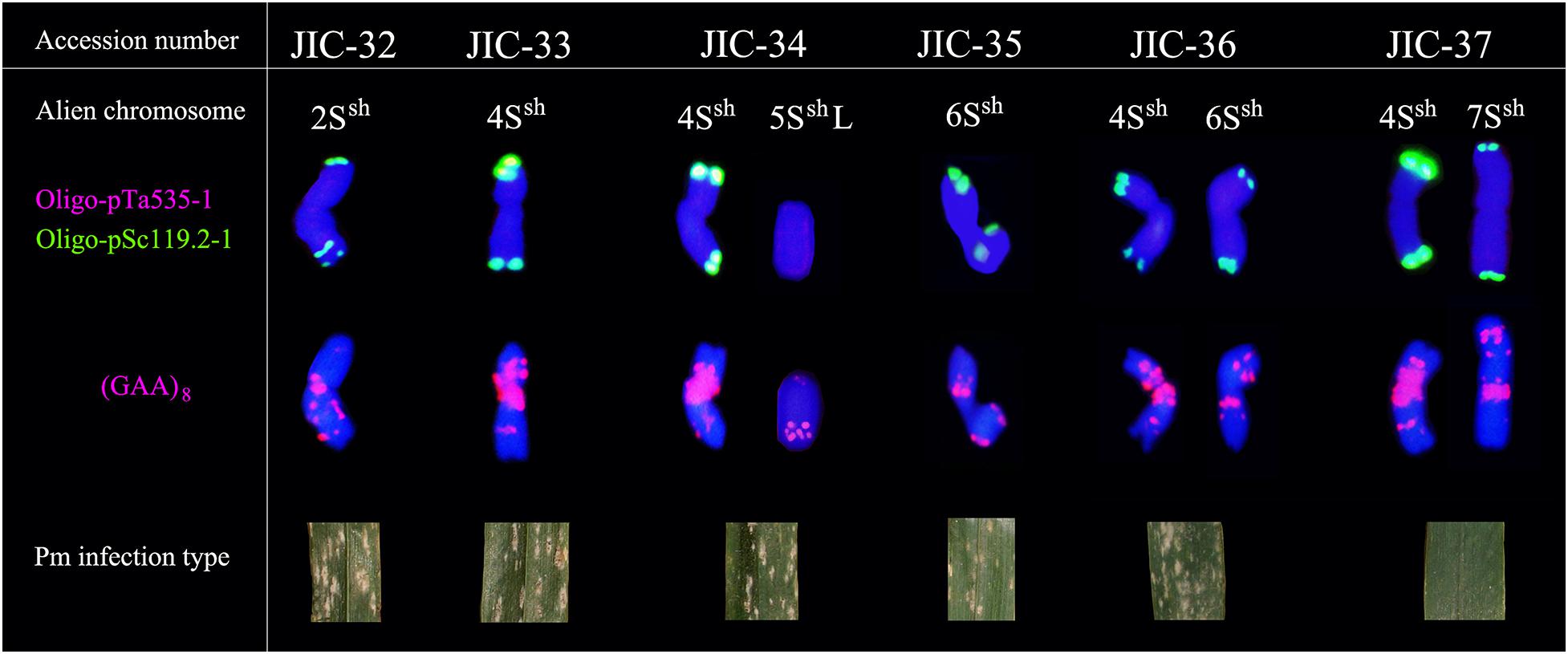
Figure 3. Standard FISH pattern of A. sharonensis chromosomes using Oligo-nucleotides as probes. Row one indicates alien chromosome; Row two indicates double-color FISH patterns using Oligo-pTa535-1 (red) and Oligo-pSc119.2-1 (green) as probes; Row three indicates FISH patterns using (GAA)8 (red) as probes after washing off the double-color FISH signals; Row four indicates photos of powdery mildew in wheat-A. sharonensis chromosome lines.
Molecular Identification of Wheat-A. sharonensis Chromosomes
In order to identify the homoeologous groups of each of the A. sharonensis chromosomes in JIC-32 to JIC-37, a total of 526 PLUG primer pairs were used to develop A. sharonensis chromosome-specific markers. As a result, fifty-one primer pairs could generate polymorphisms in A. sharonensis, the CS-A. sharonensis amphiploid, CS, JM22, and JN17. Among them, four, eight, nine, six, two, five, and seventeen belonged to chromosome homoeologous groups 1–7, respectively (Table 1, 2). The percentage of primers showing polymorphisms across the seven types of A. sharonensis chromosomes ranged from 2.6 to 15.6% (Table 1). The PCR patterns of primer pairs TNAC1137, TNAC1197, TNAC1398, TNAC1740, TNAC1867, and TNAC1924 are shown in Figure 4.
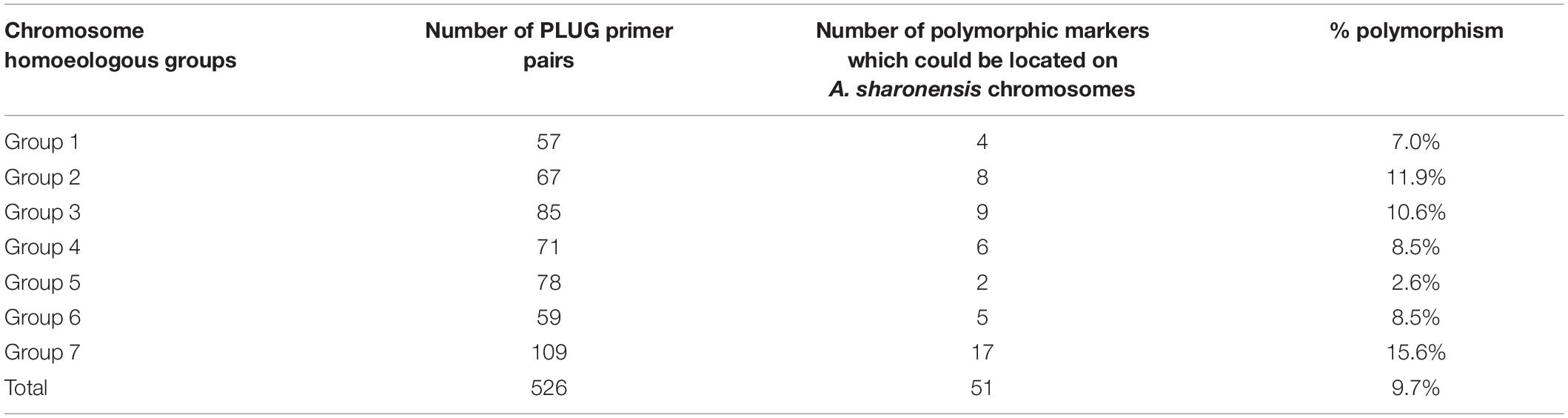
Table 1. PLUG primer pairs screened to identify specific markers of Aegilops sharonensis chromosomes.
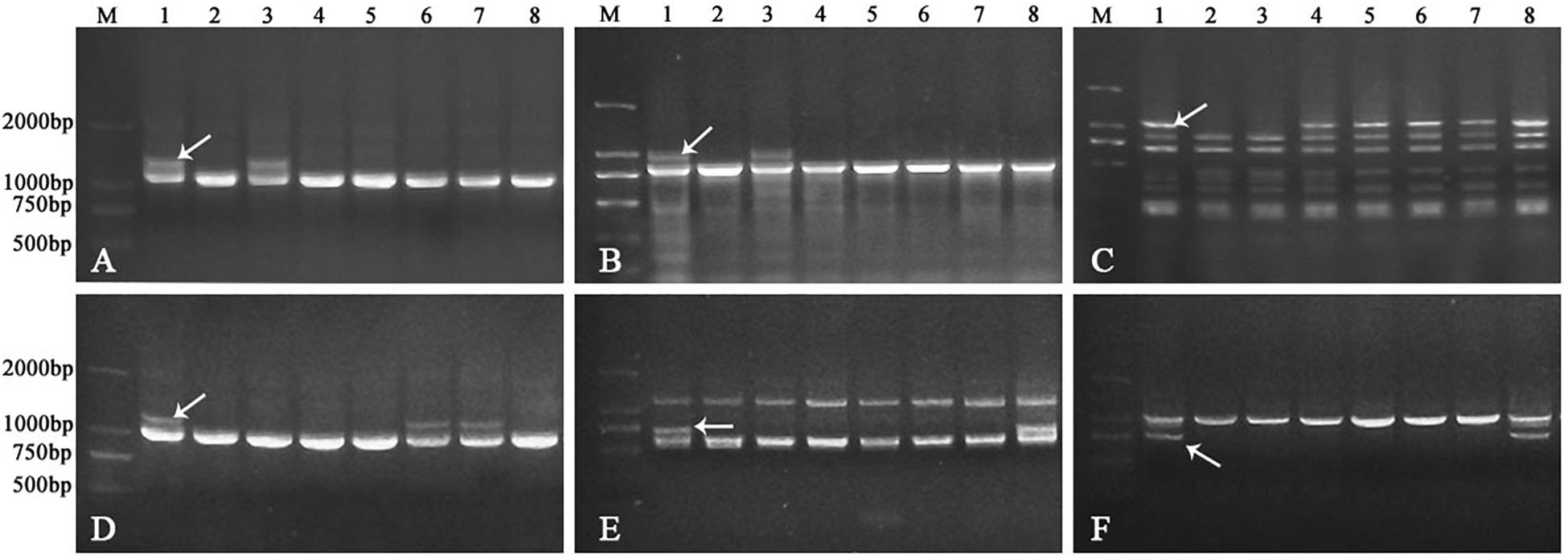
Figure 4. PCR patterns of primer pair TNAC1137 (A), TNAC1197 (B), TNAC1398 (C), TNAC1740 (D), TNAC1867 (E), and TNAC1924 (F). Lane M indicates Marker DM2000. Lanes 1-8 in panels (A–F) are CS-A. sharonensis amphiploid, CS, CS-A. sharonensis 2Ssh disomic addition, 4Ssh disomic addition, 4Ssh (4D) substitution + 5SshL monotelosomic addition, 6Ssh disomic addition, 4Ssh (4D) substitution + 6Ssh disomic addition and 4Ssh (4D) substitution + 7Ssh disomic addition lines, respectively.
TNAC1102 and another seven primer pairs specific to chromosome homoeologous group 2, could amplify polymorphisms in the A. sharonensis chromosomes of JIC-32, indicating that the pair of A. sharonensis chromosomes in that line was 2Ssh. The chromosome number of JIC-32 was 44, including the 42 complete wheat chromosomes, therefore, JIC-32 was a CS-A. sharonensis 2Ssh disomic addition (Figures 2A, B). Based on the results of molecular markers and cytological identification, the same analysis method was performed on JIC-33 to JIC-37, indicating that JIC-33 was a CS-A. sharonensis 4Ssh disomic addition (Figures 2C, D), while JIC-34 was a CS-A. sharonensis 4Ssh (4D) substitution + 5SshL monotelosomic addition (Supplementary Figures 1A, B), JIC-35 was a CS-A. sharonensis 6Ssh disomic addition (Supplementary Figures 1C, D), JIC-36 was a CS-A. sharonensis 4Ssh (4D) substitution + 6Ssh disomic addition (Supplementary Figures 1E, F) and JIC-37 was a 4Ssh (4D) substitution + 7Ssh disomic addition line (Supplementary Figures 1G, H).
Spike and Grain Characters of Wheat-A. sharonensis Chromosome Lines
Compared to spike morphologies of CS, the spikes of the six CS-A. sharonensis chromosome derivatives were all varied (Figure 5). Spikes of the CS-A. sharonensis 2Ssh disomic addition had short awns and narrow spikes. The lower inter-spikelet segments of the heads of CS-A. sharonensis 4Ssh (4D) substitution + 5SshL monotelosomic addition, 6Ssh disomic addition, 4Ssh (4D) substitution + 6Ssh disomic addition and 4Ssh (4D) substitution + 7Ssh disomic addition lines were more elongated than that of CS. The CS-A. sharonensis 4Ssh disomic addition line (JIC-33) showed slightly elongated spikelets and overall longer spikes than that of CS. The CS-A. sharonensis 4Ssh (4D) substitution + 6Ssh disomic addition (JIC-36) showed shorter spikes and fewer spikelets per head than that of CS (Figure 5).

Figure 5. Spike and grain morphologies of wheat-A. sharonensis chromosome lines. Grain and spikes from left to right are CS, CS-A. sharonensis 2Ssh disomic addition, 4Ssh disomic addition, 4Ssh (4D) substitution + 5SshL monotelosomic addition, 6Ssh disomic addition, 4Ssh (4D) substitution + 6Ssh disomic addition and 4Ssh (4D) substitution + 7Ssh disomic addition lines, respectively.
Grain morphologies of the six CS-A. sharonensis chromosome derivatives identified above were similar to that of CS, while the CS-A. sharonensis 2Ssh disomic addition (JIC-32) showed slender grains and darker pericarp color than that of CS (Figure 5) and the 4Ssh disomic addition (JIC-33) showed smaller grains than those of CS.
Disease Resistance Tests of Wheat-A. sharonensis Chromosome Lines
Stripe rust, leaf rust, stem rust, and powdery mildew resistance tests showed that all the materials were moderately to highly susceptible to stripe rust, leaf rust, and stem rust (Table 3) except that the infection reaction for leaf rust on the CS-A. sharonensis 4Ssh (4D) substitution + 7Ssh disomic addition (JIC-37) was not obtained. The CS-A. sharonensis 4Ssh (4D) substitution + 7Ssh disomic addition (JIC-37) was nearly immune to powdery mildew, while CS and other CS-A. sharonensis chromosome lines tested were highly susceptible to powdery mildew (Table 3), suggesting that chromosome 7Ssh of A. sharonensis might carry powdery mildew resistant gene(s).
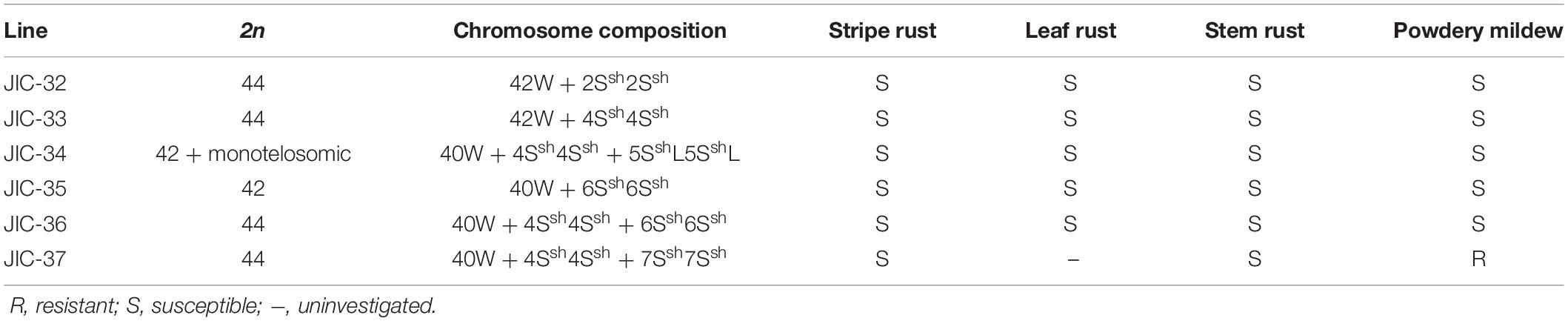
Table 3. The chromosome composition and Stripe rust, leaf rust, stem rust, and powdery mildew infection types of JIC-32 to JIC-37.
Discussion
Chromosomes Transferred From A. sharonensis Into Wheat
Transferring each pair of A. sharonensis chromosomes into wheat is difficult due to the presence of gametocidal (Gc) genes that control the preferential transmission of chromosome 4Ssh (Endo, 1982; Miller et al., 1982). Therefore, it is not easy to produce a complete set of wheat-A. sharonensis additions or substitutions (Miller et al., 1982; Olivera and Steffenson, 2009). Maan found a T. urartu-A. sharonensis amphiploid TA3398 in North Dakota in 1972 (unpublished). King et al. (1991) induced a mutation in the male fertility gene of the preferentially transmitted A. sharonensis chromosome 4S1 (some scientists defined the genome of A. sharonensis as S1S1). Friebe et al. (2003) produced a mutation of the A. sharonensis Gc2 gametocidal gene (Gc2mut), which opened a way for introgression of genes from A. sharonensis into wheat. Zhang et al. (2001) reported the production of additions 1S1, 3S1, 5S1, 6S1, and 7S1 in a 4S1 (4D) background. Antonyuk et al. (2009) studied 26 wheat-A. sharonensis introgression lines which they then separated into six groups based on different substituted chromosomes belonging to definite homoeologous groups and different numbers of translocations. Millet (2007) developed a tetraploid wheat-A. sharonensis amphiploid (genome AABBS1S1). Li X. Y. et al. (2019); Li et al. (2020) reported 24 HMW-GSs homozygous lines derived from progenies of cross wheat/A. sharonensis, and produced three 1Ssh (1A) substitution lines, two 1Ssh (1B) substitution lines, three 1Ssh (1D) substitution lines and two 1Ssh (5D) substitution lines.
So far, reports regarding the development of wheat-A. sharonensis introgression lines are very rare. Furthermore, none to date has reported the production of wheat-A. sharonensis 2Ssh introgression lines. In this research, six wheat-A. sharonensis introgression lines were identified, including a CS-A. sharonensis 2Ssh disomic addition (JIC-32), a 4Ssh disomic addition (JIC-33), a 4Ssh (4D) substitution + 5SshL monotelosomic addition (JIC-34), a 6Ssh disomic addition (JIC-35), a 4Ssh (4D) substitution + 6Ssh disomic addition (JIC-36) and a 4Ssh (4D) substitution + 7Ssh disomic addition (JIC-37). Among these six introgression lines, four possessed chromosome 4Ssh, suggesting that chromosome 4Ssh of A. sharonensis was transmitted preferentially into wheat due to the gametocidal gene, which confirms the reports of preferential transmission of gametocidal chromosomes of earlier researchers (Endo, 1982; Miller et al., 1982). These six newly identified wheat-A. sharonensis chromosome derivatives will enrich the germplasm resources available for wheat breeding.
Development of New Molecular Markers Specific for A. sharonensis Chromosomes
Previous reports regarding identification of useable molecular markers for A. sharonensis chromosomes indicated that the percentage which were polymorphic was very low, ranging from 1.3 to 11.4% (Zhang et al., 2001; Zhao et al., 2014; Wei Long, 2016; Li et al., 2020). Zhang et al. (2001) developed 21 RFLP markers to identify CS-A. sharonensis 3S1, 4S1, 5S1, 6S1, and 7S1 addition lines. Antonyuk et al. (2009) used two microsatellite primer pairs to screen wheat-A. sharonensis introgression lines. Zhao et al. (2014) used two pairs of primers from 150 SSR markers to identify the Ssh genome of A. sharonensis among F1 hybrids. Wei Long (2016) developed two molecular markers specific to the x- and y-type HMW-GSs genes of A. sharonensis, which were validated in accurately tracing and distinguishing A. sharonensis Glu-1Ssh of backcross progenies from Glu-1A, Glu-1B, and Glu-1D of wheat. Li et al. (2020) developed four molecular markers specific to the 1Ssh chromosome of A. sharonensis from 35 primer pairs.
In this study, we found that 51 PLUG markers from a total of 526 primer pairs could identify the homoeologous groups of each of the A. sharonensis chromosomes. Among these primer pairs, four, eight, nine, six, two, five, and seventeen belonged to chromosome homoeologous groups 1–7, respectively (Tables 1, 2). The percentage of each homoeologous group primers generated ranged from 2.6 to 15.6%, with an average percentage of 9.7% (Table 1).
Powdery Mildew Resistance in A. sharonensis
Wild relatives of wheat are an important gene reservoir for resistance to wheat diseases, and have been exploited extensively around the world for wheat improvement (Olivera and Steffenson, 2009). A. sharonensis, as well as other wild grasses, has co-evolved in association with many cereal pathogens, such as leaf rust, stem rust, stripe rust, and powdery mildew (Wahl et al., 1984). Among them, the highest frequency and level of resistance reported in A. sharonensis was to wheat powdery mildew (Gill et al., 1985; Valkoun et al., 1985; Dhaliwal et al., 1993; Olivera et al., 2007). Zhirov and Ternovskaya (1993) first studied a powdery mildew resistance gene in a wheat-A. sharonensis introgression line. Olivera et al. (2008) identified A. sharonensis accessions carrying major resistance genes to powdery mildew, and found different genes from accessions native to the southern and northern coastal plains of Israel.
To date, more than 70 powdery mildew resistance genes have been permanently designated (Li G. Q. et al., 2019). Among them, 19 have originated from wheat’s related species (Liu et al., 2019), such as Pm7, Pm8, Pm17, Pm20, and Pm56 from Secale cereale, Pm12, Pm32, and Pm53 from A. speltoides, Pm13 and Pm66 from A. longissima, Pm21 and Pm55 from Dasypyrum villosum, Pm19, Pm34, Pm35, and Pm58 from A. tauschii, Pm29 from A. ovata, Pm40 and Pm43 from Thinopyrum intermedium, Pm51 from T. ponticum and Pm57 from A. searsii. Among the above 19 genes mentioned, none were derived from A. sharonensis. In our present study, the CS-A. sharonensis 4Ssh (4D) substitution + 5SshL monotelosomic addition (JIC-34) and the CS-A. sharonensis 4Ssh disomic addition line (JIC-33) were highly susceptible to powdery mildew, indicating that there were no powdery mildew resistance genes on chromosomes 4Ssh and 5SshL of A. sharonensis. However, the CS-A. sharonensis 4Ssh (4D) substitution + 7Ssh disomic addition (JIC-37) was nearly immune to powdery mildew (Table 3), suggesting that the chromosome 7Ssh of A. sharonensis might carry new powdery mildew resistant gene(s).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
CL conceived and designed the experiments. XW, ZHY, HW, JBL, and RH performed the experiments. WX, GL, and JG performed disease resistance testing. YZ, FL, DC, and AL analyzed the data. XW wrote the manuscript. HL, ZJY, JJL, and CL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by Taishan Scholars Project (tsqn201812123), Natural Science Foundation of China (31971874 and 32001544), funds supported by Department of Science and Technology of Shandong Province (2019LZGC016 and ZR2020MC098), and the Modern Agricultural Industry Technology System (CARS-03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to express our appreciations to I. Dundas, The University of Adelaide, Australia, for reviewing and editing of the manuscript. We also thank Z. J. Yang, S. M. Reader, and J. Raupp for providing the materials to us.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.708551/full#supplementary-material
Supplementary Figure 1 | FISH using Oligo-nucleotides as probes on the CS- Aegilops sharonensis 4Ssh (4D) substitution + 5SshL monotelosomic addition (A,B), 6Ssh disomic addition (C,D), 4Ssh (4D) substitution + 6Ssh disomic addition (E,F), and 4Ssh (4D) substitution + 7Ssh disomic addition line (G,H). Panels (A,C,E,G) are double-color FISH patterns using Oligo-pTa535-1 (red) and Oligo-pSc119.2-1 (green) as probes; (B,D,F,H) are FISH patterns using (GAA)8 as probes. Bar indicates 10 μm.
References
Anikster, Y., Manisterski, J., Long, D. L., and Leonard, K. J. (2005). Resistance to leaf rust, stripe rust, and stem rust in Aegilops spp. In Israel. Plant Dis. 89, 303–308. doi: 10.1094/pd-89-0303
Antonyuk, M. Z., Bodylyova, M. V., and Ternovskaya, T. K. (2009). Genome structure of introgressive lines Triticum aestivum/Aegilops sharonensis. Cytol. Genet. 43, 58–67.
Danilova, T. V., Friebe, B., and Gill, B. S. (2012). Single-copy gene fluorescence in situ hybridization and genome analysis: Acc-2 loci mark evolutionary chromosomal rearrangements in wheat. Chromosoma 121, 597–611. doi: 10.1007/s00412-012-0384-7
Dhaliwal, H. S., Singh, H., Gill, K. S., and Randhawa, H. S. (1993). “Evaluation and cataloguing of wheat germplasm for disease resistanve and quality,” in Biodiversity and Wheat Improvement, ed. A. B. Damania (Chichester, UK: John Wiley and Sons), 123–140.
Endo, T. R. (1982). Gametocidal chromosome of three Aegilops species in common wheat. Canadian J. Genet. Cytol. 24, 201–206. doi: 10.1139/g82-020
Friebe, B., Zhang, P., Gill, B. S., and Nasuda, S. (2003). Characterization of a knock-out mutation at the Gc2 locus in wheat. Chromosoma 111, 509–517. doi: 10.1007/s00412-003-0234-8
Gerlach, W. L., and Bedbrook, J. R. (1979). Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleotide Acids Res. 7, 1869–1885. doi: 10.1093/nar/7.7.1869
Gill, B. S., Sharma, H. C., Raupp, W. J., Browder, L. E., Hatchett, J. H., Harvey, T. L., et al. (1985). Evaluation of Aegilops species for resistance to wheat powdery mildew, wheat leaf rust, hessian fly and greenbug. Plant Dis. 69, 314–316.
Gong, W. P., Han, R., Li, H. S., Song, J. M., Yan, H. F., Li, G. Y., et al. (2017). Agronomic traits and molecular marker identification of wheat–Aegilops caudata addition lines. Front. Plant Sci. 8:1743. doi: 10.3389/fpls.2017.01743
Gorny, A. G., and Garczynski, S. (2008). Nitrogen and phosphorus efficiency in wild and cultivated species of wheat. J. Plant Nutr. 31, 263–279. doi: 10.1080/01904160701853878
Han, R., Li, T. Y., Gong, W. P., Li, H. S., Song, J. M., Liu, A. F., et al. (2018). New resistance sources of wheat stem rust and molecular markers specific for relative chromosomes that the resistance genes are located on. Scientia Agric. Sinica 51, 1223–1232.
Ishikawa, G., Nakamura, T., Ashida, T., Saito, M., Nasuda, S., Endo, T. R., et al. (2009). Localization of anchor loci representing five hundred annotated rice genes to wheat chromosomes using PLUG markers. Theoretical Appl. Genet. 118, 499–514. doi: 10.1007/s00122-008-0916-y
Ishikawa, G., Yonemaru, J., Saito, M., and Nakamura, T. (2007). PCR-based landmark unique gene (PLUG) markers effectively assign homoeologous wheat genes to A, B and D genomes. BMC Genomics 8:135. doi: 10.1186/1471-2164-8-135
Jiang, Q. T., Zhao, Q. Z., Yang, Q., Ma, J., Zhang, X. W., Wang, C. S., et al. (2014). Amphidiploids between tetraploid wheat and Aegilops sharonensis Eig exhibit variations in high-molecular weight glutenin subunits. Genet. Resour. Crop Evol. 61, 299–305. doi: 10.1007/s10722-013-0072-3
King, I. P., Koebner, R. M. D., Reader, S. M., and Miller, T. E. (1991). Induction of a mutation in the male fertility gene of the preferentially transmitted Aegilops sharonensis chromosome 4S1 and its application for hybrid wheat production. Euphytica 54, 33–39. doi: 10.1007/bf00145628
Li, G. Q., Cowger, C., Wang, X. W., Carver, B. F., and Xu, X. Y. (2019). Characterization of Pm65, a new powdery mildew resistance gene on chromosome 2AL of a facultative wheat cultivar. Theoretical Appl. Genet. 132, 2625–2632. doi: 10.1007/s00122-019-03377-2
Li, X. Y., Wang, Q., Li, S. Y., Ma, J., Wang, J. R., Qi, P. F., et al. (2019). Stable expression and heredity of alien Glu-1Ssh in wheat-Aegilops sharonensis hybrid progenies. Genet. Resour. Crop Evol. 66, 619–632. doi: 10.1007/s10722-018-00736-8
Li, L. H., Li, X. Q., and Yang, X. M. (2006). Description Specification and Data Standard of Wheat Germplasm Resources. Beijing: China Agriculture Press.
Li, X. Y., Li, Y., Karim, H., Li, Y., Zhong, X. J., Tang, H. P., et al. (2020). The production of wheat-Aegilops sharonensis 1Ssh chromosome substitution lines harboring alien novel high-molecular-weight glutenin subunits. Genome 63, 155–167. doi: 10.1139/gen-2019-0106
Liu, C., Gong, W. P., Han, R., Guo, J., Li, G. R., Li, H. S., et al. (2019). Characterization, identification and evaluation of a set of wheat-Aegilops comosa chromosome lines. Sci. Rep. 9, 4773–4784.
Liu, C., Li, G. R., Yang, Z. J., Feng, J., Zhou, J. P., and Ren, Z. L. (2006). Specific DNA band isolation and SCAR marker construction of rye genome. Acta Botanica Boreali-Occidentalia Sinica 26, 2434–2438.
Liu, C., Qi, L. L., Liu, W. X., Zhao, W. C., Wilson, J., Friebe, B., et al. (2011). Development of a set of compensating Triticum aestivum-Dasypyrum villosum Robertsonian translocation lines. Genome 54, 836–844. doi: 10.1139/g11-051
Liu, C., Yan, H. F., Gong, W. P., Li, G. R., Liu, D. Q., and Yang, Z. J. (2013). Screening of new resistance sources of wheat leaf rust. J. Plant Genet. Resour. 14, 935–943.
Manyowa, N. M. (1989). The Genetics of Aluminum, Excess Boron, Copper and Manganese Stress Tolerance in the Tribe Triticeae and its Implications for Wheat Improvement. Ph.D. thesis. Cambridge: Cambridge University.
Miller, T. E., Hutchinson, J., and Chapman, V. (1982). Investigation of a preferentially transmitted Aegilops sharonensis chromosome in wheat. Theoretical Appl. Genet. 61, 27–33. doi: 10.1007/bf00261506
Millet, E. (2007). Exploitation of Aegilops species of section Sitopsis for wheat improvement. Israel J. Plant Sci. 55, 277–287. doi: 10.1560/ijps.55.3-4.277
Olivera, P. D., Anikster, Y., Kolmer, J. A., and Steffenson, B. J. (2007). Resistance of Sharon goatgrass (Aegilops sharonensis) to fungal diseases of wheat. Plant Dis. 91, 942–950. doi: 10.1094/pdis-91-8-0942
Olivera, P. D., Millet, E., Anikster, Y., and Steffenson, B. J. (2008). Genetics of resistance to wheat leaf rust, stem rust, and powdery mildew in Aegilops sharonensis. Phytopathology 98, 353–358. doi: 10.1094/phyto-98-3-0353
Olivera, P. D., and Steffenson, B. J. (2009). Aegilops sharonensis: origin, genetics, diversity, and potential for wheat improvement. Botany 87, 740–756. doi: 10.1139/b09-040
Slageren, M. W. V. (1994). Wild Wheats: A Monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae). Wageningen: Agricultural University Wageningen, the Netherlands.
Snyman, J. E., Pretorius, Z. A., Kloppers, F. J., and Marais, G. F. (2004). Detection of adult-plant resistance to Puccinia triticina in a collection of wild Triticum species. Genet. Resour. Crop Evol. 51, 591–597. doi: 10.1023/b:gres.0000024652.49194.ac
Tang, Z., Yang, Z. J., and Fu, S. L. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Valkoun, J., Hammer, K., Kucerova, D., and Bartos, P. (1985). Disease resistance in the genus Aegilops L. -stem rust, leaf rust, stripe rust, and powdery mildew. Kulturp Flanze 33, 1133–1153.
Wahl, I., Anikster, Y., Manisterski, J., and Segal, A. (1984). “Evolution at the center of origin,” in The Cereal Rusts. Vol. I, Origins, Specificity, Structure, and Physiology, eds A. P. Roelfs and W. R. Bushnell (Orlando, FL: Academic Press), 33–77.
Waines, J. G., Rafi, M. M., and Ehdaie, B. (1993). “Yield components and transpiration efficiency in wild wheats,” in Biodiversity and Wheat Improvement, ed. A. B. Damania (Chichester, UK: John Wiley and Sons), 173–186.
Wang, Z., Cui, Y., Chen, Y., Zhang, D., Liang, Y., Zhang, D., et al. (2014). Comparative genetic mapping and genomic region collinearity analysis of the powdery mildew resistance gene Pm41. Theoretical Appl. Genet. 127, 1741–1751. doi: 10.1007/s00122-014-2336-5
Wei Long (2016). Development of the Wheat Lines Harboring the Alien HMW-GSs from Aegilops Sharonensis. Master Dissertation. Chengdu: Sichuan Agricultural University.
Xu, X., Li, Z. S., Chen, S. Y., and Fu, J. (1992). Studies on the nullisomic backcrossing procedures for producing alien substitution lines of Triticum-Aegilops. Acta Botanica Boreali-Occidentalia Sinica 12, 9–16.
Xu, X., Monneveux, P., Damania, A. B., and Zaharieva, M. (1993). Evaluation for salt tolerance in genetic resources of Triticum and Aegilops species. Plant Genet. Resour. Newsletter 96, 11–16.
Yu, G., Champouret, N., Steuernagel, B., Olivera, P. D., Simmons, J., Williams, C., et al. (2017). Discovery and characterization of two new stem rust resistance genes in Aegilops sharonensis. Theoretical Appl. Genet. 130, 1207–1222. doi: 10.1007/s00122-017-2882-8
Zhang, H., Reader, S. M., Liu, X., Jia, J. Z., Gale, M. D., and Devos, K. M. (2001). Comparative genetic analysis of the Aegilops longissima and Ae. sharonensis genomes with common wheat. Theoretical Appl. Genet. 103, 518–525. doi: 10.1007/s001220100656
Zhao, Q. Z., Jiang, Q. T., Yang, Q., Ma, J., Wang, J. R., Chen, G. Y., et al. (2014). Characterization of intergeneric hybrid between common wheat and Aegilops sharonensis (Eig) and transfer of alien high molecular weight glutenin subunits into wheat. Cereal Res. Commun. 42, 640–647. doi: 10.1556/crc.2014.0001
Keywords: Aegilops sharonensis, chromosome derivatives, cytogenetic identification, PLUG marker, powdery mildew resistance
Citation: Wang X, Yu Z, Wang H, Li J, Han R, Xu W, Li G, Guo J, Zi Y, Li F, Cheng D, Liu A, Li H, Yang Z, Liu J and Liu C (2021) Characterization, Identification and Evaluation of Wheat-Aegilops sharonensis Chromosome Derivatives. Front. Plant Sci. 12:708551. doi: 10.3389/fpls.2021.708551
Received: 12 May 2021; Accepted: 05 July 2021;
Published: 26 July 2021.
Edited by:
István Molnár, Centre for Agricultural Research, Hungarian Academy of Sciences (MTA), HungaryReviewed by:
Ghader Mirzaghaderi, University of Kurdistan, IranMichał Tomasz Kwiatek, Poznań University of Life Sciences, Poland
Copyright © 2021 Wang, Yu, Wang, Li, Han, Xu, Li, Guo, Zi, Li, Cheng, Liu, Li, Yang, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheng Liu, bGNoNjY4ODQwN0AxNjMuY29t
 Xiaolu Wang
Xiaolu Wang Zhihui Yu4
Zhihui Yu4 Cheng Liu
Cheng Liu