- 1Department of Molecular Biology and Genetics, Institute of Plant Physiology and Genetics, Bulgarian Academy of Sciences, Sofia, Bulgaria
- 2Department of Plant Physiology, Faculty of Biology, Sofia University “St. Kliment Ohridski”, Sofia, Bulgaria
- 3Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent, Belgium
- 4VIB-UGent Center for Plant Systems Biology, Ghent, Belgium
The family of NudC proteins has representatives in all eukaryotes and plays essential evolutionarily conserved roles in many aspects of organismal development and stress response, including nuclear migration, cell division, folding and stabilization of other proteins. This study investigates an undescribed Arabidopsis homolog of the Aspergillus nidulans NudC gene, named NMig1 (for Nuclear Migration 1), which shares high sequence similarity to other plant and mammalian NudC-like genes. Expression of NMig1 was highly upregulated in response to several abiotic stress factors, such as heat shock, drought and high salinity. Constitutive overexpression of NMig1 led to enhanced root growth and lateral root development under optimal and stress conditions. Exposure to abiotic stress resulted in relatively weaker inhibition of root length and branching in NMig1-overexpressing plants, compared to the wild-type Col-0. The expression level of antioxidant enzyme-encoding genes and other stress-associated genes was considerably induced in the transgenic plants. The increased expression of the major antioxidant enzymes and greater antioxidant potential correlated well with the lower levels of reactive oxygen species (ROS) and lower lipid peroxidation. In addition, the overexpression of NMig1 was associated with strong upregulation of genes encoding heat shock proteins and abiotic stress-associated genes. Therefore, our data demonstrate that the NudC homolog NMig1 could be considered as a potentially important target gene for further use, including breeding more resilient crops with improved root architecture under abiotic stress.
Introduction
Nuclear distribution C (NudC) genes encode proteins with high structural and functional conservation from yeast to human (Osmani et al., 1990; Fu et al., 2016). NudC has been identified in the filamentous fungus Aspergillus nidulans as an essential gene required for the microtubule-dependent migration of cell nuclei and for normal colony growth (Osmani et al., 1990; Chiu et al., 1997). NudC proteins play roles in diverse cellular processes, including cell division, cell cycle progression, neuronal migration, dynein-dependent functions (Aumais et al., 2003; Zhou et al., 2003; Fu et al., 2016). Homologs of NudC from mammals, Drosophila, Caenorhabditis elegans and Arabidopsis thaliana complement nudC3 mutation in A. nidulans, and result in the normal movement of nuclei and colony growth (Cunniff et al., 1997; Morris et al., 1997; Dawe et al., 2001). These observations suggest that some functions of NudC are evolutionarily and functionally conserved in different eukaryotic organisms.
The levels of NudC mRNA and protein correlate with the proliferative activity in different cell types and tissues. The human NudC homolog is highly expressed in proliferating cells (Gocke et al., 2000) and participates in spindle formation during mitosis (Zhang et al., 2002). Depletion of NudC leads to the occurrence of multiple spindles during metaphase and induces lagging chromosomes during anaphase (Zhou et al., 2003). Either depletion or overexpression of NudC components induces cytokinesis defects in mammalian cells (Aumais et al., 2003; Zhou et al., 2003). NudC is also intimately involved in the regulation of actin dynamics by stabilizing cofilin 1, a key regulator of actin polymerization and depolymerization (Zhang et al., 2016).
The NudC homologs possess a core CS domain (a domain shared by CHORD-containing proteins and SGT1) that occurs preferably in proteins with chaperone or co-chaperone activities (Lee et al., 2004; Zheng et al., 2011). The CS domain in the NudC family has a molecular architecture similar to small heat shock chaperones, such as p23 and HSP20/alpha-crystallin proteins (Garcia-Ranea et al., 2002). One of the most important functions of p23 and HSP20 is to simultaneously interact with HSP90 and specific client proteins (Lee et al., 2004; Botër et al., 2007). The CS domain is considered as a binding module for HSP90, implying that CS domain-containing proteins are involved in recruiting heat shock proteins (HSPs) to multiprotein complexes (Lee et al., 2004). Experimental evidences support both the HSP90 co-chaperone and intrinsic chaperone functions of the NudC family. For instance, the microtubule-associated C. elegans NudC homolog, NUD-1, has shown chaperone activity in vitro, inhibiting heat-induced aggregation and precipitation of citrate synthase and luciferase (Faircloth et al., 2009). Human NudC stabilizes Lis1 (lissencephaly protein 1) through HSP90-mediated pathways and exhibits intrinsic chaperone activity in vitro preventing aggregation of citrate synthase (Yang et al., 2010; Zhu et al., 2010). There have been only few studies aimed at exploring the functions of NudC proteins in plants (Jurkuta et al., 2009; Perez et al., 2009; Ishibashi et al., 2012; Silverblatt-Buser et al., 2018; Liu et al., 2019). In A. thaliana, a gene named BOBBER1 (BOB1), encodes a small 34.5 kDa protein that interacts genetically with two 26S proteasome subunits and maintains a functional proteostasis network enabling proper plant development (Silverblatt-Buser et al., 2018). A partial loss-of-function mutation in BOB1 results in numerous developmental defects, including obstructed root growth (Perez et al., 2009). BOB1 displays features typical for canonical small heat shock proteins (sHSPs), such as in vitro chaperone activity, heat stress induction and the availability of an α-crystallin-like NudC domain (Haslbeck et al., 2005; Perez et al., 2009). At high temperatures, BOB1 colocalizes with HSP17.6 and incorporates into heat shock granules that are formed mainly in the cytoplasm of heat-exposed plant cells (Kirschner et al., 2000; Miroshnichenko et al., 2005). It has been demonstrated that heat exposure of a partial loss-of-function mutant bob1-3 during germination and at seedling stage leads to reduced thermotolerance, compared to wild-type seeds. This defect can be complemented by a functional BOB1 transgene, confirming that the phenotype is a result of the reduced BOB1 function (Perez et al., 2009).
In search of regulators of root development, we identified a previously undescribed Arabidopsis homolog of the Aspergillus NudC gene, named NMig1 (for Nuclear Migration 1), which shares structural similarity to other plant and mammalian NudC-like members. Since NMig1 is a BOB1 homolog and given the presence of the CS domain, we hypothesized the putative role of NMig1 in root development and modulation of plant defense systems under stress. Accordingly, we generated Arabidopsis plants overexpressing NMig1 and compared their root phenotypic features and responses to abiotic stress conditions (heat shock, drought, and high salinity) with those in the wild-type plants. Evaluation of the antioxidant status, accumulation of reactive oxygen species (ROS) and lipid peroxidation, as well as analysis of the expression levels of stress-related genes were undertaken. The results obtained strongly suggest a potential regulatory function of NMig1 in root development and tolerance to abiotic stress.
Materials and Methods
Bioinformatics Analysis
The NMig1 (At5g58740) sequence was used as a query to search for NMig1 homologs from the NCBI database through BLAST analysis. The amino acid sequence of NMig1 and some of its homologs were aligned using Clustal Omega (Pundir et al., 2016), and the alignment image was generated using BoxShade version 3.21 (K. Hofmann, Koeln, Germany and M. Baron, Surrey, United Kingdom). The accession numbers are as follows: Q8VXX3, NP_200682.1 (Arabidopsis thaliana); Q9LV09, NP_200152.1 (A. thaliana); Q9STN7, NP_194518.1 (A. thaliana); NP_001357629 (Zea mays); XP_002467510.1 (Sorghum bicolor); XP_015617572.1 (Oryza sativa); XP_003579589.1 (Brachypodium distachyon); NP_001236519.1 (Glycine max); XP_002525640.1 (Ricinus communis); XP_002274773.1 (Vitis vinifera), and CAL37614 (Homo sapiens). The conserved motifs and domain structure of NMig1 protein were analyzed using InterPro scan program1 (Supplementary Figure S1).
Generation of Constructs and Transformation of Arabidopsis thaliana
The recombinant plasmids were obtained through the GATEWAYTM recombination cloning technology (Invitrogen Life Technologies). The Entry Clones were generated by inserting the coding sequence of NMig1 into the pDONR221 donor vector, then the Entry Clones were recombined into the destination vectors pK7WG2 or pK7FWG2 [with C-terminal translational green fluorescent protein (GFP) fusion] under the control of CaMV 35S promoter and neomycin phosphotransferase II (nptII) gene for plant selection (Karimi et al., 2007). The generated 35S:NMig1 and 35S:NMig1-GFP constructs were introduced into Agrobacterium tumefaciens strain C58C1 (pMP90), harboring the helper plasmid pSoup. The positive clones were selected on agar solidified YEB nutrient medium supplemented with 100 mg l−1 rifampicin, 100 mg l−1 spectinomycin and 50 mg l−1 gentamicin, and subsequently transformed into wild-type Col-0 plants by the floral-dip method (Clough and Bent, 1998). The primary transformants (T0) were selected on 50 mg l−1 kanamycin and their progeny genotyped with gene specific and nptII gene primers. The positive transformants were further tested by kanamycin selection and five representative transgenic lines (L1 – L5) were generated. All analyses were carried out with homozygous T3 seedlings.
Plant Material and Growth Conditions
Arabidopsis thaliana seeds were surface sterilized, stratified for 2 days in the dark at 4°C, and grown on half-strength Murashige and Skoog (1/2MS) (Murashige and Skoog, 1962) medium (pH 5.7) solidified with 1.0% agar at 22°C. To evaluate the root phenotypes of wild-type and transgenic (T3 generation) plants, approximately 30–35 seeds from each independent transgenic line and controls were plated at least in triplicate and cultivated in a growth chamber under a 16-h light/8-h dark cycle and a light intensity of ∼150 μmol m–2 s–1. For all phenotypic analyses, plants were vertically grown on square plates (Greiner Labortechnik) for 10 days unless otherwise stated. For comparison of the root phenotype of transgenic and wild-type plants, NMig1-overexpressing lines were co-cultivated with the wild-type plants on the same agar plates divided into two equal parts. Each plate contained seedlings of one individual transgenic line and Col-0. To analyze responses to abiotic stress, 5-day-old seedlings were transferred to plates containing solid 1/2MS medium supplemented with or without 150 mM sorbitol or 150 mM NaCl and scanned every day. Heat stress was applied by exposure of 8-day-old seedlings to 42°C for 1 h and subsequently transferred into a growth chamber for 2 additional days of growth under optimal conditions. Untreated wild-type and transgenic plants were used as controls and these were grown at the same time as the individuals subjected to the stress treatments.
Morphological Characterization of Roots
Plates were scanned every day with an Epson Perfection V850 Pro scanner. Root length was quantified using the image processing software ImageJ 1.48 (National Institutes of Health, Bethesda, MD, United States), and the visible lateral roots were counted and calculated per seedling. Non-emerged lateral root primordia were quantified on the roots of 10-day-old seedlings after root clearing (Malamy and Benfey, 1997). The stress-induced inhibition of primary root growth, formation of lateral root primordia and emerging lateral roots was calculated in percentages for each genotype as a difference between growth and branching under control and stress conditions. The density of lateral root formation events was quantified by dividing the total number of emerged lateral roots plus lateral root primordia by the primary root length, and expressed as lateral root formation events per cm of primary root. In each experiment, the root system of at least 30 plants was examined under an Olympus BX51 upright microscope with a differential interference contrast (DIC) and XC50 digital microscope camera.
Confocal Microscopy and Image Analysis
Fluorescent images were acquired on an inverted laser scanning confocal microscope Zeiss LSM 710 using the ZEN software package (Carl Zeiss, Germany) after excitation by a 488 nm argon laser and detected using the bandpass 505–530 nm emission filter setting. Autofluorescence of photosynthetic pigments was observed after excitation with a 543 nm helium-neon (He-Ne) laser and detected using the bandpass 560–615 nm emission filter. The root apical meristem was imaged on an Andor Dragonfly spinning disk confocal system equipped with the Fusion software (Andor Technologies, Inc.) and iXon897 EMCCD camera at 488 nm laser excitation. Confocal images were processed with ImageJ1.52r or the CellTool software2 (Danovski et al., 2018).
Determination of Free Radical Scavenging Activity and Antioxidant Status
Fresh plant material (200 mg) was frozen in liquid nitrogen, grinded to fine powder in TissueLyser LT (QIAGEN) and mixed with 500 μl ice-cold 80% ethanol. The samples were centrifuged at 10 000 rpm for 30 min at 4°C and the supernatants were used for determination of antiradical activity by 2,2′-diphenyl-1-picrylhydrazyl (DPPH) Radical Scavenging assay, according to Brand-Williams et al. (1995), and antioxidant capacity by Ferric Reducing Antioxidant Power (FRAP) assay, as decribed by Benzie and Strain (1999).
The antiradical activity of the plant extract (40 μl) was measured by mixing it with DPPH solution (960 μl) in methanol (6.10–5M). The absorbance of the reaction solution at 515 nm was determined after 30 min of incubation at room temperature in the dark. The rate of antiradical activity was calculated by Trolox standard curve and the results were represented as μmol Trolox g–1 FW.
The antioxidant capacity assay is based on the reduction of ferric tripyridyl-s-triazine (Fe3+-TPTZ) complex to the blue colored ferrous (Fe2+) form by the antioxidants present in the plant extract. Briefly, 50 μl of the supernatant was mixed with 1.5 ml of freshly prepared FRAP reagent (0.3 M acetate buffer, 0.01 M TPTZ in 0.04 M HCl and 0.02 M FeCl3.6H2O in 10:1:1 proportion) and 150 μl distilled water. The optical density was spectrophotometrically determined after 15 min at 593 nm. Antioxidant capacity was expressed as μmol FRAP g–1 FW.
Measurement of Lipid Peroxidation and Hydrogen Peroxide Content
Fresh plant material (200 mg) was frozen in liquid nitrogen, grinded to fine powder in TissueLyser LT (QIAGEN) and mixed with 0.1% trichloroacetic acid (w/v) (TCA). The samples were centrifuged at 10 000 rpm for 30 min at 4°C and the supernatant was used for determination of lipid peroxidation and H2O2 content.
Lipid peroxidation was determined by the thiobarbituric (TBA) method, which quantifies the amount of malondialdehyde (MDA) as the end product of the lipid peroxidation (Hodges et al., 1999). Reaction mixture contained 500 μl supernatant, 500 μl 0.1 M phosphate buffer (pH 7.3) ant 1 ml TBA (0.5% w/v). The samples were placed in a boiling water bath for 45 min. After cooling on ice, the sample absorbance was read at 440, 532, and 600 nm. The MDA extinction coefficient (ε = 155 mM–1 cm–1) was used for calculations of MDA concentration and expressed as nmol g–1 FW.
Hydrogen peroxide (H2O2) content was determined according to Wolff (1994). One ml of 0.1% TCA extract was mixed with 1.9 ml reagent (composed by 100 μM xylenol orange, 250 μM ammonium ferrous sulphate, and 100 mM sorbitol in 25 mM H2SO4). The absorbance of the reactions was evaluated spectrophotometrically at 560 nm after 30 min incubation at room temperature and H2O2 extinction coefficient used for the calculations was ε = 2.24 × 105 M–1 cm–1.
In situ ROS Assays
In situ detection of reactive oxygen species (ROS) accumulation was performed by nitroblue tetrazolium (NBT) and diaminobenzidine (DAB) staining methods, according to Kumar et al. (2014) with slight modifications, as described in Nguyen et al. (2017). For tissue-specific detection of the superoxide anion (O2–) abundance, 6-day-old A. thaliana plants grown on 1/2MS, were stained in freshly prepared 0.05% NBT (w/v) in 50 mM sodium phosphate buffer (pH 7.4) for 30 min, covered with aluminum foil and incubated on a lab shaker (80 rpm) at room temperature. To detect H2O2 content, the seedlings were stained for 1 h in 1 mg ml−1 DAB dissolved in 10 mM Na2HPO4 (freshly prepared). After the incubation in NBT and DAB solutions, the seedlings were transferred into bleaching solution (ethanol: acetic acid: glycerol in ratio 3:1:1) and kept at 4°C overnight. The analyzes of the samples were performed on the following day on an Olympus BX51 upright microscope coupled to an XC50 digital microscope camera.
RNA Extraction and Gene Expression Analysis
Total RNA was extracted using the Trizol reagent (Invitrogen), followed by clean-up with RNeasy Mini Kit (Qiagen), including DNase I (Qiagen) treatment, according to the manufacturer’s protocols. The quantity and quality of RNA samples were evaluated with Thermo Fisher Scientific NanoDrop 1000 Spectrophotometer. Complementary DNA (cDNA) was synthesized from 1 mg of total RNA with the iScript cDNA Synthesis Kit (Bio-Rad), according to the manufacturer’s instructions. The quantitative PCR analysis was carried out in a BioRad CFX96 real time C1000 thermal cycler with SsoFast EvaGreen supermix (BioRad) or in a LightCycler 480 system (Roche Diagnostics) with the SYBR Green I Master kit (Roche Diagnostics), according to the manufacturer’s instructions. Conventional PCR analysis was performed with the same cDNA templates in a PCR Eppendorf Mastercycler (Eppendorf, Germany). Primer pairs were designed with Beacon Designer 4.0 (Premier Biosoft International) or Primer 3.0 (Untergasser et al., 2012). Efficiency curves were generated for each primer set and amplification efficiency between 98 and 102% was considered acceptable for a quantitative gene expression analysis (Raymaekers et al., 2009). Expression levels were normalized to those of EF1α and CDKA1;1. The average Cycle threshold (Ct) values of the reference genes EF1α and CDKA1;1 were calculated using three untreated biological replicates. Delta Ct (ΔCt) values were obtained by subtracting from the average reference Ct values the individual Ct values of each treated and untreated sample within a specific gene dataset. The relative quantity (RQ = 2^ΔCt) and the normalization factor [NF = RQ (EF1α + CDKA1;1)] were used to calculate the normalized expression of the genes of interest. In semi-quantitative analyses, EF1α was used as a reference gene. Each individual sample was analyzed in triplicate. The list of specific primer sets for the monitored genes is provided in Supplementary Table S1.
Statistical Analysis
All values reported in this study are the means of at least three independent experiments with three replicates, unless otherwise stated. Mean data from all plants on a single plate were used to generate a single replicate. The significance of the results and statistical differences were analyzed using Statgraphics Plus 5.1 software (Statistical Graphics, Warrenton, VA, United States). The data were evaluated by multifactor analysis of variance and expressed as mean ± standard error (SE). A p-value equal to or lower than 0.05 was considered statistically significant.
Accession Numbers
The Arabidopsis Information Resource (TAIR) locus identifiers for the genes mentioned in this study are At5g58740 for NMig1, At5g53400 for BOB1, At4g27890 for BOB2, At3g53990 for USP17, At4g27320 for USP21, At1g08830 for SOD1, At1g20630 for CAT1, At4g35090 for CAT2, At1g07890 for APX1, At2g33210 for HSP60, At4g24190 for HSP90, At1g74310 for HSP101, At5g52310 for RD29A, At2g01980 for SOS1, At1g07940 for EF1a, and At3g48750 for CDKA1;1.
Results
NMig1 Is a Small HSP With NudC Domain
Proteins belonging to the NudC family contain a highly conserved NudC domain of approximately 140 amino acids that share significant sequence similarity in different species (Supplementary Figure S1). The A. thaliana NMig1 (locus At5g58740) gene is located in chromosome 5 (Chr5: 23725675-23727628), and consists of five exons and four introns. The deduced protein sequence (accession number AED97093.1) is annotated in the databases as a “HSP20-like chaperones superfamily protein.” The coding sequence of NMig1 is 477 bp in length and encodes a protein of 158 amino acids with a molecular mass of 18.2 kDa (pI 5.77) (Aramemnon database; Schwacke et al., 2003).
According to the Arabidopsis developmental and root maps in the eFP Browser (electronic Fluorescent Pictograph3), NMig1 is expressed in an organ specific manner (Supplementary Figure S2). The developmental map showed that the highest expression level (expression potential) in the leaves, stem, flowers, pollen and seeds was about 510 absolute signal value (a.s.v.) (Supplementary Figure S2A). In the root, the NMig1 expression potential reached about 2766 a.s.v. depending on the growth conditions. At tissue resolution within the root, the highest level of NMig1 expression was found in the lateral root cap and columella cells (Supplementary Figure S2B).
In the A. thaliana genome, besides NMig1, two additional genes encode proteins with NudC domain, BOB1 (At5g53400) and BOB2 (At4g27890) (Supplementary Figure S1; Jurkuta et al., 2009; Perez et al., 2009). BOB1 and BOB2 show weak sequence similarity to NMig1. BOB1 displays 42.1% similarity, 27.3% identity, gaps 48.6% and overlap in 40 amino acids when compared to NMig1. BOB2 has 41.4% similarity, 30.7% identity, gaps 47% and overlap in 42 amino acids [The European Bioinformatics Institute, Pairwise Sequence Alignment, accession numbers: AAO23639.1 (BOB1), AEE85405.1 (BOB2)].
In addition, protein sequence analysis revealed the presence of a CS (CHORD-containing proteins and SGT1) domain (PF04969) that is typical for proteins functioning as independent sHSPs or co-chaperones of other HSPs (Zheng et al., 2011). The CS domain encompasses almost all of NMig1 protein and consists of approximately 100 amino acids. Comparison of NudC family members in different species displayed conservation of CS domain sequences across monocots and dicots, and also human (Supplementary Figure S1).
Expression Patterns and Root System Architecture of NMig1 Overexpressors
To investigate the biological functions of NMig1, we constructed five transgenic lines (L1, L2, L3, L4, and L5) that stably expressed NMig1 under the control of CaMV 35S promoter. In lines L4 and L5, the NMig1 gene was fused upstream of a reporter gene encoding the green fluorescent protein (GFP) (Figure 1). Quantification of NMig1 expression in the generated homozygous lines revealed strong upregulation of NMig1 transcripts, which increased between 35- and 50-fold compared to the wild-type values (Figure 2). Based on the obtained results, the lines with the highest NMig1 expression levels (L2, L3 for 35S:NMig1 and L5 for 35S:NMig1-GFP), were chosen for further phenotypic analyses and assessment of abiotic stress response.
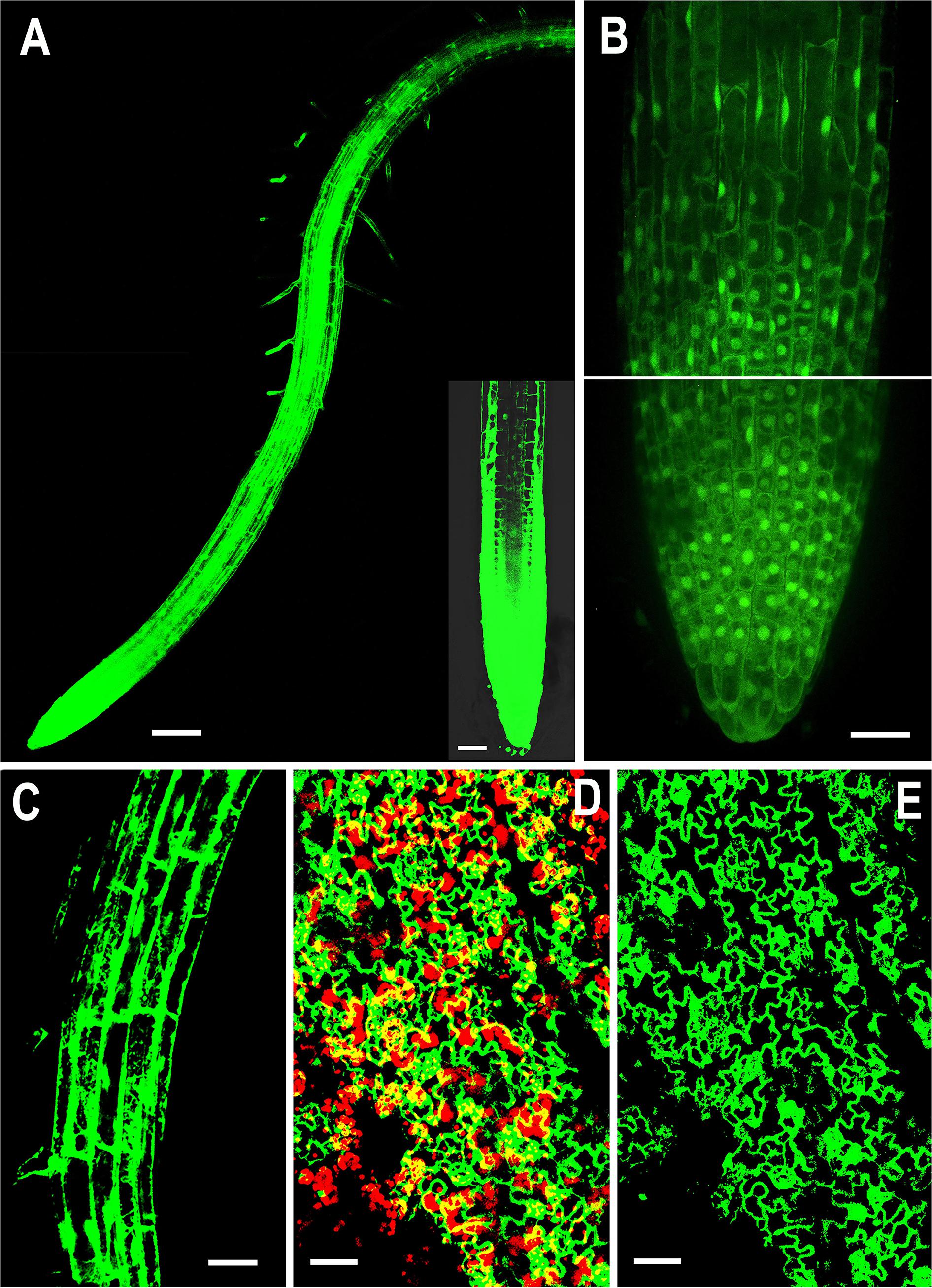
Figure 1. Representative confocal images showing localization of overexpressed NMig1 gene in the root and leaf tissues of 6-day-old Arabidopsis thaliana. Expression pattern of 35S:NMig1-GFP in: (A) primary root, (B) primary root meristem, (C) cells from root elongation zone, (D) and (E) leaf epidermal cells and chlorophyll autofluorescence (red). Scale bars: (A) 100 μm; (B), (D) and (E) 20 μm; (C) 50 μm.
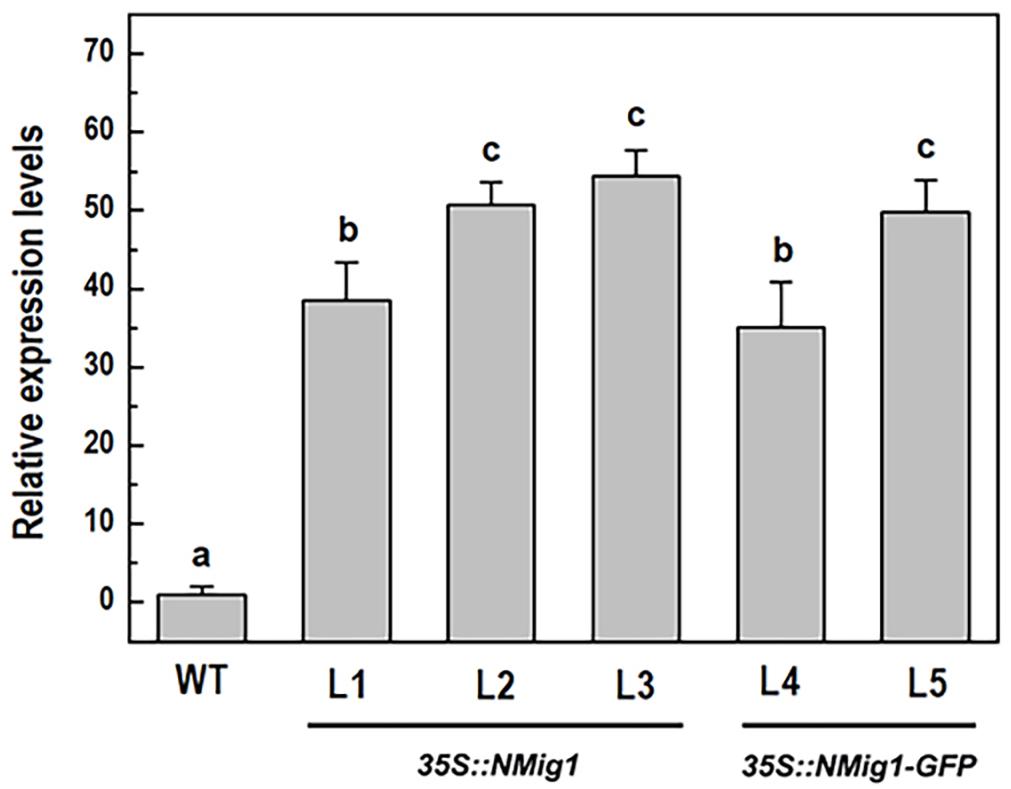
Figure 2. Expression analysis of the NMig1 gene in 10-day-old wild-type seedlings and homozygous transgenic lines overexpressing NMig1 (L1-L5). Expression levels were normalized to those of EF1a and CDKA1;1, as described in Materials and Methods. Data represents the mean values ± SE from at least three biological replicates. Different lowercase letters indicate statistically significant difference (p ≤ 0.05).
Examination of the transgenic lines harboring the 35S:NMig1-GFP construct demonstrated very strong expression in the primary roots, as the most intense signal was seen in the root apical meristem (Figure 1A). At the cellular level, high strength of the GFP signal was detected in the cytoplasm, in the nuclei, excluding the nucleolus (Figure 1B), as well as in the membrane and endomembrane systems (Figure 1C). Confocal imaging also showed intense GFP signal in the leaf epidermis, which was particularly strong in stomatal guard cells (Figures 1D,E). Overlap of NMig1-GFP and red chlorophyll autofluorescence in the leaf chloroplasts could be seen as yellow patches (Figure 1D).
After 10 days of growth, a detailed phenotypic analysis was carried out to compare root growth and branching in the lines with ectopic expression of NMig1 and the wild-type plants (Figures 3A–D). Under favorable growth conditions, all transgenic lines, overexpressing NMig1, displayed significantly greater primary root length (between 18 and 28%), as compared to the wild-type Col-0 (Figure 3C). The number of lateral roots was also enhanced, as the increase ranged between 33 and 50% in the different transgenic lines. To assess whether this increase was caused by enhanced lateral root initiation events, we additionally quantified the number of non-emerged lateral root primordia in the roots of NMig1 overexpressors and the wild-type Col-0 (Figure 3D). The results showed an increased number of lateral root primordia (between 24 and 66%) in transgenic lines, as compared to the controls. Calculation of the density of all events associated with lateral root development that include emerged lateral roots and developing non-emerged primordia estimated as a function of primary root length, revealed 26% greater density of the formative events in L2 and L5, compared to the wild-type Col-0. In L3, despite that the number of all events associated with lateral root development increased about 30%, the calculated density did not show statistically significant difference from the control (Figure 3D).
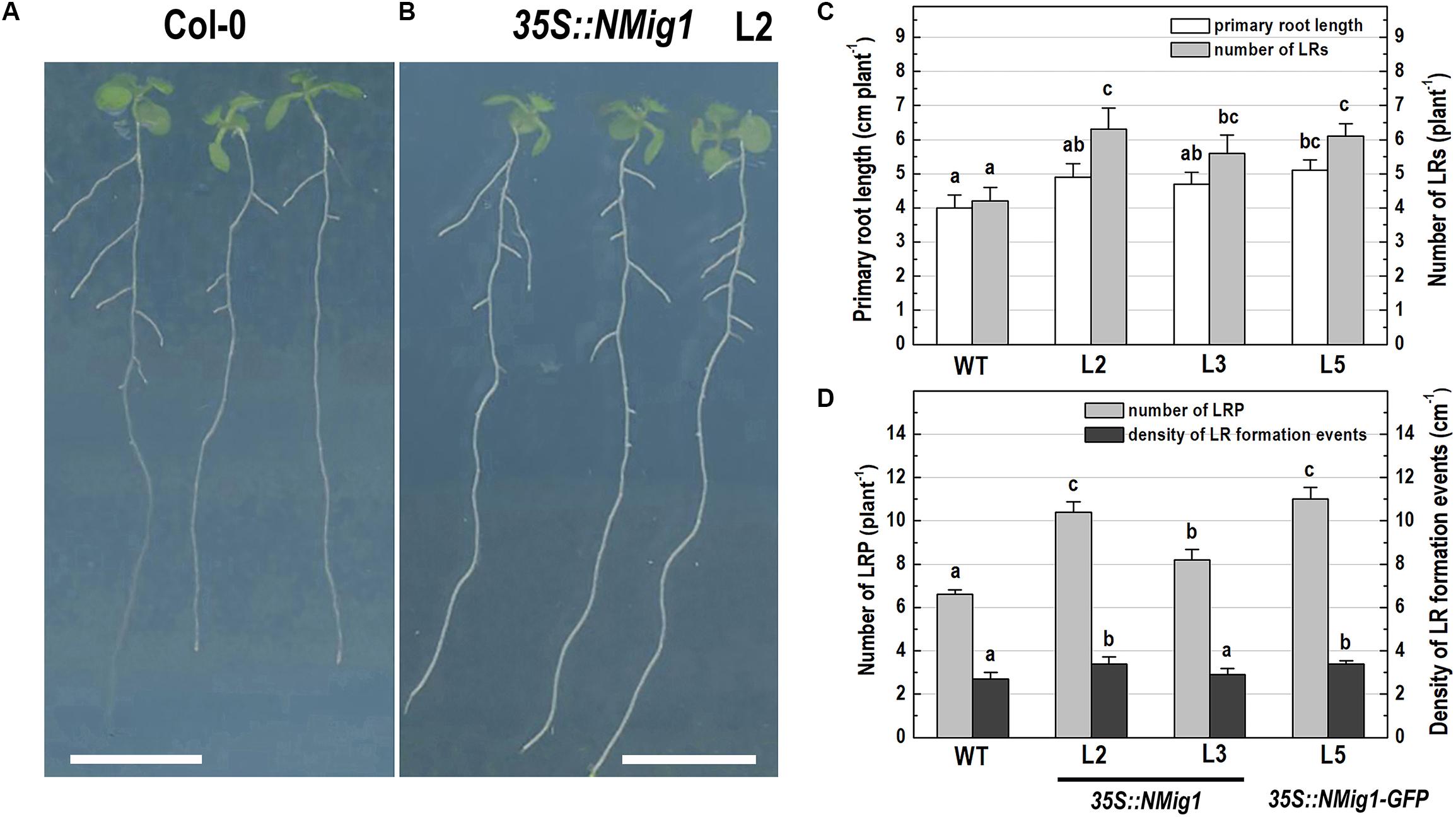
Figure 3. Primary root length and lateral root development of 10-day-old wild-type Col-0 seedlings (A), and transgenic lines overexpressing NMig1 (L2, L3, and L5) (B) under optimal growth conditions: (C) average length of the primary root and number of lateral roots; (D) number of lateral root primordia and density of lateral root formation events. Different lowercase letters indicate statistically significant difference (p ≤ 0.05). Scale bar = 1.0 cm.
Differential Expression of NMig1 Under Exposure to Abiotic Stress
Given the presence of the CS domain, which has structural homology to the HSP90 co-chaperone p23 (Botër et al., 2007), we checked the possibility of involvement of NMig1 in abiotic stress responses. To study this experimentally, we quantified the NMig1 transcript abundance after exposure of the wild-type Col-0 plants to heat shock, drought or high salinity (Figure 4). Expression of NMig1 was significantly upregulated by all the applied stress treatments between 1.5- and 2.5-fold, compared to the control (Figure 4A). The degree of the NMig1 stress induction was compared with that of two genes encoding universal stress proteins (USPs), USP17 and USP21. The USPs are involved in a wide range of stress responses, and in general, their overexpression enhances stress tolerance and alleviates the oxidative damage through the activation of oxidative stress-responsive genes and the removal of intracellular ROS (Chi et al., 2019). Both USP genes studied were upregulated by the applied stress treatments (Figures 4B–D) with increases of 2.6- to 5.6-fold for USP17 and 2.0- to 3.1-fold for USP21, compared to those of untreated controls. Thus, the stress-induced expression changes of NMig1 were of similar magnitude and direction to those found for the studied USPs. The transcript abundance of the reference gene EF1α did not change under different experimental conditions (Figure 4D).
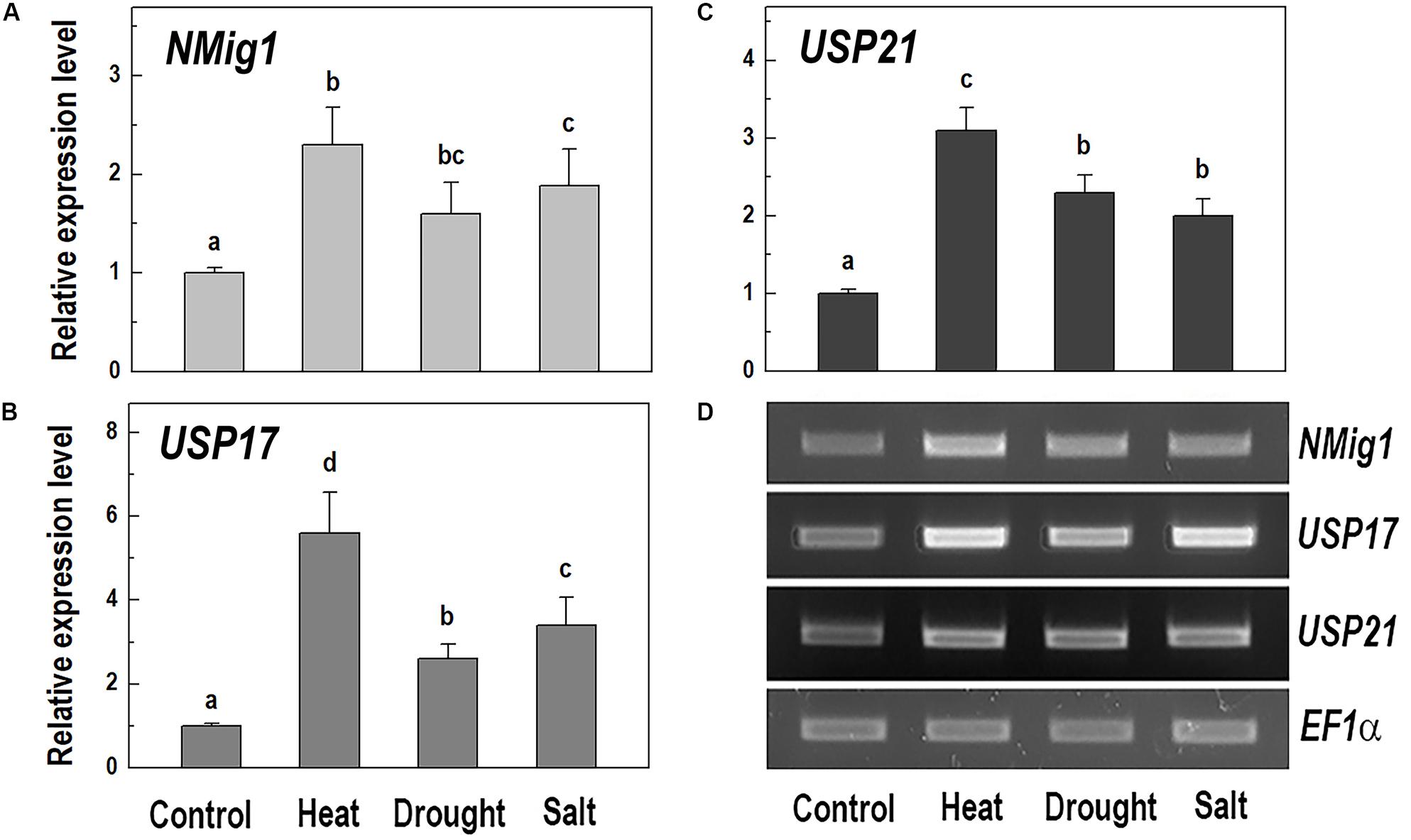
Figure 4. Quantitative RT-PCR (A–C) and semi-quantitative RT-PCR analysis (D) of the expression levels of NMig1 (A), and the universal stress proteins USP17 (B), and USP21 (C) in 10-day-old wild-type seedlings subjected to heat shock, drought and high salinity. Expression levels were normalized to those of EF1a and CDKA1;1, as described in Materials and Methods. For semi-quantitative RT-PCR analysis EF1α was used as a reference gene. Data represents the mean values ± SE from at least three biological replicates. Different lowercase letters indicate statistically significant difference (p ≤ 0.05).
Overexpression of NMig1 Promotes Root Growth and Branching Under Abiotic Stress
The response of transgenic plants to abiotic stress factors was analyzed in NMig1 overexpressors and the wild-type Col-0 grown under the above-mentioned stress conditions, heat stress, drought and high salinity (Figure 5). Evaluation of the root system morphology when compared showed that all NMig1-overexpressing lines were more tolerant toward the tested abiotic factors since the stress-mediated reduction of root growth and branching was significantly lower, compared to the wild-type Col-0 (Figures 5A,B and Supplementary Figures S3A–L), as visualized by presenting the relative inhibition of the primary root growth and branching under each stress treatment within the tested genotypes (Figures 5A,B). The most obvious beneficial effect of the NMig1 overexpression was seen for the number of lateral root primordia (Figure 5C) and density of all branching events under heat stress and salinity (Figure 5D). The heat- and salt-mediated reductions in these parameters were significantly lower in NMig1-overexpressing lines, compared to Col-0. Exposure to heat stress resulted in more than 20% inhibition in the density of all initiation and formative events associated with lateral root development in Col-0, whereas in the overexpressors L2 and L3, this parameter was inhibited by only about 6% (Figure 5D). Similarly, the inhibitory effect of drought and salinity on the density of all lateral root initiation events was at least twice lower in L2 and L3 than that observed in the wild-type.
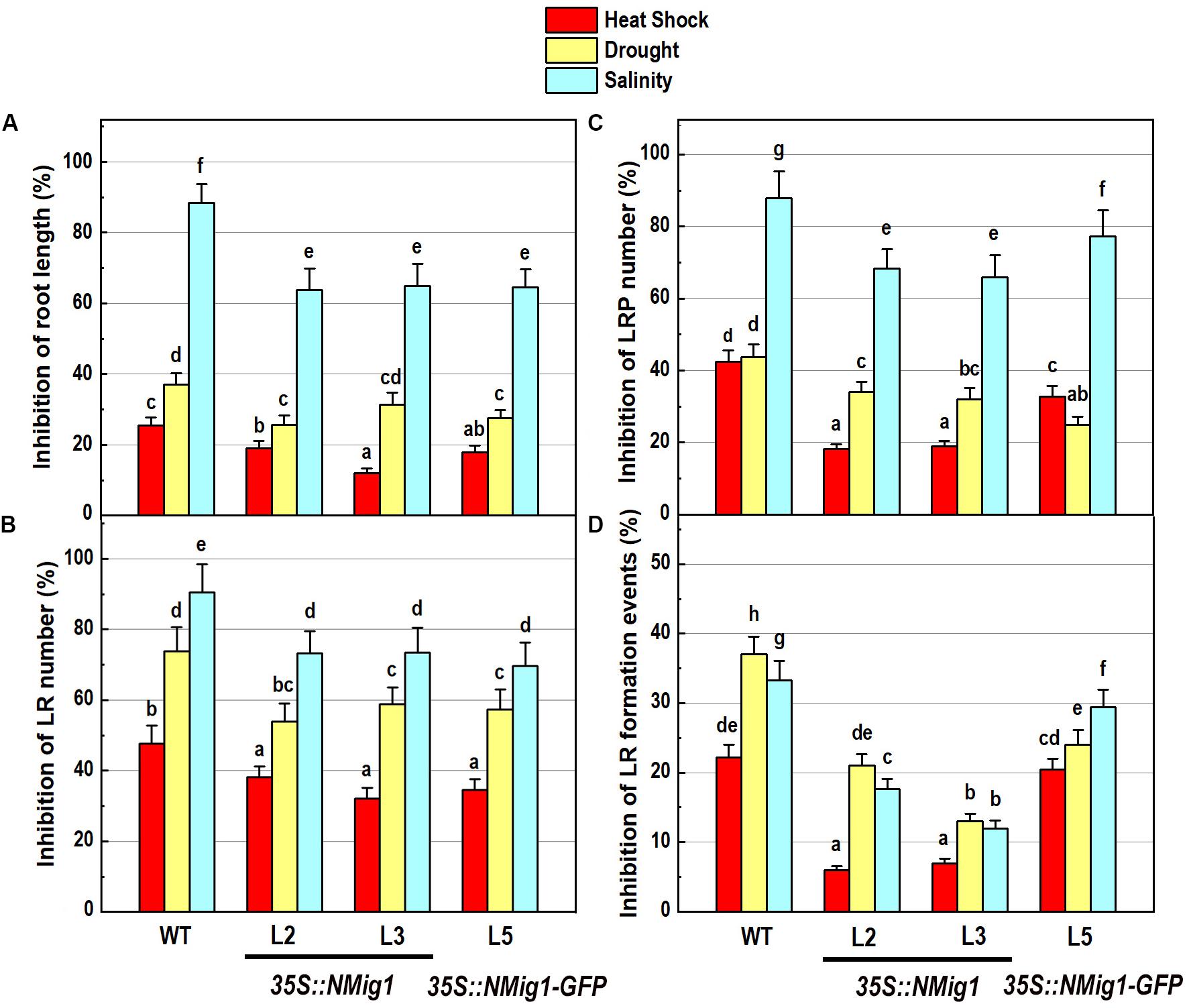
Figure 5. Inhibitory effect of various abiotic stress factors (heat shock, drought and salinity) on primary root length and lateral root development of 10-day-old wild-type seedlings and transgenic lines overexpressing NMig1 (L2, L3, and L5): (A) inhibition of the primary root length, (B) inhibition of the number of lateral roots, (C) inhibition of the number of lateral root primordia, and (D) inhibition of the density of lateral root formation events. Different lowercase letters indicate statistically significant difference (p ≤ 0.05).
NMig1 Overexpression Improves Free Radical Scavenging Activity and Antioxidant Status
To further elucidate the abiotic stress responses of the transgenic plants, the effect of NMig1 overexpression on the antioxidant status was monitored using two antioxidant assays, DPPH (Figure 6A) and FRAP (Figure 6B). Both analyses displayed that antioxidant status did not differ in unstressed transgenic and wild-type plants. The DPPH radical scavenging activity was increased by the overexpression of NMig1 under all applied stress factors (Figure 6A). The assessment of antioxidant activity also showed higher stress-induced levels in the transgenic lines, as compared to the wild-type Col-0. Thus, both antioxidant assays showed similar trends of change manifested by lower values in the stress-treated wild-type seedlings and higher values in the transgenic lines (Figures 6A,B).
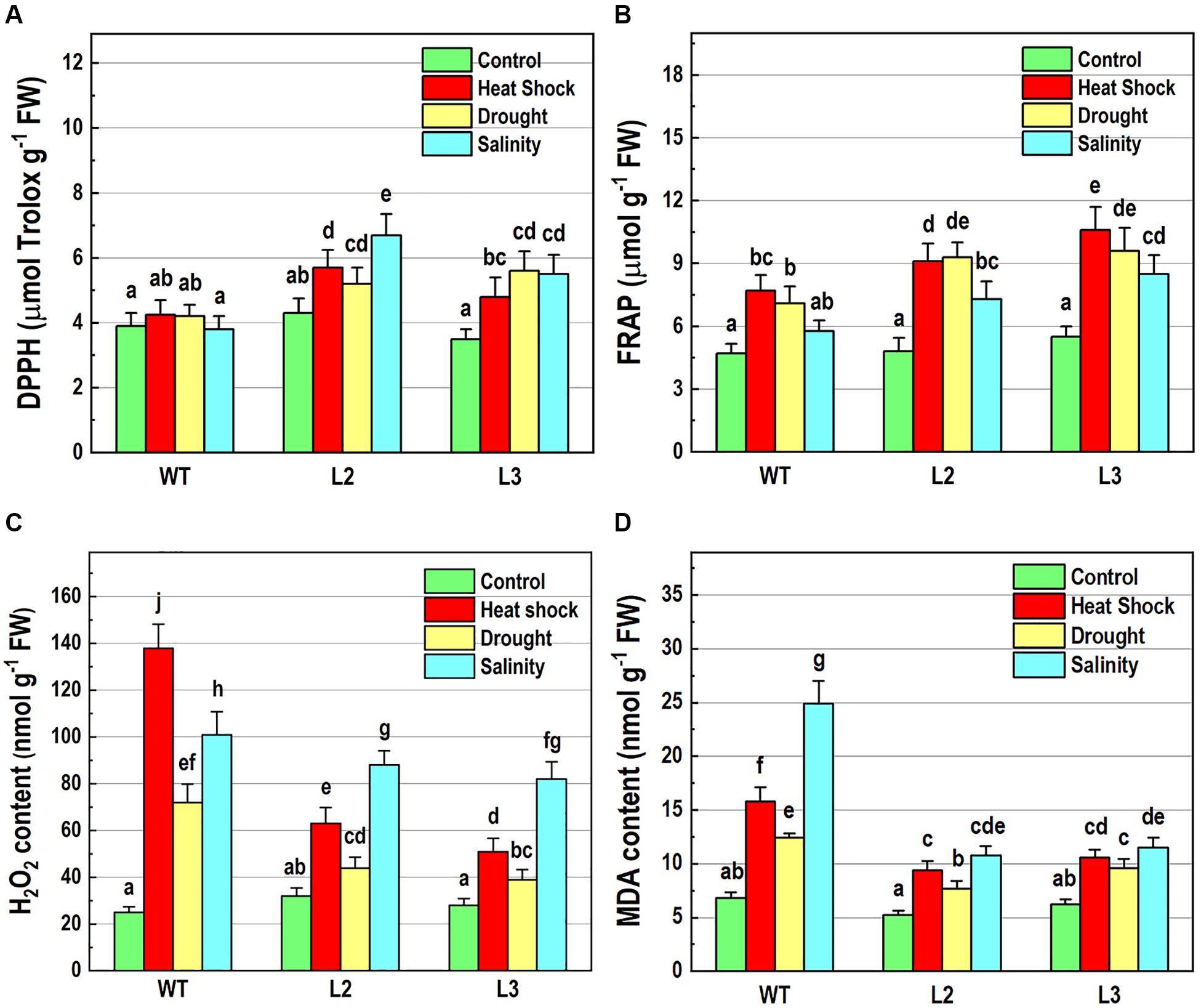
Figure 6. Determination of antioxidant-related parameters in 10-day-old wild-type seedlings and transgenic lines overexpressing NMig1 (L2 and L3) after exposure to heat shock, drought and high salinity: (A) free radical scavenging activity, (B) antioxidant capacity, (C) hydrogen peroxide (H2O2) content, and (D) malondialdehyde (MDA) level. Different lowercase letters indicate statistically significant difference (p ≤ 0.05).
Hydrogen peroxide is one of the most common and early produced ROS in stress-treated plants and comparison of H2O2 content in NMig1-overexpressing and Col-0 plants displayed considerably reduced H2O2 production in the transgenic lines (Figure 6C). The most obvious difference between NMig1-overexpressors and Col-0 plants was observed under heat stress. Exposure of Col-0 to heat shock led to more than 2-fold higher H2O2 accumulation, compared to transgenic plants. Under salinity stress, the content of O2•−H2O2 greatly increased in both the wild-type Col-0 and NMig1 overexpressors. However, this increase was less pronounced in the transgenic than in the wild-type seedlings.
Since ROS usually induce the peroxidation of membrane lipids, a quantitative assay was carried out to compare the level of lipid peroxidation, determined by the amount of MDA (Figure 6D). At control conditions, no significant difference in MDA content was detected between the NMig1-overexpressing and wild-type plants. However, the applied abiotic stresses induced greater accumulation of MDA in Col-0, as compared to that in NMig1 overexpressors. Exposure to high salinity increased about 4.0-fold the content of MDA in Col-0 and less than 2.0-fold in the transgenic lines, compared to the respective controls. Similarly, under heat shock and drought NMig1 overexpressors accumulated lower levels of MDA, compared to the wild-type Col-0.
In addition, to evaluate the effect of NMig1 overexpression on the stress-induced production of reactive oxygen species (ROS) in plant roots, the formation of superoxide anion (O2•−) and hydrogen peroxide (H2O2) was monitored using NBT (Figure 7) and DAB staining (Figure 8), respectively. Under benign growth conditions, both staining procedures showed that the O2•− and H2O2 levels in transgenic and wild-type plants exhibited no differences (Figures 7A,E, 8A,E). Abiotic stresses, however, led to a higher accumulation of O2•− and H2O2 in Col-0, where in the plant roots strong blue (Figures 7B–D) and brown (Figures 8B–D) staining was visualized. The most obvious difference between the NMig1-overexpressing plants and the wild-type Col-0 was detected under heat stress (Figures 7B,F, 8B,F). The accumulation of O2•− in the heat-treated roots of Col-0 was very strong (Figure 7B), whereas in the NMig1 overexpressors, the NBT staining intensity was compatible to the control level (Figure 7F). Exposure to drought and salinity also showed distinct staining patterns in Col-0 (Figures 7C,D, 8C,D) and in the transgenic plants (Figures 7G,H, 8G,H), but the difference in O2•− and H2O2 contents between treated and untreated seedlings was considerably lower.
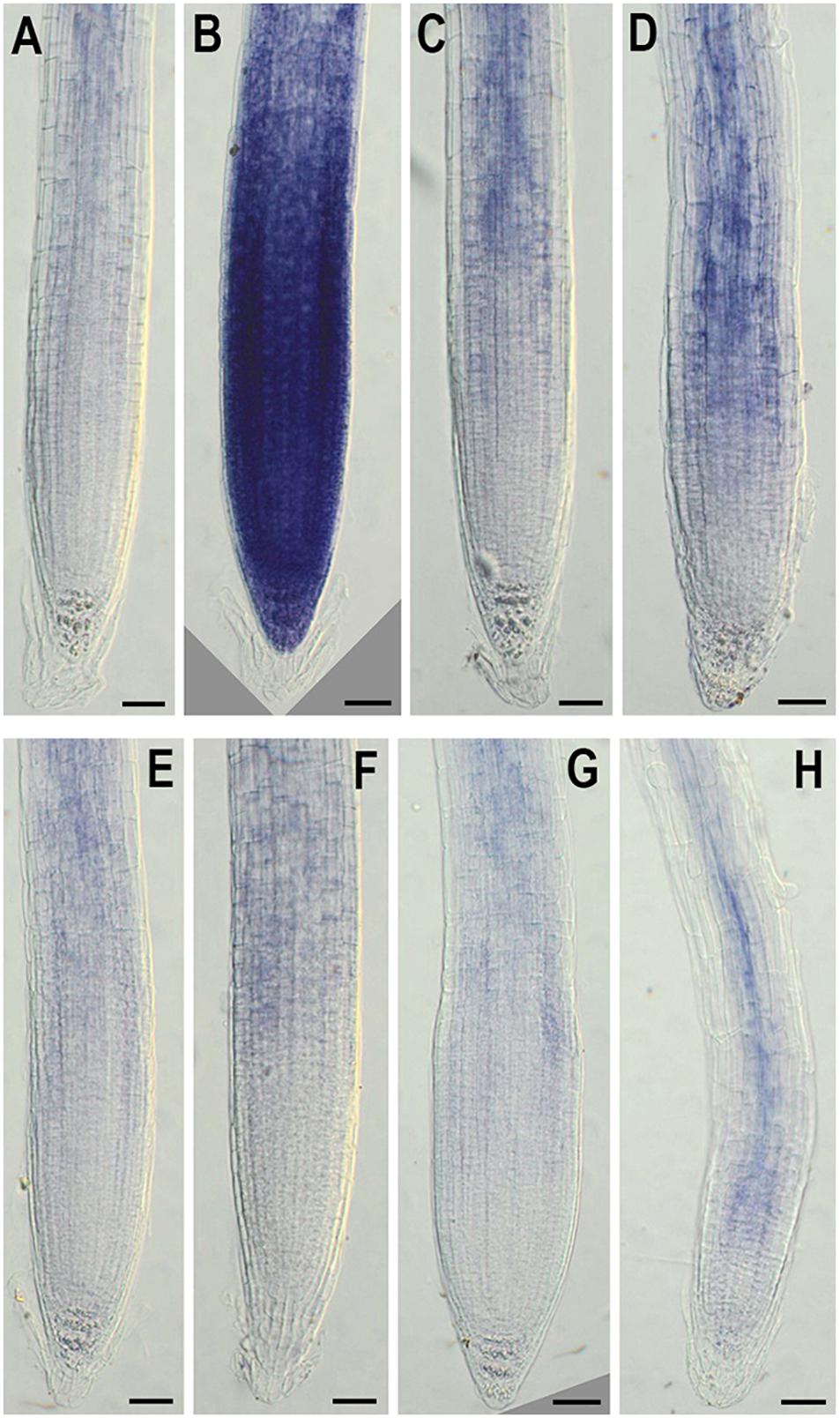
Figure 7. In situ detection of O2•− by nitroblue tetrazolium (NBT) staining in 6-day-old wild-type (A–D) and NMig1-overexpressing (E–H) seedlings under optimal growth conditions (A,E), and after exposure to heat shock (B,F), drought (C,G) and high salinity (D,H). Representative images from three independent experiments are shown. Scale bars = 50 μm.

Figure 8. In situ detection of H2O2 by 3,3-diaminobenzidine (DAB) staining in 6-day-old wild-type (A–D) and NMig1-overexpressing (E–H) seedlings under optimal growth conditions (A,E), and after exposure to heat shock (B,F), drought (C,G), and high salinity (D,H). Representative images from three independent experiments are shown. Scale bars = 50 μm.
Overexpression of NMig1 Changed the Expression of Stress-Related Genes
To explore the molecular mechanisms behind ROS detoxification under stress conditions, mediated by NMig1 overexpression, we compared the expression levels of several genes encoding major enzymes involved in antioxidant defense (Figure 9) and other stress-related maker genes (Figure 10) in the transgenic plants and the wild-type Col-0. Very low expression of the genes SOD1, CAT1, CAT2, and APX1, encoding the ROS scavenging enzymes superoxide dismutase 1, catalase 1, catalase 2 and ascorbate peroxidase 1, respectively, was detected in the control plants (Figures 9A–D). The quantitative expression analysis revealed profound stress-induced upregulation of these genes in both the wild-type and NMig1 overexpressors. More specifically, exposure of wild-type plants to heat shock, drought and salinity, increased the level of SOD1 transcripts by 3.3-, 13-, and 14.8-fold, respectively, compared to untreated controls (Figure 9A). The expression levels of CAT1 and CAT2 also showed differential responses depending of the stress type (Figures 9B,C). In both the wild-type and NMig1 overexpressors, CAT1 was maximally induced by drought and salt stress (Figure 9B), while CAT2 had the highest expression levels (more than 30-fold upregulation) under heat shock in the transgenic lines (Figure 9C). Similar expression pattern was observed for APX1 after the plant exposure to heat shock, where the APX1 expression increased by 12.5-fold in the wild-type but reached more than 25.0-fold in L2 and L3 (Figure 9D). When challenged with drought and salinity, the transgenic lines also showed higher accumulation of APX1 transcripts, but the relative increase in the gene expression was lower, as compared to the control.
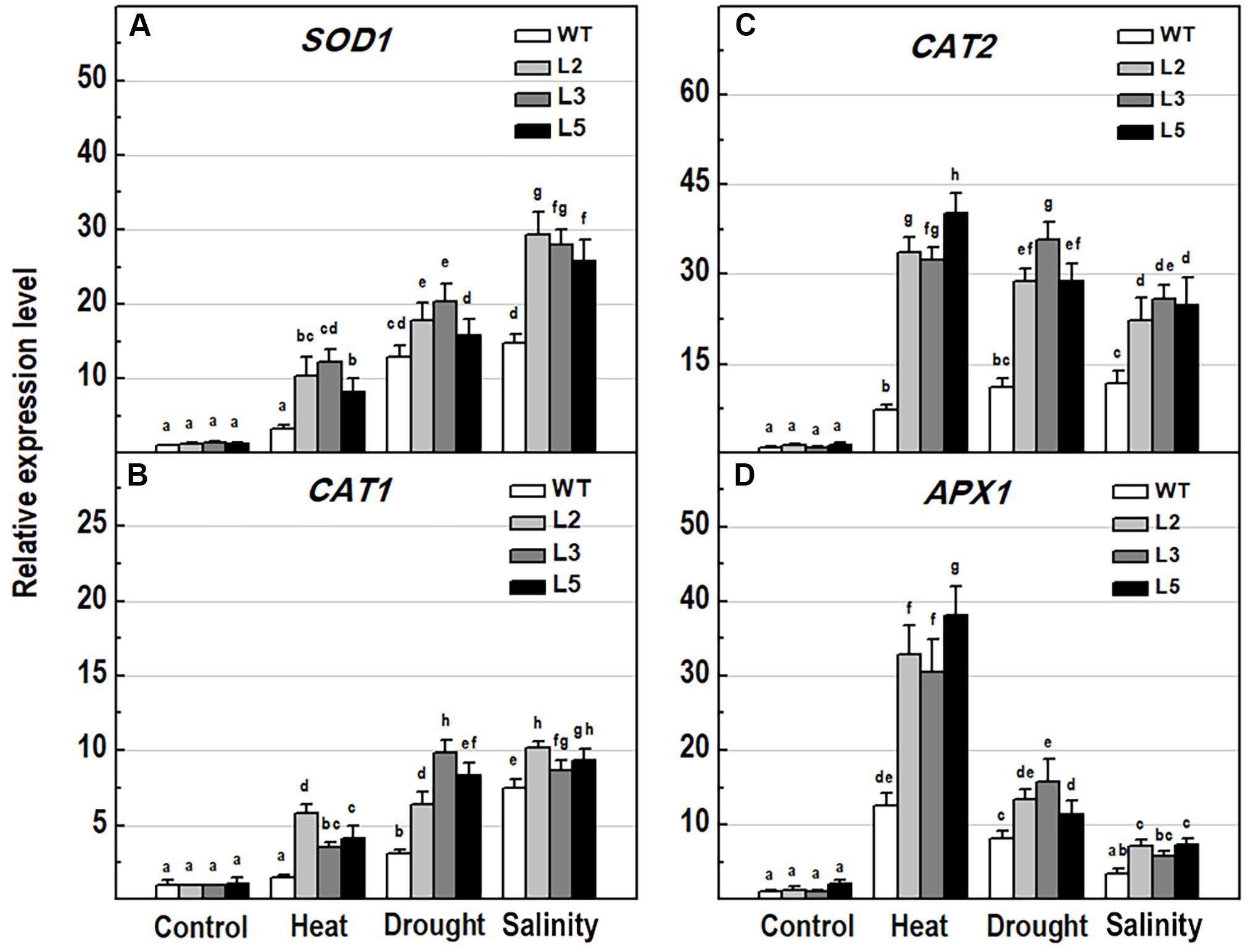
Figure 9. Analysis of the expression levels of major antioxidant genes in 10-day-old wild-type seedlings and transgenic lines overexpressing NMig1 (L2, L3, and L5) after exposure to heat shock, drought and high salinity: (A) SOD1, (B) CAT1, (C) CAT2, and (D) APX1. Expression levels were normalized to those of EF1a and CDKA1;1, as described in section “Materials and Methods.” Data represents the mean values ± SE from at least three biological replicates. Different lowercase letters indicate statistically significant difference (p ≤ 0.05).
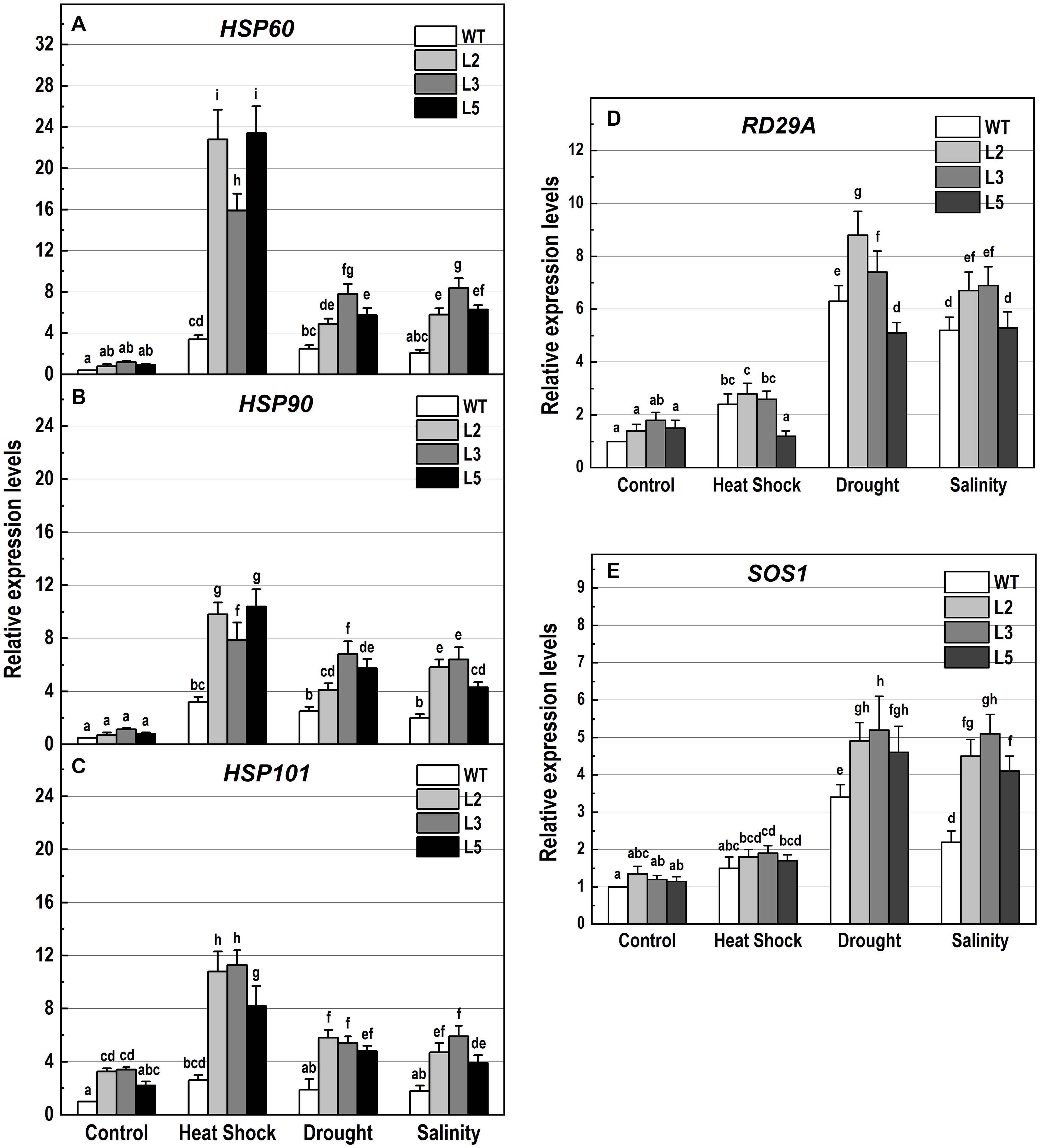
Figure 10. Analysis of the expression levels of genes encoding the heat shock proteins HSP60, HSP90 and HSP101, and the stress-responsive genes RD29A and SOS1 in 10-day-old wild-type seedlings and transgenic lines overexpressing NMig1 (L2, L3, and L5) after exposure to heat shock, drought and high salinity: (A) HSP60, (B) HSP90, (C) HSP101, (D) RD29A, and (E) SOS1. Expression levels were normalized to those of EF1a and CDKA1;1, as described in section “Materials and Methods.” Data represents the mean values ± SE from at least three biological replicates. Different lowercase letters indicate statistically significant difference (p ≤ 0.05).
Additionally, the applied stress factors led to upregulation of genes encoding heat shock proteins, HSP60, HSP90, and HSP101 (Figures 10A–C), and the abiotic stress-responsive genes RD29A and SOS1 (Figures 10D,E). Exposure to stress of the wild-type plants resulted in an increase of all studied genes, however, this increase was much lower than that observed in the transgenic lines (Figures 10A–E). In NMig1-overexpressing plants, the heat stress induced greater accumulation of HSP60 and HSP101 transcripts, between 3.3-fold and 28.5-fold, compared to the transcript abundance in the same lines under benign growth conditions (Figures 10A,C). Sorbitol-mediated drought stress and high salinity led to an increase in the expression of HSP60, RD29A and SOS1 between 3.3-fold and 7.25-fold, compared to the expression levels detected under optimal conditions.
Discussion
In the present study, we identified a heretofore uncharacterized Arabidopsis gene, designated NMig1 (for Nuclear Migration 1), which encodes NudC domain-containing protein. Overexpression of NMig1 resulted in increased primary root growth and formation of more lateral branches, demonstrating that the studied root traits positively correlated with the higher level of NMig1. Consistent with these findings are the developmental defects including obstructed root growth due to a partial loss-of-function mutation in the NudC protein BOB1 (Perez et al., 2009). The stimulating effect on lateral root formation was nearly twice stronger than that on primary root growth. These results led us to further explore how overexpression of NMig1 contributes to remodeling of root system architecture. Quantification of all branching events in the transgenic and the wild-type Col-0 plants indicated increased density of both emerging lateral roots and developing non-emerged primordia. Hence, NMig1 exerts a dual function in Arabidopsis root development, increasing lateral root initiation events and promoting lateral root emergence.
Generally, plants can adjust their root system in correlation with variable environmental conditions and it is very likely that every change in root architecture may contribute to plant performance under stress (Rellán-Álvarez et al., 2016; Motte and Beeckman, 2019). Overexpression of NMig1 positively affected root growth and branching under heat shock, drought and salinity stress manifested by longer primary roots and more lateral branches in the transgenic plants compared to the wild-type Col-0. These beneficial root traits suggest that high expression levels of NMig1 could at least partially counteract the inhibitory effect of different environmental extremes. In agreement with this observation is the transcriptional activation of NMig1 upon exposure of the wild-type Col-0 to different stress factors. The level of NMig1 induction by stress was compared to that of the genes encoding the universal stress proteins USP17 and USP21. The USPs exhibit a redox-dependent chaperone function and are involved in plant protection from abiotic stress (Jung et al., 2015; Bhuria et al., 2016). The magnitude and profile of the stress-induced expression of both USP genes were similar to those of NMig1, which indicate that molecular adjustment of Arabidopsis to stress requires transcriptional activation of these three genes at comparable levels.
A common plant response to internal and external stimuli is the generation of ROS that can potentially lead to oxidative injury of cell components (You and Chan, 2015). Plant cells control the levels of ROS by balancing ROS generation and scavenging through antioxidant enzymes and molecules, such as superoxide dismutase, catalase, peroxidase and other antioxidants that protect cells from oxidative damage (Mittler, 2017). Under benign conditions, ROS are efficiently reduced by non-enzymatic and enzymatic antioxidant components, whereas under stress the production of ROS surpasses the capacity of the antioxidant defense systems to remove them, which causes oxidative stress (Vanderauwera et al., 2005). In our experiments, the expression of four genes, SOD1, CAT1, CAT2, and APX1, encoding major antioxidant enzymes was induced by the applied stress factors with especially high expression rates in the transgenic lines. This suggests that overexpression of NMig1 largely upregulated antioxidant defense components in transgenic plants providing better protection against ROS damages. The overexpression of NMig1 was also associated with greater total antioxidant activity and ROS scavenging potential, assessed by DPPH and FRAP assays. These observations correlate well with the reduced H2O2 levels in the stress-treated transgenic plants when compared with the wild-type Col-0. Additional histochemical detection of the accumulation of O2•− and hydrogen peroxide (H2O2) specifically in the roots of the transgenic and wild-type seedlings also displayed a greater stress-induced staining in the wild-type Col-0, which was utmost under heat stress. In transgenic plants, a visible increase in staining intensities was not documented, in particular after the heat treatment. In addition, the degree of lipid peroxidation, a generally used marker for oxidative stress, quantified by the level of MDA concentration (Yadav et al., 2016) was considerably reduced in the transgenics compared to the wild-type.
Taken together, these results suggest that under stress conditions, different components of antioxidant defense system were mobilized by the overexpression of NMig1, which led to a decrease in ROS accumulation and reduction of oxidative damage in the transgenic plants. Hence, NMig1 overexpressors possess a more efficient antioxidant system compared to the wild-type plants. Since the level of antioxidants in plant tissues is closely related to plant stress tolerance (Xing et al., 2007; Sofo et al., 2015), we can suggest that overexpression of NMig1 confers tolerance to multiple abiotic stresses through optimization of antioxidant defense system (Zhang et al., 2013; Yong et al., 2017).
Furthermore, since NMig1 encodes a HSP20-like chaperone superfamily protein, it could be expected that its overexpression is associated with increased accumulation of other HSPs. In general, various HSPs are coordinately expressed and operate in chaperone networks (Haslbeck et al., 2005; Waters, 2013). Many studies have shown a correlation between HSP induction and plant adaptation to stress (Jacob et al., 2017). We quantified the transcript levels of HSP60, HSP90 and the well-known stress responsive gene HSP101 (Queitsch et al., 2000) in the transgenic plants and Col-0 under abiotic stress. Although some expression differences depending on the nature of the stress imposed were detected, all tested HSPs followed a similar expression pattern, featuring substantial increase in NMig1 overexpressors and lower detected levels in Col-0. Based on these results, we suggest that the NMig1-mediated induction of the studied HSPs could represent another mechanism contributing to abiotic stress tolerance in Arabidopsis. However, the exact molecular link between the HSP accumulation and protective role of NMig1-induced expression under various stress conditions still needs to be studied and defined.
It should be also noted that the impact of NMig1 overexpression in promoting plant tolerance varied depending on the nature of the stress imposed. Exposure to heat shock coincided with the highest expression of the studied HSPs. Drought and high salinity led to a stronger induction of the stress-related marker genes RD29A and SOS1 (Shi et al., 2000; Magwanga et al., 2019). This observation could be associated with the specific stress response mechanisms triggered by each particular stress factor and the involvement of NMig1 in the concrete molecular relays. Nevertheless, overexpression of NMig1 positively regulated the accumulation of defense gene transcripts, such as HSPs, RD29A, and SOS1, which is consistent with the higher stress tolerance of the transgenic plants.
In conclusion, the present work has provided novel insights into the involvement of plant NudC proteins in Arabidopsis root growth and in plant protection against abiotic stress factors. The proposed functions of NMig1 and the insufficient information on the precise biological roles of plant NudC proteins are challenging aspects that require further investigation.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
VVa and TB designed the experiments and wrote the manuscript. VVe, IV, GZ, MZ, MG, NV, and VVa conducted the experiments and analyzed the data. All the authors read and approved the final manuscript.
Funding
This research was financially supported by the Bulgarian National Science Fund (BNSF), Grant No. DN11/8/15.12.2017. The first steps of this work have been carried out under the joint research project between the Research Foundation – Flanders (FWO), Belgium and the Bulgarian Academy of Sciences, Grant No. VS.035.10N.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00815/full#supplementary-material
Footnotes
- ^ https://www.ebi.ac.uk/interpro/
- ^ dnarepair.bas.bg/software/CellTool
- ^ http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi
References
Aumais, J. P., Williams, S. N., Luo, W., Nishino, M., Caldwell, K. A., Caldwell, G. A., et al. (2003). Role for NudC, a dynein-associated nuclear movement protein, in mitosis and cytokinesis. J. Cell Sci. 116, 1991–2003. doi: 10.1242/jcs.00412
Benzie, I. F. F., and Strain, J.-J. (1999). Ferric reducing antioxidant power assay: direct measure of total antioxidant activity of biological fluids and modified version of simultaneous measurement of total antioxidant power and ascorbic acid concentration. Methods Enzymol. 299, 15–27. doi: 10.1016/s0076-6879(99)99005-5
Bhuria, M., Goel, P., Kumar, S., and Singh, A. K. (2016). The promoter of AtUSP is co-regulated by phytohormones and abiotic stresses in Arabidopsis thaliana. Front. Plant Sci. 7:1957. doi: 10.3389/fpls.2016.01957
Botër, M., Amigues, B., Peart, J., Breuer, C., Kadota, Y., Casais, C., et al. (2007). Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell 19, 3791–3804. doi: 10.1105/tpc.107.050427
Brand-Williams, W., Cuvelier, M. E., and Berset, C. (1995). Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 28, 25–30. doi: 10.1016/S0023-6438(95)80008-5
Chi, Y. H., Koo, S. S., Oh, H. T., Lee, E. S., Park, J. H., Phan, K. A. T., et al. (2019). The physiological functions of universal stress proteins and their molecular mechanism to protect plants from environmental stresses. Front. Plant Sci. 10:750. doi: 10.3389/fpls.2019.00750
Chiu, Y., Xiang, X., Dawe, A. L., and Morris, N. R. (1997). Deletion of nudC, a nuclear migration gene of Aspergillus nidulans, causes morphological and cell wall abnormalities and is lethal. Mol. Biol. Cell 8, 1735–1749. doi: 10.1091/mbc.8.9.1735
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cunniff, J., Chiu, Y. H., Morris, N. R., and Warrior, R. (1997). Characterization of DnudC, the Drosophila homolog of an Aspergillus gene that functions in nuclear motility. Mech. Dev. 66, 55–68. doi: 10.1016/S0925-4773(97)00085-3
Danovski, G., Dyankova, T., and Stoynov, S. (2018). CellTool: an open source software combining bio-image analysis and mathematical modelling. bioRxiv [Preprint]. doi: 10.1101/454256
Dawe, A. L., Caldwell, K. A., Harris, P. M., Morris, N. R., and Caldwell, G. A. (2001). Evolutionarily conserved nuclear migration genes required for early embryonic development in Caenorhabditis elegans. Dev. Genes Evol. 211, 434–441. doi: 10.1007/s004270100176
Faircloth, L. M., Churchill, P. F., Caldwell, G. A., and Caldwell, K. A. (2009). The microtubule-associated protein, NUD-1, exhibits chaperone activity in vitro. Cell Stress Chaperones 14, 95–103. doi: 10.1007/s12192-008-0061-1
Fu, Q., Wang, W., Zhou, T., and Yang, Y. (2016). Emerging roles of NudC family: from molecular regulation to clinical implications. Sci. China Life Sci. 59, 455–462. doi: 10.1007/s11427-016-5029-2
Garcia-Ranea, J. A., Mirey, G., Camonis, J., and Valencia, A. (2002). p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 529, 162–167. doi: 10.1016/S0014-5793(02)03321-5
Gocke, C. D., Osmani, S. A., and Miller, B. A. (2000). The human homologue of the Aspergillus nuclear migration gene nudC is preferentially expressed in dividing cells and ciliated epithelia. Histochem. Cell Biol. 114, 293–301. doi: 10.1007/s004180000197
Haslbeck, M., Franzmann, T., Weinfurtner, D., and Buchner, J. (2005). Some like it hot: the structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 12, 842–846. doi: 10.1038/nsmb993
Hodges, D. M., DeLong, J. M., Forney, C. F., and Prange, R. K. (1999). Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207, 604–611. doi: 10.1007/s004250050524
Ishibashi, N., Kanamaru, K., Ueno, Y., Kojima, S., Kobayashi, T., Machida, C., et al. (2012). ASYMMETRIC-LEAVES2 and an ortholog of eukaryotic NudC domain proteins repress expression of AUXIN-RESPONSE-FACTOR and class 1 KNOX homeobox genes for development of flat symmetric leaves in Arabidopsis. Biol. Open 1, 197–207. doi: 10.1242/bio.2012406
Jacob, P., Hirt, H., and Bendahmane, A. (2017). The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotech. J. 15, 405–414. doi: 10.1111/pbi.12659
Jung, Y. J., Melencion, S. M. B., Lee, E. S., Park, J. H., Alinapon, C. V., Oh, H. T., et al. (2015). Universal stress protein exhibits a redox-dependent chaperone function in Arabidopsis and enhances plant tolerance to heat shock and oxidative stress. Front. Plant Sci. 6:1141. doi: 10.3389/fpls.2015.01141
Jurkuta, R. J., Kaplinsky, N. J., Spindel, J. E., and Barton, M. K. (2009). Partitioning the apical domain of the Arabidopsis embryo requires the BOBBER1 NudC domain protein. Plant Cell 21, 1957–1971. doi: 10.1105/tpc.108.065284
Karimi, M., Bleys, A., Vanderhaeghen, R., and Hilson, P. (2007). Building blocks for plant gene assembly. Plant Physiol. 145, 1183–1191. doi: 10.1104/pp.107.110411
Kirschner, M., Winkelhaus, S., Thierfelder, J. M., and Nover, L. (2000). Transient expression and heat-stress-induced co-aggregation of endogenous and heterologous small heat-stress proteins in tobacco protoplasts. Plant J. 24, 397–411. doi: 10.1046/j.1365-313x.2000.00887.x
Kumar, D., Yusuf, M. A., Singh, P., Sardar, M., and Sarin, N. B. (2014). Histochemical detection of superoxide and H2O2 accumulation in Brassica juncea seedlings. Bio Protocol 4:e1108.
Lee, Y. T., Jacob, J., Michowski, W., Nowotny, M., Kuznicki, J., and Chazin, W. J. (2004). Human Sgt1 binds HSP90 through the CHORD-Sgt1 domain and not the tetratricopeptide repeat domain. J. Biol. Chem. 279, 16511–16517. doi: 10.1074/jbc.M400215200
Liu, S., Wang, J., Jiang, S., Wang, H., Gao, Y., Zhang, H., et al. (2019). Tomato SlSAP3, a member of the stress-associated protein family, is a positive regulator of immunity against Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 20, 815–830. doi: 10.1111/mpp.12793
Magwanga, R. O., Kirungu, J. N., Lu, P., Yang, X., Dong, Q., Cai, X., et al. (2019). Genome wide identification of the trihelix transcription factors and overexpression of Gh_A05G2067 (GT-2), a novel gene contributing to increased drought and salt stresses tolerance in cotton. Physiol. Plant. 167, 447–464. doi: 10.1111/ppl.12920
Malamy, J. E., and Benfey, P. N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44.
Miroshnichenko, S., Tripp, J., Nieden, U., Neumann, D., Conrad, U., and Manteuffel, R. (2005). Immunomodulation of function of small heat shock proteins prevents their assembly into heat stress granules and results in cell death at sublethal temperatures. Plant J. 41, 269–281. doi: 10.1111/j.1365-313X.2004.02290.x
Morris, S. M., Anaya, P., Xiang, X., Morris, N. R., May, G. S., and Yu-Lee, L. (1997). A prolactin-inducible T cell gene product is structurally similar to the Aspergillus nidulans nuclear movement protein NUDC. Mol. Endocrinol. 11, 229–236. doi: 10.1210/mend.11.2.9892
Motte, H., and Beeckman, T. (2019). The evolution of root branching: increasing the level of plasticity. J. Exp. Bot. 70, 785–793. doi: 10.1093/jxb/ery409
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nguyen, H. M., Sako, K., Matsui, A., Suzuki, Y., Mostofa, M. G., Ha, C. V., et al. (2017). Ethanol enhances high-salinity stress tolerance by detoxifying reactive oxygen species in Arabidopsis thaliana and rice. Front. Plant Sci. 8:1001. doi: 10.3389/fpls.2017.01001
Osmani, A. H., Osmani, S. A., and Morris, N. R. (1990). The molecular cloning and identification of a gene product specifically required for nuclear movement in Aspergillus nidulans. J. Cell. Biol. 111, 543–555. doi: 10.1083/jcb.111.2.543
Perez, D. E., Hoyer, J. S., Johnson, A. I., Moody, Z. R., Lopez, J., and Kaplinsky, N. J. (2009). BOBBER1 is a noncanonical Arabidopsis small heat shock protein required for both development and thermotolerance. Plant Physiol. 151, 241–252. doi: 10.1104/pp.109.142125
Pundir, S., Martin, M. J., and O’Donovan, C. (2016). UniProt tools. Curr. Protoc. Bioinformatics 53, 1–29. doi: 10.1002/0471250953.bi0129s53
Queitsch, C., Hong, S. W., Vierling, E., and Lindquist, S. (2000). Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12, 479–492. doi: 10.1105/tpc.12.4.479
Raymaekers, M., Smets, R., Maes, B., and Cartuyvels, R. (2009). Checklist for optimization and validation of real-time PCR assays. J. Clin. Lab. Anal. 23, 145–151. doi: 10.1002/jcla.20307
Rellán-Álvarez, R., Lobet, G., and Dinneny, J. R. (2016). Environmental control of root system biology. Annu. Rev. Plant Biol. 67, 619–642. doi: 10.1146/annurev-arplant-043015-111848
Schwacke, R., Schneider, A., Van Der Graaff, E., Fischer, K., Catoni, E., Desimone, M., et al. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26. doi: 10.1104/pp.011577
Shi, H., Ishitani, M., Kim, C., and Zhu, J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97, 6896–6901. doi: 10.1073/pnas.120170197
Silverblatt-Buser, E. W., Frick, M. A., Rabeler, C., and Kaplinsky, N. J. (2018). Genetic interactions between BOB1 and multiple 26S proteasome subunits suggest a role for proteostasis in regulating Arabidopsis development. G3 8, 1379–1390. doi: 10.1534/g3.118.300496
Sofo, A., Scopa, A., Nuzzaci, M., and Vitti, A. (2015). Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. ıInt. J. Mol. Sci. 16, 13561–13578. doi: 10.3390/ijms160613561
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3-new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Vanderauwera, S., Vandenbroucke, K., Inzé, A., van de Cotte, B., Mühlenbock, P., De Rycke, R., et al. (2005). AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 20113–20118. doi: 10.1073/pnas.1217516109
Waters, E. R. (2013). The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 64, 391–403. doi: 10.1093/jxb/ers355
Wolff, S. P. (1994). Ferrous ion oxidation in presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Methods Enzymol. 233, 182–189. doi: 10.1016/S0076-6879(94)33021-2
Xing, Y., Jia, W. S., and Zhang, J. H. (2007). AtMEK1 mediates stress-induced gene expression of CAT1 catalase by triggering H2O2 production in Arabidopsis. J. Exp. Bot. 58, 2969–2981. doi: 10.1093/jxb/erm144
Yadav, V., Arif, N., Singh, S., Srivastava, P. K., Sharma, S., Tripathi, D. K., et al. (2016). Exogenous mineral regulation under heavy metal stress: advances and prospects. Biochem. Pharmacol. 5:220. doi: 10.4172/2167-0501.1000220
Yang, Y., Yan, X., Cai, Y., Lu, Y., Si, J., and Zhou, T. (2010). NudC-like protein 2 regulates the LIS1/dynein pathway by stabilizing LIS1 with Hsp90. Proc. Natl. Acad. Sci. U.S.A. 107, 3499–3504. doi: 10.1073/pnas.0914307107
Yong, B., Wang, X., Xu, P., Zheng, H., Fei, X., Hong, Z., et al. (2017). Isolation and abiotic stress resistance analyses of a catalase gene from Ipomoea batatas (L.) Lam. BioMed Res. Internat 2017, 1–10. doi: 10.1155/2017/6847532
You, J., and Chan, Z. (2015). ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 6:1092. doi: 10.3389/fpls.2015.01092
Zhang, C., Zhang, W., Lu, Y., Yan, X., Yan, X., Zhu, X., et al. (2016). NudC regulates actin dynamics and ciliogenesis by stabilizing cofilin 1. Cell Res. 26, 239–253. doi: 10.1038/cr.2015.152
Zhang, M.-Y., Huang, N.-N., Clawson, G. A., Osmani, S. A., Pan, W., Xin, P., et al. (2002). Involvement of the fungal nuclear migration gene NudC human homolog in cell proliferation and mitotic spindle formation. Exp. Cell Res. 273, 73–84. doi: 10.1006/excr.2001.5414
Zhang, Z., Zhang, Q., Wu, J., Zheng, X., Zheng, S., Sun, X., et al. (2013). Gene knockout study reveals that cytosolic ascorbate peroxidase 2 (OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS One 8:e57472. doi: 10.1371/journal.pone.0057472
Zheng, M., Cierpicki, T., Burdette, A. J., Utepbergenov, D., Janczyk, P. Ł, Derewenda, U., et al. (2011). Structural features and chaperone activity of the NudC protein family. J. Mol. Biol. 409, 722–741. doi: 10.1016/j.jmb.2011.04.018
Zhou, T., Aumais, J. P., Liu, X., Yu-Lee, L. Y., and Erikson, R. L. (2003). A role for Plk1 phosphorylation of NudC in cytokinesis. Dev. Cell 5, 127–138. doi: 10.1016/S1534-5807(03)00186-2
Keywords: NudC gene, NMig1, abiotic stress, reactive oxygen species, heat shock proteins, Arabidopsis thaliana
Citation: Velinov V, Vaseva I, Zehirov G, Zhiponova M, Georgieva M, Vangheluwe N, Beeckman T and Vassileva V (2020) Overexpression of the NMig1 Gene Encoding a NudC Domain Protein Enhances Root Growth and Abiotic Stress Tolerance in Arabidopsis thaliana. Front. Plant Sci. 11:815. doi: 10.3389/fpls.2020.00815
Received: 15 February 2020; Accepted: 20 May 2020;
Published: 11 June 2020.
Edited by:
Manny Delhaize, Plant Industry (CSIRO), AustraliaReviewed by:
Men-Chi Chang, National Taiwan University, TaiwanVijay Pratap Singh, University of Allahabad, India
Copyright © 2020 Velinov, Vaseva, Zehirov, Zhiponova, Georgieva, Vangheluwe, Beeckman and Vassileva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tom Beeckman, tobee@psb.ugent.be; Valya Vassileva, valyavassileva@bio21.bas.bg
 Valentin Velinov
Valentin Velinov