- Department of Pomology, College of Agronomy, Jiangxi Agricultural University, Nanchang, China
Cuticular wax is closely related to plant resistance to abiotic stress. 3-Ketoacyl-CoA synthase (KCS) catalyzes the biosynthesis of very-long-chain fatty acid (VLCFA) wax precursors. In this study, a novel KCS family gene was isolated from Newhall navel orange and subsequently named CsKCS6. The CsKCS6 protein has two main domains that belong to the thiolase-like superfamily, the FAE1-CUT1-RppA and ACP_syn_III_C domains, which exist at amino acid positions 80–368 and 384–466, respectively. CsKCS6 was expressed in all tissues, with the highest expression detected in the stigma; in addition, the transcription of CsKCS6 was changed in response to drought stress, salt stress and abscisic acid (ABA) treatment. Heterologous expression of CsKCS6 in Arabidopsis significantly increased the amount of VLCFAs in the cuticular wax on the stems and leaves, but there were no significant changes in total wax content. Compared with that of the wild-type (WT) plants, the leaf permeability of the transgenic plants was lower. Further research showed that, compared with the WT plants, the transgenic lines experienced less water loss and ion leakage after dehydration stress, displayed increased survival under drought stress treatment and presented significantly longer root lengths and survival under salt stress treatment. Our results indicate that CsKCS6 not only plays an important role in the synthesis of fatty acid precursors involved in wax synthesis but also enhances the tolerance of transgenic Arabidopsis plants to abiotic stress. Thus, the identification of CsKSC6 could help to increase the abiotic stress tolerance of Citrus in future breeding programs.
Introduction
Very-long-chain fatty acids (VLCFAs), which refer mainly to a class of fatty acids with more than 18 carbon atoms, are precursor materials for the synthesis of plant cuticular waxes (Kunst and Samuels, 2003). The biosynthesis of VLCFAs involves catalysis by the multienzyme fatty acid elongase (FAE) complex. The catalytic process involves four main steps: condensation, reduction, dehydration and re-reduction (Haslam and Kunst, 2013). 3-Ketoacyl-CoA synthase (KCS) is the rate-limiting enzyme of the condensation reaction and is a key enzyme in the fatty acid extension pathway (Rossak et al., 2001).
There are many genes that encode KCS enzymes. FAE1, which is expressed only in seeds, was the first KCS family gene cloned from Arabidopsis, and its encoded protein catalyzes the synthesis of C20 and C22 fatty acids and storage lipids (James et al., 1995). To date, 21 KCS genes have been annotated in the Arabidopsis genome, and they are divided into four subfamilies, FAE1-like, KCS1-like, FDH-like, and CER6, based on their amino acid sequence homology (Costaglioli et al., 2005). Subsequently, Joubès et al. (2008) divided the existing 21 Arabidopsis KCS proteins into eight subclasses based on phylogeny. Guo et al. (2015) collected KCS genes from 26 sequenced plant genomes and reconstructed the phylogenetic tree of the KCS family genes to reveal the molecular basis of the functional divergence of different KCS genes. Furthermore, three KCS genes, including KCS1 (Todd et al., 1999; Lee and Suh, 2013), FDH (Yephremov et al., 1999; Pruitt et al., 2000) and CER6 (Millar et al., 1999; Fiebig et al., 2000; Hooker et al., 2002), have been shown to be involved in the formation of the cuticle in Arabidopsis: it has been reported that KCS1 is expressed in all tissues of Arabidopsis plants, and complete loss of function of KCS1 leads to a decrease in C26 and C30 alcohol and aldehyde compounds, respectively (Lee and Suh, 2013). FDH is expressed mainly in the flowers and young leaves of Arabidopsis; the FDH gene participates in the synthesis of VLCFAs in epidermal cells and is involved in both the regulation of plant morphological structure and the response to biotic stress (Yephremov et al., 1999; Pruitt et al., 2000; Joubès et al., 2008). KCS6/CER6/CUT1 is expressed in the aboveground tissues of Arabidopsis, especially the flowers and siliques. Overexpression of KCS6/CER6 caused an increase in the wax content of the stem epidermis of Arabidopsis. Moreover, its functional defect mutant cer6 accumulated a large quantity of C24 VLCFA derivatives, indicating that this gene is involved in the biosynthesis of C24 (or longer) VLCFAs (Millar et al., 1999; Fiebig et al., 2000; Hooker et al., 2002).
Cuticular wax is the first protective barrier that covers the surface of terrestrial plants and is in contact with the outside environment (Riederer and Schneider, 1990; Jenks et al., 1994). Therefore, cuticular wax plays an important role in plant resistance to various stresses (Krauss et al., 1997; Sieber et al., 2000). In recent years, many wax synthesis genes have been used to improve plant resistance to abiotic stresses. ECERIFERUM (CER1) genes are involved in the wax biosynthesis pathway. cer1 mutants present glossy green stems and altered wax alkane biosynthesis (Bourdenx et al., 2011; Bernard et al., 2012). Overexpression of OsGL1-2 increases the number of wax crystals of rice leaves, thereby improving the drought resistance of the plant (Islam et al., 2009). Transformation of Arabidopsis with WXP1 and WXP2 also increased the wax content of the transformants and their resistance to water stress and drought stress (Zhang et al., 2005, 2007). The apple AP2/EREBP transcription factor MdSHINE2 provides drought resistance by regulating wax biosynthesis (Zhang et al., 2019a). Similarly, the R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis (Zhang et al., 2019b). Overexpression of BnKCS1-1, BnKCS1-2, and BnCER1-2 in Brassica napus increased the wax crystal density on the leaf surface, and the water loss rate decreased together with increased drought tolerance, which was enhanced in transgenic plants (Wang et al., 2020).
Citrus is a perennial woody plant species with a high degree of heterozygosity. The relationship between wax genes and citrus resistance to abiotic stress may be more complex than that of model plant species such as Arabidopsis. In our previous studies, we analyzed the wax structure, chemical composition, and transcript levels of wax genes in Newhall navel orange (Liu et al., 2012, 2015). No wax-related genes have been identified in citrus so far; thus, the molecular mechanism underlying wax synthesis and its effect on citrus abiotic stress resistance is still unknown. In this study, a KCS6 homologous gene, which was named CsKCS6, was cloned from Newhall navel orange. The expression patterns of CsKCS6 in different tissues and in response to different abiotic stress conditions were analyzed by quantitative real-time PCR (qRT-PCR). The important role of CsKCS6 in citrus wax synthesis and its effects on abiotic tolerance were identified by overexpressing the gene in Arabidopsis. The results broadened our knowledge concerning the molecular mechanism underlying cuticular wax synthesis in citrus.
Materials and Methods
Plant Materials and Growth Conditions
Tissue samples of from adult trees of Newhall navel orange (Citrus sinensis) were collected in an orchard in Xinfeng County, Jiangxi Province, China. Young leaves of Newhall navel orange trees were collected for gene cloning, and leaves, flowers, receptacles, petals, pistils, stigmas, ovaries, flavedos, albedos, and flesh were collected from five trees per biological replicate for organ-specific expression analysis. Flavedo, albedo and flesh samples were collected from five trees per biological replicate at 60, 120, and 210 days after flowering, respectively, for analysis of expression during fruit development. Three biological replicates were used for these expression analyses.
Prior to the stress treatments, 1-year-old grafted seedlings of Newhall navel orange whose height was 30 ± 2 cm were transferred to pots that contained Hoagland’s solution in a growth chamber for 7 days to acclimate to the new conditions. The plants were then subjected to 4°C for 0, 1, 3, 6, 9, 12, and 24 h for the cold treatment. For the drought stress, salt stress and abscisic acid (ABA) treatments, we transferred the plants to Hoagland’s solution that contained 20% PEG6000, 250 mM NaCl, or 100 μM ABA, respectively, for 0, 1, 3, 6, 9, 12, and 24 h. The seedlings for each treatment were grown under a light/dark cycle of 16/8 h at 25°C. For each treatment, the leaves were sampled from 18 randomly selected seedlings at each time point, and each treatment was repeated three times for each time point.
Cloning and Sequence Analysis of CsKCS6
The KCS6 homologous cDNA sequence (Cs7g13310) from sweet orange was retrieved from the sweet orange genomic database1. A pair of gene-specific primers (P1) (Supplementary Table S1) based on the KCS6 homologous cDNA sequence were designed for CDS sequence cloning. The leaves of 1-year-old grafted seedlings of Newhall navel orange were sampled to extract the total RNA. RNA extraction method refers to TaKaRa MiniBEST Plant RNA Extraction Kit (TaKaRa, Japan) instructions. The integrity of RNA was detected by 1% agarose gel electrophoresis. The total RNA yield and quality were further determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, United States). Using the total RNA extracted above as a template, TaKaRa PrimeScriptTM RT reagent Kit with gDNA Eraser reverse transcription kit (TaKaRa, Japan) was used for cDNA synthesis. Bioinformatic analysis, which included a plant KCS6 homology analysis, multiple sequence alignment and a KCS6 phylogenetic analysis, was conducted using the methods of a previous report (Liu et al., 2017).
Expression Analysis of CsKCS6 in Navel Orange
Real-time quantitative PCR (qRT-PCR) was used for CsKCS6 transcript analysis with the specific primer P2 (Supplementary Table S1). The citrus ACTIN gene was amplified with the specific primer P3 (Supplementary Table S1) to serve as a control gene for normalizing the expression between different samples. The real-time quantitative detection is done using SYBR§ Premix Ex TaqTM kit (TaKaRa, Japan). The reactions were performed by an initial incubation at 95°C pre-denaturation for 30 s, then 95°C denaturation for 5 s, 60°C annealing for 30 s and 40 cycles. The 2–ΔΔCT method was used to calculate the relative changes in gene expression. qPCR was performed in four replicates for each sample, and data are indicated as the mean ± SD (n = 3).
Construction of the CsKCS6 Expression Vector and Its Genetic Transformation Into Arabidopsis
The full-length CDS of the Newhall navel orange CsKCS6 gene was cloned into a pCAMBIA1301 with CaMV35S promoter plant expression vector using the amplification-specific primer P4 (Supplementary Table S1) with BamHI and BstpI restriction sites. A vector-containing plasmid was introduced into Agrobacterium tumefaciens strain LBA4404, which was subsequently transformed into Arabidopsis by the Agrobacterium-mediated floral-dip method, as described by Zhou et al. (2019). Seeds of T0 transgenic plants were sown in Murashige and Skoog (MS) media to which hygromycin (10 mg L–1) was added to screen for positive T1 transgenic plants based on their resistance to hygromycin, and using the leaves of resistant seedlings as templates, PCR detection was carried out using gene specific primers of P4 and P7 (Supplementary Table S1). The expression level of positive transgenic plants was further tested by qPCR amplification of CsKCS6 and AtACTIN using two different primers: P5 and P6 (Supplementary Table S1). Seeds of the T1-positive Arabidopsis plants were grown on MS solid media that contained hygromycin (10 mg L–1) to produce T2 transgenic plants, which were used in subsequent experiments.
Extraction of Cuticular Waxes and Gas Chromatography-Mass Spectrometry (GC–MS) Analysis
Approximately 100 mg of leaves and stems of 6-week-old wild-type (WT) and transgenic Arabidopsis plants were removed. The sample is gently moistened with distilled water and then the tissue surface is tenderly drained with filter paper, after which they were placed in a reaction flask. Cuticular waxes were extracted described by immersing tissues for 1 min in 2 mL of chloroform containing 10 μg of tetracosane as an internal standard. The extracts were dried under a gentle stream of nitrogen gas. The dried wax residues were derivatized by adding 20 μL of silylation reagent (BSTFA) and 20 μL of pyridine and incubated for 40 min at 70°C. The chromatographic column used in the experiment adopts a universal (30 m × 0.25 mm × 0.25 μm) hp-5MS capillary column with polydimethylsiloxane as the stationary phase. The chromatographic conditions were the inlet temperature of 280°C, the column flow rate of 2 mL/min and a constant flow rate; the Aux-2 temperature of 280°C, helium as the carrier gas, sample injection without splitting and the injection volume of 1 μL. The mass spectrometry program is set to electron impact ionization mode, energy 70 eV, mass scanning range 50 ∼ 600 amu and full scanning mode. The temperature rise process is to hold at 50°C for 1 min, raise the temperature to 170°C at 20°C/min, and hold for 2 min; then raise the temperature to 300°C at 5°C/min and hold for 8 min. The ion peaks of various wax components are detected by GC–MS (GCMS-QP2010, Shimadzu, Agilent). According to the mass spectrum library to search and determine and compare the FID peak area data, the single wax compound was quantified relative to the internal standard. Three biological replicates were used for each sample. The specific methods used were described by Chen et al. (2011) and Liu et al. (2012).
Scanning Electron Microscopy (SEM)
Scanning electron microscopy (SEM) was used to observe the ultrastructure of the wax crystals of the stems and rosette leaves of 6-week-old Arabidopsis. The samples were fixed in 4% glutaraldehyde for 12 h and then rinsed with 0.1 mM phosphate buffer at a pH of 7.2, three times for 10 min each. Ethanol at concentrations of 30, 50, 70, 80, and 90% was dehydrated once for 15 min using a JFC-1100-type ion sputtering instrument for gold plating. The samples were ultimately observed with a JSM-T300 scanning electron microscope (JEOL, Tokyo, Japan) after they were freeze-dried and adhered to the sample table (Liu et al., 2012).
Toluidine Blue (TB) Staining and Chlorophyll Content Assays
Six-week-old healthy Arabidopsis leaves were stained in a 0.05% TB staining solution at 25°C for 10 min. The samples were then rinsed with deionized water at least three times before being images.
Healthy leaves of transgenic plants and WT plants at 6 weeks of age were taken in equal amounts, and their chlorophyll leaching content was measured according to the method of Zhang et al. (2019a).
Physiological Measurements of Arabidopsis Plants Under Drought and Salt Treatments
The water loss rate and ion leakage rate of the leaves after dehydration treatment were measured according to previously described methods (Dai et al., 2018; Xie et al., 2018).
Seeds of WT and CsKCS6-overexpressing Arabidopsis were selected and sown on MS media that were supplemented with 100 mM NaCl. Thirty seeds were sown per treatment, and the germination percentage was determined daily (Seo et al., 2009). WT and transgenic Arabidopsis seedlings that had grown on MS media for 7 days were transferred to fresh MS media supplemented with 100 and 200 mM NaCl and allowed to grow for 2 weeks, at which time they were imaged and their root length (≥5 mm) was measured.
WT and transgenic Arabidopsis that had grown on MS media for 4 weeks were transplanted into soil. After 10 days, the plants were then subjected to drought (water replenishment for 3 days after 10 days of natural drought) and salt (100/200 mM NaCl solutions were poured into the pots every 3 days, at 50 ml pot–1) treatments. Twenty plants were included per treatment, each of which included three biological replicates (Dai et al., 2018).
Data Presentation and Statistical Analysis
All quantitative data are given as the means and standard errors. Statistical analyses were conducted using Student’s t-test and Duncan’s multiple range tests in SPSS version 22. Statistical significance was considered at the P < 0.01 and P < 0.05 level for t-test and at P < 0.05 for Duncan’s multiple range tests.
Results
Cloning and Sequence Analysis of CsKCS6
In the past, a novel navel orange variety, named ‘Ganqi 3,’ was bred from a bud mutation of ‘Newhall’ navel orange. Further study revealed that the expression level of CsKCS6 in ‘Ganqi 3’ fruit peels were significant lower than that in ‘Newhall’ fruit peels, indicating the CsKCS6 might be involved in cuticular wax biosynthesis (Liu et al., 2016). Using the cDNA of the ‘Newhall Navel Orange’ leaf as a template, PCR amplification was performed using CsKCS6 specific primers (Supplementary Table S1). Sequence analysis showed that The full-length cDNA sequence of CsKCS6 (GenBank accession number MT259183) was 1750 bp, with an open reading frame (ORF) of 1491 bp. The CsKCS6 protein comprised 496 amino acids, with a putative molecular weight of 56.22 kDa and an isoelectric point of 9.18. Sequence analysis revealed that CsKCS6 is highly homologous to other KCS6 proteins in plants, including Citrus clementina CcKCS6 (98%), Hevea brasiliensis HbKCS6 (67%), Jatropha curcas JcKCS6 (68%), Glycine max GmKCS6 (67%), Malus domestica MdKCS6 (69%), Populus euphratica PeKCS6 (68%), Theobroma cacao TcKCS6 (70%), and Sesamum indicum SiKCS6 (67%). Structural analysis revealed that the FAE1-CUT1-RppA conserved domain was found at amino acid positions of 80–368 in CsKCS6 and that the ACP_syn_III_C conserved domain was found at 384–466. Both major domains belong to the thiolase-like superfamily. Among them, the Cys residue at amino acid position of 225 is the catalytic active site of the condensing enzyme and is highly conserved among different species (Figure 1). In this study, we used the maximum likelihood method of MEGA 6 software to construct a phylogenetic tree of plant KCS6 proteins. The results showed that the CsKCS6 protein formed a large branch with the KCS6 proteins of dicotyledonous plant species such as Citrus clementina, Malus domestica, Gossypium raimondii, and Glycine max. The CsKCS6 protein was most closely related to the clementine CcKCS6 and clustered onto the same small branch. In addition, the KCS6 protein of the monocotyledonous plant species Musa acuminata, Zea mays, Oryza brachyantha, and Phoenix dactylifera was classified as another large branch, and this protein was the most related to CsKCS6. The evolution of these protein sequences is essentially consistent with the process of plant evolution in nature (Figure 2).
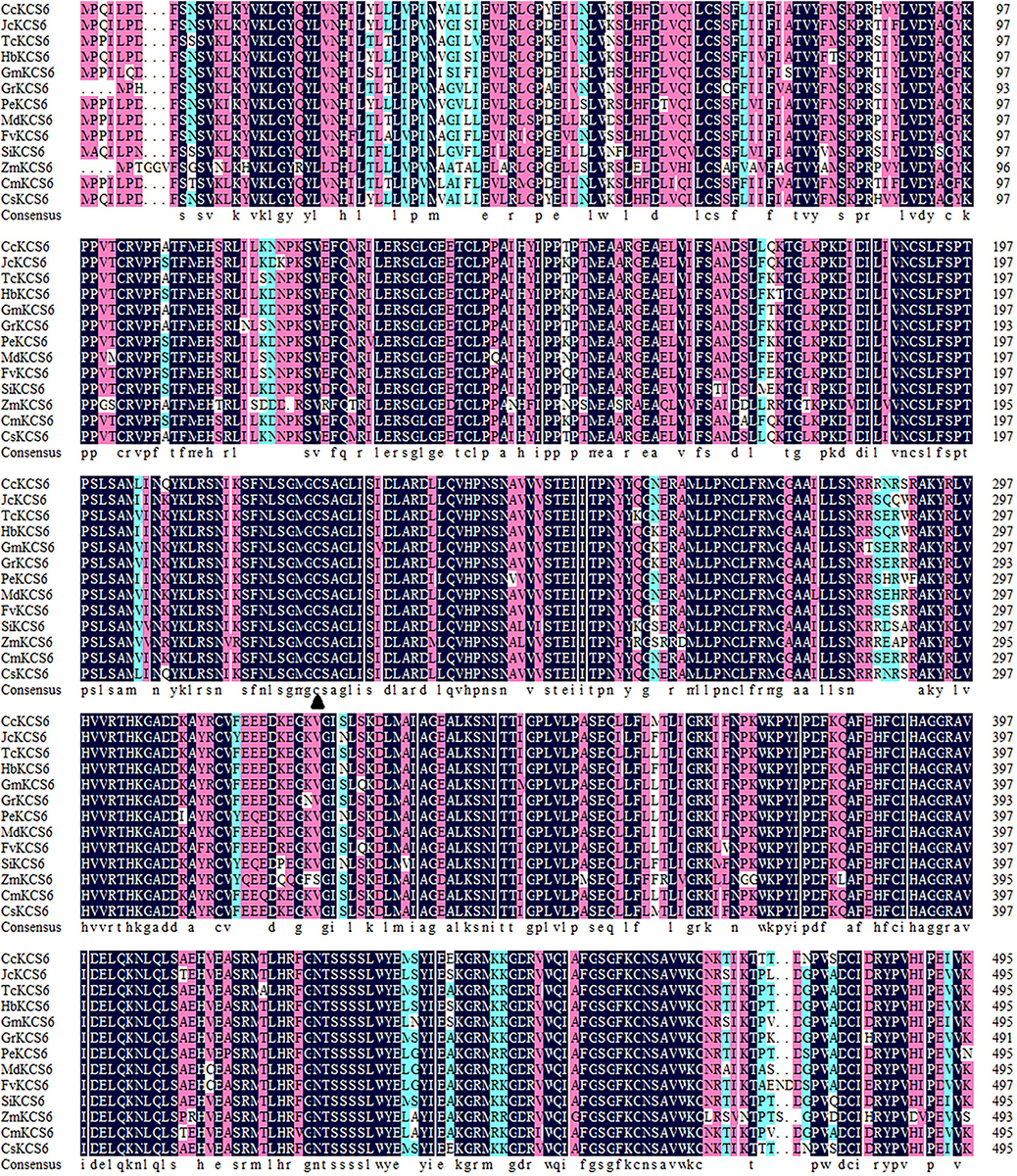
Figure 1. Multiple amino acid sequence alignment of CsKCs6 with its homologous proteins from different plant species, including Citrus clementina CcKCS6 (XP_006437664.1), Jatropha curcas JcKCS6 (XP_012082087.1), Theobroma cacao TcKCS6 (XP_007044356.2), Hevea brasiliensis HbKCS6 (XP_021637941.1), Glycine max GmKCS6 (XP_003555901.1), Gossypium raimondii GrKCS6 (XP_012467884.1), Populus euphratica PeKCS6 (XP_011007926.1), Malus domestica MdKCS6 (XP_008339680.2), Fragaria vesca subsp. Vesca FvKCS6 (XP_004299082.1), Sesamum indicum SiKCS6 (XP_011099559.1), Zea mays ZmKCS6 (NP_001310469.1), Cucumis melo CmKCS6 (XP_008446598.1), Arabidopsis thaliana AtKCS6 (AAM65060.1), Ginkgo biloba GbKCS6 (AAY52458.1), and Citrus sinensis CsKCS6. The Cys amino acid (c) site is marked with solid triangle (▲).
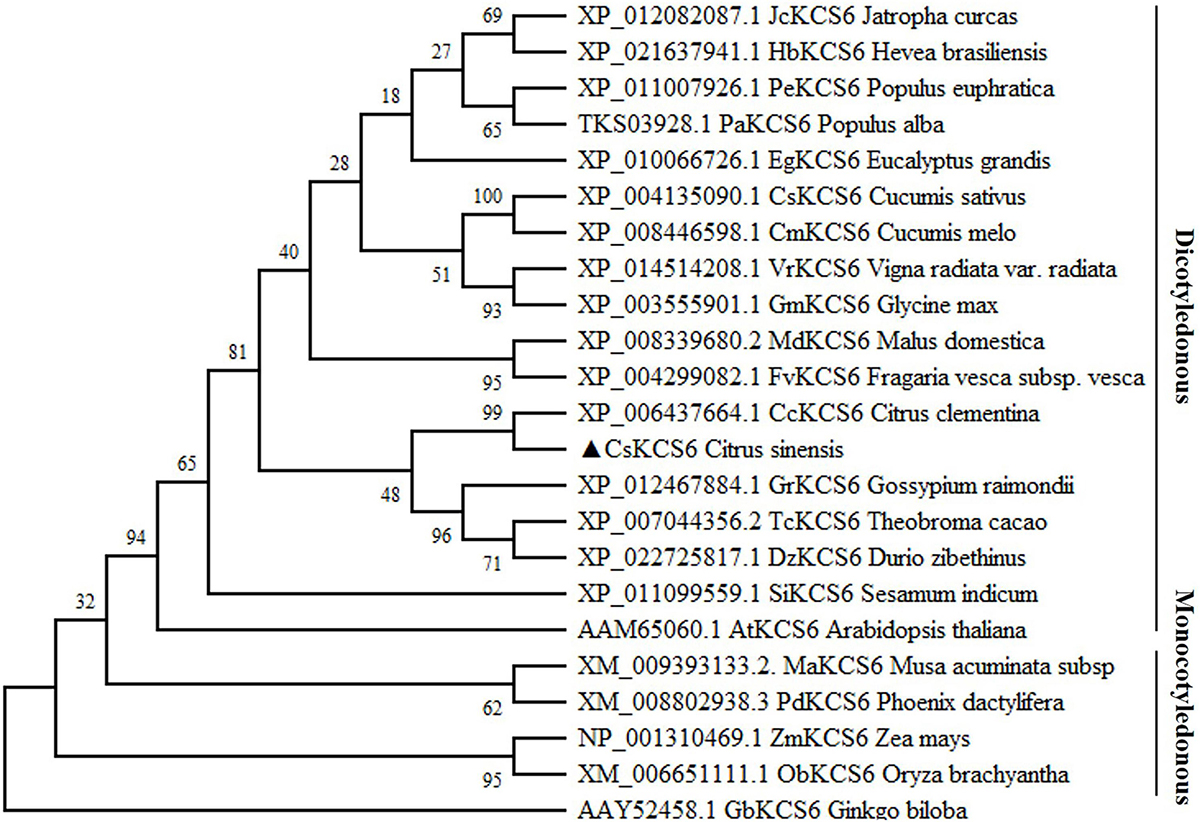
Figure 2. Phylogenetic tree of the homologs of CsKCS6 and other KCS proteins. The solid lines represent monocotyledonous and dicotyledonous plants.
Analysis of the CsKCS6 Expression Pattern in Navel Orange
The spatiotemporal expression of CsKCS6 in Newhall navel orange was determined via qPCR. The results showed that CsKCS6 Significantly high expression in the stigmas, followed by the petals and pistils. But substantially lower expression in tissues such as flavedo, albedo, flesh, leaf, receptacle, and ovary (Figure 3A). The expression level of CsKCS6 in the flavedo and flesh of Newhall navel orange increased with fruit development. However, no significant changes were observed in the albedo of Newhall navel orange during fruit development (Figures 3B–D).
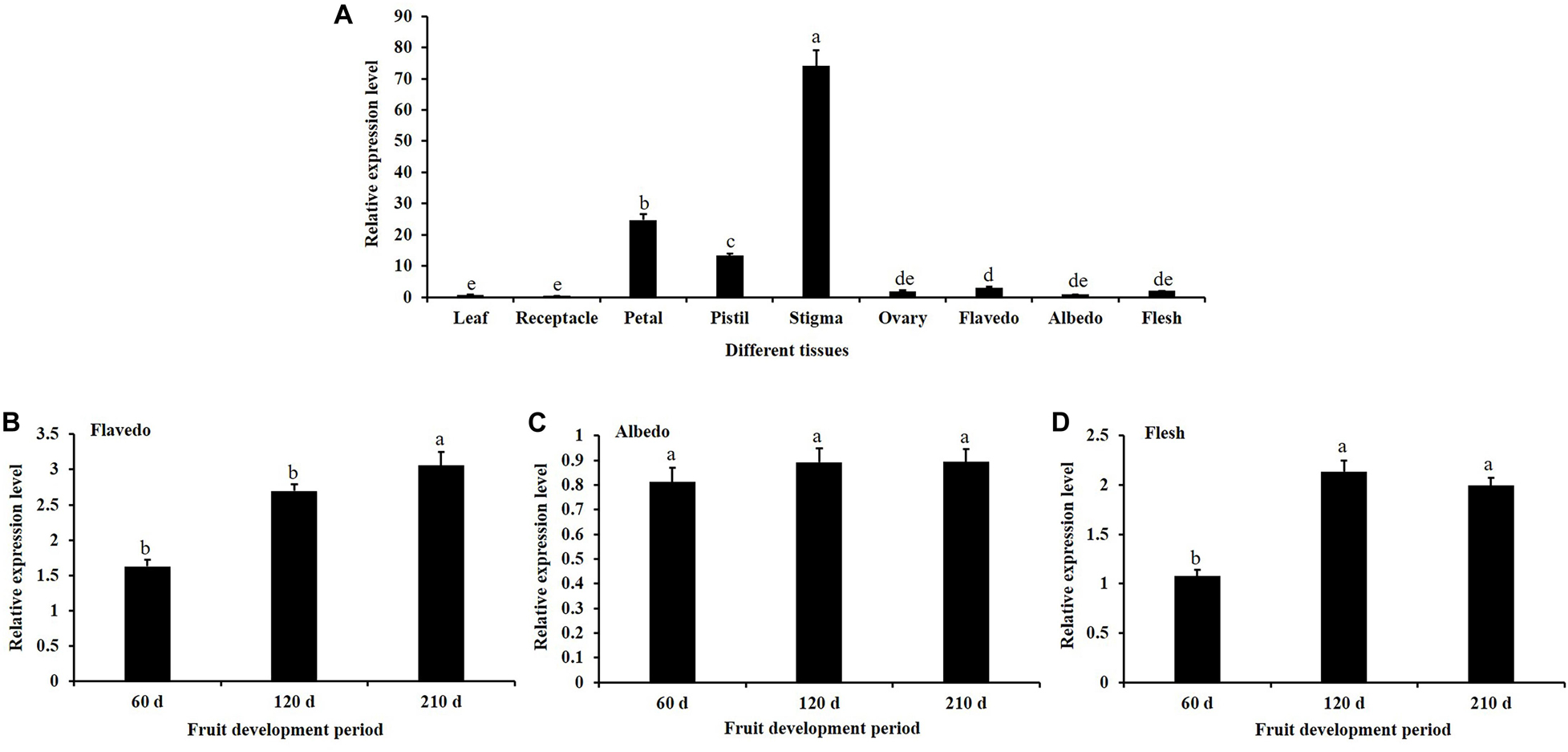
Figure 3. Expression analysis of CsKCS6 in Newhall navel orange. (A) Spatial-specific expression of CsKCS6 in different organs. (B–D) Expression pattern of CsKCS6 in flavedo, albedo and flesh tissues during the fruit development period. The y-axis shows the relative gene expression levels, as calculated by the 2–ΔΔCT method, in which CsACTIN served as the endogenous reference. Data are shown as the means ± SE calculated from three biological replicates. Different small letters represent significant differences at P < 0.05 according to Duncan’s multiple range tests.
In this study, the expression patterns of CsKCS6 gene in leaves of ‘Newhall’ navel orange under abiotic stress were investigated by qPCR. After 4°C treatment, the expression level of CsKCS6 declined significantly at 1 h (about threefold lower than 0 h), remained this level from 1 to 12 h, and then decreased again at 24 h (Figure 4A). Under the 250 mM NaCl stress, the expression level of CsKCS6 decreased significantly at 6 h, increased at 3 h, declined again at 6 h, increased to the maximum value (about 1.5-fold higher than 0 h) at 12 h, and then decreased significantly at 24 h (Figure 4B). After simulated drought treatment with 20% PEG6000, the CsKCS6 transcript decreased significantly at 1 h, increased to the maximum value (about twofold higher than 0 h) at 3 h and then decreased gradually from 3 to 24 h (Figure 4C). After 100 μM ABA treatment, the expression level of CsKCS6 decreased significantly at 1 h, peaked at 3 h (about 2.8-fold higher than 0 h), decreased again from 3 to 9 h. Surprisingly, the CsKCS6 expression increased again to a high level (about 1.5-fold higher than 0 h) at 12 h after ABA treatment, but finally declined at 24 h (Figure 4D).
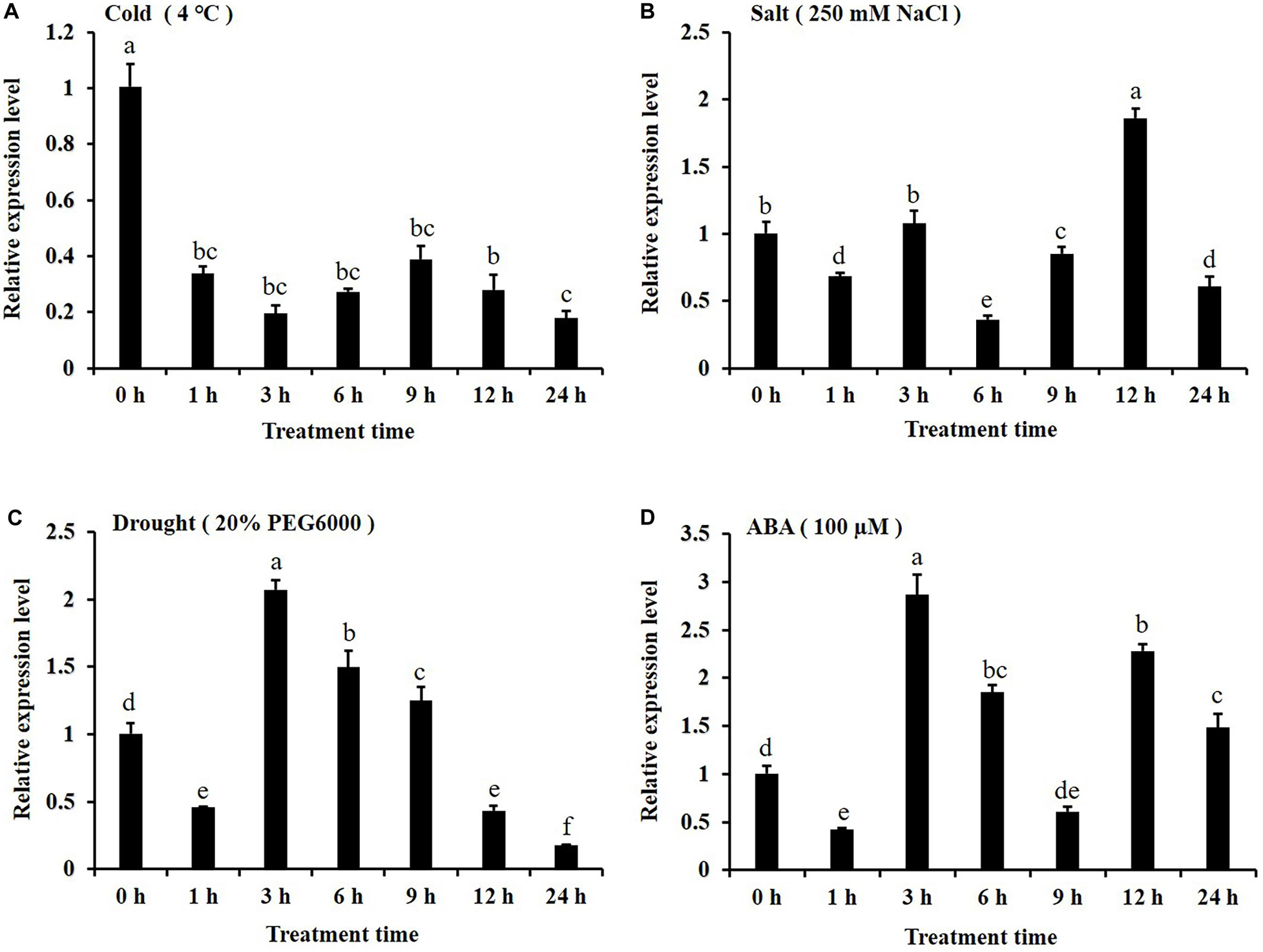
Figure 4. Expression of CsKCS6 in the leaves of Newhall navel orange plants under abiotic stress according to qRT-PCR (A–D) after cold (4°C), salt, drought and ABA treatments. The y-axis shows the relative gene expression levels, as calculated by the 2–ΔΔCT method, in which CsACTIN served as the endogenous reference. Data are shown as the means ± SE calculated from three biological replicates. Different small letters represent significant differences at P < 0.05 according to Duncan’s multiple range tests.
Phenotypic Analysis of Transgenic Arabidopsis Overexpressing CsKCS6
To investigate the function of CsKCS6 in plants, we introduced a 35S:CsKCS6-pCAMBIA1301 vector into Agrobacterium tumefaciens strain LBA4404, which were then transformed into Arabidopsis plants. The T0 generation seeds were screened on MS medium containing 10 mg/L hygromycin to obtain nine resistant seedlings. PCR detection was performed using specific primers of CsKCS6 and Hyg (Supplementary Table S1-P4, P7) after transplanting, and five positive plants were identified (Supplementary Figure S1). The cDNA was derived from the leaves of T2 generation Arabidopsis plants. Semi-quantitative PCR detection showed that CsKCS6 was strongly expressed in the leaves of five transgenic lines, while CsKCS6 was not detected in WT. Therefore, OE-CsKCS6-1 and OE-CsKCS6-2 were used for further research (Figure 5A).
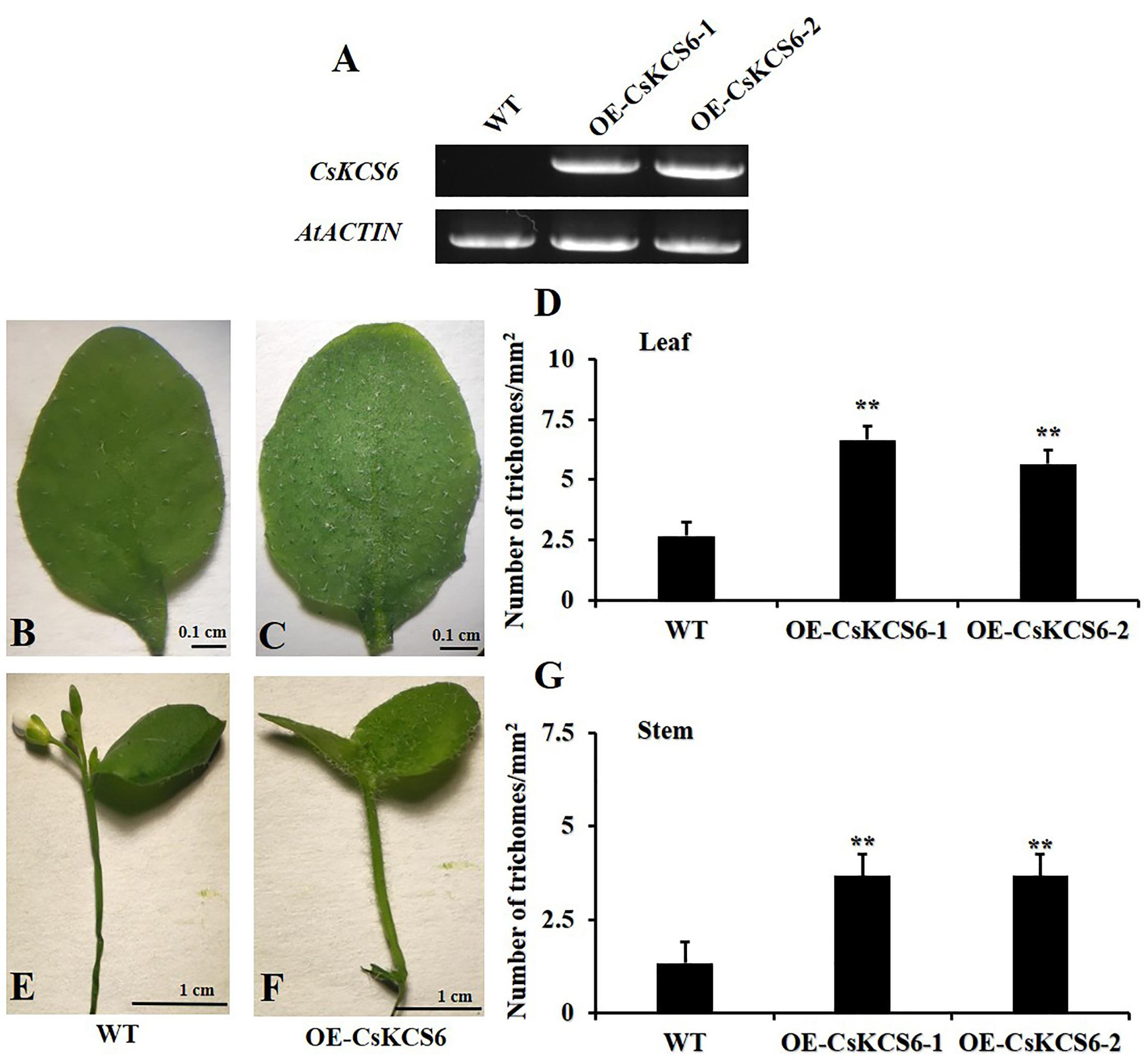
Figure 5. Phenotypes of the WT and two transgenic Arabidopsis lines (OE-KCS6-1, OE-KCS6-2). (A) Expression of CsKCS6 in different leaves of the WT and transgenic Arabidopsis lines. AtACTIN was used as a reference gene. (B) Leaves of the WT and (C) transgenic lines of 6-week-old. (D) The trichome densities in leaves of two Arabidopsis genotypes show in (B,C). (E) Stems of the WT and (F) transgenic lines. (G) The trichome densities in stems of two Arabidopsis genotypes show in (E,F). The data are shown as the means ± SE of three biological replicates. The asterisks indicate that the values of the transgenic Arabidopsis line are significantly different from the values of the WT (∗P < 0.05; ∗∗P < 0.01).
Observing the performance of transgenic Arabidopsis seedlings and wild-type seedlings of the same age, it is found that the trichome density on the surface of the leaves and stems of the transgenic plants is higher than that of the wild-type plants, and the transgenic leaves and stems of the plants are rougher than the wild-type plants (Figures 5B,C,E,F). Subsequently, we obtained photos of transgenic and wild-type Arabidopsis thaliana through stereomicroscopes, and then statistical analysis based on the pictures showed that the number of trichomes per square millimeter on the transgenic Arabidopsis leaves was more than twofold that of the WT, forming a very significant difference (Figure 5D). The number of trichomes on the stems of transgenic Arabidopsis was also significantly higher than that of the WT, with a difference of about threefold (Figure 5G).
Cuticular Wax Morphology and Composition of Wax Present on Leaves of the WT and Transgenic Arabidopsis Plants
Scanning electron microscopy was used to observe the morphology of the cuticular wax present on the leaf surfaces of the WT and transgenic Arabidopsis. The results showed that the trichomes on the leaf surfaces of the transgenic lines were more abundant than those of the WT (Figures 6A,B), this phenotypic difference is consistent with the statistics of trichomes in Figure 5D. However, no significant difference was detected in the morphology of the cuticular wax between the adaxial and abaxial sides of the leaves of the WT and transgenic Arabidopsis plants (Figures 6C–F).
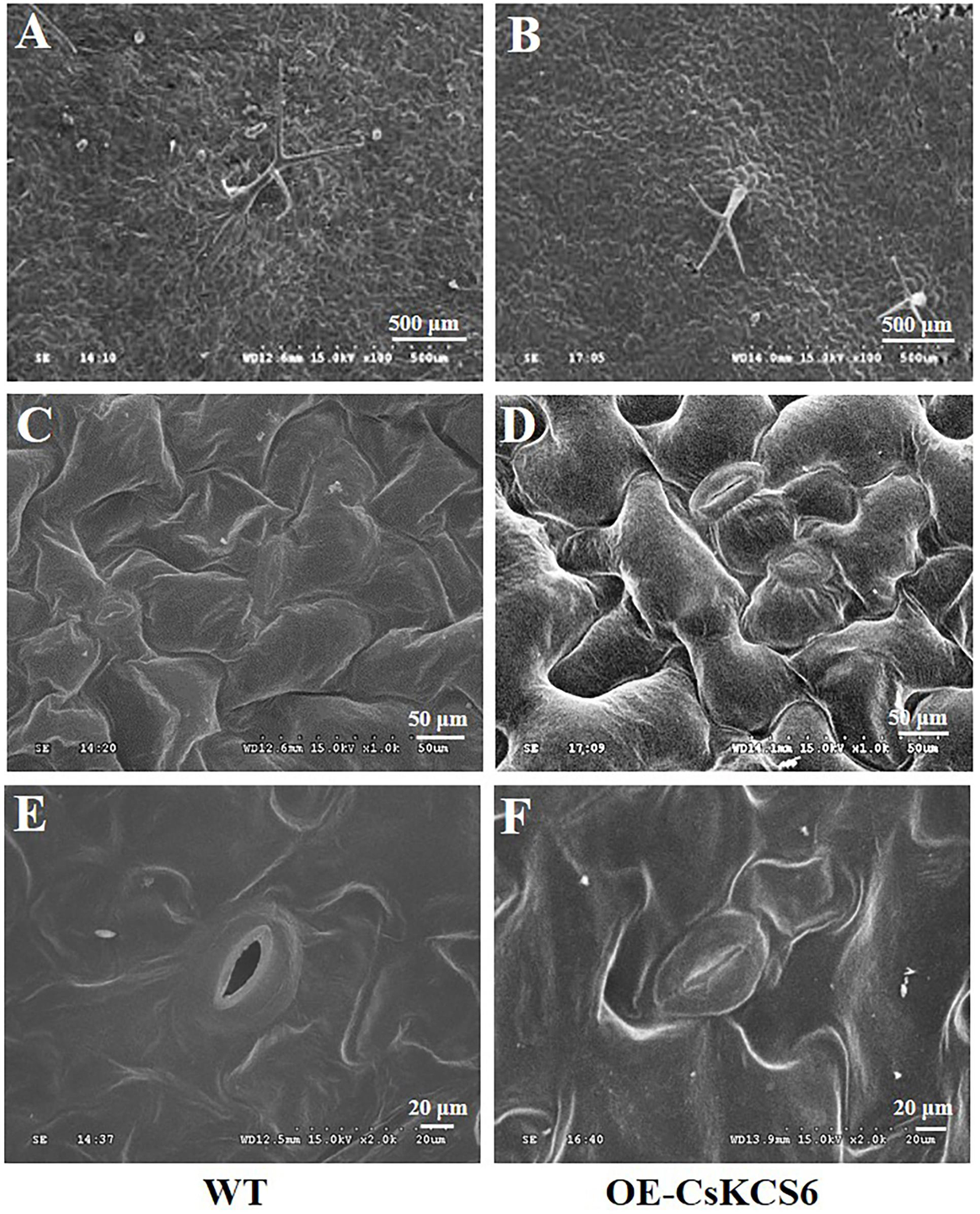
Figure 6. Morphology of the wax of the leaf surfaces of WT and transgenic plants. (A) Wax on the trichomes of WT and (B) transgenic plants (100×, Bar = 500 μm). (C) Wax on the adaxial side of the leaves of the WT and (D) transgenic plants (1000×, Bar = 50 μm). (E) Wax on the abaxial side of the leaves of WT and (F) transgenic plants (2000×, Bar = 20 μm).
GC–MS analysis revealed that the total wax content of the two transgenic lines was similar to that of the WT (Figure 7A). The amount of fatty acids in the transgenic lines was about twofold higher than that in the WT plants. However, the contents of primary alcohols and sterols in the transgenic strain lines were reduced about 2- and 1.5-fold respectively in compare with the WT. There was no significant difference in the contents of alkanes and secondary alcohols (Figure 7B). Meanwhile, the two transgenic lines had much greater amounts (increased by about 40%) of fatty acids with even chain length ≥ C24 compared to the WT plants. But the amounts of C32 and C34 primary alcohol reduced by 35% in the transgenic lines (Figure 7C).
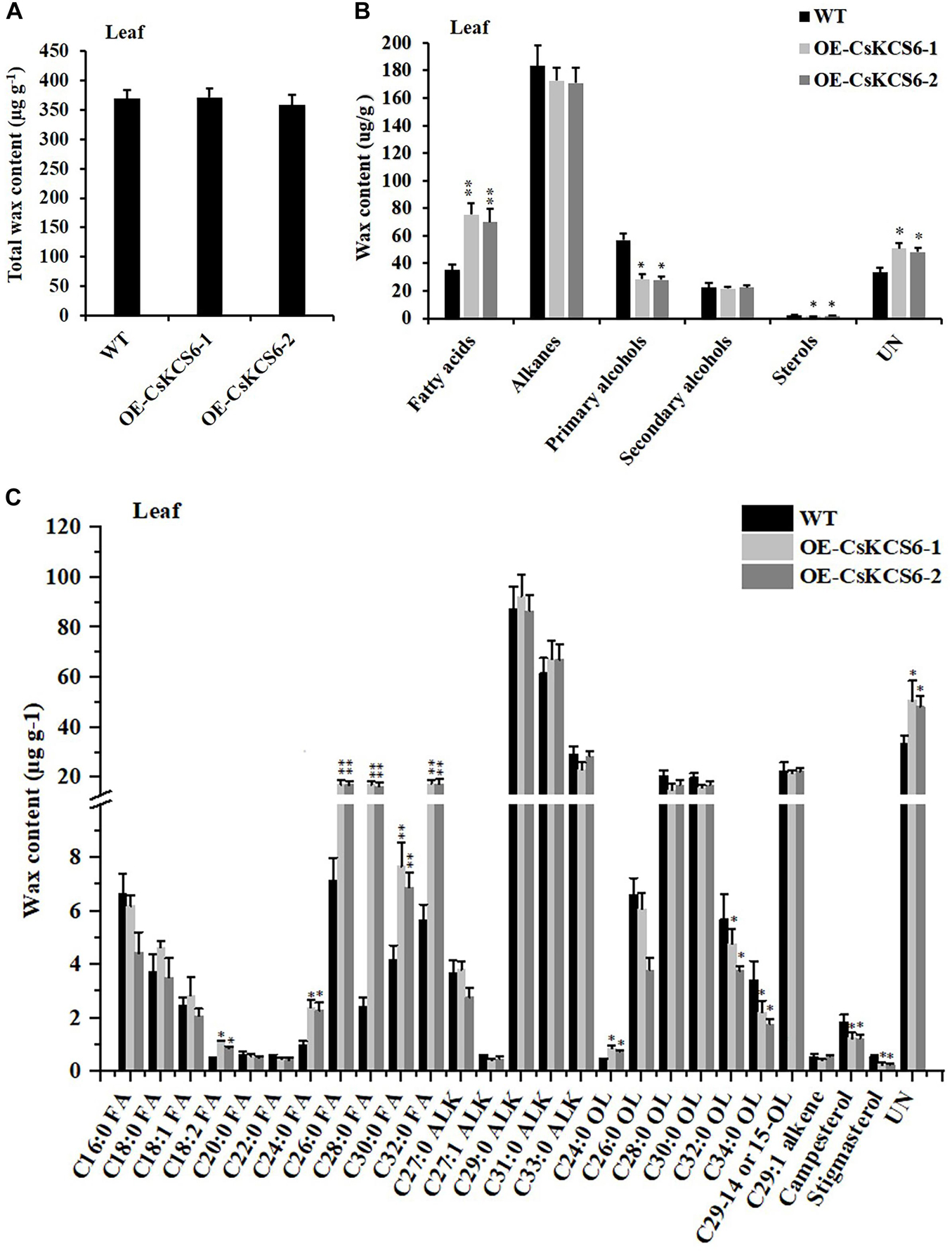
Figure 7. Cuticular wax content and composition of leaves of WT and transgenic Arabidopsis plants. (A) Total wax content, (B) wax composition, and (C) identified waxes in the leaves of the WT and two transgenic plants. FA, fatty acids; ALK, alkanes; OL, alcohols; UN, unknown component. The values are means ± SE of three biological replicates, and the asterisks indicate significant differences between WT plants and transgenic lines (*P < 0.05; **P < 0.01).
Morphology and Composition of Cuticular Wax on the Stems of WT and Transgenic Arabidopsis Plants
Scanning electron microscopy was used to observe the morphology of the cuticular wax on the surface of the stems of WT and transgenic Arabidopsis plants. The results showed that obviously more trichomes were present on the surfaces of the stems of the transgenic Arabidopsis plants than on those of the WT plants (Figures 8A,B), this phenotypic difference is consistent with the statistics of trichomes in Figure 5G. There were many granular wax crystals deposited on the surface of the stems of both genotypes (Figures 8C,D). In addition, many shallow ridges were detected on the stem surfaces of the transgenic Arabidopsis plants, which were not observed on the leaf surfaces of the WT plants.

Figure 8. Morphology of the cuticular wax on the stem surfaces of WT and transgenic Arabidopsis. (A) Trichomes of WT stems and (B) trichomes of transgenic Arabidopsis stems (100×, Bar = 500 μm). (C) Wax on the surfaces of the stems of WT and (D) transgenic Arabidopsis stems (2000×, Bar = 20 μm).
GC–MS analysis revealed that the total wax content on the stem surfaces of the two transgenic lines was similar to that of the WT (Figure 9A). The total amount of fatty acids was increased by more than twofold compared with the WT plants. The alkanes amount in the transgenic lines increased by only about 18% in comparison to the WT plants. In contrast, the amounts of primary alcohols, aldehydes, and ketones decreased significantly in the transgenic lines. The most drastic decline was observed in aldehydes, which reduced by more than 30% in the transgenic lines (Figure 9B). Notably, the increase in fatty acids content was mainly observed in even chain length chains of C16 and C26–30. And the decrease of aldehydes was mainly attribute to the reduction of C26, C28, and C30 aldehyde (Figure 9C).
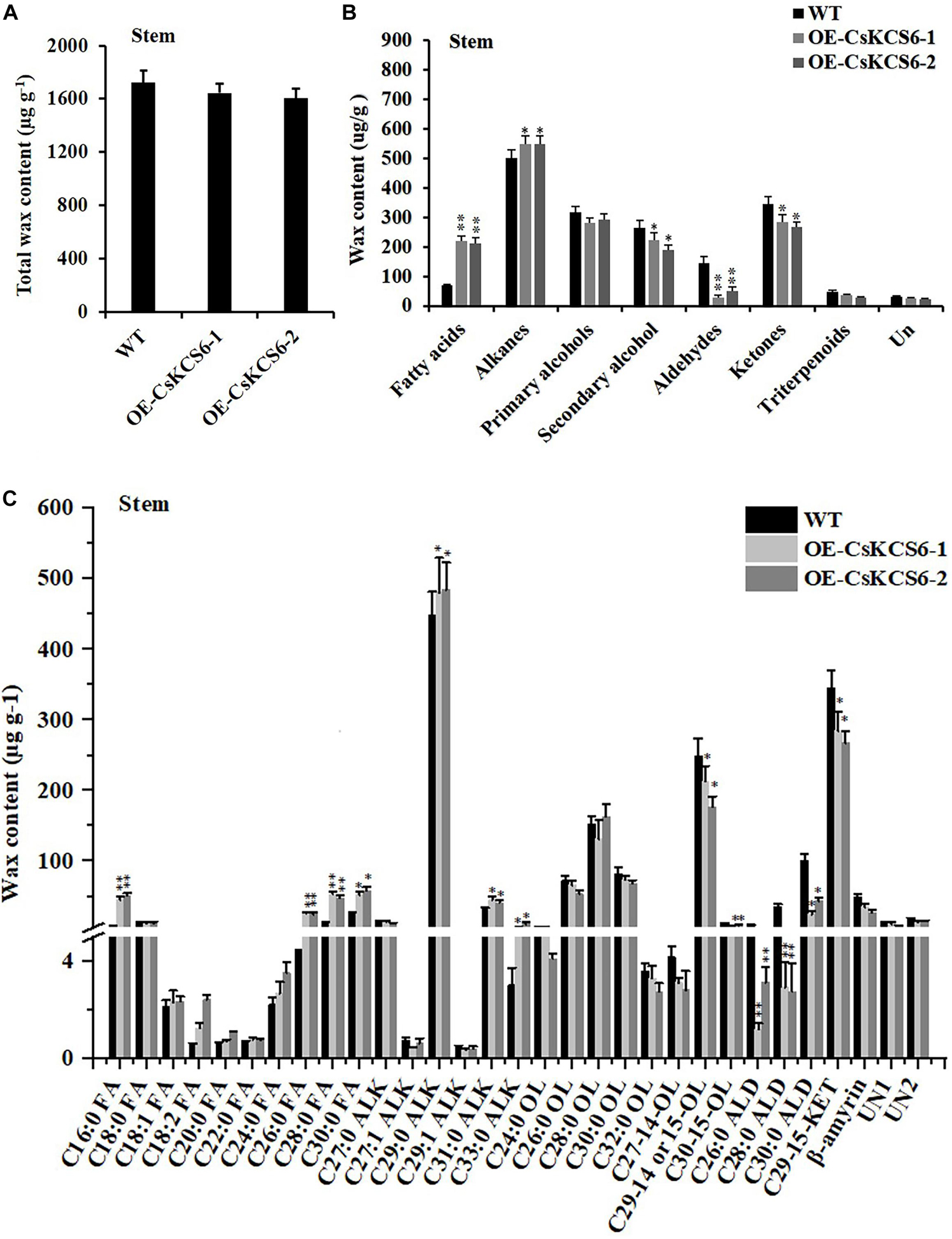
Figure 9. Content and composition of the cuticular wax of the stems of WT and transgenic Arabidopsis plants. (A) Wax content, (B) wax composition, and (C) identified waxes in the stems of the WT and two transgenic plants. FA, fatty acids; ALK, alkanes; OL, alcohols; ALD, aldehydes; KET, ketones; UN, unknown component. The values are the means ± SE of three biological replicates, and the asterisks indicate significant differences between the WT and transgenic lines (*P < 0.05; **P < 0.05).
Analysis of Cuticular Permeability of the Leaves
To explore whether the epidermal permeability was altered in CsKCS6 transgenic lines, the epidermis of leaves collected from the CsKCS6 transgenic lines and WT was stained with TB, and the chlorophyll leaching contents was measured under alcohol treatment. The results showed that both sides of the leaves of the WT were stained much deeply than were the transgenic lines (Figure 10A). Furthermore, the chlorophyll leaching contents of the WT and transgenic lines continuously increased after the alcohol treatment; however, the chlorophyll leaching contents of the transgenic lines was lower than that of the WT at all the time points of the alcohol treatment, and the difference was significant after 60 min, and the chlorophyll leaching content extracted was only about half of that of the WT at 180 min (Figure 10B).
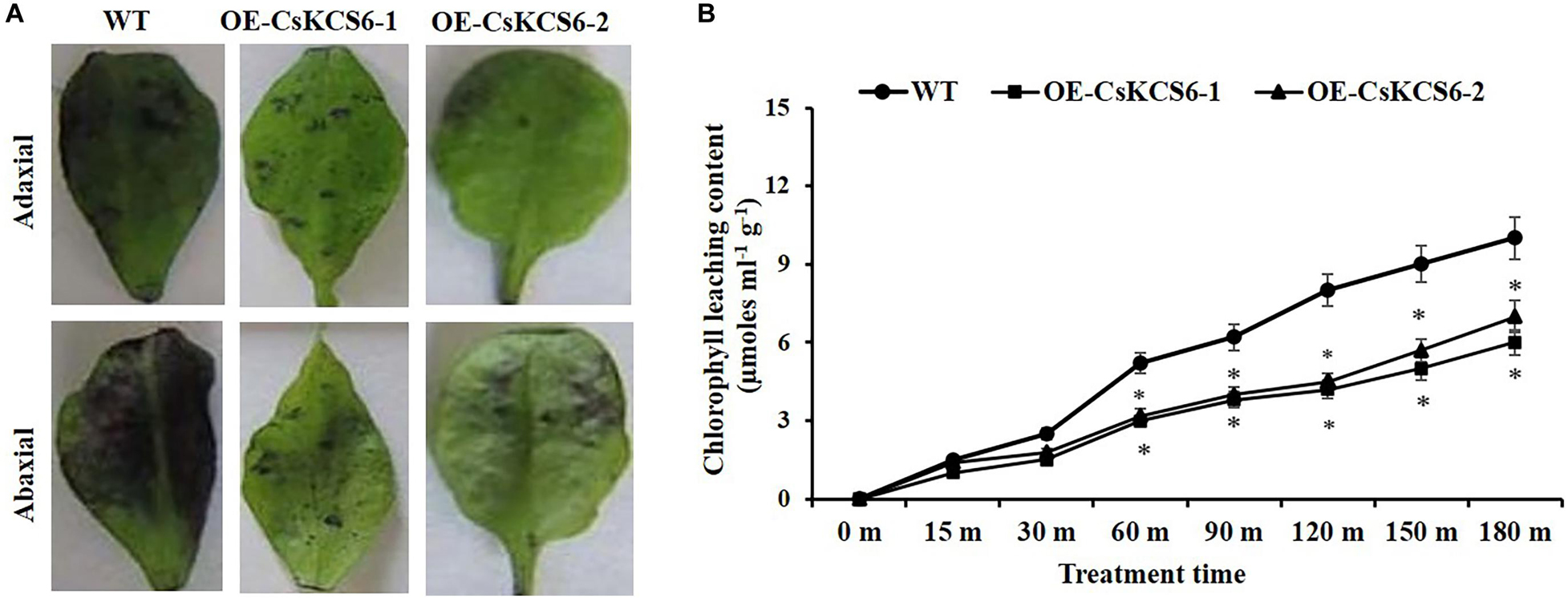
Figure 10. Alterations in cuticle permeability of CsKCS6 transgenic Arabidopsis. (A) TB staining, (B) chlorophyll leaching content of the WT and transgenic Arabidopsis plants after alcohol treatment.
Ectopic Expression of CsKCS6 in Arabidopsis Enhanced Drought Tolerance
To study whether CsKCS6 is involved in the response to osmotic stress, the plant phenotype, survival, water loss rate and ion leakage of the WT and two transgenic lines were analyzed. After drought treatment for 10 days and rewatering for 3 days, the leaf wilting of the WT plants was more severe than that of the two transgenic lines (Figures 11A,B). Moreover, the survival percentages of OE-KCS6-1 and OE-KCS6-2 were 21.66 and 23.33%, respectively, which were significantly higher than the survival rate of the WT plants (6.66%) after drought treatment (Figure 11C).
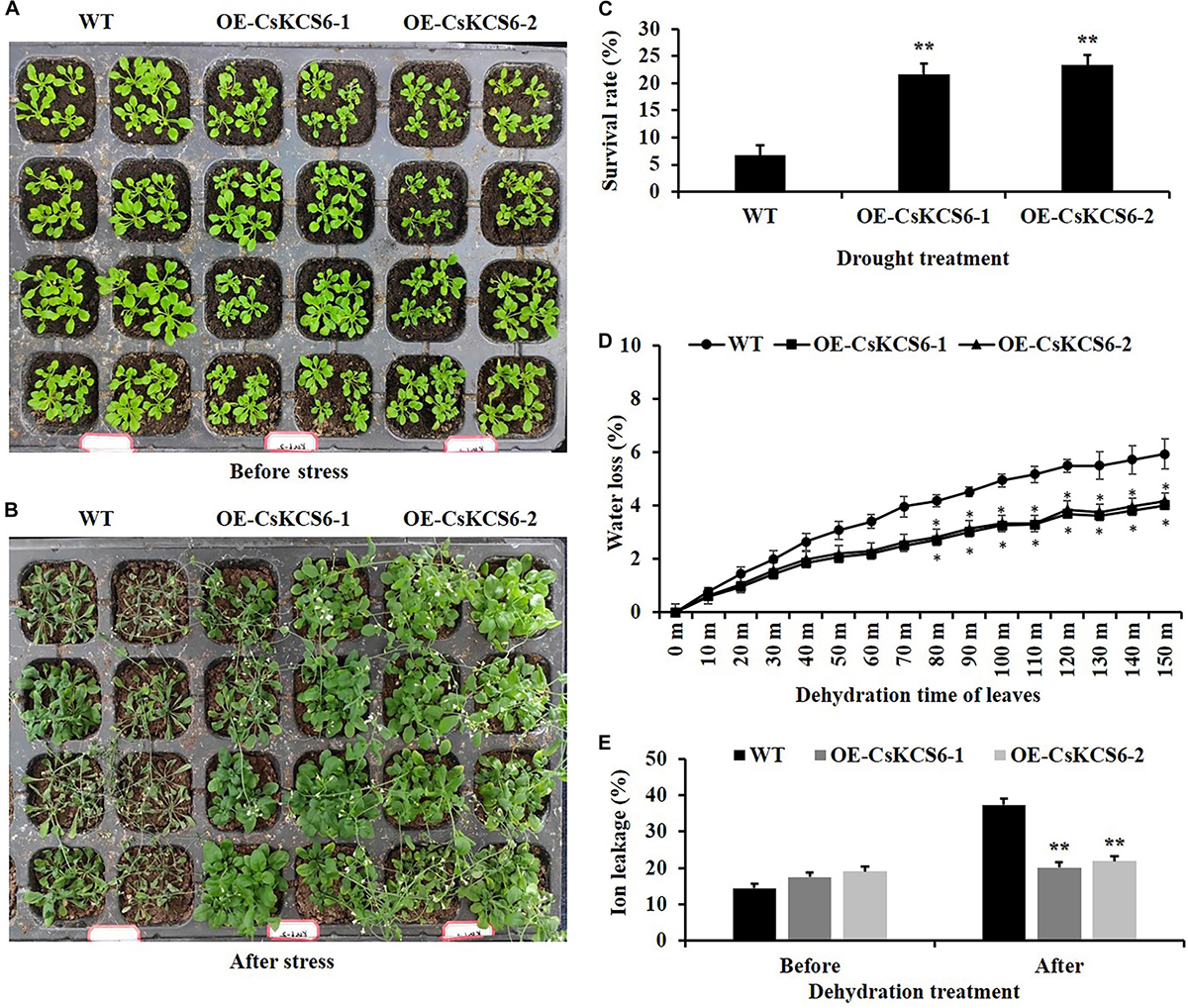
Figure 11. Phenotypes and physiological traits of WT and transgenic Arabidopsis lines after drought treatments. (A,B) Phenotypes of two genotypes after drought treatment, as well as their (C) survival rate, (D) water loss rate, and (E) ion leakage. Leaves of 6-week-old WT and transgenic plants were collected for measuring the physiological traits. The data are shown as the means ± SE of three biological replicates. The asterisks indicate that the values of the transgenic Arabidopsis line are significantly different from the values of the WT (*P < 0.05; **P < 0.01).
In addition, the transpiration rates of the leaves of the transgenic Arabidopsis plants were significantly lower than those of WT plants from 80 to 150 min after dehydration (Figure 11D). Compared with that of the WT plants, the ion leakage in the leaves of the transgenic Arabidopsis plants was also obviously reduced after the dehydration treatment. These results indicate that ectopic expression of CsKCS6 can reduce the cell membrane permeability of Arabidopsis leaves (Figure 11E). Taken together, these results suggest that ectopic expression of CsKCS6 can improve the drought tolerance of transgenic Arabidopsis.
Ectopic Expression of CsKCS6 in Arabidopsis Enhanced Salt Tolerance
To analyze the role of CsKCS6 in the plant response to salt stress, we investigated the effects of salt stress on the seed germination and root length of the WT and two transgenic lines. The seed germination rate of the two transgenic Arabidopsis lines was significantly lower than that of the WT within the first 5 days of 100 mM NaCl treatment. Interestingly, the germination rate of the two transgenic lines was similar to that of the WT plants after 8 days of 100 mM NaCl treatment (Figures 12A,B).
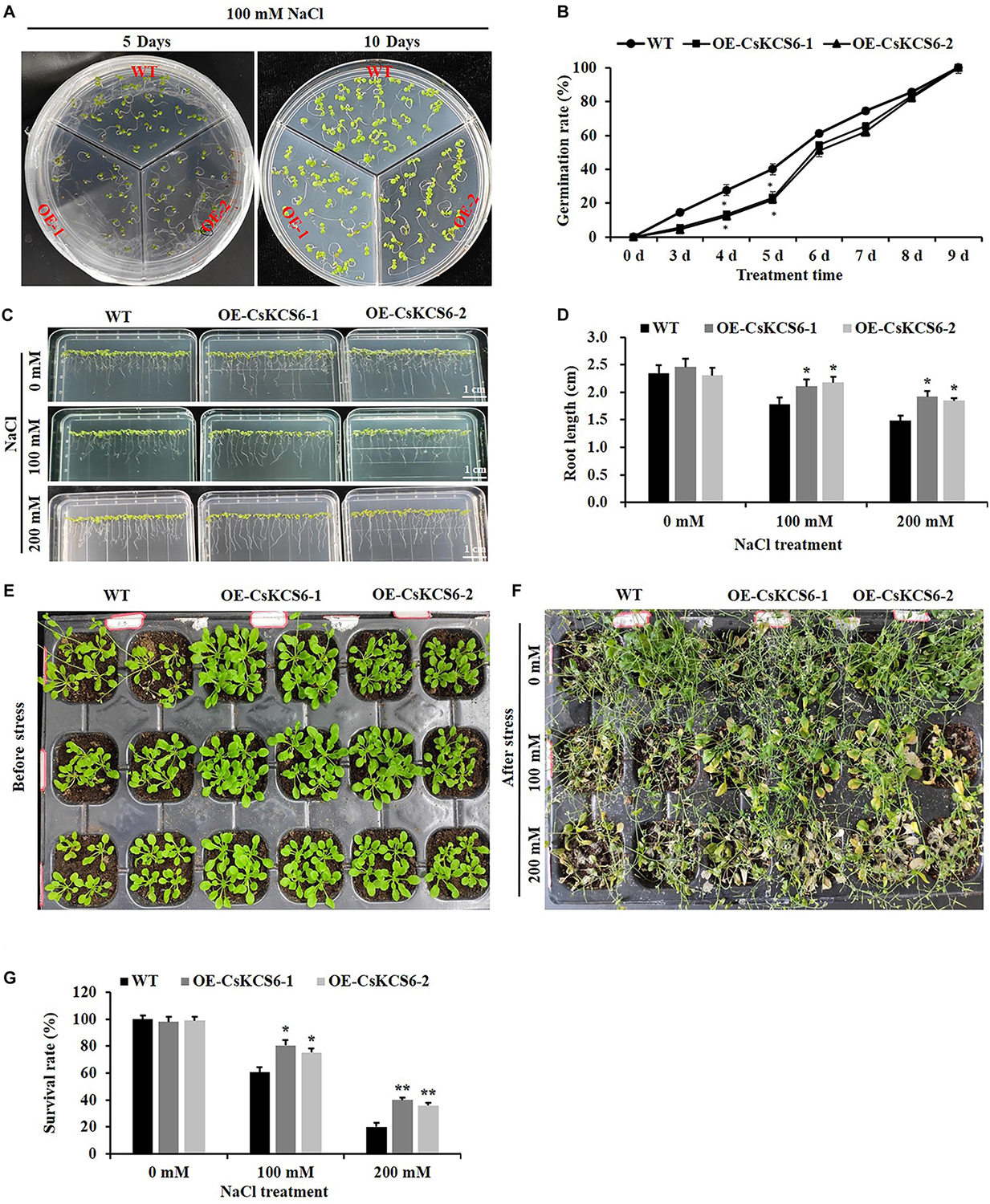
Figure 12. Phenotypes and physiological traits of the WT and transgenic Arabidopsis lines after salt treatment. (A) Seed germination. T2 and WT seeds were sown on MS media, with 30 seeds sown per line. (B) Germination rates calculated from (A), (C) root growth under salt stress treatment. (D) Root length (root length ≥ 5 mm) calculated from (C), (E,F) phenotypes of two genotype lines after salt treatment. (G) Survival. The data are shown as the mean ± SE of three biological replicates. The asterisks indicate significant differences between transgenic Arabidopsis lines and WT (*P < 0.05; **P < 0.01).
Statistical analysis showed that there was no significant difference in root length between the 2-week-old transgenic and wild-type Arabidopsis, and both could reach about 2.4 cm. However, after 100 and 200 mM NaCl treatment, the root length of the transgenic lines was about 2.2 and 1.8 cm respectively, which was significantly higher than that of the WT plants (about 1.7 and1.4 cm) (Figures 12C,D).
In addition, we compared the phenotypic differences between the WT and two transgenic lines after salt stress treatment, and more severe damage was observed in the WT plants than in the two transgenic lines after the 100 and 200 mM NaCl treatments (Figures 12E,F). Furthermore, after 100 mM NaCl treatment, the survival rate of wild-type Arabidopsis is only 60%, while the survival rate of transgenic lines reaches about 80%. When the concentration of NaCl treatment reaches 200 mM, the survival rate of the transgenic lines (40%) was about twofold higher than that of the WT (20%) (Figure 12G).
Discussion
Very-long-chain fatty acids are biosynthesized from the extension of C16 or C18 fatty acids, and the extension is catalyzed by the multienzyme complex FAE; thus, FAE components are direct precursors of cuticular wax biosynthesis. The first step of VLCFA biosynthesis is to produce β-ketoacyl-CoA, which is catalyzed by KCS (Kunst and Samuels, 2003; Yeats and Rose, 2013).
Members of the KCS family are often described as 3-ketoacyl-CoA synthases, type III polyketide synthases and FAEs, while the characteristic regions of the KCS6 protein usually contain active site residues and motifs involved in substrate binding (Funa et al., 2002). In this study, multiple sequence alignment revealed that the amino acid sequence of CsKCS6 contained two conserved domains, FAE1-CUT1-RppA and ACP_syn_III_C, which belong to the thiolase-like superfamily. The Cys residue at amino acid position of 225 is the catalytic active site of the condensing enzyme and is highly conserved among different plant species (Figure 1). These conserved sites are also present in KCS family proteins from other plant species. In addition, phylogenetic analysis revealed that CsKCS6 clustered with other KCS6 proteins from dicotyledonous plant species, especially CcKCS6, which comes from Citrus clementine and is the closest homolog of CsKCS6 (Figure 2). The structural features and phylogenetic results of this protein suggested that CsKCS6 belongs to the KCS gene family and might be involved in regulating the synthesis of VLCFAs in citrus.
The expression patterns of KCS family genes have been reported in many studies. For example, tissue-specific expression analysis of 21 KCS family genes in Arabidopsis showed that AtKCS21, AtKCS7 and AtKCS15 were underexpressed in Arabidopsis tissues; that AtKCS18/AtFAE1 and AtKCS19 were preferentially expressed in the seeds; that AtKCS10/AtFDH, AtKCS4, AtKCS9, AtKCS11, AtKCS20, AtKCS2/AtDAISY, and AtKCS1 are widely expressed in various tissues of Arabidopsis; and that these proteins may be involved in VLCFA synthesis required for growth and development (Suh et al., 2005). However, AtKCS17 is expressed only in the flowers and siliques (Joubès et al., 2008). It has been reported that AtKCS6 is highly expressed in the flowers and siliques but not in the roots (Costaglioli et al., 2005; Suh et al., 2005). In agreement with the information in these reports, CsKCS6 is highly expressed in the flowers of navel orange, especially in the stigmas (Figure 3). In our previous study, we reported that the amounts of VLCFAs continuously increased in the epicuticular wax of Newhall navel orange during fruit development. Accordingly, the expression of CsKCS6 in the flavedo and albedo of Newhall navel orange also increased during fruit development (Figure 3). Thus, CsKCS6 might be involved in the biosynthesis of VLCFAs for the cuticular wax of navel orange (Liu et al., 2015).
Cuticular wax synthesis in plants is induced by various environmental factors, such as light, water deficit and low temperature (Shepherd and Wynne Griffiths, 2006). The expression of many cuticular wax genes, including KCS family genes, also shifted under abiotic stress. For example, the expression levels of Arabidopsis AtKCS1 and AtKCS3 were downregulated under darkness and low temperature, and the expression of AtKCS10 decreased under various kinds of osmotic stress (Joubès et al., 2008). In the present study, the expression level of CsKCS6 significantly declined under low temperature stress and obviously increased at several time points after high salt, drought and ABA treatment (Figure 4). This result was in agreement with the report of Joubès et al. (2008), which indicated that the expression level of Arabidopsis AtKCS6 was decreased under low temperature stress but increased after high salt and drought treatment. Similar result also reported by Hooker et al. (2002), which suggested that osmotic stress and ABA enhance the transcription of AtKCS6/AtCER6 (Hooker et al., 2002). Interestingly, the expression levels of CsKCS6 were fluctuated after high salt, drought and ABA treatments (Figure 4). As reviewed by Grundy et al. (2015), the circadian clock controls expression of a large number of genes that are responsive to abiotic stress. Thus, many abiotic stress-responsive genes were found to be expressed rhythmically under diurnal light–dark or temperature cycles. Furthermore, rhythmic expression of abiotic stress-responsive genes under constant conditions was also reported for many plant species, such as Arabidopsis, soybean and barley (Covington et al., 2008; Habte et al., 2014; Marcolino-Gomes et al., 2014). It has been proved that dark can suppress the expression of Arabidopsis AtKCS6 (Hooker et al., 2002; Joubès et al., 2008). Thus, the CsKCS6 transcript was probably under the control of circadian clock, leading to the fluctuated changes in CsKCS6 expression after high salt, drought and ABA treatments.
To further study whether CsKCS6 participates in the biosynthesis of VLCFAs for cuticular wax and regulates the osmotic stress tolerance of navel orange, transgenic Arabidopsis lines ectopic expression CsKCS6 were generated in this study. Phenotypic observations and SEM analysis revealed that the density of trichomes on the leaves and stems of transgenic Arabidopsis was significantly higher than that of the WT plants (Figures 5, 6, 8), suggesting CsKCS6 might be involved in the development of trichomes. To our surprise, although AtKCS16 is proved to catalyze the elongation of C34 to C38 acyl-CoAs in Arabidopsis leaf trichomes (Hegebarth et al., 2017), none of the KCS6 homologous genes and other KCS family genes were reported to be related to trichome formation (Fiebig et al., 2000; Hooker et al., 2002; Serra et al., 2009; Weidenbach et al., 2015). The divergence between our result and previous reports is probably attributed to the gene function difference among plant species. Trichomes play an important role in plant resistance to biotic and abiotic stresses (Choi, 2010; Kim et al., 2012). Thus, the increase in trichome numbers in the transgenic Arabidopsis plants might enhance their tolerance to abiotic stresses.
Arabidopsis AtCER6 is also named AtCUT1 and AtKCS6 (Fiebig et al., 2000). In cer6 mutants of Arabidopsis, wax crystals are nearly undetectable in the stems and siliques due to the loss of AtCER6 function (Millar et al., 1999; Fiebig et al., 2000). Moreover, overexpression of AtCER6/AtKCS6 in Arabidopsis leads to waxless phenotype in stems of transgenic lines (Millar et al., 1999). Surprisingly, in the present study, no significant differences in the structure or total amount of wax were observed on the surfaces of the leaves and stems between the WT and transgenic Arabidopsis plants. Similar to our result, although high levels of AtCER6/AtKCS6 expression were observed in the hemizygous 35S-AtCER6/AtKCS6 plants, they did not exhibit higher wax amounts than those in WT plants (Millar et al., 1999). The total amount of cuticular wax in tuber periderms of StKCS6-silenced lines was also not significantly different from the wild-type potato (Serra et al., 2009). KCSs catalyze the rate-limiting step in VLCFA elongation and have substrate specificity (Joubès et al., 2008). Arabidopsis AtKCS6 catalyzes the elongation of C24 and C26 VLCFAs (Jenks et al., 1994; Millar et al., 1999). Tomato LeCER6 and Potato StKCS6 are both involved in the biosynthesis of VLCFAs beyond C28 (Leide et al., 2007; Serra et al., 2009). Cotton GhCER6 is responsible for the formation of C26 VLCFA (Qin et al., 2007). In agree with these reports, the amounts of VLCFAs with an even-numbered carbon chain length greater than that of C24 in the leaves and greater than that of C26 in the stems of the two transgenic lines were significantly higher than those in the WT plants, suggesting CsKCS6 is involved in the biosynthesis of VLCFAs with chain length beyond C24. In addition to the striking differences in the accumulation of VLCFAs, ectopic expression of CsKCS6 also decreased the amounts of primary and secondary alcohols, aldehydes and ketones, especially primary alcohols with chain length beyond C28 in leaves (decreased by 30%) and aldehydes with chain length longer than C26 in stems (decreased by 50%) (Figures 7, 9). The significant decreases of primary alcohols and aldehydes in transgenic plants suggested that either some wax biosynthetic genes might be suppressed by ectopic expression of CsKCS6 or that a large block in VLCFAs entering the wax biosynthetic pathway might be occurred in transgenic plants. The latter hypothesis was supported by Von Wettstein-Knowles (1979), who suggested that VLCFA channeling was probably occurred in plants so that the substrates destined for specific wax components were not freely changed.
The permeability of the cuticle plays an important role in the adaptation of plants to osmotic stress, which affects the water retention capacity of plants and delays the damage of plants from environmental stress (Yeats and Rose, 2013). Recently, several KCS family genes from different plant species were reported to play an important role in regulating cuticle permeability and plant tolerance to abiotic stress. For example, overexpression of AhKCS1 from a drought tolerant groundnut reduced the cuticle permeability and enhanced the drought tolerance in a susceptible genotype by increasing cuticular wax load in leaves (Lokesh et al., 2019). Overexpression of BnKCS1-1 and BnKCS1-2 in Brassica napus also increased drought tolerance in transgenic plants by promoting cuticular wax production (Wang et al., 2020). As reviewed by Singh et al. (2018), a number of other genes related to wax biosynthesis also have the function to enhance the resistance to abiotic stress by altering wax composition, such as WIN1/SHN1, DWA1, MYB96, CER1, CER3/WAX2, FAR family genes and so on. In agreement with these reports, a significant decrease in leaf chlorophyll permeability, TB staining, water loss rate, ion leakage and an obvious increase in the root length and survival rate were observed in the transgenic plants under drought and high salt stress (Figures 10–12), indicating that ectopic expression of CsKCS6 can reduce the cuticle permeability and enhance plant tolerance to drought and high salt. The mechanism of KCS6-like genes to influence cuticle permeability in tomato (Vogg et al., 2004; Leide et al., 2007) and potato (Serra et al., 2009) is well-established. In detail, for the tomato lecer6 mutant deficiency in KCS6, a decline in aliphatic components such as alkanes with chain length beyond C30 and an increase in cyclic triterpenoids lead to the increase in cuticle permeability (Vogg et al., 2004; Leide et al., 2007). Serra et al. (2009) reported that both the significant decrease in aliphatic compounds with chain length beyond C28 and total wax coverage in tuber periderm of StKCS6-silenced potato should be responsible for the increase of cuticle permeability. Since the wax morphology and the total wax load in the transgenic lines were similar to the WT plants, it is reasonable to deduce that the dramatic increase (about twofold) in VLCFAs with chain length beyond C24 in leaves of transgenic plants might be the major reason for the decrease in cuticle permeability. This result further supports the suggestion that cuticular wax composition rather than total wax load predominantly affects the cuticle permeability in plants (Leide et al., 2007; Isaacson et al., 2009; Parsons et al., 2012).
Conclusion
In summary, in the present study, the KCS family gene CsKCS6, which is mainly highly expressed in flowers of Newhall navel orange, was cloned from this species. Drought, salt, and ABA treatment could increase the transcript levels of CsKCS6 in leaves of Newhall navel orange. Ectopic expression of CsKCS6 in Arabidopsis promoted a significant increase in the total VLCFA content of the leaves and stems but resulted in no significant change in total wax content. Further study revealed that the drought tolerance and salt tolerance of the transgenic Arabidopsis lines were significantly enhanced compared with those of the WT plants. Our results indicate that CsKCS6 can enhance plant tolerance to drought and high salt stress by regulating the biosynthesis of VLCFAs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
Author Contributions
QW, WG, and DL originally formulated the idea. LY, WH, and YL developed methodology. WG performed statistical analyses and wrote the manuscript. All authors edited and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 31660563, 31701896, and 31860544) and by the earmarked fund for Jiangxi Agriculture Research System (Grant No. JXARS-07-cultivation post).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.564656/full#supplementary-material
Supplementary Figure S1 | Identification of positive transgenic plants by PCR. (A) The T1 generation transgenic lines of Arabidopsis were detected by specific primers of CsKCS6 and Hyg genes to obtain positive plants. (B) Semi-quantitative detection of the expression level of transgenic Arabidopsis.
Supplementary Table S1 | Primer sequences used for CsKCS6 cloning, expression analysis, vector construction, and transgenic confirmation.
Footnotes
References
Bernard, A., Domergue, F., Pascal, S., Jetter, R., and Renne, C. (2012). Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very long-chain alkane synthesis complex. Plant Cell 24, 3106–3118. ne biosynthesis by directly doi: 10.1105/tpc.112.099796
Bourdenx, B., Bernard, A., Domergue, F., Pascal, S., Léger, A., Roby, D., et al. (2011). Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol.156, 29–45.
Chen, W. W., Yu, X. H., Zhang, K. S., Shi, J. X., Sheron, D. O., Zhang, D. B., et al. (2011). Male sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in arabidopsis. Plant Physiol. 157, 842–853. doi: 10.1104/pp.111.181693
Choi, Y. E. (2010). Expression profiling of tobacco leaf trichomes identifies genes for biotic and abiotic stresses. Plant Cell Physiol. 51, 1627–1637. doi: 10.1093/pcp/pcq118
Costaglioli, P., Joubès, J., Garcia, C., Stef, M., and Arveiler, B. (2005). Profiling candidate genes involved in wax biosynthesis in Arabidopsis thaliana by microarray analysis. Biochim. Biophys. Acta1734, 247–258. doi: 10.1016/j.bbalip.2005.04.002
Covington, M. F., Maloof, J. N., Straume, M., Kay, S. A., and Harmer, S. L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9:R130. doi: 10.1186/gb-2008-9-8-r130
Dai, W. S., Wang, M., Gong, X., and Liu, J. H. (2018). The transcription factor fcwrky40 of fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating sos2 and p5cs1 homologs. New Phytol. 219, 972–989. doi: 10.1111/nph.15240
Fiebig, A., Mayfield, J. A., Miley, N. L., Chau, S., Fischer, R. L., and Preuss, D. (2000). Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12, 2001–2008. doi: 10.1105/tpc.12.10.2001
Funa, N., Ohnishi, Y., Ebizuka, Y., and Horinouchi, S. (2002). Alteration of reaction and substrate specificity of a bacterial type III polyketide synthase by site-directed mutagenesis. Biochem. J. 367, 781–789. doi: 10.1042/BJ20020953
Grundy, J., Stoker, C., and Carré, I. A. (2015). Circadian regulation of abiotic stress tolerance in plants. Front. Plant Sci. 6:48. doi: 10.3389/fpls.2015.00648
Guo, H. S., Zhang, Y. M., Sun, X. Q., Li, M. M., and Xue, J. Y. (2015). Evolution of the kcs gene family in plants: the history of gene duplication, sub/neofunctionalization and redundancy. Mol. Genet. Genomics 291, 739–752. doi: 10.1007/s00438-015-1142-3
Habte, E., Muller, L. M., Shtaya, M., Davis, S. J., and Von Korff, M. (2014). Osmotic stress at the barley root affects expression of circadian clock genes in the shoot. Plant Cell Environ. 37, 1321–1327. doi: 10.1111/pce.12242
Haslam, T. M., and Kunst, L. (2013). Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210, 93–107. doi: 10.1016/j.plantsci.2013.05.008
Hegebarth, D., Buschhaus, C., Joubès, J., Thoraval, D., Bird, D., and Jetter, R. (2017). Arabidopsis ketoacyl-CoA synthase 16 (KCS16) forms C36/C38 acyl precursors for leaf trichome and pavement surface wax. Plant Cell Environ. 40, 1761–1776. doi: 10.1111/pce.12981
Hooker, T. S., Millar, A. A., and Kunst, L. (2002). Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129, 1568–1580. doi: 10.1104/pp.003707
Isaacson, T., Kosma, D. K., Matas, A. J., Buda, G. J., and He, Y. (2009). Cutin deficiency in the tomato fruit cuticle consistently affects resistance to microbial infection and biomechanical properties, but not transpirational water loss. Plant J. 60, 363–377.
Islam, M. A., Du, H., Ning, J., Ye, H., and Xiong, L. (2009). Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol. Biol. 70, 443–456. doi: 10.1007/s11103-009-9483-0
James, D. W., Lim, E., Keller, J., Plooy, I., Ralston, E., and Dooner, H. K. (1995). Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell 7, 309–319. doi: 10.1105/tpc.7.3.309
Jenks, M. A., Joly, R. J., Peters, P. J., Rich, P. J., Axtell, J. D., and Ashworth, E. N. (1994). Chemically induced cuticle mutation affecting epidermal conductance to water vapor and disease susceptibility in Sorghum bicolor (L.) Moench. Plant Physiol. 105, 1239–1245. doi: 10.1104/pp.105.4.1239
Joubès, J., Raffaele, S., Bourdenx, B., Garcia, C., and Laroche-Traineau, J. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67:547. doi: 10.1007/s11103-008-9339-z
Kim, H. J., Seo, E. Y., Kim, J. H., Cheong, H. J., and Choi, D. I. (2012). Morphological classification of trichomes associated with possible biotic stress resistance in the genus capsicum. Plant Pathol. J. 28, 107–113. doi: 10.5423/PPJ.NT.12.2011.0245
Krauss, P., Markstädter, C., and Riederer, M. (1997). Attenuation of UV radiation by plant cuticles from woody species. Plant Cell Environ. 20, 1079–1085. doi: 10.1111/j.1365-3040.1997.tb00684.x
Kunst, L., and Samuels, A. L. (2003). Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 42, 51–80. doi: 10.1016/S0163-7827(02)00045-0
Lee, S. B., and Suh, M. C. (2013). Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Mol. Plant 6, 246–249. doi: 10.1093/mp/sss159
Leide, J., Hildebrandt, U., Reussing, K., Riederer, M., and Vogg, G. (2007). The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties: effects of a deficiency in a β-ketoacyl-coenzyme A synthase (LeCER6). Plant Physiol. 144, 1667–1679. doi: 10.1104/pp.107.099481
Liu, D., Yang, L., Wang, Y., Zhuang, X., Liu, C., Liu, S., et al. (2016). Transcriptome sequencing identified wax-related genes controlling the glossy phenotype formation of “ganqi 3,” a bud mutant derived from wild-type “newhall” navel orange. Tree Genet. Genom. 12:55. doi: 10.1007/s11295-016-1017-8
Liu, D. C., Yang, L., Luo, M., Wu, Q., Liu, S. B., and Liu, Y. (2017). Molecular cloning and characterization of Ptrzpt2-1, a zpt2 family gene encoding a cys2/his2-type zinc finger protein from trifoliate orange (poncirus trifoliata (l.) raf.) that enhances plant tolerance to multiple abiotic stresses. plant science. Plant Sci. 263, 66–78. doi: 10.1016/j.plantsci.2017.07.012
Liu, D. C., Yang, L., Zheng, Q., Wang, Y., and Wang, M. (2015). Analysis of cuticular wax constituents and genes that contribute to the formation of ‘glossy Newhall’, a spontaneous bud mutant from the wild-type ‘Newhall’ navel orange. Plant Mol. Biol. 88, 573–590. doi: 10.1007/s11103-015-0343-9
Liu, D. C., Zeng, Q., Ji, Q. X., Liu, C. F., Liu, S. B., and Liu, Y. (2012). A comparison of the ultrastructure and composition of fruits’ cuticular wax from the wild-type ‘Newhall’navel orange (Citrus sinensis [L.] Osbeck cv. Newhall) and its glossy mutant. Plant Cell Rep. 31, 2239–2246. doi: 10.1007/s00299-012-1333-x
Lokesh, U., Venkatesh, B., Kiranmai, K., Nareshkumar, A., Amarnathareddy, V., Rao, G. L., et al. (2019). Overexpression of ß-Ketoacyl CoA Synthase1 gene improves tolerance of drought susceptible groundnut (Arachis hypogaea L.) Cultivar K-6 by increased leaf epicuticular wax accumulation. Front. Plant Sci. 9:1869. doi: 10.3389/fpls.2018.01869
Marcolino-Gomes, J., Rodrigues, F. A., Fuganti-Pagliarini, R., Bendix, C., Nakayama, T. J., Celaya, B., et al. (2014). Diurnal oscillations of soybean circadian clock and drought responsive genes. PLoS One 9:e86402. doi: 10.1371/journal.pone.0086402
Millar, A. A., Clemens, S., Zachgo, S., Giblin, E. M., Taylor, D. C., and Kunst, L. (1999). CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11, 825–838. doi: 10.1105/tpc.11.5.825
Parsons, E. P., Popopvsky, S., Lohrey, G. T., Lü, S., Alkalai-Tuvia, S., and Perzelan, Y. (2012). Fruit cuticle lipid composition and fruit post-harvest water loss in an advanced backcross generation of pepper (Capsicum sp.). Physiol. Plant. 146, 15–25.
Pruitt, R. E., Vielle-Calzada, J. P., Ploense, S. E., Grossniklaus, U., and Lolle, S. J. (2000). FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc. Natl. Acad. Sci. U.S.A. 97, 1311–1316. doi: 10.1073/pnas.97.3.1311
Qin, Y. M., Pujol, F. M., Hu, C. Y., Feng, J. X., Kastaniotis, A. J., Hiltunen, J. K., et al. (2007). Genetic and biochemical studies in yeast reveal that the cotton fibre-specific GhCER6 gene functions in fatty acid elongation. J. Exp. Bot. 58, 473–481. doi: 10.1093/jxb/erl218
Riederer, M., and Schneider, G. (1990). The effect of the environment on the permeability and composition of Citrus leaf cuticles. Planta 180, 154–165. doi: 10.1007/BF00193990
Rossak, M., Smith, M., and Kunst, L. (2001). Expression of the FAE1 gene and FAE1 promoter activity in developing seeds of Arabidopsis thaliana. Plant Mol. Biol. 46, 717–725. doi: 10.1023/A:1011603923889
Seo, P. J., Xiang, F. N., Qiao, M., Park, J. Y., and Park, C. M. (2009). The myb96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151, 275–289. doi: 10.2307/40537768
Serra, O., Soler, M., Hohn, C., Franke, R., Schreiber, L., Prat, S., et al. (2009). Silencing of StKCS6 in potato periderm leads to reduced chain lengths of suberin and wax compounds and increased peridermal transpiration. J. Exp. Bot. 60, 697–707. doi: 10.1093/jxb/ern314
Shepherd, T., and Wynne Griffiths, D. (2006). The effects of stress on plant cuticular waxes. New Phytol. 171, 469–499. doi: 10.1111/j.1469-8137.2006.01826.x
Sieber, P., Schorderet, M., Ryser, U., Buchala, A., Kolattukudy, P., Métraux, J. P., et al. (2000). Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12, 721–737. doi: 10.1105/tpc.12.5.721
Singh, S., Das, S., and Geeta, R. (2018). “Role of cuticular wax in adaptation to abiotic stress: a molecular perspective,” in Abiotic Stress-Mediated Sensing and Signaling in Plants: An Omics Perspective, eds S. Zargar and M. Zargar (Singapore: Springer), 155–182.
Suh, M. C., Samuels, A. L., Jetter, R., Kunst, L., and Pollard, M. (2005). Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 139, 1649–1665. doi: 10.1104/pp.105.070805
Todd, J., Post-Beittenmiller, D., and Jaworski, J. G. (1999). KCS1 encodes a fatty acid elongase3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 17, 119–130. doi: 10.1046/j.1365-313X.1999.00352.x
Vogg, G., Fischer, S., Leide, J., Emmanuel, E., Jetter, R., Levy, A. A., et al. (2004). Tomato fruit cuticular waxes and their effects on transpiration barrier properties: functional characterization of a mutant deficient in a very-long-chain fatty acid b-ketoacyl-CoA synthase. J. Exp. Bot. 55, 1401–1410. doi: 10.1093/jxb/erh149
Von Wettstein-Knowles, P. M. (1979). “Genetics and biosynthesis of plant epicuticular waxes,” in Advances in Biochemistry and Physiology of Plant Lipids, eds L. Applequist and C. Liljenberg (Amsterdam: Elsevier), 1–26.
Wang, Y., Jin, S., Xu, Y., Li, S., and Zhang, S. (2020). Overexpression of BnKCS1-1, BnKCS1-2, and BnCER1-2 promotes cuticular wax production and increases drought tolerance in Brassica napus. Crop J. 8, 26–37. doi: 10.1016/j.cj.2019.04.006
Weidenbach, D., Jansen, M., Bodewein, T., Nagel, K. A., and Schaffrath, U. (2015). Shoot and root phenotyping of the barley mutant kcs6 (3-ketoacyl-coa synthase6) depleted in epicuticular waxes under water limitation. Plant Signal. Behav. 10:e1003752. doi: 10.1080/15592324.2014.1003752
Xie, Y. P., Chen, P., Yan, Y., Bao, C., and Li, X. (2018). An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 218, 201–218. doi: 10.1111/nph.14952
Yeats, T. H., and Rose, J. K. (2013). The formation and function of plant cuticles. Plant Physiol. 163, 5–20. doi: 10.2307/23598549
Yephremov, A., Wisman, E., Huijser, P., Huijser, C., Wellesen, K., and Saedler, H. (1999). Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11, 2187–2201. doi: 10.1105/tpc.11.11.2187
Zhang, J. Y., Broeckling, C. D., Blancaflor, E. B., Sledge, M. K., Sumner, L. W., and Wang, Z. Y. (2005). Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 42, 689–707.
Zhang, J. Y., Broeckling, C. D., Sumner, L. W., and Wang, Z. (2007). Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol. Biol. 64, 265–278. doi: 10.1007/s11103-007-9150-2
Zhang, Y. L., Zhang, C. L., Wang G. L., Wang, Y. X., and Hao, Y. J. (2019a). Apple AP2/EREBP transcription factor MdSHINE2 confers drought resistance by regulating wax biosynthesis. Planta 249, 1627–1643. doi: 10.1007/s00425-019-03115-4
Zhang, Y. L., Zhang, C. L., Wang, G. L., Wang, Y. X., and Hao, Y. J. (2019b). The R2R3 MYB transcription factor MdMYB30 modulates plant resistance against pathogens by regulating cuticular wax biosynthesis. BMC Plant Biol. 19:362. doi: 10.1186/s12870-019-1918-4
Keywords: Newhall navel orange, cuticular wax, KCS6, abiotic stresses, transgenic Arabidopsis
Citation: Guo W, Wu Q, Yang L, Hu W, Liu D and Liu Y (2020) Ectopic Expression of CsKCS6 From Navel Orange Promotes the Production of Very-Long-Chain Fatty Acids (VLCFAs) and Increases the Abiotic Stress Tolerance of Arabidopsis thaliana. Front. Plant Sci. 11:564656. doi: 10.3389/fpls.2020.564656
Received: 22 June 2020; Accepted: 14 September 2020;
Published: 06 October 2020.
Edited by:
Agnieszka Zienkiewicz, Nicolaus Copernicus University in Toruñ, PolandReviewed by:
Tegan Haslam, The University of British Columbia, CanadaJoaquín J. Salas, Instituto de la Grasa (IG), Spain
Copyright © 2020 Guo, Wu, Yang, Hu, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dechun Liu, bGRjODczMzgwODAwQDE2My5jb20=; Yong Liu, bGl1eW9uZ2p4YXVAMTYzLmNvbQ==
 Wenfang Guo
Wenfang Guo Qi Wu
Qi Wu