
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 08 May 2020
Sec. Plant Metabolism and Chemodiversity
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00557
A correction has been applied to this article in:
Corrigendum: Genetic Mapping of a Light-Dependent Lesion Mimic Mutant Reveals the Function of Coproporphyrinogen III Oxidase Homolog in Soybean
Lesion mimic mutants provide ideal genetic materials for elucidating the molecular mechanism of cell death and disease resistance. Here, we isolated a Glycine max lesion mimic mutant 2-1 (Gmlmm2-1), which displayed a light-dependent cell death phenotype. Map-based cloning revealed that GmLMM2 encods a coproporphyrinogen III oxidase and participates in tetrapyrrole biosynthesis. Knockout of GmLMM2 led to necrotic spots on developing leaves of CRISPR/Cas9 induced mutants. The GmLMM2 defect decreased the chlorophyll content by disrupting tetrapyrrole biosynthesis and enhanced resistance to Phytophthora sojae. These results suggested that GmLMM2 gene played an important role in the biosynthesis of tetrapyrrole and light-dependent defense in soybeans.
Plants have developed sophisticated defense mechanisms to protect against attack by different pathogens. The hypersensitive response (HR) is an effective resistance reaction, which induces rapid programmed cell death (PCD) in infected areas to inhibit further invasion of pathogens in normal cells (Dangl et al., 1996). HR-mediated PCD is accompanied by a burst of reactive oxygen species (ROS), triggered by the expression of pathogenesis-related (PR) genes (Qiao et al., 2010; Dickman and Fluhr, 2013). Lesion mimic mutants (lmms) are plants that spontaneously develop necrotic lesions caused by cell death, without any pathogen infection or abiotic stress, which are similar to disease symptoms or HR. Therefore, lmms are powerful tools for studying the mechanisms of HR, PCD, and disease resistance. Earlier studies showed that a number of lmms plants present enhanced disease resistance compared with their progenitor parents. Subsequently, a number of LMMs genes were mapped and cloned, and major pathways were clarified, including chlorophyll synthesis, fatty acid and lipid biosynthesis, secondary messenger, and kinase signaling pathways (Hoisington et al., 1982; Wolter et al., 1993; Takahashi et al., 1999; Lorrain et al., 2003; Tsunezuka et al., 2005; Moeder and Yoshioka, 2008; Bruggeman et al., 2015; Sun et al., 2017).
The tetrapyrrole biosynthetic pathway is a multi-branched pathway. The most of the intermediate molecules of the tetrapyrrole biosynthetic pathways are photosensitizers. Under the light, these highly photoreactive sensitizers potentially generate singlet oxygen that lead to the accumulation of copious amounts of ROS and the subsequent formation of photodynamic damage and spontaneous necrotic spots on developing leaves (Ishikawa et al., 2001). The reduced activity of some tetrapyrrole biosynthetic enzymes lead to the accumulation of photosensitized intermediates and cell death in leaves. For example, defects in uroporphyrinogen III decarboxylase (UROD) in the maize lesion mimic22 (Les22) mutant, porphobilinogen deaminase (PBGD) in the maize camouflage1 (cf1) mutant and Arabidopsis rugosa1 (rug1) mutant, and 5-aminolevulinic acid dehydratase (ALAD) in the cotton Ghlmm mutant induce spontaneous cell death in leaves (Hu et al., 1998; Huang et al., 2009; Quesada et al., 2013; Chai et al., 2017).
Many lmms exhibited up-regulation of resistance-related genes and enhanced pathogen resistance (Takahashi et al., 1999; Guo et al., 2013; Sathe et al., 2019). PR1 (Pathogensis-Related1) and NPR1 (NON-EXPRESSOR OF PATHOGENESIS-RELATED GENES1) are strongly correlated with the onset of systemic acquired resistance (SAR) (Weigel et al., 2001; van Verk et al., 2008; Zheng and Dong, 2013). Endogenous signal molecules such as jasmonic acid (JA) and salicylic acid (SA) play an important role in induced resistance against pathogen infection. The transcript levels of SA-responsive marker genes, GmPR10 (Pathogensis-Related protein10) and GmSnRK1.1 (Sucrose Non-fermenting-1-Related Protein Kinase1) were significantly increased following infection with P. sojae (Xu et al., 2014; Wang et al., 2019). AOS (Allene Oxide Synthase) generate precursors of the hormone JA, and COI1 (Coronatine Insensitive 1) is a critical component of the JA receptor. Both AOS and COI1 play an important role in JA mediated defense (Xie et al., 1998). It was also reported that OsJAMyb (Jasmine Acid induced MYB) enhanced blast resistance in transgenic rice (Cao et al., 2015).
Coproporphyrinogen III oxidase (CPO) is an enzyme in the tetrapyrrole biosynthetic pathway that catalyzes coproporphyrinogen III (Coprogen III) to protoporphyrinogen IX (Proto IX) via the oxidative decarboxylation of two propionate groups of the side chains of pyrrole rings A and B to vinyl groups (Tanaka et al., 2011). Two unrelated structural and functional CPOs, CPO/HemF and CPO/HemF, catalyze the reaction under aerobic and anaerobic conditions, respectively. CPO/HemF belongs to a family of monooxygenases and CPO/HemN belongs to the radical S-adenosylmethionine family (Ouchane et al., 2004; Phillips et al., 2004). CPO/HemF exist widely in living organisms, from lower forms of life and higher living organisms. The mutations in human CPO/HemF associated with many of the disease hereditary coproporphyria. The crystal structure of human CPO/HemF reveals critical residues for catalytic activity and substance binding sites. Disruption of CPO/HemF causes necrotic spot formation in Nicotiana tabacum, Arabidopsis, and rice (Kruse et al., 1995; Ishikawa et al., 2001; Sun et al., 2011; Wang et al., 2015).
In the present study, we carried out map-based cloning of the Glycine max lesion mimic mutant 2-1 (Gmlmm2-1) in soybean and confirmed GmLMM2 gene encodes CPO/HemF and its protein located in the chloroplast. Suppression of GmLMM2 disrupts the chloroplast structure and tetrapyrrole synthesis pathway. The lesion mimic phenotypes in Gmlmm2-1 depend on the light and might help to enhance pathogens resistance to Phytophthora sojae. Our results suggest that GmLMM2 plays an important role in the biosynthesis of tetrapyrrole and immunity in soybean.
The soybean cultivar “Williams 82” and “Hedou 12” were obtained from the Chinese Academy of Agricultural Sciences (Cheng et al., 2016). The Gmlmm2-1 mutant was isolated from an M2 population induced by ethyl methane sulfonate (EMS) (Feng et al., 2019). For further analysis, the Gmlmm2-1 mutant was backcrossed to “Williams 82” four times to purify the GmLMM2 mutation. All the plants were grown in Changchun, China. The Columbia (Col-0) ecotype of A. thaliana plants were grown in a growth chamber (Percival, IA, United States) under 150 μmol m–2s–1 16 h light/8 h dark cycles at 25°C.
The first compound leaf of the 15-day-old “Williams 82” and the Gmlmm2-1 mutant were used in a half-leaf shading experiment. Small pieces of aluminum foil were used to cover half of the Gmlmm2-1 mutant and “Williams 82” leaves to prevent light exposure; 5 days later the Gmlmm2-1 mutant and “Williams 82” leaves were photographed and staining with different reagents. Leaf trypan blue (TB) staining was performed as the method described by Yin et al. (2000). H2O2 accumulation was detected by 3, 3’-diaminobenzidine (DAB) staining (Thordal-Christensen et al., 1997).
To determine pigment content, leaves of 3-week-old Gmlmm2-1 mutant and “Williams 82” were collected and measured as previously described (Lichtenthaler and Wellburn, 1983). Net photosynthetic rate (Pn) and transpiration rate (Tr) of leaves were measured using Li-6400 photosynthesis equipment (Li-Cor, Lincoln, NE, United States) (Yamori et al., 2011). F0 (initial fluorescence) and Fm (maximal fluorescence) values were measured using a chlorophyll fluorometer OS-30p (Opti-Sciences, Hudson, NY, United States). Fv/Fm (maximum quantum efficiency of photosystem II) and Fv/F0 (maximum primary yield of photochemistry of PSII) were calculated as previously described (Genty et al., 1989). All measurements were made using three plants from 10:00 am to 12:00 noon during the R1 period with three biological replicates.
Malonyldialdehyde (MDA) levels in leaves were measured as described by Hodges et al. (1999). H2O2 content was quantified using an H2O2 detection kit according to the manufacturer’s instructions (Comin Biotechnology, Suzhou, China). The absorbance of yellow compound was measured at 415 nm using a NanoPhotometer (Implen).
Coprogen III content was measured as described by Pratibha et al. (2017). The absorbance of Coprogen III was measured at a wavelength of 402 nm. Uroporphyrinogen III (Urogen III) content was determined as described by Bogorad (1962). The absorbance of Urogen III was measured at a wavelength of 405 nm. The Proto IX, Mg-protoporphyrin IX (Mg-proto IX) and protochlorophyllide (Pchlide) contents were measured as described by Rebeiz et al. (1975). All above measurements were obtained from three biological replicates.
True leaves were cut into smaller sections of approximately 1 × 1 mm, and placed in 1.5 mL fixation solution (2.5% glutaraldehyde with phosphate, pH 7.2), and vacuumed until completely immersed in the solution. The samples were subsequently fixed with 1% osmium tetroxide at 4°C. Samples were then treated according to previously described methods (Kowalewska et al., 2016). The samples were sliced to 70 nm thickness with a MT-X (RMC, Tucson, AZ, United States) ultramicrotome and stained with uranyl acetate and lead citrate. The samples were observed using a Hitachi H-7650 electron microscope (Tokyo, Japan).
To determine the subcellular location of GmLMM2, a full-length cDNA fragment of GmLMM2 was amplified using the primer pairs listed in Supplementary Table S1 and inserted into the modified expression vector pUC19-GFP (Zheng et al., 2016). The pFL1004 construct (Supplementary Figure S8) was introduced into Arabidopsis (Col-0) mesophyll protoplasts using 20% polyethylene glycol (Meyer et al., 2006). Fluorescence was monitored using a confocal laser scanning microscope Leica SP8 (Leica, Solms, Germany). We detected the fluorescence of GFP wavelengths at 488 nm (excitation) and 495–540 nm (emission) and chlorophyll auto fluorescence wavelengths at 488 nm (excitation) and 680–700 nm (emission).
The INDEL markers for preliminary mapping were same as described previously (Song et al., 2015). New molecular markers for fine mapping are listed in Supplementary Table S1. The Gmlmm2-1 mutant was re-sequenced with a depth of approximately 30× using illumina Hiseq2000 (Song et al., 2015). The Genome Analysis Toolkit (GATK, version 3.8) was used to detect single nucleotide polymorphisms (SNPs; McKenna et al., 2010). Sequences were deposited at the National Center for Biotechnology Information (NCBI) under the accession number SRP149750.
The amino acid sequences of CPOs/HemF were downloaded from Phytozome1, Phytozome V12.0. CPOs/HemF sequences were aligned with DNAMAN 8.0. MEGA72 and used to build a phylogenetic tree via the maximum likelyhood method with 1000 bootstrap replications. CPOs/HemF motifs were analyzed using MEME3 and TBtools software (Bailey et al., 2015). Synteny was analyzed using MCScanX (Wang et al., 2012).
For mutation complementation, the 5,374 bp genomic DNA fragment of GmLMM2 (including 2,008 bp promoter region and 3,366 bp gene region) was inserted into the pCAMBIA3301 vector between Sac I and Bam HI sites. The resultant pFL1002 plasmid (Supplementary Figure S6) was then transformed into the cotyledonary explants of the Gmlmm2 mutant via Agrobacterium-mediated transformation (Zhao et al., 2016).
To knockout the GmLMM2 gene in the ‘DongNong50 (DN50)’ soybean cultivar, the modified pSC1-Cas9 was used as described by Du et al. (2016). A 20-nt single guide RNA (sgRNA) was identified using the web-based tools CRISPR-P4 and CRISPR-PLANT5, which targeted 195 to 214 position in the first exon of GMLMM2. The oligo pairs corresponding the sgRNA were annealed and ligated into a plasmid pSC1-Cas9 digesting with BspQ I. The sgRNA expression was driven under GmU6-16g-1 promoter in the recombinant plasmid pFL1003. The recombinant plasmid pFL1003 (Supplementary Figure S7) was transformed into DN50 as above (Zhao et al., 2016). The primers used for the construct are listed in Supplementary Table S1.
Total RNA was isolated from different soybean tissues using TRIzol (Invitrogen, Carlsbad, CA, United States), and genomic DNA was digested with DNase I (Takara Biotechnology, Dalian, China). cDNA was synthesized using a Fast Quant RT Kit (TIANGEN, Beijing, China). RT-qPCR was performed in a Gene Amp 5700 sequence Detection System with SYBR® premix Ex TaqTM regent (Takara). GmActin11 was used as the reference gene (Zeng et al., 2017), the data were analyzed using the 2–ΔΔCT method (Livak and Schmittgen, 2001). Three biological replicates, each with three technical replicates, were performed. The primers used in RT-qPCR are listed in Supplementary Table S1.
Phytophthora sojae strain P7076 (P. sojae P7076) was cultivated at 25°C on 10% (v/v) V8 juice agar in a polystyrene dish (Förster et al., 1994). P. sojae P7076 infection assay was carried out as previously described with some modifications (Jia et al., 2013; Hwu et al., 2017). Briefly, 20 μL 5‰ Tween 20 was dripped on middle of 10-day-old detached leaves, and then transferred 5-day-old fresh mycelial disks approximately 5.5 mm diameter with a 200 μL Eppendorf pipette tip on the middle of detached leaves. Immediately after inoculation, the leaves of soybean were plated in trays covered with plastic wrap to maintain humidity and infected for 48 and 60 h at 25°C in the dark. Infection dead cell was determined by trypan blue staining as described above. Disease resistance was evaluated by measuring the lesion area using Image J software6.
A lesion mimic mutant was isolated from the EMS-induced “Williams 82” mutant population and named Gmlmm2-1 (Glycine max lesion mimic mutant 2-1). In this mutant, small chlorotic spots were initially visible along the veins of fully expanded leaves, before spreading gradually over the entire leaf. Older leaves of mutant with irregular brown spots began to shrink and senesce rapidly. The adult leaf of mutant was yellow compared with that of “Williams 82” (Figures 1A,B). In addition to necrotic spots, the number of nodules, hundred-grain weight, number of grains per plant, and plant height were reduced in the Gmlmm2-1 mutant (Supplementary Figure S1).
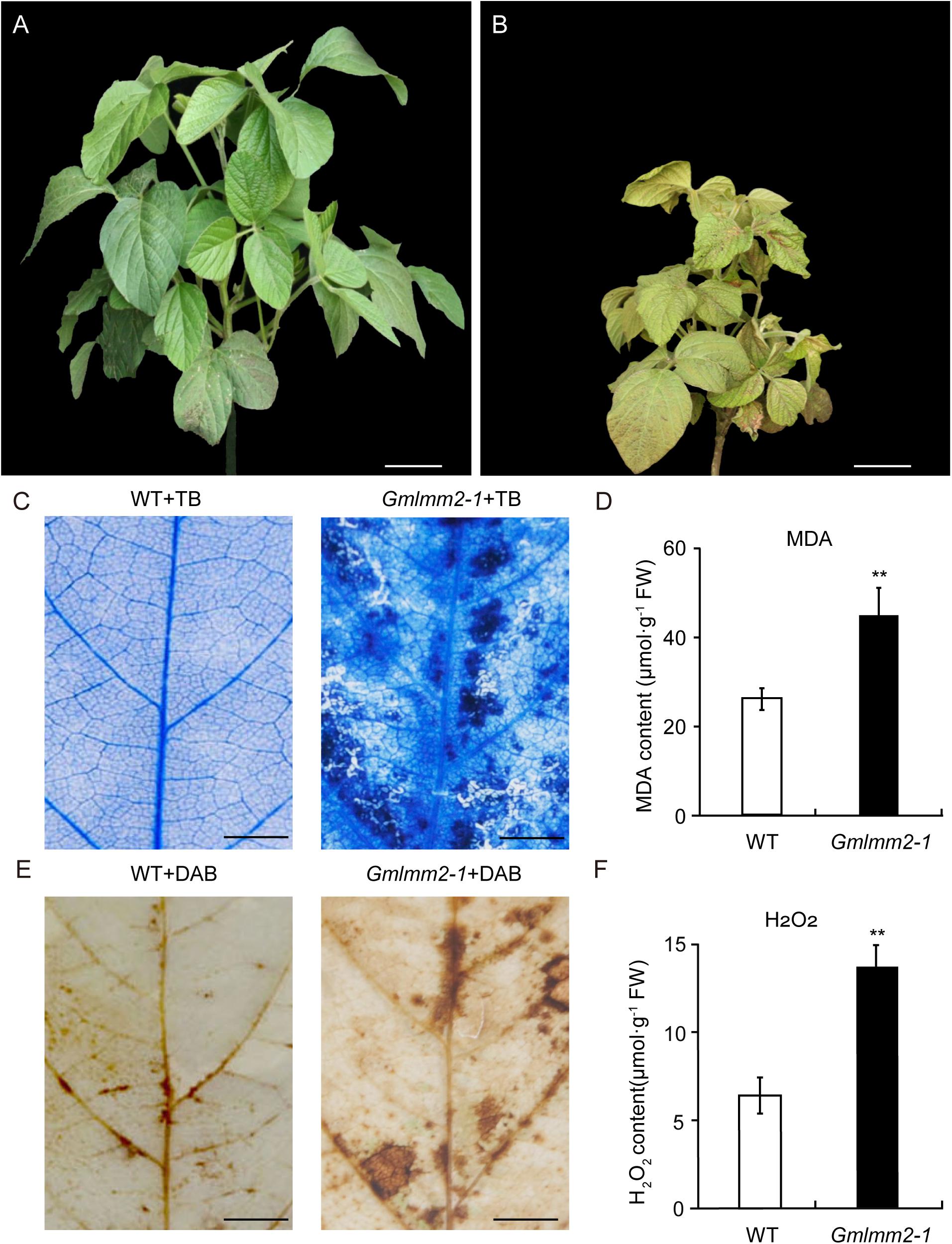
Figure 1. “Williams 82” (A) and the Gmlmm2-1 mutant (B) plants. Scale bars = 10 cm. TB staining of leaves of “Williams 82” (C) and the Gmlmm2-1 mutant. Scale bars = 1 cm. (D) MDA contents in wild type and mutant leaves. DAB staining of leaves of “Williams 82” (E) and the Gmlmm2-1 mutant. Scale bars = 1 cm. (F) H2O2 contents in wild type and mutant leaves. Asterisks indicate a significant difference by student’s t-test (**P < 0.01) and the error bars represent standard deviations.
To examine cell membrane damage situation of leaves, we performed a leaf trypan blue (TB) staining assay of both wild type and mutant at V2 stage (Figure 1C). After trypan blue staining, numerous dark blue spots appeared at the lesion sites on the Gmlmm2-1 mutant leaves; whereas the surrounding cells in the Gmlmm2-1 mutant and “Williams 82” cells were healthy (Figure 1C). The content of MDA in the Gmlmm2-1 mutant was 44.75 ± 6.42 μmol/g, which was 1.70-fold higher than that of “Williams 82” (Figure 1D). The increased level of MDA suggested that lipid peroxidation occurred in the Gmlmm2-1 mutant. The results of these experiments suggested that PCD occurred during lesion formation.
We subsequently performed DAB staining and observed red-brown polymer deposition in necrotic areas, indicating the generation of excess H2O2 (Figure 1E). The H2O2 level in the Gmlmm2-1 mutant leaves was higher than that in “Williams 82” leaves (Figure 1F), which was consistent with the DAB staining results. These findings indicated that ROS accumulation in cells was responsible for cell death and the visible necrosis in leaves.
To map the location of GmLMM2, we crossed Gmlmm2-1 mutant with “Hedou 12,” and the F1 plants exhibited the wild-type phenotype. Genetic analysis of the F2 population revealed that the necrotic phenotype of the Gmlmm2-1 mutant was controlled by a single recessive nuclear gene (WT: mutant = 618:195). The F2 population was used to identify the GmLMM2 locus. A total of 107 INDEL markers covering all 20 chromosomes were used for preliminary mapping, and the mapping result showed that GmLMM2 was restricted to the top of Chromosome 14 (Figure 2A). To fine-map the GmLMM2 locus, we developed four INDEL markers, MOL3470, MOL3552, MOL3560, and MOL4565; the GmLMM2 locus was further narrowed down to a 223 kb region between 203,085 and 426,404 bp on chromosome 14 (Chr 14) containing 32 annotated genes (Figures 2A,B). To identify the causative mutation, the Gmlmm2-1 mutant was re-sequenced with a depth of approximately 30 × using illumine Hiseq2000. We identified a A–667 to G–667 transition at the second exon of Glyma.14G003200 (Figure 2C), which caused a nonsynonymous substitution of Tyr–192 to Cys–192 in the predicted protein. No other mutations were discovered among the 32 genes in the candidate GmLMM2 genomic region (Figure 2B and Supplementary Table S2).
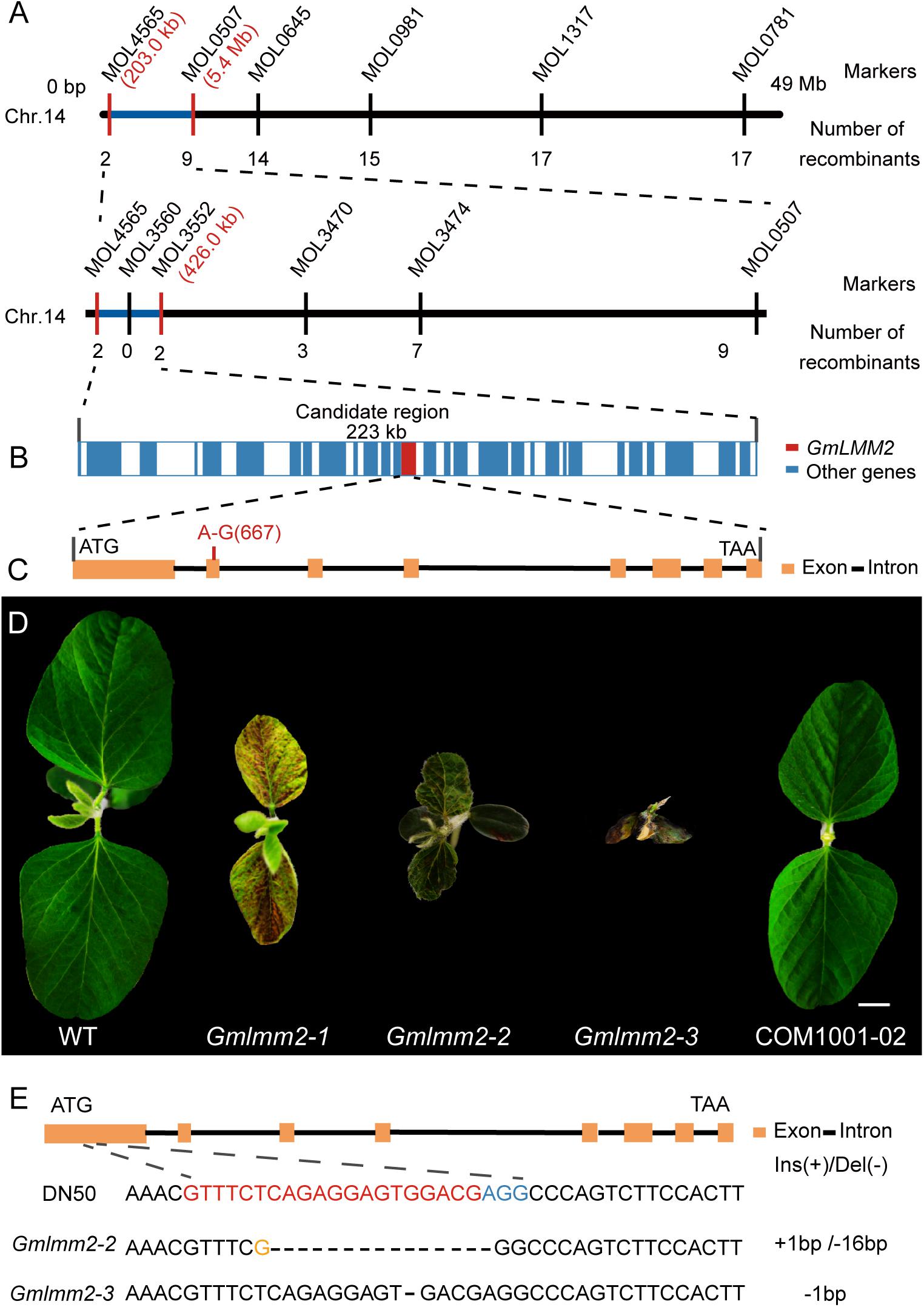
Figure 2. (A) Map-based cloning of the GmLMM2 locus. The numbers under INDELs markers indicate the number of recombinants. (B) Diagram of gene distribution in 233kb region harbored GmLMM2 gene. (C) Schematic diagram shows the structure of Glyma.14G003200. The red line denotes a A to G substitution in the second exon of GmLMM2. (D) Phenotypes of “Williams 82,” CRISPR/Cas9 plants (Gmlmm2-2 and Gmlmm2-3), complemented the Gmlmm2-1 mutant transgenic plant (COM1001-02). Scale bar = 1 cm. (E) sgRNA target sequences of “DongNong50,” Gmlmm2-2 and Gmlmm2-3. The sgRNA target sequence is shown in red and the protospacer-adjacent motif (PAM) site in blue. The yellow colored ‘G’ indicates an insertion.
A complementary vector (pFL1002, Supplementary Figure S6) was transformed into the Gmlmm2-1 mutant. Basta-resistance progenies of the transformed the Gmlmm2-1 mutant had a wild type phenotype in the T2 COM1001-02, COM1001-09, and COM1001-12 populations (Figure 2D, Supplementary Figure S3). Furthermore, we constructed a CRISPR/Cas9 expression vector (pFL1003, Supplementary Figure S7), and the construct was transformed into soybean DN50. Five heterozygous mutants were found in the T0 plants, and two independent homozygous knock-out lines, named Gmlmm2-2 and Gmlmm2-3, were found in T1 population (Figure 2D). The Gmlmm2-2 mutant contains a 16 bp deletion corresponding to CDS region of GmLMM2 gene from 200 bp to 215 bp, and a 1 bp insertion in above region. The Gmlmm2-2 mutant caused five amino acids deletion from 67th to 71th and one amino acid substitute (Glu to Arg) at 72th of GmLMM2 (Figure 2E). The Gmlmm2-3 mutant has a 1 bp deletion at 210 bp of CDS and caused a frameshift mutation to a truncated protein with 85 aa (Figure 2E). The Gmlmm2-2 and Gmlmm2-3 mutant variedly phenocopied cell death on leaf compared with the Gmlmm2-1 mutant. The Gmlmm2-2 and Gmlmm2-3 mutant also caused a lesion mimic phenotype in the stem, which was not observed in the Gmlmm2-1 mutant (Supplementary Figure S4). In addition, the Gmlmm2-2 and Gmlmm2-3 mutant exhibited pleiotropic phenotypes, including slow growth, dwarfing, and delayed development during vegetative stages, and the Gmlmm2-3 mutant was a seedling lethal mutant (Figure 2D). These data suggested the Gmlmm2-2 and Gmlmm2-3 mutants are strong alleles, whereas the Gmlmm2-1 mutant is a weak allele, and that complete loss of GmLMM2 function strongly affects soybean growth and development, and regulates cell death.
The open-reading frame (ORF) of GmLMM2 is 1158 bp in length, and the deduced protein contains 385 amino acids (Supplementary Figure S5). Phylogenetic analysis showed that GmLMM2 is more closely related to Phaseolus vulgaris and Vigna radiate homologs, which constitute an isolated branch in the phylogenetic tree. The identities of GmLMM2 and the P. vulgaris homolog are up to 91%. The results of MEME analysis showed that CPOs contain six conserved motifs (Figure 3A).
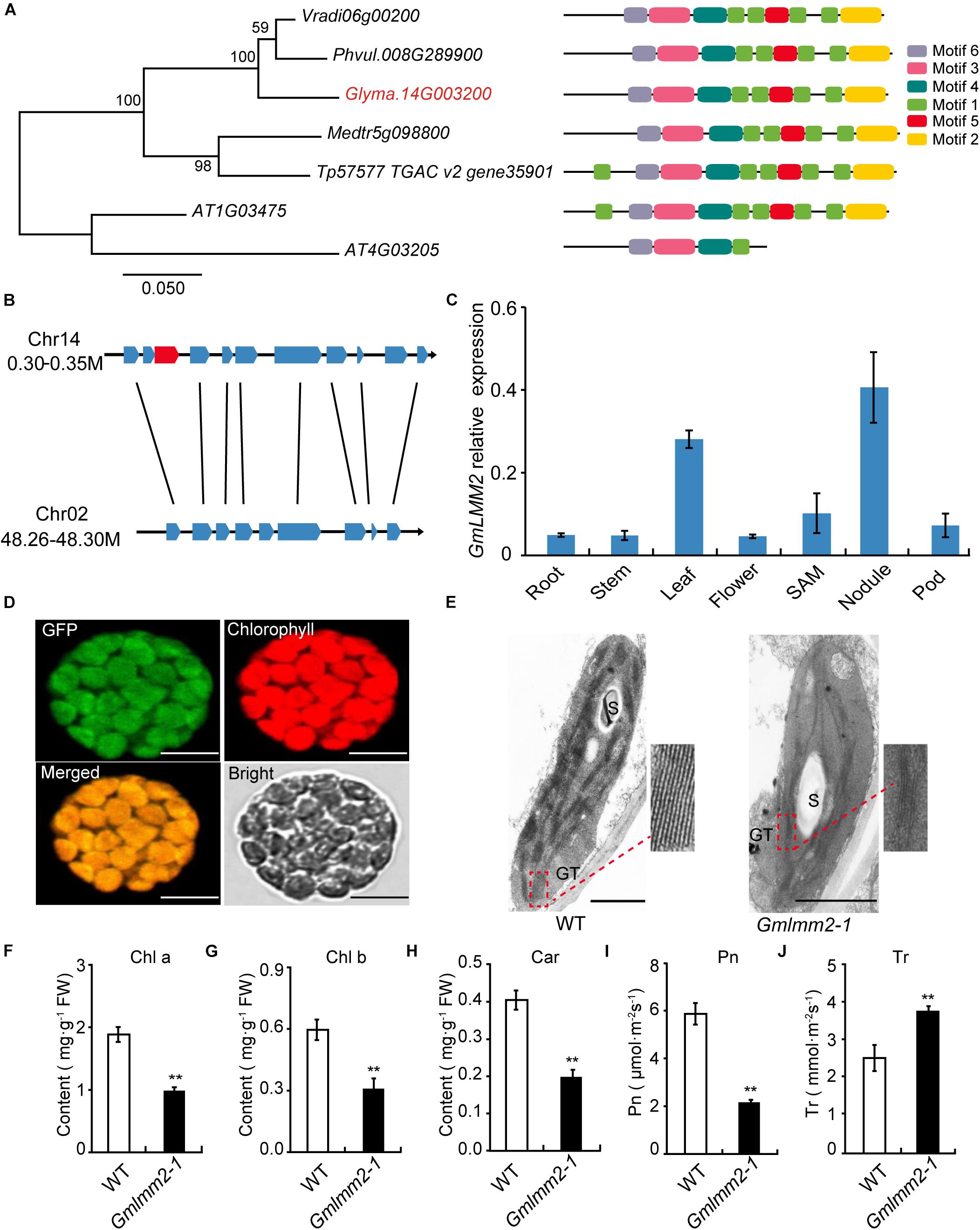
Figure 3. (A) Phylogenetic tree of CPOs/HemF in soybean and other species. Seven CPOs/HemF proteins from six species, including Vigna radiate (Vradi06g00200), Phaseolus vulgaris (Phvul.008G289900), Glycine max (Glyma.14G003200), Medicago truncatula (Medtr5g098800), Trifolium pretense (Tp57577 TGAC v2 gene35901), and Arabidopsis thaliana (AT1G03475; AT4G03205). (B) Synteny diagram plot of the genes surrounding the GmLMM2 gene. (C) Expression pattern of GmLMM2 in different tissues of “Williams 82” (WT). (D) GmLMM2 localized to the chloroplasts. GFP, GFP signal of GmLMM2; Chlorophyll, chlorophyll auto fluorescence; Merged, merged image of GFP and chlorophyll. Scale bars = 10 μm. (E) Chloroplast structure of “Williams 82” (WT) and the Gmlmm2-1 mutant. S, starch granule; GT, granal thylakoid. Scale bars = 1 μm. (F–J) Pigment contents and photosynthesis parameters of “Williams 82” and the Gmlmm2-1 mutant. (F) Chl a, chlorophyll a; (G) Chl b, chlorophyll b; (H) Car, carotenoid; (I) Pn, photosynthetic rate; (J) Tr, transpiration rate. Asterisks indications are same as Figure 1.
To find the homologs of GmLMM2 gene, we identified the 41.6 kb syntenic block on chromosome 2 from 48,264,660 bp to 48,306,267 bp, which is homologous to a 48.9 kp region to GmLMM2 located from 300,925 bp to 349,856 bp on chromosome 14 (Figure 3B, Supplementary Table S3). They shared 8 holomgous genes between these two regions. However, the putative paralog of GmLMM2 was absent from the syntenic block on chromosome 2 between Glyma.02G309600 and Glyma.02G309700 (Figure 3B). Thus, GmLMM2 is more like a single copy gene in the soybean genome.
We examined GmLMM2 expression in the root, stem, leaf, flower, nodule, SAM, and pod via quantitative real-time PCR. GmLMM2 was expressed in all organs and tissues tested (Figure 3C). The highest expression levels were detected in leaf and nodule, which may explain why the GmLMM2 mutation severely affected the growth of those tissues.
A chloroplast transit peptide in the first 40 amino acids of GmLMM2 was predicted by TargetP and ChloroP software; and GmLMM2 may be a chloroplast protein. To confirm its subcellular location, a GmLMM2-GFP fusion protein was transiently expressed in Arabidopsis protoplasts. GFP fluorescence only overlapped with the chlorophyll auto fluorescence signal (Figure 3D), which indicated that the GmLMM2 protein is localized to the chloroplast.
To confirm whether GmLMM2 is associated with chloroplast development, we examined the ultrastructure of plastids in 20-day-old seedlings in “Williams 82” and the Gmlmm2-1 mutant. The Gmlmm2-1 mutant contained fewer granal thylakoid membranes compared with the “Williams 82” leaves (Figure 3E). The abnormal chloroplast development in the Gmlmm2-1 mutant may related to the decreased pigment contents. The contents of Chl a, Chl b, and Car in the Gmlmm2-1 mutant were approximately 51, 50, and 40% of those in “Williams 82,” respectively (Figures 3F–H). The decreased chlorophyll contents also affected photosynthesis. We found that the net photosynthetic rate of “Williams 82” was 5.93 ± 0.46 μmol CO2 m–2s–1, which was 2.72-fold higher than that of the Gmlmm2-1 mutant (Figure 3I). The transpiration rate of the Gmlmm2-1 mutant was 3.75 ± 0.12 mmol H2O m–2s–1, which was 1.49-fold higher than that of “Williams 82” (Figure 3J). However, maximum chlorophyll fluorescence (Fv/Fm) and initial fluorescence ratio (Fv/F0) did not vary between the Gmlmm2-1 mutant and “Williams 82” (Supplementary Figure S2).
To further understand the impact of the GmLMM2 mutation on the tetrapyrrole biosynthesis pathway, the contents of some metabolic intermediates of tetrapyrrole biosynthesis were examined. No significant difference in Uroporphyrinogen III (Urogen III) levels was observed between wild type and mutant (Figure 4A). Whereas, the Coproporphyrinogen III (Coprogen III) level in the Gmlmm2-1 mutant was two-fold higher than that in wild type (Figure 4B). The levels of Protoporphyrinogen-IX (Proto IX), Mg-protoporphyrinogen-IX (Mg-proto IX), and Pchlide in the Gmlmm2-1 mutant were significantly lower than those in the leaves of “Williams 82” (Figures 4C–E). Therefore, light-dependent cell death in the Gmlmm2-1 mutant may be attributed to the accumulation of Coprogen III.
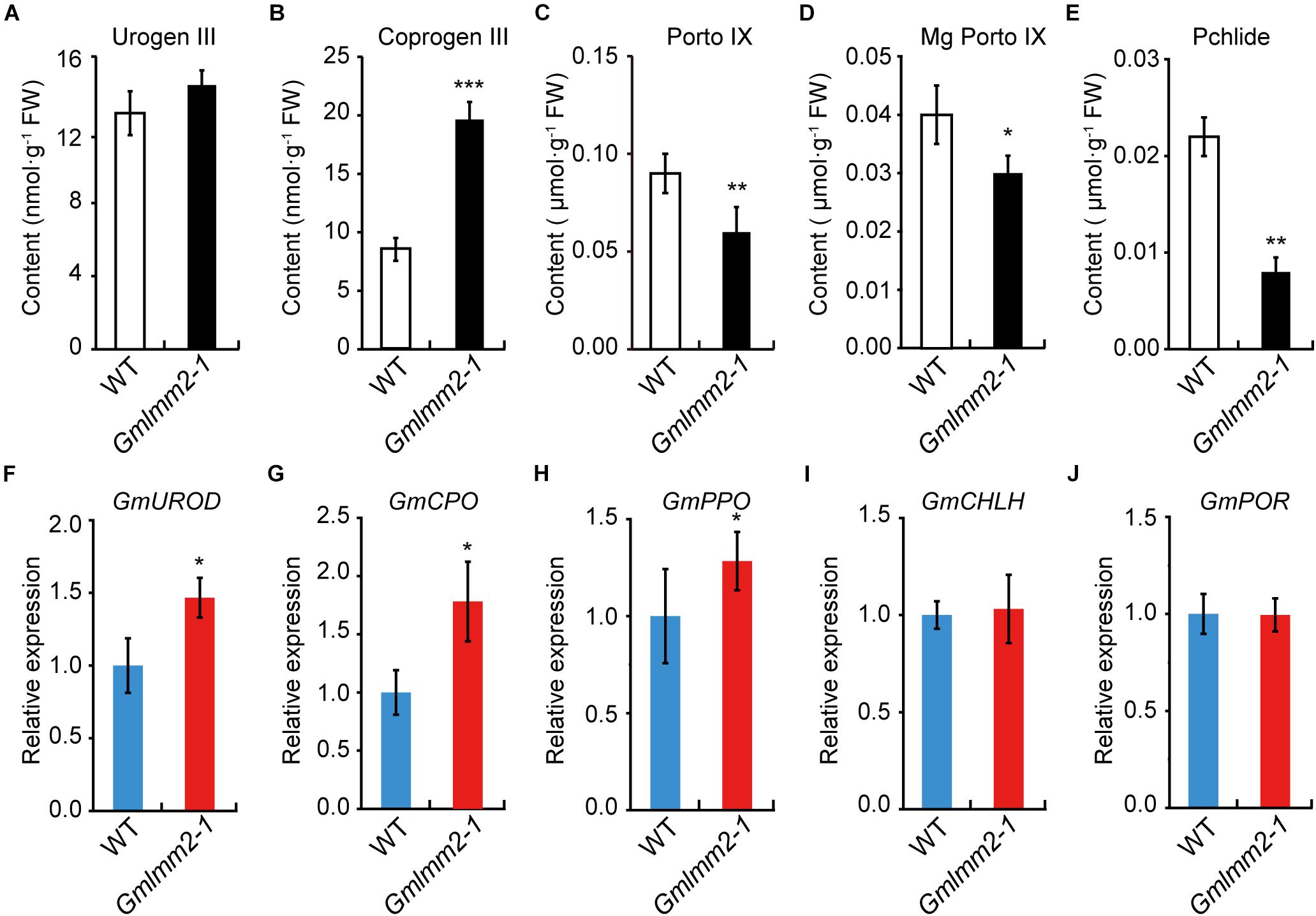
Figure 4. (A–E) Contents of metabolic intermediates in young leaves of wild type and mutant. (F–J) Expression levels of some enzymes in tetrapyrrole biosynthesis of wild type and mutant. Asterisks indicate a significant difference by student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001) and the error bars represent standard deviations.
We further analyzed the expressions of some enzymes related above intermediates, including uroporphyrinogen III decarboxylase (GmUROD, Glyma.18G021500); coproporphyrinogen III oxidase (GmCPO, Glyma.14G003200), protoporphyrinogen oxidase (GmPPO, Glyma.10G138600); Mg-chelatase subunit H (GmCHLH, Glyma.10G097800) and NADPH: Pchlide oxidoreductase (GmPOR, Glyma.12G222200). The transcript levels of GmUROD, GmCPO, and GmPPO were slightly elevated, but the transcript levels of GmCHLH and GmPOR remained substantially unchanged (Figures 4F–J). The expression raise of GmUROD and GmCPO might response to their substrates increasing in mutant plant.
To confirm whether the formation of lesions in the Gmlmm2-1 mutant leaves was light-dependent, half of the leaflets before lesion emergence were covered by aluminum foil, while the other half of the leaflets were exposed to light. Five-days later, no lesion had formed on the non-shaded or shaded leaflets of “Williams 82.” However, irregular brown lesions appeared on the light-exposed leaflets of the Gmlmm2-1 mutant, but not on the shaded part leaflets of the Gmlmm2-1 mutant (Figures 5A,B).
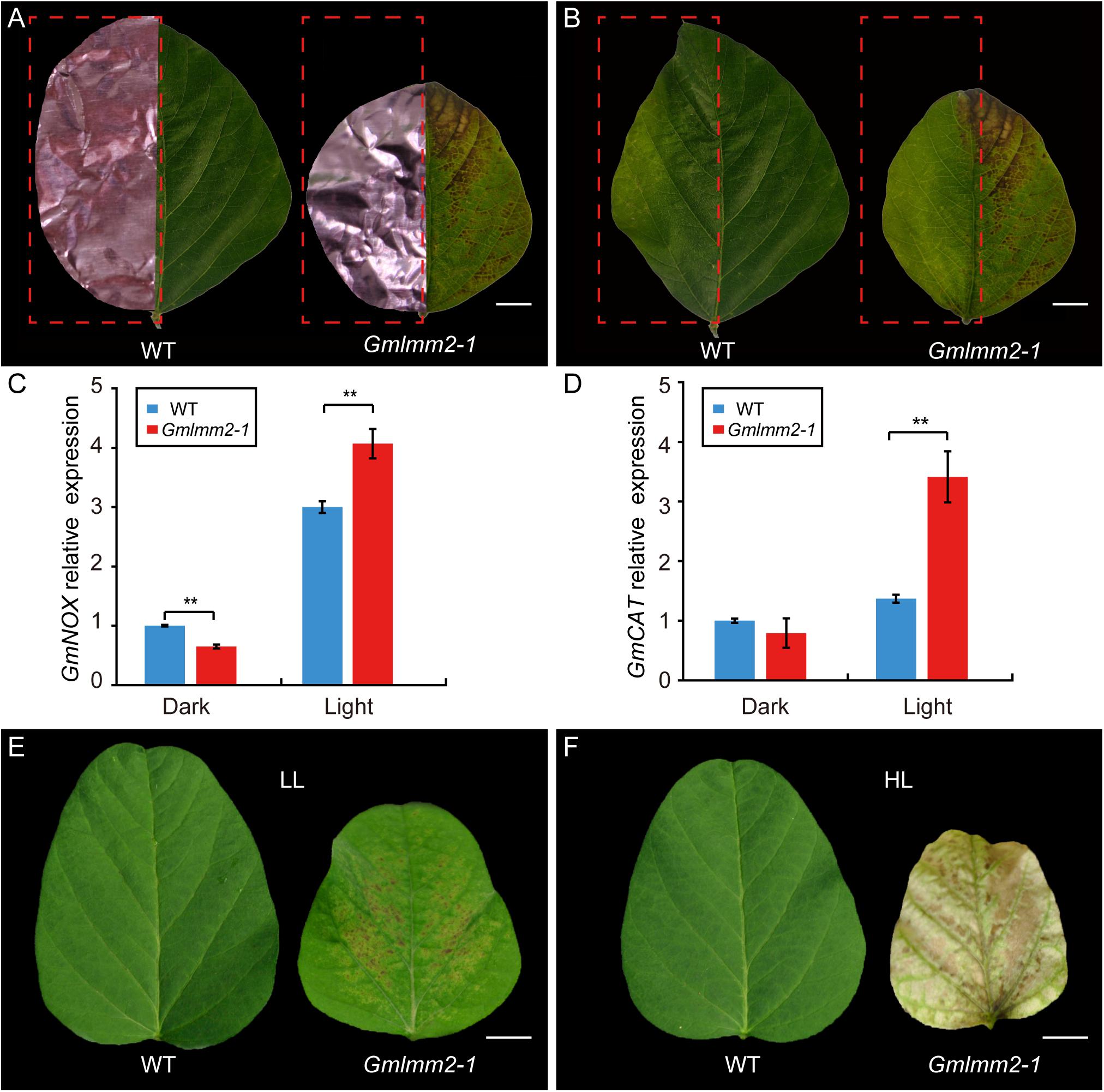
Figure 5. Light avoidance assay in leaves of “Williams 82” and the Gmlmm2-1 mutant before (A) and after (B) removing the shading foils. Scale bars = 0.5 cm. Comparison of expression levels of GmNOX (C) and GmCAT (D) between shaded (Dark) and un-shaded (Light) half leaflet. Phenotype of leaf growing under (E) lower intensity light (LL) and (F) high intensity light (HL). Scale bars = 1 cm. Asterisks’ indications are same as Figure 1.
NADPH oxidase (NOX) is related to ROS homeostasis, and catalase (CAT) is a ROS-scavenging gene (Lee et al., 2005, 2018). To determine the relationship of ROS and light condition, we compared expression of GmNOX and GmCAT of half leaflet with light covering. The transcript level of GmNOX in the Gmlmm2-1 mutant was 1.35-fold higher than that in the “Williams 82” under light. The expression of GmCAT in the Gmlmm2-1 mutant was 1.65-fold higher than that in the “Williams 82” under light. There was no significant difference in the expression of GmCAT between the Gmlmm2-1 mutant and “Williams 82” in the aluminum foil-covered leaves (Figures 5C,D). The results demonstrated that the GmLMM2 defect disrupts ROS homeostasis in the Gmlmm2-1 mutant, and that these processes are light-dependent.
The effect of light intensity on the lesion induction was also observed under different light condition. We planted “Williams 82” and Gmlmm2-1 mutant under high (20,000 Lux) and low (7,500 Lux) intensity light, respectively. We found that the higher intensity light rapidly and broadly induced severe necrosis and development, and chlorophyll deficiency compared with low intensity light (Figures 5E,F). These results suggested that light is a critical causal factor for lesion induction in the Gmlmm2-1 mutant and light intensity is correlated with the degree of cell death.
The previously study of lesion mimic mutant suggested that the Gmlmm2-1 mutant may enhanced resistance to pathogen infection. To confirm this hypothesis, the fully expanded 10-day-old true leaves of “Williams 82” and the Gmlmm2-1 mutant were inoculated with P. sojae P7076. Visible lesions were observed on the leaves of “Williams 82” and the Gmlmm2-1 mutant at 48 h post-inoculation (48 hpi) (Figure 6A). We measured the infectious area by TB staining, the infectious area in Gmlmm2-1 mutant reduced to 34.5 and 43.7%, compared with “Williams 82” at 48 and 60 hpi separately (Figures 6B–D). These results suggested that mutation of GmLMM2 inhibits the invasive of P. sojae P7076 and the Gmlmm2-1 mutant enhanced resistance to P. sojae P7076.
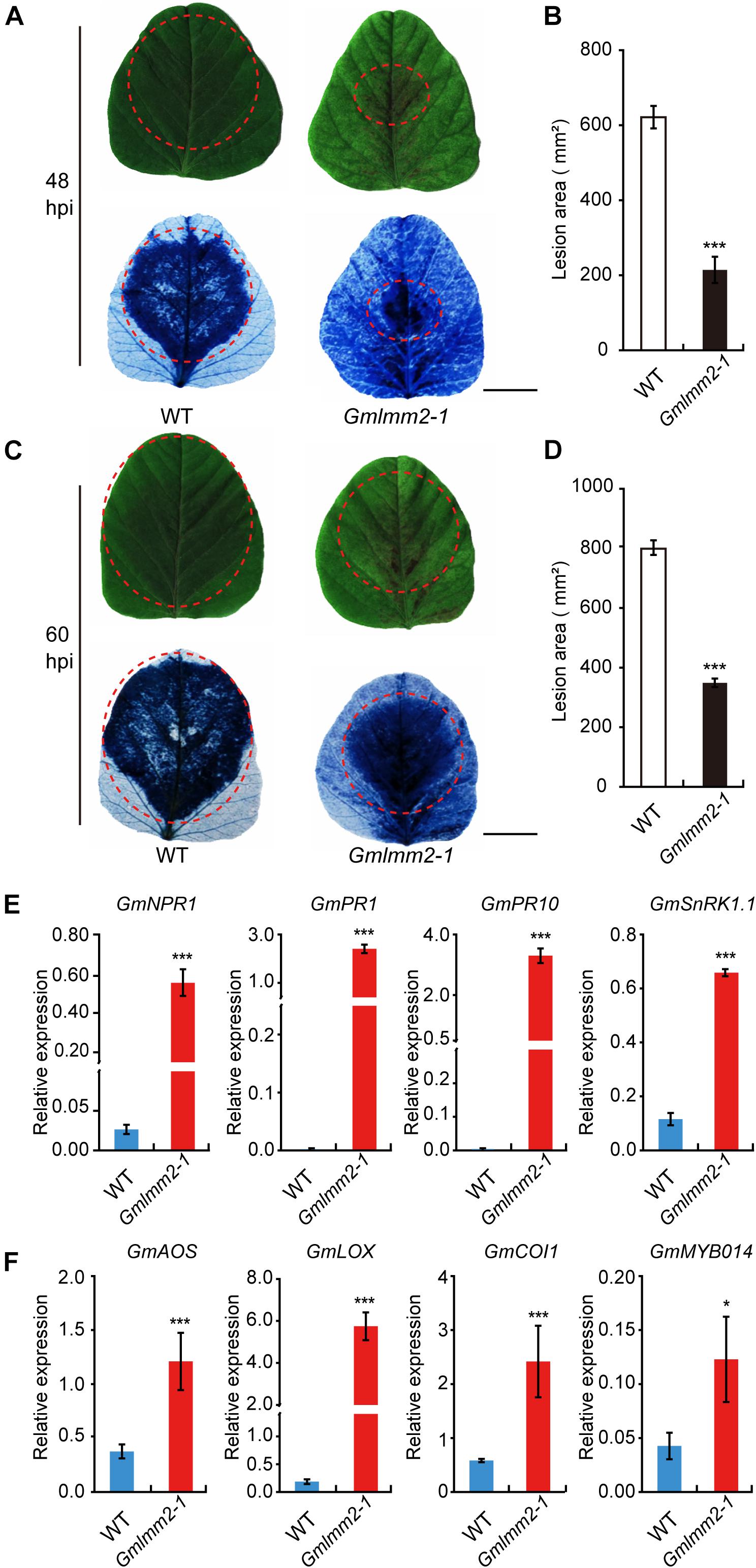
Figure 6. Up-regulation of resistance-related genes and enhanced pathogen resistance. (A–D) The infection phenotypes of “Williams82” (WT) and the Gmlmm2-1 mutant at 48 and 60 hpi with Phytophthora sojae stain P7076. Trypan blue staining demonstrated that infection levels were similar to those of the leaves shown above. Scale bars = 1 cm. The sizes of the lesions were statistically analyzed corresponding to (A–C). (E,F) The expression levels of SA-signaling and JA-signaling genes in “Williams82” (WT) and the Gmlmm2-1 mutant. The data shown are the mean values of three biological replicates and the error bars represent the standard deviation (SD). Data are presented as fold difference from the WT control after normalizing to the control gene GmActin11. Relative expression levels are presented as means ± SD. Data are from three biological replicates. *P < 0.05, **P < 0.01, ***P < 0.001 (t-test).
To test whether spontaneous lesion formation in mutant correlated with the expression of resistance-related genes, we monitored the transcript levels of these genes in “Williams 82” and the Gmlmm2-1 mutant. SA-signaling genes (GmNPR1, GmPR1, GmPR10, and GmSnRK1.1) were more highly expressed in the Gmlmm2-1 mutant compared with those in “Williams 82” (Figure 6E). The transcript level of GmSnRK in the Gmlmm2-1 mutant was 5.5-fold higher than that in “Williams 82.” The expression of GmPR10 in the Gmlmm2-1 mutant was 1373.3-fold higher than that in “Williams 82.” As shown in Figure 6F, these JA-signaling genes (GmAOS, GmLOX, GmCOI1, and GmMYB014) were activated at different levels in the Gmlmm2-1 mutant. The expression of GmCOI1 in the Gmlmm2-1 mutant was 4.17-fold higher than that in “Williams 82.” These results indicated that the expressions of defense-related genes are up-regulated during lesion development.
Coproporphyrinogen III Oxidase (CPO) catalyzes the coproporphyrinogen-III to yield protoporphyrinogen-IX in tetrapyrrole biosynthesis. Defects in CPO have been reported in Nicotiana tabacum, Arabidopsis, and rice (Kruse et al., 1995; Ishikawa et al., 2001; Sun et al., 2011; Wang et al., 2015). This study is the first report to demonstration the function of CPO in soybean. Many lmms mutants exhibit light-dependent cell death, accompanied by the upregulation of ROS-related genes (Sathe et al., 2019). In our study, the GmLMM2 defect in the Gmlmm2-1 mutant also induced a light-dependent lesion mimic phenotype, and higher light intensity led to severe necrosis and chlorophyll deficiency (Figures 5A,B,E,F). Trypan blue and DAB staining assays showed that cell death in the Gmlmm2-1 mutant was caused by abnormal H2O2 accumulation (Figure 1). The up-regulation of GmNOX led to ROS accumulation in the illuminated region of the Gmlmm2-1 mutant, and not in the dark region (Figure 5C). As a large amount of H2O2 accumulated in the illuminated region of the Gmlmm2-1 mutant, expression of the genes encoding H2O2-scavenging enzymes, such as GmCAT, was significantly upregulated (Figure 5D). ROS accumulation can trigger the expression of resistance-related genes, such as PR genes; therefore, many lmms have enhanced resistance to pathogens (Dangl and Jones, 2001; Guo et al., 2013; Wang et al., 2015; Lv et al., 2019).
Resistance-related genes may be activated in lmms and contribute to enhanced pathogen resistance (Guo et al., 2013; Wang et al., 2015). In the present study, the expressions of important genes involved in JA and SA signaling pathways were upregulated in the Gmlmm2-1 mutant and the Gmlmm2-1 mutant displayed enhanced resistance to P. sojae P7076. The SA pathway has been implicated in conferring resistance to biotrophic pathogens, such as Pseudomonas syringae and Hyaloperonospora arabidopsidis, and the JA pathway in resistance to necrotrophic pathogens, such as Alternaria brassicicola and Botrytis cinerea (Penninckx et al., 1998; Pieterse and Van Loon, 1999; Glazebrook, 2005; Spoel et al., 2007). The induction of marker genes involved in the two defense signaling pathways suggested that the Gmlmm2-1 mutant may have acquired a broad spectrum of resistance to multiple pathogens.
In summary, the Gmlmm2-1 mutant exhibits light-induced lesions. The identification of GmLMM2 improves our understanding of the molecular mechanism underlying cell death and the defense response in soybean. In addition, it helps us to further understand the complex regulation of the tetrapyrrole biosynthesis pathway in soybean.
Sequences were deposited at the National Center for Biotechnology Information (NCBI) under the accession number SRP149750.
SY and XZF conceived and designed the study. JM, DW, and XXF performed the experiments and wrote the manuscript. JM, KT, and XXF modified the images. KT analyzed the sequencing data. JM, SY, and XZF discussed the results and prepared the manuscript. All authors read and approved the manuscript.
This work was supported by a National Key R&D Project (Grant nos. 2016YFD0101900) from the Ministry of Science and Technology of China; by Programs (Grant nos. 31971901 and 31571692) from the National Natural Science Foundation of China, and by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant no. XDA0801010502).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00557/full#supplementary-material
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43:W39. doi: 10.1093/nar/gkv416
Bruggeman, Q., Raynaud, C., Benhamed, M., and Delarue, M. (2015). To die or not to die? Lessons from lesion mimic mutants. Front. Plant Sci. 6:24. doi: 10.3389/fpls.2015.00024
Cao, W. L., Chu, R. Z., Zhang, Y., Luo, J., Su, Y. Y., Xie, L. J., et al. (2015). OsJAMyb, a R2R3-type myb transcription factor, enhanced blast resistance in transgenic rice. Physiol. Mol. Plant Pathol. 92, 154–160. doi: 10.1016/j.pmpp.2015.04.008
Chai, Q., Shang, X., Wu, S., Zhu, G., Cheng, C., Cai, C., et al. (2017). 5-Aminolevulinic acid dehydratase gene dosage affects programmed cell death and immunity. Plant Physiol. 175, 511–528. doi: 10.1104/pp.17.00816
Cheng, W., Gao, J. S., Feng, X. X., Shao, Q., Yang, S. X., and Feng, X. Z. (2016). Characterization of dwarf mutants and molecular mapping of a dwarf locus in soybean. J. Integr. Agric. 15, 2228–2236.
Dangl, J. L., Dietrich, R. A., and Richberg, M. H. (1996). Death don’t have no mercy: cell death programs in plant-microbe interactions. Plant Cell 8, 1793–1807. doi: 10.2307/3870230
Dangl, J. L., and Jones, J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Dickman, M. B., and Fluhr, R. (2013). Centrality of host cell death in plant-microbe interactions. Annu. Rev. Phytopathol. 51, 543–570. doi: 10.1146/annurev-phyto-081211-173027
Du, H., Zeng, X., Zhao, M., Cui, X., Wang, Q., Hui, Y., et al. (2016). Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotechnol. 217, 90–97. doi: 10.1016/j.jbiotec.2015.11.005
Hwu, F. Y., Lai, M. W., and Liou, R. F. (2017). PpMID1 plays a role in the asexual development and virulence of Phytophthora parasitica. Front. Microbiol. 8:610. doi: 10.3389/fmicb.2017.00610
Feng, X. X., Tang, K. Q., Zhang, Y. H., Leng, J. T., Ma, J. J., Wang, Q., et al. (2019). GmPGL1, a thiamine thiazole synthase, is required for the biosynthesis of thiamine in Soybean. Front. Plant Sci. 10:1546. doi: 10.3389/fpls.2019.01546
Förster, H., Tyler, B., and Coffey, M. (1994). Phytophthora sojae races have arisen by clonal evolution and by rare outcrosses. Mol. Plant Microbe Interact. 7, 780–791.
Genty, B., Briantais, J. M., and Baker, N. R. (1989). The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta Gen. Subj. 990, 87–92.
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Guo, C. Y., Wu, G. H., Xing, J., Li, W. Q., Tang, D. Z., and Cui, B. M. (2013). A mutation in a coproporphyrinogen III oxidase gene confers growth inhibition, enhanced powdery mildew resistance and powdery mildew-induced cell death in Arabidopsis. Plant Cell Rep. 32, 687–702. doi: 10.1007/s00299-013-1403-8
Hodges, D. M., Delong, J. M., Forney, C. F., and Prange, R. K. (1999). Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207, 604–611. doi: 10.2307/23385611
Hoisington, D., Neuffer, M., and Walbot, V. (1982). Disease lesion mimics in maize: I. Effect of genetic background, temperature, developmental age, and wounding on necrotic spot formation with Les1. Dev. Biol. 93, 381–388. doi: 10.1016/0012-1606(82)90125-7
Hu, G., Yalpani, N., Briggs, S. P., and Johal, G. S. (1998). A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10, 1095–1105. doi: 10.1105/tpc.10.7.1095
Huang, M., Slewinski, T. L., Baker, R. F., Janick-Buckner, D., Buckner, B., Johal, G. S., et al. (2009). Camouflage patterning in maize leaves results from a defect in porphobilinogen deaminase. Mol. Plant 2, 773–789. doi: 10.1093/mp/ssp029
Ishikawa, A., Okamoto, H., Iwasaki, Y., and Asahi, T. (2001). A deficiency of coproporphyrinogen III oxidase causes lesion formation in Arabidopsis. Plant J. 27, 89–99. doi: 10.1046/j.1365-313x.2001.01058.x
Jia, Y., Liu, G., Park, D. S., and Yang, T. N. (2013). Inoculation and scoring methods for rice sheath blight disease. Rice Protoc. 956, 257–268. doi: 10.1007/978-1-62703-194-3_19
Kowalewska, L., Mazur, R., Suski, S., Garstka, M., and Mostowska, A. (2016). Three-dimensional visualization of the tubular-lamellar transformation of the internal plastid membrane network during runner bean chloroplast biogenesis. Plant Cell 28, 875–891. doi: 10.1105/tpc.15.01053
Kruse, E., Mock, H. P., and Grimm, B. (1995). Reduction of coproporphyrinogen oxidase level by antisense RNA synthesis leads to deregulated gene expression of plastid proteins and affects the oxidative defense system. EMBO J. 14, 3712–3720. doi: 10.1002/j.1460-2075.1995.tb00041.x
Lee, D., Lee, G., Kim, B., Jang, S., Lee, Y., Yu, Y., et al. (2018). Identification of a spotted leaf sheath gene involved in early senescence and defense response in rice. Front. Plant Sci. 9:1274. doi: 10.3389/fpls.2018.01274
Lee, D. S., Flachsová, E., Bodnárová, M., Demeler, B., Martásek, P., and Raman, C. S. (2005). Structural basis of hereditary coproporphyria. Proc. Natl. Acad. Sci. U.S.A. 102, 14232–14237. doi: 10.1073/pnas.0506557102
Lichtenthaler, H., and Wellburn, A. (1983). Determination of total carotenoids and chlorophylls a and b of leaf in different solvents. Biochem. Soc. Trans. 11, 591–592. doi: 10.1042/bst0110591
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lorrain, S., Vailleau, F., Balagué, C., and Roby, D. (2003). Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8, 263–271. doi: 10.1016/S1360-1385(03)00108-0
Lv, R. Q., Li, Z., Li, M., Dogra, V., and Kim, C. (2019). Uncoupled expression of nuclear and plastid photosynthesis-associated genes contributes to cell death in a lesion mimic mutant. Plant Cell 31, 210–230. doi: 10.1105/tpc.18.00813
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Meyer, A., Eskandari, S., Grallath, S., and Rentsch, D. (2006). AtGAT1, a high affinity transporter for gamma-aminobutyric acid in Arabidopsis thaliana. J. Biol. Chem. 281, 7197–7204. doi: 10.1074/jbc.M510766200
Moeder, W., and Yoshioka, K. (2008). Lesion mimic mutants: a classical, yet still fundamental approach to study programmed cell death. Plant Signal. Behav. 3, 764–767. doi: 10.4161/psb.3.10.6545
Ouchane, S., Steunou, A. S., Picaud, M., and Astier, C. (2004). Aerobic and anaerobic Mg-Protoporphyrin monomethyl ester cyclases in purple bacteria: a strategy adopted to bypass the repressive oxygen control system. J. Biol. Chem. 279, 6385–6394. doi: 10.1074/jbc.M309851200
Penninckx, I. A., Thomma, B. P., Buchala, A., Métraux, J. P., and Broekaert, W. F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. doi: 10.1105/tpc.10.12.2103
Phillips, J. D., Whitby, F. G., Warby, C. A., Labbe, P., Yang, C., Pflugrath, J. W., et al. (2004). Crystal structure of the oxygen-dependant coproporphyrinogen oxidase (Hem13p) of Saccharomyces cerevisiae. J. Biol. Chem. 279, 38960–38968. doi: 10.1074/jbc.M406050200
Pieterse, C. M., and Van Loon, L. C. (1999). Salicylic acid-independent plant defence pathways. Trends Plant Sci. 4, 52–58. doi: 10.1016/s1360-1385(98)01364-8
Pratibha, P., Singh, S. K., Srinivasan, R., Bhat, S. R., and Sreenivasulu, Y. (2017). Gametophyte development needs mitochondrial coproporphyrinogen III oxidase function. Plant Physiol. 174, 258–275. doi: 10.1104/pp.16.01482
Qiao, Y., Jiang, W., Lee, J. H., Park, B. S., Choi, M. S., Piao, R., et al. (2010). SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit μ1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol. 185, 258–274. doi: 10.1111/j.1469-8137.2009.03047.x
Quesada, V., Sarmiento-Mañús, R., González-Bayón, R., Hricová, A., Ponce, M. R., and Micol, J. L. (2013). PORPHOBILINOGEN DEAMINASE deficiency alters vegetative and reproductive development and causes lesions in Arabidopsis. PLoS One 8:e53378. doi: 10.1371/journal.pone.0053378
Rebeiz, C. Z., Smith, B. B., Mattheis, J. R., Rebeiz, C. C., and Dayton, D. F. (1975). Chloroplast biogenesis: biosynthesis and accumulation of Mg-protoporphyrin IX monoester and other metalloporphyrins by isolated etioplasts and developing chloroplasts. Arch. Biochem. Biophys. 167, 351–365. doi: 10.1016/0003-9861(75)90471-3
Sathe, A. P., Su, X., Chen, Z., Su, X., Chen, Z., Chen, T., et al. (2019). Identification and characterization of a spotted-leaf mutant spl40 with enhanced bacterial blight resistance in rice. Rice 12:68. doi: 10.1186/s12284-019-0326-6
Song, X., Wei, H., Cheng, W., Yang, S., Zhao, Y., Xuan, L., et al. (2015). Development of INDEL markers for genetic mapping based on whole genome resequencing in soybean. G3 5, 2793–2799. doi: 10.1534/g3.115.022780
Spoel, S. H., Johnson, J. S., and Dong, X. (2007). Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. U.S.A. 104, 18842–18847. doi: 10.1073/pnas.0708139104
Sun, C., Liu, L., Tang, J., Lin, A., Zhang, F., Fang, J., et al. (2011). RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J. Genet. Genomics 38, 29–37. doi: 10.1016/j.jcg.2010.12.001
Sun, L., Wang, Y., Liu, L. L., Wang, C., Gan, T., Zhang, Z., et al. (2017). Isolation and characterization of a spotted leaf 32 mutant with early leaf senescence and enhanced defense response in rice. Sci. Rep. 7:41846. doi: 10.1038/srep41846
Takahashi, A., Kawasaki, T., Henmi, K., Shii, K., Kodama, O., Satoh, O., et al. (1999). Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J. 17, 535–545. doi: 10.1046/j.1365-313X.1999.00405.x
Tanaka, R., Kobayashi, K., and Masuda, T. (2011). Tetrapyrrole metabolism in Arabidopsis thaliana. Arabidopsis Book 9:e0145. doi: 10.1199/tab.0145
Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Tsunezuka, H., Fujiwara, M., Kawasaki, T., and Shimamoto, K. (2005). Proteome analysis of programmed cell death and defense signaling using the rice lesion mimic mutant cdr2. Mol. Plant Microbe Interact. 18, 52–59. doi: 10.1094/MPMI-18-0052
van Verk, M. C., Pappaioannou, D., Neeleman, L., Bol, J. F., and Linthorst, H. J. M. (2008). A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 146, 1983–1995. doi: 10.1104/pp.107.112789
Wang, J., Ye, B., Yin, J., Yuan, C., Zhou, X., Li, W., et al. (2015). Characterization and fine mapping of a light-dependent leaf lesion mimic mutant 1 in rice. Plant Physiol. Biochem. 97, 44–51. doi: 10.1016/j.plaphy.2015.09.001
Wang, L., Wang, H., He, S., Meng, F., Zhang, C., Fan, S., et al. (2019). GmSnRK1. 1, a sucrose non-fermenting-1 (SNF1)-related protein kinase, promotes soybean resistance to Phytophthora sojae. Front. Plant Sci. 10:996. doi: 10.3389/fpls.2019.00996
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Weigel, R. R., Bäuscher, C., Pfitzner, A. J., and Pfitzner, U. M. (2001). NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol. Biol. 46, 143–160. doi: 10.1023/A:1010652620115
Wolter, M., Hollricher, K., Salamini, F., and Schulze-Lefert, P. (1993). The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol. Gen. Genet. 239, 122–128. doi: 10.1007/bf00281610
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., and Turner, J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Xu, P., Jiang, L., Wu, J., Li, W., Fan, S., and Zhang, S. (2014). Isolation and characterization of a pathogenesis-related protein 10 gene (GmPR10) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. Mol. Biol. Rep. 41, 4899–4909. doi: 10.1007/s11033-014-3356-6
Yamori, W., Takahashi, S., Makino, A., Price, G. D., Badger, M. R., and Von Caemmerer, S. (2011). The roles of ATP synthase and the cytochrome b6/f complexes in limiting chloroplast electron transport and determining photosynthetic capacity. Plant Physiol. 155, 956–962. doi: 10.1104/pp.110.168435
Yin, Z., Chen, J., Zeng, L., Goh, M., Leung, H., Khush, G. S., et al. (2000). Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol. Plant Microbe Interact. 13, 869–876. doi: 10.1094/MPMI.2000.13.8.869
Zeng, H., Zhang, Y., Zhang, X., Pi, E., and Zhu, Y. (2017). Analysis of EF-Hand proteins in soybean genome suggests their potential roles in environmental and nutritional stress signaling. Front. Plant Sci. 8:877. doi: 10.3389/fpls.2017.00877
Zhao, B., Dai, A., Wei, H., Yang, S., Wang, B., Jiang, N., et al. (2016). Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 90, 33–47. doi: 10.1007/s11103-015-0392-0
Zheng, K., Tian, H., Hu, Q., Guo, H., Yang, L., Cai, L., et al. (2016). Ectopic expression of R3 MYB transcription factor gene OsTCL1 in Arabidopsis, but not rice, affects trichome and root hair formation. Sci. Rep. 6:19254. doi: 10.1038/srep19254
Keywords: Glycine max, lesion mimic mutant 2, tetrapyrrole biosynthesis, coproporphyrinogen III oxidase, pathogen resistance
Citation: Ma J, Yang S, Wang D, Tang K, Feng XX and Feng XZ (2020) Genetic Mapping of a Light-Dependent Lesion Mimic Mutant Reveals the Function of Coproporphyrinogen III Oxidase Homolog in Soybean. Front. Plant Sci. 11:557. doi: 10.3389/fpls.2020.00557
Received: 01 February 2020; Accepted: 14 April 2020;
Published: 08 May 2020.
Edited by:
Chang-Jun Liu, Brookhaven National Laboratory (DOE), United StatesReviewed by:
Mingyue Gou, Henan Agricultural University, ChinaCopyright © 2020 Ma, Yang, Wang, Tang, Feng and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suxin Yang, eWFuZ3N1eGluQGlnYS5hYy5jbg==; Xian Zhong Feng, ZmVuZ3hpYW56aG9uZ0BuZWlnYWUuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.