
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 06 November 2018
Sec. Plant Development and EvoDevo
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01621
This article is part of the Research TopicGenetic Regulatory Mechanisms Underlying Developmental Shifts in Plant EvolutionView all 19 articles
Arbuscular mycorrhiza is one of the most common plant symbiotic interactions observed today. Due to their nearly ubiquitous occurrence and their beneficial impact on both partners it was suggested that this mutualistic interaction was crucial for plants to colonize the terrestrial habitat approximately 500 Ma ago. On the plant side the association is established via the common symbiotic pathway (CSP). This pathway allows the recognition of the fungal symbiotic partner, subsequent signaling to the nucleus, and initiation of the symbiotic program with respect to specific gene expression and cellular re-organization. The downstream part of the CSP is a regulatory network that coordinates the transcription of genes necessary to establish the symbiosis, comprising multiple GRAS transcription factors (TFs). These regulate their own expression as an intricate transcriptional network. Deduced from non-host genome data the loss of genes encoding CSP components coincides with the loss of the interaction itself. Here, we analyzed bryophyte species with special emphasis on the moss Physcomitrella patens, supposed to be a non-host, for the composition of the GRAS regulatory network components. We show lineage specific losses and expansions of several of these factors in bryophytes, potentially coinciding with the proposed host/non-host status of the lineages. We evaluate losses and expansions and infer clade-specific evolution of GRAS TFs.
Mycorrhiza is the most common plant–fungus symbiotic interaction we observe today. Over 80% of all extant plant species engage in this symbiotic interaction (Bonfante and Genre, 2010) which is beneficial to both partners (mutualistic). The plant provides the fungus with carbohydrates and lipids, in turn the fungus provides the plant host with nutrients like nitrate and especially phosphorus. Additionally, the fungal hyphae enlarge the rhizosphere area of the plant and seem to improve plant stress tolerance (Bago et al., 2003; Liu et al., 2007; Feddermann et al., 2010; Veresoglou et al., 2012). Several forms of mycorrhizal interactions exist, of which arbuscular mycorrhiza (AM) is the most common one. It is called ‘arbuscular’ since the fungal hyphae grow into the plant cells forming a ‘tree-like’ structure called arbuscule. In AM this structure represents the nutrient exchange zone between plant and fungus, since the plant-derived so-called peri-arbuscular membrane is loaded with transporters to facilitate the described nutrient exchange. The actual composition of the respective membrane in terms of transporters is predominantly known for plants (Harrison et al., 2002; Bonfante and Genre, 2010; Gaude et al., 2012; Luginbuehl and Oldroyd, 2017; MacLean et al., 2017).
Although beneficial for them, plants need to regulate and coordinate this symbiotic interaction because intensive cellular reprogramming is required and the plant needs to restrict the degree of colonization by the fungus in correspondence to its own nutritional status (Koide and Schreiner, 1992; Breuillin et al., 2010; Balzergue et al., 2011), e.g., to avoid carbon loss (Carbonnel and Gutjahr, 2014). Additionally, and perhaps most important, the beneficial partner needs to be distinguished from potential pathogens. Plants, and most probably already their progenitors, the streptophyte algae, evolved the so called common symbiotic pathway (CSP) (Oldroyd, 2013; Delaux et al., 2015) that enables this distinctive signaling. The pathway is called ‘common’ because a large part of the set of genes that evolved to accommodate arbuscular mycorrhiza (AM) was later recruited by the Rhizobium legume symbiosis (Kistner and Parniske, 2002). Numerous components of the CSP in AM host plants have been identified, but these analyzes were predominantly performed in seed plants (see Delaux et al., 2013b; Oldroyd, 2013 for review and Figure 1A for overview). Via this pathway the plant detects the nearby fungus by its secreted lipo-chito-oligosaccharides (LCOs) and other myc factors such as short-chain chitin oligomers (COs) (Maillet et al., 2012; Genre et al., 2013) and prepares for colonization by starting a specific transcriptional program and cellular reorganization (Gutjahr and Parniske, 2013; Pimprikar and Gutjahr, 2018). In turn, the fungus senses the plant host, predominantly through strigolactones secreted by the plant, and starts intensive hyphal growth and branching toward the symbiotic partner (Akiyama et al., 2005; Besserer et al., 2006). The fungal signals are recognized by a receptor complex at the plant plasma membrane that involves Lysine motif (LysM) receptor like kinases (RLKs). This complex seems to be more intricate than previously thought, since it becomes more and more evident that multiple signals and receptors contribute to composite signal processing (Antolín-Llovera et al., 2012; Conn and Nelson, 2015; Gutjahr et al., 2015; Sun et al., 2015; Zhang et al., 2015). The signal is transduced to the nucleus by a so far not fully characterized mechanism involving most probably mevalonate and potentially further factors (Venkateshwaran et al., 2015). Multiple ion channels in the nuclear envelope elicit a symbiotic Ca2+ oscillating signal (spiking) in the nucleus (Charpentier et al., 2008). The factors described so far make up what we will henceforth call the ‘signaling module’ of the CSP (Figure 1A). This module transduces the external signal to the nucleus where it results in calcium oscillation. This symbiosis-specific calcium spiking activates the calcium and calmodulin-dependent kinase (CCaMK), a key player of the CSP, which in turn regulates the transcription factor (TF) CYCLOPS, which is thought to initiate a transcriptional regulatory network (Singh et al., 2014;Pimprikar et al., 2016). This network of various TFs controls, together with CYCLOPS, the transcription of some additional TFs and the ‘later genes’ that encode factors which are, for example, needed for arbuscule initiation, branching and transmembrane transport (Harrison et al., 2002; Zhang et al., 2010; Takeda et al., 2011). The transcription of all those factors is tightly regulated and especially GRAS [Gibberellic acid insensitive (GAI), Repressor of GAI (RGA), and Scarecrow (SCR)] proteins are important regulators in this developmental process (Gutjahr, 2014; Xue et al., 2015). This family originated from a bacterial methylase (Zhang et al., 2012) and apparently evolved in streptophyte algae (Wilhelmsson et al., 2017), the sister lineage to land plants. GRAS proteins fulfill important regulatory roles in plant growth, response to environment and development (Peng et al., 1999; Pysh et al., 1999; Hirsch and Oldroyd, 2009; Sun et al., 2011). Recently the DNA binding capability of GRAS proteins was reported (Li et al., 2016), demonstrating that they might act as TFs. However, their mode of action as regulators is still highly debated (Hirano et al., 2017). In case of arbuscular mycorrhiza, so far, predominantly Reduced Arbuscular Mycorrhization 1 (RAM1), Required for Arbuscule Development1 (RAD1), Nodulation signaling pathway 1 (NSP1) and NSP2 were identified as prominent regulators, although NSP1 and NSP2 were previously thought to be root nodule symbiosis specific (Gobbato et al., 2012, 2013; Lauressergues et al., 2012; Maillet et al., 2012; Delaux et al., 2013a; Hohnjec et al., 2015; Park et al., 2015; Rich et al., 2015; Xue et al., 2015; Pimprikar et al., 2016) (Figure 1A). Additionally, recently further potential GRAS TFs were proposed to be involved in mycorrhizal regulation (Xue et al., 2015; Heck et al., 2016). It has been suggested that the action of the four mentioned GRAS TFs is highly interconnected or dependent of each other, thus forming a transcriptional network (Xue et al., 2015). For instance, it was shown that RAM1 interacts with RAD1 and controls several ‘later genes’ (Park et al., 2015; Xue et al., 2015). The transcription of RAM1 in turn is controlled by CYCLOPS and DELLA (Pimprikar et al., 2016). NSP1 and NSP2 were shown to interact directly in nodulation (Hirsch et al., 2009). Additionally, NSP2 was shown to interact with RAM1 by yeast-2-hybrid and bimolecular fluorescence complementation (Gobbato et al., 2012). Adding an additional layer of complexity in the control of this symbiosis, it was shown that NSP2 is regulated by the microRNA MiR171h in flowering plants (Devers et al., 2011; Lauressergues et al., 2012; Hofferek et al., 2014).
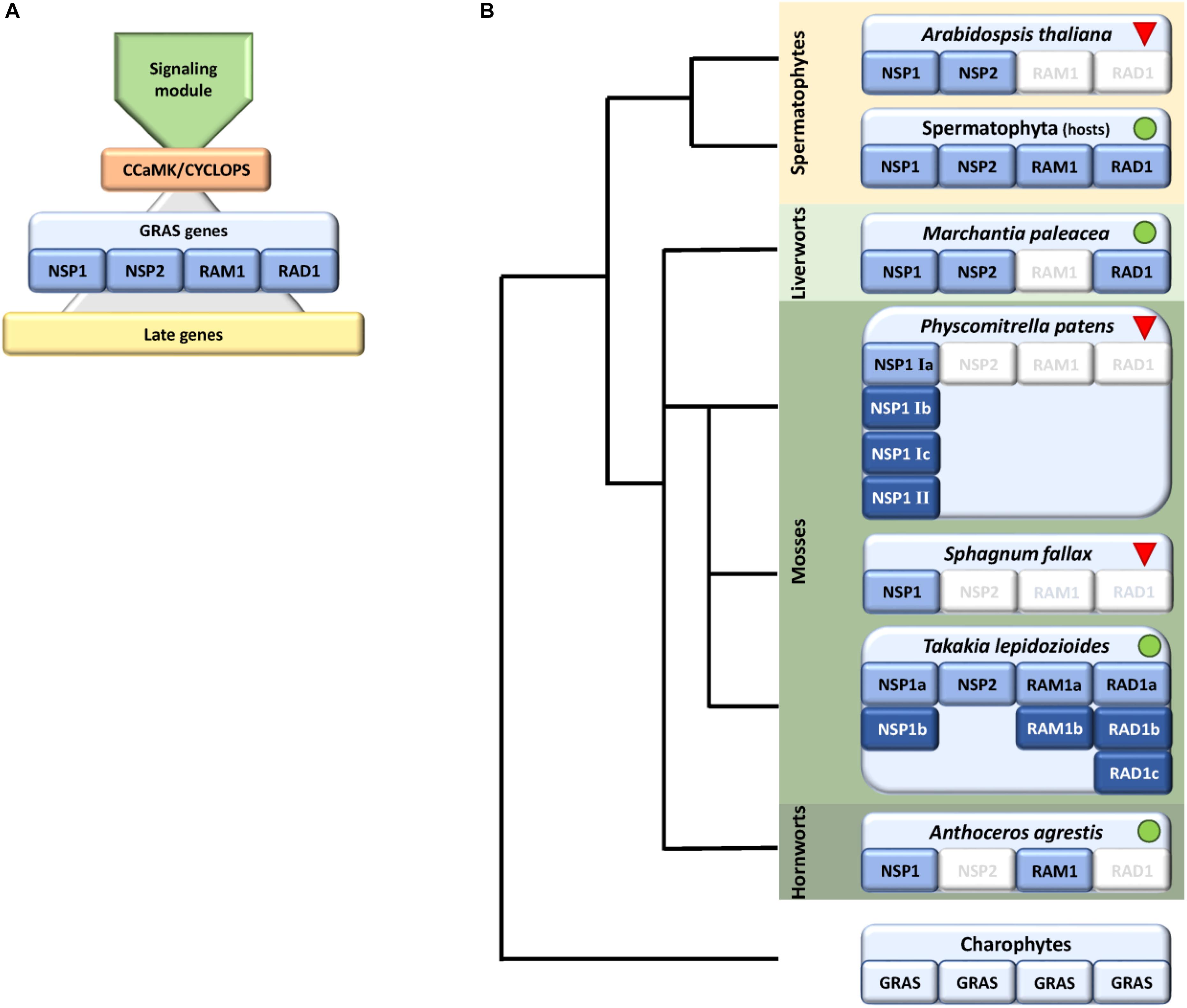
FIGURE 1. Schematic representation of identified factors in the symbiosis GRAS signaling module with a focus on bryophytes. (A) Scheme showing the main parts of the common symbiotic pathway (CSP), consisting of a signaling module, the main hub CCaMK/CYCLOPS, which controls start of the transcriptional program eventually controlled by GRAS genes (NSP1/2, RAD1, and RAM1). The GRAS module regulates the transcription of later genes. (B) Presence/absence of GRAS factors within bryophytes shown on a schematic tree, predominantly based on genomic data. We were not able to identify RAM1/RAD1 in liverworts and NSP2/RAD1 in hornworts, with the exception of RAD1 in transcriptomic data of M. paleacea. In case of mosses we observed an expansion of NSP1 in the crown group mosses (Bryophytina) but only Takakia shows the full set of symbiotic GRAS genes. Sphagnum encodes NSP1 and NSP2 whereas crown group (“true”) mosses, exemplified by P. patens, encode up to six NSP1 genes. Dark blue coloring indicates duplications of respective genes. Green dots and red triangles indicate known host and non-host status, respectively. The land plant ancestor most probably encoded all four symbiosis-specific GRAS sub families.
Due to their structure and mode of action, e.g., being functional as homo- or hetero-dimer, GRAS TFs seem not only to act as TFs but also as some kind of ‘hub proteins’ to interconnect signals from different pathways, e.g., hormone signaling, to regulate complex cellular reprogramming (Thieulin-Pardo et al., 2015; Li et al., 2016). This hub function becomes obvious in the example of DELLA proteins. These proteins are specialized GRAS TFs consisting of a GRAS domain and an additional DELLA domain. These are known to be key regulators in gibberellic acid (GA) signaling (Sun, 2011). GA presence inhibits arbuscule formation, and DELLA proteins are degraded under this condition. In turn, DELLA proteins, although not AM specific, are important for arbuscule formation (Floss et al., 2013). Very recently it was shown that DELLAs interact with CCaMK/CYCLOPS and potentially additional TFs to regulate RAM1 transcription (Pimprikar et al., 2016). Additionally, DELLAs interact with the GRAS TF DELLA interacting protein 1 (DIP1) and RAD1, which in turn interact with RAM1 (Yu et al., 2014; Park et al., 2015; Takeda et al., 2015; Xue et al., 2015; Pimprikar et al., 2016), indicating the importance of DELLA and plant hormones in AM development and regulation. Moreover, abscisic acid (ABA) has been shown to promote mycorrhizal development, possibly by stabilizing DELLA (Achard et al., 2006), and by regulating GA levels in the context of symbiosis (Martin-Rodriguez et al., 2016).
Recently, the evolution of the CSP was analyzed, covering datasets ranging from chlorophytes to spermatophytes (Delaux et al., 2014, 2015). It was shown that CSP factors are present in charophyte algae, especially those for signal perception and processing of the Ca2+ signal in the nucleus. Hence, some CSP factors were already present before the water-to-land transition (Delaux et al., 2015). However, with respect to the GRAS genes, orthologs of the symbiosis-specific GRAS TFs known from extant land plants were not detected (Delaux et al., 2015).
Symbiosis specific genes are lost in non-host plants, leading to a specific absence/presence pattern of CSP components (Delaux et al., 2014; Favre et al., 2014; Bravo et al., 2016). Hence, presence and absence of these factors may allow a conclusion on the symbiotic status of the plant analyzed. Indeed, some land plant lineages lost the ability to form a mycorrhizal partnership, among them the Brassicaceae with the prime plant model, the weed Arabidopsis thaliana. The model moss Physcomitrella patens (Funariceae) is not known to form AMF associations in nature, although intracellular growth can be occasionally detected in culture (Hanke and Rensing, 2010) and the relative Funaria hygrometrica was described to show AMF association in a companion plant assay (Parke and Linderman, 1980). While A. thaliana has lost genes required for responding to symbiotic fungi (Delaux et al., 2014), P. patens retained orthologs of these, at least for factors of the signaling module of the CSP (Wang et al., 2010; Delaux et al., 2015). Our study focuses on bryophytes, comprising mosses, hornworts, and liverworts. While liverworts and hornworts are generally considered host plants, most mosses are considered non-hosts (Field and Pressel, 2018). The crown group mosses (Bryophytina or true mosses) comprise the classes Oedipodiopsida, Polytrichopsida, Tetraphidopsida (each with a single sub class) as well as the major class Bryopsida, comprising eight sub classes. Sister lineages to the Bryophytina are the three single class comprising sub divisions Andreaeophytina, Sphagnophytina (comprising the genus Sphagnum, peat mosses) and Takakiophytina. Their branching order remains under debate, with Takakiophytina (comprising the single genus Takakia with the two species T. lepidoziodes and T. ceratophylla) probably being sister to all other mosses (Volkmar and Knoop, 2010; Ligrone et al., 2012). The only accepted evidence for host plants within the mosses is in the basal lineage represented by Takakia (Boullard, 1988). Here, we performed comprehensive phylogenomic analyzes of the GRAS transcriptional regulatory network in bryophytes and found lineage specific losses and expansions of these key symbiotic signaling pathway components. We evaluate our findings with respect to the host and non-host status of the species or lineages in question and hypothesize on the (early) clade-specific evolution of symbiotic GRAS genes.
GRAS TFs were acquired using the HMM-based (using the motif PF03514) TAPscan classification (Wilhelmsson et al., 2017) against a database of sequenced plant and algal genomes and transcriptomes (Supplementary Table S1). An initial alignment and phylogenetic tree of all GRAS TFs was constructed, and sequences from the clades representing the sub families involved in symbiosis signaling (NSP1, NSP2, RAD1, RAM1) were selected, aligned, manually curated and used to generate HMMs specific to each of the four sub families using hmmbuild from HMMer (Finn et al., 2015) 3.1b1 (HMMs available upon request). HMMsearch was used with these HMMs against all GRAS proteins in order to determine each of the sub families. To aid this selection, HMM search scores were derived of the basalmost sequences of the phylogenetic clade in question, and of the next closest phylogenetic clade in the tree. Cutoff scores were then derived to lie between these values. Because the resulting list of sequences of RAD1 and RAM1 largely overlapped, these two clades were combined in a single phylogenetic analysis. While all sequences of non-vascular plants were used, seed plants were represented by selected species to cover gymnosperms, basal angiosperms as well as mono- and di-cotyledonous flowering plants. Additionally, a putative Lunularia cruciata RAM1 sequence (Delaux et al., 2015) was added, but based on our phylogenetic analyses could not be confirmed as RAM1. Each of the three protein sets was aligned using Mafft L-INS-i (Katoh and Standley, 2013). Alignments were manually curated using Jalview (Waterhouse et al., 2009), removing identical sequences and cropping columns to only represent the GRAS domain. Sequences of non-symbiotic GRAS proteins, namely the A. thaliana DELLA proteins GAI and RGA, were added for outgroup rooting (see Supplementary Figure S1 for relationships of the GRAS clades). The best suited amino acid substitution model was determined using Prottest 3 (Darriba et al., 2011) and turned out to be JTT+G+F. Bayesian inference utilizing MrBayes 3.2 (Ronquist et al., 2012) was carried out with two hot and cold chains until the average standard deviation of split frequencies was below 0.01 and no more trend was observable. 150 trees each were discarded as burn-in. Resulting trees were visualized using FigTree 1.4.01. The three alignments that are the basis for the phylogenetic trees are provided as Supplementary Files.
Transcriptome completeness was assessed by determining the percentage of eukaryotic single copy orthologs represented as full length transcripts (Supplementary Table S1), as implemented in BUSCO (Simao et al., 2015). The 1KP transcriptome datasets are based on whole plants.
Many factors of the CSP have been identified in recent years and it has been shown that plants which do not engage in a mutualistic symbiosis with AMF lost CSP genes (Delaux et al., 2014; Favre et al., 2014; Bravo et al., 2016). Furthermore, it was shown that basic factors of the CSP, such as DMI3/CCaMK and DMI1/POLLUX, were already present in the streptophyte algae, the sister lineage of land plants (Wickett et al., 2014; Delaux et al., 2015). Most probably GRAS proteins and other TFs act in a complex way by forming homo- and hetero-dimers (or multimers) among each other which then regulate the respective target genes (Gobbato et al., 2012, 2013; Hohnjec et al., 2015; Xue et al., 2015). Since mycorrhiza establishment requires fundamental changes to cell structure and physiology, the transcriptional program needs to be tightly regulated and factors involved are key regulators of this alteration. This regulatory network was so far predominantly studied in spermatophytes (especially in the legumes lotus and medicago) leading to a somewhat biased knowledge about presence and absence of these factors. Therefore, we screened for homologs of GRAS TFs (NSP1, NSP2, RAM1, RAD1) previously described in the prime model organisms for mycorrhiza research, Medicago truncatula and/or Lotus japonicus, and further analyzed them by phylogenic inference with special emphasis on bryophytes. Through this we were able to elucidate probable presence/absence patterns for factors of the GRAS transcriptional network in bryophyte clades.
Analysis of factors downstream of the signaling module and Ca2+ spiking (CCaMK) in non-seed plants and algae has in part already been undertaken recently (Delaux et al., 2015). Analyzing specifically the GRAS TFs and creating GRAS TF trees in more depth (Figures 2–4 and Supplementary Figure S1), we found no orthologs of NSP2 (Figure 2), RAD1 and RAM1 (Figure 3) in P. patens or other crown group mosses (Bryophytina) based on fully sequenced genomes, transcriptome data (Szovenyi et al., 2010, 2014) and data from the 1,000 plant transcriptomes project 1KP (Matasci et al., 2014). We also included genomic data for the more basal lineages of Sphagnum and Takakia. In case of Sphagnum we were able to detect only NSP1 (Figures 1B, 4). In contrast to that we found all factors to be present in Takakia.
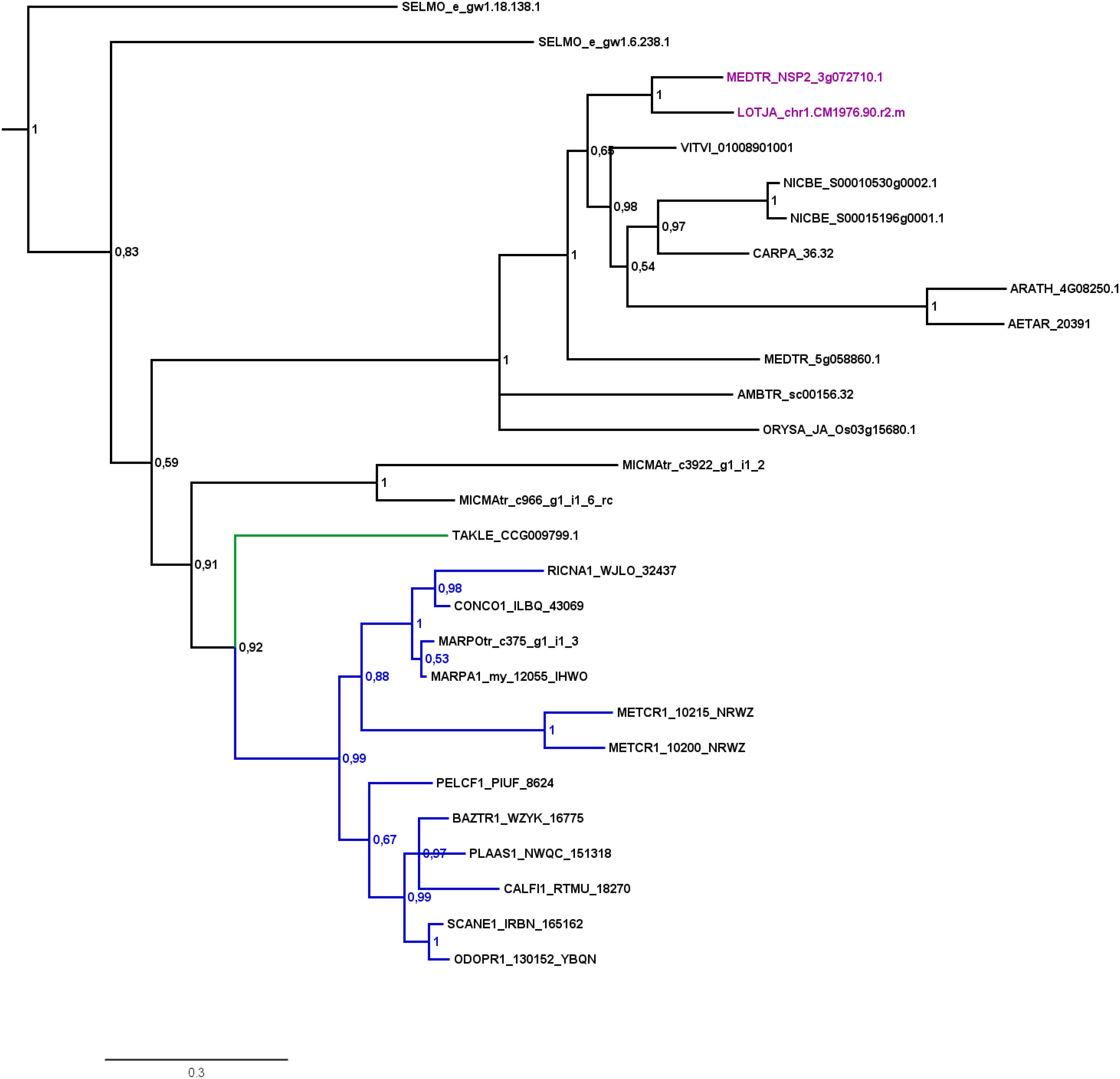
FIGURE 2. Phylogenetic tree for NSP2. Phylogenetic reconstruction of NSP2 using Bayesian inference. NSP2 was found in the basal lineage represented by the moss Takakia (green branches) and in liverworts (blue branches), but not in hornworts. Sequences of L. japonicum and M. truncatula are highlighted in purple. Five letter codes of the form MARchantia POlymorpha (MARPO) are used to abbreviate species names (Supplementary Table S1). Five letter codes followed by “1” depict 1KP sequences, those followed by “tr” represent sequences from non-1KP transcriptomes. The Marchantia paleacea transcriptome generated from mycorrhized tissue is marked by “my,” the one generated from non-mycorrhized tissue by “nm.” Note that an NSP2 transcript was found in the mycorrhized (my) 1KP library for Marchantia paleacea, as well as in the Marchantia polymorpha transcriptome. Posterior probabilities are shown at the nodes, the tree was outgroup-rooted by A. thaliana GAI and RGA (not shown).
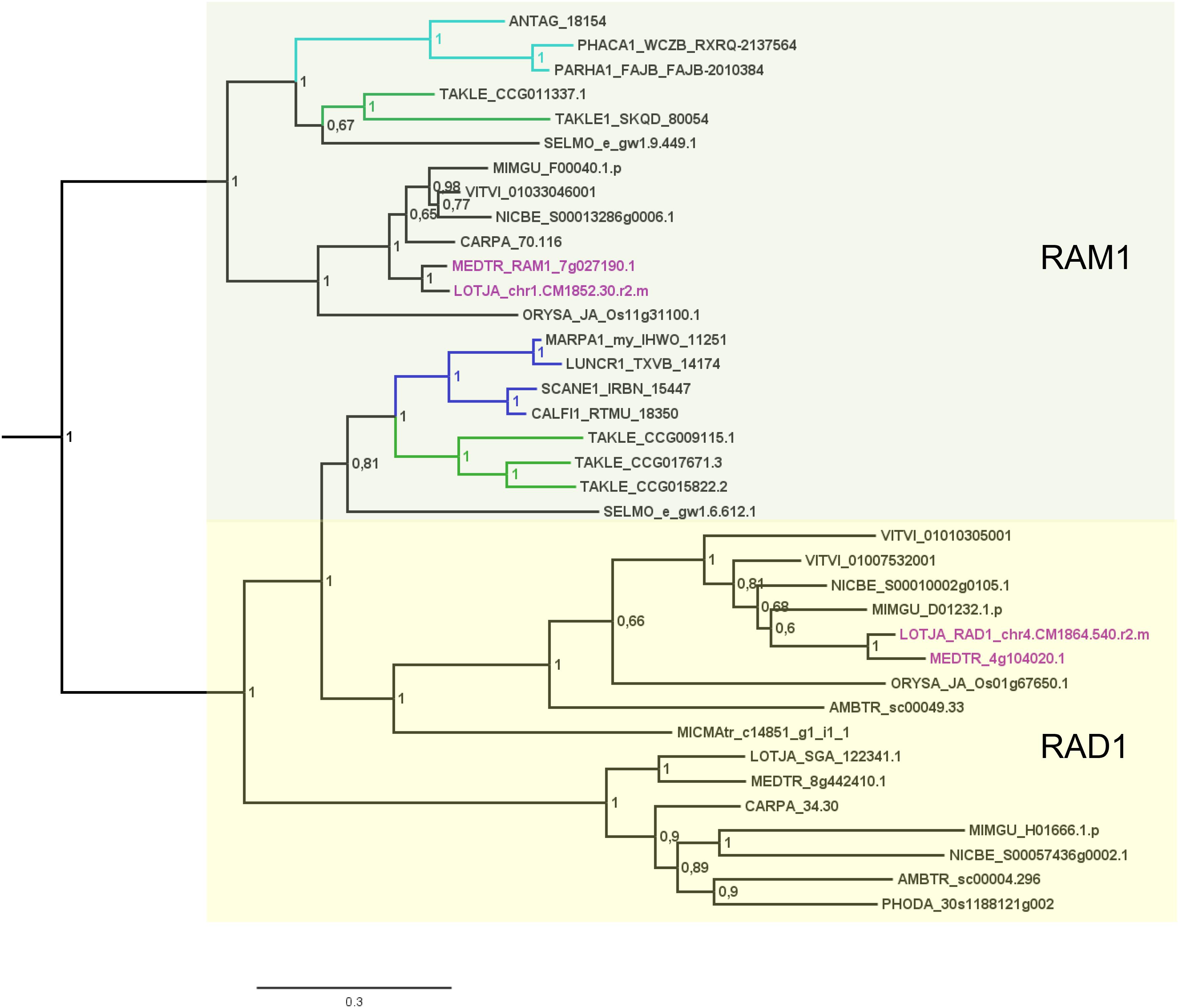
FIGURE 3. Phylogenetic tree for RAD1 and RAM1. Phylogenetic reconstruction of RAD1 and RAM1 (clades marked using boxes) using Bayesian inference. RAD1 was found in the basal moss lineage represented by Takakia (green branches) and in liverworts (blue branches), but not in hornworts. In contrast, RAM1 was found only in Takakia and hornworts (cyan branches). Sequences of L. japonicum and M. truncatula are highlighted in purple. Note that a RAD1 transcript was only found in the mycorrhized (my) 1KP library for Marchantia paleacea, and that no RAD1 gene was found in the Marchantia polymorpha genome or transcriptome. Posterior probabilities are shown at the nodes, the tree was outgroup-rooted by A. thaliana GAI and RGA (not shown); see legend to Figure 2 for explanation of naming.
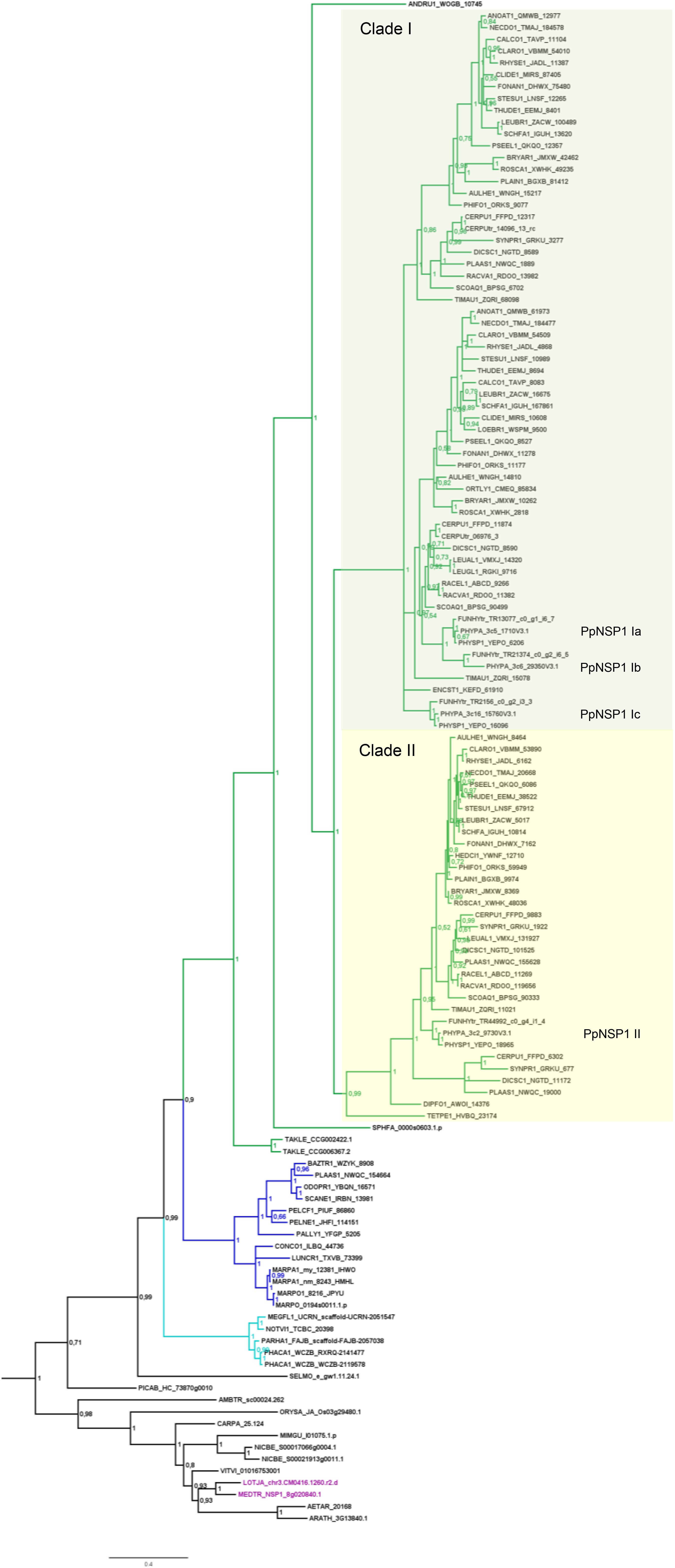
FIGURE 4. Phylogenetic tree for NSP1. Phylogenetic reconstruction of NSP1 using Bayesian inference. NSP1 was found in mosses (green branches), liverworts (blue branches), and hornworts (cyan branches). In the case of crown group mosses (Bryophytina) an expansion of NSP1 is evident due to the presence of several paralogs per species. Two main clades (marked by boxes, clades I and II) can be observed for moss NSP1. Mosses typically encode four NSP1 copies; naming of individual NSP1 genes is provided for P. patens as an example. Note that the basal lineages represented by Takakia, Sphagnum, and Andreaea do not share the diversified NSP1 clades of the other mosses. Sequences of L. japonicum and M. truncatula are shown in purple. Note that NSP1 transcripts were found in all transcriptomes of M. paleacea and M. polymorpha. Posterior probabilities are shown at the nodes, the tree was outgroup-rooted by A. thaliana GAI and RGA (not shown); see legend to Figure 2 for explanation of naming.
Our GRAS phylogenetic analyzes did not detect liverwort orthologs of RAM1, although 1KP data (Matasci et al., 2014) were included (Figure 3). Apart from that we found NSP1, NSP2, and RAD1 in the liverwort lineage. Interestingly, while NSP1 and NSP2 are detected in mycorrhizal as well as non-mycorrhizal tissue of Marchantia paleacea and Marchantia polymorpha, RAD1 is only detected in mycorrhizal M. paleacea (Figure 3). For hornworts we had access to 1KP data and preliminary sequence data for Anthoceros agrestis (kindly provided by Peter Szovenyi). We could identify NSP1 and RAM1, but not NSP2 or RAD1 in hornworts (Figures 1–4). In summary, the only full set of symbiotic GRAS TFs in bryophytes was detected in genomic data of the basal moss lineage represented by Takakia lepidozioides (kindly provided by Yikun He).
Besides the mentioned lineage-specific absence of genes, our analyzes show expansions of some GRAS sub families. Although no orthologs for NSP2, RAM1, and RAD1 were found in P. patens, we found four paralogs for NSP1 and a general expansion of this GRAS TF in crown group mosses (Bryophytina). Two main clades of NSP1s in mosses are obvious and most mosses seem to possess four NSP1 paralogs divided into the two clades (Figure 4). In case of Sphagnum and Andreaea we detected only one copy of NSP1 each, and two copies in case of Takakia. These three species represent the sister lineages to the Bryophytina and obviously did not share the later evolutionary diversification of NSP1. Interestingly, expansions of the other three GRAS sub families were only observable for Takakia, for which we identified one NSP2 but two paralogs of RAM1 and three paralogs of RAD1 (Figure 3).
Taken together, we identified a specific absence/presence pattern for the analyzed GRAS TFs involved in mycorrhiza signaling. Losses of parts of the symbiotic GRAS genes were found in all examined bryophyte lineages except for Takakia. P. patens as example for the mosses shows most losses, having lost NSP2, RAM1 and RAD1. Expansions of factors were detected in Takakia (NSP1, RAM1, RAD1) and Bryophytina (NSP1) (Figure 1B). Although data of algae were included in the database mined, and GRAS TFs are present in streptophyte algae (Bowman et al., 2017; Wilhelmsson et al., 2017), orthologs of the symbiotic GRAS TFs could not be identified in algae.
From the beginning of their conquest of land, around 500 Ma ago (Lang et al., 2010; Morris et al., 2018), land plants most probably have been in contact and/or symbiosis with fungal partners and nowadays over 80% of all extant land plants continue this mutualistic relationship (Fitter, 2005). Plants make use of a signaling module for recognition and establishment of the symbiosis, the main factors of which have already been present before the water-to-land transition of plants (Delaux et al., 2015). This might indicate that these factors are also important for microbial interactions in an aquatic environment, which are also common although so far less studied (Hempel et al., 2008; Rodriguez et al., 2009; Wurzbacher et al., 2010; Kataržytė et al., 2017). This view is supported by the fact that root nodule symbioses also make use of the CSP to establish plant–bacterial symbiosis (Oldroyd, 2013). Additionally, some microbes adopt this pathway to have parasitic access to plants; most probably parasitism is as ancient as symbiosis or predated and led to it (Corradi and Bonfante, 2012; Wang et al., 2012; Gobbato et al., 2013; Rey et al., 2015, 2017). As outlined above, components of the signaling module but not symbiosis related GRAS TFs were already present in the most recent common ancestor of land plants and charophyte algae. Indeed, we were also not able to identify sequences of charophytes orthologous to ‘symbiotic’ GRAS TFs. As proposed before (Delaux et al., 2015) the symbiotic GRAS signaling most probably evolved by duplication events from GRAS TFs already present in streptophyte algae (Bowman et al., 2017; Wilhelmsson et al., 2017).
Delaux et al. (2015) detected orthologs for symbiotic GRAS TFs in bryophytes. The full complement was detected for liverworts only (especially Lunularia cruciata, for which the transcriptome was sequenced). Furthermore, they identified two factors (RAM1 and RAD1) for Takakia, and NSP1 in hornworts and mosses. Our GRAS phylogenetic trees (Figures 2–4) expand this view in so far that we found all four factors in Takakia, added the basal moss lineage represented by Sphagnum (having NSP1 only), and found NSP2 and RAM1 in hornworts. We were not able to identify RAM1 in liverworts. The previously reported putative Lunularia cruciata transcriptomic RAM1 sequence (Delaux et al., 2015) grouped outside the RAM1/RAD1 clade in our analysis, maybe due to its fragmentary sequence. The detection of these GRAS TFs in transcriptomic data is potentially flawed because some of them are only expressed upon detection of or colonization by AM fungi, and thereby activation of the CSP (Xue et al., 2015; Pimprikar et al., 2016; Rich et al., 2017). Hence, they might not be expressed under the conditions from which the respective transcriptome was sequenced. Additionally, hornworts are unfortunately underrepresented in the 1KP data. Such problems do not apply if full genomes (with a certain quality) are available (e.g., P. patens or the liverwort M. polymorpha). The strength of our study is that we use for the first time genomic data for each of the bryophyte lineages, thus at least partially overcoming the limitations of transcriptomic data. However, it also clearly demonstrates that we need more genomic data for bryophytes and other non-seed plants (Rensing, 2017).
In the case of mosses, only NSP1 is present and clearly expanded, exemplified by, e.g., P. patens encoding four NSP1 genes. Using the M. truncatula sequences encoding NSP2, RAD1 and RAM1, no hits can be recovered in the P. patens genome assembly. Also, the best BLASTP hits of the M. truncatula genes flanking the three GRAS loci are not part of syntenic regions detected between P. patens and other plant genomes (Lang et al., 2018). Neither is any of the four NSP1 paralogs part of such a syntenic region. Hence, the regions encoding three of the four genes seem to have been lost from the genome. As outlined above, there are two moss NSP1 subclades (Figure 4, clades I and II). Most of the mosses have at least one sequence in each of these clades, and typically encode four paralogs. The topology and distribution pattern of NSP1 genes indicate a common duplication event giving rise to the two clades, observed in the crown group mosses (Bryophytina) but not shared by the sister lineages represented by Takakia, Sphagnum, and Andreaea. These duplications might be related to whole genome duplication (WGD) events observable in mosses (Lang et al., 2018), leading to subsequent neo- and/or sub-functionalization of duplicated genes (Rensing, 2014). Published expression for P. patens (Perroud et al., 2018) shows that the four paralogs show different expression levels (Nsp1 Ib lowest, Nsp1 Ia highest), but a qualitatively similar expression profile across the available developmental stages (Supplementary Table S1). NSP1 was identified as being important in root nodule symbiosis (RNS) (Catoira et al., 2000; Smit et al., 2005) and later on it was shown to also influence AM, since this TF is an important factor of the strigolactone (SL) biosynthesis pathway (Liu et al., 2011; Delaux et al., 2013b; Takeda et al., 2013; Hohnjec et al., 2015). SL biosynthesis is important for the establishment of the AM symbiosis because the fungus senses SL and the hyphal branching increases upon this stimulus (Akiyama et al., 2010). Interestingly, although P. patens does not encode an NSP2 gene, which is also necessary for SL biosynthesis in seed plants (Liu et al., 2011), it releases SLs (Proust et al., 2011). Biosynthesis of SL in P. patens is induced by phosphate starvation (a condition under which AMF association typically occurs), and leads to resistance to pathogenic fungi (Decker et al., 2017). Potentially, duplicated moss NSP1 genes sub-/neo-functionalized and compensate for the loss of NSP2. As GRAS proteins can act as hetero- and/or homodimers (Hirsch et al., 2009; Li et al., 2016) it is a feasible scenario that the four NSP1 paralogs might take over functions typically carried out by other GRAS proteins in other plants. Intriguingly, only NSP1 genes of clade I (Figure 4) seem to have been duplicated in mosses (the naming of individual NSP1 genes in the overview in Figure 1B is according to this division). This might indicate that a first sub- or neo-functionalization of NSP1 genes already occurred after the first duplication and in the second duplication event duplicated genes of group II were selected against, maybe because of unfavorable consequences due to stoichiometry of the dimer partners. However, so far, we are not able to assign certain functions to the individual NSP1 genes in P. patens.
Looking at the potential overall evolution of the symbiosis GRAS signaling genes we found support for the view of Delaux et al. (2015). Given the distribution of GRAS factors we hypothesize that the land plant ancestor encoded the full set of GRAS TFs (Figure 1). In bryophyte clades we can observe several losses (especially in mosses) and expansions (also mainly in mosses) of these GRAS TFs. Mosses have lost most factors of these GRAS genes (NSP2, RAM1, RAD1) and according to that they, including Sphagnum, are considered non-host plants (Read et al., 2000; Wang and Qiu, 2006). Although the signaling module seems to be intact, they also lost some genes which are, e.g., important for the periarbuscular membrane (Wang et al., 2010; Delaux et al., 2014, 2015). The symbiotic GRAS sub family losses might explain why a tight association or even symbiosis cannot be established in mosses. An exception at the basis of the mosses is Takakia, which encodes all four GRAS factors (plus expansions for NSP1, RAM1, and RAD1), and indeed was reported to engage in AM (Boullard, 1988). Given that Takakia represents one of the sister lineages of the Bryophytina, an evolutionary loss of NSP2, RAD1, and RAM1 during moss evolution appears the most probable scenario.
Most liver- and hornworts are considered host plants (Field and Pressel, 2018). As mentioned, we were not able to identify RAM1 in liverworts, or NSP2 and RAD1 in hornworts. This might be due to the mentioned problem with transcriptome coverage, but is also explainable by species-specific losses of AM capability exemplified by M. polymorpha, which does not show AM and lacks RAD1 (Figure 3) in contrast to its close relative M. paleacea (Humphreys et al., 2010; Bowman et al., 2017). The lack of M. polymorpha mycorrhizal association is in line with the absence of GAs and might be featured by nutrient rich habitats (Ligrone et al., 2007). While genes needed for successful mycorrhization are absent in non-host Marchantia species, other gene families are over-represented in M. polymorpha, e.g., transporters for phosphate and ammonium. These genomic adaptations might reflect the shift from mycorrhizal to non-mycorrhizal status by improving the transport capacity instead of being dependent on symbiotic organisms (Bowman et al., 2017). Nevertheless, according to our analysis both lack RAM1, which is not in line with the species’ host/non-host status, since M. paleacea, as host, should have the complete GRAS gene set. This is most probably due to the mentioned incomplete nature of the transcriptomic data. In case of NSP2 we can identify a potential coding region in the M. polymorpha genome (encoding the same protein detected in the transcriptome, Figure 2) that does not have a gene model assigned to it; updated genome versions might solve this issue. Presence of transcripts in transcriptomic data is evidence of presence of the gene, but absence of transcripts must not necessarily reflect absence of the gene (for example, the 1KP transcriptomes contain on average 84% of the conserved eukaryotic single copy gene set, Supplementary Table S1). For our overview (Figure 1B) we are thus relying mainly on genomic data in order not to represent conclusions based on transcriptomic absence of genes.
With rising morphological complexity more complex regulation and cellular reorganization is needed. Most probably we do not yet know all TFs involved in the regulation of this symbiosis, which involves tight regulation and massive cellular reorganization. Recent publications indicate that even more factors are involved, at least in seed plants (Xue et al., 2015; Heck et al., 2016). This indicates that we are only at the beginning of understanding this complex pathway and its transcriptional network (Genre and Russo, 2016; Pimprikar and Gutjahr, 2018). However, a quick survey showed that for example the GRAS TF MIG1 seems not to be present in bryophytes (data not shown). Most probably the transcriptional network to establish mycorrhizal symbiosis comprises more factors and is thereby more complex in vascular plants, due to more cell types and tissue layers. It is important to note that our current knowledge of the CSP is predominantly based on studies in spermatophytes (and here again predominantly analyzed of lotus and medicago). Maybe some GRAS genes are less important in bryophytes as compared to the situation in seed or vascular plants, for example due to their lack of roots.
If we evaluate the evolution of the CSP and its downstream components we should also be considering other plant–microbe associations and symbioses. Recently, the view was broadened since fungi belonging to the Mucoromycotina and also Ascomycotina were shown to interact in particular with liverworts and maybe hornworts (Field et al., 2014; Kowal et al., 2018), and the plant–Mucoromycotina interaction was proposed to potentially represent the ancestral state of plant–fungus interaction (Field et al., 2015). It is also known that plants interact with bacteria or even with both, fungi and bacteria (Bonfante and Anca, 2009). Foremost known is the RNS, which makes use of many factors of the CSP (Oldroyd, 2013; Genre and Russo, 2016). Interaction with cyanobacteria is also known, in particular for hornworts but also in some liverworts, mosses, ferns, and seed plants. There are more microbial/plant associations and symbioses known (e.g., Frankia, etc.) (Santi et al., 2013; Martin et al., 2017), and most probably many more we do not know yet. These interactions are important and widespread, and probably evolved already in the aquatic environment (Croft et al., 2005; Hom and Murray, 2014). How are these associations and symbioses regulated and how do the symbiotic partners identify each other? Most likely key components of the CSP (signaling module) are also involved in symbioses other than the ones they have so far been implicated in (mycorrhizal, rhizobial, and actinorhizal) (Martin et al., 2017). The signaling module apparently evolved in streptophyte algae (Delaux et al., 2015), suggesting that it may be functional in additional associations and symbioses, e.g., with cyanobacteria. The factors that process the symbiotic calcium spiking and induce a specific transcriptional program might be specific for each kind of symbiosis, leading to an association/symbiosis-specific diversification of the CSP downstream of the signaling module. More molecular studies in additional symbioses, especially in aquatic environments, are needed to unravel additional symbiosis-specific factors.
The key pathway to regulate beneficial interactions in plants seems to be the CSP (Martin et al., 2017). Furthermore, it is believed that microbial interactions enabled plants to conquer the land (Pirozynski and Malloch, 1975), highlighting the importance of this signaling pathway. Here we argue that the symbiosis related GRAS signaling genes, known to be important in regulation of AM symbiosis, were already present in the most recent common ancestor of all land plants. These genes display lineage specific losses and expansions in bryophytes, in particular in mosses. Such losses seem to reflect the non-host status. Nevertheless, the upstream CSP signaling module for symbiosis establishment seems to be intact (Wang et al., 2010; Delaux et al., 2015) and may serve in additional symbioses with a different or an extended subset of factors in the transcriptional network module. Additional studies are needed to elucidate the symbiosis-specific interplay of TFs and the functions of, e.g., the duplicated NSP1 genes in mosses.
SAR and CG conceived of the study. SAR carried out the phylogenetic analyses. CG, ACG, and SAR analyzed the data. CG and SAR wrote the paper, with contributions by ACG.
This project was funded by the German Research Foundation (DFG RE1697/6-1 to SAR), the Forschungsförderfond of the University of Marburg, the University of Freiburg/the Ministry of Science, Research and Art of the state of Baden-Württemberg (RiSC co-grant to SAR).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank M. Göttig for excellent technical assistance, C. Gutjahr for critical comments on the manuscript, and N. Fernandez-Pozo for carrying out BUSCO analyses. They would like to thank Peter Szovenyi for access to Anthoceros agrestis and Yikun He for access to Takakia lepidozioides draft genome data.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01621/full#supplementary-material
FIGURE S1 | Overview phylogeny of the GRAS family. Midpoint rooted Bayesian inference tree of selected species, including Lotus japonicus, Medicago truncatula, Arabidopsis thaliana, Physcomitrella patens, and Marchantia polymorpha. Line thickness corresponds to posterior probabilities. Colored clades depict RAD1 (red), RAM1 (purple), NSP1 (blue), NSP2 (green), and DELLA (cyan). Note that DELLA proteins were used as outgroup to root each of the trees shown in Figures 2–4.
TABLE S1 | Lists the sources of genomic and transcriptomic datasets used, five letter species abbreviations used in the Figures, BUSCO completeness percentages and expression data for the P. patens NSP1 paralogs. Alignments of NSP1, NSP2, and RAD1/RAM1 used for the phylogenetic analyses shown in Figures 2–4.
Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. doi: 10.1126/science.1118642
Akiyama, K., Matsuzaki, K., and Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827. doi: 10.1038/nature03608
Akiyama, K., Ogasawara, S., Ito, S., and Hayashi, H. (2010). Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol. 51, 1104–1117. doi: 10.1093/pcp/pcq058
Antolín-Llovera, M., Ried, M. K., Binder, A., and Parniske, M. (2012). Receptor kinase signaling pathways in plant-microbe interactions. Annu. Rev. Phytopathol. 50, 451–473. doi: 10.1146/annurev-phyto-081211-173002
Bago, B., Pfeffer, P. E., Abubaker, J., Jun, J., Allen, J. W., Brouillette, J., et al. (2003). Carbon export from arbuscular mycorrhizal roots involves the translocation of carbohydrate as well as lipid. Plant Physiol. 131, 1496–1507. doi: 10.1104/pp.102.007765
Balzergue, C., Puech-Pages, V., Bécard, G., and Rochange, S. F. (2011). The regulation of arbuscular mycorrhizal symbiosis by phosphate in pea involves early and systemic signalling events. J. Exp. Bot. 62, 1049–1060. doi: 10.1093/jxb/erq335
Besserer, A., Puech-Pages, V., Kiefer, P., Gomez-Roldan, V., Jauneau, A., Roy, S., et al. (2006). Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 4:e226. doi: 10.1371/journal.pbio.0040226
Bonfante, P., and Anca, I. A. (2009). Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu. Rev. Microbiol. 63, 363–383. doi: 10.1146/annurev.micro.091208.073504
Bonfante, P., and Genre, A. (2010). Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis. Nat. Commun. 1:48. doi: 10.1038/ncomms1046
Boullard, B. (1988). “Observations on the coevolution of fungi with hepatics,” in Coevolution of Fungi with Plants and Animals, eds K. A. Pirozynski and D. L. Hawksworth (Cambridge, MA: Academic Press).
Bowman, J. L., Kohchi, T., Yamato, K. T., Jenkins, J., Shu, S., Ishizaki, K., et al. (2017). Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304.e15. doi: 10.1016/j.cell.2017.09.030
Bravo, A., York, T., Pumplin, N., Mueller, L. A., and Harrison, M. J. (2016). Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nat. Plants 2:15208. doi: 10.1038/nplants.2015.208
Breuillin, F., Schramm, J., Hajirezaei, M., Ahkami, A., Favre, P., Druege, U., et al. (2010). Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 64, 1002–1017. doi: 10.1111/j.1365-313X.2010.04385.x
Carbonnel, S., and Gutjahr, C. (2014). Control of arbuscular mycorrhiza development by nutrient signals. Front. Plant Sci. 5:462. doi: 10.3389/fpls.2014.00462
Catoira, R., Galera, C., de Billy, F., Penmetsa, R. V., Journet, E. P., Maillet, F., et al. (2000). Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12, 1647–1666. doi: 10.1105/tpc.12.9.1647
Charpentier, M., Bredemeier, R., Wanner, G., Takeda, N., Schleiff, E., and Parniske, M. (2008). Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20, 3467–3479. doi: 10.1105/tpc.108.063255
Conn, C. E., and Nelson, D. C. (2015). Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front. Plant Sci. 6:1219. doi: 10.3389/fpls.2015.01219
Corradi, N., and Bonfante, P. (2012). The arbuscular mycorrhizal symbiosis: origin and evolution of a beneficial plant infection. PLoS Pathog. 8:e1002600. doi: 10.1371/journal.ppat.1002600
Croft, M. T., Lawrence, A. D., Raux-Deery, E., Warren, M. J., and Smith, A. G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93. doi: 10.1038/nature04056
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. doi: 10.1093/bioinformatics/btr088
Decker, E. L., Alder, A., Hunn, S., Ferguson, J., Lehtonen, M. T., Scheler, B., et al. (2017). Strigolactone biosynthesis is evolutionarily conserved, regulated by phosphate starvation and contributes to resistance against phytopathogenic fungi in a moss, Physcomitrella patens. New Phytol. 216, 455–468. doi: 10.1111/nph.14506
Delaux, P.-M., Bécard, G., and Combier, J.-P. (2013a). NSP1 is a component of the Myc signaling pathway. New Phytol. 199, 59–65. doi: 10.1111/nph.12340
Delaux, P.-M., Séjalon-Delmas, N., Bécard, G., and Ané, J.-M. (2013b). Evolution of the plant-microbe symbiotic ‘toolkit’. Trends Plant Sci. 18, 298–304. doi: 10.1016/j.tplants.2013.01.008
Delaux, P. M., Radhakrishnan, G. V., Jayaraman, D., Cheema, J., Malbreil, M., Volkening, J. D., et al. (2015). Algal ancestor of land plants was preadapted for symbiosis. Proc. Natl. Acad. Sci. U.S.A. 112, 13390–13395. doi: 10.1073/pnas.1515426112
Delaux, P. M., Varala, K., Edger, P. P., Coruzzi, G. M., Pires, J. C., and Ane, J. M. (2014). Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet. 10:e1004487. doi: 10.1371/journal.pgen.1004487
Devers, E. A., Branscheid, A., May, P., and Krajinski, F. (2011). Stars and symbiosis: microRNA- and microRNA∗-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 156, 1990–2010. doi: 10.1104/pp.111.172627
Favre, P., Bapaume, L., Bossolini, E., Delorenzi, M., Falquet, L., and Reinhardt, D. (2014). A novel bioinformatics pipeline to discover genes related to arbuscular mycorrhizal symbiosis based on their evolutionary conservation pattern among higher plants. BMC Plant Biol. 14:333. doi: 10.1186/s12870-014-0333-0
Feddermann, N., Finlay, R., Boller, T., and Elfstrand, M. (2010). Functional diversity in arbuscular mycorrhiza – the role of gene expression, phosphorous nutrition and symbiotic efficiency. Fungal Ecol. 3, 1–8. doi: 10.1016/j.funeco.2009.07.003
Field, K. J., and Pressel, S. (2018). Unity in diversity: structural and functional insights into the ancient partnerships between plants and fungi. New Phytol. doi: 10.1111/nph.15158 [Epub ahead of print].
Field, K. J., Pressel, S., Duckett, J. G., Rimington, W. R., and Bidartondo, M. I. (2015). Symbiotic options for the conquest of land. Trends Ecol. Evol. 30, 477–486. doi: 10.1016/j.tree.2015.05.007
Field, K. J., Rimington, W. R., Bidartondo, M. I., Allinson, K. E., Beerling, D. J., Cameron, D. D., et al. (2014). First evidence of mutualism between ancient plant lineages (Haplomitriopsida liverworts) and Mucoromycotina fungi and its response to simulated Palaeozoic changes in atmospheric CO. New Phytol. 205, 743–756. doi: 10.1111/nph.13024
Finn, R. D., Clements, J., Arndt, W., Miller, B. L., Wheeler, T. J., Schreiber, F., et al. (2015). HMMER web server: 2015 update. Nucleic Acids Res. 43, W30–W38. doi: 10.1093/nar/gkv397
Fitter, A. H. (2005). Darkness visible: reflections on underground ecology. J. Ecol. 93, 231–243. doi: 10.1111/j.0022-0477.2005.00990.x
Floss, D. S., Levy, J. G., Levesque-Tremblay, V., Pumplin, N., and Harrison, M. J. (2013). DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 110, E5025–E5034. doi: 10.1073/pnas.1308973110
Gaude, N., Schulze, W. X., Franken, P., and Krajinski, F. (2012). Cell type-specific protein and transcription profiles implicate periarbuscular membrane synthesis as an important carbon sink in the mycorrhizal symbiosis. Plant Signal. Behav. 7, 461–464. doi: 10.4161/psb.19650
Genre, A., Chabaud, M., Balzergue, C., Puech-Pages, V., Novero, M., Rey, T., et al. (2013). Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 198, 190–202. doi: 10.1111/nph.12146
Genre, A., and Russo, G. (2016). Does a common pathway transduce symbiotic signals in plant-microbe interactions? Front. Plant Sci. 7:96. doi: 10.3389/fpls.2016.00096
Gobbato, E., Marsh, J. F., Vernié, T., Wang, E., Maillet, F., Kim, J., et al. (2012). A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 22, 2236–2241. doi: 10.1016/j.cub.2012.09.044
Gobbato, E., Wang, E., Higgins, G., Bano, S. A., Henry, C., Schultze, M., et al. (2013). RAM1 and RAM2 function and expression during arbuscular mycorrhizal symbiosis and Aphanomyces euteiches colonization. Plant Signal. Behav. 8:e26049. doi: 10.4161/psb.26049
Gutjahr, C. (2014). Phytohormone signaling in arbuscular mycorhiza development. Curr. Opin. Plant Biol. 20, 26–34. doi: 10.1016/j.pbi.2014.04.003
Gutjahr, C., Gobbato, E., Choi, J., Riemann, M., Johnston, M. G., Summers, W., et al. (2015). Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350, 1521–1524. doi: 10.1126/science.aac9715
Gutjahr, C., and Parniske, M. (2013). Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu. Rev. Cell Dev. Biol. 29, 593–617. doi: 10.1146/annurev-cellbio-101512-122413
Hanke, S. T. and Rensing, S. A. (2010). In vitro association of non-seed plant gametophytes with arbuscular mycorrhiza fungi. Endocyt. Cell. Res. 20, 95–101.
Harrison, M. J., Dewbre, G. R., and Liu, J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14, 2413–2429. doi: 10.1105/tpc.004861
Heck, C., Kuhn, H., Heidt, S., Walter, S., Rieger, N., and Requena, N. (2016). Symbiotic fungi control plant root cortex development through the novel GRAS transcription factor MIG1. Curr. Biol. 26, 2770–2778. doi: 10.1016/j.cub.2016.07.059
Hempel, M., Blume, M., Blindow, I., and Gross, E. M. (2008). Epiphytic bacterial community composition on two common submerged macrophytes in brackish water and freshwater. BMC Microbiol. 8:58. doi: 10.1186/1471-2180-8-58
Hirano, Y., Nakagawa, M., Suyama, T., Murase, K., Shirakawa, M., Takayama, S., et al. (2017). Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD. Nat. Plants 3:17010. doi: 10.1038/nplants.2017.10
Hirsch, S., Kim, J., Munoz, A., Heckmann, A. B., Downie, J. A., and Oldroyd, G. E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21, 545–557. doi: 10.1105/tpc.108.064501
Hirsch, S., and Oldroyd, G. E. (2009). GRAS-domain transcription factors that regulate plant development. Plant Signal. Behav. 4, 698–700. doi: 10.4161/psb.4.8.9176
Hofferek, V., Mendrinna, A., Gaude, N., Krajinski, F., and Devers, E. A. (2014). MiR171h restricts root symbioses and shows like its target NSP2 a complex transcriptional regulation in Medicago truncatula. BMC Plant Biol. 14:199. doi: 10.1186/s12870-014-0199-1
Hohnjec, N., Czaja-Hasse, L. F., Hogekamp, C., and Kuster, H. (2015). Pre-announcement of symbiotic guests: transcriptional reprogramming by mycorrhizal lipochitooligosaccharides shows a strict co-dependency on the GRAS transcription factors NSP1 and RAM1. BMC Genomics 16:994. doi: 10.1186/s12864-015-2224-7
Hom, E. F., and Murray, A. W. (2014). Plant-fungal ecology. Niche engineering demonstrates a latent capacity for fungal-algal mutualism. Science 345, 94–98. doi: 10.1126/science.1253320
Humphreys, C. P., Franks, P. J., Rees, M., Bidartondo, M. I., Leake, J. R., and Beerling, D. J. (2010). Mutualistic mycorrhiza-like symbiosis in the most ancient group of land plants. Nat. Commun. 1:103. doi: 10.1038/ncomms1105
Kataržytė, M., Vaičiūtė, D., Bučas, M., Gyraitė, G., and Petkuvienė, J. (2017). Microorganisms associated with charophytes under different salinity conditions. Oceanologia 59, 177–186. doi: 10.1016/j.oceano.2016.10.002
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kistner, C., and Parniske, M. (2002). Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci. 7, 511–518. doi: 10.1016/S1360-1385(02)02356-7
Koide, R. T., and Schreiner, R. P. (1992). Regulation of the vesicular-arbuscular mycorrhizal symbiosis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 557–581. doi: 10.1146/annurev.pp.43.060192.003013
Kowal, J., Pressel, S., Duckett, J. G., Bidartondo, M. I., and Field, K. J. (2018). From rhizoids to roots? Experimental evidence of mutualism between liverworts and ascomycete fungi. Ann. Bot. 121, 221–227. doi: 10.1093/aob/mcx126
Lang, D., Ullrich, K. K., Murat, F., Fuchs, J., Jenkins, J., Haas, F. B., et al. (2018). The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 93, 515–533. doi: 10.1111/tpj.13801
Lang, D., Weiche, B., Timmerhaus, G., Richardt, S., Riano-Pachon, D. M., Correa, L. G., et al. (2010). Genome-wide phylogenetic comparative analysis of plant transcriptional regulation: a timeline of loss, gain, expansion, and correlation with complexity. Genome Biol. Evol. 2, 488–503. doi: 10.1093/gbe/evq032
Lauressergues, D., Delaux, P. M., Formey, D., Lelandais-Briere, C., Fort, S., Cottaz, S., et al. (2012). The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 72, 512–522. doi: 10.1111/j.1365-313X.2012.05099.x
Li, S., Zhao, Y., Zhao, Z., Wu, X., Sun, L., Liu, Q., et al. (2016). Crystal structure of the GRAS domain of SCARECROW-LIKE7 in Oryza sativa. Plant Cell 28, 1025–1034. doi: 10.1105/tpc.16.00018
Ligrone, R., Carafa, A., Lumini, E., Bianciotto, V., Bonfante, P., and Duckett, J. G. (2007). Glomeromycotean associations in liverworts: a molecular, cellular, and taxonomic analysis. Am. J. Bot. 94, 1756–1777. doi: 10.3732/ajb.94.11.1756
Ligrone, R., Duckett, J. G., and Renzaglia, K. S. (2012). Major transitions in the evolution of early land plants: a bryological perspective. Ann. Bot. 109, 851–871. doi: 10.1093/aob/mcs017
Liu, J., Maldonado-Mendoza, I., Lopez-Meyer, M., Cheung, F., Town, C. D., and Harrison, M. J. (2007). Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J. 50, 529–544. doi: 10.1111/j.1365-313X.2007.03069.x
Liu, W., Kohlen, W., Lillo, A., Op, den Camp, R., Ivanov, S., et al. (2011). Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell Online 23, 3853–3865. doi: 10.1105/tpc.111.089771
Luginbuehl, L. H., and Oldroyd, G. E. D. (2017). Understanding the arbuscule at the heart of endomycorrhizal symbioses in plants. Curr. Biol. 27, R952–R963. doi: 10.1016/j.cub.2017.06.042
MacLean, A. M., Bravo, A., and Harrison, M. J. (2017). Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. Plant Cell 29, 2319–2335. doi: 10.1105/tpc.17.00555
Maillet, F., Poinsot, V., André, O., Puech-Pages, V., Haouy, A., Gueunier, M., et al. (2012). Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469, 58–63. doi: 10.1038/nature09622
Martin, F. M., Uroz, S., and Barker, D. G. (2017). Ancestral alliances: plant mutualistic symbioses with fungi and bacteria. Science 356:eaad4501.
Martin-Rodriguez, J. A., Huertas, R., Ho-Plagaro, T., Ocampo, J. A., Tureckova, V., Tarkowska, D., et al. (2016). Gibberellin-abscisic acid balances during arbuscular mycorrhiza formation in tomato. Front. Plant Sci. 7:1273. doi: 10.3389/fpls.2016.01273
Matasci, N., Hung, L. H., Yan, Z., Carpenter, E. J., Wickett, N. J., Mirarab, S., et al. (2014). Data access for the 1,000 Plants (1KP) project. Gigascience 3:17. doi: 10.1186/2047-217X-3-17
Morris, J. L., Puttick, M. N., Clark, J. W., Edwards, D., Kenrick, P., Pressel, S., et al. (2018). The timescale of early land plant evolution. Proc. Natl. Acad. Sci. U.S.A. 115, E2274–E2283. doi: 10.1073/pnas.1719588115
Oldroyd, G. E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Park, H.-J., Floss, D. S., Levesque-Tremblay, V., Bravo, A., and Harrison, M. J. (2015). Hyphal branching during arbuscule development requires reduced arbuscular mycorrhiza1. Plant Physiol. 169, 2774–2788.
Parke, J. L., and Linderman, R. G. (1980). Association of vesicular-arbuscular mycorrhizal fungi with the moss Funaria hygrometrica. Can. J. Bot. 58, 1898–1904. doi: 10.1139/b80-218
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: 10.1038/22307
Perroud, P. F., Haas, F. B., Hiss, M., Ullrich, K. K., Alboresi, A., Amirebrahimi, M., et al. (2018). The Physcomitrella patens gene atlas project: large scale RNA-seq based expression data. Plant J. 95, 168–182. doi: 10.1111/tpj.13940
Pimprikar, P., Carbonnel, S., Paries, M., Katzer, K., Klingl, V., Bohmer, M. J., et al. (2016). A CCaMK-CYCLOPS-DELLA complex activates transcription of Ram1 to regulate arbuscule branching. Curr. Biol. 26, 987–998. doi: 10.1016/j.cub.2016.01.069
Pimprikar, P., and Gutjahr, C. (2018). Transcriptional regulation of arbuscular mycorrhiza development. Plant Cell Physiol. 59, 673–690. doi: 10.1093/pcp/pcy024
Pirozynski, K. A., and Malloch, D. W. (1975). The origin of land plants: a matter of mycotrophism. Biosystems 6, 153–164. doi: 10.1016/0303-2647(75)90023-4
Proust, H., Hoffmann, B., Xie, X., Yoneyama, K., Schaefer, D. G., Yoneyama, K., et al. (2011). Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development 138, 1531–1539. doi: 10.1242/dev.058495
Pysh, L. D., Wysocka-Diller, J. W., Camilleri, C., Bouchez, D., and Benfey, P. N. (1999). The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18, 111–119. doi: 10.1046/j.1365-313X.1999.00431.x
Read, D. J. D., Ducket, J. G. J., Francis, R. R., Ligron, R. R., and Russell, A. A. (2000). Symbiotic fungal associations in ‘lower’ land plants. Philos. Trans. R. Soc. B Biol. Sci. 355, 815–811. doi: 10.1098/rstb.2000.0617
Rensing, S. A. (2014). Gene duplication as a driver of plant morphogenetic evolution. Curr. Opin. Plant Biol. 17, 43–48. doi: 10.1016/j.pbi.2013.11.002
Rensing, S. A. (2017). Why we need more non-seed plant models. New Phytol. 216, 355–360. doi: 10.1111/nph.14464
Rey, T., Bonhomme, M., Chatterjee, A., Gavrin, A., Toulotte, J., Yang, W., et al. (2017). The Medicago truncatula GRAS protein RAD1 supports arbuscular mycorrhiza symbiosis and Phytophthora palmivora susceptibility. J. Exp. Bot. 68, 5871–5881. doi: 10.1093/jxb/erx398
Rey, T., Chatterjee, A., Buttay, M., Toulotte, J., and Schornack, S. (2015). Medicago truncatula symbiosis mutants affected in the interaction with a biotrophic root pathogen. New Phytol. 206, 497–500. doi: 10.1111/nph.13233
Rich, M. K., Courty, P. E., Roux, C., and Reinhardt, D. (2017). Role of the GRAS transcription factor ATA/RAM1 in the transcriptional reprogramming of arbuscular mycorrhiza in Petunia hybrida. BMC Genomics 18:589. doi: 10.1186/s12864-017-3988-8
Rich, M. K., Schorderet, M., Bapaume, L., Falquet, L., Morel, P., Vandenbussche, M., et al. (2015). The petunia GRAS transcription factor ATA/RAM1 regulates symbiotic gene expression and fungal morphogenesis in arbuscular mycorrhiza. Plant Physiol. 168, 788–797. doi: 10.1104/pp.15.00310
Rodriguez, R. J., White, J. F. Jr., Arnold, A. E., and Redman, R. S. (2009). Fungal endophytes: diversity and functional roles. New Phytol. 182, 314–330. doi: 10.1111/j.1469-8137.2009.02773.x
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Santi, C., Bogusz, D., and Franche, C. (2013). Biological nitrogen fixation in non-legume plants. Ann. Bot. 111, 743–767. doi: 10.1093/aob/mct048
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Singh, S., Katzer, K., Lambert, J., Cerri, M., and Parniske, M. (2014). CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15, 139–152. doi: 10.1016/j.chom.2014.01.011
Smit, P., Raedts, J., Portyanko, V., Debellé, F., Gough, C., Bisseling, T., et al. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308, 1789–1791. doi: 10.1126/science.1111025
Sun, J., Miller, J. B., Granqvist, E., Wiley-Kalil, A., Gobbato, E., Maillet, F., et al. (2015). Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell 27, 823–838. doi: 10.1105/tpc.114.131326
Sun, T. P. (2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21, R338–R345. doi: 10.1016/j.cub.2011.02.036
Sun, X., Xue, B., Jones, W. T., Rikkerink, E., Dunker, A. K., and Uversky, V. N. (2011). A functionally required unfoldome from the plant kingdom: intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 77, 205–223. doi: 10.1007/s11103-011-9803-z
Szovenyi, P., Perroud, P. F., Symeonidi, A., Stevenson, S., Quatrano, R. S., Rensing, S. A., et al. (2014). De novo assembly and comparative analysis of the Ceratodon purpureus transcriptome. Mol. Ecol. Resour. 15, 203–215. doi: 10.1111/1755-0998.12284
Szovenyi, P., Rensing, S. A., Lang, D., Wray, G. A., and Shaw, A. J. (2010). Generation-biased gene expression in a bryophyte model system. Mol. Biol. Evol. 28, 803–812. doi: 10.1093/molbev/msq254
Takeda, N., Haage, K., Sato, S., Tabata, S., and Parniske, M. (2011). Activation of a Lotus japonicus subtilase gene during arbuscular mycorrhiza is dependent on the common symbiosis genes and two cis-active promoter regions. Mol. Plant Microbe Interact. 24, 662–670. doi: 10.1094/MPMI-09-10-0220
Takeda, N., Handa, Y., Tsuzuki, S., Kojima, M., Sakakibara, H., and Kawaguchi, M. (2015). Gibberellins interfere with symbiosis signaling and gene expression and alter colonization by arbuscular mycorrhizal fungi in Lotus japonicus. Plant Physiol. 167, 545–557. doi: 10.1104/pp.114.247700
Takeda, N., Tsuzuki, S., Suzaki, T., Parniske, M., and Kawaguchi, M. (2013). CERBERUS and NSP1 of Lotus japonicus are common symbiosis genes that modulate arbuscular mycorrhiza development. Plant Cell Physiol. 54, 1711–1723. doi: 10.1093/pcp/pct114
Thieulin-Pardo, G., Avilan, L., Kojadinovic, M., and Gontero, B. (2015). Fairy “tails”: flexibility and function of intrinsically disordered extensions in the photosynthetic world. Front. Mol. Biosci. 2:23. doi: 10.3389/fmolb.2015.00023
Venkateshwaran, M., Jayaraman, D., Chabaud, M., Genre, A., Balloon, A. J., Maeda, J., et al. (2015). A role for the mevalonate pathway in early plant symbiotic signaling. Proc. Natl. Acad. Sci. U.S.A. 112, 9781–9786. doi: 10.1073/pnas.1413762112
Veresoglou, S. D., Chen, B., and Rillig, M. C. (2012). Arbuscular mycorrhiza and soil nitrogen cycling. Soil Biol. Biochem. 46, 53–62. doi: 10.1016/j.soilbio.2011.11.018
Volkmar, U., and Knoop, V. (2010). Introducing intron locus cox1i624 for phylogenetic analyses in Bryophytes: on the issue of Takakia as sister genus to all other extant mosses. J. Mol. Evol. 70, 506–518. doi: 10.1007/s00239-010-9348-9
Wang, B., and Qiu, Y. L. (2006). Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16, 299–363. doi: 10.1007/s00572-005-0033-6
Wang, B., Yeun, L. H., Xue, J.-Y., Liu, Y., Ané, J.-M., and Qiu, Y.-L. (2010). Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol. 186, 514–525. doi: 10.1111/j.1469-8137.2009.03137.x
Wang, E., Schornack, S., Marsh, J. F., Gobbato, E., Schwessinger, B., Eastmond, P., et al. (2012). A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr. Biol. 22, 2242–2246. doi: 10.1016/j.cub.2012.09.043
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., and Barton, G. J. (2009). Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Wickett, N. J., Mirarab, S., Nguyen, N., Warnow, T., Carpenter, E., Matasci, N., et al. (2014). Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. U.S.A. 111, E4859–E4868. doi: 10.1073/pnas.1323926111
Wilhelmsson, P. K. I., Muhlich, C., Ullrich, K. K., and Rensing, S. A. (2017). Comprehensive genome-wide classification reveals that many plant-specific transcription factors evolved in streptophyte algae. Genome Biol. Evol. 9, 3384–3397. doi: 10.1093/gbe/evx258
Wurzbacher, C. M., Bärlocher, F., and Grossart, H. P. (2010). Fungi in lake ecosystems. Aquat. Microb. Ecol. 59, 125–149. doi: 10.3354/ame01385
Xue, L., Cui, H., Buer, B., Vijayakumar, V., Delaux, P. M., Junkermann, S., et al. (2015). Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol. 167, 854–871. doi: 10.1104/pp.114.255430
Yu, N., Luo, D., Zhang, X., Liu, J., Wang, W., Jin, Y., et al. (2014). A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res. 24, 130–133. doi: 10.1038/cr.2013.167
Zhang, D., Iyer, L. M., and Aravind, L. (2012). Bacterial GRAS domain proteins throw new light on gibberellic acid response mechanisms. Bioinformatics 28, 2407–2411. doi: 10.1093/bioinformatics/bts464
Zhang, Q., Blaylock, L. A., and Harrison, M. J. (2010). Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell 22, 1483–1497. doi: 10.1105/tpc.110.074955
Keywords: bryophyte, land plant evolution, moss, mycorrhiza, Physcomitrella patens, symbiotic pathway, GRAS, transcription factor
Citation: Grosche C, Genau AC and Rensing SA (2018) Evolution of the Symbiosis-Specific GRAS Regulatory Network in Bryophytes. Front. Plant Sci. 9:1621. doi: 10.3389/fpls.2018.01621
Received: 14 May 2018; Accepted: 18 October 2018;
Published: 06 November 2018.
Edited by:
Annette Becker, Justus-Liebig-Universität Gießen, GermanyReviewed by:
Didier Reinhardt, Université de Fribourg, SwitzerlandCopyright © 2018 Grosche, Genau and Rensing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefan A. Rensing, stefan.rensing@biologie.uni-marburg.de
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.