
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 22 December 2017
Sec. Plant Abiotic Stress
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.02182
Stress-induced proline accumulation in plants is thought to result primarily from enhanced proline biosynthesis and decreased proline degradation. To identify regulatory components involved in proline transport, we screened for Arabidopsis thaliana T-DNA mutants with enhanced tolerance to toxic levels of exogenous proline (45 mM). We isolated the proline resistant 1-1 (pre1-1) mutant and map-based cloning identified PRE1 as AMINO ACID PERMEASE1 (AAP1, At1g58360), which encodes a plasma membrane-localized amino acid permease. AAP1 expression is induced by salt stress and abscisic acid, but not by proline. In pre1-1 mutants, a 19-nucleotide deletion in the AAP1 coding region produced a premature stop codon. When grown on proline-containing medium, pre1-1 mutants accumulated significantly less proline than did the wild type. Under salt stress, proline uptake decreased significantly in pre1-1 mutants. By contrast, proline uptake increased significantly in the wild type. These results suggest that AAP1 functions in the increase of proline uptake during salt stress. In addition, proline uptake promotes salt tolerance in Arabidopsis seedlings. We conclude that plants can increase proline accumulation by AtAAP1-mediated proline uptake from exogenous source, which help to improve the salt tolerance of seedlings.
Higher plants accumulate free proline in response to a wide range of environmental stresses, including salinity, drought, low temperatures, heavy metals, pathogens, nutrient deficiency, atmospheric pollution, and UV-B (Saradhi et al., 1995; Hare and Cress, 1997; Siripornadulsil et al., 2002; Ashraf and Foolad, 2007; Verbruggen and Hermans, 2008; Verslues and Sharma, 2010; Wang et al., 2015). Although the precise role of proline remains unclear, proline accumulation can improve plant adaptation to stresses, helping plant cells maintain the integrity of membranes, and affecting the scavenging of reactive oxygen species and storage of nitrogen and energy (Ashraf and Foolad, 2007; Szabados and Savoure, 2010).
In plants, increased proline accumulation following environmental stress primarily results from enhanced de novo proline biosynthesis from glutamate and reduced proline degradation (Buhl and Stewart, 1983; Rhodes et al., 1986; Kiyosue et al., 1996; Peng et al., 1996). Increased expression of PYRROLINE-5-CARBOXYLATE SYNTHASE1 (P5CS1) upon environmental stress, more than P5CS2, results in higher pyrroline-5-carboxylate synthase activity that facilitates proline biosynthesis from glutamate in Arabidopsis thaliana (Szekely et al., 2008). Arabidopsis mutants that have decreased or no expression of P5CS1 accumulate decreased amounts of proline during osmotic stress (Szekely et al., 2008). By contrast, P5CS1 overexpression enhances proline accumulation in plants (Kishor et al., 1995; Zhu et al., 1998). The down-regulation of PROLINE DEHYDROGENASE (PDH) expression reduces proline dehydrogenase activity, thus decreasing proline degradation and promoting proline accumulation upon environmental stress. Arabidopsis mutants that have decreased or no expression of PDH accumulate elevated levels of proline during environmental stress (Nanjo et al., 1999; Mani et al., 2002; Ribarits et al., 2007). These observations indicate that P5CS1 and PDH function in accumulation of proline in response to stress. Recently, it has been reported that apart from transcriptional regulation, P5CS1 and PDH1 are likely subjected to post-translational modifications (Bhaskara et al., 2015) under low water potential condition. In addition, cellular redox status has also been found to affect the level of proline accumulation under low water potential stress (Sharma et al., 2013; Shinde et al., 2016) through yet unknown mechanism. Other possible regulation of proline accumulation also include the availability of the procusor molecule, such as glutamate (Skopelitis et al., 2006; Lei et al., 2016), but the precise mechanism is not very well understood.
In addition to the regulation of proline biosynthesis and degradation, proline transport also likely plays a role in proline accumulation, possibly by redistributing proline among organs. Past studies on prokaryotes have suggested a role for proline transport in proline accumulation. For example, upon osmotic stress in Salmonella typhimurium and Escherichia coli, proline accumulated by uptake from the medium via the ProP and ProU proline transport systems, but the bacteria showed no changes in the rates of proline synthesis or degradation (Csonka, 1989). Similarly, growth of S. typhimurium on minimal medium containing high salt (0.8 M NaCl) is markedly enhanced by the addition of either proline or betaine (Booth et al., 1988), suggesting a role for the uptake of these osmolyte compounds in salt tolerance.
Reports in plants also suggest that proline transport could play an important role in proline accumulation. NaCl-induced proline biosynthesis in cell suspensions of a salt-tolerant grass (Distichlis spicata) was inhibited in growth medium containing 5.0 mM proline, however, the cells took up exogenous proline (Heyser et al., 1989). In barley (Hordeum vulgare), HvProT encodes a proline transporter and is rapidly induced in root tissue within 30 min following salt stress, implying that an increase in proline translocation occurs before proline biosynthesis (Ueda et al., 2001). In tomato (Solanum lycopersicum), proline accumulation in pollen or induced by salinity is not associated with induced P5CS expression, which made the authors to propose that proline transport, among other possibility, might be associated with changes in the intracellular proline levels (Fujita et al., 1998). In addition, LeProT1, a transporter of compatible solutes including proline, may contribute to proline accumulation in pollen (Schwacke et al., 1999). In Arabidopsis, AtProT2 expression is strongly induced under drought and salt stress (Rentsch et al., 1996). Moreover, studies using atprot knockout and AtProT overexpression lines concluded that AtProT1 and AtProT2 function in proline transport in planta because atprot2 mutants were tolerant to a normally toxic level of proline, and AtProT1 and AtProT2 overexpression lines were hypersensitive to exogenous proline (Lehmann et al., 2011). Furthermore, it was found that shoot-applied Pro could greatly enhance root elongation and large amount of proline was built up in the root apex of pdh1-2 under low water potential, suggesting that proline and/or related metabolites are transported from shoot to root (Sharma et al., 2011).
The amino acid permease (AAP) family in Arabidopsis consists of AAP1-8, which are reported to transport different amino acids (Frommer et al., 1993; Fischer et al., 1995, 2002; Okumoto et al., 2002; Lee et al., 2007; Schmidt et al., 2007; Svennerstam et al., 2008). Several AAP family members, including AAP1, can complement a yeast mutant that lacks eight amino acid transport systems; expression of these AAPs can restore growth on medium containing proline as the sole nitrogen source (Tegeder et al., 2000; Okumoto et al., 2002). In Arabidopsis, AAP1 localizes on the plasma membrane and can import neutral amino acids, including proline, into root tissue (Lee et al., 2007). Previous studies suggested that AAP1 facilitates transport of amino acids only at high concentrations, such as above 50 μM in the soil (Lee et al., 2007; Svennerstam et al., 2011). However, it was recently demonstrated that AAP1 mediates the absorption of glutamic acid and neutral amino acids (including proline) in low concentrations in the soil (Perchlik et al., 2014).
Despite considerable progress in the field, little is known concerning the role of proline uptake or redistribution in proline accumulation and stress adaptation in planta. To identify the components regulating proline transport and characterize their function in proline accumulation, we screened for proline-tolerant mutants and isolated the Arabidopsis mutant proline resistance 1-1 (pre1-1), which is tolerant to toxic levels of exogenous proline. Map-based cloning identified PRE1 as At1g58360, encoding the amino acid transporter AAP1. In this report, we show that AAP1 mediates increased proline uptake in Arabidopsis seedlings under salt stress, which contributes to improved salt tolerance.
The A. thaliana (L.) wild-type, mutant, and transgenic plants used in this study are in the Columbia-0 background. The Arabidopsis seeds were surface-sterilized with 10% sodium hypochlorite for 15 min, rinsed six times in sterile distilled water and sowed in petri dishes (9.0 cm in diameter) on Murashige and Skoog (MS) medium with 2.5% Suc and 0.8% agar (pH 5.8). After being stratified at 4°C for 3 days, seeds were germinated in a growth chamber at 22°C under a 16-h: 8-h, light: dark photoperiod (light intensity 120 μmol photons m-2 sec-1).
For treatment with various conditions, 12-day-old Arabidopsis seedlings grown on a mesh were transferred into 0.5× MS liquid medium with or without different concentration of proline, NaCl, or abscisic acid for the desired time periods with regular agitation. For measurement of proline content after proline treatment, the seedlings were washed thoroughly and quickly in liquid medium, dried between sheets of filter paper and frozen in liquid nitrogen for subsequent analysis.
For proline uptake experiments, Col and pre1-1seedlings are grown on MS medium for 2 weeks and then treated liquid MS medium supplemented with 2.5 mM unlabeled L-proline and 2.5 mM 15N-labeled L-proline under salt or control conditions for 10 h. For measurement of proline uptake after treatment, the seedlings were washed thoroughly and dried in 60°C for subsequent isotope ratio mass spectrometry analysis.
The Arabidopsis T-DNA insertional mutant pool (Zhang et al., 2005) we used is generated by transforming XVE inducible activation tagging vector (Zuo et al., 2000). For mutant screening, the seeds of the mutant population were germinated and grown horizontally on MS agar plates containing 45 mM proline. The seedlings that grew better were selected at 10 days after germination (DAG). One of the mutants was pre1-1 (proline resistant 1-1), which was allowed to grow to maturity for seed setting.
Because the T-DNA insertion does not co-segregate with the phenotype observed, map-based cloning was performed to clone the mutated gene. To this end, the homozygous pre1-1 mutant (Columbia background) was crossed with Arabidopsis Landsberg erecta to generate F2 progenies. Because the difference in root length is faster and easier to follow, F2 seeds were grown on plates with 45 mM proline and the seedlings with the longest roots were selected and used as a mapping population. The pre1-1 mutation was delimited to a 64-kb region within BAC clone X7J on chromosome 1 using a series of PCR-based simple sequence-length polymorphism markers. All eleven candidate genes in the 64-kb region were sequenced from pre1-1 mutant and this analysis identified a 19-bp deletion in the first exon of At1g58360 in pre1-1.
For pre1-1 complementation, a 6518-bp genomic fragment of AAP1/PRE1 spanning from 2140 bp upstream of the ATG to 687 bp downstream of the TGA was amplified from Arabidopsis genomic DNA using recombinant PCR with two pairs of primers (p-3f & p-3r and p-4f & p-4r) containing Sma I and Xba I restriction sites (Supplementary Table S1). The amplified fragment was digested with Sma I and Xba I and cloned into the same sites in the binary vector pCAMBIA1300 (He et al., 2012) to generate pgPRE1.
For tissue-specific analysis of PRE1 promoter activity, a 2584-bp promoter fragment of AAP1/PRE1 was amplified from Arabidopsis genomic DNA with two primers (p-5f and p-5r) containing Pst I and BamH I sites (Supplementary Table S1), digested with Pst I and BamH I and cloned into the same sites in a modified pCAMBIA1391 with a GUS reporter gene and a NOS terminator, resulting in pProAAP1-GUS.
For PRE1 subcellular localization, the PRE1 coding sequence was amplified using primers p-2f and p-2r (Supplementary Table S1), cloned into a modified pJIBIA163-1300 containing a 35S promoter, a GFP reporter gene, and a NOS terminator, resulting in p35S-PRE1:GFP. For measurement of PRE1 protein levels, a 2584-bp AAP1/PRE1 promoter fragment was amplified from pAAP1-GUS with two primers (p-5f and p-5r), and was used to replace the 35S promoter in p35S-PRE1:GFP, resulting in pProAAP1- PRE1:GFP.
The constructs were introduced into Arabidopsis plants via the Agrobacterium-mediated floral-dipping transformation method (Clough and Bent, 1998). Homozygous T3 plants with a single copy of the transgene were used in subsequent experiments.
Total RNA was extracted from ∼12-day-old seedlings as described by Hua et al. (2001), treated with DNase I to remove any remaining genomic DNA, and reverse-transcribed using SuperScript II (Invitrogen) according to the manufacturer’s recommendations. Quantitative real-time PCR was performed using a SYBR Green Real-Time PCR Master Mix (Toyobo, Tokyo, Japan) on a Mx3000P thermocycler with an initial denaturation at 95°C for 2 min followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 57°C for 15 s, and extension at 72°C for 15 s. ACTIN7 was used as an internal standard to normalize the transcript level of target genes. A melting curve was run after the PCR cycles. Reactions were repeated three times for each sample. The AAP1-specific primers used were p-6f & p-6r and the ACTIN7-specific primers were ACTIN7-F and ACTIN7-R (Supplementary Table S1).
Total protein was extracted from transgenic plants containing the construct ProAAP1-AAP1CDS-GFP using cell lysis buffer for western blot and immunoprecipitation (Beyotime P0013). Immunoblot analysis was performed using an anti-GFP antibody (Abmart) and Anti-Mouse IgG (Abmart).
Samples stored at -80°C were ground in liquid nitrogen and then measured for free proline (Pro) content by a colorimetric assay as described by Bates et al. (1973).
For measurement of proline uptake after proline treatment, the seedlings were washed thoroughly and dried for 48 h at 60°C. The dried seedlings were then ground with mixed grinding apparatus (Retsch MM400) and weighed on a microbalance, packaged in tin capsules and analyzed for 15N content by isotope ratio mass spectrometry (Thermo Finnigan MAT DELTAplus XP), using an automated nitrogen/carbon analysis system (Flash EA 1112). Un-labeled glycine (with 0.3674 at% 15N) was used as the working standard. Results are presented as atom% (N15/Ntotal) and N%. In according to N15/Ntotal = N15/(N15+N14) = atom% and mN = N14∗14+N15∗15 = N% ∗ msample, the 15N content (μg g-1 DW) in sample (mN15/msample) was calculated by atom %, N% and dry weight.
For histochemical GUS staining, ProAAP1:GUS transgenic plants were grown in MS medium for 1 week, or in soil until maturation, then whole seedlings or individual organs were immersed into staining solution with X-gluc [2 mM X-gluc; 10 mM EDTA; 100 mM NaPO4 (pH 7.2); 0.5 mM K4Fe(CN)6; 0.5 mM K3Fe(CN)6; 0.1% Trix-100], and incubated at 37°C for 2 to 6 h until blue staining was visible. De-coloration was performed in 75% ethanol for 3–5 h.
For observation of GFP fluorescence, protoplasts were isolated as described (Yoo et al., 2007) from 35S-PRE1:GFP plants grown in soil for 3–4 weeks and observed under an OLYMPUSFV1000 MPE confocal laser scanning microscope.
To identify novel components regulating proline transport, we performed a genetic screen for proline-tolerant mutants in a T-DNA insertion mutant population of Arabidopsis. This screen identified the mutant allele pre1-1 (proline resistant 1-1). Whereas there was no significant difference in root length between wild-type (Columbia) and pre1-1 seedlings grown on MS medium, wild-type root length was reduced to 33 and 42.1% of that of pre1-1 when grown on MS supplemented with 45 mM L-proline for 10 or 16 days, respectively (Figures 1A–D). In addition, the aerial tissue of pre1-1 mutants was also larger and greener compared to that of wild type. To compare proline uptake in wild type and pre1-1, the proline content in 2-week-old seedlings was quantified following treatment with 45 mM L-proline in 0.5× MS liquid medium for various time periods. For each treatment time period, the proline content in pre1-1 was significantly lower than that of wild type (Figure 1E), suggesting that the enhanced proline tolerance of pre1-1 mutants results from reduced proline uptake.
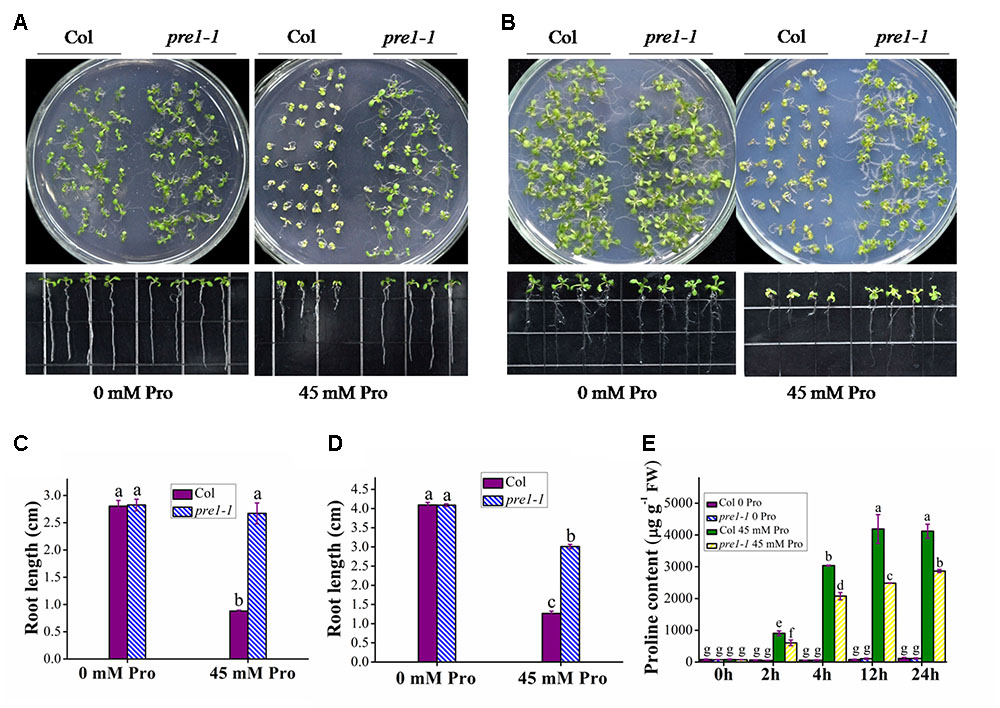
FIGURE 1. The proline-tolerant phenotype of pre1-1 mutants. Growth of wild-type (Col) and pre1-1 seedlings (A,B) and average root length (C,D) on 0.5× MS medium supplemented with 45 mM L-proline (Pro) following 10 (A,C) or 16 (B,D) days of growth. (E) The proline contents of 12-day-old wild-type and pre1-1 seedlings treated with 45 mM proline in 0.5× MS liquid medium for various time periods. Experiments were repeated a minimum of three times with similar results. Data are means ± standard error (n = 6). Different letters above columns indicate significant difference (P < 0.05, Duncan’s multiple range test).
To characterize the pre1-1 mutation, the mutant line was back-crossed with wild type (Columbia). The resulting F1 (diploid) progeny displayed moderate tolerance to proline (Supplementary Figure S1), suggesting that the pre1-1 phenotype is caused by a semi-dominant mutation. Examination of the F2 progeny revealed segregation of seedlings into longest-root, longer-root, and short-root phenotypic classes, with 70, 156, and 67 seedlines in each respective class [χ2(1:2:1) = 1.294 < χ20.05 = 5.99; P > 0.05], further demonstrating that the pre1-1 phenotype is inherited as a semi-dominant trait.
We then determined whether the pre1-1 mutation is associated with a dominant gain of function or with haplo-insufficiency by crossing homozygous pre1-1 with tetraploid wild type. The resulting F1 (triploid) progeny had a proline-sensitive phenotype similar to that of wild type (Supplementary Figure S1), suggesting that the pre1-1 phenotype is caused by haplo-insufficiency. Further analysis demonstrated that the proline-tolerant phenotype of pre1-1 is not linked to a T-DNA insertion. Therefore, individual plants with a proline-tolerant phenotype that did not carry a T-DNA insertion were isolated for map-based cloning of PRE1.
To clone PRE1, we generated an F2 population by crossing homozygous pre1-1 (Columbia) with wild type (Landsberg erecta). Since pre1-1 is a semi-dominant mutant, we obtained the mapping population by sowing F2 seeds on MS medium containing 45 mM L-proline and selecting individuals with the longest-root phenotype. Based on classical simple sequence length polymorphism markers, the PRE1 locus was mapped to the middle of chromosome 1 and was subsequently narrowed down to a 64-kb region within BAC clone X7J and F19C14 (Figure 2A). The open reading frames in this region were sequenced in pre1-1, which revealed a 19-nucleotide deletion in the first exon of AT1G58360 (Figure 2A), creating a premature stop codon that truncated the encoded protein.
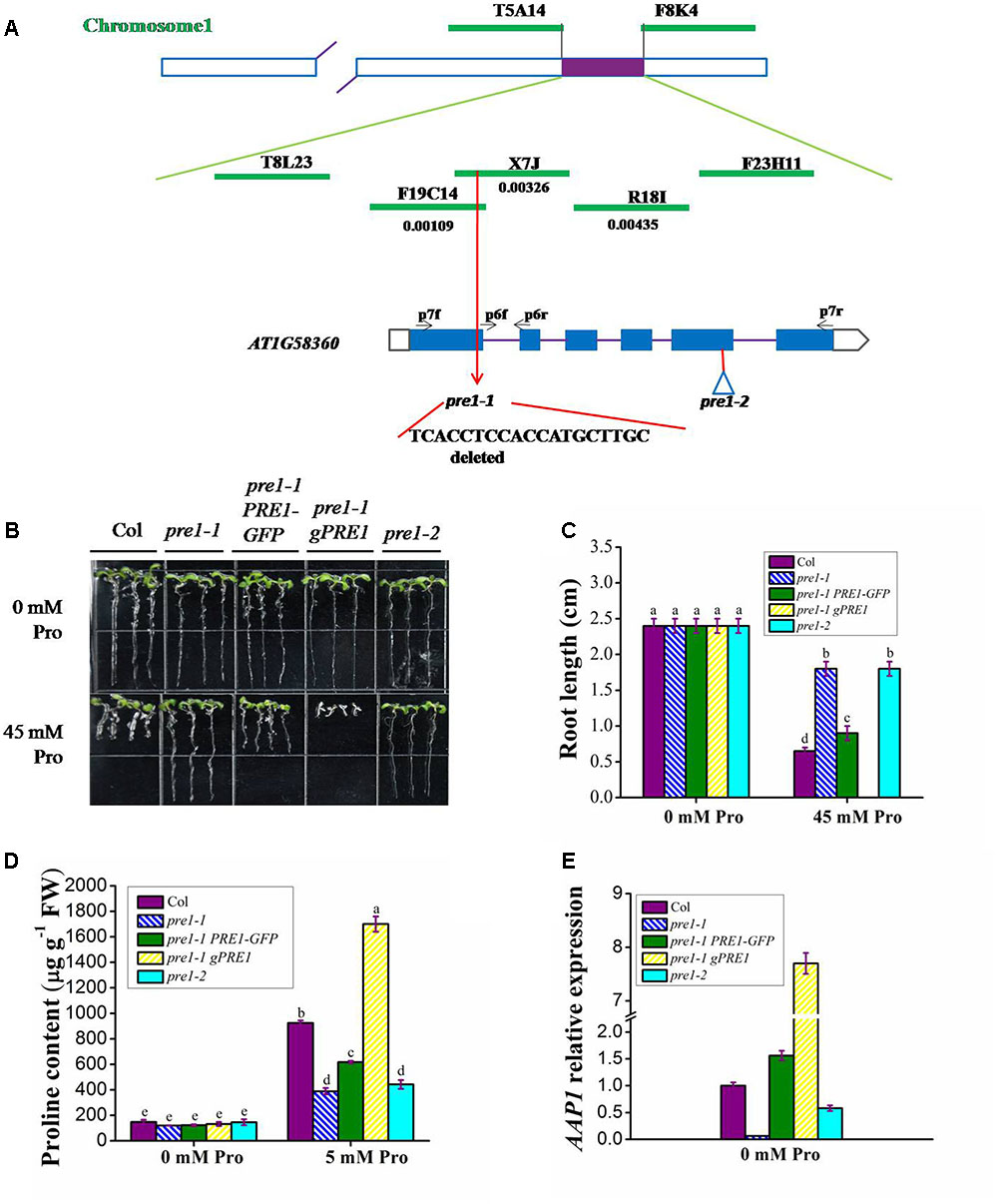
FIGURE 2. PRE1 positional cloning and pre1-1 complementation. (A) The PRE1 locus was mapped to BAC clone X7J on chromosome 1. PRE1/At1g58360/AAP1 is composed of 6 exons (black boxes) and 5 introns (lines). BAC names and their regions spanned are indicated. DNA sequencing revealed a 19-bp deletion in the first exon of PRE1/AAP1 in pre1-1, resulting in truncation of PRE1/AAP1. The T-DNA insertion site in pre1-2 is indicated with a triangle. (B) Quantification of proline sensitivity and (C) average primary root length (C) in 9-day-old seedlings of wild type, mutant, and complemented lines treated with 0.5× MS medium supplemented with 45 mM L-proline (Pro). (D) The proline content of 12-day-old seedlings of wild type, mutant, and complemented lines treated with 5 mM Pro for 24 h. (E) The relative expression of PRE1/AAP1 in 12-day-old seedlings of wild type, mutant, and complemented lines. Col, wild-type Columbia; pre1-1 PRE1-GFP, pre1-1 transformed PRE1 cDNA fused with GFP under the control of the PRE1/AAP1 promoter; pre1-1 gPRE1, pre1-1 transformed with a genomic fragment of PRE1/AAP1. Experiments were repeated a minimum of three times with similar results. The values are means ± standard error (n = 6). Different letters above columns indicate significant difference (P < 0.05, Duncan’s multiple range test).
To confirm that At1g58360 is responsible for the pre1-1 mutant phenotype, pre1-1 plants were transformed with either the AT1G58360 genomic DNA fragment from wild type (gPRE1, spanning from 2.5 kb upstream of the start codon to 687 bp downstream of the stop codon) or the AT1G58360 coding region fused with GFP under the control of the AT1G58360 promoter (PRE1-GFP). All complemented lines (pre1-1 gPRE1 and pre1-1 PRE1-GFP) displayed a proline-sensitive phenotype (Figures 2B,C) with proline sensitivity directly proportional to the level of PRE1 transcript, indicating that AT1G58360 is PRE1 and functions in proline uptake in a dose-dependent manner (Supplementary Figure S2). We have noticed that pre1-1 gPRE1 lines are generally more sensitive to proline than are pre1-1 PRE1-GFP lines, correlated with the higher level of PRE1 transcript in the former, despite of same length of the promoter is used in both constructs. The reason for this is currently unknown, but probably related to the presence of intron or 3′UTR in the former and GFP fusion in the later.
AT1G58360 has been previously annotated as AAP1, encoding an amino acid permease family member (Lee et al., 2007), which implies that pre1-1 is deficient in proline uptake. To confirm the physiological function of PRE1/AAP1, the proline contents of 12-day-old wild-type, pre1-1, pre1-1 gPRE1, and pre1-1 PRE1-GFP seedlings were measured following a 24 h treatment with 5 mM L-proline. As shown in Figure 2D, the proline content of pre1-1 gPRE1 seedlings was significantly higher than that of the other genotypes, which correlated with the significantly higher PRE1/AAP1 expression observed in pre1-1 gPRE1 (Figure 2E). The proline content of pre1-1 PRE1-GFP seedlings was slightly lower than that of wild type, but significantly higher than that of pre1-1 (Figure 2D), which agreed with the proline sensitivity of pre1-1 PRE1-GFP (Figures 2B,C). The PRE1/AAP1 transcript level in pre1-1 PRE1-GFP plants was comparable to that in wild type (Figure 2E).
To further confirm the function of PRE1/AAP1 in proline uptake, a T-DNA insertion mutant of PRE1/AAP1 (Columbia; GK-324G05-015976) was obtained from the Arabidopsis Biological Resource Center and named pre1-2. In this mutant, the T-DNA was inserted at position 1028 (the fifth exon) relative to the putative start codon of AT1G58360 (Figure 2A), and the full-length transcript of AT1G58360 could not be detected (Supplementary Figure S3). In 9-day-old seedlings, no significant difference was observed in root length between wild type and the pre1-1 and pre1-2 mutants grown on MS medium; however, the root length of pre1-2 was comparable to that of pre1-1 and approximately fourfold longer than that of wild type, following seedling growth on MS medium supplemented with 45 mM L-proline (Figures 2B,C). When treated with 5 mM L-proline for 24 h, the proline content of 12-day-old pre1-2 seedlings was significantly lower than that of wild type, similar to that observed in pre1-1 (Figure 2D). As in pre1-1, PRE1/AAP1 expression in pre1-2 was significantly lower than that in wild type (Figure 2E). The primers used here are located in the first and second exon (p6f and p6r in Figure 2A), so it seems that the partial transcript of AAP1 in front of the T-DNA insertion can still be detected in pre1-2. Since PRE1 mutation inhibits AAP1-mediated proline uptake, PRE1 is hereafter referred to as AAP1.
To characterize the AAP1 tissue-specific expression pattern, transgenic Arabidopsis plants were generated that carried the β-glucuronidase (GUS) gene driven by the AAP1 promoter (the genomic sequence approximately 2.5 kb upstream of the putative start codon of AAP1). Histochemical analysis of GUS activity showed AAP1 promoter activity in almost all plant organs. In 12-day-old seedlings, AAP1 promoter activity was observed in cotyledons, true leaves, and roots, with strong activity in the cotyledon and true leaf upper regions, root tips, and the cotyledon and root vascular tissue (Figures 3A,B). Relatively less AAP1 promoter activity was observed in root meristems. In flower buds, AAP1 promoter activity was primarily concentrated in sepals, and absent in stigmas, filaments, and anthers (Figure 3C); however, expression appeared in these tissues in mature flowers (Figures 3D,I). During the early stages of silique development, AAP1 promoter activity was detected at the extremities of siliques (Figures 3E,I) and spread throughout the silique tissue as they developed (Figure 3F). In mature siliques, strong AAP1 promoter activity was still visible in the pericarp (Figure 3G). No AAP1 expression was observed in mature seeds, aside from weak expression in the pedicel (Figure 3H). Upon silique maturation, AAP1 promoter activity became stronger in silique stalks (Figure 3I). In agreement with the observed GUS activity, the level of AAP1 transcript was high in siliques and flowers (Figure 3J).
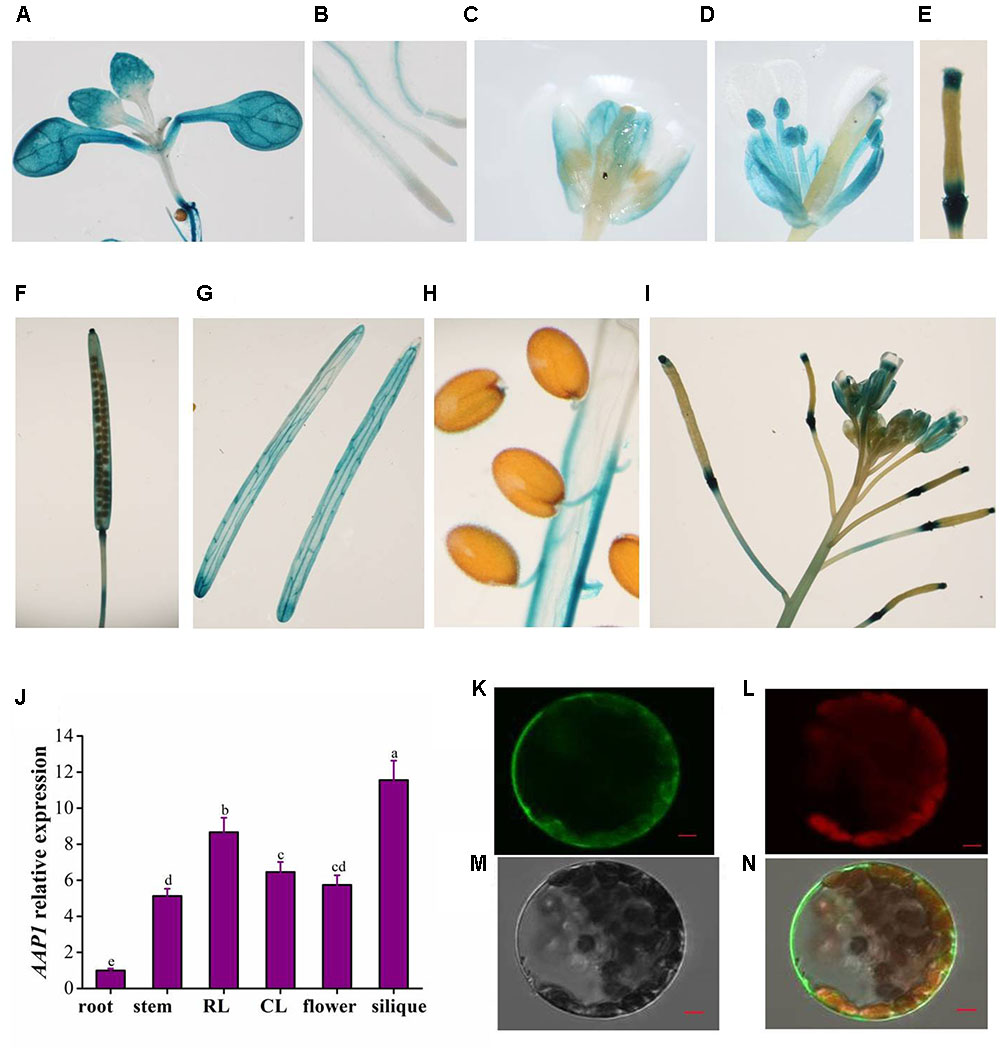
FIGURE 3. Tissue-specific expression profile of AAP1 and subcellular localization of AAP1. Histochemical staining of GUS activity directed by the AAP1 promoter observed in the (A) cotyledons and true leaves and (B) the root vascular cylinder of 12-day-old seedlings. GUS activity observed in (C) flower buds, (D) open flowers, (E) young siliques, (F) developing siliques, (G) envelope of mature siliques, (H) seed pedicels, and (I) silique stalks at the top of the main stem in mature Arabidopsis plants. (J) AAP1 transcript level in different organs of mature wild-type (Col) plants. (K) AAP1-GFP fluorescence in protoplasts. (L) Chlorophyll auto-fluorescence. (M) Bright field. (N) Merged image. Bars = 5 μm. Experiments were repeated a minimum of three times with similar results. The values are means ± standard error (n = 3). Different letters above columns indicate significant difference (P < 0.05, Duncan’s multiple range test).
To investigate the subcellular localization of AAP1, transgenic Arabidopsis plants were generated that expressed an AAP1-GFP fusion protein under the control of the 35S promoter. Protoplasts isolated from the AAP1-GFP plants were observed under a confocal microscope, which revealed GFP localized primarily on the plasma membrane (Figures 3K–N), and relatively less GFP signal in the cytoplasm.
To understand whether AAP1 expression is responsive to various abiotic stresses that induce proline accumulation, 12-day-old Arabidopsis seedlings were subjected to abiotic stress treatments and AAP1 expression levels were analyzed using quantitative real-time PCR. The AAP1 transcript level did not significantly vary over 24 h of treatment with 45 mM L-proline (Figure 4A). Similar results were seen with 5 and 10 mM proline (Supplementary Figure S4D). An increase in AAP1 transcript accumulation was detected upon addition of 50 and 100 mM NaCl (Supplementary Figure S4A). A higher increase was observed after 200 mM NaCl treatment, with an increment peak at 6 h, after which AAP1 transcript level gradually declined, remaining higher than that in control seedlings at 24 h (Figure 4B). AAP1 expression also rose rapidly following treatment with 40 μM abscisic acid (ABA) and 400 mM mannitol, and at 24 h, reached a level approximately 27- and 11-fold higher than that of control seedlings, respectively (Figures 4C,D). 4, 10 μM ABA and 100, 200 mM mannitol can also induce AAP1 expression, but to lesser extent (Supplementary Figures S4B,C).
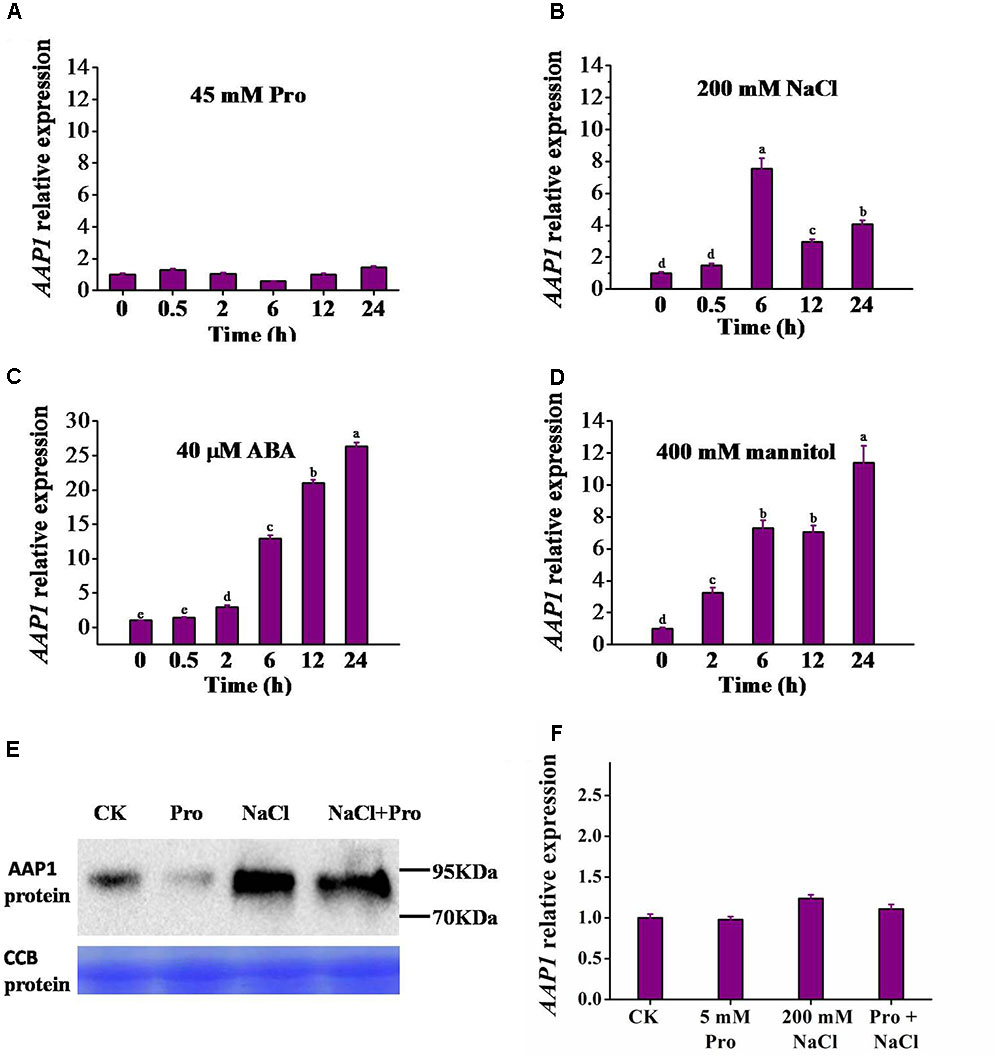
FIGURE 4. Induction of AAP1 transcript and AAP1 protein level by different treatments. AAP1 relative expression level analyzed using real-time RT-PCR in 12-day-old wild-type (Col) seedlings following treatment for different time periods with 0.5× MS liquid medium containing (A) 45 mM L-proline (Pro), (B) 200 mM NaCl, (C) 40 μM abscisic acid (ABA), or (D) 400 mM mannitol. (E) AAP1-GFP protein level in 35S-PRE1:GFP seedlings following a 10-h treatment with various compounds. (F) AAP1 relative expression level in 35S-PRE1:GFP seedlings following a 7 h treatment with 5 mM Pro, 200 mM NaCl, or 5 mM Pro and 200 mM NaCl. 35S-PRE1:GFP, pre1-1 transformed PRE1 cDNA fused with GFP driven by the 35S promoter. Experiments were repeated a minimum of three times with similar results and data are means ± standard error (n = 3). Different letters above columns indicate significant difference (P < 0.05, Duncan’s multiple range test).
Building on the AAP1 transcriptional up-regulation observed following NaCl treatment, we examined the response of AAP1-GFP fusion protein, under the 35S promoter control, to 10 h treatments with 5 mM L-proline, 200 mM NaCl, or both L-proline and NaCl. For this, we performed western blot experiments using a GFP specific antibody. The 5 mM proline is used here to be consistent with that in salt-induced proline uptake assay later. As shown in Figure 4E, the AAP1-GFP level was significantly higher following treatment with 200 mM NaCl or 200 mM NaCl and 5 mM L-proline. However, similar induction of AAP1-GFP was not observed following 5 mM L-proline treatment alone, where it appeared to be reduced compared to that in the control sample. Of note, no significant variation was observed in AAP1 transcript levels between the different treatments in 35S:AAP1-GFP seedlings (Figure 4F).
To investigate whether plants can use exogenous proline to increase their internal proline content, we measured the proline uptake in 2-week-old wild-type seedlings grown under salt stress or control conditions following treatment with 5 mM L-proline (including 2.5 mM labeled 15N L-proline). Under control conditions, a 10 h proline treatment resulted in a 571 μg g-1 DW net increase in seedling proline uptake (Figure 5A). However, under salt stress, proline treatment caused a 672 μg g-1 DW net increase in seedling proline contents, which was significantly higher than that observed under control conditions (Figure 5A).
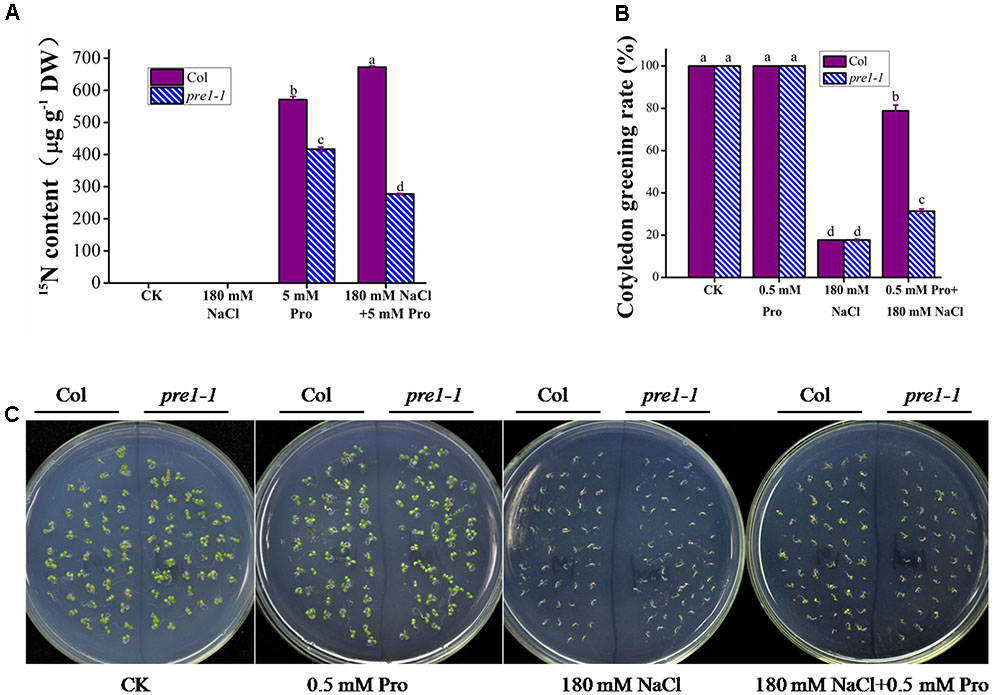
FIGURE 5. The effect of salt stress treatment on exogenous proline uptake and beneficial effects of increased proline uptake on salt tolerance in Arabidopsis seedlings. (A) The proline uptake of wild-type and pre1-1 12-day-old seedlings following a 10 h treatment with 0.5× MS liquid medium containing 5 mM L-proline (including 2.5 mM 15N labeled L-proline) with or without 180 mM NaCl. (B,C) The seeds of wild type (Col) and the pre1-1 mutant were germinated on 0.5× MS medium containing 180 mM NaCl with or without 0.5 mM L-proline (Pro), grown for 5 days. The survival rate (B) (the number of seedlings to compile statistics ≥ 200) of Col and pre1-1 seedlings and the phenotype (C) were measured and photographed, respectively. Col, wild-type Columbia. Experiments were repeated a minimum of three times with similar results. In (A,B), the data are means ± standard error (n = 3). Different letters above columns indicate significant difference (P < 0.05, Duncan’s multiple range test).
To investigate whether AAP1 plays a role in proline uptake during salt stress, pre1-1 was treated with proline as described above. Similar to the results presented in Figures 1E, 5A shows that the net proline uptake in the pre1-1 mutant (417 μg g-1 DW) was lower than that in wild type under control conditions. Under salt stress, however, the net proline uptake of the pre1-1 mutant (277 μg g-1 DW) significantly decreased and was significantly lower than that of wild type (Figure 5A).
To determine whether enhanced proline uptake improves salt stress tolerance in Arabidopsis, pre1-1 and wild-type seedlings were grown on MS medium containing 180 mM NaCl with or without 0.5 mM proline. Following a 5-day 180 mM NaCl salt stress growing, the rate of cotyledon greening was similar between wild type and pre1-1 (approximately 18%) on medium without proline, but increased to 80 and 35%, respectively, on 0.5 mM proline-containing medium (Figures 5B,C).
In this report, we identified and characterized the mutant pre1-1, which tolerates levels of exogenous proline that are toxic to wild type. Map-based cloning revealed that PRE1 encodes AMINO ACID PERMEASE 1 (AAP1) and, in pre1-1, a 19-nucleotide deletion in the first exon creates a premature stop codon that likely truncates AAP1. An AAP1-GFP fusion protein localized to the plasma membrane, consistent with previous results (Lee et al., 2007). Previous studies on AAP1 reported that it mediates the uptake of glutamic acid and other neutral amino acids, including proline (Lee et al., 2007). Our results also showed that when grown on medium supplemented with various concentrations of proline, the intracellular proline content of pre1-1 was significantly lower than that of wild-type Columbia. Furthermore, another mutant line, pre1-2, which carried a T-DNA insertion toward the 3′ end of the PRE1 coding region, displayed a similar proline-tolerant phenotype. These results support the conclusion that AAP1 is a functional proline transporter in planta. However, the semi-dominant and haplo-insufficient nature of pre1-1 indicated that the capacity of AAP1 to transport proline is relatively low. In addition, the complementation lines all showed correlation between proline sensitivity and AAP1 expression level in a dose-dependent manner. These results are consistent with a previous report that the affinity of AAP1 for proline is low (Km = 60 μM) (Lehmann et al., 2010). The pre1-1 mutant was also more tolerant to other amino acids, such as Met, Val, Phe, Cys, Thr, His, and Ile (Supplementary Figure S5), in agreement with the broad substrate specificity of AAP1 reported previously (Lee et al., 2007).
Notably, our data showed that proline uptake can increase the proline contents in Arabidopsis seedlings under salt stress if proline is available as an exogenous source. Several evidence supported this conclusion. First, in wild type, the seedling proline uptake in the presence of exogenous proline under salt stress was significantly higher than that observed under normal conditions. Second, in the pre1-1 mutant, the proline uptake under salt stress significantly decreased, suggesting that AAP1 is involved in this process. Lastly, AAP1 transcript was induced by NaCl treatment and further analyses indicated that the translation and/or the stability of AAP1 was enhanced under salt stress, suggesting that more AAP1 is present in the plasma membrane to facilitate proline transport in response to salt stress. It has been reported that in prokaryotes such as E. coli, proline accumulation during osmotic stress primarily involves increased proline uptake from the growth medium, rather than proline biosynthesis (Csonka, 1989). Our results provided evidences that in addition to de novo proline biosynthesis and reduced proline degradation, plants can also take up proline from the environment to achieve a higher internal proline content under stress, similar to prokaryotes.
Our results also demonstrate that increased proline uptake improves salt stress tolerance and survival in Arabidopsis seedlings. Improved salt tolerance in wild type compared to that in the pre1-1 mutant was evident on medium containing proline, whereas, in the absence of proline, similar salt-induced lethality was observed in both wild type and pre1-1. We have also noticed that the increase of proline uptake under salt stress is not so dramatic compared to the level of endogenous proline accumulation normally reported, yet still contribute to significant increased salt tolerance of the seedlings. However, we constantly observed this level of increase under our experimental condition. A possible explanation is that the proline taken up during salt stress was not evenly distributed. The local accumulation of proline in specific kinds of cells may exert certain protective effects. Alternatively, proline uptake usually happened faster than its de novo biosynthesis, which may be beneficial for early stage growth of the seedling under salt stress.
The protective roles of exogenous proline corroborate previous reports that exogenous application of proline may alleviate the inhibitory effects of salt stress on plant growth (Chen et al., 2011). It has been reported that pretreatment with 0.5 or 5 mM proline significantly improved the growth of Arabidopsis seedlings during a subsequent salt stress treatment (Chen et al., 2011). Exogenous proline supplied at 5–10 mM during stress treatment also increased the salinity tolerance of several plant species and cell cultures (Okuma et al., 2000; Kaya et al., 2007; Teh et al., 2015; Zheng et al., 2015). However, other amino acids have not shown similar protective effects (Okuma et al., 2000). Although a correlation was proposed between the beneficial effect of supplemental proline and both the observed reduction in Na+ accumulation (Kaya et al., 2007; Zheng et al., 2015) and the elevation of anti-oxidative capacities (Zheng et al., 2015), no causal relationship has been established.
The toxic effect of exogenous proline under non-stressed conditions has frequently been attributed to a high level of endogenous proline or its catabolic intermediate pyrroline-5-carboxylate (Hellmann et al., 2000; Mani et al., 2002; Miller et al., 2009). A high internal proline level may disrupt the ultrastructure of chloroplasts and mitochondria (Hare et al., 2002). However, the fact that exogenous proline is toxic under normal conditions does not discount its potential protective role under salt stress, since salt stress was found to ameliorate proline toxicity (Hellmann et al., 2000).
AAP1 is a transporter with a known affinity for several neutral amino acids (Lee et al., 2007). Therefore, it is likely that AAP1 mediates uptake of other neutral amino acids, in addition to proline, if they are also present exogenously. However, the salt tolerance of Arabidopsis seedlings was not improved by the presence of other neutral amino acids (Supplementary Figure S6), suggesting that proline has a specific role in alleviating the inhibitory effects of salt stress.
The redistribution of proline among plant organs, which would rely on proline transport, may also be an important salt stress tolerance mechanism. However, we found no difference between wild type and pre1-1 in proline redistribution among organs during salt-induced proline accumulation (Supplementary Figure S7), suggesting that either AAP1 is not involved in proline redistribution or other transporters may function redundantly in this process. It may be fruitful to characterize other salt-inducible amino acid transporter genes and investigate their possible roles in mediating proline redistribution among organs.
We have demonstrated that treatment with 40 μM ABA significantly increased the level of AAP1 transcript (Figure 4C) and the uptake of proline (Supplementary Figure S8) in wild type. However, in pre1-1 mutants, ABA treatment did not significantly alter proline uptake (Supplementary Figure S8), suggesting that endogenous ABA might induce proline uptake via AAP1. Hormonal regulation of genes encoding amino acid transporters has not been extensively studied, although a study in ginseng showed that expression of the transporter gene PgLHT1 is induced by ABA, salicylic acid, and methyl jasmonic acid (Zhang et al., 2013). Detailed characterization of mutants deficient in ABA synthesis or ABA responsiveness may produce a greater understanding of the involvement of ABA in AAP1 expression regulation.
To explore the contribution of proline transport to proline accumulation under salt stress, we firstly obtained an Arabidopsis mutant proline resistance 1-1 (pre1-1) in a genetic screening for more resistant to toxic level of exogenous proline and identified PRE1 as At1g58360, encoding the amino acid transporter AAP1, by map-based cloning. Our results indicated that the proline uptake in the presence of exogenous proline under salt stress was significantly higher than that observed under normal conditions in wild type. However, in the pre1-1 mutant, the proline uptake under salt stress was significantly decreased, which suggested AAP1 is involved in this process. In addition, AAP1 transcript was induced by NaCl treatment and further analyses indicated that the translation and/or the stability of AAP1 was enhanced under salt stress, suggesting that more AAP1 is present in the plasma membrane to facilitate proline transport in response to salt stress. Analysis of cotyledon greening rate under salinity with or without proline indicated that the growth of wild type seedlings was significantly better than that of pre1-1 mutant only in the presence of exogenous proline.
Taken together, we conclude that proline transport mediated by PRE1/AtAAP1 is important for proline accumulation under salt stress, which is probably a part of adaptive mechanism for improving salt tolerance.
TW, MZ, and XH designed the experiments. TW, YC, MZ, JC, JL, and HH performed the experiments. TW and XH wrote the manuscript. XH revised the manuscript.
This work was supported by the National Nature Science Foundation of China (Grant No. 31271308) project to XH.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Arabidopsis Biological Resource Center for providing the T-DNA insertional lines of aap1-2.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02182/full#supplementary-material
Ashraf, M., and Foolad, M. R. (2007). Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 59, 206–216. doi: 10.1016/j.envexpbot.2005.12.006
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/Bf00018060
Bhaskara, G. B., Yang, T. H., and Verslues, P. E. (2015). Dynamic proline metabolism: importance and regulation in water limited environments. Front. Plant Sci. 6:484. doi: 10.3389/fpls.2015.00484
Booth, I. R., Cairney, J., Sutherland, L., Stirling, D. A., and Higgins, C. F. (1988). Enteric bacteria and osmotic-stress - an integrated homeostatic system. J. Chem. Technol. Biotechnol. 42, 298–299.
Buhl, M. B., and Stewart, C. R. (1983). Effects of Nacl on proline synthesis and utilization in excised barley leaves. Plant Physiol. 72, 664–667. doi: 10.1104/pp.72.3.664
Chen, J. G., Zhang, Y. Q., Wang, C. P., Lu, W. T., Jin, J. B., and Hua, X. J. (2011). Proline induces calcium-mediated oxidative burst and salicylic acid signaling. Amino Acids 40, 1473–1484. doi: 10.1007/s00726-010-0757-2
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Csonka, L. N. (1989). Physiological and genetic responses of bacteria to osmotic-stress. Microbiol. Rev. 53, 121–147.
Fischer, W. N., Kwart, M., Hummel, S., and Frommer, W. B. (1995). Substrate-specificity and expression profile of amino-acid transporters (Aaps) in Arabidopsis. J. Biol. Chem. 270, 16315–16320. doi: 10.1074/jbc.270.27.16315
Fischer, W. N., Loo, D. D. F., Koch, W., Ludewig, U., Boorer, K. J., Tegeder, M., et al. (2002). Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. Plant J. 29, 717–731. doi: 10.1046/j.1365-313X.2002.01248.x
Frommer, W. B., Hummel, S., and Riesmeier, J. W. (1993). Expression cloning in yeast of a cdna-encoding a broad-specificity amino-acid permease from Arabidopsis-Thaliana. Proc. Natl. Acad. Sci. U.S.A. 90, 5944–5948. doi: 10.1073/pnas.90.13.5944
Fujita, T., Maggio, A., Garcia-Rios, M., Bressan, R. A., and Csonka, L. N. (1998). Comparative analysis of the regulation of expression and structures of two evolutionarily divergent genes for Δ1-pyrroline-5-carboxylate synthetase from tomato. Plant Physiol. 118, 661–674. doi: 10.1104/pp.118.2.661
Hare, P. D., and Cress, W. A. (1997). Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul. 21, 79–102. doi: 10.1023/A:1005703923347
Hare, P. D., Cress, W. A., and van Staden, J. (2002). Disruptive effects of exogenous proline on chloroplast and mitochondrial ultrastructure in Arabidopsis leaves. S. Afr. J. Bot. 68, 393–396. doi: 10.1016/S0254-6299(15)30405-1
He, J. N., Duan, Y., Hua, D. P., Fan, G. J., Wang, L., Liu, Y., et al. (2012). DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell 24, 1815–1833. doi: 10.1105/tpc.112.098707
Hellmann, H., Funck, D., Rentsch, D., and Frommer, W. B. (2000). Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiol. 123, 779–790. doi: 10.1104/pp.123.2.779
Heyser, J. W., De Bruin, D., Kincaid, M. L., Johnson, R. Y., Rodriguez, M. M., and Robinson, N. J. (1989). Inhibition of NaCl-induced proline biosynthesis by exogenous proline in halophilic Distichlis spicata suspension cultures. J. Exp. Bot. 40, 225–232. doi: 10.1093/Jxb/40.2.225
Hua, X. J., van de Cotte, B., Van Montagu, M., and Verbruggen, N. (2001). The 5′ untranslated region of the At-P5R gene is involved in both transcriptional and post-transcriptional regulation. Plant J. 26, 157–169. doi: 10.1046/j.1365-313x.2001.01020.x
Kaya, C., Tuna, A. L., Ashraf, M., and Altunlu, H. (2007). Improved salt tolerance of melon (Cucumis melo L.) by the addition of proline and potassium nitrate. Environ. Exp. Bot. 60, 397–403. doi: 10.1016/j.envexpbot.2006.12.008
Kishor, P. B. K., Hong, Z. L., Miao, G. H., Hu, C. A. A., and Verma, D. P. S. (1995). Overexpression of [delta]-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol. 108, 1387–1394. doi: 10.1104/pp.108.4.1387
Kiyosue, T., Yoshiba, Y., YamaguchiShinozaki, K., and Shinozaki, K. (1996). A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell 8, 1323–1335. doi: 10.2307/3870304
Lee, Y. H., Foster, J., Chen, J., Voll, L. M., Weber, A. P. M., and Tegeder, M. (2007). AAP1 transports uncharged amino acids into roots of Arabidopsis. Plant J. 50, 305–319. doi: 10.1111/j.1365-313X.2007.03045.x
Lehmann, S., Funck, D., Szabados, L., and Rentsch, D. (2010). Proline metabolism and transport in plant development. Amino Acids 39, 949–962. doi: 10.1007/s00726-010-0525-3
Lehmann, S., Gumy, C., Blatter, E., Boeffel, S., Fricke, W., and Rentsch, D. (2011). In planta function of compatible solute transporters of the AtProT family. J. Exp. Bot. 62, 787–796. doi: 10.1093/jxb/erq320
Lei, P., Xu, Z. Q., Liang, J. F., Luo, X. H., Zhang, Y. X., Feng, X. H., et al. (2016). Poly(gamma-glutamic acid) enhanced tolerance to salt stress by promoting proline accumulation in Brassica napus L. Plant Growth Regul. 78, 233–241. doi: 10.1007/s10725-015-0088-0
Mani, S., Van de Cotte, B., Van Montagu, M., and Verbruggen, N. (2002). Altered levels of proline dehydrogenase cause hypersensitivity to proline and its analogs in Arabidopsis. Plant Physiol. 128, 73–83. doi: 10.1104/pp.010572
Miller, G., Honig, A., Stein, H., Suzuki, N., Mittler, R., and Zilberstein, A. (2009). Unraveling Δ1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J. Biol. Chem. 284, 26482–26492. doi: 10.1074/jbc.M109.009340
Nanjo, T., Kobayashi, M., Yoshiba, Y., Kakubari, Y., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett. 461, 205–210. doi: 10.1016/S0014-5793(99)01451-9
Okuma, E., Soeda, K., Tada, M., and Murata, Y. (2000). Exogenous proline mitigates the inhibition of growth of Nicotiana tabacum cultured cells under saline conditions. Soil Sci. Plant Nutr. 46, 257–263. doi: 10.1080/00380768.2000.10408781
Okumoto, S., Schmidt, R., Tegeder, M., Fischer, W. N., Rentsch, D., Frommer, W. B., et al. (2002). High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of Arabidopsis. J. Biol. Chem. 277, 45338–45346. doi: 10.1074/jbc.M207730200
Peng, Z., Lu, Q., and Verma, D. P. S. (1996). Reciprocal regulation of delta 1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol. Gen. Genet. 253, 334–341. doi: 10.1007/Pl00008600
Perchlik, M., Foster, J., and Tegeder, M. (2014). Different and overlapping functions of Arabidopsis LHT6 and AAP1 transporters in root amino acid uptake. J. Exp. Bot. 65, 5193–5204. doi: 10.1093/jxb/eru278
Rentsch, D., Hirner, B., Schmelzer, E., and Frommer, W. B. (1996). Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. Plant Cell 8, 1437–1446. doi: 10.1105/Tpc.8.8.1437
Rhodes, D., Handa, S., and Bressan, R. A. (1986). Metabolic changes associated with adaptation of plant-cells to water-stress. Plant Physiol. 82, 890–903. doi: 10.1104/Pp.82.4.890
Ribarits, A., Abdullaev, A., Tashpulatov, A., Richter, A., Heberle-Bors, E., and Touraev, A. (2007). Two tobacco proline dehydrogenases are differentially regulated and play a role in early plant development. Planta 225, 1313–1324. doi: 10.1007/s00425-006-0429-3
Saradhi, P. P., Alia Arora, S., and Prasad, K. V. S. K. (1995). Proline accumulates in plants exposed to UV radiation and protects them against UV induced peroxidation. Biochem. Biophys. Res. Commun. 209, 1–5. doi: 10.1006/bbrc.1995.1461
Schmidt, R., Stransky, H., and Koch, W. (2007). The amino acid permease AAP8 is important for early seed development in Arabidopsis thaliana. Planta 226, 805–813. doi: 10.1007/s00425-007-0527-x
Schwacke, R., Grallath, S., Breitkreuz, K. E., Stransky, E., Stransky, H., Frommer, W. B., et al. (1999). LeProT1, a transporter for proline, glycine betaine, and gamma-amino butyric acid in tomato pollen. Plant Cell 11, 377–391. doi: 10.1105/tpc.11.3.377
Sharma, S., Shinde, S., and Verslues, P. E. (2013). Functional characterization of an ornithine cyclodeaminase-like protein of Arabidopsis thaliana. BMC Plant Biol. 13:182. doi: 10.1186/1471-2229-13-182
Shinde, S., Villamor, J. G., Lin, W. D., Sharma, S., and Verslues, P. E. (2016). Proline coordination with fatty acid synthesis and redox metabolism of chloroplast and mitochondria. Plant Physiol. 172, 1074–1088. doi: 10.1104/pp.16.01097
Sharma, S., Villamor, J. G., and Verslues, P. E. (2011). Essential role of tissue-specific proline synthesis and catabolism in growth and redox balance at low water potential. Plant Physiol. 157, 292–304. doi: 10.1104/pp.111.183210
Siripornadulsil, S., Traina, S., Verma, D. P. S., and Sayre, R. T. (2002). Molecular mechanisms of proline-mediated tolerance to toxic heavy metals in transgenic microalgae. Plant Cell 14, 2837–2847. doi: 10.1105/tpc.004853
Skopelitis, D. S., Paranychianakis, N. V., Paschalidis, K. A., Pliakonis, E. D., Delis, I. D., Yakoumakis, D. I., et al. (2006). Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 18, 2767–2781. doi: 10.1105/tpc.105.038323
Svennerstam, H., Ganeteg, U., and Nasholm, T. (2008). Root uptake of cationic amino acids by Arabidopsis depends on functional expression of amino acid permease. New Phytol. 180, 620–630. doi: 10.1111/j.1469-8137.2008.02589.x
Svennerstam, H., Jamtgard, S., Ahmad, I., Huss-Danell, K., Nasholm, T., and Ganeteg, U. (2011). Transporters in Arabidopsis roots mediating uptake of amino acids at naturally occurring concentrations. New Phytol. 191, 459–467. doi: 10.1111/j.1469-8137.2011.03699.x
Szabados, L., and Savoure, A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97. doi: 10.1016/j.tplants.2009.11.009
Szekely, G., Abraham, E., Cselo, A., Rigo, G., Zsigmond, L., Csiszar, J., et al. (2008). Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53, 11–28. doi: 10.1111/j.1365-313X.2007.03318.x
Tegeder, M., Offler, C. E., Frommer, W. B., and Patrick, J. W. (2000). Amino acid transporters are localized to transfer cells of developing pea seeds. Plant Physiol. 122, 319–325. doi: 10.1104/pp.122.2.319
Teh, C. Y., Mahmood, M., Shaharuddin, N. A., and Ho, C. L. (2015). In vitro rice shoot apices as simple model to study the effect of NaCl and the potential of exogenous proline and glutathione in mitigating salinity stress. Plant Growth Regul. 75, 771–781. doi: 10.1007/s10725-014-9980-2
Ueda, A., Shi, W. M., Sanmiya, K., Shono, M., and Takabe, T. (2001). Functional analysis of salt-inducible proline transporter of barley roots. Plant Cell Physiol. 42, 1282–1289. doi: 10.1093/pcp/pce166
Verbruggen, N., and Hermans, C. (2008). Proline accumulation in plants: a review. Amino Acids 35, 753–759. doi: 10.1007/s00726-008-0061-6
Verslues, P. E., and Sharma, S. (2010). Proline metabolism and its implications for plant-environment interaction. Arabidopsis Book 8:e0140. doi: 10.1199/tab.0140
Wang, H. Y., Tang, X. L., Wang, H. L., and Shao, H. B. (2015). Proline accumulation and metabolism-related genes expression profiles in Kosteletzkya virginica seedlings under salt stress. Front. Plant Sci. 6:792. doi: 10.3389/fpls.2015.00792
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, J., Xu, J. X., Kong, Y. Z., Ji, Z. D., Wang, X. C., An, F. Y., et al. (2005). Generation of chemical-inducible activation tagging T-DNA insertion lines of Arabidopsis thaliana. Yi Chuan Xue Bao 32, 1082–1088.
Zhang, R., Zhu, J., Cao, H. Z., Xie, X. L., Huang, J. J., Chen, X. H., et al. (2013). Isolation and characterization of LHT-type plant amino acid transporter gene from panax ginseng meyer. J. Ginseng Res. 37, 361–370. doi: 10.5142/jgr.2013.37.361
Zheng, J. L., Zhao, L. Y., Wu, C. W., Shen, B., and Zhu, A. Y. (2015). Exogenous proline reduces NaCl-induced damage by mediating ionic and osmotic adjustment and enhancing antioxidant defense in Eurya emarginata. Acta Physiol. Plant. 37:181. doi: 10.1007/s11738-015-1921-9
Zhu, B. C., Su, J., Chan, M. C., Verma, D. P. S., Fan, Y. L., and Wu, R. (1998). Overexpression of a delta (1)-pyrroline-5-carboxylate synthetase gene and analysis of tolerance to water- and salt-stress in transgenic rice. Plant Sci. 139, 41–48. doi: 10.1016/S0168-9452(98)00175-7
Keywords: Arabidopsis thaliana, amino acid permease, positional cloning, proline accumulation, proline transport, salt stress
Citation: Wang T, Chen Y, Zhang M, Chen J, Liu J, Han H and Hua X (2017) Arabidopsis AMINO ACID PERMEASE1 Contributes to Salt Stress-Induced Proline Uptake from Exogenous Sources. Front. Plant Sci. 8:2182. doi: 10.3389/fpls.2017.02182
Received: 04 August 2017; Accepted: 12 December 2017;
Published: 22 December 2017.
Edited by:
Alejandra A. Covarrubias, Universidad Nacional Autónoma de México, MexicoReviewed by:
Sandeep Sharma, Council of Scientific and Industrial Research (CSIR), IndiaCopyright © 2017 Wang, Chen, Zhang, Chen, Liu, Han and Hua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuejun Hua, xjhua@ibcas.ac.cn
†Present address: Ying Chen, VIB-UGent Center for Plant Systems Biology, Ghent University, Ghent, Belgium Min Zhang, State Key Laboratory of Plant Cell and Chromosome Engineering, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China Jiugeng Chen, National Key Laboratory of Plant Molecular Genetics, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.