- Centre for Modern Interdisciplinary Technologies, Nicolaus Copernicus University in Toruń, Toruń, Poland
In eukaryotic cells, lipids in the form of triacylglycerols (TAGs) are the major reservoir of cellular carbon and energy. These TAGs are packed into specialized organelles called lipid droplets (LDs). They can be found in most, if not all, types of cells, from bacteria to human. Recent data suggest that rather than being simple storage organelles, LDs are very dynamic structures at the center of cellular metabolism. This is also true in plants and algae, where LDs have been implicated in many processes including energy supply; membrane structure, function, trafficking; and signal transduction. Plant and algal LDs also play a vital role in human life, providing multiple sources of food and fuel. Thus, a lot of attention has been paid to metabolism and function of these organelles in recent years. This review summarizes the most recent advances on LDs degradation as a key process for TAGs release. While the initial knowledge on this process came from studies in oilseeds, the findings of the last decade revealed high complexity and specific mechanisms of LDs degradation in plants and algae. This includes identification of numerous novel proteins associated with LDs as well as a prominent role for autophagy in this process. This review outlines, systemizes, and discusses the most current data on LDs catabolism in plants and algae.
Introduction
Lipids in plants and algae can be generally divided into two major groups, storage lipids and membrane lipids. The former serve as energy and carbon reservoirs, and the latter are building blocks for photosynthetic and non-photosynthetic membranes (Li-Beisson et al., 2013). Additionally, land plants deposit a lipidic layer composed of waxes and cutin on their surface, which serves as an impermeable barrier protecting them from excessive water loss, pathogens, and toxins (Nawrath et al., 2013; Serrano et al., 2014). The storage lipids are represented mainly by triacylglycerols (TAGs) and to a much lesser degree by sterol esters, whereas lipid composition of membranes is more complex and differs between cell compartments, cell types, and organisms (Bouvier-Navé et al., 2010; Horn et al., 2011; Goold et al., 2015; Shimada et al., 2019). Broadly, galactolipids are much more abundant in plastidial membranes, whereas non-plastidial membranes are composed mostly of phospholipids (Li-Beisson et al., 2013). Synthesis of lipids in plant and algal cells starts in plastids, where fatty acids (FAs) are generated. These molecules are further used as substrates in the key cellular pathways of membrane lipid and TAGs synthesis. Before entering complex metabolic pathways FAs often undergo modifications, like desaturation or elongation. Regardless of their modification level, FAs can reside in the chloroplast, where they function as substrates for synthesis of plastidial membrane lipids, like monogalactosyldiacylglycerol (MGDG) or digalactosyldiacylglycerol (DGDG) (Wang and Benning, 2012; Lavell and Benning, 2019). When exported to the cytosol, free FAs are first exported by FATTY ACID EXPORT1 (FAX1) across the chloroplast inner envelope and then undergo vectorial acylation by the chloroplast outer envelope-localized long-chain acyl-CoA synthetase 9 (LACS9) (Schnurr et al., 2002; Li et al., 2015; Li et al., 2016; Li et al., 2019). The resulting conjugates of FAs with coenzyme A (CoA)—acyl-CoAs are transported into endoplasmic reticulum (ER) and incorporated into the cellular pool of carbon and energy in the form of TAGs (Li et al., 2016). The major pathway of TAGs synthesis is the glycerol-3-phosphate (G-3-P) (or Kennedy) pathway (Figure 1) (Ohlrogge and Browse, 1995; Chapman and Ohlrogge, 2012). In this pathway, G-3-P is successively esterified with acyl chains from acyl-CoAs. These reactions occur in a precise manner and are catalyzed by specific acyltransferases located in the endoplasmic reticulum (ER) membrane. In the first reaction, catalyzed by glycerol-3-phosphate acyltransferase (GPAT), G-3-P is linked to acyl-CoA and lysphosphatidic acid (LPA) is generated. LPA is then converted into phosphatidic acid (PA) by esterification with another acyl-CoA. This reaction is catalyzed by lysophosphatidate acyltransferase (LPAT). PA can serve as a substrate for synthesis of phospholipids or TAGs. In the latter case, dephosphorylation of PA by phosphatidic acid phosphatase (PAP) leads to formation of diacylglycerol (DAG) (Li-Beisson et al., 2013). The synthesized DAG is further converted into TAG by acyl-CoA:diacylglycerol acyltransferases (DGATs), and this reaction is considered a committed step of TAG formation (Zhang et al., 2009; Turchetto-Zolet et al., 2011; Sanjaya et al., 2013; Zienkiewicz et al., 2016; Zienkiewicz et al., 2017; Zienkiewicz et al., 2018). In addition to DGAT-mediated pathway DAG can also be acylated into TAG by the phospholipid:diacylglycerol acyltransferase (PDAT) (Figure 1). This enzyme has been shown to synthesize TAG via acyl-CoA-independent transacylation of DAG using phosphatidylcholine (PC) as acyl donor (Figure 1) (Dahlqvist et al., 2000; Zhang et al., 2009; Yoon et al., 2012).
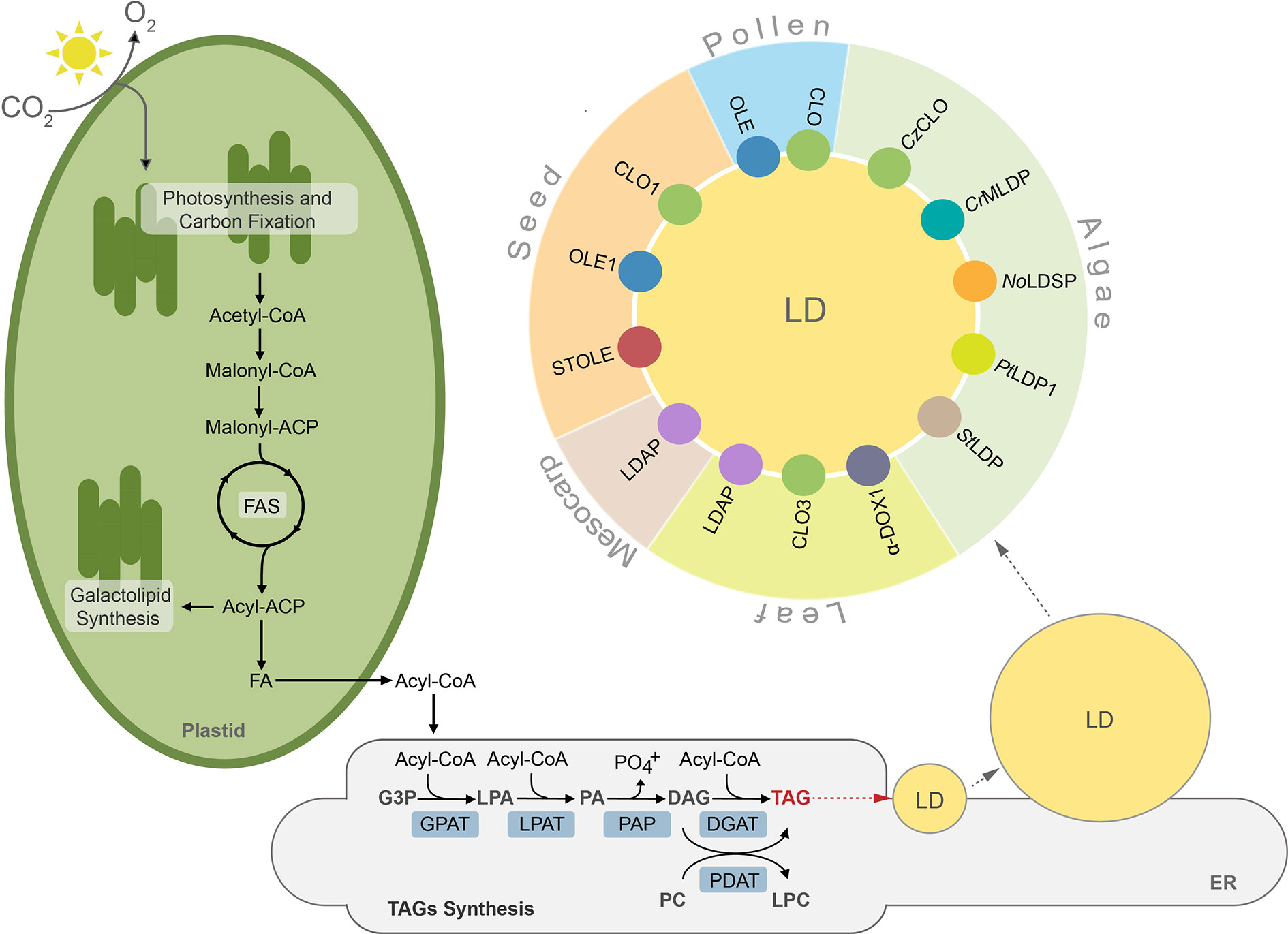
Figure 1 TAGs synthesis, lipid droplet formation and their protein equipment in plant and algal cells. ACP, acyl carrier protein; CoA, coenzyme A; CLO, caleosin; CrMLDP Chlamydomonas reinhardtii major lipid droplet protein; CzCLO, Chromochloris zofingiensis caleosin; DAG, diacylglycerol; DGAT, acylCoA:diacylglycerol acyltransferase, α-DOX1, dioxygenase 1; FA, fatty acid; FAS, fatty acid synthase, G3P, glycerol-3-phosphate; GPAT, acyl-CoA:glycerol-3-phosphate-acyltransferase; LD, lipid droplet; LDAP, lipid droplet associated protein; LPA, lysophosphatidic acid; LPAT, acyl-CoA:lysophosphatidic acid acyltransferase; LPC, lysophosphatidylcholine, NoLDSP, Nannochloropsis oceanica lipid droplet surface protein; OLE, oleosin; PA, phosphatidic acid; PAP, phosphatidic acid phosphatase, PDAT, phospholipid:diacylglycerol acyltransferase; PC, phosphatidylcholine; PtLDP1, Phaeodactylum tricornutum lipid droplet protein 1; StLDP, Stramenopile-type lipid droplet protein; STOLE, steroleosin; TAG, triacylglycerol.
TAGs synthesized in the ER membrane are continuously deposited between leaflets of the membrane bilayer, finally forming spherical organelles referred to in the literature by many terms, such as lipid droplets (thereafter LDs), oil bodies (OBs), oleosomes, or spherosomes (Figure 1). A prominent role for LDs formation at ER membrane in plants has been shown for SEIPIN proteins (Cai et al., 2015; Taurino et al., 2018; Greer et al., 2020). Indeed, loss of function Arabidopsis thaliana SEIPIN isoforms as well as their overexpression results in altered size of LDs, however, the exact role of these proteins in LDs formation remains to be clarified (Cai et al., 2015; Taurino et al., 2018). Recent studies showed also that mutation in the gene encoding AtVAP27-1 (Vesicle-Associated Membrane Protein (VAMP)–Associated Proteins (VAPs)) resulted in formation of aberrant and enlarged LDs. Moreover, a direct interaction between Arabidopsis SEIPIN isoforms (2 and 3) and AtVAP27-1 indicates that these two proteins most probably cooperate during formation of LDs (Greer et al., 2020). A putative SEIPIN ortholog has also been identified in diatom Phaeodactylum tricornutum. Overexpression of this SEIPIN in P. tricornutum resulted in biogenesis of larger LDs, suggesting similarities in the functional nature of SEIPINs between plants and algae (Lu et al., 2017).
After their complete formation, LDs separate from the ER membrane and localize to the cytosol (Chapman and Ohlrogge, 2012; Murphy, 2012; Olzmann and Carvalho, 2019). LDs have been identified in cells of diverse organisms, including bacteria (Zhang et al., 2017), yeast (Grillitsch et al., 2011), algae (Zienkiewicz et al., 2020), plants (Tzen et al., 1993), nematodes (O’Rourke et al., 2009), and mammals (Birsoy et al., 2013). This indicates their highly conserved role in cellular lipid metabolism. Until recently LDs were considered as a simple TAG storage compartment, however, intense studies in the last decade revealed that LDs represent highly dynamic structures, involved in a plethora of diverse cellular processes, like regulation of energy homeostasis, remodeling of membranes, and signaling (Chitraju et al., 2017; Welte and Gould, 2017; Yang and Benning, 2018; Fernandez-Santos et al., 2020). According to the common structural model, LDs are composed of a hydrophobic core filled mostly with TAGs surrounded by a phospholipid monolayer, and are decorated with a set of specific proteins (Figure 1). These proteins are considered essential for biogenesis, stabilization, and mobilization of LDs, and are cell-, tissue-, and organism-specific (Figure 1, Table S1) (Tzen et al., 1993; Huang, 2018).
Lipid Droplets—Multiple Variants Of The Same Organelle?
In plants, structural proteins of LDs were first and best characterized in seeds (Tzen et al., 1990; Tzen et al., 1993; Jolivet et al., 2004; Huang, 2018; Du et al., 2019). The set of major LD structural proteins found in seeds includes oleosin, caleosin, and steroleosin, however their individual isoforms are also present in non-seed tissues (Table S1) (Chapman et al., 2012; Huang, 2018). Oleosins are the most abundant integral membrane proteins of oilseed LDs. They are anchored in the phospholipid monolayer by a hydrophobic α-helical hairpin domain with a proline knot, and their C- and N- termini face the cytosol (Abell et al., 1997; Napier et al., 2001; Alexander et al., 2002). As Arabidopsis thaliana mutant of one of the seed-specific oleosins ole1 accumulates larger LDs when compared to wild type plants, it has been proposed that oleosins control the structure and size of LDs by preventing their uncontrolled fusion (Siloto et al., 2006; Shimada et al., 2008). Caleosins are much less abundant compared to oleosins in the LDs fraction from oilseeds. Their name derives from the ability to bind calcium ions (by a single EF-hand binding motif) and structural similarity to oleosins (Chen et al., 1999). Phylogenetic studies so far suggest that caleosin proteins were likely the ancestors of oleosins, as the putative genes encoding for caleosin-, but not oleosin-like proteins, are present in algae, non-vascular plants, and fungi (Jiang and Tzen, 2010; Rahman et al., 2018a; Rahman et al., 2018b). The ability to bind calcium ions, together with the presence of several phosphorylation sites and possession of peroxygenase activity suggest that caleosins mediate signaling between diverse developmental and stress signals and LDs (Hanano et al., 2006; Purkrtova et al., 2008). Unlike caleosins and oleosins, steroleosins possess only two structural motifs: an N-terminal hydrophobic region responsible for association with LD membranes through conserved proline residues, and a C-terminal domain with hydroxysteroid dehydrogenase (HSD) activity (d’Andrea et al., 2007). Steroleosins are thought to play a role in brassinosteroid-mediated cellular signaling during plant growth and development (Lin et al., 2002). Recent advances in LDs proteomics identified many new LD-associated proteins in seeds and seedlings, however their functions remain to be deciphered (Zhi et al., 2017; Kretzschmar et al., 2020).
After the seed, pollen grains are one of the most active sites in TAGs biosynthesis (Piffanelli et al., 1998; Zienkiewicz et al., 2014a). Mature pollen grains of many plants, especially oleaginous species, accumulate a high number of LDs in the cytoplasm of the vegetative cell (Zienkiewicz et al., 2014a). Similar to seeds, pollen LDs are coated with oleosin and caleosin, but no pollen steroleosin has yet been identified (Kim et al., 2002; Jiang et al., 2008; Zienkiewicz et al., 2010; Zienkiewicz K. et al., 2011). Interestingly, oleosin-like and caleosin proteins are also present in the pollen coat (Mayfield et al., 2001; Rejon et al., 2016), anther loculus, as well as in tapetal cells (Zienkiewicz K. et al., 2011; Levesque-Lemay et al., 2016). The latter tissue is directly involved in pollen development and is extremely rich in lipidic structures known as tapetosomes. They are released from degrading tapetum during anther development and targeted to the surface of developing pollen grains, eventually forming the pollen coat (Parish and Li, 2010; Levesque-Lemay et al., 2016). Interestingly, each tapetosome consists of multiple LDs clustered together and coated by oleosins (Hsieh and Huang, 2005) and/or caleosin (Zienkiewicz K. et al., 2011). This could explain the presence of both these proteins in the pollen coat (Table S1) and suggests that mature pollen grains are equipped with LD-associated proteins of both gametophytic and sporophytic origin.
Depending on developmental stage and/or physiological state of the plant, LDs can also be present in non-seed organs such as fruits, leaves, stems, and roots (Lersten et al., 2006; Shimada et al., 2015; Pyc et al., 2017a; Huang, 2018). Leaf LDs seem to be equipped with a different set of integral proteins than seeds (Table S1) (Pyc et al., 2017a; Fernandez-Santos et al., 2020). A leaf-specific isoform of caleosin, CLO3, has been shown to decorate leaf LDs and to be directly involved in triggering the oxylipin-mediated response to pathogen attack in A. thaliana (Hanano et al., 2015; Shimada et al., 2015). Infection of Arabidopsis by the pathogenic fungus Colletotrichum higginisianum leads to localization of α-dioxygenase 1 (α-DOX1) on the surface of leaf LDs and the formation of 2-hydroperoxy-octadecatrienoic acid (2-HPOT) from α-linolenic acid released from TAGs stored in LDs. 2-HPOT is then converted by CLO3 into 2-hydroxy-octadecatrienoic acid (2-HOT), which possesses anti-fungal activity (Shimada et al., 2014). Additionally, accumulation of phytoalexin deficient 3 (PAD3) protein involved in camalexin synthesis was observed on the surface of leaf LDs after Pseudomonas syringae pv tomato (Pst) DC3000 avrRpm1 infection, supporting the role of leaf LDs in the plant defense response (Fernandez-Santos et al., 2020). Besides caleosin, three isoforms of Lipid Droplet Associated Proteins (LDAPs) have been identified in the leaves of Arabidopsis. LDAPs have been shown to be essential for the maintenance and regulation of LD metabolism and to play nonredundant functions in the stress response and post-germinative growth (Gidda et al., 2016). In addition to the above mentioned LD-associated proteins, a few proteins with diverse functions were also identified in the proteome of leaf LDs, including LDAP-interacting protein (LDIP) (Pyc et al., 2017b), glycerol-3-phosphate-acyltransferase 4 (GPAT4) and 8 (GPAT8) (Fernandez-Santos et al., 2020), strictosidine synthase (STR), 2-oxoglutarate (2OG) and farnesylcysteine lyase (FCLY) (Brocard et al., 2017). These observations suggest the multifunctional nature of LDs and support the direct involvement of non-seed LDs in stress-response and defense-related pathways in plants. Among non-seed tissues, few but extremely large LDs (over 10 µm of diameter) have also been found in mesocarp cells of some oleaginous fruits, like olive and avocado (Horn et al., 2013; Bartolini et al., 2014). Similar to seed LDs, mesocarp LDs are filled with TAGs, but they mostly lack oleosin and are equipped with LDAP protein instead (Horn et al., 2013; Zhi et al., 2017).
Under favorable growth conditions, algae usually accumulate small amounts of TAGs, whereas upon stresses such as nutrient limitation (e.g. nitrogen (N) deprivation), elevated temperatures, or high light intensities they synthesize massive amounts (Goold et al., 2015; Li-Beisson et al., 2015; Zienkiewicz et al., 2016). Algal LDs are equipped with specific LD-associated proteins, different from their plant counterparts (Moellering and Benning, 2010; Vieler et al., 2012a; Zienkiewicz et al., 2016; Leyland et al., 2020). The major LD protein MLDP was first identified in green algae, Chlamydomonas reinhardtii (Moellering and Benning, 2010; Nguyen et al., 2011) and then in Dunaiella salina (Davidi et al., 2012), Scenedesmus quadricauda (Javee et al., 2016), Chromochloris zofingiensis (Wang et al., 2019), and Lobosphaera incisa (Siegler et al., 2017). MLDPs are different from oleosin in lacking the central hydrophobic region, and appear to be specific to the lineage of green algae. Similar to structural proteins covering LDs in land plants, CrMLDP also seems to be involved in the control of LD size and stabilization (Tsai et al., 2015). In turn, LDs of the oleaginous alga Nannochloropsis oceanica are decorated with Lipid Droplet Surface Protein (LDSP), which has been shown to localize on the LD surface during N deprivation (Vieler et al., 2012a; Vieler et al., 2012b; Zienkiewicz et al., 2020). NoLDSP protein, similar to plant oleosin, possesses a proline-rich hydrophobic domain in its central region, however the proline knot motif found in oleosin is absent in NoLDSP (Vieler et al., 2012a). Major LD structural proteins were also identified in the diatom P. tricornutum, including LD-associated protein (PtLDP1) (Wang et al., 2017), a homolog of oleosome-associated-protein 1 (DOAP1) from Fistulifera solaris (Maeda et al., 2014), and Stramenopile-type lipid droplet protein (StLDP) (Yoneda et al., 2016). Overexpression of both these proteins in P. tricornutum was correlated with increased TAGs content and enlarged LDs during N deprivation (Yoneda et al., 2016; Wang et al., 2017), suggesting their analogous role to plant oleosin. Recently, three caleosin-related proteins were identified by proteomic analyses of LDs from C. zofingiensis, and co-localization for two of them on LDs was confirmed in Saccharomyces cerevisiae (Wang et al., 2019). Besides the major structural proteins detected in algal LDs, multiple proteomic studies have been performed to identify new LD proteins in diverse algal strains (Siegler et al., 2017; Lupette et al., 2019; Wang et al., 2019), however their specific localization and potential role in LD homeostasis have yet to be investigated.
Regardless of LD protein and TAG composition, their mobilization is crucial for providing energy and carbon during periods of active metabolism. This process is highly coordinated with development (e.g. seed germination) as well as responses to specific environmental conditions (e.g. nutrient deprivation). The release of TAGs from LDs requires the coordinated action of molecular machineries governing protein and lipid breakdown. Below we characterize the current state of knowledge regarding these pathways and interactions.
LDs Lipolysis—Tag Lipases and Co-Workers
TAG lipases play one of the most essential roles in LDs degradation in plants and algae. These enzymes hydrolyze TAGs stored in LDs leading to the release of FAs, DAGs, MAGs, and glycerol (Graham, 2008; Kelly and Feussner, 2016). Glycerol is subsequently phosphorylated, oxygenated, and enters into glycolysis, whereas FAs undergo β-oxidation into acetyl-CoAs (Eastmond and Graham, 2001). In plants (Graham, 2008; Pracharoenwattana et al., 2010; Rinaldi et al., 2016) and green algae (Kong et al., 2018a; Kong et al., 2018b) the latter process takes place in peroxisomes (glyoxysomes), meanwhile in diatoms both mitochondrial and peroxisomal FAs β-oxidation is tough to occur (Chauton et al., 2013; Jallet et al., 2020).
So far, the TAG lipases directly involved in LD mobilization are best characterized in plants, mainly in germinating seeds and growing seedlings of A. thaliana (Eastmond, 2006; Quettier and Eastmond, 2009; Kelly et al., 2011). LDs accumulation during seed development is critical for germination and seedling growth later on (Huang, 1996; Graham, 2008; Zienkiewicz A. et al., 2011; Zienkiewicz et al., 2014b). Based on the analysis of TAGs breakdown in Arabidopsis sdp1, sdp1L, and sdp1 sdp1L mutants it was demonstrated that among these two TAG lipases only AtSDP1 (SUGAR DEPENDENT 1) plays a key role in mobilization of TAGs from LDs during seed germination (Eastmond, 2006; Kelly et al., 2011). In addition, the loss of SDP1 function caused increased TAG content in the seeds of Arabidopsis (van Erp et al., 2014), rapeseed (Brassica napus L.) (Kelly et al., 2013a), Jatropha (Jatropha curcas) (Kim et al., 2014), and soybean (Glycine max L.) (Kanai et al., 2019), suggesting that SDP1 might be involved in TAGs turnover in developing seeds as well. Indeed, a decrease in oil content has often been observed during desiccation phase of seed development in some species (Baud et al., 2002; Chia et al., 2005; Kelly et al., 2013a). It has been proposed that at early stages of post-germinative growth, the inactive form of AtSDP1 first localizes to the surface of peroxisomes and is then delivered to LDs via peroxules, initiating TAGs hydrolysis (Figure 2) (Thazar-Poulot et al., 2015; Cui et al., 2016; Esnay et al., 2020). The released FAs are then transported into peroxisomes by AtPXA1 (an ATP-binding cassette (ABC) transporter) and enter the β-oxidation process (Zolman et al., 2001). Interestingly, Arabidopsis sdp1 and sdp1 sdp1L mutants exhibit residual TAG hydrolysis on sugar-deficient medium, suggesting that additional lipases could also be involved in this process (Kelly et al., 2011). Therefore, the mobilization of TAGs was also analyzed in an Arabidopsis mutant with a lesion in ADIPOSE TRIGLYCERIDE LIPASE-LIKE (ATGL). ATGL encodes a lipase similar to human ATGL and COMPARATIVE GENE IDENTIFIER-58 (CGI-58), which possess TAG lipase, phospholipase A (PLA), and lysophosphatidic acid acyltransferase (LPAAT) activities (Eastmond, 2006; Ghosh et al., 2009; Kelly et al., 2011). However, no significant difference in the rate of TAGs breakdown was observed between wild type plants and these single mutants (Kelly et al., 2011). It was reported recently that AtOBL1, a homolog of acid lipase from Ricinus communis (RcOBL1, oil body lipase 1, Eastmond, 2004) possesses activity towards TAGs in Arabidopsis seeds (Müller and Ischebeck, 2018). Although AtOBL1 is able to cleave TAGs and is associated with LDs, seed germination and TAGs breakdown rates in the obl1 mutant were similar to those observed in wild type plants (Müller and Ischebeck, 2018). A specific LD lipoxygenase (LOX) and phospholipase A (PLA) seem to be involved in an alternative pathway of seed LDs degradation (Rudolph et al., 2011). LOX activity leads to formation of (9Z,11E,13S)-13-hydroperoxy octadeca-9,11-dienoic acid (13-HPOD) by oxygenation of linoleate moieties (18:2) of TAGs mobilized from LDs. The 13-HPOD liberated from TAGs can be later reduced to 13-HOD by the peroxygenase activity of caleosin (Rudolph et al., 2011). The LOX and PLA activities were detected in vitro on isolated LDs of cucumber (Cucumis sativus) (Rudolph et al., 2011) and olive (Olea europaea L.) (Zienkiewicz et al., 2014b) during seed germination. Thus, it was proposed that PLA might be responsible for LD membrane degradation and thereby facilitate access of LOX and lipase to TAG.
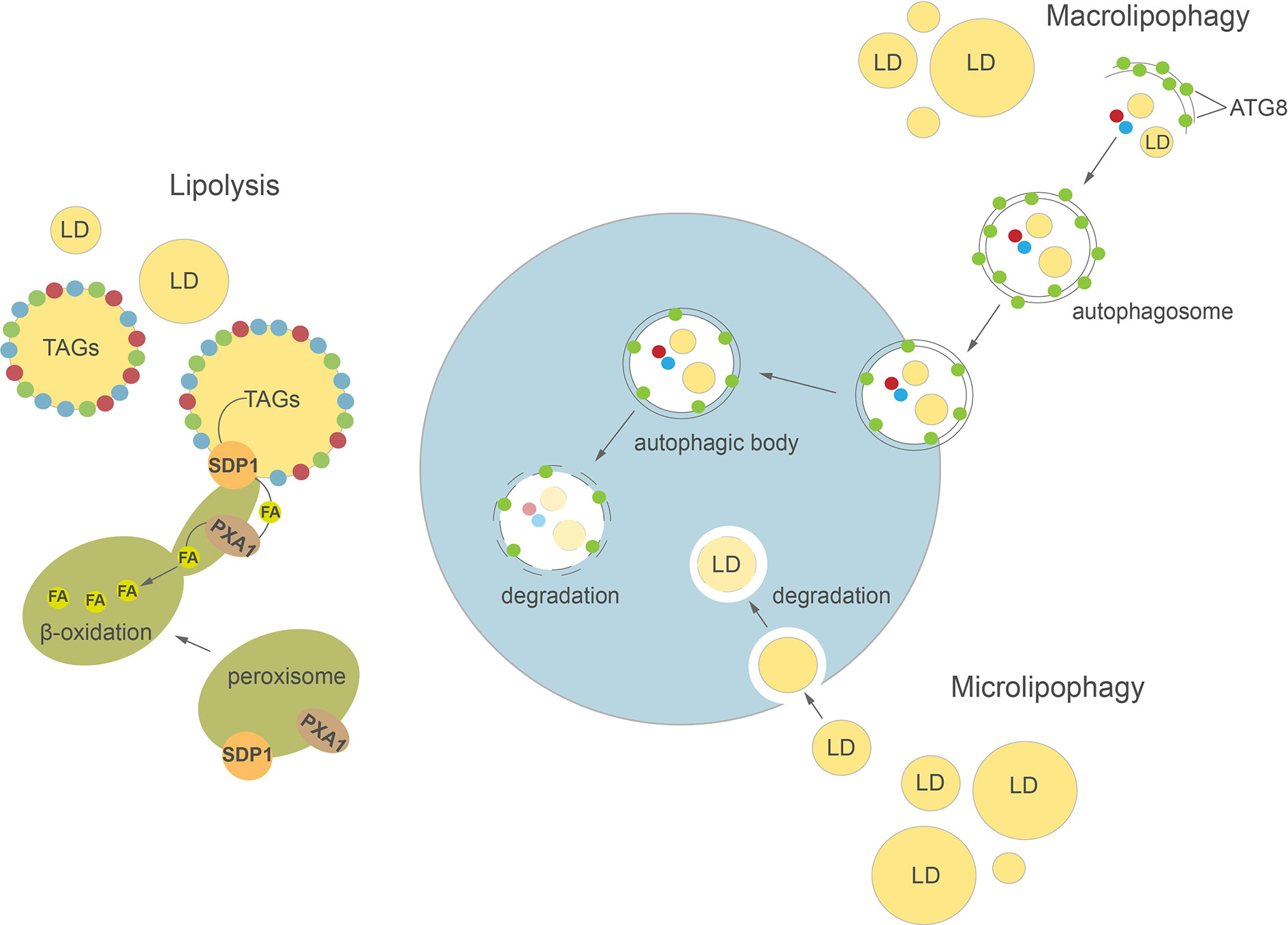
Figure 2 Cellular pathways of lipid droplets degradation in plants and algae. Detailed description in the text. ATG8, AUTOPHAGY-RELATED PROTEIN 8; FA, fatty acid; LD, lipid droplet; SDP1, SUGAR DEPENDENT 1; TAGs, triacylglycerols.
Pollen development, germination, and pollen tube growth are essential for sexual reproduction in flowering plants. During pollen hydration LDs polarize near the germinative aperture, and as the pollen grain starts to germinate, they enter the emerging pollen tube where their progressive degradation takes place (Zienkiewicz et al., 2013; Taurino et al., 2018). AtSDP1L, unlike other putative TAG lipases identified in Arabidopsis, showed exceptionally high expression in mature pollen grains (Kelly et al., 2011). This suggests that AtSDP1L may be involved in LDs breakdown during pollen germination. Two homologs of RcOBL1, TAG lipase from tobacco (NtOBL1) and AtOBL1 from Arabidopsis mentioned above, have been characterized in pollen tubes (Müller and Ischebeck, 2018). Interestingly, similar to RcOBL1 and unlike AtSDP1 in seeds, both proteins localized to LDs when ectopically expressed in growing tobacco pollen tubes. Moreover, Arabidopsis obl1 mutants showed slower pollen tube growth in vivo. The authors suggested that acyl groups released from TAGs by OBL1 might be directly channeled into the ER, where they serve as substrates for rapid membrane synthesis (Müller and Ischebeck, 2018).
LD-associated lipase activities were also found in vitro and in situ in germinating pollen and growing pollen tubes of olive (Olea europaea L.) (Rejon et al., 2012; Zienkiewicz et al., 2013). Their direct role in proper pollen tube growth has been confirmed by in vitro olive pollen germination in medium with and without sucrose, with higher lipase activity resulting in the latter case (Zienkiewicz et al., 2013). In these studies, TAG lipase activity was found only on LDs isolated from germinating pollen tubes, and not from the mature pollen grain. Similar localization pattern was also found for LOX protein (Zienkiewicz et al., 2013). In turn, PLA activity co-localized with LDs in both mature and germinating olive pollen (Zienkiewicz et al., 2013). This indicates that there are substantial differences in the temporal and spatial localization pattern of machinery governing LDs breakdown in pollen grains compared to seeds, where all the lipid degrading enzymes seem to be recruited to the LDs surface during or just after seed imbibition (Rudolph et al., 2011; Thazar-Poulot et al., 2015). Most probably, this reflects the distinct energy demands of pollen grains and seeds, which are tightly connected with their biological functions. The initiation of pollen germination and pollen tube growth occur much faster than seed imbibition and germination. This could explain the presence of some LDs degrading enzymes “on-site,” available to act as soon as pollen germination starts.
Previous studies have shown that disruption of AtSDP1 also leads to TAG accumulation in other vegetative tissues, such us leaves, stems, and roots (Kelly et al., 2013b; Fan et al., 2014). Moreover, the expression level of AtSDP1 increases during natural leaf senescence, thus it is possible that this lipase is involved in regulation of TAG homeostasis during leaf development (Troncoso-Ponce et al., 2013). The transcript levels corresponding to AtCGI-58 were up-regulated during leaf senescence as well (Troncoso-Ponce et al., 2013), and loss of function of AtCGI-58 resulted in a significant increase in TAG content of leaf mesophyll cells (James et al., 2010). While in mammals CGI-58 has been shown to act as an activator of adipose triglyceride lipase ATGL, and thus to directly regulate TAG breakdown (Lass et al., 2006), such a role has not been confirmed for plants, where CGI-58 likely regulates the activity of PXA1 but not of TAG lipase (Park et al., 2013). Consequently, the plant CGI-58 does not seem to be a bona fide TAG lipase, but rather an element of regulatory circuit of TAG homeostasis. Importantly, many putative lipases are expressed during leaf senescence (Troncoso-Ponce et al., 2013), suggesting more lipases could be involved in LD turnover in leaves.
LDs mobilization is an essential step during the transition of algal cells from quiescence to autotrophy in response to restored favorable growth conditions (Siaut et al., 2011; Tsai et al., 2018; Zienkiewicz et al., 2020). Thus, comparative transcriptomics between stress (N deprivation) and optimal (N resupply) growth conditions led to the identification of many putative TAG lipases highly expressed after triggering of the massive TAG degradation (Jaeger et al., 2017; Zienkiewicz et al., 2020). However, only a few of them have been functionally characterized to date (Table 1). Among 49 putative lipases encoded by the genome of the oleaginous marine diatom P. tricornutum, only one patatin-SDP1-like lipase, named tgl1, has been functionally characterized to date (Barka et al., 2016). The TAG lipase activity of tgl1 has been confirmed in vitro and in vivo. In the latter case, the tgl1 knockdown mutants of P. tricornutum accumulated much higher amounts of TAGs when compared to the wild type (Barka et al., 2016). In C. reinhardtii, among over 130 putative lipases encoded by its genome, only one TAG lipase has been identified and characterized recently. This TAG lipase, LIP4, shares 44% amino acid identity with AtSDP1 (Table 1) (Warakanont et al., 2019). The expression of LIP4 is downregulated in response to N deprivation and up-regulated after N resupply, whereas its mutation resulted in delay of TAG degradation, which consequently led to TAG over-accumulation in Crlip4 mutant (Warakanont et al., 2019). Two homologs of AtSDP1, named NoTGL1 and NoTGL2, have also been recently reported in N. oceanica (Nobusawa et al., 2019). Over-accumulation of TAGs was observed only in the N. oceanica knockout of NoTGL1, but not of NoTGL2, after N resupply. Moreover, NoTGL1 was found to be a specific resident of ER, thus the authors proposed its involvement in degradation of TAG de novo synthesized in the ER (Nobusawa et al., 2019). Proteomic analysis of another oleaginous green alga, L. incisa, revealed the presence of the putative TAG lipase LiSDP1. LiSDP1 shares 44% identity with AtSDP1 (Siegler et al., 2017). Ectopic expression of LiSDP1 in A. thaliana sdp1 sdp1-L mutants resulted in only partial complementation of the TAG lipase-deficient plants. LiSDP1 from L. incisa was absent in LD isolates from N-deprived cultures and did not localize to LDs when ectopically expressed in tobacco pollen tube. The authors did not exclude that LiSDP1 might be specifically recruited to the surface of LDs when needed (Siegler et al., 2017). By searching for human lipase homologs in the genome of Thalassiosira pseudonana, Trentacoste et al. (2013) identified a homolog of CGI-58, encoded by Thaps3_264297. The resulting protein has been shown to possess TAG lipase, PLA, and LPAAT activities in vitro, and a knockdown of Thaps3_264297 resulted in increased TAG content and accumulation of LDs compared to the wild type strain (Trentacoste et al., 2013). However, taking into account the proposed role of plant CGI-58 the exact role of algal CGI-58 still needs to be elucidated.
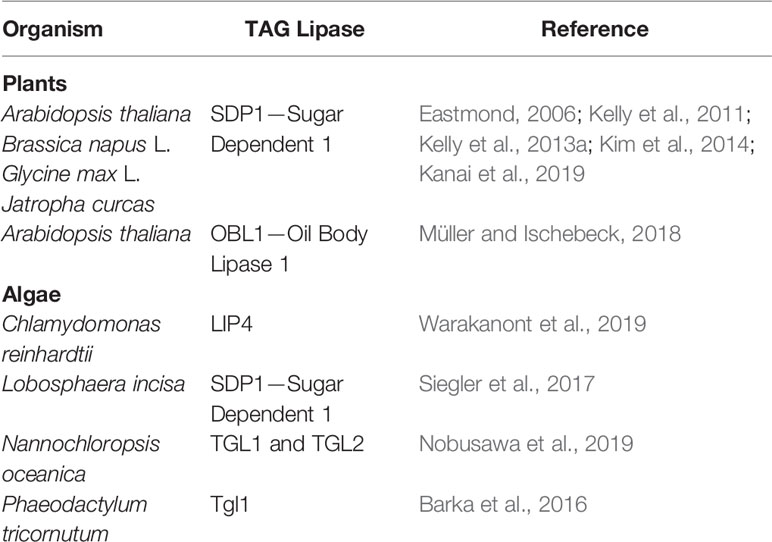
Table 1 Functionally characterized TAG lipases involved in LDs degradation identified in plants and algae.
Lipophagy—A New Player in The Field
Autophagy is a highly conserved process involved in regulation of intracellular degradation and recycling of individual molecules as well as whole organelles, via lysosomes in animals or vacuoles in yeast and plants (Avin-Wittenberg et al., 2012; Bento et al., 2016; Marshall and Vierstra, 2018; Couso et al., 2018). In plants, autophagy plays an important role in many developmental processes, such as seed development (Di Berardino et al., 2018; Sera et al., 2019) and leaf senescence (Avila-Ospina et al., 2014; Li et al., 2014). Autophagy is highly induced by various abiotic stresses, including nutrient deprivation (Sun et al., 2018; Janse van Rensburg et al., 2019), drought (Bao et al., 2020), and biotic stresses e.g. pathogen infection (Dagdas et al., 2016). Two major types of autophagy have been described in plants: macroautophagy and microautophagy (Figure 2). During macroautophagy, numerous autophagy-related (ATG) proteins participate in the induction of autophagy and formation of a double-membrane structure called an autophagosome (Yoshimoto and Ohsumi, 2018). Autophagosomes containing different cargo (macromolecules or damaged organelles) fuse with the tonoplast and subsequently enter the vacuolar lumen as autophagic bodies. The content of autophagic bodies then undergoes progressive degradation (Soto-Burgos et al., 2018). In microautophagy, cytoplasmic contents are directly captured into the vacuolar lumen by invagination of the tonoplast. Previous studies in yeast and mammalian cells demonstrated a close relationship between LD homeostasis and autophagy (van Zutphen et al., 2014; Petan et al., 2018). Indeed, autophagy may either provide FAs for LDs biogenesis (Rambold et al., 2015) or participate in their degradation in the process of lipophagy (Singh et al., 2009; van Zutphen et al., 2014). In animals, depending on the size of LDs, their degradation can be achieved either by macrolipophagy, where small, entire LDs are enclosed together with other cytoplasmic contents in an autophagosome, or by piecemeal microlipophagy, where only a small portion of a large LD is trapped in an autophagosome and then pinches off as a double membrane autolipophagosome (Singh et al., 2009; Khaldoun et al., 2014; Garcia et al., 2018). In yeast, distinct forms of microlipophagy contribute to LD degradation depending on growth conditions (van Zutphen et al., 2014; Vevea et al., 2015; Seo et al., 2017). For example, under acute glucose starvation, the molecular machinery of macroautophagy participates in the induction of microlipophagy. ATG14 is recruited to liquid-ordered membrane (Lo) domains at the surface of vacuole, where together with ATG6 it forms recruitment sites for LDs and initiates their microlipophagy (Seo et al., 2017). In contrast, under lipid and ER stress, microlipophagy depends on ESCRT (endosomal sorting complexes required for transport) machinery rather than ATG proteins (Vevea et al., 2015). Similarly, microlipophagy induced after diauxic shifts has been shown to be independent of the core ATGs but dependent on ESCRT proteins (Oku et al., 2017). Our knowledge on LDs TAG degradation inside the vacuole/lysosome is rather scarce and fragmentary. Nevertheless, one of the few reports showed that vacuolar lipase ATG15 acts in degradation of neutral lipids of LDs after their incorporation into the vacuole in yeast (Maeda et al., 2015).
Over the last decade research has also produced increasing evidence of an important role for autophagy in LDs degradation in plants (Kurusu et al., 2014; Fan et al., 2019) and algae (Zhao et al., 2014; Schwarz et al., 2017; Tsai et al., 2018). In plants, lipophagy seems to be involved among others in male reproductive development, pollen germination, and pollen tube growth (Kurusu et al., 2014; Hanamata et al., 2019; Hanamata et al., 2020; Zhao et al., 2020). The potential role of lipophagy in pollen grain maturation was demonstrated in a study on the Oryza sativa atg7 mutant (Kurusu et al., 2014). Mutation in OsATG7 was associated with a lower number of LDs in mature pollen and a higher accumulation of LDs in tapetal cells compared to wild type plants. These results, together with the observation that in wild type plants LDs were enclosed in the vacuoles of rice tapetal cells, suggest that ATG7-dependent tapetal autophagy may be responsible for LDs degradation and lipid metabolism in the tapetum (Kurusu et al., 2014). Recently, a detailed analysis of the germinating pollen lipidome in tobacco was performed using ATG-suppressed RNAi lines (Zhao et al., 2020). This study showed that silencing of ATG2 and ATG5 leads to a decrease in the number of autophagosomes at the germinative pollen aperture and is accompanied by inhibition of pollen germination and significant accumulation of TAGs and DAGs in pollen grains. Lipophagy may also contribute to the degradation of LDs during seed germination and seedling growth. The potential implication of autophagy in LDs degradation during seed germination was suggested by earlier studies of the Arabidopsis clo1 mutant (Poxleitner et al., 2006). During seed germination, LDs were commonly observed in the vacuolar lumen of wild type cotyledon cells, while loss of function of AtCLO1 resulted in the absence of LDs inside the vacuoles and slower degradation of eicosenoic acid (20:1) (Poxleitner et al., 2006). It is therefore possible that turnover of some pools of LDs is regulated by caleosin-dependent microlipophagy during seed germination. Other data showed that during in vitro germination of olive seeds, the neutral lipids stained by Sudan Black B co-localize in the area of protein bodies (known also as protein storage vacuoles), together with lipase activity, suggesting that LDs degradation takes place inside protein bodies (Zienkiewicz et al., 2014b). A significant accumulation of TAGs was observed in etiolated carbon-starved seedlings of the Arabidopsis atg5 mutant (Avin-Wittenberg et al., 2015), indicating the potential role of autophagic machinery in LDs degradation. It was demonstrated recently that LDs breakdown in senescent watermelon (Cirullus lanatus) leaves occurs via different pathways: small vacuole-associated and central vacuole-associated (Zhang et al., 2020). In leaf cells without a central vacuole, LDs interact with autophagosome-like structures before they enter small vacuoles. Meanwhile, in the cells of senescent leaves containing the central vacuole, LDs are delivered into the vacuolar lumen via a process morphologically resembling microlipophagy (Zhang et al., 2020). Lipophagy has also been implicated in LD turnover in Arabidopsis leaves during dark-induced starvation (Fan et al., 2019). Under extended darkness, DsRed-Atg8e-labeled autophagic structures were observed to be associated with LDs in the leaves of tgd1 (trigalactosyldiacylglycerol1) mutant. Moreover, ultrastructural analysis of leaf cells from dark-treated Atsdp1-4 plants, deficient in cytosolic lipolysis, demonstrated the entrance and appearance of LDs in the central vacuole. The authors suggested that this process is likely mediated by the microlipophagy pathway (Fan et al., 2019). Interestingly, disruption of molecular macroautophagy machinery in Arabidopsis double mutants atg2-1 sdp1-4 and atg5-1 sdp1-4 leads to inhibition of dark-induced lipophagy in the leaves. Taken together, the findings described by Fan et al. (2019) showed that microlipophagy observed in Arabidopsis leaves depends on core components of macroautophagy pathway, as has been described previously in yeast (van Zutphen et al., 2014).
A large number of studies on autophagy-mediated LDs degradation in algae have been performed using C. reinhardtii as a reference. Ultrastructural analysis of C. reinhardtii cells under N resupply (NR) conditions showed the appearance of small LDs in the vacuolar lumen is controlled by a process morphologically resembling microlipophagy (Tsai et al., 2018). A mutation in CrATG8, encoding one of the core ATG proteins required for the formation of the autophagosome, results in delayed degradation of TAGs after NR compared to wild type lines (Kajikawa et al., 2019). In addition, by using mCherry-ATG8 as a tool to monitor the intracellular movements of autophagosomes (Yoshimoto et al., 2004; Klionsky et al., 2007), the fusion between autophagosomes and LDs was observed at later stages of N deprivation (Tran et al., 2019). Similar to C. reinhardtii, small LDs were observed to be sequestered by the vacuole via a pathway resembling microautophagy in another green alga, Auxenochlorella protothecoides, during its heterotrophy to autotrophy transition (Zhao et al., 2014). Interestingly, the authors suggested that small portions of large LDs might also be degraded in the vacuolar lumen through a process reminiscent of the piecemeal microautophagy commonly observed in mammalian cells. Microlipophagy-like degradation of LDs was observed as well in N. oceanica under NR conditions (Zienkiewicz et al., 2020). Moreover, by using a biomolecular fluorescence complementation (BiFC) assay it was demonstrated that NoATG8 protein interacts in vivo with NoLDSP —a major LD surface protein in N. oceanica (Zienkiewicz et al., 2020). It has been proposed that this interaction might be involved in the targeting and/or fusion of LDs into the vacuole. The ATG8-interacting motif (AIM) was also identified in Stramenopile-type lipid droplet protein (StLDP), suggesting a possible link between autophagy and LD degradation in P. tricornutum (Leyland et al., 2020). The latest analyses revealed that salt stress is also associated with appearance of LDs in the vacuolar lumen in Parachlorella kessleri (You et al., 2019). Interestingly, in the unicellular alga Micrasterias denticulate, LDs degradation occurring under carbon starvation seems to be mediated by macrolipophagy, as LDs were trapped in autophagosomes and then delivered into small vacuoles (Schwarz et al., 2017).
LD Protein Turnover—The Missing Puzzle In The Crosstalk Between Lipolysis And Lipophagy?
Previous studies in animals revealed that association of ATGL with the surface of LDs depends on the degradation of the LDs structural proteins perilipin 2 (PLIN2) and perilipin 3 (PLIN3) via chaperone-mediated autophagy (CMA) (Kaushik and Cuervo, 2015). Sathyanarayan et al. (2017) proposed a model where the removal of PLINs facilitates access of ATGL to TAGs, and ATGL acts as a signal for LDs recognition by autophagosomes and thus an inducer of bulk LDs degradation via lipophagy (Sathyanarayan et al., 2017). In plants, oleosins and LDAPs have been proposed to protect LDs from the action of TAG lipases (Siloto et al., 2006; Gidda et al., 2016). Recently, two independent groups reported that ubiquitinated oleosins interact with the PUX10 protein through its UBA domain. Via its UBX domain, PUX10 in turn recruits CDC48, enabling further degradation of oleosins by the proteasome (Deruyffelaere et al., 2018; Kretzschmar et al., 2018). On the other hand, it has been found that AtOLE1 can interact with AtATG8e protein through the AIM motif (ATG8 interacting motif) (Marshall et al., 2019). This implies that autophagy can also participate in oleosin turnover during seed germination. Interestingly, ATGL and hormone-sensitive lipase (HSL) contain several putative LIR (LC3-interacting region) motifs responsible for direct interaction with microtubule associated protein 1 LC3 (ATG8 family protein) (Martinez-Lopez et al., 2016). Mutating the ATGL LIR motif blocked ATGL associations with the LD surface and lipolysis, suggesting the direct crosstalk between lipolysis and autophagy in animal cells. It remains unknown if the degradation of plant and algal TAG lipases is also controlled by autophagy machinery.
Concluding Remarks
The past decade has seen considerable progress in the identification and characterization of mechanisms governing LDs degradation in plant and algal cells. The studies reviewed here revealed the complexity of intracellular molecular networks related to LDs metabolism, and highlighted the fundamental meaning of these organelles for developmental programs and physiological responses in plants and algae. This includes essential functions of LDs structural proteins (oleosins, celeosins, and their counterparts in algal cells) as well as lipolysis and lipophagy pathways of TAGs breakdown. However, there are still many unanswered questions. For example, what is the nature of cross-talk between lipolysis and lipophagy in plant and algal cells? And what specific lipophagy receptors on the surface of LDs could be involved in activation of the autophagic machinery and lipophagy initiation? Moreover, it is still unclear whether, similar to animal cells, plant and algal TAG lipases are implicated in the induction of lipophagy, and if autophagy is responsible for their degradation. Answering these questions will help us gain a better comprehensive understanding of induction and regulation of LDs mobilization in plants and algae. Future research is needed to uncover the fascinating and multifaceted mechanisms that govern LDs turnover in plant and algal cells, as this knowledge is crucial for the development of more applied research and engineering of lipid-rich biomass production from algae and oil crops.
Author Contributions
KZ and AZ conceived, wrote the manuscript and designed the figures. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Tegan Haslam for the critical reading of the manuscript, valuable comments and English corrections.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.579019/full#supplementary-material
Supplementary Table 1 | LD-associated proteins identified in plants and algae. The corresponding references are given next to the table.
References
Abell, B. M., Holbrook, L. A., Abenes, M., Murphy, D. J., Hills, M. J., Moloney, M. M. (1997). Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell 9, 1481–1493. doi: 10.1105/tpc.9.8.1481
Alexander, L. G., Sessions, R. B., Clarke, A. R., Tatham, A. S., Shewry, P. R., Napier, J. A. (2002). Characterization and modelling of the hydrophobic domain of a sunflower oleosin. Planta 214, 546–551. doi: 10.1007/s004250100655
Avila-Ospina, L., Moison, M., Yoshimoto, K., Masclaux-Daubresse, C. (2014). Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 65, 3799–3811. doi: 10.1093/jxb/eru039
Avin-Wittenberg, T., Honig, A., Galili, G. (2012). Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma 249, 285–299. doi: 10.1007/s00709-011-0296-z
Avin-Wittenberg, T., Bajdzienko, K., Wittenberg, G., Alseekh, S., Tohge, T., Bock, R., et al. (2015). Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell 27, 306–322. doi: 10.1105/tpc.114.134205
Bao, Y., Song, W.-M., Wang, P., Yu, X., Li, B., Jiang, C., et al. (2020). COST1 regulates autophagy to control plant drought tolerance. Proc. Natl. Acad. Sci. U. S. A. 117, 7482–7493. doi: 10.1073/pnas.1918539117
Barka, F., Angstenberger, M., Ahrendt, T., Lorenzen, W., Bode, H. B., Buchel, C. (2016). Identification of a triacylglycerol lipase in the diatom Phaeodactylum tricornutum. Biochim. Biophys. Acta 1861, 239–248. doi: 10.1016/j.bbalip.2015.12.023
Bartolini, S., Leccese, A., Andreini, L. (2014). Influence of canopy fruit location on morphological, histochemical and biochemical changes in two oil olive cultivars. Plant Biosyst. - Int. J. Deal. all Asp. Plant Biol. 148, 1221–1230. doi: 10.1080/11263504.2014.980360
Baud, S., Boutin, J. P., Miquel, M., Lepiniec, L., Rochat, C. (2002). An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 40, 151–160. doi: 10.1016/S0981-9428(01)01350-X
Bento, C. F., Renna, M., Ghislat, G., Puri, C., Ashkenazi, A., Vicinanza, M., et al. (2016). Mammalian autophagy: How does it work? Annu. Rev. Biochem. 85, 685–713. doi: 10.1146/annurev-biochem-060815-014556
Birsoy, K., Festuccia, W. T., Laplante, M. (2013). A comparative perspective on lipid storage in animals. J. Cell Sci. 126, 1541–1552. doi: 10.1242/jcs.104992
Bouvier-Navé, P., Berna, A., Noiriel, A., Compagnon, V., Carlsson, A. S., Banas, A., et al. (2010). Involvement of the phospholipid sterol acyltransferase1 in plant sterol homeostasis and leaf senescence. Plant Physiol. 152, 107–119. doi: 10.1104/pp.109.145672
Brocard, L., Immel, F., Coulon, D., Esnay, N., Tuphile, K., Pascal, S., et al. (2017). Proteomic analysis of lipid droplets from Arabidopsis aging leaves brings new insight into their biogenesis and functions. Front. Plant Sci. 8:894:894. doi: 10.3389/fpls.2017.00894
Cai, Y., Goodman, J. M., Pyc, M., Mullen, R. T., Dyer, J. M., Chapman, K. D. (2015). Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell. 27, 2616–2636. doi: 10.1105/tpc.15.00588
Chapman, K. D., Ohlrogge, J. B. (2012). Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 287, 2288–2294. doi: 10.1074/jbc.R111.290072
Chapman, K. D., Dyer, J. M., Mullen, R. T. (2012). Biogenesis and functions of lipid droplets in plants: Thematic Review Series: Lipid Droplet Synthesis and Metabolism: from Yeast to Man. J. Lipid Res. 53, 215–226. doi: 10.1194/jlr.R021436
Chauton, M. S., Winge, P., Brembu, T., Vadstein, O., Bones, A. M. (2013). Gene regulation of carbon fixation, storage, and utilization in the diatom Phaeodactylum tricornutum acclimated to light/dark cycles. Plant Physiol. 161, 1034–1048. doi: 10.1104/pp.112.206177
Chen, J. C., Tsai, C. C., Tzen, J. T. (1999). Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil bodies of plant seeds. Plant Cell Physiol. 40, 1079–1086. doi: 10.1093/oxfordjournals.pcp.a029490
Chia, T. Y., Pike, M. J., Rawsthorne, S. (2005). Storage oil breakdown during embryo development of Brassica napus (L.). J. Exp. Bot. 5, 1285–1296. doi: 10.1093/jxb/eri129
Chitraju, C., Mejhert, N., Haas, J. T., Diaz-Ramirez, L. G., Grueter, C. A., Imbriglio, J. E., et al. (2017). Triglyceride synthesis by DGAT1 protects adipocytes from lipid-induced ER stress during lipolysis. Cell Metab. 26, 407–418.e3. doi: 10.1016/j.cmet.2017.07.012
Couso, I., Pérez-Pérez, M. E., Martínez-Force, E., Kim, H. S., He, Y., Umen, J. G., et al. (2018). Autophagic flux is required for the synthesis of triacylglycerols and ribosomal protein turnover in Chlamydomonas. J. Exp. Bot. 69, 1355–1367. doi: 10.1093/jxb/erx372
Cui, S., Hayashi, Y., Otomo, M., Mano, S., Oikawa, K., Hayashi, M., et al. (2016). Sucrose production mediated by lipid metabolism suppresses the physical interaction of peroxisomes and oil bodies during germination of. Arabidopsis thaliana J. Biol. Chem. 291, 19734–19745. doi: 10.1074/jbc.M116.748814
Dagdas, Y. F., Belhaj, K., Maqbool, A., Chaparro-Garcia, A., Pandey, P., Petre, B., et al. (2016). An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. Elife 5, e10856. doi: 10.7554/eLife.10856
Dahlqvist, A., Stahl, U., Lenman, M., Banas, A., Lee, M., Sandager, L., et al. (2000). Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. U. S. A. 97, 6487–6492. doi: 10.1073/pnas.120067297
Davidi, L., Katz, A., Pick, U. (2012). Characterization of major lipid droplet proteins from Dunaliella. Planta 236, 19–33. doi: 10.1007/s00425-011-1585-7
Deruyffelaere, C., Purkrtova, Z., Bouchez, I., Collet, B., Cacas, J.-L., Chardot, T., et al. (2018). PUX10 is a CDC48A adaptor protein that regulates the extraction of ubiquitinated oleosins from seed lipid droplets in Arabidopsis. Plant Cell 30, 2116–2136. doi: 10.1105/tpc.18.00275
Di Berardino, J., Marmagne, A., Berger, A., Yoshimoto, K., Cueff, G., Chardon, F., et al. (2018). Autophagy controls resource allocation and protein storage accumulation in Arabidopsis seeds. J. Exp. Bot. 69, 1403–1414. doi: 10.1093/jxb/ery012
d’Andrea, S., Canonge, M., Beopoulos, A., Jolivet, P., Hartmann, M. A., Miquel, M., et al. (2007). At5g50600 encodes a member of the short-chain dehydrogenase reductase superfamily with 11beta- and 17beta-hydroxysteroid dehydrogenase activities associated with Arabidopsis thaliana seed oil bodies. Biochimie 89, 222–229. doi: 10.1016/j.biochi.2006.09.013
Du, C., Liu, A., Niu, L., Cao, D., Liu, H., Wu, X., et al. (2019). Proteomic identification of lipid-bodies-associated proteins in maize seeds. Acta Physiol. Plant 41, 70. doi: 10.1007/s11738-019-2854-5
Eastmond, P. J., Graham, I. A. (2001). Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 6, 72–78. doi: 10.1016/s1360-1385(00)01835-5
Eastmond, P. J. (2004). Cloning and characterization of the acid lipase from castor beans. J. Biol. Chem. 279, 45540–45545. doi: 10.1074/jbc.M408686200
Eastmond, P. J. (2006). SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18, 665–675. doi: 10.1105/tpc.105.040543
Esnay, N., Dyer, J. M., Mullen, R. T., Chapman, K. D. (2020). Lipid droplet–peroxisome connections in plants. Contact 3:2515256420908765. doi: 10.1177/2515256420908765
Fan, J., Yan, C., Roston, R., Shanklin, J., Xu, C. (2014). Arabidopsis lipins, PDAT1 acyltransferase, and SDP1 triacylglycerol lipase synergistically direct fatty acids toward β-oxidation, thereby maintaining membrane lipid homeostasis. Plant Cell 26, 4119–4134. doi: 10.1105/tpc.114.130377
Fan, J., Yu, L., Xu, C. (2019). Dual role for autophagy in lipid metabolism in Arabidopsis. Plant Cell 31, 1598–1613. doi: 10.1105/tpc.19.00170
Fernandez-Santos, R., Izquierdo, Y., Lopez, A., Muniz, L., Martinez, M., Cascon, T., et al. (2020). Protein profiles of lipid droplets during the hypersensitive defense response of Arabidopsis against Pseudomonas infection. Plant Cell Physiol. 61, 1144–1157. doi: 10.1093/pcp/pcaa041
Garcia, E. J., Vevea, J. D., Pon, L. A. (2018). Lipid droplet autophagy during energy mobilization, lipid homeostasis and protein quality control. Front. Biosci. (Landmark Ed. 23, 1552–1563. doi: 10.2741/4660
Ghosh, A. K., Chauhan, N., Rajakumari, S., Daum, G., Rajasekharan, R. (2009). At4g24160, a soluble acyl-coenzyme A-dependent lysophosphatidic acid acyltransferase. Plant Physiol. 151, 869–881. doi: 10.1104/pp.109.144261
Gidda, S. K., Park, S., Pyc, M., Yurchenko, O., Cai, Y., Wu, P., et al. (2016). Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol. 170, 2052–2071. doi: 10.1104/pp.15.01977
Goold, H., Beisson, F., Peltier, G., Li-Beisson, Y. (2015). Microalgal lipid droplets: composition, diversity, biogenesis and functions. Plant Cell Rep. 34, 545–555. doi: 10.1007/s00299-014-1711-7
Graham, I. A. (2008). Seed storage oil mobilization. Annu. Rev. Plant Biol. 59, 115–142. doi: 10.1146/annurev.arplant.59.032607.092938
Greer, M. S., Cai1, Y., Gidda, S. K., Esnay, N., Kretzschmar, F. K., Seay, D., et al. (2020). SEIPIN isoforms interact with the membrane-tethering protein VAP27-1 for lipid droplet formation. Plant Cell. doi: 10.1105/tpc.19.00771
Grillitsch, K., Connerth, M., Kofeler, H., Arrey, T. N., Rietschel, B., Wagner, B., et al. (2011). Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim. Biophys. Acta 1811, 1165–1176. doi: 10.1016/j.bbalip.2011.07.015
Hanamata, S., Sawada, J., Toh, B., Ono, S., Ogawa, K., Fukunaga, T., et al. (2019). Monitoring autophagy in rice tapetal cells during pollen maturation. Plant Biotechnol. (Tokyo Jpn.) 36, 99–105. doi: 10.5511/plantbiotechnology.19.0417a
Hanamata, S., Sawada, J., Ono, S., Ogawa, K., Fukunaga, T., Nonomura, K.-I., et al. (2020). Impact of autophagy on gene expression and tapetal programmed cell death during pollen development in rice. Front. Plant Sci. 11:172:172. doi: 10.3389/fpls.2020.00172
Hanano, A., Burcklen, M., Flenet, M., Ivancich, A., Louwagie, M., Garin, J., et al. (2006). Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. J. Biol. Chem. 281, 33140–33151. doi: 10.1074/jbc.M605395200
Hanano, A., Bessoule, J.-J., Heitz, T., Blee, E. (2015). Involvement of the caleosin/peroxygenase RD20 in the control of cell death during Arabidopsis responses to pathogens. Plant Signal. Behav. 10, e991574. doi: 10.4161/15592324.2014.991574
Horn, P. J., Ledbetter, N. R., James, C. N., Hoffman, W. D., Case, C. R., Verbeck, G. F., et al. (2011). Visualization of lipid droplet composition by direct organelle mass spectrometry. J. Biol. Chem. 286, 3298–3306. doi: 10.1074/jbc.M110.186353
Horn, P. J., James, C. N., Gidda, S. K., Kilaru, A., Dyer, J. M., Mullen, R. T., et al. (2013). Identification of a new class of lipid droplet-associated proteins in plants. Plant Physiol. 162, 1926–1936. doi: 10.1104/pp.113.222455
Hsieh, K., Huang, A. H. C. (2005). Lipid-rich tapetosomes in Brassica tapetum are composed of oleosin-coated oil droplets and vesicles, both assembled in and then detached from the endoplasmic reticulum. Plant J. 43, 889–899. doi: 10.1111/j.1365-313X.2005.02502.x
Huang, A. H. (1996). Oleosins and oil bodies in seeds and other organs. Plant Physiol. 110, 1055–1061. doi: 10.1104/pp.110.4.1055
Huang, A. H. C. (2018). Plant lipid droplets and their associated proteins: potential for rapid advances. Plant Physiol. 176, 1894–1918. doi: 10.1104/pp.17.01677
Jaeger, D., Winkler, A., Mussgnug, J. H., Kalinowski, J., Goesmann, A., Kruse, O. (2017). Time-resolved transcriptome analysis and lipid pathway reconstruction of the oleaginous green microalga Monoraphidium neglectum reveal a model for triacylglycerol and lipid hyperaccumulation. Biotechnol. Biofuels 10, 197. doi: 10.1186/s13068-017-0882-1
Jallet, D., Xing, D., Hughes, A., Moosburner, M., Simmons, M. P., Allen, A. E., et al. (2020). Mitochondrial fatty acid β-oxidation is required for storage-lipid catabolism in a marine diatom. New Phytol. doi: 10.1111/nph.16744
James, C. N., Horn, P. J., Case, C. R., Gidda, S. K., Zhang, D., Mullen, R. T., et al. (2010). Disruption of the Arabidopsis CGI-58 homologue produces Chanarin-Dorfman-like lipid droplet accumulation in plants. Proc. Natl. Acad. Sci. U. S. A. 107, 17833–17838. doi: 10.1073/pnas.0911359107
Janse van Rensburg, H. C., Van den Ende, W., Signorelli, S. (2019). Autophagy in plants: both a puppet and a puppet master of sugars. Front. Plant Sci. 10:14:14. doi: 10.3389/fpls.2019.00014
Javee, A., Sulochana, S. B., Pallissery, S. J., Arumugam, M. (2016). Major lipid body protein: a conserved structural component of lipid body accumulated during abiotic stress in S. quadricauda CASA-CC202. Front. Energy Res. 4:2016.00037:37. doi: 10.3389/fenrg.2016.00037
Jiang, P.-L., Tzen, J. T. C. (2010). Caleosin serves as the major structural protein as efficient as oleosin on the surface of seed oil bodies. Plant Signal. Behav. 5, 447–449. doi: 10.4161/psb.5.4.10874
Jiang, P.-L., Jauh, G.-Y., Wang, C.-S., Tzen, J. T. C. (2008). A unique caleosin in oil bodies of lily pollen. Plant Cell Physiol. 49, 1390–1395. doi: 10.1093/pcp/pcn103
Jolivet, P., Roux, E., D’Andrea, S., Davanture, M., Negroni, L., Zivy, M., et al. (2004). Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. PPB 42, 501–509. doi: 10.1016/j.plaphy.2004.04.006
Kajikawa, M., Yamauchi, M., Shinkawa, H., Tanaka, M., Hatano, K., Nishimura, Y., et al. (2019). Isolation and characterization of Chlamydomonas autophagy-related mutants in nutrient-deficient conditions. Plant Cell Physiol. 60, 126–138. doi: 10.1093/pcp/pcy193
Kanai, M., Yamada, T., Hayashi, M., Mano, S., Nishimura, M. (2019). Soybean (Glycine max L.) triacylglycerol lipase GmSDP1 regulates the quality and quantity of seed oil. Sci. Rep. 9, 8924. doi: 10.1038/s41598-019-45331-8
Kaushik, S., Cuervo, A. M. (2015). Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 17, 759–770. doi: 10.1038/ncb3166
Kelly, A. A., Feussner, I. (2016). Oil is on the agenda: Lipid turnover in higher plants. Biochim. Biophys. Acta 1861, 1253–1268. doi: 10.1016/j.bbalip.2016.04.021
Kelly, A. A., Quettier, A. L., Shaw, E., Eastmond, P. J. (2011). Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol. 157, 866–875. doi: 10.1104/pp.111.181784
Kelly, A. A., Shaw, E., Powers, S. J., Kurup, S., Eastmond, P. J. (2013a). Suppression of the SUGAR-DEPENDENT1 triacylglycerol lipase family during seed development enhances oil yield in oilseed rape (Brassica napus L.). Plant Biotechnol. J. 11, 355–361. doi: 10.1111/pbi.12021
Kelly, A. A., van Erp, H., Quettier, A.-L., Shaw, E., Menard, G., Kurup, S., et al. (2013b). The sugar-dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiol. 162, 1282–1289. doi: 10.1104/pp.113.219840
Khaldoun, S. A., Emond-Boisjoly, M.-A., Chateau, D., Carriere, V., Lacasa, M., Rousset, M., et al. (2014). Autophagosomes contribute to intracellular lipid distribution in enterocytes. Mol. Biol. Cell 25, 118–132. doi: 10.1091/mbc.E13-06-0324
Kim, H. U., Hsieh, K., Ratnayake, C., Huang, A. H. C. (2002). A novel group of oleosins is present inside the pollen of Arabidopsis. J. Biol. Chem. 277, 22677–22684. doi: 10.1074/jbc.M109298200
Kim, M. J., Yang, S. W., Mao, H.-Z., Veena, S. P., Yin, J.-L., Chua, N.-H. (2014). Gene silencing of Sugar-dependent 1 (JcSDP1), encoding a patatin-domain triacylglycerol lipase, enhances seed oil accumulation in Jatropha curcas. Biotechnol. Biofuels 7:36. doi: 10.1186/1754-6834-7-36
Klionsky, D. J., Cuervo, A. M., Seglen, P. O. (2007). Methods for monitoring autophagy from yeast to human. Autophagy 3, 181–206. doi: 10.4161/auto.3678
Kong, F., Burlacot, A., Liang, Y., Légeret, B., Alseekh, S., Brotman, Y., et al. (2018a). Interorganelle communication: peroxisomal MALATE DEHYDROGENASE2 connects lipid catabolism to photosynthesis through redox coupling in Chlamydomonas. Plant Cell. 30, 1824–1847. doi: 10.1105/tpc.18.00361
Kong, F., Romero, I. T., Warakanont, J., Li-Beisson, Y. (2018b). Lipid catabolism in microalgae. New Phytol. 218, 1340–1348. doi: 10.1111/nph.15047
Kretzschmar, F. K., Mengel, L. A., Müller, A. O., Schmitt, K., Blersch, K. F., Valerius, O., et al. (2018). PUX10 is a lipid droplet-localized scaffold protein that interacts with CELL DIVISION CYCLE48 and is involved in the degradation of lipid droplet proteins. Plant Cell 30, 2137–2160. doi: 10.1105/tpc.18.00276
Kretzschmar, F. K., Doner, N. M., Krawczyk, H. E., Scholz, P., Schmitt, K., Valerius, O., et al. (2020). Identification of low-abundance lipid droplet proteins in seeds and seedlings. Plant Physiol. 182, 1326–1345. doi: 10.1104/pp.19.01255
Kurusu, T., Koyano, T., Hanamata, S., Kubo, T., Noguchi, Y., Yagi, C., et al. (2014). OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10, 878–888. doi: 10.4161/auto.28279
Lass, A., Zimmermann, R., Haemmerle, G., Riederer, M., Schoiswohl, G., Schweiger, M., et al. (2006). Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 3, 309–319. doi: 10.1016/j.cmet.2006.03.005
Lavell, A. A., Benning, C. (2019). Cellular organization and regulation of plant glycerolipid metabolism. Plant Cell Physiol. 60, 1176–1183. doi: 10.1093/pcp/pcz016
Lersten, N. R., Czlapinski, A. R., Curtis, J. D., Freckmann, R., Horner, H. T. (2006). Oil bodies in leaf mesophyll cells of angiosperms: overview and a selected survey. Am. J. Bot. 93, 1731–1739. doi: 10.3732/ajb.93.12.1731
Levesque-Lemay, M., Chabot, D., Hubbard, K., Chan, J. K., Miller, S., Robert, L. S. (2016). Tapetal oleosins play an essential role in tapetosome formation and protein relocation to the pollen coat. New Phytol. 209, 691–704. doi: 10.1111/nph.13611
Leyland, B., Boussiba, S., Khozin-Goldberg, I. (2020). A review of diatom lipid droplets. Biol. (Basel) 9, 38. doi: 10.3390/biology9020038
Li, F., Chung, T., Vierstra, R. D. (2014). AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26, 788–807. doi: 10.1105/tpc.113.120014
Li, N., Gügel, I. L., Giavalisco, P., Zeisler, V., Schreiber, L., Soll, J., et al. (2015). FAX1, a novel membrane protein mediating plastid fatty acid export. PloS Biol. 3, e1002053. doi: 10.1371/journal.pbio.1002053
Li, N., Xu, C., Li-Beisson, Y., Philippar, K. (2016). Fatty acid and lipid transport in plant cells. Trends Plant Sci. 21, 145–158. doi: 10.1016/j.tplants.2015.10.011
Li, N., Zhang, Y., Meng, H., Li, S., Wang, S., Xiao, Z., et al. (2019). Characterization of Fatty Acid Exporters involved in fatty acid transport for oil accumulation in the green alga Chlamydomonas reinhardtii. Biotechnol. Biofuels 12, 14. doi: 10.1186/s13068-018-1332-4
Li-Beisson, Y., Shorrosh, B., Beisson, F., Andersson, M. X., Arondel, V., Bates, P. D., et al. (2013). Acyl-lipid metabolism. Arab. B. 11, e0161. doi: 10.1199/tab.0161
Li-Beisson, Y., Beisson, F., Riekhof, W. (2015). Metabolism of acyl-lipids in Chlamydomonas reinhardtii. Plant J. 82, 504–522. doi: 10.1111/tpj.12787
Lin, L.-J., Tai, S. S. K., Peng, C.-C., Tzen, J. T. C. (2002). Steroleosin, a sterol-binding dehydrogenase in seed oil bodies. Plant Physiol. 128, 1200–1211. doi: 10.1104/pp.010928
Lu, Y., Wang, X., Balamurugan, S., Yang, W.-D., Liu, J.-S., Dong, H.-P., et al. (2017). Identification of a putative seipin ortholog involved in lipid accumulation in marine microalga Phaedactylum tricornutum. J. App. Phycol. 29, 2821–2829. doi: 10.1007/s10811-017-1173-8
Lupette, J., Jaussaud, A., Seddiki, K., Morabito, C., Brugière, S., Schaller, H., et al. (2019). The architecture of lipid droplets in the diatom Phaeodactylum tricornutum. Algal. Res. 38:101415. doi: 10.1016/J.ALGAL.2019.101415
Maeda, Y., Sunaga, Y., Yoshino, T., Tanaka, T. (2014). Oleosome-associated protein of the oleaginous diatom Fistulifera solaris contains an endoplasmic reticulum-targeting signal sequence. Mar. Drugs 12, 3892–3903. doi: 10.3390/md12073892
Maeda, Y., Oku, M., Sakai, Y. (2015). A defect of the vacuolar putative lipase Atg15 accelerates degradation of lipid droplets through lipolysis. Autophagy 11, 1247–1258. doi: 10.1080/15548627.2015.1056969
Marshall, R. S., Vierstra, R. D. (2018). Autophagy: the master of bulk and selective recycling. Annu. Rev. Plant Biol. 69, 173–208. doi: 10.1146/annurev-arplant-042817-040606
Marshall, R. S., Hua, Z., Mali, S., McLoughlin, F., Vierstra, R. D. (2019). ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 177, 766–781.e24. doi: 10.1016/j.cell.2019.02.009
Martinez-Lopez, N., Garcia-Macia, M., Sahu, S., Athonvarangkul, D., Liebling, E., Merlo, P., et al. (2016). Autophagy in the CNS and periphery coordinate lipophagy and lipolysis in the brown adipose tissue and liver. Cell Metab. 23, 113–127. doi: 10.1016/j.cmet.2015.10.008
Mayfield, J. A., Fiebig, A., Johnstone, S. E., Preuss, D. (2001). Gene families from the Arabidopsis thaliana pollen coat proteome. Science 292, 2482–2485. doi: 10.1126/science.1060972
Moellering, E. R., Benning, C. (2010). RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell 9, 97–106. doi: 10.1128/ec.00203-09
Müller, A. O., Ischebeck, T. (2018). Characterization of the enzymatic activity and physiological function of the lipid droplet-associated triacylglycerol lipase AtOBL1. New Phytol. 217, 1062–1076. doi: 10.1111/nph.14902
Murphy, D. J. (2012). The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 249, 541–585. doi: 10.1007/s00709-011-0329-7
Napier, J. A., Beaudoin, F., Tatham, A. S., Alexander, L. G., Shewry, P. R. (2001). The seed oleosins: structure, properties and biological role. Adv. Bot. Res. 35, 111–138. doi: 10.1016/S0065-2296(01)35005-X
Nawrath, C., Schreiber, L., Franke, R. B., Geldner, N., Reina-Pinto, J. J., Kunst, L. (2013). Apoplastic diffusion barriers in Arabidopsis. Arab. B. 11, e0167. doi: 10.1199/tab.0167
Nguyen, H. M. ,., Baudet, M., Cuiné, S., Adriano, J., Barthe, D., Billon, E., et al. (2011). Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics 11, 4266–4273. doi: 10.1002/pmic.201100114
Nobusawa, T., Yamakawa-Ayukawa, K., Saito, F., Nomura, S., Takami, A., Ohta, H. (2019). A homolog of Arabidopsis SDP1 lipase in Nannochloropsis is involved in degradation of de novo-synthesized triacylglycerols in the endoplasmic reticulum. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864, 1185–1193. doi: 10.1016/j.bbalip.2019.05.013
Ohlrogge, J., Browse, J. (1995). Lipid biosynthesis. Plant Cell 7, 957–970. doi: 10.1105/tpc.7.7.957
Oku, M., Maeda, Y., Kagohashi, Y., Kondo, T., Yamada, M., Fujimoto, T., et al. (2017). Evidence for ESCRT- and clathrin-dependent microautophagy. J. Cell Biol. 216, 3263–3274. doi: 10.1083/jcb.201611029
Olzmann, J. A., Carvalho, P. (2019). Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155. doi: 10.1038/s41580-018-0085-z
O’Rourke, E. J., Soukas, A. A., Carr, C. E., Ruvkun, G. (2009). C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 10, 430–435. doi: 10.1016/j.cmet.2009.10.002
Parish, R. W., Li, S. F. (2010). Death of a tapetum: A programme of developmental altruism. Plant Sci. 178, 73–89. doi: 10.1016/j.plantsci.2009.11.001
Park, S., Gidda, S. K., James, C. N., Horn, P. J., Khuu, N., Seay, D. C., et al. (2013). The α/β hydrolase CGI-58 and peroxisomal transport protein PXA1 coregulate lipid homeostasis and signaling in Arabidopsis. Plant Cell. 25, 1726–1739. doi: 10.1105/tpc.113.111898
Petan, T., Jarc, E., Jusovic, M. (2018). Lipid droplets in cancer: guardians of fat in a stressful world. Molecules 23, 1941. doi: 10.3390/molecules23081941
Piffanelli, P., Ross, J. H. E., Murphy, D. J. (1998). Biogenesis and function of the lipidic structures of pollen grains. Sex Plant Reprod. 11, 65–80. doi: 10.1007/s004970050122
Poxleitner, M., Rogers, S. W., Lacey Samuels, A., Browse, J., Rogers, J. C. (2006). A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J. 47, 917–933. doi: 10.1111/j.1365-313X.2006.02845.x
Pracharoenwattana, I., Zhou, W., Smith, S. M. (2010). Fatty acid beta-oxidation in germinating Arabidopsis seeds is supported by peroxisomal hydroxypyruvate reductase when malate dehydrogenase is absent. Plant Mol. Biol. 72, 101–109. doi: 10.1007/s11103-009-9554-2
Purkrtova, Z., Le Bon, C., Kralova, B., Ropers, M.-H., Anton, M., Chardot, T. (2008). Caleosin of Arabidopsis thaliana : effect of calcium on functional and structural properties. J. Agric. Food Chem. 56, 11217–11224. doi: 10.1021/jf802305b
Pyc, M., Cai, Y., Greer, M. S., Yurchenko, O., Chapman, K. D., Dyer, J. M., et al. (2017a). Turning over a new leaf in lipid droplet biology. Trends Plant Sci. 22, 596–609. doi: 10.1016/j.tplants.2017.03.012
Pyc, M., Cai, Y., Gidda, S. K., Yurchenko, O., Park, S., Kretzschmar, F. K., et al. (2017b). Arabidopsis lipid droplet-associated protein (LDAP) - interacting protein (LDIP) influences lipid droplet size and neutral lipid homeostasis in both leaves and seeds. Plant J. 92, 1182–1201. doi: 10.1111/tpj.13754
Quettier, A.-L., Eastmond, P. J. (2009). Storage oil hydrolysis during early seedling growth. Plant Physiol. Biochem. PPB 47, 485–490. doi: 10.1016/j.plaphy.2008.12.005
Rahman, F., Hassan, M., Hanano, A., Fitzpatrick, D. A., McCarthy, C. G. P., Murphy, D. J. (2018a). Evolutionary, structural and functional analysis of the caleosin/peroxygenase gene family in the Fungi. BMC Genomics 19, 976. doi: 10.1186/s12864-018-5334-1
Rahman, F., Hassan, M., Rosli, R., Almousally, I., Hanano, A., Murphy, D. J. (2018b). Evolutionary and genomic analysis of the caleosin/peroxygenase (CLO/PXG) gene/protein families in the Viridiplantae. PloS One 13, e0196669. doi: 10.1371/journal.pone.0196669
Rambold, A. S., Cohen, S., Lippincott-Schwartz, J. (2015). Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678–692. doi: 10.1016/j.devcel.2015.01.029
Rejon, J. D., Zienkiewicz, A., Rodriguez-Garcia, M., II, Castro, A. J. (2012). Profiling and functional classification of esterases in olive (Olea europaea) pollen during germination. Ann. Bot. 110, 1035–1045. doi: 10.1093/aob/mcs174
Rejon, J. D., Delalande, F., Schaeffer-Reiss, C., Alche, J., de, D., Rodriguez-Garcia, M., II, et al. (2016). The pollen coat proteome: at the cutting edge of plant reproduction. Proteomes 4, 5. doi: 10.3390/proteomes4010005
Rinaldi, M. A., Patel, A. B., Park, J., Lee, K., Strader, L. C., Bartel, B. (2016). The Roles of β-oxidation and cofactor homeostasis in peroxisome distribution and function in Arabidopsis thaliana. Genetics 204, 1089–1115. doi: 10.1534/genetics.116.193169
Rudolph, M., Schlereth, A., Korner, M., Feussner, K., Berndt, E., Melzer, M., et al. (2011). The lipoxygenase-dependent oxygenation of lipid body membranes is promoted by a patatin-type phospholipase in cucumber cotyledons. J. Exp. Bot. 62, 749–760. doi: 10.1093/jxb/erq310
Sanjaya, M. R., Durrett, T. P., Kosma, D. K., Lydic, T. A., Muthan, B., Bagyalakshmi, M., et al. (2013). Altered lipid composition and enhanced nutritional value of Arabidopsis leaves following introduction of an algal diacylglycerol acyltransferase 2. Plant Cell. 25, 677–693. doi: 10.1105/tpc.112.104752
Sathyanarayan, A., Mashek, M. T., Mashek, D. G. (2017). ATGL promotes autophagy/lipophagy via SIRT1 to control hepatic lipid droplet catabolism. Cell Rep. 19, 1–9. doi: 10.1016/j.celrep.2017.03.026
Schnurr, J. A., Shockey, J. M., de Boer, G. J., Browse, J. A. (2002). Fatty acid export from the chloroplast. Molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis. Plant Physiol. 129, 1700–1709. doi: 10.1104/pp.003251
Schwarz, V., Andosch, A., Geretschlager, A., Affenzeller, M., Lutz-Meindl, U. (2017). Carbon starvation induces lipid degradation via autophagy in the model alga Micrasterias. J. Plant Physiol. 208, 115–127. doi: 10.1016/j.jplph.2016.11.008
Seo, A. Y., Lau, P.-W., Feliciano, D., Sengupta, P., Gros, M. A., Cinquin, B., et al. (2017). AMPK and vacuole-associated Atg14p orchestrate mu-lipophagy for energy production and long-term survival under glucose starvation. Elife 6, e21690. doi: 10.7554/eLife.21690
Sera, Y., Hanamata, S., Sakamoto, S., Ono, S., Kaneko, K., Mitsui, Y., et al. (2019). Essential roles of autophagy in metabolic regulation in endosperm development during rice seed maturation. Sci. Rep. 9, 18544. doi: 10.1038/s41598-019-54361-1
Serrano, M., Coluccia, F., Torres, M., L’Haridon, F., Metraux, J.-P. (2014). The cuticle and plant defense to pathogens. Front. Plant Sci. 5:274:274. doi: 10.3389/fpls.2014.00274
Shimada, T. L., Shimada, T., Takahashi, H., Fukao, Y., Hara-Nishimura, I. (2008). A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 55, 798–809. doi: 10.1111/j.1365-313X.2008.03553.x
Shimada, T. L., Takano, Y., Shimada, T., Fujiwara, M., Fukao, Y., Mori, M., et al. (2014). Leaf oil body functions as a subcellular factory for the production of a phytoalexin in Arabidopsis. Plant Physiol. 164, 105–118. doi: 10.1104/pp.113.230185
Shimada, T. L., Takano, Y., Hara-Nishimura, I. (2015). Oil body-mediated defense against fungi: From tissues to ecology. Plant Signal. Behav. 10, e989036. doi: 10.4161/15592324.2014.989036
Shimada, T. L., Shimada, T., Okazaki, Y., Higashi, Y., Saito, K., Kuwata, K., et al. (2019). HIGH STEROL ESTER 1 is a key factor in plant sterol homeostasis. Nat. Plants 5, 1154–1166. doi: 10.1038/s41477-019-0537-2
Siaut, M., Cuiné, S., Cagnon, C., Fessler, B., Nguyen, M., Carrier, P., et al. (2011). Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 2011 11:7. doi: 10.1186/1472-6750-11-7
Siegler, H., Valerius, O., Ischebeck, T., Popko, J., Tourasse, N. J., Vallon, O., et al. (2017). Analysis of the lipid body proteome of the oleaginous alga Lobosphaera incisa. BMC Plant Biol. 17, 98. doi: 10.1186/s12870-017-1042-2
Siloto, R. M. P., Findlay, K., Lopez-Villalobos, A., Yeung, E. C., Nykiforuk, C. L., Moloney, M. M. (2006). The accumulation of oleosins determines the size of seed oil bodies in Arabidopsis. Plant Cell 18, 1961–1974. doi: 10.1105/tpc.106.041269
Singh, R., Kaushik, S., Wang, Y., Xiang, Y., Novak, I., Komatsu, M., et al. (2009). Autophagy regulates lipid metabolism. Nature 458, 1131–1135. doi: 10.1038/nature07976
Soto-Burgos, J., Zhuang, X., Jiang, L., Bassham, D. C. (2018). Dynamics of autophagosome formation. Plant Physiol. 176, 219–229. doi: 10.1104/pp.17.01236
Sun, X., Jia, X., Huo, L., Che, R., Gong, X., Wang, P., et al. (2018). MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 41, 469–480. doi: 10.1111/pce.13110
Taurino, M., Costantini, S., De Domenico, S., Stefanelli, F., Ruano, G., Delgadillo, M. O., et al. (2018). SEIPIN proteins mediate lipid droplet biogenesis to promote pollen transmission and reduce seed dormancy. Plant Physiol. 176, 1531–1546. doi: 10.1104/pp.17.01430
Thazar-Poulot, N., Miquel, M., Fobis-Loisy, I., Gaude, T. (2015). Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Proc. Natl. Acad. Sci. U. S. A. 112, 4158–4163. doi: 10.1073/pnas.1403322112
Tran, Q.-G., Yoon, H. R., Cho, K., Lee, S.-J., Crespo, J. L., Ramanan, R., et al. (2019). Dynamic interactions between autophagosomes and lipid droplets in Chlamydomonas reinhardtii. Cells 8, 992. doi: 10.3390/cells8090992
Trentacoste, E. M., Shrestha, R. P., Smith, S. R., Gle, C., Hartmann, A. C., Hildebrand, M., et al. (2013). Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc. Natl. Acad. Sci. U. S. A. 110, 19748–19753. doi: 10.1073/pnas.1309299110
Troncoso-Ponce, M. A., Cao, X., Yang, Z., Ohlrogge, J. B. (2013). Lipid turnover during senescence. Plant Sci. 205–206, 13–19. doi: 10.1016/J.PLANTSCI.2013.01.004
Tsai, C.-H., Zienkiewicz, K., Amstutz, C. L., Brink, B. G., Warakanont, J., Roston, R., et al. (2015). Dynamics of protein and polar lipid recruitment during lipid droplet assembly in Chlamydomonas reinhardtii. Plant J. 83, 650–660. doi: 10.1111/tpj.12917
Tsai, C. H., Uygun, S., Roston, R., Shiu, S. H., Benning, C. (2018). Recovery from N deprivation is a transcriptionally and functionally distinct state in Chlamydomonas. Plant Physiol. 176, 2007–2023. doi: 10.1104/pp.17.01546
Turchetto-Zolet, A., Maraschin, F. S., de Morais, G. L., Cagliari, A., Andrade, C. M., Margis-Pinheiro, M., et al. (2011). Evolutionary view of acyl-CoA diacylglycerol acyltransferase (DGAT), a key enzyme in neutral lipid biosynthesis. BMC Evol. Biol. 11:263. doi: 10.1186/1471-2148-11-263
Tzen, J. T., Lai, Y. K., Chan, K. L., Huang, A. H. (1990). Oleosin isoforms of high and low molecular weights are present in the oil bodies of diverse seed species. Plant Physiol. 94, 1282–1289. doi: 10.1104/pp.94.3.1282
Tzen, J. T. C., Cao, Y., Laurent, P., Ratnayake, C., Huang, A. H. C. (1993). Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiol. 101, 267–276. doi: 10.1104/pp.101.1.267
van Erp, H., Kelly, A. A., Menard, G., Eastmond, P. J. (2014). Multigene engineering of triacylglycerol metabolism boosts seed oil content in Arabidopsis. Plant Physiol. 165, 30–36. doi: 10.1104/pp.114.236430
van Zutphen, T., Todde, V., de Boer, R., Kreim, M., Hofbauer, H. F., Wolinski, H., et al. (2014). Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 25, 290–301. doi: 10.1091/mbc.E13-08-0448
Vevea, J. D., Garcia, E. J., Chan, R. B., Zhou, B., Schultz, M., Di Paolo, G., et al. (2015). Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev. Cell 35, 584–599. doi: 10.1016/j.devcel.2015.11.010
Vieler, A., Brubaker, S. B., Vick, B., Benning, C. (2012a). A lipid droplet protein of Nannochloropsis with functions partially analogous to plant oleosins. Plant Physiol. 158, 1562–1569. doi: 10.1104/pp.111.193029
Vieler, A., Wu, G., Tsai, C. H., Bullard, B., Cornish, A. J., Harvey, C., et al. (2012b). Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PloS Genet. 8, e1003064. doi: 10.1371/journal.pgen.1003064
Wang, Z., Benning, C. (2012). Chloroplast lipid synthesis and lipid trafficking through ER-plastid membrane contact sites. Biochem. Soc Trans. 40, 457–463. doi: 10.1042/BST20110752
Wang, X., Hao, T.-B., Balamurugan, S., Yang, W.-D., Liu, J.-S., Dong, H.-P., et al. (2017). A lipid droplet-associated protein involved in lipid droplet biogenesis and triacylglycerol accumulation in the oleaginous microalga Phaeodactylum tricornutum. Algal. Res. 26, 215–224. doi: 10.1016/J.ALGAL.2017.07.028
Wang, X., Wei, H., Mao, X., Liu, J. (2019). Proteomics analysis of lipid droplets from the oleaginous alga Chromochloris zofingiensis reveals novel proteins for lipid metabolism. Genomics Proteomics Bioinf. 17, 260–272. doi: 10.1016/J.GPB.2019.01.003
Warakanont, J., Li-Beisson, Y., Benning, C. (2019). LIP4 is involved in triacylglycerol degradation in Chlamydomonas reinhardtii. Plant Cell Physiol. 60, 1250–1259. doi: 10.1093/pcp/pcz037
Welte, M. A., Gould, A. P. (2017). Lipid droplet functions beyond energy storage. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1260–1272. doi: 10.1016/j.bbalip.2017.07.006
Yang, Y., Benning, C. (2018). Functions of triacylglycerols during plant development and stress. Curr. Opin. Biotechnol. 49, 191–198. doi: 10.1016/j.copbio.2017.09.003
Yoneda, K., Yoshida, M., Suzuki, I., Watanabe, M. M. (2016). Identification of a major lipid droplet protein in a marine diatom Phaeodactylum tricornutum. Plant Cell Physiol. 57, 397–406. doi: 10.1093/pcp/pcv204
Yoon, K., Han, D., Li, Y., Sommerfeld, M., Hu, Q. (2012). Phospholipid:diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell. 24, 3708–3724. doi: 10.1105/tpc.112.100701
Yoshimoto, K., Ohsumi, Y. (2018). Unveiling the molecular mechanisms of plant autophagy-from autophagosomes to vacuoles in plants. Plant Cell Physiol. 59, 1337–1344. doi: 10.1093/pcp/pcy112
Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, T., et al. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967–2983. doi: 10.1105/tpc.104.025395
You, Z., Zhang, Q., Peng, Z., Miao, X. (2019). Lipid droplets mediate salt stress tolerance in Parachlorella kessleri. Plant Physiol. 181, 510–526. doi: 10.1104/pp.19.00666
Zhang, M., Fan, J., Taylor, D. C., Ohlrogge, J. B. (2009). DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell. 21, 3885–3901. doi: 10.1105/tpc.109.071795
Zhang, C., Yang, L., Ding, Y., Wang, Y., Lan, L., Ma, Q., et al. (2017). Bacterial lipid droplets bind to DNA via an intermediary protein that enhances survival under stress. Nat. Commun. 8:15979. doi: 10.1038/ncomms15979
Zhang, C., Qu, Y., Lian, Y., Chapman, M., Chapman, N., Xin, J., et al. (2020). A new insight into the mechanism for cytosolic lipid droplet degradation in senescent leaves. Physiol. Plant 168, 835–844. doi: 10.1111/ppl.13039
Zhao, L., Dai, J., Wu, Q. (2014). Autophagy-like processes are involved in lipid droplet degradation in Auxenochlorella protothecoides during the heterotrophy-autotrophy transition. Front. Plant Sci. 5:400:400. doi: 10.3389/fpls.2014.00400
Zhao, P., Zhou, X.-M., Zhao, L.-L., Cheung, A. Y., Sun, M.-X. (2020). Autophagy-mediated compartmental cytoplasmic deletion is essential for tobacco pollen germination and male fertility. Autophagy 1–13. doi: 10.1080/15548627.2020.1719722
Zhi, Y., Taylor, M. C., Campbell, P. M., Warden, A. C., Shrestha, P., El Tahchy, A., et al. (2017). Comparative lipidomics and proteomics of lipid droplets in the mesocarp and seed tissues of Chinese Tallow (Triadica sebifera). Front. Plant Sci. 8:1339:1339. doi: 10.3389/fpls.2017.01339
Zienkiewicz, K., Castro, A. J., Alche, J., de, D., Zienkiewicz, A., Suarez, C., et al. (2010). Identification and localization of a caleosin in olive (Olea europaea L.) pollen during in vitro germination. J. Exp. Bot. 61, 1537–1546. doi: 10.1093/jxb/erq022
Zienkiewicz, A., Jimenez-Lopez, J. C., Zienkiewicz, K., de Dios Alche, J., Rodriguez-Garcia, M., II (2011). Development of the cotyledon cells during olive (Olea europaea L.) in vitro seed germination and seedling growth. Protoplasma 248, 751–765. doi: 10.1007/s00709-010-0242-5
Zienkiewicz, K., Zienkiewicz, A., Rodriguez-Garcia, M., II, Castro, A. J. (2011). Characterization of a caleosin expressed during olive (Olea europaea L.) pollen ontogeny. BMC Plant Biol. 11:122. doi: 10.1186/1471-2229-11-122
Zienkiewicz, A., Zienkiewicz, K., Rejon, J. D., Rodriguez-Garcia, M., II, Castro, A. J. (2013). New insights into the early steps of oil body mobilization during pollen germination. J. Exp. Bot. 64, 293–302. doi: 10.1093/jxb/ers332
Zienkiewicz, A., Krzysztof, Z., Rodriguez-Garcia, M., II (2014a). “Storage lipids in developing and germinating pollen grain of flowering plants”, in Reproductive Biology of Plants. Eds. Ramawat, K., Merillon, J. M., and Shivanna, K. (Boca Raton: CRC Press), 133–146. doi: 10.1201/b16535
Zienkiewicz, A., Zienkiewicz, K., Rejon, J. D., Alche, J., de, D., Castro, A. J., et al. (2014b). Olive seed protein bodies store degrading enzymes involved in mobilization of oil bodies. J. Exp. Bot. 65, 103–115. doi: 10.1093/jxb/ert355
Zienkiewicz, K., Du, Z. Y., Ma, W., Vollheyde, K., Benning, C. (2016). Stress-induced neutral lipid biosynthesis in microalgae - Molecular, cellular and physiological insights. Biochim. Biophys. Acta. 1861, 1269–1281. doi: 10.1016/j.bbalip.2016.02.008
Zienkiewicz, K., Zienkiewicz, A., Poliner, E., Du, Z. Y., Vollheyde, K., Herrfurth, C., et al. (2017). Nannochloropsis, a rich source of diacylglycerol acyltransferases for engineering of triacylglycerol content in different hosts. Biotechnol. Biofuels 10:8. doi: 10.1186/s13068-016-0686-8
Zienkiewicz, K., Benning, U., Siegler, H., Feussner, I. (2018). The type 2 acyl CoA:diacylglycerol acyltransferase family of the oleaginous microalga Lobosphaera incisa. BMC Plant Biol. 18, 98. doi: 10.1186/s12870-018-1510-3
Zienkiewicz, A., Zienkiewicz, K., Poliner, E., Pulman, J. A., Du, Z.-Y., Stefano, G., et al. (2020). The microalga Nannochloropsis during transition from quiescence to autotrophy in response to nitrogen availability. Plant Physiol. 182, 819–839. doi: 10.1104/pp.19.00854
Keywords: lipid droplets (LDs), triacylglycerols (TAGs), lipid droplet degradation, lipolysis, lipase, autophagy, lipophagy
Citation: Zienkiewicz K and Zienkiewicz A (2020) Degradation of Lipid Droplets in Plants and Algae—Right Time, Many Paths, One Goal. Front. Plant Sci. 11:579019. doi: 10.3389/fpls.2020.579019
Received: 01 July 2020; Accepted: 24 August 2020;
Published: 09 September 2020.
Edited by:
Kazufumi Yazaki, Kyoto University, JapanReviewed by:
Takashi L. Shimada, Chiba University, JapanYonghua Li-Beisson, Commissariat à l’Energie Atomique et aux Energies Alternatives (CEA), France
Copyright © 2020 Zienkiewicz and Zienkiewicz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Agnieszka Zienkiewicz, YWdhemV0QHVtay5wbA==
 Krzysztof Zienkiewicz
Krzysztof Zienkiewicz Agnieszka Zienkiewicz
Agnieszka Zienkiewicz