- 1Department of Molecular Biology, Sejong University, Seoul, South Korea
- 2Department of Bioindustry and Bioresource Engineering, Sejong University, Seoul, South Korea
- 3Plant Engineering Research Institute, Sejong University, Seoul, South Korea
Root hairs are tubular outgrowths that originate from epidermal cells. Exposure of Arabidopsis to cadmium (Cd) and arsenic [arsenite, As(III)] increases root hair density and length. To examine the underlying mechanism, we measured the expression of genes involved in fate determination and morphogenesis of root hairs. Cd and As(III) downregulated TTG1 and GL2 (negative regulators of fate determination) and upregulated GEM (positive regulator), suggesting that root hair fate determination is stimulated by Cd and As(III). Cd and As(III) increased the transcript levels of genes involved in root hair initiation (RHD6 and AXR2) and root hair elongation (AUX1, AXR1, ETR1, and EIN2) except CTR1. DR5::GUS transgenic Arabidopsis showed a higher DR5 expression in the root tip, suggesting that Cd and As(III) increased the auxin content in the root tip. Knockdown of TTG1 in Arabidopsis resulted in increased root hair density and decreased root hair length compared with the control (Col-0) on 1/2 MS media. This phenotype may be attributed to the downregulation of GL2 and CTR1 and upregulation of RHD6. By contrast, gem mutant plants displayed a decrease in root hair density and length with reduced expression of RHD6, AXR2, AUX1, AXR1, ETR1, CTR1, and EIN2. Taken together, our results indicate that fate determination, initiation, and elongation of root hairs are stimulated in response to Cd and As(III) through the modulation of the expression of genes involved in these processes in Arabidopsis.
Introduction
The metalloid arsenic (As) and the heavy metal cadmium (Cd) are highly toxic elements present in the environment, and they pose a considerable risk to living organisms owing to their accumulation in the soil and water (Godt et al., 2006; Zhao et al., 2009). Typical toxic effects of Cd on plants are the inhibition of primary root growth and increased lateral root density (Hu et al., 2013). Although the influence of Cd on root hairs has not been researched in detail, studies have shown that Cd at specific concentrations (that inhibit root growth by about 50%) increases root hair density and length in various plant species including radish (Vitoria et al., 2003), barley (Durcekova et al., 2007), sorghum (Kuriakose and Prasad, 2008), and Rhode grass (Kopittke et al., 2010). However, the molecular mechanism of root hair enhancement in response to Cd and As has not been reported to date.
Root hairs are tubular outgrowths from specific epidermal cells that function in nutrient and water absorption (Peterson and Farquhar, 1996). In Arabidopsis (Arabidopsis thaliana), root anatomy is well determined, and the different steps of root hair development including fate determination, initiation, bulge formation, and tip growth have been defined (Schiefelbein and Somerville, 1990; Dolan et al., 1993; Galway et al., 1994). By definition, epidermal cells located over the anticlinal junction between two underlying cortical cells differentiate into trichoblast cells that develop root hairs, while epidermal cells that lie above the outer periclinal cortical cells become non-hair cell atrichoblasts (Masucci et al., 1996; Dolan, 2006).
In Arabidopsis, several genes and transcription factors are involved in epidermal cell fate determination. Previous studies showed that root epidermal cells differentiate into non-hair cells through the action of the transcriptional complex WER-GL3/EGL3-TTG1, which consists of the R2-R3 MYB transcription factor WER (WEREWOLF), the basic Helix-Loop-Helix (bHLH) transcription factors GL3 (GLABRA3) and EGL3 (ENHANCER OF GLABRA3), and TTG1 encoding a protein of 341 amino acid residues containing four WD-40 repeats. By contrast, a root hair cell is produced if the transcriptional complex CPC/TRY/ETC1 (CAPRICE/TRIPTYCHON/ENHANCER OF TRY AND CPC1, one-repeat Myb protein)-GL3/EGL3-TTG1 is present in the epidermal cell (Wada et al., 1997; Lee and Schiefelbein, 2002; Schellmann et al., 2002; Kirik et al., 2004; Simon et al., 2007; Benitez and Alvarez-Buylla, 2010). The generation of each complex depends on the relative abundance of two competing transcription factors, WER/MYB23 and CPC/TRY/ETC1 (Wada et al., 1997; Lee and Schiefelbein, 1999). In non-hair cells, high expression levels of WER make the complex to be mainly an active form, which promotes the expression of GL2 (GLABRA2, homeodomain transcription factor) and the repression of RHD6 (ROOT HAIR DEFECTIVE 6, bHLH transcription factor) transcription, suppressing the expression of atrichoblast-specific genes and preventing root hair formation (Masucci and Schiefelbein, 1994, 1996; Rerie et al., 1994; Lee and Schiefelbein, 1999; Menand et al., 2007; Bruex et al., 2012). On the other hand, in root hair cells, the perception of a JACKDAW (JKD, zinc finger transcription factor)-induced positional signal in the cortex by SCM (SCRAMBLED, leucine-rich repeat receptor-like kinase) inhibits WER expression (Kwak et al., 2005; Hassan et al., 2010). Eventually, reduced WER expression as well as competitive inhibition of WER binding to GL3/EGL3-TTG1 by CPC/TRY/ETC1 (Lee and Schiefelbein, 2002) lead to the formation of an inactive complex, CPC-GL3/EGL3-TTG1, and the repression of GL2 and root hair cell differentiation. The WER-GL3/EGL3-TTG1 complex positively regulates the transcription of CPC/TRY/ETC1 and negatively regulates that of GL3/EGL3 in atrichoblast cells. Therefore, it was proposed that CPC/TRY/ETC1 translocates from non-hair cells to hair cells and GL3/EGL3 in the opposite direction via plasmodesmata (Kurata et al., 2005; Kang et al., 2013).
Genetic analysis of root hair mutants demonstrated that the phytohormones ethylene and auxin regulate root hair formation independently or downstream of the WER-GL3/EGL3-TTG1 or CPC-GL3/EGL3-TTG1 pathway (Schiefelbein, 2000; Muller and Schmidt, 2004). For instance, ROOT HAIR DEFECTIVE6 (RHD6), a downstream gene of GL2, encodes a bHLH transcription factor (Qing and Aoyama, 2012) and its mutation results in a decrease of root hair number as well as a shift in the position of root hair emergence (Masucci and Schiefelbein, 1994; Menand et al., 2007). However, this defect can be reversed by the application of auxin (IAA) or ACC (ethylene precursor), implying that RHD6 plays a role in root hair formation (probably initiation) via auxin and/or ethylene related pathways. Mutations in the AXR2 (AUXIN RESISTANT2) gene, which encodes the Aux/IAA protein, result in a decline in root hair number (Pitts et al., 1998; Nagpal et al., 2000; Rigas et al., 2013). AXR2 represents a converging point of the auxin and ethylene pathways, since treatment with IAA or ACC does not recover root hair numbers in axr2 mutants (Masucci and Schiefelbein, 1996). Mutants of the AUX1 gene, which encodes an auxin influx carrier, display shorter root hairs (Pitts et al., 1998; Schmidt and Schikora, 2001; Yu et al., 2015). Mutants of AXR1 (AUXIN RESISTANT1), which encodes a ubiquitin-activating enzyme E1, have shorter root hairs than the WT (Cernac et al., 1997; Pitts et al., 1998). Regarding ethylene, root hair formation is suppressed by the ethylene biosynthesis inhibitor AVG (Tanimoto et al., 1995; Masucci and Schiefelbein, 1996) and enhanced by ACC (ethylene precursor) treatment (Tanimoto et al., 1995). Nevertheless, the role of ethylene in root hair formation remains unclear, since the ethylene signaling mutants etr1 (ETHYLENE RESPONSE1) and ein2 (ETHYLENE INSENSITIVE2) show normal root hair density (Masucci and Schiefelbein, 1996). However, ein2 mutants have shorter root hairs (Rahman et al., 2002). In addition, the ethylene response mutant ctr1 (CONSTITUTIVE TRIPLE RESPONSE1), which is constitutively active in ethylene signal transduction, possesses a greater root hair density (Masucci and Schiefelbein, 1996), and the ethylene overproducing mutant eto1 exhibits longer hairs (Strader et al., 2010) than the WT. Furthermore, ethylene promotes auxin biosynthesis and transport, implying that ethylene may be involved in root hair formation through the auxin pathway (Ruzicka et al., 2007; Swarup et al., 2007).
In the present work, we examined the enhancement of root hair density and length by Cd and As(III) based on the hypothesis that root hair fate determination and morphogenesis are affected by cadmium and arsenic. Our results suggested that cadmium and arsenic increase root hair density and length by modulating the expression of genes involved in fate determination and the initiation and elongation of root hairs. Based on our results, we proposed a model in which cadmium and arsenic enhance root hair density and length by promoting the convergence of developmental (TTG1/GL2/GEM) and hormonal (auxin and ethylene) signaling pathways.
Materials and Methods
Plant Material and Heavy Metal Treatment
Seeds of Arabidopsis (A. thaliana L. Heynh) ‘Columbia’ ecotype, ttg1 (SALK_054584), and gem (SALK_145846C) were obtained from the Arabidopsis Biological Resource Center (Ohio State University) and used in the study. Seeds were surface-sterilized by immersion in 95% ethanol for 3 min and a 20% bleach solution containing 0.1% Tween 20 for 5 min, followed by at least three rinses in ddH2O (sterile Milli-Q water). Then, seeds were germinated and grown on agar plates containing 1/2 MS nutrient medium (Duchefa) and 1% sucrose, with pH adjusted to 5.7 by KOH, and 0.5% agar in a growth chamber at 23°C with 60% relative humidity in a long day photoperiod (16 h light/8 h dark) in a vertical position. In each experiment, 10 seeds were cultured on a 90-mm Petri dish containing 20 mL of medium. Aliquots of 100 mM CdSO4 and AsNaO2 stock solution for cadmium and arsenic media were added to the 1/2 MS media after autoclaving to result in final concentrations of 40 μM Cd and 10 μM As(III).
Morphometric Analysis
After 1 week of growth, Arabidopsis seedlings were viewed under a Leica stereoscopic microscope (MZ10F, Germany). Photographs were taken using a DFC450 C (Leica, Germany) camera under appropriate magnification (5–40×). Root hair density was determined as the number of hairs in a 5 mm section from the starting point of the root differentiation zone under a microscope (40×). Root hair length was measured in a 5 mm section from the starting point of the root differentiation zone and quantified using Image J software (National Institutes of Health1). Results were expressed as the mean ± standard error (SE). All measurements indicate the average of three independent experiments, each involving 10 seedlings.
Quantitative RT-PCR Analysis
For gene expression analysis, total RNA was isolated from Arabidopsis plants grown on 1/2 MS agar plates supplemented with or without Cd and As(III) for 1 week using the TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. The first strand cDNA was synthesized from 2 μg of each RNA sample using the cDNA Synthesis Kit (Wizbio, Korea). qRT-PCR was performed using a CFX ConnectTM Real-Time PCR Detection System (Bio-Rad Laboratories Inc., Hercules, CA, USA). Each 20 μL reaction mixture consisted of 10 μL of SYBR Supermix (SsoAdvancedTM Universal SYBR® Green Supermix), 1 μL of cDNA, 7 μL of nuclease-free water, and 1 μL of each primer. The qRT-PCR reaction thermal conditions were as follows: 95°C for 30 s, 40 cycles at 95°C for 15 s, and 60°C for 20 s. All samples were run in triplicate (three biological repeats) for cDNA and each primer set. Relative transcript levels were assessed by normalization to actin transcript abundance as an endogenous control. The gene specific primers are listed in Supplementary Table S1.
Semi-Quantitative RT-PCR
Total RNA was isolated from Arabidopsis plants grown on 1/2 MS agar plates with and without Cd and As(III) for 1 week using the TRIzol reagent (Invitrogen), and RT-PCR was performed using 250 ng of total RNA with an RT-PCR kit (Takara) as follows: 30 cycles of 30 s at 95°C, 30 s at different annealing temperatures, 120 s at 72°C. Annealing temperatures were as follows: 60°C for AtActin; 58°C for AtTTG1, AtGEM, and AtAXR2; 51°C for AtRHD6, AtAUX1, and AtAXR1; 52°C for AtGL2; and 50°C for AtEIN2, AtCTR1, and AtETR1. The RT-PCR analysis was repeated at least five times using separately harvested samples. For semi-quantitative RT-PCR analysis, RT-PCR products were analyzed using ImageJ software to quantify the expression level of each gene. The gene-specific primers used for semi-quantitative RT-PCR are listed in Supplementaty Table S2.
β-Glucuronidase Assay
GUS histochemical staining of Arabidopsis plants harboring AtGL2::GUS and AtDR5::GUS fusion constructs was performed as previously described (Prasinos et al., 2005) with minor modifications. Briefly, 5-day-old seedlings grown vertically on 1/2 MS agar medium supplemented with or without Cd (40 μM) and As(III) (10 μM) were immersed in GUS staining solution (50 mM NaPO4 buffer, pH 6.8, 0.5 M EDTA pH 8.0, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 20% triton X-100, and 2 mM X-gluc), and incubated overnight at 37°C. Photographs were taken with a microscope (Leica MZ10F, Germany) equipped with a digital camera (DFC450 C). GUS stained tissues represent the results of at least five independent seedlings.
Statistical Analysis
One-way ANOVA was used to analyze the data using SAS software (version 9.1), and means were compared using Tukey’s multiple comparison test, with significant differences at P ≤ 0.05 or P ≤ 0.01.
Results
Root Hair Density and Length Were Increased in Response to Cd and As(III) in Arabidopsis
Arabidopsis plants grown on 1/2 MS agar medium containing Cd (CdSO4, 40 μM) and AsIII (AsNaO2, 10 μM) for 1 week showed a 50% decrease in primary root length (Figures 1A,B). By contrast, root hair length and density increased significantly in Arabidopsis plants exposed to Cd and As(III) compared with those in control plants (Figure 2A). Exposure to Cd at 20 and 40 μM increased the number of root hairs in a 5 mm section from the starting point of the root differentiation zone by 22 and 50%, respectively, compared with control plants (Figure 2B). Root hair number and length could not be measured at 60 μM Cd because germination was completely inhibited (data not shown). As(III) treatment increased the number of root hairs by 23% at 5 μM, 39% at 10 μM, and 19% at 30 μM (Figure 2C). Cd-treated plants showed an increase in root hair length of 38% at 20 μM and 77% at 40 μM compared with that of the control (1/2 MS) (Figure 2B). As(III) treatment also increased root hair length by 32% at 5 μM, 63% at 10 μM, and 62% at 30 μM (Figure 2C).
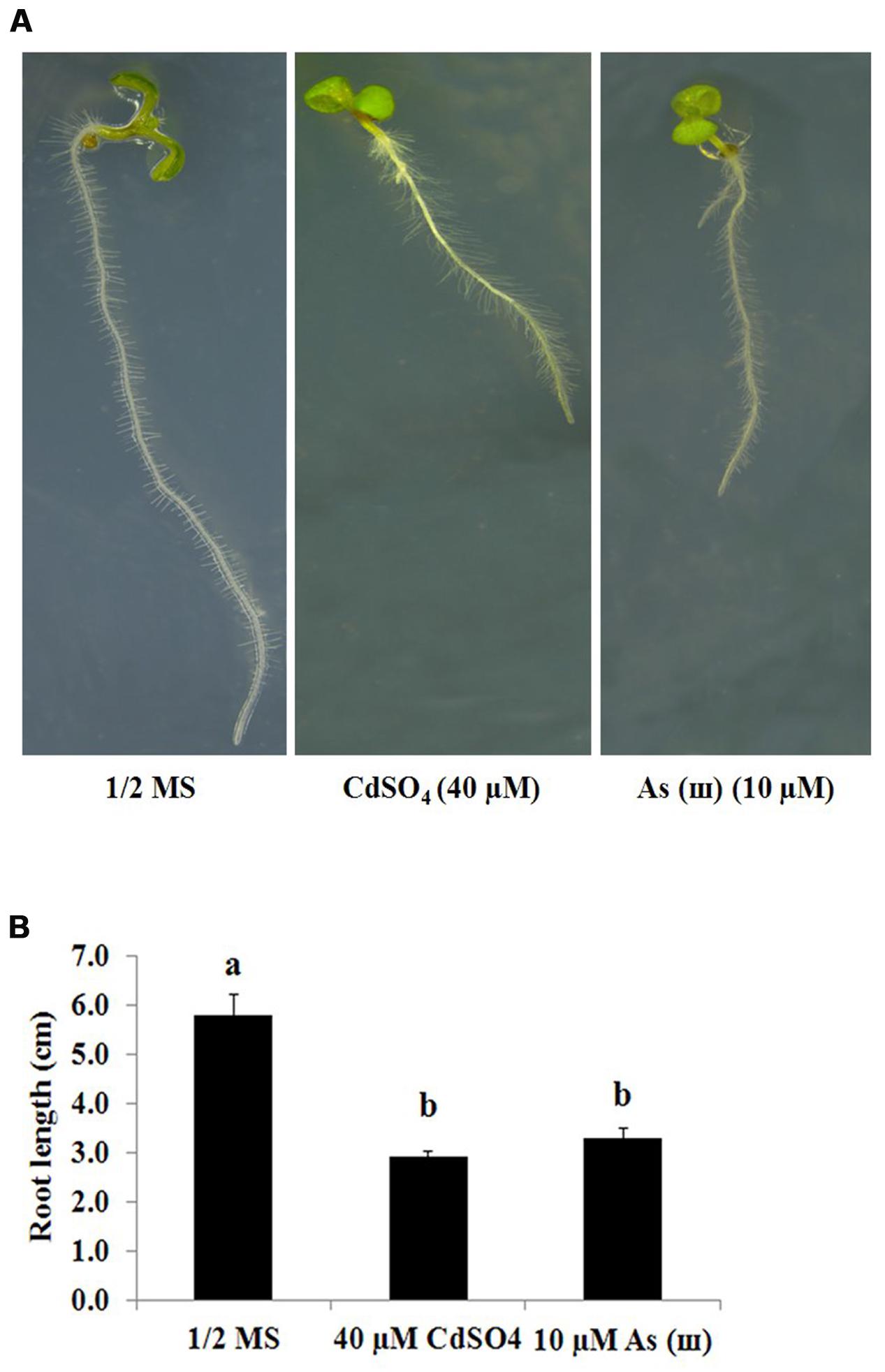
FIGURE 1. Inhibitory effect of Cd (CdSO4) and As(III) (AsNaO2) stress on primary root growth in Arabidopsis. (A) Morphology of 1-week-old seedlings germinated and grown on 1/2 MS agar plates containing no metal (left), 40 μM CdSO4 (center), and 10 μM AsNaO2 (right). (B) Quantification of the primary root length of Arabidopsis shown in (A). Values are average (±SE) of three independent experiments, each involving 10 seedlings. Different letters in each column indicate a significant difference according to Tukey’s multiple comparison test (p ≤ 0.01).
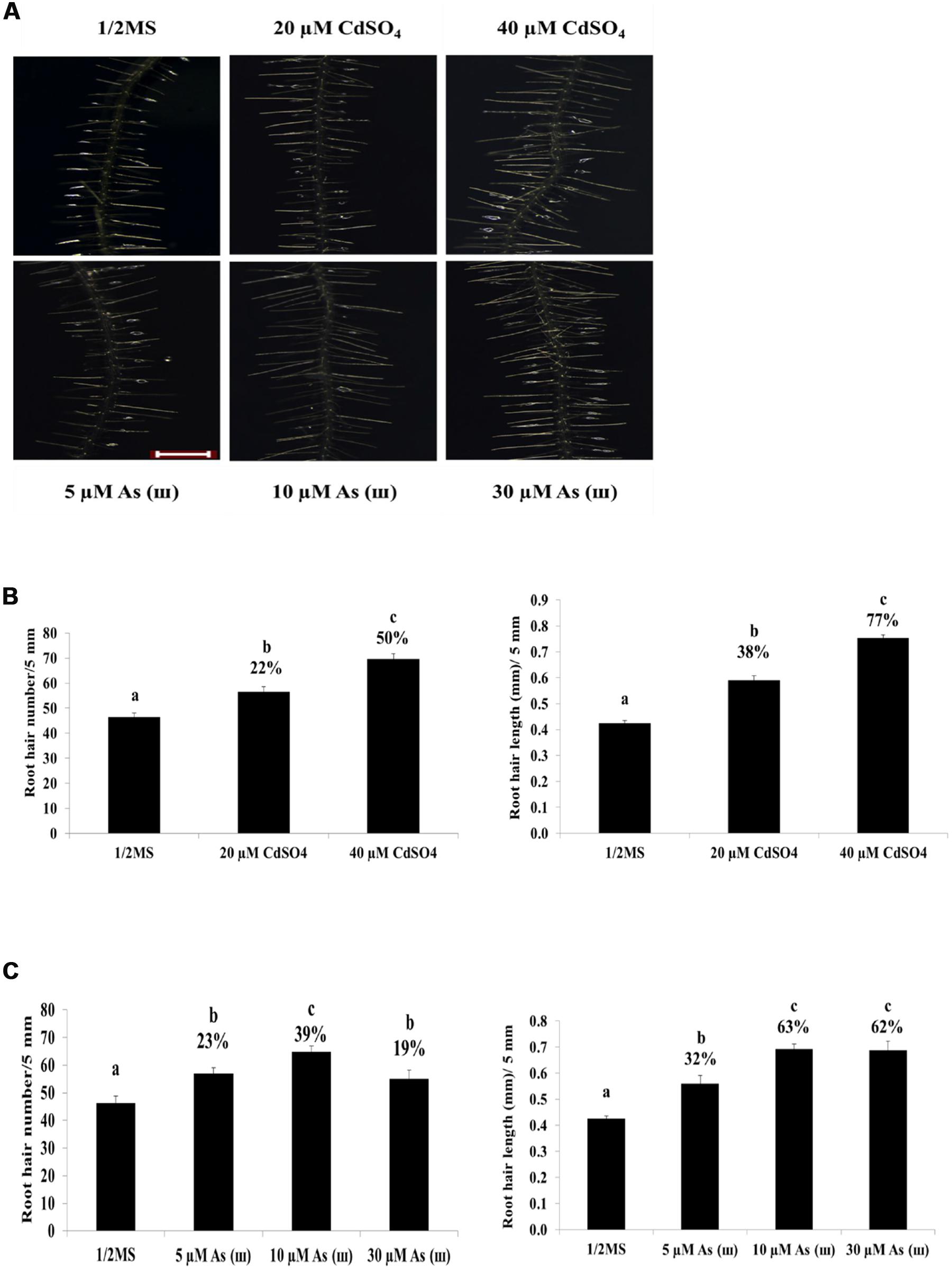
FIGURE 2. Effect of Cd and As(III) on the length and density of root hairs in Arabidopsis. (A) Root hair morphology of Arabidopsis Col-0 seedlings germinated and grown for a week on 1/2 MS agar media treated with no metal, 20, 40 μM CdSO4, and 5, 10, 30 μM As(III). Roots were observed at 40× magnification; scale bars = 0.2 mm. Photographs show plants representative of the 10 plants examined in each experiment. (B) Density and length of root hair treated with Cd shown in (A). Root hair density was expressed as the number counted in a 5 mm section from the starting point of differentiation zone under a microscope. Root hair length was measured in a 5 mm section of differentiation zone. Root hairs were photographed with Leica Stereoscopic microscope (MZ10F, Germany), and their length was quantified using Image J software as described in Materials and methods. Values are average (±SE) of three independent experiments, each involving 10 seedlings. (C) Density and length of root hairs treated with As(III) shown in (A). Root hair density and length were measured as in (B). Different letters in each column indicate a significant difference according to Tukey’s multiple comparison test (p ≤ 0.01).
To determine whether the metal-induced increase in root hair density was related to the reduction of root cell length, we measured root epidermal cell length in control and metal-treated plants. The root cell length of control plants was 151.7 μm, whereas that of Cd and As(III)-treated plants was 140.38 and 144.2 μm, respectively (Supplementary Figure S4). These results indicated that the increase in root hair number was not associated with changes in root cell length.
Cd and As(III) Modulated the Expression of Genes Involved in Root Hair Fate Determination (Differentiation)
Our findings that exposure to metals increased root hair density and length suggested that Cd and As(III) promoted the differentiation and elongation of root hairs. Developmental pathways regulated by the TTG1/GL2 interaction determine root epidermal cell fate (Masucci and Schiefelbein, 1996). We therefore measured the expression levels of TTG1 and GL2 using semi-quantitative RT-PCR (Figure 3A, graphs in Supplementary Figure S1), qRT-PCR (Figure 3B), and GUS staining (Figures 4A,B). Cd and As(III) markedly downregulated TTG1 expression compared with that in the control, providing an explanation for the increment in root hair density induced by Cd and As(III), since TTG1 is a negative regulator of root hair differentiation (Figures 3A,B). The expression of GL2, another negative regulator of root hair differentiation, was also repressed by Cd and As(III). In addition, GUS activity in Arabidopsis plants expressing the GL2::GUS construct which were germinated and grown for 5 days on medium containing Cd and As(III) decreased compared with that in controls, suggesting that GL2 expression is downregulated by Cd and As(III) (Figures 4A,B). This finding is contrary to previous studies showing that GL2-GUS is preferentially expressed in developing atrichoblast cells within the meristematic and elongation zones of the Arabidopsis root (Masucci et al., 1996). Taken together, these results indicated that the Cd or As(III) induced increase in root hair density was probably caused by the inhibition of GL2 and TTG1 gene expression. Because GEM plays a positive role in root hair fate determination (Caro et al., 2007), we examined the expression of GEM in Cd and As(III) treated plants. We found that the two metals significantly increased GEM mRNA levels compared with those in control untreated plants (Figures 3A,B). Taken together, our results suggested that Cd and As(III) increased root hair density by promoting root hair differentiation through the repression of TTG1 and GL2 (negative regulators of root hair differentiation) and the upregulation of GEM expression (positive regulator).
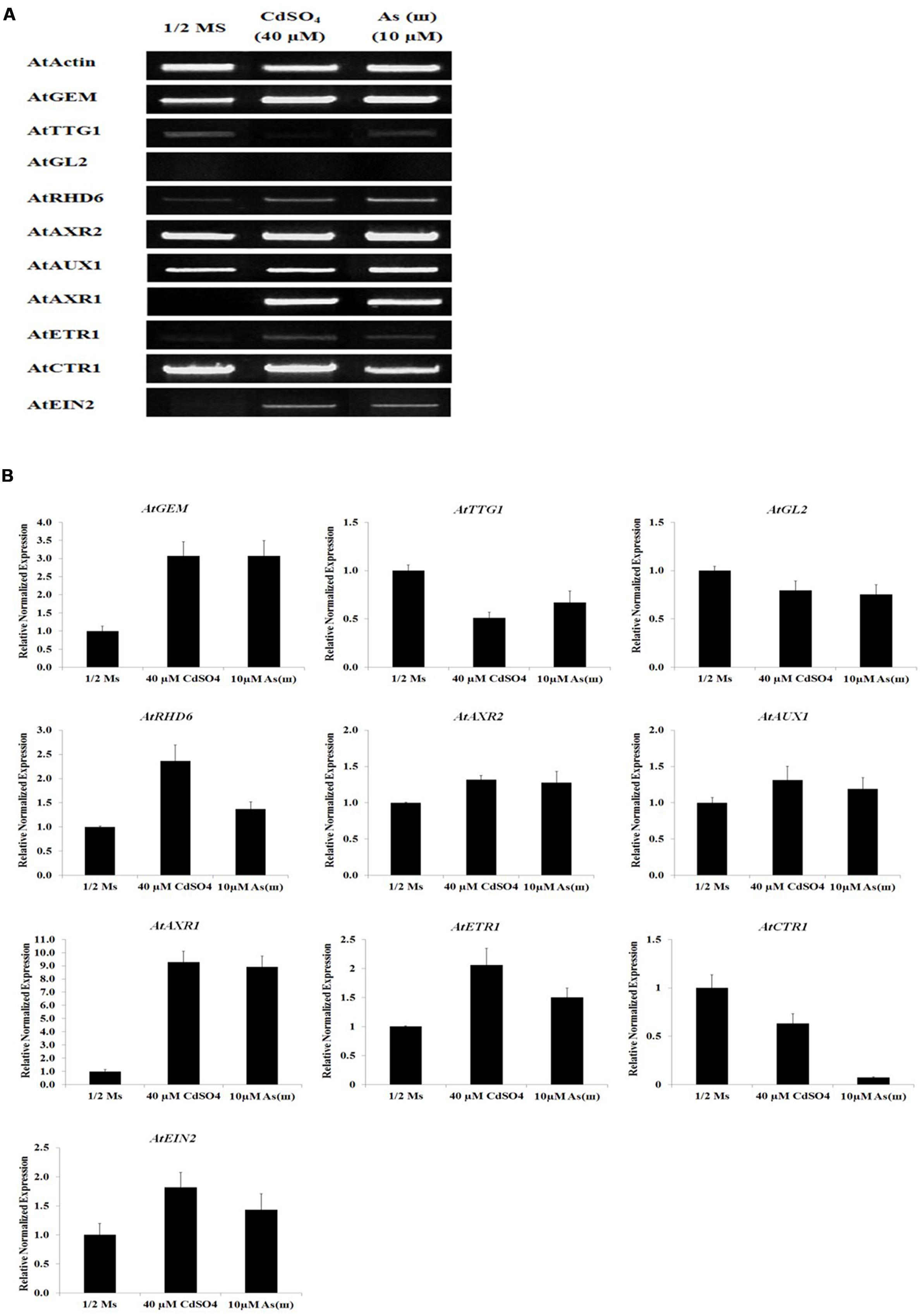
FIGURE 3. Effect of Cd and As(III) on expressions of genes involved in the differentiation (fate determination) and elongation of root hairs in Arabidopsis. (A) Semi-quantitative RT-PCR results showing relative transcript levels of various genes involved in the differentiation (GEM, TTG1, and GL2), initiation (RHD6 and ACXR2) and elongation (AUX1, AXR1, ETR1, CTR1, and EIN2) of root hairs in response to Cd and As(III) in Arabidopsis. RT-PCR was performed using total RNA isolated from 1-week-old plants which were germinated and grown on 1/2 MS agar media treated with no metal, 40 μM CdSO4 and 10 μM As(III).The gel photography represents 3 experimental replicates. (B) Quantitative real-time PCR results of various genes involved in root hair differentiation and elongation shown in (A). Transcript levels were normalized to the transcript level of Actin.
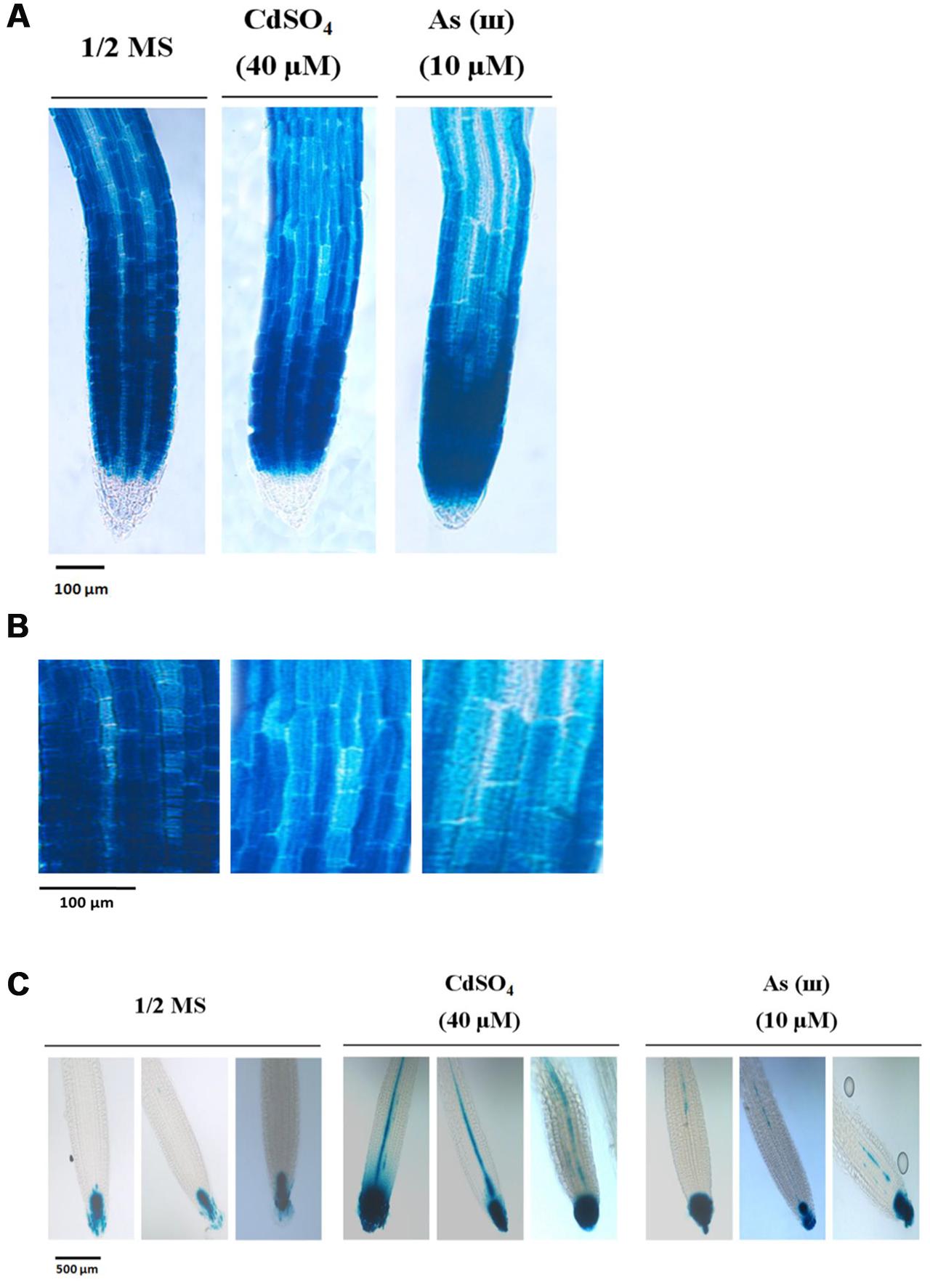
FIGURE 4. Down-regulation of GL2 and up-regulation of DR5 by cadmium and arsenite in Arabidopsis root. (A) GUS activity of primary root tips (2 mm) of 5-day-old transgenic Arabidopsis Col-0 seedlings carrying the GL2-GUS construct. Plants were germinated and grown in 1/2 MS media supplemented with or without cadmium and arsenite, and were stained to visualize GUS activity (20× magnification). (B) GUS activity at higher magnification (40×). Left panel, 1/2 MS media; central panel, 1/2 MS media with 40 μM CdSO4; right panel, 1/2 MS media with 10 μM As(III). Scale bar indicates a 200 μm in size. Each photograph represents five plants. (C) GUS activity of primary root tips (2.3 mm) of 5-day-old transgenic Arabidopsis Col-0 seedlings carrying the DR5-GUS construct. Plants were germinated and grown in 1/2 MS media supplemented with or without cadmium and arsenite, and were stained to visualize GUS activity (20× magnification). Left panel, 1/2MS media; central panel, 1/2 MS media with 40 μM CdSO4; right panel, 1/2 MS media with 10 μM As(III). Scale bar indicates a 500 μm in size. Each photograph represents five plants.
Cd and As(III) Modulated the Expression of Genes Involved in Root Hair Initiation and Elongation (Auxin/Ethylene Signaling Pathway)
Several plant hormones are involved in root hair development (Lee and Cho, 2013). Among these, auxin and ethylene have been studied extensively (Masucci and Schiefelbein, 1996; Pitts et al., 1998; Cho and Cosgrove, 2002). Given the important role of auxin and ethylene in root hair initiation and elongation, we examined whether the expression of genes related to these hormones was affected by Cd and As(III).
The RHD6 gene, a downstream gene of GL2, plays a key role in root hair initiation, as shown by the decreased root hair density and shift in the root hair initiation site observed in rhd6 mutants (Masucci and Schiefelbein, 1994; Menand et al., 2007). Assessment of RHD6 transcript levels in Col-0 Arabidopsis showed that Cd and As(III) upregulated RHD6 expression (Figures 3A,B), suggesting that root hair initiation is promoted by Cd and As(III) through the enhancement of RHD6 expression.
Next, we analyzed the expression of auxin-related genes in plants grown in media containing Cd and As(III). As shown in Figures 3A,B, AXR2, which functions in root hair initiation, and AUX1 and AXR1, which are involved in root hair elongation, were upregulated in response to Cd and As(III). In addition, DR5::GUS transgenic Arabidopsis showed that auxin-inducible DR5 expression was enhanced by Cd and As(III), indicating that the auxin level at root tip is increased by Cd and As(III) (Figure 4C). The expression of DR5-GUS around the vascular cylinder indicates that auxin is translocating to the root tip. Taken together, these results suggested that the auxin signaling pathway is activated in response to Cd and As(III), leading to root hair elongation.
The result that Cd and As(III) upregulated the expression of the auxin-resistant gene AXR1 indicated that crosstalk between auxin and ethylene may be involved in metal-induced root hair elongation, since AXR1 is upregulated by ethylene (Swarup et al., 2007). This raised the question of whether the change in transcript levels also occurs in ethylene signaling genes. To address this question, the mRNA levels of ETR1, CTR1, and EIN2 were analyzed in Cd- and As(III)-treated plants. ETR1 and EIN2 were upregulated, whereas CTR1, a negative regulator of ethylene signaling, was downregulated by Cd and As(III). This indicated that the increase in root hair elongation in response to Cd and As(III) was mediated by the modulation of the expression of ethylene signaling genes (Figures 3A,B).
Root Hair Density and RHD6 Expression Were Enhanced, and Root Hair Length was Reduced in ttg1 Mutants
The effects of Cd and As(III) on root hair development were also examined in the ttg1 (negative regulator of root hair differentiation) mutant (Figure 5). ttg1 Arabidopsis (SALK_ 054584, Figure 5A) showed extremely low expression of AtTTG1 (Figure 5B) and a decrease in root length in response to Cd and As(III) (Figures 5C,D). As shown in Figures 5E,F, root hair density was higher in ttg1 Arabidopsis plants than in control plants in 1/2 MS media, which is consistent with the role of TTG1 as a negative regulator of root hair fate determination. However, root hair density was not enhanced by Cd and As(III), suggesting that the root hairs of the ttg1 mutant were already fully differentiated to the level of metal-treated Col-0. By contrast, root hair length was slightly shorter in the ttg1 mutant than in Col-0 in 1/2 MS medium, and it was increased by Cd and As(III) to the level of metal-treated Col-0. This implied that root hair differentiation was promoted, whereas root hair elongation was not enhanced in ttg1, confirming that TTG1 is a negative regulator of root hair differentiation but not of root hair elongation.
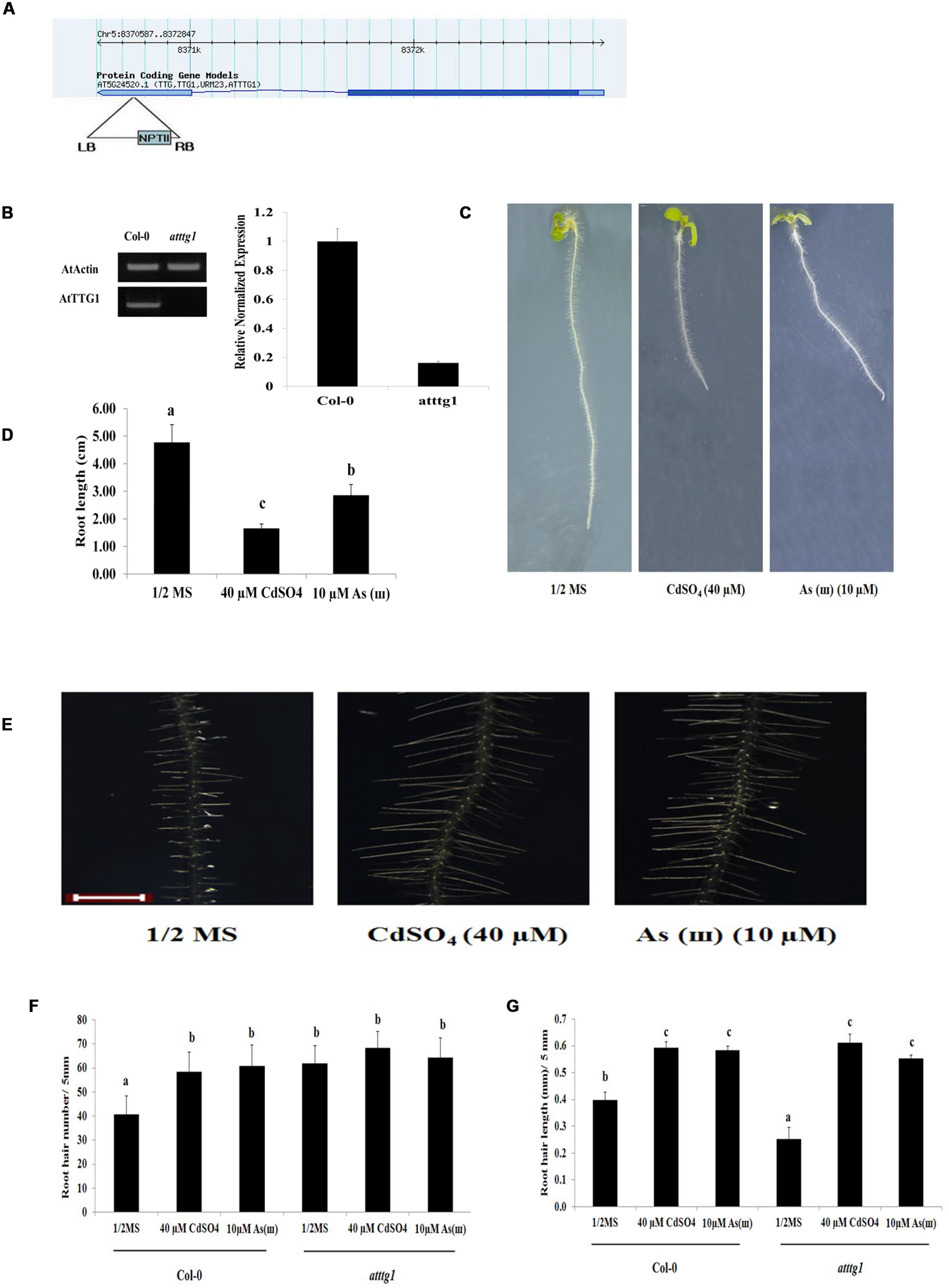
FIGURE 5. Root hair length and density in response to Cd and As(III) in ttg1 Arabidopsis. (A) Diagram showing T-DNA insertion position in ttg1 Arabidopsis. (B) Left, semi-quantitative RT-PCR products (transcript levels) of AtTTG1 in control (Col-0) and ttg1 Arabidopsis. PCR product of actin (AtActin) was used as a loading control; Right, real-time PCR results. (C) Morphology of ttg1 Arabidopsis (1-week-old) seedlings germinated and grown on 1/2 MS agar plates containing no metal (left), 40 μM CdSO4 (center), and 10 μM AsNaO2 (right). (D) Quantification of the primary root length of ttg1 Arabidopsis shown in (C). (E) Root hair morphology of ttg1 Arabidopsis germinated and grown on 1/2 MS with no metal (left), 40 μM CdSO4 (center) and 10 μM As(ø) (right). Roots were observed at 40× magnification; scale bars = 0.2 mm. (F) Graphs representing root hair density in a 5 mm section from the starting point of differentiation zone in ttg1 Arabidopsis shown in (E). (G) Graphs representing root hair length in a 5 mm section from the starting point of differentiation zone in ttg1 Arabidopsis shown in (E). Values are average (±SE) of three independent experiments, each involving 10 seedlings. Different letters over each column indicate a significant difference according to Tukey’s multiple comparison test (p ≤ 0.01).
To understand the molecular mechanism underlying the enhanced root hair density and slightly reduced root hair length in ttg1 plants, the expression of genes involved in fate determination, initiation, and elongation of root hairs was examined (Figure 6 and Supplementary Figure S2). The expression of GL2, which was reported as a negative regulator of root hair fate determination, was repressed and not induced by Cd and As(III). This result supports previous reports that GL2 is induced by TTG1, resulting in the suppression of root hair differentiation (Hung et al., 1998). In addition, the expression of RHD6, which is inhibited by GL2 and involved in root hair initiation (Masucci and Schiefelbein, 1996), was upregulated in ttg1 plants and this effect was enhanced by Cd and As(III). However, AXR2 expression was not increased in ttg1 plants, despite the fact that the axr2 mutant has a lower number of root hairs (Masucci and Schiefelbein, 1996), implying that AXR2 is involved in root hair initiation similar to RHD6. The expression of auxin/ethylene signaling genes (AUX1, AXR1, ETR1, CTR1, and EIN2) involved in root hair elongation in ttg1 plants was similar to that in Col-0 plants, confirming that auxin/ethylene signaling genes are not controlled by TTG1/GL2. However, it was not clear why root hair length was shorter in ttg1 than in Col-0, since the expression of root hair elongation genes was not decreased. Regarding root hair initiation and elongation in response to Cd and As(III), similar to Col-0, the expression of genes involved in root hair initiation (RHD6 and AXR2) and elongation (AUX1, AXR1, ETR1, and EIN2) was increased, while CTR1 expression was reduced.
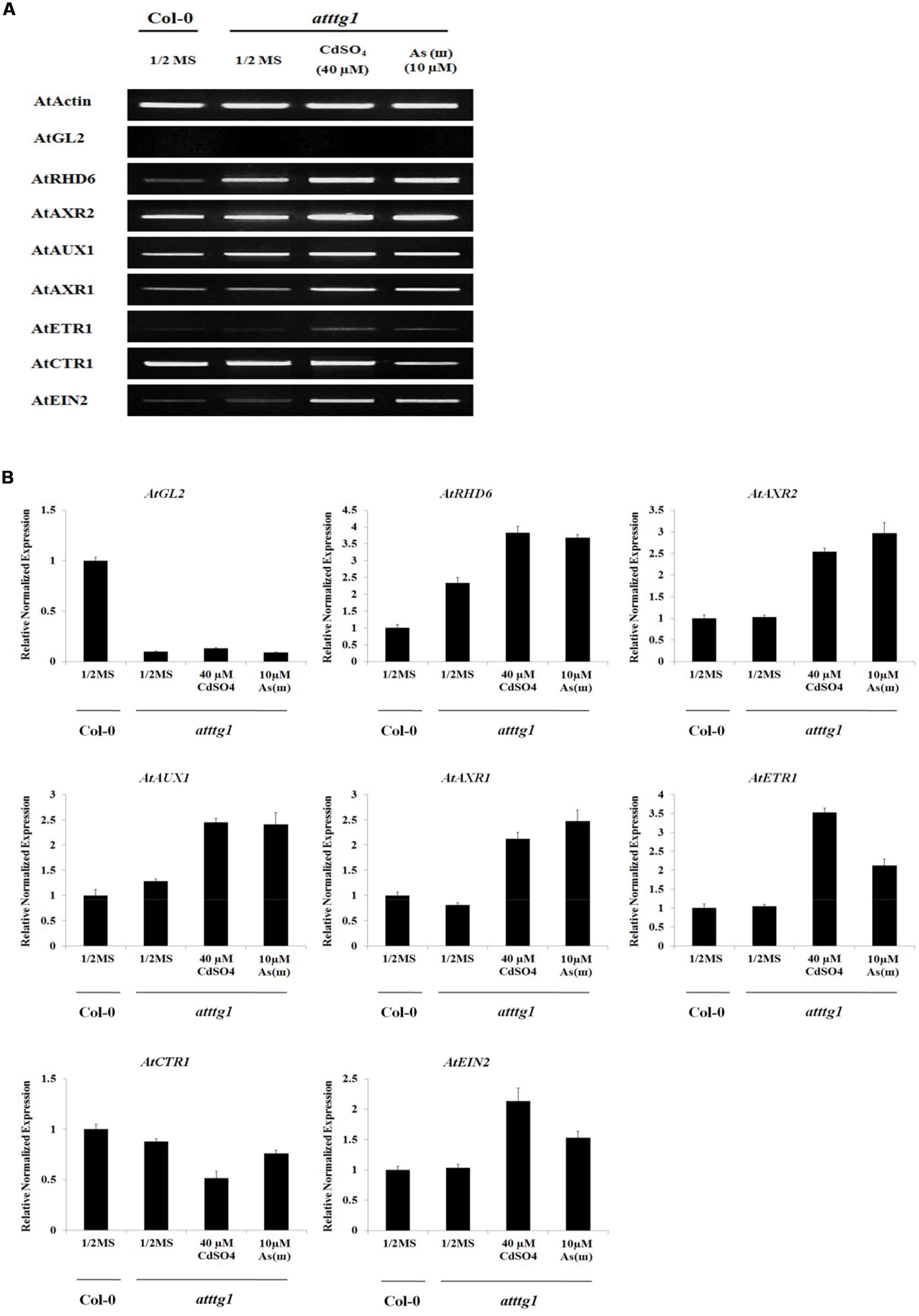
FIGURE 6. Effect of Cd and As(III) on expressions of genes participated in the differentiation (fate determination) and elongation of root hairs in ttg1 Arabidopsis. (A) Semi-quantitative RT-PCR results showing relative transcript levels of various genes involved in the differentiation (GL2), initiation (RHD6 and AXR2) and elongation (AUX1, AXR1, ETR1, CTR1, and EIN2) of root hairs in Col-0 and ttg1 Arabidopsis. RT-PCR was performed using total RNA isolated from 1-week old plants which were germinated and grown on 1/2 MS agar media treated with no metal, 40 μM CdSO4 and 10 μM As(III). The gel photography represents three experimental replicates. (B) Quantitative real-time PCR results of various genes involved in root hair differentiation and elongation shown in (A). Transcript levels were normalized to the transcript level of Actin.
Root Hair Density and Length and the Expression of Genes Involved in Root Hair Initiation and Elongation Were Decreased in gem Mutants
Root hair density and length were also examined in gem (positive regulator of root hair fate determination) Arabidopsis mutants. As shown in Figure 7, both root hair density and length were decreased in gem plants (SALK_145846C) compared with those in control Col-0, whereas both were enhanced to the level of the control in response to Cd and As(III). This suggested that Cd and As(III) promoted root hair development in gem mutants by stimulating root hair differentiation (fate determination), initiation, and elongation.
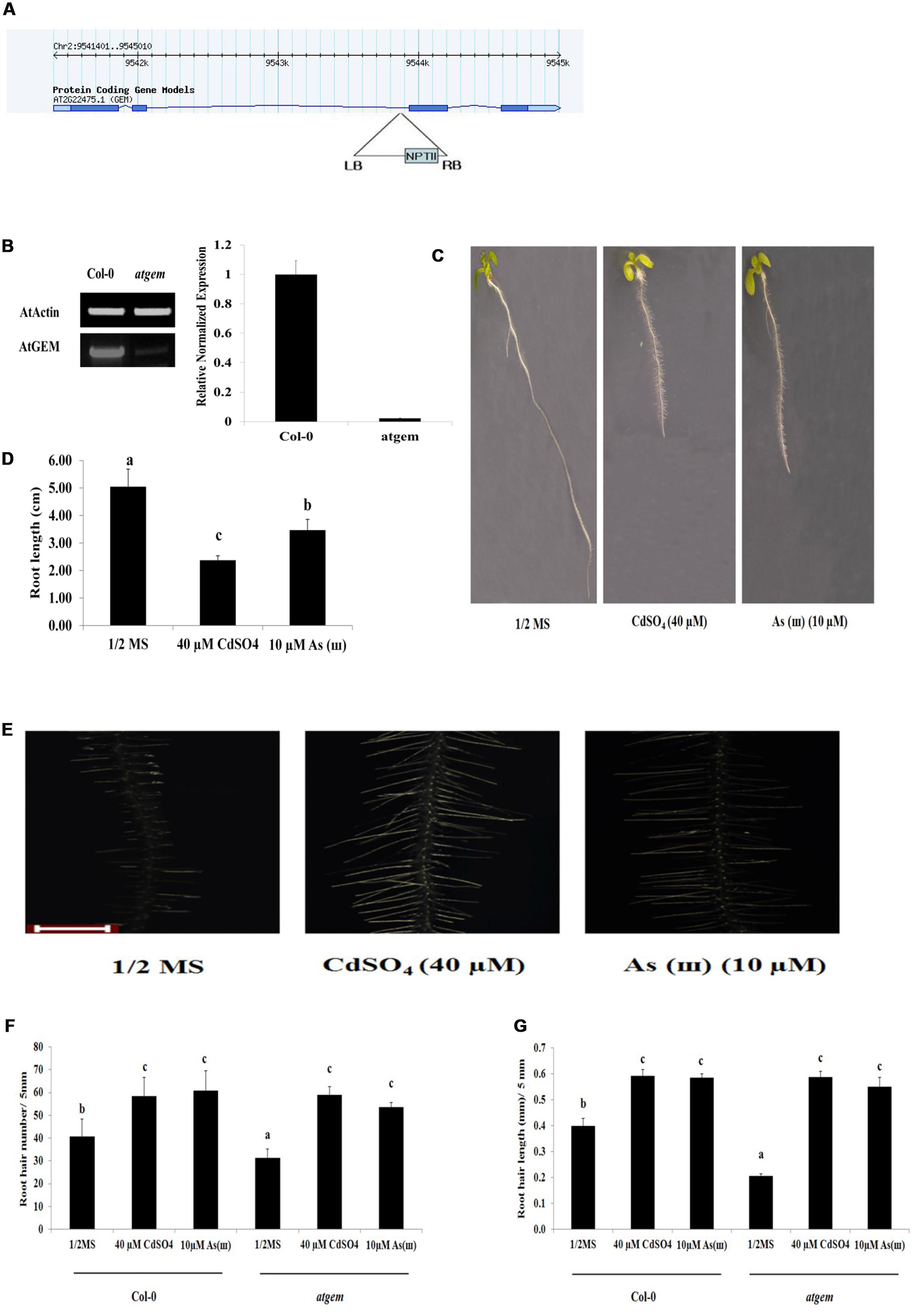
FIGURE 7. Root hair length and density in response to Cd and As(III) in gem Arabidopsis. (A) Diagram showing T-DNA insertion position in gem Arabidopsis. (B) Right, transcript levels (semi-quantitative RT-PCR) of AtGEM in control (Col-0) and gem Arabidopsis. A PCR product of actin (AtActin) was used as a loading control; Right, real-time PCR result. (C) Morphology of gem Arabidopsis (1-week-old) seedlings germinated and grown on 1/2 MS agar plates containing no metal (left), 40 μM CdSO4 (center), and 10 μM AsNaO2 (right). (D) Quantification of root length of gem Arabidopsis shown in (C). (E) Root hair morphology of gem Arabidopsis germinated and grown on 1/2 MS with no metal (left), 40 μM CdSO4 (center), and 10 μM As(III) (right). Roots were observed at 40× magnification; scale bars = 0.2 mm. (F) Graphs representing root hair density in a 5 mm section from the starting point of differentiation zone in gem Arabidopsis root shown in (E). (G) Graphs representing root hair length in a 5 mm section from the starting point of differentiation zone in gem Arabidopsis root shown in (E). Values are average (±SE) of three-independent experiments, each involving 10 seedlings. Different letters in each column indicate a significant difference according to Tukey’s multiple comparison test (p ≤ 0.01).
To explain the Cd/As(III)-induced root hair development in gem plants, we examined the expression of genes involved in differentiation, initiation, and elongation of root hairs (Figure 8 and Supplementary Figure S3). The expression of TTG1, a negative regulator of root hair differentiation, was lower in gem plants than in Col-0 and further downregulated by Cd and As(III). However, the expression of another negative regulator, GL2, was not altered. These results suggested that GEM is not a negative regulator of TTG1 and GL2, which is in disagreement with previous reports indicating that GEM is a negative regulator of TTG1/GL2 and promotes root hair differentiation (Caro et al., 2007). Furthermore, the lower expression of TTG1 did not correlate with the reduced root hair density in gem plants, since ttg1 plants displayed higher root hair density (Figure 5F). In addition, the downregulation of RHD6 and AXR2, which function in root hair initiation, also contributed to the decrease of root hair density in gem plants.
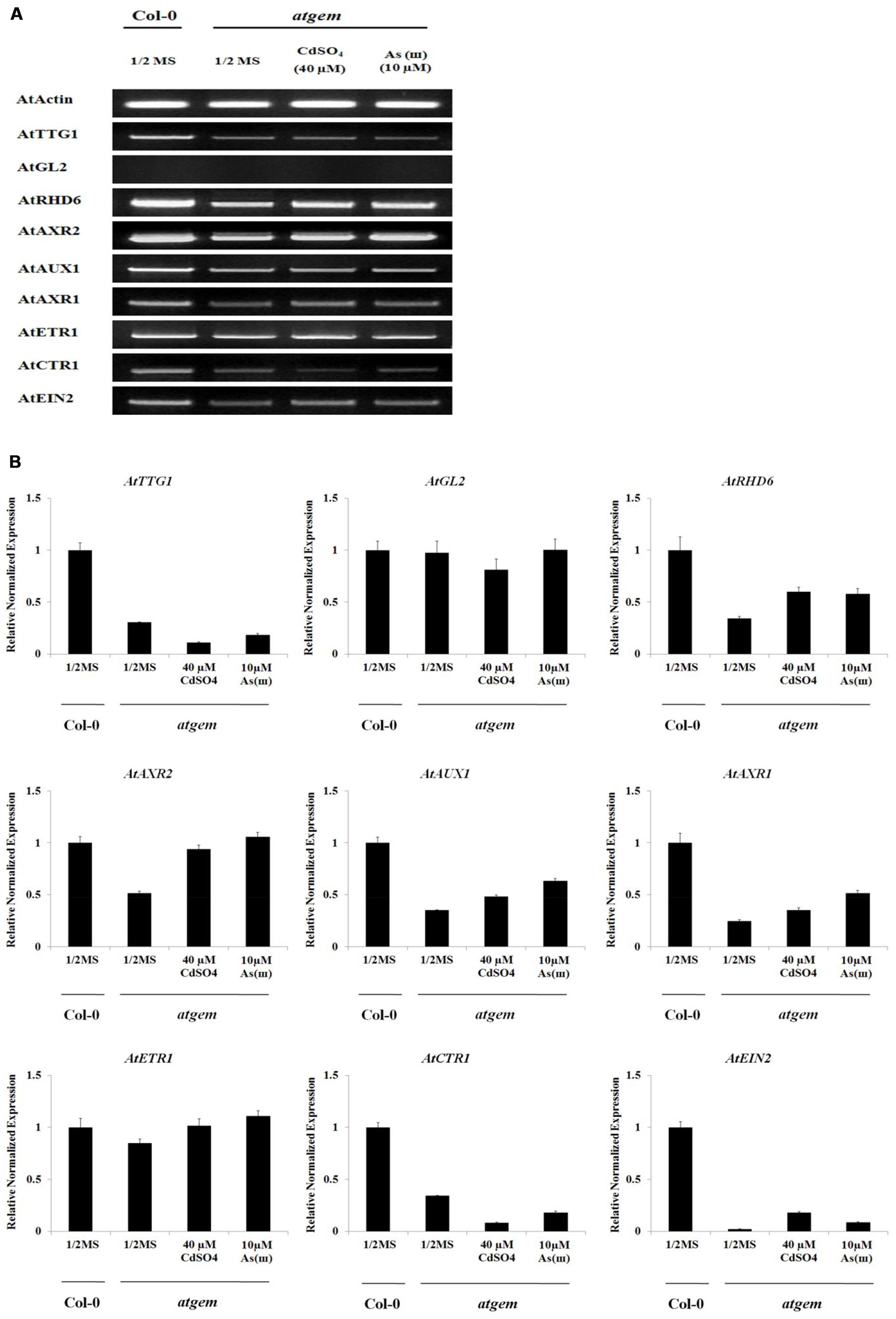
FIGURE 8. Effect of Cd and As(III) on expressions of genes engaged in the differentiation (fate determination) and elongation of root hairs in gem Arabidopsis. (A) Semi-quantitative RT-PCR results showing relative transcript levels of various genes involved in the differentiation (TTG1 and GL2), initiation (RHD6 and ACXR2) and elongation (AUX1, AXR1, ETR1, CTR1, and EIN2) of root hairs in Col-0 and gem Arabidopsis. RT-PCR was performed using total RNA isolated from 1-week old plants which were germinated and grown on 1/2 MS agar media treated with no metal, 40 μM CdSO4 and 10 μM As(III). The gel photography represents three experimental replicates. (B) Quantitative real-time PCR results of various genes involved in root hair differentiation and elongation shown in (A). Transcript levels were normalized to the transcript level of Actin.
Consistent with the decreased root hair length, the expression of genes involved in root hair elongation (AUX1, AXR1, ETR1, and EIN2) was inhibited in gem plants. Although Cd and As(III) upregulated these repressed genes, their expression did not recover to the level of Col-0 except AXR2 and ETR1. It is beyond our expectation that CTR1 expression was also repressed in gem plants, since CTR1 is a blocker of ethylene signal transduction involved in root hair elongation.
Discussion
In the present study, we showed that Cd and As(III) increased root hair density probably by converting non-hairy cells to hairy cells through the modulation of the expression of genes involved in root hair fate determination (Figures 3 and 9). First, Cd and As(III) downregulated TTG1, which is essential for generating the WER-GL3/EGL3-TTG1 complex; this impaired its ability to upregulate GL2, which resulted in cells entering the root hair cell fate pathway. Second, Cd and As(III) may inhibit GL2 expression directly and render it unable to inhibit RHD6 transcription, which is involved in hair initiation, as well as impairing the expression of atrichoblast-specific genes. Third, Cd and As(III) promoted GEM expression, positively regulating RHD6 expression, which was downregulated in gem Arabidopsis (Figure 8); this led to the upregulation of root hair cell differentiation genes and the determination of hair cell fate.
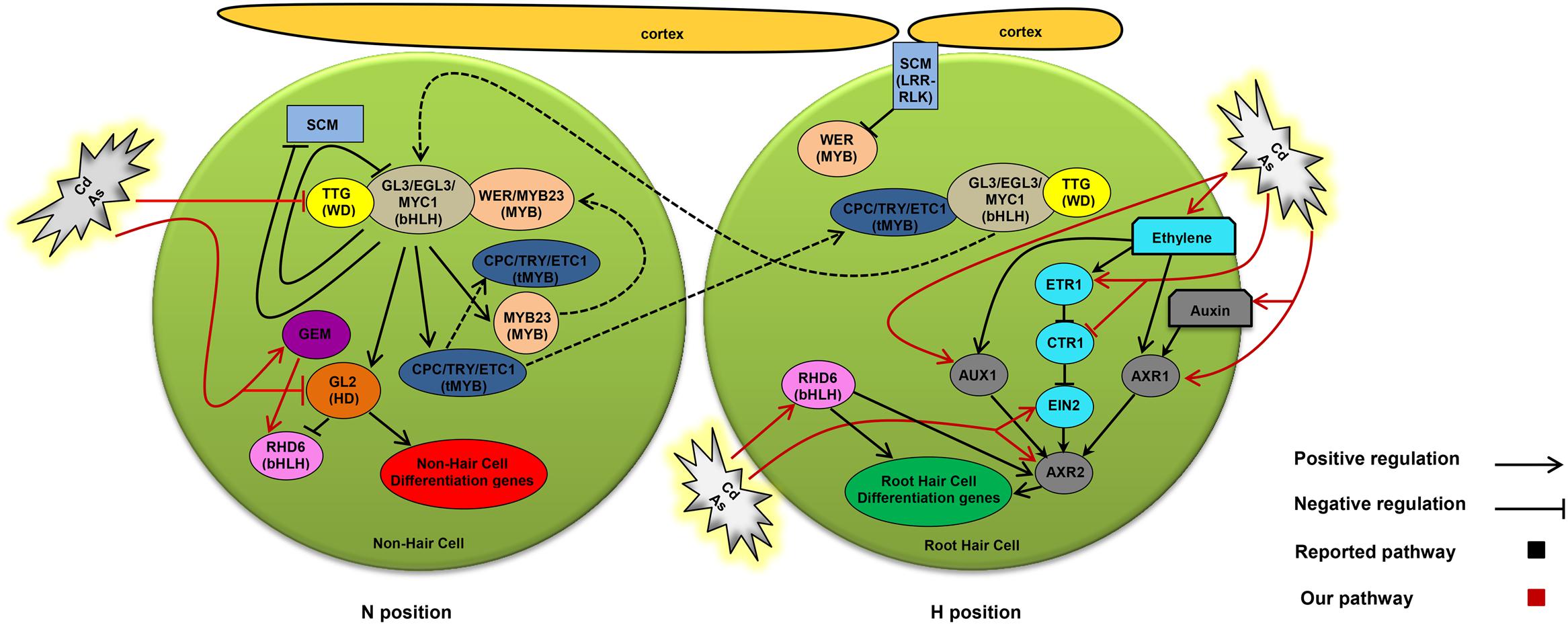
FIGURE 9. Proposed model showing how cadmium and arsenic (As III) promote root hair differentiation and elongation through modulating intrinsic developmental and hormonal pathways in Arabidopsis. Arrows show positive regulation, blunted lines show negative regulation, and broken lines illustrate intercellular or intracellular protein translocation. This model is adapted from Bruex et al. (2012).
The next step of root hair development may be root hair morphogenesis, including initiation and elongation. RHD6 and AXR2 are downstream components of fate determination that initiate root hair morphogenesis, since rhd6 and axr2 mutants have reduced root hair density (Masucci and Schiefelbein, 1994; Rigas et al., 2013). Our data showed that RHD6 expression was increased by Cd and As(III), resulting in increased root hair density and length. RHD6 expression was higher in the ttg1 mutant, in which root hair density is increased, and was lower in the gem mutant, in which root hair number is reduced, than in the control. AXR2 expression was enhanced by Cd and As(III), remained unchanged in ttg1, and was reduced in gem plants. Auxin and ethylene promote root hair initiation via AXR2 independently of RHD6, since they rescue low root hair numbers in the rhd6 mutant but not in the axr2 mutant (Masucci and Schiefelbein, 1996). Although the ctr1 mutant has a higher number of root hairs (Masucci and Schiefelbein, 1996), it is not clear whether it functions in hair initiation, since supporting data was not reported thereafter.
The auxin signaling components AUX1 and AXR1 are involved in root hair elongation since aux1 and axr1 mutants have shorter hairs (Pitts et al., 1998). Consistent with this result, AUX1 and AXR1 were upregulated at concentrations of Cd and As(III) that promoted root hair elongation (Figure 3). In addition, their expression was decreased in gem plants, which have shorter root hairs (Figure 8), and not affected in ttg1 Arabidopsis, which has a higher root hair number (Figure 6). These results confirmed that GEM is also involved in root hair elongation. In accordance with our results regarding AUX1, a previous report indicated that Cd stress induces AUX1 expression in rice root hairs, and the root hair length in osaux1 mutant rice is shorter than that of the WT (Yu et al., 2015). The involvement of auxin in root hair elongation is supported by our results showing increased levels of auxin in the root tip in parallel with the increased root hair length caused by Cd and As(III) (Figure 4C).
Ethylene signaling components also seem to play a role in root hair elongation, since the ethylene insensitive mutant ein2 has shorter hairs (Rahman et al., 2002), while the ethylene overproducing mutant eto1 has longer root hairs (Strader et al., 2010). In support of this, the ethylene signaling components ETR1 and EIN2 were upregulated in response to Cd and As(III), stimulating root hair elongation (Figure 3). Consistent with this result, the negative regulator of ethylene signaling CTR1 was downregulated by Cd and As(III). In the case of ttg1 plants showing an increase in root hair density, the expression of ethylene signaling genes was not altered (Figure 6). By contrast, ethylene signaling genes were repressed in gem plants, which display a reduction in root hair density and length (Figure 8). Regarding the relationship between ethylene and auxin, ethylene is able to enhance auxin synthesis and transport (Ruzicka et al., 2007; Swarup et al., 2007), and ethylene promotes root hair elongation via auxin-induced microtubule randomization (Takahashi et al., 2003). In addition, ethylene may be involved in root hair initiation, since ACC (ethylene precursor) enhances root hair number (Tanimoto et al., 1995) and AVG (ethylene biosynthesis inhibitor) reduces root hair number (Masucci and Schiefelbein, 1996).
It is also possible that Cd promotes root hair elongation by increasing ROS production, which activates root hair cell elongation (Takeda et al., 2008). This hypothesis is supported by reports that root hair length was induced by Cu (Pasternak et al., 2005) and Cr (Castro et al., 2007), which are associated with changes in ROS metabolism and auxin homeostasis.
It is also suggested that Cd enhances root hair formation by inducing Fe deficiency through the inhibition of Fe uptake, since Fe deficiency promotes root hair production and elongation by enhancing ethylene production (Schmidt and Schikora, 2001). Cd reduces Fe uptake by competing for the iron transporter IRT1 (Vert et al., 2002).
During the last decades, dynamic gene regulatory network (GRN) models have been used extensively to investigate cell fate determination (von Dassow et al., 2000; Albert and Othmer, 2003; Espinosa-soto et al., 2004; Almaas, 2007). These models integrate experimental data and produce a dynamic model to explore developmental patterns. Such models are potentially useful and can improve our knowledge regarding biological developmental systems, since they provide powerful tools to interpret contrasting data. In addition, new hypotheses can arise from these models that can be experimentally validated. For instance, Benitez et al. (2007) used a dynamic model to show that positional SCM signals in the cortex can function either positively or negatively on WER and CPC expression within the active or inactive complex, respectively. Although WER expression is proposed to be repressed by the SCM signal (Kwak et al., 2005), the possibility of a direct effect on CPC could not be excluded (Wada et al., 1997; Lee and Schiefelbein, 2002; Costa and Dolan, 2003; Benitez et al., 2007). In the model presented here, we attempted to show how Cd and As (III) affect the GRN in root epidermal cells based on the transcriptional levels of the genes examined. Our model (Figure 9) is only a schematic integration of the active molecules in each cell type. Moreover, this does not include the complete GRN model components within each cell type, in contrast to dynamic models presented in recent studies (Benitez et al., 2008; Kwak and Schiefelbein, 2008; Savage et al., 2008). Information derived from dynamic GRN studies (Benitez et al., 2007, 2008; Kwak and Schiefelbein, 2008; Benitez and Alvarez-Buylla, 2010) can improve our understanding of cell fate determination and patterning, as well as possible interactions within the GRN and cell to cell communication in Arabidopsis root epidermal cells. In this context, the GRN model can not only predict and provide novel insights into GRN component interactions involved in root hair patterning in response to Cd and As, but also help link heavy metal signaling pathways and genetic regulatory network components under different experimental condition in future studies.
For further experiments, the conversion of non-hair cell to hair cells (change of fate) induced by Cd and As(III) should be examined using morphological analyses. In addition, the expression of various fate-determining genes in response to metals, such as WER, GL3, EGL3, CPC1, TRY, and ETC1, should be examined. Analysis of the effects of metals on recently identified genes expressed in hairy and non-hairy cells is also meaningful (Bruex et al., 2012).
Conclusion
The density and length of root hairs are increased in response to Cd and As(III) in Arabidopsis by the following mechanisms: First, root hair fate determination (differentiation) is stimulated through the upregulation of GEM (positive regulator) and the downregulation of TTG1/GL2 (negative regulators). Second, the initiation of root hair is promoted by the upregulation of RHD6. Third, the elongation of root hair is promoted by increasing auxin content and the expression of the auxin signaling genes AXR2, AUX1, and AXR1. Finally, root hair elongation is stimulated by the upregulation of the ethylene signaling genes ETR1 and EIN2, and the downregulation of CTR1.
Author Contributions
SH designed and supervised whole research, and wrote the article with contributions of all the authors; RB and DK equally performed most of the experiments; JK performed an initial experiment.
Funding
This work was supported by the Bio-industry Technology Development Program funded by the Ministry for Food, Agriculture, Forestry and Fisheries [312033-5]; and the Civil research projects for solving social problems through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning [NRF-2015M3C8A6A06014500], KOREA.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. MyoungMin Lee (Yonsei U., Korea) for the supply of transgenic Arabidopsis containing GL2::GUS construct; Prof. Jun Lim (Konkuk U., Korea) for transgenic Arabidopsis containing DR5::GUS construct.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01763/full#supplementary-material
Abbreviations
ACC, 1-aminocyclopropane-1-carboxylic acid; ANOVA, analysis of variance; AsNaO2, Arsenate sodium; AVG, aminoethoxyvinylglycine; CdSO4, Cadmium sulphate; EDTA, Ethylene diamine tetra acetic acid; GUS, β-glucuronidase; MS, Murashige and Skoog; qRT-PCR, quantitative RT-PCR; ROS, Reactive oxygen species; WT, Wild type; X-gluc, 5-bromo-4-chloro-3-indolyl-β-D-glucuronidase.
Footnotes
References
Albert, R., and Othmer, H. G. (2003). The topology of the regulatory interactions predicts the expression pattern of the segment polarity genes in Drosophila melanogaster. J. Theor. Biol. 223, 1–18. doi: 10.1016/S0022-5193(03)00035-3
Almaas, E. (2007). Biological impacts and context of network theory. J. Exp. Biol. 210, 1548–1558. doi: 10.1242/jeb.003731
Benitez, M., and Alvarez-Buylla, E. R. (2010). Dynamic-module redundancy confers robustness to the gene regulatory network involved in hair patterning of Arabidopsis epidermis. Biosystems 102, 11–15. doi: 10.1016/j.biosystems.2010.07.007
Benitez, M., Espinosa-Soto, C., Padilla-Longoria, P., and Alvarez-Buylla, E. R. (2008). Interlinked nonlinear subnetworks underlie the formation of robust cellular patterns in Arabidopsis epidermis: a dynamic spatial model. BMC Syst. Biol. 2:98. doi: 10.1186/1752-0509-2-98
Benitez, M., Espinosa-Soto, C., Padilla-Longoria, P., Diaz, J., and Alvarez-Buylla, E. R. (2007). Equivalent genetic regulatory networks in different contexts recover contrasting spatial cell patterns that resemble those in Arabidopsis root and leaf epidermis: a dynamic model. Int. J. Dev. Biol. 51, 139–155. doi: 10.1387/ijdb.062183mb
Bruex, A., Kainkaryam, R. M., Wieckowski, Y., Kang, Y. H., Bernhardt, C., Xia, Y., et al. (2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLos Genet. 8:e1002446. doi: 10.1371/journal.pgen.1002446
Caro, E., Castellano, M. M., and Gutierrez, C. (2007). A chromatin link that couples cell division to root epidermis patterning in Arabidopsis. Nature 447, 213–217. doi: 10.1038/nature05763
Castro, R. O., Trujillo, M. M., Bucio, J. L., and Cervantes, C. (2007). Effects of dichromate on growth and root system architecture of Arabidopsis thaliana seedlings. Plant Sci. 172, 684–691. doi: 10.1016/j.plantsci.2006.11.021
Cernac, A., Lincoln, C., Lammer, D., and Estelle, M. (1997). The SAR1 gene of Arabidopsis acts downstream of the AXR1 gene in auxin response. Development 124, 1583–1591.
Cho, H. T., and Cosgrove, D. J. (2002). Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14, 3237–3253. doi: 10.1105/tpc.006437
Costa, S., and Dolan, L. (2003). Epidermal patterning genes are active during embryogenesis in Arabidopsis. Development 130, 2893–2901. doi: 10.1242/dev.00493
Dolan, L. (2006). Positional information and mobile transcriptional regulators determine cell pattern in the Arabidopsis root epidermis. J. Exp. Bot. 57, 51–54. doi: 10.1093/jxb/erj037
Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., et al. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84.
Durcekova, K., Huttova, J., Mistrik, I., Olle, M., and Tamas, L. (2007). Cadmium induces premature xylogenesis in barley roots. Plant Soil 290, 61–68. doi: 10.1007/s11104-006-9111-6
Espinosa-soto, C., Padilla-Longoria, P., and Alvarez-Buylla, E. R. (2004). A gene regulatory network model for cell-fate determination during Arabidopsis thaliana flower development that is robust and recovers experimental gene expression profiles. Plant Cell 16, 2923–2939. doi: 10.1105/tpc.104.021725
Galway, M. E., Masucci, J. D., Lloyd, A. M., Walbot, V., Davis, R. W., and Schiefelbein, J. W. (1994). The Ttg gene is required to specify epidermal-cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166, 740–754. doi: 10.1006/dbio.1994.1352
Godt, J., Scheidig, F., Grosse-Siestrup, C., Esche, V., Brandenburg, P., Reich, A., et al. (2006). The toxicity of cadmium and resulting hazards for human health. J. Occup. Med. Toxicol. 1:22. doi: 10.1186/1745-6673-1-22
Hassan, H., Scheres, B., and Blilou, I. (2010). JACKDAW controls epidermal patterning in the Arabidopsis root meristem through a non-cell-autonomous mechanism. Development 137, 1523–1529. doi: 10.1242/dev.048777
Hu, Y. F., Zhou, G. Y., Na, X. F., Yang, L. J., Nan, W. B., Liu, X., et al. (2013). Cadmium interferes with maintenance of auxin homeostasis in Arabidopsis seedlings. J. Plant Physiol. 170, 965–975. doi: 10.1016/j.jplph.2013.02.008
Hung, C. Y., Lin, Y., Zhang, M., Pollock, S., Marks, M. D., and Schiefelbein, J. (1998). A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiol. 117, 73–84. doi: 10.1104/Pp.117.1.73
Kang, Y. H., Song, S. K., Schiefelbein, J., and Lee, M. M. (2013). Nuclear trapping controls the position-dependent localization of CAPRICE in the root epidermis of Arabidopsis. Plant Physiol. 163, 193–204. doi: 10.1104/pp.113.221028
Kirik, V., Simon, M., Huelskamp, M., and Schiefelbein, J. (2004). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268, 506–513. doi: 10.1016/j.ydbio.2003.12.037
Kopittke, P. M., Blamey, F. P. C., and Menzies, N. W. (2010). Toxicity of Cd to signal grass (Brachiaria decumbens Stapf.) and Rhodes grass (Chloris gayana Kunth.). Plant Soil 330, 515–523. doi: 10.1007/s11104-009-0224-6
Kurata, T., Ishida, T., Kawabata-Awai, C., Noguchi, M., Hattori, S., Sano, R., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132, 5387–5398. doi: 10.1242/dev.02139
Kuriakose, S. V., and Prasad, M. N. V. (2008). Cadmium stress affects seed germination and seedling growth in Sorghum bicolor (L.) Moench by changing the activities of hydrolyzing enzymes. Plant Growth Regul. 54, 143–156. doi: 10.1007/s10725-007-9237-4
Kwak, S. H., Shen, R., and Schiefelbein, J. (2005). Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307, 1111–1113. doi: 10.1126/science.1105373
Kwak, S. H., and Schiefelbein, J. (2008). A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern in Arabidopsis. Curr. Biol. 18, 1949–1954. doi: 10.1016/j.cub.2008.10.064
Lee, M. M., and Schiefelbein, J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. doi: 10.1016/S0092-8674(00)81536-6
Lee, M. M., and Schiefelbein, J. (2002). Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14, 611–618. doi: 10.1105/tpc.010434
Lee, R. D., and Cho, H. T. (2013). Auxin, the organizer of the hormonal/environmental signals for root hair growth. Front. Plant Sci. 4:448. doi: 10.3389/fpls.2013.00448
Masucci, J. D., Rerie, W. G., Foreman, D. R., Zhang, M., Galway, M. E., Marks, M. D., et al. (1996). The homeobox gene GLABRA 2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260.
Masucci, J. D., and Schiefelbein, J. W. (1994). The rhd6 Mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 106, 1335–1346. doi: 10.1104/pp.106.4.1335
Masucci, J. D., and Schiefelbein, J. W. (1996). Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8, 1505–1517. doi: 10.1105/tpc.8.9.1505
Menand, B., Yi, K. K., Jouannic, S., Hoffmann, L., Ryan, E., Linstead, P., et al. (2007). An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316, 1477–1480. doi: 10.1126/science.1142618
Muller, M., and Schmidt, W. (2004). Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol. 134, 409–419. doi: 10.1104/pp.103.029066
Nagpal, P., Walker, L. M., Young, J. C., Sonawala, A., Timpte, C., Estelle, M., et al. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–574. doi: 10.1104/pp.123.2.563
Pasternak, T., Rudas, V., Potters, G., and Jansen, M. A. K. (2005). Morphogenic effects of abiotic stress: reorientation of growth in Arabidopsis thaliana seedlings. Environ. Exp. Bot. 53, 299–314. doi: 10.1016/j.envexpbot.2004.04.009
Peterson, R. L., and Farquhar, M. L. (1996). Root hairs: specialized tubular cells extending root surfaces. Bot. Rev. 62, 1–40. doi: 10.1007/Bf02868919
Pitts, R. J., Cernac, A., and Estelle, M. (1998). Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16, 553–560. doi: 10.1046/j.1365-313x.1998.00321.x
Prasinos, C., Krampis, K., Samakovli, D., and Hatzopoulos, P. (2005). Tight regulation of expression of two Arabidopsis cytosolic Hsp90 genes during embryo development. J. Exp. Bot. 56, 633–644. doi: 10.1093/jxb/eri035
Qing, L., and Aoyama, T. (2012). Pathways for epidermal cell differentiation via the homeobox gene GLABRA2: update on the roles of the classic regulator. J. Integr. Plant Biol. 54, 729–737. doi: 10.1111/j.1744-7909.2012.01159.x
Rahman, A., Hosokawa, S., Oono, Y., Amakawa, T., Goto, N., and Tsurumi, S. (2002). Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiol. 130, 1908–1917. doi: 10.1104/pp.010546
Rerie, W. G., Feldmann, K. A., and Marks, M. D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8, 1388–1399. doi: 10.1101/gad.8.12.1388
Rigas, S., Ditengou, F. A., Ljung, K., Daras, G., Tietz, O., Palme, K., et al. (2013). Root gravitropism and root hair development constitute coupled developmental responses regulated by auxin homeostasis in the Arabidopsis root apex. New Phytol. 197, 1130–1141. doi: 10.1111/nph.12092
Ruzicka, K., Ljung, K., Vanneste, S., Podhorska, R., Beeckman, T., Friml, J., et al. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19, 2197–2212. doi: 10.1105/tpc.107.052126
Savage, N. S., Walker, T., Wieckowski, Y., Schiefelbein, J., Dolan, L., and Monk, N. A. M. (2008). A mutual support mechanism through intercellular movement of CAPRICE and GLABRA3 can pattern the Arabidopsis root epidermis. PLoS Biol. 6:e235. doi: 10.1371/journal.pbio.0060235
Schellmann, S., Schnittger, A., Kirik, V., Wada, T., Okada, K., Beermann, A., et al. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21, 5036–5046. doi: 10.1093/emboj/cdf524
Schiefelbein, J. W. (2000). Constructing a plant cell. The genetic control of root hair development. Plant Physiol. 124, 1525–1531. doi: 10.1104/pp.124.4.1525
Schiefelbein, J. W., and Somerville, C. (1990). Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2, 235–243. doi: 10.1105/tpc.2.3.235
Schmidt, W., and Schikora, A. (2001). Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiol. 125, 2078–2084. doi: 10.1104/pp.125.4.2078
Simon, M., Lee, M. M., Lin, Y., Gish, L., and Schiefelbein, J. (2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 311, 566–578. doi: 10.1016/j.ydbio.2007.09.001
Strader, L. C., Chen, G. L., and Bartel, B. (2010). Ethylene directs auxin to control root cell expansion. Plant J. 64, 874–884. doi: 10.1111/j.1365-313X.2010.04373.x
Swarup, R., Perry, P., Hagenbeek, D., Van Der Straeten, D., Beemster, G. T. S., Sandberg, G., et al. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19, 2186–2196. doi: 10.1105/tpc.107.052100
Takahashi, H., Kawahara, A., and Inoue, Y. (2003). Ethylene promotes the induction by auxin of the cortical microtubule randomization required for low-pH-induced root hair initiation in lettuce (Lactuca sativa L.) seedlings. Plant and Cell Physiology 44, 932–940. doi: 10.1093/Pcp/Pcg119
Takeda, S., Gapper, C., Kaya, H., Bell, E., Kuchitsu, K., and Dolan, L. (2008). Local positive feedback regulation determines cell shape in root hair cells. Science 319, 1241–1244. doi: 10.1126/science.1152505
Tanimoto, M., Roberts, K., and Dolan, L. (1995). Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J. 8, 943–948. doi: 10.1046/j.1365-313X.1995.8060943.x
Vert, G., Grotz, N., Dedaldechamp, F., Gaymard, F., Guerinot, M. L., Briat, J. F., et al. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14, 1223–1233. doi: 10.1105/tpc.001388
Vitoria, A. P., Rodriguez, A. P. M., Cunha, M., Lea, P. J., and Azevedo, R. A. (2003). Structural changes in radish seedlings exposed to cadmium. Biol. Plant. 47, 561–568. doi: 10.1023/B:BIOP.0000041062.00539.7a
von Dassow, G., Meir, E., Munro, E. M., and Odell, G. M. (2000). The segment polarity network is a robust development module. Nature 406, 188–192. doi: 10.1038/35018085
Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116.
Yu, C. L., Sun, C. D., Shen, C. J., Wang, S. K., Liu, F., Liu, Y., et al. (2015). The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in Cd stress responses in rice (Oryza sativa L.). Plant J. 83, 818–830. doi: 10.1111/tpj.12929
Keywords: root hair, cadmium, arsenic, gene expressions, Arabidopsis
Citation: Bahmani R, Kim DG, Kim JA and Hwang S (2016) The Density and Length of Root Hairs Are Enhanced in Response to Cadmium and Arsenic by Modulating Gene Expressions Involved in Fate Determination and Morphogenesis of Root Hairs in Arabidopsis. Front. Plant Sci. 7:1763. doi: 10.3389/fpls.2016.01763
Received: 01 July 2016; Accepted: 08 November 2016;
Published: 23 November 2016.
Edited by:
Wim Van den Ende, KU Leuven, BelgiumReviewed by:
Iwona Małgorzata Morkunas, University of Life Sciences in Poznañ, PolandElena R. Alvarez-Buylla, Universidad Nacional Autónoma de Mexico, Mexico
Copyright © 2016 Bahmani, Kim, Kim and Hwang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seongbin Hwang, c2Jod2FuZ0BzZWpvbmcuYWMua3I=
†These authors have contributed equally to this work.
 Ramin Bahmani
Ramin Bahmani Dong G. Kim
Dong G. Kim Jin A. Kim
Jin A. Kim Seongbin Hwang
Seongbin Hwang