- Department of Genetics and Genome Biology, University of Leicester, Leicester, United Kingdom
The amphipod Parhyale hawaiensis is a model organism of growing importance in the fields of evolutionary development and regeneration. A small, hardy marine crustacean that breeds year-round with a short generation time, it has simple lab culture requirements and an extensive molecular toolkit including the ability to generate targeted genetic mutant lines. Here we identify canonical core and regulatory clock genes using genomic and transcriptomic resources as a first step in establishing this species as a model in the field of chronobiology. The molecular clock of P. hawaiensis lacks orthologs of the canonical circadian genes cryptochrome 1 and timeless, in common with the mammalian system but in contrast to many arthropods including Drosophila melanogaster. Furthermore the predicted CLOCK peptide is atypical and CRY2 shows an extended 5′ region of unknown function. These results appear to be shared by two other amphipod species.
Introduction
The subphylum Crustacea has a rich history of chronobiological research. One of the earliest recorded examples of a persistent circadian phenomenon in animals was the color changes seen in the prawn Hippolyte varians (Gamble and Keeble, 1900), while investigations on the circadian and circatidal rhythms of the fiddler crab Uca pugnax helped lay the basis for modern chronobiology (Palmer, 1991), with published works on the persistence of chromatophore and locomotor activity rhythms in constant conditions; temperature compensation; persistance of rhythms in isolated tissue; and the phase-shifting effect of light pulses, this latter observation coming a decade before the phase response curve (PRC) was incorporated into circadian concepts (De Coursey, 1960). A variety of endogenously controlled rhythmic phenomena have been documented in wide range of crustaceans, not only circadian rhythms but also tidal, lunar and semi-lunar (Strauss and Dircksen, 2010), under the control of zeitgebers such as light, hydrostatic pressure, immersion, wave motion and salinity. Where the clade has historically proved lacking, however, is in providing an organism with a molecular toolkit comparable to that of the fruit fly Drosophila melanogaster, in which transgenics, reporter genes and binary expression systems have been used to characterize, locate and dissect the endogenous biological clock at a neurological level (Helfrich-Förster, 2005).
At the center of the Drosophila clock are the bHLH-PAS proteins CLOCK (Allada et al., 1998; Gekakis et al., 1998, CLK) and CYCLE (Rutila et al., 1998, CYC), which form a heterodimer that drives transcription of clock-controlled genes including period (Konopka and Benzer, 1971, per) and timeless (Sehgal et al., 1994, tim), whose protein products dimerise and interact with CLK:CYC to inhibit its transcription activity and thus their own expression. TIM is degraded by the light-activated CRYPTOCHROME (Emery et al., 1998, CRY), exposing PER to degradation in turn and ending CLK:CYC repression. The transcription-translation feedback loop enacted by these core clock genes is modified to achieve a circadian periodicity of 24 h through the contributions of numerous regulatory genes such as doubletime, vrille, shaggy and clockwork orange. In mammals the clock is similarly well understood thanks to work on Mus musculus (Reppert and Weaver, 2001), showing that the molecular system is remarkably conserved but not identical. In mammals the CLK:BMAL1 heterodimer (where BMAL1 is an ortholog of CYC) is inhibited by multiple PER and CRY proteins, the latter notably different to the Drosophila CRY in lacking photosensitivity. A number of species (Zhu et al., 2005) have been found to have both a Drosophila-like CRY (often termed CRY1) and a mammalian-like CRY (CRY2).
In crustaceans, the first core clock gene to be identified was clock in the prawn Macrobrachium rosenbergii (Yang et al., 2006). The falling cost of next generation sequencing has subsequently brought a wave of such data in crustacean species including the water flea Daphnia pulex (Tilden et al., 2011), the copepods Calanus finmarchicus (Christie et al., 2013) and Tigriopus californicus (Nesbit and Christie, 2014), the lobster Nephrops norvegicus (Sbragaglia et al., 2015), the amphipod Talitrus saltator (O'Grady et al., 2016) and the Antarctic krill Euphausia superba (Hunt et al., 2017), in some cases including identification of rhythmic transcription. The clock of E. superba has been further characterized through analysis of the interactions of its protein components (Biscontin et al., 2017), as has that of the isopod Eurydice pulchra (Zhang et al., 2013), while tissue-specific identification of clock components has been performed in the American lobster Homarus americanus (Christie et al., 2018a,b).
Nevertheless much work remains to be done in determining how these genes generate rhythms, and in what tissues and cells they are active and when, and with which genes they interact. More fine-grained investigation into the crustacean clock requires a model organism of similar power to D. melanogaster, and the establishment of such a model also promises to significantly advance our understanding of the animal clock through comparative analysis.
The amphipod Parhyale hawaiensis was first isolated for lab culture in 1997 from the filtration system of the John G. Shedd Aquarium, “preselected [for] minimal care,” in the words of the collectors (Browne et al., 2005). A circumtropical marine detritivore found in shallow water mangrove zones, it is a robust, fecund organism that is cheap to maintain and breeds year-round with a quick generation time, making it well-suited to lab culture. Thanks to a reproductive system that permits the collection and manipulation of embryos from a single cell stage this species has become established as a model organism of some significance in the field of development. It has further proven itself to be amenable to molecular analysis and manipulation, including in situ hybridization, RNAi-mediated knockdown, transposon and integrase-based genetic transformation and CRISPR/Cas9 mutagenesis, and recently the 3.6 Gb genome was published (Kao et al., 2016).
Here we describe the creation of a head transcriptome to identify core and regulatory clock genes and use the genome to gain independent verification of our findings, which include the apparent loss of a number of major clock genes and the identification of a peptide feature that may be unique to amphipods.
Materials and Methods
Animal Husbandry
Animals were maintained in polypropylene containers with lids 3/4 on, with a layer of crushed coral approximately 1 cm deep and filled with 1 L artificial seawater prepared using 30g Tetra Marine Sea Salt per liter of distilled tap water to give a specific gravity of 1.022. Cultures were maintained on a 12:12 light:dark cycle with lights on at 9am, illumination coming from fluorescent room lighting at 300 lux, at 20° C. Animals were fed twice weekly, once with shredded carrot and once with a pinch of standard fish flakes.
Head Transcriptome
At eight time-points across 24 h beginning at 9am (lights on, zeitgeber time (ZT) 0) and every 3 h thereafter, a total of 20 adult males were collected per time-point and snap-frozen on a metal plate atop dry ice, whereupon the heads were dissected and stored at -80°C until use. Total RNA was extracted from five heads per time-point using a micropestle to homogenize the tissue in TRIzol reagent (Invitrogen), cleaned using the PureLink Micro Kit (ThermoFisher Scientific) with on-column DNase treatment to remove genomic DNA contamination and eluted with RNase free water. RNA extractions were quality assessed using Nanodrop and Agilent 2100 Bioanalyzer. Samples were required to meet a minimum quality of > 50 ng μL-1 concentration; 260/280 ratio 1.8 –2.2; 260/230 ratio 2.0–2.4 for sequencing use, and those that failed were discarded and the extraction process repeated on another five heads from storage.
One sample per time-point was delivered to Glasgow Polyomics (University of Glasgow) and subject to polyA mRNA enrichment with the TruSeq Stranded mRNA Library Prep Kit (Illumina). The resultant libraries were sequenced using the Illumina NextSeq 500 platform to generate 75 bp paired-end strand-specific reads, which were subject to gentle quality control comprising low quality read removal < Q10 and adapter trimming by Glasgow Polyomics. After download of this dataset FastQC 0.11.2 (Andrews, 2010) was used to assess read quality and Cutadapt 1.9.1 (Martin, 2011) used to remove remaining traces of the TruSeq Index and Universal adapters using the sequence “GATCGGAAGAGC” common to both adapter types as criteria. Reads were then filtered using Trimmomatic 0.32 (Bolger et al., 2014) to remove reads with an average quality of less than 20 and/or shorter than 72 bases.
A multi-k-mer, multi-assembler approach was adopted for assembly. Trans-ABySS 1.5.5 (Robertson et al., 2010), Bridger 2014-12-01 (Chang et al., 2015) and Trinity r20140717 (Haas et al., 2013) were used to generate individual assemblies using k-mers ranging from 19 to 71 with a minimum contig length of 200 bp. These were subsequently merged and duplicates and containments were removed at 99% identity using CD-HIT-EST 4.6.6 (Li and Godzik, 2006). TransRate (Smith-Unna et al., 2016) was used to identify well-assembled contigs based on read-mapping evidence. TransDecoder 3.0.0 (Haas et al., 2013) was used to identify predicted peptides from the well-assembled contigs (single_best_orf, -m 90), and CD-HIT used at 90% peptide identity to produce a final list of well-assembled, non-redundant coding contigs that were extracted from the original merged transcriptome assembly and used for all downstream purposes. Transcriptome completeness was assessed using BUSCO (Waterhouse et al., 2018) against the arthropoda lineage dataset.
Identification of Clock Genes
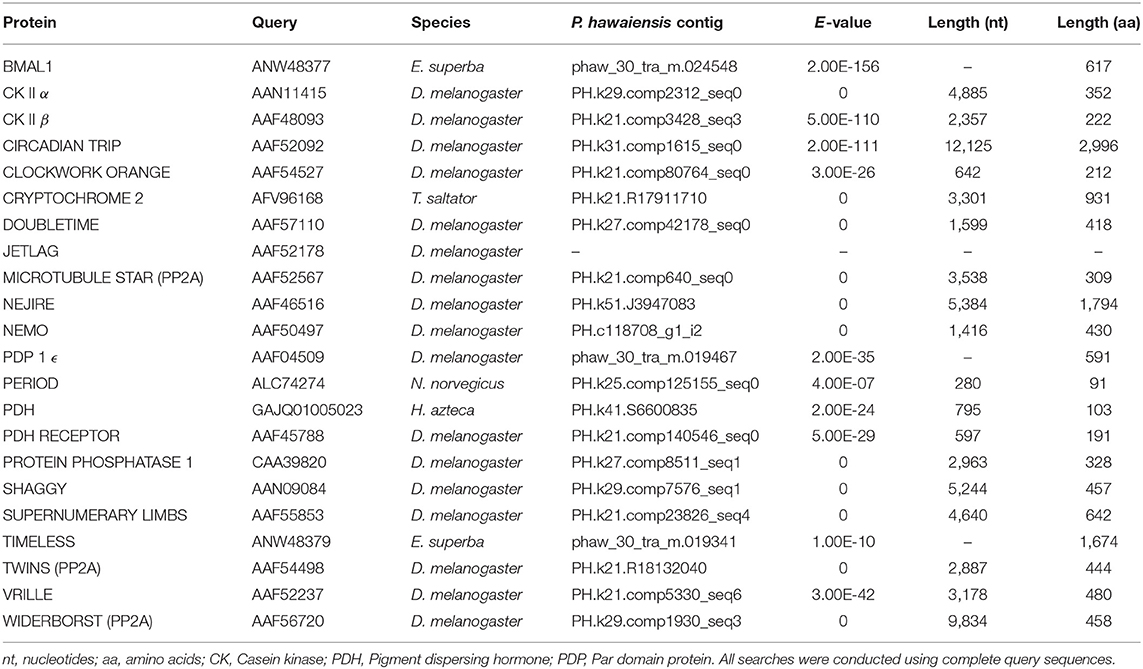
The head transcriptome was searched for core and regulatory circadian genes using the tblastn function of BLAST+ with an E-value cutoff of 1E-5 (Table 1). The protein sequences inferred from the genome (Kao et al., 2016, see https://doi.org/10.6084/m9.figshare.3498104.v4) were searched in the same manner using blastp. Genes that returned little or no evidence through this approach were searched for in the genome using tblastn and, where evidence was subsequently obtained, the scaffold and coordinates of the alignments were noted. The head transcriptome reads were aligned to the genome using HISAT2 2.1.0 (Kim et al., 2015) and visualized with IGV 2.3.82 (Robinson et al., 2011), and Stringtie 1.3.3b (Pertea et al., 2015) was used to assemble transcripts of interest from these read alignments. Gene sequences identified through BLAST+ searches were extended using these alignments and transcripts where appropriate. Sequence data from two other amphipods, Hyalella azteca (GCA_000764305.2) and Talitrus saltator (GDUJ00000000) was further used to extend partial transcripts and lend support to atypical results. RNA-seq reads for H. azteca (PRJNA312414) were downloaded, aligned and assembled as above. P. hawaiensis sequences obtained from BLAST+ searches and contigs identified as circadian genes in T. saltator (O'Grady et al., 2016) were used to search the genomes/transcriptomes of all three species, with results expanded using read mapping evidence and then these extended sequences used to search all resources again until no further extension was possible. Sequences returned by these processes were translated to peptides and subject to validation using the NCBI BLAST web interface (blastp, Database “nr,” Entrez Query “all[filter] NOT predicted[title] NOT hypothetical[title] NOT putative[title]”) to ascertain their closest match in other species.
Peptide domain analysis was conducted using SMART (Letunic et al., 2015). Peptide sequences were visualized using DOG 2.0 (Ren et al., 2009). Phylogenetic trees were created using MEGA 7.0.26 (Kumar et al., 2016); peptide sequences (Table S1) were aligned using MUSCLE at default settings, and neighbor-joining trees generated using 1,000 bootstrap replications and complete deletion.
Results
Transcriptome Assembly and Mining
After quality control the RNA-seq library for each head timepoint sample was approximately 22 million read pairs, with a total of 178,028,148 used in a final assembly of 86,686 contigs with an N50 of 2,797 and contig length ranging from 267 – 35,494. BUSCO analysis indicated 91.1% complete BUSCOs (81.1% single copy, 10% duplicated), 5.1% present but fragmented and 3.8% missing from a dataset of 1,066 groups searched. This Transcriptome Shotgun Assembly project has been deposited at GenBank under the accession GFVL00000000.
Evidence was found for the existence of the core clock genes bmal1, period and the mammalian-like cryptochrome 2 in this assembly. Also identified were 16 regulatory clock genes (Tables 1, 2).
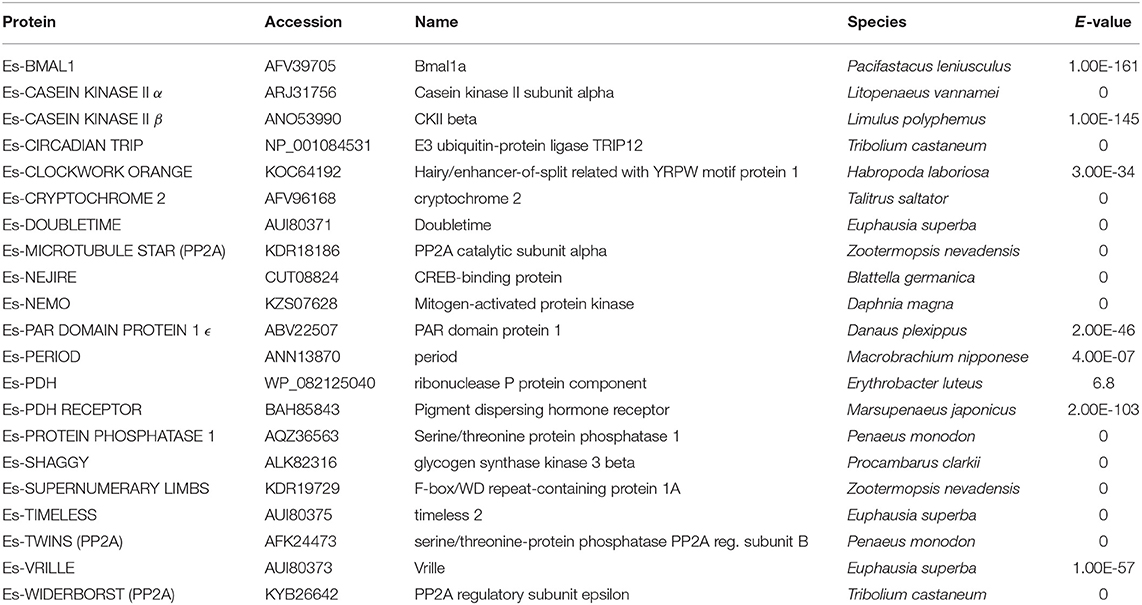
Table 2. BLAST search results using P. hawaiensis clock gene candidates vs. NCBI non-redundant protein database.
BMAL1/CYCLE
P. hawaiensis bmal1 (Ph-bmal1), found in complete form in the genome-inferred peptides (phaw_30_tra_m.024548) and in fragmentary form in the head transcriptome assembly, shows the typical complement of domains when translated (Figure 1A), with a bHLH domain, two PAS domains, a PAC domain immediately following the second PAS, and a BMAL1 C-terminal Region (BCTR), a transactivation domain characteristic of this gene (except in D. melanogaster CYCLE). The peptide returned Bmal1a in Pacifastacus leniusculus (accession AFV39705; 1E-161) from a BLAST confirmation query. Ph-bmal1 showed very low levels of expression in the head transcriptome (Table S3).
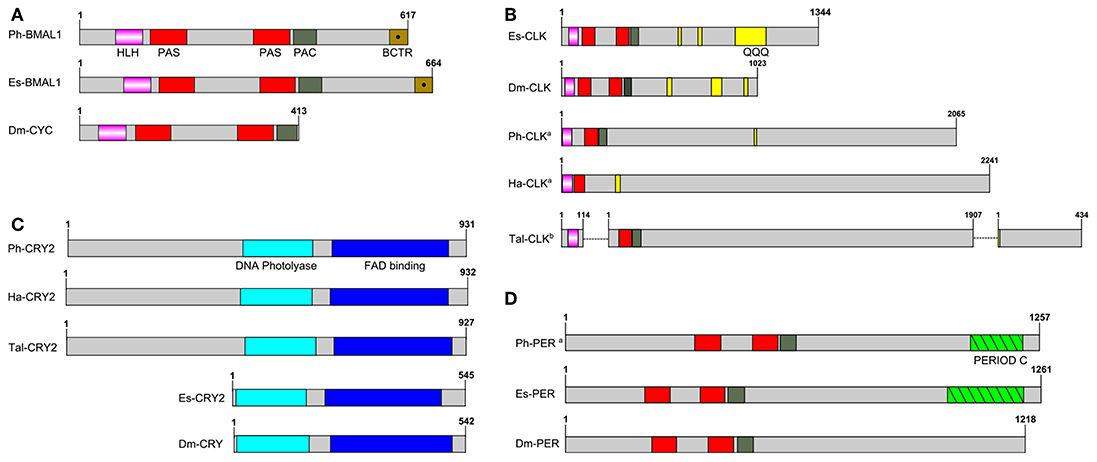
Figure 1. Schematics of predicted P. hawaiensis core clock peptides aligned with examples from other species. (A) BMAL1/CYCLE, (B) CLOCK, (C) CRYPTOCHROME and (D) PERIOD. Ph, Parhayle hawaiensis; Dm, Drosophila melanogaster; Es, Euphausia superba; Tal, Talitrus saltator; Ha, Hyalella azteca; QQQ, polyglutamine domain (defined as a run of 10 or more amino acids in which 60% or more are glutamine).apeptide is predicted on the basis of BLAST alignments and read mapping.bpeptide is a construct of multiple separate de novo assembled contigs. Both Tal-CRY2 and Tal-CLK have been previously reported in a shorter form (O'Grady et al., 2016).
CLOCK
No evidence was found of a P. hawaiensis clock gene (Ph-clock/Ph-clk) through BLAST mining of the head transcriptome or genome-inferred peptides. Searching the genome using Eurydice pulchra CLK 5 (AGV28720) identified orthologous sequences on scaffold LQNS02278184.1 (Supplementary Section A). When translated and combined, these sequences returned clock in the American lobster Homarus americanus (AWC08577; 3E-69), while SMART domain analysis identified bHLH, PAS and PAC domains. A small number of reads mapped to these fragments, indicating very low levels of expression in the head. Through iterative searches of the P. hawaiensis and H. azteca genomes using the CLK protein reported for the amphipod T. saltator (O'Grady et al., 2016), and informed by read mapping evidence, this was further extended to a putative final sequence encoding a 2,065 aa peptide. A putative Ha-CLK was also identified in this manner (Figure 1B).
Searching the T. saltator transcriptome with Ph-CLK and Ha-CLK identified two additional contigs of interest beyond Tal-CLK (Figure 1B): GDUJ01041853 encodes a truncated peptide with a bHLH domain that returns clock in Euphausia superba (ANW48378; 6E-24), while GDUJ01061398 shows homology (BLAST alignment 41% identity, 1E-40) to the 3′ end of Ph-CLK and Ha-CLK.
CRYPTOCHROME 2
P. hawaiensis cryptochrome 2 (Ph-cryptochrome 2/Ph-cry2) was identified in complete form in the head transcriptome, returning cryptochrome 2 in T. saltator (AFV96168; e-value 0.0) in a BLAST confirmation query. SMART analysis of the translated 931 aa peptide identified a DNA photolyase and FAD binding domain (Figure 1C), while an extended N-terminus of 357 amino acids had no identifiable functional domain or motif. A neighbor-joining phylogenetic tree confirmed the nature of the peptide as a mammalian-like CRY2 rather than a Drosophila-like CRY1 (Figure 2A). This coding sequence was supported by genome search and read mapping evidence: Stringtie assembled an 11-exon transcript (Supplementary Section A) located on scaffold LQNS02278089.1 that shows 99.5% identity to the de novo assembled contig, with the first exon incorporating both the N-terminus and a partial DNA photolyase domain in one contiguous sequence, indicating that the de novo assembled contig is unlikely to be a chimeric construct. Further to this the genome-inferred peptides, which were generated with reference to an independently assembled de novo transcriptome from embryonic and limb tissue (Kao et al., 2016), include a contig (phaw_30_tra_m.017772) covering 189 amino acids of the N-terminus and the entire DNA photolyase domain, and another (phaw_30_tra_m.017773) covering the first 160 amino acids of the N-terminus. Of the core clock genes identified Ph-cry2 showed the highest levels of expression (Table S3).
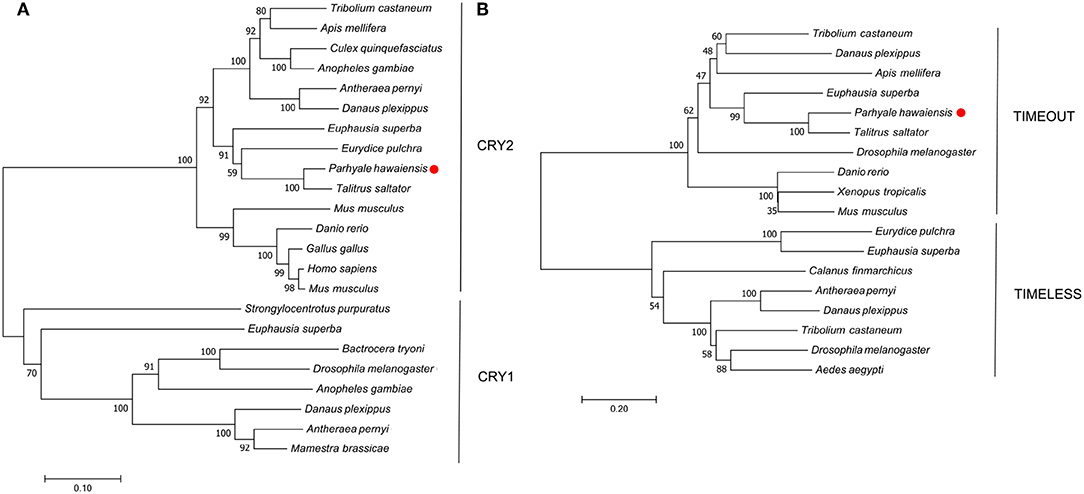
Figure 2. Optimal neighbor-joining trees depicting the evolutionary relationships of (A) CRYPTOCHROME and (B) TIMELESS/TIMEOUT peptides.
No evidence was found in any P. hawaiensis, T. saltator or H. azteca resource for a Drosophila-like cryptochrome.
PERIOD
A small 3′ fragment of P. hawaiensis period (Ph-period/Ph-per) was found in the head transcriptome, the peptide of which returned period in Macrobrachium nipponense from a BLAST confirmation query (ANN13870; e-value 4E-07). Searching the genome using this sequence and the predicted PER protein of Hyalella azteca (XP_018012603) returned multiple alignments on scaffold LQNS02276498.1 (Supplementary Section A). A small number of head transcriptome reads mapped to 14 of these regions. Translated and combined to form a fragmented putative peptide sequence 1,257 amino acids in length, SMART domain analysis revealed two PAS domains, a PAC domain and a PERIOD C domain (Figure 1D). This construct returned period isoform 1 in Homarus americanus (AWC08578; 3E-99) from a BLAST confirmation query. Ph-per showed very low levels of expression in the head transcriptome (Table S3).
TIMELESS
A mammalian-like TIMELESS was identified in the genome-inferred peptides resource. This is an ortholog of the D. melanogaster timeless paralog timeout (Figure 2B), a gene crucial to early development that is seemingly ubiquitous in animals (Sandrelli et al., 2008) and may have only peripheral involvement with the endogenous clock. No true TIMELESS could be identified in any P. hawaiensis, H. azteca or T. saltator resource.
Regulatory and Clock-Controlled Genes
Putative orthologs were identified for the regulatory genes casein kinase II α and β, circadian trip, clockwork orange, doubletime, microtubule star, nejire, nemo, par domain protein ϵ, protein phosphatase I, shaggy, supernumerary limbs, twins, vrille and widerborst, and for the clock-controlled genes pigment dispersing hormone (pdh) and pdh receptor (see Supplementary Section B). Each sequence was supported through its presence in an independently assembled leg/embryo transcriptome, the genome, or both.
Discussion
In silico mining of a head transcriptome resulted in the identification and characterization of four putative core clock proteins in Parhyale hawaiensis; Ph-BMAL1, Ph-CLK, Ph-PER, and Ph-CRY2. BUSCO analysis of this transcriptome indicated a high level of completeness and despite searching multiple additional resources including the recently published genome, we were unable to identify an ortholog of TIM or a Drosophila-like CRY. This suggests that P. hawaiensis has evolved what might be termed a mammalian-style molecular clock—a system lacking a TIM or CRY1, as originally reported in M. musculus (Reppert and Weaver, 2000). This system is found in vertebrates, hymenopterans and now in crustaceans. The gene reported as T. saltator timeless (O'Grady et al., 2016) appears to be a timeout (Figure 2B) and no sequence encoding a CRY1 or true TIM could be found in the various transcriptomic and genomic resources available for T. saltator and H. azteca, indicating that this system may be common in amphipods. This contrasts with other crustaceans studied so far - Daphnia pulex possesses the full complement of core clock proteins including a CRY1 and CRY2 (Tilden et al., 2011), as does Calanus finmarchicus (Christie et al., 2013), Trigriopus californicus (Nesbit and Christie, 2014), Limulus polyphemus (Chesmore et al., 2016) and Euphausia superba (Hunt et al., 2017). Eurydice pulchra lacks only a CRY1 (Zhang et al., 2013), as does Nephrops norvegicus (Sbragaglia et al., 2015).
This is not the only interesting aspect of the P. hawaiensis circadian system. We were unable to identify a complete coding sequence for clock, relying on alignments and read mapping to generate an incomplete construct that is, aside from the other amphipods analyzed here, notably different from the orthologs of other species. The putative amphipod CLK peptides appear to lack a second PAS domain and an extensive polyglutamine region that is characteristic of the gene (Saleem et al., 2001), with only very small glutamine-rich regions identified (Figure 1B). They are also extremely long, each over 2,000 aa in partial form, far surpassing the complete E. superba CLK (1,344 aa) one of the longest complete CLK peptides identified to date. If the two additional T. saltator contigs identified in the Results are part of the full T. saltator clock gene this would result in a peptide at least 2,455 aa in length. In H. azteca and T. saltator there is evidence of fragmentation in this gene, with bHLH-encoding sequences present in separate scaffolds/contigs. This fragmentation was also present in the first draft of the P. hawaiensis genome but was resolved with the second draft, and it is likely that the fragmentation is in silico rather than in vivo for all three species. The default assumption should be that Ph-CLK forms a heterodimer with Ph-BMAL1 to drive clock gene expression as is usual in animals, but the atypical nature of the amphipod CLK peptides - in particular the apparent lack of a second PAS domain which, if accurate, may impact dimerisation - encourages speculation about alternative partners for Ph-BMAL1. In mice BMAL1 is a promiscuous binding protein (Hogenesch et al., 1998) and other bHLH-PAS proteins have substituted for CLK in other species, for example methoprene-tolerent (MET) in the mosquito Aedes aegypti (Shin et al., 2012), a putative ortholog of which can be found in fragmentary form in the head transcriptome and complete in the genome-inferred peptides (phaw_30_tra_m.006390).
In this context we consider the cryptochrome Ph-CRY2, which shows an extraordinary extension at the N-terminus, with various lines of evidence suggesting the same may beis true for H. azteca and T. saltator. In H. azteca a predicted cryptochrome has been reported (accession XM_018166003), and while somewhat extended at the 5′ end it does not include an N-terminus comparable to that of P. hawaiensis. However a BLAST search of the genome using the Ph-CRY2 peptide returned alignments on scaffold KV721600.1 suggesting the existence of such a feature upstream of the reported sequence. Read alignment and assembly generated an 11-exon transcript (Supplementary Section A), the second exon coding for the N-terminus and partial DNA photolyase domain and terminating at the same point as the first exon of the Stringtie Ph-cry2 transcript. When translated, this transcript encodes a peptide 932 amino acids long (Ha-CRY2) incorporating the extended N-terminus with the previously predicted H. azteca CRY. In the paper detailing the molecular clock of T. saltator (O'Grady et al., 2016) two contigs were identified as representing cry2. An assumed complete peptide of 565 amino acids was reported, but one of the de novo assembled transcriptome contigs (accession GDUJ01076706.1, reported as comp100937_c0_seq1) includes an N-terminus of 362 amino acids that is not considered part of the CRY2 peptide by the authors, who verified the sequence using RACE PCR and suggest the contig may have been misassembled. However this sequence shows 55.9% identity and 71.3% similarity to the P. hawaiensis N-terminus, and considering the lines of evidence from P. hawaiensis and H. azteca the possibility of incomplete RACE PCR or splice variants should not be discounted. Whether this N-terminus serves a function, and what that might be, is again speculative at this point, but an unusual transcriptional heterodimer may require an unusual repressor. Perhaps then Ph-CRY2 serves in the repressive limb of the P. hawaiensis circadian feedback loop as does CRY2 in many other species, and it has evolved this N-terminus as a means to interact with, for example, a MET:BMAL1 heterodimer, or an atypical CLK:BMAL1.
We identified putative orthologs of all regulatory genes searched for, with the exception of jetlag, results that can form the basis for future work investigating if, and how, these genes curate the central feedback loops. A candidate for Ph-PDH proved difficult to identify due to its divergent sequence including at the conserved NSELINS motif. Ph-pdh was found only through the use of the putative Hyalella azteca preprohormone Hyaaz-prepro-PDH (Christie, 2014) as a query term, and while a BLAST confirmation query returned only one unrelated result (2) Ph-PDH shares 47.6% identity and 62.1% similarity with Hyaaz-prepro-PDH. A signal peptide with a predicted cleavage site between residues 36 and 37 was detected using SignalP 5.0 (Armenteros et al., 2019) and through alignment with Hyaaz-prepro-PDH a 23 amino acid mature peptide was identified with 91.3% identity and 100% similarity. T. saltator is reported to possess two PDH peptides, the first (Tal-PDH I) unusual with regards to its long mature peptide form but showing the NSELINS motif and the second (Tal-PDH II) identified using relaxed search criteria. This latter PDH sequence and that of H. azteca—itself described as atypical (Christie, 2014)—were the only search terms from dozens used that returned a candidate contig. In insects the homolog of PDH, called Pigment dispersing factor (PDF) is a neuropeptide and neurohormone that is crucial for the synchronization among the different circadian neurons (Renn et al., 1999). In crustaceans PDH has two distinct types, α-PDH and β-PDH, which are found in multiple isoforms (Strauss and Dircksen, 2010), and the neuroanatomy of PDH-expressing cells has been described in numerous species, with evidence of PDH involvement in circadian rhythmicity in several of them, such as the crab Cancer productus in which only one of two β-PDH isoforms was found to be expressed in the brain and was able to substitute for D. melanogaster PDF in pdf01 flies (Beckwith et al., 2011). While we have confirmation of expression in the head, the novel nature of the putative Ph-PDH requires further work to confirm its identity and to ascertain whether the sequence divergence has any impact on its functioning in the circadian pathways.
Given a lack of timepoint replicates in our head transcriptome data no firm conclusions can be drawn regarding rhythmic gene expression, though the data is suggestive of a bimodal pattern in a number of clock-related genes (Figure S1, Tables S2, S3). This transcriptomic data also suggests very low expression levels for the core clock genes in head tissue, low enough that only short fragments were assembled of Ph-bmal1 and Ph-period, and nothing of Ph-clock. This result is consistent with the isopod E. pulchra in which it has been determined that Ep-PERIOD is present in only 10 cells in the brain (Zhang et al., 2013).
This in silico analysis of these amphipod circadian systems has generated plenty of avenues for future enquiry. These sequences need to be confirmed through further sequencing, and their expression patterns more stringently characterized, both temporally and in terms of location using more extensive RNA-seq and/or RT-PCR. If it is found that Ph-CLK has been accurately described here, its ability to bind with Ph-BMAL1 and drive gene expression needs to be confirmed. The possibility of other partners for Ph-BMAL1 should also be investigated, and the functional significance of the N-terminus of Ph-CRY2 determined. Finally, further to the molecular work required it will be vital to develop behavioral and physiological assays that can be used to study the effects of clock gene manipulation. In Drosophila melanogaster a number of rhythmic phenomena have been identified including locomotor activity, eclosion, egg deposition, eye sensitivity, and oxygen consumption (Helfrich-Förster, 1996), while as a model organism P. hawaiensis has been largely exploited at a molecular level and often at early developmental stages, and behavioral observations are lacking. We have gathered encouraging preliminary locomotor activity results (Hunt, 2016), albeit compromised by issues with temperature control, hunger, age-matching and water evaporation, aeration and fouling. These results align with the observations of Poovachiranon et al. (1986) in indicating a nocturnal peak in activity that persists in constant darkness, with some individuals exhibiting 12 h periodic components, a finding that may be of interest to researchers studying circa-tidal rhythms given that P. hawaiensis is found in inter-tidal zones. We urge interested parties to work toward optimizing such behavioral assays and also to undertake the identification of other relevant characteristics that may be experimentally tractable. Only with this information in hand can one begin the task of employing P. hawaiensis to bring about new insights in crustacean chronobiology, a field with a rich research history that is ripe for significant advancement through the exploitation of a model with a diverse and powerful genetic toolkit.
Data Availability Statement
The P. hawaiensis head transcriptome is archived at the NCBI SRA under project PRJNA399131. Newly identified/expanded putative coding and peptide sequences of core clock genes are detailed in Supplementary Material.
Author Contributions
BH performed RNA extraction, de novo assembly, genome and transcriptome analysis, and drafted the manuscript. ER and EM conceived the study. All authors approved the final manuscript.
Funding
This work was funded by the Natural Environment Research Council (NERC) UK through an Antarctic Funding Initiative grant NE/D008719/1 (ER) and a Ph.D. studentship (BH). EM was supported by research grant NE/N010019/1 from the Natural Environment Research Council, UK.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are deeply grateful to Ana Patricia Ramos and Michalis Averof for supplying us with animals and husbandry advice and for permitting us to search an unpublished leg and embryo tissue assembly which proved vital in identifying the PER fragment in our own head assembly. Thanks are also due to Damian Kao, Aziz Aboobaker, Anastasios Pavlopoulos and all other authors of Kao et al. (2016) for granting us access to the then unpublished P. hawaiensis genome. Parts of this research first appeared in the thesis of BH (Hunt, 2016). This research used the ALICE High Performance Computing Facility at the University of Leicester.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.01325/full#supplementary-material
References
Allada, R., White, N. E., So, W., Hall, J. C., and Rosbash, M. (1998). A mutant Drosophila momolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93, 791–804. doi: 10.1016/S0092-8674(00)81440-3
Andrews, S. (2010). FastQC a Quality Control Tool for High Throughput Sequence Data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Armenteros, J. J. A., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Beckwith, E. J., Lelito, K. R., Hsu, Y.-W. A., Medina, B. M., Shafer, O., Ceriani, M. F., et al. (2011). Functional conservation of clock output signaling between flies and intertidal crabs. J. Biol. Rhyth. 26, 518–529. doi: 10.1177/0748730411420242
Biscontin, A., Wallach, T., Sales, G., Grudziecki, A., Janke, L., Sartori, E., et al. (2017). Functional characterization of the circadian clock in the Antarctic krill, Euphausia superba. Sci. Rep. 7:17742. doi: 10.1038/s41598-017-18009-2
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Browne, W. E., Price, A. L., Gerberding, M., and Patel, N. H. (2005). Stages of embryonic development in the amphipod crustacean, Parhyale hawaiensis. Genesis 42, 124–149. doi: 10.1002/gene.20145
Chang, Z., Li, G., Liu, J., Zhang, Y., Ashby, C., Liu, D., et al. (2015). Bridger: a new framework for de novo transcriptome assembly using RNA-seq data. Genome Biol. 16:30. doi: 10.1186/s13059-015-0596-2
Chesmore, K. N., Watson, W. H., and Chabot, C. C. (2016). Identification of putative circadian clock genes in the American horseshoe crab, Limulus polyphemus. Comparat. Biochem. Physiol. D 19, 45–61. doi: 10.1016/j.cbd.2016.06.001
Christie, A. E. (2014). Identification of the first neuropeptides from the Amphipoda (Arthropoda, Crustacea). Gen. Comparat. Endocrinol. 206, 96–110. doi: 10.1016/j.ygcen.2014.07.010
Christie, A. E., Fontanilla, T. M., Nesbit, K. T., and Lenz, P. H. (2013). Prediction of the protein components of a putative Calanus finmarchicus (Crustacea, Copepoda) circadian signaling system using a de novo assembled transcriptome. Compar. Biochem. Physiol. D Genom. Proteom. 8, 165–193. doi: 10.1016/j.cbd.2013.04.002
Christie, A. E., Yu, A., Pascual, M. G., Roncalli, V., Cieslak, M. C., Warner, A. N., et al. (2018a). Circadian signaling in Homarus americanus: region-specific de novo assembled transcriptomes show that both the brain and eyestalk ganglia possess the molecular components of a putative clock system. Mar. Genom. 40, 25–44. doi: 10.1016/j.margen.2018.03.002
Christie, A. E., Yu, A., Roncalli, V., Pascual, M. G., Cieslak, M. C., Warner, A. N., et al. (2018b). Molecular evidence for an intrinsic circadian pacemaker in the cardiac ganglion of the American lobster, Homarus americanus - Is diel cycling of heartbeat frequency controlled by a peripheral clock system? Mar. Genom. 41, 19–30. doi: 10.1016/j.margen.2018.07.001
De Coursey, P. J. (1960). Daily light sensitivity rhythm in a rodent. Science 131, 33–35. doi: 10.1126/science.131.3392.33
Emery, P., So, W., Kaneko, M., Hall, J. C., and Rosbash, M. (1998). CRY, a < i>Drosophila < \i> clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95, 669–679. doi: 10.1016/S0092-8674(00)81637-2
Gamble, F. W., and Keeble, F. W. (1900). Hippolyte varians: a study in colour-change. Q. J. Microse Sci. 43, 589–703.
Gekakis, N., Staknis, D., Nguyen, H. B., Davis, F. C., Wilsbacher, L. D., King, D. P., et al. (1998). Role of the CLOCK protein in the mammalian circadian mechanism. Science (New York, N.Y.) 280, 1564–1569. doi: 10.1126/science.280.5369.1564
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Helfrich-Förster, C. (1996). Drosophila rhythms: from brain to behavior. Semin. Cell Dev. Biol. 7, 791–802. doi: 10.1006/scdb.1996.0097
Helfrich-Förster, C. (2005). Neurobiology of the fruit fly's circadian clock. Genes Brain Behav. 4, 65–76. doi: 10.1111/j.1601-183X.2004.00092.x
Hogenesch, J. B., Gu, Y. Z., Jain, S., and Bradfield, C. A. (1998). The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. U.S.A. 95, 5474–5479. doi: 10.1073/pnas.95.10.5474
Hunt, B. J. (2016). Advancing molecular crustacean chronobiology through the characterisation of the circadian clock in two malacostracan species, Euphausia superba and Parhyale hawaiensis. (Thesis). Leicester, UK: University of Leicester.
Hunt, B. J., Özkaya, Ö., Davies, N. J., Gaten, E., Seear, P., Kyriacou, C. P., et al. (2017). The Euphausia superba transcriptome database, SuperbaSE: An online, open resource for researchers. Ecol. Evol. 7, 6060–6077. doi: 10.1002/ece3.3168
Kao, D., Lai, A. G., Stamataki, E., Rosic, S., Konstantinides, N., Jarvis, E., et al. (2016). The genome of the crustacean Parhyale hawaiensis, a model for animal development, regeneration, immunity and lignocellulose digestion. 5:e20062. doi: 10.7554/eLife.20062
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Konopka, R. J., and Benzer, S. (1971). Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 68, 2112–2116. doi: 10.1073/pnas.68.9.2112
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucl. Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Nesbit, K. T., and Christie, A. E. (2014). Identification of the molecular components of a Tigriopus californicus (Crustacea, Copepoda) circadian clock. Compar. Biochem. Physiol. D 12, 16–44. doi: 10.1016/j.cbd.2014.09.002
O'Grady, J. F., Hoelters, L. S., Swain, M. T., and Wilcockson, D. C. (2016). Identification and temporal expression of putative circadian clock transcripts in the amphipod crustacean Talitrus saltator. PeerJ 4:e2555. doi: 10.7717/peerj.2555
Palmer, J. D. (1991). Contributions made to chronobiology by studies of fiddler crab rhythms. Chronobiol. Int. 8, 110–130. doi: 10.3109/07420529109059163
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Poovachiranon, S., Boto, K., and Duke, N. (1986). Food preference studies and ingestion rate measurements of the mangrove amphipod Parhyale hawaiensis (dana). J. Exp. Mar. Biol. Ecol. 98, 129–140. doi: 10.1016/0022-0981(86)90078-X
R Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Renn, S. C., Park, J. H., Rosbash, M., Hall, J. C., and Taghert, P. H. (1999). A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802. doi: 10.1016/S0092-8674(00)81676-1
Reppert, S. M., and Weaver, D. R. (2000). Comparing clockworks: mouse versus fly. J. Biol. Rhyth. 15, 357–364. doi: 10.1177/074873000129001459
Reppert, S. M., and Weaver, D. R. (2001). Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63, 647–676. doi: 10.1146/annurev.physiol.63.1.647
Robertson, G., Schein, J., Chiu, R., Corbett, R., Field, M., Jackman, S. D., et al. (2010). De novo assembly and analysis of RNA-seq data. Nat. Methods 7, 909–912. doi: 10.1038/nmeth.1517
Robinson, J. T., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. doi: 10.1038/nbt.1754
Robinson, M. D., and Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25–R25. doi: 10.1186/gb-2010-11-3-r25
Rutila, J. E., Suri, V., Le, M., So, W. V., Rosbash, M., and Hall, J. C. (1998). Cycle is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93, 805–814. doi: 10.1016/S0092-8674(00)81441-5
Saleem, Q., Anand, a., Jain, S., and Brahmachari, S. K. (2001). The polyglutamine motif is highly conserved at the Clock locus in various organisms and is not polymorphic in humans. Hum. Genet. 109, 136–142. doi: 10.1007/s004390100550
Sandrelli, F., Costa, R., Kyriacou, C. P., and Rosato, E. (2008). Comparative analysis of circadian clock genes in insects. Insect Mol. Biol. 17, 447–463. doi: 10.1111/j.1365-2583.2008.00832.x
Sbragaglia, V., Lamanna, F., M. Mat, A., Rotllant, G., Joly, S., Ketmaier, V., et al. (2015). Identification, characterization, and diel pattern of expression of canonical clock genes in Nephrops norvegicus (Crustacea: Decapoda) eyestalk. PLoS ONE 10:e0141893. doi: 10.1371/journal.pone.0141893
Sehgal, A., Price, J. L., Man, B., and Young, M. W. (1994). Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 263, 1603–1606. doi: 10.1126/science.8128246
Shin, S. W., Zou, Z., Saha, T. T., and Raikhel, A. S. (2012). bHLH-PAS heterodimer of methoprene-tolerant and Cycle mediates circadian expression of juvenile hormone-induced mosquito genes. Proc. Natl. Acad. Sci. U.S.A. 109, 16576–16581. doi: 10.1073/pnas.1214209109
Smith-Unna, R., Boursnell, C., Patro, R., Hibberd, J. M., and Kelly, S. (2016). TransRate: reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 26, 1134–1144. doi: 10.1101/gr.196469.115
Strauss, J., and Dircksen, H. (2010). Circadian clocks in crustaceans: identified neuronal and cellular systems. Front. Biosci. 15, 1040–1074. doi: 10.2741/3661
Tilden, A. R., McCoole, M. D., Harmon, S. M., Baer, K. N., and Christie, A. E. (2011). Genomic identification of a putative circadian system in the cladoceran crustacean Daphnia pulex. Comparat. Biochem. Physiol. D 6, 282–309. doi: 10.1016/j.cbd.2011.06.002
Waterhouse, R. M., Seppey, M., Simão, F. A., Manni, M., Ioannidis, P., Klioutchnikov, G., et al. (2018). BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35, 543–548. doi: 10.1093/molbev/msx319
Yang, J. S., Dai, Z. M., Yang, F., and Yang, W. J. (2006). Molecular cloning of Clock cDNA from the prawn, Macrobrachium rosenbergii. Brain Res. 1067, 13–24. doi: 10.1016/j.brainres.2005.10.003
Zhang, L., Hastings, M. H., Green, E. W., Tauber, E., Sladek, M., Webster, S. G., et al. (2013). Dissociation of circadian and circatidal timekeeping in the marine crustacean Eurydice pulchra. Curr. Biol. 23, 1863–1873. doi: 10.1016/j.cub.2013.08.038
Keywords: crustacean, amphipod, parhyale, talitrus, hyalella, circadian, clock, genetics
Citation: Hunt BJ, Mallon EB and Rosato E (2019) In silico Identification of a Molecular Circadian System With Novel Features in the Crustacean Model Organism Parhyale hawaiensis. Front. Physiol. 10:1325. doi: 10.3389/fphys.2019.01325
Received: 16 July 2019; Accepted: 03 October 2019;
Published: 18 October 2019.
Edited by:
Nicholas Simon Foulkes, Karlsruhe Institute of Technology (KIT), GermanyReviewed by:
David Wilcockson, Aberystwyth University, United KingdomJoe Hull, Agricultural Research Service, United States Department of Agriculture, United States
Maria Fernanda Ceriani, Leloir Institute Foundation (FIL), Argentina
Copyright © 2019 Hunt, Mallon and Rosato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ezio Rosato, ZXI2QGxlLmFjLnVr
 Benjamin James Hunt
Benjamin James Hunt Eamonn B. Mallon
Eamonn B. Mallon Ezio Rosato
Ezio Rosato