- 1Department of Gastrointestinal Surgery, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, China
- 2The Second Clinical College, Guangdong Provincial Hospital of Chinese Medicine, Guangzhou University of Chinese Medicine, Guangzhou, China
- 3The First Clinical College, Guangzhou University of Chinese Medicine, Guangzhou, China
- 4School of Basic Medical Science, Guangzhou University of Chinese Medicine, Guangzhou, China
- 5Department of Laboratory Medicine, Zhongnan Hospital of Wuhan University, Wuhan University, Wuhan, China
Background: There is debate as to whether c-Myc predicts prognosis in colorectal cancer (CRC). In this study, we aimed to review the association between c-Myc and CRC prognosis.
Methods: Pertinent studies were identified by searching electronic databases and carefully reviewing the reference lists of pertinent studies until March 2016. The summary hazard ratio (HR) and corresponding 95% confidence interval (CI) were calculated to study the association between c-Myc and CRC prognosis.
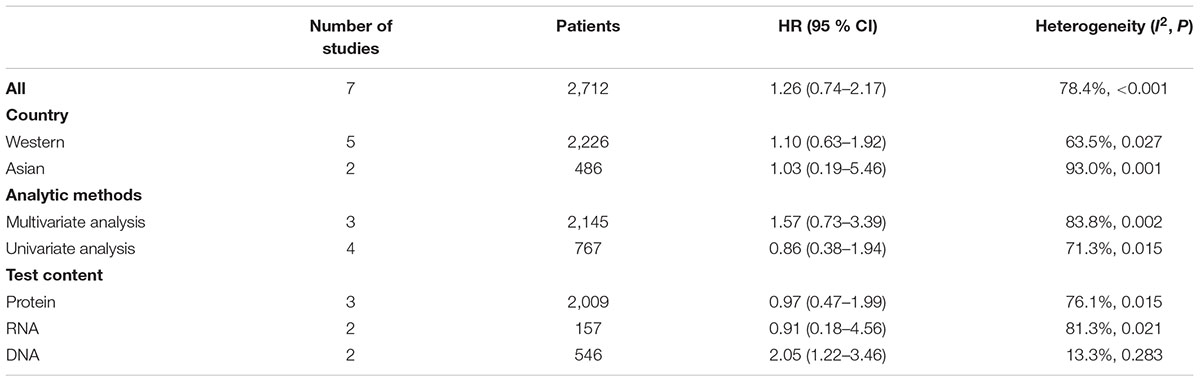
Results: Eight cohort studies (including seven studies about overall survival [OS] and one study about disease free survival [DFS]) were included. The pooled HR of OS was 1.13 (95% CI: 0.66–1.95). In subgroup analysis, no significant association between c-Myc and CRC prognosis was found in the studies either from Western countries (HR: 0.87, 95% CI: 0.68–1.10) or Asian countries (HR: 1.89, 95% CI: 0.62–5.77). HRs were 0.86 (95% CI: 0.38–1.94) and 1.57 (95% CI: 0.73–3.39) for the studies using univariate analysis and multivariate analysis, respectively. HR from the studies that examined DNA level was significantly different (HR: 2.05, 95% CI: 1.22–3.46); while that about RNA level or protein level was not significantly different.
Conclusion: c-Myc was not associated with CRC prognosis in this meta-analysis. However, the conclusion is preliminary and should be examined in future studies.
Introduction
Colorectal cancer (CRC) is the third most frequent cancer worldwide and the fourth most common cause of cancer-related death (Torre et al., 2015). There are approximately 1.4 million new cases and 700,000 deaths from CRC each year (Torre et al., 2015). CRC is a heterogeneous and complicated disease affected by both environmental and genetic factors. A number of cancer-related genes are correlated with CRC prognosis, but the survival benefit associated with targeted therapies is only 4–5 months (Bokemeyer et al., 2012), indicating that the precise molecular mechanisms of CRC are unclear.
The Myc family encodes three highly related nuclear phosphoproteins: c-Myc, l-Myc, and n-Myc (Mukherjee et al., 1992). c-Myc functions as an oncogene, participating in cell growth, death, transformation, and therapy sensitivity (Hermeking and Eick, 1994; Lee K.B. et al., 2016; Wang et al., 2016). The c-Myc protein occupies regulatory regions of up to 15% of all genes and can both activate or suppress various target genes (Dang et al., 2006; Feng et al., 2016). The target genes of c-Myc are involved in various cellular functions, including survival, cell cycle, protein synthesis, cell adhesion, and non-coding RNA expression (Dang et al., 2006).
Aberrant expression of c-Myc was observed in many human cancers and was elevated in up to 70–80% of CRC (Erisman et al., 1985). Several studies have focused on the association between c-Myc and CRC prognosis. Bhatavdekar et al. (1997) reported that measurement of c-Myc expression in primary CRC tissue did not predict prognosis (Erisman et al., 1988). However, some studies showed that positive c-Myc expression had the strongest association with poor survival in CRC patients (Rowley et al., 1990; Bhatavdekar et al., 1997; Kakisako et al., 1998; Lee et al., 2015). For example, Lee et al. (2015) indicated that c-Myc was an independent factor for poor prognosis in consecutive CRC patients according to multivariate analysis. On the other hand, several studies found that c-Myc was correlated with a favorable prognosis of CRC patients (Smith and Goh, 1996; Bockleman et al., 2012; Toon et al., 2014). For example, Smith and Goh (1996) demonstrated that overexpression of c-Myc mRNA in CRC tumors was associated with a better prognosis. All these findings suggested that the prognostic value of c-Myc in CRC remained controversial and inconclusive. Therefore, we conducted a meta-analysis to evaluate the association between c-Myc and CRC prognosis.
Materials and Methods
Literature Search
The Research Ethics Committee of Guangzhou University of Chinese Medicine provided ethical approval. PubMed, EMBASE, ISI Web of Knowledge, and the Cochrane Database were searched for eligible studies up to March 14th, 2016. The search strategy was carried out using the following words: “colorectal” (large intestine, large bowel, colon, colonic, rectal or rectum), “cancer” (carcinoma, tumor, neoplasm or cancers), “c-Myc” and “prognosis” (prognoses, prognostic, predictive, biomarker, marker, survival, survive, cox, log-rank or Kaplan-Meier). The search strategy for the Pubmed database was shown in Appendix 1. The reference lists of pertinent publications were also checked for the eligible studies. Only studies published in English were included. In case of duplicate reports or of studies obviously reporting results from the same study, only the latest published studies were selected. This meta-analysis was performed according to the preferred reporting items for systematic reviews and meta-analysis (PRISMA statement) (Moher et al., 2009). The PRISMA 2009 Checklist was shown in Appendix 2.
Selection Criteria
The inclusion and exclusion criteria consisted of the following three aspects: (1) studies of colorectal cancer (including colon cancer, or rectal cancer) were included; (2) the relationship between c-Myc and patients’ prognosis [i.e., overall survival (OS), disease free survival (DFS), or relapse free survival (RFS)] was studied; and the hazard ratio (HR) and its 95% confidence interval (CI) were provided; and (3) studies were published in the English language. The eligible studies included cohort studies and randomized control trials.
Data Extraction
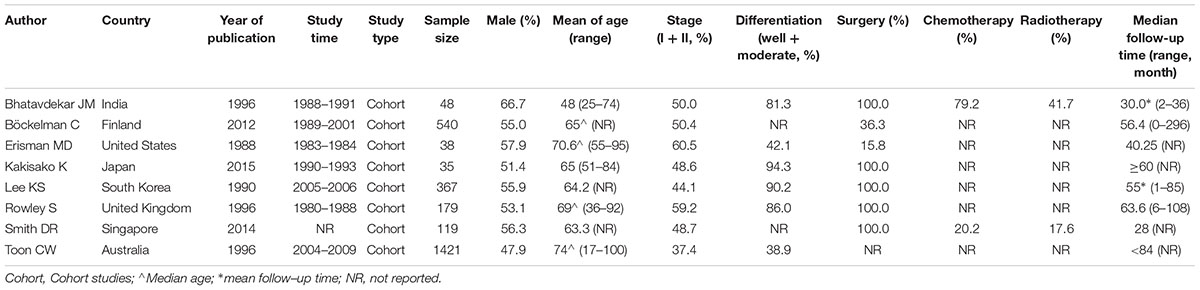
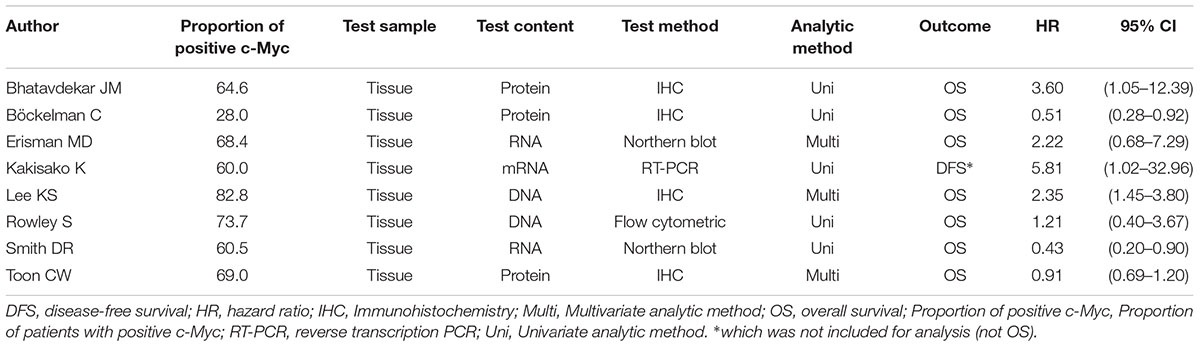
The titles and abstracts of all the studies were screened by two of three reviewers independently (X-tW, J-lW, and Y-kL). The eligible or uncertain studies were retrieved for the full texts. Two of three reviewers (X-tW, J-lW and Y-kL) read the full texts and identified the eligible publications. For each eligible study, the following information was extracted: first author, year of publication, country of origin, study time, study type, sample sizes, the characteristics of the patients (gender, stage, differentiation, and treatment method), median follow-up time, the c-Myc information (proportion of positive c-Myc, test sample, test content, and analytic method) and prognosis. Country of origin was categorized as Western countries and Asian countries. Disagreements in data collection were resolved by consensus.
Statistical Analysis
The association between c-Myc and CRC survival was examined using HR with its 95% CI. DFS and OS were analyzed separately.
The heterogeneity of the individual HR was calculated using Chi-square tests. A heterogeneity test with inconsistency index statistic (I2) and Q statistic were carried out (Handoll, 2006). The Q test suggested lack of heterogeneity when P > 0.10, and summary HR was examined using fixed-effect model (Mantel and Haenszel, 1959). Otherwise, random-effect model was executed (DerSimonian and Laird, 1986). Subgroup analysis were conducted according to different countries (West [Europe and America], and Asia), analytic methods (univariate analysis, multivariate analysis) and test content (Protein, DNA, RNA). Meta-regression was performed to find out the factors related with the heterogeneity of the HRs. A sensitivity analysis was carried out to evaluate the stability of the results. In addition, Egger’s test and funnel plots were utilized to evaluate publication bias. All statistical analyses were conducted using STATA software (version 12.0).
Results
Characteristics of Studies
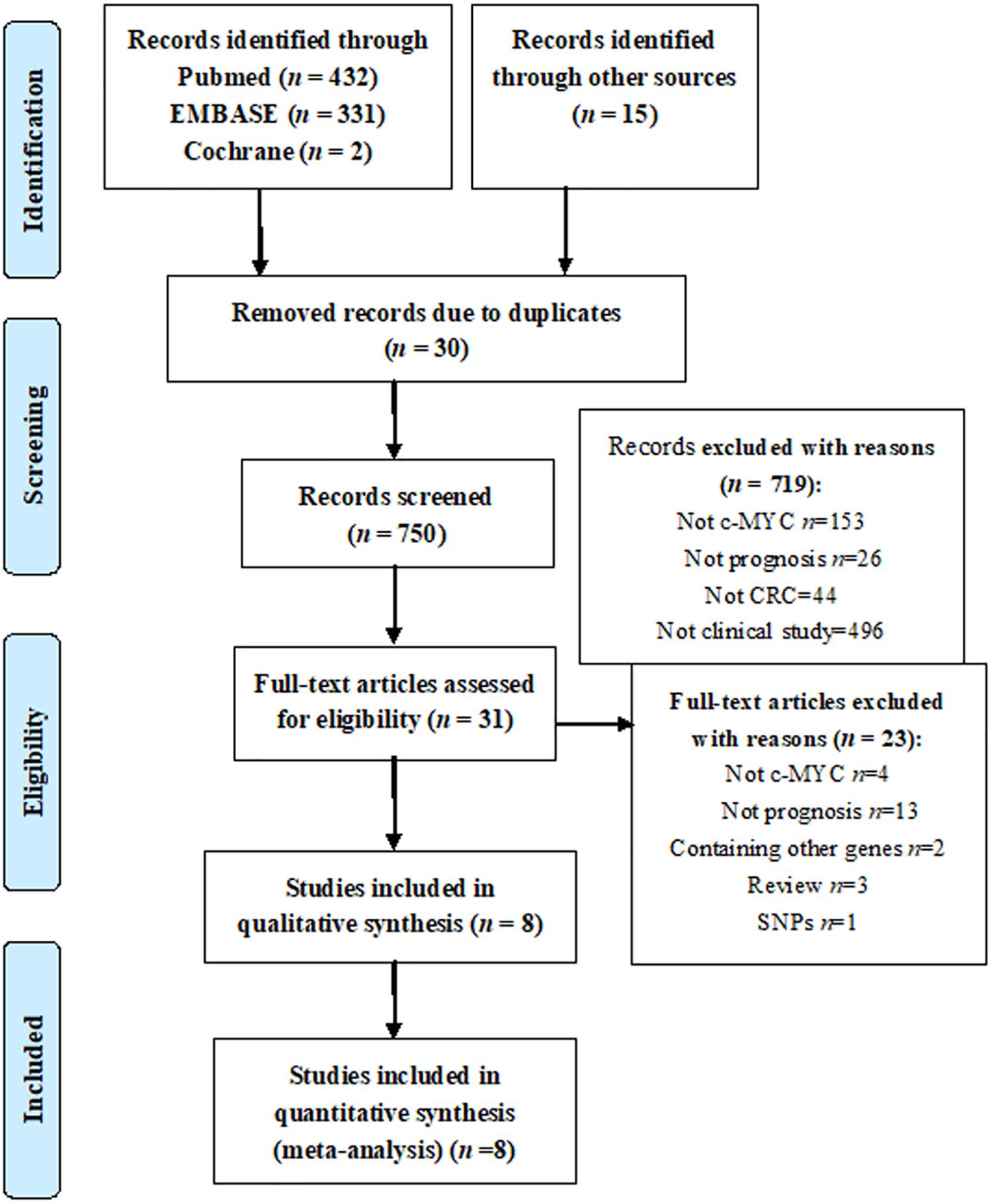
The initial search strategy identified 780 potentially eligible studies. Thirty studies were excluded because of duplication. We excluded 719 studies after detailed review of the abstract. The remaining 31 studies were evaluated for the full texts. Four studies did not involve c-Myc, thirteen studies did not deal with prognosis, two included other genes, three were review articles, and one was about single-nucleotide polymorphism and was therefore excluded. Eventually, we included eight studies in our meta-analysis (Figure 1; Erisman et al., 1988; Rowley et al., 1990; Smith and Goh, 1996; Bhatavdekar et al., 1997; Kakisako et al., 1998; Bockleman et al., 2012; Toon et al., 2014; Lee et al., 2015).
Three studies were from Asian countries (Smith and Goh, 1996; Bhatavdekar et al., 1997; Kakisako et al., 1998; Lee et al., 2015), and others were from Western countries. A total of 2,947 patients were included (Table 1). All of the eligible studies were cohort studies. The proportion of patients with positive c-Myc was ≥60%, except the study by Bockleman et al. (2012) (Table 2). One study reported DFS, while others reported OS (Table 2). The HR from the only one study about DFS of c-Myc was 5.81 (95% CI: 1.02–32.96; 35 patients). The following results were based on OS.
Meta-Analysis About OS of c-Myc in CRC Patients
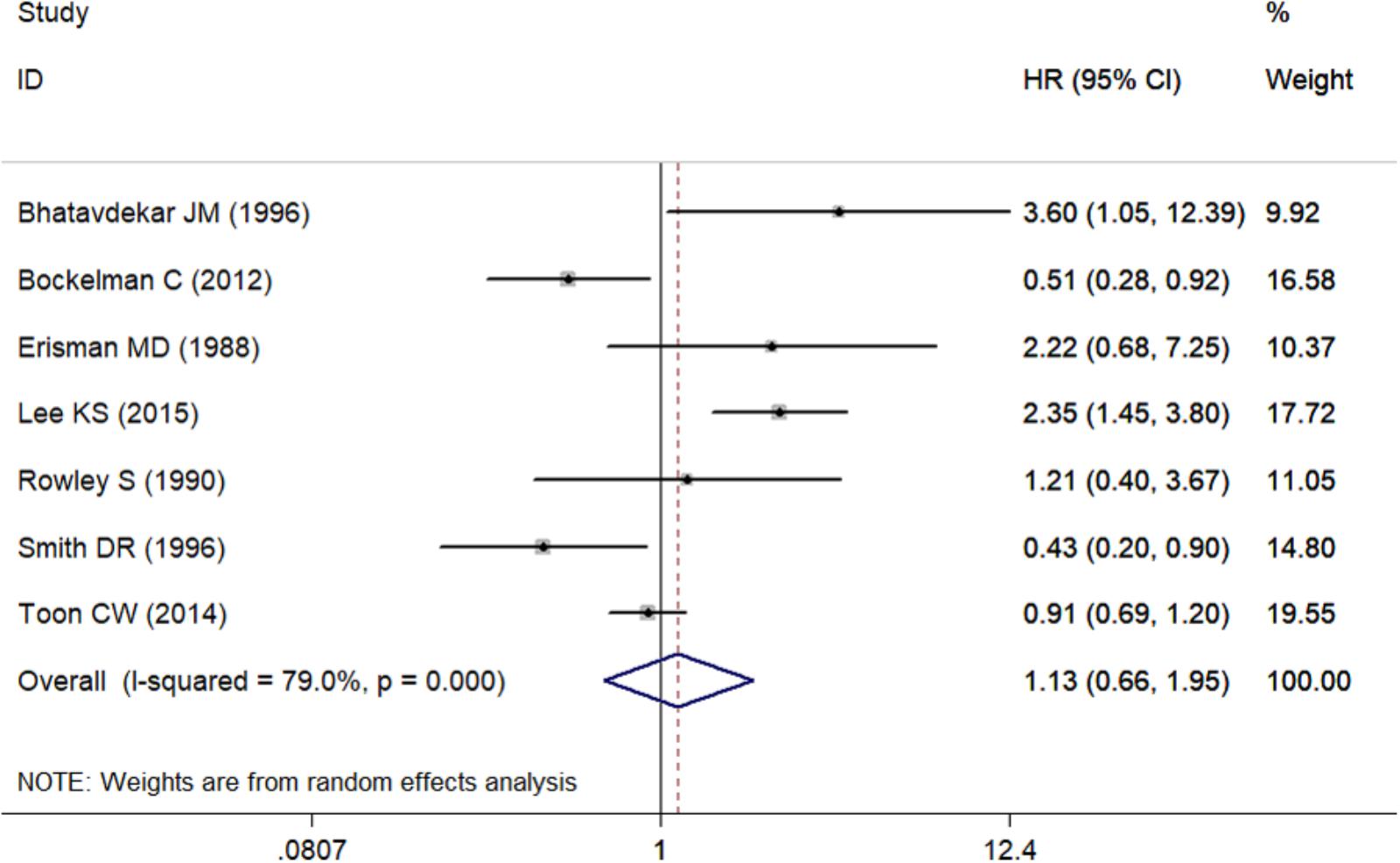
Seven studies including 2,712 CRC patients were involved (Table 2). The prognostic roles of c-Myc in CRC were summarized in Figure 2. Inconsistent HRs were observed among studies, suggesting either favorable or poor prognostic roles of c-Myc in CRC. A random-effects model was executed to obtain an unadjusted pooled HR of 1.13 (95% CI: 0.66–1.96, I2 = 79.0%, P < 0.001).
Subgroup Analysis
The pooled HR for studies from Western countries was 1.10 (95% CI: 0.63–1.92; I2 = 63.5%, P = 0.027, Figure 3 and Table 3). For studies from Asian countries, the pooled HR was 1.03 (95% CI: 0.19–5.46; I2 = 93.0%, P < 0.001, Figure 3 and Table 3).
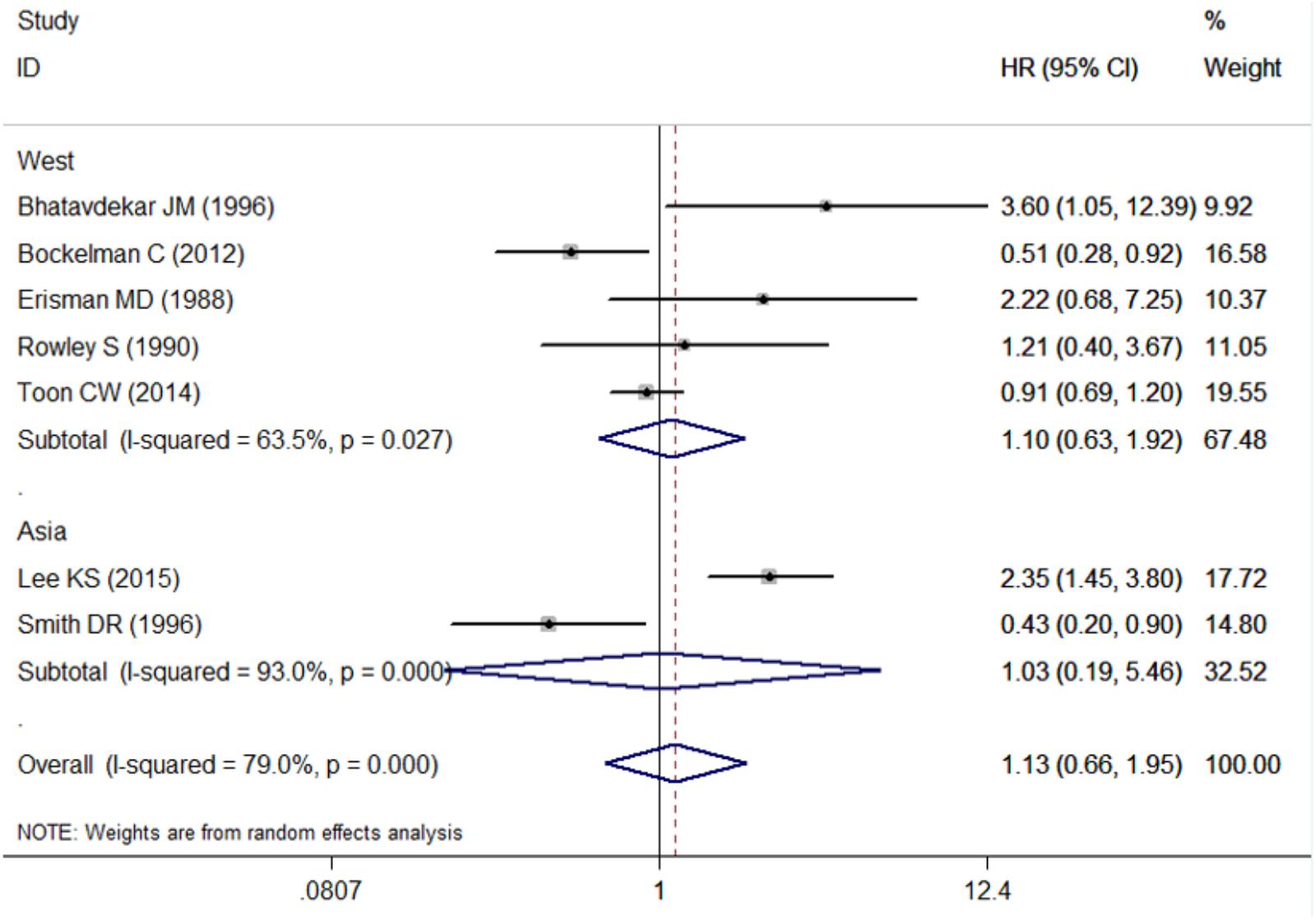
FIGURE 3. Subgroup analysis for the association between c-Myc and overall survival in the studies from different countries. West, western countries; Asia, Asian countries.
Pooled HR was 1.57 (95% CI: 0.73–3.39) by combining three studies that provided multivariate analysis (P = 0.002, I2 = 83.8%, Figure 4 and Table 3). In addition, the pooled HR from four studies providing univariate analysis was 0.86 (95% CI: 0.38–1.94) based on the result of random-effect model (P = 0.015, I2 = 71.3%, Figure 4 and Table 3).
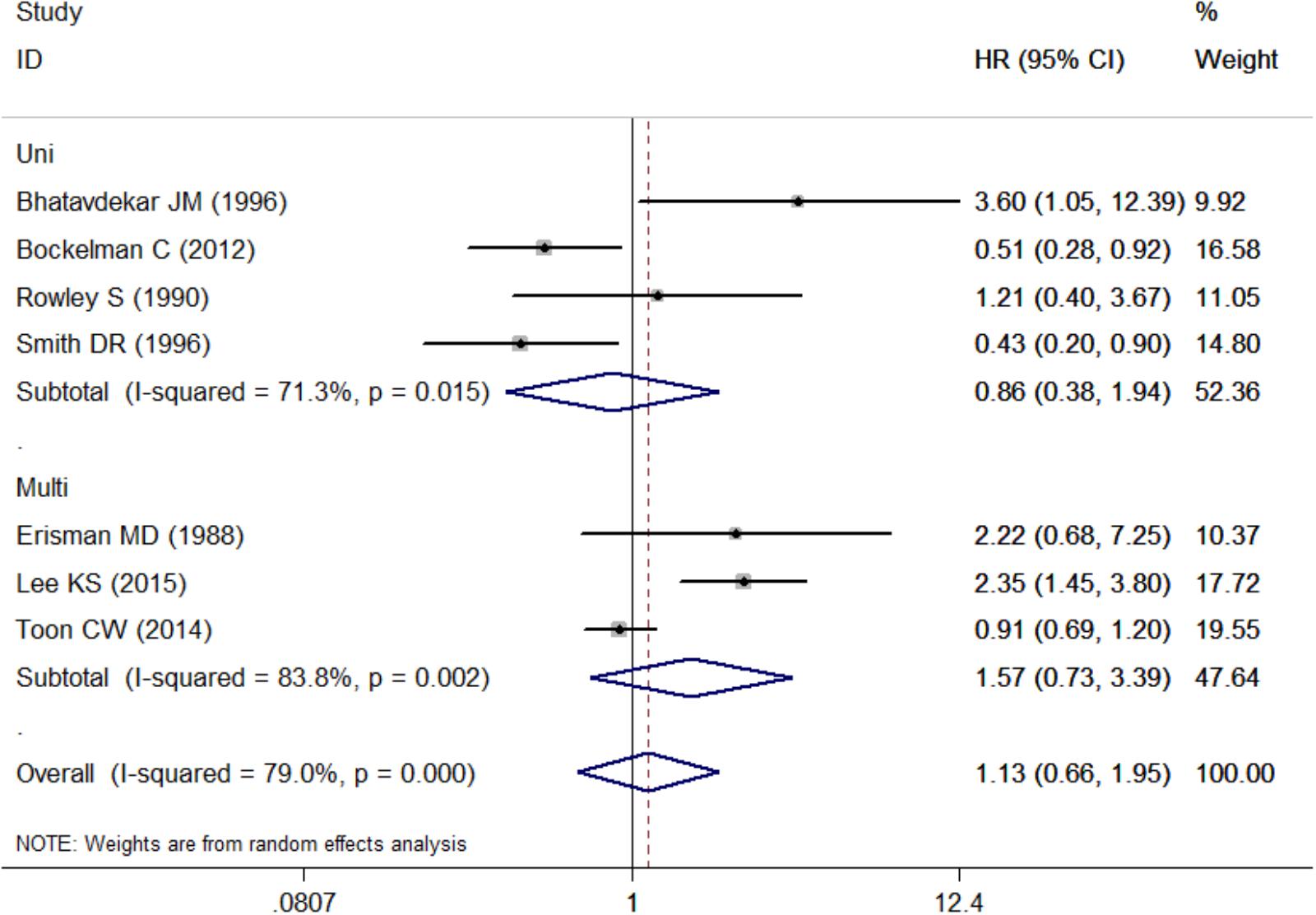
FIGURE 4. Subgroup analysis for the association between c-Myc and overall survival in the studies using different analytic methods. Uni, univariate analysis; Multi, multivariate analysis.
Three studies examined protein level of c-Myc, two studies examined RNA level, while two studies examined DNA level. Pooled HR was 0.97 (95% CI: 0.47–1.99, Figure 5 and Table 3) for protein level of c-Myc, and 0.91 (95% CI: 0.18–4.56) for RNA level. HR from three studies that examined DNA level was 2.05 (95% CI: 1.22–3.46).
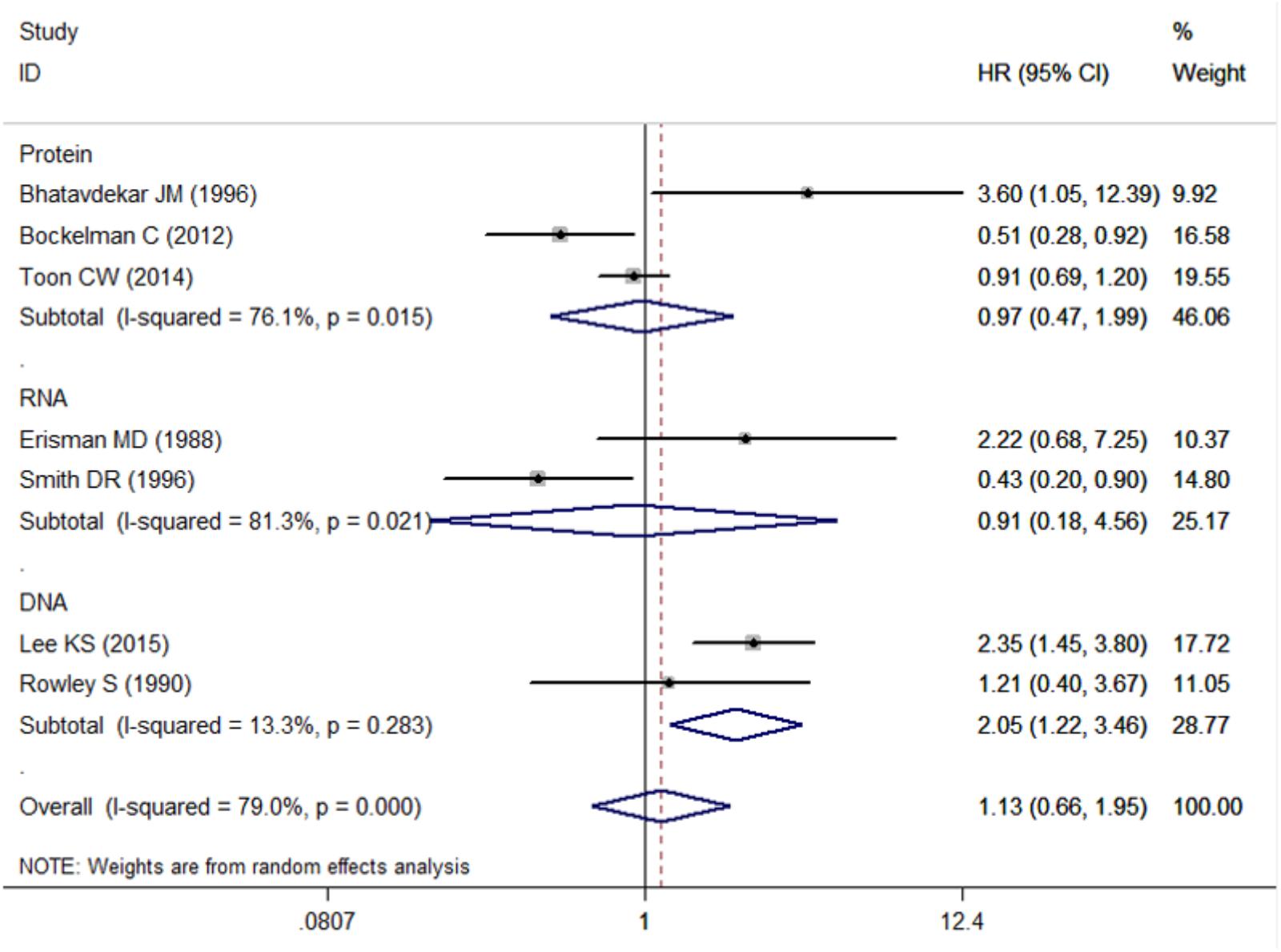
FIGURE 5. Subgroup analysis for the association between c-Myc and overall survival in the studies using different test content (including Protein, DNA, RNA).
Analysis of Heterogeneity
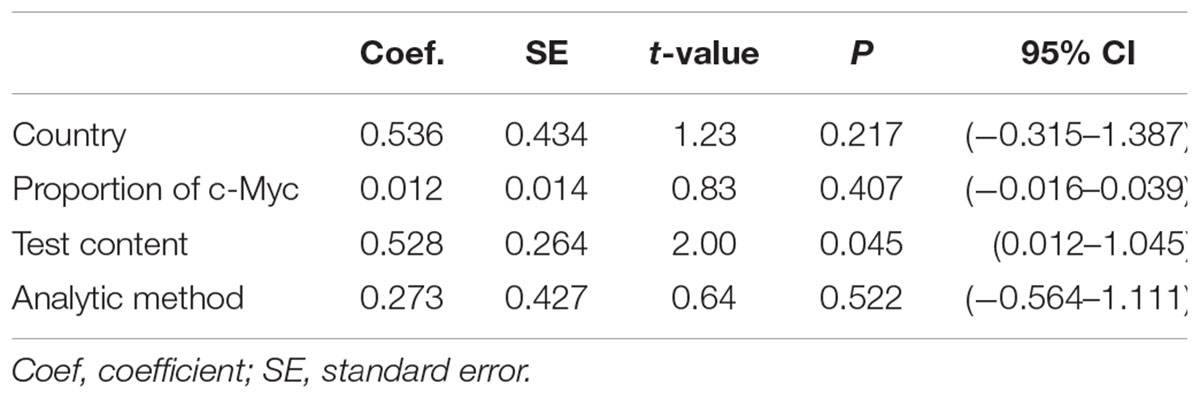
There was significant heterogeneity for OS among seven studies (Figure 2). Meta-regression was performed. The variable “Test content” was related with the heterogeneity of the HRs (Table 4). Sensitivity analysis (Figure 6A) and funnel plot (Figure 6B) were carried out to evaluate the influence of potential publication bias. We did not observe significantly publication bias from egger’s test (P = 0.368). However, the shape of the funnel plot indicated some studies were out of the reference line (Figure 6B). Each study in sensitivity analysis was successively removed to evaluate the effect of individual study on the pooled HR (Figure 6A). The results showed that the studies conducted by Bockleman et al. (2012); Toon et al. (2014) were out of the reference line, which demonstrated that there might be publication bias for OS.
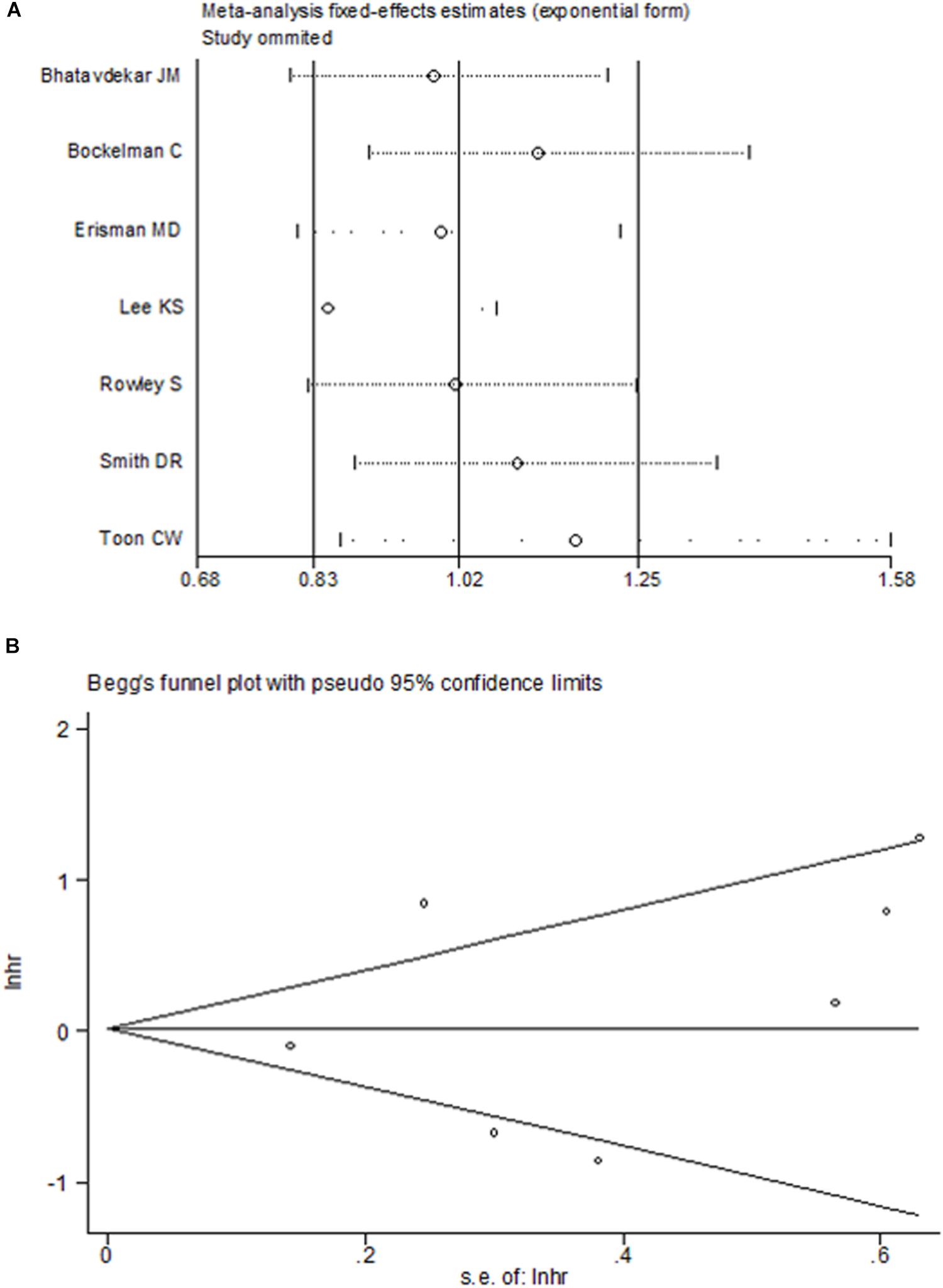
FIGURE 6. Sensitive analysis (A) and Begg’s funnel plot (B) for the assessment of included studies in overall survival.
Discussion
This study is the first meta-analysis to examine the association between c-Myc and CRC prognosis. We found that c-Myc was not significantly associated with CRC prognosis.
c-Myc participates in cell proliferation, differentiation, metabolism, survival, and apoptosis by regulating human genes (Guo et al., 2016; Su et al., 2016; Subramaniam et al., 2016). The c-Myc gene can promote tumourigenesis in many types of cancers (Aprelikova et al., 2016; Richart et al., 2016) and plays an important role in the progression of CRC (Smith and Goh, 1996; Kriegl et al., 2012).
Several studies have reported c-Myc status in many cancers, including prostate cancer (Zeng et al., 2015), breast cancer (Elster et al., 2016), and CRC (Lee K.S. et al., 2016). Some cancers with c-Myc overexpression, including oesophageal squamous cell carcinoma, gastric carcinoma, and soft tissue leiomyosarcoma, are correlated with poor survival (Ninomiya et al., 1991; Tsiatis et al., 2009; Wang et al., 2011). Likewise, several cancers with c-Myc gene amplification were associated with poor survival (Dimova et al., 2006; Choi et al., 2012; Seo et al., 2014). However, the prognostic value of c-Myc in CRC patients is quite controversial. It was reported that overexpression of either c-Myc mRNA or c-Myc protein in CRC patients was associated with favorable survival (Smith and Goh, 1996; Toon et al., 2014), but these were opposite results to previous studies that showed that high expression of c-Myc in CRC predicted worse survival outcome (Erisman et al., 1988). The association between c-Myc expression and CRC patients’ prognosis remains debatable. Therefore, it is required to further estimate c-Myc expression in CRC to obtain a conclusion regarding its prognostic value. Therefore, a meta-analysis including 2,947 CRC patients was performed. It was demonstrated that the c-Myc was not significantly associated with CRC prognosis in the overall investigated populations.
In subgroup analysis by ethnicity, we did not detect significant association between c-Myc and survival in either Europeans or Asians, indicating that ethnic differences in genetic backgrounds and the lifestyle context do not influence the association between c-Myc and CRC prognosis.
Nevertheless, there were some limitations in our study. First, adjusted confounding factors, including BMI and environmental factors, varied among studies. What was more, the method of therapy greatly affected the survival time of the CRC patients. Although all of the included patients were diagnosed as CRC, the use of specific therapy differed among the included studies. Thus, the confounding effects of different therapies remain unclear. Second, publication bias was observed among the studies, it might be inevitable due to unpublished studies or original data. Third, test content and evaluation criteria of c-Myc varied among studies, possibly giving rise to significant heterogeneity. HR from three studies that examined DNA level was significantly different, while those about RNA level or protein level were not significantly different. Fourth, only eight studies were enrolled in the meta-analysis, and each study included a relatively small sample size.
Overall, the meta-analysis indicates that c-Myc is not associated with CRC prognosis. However, due to the potential limitations, conclusions must be drawn with caution, and additional larger studies, particularly studies with sub-groups for environmental-genetic interactions, should be performed to validate our findings.
Author Contributions
W-LH and YP wrote the draft of the paper. X-TW extracted the data and helped to modify the manuscript. J-LW and Y-KL extracted the data. T-WL searched the databases. Q-YZ and YH analyzed the data. X-LC conceived the study, searched the databases and modified the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81774451, 81701834, and 81871994), the Outstanding Youth Foundation of Guangdong Province Colleges and Universities (YQ2015041), the Young Talents Foundation of Guangzhou University of Chinese Medicine (QNYC20140101), the Natural Science Foundation of Guangdong Province (2017A030313827, 2015A030313036), and Guangdong high level universities program of Guangzhou University of Chinese Medicine.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank JiaXing from the First Affiliated Hospital of Sun Yat-sen University for his suggestions about modifying the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01549/full#supplementary-material
References
Aprelikova, O., Chen, K., El Touny, L. H., Brignatz-Guittard, C., Han, J., Qiu, T., et al. (2016). The epigenetic modifier JMJD6 is amplified in mammary tumors and cooperates with c-Myc to enhance cellular transformation, tumor progression, and metastasis. Clin. Epigenetics 8:38. doi: 10.1186/s13148-016-0205-6
Bhatavdekar, J. M., Patel, D. D., Ghosh, N., Chikhlikar, P. R., Trivedi, T. I., Suthar, T. P., et al. (1997). Coexpression of Bcl-2, c-Myc, and p53 oncoproteins as prognostic discriminants in patients with colorectal carcinoma. Dis. Colon. Rectum 40, 785–790. doi: 10.1007/BF02055433
Bockleman, C., Koskensalo, S., Hagstrom, J., Lundin, M., Ristimaki, A., and Haglund, C. (2012). CIP2A overexpression is associated with c-Myc expression in colorectal cancer. Cancer Biol. Ther. 13, 289–295. doi: 10.4161/cbt.18922
Bokemeyer, C., Van Cutsem, E., Rougier, P., Ciardiello, F., Heeger, S., Schlichting, M., et al. (2012). Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur. J. Cancer 48, 1466–1475. doi: 10.1016/j.ejca.2012.02.057
Choi, J. S., Seo, J., Jung, E. J., Kim, E. J., Lee, G. K., and Kim, W. H. (2012). c-MYC amplification in mucinous gastric carcinoma: a possible genetic alteration leading to deeply invasive tumors. Anticancer Res. 32, 5031–5037.
Dang, C. V., O’donnell, K. A., Zeller, K. I., Nguyen, T., Osthus, R. C., and Li, F. (2006). The c-Myc target gene network. Semin. Cancer Biol. 16, 253–264. doi: 10.1016/j.semcancer.2006.07.014
DerSimonian, R., and Laird, N. (1986). Meta-analysis in clinical trials. Control Clin. Trials 7, 177–188. doi: 10.1016/0197-2456(86)90046-2
Dimova, I., Raitcheva, S., Dimitrov, R., Doganov, N., and Toncheva, D. (2006). Correlations between c-myc gene copy-number and clinicopathological parameters of ovarian tumours. Eur. J. Cancer 42, 674–679. doi: 10.1016/j.ejca.2005.11.022
Elster, D., Jaenicke, L. A., Eilers, M., and Von Eyss, B. (2016). TEAD activity is restrained by MYC and stratifies human breast cancer subtypes. Cell Cycle 15, 2551–2556. doi: 10.1080/15384101.2016.1207837
Erisman, M. D., Litwin, S., Keidan, R. D., Comis, R. L., and Astrin, S. M. (1988). Noncorrelation of the expression of the c-myc oncogene in colorectal carcinoma with recurrence of disease or patient survival. Cancer Res. 48, 1350–1355.
Erisman, M. D., Rothberg, P. G., Diehl, R. E., Morse, C. C., Spandorfer, J. M., and Astrin, S. M. (1985). Deregulation of c-myc gene expression in human colon carcinoma is not accompanied by amplification or rearrangement of the gene. Mol. Cell. Biol. 5, 1969–1976. doi: 10.1128/MCB.5.8.1969
Feng, X. H., Liang, Y. Y., Liang, M., Zhai, W., and Lin, X. (2016). Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitor p15(Ink4B). Mol. Cell 63:1089. doi: 10.1016/j.molcel.2016.08.027
Guo, J., Hao, J., Jiang, H., Jin, J., Wu, H., Jin, Z., et al. (2016). Proteasome activator subunit 3 promotes pancreatic cancer growth via c-Myc-glycolysis signaling axis. Cancer Lett. 386, 161–167. doi: 10.1016/j.canlet.2016.08.018
Handoll, H. H. (2006). Systematic reviews on rehabilitation interventions. Arch. Phys. Med. Rehabil. 87:875. doi: 10.1016/j.apmr.2006.04.006
Hermeking, H., and Eick, D. (1994). Mediation of c-Myc-induced apoptosis by p53. Science 265, 2091–2093. doi: 10.1126/science.8091232
Kakisako, K., Miyahara, M., Uchino, S., Adachi, Y., and Kitano, S. (1998). Prognostic significance of c-myc mRNA expression assessed by semi-quantitative RT-PCR in patients with colorectal cancer. Oncol. Rep. 5, 441–445. doi: 10.3892/or.5.2.441
Kriegl, L., Vieth, M., Kirchner, T., and Menssen, A. (2012). Up-regulation of c-MYC and SIRT1 expression correlates with malignant transformation in the serrated route to colorectal cancer. Oncotarget 3, 1182–1193. doi: 10.18632/oncotarget.628
Lee, K. B., Jin, H., Ye, S., Park, B. H., and Kim, S. M. (2016). Recombinant human bone morphogenetic protein-2 inhibits gastric cancer cell proliferation by inactivating Wnt signaling pathway via c-Myc with aurora kinases. Oncotarget 7, 73473–73485. doi: 10.18632/oncotarget.11969
Lee, K. S., Kwak, Y., Nam, K. H., Kim, D. W., Kang, S. B., Choe, G., et al. (2016). Favorable prognosis in colorectal cancer patients with co-expression of c-MYC and ss-catenin. BMC Cancer 16:730. doi: 10.1186/s12885-016-2770-7
Lee, K. S., Kwak, Y., Nam, K. H., Kim, D. W., Kang, S. B., Choe, G., et al. (2015). c-MYC copy-number gain is an independent prognostic factor in patients with colorectal cancer. PLoS One 10:e0139727. doi: 10.1371/journal.pone.0139727
Mantel, N., and Haenszel, W. (1959). Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 22, 719–748.
Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G., and Prisma Group. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6:e1000097. doi: 10.1371/journal.pmed.1000097
Mukherjee, B., Morgenbesser, S. D., and Depinho, R. A. (1992). Myc family oncoproteins function through a common pathway to transform normal cells in culture: cross-interference by max and trans-acting dominant mutants. Genes Dev. 6, 1480–1492. doi: 10.1101/gad.6.8.1480
Ninomiya, I., Yonemura, Y., Matsumoto, H., Sugiyama, K., Kamata, T., Miwa, K., et al. (1991). Expression of c-myc gene product in gastric carcinoma. Oncology 48, 149–153. doi: 10.1159/000226915
Richart, L., Carrillo-De Santa Pau, E., Rio-Machin, A., De Andres, M. P., Cigudosa, J. C., Lobo, V. J., et al. (2016). BPTF is required for c-MYC transcriptional activity and in vivo tumorigenesis. Nat Commun 7:10153. doi: 10.1038/ncomms10153
Rowley, S., Newbold, K. M., Gearty, J., Keighley, M. R., Donovan, I. A., and Neoptolemos, J. P. (1990). Comparison of deoxyribonucleic acid ploidy and nuclear expressed p62 c-myc oncogene in the prognosis of colorectal cancer. World J Surg. 14, 545–550; discussion 551. doi: 10.1007/BF01658688
Seo, A. N., Yang, J. M., Kim, H., Jheon, S., Kim, K., Lee, C. T., et al. (2014). Clinicopathologic and prognostic significance of c-MYC copy number gain in lung adenocarcinomas. Br. J. Cancer 110, 2688–2699. doi: 10.1038/bjc.2014.218
Smith, D. R., and Goh, H. S. (1996). Overexpression of the c-myc proto-oncogene in colorectal carcinoma is associated with a reduced mortality that is abrogated by point mutation of the p53 tumor suppressor gene. Clin. Cancer Res 2, 1049–1053.
Su, R., Gong, J. N., Chen, M. T., Song, L., Shen, C., Zhang, X. H., et al. (2016). c-Myc suppresses miR-451 dash, verticalYWTAZ/AKT axis via recruiting HDAC3 in acute myeloid leukemia. Oncotarget 7, 77430–77443. doi: 10.18632/oncotarget.12679
Subramaniam, K. S., Omar, I. S., Kwong, S. C., Mohamed, Z., Woo, Y. L., Mat Adenan, N. A., et al. (2016). Cancer-associated fibroblasts promote endometrial cancer growth via activation of interleukin-6/STAT-3/c-Myc pathway. Am. J. Cancer Res. 6, 200–213.
Toon, C. W., Chou, A., Clarkson, A., Desilva, K., Houang, M., Chan, J. C., et al. (2014). Immunohistochemistry for myc predicts survival in colorectal cancer. PLoS One 9:e87456. doi: 10.1371/journal.pone.0087456
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108. doi: 10.3322/caac.21262
Tsiatis, A. C., Herceg, M. E., Keedy, V. L., Halpern, J. L., Holt, G. E., Schwartz, H. S., et al. (2009). Prognostic significance of c-Myc expression in soft tissue leiomyosarcoma. Mod. Pathol. 22, 1432–1438. doi: 10.1038/modpathol.2009.113
Wang, L., Xue, M., and Chung, D. C. (2016). c-Myc is regulated by HIF-2alpha in chronic hypoxia and influences sensitivity to 5-FU in colon cancer. Oncotarget 7, 78910–78917. doi: 10.18632/oncotarget.12911
Wang, W., Xue, L., and Wang, P. (2011). Prognostic value of beta-catenin, c-myc, and cyclin D1 expressions in patients with esophageal squamous cell carcinoma. Med. Oncol. 28, 163–169. doi: 10.1007/s12032-010-9436-0
Keywords: c-Myc, colorectal cancer, prognosis, biomarker, meta-analysis
Citation: He W-L, Weng X-T, Wang J-L, Lin Y-K, Liu T-W, Zhou Q-Y, Hu Y, Pan Y and Chen X-L (2018) Association Between c-Myc and Colorectal Cancer Prognosis: A Meta-Analysis. Front. Physiol. 9:1549. doi: 10.3389/fphys.2018.01549
Received: 21 November 2017; Accepted: 16 October 2018;
Published: 13 November 2018.
Edited by:
Angelica Merlot, The University of New South Wales, AustraliaReviewed by:
Antonio Longo, Università degli Studi di Catania, ItalyGregory Pond, McMaster University, Canada
Copyright © 2018 He, Weng, Wang, Lin, Liu, Zhou, Hu, Pan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunbao Pan, cGFueXVuYmFvQG91dGxvb2suY29t Xin-Lin Chen, Y2hlbnhsc3Vtc0AxMjYuY29t
 Wei-Ling He1
Wei-Ling He1 Yunbao Pan
Yunbao Pan Xin-Lin Chen
Xin-Lin Chen