- 1Center of Ophthalmology, The Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, Zhejiang, China
- 2Department of Computer Science, Hangzhou Dianzi University, Hangzhou, Zhejiang, China
Longitudinal disease progression evaluation between follow-up examinations relies on precise registration of medical images. Compared to other medical imaging methods, color fundus photograph, a common retinal examination, is easily affected by eye movements while shooting, for which we think it is necessary to develop a reliable longitudinal registration method for this modality. Thus, the purpose of this study was to propose a robust registration method for longitudinal color fundus photographs and establish a longitudinal retinal registration dataset. In the proposed algorithm, radiation-variation insensitive feature transform (RIFT) feature points were calculated and aligned, followed by further refinement using a normalized total gradient (NTG). Experiments and ablation analyses were conducted on both public and private datasets, using the mean registration error and registration success plot as the main evaluation metrics. The results showed that our proposed method was comparable to other state-of-the-art registration algorithms and was particularly accurate for longitudinal images with disease progression. We believe the proposed method will be beneficial for the longitudinal evaluation of fundus images.
Introduction
Diabetic retinopathy (DR) is one of the major diseases that can cause blindness. It is estimated that about 600 million people will have diabetes by 2040 [1], a third of whom will be affected by DR [2]. Regular follow-up and accurate analysis of longitudinal examinations play an important part in the management of DR [3]. However, the quantitative analysis of longitudinal images is still challenging, due to the tremendous discrepancies between the images caused by vastly different photographing conditions, involuntary eye movements, and pathological changes [4], which can disturb the observation and influence the evaluation of retinal image biomarkers [5]. Registration, which means the process of establishing pixel-to-pixel correspondence between two images, grants us the chance to eliminate these discrepancies before longitudinal assessment [6]. Therefore, a preliminary registration of two retinal images is required to reduce these effects and generate a reliable disease progression conclusion.
As a necessary work of retinal image analysis, the registration of retinal fundus images is a classic topic, in which tremendous efforts have been put into this area during past decades. From a methodological point of view, retinal image registration methods can be classified into three groups: feature-based, intensity-based, and hybrid methods. In feature-based registration methods, invariant features of the retinal images are extracted and utilized for seeking the best geometric transformation between two images. Retinal vessel bifurcations [7–11], optic disc, and fovea [12, 13] are previously commonly used features. However, some of these features rely on the segmentation of retinal structures and are sensitive to image quality. Therefore, easily-obtained and stable key-point detection is the premise for robust registration through feature-based methods, for example, Harris corner [14], scale invariant feature transform (SIFT) [15], and Speeded-Up Robust Features (SURF) [16] are classic feature points that have been extensively studied. Hernandez-Matas et al. [17, 18] introduced a feature-based registration framework exploiting the spherical eye model and pose estimation. In intensity-based methods, the intensity information is calculated and used to measure the similarity of the images and the registration performance, such as cross-correlation [19], mutual information [20], and phase correlation [21]. Hybrid registration methods combine feature-based and intensity-based methods together to seek better performance [4, 22]. Compared to single feature-based or intensity-based methods, hybrid methods have great potential for more accurate and practical image alignment, but it is less investigated. Although the registration of color fundus photographs has been intensively studied, the steps of seeking higher and more robust performance have never stopped.
Although there has been intensive research work in registration, further research is still needed. First, novel registration methods developed on other modalities should be applied to retinal images to seek better performance. Second, instead of paying attention to the improvement and development of registration methods, researchers should focus more on the clinical applicability of the proposed methods, which is extremely important for longitudinal follow-up examinations. Third, the development and evaluation of registration methods rely on the publication of open-access datasets. To the best of our knowledge, the Fundus Image Registration (FIRE) dataset is the only registration dataset that has been made publicly available [23]. We thought it would be useful to develop a registration dataset made up of longitudinal images with clarified medical diagnoses. Therefore, developing registration methods in clinical settings and establishing registration datasets would be greatly beneficial for interdisciplinary cooperation and clinical transformation of computation methods.
In this study, a robust registration method for longitudinal color fundus photographs based on both feature and intensity is proposed. An ablation study showed the necessity of combining the two main parts. A comprehensive comparison between the proposed algorithm and other state-of-the-art methods was conducted to investigate its features. The dataset will be available for registration research. We believe this work will be beneficial to follow up retinal image analysis and disease progression assessment.
Materials and methods
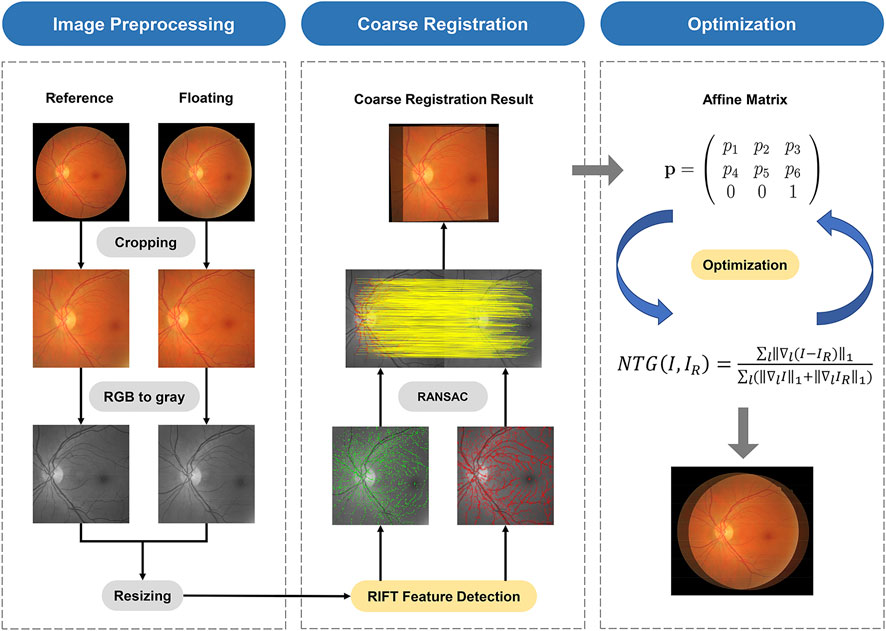
The proposed registration framework is a combination of feature-based and intensity-based methods. The flow of this work is shown in Figure 1.
Retinal image datasets
For the evaluation of the proposed registration method, we use two datasets, FIRE and FI-LORE, consisting of color fundus image pairs different from each other in terms of actual photographing and patient conditions. These datasets are described in detail hereinafter.
The Fundus Image Registration (FIRE) dataset [23] comprises 134 image pairs, which are further classified into three categories according to their characteristics. Category S contains 71 image pairs with more than 75% overlap area and super-resolution but no anatomical changes, while category P contains 49 image pairs with less than 75% overlap area and no anatomical changes. Category A contains 14 image pairs with high overlap and large anatomical changes due to retinopathy, which can be used to mimic practical longitudinal examinations. All the images have a resolution of 2,912 × 2,912 pixels. FIRE provides ground truths for the calculation of registration errors.
The Fundus Image for Longitudinal Registration (FI-LORE) dataset consists of 83 color fundus image pairs from 78 eyes of 54 diabetic retinopathy patients who underwent longitudinal examinations at the Second Affiliated Hospital of Zhejiang University, School of Medicine, from May 2020 to July 2020. Photograph conditions, involuntary movements of the eye, and disease progression and treatments, such as laser scars, all contribute to the differences of each image in a pair. Additionally, some of them are of low image quality because of complications, such as cataracts. FI-LORE can fully reflect practical conditions of the follow-up in clinics and test the robustness of the proposed method. All the images have a resolution of 1,500 × 1,500 pixels. Finally, to compute the registration error, we follow the annotation rule of the FIRE dataset [23], carefully choosing 10 corresponding points and repeatedly correcting the exact location of the points to guarantee the reliability of the ground truths. The FI-LORE dataset will be made publicly available.
Proposed registration framework
To normalize the images in different datasets taken at different examinations, preprocessing is the first step in the algorithm. First, the mask provided by the FIRE dataset was utilized to delete the blank margin of the original images. Second, the cropped images are further resized to 1,500 × 1,500 pixels to reach unity of the whole dataset. Furthermore, to simplify the calculation process, the RGB images are transformed into grayscale images.
Radiation-variation insensitive feature transform (RIFT) is a feature-based registration method with great robustness to non-linear radiation distortion (NRD) [24]. NRD is a rather common phenomenon that can be caused by involuntary movements of the eye. Therefore, we think it can be used for retinal image alignment tasks. The detail of the RIFT calculation can be found in the original article [24]. The alignment process is realized by the RANdom SAmple Consensus (RANSAC) algorithm [25]. Then, an affine transformation matrix
Registration evaluation
To quantitatively assess the performance of the registration result, we adopt a widely accepted registration error calculation method [23], which requires the ground truths of image pairs. Given the sets of reference points,
The
Results
Results of the ablation study
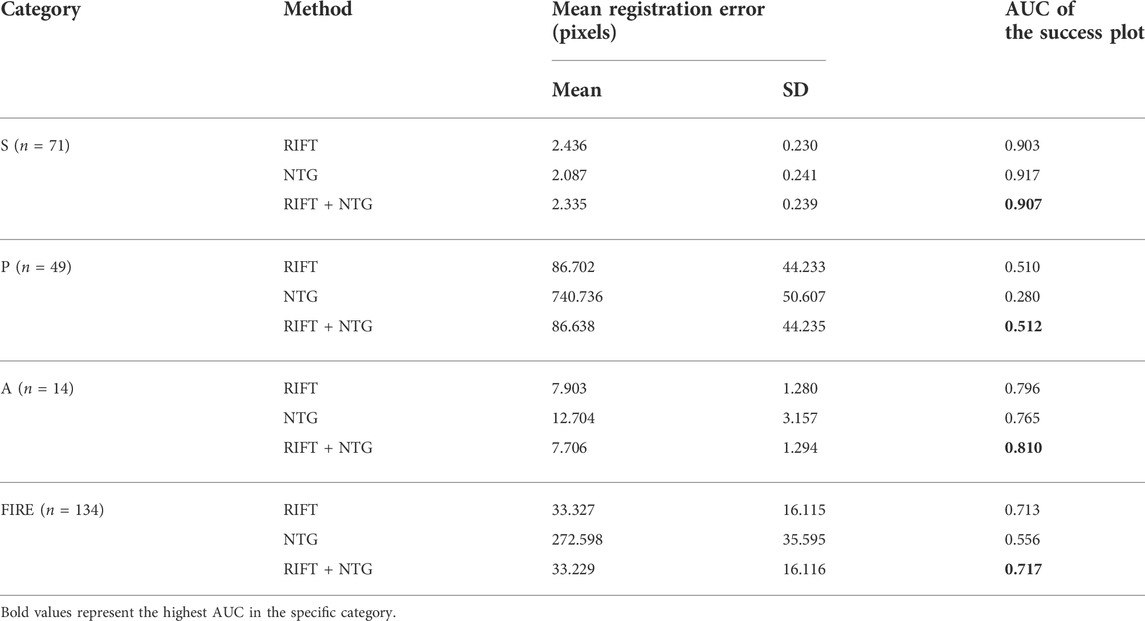
To better understand the contributions of each part of the registration framework and validate the effectiveness of the combination of RIFT and NTG, we conducted an ablation study to see the registration performance of these two procedures themselves. Table 1 shows the registration results of the ablation study in the FIRE dataset. Figure 2 is the success plot of the ablation study. From the results, we can see that the NTG performs better in image pairs with high overlap, but once the overlap is small, the NTG method performance is relatively poor. However, RIFT is on the opposite side of the NTG. The combination of the algorithms outperforms each one of them. Therefore, the combination of RIFT and NTG grants algorithm robustness to image pairs of different overlap areas.
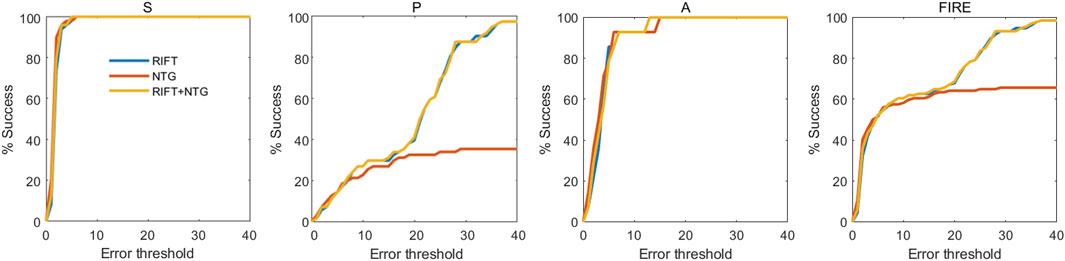
FIGURE 2. Registration success plot of the ablation study. The x-axis marks, in pixels, the registration error threshold under which registration is considered to be successful. The y-axis marks the percentage of successfully registered image pairs for a given threshold.
Comparison to other registration methods
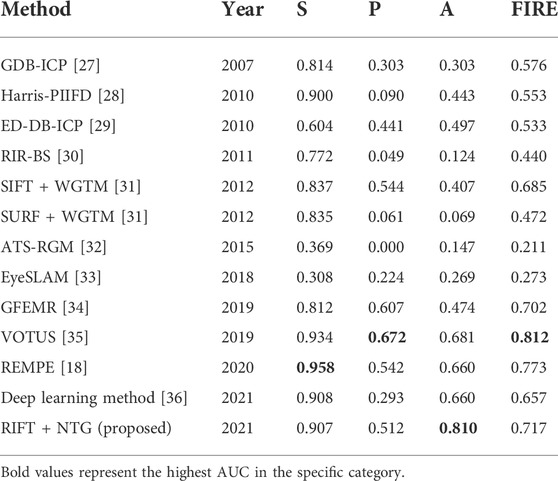
To further assess the accuracy of the proposed method in color fundus image registration, we compare our results to other state-of-the-art image registration methods which are already utilized in the FIRE dataset, including GDB-ICP [27], Harris-PIIFD [28], ED-DB-ICP [29], RIR-BS [30], SIFT + WGTM [31], SURF + WGTM [31], ATS-RGM [32], EyeSLAM [33], GFEMR [34], VOTUS [35], REMPE [18], and a deep-learning based registration method proposed by Rivas-Villar et al [36]. Figure 3 is the qualitative illustration of the proposed method registration results. Table 2 lists the methods used for comparison and the AUC of the success plot. Figure 4 contains the success plot of the proposed method and some other methods whose results are publicly available online. From the aforementioned results, one can conclude that the proposed method is competitive to the leading registration methods in category S. The AUC of category P clearly underperforms some algorithms. But the proposed method outperforms all the methods in category A, which stands for the longitudinal study. Therefore, we think the proposed method can remain robust under the anatomical changes and disease progression and is well worth further study.

FIGURE 3. Registration results of the proposed algorithm in the FIRE dataset. The overlap decreases from the top row to bottom. (A) and (B) are image pairs without registration. (C) Image pairs shown in an overlaying form after registration. (D) Checkerboard comparisons of the proposed method.
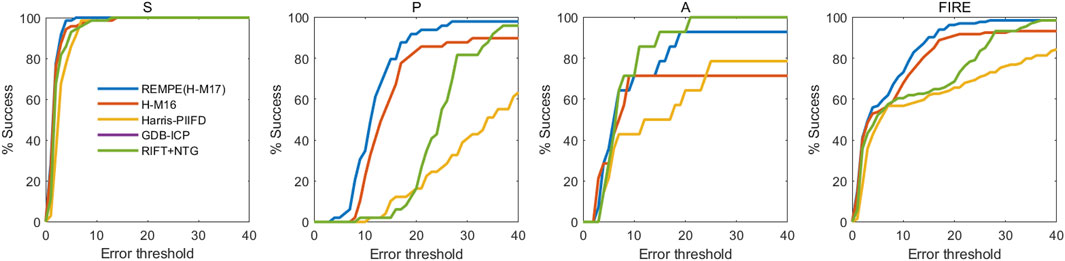
FIGURE 4. Registration success plot of the comparisons between the proposed and other registration methods. The x-axis marks, in pixels, the registration error threshold under which registration is considered to be successful. The y-axis marks the percentage of successfully registered image pairs for a given threshold.
Results in the FI-LORE dataset
Retinal images in real clinical conditions may be of poor quality due to complications such as cataracts, loss of focus, and various light conditions, posing great problems to the practical use of registration. As described earlier, FI-LORE is a collection from real ophthalmologic practice, which can be used to validate the utility of the proposed method. Figure 5 shows some image pairs before and after registration. We can see that despite these image pairs having dissimilar illumination conditions, pathological changes, and disease progression, our method can robustly align the longitudinal images to a satisfying extent. Figure 6 is the success plot of the proposed method and other state-of-the-art color fundus image registration methods in FI-LORE. The AUCs of RIFT, NTG, RITF + NTG (proposed), REMPE, and GFEMR are 0.841, 0.755, 0.850, 0.840, and 0.808, respectively. Through the quantitative analysis of the registration results in FI-LORE, we have validated the superior performance of the registration method in real clinical conditions.
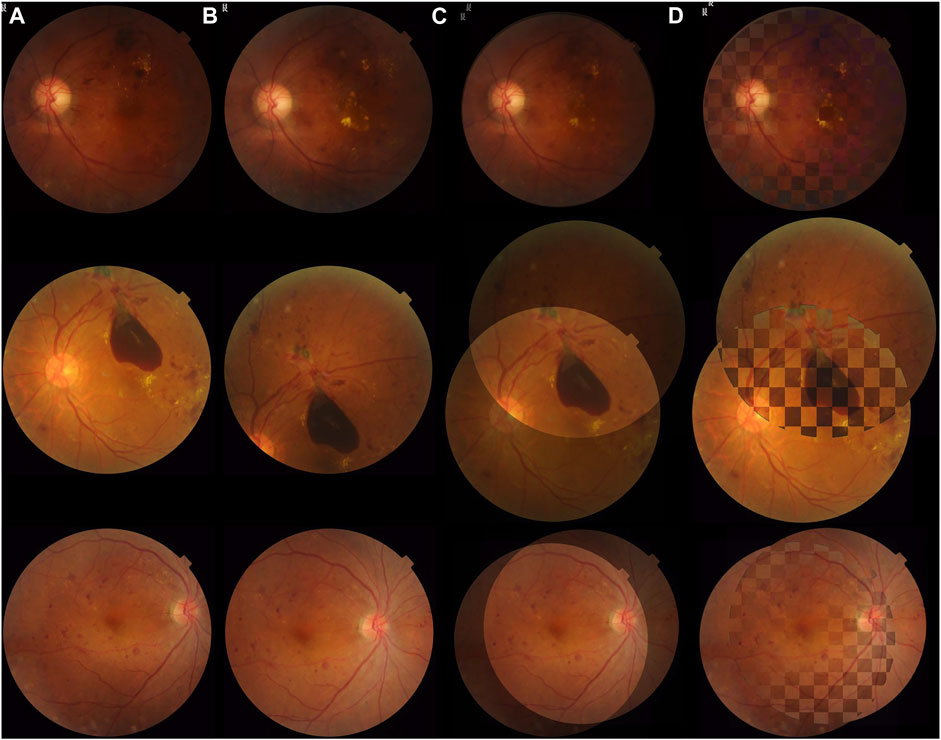
FIGURE 5. Registration results of the proposed algorithm in FI-LORE. The pairs listed, respectively, represent poor illumination quality, pathological change, and disease progression, which are common conditions in longitudinal examinations. (A) and (B) are image pairs without registration. (C) Image pairs shown in an overlaying form after registration. (D) Checkerboard comparisons of the proposed method.
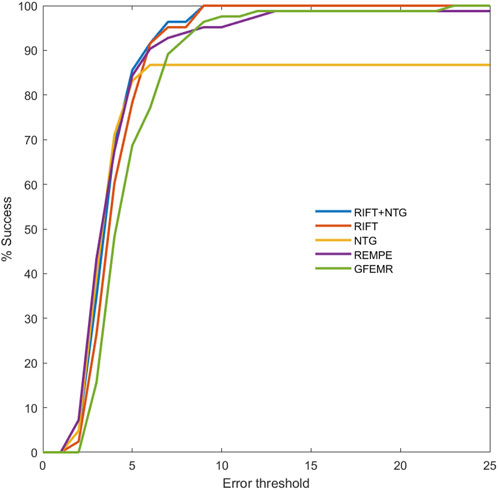
FIGURE 6. Registration success plot of the registration results in FI-LORE. The x-axis marks, in pixels, the registration error threshold under which registration is considered to be successful. The y-axis marks the percentage of successfully registered image pairs for a given threshold. The AUCs of RIFT, NTG, RITF + NTG (proposed), REMPE, and GFEMR are 0.841, 0.755, 0.850, 0.840, and 0.808, respectively.
Discussion
Longitudinal assessment of DR retinal images is of great importance, and longitudinal registration is an important and fundamental task which has often been neglected in clinical situations, especially for follow-up examinations. Precise alignment of different examination images is the premise for accurate detection and analysis of pathological changes, which has already been adopted in some automated retinal image analyzing devices [37]. In this study, we proposed a hybrid registration method with comprehensive experiments showing its excellent performance in longitudinal images and established a color fundus photograph dataset with pixel-wise annotation ground truth. In Table 1, the NTG shows the best performance in category S, and RIFT registered better in category P. We can conclude that the intensity-based registration method, NTG, is more precise in image pairs with large overlap while RIFT is the opposite. Thus, the combination of RIFT and NTG is reasonable and has been proved to be the best on the whole FIRE. Results from the intensive comparison experiments showed that our method is comparable to state-of-the-art image registration methods, such as GFEMR [34], VOTUS [35], and REMPE [18]. For longitudinal images in category A, the proposed method outperformed other state-of-the-art methods, for which we think it is suitable for the clinical evaluation of disease progression in follow-up examinations. This conclusion is further validated using a private dataset, FI-LORE, with more longitudinal images. Taking all of these into consideration, we believe the proposed method is good at registering longitudinal retinal images and will be beneficial in clinical use. It should be noted that in category P, which is made up of images with partial overlap, the MRE and AUC are far less than those in the other two classes. The main source of error came from several misregistered image pairs which show an MRE of nearly a thousand pixels. The same trends can also be observed in some state-of-the-art methods. The private dataset also contains images with less overlap, and the proposed method can also register them, as shown in Figure 6. Further research is needed to validate its performance and investigate the reason why these algorithms did not perform well in category P. During image pre-processing, we thought resizing might affect the final results. In the current study, the size of 500 × 500 pixels is recommended. We conducted experiments on 250 × 250 and 750 × 750 pixels, results of which can be found in Supplementary Material. Also, mutual information was evaluated to confirm the performance of the NTG, and relevant results are given in Supplementary Material.
Most of the development of registration methods focuses on either feature-based or intensity-based registration. As far as we know, only several studies adopted hybrid methods. In 2016, Saha et al. proposed a hybrid method using Speeded-Up Robust Features (SURF) and Binary Robust Independent Elementary Features (BRIEF) [4], which are both well-known registration methods and their combination generated better performance. In our study, two registration methods, RIFT and NTG, were adopted, and further investigation revealed their own characteristics in image registration. To the best of our knowledge, this is the first study that used RIFT and NTG in ophthalmic imaging and investigated their applicability to different image overlaps. We think that the two-step hybrid registration method is promising in retinal imaging.
Deep learning has been showing great potential in medical image processing, including segmentation and registration. Several methods have been proposed to attempt to utilize deep learning in retinal image registration [36, 38–45]. However, to the best of our knowledge, there are two inherent problems limiting the development of deep learning-based registration. On the one hand, unlike other image processing issues (segmentation, enhancement, etc.), registration contains two steps theoretically, feature recognition and feature alignment. In retinal image registration, these usually mean retinal feature extraction (feature points, vessel network, etc.) and retinal feature alignment. Therefore, an inevitable question comes up, that is, when and where to adopt deep learning in the registration workflow. Different researchers provided various solutions. Some researchers adopted deep learning in the feature detection process and further aligned the feature points using conventional image alignment methods, such as RANSAC[40, 41], while some work constructed an outlier-rejection network to compute the image transformation matrix [45, 46]. There is no consensus on how deep learning should be added to the registration pipeline [46]. Moreover, in most deep learning-based registration algorithms, accurate registration relies on accurate segmentation, which is still an ongoing research topic in medical image processing. On the other hand, training and validation of deep learning networks rely on massive labeled data. In the specific topic of image registration, ground truth annotation is labor-intensive and time-consuming. For some deep learning methods using vessel segmentation, the networks also need large annotated vessel segmentation datasets. From these two perspectives, we tend to believe that although deep learning has shed light on medical image processing and analysis, it is still in the exploration stage for image registration. In the current study, we compare a state-of-the-art registration method with the proposed method, and the results showed that for longitudinal retinal image registration, our proposed method still stood out. Deep learning-assisted retinal image registration should be paid more attention to find out whether it is actually superior to conventional algorithms.
The development of retinal image registration methods is limited due to the lack of registration datasets. As far as we know, FIRE is the only dataset that focuses on retinal image registration and proposes pixel-level ground truth which can be used for the development and evaluation of registration methods. However, the longitudinal category contains only 14 image pairs, significantly small when compared to other categories. Taking this situation and the clinical use of registration methods into account, we collected and annotated 83 image pairs, especially for longitudinal image registration tasks. These image pairs are different in photograph conditions, involuntary movements of the eye, and disease progression and treatments. We believe that the adoption of this dataset can greatly benefit the study of retinal image registration.
Because of the specialty of registration in retinal image analysis, some methods have been claimed to be put into clinical use. To the best of our knowledge, there is one registration software, the DualAlign i2k software package (Clifton Park, NY), that has been made commercial. The software was developed based on the GDB-ICP algorithm, which has been compared in our work [27]. With growing interest in image registration, more novel and efficient methods have been proposed to ensure better and swifter registration performance. These methods show promise for medical image registration tasks. However, due to the lack of interdisciplinary cooperation of medical and computer science researchers, the study of these novel methods for medical use is limited. In this study, we focus on two methods and validate their performance. We believe more research is needed to provide more possibilities for more precise and swifter image analysis in real clinical use.
There are some limitations to our current study. First, the proposed algorithm performed relatively poorly in the category which stands for images with small overlay. However, there are also some similar image pairs in the FI-LORE dataset, but the proposed methods did not perform like that, which is confusing. More image pairs are needed to further test this method. Second, in the current study, we still focus on unimodality registration tasks. The performance of this method in multi-modal tasks needs more examination. Third, some state-of-the-art methods should be compared with the local dataset FI-LORE, but due to the lack of reliable source codes and our inability to completely repeat the methods, we failed to put them into further comparison. Finally, there are some artificial intelligence algorithms that have been developed for retinal image registration tasks [22, 47, 48]. We have compared one deep learning algorithm, but further studies are needed to investigate deep learning in the context of retinal image alignment.
Conclusion
RIFT can better align images with small overlap, while the NTG is more precise with large overlap image pairs. Thus, the combination of RIFT and NTG was reasonable and outperformed single RIFT or NTG. The proposed method was comparable to other state-of-the-art registration algorithms and was especially accurate for longitudinal images with disease progression. We believe that the proposed method will be beneficial for the longitudinal evaluation of fundus images.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author contributions
JZ and KJ contributed to the idea, performed the experiments, analyzed the results, and wrote the manuscript. RG helped with the experiments and gave meaningful advice in the algorithm part. YY and YZ helped perform the analysis. YS helped with the preparatory work. JY contributed to the conception of the study and supervised the whole process of the experiment and writing.
Funding
The study was supported in part by the National Key Research and Development Program of China (2019YFC0118400), the Key Research and Development Program of Zhejiang Province (2019C03020), the Clinical Medical Research Center for Eye Diseases of Zhejiang Province (2021E50007), and the Natural Science Foundation of Zhejiang Province (grant number LQ21H120002).
Acknowledgments
The authors would like to thank C. Hernandez-Matas et al. for providing with the data from the FIRE database (see https://projects.ics.forth.gr/cvrl/fire/).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphy.2022.978392/full#supplementary-material
References
1. Ogurtsova K, da Rocha Fernandes JD, Huang Y, Linnenkamp U, Guariguata L, Cho NH, et al. IDF Diabetes Atlas: Global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Res Clin Pract (2017) 128:40–50. doi:10.1016/j.diabres.2017.03.024
2. Yau JW, Rogers SL, Kawasaki R, Lamoureux EL, Kowalski JW, Bek T, et al. Global prevalence and major risk factors of diabetic retinopathy. Diabetes Care (2012) 35(3):556–64. eng. Epub 2012/02/04. doi:10.2337/dc11-1909
3. Wong TY, Sun J, Kawasaki R, Ruamviboonsuk P, Gupta N, Lansingh V, et al. Guidelines on diabetic eye care. Ophthalmology (2018) 125(10):1608–22. EnglishCited in: Pubmed; PMID WOS:000445012100028. doi:10.1016/j.ophtha.2018.04.007
4. Saha SK, Xiao D, Frost S, Kanagasingam Y. A two-step approach for longitudinal registration of retinal images. J Med Syst (2016) 40(12):277. Epub 2016/10/28Cited in: Pubmed; PMID 27787783. doi:10.1007/s10916-016-0640-0
5. Ting DSW, Peng L, Varadarajan AV, Keane PA, Burlina PM, Chiang MF, et al. Deep learning in ophthalmology: The technical and clinical considerations. Prog Retin Eye Res (2019) 72:100759. EnglishCited in: Pubmed; PMID WOS:000488311400001. doi:10.1016/j.preteyeres.2019.04.003
6. Zitová B, Flusser J. Image registration methods: A survey. Image Vis Comput (2003) 21(11):977–1000. doi:10.1016/s0262-8856(03)00137-9
7. Zana F, Klein JC. A multimodal registration algorithm of eye fundus images using vessels detection and Hough transform. IEEE Trans Med Imaging (1999) 18(5):419–28. doi:10.1109/42.774169
8. Can AS, Charles V. A feature-based, robust, hierarchical algorithm for registering pairs of images of the curved human retina. IEEE Transactions on Pattern Analysis & Machine Intelligence 24 (2002).doi:10.1109/34.990136
9. Laliberte F, Gagnon L, Sheng Y. Registration and fusion of retinal images--an evaluation study. IEEE Trans Med Imaging (2003) 22(5):661–73. Epub 2003/07/09. doi:10.1109/TMI.2003.812263Cited in: Pubmed; PMID 12846435
10. Matsopoulos GK, Asvestas PA, Mouravliansky NA, Delibasis KK. Multimodal registration of retinal images using self organizing maps. IEEE Trans Med Imaging (2004) 23:1557–63. doi:10.1109/tmi.2004.836547
11. Fang B, Tang YY. Elastic registration for retinal images based on reconstructed vascular trees. IEEE Trans Biomed Eng (2006) 53(6):1183–7. Epub 2006/06/10. doi:10.1109/TBME.2005.863927
12. Hart WE, Goldbaum MH. Registering retinal images using automatically selected control point pairs, Proceedings of the Image processing, icip-94. IEEE International Conference (1994). doi:10.1109/icip.1994.413740
13. Nunes JC, Bouaoune Y, Delechelle E, Bunel P. A multiscale elastic registration scheme for retinal angiograms. Comput Vis Image Underst (2004) 95(2):129–49. doi:10.1016/j.cviu.2004.03.007
14. Harris C, Stephens M. A combined corner and edge detector. Proceedings of the Alvey vision conference (1988). doi:10.5244/c.2.23
15. Lowe DG. Distinctive image features from scale-invariant keypoints. Int J Comp Vis (2004) 60(2):91–110. doi:10.1023/B:VISI.0000029664.99615.94
16. Bay H, Ess A, Tuytelaars T, Van Gool L. Speeded-up robust features (SURF). Computer Vis Image Understanding (2008) 110(3):346–59. doi:10.1016/j.cviu.2007.09.014
17. Hernandez-Matas C, Zabulis X, Argyros AA. Retinal image registration through simultaneous camera pose and eye shape estimation. Annu Int Conf IEEE Eng Med Biol Soc (2016) 2016:3247–51. eng. Epub 2017/03/09. doi:10.1109/embc.2016.7591421Cited in: Pubmed; PMID 28269000
18. Hernandez-Matas C, Zabulis X, Argyros A. Rempe: Registration of retinal images through eye modelling and pose estimation. IEEE J Biomed Health Inform (2020) 24:3362–73. eng. Epub 2020/04/06. doi:10.1109/jbhi.2020.2984483Cited in: Pubmed; PMID 32248134
19. Cideciyan AV. Registration of ocular fundus images: An algorithm using cross-correlation of triple invariant image descriptors. IEEE Eng Med Biol Mag (2002) 14(1):52–8. doi:10.1109/51.340749
20. Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging (1997) 16(2):187–98. doi:10.1109/42.563664
21. Suthaharan S, Rossi EA, Snyder V, Chhablani J, Lejoyeux R, Sahel JA, et al. Laplacian feature detection and feature alignment for multimodal ophthalmic image registration using phase correlation and Hessian affine feature space. Signal Processing (2020) 177:107733. Epub 2020/09/19Cited in: Pubmed; PMID 32943806. doi:10.1016/j.sigpro.2020.107733
22. ÁlvaroHervella SJR, Novo J, Ortega M. Multimodal registration of retinal images using domain-specific landmarks and vessel enhancement. Proced Comp Sci (2018) 126:97–104. doi:10.1016/j.procs.2018.07.213
23. Hernandez-Matas C, Zabulis X, Triantafyllou A, Anyfanti P, Douma S, Argyros AA. Fire: Fundus image registration dataset. Model Artif Intelligence Ophthalmol (2017) 1(4):16–28. doi:10.35119/maio.v1i4.42
24. Li J, Hu Q, Ai M. Rift: Multi-Modal image matching based on radiation-variation insensitive feature transform. IEEE Trans Image Process (2019) 29:3296–310. Epub 2019/12/24. doi:10.1109/TIP.2019.2959244Cited in: Pubmed; PMID 31869789
25. Hossein-Nejad Z, Nasri M, . A-RANSAC: Adaptive random sample consensus method in multimodal retinal image registration. Biomed Signal Process Control (2018) 45:325–38. doi:10.1016/j.bspc.2018.06.002
26. Shu-Jie C, Hui-Liang S, Chunguang L, Xin JH. Normalized total gradient: A new measure for multispectral image registration. IEEE Trans Image Process (2018) 27(3):1297–310. Epub 2018/07/11. doi:10.1109/TIP.2017.2776753Cited in: Pubmed; PMID 29990251
27. Yang G, Stewart CV, Sofka M, Tsai CL. Registration of challenging image pairs: Initialization, estimation, and decision. IEEE Trans Pattern Anal Mach Intell (2007) 29(11):1973–89. eng. Epub 2007/09/13. doi:10.1109/tpami.2007.1116in: Pubmed; PMID 17848778
28. Chen J, Tian J, Lee N, Zheng J, Smith RT, Laine AF. A partial intensity invariant feature descriptor for multimodal retinal image registration. IEEE Trans Biomed Eng (2010) 57(7):1707–18. eng. Epub 2010/02/18. doi:10.1109/TBME.2010.2042169Cited in: Pubmed; PMID 20176538
29. Tsai CL, Li CY, Yang G, Lin KS. The edge-driven dual-bootstrap iterative closest point algorithm for registration of multimodal fluorescein angiogram sequence. IEEE Trans Med Imaging (2010) 29(3):636–49. Epub 2009/08/28. doi:10.1109/TMI.2009.2030324Cited in: Pubmed; PMID 19709965
30. Chen L, Xiang Y, Chen Y, Zhang X. Retinal image registration using bifurcation structures. 2011 18th IEEE International Conference on Image Processing (2011). p. 2169–72. doi:10.1109/icip.2011.6116041
31. Izadi M, Saeedi P. Robust weighted graph transformation matching for rigid and nonrigid image registration. IEEE Trans Image Process (2012) 21(10):4369–82. doi:10.1109/TIP.2012.2208980
32. Serradell E, Pinheiro MA, Sznitman R, Kybic J, Moreno-Noguer F, Fua P. Non-rigid graph registration using active testing search. IEEE Trans Pattern Anal Mach Intell (2015) 37(3):625–38. Epub 2015/09/10. doi:10.1109/TPAMI.2014.2343235Cited in: Pubmed; PMID 26353266
33. Braun D, Yang S, Martel JN, Riviere CN, Becker BC. EyeSLAM: Real-time simultaneous localization and mapping of retinal vessels during intraocular microsurgery. Int J Med Robot (2018) 14(1). Epub 2017/07/19. doi:10.1002/rcs.1848
34. Wang J, Chen J, Xu H, Zhang S, Mei X, Huang J, et al. Gaussian field estimator with manifold regularization for retinal image registration. Signal Processing (2019) 157:225–35. doi:10.1016/j.sigpro.2018.12.004
35. Motta D, Casaca W, Paiva A. Vessel optimal transport for automated alignment of retinal fundus images. IEEE Trans Image Process (2019) 28(12):6154–68. Epub 2019/07/10. doi:10.1109/TIP.2019.2925287
36. Rivas-Villar D, Hervella AS, Rouco J, Novo J. Color fundus image registration using a learning-based domain-specific landmark detection methodology. Comput Biol Med (2021) 140:105101. Epub 2021/12/08. doi:10.1016/j.compbiomed.2021.105101
37. Grzybowski A, Brona P, Lim G, Ruamviboonsuk P, Tan GSW, Abramoff M, et al. Artificial intelligence for diabetic retinopathy screening: A review. Eye (London, England) (2020) 34(3):451–60. Epub 2019/09/07Cited in: Pubmed; PMID 31488886. doi:10.1038/s41433-019-0566-0
38. Mahapatra D, Antony B, Sedai S, Garnavi R. Deformable medical image registration using generative adversarial networks, Proceedings of the IEEE 15th international symposium on biomedical imaging (ISBI 2018). p. 1449–53. doi:10.1109/isbi.2018.8363845
39. Lee J, Liu P, Cheng J, Fu H. A deep step pattern representation for multimodal retinal image registration. IEEE/CVF International Conference on Computer Vision (ICCV) (2019). doi:10.1109/iccv.2019.00518
40. De Silva T, Chew EY, Hotaling N, Cukras CA. Deep-learning based multi-modal retinal image registration for the longitudinal analysis of patients with age-related macular degeneration. Biomed Opt Express (2021) 12(1):619–36. eng. Epub 2021/02/02. doi:10.1364/boe.408573. Cited in: Pubmed; PMID 33520392
41. Ding L, Kuriyan AE, Ramchandran RS, Wykoff CC, Sharma G. Weakly-supervised vessel detection in ultra-widefield fundus photography via iterative multi-modal registration and learning. IEEE Trans Med Imaging (2020) 40:2748–58. eng. Epub 2020. doi:10.1109/tmi.2020.3027665./09/30Cited in: Pubmed; PMID 32991281
42. Luo G, Chen X, Shi F, Peng Y, Xiang D, Chen Q, et al. Multimodal affine registration for ICGA and MCSL fundus images of high myopia. Biomed Opt Express (2020) 11(8):4443–57. eng. Epub 2020/09/15. doi:10.1364/boe.393178. Cited in: Pubmed; PMID 32923055
43. Tian Y, Hu Y, Ma Y, Hao H, Mou L, Yang J, et al. Multi-scale U-net with edge guidance for multimodal retinal image deformable registration. Annu Int Conf IEEE Eng Med Biol Soc (2020) 2020:1360–3. eng. Epub 2020/10/07. doi:10.1109/embc44109.2020.9175613. Cited in: Pubmed; PMID 33018241
44. Zhang J, An C, Dai J, Amador M, Bartsch D, Borooah S, et al. Joint vessel segmentation and deformable registration on multi-modal retinal images based on style transfer Proceedings of the IEEE international conference on image processing New Jersey, United States: IEEE (2019). p. 839–43. doi:10.1109/icip.2019.8802932
45. Wang Y, Zhang J, An C, Cavichini M, Jhingan M, Amador-Patarroyo MJ, et al. A segmentation based robust deep learning framework for multimodal retinal image registration Proceedings of the Icassp 2020 - 2020 IEEE international conference on acoustics, speech and signal processing (ICASSP) (2020). p. 1369–73. doi:10.1109/icassp40776.2020.9054077
46. Zhang J, Wang Y, Dai J, Cavichini M, Bartsch DUG, Freeman WR, et al. Two-step registration on multi-modal retinal images via deep neural networks. IEEE Trans Image Process (2022) 31:823–38. doi:10.1109/TIP.2021.3135708
47. Wang Y, Zhang J, Cavichini M, Bartsch DG, Freeman WR, Nguyen TQ, et al. Robust content-adaptive global registration for multimodal retinal images using weakly supervised deep-learning framework. IEEE Trans Image Process (2021) 30:3167–78. eng. Epub 2021. doi:10.1109/tip.2021.3058570./02/19Cited in: Pubmed; PMID 33600314
Keywords: registration, color fundus photograph, retinal imaging, diabetic retinopathy, disease progression
Citation: Zhou J, Jin K, Gu R, Yan Y, Zhang Y, Sun Y and Ye J (2022) Color fundus photograph registration based on feature and intensity for longitudinal evaluation of diabetic retinopathy progression. Front. Phys. 10:978392. doi: 10.3389/fphy.2022.978392
Received: 26 June 2022; Accepted: 06 September 2022;
Published: 27 September 2022.
Edited by:
Liwei Liu, Shenzhen University, ChinaCopyright © 2022 Zhou, Jin, Gu, Yan, Zhang, Sun and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Ye, eWVqdWFuQHpqdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Jingxin Zhou
Jingxin Zhou Kai Jin
Kai Jin Renshu Gu2
Renshu Gu2 Yiming Sun
Yiming Sun