- 1National Key Laboratory for Innovation and Transformation of Luobing Theory, The Key Laboratory of Cardiovascular Remodeling and Function Research, Chinese Ministry of Education, Chinese National Health Commission and Chinese Academy of Medical Sciences, Department of Cardiology, Qilu Hospital of Shandong University, Jinan, China
- 2Department of Gerontology, The First Affiliated Hospital of Shandong First Medical University and Shandong Provincial Qianfoshan Hospital, Jinan, China
- 3Department of Cardiology, Qilu Hospital (Qingdao), Cheeloo College of Medicine, Shandong University, Qingdao, China
Myocardial infarction (MI) imposes a huge medical and economic burden on society, and cardiac repair after MI involves a complex series of processes. Understanding the key mechanisms (such as apoptosis, autophagy, inflammation, and fibrosis) will facilitate further drug development and patient treatment. Presently, a substantial body of evidence suggests that the regulation of epigenetic processes contributes to cardiac repair following MI, with DNA methylation being among the notable epigenetic factors involved. This article will review the research on the mechanism of DNA methylation regulation after MI to provide some insights for future research and development of related drugs.
1 Introduction
Cardiovascular disease (CVD) is the leading cause of death worldwide and poses a significant medical and economic burden on countries worldwide (Han, 2019; Sturgeon et al., 2019; Zhao et al., 2019; Townsend et al., 2022). Among CVD events, myocardial infarction (MI) has been shown to contribute significantly to disease burden and mortality (Mathers and Loncar, 2006; White and Chew, 2008; Heidenreich et al., 2013). Following MI, cardiac repair involves a complex series of events, including an inflammatory phase, repair and proliferation phase, and mature phase (Prabhu and Frangogiannis, 2016). The inflammatory phase, initiated by tissue damage such as cell necrosis, triggers the recruitment of immune cells that generate a robust inflammatory response to remove damaged cells and extracellular matrix components (Frangogiannis, 2012). Transitioning from the inflammatory phase to the repair and proliferation phase requires the activation of multiple inhibition pathways to reduce post-infarction inflammation, involving a variety of cell types and extracellular matrix components such as neutrophils, monocyte-macrophage systems, and endothelial cells. The success of this transition is critical for effective repair of the infarcted area (Frangogiannis, 2012; Kain et al., 2014). During the repair and proliferation phase, inflammation gradually subsides, and fibroblasts transform into myofibroblast phenotypes, leading to a series of fibrotic reactions that culminate in scar formation and neovascularization in the mature phase (Nahrendorf et al., 2010). Despite significant progress in understanding the pathogenesis of MI and the development of novel treatments such as reperfusion therapy and drug therapy, treating MI patients remains challenging (Saleh and Ambrose, 2018). Identifying key molecular targets, such as those involved in apoptosis, autophagy, inflammation, and fibrosis, that occur successively after MI may lead to the development of more effective treatment strategies in the future.
DNA methylation is a well-known epigenetic modification in mammalian genomes that is regulated by a family of DNA methyltransferases (DNMTs). DNMTs catalyze the transfer of a methyl group from s-adenine methionine to a cytosine residue at position 5C to form 5 mC (Moore et al., 2013). Dnmt3a and DNMT3b are responsible for de novo methylation (Figure 1) during development by establishing a novel methylation pattern on unmodified DNA (Okano et al., 1999). DNMT1 conveys DNA methylation patterns during DNA replication of the newly synthesized daughter strand of the parental strand and plays a role in maintaining post-replicative methylation in cells (Li et al., 1992; Sen et al., 2010; Moore et al., 2013). However, some studies argue for a cooperative role of the three DNMTs during DNA methylation (Fatemi et al., 2002; Kim et al., 2002). DNA methylation predominantly occurs at palindromic cytosine phosphate guanine dinucleotides (CpGs) in the human genome, with 60%–80% of approximately 28 million CpGs usually being methylated (Smith and Meissner, 2013). CpG islands (CpGis), known as CG dense regions, account for less than 10% of CpGs and are located in promoter regions of approximately 70% of annotated genes, including nearly all housekeeping genes, tissue-specific genes, and developmentally regulated genes, to regulate transcription (Larsen et al., 1992; Saxonov et al., 2006; Zhu et al., 2008; Deaton and Bird, 2011). DNA methylation can directly interfere with the binding of specific transcription factors or indirectly bind specific transcriptional repressors, such as methyl CpG binding protein (MeCP) and methyl CpG binding domain protein, to repress chromatin transcriptional states and mediate gene silencing (Iguchi-Ariga and Schaffner, 1989; Comb and Goodman, 1990; Falzon and Kuff, 1991; Prendergast et al., 1991; Hendrich and Tweedie, 2003; Das and Singal, 2004; Russell-Hallinan et al., 2018). However, DNA methylation may play different roles depending on the genomic region sequences (Moore et al., 2013). Intergenic regions, for instance, serve to suppress potentially harmful genetic factors (Walsh et al., 1998; Gaudet et al., 2004), while DNA hypermethylation within gene bodies is linked to increased gene expression (Hellman and Chess, 2007; Ball et al., 2009; Aran et al., 2011).
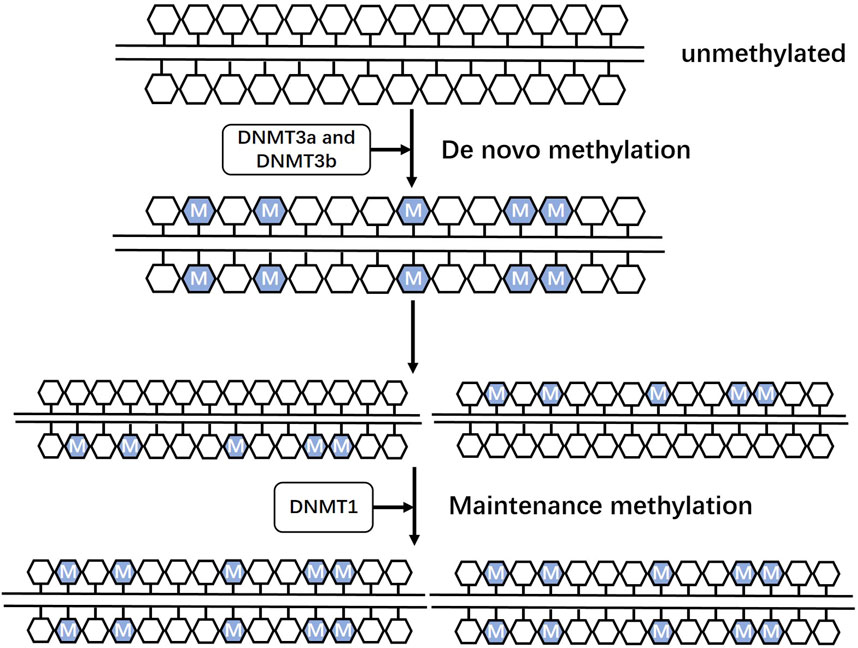
FIGURE 1. Roles of three DNA methyltransferases Dnmt3a and DNMT3b are responsible for de novo methylation during development by establishing a novel methylation pattern on unmodified DNA. DNMT1 conveys DNA methylation patterns during DNA replication of the newly synthesized daughter strand of the parental strand and plays a role in maintaining post-replicative methylation in cells.
While DNA methylation was initially considered a long-lasting and stable epigenetic modification, current research indicates that it is a dynamic and reversible biological process (Fardi et al., 2018; Russell-Hallinan et al., 2018). The process may be mediated by ten-eleven translocation (Tet) enzymes, namely Tet1, Tet2, and Tet3. Tet1, in particular, introduces a hydroxyl group to the methyl group at the 5 mC site, resulting in the formation of 5 hmC (Tahiliani et al., 2009; Ito et al., 2010). Once formed, 5 hmC may be converted to 5 fC and 5 caC under Tet enzyme catalyzed oxidation, followed by decarboxylation to cytosine (Ito et al., 2011). 5 fC and 5 caC can also be excised by thymine DNA glycosylase and subsequently subjected to pathways such as base excision repair to remove methylation modifications from cytosine (Kohli and Zhang, 2013). There is also supporting evidence indicating that akin to methylation, 5 hmC can influence gene expression, leading to notable decreases in the binding affinity of MeCP2 (Valinluck et al., 2004).
Research has demonstrated alterations in the degree of methylation of certain genes in individuals with myocardial infarction (Ek et al., 2016; Rask-Andersen et al., 2016; Ward-Caviness et al., 2018; Thunders et al., 2019), which are associated with cardiac function, cardiogenesis, and recovery after ischemic injury. To understand the early dynamic changes of gene expression after MI, RNA-seq and MeDIP-seq were used on heart tissues from AMI mice at different time points (Han et al., 2022; Luo et al., 2022), and the results showed that DNA methylation plays an important role in the pathophysiological progression after MI. We reviewed the existing research and elucidated the role of DNA methylation regulation in inflammation, fibrosis and other aspects after myocardial infarction.
2 Roles of DNA methylation after myocardial infarction
2.1 Inflammation and DNA methylation
After myocardial infarction, hypoxia often leads to endothelial cell barrier impairment and increased vascular permeability due to decreased adenylate cyclase activity and intracellular cAMP levels. This promotes leukocyte infiltration and activates the death process of myocardial cells, primarily cell necrosis, accompanied by apoptosis and autophagy (Ogawa et al., 1992; Hotchkiss et al., 2009; Nahrendorf et al., 2010; Eltzschig and Eckle, 2011; Kain et al., 2014; Prabhu and Frangogiannis, 2016). Immediate reperfusion treatment releases soluble inflammatory mediators that recruit neutrophils, which directly damage endothelial cells by producing reactive oxygen species (ROS), cytokines, proteases, and lipids. These further increases leukocyte adhesion and damages the tight junctions between cells, exacerbating endothelial cell barrier dysfunction (Ma et al., 1993; Weyrich et al., 1993; Vinten-Johansen, 2004; Arslan et al., 2011; Timmers et al., 2012). Endogenous molecules released by damaged or dead cells and extracellular matrix, called danger-associated molecular patterns (DAMPs), initiate immune responses after MI, such as high-mobility group box-1 (HMGB1), heat shock proteins (HSPs), and fibronectin fragments (Fan et al., 2005; Timmers et al., 2012; Mi et al., 2019; Gong et al., 2020). These molecules continuously activate pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), in monocytes and neutrophils of innate immunity. The activation of these molecules initiates intracellular signal transduction pathways, such as mitogen-activated protein kinase (MAPK), Janus kinase (JAK), and calcineurin pathway. This induces the cascade amplification of nuclear factor-kappaB (NF-κB) and activating protein AP-1 to downstream inflammatory mediators (Mann, 2011; Newton and Dixit, 2012; Ghigo et al., 2014). An excessive inflammatory response may lead to pathological phenomena, such as maladaptive remodeling, interstitial fibrosis, and impaired myocardial contractility. Therefore, the recruitment of inflammatory-related cells must be strictly controlled to ensure functional cardiac healing. For instance, secreting growth differentiation factor-15, transforming growth factor-β (TGF-β), and other anti-inflammatory signals can limit the infiltration range of immune cells (Wilson et al., 2004; Wrigley et al., 2011; Ghigo et al., 2014).
The initial response of immune cells to damage is regulated by immediate epigenetic modifications, such as DNA methylation-mediated key activation. However, limited research has been conducted on the epigenetic modification of immune cells during the later stages, particularly during the repair regression phase (Busslinger and Tarakhovsky, 2014; Phan et al., 2017; Placek et al., 2019). In an experiment, rats with MI were administered with 5-azacytidine (5-AZ) as a DNA methyltransferase inhibitor (DNMTi). This intervention led to a reduction in the count of M1 phenotype macrophages expressing iNOS and an elevation in the presence of anti-inflammatory M2 phenotype macrophages in the infarcted myocardium (Figure 2). The expression frequency of Arg1 increased, improving the myocardial diastolic function after MI (Stresemann and Lyko, 2008; Kim et al., 2014). Dynamic DNA methylation modification plays a significant role in the self-renewal and differentiation of immune cells, presenting itself as a potential target for addressing inflammatory processes in diseases (Luo et al., 2018; Morales-Nebreda et al., 2019).
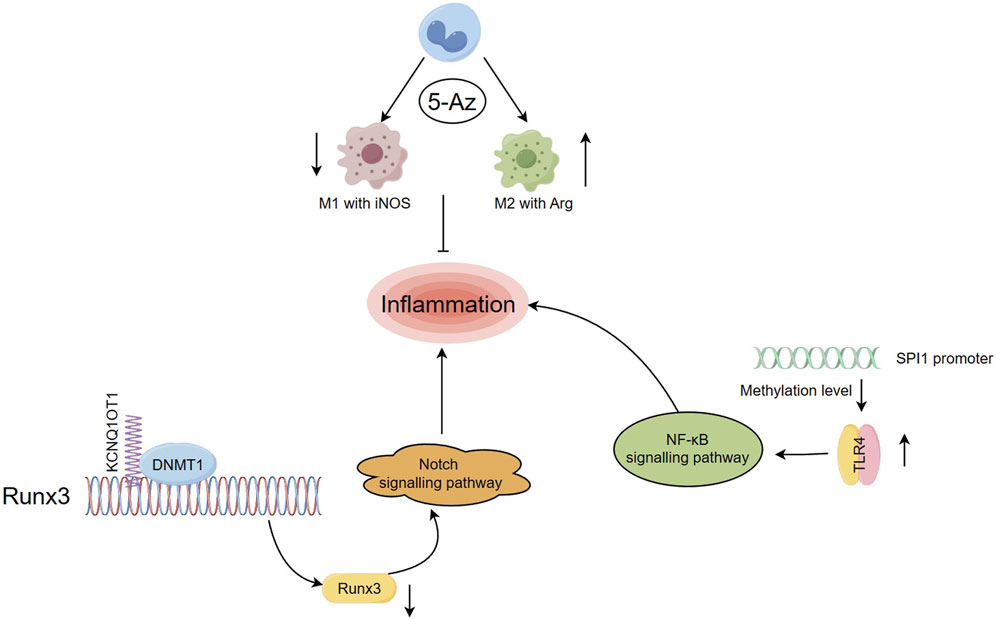
FIGURE 2. Regulation of DNA methylation on inflammation after myocardial infarction (Han, 2019) Rats with MI were administered with 5-Az, resulting in a decrease in the number of M1 phenotype macrophages expressing iNOS and an increase in the level of anti-inflammatory M2 phenotype macrophages. (Zhao et al., 2019). lncRNA KCNQ1OT1 mediated imprinted gene silencing by recruiting DNMT1 and elevating the CpGi methylation level in the promoter region of RUNX3, which in turn suppressed RUNX3 expression levels and affected inflammatory responses through the Notch pathway (Townsend et al., 2022). After MI, the levels of DNA methylation at the SPI1 promoter CpGi were significantly decreased, upregulated SPI1 expression levels bound to the TLR4 promoter and activated the TLR4/NF-κB pathway, exacerbating inflammation.
Studies have demonstrated that, in addition to coding genes, long noncoding RNAs (lncRNAs) play a role in the progression of genetic variations following MI. They are involved in regulating cardiac development and pathological conditions, such as myocardial ischemia (Saddic et al., 2017). For example, the lncRNA potassium voltage-gated channel subfamily Q member 1 overlapping transcript 1 (KCNQ1OT1) is expressed at significantly higher levels in MI patients compared to healthy individuals (Vausort et al., 2014). KCNQ1OT1 is responsible for silencing a gene cluster in cis and is associated with epigenetic modifications such as DNA methylation and histone modifications that play a role in colorectal carcinogenesis (Nakano et al., 2006; Wan et al., 2013). Previous research has shown that knockdown of KCNQ1OT1 can modulate adiponectin receptor 1 and attenuate ischemia-reperfusion (I/R) injury after acute myocardial infarction (AMI) by affecting the p38 MAPK/NF-κB signaling pathways (Li et al., 2017). In a study on MI mice, it was found that lncRNA KCNQ1OT1 mediated imprinted gene silencing by recruiting DNMT1 and elevating the CpGi methylation level in the promoter region of runt related transcription factor 3 (RUNX3), which in turn suppressed RUNX3 expression levels. This affected cardiac microvascular endothelial cells activity and inflammatory responses in post-MI mice through the Notch1 pathway (Figure 2), and the dysfunction of endothelial cells was associated with adverse cardiac remodeling after MI (Mohammad et al., 2010; Wang Y. et al., 2019). Furthermore, when RUNX3 is knocked down as a common downstream target of TGF-β and Notch signal pathways in cardiovascular disease, it can attenuate hypoxia-induced endothelial-mesenchymal transition (EndMt) and reverse human cardiac microvascular endothelial cells function. This suggests that targeting RUNX3 may be a novel approach to mediating EndMt for the treatment of CVD (Liu et al., 2017).
The proto-oncogene SPI1 is considered a key player in the progression to heart failure (HF) after MI. It is tightly associated with inflammation, immune activity, and apoptosis, and its possible target TLR4 plays an equally important role in mediating tissue damage and inflammatory responses (Niu et al., 2019; Ramirez-Carracedo et al., 2020). A recent study showed that in MI mice and hypoxia-exposed HL-1 cells, the levels of DNA methylation at the SPI1 promoter CpGi were significantly decreased, leading to the activation of SPI1 transcription. Upregulated SPI1 expression levels then bound to the TLR4 promoter and activated the TLR4/NF-κB pathway, exacerbating inflammation and cardiomyocyte apoptosis (Liu and Huang, 2022) (Figure 2). Previous studies have confirmed that the activation of the TLR4/NF-κB signaling pathway is involved in inducing tissue injury and inflammation after MI. Inhibition of this pathway is a promising and controllable target for anti-inflammatory treatment after MI (Li et al., 2018; Wang XZ. et al., 2019; Liu et al., 2019; Li et al., 2020).
During different stages after MI, immune cells exhibit functional heterogeneity. In the early stages of myocardial injury, functional changes involving macrophages are accompanied by infiltration of neutrophils and monocytes that promote inflammation. Later on, repair-predominant subpopulations gradually replace these cells (Bajpai et al., 2019; Dick et al., 2019). Further exploration of cell-specific functional type changes during inflammation, linked to spatiotemporally specific epigenetic modifications such as DNA methylation, is necessary to identify the best targets for clinical treatment after MI.
2.2 Autophagy and DNA methylation
As a dynamic circulatory system, autophagy primarily transports long-lived proteins and damaged organelles into lysosomes for degradation (Yang and Klionsky, 2010; Mizushima and Komatsu, 2011). Autophagy is often associated with ischemic heart disease (Bravo-San Pedro et al., 2017) and can be further enhanced by I/R (Valentim et al., 2006). During acute ischemia, autophagy may play a protective role in maintaining energy supply and reducing infarct size (Matsui et al., 2007; Zhu et al., 2007; Kanamori et al., 2011; Chen et al., 2013), while also endowing the body with some anti-stress ability (Yan et al., 2006). However, excessive activation of autophagy by oxidative stress during reperfusion (Liu et al., 2018), which clears damaged organelles intracellularly, can be harmful to the organism. Autophagy is a controversial topic, as many studies have shown that autophagy, as a stress response mechanism, is upregulated in myocardial reperfusion injury, and the impairment of autophagy clearance ability can lead to cell death (Ma et al., 2012). Therefore, balancing autophagy activity in a timely manner to keep it within a safe range is crucial to maintain the stability of myocardial cells and overall cardiac structure and function (Nakai et al., 2007; Xu et al., 2015) and prevent cardiac dysfunction after MI. Currently, there is no effective intervention strategy for regulating autophagy-related cardiovascular diseases in clinical practice (Zhou et al., 2019). However, epigenetic modifications such as DNA methylation have shown some therapeutic prospects in regulating autophagy after MI.
Poorly regulated autophagy is associated with cardiovascular diseases, including myocardial infarction (Tanaka et al., 2000; Yamamoto et al., 2000; Maejima et al., 2013). PTEN-induced putative kinase 1 (Pink1) is downregulated in advanced stages of human HF and is believed to be crucial for maintaining normal cardiac function (Billia et al., 2011). Family with sequence similarity 65 member B (FAM65B) is involved in several cellular processes, such as cell differentiation, but its role in regulating autophagy in cardiomyocytes was previously unknown (Balasubramanian et al., 2014; Diaz-Horta et al., 2014). Recently, a study revealed that autophagy-related circular RNA (ACR) directly binds to DNMT3b and prevents DNA methylation of the Pink1 promoter, a process regulated by DNMT3b. ACR activates the expression of the Pink1 promoter by targeting a reduction in its DNA methylation level. Additionally, it facilitates the phosphorylation of FAM65B, leading to the inhibition of autophagy and cell death. This process reduces ischemia/reperfusion injury and the area of myocardial infarction (Figure 3). The existing evidence demonstrates that the ACR-Pink1-FAM65B axis, which is regulated by the DNA methylation pathway, can be utilized as a regulator of cardiac autophagy (Zhou et al., 2019). Circular RNA interacts with DNMT and participates in the regulation of DNA methylation, which highlights its potential as a therapeutic target for MI and I/R injury.
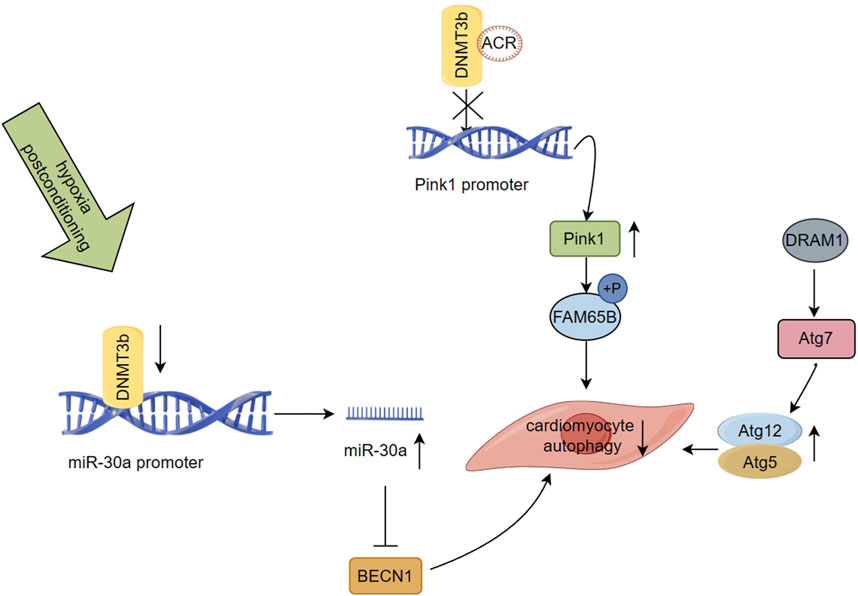
FIGURE 3. Regulation of DNA methylation on autophagy after myocardial infarction (Han, 2019) ACR binds to DNMT3b and prevents DNA methylation of the Pink1 promoter, which activates the expression of the Pink1, and it mediates FAM65B phosphorylation to inhibit autophagy and cell death (Zhao et al., 2019). Suppressed DNMT3b activity leads to miR-30a hypomethylation and increases its expression, which results in the targeting of the autophagy related gene BECN1 by miR-30a, inhibiting autophagy induction in aging cardiomyocytes (Townsend et al., 2022). DRAM1 was found to be negatively regulated in the rat model of AMI. Regulating the autophagic flow of DRAM1-Atg7-Atg12/Atg5 under the stress condition of myocardial ischemia can alleviate autophagic flow and regulating the mechanism of myocardial cell protection.
Ischemic postconditioning (IPostC) is a potential strategy for protecting cardiomyocytes during early reperfusion by intermittently interrupting coronary blood flow, thereby reducing I/R injury and final infarct size (Zhao et al., 2003; Heusch et al., 2011). IPostC has been implicated in regulating autophagy and may offer myocardial protection in I/R injury (Ma et al., 2011; Wang et al., 2016). The methylation status of miRNA promoters may serve as potential epigenetic biomarkers for clinical applications and directly regulate their expression levels (Vera et al., 2017; Ortiz et al., 2018). A study investigating hypoxic postconditioning (HPostC) in aging cardiomyocytes exposed to hypoxia reoxygenation (H/R) injury revealed that DNMT3b mediates hypomethylation of the miR-30a promoter, leading to an increase in the expression levels of miR-30a. This results in the targeting of the autophagy related gene BECN1 by miR-30a, inhibiting autophagy induction in aging cardiomyocytes, and promoting HPostC to exert cardioprotective effects (Figure 3). Suppressed DNMT3b activity leads to miR-30a hypomethylation and increases its expression, providing an additional epigenetic regulatory pathway (Wang et al., 2020). This study not only uncovers the protective effect of IPostC against I/R injury in aging cardiomyocytes but also sheds light on the underlying molecular mechanism of I/R injury via autophagy regulation. It provides a highly promising DNA methylation-related therapeutic target for achieving precise treatment and improving myocardial injury in elderly patients with MI, in agreement with previous clinical reports (Wang et al., 2016; Zhang et al., 2018).
Damage-regulated autophagy modulator 1 (DRAM1) is a lysosomal membrane protein that is be beneficial in attenuating autophagic flux impairment and cardiac functional maladaptation in acute myocardial infarction (AMI) due to ischemic stress, ultimately improving cardiac outcomes in patients (Crighton et al., 2006; Wu et al., 2014; Wu et al., 2018). A recent study using an AMI rat model demonstrated that DRAM1 upregulates the mRNA level of Atg12 in cardiomyocytes in vitro and in vivo, while decreasing the levels of free Atg12 protein. Additionally, an increase in the formation of Atg12-Atg5 conjugates was identified. DRAM1 showed a significant direct interaction only with Atg7, which regulated the autophagic flow of DRAM1-Atg7-Atg12/Atg5 under the stress condition of myocardial ischemia, thereby alleviating autophagic flow and regulating the mechanism of myocardial cell protection (Wu et al., 2018) (Figure 3). Moreover, DRAM1 was found to be negatively regulated in the rat model of AMI, where ischemia-induced aberrant DNA hypermethylation status at multiple promoter CpG sites of DRAM1 was associated with decreased DRAM1 expression levels in the infarct border zone. In summary, these findings suggest that targeting DNA methylation and autophagy pathways via DRAM1 could be a promising therapeutic strategy to regulate autophagy flux and improve outcomes in patients with AMI.
In recent years, epigenetic regulation such as DNA methylation and histone modification has been extensively investigated in the context of autophagy. These modifications can not only directly modify autophagy-related genes but also influence signaling molecules that regulate these genes (Klionsky et al., 2021). In cancer research, the methylation status of autophagy-regulating effector molecules has been found to play a significant role in exhibiting biphasic regulatory effects (Liu H. et al., 2013; Liao et al., 2014; Nihira et al., 2014; Zhang et al., 2016; Bhol et al., 2020). Similarly, DNA methylation in conjunction with autophagy in the context of MI and I/R injury presents a promising avenue for research. Targeted therapy strategies based on the aberrant DNA methylation status of autophagy-related molecules in cardiomyocytes may become a crucial clinical direction in the near future.
2.3 Fibrosis and DNA methylation
In tissues affected by myocardial infarction, ongoing ischemia, hypoxia, inflammation, and other stimuli induce cardiac fibrosis and abnormal cardiac remodeling, which are inevitable pathological changes in MI progression (van den Borne et al., 2010; Barallobre-Barreiro et al., 2012; Opie et al., 2006; Brown et al., 2005). Interstitial fibroblasts respond to elevated TGF-β and changes in the extracellular matrix by transforming into myofibroblasts, leading to further increases in proliferative activity (Willems et al., 1994; Frangogiannis et al., 2000; Souders et al., 2009; Turner and Porter, 2013). These pathways serve as markers for the proliferative phase of repair after MI and are highly involved in regulating cardiac fibrosis-related remodeling, which has a major impact on cardiac function (Hinz et al., 2007). Under stress conditions, cardiac fibroblasts express α-smooth muscle actin (α-SMA) and the smooth muscle dedifferentiation marker myosin heavy chain embryonic isoform (Frangogiannis et al., 2000). This disrupts the equilibrium between matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases, leading to the production of an extracellular matrix (ECM) that distinguishes fibroblasts (Berk et al., 2007). The reparative proliferation process of the myocardium in the infarcted area involves the regulated deposition and breakdown of ECM, particularly collagen, which is controlled by regulatory elements such as MMPs (Watson et al., 2014; Olsen et al., 2017). Although fibrosis initially reflects adaptive protective mechanisms, dysregulation of the balance can lead to excessive ECM deposition, resulting in pathological reactive fibrosis that increases myocardial stiffness, disrupts tissue architecture, and impairs cardiac function (Levick et al., 2009; Qi et al., 2014; Murtha et al., 2017). Accumulating evidence suggests that the hyperactive fibrotic phenotype may be attributed to the accumulation of gene expression within the interstitium, and the causal role that epigenetics plays in fibrosis is gradually being recognized. DNA methylation may play an important role in the progression of cardiac fibrosis (Gabrielsen et al., 2007; Mann and Mann, 2013; Watson et al., 2014; Yang and Schwartz, 2015; Kale et al., 2021).
NEIL3 is a mammalian oxidized base-specific DNA glycosylase in the base excision repair pathway. It is elevated in the myocardium of patients with HF, and its expression increases significantly in fibroblast enriched fractions after myocardial infarction in mice (Liu M. et al., 2013; Olsen et al., 2017). NEIL3 regulates cell proliferation in various human tumor tissues, murine hematopoietic tissues, and various stem cell populations in mice (Torisu et al., 2005; Hildrestrand et al., 2009; Regnell et al., 2012; Reis and Hermanson, 2012; Rolseth et al., 2013). Some studies suggest that NEIL3 affects the balance between methylation and oxidative demethylation of cytosine related epigenetic modifications in the heart. These modifications include differential regulation of both 5 mC and 5 hmc. Alternatively, NEIL3 may regulate the proliferation and differentiation of fibroblast-like cells during post-MI repair by affecting DNA methylation sites. The results show that NEIL3 deficient mice have a decreased ability to regulate cell proliferation after MI, implying a deregulation of fibroblast-like cells proliferation and differentiation. This leads to a disruption of the extracellular matrix balance, including increased MMP2 production and consequent fatal consequences such as myocardial rupture. Therefore, understanding the regulatory mechanisms of fibroblast-like cell proliferation and differentiation and ECM balance is important for developing clinical antifibrotic drugs. NEIL3 could be a potential therapeutic target for this purpose.
After MI, ischemic hypoxic injuries commonly occur and may require long-term adaptation involving epigenetic modifications to maintain the fibrotic phenotype. A study quantified the expression levels of the CAIX gene, regulated by hypoxia-inducible factor-1 (HIF-1), as a means to assess the degree of myocardial tissue hypoxia. Prolonged exposure of human cardiac fibroblasts to 1% hypoxia resulted in a profibrotic state, and the degree of hypoxia correlated with collagen 1 (Col1) and α-SMA expression levels and collagen deposition (Holotnakova et al., 2008). HIF-1a plays a role in regulating global DNA hypermethylation and a profibrotic state, potentially through the binding of hypoxia response elements (HRE) to DNMT1 and DNMT3b promoter sites, resulting in a significant increase in DNMT1 and DNMT3b expression levels (Watson et al., 2014). Moreover, under hypoxic conditions, the profibrotic effect of TGF-β, a profibrotic agonist, can be inhibited by the DNMTi 5-aza-2′- deoxycytidine (5-aza), which significantly reduces the expression levels of α-SMA and Col1 (Figure 4). However, previous studies have shown that TGF-β1 may inhibit DNMT expression levels and overall activity, while upregulating Col1A1 in cardiac fibroblasts. The role of DNA methylation in the fibrotic process in vivo during hypoxia induction requires further investigation, but the strong correlation of DNMT1 and DNMT3b in hypoxic injury and fibrosis suggests the potential for investigating basic and fibrosis-targeted therapies (Watson et al., 2014).
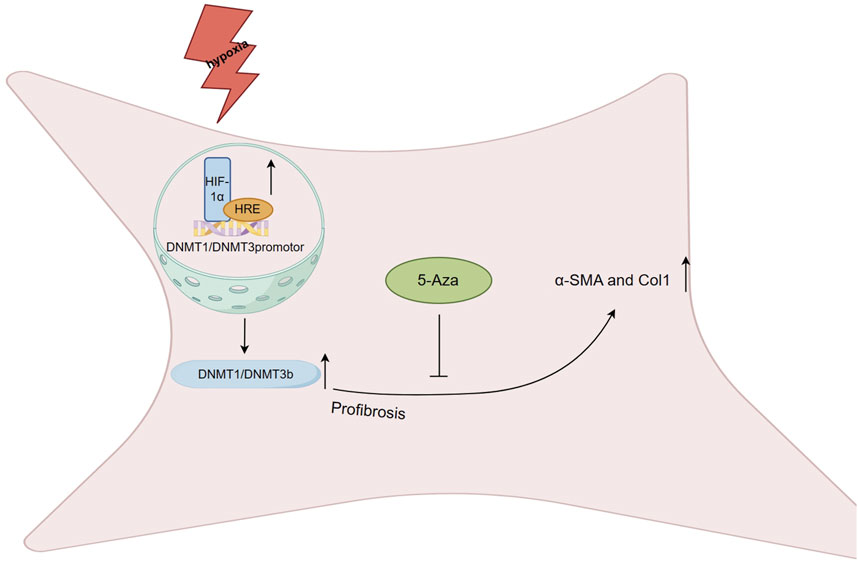
FIGURE 4. Regulation of DNA methylation on fibrosis after myocardial infarction HIF-1a plays a role in regulating global DNA hypermethylation and a profibrotic state, potentially through the binding of hypoxia response elements (HRE) to DNMT1 and DNMT3b promoter sites, resulting in a significant increase in DNMT1 and DNMT3b expression levels. Under hypoxic conditions, the profibrotic effect of TGF-β can be inhibited by the 5-Aza, which significantly reduces the expression levels of α-SMA and Col1.
The differentiation of cardiac fibroblasts into α-SMA expressing myofibroblasts is a crucial aspect of the fibrotic process after MI. Recent studies have suggested that post-MI α-SMA expression is regulated by DNA methylation, highlighting the importance of both in cardiac fibroblast differentiation (Hinz et al., 2007; Souders et al., 2009; Williams et al., 2014; He et al., 2019). In the infarcted area of MI rats, α-SMA is overexpressed, but DNMT1 expression levels are decreased. In vitro treatment of cardiac fibroblasts with TGF-β1 has been shown to induce upregulation of α-SMA expression by significantly inhibiting DNMT1-mediated DNA methylation at multiple CpG sites in the α-SMA promoter. The Smad and MAPK pathways also play a role in regulating DNMT1 expression in cardiac fibroblasts (He et al., 2019).
Although there are still controversies regarding cardiac fibroblast differentiation, it is undeniable that DNA methylation plays an essential role in maintaining the fibrotic phenotype during the profibrotic process after MI. Further research is needed to resolve the existing contradictions and to investigate the specific manifestations of post-MI epigenetic modifications in the fibrotic process (Pan et al., 2013; Watson et al., 2014; He et al., 2019). DNMT-related drug targets hold great potential for regulating expression in both fibrosis and the control of cardiac fibroblast differentiation, making them highly valuable as clinical therapeutic properties (Tao et al., 2014; Watson et al., 2014).
2.4 Cardiomyocyte proliferation and DNA methylation
The imbalance of cardiomyocyte death and regeneration is one of the factors contributing to adverse myocardial remodeling and cardiac dysfunction (Gill et al., 2002; van Empel et al., 2005). The heart, being one of the least regenerative organs in the mammalian body, has limited regenerative capacity in the adult period, as cardiomyocytes essentially exit the mitotic cell cycle and stop cell proliferation (Drenckhahn et al., 2008; Walsh et al., 2010; Laflamme and Murry, 2011). This lack of regenerative capability is the basis for functional deficits in the regeneration and repair of the adult heart in response to pathological insults, such as MI, ultimately leading to cardiac dysfunction and HF (Ponnusamy et al., 2019). However, non-mammals such as amphibians and teleosts maintain significant cardiac regenerative capacity throughout life (Oberpriller and Oberpriller, 1974; Jopling et al., 2010; Kikuchi et al., 2010), and adult zebrafish can induce cardiomyocyte proliferation and overcome scar formation after removing 20% of their ventricles, achieving complete cardiac regeneration (Poss et al., 2002). Recent studies have shown that humans and other mammals still maintain some renewal capacity throughout their lifespan, including after MI (Beltrami et al., 2001; Bergmann et al., 2009). A small number of cardiomyocytes undergoing mitotic proliferation have been observed in the infarct border zone after MI in patients (Beltrami et al., 2001). However, eventually, the myocardium does not regenerate appreciably but instead replaces it with a fibrotic scar (Nag et al., 1983). In ischemic heart disease, 152 cardiomyocytes per million undergo proliferation, a tenfold elevation compared with the normal left ventricle (Kajstura et al., 1998). Achieving therapeutic cardiac regeneration, such as activating cardiomyocyte proliferation in situ, is currently an important biomedical goal of therapeutic strategies after MI (Karra and Poss, 2017). Nonetheless, strong and precise targeting of cardiomyocyte proliferation is necessary to prevent unintended proliferative events in non-cardiomyocytes and to avoid any neoplastic effects, ensuring safety (Lin and Pu, 2014; Gabisonia and Recchia, 2018).
Cardiac progenitor cells (CPCs) are crucial resident cells of the heart with multipotent, clonogenic, and self-renewal abilities for cardiac regeneration (Urbanek et al., 2006; Sanada et al., 2014; Leri et al., 2015). These cells can differentiate into cardiomyocytes, endothelial cells, and vascular smooth muscle cells, playing a significant role in protecting the heart and generating blood vessels to prevent adverse cardiac remodeling after MI (Beltrami et al., 2003; Matsuura et al., 2009; van Berlo et al., 2014; Liu et al., 2015). However, after MI, the population of distinct cardiac side-population cells is reduced to less than half of their original levels 1 day after the injury, and only a few CPCs remain once large numbers are transplanted to the injured heart (Mouquet et al., 2005; Su et al., 2018). Moreover, patients who suffered from MI showed higher levels of serum HMGB1, a protein closely related to inflammatory response, cell proliferation, and apoptosis (Bell et al., 2006; Goldstein et al., 2006; Gwak et al., 2012; Kang et al., 2014). Studies have reported that CPCs treated with hypoxia significantly increased the level of expressed HMGB1, and knockdown of HMGB1 could attenuate hypoxia-induced apoptosis in these cells. This mechanism was found to be due to the hypoxic stimulus inhibiting the expression of DNMT1. This reduction in DNMT1 led to a decrease in the methylation of CpGi at HMGB1 promoter in CPCs, subsequently upregulating the HMGB1 expression level. Additionally, the MAPK signaling pathway is also involved in regulating the mechanism of post-hypoxic apoptosis of CPCs mediated by HMGB1. This study identified an important role of the MAPKs/DNMT1/HMGB1 signaling axis in regulating post-hypoxic apoptosis of CPCs and provided a new direction for CPCs apoptosis and proliferation, contributing to the development of valuable targets for stem cell therapy after MI (Su et al., 2018).
Numerous studies have demonstrated the involvement of the Notch signaling pathway in processes relevant to the regulation of cardiomyocyte proliferation during heart development (Pedrazzini, 2007; Niessen and Karsan, 2008; de la Pompa and Epstein, 2012; MacGrogan et al., 2018; Wang et al., 2021). In particular, Notch1 is highly expressed in immature proliferating cardiomyocytes during the early neonatal period to protect them from apoptosis (Collesi et al., 2008). Reactivation of the Notch signaling pathway has been shown to induce embryonic stem cell-derived ventricular myocytes to re-enter the cell cycle (Campa et al., 2008). Studies have demonstrated that DNA methylation serves as an irreversible marker of transcriptional repression (Cedar and Bergman, 2009), and after birth in neonatal rats, cardiomyocyte proliferation is reduced coincident with a significant reduction in Notch1 signaling levels (Felician et al., 2014). Yet, utilizing adeno-associated virus vectors for gene transfer to activate the Notch1 signaling pathway after MI in adult mice proves ineffective. Additionally, deliberately activating the Notch1 pathway does not succeed in inducing cardiomyocyte proliferation in adult mice after MI. It has been proposed that the terminal differentiation state of cardiomyocytes is associated with the temporally progressive DNA methylation of Notch1 promoters and its target genes (Hes1, Hey1, and Hey2). Treatment with DNMTi 5-aza has been confirmed to correlate with the repressive role of Notch gene promoter methylation. This study argues that induction of adult cardiomyocyte proliferation by Notch pathway stimulation is not a suitable strategy to promote cardiac regenerative responses in adult mice. However, as research progresses, DNA demethylation-related epigenetic modifications and epigenetic drugs such as DNMTi advance clinical experiments in the field of cancer (Jones et al., 2016; Wu and Zhang, 2017; Soler-Botija et al., 2020). The methylation sites of Notch and its target genes may become highly promising therapeutic targets soon.
A long non-coding RNA named cardiomyocyte proliferation regulator (CPR) has potential regulatory effects on cardiomyocyte proliferation and cardiac repair. Studies have found that CPR negatively regulates cardiomyocyte proliferation and regeneration, whereas silencing CPR significantly increases cardiomyocyte proliferation in postnatal and adult hearts and restores cardiac function after MI (Ponnusamy et al., 2019). The interaction of CPR with DNMT3a guides DNMT3a to bind CpGi sites of the minichromosome maintenance protein 3 (Mcm3) promoter region, an initiator of eukaryotic genome replication and cell cycle progression. This targeted binding leads to an increase in methylation levels, subsequently diminishing the expression of Mcm3 and inhibiting cardiomyocyte proliferation (Figure 5). Loss of cardiomyocyte cell cycle activity induces robust fibrosis response and scar formation in the adult heart after injury (Porrello et al., 2011). Aberrant fibrosis, scarring, and loss of cardiac regenerative capacity after MI are all critical contributors to adverse cardiac remodeling. The reduction of methylation levels of the Mcm3 promoter region by targeted inhibition of CPR expression or using epigenetic modifying drugs such as DNMTi may be a constructive view to promote cardiomyocyte proliferation after MI and alleviate adverse cardiac remodeling (Ponnusamy et al., 2019).
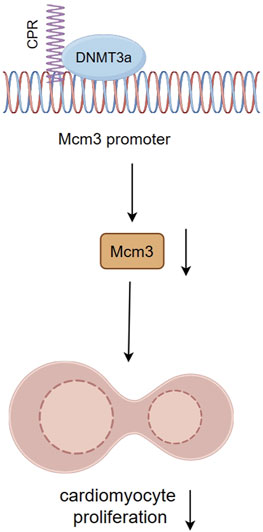
FIGURE 5. The interaction between CPR and DNMT3a inhibits cardiomyocyte proliferation by inhibiting Mcm3. The interaction of CPR with DNMT3a guides DNMT3a to bind CpGi sites of Mcm3 promoter region, directionally elevating methylation levels, thereby reducing Mcm3 expression and inhibiting cardiomyocyte proliferation.
Epigenetic reprogramming exhibits great potential after MI, such as transplantation to treat MI by in vitro DNA methylation modification, converting bone marrow progenitor cells into cardiac progenitor cells (Rajasingh et al., 2011), and reprogramming of fibroblasts into functional cardiomyocyte-like cells or inducing cardiomyocyte proliferation (Ieda et al., 2010; Liu and Schwartz, 2012; Qian et al., 2012). Through continued research progress, we can bridge the gap in clinical trials related to epigenetic drugs for cardiomyocyte proliferation in cardiovascular diseases and identify the most effective targets to promote cardiac tissue regeneration after MI.
2.5 Drug treatment and DNA methylation
Currently, medications such as aspirin, warfarin, tissue plasminogen activator, and interventional therapy, such as percutaneous coronary intervention (PCI), are the mainstay of treatment for AMI (Lu et al., 2015; Doenst et al., 2019; Sabatine and Braunwald, 2021). However, unpredictable complications such as bleeding, ischemia/reperfusion injury, and coronary restenosis may occur, highlighting the need for safer and more innovative therapeutic strategies to optimize clinical outcomes (McCarthy et al., 2018; Doenst et al., 2019; Mackman et al., 2020; Zhang et al., 2022).Several studies have observed significant individual methylation differences in patients with MI (Ek et al., 2016; Rask-Andersen et al., 2016; Thunders et al., 2019; Han et al., 2022; Luo et al., 2022; Ren et al., 2022). Further exploration of the regulatory mechanisms of DNA methylation and its potential impact on metabolism and vascular physiology after MI may help reduce the incidence of post-MI complications (Ward-Caviness et al., 2018). Epigenetic modification-related drugs, particularly DNMT inhibitors, have demonstrated efficacy in the treatment of acute myeloid leukemia and myelodysplastic syndrome (Sudan et al., 2006; Cashen et al., 2010; Lübbert et al., 2012; Saunthararajah et al., 2012). Although the research on DNA methylation-related drugs in cardiovascular diseases is relatively lacking, the existing achievements reflect considerable value and promise, given the success of epigenetic drugs in other fields such as cancer.
Clopidogrel is a medication that significantly reduces the risk of adverse ischemic events after PCI compared to aspirin (Steinhubl et al., 2002). However, resistance to clopidogrel remains an important factor contributing to the recurrence of ischemic events following antiplatelet therapy (Mehta et al., 2001). Recent studies have shown that clopidogrel resistance is associated with increased DNA methylation levels at the promoters of genes such as ABCB1, P2Y12, and PON1 (Su et al., 2014; Su et al., 2017; Su et al., 2019). Furthermore, CYP2C19, a key enzyme in the biotransformation of clopidogrel, and its reduction in DNA methylation have been found to increase the risk of resistance and worsen clinical outcomes in STEMI patients undergoing clopidogrel preconditioning after PCI (Sukmawan et al., 2021). All 14 CpGis of the CYP2C19 gene are located in the gene body, which, according to the methylation paradox, suggests that reduced DNA methylation levels in the CYP2C19 gene body are responsible for its reduced transcriptional expression, according to the methylation paradox (Jones, 1999). The reduction in CYP2C19 levels blocks the biotransformation of clopidogrel, reducing active metabolites, which leads to resistance. Genetic polymorphisms of CYP2C19 have also been found to increase the risk of resistance to clopidogrel by 4.2 to 5.3-fold (Sukmawan et al., 2021). Recent advances in CYP2C19 DNA methylation and clopidogrel resistance further point to epigenetic modifications playing a key role in post-MI pharmacotherapy.
Effective mitigation strategies are urgently needed for H/R injury caused by post-MI treatment (Hausenloy and Yellon, 2013). Dexmedetomidine (DEX), a highly selective α2-adrenoceptor agonist commonly used as a sedative and anesthetic agent in clinical settings, has been extensively studied for its protective effects against H/R-mediated organ injuries, including those affecting the brain, liver, and kidney (Yuan et al., 2019; Yu et al., 2020; Zhang B. et al., 2021; Wu et al., 2021). DEX has been shown to inhibit cardiomyocyte apoptosis triggered by H/R injury by upregulating SIRT1 via the SIRT1/CHOP pathway (Zhang Y. et al., 2021). Overexpression of SIRT1 can hinder NF-κB acetylation, thus improving cardiac function (Lin et al., 2020). Moreover, a recent study has refined and expanded on these findings by exploring the link between DNA methylation and H/R injury. By increasing SIRT1 expression levels through DNMTi-mediated demethylation of the SIRT1 promoter, DEX can inhibit NF-κB activation and ameliorate H/R-triggered myocardial injury (Wang et al., 2022). Given that Tet1-mediated DNA demethylation is critical to DEX’s mechanism of action in alleviating H/R injury, this approach represents a promising new pharmacological strategy based on DNA methylation for mitigating H/R injury after MI.
Although progress in epigenetic drug applications for MI-related cardiovascular diseases in clinical trials has been slow, preclinical studies in animal models have shown promising results (Soler-Botija et al., 2020). For instance, in a rat MI model, the DNA methylation inhibitor 5-AZ was found to shift macrophages towards an M2 anti-inflammatory phenotype and prevent cardiac contractile decompensation by inhibiting the promoter activity of inducible nitric oxide synthase (iNOS). Furthermore, 5-AZ was found to activate ubiquitin-conjugating enzyme 9 to mediate the sumoylation of interferon regulatory factor 1 in macrophages, which circumvented rapid degradation by ubiquitination in the proteasome and enabled its accumulation in cells (Nakagawa and Yokosawa, 2000). By altering its effect and incapacitating iNOS, 5-AZ promotes macrophages to switch to an M2 anti-inflammatory phenotype (Jeong et al., 2015). Given the clinical availability of 5-AZ as an epigenetic drug, it may offer a promising new direction for post-MI inflammation therapy. In an MI rat model with reperfusion injury, treatment with another DNMTi, epigallocatechin-3-gallate (EGCG), reduced plasma interleukin-6 levels, and post-ischemic neutrophil infiltration. EGCG suppressed IκB kinase (IKK) activity after reperfusion and markedly weakened the degradation of inhibitor κB-α, thereby significantly reducing NF-κB’s bound state to DNA. Moreover, EGCG attenuated c-Jun phosphorylation at all time points post-MI reperfusion, affecting the activation process of AP-1 (Aneja et al., 2004). These findings suggest that EGCG may inhibit the IKK/NF-κB signal transduction pathway directly or indirectly by altering the redox status at the site of inflammation. By leveraging its properties as an epigenetic drug and understanding its underlying molecular mechanism, EGCG may offer a promising clinical therapeutic strategy for modulating the associated inflammation during blood flow reconstruction after MI.
We summarize the DNA methylation drugs mentioned above and the DNA methylation targets involved in the pathophysiological processes after myocardial infarction in Table 1; Supplementary Table S1, respectively.
3 Conclusion
Considerable in vivo and in vitro evidence now supports the potential use of epigenetic drugs as therapeutic tools to improve functional recovery after MI, however, there is currently no corresponding clinical evidence. Conducting clinical trials with a limited number of patients can lay the groundwork for identifying safe and efficacious epigenetic drugs, paving the way for their broader application in a larger patient population. Currently, epigenetic drugs in clinical trials are broadly divided into two classes: genomic drugs, such as the DNMTi mentioned, for extensive reprogramming, and targeted precision medicine for the treatment of specific diseases. DNMTi, in particular, have shown great promise in post-MI treatment, suggesting their potential for further development in clinical applications. We believe that drugs targeting the prevention of cardiomyocyte death and promotion of cardiomyocyte regeneration during the process of myocardial infarction may have promising clinical prospects. However, many drugs, such as DNMTi, are administered systemically, and their side effects require further evaluation. With the development of targeted drug delivery technologies, these drugs may be widely applied in the future.
Author contributions
WH: Methodology, Writing–original draft, Writing–review and editing. WW: Investigation, Writing–original draft, Writing–review and editing. QW: Writing–review and editing. KM: Writing–review and editing. LH: Writing–review and editing. JZ: Funding acquisition, Supervision, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82270331 and 82000235), Qingdao Key Health Discipline Development Fund and Qingdao Key Clinical Specialty Elite Discipline (QDZDZK-2022008). The funding sources had no roles in the design and conduct of this work.
Acknowledgments
Figures 2–5 in the manuscript were drawn in Figdraw.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1267585/full#supplementary-material
References
Aneja, R., Hake, P. W., Burroughs, T. J., Denenberg, A. G., Wong, H. R., and Zingarelli, B. (2004). Epigallocatechin, a green tea polyphenol, attenuates myocardial ischemia reperfusion injury in rats. Mol. Med. 10 (1-6), 55–62. doi:10.2119/2004-00032.aneja
Aran, D., Toperoff, G., Rosenberg, M., and Hellman, A. (2011). Replication timing-related and gene body-specific methylation of active human genes. Hum. Mol. Genet. 20 (4), 670–680. doi:10.1093/hmg/ddq513
Arslan, F., de Kleijn, D. P., and Pasterkamp, G. (2011). Innate immune signaling in cardiac ischemia. Nat. Rev. Cardiol. 8 (5), 292–300. doi:10.1038/nrcardio.2011.38
Bajpai, G., Bredemeyer, A., Li, W., Zaitsev, K., Koenig, A. L., Lokshina, I., et al. (2019). Tissue resident CCR2-and CCR2+ cardiac macrophages differentially orchestrate monocyte recruitment and fate specification following myocardial injury. Circulation Res. 124 (2), 263–278. doi:10.1161/CIRCRESAHA.118.314028
Balasubramanian, A., Kawahara, G., Gupta, V. A., Rozkalne, A., Beauvais, A., Kunkel, L. M., et al. (2014). Fam65b is important for formation of the HDAC6-dysferlin protein complex during myogenic cell differentiation. FASEB J. official Publ. Fed. Am. Soc. Exp. Biol. 28 (7), 2955–2969. doi:10.1096/fj.13-246470
Ball, M. P., Li, J. B., Gao, Y., Lee, J. H., LeProust, E. M., Park, I. H., et al. (2009). Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 27 (4), 361–368. doi:10.1038/nbt.1533
Barallobre-Barreiro, J., Didangelos, A., Schoendube, F. A., Drozdov, I., Yin, X., Fernández-Caggiano, M., et al. (2012). Proteomics analysis of cardiac extracellular matrix remodeling in a porcine model of ischemia/reperfusion injury. Circulation 125 (6), 789–802. doi:10.1161/CIRCULATIONAHA.111.056952
Bell, C. W., Jiang, W., Reich, C. F., and Pisetsky, D. S. (2006). The extracellular release of HMGB1 during apoptotic cell death. Am. J. physiology Cell physiology 291 (6), C1318–C1325. doi:10.1152/ajpcell.00616.2005
Beltrami, A. P., Barlucchi, L., Torella, D., Baker, M., Limana, F., Chimenti, S., et al. (2003). Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 114 (6), 763–776. doi:10.1016/s0092-8674(03)00687-1
Beltrami, A. P., Urbanek, K., Kajstura, J., Yan, S. M., Finato, N., Bussani, R., et al. (2001). Evidence that human cardiac myocytes divide after myocardial infarction. N. Engl. J. Med. 344 (23), 1750–1757. doi:10.1056/NEJM200106073442303
Bergmann, O., Bhardwaj, R. D., Bernard, S., Zdunek, S., Barnabé-Heider, F., Walsh, S., et al. (2009). Evidence for cardiomyocyte renewal in humans. Sci. (New York, NY) 324 (5923), 98–102. doi:10.1126/science.1164680
Berk, B. C., Fujiwara, K., and Lehoux, S. (2007). ECM remodeling in hypertensive heart disease. J. Clin. investigation 117 (3), 568–575. doi:10.1172/JCI31044
Bhol, C. S., Panigrahi, D. P., Praharaj, P. P., Mahapatra, K. K., Patra, S., Mishra, S. R., et al. (2020). Epigenetic modifications of autophagy in cancer and cancer therapeutics. Seminars cancer Biol. 66, 22–33. doi:10.1016/j.semcancer.2019.05.020
Billia, F., Hauck, L., Konecny, F., Rao, V., Shen, J., and Mak, T. W. (2011). PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc. Natl. Acad. Sci. U. S. A. 108 (23), 9572–9577. doi:10.1073/pnas.1106291108
Bravo-San Pedro, J. M., Kroemer, G., and Galluzzi, L. (2017). Autophagy and mitophagy in cardiovascular disease. Circulation Res. 120 (11), 1812–1824. doi:10.1161/CIRCRESAHA.117.311082
Brown, R. D., Ambler, S. K., Mitchell, M. D., and Long, C. S. (2005). The cardiac fibroblast: therapeutic target in myocardial remodeling and failure. Annu. Rev. Pharmacol. Toxicol. 45, 657–687. doi:10.1146/annurev.pharmtox.45.120403.095802
Busslinger, M., and Tarakhovsky, A. (2014). Epigenetic control of immunity. Cold Spring Harb. Perspect. Biol. 6 (6), a019307. doi:10.1101/cshperspect.a019307
Campa, V. M., Gutiérrez-Lanza, R., Cerignoli, F., Díaz-Trelles, R., Nelson, B., Tsuji, T., et al. (2008). Notch activates cell cycle reentry and progression in quiescent cardiomyocytes. J. Cell Biol. 183 (1), 129–141. doi:10.1083/jcb.200806104
Cashen, A. F., Schiller, G. J., O'Donnell, M. R., and DiPersio, J. F. (2010). Multicenter, phase II study of decitabine for the first-line treatment of older patients with acute myeloid leukemia. J. Clin. Oncol. official J. Am. Soc. Clin. Oncol. 28 (4), 556–561. doi:10.1200/JCO.2009.23.9178
Cedar, H., and Bergman, Y. (2009). Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10 (5), 295–304. doi:10.1038/nrg2540
Chen, H. H., Mekkaoui, C., Cho, H., Ngoy, S., Marinelli, B., Waterman, P., et al. (2013). Fluorescence tomography of rapamycin-induced autophagy and cardioprotection in vivo. Circ. Cardiovasc. imaging 6 (3), 441–447. doi:10.1161/CIRCIMAGING.112.000074
Collesi, C., Zentilin, L., Sinagra, G., and Giacca, M. (2008). Notch1 signaling stimulates proliferation of immature cardiomyocytes. J. Cell Biol. 183 (1), 117–128. doi:10.1083/jcb.200806091
Comb, M., and Goodman, H. M. (1990). CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic acids Res. 18 (13), 3975–3982. doi:10.1093/nar/18.13.3975
Crighton, D., Wilkinson, S., O'Prey, J., Syed, N., Smith, P., Harrison, P. R., et al. (2006). DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126 (1), 121–134. doi:10.1016/j.cell.2006.05.034
Das, P. M., and Singal, R. (2004). DNA methylation and cancer. J. Clin. Oncol. official J. Am. Soc. Clin. Oncol. 22 (22), 4632–4642. doi:10.1200/JCO.2004.07.151
Deaton, A. M., and Bird, A. (2011). CpG islands and the regulation of transcription. Genes & Dev. 25 (10), 1010–1022. doi:10.1101/gad.2037511
de la Pompa, J. L., and Epstein, J. A. (2012). Coordinating tissue interactions: notch signaling in cardiac development and disease. Dev. Cell 22 (2), 244–254. doi:10.1016/j.devcel.2012.01.014
Diaz-Horta, O., Subasioglu-Uzak, A., Grati, M., DeSmidt, A., Foster, J., Cao, L., et al. (2014). FAM65B is a membrane-associated protein of hair cell stereocilia required for hearing. Proc. Natl. Acad. Sci. U. S. A. 111 (27), 9864–9868. doi:10.1073/pnas.1401950111
Dick, S. A., Macklin, J. A., Nejat, S., Momen, A., Clemente-Casares, X., Althagafi, M. G., et al. (2019). Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat. Immunol. 20 (1), 29–39. doi:10.1038/s41590-018-0272-2
Doenst, T., Haverich, A., Serruys, P., Bonow, R. O., Kappetein, P., Falk, V., et al. (2019). PCI and CABG for treating stable coronary artery disease: JACC review topic of the week. J. Am. Coll. Cardiol. 73 (8), 964–976. doi:10.1016/j.jacc.2018.11.053
Drenckhahn, J. D., Schwarz, Q. P., Gray, S., Laskowski, A., Kiriazis, H., Ming, Z., et al. (2008). Compensatory growth of healthy cardiac cells in the presence of diseased cells restores tissue homeostasis during heart development. Dev. Cell 15 (4), 521–533. doi:10.1016/j.devcel.2008.09.005
Ek, W. E., Hedman Å, K., Enroth, S., Morris, A. P., Lindgren, C. M., Mahajan, A., et al. (2016). Genome-wide DNA methylation study identifies genes associated with the cardiovascular biomarker GDF-15. Hum. Mol. Genet. 25 (4), 817–827. doi:10.1093/hmg/ddv511
Eltzschig, H. K., and Eckle, T. (2011). Ischemia and reperfusion--from mechanism to translation. Nat. Med. 17 (11), 1391–1401. doi:10.1038/nm.2507
Falzon, M., and Kuff, E. L. (1991). Binding of the transcription factor EBP-80 mediates the methylation response of an intracisternal A-particle long terminal repeat promoter. Mol. Cell. Biol. 11 (1), 117–125. doi:10.1128/mcb.11.1.117
Fan, G. C., Ren, X., Qian, J., Yuan, Q., Nicolaou, P., Wang, Y., et al. (2005). Novel cardioprotective role of a small heat-shock protein, Hsp20, against ischemia/reperfusion injury. Circulation 111 (14), 1792–1799. doi:10.1161/01.CIR.0000160851.41872.C6
Fardi, M., Solali, S., and Farshdousti Hagh, M. (2018). Epigenetic mechanisms as a new approach in cancer treatment: an updated review. Genes & Dis. 5 (4), 304–311. doi:10.1016/j.gendis.2018.06.003
Fatemi, M., Hermann, A., Gowher, H., and Jeltsch, A. (2002). Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur. J. Biochem. 269 (20), 4981–4984. doi:10.1046/j.1432-1033.2002.03198.x
Felician, G., Collesi, C., Lusic, M., Martinelli, V., Ferro, M. D., Zentilin, L., et al. (2014). Epigenetic modification at Notch responsive promoters blunts efficacy of inducing notch pathway reactivation after myocardial infarction. Circulation Res. 115 (7), 636–649. doi:10.1161/CIRCRESAHA.115.304517
Frangogiannis, N. G. (2012). Regulation of the inflammatory response in cardiac repair. Circulation Res. 110 (1), 159–173. doi:10.1161/CIRCRESAHA.111.243162
Frangogiannis, N. G., Michael, L. H., and Entman, M. L. (2000). Myofibroblasts in reperfused myocardial infarcts express the embryonic form of smooth muscle myosin heavy chain (SMemb). Cardiovasc. Res. 48 (1), 89–100. doi:10.1016/s0008-6363(00)00158-9
Gabisonia, K., and Recchia, F. A. (2018). Gene therapy for heart failure: new perspectives. Curr. heart Fail. Rep. 15 (6), 340–349. doi:10.1007/s11897-018-0410-z
Gabrielsen, A., Lawler, P. R., Yongzhong, W., Steinbrüchel, D., Blagoja, D., Paulsson-Berne, G., et al. (2007). Gene expression signals involved in ischemic injury, extracellular matrix composition and fibrosis defined by global mRNA profiling of the human left ventricular myocardium. J. Mol. Cell. Cardiol. 42 (4), 870–883. doi:10.1016/j.yjmcc.2006.12.016
Gaudet, F., Rideout, W. M., Meissner, A., Dausman, J., Leonhardt, H., and Jaenisch, R. (2004). Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol. Cell. Biol. 24 (4), 1640–1648. doi:10.1128/mcb.24.4.1640-1648.2004
Ghigo, A., Franco, I., Morello, F., and Hirsch, E. (2014). Myocyte signalling in leucocyte recruitment to the heart. Cardiovasc. Res. 102 (2), 270–280. doi:10.1093/cvr/cvu030
Gill, C., Mestril, R., and Samali, A. (2002). Losing heart: the role of apoptosis in heart disease--a novel therapeutic target? FASEB J. official Publ. Fed. Am. Soc. Exp. Biol. 16 (2), 135–146. doi:10.1096/fj.01-0629com
Goldstein, R. S., Gallowitsch-Puerta, M., Yang, L., Rosas-Ballina, M., Huston, J. M., Czura, C. J., et al. (2006). Elevated high-mobility group box 1 levels in patients with cerebral and myocardial ischemia. Shock (Augusta, Ga) 25 (6), 571–574. doi:10.1097/01.shk.0000209540.99176.72
Gong, T., Liu, L., Jiang, W., and Zhou, R. (2020). DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 20 (2), 95–112. doi:10.1038/s41577-019-0215-7
Gwak, G. Y., Moon, T. G., Lee, D. H., and Yoo, B. C. (2012). Glycyrrhizin attenuates HMGB1-induced hepatocyte apoptosis by inhibiting the p38-dependent mitochondrial pathway. World J. gastroenterology 18 (7), 679–684. doi:10.3748/wjg.v18.i7.679
Han, Y. (2019). Chinese journal of cardiology in EHJ CardioPulse. Eur. heart J. 40 (39), 3220–3221. doi:10.1093/eurheartj/ehz689
Han, Y., Duan, B., Wu, J., Zheng, Y., Gu, Y., Cai, X., et al. (2022). Analysis of time series gene expression and DNA methylation reveals the molecular features of myocardial infarction progression. Front. Cardiovasc. Med. 9, 912454. doi:10.3389/fcvm.2022.912454
Hausenloy, D. J., and Yellon, D. M. (2013). Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J. Clin. investigation 123 (1), 92–100. doi:10.1172/JCI62874
He, Y., Ling, S., Sun, Y., Sheng, Z., Chen, Z., Pan, X., et al. (2019). DNA methylation regulates α-smooth muscle actin expression during cardiac fibroblast differentiation. J. Cell. physiology 234 (5), 7174–7185. doi:10.1002/jcp.27471
Heidenreich, P. A., Albert, N. M., Allen, L. A., Bluemke, D. A., Butler, J., Fonarow, G. C., et al. (2013). Forecasting the impact of heart failure in the United States: a policy statement from the American Heart Association. Circ. Heart Fail 6 (3), 606–619. doi:10.1161/HHF.0b013e318291329a
Hellman, A., and Chess, A. (2007). Gene body-specific methylation on the active X chromosome. Sci. (New York, NY) 315 (5815), 1141–1143. doi:10.1126/science.1136352
Hendrich, B., and Tweedie, S. (2003). The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. TIG 19 (5), 269–277. doi:10.1016/S0168-9525(03)00080-5
Heusch, G., Musiolik, J., Gedik, N., and Skyschally, A. (2011). Mitochondrial STAT3 activation and cardioprotection by ischemic postconditioning in pigs with regional myocardial ischemia/reperfusion. Circulation Res. 109 (11), 1302–1308. doi:10.1161/CIRCRESAHA.111.255604
Hildrestrand, G. A., Neurauter, C. G., Diep, D. B., Castellanos, C. G., Krauss, S., Bjørås, M., et al. (2009). Expression patterns of Neil3 during embryonic brain development and neoplasia. BMC Neurosci. 10, 45. doi:10.1186/1471-2202-10-45
Hinz, B., Phan, S. H., Thannickal, V. J., Galli, A., Bochaton-Piallat, M. L., and Gabbiani, G. (2007). The myofibroblast: one function, multiple origins. Am. J. pathology 170 (6), 1807–1816. doi:10.2353/ajpath.2007.070112
Holotnakova, T., Ziegelhoffer, A., Ohradanova, A., Hulikova, A., Novakova, M., Kopacek, J., et al. (2008). Induction of carbonic anhydrase IX by hypoxia and chemical disruption of oxygen sensing in rat fibroblasts and cardiomyocytes. Pflugers Archiv Eur. J. physiology 456 (2), 323–337. doi:10.1007/s00424-007-0400-6
Hotchkiss, R. S., Strasser, A., McDunn, J. E., and Swanson, P. E. (2009). Cell death. N. Engl. J. Med. 361 (16), 1570–1583. doi:10.1056/NEJMra0901217
Ieda, M., Fu, J. D., Delgado-Olguin, P., Vedantham, V., Hayashi, Y., Bruneau, B. G., et al. (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142 (3), 375–386. doi:10.1016/j.cell.2010.07.002
Iguchi-Ariga, S. M., and Schaffner, W. (1989). CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes & Dev. 3 (5), 612–619. doi:10.1101/gad.3.5.612
Ito, S., D'Alessio, A. C., Taranova, O. V., Hong, K., Sowers, L. C., and Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466 (7310), 1129–1133. doi:10.1038/nature09303
Ito, S., Shen, L., Dai, Q., Wu, S. C., Collins, L. B., Swenberg, J. A., et al. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Sci. (New York, NY) 333 (6047), 1300–1303. doi:10.1126/science.1210597
Jeong, H. Y., Kang, W. S., Hong, M. H., Jeong, H. C., Shin, M. G., Jeong, M. H., et al. (2015). 5-Azacytidine modulates interferon regulatory factor 1 in macrophages to exert a cardioprotective effect. Sci. Rep. 5, 15768. doi:10.1038/srep15768
Jones, P. A. (1999). The DNA methylation paradox. Trends Genet. TIG 15 (1), 34–37. doi:10.1016/s0168-9525(98)01636-9
Jones, P. A., Issa, J. P., and Baylin, S. (2016). Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 17 (10), 630–641. doi:10.1038/nrg.2016.93
Jopling, C., Sleep, E., Raya, M., Martí, M., Raya, A., and Izpisúa Belmonte, J. C. (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464 (7288), 606–609. doi:10.1038/nature08899
Kain, V., Prabhu, S. D., and Halade, G. V. (2014). Inflammation revisited: inflammation versus resolution of inflammation following myocardial infarction. Basic Res. Cardiol. 109 (6), 444. doi:10.1007/s00395-014-0444-7
Kajstura, J., Leri, A., Finato, N., Di Loreto, C., Beltrami, C. A., and Anversa, P. (1998). Myocyte proliferation in end-stage cardiac failure in humans. Proc. Natl. Acad. Sci. U. S. A. 95 (15), 8801–8805. doi:10.1073/pnas.95.15.8801
Kale, A., Sankrityayan, H., Anders, H. J., and Gaikwad, A. B. (2021). Epigenetic and non-epigenetic regulation of Klotho in kidney disease. Life Sci. 264, 118644. doi:10.1016/j.lfs.2020.118644
Kanamori, H., Takemura, G., Goto, K., Maruyama, R., Ono, K., Nagao, K., et al. (2011). Autophagy limits acute myocardial infarction induced by permanent coronary artery occlusion. Am. J. physiology Heart circulatory physiology 300 (6), H2261–H2271. doi:10.1152/ajpheart.01056.2010
Kang, R., Chen, R., Zhang, Q., Hou, W., Wu, S., Cao, L., et al. (2014). HMGB1 in health and disease. Mol. aspects Med. 40, 1–116. doi:10.1016/j.mam.2014.05.001
Karra, R., and Poss, K. D. (2017). Redirecting cardiac growth mechanisms for therapeutic regeneration. J. Clin. investigation 127 (2), 427–436. doi:10.1172/JCI89786
Kikuchi, K., Holdway, J. E., Werdich, A. A., Anderson, R. M., Fang, Y., Egnaczyk, G. F., et al. (2010). Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464 (7288), 601–605. doi:10.1038/nature08804
Kim, G. D., Ni, J., Kelesoglu, N., Roberts, R. J., and Pradhan, S. (2002). Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 21 (15), 4183–4195. doi:10.1093/emboj/cdf401
Kim, Y. S., Kang, W. S., Kwon, J. S., Hong, M. H., Jeong, H. Y., Jeong, H. C., et al. (2014). Protective role of 5-azacytidine on myocardial infarction is associated with modulation of macrophage phenotype and inhibition of fibrosis. J. Cell. Mol. Med. 18 (6), 1018–1027. doi:10.1111/jcmm.12248
Klionsky, D. J., Petroni, G., Amaravadi, R. K., Baehrecke, E. H., Ballabio, A., Boya, P., et al. (2021). Autophagy in major human diseases. EMBO J. 40 (19), e108863. doi:10.15252/embj.2021108863
Kohli, R. M., and Zhang, Y. (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502 (7472), 472–479. doi:10.1038/nature12750
Laflamme, M. A., and Murry, C. E. (2011). Heart regeneration. Nature 473 (7347), 326–335. doi:10.1038/nature10147
Larsen, F., Gundersen, G., Lopez, R., and Prydz, H. (1992). CpG islands as gene markers in the human genome. Genomics 13 (4), 1095–1107. doi:10.1016/0888-7543(92)90024-m
Leri, A., Rota, M., Pasqualini, F. S., Goichberg, P., and Anversa, P. (2015). Origin of cardiomyocytes in the adult heart. Circulation Res. 116 (1), 150–166. doi:10.1161/CIRCRESAHA.116.303595
Levick, S. P., McLarty, J. L., Murray, D. B., Freeman, R. M., Carver, W. E., and Brower, G. L. (2009). Cardiac mast cells mediate left ventricular fibrosis in the hypertensive rat heart. Hypertension 53 (6), 1041–1047. doi:10.1161/HYPERTENSIONAHA.108.123158
Li, E., Bestor, T. H., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69 (6), 915–926. doi:10.1016/0092-8674(92)90611-f
Li, H., Yang, H., Wang, D., Zhang, L., and Ma, T. (2020). Peroxiredoxin2 (Prdx2) reduces oxidative stress and apoptosis of myocardial cells induced by acute myocardial infarction by inhibiting the TLR4/nuclear factor kappa B (NF-κB) signaling pathway. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 26, e926281. doi:10.12659/MSM.926281
Li, X., Dai, Y., Yan, S., Shi, Y., Han, B., Li, J., et al. (2017). Down-regulation of lncRNA KCNQ1OT1 protects against myocardial ischemia/reperfusion injury following acute myocardial infarction. Biochem. biophysical Res. Commun. 491 (4), 1026–1033. doi:10.1016/j.bbrc.2017.08.005
Li, Y., Wang, J., Sun, L., and Zhu, S. (2018). LncRNA myocardial infarction-associated transcript (MIAT) contributed to cardiac hypertrophy by regulating TLR4 via miR-93. Eur. J. Pharmacol. 818, 508–517. doi:10.1016/j.ejphar.2017.11.031
Liao, Y. P., Chen, L. Y., Huang, R. L., Su, P. H., Chan, M. W., Chang, C. C., et al. (2014). Hypomethylation signature of tumor-initiating cells predicts poor prognosis of ovarian cancer patients. Hum. Mol. Genet. 23 (7), 1894–1906. doi:10.1093/hmg/ddt583
Lin, B., Zhao, H., Li, L., Zhang, Z., Jiang, N., Yang, X., et al. (2020). Sirt1 improves heart failure through modulating the NF-κB p65/microRNA-155/BNDF signaling cascade. Aging 13 (10), 14482–14498. doi:10.18632/aging.103640
Lin, Z., and Pu, W. T. (2014). Strategies for cardiac regeneration and repair. Sci. Transl. Med. 6 (239), 239rv1. doi:10.1126/scitranslmed.3006681
Liu, C. Y., Zhang, Y. H., Li, R. B., Zhou, L. Y., An, T., Zhang, R. C., et al. (2018). LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat. Commun. 9 (1), 29. doi:10.1038/s41467-017-02280-y
Liu, H., He, Z., von Rütte, T., Yousefi, S., Hunger, R. E., and Simon, H. U. (2013a). Down-regulation of autophagy-related protein 5 (ATG5) contributes to the pathogenesis of early-stage cutaneous melanoma. Sci. Transl. Med. 5 (202), 202ra123. doi:10.1126/scitranslmed.3005864
Liu, M., Doublié, S., and Wallace, S. S. (2013b). Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage. Mutat. Res. 743-744, 4–11. doi:10.1016/j.mrfmmm.2012.12.003
Liu, M., Yin, L., Li, W., Hu, J., Wang, H., Ye, B., et al. (2019). C1q/TNF-related protein-9 promotes macrophage polarization and improves cardiac dysfunction after myocardial infarction. J. Cell. physiology 234 (10), 18731–18747. doi:10.1002/jcp.28513
Liu, X., Hall, S. R. R., Wang, Z., Huang, H., Ghanta, S., Di Sante, M., et al. (2015). Rescue of neonatal cardiac dysfunction in mice by administration of cardiac progenitor cells in utero. Nat. Commun. 6, 8825. doi:10.1038/ncomms9825
Liu, Y., and Schwartz, R. J. (2012). Reprogrammed cardiac fibroblasts to the rescue of heart failure. Circulation Res. 111 (7), 831–832. doi:10.1161/CIRCRESAHA.112.279745
Liu, Y., Zou, J., Li, B., Wang, Y., Wang, D., Hao, Y., et al. (2017). RUNX3 modulates hypoxia-induced endothelial-to-mesenchymal transition of human cardiac microvascular endothelial cells. Int. J. Mol. Med. 40 (1), 65–74. doi:10.3892/ijmm.2017.2998
Liu, Z., and Huang, S. (2022). Upregulation of SPI1 during myocardial infarction aggravates cardiac tissue injury and disease progression through activation of the TLR4/NFκB axis. Am. J. Transl. Res. 14 (4), 2709–2727.
Lu, L., Liu, M., Sun, R., Zheng, Y., and Zhang, P. (2015). Myocardial infarction: symptoms and treatments. Cell Biochem. biophysics 72 (3), 865–867. doi:10.1007/s12013-015-0553-4
Lübbert, M., Rüter, B. H., Claus, R., Schmoor, C., Schmid, M., Germing, U., et al. (2012). A multicenter phase II trial of decitabine as first-line treatment for older patients with acute myeloid leukemia judged unfit for induction chemotherapy. Haematologica 97 (3), 393–401. doi:10.3324/haematol.2011.048231
Luo, C., Hajkova, P., and Ecker, J. R. (2018). Dynamic DNA methylation: in the right place at the right time. Sci. (New York, NY) 361 (6409), 1336–1340. doi:10.1126/science.aat6806
Luo, X., Hu, Y., Shen, J., Liu, X., Wang, T., Li, L., et al. (2022). Integrative analysis of DNA methylation and gene expression reveals key molecular signatures in acute myocardial infarction. Clin. epigenetics 14 (1), 46. doi:10.1186/s13148-022-01267-x
Ma, H., Guo, R., Yu, L., Zhang, Y., and Ren, J. (2011). Aldehyde dehydrogenase 2 (ALDH2) rescues myocardial ischaemia/reperfusion injury: role of autophagy paradox and toxic aldehyde. Eur. heart J. 32 (8), 1025–1038. doi:10.1093/eurheartj/ehq253
Ma, X., Liu, H., Foyil, S. R., Godar, R. J., Weinheimer, C. J., Hill, J. A., et al. (2012). Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation 125 (25), 3170–3181. doi:10.1161/CIRCULATIONAHA.111.041814
Ma, X. L., Weyrich, A. S., Lefer, D. J., Buerke, M., Albertine, K. H., Kishimoto, T. K., et al. (1993). Monoclonal antibody to L-selectin attenuates neutrophil accumulation and protects ischemic reperfused cat myocardium. Circulation 88 (2), 649–658. doi:10.1161/01.cir.88.2.649
MacGrogan, D., Münch, J., and de la Pompa, J. L. (2018). Notch and interacting signalling pathways in cardiac development, disease, and regeneration. Nat. Rev. Cardiol. 15 (11), 685–704. doi:10.1038/s41569-018-0100-2
Mackman, N., Bergmeier, W., Stouffer, G. A., and Weitz, J. I. (2020). Therapeutic strategies for thrombosis: new targets and approaches. Nat. Rev. Drug Discov. 19 (5), 333–352. doi:10.1038/s41573-020-0061-0
Maejima, Y., Kyoi, S., Zhai, P., Liu, T., Li, H., Ivessa, A., et al. (2013). Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat. Med. 19 (11), 1478–1488. doi:10.1038/nm.3322
Mann, D. L. (2011). The emerging role of innate immunity in the heart and vascular system: for whom the cell tolls. Circulation Res. 108 (9), 1133–1145. doi:10.1161/CIRCRESAHA.110.226936
Mann, J., and Mann, D. A. (2013). Epigenetic regulation of wound healing and fibrosis. Curr. Opin. rheumatology 25 (1), 101–107. doi:10.1097/BOR.0b013e32835b13e1
Mathers, C. D., and Loncar, D. (2006). Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 3 (11), e442. doi:10.1371/journal.pmed.0030442
Matsui, Y., Takagi, H., Qu, X., Abdellatif, M., Sakoda, H., Asano, T., et al. (2007). Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circulation Res. 100 (6), 914–922. doi:10.1161/01.RES.0000261924.76669.36
Matsuura, K., Honda, A., Nagai, T., Fukushima, N., Iwanaga, K., Tokunaga, M., et al. (2009). Transplantation of cardiac progenitor cells ameliorates cardiac dysfunction after myocardial infarction in mice. J. Clin. investigation 119 (8), 2204–2217. doi:10.1172/JCI37456
McCarthy, C. P., Vaduganathan, M., McCarthy, K. J., Januzzi, J. L., Bhatt, D. L., and McEvoy, J. W. (2018). Left ventricular thrombus after acute myocardial infarction: screening, prevention, and treatment. JAMA Cardiol. 3 (7), 642–649. doi:10.1001/jamacardio.2018.1086
Mehta, S. R., Yusuf, S., Peters, R. J., Bertrand, M. E., Lewis, B. S., Natarajan, M. K., et al. (2001). Effects of pretreatment with clopidogrel and aspirin followed by long-term therapy in patients undergoing percutaneous coronary intervention: the PCI-CURE study. Lancet 358 (9281), 527–533. doi:10.1016/s0140-6736(01)05701-4
Mi, L., Zhang, Y., Xu, Y., Zheng, X., Zhang, X., Wang, Z., et al. (2019). HMGB1/RAGE pro-inflammatory axis promotes vascular endothelial cell apoptosis in limb ischemia/reperfusion injury. Biomed. Pharmacother. = Biomedecine Pharmacother. 116, 109005. doi:10.1016/j.biopha.2019.109005
Mizushima, N., and Komatsu, M. (2011). Autophagy: renovation of cells and tissues. Cell 147 (4), 728–741. doi:10.1016/j.cell.2011.10.026
Mohammad, F., Mondal, T., Guseva, N., Pandey, G. K., and Kanduri, C. (2010). Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Dev. Camb. Engl. 137 (15), 2493–2499. doi:10.1242/dev.048181
Moore, L. D., Le, T., and Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacol. official Publ. Am. Coll. Neuropsychopharmacol. 38 (1), 23–38. doi:10.1038/npp.2012.112
Morales-Nebreda, L., McLafferty, F. S., and Singer, B. D. (2019). DNA methylation as a transcriptional regulator of the immune system. Transl. Res. J. laboratory Clin. Med. 204, 1–18. doi:10.1016/j.trsl.2018.08.001
Mouquet, F., Pfister, O., Jain, M., Oikonomopoulos, A., Ngoy, S., Summer, R., et al. (2005). Restoration of cardiac progenitor cells after myocardial infarction by self-proliferation and selective homing of bone marrow-derived stem cells. Circulation Res. 97 (11), 1090–1092. doi:10.1161/01.RES.0000194330.66545.f5
Murtha, L. A., Schuliga, M. J., Mabotuwana, N. S., Hardy, S. A., Waters, D. W., Burgess, J. K., et al. (2017). The processes and mechanisms of cardiac and pulmonary fibrosis. Front. physiology 8, 777. doi:10.3389/fphys.2017.00777
Nag, A. C., Carey, T. R., and Cheng, M. (1983). DNA synthesis in rat heart cells after injury and the regeneration of myocardia. Tissue & Cell 15 (4), 597–613. doi:10.1016/0040-8166(83)90010-1
Nahrendorf, M., Pittet, M. J., and Swirski, F. K. (2010). Monocytes: protagonists of infarct inflammation and repair after myocardial infarction. Circulation 121 (22), 2437–2445. doi:10.1161/CIRCULATIONAHA.109.916346
Nakagawa, K., and Yokosawa, H. (2000). Degradation of transcription factor IRF-1 by the ubiquitin-proteasome pathway. The C-terminal region governs the protein stability. Eur. J. Biochem. 267 (6), 1680–1686. doi:10.1046/j.1432-1327.2000.01163.x
Nakai, A., Yamaguchi, O., Takeda, T., Higuchi, Y., Hikoso, S., Taniike, M., et al. (2007). The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 13 (5), 619–624. doi:10.1038/nm1574
Nakano, S., Murakami, K., Meguro, M., Soejima, H., Higashimoto, K., Urano, T., et al. (2006). Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer Sci. 97 (11), 1147–1154. doi:10.1111/j.1349-7006.2006.00305.x
Newton, K., and Dixit, V. M. (2012). Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 4 (3), a006049. doi:10.1101/cshperspect.a006049
Niessen, K., and Karsan, A. (2008). Notch signaling in cardiac development. Circulation Res. 102 (10), 1169–1181. doi:10.1161/CIRCRESAHA.108.174318
Nihira, K., Miki, Y., Iida, S., Narumi, S., Ono, K., Iwabuchi, E., et al. (2014). An activation of LC3A-mediated autophagy contributes to de novo and acquired resistance to EGFR tyrosine kinase inhibitors in lung adenocarcinoma. J. pathology 234 (2), 277–288. doi:10.1002/path.4354
Niu, X., Zhang, J., Zhang, L., Hou, Y., Pu, S., Chu, A., et al. (2019). Weighted gene Co-expression network analysis identifies critical genes in the development of heart failure after acute myocardial infarction. Front. Genet. 10, 1214. doi:10.3389/fgene.2019.01214
Oberpriller, J. O., and Oberpriller, J. C. (1974). Response of the adult newt ventricle to injury. J. Exp. zoology 187 (2), 249–253. doi:10.1002/jez.1401870208
Ogawa, S., Koga, S., Kuwabara, K., Brett, J., Morrow, B., Morris, S. A., et al. (1992). Hypoxia-induced increased permeability of endothelial monolayers occurs through lowering of cellular cAMP levels. Am. J. physiology 262 (3 Pt 1), C546–C554. doi:10.1152/ajpcell.1992.262.3.C546
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99 (3), 247–257. doi:10.1016/s0092-8674(00)81656-6
Olsen, M. B., Hildrestrand, G. A., Scheffler, K., Vinge, L. E., Alfsnes, K., Palibrk, V., et al. (2017). NEIL3-Dependent regulation of cardiac fibroblast proliferation prevents myocardial rupture. Cell Rep. 18 (1), 82–92. doi:10.1016/j.celrep.2016.12.009
Opie, L. H., Commerford, P. J., Gersh, B. J., and Pfeffer, M. A. (2006). Controversies in ventricular remodelling. Lancet 367 (9507), 356–367. doi:10.1016/S0140-6736(06)68074-4
Ortiz, I., Barros-Filho, M. C., Dos Reis, M. B., Beltrami, C. M., Marchi, F. A., Kuasne, H., et al. (2018). Loss of DNA methylation is related to increased expression of miR-21 and miR-146b in papillary thyroid carcinoma. Clin. epigenetics 10 (1), 144. doi:10.1186/s13148-018-0579-8
Pan, X., Chen, Z., Huang, R., Yao, Y., and Ma, G. (2013). Transforming growth factor β1 induces the expression of collagen type I by DNA methylation in cardiac fibroblasts. PloS one 8 (4), e60335. doi:10.1371/journal.pone.0060335
Pedrazzini, T. (2007). Control of cardiogenesis by the notch pathway. Trends Cardiovasc. Med. 17 (3), 83–90. doi:10.1016/j.tcm.2007.01.003
Phan, A. T., Goldrath, A. W., and Glass, C. K. (2017). Metabolic and epigenetic coordination of T cell and macrophage immunity. Immunity 46 (5), 714–729. doi:10.1016/j.immuni.2017.04.016
Placek, K., Schultze, J. L., and Aschenbrenner, A. C. (2019). Epigenetic reprogramming of immune cells in injury, repair, and resolution. J. Clin. investigation 129 (8), 2994–3005. doi:10.1172/JCI124619
Ponnusamy, M., Liu, F., Zhang, Y. H., Li, R. B., Zhai, M., Liu, F., et al. (2019). Long noncoding RNA CPR (cardiomyocyte proliferation regulator) regulates cardiomyocyte proliferation and cardiac repair. Circulation 139 (23), 2668–2684. doi:10.1161/CIRCULATIONAHA.118.035832
Porrello, E. R., Mahmoud, A. I., Simpson, E., Hill, J. A., Richardson, J. A., Olson, E. N., et al. (2011). Transient regenerative potential of the neonatal mouse heart. Sci. (New York, NY) 331 (6020), 1078–1080. doi:10.1126/science.1200708
Poss, K. D., Wilson, L. G., and Keating, M. T. (2002). Heart regeneration in zebrafish. Sci. (New York, NY) 298 (5601), 2188–2190. doi:10.1126/science.1077857
Prabhu, S. D., and Frangogiannis, N. G. (2016). The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circulation Res. 119 (1), 91–112. doi:10.1161/CIRCRESAHA.116.303577
Prendergast, G. C., Lawe, D., and Ziff, E. B. (1991). Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell 65 (3), 395–407. doi:10.1016/0092-8674(91)90457-a
Qi, G. M., Jia, L. X., Li, Y. L., Li, H. H., and Du, J. (2014). Adiponectin suppresses angiotensin II-induced inflammation and cardiac fibrosis through activation of macrophage autophagy. Endocrinology 155 (6), 2254–2265. doi:10.1210/en.2013-2011
Qian, L., Huang, Y., Spencer, C. I., Foley, A., Vedantham, V., Liu, L., et al. (2012). In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485 (7400), 593–598. doi:10.1038/nature11044
Rajasingh, J., Thangavel, J., Siddiqui, M. R., Gomes, I., Gao, X. P., Kishore, R., et al. (2011). Improvement of cardiac function in mouse myocardial infarction after transplantation of epigenetically-modified bone marrow progenitor cells. PloS one 6 (7), e22550. doi:10.1371/journal.pone.0022550
Ramirez-Carracedo, R., Tesoro, L., Hernandez, I., Diez-Mata, J., Piñeiro, D., Hernandez-Jimenez, M., et al. (2020). Targeting TLR4 with ApTOLL improves heart function in response to coronary ischemia reperfusion in pigs undergoing acute myocardial infarction. Biomolecules 10 (8), 1167. doi:10.3390/biom10081167
Rask-Andersen, M., Martinsson, D., Ahsan, M., Enroth, S., Ek, W. E., Gyllensten, U., et al. (2016). Epigenome-wide association study reveals differential DNA methylation in individuals with a history of myocardial infarction. Hum. Mol. Genet. 25 (21), 4739–4748. doi:10.1093/hmg/ddw302
Regnell, C. E., Hildrestrand, G. A., Sejersted, Y., Medin, T., Moldestad, O., Rolseth, V., et al. (2012). Hippocampal adult neurogenesis is maintained by Neil3-dependent repair of oxidative DNA lesions in neural progenitor cells. Cell Rep. 2 (3), 503–510. doi:10.1016/j.celrep.2012.08.008
Reis, A., and Hermanson, O. (2012). The DNA glycosylases OGG1 and NEIL3 influence differentiation potential, proliferation, and senescence-associated signs in neural stem cells. Biochem. biophysical Res. Commun. 423 (4), 621–626. doi:10.1016/j.bbrc.2012.04.125
Ren, J., Jiang, L., Liu, X., Liao, Y., Zhao, X., Tang, F., et al. (2022). Heart-specific DNA methylation analysis in plasma for the investigation of myocardial damage. J. Transl. Med. 20 (1), 36. doi:10.1186/s12967-022-03234-9
Rolseth, V., Krokeide, S. Z., Kunke, D., Neurauter, C. G., Suganthan, R., Sejersted, Y., et al. (2013). Loss of Neil3, the major DNA glycosylase activity for removal of hydantoins in single stranded DNA, reduces cellular proliferation and sensitizes cells to genotoxic stress. Biochimica biophysica acta 1833 (5), 1157–1164. doi:10.1016/j.bbamcr.2012.12.024
Russell-Hallinan, A., Watson, C. J., and Baugh, J. A. (2018). Epigenetics of aberrant cardiac wound healing. Compr. Physiol. 8 (2), 451–491. doi:10.1002/cphy.c170029
Sabatine, M. S., and Braunwald, E. (2021). Thrombolysis in myocardial infarction (TIMI) study group: JACC focus seminar 2/8. J. Am. Coll. Cardiol. 77 (22), 2822–2845. doi:10.1016/j.jacc.2021.01.060
Saddic, L. A., Sigurdsson, M. I., Chang, T. W., Mazaika, E., Heydarpour, M., Shernan, S. K., et al. (2017). The long noncoding RNA landscape of the ischemic human left ventricle. Circ. Cardiovasc. Genet. 10 (1), e001534. doi:10.1161/CIRCGENETICS.116.001534
Saleh, M., and Ambrose, J. A. (2018). Understanding myocardial infarction. F1000Research 7, F1000 Faculty Rev-1378. doi:10.12688/f1000research.15096.1
Sanada, F., Kim, J., Czarna, A., Chan, N. Y., Signore, S., Ogórek, B., et al. (2014). c-Kit-positive cardiac stem cells nested in hypoxic niches are activated by stem cell factor reversing the aging myopathy. Circulation Res. 114 (1), 41–55. doi:10.1161/CIRCRESAHA.114.302500
Saunthararajah, Y., Triozzi, P., Rini, B., Singh, A., Radivoyevitch, T., Sekeres, M., et al. (2012). p53-Independent, normal stem cell sparing epigenetic differentiation therapy for myeloid and other malignancies. Seminars Oncol. 39 (1), 97–108. doi:10.1053/j.seminoncol.2011.11.011
Saxonov, S., Berg, P., and Brutlag, D. L. (2006). A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. U. S. A. 103 (5), 1412–1417. doi:10.1073/pnas.0510310103
Sen, G. L., Reuter, J. A., Webster, D. E., Zhu, L., and Khavari, P. A. (2010). DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463 (7280), 563–567. doi:10.1038/nature08683
Smith, Z. D., and Meissner, A. (2013). DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14 (3), 204–220. doi:10.1038/nrg3354
Soler-Botija, C., Forcales, S. V., and Bayés Genís, A. (2020). Spotlight on epigenetic reprogramming in cardiac regeneration. Seminars Cell & Dev. Biol. 97, 26–37. doi:10.1016/j.semcdb.2019.04.009
Souders, C. A., Bowers, S. L., and Baudino, T. A. (2009). Cardiac fibroblast: the renaissance cell. Circulation Res. 105 (12), 1164–1176. doi:10.1161/CIRCRESAHA.109.209809
Steinhubl, S. R., Berger, P. B., Mann, J. T., Fry, E. T., DeLago, A., Wilmer, C., et al. (2002). Early and sustained dual oral antiplatelet therapy following percutaneous coronary intervention: a randomized controlled trial. Jama 288 (19), 2411–2420. doi:10.1001/jama.288.19.2411
Stresemann, C., and Lyko, F. (2008). Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. cancer 123 (1), 8–13. doi:10.1002/ijc.23607
Sturgeon, K. M., Deng, L., Bluethmann, S. M., Zhou, S., Trifiletti, D. M., Jiang, C., et al. (2019). A population-based study of cardiovascular disease mortality risk in US cancer patients. Eur. heart J. 40 (48), 3889–3897. doi:10.1093/eurheartj/ehz766
Su, J., Fang, M., Tian, B., Luo, J., Jin, C., Wang, X., et al. (2018). Hypoxia induces hypomethylation of the HMGB1 promoter via the MAPK/DNMT1/HMGB1 pathway in cardiac progenitor cells. Acta biochimica biophysica Sinica 50 (11), 1121–1130. doi:10.1093/abbs/gmy118
Su, J., Li, J., Yu, Q., Xu, X., Wang, J., Yang, J., et al. (2019). Association of PON1 gene promoter DNA methylation with the risk of Clopidogrel resistance in patients with coronary artery disease. J. Clin. laboratory analysis 33 (5), e22867. doi:10.1002/jcla.22867
Su, J., Li, X., Yu, Q., Liu, Y., Wang, Y., Song, H., et al. (2014). Association of P2Y12 gene promoter DNA methylation with the risk of clopidogrel resistance in coronary artery disease patients. BioMed Res. Int. 2014, 450814. doi:10.1155/2014/450814
Su, J., Yu, Q., Zhu, H., Li, X., Cui, H., Du, W., et al. (2017). The risk of clopidogrel resistance is associated with ABCB1 polymorphisms but not promoter methylation in a Chinese Han population. PloS one 12 (3), e0174511. doi:10.1371/journal.pone.0174511
Sudan, N., Rossetti, J. M., Shadduck, R. K., Latsko, J., Lech, J. A., Kaplan, R. B., et al. (2006). Treatment of acute myelogenous leukemia with outpatient azacitidine. Cancer 107 (8), 1839–1843. doi:10.1002/cncr.22204
Sukmawan, R., Hoetama, E., Suridanda Danny, S., Giantini, A., Listiyaningsih, E., Gilang Rejeki, V., et al. (2021). Increase in the risk of clopidogrel resistance and consequent TIMI flow impairment by DNA hypomethylation of CYP2C19 gene in STEMI patients undergoing primary percutaneous coronary intervention (PPCI). Pharmacol. Res. Perspect. 9 (2), e00738. doi:10.1002/prp2.738
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Sci. (New York, NY) 324 (5929), 930–935. doi:10.1126/science.1170116
Tanaka, Y., Guhde, G., Suter, A., Eskelinen, E. L., Hartmann, D., Lüllmann-Rauch, R., et al. (2000). Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406 (6798), 902–906. doi:10.1038/35022595
Tao, H., Yang, J. J., Chen, Z. W., Xu, S. S., Zhou, X., Zhan, H. Y., et al. (2014). DNMT3A silencing RASSF1A promotes cardiac fibrosis through upregulation of ERK1/2. Toxicology 323, 42–50. doi:10.1016/j.tox.2014.06.006
Thunders, M., Holley, A., Harding, S., Stockwell, P., and Larsen, P. (2019). Using NGS-methylation profiling to understand the molecular pathogenesis of young MI patients who have subsequent cardiac events. Epigenetics 14 (6), 536–544. doi:10.1080/15592294.2019.1605815
Timmers, L., Pasterkamp, G., de Hoog, V. C., Arslan, F., Appelman, Y., and de Kleijn, D. P. (2012). The innate immune response in reperfused myocardium. Cardiovasc. Res. 94 (2), 276–283. doi:10.1093/cvr/cvs018
Torisu, K., Tsuchimoto, D., Ohnishi, Y., and Nakabeppu, Y. (2005). Hematopoietic tissue-specific expression of mouse Neil3 for endonuclease VIII-like protein. J. Biochem. 138 (6), 763–772. doi:10.1093/jb/mvi168
Townsend, N., Kazakiewicz, D., Lucy Wright, F., Timmis, A., Huculeci, R., Torbica, A., et al. (2022). Epidemiology of cardiovascular disease in Europe. Nat. Rev. Cardiol. 19 (2), 133–143. doi:10.1038/s41569-021-00607-3
Turner, N. A., and Porter, K. E. (2013). Function and fate of myofibroblasts after myocardial infarction. Fibrogenesis tissue repair 6 (1), 5. doi:10.1186/1755-1536-6-5
Urbanek, K., Cesselli, D., Rota, M., Nascimbene, A., De Angelis, A., Hosoda, T., et al. (2006). Stem cell niches in the adult mouse heart. Proc. Natl. Acad. Sci. U. S. A. 103 (24), 9226–9231. doi:10.1073/pnas.0600635103
Valentim, L., Laurence, K. M., Townsend, P. A., Carroll, C. J., Soond, S., Scarabelli, T. M., et al. (2006). Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. J. Mol. Cell. Cardiol. 40 (6), 846–852. doi:10.1016/j.yjmcc.2006.03.428
Valinluck, V., Tsai, H. H., Rogstad, D. K., Burdzy, A., Bird, A., and Sowers, L. C. (2004). Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic acids Res. 32 (14), 4100–4108. doi:10.1093/nar/gkh739
van Berlo, J. H., Kanisicak, O., Maillet, M., Vagnozzi, R. J., Karch, J., Lin, S. C., et al. (2014). c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature 509 (7500), 337–341. doi:10.1038/nature13309
van den Borne, S. W., Diez, J., Blankesteijn, W. M., Verjans, J., Hofstra, L., and Narula, J. (2010). Myocardial remodeling after infarction: the role of myofibroblasts. Nat. Rev. Cardiol. 7 (1), 30–37. doi:10.1038/nrcardio.2009.199
van Empel, V. P., Bertrand, A. T., Hofstra, L., Crijns, H. J., Doevendans, P. A., and De Windt, L. J. (2005). Myocyte apoptosis in heart failure. Cardiovasc. Res. 67 (1), 21–29. doi:10.1016/j.cardiores.2005.04.012
Vausort, M., Wagner, D. R., and Devaux, Y. (2014). Long noncoding RNAs in patients with acute myocardial infarction. Circulation Res. 115 (7), 668–677. doi:10.1161/CIRCRESAHA.115.303836
Vera, O., Jimenez, J., Pernia, O., Rodriguez-Antolin, C., Rodriguez, C., Sanchez Cabo, F., et al. (2017). DNA methylation of miR-7 is a mechanism involved in platinum response through MAFG overexpression in cancer cells. Theranostics 7 (17), 4118–4134. doi:10.7150/thno.20112
Vinten-Johansen, J. (2004). Involvement of neutrophils in the pathogenesis of lethal myocardial reperfusion injury. Cardiovasc. Res. 61 (3), 481–497. doi:10.1016/j.cardiores.2003.10.011
Walsh, C. P., Chaillet, J. R., and Bestor, T. H. (1998). Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20 (2), 116–117. doi:10.1038/2413
Walsh, S., Pontén, A., Fleischmann, B. K., and Jovinge, S. (2010). Cardiomyocyte cell cycle control and growth estimation in vivo--an analysis based on cardiomyocyte nuclei. Cardiovasc. Res. 86 (3), 365–373. doi:10.1093/cvr/cvq005
Wan, J., Huang, M., Zhao, H., Wang, C., Zhao, X., Jiang, X., et al. (2013). A novel tetranucleotide repeat polymorphism within KCNQ1OT1 confers risk for hepatocellular carcinoma. DNA Cell Biol. 32 (11), 628–634. doi:10.1089/dna.2013.2118
Wang, J., Han, D., Sun, M., and Feng, J. (2016). A combination of remote ischemic perconditioning and cerebral ischemic postconditioning inhibits autophagy to attenuate plasma HMGB1 and induce neuroprotection against stroke in rat. J. Mol. Neurosci. MN 58 (4), 424–431. doi:10.1007/s12031-016-0724-9
Wang, L., Wang, S., Jia, T., Sun, X., Xing, Z., Liu, H., et al. (2022). Dexmedetomidine prevents cardiomyocytes from hypoxia/reoxygenation injury via modulating tetmethylcytosine dioxygenase 1-mediated DNA demethylation of Sirtuin1. Bioengineered 13 (4), 9369–9386. doi:10.1080/21655979.2022.2054762
Wang, W., Hu, Y. F., Pang, M., Chang, N., Yu, C., Li, Q., et al. (2021). BMP and notch signaling pathways differentially regulate cardiomyocyte proliferation during ventricle regeneration. Int. J. Biol. Sci. 17 (9), 2157–2166. doi:10.7150/ijbs.59648
Wang, X. Z., Yu, Z. X., Nie, B., and Chen, D. M. (2019b). Perindopril inhibits myocardial apoptosis in mice with acute myocardial infarction through TLR4/NF-κB pathway. Eur. Rev. Med. Pharmacol. Sci. 23 (15), 6672–6682. doi:10.26355/eurrev_201908_18558
Wang, Y., Hao, Y., Zhang, H., Xu, L., Ding, N., Wang, R., et al. (2020). DNA hypomethylation of miR-30a mediated the protection of hypoxia postconditioning against aged cardiomyocytes hypoxia/reoxygenation injury through inhibiting autophagy. Circulation J. official J. Jpn. Circulation Soc. 84 (4), 616–625. doi:10.1253/circj.CJ-19-0915
Wang, Y., Yang, X., Jiang, A., Wang, W., Li, J., and Wen, J. (2019a). Methylation-dependent transcriptional repression of RUNX3 by KCNQ1OT1 regulates mouse cardiac microvascular endothelial cell viability and inflammatory response following myocardial infarction. FASEB J. official Publ. Fed. Am. Soc. Exp. Biol. 33 (12), 13145–13160. doi:10.1096/fj.201900310R
Ward-Caviness, C. K., Agha, G., Chen, B. H., Pfeiffer, L., Wilson, R., Wolf, P., et al. (2018). Analysis of repeated leukocyte DNA methylation assessments reveals persistent epigenetic alterations after an incident myocardial infarction. Clin. epigenetics 10 (1), 161. doi:10.1186/s13148-018-0588-7
Watson, C. J., Collier, P., Tea, I., Neary, R., Watson, J. A., Robinson, C., et al. (2014). Hypoxia-induced epigenetic modifications are associated with cardiac tissue fibrosis and the development of a myofibroblast-like phenotype. Hum. Mol. Genet. 23 (8), 2176–2188. doi:10.1093/hmg/ddt614
Weyrich, A. S., Ma, X. Y., Lefer, D. J., Albertine, K. H., and Lefer, A. M. (1993). In vivo neutralization of P-selectin protects feline heart and endothelium in myocardial ischemia and reperfusion injury. J. Clin. investigation 91 (6), 2620–2629. doi:10.1172/JCI116501
White, H. D., and Chew, D. P. (2008). Acute myocardial infarction. Lancet 372 (9638), 570–584. doi:10.1016/S0140-6736(08)61237-4
Willems, I. E., Havenith, M. G., De Mey, J. G., and Daemen, M. J. (1994). The alpha-smooth muscle actin-positive cells in healing human myocardial scars. Am. J. pathology 145 (4), 868–875.
Williams, S. M., Golden-Mason, L., Ferguson, B. S., Schuetze, K. B., Cavasin, M. A., Demos-Davies, K., et al. (2014). Class I HDACs regulate angiotensin II-dependent cardiac fibrosis via fibroblasts and circulating fibrocytes. J. Mol. Cell. Cardiol. 67, 112–125. doi:10.1016/j.yjmcc.2013.12.013
Wilson, E. M., Diwan, A., Spinale, F. G., and Mann, D. L. (2004). Duality of innate stress responses in cardiac injury, repair, and remodeling. J. Mol. Cell. Cardiol. 37 (4), 801–811. doi:10.1016/j.yjmcc.2004.05.028
Wrigley, B. J., Lip, G. Y., and Shantsila, E. (2011). The role of monocytes and inflammation in the pathophysiology of heart failure. Eur. J. heart Fail. 13 (11), 1161–1171. doi:10.1093/eurjhf/hfr122
Wu, X., He, L., Chen, F., He, X., Cai, Y., Zhang, G., et al. (2014). Impaired autophagy contributes to adverse cardiac remodeling in acute myocardial infarction. PloS one 9 (11), e112891. doi:10.1371/journal.pone.0112891
Wu, X., Qin, Y., Zhu, X., Liu, D., Chen, F., Xu, S., et al. (2018). Increased expression of DRAM1 confers myocardial protection against ischemia via restoring autophagy flux. J. Mol. Cell. Cardiol. 124, 70–82. doi:10.1016/j.yjmcc.2018.08.018
Wu, X., and Zhang, Y. (2017). TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18 (9), 517–534. doi:10.1038/nrg.2017.33
Wu, Y., Qiu, G., Zhang, H., Zhu, L., Cheng, G., Wang, Y., et al. (2021). Dexmedetomidine alleviates hepatic ischaemia-reperfusion injury via the PI3K/AKT/Nrf2-NLRP3 pathway. J. Cell. Mol. Med. 25 (21), 9983–9994. doi:10.1111/jcmm.16871
Xu, Q., Li, X., Lu, Y., Shen, L., Zhang, J., Cao, S., et al. (2015). Pharmacological modulation of autophagy to protect cardiomyocytes according to the time windows of ischaemia/reperfusion. Br. J. Pharmacol. 172 (12), 3072–3085. doi:10.1111/bph.13111
Yamamoto, S., Sawada, K., Shimomura, H., Kawamura, K., and James, T. N. (2000). On the nature of cell death during remodeling of hypertrophied human myocardium. J. Mol. Cell. Cardiol. 32 (1), 161–175. doi:10.1006/jmcc.1999.1064
Yan, L., Sadoshima, J., Vatner, D. E., and Vatner, S. F. (2006). Autophagy: a novel protective mechanism in chronic ischemia. Cell Cycle 5 (11), 1175–1177. doi:10.4161/cc.5.11.2787
Yang, I. V., and Schwartz, D. A. (2015). Epigenetics of idiopathic pulmonary fibrosis. Transl. Res. J. laboratory Clin. Med. 165 (1), 48–60. doi:10.1016/j.trsl.2014.03.011
Yang, Z., and Klionsky, D. J. (2010). Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12 (9), 814–822. doi:10.1038/ncb0910-814
Yu, W., Lyu, J., Jia, L., Sheng, M., Yu, H., and Du, H. (2020). Dexmedetomidine ameliorates Hippocampus injury and cognitive dysfunction induced by hepatic ischemia/reperfusion by activating SIRT3-mediated mitophagy and inhibiting activation of the NLRP3 inflammasome in young rats. Oxidative Med. Cell. Longev. 2020, 7385458. doi:10.1155/2020/7385458
Yuan, M., Meng, X. W., Ma, J., Liu, H., Song, S. Y., Chen, Q. C., et al. (2019). Dexmedetomidine protects H9c2 cardiomyocytes against oxygen-glucose deprivation/reoxygenation-induced intracellular calcium overload and apoptosis through regulating FKBP12.6/RyR2 signaling. Drug Des. Dev. Ther. 13, 3137–3149. doi:10.2147/DDDT.S219533
Zhang, B., Zhang, J., Ainiwaer, Y., He, B., Geng, Q., Lin, L., et al. (2021a). Dexmedetomidine attenuates myocardial injury induced by renal ischemia/reperfusion by inhibiting the HMGB1-TLR4-MyD88-NF-κb signaling pathway. Ann. Clin. laboratory Sci. 51 (3), 376–384.
Zhang, J., Zhang, X., Cui, Y., Ferdous, M., Cui, L., and Zhao, P. (2018). Different postconditioning cycles affect prognosis of aged patients undergoing primary percutaneous coronary intervention. Cardiol. J. 25 (6), 666–673. doi:10.5603/CJ.a2017.0083
Zhang, Q., Wang, L., Wang, S., Cheng, H., Xu, L., Pei, G., et al. (2022). Signaling pathways and targeted therapy for myocardial infarction. Signal Transduct. Target. Ther. 7 (1), 78. doi:10.1038/s41392-022-00925-z
Zhang, X., Li, C., Wang, D., Chen, Q., Li, C. L., and Li, H. J. (2016). Aberrant methylation of ATG2B, ATG4D, ATG9A and ATG9B CpG island promoter is associated with decreased mRNA expression in sporadic breast carcinoma. Gene 590 (2), 285–292. doi:10.1016/j.gene.2016.05.036
Zhang, Y., Zhao, Q., Li, X., and Ji, F. (2021b). Dexmedetomidine reversed hypoxia/reoxygenation injury-induced oxidative stress and endoplasmic reticulum stress-dependent apoptosis of cardiomyocytes via SIRT1/CHOP signaling pathway. Mol. Cell. Biochem. 476 (7), 2803–2812. doi:10.1007/s11010-021-04102-8
Zhao, D., Liu, J., Wang, M., Zhang, X., and Zhou, M. (2019). Epidemiology of cardiovascular disease in China: current features and implications. Nat. Rev. Cardiol. 16 (4), 203–212. doi:10.1038/s41569-018-0119-4
Zhao, Z. Q., Corvera, J. S., Halkos, M. E., Kerendi, F., Wang, N. P., Guyton, R. A., et al. (2003). Inhibition of myocardial injury by ischemic postconditioning during reperfusion: comparison with ischemic preconditioning. Am. J. physiology Heart circulatory physiology 285 (2), H579–H588. doi:10.1152/ajpheart.01064.2002
Zhou, L. Y., Zhai, M., Huang, Y., Xu, S., An, T., Wang, Y. H., et al. (2019). The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/FAM65B pathway. Cell death Differ. 26 (7), 1299–1315. doi:10.1038/s41418-018-0206-4
Zhu, H., Tannous, P., Johnstone, J. L., Kong, Y., Shelton, J. M., Richardson, J. A., et al. (2007). Cardiac autophagy is a maladaptive response to hemodynamic stress. J. Clin. investigation 117 (7), 1782–1793. doi:10.1172/JCI27523
Keywords: myocardial infarction, DNA methylation, inflammation, autophagy, proliferation, fibrosis
Citation: Han W, Wang W, Wang Q, Maduray K, Hao L and Zhong J (2024) A review on regulation of DNA methylation during post-myocardial infarction. Front. Pharmacol. 15:1267585. doi: 10.3389/fphar.2024.1267585
Received: 26 July 2023; Accepted: 25 January 2024;
Published: 13 February 2024.
Edited by:
Eliot Ohlstein, Drexel University, United StatesReviewed by:
Sujith Dassanayaka, University of Louisville, United StatesJi-Xin Tang, Guangdong Medical University, China
Copyright © 2024 Han, Wang, Wang, Maduray, Hao and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingquan Zhong, MTk4NzYyMDAwNzc4QGVtYWlsLnNkdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Wenqiang Han1†
Wenqiang Han1† Jingquan Zhong
Jingquan Zhong