- 1Department of Pathology, The Second Hospital of Jilin University, Changchun, Jilin, China
- 2Department of Urology, The Second Hospital of Jilin University, Changchun, Jilin, China
- 3Department of Orthopedics, The Second Hospital of Jilin University, Changchun, Jilin, China
- 4Department of Anesthesiology, The Second Hospital of Jilin University, Changchun, Jilin, China
The integrated application of artificial intelligence (AI) and digital pathology (DP) technology has opened new avenues for advancements in oncology and molecular pathology. Consequently, studies in renal cell carcinoma (RCC) have emerged, highlighting potential in histological subtype classification, molecular aberration identification, and outcome prediction by extracting high-throughput features. However, reviews of these studies are still rare. To address this gap, we conducted a thorough literature review on DP and AI applications in RCC through database searches. Notably, we found that AI models based on deep learning achieved area under the curve (AUC) of over 0.93 in subtype classification, 0.89-0.96 in grading of clear cell RCC, 0.70-0,89 in molecular prediction, and over 0.78 in survival prediction. This review finally discussed the current state of researches and potential future directions.
Introduction
Renal cell carcinoma
Renal cell carcinoma (RCC) refers to a diverse group of malignant tumors originating from the epithelial cells of the renal tubules, with an increasing incidence (1). The 2022 Fifth World Health Organization (WHO) Blue Book divided RCC into morphologically and molecularly defined RCCs, classifying RCCs into three major subtypes: clear cell RCC (ccRCC), papillary RCC (pRCC), and chromophobe RCC (chRCC), which together constitute over 90% of RCC cases (2). CcRCC is the most common subtype (70%-80% of cases), which is graded using the Fuhrman classification, now replaced by the World Health Organization/International Society of Urology (WHO/ISUP) grading system (3). In addition to histological classifications, the Fifth WHO update introduced molecular-driven categorizations (4), emphasizing the importance of molecular profiling for RCC diagnosis and prognosis.
Digital pathology, WSI, and pathomics
Digital pathology (DP) is a technique that utilizes digital technology and computer-aided tools to convert images from traditional glass slides into high-resolution digital formats (5). Initially, DP focused on digital image acquisition. With the advancements in artificial intelligence (AI), DP has now integrated AI to further enhance its capabilities. This integration enables advanced data storage and retrieval, while offering automated image analysis and diagnostic support (5).
Whole-slide imaging (WSI) is a significant technology in DP that utilizes automated microscopes to capture high-resolution detailed images (6). It employs computer algorithms for precise stitching and processing, enabling the quantification of features such as shape, size, and color in pathological images (6). A randomized controlled trial compared six pathologists’ diagnostic accuracy using glass slides versus WSIs for the same cases. The results showed no significant differences, confirming diagnostic equivalence (7).
Pathomics is an emerging subject, which is the integration of pathology and omics, aiming to utilize computational techniques to process and interpret pathology image data (8). Through high-throughput feature extraction, visualization, and quantification, it aims to develop predictive models that uncover potential biomarkers (9).
Artificial intelligence
In the medical milieu, AI employs advanced algorithms like machine learning (ML) and deep learning (DL), to analyze complex medical data, assist diagnosis and treatment planning, and optimize patient outcomes (9, 10). It has been integrated with diagnostic technologies such as X-rays, CT scans, magnetic resonance imaging, ultrasounds, and gene sequencing, achieving satisfactory results in predicting diagnosis, prognosis, and treatment response (9).
In pathology, the application of AI covers many aspects, such as cytological screening, morphological quantification analysis, tissue pathology diagnosis, and prognosis assessment (11, 12). M. Giulietti et al. (13) compared the prediction performance of AI-based and non-AI-based predictors, with the former performing slightly better. A. Distante et al. (14) assessed the precision, efficiency, and objectivity of histopathological analysis, suggesting that AI can overcome intra and interobserver variability and time consumption.
One of the bottlenecks in the application of WSI integrated with AI is interpretability, also known as black box features (15). The features automatically extracted by the initial AI model during training are often high dimensional and abstract. Although these features are effective in optimizing model performance, they lack biological significance, making it difficult to explain the relationship between these features and prediction tasks (15). In response to the growing demand for interpretability in medical fields, researchers primarily focus on improving the steps of feature extraction and prediction models to enhance this aspect (16). For example, Zhang et al. (17) proposed a Structural Priors Guided Network (SPG-Net) that not only achieves high segmentation accuracy but also incorporates prior structural knowledge, making the model’s predictions more interpretable.
The previous reviews summarized the applications of AI and DP in the diagnosis and prediction of pathology and RCC (18, 19). However, they did not focus on the extracted features. This study aimed to address this gap by providing a comprehensive overview of methods for enhancing the interpretability of AI and mitigating its black box nature in future research. We focused on summarizing and analyzing the existing literature related to feature extraction techniques in AI for RCC, offering insights into how these methods can improve model interpretability.
Materials and methods
A literature search was performed on the PubMed and the Web of Science database platforms in January 2024. The search was carried out using the following terms: “((renal cell carcinoma) AND ((artificial intelligence) OR (machine learning) OR (deep learning))).” In addition, a manual search was performed to identify additional potentially relevant articles.
English publications were included if published between 2017 and January 2024, due to advancements in AI, particularly DL and CNNs, which began impacting DP and improving model accuracy. Nonpeer reviewed articles, case reports, comments, and conference summaries were excluded. A duplicate search was performed in Endnote software (version 20), together with the manual screening of titles and abstracts. Literature screening and evaluation were conducted following the PRISMA2020 checklist (20).
Results and discussion
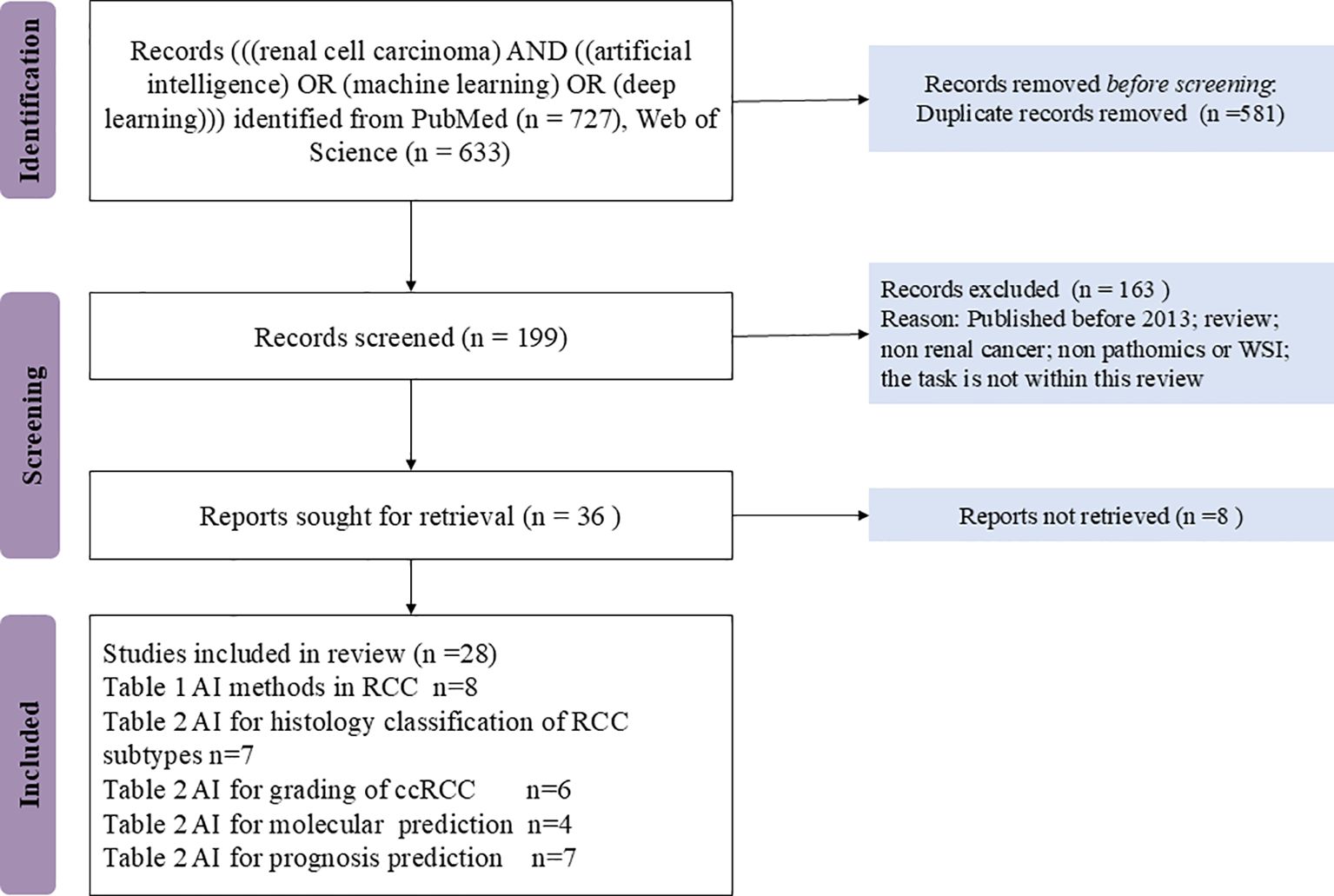
Figure 1 provided the flow diagram for the PRISMA strategy. In all, 1032 records were excluded, and the 28 remaining full texts were checked for suitability.
The text mining software programs SATI (http://sationline.cn/) and Voyant (https://voyanttools.org/) were used to read and analyze full records and cited references. Of the 28 records in the study, 2 were not in the Web of Science core collection, and 26 records were included in the text mining analysis. The results of the analyses of the keywords, authors, institutions, and citations were shown in Supplementary Materials 1–4.
Although the results based on text mining were only objective index statistics, and did not involve the overall scientific evaluation of the literature, they can help us to explore the rich information of research in this field.
AI algorithms in RCC
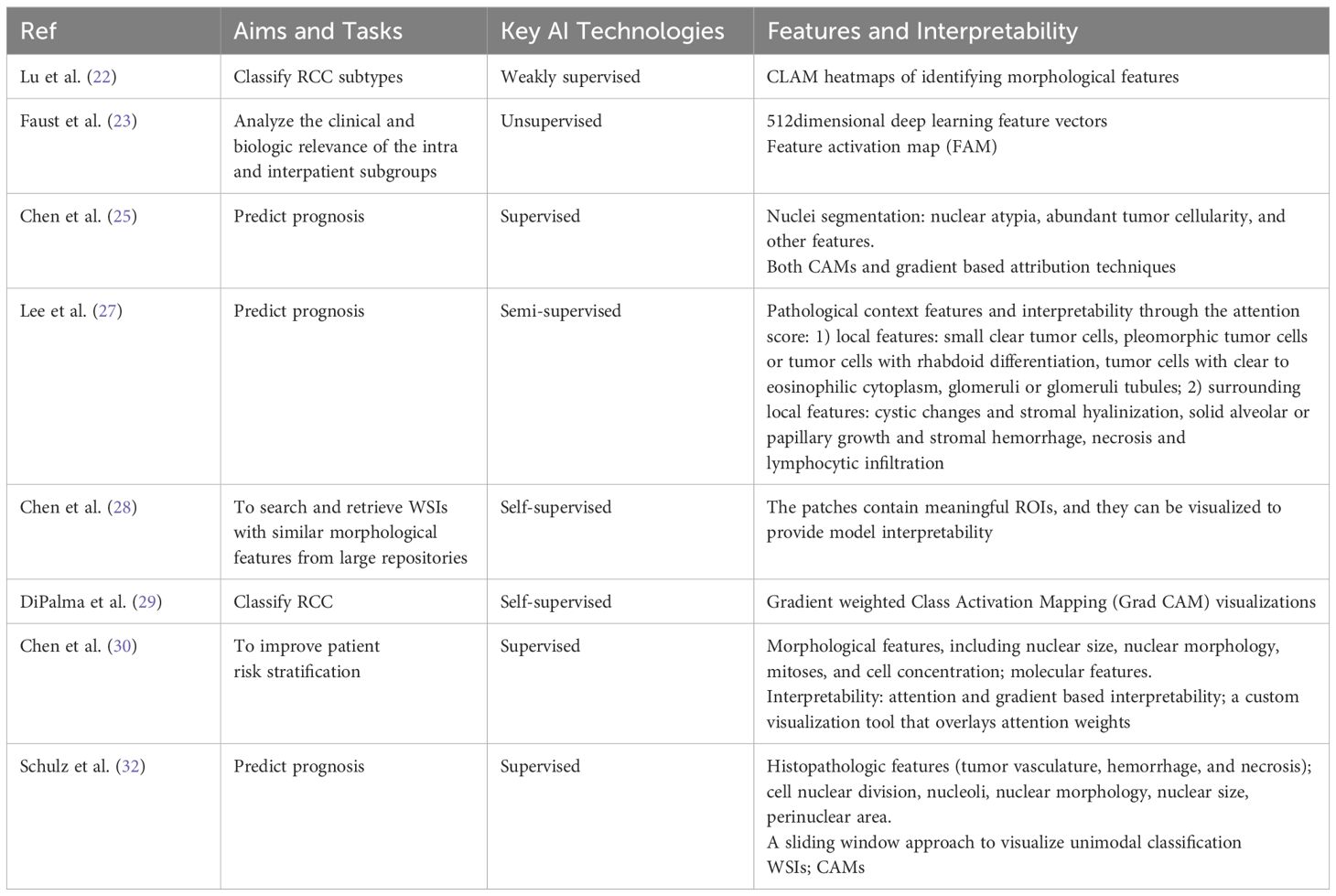
The current AI approaches used for classification and prediction include supervised, weakly supervised, self-supervised, and unsupervised learning (21). Supervised learning involves fully labeled data, which provides broader context and clearer relationships within the data, potentially enhancing the model’s ability to explain its predictions. However, its scalability is limited by the time-consuming, labor-intensive, and error-prone labeling process (21). In weakly supervised learning, image-level (WSI-level) labels are commonly used, which offer coarse annotations for entire images. These methods typically provide less detailed contextual information compared to fully supervised methods, which use pixel-level labels or bounding boxes (22). Unsupervised learning methods depend solely on the intrinsic properties of the data, such as similarity, consistency, or relationships, to uncover underlying structures or representations, often applied in medical image analysis Meanwhile, self-supervised learning, which falls between supervised and unsupervised approaches, leverages unlabeled data by creating pretext tasks that help the model learn useful representations without explicit labels (23). A summary of AI algorithms in RCC is provided in Table 1.
Supervised learning
In supervised learning, one of the most widely used deep learning (DL) architectures is the Convolutional Neural Network (CNN), which consists of multiple layers of convolutional filters that can automatically extract features from images (24). CNNs have demonstrated superior performance in image segmentation, detection, and generation tasks (24). However, these methods also require either manual annotation of gigapixel WSIs or large datasets of WSIs with slide level labels (25). The input to a CNN is a small patch image obtained from segmenting the WSI, focusing on local features, such as morphological changes or different growth patterns of tumor cells, but often failing to pay attention to contextual background features (25). For example, Chen et al. (25) proposed Supervised Multimodal Fusion, an interpretable strategy for end-to-end multimodal fusion of histology images and genomic features. This approach not only utilizes CNNs but also incorporates other methods, such as: 1) graph CNNs for learning cell graph features, 2) Graph Convolutional Networks (GCNs) for survival outcome prediction from histology, and 3) the Kronecker product of gated feature representations along with a gating-based attention mechanism.
Weakly supervised learning
In weakly supervised learning, the researchers implemented Graph Neural Network (GNN) to extract these features using attention and integrated gradients (IG) for model interpretation to fully exploit the contextual features of WSI (26). However, these methods overlook the spatial interactions of local pathological features and lack interpretability for contextual features in WSI (26). To address these limitations, Lee et al. (27) proposed a semi-supervised tumor environment related context learning using graph deep learning (TEA graph), which is a GNN based method that represents WSI through super patches and can analyze the context of spatial interactions of histopathological features. Additionally, in study (22), a weakly supervised clustering-constrained attention-based multiple instance learning (CLAM) method was introduced. CLAM utilizes attention-based learning to identify subregions of high diagnostic value while applying instance-level clustering to refine and constrain the feature space, eliminating the need for pixel-level annotations, ROI extraction, or sampling.
Unsupervised learning
Several approaches have been proposed in both unsupervised and self-supervised learning. For instance, in study (23), an unsupervised pretrained CNN was used as a feature extractor for histopathology, generating all groupings through unsupervised learning techniques such as dimensionality reduction and clustering. In study (28), a self-supervised image search method for histology pipelines was introduced, utilizing a Vector Quantized Variational Autoencoder (VQVAE) trained on a large dataset to improve feature extraction. Additionally, in study (29), a self-supervised deep learning method, known as Resolution-Based Distillation, was proposed. This technique distills the learned representation knowledge from a teacher model into a student model trained at a lower resolution, while minimizing the impact on classification performance.
Both weakly supervised and unsupervised methods face greater challenges in interpretability compared to supervised algorithms that use more comprehensive annotations (21). These challenges include obtaining reliable and robust weak labels, addressing ambiguities and uncertainties in the data, and evaluating and interpreting results (21). Nonetheless, there is growing interest in developing weakly supervised and unsupervised learning methods for medical image analysis, as they can leverage rich unlabeled or partially labeled data, thereby reducing the annotation burden.
Model fusion techniques
Model fusion techniques are commonly used to deal with multimodal data, including pathomics, radiomics, genomics, and proteomics data and other clinical medical record data, as these diverse data sources provide complementary information that can enhance performance in prediction tasks (30). Model fusion can be performed at different stages of an algorithm pipeline, in early fusion, late fusion, or hybrid fusion (21, 31). Model fusion can be achieved through ensemble learning, or multimodal deep learning methods, which use neural networks to identify joint representations from different data modalities (28). For example, in study (30), a supervised multimodal fusion DL model was designed to integrate whole-slide images and molecular profile data using a weakly supervised multimodal deep learning algorithm. Schulz et al. (32) developed a supervised multimodal model that can integrate multiple medical images (WSI, CT, MRI) and genomic data and fuses the multi-source output information through the attention layer. Chen et al. (28) proposed an image search method based on self-supervised learning to optimize the histological analysis process. They enhanced the interpretability of the model by visualizing the ROIs, providing an intuitive tool for pathologists to evaluate the basis of model decision-making.
Application of AI in RCC
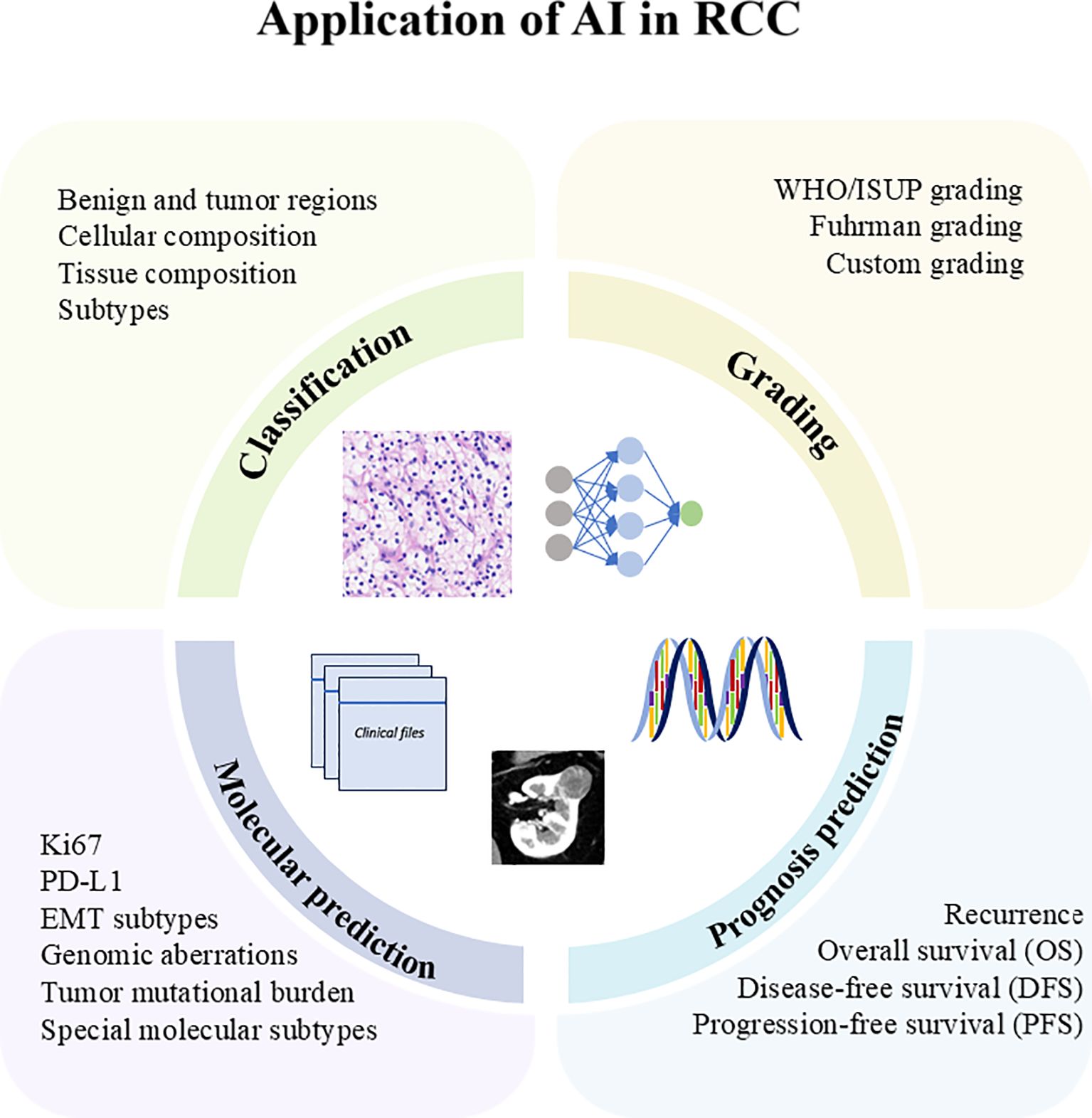
The application of AI in RCC has enhanced classification, grading, molecular prediction, and prognosis prediction, as shown in Figure 2. In the following sections, we summarized and elaborated on these applications in Table 2.
AI for classification
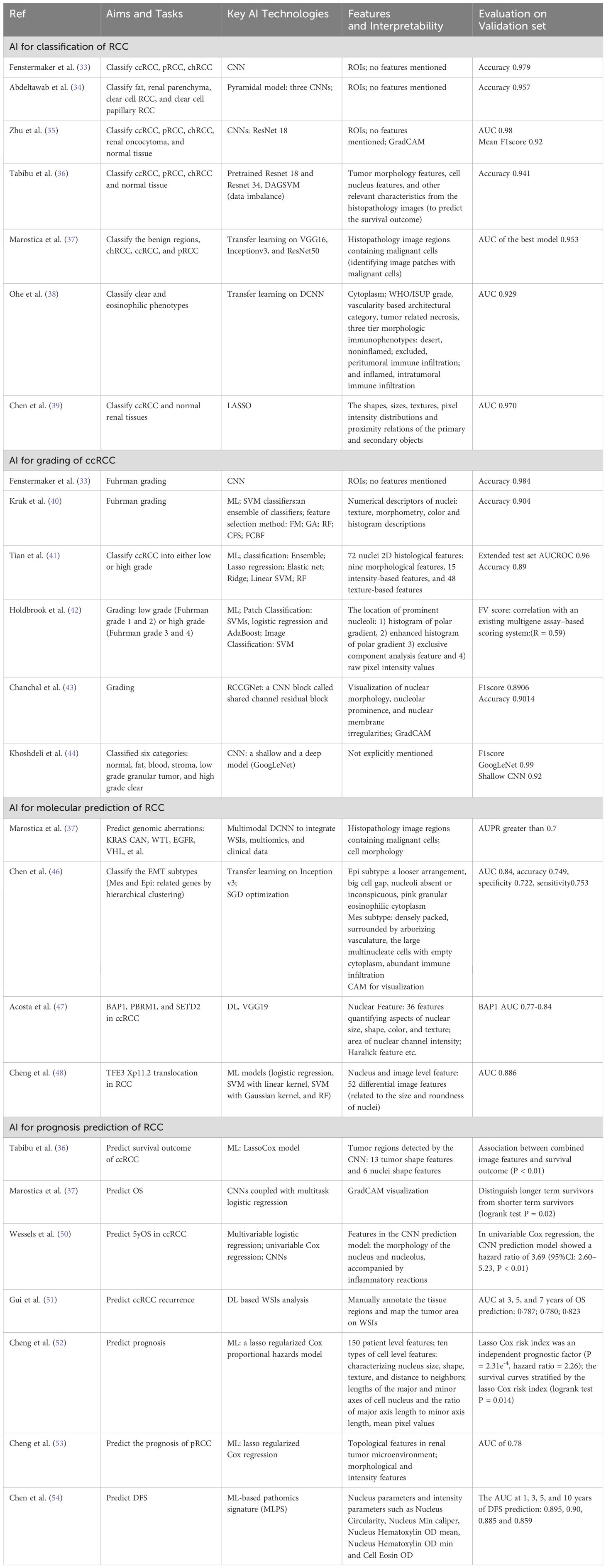
Supervised CNN architectures and techniques are commonly employed for the classification of RCC (33–36). The working pipeline begins with pathologists manually annotating regions of interest (ROIs) on the pixel level in WSIs. These ROIs are then segmented into reasonably sized patches for classification at the patch level. During this process, data augmentation techniques are applied to preprocess the WSIs and address the issue of small datasets (33, 35).
For example, in study (35), the model not only achieves whole-slide classification but also visualizes key indicative regions and features on the slides, thereby enhancing its explainability. To further improve interpretability, the Gradient-weighted Class Activation Mapping (Grad-CAM) method is utilized with (AUC) values of 0.97 (95% CI: 0.96–0.98) for external set. E. Marostica et al. (37) integrated VGG16, Inceptionv3, and ResNet50 with multiomics and clinical data, and extracted features like cell morphology to diagnose the RCC subtypes, predict patients’ survival outcomes, Malignant cells are divided into ROI to identify malignant cells and complete the above tasks. However, the specific features extracted were not explicitly mentioned in the subsequent task of linking histopathological images with genomic data to reveal molecular morphology. Y. Yasukochi, et al. (38) employed a deep convolutional neural network (DCNN) trained via transfer learning on WSIs to predict the eosinophilic phenotype of RCC in a high AI score group. They extracted features related to clear and eosinophilic phenotypes and angiogenesis gene signatures. The independent validation set achieved an AUC of 0.929 for predicting clear or mixed/eosinophilic phenotypes, though external validation results were not provided. Chen et al. (39) extracted a total of 346 useful quantitative image features, including shapes, sizes, textures, pixel intensity distributions, and proximity relations in WSI for diagnosis of ccRCC patients. The AUC value of the model was 0.970 in the test cohort, and 0.814 in the external validation cohort.
AI for grading of ccRCC
The classic WHO/ISUP grading system for ccRCC relies on the morphological characteristics of the nucleus, in particular on the presence of nucleoli at different magnifications (3). Key steps in the structured pipeline of automated grading systems based on ML technology include identification of ROIs, segmentation of nuclei, numerical descriptors pertaining to nuclei features, feature selection, and classification (40–42). Most studies utilize fivefold or tenfold cross validation techniques and image enhancement strategies to reduce the impact of small datasets (42).
For instance (Table 2), studies (40–42), have extracted cellular features such as cellular information included texture, morphometry, color, intensity, histogram descriptions and pleomorphic features. When building an automated classification system, selecting and improving classifiers is a crucial step, and some studies have proposed improved classifiers based on traditional SVM, RF, such as ensemble learners (40). In studies (33, 43, 44), classic architectures of DL, such as ResNet, Inception, and DL algorithms improved based on classic architectures are also used for the grading task. Chanchal et al. (43) proposed a novel shared channel residual (SCR) block to share the information between different layers and strengthens the local semantic features. Satisfactory accuracy and F1 scores were obtained by pathologists marking the tumor region ROI and extracting nuclear morphology visualization, nucleolar protrusion and nuclear membrane irregularities.
AI for molecular prediction
Molecular driven categorizations of RCC, such as TFE3/TFEB rearranged RCC, and ALK rearranged RCC, have been added into the Fifth WHO, due to the importance of molecular profiling in RCC tumorigenesis, prevalence rates, diagnosis and prognosis (2). However, traditional morphological diagnosis based on microscopic pathological sections often falls short in accurately identifying tumor molecular heterogeneity. Additionally, gene expression analysis is not readily implemented in routine clinical practice due to cost and technical challenges (45). In response, AI technology utilizes DL algorithms to explore the correlation between morphological features and molecular characteristics extracted from tumor tissue images, consistently inferring molecular tumor subtypes from conventional histology.
For example (Table 2), Q. Chen et al. (46) developed an EMT gene signature that was used to classify ccRCC into epithelial and mesenchymal subtypes through DCNN, but the sample size used in that study was relatively small, and the neural network was trained on a single data set. Acosta et al. (47) developed a DL model to identify mutations in BAP1, PBRM1, and SETD2, achieving commendable performance. Cheng et al. (48) designed differential image features that are closely related to the size and roundness of the nucleus. Through in-depth study and analysis of these features, a variety of ML models have shown good diagnostic performance in distinguishing TFE3 Xp11.2 translocation-associated RCC from other types of RCC.
The use of AI technology to analyze pathological sections, extract features, and predict molecular subtypes offers pathologists a new perspective (47). However, this field is still in its early stages, and more data and in-depth studies are needed to validate the accuracy and reliability of AI’s association with RCC molecular characteristics.
AI for prognosis prediction
Accurate prognostic predictions are crucial for clinical decision-making and evaluating therapeutic effects in patients with renal cell carcinoma (RCC), leading researchers to focus on developing clinical risk models (49). In prognostic prediction tasks, regression models in machine learning, such as Cox regression, Lasso-regularized Cox regression, and logistic regression (which is also employed for classification tasks), are commonly utilized (36). The typical workflow involves first extracting relevant features from WSIs, followed by using machine learning classifiers for risk scoring and classification (sometimes for feature transformation or further analysis rather than direct classification). Finally, survival outcomes are predicted using regression models (36, 50).
For example, F. Wessels et al. (50) established a univariate logistic regression model based on CNN prediction model and a multivariate logistic regression model combining CNNs prediction and clinicopathological parameters (we only showed univariate logistic regression results in Table 2). The results in the validation group showed that the AUROC of both models was 0.88. Gui et al. (51) combined the WSI score with a score based on six single nucleotide polymorphisms (SNPs) and the Leibovich score based on clinical pathological risk factors, to construct a multimodal recurrence scoring system. They found that the multimodal recurrence scoring system had higher predictive accuracy than single modality scores and clinical pathological risk factors, and could more accurately predict the recurrence free interval (RFI) for localized renal cell carcinoma patients. Cheng et al. (52) extracted nucleus features and topological features in the renal tumor microenvironment and eigengenes from functional genomics data to predict ccRCC prognosis. This team conducted another study (53) to predict patient prognosis in pRCC by extracting topological features included the histogram of cooccurrence of nucleus patterns and bag of edge histogram (BOEH) features spatial arrangement of different cell patterns in the tumor microenvironment. The author mentioned that the prediction effect of the integrated model is better than that of the model considering a single factor (52) and the proposed topological features were superior to traditional clinical features and cell morphology and intensity features in predicting patient outcomes (53). The group led by Shanghai Jiao Tong University School of Medicine (54) used QuPath digital pathology software (55) (a commonly used digital pathological analysis software) to carry out cell and nuclear segmentation and detected pathological signatures as machine learning based pathomics signature (MLPS) to predict the clinical outcomes of ccRCC patients. There are a total of 43 different pathological features, mainly nuclear and intensity parameters, used in the MLPS classification system, and satisfactory survival prediction based on MLPS classification has been obtained.
Conclusions and future prospects
This article summarized relevant literatures on the application of AI and DP in RCC and outlined the methods currently employed to enhance interpretability, which is especially critical in the medical field. Visualization techniques are often used to visually highlight meaningful features and regions recognizable by pathologists to increase interpretability (23, 43). Features extracted based on WSI include but are not limited to color, texture, shape features, topology, etc. Tumor cell morphological (36, 37), and tumor microenvironment features (25, 27, 39) are common in RCC researches. The prediction tasks based on the above extracted features can achieve satisfactory results, while studies that do not provide extracted features often need to increase credibility by improving the interpretability of the algorithm (33, 36).
Although great progress has been made in the application of AI technology to the medical field, many challenges remain. A conflict is seen between the limitations of computing resources, the extent of original research materials, and the requirements for computing power and size of the data set for the use of robust AI models (56). Therefore, improving computing power and upgrading scanning equipment are issues that computer engineering should continue to address. Although the use of large databases has made up somewhat for the problem of insufficient data set in a single research center, the following issues still exist. 1. Extracting meaningful information from huge databases is still an arduous task. 2. The curse of dimensionality for high-throughput sequencing data leads to serious overfitting problems. 3. The quality of these data cannot be fully guaranteed, and the lack of multicenter external validation affects research reliability. The application of AI to the medical field also involves ethical issues, the optimizing of workflow, acceptance by pathologists, providing real assistance clinically, and delineating responsibilities for the resolution of clinical problems that need to be addressed.
Targeted therapy, chemotherapy, and immunotherapy are important adjuvants to surgical intervention in RCC (57). Therefore, the diagnosis and prognosis prediction of RCC based on clinical data, imaging data, pathological characteristics, and genes information are worth studying further. However, based on the findings of this review, researches on RCC primarily focus on three major histological subtypes, effectively identifying molecular aberrations and accurately predicting prognosis remain key challenges for future studies. Given the rarity of cases with specific molecular subtypes, the necessity for multicenter and collaborative studies is particularly pronounced. Furthermore, the integration of multimodal data from pathology, radiomics, genomics, and proteomics (58), along with the application of advanced AI technologies such as Large Language Models (59) is expected to make significant contributions to this field.
Our study has some limitations. The number of collected literature is limited, and only English articles and reviews on Web of Science and PubMed database are included, without articles of other languages or types. Our follow up plan is to develop systematic research to find more scientific and unified valuable feature extraction methods to break through the bottleneck of interpretability.
Data availability statement
The data sets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Author contributions
M-YL: Data curation, Funding acquisition, Methodology, Writing – original draft, Writing – review & editing. YP: Data curation, Methodology, Writing – original draft. YL: Formal analysis, Software, Validation, Writing – review & editing. HM: Formal analysis, Software, Writing – original draft. P-LS: Funding acquisition, Project administration, Writing – review & editing. H-WG: Conceptualization, Project administration, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China (81902342); Natural Science Foundation of Jilin Province (YDZJ202401458ZYTS/ YDZJ202401240ZYTS/ YDZJ202401451ZYTS); Jilin Province Medical and Health Talent Special Project (2024WSZX-A01); Bureau of Jilin Province Department(2024WSZX-B06); Jilin Province Key Laboratory (20170622006JC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2025.1516264/full#supplementary-material
Abbreviations
AI, artificial intelligence; API, application programming interface; AUC, area under the curve; ccRCC, clear cell renal cell carcinoma; CFS, correlation feature selection; chRCC, chromophobe renal cell carcinoma; CNN, convolutional neural network; DAG, Directed Acyclic Graph; DCNN, deep convolutional neural network; DFS, disease free survival; DL, deep learning; DP, digital pathology; EHPG, enhanced histogram of polar gradient; FCBF, fast correlation-based filter selection method; FM, Fisher selection; GA, genetic algorithm selection; GGMRF, generalized Gauss Markov random field; GNN, Graph Neural Network; Grad CAM, Gradient weighted Class Activation Mapping; HPG, histogram of polar gradient; IG, integrated gradient; IHC, immunohistochemistry; ISUP, International Society of Urological Pathology; LASSO, least absolute shrinkage and selection operator; ML, machine learning; OS, overall survival; PFS, progression free survival; pRCC, papillary renal cell carcinoma; RCC, Renal cell carcinoma; RF, random forest selection; ROC, Receiver Operating Characteristic; ROI, region of interest; SGD, Stochastic gradient descent; TMB, tumor mutation burden; WSI, whole slide imaging; XCA, exclusive component analysis feature.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. (2016) 66:7–30. doi: 10.3322/caac.21332
2. Moch H, Amin MB, Berney DM, Comperat EM, Gill AJ, Hartmann A, et al. The 2022 world health organization classification of tumours of the urinary system and male genital organs-part A: renal, penile, and testicular tumours. Eur Urol. (2022) 82:458–68. doi: 10.1016/j.eururo.2022.06.016
3. Delahunt B, Cheville JC, Martignoni G, Humphrey PA, Magi-Galluzzi C, McKenney J, et al. The International Society of Urological Pathology (ISUP) grading system for renal cell carcinoma and other prognostic parameters. Am J Surg Pathol. (2013) 37:1490–504. doi: 10.1097/PAS.0b013e318299f0fb
4. Smith SC, Wobker SE. The new WHO 2022 category of molecularly defined renal carcinomas: Accessible to practicing pathologists. Am J Clin Pathol. (2023) 160(6):545–8. doi: 10.1093/ajcp/aqad101
5. Jahn SW, Plass M, Moinfar F. Digital pathology: advantages, limitations and emerging perspectives. J Clin Med. (2020) 9:3697. doi: 10.3390/jcm9113697
6. Hanna MG, Parwani A, Sirintrapun SJ. Whole slide imaging: technology and applications. Adv Anat Pathol. (2020) 27:251–9. doi: 10.1097/PAP.0000000000000273
7. Hanna MG, Reuter VE, Hameed MR, Tan LK, Chiang S, Sigel C, et al. Whole slide imaging equivalency and efficiency study: experience at a large academic center. Mod Pathol. (2019) 32:916–28. doi: 10.1038/s41379-019-0205-0
8. Schuettfort VM, Pradere B, Rink M, Comperat E, Shariat SF. Pathomics in urology. Curr Opin Urol. (2020) 30:823–31. doi: 10.1097/MOU.0000000000000813
9. Buch VH, Ahmed I, Maruthappu M. Artificial intelligence in medicine: current trends and future possibilities. Br J Gen Pract. (2018) 68:143–4. doi: 10.3399/bjgp18X695213
10. Koteluk O, Wartecki A, Mazurek S, Kołodziejczak I, Mackiewicz A. How do machines learn? Artificial intelligence as a new era in medicine. J Pers Med. (2021) 11:32. doi: 10.3390/jpm11010032
11. Marletta S, Eccher A, Martelli FM, Santonicco N, Girolami I, Scarpa A, et al. Artificial intelligence-based algorithms for the diagnosis of prostate cancer: A systematic review. Am J Clin Pathol. (2024) 161:526–34. doi: 10.1093/ajcp/aqad182
12. Marletta S, L'Imperio V, Eccher A, Antonini P, Santonicco N, Girolami I, et al. Artificial intelligence-based tools applied to pathological diagnosis of microbiological diseases. Pathol Res Pract. (2023) 243:154362. doi: 10.1016/j.prp.2023.154362
13. Giulietti M, Cecati M, Sabanovic B, Scire A, Cimadamore A, Santoni M, et al. The role of artificial intelligence in the diagnosis and prognosis of renal cell tumors. Diagnostics (Basel). (2021) 11:206. doi: 10.3390/diagnostics11020206
14. Distante A, Marandino L, Bertolo R, Ingels A, Pavan N, Pecoraro A, et al. Artificial intelligence in renal cell carcinoma histopathology: current applications and future perspectives. Diagnostics (Basel). (2023) 13:2294. doi: 10.3390/diagnostics13132294
15. Dara S, Tumma P eds. (2018). “Feature extraction by using deep learning: A survey”, in: 2018 Second International Conference on Electronics, Communication and Aerospace Technology (ICECA), 29-31 March 2018. United States: IEEE, Institute of Electrical and Electronics Engineers.
16. Duan Y, Lu J, Zheng W, Zhou J. Deep adversarial metric learning. IEEE Trans Image Process. (2020) 29:2037–51. doi: 10.1109/TIP.83
17. Zhang Y, Xi R, Zeng L, Towey D, Bai R, Higashita R, et al. Structural priors guided network for the corneal endothelial cell segmentation. IEEE Trans Med Imaging. (2024) 43:309–20. doi: 10.1109/TMI.2023.3300656
18. Khene ZE, Kammerer-Jacquet SF, Bigot P, Rabilloud N, Albiges L, Margulis V, et al. Clinical application of digital and computational pathology in renal cell carcinoma: A systematic review. Eur Urol Oncol. (2024) 7:401–11. doi: 10.1016/j.euo.2023.10.018
19. Wessels F, Kuntz S, Krieghoff-Henning E, Schmitt M, Braun V, Worst TS, et al. Artificial intelligence to predict oncological outcome directly from hematoxylin and eosin-stained slides in urology. Minerva Urol Nephrol. (2022) 74:538–50. doi: 10.23736/S2724-6051.22.04758-9
20. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Rev Panam Salud Publ. (2022) 46:89. doi: 10.1186/s13643-021-01626-4
21. Lipkova J, Chen RJ, Chen B, Lu MY, Barbieri M, Shao D, et al. Artificial intelligence for multimodal data integration in oncology. Cancer Cell. (2022) 40:1095–110. doi: 10.1016/j.ccell.2022.09.012
22. Lu MY, Williamson DFK, Chen TY, Chen RJ, Barbieri M, Mahmood F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat BioMed Eng. (2021) 5:555–70. doi: 10.1038/s41551-020-00682-w
23. Faust K, Roohi A, Leon AJ, Leroux E, Dent A, Evans AJ, et al. Unsupervised resolution of histomorphologic heterogeneity in renal cell carcinoma using a brain tumor-educated neural network. JCO Clin Cancer Inform. (2020) 4:811–21. doi: 10.1200/CCI.20.00035
24. Sarker IH. Deep learning: A comprehensive overview on techniques, taxonomy, applications and research directions. SN Comput Science. (2021) 2:420. doi: 10.1007/s42979-021-00815-1
25. Chen RJ, Lu MY, Wang J, Williamson DFK, Rodig SJ, Lindeman NI, et al. Pathomic fusion: an integrated framework for fusing histopathology and genomic features for cancer diagnosis and prognosis. IEEE Trans Med Imaging. (2022) 41:757–70. doi: 10.1109/TMI.2020.3021387
26. Xie Y, Xu Z, Zhang J, Wang Z, Ji S. Self-supervised learning of graph neural networks: A unified review. IEEE Trans Pattern Anal Mach Intell. (2023) 45:2412–29. doi: 10.1109/TPAMI.2022.3170559
27. Lee Y, Park JH, Oh S, Shin K, Sun J, Jung M, et al. Derivation of prognostic contextual histopathological features from whole-slide images of tumours via graph deep learning. Nat BioMed Eng. (2022). doi: 10.1038/s41551-022-00923-0
28. Chen C, Lu MY, Williamson DFK, Chen TY, Schaumberg AJ, Mahmood F. Fast and scalable search of whole-slide images via self-supervised deep learning. Nat BioMed Eng. (2022) 6:1420–34. doi: 10.1038/s41551-022-00929-8
29. DiPalma J, Suriawinata AA, Tafe LJ, Torresani L, Hassanpour S. Resolution-based distillation for efficient histology image classification. Artif Intell Med. (2021) 119:102136. doi: 10.1016/j.artmed.2021.102136
30. Chen RJ, Lu MY, Williamson DFK, Chen TY, Lipkova J, Noor Z, et al. Pan-cancer integrative histology-genomic analysis via multimodal deep learning. Cancer Cell. (2022) 40:865–78 e6. doi: 10.1016/j.ccell.2022.07.004
31. Shi M, Li X, Li M, Si Y. Attention-based generative adversarial networks improve prognostic outcome prediction of cancer from multimodal data. Brief Bioinform. (2023) 24:bbad329. doi: 10.1093/bib/bbad329
32. Schulz S, Woerl AC, Jungmann F, Glasner C, Stenzel P, Strobl S, et al. Multimodal deep learning for prognosis prediction in renal cancer. Front Oncol. (2021) 11:788740. doi: 10.3389/fonc.2021.788740
33. Fenstermaker M, Tomlins SA, Singh K, Wiens J, Morgan TM. Development and validation of a deep-learning model to assist with renal cell carcinoma histopathologic interpretation. Urology. (2020) 144:152–7. doi: 10.1016/j.urology.2020.05.094
34. Abdeltawab H, Khalifa F, Ghazal M, Cheng L, Gondim D, El-Baz A. A pyramidal deep learning pipeline for kidney whole-slide histology images classification. Sci Rep. (2021) 11:20189. doi: 10.1038/s41598-021-99735-6
35. Zhu M, Ren B, Richards R, Suriawinata M, Tomita N, Hassanpour S. Development and evaluation of a deep neural network for histologic classification of renal cell carcinoma on biopsy and surgical resection slides. Sci Rep. (2021) 11:7080. doi: 10.1038/s41598-021-86540-4
36. Tabibu S, Vinod PK, Jawahar CV. Pan-Renal Cell Carcinoma classification and survival prediction from histopathology images using deep learning. Sci Rep. (2019) 9:10509. doi: 10.1038/s41598-019-46718-3
37. Marostica E, Barber R, Denize T, Kohane IS, Signoretti S, Golden JA, et al. Development of a histopathology informatics pipeline for classification and prediction of clinical outcomes in subtypes of renal cell carcinoma. Clin Cancer Res. (2021) 27:2868–78. doi: 10.1158/1078-0432.CCR-20-4119
38. Ohe C, Yoshida T, Amin MB, Uno R, Atsumi N, Yasukochi Y, et al. Deep learning-based predictions of clear and eosinophilic phenotypes in clear cell renal cell carcinoma. Hum Pathol. (2023) 131:68–78. doi: 10.1016/j.humpath.2022.11.004
39. Chen S, Zhang N, Jiang L, Gao F, Shao J, Wang T, et al. Clinical use of a machine learning histopathological image signature in diagnosis and survival prediction of clear cell renal cell carcinoma. Int J Cancer. (2021) 148:780–90. doi: 10.1002/ijc.v148.3
40. Kruk M, Kurek J, Osowski S, Koktysz R, Swiderski B, Markiewicz T. Ensemble of classifiers and wavelet transformation for improved recognition of Fuhrman grading in clear-cell renal carcinoma. Biocybernetics Biomed Engineering. (2017) 37:357–64. doi: 10.1016/j.bbe.2017.04.005
41. Tian K, Rubadue CA, Lin DI, Veta M, Pyle ME, Irshad H, et al. Automated clear cell renal carcinoma grade classification with prognostic significance. PloS One. (2019) 14:e0222641. doi: 10.1371/journal.pone.0222641
42. Holdbrook DA, Singh M, Choudhury Y, Kalaw EM, Koh V, Tan HS, et al. Automated renal cancer grading using nuclear pleomorphic patterns. JCO Clin Cancer Inform. (2018) 2:1–12. doi: 10.1200/CCI.17.00100
43. Chanchal AK, Lal S, Kumar R, Kwak JT, Kini J. A novel dataset and efficient deep learning framework for automated grading of renal cell carcinoma from kidney histopathology images. Sci Rep. (2023) 13:5728. doi: 10.1038/s41598-023-31275-7
44. Khoshdeli M, Borowsky A, Parvin B. Deep learning models differentiate tumor grades from H&E stained histology sections. Annu Int Conf IEEE Eng Med Biol Soc. (2018) 2018:620–3. doi: 10.1109/EMBC.2018.8512357
45. Udager AM, Mehra R. Morphologic, molecular, and taxonomic evolution of renal cell carcinoma: A conceptual perspective with emphasis on updates to the 2016 world health organization classification. Arch Pathol Lab Med. (2016) 140:1026–37. doi: 10.5858/arpa.2016-0218-RA
46. Chen Q, Kuai Y, Wang S, Zhu X, Wang H, Liu W, et al. Deep learning-based classification of epithelial-mesenchymal transition for predicting response to therapy in clear cell renal cell carcinoma. Front Oncol. (2021) 11:782515. doi: 10.3389/fonc.2021.782515
47. Acosta PH, Panwar V, Jarmale V, Christie A, Jasti J, Margulis V, et al. Intratumoral resolution of driver gene mutation heterogeneity in renal cancer using deep learning. Cancer Res. (2022) 82:2792–806. doi: 10.1158/0008-5472.CAN-21-2318
48. Cheng J, Han Z, Mehra R, Shao W, Cheng M, Feng Q, et al. Computational analysis of pathological images enables a better diagnosis of TFE3 Xp11. 2 translocation Renal Cell carcinoma Nat Commun. (2020) 11:1778. doi: 10.1038/s41467-020-15671-5
49. Wei JH, Feng ZH, Cao Y, Zhao HW, Chen ZH, Liao B, et al. Predictive value of single-nucleotide polymorphism signature for recurrence in localised renal cell carcinoma: a retrospective analysis and multicentre validation study. Lancet Oncol. (2019) 20:591–600. doi: 10.1016/S1470-2045(18)30932-X
50. Wessels F, Schmitt M, Krieghoff-Henning E, Kather JN, Nientiedt M, Kriegmair MC, et al. Deep learning can predict survival directly from histology in clear cell renal cell carcinoma. PloS One. (2022) 17:e0272656. doi: 10.1371/journal.pone.0272656
51. Gui CP, Chen YH, Zhao HW, Cao JZ, Liu TJ, Xiong SW, et al. Multimodal recurrence scoring system for prediction of clear cell renal cell carcinoma outcome: a discovery and validation study. Lancet Digit Health. (2023) 5:e515–e24. doi: 10.1016/S2589-7500(23)00095-X
52. Cheng J, Zhang J, Han Y, Wang X, Ye X, Meng Y, et al. Integrative analysis of histopathological images and genomic data predicts clear cell renal cell carcinoma prognosis. Cancer Res. (2017) 77:e91–e100. doi: 10.1158/0008-5472.CAN-17-0313
53. Cheng J, Mo X, Wang X, Parwani A, Feng Q, Huang K. Identification of topological features in renal tumor microenvironment associated with patient survival. Bioinformatics. (2018) 34:1024–30. doi: 10.1093/bioinformatics/btx723
54. Chen S, Jiang L, Gao F, Zhang E, Wang T, Zhang N, et al. Machine learning-based pathomics signature could act as a novel prognostic marker for patients with clear cell renal cell carcinoma. Br J Cancer. (2022) 126:771–7. doi: 10.1038/s41416-021-01640-2
55. Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. (2017) 7:16878. doi: 10.1038/s41598-017-17204-5
56. Kalra S, Tizhoosh HR, Shah S, Choi C, Damaskinos S, Safarpoor A, et al. Pan-cancer diagnostic consensus through searching archival histopathology images using artificial intelligence. NPJ Digit Med. (2020) 3:31. doi: 10.1038/s41746-020-0238-2
57. Khodabakhshi Z, Amini M, Mostafaei S, Haddadi Avval A, Nazari M, Oveisi M, et al. Overall survival prediction in renal cell carcinoma patients using computed tomography radiomic and clinical information. J Digital Imaging. (2021) 34:1086–98. doi: 10.1007/s10278-021-00500-y
58. Vanguri RS, Luo J, Aukerman AT, Egger JV, Fong CJ, Horvat N, et al. Multimodal integration of radiology, pathology and genomics for prediction of response to PD-(L)1 blockade in patients with non-small cell lung cancer. Nat Cancer. (2022) 3:1151–64. doi: 10.1038/s43018-022-00416-8
Keywords: digital pathology, artificial intelligence, deep learning, WSI, RCC, prediction, diagnosis, prognosis
Citation: Li M-Y, Pan Y, Lv Y, Ma H, Sun P-L and Gao H-W (2025) Digital pathology and artificial intelligence in renal cell carcinoma focusing on feature extraction: a literature review. Front. Oncol. 15:1516264. doi: 10.3389/fonc.2025.1516264
Received: 24 October 2024; Accepted: 06 January 2025;
Published: 24 January 2025.
Edited by:
Francesca Sanguedolce, University of Foggia, ItalyCopyright © 2025 Li, Pan, Lv, Ma, Sun and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Wen Gao, Z2FvaG9uZ3dlbkBqbHUuZWR1LmNu; Ping-Li Sun, cGluZ2xpc3VuQGpsdS5lZHUuY24=
 Ming-Yue Li1
Ming-Yue Li1 Ping-Li Sun
Ping-Li Sun Hong-Wen Gao
Hong-Wen Gao