- 1Department of Pathology and Laboratory Medicine, The Ohio State University Medical Center, Columbus, OH, United States
- 2Comprehensive Cancer Center, The Ohio State University Medical Center, Columbus, OH, United States
- 3Department of Surgery, Division of Surgical Oncology, The Ohio State University Medical Center, Columbus, OH, United States
Liposarcoma is the most prevalent sarcoma in adults representing 20% of all sarcomas with well-differentiated/dedifferentiated among the most common subtypes represented. Despite multimodality treatment approaches, there has not been any appreciable change in survival benefit in the past 10 years. The future of targeted therapy for WD/DDLPS is promising with the intention to spare multi-visceral removal due to radical surgical resection. Therefore, there is a need to expand upon the molecular landscape of WDLPS and DDLPS which can help identify potential therapeutic targets for the treatment of this disease. Targeted transcriptome analysis using the NanoString tumor signaling 360 panel revealed a dysregulation in glucose metabolism and HIF1 signaling pathways in both WDLPS and DDLPS when compared to normal fat controls. WDLPS, however, demonstrated upregulation of HIF-1A and TGF-β when compared to DDLPS by targeted transcriptome analysis and orthogonal validation by RT-qPCR suggesting activation of EMT pathway in WDLPS when compared to DDLPS. Our findings implicate a putative role for dysregulation in glucose metabolism, TGF-β and HIF1 signaling in the pathogenesis of both WD/DDLPS suggesting a possible proinflammatory tumor environment within WDLPS and subsequent activation of the TGF-β signaling pathway.
Introduction
Liposarcoma is one of the prevalent mesenchymal neoplasms in adults, representing 20% of all sarcomas and is divided into 5 main subtypes including atypical lipomatous tumor (ALT)/well-differentiated liposarcoma (WDLPS), dedifferentiated liposarcoma (DDLPS), myxoid liposarcoma, pleomorphic liposarcoma, and atypical spindle cell/pleomorphic lipomatous tumor. Unique molecular and biological differences exist that define the histopathologic/clinical subtypes. The largest subgroup includes both ALT/WDLPS and DDLPS representing approximately 45% of all liposarcomas (1–3). When localized to the extremities, the term atypical lipomatous tumor is used, whilst liposarcoma is used for retroperitoneal and central body cavity locations.
The morphology and clinical behavior of WDLPS and DDLPS differ drastically, and the biological behavior of DDLPS portends a worse prognosis due to its aggressive nature, high local recurrence rate, and ability to metastasize (4–7). We also recognize that 90% of DDLPS arises de novo in the absence of a WDLPS precursor lesion. (8) The non-adipogenic morphology of DDLPS is distinct from WDLPS, often consisting of a high-grade non-lipogenic spindle cell sarcoma (7). In cases of secondary DDLPS the dedifferentiation of WDLPS is not well understood. Moreover, both WDLPS and DDLPS are among the most common retroperitoneal sarcoma establishing a critical need to further elucidate the molecular landscape that defines tumorigenesis in retroperitoneal liposarcoma and to understand the underlying mechanisms that distinguish the unique biological behavior of WDLPS and DDLPS. For the purposes of uniformity, we will restrict our terminology to WDLPS rather than ALT.
Previous studies on molecular profiling of lipomatous tumors have established amplification of MDM2 as a diagnostic marker distinguishing WDLPS and DDLPS from benign lipomatous tumors as well as other subtypes of liposarcoma (9, 10). Both WDLPS and DDLPS harbor supernumerary ring and giant marker chromosomes resulting from amplification of the 12q13-15 region containing the MDM2 locus. The MDM2-p53 axis has been well established in summarizing the regulatory mechanisms by which MDM2 functions to facilitate p53 degradation by E3 ubiquitin ligase thereby inhibiting the tumor suppressor activity of p53 (11, 12). Further, overexpression of neighboring genes, CDK4 and HMGAI, have been identified in WDLPS and DDLPS as well as exploited as potential molecular targets for treatment (10). However, the molecular genetic determinants of transformation of WDLPS to DDLPS are not known.
The landscape of systemic and molecular-based therapies for WDLPS and DDLPS are limited to CDK4 and PDL1 inhibitors. Small molecule MDM2 (i.e. nutlins, Siremadlin) and cdk4 inhibitors (ribociclib) have shown limited efficacy in phase I clinical trials (13, 14). Novel targets of the EMT pathway are emerging as potential therapeutics for the treatment of DDLPS. For example, CCNDBP1, a tumor suppressor, has been shown to suppress gene expression and protein levels of key regulators of the EMT in vivo. (15).
Surgical resection is currently the standard treatment. (16) Chemotherapy used in the adjuvant setting for DDLPS has shown no significant improvement in clinical outcomes regarding local recurrence. More so, the overall 5-year survival rate of DDLPS is approximately 40-60% and no significant improvement in survival rates has been achieved in the past 15 years. (17) Despite surgical resection, retroperitoneal WDLPS recurs in approximately 20% of cases and has a higher incidence of dedifferentiation to DDLPS with an increased 20-year mortality rate between 30-40% (2, 18). Cytotoxic chemotherapy is not routinely utilized in patients with retroperitoneal WDLPS indicating a critical need to identify therapeutic targets for the treatment of this aggressive subtype of WDLPS. Importantly, the cellular pathways that define the sarcomagenesis of WDLPS and DDLPS and distinguish each entity continue to remain elusive. Thus, there remains an urgent need for more targeted therapeutic options for WD/DDLPS patients.
Using the targeted NanoString 360 tumor signaling transcriptomic panel, we further explore the molecular underpinnings of WDLPS and DDLPS and identify key cellular pathways unique to the two liposarcoma subtypes. Expanding our understanding of the molecular drivers in WDLPS may lead to the identification of therapeutic targets for its treatment to prevent recurrence.
Materials and methods
Sample selection
Snap-frozen tissue samples were retrieved from tissue biorepository, obtained at the time of radical surgical resections. All the patients were chemoradiation therapy naïve, and the nature of the surgical resections were R1.
Selection criteria included a known diagnosis of retroperitoneal WDLPS [n=12 (males = 7; females = 5); median age = 61.5 years] and DDLPS [n=12 (males = 8; females = 4); median age = 61.5 years]. Normal fat was collected as normal controls [n = 4 (males =3; females = 1); median age = 59 years] (Supplementary Table 1).
Quantitative real-time PCR analysis
Total RNA was isolated from snap-frozen human tissue samples using RNeasy Lipid Tissue Mini Kit according to manufacturer’s protocol (Qiagen). Preparation of cDNA was achieved using the high-capacity cDNA reverse transcription kit with RNase Inhibitor (Applied Biosystems, Thermo Fischer Scientific). SYBR Green (Bio-Rad) biochemistries was used to perform qRT-PCR to quantify gene expression with GAPDH or ACTB as reference genes.
NanoString nCounter gene expression assay
NanoString nCounter Tumor Signaling 360 panel was used for direct detection of target molecules using color-coded molecular barcodes, providing a digital simultaneous quantification of the number of target molecules. The panel included standard probes for the Tumor Signaling 360 panel without customization. A concentration of 100 ng of total RNA was hybridized overnight with nCounter Reporter (8 μL) probes in hybridization buffer and in excess of nCounter Capture probes (2 μL) at 65°C for 18 h. After overnight hybridization, probe excess was removed using two-step magnetic bead-based purification on an automated fluidic handling system (nCounter Prep Station). Biotinylated capture probe-bound samples were immobilized and recovered on a streptavidin-coated cartridge. The abundance of specific target molecules was then quantified using the nCounter digital analyzer. Individual fluorescent barcodes and target molecules present in each sample were recorded with a CCD camera by performing a high-density scan (325 fields of view). Images were processed internally into a digital format and exported as Reporter Code Count (RCC).
Western Blot
A total of 20 µg of protein was loaded into a 10% Tris-HCL gel. Proteins were transferred to a PVDF membrane (0.2 µm). The PVDF membrane was blocked with 5% nonfat dry milk in 1X TBS-T (1% polysorbate 20) for 1 hour and incubated with the primary antibody, rabbit anti-human TGF-β (Cell Signaling, Danvers, MA, USA, #3711) or rabbit anti-human GAPDH (Cell Signaling, 2118S) overnight at 4°C. Membranes were incubated in a secondary goat anti-rabbit HRP-linked antibody (Thermo Fisher Scientific, 31460, Rockford, IL, USA) for 2 hours. Membranes were incubated with chemiluminescence ECL Western Blotting substrate (ThermoScientific, Waltham, MA, USA). ImageJ was used to quantify the protein intensity of TGF-β relative to GAPDH.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 9.5.1 (GraphPad Software, San Diego, CA, USA). Statistical differences were determined using a one-way ANOVA followed by a Tukey-Kramer or Bonferroni test for post hoc comparison. Nanostring gene expression was performed by using ROSILAND® software. In brief, ROSILAND® software follows the nCounter® Advanced Analysis protocol for normalization. Fold changes and p-values were calculated using the nCounter® Advanced Analysis 2.0. The adjusted p-value was calculated using the Benjamini-Hochberg method of estimating the false discovery rate (FDR).
Results
WDLPS and DDLPS demonstrate a global differential gene expression and downregulation in genes associated with glucose metabolism compared to normal fat
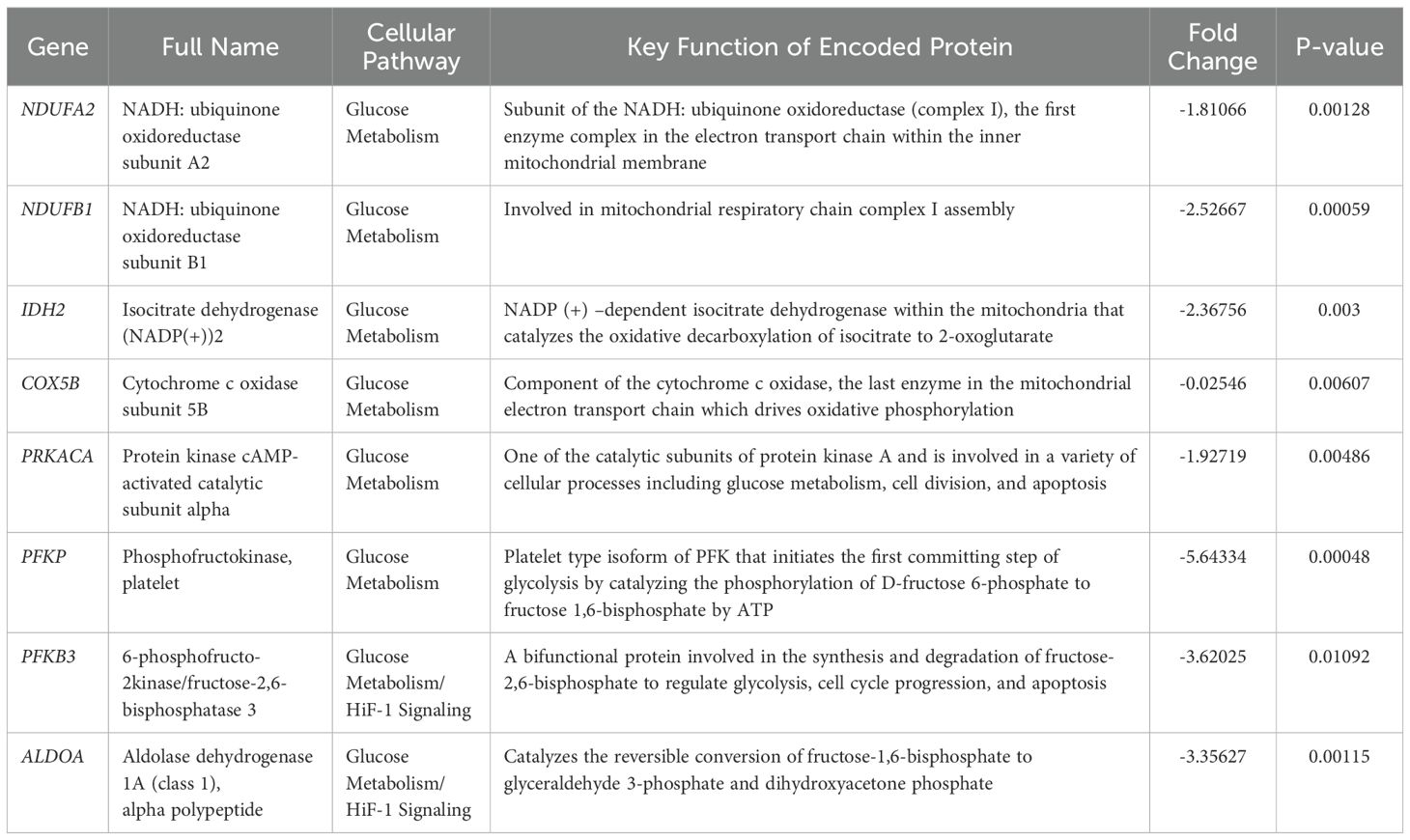
The NanoString nCounter Tumor Signaling 360 Panel was used to characterize the gene expression profiles of WDLPS, DDLPS, and the normal fat control. The heat-map (Figure 1A) and principal component analysis (Figure 1B) show global differential gene expression and clustering between well differentiated liposarcoma, dedifferentiated liposarcoma and normal fat. The panel analyzed 760 genes involved in global pathways including tumorigenesis, invasion, metastasis, angiogenesis, immune evasion, inflammation and glucose metabolism (Figures 1C, D). Our initial evaluation revealed 352 differentially expressed genes involved in a variety of biochemical pathways including signal transduction, metabolism, autophagy, and epigenetic and transcriptional regulation (Figure 1C). Of the 352 differentially expressed genes between both the liposarcoma groups (WDLPS and DDLPS) and normal fat, 147 genes were involved in signal transduction pathways that function to regulate cell cycle progression, immune response, autophagy, and oxidative stress (Figures 1C, D). Glucose metabolism demonstrated a global downregulation in both WDLPS and DDLPS when compared to normal fat (global significance score >2.0) as highlighted in Figure 1D. Additionally, glutamine metabolism and HIF-1 signaling pathways were upregulated in both WDLPS and DDLPS thereby suggesting a metabolic shift in WDLPS and DDLPS when compared to the nonneoplastic normal fat control (Figure 1D).
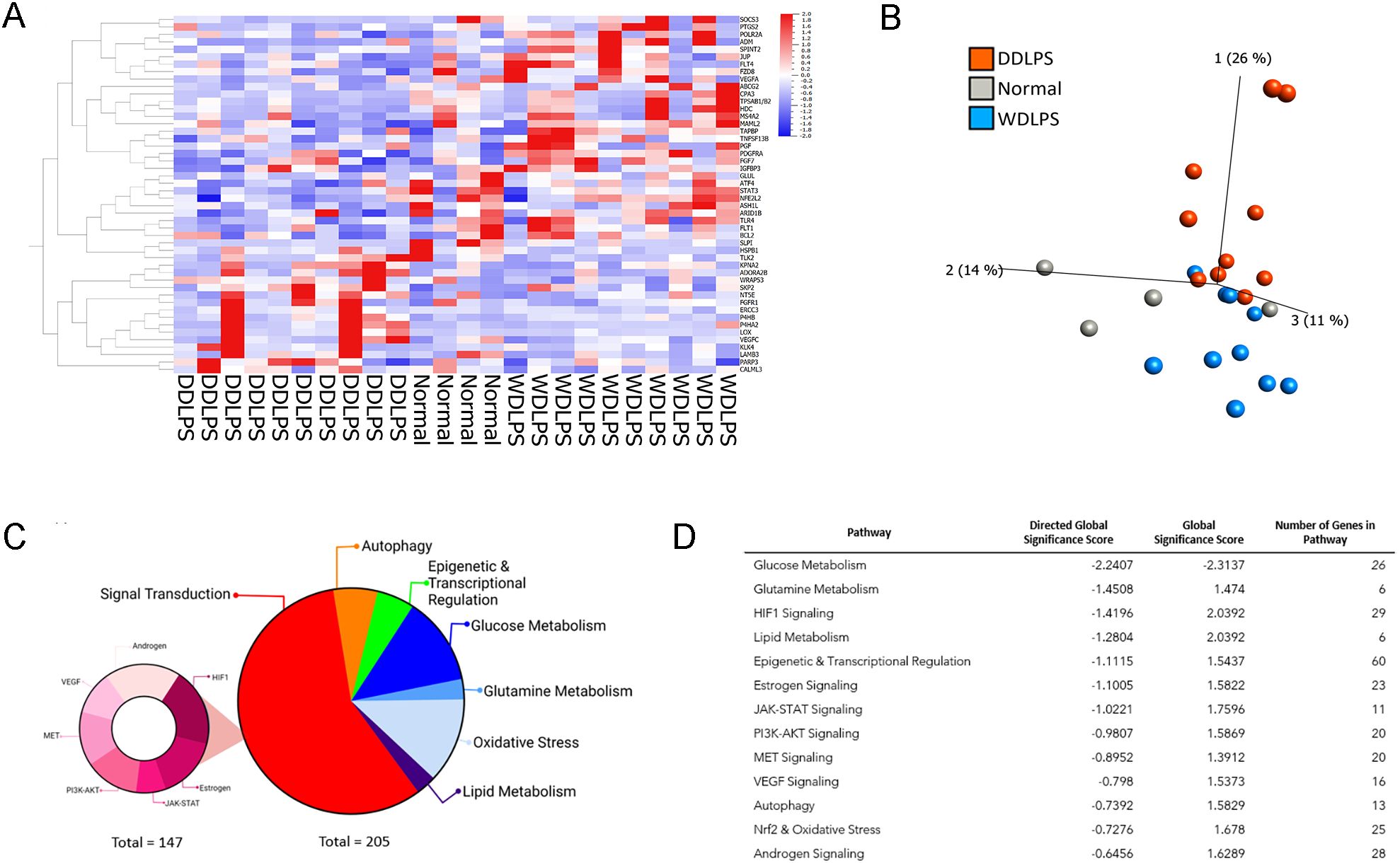
Figure 1. NanoString nCounter Tumor Signaling 360 Panel was used to characterize the gene expression profiles and pathway analysis of WDLPS, DDLPS, and the normal fat control. (A, B) Heat map (A) Principal component analysis (B) generated by hierarchical clustering analysis to represent the unique expression profiles for WDLPS, DDLPS, and normal fat. Global pathway analysis and differential gene expression show differentially expressed genes involved in a variety of biochemical pathways including signal transduction, metabolism, autophagy, and epigenetic and transcriptional regulation (Global significance cutoff scores of 1.0 and -1.0) (C, D).
Targeted transcriptomic analysis of WDLPS and normal fat tissue identified 82 differentially expressed genes representing significantly altered cellular pathways. Among the pathways with the greatest significance score include glucose metabolism and HIF-1-signaling (downregulated) as well as antigen presentation (upregulated) with significance score >2.0 (Figures 2A–C). Our initial comparison between DDLPS and normal fat in Figures 2D–F revealed 118 differentially expressed genes representing significantly altered cellular pathways (significance score >2.0) including glucose metabolism (downregulated) as well as HIF-1signaling, T-cell exhaustion, hedgehog (upregulated). The comparison between WDLPS and DDLPS (Figures 2G–I) shows differential gene expressions involved in the Notch, TNF and VEGF-signaling pathways, though the significance scores are <2.0.
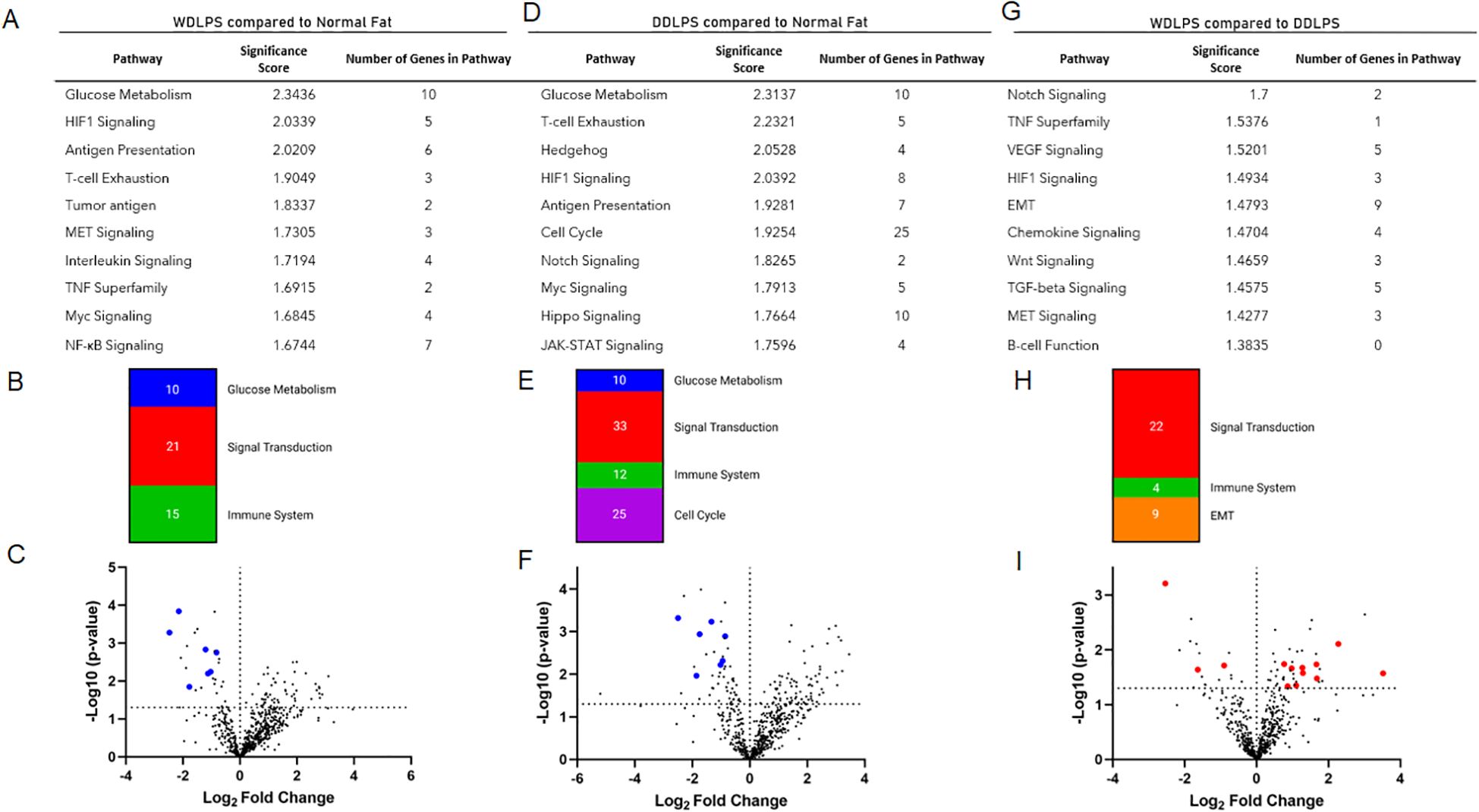
Figure 2. NanoString nCounter Tumor Signaling 360 Panel was used to characterize the gene expression profiles and pathway analysis of WDLPS, DDLPS, and the normal fat control. (A, B) Significantly altered pathways in WDLPS compared to normal fat (significance score cutoff 1.0). (C) Volcano plot demonstrating the distribution of differential gene expression for WDLPS and normal fat. (D, E) Significantly altered pathways in DDLPS compared to normal fat (significance score cutoff 1.0). (F) Volcano plot demonstrating the distribution of differential gene expression for DDLPS and normal fat. (G, H) Significantly altered pathways in WDLPS compared to DDLPS (significance score cutoff 1.0). (I) Volcano plot demonstrating the distribution of differential gene expression for WDLPS and DDLPS; blue and red dots correspond to genes associated with glucose metabolism and signal transduction, respectively.
Significantly downregulated genes in both WDLPS and DDLPS when compared to normal fat were PFKP, ALDOA, PKM, PFKFB3, LDHA, NDUFB1, IDH2, COX5B, NDUFA2, and PRKACA which share key modulators of glucose metabolism, specifically aerobic respiration including glycolysis and the electron transport chain (Table 1; Figures 3A–C).
Figure 3. WDLPS and DDLPS demonstrate a global downregulation in genes associated with glucose metabolism and upregulated HIF1 signaling. (A) Overview of glycolysis. (B) Differentially expressed genes involved in glucose metabolism (padj < 0.05; log2 fold change > 1 & < -1). (C) Real-time PCR demonstrating a downregulation of NDUFA2, IDH2, PRKCAC in both WDLPS and DDLPS when compared to normal fat as well as downregulation of GABPA in DDLPS when compared to WDLPS and normal fat. (D) HIF1A expression is upregulated in WDLPS when compared to DDLPS and normal fat. (E) Differential expression of genes associated with HIF1-signaling pathway (padj < 0.05; log2 fold change > 1 & < -1). * represents p<0.05; ** represents p<0.01; and *** represents p<0.001.
Both WDLPS and DDLPS displayed reduced aerobic glucose metabolism when compared to normal fat. Downregulated genes involved in glucose metabolism identified in the DDLPS group were similar to genes identified in WDLPS which included PFKP, PFKFB3, IDH2, PRKACA, ALDOA, COX5B, NDUFB1, and NDUFA2 (Figures 3A–C). Real-time PCR was performed to validate differential gene expression in WDLPS, DDLPS, and normal fat. Figures 3B, C highlights decreased gene expression of NDUFA2, IDH2, and PRKCAC in both WDLPS and DDLPS when compared to normal fat tissue. Among the genes involved in glucose metabolism include PFKP, PFKFB3, IDH2, and PRKACA which are known to serve multifactorial roles in carcinogenesis including differentiation, cell-cycle regulation, and apoptosis however, the role of these genes in sarcomagenesis of WDLPS and DDLPS is not well elucidated.
Glutamine synthesis is altered in DDLPS when compared to WDLPS and normal fat
One means by which cancer metabolism is altered is by utilizing a variety of different biosynthetic substrates including glutamine. Glutamine synthesis was reduced in both WDLPS and DDLPS when compared to normal fat potentially supporting a metabolic shift toward glutaminolysis. Interestingly, GA binding protein transcription factor subunit alpha, GABPA, was downregulated in DDLPS when compared to normal fat and WDLPS and confirmed by RT-PCR (Figure 3C). However, the role of GABPA in the pathogenesis of DDLPS is still unknown.
The HIF-1 signaling pathway is upregulated in WDLPS
The HIF-1 pathway was significantly altered in both WDLPS and DDLPS when compared to the normal fat control (Figures 3D, E). Expression of HIF-1A was significantly upregulated in WDLPS and DDLPS compared to normal fat (Figure 3D) and confirmed by RT-PCR. Further exploration of the HIF-1A pathway revealed differences in the transcriptome profile of WDLPS and DDLPS (Figure 3E). Expression of ADM and FLT1, genes also involved in angiogenesis, were increased in WDLPS when compared to DDLPS (Figure 3E). In contrast, DDLPS showed an upregulation of P4HA2, a key mediator of collagen synthesis when compared to WDLPS (Figure 3E).
Profiling of resident immune cells reveals an increased population of T-cells in WDLPS
Resident immune cells serve a predominantly anti-inflammatory role within normal adipose tissue as well as indirectly regulating adipocyte metabolism. Therefore, as an additional metric, we evaluated the composition of the immune fraction within the tumor microenvironments of WDLPS and DDLPS as well as normal adipose tissue using Nanostring cell profiling capabilities (Figure 4). The population of neutrophils was elevated in both WDLPS and DDLPS when compared to the normal fat control (Figure 4A). Immune profiling revealed increased lymphocytes, specifically T-cells and cytotoxic T-cells, in the WDLPS samples when compared to both DDLPS and normal adipose tissue (Figure 4A). Populations of the remaining immune cells including dendritic cells, macrophages, and mast cells remained unchanged (Figure 4A).
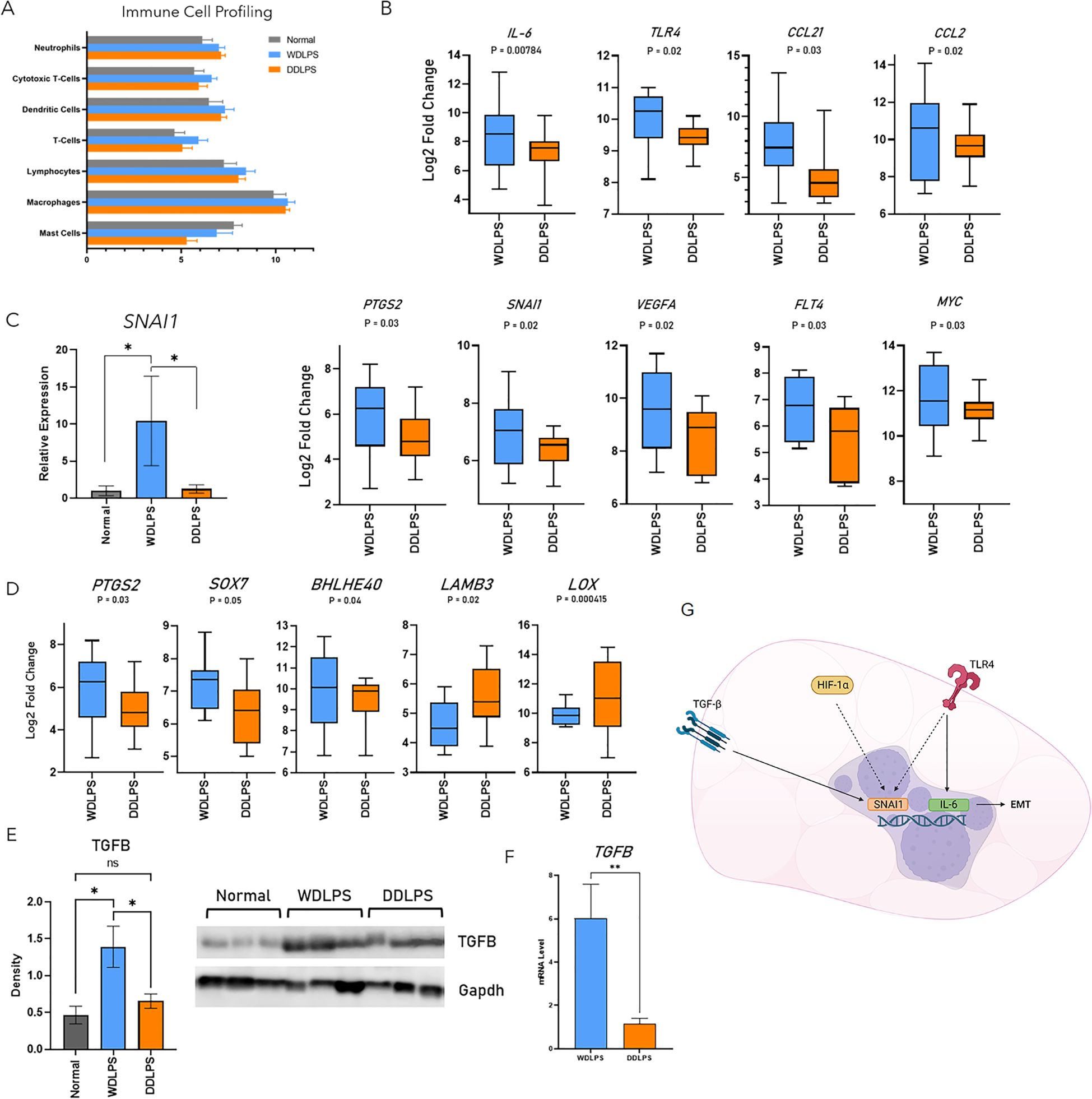
Figure 4. Profiling of resident immune cells reveals an increased population T-cells and upregulation of the EMT pathway in WDLPS. (A) Cell profiling of resident immune populations based on NanoString nCounter Tumor Signaling 360 Panel capture probes demonstrates increased lymphocytes, specifically T-cell and cytotoxic T-cells in WDLPS compared to DDLPS and normal fat. Neutrophils are increased in WDLPS and DDLPS compared to normal fat. (B) Differential expression of genes involved in the EMT pathway demonstrates upregulation of IL-6, TLR4, CCL21, CCL2, PTGS2, SNAI1, VEGFA, FLT4, and MYC when compared to DDLPS (padj < 0.05; log2 fold change > 1 & < -1). (C) SNAI1 is upregulated in WDLPS when compared to DDLPS and normal fat by real-time PCR. (D) Genes (PTGS2, SOX7, BHLHE40, LAMB3, LOX) downstream of TGF-β signaling are upregulated in WDLPS when compared to DDLPS. (E, F) TGF-β protein and mRNA levels (E) are increased in WDLPS confirmed by Western blot and real-time qPCR (*p-value < 0.05). (G) Summary of EMT-related pathways activated in WDLPS. ** represents p<0.01.
Epithelial-mesenchymal transition and TGF-β pathways are upregulated in WDLPS
Our analysis established that WDLPS and DDLPS share similar metabolic profiles, specifically a shift from glucose metabolism to glutaminolysis thus exhibiting a cancer-promoting metabolic phenotype. Of the 760 differentially expressed genes, 47 genes were significantly altered in WDLPS when compared to DDLPS (Figure 1). Interestingly, the corresponding cellular pathways significantly altered in WDLPS when compared to DDLPS are predominantly associated with metastasis including inflammation, angiogenesis, and extracellular matrix remodeling (Figures 4B–D). Our data revealed that genes involved in metastasis, specifically pathways that comprise the epithelial-mesenchymal transition, EMT, pathway were differentially expressed in the WDLPS and DDLPS (Figures 4B–D). It is well known that the biological behavior of DDLPS is more aggressive than WDLPS and can metastasize to distant sites. The behavior of WDLPS, however, is more insidious as it tends to expand in the peritoneal and retroperitoneal cavities engulfing visceral organs. Genes involved in the EMT pathway including proinflammatory cytokines/chemokines IL-6, CCL21, CCL2, as well as the innate immune receptor, TLR4, were upregulated in WDLPS when compared to DDLPS (Figure 4B). Additionally, genes involved in angiogenesis and vascular permeability including VEGFA, FLT4, FLT1, and PTGS2 were upregulated in WDLPS when compared to DDLPS with the exception of VEGFC which showed increased mRNA levels in DDLPS when compared to WDLPS (Figure 4B). Snail family transcriptional repressor 1, SNAI1, is known to be involved in promoting the EMT pathway in human cancer and is a direct downstream target of the HIF-1 pathway as well as the TGF-β signaling pathway. Evaluation of differentially expressed genes involved in the EMT pathway revealed an upregulation of SNAI1 in the WDLPS when compared to DDLPS (Figure 4B). Measurement of SNAI1 mRNA levels by real-time PCR confirmed that SNAI1 is upregulated in WDLPS when compared to DDLPS as well as normal fat (Figure 4C).
The TGF-β signaling pathway plays an integral role in adipogenesis by promoting the differentiation from preadipocytes to mature adipocytes programmed to store lipids primarily in the form of triglyceride. The TGF-β signaling also plays a role in cell proliferation and apoptosis and has been implicated in a variety of human cancers serving as a therapeutic target. The gene expression profiles of both WDLPS and DDLPS demonstrated that genes involved in the TGF-β signaling pathway by direct or indirect downstream transcriptional regulation are predominately up-regulated in WDLPS when compared to DDLPS (Figure 4D). Gene expression of TGF-β has been shown to be upregulated in the DDLPS by in vitro and in vivo studies. It is conceivable that the upregulation of TGF-β may implicate proinflammatory cytokines and other activators of EMT that drive the tumorigenesis of WDLPS (Figure 4G). However, our data revealed that both TGF- β protein and mRNA levels were elevated in WDLPS when compared to DDLPS (Figures 4E, F) as well as several genes involved in the TGF-β pathway including PTGS2, and SOX7 apart from BHLHE40, LAMB3, and LOX which were unchanged or down-regulated in WDLPS, respectively (Figure 4D).
Discussion
The NanoSting nCounter panel has also been widely accepted as an efficient screening modality for various tumor types including soft tissue sarcomas with the goal of identifying key tumorigenic pathways to aid in the further understanding of tumor biology. (19–22) NanoString has demonstrated efficacy and cost-effectiveness for the identification and diagnosis of WDLPS and DDLPS in a predominantly retrospective series of low-grade adipogenic tumors when compared to MDM2 FISH. (23) More recently, the NanoString nCounter platform has been employed to evaluate the immune profile of DDLPS for targeted immunotherapy. (24) This the first pilot study to identify key metabolic signatures between WDLPS and DDLPS utilizing the NanoString nCounter Tumor Signaling 360 Panel.
WDLPS and DDLPS share the cytogenetic abnormality, specifically amplification of the 12q13-15 region that leads to subsequent diagnostic amplification of MDM2 and CDK4. Previous cytogenetic studies have identified alternative amplification of 1p32 and 6q23 in DDLPS and not identified in WDLPS. (25) Located within the 1p32 and 6q23 loci, are the genes jun proto-oncogene (JUN) and mitogen-activated protein kinase kinase5 (MAP3K5). (26–28) Lineage-defining molecular mechanisms of WDLPS and DDLPS, however, remain unknown.
The NanoString nCounter Tumor Signaling 360 Panel was selected for targeted transcriptomic analysis due to the comprehensive coverage of genes involved in tumorigenesis, immune signaling, and microenvironment. Results of the current study have provided insight into additional molecular pathways that can be further explored to develop enhanced therapeutic modalities for retroperitoneal WDLPS and DDLPS including molecular pathways that are somewhat surprising in distinguishing WDLPS from DDLPS such as HIF-1 signaling, TGF-β signaling, and the epithelial-mesenchymal transition (EMT) pathways that are indicators of aggressive tumor behavior and metastasis.
We have established that both WDLPS and DDLPS have metabolic profiles generated from our targeted analysis that differ significantly from normal fat including both notably glucose and glutamine metabolism. Glucose is a primary focus in cancer metabolism, especially in the setting of decreased nutrient precursors in the tumor microenvironment. Cancer cells maintain high rates of glucose metabolism and oxidative phosphorylation to fulfill the high anabolic demand. (29, 30) One mechanism of particular interest is the Warburg effect in which a state of hypoxia or pseudohypoxia induced by the tumor microenvironment and rapid growth reduces oxidative phosphorylation while increasing glucose metabolism. (31, 32) The tricarboxylic acid (TCA) cycle remains intact to support tumor cell growth and is thought to be rewired to provide the building blocks for cancer metabolism. (33, 34)
Glutamine is involved in a variety of energy-generating biosynthetic pathways including cancer metabolism which has generated special interest to utilize glutamine metabolism as a potential therapeutic target. (35) Glutamine is the most abundant amino acid in circulation and provides a readily accessible carbon and nitrogen source for cellular metabolism. Cancer cells can exploit glutamine for anaplerotic pathways by inducing glutaminolysis which converts glutamine to the byproduct α-ketoglutarate for entry to the TCA cycle. (36) Targeting glutamine metabolism has been shown to slow growth in soft tissue sarcomas. (37) Inhibition of glutamine synthetase induced antiproliferative effects in sarcoma cell lines (rhabdomyosarcoma, fibrosarcoma, osteosarcoma) including the liposarcoma derived cell line (SW872) In particular, liposarcoma cells were highly sensitive to the cytotoxic effects of antitumor enzyme L-asparaginase increasing glutamine synthetase levels and reducing cell proliferation. (38) However, the effects of glutamine metabolism are not well-elucidated in human liposarcoma. Results from the targeted transcriptome analysis revealed reduced glutamine synthesis in both the WDLPS and DDLPS samples when compared to the normal fat controls suggesting a tumor-induced metabolic shift from glutamine synthesis to glutaminolysis. Inhibitors of glutamine synthesis may be potential therapeutic options in the treatment of sarcomas that express high levels of glutaminase and are dependent on glutamine metabolism.
The HIF-1 signaling pathway is crucial for cancer cells to adapt to hypoxic stress, specifically intratumoral hypoxia and pseudohypoxia, and is considered a risk factor for poor prognosis in a variety of cancer types including retroperitoneal sarcomas (leiomyosarcoma, malignant peripheral nerve sheath tumor, WDLPS and DDLPS). HIF-1 is a heterodimer consisting of α and β subunits. (39) Downstream targets of HIF-1 include genes involved in angiogenesis (i.e. vascular endothelial growth factor, VEGF) (38)
Transforming growth factor β (TGF-β) has long been considered a multifactorial regulator of embryonic development and homeostasis by stimulating or inhibiting cell proliferation. Dysregulation of TGF-β has been implicated in metabolic disease promoting fibrosis and inflammation, and cancer. (40–42) In early tumorigenesis, TGF-β signaling is shifted toward tumor suppression. However, unregulated TGF-β expression in cancer cells overcomes the apoptotic/cell cycle control of TGF-β promoting tumor progression. The non-canonical pathway activates various pathways including PI3K, JNK, and ERK MAP kinases involved in transcriptional regulation. (43) The absence of key components of the TGF-β pathway suggest reduced invasive potential. (44, 45) Several genes involved in the TGF- β pathway were found to be elevated in DDLPS when compared to WDLPS suggesting a potential role in the progression of DDLPS. The therapeutic potential of TGF-β in liposarcoma was investigated to show that inhibition of TGF- β signaling reduced the growth and metastasis of liposarcoma cells in mice.
Activation of the TGF-β pathway can promote epithelial to mesenchymal transition (EMT) promoting a motile mesenchymal phenotype which is a characteristic of invasion and metastasis. (46, 47) Downstream targets of TGF-β such as SNAI1 mediate the EMT pathway in a SMAD-dependent manner. (48) Nine genes involved in the EMT pathway were upregulated in WDLPS when compared to DDLPS including proinflammatory cytokines/chemokines (IL-6, TLR4, CCL21, CCL2, PTGS2), cell proliferation (MYC), and angiogenesis (VEGFA, FLT1, and FLT4). The expression of SNAI1 was upregulated in WDLPS when compared to DDLPS. Although WDLPS demonstrates a predominantly adipose-like morphology in contrast to the high-grade spindle morphology of DDLPS, both WDLPS and DDLPS can show focal to extensive myxoid stroma, sclerosis, or inflammatory infiltrate. A study evaluating the degree of sclerosis in cases of retroperitoneal WDLPS showed that the degree of sclerosis and not myxoid or inflammation had the greatest impact on prognosis. Cases of minimally sclerotic WDLPS have a more favorable outcome when compared to cases with advanced sclerosis. (44) Although WDLPS is not considered to possess metastatic potential, these findings suggest a possible proinflammatory tumor environment within WDLPS and subsequent activation of the TGF-β signaling pathway (Figure 4G). Additional studies are warranted to understand the role of TGF-β in the pathogenesis of WDLPS.
In conclusion, our data suggests that WDLPS and DDLPS demonstrate diverse molecular and metabolic profiles. In an era of personalized medicine, some questions remain; what additional molecular aberrations account for the different behavior and prognosis of WDLPS and DDLPS, and how can these molecular pathways be targeted for therapeutic alternatives to surgical resection? Therapeutic agents have been developed and are currently in preclinical or clinical phases for pathways including glutamine metabolism and TGF-β signaling which have demonstrated efficacy, at least, in vivo in liposarcoma-derived cell lines. The future of targeted therapy for WDLPS is promising with the goal to spare multi-visceral removal as a consequence of radical surgical resection and prevention of dedifferentiation to DDLPS.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by The Ohio State Wexner Medical Center IRB#2022C0037. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because This was a study involving archived human tissue with minimal impact to the participants. The requirement of ethical approval was waived by The Ohio State Wexner Medical Center IRB#2022C0037 for the studies involving animals because patient consent was not deemed necessary for the study and was waived by the IRB. The studies were conducted in accordance with the local legislation and institutional requirements.
Author contributions
AP: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. NH: Data curation, Formal analysis, Writing – review & editing. PU: Formal analysis, Writing – review & editing, Supervision, Validation, Writing – original draft. PS: Writing – review & editing, Resources. RP: Resources, Writing – review & editing, Investigation. SO: Writing – review & editing, Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Supervision, Validation, Writing – original draft. OI: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing, Funding acquisition, Investigation, Resources, Visualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded in part by NIH grants R01DE033906 and DP1DA054344 awarded to SO, and a pilot grant from NanoString Inc., awarded to OI. The authors declare that this study received funding per the funding agencies listed in manuscript. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Acknowledgments
We would like to acknowledge the Ohio State University Comprehensive Cancer Center Genomics Shared Resource P30CA016058 and its laboratory supervisor, Paolo Fadda, Pharm.D., for performing the NanoString assays and protocols. Nanostring nCounter.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1456071/full#supplementary-material
Supplementary Table 1 | Sample information and collection criteria.
References
1. Henricks WH, Chu YC, Goldblum JR, Weiss SW. Dedifferentiated liposarcoma: A clinicopathological analysis of 155 cases with a proposal for an expanded definition of dedifferentiation. Am J Surg Pathol. (1997) 21:271–81. doi: 10.1097/00000478-199703000-00002
2. Weiss SW, Rao VK. Well-differentiated liposarcoma (Atypical lipoma) of deep soft tissue of the extremities, retroperitoneum, and miscellaneous sites. A follow-up study of 92 cases with analysis of the incidence of ‘Dedifferentiation. Am J Surg Pathol. (1992) 16:1051–58. doi: 10.1097/00000478-199211000-00003
3. Boland JM, Weiss SW, Oliveira AM, Erickson-Johnson ML, Folpe AL. Liposarcomas with mixed well-differentiated and pleomorphic features: A clinicopathologic study of 12 cases. Am J Surg Pathol. (2010) 34:837–435. doi: 10.1097/PAS.0b013e3181dbf2f7
4. Thway K. Well-differentiated liposarcoma and dedifferentiated liposarcoma: an updated review. Semin Diagn Pathol. (2019) 36:112–21. doi: 10.1053/j.semdp.2019.02.006
5. Gahvari Z, Parkes A. Dedifferentiated liposarcoma: systemic therapy options. Curr Treat Options Oncol. (2020) 21:155. doi: 10.1007/s11864-020-0705-7
6. Crago AM, Dickson MA. Liposarcoma. Surg Oncol Clinics North America. (2016) 25:761–35. doi: 10.1016/j.soc.2016.05.007
7. Lee ATJ, Thway K, Huang PH, Jones RL. Clinical and molecular spectrum of liposarcoma. J Clin Oncology: Off J Am Soc Clin Oncol. (2018) 36:151–595. doi: 10.1200/JCO.2017.74.9598
8. Mori T, Yamada Y, Kinoshita I, Kohashi K, Yamamoto H, Ito Y, et al. Clinicopathological and histopathological review of dedifferentiated liposarcoma: A comprehensive study of 123 primary tumours. Histopathology. (2022) 80:538–75. doi: 10.1111/his.14588
9. Cassinelli G, Pasquali S, Lanzi C. Beyond targeting amplified MDM2 and CDK4 in well differentiated and dedifferentiated liposarcomas: from promise and clinical applications towards identification of progression drivers. Front Oncol. (2022) 12:965261. doi: 10.3389/fonc.2022.965261
10. Assi T, Kattan J, Rassy E, Nassereddine H, Farhat F, Honore C, et al. Targeting CDK4 (Cyclin-dependent kinase) amplification in liposarcoma: A comprehensive review. Crit Rev Oncology/Hematology. (2020) 153:103029. doi: 10.1016/j.critrevonc.2020.103029
11. Marino-Enriquez A. Advances in the molecular analysis of soft tissue tumors and clinical implications. Surg Pathol Clinics. (2015) 8:525–37. doi: 10.1016/j.path.2015.06.001
12. Sirvent N, Coindre J-M, Maire G, Hostein I, Keslair F, Guillou L, et al. Detection of MDM2-CDK4 amplification by fluorescence in situ hybridization in 200 paraffin-embedded tumor samples: utility in diagnosing adipocytic lesions and comparison with immunohistochemistry and real-time PCR. Am J Surg Pathol. (2007) 31:1476–895. doi: 10.1097/PAS.0b013e3180581fff
13. Traweek RS, Cope BM, Roland CL, Keung EZ, Nassif EF, Erstad DJ. Targeting the MDM2-P53 pathway in dedifferentiated liposarcoma. Front Oncol. (2022) 12:1006959. doi: 10.3389/fonc.2022.1006959
14. Tripathy D, Bardia A, Sellers WR. Ribociclib (LEE011): mechanism of action and clinical impact of this selective cyclin-dependent kinase 4/6 inhibitor in various solid tumors. Clin Cancer Research: Off J Am Assoc Cancer Res. (2017) 23:3251–625. doi: 10.1158/1078-0432.CCR-16-3157
15. Yang L, Wu Z, Sun W, Luo P, Chen S, Chen Y, et al. CCNDBP1, a prognostic marker regulated by DNA methylation, inhibits aggressive behavior in dedifferentiated liposarcoma via repressing epithelial mesenchymal transition. Front Oncol. (2021) 11:687012. doi: 10.3389/fonc.2021.687012
16. De Vita A, Mercatali L, Recine F, Pieri F, Riva N, Bongiovanni A, et al. Current classification, treatment options, and new perspectives in the management of adipocytic sarcomas. OncoTargets Ther. (2016) 9:6233–46. doi: 10.2147/OTT.S112580
17. Dickson MA, Schwartz GK, Keohan ML, D’Angelo SP, Gounder MM, Chi P, et al. Progression-free survival among patients with well-differentiated or dedifferentiated liposarcoma treated with CDK4 inhibitor palbociclib: A phase 2 clinical trial. JAMA Oncol. (2016) 2:937. doi: 10.1001/jamaoncol.2016.0264
18. Gootee JM, Curtin CE, Aurit SJ, Randhawa SE, Kang BY, Silberstein PT. Treatment facility: an important prognostic factor for dedifferentiated liposarcoma survival. Federal Practitioner: For Health Care Professionals VA DoD PHS. (2019) 36:S34–41.
19. Núñez KG, Sandow T, Lakey MA, Fort D, Cohen AJ, Thevenot PT. Distinct gene expression profiles in viable hepatocellular carcinoma treated with liver-directed therapy. Front Oncol. (2022) 12:809860. doi: 10.3389/fonc.2022.809860
20. Amaddeo G, Nguyen CT, Maillé P, Mulé S, Luciani A, Machou C, et al. Intrahepatic Immune Changes after Hepatitis c Virus Eradication by Direct-Acting Antiviral Therapy. Liver International: Off J Int Assoc Study Liver. (2020) 40:74–82. doi: 10.1111/liv.14226
21. Cheng Y, Zhang X, Wang Z, Wang J. Reconstruction of immune microenvironment and signaling pathways in endometrioid endometrial adenocarcinoma during formation of lymphovascular space involvement and lymph node metastasis. Front Oncol. (2020) 10:595082. doi: 10.3389/fonc.2020.595082
22. Chang KTE, Goytain A, Tucker T, Karsan A, Lee C-H, Nielsen TO, et al. Development and evaluation of a pan-sarcoma fusion gene detection assay using the nanoString nCounter platform. J Mol Diagnostics: JMD. (2018) 20:63–775. doi: 10.1016/j.jmoldx.2017.09.007
23. Wang XQ, Wang XQ, Hsu ATYW, Goytain A, Ng TLT, Nielsen TO. A rapid and cost-effective gene expression assay for the diagnosis of well-differentiated and dedifferentiated liposarcomas. J Mol Diagnostics: JMD. (2021) 23:274–845. doi: 10.1016/j.jmoldx.2020.11.011
24. Jirovec A, Flaman A, Godbout E, Serrano D, Werier J, Purgina B, et al. Immune profiling of dedifferentiated liposarcoma and identification of novel antigens for targeted immunotherapy. Sci Rep. (2024) 14:112545. doi: 10.1038/s41598-024-61860-3
25. Schmidt H, Bartel F, Kappler M, Würl P, Lange H, Bache M, et al. Gains of 13q are correlated with a poor prognosis in liposarcoma. Modern Pathology: Off J United States Can Acad Pathology Inc. (2005) 18:638–445. doi: 10.1038/modpathol.3800326
26. Haluska FG, Huebner K, Isobe M, Nishimura T, Croce CM, Vogt PK. Localization of the human JUN protooncogene to chromosome region 1p31-32. Proc Natl Acad Sci United States America. (1988) 85:2215–18. doi: 10.1073/pnas.85.7.2215
27. Coindre J-M, Mariani O, Chibon F, Mairal A, De Saint Aubain Somerhausen N, Favre-Guillevin E, et al. Most Malignant fibrous histiocytomas developed in the retroperitoneum are dedifferentiated liposarcomas: A review of 25 cases initially diagnosed as Malignant fibrous histiocytoma. Modern Pathology: Off J United States Can Acad Pathology Inc. (2003) 16:256–625. doi: 10.1097/01.MP.0000056983.78547.77
28. Snyder EL, Sandstrom DJ, Law K, Fiore C, Sicinska E, Brito J, et al. C-jun amplification and overexpression are oncogenic in liposarcoma but not always sufficient to inhibit the adipocytic differentiation programme. J Pathol. (2009) 218:292–300. doi: 10.1002/path.2564
29. Bose S, Le A. Glucose metabolism in cancer. Adv Exp Med Biol. (2018) 1063:3–12. doi: 10.1007/978-3-319-77736-8_1
30. Lin X, Xiao Z, Chen T, Liang SH, Guo H. Glucose metabolism on tumor plasticity, diagnosis, and treatment. Front Oncol. (2020) 10:317. doi: 10.3389/fonc.2020.00317
31. Garber K. Energy boost: the warburg effect returns in a new theory of cancer. J Natl Cancer Institute. (2004) 96:1805–6. doi: 10.1093/jnci/96.24.1805
32. Koppenol WH, Bounds PL, Dang CV. Otto warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. (2011) 11:325–75. doi: 10.1038/nrc3038
33. Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci. (2016) 73:377–925. doi: 10.1007/s00018-015-2070-4
34. Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. (2017) 551:115–18. doi: 10.1038/nature24057
35. Altman BJ, Stine ZE, Dang CV. From krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. (2016) 16:619–345. doi: 10.1038/nrc.2016.71
36. Yang W-H, Qiu Y, Stamatatos O, Janowitz T, Lukey MJ. Enhancing the efficacy of glutamine metabolism inhibitors in cancer therapy. Trends Cancer. (2021) 7:790–8045. doi: 10.1016/j.trecan.2021.04.003
37. Lee P, Malik D, Perkons N, Huangyang P, Khare S, Rhoades S, et al. Targeting glutamine metabolism slows soft tissue sarcoma growth. Nat Commun. (2020) 11:498. doi: 10.1038/s41467-020-14374-1
38. Tardito S, Uggeri J, Bozzetti C, Bianchi MG, Rotoli BM, Franchi-Gazzola R, et al. The inhibition of glutamine synthetase sensitizes human sarcoma cells to L-asparaginase. Cancer Chemotherapy Pharmacol. (2007) 60:751–58. doi: 10.1007/s00280-007-0421-z
39. Rashid M, Zadeh LR, Baradaran B, Molavi O, Ghesmati Z, Sabzichi M, et al. Up-down regulation of HIF-1α in cancer progression. Gene. (2021) 798:145796. doi: 10.1016/j.gene.2021.145796
40. Grafe I, Alexander S, Peterson JR, Snider TN, Levi B, Lee B, et al. TGF-β Family signaling in mesenchymal differentiation. Cold Spring Harbor Perspect Biol. (2018) 10:a022202. doi: 10.1101/cshperspect.a022202
41. Neuzillet C, Tijeras-Raballand A, Cohen R, Cros J, Faivre S, Raymond E, et al. Targeting the TGFβ Pathway for cancer therapy. Pharmacol Ther. (2015) 147:22–31. doi: 10.1016/j.pharmthera.2014.11.001
42. Peng D, Fu M, Wang M, Wei Y, Wei X. Targeting TGF-β Signal transduction for fibrosis and cancer therapy. Mol Cancer. (2022) 21:1045. doi: 10.1186/s12943-022-01569-x
43. Li S-N, Wu J-F. TGF-β/SMAD signaling regulation of mesenchymal stem cells in adipocyte commitment. Stem Cell Res Ther. (2020) 11:415. doi: 10.1186/s13287-020-1552-y
44. Dehner CA, Moon T, Lyu Y, Zhang X, Zhou Z, Yang K, et al. Mutations involving TGFB and MAPK may be associated with Malignancy in granular cell tumors. Genes Chromosomes Cancer. (2023) 62:301–7. doi: 10.1002/gcc.23123
45. Valkov A, Sorbye SW, Kilvaer TK, Donnem T, Smeland E, Bremnes RM, et al. The prognostic impact of TGF-Β1, fascin, NF-κB and PKC-ζ Expression in soft tissue sarcomas. PloS One. (2011) 6:e175075. doi: 10.1371/journal.pone.0017507
46. Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. (2009) 119:1429–375. doi: 10.1172/JCI36183
47. Lin Y-T, Wu K-J. Epigenetic regulation of epithelial-mesenchymal transition: focusing on hypoxia and TGF-β Signaling. J Biomed Sci. (2020) 27:395. doi: 10.1186/s12929-020-00632-3
Keywords: dedifferentiated liposarcoma, well-differentiated liposarcoma, NanoString, TGF-β, transcriptome, glucose metabolism, HIF, signaling pathways
Citation: Patton A, Horn N, Upadhaya P, Sarchet P, Pollock RE, Oghumu S and Iwenofu OH (2024) Targeted transcriptomic analysis of well-differentiated and dedifferentiated liposarcoma reveals multiple dysregulated pathways including glucose metabolism, TGF-β, and HIF-1 signaling. Front. Oncol. 14:1456071. doi: 10.3389/fonc.2024.1456071
Received: 27 June 2024; Accepted: 16 October 2024;
Published: 26 November 2024.
Edited by:
Joanna Kitlinska, Georgetown University, United StatesReviewed by:
Alessandro De Vita, Scientific Institute of Romagna for the Study and Treatment of Tumors (IRCCS), ItalyToru Wakamatsu, Osaka International Cancer Institute, Japan
Copyright © 2024 Patton, Horn, Upadhaya, Sarchet, Pollock, Oghumu and Iwenofu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steve Oghumu, U3RldmUuT2dodW11QG9zdW1jLmVkdQ==; Obiajulu Hans Iwenofu, SGFucy5Jd2Vub2Z1QG9zdW1jLmVkdQ==
 Ashley Patton1
Ashley Patton1 Raphael E. Pollock
Raphael E. Pollock Steve Oghumu
Steve Oghumu Obiajulu Hans Iwenofu
Obiajulu Hans Iwenofu