- 1Department of Thoracic Cardiovascular Surgery, The Eighth Affiliated Hospital of Sun Yat−sen University, Shenzhen, Guangdong, China
- 2Community Health Center, The Eighth Affiliated Hospital of Sun Yat-sen University, Shenzhen, Guangdong, China
- 3Hangzhou Institute of Medicine (HIM), Chinese Academy of Sciences, Hangzhou, Zhejiang, China
- 4BGI Genomics, BGI-Shenzhen, Shenzhen, China
- 5Beijing Etown Academy, Beijing, China
Introduction: Adenocarcinoma in situ (AIS) and minimally invasive adenocarcinoma (MIA) are considered pre-invasive forms of lung adenocarcinoma (LUAD) with a 5-year recurrence-free survival of 100%. We investigated genomic profiles in early tumorigenesis and distinguished mutational features of preinvasive to invasive adenocarcinoma (IAC) for early diagnosis.
Methods: Molecular information was obtained from a 689-gene panel in the 90 early-stage LUAD Chinese patients using next-generation sequencing. Gene signatures were identified between pathology subtypes, including AIS/MIA (n=31) and IAC (n=59) in this cohort. Mutational and clinicopathological information was also obtained from the Cancer Genome Atlas (TCGA) as a comparison cohort.
Results: A higher mutation frequency of TP53, RBM10, MUC1, CSMD, MED1, LRP1B, GLI1, MAP3K, and RYR2 was observed in the IAC than in the AIS/MIA group. The AIS/MIA group showed higher mutation frequencies of ERBB2, BRAF, GRIN2A, and RB1. Comparable mutation rates for mutually exclusive genes (EGFR and KRAS) across cohorts highlight the critical transition to invasive LUAD. Compared with the TCGA cohort, EGFR, KRAS, TP53, and RBM10 were frequently mutated in both cohorts. Despite limited gene mutation overlap between cohorts, we observed variant mutation types in invasive LUAD. Additionally, the tumor mutation burden (TMB) values were significantly lower in the AIS/MIA group than in the IAC group in both the Chinese cohort (P=0.0053) and TCGA cohort (P<0.01).
Conclusion: These findings highlight the importance of distinguishing preinvasive from invasive LUAD in the early stages of LUAD and both pathology and molecular features in clinical practice, revealing genomic tumor heterogeneity and population differences.
1 Introduction
Lung cancer remains the leading cause of cancer-related deaths and ranks second in newly diagnosed carcinoma worldwide, with 2.2 million new cases and 1.8 million new deaths per year (1). Non-small-cell lung cancer (NSCLC) accounts for 80%~85% of all lung cancers, and it is composed of squamous-cell carcinoma, adenocarcinoma, and large-cell carcinoma (1). LUAD is a major subtype of lung cancer. Lung adenocarcinoma (LUAD) accounts for approximately 40% of all lung carcinomas (1). If diagnosed and treated in the early stages, the survival rate for lung cancer participants who undergo complete surgical resection within 1 month after diagnosis can reach 92% (2). Furthermore, the overall survival (OS) rate varies among different stages of primary lung cancer; stage IA (tumor size < 1 cm) has a 5-year OS rate of 82%, whereas stages beyond IIB have a 5-year OS rates of less than 50% (3).
Most cases of early-stage lung cancer are treated with surgery alone, whereas combination therapy may be used in other scenarios (4). The 2015 World Health Organization (WHO) classification of lung tumors introduces new concepts for LUAD, such as adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA) (5); Invasive adenocarcinoma (IAC) also recommended by the WHO to be classified by comprehensive histologic pattern, which includes five predominant adenocarcinoma patterns: lepidic, acinar, papillary, solid, and micropapillary (5). Others include invasive mucinous adenocarcinoma, colloid adenocarcinoma, fetal adenocarcinoma, and enteric adenocarcinoma (5). The recurrence-free survival (RFS) rate varies among different pathology subtypes, with AIS and MIA having a 100% RFS rate (6). There is a 40% recurrence rate in NSCLC cases with all pathology subtypes after complete primary tumor resection and systematic lymph node dissection within 5 years (7). Clinical judgment and treatment therapy for early-stage lung cancer is crucial in clinical practice. Early diagnosis of pathology subtypes, along with an understanding of the genetic landscape, can help to prolong the survival of lung cancer patients.
In recent years, mutation profiles of NSCLC have been clarified, with frequent mutations identified in driver genes such as epidermal growth factor receptor (EGFR), and erb-b2 receptor tyrosine kinase 2 (ERBB2) (8, 9). Various treatment options are available for patients with NSCLC, including targeted therapy, immunotherapy, chemoradiotherapy (CRT), and complete resection (4, 10). However, the crucial predictive biomarkers for early-stage NSCLC remain largely unknown and require validation and reproducibility (11). Genomic intratumor heterogeneity has been identified in early-stage LUAD, and different pathology subtypes, including AIS (12). In preinvasive and invasive LUAD, EGFR mutation is the most common driver alterations across AIS, MIA, and IAC (13). In addition, other canonical cancer gene mutations such as ERBB2, NRAS proto-oncogene, GTPase (NRAS), and B-Raf proto-oncogene (BRAF) are early trunk mutations during the carcinogenesis of LUAD (13, 14). Meanwhile, mutations in the tumor protein p53 gene (TP53) and cell mobility, gap junction, and metastasis-related genes may be late events associated with subclonal diversification and neoplastic progression (14).
Targeted exome sequencing has been widely used for mutation detection in cancer therapy (15). In one retrospective study, four gene mutations were identified, including MAP2K1 insertion-deletions (indels), BRAF non-V600E kinase mutations, and exon 20 insertions (20ins) in both EGFR and ERBB2, which were enriched in pre-invasive tumors among 3,254 Chinese patients with LUAD (16). The dataset from TCGA has also been used for comparison studies or validation of the gene signature results (17). Despite these observations, genomic characteristics and ethnic differences in the WHO 2015 pathology classifications of LUAD genomics have yet to be systematically elucidated because of the lack of a sufficiently large cohort.
To address the challenges mentioned above, our study aimed to identify the unique mutational features that distinguish preinvasive from invasive LUAD in both our Chinese cohort and the TCGA cohort. To achieve this, we collected epidemiological and clinicopathological information from early-stage LUAD patients with AIS/MIA or IAC from both our Chinese cohort and the TCGA cohort. In addition, surgically resected specimens were subjected to targeted exome sequencing to reveal the molecular status differences between AIS/MIA and IAC.
2 Materials and methods
2.1 Patients and sample collection
The resected tissue samples were collected from 90 primary LUAD patients who underwent resection surgery from July 2020 to December 2021. Gene mutation data was obtained using a capture-based targeted sequencing approach with a 689-gene panel. Relevant epidemiological and clinicopathologic information records were also obtained. The public dataset of TCGA was derived from the MSK study with histologic data in cBioportal for Cancer Genomics (18). Only patients data from stages I and II were included and the clinical data was selected following the 2015 WHO criteria (5).
2.2 DNA Extraction, library preparation, and target capture sequencing
Genomic DNA (50-200 ng) was extracted from formalin-fixed paraffin-embedded (FFPE) samples using QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). The extracted tissue DNA samples were qualified and used to construct cDNA libraries (Integrated DNA Technologies, Coralville, IA, USA). Custom-designed 689-gene probe panels (Integrated DNA Technologies, Coralville, IA, USA) were used to capture their respective, as listed in Supplementary Table S1. To eliminate germline mutations, a control library was created using DNA extracted from peripheral blood samples. The cDNA libraries were sequenced using the MGISEQ-2000 platform (MGI, Shenzhen, China) by the manufacturer’s recommendations. The workflow for potentially actionable variants was introduced following a previous pipeline (19).
2.3 Sequencing data analysis and immunohistochemistry
The raw fastq data generated by the MGISEQ-2000 sequencer underwent filtering by SOAPnuke (RRID : SCR_015025, https://scicrunch.org/resolver/SCR_015025) to remove reads with low quality. The reference human genome used was GRCh37/hg19, and clean reads were mapped to this genome using the UCSC genome browser (RRID : SCR_005780, https://scicrunch.org/resolver/SCR_005780). Single nucleotide variants (SNVs) and small insertions/deletions (Ins/Del) were identified using the Genome Analysis Tool kit (GATK) with parameters adapted to HaloPlex-generated sequences. Copy number variants (CNVs) were called using the CNVnator read-depth algorithm. Tumor mutation burden (TMB) was assessed through targeted sequencing of approximately 1.25Mb, which broadly recapitulated previous results of whole exome TMB analysis. Tumor mutation burden was calculated as the number of all nonsynonymous mutations per 0.7 Mb of the targeted coding region. TMB was obtained by calculating the number of mutations (allele frequency > 1.5%) in non-driver genes per Mb in each sample. MSIsensor and MANTIS were used to detect the status of microsatellite instability (MSI) (20, 21).
Programmed death ligand 1 (PD-L1) expression was conducted by immunohistochemistry in the central laboratory. The PD-L1 expression was determined by the Tumor Proportion Score (TPS) method with the IHC 22C3 pharmDx kit (Agilent Technologies, Santa Clara, California, USA).
2.4 Statistical analysis
Visualization and statistical analyses were performed using R (v4.1.0) and prism graphpad (RRID : SCR_000306, https://scicrunch.org/resolver/SCR_000306). All distributed data were presented as mean ± standard deviation. Chi-square test was employed to analyze the differences between AIS/MIA and IAC groups. P < 0.05 is regarded as statistically significant. GO and KEGG were performed using Database for Annotation, Visualization, and Integrated Discovery v6.8 (DAVID, https://david.ncifcrf.gov/) (22). DAVID v6.8 (https://david.ncifcrf.gov/tools.jsp), an online set of functional annotation tools, was used to analyze biological processes, cellular components, molecular functions, and pathways for DEGs. GO terms and KEGG pathways (RRID : SCR_012773, https://scicrunch.org/resolver/SCR_012773) with P value <0.05 were considered statistically significant.
3 Results
3.1 Clinical characteristics and mutation spectrum of LUAD patients
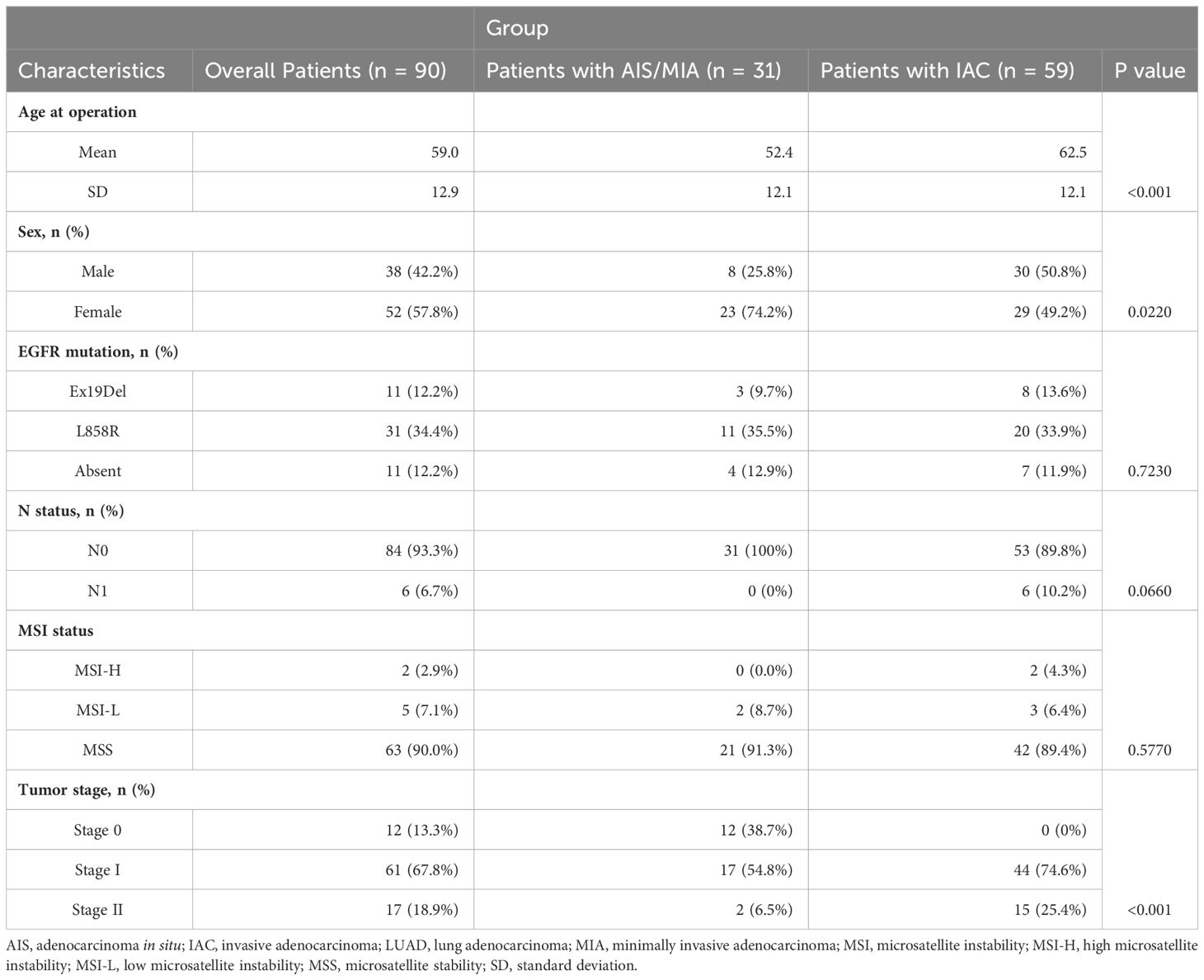
Clinical data were employed for 90 primary LUAD patients. The mean age was 59 years (standard deviation [SD] = 12.9 years) for this cohort, with 42.2% (38/90) male and 58.7% (52/90) female. 13.3% (12/90) were adenocarcinoma in stage 0, Others were 67.8% (61/90) in stage I, and 18.9% (17/90) in stage II (Supplementary Table S2). The detailed clinical characteristics of the patients are shown in Table 1 and Supplementary Table S2. In brief, 12.2% (11/90) had Ex19Del mutant EGFR, 34.4% (31/90) were L858R mutant EGFR, others 12.2% (11/90) were absent of these two classical EGFR activating mutations. The whole LUAD patients were also divided into subgroups of AIS, MIA, and IAC. The age of the AIS/MIA group was significantly younger than the IAC group (52.4 versus 62.5 years, P < 0.001). Besides, the composition of sex and tumor stage were also significantly different between AIS/MIA and IAC group patients. The AIS/MIA group had more females and more cancers in the early stages. Other factors such as N stage, MSI, and EGFR mutation status (Ex19Del, L858R, Absent) had no difference between the AIS/MIA and IAC groups (Table 1).
A panel of 689 cancer-related genes was applied to 90 primary lung tissue samples. Mutation genes were also revealed in this cohort (Supplementary Table S3). All 90 patients had at least one potentially actionable variant. Full details of variants for all patients are available in Supplementary Table S3. In summary, gene landscape was different between AIS/MIA and IAC groups (Figure 1A, Supplementary Figures S1A, B). The most frequently mutated genes with various pathology subtypes (AIS/MIA/IAC) are displayed. The mutation frequencies of TP53 (31% versus 6%), RNA binding motif protein 10 (RBM10, 24% versus 6%), mucin-16 (MUC16, 14% versus 0%), CUB and sushi multiple domains 3 (CSMD3, 8% versus 3%), mediator complex subunit 12 (MED12, 8% versus 6%), low-density lipoprotein receptor-related protein 1b (LRP1B, 8% versus 3%), GLI family zinc finger 1 (GLI1, 8% versus 0%), mitogen-activated protein kinase kinase 4 (MAP3K4, 7% versus 3%), and ryanodine receptor 2 (RYR2, 7% versus 3%) were higher in the IAC group than in the AIS/MIA group. The AIS/MIA group showed higher mutation frequencies of ERBB2 (16% versus 3%), BRAF (10% versus 3%), glutamate ionotropic receptor NMDA type subunit 2A (GRIN2A, 6% versus 2%), and RB transcriptional corepressor 1 (RB1, 6% versus 0%) than the IAC groups (Figure 1A, Supplementary Figures S1A, B). The mutation frequencies were similar between AIS/MIA and IAC groups for the two top driver genes of EGFR (55% versus 54%) and KRAS proto-oncogene, GTPase, (KRAS, 10% versus 10%) (Figure 1A, Supplementary Figures S1A, B). The data suggest that ERBB2, BRAF, GRIN2A, RB1 were early-stage events for LUAD. The late events were mutations for TP53, RBM10, MUC1, CSMD3, MED1, LRP1B, GLI1, RYR2 mutations. Besides, EGFR and KRAS mutations accompanied all the events. In addition, by analyzing the mutation types of 15 high-frequent mutated genes between AIS/MIA and IAC (Figure 1B), we identified that the mutation type of EGFR varied in IAC, with genes such as RYR2, GLI1, LRP1B, MED12, CSMD3, MUC16, RBM10, TP53 having higher mutation frequency and more mutation types, such as structure change for genes in invasive LUAD.
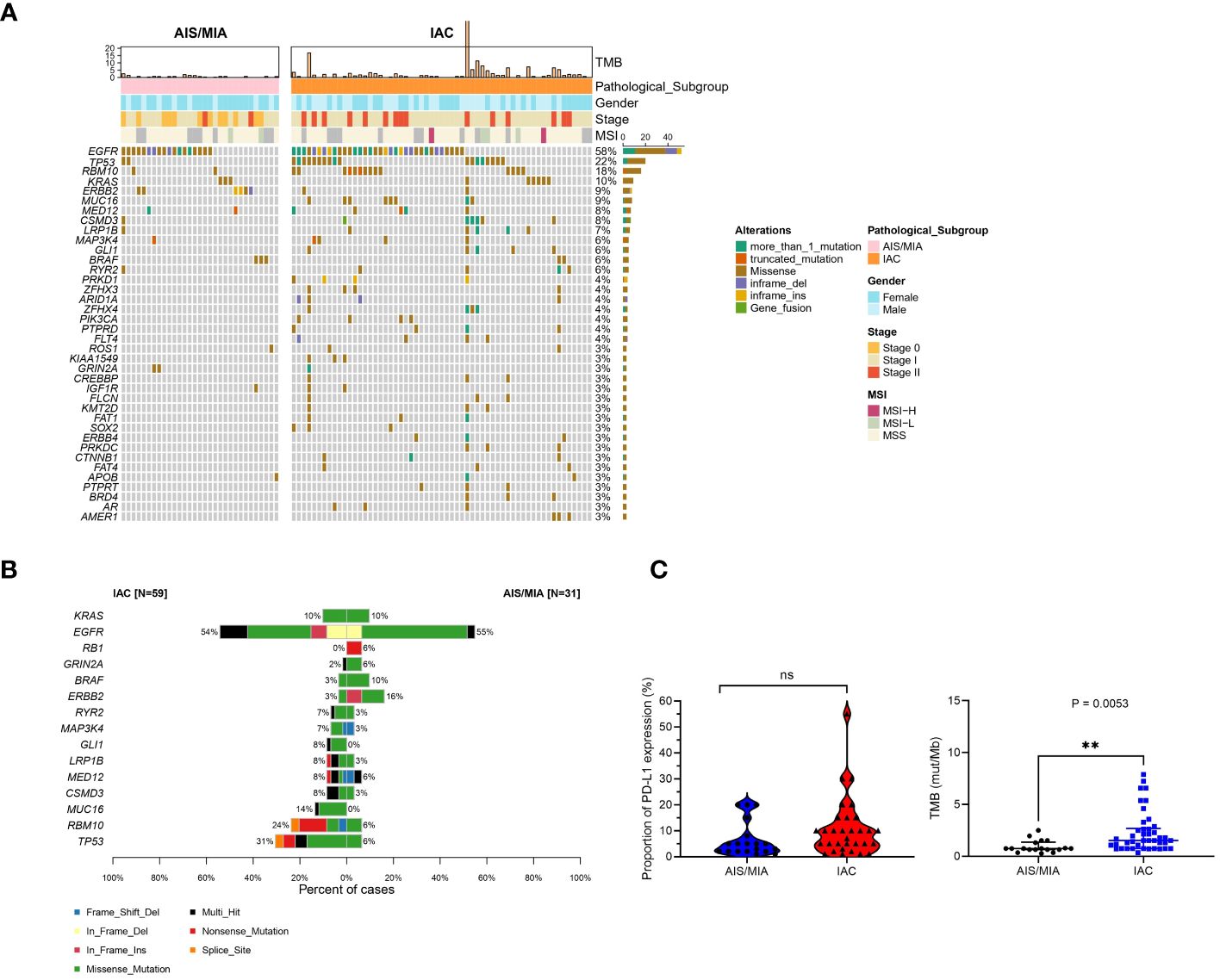
Figure 1 Distinct mutation spectrum of preinvasive and invasive LUAD patients. (A) Waterfall plot with top 38 genes for LUAD patients in AIS/MIA and IAC group; (B) The mutation types of 15 high-frequent mutated genes between AIS/MIA and IAC; (C) Proportion of PD-L1 expression level in AIS/MIA and IAC group (left), Proportion of TMB values in AIS/MIA and IAC group (right). AIS: adenocarcinoma in situ, MIA: minimally invasive adenocarcinoma, IAC: Invasive adenocarcinoma, MSI: microsatellite instability, TMB: tumor mutation burden, ns, not significant, **: P < 0.01.
3.2 Identification of core genes in tumors originating from the lung
In Figure 1C, tumor mutation burden (TMB) values of the AIS/MIA group were significantly lower than those of the IAC group (3.92 versus 0.94 mut/Mb, P = 0.0053). The proportion of PD-L1 expression in the IAC group was slightly higher than that in the AIS/MIA group, but this difference was not significant. Furthermore, as shown in Figure 2A, these two groups shared 26 genes in common, including EGFR, ERBB2, MED12, TP53, insulin-like growth factor 1 receptor (IGF1R), mutS homolog 5 (MSH5), BRAF, CSMD3, RYR2, estrogen receptor 1 (ESR1), KRAS, LRP1B, notch receptor 4 (NOTCH4), IKAROS family zinc finger 1 (IKZF1), laminin subunit alpha 2 (LAMA2), RBM10, KIT proto-oncogene, receptor tyrosine kinase (KIT), fibroblast growth factor receptor 4 (FGFR4), ROS proto-oncogene 1, receptor tyrosine kinase (ROS1), MAP3K4, GRIN2A, phosphatase and tensin homolog (PTEN), adhesion G protein-coupled receptor A2 (ADGRA2), apolipoprotein B (APOB), XPC complex subunit, DNA damage recognition and repair factor (XPC), neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 (NYAP2). In addition, EGFR, ERBB2, MED12, TP53, BRAF, and KRAS are six genes shared across AIS/MIA/IAC groups (Supplementary Figure S2A), all of which play an important role during tumorigenesis. There were 15 genes unique to MIA/AIS rather than AIS. More detailed information can be found in Supplementary Table S3.
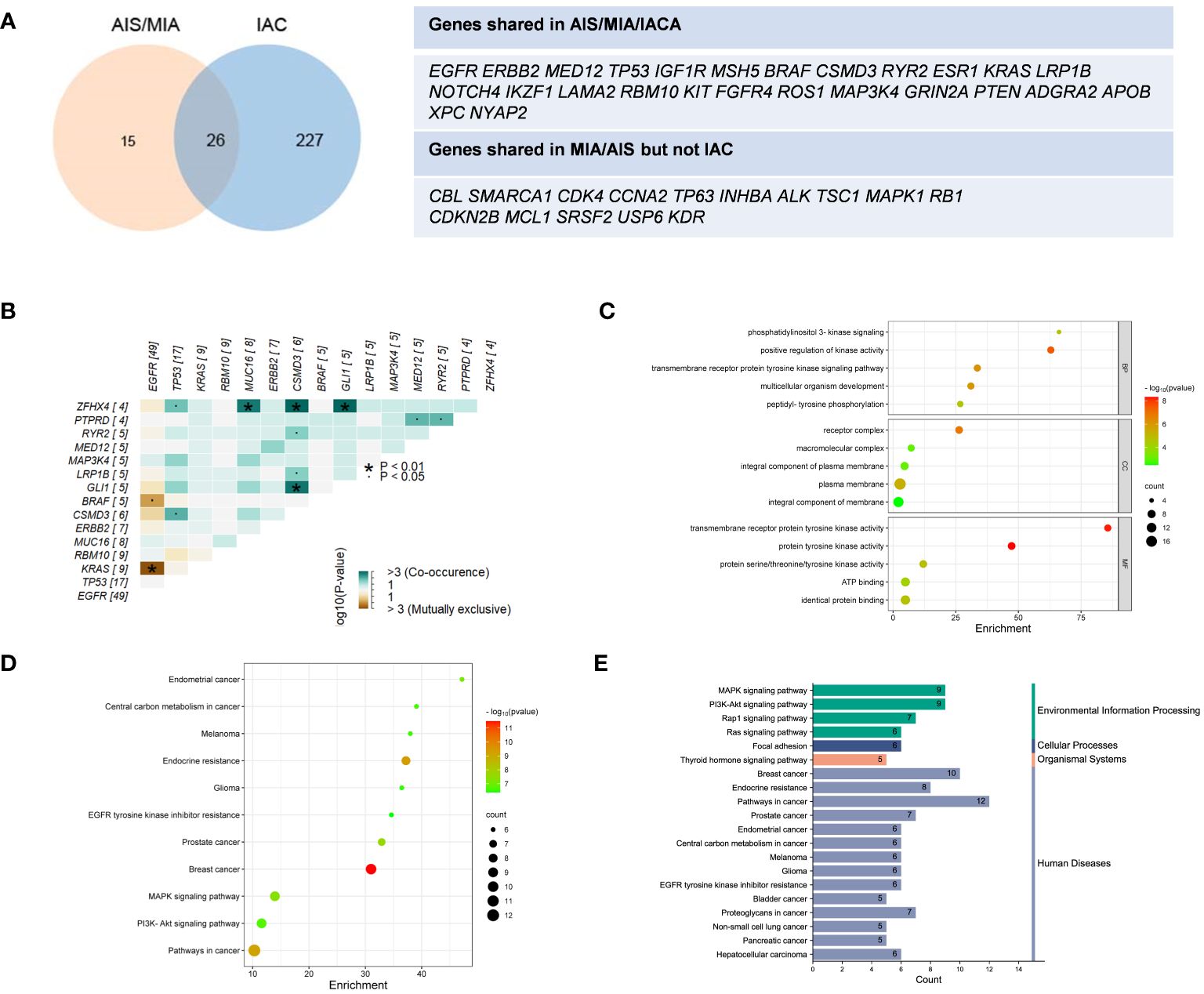
Figure 2 Distinct distribution of genes and pathway analysis. (A) Shared genes between IAS/MIA and IAC groups; (B) Gene co-occurrence figure for the top 15 frequently mutated genes; (C–E) The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment for the shared genes between IAS/MIA and IAC groups. ▪: P < 0.05. *: P < 0.01.
According to the gene co-occurrence figure (Figure 2B), there was a significant exclusivity between mutations in KRAS (P < 0.01), BRAF (P < 0.05), and EGFR. Additionally, CSMD3 was found to co-occur with TP53, ZFHX4, RYR2, LRP1B and GLI1. Previous studies have shown that high expression of CSMD3 is associated with poor outcomes (23). In our results, we found that zinc finger homeobox 4 (ZFHX4) co-occurs with TP53, MUC16, CSMD3, and GL1. It is worth noting that ZFHX4 was found to be upregulated in lung adenocarcinoma (LUAD) and was associated with a poor prognosis, as indicated by previous research (24).
In the analysis of core genes and related signaling pathways, we adopted the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (25). A total of 20 important pathways in the lung cancer population were grouped (Figures 2C–E). The genes shared between preinvasive and invasive LUAD play important roles in the cellular component of the receptor complex, positive regulation of kinase activity for biological processes, and transmembrane receptor protein kinase activity function. Moreover, these genes function in focal adhesion and pathways in cancer, which are crucial for neoplastic progression. Thus, membrane-related kinase activity could be an early event from preinvasive to invasive LUAD. The differentially mutated genes suggest a better understanding of early lung cancer progression.
3.3 Clinical implications and comparison with the TCGA cohort
To further compare the clinical significance in multiple dimensions, we used the public database from TCGA. Full details of variants for all patients are available in Supplementary Table S4. From our Chinese cohort and TCGA cohort, we identified that our Chinese group contained 34% (31/90) AIS/MIA patients, compared with 12% (64/524) in the TCGA cohort. In our Chinese cohort, 66% (59/90) of the patients had IAC, whereas a higher percentage of patients (88%, 460/524) had IAC in the TCGA cohort (Figure 3A). Moreover, the tumor stages we analyzed were also different between our cohort and the TCGA cohort. The percentages were 13% (12/90), 68% (61/90), 19% (17/90) for Stage 0/I/II in our cohort, compared with 0% (0/524), 82% (432/524), 18% (92/524) in the TCGA group (Figure 3A). TMB values were also analyzed for the Chinese group and the TCGA group (Figure 3B, up panel). In the TCGA cohort, the TMB values were significantly higher in the IAC group than in the MIA group (7.625 versus 4.450, P < 0.01). Additionally, it seems that our group had a lower TMB of the AIS/MIA group than the TCGA (0.939 versus 4.450, P < 0.0001). This might be because our group had 13% Stage 0 and we had 12 AIS patients, while the TCGA group was in higher stages with MIA.
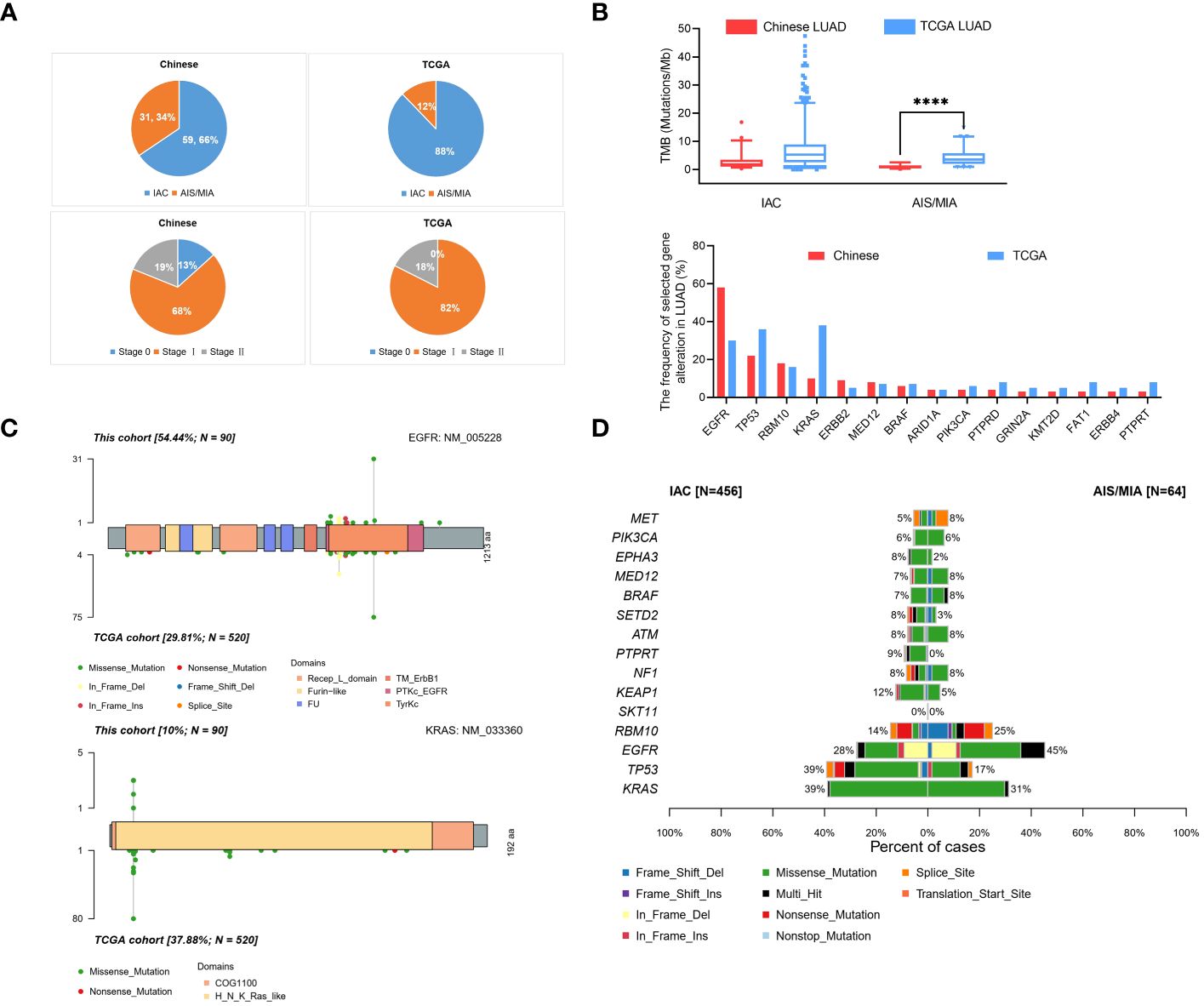
Figure 3 Comparison of data in Chinese LUAD and the TCGA LUAD cohorts. (A) Pathology and tumor stage status in our cohort with Chinese patients and TCGA cohort; (B) Proportion of TMB values in AIS/MIA and IAC group in our group and TCGA group (up panel), gene alternations in our group and TCGA group (lower panel); (C) Distribution and comparison of deleterious mutations detected in EGFR gene (up panel) and KRAS gene (lower panel) both in Chinese group and TCGA group; (D) The comparative bar plot of the top 15 frequently mutated genes in the TCGA cohort. ****: P < 0.0001.
By analyzing the mutation spectrum of the top 35 genes in our Chinese cohort and the TCGA cohort, we identified that different genes varied significantly under different circumstances (Supplementary Figure S2B and Figure 3B, lower panel). We also compared two driver genes, EGFR and KRAS, in the whole population. The variation frequency of EGFR was 54.44% in our cohort, compared with 29.81% in the TCGA cohort (Figure 3C, up panel). Additionally, the variation frequency of KRAS was 10% in our cohort, compared with 37.88% in the TCGA cohort (Figure 3C, lower panel). In Figure 3D, the comparative bar plot shows the 10 mutation types of the top 15 genes in TCGA cohort. Although the overlapped genes between our Chinese cohort and TCGA cohort varied, the mutation types increased in invasive LUAD in the TCGA cohort.
4 Discussion
We profiled this functional study on 90 early-stage LUAD patients, combining the AIS and MIA groups due to their high 5-year RFS of 100% (6). This RFS was higher than NSCLC cases with all pathology subtypes, which experienced a recurrence rate of 40% within 5 years (7). We analyzed gene mutational profiles using capture-based targeted sequencing and compared the most frequently mutated genes within AIS/MIA and IAC groups. Our results showed that the gene landscape was different between the two groups. The mutation frequency of TP53, RBM10, MUC16, CSMD3, MED12, LRP1B, GLI1, MAP3K4, RYR2 was higher in the IAC group than in the AIS/MIA group. Additionally, during the follow-up, there were only two recurrence events in the IAC group (2/25, 8%) and no events in the preinvasive group (0/9, 0%), indicating a difference in recurrence rates between preinvasive and invasive LUAD groups. Although the difference was not significant, this may be due to the short follow-up time of less than 3 years, which was too short for early-stage LUAD.
Genomic intratumor heterogeneity has been identified in early-stage LUAD with different pathology subtypes, including AIS (12, 13). Mutations including MAP2K1, BRAF, EGFR, and ERBB2, were identified as enriched in pre-invasive tumors (16). Additionally, ERBB2, NRAS, and BRAF are considered to be early trunk mutations during LUAD carcinogenesis (13, 14). Our study also identified that ERBB2 and BRAF were early-stage events for LUAD with higher mutation frequency in the preinvasive group. ERBB2 is a proto-oncogene, and it’s alternation is associated with poor survival and worse outcomes (26). Our results also showed that mutations in BRAF (P < 0.05) with EGFR were significantly mutually exclusive. This has also been recently confirmed in another Chinese cohort (27). In our study, GRIN2A and RB1 were also highly mutated in the preinvasive group and were considered to be early-stage events for LUAD in our Chinese cohort. The mutation frequency of ERBB2, BRAF, GRIN2A, and RB1 was found to be low in the TCGA cohort, and there was no significant difference in mutation frequency between the preinvasive and invasive LUAD groups in TCGA.
In our study, TP53, RBM10, MUC16, CSMD3, MED1, LRP1B, GLI1, and RYR2 were identified as late events with a higher mutation frequency in the invasive LUAD group. TP53 mutations were the most enriched alternations in invasive LUAD, suggesting that it may play a more important role in the acquisition of invasiveness. It has also been revealed that TP53 and cell mobility, gap junction, and metastasis-related genes may be late events associated with subclonal diversification and neoplastic progression (13, 14). One study identified that RBM10 plays an important role in invasive LUAD, whereas another study showed that this gene is significantly mutated in the pre/minimally invasive group (14). It has been reported that as a tumor suppressor gene, RBM10 can also promote lung cancer (28). For the TCGA cohort results, the mutation frequency of RBM10 was 16.1%, and it was more highly mutated in the preinvasive group than in the invasive LUAD group (25% versus 14%). Interestingly, MUC16 (CA125) is cleaved and shed into the bloodstream, and its serum level is thought to be a key indicator of lung cancer metastasis to the liver (29). Additionally, MUC16 plays crucial roles in lung cancer pathogenesis, progression, and chemoresistance (30). In general, MUC16 might play an important role during the entire process from preinvasive to invasive LUAD.
In both preinvasive and early invasive LUAD, EGFR mutation is the most common driver alterations across AIS, MIA, and IAC (13). EGFR is a transmembrane protein that can transduce important growth factor signaling from the extracellular milieu to the cell, thus regulating downstream events including increased proliferation, angiogenesis, metastasis, and decreased apoptosis (31). KRAS alterations occur early in the carcinogenesis process and promote cancer cell survival, invasion, and migration (32). Our results showed that mutations in KRAS and EGFR were mutually exclusive (P < 0.01). This has been validated in another study that EGFR mutations are never found in LUAD tumors with KRAS mutations (33). In our study, for driver genes such as EGFR and KRAS, the mutation was similar between the AIS/MIA and IAC groups, indicating that EGFR and KRAS mutations were separately occurring and accompanying all events. It has also been reported that EGFR and KRAS mutations are significantly mutated genes in the pre/minimally invasive group (14). In addition, the EGFR mutation type varies greatly in IAC compared with AIS/MIA in our Chinese cohort. The distribution and comparison of deleterious mutations detected for EGFR and KRAS exhibited some overlap between the Chinese group and TCGA group but also showed variations. In our study, the pre-invasive LUAD exhibited a high frequency of driver mutations, including those in EGFR, KRAS, ALK, and ERBB2 genes, which was also found in other publications (14, 34). The population for TCGA cohort also yielded a high incidence of EGFR mutations, which might due to the background of the United States (35).
In conclusion, we demonstrated that ERBB2, BRAF, GRIN2, RB1, TP53, RBM10, MUC1, CSMD3, MED1, LRP1B, GLI1, MAP3K4, RYR2, EGFR, and KRAS genes all play crucial roles in carcinogenesis from preinvasive to invasive LUAD in our Chinese cohort. The usability of targeted sequencing for biomarker detection has strong clinical significance for LUAD. Furthermore, in comparison of our findings with the TCGA database, EGFR, KRAS, TP53, and RBM10 were the most frequently mutated genes in the two cohorts. In addition, we also observed an increase in mutation types in IAC in both cohorts. Overall, our current understanding of pathological and molecular features in clinical practice reveals genomic tumor heterogeneity of resectable LUAD.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by Ethics Committee of The Eighth Affiliated Hospital of Sun Yat-sen University and informed consents from all patient. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by Ethics Committee of The Eighth Affiliated Hospital of Sun Yat-sen University and informed consents from all patient. The study was conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
YL: Writing – original draft. DL: Writing – original draft. HH: Methodology, Writing – review & editing. HM: Data curation, Formal analysis, Writing – original draft. ML: Data curation, Formal analysis, Writing – original draft. BR: Writing – review & editing. BC: Data curation, Formal analysis, Writing – original draft. WZ: Data curation, Formal analysis, Writing – review & editing. DS: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. DM: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. YJ: Writing – original draft, Writing – review & editing. FQ: Conceptualization, Writing – original draft. HL: Conceptualization, Writing – review & editing, Supervision. BJ: Conceptualization, Funding acquisition, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The present study was supported by the Futian Healthcare Research Project (No. FTWS2021012) and the Guangdong Medical Science and Technology Research Foundation (A2021177).
Acknowledgments
We would like to first acknowledge all the patients and their families who participated in this study. We’d also express our gratitude to Dr. Fan Qiu, Huaming Li, Bo Jiang, and Binbin Chen for their helpful suggestions.
Conflict of interest
Authors BC, WZ, DS and DM were employed by the company BGI Genomics Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2024.1389618/full#supplementary-material
Supplementary Figure 1 | Mutation spectrum of the LUAD patients in a Chinese cohort. (A) Waterfall plot with top frequently mutated genes in LUAD patients with AIS/MIA; (B) Waterfall plot with top frequently mutated genes in LUAD patients with IAC.
Supplementary Figure 2 | Comparison of data in Chinese LUAD and the TCGA cohorts. (A) Shared genes between IAS, MIA and IAC groups in a Chinese cohort; (B) Top 35 frequently mutated genes in our Chinese cohort and TCGA cohort.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Viviani S. Survival of patients with stage I lung cancer detected on CT screening. N Engl J Med. (2008) 358:1862. doi: 10.1056/NEJMx080010
3. Goldstraw P, Chansky K, Crowley J, Rami-Porta R, Asamura H, Eberhardt WEE, et al. The IASLC lung cancer staging project: Proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM Classification for lung cancer. J Thorac Oncol. (2016) 11:39–51. doi: 10.1016/j.jtho.2015.09.009
4. Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A, et al. Non-small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. (2022) 20:497–530. doi: 10.6004/jnccn.2022.0025
5. Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. (2015) 10:1243–60. doi: 10.1097/JTO.0000000000000630
6. Yotsukura M, Asamura H, Motoi N, Kashima J, Yoshida Y, Nakagawa K, et al. Long-term prognosis of patients with resected adenocarcinoma in situ and minimally invasive adenocarcinoma of the lung. J Thorac Oncol. (2021) 16:1312–20. doi: 10.1016/j.jtho.2021.04.007
7. Maeda R, Yoshida J, Hishida T, Aokage K, Nishimura M, Nishiwaki Y, et al. Late recurrence of non-small cell lung cancer more than 5 years after complete resection: Incidence and clinical implications in patient follow-up. Chest. (2010) 138:145–50. doi: 10.1378/chest.09-2361
8. Zhu QG, Zhang SM, Ding XX, He B, Zhang HQ. Driver genes in non-small cell lung cancer: Characteristics, detection methods, and targeted therapies. Oncotarget. (2017) 8:57680–92. doi: 10.18632/oncotarget.17016
9. Chang YS, Tu SJ, Chen YC, Liu TY, Lee YT, Yen JC, et al. Mutation profile of non-small cell lung cancer revealed by next generation sequencing. Respir Res. (2021) 22:1–10. doi: 10.1186/s12931-020-01608-5
10. Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. (2021) 398:535–54. doi: 10.1016/S0140-6736(21)00312-3
11. Rodríguez M, Ajona D, Seijo LM, Sanz J, Valencia K, Corral J, et al. Molecular biomarkers in early stage lung cancer. Transl Lung Cancer Res. (2021) 10:1165–85. doi: 10.21037/tlcr-20-750
12. Qiu Y, Liu L, Yang H, Chen H, Deng Q, Xiao D, et al. Intratumor heterogeneity of driver mutations and TMB distribution in 30 early-stage LUAD patients with multiple lesions. Front Oncol. (2022) 12:952572. doi: 10.3389/fonc.2022.952572
13. Zhang C, Zhang J, Xu FP, Wang YG, Xie Z, Su J, et al. Genomic landscape and immune microenvironment features of preinvasive and early invasive lung adenocarcinoma. J Thorac Oncol. (2019) 14:1912–23. doi: 10.1016/j.jtho.2019.07.031
14. Chen H, Carrot-Zhang J, Zhao Y, Hu H, Freeman SS, Yu S, et al. Genomic and immune profiling of pre-invasive lung adenocarcinoma. Nat Commun. (2019) 10:1–6. doi: 10.1038/s41467-019-13460-3
15. Zhou Yj, Zheng W, Zeng Qh, Ye Y, Wang C, Fang C, et al. Targeted exome sequencing identifies mutational landscape in a cohort of 1500 Chinese patients with non-small cell lung carcinoma (NSCLC). Hum Genomics. (2021) 15:1–10. doi: 10.1186/s40246-021-00320-9
16. Xiang C, Ji C, Cai Y, Teng H, Wang Y, Zhao R, et al. Distinct mutational features across preinvasive and invasive subtypes identified through comprehensive profiling of surgically resected lung adenocarcinoma. Mod Pathol. (2022) 35:1181–92. doi: 10.1038/s41379-022-01076-w
17. Vinayanuwattikun C, Le Calvez-Kelm F, Abedi-Ardekani B, Zaridze D, Mukeria A, Voegele C, et al. Elucidating genomic characteristics of lung cancer progression from in situ to invasive adenocarcinoma. Sci Rep. (2016) 6:1–10. doi: 10.1038/srep31628
18. Caso R, Sanchez-Vega F, Tan KS, Mastrogiacomo B, Zhou J, Jones GD, et al. The underlying tumor genomics of predominant histologic subtypes in lung adenocarcinoma. J Thorac Oncol. (2020) 15:1844–56. doi: 10.1016/j.jtho.2020.08.005
19. Shao D, Lin Y, Liu J, Wan L, Liu Z, Cheng S, et al. A targeted next-generation sequencing method for identifying clinically relevant mutation profiles in lung adenocarcinoma. Sci Rep. (2016) 6:1–9. doi: 10.1038/srep22338
20. Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. BIOINFORMATICS. (2014) 30:1015–6. doi: 10.1093/bioinformatics/btt755
21. Kautto EA, Bonneville R, Miya J, Yu L, Krook MA, Reeser JW, et al. Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget. (2017) 8:7452–63. doi: 10.18632/oncotarget.13918
22. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. (2009) 4:44–57. doi: 10.1038/nprot.2008.211
23. Zhang S, Zeng X, Lin S, Liang M, Huang H. Identification of seven-gene marker to predict the survival of patients with lung adenocarcinoma using integrated multi-omics data analysis. J Clin Lab Anal. (2022) 36:1–14. doi: 10.1002/jcla.24190
24. Xia W, Mao Q, Chen B, Wang L, Ma W, Liang Y, et al. The TWIST1-centered competing endogenous RNA network promotes proliferation, invasion, and migration of lung adenocarcinoma. Oncogenesis. (2019) 8:62. doi: 10.1038/s41389-019-0167-6
25. Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. (2023) 51:D587–92. doi: 10.1093/nar/gkac963
26. Sharma M, Dewan A, Diwan H, Nathany S BU. A narrative review of ERBB2 in non-small cell lung carcinoma. Cancer Res Stat Treat. (2022) 5:97–104. doi: 10.4103/crst.crst_323_21
27. Peng P, Lv G, Hu J, Wang K, Lv J, Guo G. Co-mutations of epidermal growth factor receptor and BRAF in Chinese non-small cell lung cancer patients. Ann Transl Med. (2021) 9:1321–1. doi: 10.21037/atm-21-3570
28. Cao Y, Di X, Zhang Q, Li R, Wang K. RBM10 regulates tumor apoptosis, proliferation, and metastasis. Front Oncol. (2021) 11:603932. doi: 10.3389/fonc.2021.603932
29. Saad HM, Tourky GF, Al-kuraishy HM, Al-Gareeb AI, Khattab AM, Elmasry SA, et al. The potential role of MUC16 (CA125) biomarker in lung cancer: A magic biomarker but with adversity. Diagnostics. (2022) 12:1–20. doi: 10.3390/diagnostics12122985
30. Chen Y, Huang Y, Kanwal M, Li G, Yang J, Niu H, et al. MUC16 in non-small cell lung cancer patients affected by familial lung cancer and indoor air pollution: Clinical characteristics and cell behaviors. Transl Lung Cancer Res. (2019) 8:476–88. doi: 10.21037/tlcr.2019.07.10
31. Da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol Mech Dis. (2011) 6:49–69. doi: 10.1146/annurev-pathol-011110-130206
32. Jang TW, Oak CH, Chang HK, Suo SJ, Jung MH. EGFR and KRAS mutations in patients with adenocarcinoma of the lung. Korean J Intern Med. (2009) 24:48–54. doi: 10.3904/kjim.2009.24.1.48
33. Reck M, Carbone DP, Garassino M, Barlesi F. Targeting KRAS in non-small-cell lung cancer: recent progress and new approaches. Ann Oncol. (2021) 32:1101–10. doi: 10.1016/j.annonc.2021.06.001
34. Liu L, Liu J, Shao D, Deng Q, Tang H, Liu Z, et al. Comprehensive genomic profiling of lung cancer using a validated panel to explore therapeutic targets in East Asian patients. Cancer Sci. (2017) 108:2487–94. doi: 10.1111/cas.13410
Keywords: adenocarcinoma in situ, minimally invasive adenocarcinoma, invasive adenocarcinoma, genomic landscape, targeted exome sequencing, lung adenocarcinoma
Citation: Lin Y, Li D, Hui H, Miao H, Luo M, Roy B, Chen B, Zhang W, Shao D, Ma D, Jie Y, Qiu F, Li H and Jiang B (2024) Genomic landscape and tumor mutational features of resected preinvasive to invasive lung adenocarcinoma. Front. Oncol. 14:1389618. doi: 10.3389/fonc.2024.1389618
Received: 21 February 2024; Accepted: 24 April 2024;
Published: 13 May 2024.
Edited by:
Melanie Kucherlapati, Harvard University, United StatesReviewed by:
Alina Basnet, Upstate Medical University, United StatesFengbiao Mao, Peking University Third Hospital, China
Copyright © 2024 Lin, Li, Hui, Miao, Luo, Roy, Chen, Zhang, Shao, Ma, Jie, Qiu, Li and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huaming Li, bGlobTY4QG1haWwuc3lzdS5lZHUuY24=; Bo Jiang, amlhbmdiNTVAbWFpbC5zeXN1LmVkdS5jbg==; Fan Qiu, cWl1ZjdAbWFpbC5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
 Yangui Lin1†
Yangui Lin1† Bhaskar Roy
Bhaskar Roy Binbin Chen
Binbin Chen Wei Zhang
Wei Zhang Di Shao
Di Shao