
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 30 October 2023
Sec. Gastrointestinal Cancers: Colorectal Cancer
Volume 13 - 2023 | https://doi.org/10.3389/fonc.2023.1270991
This article is part of the Research TopicGut Microbiota in Health and DiseaseView all 19 articles
Colorectal cancer (CRC) ranks third in terms of incidence among all kinds of cancer. The main cause of death is metastasis. Recent studies have shown that the gut microbiota could facilitate cancer metastasis by promoting cancer cells proliferation, invasion, dissemination, and survival. Multiple mechanisms have been implicated, such as RNA-mediated targeting effects, activation of tumor signaling cascades, secretion of microbiota-derived functional substances, regulation of mRNA methylation, facilitated immune evasion, increased intravasation of cancer cells, and remodeling of tumor microenvironment (TME). The understanding of CRC metastasis was further deepened by the mechanisms mentioned above. In this review, the mechanisms by which the gut microbiota participates in the process of CRC metastasis were reviewed as followed based on recent studies.
Colorectal cancer (CRC) has the third highest incidence rate among all types of cancers globally (1). The main cause of death of CRC patients is metastasis, which is also a clinical challenge (2, 3). Metastasis is a multi-step and multi-factor process including the separation of tumor cells from each other, invasion into surrounding tissues, adhesion to endothelial cells, and migration from the primary site to secondary site. Several mechanisms have been implicated, such as epithelial-mesenchymal transition (EMT) (4, 5), changes in expression of intercellular adhesion molecules (6), loss of structural integrity of the basement membrane (7), remodeling of the pre-metastatic niche (8), and induction of angiogenesis (9). Nonetheless, it is worth noting that current understanding of CRC couldn’t fully illuminate the role of systematic factors like exercise, diet and aging in CRC metastasis.
Gut microbiota located within the intestinal tract comprises a large and diverse community including bacteria, yeasts, fungi viruses and parasites, which are referred to as the second gene pool of the human body (10). As one of the earliest encountered foreign antigens in the human body, gut microbiota plays essential roles in various physiological and pathological processes. Previously, the main roles attributed to gut microbiota were the synthesis of essential amino acids and vitamins, the digestion of polysaccharides that are difficult to assimilate, and contribution to human metabolic processes (11). Additionally, gut microbiota provides essential signals for the development and functioning of immune system (12). In recent years, numerous studies have suggested that the gut microbiota also participates in oncogenesis and progression of cancer, particularly in the process of metastasis (13–15). On the one hand, the gut microbiota secretes various metabolites or virulence factors that damage host DNA (16) and contributes to a pro-inflammatory environment (17), leading to pre-cancerous lesions. On the other hand, the gut microbiota directly interacts with cancer cells, thereby increasing invasion and proliferation of cancer cells (18). Furthermore, several studies have indicated that the gut microbiota may facilitate metastasis by affecting the recruitment of immune cells and remodeling the tumor microenvironment (TME) (19–23). The mechanisms by which the gut microbiota participates in the process of CRC metastasis were reviewed as followed based on recent studies.
The progression of CRC involves multiple signaling pathways (24, 25). The disruption of cell cycle and the acquisition of unlimited proliferative capacity are key steps in cancer progression. Researches have indicated that Fusobacterium, a specific type of bacteria, has a significantly higher relative abundance in CRC tissue compared to normal one (26–28). The quantity of Fusobacterium also exhibits statistical differences between different stages of cancer progression. Furthermore, during the transition from adenoma to malignant tumor, the abundance of Fusobacterium gradually increases (29). Recent studies have demonstrated that the gut microbiota may promote cancer cell proliferation through mechanisms as follows.
RNA-mediated targeting effects are important mechanisms of epigenetic regulation (30), including the synthesis of various non-coding RNAs and their impact on downstream genes (31, 32). MicroRNAs (miRNAs) are one of the key players in this process, regulating various biological processes such as tumorigenesis. Recent studies have indicated that the gut microbiota is involved in RNA-mediated targeting effects that regulate cancer cell proliferation.
Fusobacterium not only facilitated the proliferation and invasiveness of co-cultured CRC cell lines but also promoted tumor formation in APCMin/+ mice. Fusobacterium activated the TLR4/MYD88 receptors on the surface of cancer cells, leading to the activation of NFκB. NFκB then binds to the upstream region of the transcription start site (TSS) of miR21, upregulating its expression. MiR21, in turn, bound to the 3’ end binding site of RASA1, inhibiting its expression (33). RASA1 is a member of the RAS GTPase-activating protein (RAS-GAP) family, and its binding to the well-known oncogenic protein RAS can inhibit RAS activity (34). Some studies have suggested that mutations or loss of function in RASA1 in CRC leads to activation of the RAS-MAPK cascade (35–37). The MAPK pathway is reported to induce the synthesis of cycling D1, promoting cell division (38). The MAPK pathway has also been shown to participate in the proliferation of cancer cells in multiple studies (39, 40).
Peptostreptococcus micros (P. micros) is an opportunistic pathogen found in the oral cavity that is closely associated with periodontitis (41). It can also cause suppurative infections in various organs throughout the body (41). Chang et al. found that P. micros could significantly foster the proliferation of LoVo and HT-29 cell lines in vitro (42). To unveil the underlying mechanism, they constructed xenograft models. It came out that tumors derived from cancer cells co-cultured with P. micros had larger volume and weight (42). Further investigations revealed that P. micros suppressed the expression of protein tyrosine phosphatase receptor R (PTPRR) by upregulating miR-218-5p, ultimately activating the Ras/ERK/cFos signaling pathway (42). The Ras/ERK signaling pathway is part of MAPK pathway and also participates in the proliferation of CRC cells (43).
Significantly associated with inflammatory bowel disease (IBD) and CRC, Enterotoxigenic Bacteroides fragilis (ETBF) is a molecular subtype of Bacteroides fragilis (44, 45). ETBF could downregulate the expression of miR-149-3p in cancer cell lines and influences the selective splicing of the KAT2A gene through PHF5A. Ultimately, KAT2A directly binds to the promoter region of SOD2, activating the SOD2 gene (46). SOD2 has been shown to modulate energy metabolism and promote proliferation of CRC (47).
The Wnt/β-catenin signaling pathway plays a crucial role in physiological processes such as cell proliferation and differentiation, stem cell renewal, embryonic development, and tissue homeostasis (48). Dysregulation of this pathway is widely considered a key oncogenic signal and is of significant importance in the development of different kinds of cancers (49). Certain bacteria, such as Fusobacterium nucleatum (F. nucleatum), could facilitate cancer cell proliferation through the Wnt/β-catenin pathway (18, 50). For example, F. nucleatum produces a virulence factor called FadA (51), which binds to the E-cadherin domain EC5 on the surface of CRC cells. This interaction leads to the dephosphorylation of β-catenin, accumulation of β-catenin in the cytoplasm, and translocation of β-catenin to the cell nucleus. Subsequently, the expression of transcription factors lymphoid enhancer-binding factor (LEF)/T-cell factor (TCF), NFκB, and oncogenes such as Myc and Cyclin D1 is upregulated, promoting CRC cell proliferation (18). Additionally, FadA could promote the expression of chk2 through the E-cadherin/β-catenin pathway, leading to increased DNA damage and elevated proliferative capacity in CRC cells (50). Furthermore, some studies have reported that probiotics have the ability to inhibit cancer cell proliferation and promote apoptosis (52–55). qPCR and western blot results have shown that during this process, the gene expression and protein content of β-catenin in CRC decrease, suggesting that probiotics may inhibit CRC cell proliferation by regulating β-catenin-related pathways (52). Nonetheless, the underlying mechanisms of these effects are still need to be explored.
The PI3K-Akt pathway is widely activated in various tumors and is closely associated with tumor development (56–59). Gram-positive anaerobic bacteria, such as Peptostreptococcus anaerobius (P. anaerobius), present in the oral cavity and intestines (60), could bind to integrin α2β1 on the surface of CRC cells through its surface protein called putative cell wall binding repeat 2 (PCWBR2). This interaction activates the PI3K-Akt signaling pathway through Focal Adhesion Kinase 9 (FAK9), ultimately promoting cancer cell proliferation (61).
The MAPK-ERK pathway, a cell proliferation signaling pathway located on the cell surface and extended to the nucleus, plays a crucial role in cell proliferation (62). Activation of the MAPK-ERK pathway is increasingly implicated in the occurrence and progression of CRC (63). The oral pathogenic bacterium Porphyromonas gingivalis (P. gingivalis), once colonizing the colon, can selectively invade CRC cells and activate the MAPK-ERK pathway, thereby promoting tumor proliferation (64).
Not only individual bacterial species but also the overall balance of the gut microbiota is crucial in regulating cancer proliferation. Bai et al. have found that smoking induced gut microbiota dysbiosis altered gut metabolites and impaired gut barrier function, ultimately activating the oncogenic MAPK-ERK signaling and enhancing cancer cell proliferation (65). Portulaca oleracea, a medicinal plant and a member of the Portulacaceae family, is well-known for its resistance against microbiota, inflammation, and cancer (66). Portulaca oleracea extract (POE) has been found to reduce tumor quantity and improve survival rate in carcinogen-induced mouse models through restoring the balance of gut microbiota. Further results have shown that POE upregulates the expression of TP53, inhibits the Wnt/β-catenin signaling pathway and reduces the expression of c-Myc and Cyclin D1, ultimately suppressing cancer cell proliferation (67).
In addition to unlimited proliferative capacity, invasive growth into surrounding tissues is another characteristic of malignant tumors. Breaking through the basement membrane is the first step for distant metastasis (68). It has been shown that a positive correlation between the gut microbiota and tumor progression stages exists (29). Since one of the defining criteria for tumor progression stages is the depth of tumor infiltration (69), the gut microbiota has the potential to regulate the invasive properties of cancer cells.
The metabolic products derived from microorganisms, such as l-2-hydroxyglutarate, succinate, fumarate, d-2-hydroxyglutarate, and lactate, can accumulate in tumor lesions and exacerbate the malignancy of the tumor (70). Furthermore, some metabolites could hijack signaling pathways related to tumor metastasis through gene regulation (71). Formate, a major metabolic product of F. nucleatum, can activate the AhR signaling pathway in CRC, enhancing its cancer stem cell properties and increasing the invasiveness of CRC, ultimately promoting cancer metastasis (72).
EMT is a cellular biological process (73, 74) that endows cancer cells with invasive and anti-apoptotic capabilities (75, 76). EMT triggers the process of dissemination and invasion, ultimately leading the formation of metastases (77, 78). Certain strains of Escherichia coli can produce a virulence protein called cytotoxic necrotizing factor 1 (CNF1) (79). CNF1 induces the recruitment of mTOR to lysosomes, consequently increasing invasiveness of CRC cell lines and inducing the expression of EMT markers (80). These findings suggest that gut microbiota has the potential to induce EMT in cancer cells.
Hydrogen sulfide (H2S) has been identified as the third gasotransmitter after nitric oxide (NO) and carbon monoxide (CO) and participates in a variety of biological processes (81). There are two sources of luminal H2S: the inorganic and organic metabolism of intestinal bacteria (82) and endogenously synthesized in the mammal cells (83). Endogenous H2S fosters metastasis, partly through induction of ATP citrate lyase (ACLY) to facilitate EMT (84, 85). Since the luminal H2S mainly originate from bacterial metabolism and directly contact with intestinal epithelial cells (86), the intestinal flora has the potential to facilitate CRC metastasis through modulating endogenous H2S synthesis and related pathways.
The presence of the microbiota has been shown to induce epigenetic changes in mouse tissues at transcriptional level (87, 88). N6-methyladenosine (m6A), one of the epigenetic modification mechanisms of mRNA, could influence various fundamental biological processes (89). METTL3, the main m6A methyltransferase, is involved in the progression of several types of cancers, including acute myeloid leukemia (90), hepatocellular carcinoma (91), and lung cancer (92). In CRC, F. nucleatum has been shown to inhibit the Hippo pathway and activate the YAP signaling, leading to the suppression of METTL3 expression through the transcription factor FOXD3. The inhibition of METTL3 resulted in decreased m6A methylation of KIF26B mRNA, a gene associated with cell-cell adhesion and important for cancer cell invasion. Consequently, the expression of KIF26B were promoted, leading to enhanced tumor cell invasiveness. Therefore, F. nucleatum could induce epigenetic modifications in the KIF26B gene at transcriptional level through the YAP/FOXD3/METTL3 axis, ultimately facilitating the invasiveness of cancer cells (93).
In conclusion, gut microbiota is capable of promoting proliferation and invasiveness of CRC cells via multiple mechanisms (see Figure 1 for details).
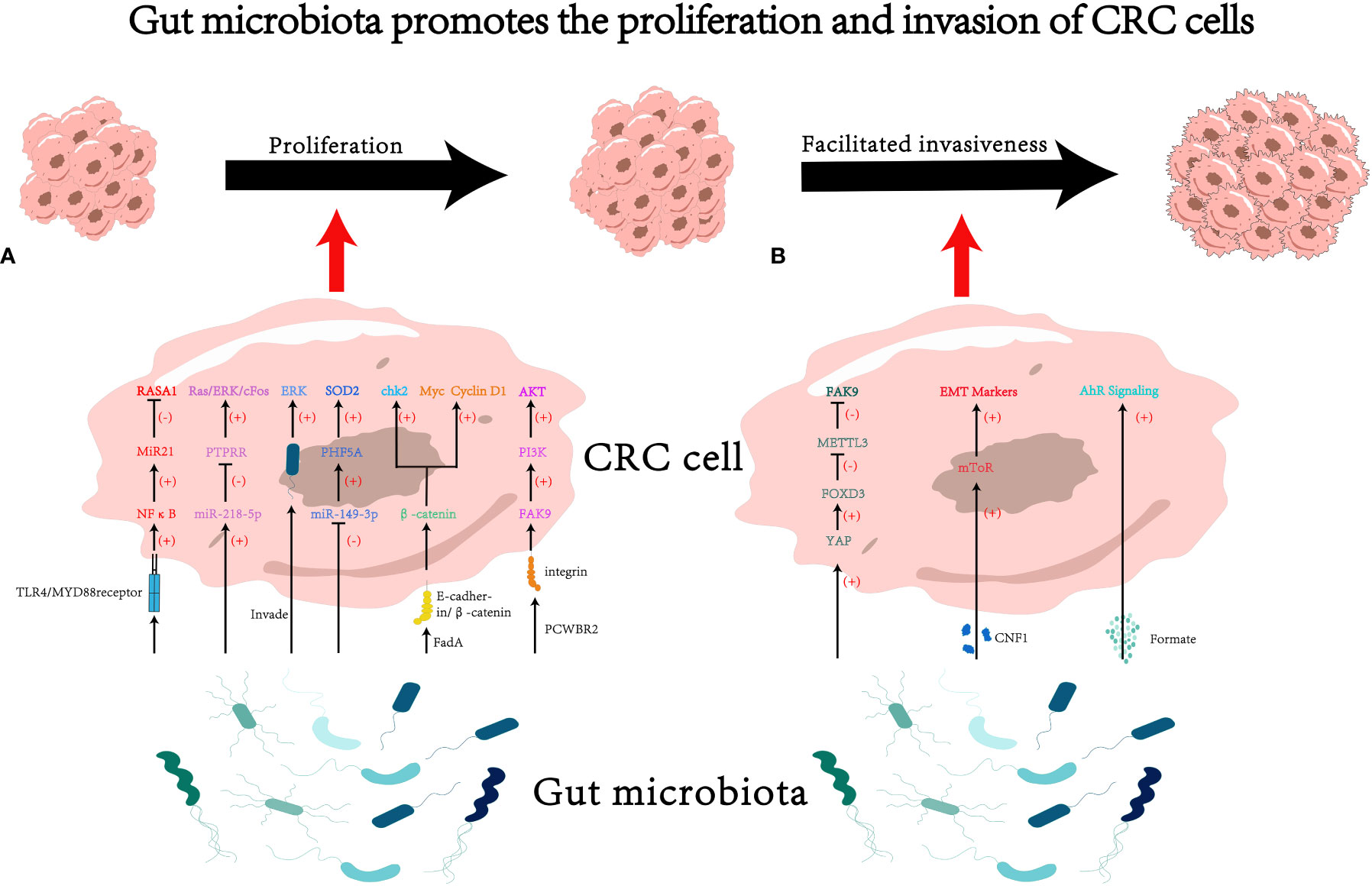
Figure 1 The impact of gut microbiota on proliferation and invasion of colorectal cancer (CRC) cells. (A) Gut microbiota may influence cancer cell proliferation through various pathways, such as modulating RNA-mediated targeting effects (33, 42, 46) and activating the cascades of cancer signaling (18, 50, 60, 64). (B) Gut microbiota may influence cancer cell invasiveness through various pathways, such as secreting microbiota-derived functional substances (70, 79), and regulating mRNA methylation (93). PCWBR2, putative cell wall binding repeat 2. CNF1, cytotoxic necrotizing factor 1.
Most kinds of cancers rely on blood vessels, lymphatic vessels, and other channels for metastasis. Survival pressure like anoikis, shear forces, and immune attacks are exerted on cancer cells once they enter the circulatory system (68). Therefore, dissemination and survival are crucial prerequisites for cancer cells to complete metastasis. In recent years, studies have found that gut microbiota not only promotes cancer cell proliferation and increases their invasiveness but also facilitates cancer cell dissemination and survival (94–96). The mechanisms behind this include regulating intravasation of cancer cells to facilitate dissemination, participating in immune evasion to promote cancer cell survival, and modulating the tumor microenvironment to facilitate the formation of metastatic lesions.
Structural and functional disruptions of vascular basement membrane (97), as well as tumor cell reprogramming (98), are two important processes involved in hematogenous metastasis of tumors. The former provides a physical basis for cancer cells to breach blood vessels and enter the bloodstream, while the latter enhances the intravasation and migration capabilities of tumor cells.
Under a high-fat diet, elevated levels of deoxycholic acid (DCA) in the host gut was detected, which enhanced vasculogenic mimicry in tumor tissues (99) —— the formation of structures that lack endothelial cells but possess normal vascular functions (100). This study suggests that the gut microbiota’s regulation of host metabolism may contribute to vasculogenic mimicry and promote tumor metastasis. However, the direct association between bile salt-hydrolyzing bacteria and intestinal DCA levels requires further investigation. Therefore, the mechanisms by which the gut microbiota regulates tumor vasculogenic mimicry through DCA still need to be further validated.
The adhesion of circulating tumor cells to endothelial cells and extravasation into pre-metastatic sites is an important process in tumor metastasis (101). Intercellular adhesion molecule 1 (ICAM1), a member of the immunoglobulin superfamily, has been shown to promote tumor cell adhesion to endothelial cells and facilitate metastasis (102). Its expression levels also positively correlate with tumor progression and metastasis in clinical settings (103). F. nucleatum could activate the NF-κB pathway by acting on the pattern recognition receptor ALPK1 on cells, thereby upregulating ICAM1 expression and promoting CRC cell adhesion to endothelial cells (96), ultimately facilitating metastasis of CRC.
Immune surveillance imposes strong selective pressure on cancer cells (104). The gut microbiota can directly or indirectly inhibit the function of immune cells, thus mediating immune evasion of tumor cells (61, 105–107).
The TIGIT (T cell immunoglobulin and ITIM domain) receptor is expressed on all NK cells and some other types of lymphocytes (108). F. nucleatum can directly interact with the TIGIT receptor through its surface virulence protein Fap2, thereby inhibiting the cytotoxicity of NK cells against cancer cells and ultimately inducing immune evasion of tumor cells (105). In addition to affecting the host’s innate immunity, the gut microbiota also regulates host adaptive immunity. Research by Jiang et al. has shown that succinate produced by F. nucleatum could inhibit the cGAS-IFNβ pathway, leading to reduced levels of chemokines CCL5 and CXCL10 in the tumor, thereby limiting the migration of CD8+ T cells to TME and suppressing the anti-tumor response of CD8+ T cells (109).
Myeloid-derived suppressor cells (MDSCs) from the bone marrow exert immunosuppressive effects through the depletion of amino acids and the expression of TGFβ and PD-L1 (110). Certain specific pathogens such as F. nucleatum and P. anaerobius can induce tumor-derived chemokine CXCL1 to recruit the MDSCs, thereby suppressing anti-tumor immunity (61, 107). The gut microbiota can also activate the TLR-calcineurin-NFAT-IL-6 signaling cascade on MDSCs, leading to the STAT3-dependent induction of the inhibitory protein B7H3/4, resulting in functional inhibition of cytotoxic T cells and ultimately promoting tumor immune evasion (111).
It’s worth noting that the effects imposed on the anti-tumor immunity by gut microbiota is a double-edged sword. A consortium of 11 bacterial strains was found to induce a strong CD8+ T cell response that boosted the efficacy of immune checkpoint blockade in mice (112). Other species such as Enterococus hirae could facilitate anti-tumor immunity in mice by enhancing CD8+ T cell anti-tumor responses when used in combination with cyclophosphamide chemotherapy (113). Bachem et al. discovered that butyrate, a microbiota-derived short-chain fatty acid (SCFA), enhances CD8+ T cell metabolism and promotes their differentiation into memory T cells (114). Similarly, microbiome-derived inosine could facilitate the differentiation of TH1 cells in an adenosine 2A receptor-dependent manner and consequently improve the antitumor effect induced by the ICB therapy (115). Since the adenosine 2A receptor has been demonstrated to inhibit TH1 differentiation in vitro as well as antitumor immunity in vivo (116–119) and only a few has reported that adenosine 2A receptor signaling can sustain TH1 and antitumor immunity (120, 121), the crosstalk between microbiota-derived metabolites, adenosine 2A receptor signaling and host immunity needs to be further investigated. In terms of clinical practice, a phase I clinical trial enrolling 20 patients have shown that fecal microbiota transportation (FMT) in combination with anti PD-1therapy could lead to a promoted immune status in patients with melanoma (122). These researches indicate that the correlation between gut microbiota and host immunity could be far more complicated and worth further investigation.
Besides regulating the anti-tumor immunity, the gut microbiota plays an important role in the development and maturation of the host immune function. Germ-free mice are unable to develop mature isolated lymphoid follicles (123). Additionally, the gut microbiota can regulate the function of different types of immune cells such as Treg cells, DC cells, and T cells, thereby establishing a normal intestinal immune homeostasis during early host development by balancing local pro-inflammatory and anti-inflammatory responses (124–126). Similarly, changes in the functional status of the immune system can change the composition of the gut microbiota. Activation of the AhR pathway in Th17/Th22 cells can induce the production of IL-22 and IL-17, which in turn can stimulate intestinal epithelial cells to secrete antimicrobial peptides, ultimately limiting the proliferation of pathogenic microbial communities (125). Individuals with immune deficiencies are more prone to dysbiosis of the gut microbiota, leading to various chronic inflammations (127).
These facts indicate that the gut microbiota-immunity axis is a complex bidirectional process. During the occurrence and development of tumors, changes in the gut microbiota are accompanied by immune dysregulation. The aforementioned studies have revealed various mechanisms by which the gut microbiota participates in immune evasion, providing a new perspective for a deeper understanding of the correlation between gut microbiota and the host immunity.
TME consists of various cell components (128, 129), and its complexity has made it a tendency to view the TME as an organ itself (128). In certain situations, these components can produce bioactive factors and release them into the TME, thereby promoting tumor angiogenesis, invasion, and metastasis (130–133). Recent studies have found that there could be multiple kinds of bacteria with regulation effect in the TME besides the cell component. For instance, Xu and colleagues found that F. nucleatum could facilitate tumor metastasis in a CCL20-dependent manner (94). Although the only known receptor for CCL20—CCR6 is mainly expressed in immature dendritic cells, innate lymphoid cells, regulatory CD4 T cells, Th17 cells and B cells (134), a positive correlation between F. nucleatum-induced CCL20 expression and F4/80+ CCR6+ macrophage in lung metastasis tissues was observed (94). And they also found that F. nucleatum could directly promote the polarization of M2 macrophages in tumor tissues (94). Current researches have shown that M2 macrophages play important roles in immune suppression, tumor angiogenesis, and EMT (135–137). There are also studies showing that several bacteria such as segmented filamentous bacterium (SFB; Candidatus Savagella), ETBF, Bifidobacterium spp., F. nucleatum could modify the polarization of CD4+T cells into TH17 cells (138–141). TH17 cells has been indicated to foster an inflamed and tolerogenic TME (142, 143), providing a potential mechanism by which gut microbiota facilitates the future process of metastasis.
Not only is the microenvironment of the primary tumor important for tumor metastasis, but also the remodeling of the microenvironment in the metastatic foci plays a crucial role in the process of tumor metastasis. According to the “seed and soil” hypothesis, certain tumor cells can selectively settle in organs with suitable growth environments (144). Since the microenvironments of different organs vary, a particular type of tumor cells tends to preferentially colonize a specific organ (145). This preference may originate from the selective remodeling of the target organ by the primary tumor before metastasis occurs (146). Proteus mirabilis (P. mirabilis) and Bacteroides vulgatus (B. vulgatus) can modulate the hepatic immune niche by regulating the proliferation of Kupffer cells and inhibiting their phagocytic ability, ultimately fostering liver metastasis of CRC (19).
In conclusion, gut microbiota can facilitate the dissemination and survival of CRC cells via different ways(see Figure 2 for details).
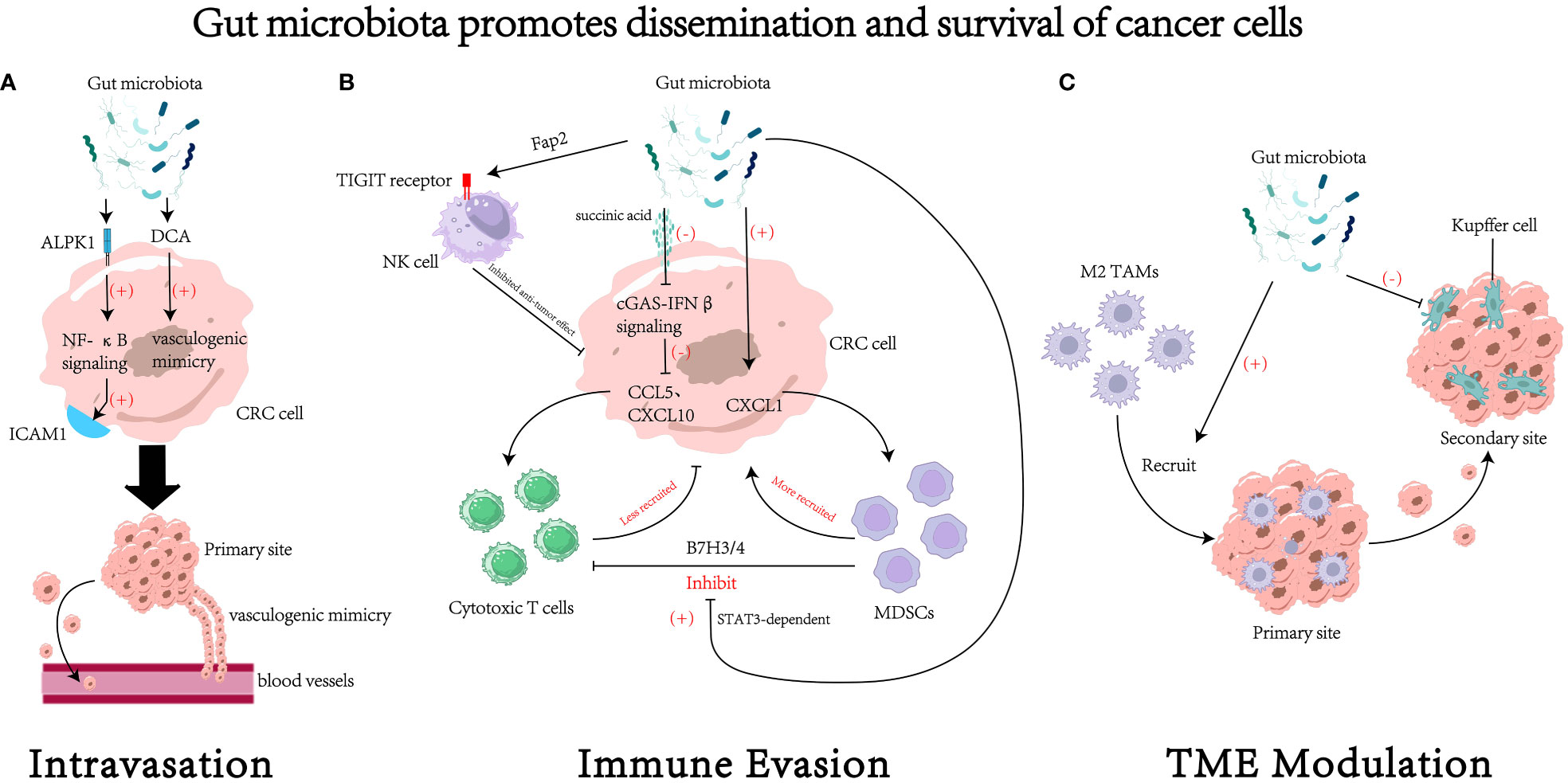
Figure 2 The impact of gut microbiota on the dissemination and survival of colorectal cancer (CRC) cell. (A) Gut microbiota could regulate the process of intravasation through various mechanisms, such as increasing the expression of intercellular adhesion molecule 1 (96) and promoting the formation of vasculogenic mimicry (99). (B) Gut microbiota could regulate survival of cancer cells by modulating tumor immune evasion through various pathways. For example, gut microbiota can inhibit the cytotoxic activity of natural killer (NK) cells through receptor-ligand interactions (105), reduce the recruitment of cytotoxic T cells (109), increase the recruitment of myeloid-derived suppressor cells (MDSCs) (61, 107), and foster the suppressive effects of MDSCs on cytotoxic T cells (111). (C) Gut microbiota may remodel the tumor microenvironment (TME) to facilitate the process of dissemination and colonization. For instance, it could recruit M2 tumor associated macrophages (M2 TAMs) to the primary site (94) and inhibit the function of Kupffer cells in the secondary site (19). DCA, deoxycholic acid. ICAM1, intercellular adhesion molecule 1. MDSCs, myeloid-derived suppressor cells. TAMs, tumor associated macrophages.
Studies on the correlation between gut microbiota and CRC can be traced back to 1951. Subsequent advancements in techniques such as 16S rRNA sequencing and metagenomic sequencing have made it possible to identify gut microbiota that are significantly associated with CRC. Multiple mechanisms have been proposed regarding how the gut microbiota regulates anti-tumor effects and participates in pathological processes especially metastasis of CRC in the following years (see Table 1 for details). Consequently, CRC is a suitable model disease to investigate novel strategies for early cancer detection. Stool-based screening such as 16s rRNA sequencing is considered as a promising, non-invasive approach compared with colonoscopies (155). For bacteria widely participated in the initiation and progression of CRC, high-specific therapy strategies such as targeted antibiotic (156) and bioinorganic hybrid bacteriophage (157) has presented an attractive prospect for prevention and curation.
Recent studies have implicated that oncogenesis and progression of cancers could be consequences of the dysregulated immunologic function. Mechanisms like immune checkpoint shed a light on the complicated networks between cancer and immunity. Still, such theories cannot fully illuminate the role of systematic factors, such as exercise, diet and aging, in crosstalk between cancer and immunity. As one of the earliest encountered environmental antigens in the human body, gut microbiota could facilitate cancer metastasis and modulate immune response through mechanisms mentioned afore, which may explain the role of systematic factors in cancer and immunologic function. Nonetheless, more efforts should be dedicated to further unveil the mechanisms by which systematic factors such as gut microbiota regulate the process of cancer oncogenesis and progression.
SY: Writing – original draft. SW: Writing – review & editing. BX: Writing – review & editing. CP: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Supported by the National Natural Science Foundation of China, Zhongnan Hospital of Wuhan University (No. 81401515), Science, Technology and Innovation Seed Fund (No. znpy2018030), the Program of Excellent Doctoral (Postdoctoral) of Zhongnan Hospital of Wuhan University (No. ZNYB2021013), “351 talent project (Luojia Young Scholars)” of Wuhan University, Training project for young and middle-aged medical key personnel of Wuhan City and Wuhan Peritoneal Cancer Clinical Medical Center, the Health Commission of Hubei Province Scientific Research Project (No. WJ2019H012), Improvement Project for Theranostic ability on Difficulty miscellaneous disease (Tumor) (No. ZLYNXM202018), National Key Clinical Specialty Construction Project and National Natural Science Fund Youth Fund of China (No. 81702411).
Sincerely thanks to the irreplaceable contribution by the authors who helped edit and revise the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer. JAMA-J Am Med Assoc (2021) 325(7):669. doi: 10.1001/jama.2021.0106
3. Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal TRANSDUCT TAR (2020) 5(1):28. doi: 10.1038/s41392-020-0134-x
4. Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu Rev Pathol: Mech Dis (2018) 13(1):395–412. doi: 10.1146/annurev-pathol-020117-043854
5. Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. CELL (2016) 166(1):21–45. doi: 10.1016/j.cell.2016.06.028
6. Su JX, Li SJ, Zhou XF, Zhang ZJ, Yan Y, Liu SL, et al. Chemotherapy-induced metastasis: molecular mechanisms and clinical therapies. Acta Pharmacol Sin (2023) 44(9):1725–36. doi: 10.1038/s41401-023-01093-8
7. Ray A, Callaway MK, Rodríguez-Merced NJ, Crampton AL, Carlson M, Emme KB, et al. Stromal architecture directs early dissemination in pancreatic ductal adenocarcinoma. JCI Insight (2022) 7(3):e150330. doi: 10.1172/jci.insight.150330
8. Wang G, Li J, Bojmar L, Chen H, Li Z, Tobias GC, et al. Tumour extracellular vesicles and particles induce liver metabolic dysfunction. NATURE (2023) 618(7964):374–82. doi: 10.1038/s41586-023-06114-4
9. Liu Z, Chen H, Zheng L, Sun L, Shi L. Angiogenic signaling pathways and anti-angiogenic therapy for cancer. Signal TRANSDUCT TAR (2023) 8(1):198. doi: 10.1038/s41392-023-01460-1
10. Ferreira RDS, Mendonça LABM, Ribeiro CFA, Calças NC, Guimarães RCA, Nascimento VA, et al. Relationship between intestinal microbiota, diet and biological systems: an integrated view. Crit Rev Food Sci (2022) 62(5):1166–86. doi: 10.1080/10408398.2020.1836605
11. Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. SCIENCE (2006) 312(5778):1355–9. doi: 10.1126/science.1124234
12. Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev (2017) 279(1):70–89. doi: 10.1111/imr.12567
13. Matsushita M, Fujita K, Hayashi T, Kayama H, Motooka D, Hase H, et al. Gut microbiota–derived short-chain fatty acids promote prostate cancer growth via IGF1 signaling. Cancer Res (2021) 81(15):4014–26. doi: 10.1158/0008-5472.CAN-20-4090
14. Bertocchi A, Carloni S, Ravenda PS, Bertalot G, Spadoni I, Cascio AL, et al. Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell (2021) 39(5):708–24. doi: 10.1016/j.ccell.2021.03.004
15. Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. GASTROENTEROLOGY (2017) 152(6):1419–33. doi: 10.1053/j.gastro.2017.01.009
16. Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carrá A, et al. The human gut bacterial genotoxin colibactin alkylates DNA. SCIENCE (2019) 363(6428):eaar7785. doi: 10.1126/science.aar7785
17. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini GA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe (2013) 14(2):207–15. doi: 10.1016/j.chom.2013.07.007
18. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via its FadA Adhesin. Cell Host Microbe (2013) 14(2):195–206. doi: 10.1016/j.chom.2013.07.012
19. Yuan N, Li X, Wang M, Zhang Z, Qiao L, Gao Y, et al. Gut microbiota alteration influences colorectal cancer metastasis to the liver by remodeling the liver immune microenvironment. GUT LIVER (2022) 16(4):575–88. doi: 10.5009/gnl210177
20. Nosho K. Association ofFusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J GASTROENTERO (2016) 22(2):557. doi: 10.3748/wjg.v22.i2.557
21. Borowsky J, Haruki K, Lau MC, Costa AD, Väyrynen JP, Ugai T, et al. Association ofFusobacterium nucleatum with Specific T-cell Subsets in the Colorectal Carcinoma Microenvironment. Clin Cancer Res (2021) 27(10):2816–26. doi: 10.1158/1078-0432.CCR-20-4009
22. Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol (2015) 1(5):653. doi: 10.1001/jamaoncol.2015.1377
23. Sakamoto Y, Mima K, Ishimoto T, Ogata Y, Imai K, Miyamoto Y, et al. Relationship betweenFusobacterium nucleatum and antitumor immunity in colorectal cancer liver metastasis. Cancer Sci (2021) 112(11):4470–7. doi: 10.1111/cas.15126
24. Dorard C, Madry C, Buhard O, Toifl S, Didusch S, Ratovomanana T, et al. RAF1 contributes to cell proliferation and STAT3 activation in colorectal cancer independently of microsatellite and KRAS status. ONCOGENE (2023) 42(20):1649–60. doi: 10.1038/s41388-023-02683-w
25. Pilat JM, Brown RE, Chen Z, Berle NJ, Othon AP, Washington MK, et al. SELENOP modifies sporadic colorectal carcinogenesis and WNT signaling activity through LRP5/6 interactions. J Clin Invest (2023) 133(13):e165988. doi: 10.1172/JCI165988
26. Yamaoka Y, Suehiro Y, Hashimoto S, Hoshida T, Fujimoto M, Watanabe M, et al. Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population. J Gastroenterol (2018) 53(4):517–24. doi: 10.1007/s00535-017-1382-6
27. Gao R, Kong C, Huang L, Li H, Qu X, Liu Z, et al. Mucosa-associated microbiota signature in colorectal cancer. Eur J Clin Microbiol (2017) 36(11):2073–83. doi: 10.1007/s10096-017-3026-4
28. Li Y, Ge QK, Cao J, Zhou YJ, Du YL, Shen B, et al. Association ofFusobacterium nucleatum infection with colorectal cancer in Chinese patients. World J GASTROENTERO (2016) 22(11):3227. doi: 10.3748/wjg.v22.i11.3227
29. Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol (2014) 33(8):1381–90. doi: 10.1007/s10096-014-2081-3
30. Babar Q, Saeed A, Tabish TA, Pricl S, Townley H, Thorat N. Novel epigenetic therapeutic strategies and targets in cancer. Biochim Biophys Acta (BBA) - Mol Basis Dis (2022) 1868(12):166552. doi: 10.1016/j.bbadis.2022.166552
31. Withers JB, Mondol V, Pawlica P, Rosa-Mercado NA, Tycowski KT, Ghasempur S, et al. Idiosyncrasies of viral noncoding RNAs provide insights into host cell biology. Annu Rev Virol (2019) 6(1):297–317. doi: 10.1146/annurev-virology-092818-015811
32. Pan X, Hong X, Li S, Meng P, Xiao F. METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. Exp Mol Med (2021) 53(1):91–102. doi: 10.1038/s12276-020-00510-w
33. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor–κB, and up-regulating expression of microRNA-21. GASTROENTEROLOGY (2017) 152(4):851–66. doi: 10.1053/j.gastro.2016.11.018
34. Jia Z, An J, Liu Z, Zhang F. Non-coding RNAs in colorectal cancer: their functions and mechanisms. Front Oncol (2022) 12:783079. doi: 10.3389/fonc.2022.783079
35. Kent OA, Mendell JT, Rottapel R. Transcriptional regulation of miR-31 by oncogenic KRAS mediates metastatic phenotypes by repressing RASA1. Mol Cancer Res (2016) 14(3):267–77. doi: 10.1158/1541-7786.MCR-15-0456
36. Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J, et al. MicroRNA-31 activates the RAS pathway and functions as an oncogenic MicroRNA in human colorectal cancer by repressing RAS p21 GTPase activating protein 1 (RASA1). J Biol Chem (2013) 288(13):9508–18. doi: 10.1074/jbc.M112.367763
37. Sun D, Wang C, Long S, Ma Y, Guo Y, Huang Z, et al. C/EBP-β-activated microRNA-223 promotes tumour growth through targeting RASA1 in human colorectal cancer. BRIT J Cancer (2015) 112(9):1491–500. doi: 10.1038/bjc.2015.107
38. Ou WB, Lundberg MZ, Zhu S, Bahri N, Kyriazoglou A, Xu L, et al. YWHAE-NUTM2 oncoprotein regulates proliferation and cyclin D1 via RAF/MAPK and Hippo pathways. ONCOGENESIS (2021) 10(5):37. doi: 10.1038/s41389-021-00327-w
39. Hassan A, Oh YI, Lee CH, Kim YJ, Cho SB, Alam MM, et al. Design, synthesis, and study of novel phenethyl-based antitumor phospholipids downregulating p38 mitogen-activated protein kinase. J ENZYM INHIB Med CH (2023) 38(1):2217695. doi: 10.1080/14756366.2023.2217695
40. Lee CL, Cremona M, Farrelly A, Workman JA, Kennedy S, Aslam R, et al. Preclinical evaluation of the CDK4/6 inhibitor palbociclib in combination with a PI3K or MEK inhibitor in colorectal cancer. Cancer Biol Ther (2023) 24(1):2223388. doi: 10.1080/15384047.2023.2223388
41. Yu Q, Sun L, Xu Z, Fan L, Du Y. Severe pneumonia caused by Parvimonas micra: a case report. BMC Infect Dis (2021) 21(1):364. doi: 10.1186/s12879-021-06058-y
42. Chang Y, Huang Z, Hou F, Liu Y, Wang L, Wang Z, et al. Parvimonas micra activates the Ras/ERK/c-Fos pathway by upregulating miR-218-5p to promote colorectal cancer progression. J Exp Clin CANC Res (2023) 42(1):13. doi: 10.1186/s13046-022-02572-2
43. Li H, Wang Y, Rong SK, Li L, Chen T, Fan YY, et al. Integrin alpha1 promotes tumorigenicity and progressive capacity of colorectal cancer. Int J Biol Sci (2020) 16(5):815–26. doi: 10.7150/ijbs.37275
44. Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, et al. The bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis (2015) 60(2):208–15. doi: 10.1093/cid/ciu787
45. Grellier N, Suzuki MT, Brot L, Rodrigues AMS, Humbert L, Escoubeyrou K, et al. Impact of IBD-associated dysbiosis on bacterial quorum sensing mediated by acyl-homoserine lactone in human gut microbiota. Int J Mol Sci (2022) 23(23):15404. doi: 10.3390/ijms232315404
46. Cao Y, Wang Z, Yan Y, Ji L, He J, Xuan B, et al. Enterotoxigenic bacteroidesfragilis promotes intestinal inflammation and Malignancy by inhibiting exosome-packaged miR-149-3p. GASTROENTEROLOGY (2021) 161(5):1552–66. doi: 10.1053/j.gastro.2021.08.003
47. Zhou C, Lyu LH, Miao HK, Bahr T, Zhang QY, Liang T, et al. Redox regulation by SOD2 modulates colorectal cancer tumorigenesis through AMPK-mediated energy metabolism. Mol CARCINOGEN (2020) 59(5):545–56. doi: 10.1002/mc.23178
48. Zhang Y, Wang X. Targeting the Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol (2020) 13(1):165. doi: 10.1186/s13045-020-00990-3
49. Peng Y, Xu Y, Zhang X, Deng S, Yuan Y, Luo X, et al. A novel protein AXIN1-295aa encoded by circAXIN1 activates the Wnt/beta-catenin signaling pathway to promote gastric cancer progression. Mol Cancer (2021) 20(1):158. doi: 10.1186/s12943-021-01457-w
50. Guo P, Tian Z, Kong X, Yang L, Shan X, Dong B, et al. FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J Exp Clin CANC Res (2020) 39(1):1–202. doi: 10.1186/s13046-020-01677-w
51. Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J BACTERIOL (2005) 187(15):5330–40. doi: 10.1128/JB.187.15.5330-5340.2005
52. Ghanavati R, Akbari A, Mohammadi F, Asadollahi P, Javadi A, Talebi M, et al. Lactobacillus species inhibitory effect on colorectal cancer progression through modulating the Wnt/beta-catenin signaling pathway. Mol Cell Biochem (2020) 470(1-2):1–13. doi: 10.1007/s11010-020-03740-8
53. Ghanavati R, Asadollahi P, Shapourabadi MB, Razavi S, Talebi M, Rohani M. Inhibitory effects of Lactobacilli cocktail on HT-29 colon carcinoma cells growth and modulation of the Notch and Wnt/β-catenin signaling pathways. Microb PATHOGEN (2020) 139:103829. doi: 10.1016/j.micpath.2019.103829
54. Wei L, Lin J, Chu J, Chen H, Li Q, Peng J. Scutellaria barbata D. Don inhibits colorectal cancer growth via suppression of Wnt/β-catenin signaling pathway. Chin J Integr Med (2017) 23(11):858–63. doi: 10.1007/s11655-017-2775-3
55. Karimi AS, Tafvizi F, Tajabadi EM. Heat-killed probiotic bacteria induce apoptosis of HT-29 human colon adenocarcinoma cell line via the regulation of Bax/Bcl2 and caspases pathway. Hum Exp Toxicol (2019) 38(9):1069–81. doi: 10.1177/0960327119851255
56. Tan M, Xu J, Siddiqui J, Feng F, Sun Y. Depletion of SAG/RBX2 E3 ubiquitin ligase suppresses prostate tumorigenesis via inactivation of the PI3K/AKT/mTOR axis. Mol Cancer (2016) 15(1):81. doi: 10.1186/s12943-016-0567-6
57. Zhang Q, Tang X, Zhou Y, Chen X, Peng K, Jiang R, et al. LINC01060 knockdown inhibits osteosarcoma cell Malignant behaviorsin vitro and tumor growth and metastasisin vivo through the PI3K/Akt signaling. Cancer Biol Ther (2023) 24(1):2198904. doi: 10.1080/15384047.2023.2198904
58. Long J, Pi X. Polyphyllin I promoted melanoma cells autophagy and apoptosis via PI3K/akt/mTOR signaling pathway. BioMed Res Int (2020) 2020:1–9. doi: 10.1155/2020/5149417
59. Zhou J, Jiang YY, Chen H, Wu YC, Zhang L. Tanshinone I attenuates the Malignant biological properties of ovarian cancer by inducing apoptosis and autophagy via the inactivation of PI3K/AKT/mTOR pathway. Cell PROLIFERAT (2020) 53(2):e12739. doi: 10.1111/cpr.12739
60. Zhang X, Yu D, Wu D, Gao X, Shao F, Zhao M, et al. Tissue-resident Lachnospiraceae family bacteria protect against colorectal carcinogenesis by promoting tumor immune surveillance. Cell Host Microbe (2023) 31(3):418–32. doi: 10.1016/j.chom.2023.01.013
61. Long X, Wong CC, Tong L, Chu ESH, Szeto CH, Go MYY, et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol (2019) 4(12):2319–30. doi: 10.1038/s41564-019-0541-3
62. Farooqi AA, Attar R, Xu B. Anticancer and anti-metastatic role of thymoquinone: regulation of oncogenic signaling cascades by thymoquinone. Int J Mol Sci (2022) 23(11):6311. doi: 10.3390/ijms23116311
63. Saeed H, Leibowitz BJ, Zhang L, Yu J. Targeting Myc-driven stress addiction in colorectal cancer. Drug Resist Update (2023) 69:100963. doi: 10.1016/j.drup.2023.100963
64. Mu W, Jia Y, Chen X, Li H, Wang Z, Cheng B. Intracellular Porphyromonas gingivalis Promotes the Proliferation of Colorectal Cancer Cells via the MAPK/ERK Signaling Pathway. Front Cell Infect MI (2020) 10:584798. doi: 10.3389/fcimb.2020.584798
65. Bai X, Wei H, Liu W, Coker OO, Gou H, Liu C, et al. Cigarette smoke promotes colorectal cancer through modulation of gut microbiota and related metabolites. GUT (2022) 71(12):2439–50. doi: 10.1136/gutjnl-2021-325021
66. Kim KH, Park EJ, Jang HJ, Lee SJ, Park CS, Yun BS, et al. 1-carbomethoxy-β-carboline, derived from portulaca oleracea L., ameliorates LPS-mediated inflammatory response associated with MAPK signaling and nuclear translocation of NF-κB. Molecules (Basel Switzerland) (2019) 24(22):4042. doi: 10.3390/molecules24224042
67. Yi S, Jin X, Liu B, Wu P, Xiao W, Chen W. Portulaca oleracea extract reduces gut microbiota imbalance and inhibits colorectal cancer progression via inactivation of the Wnt/β-catenin signaling pathway. Phytomed (Stuttgart) (2022) 105:154279. doi: 10.1016/j.phymed.2022.154279
68. Gerstberger S, Jiang Q, Ganesh K. Metastasis. CELL (2023) 186(8):1564–79. doi: 10.1016/j.cell.2023.03.003
69. Delattre JF, Erdogan ASO, Cohen R, Shi Q, Emile JF, Taieb J, et al. A comprehensive overview of tumour deposits in colorectal cancer: Towards a next TNM classification. Cancer Treat Rev (2022) 103:102325. doi: 10.1016/j.ctrv.2021.102325
70. Ternes D, Karta J, Tsenkova M, Wilmes P, Haan S, Letellier E. Microbiome in colorectal cancer: how to get from meta-omics to mechanism? Trends Microbiol (Regular ed.) (2020) 28(8):698. doi: 10.1016/j.tim.2020.05.013
71. Wei Q, Qian Y, Yu J, Wong CC. Metabolic rewiring in the promotion of cancer metastasis: mechanisms and therapeutic implications. ONCOGENE (2020) 39(39):6139–56. doi: 10.1038/s41388-020-01432-7
72. Ternes D, Tsenkova M, Pozdeev VI, Meyers M, Koncina E, Atatri S, et al. The gut microbial metabolite formate exacerbates colorectal cancer progression. Nat Metab (2022) 4(4):458–75. doi: 10.1038/s42255-022-00558-0
73. Varga J, De Oliveira T, Greten FR. The architect who never sleeps: tumor-induced plasticity. FEBS Lett (2014) 588(15):2422–7. doi: 10.1016/j.febslet.2014.06.019
74. Marconi GD, Fonticoli L, Rajan TS, Pierdomenico SD, Trubiani O, Pizzicannella J, et al. Epithelial-mesenchymal transition (EMT): the type-2 EMT in wound healing, tissue regeneration and organ fibrosis. Cells (Basel Switzerland) (2021) 10(7):1587. doi: 10.3390/cells10071587
75. Mitra A, Mishra L, Li SEMT. CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget (2015) 6(13):10697–711. doi: 10.18632/oncotarget.4037
76. Kim HS, Lee SI, Choi YR, Kim J, Eun JW, Song KS, et al. GNAQ-regulated ZO-1 and ZO-2 act as tumor suppressors by modulating EMT potential and tumor-repressive microenvironment in lung cancer. Int J Mol Sci (2023) 24(10):8801. doi: 10.3390/ijms24108801
77. Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell (2012) 22(6):709–24. doi: 10.1016/j.ccr.2012.10.012
78. Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell (2012) 22(6):725–36. doi: 10.1016/j.ccr.2012.09.022
79. Meda LLT, Landraud L, Petracchini S, Descorps-Declere S, Perthame E, Nahori MA, et al. The cnf1 gene is associated with an expanding Escherichia coli ST131 H30Rx/C2 subclade and confers a competitive advantage for gut colonization. GUT Microbes (2022) 14(1):2121577. doi: 10.1080/19490976.2022.2121577
80. Fabbri A, Travaglione S, Rosadi F, Ballan G, Maroccia Z, Giambenedetti M, et al. The Escherichia coli protein toxin cytotoxic necrotizing factor 1 induces epithelial mesenchymal transition. Cell Microbiol (2020) 22(2):e13138. doi: 10.1111/cmi.13138
81. Lin H, Yu Y, Zhu L, Lai N, Zhang L, Guo Y, et al. Implications of hydrogen sulfide in colorectal cancer: Mechanistic insights and diagnostic and therapeutic strategies. Redox Biol (2023) 59:102601. doi: 10.1016/j.redox.2023.102601
82. Wolf PG, Cowley ES, Breister A, Matatov S, Lucio L, Polak P, et al. Diversity and distribution of sulfur metabolic genes in the human gut microbiome and their association with colorectal cancer. MICROBIOME (2022) 10(1):64. doi: 10.1186/s40168-022-01242-x
83. Cirino G, Szabo C, Papapetropoulos A. Physiological roles of hydrogen sulfide in mammalian cells, tissues, and organs. Physiol Rev (2023) 103(1):31–276. doi: 10.1152/physrev.00028.2021
84. Ascenção K, Dilek N, Augsburger F, Panagaki T, Zuhra K, Szabo C. Pharmacological induction of mesenchymal-epithelial transition via inhibition of H2S biosynthesis and consequent suppression of ACLY activity in colon cancer cells. Pharmacol Res (2021) 165:105393. doi: 10.1016/j.phrs.2020.105393
85. Phillips CM, Zatarain JR, Nicholls ME, Porter C, Widen SG, Thanki K, et al. Upregulation of cystathionine-β-synthase in colonic epithelia reprograms metabolism and promotes carcinogenesis. Cancer Res (Chicago Ill.) (2017) 77(21):5741–54. doi: 10.1158/0008-5472.CAN-16-3480
86. Blachier F, Andriamihaja M, Larraufie P, Ahn E, Lan A, Kim E. Production of hydrogen sulfide by the intestinal microbiota and epithelial cells and consequences for the colonic and rectal mucosa. Am J physiology: Gastrointest liver Physiol (2021) 320(2):G125–35. doi: 10.1152/ajpgi.00261.2020
87. Wang X, Li Y, Chen W, Shi H, Eren AM, Morozov A, et al. Transcriptome-wide reprogramming of N6-methyladenosine modification by the mouse microbiome. Cell Res (2018) 29(2):167–70. doi: 10.1038/s41422-018-0127-2
88. Jabs S, Biton A, Becavin C, Nahori MA, Ghozlane A, Pagliuso A, et al. Impact of the gut microbiota on the m(6)A epitranscriptome of mouse cecum and liver. Nat Commun (2020) 11(1):1344. doi: 10.1038/s41467-020-15126-x
89. Patil DP, Pickering BF, Jaffrey SR. Reading m(6)A in the Transcriptome: m(6)A-Binding Proteins. Trends Cell Biol (2018) 28(2):113–27. doi: 10.1016/j.tcb.2017.10.001
90. Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med (2017) 23(11):1369–76. doi: 10.1038/nm.4416
91. Chen M, Wei L, Law CT, Tsang FHC, Shen J, Cheng CLH, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. HEPATOLOGY (2018) 67(6):2254–70. doi: 10.1002/hep.29683
92. Yin H, Zhang X, Yang P, Zhang X, Peng Y, Li D, et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat Commun (2021) 12(1):1394. doi: 10.1038/s41467-021-21514-8
93. Chen S, Zhang L, Li M, Zhang Y, Sun M, Wang L, et al. Fusobacterium nucleatum reduces METTL3-mediated m(6)A modification and contributes to colorectal cancer metastasis. Nat Commun (2022) 13(1):1248. doi: 10.1038/s41467-022-28913-5
94. Xu C, Fan L, Lin Y, Shen W, Qi Y, Zhang Y, et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. GUT Microbes (2021) 13(1):1980347. doi: 10.1080/19490976.2021.1980347
95. Yin H, Miao Z, Wang L, Su B, Liu C, Jin Y, et al. Fusobacterium nucleatum promotes liver metastasis in colorectal cancer by regulating the hepatic immune niche and altering gut microbiota. Aging (2022) 14(4):1941–58. doi: 10.18632/aging.203914
96. Zhang Y, Zhang L, Zheng S, Li M, Xu C, Jia D, et al. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-κB/ICAM1 axis. GUT Microbes (2022) 14(1):2038852. doi: 10.1080/19490976.2022.2038852
97. Arwert EN, Harney AS, Entenberg D, Wang Y, Sahai E, Pollard JW, et al. A unidirectional transition from migratory to perivascular macrophage is required for tumor cell intravasation. Cell Rep (2018) 23(5):1239–48. doi: 10.1016/j.celrep.2018.04.007
98. Cabrera RM, Mao S, Surve CR, Condeelis JS, Segall JE. A novel neuregulin - jagged1 paracrine loop in breast cancer transendothelial migration. Breast Cancer Res (2018) 20(1):24. doi: 10.1186/s13058-018-0960-8
99. Song X, An Y, Chen D, Zhang W, Wu X, Li C, et al. Microbial metabolite deoxycholic acid promotes vasculogenic mimicry formation in intestinal carcinogenesis. Cancer Sci (2022) 113(2):459–77. doi: 10.1111/cas.15208
100. Qiao K, Liu Y, Xu Z, Zhang H, Zhang H, Zhang C, et al. RNA m6A methylation promotes the formation of vasculogenic mimicry in hepatocellular carcinoma via Hippo pathway. ANGIOGENESIS (2021) 24(1):83–96. doi: 10.1007/s10456-020-09744-8
101. Strilic B, Offermanns S. Intravascular survival and extravasation of tumor cells. Cancer Cell (2017) 32(3):282–93. doi: 10.1016/j.ccell.2017.07.001
102. Alsabbagh R, Ahmed M, Alqudah M, Hamoudi R, Harati R. Insights into the molecular mechanisms mediating extravasation in brain metastasis of breast cancer, melanoma, and lung cancer. CANCERS (2023) 15(8):2258. doi: 10.3390/cancers15082258
103. Tung SY, Chang SF, Chou MH, Huang WS, Hsieh YY, Shen CH, et al. CXC chemokine ligand 12/stromal cell-derived factor-1 regulates cell adhesion in human colon cancer cells by induction of intercellular adhesion molecule-1. J BioMed Sci (2012) 19(1):91. doi: 10.1186/1423-0127-19-91
104. Martin TD, Patel RS, Cook DR, Choi MY, Patil A, Liang AC, et al. The adaptive immune system is a major driver of selection for tumor suppressor gene inactivation. Sci (Am Assoc Adv Sci) (2021) 373(6561):1327–35. doi: 10.1126/science.abg5784
105. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the fap2 protein of fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. IMMUNITY (2015) 42(2):344–55. doi: 10.1016/j.immuni.2015.01.010
106. Gur C, Maalouf N, Shhadeh A, Berhani O, Singer BB, Bachrach G, et al. Fusobacterium nucleatum supresses anti-tumor immunity by activating CEACAM1. ONCOIMMUNOLOGY (2019) 8(6):e1581531. doi: 10.1080/2162402X.2019.1581531
107. Zhang Q, Ma C, Duan Y, Heinrich B, Rosato U, Diggs LP, et al. Gut microbiome directs hepatocytes to recruit MDSCs and promote cholangiocarcinoma. Cancer Discovery (2021) 11(5):1248–67. doi: 10.1158/2159-8290.CD-20-0304
108. Chu X, Tian W, Wang Z, Zhang J, Zhou R. Co-inhibition of TIGIT and PD-1/PD-L1 in cancer immunotherapy: mechanisms and clinical trials. Mol Cancer (2023) 22(1):93. doi: 10.1186/s12943-023-01800-3
109. Jiang SS, Xie YL, Xiao XY, Kang ZR, Lin XL, Zhang L, et al. Fusobacterium nucleatum-derived succinic acid induces tumor resistance to immunotherapy in colorectal cancer. Cell Host Microbe (2023) 31(5):781–97. doi: 10.1016/j.chom.2023.04.010
110. Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun (2016) 7(1):12150. doi: 10.1038/ncomms12150
111. Peuker K, Strigli A, Tauriello DVF, Hendricks A, von Schönfels W, Burmeister G, et al. Microbiota-dependent activation of the myeloid calcineurin-NFAT pathway inhibits B7H3- and B7H4-dependent anti-tumor immunity in colorectal cancer. Immun (Cambridge Mass.) (2022) 55(4):701–17. doi: 10.1016/j.immuni.2022.03.008
112. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. NATURE (2019) 565(7741):600–5. doi: 10.1038/s41586-019-0878-z
113. Daillère R, Vétizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. IMMUNITY (2016) 45(4):931–43. doi: 10.1016/j.immuni.2016.09.009
114. Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K, et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. IMMUNITY (2019) 51(2):285–97. doi: 10.1016/j.immuni.2019.06.002
115. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. SCIENCE (2020) 369(6510):1481–9. doi: 10.1126/science.abc3421
116. Hasko G, Kuhel DG, Nemeth ZH, Mabley JG, Stachlewitz RF, Virág L, et al. Inosine inhibits inflammatory cytokine production by a posttranscriptional mechanism and protects against endotoxin-induced shock. J Immunol (2000) 164(2):1013–9. doi: 10.4049/jimmunol.164.2.1013
117. He B, Hoang TK, Wang T, Ferris M, Taylor CM, Tian X, et al. Resetting microbiota by Lactobacillus reuteri inhibits T reg deficiency-induced autoimmunity via adenosine A2A receptors. J Exp Med (2017) 214(1):107–23. doi: 10.1084/jem.20160961
118. Csóka B, Himer L, Selmeczy Z, Vizi ES, Pacher P, Ledent C, et al. Adenosine A2A receptor activation inhibits T helper 1 and T helper 2 cell development and effector function. FASEB J (2008) 22(10):3491–9. doi: 10.1096/fj.08-107458
119. Ohta A, Gorelik E, Prasad SJ, Ronchese F, Lukashev D, Wong MKK, et al. A2A adenosine receptor protects tumors from antitumor T cells. Proc Natl Acad Sci - PNAS (2006) 103(35):13132–7. doi: 10.1073/pnas.0605251103
120. Cekic C, Linden J. Adenosine A2A receptors intrinsically regulate CD8+ T cells in the tumor microenvironment. Cancer Res (2014) 74(24):7239–49. doi: 10.1158/0008-5472.CAN-13-3581
121. Lioux T, Mauny MA, Lamoureux A, Bascoul N, Hays M, Vernejoul F, et al. Design, synthesis, and biological evaluation of novel cyclic adenosine-inosine monophosphate (cAIMP) analogs that activate stimulator of interferon genes (STING). J Med Chem (2016) 59(22):10253–67. doi: 10.1021/acs.jmedchem.6b01300
122. Routy B, Lenehan JG, Miller WH, Jamal R, Messaoudene M, Daisley BA, et al. Fecal microbiota transplantation plus anti-PD-1 immunotherapy in advanced melanoma: a phase I trial. Nat Med (2023) 29(8):2121–32. doi: 10.1038/s41591-023-02453-x
123. Sommer F, Bäckhed F. The gut microbiota — masters of host development and physiology. Nat Rev Microbiol (2013) 11(4):227–38. doi: 10.1038/nrmicro2974
124. Breyner NM, Michon C, de Sousa CS, Boas PBV, Chain F, Azevedo VA, et al. Microbial Anti-Inflammatory Molecule (MAM) from Faecalibacterium prausnitzii Shows a Protective Effect on DNBS and DSS-Induced Colitis Model in Mice through Inhibition of NF-κB Pathway. Front Microbiol (2017) 8:114. doi: 10.3389/fmicb.2017.00114
125. Corrêa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MAR. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol (2016) 5(4):e73. doi: 10.1038/cti.2016.17
126. Gurav A, Sivaprakasam S, Bhutia YD, Boettger T, Singh N, Ganapathy V. Slc5a8, a Na+-coupled high-affinity transporter for short-chain fatty acids, is a conditional tumour suppressor in colon that protects against colitis and colon cancer under low-fibre dietary conditions. Biochem J (2015) 469(2):267–78. doi: 10.1042/BJ20150242
127. Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Costa GD, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med (2016) 22(6):598–605. doi: 10.1038/nm.4102
128. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. CELL (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
129. Gabriele Multhoff MMAJ. Chronic inflammation in cancer development. Front Immunol (2012) 2:98. doi: 10.3389/fimmu.2011.00098
130. Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. CELL (2010) 141(1):39–51. doi: 10.1016/j.cell.2010.03.014
131. Reis S, Da CL, Peluzio M. Intestinal microbiota and colorectal cancer: changes in the intestinal microenvironment and their relation to the disease. J Med Microbiol (2019) 68(10):1391–407. doi: 10.1099/jmm.0.001049
132. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell (2010) 140(6):883–99. doi: 10.1016/j.cell.2010.01.025
133. DeNardo DG, Andreu P, Coussens LM. Interactions between lymphocytes and myeloid cells regulate pro- versus anti-tumor immunity. Cancer METAST Rev (2010) 29(2):309–16. doi: 10.1007/s10555-010-9223-6
134. Meitei HT, Jadhav N, Lal G. CCR6-CCL20 axis as a therapeutic target for autoimmune diseases. AUTOIMMUN Rev (2021) 20(7):102846. doi: 10.1016/j.autrev.2021.102846
135. Jayasingam SD, Citartan M, Thang TH, Mat Zin AA, Ang KC, Ch'Ng ES. Evaluating the polarization of tumor-associated macrophages into M1 and M2 phenotypes in human cancer tissue: technicalities and challenges in routine clinical practice. Front Oncol (2020) 9:1512. doi: 10.3389/fonc.2019.01512
136. Zhang Q, Huang F, Yao Y, Wang J, Wei J, Wu Q, et al. Interaction of transforming growth factor-β-Smads/microRNA-362-3p/CD82 mediated by M2 macrophages promotes the process of epithelial-mesenchymal transition in hepatocellular carcinoma cells. Cancer Sci (2019) 110(8):2507–19. doi: 10.1111/cas.14101
137. DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol (2019) 19(6):369–82. doi: 10.1038/s41577-019-0127-6
138. Ivanov II, Frutos RL, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe (2008) 4(4):337–49. doi: 10.1016/j.chom.2008.09.009
139. Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. CELL (2015) 163(2):367–80. doi: 10.1016/j.cell.2015.08.058
140. Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci (2016) 113(50):E8141–50. doi: 10.1073/pnas.1617460113
141. Brennan CA, Clay SL, Lavoie SL, Bae S, Lang JK, Fonseca-Pereira D, et al. Fusobacterium nucleatum drives a pro-inflammatory intestinal microenvironment through metabolite receptor-dependent modulation of IL-17 expression. GUT Microbes (2021) 13(1):1987780. doi: 10.1080/19490976.2021.1987780
142. Calcinotto A, Brevi A, Chesi M, Ferrarese R, Perez LG, Grioni M, et al. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat Commun (2018) 9(1):4832. doi: 10.1038/s41467-018-07305-8
143. Overacre-Delgoffe AE, Bumgarner HJ, Cillo AR, Burr AHP, Tometich JT, Bhattacharjee A, et al. Microbiota-specific T follicular helper cells drive tertiary lymphoid structures and anti-tumor immunity against colorectal cancer. IMMUNITY (2021) 54(12):2812–24. doi: 10.1016/j.immuni.2021.11.003
144. Liu Q, Zhang H, Jiang X, Qian C, Liu Z, Luo D. Factors involved in cancer metastasis: a better understanding to "seed and soil" hypothesis. Mol Cancer (2017) 16(1):176. doi: 10.1158/1538-7445.EPSO16-B17
145. Akhtar M, Haider A, Rashid S, Al-Nabet ADMH. Paget’s “Seed and soil” Theory of cancer metastasis: an idea whose time has come. Adv ANAT Pathol (2019) 26(1):69–74. doi: 10.1097/PAP.0000000000000219
146. Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer (2017) 17(5):302–17. doi: 10.1038/nrc.2017.6
147. McCOY WC, Mason JM. Enterococcal endocarditis associated with carcinoma of the sigmoid; report of a case. J Med Assoc State Ala (1951) 21(6):162–6.
148. Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res (2012) 22(2):292–8. doi: 10.1101/gr.126573.111
149. Uribe-Herranz M, Rafail S, Beghi S, Gil-de-Gómez L, Verginadis I, Bittinger K, et al. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy-induced antitumor immune response. J Clin Invest (2020) 130(1):466–79. doi: 10.1172/JCI124332
150. Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med (2019) 25(4):679–89. doi: 10.1038/s41591-019-0406-6
151. Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med (2019) 25(4):667–78. doi: 10.1038/s41591-019-0405-7
152. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. SCIENCE (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
153. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin JM, Morrison RM, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. SCIENCE (2021) 371(6529):595–602. doi: 10.1126/science.abf3363
154. Spencer CN, McQuade JL, Gopalakrishnan V, McCulloch JA, Vetizou M, Cogdill AP, et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. SCIENCE (2021) 374(6575):1632–40. doi: 10.1126/science.aaz7015
155. Navarro M, Nicolas A, Ferrandez A, Lanas A. Colorectal cancer population screening programs worldwide in 2016: An update. World J GASTROENTERO (2017) 23(20):3632–42. doi: 10.3748/wjg.v23.i20.3632
156. Wang M, Rousseau B, Qiu K, Huang G, Zhang Y, Su H, et al. Killing tumor-associated bacteria with a liposomal antibiotic generates neoantigens that induce anti-tumor immune responses. Nat Biotechnol (2023). doi: 10.1038/s41587-023-01957-8
Keywords: gut microbiota, colorectal cancer, metastasis, tumor progression, immune evasion
Citation: Yu S, Wang S, Xiong B and Peng C (2023) Gut microbiota: key facilitator in metastasis of colorectal cancer. Front. Oncol. 13:1270991. doi: 10.3389/fonc.2023.1270991
Received: 08 August 2023; Accepted: 09 October 2023;
Published: 30 October 2023.
Edited by:
Diogo Alpuim Costa, CUF Oncologia, PortugalReviewed by:
Ana Raimundo, CUF Institute of Oncology, PortugalCopyright © 2023 Yu, Wang, Xiong and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunwei Peng, d2h1cGVuZ2N3QHdodS5lZHUuY24=; Bin Xiong, YmlueGlvbmcxOTYxQHdodS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.