- 1Department of Clinical Pharmacy, School of Pharmacy, Taipei Medical University, Taipei, Taiwan
- 2Department of General Surgery, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan
- 3Department of Anesthesiology, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan
- 4Department of Anesthesiology, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan
- 5Master Program in Clinical Genomics and Proteomics, School of Pharmacy, Taipei Medical University, Taipei, Taiwan
- 6Department of Pharmacy, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan
- 7Integrative Research Center for Critical Care, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan
Introduction: Cytoreductive surgery (CRS) and hyperthermic intraperitoneal chemotherapy (HIPEC) are considered for patients with peritoneal metastasis (PM). However, patients selection that relies on conventional prognostic factors is not yet optimal. In this study, we performed whole exome sequencing (WES) to establish tumor molecular characteristics and expect to identify prognosis profiles for PM management.
Methods: In this study, blood and tumor samples were collected from patients with PM before HIPEC. Tumor molecular signatures were determined using WES. Patient cohort was divided into responders and non-responders according to 12-month progression-free survival (PFS). Genomic characteristics between the two cohorts were compared to study potential targets.
Results: In total, 15 patients with PM were enrolled in this study. Driver genes and enriched pathways were identified from WES results. AGAP5 mutation was found in all responders. This mutation was significantly associated with better OS (p = 0.00652).
Conclusions: We identified prognostic markers that might be useful to facilitate decision-making before CRS/HIPEC.
1 Introduction
Peritoneal metastases (PMs) are aggressive advanced-stage manifestations that predominantly originate from intra-abdominal malignancies such as gastric, colorectal, appendiceal, and ovarian cancers. Patients with PM showed limited overall survival (OS) of approximately 3.1 months (1). Cytoreductive surgery and systemic chemotherapy are still the standard treatments. Over the last two decades, marginal efficacy of systemic palliative chemotherapy in PM management (2–4) has led to an alternative approach: cytoreductive surgery (CRS) with hyperthermic intraperitoneal chemotherapy (HIPEC) (5).
Poor distribution of systemic chemotherapy to the peritoneal cavity is due to the plasma-peritoneal barrier and high interstitial pressure of tumors. Therefore, intraperitoneal infusion offers preferentially higher efficacy in delivering chemotherapeutic drugs to tumor nodules while localizing the diffusion of these drugs in the peritoneum to decrease their systemic toxicity (5, 6). Furthermore, hyperthermia with the range of temperature from 41 to 43°C augments the cytotoxic effect of intraperitoneal chemotherapeutic agents via DNA repair inhibition, promotion of heat shock proteins, immune cell recruitment (e.g., natural killer cells, dendritic cells, T cells) and apoptosis (7, 8).
While systemic chemotherapy increases the median OS of patients with PM to roughly 1 year (9-15 months) (6, 9, 10), some results from CRS/HIPEC have shown superior efficacy to improve the survival duration. Median survival times varied according to the origin of the primary cancer. For colorectal origin, the median OS of PM patients receiving CRS/HIPEC was 32-41 months (11, 12); patients with epithelial ovarian cancer achieved a median OS of 45.7 months (13); and the duration of survival for patients with mesothelioma was up to 53 months (14).
Despite these promising results, a considerable group of patients were reported to have relapsed within 12 months following CRS/HIPEC (15–17). In addition, recent randomized clinical trials failed to confirm the favorable outcome of CRS/HIPEC over mono-modal systemic chemotherapy or palliative surgery in colorectal PM and recurrent epithelial ovarian cancer (18, 19). The controversial results highlighted shortcomings of current guidelines for CRS/HIPEC. First, the protocols vary across institutes regarding open or closed abdominal perfusion, chemotherapeutic drugs used, administration duration, and the optimal temperature range, making it difficult to extrapolate results to all patients (20, 21). Second, conventional prognostic factors for CRS/HIPEC prominently depend on clinical and pathological data such as patient’s clinical performance status (Eastern Cooperative Oncology Group (ECOG) score, peritoneal carcinomatosis index (PCI), histologic tumor grade, lymph node status, signet ring cell differentiation, the completeness of cytoreduction, and the experience of the operative team (22, 23). Nonetheless, the biological heterogeneity of peritoneal tumors in different patients raises concerns that the generally standardized regimen of HIPEC might not be effective in all cases and plausibly necessitates a personalized approach to optimize clinical decision-making. Thus, insights of tumor molecular characteristics are of great value to improve patient selection for CRS/HIPEC.
Recently, high-throughput sequencing technology has shed light on genomic profiles in individual cancer therapy. Indeed, genomic application for prognostic significance after CRS/HIPEC therapy was reported (24–26). However, the current information is still not able to be translated into clinical applications (27). Herein, we sought to unravel the tumor-specific genetic alterations, thereby providing predictive genetic-guided factors associated with HIPEC responsiveness.
2 Materials and methods
2.1 Patient cohort
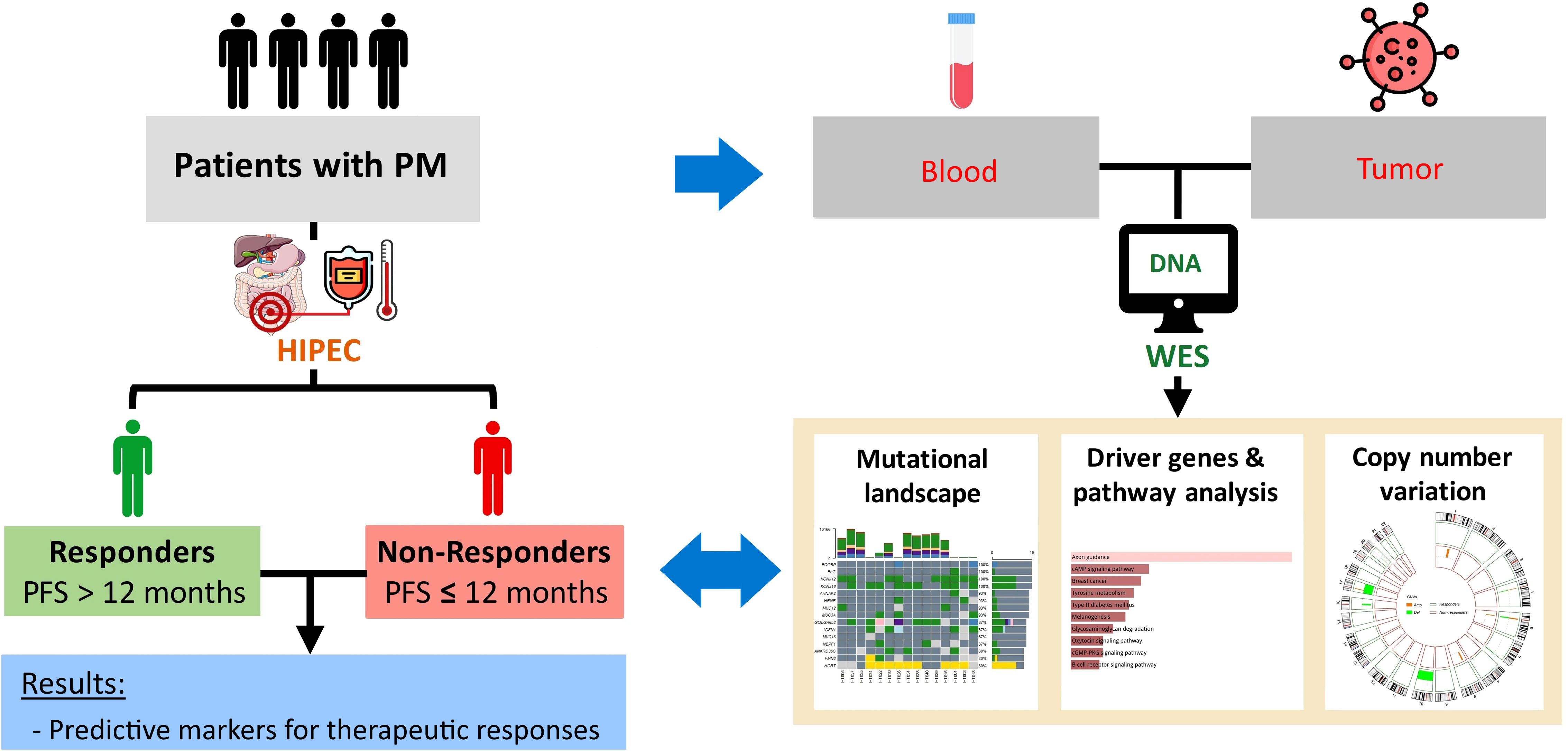
This study was conducted with the approval of the Joint Institutional Review Board of Taipei Medical University (approval no. N201807067). The study design is demonstrated in Figure 1. Patients diagnosed with AJCC (American Joint Committee on Cancer) stage IV peritoneal metastasis based on computerized tomography (CT) scans, no comorbidities such as heart disease, epilepsy, severe kidney, or liver dysfunction, aged 20–75 years, and with a body weight of 50–100 kilograms were recruited for this study. All eligible patients have signed informed consent forms. After surgery, patients with at least 12 months of follow-up were stratified into two groups based on the 12-month progression-free survival (PFS) (or early recurrence): responders and non-responders.
PFS was defined as the time interval from CRS/HIPEC to the first occurrence of the following events: local or systemic recurrence evaluated by imaging assessment and/or histological confirmation by a laparotomy/laparoscopy; and death from any cause. OS was also investigated from the date of surgery to the date of death from any cause or censored on the date of last follow-up for alive patients.
2.2 HIPEC procedure and specimen collection
HIPEC was performed with closed technique for 1 to 2 hours. The PerformerHT (RanD, Medolla, Italy) was used to assure the target temperature ranging from 42.0 to 43.0°C. Chemotherapeutic agents were used in HIPEC as monotherapy or in combination according to primary cancer types, including mitomycin C, docetaxel, cisplatin, and others (etoposide, doxorubicin, mesna, Ifosfamide).
Tumor and matched peripheral blood samples of all patients were collected prior to the HIPEC regimen in the operating room. The collected tumor tissues were stored in RNAlater and kept in liquid nitrogen. The tumor tissues were ground with tissue homogenizers for DNA isolation. Peripheral blood mononuclear cells (PBMCs) were extracted from peripheral blood using Ficoll. DNA was isolated from the tumor tissue and PBMCs with Allprep DNA/RNA kit (Qiagen) according to the manufacturer’s protocol. The DNA concentration per sample was assessed by absorbance measures using the NanoDrop Spectrophotometer (Thermo Scientific, Waltham, MA, USA) and by fluorometric method with the Qubit 2.0 Fluorometer (Thermo Scientific).
2.3 Whole exome sequencing and bioinformatic analysis
WES was performed with the Illumina NovaSeq 6000 platform at 150-bp pair-end reads. The average sequencing coverage was 200× for the samples. To assess raw sequencing reads, quality control was performed using FastQC and trimming was performed using Trimmomatic. The Genome Analysis Toolkit version 4.0 (GATK4) was applied for variant calling according to the GATK best practice (28).
The processed reads were mapped and aligned to the reference human genome (GRCh38/hg38) using Burrows-Wheeler Aligner (BWA) (29). BAM files were sorted using SAMtools; polymerase chain reaction (PCR) duplicates were omitted with MarkDuplicates (Picard). Somatic variants (single nucleotide variants (SNVs) and insertions/deletions (INDELs)) were called using GATK4 Mutect2. Subsequently, Ensembl’s Variant Effect Predictor (version 104.3) was used to annotate the variants (30).
The tumor mutational burden (TMB) was calculated as the total number of non-synonymous somatic mutations per mega-base (Mb) of WES data. Mutational data were then analyzed and visualized using the Maftools package (31). Driver genes were identified from the oncodriveCLUST pipeline integrated in the Maftools package. This tool takes into account genes with mutations that are clustered in particular regions of the protein, which are potentially functionally deleterious (32). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was subsequently carried out using the web-based tool, Enrichr, for this gene list (33).
2.4 Copy number variation analysis
To call the allele-specific copy number for each sample, BAM files were subjected to the Sequenza pipeline, and the GRCh38/hg38 human reference genome assembly was used for annotation (34). Segmentation output files from Sequenza were used as the input for Genome Identification of Significant Targets in Cancer (GISTIC) version 2.0 to detect CNV events at gene level and identify regions with significant amplification or deletion in each cohort (i.e., responders and non-responders) (35). To avoid gender bias, the X and Y chromosomes were excluded from the CNV analysis. Significantly recurrent focal CNV regions in each cohort were determined by GISTIC 2.0 with a false discovery rate (FDR) q-value of < 0.25. The CNV event of a gene was defined as amplification, gain, loss, or deletion based on the GISTIC score.
2.5 Statistical analysis
Pre-, peri-, and postoperative clinicopathological data were compared between responders and non-responders. Chi-squared test or Fisher’s exact test was performed for categorical variables. For continuous variables, Student’s t-test and Mann Whitney-U test were respectively used for normally distributed and non-normally distributed data. Survival analyses were conducted using the Kaplan-Meier method, and the log-rank test was used to compare survival curves across different groups of interest. A p value of < 0.05 was considered statistically significant. All statistical analyses were performed in R version 4.0.5.
3 Results
3.1 Patient characteristics
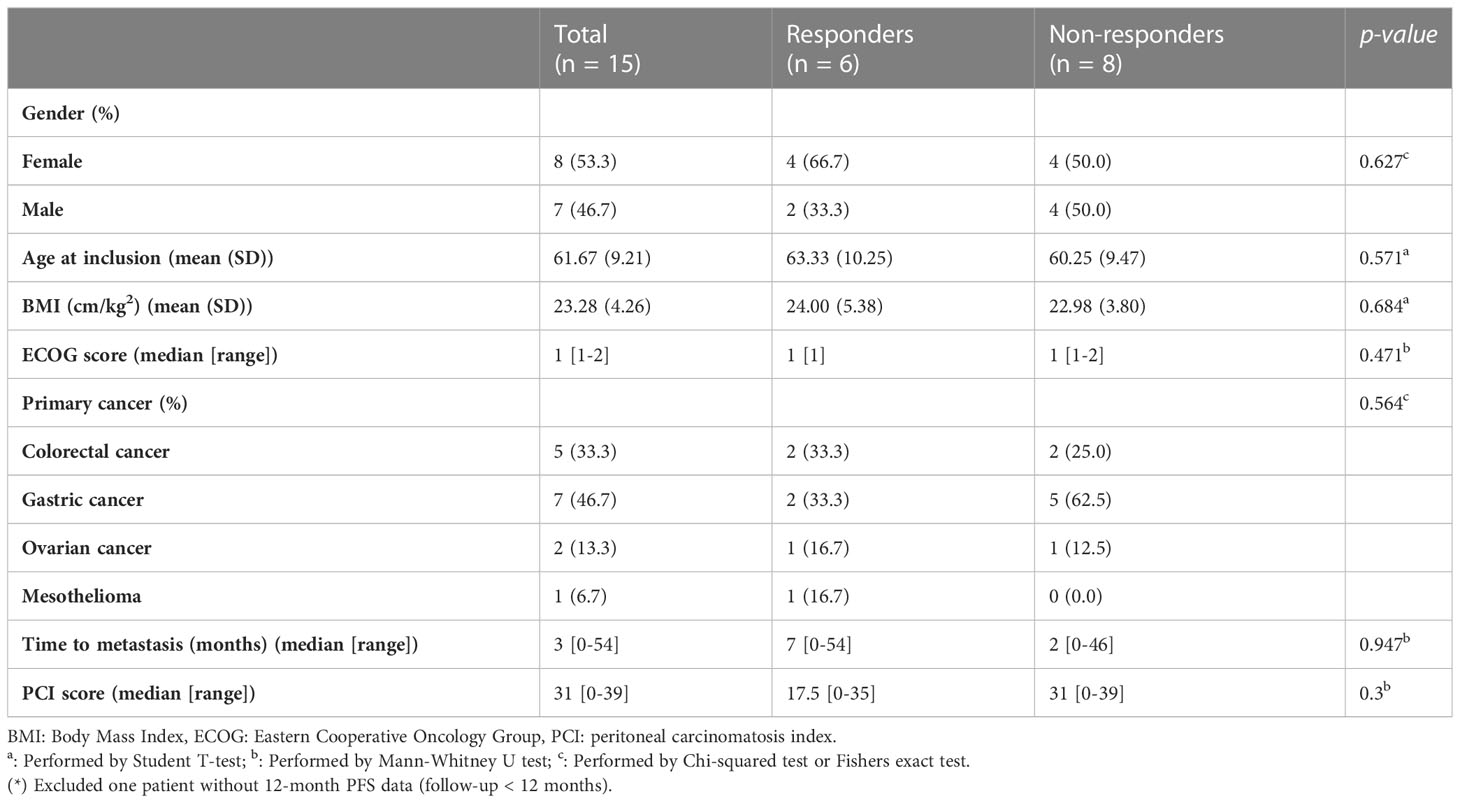
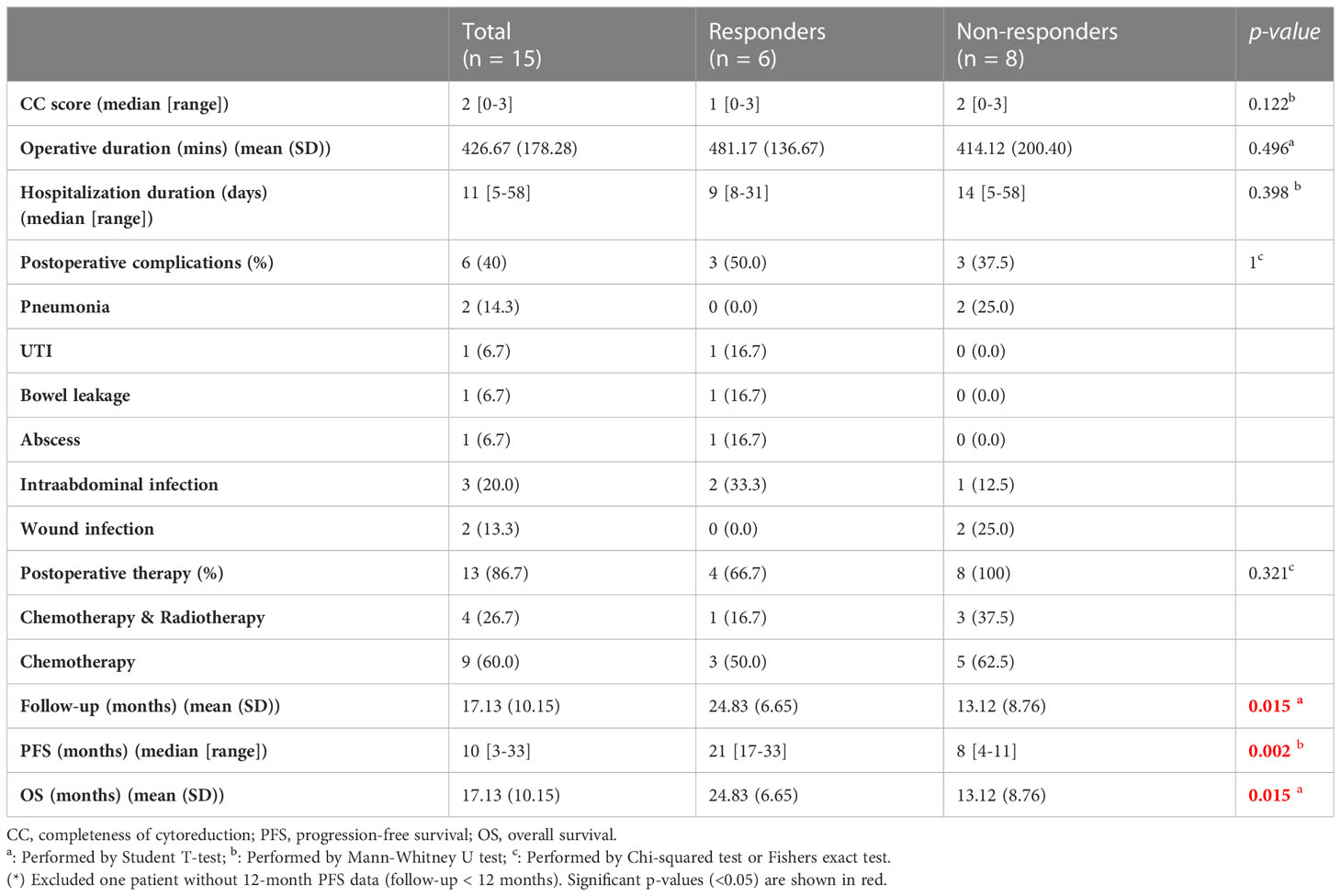
In total, 15 patients with PM receiving CRS/HIPEC at Wanfang Hospital (Taipei, Taiwan) were enrolled in this study. Preoperative-, intraoperative-, and postoperative-related clinical data were recorded for analysis (Tables 1, 2). Five patients (33.3%) had primary colorectal cancer (CRC), seven patients (46.7%) had primary gastric cancer, two patients (13.3%) had primary ovarian cancer, and one patient (6.7%) had mesothelioma. This cohort was divided into responders (n = 6) and non-responders (n = 8) based on their 12-month PFS after CRS/HIPEC. One patient was excluded due to loss of follow-up at three months after CRS/HIPEC. No statistically significant difference was observed between the two groups in any of the patients’ baseline or operative characteristics (Tables 1, 2). As to clinical outcomes, responders achieved a median PFS of 21 (range, 17–33) months and an average OS of 24.83 months, whereas non-responders had a median PFS of 8 (range, 4–11) months and an average OS of 13.12 months (p = 0.002 and 0.015, respectively).
3.2 Somatic mutational profiles in tumor samples of all patients (n = 15)
There were 15 tumor and matched blood samples obtained prior to the HIPEC regimen for WES. Missense mutations were the highest among the mutational classifications (Figures 2A, B), and SNVs dominated all mutational types (Figure 2C). The T>C and C>T transitions constituted the highest proportion of SNVs, at approximately 50% of events (Figure 2D). Overall, the median total number of mutations per tumor sample was 6350 (range, 253–10,166) (Figure 2E). The top 10 genes with the highest rate of mutation were MUC16 (87%), FLG (100%), MUC3A (93%), AHNAK2 (93%), HRNR (93%), MUC12 (93%), FCGBP (100%), IGFN1 (87%), KCNJ18 (100%), and KCNJ12 (100%) (Figure 2F). Alterations in FCGBP, FLG, KCNJ12, and KCNJ18 were observed in all tumor samples. Detailed classifications of variants harbored in the top 15 mutated genes are presented in a waterfall plot (Figure 2G).
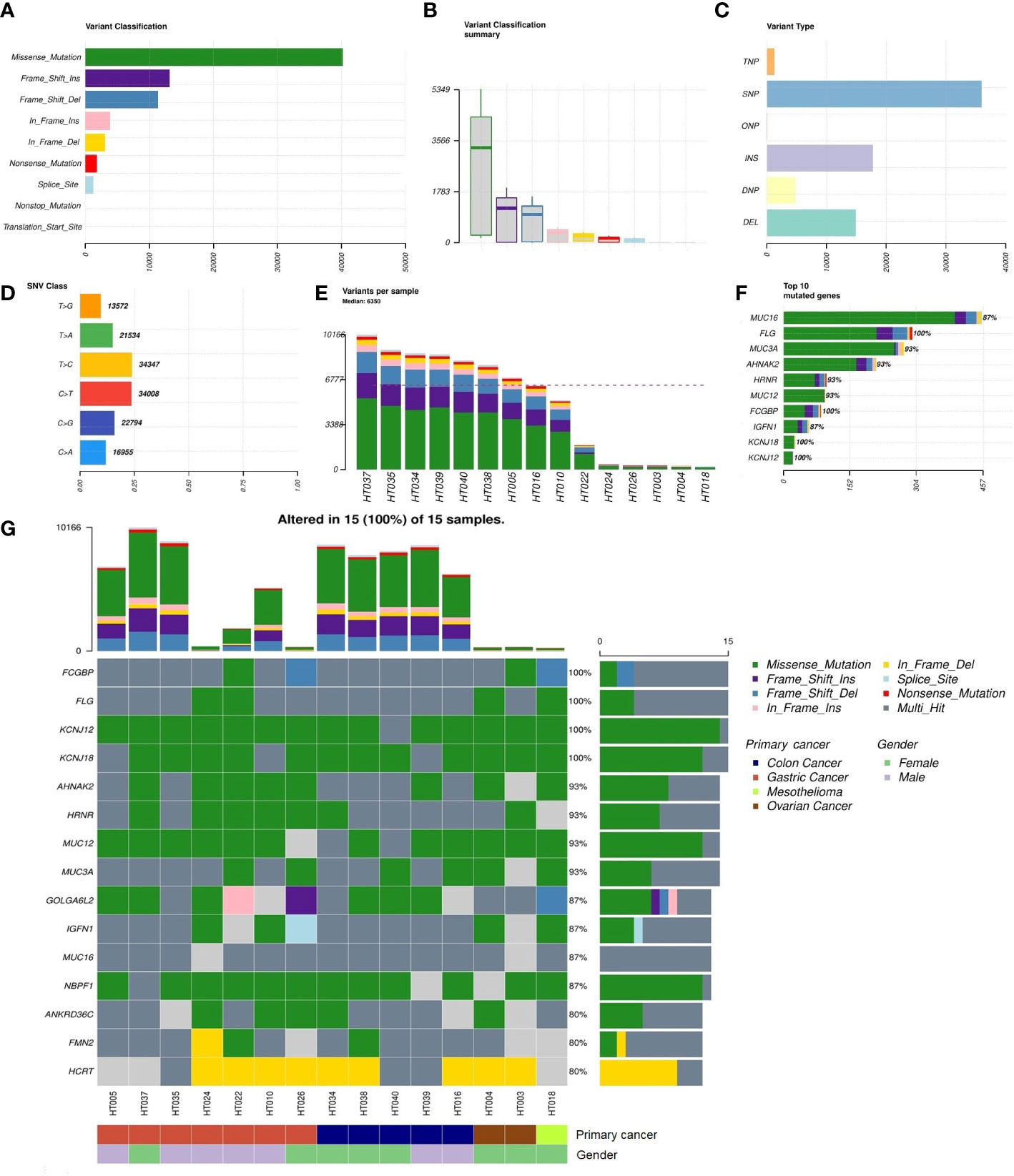
Figure 2 Summary of the non-synonymous mutational landscape (n = 15 patients). (A, B) Classifications of non-synonymous mutations (i.e., single-nucleotide variants (SNVs) and insertions/deletions (INDELs)). (C) Types of non-synonymous mutations. (D) Frequencies of transition and transversion events in SNVs. (E) Total mutational loads in each tumor sample. (F) The top 10 genes with the highest rate of mutation. (G) Oncoplot showing the top 15 most frequently mutated genes with detailed annotations in each tumor sample.
3.3 Driver genes and pathway analysis across all tumor samples (n = 15)
In total, 1904 potential driver genes were annotated in this cohort using OncodriveCLUST (p < 0.05, FDR < 0.05) (Table S1). These genes were used as input for the functional gene enrichment analysis (i.e., the KEGG pathway analysis) in the Enrichr web-based tool, which revealed significantly enriched pathways related to signaling, metabolism, tumorigenesis, and immune response (Figures 3A, B). Among those, ‘axon guidance’ was a significantly enriched pathway in this cohort. Molecules involved in this pathway are listed in Table S2. As shown in the Figure 3C, mitogen-activated protein kinase 1 (MAPK1) and MAPK3 were the most frequently clustered genes in the top annotated enriched pathways.
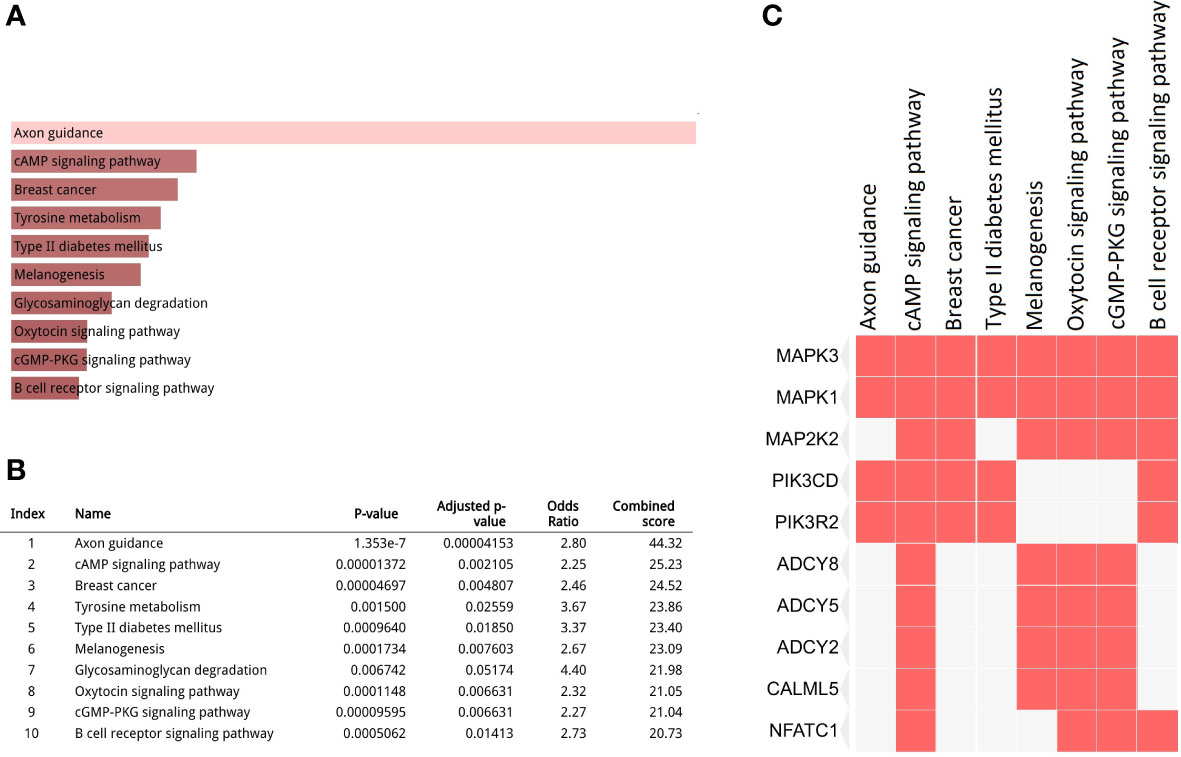
Figure 3 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis for the driver gene list. (A, B) Top 10 enriched pathways (p < 0.05, false discovery rate (FDR) < 0.05); (C) Clustergram representing genes involved in the enriched pathways.
3.4 Comparisons of tumor molecular characteristics between responders and non-responders
We further compared the TMB between responders and non-responders. Results revealed higher TMB in pretreatment tumors of non-responders; however, the difference did not reach statistical significance (median TMB of 133.04 mutations/Mb vs. 71.14 mutations/Mb, p = 0.4316) (Figure S1). Likewise, while the total number of non-synonymous (ns)SNVs was relatively higher in non-responders than in responders (median of 3147 vs. 1892, p = 0.4908), the impact of nsSNV loads on patient outcomes was not clearly established (Figure S2).
In the CNV analysis, fewer amplification than deletion events were observed in responders at the chromosome-arm level, while there were only amplified chromosomal events in non-responders (Figures 4A, C). Specifically, gains of 4p, 5q, 10q, 17q, 20q, and gains of 1q, 5q, and 8p were respectively reported for responders and non-responders. Meanwhile, chromosomal losses of 4p, 5q, 6q, 10q, 16q, 17p, and 20q were exclusively observed in responders. In total, eight recurrent focal amplification and nine deletion peaks were also identified across cohorts (q value of < 0.25) (Figure 4B). Among them, amplification at 4p11 (67%), 17q11.1 (67%), and 20q11.21 (67%) and deletion at 6q27 (50%) and 17p11.2 (50%) were the most frequent events in responders. On the other hand, amplification of 5q11.1 (75%) was mostly observed in non-responders. The amplification and deletion events at gene level are summarized in Table S3.
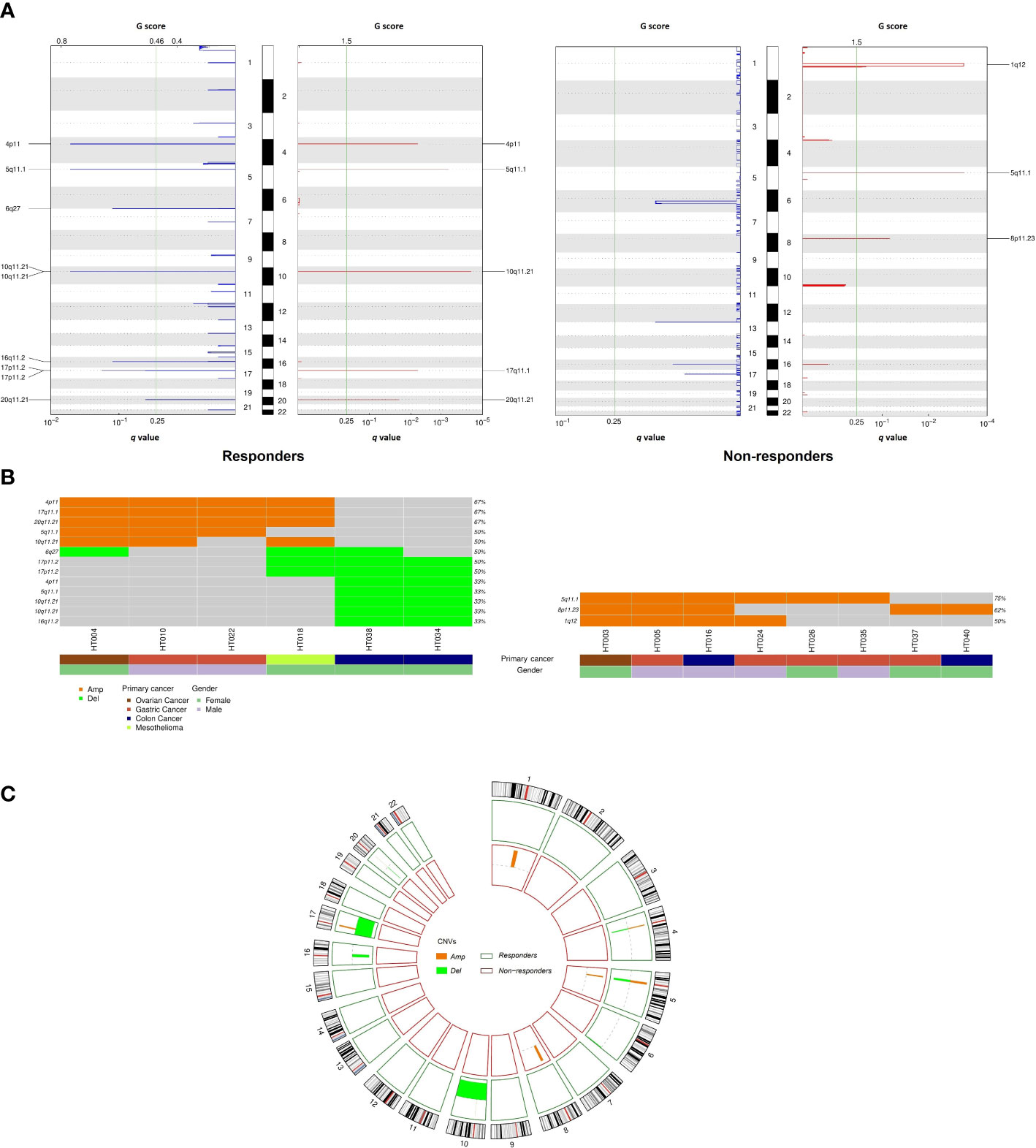
Figure 4 Comparison of copy number variation between responders and non-responders. (A) Amplifications and deletion peaks identified by GISTIC2.0 (q < 0.25); (B) Recurrent focal amplification and deletion regions detected for each tumor; (C) Circos plot summarizing copy number variation distribution in the two cohorts.
3.5 Mutations of AGAP5 associated with prognostic significance
To identify differentially mutated genes, we conducted Fisher’s exact test for all genes with alterations between responders and non-responders. Fourteen genes were recurrently and preferentially mutated in either responders or non-responders (p < 0.05) (Figure 5A). Among those, CFAP46, TRIM28, TOP3B, TAS1R2, POU4F1, MAP3K21, MAP3K13, HPS1, DYSF, DIDO1, and CACNA1A were found as unique in tumors from non-responders. Notably, AGAP5 was the top differentially mutated gene, with missense mutations reported in all responding tumors (100%) and one of eight samples in non-responders (12.5%). The pattern of missense mutations on different functional domains of AGAP5 is demonstrated with a lollipop plot (Figure 5B). In the survival analysis, mutated AGAP5 was significantly correlated with better OS following CRS/HIPEC compared to the wild-type gene (p = 0.00652, hazard ratio (HR) = 3.75 × 10-10) (Figure 6). Besides AGAP5, results of the survival analyses for the remaining genes are shown in Figure S3. No significant associations were identified.
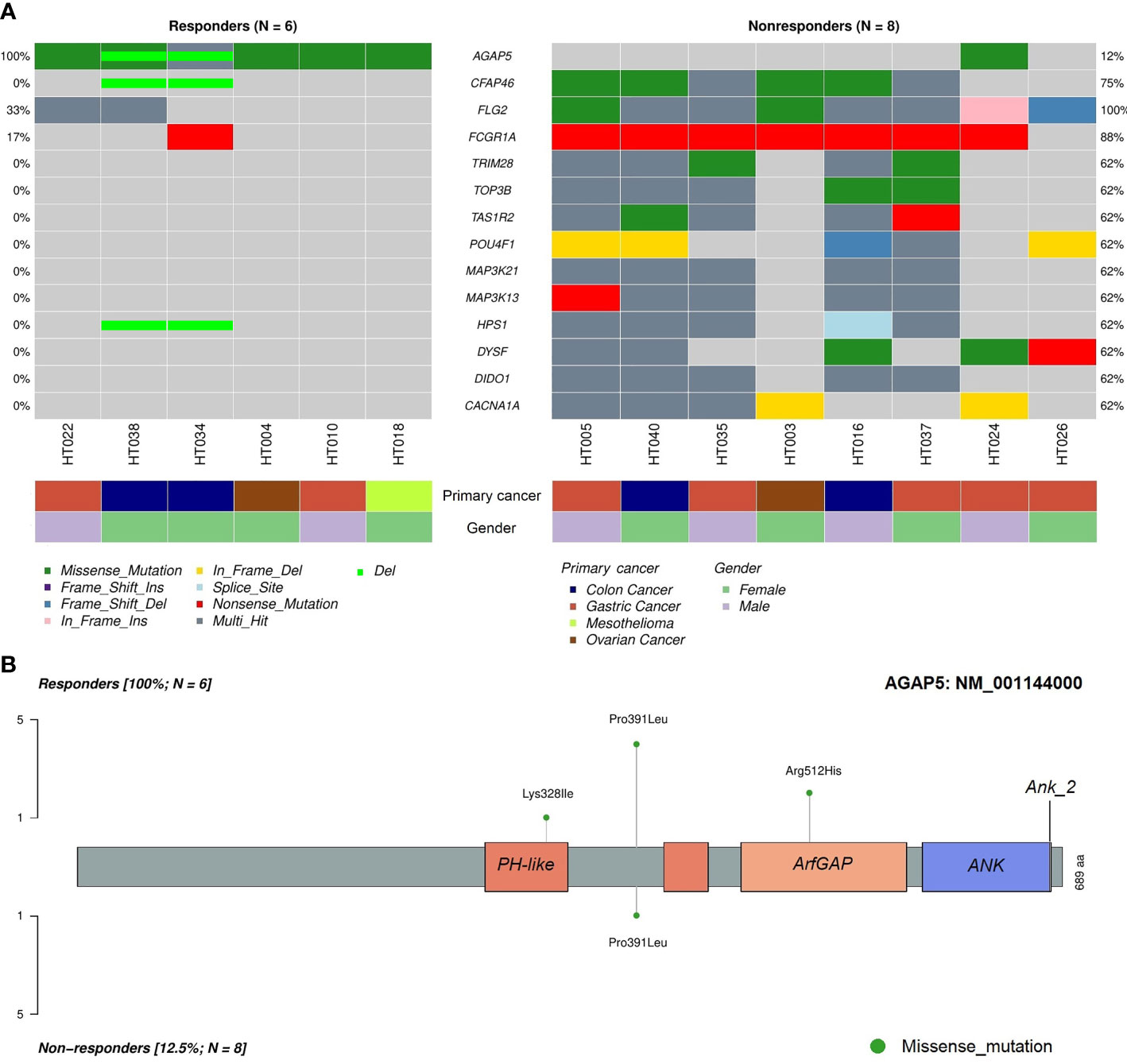
Figure 5 Differentially mutated genes in tumor samples between responders and non-responders. (A) Differentially mutated genes between the two cohorts with frequencies and classifications of mutations. (B) Lollipop plot demonstrating mutated amino acids on different functional domains of AGAP5. ANK, ankyrin repeat; PH, pleckstrin homology.
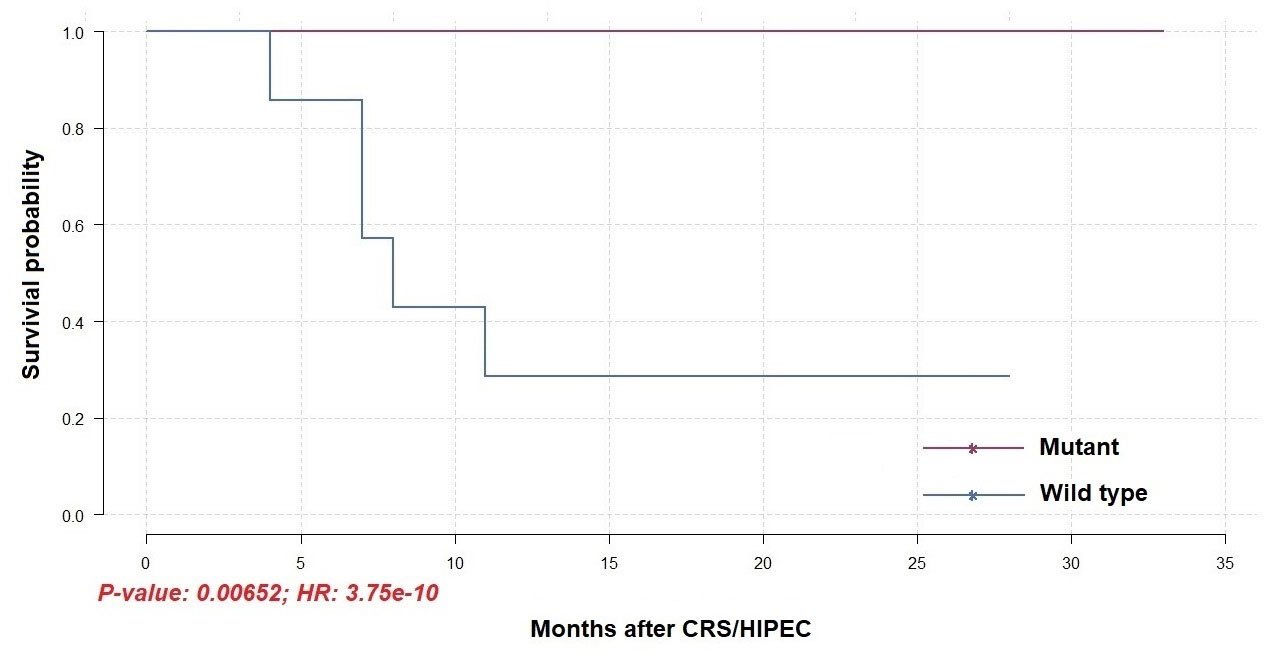
Figure 6 Overall survival of patients receiving CRS/HIPEC with or without a mutated AGAP5 gene (N = 14).
4 Discussion
In this study, molecular signatures from WES may facilitate clinical decision-making for PM management. First, alterations in FCGBP, FLG, KCNJ12, and KCNJ18 were observed in all tumor samples, regardless of the responsiveness towards HIPEC. FCGBP is involved in the intestinal immune defense against inflammation and carcinogenesis. Reduced expression of FCGBP was frequently observed in colorectal adenoma and CRC (36). FLG acts as a defense line (i.e., establishment of a skin barrier (GO:0061436)) that exhibits mutations in several cancer types (e.g., non-melanoma skin cancer, CRC, cervical cancer, and prostate cancer) (37), whereas KCNJ12 and KCNJ18 are involved in regulating transmembrane transport (GO:0034762), which is important in cell proliferation, differentiation, and apoptosis (38). Acquired mutations may imply impaired functions of these genes in primary tumorigenesis as well as the development of PM.
Importantly, ‘axon guidance’ was annotated as the most important pathway in our cohort. The ‘axon guidance’ pathway modulates a variety of key biological functions including cell survival, proliferation, differentiation, migration, and invasion in different cancer types (39). Dysregulation of axon guidance molecules has frequently been reported in cancers. EPHB6 was down-regulated in several cancer types such as metastatic lung cancer, melanoma, and CRC (40–42), while high expression of SEMA4D promoted angiogenesis in many tumors (43, 44). Pepinemab, an anti-SEMA4D has been developed and tested in several solid tumors (45, 46). An early phase clinical trial has demonstrated that Pepinemab combined with other immune checkpoint inhibitors was well-tolerated and showed anti-tumor activity in immune-resistant tumors (46). Recently, Yu et al. reported that suppression of ERBB2 (Erb-B2 Receptor Tyrosine Kinase 2) was observed with the negative regulatory activity on MAPK1/MAPK3 signaling pathway that led to the hinderance of tumor progression in ovarian cancer cells (47). Taken together, these findings revealed potential therapeutic targets for PM.
Our results indicated that TMB was higher in non-responders than that in responders, which is consistent with a previous finding of colorectal PM treated with CRS/HIPEC (48). Conversely, the correlation between high TMB and better responses to HIPEC was reported in gastric PM patients (49). The inconsistent results might be due to the tumor heterogeneity. In addition, the CNV analysis revealed targets with prognostic potential for therapeutic response and survival outcomes. For example, ABCC3 (17p11.2) and ABCC2 (10q11.21) were found to be deleted in responders. These ATP-binding cassette (ABC) transporters regulate the efflux of drugs from cancer cells; therefore, deletions in these gene loci were previously described as producing favorable responses to neoadjuvant chemotherapy (50). In contrast, five of eight non-responding samples exhibited amplified ANK1 and FGFR1 in the 8p11.23 region. High expression of ANK1 and FGFR1 are associated with poor outcomes in several cancer types (51, 52).
Here, we identified 11 exclusively mutated genes in non-responders, namely CFAP46, TRIM28, TOP3B, TAS1R2, POU4F1, MAP3K21, MAP3K13, HPS1, DYSF, DIDO1, and CACNA1A. Importantly, we found that mutated AGAP5 was observed in all responders and was associated with increased OS after CRS/HIPEC. In addition, deletion of AGAP5 was also detected in responders HT034 and HT038 (Figure 5A).
AGAP5 is a protein-coding gene with the function of a GTPase activity activator. AGAP5 consists of an ADP-ribosylation factor (Arf) GAP domain, ankyrin repeat (ANK) domains, and pleckstrin homology (PH) domain 5 (Figure 5B). The PH domain is a critical protein-binding site for catalyzing GTPase activity, and the PH-Arf GAP domain interaction is necessary to activate AGAP function (53). Arfs are proteins that belong to a subfamily of Ras small GTPases without intrinsic GTPase activities. Arfs are activated by GTP exchange factors (Arf GEFs) that assist the conversion of GDP to GTP and are terminated by GTPase-activating proteins (Arf GAPs) (54). In addition, a growing body of evidence suggests the functions of Arf GAPs through an Arf-independent manner (55–57). Under normal physiological circumstances, Arf proteins play important roles in regulating key cellular functions, including membrane trafficking, lipid metabolism, energy utility, cell motility, mitosis, apoptosis, and transcription (54, 58). Upregulation of Arf1, Arf4, and Arf6 were found in breast, gastric, prostate, or lung cancer. Furthermore, overexpression of Arf GAPs (i.e., AGAP1 and AGAP2) was identified in different cancer types such as breast, colon, lung, ovarian, and hepatocellular carcinoma (57). AGAP2 with regulatory activity on Arf1 and Arf5 enhances cancer cell survival, migration, and invasion in glioblastoma (59). Amplified AGAP2 and ASAP1 were associated with impaired OS and progression-free survival in uveal melanoma (60).
The limitation of this study is the small sample size. To establish a better signature to predict the therapeutic outcomes, more clinical samples for further analysis are needed. In addition, our cohort comprised patients with PM derived from different origins (i.e., CRC, gastric cancer, ovarian cancer, and mesothelioma); thus, sub-group analyses according to primary cancer types should be considered to generate more stringent conclusions. Despite these shortcomings, we successfully identified potential shared genetic markers with prognostic value for CRS/HIPEC. These exploratory findings may provide a rationale for clinical decision-making.
In conclusion, we uncovered the molecular characteristics from PM patients and reported a list of driver genes as well as enriched signaling pathways. The results might be helpful for further drug discovery. Importantly, AGAP5 was found to be significantly associated with the better OS. This finding is helpful to detect the good responders after CRS/HIPEC. In summary, our study provides a tailor prognostic signature for CRS/HIPEC therapy.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA891798, accession number PRJNA891798.
Ethics statement
The studies involving human participants were reviewed and approved by The Joint Institutional Review Board of Taipei Medical University (approval no. N201807067). The patients/participants provided their written informed consent to participate in this study.
Author contributions
Q-AN, W-HC, Y-TT, M-CH and W-CC conceptualized, conceived, and designed the study. Y-TT, M-CH performed clinical diagnosis and sample collection. Q-AN, C-MC, W-TL prepared samples for sequencing. Q-AN, W-HC, W-CC analyzed data. Q-AN, W-HC, Y-TT, M-CH and W-CC interpreted results. Q-AN, W-HC wrote the original draft. Y-TT, M-CH and W-CC provided the funding. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Ministry of Science and Technology, Taiwan (MOST 110-2628-B-038-020; MOST 110-2314-B-038-161, and MOST 110-2321-B-038-002 - Establishing A Translational Female Cancer Biomedical Big Data Bank and Developing Precision Medicine Healthcare System).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1054406/full#supplementary-material
Supplementary Table 1 | List of driver genes.
Supplementary Table 2 | KEGG pathway analysis of driver genes.
Supplementary Table 3 | Genes with an amplification or deletion event.
References
1. Sadeghi B, Arvieux C, Glehen O, Beaujard AC, Rivoire M, Baulieux J, et al. Peritoneal carcinomatosis from non-gynecologic malignancies: Results of the evocape 1 multicentric prospective study. Cancer (2000) 88(2):358–63. doi: 10.1002/(sici)1097-0142(20000115)88:2<358::aid-cncr16>3.0.co;2-o
2. Verwaal VJ, van Ruth S, de Bree E, van Sloothen GW, van Tinteren H, Boot H, et al. Randomized trial of cytoreduction and hyperthermic intraperitoneal chemotherapy versus systemic chemotherapy and palliative surgery in patients with peritoneal carcinomatosis of colorectal cancer. J Clin Oncol (2003) 21(20):3737–43. doi: 10.1200/JCO.2003.04.187
3. van Oudheusden TR, Razenberg LG, van Gestel YR, Creemers GJ, Lemmens VE, de Hingh IH. Systemic treatment of patients with metachronous peritoneal carcinomatosis of colorectal origin. Sci Rep (2015) 5:18632. doi: 10.1038/srep18632
4. Franko J, Shi Q, Meyers JP, Maughan TS, Adams RA, Seymour MT, et al. Prognosis of patients with peritoneal metastatic colorectal cancer given systemic therapy: An analysis of individual patient data from prospective randomised trials from the analysis and research in cancers of the digestive system (Arcad) database. Lancet Oncol (2016) 17(12):1709–19. doi: 10.1016/S1470-2045(16)30500-9
5. Sugarbaker PH. Peritoneal carcinomatosis: Natural history and rational therapeutic interventions using intraperitoneal chemotherapy. Cancer Treat Res (1996) 81:149–68. doi: 10.1007/978-1-4613-1245-1_13
6. Coccolini F, Gheza F, Lotti M, Virzi S, Iusco D, Ghermandi C, et al. Peritoneal carcinomatosis. World J Gastroenterol (2013) 19(41):6979–94. doi: 10.3748/wjg.v19.i41.6979
7. Witkamp AJ, de Bree E, Van Goethem R, Zoetmulder FA. Rationale and techniques of intra-operative hyperthermic intraperitoneal chemotherapy. Cancer Treat Rev (2001) 27(6):365–74. doi: 10.1053/ctrv.2001.0232
8. Ahmed K, Tabuchi Y, Kondo T. Hyperthermia: An effective strategy to induce apoptosis in cancer cells. Apoptosis (2015) 20(11):1411–9. doi: 10.1007/s10495-015-1168-3
9. Pyrhonen S, Kuitunen T, Nyandoto P, Kouri M. Randomised comparison of fluorouracil, epidoxorubicin and methotrexate (Femtx) plus supportive care with supportive care alone in patients with non-resectable gastric cancer. Br J Cancer (1995) 71(3):587–91. doi: 10.1038/bjc.1995.114
10. Glimelius B, Ekstrom K, Hoffman K, Graf W, Sjoden PO, Haglund U, et al. Randomized comparison between chemotherapy plus best supportive care with best supportive care in advanced gastric cancer. Ann Oncol (1997) 8(2):163–8. doi: 10.1023/a:1008243606668
11. Elias D, Lefevre JH, Chevalier J, Brouquet A, Marchal F, Classe JM, et al. Complete cytoreductive surgery plus intraperitoneal chemohyperthermia with oxaliplatin for peritoneal carcinomatosis of colorectal origin. J Clin Oncol (2009) 27(5):681–5. doi: 10.1200/JCO.2008.19.7160
12. Kuijpers AM, Mirck B, Aalbers AG, Nienhuijs SW, de Hingh IH, Wiezer MJ, et al. Cytoreduction and hipec in the Netherlands: Nationwide long-term outcome following the Dutch protocol. Ann Surg Oncol (2013) 20(13):4224–30. doi: 10.1245/s10434-013-3145-9
13. Bakrin N, Bereder JM, Decullier E, Classe JM, Msika S, Lorimier G, et al. Peritoneal carcinomatosis treated with cytoreductive surgery and hyperthermic intraperitoneal chemotherapy (Hipec) for advanced ovarian carcinoma: A French multicentre retrospective cohort study of 566 patients. Eur J Surg Oncol (2013) 39(12):1435–43. doi: 10.1016/j.ejso.2013.09.030
14. Yan TD, Deraco M, Baratti D, Kusamura S, Elias D, Glehen O, et al. Cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for malignant peritoneal mesothelioma: Multi-institutional experience. J Clin Oncol (2009) 27(36):6237–42. doi: 10.1200/JCO.2009.23.9640
15. Konigsrainer I, Horvath P, Struller F, Forkl V, Konigsrainer A, Beckert S. Risk factors for recurrence following complete cytoreductive surgery and hipec in colorectal cancer-derived peritoneal surface malignancies. Langenbecks Arch Surg (2013) 398(5):745–9. doi: 10.1007/s00423-013-1065-6
16. Braam HJ, van Oudheusden TR, de Hingh IH, Nienhuijs SW, Boerma D, Wiezer MJ, et al. Patterns of recurrence following complete cytoreductive surgery and hyperthermic intraperitoneal chemotherapy in patients with peritoneal carcinomatosis of colorectal cancer. J Surg Oncol (2014) 109(8):841–7. doi: 10.1002/jso.23597
17. Tan GHC, Chia CS, Tan SH, Soo KC, Teo MCC. Early recurrence after cytoreductive surgery (Crs) and hyperthermic intraperitoneal chemotherapy (Hipec). Int J Clin Oncol (2018) 23(5):989–98. doi: 10.1007/s10147-018-1301-8
18. Quenet F, Elias D, Roca L, Goere D, Ghouti L, Pocard M, et al. Cytoreductive surgery plus hyperthermic intraperitoneal chemotherapy versus cytoreductive surgery alone for colorectal peritoneal metastases (Prodige 7): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol (2021) 22(2):256–66. doi: 10.1016/S1470-2045(20)30599-4
19. Zivanovic O, Chi DS, Zhou Q, Iasonos A, Konner JA, Makker V, et al. Secondary cytoreduction and carboplatin hyperthermic intraperitoneal chemotherapy for platinum-sensitive recurrent ovarian cancer: An msk team ovary phase ii study. J Clin Oncol (2021) 39(23):2594–604. doi: 10.1200/JCO.21.00605
20. Auer RC, Sivajohanathan D, Biagi J, Conner J, Kennedy E, May T. Indications for hyperthermic intraperitoneal chemotherapy with cytoreductive surgery: A clinical practice guideline. Curr Oncol (2020) 27(3):146–54. doi: 10.3747/co.27.6033
21. Dodson RM, Kuncewitch M, Votanopoulos KI, Shen P, Levine EA. Techniques for cytoreductive surgery with hyperthermic intraperitoneal chemotherapy. Ann Surg Oncol (2018) 25(8):2152–8. doi: 10.1245/s10434-018-6336-6
22. Froysnes IS, Larsen SG, Spasojevic M, Dueland S, Flatmark K. Complete cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for colorectal peritoneal metastasis in Norway: Prognostic factors and oncologic outcome in a national patient cohort. J Surg Oncol (2016) 114(2):222–7. doi: 10.1002/jso.24290
23. Kwakman R, Schrama AM, van Olmen JP, Otten RH, de Lange-de Klerk ES, de Cuba EM, et al. Clinicopathological parameters in patient selection for cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for colorectal cancer metastases: A meta-analysis. Ann Surg (2016) 263(6):1102–11. doi: 10.1097/SLA.0000000000001593
24. Hulshof EC, Lim L, de Hingh I, Gelderblom H, Guchelaar HJ, Deenen MJ. Genetic variants in DNA repair pathways as potential biomarkers in predicting treatment outcome of intraperitoneal chemotherapy in patients with colorectal peritoneal metastasis: A systematic review. Front Pharmacol (2020) 11:577968. doi: 10.3389/fphar.2020.577968
25. Solomon D, Leigh N, Bekhor E, Feferman Y, Dhorajiya P, Feingold D, et al. The role of molecular biomarkers in outcomes and patient selection for cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for peritoneal metastases of colorectal origin. Surgeon (2021) 19(6):e379–e85. doi: 10.1016/j.surge.2020.11.002
26. Shannon NB, Tan JW, Tan HL, Wang W, Chen Y, Lim HJ, et al. A set of molecular markers predicts chemosensitivity to mitomycin-c following cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for colorectal peritoneal metastasis. Sci Rep (2019) 9(1):10572. doi: 10.1038/s41598-019-46819-z
27. Lund-Andersen C, Torgunrud A, Fleten KG, Flatmark K. Omics analyses in peritoneal metastasis-utility in the management of peritoneal metastases from colorectal cancer and pseudomyxoma peritonei: A narrative review. J Gastrointest Oncol (2021) 12(Suppl 1):S191–203. doi: 10.21037/jgo-20-136
28. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res (2010) 20(9):1297–303. doi: 10.1101/gr.107524.110
29. Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics (2009) 25(14):1754–60. doi: 10.1093/bioinformatics/btp324
30. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The ensembl variant effect predictor. Genome Biol (2016) 17(1):122. doi: 10.1186/s13059-016-0974-4
31. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res (2018) 28(11):1747–56. doi: 10.1101/gr.239244.118
32. Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. Oncodriveclust: Exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics (2013) 29(18):2238–44. doi: 10.1093/bioinformatics/btt395
33. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res (2016) 44(W1):W90–7. doi: 10.1093/nar/gkw377
34. Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, et al. Sequenza: Allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol (2015) 26(1):64–70. doi: 10.1093/annonc/mdu479
35. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. Gistic2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol (2011) 12(4):R41. doi: 10.1186/gb-2011-12-4-r41
36. Lee S, Bang S, Song K, Lee I. Differential expression in normal-Adenoma-Carcinoma sequence suggests complex molecular carcinogenesis in colon. Oncol Rep (2006) 16(4):747–54. doi: 10.3892/or.16.4.747
37. Skaaby T, Husemoen LL, Thyssen JP, Meldgaard M, Thuesen BH, Pisinger C, et al. Filaggrin loss-of-Function mutations and incident cancer: A population-based study. Br J Dermatol (2014) 171(6):1407–14. doi: 10.1111/bjd.12969
38. Lee I, Lee SJ, Kang TM, Kang WK, Park C. Unconventional role of the inwardly rectifying potassium channel Kir2.2 as a constitutive activator of rela in cancer. Cancer Res (2013) 73(3):1056–62. doi: 10.1158/0008-5472.CAN-12-2498
39. Chedotal A, Kerjan G, Moreau-Fauvarque C. The brain within the tumor: New roles for axon guidance molecules in cancers. Cell Death Differ (2005) 12(8):1044–56. doi: 10.1038/sj.cdd.4401707
40. Bailey CM, Kulesa PM. Dynamic interactions between cancer cells and the embryonic microenvironment regulate cell invasion and reveal Ephb6 as a metastasis suppressor. Mol Cancer Res (2014) 12(9):1303–13. doi: 10.1158/1541-7786.MCR-13-0673
41. Mateo-Lozano S, Bazzocco S, Rodrigues P, Mazzolini R, Andretta E, Dopeso H, et al. Loss of the eph receptor B6 contributes to colorectal cancer metastasis. Sci Rep (2017) 7:43702. doi: 10.1038/srep43702
42. Muller-Tidow C, Diederichs S, Bulk E, Pohle T, Steffen B, Schwable J, et al. Identification of metastasis-associated receptor tyrosine kinases in non-small cell lung cancer. Cancer Res (2005) 65(5):1778–82. doi: 10.1158/0008-5472.CAN-04-3388
43. Zhou H, Binmadi NO, Yang YH, Proia P, Basile JR. Semaphorin 4d cooperates with vegf to promote angiogenesis and tumor progression. Angiogenesis (2012) 15(3):391–407. doi: 10.1007/s10456-012-9268-y
44. Basile JR, Castilho RM, Williams VP, Gutkind JS. Semaphorin 4d provides a link between axon guidance processes and tumor-induced angiogenesis. Proc Natl Acad Sci U.S.A. (2006) 103(24):9017–22. doi: 10.1073/pnas.0508825103
45. Rossi AJ, Khan TM, Hong H, Lesinski GB, Wu C, Hernandez JM. Pepinemab (Anti-Sema4d) in combination with ipilimumab or nivolumab for patients with resectable pancreatic and colorectal cancer. Ann Surg Oncol (2021) 28(8):4098–9. doi: 10.1245/s10434-021-10111-0
46. Shafique MR, Fisher TL, Evans EE, Leonard JE, Pastore DRE, Mallow CL, et al. A phase Ib/Ii study of pepinemab in combination with avelumab in advanced non-small cell lung cancer. Clin Cancer Res an Off J Am Assoc Cancer Res (2021) 27(13):3630–40. doi: 10.1158/1078-0432.Ccr-20-4792
47. Yu TT, Wang CY, Tong R. Erbb2 gene expression silencing involved in ovarian cancer cell migration and invasion through mediating Mapk1/Mapk3 signaling pathway. Eur Rev Med Pharmacol Sci (2020) 24(10):5267–80. doi: 10.26355/eurrev_202005_21309
48. Hallam S, Stockton J, Bryer C, Whalley C, Pestinger V, Youssef H, et al. The transition from primary colorectal cancer to isolated peritoneal malignancy is associated with an increased tumour mutational burden. Sci Rep (2020) 10(1):18900. doi: 10.1038/s41598-020-75844-6
49. Zeng L, Huang X, Tian Y, Huang J, Liu H, Wen J, et al. Tumor mutational burden associated with response to hyperthermic intraperitoneal chemotherapy. Front Oncol (2022) 12:796263. doi: 10.3389/fonc.2022.796263
50. Litviakov NV, Cherdyntseva NV, Tsyganov MM, Slonimskaya EM, Ibragimova MK, Kazantseva PV, et al. Deletions of multidrug resistance gene loci in breast cancer leads to the down-regulation of its expression and predict tumor response to neoadjuvant chemotherapy. Oncotarget (2016) 7(7):7829–41. doi: 10.18632/oncotarget.6953
51. Hall AE, Lu WT, Godfrey JD, Antonov AV, Paicu C, Moxon S, et al. The cytoskeleton adaptor protein ankyrin-1 is upregulated by P53 following DNA damage and alters cell migration. Cell Death Dis (2016) 7:e2184. doi: 10.1038/cddis.2016.91
52. Krook MA, Reeser JW, Ernst G, Barker H, Wilberding M, Li G, et al. Fibroblast growth factor receptors in cancer: Genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. Br J Cancer (2021) 124(5):880–92. doi: 10.1038/s41416-020-01157-0
53. Luo R, Miller Jenkins LM, Randazzo PA, Gruschus J. Dynamic interaction between arf gap and ph domains of Asap1 in the regulation of gap activity. Cell Signal (2008) 20(11):1968–77. doi: 10.1016/j.cellsig.2008.07.007
54. Sztul E, Chen PW, Casanova JE, Cherfils J, Dacks JB, Lambright DG, et al. Arf gtpases and their gefs and gaps: Concepts and challenges. Mol Biol Cell (2019) 30(11):1249–71. doi: 10.1091/mbc.E18-12-0820
55. Luo R, Reed CE, Sload JA, Wordeman L, Randazzo PA, Chen PW. Arf gaps and molecular motors. Small GTPases (2019) 10(3):196–209. doi: 10.1080/21541248.2017.1308850
56. East MP, Kahn RA. Models for the functions of arf gaps. Semin Cell Dev Biol (2011) 22(1):3–9. doi: 10.1016/j.semcdb.2010.07.002
57. Casalou C, Faustino A, Barral DC. Arf proteins in cancer cell migration. Small GTPases (2016) 7(4):270–82. doi: 10.1080/21541248.2016.1228792
58. Donaldson JG, Jackson CL. Arf family G proteins and their regulators: Roles in membrane transport, development and disease. Nat Rev Mol Cell Biol (2011) 12(6):362–75. doi: 10.1038/nrm3117
59. Qi Q, Kang SS, Zhang S, Pham C, Fu H, Brat DJ, et al. Co-Amplification of phosphoinositide 3-kinase enhancer a and cyclin-dependent kinase 4 triggers glioblastoma progression. Oncogene (2017) 36(32):4562–72. doi: 10.1038/onc.2017.67
Keywords: hyperthermic intraperitoneal chemotherapy (HIPEC), peritoneal metastasis, whole exome sequencing (WES), somatic mutation, AGAP5
Citation: Nguyen Q-A, Chou W-H, Hsieh M-C, Chang C-M, Luo W-T, Tai Y-T and Chang W-C (2023) Genetic alterations in peritoneal metastatic tumors predicted the outcomes for hyperthermic intraperitoneal chemotherapy. Front. Oncol. 13:1054406. doi: 10.3389/fonc.2023.1054406
Received: 26 September 2022; Accepted: 27 March 2023;
Published: 26 April 2023.
Edited by:
Wim Ceelen, Ghent University, BelgiumReviewed by:
Shiwei Liu, Harvard University, United StatesChukwuemeka Ihemelandu, MedStar Washington Hospital Center, United States
Aruna Prabhu, Thangam Cancer Center, India
Copyright © 2023 Nguyen, Chou, Hsieh, Chang, Luo, Tai and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Chiao Chang, d2NjQHRtdS5lZHUudHc=; Yu-Ting Tai, eXR0YWlAdy50bXUuZWR1LnR3
†These authors have contributed equally to this work
 Quynh-Anh Nguyen
Quynh-Anh Nguyen Wan-Hsuan Chou
Wan-Hsuan Chou Mao-Chih Hsieh
Mao-Chih Hsieh Che-Mai Chang
Che-Mai Chang Wei-Tzu Luo
Wei-Tzu Luo Yu-Ting Tai
Yu-Ting Tai Wei-Chiao Chang
Wei-Chiao Chang