- 1Women’s College Research Institute, Women’s College Hospital, University of Toronto, Toronto, ON, Canada
- 2Institute of Medical Science, Faculty of Medicine, University of Toronto, Toronto, ON, Canada
- 3Nottingham Biodiscovery Institute, School of Medicine, University of Nottingham, Nottingham, United Kingdom
- 4Department of Laboratory Medicine and Pathology, Faculty of Medicine, University of Toronto, Toronto, ON, Canada
- 5International Hereditary Cancer Center, Department of Genetics and Pathology, Pomeranian Medical University in Szczecin, Szczecin, Poland
- 6Sunnybrook Health Science Centre, University of Toronto, Toronto, ON, Canada
- 7Genome Stability Laboratory, Centre Hospitalier Universitaire (CHU) de Québec Research Center, Oncology Axis, Department of Molecular Biology, Medical Biochemistry and Pathology, Laval University Cancer Research Center, Québec, QC, Canada
- 8Department of Biochemistry and Molecular Biology, College of Medicine, Howard University, Washington, DC, United States
- 9National Human Genome Center, College of Medicine, Howard University, Washington, DC, United States
- 10Humber River Hospital, University of Toronto, Toronto, ON, Canada
- 11Dalla Lana School of Public Health, University of Toronto, Toronto, ON, Canada
Background: RECQL (also known as RECQ1 and RECQL1) is a gene of recent interest in breast cancer and an association between high levels of RECQL protein in breast cancer tumour cells and good survival of patients has been reported.
Methods: To validate this association, we measured the RECQL protein levels in tumours of 933 breast cancer patients using immunohistochemistry analysis and followed the patients for death from breast cancer.
Results: Women with a level of RECQL protein above the 75th percentile had better 15-year disease-specific survival among ER-positive patients (62.5% vs. 48.7%, HR= 0.72, 95%CI= 0.52-0.98, p-value = 0.04), but not among ER- patients (48.9% vs. 48.0%, HR= 1.07, 95%CI= 0.67-1.69, p-value= 0.79). Among the ER-negative patients, high RECQL protein levels were associated with better survival among women who received tamoxifen treatment (67.0% vs. 51.5%, HR= 0.64, 95%CI= 0.41-0.99, p-value= 0.04).
Conclusion: RECQL might be a new predictive marker for tamoxifen treatment among ER-positive patients.
Introduction
Altered expression levels of several genes in breast cancer predict patient prognosis. A correlation has been observed between poor breast cancer prognosis and lower levels of RECQL (also known as RECQ1 and RECQL1) mRNA or RECQL protein expression (1, 2). RECQL is the smallest and most abundant member of the RecQ family of DNA helicases (3, 4). It has two main domains; the core helicase domain, involved in ATP binding and hydrolysis, and the RecQ C-terminal domain (RQC) which play an essential role in the unwinding of DNA (4–6). RECQL performs its helicase activity in an ATP-dependent manner in a 3’ to 5’ direction (7). As a helicase, it has many essential functions in DNA replication, such as maintaining the DNA replication fork progression and restarting stalled replication forks (8–11). It is also involved in maintaining genome stability, double-strand break repair, and telomere maintenance (4, 6, 12–14). In addition to its roles as a DNA helicase, RECQL is involved in branch migration of Holiday junctions and strand annealing (7). The depletion of this helicase could impair normal cellular function and lead to increased DNA damage and compromised genome stability.
An association between mutations in RECQL and breast cancer susceptibility was first reported by Cybulski et al. (15). Other studies have supported this association (16–21) but there have been negative studies as well (22–26). It has also been suggested that RECQL is a susceptibility gene for familial colorectal cancer (27).
In addition to the cancer susceptibility role, RECQL was suggested to be a prognostic marker. In a study by Arora et al. low RECQL mRNA and protein levels in breast tumours were associated with poor breast cancer prognosis (1). This correlation was observed in a second study which showed low RECQL protein levels were correlated with poor survival (HR: 2.12, p-value: 0.015) (2). Furthermore, in a recent study, the cellular mechanism by which RECQL affects ER-positive cells was proposed, wherein RECQL in cooperation with FOXA1, directly regulates the expression of the ESR1 gene which encodes the ERα protein (28).
To investigate this association further, we measured the RECQL protein levels in tumours from 933 breast cancer patients by immunohistochemistry (IHC) and analyzed their 15-year survival.
Methods
Study Population
This study was conducted by analyzing patients from the Banting study (29). Total of 1,601 breast cancer patients were diagnosed and enrolled between 1987 to 1999 in the Banting study and their therapy reflected the commonly used treatments during that time period. Data files and tumour samples from 933 breast cancer patients were available and analyzed in this study. Patients were followed from the date of diagnosis to the date of death from breast cancer or the date of last follow up (if alive). The average follow-up was 12.1 years (range: 0 to 31.3 years). The age at diagnosis ranged from 24 to 93 (mean 55.4 years). 55.1% of subjects were post-menopausal at diagnosis and 77.0% were ER-positive. The majority of ER-positive patients received tamoxifen (60.6%). No other endocrine therapy was used.
Tissue Microarray and Immunohistochemistry
Tissue microarrays (TMA) were made from 0.6-mm cores sampled from the formalin-fixed paraffin-embedded tumour blocks. Each TMA contains three cores from each tumour block. Immunohistochemical staining for the RECQL protein was conducted based on a previously described method (30–33) by using a combination of Thermo Scientific Shandon Sequenza chamber system (REF: 72110017), Novolink Max Polymer Detection System (RE7280-K: 1,250 tests), and the Leica Bond Primary Antibody Diluent (AR9352), according to the manufacturer’s instructions (Leica Microsystems). The slides were dewaxed and dehydrated by Leica Autostainer XL machine. TMA sections were pretreated with sodium citrate buffer (pH 6.0) and heated for 20 minutes at 95 °C in a microwave (Whirlpool JT359 Jet Chef 1000W) for antigen retrieval. Each set of slides was incubated with primary anti-RECQL antibody (Bethyl Laboratories, catalog no. A300-450A) at a dilution of 1:1,000 for 60 minutes. For each run, positive and negative controls were included. Negative control was utilized to reassure all the staining was because of specific antibody-antigen interaction. The negative control slide had only breast tissue without adding any antibody to it. The positive control slide was a liver tissue with known expression of RECQL to control for the reactivity of the RECQL antibody and also breast cancer tissue stained with an antibody for β-globulin to control for the reactivity of immunohistochemistry enzyme.
IHC Evaluation
After scoring whole-field inspection of the cores, intensities of the nuclear stainings were classified (0 = no staining, 1 = weak staining, 2 = moderate staining, and 3 = strong staining; Supplementary Figure 1). For each intensity classification, the percentage of the stained nuclei was estimated. An H-index (range 0-300) was calculated by multiplying the staining intensity score by the percentage of stained nuclei for each core. For each tumour sample, the median H-index of the three core replicates was used for the data analysis. The distribution of the median H-index for the entire cohort was shown in Supplementary Table 1. The mean and median of the coefficient of variation (CV) of H-index for the three cores across all samples were 10.4% and 6.9%, respectively.
Statistical Analysis
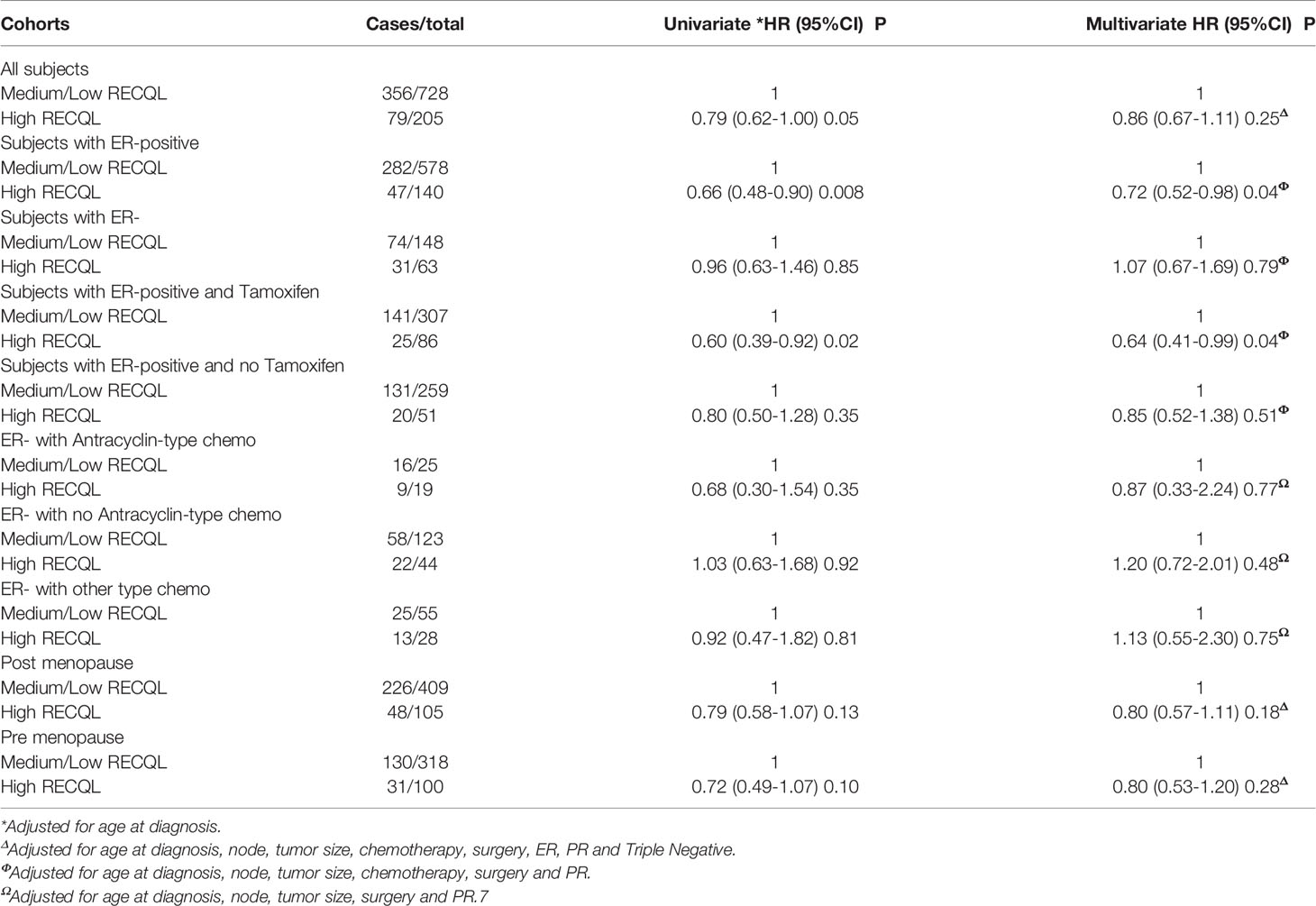
Subjects were divided into two groups based on their RECQL protein levels (Table 1). We considered a high RECQL level to be one in the highest quartile (n = 205) and a medium/low RECQL level to be one in the bottom three quartiles (n = 728). This classification had the best performance in terms of distinguishing patients with poor from good survival compared to using the first quartile or median as the cut-off point. Student t tests and Fisher exact tests were used as appropriate. Estimation of cumulative survival probabilities was conducted by the Kaplan–Meier method. A log-rank test was performed for analyzing the difference between survival. A Cox proportional hazards model was used to conduct a multivariate survival analysis. Hazard ratios and 95% CIs (95% confidence intervals) for each variable were estimated. All statistical tests were two-sided. A p-value < 0.05 was considered to be significant. SAS (version 9.4) was used for the data analysis
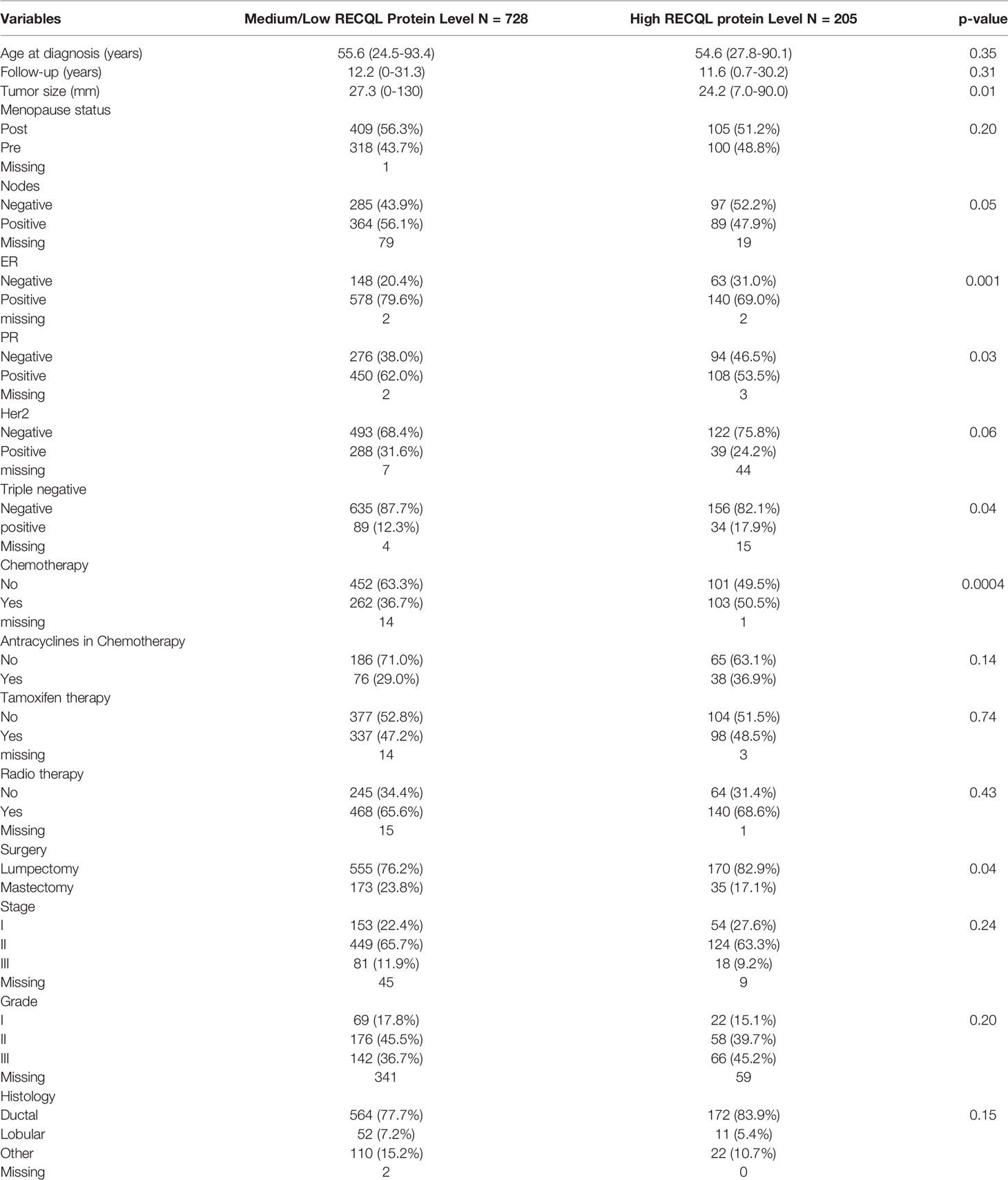
Table 1 Comparison of clinicopathological characteristics between patients with medium/low versus high RECQL protein levels.
Results
Clinicopathological Characteristics
Individuals with a tumour with a medium/low RECQL protein levels had a higher prevalence of lymph node positivity (56.1% vs. 47.9%, p-value= 0.05), a larger mean tumour size (27.3 mm vs 24.2 mm, p-value = 0.01), a higher proportion of ER-positive tumours (79.6% vs. 69.0%, p-value = 0.001) and a smaller proportion of triple-negative tumours (12.3% vs. 17.9%, p-value = 0.04) compared to those with a high level of RECQL protein. There were no significant differences in HER2 status or tumour grade.
Association of RECQL Protein Expression With Survival Among ER-Positive Patients
Of the 933 tumour samples in the study, 78% (n = 728) had a medium/low RECQL protein level, and 22% (n = 205) had a high level. Patients with a high RECQL levels had superior 15-year survival compared to patients with medium/low RECQL levels (58.3% vs. 48.7%, HR = 0.79, 95%CI = 0.62-1.00, p-value = 0.05) (Figure 1A; Table 2). After adjustment for age at diagnosis, lymph node status, tumor size, chemotherapy, surgery, ER, PR and Triple Negative, a similar association was observed (58.3% vs. 48.7%, HR= 0.86, 95%CI= 0.67-1.11) but it was not significant (p-value= 0.25) (Table 2).
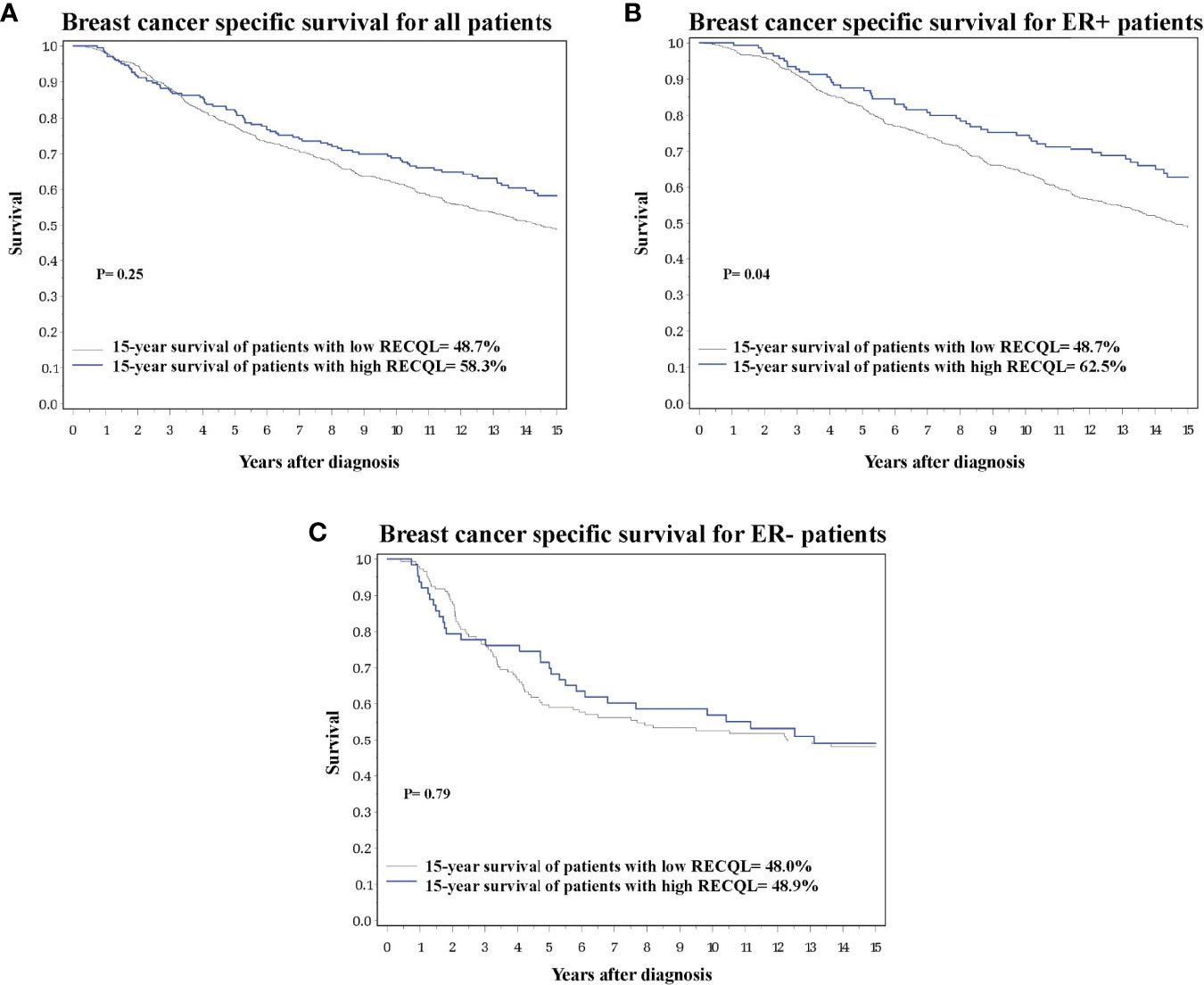
Figure 1 Breast cancer specific survival based on RECQL protein expression. (A) Breast cancer specific survival in the entire dataset based on RECQL protein expression. (B) Breast cancer specific survival in ER-positive patients based on RECQL protein expression. (C) Breast cancer specific survival in ER- patients based on RECQL protein expression.
Multivariate survival analysis was conducted separately for ER-positive and ER-negative patients. Among ER-positive patients a higher 15-year survival rate was observed for those with a high level of RECQL protein (62.5% vs. 48.7%, HR = 0.72, 95%CI = 0.52-0.98, p-value = 0.04) (Figure 1B; Table 2). Among the ER-negative patients no difference was seen (48.9% vs. 48.0%, HR = 1.07, 95%CI = 0.67-1.69, p-value = 0.79) (Figure 1C; Table 2).
We next investigated the effect of RECQL levels on ER-positive breast cancer patients subdivided by tamoxifen therapy. Among the ER-positive patients who received tamoxifen treatment, those who had higher RECQL protein levels had a better survival rate than patients with low RECQL levels (67.0% vs. 51.5%, HR = 0.64, 95%C I = 0.41-0.99, p = 0.04) (Figure 2A; Table 2). Among ER-positive patients who did not receive tamoxifen therapy, there was a smaller, non-significant, association between RECQL protein levels and survival (57.2% vs. 47.1%, HR = 0.85, 95%CI = 0.52-1.38; p = 0.5) (Figure 2B; Table 2).
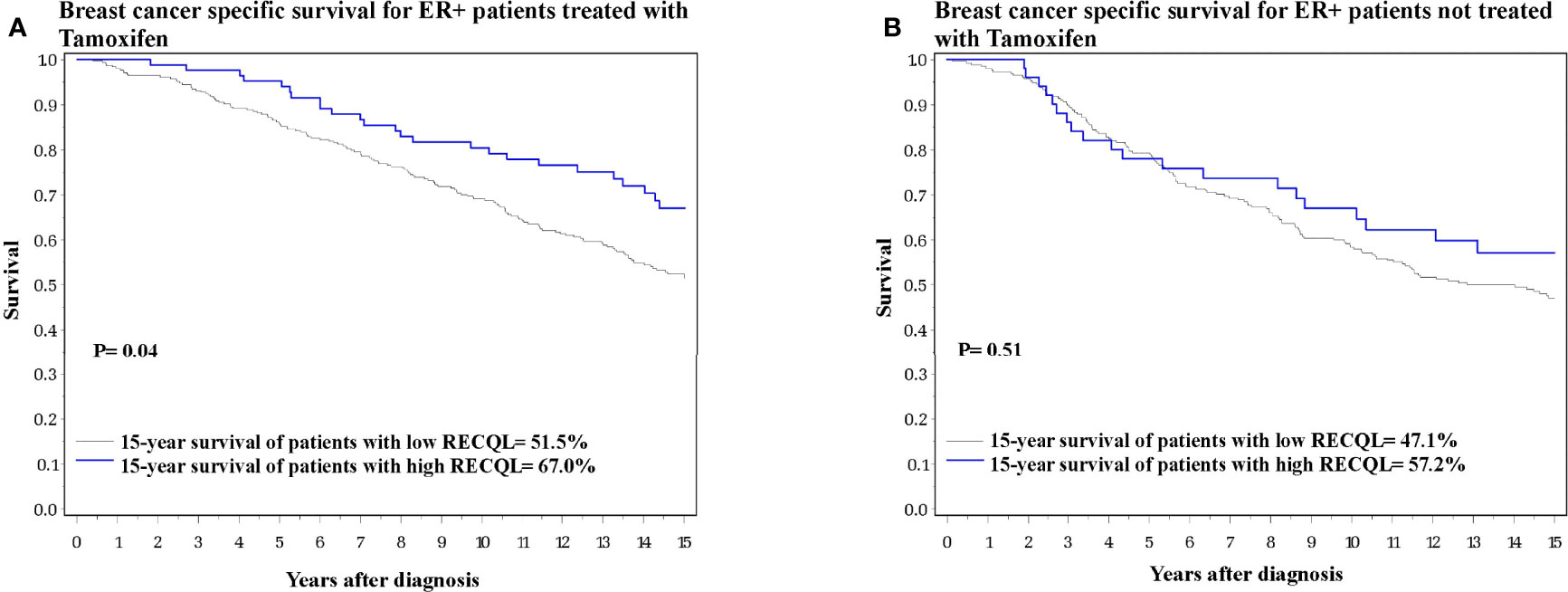
Figure 2 Breast cancer specific survival among ER-positive patients based on RECQL protein expression. (A) Breast cancer specific survival among ER-positive patients who received Tamoxifen based on RECQL protein expression. (B) Breast cancer specific survival among ER-positive patients who did not receive Tamoxifen based on RECQL protein expression.
In terms of 15-year survival, the benefit of tamoxifen treatment was greater in ER-positive patients with a high RECQL level (76.6% vs. 62.1%, HR = 0.37, 95%CI = 0.18-0.74, P = 0.005) than among ER-positive patients with a low/medium RECQ level (55.0% vs. 52.0%, HR = 0.73, 95%CI = 0.55-0.96, P = 0.02) (Figure 3; Table 3).
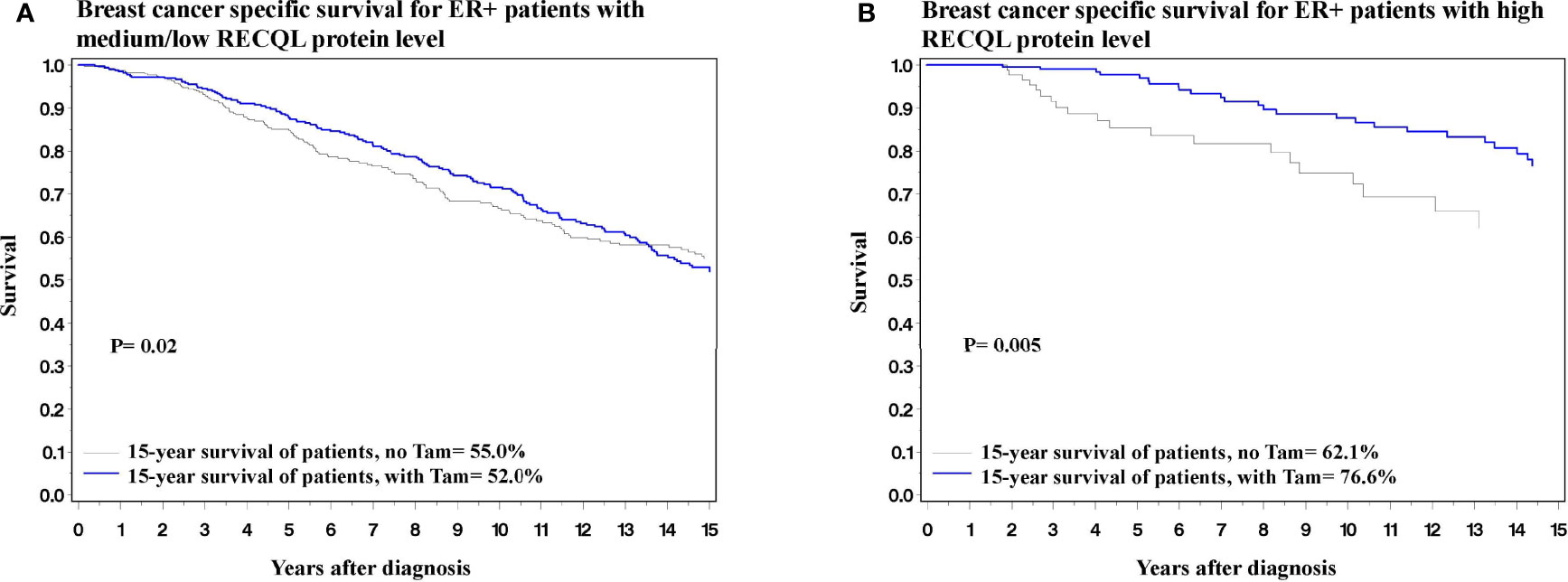
Figure 3 Breast cancer specific survival among ER-positive patients based on Tamoxifen treatment. (A) Breast cancer specific survival among ER-positive patients with medium/low level of RECQL protein based on Tamoxifen treatment. (B) Breast cancer specific survival among ER-positive patients with high level of RECQL protein based on Tamoxifen treatment.
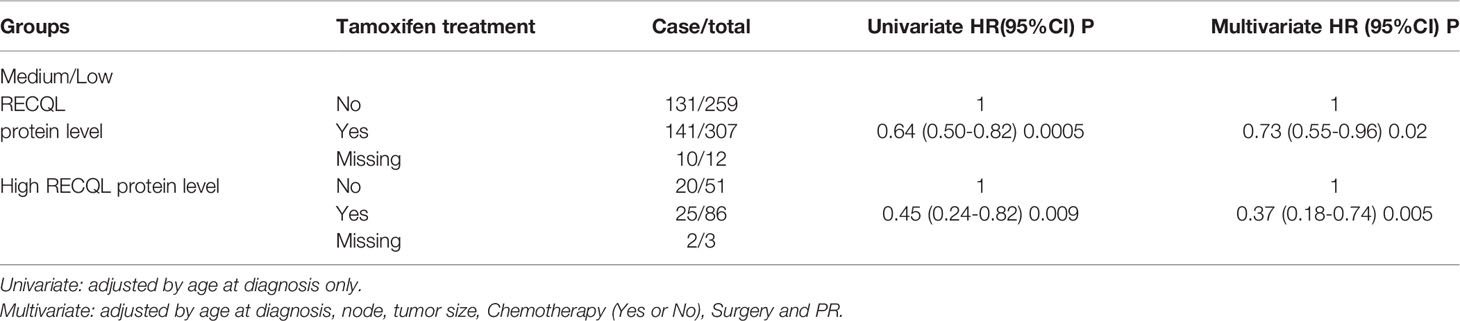
Table 3 Analysis of tamoxifen treatment influence on survival of ER-positive patients with medium/low and high RECQL protein levels.
Discussion
We analyzed the association between RECQL protein levels and breast cancer survival among 933 breast cancer patients diagnosed from 1987 to 1999 in Toronto, Canada. Among unselected breast cancer patients medium/low levels of RECQL protein were associated with inferior disease-specific survival. Additional analysis revealed that this association was seen only among ER-positive patients, in particular among those who received endocrine therapy (Tamoxifen). Together, these data show that RECQL protein level is a prognostic factor for realizing a benefit from endocrine therapy.
Three other studies have investigated the association between RECQL protein or mRNA levels and breast cancer survival (1, 2, 34). The first study showed that in separate cohorts of 848 and 1977 breast cancer patients, lower protein and mRNA levels of RECQL were associated with worse prognosis; further analysis revealed that this association only held among the ER-positive patients (1). The second study also observed that among 774 breast cancer patients, lower mRNA levels of RECQL were associated with poor distant recurrence-free survival (HR: 2.77, p-value <0.001) and disease-specific survival (HR: 3.10, p-value <0.001). In a cohort of 322 breast cancer patients, low RECQL protein levels correlated with poor survival (HR: 2.12, p-value: 0.015); however, the authors did not compare this association among ER-positive versus ER- individuals (2). These studies also showed that lower RECQL protein levels were associated with poor clinicopathological characteristics (1, 2). We also observed that medium/low RECQL levels were associated with poor clinicopathological characteristics, such as a higher proportion of lymph node-positive tumours (56.1% vs. 47.9%, p-value= 0.05) and larger tumour sizes (27.3 vs 24.2, p-value= 0.01). However, other clinicopathological characteristics were better among the medium/low RECQL patients of our cohort; medium/low RECQL levels were associated with a larger proportion of ER-positive tumours (79.6% vs. 69.0%) and a smaller proportion of triple-negative tumour (12.3% vs. 17.9%), while there was no difference between the medium/low versus high RECQL patients in HER2 status, tumour stage, tumour grade, and tumour histology. Lastly, the third study was only focused on RECQL mRNA levels (34). They observed that a higher expression of RECQL mRNA was correlated with shorter relapse-free survival (RFS) (HR: 1.28, p-value <0.001, n= 3955) and post-progression survival (PPS) (HR: 1.32, p-value: 0.027, n= 414) in all breast cancers. However, higher expression of RECQL mRNA did not affect overall survival (OS) (HR: 1.04, p-value: 0.74, n= 1402) or distant metastasis-free survival (DMFS) (HR: 1.06, p-value: 0.57, n= 1747). The current body of evidence suggests that higher RECQL protein levels are associated with higher survival rates among breast cancer patients with ER-positive tumours.
The cellular mechanism of RECQL effect on the prognosis of ER-positive tumours might be explained by a recent study that explored the role RECQL plays in regulating ERα expression. According to this newly published study, in a helicase dependant manner, RECQL in cooperation with FOXA1 increases the chromatin accessibility at the regulatory site of the ESR1 gene (the gene encoding ERα), and a group of other ERα target genes (28). Therefore, higher levels of RECQL protein would increase ERα expression and its downstream effect. This is an important observation in breast cancer biology for two main reasons. First, higher ERα levels are associated with a better prognosis because ERα inhibits tumour invasiveness (35–38). Second, higher levels of ERα would enable a more desirable response to endocrine therapy (38–41). Therefore, higher RECQL level directly increases ERα level, and in turn could reduce tumour invasiveness and improve the response to endocrine therapy. Unfortunately, we did not have ER expression values in our cohort to compare with the RECQL data to confirm their positive correlation. The relationship between RECQL levels and ER expression, tumour invasiveness as well as endocrine treatment efficiency could be the subject of further research. More importantly, future studies should investigate how to induce RECQL expression in ER-positive patients to improve their prognosis, especially in patients with lower RECQL protein levels. The findings of such studies could have crucial clinical implications, especially considering that more than 70% of breast cancer cases are ER-positive (35, 42).
One question that remains to be answered is why patients with lower RECQL levels had lower survival, while lower RECQL levels result in lower levels of ERα, and as a result the mitogenic effects of estrogen and ERα should be reduced. Three factors should be considered in answering this question. First, lower ERα levels would result in higher invasiveness, reducing the survival rate (35, 36). Second, we observed a larger tumour size in patients with lower levels of RECQL protein, so either lower RECQL levels do not cause enough reduction in ERα levels to dampen the mitogenic effects of ERα, or there are other factors involved that not only compensated for the reduced mitogenic effects of ERα due to its reduction, but caused increased mitogenic effects and a larger tumour size. Third, it is likely that RECQL also impacts breast cancer patient survival through non-ERα dependent effects and through its role in maintaining the chromosomal stability and DNA damage response. Additional studies should be conducted to investigate if such factors exist, and if they do, what the mechanism by which they act as mitogens is, and what implications do they have for breast cancer and possibly other neoplasms.
Conclusion
We have shown that higher RECQL protein levels are associated with improved ER-positive breast cancer-specific survival and better response to endocrine therapy with tamoxifen. Therefore, RECQL could be a prognostic and predictive candidate biomarker in ER-positive breast cancer patients responding to tamoxifen’s endocrine treatment.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
This study has been approved by the ethics board committee at Women’s College Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
AM wrote the manuscript draft, AS did the laboratory assays, PS and VG did the statistical analysis, CC, SN-M, SS, SM, SN, and MA worked together in conceptualizing the study idea, contributing laboratory and biospecimen resources required for the study and writing the manuscript, AAS reviewed the IHC slides and helped with the revision of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The study was funded by Canadian Institute for Health Research, grant# 152939.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge the pathology lab at Sunnybrook Health Science Centre, Toronto, Canada for making the tissue microarrays used in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.877617/full#supplementary-material
Abbreviations
ER, Estrogen receptor; HR, Hazard ratio; IHC, Immunohistochemistry; TMA, Tissue microarrays.
References
1. Arora A, Parvathaneni S, Aleskandarany MA, Agarwal D, Ali R, Abdel-Fatah T, et al. Clinicopathological and Functional Significance of RECQL1 Helicase in Sporadic Breast Cancers. Mol Cancer Ther (2017) 16(1):239–50. doi: 10.1158/1535-7163.MCT-16-0290
2. Xu H, Xu Y, Ouyang T, Li J, Wang T, Fan Z, et al. Low Expression of RECQL is Associated With Poor Prognosis in Chinese Breast Cancer Patients. BMC cancer (2018) 18(1):1–0. doi: 10.1186/s12885-018-4585-1
3. Sami F, Sharma S. Probing Genome Maintenance Functions of Human RECQ1. Comput Struct Biotechnol J (2013) 6(7):e201303014. doi: 10.5936/csbj.201303014
4. Debnath S, Sharma S. RECQ1 Helicase in Genomic Stability and Cancer. Genes (2020) 11(6):622. doi: 10.3390/genes11060622
5. Mojumdar A. Mutations in Conserved Functional Domains of Human RecQ Helicases Are Associated With Diseases and Cancer: A Review. Biophys Chem (2020) 16:106433. doi: 10.1016/j.bpc.2020.106433
6. Lu H, Davis AJ. Human RecQ Helicases in DNA Double-Strand Break Repair. Front Cell Dev Biol (2021) 9:279. doi: 10.3389/fcell.2021.640755
7. Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, et al. Biochemical Analysis of the DNA Unwinding and Strand Annealing Activities Catalyzed by Human RECQ1*[boxs]. J Biol Chem (2005) 280(30):28072–84. doi: 10.1074/jbc.M500264200
8. Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, et al. Human RECQ1 and RECQ4 Helicases Play Distinct Roles in DNA Replication Initiation. Mol Cell Biol (2010) 30(6):1382–96. doi: 10.1128/MCB.01290-09
9. Lu X, Parvathaneni S, Hara T, Lal A, Sharma S. Replication Stress Induces Specific Enrichment of RECQ1 at Common Fragile Sites FRA3B and FRA16D. Mol cancer (2013) 12(1):1–2. doi: 10.1186/1476-4598-12-29
10. Berti M, Chaudhuri AR, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, et al. Human RECQ1 Promotes Restart of Replication Forks Reversed by DNA Topoisomerase I Inhibition. Nat Struct Mol Biol (2013) 20(3):347–54. doi: 10.1038/nsmb.2501
11. Popuri V, Croteau DL, Brosh C.OMMAJr RM, Bohr VA. RECQ1 is Required for Cellular Resistance to Replication Stress and Catalyzes Strand Exchange on Stalled Replication Fork Structures. Cell Cycle (2012) 11(22):4252–65. doi: 10.4161/cc.22581
12. Brosh RM Jr., Sharma S, Sommers JA. Processing of DNA Replication and Repair Intermediates by the Concerted Action of RecQ Helicases and Rad2 Structure-Specific Nucleases. Protein Pept letters (2008) 15(1):89–102. doi: 10.2174/092986608783330369
13. Sharma S, Stumpo DJ, Balajee AS, Bock CB, Lansdorp PM, Brosh RM Jr., et al. RECQL, a Member of the RecQ Family of DNA Helicases, Suppresses Chromosomal Instability. Mol Cell Biol (2007) 27(5):1784–94. doi: 10.1128/MCB.01620-06
14. Sharma S, Brosh RM Jr. Human RECQ1 is a DNA Damage Responsive Protein Required for Genotoxic Stress Resistance and Suppression of Sister Chromatid Exchanges. PloS One (2007) 2(12):e1297. doi: 10.1371/journal.pone.0001297
15. Cybulski C, Carrot-Zhang J, Rivera B, Kashyap A, Wokołorczyk D, Giroux S, et al. Germline RECQL Mutations Are Associated With Breast Cancer Susceptibility. Nat Genet (2015) 47(6):643–6. doi: 10.1038/ng.3284
16. Sun J, Wang Y, Xia Y, Xu Y, Ouyang T, Li J, et al. Mutations in RECQL Gene Are Associated With Predisposition to Breast Cancer. PloS Genet (2015) 11(5):e1005228. doi: 10.1371/journal.pgen.1005228
17. Kwong A, Shin VY, Cheuk IW, Chen J, Au CH, Ho DN, et al. Germline RECQL Mutations in High Risk Chinese Breast Cancer Patients. Breast Cancer Res Treat (2016) 157(2):211–5. doi: 10.1007/s10549-016-3784-1
18. Sun J, Meng H, Yao L, Lv M, Bai J, Zhang J, et al. Germline Mutations in Cancer Susceptibility Genes in a Large Series of Unselected Breast Cancer Patients. Clin Cancer Res (2017) 23(20):6113–9. doi: 10.1158/1078-0432.CCR-16-3227
19. Tervasmäki A, Mantere T, Hartikainen JM, Kauppila S, Lee HM, Koivuluoma S, et al. Rare Missense Mutations in RECQL and POLG Associate With Inherited Predisposition to Breast Cancer. Int J cancer (2018) 142(11):2286–92. doi: 10.1002/ijc.31259
20. Cybulski C, Kluźniak W, Huzarski T, Wokołorczyk D, Kashyap A, Rusak B, et al. The Spectrum of Mutations Predisposing to Familial Breast Cancer in Poland. Int J cancer (2019) 145(12):3311–20. doi: 10.1002/ijc.32492
21. Palmer JR, Polley EC, Hu C, John EM, Haiman C, Hart SN, et al. Contribution of Germline Predisposition Gene Mutations to Breast Cancer Risk in African American Women. JNCI: J Natl Cancer Institute (2020) 112(12):1213–21. doi: 10.1093/jnci/djaa040
22. Bogdanova N, Pfeifer K, Schürmann P, Antonenkova N, Siggelkow W, Christiansen H, et al. Analysis of a RECQL Splicing Mutation, C. 1667_1667+ 3delagta, in Breast Cancer Patients and Controls From Central Europe. Familial Cancer (2017) 16(2):181–6. doi: 10.1007/s10689-016-9944-y
23. Li N, Rowley SM, Goode DL, Amarasinghe KC, McInerny S, Devereux L, et al. Mutations in RECQL Are Not Associated With Breast Cancer Risk in an Australian Population. Nat Genet (2018) 50(10):1346–8. doi: 10.1038/s41588-018-0206-9
24. Rashid MU, Muhammad N, Khan FA, Shehzad U, Naeemi H, Malkani N, et al. Prevalence of RECQL Germline Variants in Pakistani Early-Onset and Familial Breast Cancer Patients. Hereditary Cancer Clin Practice (2020) 18(1):1–9. doi: 10.1186/s13053-020-00159-6
25. Hu C, Hart SN, Gnanaolivu R, Huang H, Lee KY, Na J, et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. New Engl J Med (2021) 384(5):440–51. doi: 10.1056/NEJMoa2005936
26. Dorling L, Carvalho S, Allen J, González-Neira A, Luccarini C, Wahlström C, et al. Breast Cancer Risk Genes-Association Analysis in More Than 113,000 Women. N Engl J Med (2021) 384(5):428–39. doi: 10.1056/nejmoa1913948
27. Díaz-Gay M, Franch-Expósito S, Arnau-Collell C, Park S, Supek F, Muñoz J, et al. Integrated Analysis of Germline and Tumor DNA Identifies New Candidate Genes Involved in Familial Colorectal Cancer. Cancers (2019) 11(3):362. doi: 10.3390/cancers11030362
28. Lu X, Redon CE, Tang W, Parvathaneni S, Bokhari B, Debnath S, et al. Genome-Wide Analysis Unveils DNA Helicase RECQ1 as a Regulator of Estrogen Response Pathway in Breast Cancer Cells. Mol Cell Biol (2021) 41(4):e00515–20. doi: 10.1128/MCB.00515-20
29. Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-Negative Breast Cancer: Clinical Features and Patterns of Recurrence. Clin Cancer Res (2007) 13(15):4429–34. doi: 10.1158/1078-0432.CCR-06-3045
30. Abdel-Fatah TM, Middleton FK, Arora A, Agarwal D, Chen T, Moseley PM, et al. Untangling the ATR-CHEK1 Network for Prognostication, Prediction and Therapeutic Target Validation in Breast Cancer. Mol Oncol (2015) 9(3):569–85. doi: 10.1016/j.molonc.2014.10.013
31. Abdel-Fatah TM, Arora A, Alsubhi N, Agarwal D, Moseley PM, Perry C, et al. Clinicopathological Significance of ATM-Chk2 Expression in Sporadic Breast Cancers: A Comprehensive Analysis in Large Cohorts. Neoplasia (2014) 16(11):982–91. doi: 10.1016/j.neo.2014.09.009
32. Abdel-Fatah TM, Perry C, Arora A, Thompson N, Doherty R, Moseley PM, et al. Is There a Role for Base Excision Repair in Estrogen/Estrogen Receptor-Driven Breast Cancers? Antioxid Redox Signal (2014) 21(16):2262–8. doi: 10.1089/ars.2014.6077
33. Abdel-Fatah TM, Russell R, Agarwal D, Moseley P, Abayomi MA, Perry C, et al. DNA Polymerase β Deficiency is Linked to Aggressive Breast Cancer: A Comprehensive Analysis of Gene Copy Number, mRNA and Protein Expression in Multiple Cohorts. Mol Oncol (2014) 8(3):520–32. doi: 10.1016/j.molonc.2014.01.001
34. Zhu X, Chen H, Yang Y, Xu C, Zhou J, Zhou J, et al. Distinct Prognosis of mRNA Expression of the Five RecQ DNA-Helicase Family Members–RECQL, BLM, WRN, RECQL4, and RECQL5–in Patients With Breast Cancer. Cancer Manage Res (2018) 10:6649. doi: 10.2147/CMAR.S185769
35. Maynadier M, Nirdé P, Ramirez JM, Cathiard AM, Platet N, Chambon M, et al. Role of Estrogens and Their Receptors in Adhesion and Invasiveness of Breast Cancer Cells. InHormonal Carcinogenesis V (2008) 617:485–91. Springer, New York, NY. doi: 10.1007/978-0-387-69080-3_48
36. Chimge NO, Baniwal SK, Little GH, Chen YB, Kahn M, Tripathy D, et al. Regulation of Breast Cancer Metastasis by Runx2 and Estrogen Signaling: The Role of SNAI2. Breast Cancer Res (2011) 13(6):1–3. doi: 10.1186/bcr3073
37. McGuire WL. Hormone Receptors: Their Role in Predicting Prognosis and Response to Endocrine Therapy. InSeminars Oncol (1978) 5:428–33.
38. Clarke R, Liu MC, Bouker KB, Gu Z, Lee RY, Zhu Y, et al. Antiestrogen Resistance in Breast Cancer and the Role of Estrogen Receptor Signaling. Oncogene (2003) 22(47):7316–39. doi: 10.1038/sj.onc.1206937
39. Musgrove EA, Sutherland RL. Biological Determinants of Endocrine Resistance in Breast Cancer. Nat Rev Cancer (2009) 9(9):631–43. doi: 10.1038/nrc2713
40. Muss HB. Endocrine Therapy for Advanced Breast Cancer: A Review. Breast Cancer Res Treat (1992) 21(1):15–26. doi: 10.1007/BF01811960
41. Madeira M, Mattar A, Logullo ÂF, Soares FA, Gebrim LH. Estrogen Receptor Alpha/Beta Ratio and Estrogen Receptor Beta as Predictors of Endocrine Therapy Responsiveness–a Randomized Neoadjuvant Trial Comparison Between Anastrozole and Tamoxifen for the Treatment of Postmenopausal Breast Cancer. BMC cancer (2013) 13(1):1–2. doi: 10.1186/1471-2407-13-425
Keywords: breast cancer, RECQL, survival, ER-positive, expression
Citation: Mahmoodi A, Shoqafi A, Sun P, Giannakeas V, Cybulski C, Nofech-Mozes S, Masson JY, Sharma S, Samani AA, Madhusudan S, Narod SA and Akbari MR (2022) High Expression of RECQL Protein in ER-Positive Breast Tumours Is Associated With a Better Survival. Front. Oncol. 12:877617. doi: 10.3389/fonc.2022.877617
Received: 16 February 2022; Accepted: 28 April 2022;
Published: 31 May 2022.
Edited by:
Adayabalam Sambasivan Balajee, Oak Ridge Institute for Science and Education (ORISE), United StatesReviewed by:
Pete Simpson, The University of Queensland, AustraliaLawrence Panasci, Segal Cancer Centre, Canada
Copyright © 2022 Mahmoodi, Shoqafi, Sun, Giannakeas, Cybulski, Nofech-Mozes, Masson, Sharma, Samani, Madhusudan, Narod and Akbari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad R. Akbari, bW9oYW1tYWQuYWtiYXJpQHV0b3JvbnRvLmNh
 Ardalan Mahmoodi
Ardalan Mahmoodi Ahmed Shoqafi3
Ahmed Shoqafi3 Vasily Giannakeas
Vasily Giannakeas Sudha Sharma
Sudha Sharma Srinivasan Madhusudan
Srinivasan Madhusudan Steven A. Narod
Steven A. Narod Mohammad R. Akbari
Mohammad R. Akbari