- 1Division of Digestive Surgery, Xijing Hospital of Digestive Diseases, Air force Medical University, Xi’an, China
- 2School of Clinical Medicine, Xi’an Medical University, Xi’an, China
- 3Department of Neurosurgery, Xijing Hospital, Air Force Medical University, Xi’an, China
- 4State Key Laboratory of Cancer Biology, Institute of Digestive Diseases, Xijing Hospital, The Fourth Military Medical University, Xi’an, China
- 5Xi’an Institute of Flight of the Air Force, Ming Gang Station Hospital, Minggang, China
- 6Department of General Surgery, The Southern Theater Air Force Hospital, Guangzhou, China
- 7Department of Experiment Surgery, Xijing Hospital, Fourth Military Medical University, Xi’an, China
Simple summary: Detecting deficient mismatch repair (dMMR) in patients with colorectal cancer is essential for clinical decision-making, including evaluation of prognosis, guidance of adjuvant chemotherapy and immunotherapy, and primary screening for Lynch syndrome. However, outside of tertiary care centers, existing detection methods are not widely disseminated and highly depend on the experienced pathologist. Therefore, it is of great clinical significance to develop a broadly accessible and low-cost tool for dMMR prediction, particularly prior to surgery. In this study, we developed a convenient and reliable model for predicting dMMR status in CRC patients on routine preoperative characterization utilizing multiple machine learning algorithms. This model will work as an automated screening tool for identifying patients suitable for mismatch repair testing and consequently for improving the detection rate of dMMR, while reducing unnecessary labor and cost in patients with proficient mismatch repair.
Background: Deficient mismatch repair (dMMR) indicates a sustained anti-tumor immune response and has a favorable prognosis in patients with colorectal cancer (CRC). Although all CRC patients are recommended to undergo dMMR testing after surgery, current diagnostic approaches are not available for all country hospitals and patients. Therefore, efficient and low-cost predictive models for dMMR, especially for preoperative evaluations, are warranted.
Methods: A large scale of 5596 CRC patients who underwent surgical resection and mismatch repair testing were enrolled and randomly divided into training and validation cohorts. The clinical features exploited for predicting dMMR comprised the demographic characteristics, preoperative laboratory data, and tumor burden information. Machine learning (ML) methods involving eight basic algorithms, ensemble learning methods, and fusion algorithms were adopted with 10-fold cross-validation, and their performance was evaluated based on the area under the receiver operating characteristic curve (AUC) and calibration curves. The clinical net benefits were assessed using a decision curve analysis (DCA), and a nomogram was developed to facilitate model clinical practicality.
Results: All models achieved an AUC of nearly 0.80 in the validation cohort, with the stacking model exhibiting the best performance (AUC = 0.832). Logistical DCA revealed that the stacking model yielded more clinical net benefits than the conventional regression models. In the subgroup analysis, the stacking model also predicted dMMR regardless of the clinical stage. The nomogram showed a favorable consistence with the actual outcome in the calibration curve.
Conclusion: With the aid of ML algorithms, we developed a novel and robust model for predicting dMMR in CRC patients with satisfactory discriminative performance and designed a user-friendly and convenient nomogram.
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second most common cause of cancer-related death worldwide, posing an ongoing threat to public health (1). As a molecular subtype of CRC, microsatellite instability (MSI) or deficient mismatch repair (dMMR) plays a prominent role in the formation of tumors and development of cancer and is therefore a potential therapeutic target (2). dMMR leads to the accumulation of multiple mutations and MSI to further induces tumorigenesis (3), which is observed in approximately 15% of CRC cases and incorporated in the universal screening protocol for Lynch syndrome (4).
MSI or dMMR has a favorable prognosis but gains no benefit from neoadjuvant chemotherapy with fluorouracil in CRC patients with stage II (5). More importantly, recent studies elucidate that immune checkpoint blockade (ICB) treatment in patients with MSI or dMMR can yield remarkable clinical benefits (6). Further investigations suggest that the benefit of ICB treatment in these patients is not limited to CRC but encompasses all solid tumors (7). Now, MSI or dMMR has been approved by the FDA as the first pan-cancer biomarker for immunotherapy response (8). Given the extensive clinical applications, MSI or dMMR testing has been recommended by multiple international guidelines for all CRC patients (9–11). However, in clinical practice, not all CRC patients are tested for MSI or dMMR, especially those in developing cities and hospitals; this is because testing requires particular genetic or immunohistochemical examinations, which are costly and time-consuming and rely on excellent pathology laboratories and doctors (12). Therefore, a low-cost tool that can be used in all CRC patients from different cities and hospitals is essential.
Machine learning (ML) has shown great potential in identifying features of disease subtypes and outcomes, with successful application in disease screening (13), prognosis evaluation (14), and efficacy prediction (15). Previous studies have suggested that ML can recognize pathomorphological characteristics that contribute to MSI or dMMR from hematoxylin and eosin-stained tissue sections. Although these results are promising, the performance of ML is poorer when tissue areas are smaller than surgically resected specimens and is subject to inter-hospital variability in relation to the quality of histological sections (16, 17). Meanwhile, researchers have discovered susceptibility genes associated with MSI or dMMR from genomic sequencing data based on ML algorithms. However, genetic testing is expensive, making it difficult to test all patients (18).
In this study, we aimed to develop an ML-based model for predicting dMMR that is easily accessible and consistent among different regions and hospitals based on routine clinical data. As an automated screening tool, the model is expected to complement further confirmatory testing, while reducing unnecessary labor and cost in patients with proficient mismatch repair (pMMR).
Methods
Patients
This retrospective study included a primary cohort of CRC patients who underwent surgical resection and dMMR testing between January 2015 and August 2021 in the Xijing Hospital of Digestive Diseases, Air Force Military Medical University (Shaanxi, China). The inclusion criteria were as follows: (1) primary colorectal cancer confirmed via cytological and histological examinations; (2) dMMR testing; and (3) complete clinical and pathological data. In contrast, the exclusion criteria were as follows: (1) coexistence of other primary malignant tumors and (2) chemoradiotherapy, immunotherapy, and other related anti-tumor therapies before surgery. In total, 5596 patients and 80 clinical features, including primary demographic characteristics, tumor information, and routine laboratory data, were investigated in our study. All data retrieved from electronic patient records were further transformed and normalized to facilitate feature selection and model building, and all clinical data were explicitly scrutinized. The study was approved by the Medical Ethics Committee of the First Affiliated Hospital of the Air Force Military Medical University (No. KY20112170-C-1).
Model development
Traditional models depend largely on human-selected features, while ML can learn features from data, which allows researchers to obtain untapped information and detect difficult-to-discern patterns (19). Therefore, we constructed a predictive model based on ML. In this study, the cohort was split into separate training and validation sets at an 8:2 ratio using the light gradient boosting machine (LGBM) (random state = 3), which could maintain the same ratio of positive-to-negative samples in the training and validation sets. As a classical approach, cross-validation is typically exploited to avoid overfitting of training data in ML (20). Thus, in the training set, we employed k-fold cross-validation (k = 10), a procedure that uses subsets of data to iteratively train and validate the predictive performance of a model. In this procedure, a cohort is randomly divided into 10 subsets, of which nine subsets are included in the training set and the remaining subset in the testing set; the optimal hyperparameters arise from the combination of the best cross-validation results and the model that will provide the best predictive performance on a new sample (21). For model development, we trained eight basic models based on eight different ML pipelines. The LGBM is a highly efficient gradient boosting decision tree suitable for scenarios with large amounts of data and high-dimensional features (22). Random forest (RF) is an ensemble learning algorithm that builds and merges diverse decision trees to obtain the optimal classification performance (23). Gaussian Naive Bayesian (GNB), a supervised learning method, approaches the classification task with the naive assumption of independence between every pair of features (24). The K-nearest neighbor (KNN) is a simple and effective classification algorithm using the k-nearest points of inputs to predict responses (25). The multilayer perceptron (MLP) is a feedforward neuronal network consisting of an input layer, an output layer, and one or more hidden layers that are closely connected; it is widely used in distinguishing data that are not linearly separable (26, 27). A classification and regression tree (CART) is a modeling approach for classification (binary response) and regression (continuous response) that has been successfully utilized in clinical practice (28). The support vector machine (SVM) is currently the mainstream classifier in ML, which has been intensively applied for pattern recognition and data classification (29). Logistic regression (LR) is a generalized linear model used to solve binary classification problems, which can fit binary or multinomial LR (30). To further optimize the model, we applied bagging bootstrap aggregation, an ensemble technique used for classification or regression, to determine the hyperparameters of the basic models (31). Next, we built a fusion model with hard voting (based on the average of the bagging model) or soft voting (based on the weighted average of the predicted class probabilities) (32).
Feature importance
Shapley additive explanation (SHAP), one of the most optimal ML explication methods based on the game theory, allows both local and global interpretabilities of the output of any ML-based model (33). To determine which features contributed most to the model predictions, we calculated and visualized the SHAP values on the stacking predictions. Each SHAP value measured how much each feature contributed, either positively or negatively, to the risk of dMMR assigned by the model.
Model performance
We adopted the area under the receiver operating characteristic curve (AUC) as the mainstay parameter of model performance. Furthermore, we calculated and compared the sensitivity, specificity, precision, negative predictive value (NPV), false discovery rate (FDR), accuracy, average precision (AP), and confusion matrix to further assess the comprehensive performance of the ML-based models. In addition, a decision curve analysis (DCA) was employed to evaluate the clinical net benefit of the models (34).
Statistical analysis
Statistical analysis was conducted using R (version 4.1.2; https://www.Rproject.org), and ML modeling was performed using Python (version 3.6.5; https://www.python.org). The following Python packages were used: “imblearn,” “sklearn,” “lightgbm,” “randomForest,” and “mlxtend.” Meanwhile, the following R packages were utilized: “tableone,” “survival,” “mice,” and “rms.” Qualitative data were compared between the two groups using the χ2 test or Fisher’s exact test. P < 0.05 was considered statistically significant.
Results
Baseline clinical data
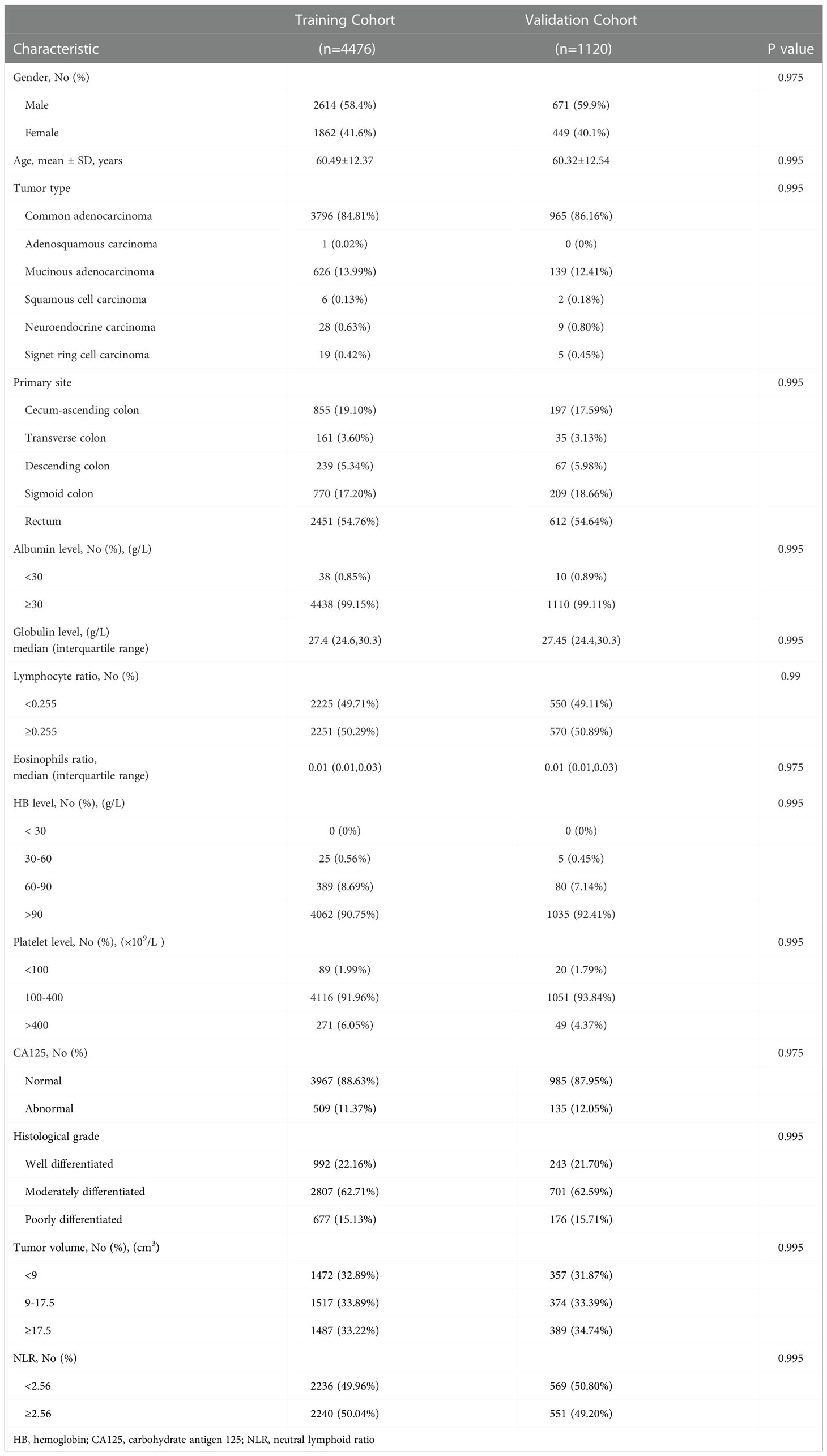
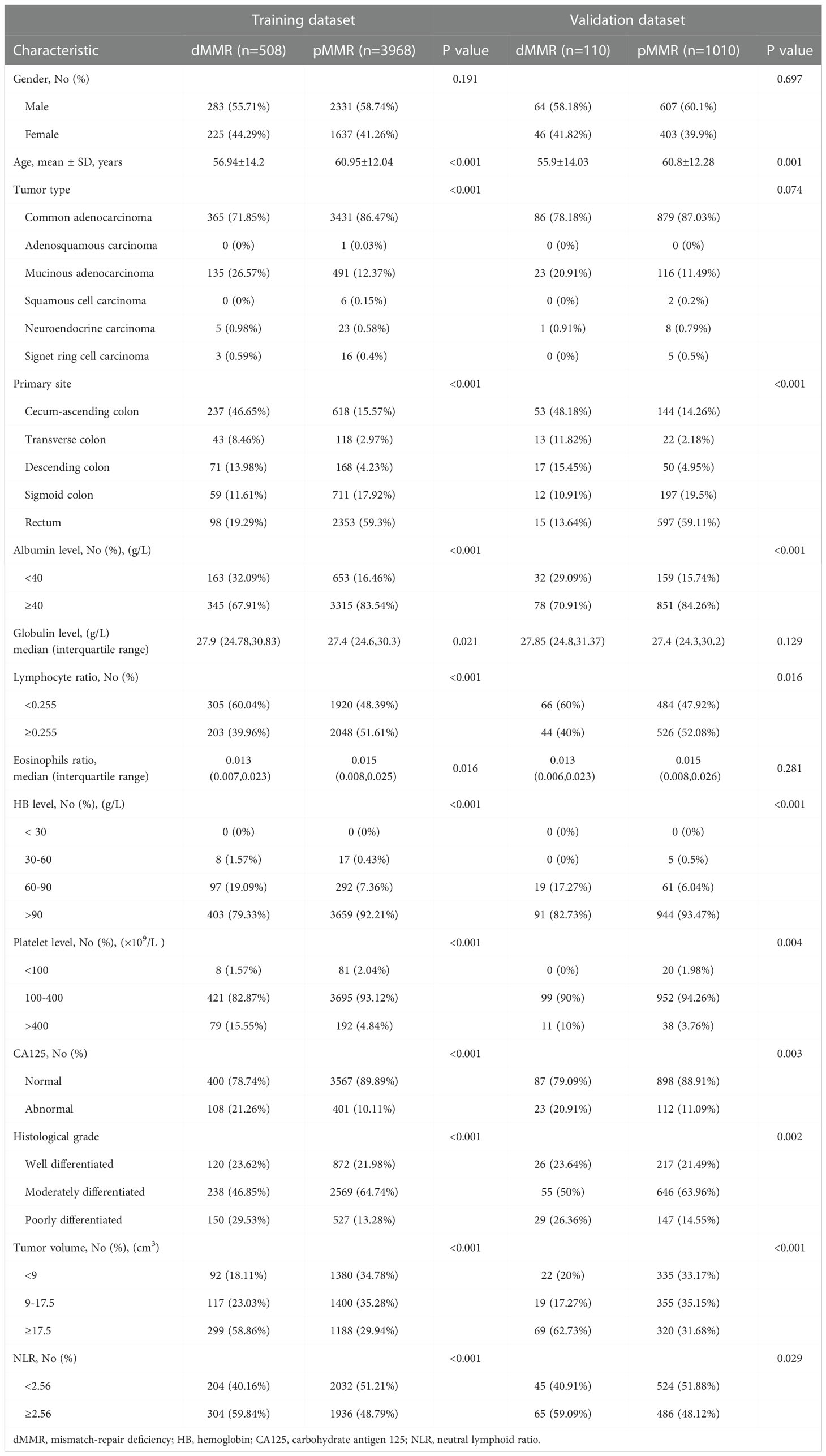
The flowchart of this study is shown in Figure 1. The primary data were extracted from electronic medical records between January 2015 and August 2021. According to the inclusion and exclusion criteria, 5596 patients were enrolled as the study population: 4476 patients included in the training cohort and 1120 in the validation cohort. No significant differences were found in the prevalence of dMMR between the two cohorts. dMMR was found in 508 (11.3%) and 110 (9.82%) patients in the training and validation cohorts, respectively. There was no significant difference between the difference of clinical characteristics between the training cohorts and testing cohorts, including gender, age, tumor type, primary site, histological grade, tumor size and other experimental indicators (P >0.05). The baseline characteristics of the cohorts are shown in Table 1.
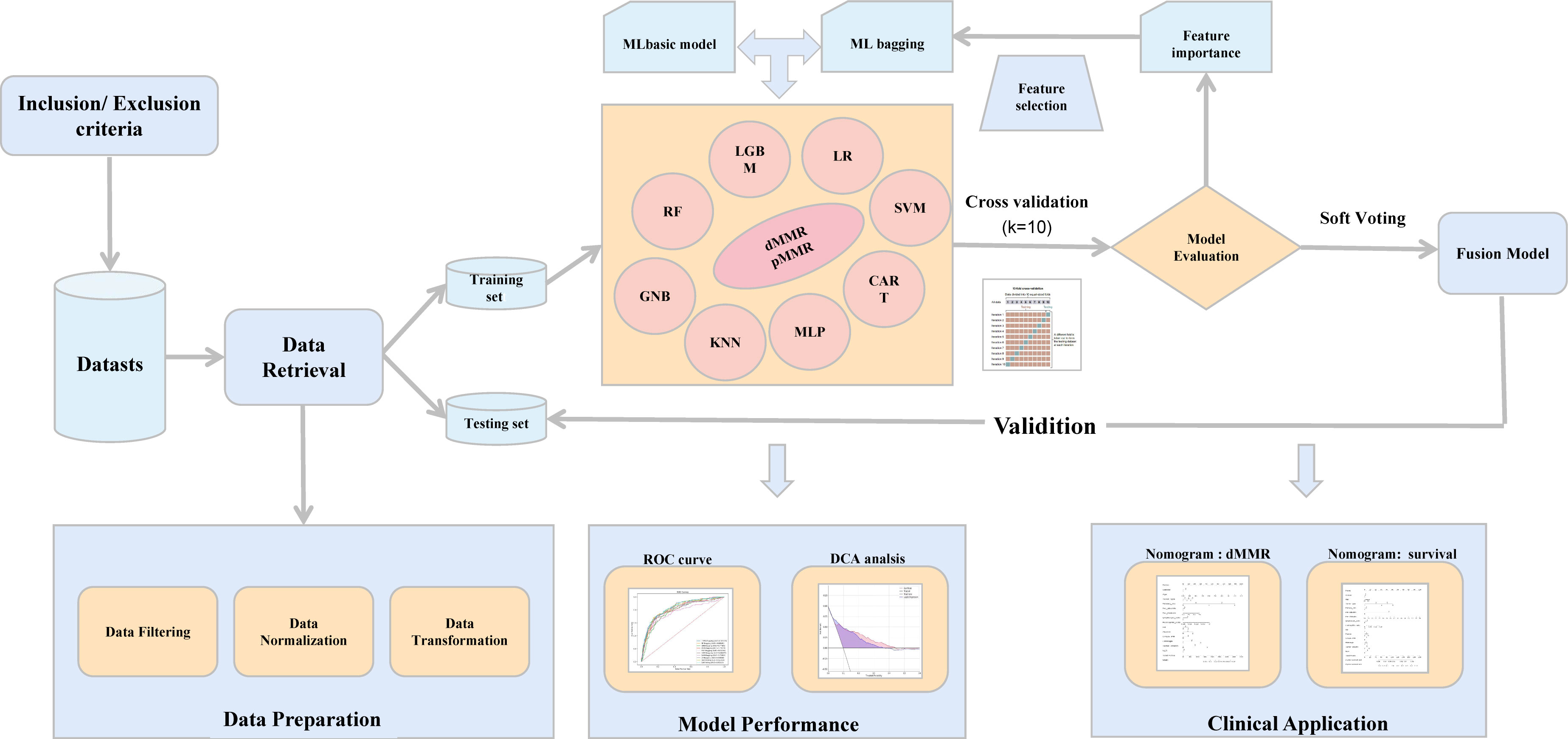
Figure 1 The workflow of the study. ML, machine learning; ROC, receiver operating characteristic; DCA, decision curve analysis.
Fourteen clinical features with better performance were eventually employed to build the final model: age at diagnosis, sex, hemoglobin (HB) level, platelet count, albumin level, globulin level, lymphocyte ratio, eosinophil ratio, neutral lymphoid ratio (NLR), CA125 level, primary site, histologic tumor grade, tumor type, and tumor volume. The baseline characteristics of the dMMR and pMMR groups are shown in detail in Table 2.
Parameters
We trained the LGBM with a depth of 5, a learning rate of 0.012, basic learners of 230, a leaf size of 8, and maximum bins of 256. For the RF and CART, the maximum depths of the basic trees were 10, and the basic learners were 500. For the KNN, the leaf size was 30, and the optimum number of neighbors was 400. For the MLP, we used three hidden layers with a size of 50, 30, and 10, respectively; a learning rate of 0.08; and the Adam optimizer and ReLU activation function. For the SVM, we combined a C value of 0.01 with a kernel smoothing parameter of 0.01. Each bagging model was also linked with 10 datasets generated by random sampling for 10 times, allowing the construction of the predictive models based on identical basic algorithms and evaluation of their performance using the 10-fold cross-validation test. The eventual stacking model consisted of eight bagging models, with final weight values of 42 (LGBM), 12 (RF), 2 (GNB), 4 (KNN), 4 (MLP), 10 (DT), 34 (SVM), and 24 (LR).
Feature importance
To further determine the association of each feature with the outcome of mismatch repair (MMR), we employed the SHAP values. Meanwhile, we calculated and visualized the significance of each feature by analyzing the SHAP values in all individual models (Supplementary Table S1) and stacking model (Figure 2). The 10 most important features in the stacking model were the primary tumor site, tumor volume, age at diagnosis, histological tumor grade, tumor type, and preoperative HB level, albumin level, platelet count, NLR, and eosinophil ratio. Notably, a larger tumor volume, younger age, higher histological tumor grade, higher platelet count, and higher NLR ratio were associated with a higher risk of dMMR, while a higher preoperative HB level, albumin level, and eosinophil ratio were related to a lower risk of dMMR.
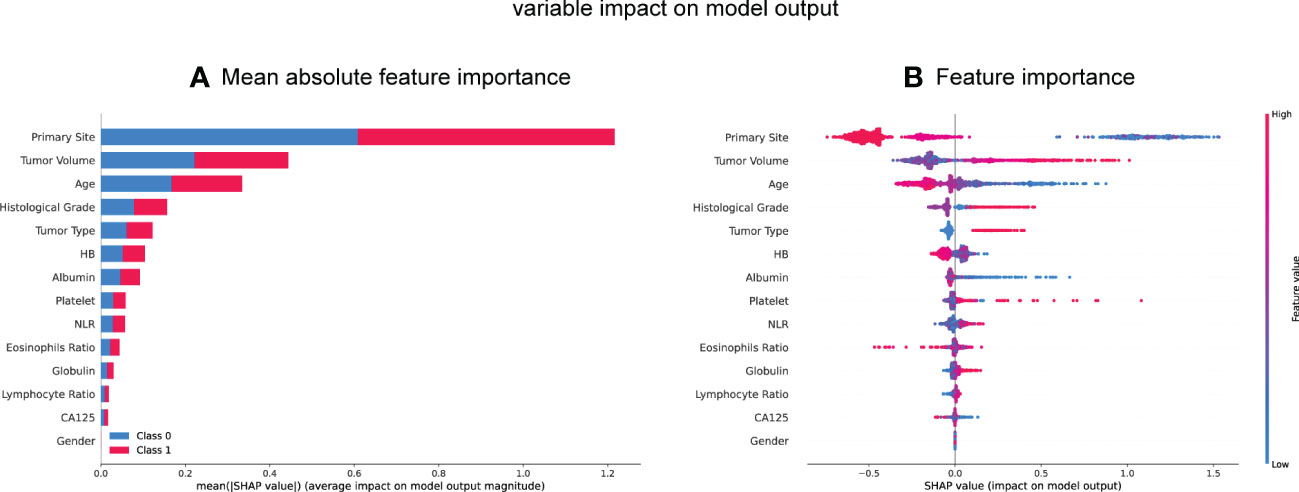
Figure 2 SHAP summary plot of the 14 feature of the stacking model. (A, B) Each dot corresponds to each feature attribution value for the model of each patient, with positive values indicating a contribution that increase the probability of dMMR development while negative values indicating a contribution that decreases the probability. Dots are colored according to the values of features for the respective patient and accumulate vertically to depict density. Red represents higher feature values, and blue respective lower feature values. HB, hemoglobin; NLR, neutral lymphoid ratio; CA125, carbohydrate antigen 125.
Model performance
Expectedly, all individual models demonstrated optimal predictive performance in the internal validation set (Figure 3A). Additionally, the stacking model exhibited an outstanding predictive performance, with a relatively high AUC of 0.831 and NPV of 97.03%. The AUC, sensitivity, specificity, precision, NPV, FDR, accuracy, AP, F1-sore, and MCC of each model in the internal validation set are listed in Supplementary Table S2. An intuitive and concise confusion matrix was also employed to evaluate the performance of the models; the detailed outcomes of model prediction in the internal testing set are shown in (Supplementary Table S3).
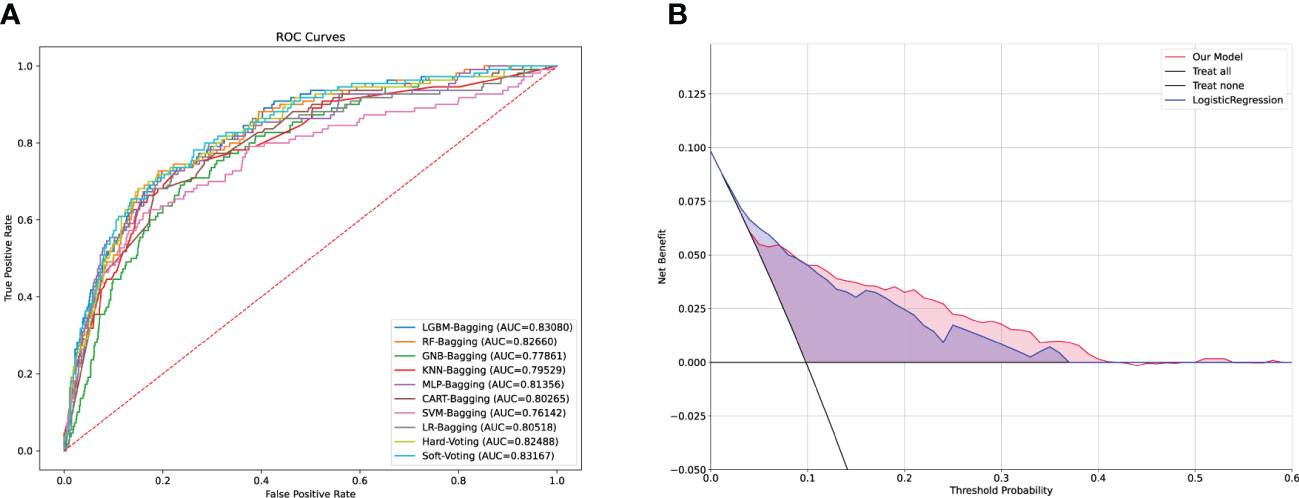
Figure 3 Comparison of the performance among machine learning models and DCA analysis. (A) ROC curves of the machine learning in the validation cohort. (B) Logistic DCA analysis for two models in the validation cohort. Blue shading (LR model) represents the traditional model only based on logistic regression algorithm, and red shading (stacking model) means the machine model based on ensemble learning strategies and machine learning fusion algorithms in our study. ROC, receiver operating characteristic; DCA, decision curve analysis; AUC, area under curve; LR, logistic regression.
To further appraise whether our ML-based models performed better than did the classical LR strategy, we fit our data in the LR models. The stacking model had a higher AUC and NPV than the LR model (Supplementary Table S4). Considering the clinical implications and with the guidance of the models, we drew a DCA curve by performing a logistic DCA analysis. The corresponding results revealed a favorable net clinical benefit of both the stacking and LR models, although the stacking model had a stronger effect (Figure 3B).
Subgroup analysis
To further confirm the comprehensive performance of the stacking model in complicated clinical scenarios, we stratified our cohort into four subgroups according to the AJCC stage. The analysis showed that the stacking model achieved a promising discriminative capacity, irrespective of the tumor stage (Figure 4).
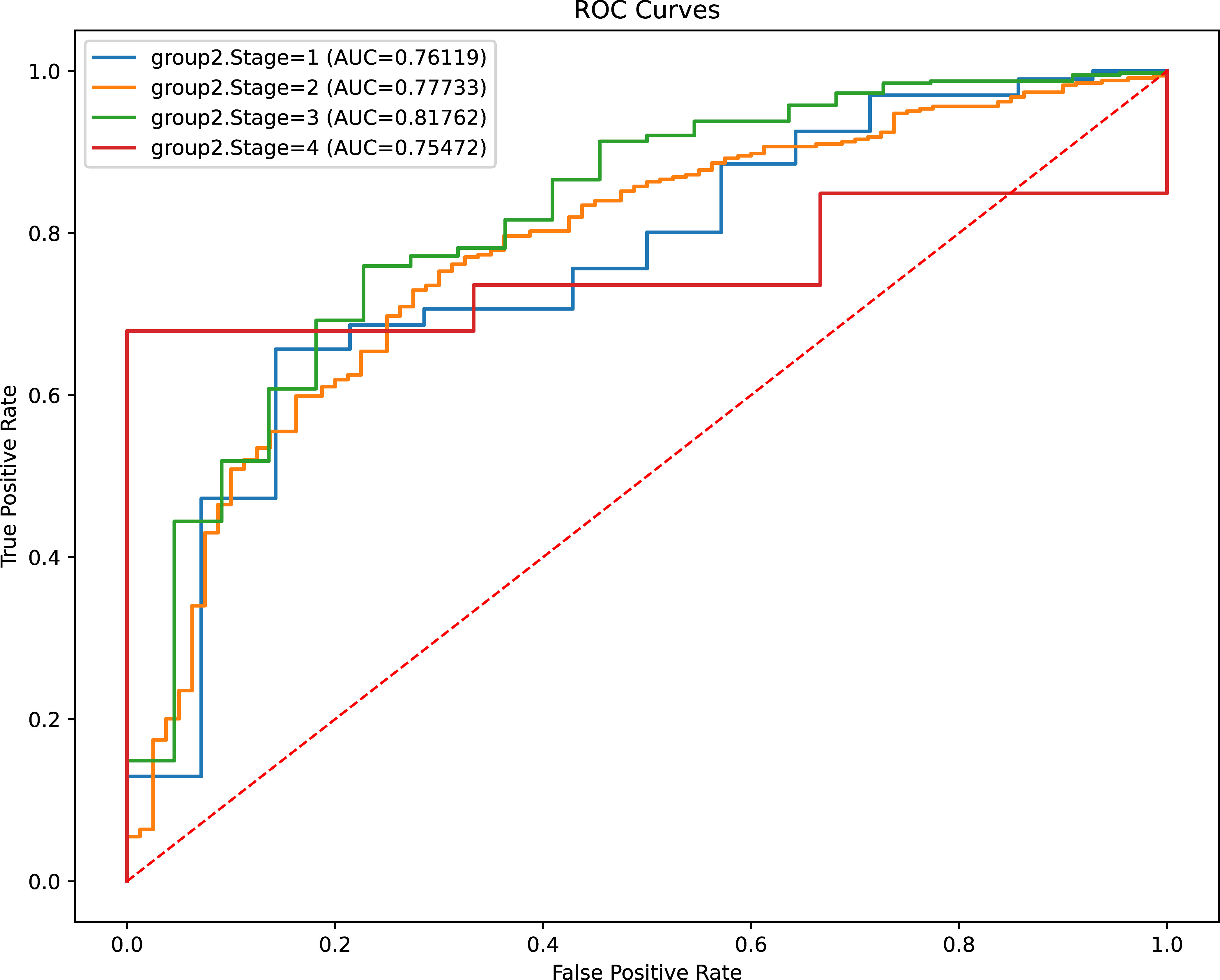
Figure 4 Evaluation of models’ discriminant capability for CRC patients with different clinical stage. ROC, receiver operating characteristic; AUC, area under curve.
Clinical application
To facilitate the clinical application of the model, we further developed a concise and accessible nomogram based on 14 crucial features that can be used to accurately assess the MMR status of CRC patients (Figure 5A). Thereafter, a calibration curve was adopted to evaluate the predictive power of the nomogram. The calibration curve indicated that the error between the actual and predicted dMMR rates was very small, suggesting that the nomogram possesses a preferable accuracy in predicting dMMR (Figure 5B). The features could also well predict the prognosis of CRC patients. Similarly, we constructed a prognostic nomogram and calibration curve, which showed that the predictive power was close to the ideal curve, indicating that the prognostic nomogram has a great predictive capability (Figure 5C, D).
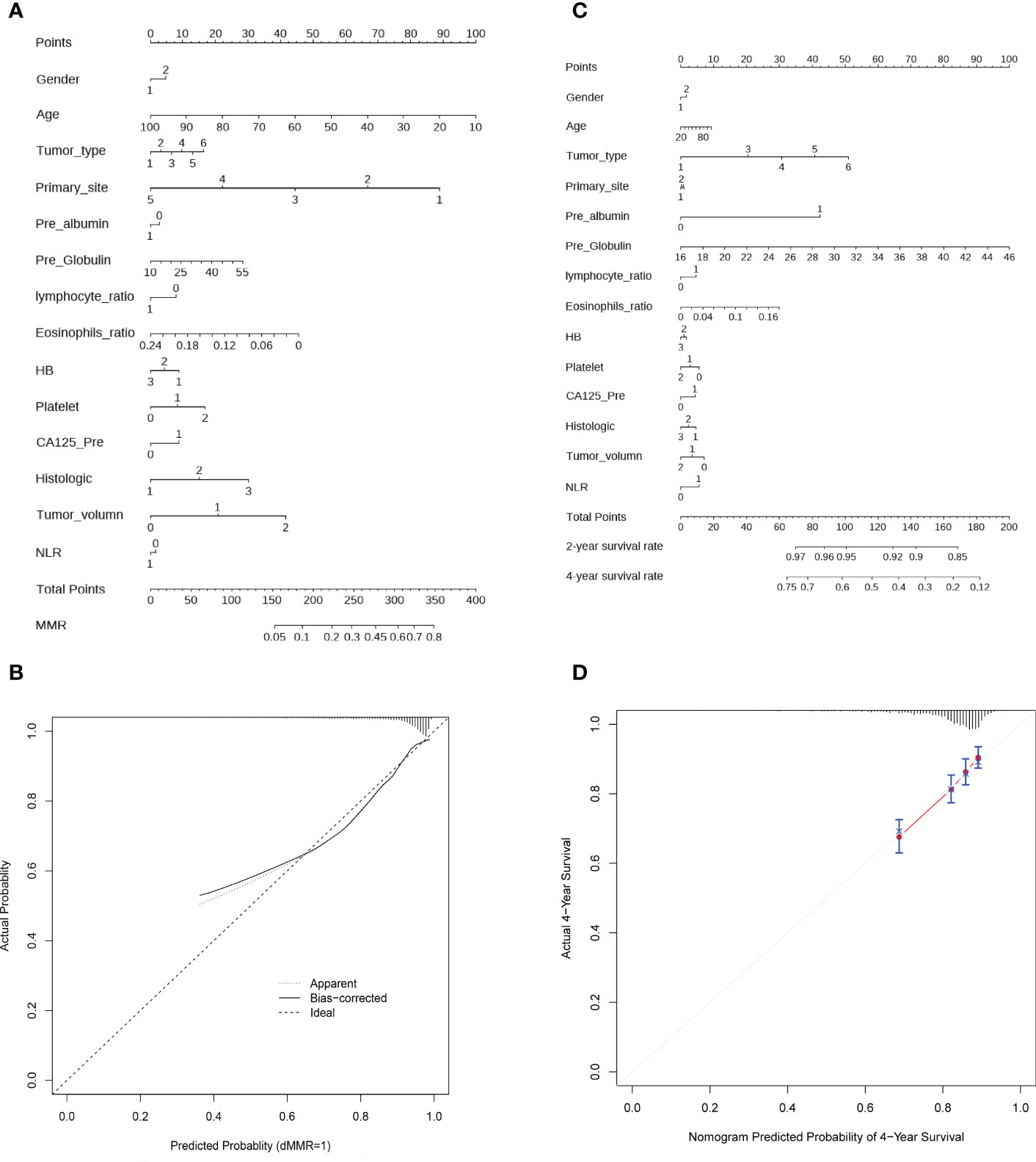
Figure 5 Construction of nomogram and calibration diagram. (A) A nomogram to predict the dMMR probability of CRC patients with preoperative routine indexes. In the nomogram, the total points are the sum of individual point for each feature, with larger total points reflecting greater probability of dMMR. (B) Calibration curve to evaluate the predictive power of the nomogram. (C) Nomogram to predict the 2- and 4- year OS of CRC patient, with higher total points denoting worse prognosis. (D) Calibration curve for the estimation of 4-year OS predicted by the nomogram. The diagonal dotted line represents theoretical response of perfect nomogram, the red solid line indicates the performance of nomogram. Abbreviations: HB, hemoglobin; NLR, neutral lymphoid ratio; CA125, carbohydrate antigen 125; dMMR, mismatch repair deficiency.
Discussion
dMMR or MSI in CRC patients can serve as a cardinal parameter for clinical decision-making, as it identifies a distinct patient subset with a favorable prognosis, in whom treatment with standard fluorouracil-based chemotherapy has no clinical benefit and in whom ICB treatment might be of remarkable benefit (35). However, in clinical practice, only a portion of CRC patients is tested for dMMR or MSI owing to the associated high costs and dependence on the operator. Therefore, a low-cost, broadly accessible, and robust predictive model for dMMR or MSI that can guide diagnostic and therapeutic strategies is urgently needed.
Recently, an increasing number of researchers harnessed ML as a breakthrough tool for sophisticated medical issues, which has made great clinical contributions, including disease prediction, prognosis evaluation, and new drug development. ML-based models can achieve considerably better accuracy, stability, and interpretability compared with traditional regression models. Thus, we developed and validated an innovative and cost-efficient model to predict dMMR by utilizing eight ML algorithms and combining 14 parameters that are ubiquitously available in clinical practice. Although partial parameters were found to have relatively weak contribution to the model output, they were essential for overall model performance (36). In order to get a higher sensitivity, specificity and prediction power model, we retained all parameters filtered by multiple ML algorithms. Notably, all tests for these parameters were completely standardized and consistent among different regions and hospitals. Thus, the findings could be easily generalized to the complicated clinical environment. The receiver operating characteristic curve analysis showed that both the final fusion model and individual models had a satisfactory performance, while the DCA analysis revealed more net benefits of the fusion model than of the conventional regression model. Most importantly, our model showed a prominent capability in recognizing dMMR in CRC patients regardless of the clinical stage. Ultimately, we constructed a user-friendly nomogram comprising the features of the fusion model that can be used in identifying dMMR in CRC patients and treating them with a personalized strategy.
We established an ML-based model based on adequate clinical data to predict dMMR in CRC patients. Compared with previous analogous studies discerning dMMR or MSI with the combination of histomorphological patterns with features associated with dMMR or MSI from hematoxylin and eosin-stained slides of CRC patients and combination of susceptible genes related to dMMR or MSI with transcriptome data, our study had several considerable strengths. First, we focused on incorporating dMMR and adequate clinical information into our model, which was easily accessible and closely associated with the comprehensive state of the patients. Meanwhile, the detection methods of all features were objective and normalized, which eliminated the variation between hospitals at different levels and between different locations. Notably, all data were derived from preoperative examinations, which could be useful in identifying patients with dMMR early, especially those who could not undergo surgical resection or those who developed distant metastases. Second, all ML-based models achieved a satisfactory discriminative power. More importantly, both the individual and stacking models yielded a crucial NPV (> 0.97), indicating that our model could furnish an additional value as an automatic screening tool to assist in clinical decision-making before confirmatory MMR testing. Specifically, if all patients predicted to have pMMR are excluded from confirmatory detection, this will dramatically decrease the number of patients undergoing MMR testing, yielding substantial test-related labor and cost savings. Third, several previous studies only used a single ML algorithm, while our study utilized ensemble learning strategies and ML fusion algorithms, which generated comprehensive learning with strong robustness and generalizability. Finally, previous studies have suggested that concordance between MMR and MSI analysis is quite high especially in CRC patients (37), so our model may be useful for MSI assessment.
Our findings on the clinicopathological characteristics of dMMR are consistent with previously published data. dMMR was more frequently associated with mucinous adenocarcinoma, poorly differentiated, and mostly located in the right hemicolon (38, 39). Moreover, younger CRC patients were more likely to have dMMR than older patients, which might be related to their higher metabolism rate (40). Notably, beyond these classical risk factors, our research also identified several risk factors particularly relevant for dMMR by adopting multiple ML methods; these factors included a larger tumor volume; lower HB level, albumin level, lymphocyte ratio, and eosinophil ratio; and higher platelet count, globulin level, NLR, and CA125 level. Numerous studies have demonstrated that inflammatory mediators, important promoters of genetic alterations, play an important role in tumor initiation and progression (41). Specifically, inflammatory immune cells, such as macrophages and neutrophils, produce a wide variety of reactive substances, which could directly trigger DNA damage of nonimmune cells to further increase mutation frequency and genomic instability (42). When DNA sustains damage, specific proteins and enzymes are activated to repair such damage (43). During this process, HB and albumin as the primary vectors of nutrients could reduce DNA injury and strengthen the ability to repair DNA damage. Conversely, if the levels of HB and albumin decrease, the capacity to repair DNA damage will distinctly increase (44, 45). These findings coincide with our data on the lower levels of HB and albumin and higher NLR. Accumulating evidence also suggests that platelets play a critical role in angiogenesis modulation, tumor immune microenvironment maintenance, and cancer progression (46). An increased platelet count might be strongly associated with carcinogenesis and prognosis of CRC patients with dMMR (47). Interestingly, eosinophils could have both positive and negative effects: They could both promote tumor progression and exert anti-tumor activities by secreting cytokines (48, 49). Previous studies have also indicated that CA125 could serve as an important diagnostic and prognostic marker in CRC (50). Although several studies reported that these features are closely associated with dMMR in CRC patients, the biological explanations for the underlying mechanisms are not well understood, which is worthy of further exploration.
Because the immune microenvironment and clinicopathological features were varied greatly in different tumors, our model may not be applicable to the assessment of MSI or MMR in other tumors. However, the insights and methods of this study were completely generalized on other tumors. Additionally, this study had several limitations. First, our analysis was conducted at a single medical center; more external test cohorts from different hospitals and regions are of great necessity to enhance the robustness and generalizability of our models. Second, this study had a retrospective design; a large prospective clinical trial is necessary before MMR testing could be routinely performed in clinical practice. Third, although we utilized SHAP values and previous data to interpret the feature performance in our model, further basic science studies are required to determine the underlying mechanisms.
Conclusions
In conclusion, we successfully confirmed 14 features closely related to dMMR in CRC patients: age, sex, HB level, platelet count, albumin level, globulin level, lymphocyte ratio, eosinophil ratio, NLR, CA125 level, primary site, histologic tumor grade, tumor type, and tumor volume. An innovative and universal fusion model based on multiple ML algorithms was constructed to predict dMMR in CRC patients, which achieved a satisfactory predictive performance. A user-friendly nomogram was also adopted, which yielded benefits and demonstrated prospects for clinical application.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
Written informed consent was not obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
JL and JZ designed the study. DX, RC, and YJ contributed to the conception of the study and completed the manuscript together. SW, ZL and XC contributed significantly to statistical analysis and manuscript preparation. XF and JZ helped perform the analysis with constructive discussions. All authors contributed to the article and approved the submitted version.
Funding
This research was founding in part by grants from the National Natural Science Foundation of China (82172781) and the Key Research and Development Program of Shaanxi (2019SF-010).
Acknowledgments
We are thankful to Air Force Military Medical University first affiliation Xijing digestive hospital (Shaanxi, China) for supporting this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.1049305/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med (2015) 21(11):1350–6. doi: 10.1038/nm.3967
3. Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med (2016) 22(11):1342–50. doi: 10.1038/nm.4191
4. Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, et al. Feasibility of screening for lynch syndrome among patients with colorectal cancer. J Clin Oncol (2008) 26(35):5783–8. doi: 10.1200/JCO.2008.17.5950
5. Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol (2010) 28(20):3219–26. doi: 10.1200/JCO.2009.27.1825
6. Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-Deficient/Microsatellite instability-high metastatic colorectal cancer. J Clin Oncol (2018) 36(8):773–9. doi: 10.1200/JCO.2017.76.9901
7. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science (2017) 357(6349):409–13. doi: 10.1126/science.aan6733
8. Prasad V, Kaestner V, Mailankody S. Cancer drugs approved based on biomarkers and not tumor type-FDA approval of pembrolizumab for mismatch repair-deficient solid cancers. JAMA Oncol (2018) 4(2):157–8. doi: 10.1001/jamaoncol.2017.4182
9. Dong C, Ding Y, Weng S, Li G, Huang Y, Hu H, et al. Update in version 2021 of CSCO guidelines for colorectal cancer from version 2020. Chin J Cancer Res (2021) 33(3):302–7. doi: 10.21147/j.issn.1000-9604.2021.03.02
10. Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, et al. Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw (2021) 19(3):329–59. doi: 10.6004/jnccn.2021.0012
11. Yoshino T, Argilés G, Oki E, Martinelli E, Taniguchi H, Arnold D, et al. Pan-Asian adapted ESMO clinical practice guidelines for the diagnosis treatment and follow-up of patients with localised colon cancer. Ann Oncol (2021) 32(12):1496–510. doi: 10.1016/j.annonc.2021.08.1752
12. Shia J. The diversity of tumours with microsatellite instability: molecular mechanisms and impact upon microsatellite instability testing and mismatch repair protein immunohistochemistry. Histopathology (2021) 78(4):485–97. doi: 10.1111/his.14271
13. Lu MT, Raghu VK, Mayrhofer T, Aerts H, Hoffmann U. Deep learning using chest radiographs to identify high-risk smokers for lung cancer screening computed tomography: Development and validation of a prediction model. Ann Intern Med (2020) 173(9):704–13. doi: 10.7326/M20-1868
14. Poirion OB, Jing Z, Chaudhary K, Huang S, Garmire LX. DeepProg: an ensemble of deep-learning and machine-learning models for prognosis prediction using multi-omics data. Genome Med (2021) 13(1):112. doi: 10.1186/s13073-021-00930-x
15. Feng L, Liu Z, Li C, Li Z, Lou X, Shao L, et al. Development and validation of a radiopathomics model to predict pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer: a multicentre observational study. Lancet Digit Health (2022) 4(1):e8–e17. doi: 10.1016/S2589-7500(21)00215-6
16. Echle A, Grabsch HI, Quirke P, van den Brandt PA, West NP, Hutchins GGA, et al. Clinical-grade detection of microsatellite instability in colorectal tumors by deep learning. Gastroenterology (2020) 159(4):1406–1416.e1411. doi: 10.1053/j.gastro.2020.06.021
17. Yamashita R, Long J, Longacre T, Peng L, Berry G, Martin B, et al. Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet Oncol (2021) 22(1):132–41. doi: 10.1016/S1470-2045(20)30535-0
18. Bao X, Zhang H, Wu W, Cheng S, Dai X, Zhu X, et al. Analysis of the molecular nature associated with microsatellite status in colon cancer identifies clinical implications for immunotherapy. J Immunother Cancer (2020) 8(2):e001437. doi: 10.1136/jitc-2020-001437
19. Churpek MM, Yuen TC, Winslow C, Meltzer DO, Kattan MW, Edelson DP. Multicenter comparison of machine learning methods and conventional regression for predicting clinical deterioration on the wards. Crit Care Med (2016) 44(2):368–74. doi: 10.1097/CCM.0000000000001571
20. Ferdinandy B, Gerencsér L, Corrieri L, Perez P, Újváry D, Csizmadia G, et al. Challenges of machine learning model validation using correlated behaviour data: Evaluation of cross-validation strategies and accuracy measures. PloS One (2020) 15(7):e0236092. doi: 10.1371/journal.pone.0236092
21. Moreno-Torres JG, Saez JA, Herrera F. Study on the impact of partition-induced dataset shift on k-fold cross-validation. IEEE Trans Neural Netw Learn Syst (2012) 23(8):1304–12. doi: 10.1109/TNNLS.2012.2199516
22. Ahn BC, So JW, Synn CB, Kim TH, Kim JH, Byeon Y, et al. Clinical decision support algorithm based on machine learning to assess the clinical response to anti-programmed death-1 therapy in patients with non-small-cell lung cancer. Eur J Cancer (2021) 153:179–89. doi: 10.1016/j.ejca.2021.05.019
23. Ubels J, Schaefers T, Punt C, Guchelaar HJ, de Ridder J. RAINFOREST: a random forest approach to predict treatment benefit in data from (failed) clinical drug trials. Bioinformatics (2020) 36(Suppl_2):i601–9. doi: 10.1093/bioinformatics/btaa799
24. Garcia-Vidal C, Puerta-Alcalde P, Cardozo C, Orellana MA, Besanson G, Lagunas J, et al. Machine learning to assess the risk of multidrug-resistant gram-negative bacilli infections in febrile neutropenic hematological patients. Infect Dis Ther (2021) 10(2):971–83. doi: 10.1007/s40121-021-00438-2
25. Abu Alfeilat HA, Hassanat ABA, Lasassmeh O, Tarawneh AS, Alhasanat MB, Eyal Salman HS, et al. Effects of distance measure choice on K-nearest neighbor classifier performance: A review. Big Data (2019) 7(4):221–48. doi: 10.1089/big.2018.0175
26. Alkadri S, Ledwos N, Mirchi N, Reich A, Yilmaz R, Driscoll M, et al. Utilizing a multilayer perceptron artificial neural network to assess a virtual reality surgical procedure. Comput Biol Med (2021) 136:104770. doi: 10.1016/j.compbiomed.2021.104770
27. Tang J, Deng C, Huang GB. Extreme learning machine for multilayer perceptron. IEEE Trans Neural Netw Learn Syst (2016) 27(4):809–21. doi: 10.1109/TNNLS.2015.2424995
28. Nunn ME, Fan J, Su X, Levine RA, Lee HJ, McGuire MK. Development of prognostic indicators using classification and regression trees for survival. Periodontol 2000 (2012) 58(1):134–42. doi: 10.1111/j.1600-0757.2011.00421.x
29. Nedaie A, Najafi AA. Support vector machine with dirichlet feature mapping. Neural Netw (2018) 98:87–101. doi: 10.1016/j.neunet.2017.11.006
30. LaValley MP. Logistic regression. Circulation (2008) 117(18):2395–9. doi: 10.1161/CIRCULATIONAHA.106.682658
31. Hsiao YW, Tao CL, Chuang EY, Lu TP. A risk prediction model of gene signatures in ovarian cancer through bagging of GA-XGBoost models. J Adv Res (2021) 30:113–22. doi: 10.1016/j.jare.2020.11.006
32. Jones IA, Van Oyen MP, Lavieri MS, Andrews CA, Stein JD. Predicting rapid progression phases in glaucoma using a soft voting ensemble classifier exploiting kalman filtering. Health Care Manag Sci (2021) 24(4):686–701. doi: 10.1007/s10729-021-09564-2
33. Nohara Y, Matsumoto K, Soejima H, Nakashima N. Explanation of machine learning models using shapley additive explanation and application for real data in hospital. Comput Methods Prog BioMed (2022) 214:106584. doi: 10.1016/j.cmpb.2021.106584
34. Vickers AJ, Holland F. Decision curve analysis to evaluate the clinical benefit of prediction models. Spine J (2021) 21(10):1643–8. doi: 10.1016/j.spinee.2021.02.024
35. Zaanan A, Meunier K, Sangar F, Fléjou JF, Praz F. Microsatellite instability in colorectal cancer: from molecular oncogenic mechanisms to clinical implications. Cell Oncol (Dordr) (2011) 34(3):155–76. doi: 10.1007/s13402-011-0024-x
36. Tseng PY, Chen YT, Wang CH, Chiu KM, Peng YS, Hsu SP, et al. Prediction of the development of acute kidney injury following cardiac surgery by machine learning. Crit Care (2020) 24(1):478. doi: 10.1186/s13054-020-03179-9
37. Bartley AN, Luthra R, Saraiya DS, Urbauer DL, Broaddus RR. Identification of cancer patients with lynch syndrome: clinically significant discordances and problems in tissue-based mismatch repair testing. Cancer Prev Res (Phila) (2012) 5(2):320–7. doi: 10.1158/1940-6207.CAPR-11-0288
38. Jenkins MA, Hayashi S, O’Shea AM, Burgart LJ, Smyrk TC, Shimizu D, et al. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: a population-based study. Gastroenterology (2007) 133(1):48–56. doi: 10.1053/j.gastro.2007.04.044
39. Song Y, Wang L, Ran W, Li G, Xiao Y, Wang X, et al. Effect of tumor location on clinicopathological and molecular markers in colorectal cancer in Eastern China patients: An analysis of 2,356 cases. Front Genet (2020) 11:96. doi: 10.3389/fgene.2020.00096
40. Law JH, Koh FH, Tan KK. Young colorectal cancer patients often present too late. Int J Colorectal Dis (2017) 32(8):1165–9. doi: 10.1007/s00384-017-2837-1
41. Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol (2015) 12(10):584–96. doi: 10.1038/nrclinonc.2015.105
42. Uyanik B, Goloudina AR, Akbarali A, Grigorash BB, Petukhov AV, Singhal S, et al. Inhibition of the DNA damage response phosphatase PPM1D reprograms neutrophils to enhance anti-tumor immune responses. Nat Commun (2021) 12(1):3622. doi: 10.1038/s41467-021-23330-6
43. Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen (2017) 58(5):235–63. doi: 10.1002/em.22087
44. Bristow RG, Hill RP. Hypoxia and metabolism. hypoxia, DNA repair and genetic instability. Nat Rev Cancer (2008) 8(3):180–92. doi: 10.1038/nrc2344
45. Simunkova M, Lauro P, Jomova K, Hudecova L, Danko M, Alwasel S, et al. Redox-cycling and intercalating properties of novel mixed copper(II) complexes with non-steroidal anti-inflammatory drugs tolfenamic, mefenamic and flufenamic acids and phenanthroline functionality: Structure, SOD-mimetic activity, interaction with albumin, DNA damage study and anticancer activity. J Inorg Biochem (2019) 194:97–113. doi: 10.1016/j.jinorgbio.2019.02.010
46. Haemmerle M, Stone RL, Menter DG, Afshar-Kharghan V, Sood AK. The platelet lifeline to cancer: Challenges and opportunities. Cancer Cell (2018) 33(6):965–83. doi: 10.1016/j.ccell.2018.03.002
47. Wang W, Wang G, Fu S, Zhang B, Liu Z, Wang R. Decreased mean platelet volume is associated with microsatellite instability in colorectal cancer: A propensity score-matched analysis. Cancer biomark (2021) 31(4):351–9. doi: 10.3233/CBM-203250
48. Liu LY, Bates ME, Jarjour NN, Busse WW, Bertics PJ, Kelly EA. Generation of Th1 and Th2 chemokines by human eosinophils: evidence for a critical role of TNF-alpha. J Immunol (2007) 179(7):4840–8. doi: 10.4049/jimmunol.179.7.4840
49. Grisaru-Tal S, Itan M, Klion AD, Munitz A. A new dawn for eosinophils in the tumour microenvironment. Nat Rev Cancer (2020) 20(10):594–607. doi: 10.1038/s41568-020-0283-9
Keywords: colorectal cancer, deficient mismatch repair, real-world research, machine learning, routine preoperative characterization
Citation: Xu D, Chen R, Jiang Y, Wang S, Liu Z, Chen X, Fan X, Zhu J and Li J (2022) Application of machine learning in the prediction of deficient mismatch repair in patients with colorectal cancer based on routine preoperative characterization. Front. Oncol. 12:1049305. doi: 10.3389/fonc.2022.1049305
Received: 21 September 2022; Accepted: 07 December 2022;
Published: 22 December 2022.
Edited by:
Zexian Liu, Sun Yat-sen University Cancer Center (SYSUCC), ChinaReviewed by:
Pouria Samadi, Hamadan University of Medical Sciences, IranFrancesco Pepe, University of Naples Federico II, Italy
Copyright © 2022 Xu, Chen, Jiang, Wang, Liu, Chen, Fan, Zhu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jipeng Li, amlwZW5nTGkxOTc0QGFsaXl1bi5jb20=; Jun Zhu, empzdHlAZm1tdS5lZHUuY24=
†These authors have contributed equally to this work
 Dong Xu1,2†
Dong Xu1,2† Jun Zhu
Jun Zhu Jipeng Li
Jipeng Li