- 1Department of Hepatology, Tianjin Second People’s Hospital, Tianjin Institute of Hepatology, Tianjin, China
- 2Department of Hepatobiliary Oncology, Liver Cancer Center, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Key Laboratory of Cancer Prevention and Therapy, Tianjin’s Clinical Research Center for Cancer, Tianjin Medical University, Tianjin, China
Emerging studies have revealed the role of microbiota in regulating tumorigenesis, development, and response to antitumor treatment. However, most studies have focused on gut microbiota, and little is known about the intratumoral microbiome. To date, the latest research has indicated that the intratumoral microbiome is a key component of the tumor microenvironment (TME), and can promote a heterogeneous immune microenvironment, reprogram tumor metabolism to affect tumor invasion and metastasis. In this review, we will summarize existing studies on the intratumoral microbiome of gastrointestinal cancers and reveal their crosstalk. This will provide a better understanding of this emerging field and help to explore new therapeutic approaches for cancer patients by targeting the intratumoral microbiome.
Introduction
Interactions between the microbiome and human body are well known to be complex (1, 2). The microbiome can affect different physical processes in several ways, most importantly through metabolism and immunity. Approximately 20% of human malignant tumors are associated with the microbiome (3), of which gastrointestinal cancers, including Helicobacter pylori-associated gastric cancer, hepatitis B virus-associated hepatocellular carcinoma (HCC), and Fusobacterium nucleatum-associated colorectal cancer (CRC), account for a vast proportion.
The commensal microbiota mainly resides in the gut (4). Emerging evidence has indicated an association between gut dysbiosis and various tumors (5), as well as revealed the potential of gut microbiota as a non-invasive diagnostic marker for tumors. For example, Ren et al. reported a decrease of butyrate-producing bacteria but increased lipopolysaccharide-producing bacteria in early HCC (6). Similarly, a decrease of methanogenic archaea, Saccharomycetes, and Pneumocystidomycetes, but enrichment of halophilic archaea, Malasseziomycetes in fecal samples of patients with CRC were also found (7). In addition, a series of microbial prognostic models are established to discriminate cases from control individuals (6, 8). Moreover, the dysbiosis of microbiome is stage-specific and specific microbial markers are associated with the survival of patients, independently of tumor stage, lymph node metastases, or clinical parameters (8). Gut dysbiosis can lead to alterations in key proteases and metabolites, such as toll-like receptor, nuclear factor-kappa B (NF-κB), and short-chain fatty acids, regulate immunity and metabolism so as to induce tumorigenesis and development (9). Furthermore, the key role of gut microbiota in mediating tumor responses to chemotherapy and immunotherapy has also been highlighted (10, 11).
Considering the outstanding progression of gut microbiota, studies on intratumoral microbiome have also substantially advanced in recent years. The intratumoral microbiome is reported to interact with TME and play important roles in regulating tumorigenesis, development, and response to antitumor treatments (1, 12, 13). Here, we will review studies on the intratumoral microbiome of gastrointestinal cancers.
Intratumoral microbiome of gastrointestinal cancer
As early as in the last century, scientists have detected the presence of bacteria in tumor tissues (14). However, characterizing the intratumoral microbiome remains difficult because of the extremely low microbial biomass of tumors and limited detection technology. With the development of next-generation sequencing technology, emerging studies have begun to explore the composition of the intratumoral microbiome and its role in tumorigenesis and progression. A recent study detected intratumoral bacteria in 1526 tumor tissues of melanoma, pancreatic cancer, lung cancer, ovarian cancer, glioblastoma, bone cancer, and breast cancer using multiple technologies (e.g., 16S rRNA sequencing, immunohistochemistry, immunofluorescence hybridization, and bacterial culture) (15). Intratumoral bacteria were found to be tumor-specific and associated with smoking history and immunotherapy response. Moreover, they may affect tumor occurrence, development and their therapeutic responses by regulating inflammation and immunity, participating in metabolic processes, and destroying DNA stability (16). In the next section, we will elaborate on the research progress about intratumoral microbiome in different types of gastrointestinal cancers.
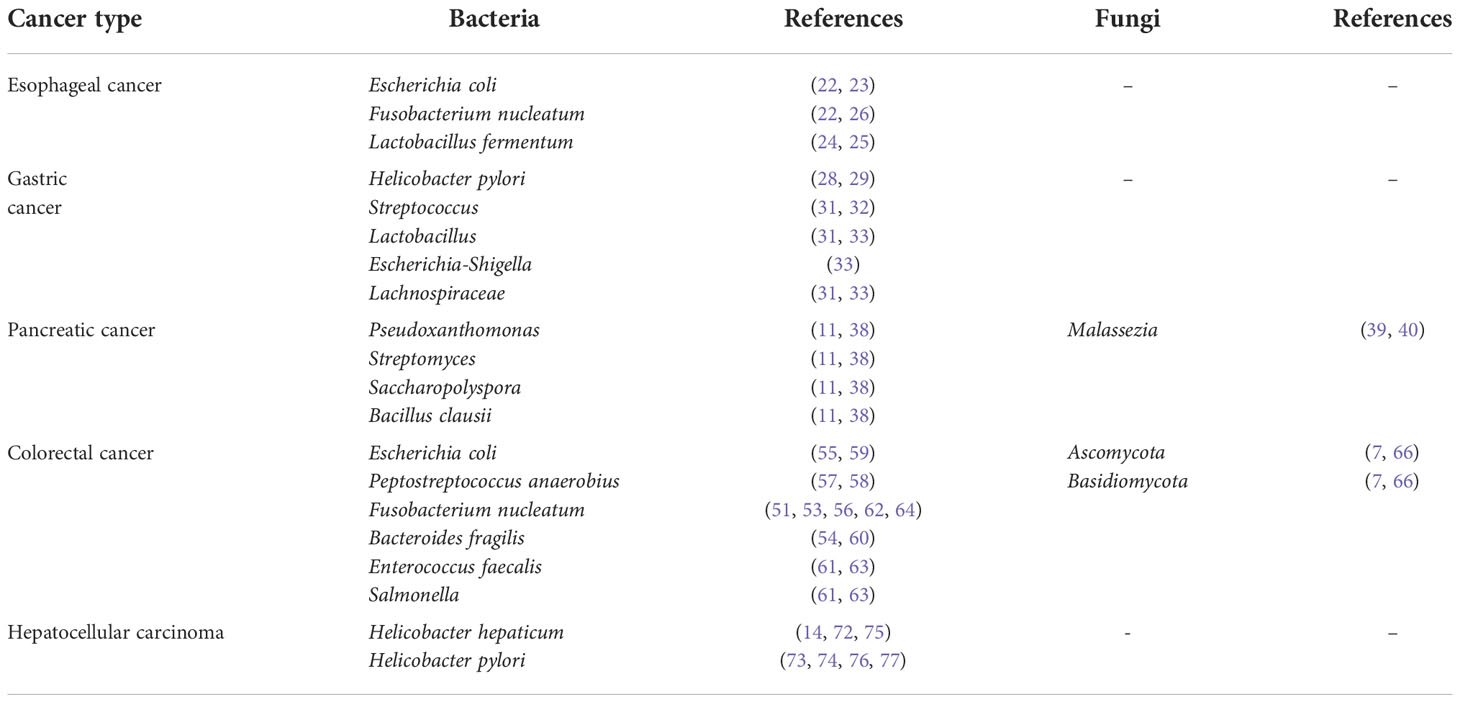
Esophageal cancer
Esophageal cancer is the fourth leading cause of cancer-related death in China with 5-year survival rate less than 20%. It is associated with squamous dysplasia, alcohol consumption, cigarette smoking, and dietary habits (17, 18). Epidemiologic studies have shown the association between variations of the esophageal microbiota and esophageal disease (19, 20). For example, normal esophagus was found to be with higher abundance of Streptococcus, while esophagitis and Barrett’s esophagus is enriched in gram-negative bacteria, such as Veillonella, Prevotella, Haemophilus, Neisseria, Granulicatella, and Fusobacterium (21). In contrast, esophageal cancer is associated with specific Gram-negative bacteria (e.g., Escherichia coli and Fusobacterium nucleatum) (22, 23). Similarly, a decrease of Veillonella and Granulicatella while an enrichment of Lactobacillus fermentum were found in esophageal adenocarcinoma (EAC) patients compared to controls and Barrett’s esophagus patients (24, 25). Attentionally, the relative abundance of Fusobacterium spp. was also gradually increased from physiological normal esophagus to esophageal squamous cell carcinoma (ESCC) (26), associated with advanced tumor stage and poor overall survival (27), while the abundance of Proteobacteria was decreased. In a word, esophageal cancers of various pathological types are all complicated with microbial dysbiosis, meanwhile, with decreased microbial diversity compared with control individuals (24, 26).
Although the composition and diversity of the esophageal microbiota correlate with esophageal disease (22), while most data on esophageal microbiota are derived from small-scale cross-sectional studies and the evidence is insufficient to assume causality.
Gastric cancer
Gastric cancer is a common gastrointestinal cancer and a leading cause of cancer-related death. Proteobacteria (e.g., H. pylori) are primarily detected in gastric cancer (28, 29) and have been reported to promote precancerous lesions, such as gastric atrophy, intestinal metaplasia, and atypical hyperplasia, which can eventually lead to gastric cancer (30). Previous hypotheses suggest that an acidic microenvironment in the stomach causes a lack of bacterial diversity. While the microbial diversity in gastric cancer have been found significantly increased as compared with that in chronic gastritis and intestinal metaplasia, with higher relative abundance of Streptococcus (31, 32). Microbial alpha-diversity increases with disease severity, that is, chronic gastritis has the lowest microbial diversity, whereas gastric cancer has the highest (31). As reported, Proteobacteria, Firmicutes, Bacteroidetes, Fusobacteria, and Actinobacteria are the dominant bacteria at the phylum level, and potential cancer-promoting bacteria (e.g., Lactobacillus, Escherichia-Shigella, Lachnospiraceae) are enriched in gastric cancer at the genus level (33). This is consistent with the discovery in other types of tumors (34–36). Altered microbiota in gastric cancer (e.g., bacterial overgrowth and diversified microbial community) might potentially promote inflammation and carcinogenesis (33).
The community structure and microbial diversity of gastric cancer remain poorly understood. Nonetheless, bacterial overgrowth is potentially associated with the development of gastric cancer, and the microbial community construct in gastric cancer and its potential role in carcinogenesis also remain to be further explored.
Pancreatic cancer
Pancreatic cancer is one of the most aggressive human malignancies with a five-year survival rate of 8%. The pancreas used to be considered a sterile organ. However, emerging studies have demonstrated the presence of bacterial species in the pancreas. A variety of bacteria were detected in 76% of pancreatic ductal adenocarcinoma (PDAC) tissues with a higher proportion in pancreatic cancer tissues compared with normal pancreatic tissues (12, 37). Proteobacteria, Bacteroidetes, and Firmicutes were found to be the dominant phyla in tumor tissues (12). Higher alpha-diversity of tumor microbiome and high abundances of a microbial signature (Pseudoxanthomonas-Streptomyces-Saccharopolyspora-Bacillus clausii) were correlated with longer survival, which could contribute to the anti-tumor immune response by favoring recruitment and activation of CD8+ T cells (11, 38). In addition, bacterial species cultured from fresh PDAC tumor tissues were found to induce resistance to chemotherapeutic drug gemcitabine by preclinical study (37). Beyond intratumoral bacteria, Aykut et al. explored the fungal community of PDAC (39, 40). They showed that fungi migrated from gut lumen to the pancreas, and PDAC showed an alarming increase in fungi compared to normal pancreatic tissue both in humans and mouse models. Specifically, patients with PDAC could be distinguished from healthy individuals by the abundance of markedly enriched Malassezia spp.
Attentionally, the origin of tumor-associated bacteria is also a research hotspot. Considering constant interactions between the pancreas and gut, the development of pancreatic cancer is closely related to the dysregulation and mislocation of gut microbiota (12, 41). Consistent with the above, studies have estimated that PDAC-associated bacteria can be translocated from the gastrointestinal tract in a retrograde manner (12, 37). These findings provide the possibility for further exploration of intratumoral microbiome and for development of new strategies for the diagnosis and treatment of pancreatic cancer (42).
CRC
CRC is a common malignant tumor of the digestive tract with poor prognosis (43). The etiology of CRC involves genetic and environmental factors (44), in which dietary habits, obesity, and heavy drinking play important roles in the occurrence of sporadic CRC. Furthermore, emerging studies have shown the potential role of gut microbiota in the development, diagnosis and treatment of CRC (7, 45–49).
Compared with control individuals, there is no doubt that the gut microbiota in CRC patients is dysbiosis (50–53). Gut commensal bacteria (e.g., E. coli, Fusobacteria, enterotoxin-producing Bacteroides fragilis, and Peptostreptococcus anaerobius) have been found to be increased in patients with CRC than in healthy individuals (54–60). Enterococcus faecalis, Salmonella (61) and F. nucleatum are also revealed with significant associations with CRC (56, 62, 63). Special species have been shown to promote tumor cell proliferation in vitro and in vivo (64), and even predict a poor prognosis (51, 65). Although studies on fungal communities of CRC are limited, but some new findings have been obtained. Coker et al. revealed higher Basidiomycota: Ascomycota ratio in CRC patients and different clusters of fungal components in early-stage and late-stage CRC patients, indicating that mycobiome profiles were stage-specific (7). Ascomycota, Glomeromycota, and Basidiomycota were found to be the dominant phyla by characterized fungal microbiota profiles in 27 cases of colorectal adenomas and adjacent tissues (66). Adenoma size and disease stage were closely associated with fungal microbiota. Furthermore, a series of intestinal microbial biomarkers were identified to distinguish CRC patients from controls (7, 66). This indicates the potential of intestinal microbiome as a tool towards targeted non-invasive biomarkers for CRC.
Many studies on the intestinal microbiome of CRC are performed using fecal samples because of its easy and non-invasive procedure. However, tissue samples from colonic mucosa are more valuable to disentangle the physiopathology of CRC disease and cumulative studies have shown different microbiome profiles between mucosal and fecal samples (67–71). So far, the unified microbial community structure associated with CRC has not been determined and sample collection is another challenging in microbiome studies of CRC. Accordingly, further studies are warranted to determine which is more representative of the real microbial structure change.
HCC
Hepatitis virus is well known to be closely related to HCC, but the role of bacteria in the occurrence and development of HCC remains unclear. In 1992, the Frederick Cancer Research Center identified a spiral bacterium-Helicobacter hepaticus from the liver tissue, and considered it as a cause of hepatitis and liver tumors in mice (14). Since then, scientists have carried out a series of studies on the association between Helicobacter spp. and chronic liver diseases (72–75). The Helicobacter 16S rRNA gene was sequentially detected in liver tissues of patients with chronic liver diseases and HCC (76–78). In addition, an association between H. pylori infection and mortality of patients with HCC has been observed (79). However, studies on the intratumoral microbiota of HCC are scarce and further investigations on the role of other bacteria in HCC are needed.
The above are reported researches on intratumoral microbiome of gastrointestinal cancers, and we summarize the intratumoral microbiome in Table 1.
Crosstalk between intratumoral microbiome and TME
Emerging studies have demonstrated intratumoral microbiome as a component of TME, and the crosstalk between intratumoral microbiome and TME is mutual and highly dynamic (1, 15). Specifically, the intratumoral microbiome can induce immunosuppression or immunoactivation, reprogram tumor metabolism, and form a heterogeneity TME so as to promote or inhibit tumor development (80, 81). The potential crosstalk reported between the microbiome and TME is shown in Figure 1.
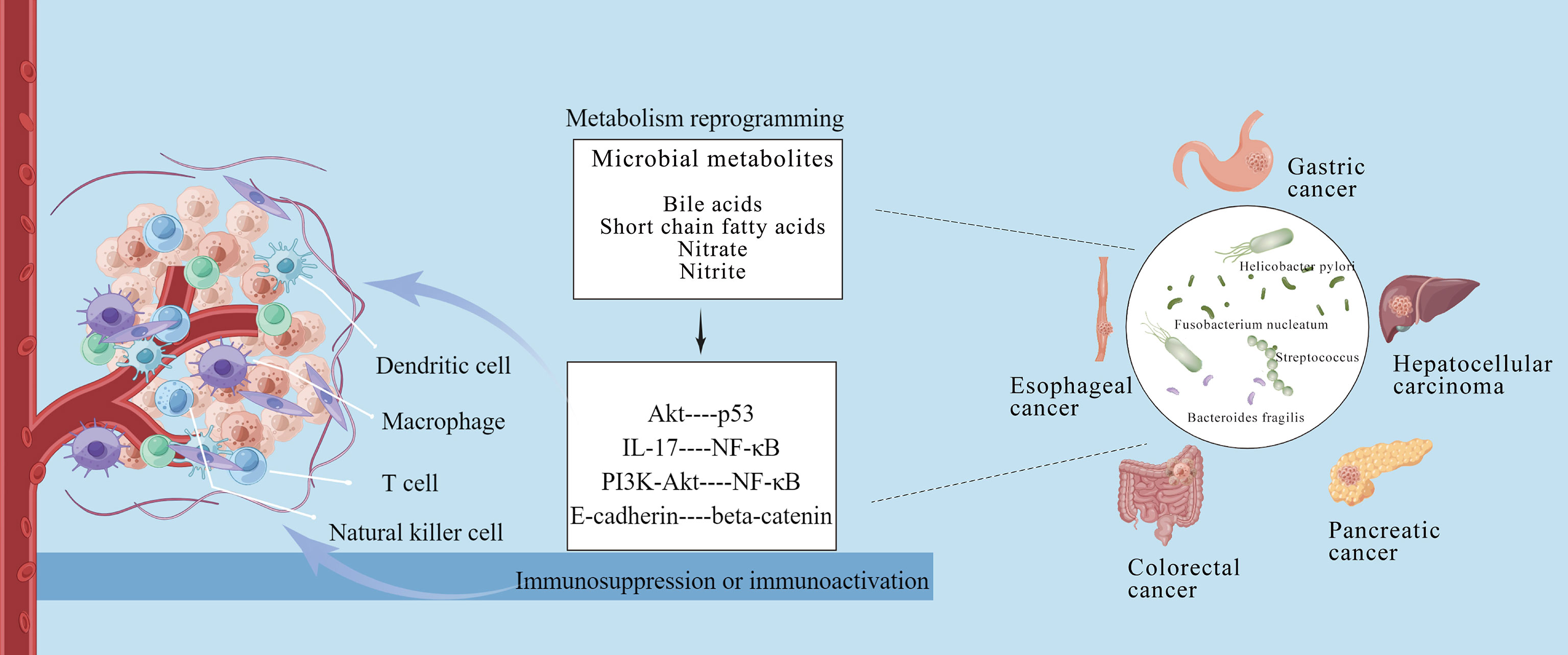
Figure 1 Crosstalk between intratumoral microbiome of gastrointestinal cancers and tumor microenvironment. The intratumoral microbiota can induce immunosuppression or immunoactivation via metabolism reprogramming, and form a heterogeneity tumor microenvironment to affect tumor occurrence and development. This figure is drawn by Figdraw.
The reprogramming of immune infiltration plays important roles in the crosstalk between the intratumoral microbiome and TME (2, 82). A recent research revealed that microbial components (e.g., DNA, RNA, bacterial peptides, and lipopolysaccharides) could be detected in tumor and immune cells, indicating that intratumoral microbiome might affect immune infiltration in TME (15). Similarly, bacterial polypeptide fragments were found to be directly presented on the surface of tumor cells or antigen-presenting cells through human leukocyte antigens. They could promote T cell activation and potential tumor immune response (83). Tumor monocytes can be induced to produce interferon-I, regulate macrophage polarization, promote communication of natural killer and dendritic cells, and reprogram the TME (84). In addition, the microbiota may regulate immune invasion in different tumors in different ways. Proteus bacteria (mainly E. coli) in CRC may destroy the intestinal barrier, migrate and colonize the liver through the damaged intestinal barrier, and promote immune cell recruitment in the liver (e.g., macrophages, neutrophils, and monocytes) to induce liver metastasis (85). F. nucleatum is reported to bind to the immunosuppressive receptor T cell immunoglobulin through its surface adhesin, thereby inhibiting T cell activation and natural killer cell lethality (86). Pushalkar et al. demonstrated that intratumoral bacteria in pancreatic cancer could promote oncogenesis by inducing innate and adaptive immune suppression (12). While bacterial ablation induced immunogenic reprogramming, including reduction of myeloid-derived suppressor cells, increase of M1 macrophage differentiation, promotion of Th1 differentiation of CD4+ T cells, and activation of CD8+ T cells (12). Mechanistically, the immune-suppression characteristic of PDAC were generated by bacteria through differentially activating select toll-like receptors in monocytic cells. Furthermore, special bacterial species were found to favor recruitment and activation of CD8+ T cells, contribute to the anti-tumor immune response and affect clinical outcomes of pancreatic cancer (11).
Many of the above findings are still arguable and need further research. While, some preclinical models have clarified potential mechanisms by which microbiota can contribute to tumorigenesis and established a more causative role. P. gingivalis infection was found to enhance proliferation of PDAC cells by cell lines and a xenograft model (87). This was independent of TLR2 signaling and associated with augmentation of the Akt signaling pathway (87). Similarly, Akt signaling pathway activated by H pylori can also result in subsequent degradation of tumor suppressor p53 in gastric epithelial cells and increase survival of gastric epithelial cells with sustained DNA damage (88). Enterotoxigenic B. fragilis can lead to colitis that triggers IL-17 production and inflammatory pathway activation, such as NF-κB, to facilitate tumor growth (54, 89). While PI3K-Akt pathway activated in CRC can lead to increased cell proliferation and NF-κB activation (58). F. nucleatum may selectively bind and activate E-cadherin/beta-catenin signaling via its FadA adhesin, inducing inflammation and CRC (90). Enterococcus faecalis in CRC induces superoxide production, thus damaging the DNA of epithelial cells (91, 92).
Additionally, tumor microbiome is reported to directly affect the response to antitumor therapies (93, 94). The enrichment of Firmicutes, reciprocal changes in abundance of Verrucomicrobia and Proteobacteria, were found to be correlated with better immunocheckpoint inhibitor (ICI) response across various tumors (93, 94). In addition, F. nucleatum is found to promote the resistance of colorectal cancer to chemotherapy (95). Mechanistically, F. nucleatum activated the autophagy pathway by targeting at TLR4 and MYD88 innate immune signaling and specific microRNAs to promote chemoresistance (95). A study about CD47-based cancer immunotherapy revealed that accumulative Bifidobacterium in TME facilitates local anti-CD47 treatment by a stimulator of interferon genes (STING)- and interferon-dependent fashion (96). Besides, disruption of gut microbiota is estimated to affect the response of tumors to immunotherapy and chemotherapy both in preclinical models and cancer patients (97–100). Antibiotic treatment can reduce response to ICI, while the presence of certain bacteria strains correlates with better outcomes (10, 101). Mechanistically, gut microbiota could modulate the expression of immune checkpoints, the function of dendritic cell, the homing and recruitment of lymphocyte (82, 83), as well as the production of critical metabolites, such as short chain fatty acids (SCFA) (102, 103).
The microbiome participates in the modulation of human metabolism and microbial dysbiosis can induce systemic metabolic alterations (104, 105). To date, intratumoral microbiome has been shown to reprogram tumor metabolism through derived metabolites, and a differential enrichment of metabolic functional pathways induced by microbiota may correlate with clinical outcome. As reported, patients with PDAC who are enrichment in xenobiotics biodegradation and lipids metabolism pathways have shown better outcomes (11). Metabolites, such as the secondary bile acids (e.g., deoxycholic acid, lithocholic acid) and SCFA (e.g., butyrate), have been reported to regulate inflammation and T cell differentiation (106–108) and implicated in the tumorigenesis (109, 110). HCC oncogenesis is found to be closely associated with intestinal flora, bile acid metabolism, and tumor immunity (111). Gut microbiota-mediated bile acid metabolism regulates the occurrence and progression of HCC by affecting the accumulation of hepatic CXCR6+ natural killer T (NKT) cells through mediating the expression of CXCL16 in liver sinusoidal endothelial cells (111). SCFAs, mainly butyrate, can directly modulate CD8(+) cytotoxic T lymphocytes and Tc17 cells, as well as induce senescence-like phenotypes and the development of CRC (106, 107). The secondary metabolites produced by microbiota, such as reactive nitrate and nitrite, can lead to accumulation of carcinogenic N-nitroso compounds and associate with tumor development (26, 112). These studies shed light on a new theoretical basis for tumor treatment by regulating microbial metabolism and the microbiota.
Conclusions
Despite decades of research, gastrointestinal cancers remain the most lethal malignant tumor with limited treatment options and poor clinical outcomes. Emerging understanding of microbiome in gastrointestinal cancers will potentially provide new insights and opportunities for the development of novel biomarkers and treatment strategies. Current studies have revealed the role of microbiota in regulating tumorigenesis, development, and response to antitumor treatment, while challenges remain in our understanding of intratumoral microbiome. For example, how are the mechanisms of action of intratumoral microbiome in various tumor types? How are they different? What are the effects of microbial metabolites on tumors? How does intratumoral microbiota regulate the inflammatory carcinogenic pathway and immune response? Moreover, research discoveries on the various types of tumors have been discrepant, and this may be due to several factors, such as the tumor type, research methods, and microbial complexity. Moreover, almost no good clinical data exists for the claims. Therefore, the diversity, origin, and mechanism of action of intratumoral microbiome require further clarification. However, intratumoral microbiome has the potential to be used as diagnostic tools for tumor typing and to be considered as a potential new therapeutic target. Further in-depth researches on the intratumoral microbiome are expected to reveal the complex correlation between intratumoral microbiome and gastrointestinal cancers. Overall, this exciting field will allow us to explore the mystery of carcinogenesis from another perspective and to discover novel targets for precision medicine in the management of gastrointestinal cancers.
Author contributions
SL: Data curation, Writing-original draft preparation. WL, QL: Writing- reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Key project of Science and Technology, Tianjin Municipal Science and Technology Bureau (19YFZCSY00020).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science (2021) 371(6536):eabc4552. doi: 10.1126/science.abc4552
2. Poutahidis T. Erdman s E.Commensal bacteria modulate the tumor microenvironment. Cancer Lett (2016) 380(1):356–8. doi: 10.1016/j.canlet.2015.12.028
3. De Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol (2012) 13(6):607–15. doi: 10.1016/S1470-2045(12)70137-7
4. Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature (2012) 486(7402):207–14. doi: 10.1038/nature11234
5. Chen D, Wu J, Jin D, Wang B, Cao H. Fecal microbiota transplantation in cancer management: Current status and perspectives. Int J Cancer (2019) 145(8):2021–31. doi: 10.1002/ijc.32003
6. Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut (2019) 68(6):1014–23. doi: 10.1136/gutjnl-2017-315084
7. Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, et al. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut (2019) 68(4):654–62. doi: 10.1136/gutjnl-2018-317178
8. Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology (2018) 155(2):529–541.e5. doi: 10.1053/j.gastro.2018.04.018
9. Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer (2013) 13(11):759–71. doi: 10.1038/nrc3611
10. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
11. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell (2019) 178(4):795–806.e12. doi: 10.1016/j.cell.2019.07.008
12. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discovery (2018) 8(4):403–16. doi: 10.1158/2159-8290.CD-17-1134
13. Sepich-Poore GD, Carter H. Knight R.Intratumoral bacteria generate a new class of therapeutically relevant tumor antigens in melanoma. Cancer Cell (2021) 39(5):601–3. doi: 10.1016/j.ccell.2021.04.008
14. Fox JG, Dewhirst FE, Tully JG, Paster BJ, Yan L, Taylor NS, et al. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol (1994) 32(5):1238–45. doi: 10.1128/jcm.32.5.1238-1245.1994
15. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science (2020) 368(6494):973–80. doi: 10.1126/science.aay9189
16. Ramirez-Labrada AG, Isla D, Artal A, Arias M, Rezusta A, Pardo J, et al. The influence of lung microbiota on lung carcinogenesis, immunity, and immunotherapy. Trends Cancer (2020) 6(2):86–97. doi: 10.1016/j.trecan.2019.12.007
17. Oze I, Matsuo K, Wakai K, Nagata C, Mizoue T, Tanaka K, et al. Alcohol drinking and esophageal cancer risk: an evaluation based on a systematic review of epidemiologic evidence among the Japanese population. Jpn J Clin Oncol (2011) 41(5):677–92. doi: 10.1093/jjco/hyr026
18. Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y, Ueda J, et al. Epidemiology of esophageal cancer in Japan and China. J Epidemiol (2013) 23(4): 233–42. doi: 10.2188/jea.JE20120162
19. Snider EJ, Compres G, Freedberg DE, Khiabanian H, Nobel YR, Stump S, et al. Alterations to the esophageal microbiome associated with progression from barrett's esophagus to esophageal adenocarcinoma. Cancer Epidemiol Biomarkers Prev (2019) 28(10):1687–93. doi: 10.1158/1055-9965.EPI-19-0008
20. Plaza-Diaz J, Alvarez-Mercado AI, Ruiz-Marin CM, Reina-Pérez I, Pérez-Alonso AJ, Sánchez-Andujar MB, et al. Association of breast and gut microbiota dysbiosis and the risk of breast cancer: a case-control clinical study. BMC Cancer (2019) 19(1):495. doi: 10.1186/s12885-019-5660-y
21. Yang L, Lu X, Nossa CW, Francois F, Peek RM, Pei Z. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology (2009) 137(2):588–97. doi: 10.1053/j.gastro.2009.04.046
22. Ajayi TA, Cantrell S, Spann A, Garman KS. Barrett's esophagus and esophageal cancer: Links to microbes and the microbiome. PLos Pathog (2018) 14(12):e1007384. doi: 10.1371/journal.ppat.1007384
23. Zaidi AH, Kelly LA, Kreft RE, Barlek M, Omstead AN, Matsui D, et al. Associations of microbiota and toll-like receptor signaling pathway in esophageal adenocarcinoma. BMC Cancer (2016) 16:52. doi: 10.1186/s12885-016-2093-8
24. Elliott DRF, Walker AW, O'donovan M, Parkhill J, Fitzgerald RC. A non-endoscopic device to sample the oesophageal microbiota: a case-control study. Lancet Gastroenterol Hepatol (2017) 2(1):32–42. doi: 10.1016/S2468-1253(16)30086-3
25. Lv J, Guo L, Liu JJ, Zhao HP, Zhang J, Wang JH. Alteration of the esophageal microbiota in barrett's esophagus and esophageal adenocarcinoma. World J Gastroenterol (2019) 25(18):2149–61. doi: 10.3748/wjg.v25.i18.2149
26. Yang W, Chen CH, Jia M, Xing X, Gao L, Tsai HT, et al. Tumor-associated microbiota in esophageal squamous cell carcinoma. Front Cell Dev Biol (2021) 9:641270. doi: 10.3389/fcell.2021.641270
27. Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, et al. Human microbiome fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res (2016) 22(22):5574–81. doi: 10.1158/1078-0432.CCR-16-1786
28. Cai H, Ye F, Michel A, Murphy G, Sasazuki S, Taylor PR, et al. Helicobacter pylori blood biomarker for gastric cancer risk in East Asia. Int J Epidemiol (2016) 45(3):774–81. doi: 10.1093/ije/dyw078
29. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
30. Correa P, Houghton J. Carcinogenesis of helicobacter pylori. Gastroenterology (2007) 133(2):659–72. doi: 10.1053/j.gastro.2007.06.026
31. Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY, et al. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter (2014) 19(6):407–16. doi: 10.1111/hel.12145
32. Dai D, Yang Y, Yu J, Dang T, Qin W, Teng L, et al. Interactions between gastric microbiota and metabolites in gastric cancer. Cell Death Dis (2021) 12(12):1104. doi: 10.1038/s41419-021-04396-y
33. Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, et al. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol (2016) 28(3):261–6. doi: 10.1097/MEG.0000000000000542
34. Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol (2009) 58(Pt 4):509–16. doi: 10.1099/jmm.0.007302-0
35. Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep (2014) 4:4202. doi: 10.1038/srep04202
36. Leung A, Tsoi H. Yu J.Fusobacterium and escherichia: models of colorectal cancer driven by microbiota and the utility of microbiota in colorectal cancer screening. Expert Rev Gastroenterol Hepatol (2015) 9(5):651–7. doi: 10.1586/17474124.2015.1001745
37. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science (2017) 357(6356):1156–60. doi: 10.1126/science.aah5043
38. Bradley CA. Tumour microbiome defines outcomes. Nat Rev Cancer (2019) 19(10):545. doi: 10.1038/s41568-019-0201-1
39. Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature (2019) 574(7777):264–7. doi: 10.1038/s41586-019-1608-2
40. Lee SC. mSphere of influence: the mycobiota in human health and disease. mSphere (2020) 5(1):e00974–19. doi: 10.1128/mSphere.00974-19
41. Vitiello GA, Cohen DJ. Miller G.Harnessing the microbiome for pancreatic cancer immunotherapy. Trends Cancer (2019) 5(11):670–6. doi: 10.1016/j.trecan.2019.10.005
42. Li JJ, Zhu M, Kashyap PC, Chia N, Tran NH, McWilliams RR, et al. The role of microbiome in pancreatic cancer. Cancer Metastasis Rev (2021) 40(3):777–89. doi: 10.1007/s10555-021-09982-2
43. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
44. Louis P, Hold GL. Flint h J.The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol (2014) 12(10):661–72. doi: 10.1038/nrmicro3344
45. Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu William KK, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun (2015) 6:8727. doi: 10.1038/ncomms9727
46. Chiu DK, Tse AP, Xu IM, Di CJ, Lai RK, Li LL, et al. Hypoxia inducible factor HIF-1 promotes myeloid-derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat Commun (2017) 8(1):517. doi: 10.1038/s41467-017-00530-7
47. Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut (2018) 67(6):1024–32. doi: 10.1136/gutjnl-2017-314281
48. Jobin C. Colorectal cancer: looking for answers in the microbiota. Cancer Discovery (2013) 3(4):384–7. doi: 10.1158/2159-8290.CD-13-0042
49. Tlaskalova-Hogenova H, Vannucci L, Klimesova K, Stepankova R, Krizan J, Kverka M. Microbiome and colorectal carcinoma: insights from germ-free and conventional animal models. Cancer J (2014) 20(3):217–24. doi: 10.1097/PPO.0000000000000052
50. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe (2013) 14(2):207–15. doi: 10.1016/j.chom.2013.07.007
51. Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y, et al. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin Transl Gastroenterol (2016) 7(11):e200. doi: 10.1038/ctg.2016.53
52. Chen D, Zhang Y, Wang W, Chen H, Ling T, Yang R, et al. Identification and characterization of robust hepatocellular carcinoma prognostic subtypes based on an integrative metabolite-protein interaction network. Adv Sci (Weinh) (2021) 8(17):e2100311. doi: 10.1002/advs.202100311
53. Bashir A, Miskeen AY, Bhat A, Fazili KM, Ganai BA. Fusobacterium nucleatum: an emerging bug in colorectal tumorigenesis. Eur J Cancer Prev (2015) 24(5):373–85. doi: 10.1097/CEJ.0000000000000116
54. Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science (2018) 359(6375):592–7. doi: 10.1126/science.aah3648
55. Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, et al. Colonization of the human gut by e. coli and colorectal cancer risk. Clin Cancer Res (2014) 20(4):859–67. doi: 10.1158/1078-0432.CCR-13-1343
56. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res (2012) 22(2):299–306. doi: 10.1101/gr.126516.111
57. Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC, et al. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology (2017) 152(6):1419–1433.e5. doi: 10.1053/j.gastro.2017.01.009
58. Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol (2019) 4(12):2319–30. doi: 10.1038/s41564-019-0541-3
59. Veziant J, Gagniere J, Jouberton E, Bonnin V, Sauvanet P, Pezet D, et al. Association of colorectal cancer with pathogenic escherichia coli: Focus on mechanisms using optical imaging. World J Clin Oncol (2016) 7(3):293–301. doi: 10.5306/wjco.v7.i3.293
60. Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe (2018) 23(2):203–214.e5. doi: 10.1016/j.chom.2018.01.007
61. Li S, Liu J, Zheng X, Ren L, Yang Y, Li W, et al. Tumorigenic bacteria in colorectal cancer: mechanisms and treatments. Cancer Biol Med (2021). doi: 10.20892/j.issn.2095-3941.2020.0651
62. Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of fusobacterium with colorectal carcinoma. Genome Res (2012) 22(2):292–8. doi: 10.1101/gr.126573.111
63. Henstra C, Van Praagh J, Olinga P, Nagelkerke A. The gastrointestinal microbiota in colorectal cancer cell migration and invasion. Clin Exp Metastasis (2021) 38(6):495–510. doi: 10.1007/s10585-021-10130-x
64. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of fusobacterium persistence and antibiotic response in colorectal cancer. Science (2017) 358(6369):1443–8. doi: 10.1126/science.aal5240
65. Andzinski L, Kasnitz N, Stahnke S, Wu CF, Gereke M, von Köckritz-Blickwede M, et al. Type I IFNs induce anti-tumor polarization of tumor associated neutrophils in mice and human. Int J Cancer (2016) 138(8):1982–93. doi: 10.1002/ijc.29945
66. Luan C, Xie L, Yang X, Miao H, Lv N, Zhang R, et al. Dysbiosis of fungal microbiota in the intestinal mucosa of patients with colorectal adenomas. Sci Rep (2015) 5:7980. doi: 10.1038/srep07980
67. Yu LC, Wei SC, Ni YH. Impact of microbiota in colorectal carcinogenesis: lessons from experimental models. Intest Res (2018) 16(3):346–57. doi: 10.5217/ir.2018.16.3.346
68. Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst (2013) 105(24):1907–11. doi: 10.1093/jnci/djt300
69. Zackular JP, Rogers MA, Ruffin MTT, Schloss PD. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila) (2014) 7(11):1112–21. doi: 10.1158/1940-6207.CAPR-14-0129
70. Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun (2015) 6:6528. doi: 10.1038/ncomms7528
71. Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med (2019) 25(4):679–89. doi: 10.1038/s41591-019-0406-6
72. Dore MP, Realdi G, Mura D, Graham DY, Sepulveda AR. Helicobacter infection in patients with HCV-related chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Dig Dis Sci (2002) 47(7):1638–43. doi: 10.1023/A:1015848009444
73. Pellicano R, Leone N, Berrutti M, Cutufia MA, Fiorentino M, Rizzetto M, et al. Helicobacter pylori seroprevalence in hepatitis c virus positive patients with cirrhosis. J Hepatol (2000) 33(4):648–50. doi: 10.1016/S0168-8278(00)80018-5
74. Avenaud P, Marais A, Monteiro L, Le Bail B, Bioulac Sage P, Balabaud C, et al. Detection of helicobacter species in the liver of patients with and without primary liver carcinoma. Cancer (2000) 89(7):1431–9. doi: 10.1002/1097-0142(20001001)89:7<1431::AID-CNCR4>3.0.CO;2-5
75. Yang J, Ji S, Zhang Y, Wang J. Helicobacter hepaticus infection in primary hepatocellular carcinoma tissue. Singapore Med J (2013) 54(8):451–7. doi: 10.11622/smedj.2013153
76. Nilsson HO, Mulchandani R, Tranberg KG, Stenram U, Wadström T. Helicobacter species identified in liver from patients with cholangiocarcinoma and hepatocellular carcinoma. Gastroenterology (2001) 120(1):323–4. doi: 10.1053/gast.2001.21382
77. Fan XG, Zou YY, Wu AH, Li TG, Hu GL, Zhang Z. Seroprevalence of helicobacter pylori infection in patients with hepatitis b. Br J BioMed Sci (1998) 55(3):176–8. doi: 10.1097/00022744-199809000-00014
78. Rocha M, Avenaud P, Menard A, Le Bail B, Balabaud C, Bioulac-Sage P, et al. Association of helicobacter species with hepatitis c cirrhosis with or without hepatocellular carcinoma. Gut (2005) 54(3):396–401. doi: 10.1136/gut.2004.042168
79. Yang JD, Mohamed EA, Aziz AO, Shousha HI, Hashem MB, Nabeel MM, et al. Characteristics, management, and outcomes of patients with hepatocellular carcinoma in Africa: a multicountry observational study from the Africa liver cancer consortium. Lancet Gastroenterol Hepatol (2017) 2(2):103–11. doi: 10.1016/S2468-1253(16)30161-3
80. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell (2014) 157(1):121–41. doi: 10.1016/j.cell.2014.03.011
81. Dzutsev A, Badger JH, Perez-Chanona E, Roy S, Salcedo R, Smith CK, et al. Microbes and cancer. Annu Rev Immunol (2017) 35:199–228. doi: 10.1146/annurev-immunol-051116-052133
82. Erdman SE, Poutahidis T. The microbiome modulates the tumor macroenvironment. Oncoimmunology (2014) 3:e28271. doi: 10.4161/onci.28271
83. Cullin N, Azevedo Antunes C, Straussman R, Stein-Thoeringer CK, Elinav E. Microbiome and cancer. Cancer Cell (2021) 39(10):1317–41. doi: 10.1016/j.ccell.2021.08.006
84. Lam KC, Araya RE, Huang A, Chen Q, Di Modica M, Rodrigues RR, et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell (2021) 184(21):5338–5356.e21. doi: 10.1016/j.cell.2021.09.019
85. Bertocchi A, Carloni S, Ravenda PS, Bertalot G, Spadoni I, Lo Cascio A, et al. Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell (2021) 39(5):708–724.e11. doi: 10.1016/j.ccell.2021.03.004
86. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity (2015) 42(2):344–55. doi: 10.1016/j.immuni.2015.01.010
87. Gnanasekaran J, Binder Gallimidi A, Saba E, Pandi K, Eli Berchoer L, Hermano E, et al. Intracellular porphyromonas gingivalis promotes the tumorigenic behavior of pancreatic carcinoma cells. Cancers (Basel) (2020) 12(8):2331. doi: 10.3390/cancers12082331
88. Wei J, Nagy TA, Vilgelm A, Zaika E, Ogden SR, Romero-Gallo J, et al. Regulation of p53 tumor suppressor by helicobacter pylori in gastric epithelial cells. Gastroenterology (2010) 139(4):1333–43. doi: 10.1053/j.gastro.2010.06.018
89. Cheng WT, Kantilal HK, Davamani F. The mechanism of bacteroides fragilis toxin contributes to colon cancer formation. Malays J Med Sci (2020) 27(4):9–21. doi: 10.21315/mjms2020.27.4.2
90. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating e-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe (2013) 14(2):195–206. doi: 10.1016/j.chom.2013.07.012
91. Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, desulfovibrio and enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol (2008) 23(8 Pt 1):1298–303. doi: 10.1111/j.1440-1746.2008.05490.x
92. Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke MM. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res (2008) 68(23):9909–17. doi: 10.1158/0008-5472.CAN-08-1551
93. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell (2018) 33(4):570–80. doi: 10.1016/j.ccell.2018.03.015
94. Huang C, Li M, Liu B, Zhu H, Dai Q, Fan X, et al. Relating gut microbiome and its modulating factors to immunotherapy in solid tumors: A systematic review. Front Oncol (2021) 11:642110. doi: 10.3389/fonc.2021.642110
95. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell (2017) 170(3):548–563.e16. doi: 10.1016/j.cell.2017.07.008
96. Shi Y, Zheng W, Yang K, Harris KG, Ni K, Xue L, et al. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J Exp Med (2020) 217(5):e20192282. doi: 10.1084/jem.20192282
97. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science (2013) 342(6161):967–70. doi: 10.1126/science.1240527
98. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (2015) 350(6264): 1084–9. doi: 10.1126/science.aac4255
99. Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
100. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science (2018) 359(6371):97–103. doi: 10.1126/science.aan4236
101. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science (2018) 359(6371):104–8. doi: 10.1126/science.aao3290
102. Yi M, Yu S, Qin S, Liu Q, Xu H, Zhao W, et al. Gut microbiome modulates efficacy of immune checkpoint inhibitors. J Hematol Oncol (2018) 11(1):47. doi: 10.1186/s13045-018-0592-6
103. Yi M, Jiao D, Qin S, Chu Q, Li A, Wu K. Manipulating gut microbiota composition to enhance the therapeutic effect of cancer immunotherapy. Integr Cancer Ther (2019) 18:1–13. doi: 10.1177/1534735419876351
104. Tremaroli V. Backhed F.Functional interactions between the gut microbiota and host metabolism. Nature (2012) 489(7415):242–9. doi: 10.1038/nature11552
105. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science (2012) 336(6086):1262–7. doi: 10.1126/science.1223813
106. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature (2013) 504(7480):451–5. doi: 10.1038/nature12726
107. Luu M, Weigand K, Wedi F, Breidenbend C, Leister H, Pautz S, et al. Regulation of the effector function of CD8(+) T cells by gut microbiota-derived metabolite butyrate. Sci Rep (2018) 8(1):14430. doi: 10.1038/s41598-018-32860-x
108. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science (2013) 341(6145):569–73. doi: 10.1126/science.1241165
109. Rossi T, Vergara D, Fanini F, Maffia M, Bravaccini S, Pirini F. Microbiota-Derived Metabolites Tumor Progression and Metastasis. Int J Mol Sci (2020) 21(16). doi: 10.3390/ijms21165786
110. Okumura S, Konishi Y, Narukawa M, Sugiura Y, Yoshimoto S, Arai Y, et al. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat Commun (2021) 12(1):5674. doi: 10.1038/s41467-021-25965-x
111. Jia B. Commentary: Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Front Immunol (2019) 10:282. doi: 10.3389/fimmu.2019.00282
Keywords: tumor microenvironment, intratumoral microbiome, gastrointestinal cancer, commensal microbiota, tumor metabolism
Citation: Li S, Li Q and Lu W (2022) Intratumoral microbiome and gastrointestinal cancers. Front. Oncol. 12:1047015. doi: 10.3389/fonc.2022.1047015
Received: 17 September 2022; Accepted: 11 November 2022;
Published: 29 November 2022.
Edited by:
Aditi Banerjee, University of Maryland, Baltimore, United StatesReviewed by:
Ho Tsoi, The University of Hong Kong, Hong Kong SAR, ChinaSanjit Roy, University of Maryland, Baltimore, United States
Copyright © 2022 Li, Li and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Lu, bHV3ZWkxOTY2QDEyNi5jb20=; Qian Li, Y3JiaWN1QDE2My5jb20=
 Shengnan Li
Shengnan Li Qian Li1*
Qian Li1*